Abstract
Glandular epithelia, including the mammary and prostate glands, are composed of basal cells (BCs) and luminal cells (LCs)1,2. Many glandular epithelia develop from multipotent basal stem cells (BSCs) that are replaced in adult life by distinct pools of unipotent stem cells1,3–8. However, adult unipotent BSCs can reactivate multipotency under regenerative conditions and upon oncogene expression3,9–13. This suggests that an active mechanism restricts BSC multipotency under normal physiological conditions, although the nature of this mechanism is unknown. Here we show that the ablation of LCs reactivates the multipotency of BSCs from multiple epithelia both in vivo in mice and in vitro in organoids. Bulk and single-cell RNA sequencing revealed that, after LC ablation, BSCs activate a hybrid basal and luminal cell differentiation program before giving rise to LCs—reminiscent of the genetic program that regulates multipotency during embryonic development7. By predicting ligand–receptor pairs from single-cell data14, we find that TNF—which is secreted by LCs—restricts BC multipotency under normal physiological conditions. By contrast, the Notch, Wnt and EGFR pathways were activated in BSCs and their progeny after LC ablation; blocking these pathways, or stimulating the TNF pathway, inhibited regeneration-induced BC multipotency. Our study demonstrates that heterotypic communication between LCs and BCs is essential to maintain lineage fidelity in glandular epithelial stem cells.
Genetic lineage tracing of BCs of the mammary gland (MG) under physiological conditions demonstrates that BCs are multipotent during embryonic development, giving rise to BCs and LCs that become lineage-restricted at the end of embryonic development and sustain only the BC lineage postnatally1,3,7,8. Transplantation of adult unipotent MG BCs alone results in MG outgrowth composed of BCs and LCs3,9–11, demonstrating the multipotent potential of BCs when they are transplanted without LCs, and illustrating the discrepancy between the fate of BCs under physiological conditions and their potential under regenerative conditions. By contrast, when BCs are transplanted together with LCs, BCs maintain their unipotent basal fate3, suggesting that LCs might signal to BCs to restrict their multilineage differentiation.
LC ablation drives multipotency of MG BSCs
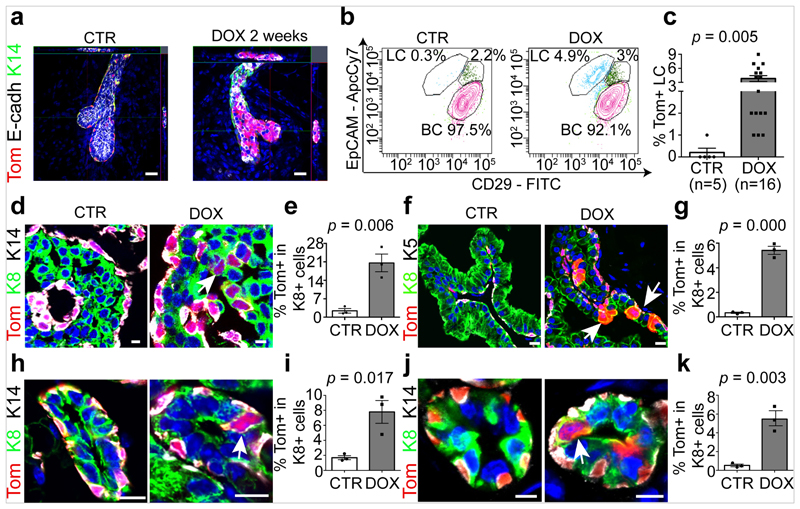
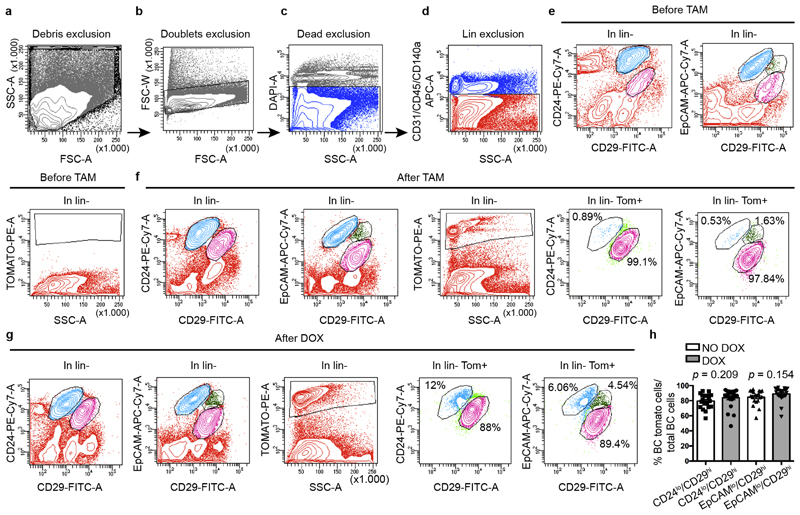
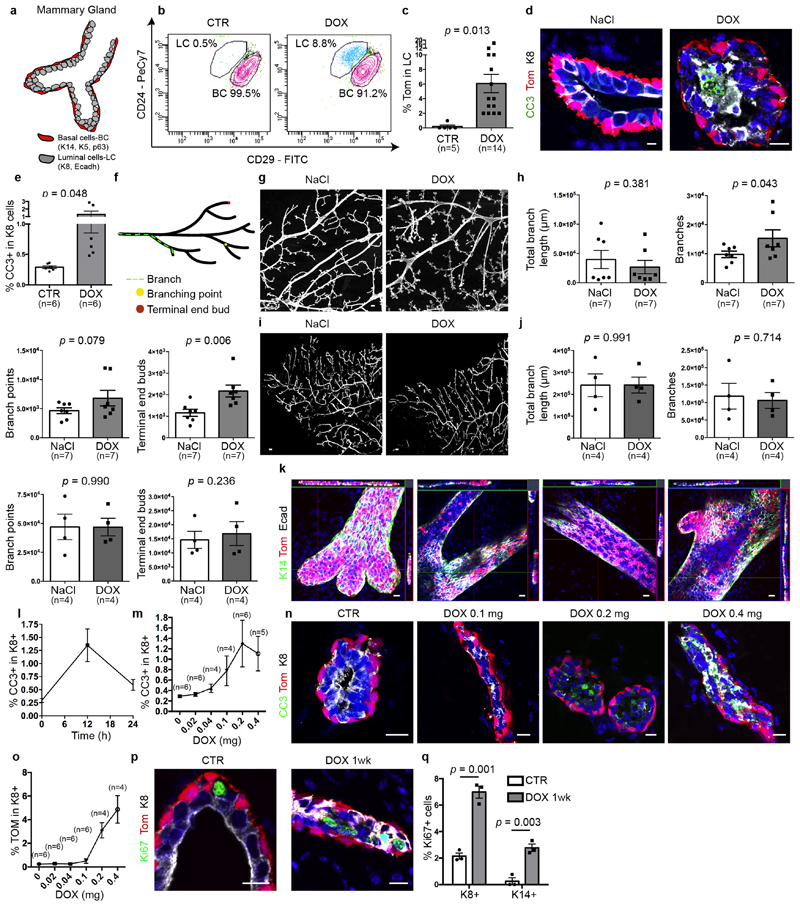
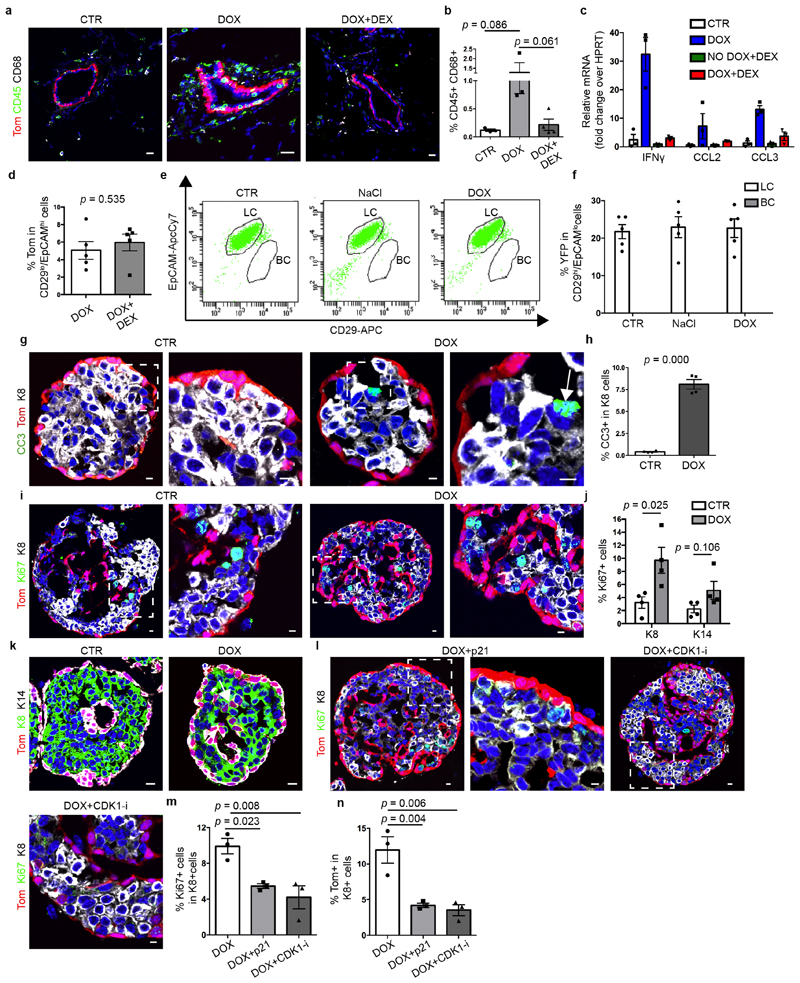
To test whether LCs restrict the multipotency of BCs, we developed a genetic mouse model that enabled us to lineage-trace BCs and lineage-ablate LCs in a temporally regulated manner. The administration of tamoxifen to eight-week-old K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA female mice led to the labelling of most of the BCs, as demonstrated by the high proportion of tdTomato expression in CD29high CD24low or CD29high EpCAMlow BCs when analysed by fluorescence-activated cell sorting (FACS) (Extended Data Fig. 1). One week after BC labelling, we administered doxycycline (DOX) to ablate LCs or saline solution (NaCl) as control by intraductal injection (IDI) into the contralateral gland, and one week later we assessed the fate of BCs by FACS and confocal microscopy. Consistent with previous studies1,3,11 in the absence of LC ablation, BCs gave rise only to BCs. By contrast, upon DOX administration—which led to apoptosis of LCs, as visualized by the presence of cleaved caspase 3 (CC3) and by the remodelling of the MG—tdTomato was observed in both BCs and LCs. FACS analysis for CD24 and CD29, which enables discrimination between BCs and LCs3,9,10, showed the appearance of tdTomato-labelled cells in the LC (CD29low CD24high) population. FACS analysis for EpCAM and CD29 revealed—in addition to CD29high EpCAMlow BCs—the appearance of tdTomato-labelled cells in two distinct populations: an intermediate CD29high EpCAMhigh population and CD29low EpCAMhigh LCs (Fig. 1a–c, Extended Data Fig. 2a–k). Increasing the dose of DOX led to increased apoptosis of LCs and it was found that the presence of 1% CC3+ LCs, 12 h after the administration of 0.2 mg DOX, was the critical threshold required to stimulate the replacement of LCs by BCs. LC ablation and activation of BC multipotency was associated with increased BC and LC proliferation (Extended Data Fig. 2l–q). Similar to what has been reported after bacterial infection of the prostate gland, which promotes prostate cell death leading to the recruitment of inflammatory cells and the stimulation of BC multipotency15, we found that LC ablation led to the recruitment of CD45+ and CD68+ inflammatory cells. Administration of dexamethasone—a potent anti-inflammatory steroid—decreased macrophage infiltration and the expression of cytokines and chemokines and did not inhibit BC multipotency after LC ablation (Extended Data Fig. 3a–d); this suggests that inflammation is not essential for BC multipotency.
Fig. 1. LC ablation promotes BC multipotency in glandular epithelia.
a, Whole-mount confocal imaging of immunostaining for tdTomato (Tom), K14 and E-cadherin (E-cadh) in the mammary glands of K5CreER/tdTomato/K8rtTA/TetO-DTA mice after IDI with NaCl (Control, n = 3 independent experiments) or 0.2 mg DOX (n = 3 independent experiments) and chasing for two weeks. Scale bars, 20 μm. b, Representative FACS plot of CD29 and EpCAM expression in Lin−tdTomato+ epithelial cells from control mice (n = 5) and from mice 1 week after DOX administration (n = 16). The percentage of the gated population out of all epithelial cells is shown. c, Quantification of tdTomato expression in CD29low EpCAMhigh LCs from control (n = 5) and DOX-treated (n = 16) mice. The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from unpaired two-sided t-tests. d–k, Confocal imaging of immunostaining for tdTomato, K8 and K14 or tdTomato, K8 and K5 in the MG organoid (d), prostate (f), salivary gland (h) and sweat gland (j) tissues and the respective quantification of tdTomato+ cells in K8+ LCs (e, g, i, k) in K5CreER/tdTomato/K8rtTA/TetO-DTA organoids (d, e) or in mice (f–k) treated with DOX and analysed 72 h later for organoids and 1 week later for in vivo mouse experiments. Scale bars, 10 μm (organoid); 5 μm (mouse tissues). Hoechst nuclear staining is shown in blue in immunofluorescence images. n = 3 independent experiments. In e, g, i, k, the bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from unpaired two-sided t-test.
To assess the plasticity of LCs under regenerative conditions, we generated another mouse model enabling the ablation of BCs after LC lineage tracing (K14rtTA/TetO-DTA/K8CreER/Rosa-YFP). We did not observe replacement of BCs by LCs after the administration of DOX (Extended Data Fig. 3e, f), suggesting that BCs are more plastic than LCs under regenerative conditions—similar to the situation observed after cell transplantation3.
To assess whether the promotion of BSC multipotency is a basal intrinsic mechanism, we performed lineage tracing and lineage ablation using MG organoids that were cultured in vitro and composed of BCs and LCs only. Similar to what was observed in vivo, DOX-induced diphtheria toxin A (DTA) led to the apoptosis of LCs and to increased proliferation of both BCs and LCs (Extended Data Fig. 3g–j). Seventy-two hours after the generation of DOX-induced DTA, we found that about 20% of LCs had been replaced by tdTomato+ BCs, showing that LC ablation in the absence of stroma and inflammatory cells is sufficient to rapidly promote BC multipotency in a reductionist in vitro assay (Fig. 1d, e, Extended Data Fig. 3g–k). These findings further suggest that LCs restrict BC multipotency, and that the promotion of BC multipotency is an intrinsic mechanism that does not depend on stromal or immune cells.
To assess whether proliferation is a necessary step for the generation of LCs by BCs, we inhibited cell proliferation in MG organoids derived from K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA mice by adenovirus-mediated p21 overexpression or by using CDK1 inhibitor. We found that decreasing cell proliferation reduced BC multipotency after LC ablation (Extended Data Fig. 3l–n), suggesting that cell proliferation contributes to de novo generation of LCs from BCs.
BSC multipotency in glandular epithelia
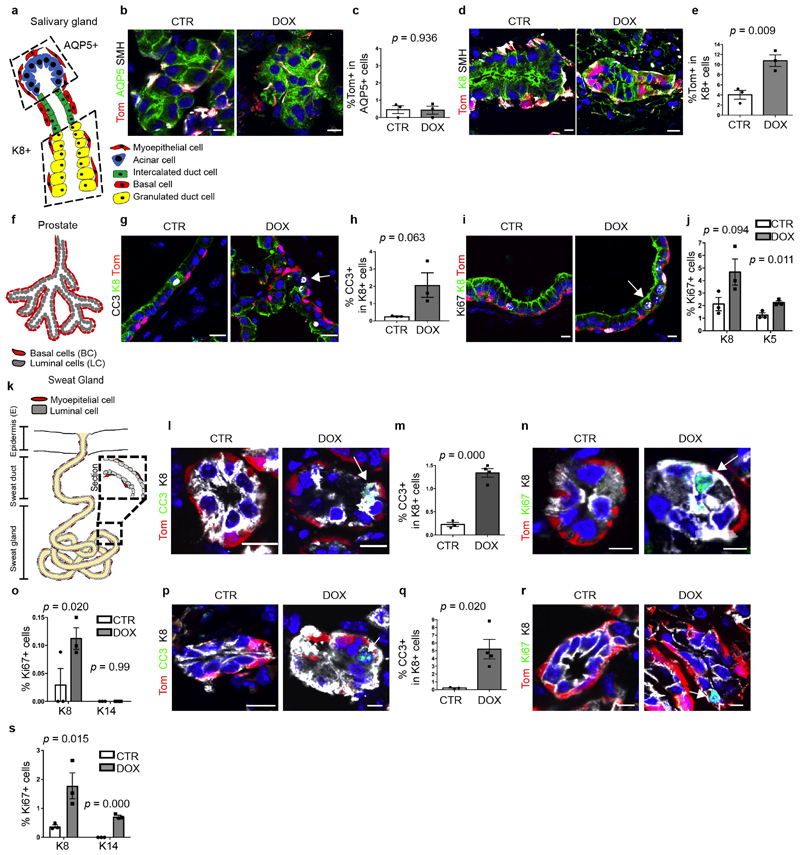
Similar to the MG, the prostate gland is also a branched epithelium composed of BCs and LCs2,16. Transplantation of adult prostate BCs without LCs into the kidney capsule can also lead to the formation of prostate outgrowth2,16, whereas under physiological conditions the adult prostate BCs mostly give rise to BCs as demonstrated by lineage tracing4,17, showing that the regenerative conditions associated with transplantation stimulate plasticity and multipotency of prostate BSCs. To test whether cell–cell interactions also regulate BC fate in the prostate, as was proposed after deletion of E-cadherin in LCs17, we assessed whether LC ablation also stimulates multipotency of prostate BCs. In the absence of LC ablation, very few LCs were tdTomato+ after BC lineage tracing, consistent with the presence of rare multipotent BCs during adult prostate homeostasis2,16. By contrast, one week after LC ablation, many more tdTomato-labelled LCs were observed throughout the prostate (Fig. 1f, g), demonstrating that LC ablation stimulates BSC multipotency in the adult prostate—consistent with previous findings18.
To determine whether this paradigm of cell fate restriction occurs in other glandular epithelia, we assessed whether LC ablation stimulates multipotency in salivary glands and sweat glands, which are also composed of basal or myoepithelial cells and intercalated ductal or luminal cells that are maintained by their lineage-restricted stem cells during homeostasis6,19,20. The salivary gland is composed of three regions: the acinar, the intercalated ductal and the granulated ductal regions. Conflicting results have been reported as to whether irradiation stimulates multipotency of BCs in the acinar compartment of the salivary glands20,21. Multipotency of BCs towards granulated cells has been described both in homeostatic conditions and upon irradiation21. In sweat glands, lineage ablation of myoepithelial cells increased the proliferation of myoepithelial cells, whereas ablation of LCs increased the proliferation of LCs. This suggests that unipotent stem cells contribute to sweat gland regeneration upon injury, but does not rule out the contribution of multipotent stem cells to sweat gland repair7. To our knowledge, lineage tracing of BCs followed by LC ablation has not been performed in either salivary or sweat glands.
In the absence of LC ablation, K5CreER lineage tracing in the sweat glands and salivary glands led to the labelling of BCs and myoepithelial cells and not of ductal cells or LCs. However, after LC ablation, tdTomato+ cells were observed in the K8+ intercalated ductal cells (but not in the acinar cells) of the salivary gland, and in the LCs of the sweat gland (Fig. 1h–k, Extended Data Fig. 4a–e), demonstrating that LC ablation stimulates BC multipotency in these glandular epithelia. Moreover, in these three glandular epithelia, DOX-induced DTA led to apoptosis of LCs and increased proliferation of both BCs and LCs (Extended Data Fig. 4f–s). Taken together, these data indicate that in four distinct glandular epithelia, maintained by lineage-restricted stem cells during physiological conditions, LC ablation stimulates BC multipotency.
BSC multipotency and hybrid state
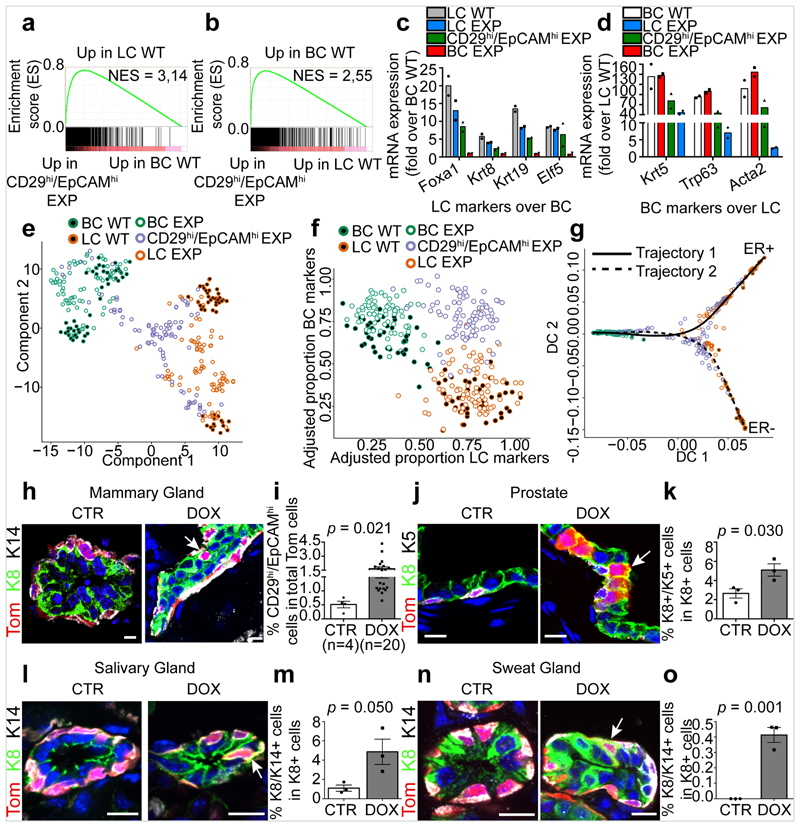
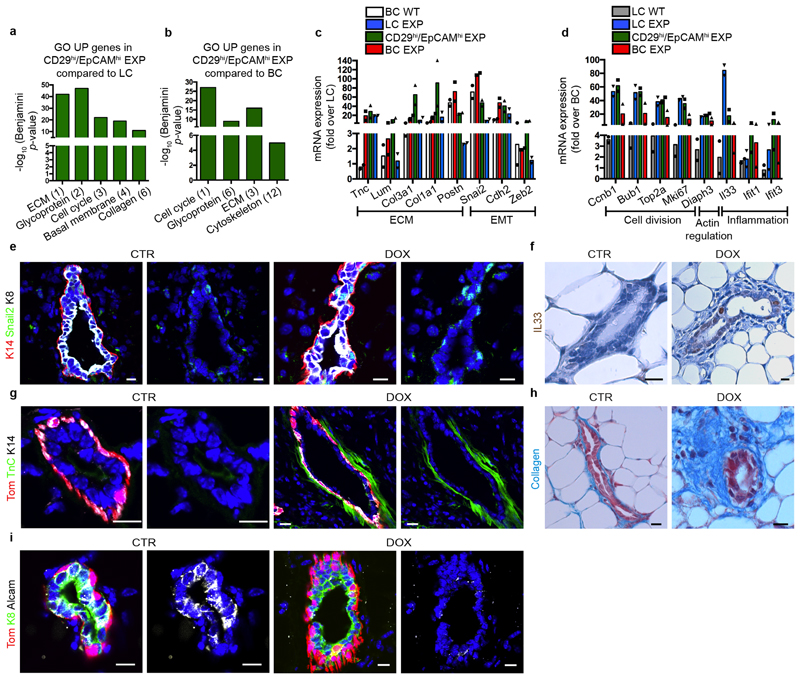
To elucidate the molecular mechanisms by which LC ablation induces BC multipotency, we first performed bulk RNA sequencing (RNA-seq) of FACS-isolated BCs and LCs from the control MG and after LC ablation—including the intermediate CD29high EpCAMhigh population that appeared after LC ablation. Gene set enrichment analysis (GSEA) comparing the transcriptome of the CD29high EpCAMhigh population with the basal and luminal signatures show that this new population expressed a hybrid basal (Krt5, Trp63, Acta2/SMA (which encodes smooth muscle actin)) and luminal (Foxa1, Krt8, Krt19, Elf5) gene signature, suggesting that this population represents a transition state between BCs and LCs. After LC ablation, Gene Ontology of the differentially regulated genes showed that BCs and LCs upregulated many genes controlling cell division, the extracellular matrix, actin regulators, inflammation and the epithelial–mesenchymal transition, which was confirmed by immunostaining and in situ characterization (Fig. 2a–d, Extended Data Fig. 5).
Fig. 2. BSC multipotency is associated with a hybrid basal and luminal signature.
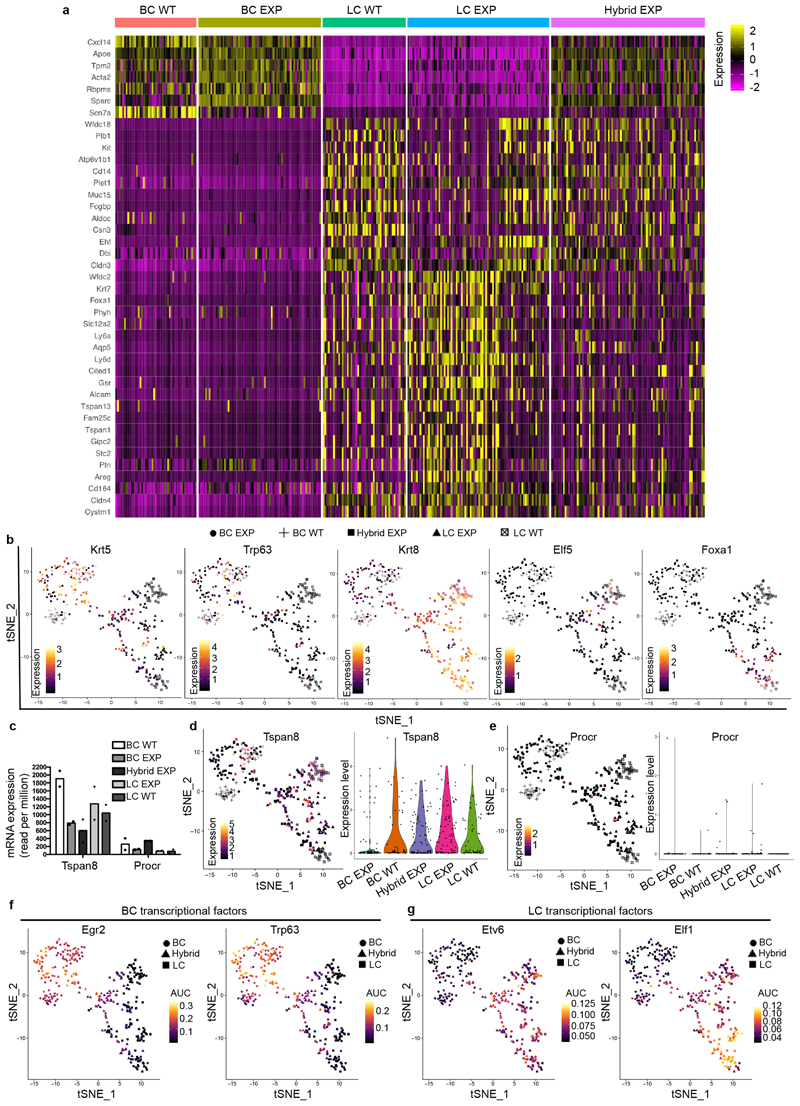
a, b, GSEA of the upregulated genes in LCs (a, compared with BCs) or in BCs (b, compared with LCs). Normalized enrichment scores (NES) are indicated. The designations ‘up in LCs’ and ‘up in BCs’ are derived from the RNA-seq analysis of FACS-isolated wild-type (WT) BCs and LCs (>2.5-fold). c, d, Relative mRNA expression of luminal (c, fold over wild-type BCs) and basal (d, fold over wild-type LCs) genes measured by RNA-seq in FACS-isolated wild-type LCs, LCs under experimental conditions (EXP LCs), CD29high EpCAMhigh EXP, EXP BCs and wild-type BCs. Mean of 2 independent experiments. e, Dimensionality reduction using t-SNE of scRNA-seq data of FACS-isolated wild-type BCs, wild-type LCs, EXP BCs, CD29high EpCAMhigh EXP and EXP LCs. f, Adjusted proportion of LC and BC marker genes detected in each cell of the different cell populations. g, Diffusion map of scRNA-seq data showing the lineage trajectories, obtained using Slingshot analysis. For scRNA-seq data, each circle (n = 337) represents one cell. h, Confocal imaging of the MG after immunostaining for tdTomato, K8 and K14 in K5CreER/tdTomato/K8rtTA/TetO-DTA mice, 1 week after IDI with NaCl or DOX. n = 3 mice per condition. i, FACS quantification of tdTomato in CD29high EpCAMhigh hybrid population. Control, n = 4 mice; DOX treatment, n = 20 mice. The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from unpaired two-sided t-tests. j–o, Confocal imaging of prostate (j), salivary gland (l) and sweat gland (n) after immunostaining for tdTomato, K8 and K5 or tdTomato, K8 and K14 from K5CreER/tdTomato/K8rtTA/TetO-DTA mice treated with DOX for 5 days and chased for 1 week, and quantification of the K8+ K5+ (k) or K8+ K14+ (m, o) hybrid cells in the immunostaining of prostate (k), salivary gland (m) and sweat gland (o). n = 3 mice per condition. The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from unpaired two-sided t-test. Hoechst nuclear staining is shown in blue. Scale bars, 5 μm.
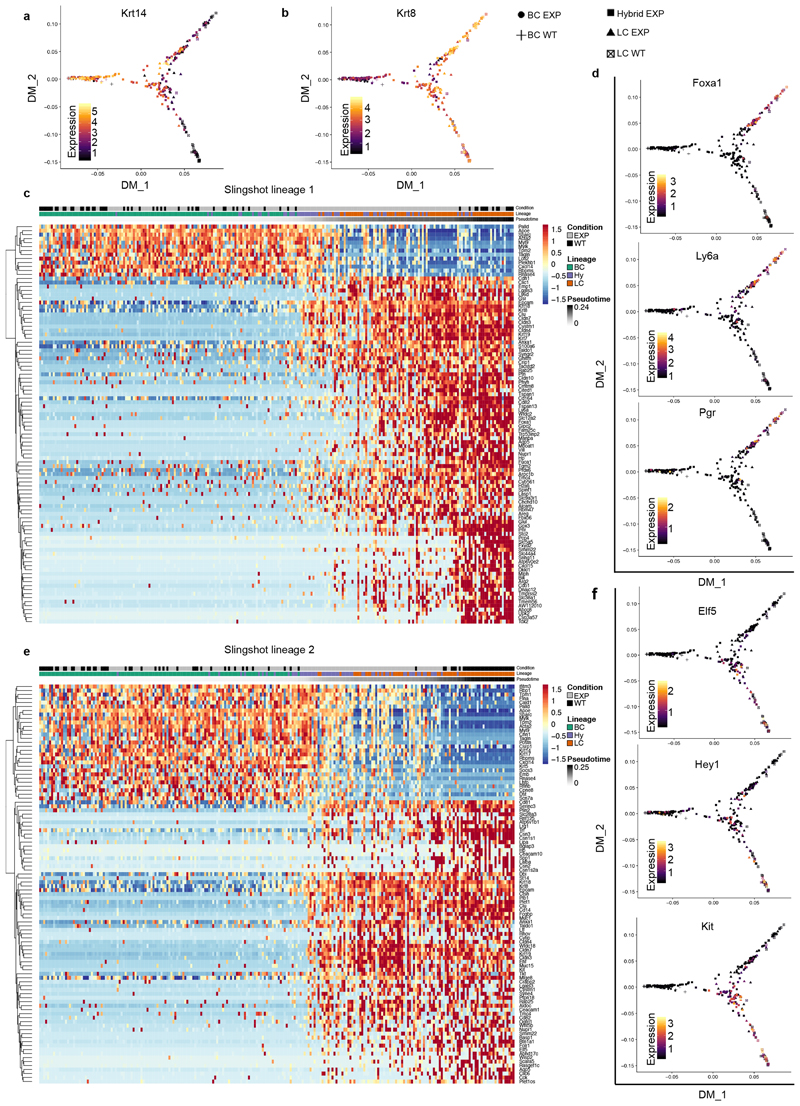
Dimensionality reduction analysis using t-distributed stochastic neighbour embedding (t-SNE) and unsupervised clustering of single-cell RNA sequencing (scRNA-seq) data showed that adult mouse BCs and LCs are molecularly distinct, and that LCs can be subdivided into two populations—as previously reported7,22–24—that remain transcriptionally very similar to the respective control BCs and LCs upon lineage ablation. Notably, the new CD29high EpCAMhigh population that arises after LC ablation represents a cell state that presents a continuum of gene expression between the BC and the LC lineages. These intermediate hybrid cells express high levels of both basal and luminal markers, such as Krt5 and Krt8, and show a progressive decrease of Trp63, a master regulator of MG development and BC fate7,25–27. An important fraction of these cells expressed either Foxa1, a master regulator of the LC oestrogen receptor (ER)+ fate28, or Elf5, a transcription factor associated with LC ER– and alveolar fate29 (Fig. 2e, f, Extended Data Fig. 6a, b). To determine whether the hybrid population emerges from a subpopulation of quiescent multipotent BSCs expressing Tspan8 or Procr30,31, we assessed the expression of these markers in our bulk and single-cell RNA-seq data. The hybrid cells do not higher levels of any of these markers compared with normal basal or luminal cells (Extended Data Fig. 6c–e), suggesting that hybrid cells are not derived from the putative quiescent basal population expressing Tspan8 or Procr. We then analysed the scRNA-seq data using SCENIC, a bioinformatic method adapted to scRNA-seq data that enables the identification of regulatory modules by inferring co-expression between transcription factors and their target genes containing the respective transcription-factor-binding motif in their regulatory regions in each cell32. The analysis suggested that the transition from the basal towards the hybrid cells is controlled by different BC transcription factors—such as those encoded by Egr2 and Trp63—together with the activation of the pro-luminal program mediated by transcription factors encoded by Etv6 and Elf1, which have been found to be active in MG multipotent embryonic progenitors7 (Extended Data Fig. 6f, g). Lineage trajectory inference using Slingshot33 further shows that the transition from BCs to LCs upon LC ablation is characterized by the presence of the CD29high EpCAMhigh hybrid state, intermediate between BCs and LCs. Two trajectories were found, heading towards either ER+ or ER– LCs, with the branching point occurring in the hybrid population (Fig. 2g, Extended Data Fig. 7). This suggests that the cell fate choice between ER+ and ER– is already occurring in the hybrid state. Taken together, the scRNA-seq analysis after LC ablation indicates that the reactivation of multipotency in BCs is a progressive process that is associated with the transient expression of a hybrid signature in the activated BCs, which then move towards the luminal compartment and become reprogrammed into ER+ or ER– LCs while losing their basal identity.
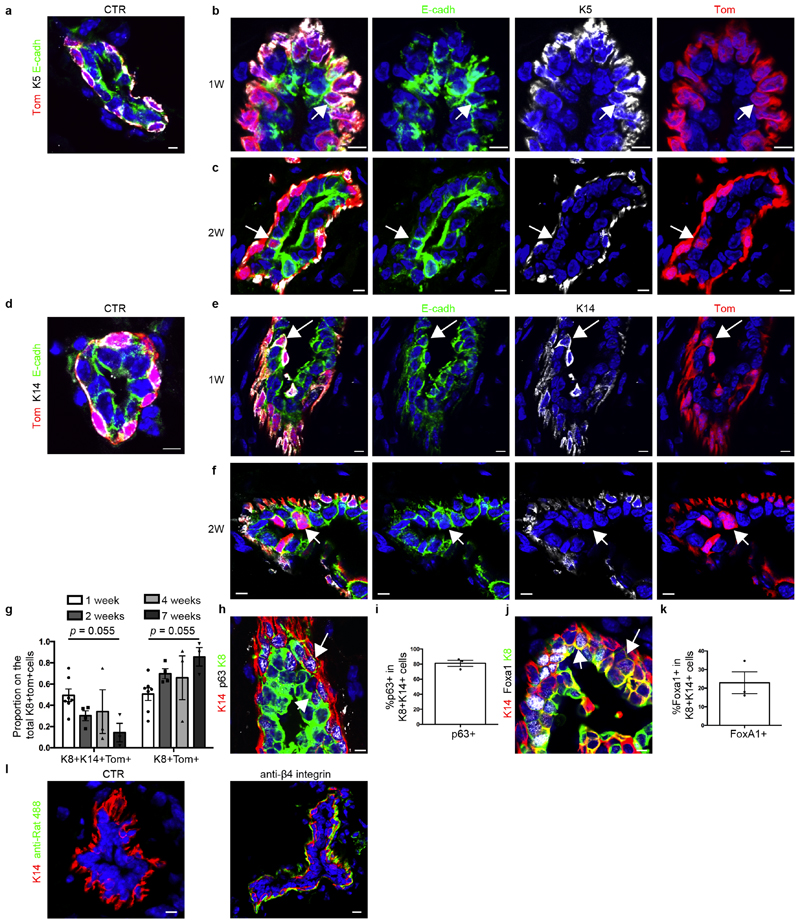
Immunostaining of basal and luminal markers at different time points after LC ablation confirmed the formation of hybrid basal and luminal cells and showed a progressive reduction in the proportion of hybrid cells in favour of luminal K8+tdTomato+ cells over time (Fig. 2h, i, Extended Data Fig. 8a–k). Activation of BC multipotency in the prostate, sweat glands and salivary glands was also associated with the transient appearance of hybrid basal and luminal cells (Fig. 2j–o), suggesting that the emergence of hybrid state is a conserved mechanism during the reactivation of multipotency in glandular epithelia.
TNF restricts BSC multipotency
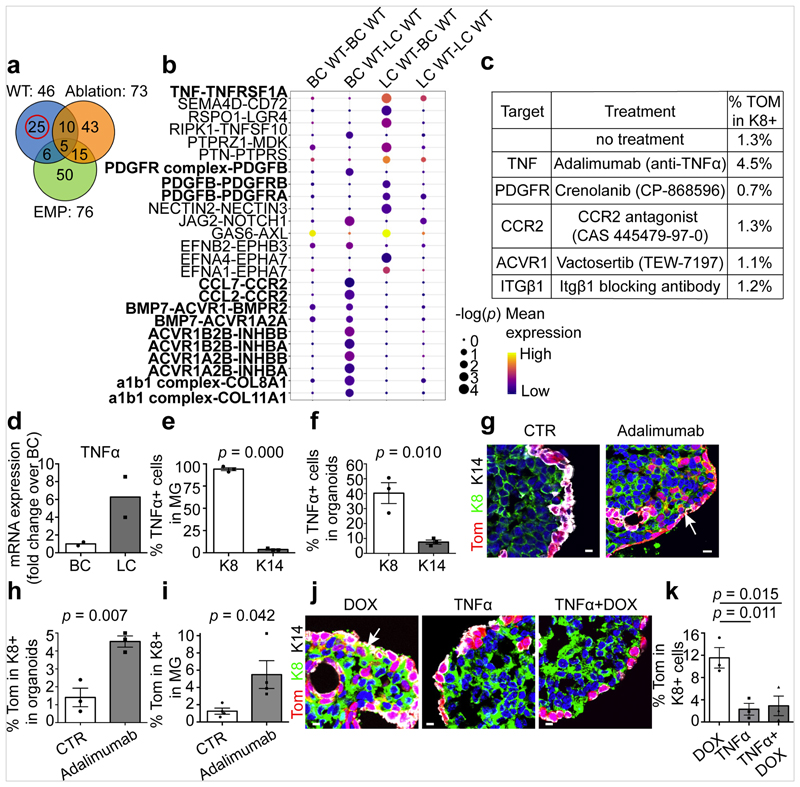
To define the mechanisms that restrict multipotency in the absence of injury we used CellPhone-DB, software that enables the definition of highly significant ligand–receptor pair interactions between the different cell types presented in single-cell data14. We assessed the ligand–receptor interaction pairs identified in control adult BCs and LCs, embryonic multipotent progenitors and upon LC ablation. The comparison between these pairs showed the presence of different common ligand–receptor pairs among the different conditions. The ligand–receptor pairs found exclusively in adult BCs and LCs under homeostatic conditions were those thought to regulate the communication between BCs and LCs during homeostasis (Fig. 3a, b). We studied the role of the most significant ligand–receptor pairs by using inhibitors of either the receptor or the ligand and then assessing the activation of BSC multipotency in MG organoids in the absence of injury (Fig. 3c). Among the different inhibitor molecules tested, the inhibition of TNF (previously known as TNFα)—which is expressed exclusively by LCs both in vivo and in organoids in vitro (Fig. 3d–f)—using the anti-TNF blocking antibody adalimumab, was found to stimulate BC multipotency in the absence of injury. This suggests that the expression of TNF by LCs restricts BSC multipotency under normal physiological conditions. To assess the relevance of this mechanism in vivo, we injected adalimumab intraductally into the MG of adult mice; this was also found to stimulate BSC multipotency (Fig. 3g–i, Extended Data Fig. 8l). Finally, we showed that the addition of TNF decreases BC multipotency after LC ablation (Fig. 3j, k). These data provide direct evidence that basal–luminal interaction, through the secretion of TNF by LCs, restricts the multipotency of BSCs.
Fig. 3. TNF expression by LCs restricts BC multipotency under homeostatic conditions.
a, Venn diagram representing the interaction between the significant ligand–receptor pairs identified by CellPhone-DB analysis on scRNa-seq data from wild-type, ablation and embryonic mammary progenitors (EMP) (n = 95 cells). b, The mean expression and P values of the ligand–receptor interaction exclusively significant for wild-type cells (n = 25 pairs). For CellPhone-DB analysis, P values are derived from one-sided permutation tests (significant P < 0.05). c, The percentage of K8+tdTomato+ cells arising from BCs in K5CreER/tdTomato organoids treated with inhibitors of the ligand–receptor pairs tested. Mean of 2 independent experiments. d–f, mRNA (d) and protein (e, f) expression of TNF in MG (d, e) and MG organoids (f). Mean of 2 independent experiments normalized over wild-type BCs (d). Quantification of TNF K8+ and K14+ cells from the immunofluorescence after cytospin of MG cells (e) or organoids (f). n = 3 independent experiments. The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from unpaired two-sided t-tests. g, h, Confocal imaging of control or adalimumab-treated K5CreER/tdTomato organoids after immunostaining for tdTomato, K8 and K14 (g) and quantification of tdTomato+K8+ cells in these organoids (h). For adalimumab treatment, organoids were treated with adalimumab for 48 h and analysed after 72 h. The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from paired two-sided t-tests. n = 3 independent experiments. i, Quantification of tdTomato+K8+ cells in the MG of control K5CreER/tdTomato mice or mice analysed one week after IDI of adalimumab (n = 4 mice per condition). The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from unpaired two-sided t-test. j, k, Confocal imaging of immunostaining for tdTomato, K8 and K14 in K5CreER/tdTomato organoids treated with DOX, TNF and DOX+TNF for 48 h and analysed after 72 h (j) and quantification of tdTomato+ K8+ cells in these organoids (k). n = 3 independent experiments. The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from ANOVA followed by two-sided Dunnett’s test. Hoechst nuclear staining is shown in blue. Scale bars, 10 μm.
Mechanisms promoting BSC multipotency
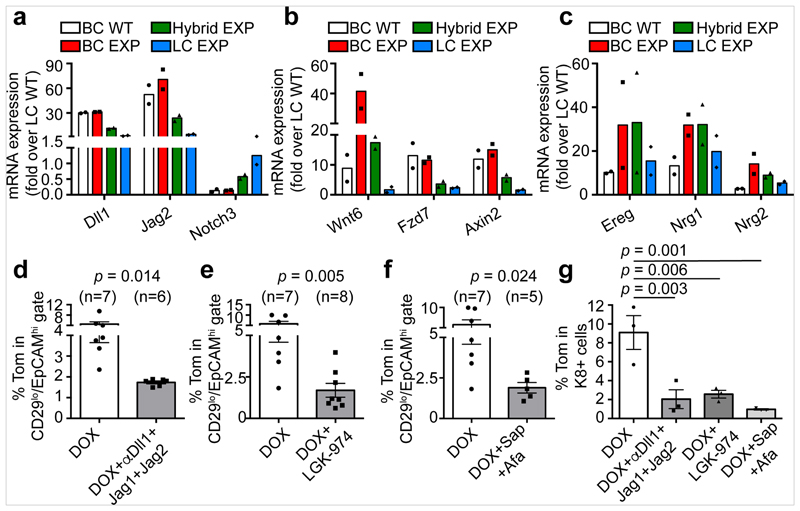
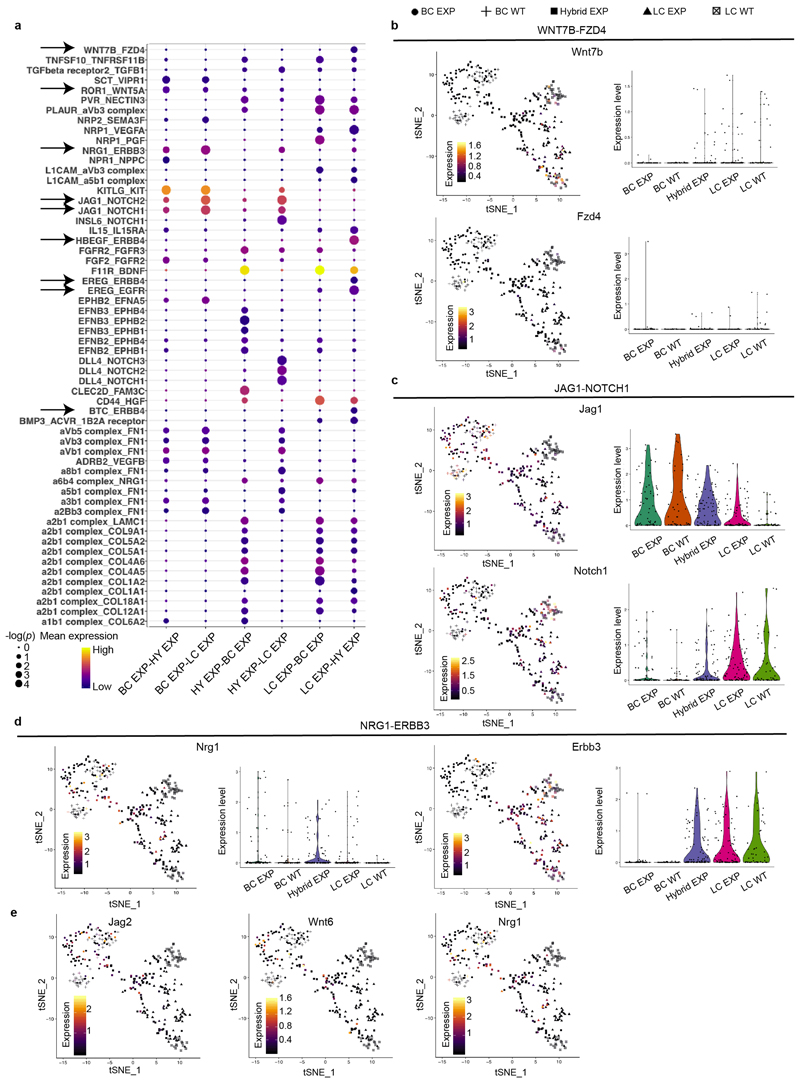
Analysis using CellPhone-DB after LC ablation revealed strong statistical enrichment in the following ligand–receptor pairs: WNT7B–FZD4, ROR1–WNT5A, NRG1–ERBB3, EREG–EGFR, JAG1–NOTCH1 and JAG1–NOTCH2, suggesting that these signalling pathways might regulate BSC multipotency after LC ablation. Bulk and scRNA-seq of BCs, hybrid cells and LCs after ablation showed that Notch ligands (Dll1 and Jag2) that activate Notch signalling and LC fate8,27,34,35, genes encoding members of the Wnt signalling pathway (Wnt6, Fzd7, Axin2) and genes encoding proteins of the EGFR signalling pathway (Ereg, Nrg1, Nrg2)36–40 were upregulated in BCs and hybrid cells after LC ablation compared to their expression under physiological conditions (Fig. 4a–c, Extended Data Fig. 9).
Fig. 4. Notch, Wnt and ErbB signalling pathways regulate BSC multipotency after LC ablation.
a–c, mRNA expression of key genes involved in Notch (a), Wnt (b) and ErbB (c) pathways that were upregulated after LC ablation in BCs and/or hybrid cells (CD29high EpCAMhigh). Results are normalized over wild-type LC expression. Replicates from n = 2 independent experiments. d–f, Quantification of the expression of tdTomato in CD29low EpCAMhigh LCs in mice treated with DOX and with anti-Notch ligand antibodies (d), Wnt inhibitor (e) or anti-ErbB inhibitors (f). Afa, afatinib; sap, sapitinib. The number of mice analysed is shown in parentheses. The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from unpaired two-sided t-tests. g, Quantification of tdTomato+K8+ cells in MG organoids first treated with tamoxifen and then with DOX and the different inhibitors for 72 h. n = 3 independent experiments. The bar height and error bars are mean ±s.e.m., with individual data points shown. P values are derived from ANOVA followed by two-sided Dunnett’s tests.
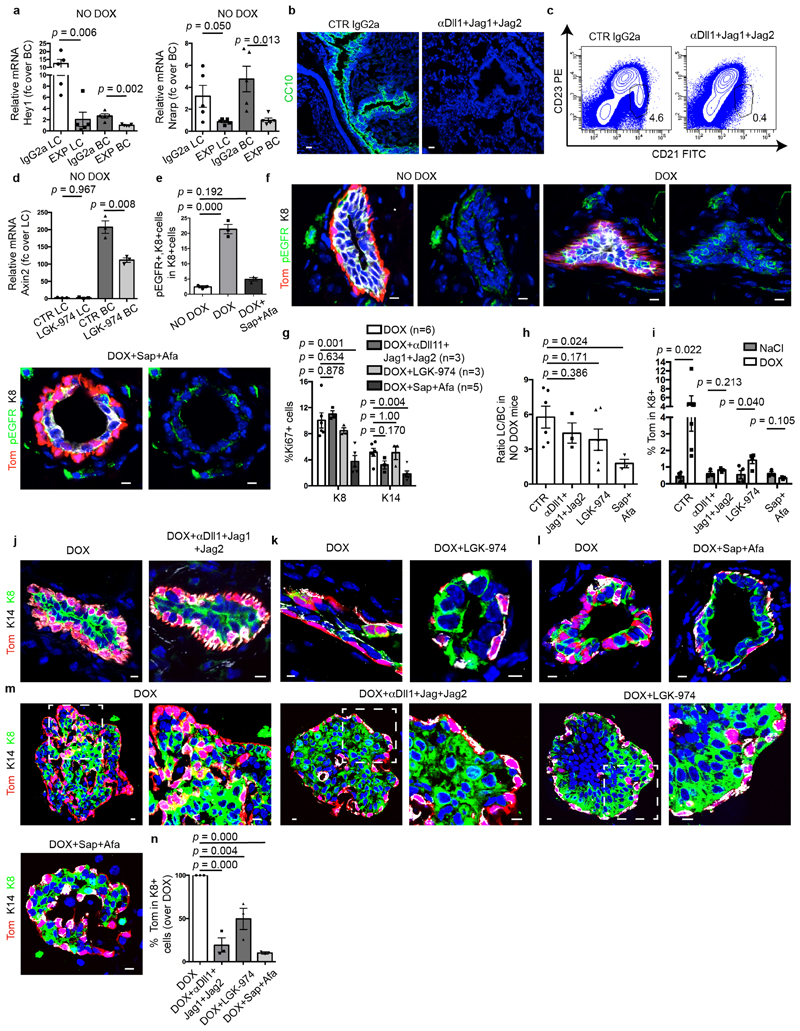
To functionally characterize the role of the Notch, Wnt and EGFR pathways in the activation of BC multipotency, we assessed how pharmacological inhibition of these signalling pathways in the MG in vivo and in organoids in vitro affects BSC fate after LC ablation. Concomitant administration of the antibodies anti-Dll1, anti-Jag1 and anti-Jag241—which inhibit the Notch pathway, as shown by the decrease in expression of the Notch target genes Hey1 and Nrarp—decreased the activation of multipotency in BCs after LC ablation (Fig. 4d, Extended Data Fig. 10a–c, j). These data show that Notch signalling, in addition to regulating LC differentiation during development and cell fate reprogramming after the overexpression of the active form of Notch1 in BCs8,27,34,35, is also required to activate multipotency and generate newly formed LCs from BCs after LC ablation. To assess whether Wnt signalling controls BSC multipotency, we administered LGK-974—an inhibitor of Porcupine that prevents the secretion of Wnt ligands42—to mice and assessed the activation of BC fate transition after LC ablation. Administration of LGK-974 inhibited Wnt signalling, as shown by the decrease in expression of the Wnt target gene Axin2, and decreased de novo formation of LCs from BCs after LC ablation (Fig. 4e, Extended Data Fig. 10d, k). To assess the functional importance of ErbB signalling on BC multipotency, we treated mice with afatinib (an inhibitor of EGFR, ErbB-2 and ErbB-4) and sapitinib (a pan-ErbB inhibitor) and assessed the effect of these inhibitors on BC fate after LC ablation. Administration of afatinib and sapitinib efficiently inhibited the EGFR pathway, as shown by the decrease of phospho-EGFR, and strongly decreased BC multipotency after LC ablation (Fig. 4f, Extended Data Fig. 10e, f, l). In contrast to Notch and Wnt inhibition—which did not affect the proliferation of BCs and LCs—afatinib and sapitinib decreased cell proliferation, in particular of that of LCs, and decreased the proportion of LCs in the MG. Treatment of control (NaCl-injected) glands with the various inhibitors did not activate BC multipotency (Extended Data Fig. 10g–i). These data show that EGFR signalling—mediated by EREG, NRG1 and NRG2—regulates the activation of BC multipotency after LC ablation, possibly by reducing cell proliferation. The inhibition of these different signalling pathways also decreased the activation of BC multipotency after LC ablation in MG organoids in vitro, showing that these inhibitors regulate BC multipotency directly and do not act indirectly through the immune cells or the stromal cells (Fig. 4g, Extended Data Fig. 10 m, n).
Discussion
Lineage ablation of tracheal BSCs has revealed that luminal progenitors that are committed to terminal differentiation can revert back to a BSC state in the absence of BCs and can contribute to tissue repair—this highlights the important plasticity of luminal progenitors and their ability to de-differentiate into BSCs after injuries43. Severe tracheal injuries can lead to the activation of facultative myoepithelial stem cells from the submucosal gland that acquire multipotent BSC-like properties44,45. Our study shows that the regulation of multipotent versus unipotent BC fate and the lineage restriction that occurs in most adult glandular epithelia are regulated by cell–cell communications that control the fate of stem cells, instructing them in real time of the need to generate the different cell lineages required in the tissue. We identify TNF—a cytokine that has previously been shown to contribute to mammary epithelial proliferation, branching morphogenesis and alveolar differentiation46,47—as a key factor secreted by LCs that restricts the multipotency of BSCs under physiological conditions. We also demonstrate the importance of the Notch, Wnt and ErbB signalling pathways for the activation of BSC multipotency upon LC ablation. Further studies will be required to better understand how these signalling pathways restrict or promote BSC multipotency and whether all BCs or only a restricted population of BSCs can reactivate multipotency, and to define the conserved and tissue-specific mechanisms that promote multipotency in the different glandular epithelia. The mechanism that regulates stem cell multipotency in adult glandular tissues is likely to be relevant for tumorigenesis, because reactivation of multipotency4,12,13, cell fate change48 and lineage infidelity49 have all been associated with tumour heterogeneity.
Methods
Mice
The generation of K5CreER and K8rtTA mice was previously described3,50. TetO-DTA mice51 were provided by J. Rajagopal. Rosa-tdTomato mice52 were purchased from the Jackson Laboratory. Mouse colonies were maintained in a certified animal facility in accordance with European guidelines. The experiments were approved by the local ethical committee (CEBEA) under protocols 671N and 673N. The study is compliant with all relevant ethical regulations regarding animal research. Mice were analysed at adult age (over 8 weeks old), as indicated in the figure legends.
Sample size, randomization and blinding
The sample size was chosen based on previous experience in the laboratory, for each experiment to yield high power to detect specific effects. No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Organoid culture
For organoid culture, we used a previously published protocol53. In brief, fat pads of 8–9-week-old virgin female mice were dissected and the lymph nodes removed. Tissues were briefly washed in 70% ethanol and manually chopped into 1 mm3 pieces. The finely minced tissue was transferred to a digestion mix consisting of serum-free Leibovitz’s L15 medium (Gibco) containing 3 mg ml−1 collagenase A (Sigma) and 1.5 mg ml−1 trypsin (Sigma). This was incubated for 1 h at 37 °C to liberate epithelial tissue fragments (‘organoids’). Isolated organoids were mixed with 50 μl of phenol-red-free Matrigel (BD Biosciences) and seeded in 24-well plates. The basal culture medium contained phenol-red-free DMEM/F-12 with penicillin/streptomycin, 10 mM HEPES (Invitrogen), Glutamax (Invitrogen), N2 (Invitrogen) and B27 (Invitrogen). The basal medium was supplemented with Nrg1 (100 ng ml−1, R&D), Noggin (100 ng ml−1, Peprotech) and R-spondin 1 (100 ng ml−1, R&D). Then, 500 μl supplemented basal culture medium was added per well and organoids were maintained in a 37 °C humidified atmosphere under 5% CO2. After one week in culture, mammary organoids were released from the Matrigel by breaking the matrix with a P1000 pipette on ice. After 2–3 passages of washing and centrifugation at 1,500 rpm (140g) for 5 min at 4 °C, mammary cells were resuspended in Matrigel, seeded in 24-well plates and exposed to the previously described culture conditions. To induce the expression of tdTomato, mammary organoids were treated with 1 μM of 4-hydroxytamoxifen for 24 h, and to promote LC ablation the organoids were treated with 10 μg ml−1 of DOX for 48 h. For the treatment of organoids, we added to the medium described above 1 μM LGK-974, 1 μM sapitinib + 1 μM afatinib, and 5 μg ml−1 for the combination of blocking antibodies for Dll1, Jag1 and Jag2. LGK-974 (S7143), sapitinib (AZD8931, S2192) and afatinib (BIBW2992, S1011) were purchased from Selleckchem. Anti-DLL1, anti-J1.b70 and anti-J2.b33 blocking antibodies were provided by the laboratory of C. Siebel (Genentech). Organoids were treated with DOX + inhibitors for 48 h, then the media changed and the organoids kept under treatments for 72 h or for 9 days. CDK1 inhibitor (Merck, RO-3306) was added in the organoid culture medium to a concentration of 1 μM 24 h before DOX administration. The treatments were renewed every 2 days. To study the role of different ligand–receptor couples in restriction of multipotency, we used the following molecules: TNFα blocking antibody 2 μg ml−1 (adalimumab, A2010, Selleckchem), CCR2 antagonist 10 nM (CAS 445479-97-0, Calbiochem, 227016), PDFGR inhibitor 20 nM (crenolanib, S2730, Selleckchem), Activin receptor inhibitor 50 nM (vactosertib, TEW-7197, Selleckchem), ITGβ1 blocking antibody 10 μg ml−1 (LEAF purified anti-mouse/rat CD29 antibody, 102209, BioLegend).
Organoid adenovirus infection
Cultured mammary organoids were collected from Matrigel as mentioned in the section ‘Organoid culture’ by breaking the matrix on ice followed by 2–3 passages of washing and centrifugation at 1,500 rpm (140g) for 5 min at 4 °C. The structured organoids were broken into single cells by pipetting the suspension with 200 μl tips 60 times. After centrifugation at 1,500 rpm (140g) for 5 min at room temperature, the pellet was resuspended with 250 μl Opti-MEM (Gibco). The cell suspension was then mixed with adenovirus CTR or P21 (abm, 000218A) and 5 μg polybrene (Sigma- TR-1003-G), and the mix seeded into a 48-well plate. The plate was covered with Parafilm and centrifuged for 1 h at 600g at 37 °C. After the centrifugation, the Parafilm was removed and the plate incubated for 3 h at 37 °C in humidified atmosphere under 5% CO2. All the mixtures were collected and centrifuged at 600g for 5 min at room temperature. The cell pellet was resuspended with Matrigel and seeded into 24-well plate, with 500 μl cell suspension in each well. Plates were incubated for 15 min in a 37 °C humidified atmosphere under 5% CO2, then the organoids were added to the culture medium. The medium was changed every two days.
Targeting tdTomato expression in the MG, prostate, salivary gland and paw skin
For MG, adult female K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA mice were induced by intraperitoneal (IP) injection with 2.5 mg of tamoxifen per day (diluted in sunflower seed oil, Sigma, T5648) for 6 days. For prostate, salivary gland and paw skin, adult male K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA mice were induced by IP injection with 2.5 mg of tamoxifen per day (diluted in sunflower seed oil, Sigma, T5648) for 5 days.
Administration of DOX in the MG, prostate, salivary gland and paw skin
For MG, 1 week after tamoxifen administration, 0.2 mg per gland DOX diluted in 0.9% NaCl solution (physiological serum) was injected intraductally (IDI)54 in K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA female mice and, to avoid side effects due to the acidity of DOX, the pH was adjusted to 5–5.5. For prostate, salivary gland and paw skin, 1 week after tamoxifen administration, K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA male mice were administered 2 mg DOX per day (diluted in PBS) by IP injection for 5 days. All mice were killed 7 days after the last DOX treatment.
MG whole-mount processing and immunostaining
For MG processing at 8–11 weeks, inguinal glands were dissected and enzymatically digested in HBSS (Gibco) + 300 U ml−1 collagenase (Sigma, C0130) + 300 μg ml−1 hyaluronidase (Sigma, 4272) for 30 min at 37 °C under shaking. Glands were later fixed in 4% paraformaldehyde (PFA) for 2 h at room temperature (RT), followed by three 10-min washes in PBS. For whole-mount (WM) immunostaining, all samples were incubated in blocking buffer (BSA 1%, horse serum (HS) 5%, Triton X 0.8% in PBS) for 3 h at RT. The different primary antibodies were incubated overnight at RT and washed for 1 h at RT with PBS 0.2% Tween20 before incubation with secondary antibodies diluted in blocking buffer at 1:800 for 5 h at RT. The following primary antibodies were used: rabbit anti-K14 (1:800, Thermo), rat anti-E-cadherin (1:800, eBioscience, 14-3249-82). The following secondary antibodies were used: anti-rabbit, anti-rat, conjugated to Alexa Fluor 488 (Molecular Probes) or Cy5 (Jackson ImmunoResearch). Nuclei were stained with a Hoechst solution (1:1,000 in PBS 0.2% Tween20) for 30 min and washed for another hour in PBS 0.2% Tween20 before mounting on slides in DAKO mounting medium supplemented with 2.5% DABCO (Sigma).
Immunofluorescence and immunohistochemistry
Dissected MGs, salivary gland and paw skin were pre-fixed in 4% PFA for 2 h at RT or directly embedded in OCT compound (Tissue Tek) and kept at −80°C. Pre-fixed tissues were washed in PBS, incubated overnight in 30% sucrose in PBS at 4 °C and embedded in OCT and kept at −80°C. Sections of 4 μm were cut using a HM560 Microm cryostat (Mikron Instruments). Non-pre-fixed tissue sections were fixed in PFA 4% for 10 min at RT. Tissue sections were incubated in blocking buffer (BSA 1%, HS 5%, Triton-X 0.2% in PBS) for 1 h at RT. The different primary antibodies were incubated overnight at 4 °C. Sections were then rinsed in PBS and incubated with the corresponding secondary antibodies diluted at 1:400 in blocking buffer for 1 h at RT. The following primary antibodies were used: rat anti-K8 (1:1,000, Troma-I, Developmental Studies Hybridoma Bank, University of Iowa), rabbit or chicken anti-K14 (1:1,000, Thermo), rabbit anti-K5 (1:1,000, PRB-160P-0100, lot number 3260860, Covance), rabbit anti-p63 (1:500, clone EPR5701, ab124762, Abcam), rabbit anti-smMHC (1:100, BT562, lot no 5620305, Biomedical Technologies), rabbit anti-FoxA1 (1:100, clone EPR10881, ab170933, Abcam), rat anti-E-cadherin (1:500, clone DECMA-1, 14-3249-82, eBioscience), rat anti-integrin β1 (1:50, clone MB1.2, MAB1997, lot number 636365, Millipore), rat anti-AQP5 (1:500, AB3559, Millipore), goat anti-TNFα (1:100, AF-410, R&D), rabbit anti-cleaved caspase 3 (1:100, clone 5A1E, 9664S, Cell Signaling), rabbit anti-Ki67 (1:200, ab15580, Abcam), rat anti-CD45 (1:500, clone 30-F11, 553079, BD), rabbit anti-CD68 (1:500, ab125212, Abcam), rabbit anti-snail2 (1:100, clone C19G7, 9585, Cell Signaling), rat anti-TnC (1:500, clone EPR4219, ab108930, Abcam), goat anti-Alcam (1:200, FAB1172A, R&D), goat anti-IL33 (1:100, AF3626, R&D), rabbit anti-pEGFR(Y1092) (1:200, clone EP774Y, ab40815, Abcam). The following secondary antibodies, diluted 1:400, were used: anti-rabbit (A21206), anti-rat (A21208), anti-chicken (A11039) conjugated to Alexa Fluor 488 (Molecular Probes), anti-rabbit (711-295-152), rhodamine Red-X or anti-rabbit (711-605-152), anti-rat (712-605-153), anti-chicken (703-605-155) Cy5 (Jackson ImmunoResearch). For intraductal injection the rat anti-β4 integrin (clone 346-11A, 553745, BD) was used. Nuclei were stained with Hoechst solution (1:2,000) and slides were mounted in DAKO mounting medium supplemented with 2.5% DABCO (Sigma). Dissected lung tissue was pre-fixed in PFA 4% for 2 h. Pre-fixed tissues were washed in PBS, incubated overnight in 30% sucrose in PBS at 4 °C and embedded in OCT and kept at −80°C. Sections of 4 μm were cut using a HM560 Microm cryostat (Mikron Instruments). Tissue sections were incubated in blocking buffer (BSA 1%, HS 5%, Triton-X 0.2% in PBS) for 1 h at RT. The anti-CC10 (1:1,000; sc9772, Santa Cruz) primary antibody was incubated overnight at 4 °C. Sections were then rinsed in PBS and incubated with the anti-goat secondary antibody (A11055) diluted at 1:400 in blocking buffer for 1 h at RT. Nuclei were stained with Hoechst solution (1:2,000) and slides were mounted in DAKO mounting medium supplemented with 2.5% DABCO (Sigma). For paraffin samples, paraffin sections were deparaffinized and rehydrated. The antigen unmasking procedure was performed for 20 min at 95 °C in EDTA (pH 9). Slides were incubated with anti-IL33 antibody at 4 °C overnight, followed by a linker rabbit anti-goat (Abcam 1:250) for 30 min at RT. Finally, slides were incubated with the OmniMap HRP-conjugated anti-rabbit antibody (Ventana) for 30 min. Standard ABC kit, and ImmPACT DAB (Vector Laboratories) were used for the detection of HRP activity. Nuclei staining was done with Mayer’s Haematoxylin (Labonord), followed by dehydration and mounting with SafeMount (Labonord).
Histology and immunostaining on prostate sections
Prostate tissue of adult mice was micro-dissected under a stereoscope to separate the different lobes. Dissected prostates were pre-fixed in 4% PFA for 2 h at RT. After three 5-min washes with PBS, samples were incubated overnight in 30% sucrose in PBS at 4 °C. Tissues were embedded in OCT and kept at −80°C. Sections of 5 μm were cut using a HM560 Microm cryostat (Mikron Instrument). Sections were incubated in blocking buffer (1% BSA, 5% HS, 0.2% TritonX-100 in PBS) for 1 h at RT. Primary antibodies diluted in blocking buffer were incubated overnight at 4 °C. Tissues were rinsed three times in PBS, 5 min each, and incubated with secondary antibodies diluted in blocking buffer for 1 h at RT. Chicken anti-K5 (1:1,000, 905901, Covance) and rat anti-K8 TROMA-1 (1:1,000, Developmental Studies Hybridoma Bank) were used as primary antibodies. To stain proliferating cells, rabbit anti-Ki67 (1:100, ab15580, AbCam) diluted in blocking buffer was incubated for 1 h at RT. To stain apoptotic cells, rabbit anti-cleaved caspase3 (Asp175) (1:150, 9664S, Cell Signaling) diluted in blocking buffer (1% BSA, 5% HS, 0.3% TritonX-100 in PBS) was incubated for 1.5 h at RT. Anti-chicken, anti- rabbit and anti-rat conjugated to Alexa Fluor 488 (1:400, Molecular Probes; A11039, A21206, A21208), and anti-chicken, anti-rabbit and anti-rat conjugated to Cy5 (1:400, Jackson ImmunoResearch; 703-605-155, 711-605-152 712-605-153). Nuclei were stained with Hoechst 33342 dye (Sigma) (diluted 1:1,000 with the secondary antibodies) and slides were mounted in glycergel mounting medium (DAKO) supplemented with 2.5% DABCO (Sigma).
Microscope image acquisition
Confocal images were acquired at RT using a Zeiss LSM780 confocal microscope fitted on an Axiovert M200 inverted microscope equipped with a C-Apochromat (40×, numerical aperture (NA) = 1.2) water immersion objective (Carl Zeiss). Optical sections of 1,024 × 1,024 pixels or 512 × 512 pixels were collected sequentially for each fluorochrome. Images of maps reconstructing the MG whole-mounts were obtained at RT using an EC Plan Neofluar (5×, NA = 0.16). Optical sections of 512 × 512 pixels were collected sequentially for each fluorochrome. The datasets generated were merged and displayed with the ZEN 2 software.
MG whole-mount branching analysis
Maps of the fourth and fifth MG from the contralateral gland of the same mouse (NaCl injected/DOX injected, oestrus not checked) were obtained as described in the section ‘Microscope image acquisition’ and images were analysed with Fiji software. In brief, the entire map was treated in order with flowing plug-ins:3D projection > Split channels > threshold>skeletonize (3D)>analysis skeleton (3D). Only the K14 immunostaining was analysed.
Mammary cell preparation
Adult MGs were dissected and the lymph nodes removed. Tissues were briefly washed in HBSS and chopped in 1 mm3 pieces. Chopped tissues were digested in HBSS + 300 U ml−1 collagenase (Sigma, C0130) + 300 μg ml−1 hyaluronidase (Sigma, 4272) for 2 h at 37 °C under agitation. Physical dissociation using a P1000 pipette was performed every 15 min throughout the enzymatic digestion. EDTA (5 mM) was added for 2 min, followed by 0.25% trypsin-EDTA for 2 min before filtration through a 70-μm mesh, and 2 successive washes in 2% FBS/PBS.
Cell labelling, flow cytometry and sorting
Samples were incubated in 250 μl of 2% FBS/PBS with fluorochrome-conjugated primary antibodies for 30 min, with shaking every 10 min. Primary antibodies were washed with 2% FBS/PBS, and cells were resuspended in 2.5 mg ml−1 DAPI (Invitrogen, D1306) before analysis. The following primary antibodies were used: APC-conjugated anti-CD45 (1:100, clone 30-F11, eBiosciences), APC-conjugated anti-CD31 (1:100, clone 390, eBiosciences), APC-conjugated anti-CD140a (1:100, clone APA5, eBiosciences), PE-Cy7-conjugated anti-CD24 (1:100, clone M1/69, BD Biosciences), FITC-conjugated anti-CD29 (1:100, clone Ha2/5, BD Biosciences), APC-Cy7-conjugated anti-EpCAM (1:100, clone G8.8, BioLegend). Data analysis and cell sorting were performed on a FACSAria sorter using the FACS DiVa software (BD Biosciences). Dead cells were excluded with DAPI; CD45+, CD31+ and CD140a+ cells were excluded (Lin+) before analysis of the tdTomato+ cells.
RNA-seq and analysis of bulk samples
Wild-type LCs (150,000), wild-type BCs (150,000), EXP Tomato LCs (10,000), EXP Tomato BCs (150,000) and EXP hybrid cells (5,000) were isolated by FACS as described in the section ‘Cell labelling, flow cytometry and sorting’ and collected into kit lysis buffer. RNA was extracted with Qiagen RNeasy Micro Kit. RNA quality was checked using a Bioanalyzer 2100 (Agilent Technologies). Indexed cDNA libraries were obtained using the Ovation Solo RNA-Seq System (NuGen) following the manufacturer’s recommendations. The multiplexed libraries (18 pM) were loaded on flow cells and sequences were produced using a NovaSeq 6000 S2 Reagent Kit (200 cycles from a NovaSeq 6000 System (Illumina). Approximately 19 million paired-end reads per sample were mapped against the mouse reference genome (GRCm38/mm10) using STAR software to generate read alignments for each sample. Annotations for Mus_musculus.GRCm38.87.gtf were obtained from ftp.Ensembl.org. After transcripts assembly, gene-level counts were obtained using HTSeq. Genes with expression levels lower than 5 were filtered out. The fold changes of mean gene expression for the duplicates were used to calculate the level of differential gene expression.
GSEA analysis
For Fig. 2, GSEA analysis was performed using ranked fold change of probe expression values between EpCAMhigh CD29high cells and wild-type BCs or wild-type LCs and genes upregulated in BCs or LCs for the displayed dataset55.
Gene ontology analysis of single population signatures
Genes upregulated in each signature were tested for enrichment in each Gene Ontology (GO) class using the DAVID v.6.8 web server56,57. Statistically significant enrichments correspond to those presenting a Benjamini-corrected P value ≤ 0.05, although some genes involved in non-statistically-significant GO classes were plotted for their biological relevance.
scRNA-seq
scRNA-seq was performed using a modified Smartseq-2 protocol58. Lysis buffer (1 μl) in 384-well PCR plates for cell sorting was prepared with 0.4% v/v Triton-X lysis buffer, 2.5 mM dNTPs and 2.5 μM oligo-dT30-VN. Reverse transcription and PCR was performed according to the Smartseq-2 protocol with reduced volumes: 1 μl of reverse transcription mix instead of 5.7 μl and 3 μl PCR Master mix instead of 15 μl. cDNA was amplified for 23 cycles and cleaned using HighPrep PCR beads (MagBio Genomics) at a 0.8× ratio on a Hamilton STAR (Hamilton Germany GmbH) liquid handler, eluted in 30 μl buffer EB (Qiagen) and transferred to 384_PP acoustic plates (LabCyte). DNA quantification was performed with Picogreen assay (Thermo Fisher) and a subset of samples were selected for quality control using an Agilent 2100 BioAnalyser (Agilent Technologies) using a High Sensitivity DNA kit.
Library preparation continued from Smartseq-2
Next-generation sequencing library preparation was performed using a Nextera XT DNA library preparation kit with volumes reduced by one-tenth (Illumina) using an acoustic dispenser. In brief, 100 pg of cDNA in a volume of 500 nl was tagmented by adding 1.5 μl Tn5-buffer mix and incubating for 10 min at 55 °C. Tagmented samples were barcoded with Nextera index sets A–D and amplified with 12 cycles of PCR. After PCR, all samples were pooled and cleaned using HighPrep PCR beads at a 0.6× ratio. Library pools were eluted in buffer EB and quality control was performed using an Agilent 2100 BioAnalyser and High Sensitiviy DNA chip before adjusting to a concentration of 4 nM. The diluted pools were quantified using a KAPA qPCR library quantification kit on a LightCycler 480 (Roche) before a final dilution to 2 nM. The pool of 384 samples was sequenced on 2 lanes of a HiSeq4000 with 1×51bp read length.
Single-cell bioinformatics analysis
Sequencing reads were trimmed for adaptor sequences using cutadapt (v.1.13) and reads were aligned to the GRCm38 reference genome using STAR with default parameters (v.2.5.2b)59. The expression count matrix was generated using HTSeq (v.0.6.0)60 on GENCODE M12 transcript annotations and counts for each protein-coding gene were collapsed. Quality control was performed using the scater R package (v.1.8.0)61. Cells that complied with one of the following conditions were excluded: had fewer than 5 × 104 counts, showed expression of fewer than 3,000 unique genes or had more than 8% counts belonging to mitochondrial sequences. Out of the 384 samples that passed sequencing, 337 passed quality control. Genes for which less than 20 counts were observed across the complete dataset were excluded from further analysis. Read counts were normalized using scran with default parameters (v.1.8.4). t-SNE was performed using the SEURAT62 package (v.2.3.3) RunTSNE function in R, plots were generated using the ggplot2 R package (v.3.0.0). For lineage marker gene discovery, we set the thresholds to all genes with an area under the receiver operating characteristic curve greater than 0.8 and a P value lower than 0.01. BC or LC specific markers were obtained from a previously published paper7. The adjusted proportion of specific markers for each cell was computed by counting the number of specific LC or BC markers over the total number of LC or BC specific markers and correcting for differences in sensitivity by modelling the linear relationship between the adjusted proportion of markers detected and the total number of genes detected for each cell. Gene regulatory network analysis was performed using pySCENIC (v.0.9.3)32 using default parameters. SCENIC59 is a tool to infer gene regulatory networks from single-cell RNA-seq data32.The method consists of three steps: first, modules of genes that are co-expressed with transcription factors (TFs) are identified from the counts matrix by the GENIE3 algorithm63. These modules are then pruned by performing cis-regulatory motif detection on the putative target genes using RcisTarget32, whereby only genes that contain the binding motif for the TF are kept in the module. This set of a TF with its putative target genes is then referred to as a regulon. Finally, the ‘activity’ of each regulon is measured in each cell using a recovery analysis, whereby all genes in a particular cell are ranked from low to high expression and plotted against the number of genes in a particular regulon that can be recovered in that cell. The area under the recovery curve (AUC) is then used as a measure of the activity for the regulon in that cell. Differentially activated regulons were determined by performing a two-sided Kolmogorov–Smirnov non-parametric rank sum test on the regulon AUC values of cells in the various clusters, P values were false discovery rate (FDR)-adjusted using the Benjamini–Hochberg method and regulons with an adjusted P value less than 0.01 were considered differentially activated.
Lineage trajectory inference
Lineage trajectory inference was performed by applying slingshot (v.1.1.0)33 to a diffusion map representation of the single-cell RNA-seq data. The diffusion map was computed using the destiny package64 (v.2.12.0). k-Means clustering (k = 4) was applied to the first two components of the diffusion map embedding to obtain cluster assignments. Lineage trajectories were calculated by slingshot, using the diffusion map and k-means clusters as input. Genes with significantly varying expression along a given trajectory were detected by fitting a generalized additive model (GAM) to the expression data with a LOESS term for the pseudotime variable of the trajectory, using the R package gam (v.1.16, https://CRAN.R-project.org/package=gam). Resulting P values were FDR-adjusted using the Benjamini–Hochberg method and genes with an adjusted P value of less than 0.01 were considered differentially expressed along the lineage trajectory.
CellPhone-DB analysis
The CellPhone-DB method was developed14 to predict cell–cell interaction based on single-cell transcriptomics data in a statistic framework. In brief, a curated database of ligand–receptor interactions was created on the basis of literature searching and known databases. This database is then used to look for enriched expression of a receptor by one cell type and of its corresponding ligand by another cell type. The input data consists of the normalized gene counts matrix and a metadata file containing the cell type label for each cell. First, the cell type labels of all cells are randomly permuted and the mean of the average receptor expression and the average ligand expression for each pair of cell types is calculated. This is repeated 1,000 times to construct a null distribution for each ligand–receptor pair in each pair of cell types. A P value for the likelihood that a given interaction is specific for a certain cell type pair can then be obtained by taking the proportion of means from the null distribution that are greater than or equal to the observed mean. Significant interactions can then be prioritized on the basis of their P value. Biologically relevant interactions can then be chosen on the basis of their average expression level and previous knowledge. The CellPhoneDB database is publicly available at https://www.cellphonedb.org/ and the software package to perform the statistical tests is available at https://github.com/Teichlab/cellphonedb. We ran CellPhoneDB on our Smart-seq2 dataset, split between wild-type and ablation conditions, using default parameters as described above. However, because the database currently only contains information on human receptor–ligand interactions, the mouse genes from our dataset were first converted to their human orthologues before analysis.
RNA extraction and quantitative real-time PCR
The protocol used for RNA extraction on FACS-isolated cells has been previously described12. In brief, RNA extraction was performed using the RNeasy micro kit (Qiagen) according to the manufacturer’s recommendations and DNase treatment. After nanodrop RNA quantification and analysis of RNA integrity, purified RNA was used to synthesize the first-strand cDNA in a 50 ml final volume, using Superscript II (Invitrogen) and random hexamers (Roche). Genomic contamination was detected by performing the same procedure without reverse transcriptase. Quantitative PCR analyses were performed with 1 ng of cDNA as template, using FastStart Essential DNA green master (Roche) and a Light Cycler 96 (Roche) for real-time PCR system. Relative quantitative RNA was normalized using the housekeeping geneGapdh. Primers were designed using the PrimerBank database (http://pga.mgh.harvard.edu/primerbank/) and are listed in Supplementary Table 1. Analysis of the results was performed using Light Cycler 96 software (Roche) and relative quantification was performed using the ddCt method using HPRT as a reference.
Adalimumab (anti-TNF) intraductal injection
For intraductal injection of adalimumab (A2010, Selleckchem), mice were prepared as described54. The controlateral fourth gland of the same mouse K5CreER/Rosa-tdTomato was injected either with PBS or with 0.15 mg of blocking antibody. Mice were euthanized after one week. To check the antibody penetration into the basal layer, the fourth gland of CD1 mice was injected intraductally with 15 μl of rat anti-β4 integrin (clone 346-11A, 553745, BD Biosciences), and after 2 days the gland was stained with anti-rat conjugated to Alexa Fluor 488 secondary antibody. Tissue sections were fixed in 4% PFA for 10 min, rinsed in PBS, and then incubated in blocking buffer (BSA 1%, HS 5%, Triton-X 0.2% in PBS) for 1 h at RT. The anti-rat secondary antibody (1:500, Molecular Probes, A21208) was incubated overnight at 4 °C. Sections were then rinsed in PBS and nuclei were stained with Hoechst solution (1:2,000). Slides were mounted in DAKO mounting medium supplemented with 2.5% DABCO (Sigma).
Blocking antibodies and small-molecule treatments
Anti-DLL1, anti-J1.b70 and anti-J2.b33 blocking antibodies were provided by the laboratory of C. Siebel (Genentech). During treatment with blocking antibodies, K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA mice were injected intraperitoneally for the first time with 15 mg kg−1 of each blocking antibody or 30 mg kg−1 of control antibody two days before intraductal DOX administration, and the second time with 7.5 mg kg−1 of each blocking antibody or 15 mg kg−1 of control antibody two days after intraductal DOX administration. LGK-974 (catalogue number S7143), sapitinib (AZD8931, S2192) and afatinib (BIBW2992, S1011) were purchased from Selleckchem. LGK-974 was dissolved in 0.5% methylcellulose (Sigma-Aldrich, M0430), 0.4% Tween 80 (Sigma-Aldrich, P8074) solution in distilled water. During LGK-974 treatment, K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA mice received daily for 9 days 10 mg kg−1 LGK-974 by oral gavage, the IDI of DOX was performed on the third day of treatment and mice were killed 7 days after DOX administration. Sapitinib was dissolved in 1% Tween 80 solution in distilled water and afatinib in 0.5% methylcellulose, 0.4% Tween 80 solution in distilled water. During sapitinib and afatinib treatment, mice received daily for 8 days 20 mg kg−1 sapitinib and 10 mg kg−1 afatinib by oral gavage, the intraductal injection of DOX was performed on the third day of treatment and mice were killed 7 days after. For both treatments, control K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA mice received 0.5% methylcellulose, 0.4% Tween 80 solution in distilled water by oral gavage for 9 days.
Statistics and reproducibility
All experiments were repeated at least three times with similar results unless a different number of repeats is stated in the legend or in the figure. Two-tailed Student’s t-tests were performed using GraphPad Prism v.6.0 (GraphPad Software). ANOVA was followed in case of statistical significance by two-sided Dunnett’s tests; many-to-one comparisons were performed using IBM-SPSS v.26.0 statistical software (IBM). P and n values are indicated in the figure legends or in the figures themselves. No statistical method was used to predetermine sample size. All experimental mice used in this study were females or males of mixed genetic backgrounds. No mice were excluded from the study. No method of randomization was used. The investigators were not blinded to allocation during experiments or outcome assessment.
Extended Data
Extended Data Fig. 1. FACS analysis of MG epithelial cells.
a–d, Unicellular suspension of MG cells from adult K5CreER/tdTomato/K8rtTA/TetO-DTA mice induced at 8 weeks stained for endothelial, immune and fibroblast markers (Lin+) (CD31, CD45, CD140a) in APC, EpCAM in Apc-Cy7 and CD29 in FITC, were gated to eliminate debris (a), doublets were discarded with gates (b), the living cells were gated by DAPI dye exclusion (c) and the non-epithelial Lin+ cells were discarded (d). CD24/CD29 and EpCAM/CD29 expression was studied in Lin−. e–g, CD24/CD29 gates, EpCAM/CD29 gates and the absence of leakiness of fluorescent reporter recombination before tamoxifen injection (e), after tamoxifen injection (f) and after DOX injection (g). The CD29low CD24high and the CD29low EpCAMhigh gate corresponds to LCs, the CD29high CD24low and the CD29high EpCAMlow gate corresponds to BCs, the CD29high EpCAMhigh correspond to the new population described in this manuscript. A total of 400,000 events is shown in the graphs. Percentages are calculated on the total LCs and BCs or LCs, CD29high EpCAMhigh and BCs shown. n = 3 independent experiments per condition. h, Graph showing the percentage of BCs that were tdTomato-labelled on the total number of BCs gated on CD24/CD29 before DOX injection (79.5 ± 2.2%; n = 20 mice, data are mean ± s.e.m.) after DOX injection (CD24 83.9 ± 2.6%; n = 23 mice, data are mean ± s.e.m.) and EpCAM/CD29 before DOX injection (84.8 ± 2.2%; n = 20 mice, data are mean ± s.e.m.) and after DOX injection (88.9 ± 1.9%; n = 23 mice, data are mean ± s.e.m.). P values are derived from unpaired two-sided t-tests.
Extended Data Fig. 2. DOX administration to K5CreER/tdTomato/K8rtTA/TetO-DTA mice promotes LC death, MG remodelling and proliferation.
a, Scheme summarizing the histology of the MG and its different lineages. b, c, FACS plot of CD29/CD24 expression in Lin−tdTomato+ epithelial cells (b) and quantification of tdTomato+ LC (c) in CTR mice and 1 week after DOX administration. The percentage of the gated population from all epithelial cells is shown. Number of mice analysed is shown in parentheses. P values are derived from unpaired two-sided t-test. d, e, Confocal imaging (d) and FACS quantification (e) of immunostaining for tdTomato, cleaved caspase 3 (CC3) and K8 in K5CreER/tdTomato/K8rtTA/TetO-DTA MG after IDI with NaCl (0.29 ± 0.02 CC3+ cells) or 0.2 mg DOX (1.3 ± 0.4 CC3+ cells) and chased for 12 h (n = 6 mice per condition). Scale bar (d), 5 μm. P values are derived from unpaired two-sided t-tests. f–j, Scheme of the different parameters measured to quantify MG remodelling (f), quantification and representation after ImageJ analysis of branch length, number of branches, branch points and terminal end buds of K5CreER/Rosa-tdTomato/K8rtTA/tet-O-DTA (g, h) or K5CreER/Rosa-tdTomato/K8rtTA contralateral MG (i, j) injected either with NaCl or DOX. Number of mice analysed is shown in parentheses. Scale bars, 100 μm. P values are derived from paired two-sided t-tests. k, Confocal imaging of immunostaining for tdTomato, E-cadherin and K14 of terminal end buds, branching points and ducts where LC replacement is occurring in a patchy and focal manner throughout the gland (n = 3 mice). Scale bars, 20 μm. l, m, Graphs showing the percentage of K8+ CC3+ cells at 0 h, 12 h and 24 h (n = 3 mice per condition) after DOX injection (l) and FACS quantification of K8+ CC3+ cells 12 h after different DOX doses (m). Number of mice analysed is shown in parentheses. n, o, Confocal imaging of immunostaining for tdTomato, K8 and CC3 (n) and FACS quantification of K8+tdTomato+ cells (o) in mice after intraductal injection with NaCl or DOX at different doses after 12 h. Number of mice analysed is shown in parentheses. p, q, Confocal imaging (p) and quantification (q) of immunostaining for tdTomato, Ki67 and K8 in K5CreER/tdTomato/K8rtTA/TetO-DTA MG after IDI with NaCl or 0.2 mg DOX and chased for 1 week. Scale bars, 5 μm. n = 3 mice per condition. P values are derived from unpaired two-sided t-tests. Data are mean ± s.e.m. For the immunofluorescence data, Hoechst nuclear staining is shown in blue.
Extended Data Fig. 3. DOX-induced DTA in LCs promotes inflammation, LC death, proliferation and activation of BC multipotency in MG in vivo and organoids in vitro.
a, b, Confocal imaging of immunostaining for tdTomato, CD68 and CD45 (a) and FACS quantification of CD45+ CD68+ cells in MG cells from K5CreER/Rosa-tdTomato/K8rtTA/tet-O-DTA CTR mice or DOX or DOX+DEX treated (DEX, dexamethasone) (b). n = 3 mice. P values are derived from unpaired two-sided t-tests. c, qRT–PCR analysis of the whole MG of K5CreER/Rosa-tdTomato/K8rtTA/tet-O-DTA mice CTR, DOX, No DOX+DEX or DOX+DEX (n = 3 mice for each group), showing the upregulation of inflammation markers upon LC ablation and its normalization after DEX treatment. d, FACS quantification of tdTomato+ LCs in mice after DOX or DOX+DEX treatment (n = 5 mice). P values are derived from unpaired two-sided t-tests. e, f, Representative FACS plot of CD29/EpCAM expression in Lin− YFP+ epithelial cells of MG from K14rtTA/TetO-DTA/K8CreER/Rosa-YFP mice in CTR or one week after IDI of NaCl or DOX (e) and quantification of YFP+ cells in LCs and BCs (f) (n = 5 mice). g, h, Confocal imaging (g) and quantification of immunostaining (h) for tdTomato, CC3 and K8 in K5CreER/tdTomato/K8rtTA/TetO-DTA MG organoids without DOX (CTR) or 6 h after DOX. n = 4 independent experiments. P values are derived from unpaired two-sided t-tests. i, j, Confocal imaging and quantification of immunostaining for tdTomato, Ki67 and K8 in K5CreER/tdTomato/K8rtTA/TetO-DTA in organoids without DOX (CTR) or 24 h after DOX. n = 4 independent experiments. P values are derived from unpaired two-sided t-tests. k, Confocal imaging of immunostaining for tdTomato, K8 and K14 in K5CreER/tdTomato/K8rtTA/TetO-DTA CTR organoids or after DOX. Lower magnification of the images shown in Fig. 1d. n = 3 independent experiments. l, m, Confocal imaging (l) and quantification (m) of immunostaining for tdTomato, Ki67 and K8 in K5CreER/tdTomato/K8rtTA/TetO-DTA organoids treated with DOX, DOX+adeno-P21 or DOX+CDK1 inhibitor (CDK1-i) for 48 h and chased for 24 h. n, Quantification of tdTomato+ K8+ cells in K5CreER/tdTomato/K8rtTA/TetO-DTA organoids treated with DOX, DOX+adeno-P21 or DOX+CDK1 inhibitor. For l–n, n = 3 independent experiments. P values are derived from ANOVA followed by two-sided Dunnett’s tests. Data are mean ± s.e.m. For the immunofluorescence data, Hoechst nuclear staining is shown in blue. Scale bars, 10 μm.
Extended Data Fig. 4. DOX administration in adult K5CreER/tdTomato/K8rtTA/TetO-DTA mice promotes LC death and proliferation in the prostate, salivary gland and sweat gland.
a, Schematic representation of the histology of the salivary gland and its lineages. b, c, Confocal imaging (b) and quantification (c) of immunostaining for tdTomato, AQP5 and smooth muscle heavy chain myosin (SMH) of salivary gland acinar cells. n = 3 mice per condition. P values are derived from unpaired two-sided t-tests. d, e, Confocal imaging (d) and quantification (e) of immunostaining for tdTomato, K8 and SMH in the granulated duct showing the contribution of BCs to replenishing granulated ductal cells during homeostasis. n = 3 mice per condition. P values are derived from unpaired two-sided t-tests. f, Schematic representation of the histology of prostate and its lineages. g–j, Confocal imaging (g) and quantification (h) of prostate immunostaining for tdTomato, K8 and CC3 or Ki67 in CTR or K5CreER/tdTomato/K8rtTA/TetO-DTA mice treated with DOX for 5 days and chased for 1 day. P values are derived from unpaired two-sided t-tests. n = 3 mice per condition. k, Schematic representation of the histology of the sweat gland and its different lineages. l–o, Confocal imaging (l, n) and quantification (m, o) of sweat gland immunostaining for tdTomato, K8 and CC3 (l, m) or tdTomato, K8 and Ki67 (n, o) in CTR or K5CreER/tdTomato/K8rtTA/TetO-DTA mice treated with DOX for 5 days and chased for 1 day. P values are derived from unpaired two-sided t-tests. n = 3 mice per condition. p–s, Confocal imaging (p, r) and quantification (q, s) of the intercalated duct of salivary gland immunostaining for tdTomato, K8 and CC3 (p, q) or tdTomato, K8 and Ki67 (r, s) in CTR or K5CreER/tdTomato/K8rtTA/TetO-DTA mice treated with DOX for 5 days and chased for 1 day. n = 4 for CC3 and n = 3 for Ki67 mice per condition. P values are derived from unpaired two-sided t-tests. Data are mean ± s.e.m. For the immunofluorescence data, Hoechst nuclear staining is shown in blue. Scale bars, 5 μm.
Extended Data Fig. 5. Transcriptional profiling associated with BC multipotency induced by LC ablation.
a, b, Gene ontology (GO) analysis of genes upregulated more than 2.5-fold in CD29high EpCAMhigh cells compared to LCs (a) or BCs (b). Histograms represent –log10 of the Benjamini P value. n = 2 independent experiments. c, d, Graphs representing mRNA expression measured by bulk population RNA sequencing of basal (c) and luminal (d) upregulated genes in FACS-isolated wild-type BCs, LC EXP, BC EXP, CD29high EpCAMhigh EXP. Fold change over wild-type LCs (c) or over wild-type BCs (d) of genes involved in different biological processes. n = 2 independent experiments. e, Confocal images of MGs from CTR mice or mice with IDI DOX.Immunostaining for K14, Snail2 and K8. f, Immunohistochemistry of Il33 in CTR MGs or MG after DOX IDI. g, Immunostaining for tdTomato, TnC and K14. h, Masson’s trichrome staining for collagen in CTR MGs or MG after DOX IDI. i, Immunostaining for tdTomato, Alcam and K14. n = 3 mice per condition. Hoechst nuclear staining in blue. Scale bars, 5 μm.
Extended Data Fig. 6. Single-cell transcriptional profiling reveals the hybrid features of CD29high EpCAMhigh cells associated with LC-ablation-induced BC multipotency.
a Heat map of normalized scRNa-seq expression. Rows represent marker genes (ROC AUC >0.7 and log fold change >0.25) for clusters discovered through unsupervised clustering (resolution = 0.2) and columns represent individual cells, grouped by lineage and treatment condition. Colour values in the heat map represent normalized and scaled expression values. These data show the hybrid basal and luminal signature of the CD29high EpCAMhigh cells (Hybrid EXP) after LC ablation. b, t-SNE plots illustrating the expression of markers for BC and LC lineages (n = 337 cells). c, Graph showing the mRNA expression (read per million) of Tspan8 and Procr. n = 2 independent experiments. d, t-SNE and violin plots illustrating the expression for Tspan8. These data show that Tspan8 is expressed at similar levels in the different MG populations under physiological conditions and after LC ablation (n = 337 cells). e, t-SNE and violin plots illustrating the expression for Procr. These data show that Procr is expressed at very low levels in the different MG populations under physiological conditions and is not increased after LC ablation (n = 337 cells). For t-SNE plots, data points represent individual cells, the colour scaling represents the expression level of the respective marker gene (high, yellow; low, black). For violin plots the minima, maxima, centre and percentiles are provided in the Source Data. f, g, t-SNE plots for differentially activated regulons (FDR-corrected P value <0.01 from one-sided Kolmogorov–Smirnov test) as computed by SCENIC, data points represent individual cells (n = 337), the colour scaling represents the regulon AUC value as a measure of the number of transcription factor target genes being expressed (high yellow, low black).
Extended Data Fig. 7. Lineage trajectory inference using Slingshot.
a, b, Diffusion maps showing normalized expression of Krt14 (a, BC) and Krt8 (b, LC). c, Heat map representing the top 100 most significant (lowest adjusted P value) genes differentially expressed along trajectory 1 (n = 337 cells). FDR-corrected P values from F-test on Loess-term for pseudotime in the GAM fit. d, Diffusion maps showing normalized expression of markers for ER+ LC (Foxa1, Ly6a/Sca1 and Pgr). e, Heat map representing the top 100 most significant (lowest adjusted P value) genes differentially expressed along trajectory 2 (n = 337 cells). FDR-corrected P values from F-test on Loess-term for pseudotime in the GAM fit. f, Diffusion maps showing normalized expression of markers for ER- LC (Elf5, Hey1 and Kit). For the diffusion maps the colour bars represent normalized expression levels, going from black (low expression) to yellow (high expression). For the heat maps rows represent the genes, ordered by hierarchical clustering. Colours correspond to the Z-score scaled expression of a gene. Columns represent cells, which are ordered by their pseudotime value in the respective trajectory and their types (BC, hybrid or LC) in control or experimental condition (wild-type or EXP) are indicated at the top of the heat map.
Extended Data Fig. 8. Immunostaining of basal and luminal markers after LC ablation and antibody permeability.
a–f, Confocal imaging of immunostaining for E-cadherin, K5 and tdTomato (a–c) or E-cadherin, K14 and tdTomato (d–f) in CTR (a, d) or K5CreER/tdTomato/K8rtTA/TetO-DTA mice after IDI of DOX and chased for 1 week (b, e) or 2 weeks (c, f) (n = 3 mice per condition). g, Quantification of the proportion of hybrid K8+ K14+tdTomato+ cells and K8+tdTomato+ cells on the total K8+tdTomato+ cells at 1 week (n = 7 mice), 2 weeks (n = 4 mice), 4 weeks (n = 3 mice) and 7 weeks (n = 3 mice) after DOX treatment. The P value between 1 week and 7 weeks, derived from ANOVA followed by two-sided Dunnett’s test, is shown. h, i, Confocal imaging of immunostaining for K14, p63 and K8 (h) and quantification of p63+ cells in K8+ K14+ cells (i) in CTR or K5CreER/tdTomato/K8rtTA/TetO-DTA mice after DOX IDI and chased for 1 week. n = 3 mice per condition. j, k, Confocal imaging of immunostaining for K14, Foxa1 and K8 (j) and quantification of Foxa1+ cells in K8+ K14+ cells (k) in CTR or K5CreER/tdTomato/K8rtTA/TetO-DTA mice after DOX IDI and chased for 1 week. n = 3 mice per condition. l, Confocal imaging of immunostaining for K14 and anti-rat Alexa Fluor 488 (green) in CTR MG not injected or MG after IDI of rat anti-β4 integrin (CD104) and chased for 2 days. n = 3 mice per condition. Data are mean ± s.e.m. For the immunofluorescence data, Hoechst nuclear staining is shown in blue. Scale bars, 5 μm.
Extended Data Fig. 9. CellPhone-DB and scRNA-seq on the heterotypic interaction significant upon LC ablation.
a, Graph representing the mean expression and P values of the heterotypic ligand–receptor interaction significant only after LC ablation compared to the wild type condition (n = 242 cells). P values are derived from a one-sided permutation test. b–d, t-SNE and violin plots illustrating the expression for the ligand–receptor couples WNT7B–FZD4 (b), JAG1–NOTCH1 (c) and NRG1–ERBB3 (d) in the different cell populations and experimental conditions (n = 337 cells). For violin plots the minima, maxima, centre and percentiles are provided in Source Data. e, t-SNE plots illustrating the expression of Jag2, Wnt6 and Nrg1 (n = 337 cells). Data points represent individual cells. The colour scaling represents the expression level of the respective marker gene (high, yellow; low, black).
Extended Data Fig. 10. Effect of administration of anti Dll1, Jag1 and Jag2 blocking antibodies; LGK-974; sapitinib and afatinib on MG and MG organoids.
a, qRT–PCR analysis of Notch target gene Hey1 and Nrarp of FACS-isolated LCs and BCs of K5CreER/tdTomato/K8rtTA/TetO-DTA mice treated with IgG2a (CTR Ab) or anti-Dll1, Jag1, Jag2 blocking antibodies (n = 5 mice per condition), showing the efficient inhibition of Notch target genes after blocking antibody administration. P values are derived from unpaired two-sided t-tests. b, Immunostaining for CC10 in lung sections of K5CreER/tdTomato/K8rtTA/TetO-DTA mice treated with CTR IgG2a or anti-Dll1, Jag1 and Jag2 blocking antibodies, showing the disappearance of the goblet cells demonstrating the efficiency of Notch inhibition (n = 3 mice per condition). c, Representative FACS plot of CD21/CD23 in CD45+ CD5− spleen cells in mice treated with CTR IgG2a or anti-Dll1, Jag1 and Jag2 blocking antibodies showing the disappearance of the marginal zone B cells (CD21high CD23−) demonstrating the efficiency of Notch inhibition (n = 3 mice per condition). d, qRT–PCR analysis of Wnt target gene Axin2 of FACS-isolated LCs and BCs of K5CreER/tdTomato/K8rtTA/TetO-DTA CTR mice or treated with LGK-974, showing the efficient inhibition of Wnt target genes by LGK-974 (n = 3 mice per condition). P values are derived from unpaired two-sided t-tests. e, f, Confocal imaging (e) and quantification (f) of immunostaining for tdTomato, phospho-EGFR (p-EGFR) and K8 in CTR and K5CreER/tdTomato/K8rtTA/TetO-DTA MG after DOX IDI and chased for 1 week and mice after DOX IDI and treated with sapitinib (Sap) and afatinib (Afa) and chased for 1 week (n = 3 mice per condition). P values are derived from ANOVA followed by two-sided Dunnett’s tests. g, Quantification of the expression of Ki67 in MG after IDI of DOX in the absence or in the presence of different inhibitors. The number of independent mice analysed is shown in parentheses. P values are derived from ANOVA followed by two-sided Dunnett’s test. h, FACS analysis of the LC/BC ratio of MG in control mice (n = 6 mice); mice treated with anti-Notch ligand antibodies (n = 3 mice); with LGK-974 (n = 4 mice) and anti-ErbB inhibitors (n = 3 mice) for 9 days showing that the inhibition of Notch or Wnt signalling does not affect the ratio between LCs and BCs whereas Erbb inhibitors decreased the proportion of LCs. P values are derived from ANOVA followed by two-sided Dunnett’s test. i, FACS analysis of MG showing tdTomato+ cells in LCs after NaCl IDI (n = 4 mice); DOX IDI (n = 5 mice); NaCl or DOX IDI and anti-Notch ligand antibodies (n = 3 mice); NaCl or DOX IDI and Wnt inhibitor (n = 4 mice); NaCl or DOX IDI and anti-ErbB inhibitors (n = 3 mice); for 9 days. P values are derived from unpaired two-sided t-tests. j–m, Confocal imaging of immunostaining for tdTomato, K8 and K14 of MG (j–l) and organoids (m) after DOX or DOX+anti-Notch ligand antibodies (j); DOX+Wnt inhibitor (k); DOX+sapitinib/afatinib (l). n, Quantification of the expression of tdTomato in K8+ cells in MG organoids treated with DOX, DOX+anti-Notch ligand antibodies, DOX+LGK-974 or DOX+sapitinib/afatinib for 48 h and chased for 10 days with media and the corresponding inhibitor. Data are normalized over DOX. For m, n, n = 3 independent experiments. Data are mean ± s.e.m. P values are derived from ANOVA followed by two-sided Dunnett’s test. Data are mean ± s.e.m. For the immunofluorescence data, Hoechst nuclear staining is shown in blue. Scale bars, 5 μm.
Supplementary Material
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Acknowledgements
We thank the ULB animal facility and ULB genomic core facility (F. Libert and A. Lefort); J.M.Vanderwinden and LiMif for the help with confocal microscopy, J. Rajagopal and P. R. Tata for their suggestions concerning the TetO-DTA mice; and B. Lloyd-Lewis for help with the organoids culture. This work was supported by the ERC and the FNRS. A.C. is supported by the FNRS/FRIA. S.L. is a long-term EMBO fellow. C.B. is supported by WELBIO, FNRS, TELEVIE, Fond Erasme, FondationContre le Cancer, ULB Foundation, European Research Council, the Foundation BailletLatour. A.Sifrim., J.V.H. and T.V. are supported by KULeuven (SymBioSys – C14/18/092), the FondationContre le Cancer (2015-143), and FWO postdoctoral fellowships 12W7318N and I001818N.
Footnotes
Author contributions A.C., S.L. and C.B. designed the experiments and performed data analysis. A.C. and S.L. performed most of the biological experiments. E.T. performed the experiments and data analysis on prostate glands. A.Sifrim., M.M., Y.S., J.V.H. and T.V. performed the bioinformatic analysis. A.D. and G.B. provided technical help. C.D. performed FACS experiments. N.D. provided technical help with single-cell RNA sequencing. A.C., S.L., M.F., A.W. and A.V.K. performed immunostainings, blocking antibodies and small-molecule treatments and experiments with follow-up mice. A.Sahay contributed genetic tools. V.d.M. performed statistical analysis. C.W.S. provided the Notch antibodies. A.C., A.V.K. and C.B. wrote the manuscript. All authors read and approved the final manuscript.
Competing interestsThe authors declare no competing interests.
Additional information
Supplementary information is available for this paper at
Peer review information Nature thanks Jacco van Rheenen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Online content Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-020-2632-y
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
Bulk RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE127975. Single-cell RNA-seq data have been deposited in the Gene Expression Omnibus under accession number GSE148791. Data supporting the findings of this study are available within the article. Source data are provided with this paper.
References
- 1.Lloyd-Lewis B, Harris OB, Watson CJ, Davis FM. Mammary stem cells: premise, properties, and perspectives. Trends Cell Biol. 2017;27:556–567. doi: 10.1016/j.tcb.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Toivanen R, Shen MM. Prostate organogenesis: tissue induction, hormonal regulation and cell type specification. Development. 2017;144:1382–1398. doi: 10.1242/dev.148270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 4.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ousset M, et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- 6.Lu CP, et al. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell. 2012;150:136–150. doi: 10.1016/j.cell.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wuidart A, et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat Cell Biol. 2018;20:666–676. doi: 10.1038/s41556-018-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lilja AM, et al. Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat Cell Biol. 2018;20:677–687. doi: 10.1038/s41556-018-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 10.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 11.Prater MD, et al. Mammary stem cells have myoepithelial cell properties. Nat Cell Biol. 2014;16:942–950. doi: 10.1038/ncb3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 13.Koren S, et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature. 2015;525:114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- 14.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon OJ, Zhang L, Ittmann MM, Xin L. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc Natl Acad Sci USA. 2014;111:E592–E600. doi: 10.1073/pnas.1318157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JJ, Shen MM. Prostate stem cells and cancer stem cells. Cold Spring Harb Perspect Med. 2019;9 doi: 10.1101/cshperspect.a030395. a030395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toivanen R, Mohan A, Shen MM. Basal progenitors contribute to repair of the prostate epithelium following induced luminal anoikis. Stem Cell Reports. 2016;6:660–667. doi: 10.1016/j.stemcr.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton C, Liu Y, Yu C, Xie Q, Wang ZA. Luminal-contact-inhibition of epithelial basal stem cell multipotency in prostate organogenesis and homeostasis. Biol Open. 2019;8 doi: 10.1242/bio.045724. bio045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emmerson E, Knox SM. Salivary gland stem cells: A review of development, regeneration and cancer. Genesis. 2018;56:e23211. doi: 10.1002/dvg.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng PL, Aure MH, Maruyama T, Ovitt CE. Limited regeneration of adult salivary glands after severe injury involves cellular plasticity. Cell Rep. 2018;24:1464–1470.e3. doi: 10.1016/j.celrep.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May AJ, et al. Diverse progenitor cells preserve salivary gland ductal architecture after radiation-induced damage. Development. 2018;145 doi: 10.1242/dev.166363. dev166363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal B, et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01560-x. 1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach K, et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02001-5. 2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraddi RR, et al. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep. 2018;24:1653–1666.e7. doi: 10.1016/j.celrep.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurita T, Medina RT, Mills AA, Cunha GR. Role of p63 and basal cells in the prostate. Development. 2004;131:4955–4964. doi: 10.1242/dev.01384. [DOI] [PubMed] [Google Scholar]
- 26.Signoretti S, et al. p63 regulates commitment to the prostate cell lineage. Proc Natl Acad Sci USA. 2005;102:11355–11360. doi: 10.1073/pnas.0500165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yalcin-Ozuysal O, et al. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 2010;17:1600–1612. doi: 10.1038/cdd.2010.37. [DOI] [PubMed] [Google Scholar]
- 28.Bernardo GM, et al. FOXA1 is an essential determinant of ERα expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi YS, Chakrabarti R, Escamilla-Hernandez R, Sinha S. Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev Biol. 2009;329:227–241. doi: 10.1016/j.ydbio.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 30.Fu NY, et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat Cell Biol. 2017;19:164–176. doi: 10.1038/ncb3471. [DOI] [PubMed] [Google Scholar]
- 31.Wang D, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- 32.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buono KD, et al. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 35.Bouras T, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 36.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 37.van Genderen C, et al. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 38.Chu EY, et al. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- 39.Forster N, et al. Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev Cell. 2014;28:147–160. doi: 10.1016/j.devcel.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balko JM, et al. The receptor tyrosine kinase ErbB3 maintains the balance between luminal and basal breast epithelium. Proc Natl Acad Sci USA. 2012;109:221–226. doi: 10.1073/pnas.1115802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lafkas D, et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature. 2015;528:127–131. doi: 10.1038/nature15715. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci USA. 2013;110:20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tata PR, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lynch TJ, et al. Submucosal gland myoepithelial cells are reserve stem cells that can regenerate mouse tracheal epithelium. Cell Stem Cell. 2018;22:653–667.e5. doi: 10.1016/j.stem.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tata A, et al. Myoepithelial cells of submucosal glands can function as reserve stem cells to regenerate airways after injury. Cell Stem Cell. 2018;22:668–683.e6. doi: 10.1016/j.stem.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varela LM, Ip MM. Tumor necrosis factor-alpha: a multifunctional regulator of mammary gland development. Endocrinology. 1996;137:4915–4924. doi: 10.1210/endo.137.11.8895364. [DOI] [PubMed] [Google Scholar]
- 47.Lee PP, Hwang JJ, Murphy G, Ip MM. Functional significance of MMP-9 in tumor necrosis factor-induced proliferation and branching morphogenesis of mammary epithelial cells. Endocrinology. 2000;141:3764–3773. doi: 10.1210/endo.141.10.7697. [DOI] [PubMed] [Google Scholar]
- 48.Youssef KK, et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat Cell Biol. 2012;14:1282–1294. doi: 10.1038/ncb2628. [DOI] [PubMed] [Google Scholar]
- 49.Ge Y, et al. Stem cell lineage infidelity drives wound repair and cancer. Cell. 2017;169:636–650.e14. doi: 10.1016/j.cell.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watson JK, et al. Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Rep. 2015;12:90–101. doi: 10.1016/j.celrep.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber T, et al. Inducible gene expression in GFAP+ progenitor cells of the SGZ and the dorsal wall of the SVZ—a novel tool to manipulate and trace adult neurogenesis. Glia. 2011;59:615–626. doi: 10.1002/glia.21129. [DOI] [PubMed] [Google Scholar]
- 52.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jardé T, et al. Wnt and Neuregulin1/ErbB signalling extends 3D culture of hormone responsive mammary organoids. Nat Commun. 2016;7 doi: 10.1038/ncomms13207. 13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krause S, Brock A, Ingber DE. Intraductal injection for localized drug delivery to the mouse mammary gland. J Vis Exp. 2013;80:e50692. doi: 10.3791/50692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad.Sci. USA; 2005. pp. 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 57.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 59.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. Inferring regulatory networks from expression data using tree-based methods. PLoS One. 2010;5:e12776. doi: 10.1371/journal.pone.0012776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Angerer P, et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE127975. Single-cell RNA-seq data have been deposited in the Gene Expression Omnibus under accession number GSE148791. Data supporting the findings of this study are available within the article. Source data are provided with this paper.