Abstract
Understanding the population structure of tropical anguillids residing in the Pacific is vital for their conservation management. Here, the population genetic structure of five sympatric freshwater eels (Anguilla marmorata Quoy & Gaimard, A. megastoma Kaup, A. obscura Steindachner, A. reinhardtii Günther and A. australis Richardson) across 11 western South Pacific (WSP) islands was investigated based on partial nucleotide sequences of the mtDNA control region and the nuclear GTH2b genes of 288 newly collected samples jointly with existing sequences. WSP anguillids are characterised by overall high levels of genetic diversity. Both mtDNA and nuclear sequences provided no evidence for distinct geographic clines or barriers in any of the species across the WSP. The occurrence of admixed individuals between A. marmorata and A. megastoma was confirmed, and a new possible occurrence of a further species was revealed (A. interioris Whitley on Bougainville Island). All species showed evidence for demographic population growth in the Pleistocene, and a subsequent population reduction for A. megastoma. Common spawning grounds and mixing of larvae by ocean currents could promote the lack of pronounced isolation by distance, a finding that has significant implications for the future management of anguillids in the area.
Keywords: genetic homogeneity, hybridisation, recruitment, spawning, sympatry, tropical eels
1. Introduction
Sympatric and closely related organisms are excellent models to infer the underpinning mechanisms behind genetic diversity and connectivity in marine ecosystems (Dawson, 2012; Palumbi, 1994). Moreover, multispecies comparative studies examining gene flow have important implications for conservation and management strategies, as they offer a more comprehensive assessment of species biodiversity and evolution (Weber, Mérigot, Valière & Chenuil, 2015). Patterns of population structure and phylogeography of economically important species with similar life-history characteristics may be concordant or disparate (Husemann, Ray, King, Hooser & Danley, 2012; Mcmillen-Jackson & Bert, 2003), providing insights into ecological and anthropogenic factors that might influence them. Interestingly, sympatric species that share large distribution areas are possibly less prone to extinctions due to fishing pressure. However, uncontrolled fisheries and habitat loss due to climate change and fluctuation of sea water levels can have deleterious effects on the habitat of particularly tropical fish, resulting in contracted distributions and reductions in population size (Nicholls & Cazenave, 2010).
The western South Pacific islands (WSP—including Samoa and archipelagos to the west) host six (Anguilla australis Richardson, A. dieffenbachia Gray, A. marmorata Quoy & Gaimard, A. megastoma Kaup, A. obscura Steindachner, A. reinhardtii Günther) of the 19 global species and subspecies of Anguilla eels (Jellyman, 2003; not including A. bicolor pacifica McClelland and A. interioris Whitley that occur at the border of the Bismarck Sea). They share catadromous life histories, with growth occurring in freshwater or estuarine areas and spawning in tropical ocean waters (Aida, Tsukamoto & Yamauchi, 2003; Schmidt, 1923; Tesch, 1977; Watanabe, Aoyama, Nishida & Tsukamoto, 2005). All species except A. bicolor (Near Threatened) and A. marmorata (Least Concern) are classified as Data Deficient or have not been assessed by the International Union for Conservation of Nature (IUCN; Jacoby & Gollock, 2014). As the majority of studies on eels have focused on temperate species, large knowledge gaps currently exist on their ecology, population genetic structure and spawning grounds. Based on differences in the number of vertebrae, there may be four spawning populations for A. marmorata in the Pacific (North Pacific, Micronesia, WSP and central South Pacific Ocean (CSP—islands east of Samoa)), two for A. megastoma (WSP, CSP) and A. australis (Australia/Tasmania, New Zealand/New Caledonia), and single panmictic populations for A. reinhardtii, A. obscura and A. dieffenbachii (Ege, 1939; Watanabe, Miller, Aoyama & Tsukamoto, 2011; Watanabe et al., 2008). Molecular genetic data also confirm the existence of three to four populations of A. marmorata (Gagnaire et al., 2011; Ishikawa, Tsukamoto & Nishida, 2004; Minegishi, Aoyama & Tsukamoto, 2008). Moreover, spatial introgression into the South Pacific lineage of A. marmorata from Indian and North Pacific populations implies a subtle population structure in this area (Gagnaire et al., 2011).
Demographic assessment also requires that species delimitation is taken into account. Natural hybrids have been previously reported between Atlantic eel species (Avise et al., 1990; Boëtius, 1980; Williams, Koehn & Thorsteinsson, 1984), whereas hybrids of, for example, short-finned and long-finned eels have been produced under laboratory conditions only (Burgerhout et al., 2011; Lokman & Young, 2000; Okamura et al., 2004). In the Pacific, relatively high levels of hybridisation were recently revealed between the two long-finned eels A. marmorata and A. megastoma from Gaua Island, Vanuatu, and linked with reproductive sympatry of the species as suggested by satellite tracking (Schabetsberger et al., 2015).
An increasing economic interest in Pacific tropical anguillid species has been stimulated by sharp declines in temperate eel species recruitment (Aoyama, Shinoda, Yoshinaga & Tsukamoto, 2012; Arai, 2016; Briand, Fatin, Fontenelle & Feunteun, 2003; Iglesias & Lobón-Cerviá, 2012). East Asian countries are the main consumers of eels and eel products, with more than 90% of Anguilla production based on farming after significant declines of wild stocks due to overfishing (Arai, 2014; Limburg & Waldman, 2009; Shiraishi & Crook, 2015). High demand and exploitation of eels resulted in the classification of European, Japanese and American eels as Critically Endangered or Endangered by the IUCN Red List (Jacoby & Gollock, 2014), with the European eel also listed in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). Until recently, the Philippines and Indonesia have been the main supplier of wild tropical glass eels for aquaculture in East Asia (Crook & Nakamura, 2013), although attempts for the commercial exploitation of wild eel stocks from WSP countries have also been reported (Pickering & Sasal, 2017; S. Tiitii, Ministry of Agriculture and Fisheries of Western Samoa and D. Kalfatak, Department of Environmental Protection and Conservation, Vanuatu, pers. comm.). However, the lack of natural history information and formal stock assessments of eels in this area impedes their management and conservation, and drastic temporal changes in local species compositions attributed to different seasonal spawning dynamics prevent the selective glass eel catch of commercially preferred species (Aoyama et al., 2015). In line with recent discoveries of cryptic species (A. luzonensis Watanabe, Aoyama & Tsukamoto in the Philippines; Watanabe, Aoyama & Tsukamoto, 2009), there is an urgent need for more information on the distribution, ecology and life history of freshwater eels in the WSP. To this end, this study samples new WSP geographic regions to provide novel insights into processes determining spatial levels of genetic variation, resulting in important conservation and management implications for five understudied but important species.
2. Methods
2.1. Study area and PCR amplification
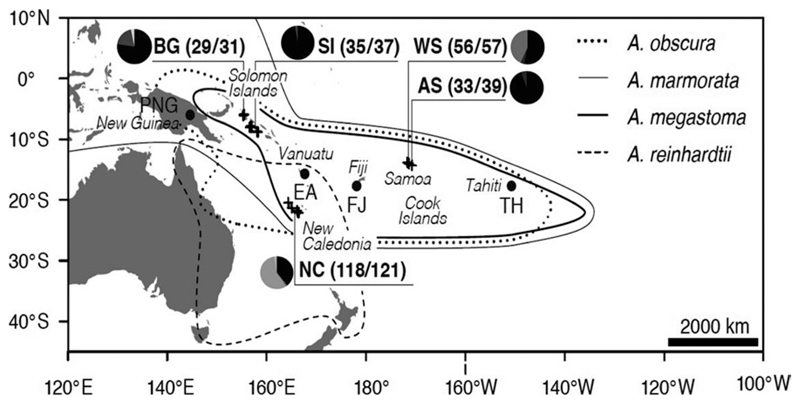
A total of 288 adult or juvenile Anguilla specimens were collected during 2015–2016 in rivers or lakes of islands in the WSP, by electrofishing and hand nets in collaboration with local fishermen (Supporting information Table S1). Sampling localities included Bougainville Island (BG: n = 31), Solomon Islands (SI: n = 37), New Caledonia (NC: n = 124), Western Samoa (WS: n = 57) and American Samoa (AS: n = 39; Figure 1, Supporting information Table S1). Samples were treated according to each permit’s requirements, as issued by local authorities. Eels were sacrificed or anaesthetised in a freshwater bath containing 40 mg/L metomidate (Marinil™, Wildlife Labs) or clove oil until motionless. They were subsequently measured, genetically sampled and released (except all individuals from Bougainville). Total length, and the distance from the lower jaw to the anus and to the dorsal fin, was measured to the nearest mm. All individuals were morphologically assigned to a species by assessment of body proportions, coloration and dentition of the upper jaw following Ege (1939). Small pieces of tissue and fin clips were stored in 95% ethanol until genomic DNA extraction was conducted using the DNeasy Blood and Tissue Kit (Qiagen) as per the manufacturer’s instructions or a standard phenol chloroform procedure (Sambrook, Fritsch & Maniatis, 1989).
Figure 1.
Sample collection locations, with numbers of individuals analysed (mitochondrial/nuclear markers). Sampling localities included Bougainville Island (BG), Solomon Islands (SI), New Caledonia (NC), Western Samoa (WS) and American Samoa (AS) and are represented with a cross (+). Sampling sites from previous studies included Papua New Guinea (PNG), Vanuatu (EA), Fiji (FJ) and Tahiti (TH) and are depicted with a dot (.). Species included: Anguilla marmorata (●); A. megastoma (●); A. obscura (●); A. reinhardtii (●); A. australis (●); A. interioris (●).
2.2. Molecular species identification
To assign all individuals genetically to known species, mitochondrial (control region, CR) and nuclear (gonadotropin II-beta subunit, GTH2b) sequence variation was examined using MTF-MTR and GTH2bF-GTH2bR primers in 20 μl reaction volumes (see Schabetsberger et al., 2015 for details). Briefly, polymerase chain reactions (PCRs) were carried out with GoTaq × 5 reaction buffer (1.5 mM MgCl2 in × 1 concentration), 200–250 μM of each deoxyribonucleotide triphosphate (dNTP; Bioline), 0.2–1.0 μM of each primer (MWG-Biotech), 1.0–2.5 U GoTaq polymerase (Promega) and thermal PCR profiles for the CR and GTH2b genes as described in Schabetsberger et al. (2015). PCR products were sequenced commercially (Macrogen, Netherlands).
All sequences were checked with Proseq 3.2 (Filatov, 2009), and all mtDNA (CR) sequences (from 498 bp—A. australis, to 526 bp—A. marmorata) were compared with those available in GenBank using the standard nucleotide BLAST (blastn) against the nucleotide collection (nr/nt) database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The 268 bp nuclear sequences validated the morphological species identification at two species-specific single nucleotide polymorphisms (SNPs) for some of the studied species (C/T at base position 86, and T/C at position 169; A. megastoma and A. obscura show genotype C and T, whereas A. marmorata has the genotype T and C, respectively; see Schabetsberger et al., 2015 for details); admixed individuals possess heterozygote genotypes at the two species-specific nuclear SNPs and were excluded from further species-specific analyses presented herein. The newly acquired samples were further combined with 83 sequences already available from Gaua Island (GA, Schabetsberger et al., 2015), as well as mtDNA sequences provided in Minegishi et al. (2008; Fiji, FJ: 20 individuals; Tahiti, ΤΗ: 19 individuals; New Caledonia, NC: 9 individuals; Papua New Guinea, PNG: 15 individuals).
2.3. Population genetic analyses
Number of haplotypes (H), polymorphic sites (s), haplotype (h) and nucleotide (π) diversities (Nei, 1987) were calculated at the level of each species for each marker and sampling site with Arlequin 3.5.2.2 (Excoffier, Laval & Schneider, 2005). The nuclear phased haplotypes were estimated using Bayesian methods in Phase 2.1 (Stephens & Donnelly, 2003). Gametic phases with posterior probabilities equal to or higher than 0.7 were considered resolved (Harrigan, Mazza & Sorenson, 2008). Past recombination for all data sets was investigated using a PHI Test (Bruen, Philippe & Bryant, 2006) in SplitsTree 4.14.4 (Huson & Bryant, 2006). No significant signals of recombination were detected, and the full nonrecombining sequence was used. Average intraspecific genetic p-distances for CR within and between sampling locations were calculated in MEGA 7.0.21 (Kumar, Stecher & Tamura, 2015), with pairwise deletion for the gaps/missing data treatment. In addition, the degree of genetic differentiation among localities was estimated using genetic distance-based ΦST, with 10,000 permutations in Arlequin. For A. marmorata, temporal genetic variation across temporally adjacent years was also assessed by calculating pairwise Φ ST values between the newly acquired samples from New Caledonia and the data presented in Minegishi et al. (2008) in Arlequin. Sequential Bonferroni corrections to the significance level of 0.05 were applied (Rice, 1989). A hierarchical analysis of molecular variance (AMOVA) was performed in Arlequin, to test for significant differences between groups of A. marmorata samples. Data were grouped according to geographical location as follows: north of New Caledonia (Papua New Guinea, Bougainville, Solomon Islands, Vanuatu), New Caledonia (this study and data from Minegishi et al., 2008), Fiji, Samoa Islands (Western Samoa and American Samoa) and Tahiti. In addition, a Mantel test was used to test for significant correlations between genetic distances and geographical distances in GenAlEx 6.5 (Peakall & Smouse, 2012).
Haplotype networks for each marker were constructed in PopART 1.7 (Leigh & Bryant, 2015) using statistical parsimony (Clement, Snell, Walker, Posada & Crandall, 2002) to visualise the genealogical relationships between haplotypes across the region. To determine whether the two New Caledonian A. australis individuals group with those caught in Australia or New Zealand (Accession Numbers: AB278891-AB278942), phylogenetic maximum-likelihood (ML) analysis of the CR sequences was performed on RAxML 8.0 (Stamatakis, Hoover, Rougemont & Renner, 2008). The GTRGAMMA model and 1,000 replications for the final tree calculations were used. Sequences of A. obscura were used as outgroup.
2.4. Demographic analysis
Recent demographic change, gene flow–drift or selection and fluctuations in population size were assessed using Tajima’s D (Tajima, 1989) and Fu’s FS (Fu, 1997) for both markers in Arlequin for the species with sufficient sample size. Furthermore, for the mitochondrial data set, a Bayesian skyline plot (BSP) model was generated in BEAUti and Beast 1.8.0 (Drummond, Rambaut, Shapiro & Pybus, 2005) and plotted using the 95% highest posterior density. Program default parameters were used, and different mutation models per species were chosen by jModeltest 2.1.9 (Darriba, Taboada, Doallo & Posada, 2012; Guindon & Gascuel, 2003) using the Bayesian information criterion (BIC). BSP analyses were conducted using a strict molecular clock with a substitution rate of 5% between lineages per one million years (Minegishi et al., 2011). The Markov chain Monte Carlo (MCMC) sampling schemes were set to 50–300 million iterations depending on the length required for convergence. All runs were sampled every 1,000 iterations, after discarding the first 10% as burn-in. The BSP outputs were analysed using Tracer 1.5 with 10% burn-in (Rambaut, Suchard, Xie & Drummond, 2013).
3. Results
Overall, 275 and 281 partial mitochondrial (CR) and nuclear (GTH2b) sequences were aligned, respectively (Accession numbers: MG977152-MG977423 and MH251344-MH251612). The CR sequences comprised 37 individuals from the Solomon Islands, 31 from Bougainville Island, 118 from New Caledonia, 56 from Western Samoa and 33 from American Samoa (Supporting information Table S1). The GTH2b sequences originated from 35 samples from the Solomon Islands, 29 samples from Bougainville Island, 121 samples from New Caledonia, 57 samples from Western Samoa and 39 samples from American Samoa (Supporting information Table S1). After combining morphological evidence with sequence data (mtDNA and nuclear markers), 175 individuals were assigned to A. marmorata, 10 individuals to A. megastoma, 26 individuals to A. obscura, 69 individuals to A. reinhardtii, three individuals to A. australis and one individual to A. interioris. Two individuals exhibited morphological and mtDNA disagreement combined with nuclear heterozygosity. Both specimens (one from Bougainville Island and one from American Samoa) were morphologically determined as A. megastoma, whereas sequence analysis assigned them to A. marmorata (Supporting information Table S1). Therefore, they were considered as admixed individuals and removed from further population genetic analyses presented in this study. The A. interioris sample as identified by its mtDNA was recorded in Bougainville Island, representing a range expansion of the species. However, it possessed ambiguous bases in both positions (86 and 169) of the nuclear GTH2b sequence, which were assumed to be uninformative with respect to species status due to the absence of reference sequences.
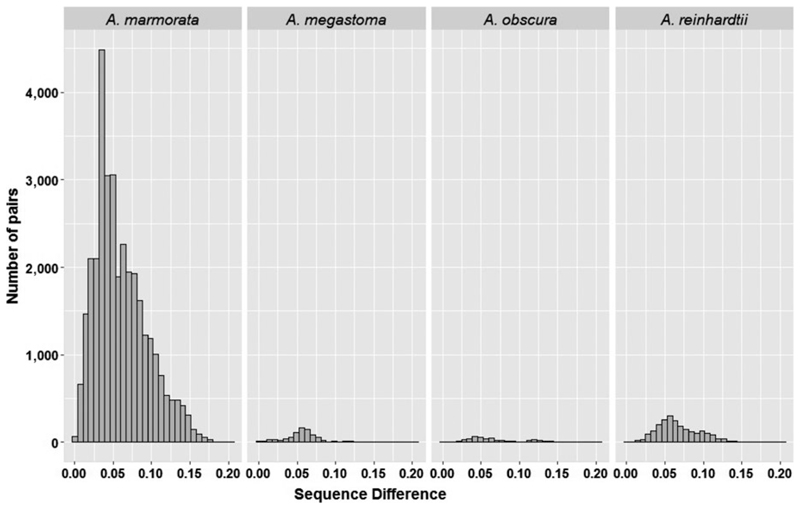
For the mtDNA CR data set, interspecies haplotype diversity ranged from 0.986 (A. megastoma) to 1.000 (A. obscura and A. australis), and corresponding nucleotide diversity values ranged from 0.038 (A. australis) to 0.069 (A. obscura; Table 1). The number of haplotypes and variable sites ranged from 2 and 19 in A. australis to 218 and 371 in A. marmorata, respectively (Table 1), with intraspecific diversity values exhibiting similar ranges (Supporting information Table S2a-c). Three of the five variable number of tandem repeat (VNTR) regions firstly reported by Ishikawa et al. (2004) were observed only in A. marmorata. Following the terminology of Minegishi et al. (2008), VNTR types 1 and 4 were found in all locations; one individual from Tahiti had Type 3, whereas Types 2 and 5 were absent. The lowest mean CR intraspecific difference was detected in A. megastoma (5.6%; Supporting information Table S3a), whereas A. marmorata exhibited the highest intraspecific variability (from 0% to 20.4%; Figure 2; Supporting information Table S3a). Interspecific variation ranged from 24.2% (between A. megastoma and A. australis; Supporting information Table S3b) to 34.4% (between A. marmorata and A. megastoma; Supporting information Table S3b). The corresponding summary statistic values for GTH2b were overall lower than for CR (Table 1).
Table 1.
Summary statistics for the total mtDNA control region and nuclear GTH2b partial gene sequences of six Anguilla species (putative admixed individuals were excluded from this analysis), n: number of individuals; h: number of different haplotypes; h: haplotype diversity; π: nucleotide diversity; s: number of polymorphic sites. D: Tajima’s D; F: Fu’s F s; standard deviations are in brackets. *p < 0.05; **p < 0.01; ***p < 0.001. Two sequences per individual eel for GTH2b were included for the analysis
| Species | Control region | GTH2b | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | H | h | π | s | D | F | n | H | h | π | s | D | F | |
| A. marmorata | 259 | 218 | 0.998 (±0.001) | 0.065 (±0.032) | 371 | −1.492* | −23.631* | 390 | 29 | 0.239 (±0.029) | 0.002 (±0.002) | 25 | −2.261*** | −29.456*** |
| A. megastoma | 42 | 32 | 0.986 (±0.008) | 0.056 (±0.028) | 149 | −0.735 | −3.595 | 72 | 8 | 0.421 (±0.068) | 0.004 (±0.003) | 7 | −0.584 | −2.081 |
| A. reinhardtii | 68 | 67 | 0.999 (±0.003) | 0.065 (±0.032) | 218 | −1.135 | −24.098*** | 136 | 7 | 0.292 (±0.050) | 0.002 (±0.002) | 6 | −1.088 | −3.161 |
| A. obscura | 33 | 33 | 1.000 (±0.008) | 0.069 (±0.034) | 157 | −0.538 | −12.283*** | 66 | 9 | 0.580 (±0.062) | 0.006 (±0.004) | 8 | −0.313 | −1.906 |
| A. australis | 2 | 2 | 1.000 (±0.500) | 0.038 (±0.039) | 19 | 0.000 | 2.944 | 6 | 3 | 0.733 (±0.155) | 0.003 (±0.003) | 2 | −0.050 | −0.427 |
| A. interioris | 1 | 1 | NA | NA | NA | NA | NA | 1 | 1 | NA | NA | NA | NA | NA |
Figure 2.
Intraspecific pairwise sequence differences in the mtDNA control region between individuals of four Anguilla species (A. marmorata, A. megastoma, A. obscura and A. reinhardtii)
The CR pairwise comparisons for the temporal sampling in New Caledonia (new data presented herein compared to data presented in Minegishi et al., 2008) revealed a lack of differentiation (ΦST = 0.0739, nonsignificant after Bonferroni corrections), and all samples were therefore pooled for subsequent analyses (Table 2a). For A. marmorata, pairwise genetic distances between islands ranged from −0.0506 to 0.1627 and −0.0038 to 0.1287 for CR and GTH2b, respectively (Table 2a). In total, 25 of 45 pairwise comparisons showed significant differences in Φ ST after Bonferroni correction. However, neither a significant association between genetic and geographic distances across sample collections (R 2 = 0.0398, p = 0.0800) nor sharp geographic clines within the WSP (Table 2a) were detected. In addition, the hierarchical AMOVA neither showed significant variation among geographically predefined groups (FCT = 0.0505, p = 0.0574, 5.05%) nor significant variation among populations within the main regions of A. marmorata occurrences (FSC = 0.0482, p < 0.0001, 4.58%), although within-population genetic variation was highest for this species (FST = 0.0963, p < 0.0001, 90.37%). Bougainville Island was the only locality with significant pairwise differentiation for GTH2b. No significant genetic structure among sites was found for both CR and GTH2b for the other two species with sufficient sample sizes (A. megastoma and A. obscura, Table 2b-c).
Table 2.
Pairwise measures of population structure. Values in lower diagonal relate to the mtDNA control region, and upper diagonal values are from the nuclear GTH2b locus. Italic and bold Φ ST values are significant before and after sequential Bonferroni correction, respectively; p values are included in brackets. (a) Anguilla marmorata, (b) A. megastoma and (c) A. obscura
| (a) | Papua New Guinea | Bougainville | Solomon Islands | Vanuatu | New Caledonia | New Caledonia Min | Fiji | Western Samoa | American Samoa | Tahiti |
|---|---|---|---|---|---|---|---|---|---|---|
| Papua New Guinea | – | |||||||||
| Bougainville | 0.1202 (0.0001) | – | ||||||||
| Solomon Islands | 0.0553 (0.0126) | 0.0930 (0.0000) | – | |||||||
| Vanuatu | 0.0862 (0.0065) | 0.0994 (0.0001) | −0.0026 (0.4827) | – | ||||||
| New Caledonia | 0.1016 (0.0000) | 0.0328 (0.0067) | 0.0671 (0.0000) | 0.0650 (0.0000) | – | |||||
| New Caledonia Min | −0.0506 (0.9682) | 0.0930 (0.0055) | 0.0440 (0.0666) | 0.0613 (0.0552) | 0.0739 (0.0101) | – | ||||
| Fiji | 0.0363 (0.0890) | 0.1238 (0.0001) | 0.0630 (0.0066) | 0.0512 (0.0228) | 0.0805 (0.0002) | 0.0158 (0.2265) | – | |||
| Western Samoa | 0.1527 (0.0000) | 0.0107 (0.1205) | 0.1477 (0.0000) | 0.1433 (0.0000) | 0.0482 (0.0003) | 0.1223 (0.0004) | 0.1533 (0.0000) | – | ||
| American Samoa | 0.1523 (0.0000) | 0.0354 (0.0070) | 0.1369 (0.0000) | 0.1267 (0.0000) | 0.0376 (0.0007) | 0.1141 (0.0009) | 0.1297 (0.0000) | 0.0108 (0.0936) | – | |
| Tahiti | 0.1296 (0.0015) | 0.1560 (0.0000) | 0.1157 (0.0001) | 0.0821 (0.0040) | 0.0933 (0.0002) | 0.0911 (0.0230) | 0.0180 (0.1408) | 0.1627 (0.0000) | 0.1283 (0.0000) | – |
| (b) | Bougainville | Solomon Islands | Vanuatu | Western Samoa | American Samoa | |||||
| Bougainville | – | −0.1707 (0.9999) | 0.0373 (0.2560) | −0.1707 (0.9999) | 0.0895 (0.1582) | |||||
| Solomon Islands | 0.0125(0.9999) | – | −0.1795 (0.9999) | 0.0000 (0.9999) | 0.2859 (0.3312) | |||||
| Vanuatu | 0.0510 (0.1195) | −0.0610 (0.9999) | – | −0.1795 (0.9999) | 0.2672 (0.0373) | |||||
| Western Samoa | −0.1236(0.9999) | 1.0000(0.9999) | 0.1426 (0.9999) | – | 0.2859 (0.3338) | |||||
| American Samoa | 0.1706(0.2119) | −0.2938(0.9999) | 0.2013 (0.0507) | −0.2061 (0.9999) | – | |||||
| (c) | Vanuatu | New Caledonia | Western Samoa | |||||||
| Vanuatu | – | −0.0938 (0.9999) | 0.0012 (0.3400) | |||||||
| New Caledonia | −0.1095 (0.8712) | – | −0.1312 (0.7624) | |||||||
| Western Samoa | −0.3262 (0.9618) | 0.0479 (0.0859) | – | |||||||
For CR, haplotypes did not assort by sample location for any of the studied species (Supporting information Figure S1I). While a similar lack of geographic structure was evident in the GTH2b networks (Supporting information Figure S1II), a number of haplotypes were unique to Bougainville Island for A. marmorata, supporting the distinctiveness of this region (Supporting information Figure S1IIa). The ML topology based on CR revealed two lineages for A. australis, with very low support values for individuals from both Australia and New Zealand (Supporting information Figure S2). Each New Caledonian sample grouped with different lineages.
3.1. Demographic history
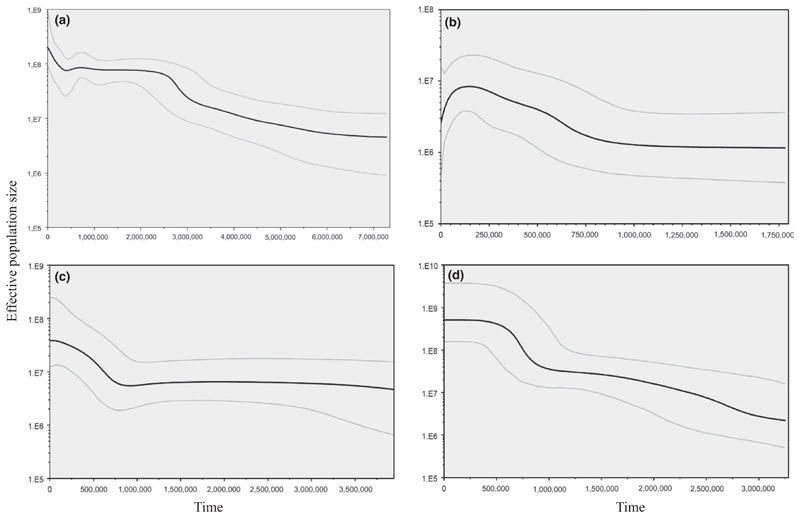
Demographic analyses revealed contrasting findings across species. Anguilla marmorata was the only species with significantly negative Tajima’s D and Fu’s FS values for both markers, conforming to a model of population expansion (Table 1), while these values were not significant for A. megastoma for both markers. For the mtDNA data sets of A. obscura and A. reinhardtii, Tajima’s D was not significant despite significantly negative Fu’s FS values, providing evidence of population expansion; however, both indices were not significant for GTH2b. The BSP analysis based on mtDNA data indicated increasing population sizes for in all species over the past 750,000 years (A. reinhardtii, A. obscura and A. marmorata), with A. megastoma, however, also showing a decline in the last 100,000 years (Figure 3).
Figure 3.
Bayesian skyline plots for each species collection showing the maternal effective population size (mean and 95% confidence interval) back in time (years) since present day. (a) A. marmorata, (b) A. megastoma, (c) A. obscura and (d) A. reinhardtii
4. Discussion
Each species of freshwater eel was characterised by overall low levels of population structure in our study area. Previous studies revealed the clear existence of distinct genetic groups separated by the Indian and Pacific Oceans in A. marmorata (Gagnaire et al., 2011; Ishikawa et al., 2004; Minegishi et al., 2008; Watanabe et al., 2008). Within the WSP, fast-evolving nuclear markers such as microsatellites and AFLPs revealed either one stock (A. marmorata, Ishikawa et al., 2004; Minegishi et al., 2008), a lack (A. reinhardtii, Shen & Tzeng, 2007a), weak (A. marmorata, Gagnaire et al., 2011) or a distinct population structure (A. australis, Shen & Tzeng, 2007b), whereas no structure was detected with mtDNA markers for A. australis (Watanabe, 2001). Gagnaire et al. (2011) suggested the presence of multiple spawning grounds in the South Pacific, whereas other evidence suggests a single spawning area in the Southern Hemisphere and in the South Equatorial Current (Aoyama et al., 1999). In the present study, both mtDNA and nuclear sequences provided no evidence for distinct geographic clines in any of the five study species throughout the WSP. For A. marmorata, A. megastoma, A. obscura and A. reinhardtii, this is in accordance with analyses on the number of vertebrae within the WSP and the lack of differences among sampling localities (Watanabe et al., 2008, 2011), although differences in morphology suggest the existence of two spawning stocks for A. australis (Watanabe, Aoyama & Tsukamoto, 2006).
It is necessary to acknowledge the limitations imposed by sampling restrictions and the markers used. Uneven sampling across study species may have partially influenced the outcome of the current study. Particularly for A. megastoma and A. obscura, sampling was rather incomplete and underrepresented, whereas both A. australis and A. reinhardtii were covered at the northern limit of their distribution. Both CR and GTH2b have previously been successfully applied to anguillid studies (Ishikawa et al., 2004; Minegishi et al., 2008, 2011; Schabetsberger et al., 2015), although particularly the GTH2b locus does not exhibit the level of variation required to detect subtle population structure. Compared to the present study, which is limited to one nuclear and mitochondrial gene each, next-generation sequencing techniques could characterise a comprehensive suite of nuclear single nucleotide polymorphisms (SNPs), providing more power in the evaluation of population structure and gene flow in tropical anguillids.
In general, this study corroborates that freshwater eels exhibit low levels of genetic structure, which might be credited to the migration of adults from different archipelagos to shared spawning grounds. Alternatively, different spawning areas may exist, but larval mixing in ocean gyres could lead to lack of spatially distinct demes in the WSP. Two major current systems disperse leptocephali within the WSP: the westward South Equatorial Current and the eastward South Equatorial Counter Current. Both are characterised by intra- and interannual variability and could potentially transport leptocephali to both directions (Schabetsberger et al., 2016). It remains unknown where A. obscura, A. reinhardtii and A. australis are spawning in the WSP (see Jellyman & Bowen, 2009), although their offspring get dispersed by these two major currents running into opposite direction.
Analyses of eel spatial population structure can be confounded by temporal genetic variation (previously evidenced in A. anguilla; Dannewitz et al., 2005; Pujolar, Maes & Volckaert, 2006). In this study, no temporal genetic differentiation for A. marmorata in New Caledonia was found, whereas temporal genetic heterogeneity was, for example, detected in the Indian Ocean by Gagnaire et al. (2009). The mixing of individuals can also be attributed to the prolonged spawning and recruitment of tropical anguillids, which has been suggested to last nearly the entire year, facilitating spawning groups that originate from different generations (Arai & Abdul Kadir, 2017; Kuroki et al., 2009; Shen & Tzeng, 2007a). The spawning ecology of tropical eels differs significantly from those found in temperate waters, enabling interbreeding between silver eels from different areas and cohorts (Acou, Laffaille, Legault & Feunteun, 2008; Kotake et al., 2007; Verreault, Mingelbier & Dumont, 2012). A year-round spawning ability may help to maintain high population sizes in the Indian Ocean (Arai & Abdul Kadir, 2017). However, the continuous and targeted fisheries, the unregulated removal of recruits for eel farming and the lack of population assessment may nevertheless lead to stock depletion. Therefore, assessing the consequences of overexploitation of WSP tropical eels requires further information on their spawning ecology and life history.
Using the same set of genetic markers as applied in the present study, the high levels of admixture between A. marmorata and A. megastoma have previously been recorded from Vanuatu, with evidence for different levels of admixture (about 15% of individuals; Schabetsberger et al., 2015). In the present study, admixed individuals between these two species were also detected in American Samoa (one individual, 2.56%) and in Bougainville (one individual, 3.13%). Comparable levels of hybridisation have also been detected in Iceland between A. rostrata and A. anguilla, with proportions of admixed individuals ranging between 10.7% and 15.5% of examined samples depending on year and sampling location (Albert, Jnsson & Bernatchez, 2006; Pujolar et al., 2014). Moreover, the high occurrence of admixed individuals in Iceland could be related to the intermediate duration and development time of larvae and/or their transport from one branch of the North Atlantic Current to Iceland (Pujolar et al., 2014). Similarly, the high occurrence of admixed individuals in Vanuatu could be linked to the geographic proximity to the hypothetical spawning area north-west of Fiji (Schabetsberger et al., 2015); a more detailed analysis of patterns of hybridisation is currently ongoing (Gubili et al. unpublished). One individual from Bougainville morphologically classified as A. megastoma was identified as A. interioris by mtDNA. Considering the small number of A. interioris caught in the wild and a morphological distinction based on the number of vertebrae (A. interioris typically has between 100 and 106 vertebrae and A. megastoma between 108 and 116; Froese & Pauly, 2011), an initial misclassification is not overly surprising.
The WSP hosts the largest number of sympatric anguillid species globally (Jellyman, 2003). All four species investigated for their population history showed patterns of population expansion around 750,000 years before present, that is during the Pleistocene period. However, A. megastoma also appeared to decline over the last around 100,000 years. The time estimates of these demographic events critically depend on assumed mutation rates and thus should be treated cautiously. Nonetheless, this period coincides with the glacial and interglacial cycles of the Pleistocene (1,600,000 to 10,000 years before present; Imbrie et al., 1992), which are known to have had profound effects on the genetic architecture of many marine and freshwater species (Grant & Waples, 2000). The simultaneous range expansion of WSP anguillids might have been triggered by climate oscillations that became more pronounced after the mid-Pleistocene (<780,000 years ago; Lowe & Walker, 2014). High levels of haplotype diversity combined with relatively low nucleotide diversity are generally indicative of drastic reduction in population size followed by sudden expansion (Grant & Bowen, 1998). The A. megastoma population decline in the last 100,000 years could be linked to the species residing in rivers of higher altitude. Hence, habitat alterations such as erosion, deposition and sea level changes could have negatively impacted the species’ demographic history.
The assessment and management of eel stocks in the WSP is currently hampered by a lack of information on migration and population structure. The main finding of this study is that each species comprises populations without pronounced isolation by distance or geographic clines, suggesting high levels of at least connectivity among locations. Moreover, hybridisation between long-finned eels has been documented across the WSP due to sympatric spawning, with the occurrence of hybrids being more restricted outside Vanuatu. Finally, tropical eel populations also exhibit different demographic histories that appear to have initially undergone expansion due to climatic oscillations in mid-Pleistocene. These findings have important implications for sustainable management required to ensure future stock viability, particularly as trade shifts towards tropical species to satisfy current and future demand.
Supplementary Material
Acknowledgments
Funding for this study was provided by the Austrian Science Fund (FWF): P28381-B29 and the University of Salford. We would like to thank the members of staff of the Ministry of Agriculture and Fisheries (MAF) and the Marine Conservation Section of the Ministry of Natural Resources & Environment (Western Samoa) for all their help and support. We are extremely thankful to Ruben Sommaruga for his support of the project and Finn Okland, Meelis Tambets, Donna Kalfatak, Aurelie Dessier, Kay Hnauane and Nicolas Charpin for their help during sampling. We are also indebted to Michael Matschiner, Bernd Egger, Julia M.I. Bath and Peter H. Comes for their constructive comments. Collecting permits were issued by Direction de l’environement (New Caledonia: Province Nord-60912-1286-2016/JJC and Province Sud-APA_NCPS_2016.004), Department of Marine and Wildlife Resources (American Samoa: 2015/010), Ministry of Natural Resources and Environment (Western Samoa: 1041072), Solomon Islands (RP/2014/007) and Bougainville (P6025227 9AUT6307158M1506303).
Funding information
Austrian Science Fund (FWF), Grant/Award Number: P28381-B29; University of Salford
References
- Acou A, Laffaille P, Legault A, Feunteun E. Migration pattern of silver eel Anguilla anguilla L.) in an obstructed river system. Ecology of Freshwater Fish. 2008;17:432–442. [Google Scholar]
- Aida K, Tsukamoto K, Yamauchi K. Eel biology. Tokyo, Japan: Springer Verlag; 2003. [Google Scholar]
- Albert V, Jnsson B, Bernatchez L. Natural hybrids in Atlantic eels Anguilla anguilla A rostrata): Evidence for successful reproduction and fluctuating abundance in space and time. Molecular Ecology. 2006;15:1903–1916. doi: 10.1111/j.1365-294X.2006.02917.x. [DOI] [PubMed] [Google Scholar]
- Aoyama J, Mochioka N, Otake T, Ishikawa S, Kawakami Y, Castle P, et al. Tsukamoto K. Distribution and dispersal of anguillid leptocephali in the western Pacific Ocean revealed by molecular analysis. Marine Ecology Progress Series. 1999;188:193–200. [Google Scholar]
- Aoyama J, Shinoda A, Yoshinaga T, Tsukamoto K. Late arrival of the Japanese glass eel Anguilla japonica at the Sagami River estuary in recent consecutive two-year classes: Ecology and socioeconomic impacts. Fisheries Science. 2012;78:1195–1204. [Google Scholar]
- Aoyama J, Yoshinaga T, Shinoda A, Shirotori F, Yambot AV, Han Y-S. Seasonal changes in species composition of glass eels of the genus Anguilla (Teleostei: Anguillidae) recruiting to the Cagayan River, Luzon Island, the Philippines. Pacific Science. 2015;69:263–270. [Google Scholar]
- Arai T. Do we protect freshwater eels or do we drive them to extinction? SpringerPlus. 2014;3:534. doi: 10.1186/2193-1801-3-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T. Current status and concerns on tropical anguillid eel stocks. Scientia Bruneiana. 2016;15:33–43. [Google Scholar]
- Arai T, Abdul Kadir SR. Opportunistic spawning of tropical anguillid eels Anguilla bicolor bicolor and A bengalensis bengalensis . Scientific Reports. 2017;7:41649. doi: 10.1038/srep41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Nelson WS, Arnold J, Koenh RK, Williams GC, Thorsteinsson V. The evolutionary genetic status of Icelandic eels. Evolution. 1990;44:1254–1262. doi: 10.1111/j.1558-5646.1990.tb05229.x. [DOI] [PubMed] [Google Scholar]
- Boëtius J. Atlantic Anguilla. A presentation of old and new data of total numbers of vertebrae with special reference to the occurrence of Anguilla rostrata in Europe. Dana. 1980;1:93–112. [Google Scholar]
- Briand C, Fatin D, Fontenelle G, Feunteun E. Estuarine and fluvial recruitment of the European glass eel, Anguilla anguilla in an exploited Atlantic estuary. Fisheries Management and Ecology. 2003;10:377–384. [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgerhout E, Brittijn SA, Kurwie T, Decker P, Dirks RP, Palstra AP, et al. Van den Thillart GEEJM. First artificial hybrid of the eel species Anguilla australis and Anguilla anguilla . BMC Developmental Biology. 2011;11:16. doi: 10.1186/1471-213X-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Snell Q, Walker P, Posada D, Crandall K. TCS: Estimating gene genealogies. Parallel and Distributed Processing Symposium, International Proceedings; 2002. p. 184. [Google Scholar]
- Crook V, Nakamura M. Glass eels: Assessing supply chain and market impacts of a CITES listing on Anguilla species. TRAFFIC Bulletin. 2013;25:24–30. [Google Scholar]
- Dannewitz J, Maes GE, Johansson L, Wickström H, Volckaert FAM, Jarvi T. Panmixia in the European eel: A matter of time. Proceedings of the Royal Society London B. 2005;272:1129–1137. doi: 10.1098/rspb.2005.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nature Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MN. Parallel phylogeographic structure in ecologically similar sympatric sister taxa. Molecular Ecology. 2012;21:987–1004. doi: 10.1111/j.1365-294X.2011.05417.x. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Ege V. A revision of the genus Anguilla Shaw. Dana Report. 1939;16:8–256. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Filatov DA. Processing and population genetic analysis of multigenic datasets with ProSeq3 software. Bioinformatics. 2009;25:3189–3190. doi: 10.1093/bioinformatics/btp572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froese R, Pauly D. FishBase. [Accessed 03 May 2017];World Wide Web electronic publication. 2011 http://www.fishbase.org. [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire PA, Minegishi Y, Aoyama J, Réveillac E, Robinet T, Bosc P, et al. Berrebi P. Ocean currents drive secondary contact between Anguilla marmorata populations in the Indian Ocean. Marine Ecology Progress Series. 2009;379:267–278. [Google Scholar]
- Gagnaire PA, Minegishi Y, Zenboudgi S, Valade P, Aoyama J, Berrebi P. Within-population structure highlighted by differential introgression across semipermeable barriers to gene flow in Anguilla marmorata . Evolution. 2011;65:3413–3427. doi: 10.1111/j.1558-5646.2011.01404.x. [DOI] [PubMed] [Google Scholar]
- Grant WS, Bowen BW. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. Journal of Heredity. 1998;89:415–426. [Google Scholar]
- Grant WS, Waples RS. Spatial and temporal scales of genetic variability in marine and anadromous species: Implications for fisheries oceanography. In: Pitcher TJ, editor. Fish and Aquatic Resources Series. Oxford: Blackwell Science; 2000. ISSN 1746-2606. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Harrigan RJ, Mazza ME, Sorenson MD. Computation vs cloning: Evaluation of two methods for haplotype determination. Molecular Ecology Resources. 2008;8:1239–1248. doi: 10.1111/j.1755-0998.2008.02241.x. [DOI] [PubMed] [Google Scholar]
- Husemann M, Ray JW, King RS, Hooser EA, Danley PD. Comparative biogeography reveals differences in population genetic structure of five stream fishes. Biological Journal of the Linnean Society. 2012;107:867–885. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Iglesias T, Lobón-Cerviá J. Size and number of male silver eels Anguilla anguilla (L.) in a Cantabrian river over two decades (1990–2011) Fundamental and Applied Limnology. 2012;181:229–239. [Google Scholar]
- Imbrie J, Boyle EA, Clemens SC, Duffy A, Howard WR, Kukla G, et al. Toggweiler JR. On the structure and origin of major glaciations cycles 1 Linear responses to Milankovitch forcing. Paleoceanography. 1992;7:701–738. [Google Scholar]
- Ishikawa S, Tsukamoto K, Nishida M. Genetic evidence for multiple geographic populations of the giant mottled eel Anguilla marmorata in the Pacific and Indian oceans. Icthyological Research. 2004;51:343–353. [Google Scholar]
- Jacoby D, Gollock M. Anguilla anguilla The IUCN red list of threatened species. Version 2014.3. 2014 www.iucnredlist.org. [Google Scholar]
- Jellyman DJ. The distribution and biology of the South Pacific species of Anguilla 1789. In: Aida K, Tsukamoto K, Yamauchi K, editors. Eel biology. Tokyo, Japan: Springer; 2003. pp. 275–292. [Google Scholar]
- Jellyman DJ, Bowen MM. Modelling larval migration routes and spawning areas of anguillid eels of New Zealand and Australia. American Fisheries Society Symposium; 2009. pp. 255–274. [Google Scholar]
- Kotake A, Arai T, Okamura A, Yamada Y, Utoh T, Oka HP, et al. Tsukamoto K. Ecological aspects of Japanese eels, Anguilla japonica collected from coastal areas of Japan. Zoological Science. 2007;24:1213–1221. doi: 10.2108/zsj.24.1213. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0. Molecular Biology and Evolution. 2015;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki M, Aoyama J, Miller MJ, Yoshinaga T, Shinoda A, Hagihara S, Tsukamoto K. Sympatric spawning of Anguilla marmorata and Anguilla japonica in the western North Pacific Ocean. Journal of Fish Biology. 2009;74:1853–1865. doi: 10.1111/j.1095-8649.2009.02299.x. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. Popart: Full-feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6:1110–1116. [Google Scholar]
- Limburg KE, Waldman JR. Dramatic declines in North Atlantic diadromous fishes. BioScience. 2009;59:955–965. [Google Scholar]
- Lokman PM, Young G. Induced spawning and early ontogeny of New Zealand freshwater eels Anguilla dieffenbachia and A australis) New Zealand Journal of Marine and Freshwater Research. 2000;34:135–145. [Google Scholar]
- Lowe J, Walker M. Reconstructing quaternary environments. London, New York: Routledge; 2014. [Google Scholar]
- Mcmillen-Jackson AL, Bert TM. Disparate patterns of population genetic structure and population history in two sympatric penaeid shrimp species Farfantepenaeus aztecus and Litopenaeus setiferus) in the eastern United States. Molecular Ecology. 2003;12:2895–2902. doi: 10.1046/j.1365-294x.2003.01955.x. [DOI] [PubMed] [Google Scholar]
- Minegishi Y, Aoyama J, Tsukamoto K. Multiple population structure of the giant mottled eel, Anguilla marmorata . Molecular Ecology. 2008;17:3109–3122. doi: 10.1111/j.1365-294X.2008.03822.x. [DOI] [PubMed] [Google Scholar]
- Minegishi Y, Gagnaire PA, Aoyama J, Bosc P, Feunteun E, Tsukamoto K, Berrebi P. Present and past genetic connectivity of the Indo-Pacific tropical eel Anguilla bicolor . Journal of Biogeography. 2011;39:408–420. [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY: Columbia University Press; 1987. [Google Scholar]
- Nicholls RJ, Cazenave A. Sea-level rise and its impact on coastal zones. Science. 2010;328:1517–1520. doi: 10.1126/science.1185782. [DOI] [PubMed] [Google Scholar]
- Okamura A, Zhang H, Utoh T, Akazawa A, Yamada Horie NY, Mikawa N, et al. Oka HP. Artificial hybrid between Anguilla anguilla and A. japonica . Journal of Fish Biology. 2004;64:1450–1454. [Google Scholar]
- Palumbi SR. Genetic divergence, reproductive isolation, and marine speciation. Annual Review in Ecology and Systematics. 1994;25:547–572. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: Genetic analysis in Excel Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering TD, Sasal P. Workshop on South Pacific freshwater eels: current knowledge and future research Suva, Fiji. 13-15 June 2016; Noumea, New Caledonia: Pacific Community; 2017. p. 36. [Google Scholar]
- Pujolar JM, Jacobsen MW, Als TD, Frydenberg J, Magnussen E, Jónsson B, et al. Hansen MM. Assessing patterns of hybridization between North Atlantic eels using diagnostic single-nucleotide polymorphisms. Heredity. 2014;112:627–637. doi: 10.1038/hdy.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujolar JM, Maes GE, Volckaert FAM. Genetic patchiness among recruits in the European eel Anguilla anguilla . Marine Ecology Progress Series. 2006;307:209–217. [Google Scholar]
- Rambaut A, Suchard MA, Xie W, Drummond AJ. Tracer v1.6. 2013 Available: http://tree.bio.ed.ac.uk/software/tracer. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schabetsberger R, Miller MJ, Dall’Olmo G, Kaiser R, Watanabe S, Aerestrup K, Tsukamoto K. Hydrographic features of anguillid spawning areas: Potential signposts for migrating eels. Marine Ecology Progress Series. 2016;554:141–155. doi: 10.3354/meps11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabetsberger R, Økland F, Kalfatak D, Sichrowsky U, Tambets M, Aarestrup K, et al. Quartly G. Genetic and migratory evidence for sympatric spawning of tropical Pacific eels from Vanuatu. Marine Ecology Progress Series. 2015;521:171–187. [Google Scholar]
- Schmidt J. The breeding places of the eel. Philosophical Transactions of the Royal Society B. 1923;211:179–208. [Google Scholar]
- Shen KN, Tzeng WN. Population genetic structure of the year-round spawning tropical eel, Anguilla reinhardtii in Australia. Zoological Studies. 2007a;46:441–453. [Google Scholar]
- Shen KN, Tzeng WN. Genetic differentiation among populations of the short finned eel Anguilla australis from the East Australia and New Zealand. Journal of Fish Biology. 2007b;70:177–190. [Google Scholar]
- Shiraishi H, Crook V. Eel market dynamics: An analysis of Anguilla production, trade and consumption in East Asia. Tokyo, Japan: TRAFFIC; 2015. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J, Renner S. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. The American Journal of Human Genetics. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesch FW. The Eel. London, UK: Chapman Hall; 1977. [Google Scholar]
- Verreault G, Mingelbier M, Dumont P. Spawning migration of American eel Anguilla rostrata from pristine 1843 1872) to contemporary 1963 1990) periods in the St Lawrence Estuary, Canada. Journal of Fish Biology. 2012;81:387–407. doi: 10.1111/j.1095-8649.2012.03366.x. [DOI] [PubMed] [Google Scholar]
- Watanabe S. Unpublished PhD thesis. University of Tokyo; Japan: 2001. Taxonomic study of the freshwater eels, the genus Anguilla Schrank 1798. [Google Scholar]
- Watanabe S, Aoyama J, Miller MJ, Ishikawa S, Feunteun E, Tsukamoto K. Evidence of population structure in the giant mottled eel, Anguilla marmorata using total number of vertebrae. Copeia. 2008;2008:680–688. [Google Scholar]
- Watanabe S, Aoyama J, Nishida M, Tsukamoto K. A molecular genetic evaluation of the taxonomy of eels of the genus Anguilla (Pisces: Anguilliformes) Bulletin of Marine Science. 2005;76:675–690. [Google Scholar]
- Watanabe S, Aoyama J, Tsukamoto K. Confirmation of morphological differences between Anguilla australis australis and A australis schmidtii . New Zealand Journal or Marine and Freshwater Research. 2006;40:325–331. [Google Scholar]
- Watanabe S, Aoyama J, Tsukamoto K. A new species of freshwater eel Anguilla luzonensis (Teleostei: Anguillidae) from Luzon Island of the Philippines. Fisheries Science. 2009;75:387–392. [Google Scholar]
- Watanabe S, Miller MJ, Aoyama J, Tsukamoto K. Analysis of vertebral counts of the tropical anguillids Anguilla megastoma A obscura and A reinhardtii in the western South Pacific in relation to their possible population structure and phylogeny. Environmental Biology of Fishes. 2011;91:353–360. [Google Scholar]
- Weber AAT, Mérigot B, Valière S, Chenuil A. Influence of the larval phase on connectivity: Strong differences in the genetic structure of brooders and broadcasters in the Ophioderma longicauda species complex. Molecular Ecology. 2015;24:6080–6094. doi: 10.1111/mec.13456. [DOI] [PubMed] [Google Scholar]
- Williams GC, Koehn RK, Thorsteinsson V. Icelandic eels: Evidence for a single species of Anguilla in the North Atlantic. Copeia. 1984;1:221–223. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.