Abstract
Background
Over 28 000 individuals were infected with Ebola virus during the West Africa (2013–2016) epidemic, yet there has been criticism of the lack of robust clinical descriptions of Ebola virus disease (EVD) illness from that outbreak.
Objectives
To perform a meta-analysis of published data from the epidemic to describe the clinical presentation, evolution of disease, and predictors of mortality in individuals with EVD. To assess the quality and utility of published data for clinical and public health decision-making.
Data sources
Primary articles available in PubMed and published between January 2014 and May 2017.
Eligibility
Studies that sequentially enrolled individuals hospitalized for EVD and that reported acute clinical outcomes.
Methods
We performed meta-analyses using random-effect models and assessed heterogeneity using the I 2 method. We assessed data representativeness by comparing meta-analysis estimates with WHO aggregate data. We examined data utility by examining the availability and compatibility of data sets.
Results
In all, 3653 articles were screened and 34 articles were included, representing 16 independent cohorts of patients (18 overlapping cohorts) and at least 6168 individuals. The pooled estimate for case fatality rate was 51% (95% CI 46%–56%). However, pooling of estimates for clinical presentation, progression, and predictors of mortality in individuals with EVD were hampered by significant heterogeneity, and inadequate data on clinical progression. Our assessment of data quality found that heterogeneity was largely unexplained, and data availability and compatibility were poor.
Conclusions
We have quantified a missed opportunity to generate reliable estimates of the clinical manifestations of EVD during the West Africa epidemic. Clinical data standards and data capture platforms are urgently needed.
Keywords: Ebola, Ebola virus disease, emerging infection, epidemic, outbreak, viral hemorrhagic
Introduction
The Ebola virus disease (EVD) epidemic in West Africa (2013–2016) was unprecedented, with at least 28 000 individuals infected [1]. Over the course of the epidemic there were gains in clinical understanding and clinical care despite the challenging circumstances–the estimated case fatality rate in the most affected countries fell from 70% to 39% [2] and was 18.5% in individuals evacuated to high-income countries [3]. Case reports detailed previously unseen or unrecognized clinical manifestations ranging from disease relapse [4], through to survival following in utero infection [5], and sustained virus sequestration in immune privileged sites [4,6,7].
It is critical to translate the personal expertise gained by clinicians during the outbreak into an evidence base to inform the treatment of individuals in future outbreaks, and to provide reliable data on clinical presentation and natural history with which to design clinical trials. Before the West Africa outbreak there were variations in clinical data estimates (including case fatality rates) due to case ascertainment biases, inconsistencies in data collection methods, and the temporal and spatial distribution of outbreaks. Although it was hoped that clear and actionable data would become available due to the scale and duration of the West Africa outbreak, when the WHO released their most recent guidelines for clinical care they concluded that their recommendations were still limited by the scarcity of robust clinical descriptions [8]. This is despite over 2800 articles being published during the outbreak [9]. Why this considerable academic output did not translate into robust data to inform clinical care has not been evaluated.
The first objective of this work is to determine the extent to which the literature can be synthesized by meta-analysis to describe the evolution of patient signs and symptoms during the course of EVD, and to identify clinical predictors of mortality with greater confidence. The second aim is to determine the extent to which the literature constitutes ‘actionable data’ [10]. This is the extent to which the findings are accessible, objective and sufficient to inform clinical, public health and research decision-making for future outbreaks [10]. This work is the first systematic analysis of clinical data accumulated during the outbreak.
Methods
This systematic review was conducted in accordance with PRISMA guidelines [11] and prospectively registered on the PROS-PERO database (registration CRD42017070150).
Data sources and searches
We conducted a systematic search of PubMed for articles pub-lished between 1 January 2014 and 31 May 2017, updating a search published previously [9]. The search term was ‘Ebola’ We searched for epidemiological data (case number and mortality data) on the WHO website.
Study selection and criteria
We included articles describing hospitalized individuals with laboratory-confirmed EVD who were treated in the three most affected countries (Liberia, Sierra Leone, Guinea) during the epidemic in West Africa (2013–2016).
We included articles that described one or more clinical aspects of disease (defined as a symptom, examination finding, or laboratory investigation finding) and an acute patient outcome (at a minimum, death up to or within 28 days of discharge). Hospitalization was defined as admission to an Ebola Treatment Unit/Centre (ETC) or a pre-existing hospital. A post hoc addition was made to include several sites that were originally termed Ebola holding centres but admitted patients and provided supportive medical therapy (e.g. intravenous fluids) and that were retrospectively retermed as ETCs by national governments. To identify representative clinical data sets, articles must have included all EVD patients in their facility for the defined period and so articles that reported a subset of patients (usually on the basis of age or pregnancy status) were excluded, as were articles that included additional enrolment criteria (usually clinical trials).
We excluded articles that described fewer than ten patients. Language was restricted to articles with English and French abstracts.
Data extraction and quality assessment
We reviewed manuscript titles, abstracts and full text in duplicate based on pre-agreed selection criteria. For eligible articles, data were extracted in duplicate into an electronic data form, with two independent reviewers following the study data management standard operating procedure. The senior author made final determination of article selection or data extraction where there was disagreement. Quality assessment was performed only by the senior author.
We extracted study characteristics including the number of patients included, selection criteria, dates of patient enrolment and dates of publication. Where hospitalized and non-hospitalized patients were included, we extracted only data on hospitalized patients.
We extracted patient information including demographics, temporal characteristics of disease, clinical manifestations of disease (based on the WHO case definition used during the outbreak [12]), patient co-morbidities, vital signs, predictors of mortality, mortality and patient treatment. Data extraction was recorded by survival status of the patient where that information was available. We recorded symptom data at three time-points (on admission (defined as <24 h), during hospitalization (subsequent days) or at any time (for when an article did not describe the time of data collection, or this could not be deduced). Vital sign data were abstracted on admission only because there was little longitudinal data on pilot testing.
For all variables, we extracted numerator and denominator data for dichotomous variables, and summary statistics for continuous variables (mean or median, and standard deviation, or interquartile range, or range, or proportion high or low). For predictors of survival we extracted unadjusted and adjusted odds or risk ratios with 95% confidence intervals. Data were reported as missing for each variable when it was not available in the manuscript. Manuscript authors were not contacted for missing data.
Quality assessment
An existing observational data quality assessment scale [13] was piloted but found to be unsuitable so we devised a study-specific scale where assessment criteria were based on parameters independently identified as critical for outbreak evaluation [14]. These include that the data source is defined; the timeframe of data collection is defined; and there is transparency regarding the proportion of cases for which an outcome is known. Clinical interpretability of data was assessed based on whether each article provided an estimate of clinical descriptors that were known or suspected co-variates of outcome at the time the West Africa outbreak commenced (age, pregnancy status, viral load and time from symptom onset to patient admission) [15–17].
Treatment of duplicate data
Duplicate or overlapping data sets were first identified on the basis of replication of authors, facilities, enrolment periods, sample size or statement of previous publication. Because some facility names and affiliated organizations changed during the outbreak, articles were then manually mapped by the senior author who was familiar with ETC operations during the outbreak.
When duplicate or overlapping data sets occurred, they were ranked from that which included the most patients, to that with the fewest number of patients. The first ranked (largest data set) was always used for reporting patient variables when data were available. However, because more detailed clinical description could be included in one or more lower ranked data sets, these were not excluded outright. Instead, a variable of interest was searched in a step-wise manner down the ranking scale and then reported for the highest ranked article where it was contained. We refer to the first ranked data sets collectively as the ‘primary’ data set.
Statistical analysis
Descriptive statistics are presented as frequencies for categorical variables, means and standard deviations for normally distributed data, and median with range for other continuous variables. Relative risks were calculated from the number of events and participants in each group. All p-values are calculated from two-tailed tests of statistical significance with a type I error rate of 5%.
Meta-analysis
Individual proportion or relative risk estimates are displayed graphically with forest plots, and on summary forest plots when variables are clinically related. Meta-analysis was performed using a random effects model. A binomial-specific method was used for meta-analysis of proportions [18]. Heterogeneity for all meta-analysis was assessed using the χ 2 test for assessment of heterogeneity and quantified with the I 2 statistic. Estimates of publication bias in meta-analysis (such as Egger's test) were not included because of their limited utility when there are a small number of publications [19]. Meta-analysis was only performed when three or more articles reported data for the variable.
Actionable data analysis
Three criteria were used to perform an actionable data analysis: the representativeness of the data to the underlying population, the ‘transparency’ of reporting [10], and the ability for the data to be quantitatively synthesized (compatibility) [10].
We assessed data representativeness by comparison with WHO data sets. For symptom data, this was aggregate data from the first year of the outbreak (data stratified by hospitalization status was not available) [20]. There are no aggregated estimates available for vital sign data. The comparative estimate for case fatality rate (CFR) was based on individual patient data (IPD) on hospitalized individuals during the entire outbreak [2].
We assessed data transparency by reporting whether anonymized IPD were available for an article, and whether publication of the same cohort in another manuscript was acknowledged when applicable.
We assessed data compatibility for synthesis by comparing the consistency of data reporting for two key co-variates of outcome that could be used in meta-regression (patient age and estimate of viral load) and by mapping predictors of patient outcomes.
We performed all analyses with stata/MP version 15.0.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation or writing of the report.
Results
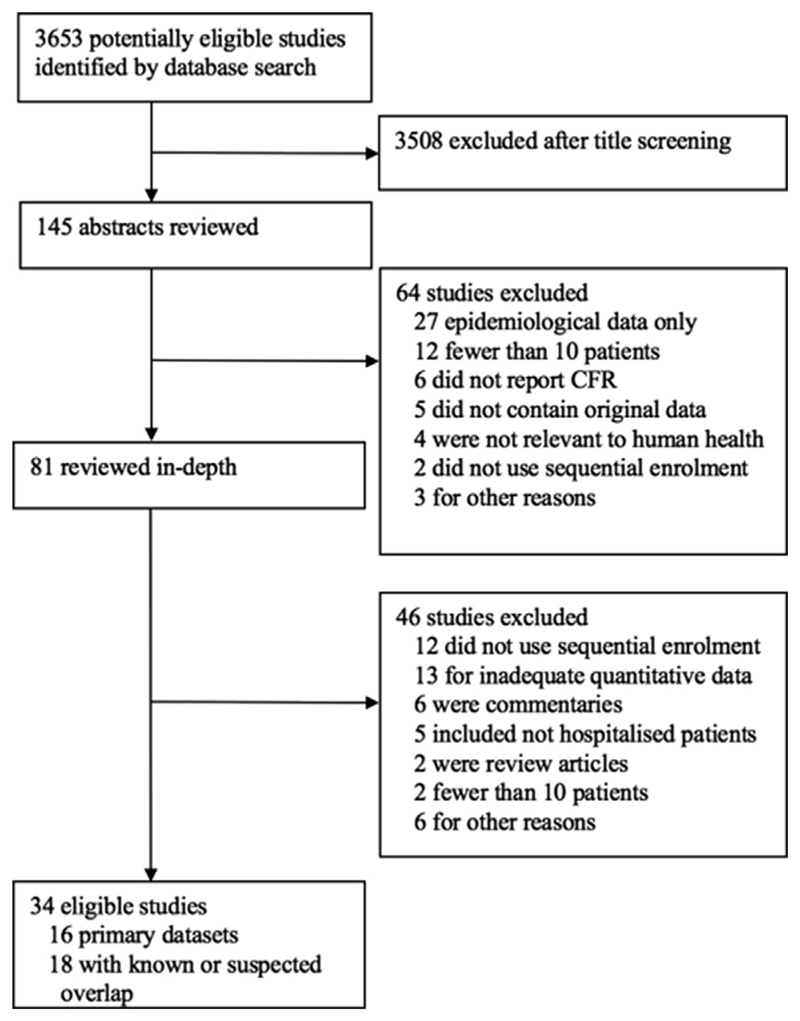
A total of 3653 articles were screened (Fig. 1), and 34 articles were eligible for inclusion (see Supplementary material, Appendix S1) [21–54]. Out of these, 16 articles represent the largest, or only, representation of patients (termed the primary data set) and 18 articles duplicate or overlap with these data and are only used when data are missing from the primary data set. The total number of patients included in the primary data set is 6168. For this data set the median number of patients per article was 207 (interquartile range 108–607).
Fig. 1.
Summary of article selection process.
Quality of evidence
Appendix S2 (see Supplementary material) details the quality of included evidence. Articles reported the data source and time of collection in 47% (16/34) of articles, the exact dates of the study in 79% (27/34) of articles, and provided specific details of laboratory confirmation in 59% (20/34) of articles. Most articles (53%, 18/34) described if patients lost to follow up were excluded from analysis, and detailed the proportion of cases for whom the clinical outcome was known (97%, 33/34). Fewer articles (35%, 12/34) detailed the extent of missing data for other variables, or described statistical management of missing data. Reporting of covariates for estimating mortality varied by category–88% (30/34) reported age, 68% (23/34) time from symptom onset to hospitalization, 59% (20/34) viral load (or cycle threshold), and 9% (3/34) pregnancy status.
Case fatality rate
The pooled CFR of the primary data set was 51% (95% CI 46%–56%); however, heterogeneity was significant (I 2 = 92.8%, p < 0.01). The forest plot for CFR is provided in the Supplementary material (Appendix S3).
Symptoms of EVD infection
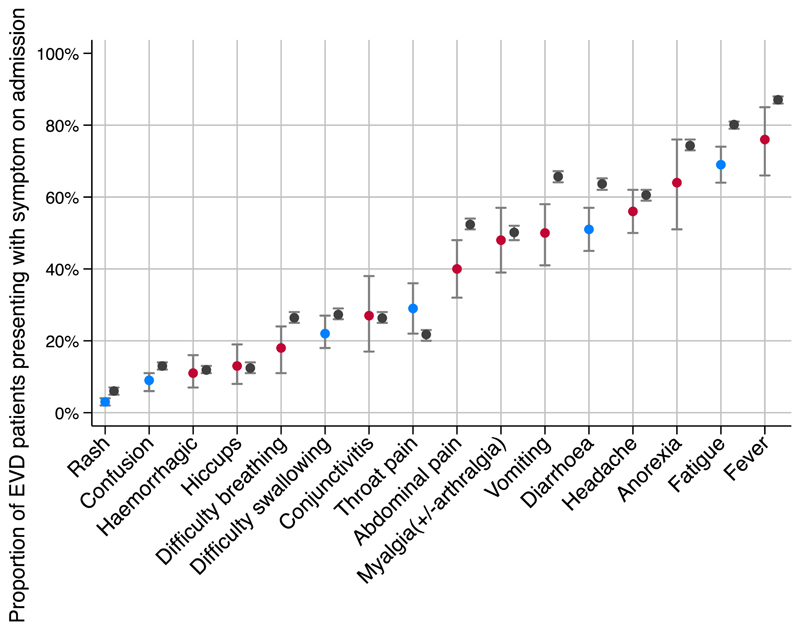
In pooled analysis of the primary data set, the most common presenting symptoms were fever (76%, 95% CI 66%–85%), fatigue (71%, 95% CI 64%–74%) and anorexia (64%, 95% CI 51%–76%) (Fig. 2). There was high (defined as I 2 value > 75% [55]) heterogeneity for symptoms of haemorrhagic manifestations, hiccups, difficulty breathing, conjunctivitis, abdominal pain, myalgia, vomiting, headache, anorexia and fever. There was low to moderate hetero-geneity for all other presented symptoms based on I 2 estimate, although the χ 2 for heterogeneity was still significant for diarrhoea (p < 0.01) and lethargy (p 0.01) (see Supplementary material, Appendix S4).
Fig. 2.
Meta-analysis of proportion of individuals with Ebola virus disease (EVD) presenting with a symptom compared with reference data. Blue represents a meta-analysis estimate with low or moderate heterogeneity for the pooled estimate, red represents a meta-analysis estimate with high heterogeneity. Grey is WHO reference data. All estimates are shown with their 95% CI.
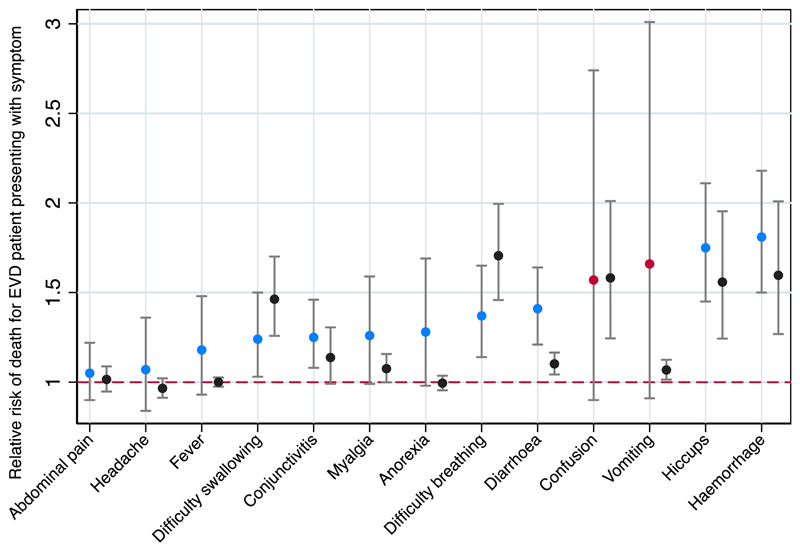
The pooled relative risk of death was significantly >1 in patients presenting with the symptoms of haemorrhage, hiccups, diarrhoea, breathing difficulty, conjunctivitis and difficulty swallowing compared with those presenting without the symptom (Fig. 3). Heterogeneity was low to moderate for all estimates except for confusion and vomiting, although the χ 2 test for heterogeneity was significant for headache (p 0.046) (see Supplementary material, Appendix S5).
Fig. 3.
Meta-analysis of the relative risk of death in individuals with Ebola virus disease (EVD) presenting with a symptom on admission, compared with symptom absence, and showing comparison reference data. Blue represents a meta-analysis estimate with low or moderate heterogeneity for the pooled estimate, red represents a meta-analysis estimate with high heterogeneity. Grey is WHO reference data. All estimates are shown with their 95% CI.
Systematic review was not feasible for symptoms during hos-pitalization. Only one article systematically reported patient symptoms in the period following admission [31], although sporadic reporting of some other symptoms occurred in a further two articles.
Meta-analysis of vital sign data was not feasible, because only two articles systematically reported data [38,52], with some detail in one further article [42].
Supportive care for EVD infection
Only one article differentiated between medications that were prescribed (as part of a protocol) and those that were actually administered to patients. Five articles detailed the exact number of patients receiving antibiotics and antimalarials (and one article detailed route of admission) and six articles described intravenous fluid use (with an estimate of fluid volume provided in half of these manuscripts).
Actionable data analysis
Data representativeness
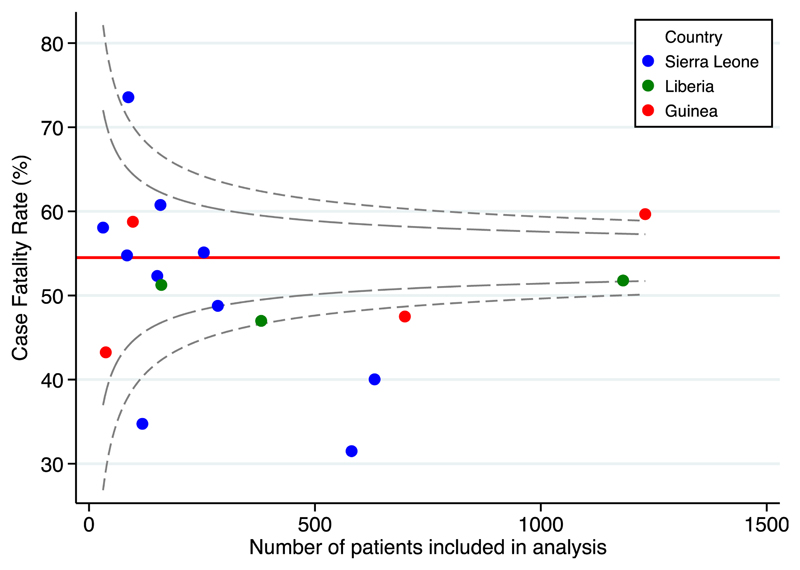
Fig. 4 depicts the CFR reported in the published articles compared with reference data from WHO (CFR from all hospitalized patients during the outbreak) [2]. Our estimate of CFR was 51%, the WHO estimate was 54.5%.
Fig. 4.
Funnel plot of case fatality rate (CFR) by country. The horizontal red line is a reference value that depicts the overall CFR for hospitalized patients during the outbreak reported by WHO, with 95% and 99.8% Cl.
Fig. 2 shows the meta-analysis estimates for the proportion of patients presenting with a symptom compared with the reference data (WHO data for all confirmed and probable patients presenting during the first year of the outbreak).
Fig. 3 depicts a comparison of our meta-analysis estimates for the relative risk of death for a patient presenting with a symptom at the time of admission, in comparison with reference data (as outlined above).
Data transparency
Eighty-five per cent (29/34) of articles made no statement on IPD availability, and data were only available for two articles where a statement was made. Eighty-two per cent (28/34) of articles re-ported duplicate or overlapping data (although this may be an underestimation, as we did not systematically review articles that did not meet our inclusion criteria). In only one of these articles was there acknowledgement of duplicate presentation (see Supplementary material, Appendix S6).
Data compatibility
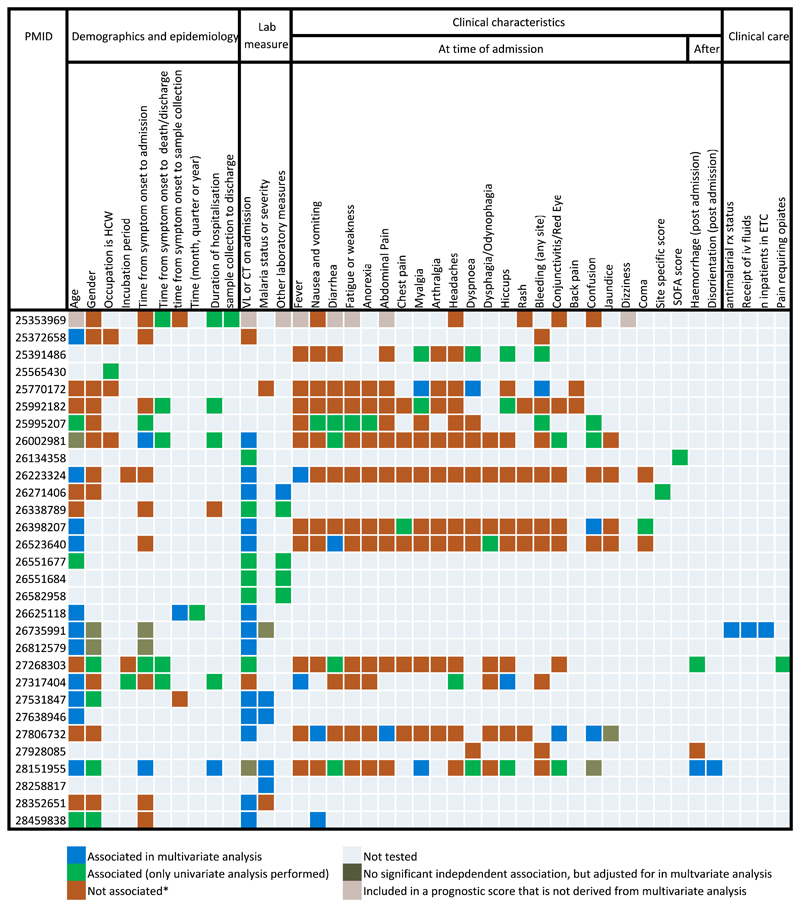
Of the 15 manuscripts presenting data according to age categories, the categories selected for stratification were the same for only two articles (see Supplementary material, Appendix S7). Between the 15 manuscripts that stratified by viral load for analysis, ten different categorization systems were used (see Supplementary material, Appendix S7). The diversity of variables found to be predictive of case fatality are shown in Fig. 5.
Fig. 5.
Reporting of predictors of mortality for patients with laboratory confirmed Ebola virus disease (EVD). Cells with a red tab reflect a complex relationship (such as statistical significance for only a subset of data). Asterisk indicates the it includes if association on univariate analysis is not supported by multivariate analysis. VL, viral load; Ct, cycle threshold; Rx, treatment.
Discussion
In this analysis of the clinical presentation of EVD, we report relatively comprehensive data for symptoms on admission (and their association with CFR). We planned to describe the evolution of patient symptoms and predictors for death because this information is not available in epidemiology data sets and is of significant utility for clinical decision-making. However, we found the literature inadequate to perform such analyses. In some ETCs, the lack of data reflects the clinical reality of working in a humanitarian setting, where at times, careful clinical observation was not feasible. However, in many other ETCs, especially those reporting data later during the outbreak, some data are expected to exist.
Meta-analysis indicated substantial heterogeneity for analysis of the proportion of patients presenting with a symptom, and for analysis of CFR. We attempted to limit clinical diversity by only including articles that enrolled all patients in a sequential manner presenting for treatment. However, populations may have differed between studies due to ETCs using different admission routes. Moreover, selection bias may have occurred because early during the outbreak some overwhelmed ETCs turned away patients and in other cases fears or rumours regarding ETCs may have altered admission patterns. The representativeness of estimates is further diminished by limited reporting of covariates for mortality (including time to hospitalization, cycle threshold and pregnancy status).
When we compared with WHO estimates, the limitations of clinical decision-making based on the literature became evident. For example, our estimates of the proportion of patients presenting with vomiting (50%, 95% CI 41%–58%) and diarrhoea (51%, 95% CI 45%–57%) are substantially lower than WHO estimates (66%, 95% CI 64%–67%; and 64%, 95% CI 62%–65%, respectively) with nonoverlapping confidence intervals, and our point estimates of the relative risk of death when presenting with each of these symptoms are higher than WHO estimates–diarrhoea 1.4 (95% CI 1.2–1.6) compared with 1.1 (95% 1.0–1.2); vomiting 1.7 (95% CI 0.9–3.0) compared with 1.1 (95% CI 1.0–1.1). Whether gastrointestinal manifestations are a common manifestation of EVD among all patients, or is relatively rarer but more discriminative of patients at risk of death, is unclear, which limits the utility of this information for clinical triage. The meta-analysis estimate of CFR is lower than for the WHO estimate in all hospitalized individuals with known outcomes. This might represent publication bias, where ETCs with high survival rates are more likely to publish their findings.
Two issues identified from the actionable data analysis have particular clinical relevance. First, the lack of available IPD represents a significant impediment to pooling or comparison of data, especially given the lack of consistency in reporting of summary measures. Second, overlapping cohorts of patients often only became evident due to author experience with the outbreak, and only one article declared previous publication (although we recognize that authors of some manuscripts may not be aware of duplicate publication). The consequence is that those relying on the literature as an evidence base may see consensus in the literature where it does not exist.
Despite poorly standardized reporting of longitudinal clinical data, there was near universal consistency in the reporting of symptoms on admission, no doubt facilitated by use of WHO case reporting forms. Therefore, a solution is in adoption of such best practice standardized case reporting forms for clinical data. These provide clarity regarding objective data (e.g. discriminating between lack of a symptom, with lack of collection of data on a symptom), provide data dictionaries (that provide clear agreed definitions for subjective terms such as ‘fatigue’) and can provide options to allow for different complexities of data collection depending on resources (ranging from symptom reporting only through to comprehensive laboratory and treatment reporting). These forms are increasingly being produced for reporting the natural history of other emerging infectious diseases, such as the WHO protocol for Zika virus infection [56]; the findings of this work further support their use.
Our findings demonstrate that clinical data standards and patient registries, akin to the value of cancer registries, are needed. The aggregated epidemiological data sets curated by WHO are clearly valuable (although not without data collection and analysis constraints themselves) [2]. Due to these, confidence in estimates of the epidemiology of EVD have improved markedly and have been instrumental in directing public health control. An evidence base of this calibre is just as important for informing the clinical care of patients but does not exist. Such a data set is feasible given that sufficient data for publication of a clinical data set exist for at least 6168 patients. This approach has been used previously for aggre-gation of clinical trial data for malaria, and in this instance has allowed IPD pooled meta-analysis to inform treatment guidelines with greater statistical power than would otherwise be possible [57]. Centralized patient registries have also been suggested for clinical trials of other tropical diseases [58], rare diseases [10] and resistant pathogens [59], where fragmented data reporting also occurs. Importantly, in outbreak settings, centralized databases should also be used to facilitate and advocate for ethical data collection and storage practices. Some of this work must be undertaken in advance of outbreaks, to test and streamline data collection tools that are feasible to use in the early stages of outbreaks when resources are stretched. Therefore, to guide these patient registries, we suggest that a systematic assessment of clinical knowledge gaps for epidemic-prone infections should be prioritized. In the first instance, this should be undertaken for WHO research and development blueprint priority pathogens.
There are several limitations to our analysis. We do not wish for our comparison with WHO data to be misconstrued as comparison with reference standard data, but rather that it constitutes the best available comparison to approximate the representativeness of data included. In particular, our comparisons for symptom data are limited as the WHO data include non-hospitalized cases. For pragmatic reasons we limited analysis to clinical data available in standardized formats (e.g. mean, median, categorical data) although clinically meaningful data may exist in other formats. Planned subgroup analysis by admission route was not possible due poor consistency of reporting. Subgroup analysis for CFR stratified by date was not possible because the distribution of date of admissions within each study was not available.
Conclusions
This systematic review summarizes clinical data for 6168 individuals with EVD. Synthesis of data largely failed due to lack of standardization and transparency in reporting of clinical data. Given the volume of clinical data that was collected, more profound gains in our knowledge regarding the natural history of EVD and factors influencing patient outcomes could have been achieved if the data had been more thoughtfully defined, assembled and shared. The most appropriate method of achieving this is through data-standardization initiatives, patient registries and structured assessment of data priorities. Although not diminishing the practical difficulties of doing so, we advocate strongly for a patient-centred research response to be embedded in the clinical response to EVD outbreaks as a means to improve evidence-based care and patient survival.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2019.06.032.
Acknowledgements
This report is independent research funded and undertaken by the UK Public Health Rapid Support Team, which is funded by the UK Government. The views expressed in this publication are those of the author(s) and not necessarily those of the National Health Service, the National Institute for Health Research or the Department of Health. This work was supported by the Wellcome Trust of Great Britain (grant numbers 107834/Z/15/Z and 106491/Z/14/Z). AR was funded by a Rhodes Scholarship. The funders had no role in the study design, the collection, analysis and interpretation of data, in the writing of the report, or in the decision to submit the paper for publication.
Footnotes
Transparency declaration
The authors have no competing interests to declare.
Author contributions
AR, AS and PH conceived of the manuscript; AR, AS, RR, EL, AE, AC, MB, NK and LC contributed to data collection; AR, PH, LO and KS contributed to data analysis and interpretation; and AR and PH drafted the document. All authors agree with the contents.
References
- [1].World Health Organization. Ebola situation reports: archive. 2016 Available at: http://www.who.int/csr/disease/ebola/situation-reports/archive/en/
- [2].Garske T, Cori A, Ariyarajah A, Blake IM, Dorigatti I, Eckmanns T, et al. Het-erogeneities in the case fatality ratio in the West African Ebola outbreak 2013–2016. Phil Trans R Soc B. 2017;372 doi: 10.1098/rstb.2016.0308. 20160308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Uyeki TM, Mehta AK, Davey RTJ, Liddell AM, Wolf T, Vetter P, et al. Clinical management of Ebola virus disease in the United States and Europe. N Engl J Med. 2016;374:636–46. doi: 10.1056/NEJMoa1504874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jacobs M, Rodger A, Bell DJ, Bhagani S, Cropley I, Filipe A, et al. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet. 2016;388:498–503. doi: 10.1016/S0140-6736(16)30386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].World Health Organization. Technical Report. Geneva: World Health Organization; 2016. Clinical care for survivors of Ebola virus disease. Available at: http://apps.who.int/iris/bitstream/10665/204235/1/WHO_EVD_OHE_PED_16.1_eng.pdf. [Google Scholar]
- [6].Varkey JB, Shantha JG, Crozier I, Kraft CS, Lyon GM, Mehta AK, et al. Persistence of Ebola virus in ocular fluid during convalescence. N Engl J Med. 2015;372:2423–7. doi: 10.1056/NEJMoa1500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chughtai AA, Barnes M, Macintyre CR. Persistence of Ebola virus in various body fluids during convalescence: evidence and implications for disease transmission and control. Epidemiol Infect. 2016;144:1652–60. doi: 10.1017/S0950268816000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lamontagne F, Fowler RA, Adhikari NK, Murthy S, Brett-Major DM, Jacobs M, et al. Evidence-based guidelines for supportive care of patients with Ebola virus disease. Lancet. 2017;391:701–8. doi: 10.1016/S0140-6736(17)31795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rojek A, Horby P, Dunning J. Insights from clinical research completed during the west Africa Ebola virus disease epidemic. Lancet Infect Dis. 2017;17:e280–92. doi: 10.1016/S1473-3099(17)30234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Frieden TR. Evidence for health decision making–beyond randomized, controlled trials. N Engl J Med. 2017;377:465–75. doi: 10.1056/NEJMra1614394. [DOI] [PubMed] [Google Scholar]
- [11].Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].World Health Organization and others. Case definition recommendations for Ebola or Marburg virus diseases: interim guideline. 2014 Available at: http://apps.who.int/iris/bitstream/10665/146397/1/WHO_EVD_CaseDef_14.1_eng.pdf?ua=1&ua=1.
- [13].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- [14].Cori A, Donnelly CA, Dorigatti I, Ferguson NM, Fraser C, Garske T, et al. Key data for outbreak evaluation: building on the Ebola experience. Phil Trans R Soc B. 2017;372 doi: 10.1098/rstb.2016.0371. 20160371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mupapa K, Mukundu W, Bwaka MA, Kipasa M, De Roo A, Kuvula K, et al. Ebola hemorrhagic fever and pregnancy. J Infect Dis. 1999;179:S11–2. doi: 10.1086/514289. [DOI] [PubMed] [Google Scholar]
- [16].MacNeil A, Farnon EC, Wamala J, Okware S, Cannon DL, Reed Z, et al. Proportion of deaths and clinical features in Bundibugyo Ebola virus infection, Uganda. Emerg Infect Dis. 2010;16:1969. doi: 10.3201/eid1612.100627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sanchez A, Lukwiya M, Bausch D, Mahanty S, Sanchez AJ, Wagoner KD, et al. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J Virol. 2004;78:10370–7. doi: 10.1128/JVI.78.19.10370-10377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nyaga VN, Arbyn M, Aerts M. Metaprop: a stata command to perform meta-analysis of binomial data. Arch Public Health. 2014;72:39. doi: 10.1186/2049-3258-72-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].WHO Ebola Response Team. Agua-Agum J, Ariyarajah A, Aylward B, Blake IM, Brennan R, Cori A, et al. West African Ebola epidemic after one year–slowing but not yet under control. N Engl J Med. 2015;372:584–7. doi: 10.1056/NEJMc1414992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Faye O, Andronico A, Faye O, Salje H, Boelle PY, Magassouba N, et al. Use ofviremia to evaluate the baseline case fatality ratio of Ebola virus disease and inform treatment studies: a retrospective cohort study. PLoS Med. 2015;12:e1001908. doi: 10.1371/journal.pmed.1001908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cotte J, Cordier PY, Bordes J, Janvier F, Esnault P, Kaiser E, et al. Fluid resus-citation in Ebola virus disease: a comparison of peripheral and central venous accesses. Anaesth Crit Care Pain Med. 2015;34:317–20. doi: 10.1016/j.accpm.2015.06.010. [DOI] [PubMed] [Google Scholar]
- [23].Janvier F, Foissaud V, Cotte J, Team HWETCM, Aletti M, Savini H, et al. Monitoring of prognostic laboratory markers in Ebola virus disease. J Infect Dis. 2015;213:1049. doi: 10.1093/infdis/jiv546. [DOI] [PubMed] [Google Scholar]
- [24].Vernet MA, Reynard S, Fizet A, Schaeffer J, Pannetier D, Guedj J, et al. Clinical, virological, and biological parameters associated with outcomes of Ebola virus infection in Macenta, Guinea. JCI Insight. 2017:2. doi: 10.1172/jci.insight.88864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Loubet P, Palich R, Kojan R, Peyrouset O, Danel C, Nicholas S, et al. Development of a prediction model for Ebola virus disease: a retrospective study in Nzerekore Ebola treatment center, Guinea. Am J Trop Med Hyg. 2016;95:1362–7. doi: 10.4269/ajtmh.16-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ji YJ, Duan XZ, Gao XD, Li L, Li C, Ji D, et al. Clinical presentations and outcomes of patients with Ebola virus disease in Freetown, Sierra Leone. Infect Dis Poverty. 2016;5:101. doi: 10.1186/s40249-016-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xu Z, Jin B, Teng G, Rong Y, Sun L, Zhang J, et al. Epidemiologic characteristics, clinical manifestations, and risk factors of 139 patients with Ebola virus disease in western Sierra Leone. Am J Infect Control. 2016;44:1285–90. doi: 10.1016/j.ajic.2016.04.216. [DOI] [PubMed] [Google Scholar]
- [28].Hartley MA, Young A, Tran AM, Okoni-Williams HH, Suma M, Mancuso B, et al. Predicting Ebola severity: a clinical prioritization score for Ebola virus disease. PLoS Negl Trop Dis. 2017;11:e0005265. doi: 10.1371/journal.pntd.0005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Waxman M, Aluisio AR, Rege S, Levine AC. Characteristics and survival of patients with Ebola virus infection, malaria, or both in Sierra Leone: a retrospective cohort study. Lancet Infect Dis. 2017;17:654–60. doi: 10.1016/S1473-3099(17)30112-3. [DOI] [PubMed] [Google Scholar]
- [30].Haaskjold YL, Bolkan HA, Krogh Kø, Jongopi J, Lundeby KM, Mellesmo S, et al. Clinical features of and risk factors for fatal Ebola virus disease, Moyamba district, Sierra Leone, December 2014-February 2015. Emerg Infect Dis. 2016;22:1537. doi: 10.3201/eid2209.151621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Arranz J, Lundeby KM, Hassan S, Fuentes LMZ, Garces PSJ, Haaskjold YL, et al. Clinical features of suspected Ebola cases referred to the Moyamba ETC, Sierra Leone: challenges in the later stages of the 2014 outbreak. BMC Infect Dis. 2016;16:308. doi: 10.1186/s12879-016-1609-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Crowe SJ, Maenner MJ, Kuah S, Erickson BR, Coffee M, Knust B, et al. Prognostic indicators for Ebola patient survival. Emerg Infect Dis. 2016;22:217. doi: 10.3201/eid2202.151250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].de La Vega MA, Caleo G, Audet J, Qiu X, Kozak RA, Brooks JI, et al. Ebola viral load at diagnosis associates with patient outcome and outbreak evolution. J Clin Invest. 2015;125:4421. doi: 10.1172/JCI83162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Theocharopoulos G, Danis K, Greig J, Hoffmann A, De Valk H, Jimissa A, et al. Ebola management centre proximity associated with reduced delays of healthcare of Ebola virus disease (EVD) patients, Tonkolili, Sierra Leone, 2014–15. PloS One. 2017;12:e0176692. doi: 10.1371/journal.pone.0176692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rosenke K, Adjemian J, Munster VJ, Marzi A, Falzarano D, Onyango CO, et al. Plasmodium parasitemia associated with increased survival in Ebola virus-infected patients. Clin Infect Dis. 2016;63:1026–33. doi: 10.1093/cid/ciw452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gignoux E, Azman AS, de Smet M, Azuma P, Massaquoi M, Job D, et al. Effect of artesunate-amodiaquine on mortality related to Ebola virus disease. N Engl J Med. 2016;374:23–32. doi: 10.1056/NEJMoa1504605. [DOI] [PubMed] [Google Scholar]
- [37].Bordes J, Janvier F, Aletti M, de Greslan T, Gagnon N, Cotte J, et al. Organ failures on admission in patients with Ebola virus disease. Intens Care Med. 2015;41:1504–5. doi: 10.1007/s00134-015-3912-0. [DOI] [PubMed] [Google Scholar]
- [38].Cournac JM, Karkowski L, Bordes J, Aletti M, Duron S, Janvier F, et al. Rhab-domyolysis in Ebola virus disease. Results of an observational study in a treatment center in Guinea. Clin Infect Dis. 2016;62:19–23. doi: 10.1093/cid/civ779. [DOI] [PubMed] [Google Scholar]
- [39].Barry M, Traore FA, Sako FB, Kpamy DO, Bah EI, Poncin M, et al. Ebola outbreak in Conakry, Guinea: epidemiological, clinical, and outcome features. Med Malad Infect. 2014;44:491–4. doi: 10.1016/j.medmal.2014.09.009. [DOI] [PubMed] [Google Scholar]
- [40].Barry M, Toure A, Traore FA, Sako FB, Sylla D, Kpamy DO, et al. Clinical pre-dictors of mortality in patients with Ebola virus disease. Clin Infect Dis. 2015;60:1821–4. doi: 10.1093/cid/civ202. [DOI] [PubMed] [Google Scholar]
- [41].Qureshi AI, Chughtai M, Bah EI, Barry M, Beavogui K, Loua TO, et al. High survival rates and associated factors among Ebola virus disease patients hospitalized at Donka National Hospital, Conakry, Guinea. J Vasc Interv Neurol. 2015;8:S4–11. [PMC free article] [PubMed] [Google Scholar]
- [42].Bah EI, Lamah MC, Fletcher T, Jacob ST, Brett-Major DM, Sall AA, et al. Clinical presentation of patients with Ebola virus disease in Conakry, Guinea. N Engl J Med. 2014;372:40–7. doi: 10.1056/NEJMoa1411249. [DOI] [PubMed] [Google Scholar]
- [43].Kerber R, Krumkamp R, Diallo B, Jaeger A, Rudolf M, Lanini S, et al. Analysis of diagnostic findings from the European mobile laboratory in gueckedou, Guinea, March 2014 through March 2015. J Infect Dis. 2016;214(3):S250–7. doi: 10.1093/infdis/jiw269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Li J, Duan HJ, Chen HY, Ji YJ, Zhang X, Rong YH, et al. Age and Ebola viral load correlate with mortality and survival time in 288 Ebola virus disease patients. Int J Infect Dis. 2016;42:34–9. doi: 10.1016/j.ijid.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yan T, Mu J, Qin E, Wang Y, Liu L, Wu D, et al. Clinical characteristics of 154 patients suspected of having Ebola virus disease in the Ebola holding center of Jui Government Hospital in Sierra Leone during the 2014 Ebola outbreak. Eur J Clin MicrobiolI Infect Dis. 2015;34:2089–95. doi: 10.1007/s10096-015-2457-z. [DOI] [PubMed] [Google Scholar]
- [46].Zhang X, Rong Y, Sun L, Liu L, Su H, Zhang J, et al. Prognostic analysis of patients with Ebola virus disease. PLoS Negl Trop Dis. 2015;9:e0004113. doi: 10.1371/journal.pntd.0004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Qin E, Bi J, Zhao M, Wang Y, Guo T, Yan T, et al. Clinical features of patients with Ebola virus disease in Sierra Leone. Clin Infect Dis. 2015;61:491–5. doi: 10.1093/cid/civ319. [DOI] [PubMed] [Google Scholar]
- [48].Lanini S, Portella G, Vairo F, Kobinger GP, Pesenti A, Langer M, et al. Blood kinetics of Ebola virus in survivors and nonsurvivors. J Clin Invest. 2015;125:4692–8. doi: 10.1172/JCI83111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fitzpatrick G, Vogt F, Moi Gbabai O, Black B, Santantonio M, Folkesson E, et al. Describing readmissions to an Ebola case management centre (CMC), Sierra Leone, 2014. Euro Surveill. 2014:19. [PubMed] [Google Scholar]
- [50].Dallatomasina S, Crestani R, Sylvester Squire J, Declerk H, Caleo GM, Wolz A, et al. Ebola outbreak in rural West Africa: epidemiology, clinical features and outcomes. Trop Med Int Health. 2015;20:448–54. doi: 10.1111/tmi.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hunt L, Gupta-Wright A, Simms V, Tamba F, Knott V, Tamba K, et al. Clinical presentation, biochemical, and haematological parameters and their association with outcome in patients with Ebola virus disease: an observational cohort study. Lancet Infect Dis. 2015;15:1292–9. doi: 10.1016/S1473-3099(15)00144-9. [DOI] [PubMed] [Google Scholar]
- [52].Schieffelin JS, Shaffer JG, Goba A, Gbakie M, Gire SK, Colubri A, et al. Clinical illness and outcomes in patients with Ebola in Sierra Leone. N Engl J Med. 2014;371:2092–100. doi: 10.1056/NEJMoa1411680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ansumana R, Bonwitt J, Stenger DA, Jacobsen KH. Ebola in Sierra Leone: a call for action. Lancet. 2014;384:303. doi: 10.1016/S0140-6736(14)61119-3. [DOI] [PubMed] [Google Scholar]
- [54].Levine AC, Shetty PP, Burbach R, Cheemalapati S, Glavis-Bloom J, Wiskel T, et al. Derivation and internal validation of the Ebola prediction score for risk stratification of patients with suspected Ebola virus disease. Ann Emerg Med. 2015;66:285–293.e1. doi: 10.1016/j.annemergmed.2015.03.011. [DOI] [PubMed] [Google Scholar]
- [55].Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. bMj. 2003;327:557. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].World Health Organization. Standardized protocol: natural history protocol for zikavirus infection. 2017 Availableat: http://www.who.int/reproductivehealth/zika/Natural-history-protocol-complete.pdf.
- [57].Merson L, Gaye O, Guerin PJ. Avoiding data dumpsters–toward equitable and useful data sharing. N Engl J Med. 2016;374:2414–5. doi: 10.1056/NEJMp1605148. [DOI] [PubMed] [Google Scholar]
- [58].Bush JT, Wasunna M, Alves F, Alvar J, Olliaro PL, Otieno M, et al. Systematic review of clinical trials assessing the therapeutic efficacy of visceral leishmaniasis treatments: a first step to assess the feasibility of establishing an individual patient data sharing platform. PLoS Negl Trop Dis. 2017;11:e0005781. doi: 10.1371/journal.pntd.0005781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].McCarthy MW, Walsh TJ. Drug development challenges and strategies to address emerging and resistant fungal pathogens. Exp Rev Anti-infect Ther. 2017;15:577–84. doi: 10.1080/14787210.2017.1328279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.