Abstract
Little is known about the genetic architecture of traits affecting educational attainment other than cognitive ability. We used Genomic Structural Equation Modeling and prior genome-wide association studies (GWAS) of educational attainment (n = 1,131,881) and cognitive test performance (n = 257,841) to estimate SNP associations with educational attainment variation that is independent of cognitive ability.We identified 157 genome-wide significant loci and a polygenic architecture accounting for 57% of genetic variance in educational attainment. Non-cognitive genetics were enriched in the same brain tissues and cell types as cognitive performance but showed different associations with gray-matter brain volumes. Non-cognitive genetics were further distinguished by associations with personality traits, less risky behavior,and increased risk for certain psychiatric disorders.For socioeconomic success and longevity, non-cognitive and cognitive-performance genetics demonstrated similar-magnitude associations. By conducting a GWAS of a phenotype that was not directly measured, we offer a first view of genetic architecture of non-cognitive skills influencing educational success.
“It takes something more than intelligence to act intelligently.”
– Fyodor Dostoyevsky, Crime and Punishment
Success in school—and life—depends on skills beyond cognitive ability1–4. Randomized trials of early-life education interventions find substantial benefits to educational outcomes, employment, and adult health, even though the interventions have no lasting effects on children’s cognitive functions5,6. These results have captured attention of educators and policy makers, motivating interest in so-called “non-cognitive skills”7–9. Non-cognitive skills suspected to be important for educational success include motivation, curiosity, persistence, and self-control1,10–13. However, questions have been raised about the substance of these skills and the magnitudes of their impacts on life outcomes14.
Twin studies find evidence that non-cognitive skills are heritable3,15–18. Genetic analysis could help clarify the contribution of these skills to educational attainment and elucidate their connections with other traits. However, lack of consistent and reliable measurements of non-cognitive skills in existing genetic datasets pose challenges19.
To overcome these challenges, we designed a GWAS of a latent trait, i.e. a trait not measured in any of the genotyped subjects20. We borrowed the strategy used in the original analysis of non-cognitive skills within the discipline of economics21,22: we defined genetic influences on non-cognitive skills as the genetic variation in educational attainment that was not explained by cognitive skills. We then performed GWAS on this residual “non-cognitive” genetic variation in educational attainment. This approach is a necessarily imperfect representation of the true relationship between cognitive and non-cognitive skills; in human development, cognitive abilities and other skills relevant for educational attainment likely interact dynamically, each influencing the other23. Our analysis excludes genetic influences on education-relevant skills that also influence measured cognitive abilities. The value of this imperfect approach is to make a quantity otherwise difficult to study tractable for analysis.
We conducted analysis using Genomic Structural Equation Modeling (Genomic-SEM)24 applied to published GWAS summary statistics for educational attainment and cognitive performance25. Our analysis used these summary statistics to “subtract” genetic influence on cognitive performance from the association of each single-nucleotide polymorphism (SNP) with educational attainment. The remaining associations of each SNP with educational attainment formed a new GWAS of a non-cognitive skills phenotype that was never directly measured. We call this novel statistical approach GWAS-by-subtraction.
We used results from the GWAS-by-subtraction of non-cognitive skills to conduct two sets of analyses. First, we conducted hypothesis-driven analysis using the phenotypic annotation approach26. We used genetic correlation and polygenic score analysis to test the hypothesis that non-cognitive skills influence educational and economic attainments and longevity and to investigate traits and behaviors that constitute non-cognitive skills. Second, we conducted hypothesis-free bioinformatic annotation analysis to explore the tissues, cell-types, and brain structures that might distinguish the biology of non-cognitive skills from the biology mediating cognitive influences on educational attainment.
Results
GWAS-by-subtraction identifies genetic associations with non-cognitive variance in educational attainment
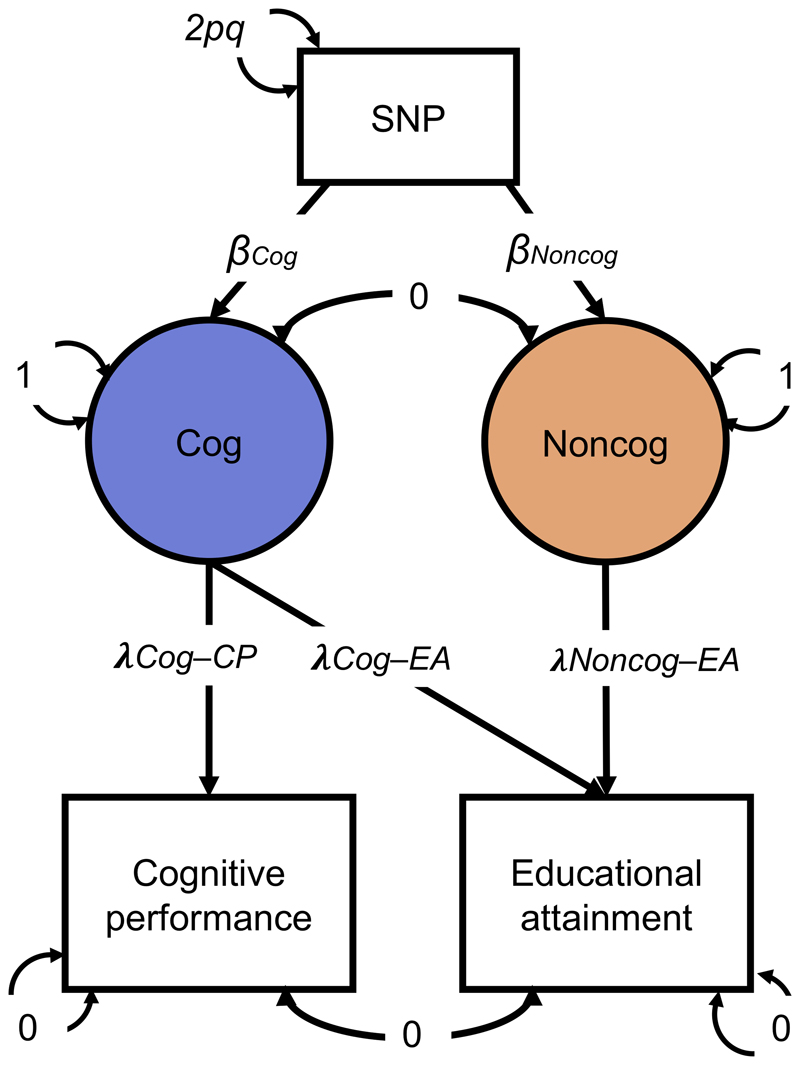
The term “non-cognitive skills” was originally coined by economists studying individuals who were equivalent in cognitive ability but who differed in educational attainment22. Our analysis of non-cognitive skills was designed to mirror this original approach: we focused on genetic variation in educational outcomes not explained by genetic variation in cognitive ability. Specifically, we applied Genomic Structural Equation Modeling (Genomic-SEM)24 to summary statistics from GWASs of educational attainment25 and cognitive performance25. Both phenotypes were regressed on a latent factor representing genetic variance in cognitive performance (hereafter “Cog”). Educational attainment was further regressed on a second latent factor representing the residual genetic variance in educational attainment left over after regressing-out variance related to cognitive performance (hereafter “NonCog”). By construction, NonCog genetic variance was independent of Cog genetic variance (r g = 0). In other words, the NonCog factor represents genetic variation in educational attainment that is not accounted for by the Cog factor. These two latent factors were then regressed on individual SNPs, yielding a GWAS of the latent constructs NonCog and Cog. A graphical representation of the model is presented in Figure 1. Parameters are derived in terms of the observed moments of the joint distribution of educational attainment, cognitive performance, and a SNP (see Supplementary Note).
Figure 1. GWAS-by-subtraction Genomic-SEM model.
Cholesky model as fitted in Genomic SEM, with path estimates for a single SNP included as illustration. SNP, cognitive performance (CP), and educational attainment (EA) are observed variables based on GWAS summary statistics. The genetic covariance between CP and EA is estimated based on GWAS summary statistics for CP and EA. The model is fitted to a 3 x 3 observed variance-covariance matrix (i.e. SNP, CP, EA). Cog and NonCog are latent (unobserved) variables. The covariances between CP and EA and between Cog and NonCog are fixed to 0. The variance of the SNP is fixed to the value of 2pq (p = reference allele frequency, q = alternative allele frequency, based on 1000 Genomes phase 3). The residual variances of CP and EA are fixed to 0, so that all variance is explained by the latent factors. The variances of the latent factors are fixed to 1. The observed variables CP and EA were regressed on the latent variables resulting in the estimates for the path loadings: λCog-CP = 0.4465; λCog-EA = 0.2237; λNonCog-EA = 0.2565. The latent variables were then regressed on each SNP that met QC criteria.
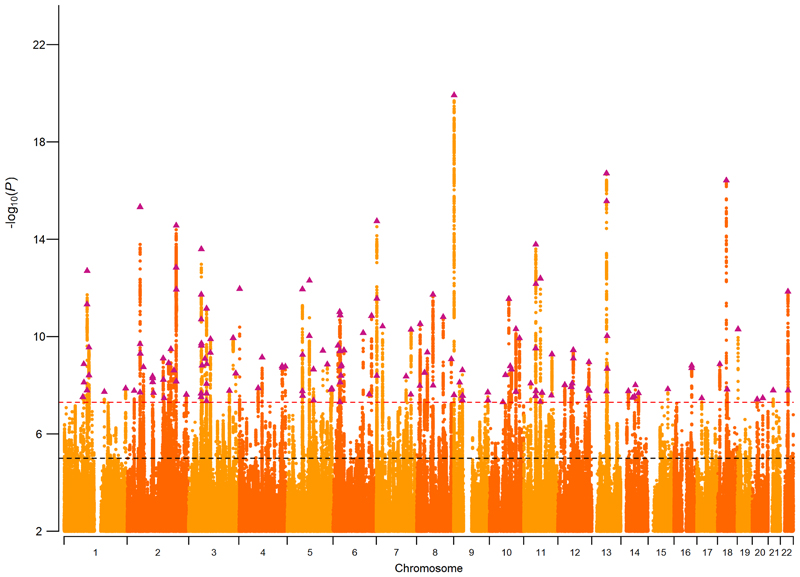
The NonCog latent factor accounted for 57% of total genetic variance in educational attainment. Using LD Score regression27, we estimated SNP-heritability for NonCog to be h 2 NonCog = 0.0637 (SE = 0.0021). After conventional GWAS significance threshold correction, GWAS of NonCog identified 157 independent genome-wide significant lead SNPs (independent SNPs defined as outside a 250-kb window, or within a 250-kb window and r 2 < 0.1). The results from the NonCog GWAS are graphed as a Manhattan plot in Figure 2. NonCog and Cog GWAS details are reported in Supplementary Tables 1-4, Supplementary Figure 1, and the Supplementary Note. In addition, we report a series of sensitivity analyses as follows: analysis of potential biases due to cohort differences (Supplementary Table 5 and Supplementary Figs. 2-4); analysis of impact of allowing for positive genetic correlations between NonCog and Cog (Supplementary Tables 6 and 7, and Supplementary Figs. 5 and 6; analysis of impact of allowing for a moderate causal effect of educational attainment on cognitive performance28 (Supplementary Table 8 and Supplementary Figs. 7-9).
Figure 2. Manhattan plot of SNP associations with NonCog.
Plot of the -log10(P-value) associated with the Wald test (two-sided) of βNonCog for all SNPs, ordered by chromosome and base position. Purple triangles indicate genome-wide significant (P < 5 × 10-8) and independent (within a 250-kb window and r 2 < 0.1) associations. The red dashed line marks the threshold for genome-wide significance (P = 5 × 10−8), and the black dashed line the threshold for nominal significance (P = 1 × 10−5).
Phenotypic annotation analysis elucidates behavioral, psychological and psychiatric correlates of non-cognitive skills genetics
Our phenotypic annotation analyses proceeded in two steps. First, we conducted polygenic score (PGS) and genetic correlation (rG) analysis to test whether our GWAS-by-subtraction succeeded in identifying genetic influences that were important to educational attainment and also distinct from genetic influences on cognitive ability. Second, we conducted PGS and rG analyses to explore how NonCog related to a network of phenotypes that psychology and economics research suggests might form the basis of non-cognitive influences on educational attainment.
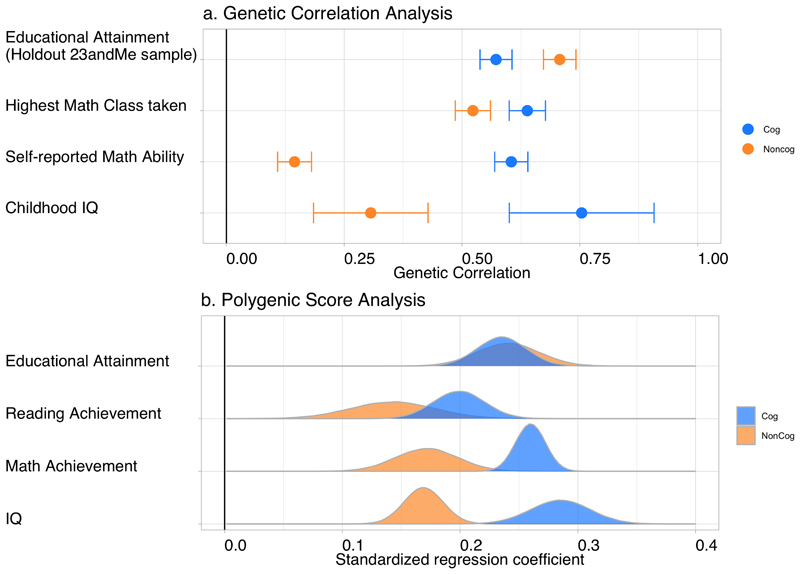
NonCog genetics are distinct from cognitive performance and are important to education, socioeconomic attainment, and longevity. To establish whether the Genomic-SEM GWAS-by-subtraction succeeded in isolating genetic variance in education that was independent of cognitive function, we compared genetic associations of NonCog and Cog with educational attainment and cognitive test performance. Results for analysis of education and cognitive test phenotypes are graphed in Figure 3.
Figure 3. Polygenic prediction and genetic correlations with IQ and educational achievement.
a, Genetic correlations of NonCog and Cog with educational attainment, highest math class taken, self-reported math ability, and childhood IQ. The dots represent genetic correlations estimated using Genomic SEM. Correlations with NonCog are in orange, and with Cog in blue. Error bars represent 95% CIs. Exact estimates and P-values are reported in Supplementary Table 14. For analysis of genetic correlations with educational attainment, we re-ran the Genomic-SEM model to compute NonCog and Cog using summary statistics that omitted the 23andMe sample from the educational attainment GWAS. We then used the 23andMe sample to run the GWAS of educational attainment. Thus, there is no sample overlap in this analysis. b, Effect-size distributions from meta-analysis of NonCog and Cog polygenic score associations with cognitive test performance and educational attainment. Outcomes were regressed simultaneously on NonCog and Cog polygenic scores. Effect-sizes entered into the meta-analysis were standardized regression coefficients interpretable as Pearson r. Exact estimates and P-values are reported in Supplementary Table 12. Samples and measures are detailed in Supplementary Tables 9 and 10. Traits were measured in different samples: educational attainment was measured in the AddHealth, Dunedin, E-Risk, NTR, and WLS samples (n = 24,056); reading achievement and mathematics achievement were measured in the AddHealth, NTR, and Texas-Twin samples (n = 9,274 for reading achievement; n = 10,747 for mathematics achievement); cognitive test performance (IQ) was measured in the Dunedin, E-Risk, NTR, Texas Twins, and WLS samples (n = 11,351). The densities were obtained by randomly generating normal distributions where the meta-analytic estimate was included as the mean and the meta-analytic standard error as the standard deviation.
We conducted PGS analysis of educational attainment in the Netherlands Twin Register29 (NTR), National Longitudinal Study of Adolescent to Adult Health30 (AddHealth), Dunedin Longitudinal Study31, E-Risk32, and Wisconsin Longitudinal Study33 (WLS) cohorts (meta-analysis n = 24,056; cohorts descriptions in Supplementary Tables 9 and 10 and Supplementary Note). PGS effect-sizes were the same for NonCog and Cog (NonCog β = 0.24 (SE = 0.03), Cog β = 0.24 (SE = 0.02), P diff = 0.702; all PGS results are reported in Supplementary Tables 11 and 12). We conducted complementary genetic correlation analysis using Genomic SEM and GWAS summary statistics from a hold-out-sample GWAS of educational attainment (Supplementary Note). This analysis allowed us to compute an out-of-sample genetic correlation of NonCog with educational attainment. NonCog showed a stronger genetic correlation with educational attainment as compared to Cog (NonCog r g = 0.71 (SE = 0.02), Cog r g = 0.57 (SE = 0.02), P diff < 0.0001; all genetic correlation results are reported in Supplementary Tables 13 and 14).
We conducted PGS analysis of cognitive test performance in the NTR, Texas Twin Project34, Dunedin, E-Risk, and WLS cohorts (combined n = 11,351). The goal of our GWAS-by-subtraction analysis was to exclude, as much as possible, genetic variance in cognitive ability from genetic variance in skills relevant for education. Consistent with this goal, effect-sizes for NonCog PGS associations with full-scale IQ were smaller by half as compared to Cog PGS associations (NonCog β = 0.17 (SE = 0.02), Cog β = 0.29 (SE = 0.03); P diff < 0.0001). However, the non-zero correlation between the NonCog PGS and full-scale IQ is a reminder that the cognitive performance GWAS used in our GWAS-by-subtraction analyses does not capture the entirety of genetic influences on all forms of cognitive tests measured at all points in the lifespan. Additional PGS analyses of IQ subscales are reported in Supplementary Figure 10 and Supplementary Tables 11 and 12.
We conducted complementary genetic correlation analysis using results from a published GWAS of childhood IQ35. Parallel to PGS analysis, the NonCog genetic correlation with childhood IQ was smaller by more than half as compared to the Cog genetic correlation (NonCog r g = 0.31 (SE = 0.06), Cog r g = 0.75 (SE = 0.08), P diff_fdr < 0.0001). Of the total genetic correlation between childhood IQ and educational attainment, 31% of the covariance was explained by NonCog and 69% by Cog.
We next examined downstream economic and health outcomes associated with greater educational attainment36,37. In PGS analysis in the AddHealth and Dunedin cohorts (n = 6,358), NonCog and Cog PGSs showed similar associations with occupational attainment (NonCog β = 0.21 (SE = 0.01), Cog β = 0.21 (SE = 0.01), P diff = 0.902). In genetic correlation analysis, NonCog showed a similar relationship to income38 as Cog (NonCog r g = 0.62, (SE = 0.04), Cog r g = 0.62 (SE = 0.04), P diff_fdr = 0.947) and a stronger relationship with neighborhood deprivation38, a measure related to where a person can afford to live (NonCog r g = -0.51 (SE = 0.05), Cog r g = -0.32 (SE = 0.04), P diff_fdr = 0.001). In Genomic-SEM analysis, NonCog explained 53% of the genetic correlation between educational attainment and income and 65% of the genetic correlation between educational attainment and neighborhood deprivation (Supplementary Table 15).
We conducted genetic correlation analysis of longevity based on GWAS of parental lifespan39. Genetic correlations were stronger for NonCog as compared to Cog (NonCog r g = 0.37 (SE = 0.03); Cog r g = 0.27 (SE = 0.03); P diff_fdr = 0.024). In Genomic-SEM analysis, NonCog explained 61% of the genetic correlation between educational attainment and longevity.
In sum, NonCog and Cog genetics showed similar relationships with educational attainment and its long-term outcomes, despite NonCog genetic having a much weaker relationship to measured cognitive test performance than Cog genetics. These findings broadly support the hypothesis that non-cognitive skills distinct from cognitive abilities are an important contributor to success across the life course.
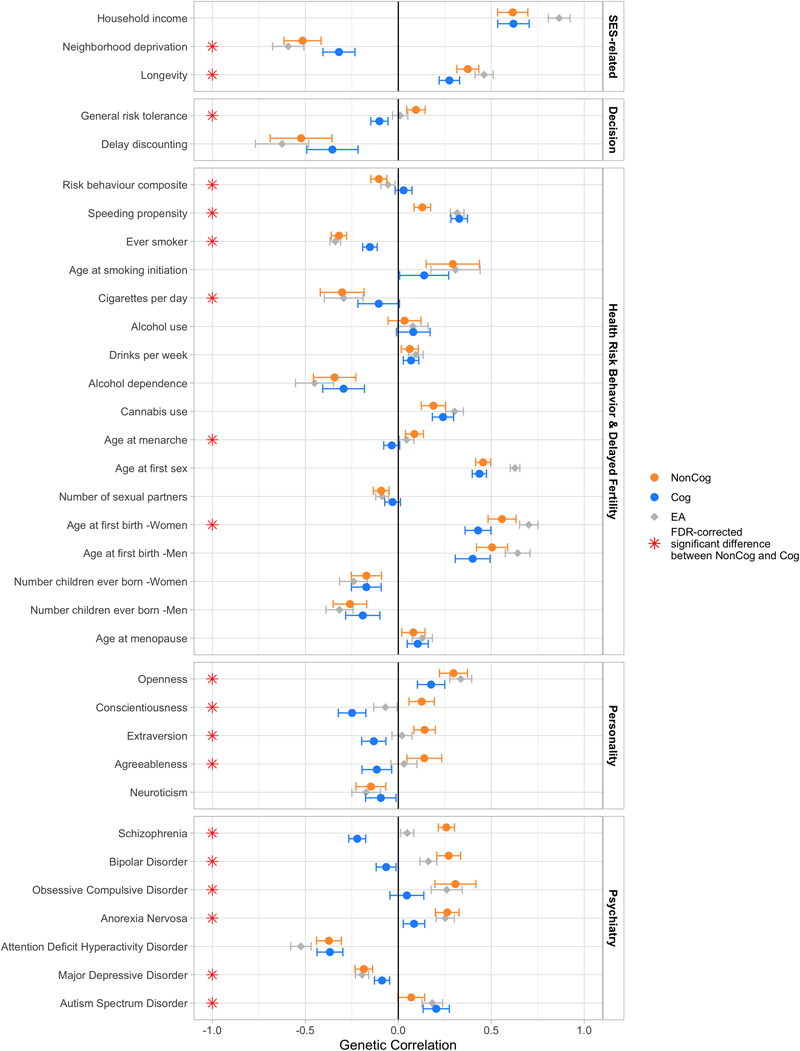
We next conducted a series of genetic correlation analyses to explore the network of phenotypes to which NonCog was genetically correlated. To develop understanding of the substance of non-cognitive skills, we tested where in that network of phenotypes genetic correlations with NonCog diverged from genetic correlations with Cog. Our analysis was organized around four themes: decision-making preferences, health-risk and fertility behaviors, personality traits, and psychiatric disorders. Results of genetic correlation analyses are graphed in Figure 4 and Supplementary Figure 11. Results are reported in Supplementary Table 14.
Figure 4. Estimates of genetic correlations with NonCog, Cog, and educational attainment.
Genetic correlations of NonCog, Cog, and educational attainment with selected phenotypes. The dots represent genetic correlations estimated in Genomic SEM. Correlations with NonCog are in orange, with Cog in blue, and with educational attainment in gray. Error bars represent 95% CIs. Red stars indicate a statistically significant (FDR corrected P < 0.05, two-tailed test) difference in the magnitude of the correlation with NonCog versus Cog. Exact P-values for all associations are reported in Supplementary Table 14. The FDR correction was applied based on all genetic correlations tested (including in Supplementary Fig. 11). The difference test is based on a chi-squared test associated with a comparison between a model constraining these two correlations to be identical versus a model where the correlations are freely estimated. Source GWAS are listed in Supplementary Table 13.
NonCog genetics were associated with decision-making preferences. In economics, non-cognitive influences on achievement and health are often studied in relation to decision-making preferences40–43. NonCog was genetically correlated with higher tolerance of risks44 (r g = 0.10 (SE = 0.03)) and willingness to forego immediate gratification in favor of a larger reward at a later time45 (delay discounting r g = -0.52 (SE = 0.08)). In contrast, Cog was genetically correlated with generally more cautious decision-making characterized by lower levels of risk tolerance (r g = -0.35 (SE = 0.07), P diff_fdr < 0.0001) and delay discounting (r g = -0.35 (SE = 0.07), P diff_fdr = 0.082).
NonCog genetics were associated with less health-risk behavior and delayed fertility. An alternative approach to studying specific non-cognitive skills is to infer individual differences in non-cognitive skills from patterns of health-risk behavior. NonCog was genetically correlated with less health-risk behavior as indicated by analysis of obesity46, substance use44,47–50, and sexual behaviors and early fertility44,51,52 (r g range 0.2-0.5), with the exception that the r g with alcohol use was not different from zero and r g with cannabis use was positive. Genetic correlations for Cog were generally in the same direction but of smaller magnitude.
NonCog genetics were associated with a broad spectrum of personality characteristics linked with social and professional competency. In psychology, non-cognitive influences on achievement are conceptualized as personality traits, i.e. patterns of stable individual differences in emotion and behavior. The model of personality that has received the most attention in genetics is a five-factor model referred to as the Big Five. Genetic correlation analysis of the Big Five personality traits53–55 revealed NonCog genetics were most strongly associated with Openness to Experience (being curious and eager to learn; r g = 0.30 (SE = 0.04)) and were further associated with a pattern of personality characteristic of changes that occur as people mature in adulthood56. Specifically, NonCog showed a positive r g with Conscientiousness (being industrious and orderly; r g = 0.13 (SE = 0.03)), Extraversion (being enthusiastic and assertive; r g = 0.14 (SE = 0.03)), and Agreeableness (being polite and compassionate; r g = 0.14 (SE = 0.05)), and negative r g with Neuroticism (being emotionally volatile; r g = -0.15 (SE = 0.04)). Genetic correlations of Cog with Openness to Experience and Neuroticism were similar to those for NonCog (P diff_fdr-Openness = 0.040, P diff_fdr-Neuroticism = 0.470). In contrast, genetic correlations of Cog with Conscientiousness, Extraversion, and Agreeableness were in the opposite direction (r g = -0.25 to -0.12, P diff_fdr < 0.0005). PGS analysis of personality traits is reported in Supplementary Table 12, Supplementary Figure 12, and the Supplementary Note.
NonCog genetics were associated with higher risk for multiple psychiatric disorders. In clinical psychology and psychiatry, research is focused on mental disorders. Mental disorders are generally associated with impairments in academic achievement and social role functioning57,58. However, positive genetic correlations with educational attainment and creativity have been reported for some disorders59,60. We therefore tested NonCog r g with psychiatric disorders based on published case-control GWAS of mental disorders61–67. NonCog was associated with higher risk for multiple clinically defined disorders, including anorexia nervosa (r g = 0.26 (SE = 0.04)), obsessive-compulsive disorder (r g = 0.31 (SE = 0.06)), bipolar disorder (r g = 0.27 (SE = 0.03)), and schizophrenia (r g = 0.26 (SE = 0.02)). Genetic correlations between Cog and psychiatric disorders were either smaller in magnitude (anorexia nervosa r g = 0.08 (SE = 0.03), P diff_fdr < 0.001; obsessive-compulsive disorder r g = 0.05 (SE = 0.05), P diff_fdr = 0.002) or in the opposite direction (bipolar disorder r g = -0.07 (SE = 0.03), P diff_fdr < 0.001; schizophrenia r g = -0.22 (SE = 0.02), P diff_fdr < 0.001). Both NonCog and Cog showed negative genetic correlations with attention-deficit/hyperactivity disorder (NonCog r g = -0.37 (SE = 0.03), Cog r g = -0.37 (SE = 0.04), P diff_fdr = 0.947).
In sum, NonCog genetics were associated with phenotypes from economics and psychology thought to mediate non-cognitive influences on educational success. These associations contrasted with associations for Cog genetics, supporting distinct pathways of influence on achievement in school and later in life. Opposing patterns of association were also observed for psychiatric disorders, suggesting that the unexpected positive genetic correlation between educational attainment and mental health problems uncovered in previous studies60,68,69 arises from non-cognitive genetic influences on educational attainment.
Biological annotation analyses reveal shared and specific neurobiological correlates
The goal of biological annotation of GWAS discoveries is to elucidate molecular mechanisms mediating genetic influences on the phenotype of interest. Our biological annotation analysis proceeded in two steps. First, we conducted enrichment analysis to test whether some tissues and cell-types were more likely to mediate NonCog and Cog heritabilities than others. Second, we conducted genetic correlation analysis to explore how NonCog and Cog genetics related to different brain structures.
NonCog and Cog genetics were enriched in similar tissues and cells. We tested whether common variants in genes specifically expressed in 53 GTEx tissues70 or in 152 tissues captured in a previous aggregation of RNA-seq studies71,72 were enriched in their effects on Cog or NonCog. Genes predominantly expressed in the brain rather than peripheral tissues were enriched in both NonCog and Cog (Supplementary Table 16).
To examine expression patterns at a more granular level of analysis, we used MAGMA73 and stratified LD score regression74 to test enrichment of common variants in 265 nervous system cell-type-specific gene-sets75 (Supplementary Table 17). In MAGMA analysis, common variants in 95 of 265 gene-sets were enriched for association with NonCog. The enriched cell-types were predominantly neurons (97%), with enrichment most pronounced for telencephalon-projecting neurons, di- and mesencephalon neurons, and to a lesser extent, telencephalon interneurons (Supplementary Fig. 13 and Supplementary Table 18). Enrichment for Cog was similar to NonCog (correlation between Z-statistics Pearson’s r = 0.85), and there were no differences in cell-type-specific enrichment, suggesting that the same types of brain cells mediate genetic influences on NonCog and Cog (Supplementary Fig. 14). Stratified LDSC results were similar to results from MAGMA (Supplementary Note, Supplementary Fig. 15, and Supplementary Table 19).
The absence of differences in cell-type specific enrichment is surprising given that NonCog and Cog are genetically uncorrelated. We therefore used the TWAS/Fusion tool76 to conduct gene-level analysis. This analysis revealed a mixture of concordant and discordant gene effects on NonCog and Cog consistent with the genetic correlation of zero (Supplementary Note, Supplementary Fig. 16, and Supplementary Table 20).
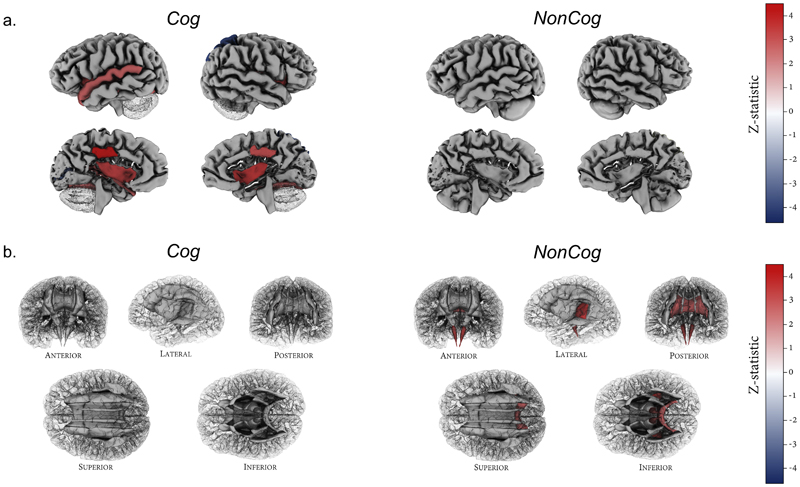
NonCog and Cog genetics show diverging associations with total and regional brain volumes. Educational attainment has previously been found to be genetically correlated with greater total brain volume77,78. We therefore used a GWAS of regional brain volume to compare the r g of NonCog and Cog with total brain volume and with 100 regional brain volumes (99 gray matter volumes and white matter volume) controlling for total brain volume (Supplementary Table 21)79. For total brain volume, genetic correlation was stronger for Cog as compared to NonCog (Cog r g = 0.22 (SE = 0.04), NonCog r g = 0.07 (SE = 0.03), P diff = 0.005). Total gray matter volume, controlling for total brain volume, was not associated with either NonCog or Cog (NonCog: r g = 0.07 (SE = 0.04); Cog: r g = 0.06 (SE = 0.04)). For total white matter volume, conditional on total brain volume, genetic correlation was weakly negative for NonCog as compared to Cog (NonCog r g = -0.12 (SE = 0.04), Cog (r g = -0.01 (SE = 0.04), P diff = 0.04).
NonCog was not associated with any of the regional gray-matter volumes after FDR correction. In contrast, Cog was significantly associated with regional gray-matter volumes for the bilateral fusiform, insula and posterior cingulate (r g range 0.11-0.17), as well as left superior temporal (r g = 0.11 (SE = 0.04)), left pericalcarine (r g = -0.16 (SE = 0.05)) and right superior parietal volumes (r g = -0.22 (SE = 0.06)) (Fig. 5).
Figure 5. Genetic correlations with regional gray matter volumes and white matter tracts.
a, Cortical patterning of FDR-corrected significant genetic correlations with regional gray matter volumes for Cog versus NonCog, after correction for total brain volume. Regions of interest are plotted according to the Desikan-Killiany-Tourville atlas97, shown on a single manually-edited surface (http://mindboggle.info)98. Exact estimates and P-values are reported in Supplementary Table 21. Cog showed significant associations with gray matter volume for the bilateral fusiform, insula and posterior cingulate, the left superior temporal and left pericalcarine and right superior parietal volumes. NonCog was not associated with any of the regional brain volumes. b, White matter tract patterning of FDR-corrected significant genetic correlations with regional mode of anisotropy (MO) for Cog versus NonCog. White matter tract probability maps are plotted according to the Johns Hopkins University DTI atlas (https://identifiers.org/neurovault.image:1401)99. Exact estimates and P-values are reported in Supplementary Table 21. Cog was not associated with regional MO. NonCog showed significant associations with MO in the corticospinal tract, the retrolenticular limb of the internal capsule and the splenium of the corpus callosum.
Finally, we tested genetic correlation of NonCog and Cog with white matter tract integrity as measured using diffusion tensor imaging (DTI)80. Analyses included 5 DTI parameters in each of 22 white matter tracts (Supplementary Table 22). NonCog was positively associated with the mode of anisotropy parameter (which denotes a more tubular, as opposed to planar, water diffusion) in the corticospinal tract, retrolenticular limb of the internal capsule, and splenium of the corpus callosum (Fig. 5). However, all correlations were small (0.10 < r g < 0.14), and we detected no genetic correlations that differed between NonCog and Cog (Supplementary Note).
Discussion
GWAS of non-cognitive influences on educational attainment identified 157 independent loci and polygenic architecture accounting for more than half the genetic variance in educational attainment. In genetic correlation and PGS analysis, these non-cognitive (NonCog) genetics showed similar magnitude of associations with educational attainment, economic attainment, and longevity to genetics associated with cognitive influences on educational attainment (Cog). As expected, NonCog genetics had much weaker associations with cognition phenotypes as compared to Cog genetics. These results contribute new GWAS evidence in support of the hypothesis that heritable non-cognitive skills influence educational attainment and downstream life-course economic and health outcomes.
Phenotypic and biological annotation analyses shed light on the substance of heritable non-cognitive skills influencing education. Economists hypothesize that preferences that guide decision-making in the face of risk and delayed rewards represent non-cognitive influences on educational attainment. Consistent with this hypothesis, NonCog genetics were associated with higher risk tolerance and lower time discounting. These decision-making preferences are associated with financial wealth, whereas opposite preferences are hypothesized to contribute to a feedback loop perpetuating poverty81. Consistent with results from analysis of decision-making preferences, NonCog genetics were also associated with healthier behavior and later fertility.
Psychologists hypothesize that the Big Five personality characteristics of conscientiousness and openness are the two “pillars of educational success”2,3,82. Our results provide some support for this hypothesis, with the strongest genetic correlation evident for openness. However, they also show that non-cognitive skills encompass the full range of personality traits, including agreeableness, extraversion, and the absence of neuroticism. This pattern mirrors the pattern of personality change that occurs as young people mature into adulthood56. Thus, non-cognitive skills share genetic etiology with what might be termed as “mature personality”. The absolute magnitudes of genetic correlations between NonCog and individual personality traits are modest. This result suggests that the personality traits described by psychologists capture some, but not all, genetic influence on non-cognitive skills.
Although the general pattern of findings in our phenotypic annotation analysis indicated non-cognitive skills were genetically related to socially desirable characteristics and behaviors, there was an important exception. Genetic correlation analysis of psychiatric disorder GWAS revealed positive associations of NonCog genetics with schizophrenia, bipolar disorder, anorexia nervosa, and obsessive-compulsive disorder. Previously, these psychiatric disorders have been shown to have a positive r g with educational attainment, a result that has been characterized as paradoxical given the impairments in educational and occupational functioning typical of serious mental illness. Our results clarify that these associations are driven by non-cognitive factors associated with success in education. These results align with the theory that clinically defined psychiatric disorders represent extreme manifestations of dimensional psychological traits, which might be associated with adaptive functioning within the normal range83–85.
Finally, biological annotation analyses suggested that genetic variants contributing to educational attainment not mediated through cognitive abilities are enriched in genes expressed in the brain, specifically in neurons. Even though NonCog and Cog were genetically uncorrelated, variants in the same neuron-specific gene-sets were enriched for both traits. Although we found some evidence of differences between NonCog and Cog in associations with gray matter volumes, moderate sample sizes in neuroimaging GWAS mean these results must be treated as preliminary, requiring replication with data from larger-scale GWAS of white-matter and gray-matter phenotypes. Limited differentiation of NonCog and Cog in biological annotation analyses focused at the levels of tissue and cell type highlights need for finer-grained molecular data resources to inform these analyses and the complementary value of phenotypic annotation analyses focused at the level of psychology and behavior.
We acknowledge limitations. Cognitive and non-cognitive skills develop in interaction with one another. For example, the dynamic mutualism hypothesis86 proposes that non-cognitive characteristics shape investments of time and effort, leading to differences in the pace of cognitive development87,88. However, in Genomic-SEM analysis, the NonCog factor is, by construction, uncorrelated with genetic influences on adult cognition as measured in the Cog GWAS. Our statistical separation of NonCog from cognition is thus a simplified representation of development. Longitudinal studies with repeated measures of cognitive and candidate non-cognitive skills are needed to study their reciprocal relationships across development89,90. Our statistical separation of NonCog from cognition is also incomplete. The ability to control statistically for any variable, genetic or otherwise, depends on how well and comprehensively that variable is measured91. The tests of cognitive performance included in the Cog GWAS likely do not capture all genetic influences on all forms of cognitive ability across the lifespan92,93. Despite these limitations, our simplified and incomplete statistical separation of NonCog from Cog allowed us to test whether heritable traits other than cognitive ability influenced educational attainment and to explore what those traits might be.
Because our analysis was based on GWAS of educational attainment, non-cognitive genetics identified here may differ from non-cognitive genetics affecting other socioeconomic attainments like income, or traits and behaviors that mediate responses to early childhood interventions, to the extent that those genetics do not affect educational attainment. Parallel analysis of alternative attainment phenotypes will clarify the specificity of discovered non-cognitive genetics.
In the case of GWAS of educational attainment, the included samples were drawn mainly from Western Europe and the U.S., and participants completed their education in the late 20th and early 21st centuries. The phenotype of educational attainment reflects an interaction between an individual and the social system in which they are educated. Differences across social systems, including education policy, culture, and historical context, may result in different heritable traits influencing on educational attainment94. Results therefore may not generalize beyond the times and places GWAS samples were collected.
Generalization of the NonCog factor is also limited by restriction of included GWAS to individuals of European ancestry. Lack of methods for integrating genome-scale genetic data across populations with different ancestries95,96 requires this restriction, but raises threats to external validity. GWAS of other ancestries and development of methods for trans-ancestry analysis can enable analysis of (Non)Cog in non-European populations.
Within the bounds of these limitations, results illustrate the application of Genomic-SEM to conduct GWAS of a phenotype not directly measured in GWAS databases. This application could have broad utility beyond the genetics of educational attainment. The GWAS-by-subtraction method allowed us to study a previously hard-to-interpret residual value. Our analysis provides a first view of the genetic architecture of non-cognitive skills influencing educational success. These skills are central to theories of human capital formation within the social and behavioral sciences and are increasingly the targets of social policy interventions. Our results establish that non-cognitive skills are central to the heritability of educational attainment and illuminate connections between genetic influences on these skills and social and behavioral science phenotypes.
Methods
Meta-analysis of educational attainment GWAS
We reproduced the Social Science Genetic Association Consortium (SSGAC) 2018 GWAS of educational attainment25 by meta-analyzing published summary statistics for n = 766,345 (www.thessgac.org/data) with summary statistics obtained from 23andMe, Inc. (n = 365,538). We included SNPs with sample size > 500,000 and MAF > 0.005 in the 1000 Genomes reference set (10,101,243 SNPs). We did not apply genomic control, as standard errors of publicly available and 23andMe summary statistics were already corrected25. Meta-analysis was performed using METAL100.
GWAS-by-subtraction
The objective of our GWAS-by-subtraction analysis was to estimate, for each SNP, the association with educational attainment that was independent of that SNP’s association with cognition (hereafter, the NonCog SNP effect). We used Genomic-SEM24 in R 3.4.3 to analyze GWAS summary statistics for the educational attainment and cognitive performance phenotypes in the SSGAC’s 2018 GWAS25. The model regressed the educational-attainment and cognitive-performance summary statistics on two latent variables, Cog and NonCog (Fig. 1). Cog and NonCog were then regressed on each SNP in the genome. This analysis allowed for two paths of association with educational attainment for each SNP. One path was fully mediated by Cog. The other path was independent of Cog and measured the non-cognitive SNP effect, NonCog. To identify independent hits with P < 5 × 10-8 (the customary P-value threshold to approximate an alpha value of 0.05 in GWAS), we pruned the results using a radius of 250 kb and an LD threshold of r 2 < 0.1 (Supplementary Tables 1-3). We explore alternative lead SNPs and loci definition in Supplementary Table 4. The parameters estimated in a GWAS-by-subtraction and their derivation in terms of the genetic covariance are described in the Supplementary Note (model specification), and practical analysis steps are further described in the Supplementary Note (SNP filtering). The effective sample size of the NonCog and Cog GWAS was estimated to 510,795 and 257,700, respectively (see Supplementary Note). We investigated biases from unaccounted-for heterogeneity in overlap across SNPs in the educational attainment and cognitive performace GWAS and describe possible strategy to deal with it (Supplementary Note). We investigated potential biases due to cohort differences in SNP heritability in the Supplementary Note. We evaluated the consequences of modifying r g (NonCog, Cog) = 0 by evaluating r g = 0.1, 0.2 or 0.3, and we investigated the consequences of a violation of the assumed causation between cognitive performance and educational attainment in the Supplementary Note.
Genetic correlations
We used Genomic-SEM to compute genetic correlations of Cog and NonCog with other education-linked traits for which well-powered GWAS data were available (SNP-h 2 z-statistics > 2; Supplementary Table 13) and to test whether genetic correlations with these traits differed between Cog and NonCog. Specifically, models tested the null hypothesis that trait genetic correlations with Cog and NonCog could be constrained to be equal using a chi-squared test with FDR adjustment to correct for multiple testing. The FDR adjustment was conducted across all genetic correlation analyses reported in the article, excluding the analyses of brain volumes described below. Finally, we used Genomic-SEM analysis of genetic correlations to estimate the percentage of the genetic covariance between educational attainment and the target traits that was explained by Cog and NonCog using the model illustrated in Supplementary Figure 17.
Polygenic score analysis
Polygenic score analyses were conducted in data drawn from six population-based cohorts from the Netherlands, the U.K., the U.S., and New Zealand: (1) the Netherlands Twin Register (NTR)29,101, (2) E-Risk32, (3) the Texas Twin Project34, (4) the National Longitudinal Study of Adolescent to Adult Health (AddHealth)30,102, dbGaP accession phs001367.v1.p1; (5) Wisconsin Longitudinal Study on Aging (WLS)33, dbGaP accession phs001157.v1.p1; and (6) the Dunedin Multidisciplinary Health and Development Study31. Supplementary Tables 9 and 10 describe cohort-specific metrics, and we include a short description of the cohorts’ populations and recruitment in Supplementary Note. Only participants with European ancestry were included in the analysis, due to the low portability of PGS between different ancestry populations. Polygenic scores were computed with PLINK based on weights derived using the LD-pred103 software with an infinitesimal prior and the 1000 Genomes phase 3 sample as a reference for the LD structure. LD-pred weights were computed in a shared pipeline to ensure comparability between cohorts. Each outcome (e.g., IQ score) was regressed on the Cog and NonCog polygenic scores and a set of control variables (sex, 10 principal components derived from the genetic data and, for cohorts in which these quantities varied, genotyping chip and age), using Stata 14 for WLS, Stata 15 for E-Risk and the Dunedin Study, and R (versions 3.4.3 and newer) for NTR, AddHealth, and the Texas Twin Project. In cohorts containing related individuals, non-independence of observations from relatives was accounted for using generalized estimation equations (GEE) or by clustering of standard errors at the family level. We used a random effects meta-analysis to aggregate the results across the cohorts. This analysis allows a cohort-specific random intercept. Individual cohort results are in Supplementary Table 11 and meta-analytic estimates in Supplementary Table 12.
Biological annotation
Enrichment of tissue-specific gene expression. We used gene-sets defined in Finucane et al.104 to test for the enrichment of genes specifically expressed in one of 53 GTEx tissues70, or 152 tissues captured by the Franke et al. aggregation of RNA-seq studies71,72. This analysis seeks to confirm the role of brain tissues in mediating Cog and NonCog influences on educational attainment. The exact analysis pipeline used is available online (https://github.com/bulik/ldsc/wiki/Cell-type-specific-analyses).
Enrichment of cell-type specific expression. We leveraged single cell RNA sequencing (scRNA-seq) data of cells sampled from the mouse nervous system75 to identify cell-type specific RNA expression. Zeisel et al.75 sequenced cells obtained from 19 regions in the contiguous anatomical regions in the peripheral sensory, enteric, and sympathetic nervous system. After initial QC, they retained 492,949 cells, which were sampled down to 160,796 high quality cells. These cells were further grouped into clusters representing 265 broad cell-types. We analyzed the dataset published by Zeisel et al. containing mean transcript counts for all genes with count >1 for each of the 265 clusters (Supplementary Table 17). We restricted analysis to genes with expression levels above the 25th percentile. For each gene in each cell-type, we computed the cell-type specific proportion of reads for the gene (normalizing the expression within cell-type). We then computed the proportion of proportions over the 265 cell-types (computing the specificity of the gene to a specific cell-type). We ranked the 12,119 genes retained in terms of specificity to each cell-type and then retained the 10% of genes most specific to a cell-type as the “cell-type specific” gene-set. We then tested whether any of the 265 cell-type specific gene-sets were enriched in the Cog or NonCog GWAS. This analysis sought to identify specific cell-types and specific regions in the brain involved in the etiology of Cog and NonCog. We further computed the difference in enrichment for Cog and NonCog to test whether any cell types were specific to either trait. For these analyses, we leveraged two widely used enrichment analysis tools: MAGMA73 and stratified LD score regression74 with the European reference panel from 1000 Genomes Project Phase 3 as SNP location and LD structure reference, Gencode release 19 as gene location reference and the human-mouse homology reference from MGI (http://www.informatics.jax.org/downloads/reports/HOM_MouseHumanSequence.rpt).
MAGMA. We used MAGMA (v1.07b73), a program for gene-set analysis based on GWAS summary statistics. We computed gene-level association statistics using a window of 10 kb around the gene for both Cog and NonCog. We then used MAGMA to run a competitive gene-set analysis, using the gene P-values and gene correlation matrix (reflecting LD structure) produced in the gene-level analysis. The competitive gene-set analysis tests whether the genes within the cell-type-specific gene-set described above are more strongly associated with Cog/NonCog than other genes.
Stratified LD-score regression. We used LD-score regression to compute LD scores for the SNPs in each of our “cell-type specific” gene-sets. Parallel to MAGMA analysis, we added a 10-kb window around each gene. We ran partitioned LD-score regression to compute the contribution of each gene-set to the heritability of Cog and NonCog. To guard against inflation, we used LD score best practices, and included the LD score baseline model (baselineLD.v2.2) in the analysis. We judged the statistical significance of the enrichment based on the P-value associated with the tau coefficient.
Difference in enrichment between Cog and NonCog. To compute differences in enrichment, we compute a standardized difference between the per-annotation enrichment for Cog and NonCog as:
| (Equation 1) |
where e Cog is the enrichment of a particular gene-set for Cog, eNonCog is the enrichment for the same gene-set for NonCog, seCog is the standard error of the enrichment for Cog, seNonCog is the standard error of the enrichment for NonCog, and CTI is the LD score cross-trait intercept, a metric of dependence between the GWASs of Cog and NonCog.
We investigated the significance of the difference between Cog and NonCog tau coefficient with Equation 1 as well as by computing jackknifed standard errors. From the jackknifed estimates of the coefficient output by the LDSC software, we computed the jackknifed estimates and standard errors of the difference between Cog and NonCog tau coefficients, as well as a z-statistic for each annotation.
Enrichment of gene expression in the brain. We performed a transcriptome-wide association study (TWAS) using FUSION76 (http://gusevlab.org/projects/fusion/). We used pre-computed brain-gene-expression weights available on the FUSION website, generated from 452 human individuals as part of the CommonMind Consortium. We then superimposed the bivariate distribution of the results of the TWAS for Cog and NonCog over the bivariate distribution expected given the sample overlap between educational attainment and cognitive performance (the GWAS on which our GWAS of Cog and NonCog are based, see Supplementary Note).
Brain modalities
Brain volumes. We conducted genetic correlation analysis of brain volumes using GWAS results published by Zhao et al.79, who performed GWAS of total brain volume and 100 regional brain volumes, including 99 gray matter volumes and total white matter volume (Supplementary Table 21). Analyses included covariate adjustment for sex, age, their square interaction and 20 principle components. Analyses of regional brain volumes additionally included covariate adjustment for total brain volume. GWAS summary statistics for these 101 brain volumes were obtained from https://med.sites.unc.edu/bigs2/data/gwas-summary-statistics/. Summary statistics were filtered and pre-processed using Genomic-SEM’s “munge” function, retaining all HapMap3 SNPs with allele frequency > 0.01 outside the MHC region. We used Genomic-SEM to compute the genetic correlations between Cog, NonCog and brain volumes. Analyses of regional volumes controlled for total brain volume. For each volume, we tested whether correlations differed between Cog and NonCog. Specifically, we used a chi-squared test to evaluate the null hypothesis that the two genetic correlations were equal. We used FDR adjustment to correct for multiple testing. The FDR adjustment is applied to the results for all gray matter volumes for Cog and NonCog separately.
White matter structures. We conducted genetic-correlation analysis of white-matter structures using GWAS results published by Zhao et al.80, who performed GWAS of diffusion tensor imaging (DTI) measures of the integrity of white-matter tracts. DTI parameters were derived for fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD), radial diffusivity (RD), and mode of anisotropy (MO). Each of these parameters was measured for 22 white matter tracts of interests (Supplementary Table 22), resulting in 110 GWAS. GWAS summary statistics for these 110 GWAS were obtained from https://med.sites.unc.edu/bigs2/data/gwas-summary-statistics/. Summary statistics were filtered and processed using Genomic-SEM’s “munge” function, retaining all HapMap3 SNPs with allele frequency > 0.01 outside the MHC region. For each white matter structure, we tested whether genetic correlations differed between Cog and NonCog. Specifically, we used a chi-squared test to evaluate the null hypothesis that the two genetic correlations were equal. We used FDR adjustment to correct for multiple testing. As these different diffusion parameters are statistically and logically interdependent, having been derived from the same tensor, FDR adjustment was applied to the results for each type of white matter diffusion parameter separately. FDR correction was applied separately for Cog and NonCog.
Additional Resources
A FAQ on why, how and what we studied is available here: https://medium.com/@kph3k/investigating-the-genetic-architecture-of-non-cognitive-skills-using-gwas-by-subtraction-b8743773ce44
A tutorial on how to perform GWAS-by-subtraction: http://rpubs.com/MichelNivard/565885
Additional resources to Genomic SEM software:
-
-
A wiki including numerous tutorials: https://github.com/MichelNivard/GenomicSEM/wiki
-
-
A Genomic SEM user group for specific questions relating to models and software: https://groups.google.com/g/genomic-sem-users
-
-
A venue to report technical issues: https://github.com/MichelNivard/GenomicSEM/issues
Supplementary Material
Acknowledgements
This study was developed with support from the Jacobs Foundation at a meeting organized by D.W.B. and K.P.H. with support from E.M.T.-D. and C.M. and also attended by co-authors P.B., B.W.D., and J.W. We gratefully acknowledge contributions to the meeting from Katrin Mannik and Felix Tropf, and the Jacobs Foundation Fellowship team who made the meeting possible. D.W.B., K.P.H., M.G.N., E.M.T.-D., and C.M. are fellows of the Foundation. J.W. is a Jacobs Foundation Young Scholar.
This study used GWAS summary statistics published by the Social Science Genetic Association Consortium (SSGAC) and additional data obtained from 23andMe. We thank the research participants and employees of 23andMe for making this work possible. We thank the SSGAC and COGENT consortia for sharing their summary statistics of the GWASs of educational attainment and cognitive performance, especially Aysu Okbay for her quick and repeated help with providing these data. This study used data from the Netherlands Twin Register (NTR), the Texas Twin Study, the National Longitudinal Study of Adolescent to Adult Health (Add Health), the Dunedin Longitudinal Study, the E-Risk Study, and the Wisconsin Longitudinal Study (WLS).
NTR is supported by: ‘Twin-family database for behavior genetics and genomics studies’ (NWO 480-04-004), Longitudinal data collection from teachers of Dutch twins and their siblings (NWO-481-08-011); Twin-family study of individual differences in school achievement (NWO 056-32-010) and Gravitation program of the Dutch Ministry of Education, Culture and Science and the Netherlands Organization for Scientific Research (NWO 0240-001-003); NWO Groot (480-15-001/674): Netherlands Twin Registry Repository: researching the interplay between genome and environment; NWO- Spi-56-464-14192 Biobanking and Biomolecular Resources Research Infrastructure (BBMRI – NL, 184.021.007 and 184.033.111); European Research Council (ERC-230374); the Avera Institute for Human Genetics, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH, R01D0042157-01A); the NIMH Grand Opportunity grants (1RC2MH089951-01 and 1RC2 MH089995-01). The Texas Twin Project is supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development grants R01HD083613 and R01HD092548. Add Health is supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development grant P01HD31921, and GWAS grants R01HD073342 and R01HD060726, with cooperative funding from 23 other federal agencies and foundations. The Dunedin Multidisciplinary Health and Development Study is supported by the NZ HRC, NZ MBIE, National Institute on Aging grant R01AG032282, and UK Medical Research Council grant MR/P005918/1. The E-Risk Study is supported by the UK Medical Research Council grant G1002190 and Eunice Kennedy Shriver National Institute of Child Health and Human Development grant R01HD077482. The Wisconsin Longitudinal Study is supported by National Institute on Aging grants R01AG041868 and P30AG017266.
Some of the work used a high-performance computing facility partially supported by grant 2016-IDG-1013 from the North Carolina Biotechnology Center. The Population Research Center at the University of Texas at Austin is supported by NIH grant P2CHD042849.
P.A.D. is supported by the grant 531003014 from The Netherlands Organisation for Health Research and Development (ZonMW). P.B. is supported by the NORFACE-DIAL grant number 462-16-100. S.R.C. is supported by the UK Medical Research Council grant MR/R024065/1 and NIH grant R01AG054628. EMTD is supported by NIH grants R01AG054628 and R01HD083613. A.A. is supported by the Foundation Volksbond Rotterdam and by ZonMw grant 849200011. E.v.B. is supported by NWO VENI grant 451-15-017. D.I.B. acknowledges the Royal Netherlands Academy of Science (KNAW) Professor Award (PAH/6635). B.W.D. is supported by award # 96-17-04 from the Russell Sage Foundation and the Ford Foundation. H.F.I. was supported by the “Aggression in Children: Unraveling gene-environment interplay to inform Treatment and InterventiON strategies” project (ACTION). ACTION received funding from the European Union Seventh Framework Program (FP7/2007-2013) under grant agreement no 602768. J.W. is supported by a postdoctoral fellowship by the AXA Research Fund. D.W.B. is a fellow of the Canadian Institute for Advanced Research Child Brain Development Network. K.P.H. and E.M.T.-D. are Faculty Research Associates of the Population Research Center at the University of Texas at Austin, which is supported by grant, 5-R24-HD042849, from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). M.G.N. is supported by ZonMW grants 849200011 and 531003014 from The Netherlands Organisation for Health Research and Development, a VENI grant awarded by NWO (VI.Veni.191G.030), and NIH grant R01MH120219.
Footnotes
Author Contributions
Conceived and designed the experiment: D.W.B., K.P.H., M.G.N., P.A.D., and M.M. conceived the idea for the study with assistance from E.M.T.-D., B.W.D., P.B., C.M., and J.W. Analyzed the data: P.A.D., M.M., T.T.M., P.B., B.W.D., D.W.B., D.L.C., K.S., S.R.C., M.G.N., A.A., and H.F.I. Wrote the paper: D.W.B., K.P.H., M.G.N., M.M., P.A.D., and E.M.T.-D. with helpful contributions from P.B., B.W.D., and S.R.C. A.D.G., L.A., E.v.B., D.I.B., A.C., K.M.H., T.E.M., R.P., J.A.P., B.S.W., E.L.d.Z., and previously mentioned authors contributed to interpretation of data, provided critical feedback on manuscript drafts, and approved the final draft.
Competing Interests
The authors declare no competing interests.
Code availability
Code used to run the analyses is available at: https://github.com/PerlineDemange/non-cognitive
A tutorial on how to perform GWAS-by-subtraction: http://rpubs.com/MichelNivard/565885
All additional software used to perform these analyses are available online.
Data availability
GWAS summary data for NonCog and Cog (excluding 23andMe) have been deposited in the GWAS Catalog with accession numbers GCST90011874 and GCST90011875, respectively (NonCog GWAS: ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST90011874, Cog GWAS: ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST90011875).
For 23andMe dataset access, see https://research.23andme.com/dataset-access/.
Part of the National Longitudinal Study of Adolescent to Adult Health (Add Health) data is publicly available and can be downloaded at the following link: https://data.cpc.unc.edu/projects/2/view#public_li. For restricted access data, details of the data sharing agreement and data access requirements can be found at the following link: https://data.cpc.unc.edu/projects/2/view
The Dunedin study datasets reported in the current article are not publicly available due to lack of informed consent and ethical approval, but are available on request by qualified scientists. Requests require a concept paper describing the purpose of data access, ethical approval at the applicant’s university, and provision for secure data access. We offer secure access on the Duke, Otago and King's College campuses. All data analysis scripts and results files are available for review (https://moffittcaspi.trinity.duke.edu/research-topics/dunedin).
The E-Risk Longitudinal Twin Study datasets reported in the current article are not publicly available due to lack of informed consent and ethical approval, but are available on request by qualified scientists. Requests require a concept paper describing the purpose of data access, ethical approval at the applicant’s university, and provision for secure data access. We offer secure access on the Duke and King's College campuses. All data analysis scripts and results files are available for review (https://moffittcaspi.trinity.duke.edu/research-topics/erisk).
Netherlands Twin Register data may be accessed, upon approval of the data access committee (email: ntr.datamanagement.fgb@vu.nl).
Researchers will be able to obtain Texas Twins data through managed access. Requests for managed access should be sent to Dr. Elliot Tucker-Drob (tuckerdrob@utexas.edu) and Dr. Paige Harden (harden@utexas.edu), joint principal investigators of the Texas Twin Project.
Wisconsin Longitudinal study data can be requested following this form: https://www.ssc.wisc.edu/wlsresearch/data/Request_Genetic_Data_28_June_2017.pdf
References
- 1.Moffitt TE, et al. A gradient of childhood self-control predicts health, wealth, and public safety. Proc Natl Acad Sci USA. 2011;108:2693–2698. doi: 10.1073/pnas.1010076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Stumm S, Hell B, Chamorro-Premuzic T. The hungry mind: intellectual curiosity is the third pillar of academic performance. Perspect Psychol Sci. 2011;6:574–588. doi: 10.1177/1745691611421204. [DOI] [PubMed] [Google Scholar]
- 3.Tucker-Drob EM, Briley DA, Engelhardt LE, Mann FD, Harden KP. Genetically-mediated associations between measures of childhood character and academic achievement. J Pers Soc Psychol. 2016;111:790–815. doi: 10.1037/pspp0000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heckman JJ, Stixrud J, Urzua S. The effects of cognitive and noncognitive abilities on labor market outcomes and social behavior. J Labor Econ. 2006;24:411–482. [Google Scholar]
- 5.Heckman JJ, Moon SH, Pinto R, Savelyev PA, Yavitz A. The rate of return to the HighScope Perry Preschool Program. J Public Econ. 2010;94:114–128. doi: 10.1016/j.jpubeco.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conti G, Heckman JJ, Pinto R. The effects of two influential early childhood interventions on health and healthy behaviour. Econ J. 2016;126:F28–F65. doi: 10.1111/ecoj.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutman LM, Schoon I. The impact of non-cognitive skills on outcomes for young people. Educ Endow Found. 2013;59:2019. [Google Scholar]
- 8.Garcia E. The Need to Address Noncognitive Skills in the Education Policy Agenda. 2014 https://www.epi.org/publication/the-need-to-address-noncognitive-skills-in-the-education-policy-agenda/
- 9.Kautz T, Heckman JJ, Diris R, Ter Weel B, Borghans L. OECD Education Working Papers, No. 110. OECD Publishing; Paris: 2014. Fostering and measuring skills: improving cognitive and non-cognitive skills to promote lifetime success. [Google Scholar]
- 10.Heckman JJ. Skill formation and the economics of investing in disadvantaged children. LIFE CYCLES. 2006;312:4. doi: 10.1126/science.1128898. [DOI] [PubMed] [Google Scholar]
- 11.Heckman JJ, Kautz T. Hard evidence on soft skills. Labour Econ. 2012;19:451–464. doi: 10.1016/j.labeco.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimfeld K, Kovas Y, Dale PS, Plomin R. True grit and genetics: Predicting academic achievement from personality. J Pers Soc Psychol. 2016;111:780–789. doi: 10.1037/pspp0000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson M, Abraham C, Bond R. Psychological correlates of university students’ academic performance: a systematic review and meta-analysis. Psychol Bull. 2012;138:353–387. doi: 10.1037/a0026838. [DOI] [PubMed] [Google Scholar]
- 14.Smithers LG, et al. A systematic review and meta-analysis of effects of early life non-cognitive skills on academic, psychosocial, cognitive and health outcomes. Nat Hum Behav. 2018;2:867–880. doi: 10.1038/s41562-018-0461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovas Y, et al. Why children differ in motivation to learn: Insights from over 13,000 twins from 6 countries. Personal Individ Differ. 2015;80:51–63. doi: 10.1016/j.paid.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loehlin JC. Genes and environment in personality development. Sage Publications; 1992. [Google Scholar]
- 17.Tucker-Drob EM, Harden KP. Learning motivation mediates gene-by-socioeconomic status interaction on mathematics achievement in early childhood. Learn Individ Differ. 2012;22:37–45. doi: 10.1016/j.lindif.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malanchini M, Engelhardt LE, Grotzinger AD, Harden KP, Tucker-Drob EM. “Same but different”: associations between multiple aspects of self-regulation, cognition, and academic abilities. J Pers Soc Psychol. 2019;117:1164–1188. doi: 10.1037/pspp0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris TT, Smith GD, van Den Berg G, Davies NM. Investigating the longitudinal consistency and genetic architecture of non-cognitive skills, and their relation to educational attainment. 2018 doi: 10.1101/470682. [DOI]
- 20.Liu JZ, Erlich Y, Pickrell JK. Case–control association mapping by proxy using family history of disease. Nat Genet. 2017;49:325–331. doi: 10.1038/ng.3766. [DOI] [PubMed] [Google Scholar]
- 21.Bowles S, Gintis H. Schooling In Capitalist America: Educational Reform And The Contradictions Of Economic Life. Basic Books; 1977. [Google Scholar]
- 22.Heckman JJ, Rubinstein Y. The importance of noncognitive skills: lessons from the GED Testing Program. Am Econ Rev. 2001;91:145–149. [Google Scholar]
- 23.Ackerman PL, Kanfer R, Goff M. Cognitive and noncognitive determinants and consequences of complex skill acquisition. J Exp Psychol Appl. 1995;1:270–304. [Google Scholar]
- 24.Grotzinger AD, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belsky DW, Harden KP. Phenotypic annotation: using polygenic scores to translate discoveries from genome-wide association studies from the top down. Curr Dir Psychol Sci. 2019;28:82–90. doi: 10.1177/0963721418807729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie SJ, Tucker-Drob EM. How much does education improve intelligence? A meta-analysis. Psychol Sci. 2018;29:1358–1369. doi: 10.1177/0956797618774253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ligthart L, et al. The Netherlands Twin Register: longitudinal research based on twin and twin-family designs. Twin Res Hum Genet. 2019;22:623–636. doi: 10.1017/thg.2019.93. [DOI] [PubMed] [Google Scholar]
- 30.Harris KM, et al. Cohort profile: The National Longitudinal Study of Adolescent to Adult Health (Add Health) Int J Epidemiol. 2019;48:1415–1415k. doi: 10.1093/ije/dyz115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulton R, Moffitt TE, Silva PA. The Dunedin Multidisciplinary Health and Development Study: overview of the first 40 years, with an eye to the future. Soc Psychiatry Psychiatr Epidemiol. 2015;50:679–693. doi: 10.1007/s00127-015-1048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffitt TE, E-risk Team. Teen-aged mothers in contemporary Britain. J Child Psychol Psychiatry. 2002;43:727–742. doi: 10.1111/1469-7610.00082. [DOI] [PubMed] [Google Scholar]
- 33.Herd P, Carr D, Roan C. Cohort profile: Wisconsin longitudinal study (WLS) Int J Epidemiol. 2014;43:34–41. doi: 10.1093/ije/dys194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harden KP, Tucker-Drob EM, Tackett JL. The Texas Twin Project. Twin Res Hum Genet. 2013;16:385–390. doi: 10.1017/thg.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benyamin B, et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry. 2014;19:253–258. doi: 10.1038/mp.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chetty R, et al. The association between income and life expectancy in the United States, 2001-2014. JAMA. 2016;315:1750–1766. doi: 10.1001/jama.2016.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Case A, Deaton A. Mortality and morbidity in the 21st century. Brook Pap Econ Act. 2017;2017:397–476. doi: 10.1353/eca.2017.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill WD, et al. Molecular genetic contributions to social deprivation and household income in UK Biobank. Curr Biol. 2016;26:3083–3089. doi: 10.1016/j.cub.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timmers PR, et al. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. eLife. 2019;8:e39856. doi: 10.7554/eLife.39856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almlund M, Duckworth AL, Heckman J, Kautz T. Handbook of the Economics of Education. Vol. 4. Elsevier; 2011. Personality psychology and economics; pp. 1–181. [Google Scholar]
- 41.Borghans L, Duckworth AL, Heckman JJ, terWeel B. The economics and psychology of personality traits. J Hum Resour. 2008;43:972–1059. [Google Scholar]
- 42.Rabin M. A perspective on psychology and economics. Eur Econ Rev. 2002;29 [Google Scholar]
- 43.Becker A, Deckers T, Dohmen T, Falk A, Kosse F. The relationship between economic preferences and psychological personality measures. Annu Rev Econ. 2012;4:453–478. [Google Scholar]
- 44.Linnér RK, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–257. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez-Roige S, et al. Genome-wide association study of delay discounting in 23,217 adult research participants of European ancestry. Nat Neurosci. 2018;21:16–18. doi: 10.1038/s41593-017-0032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yengo L, et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tobacco, Consortium Genetics. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters RK, et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21:1656–1669. doi: 10.1038/s41593-018-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schumann G, et al. KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci USA. 2016;113:14372–14377. doi: 10.1073/pnas.1611243113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasman JA, et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal effect of schizophrenia liability. Nat Neurosci. 2018;21:1161–1170. doi: 10.1038/s41593-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linnér RK, et al. Multivariate genomic analysis of 1.5 million people identifies genes related to addiction, antisocial behavior, and health. bioRxiv. 2020 doi: 10.1101/2020.10.16.342501. 2020.10.16.342501. [DOI] [Google Scholar]
- 52.Barban N, et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat Genet. 2016;48:1462–1472. doi: 10.1038/ng.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lo M-T, et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat Genet. 2017;49:152–156. doi: 10.1038/ng.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.John OP, Naumann LP, Soto CJ. Paradigm shift to the integrative Big Five Trait taxonomy. Handb Personal Theory Res. 2008:114–158. doi: 10.1016/S0191-8869(97)81000-8. [DOI] [Google Scholar]
- 55.de Moor MHM, et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry. 2012;17:337–349. doi: 10.1038/mp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caspi A, Roberts BW, Shiner RL. Personality development: stability and change. Annu Rev Psychol. 2005;56:453–484. doi: 10.1146/annurev.psych.55.090902.141913. [DOI] [PubMed] [Google Scholar]
- 57.Kessler RC, et al. Social consequences of psychiatric disorders, I: educational attainment. Am J Psychiatry. 1995;152:1026–1032. doi: 10.1176/ajp.152.7.1026. [DOI] [PubMed] [Google Scholar]
- 58.Breslau J, Lane M, Sampson N, Kessler RC. Mental disorders and subsequent educational attainment in a US national sample. J Psychiatr Res. 2008;42:708–716. doi: 10.1016/j.jpsychires.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Power RA, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18:953–955. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 60.Bansal V, et al. Genome-wide association study results for educational attainment aid in identifying genetic heterogeneity of schizophrenia. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05510-z. 3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruderfer DM, et al. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 2018;173:1705–1715. doi: 10.1016/j.cell.2018.05.046. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jansen PR, et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet. 2019;51:394–403. doi: 10.1038/s41588-018-0333-3. [DOI] [PubMed] [Google Scholar]
- 64.Duncan L, et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am J Psychiatry. 2017;174:850–858. doi: 10.1176/appi.ajp.2017.16121402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grove J, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arnold PD, et al. Revealing the complex genetic architecture of obsessive–compulsive disorder using meta-analysis. Mol Psychiatry. 2018;23:1181–1188. doi: 10.1038/mp.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ripke S, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nieuwboer HA, Pool R, Dolan CV, Boomsma DI, Nivard MG. GWIS: genome-wide inferred statistics for functions of multiple phenotypes. Am J Hum Genet. 2016;99:917–927. doi: 10.1016/j.ajhg.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6 doi: 10.1038/ncomms6890. 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fehrmann RSN, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 73.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:1–19. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeisel A, et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014. doi: 10.1016/j.cell.2018.06.021. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gusev A, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nave G, Jung WH, Karlsson Linnér R, Kable JW, Koellinger PD. Are bigger brains smarter? Evidence from a large-scale preregistered study. Psychol Sci. 2019;30:43–54. doi: 10.1177/0956797618808470. [DOI] [PubMed] [Google Scholar]
- 78.Elliott ML, et al. A polygenic score for higher educational attainment is associated with larger brains. Cereb Cortex. 2019;29:3496–3504. doi: 10.1093/cercor/bhy219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao B, et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat Genet. 2019;51:1637–1644. doi: 10.1038/s41588-019-0516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao B, et al. Large-scale GWAS reveals genetic architecture of brain white matter microstructure and genetic overlap with cognitive and mental health traits (n=17,706) Mol Psychiatry. doi: 10.1038/s41380-019-0569-z. published online 30 October 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haushofer J, Fehr E. On the psychology of poverty. Science. 2014;344:862–867. doi: 10.1126/science.1232491. [DOI] [PubMed] [Google Scholar]
- 82.Briley DA, Domiteaux M, Tucker-Drob EM. Achievement-relevant personality: relations with the Big Five and validation of an efficient instrument. Learn Individ Differ. 2014;32:26–39. doi: 10.1016/j.lindif.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smoller JW, et al. Psychiatric genetics and the structure of psychopathology. Mol Psychiatry. 2019;24:409–420. doi: 10.1038/s41380-017-0010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nat Rev Genet. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 85.Meehl PE. Schizotaxia, schizotypy, schizophrenia. Am Psychol. 1962;17:827–838. [Google Scholar]
- 86.von Stumm S, Ackerman PL. Investment and intellect: a review and meta-analysis. Psychol Bull. 2013;139:841–869. doi: 10.1037/a0030746. [DOI] [PubMed] [Google Scholar]
- 87.Tucker-Drob EM, Harden KP. A behavioral genetic perspective on non-cognitive factors and academic achievement. In: Grigorenko EL, Tan M, Latham SR, Bouregy S, editors. Genetics, Ethics and Education. Cambridge University Press; 2017. pp. 134–158. [DOI] [Google Scholar]
- 88.Tucker-Drob EM. Handbook of competence and motivation: Theory and application. 2nd. The Guilford Press; 2017. Motivational factors as mechanisms of gene-environment transactions in cognitive development and academic achievement; pp. 471–486. [Google Scholar]
- 89.Tucker-Drob EM, Harden KP. Intellectual interest mediates gene × socioeconomic status interaction on adolescent academic achievement: intellectual interest and G×E. Child Dev. 2012;83:743–757. doi: 10.1111/j.1467-8624.2011.01721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Malanchini M, et al. Reading self-perceived ability, enjoyment and achievement: A genetically informative study of their reciprocal links over time. Dev Psychol. 2017;53:698–712. doi: 10.1037/dev0000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Westfall J, Yarkoni T. Statistically controlling for confounding constructs is harder than you think. PLoS One. 2016;11:e0152719. doi: 10.1371/journal.pone.0152719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.de la Fuente J, Davies G, Grotzinger AD, Tucker-Drob EM, Deary IJ. Genetic “General Intelligence,” Objectively Determined and Measured. 2019 doi: 10.1101/766600. [DOI] [Google Scholar]
- 93.Tucker-Drob EM, Briley DA. Continuity of genetic and environmental influences on cognition across the life span: a meta-analysis of longitudinal twin and adoption studies. Psychol Bull. 2014;140:949–979. doi: 10.1037/a0035893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tropf FC, et al. Hidden heritability due to heterogeneity across seven populations. Nat Hum Behav. 2017;1:757–765. doi: 10.1038/s41562-017-0195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duncan L, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10 doi: 10.1038/s41467-019-11112-0. 3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin AR, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein A, Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci. 2012;6:171. doi: 10.3389/fnins.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klein A. Mindboggle-101 manually labeled individual brains. 2016 doi: 10.7910/DVN/HMQKCK. [DOI] [Google Scholar]
- 99.Gorgolewski KJ, et al. NeuroVault.org: a web-based repository for collecting and sharing unthresholded statistical maps of the human brain. Front Neuroinformatics. 2015;9:8. doi: 10.3389/fninf.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Willemsen G, et al. The Adult Netherlands Twin Register: twenty-five years of survey and biological data collection. Twin Res Hum Genet. 2013;16:271–281. doi: 10.1017/thg.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Highland HM, Avery CL, Duan Q, Li Y, Harris KM. Quality control analysis of Add Health GWAS data. 2018 https://www.cpc.unc.edu/projects/addhealth/documentation/guides/AH_GWAS_QC.pdf.
- 103.Vilhjálmsson BJ, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Finucane HK, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code used to run the analyses is available at: https://github.com/PerlineDemange/non-cognitive
A tutorial on how to perform GWAS-by-subtraction: http://rpubs.com/MichelNivard/565885
All additional software used to perform these analyses are available online.
GWAS summary data for NonCog and Cog (excluding 23andMe) have been deposited in the GWAS Catalog with accession numbers GCST90011874 and GCST90011875, respectively (NonCog GWAS: ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST90011874, Cog GWAS: ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST90011875).
For 23andMe dataset access, see https://research.23andme.com/dataset-access/.
Part of the National Longitudinal Study of Adolescent to Adult Health (Add Health) data is publicly available and can be downloaded at the following link: https://data.cpc.unc.edu/projects/2/view#public_li. For restricted access data, details of the data sharing agreement and data access requirements can be found at the following link: https://data.cpc.unc.edu/projects/2/view
The Dunedin study datasets reported in the current article are not publicly available due to lack of informed consent and ethical approval, but are available on request by qualified scientists. Requests require a concept paper describing the purpose of data access, ethical approval at the applicant’s university, and provision for secure data access. We offer secure access on the Duke, Otago and King's College campuses. All data analysis scripts and results files are available for review (https://moffittcaspi.trinity.duke.edu/research-topics/dunedin).
The E-Risk Longitudinal Twin Study datasets reported in the current article are not publicly available due to lack of informed consent and ethical approval, but are available on request by qualified scientists. Requests require a concept paper describing the purpose of data access, ethical approval at the applicant’s university, and provision for secure data access. We offer secure access on the Duke and King's College campuses. All data analysis scripts and results files are available for review (https://moffittcaspi.trinity.duke.edu/research-topics/erisk).
Netherlands Twin Register data may be accessed, upon approval of the data access committee (email: ntr.datamanagement.fgb@vu.nl).
Researchers will be able to obtain Texas Twins data through managed access. Requests for managed access should be sent to Dr. Elliot Tucker-Drob (tuckerdrob@utexas.edu) and Dr. Paige Harden (harden@utexas.edu), joint principal investigators of the Texas Twin Project.
Wisconsin Longitudinal study data can be requested following this form: https://www.ssc.wisc.edu/wlsresearch/data/Request_Genetic_Data_28_June_2017.pdf