Abstract
Neural stem cells (NSCs) generate neurons throughout life in the mammalian hippocampus. However, the potential for long-term self-renewal of individual NSCs within the adult brain remains unclear. We used 2-photon microscopy and followed NSCs that were genetically labeled through conditional recombination driven by the regulatory elements of the stem cell-expressed genes GLI Family Zinc Finger 1 (Gli1) or Achaete-scute homolog 1 (Ascl1). Through intravital imaging of NSCs and their progeny, we identify a population of Gli1-targeted NSCs showing long-term self-renewal in the adult hippocampus. In contrast, once activated, Ascl1-targeted NSCs undergo limited proliferative activity before they become exhausted. Using single-cell RNA sequencing, we show that Gli1- and Ascl1-targeted cells have highly similar yet distinct transcriptional profiles, supporting the existence of heterogeneous NSC populations with diverse behavioral properties. Thus, we here identify long-term self-renewing NSCs that contribute to the generation of new neurons in the adult hippocampus.
Introduction
In contrast to many organs the adult mammalian brain shows poor regenerative capacity, and fails to replace neurons that become dysfunctional or those lost through acute or chronic injury. However, the discovery of neural stem cells (NSCs) within discrete regions of the adult brain opened up the possibility to harness these cells for endogenous brain repair1. Despite some conflicting results regarding the human brain, the hippocampal dentate gyrus (DG) seems to be among the brain areas that generate new neurons throughout life2–8. Within the DG, NSCs generate new excitatory granule cells that are important for a variety of hippocampal functions3. Previous work identified the cellular and molecular identity of neurogenic cells in the DG with largely quiescent radial glia-like cells (R or type I cells) that give rise to more rapidly cycling non-radial glia-like cells (NR or type II cells)9–11. However, it remained controversial, due to ambiguity when recovering lineage information from static pulse-chase lineage tracing assays, whether NSCs exist in the DG that possess long-term self-renewal potential (i.e., renewing through cell division while giving rise to differentiated progeny) or if activation of NSCs leads to their rapid depletion from the stem cell pool via terminal differentiation12–16. Exhaustion of NSCs upon initial activation may explain the dramatic decrease of hippocampal neurogenesis that drops around 80% from 2 to 8 months of age in the rodent brain3, 17–19. However, neurogenesis plateaus and continues throughout the entire life span, albeit at low levels, which may be due to increased quiescence of aged NSCs compared to NSCs in the young hippocampus20. Theoretical modelling approaches suggest that long-term self-renewal may be required to sustain neurogenesis throughout life, but experimental evidence remains scant12, 21. To probe directly the potential for long-term self-renewal of individual NSCs in the adult DG we used intravital imaging together with single-cell RNA sequencing (scRNA-seq) to analyze the molecular and behavioral properties of GLI Family Zinc Finger 1 (Gli1)-targeted NSCs and compared them to Achaete-scute homolog 1 (Ascl1)-targeted neurogenic cells16.
Results
Gli1-lineages contain long-term self-renewing clones
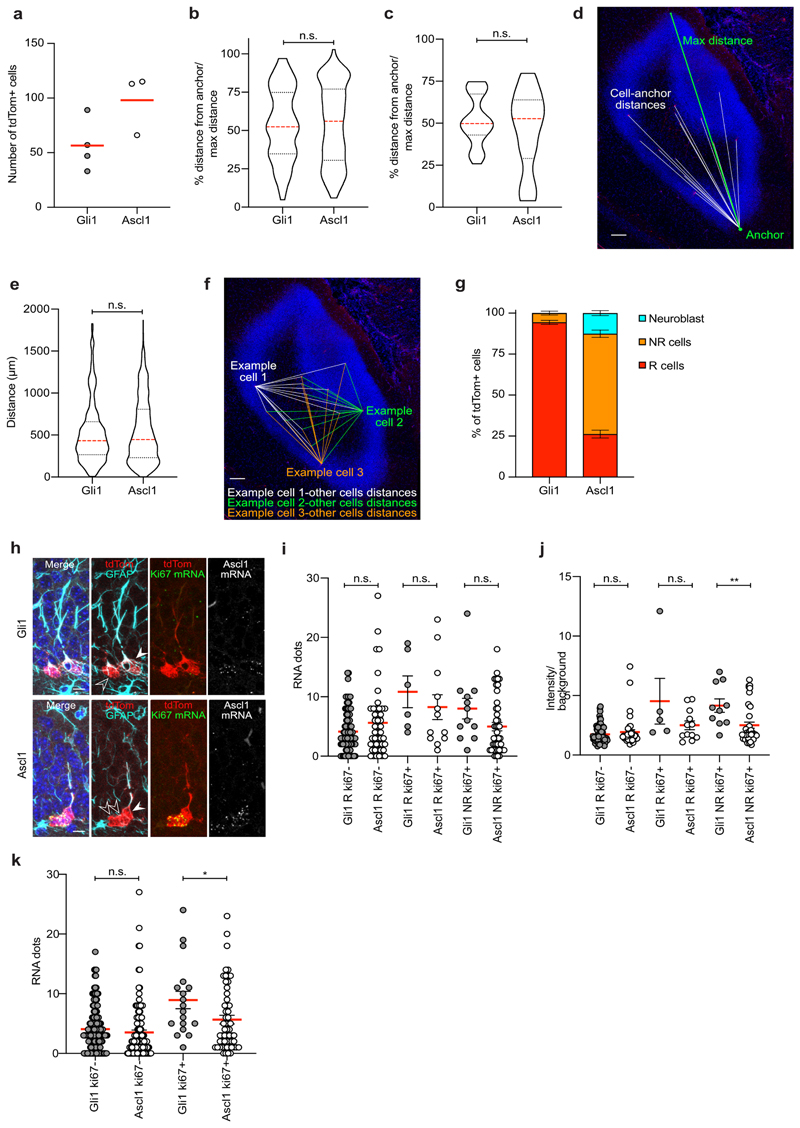
Using static analyses of individual time points we found that low-dose Tamoxifen (Tam)-induced recombination in Gli1-CreERT2 mice crossed with a tdTomato reporter mouse line sparsely labeled neurogenic cells in the subgranular zone of the DG (Fig. 1a)22. The number and spatial distribution of recombined cells was comparable between Gli1- and Ascl1CreERT2 lines (Extended Data Fig. 1a-f and Supplementary Movie 1). Soon after Tam administration, recombined cells showed the morphology of radial glia-like NSCs (hereafter called R cells)10, 11. In contrast to Ascl1-targeted cells, only a small fraction (approx. 5%) of Gli1-targeted cells in the DG showed morphological features and marker expression of non-radial precursors, NR cells, soon after Tam administration (Fig. 1a, Extended Data Fig. 1g). By using single-molecule RNAscope technology, we corroborated previous results23 showing that the pro-activation factor Ascl1 exhibits comparable mRNA levels in activated (ki67+) and quiescent (ki67-) Gli1- and Ascl1-targeted R cells. Additionally, we found slightly higher Ascl1 expression levels in Gli1- compared to Ascl1-targeted dividing NR cells and in ki67+ Gli1- compared to ki67+ Ascl1-targeted cells (Extended Data Fig. 1h-k). Using immunohistochemistry, we found that Gli1-targeted R and NR cells expressed a previously described set of marker proteins (Fig. 1a).
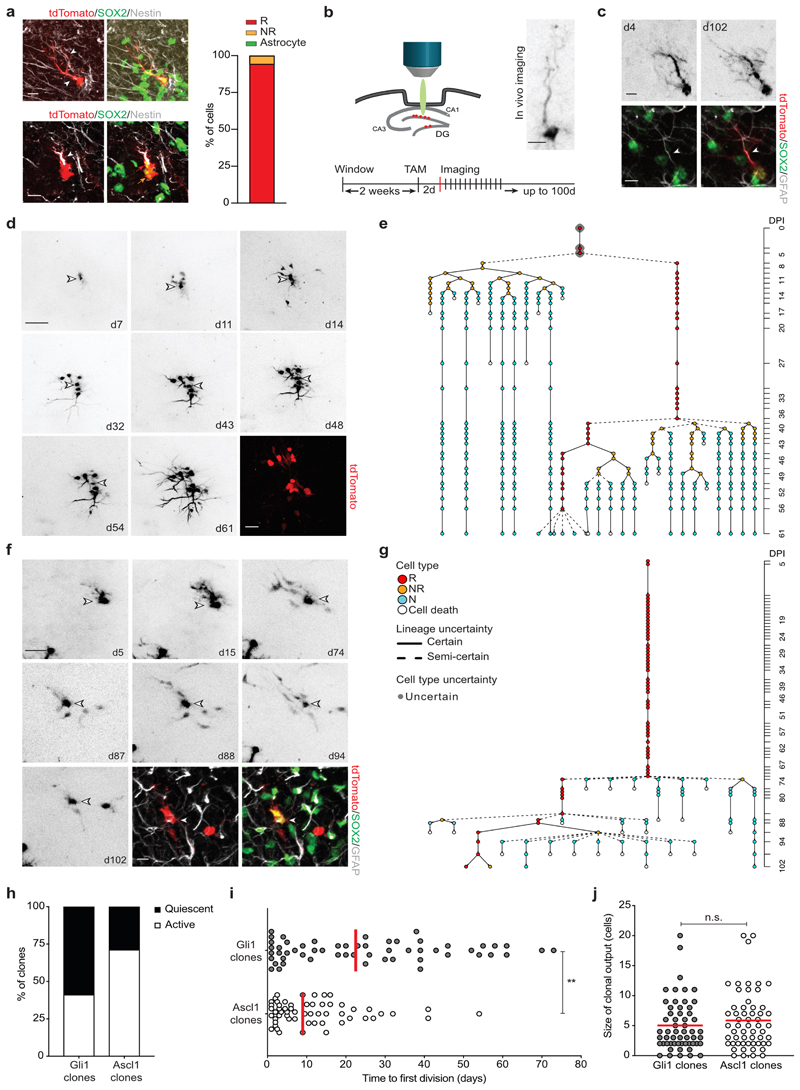
Fig. 1. Gli1-targeted R cells contain long-term self-renewing hippocampal stem cells.
a, Representative confocal images of R (tdTom+/red, SOX2+/green, Nestin+/white; red arrow; white arrowheads indicate major radial R cell process) and NR cells (tdTom+/red, SOX2+/green, Nestin-/white; orange arrow) after recombination with imaging dose. Quantification of tdTom+ cell types in the SGZ 2d after recombination. b, Experimental setup for chronic in vivo imaging of Gli1-tdTom NSCs over up to 100d. Shown is a representative R cell. c, Representative in vivo images of a Gli1-targeted quiescent R cell at 4 and 102 dpi and post hoc immunohistochemical analysis of the same R cell (tdTom+/red, SOX2+/green, GFAP+/white; white arrowhead indicates major radial process) at 102 dpi. The horizontal view of the cell corresponds to the view obtained during in vivo imaging. d, Selected pictures (collapsed z-stacks) of a Gli1-targeted R cell (white arrowhead) and its progeny imaged over the course of 2 months exhibiting long-term self-renewal (51d). Post hoc immunohistochemical analysis of the same clone (tdTom+/red) at 61dpi. e, Lineage tree deduced from tracking the R cell in D. Identified cell types are color-coded, and lineage transitions are depicted depending on their certainty (see methods). f, Example of a Gli1-targeted R cell clone showing long R cell persistence. Post hoc immunohistochemical analysis of the same clone (tdTom+/red, SOX2+/green, GFAP+/white) at 102dpi shows the presence of a R cell (white arrowhead, GFAP+/SOX2+), a NR cell (GFAP-/SOX2+) and a neuron (GFAP-/SOX2-). g, Lineage tree deduced from tracking the R cell in F and its progeny. h, Percentage of active and quiescent clones in Gli1 and Ascl1-targeted cells (total Gli1: 136; Gli1 active: 41.1%; Gli1 quiescent: 58.8 %; total Ascl1: 80; Ascl1 active: 71.2 %; Ascl1 quiescent: 28.75 %. Ascl1 cell data consist of 19 new lineages and 61 previously described lineages (see Methods). i, Time until the first division (in days) of Gli1 and Ascl1 R cells calculated from the start of the imaging 2dpi (Gli1: 24.98 ± 2.8, n = 56 R cells; Ascl1: 11.85 ± 1.5, n = 57 R cells; Mann Whitney test: Mann-Whitney U = 1055; **p = 0.0017, two-tailed). j, Final number of cells per active clone (Gli1: 5.03 ± 0.57, n = 56 clones; Ascl1: 5.86 ± 0.64, n = 57 clones; Mann Whitney test: Mann-Whitney U = 1440; p = 0.3699, two-tailed). Circles represent individual clones. Values are shown as mean ± s.e.m. Scale bars represent 10 μm (a, c, staining f), 20 μm (staining d), 25 μm (f), 50 μm (d). For detailed statistics, see Supplementary Table 5.1
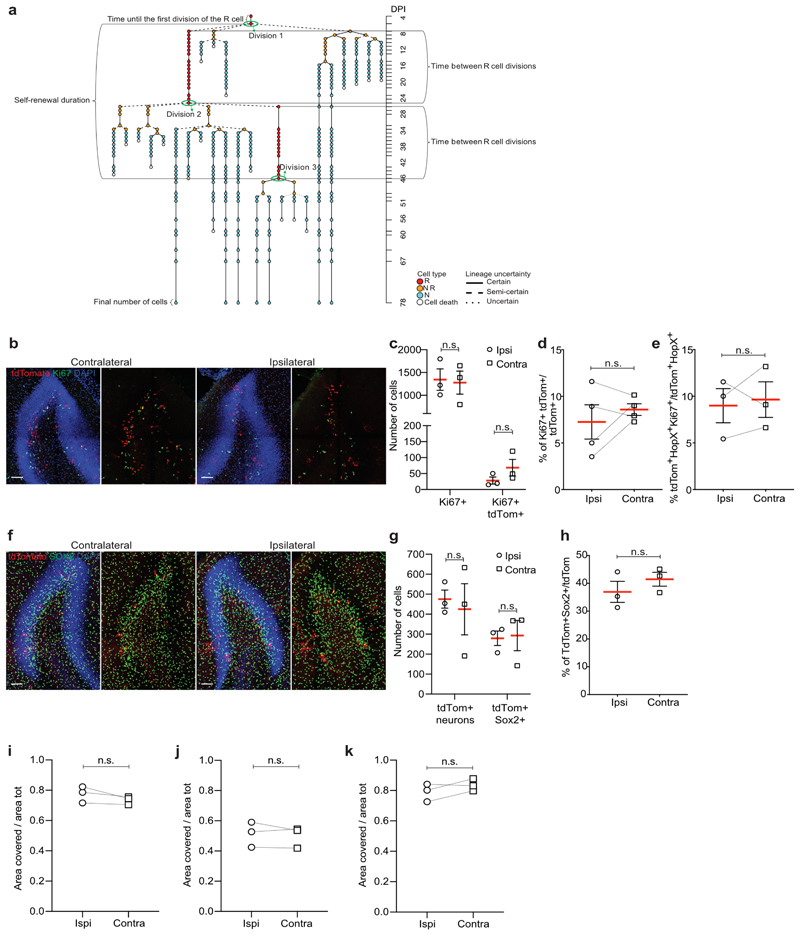
To follow individual Gli1-targeted R cells over time, we used chronic intravital imaging of sparsely labeled hippocampal NSCs (Fig. 1b, c)16. Gli1-targeted R cells became recruited into the proliferating pool of neurogenic cells over time (Fig. 1d-g, Supplementary Fig. 1 and Supplementary Movies 2, 3). The fraction of R cells remaining quiescent over the time course was higher in Gli1-targeted R cells compared to Ascl1-targeted R cells, while entry into cell cycle occurred throughout the imaging period (Fig. 1h, i and Extended Data Fig. 2a). Hippocampal window implantation and imaging did not substantially affect proliferation, R cell activation or cellular output of sparsely labeled Gli1-targeted cells (Extended Data Fig. 2b-h). Moreover, it did not cause significant gliosis and inflammation in the ipsilateral side of the DG compared to the contralateral side (Extended Data Fig. 2i-k). Once Gli1-targeted R cells entered cell cycle, they produced an average of five granule neurons (Fig. 1j and Extended Data Fig. 2a). Most strikingly, we found that some Gli1-targeted R cells returned to quiescence after producing progeny (Fig. 1d, e and Supplementary Fig. 1).
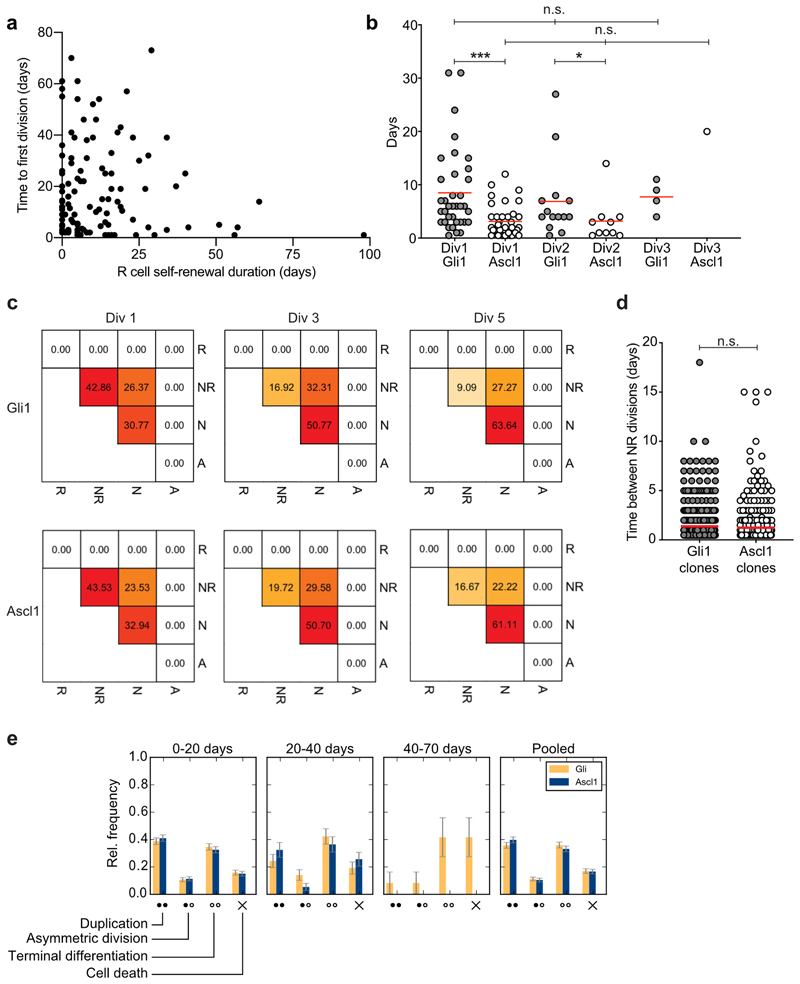
The fraction of long-term self-renewing R cells was substantially higher in Gli1-targeted clones compared to Ascl1-targeted clones, with no unambiguously identifiable Ascl1-targeted R cells surviving over a 30-day time period following activation (Fig. 2a-c and Extended Data Fig. 2a). By contrast, a fraction of Gli1-targeted R cells produced progeny and persisted for more than 56 days upon activation, with one clone showing at least 100 days of self-renewal (Fig. 2a), indicating that the adult DG harbors NSCs with long-term self-renewal capacity. Thus, Gli1-mediated recombination labels an adult NSC population that retains the ability to self-renew over extended times, whereas Ascl1-targeted clones label R cells that become exhausted upon activation (Fig. 2a, b, d). Despite the fact that the average activation time of Gli1-targeted R cells was also significantly longer than that of Ascl1-targeted R cells (Fig. 1i), there was no correlation between the time to first division and self-renewal duration of R cells (Extended Data Fig. 3a).
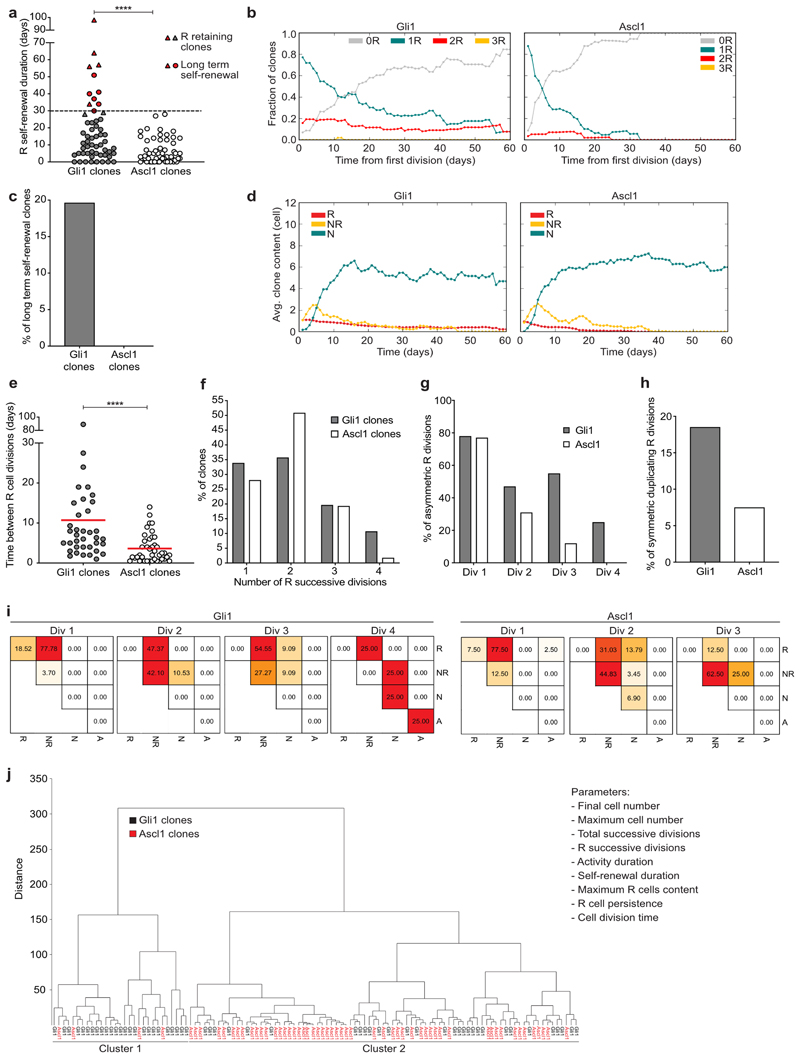
Fig. 2. Diverse behavioral features of Gli1- and Ascl1-targeted NSCs.
a, Self-renewal duration (time in days between first and last division in each lineage) of Gli1 and Ascl1- targeted R cells (Gli1: 17.80 ± 2.5, n = 56 clones; Ascl1: 6.5 ± 0.9, n = 57 clones; Mann Whitney test: Mann-Whitney U = 931; ****p < 0.0001, two-tailed). Triangles depict clones where R cell was still present at the end of the imaging (reported only for the clones with observed self-renewal duration ≥ 28d). The described self-renewal duration of these clones is an underestimation of their actual capacity. Red dots depict R cells showing long-term self-renewal. The dashed line represents a long-term self-renewal threshold (30d). b, Fraction of clones with different R content (0R, 1R, 2R, 3R cells) over time, normalised to the surviving population. All (certain, semi-certain and uncertain) R cells are considered. Lineage trees shifted to the first R cell division time point. c, Percentage of clones with R cell self-renewal ≥30d (Gli1: 19.64%; n = 56 clones; Ascl1: 0%; n = 57 clones). Only R cells that were classified as certain are considered. d, Average clone content for Gli1 and Ascl1 lineages over time. R, radial glia like cell; NR, proliferating progenitor; N, neuron. e, Average time (in days) between R cell divisions (Gli1: 10.71 ± 2.4, n = 37 clones; Ascl1: 3.64 ± 0.5, n = 41 clones; Mann Whitney test: Mann-Whitney U = 337.5; ****p < 0.0001, two-tailed). f, Number of successive R cell divisions in Gli1 and Ascl1 lineages (Gli1; 1.div: 33.93%, 2.div: 35.71%, 3.div: 19.64%, 4.div: 10.71%, n = 56 clones; Ascl1 1.div: 28.07%, 2.div: 50.88%, 3 div: 19.30%, 4 div: 1.75%, n = 57 clones). g, Percentage of asymmetric proliferative R divisions (R mother cell generates one R daughter cell and one NR daughter cell) in division rounds (div) 1 to 4 (Gli1 div 1: 77.78%; n = 27 divisions; Gli1 div 2: 47.37%; n = 19 divisions; Gli1 div 3: 54.55%; n = 11 divisions; Gli1 div 4: 25.0%; n = 4 divisions. Ascl1 div 1: 77.5%; n = 40 divisions; Ascl1 div 2: 31.03%; n = 29 divisions; Ascl1 div 3: 12.5%; n = 8 divisions). h, Percentage of symmetric duplicating R cell divisions in the first division (Gli1 = 18.52%, n = 27 divisions; Ascl1 = 7.50%, n = 40 divisions). i, Heat map representing the frequencies of modes of division of Gli1- targeted R cells (division rounds (Div) 1 to 4. Div1 n = 27 divisions; Div2 n = 19 divisions; Div3 n = 11 divisions; Div4 n = 4 divisions) and Ascl1- targeted R cells (division rounds (Div) 1 to 3. Div1 n = 40 divisions; Div2 n = 29 divisions; Div3 n = 8 divisions). R, radial glia-like cell; NR, proliferating progenitor; N, neuron; A, astrocyte. j, Lineage tree clustering using the Ward’s Hierarchical Clustering Method, Euclidean distances. Listed are the different parameters used. Gli1 lineages are contained in Cluster1 (45.4%) and Cluster2 (54.6%). Ascl1 lineages fall in Cluster1 (7.0%) and Cluster2 (93.0%). Cluster1 consists of 86.2% Gli1 lineages and 13.8% Ascl1 lineages. Cluster2 consists of 36.1% Gli1 derived lineages and 63.9% Ascl1 derived lineages. Circles represent individual clones. Values are shown as mean ± s.e.m. For detailed statistics, see Supplementary Table 5.2
As further evidence of distinct behavioral features of NSC populations in the adult DG, we found that Gli1-targeted R cells showed a higher abundance of symmetric (duplicative) divisions and more asymmetric (renewing) divisions over consecutive cell divisions as compared to Ascl1-targeted cells (Fig. 2f-i)16. This was also reflected in distinct cell division characteristics: Gli1-targeted R cells had significantly longer time intervals between consecutive R cell divisions, but showed higher total numbers of cell divisions (Fig. 2e, f and Supplementary Fig. 1). The comparison of the time between divisions among different successive R divisions (counted from 1st asymmetric R division) in each mouse line revealed no differences either in Gli1- or Ascl1-targeted R cells, suggesting that this time scale does not change according to the division history of the cell (Extended Data Fig. 3b). However, the average time between divisions was significantly higher in Gli1- compared to Ascl1-targeted R cells, in accordance with the average time between R cell divisions per clone shown in Figure 2e (Extended Data Fig. 3b). Moreover, by clustering cell lineages using a set of pre-defined core characteristics (see Methods), we found a preferential clustering of Gli1- versus Ascl1-targeted clones, supporting the partial behavioral diversity of these two genetically-targeted lineages (Fig. 2j).
In contrast to R cells, we found that the behavior of NR cells was remarkably similar between Gli1- and Ascl1-derived lineages regarding cell division kinetics and lineage choices (Fig. 2i, Extended Data Fig. 3c-e). Thus, long-term intravital imaging-derived data indicate functional heterogeneity of radial glial cells within the adult hippocampus, identifying a long-term self-renewing NSC population targeted by the Gli1-promoter and a second population, targeted by the Ascl1-promoter, with only short-term renewal potential.
Molecular diversity of Gli1- and Ascl1-targeted lineages
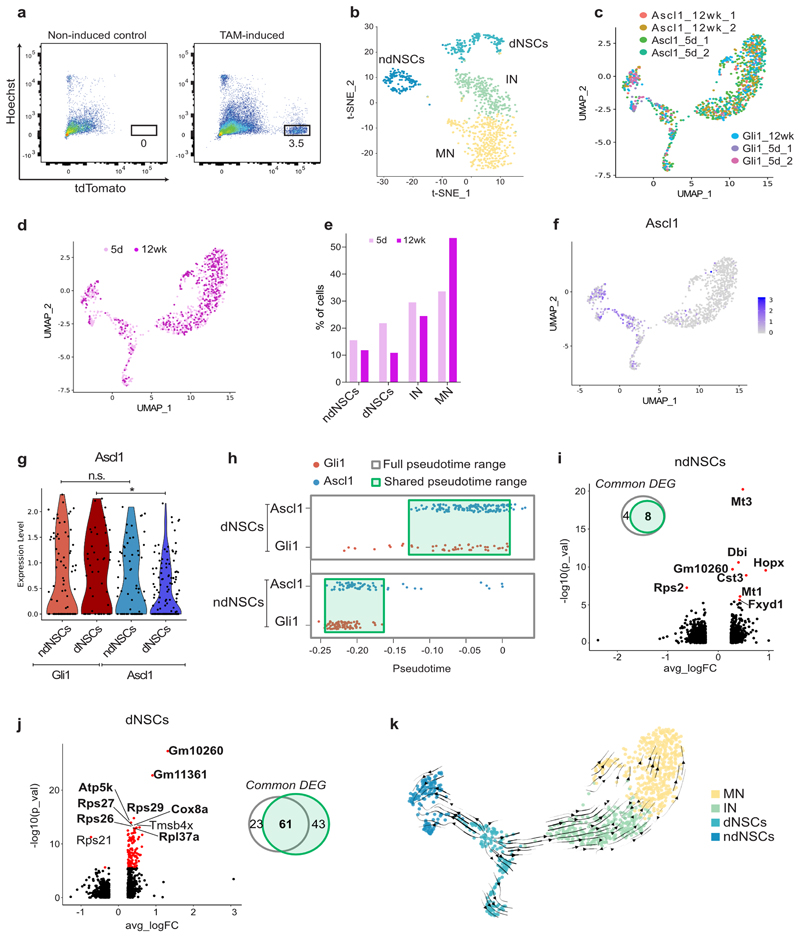
The observed differential behavior of Gli1- versus Ascl1-targeted cell lineages may be explained i) by diverse sets of NSC populations, ii) by a hierarchical organization in which Gli1-targeted NSCs with long-term renewal potential give rise to Ascl1-targeted NSCs with limited renewal potential, or iii) by the alignment of NSCs along a spectrum of diminishing renewal potential. To seek independent evidence for potential sublineage relationships, we used fluorescence activated cell sorting (FACS) to isolate Gli1- and Ascl1-targeted cells directly from the adult hippocampus at different time points after Tam-induced recombination followed by deep single-cell RNA-seq using SmartSeq2 (Fig. 3a and Extended Data Fig. 4a). As expected, we found distinct populations of cells using Uniform Manifold Approximation and Projection (UMAP) and t-Distributed Stochastic Neighbor Embedding (t-SNE) that segregated robustly into clusters of non-dividing NSCs (ndNSCs), dividing NSCs (dNSCs), and immature (IN) and mature granule cells (MN) for both Gli1- and Ascl1-targeted populations (Fig. 3b, c and Extended Data Fig. 4b). All four clusters could be identified 5 days and 12 weeks after Tam-induced recombination (Fig. 3d and Extended Data Fig. 4c), albeit the relative numbers of cell-specific profiles shifted with time, consistent with the in vivo live-imaging data (Fig. 3d, Extended Data Fig. 4d-e). In line with the RNAscope data (Extended Data Fig. 1h-k), Ascl1 mRNA was expressed at the same level in Gli1- and Ascl1-targeted non-dividing NSCs, but showed moderately higher expression levels in Gli1- compared to Ascl1-targeted dividing NSCs (Extended Data Fig. 4f-g). Since the dividing NSC cluster likely encompasses R and NR cells, we sought to investigate the heterogeneity of this cluster. Despite regressing out cell cycle genes, we were unable to further sub-cluster the dividing NSCs based on the expression patterns of key marker genes for radial (e.g. Hopx, Nestin, Gfap, Spot14/Thrsp, Sox2) vs non-radial NSCs (Tbr2/Eomes, Sox2) (data not shown).
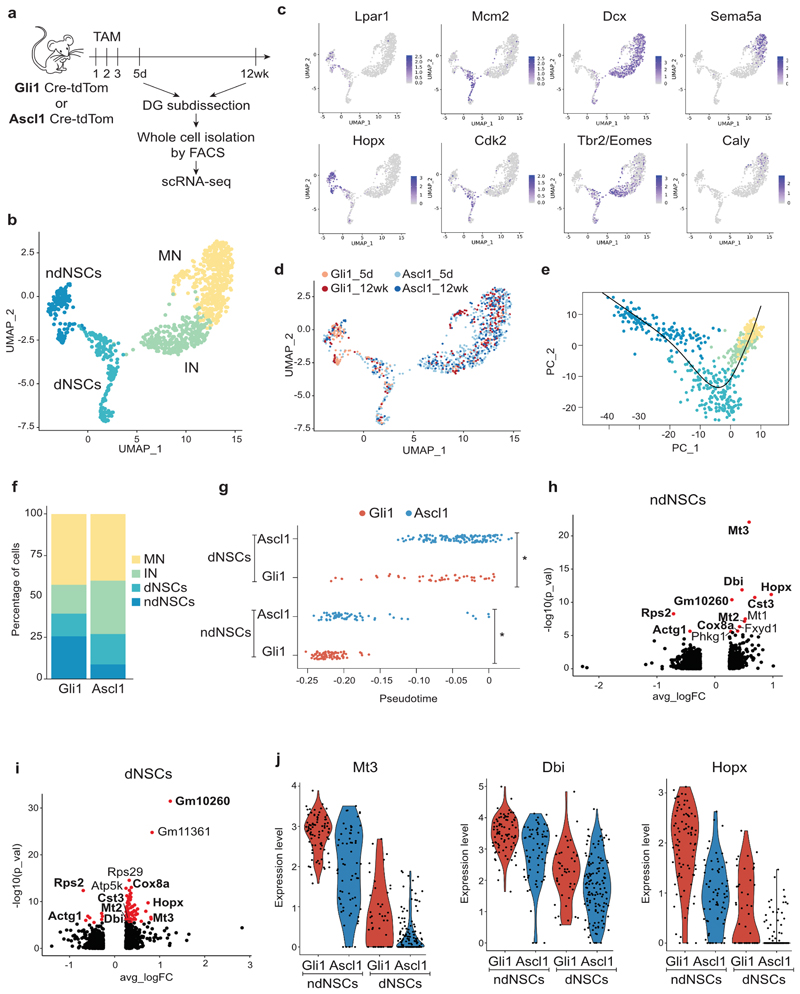
Fig. 3. scRNA-seq of Gli1- and Ascl1-targeted cells identifies NSCs with self-renewal potential.
a, Overview of the experimental approach. b, Uniform Manifold Approximation and Projection (UMAP) visualization of the 4 clusters (non-dividing NSCs (ndNSCs), dividing NSCs (dNSCs), Immature Neurons (IN), Mature Neurons (MN)) identified in Gli1- and Ascl1 targeted tdTomato+ neuronal cells. c, Expression pattern of two cluster-specific genes (top and bottom) for ndNSCs (far left), dNSCs (left), IN (right), MN (far right). d, Gli1- and Ascl1-targeted tdTomato+ cells isolated 5 days (Gli1_5d, Ascl1_5d) or 12 weeks (Gli1_12wk, Ascl1_12wk) after the last tamoxifen injection displayed on the UMAP plot from panel (b). e, Lineage inference using Slingshot. f, Percentages of ndNSCs, dNSCs, IN, MN among all Gli1- or Ascl1- targeted cells. g, Position of Gli1 (red) and Ascl1 (blue) ndNSCs and dNSCs along the pseudotime axis as calculated by Monocle. Unpaired t-test, ndNSCs p = 2.55e-06, dNSCs p = 0.0004042, *p < 0.05. h-i, Volcano plots showing significantly differentially expressed genes (DEGs) (red, padj < 0.05) between Gli1 or Ascl1-targeted ndNSCs (h) or dNSCs (i). Bolded gene names highlight DEGs found in both ndNSCs (h) and dNSCs (i) comparisons. j, Violin plots showing the top 3 common DEGs. For detailed statistics, see Supplementary Table 5.3
We next performed lineage inference analysis using Slingshot 24, which confirmed a developmental trajectory from ndNSCs via dNSCs to mature neurons (Fig. 3e). In accordance with the imaging-based data, we found that the relative number of sequenced non-dividing NSCs was substantially higher in Gli1-targeted cells compared to Ascl1-targeted cells (Fig. 3f). We then asked whether NSC populations may differ in their developmental maturity using pseudotime-based alignment: indeed, we found that Gli1-targeted non-dividing NSCs appeared to be further distant from neuronal progeny compared to Ascl1-targeted non-dividing R cells, suggesting that at least a fraction of Gli1-targeted R cells may be in a more quiescent/undifferentiated state compared to Ascl1-targeted R cells (Fig. 3g). Despite the high level of similarity in the gene expression pattern between Gli1- and Ascl1-targeted cells, some genes were differentially expressed between ndNSCs and dNSCs derived from Gli1- or Ascl1-targeted cells (Fig. 3h-j, Supplementary Tables 1-2). To verify that these differentially expressed genes did not only result from the small proportion of outlier cells occupying earlier (Gli1) or later (Ascl1) areas of the pseudotime, we performed a differential expression analysis only between Gli1 and Ascl1 cells that shared the same pseudotime range (Extended Data Fig. 4h). Comparison between DE genes identified in the presence or absence of the pseudotime outliers only revealed a minor influence of the outliers on the top DE genes (Extended Data Fig. 4i-j). While Gli1 and Ascl1 lines do not label entirely distinct NSC populations, our results suggest that the DE genes identified between Gli1- and Ascl1-targeted NSCs are not only driven by a small proportion of pseudotime outliers, but rather reflect inherent transcriptional differences between these two lines. Notably, we identified several genes that had been previously implicated in NSC quiescence and self-renewal to be differentially expressed between Gli1- and Ascl1-targeted NSCs, among others including Metallothionein-3 (Mt3), diazepam binding inhibitor (Dbi) and Hopx25–27. These findings suggest that the identified gene set expressed at high levels in Gli1-targeted NSCs may be highly relevant in governing NSC self-renewal.
Based on the intravital imaging data, we expected to detect transcriptional profiles that may indicate the return to quiescence after activation in Gli1-targeted cells. To probe for this we analyzed the relative abundance of nascent and mature mRNA in each cell as an indicator of the future state of the cell using RNA velocity in Gli1- and Ascl1-targeted cells28. While most dividing NSCs showed a positive velocity going towards immature neurons, a fraction of dividing NSCs showed a negative velocity pointing toward non-dividing NSCs. We could confirm this velocity pattern with a different RNA velocity method which takes into account the full transcriptional dynamics of splicing kinetics using a likelihood-based dynamical model scVelo (###PMID: 32747759) (Extended Data Fig. 4k). Using an independent dataset29, Bergen and colleagues also found that radial-glial cells harbor a velocity vector pointing away from the neuronal differentiation path (###PMID: 32747759) further supporting our observations. Interestingly, activated Gli1-targeted NSCs showed a higher fraction of cells with such negative velocities as compared to Ascl1-targeted cells (Fig. 4a, b). We next compared differentially expressed genes between dividing NSCs with negative versus positive velocities and found that those with negative velocities upregulated a set of genes previously implicated in NSC quiescence, such as Thrsp (Spot14) and Apoe, and down-regulated genes associated with cell division such as the cell division cycle-associated protein 4 (Cdca4) (Fig. 4c, Supplementary Table 3)9, 30, 31. These findings confirm the presence of NSCs, enriched in the Gli1-targeted population that shows transcriptional dynamics indicative of a return to a quiescent state.
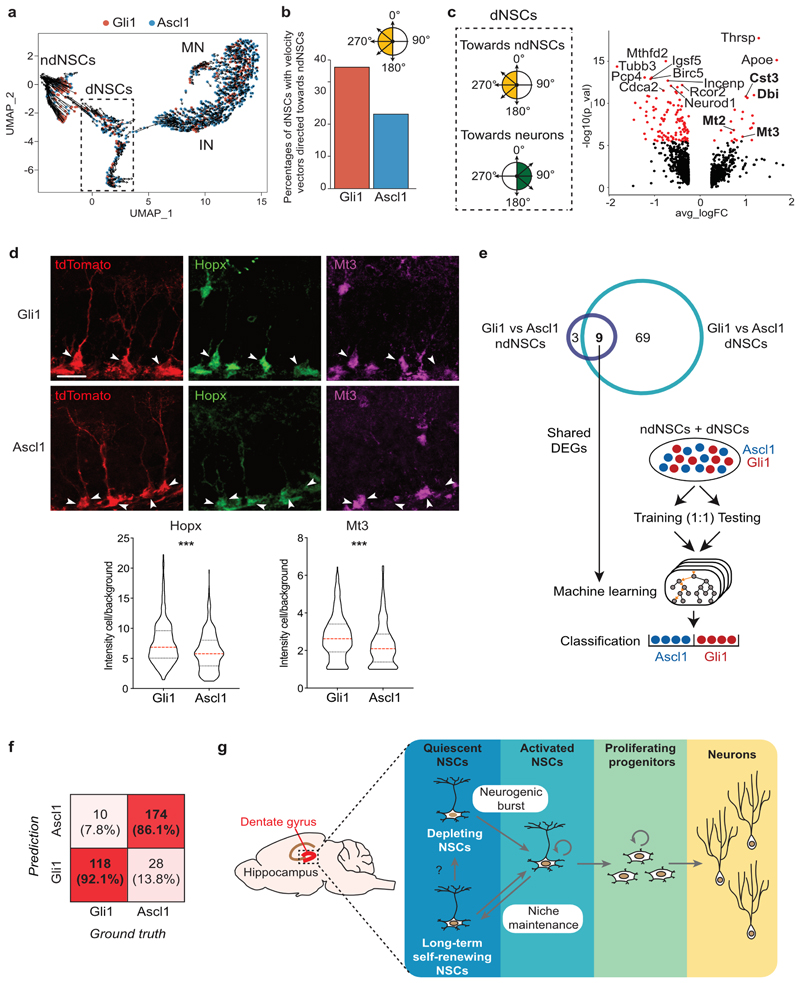
Fig. 4. Molecular profiling reveals distinct features of Gli1- vs. Ascl1-targeted NSCs.
a, The observed and the extrapolated future states (arrows) of Gli1 (red) or Ascl1 (blue) cells was calculated using RNA velocity. The velocities are visualized on the UMAP plot from panel (Fig. 3b). Velocity estimates based on nearest-cell pooling (k=300) were used. b, Percentages of dNSCs Gli1 (red) or Ascl1 (blue) with a velocity vector directed towards ndNSCs (yellow quadrants, vector angles between 180° and 380°). c, Volcano plot showing significantly DEGs (red, padj < 0.05) between dNSCs displaying an RNA velocity vector directed towards ndNSCs (yellow quadrants, vector angles between 180° and 380°), or towards IN (green quadrants, vector angles between >0° and <180°). Bolded gene names highlight common DEG with volcano plots (Fig. 3h) and (Fig. 3i). d, Top: representative examples of Gli1 and Ascl1 R cells stained using 4i protocol, 5 days post tamoxifen injection. Visualization of single channels: tdTom/red, Hopx/green, Mt3/magenta. Bottom: quantification of Hopx (Gli1 = 7.62 ± 0.197, n = 337 cells; Ascl1 = 6.14 ± 0.177, n = 316 cells; Unpaired t-test: t = 5.532; df = 651; ***p < 0.0001, two-tailed) and Mt3 (Gli1 = 2.72 ± 0.064, n = 337 cells; Ascl1 = 2.34 ± 0.065, n = 318 cells; Unpaired t-test: t = 4.201; df = 653; ***p < 0.0001, two-tailed) protein levels (fluorescence intensity of the cell/background) in Gli1- and Ascl1- targeted R cells. e, Schematic of machine learning approach. ndNSCs and dNSCs from Gli1- and Ascl1- targeted cells were divided into a training and a testing set (1:1). The 9 DEGs found in both ndNSCs (Fig. 3h) and dNSCs (Fig. 3i) comparisons were used to build three different machine learning models for classification: Random Forest Classifier (RFC), k Nearest Neighbor (k-NN) and Generalized Linear Models (GLM). The genotype (Gli1 or Ascl1) of ndNSCs and dNSCs was than predicted using the three classification models. f, Numbers and prediction accuracy rates to classify Gli1- vs. Ascl1-targeted cells using RFC, k-NN and GLM models together. Shades of red represent the accuracy of the prediction. g, Schematic showing the differential behavior of the two NSC populations in the DG: the first one gives rise to a burst of neurogenic activity followed by the depletion of the NSC. The second one is able to perform long-term self-renewal contributing to stem cell niche maintenance. Values are reported as mean ± s.e.m. Bars in violin plots represent median and quartiles. Scale bars represent 20 μm (d). For detailed statistics, see Supplementary Table 5.4
Hopx/Mt3 protein levels differentiate between Gli1- and Ascl1-targeted R cells
We next asked if the identified differentially expressed genes also showed distinct expression profiles at the protein level. We established a modified 4i technology to be used on tissue sections, allowing for the consecutive antibody-based analyses of up to 20 different proteins32. We first confirmed that both Gli1- and Ascl1-targeted cells expressed commonly used markers of R cells such as SOX2, GFAP and Id4 (Extended Data Fig. 5a, b). We next analyzed the expression of Hopx, recently associated with stem cell activity and self-renewal capacity of postnatal NSCs9, 26, 33. In accordance with the mRNA expression profiles, we found that Hopx protein was expressed in virtually all R cells analyzed (that were targeted by the Gli1- or Ascl1-promoter) (Fig. 4d and Extended Data Fig. 5b, d). However, Hopx protein levels were significantly higher in Gli1-targeted cells compared to Ascl1-targeted cells, indicating that differences in Hopx mRNA expression were also translated into distinct protein expression differences between Gli1- and Ascl1-targeted R cells. Protein expression levels of other stem cell associated proteins such as SOX2 were comparable between Gli1- and Ascl1-targeted NSCs (Extended Data Fig. 5e). To further substantiate these findings, we found that Metallothionein-3 (Mt3), showing differentially expressed mRNA levels (Fig. 3h-i), was also differentially expressed on the protein level (Fig. 4d and Extended Data Fig. 5b). Moreover, negative cells for Mt3 protein expression also showed lower levels of Hopx protein (Extended Data Fig. 5f).
Single cell gene expression predicts molecular origin of hippocampal R cells
Based on the differential expression of a subset of core genes in Gli1- and Ascl1-targeted cells in both ndNSCs and dNSCs (Fig. 3h, i), we next asked if commonly used machine learning strategies, such as Random Forest Classifier (RFC), k Nearest Neighbor (k-NN) and Generalized Linear Models (GLM), may allow for prediction of cellular origin (Fig. 4e). Strikingly, all three models were able to predict accurately Gli1- versus Ascl1-derived non-dividing and dividing R cells (Fig. 4f). These findings suggest that, despite the high transcriptional similarity overall and the partial overlap in terms of behavior, significant differences do exist in the targeted populations that allow for predicting the cellular origin of profiled cells. Thus, manipulating their expression levels in adult NSCs may represent a future entry point to alter the functional behavior of neurogenic cells in the adult brain.
Discussion
We used chronic intravital imaging and mRNA/protein analyses to define the functional and molecular properties of individual NSCs and their progeny targeted through Gli1- and Ascl1-promoters within the adult hippocampus. We find that a fraction of Gli1-targeted NSCs show evidence of long-term self-renewal, producing progeny for months following initial activation. In contrast, once activated, Ascl1-targeted R cells produce a burst of neurogenic activity before they terminally differentiate within weeks (Fig. 4g). Thus, the data presented here reconcile previously conflicting results regarding the self-renewal capacity of hippocampal NSCs, identifying functional diversity of neurogenic NSCs within the adult DG15, 34. We show that behavioral states of NSCs in the adult hippocampus are heterogeneous and consist of R cells that become exhausted upon activation and R cells that retain the ability to self-renew over the longer term. These imaging-based findings, which are consistent with previous interpretations of snap-shot based static data12, may be explained by three alternative scenarios: i) diverse types of NSC exist in the adult DG, one showing long-term self-renewal and the other rapid exhaustion upon activation, ii) Gli1-targeted cells may be developmentally upstream of Ascl1-targeted cells, or iii) NSCs’ behavior distributes along a spectrum of self-renewal potential with Gli1 targeting NSCs mostly at one end and Ascl1 targeting NSCs largely at the opposing end of the spectrum.
Some observations support a bona fide diversity of NSCs in the adult DG: First, cell division kinetics and patterns of intermediate quiescence between subsequent divisions (Fig. 2e-i and Extended Data Fig. 3b) are distinct between Ascl1- and Gli1-targeted cells, indicating that Ascl1-lineages cannot be straightforwardly superimposed onto Gli1-targeted lineages. Second, deep sequencing of single Gli1- and Ascl1-targeted R cells showed subtle but decisive differences in gene expression. Although there is a certain overlap at the single-cell level, these differential gene expression results are not driven by a small proportion of outlier cells in each group and are also reflected at the protein level (i.e., Hopx, Mt3) allowing for the separation of Gli1- vs. Ascl1-targeted cells (Fig. 3h-j, Fig. 4d-f). However, other findings are consistent with the hypothesis that Gli1-targeted R cells are upstream of Ascl1-targeted cells: We found that Gli1-targeted R cells appeared to be more undifferentiated based on pseudotiming analyses (Fig. 3g). Furthermore, we found that the behavior of NR cells was remarkably similar between Gli1- and Ascl1-targeted cells (Extended Data Fig. 3c-e).
Finally, further aspects of our data suggest that NSCs’ behavior aligns along a spectrum of stem cell potential: Not only Gli1-targeted R cells but also a small fraction of Ascl1-targeted R cells (approx. 8%) show symmetrical cell division following initial activation (i.e., generating two daughter R cells through duplication). However, in the 56 active lineages analyzed, we never observed symmetric (duplicative) divisions in the second or later rounds of division of Gli1-targeted R cells, as would be expected if Gli1-targeted R cells give rise to Ascl1-targeted R cells. Secondly, despite the finding that individual NSCs can exhibit distinct and diverse behaviors once activated, Ascl1- and Gli1-mediated targeting do not label two completely distinct NSC populations as suggested by the partial overlap in molecular profiles and behaviors. For example, the hierarchical clustering (Fig. 2j) shows a mixture of Gli1 and Ascl1 clones in both clusters, although in different proportions. Therefore, it might be plausible that each line biases toward one behavior and away from another along a spectrum of renewal potential, and that factors such as local microenvironmental cues may determine where a given NSC falls along this spectrum.
To discriminate among these scenarios, future experiments will need to combine intravital imaging with intersectional genetic lineage-tracing approaches and/or targeted ablation of Gli1- versus Ascl1-targeted NSCs. However, the application of these approaches in combination is extremely challenging and currently not feasible. Despite distinct behavioral properties of Gli1-targeted R cells compared to Ascl1-targeted R cells, mRNA expression profiles were remarkably similar albeit not identical between Gli1- and Ascl1-targeted populations, and we confirmed the differential expression levels of candidate genes on the protein level (i.e., Mt3, Hopx). Clearly, cell fate may be directed through mechanisms independent of gene transcription/expression, but rather by modifying signaling activity through transcription-independent mechanisms12–14. In addition, Ascl1- and Gli1-targeted populations are not discrete but overlap. Thus, future studies using more extensive single-cell deep sequencing and/or unbiased expression analyses of not only transcripts but also translated proteins on a single cell level will be needed to identify the detailed molecular differences mediating behavioral diversity of adult hippocampal NSCs. Future experiments will also aim to modify the imaging protocol to fully rule out the possibility that chronic exposure to gas anesthesia, surgery or window implantation may have some effect on neurogenesis. However, the identification of long-term self-renewal in Gli1-targeted and not in Ascl1-targeted NSCs supports the validity of our observations and approach. Using chronic in vivo imaging and single cell molecular analyses, our data resolve functional heterogeneity of NSCs within the adult brain. Thus, the data presented here identify long-term self-renewing NSCs in the hippocampus and characterize the molecular and cellular framework to direct distinct behaviors of R cells with the aim to enhance neurogenesis for regenerative repair in the adult brain.
Methods
Transgenic animals and labeling paradigm for NSCs
Animal experiments were approved by the Cantonal Commission for Animal Experimentation of the Canton of Zurich, Switzerland in accordance with national and cantonal regulations. To generate Gli1CreERT2/tdTom mice (Gli1Cre +/- Ai14+/-), Gli1CreERT2 mice (Gli1tm3(cre/ERT2)Alj, Jackson lab strain 007913) are crossed to homozygous CAG tdTomato (Ai14; B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze; JacksonLab strain 007914) reporter strain. Ascl1CreERT2 mice (Ascl1CreERT2 +/- Ai14+/-, Ascl1Cre ERT2,Ascl1 tm1.1(Cre/ERT2)Jejo; JacksonLab strain 012882) were bred with the CAG tdTomato line (Ai14; B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze ; JacksonLab strain 007914) to obtain Ascl1CreERT2/tdTom mice for imaging. Mice were group housed in ventilated cages (21-23 0C) under a 12h dark/light cycle with ad libitum access to food and water. The genetic background of the two mouse lines used in this study was confirmed by sequencing analysis of SNPs in the genome, using a panel of 2050 SNPs (Biolytix) and compared to results from C57BL6 and 129S control animals. Ascl1CreERT2/tdTom mice (n=3) exhibited an overlap of 99,1% (C57BL6) and 43,6% (129S) with control animals. Gli1CreERT2/tdTom mice (n=3) showed an overlap of 91,5% (C57BL6) and 46,7% (129S) with controls. At the age of 6-7 weeks mice of mixed sex underwent the implantation of the hippocampal window. Two weeks after the placement of the window sparse labeling of NSCs is achieved by single i.p. injection of Tamoxifen (Tam; 60-70mg/kg bodyweight for Gli1CreERT2/tdTom mice and 70-80mg/kg bodyweight for Ascl1CreERT2/tdTom mice; Sigma). Imaging fields of view (SPOTs) containing identified R cells were selected to obtain 10-20 SPOTs per mouse.
Chronic hippocampal window implantation
The surgical intervention for placing of the hippocampal window was carried out in mice aged 6-7 weeks as explained in previous studies16, 35. Briefly, above the dorsal DG (–2.0 posterior /–1.5 lateral mm from Bregma) the skin is opened and the cranial bone is locally removed (circle 3mm Ø). A cylinder (3mm Ø) of cortical tissue that has been punched out using biopsy punches (Miltex) until a defined depth (1.5mm, level of the corpus callosum) is aspirated using a blunt needle (22g) connected to an air pump until white matter tracks are visible. As soon as no bleeding occurs anymore, the hippocampal window (3mm Ø stainless steel cannula, 1.5mm height, covered by a 3mm Ø glass coverslip, Warner Instruments) is inserted, secured in place using the stereotactic arm and stably fixed to the cranial bone with UV curable dental cement (Ivoclar Vivadent).
In vivo 2-photon imaging in the hippocampus
Chronic 2-photon imaging was performed from 2 weeks after hippocampal window surgery, as described previously16, 35. In brief, after installation of an aluminum headpost on the contralateral side of the mouse head for repeated placement of mice in a frame, mice were anaesthetized with isoflurane (1.5–2% in O2) and the body temperature was measured and kept at 37°C with a heating pad. The imaging was performed on custom-built 2-photon microscope (Sutter Instrument Movable Objective Microscope type) using a long-working distance objective (water immersion, 16x magnification, 0.8NA, Nikon) and equipped with a ytterbium-doped laser system at 1045nm and 200fs (Femtotrain, High-Q lasers) or a fiber oscillator laser at 1070nm (Fidelity-2, Coherent) to excite tdTomato labeled cells in the DG. Emission light was detected using a photomultiplier tube (Hamamatsu) after passing a red emission filter (610/75 nm; AHF). Individual fields of view (SPOTs) containing R cells were selected and could be revisited in subsequent sessions using a coordinate system with the windows cannula being the reference (zero position, x=0, y=0, z=0). Every SPOT was imaged repeatedly by acquisition of a z-stack (512x512 pixel resolution, 2x zoom, 5μm step-size) carefully considering and checking for all cells of the clone. All SPOTs were checked on a daily basis, and no z-stack was acquired if no changes occurred. The time spent under the microscope for the mouse was kept minimal (<1h per day).
Identification and coding of lineages after processing imaging raw data
For detailed description of the criteria for coding of cells within clonal lineages, please refer to the Material and Methods section & Supplemental table with cell type characteristics in a previously published study16. Briefly, all timepoints and z-levels of a chronically imaged SPOT were compiled into a single file and the structural information from all z-levels were considered in coding cell types, cellular behavior and lineage relationships. The ROI manager in FIJI was used to code each individual cell at every timepoint throughout the compiled imaging file. Every lineage was annotated with a code by an individual researcher and double checked by a second researcher. The coding parameters were: CellID, CellType, Uncertainty CellType, Timepoint, MotherID, Uncertainty MotherCell, SisterID, Uncertainty SisterCell and CellDeath. Only if both agreed on the coding of cell type characterization or lineage transitions, this was coded as certain, otherwise as semi-certain or uncertain. The resulting compiled lineage trees were again double checked for inconsistencies with the imaging raw data.
Analysis of the lineage trees
The assembling of the tree annotations and part of the data analysis was performed in R software 36 using custom made scripts 16. The dataset consists of 216 (136 Gli1 and 80 Ascl1) lineages. 19 Ascl1 new lineages (from 2 additional mice) have been added to the Ascl1 dataset shown in Pilz et al., 2018. Two lineages from the previous dataset were excluded because of uncertainty due to imaging quality. The observed lineage behavior of newly added Ascl1-targeted cells was fully consistent to the one described before16. In total 17 (6 Gli1 and 11 Ascl1) animals have been used. The Gli1 dataset consists of 1525 cells and 1390 lineage transitions. Multiple parameters (listed below) were used to describe the behavior of the clones and to perform the hierarchical clustering of the lineage trees. The value of some parameters was calculated using the R script and then double checked and corrected manually; the value of other parameters was calculated manually after inspection of the lineage trees. For the majority of the analysis all the active clones are considered. In some cases (specified in the figure legend) some clones are excluded. The Ward’s Hierarchical Clustering Method using the Euclidean similarity index was used to perform the hierarchical clustering analysis37.
Parameters:
-
-
Maximum cell number: it represents the maximum cell number reached in the clone at one point (the time point can be any).
-
-
Final cell number: it represents the number of cells present in the clone at the last time point of the imaging (Extended Data Fig. 2a).
-
-
Number of successive divisions: it represents the maximum number of successive divisions in the clone (both R and NR cells are considered).
-
-
Self-renewal duration of R cell: it represents the time (in days) from the first R division until the last R in the clone is seen. Only certain R cells are considered. If the R cell disappears after the first division the self-renewal time is 0 (Extended Data Fig. 2a).
-
-
Activity duration of the clone: it represents the time (in days) from the first R division until the last division of any cell (R or NR) in the clone. If the clone does only 1 division the activity duration is 0.
-
-
Number of R successive divisions: it represents the maximum number of R successive divisions in the clone. Only certain R cells are considered (Extended Data Fig. 2a).
-
-
Maximum R clone content: it represents the maximum number of R cells that are present at the same time (at any time point) in the clone. Only certain R cells are considered.
-
-
Persistence of the R cell in the clone: it represents the time (in days) from the beginning of the imaging to the last time point the last R cell is seen in the clone. Only certain R cells are considered.
-
-
Time between R divisions: it represents the average time (in days) between R divisions in the clone (excluding the first R root cell). All certain R cells are considered. If the R cell is depleted after the first division of the R root cell, the time between R divisions is 0 (Extended Data Fig. 2a).
-
-
Time until division of R cells in successive divisions: it represents the time (in days) between divisions among different consecutive R divisions. The divisions were counted from the first neurogenic division (Div1 = first asymmetric R division in the tree).
-
-
Time until the first division of the R cell: it represents the time (in days) from the beginning of the imaging to the last time point the single R cell is seen before the first division. If the clone is observed with 2 cells at the first imaging session (2dpi) the value 1 is assigned. This parameter was not used for the hierarchical clustering analysis.
To determine the time-dependent fraction of clones by R content and the average clone content (Fig. 2b, d), all lineage trees were first shifted to the time point of the first R cell division to align them to a common reference time point related to cell fate. The fraction of clones by R content was obtained by determining the number of clones with the respective R content and normalizing the result by the number of observed clones. This accounted for the fact that different lineage trees had different total observation times. Likewise, the average clone content was obtained by computing the average number of the respective cell type within the observed population at each time point. The same conventions were used to obtain the results shown in Extended Data Fig. 3e. There, when determining division types, uncertain lineage relations were included when they were binary, i.e., when they associated one mother cell with exactly two daughter cells.
For the analysis of the modes of R and NR cell divisions (heat maps) only certain R and NR mother and daughter cells and certain transitions were considered. If a cell undergoes cell death, the cell fate at the last time point before cell death is taken.
The time (in days) between NR divisions takes into account only certain NR cells that undergo certain and semi-certain transitions. The tree visualization was performed using the igraph package of R38. Pheatmap package of R was used for heat map visualization39. PAST3 software was used for hierarchical clustering visualization. Quantification graphs were visualized using Graph pad Prism (version 8).
Immunohistochemical identification of imaged clones and confocal microscopy
After the in vivo imaging experiments were terminated, animals were put to deep anesthesia (Burprenophine) and then perfused with first cold saline and then paraformaldehyde (4% PFA in phosphate buffer). Brains were post-fixed overnight and cryo-protected (30% sucrose) before being cut horizontally in 60μm thickness on a cryotom (Leica SM2010R). Post-imaging immunohistochemical stainings were performed with antibodies against Sox2 (1:500, goat, Santa Cruz), GFAP (1:500, mouse, Sigma), Hopx (1:500, mouse, Santa Cruz) and Ki67 (1:250, rabbit, Abcam). For further quantifications Nestin (1:250, mouse, BD), Sox2, GFAP and Iba1 (1:500, rabbit, WAKO) were stained on 60μm horizontal sections (not previously imaged in vivo). Images were taken on confocal laser scanning microscopes (Olympus FV1000 using FluoView FV1000 or Zeiss LSM800 using ZEN Pro software). A detailed list of antibodies is provided in Supplementary Table 4.
Distribution of R cells and clones within the DG
For the analysis of the distribution of recombined R cells and imaged clones within the DG, mice received a single i.p. Tamoxifen injection (60-70mg/kg bodyweight for Gli1CreERT2/tdTom mice and 70-80mg/kg bodyweight for Ascl1CreERT2/tdTom mice). Two days following the induction (2dpi) or after the in vivo imaging experiments were terminated (two months after the induction) the mice were perfused with cold saline followed by paraformaldehyde (4% PFA in phosphate buffer). Brains were post-fixed overnight and cryo-protected (30% sucrose) before being cut horizontally in 60μm thickness on a cryotom (Leica SM2010R).
For the analysis of the distribution of recombined R cells 2dpi, immunohistochemical stainings were performed with the antibody against Sox2 (1:500, goat, Santa Cruz) and recombined R cells were identified for being Sox2+/tdTomato+ and showing a radial-glia morphology. The position of the recombined R cells was registered with X and Y coordinates in the first 5 slices containing the DG (300μm considered on Z axis).
For the analysis of the distribution of imaged clones, the clones were recognized in slices and their position was registered taking into account the center of the clone.
Additionally, a reference point (anchor) position was registered at the bottom tip of the DG in each slice. The distance between the anchor and the top tip of the DG was calculated in each slice (max distance). Finally, the distance between the anchor and each recombined R cell or imaged clone was calculated in each slice and normalized for the max distance (Extended Data Fig. 1b-d).
Furthermore, using the same registered X and Y coordinates the distance between each recombined R cell and all the others recombined R cells in each slice was measured (Extended Data Fig. 1e, f) at 2dpi.
Multiplexed immunostaining
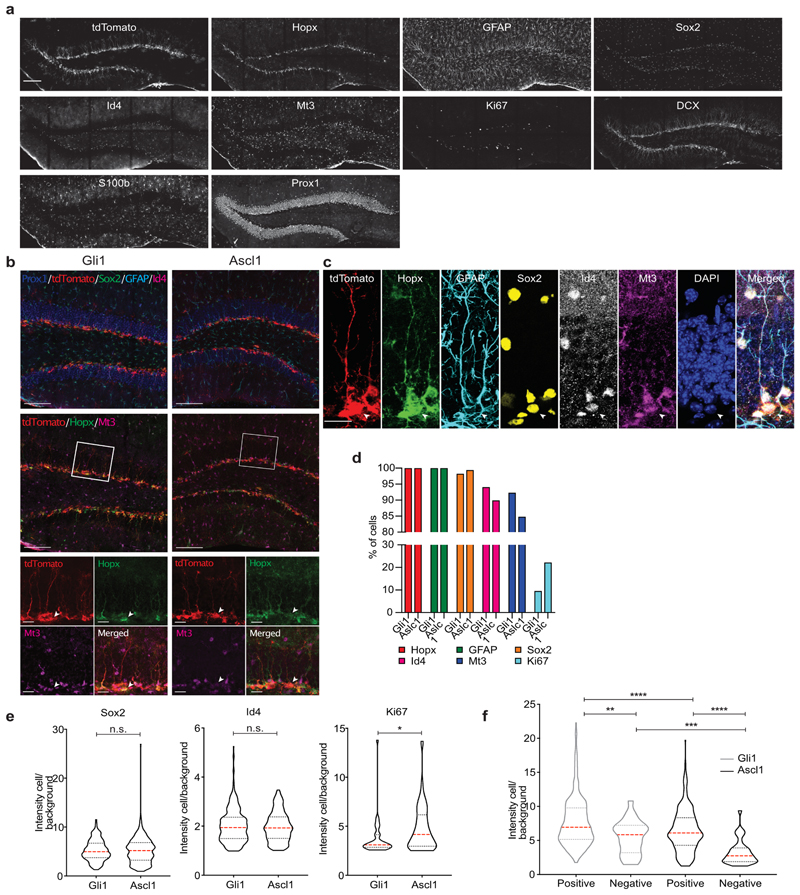
For analysis of protein expression of the Gli1 and Ascl1-labelled NSCs, mice received Tamoxifen injections (180mg/kg bodyweight) on three consecutive days to achieve high rates of recombination. Five days following the first induction, the mice were perfused with cold saline followed by paraformaldehyde (4% PFA in phosphate buffer). The brains were post-fixed in PFA overnight, then cryo-protected for two days. Coronal sections were cut at 40μm on a cryotome (Leica SM2010R). Prior to staining, sections were mounted in glass-bottomed 24 well-plates (Cellvis P24-1.5H-N) coated with poly-D-lysine. Using iterative indirect immunofluorescence imaging (4i) technique, adapted for fresh-frozen tissue32, the tissue was stained for tdTomato (1:500, goat, Sicgen), Hopx (1:500, mouse, Santa Cruz), Ki67 (1:250, rat, eBioscience), Sox2 (1:250, rat, Invitrogen), DCX (1:300, goat, Santa Cruz), s100β (1:500, rabbit, abcam), Id4 (1:250, rabbit, Biocheck), Mt3 (1:300, rabbit, abcam), GFAP (1:750, chicken, Novus), and Prox1 (1:500, rabbit, Millipore). Images were acquired with a confocal laser-scanning microscope (Zeiss LSM 800 using ZEN Pro software). Between iterations of staining, antibodies were eluted from the tissue using a 0.5M glycine-based buffer (pH 2.5) containing 3M Urea (Sigma-Aldrich), 3M Guanidinium Chloride (Sigma-Aldrich), and 0.07M TCEP-HCl (Sigma-Aldrich). During imaging, the tissue was stored in a phosphate buffer containing 0.7M of the free radical scavenger N-Acetyl-Cysteine to prevent photo-induced crosslinking of the antibodies to their respective epitopes. A detailed list of antibodies is provided in Supplementary Table 4.
RNA-Scope
For the analysis of Ascl1 mRNA levels using RNA-Scope, mice received a single i.p. Tamoxifen injection (80mg/kg bodyweight for Gli1CreERT2/tdTom mice and 90mg/kg bodyweight for Ascl1CreERT2/tdTom mice). Five days following the induction (5dpi) mice were perfused with 0.9% saline followed by 10% formalin. Brains were then put in 70% EtOH, embedded in paraffin and sectioned in a coronal plane at 5 µm thickness. The sections were then stained according to the company’s recommended protocol with probes against Ascl1 and Ki67 (RNAscope Probe Mm-Ascl1-C2 and Mm-Mki67), with target retrieval performed for 15 min and Protease Plus treatment for 30 min. The HRP-C1 label (Ki67) was visualized with TSA based fluorophore Cy5 (1:1500, Akoya Bioscience) whereas the HRP-C2 label (Ascl1) was detected with FITC (1:1000, Akoya Bioscience). Slides were then processed for immunofluorescence using antibodies against GFAP (1:750, rat, Millipore) and tdTomato (1:250, goat, Sicgen) and counterstained with DAPI. Images were acquired using a 40X objective on Leica SP8 confocal. For the quantification of the RNA-Scope signal two methods were used: i) the number of dots in each cell were counted; ii) the mean intensity of the cells was taken.
Quantification, statistical analysis and reproducibility
The quantifications of: i) immunohistochemical stainings, ii) distribution of R cells and clones within the DG, iii) different markers in the multiplexed immunostaining experiments and iv) RNA-Scope experiments were performed blinded. The quantifications of the immunohistochemical stainings and the distribution of R cells and clones in the DG were performed on a minimum of 3 animals, using 5-6 DG sections per brain (dorsal DG). The quantifications of markers in multiplexed immunostaining and RNA-Scope experiments were performed on a minimum of 3 animals, using 3 representative DG sections per brain. All the applied statistics were conducted on data from three or more biologically independent experimental replicates.
The statistical analysis of the data was performed using Graph pad Prism (version 8). A detailed report of statistical tests used, test results, sample sizes and p-values can be found in Supplementary Table 5.1-5.4 and in figure legends. First, normality was tested and when data did not follow a normal distribution according to the Shapiro-Wilk test, the non-parametric Mann Whitney test was performed. Alternatively, a paired or an unpaired two-tailed t-test was used for mean comparison depending on the nature of the analysis (within or between subjects). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 were considered significant. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications16.
Single-cell isolation
Gli1CreERT2/tdTom or Ascl1 CreERT2/tdTom mice were injected with Tamoxifen (180mg/kg bodyweight; 3x 5d timepoint; 2x 12wk timepoint). 5 days or 12 weeks after the first Tam injection, the dentate gyrus (DG) was sub-dissected in ice-cold HBSS. DG dissociation was performed using the Neural Tissue Dissociation Kit (P) (Miltenyi Biotecj cat #: 130-092-628) and Myelin removal beads II (Miltenyi Biotec cat #: 130-096-733) according to the manufacturer’s protocol. Hoechst 33342 (Life Technologies) was added for live cell discrimination. The gating strategy started with a gate on the clearly identifiable population of whole cells that separates from the debris in FSC-A/SSC-A, doublets were then eliminated using a FSC-A/FSC-H gate followed by a SSC-A/SSC-H gate, finally live Hoechst- tdTomato+ cells were selected for sorting (Extended Data Fig. 4a). Across all sorting experiments we identified 2-3 % of whole cells among which >97% were singlets containing between 2.5 and 5 % of live Hoechst- tdTomato+ cells. scRNA-seq of individual cells confirmed a purity of >99%. Individual Hoechst- tdTom+ cells were sorted into 384 well plates containing lysis buffer using a FACSAria III sorter (BD Biosciences). Flow cytometry data were analyzed using FlowJo v10. Sorted plates were stored at -80°C until library preparation.
Single-cell library preparation and sequencing
Library preparation was performed using a mosquito robot HV genomics (TTP Labtech Ltd, UK) following the Smart-seq2 protocol40. Briefly, 384 well plates containing sorted single nuclei in lysis buffer were thawed and reverse transcription with Superscript II (Life Technologies, cat #: 18064014) and PCR using KAPA Hifi HotStart ReadyMix (Kapa cat #: KK2602) were performed with the following biotinylated primers (Qiagen): Oligodt (AA GCA GTG GTA TCA ACG CAG AGT ACT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTV N), TSO (AAG CAG TGG TAT CAA CGC AGA GTA CATr GrG+G) and ISPCR primers (AA GCA GTG GTA TCA ACG CAG AGT). Following RT-PCR, clean up with Agencourt AMPure XP beads (Beckman Coulter cat #: B37419AA) was carried out and sample concentrations were measured using Bioanalyzer (Agilent Technologies) and normalized at a concentration of 0.3 ng/µl. The Nextera XT DNA library prep kit (Illumina cat #: FC-131-1096) was used for subsequent sample preparation. Samples were subjected to a tagmentation reaction, indexing, and PCR amplified. Libraries were then mixed in 384-sample pools and purified with Agencourt AMPure XP beads. Ready DNA libraries were quality controlled using D1000 Screen Tape Assay (Agilent Technologies). Samples were sequenced at the Functional Genomics Center Zurich on Illumina HiSeq 2500 or HiSeq4000 sequencers with single-end 125bp reads.
Single-cell RNA-seq analysis
Single-end 126nt-long reads were adaptor removed and trimmed using cutadapt v1.16 and sickle v1.33 with default parameters. Reads were mapped against the mouse GRCm38.90 primary assembly 126 with the Ensembl GRCm38.90 GTF annotation using STAR v2.6.0c41 in `alignReads` mode. Gene count matrices were quantified at the exon level ignoring multimappers using featurecounts from subread v1.6.2. RNA velocity loom files were generated with velocyto v0.17.17 28 in `run-smartseq2` mode using the Ensembl_GRCm38.90 GTF annotation. Cells were quality checked using scater v1.12.242. Only cells with 1000-6000 genes detected, less than 8% of mitochondrial reads and more than 50’000 reads were kept. In each dataset (i.e 384 well-plate), cells with more or less than 1.5 fold the median read content per cell were excluded. A total of 1465 cells passed QC filtering. 410 cells were filtered out including: s100b+ Aqp4+ Aldh1l1+ astrocytes (only detected in Gli1CreERT2/tdTom cells), tmem119+ microglia and gad1+ gad2+ interneurons. A total of 1055 cells were retained for final analysis. Seurat v3.1.1 was used to normalize read counts, regress out the library size and mitochondrial reads proportion per cell, find variable features with the `vst` method, integrate multiple plates as in 43, dimensionality reduce the data (PCA, t-SNE, UMAP), cluster cells and test for differentially expressed genes. Differential gene expression by cell type (e.g. Ascl vs Gli contrast) was tested only for genes detected in at least 20% of the cells in either of the populations. Logistic regression fits with batch as latent variable (null model) and cell type as predictor were run using Seurat's `FindMarkers`. P-values were adjusted for multiple testing using the Benjamini–Hochberg method within each contrast; genes with a False Discovery Rate (FDR)-adjusted p-value below 0.05 were deemed significant. Batch-affected genes were similarly tested using logistic regression with cell type as latent variable and batch as predictor (Batch1: plates Ascl1_12wk_1, Ascl1_5day_1, Gli1_12wk, Gli1_5day_1 and Gli1_5day_2; batch2: plates Ascl1_12wk_2 and Ascl1_5day_2). Genes with no detectable effects between the two batches (FDR-adjusted p-value greater or equal to 0.05) were subjected to downstream RNA velocity analysis. RNA velocity was run with velocyto.R v0.6 using default parameters. RNA velocities were plotted on top of Seurat's UMAP embedding; cells were colored according to Seurat's clustering results. Lineage inference analysis was performed using Slingshot v1.3.224. For pseudotime, independent component analysis was computed with fastICA package v1.2.2. Data analysis was carried out in R v3.6.1.
Prediction algorithm
DEGs used for the prediction of Gli1 and Asc1 genotype were obtained by taking the common genes between DEG calculated for ndNSC and DEG calculated for dNSC using edgeR package (9 genes in total). The data, containing ndNSC and dNSC cells, were split into training and test set. The training set consisted of 50% of cells from the least populated class and the corresponding number of cells was taken from the more populated class. The prediction consists of an Ensemble pairwise classification consisting of three models - Generalized linear model, k-nearest neighbors and random forest (implemented in Caret package). For each model the cross-validation was performed 10 times with 10% of the training data for the parameter tuning. Each model was used to perform classification on the test set using the stats package predict function. In order to ensure a better reproducibility of the prediction the process of fitting and prediction was done 1000 times (seeds) for each of the three models. For each seed, the data was randomly split by the same means as above to ensure that different cells are used in different seed. The accuracy was computed by assigning a score of -1 to the cells that were wrongly classified in each seed, per model, and + 1 to the cells that were correctly classified. The final accuracy was taken by comparing the proportion of correctly and wrongly classified cells according to the score system described above.
Reporting summary
Further information is available in the Nature Research Reporting Summary.
Extended Data
Extended Data Fig. 1. Characterization of Gli1 and Ascl1-targeted cells.
a, Number of tdTom+ cells in the SGZ 2d after recombination in Gli1 (n = 4 mice) and Ascl1 (n = 3 mice). b, Percentage of the distance of tdTom+ R cells from the anchor/max distance in Gli1 and Ascl1 at 2dpi. Horizontal sections, first 300 μm of DG are considered (Gli1: 53.93 ± 2.27, n = 106 cells; Ascl1: 54.35 ± 1.83, n = 199 cells; Mann Whitney test: Mann-Whitney U = 10407; p = 0.849, two-tailed). c, Percentage of the distance of imaged clones from the anchor/max distance in Gli1 and Ascl1 at 2mpi. Horizontal sections (Gli1: 51.6 ± 2.31, n = 40 clones; Ascl1: 45.6 ± 5.63, n = 18 clones; Mann Whitney test: Mann-Whitney U = 337; p = 0.708, two-tailed). d, Representation of the method used to quantify the distances between R cells and the anchor. Horizontal view of the DG. e, Quantification of the distance between pairs of tdTom+ R cells in Gli1 and Ascl1 at 2dpi. Horizontal sections, first 300 μm of DG are considered (Gli1: 523.5 ± 14.86, n = 608 pair of cells; Ascl1: 546.1 ± 9.25, n = 1696 pair of cells; Mann Whitney test: Mann-Whitney U = 505390; p = 0.469, two-tailed). f, Representation of the method used to quantify the distances between pair of R cells (only the pairings of 3 R cells with all the other R cells in the section are shown as examples). Horizontal view of the DG. g, Quantification of tdTom+ cell types in the SGZ 2d after recombination in Gli1 (n = 4 mice) and Ascl1 (n = 3 mice). h, Representative pictures of RNA-Scope with Ki67 probe (green) and Ascl1 probe (grey) and immunostaining for tdTomato (red) and GFAP (cyan) of Gli1 and Ascl1-targeted cells at 5dpi. Filled arrowheads point to R cells; empty arrowheads point to NR cells. i, Quantification of the Ascl1 mRNA dots of: R ki67- cells (Gli1: 4.136 ± 0.366, n = 88 cells; Ascl1: 5.617 ± 0.851, n = 47 cells; Unpaired t-test: t = 1.855; df = 133; p = 0.065, two-tailed), R ki67+ cells (Gli1: 10.83 ± 2.676, n = 6; Ascl1: 8.250 ± 2.089, n = 12; Unpaired t-test: t = 0.734; df = 16; p = 0.473, two-tailed), NR ki67+ cells (Gli1: 8.000 ± 1.741, n = 12; Ascl1: 4.978 ± 0.7160, n= 45; Unpaired t-test: t = 1.834; df = 55; p = 0.0721, two-tailed). j, Quantification of the Ascl1 mRNA levels (fluorescence intensity of the cell/background) of: R ki67- cells (Gli1: 1.739 ± 0.083, n = 77 cells; Ascl1: 1.933 ± 0.198, n = 40 cells; Unpaired t-test: t = 1.055; df = 115; p = 0.293, two-tailed), R ki67+ cells (Gli1: 4.528 ± 1.919, n = 5; Ascl1: 2.502 ± 0.369, n = 12; Unpaired t-test: t = 1.54; df = 15; p = 0.144, two-tailed), NR ki67+ cells (Gli1: 4.157 ± 0.566, n = 10; Ascl1: 2.510 ± 0.258, n= 39; Unpaired t-test: t = 2.815; df = 47; **p = 0.007, two-tailed). k, Quantification of the Ascl1 mRNA dots of: ki67- cells (Gli1: 4.067 ± 0.295, n = 149 cells; Ascl1: 3.516 ± 0.405, n = 124 cells; Unpaired t-test: t = 1.121; df = 271; p = 0.263, two-tailed), ki67+ cells (Gli1: 8.944 ± 1.454, n = 18; Ascl1: 5.667 ± 0.728, n = 57; Unpaired t-test: t = 2.141; df = 73; *p = 0.035, two-tailed). Values are shown as mean ± s.e.m. Bars in violin plots represent median and quartiles. Scale bars represent 100 μm (d,f) and 10 μm (h). For detailed statistics, see Supplementary Table 5.1
Extended Data Fig. 2. Intravital 2-photon imaging does not affect proliferation or lineage commitment of Gli1-targeted NSCs.
a, Explanatory scheme of the different parameters extracted from lineage data of the imaged clones. b, Comparison of cell proliferation in contralateral and ipsilateral side in Gli1 mice after 2 months of 2-photon imaging (dorsal DG, horizontal sections). c, Quantification of Ki67+ cells and Ki67+ tdTom+ cells in contra- and ipsilateral side. Horizontal sections, first 360 μm of DG are considered (Ki67+ ipsilateral: 1345 ± 235.7, n = 3 mice; Ki67+ contralateral: 1278 ± 252.2, n = 3 mice; Paired t-test: t = 0.2631; df = 2; p = 0.817, two-tailed. Ki67+/tdTom+ ipsilateral: 28 ± 11.06, n = 3 mice; Ki67+/tdTom+ contralateral: 68.33 ± 26.1, n = 3 mice; Paired t-test: t = 2.678; df = 2; p = 0.116, two-tailed). d, Percentage of Ki67+ tdTom+/tdTom+ cells in contra- and ipsilateral side (ipsilateral: 7.26 ± 1.84, n = 4 mice; contralateral: 8.58 ± 0.63, n = 4 mice; Paired t-test: t = 0.895; df = 3; p = 0.436, two-tailed). e, Percentage of tdTom+ Hopx+ Ki67+/tdTom+ Hopx+ cells in contra- and ipsilateral side (ipsilateral: 9.0 ± 1.83, n = 3 mice; contralateral: 9.63 ± 1.91, n = 3 mice; Paired t-test: t = 0.392; df = 2; p = 0.733, two-tailed). f, Comparison of the number and identity of tdTom+ cells in contralateral and ipsilateral side in Gli1 mice after 2 months of 2-photon imaging (dorsal DG, horizontal sections). g, Quantification of tdTom+ neurons and Sox2+ tdTom+ glial cells in contra- and ipsilateral side. Horizontal sections, first 360 μm of DG are considered (tdTom+ ipsilateral: 475.3 ± 45.16, n = 3 mice; tdTom+ contralateral: 424.7 ± 128.2, n = 3 mice; Paired t-test: t = 0.475; df = 2; p = 0.681, two-tailed. tdTom+ SOX2+ ipsilateral: 279.3 ± 36.5, n=3 mice; tdTom+ Sox2+ contralateral: 293 ± 75.5, n = 3 mice; Paired t-test: t = 0.345; df = 2; p = 0.762, two-tailed). h, Percentage of Sox2+ tdTom+/tdTom+ in contra- and ipsilateral side (ipsilateral: 36.93 ± 3.78, n = 3 mice; contralateral: 41.49 ± 2.49, n = 3 mice; Paired t-test: t = 1.347; df = 2; p = 0.31, two-tailed). i, Quantification of the DG area covered by GFAP+ cells/area tot in Gli1 contra- and ipsilateral side at 2 weeks after surgery (ipsilateral: 0.77 ± 0.03, n = 3 mice; contralateral: 0.73 ± 0.01, n = 3 mice; Paired t -test: t = 2.004; df = 2; p = 0.183, two-tailed). j, Quantification of the DG area covered by Iba1+ cells/area tot in Gli1 contra- and ipsilateral side at 2 weeks after surgery (ipsilateral: 0.51 ± 0.05, n = 3 mice; contralateral: 0.5 ± 0.04, n = 3 mice; Paired t-test: t = 0.677; df = 2; p = 0.568, two-tailed). k, Quantification of the DG area covered by GFAP+ cells/area tot in Gli1 contra- and ipsilateral side at 2.5 months after surgery (ipsilateral: 0.79 ± 0.03, n = 3 mice; contralateral: 0.83 ± 0.02, n = 3 mice; Paired t-test: t = 1.654; df = 2; p = 0.24, two-tailed). Values are shown as mean ± s.e.m. Scale bars represent 100 μm (b, f). For detailed statistics, see Supplementary Table 5.1
Extended Data Fig. 3. NR cells behavior is similar between Gli1- and Ascl1- derived lineages.
a, Relationship between time to first division and R cell self-renewal duration. Gli1 and Ascl1-targeted R cells are shown as dots in the XY correlation graph (correlation analysis: Spearman r = -0.03, n= 113 XY pairs; p (two-tailed) = 0.744). b, Time until division of R cells (in days) in successive divisions. Div1: first division after the first neurogenic division in the clone. Comparison among consecutive divisions in each mouse line (Gli1 Div1: 8.49 ± 1.29, n = 36 divisions; Gli1 Div2: 6.89 ± 1.95, n = 14 divisions; Gli1 Div3: 7.75 ± 1.49, n = 4 divisions; p = 0.511. Ascl1 Div1: 3.12 ± 0.49, n = 37 divisions; Ascl1 Div2: 3.2 ± 1.28, n = 10 divisions; Ascl1 Div3: 20, n = 1 divisions; p = 0.225. Statistical test used: Kruskal Wallis) and comparison between Gli1 and Ascl1 (Div1: Mann Whitney test: Mann-Whitney U = 302; ***p < 0.0001, two-tailed; Div2: Mann Whitney test: Mann-Whitney U = 34.5; *p = 0.034, two-tailed). c, Heat maps representing the frequencies of division modes of Gli1- and Ascl1- targeted NR cells (division rounds (Div) 1, 3 and 5 counting from the first NR division. Gli1 Div1 n = 91 divisions; Gli1 Div3 n = 65 divisions; Gli1 Div5 n = 11 divisions. Ascl1 Div1 n = 85 divisions; Ascl1 Div3 n = 71 divisions; Ascl1 Div5 n = 18 divisions). R, radial glia-like cell; NR, proliferating progenitor cell; N, neuron; A, astrocyte. Division events contain only certain lineage relations. d, Time (in days) between NR cell divisions (Gli1: 1.38 ± 0.08, n = 460 cells; Ascl1: 1.24 ± 0.08, n = 461 cells; Mann Whitney test: Mann-Whitney U = 100475; p = 0.1207, two-tailed). e, Relative frequency of the NR cells cell fate distribution. The lineage trees are shifted to the first R cell division and NR divisions are pooled according to the time windows in which they take place (Gli1 n in 0-20 days interval: 410, Gli1 n in 20-40 days interval: 78, Gli1 n in 40-70 days interval: 12; Ascl1 n in 0- 20 days interval: 424, Ascl1 n in 20-40 days interval: 74, Ascl1 n in 40-70 days interval: 0). Error bars represent the standard error of the proportion. Division events contain all binary lineage relations (certain and uncertain). Values are shown as mean ± s.e.m. For detailed statistics, see Supplementary Table 5.2
Extended Data Fig. 4. Transcriptional analysis of Gli1- vs Ascl1-targeted cells.
a, Representative FACS plots showing gating for live (Hoechst-) tdTomato+ DG cells sorted for scRNA-seq. b, t-Distributed Stochastic Neighbor Embedding (t-SNE) visualization of the 4 clusters (non-dividing NSCs (ndNSCs), dividing NSCs (dNSCs), Immature Neurons (IN), Mature Neurons (MN)) identified in Gli1- and Ascl1 targeted tdTomato+ neuronal cells. c, Uniform Manifold Approximation and Projection (UMAP) visualization of the seven individual datasets used for analysis. _1 and _2 indicate duplicates for a given data point. d, UMAP visualization of the cell isolated 5 days or 12 weeks after tamoxifen injection. e, Distribution of the cells among the 4 clusters, 5 days or 12 weeks after tamoxifen injection. f, Expression pattern of Ascl1 mRNA. g, Violin plots showing Ascl1 mRNA levels in Gli1- and Ascl1-derived ndNSCs and dNSCs (Wilcoxon text, *p < 0.05). h, Position of Gli1 (red) and Ascl1 (blue) ndNSCs and dNSCs along the pseudotime axis as calculated by Monocle. Cells present along the full pseudotime range (grey rectangle) or sharing the same pseudotime range (green rectangle) are depicted. i-j, Volcano plots showing significantly differentially expressed genes (DEGs) (red, padj < 0.05) between Gli1 or Ascl1-targeted ndNSCs (i) or dNSCs (j) when comparing only cells with shared pseudotime range. Venn diagrams indicate the overlap between DEGs found when comparing Gli1 or Ascl1-targeted ndNSCs (i) or dNSCs (j) using the full pseudotime range (grey circles) or the shared pseudotime range (green circles). Among the top 10 DEGs, bolded gene names highlight the DEGs found in both comparisons. k, UMAP visualization of RNA velocities calculated using scVelo. For detailed statistics, see Supplementary Table 5.3
Extended Data Fig. 5. Stem cell markers associated with quiescence are differentially expressed between Gli1- and Ascl1-targeted NSCs.
a, Representative example of a Gli1- tdTom DG section (5 days post tamoxifen injection) stained with multiple antibodies using the 4i protocol. Visualization of 9 different cellular markers in addition to tdTomato. Single channels are shown. b, Representative images of a Gli1 and an Ascl1 DG section stained using 4i protocol, 5 days post tamoxifen injection. Visualization of six different cellular markers plus tdTomato. (Top) Prox1/blue, tdTom/red, SOX2/green, GFAP/light blue, Id4/magenta. (Middle) tdTom/red, Hopx/green, Mt3/magenta. (Bottom) single channels at higher magnification (regions in the white squares). c, Example of a Gli1-targeted R cell stained using the 4i protocol (5 days post tamoxifen injection). Visualization of 5 different R cell markers in addition to tdTomato and DAPI. Displayed are single channels and a merged picture. d, Percentage of cells that express different NSC and proliferation markers in Gli1- and Ascl1-targeted R cells (Gli1 n = 330 cells; Ascl1 n = 316 cells). e, Quantification of Sox2 (Gli1 = 5.24 ± 0.11 n = 330 cells; Ascl1 = 5.22 ± 0.14, n = 316 cells; Mann Whitney test: Mann-Whitney U = 51100; p = 0.661, two-tailed), Id4 (Gli1 = 2.02 ± 0.03, n = 317 cells; Ascl1 = 1.98 ± 0.03, n = 285 cells; Mann Whitney test: Mann-Whitney U = 44567; p = 0.776, two-tailed) and Ki67 (Gli1 = 3.78 ± 0.36, n = 32 cells; Ascl1 = 4.82 ± 0.28, n = 69 cells; Mann Whitney test: Mann-Whitney U = 760; p = *0.011, two-tailed) protein levels (fluorescence intensity of the cell/background) in Gli1- and Ascl1-targeted R cells. f, Quantification of Hopx protein levels (fluorescence intensity of the cell/background) in Gli1- and Ascl1- targeted R cells that are either positive or negative for Mt3 (Gli1 positive: 7.77 ± 0.20, n = 313 cells; Gli1 negative: 5.47 ± 0.5, n = 24 cells; Whitney test: Mann-Whitney U = 2394; **p = 0.003, two-tailed. Ascl1 positive: 6.46±0.18, n = 284 cells; Ascl1 negative: 3.28 ± 0.32, n = 32 cells; Whitney test: Mann-Whitney U = 1651; ****p < 0.0001, two-tailed. Gli1 positive vs Ascl1 positive: Mann Whitney test: Mann-Whitney U = 35578; ****p < 0.0001, two-tailed. Gli1 negative vs Ascl1 negative: Mann Whitney test: Mann-Whitney U = 182; ***p = 0.0006, two-tailed). Values are shown as mean ± s.e.m. Bars in violin plots represent median and quartiles. Scale bars represent 400 μm (a), 100 μm (b) and 20 μm (c, high magnification and b). For detailed statistics, see Supplementary Table 5.4
Supplementary Material
Acknowledgments
This work was supported by the European Research Council (STEMBAR to S. Jessberger and BRAINCOMPATH to F. Helmchen), the Swiss National Science Foundation (BSCGI0_157859 and 310030_196869 to S. Jessberger), the Zurich Neuroscience Center, the University of Zurich (UZH) Forschungskredit fellowship (B.N. Jaeger) and the Wellcome Trust (098357/Z/12/Z to B.D. Simons). L. Harris was supported by a fellowship from the Francis Crick Institute. Work in F. Guillemot’s lab is supported by the Francis Crick Institute, which receives its funding from Cancer Research UK (FC0010089), the UK Medical Research Council (FC0010089) and the Wellcome Trust (FC0010089), and by the Wellcome Trust (Investigator Award 106187/Z/14/Z to F. Guillemot). We thank P. Bethge for experimental help and D. Chichung Lie for comments on the manuscript. We thank E. Yángüez López-Cano from the Functional Genomics Center Zurich (UZH and ETHZ) and the Cytometry Facility of UZH. J. Sarabia del Castillo, G. Gut, and L. Pelkmans contributed to 4i experiments.
Footnotes
Author contributions
S.B. performed imaging, analyzed data, and co-wrote the manuscript. B.N.J. performed scRNA-seq, analyzed data, and co-wrote the manuscript. G.A.P. performed imaging, analyzed data, and co-wrote the manuscript. J.D.C. performed 4i analyses. M.K. performed scRNA-seq. L.H. performed the RNAscope experiments. I.M. performed computational analyses of scRNA-seq data. V.I.K. performed computational analyses of scRNA-seq data. D.J.J. and B.D.S. contributed to the concept, performed data analyses, and co-wrote the manuscript. F.G. and F.H. revised the manuscript. S.J. developed the concept and wrote the manuscript.
Competing interests statement
The authors declare no competing interests.
Data availability
All data are available from the authors at request. scRNA-seq data have been submitted into GEO (GSE138941).
Code availability
Source code for next-generation sequencing data processing and single-cell RNA-seq are available at https://github.com/imallona/stem_cells_hippocampus_jessberger_lab
References
- 1.Gage FH. Science. Vol. 364. New York, N.Y: 2019. Adult neurogenesis in mammals; pp. 827–828. [DOI] [PubMed] [Google Scholar]
- 2.Sorrells SF, et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018 doi: 10.1038/nature25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goncalves JT, Schafer ST, Gage FH. Adult Neurogenesis in the Hippocampus: From Stem Cells to Behavior. Cell. 2016;167:897–914. doi: 10.1016/j.cell.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Eriksson PS, et al. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 5.Knoth R, et al. Murine features of neurogenesis in the human hippocampus across the lifespan from 0 to 100 years. PLoS ONE. 2010;5:e8809. doi: 10.1371/journal.pone.0008809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreno-Jimenez EP, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer's disease. Nature medicine. 2019;25:554–560. doi: 10.1038/s41591-019-0375-9. [DOI] [PubMed] [Google Scholar]
- 7.Tobin MK, et al. Human Hippocampal Neurogenesis Persists in Aged Adults and Alzheimer's Disease Patients. Cell Stem Cell. 2019;24:974–982 e973. doi: 10.1016/j.stem.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spalding KL, et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin J, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suh HK, et al. In Vivo Fate Analysis Reveals the Multipotent and Self-Renewal Capacities of Sox2+ Neural Stem Cells in the Adult Hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seri B, Garcia-Verdugo JM, McEwen BS, Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci. 2001;21:7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urban N, et al. Science. Vol. 353. New York, N.Y: 2016. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein; pp. 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonaguidi MA, et al. In Vivo Clonal Analysis Reveals Self-Renewing and Multipotent Adult Neural Stem Cell Characteristics. Cell. 2011 doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Encinas JM, et al. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kempermann G. The pessimist's and optimist's views of adult neurogenesis. Cell. 2011;145:1009–1011. doi: 10.1016/j.cell.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Pilz GA, et al. Science. Vol. 359. New York, N.Y: 2018. Live imaging of neurogenesis in the adult mouse hippocampus; pp. 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsimpardi L, Lledo PM. Regulation of neurogenesis in the adult and aging brain. Current opinion in neurobiology. 2018;53:131–138. doi: 10.1016/j.conb.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Ben Abdallah NM, Slomianka L, Vyssotski AL, Lipp HP. Early age-related changes in adult hippocampal neurogenesis in C57 mice. Neurobiology of aging. 2010;31:151–161. doi: 10.1016/j.neurobiolaging.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalamakis G, et al. Quiescence Modulates Stem Cell Maintenance and Regenerative Capacity in the Aging Brain. Cell. 2019;176:1407–1419. doi: 10.1016/j.cell.2019.01.040. e1414. [DOI] [PubMed] [Google Scholar]
- 21.Ziebell F, Dehler S, Martin-Villalba A, Marciniak-Czochra A. Development. Vol. 145. Cambridge, England: 2018. Revealing age-related changes of adult hippocampal neurogenesis using mathematical models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- 23.Blomfield IM, et al. Id4 promotes the elimination of the pro-activation factor Ascl1 to maintain quiescence of adult hippocampal stem cells. Elife. 2019;8 doi: 10.7554/eLife.48561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumitru I, Neitz A, Alfonso J, Monyer H. Diazepam Binding Inhibitor Promotes Stem Cell Expansion Controlling Environment-Dependent Neurogenesis. Neuron. 2017;94:125–137 e125. doi: 10.1016/j.neuron.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Berg DA, et al. A Common Embryonic Origin of Stem Cells Drives Developmental and Adult Neurogenesis. Cell. 2019;177:654–668 e615. doi: 10.1016/j.cell.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuzwa SA, et al. Developmental Emergence of Adult Neural Stem Cells as Revealed by Single-Cell Transcriptional Profiling. Cell reports. 2017;21:3970–3986. doi: 10.1016/j.celrep.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 28.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochgerner H, Zeisel A, Lonnerberg P, Linnarsson S. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat Neurosci. 2018;21:290–299. doi: 10.1038/s41593-017-0056-2. [DOI] [PubMed] [Google Scholar]
- 30.Yang CP, Gilley JA, Zhang G, Kernie SG. Development. Vol. 138. Cambridge, England: 2011. ApoE is required for maintenance of the dentate gyrus neural progenitor pool; pp. 4351–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knobloch M, et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 2013;493:226–230. doi: 10.1038/nature11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gut G, Herrmann MD, Pelkmans L. Science. Vol. 361. New York, N.Y: 2018. Multiplexed protein maps link subcellular organization to cellular states. [DOI] [PubMed] [Google Scholar]
- 33.Zweifel S, et al. HOPX Defines Heterogeneity of Postnatal Subventricular Zone Neural Stem Cells. Stem Cell Reports. 2018;11:770–783. doi: 10.1016/j.stemcr.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonaguidi MA, Song J, Ming GL, Song H. A unifying hypothesis on mammalian neural stem cell properties in the adult hippocampus. Current opinion in neurobiology. 2012;22:754–761. doi: 10.1016/j.conb.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pilz GA, et al. Functional Imaging of Dentate Granule Cells in the Adult Mouse Hippocampus. J Neurosci. 2016;36:7407–7414. doi: 10.1523/JNEUROSCI.3065-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2017 http://igraph.org/
- 37.Picardo MA, et al. Pioneer GABA cells comprise a subpopulation of hub neurons in the developing hippocampus. Neuron. 2011;71:695–709. doi: 10.1016/j.neuron.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal, Complex Systems. 2006 [Google Scholar]
- 39.Kolde R. pheatmap: Pretty Heatmaps. R package version 1.0.8. 2015 [Google Scholar]
- 40.Jaeger BN, et al. Miniaturization of Smart-seq2 for Single-Cell and Single-Nucleus RNA Sequencing. STAR Protoc. 2020;1:100081. doi: 10.1016/j.xpro.2020.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the authors at request. scRNA-seq data have been submitted into GEO (GSE138941).
Source code for next-generation sequencing data processing and single-cell RNA-seq are available at https://github.com/imallona/stem_cells_hippocampus_jessberger_lab