Abstract
Three classes of ion-driven protein motors have been identified to date: ATP synthase, the bacterial flagellar motor, and a proton-driven motor that powers gliding motility and the Type 9 protein secretion system (T9SS) in Bacteroidetes bacteria. Here, we present cryo-EM structures of the gliding motility/T9SS motors GldLM from Flavobacterium johnsoniae and PorLM from Porphyromonas gingivalis. The motor is an asymmetric inner membrane protein complex in which the single transmembrane helices of two periplasm-spanning GldM/PorM proteins are positioned inside a ring of five GldL/PorL proteins. Mutagenesis and single-molecule tracking identifies protonatable amino acid residues in the transmembrane domain of the complex that are important for motor function. Our data provide evidence for a mechanism in which proton flow results in rotation of the periplasm-spanning GldM/PorM dimer inside the intra-membrane GldL/PorL ring to drive processes at the bacterial outer membrane.
Introduction
Three classes of ion-driven machines have been identified in biological systems. In ATP synthases the ion-motive gradient drives a rotary movement that results in the release of ATP from the catalytic sites of the enzyme1, 2. In the bacterial flagellum, ion flow through stator units linked to the cell wall results in a mechanical force that causes the flagellum to rotate3, 4, 5. The third type of ion-driven motor is found in certain Bacteroidetes where it is used to power gliding motility across solid surfaces6 using the proton motive force (PMF) across the inner membrane (IM) as the energy source7, 8, 9. The gliding motility motor generates a rotary motion at the cell surface9 that results in a helical flow of surface adhesin proteins7, 10, 11, possibly by driving a mobile track to which the adhesins are attached12.
In the model Bacteriodetes gliding bacterium Flavobacterium johnsoniae there is genetic evidence that two conserved inner membrane proteins GldL and GldM form a PMF-transducing gliding motility motor13 (Fig. 1a). Neither of these proteins has sequence similarity to the components of the other classes of ion-driven motors. GldL is composed of two predicted transmembrane helices (TMH) followed by along cytoplasmic tail14. GldM is mainly periplasmic and anchored to the inner membrane by a N-terminal TMH14. A recent structure of the periplasmic portion of GldM reveals that it is a homodimer that folds into four pairs of domains15. These paired domains are arranged end to end to form a rod that is long enough (180Å) to span the periplasm and interact with the gliding motility apparatus present at the outer membrane (OM).
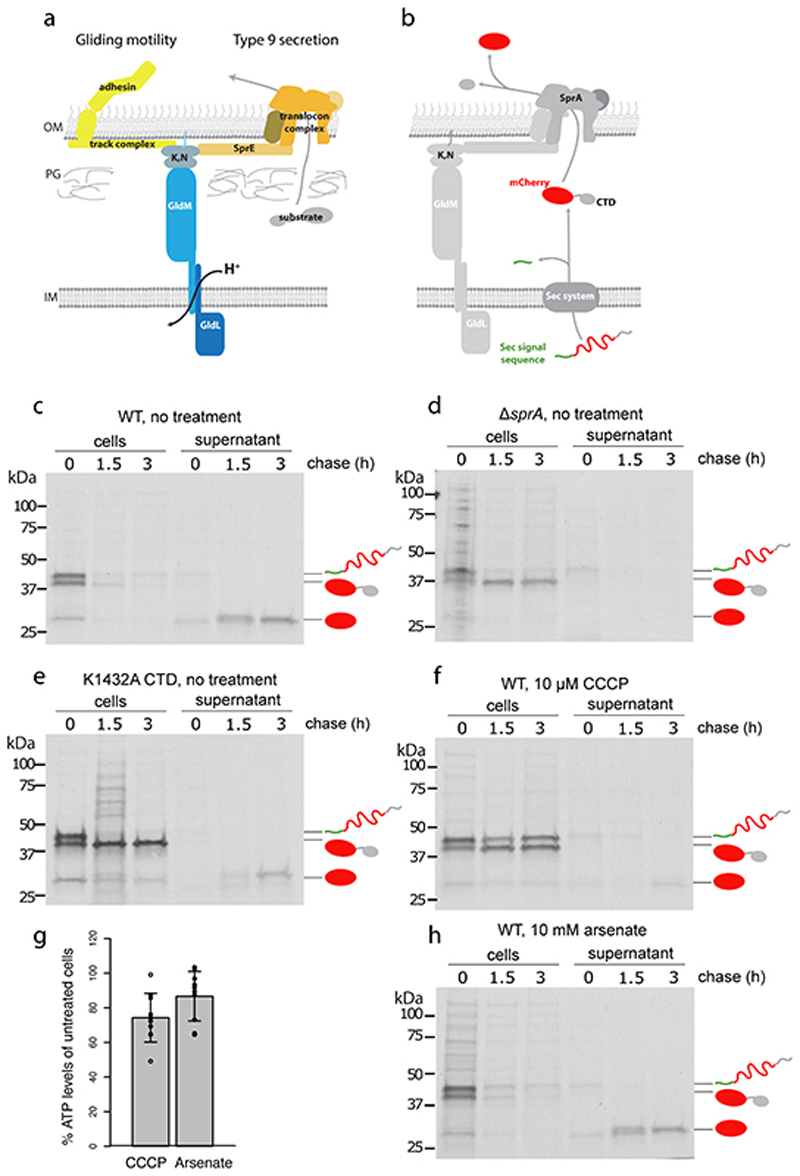
Fig. 1. Both gliding motility and Type 9 protein export require GldLM and the protonmotive force.
a, Schematic showing the proposed relationship of the GldLM complex to the major components of the gliding motility (left) and Type 9 protein transport (right) systems. OM, outer membrane; IM inner membrane; PG peptidoglycan; K, N indicates the complex of GldK and GldN proteins.
b, Schematic showing the two-stage export process of the model T9SS substrate protein SSRemA-mCherry-CTDRemA. CTD, C-terminal T9SS-targeting domain.
c-f,h, Pulse-chase analysis of the export of the SSRemA-mCherry-CTDRemA fusion protein from F. johnsoniae. Cells were labelled with [35S]methionine for 30 min, chased with cold methionine for 0 to 3 h (as indicated), and then separated into cell and supernatant (culture medium) fractions. Fusion protein was enriched by anti-mCherry immunoprecipitation and then analysed by SDS-PAGE and autoradiography. The successively processed forms of the fusion protein are marked using the cartoon representations from b. Trace amounts of free mCherry at t=0 h is background likely due to proteolytic clipping during sample workup. Similar data were obtained for two biological repeats. Export of the fusion protein in wild-type (WT) cells (panel c) was blocked by treatment with the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP, panel f) but not by the ATP synthase inhibitor arsenate (panel, h). Control experiments confirm that the observed export of the mCherry fusion requires the T9SS (ΔsprA strain Fl_00420, panel d) and a functional CTD (poorly-functional K1432A CTD19, panel e). Note, that transport of the fusion across the cytoplasmic membrane by the Sec apparatus is also inhibited by CCCP (panel f) in agreement with previous observations in Escherichia coli 21. The trace transport at 3 h observed in (f) is eliminated by increasing the CCCP concentration to 50μM CCCP. This concentration of protonophore was not used here as it has significant effects on cellular ATP levels. g, Measurements of whole cell ATP levels confirm that the effects of CCCP on protein transport are not an indirect effect of decreased cellular ATP. Bioluminescence measurements of ATP levels from treated cells were normalised to those of untreated cells from the same starting culture. CCCP and arsenate concentrations were as in panels f and h. Error bars represent 1 SD (10 biological repeats) about the mean. The datasets were tested for normality using the Shapiro-Wilk test and also inspected visually using Q-Q plots. The Bartlett test was used to establish that the variances of the two datasets were likely homogeneous. An independent two-tailed t-test indicated that the ATP levels in CCCP- and arsenate-treated cells were not significantly different (n.s.; t-value = -1.9619, df = 17.994, p-value = 0.06543>0.05) even though only CCCP treatment blocks T9SS protein export.
During the biogenesis of the gliding motility apparatus the adhesin proteins are trafficked to the cell surface by the Type IX secretion system (T9SS)13, 16(Fig. 1a). The T9SS is a Bacteroidetes-specific OM protein transporter. It which in the dental pathogen Porphyromonas gingivalis it is an essential virulence determinant.17. Unexpectedly, the gliding motility motor proteins GldL and GldM, are essential for Type 9 protein secretion13, 16 even in Bacteroidetes species, that do not glide16, such as P. ginigivalis where they are named PorL and PorMHere, we report molecular and structural analysis of the GldLM and PorLM complexes involved in gliding motility and Type 9 secretion..
Results
Type 9 secretion uses theprotonmotive force
In gliding motility the GldLM complex powers the movement of cell surface adhesins. The role of GldLM in T9SS is unclear. However, the known motor function of GldLM suggests that it might power the movement of substrates through the T9SS translocon. To test this hypothesis we assessed the energy requirements of the T9SS.
Type 9 substrates are first transported an across the IM using the Sec apparatus. A conserved C-terminal domain (CTD) then targets the protein to the T9SS translocon in the OM18, 19, 20 (Fig. 1a,b). A challenge in investigating the energetics of T9SS transport across the OM is that the preceding Sec-dependent stepuses both ATP and the PMF21. In order to disentangle the energy requirements of T9SS and Sec we used pulse-chase assays to follow the export of a model T9SS substrate protein19 from F. johnsoniae cells(Fig. 1b). Successive removal of the Sec signal sequence and of the CTD mark transport across the IM and OM, respectively (Fig. 1b,c). Control experiments confirmed that export of the substrate protein and CTD processing depend on the core T9SS translocon protein SprA (Fig. 1d) and on a functional CTD (Fig. 1e).
Addition of the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to collapse the PMF blocked transport of the periplasmic intermediate across the OM (Fig. 1f) which suggests that Type 9 transport is PMF-dependent. CCCP treatment was also associated with a small reduction in cellular ATP levels (Fig. 1g), raising the possibility that the protonophore was acting indirectly on transport by perturbing cytoplasmic ATP concentrations. To investigate this possibility we reduced the cytoplasmic ATP level using the ATP synthase inhibitor arsenate and found that a concentration of arsenate that alters cellular ATP levels to the same extent as CCCP (Fig. 1g) had no effect on export of the Type 9 substrate protein (Fig. 1h). This means that the effects of the protonophore on Type 9 transport are not mediated through alterations in ATP levels. We conclude that the PMF across the IMdrives Type 9 protein secretion across the OM, explaining why the PMF-driven GldLM motor is required for Type 9 protein transport. We note that the T9SS contains no other IM proteins that could utilise the PMF.
These data support a model in which the GldLM motor transduces the IM PMF to power both gliding motility and Type 9 protein export. In P. gingivalis the distal end of the GldM homologue PorM (domain pair 4) contacts a large ring structure on the inner face of the outer membrane composed of the proteins PorK and PorN14, 15, 22, 23. Recent data suggest that these PorKN rings are connected to the T9SS translocon via the T9SS-specific lipoprotein PorW23. In F. johnsoniae the PorK, PorN, and PorW homologues GldK, GldN, and SprE (are essential for Type 9 protein transport13, 24 and may function like their P. gingivalis counterparts (Fig. 1a). Because GldK and GldN in F. johnsoniae are inferred to link the GldLM motor to the tracks on which the gliding adhesins run6 (Fig. 1a), it is likely that energy transfer from the GldLM motor to Type 9 protein transport and gliding motility bifurcates after the shared GldKN component (Fig. 1a).
The GldLM motor is an asymmetric 5:2 subunit complex
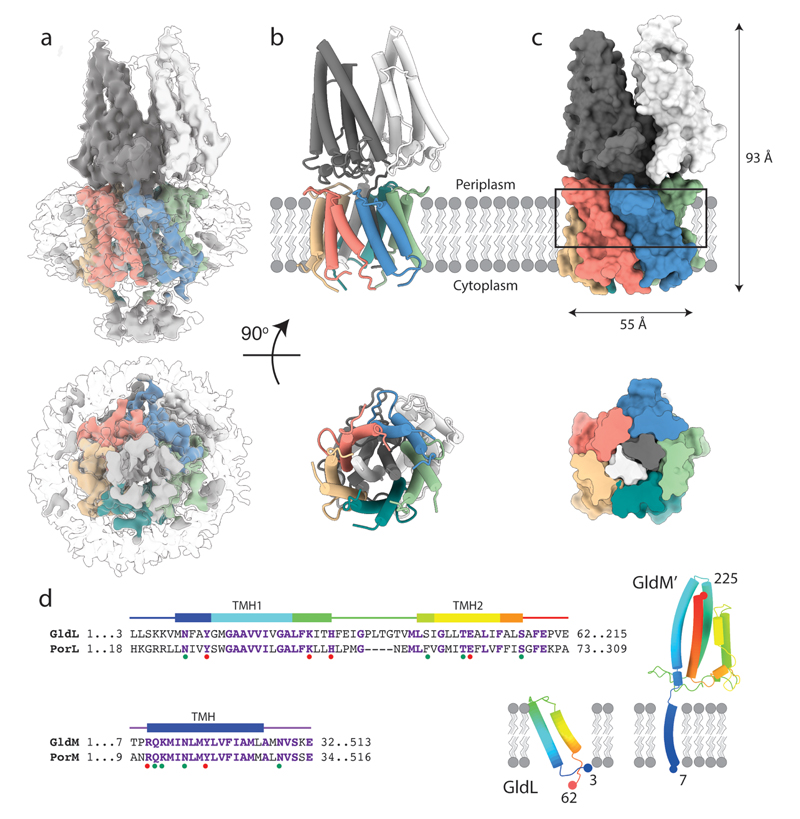
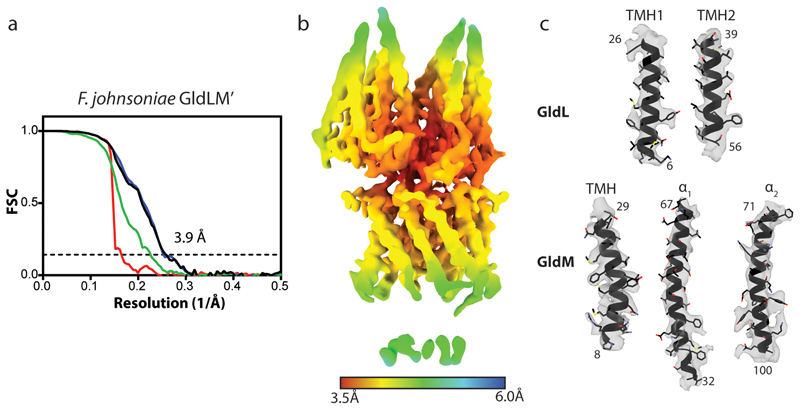
We screened GldLM homologues from multiple organisms for heterologous expression in Escherichia coli and suitability for single particle cryoEM. Intact GldLM motor complexes proved unsuitable for high resolution structure determination by cryoEM because the anisotropic shape of the molecule led to a poor distribution of views. However, imaging a F. johnsoniae GldLM complex in which the GldM protein was C-terminally truncated after the first folded periplasmic domain (hereafter referred to as GldLM′) allowed the calculation of an EM volume at 3.9 Å resolution and de novo building of a full atomic model (Fig. 2, Extended Data Figs. 1–5). The structure of GldLM′ is among the smallest protein complexes resolved by cryoEM, with less than 80kDa of ordered protein in the complex. The transmembrane domain of the complex is fully defined in the model (Fig. 2a-c). GldL is ordered between residue 3 and 62 (Fig. 2d) with 153 residues of the cytoplasmic C-terminus disordered. The ordered portion of GldL forms a pair of TMHs arranged at ~25° to the membrane normal with a well-structured loop joining them on the periplasmic face of the membrane (Fig. 2d). Almost the entirety of the truncated GldM construct could be modelled (residue 7 to 225, Fig. 2d). The single-pass TMH is resolved and is topped by the periplasmically-located helical domain (0.7Å RMSD to the previously reported X-ray structure of the domain, pdb 6ey415) (Fig. 2d). The overall complex is formed from five copies of GldL and two copies of GldM (Fig. 2b,c). The GldL TMH pairs form a distorted pentameric cage enclosing the two copies of the GldM TMH, and shielding them from the lipid bilayer (Fig. 2a-c). The periplasmic domains of the GldM subunits pack against the top of the ring of GldL TMHs (Fig. 2a-c). Only a single conformation of the GldLM′ complex is seen in our current data.
Fig. 2. Structure of the F. johnsoniae GldLM′ complex.
a, 3D cryoEM reconstruction of the detergent-solubilized GldLM′ complex at high (coloured by protein chain) and low (semi-transparent) contours. The detergent micelle is seen at the low contour level and is used to estimate the location of the membrane bilayer shown in b,c.
b,c, Cartoon (b) and space-filling (c) representations of the GldLM′ complex. The GldL subunits are coloured salmon, blue, green, teal, and yellow, and the GldM subunits are coloured white and dark grey. In c the lower panel shows a slab through the region indicated by the box in the upper panel. In b,c the lower panels show the complex viewed from the cytoplasm. Approximate dimensions of the complex are indicated.
d, (Left) Sequence alignment of the transmembrane regions of F. johnsoniae and P. gingivalis gliding motility/T9SS motor protein homologues. Secondary structure elements seen in the GldLM′ structure are shown above the alignments. Dots below the alignment indicate positions substituted in the functional studies shown in Fig. 4. The dots are coloured according to the whether the substitutions affect (red) or do not affect (green) gliding motility and/or Type 9 protein secretion. (Right) Structures of individual GldL and GldM′ subunits extracted from the complex. Numbers give the first and last residues of the subunits seen in the structure.
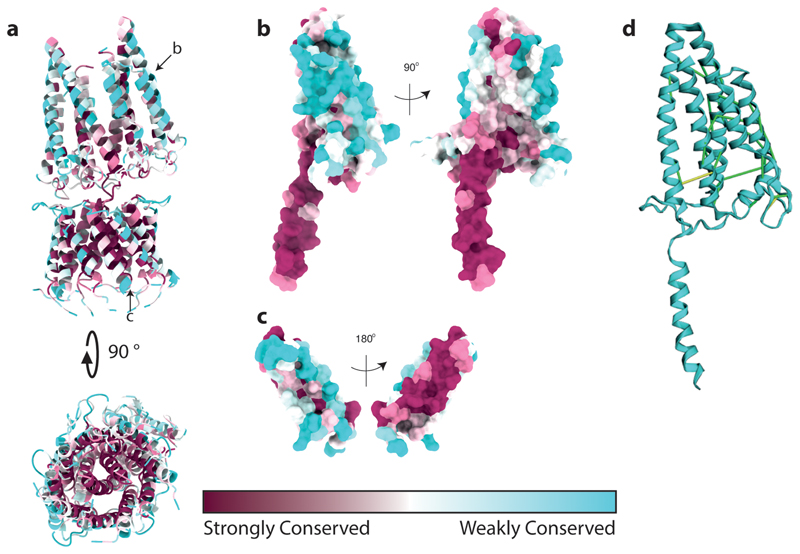
The overall structure of the GldLM′ complex is asymmetric. In the plane of the membrane this is due to the stoichiometry mismatch of the two types of subunit in the transmembrane helix bundle. In addition, the periplasmic domains of the two GldM chains adopt different tilts relative to the top of their TMH anchor (Fig. 2b,c). The result is that in the membrane domain, and at the periplasmic subunit interface, each of the five copies of GldL make different contacts with GldM (Extended Data Fig. 6). In the GldLM′ complex the TMHs are closely packed (Fig. 2c), implying that conformational change in the TMH of one subunit of the complex will only be possible if there are concerted conformational changes in the other subunits. As expected, the surfaces buried between the subunits of the GldLM′ complex are highly conserved and the exposed surfaces highly variable (Extended Data Fig. 7).
Orientation of the GldM periplasmic domains
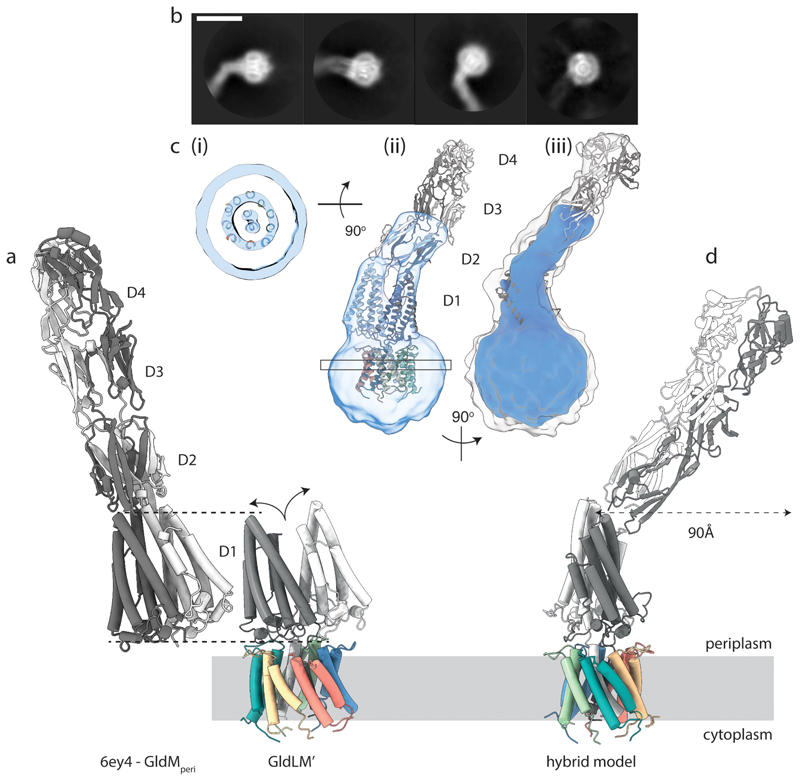
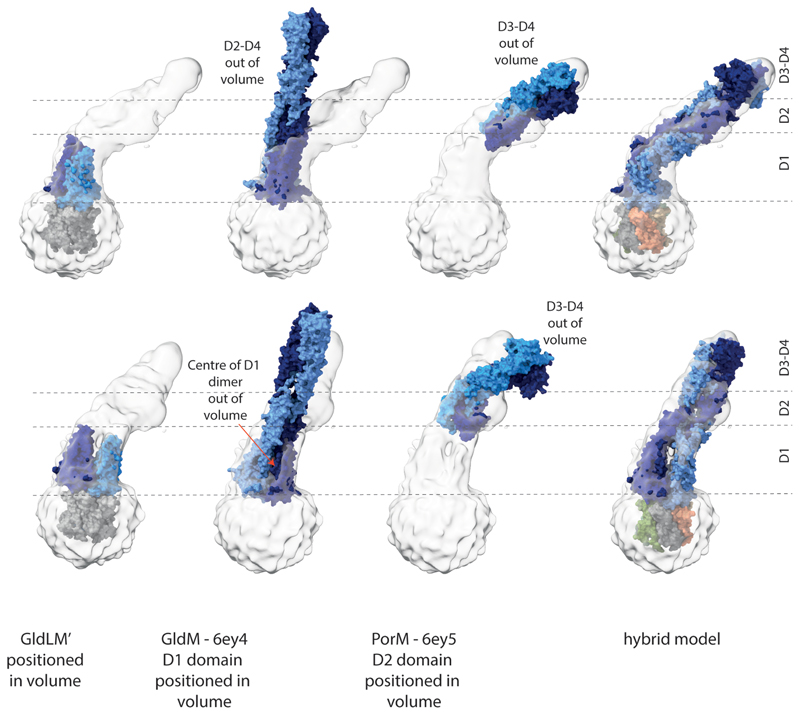
An overlay of the first periplasmic domains of GldM in our cryoEM structure of the GldLM′ complex with the equivalent domains in the previously reported crystal structures of the dimeric GldM periplasmic region15 reveals that the two copies of the domain are splayed apart in the cryoEM structure compared with the crystal structure (Fig. 3a). Splaying may be due in part to packing of the GldM TMHs in the GldL cage resulting in the two GldM helices being displaced vertically with respect to each other, thereby precluding formation of the dimer present in the crystal structure. The splaying of these domains is likely also a result of the interactions they make with the tops of the GldL subunits. The splayed arrangement of the two copies of the domain would be consistent with the lack of evidence from co-variance or sequence conservation for a strong dimerisation interface between the first periplasmic domains (Extended Data Fig. 7) and with the observation that the equivalent domain from the P. gingivalis homologue PorM crystalizes as a monomer15. We were, nevertheless, concerned that this difference between the cryoEM and crystal structures might have arisen due to truncation of the periplasmic region of GldM. We tried to resolve this issue by examining a lower resolution cryoEM volume derived from images of P. gingivalis PorLM. The data resolved the two TMH helices of PorM but not thesecondary structure elements in the periplasmic domains. Nevertheless, we were able to position our atomic model for the F. johnsoniae GldLM′ complex in the lower resolution P. gingivalis PorLM EM volume (Fig. 3b,c). Interpretation of the lower resolution volume in this way shows that the first periplasmic domains are also splayed apart in full length PorM (Fig. 3c, Extended Data Fig. 8). This splaying of the first domains is also consistent with a recent crystal structure of the entire periplasmic portion of PorM25 Density for the D2 domains is visible in the PorLM EM volume and is compatible with previously determined crystal structures15. PorM domains 3 and 4 were only present in the PorLM complex volume at very low contour levels, Nevertheless, it is apparent that the approximately linear arrangement of domains 2 to 4 seen in the crystal structure of F. johnsoniae GldM is a better model for this region in the intact PorLM complex than the bent arrangement seen in the crystal structure of the PorM domains 2 to 4 construct noted above (Fig. 3c, Extended Data Fig. 8).
Fig. 3. Structure of the complete GldLM motor complex.
a, Comparison of the crystal structure of the periplasmic domains of GldM (PDB 6ey4, GldMperi) with the cryoEM structure of the periplasmically-truncated GldLM′ complex. The structures are displayed with the first periplasmic domain of the dark grey GldM chain aligned (dotted lines) and demonstrate the splaying (curved arrows) of the GldM chains in the context of the truncated complex. The GldL subunits are individually coloured as in Fig. 2a-c.
b, Representative 2D classes from micrographs of the full-length P. gingivalis motor (PorLM) showing detail in the transmembrane region (all 4 panels), a large bend between the first two periplasmic domains of PorM (left three panels), and splaying of the first periplasmic domains of the two copies of PorM (second panel). Scale bar 100 Å.
c, Different views of the 3D cryoEM volume of the P. gingivalis PorLM motor at a high (blue) and low (white in iii) contour level. (i) The PorM TMHs can be resolved allowing confident placement of the F. johnsoniae GldLM′ model in the volume. (ii) Splaying of the PorM first periplasmic domain pair is seen as in the F. johnsoniae GldLM′ complex. (iii) The extension to the volume seen at a low contour level matches well to the length and shape of the dimeric GldM periplasmic domains 2-4 seen in the crystal structure. See also Extended Data Fig. 8.
d, Composite model for the GldLM complex produced by combining the cryoEM structure of the GldLM′ complex with the crystallographic structures of the GldM periplasmic domains D2-D4. The bend between periplasmic domains 1 and 2 seen in the low resolution cryoEM volume would allow the tip of a rotating GldM dimer to define a large track at the outer membrane. The subunits are coloured as in panel a.
We combined our structural data with existing crystallographic data to produce a composite model for the GldLM complex in which the rod-like periplasmic arm of PorM/GldM projects into the periplasm at an appreciable angle away from the membrane normal (Fig. 3d).
Functionally important residues in the GldLM transmembrane domain
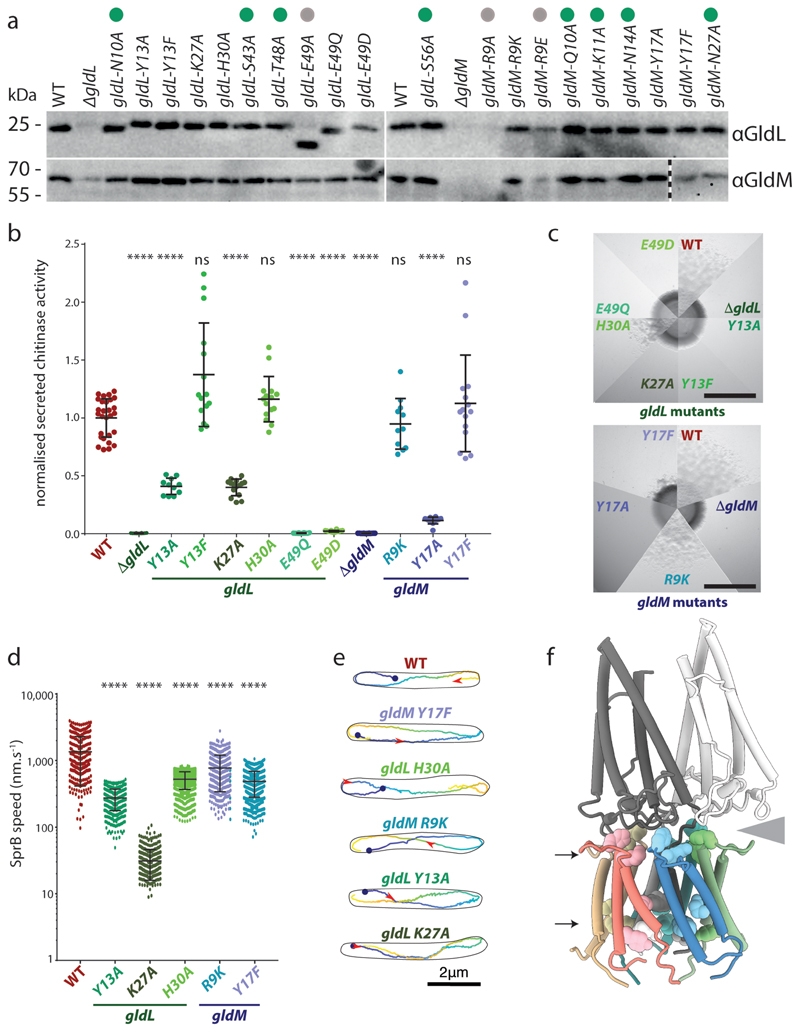
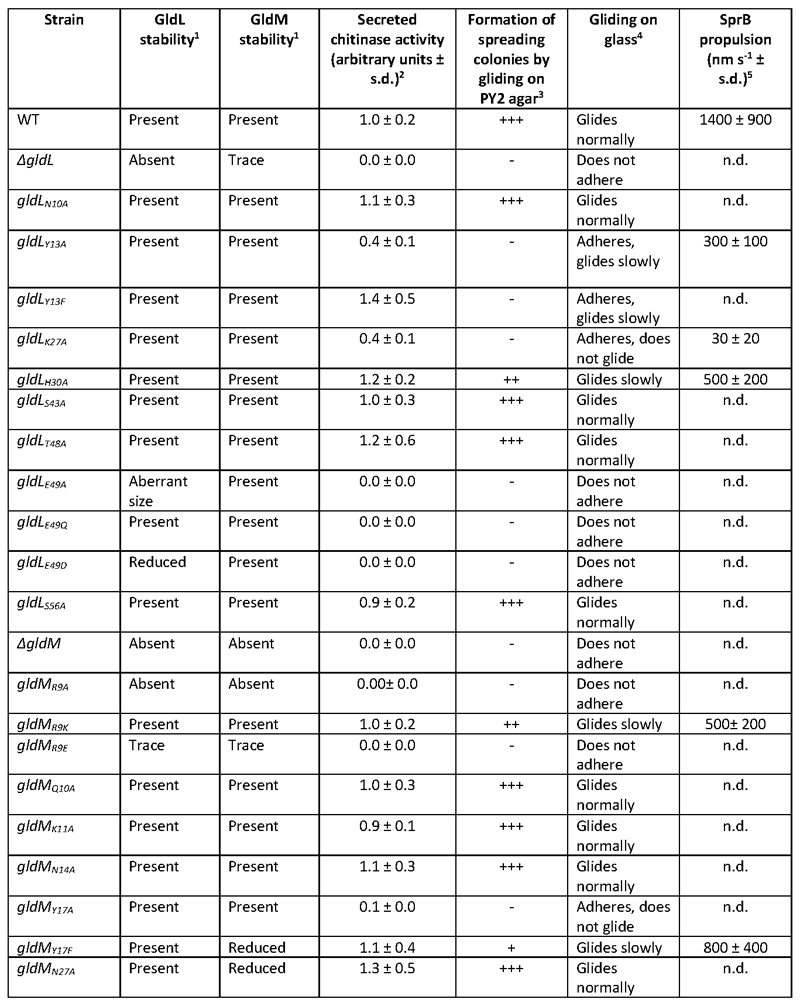
To identify residues within GldLM that might be important in transducing proton flow to mechanical motion we individually substituted all polar residues in the transmembrane domain (identified in Fig. 2d) by mutagenesis of the chromosomal gldL or gldM genes of F. johnsoniae. The resulting variant proteins were stably expressed with the exceptions of GldLE49A (proteolytically clipped) and GldMR9A and GldMR9E (that likely remove a topogenic signal for membrane insertion26) which were not analysed further. GldL and GldM are unstable when expressed individually (Fig. 4a), so any substitutions that disrupt GldLM complex formation will likely destabilise both proteins. Based on this criterion, all remaining variants were able to form GldLM complexes.
Fig. 4. Functional analysis of protonatable residues in the transmembrane domain of GldLM.
a-e, Phenotypic comparisons between F. johnsoniae wild type (WT) cells and gldLM single codon mutants.
a, Whole cell immunoblotting of the indicated strains for the presence of GldL and GldM. Variants that were not taken forward to phenotypic analysis due to aberrant expression are labelled with a grey dot. Variants that were subsequently found (Extended Data Fig. 9) to be have Type 9 protein transport and gliding motility phenotypes that were indistinguishable from the WT are labelled with a green dot and are omitted from panels b-e. Similar data were obtained for three independent preparations. The vertical dotted line indicates where an irrelevant lane was excised from the bottom right hand blot (original blot in source data file gels.pdf).
b, Type 9 protein transport activity assessed from the extracellular levels of the T9SS-secreted enzyme chitinase. 5ml cultures were grown overnight and diluted to the same OD, 10 μl was then assayed for activity. Error bars show 1 standard deviation about the mean. Significance of differences relative to WT were assessed with one-way Annova followed by Dunn’s multiple comparisons test (non-normally distributed) or Dunnet’s multiple comparison test. **** = p < 0.0001, ns = not significant. At least 3 technical replicates of at least 3 biological repeats were performed for each strain.
c, Gliding motility assessed by spreading on agar plates. Scale bar 5 mm.
d,e, Cell surface movement of single fluorophore-labelled SprB adhesin molecules. d, Plot showing the speed of helically moving adhesins in a WT (321 tracks), gldL(Y13A) (499 tracks), gldL(K27A) (369 tracks), gldL(H30A) (2,259 tracks), gldM(R9K) (429 tracks), or gldM(Y17F) (1519 tracks) background. For each strain the tracks are pooled data from the analysis of 3 independent cultures. Error bars show 1 standard deviation about the mean. Significance of differences relative to the WT were assessed with one-way Annova followed by Dunn’s multiple comparisons test (non-normally distributed) or Dunnet’s multiple comparison test. **** = p < 0.0001. e, Examples of adhesin helical tracks extracted from Supplemental Videos 2–7 and coloured from blue (start) to yellow (end). The red arrowhead marks the position along the track that the adhesin has reached at 3.5s after the start point (blue dot).
f, Protonatable residues in the GldLM transmembrane domain that exhibit reduced function when substituted. The residues are show in space-filling representation on a cartoon model of GldLM′ and fall in two bands (indicated with arrows) located at the periplasmic GldL-GldM interface and close to the cytoplasmic end of the TMH bundle. The grey wedge indicates the space between one of the GldM subunits and the top of the GldL ring.
The effects of the GldLM substitutions on Type 9 protein transport activity were assessed by determining the amount of the T9SS substrate chitinase secreted by cells (Fig. 4b). Transport activity was completely blocked in the gldL E49Q and gldL E49D strains, reduced in the gldL Y13A, gldL K27A, and gldM Y17A mutants, but not detectably compromised by the other substitutions
The effects of the gldLM mutations on gliding motility were assessed by monitoring the ability of F. johnsoniae colonies to spread on agar plates (Fig. 4c) and by microscopy of cells gliding on glass (Extended Data Fig. 9 and Supplemental Video 1). All mutants that had Type 9 protein transport defects were partially or completely defective in gliding. However, additional mutants that were not defective for protein secretion (gldL Y13F, gldL H30A, gldM R9K, and gldM Y17F - though the reduced levels of protein in the latter may explain the motility phenotype) were partially defective in gliding motility. The wider range of mutants that are affected in gliding relative to T9SS function suggests that motility requires a higher level of motor function than protein transport.
The adhesin proteins required for gliding are exported by the T9SS. Consequently,we found that gldLM mutants defective for Type 9 protein transport (gldL E49Q and gldL E49D) cannot adhere to glass slides or glide because they do not have surface adhesins (Extended Data Table 3). This raises the possibility that the gliding defects in other gldLM mutants might be due to effects of mutations on adhesin export by the T9SS. To determine the effects of the gldLM mutations on gliding motor function independent of their effects on Type 9 transport we directly measured adhesin flow. We tracked the movement of individual adhesin molecules using a HaloTag domain fused to the N-terminal end of the major gliding adhesin SprB and labelled with a photoactivatable fluorophore. In the wild-type strain the fluorescently labelled SprB molecules moved in helical trajectories along the cell body with an average velocity of ~1.5μm s-1, consistent with previous studies7–10 (Fig. 4d,e and Supplemental Video 2). In gldLM strains that were competent for secretion but had gliding defects, SprB still moved in helical patterns but the speed of adhesin movement was much slower than in the wild-type (Fig. 4d,e and Supplemental Videos 3–7). Thus, the GldLM substitutions in these strains directly affect the mechanical force driving adhesin movement..
The results of our amino acid substitution experiments (Fig. 4a-e, Extended Data Table 3) show that functionally important protonatable amino acids are clustered in the membrane core of the GldLM complex (GldLY13, GldLE49, GldMR9, and GldMY17) and at the periplasmic GldL/GldM interface (GldLK27 and GldLH30) (Fig. 4f). The transmembrane helix bundle is tightly packed with no obvious water-filled cavities (Fig. 2c) so the route of proton movement between these two layers of residues cannot be deduced from our structural model.
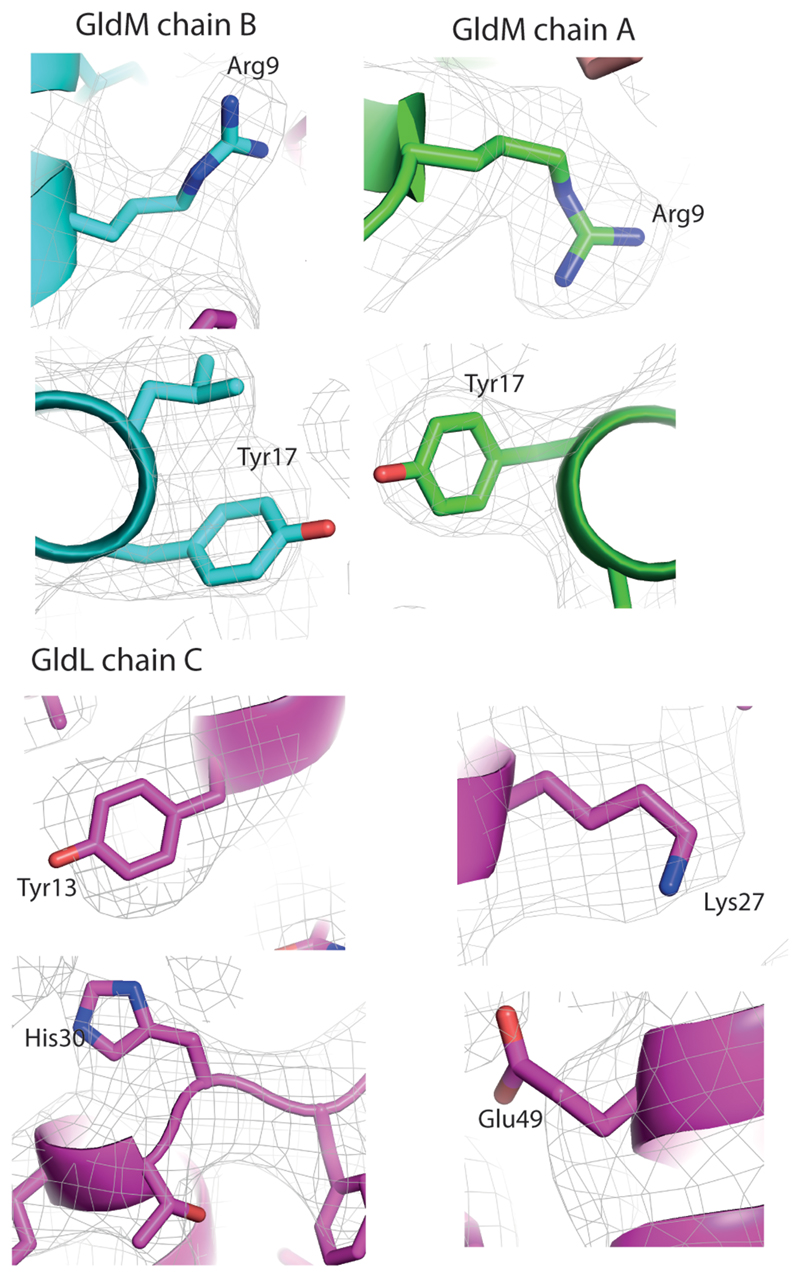
The side chains of most of the functionally important protonatable amino acids are large (GldLY13, GldLK27, GldLH30, GldMR9, and GldMY17) and their position can be modelled from the EM density even at the moderate resolution of the structure (Extended Data Figs 4, 10). The exception is GldLE49 for which the full side chain EM density is not seen, most likely because glutamic acid residues are rapidly decarboxylated in the electron beam.
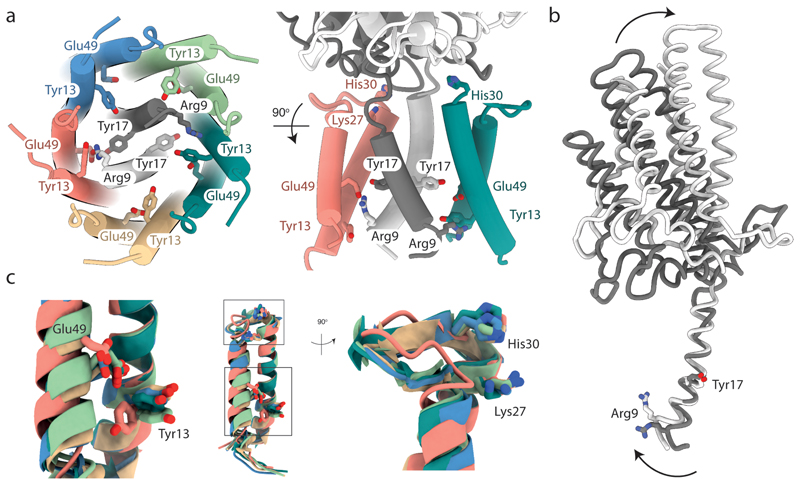
The symmetry mismatch in the GldLM′ complex means that within the membrane core the functionally important protonatable amino acids are in a different environment in each copy of the subunits. Most notably this positions GldMR9 in chain A (coloured white in the figures) to form a salt bridge with GldLE49 in chain C (coloured salmon in the figures) (Fig. 5a). This interaction can be confidently inferred even in the absence of EM density for the glutamate side chain due to the observed ordering and orientation of the arginine side chain, as well as the distortion of the N-terminus of the GldM TMH caused by the interaction (Fig. 5b, and discussed further below). The necessity to form this salt bridge would explain why GldLE49 cannot be functionally substituted with the shorter side chain of aspartate (GldLE49D variant, Fig. 4). The putative ion pair is bracketed by the functionally important GldMY17 from the other GldM molecule (chain B, coloured grey in the figures) and GldLY13 from the same chain C as the ion-paired glutamate (Fig. 5a). Due to the asymmetric nature of the transmembrane domain, GldMR9 in the other copy of GldM (chain B) is no longer positioned to interact with GldLE49 on the nearest GldL subunit, and so no ion pair is formed between these residues (Fig. 5a).
Fig. 5. The structural asymmetry of the GldLM′ complex leads to important differences in residue environments between chains.
The side chains of functionally important resides within the transmembrane domain of GldLM are shown in stick representation with oxygen atoms red and nitrogen atoms blue. Note that the side chains of Glu49 are absent from the EM density due to decarboxylation. The side chain of Glu49 in the salmon chain (chain C) is modelled on the basis that it is inferred to form an ion pair with Arg9 in the white GldM chain (chain A). The modelled side chain positions of Glu49 in the other GldL chains are arbitrary. Proteins chains are coloured as in Fig. 2 a-c.
a, The side chains of functionally important resides. For clarity, only two GldL subunits are shown in the right hand view.
b, Overlay of the two copies of GldM′ by superimposition of the TMH. Arg9 in the white chain (chain A) is involved in the ion pair with GldL chain C Glu49 shown in a. Arrows indicate the conformational changes between the two GldLM chains.
c, Superimposition of the five copies of GldL by overlaying TMH2. The boxes in the central panel indicating the positions of the two regions magnified on either side.
Structural asymmetry in the GldLM complex
An overlay of the two GldM′ subunits shows that the cytoplasmic end of the TMH must be bent in order for GldMR9 to ion pair with GldLE49 (Fig. 5b). In the putatively ion-paired copy of GldM the angle between the TMH and the periplasmic domain is less acute than in the other GldM chain, and so the periplasmic domain is less tilted relative to the GldL ring (Figs. 4f and 5b). The periplasmic domain of the GldM subunit that is not participating in the putative ion pair within the transmembrane domain tilts to make extensive interactions with the top of the ion-paired GldL subunit (Figs. 4f and 5b).
An overlay of the five copies of the GldL protein found in the GldLM′ complex reveals that the helix in which GldLE49 ion pairs with GldMR9 is shifted compared to the equivalent helix in the other copies of GldL (Fig. 5c). This helix movement is accompanied by a reorientation of GldLY13 located on the second TMH of the same chain and a remodelling of the periplasmic loop that is packed against the GldM periplasmic domain (Fig. 5c). In this context it is notable that substitution of two invariant residues (GldLK27 and GldLH30) within the GldL periplasmic loop that appear to mediate interactions with the GldM periplasmic domain significantly impair GldLM function (Figs. 4b,d,e and 5c). This implies that there are contacts in the transmembrane region and between the subunits in the periplasm that are required for proper coupling and full activity.
Discussion
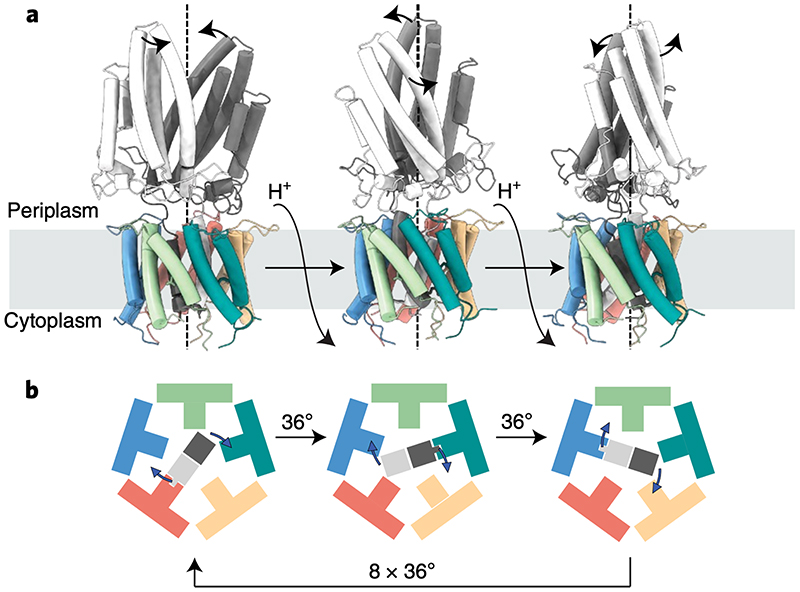
Sequence and structural conservation between GldLM and PorLM suggest all T9SS motors will share the same mechanism of action. Hereafter we use GldLM to denote the homologous complexes across all Bacteriodetes. Proton flow through the GldLM complex needs to move the GldM periplasmic arm in order to drive gliding motility and protein transport at the OM. The structure of the GldLM complex is asymmetric in the IM due to the symmetry mismatch created by placement of two GldM chains in the center of five GldL subunits. The result of this mismatch is that the two GldM chains cannot make identical interactions with the GldL ring. Specifically, only one of the GldM chains can form an ion pairing interaction to a GldL subunit using the mechanistically essential residues GldMR9 and GldLE49. In order for the GldM dimer to move relative to the GldL ring this ion pair must be broken. We propose that this is achieved by proton uptake from the periplasmic side of the membrane thereby linking transmembrane proton flow to mechanical change in the complex. We then envisage that the GldM dimer rotates by 36° (i.e. one tenth of a full circle) to bring the second GldM monomer into the correct register to ion pair with a new GldL subunit with concomitant proton loss to the cytoplasmic side of the IM. Iteration of this motion ten times would cause the GldM dimer to rotate fully back to its original position with the identity of the ion-paired GldM subunit alternating at each step (Fig. 6). Thus, the caging of GldM within GldL combined with a lack of shared symmetry between protein chains leads to the hypothesis that GldLM is a rotary motor (see Supplemental Video 8 for ananimation to illustrate the approximate molecular changes that would occur in the GldLM′ complex on rotation of the GldM dimer within GldL). We have assumed in our model that GldM rotates unidirectionally. We cannot exclude the possibility that GldM rotates in opposite directions in successive steps resulting in an oscillatory rather than rotary motion of the periplasmic arm with one direction representing a mechanistically important ‘power-stroke’ and the other a ‘slip’ to restore the starting state. Such a motion would require the flow of twice as many protons per power stroke relative to the unidirectional rotation model.
Fig. 6. Model for the mechanism of coupling proton flow to GldM rotation in the GldLM motor.
a, GldLM′ is shown in a cartoon representation, coloured as in earlier Figures. Panels represent the model after successive 36° rotations of the GldM′ subunits driven by net flow of a single proton down the transmembrane proton electrochemical gradient.
b, The transmembrane domain of the motor is shown schematically viewed from the cytoplasm and coloured as in a. The central GldM dimer rotates within the GldL pentamer in 36° steps driven by proton flow. 10 steps in total are required to bring the GldM dimer back to the starting position. At each step a salt bridge is present between one GldM chain and one GldL chain. This binding interaction is represented by the fit of the GldM chain into a GldM-binding box on the GldL subunit. The direction of rotation shown is arbitrary.
Experimental support for the idea that protonation state changes at the GldMR9 – GldLE49 ion pair are crucial for motor function comes from the observation that replacement of the glutamic acid side chain of GldLE49 by the non-protonatable isomorph glutamine abolishes motor function (GldLE49Q variant in Fig. 4). GldMY17 and GldLY13 are positioned to act as potential proton donors and acceptors, respectively, to the ion pair (Fig. 5a). Consistent with this idea, the substitution data indicate that protonation of these side chains is functionally important (GldLY13F and GldMY17F variants, Fig. 4). However, the substitution data also show the importance of the aromatic nature of GldLY13 and GldMY17, since GldLM functions are more severely compromised when they are replaced by alanine than when they are substituted by phenylalanine (compare GldLY13A with GldLY13F and GldMY17A with GldMY17F variants, Fig. 4).
The close packing of subunits in the transmembrane bundle and the contacts between the transmembrane bundle and the base of the periplasmic domains in the GldLM′ structure indicates that the motions of the motor are likely to be coupled through the complex (Figs 2a-c and 4f). The Co-operative movements of the two GldM proteins will be reinforced because it is the periplasmic domain of the GldM subunit not involved in the ion pair that contacts the periplasmic loops of the ion-paired GldL subunit (Figs. 4f, 5a).
Structural analysis of the intact P. gingivalis PorLM complex demonstrates that the periplasmic region of PorM is bent outwards at the junction between the first and second periplasmic domains (Fig. 3b,c). This bending means that rotation of PorM would cause the tip of the periplasmic arm to move within a circle of around 200 Å in diameter at the outer membrane (Fig. 3d). Although very wide, this circle is still considerably smaller than the 500 Å diameter of the PorKN ring with which PorM interacts15, 22, 23 (Fig. 1a). It is unlikely, therefore, that the orbit of PorM is matched to the circumference of PorKN. Within the periplasm the PorM arm must cross the cell wall. For the cell wall not to impede rotation of the arm, the arm would need to sit in a gap in the peptidoglycan of close to 200 Å in diameter. Since this is larger than the normal peptidoglycan pore size27, 28 it is likely that the cell wall is remodelled to accommodate the PorLM motor as occurs with other large trans-envelope proteins29, 30. Similar considerations are likely to apply to the GldLM motor in gliding bacteria.
In order for movement of the distal end of the GldM arm to drive processes at the outer membrane, the GldL ring needs to remain stationary in the inner membrane to act as a stator to the rotating GldM protein. Because GldL is embedded in a fluid lipid bilayer, this stator function must be achieved by attaching GldL to a stationary cellular component. Many other bacterial machines use the cell wall as an anchor point. However, GldL cannot be directly immobilized in this way since GldM covers the periplasm-exposed parts of GldL. No other conserved IM proteins that could indirectly link GldL to the cell wall have been identified in the T9SS and gliding motility systems. Instead, it is most likely that GldL is held in position through interactions between the currently unvisualized cytoplasmic tail and a relatively static large cytoplasmic structure such as a cytoskeletal element or the chromosome. In P. gingivalis the cytoplasmic tail of the GldL homologue PorL binds the chemotaxis response regulator protein homologue PorX31 which raises the possibility that the activity of the motor protein could be regulated by modulating the anchoring of the PorL/GldL subunit.
Comparison of GldLM with the other classes of ion-driven motors reveal that the gliding motility/T9SS motor shares no structural or mechanistic properties with ATP synthases, but that the arrangement of ten GldL TMHs around two central GldM TMHs is similar to the structural organization of the transmembrane core of MotAB motors of the flagellar stator class5, 32 despite their lack of sequence similarity.
Importantly, the flagellar stator units are also rotary motors5. The overall structural similarity between the transmembrane domains of GldLM and MotAB suggests that the fundamental rotary mechanism is the same in the two classes of motor. Given the lack of sequence similarity between GldLM and MotAB it is not possible to determine whether the two classes of motor arose by divergence from a common ancestor, or convergent evolution to the smallest structural framework with which it is possible to build an asymmetric rotor mechanism.
In other respects the gliding motility/T9SS motor and flagellar stator units are different. In the gliding motility/T9SS motor the central units (GldM) form the rotor and the surrounding subunits (GldL) are stationary, but in the flagellar stator units the peripheral subunits (MotA) rotate around the fixed central subunits (MotB). In the gliding motility/T9SS motor work is transduced to the periplasmic side of the membrane. By contrast, the flagellar stator units transfer energy to cytoplasmic components. Additionally, the details of the ion coupling steps are likely to differ between the two motor classes given that the flagellar stator family does not form an ion pair and the crucial protonatable aspartate residue in the stator motor is on the central (MotB) rather than surrounding (MotA) subunits.
In conclusion, the structure of the GldLM complex reveals the architecture of a third class of ion-driven molecular motors and provides a molecular basis for understanding how the gliding motility/T9SS motor transduces the energy of the inner membrane PMF across the bacterial cell envelope.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Bacterial strains and growth conditions
All strains and plasmids used in this work are listed in Tables S1 and S2. F. johnsoniae was routinely grown in Casitone Yeast Extract (CYE) medium33 at 30 °C with shaking. To assess motility on glass and for chitinase secretion measurements cells were grown in Motility Medium34 (MM). PY2 medium35 was used to assess motility on agar plates. E. coli cells were routinely grown in Luria Bertani (LB) medium at 37 °C with shaking. When required, antibiotics were added at the following concentrations: erythromycin, 100 μg ml-1; spectinomycin, 100 μg ml-1; ampicillin, 100 μg ml-1; kanamycin, 30 μg ml-1.
Genetic constructs
All primers used in this work are described in Table S3 All plasmid constructs were verified by sequencing.
A vector directing the co-expression of F. johnsoniae GldL and a C-terminally truncated and Twin-Strep tagged GldM under the control of a rhamnose-inducible promoter was produced as follows. The chromosomal region encoding GldL and the first 225 amino acids of GldM was amplified using primers RHJ164 and RHJ165. The plasmid pT1236 (map shown in Supplementary Fig. 1) was linearized by amplification with primers RHJ162 and RHJ163. The two fragments were assembled by Gibson cloning to yield plasmid pRHJ007. A vector for expression of F. johnsoniae GldL and the first 232 amino acids of GldM was produced similarly, using primer pair RHJ166 & RHJ167 to create pRHJ008.
A vector directing the co-expression of P. gingivalis ATCC 33277 PorL and PorM in E. coli was produced as follows. pWALDO-sfGFP37 was digested with BamHI and HindIII and the resulting fragment coding for the TEV cleavage site, superfolder GFP, and a His8 tag was ligated into the corresponding sites in the first multiple cloning site of pCDFDuet-1 (Novagen), yielding plasmid pRHJ001. A C-terminal Twin-Strep tag-coding sequence was added to the second multi-cloning site of pRHJ001 by Q5 site-directed mutagenesis (New England Biolabs) using primers RHJ025 and RHJ026, giving plasmid pRHJ002. The NcoI site in the superfolder GFP-coding sequence was removed by QuikChange mutagenesis (Agilent) using primers RHJ046 and RHJ047, yielding plasmid pRHJ003. pRHJ003 was digested with NcoI and XhoI and the resulting fragment ligated into the corresponding sites of pETDuet-1 to give pRHJ004. P. gingivalis porL was amplified from genomic DNA with primers RHJ051 and RHJ052. The resulting 0.9 kb fragment was inserted between the NcoI and BamHI sites of pRHJ004 to give pRHJ005. P. gingivalis porM was amplified with primers RHJ057 and RHJ058. The resulting 1.6 kb fragment was inserted between the NdeI and KpnI sites of pRHJ005 to give pRHJ006.
A suicide vector to produce an in-frame unmarked deletion of F. johnsoniae gldL was produced as follows. A 2.6 kbp region corresponding to the first 36 bp of gldL together with the directly upstream region was amplified with primers FL309 and FL310. A 2.5 kbp region corresponding to the final 36 bp of gldL together with the directly downstream region was amplified with primers FL311 and FL312. The vector pYT31338 was linearized by amplification with primers FL313 and FL314. These three fragments were then assembled by Gibson cloning to give plasmid pFL89.
A suicide vector to produce an in-frame unmarked deletion of F. johnsoniae gldM was produced as follows. A 2.7 kb region corresponding to the first 31 bp of gldM together with the directly upstream region was amplified with primers RHJ148 and RHJ160. A 2.5 kb region corresponding to the final 38 bp of gldM together with the directly downstream region was amplified using primers RHJ161 and RHJ147. The vector pGEM-T was linearized by amplification using primer RHJ144 and RHJ145. These three fragments were assembled by Gibson cloning to yield pRHJ010. The fragment containing the two F. johnsoniae chromosomal regions was then amplified from pRHJ010 with primers RHJ110 and RHJ113. The resulting 5.2 kb fragment was inserted between the BamHI and KpnI sites of pYT35438 to give plasmid pRHJ011.
The strategy to generate point mutations in gldL involved the use of an intermediate cloning vector that was produced as follows. A 5.3 kb region including the gldL gene sequence and surrounding chromosomal regions was amplified using primers RHJ146 and RHJ149. pGEM-T was linearized by amplification with primers RHJ144 and RHJ145. The two fragments were assembled by Gibson cloning to give pRHJ012. An intermediate cloning vector for generating mutations in gldM was produced in an analogous manner using primers RHJ147 and RHJ148 and yielding plasmid pRHJ013.
A suicide vector for the introduction of the gldL(N10A) point mutation was produced as follows. pRHJ012 was linearized by amplification with primers RHJ310 and RHJ311, which introduce the desired 2 bp point mutation in codon N10. The resulting fragment was re-circularised by in vivo assembly39, yielding plasmid pRHJ014. The fragment containing the mutated gldL sequence and adjacent regions was then amplified from pRHJ014 with primers RHJ341 and RHJ342. The vector pYT354 was linearized by digestion with BamHI and SalI. The two fragments were then assembled by in vivo Gibson cloning to give plasmid pRHJ036. Other point mutations were generated similarly, using pRHJ012 as a template for mutations in gldL and pRHJ013 as a template for mutations in gldM, using the primers described in Table S3.
A suicide vector for the construction of halotag-sprB strains (placing Twin-Strep-HaloTag directly after the Sec signal peptide of SprB, between residues Ser29 and Phe30) was produced by Gibson assembly of the following four fragments yielding plasmid pAK021: pYT313 linearized with primers AK41 and AK62; nucleotide 88 to 2647 of sprB amplified from the F. johnsoniae chromosome with primers AK59 and AK60; twinstrep-halotag amplified from plasmid pET21-ts-halo-RemA97CTD with primers AK37 and AK61; a genomic fragment extending from nucleotide 2543 of sprD through to nucleotide 87 of sprB was amplified from the F. johnsoniae chromosome with primers AK39 and AK40.
For pulse-chase experiments, a plasmid directing production of a tripartite fusion protein consisting of the F. johnsoniae RemA signal sequence, mCherry, and the RemA C-terminal domain under the control of the remA promoter was constructed as follows. A region encompassing 361 nucleotides immediately upstream of remA together with the first 150 nucleotides of remA was amplified from genomic DNA using primers PG001 and PG002, then cloned into the XbaI-SpeI restriction sites of pCP1133. Between the SpeI and SacI sites of the resulting plasmid was inserted the coding sequence for mCherry with no stop codon amplified from plasmid pRVCHYC-540 using primers PG003 and PG004. Finally, a region coding for the 97 C-terminal residues of RemA was amplified with primers PG005 and PG006 and inserted between the SacI and SalI sites to produce plasmid pCP-remAus-mch-CTD97remA. A plasmid coding for the fusion protein with a K1432A substitution in the RemA CTD was made by the QuikChange mutagenesis (Agilent) using primers PG007 and PG008.
Suicide and expression plasmids were introduced into F. johnsoniae strains by biparental mating using E. coli S17-141 as the donor strain, as previously described33. Point mutations in gldL were introduced into the chromosome by using the ΔgldL strains Fl_082 or Ak_205 as the recipient. Point mutations in gldM were introduced into the chromosome by using the ΔgldM strains Rhj_006 or Ak_203 as the recipient. Erythromycin resistance was used to select cells with a chromosomally-integrated suicide plasmid. One of the resulting clones was grown overnight in CYE without antibiotic to allow for loss of the plasmid backbone, and then plated onto CYE agar containing 5 % sucrose. Sucrose-resistant colonies were screened by PCR for the presence of the desired chromosomal modification and then verified by sequencing.
Purification of PorLM and GldLM’ complexes
GldLM′ proteins were overproduced from plasmids pRHJ007 and pRHJ008 as follows. Colonies of BL21(DE3) cells carrying the appropriate plasmid were inoculated into 50 ml 2xYT medium and cultured at 37°C with shaking for 6-8 h. The cells were diluted to OD600 = 0.02 in TB supplemented with 0.2 % L-rhamnose and then grown at 37°C with shaking for 14 h. Cells were harvested by centrifugation at 5,000g for 15 min at 4 °C. Cells were washed once in Dulbecco A phosphate buffered saline (PBS) and stored at -20°C until further use.
The frozen cell pellet was resuspended in 3.3 ml per g of cells of Lysis Buffer (PBS supplemented with 1 mM EDTA, 30 μg ml-1 Dnase I, 400 μg ml-1 lysozyme, and 1 tablet per 100 ml SIGMAFAST™ protease inhibitor cocktail). The cells were then disrupted using an Emulsiflex homogeniser operated at 15,000 PSI. The resulting lysate was centrifuged at 27,000g for 30 min at 4 °C to remove cellular debris before the membrane fraction was recovered by centrifugation at 210,000g for 1 h at 4 °C. The membrane pellets were stored overnight at 4 °C before being resuspended in 8 ml Resuspension Buffer (PBS, 1 mM EDTA) per g membrane. 1 ml 10 % (w/v) lauryl maltose neopentyl glycol (LMNG; Anatrace) solution was added per g of membranes and the suspension was gently stirred at 4 °C for 2 h. Non-solubilised material was removed by centrifugation at 75,000g at 10 °C for 30 min. The resulting supernatant solution was passed through a 5 ml StrepTactin XT cartridge (IBA). The column was washed with 10 column volumes (CV) StrepTactin Wash Buffer (PBS, 1 mM EDTA, 0.02 % LMNG). Protein was eluted in 2 CV StrepTactin Elution Buffer (PBS, 0.02 % LMNG, 1 mM EDTA, 50 mM D-biotin). The elution fractions were concentrated to 500 μl using a 100 kDa MWCO Amicon Ultra–15 centrifugal filter, and injected on to a Superose 6 10/300 Increase GL size-exclusion chromatography column (GE Healthcare) equilibrated in 20 mM HEPES pH 7.5, 150 mM NaCl, 0.02 % LMNG. Protein purity was assessed by SDS-PAGE and GldLM′-containing fractions were pooled, concentrated using a GE Healthcare 100 kDa MWCO Vivaspin 500 concentrator, and stored at 4°C until use. Protein concentrations were determined spectrophotometrically using A280nm 1 = 1 mg ml-1.
Production of recombinant PorLM complexes was carried out as follows. An overnight culture of E. coli Mt56(DE3)42 carrying pRHJ006 in 2xYT medium43 was diluted 100-fold into fresh Terrific Broth medium44 containing 100 μg ml-1 ampicillin, and grown at 37°C with shaking to OD600 = 4.0. Protein production was then induced by addition of 0.1 mM IPTG and the cells cultured for 15 h at 24°C with shaking. Cells were harvested and proteins purified as described above for the GldLM′ complex, with the following differences. 1 mM EDTA was omitted from the StrepTactin Elution Buffer. Following elution from the StrepTactin XT cartridge, the eluted fraction was subsequently passed through a 5 ml Ni-NTA Superflow cartridge (Qiagen). The column was washed with 10 CV Ni-NTA Wash Buffer 1 (PBS, 0.02 % LMNG, 20 mM imidazole), followed by 10 CV Ni-NTA Wash Buffer 2 (PBS, 0.02 % LMNG, 40 mM imidazole). Protein was eluted in 1.2 CV Ni-NTA Elution Buffer (PBS, 0.02 % LMNG, 250 mM imidazole). 1mg of TEV-His6 protease was added per 10 mg eluted protein and the sample dialysed overnight against Dialysis Buffer (PBS, 0.02 % LMNG) in 3.5 kDa molecular weight cutoff (MWCO) SnakeSkin® dialysis tubing (Thermo Scientific) at 4°C. The dialysate was centrifuged at 3,220g for 15 min at 4°C. Imidazole from a 2M stock was added to the supernatant to achieve a final concentration of 20 mM. The supernatant was passed over a Ni-NTA column. The flowthrough containing PorLM was collected and then further processed by SEC chromatography as described for the GldLM′ purification above.
Cryo-EM sample preparation and imaging
Four microlitre aliquots of purified GldLM′ (A280nm = 0.4-0.5) or PorLM (A280nm = 0.2-0.5) complexes were applied onto glow-discharged holey carbon coated grids (Quantifoil 300 mesh, Au R1.2/1.3), adsorbed for 10 s, blotted for 2 s at 100% humidity at 4 °C and plunge frozen in liquid ethane using a Vitrobot Mark IV (FEI).
Data were collected in counting mode on a Titan Krios G3 (FEI) operating at 300 kV with a GIF energy filter (Gatan) and K2 Summit detector (Gatan) using a pixel size of 0.822 Å and a total dose of 48 e−/Å2 spread over 20 fractions.
Cryo-EM data processing
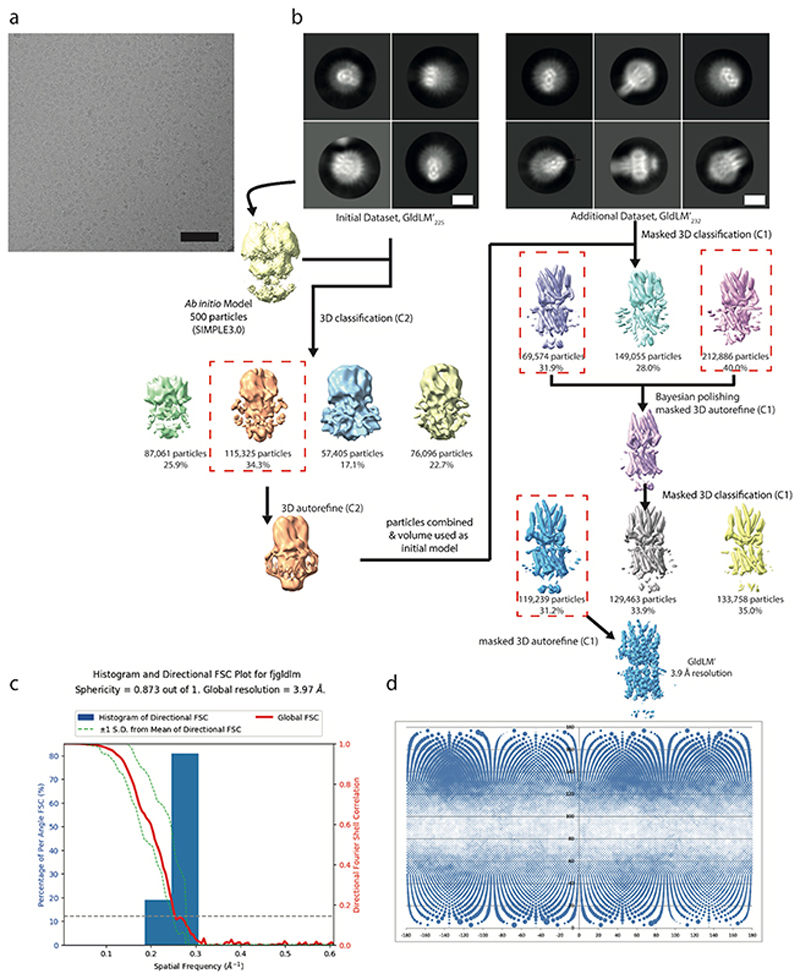
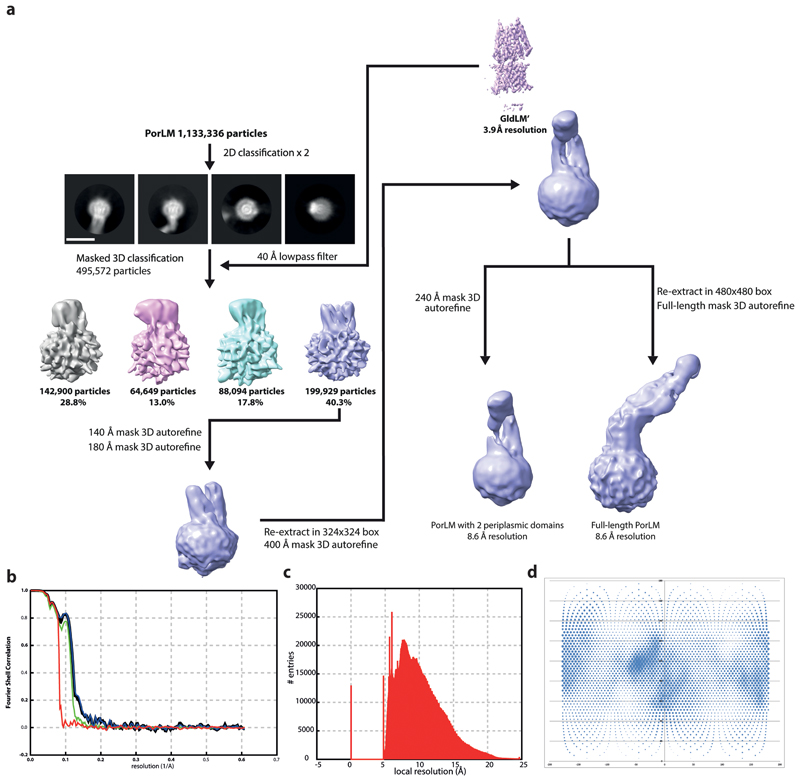
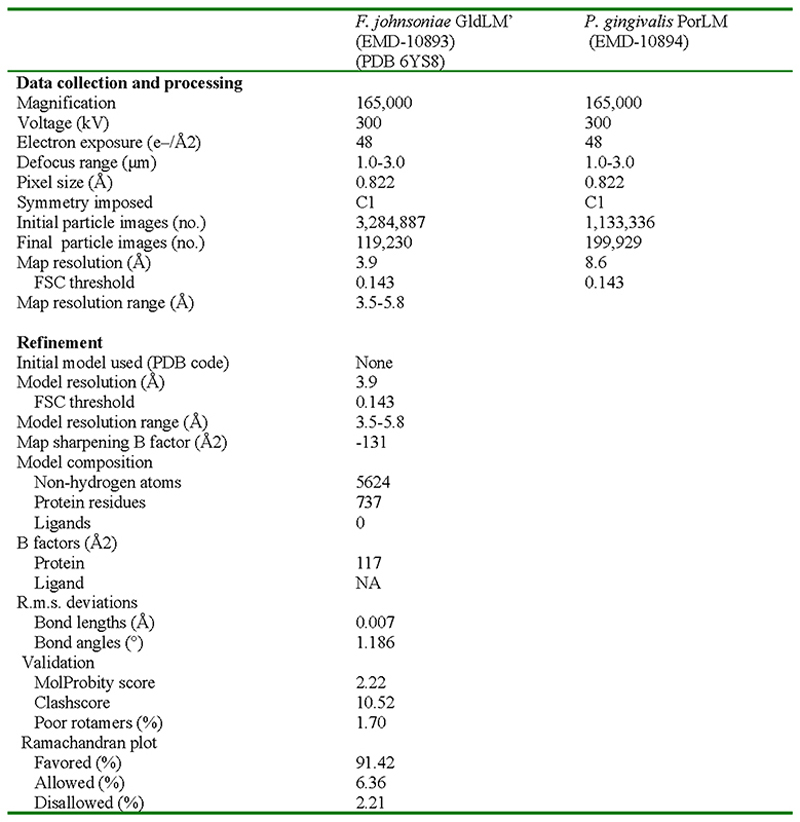
A workflow summarising the cryoEM data processing is shown in Extended Data Fig. 2. Briefly, data sets were collected from GldLM′225 and GldLM′232 samples and combined. Motion correction and dose weighting were performed using MotionCor implemented in Relion 3.045. Contrast transfer functions were calculated using CTFFIND446. Particles were picked in Simple47 and processed in Relion 3.045. Gold standard Fourier shell correlations using the 0.143 criterion and local resolution estimations were calculated within Relion 3.045 (Extended Data Fig. 3). In total 3,284,887 GldLM′ particles were extracted from 9,858 movies in 256 x 256 pixel boxes and subjected to rounds of reference-free 2D classification, from which 531,515 particles were recovered. An ab initio initial model was generated from a subset of 500 particles using SIMPLE3.047. This model was low-pass filtered to 60 Å and used as reference for 3D classification (4 classes, 7.5° sampling) against a 335,887 particle subset followed by refinement which yielded a 6.8 Å map. This map was used as initial reference (40 Å low-pass filtered) and mask for 3D classification (3 classes, 15 iterations at 7.5° sampling then 10 iterations at 3.75° sampling) against the entire dataset and further refined to 4.0 Å. Bayesian particle polishing followed by another round of 3D classification with local angular searches yielded a 3.9 Å map from 119,230 particles after refinement. A directional FSC plot for GldLM′ was calculated using the 3DFSC server48 and the resulting particle orientation distributions are shown in Extended Data Fig. 2c,d. Refinement of other of the initial 3D classes or more complex processing strategies failed to generate any improvement of this volume or other usable volumes.
PorLM particles (1,133,336) were extracted from 14,135 movies in 324 x 324 boxes and subjected to two rounds of reference-free 2D classification, from which 495,572 particles were recovered. After re-centring and re-extraction in a smaller box (256 x 256), particles were subjected to 3D classification (4 classes, 15 iterations at 7.5° sampling then 10 iterations at 3.75° sampling) against a 40 Å low-pass filtered GldLM’ map (Extended Data Fig. 3). Selected particles (199,929) were refined against the corresponding map (low-pass filtered to 40 Å) first using a soft spherical mask of 140 Å and then with a 180 Å mask. Re-centered particles, now in a 324 x 324 pixel box, were refined using a spherical mask of 400 Å. Particle re-centring and re-extraction in 480 x 480 pixel boxes followed by refinement using local searches with a mask covering the stalk and base of PorLM yielded an ~9 Å map which allows rigid body placement of prior crystal structures within the envelope but does not allow de novo modelling.
GldLM’ model building and refinement
The first periplasmic domains (residues 36-224) of the F. johnsoniae GldM crystal structure15 (PDB 6ey4) were rigid body fitted into the GldLM′ cryoEM density map using Coot49. All other residues were built de novo using Coot49 guided by TMH predictions from TMHMM50. Multiple rounds of rebuilding in both the globally sharpened and local-resolution filtered maps and real-space refinement in Phenix51 using secondary structure, rotamer, and Ramachandran restraints yielded the final model described in Extended Data Fig. 5. Validation was performed using Molprobity52. Conservation analysis was carried out using the Consurf server53.
The GldLM′ model and the C-terminal periplasmic domains (residues 225-515) of PorM (PDB 6ey5) were docked into the PorLM map using Chimera54. Figures were prepared using UCSF ChimeraX54 and Pymol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Antibody production
To produce GldL antibodies a 462-bp fragment of gldL spanning the cytoplasmic domain (GldLC, residues 66-215) was amplified from genomic DNA using primers FL125 and FL126. This fragment was inserted between the NdeI and BamHI sites of pWALDO-sfGFPd. The resulting vector, pFL43, produces a GldLC-TEV-sfGFP-His8 fusion protein.
To produce GldM antibodies a 1305-bp fragment of gldM spanning the periplasmic domain (GldMP, residues 78-513) was amplified from genomic DNA using primers FL128 and FL129. This fragment was inserted between the NdeI and BamHI sites of pWALDO-sfGFPd. The resulting vector, pFL44, produces a GldMP-TEV-sfGFP-His8 fusion protein.
E. coli BL21 Star (DE3) cells containing either pFL43 or pFL44 were grown in 1.2 l LB medium at 37 °C to OD600 = 0.5 mid-log phase and protein expression induced by addition of 400 μM IPTG. The cells were then cultured for an additional 5 h at 20 °C. Cells were harvested by centrifugation at 6,000g for 25 min and stored at -20°C until further use. All purification steps were carried out at 4°C. Cell pellets were resuspended in PBS containing 30 μg ml-1 DNase I, 400 μg ml-1 lysozyme and 1 mM phenylmethylsulfonyl fluoride at a ratio of 5 ml of buffer to 1 g of cell pellet. Cells were incubated on ice for 30 min before being lysed by two passages through a TS series 1.1kW cell disruptor (Constant System Ltd) at 30,000 PSI. Cell debris was removed by centrifugation at 20,000g for 25 min. The supernatant was then clarified using a 0.22 μm syringe filter unit (Millipore) and circulated through a 5 ml HisTrap HP column (GE Healthcare) for 2 h. The column was washed with 10 CV of PBS containing 10 mM imidazole and bound proteins were eluted with a 10-500 mM linear imidazole in 10 CV of PBS. Peak fractions were collected, diluted with an equal volume of PBS, pH 8.0, containing 0.5 mM EDTA, and dialyzed for 1 h at 4 °C against 1 l of the same buffer. TEV-His6 protease was added to the pooled fractions at a 1 to 100 protein mass ratio and dialysis was continued overnight at 4°C against 1 l of PBS containing 0.5 mM EDTA and 1 mM DTT. The cleavage reaction was then circulated through a HisTrap HP column (GE Healthcare) for 2 h and the flow through collected. This preparation was subjected to SDS-PAGE followed by Coomassie Blue staining. The band corresponding to the recombinant protein domain was excised from the gel and used by Davids Biotechnologie GmbH (Regensburg, DE) to raise polyclonal antibodies.
Preparation of samples for whole cell immunoblotting analysis of GldL and GldM
Strains were grown in CYE medium for 22 h, reaching OD600 = 5.5-6.5. The cells in 1 ml samples of the culture were harvested by centrifugation at 9,000g for 10 min, resuspended and washed once in 1 ml PBS, and finally resuspended in PBS to an OD600 = 5.0. These samples were then diluted ten-fold in PBS, subject to SDS-PAGE, and analysed by immunoblotting with GldLC or GldMP primary antibodies, and anti-Rabbit IgG HRP Conjugate (Promega) secondary antibodies.
Measurement of chitinase secretion
Cells were grown in MM for 15.5 h, reaching OD600 = 0.75-1.25. 5 ml culture samples were subject to centrifugation at 3,720g for 10 min to remove cells. Chitinase activity in the resulting supernatants was determined using a fluorometric chitinase assay kit (Sigma) with the synthetic substrate 4-methylumbelliferyl N,N’-diacetyl-β-D-chitobioside. Statistical analysis of the results was carried out using GraphPad Prism version 7.03 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). Normality of datasets was assessed by the Kolmogorov-Smirnov test. Normally distributed datasets were compared to the parental strain using one-way ANOVA followed by Dunn’s multiple comparison test. Non-normally distributed datasets were compared to WT using a Kruskal-Wallis test followed by Dunnett’s multiple comparisons test.
Measurement of gliding motility on agar
Strains were grown overnight in PY2 medium, washed once in PY2 medium, then resuspended in PY2 medium to an OD600 = 0.1. A 2 μl sample was then spotted on PY2 agar plates. Plates were incubated at 25°C for 48 h before imaging with a Zeiss AXIO Zoom MRm CCD camera and Zeiss software (ZenPro 2012, version 1.1.1.0).
Microscopic observation of gliding motility on glass
Strains were grown overnight in MM, diluted 40-fold into fresh medium and grown for 5 h at 25°C with 50 rpm shaking. An aliquot of the culture was introduced to a tunnel slide, incubated for 5 min, washed twice with 100 μl fresh medium and imaged in phase contrast on a Zeiss Axoivert 200 microscope fitted with a Photometrics Coolsnap HQ Camera using a 63x 1.4NA Plan-Apochromat lens. Tif images were captured using Metamorph software (Molecular Devices) and then modified in Image J55. Videos were collected at 20 frames per second with a 15 ms exposure time using 2x2 binning.
Single molecule fluorescence imaging of SprB mobility
Strains expressing the Halo Tag-SprB fusion were grown overnight in CYE medium, diluted 40-fold into fresh medium and grown at 30°C with 180 rpm shaking to early stationary phase (OD600 = 0.8-1.0). The HaloTag-SprB fusion was labelled by mixing 1 ml of this culture with 1 μl of a 10 μM stock solution of PA Janelia Fluor 646 HaloTag ligand56, 57 in dimethyl sulfoxide (DMSO) and cultured for a further 30 min. The cells were harvested at 9000g for 3 min, washed 3 times with 650 μl PY2 medium, and then 2 μl cells were spotted onto a 1 % agarose pad containing 50 % PY2 medium.
Fluorescence images were acquired at 25°C using a Nanoimager (Oxford Nanoimaging) equipped with 405nm and 640nm 1W DPSS lasers. Optical magnification was provided by a 100x oil-immersion objective (Olympus, numerical aperture (NA) 1.4) and images were acquired using an ORCA-Flash4.0 V3 CMOS camera (Hamamatsu). Cells were imaged using a 20 ms exposure time, with the 405 nm photoactivation laser at 10% power and the 640 nm measurement laser at 20% power. Different strains were imaged at different strobing frequencies to accommodate the large differences in adhesin velocities between the different gldLM mutants. Specifically, strain Ak_73 (wild type gldLM) was imaged without strobing, strains Ak_196 (gldL(Y13A)), Ak_197 (gldL(H30A)), and Ak_199 (gldM(Y17F)) were imaged with a 60 ms dark interval, whilst Ak_198 (gldL(K27A)) was imaged with a 480 ms dark interval.
Fluorescent foci were tracked using the Nanoimager software. Helical tracks were exported as csv files and the average frame to frame displacement calculated. The resulting data were analysed using GraphPad Prism version 7.03 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com), using a Kruskal-Wallis test followed by Dunn’s multiple comparisons test. The data were plotted using Prism.
Pulse-chase assay of Type 9 protein export
The required F. johnsoniae strain was inoculated from a freshly-streaked plate into 5 ml SDY medium (SD medium58 containing 0.01% yeast extract) and cultured for 24 h at 30 °C with shaking. The cultures were harvested by centrifugation, resuspended in the same volume of SDAC (SD medium with 0.04 mg ml-1 of all L-amino acids except methionine plus 1.5% CYE medium), then diluted one in five in fresh SDAC. These cultures were grown for 2.5 h at 30 °C and 30 rpm shaking. 40 μCi ml-1 EasyTag L-[35S]-methionine (Perkin-Elmer) was then added and growth was continued for 30 min. Cultures were harvested by centrifugation and resuspended in the same volume of unlabelled SDAC containing 0.4 mg ml-1 unlabelled L-methionine, and where required, either 10 μM carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or 10 mM sodium arsenate. The cultures were further incubated with shaking at 30 °C. At appropriate time points 1 ml samples were removed from each culture and the cells pelleted by centrifugation. Cells were resuspended in RIPA buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 0.09% sodium azide). Both the resuspended pellets and culture supernatants stored at 4 °C until the end of the time course, at which point all samples were incubated for a further 1 hour at 4 °C. The cell pellet samples were then diluted with 1.5 volumes of RND buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA), centrifuged for 10 min at 13,000g and the supernatant retained. Both these cell pellet-derived samples and the culture supernatant samples were incubated with 10 μl RFP-trap MA (Chromotek) for one hour at 24 °C with constant mixing. The RFP-trap resin was isolated on a magnetic rack and washed with 2 x 800 μl RND buffer, then resuspended in SDS-PAGE sample buffer. Samples were separated by SDS-PAGE in a Bio-rad TGX minigel system using precast 4-20% polyacrylamide gels run until the 20kDa marker was at the bottom of the gel. The gels were fixed, dried and exposed to radiographic film (GE Healthcare).
Measurement of cellular ATP levels
Strains were cultured as for the pulse-chase experiments to the point at which the cells were transferred to SDAC medium. The cells were then incubated for 3 h at 30 ˚C and 110 rpm. 0.5 ml samples were treated with 10 μM CCCP, or 10 mM sodium arsenate, or left untreated for 20 min at 24 ˚C. Cellular ATP levels were then determined using an ATP Bioluminescence Assay Kit HS II (Roche). Cells were diluted to OD600 = 0.1 with dilution buffer from the kit. 200 μl aliquots of diluted cells were combined with 200 μl lysis buffer from the kit and incubated at 70 ˚C for 5 min. Lysates were immediately transferred to ice and then clarified by centrifugation. The ATP content in the lysates was determined according to the kit manufacturer’s instructions and using a CLARIOstar Plus plate reader (BMG Labtech). Statistical analysis of cellular ATP levels was carried out using R59.
Extended Data
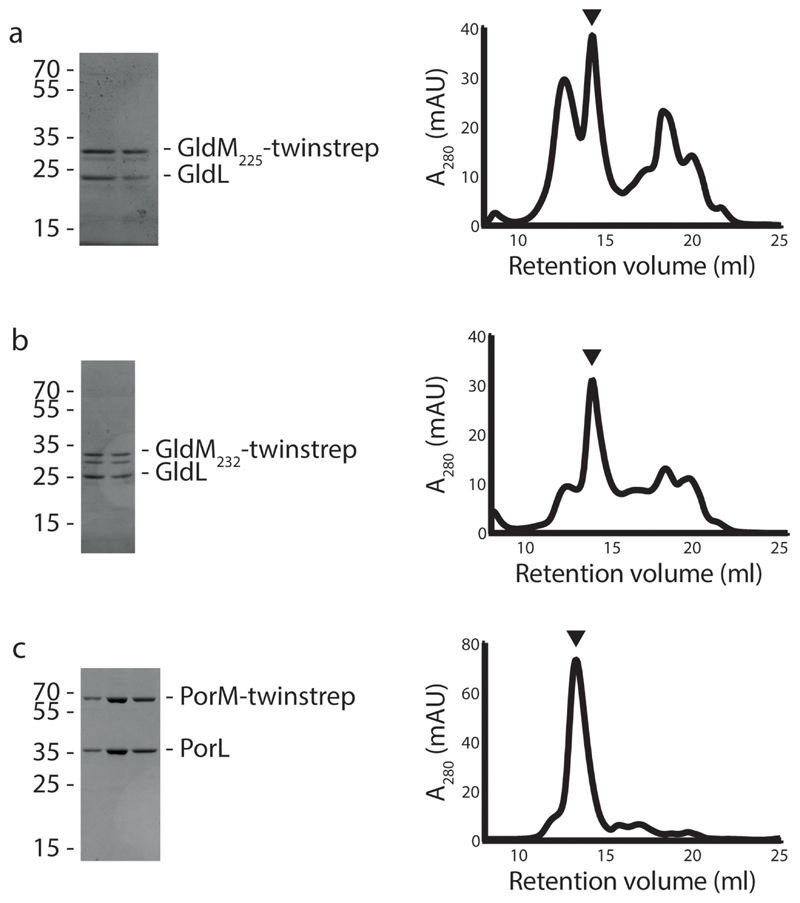
Extended Data Fig. 1. Purification of the GldLM’ and PorLM complexes.
Coomassie Blue-stained SDS-PAGE gels and size-exclusion column traces of complexes used for cryoEM structure determination. In each case the size exclusion column fractions that were pooled to make the cryoEM grids are denoted by an arrow and correspond to the fractions analysed on the gels. A280 is the absorption at 280nm in arbitrary units.
a, GldLM225(produced from pRHJ007). b, GldLM232 (produced from pRHJ008). c, PorLM (produced from pRHJ006).
Extended Data Fig. 2. Cryo-EM workflow for GldLM′.
a, Exemplar micrograph collected at a defocus value of approximately -2.5 μm, Scale bar is 200 Å.
b, Image processing workflow for GldLM’. Scale bar is 100 Å.
c, Directional Fourier Shell Correlation (FSC) plot for GldLM′ calculated using the 3DFSC server52.
d, Particle Orientation Distributions from final refined volume. Rotation (phi) is plotted along the x axis and tilt (theta) on the y axis. The area of the dot at the angular position is proportional to the number of particles assigned to that view.
Extended Data Fig. 3. Cryo-EM workflow for PorLM.
a, Image processing workflow for PorLM. Scale bar is 100 Å.
b, Fourier Shell Correlation (FSC) plot for PorLM Red, phase randomised; Green, unmasked; Blue, masked; Black, corrected.
c, Local resolution histogram of PorLM volume
d, Particle Orientation Distributions from final refined volume. Rotation (phi) is plotted along the x axis and tilt (theta) on the y axis. The area of the dot at the angular position is proportional to the number of particles assigned to that view.
Extended Data Fig. 4. Cryo-EM map quality and resolution estimates.
a, Fourier Shell Correlation (FSC) plot for the GldLM′ structure. The resolution at the gold-standard cutoff (FSC = 0.143) is indicated by the dashed line. Curves: Red, phase-randomized; Green, unmasked; blue, masked; black, MTF-corrected.
b, Local resolution estimates (in Å) for the sharpened GldLM′ map.
c, Representative modelled densities.
Extended Data Fig. 5. Cryo-EM data collection, refinement and validation statistics.
Extended Data Fig. 6. Contacts between the periplasmic loops of GldL and GldM.
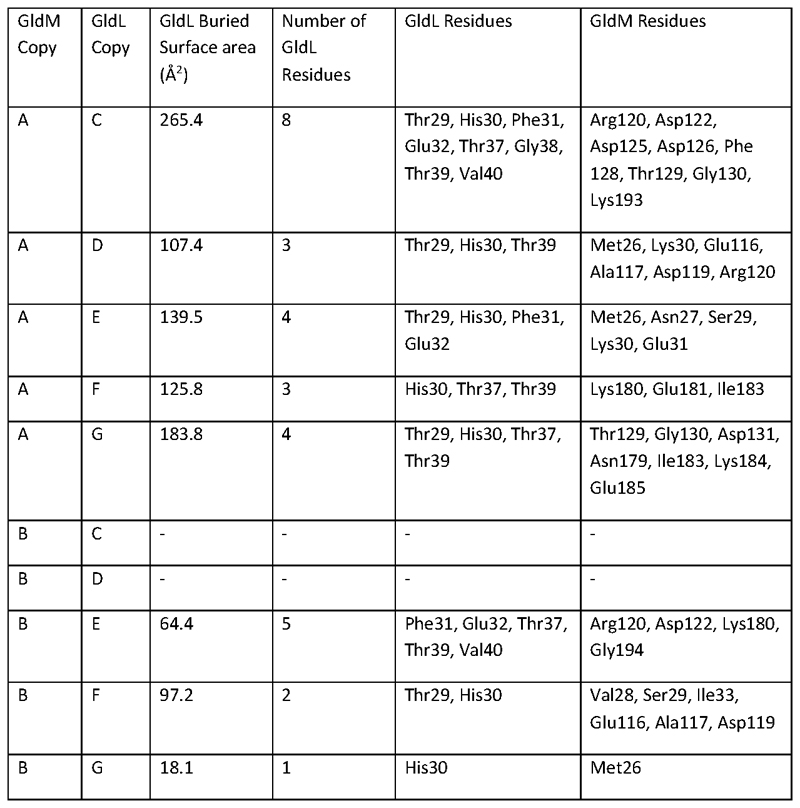
The PDBePISA server (http://www.ebi.ac.uk/pdbe/pisart.html)65 was used to calculate the sizes of the interfaces between the periplasmic loops of each copy of GldL and the periplasmic domains of GldM. The periplasmic loops of GldL were taken to include residues 29-41.
Extended Data Fig. 7. Conservation analysis of the GldLM′ complex.
a-c, Sequence conservation assessed using Consurf57.
a, The whole complex in cartoon representation. The chains shown individually in b and c are indicated with black arrows.
b,c, GldM (chain B) and GldL (chain F) in surface representation. The left hand panels show the chains in the same orientation as the upper panel of a.
d, Co-varying residue pairs in the first periplasmic domain of GldM identified using the Gremlin server64. The highest-scoring pairs (score cut-off 1.63) are shown on the structure of the GldLM′ complex. Contacts with a minimum atom-atom distance of <5 Å are shown in green, and 5-10 Å in yellow. Note, that no high-scoring GldM-GldM intermolecular pairs were observed for D1. Conversely dimerization of the other periplasmic domains is supported by co-variance with the following strongly co-varying pairs identified in the other domains D2:Val253-Ile284, Phe248-Glu-293, Tyr236-Arg261; D3: Thr327-Ala349, Asp331-Lys371; D4: Ala456-Asp483.
Extended Data Fig. 8. The full length PorLM volume is compatible with the shape of GldLM′ but not with the domain arrangements of earlier crystallographic structures of GldM (6ey4,) or PorM (6ey5) fragments.
The PorLM volume is contoured at 2.5s and the upper and lower panels are related by a 90° rotation about the y axis. The approximate domain boundaries are indicated by the dashed lines and domains in these regions are labelled on the right hand side of the figure.
The cryoEM structure of GldLM′ is positioned by docking the GldM′ TMHs onto the PorM TMHs as described in the text and Fig. 3.
The D1 dimer of GldM 6ey415 is docked into the D1 portion of the volume and shows (1) poor fit within D1 because the centre of the closed crystallographic dimer protrudes from the volume and (2) poor fit in the D2-D4 region when no bend is introduced between D1-D2.
PorM 6ey515 is positioned by placement of D2 into its portion of the volume and shows a poor fit for D3-4 which protrude from the volume.
The hybrid model presented in Figure 3 is shown for comparison, with GldM coloured as in other panels and GldL chains coloured individually.
Extended Data Fig. 9. Gliding motility and T9SS phenotypes of mutant strains.
Footnotes for table:1 Data from Fig. 4a.
2 Activity of the major T9SS substrate chitinase in the extracellular medium. Includes data shown in Fig. 4b.
3 +++, spreads as well as the parental strain; ++ spreads less well than the parental strain; + residual spreading; - does not spread. Includes data shown in Fig. 4c.
4 Includes data from Supplementary Movie 1. Note that only strains possessing cell surface adhesins are able to adhere to glass slides, and that adhesin export to the cell surface requires the T9SS which in turn depends on functional GldLM.
5 Data from Fig. 4d.
Extended Data Fig. 10. Closeup views of density around side chains discussed in manuscript.
Cryo-EM volumes contoured at 3 sigma. Key side chains are shown for both copies of GldM and for the GldL chain involved in the putative GldLE49 – GldMR9 salt-bridge.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank Mark McBride and Yongtao Zhu for providing reagents for the genetic manipulation of F. johnsoniae. We thank Luke Lavis for supplying the Janelia Fluor ligands, Sam Hickman for advice on fluorescence imaging, and Alan Wainman for assistance with phase contrast imaging. We thank Paul Guy for his involvement in producing the pulse-chase mCherry-CTD fusion construct and Richard Berry for discussions and comments on the manuscript. We acknowledge the use of the Central Oxford Structural Microscopy and Imaging Centre (COSMIC), the Oxford Micron Advanced Imaging Facility, and the Oxford Advanced Proteomics Facility. We thank Kevin Foster for providing additional imaging facilities. This work was supported by Wellcome Trust studentships to R.H.J. and A.S. (1009136), a Biotechnology and Biological Sciences Research Council studentship to A.K., and Wellcome Trust Investigator Awards 107929/Z/15/Z and 100298/Z/12/Z. COSMIC was supported by a Wellcome Trust Collaborative Award 201536/Z/16/Z, the Wolfson Foundation, a Royal Society Wolfson Refurbishment Grant, the John Fell Fund, and the EPA and Cephalosporin Trusts.
Footnotes
Author contributions
R.H.J carried out all genetic and biochemical work except as credited otherwise. J.C.D. and S.M.L. collected EM data and determined the structure. A.K. developed and carried out the SprB tracking experiments including strain construction. F.A. carried out the pulse chase analysis of protein export, A.S. assayed cellular ATP levels, F.L. constructed the ΔgldL strain and produced the GldL and GldM antibodies. B.C.B. and S.M.L. conceived the project. All authors interpreted data and wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Author Information
Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
Data availability
The cryoEM volumes have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-10893 and EMD-10894, and the coordinates have been deposited in the Protein Data Bank (PDB) with accession code 6YS8. Source Data for gels are provided with paper.
References
- 1.Guo H, Rubinstein JL. Cryo-EM of ATP synthases. Curr Opin Struct Biol. 2018;52:71–79. doi: 10.1016/j.sbi.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 3.Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008;41:103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- 4.Kojima S. Dynamism and regulation of the stator, the energy conversion complex of the bacterial flagellar motor. Curr Opin Microbiol. 2015;28:66–71. doi: 10.1016/j.mib.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Deme JC, et al. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBride MJ. Bacteroidetes Gliding Motility and the Type IX Secretion System. Microbiol Spectr. 2019;7 doi: 10.1128/microbiolspec.psib-0002-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A. 2013;110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridgway HF. Source of energy for gliding motility in Flexibacter polymorphus: effects of metabolic and respiratory inhibitors on gliding movement. J Bacteriol. 1977;131:544–556. doi: 10.1128/jb.131.2.544-556.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrivastava A, Lele PP, Berg HC. A rotary motor drives Flavobacterium gliding. Curr Biol. 2015;25:338–341. doi: 10.1016/j.cub.2014.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrivastava A, Roland T, Berg HC. The Screw-Like Movement of a Gliding Bacterium Is Powered by Spiral Motion of Cell-Surface Adhesins. Biophys J. 2016;111:1008–1013. doi: 10.1016/j.bpj.2016.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson SS, Bollampalli S, McBride MJ. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol. 2008;190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrivastava A, Berg HC. A molecular rack and pinion actuates a cell-surface adhesin and enables bacterial gliding motility. Science Advances. 2020;6:1–6. doi: 10.1126/sciadv.aay6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J Bacteriol. 2013;195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vincent MS, et al. Characterization of the Porphyromonas gingivalis Type IX Secretion Trans-envelope PorKLMNP Core Complex. J Biol Chem. 2017;292:3252–3261. doi: 10.1074/jbc.M116.765081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leone P, et al. Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space. Nat Commun. 2018;9:429. doi: 10.1038/s41467-017-02784-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato K, et al. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A. 2010;107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasica AM, Ksiazek M, Madej M, Potempa J. The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function. Front Cell Infect Microbiol. 2017;7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Diego I, et al. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain. Sci Rep. 2016;6:23123. doi: 10.1038/srep23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni SS, Zhu Y, Brendel CJ, McBride MJ. Diverse C-Terminal Sequences Involved in Flavobacterium johnsoniae Protein Secretion. J Bacteriol. 2017;199 doi: 10.1128/JB.00884-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauber F, Deme JC, Lea SM, Berks BC. Type 9 secretion system structures reveal a new protein transport mechanism. Nature. 2018;564:77–82. doi: 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nijeholt JALA, Driessen AJM. The bacterial Sec-translocase: structure and mechanism. Philos T R Soc B. 2012;367:1016–1028. doi: 10.1098/rstb.2011.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorasia DG, et al. Structural Insights into the PorK and PorN Components of the Porphyromonas gingivalis Type IX Secretion System. PLoS Pathog. 2016;12:e1005820. doi: 10.1371/journal.ppat.1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorasia DG, et al. structure and organisation of the type IX secretion system. BioRxiv. 2020 doi: 10.1101/2020.05.13.094771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes RG, Samarasam MN, Van Groll EJ, McBride MJ. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J Bacteriol. 2011;193:5322–5327. doi: 10.1128/JB.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato K, Okada K, Nakayama K, Imada K. PorM, a core component of bacterial type IX secretion system, forms a dimer with a unique kinked-rod shape. Biochem Biophys Res Commun. 2020 doi: 10.1016/j.bbrc.2020.08.018. [DOI] [PubMed] [Google Scholar]
- 26.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 27.Turner RD, Hurd AF, Cadby A, Hobbs JK, Foster SJ. Cell wall elongation mode in Gram-negative bacteria is determined by peptidoglycan architecture. Nat Commun. 2013;4:1496. doi: 10.1038/ncomms2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner RD, Mesnage S, Hobbs JK, Foster SJ. Molecular imaging of glycan chains couples cell-wall polysaccharide architecture to bacterial cell morphology. Nat Commun. 2018;9:1263. doi: 10.1038/s41467-018-03551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koraimann G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci. 2003;60:2371–2388. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheurwater EM, Burrows LL. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol Lett. 2011;318:1–9. doi: 10.1111/j.1574-6968.2011.02228.x. [DOI] [PubMed] [Google Scholar]
- 31.Vincent MS, Durand E, Cascales E. The PorX Response Regulator of the Porphyromonas gingivalis PorXY Two-Component System Does Not Directly Regulate the Type IX Secretion Genes but Binds the PorL Subunit. Front Cell Infect Microbiol. 2016;6:96. doi: 10.3389/fcimb.2016.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celia H, et al. Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry. Commun Biol. 2019;2:358. doi: 10.1038/s42003-019-0604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride MJ, Kempf MJ. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J Bacteriol. 1996;178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, McBride MJ, Subramaniam S. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol. 2007;189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal S, Hunnicutt DW, McBride MJ. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci U S A. 1997;94:12139–12144. doi: 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dietsche T, et al. Structural and Functional Characterization of the Bacterial Type III Secretion Export Apparatus. PLoS Pathog. 2016;12:e1006071. doi: 10.1371/journal.ppat.1006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. Optimization of membrane protein overexpression and purification using GFP fusions. Nat Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, et al. Genetic analyses unravel the crucial role of a horizontally acquired alginate lyase for brown algal biomass degradation by Zobellia galactanivorans. Environ Microbiol. 2017;19:2164–2181. doi: 10.1111/1462-2920.13699. [DOI] [PubMed] [Google Scholar]
- 39.Jacobus AP, Gross J. Optimal cloning of PCR fragments by homologous recombination in Escherichia coli. PloS one. 2015;10:e0119221. doi: 10.1371/journal.pone.0119221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon R, Priefer U, Puhler A. A Broad Host Range Mobilization System for Invivo Genetic-Engineering - Transposon Mutagenesis in Gram-Negative Bacteria. Bio-Technol. 1983;1:784–791. [Google Scholar]
- 42.Baumgarten T, et al. Isolation and characterization of the E. coli membrane protein production strain Mutant56(DE3) Sci Rep. 2017;7:45089. doi: 10.1038/srep45089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller J. Experiments in Molecular Genetics. Cold Spring Harbor; New York: 1972. [Google Scholar]
- 44.Tartof KD, aH CA. Improved media for growing plasmid and cosmid clones. Focus®. 1987;9:12. [Google Scholar]
- 45.Zivanov J, Nakane T, Scheres SHW. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reboul CF, Eager M, Elmlund D, Elmlund H. Single-particle cryo-EM-Improved ab initio 3D reconstruction with SIMPLE/PRIME. Protein Sci. 2018;27:51–61. doi: 10.1002/pro.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan YZ, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nature methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 51.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta crystallographica Section D, Structural biology. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams CJ, et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein science: a publication of the Protein Society. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashkenazy H, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grimm JB, Brown TA, English BP, Lionnet T, Lavis LD. Synthesis of Janelia Fluor HaloTag and SNAP-Tag Ligands and Their Use in Cellular Imaging Experiments. Methods Mol Biol. 2017;1663:179–188. doi: 10.1007/978-1-4939-7265-4_15. [DOI] [PubMed] [Google Scholar]
- 57.Grimm JB, et al. Bright photoactivatable fluorophores for single-molecule imaging. Nat Methods. 2016;13:985–988. doi: 10.1038/nmeth.4034. [DOI] [PubMed] [Google Scholar]
- 58.Chang LYE, Pate JL. Nutritional-Requirements of Cytophaga-Johnsonae and Some of Its Auxotrophic Mutants. Curr Microbiol. 1981;5:235–240. [Google Scholar]
- 59.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 60.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci U S A. 2013;110:15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryoEM volumes have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-10893 and EMD-10894, and the coordinates have been deposited in the Protein Data Bank (PDB) with accession code 6YS8. Source Data for gels are provided with paper.