Summary
De novo mutations (DNMs) in protein-coding genes are a well-established cause of developmental disorders (DD)1. However, known DD-associated genes only account for a minority of the observed excess of such DNMs1,2. To identify novel DD-associated genes, we integrated healthcare and research exome sequences on 31,058 DD parent-offspring trios, and developed a simulation-based statistical test to identify gene-specific enrichments of DNMs. We identified 285 significantly DD-associated genes, including 28 not previously robustly associated with DDs. Despite detecting more DD-associated genes, much of the excess of DNMs of protein-coding genes remains unaccounted for. Modelling suggests that over 1,000 novel DD-associated genes await discovery, many of which are likely to be less penetrant than the currently known genes. Research access to clinical diagnostic datasets will be critical for completing the map of dominant DDs.
Introduction
It has previously been estimated that ~42-48% of patients with a severe developmental disorder (DD) have a pathogenic de novo mutation (DNM) in a protein coding gene1,2. However, most of these patients remain undiagnosed despite the identification of hundreds of DD-associated genes. This implies that there are more DD relevant genes to find. Existing methods to detect gene-specific enrichments of damaging DNMs ignore much prior information about which variants are more likely to be disease-associated; missense variants and protein-truncating variants (PTVs) vary in their impact on protein function3–6. Known dominant DD-associated genes are strongly enriched in the minority of genes that exhibit strong selective constraint on heterozygous PTVs7. To identify additional DD-associated genes, we need to increase our power to detect gene-specific enrichments for damaging DNMs by both increasing sample sizes and improving our statistical methods. In previous studies of pathogenic Copy Number Variation, utilising healthcare data has been key to achieve larger sample sizes than would be possible in a research setting alone8,9.
Identification of 285 DD-associated genes
Following clear consent practices and only using aggregate, de-identified data, we pooled DNMs in DD patients from three centres: GeneDx (a US-based diagnostic testing company), the Deciphering Developmental Disorders study, and Radboud University Medical Center. We performed stringent quality control on variants and samples to obtain 45,221 coding and splicing DNMs in 31,058 individuals (Supplementary Fig. 1; Supplementary Table 1), including data on 24,348 trios not previously published. These DNMs included 40,992 single nucleotide variants (SNVs) and 4,229 indels. The three cohorts have similar clinical characteristics, male/female ratios, enrichments of DNMs by mutational class, and prevalences of known disorders (Supplementary Fig. 2).
To detect gene-specific enrichments of damaging DNMs, we developed a method named DeNovoWEST (De Novo Weighted Enrichment Simulation Test, https://github.com/queenjobo/DeNovoWEST). DeNovoWEST scores all classes of sequence variants on a unified severity scale based on empirically-estimated positive predictive values of being pathogenic (Supplementary Fig. 3–4). We perform two tests per gene: an enrichment test on all nonsynonymous DNMs and a test designed to detect genes likely acting via an altered-function mechanism, which combines a missense enrichment test with a missense clustering test. We then applied a Bonferroni multiple testing correction accounting for the number of genes (n=18,762) and two tests per gene.
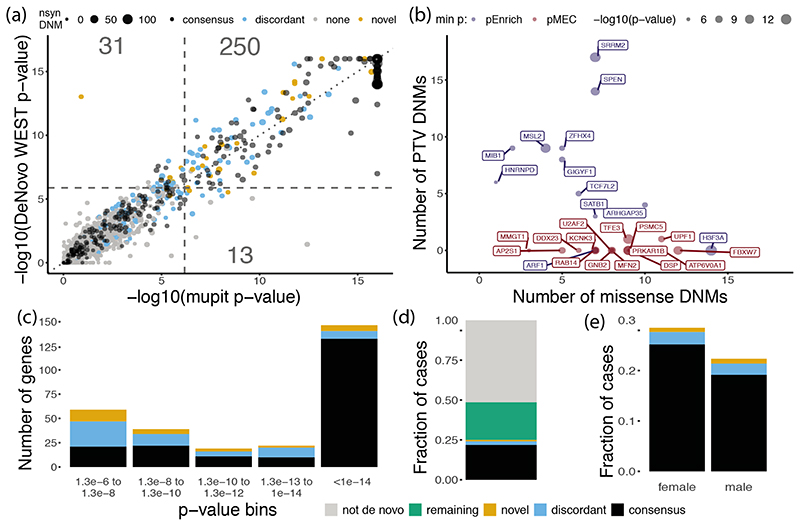
We first applied DeNovoWEST to all individuals in our cohort and identified 281 significantly enriched genes, 18 more than when using our previous method1 (Supplementary Fig. 5; Fig. 1a). The majority (196/281; 70%) of these significant genes already had sufficient evidence of DD-association to be considered of diagnostic utility (as of late 2019) by all three centres, and we refer to them as “consensus” genes. 54/281 of these significant genes were previously considered diagnostic by one or two centres (“discordant” genes). Applying DeNovoWEST to synonymous DNMs, as a negative control analysis, identified no significantly enriched genes (Supplementary Fig. 6).
Figure 1. Results of DeNovoWEST analysis.
(a) Comparison of p-values using the new method (DeNovoWEST) versus the previous method (mupit)1, run on the full cohort. Dashed lines indicate the threshold for genome-wide significance (one sided, Bonferroni correction). Point size is proportional to the number of nonsynonymous DNMs in our cohort (nsyn). The number of genes that fall into each quadrant are annotated. (b) The number of missense and PTV DNMs in the novel genes. Point size is proportional to the log10(-p-value) from analysis of the undiagnosed subset. Point colour corresponds to which test p-value was more significant: non-synonymous enrichment test in blue (pEnrich), missense enrichment and clustering test in red (pMEC). (c) The distribution of significant p-values from analysis of the undiagnosed subset for discordant and novel genes; p-values for consensus genes come from the full cohort analysis. The number of genes in each p-value bin is coloured by diagnostic gene group (n = 285 significant genes; one-sided p-values, Bonferroni corrected). Green represents the remaining fraction of cases expected to have a pathogenic de novo coding mutation and grey is the fraction of cases that are likely to be explained by other factors. (d) The fraction of cases (n = 31,058) with a nonsynonymous mutation in each diagnostic gene group. (e) The fraction of cases with a nonsynonymous mutation in each diagnostic gene group split by sex (n = 13,636 female and 17,422 male). In all panels, black, blue and orange represents consensus, discordant and novel genes respectively.
To discover novel DD-associated genes with greater power, we applied DeNovoWEST to DNMs in patients without damaging DNMs in consensus genes (we refer to this subset as ‘undiagnosed’ patients) and identified 94 significant genes (Supplementary Fig. 7; Supplementary Table 2), of which 33 were putative ‘novel’ DD-associated genes. To ensure robustness to potential mutation rate variation between genes, we determined whether any of the putative novel DD-associated genes had significantly more synonymous variants in the Genome Aggregation Database6 (gnomAD) of population variation than expected under our null mutation model (Supplementary Note). We identified 11/33 genes with a significant excess of synonymous variants. For these 11 genes we repeated the DeNovoWEST test, increasing the null mutation rate by the ratio of observed to expected synonymous variants in gnomAD. Five of these genes fell below our exome-wide significance threshold and were removed, leaving 28 novel genes, with a median of 10 nonsynonymous DNMs (Fig. 1b–c; Supplementary Table 3). There were 314 patients with nonsynonymous DNMs in these 28 genes (1.0% of our cohort); all these DNMs were inspected in IGV10 and, of 198 for which experimental validation was attempted, all were confirmed as DNMs. The DNMs in these novel genes were distributed randomly across the three datasets (no genes with p < 0.001, heterogeneity test). Six of the 28 novel DD-associated genes are corroborated by OMIM entries or publications, including TFE3 11,12 which was described in two recent publications.
We also investigated whether some synonymous DNMs might be pathogenic by disrupting splicing. We identified a small but significant enrichment of synonymous DNMs with high values of the splicing pathogenicity score SpliceAI13 (≥ 0.8, 1.56-fold enriched, p = 0.0037, Poisson test; Supplementary Table 4). This enrichment corresponds to an excess of ~15 splice-disrupting synonymous DNMs in our cohort, of which six are accounted for by a recurrent synonymous DNM in KAT6B known to disrupt splicing14.
Taken together, 25.0% of our cohort has a nonsynonymous DNM in one of the consensus or significant DD-associated genes (Fig. 1d). We noted significant sex differences in the autosomal burden of nonsynonymous DNMs (Supplementary Fig. 8). The rate of nonsynonymous DNMs in consensus autosomal genes was significantly higher in females than males (OR = 1.16, p = 4.4 × 10-7, Fisher’s exact test; Fig. 1e), as noted previously1. However, the exome-wide burden of autosomal nonsynonymous DNMs in all genes was not significantly different between undiagnosed males and females (OR = 1.03, p = 0.29, Fisher’s exact test).
This suggests the existence of subtle sex differences in the genetic architecture of DD, especially with regard to known and undiscovered disorders. This could include sex-biased contribution of polygenic, oligogenic and/or environmental modifiers of phenotypic variation and thus clinical ascertainment.
Characteristics novel DD-associated genes
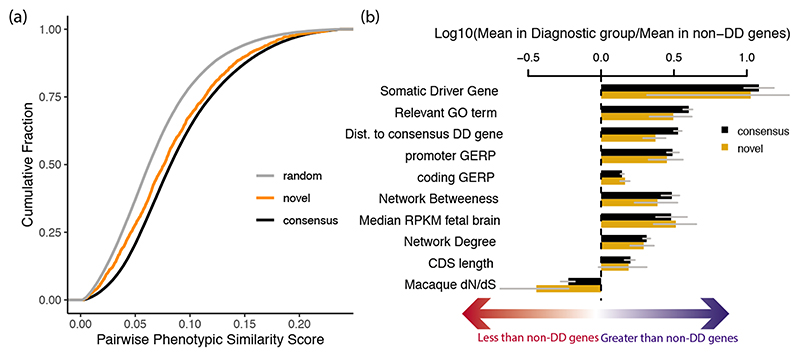
Based on semantic similarity15 between Human Phenotype Ontology terms, patients with DNMs in the same novel DD-associated gene were less phenotypically similar to each other, on average, than patients with DNMs in a consensus gene (p = 2.3 × 10-11, Wilcoxon rank-sum test; Fig. 2a; Supplementary Figure 9). This suggests that these novel disorders less often result in distinctive and consistent clinical presentations, which may have made these disorders harder to discover via a phenotype-driven approach. Each of these novel disorders requires genotype-phenotype characterisation, which is beyond the scope of this study.
Figure 2. Properties of novel genes.
(a) The phenotypic similarity of patients with DNMs in novel and consensus genes. Random phenotypic similarity was calculated from random pairs of patients. Cases with DNMs in the same novel gene were less phenotypically similar than cases with DNMs in the same consensus gene (p = 2.3 × 10-11, two-sided Wilcoxon rank-sum test). (b) Comparison of properties of consensus (n = 380) and novel (n = 28) DD genes known to be differential between consensus and non-DD genes (95% bootstrapped confidence intervals shown).
Overall, novel DD-associated genes encode proteins that have very similar functional and evolutionary properties to consensus genes (Fig. 2b; Supplementary Table 5). Despite the high-level functional similarity between known and novel DD-associated genes, nonsynonymous DNMs in the more recently discovered DD-associated genes are much more likely to be missense DNMs, and less likely to be PTVs (discordant and novel; p = 1.2 × 10-25, chi-squared test). Fifteen (54%) of the 28 novel genes only had missense DNMs. Consequently, we expect that a greater proportion of the novel genes will act via altered-function mechanisms (e.g. dominant negative or gain-of-function). For example, the novel gene PSMC5 (DeNovoWEST p = 2.6 × 10-15) had one inframe deletion and nine missense DNMs, eight of which altered two structurally important amino acids in the AAA+ ATPase domain, and so is likely to operate via an altered-function mechanism (Supplementary Fig. 10a–b). None of the novel genes exhibited significant clustering of de novo PTVs.
We observed that missense DNMs were more likely to affect functional protein domains than other coding regions. We observed a 2.63-fold enrichment (p = 2.2 × 10-68, G-test) of missense DNMs residing in protein domains among consensus genes and a 1.80-fold enrichment (p = 8.0 × 10-5, G-test) in novel DD-associated genes, but no enrichment for synonymous DNMs (Supplementary Table 6). Four protein domain families in consensus genes were enriched for missense DNMs (Supplementary Table 7): ion transport protein (PF00520, p = 6.9 × 10-4, G-test Bonferroni corrected), ligand-gated ion channel (PF00060, p = 4.0 × 10-6), protein kinase domain (PF00069, p = 0.043), and kinesin motor domain (PF00225, p = 0.027). Missense DNMs in all four enriched domain families have previously been associated with DD (Supplementary Table 8)16–18.
We observed a significant overlap between the 285 DNM-enriched DD-associated genes and a set of 369 previously described cancer driver genes19 (overlap of 70 genes; p = 1.7 × 10-49, logistic regression correcting for shet), as observed previously20,21, as well as a significant enrichment of nonsynonymous DNMs in both overlapping and non-overlapping cancer genes (Supplementary Table 9). We observe 117 DNMs at 76 recurrent somatic mutations observed in at least three patients in The Cancer Genome Atlas (TCGA)22. By modelling the germline mutation rate at these somatic driver mutations, we found that recurrent nonsynonymous mutations in TCGA are enriched 21-fold in our cohort (p < 10-50, Poisson test, Supplementary Fig. 11), whereas recurrent synonymous mutations in TCGA are not significantly enriched (2.4-fold, p = 0.13, Poisson test). This suggests that this observation is driven by the pleiotropic effects of these mutations in development and tumourigenesis, rather than hypermutability.
Recurrent mutations
We identified 773 recurrent DNMs (736 SNVs and 37 indels), observed in 2-36 individuals, which allowed us to interrogate systematically the factors driving recurrent germline mutation. We considered three potential contributory factors: (i) clinical ascertainment enriching for pathogenic mutations, (ii) greater mutability at specific sites, and (iii) positive selection conferring a proliferative advantage in the male germline23. We observed evidence that all three factors contribute, but not mutually exclusively. Clinical ascertainment drives the observation that 65% of recurrent DNMs were in consensus genes, a 5.4-fold enrichment compared to DNMs only observed once (p < 10-50, proportion test). Hypermutability underpins the observation that 64% of recurrent de novo SNVs occurred at hypermutable CpG dinucleotides24, a 2.0-fold enrichment over DNMs only observed once (p = 3.3 × 10-68, chi-square test).
To assess the contribution of germline selection to recurrent DNMs, we initially focused on the 12 known germline selection genes, which all operate through activation of the RAS-MAPK signalling pathway25,26. We identified 39 recurrent DNMs in 11 of these genes, 38 of which are missense and all of which are known to be activating in the germline (see Supplement). As expected, given that hypermutability is not the driving factor for recurrent mutation in these genes, these 39 recurrent DNMs were depleted for CpGs relative to other recurrent mutations (6/39 vs 425/692, p = 3.4 × 10-8, chi-squared test).
Positive germline selection can increase the apparent mutation rate more strongly23 than either clinical ascertainment (10-100X in our dataset) or hypermutability (~10X for CpGs). However, only a minority of the most highly recurrent mutations in our dataset are in genes that have been previously associated with germline selection. Nonetheless, several lines of evidence suggested that the majority of these most highly recurrent mutations are likely to confer a germline selective advantage. Based on the observations above, DNMs under germline selection should be more likely to be activating missense mutations, and should be less enriched for CpG dinucleotides. Extended Data Table 1 shows the 16 de novo SNVs observed nine or more times in our cohort, only two of which are in known germline selection genes. All but two of these 16 de novo SNVs cause missense changes, all but two of these genes cause disease by an altered-function mechanism, and these DNMs were depleted for CpGs relative to all recurrent mutations. Two of these genes with highly recurrent de novo SNVs, SHOC2 and PPP1CB, encode interacting proteins that regulate the RAS-MAPK pathway, and pathogenic variants in these genes are associated with a Noonan-like syndrome27. Moreover, two of these recurrent DNMs are in the same gene SMAD4, which encodes a key component of the TGF-beta signalling pathway, potentially expanding the pathophysiology of germline selection beyond the RAS-MAPK pathway. Confirming germline selection of these mutations will require deep sequencing of testes and/or sperm26.
Incomplete penetrance and pre/perinatal death
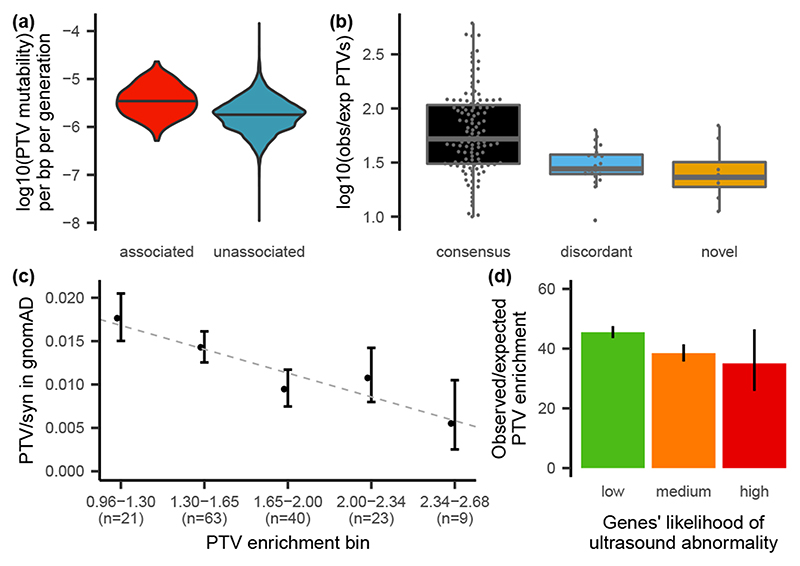
Nonsynonymous DNMs in consensus or significant DD-associated genes accounted for half of the exome-wide nonsynonymous DNM burden associated with DD (Fig. 1b). Despite our identification of 285 significantly DD-associated genes, there remains a substantial burden of both missense and protein-truncating DNMs in unassociated genes (those that are neither significant in our analysis nor on the consensus gene list). This residual burden of protein-truncating DNMs is greatest in genes that are intolerant of PTVs in the general population (Supplementary Fig. 12) suggesting that more haploinsufficient (HI) disorders await discovery. We observed that PTV mutability (estimated from a null germline mutation model) was significantly lower in unassociated genes compared to DD-associated genes (p = 4.5 × 10-68, Wilcox rank-sum test Fig. 3a), which leads to reduced statistical power to detect DNM enrichment in unassociated genes, consistent with our hypothesis that many more HI disorders await discovery.
Figure 3. Factors influencing power.
(a) PTV mutability is significantly lower (p = 4.6 × 10-68, two-sided Wilcox rank sum test) in genes that are not significantly DD-associated (blue) than in DD-associated genes (red). Median depicted with a black horizontal line. (b) Distribution of PTV enrichment in significant, likely haploinsufficient, genes by category (118 consensus, 23 discordant, 8 novel genes). Lower and upper hinges correspond to first and third quantiles. Median depicted by a horizontal grey line. The upper and lower whiskers extend 1.5 times the inter-quartile range. (c) Comparison of PTV enrichment in our cohort vs the PTV to synonymous ratio in gnomAD, for genes that are significantly PTV-enriched in our cohort (without variant weighting; n = 156 genes). PTV enrichment bins labelled with log10(enrichment). Dashed line indicates regression. Confidence intervals are 95% of the rate ratio. (d) Overall PTV enrichment across genes grouped by likelihood of presenting with a structural malformation on prenatal ultrasound (145 low, 65 medium, 6 low genes). PTV enrichment is significantly higher for genes with a low likelihood compared to other genes (p = 4.6 × 10-5, two-sided Poisson test). Poisson 95% confidence intervals shown.
A key parameter in estimating statistical power to detect novel HI disorders is the fold-enrichment of de novo PTVs expected in undiscovered HI disorders. We observed that novel DD-associated HI genes had significantly lower PTV enrichment compared to the consensus HI genes (p = 0.005, Wilcox rank-sum test; Fig. 3b). Two additional factors that could lower DNM enrichment, and thus power to detect a novel DD-association, are reduced penetrance and increased pre/perinatal death (due to spontaneous fetal loss, termination of pregnancy for fetal anomaly, stillbirth, or early neonatal death). To evaluate incomplete penetrance, we investigated whether HI genes with a lower enrichment of de novo PTVs in our cohort are associated with greater prevalences of PTVs in the general population. We observed a significant negative correlation (p = 0.031, weighted linear regression) between PTV enrichment in our cohort and the ratio of PTV to synonymous variants in gnomAD6, suggesting that incomplete penetrance does lower de novo PTV enrichment in our cohort (Fig. 3c).
Additionally, we observed that the fold-enrichment of de novo PTVs in consensus HI DD-associated genes in our cohort was significantly higher for genes with a low likelihood of presenting with a prenatal structural malformation (p = 4.6 × 10-5, Poisson test, Fig. 3d), suggesting that pre/perinatal death decreases our power to detect some novel disorders (see supplement for details).
Hundreds of DD genes not yet discovered
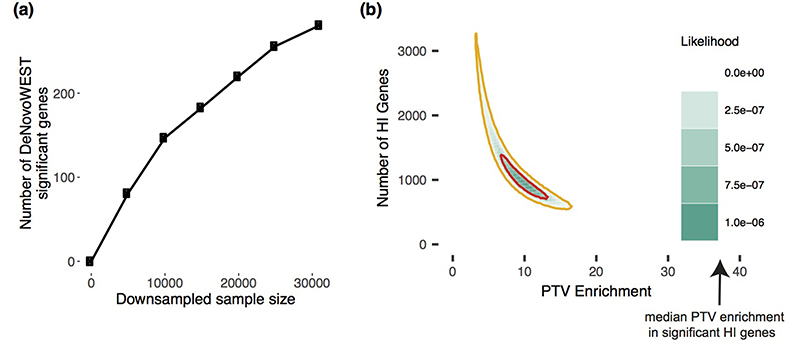
Downsampling of our cohort and repeating enrichment analyses showed that the discovery of DD-associated genes has not plateaued (Extended Data Fig 1a). Increasing sample sizes should result in the discovery of many novel DD-associated genes. To estimate how many haploinsufficient genes might await discovery, we modelled the likelihood of the observed distribution of de novo PTVs among genes as a function of varying numbers of undiscovered HI DD-associated genes and fold-enrichments of de novo PTVs in those genes. We found that the remaining PTV burden is most likely spread across ~1,000 genes with ~10-fold PTV enrichment (Extended Data Fig 1b). This fold enrichment is three times lower than in known HI DD-associated genes, suggesting that incomplete penetrance and/or pre/perinatal death is more prevalent among undiscovered HI genes. We modelled the missense DNM burden separately and also observed that the most likely architecture of undiscovered DD-associated genes is one that comprises over 1,000 genes with a substantially lower fold-enrichment than in currently known DD-associated genes (Supplementary Fig. 13).
We calculated that a sample size of ~350,000 parent-offspring trios would be needed to have 80% power to detect a 10-fold enrichment of de novo PTVs for an average gene. Using this inferred 10-fold enrichment among undiscovered HI genes, from our current data we can evaluate the likelihood that any gene i is an undiscovered HI gene, by comparing the likelihood of the number of de novo PTVs observed in each gene to have arisen from the null mutation rate or from a 10-fold increased PTV rate. Among the ~19,000 non-DD-associated genes, ~1,200 were more than three times more likely to have arisen from a 10-fold increased PTV rate, whereas ~7,000 were three times more likely to have no de novo PTV enrichment.
Discussion
In this study, we have presented evidence for 28 novel developmental disorders by developing an improved statistical test for mutation enrichment and applying it to a dataset of exome sequences from 31,058 parent-offspring trios. Most of the increased power to detect novel disorders comes from the increase in sample size, rather than the improved statistical test. These 28 novel genes account for 1.0% of our cohort, and their inclusion in diagnostic workflows will catalyse increased diagnosis of similar patients globally. The value of this study for improving diagnostic yield extends beyond these 28 novel genes; the total number of genes added to diagnostic workflows of the three participating centres (including newly validated discordant genes) ranged from 48-65 genes. We have shown that both incomplete penetrance and pre/perinatal death reduce our power to detect novel DDs postnatally, and hypothesise that one or both of these factors are operating more strongly among undiscovered DD-associated genes. In addition, we have identified a set of highly recurrent mutations that are strong candidates for novel germline selection mutations, which should result in a higher than expected disease incidence that increases dramatically with increased paternal age.
Our study is approximately three times larger than a recent meta-analysis of DNMs from a collection of individuals with autism spectrum disorder, intellectual disability, and/or a developmental disorder28. We identified ~2.3 times as many significantly DD-associated genes as this previous study when using Bonferroni-corrected exome-wide significance (285 vs 124). In contrast to meta-analyses of published DNMs, the harmonised filtering of candidate DNMs across cohorts in this study should be more robust to cohort-specific differences in the sensitivity and specificity of detecting DNMs.
We inferred indirectly that developmental disorders with higher rates of detectable prenatal structural abnormalities had greater pre/perinatal death. The potential size of this effect can be quantified from the recently published PAGE study of genetic diagnoses in a cohort of fetal structural abnormalities29. In this latter study, genetic diagnoses were not returned to participants during the pregnancy, and so genetic diagnostic information could not influence pre/perinatal death. In the PAGE study data, 69% of fetal abnormalities with a genetically diagnosable cause died perinatally or neonatally. This emphasises the substantial impact that pre/perinatal death can have on reducing the ability to discover novel DDs from postnatal recruitment alone, and motivates the integration of genetic data from prenatal, neonatal and postnatal studies in future studies.
To empower our mutation enrichment testing, we estimated positive predictive values (PPV) of each DNM being pathogenic on the basis of their predicted protein consequence, CADD score3, selective constraint against heterozygous PTVs across the gene (shet)30, and, for missense variants, presence in a region under selective missense constraint4. These PPVs should also be informative for variant prioritisation in the diagnosis of dominant developmental disorders. Further work is needed to see whether these PPVs might be informative for recessive developmental disorders, and in other types of dominant disorders. More generally, we hypothesise that empirically-estimated PPVs based on variant enrichment in large datasets will be similarly informative in many other disease areas.
We adopted a conservative statistical approach to identifying DD-associated genes. In two previous studies using the same significance threshold, we identified 26 novel DD-associated genes1,31. All 26 are now regarded as being diagnostic, and have entered routine clinical diagnostic practice. Had we used a significance threshold of FDR < 10% as used in Satterstrom, Kosmicki, Wang et al32, we would have identified 770 DD-associated genes. The FDR of individual genes depends on the significance of other genes being tested, so are not appropriate for assessing the significance of individual genes, but rather for defining gene-sets. There are 184 consensus genes that did not cross our significance threshold in this study. It is likely that many of these cause disorders that were under-represented in our study due to the ease of clinical diagnosis on the basis of distinctive clinical features or targeted diagnostic testing. These ascertainment biases will not impact the representation of novel DDs in our cohort.
Our modelling suggested that likely over 1,000 DD-associated genes remain to be discovered, and that reduced penetrance and pre/perinatal death will reduce our power to identify these genes through DNM enrichment. Identifying these genes will require both improved analytical methods and greater sample sizes. As sample sizes increase, accurate modelling of gene-specific mutation rates becomes more important. In our analyses of 31,058 trios, we observed evidence that mutation rate heterogeneity among genes can lead to over-estimating the statistical significance of mutation enrichment based on an exome-wide mutation model. We advocate the development of more granular mutation rate models, based on large-scale population variation resources, that correct for all technical and biological complexities, to ensure that larger studies are robust to mutation rate heterogeneity.
We anticipate that the variant-level weights used by DeNovoWEST will improve over time. As reference population samples, such as gnomAD6, increase in size, weights based on selective constraint metrics (e.g. shet, regional missense constraint) will improve. Weights could also incorporate more functional information, such as expression in disease-relevant tissues. For example, we observe that DD-associated genes are significantly more likely to be expressed in fetal brain (Supplementary Fig. 14). Furthermore, novel metrics based on gene co-regulation networks can predict whether genes function within a disease-relevant pathway33. As a cautionary note, including more functional information may increase power to detect some novel disorders while decreasing power for disorders with pathophysiology different from known disorders. Our analyses also suggest that variant-level weights could be further improved by incorporating other variant prioritisation metrics, such as upweighting variants predicted to impact splicing, variants in particular protein domains, or variants that are somatic driver mutations during tumorigenesis. In developing DeNovoWEST, we explored applying both variant-level weights and gene-level weights in separate stages of the analysis, however, subtle but pervasive correlations between gene-level metrics (e.g. shet) and variant-level metrics (e.g. regional missense constraint, CADD) presents statistical challenges to implementation. Finally, the discovery of less penetrant disorders can be empowered by analytical methodologies that integrate both DNMs and rare inherited variants, such as TADA34. Nonetheless, using current methods focused on DNMs alone, we estimated that ~350,000 parent-child trios would need to be analysed to have ~80% power to detect HI genes with a 10-fold PTV enrichment. Discovering non-HI disorders will need even larger sample sizes. Reaching this number of sequenced families will be impossible for an individual research study or clinical centre, therefore it is essential that genetic data generated as part of routine diagnostic practice is shared with the research community such that it can be aggregated to drive discovery of novel disorders and improve diagnostic practice.
Extended Data
Extended Data Figure 1. Exploring the remaining number of DD genes.
(a) Number of significant genes from downsampling full cohort and running DeNovoWEST’s enrichment test. (b) Results from modelling the likelihood of the observed distribution of de novo PTV mutations. This model varies the numbers of remaining haploinsufficient (HI) DD genes and PTV enrichment in those remaining genes. The 50% credible interval is shown in red and the 90% credible interval is shown in orange. Note that the median PTV enrichment in genes that are significant and known to operate via a loss-of-function mechanism (shown with an arrow) is 39.7.
Extended Data Table 1. Recurrent Mutations.
De novo single nucleotide variants with more than 9 recurrences in our cohort annotated with relevant information, such as CpG status, whether the impacted gene is a known somatic driver or germline selection gene, and diagnostic gene group (e.g. consensus). “Recur” refers to the number of recurrences. “Likely mechanism” refers to mechanisms attributed to this gene in the published literature.
| Symbol | Chr | Position | Ref | Alt | Consequence | Recur | Likely mechanism | CpG | Somatic Driver Gene | Germline Selection Gene | DD status |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PACS1 | 11 | 65978677 | C | T | missense | 36 | activating | Yes | - | - | consensus |
| PPP2R5D | 6 | 42975003 | G | A | missense | 22 | dominant negative | - | - | - | consensus |
| SMAD4 | 18 | 48604676 | A | G | missense | 21 | activating | - | Yes | - | consensus |
| PACS2 | 14 | 105834449 | G | A | missense | 13 | dominant negative | Yes | - | - | discordant |
| MAP2K1 | 15 | 66729181 | A | G | missense | 11 | activating | - | Yes | Yes | consensus |
| PPP1CB | 2 | 28999810 | C | G | missense | 11 | all missense/in frame | - | - | - | consensus |
| NAA10 | X | 153197863 | G | A | missense | 11 | all missense/in frame | Yes | - | - | consensus |
| MECP2 | X | 153296777 | G | A | stop gain | 11 | loss of function | Yes | - | - | consensus |
| CSNK2A1 | 20 | 472926 | T | C | missense | 10 | activating | - | - | - | consensus |
| CDK13 | 7 | 40085606 | A | G | missense | 10 | all missense/in frame | - | - | - | consensus |
| SHOC2 | 10 | 112724120 | A | G | missense | 9 | activating | - | - | - | consensus |
| PTPN11 | 12 | 112915523 | A | G | missense | 9 | activating | - | Yes | Yes | consensus |
| SMAD4 | 18 | 48604664 | C | T | missense | 9 | activating | Yes | Yes | - | consensus |
| SRCAP | 16 | 30748664 | C | T | stop gain | 9 | dominant negative | Yes | - | - | consensus |
| FOXP1 | 3 | 71021817 | C | T | missense | 9 | loss of function | Yes | - | - | consensus |
| CTBP1 | 4 | 1206816 | G | A | missense | 9 | dominant negative | Yes | - | - | discordant |
Supplementary Material
Acknowledgements
We thank the families and their clinicians for their participation and engagement. We are very grateful to our colleagues who assisted in the generation and processing of data. Inclusion of RadboudUMC data was in part supported by the Solve-RD project that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 779257. This work was in part financially supported by grants from the Netherlands Organization for Scientific Research: 917-17-353 to CG. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003]. This study makes use of DECIPHER which is funded by Wellcome. See www.ddduk.org/access.html for full acknowledgement. The DDD study would like to acknowledge the tireless work of Rosemary Kelsell. Finally we acknowledge the contribution of an esteemed DDD clinical collaborator, M. Bitner-Glindicz, who died during the course of the study.
Footnotes
Author contributions
J.K., K.E.S., L.W., K.J.A., M.E.H., C.G., and K.R. contributed to the generation of figures and writing of the manuscript. J.K., K.E.S., L.W., Z.Z., K.J.A., R.Y.E., G.G., S.H.L., H.C.M., J.F.M., E.B., R.P., M.R.F.R., and H.G.Y. contributed to the generation and quality control of data. J.K., K.E.S., L.W., Z.Z., K.J.A., R.I.T., J.F.M., P.J.S., P.D., E.J.G., N.H., J.L., I.M., A.Y., and K.R. developed methods, contributed data, or performed analyses. H.C.M., L.E.L.M.V., J.J., C.F.W., H.G.B., H.V.F., D.R.F., J.C.B., M.E.H., C.G., and K.R. provided experimental and analytical supervision. M.E.H., C.G., and K.R. provided project supervision.
Competing interests
Z.Z., K.J.A., R.I.T., J.J., and K.R. are employees of GeneDx. J.J. and K.R. are shareholders of OPKO. M.E.H. is a co-founder of, consultant to, and holds shares in, Congenica Ltd, a genetics diagnostic company.
Contributor Information
DDD Study consortium authors:
Silvia Borras, Caroline Clark, John Dean, Zosia Miedzybrodzka, Alison Ross, Stephen Tennant, Tabib Dabir, Deirdre Donnelly, Mervyn Humphreys, Alex Magee, Vivienne McConnell, Shane McKee, Susan McNerlan, Patrick J. Morrison, Gillian Rea, Fiona Stewart, Trevor Cole, Nicola Cooper, Lisa Cooper-Charles, Helen Cox, Lily Islam, Joanna Jarvis, Rebecca Keelagher, Derek Lim, Dominic McMullan, Jenny Morton, Swati Naik, Mary O’Driscoll, Kai-Ren Ong, Deborah Osio, Nicola Ragge, Sarah Turton, Julie Vogt, Denise Williams, Simon Bodek, Alan Donaldson, Alison Hills, Karen Low, Ruth Newbury-Ecob, Andrew M. Norman, Eileen Roberts, Ingrid Scurr, Sarah Smithson, Madeleine Tooley, Steve Abbs, Ruth Armstrong, Carolyn Dunn, Simon Holden, Soo-Mi Park, Joan Paterson, Lucy Raymond, Evan Reid, Richard Sandford, Ingrid Simonic, Marc Tischkowitz, Geoff Woods, Lisa Bradley, Joanne Comerford, Andrew Green, Sally Lynch, Shirley McQuaid, Brendan Mullaney, Jonathan Berg, David Goudie, Eleni Mavrak, Joanne McLean, Catherine McWilliam, Eleanor Reavey, Tara Azam, Elaine Cleary, Andrew Jackson, Wayne Lam, Anne Lampe, David Moore, Mary Porteous, Emma Baple, Júlia Baptista, Carole Brewer, Bruce Castle, Emma Kivuva, Martina Owens, Julia Rankin, Charles Shaw-Smith, Claire Turner, Peter Turnpenny, Carolyn Tysoe, Therese Bradley, Rosemarie Davidson, Carol Gardiner, Shelagh Joss, Esther Kinning, Cheryl Longman, Ruth McGowan, Victoria Murday, Daniela Pilz, Edward Tobias, Margo Whiteford, Nicola Williams, Angela Barnicoat, Emma Clement, Francesca Faravelli, Jane Hurst, Lucy Jenkins, Wendy Jones, V. K. Ajith Kumar, Melissa Lees, Sam Loughlin, Alison Male, Deborah Morrogh, Elisabeth Rosser, Richard Scott, Louise Wilson, Ana Beleza, Charu Deshpande, Frances Flinter, Muriel Holder, Melita Irving, Louise Izatt, Dragana Josifova, Shehla Mohammed, Aneta Molenda, Leema Robert, Wendy Roworth, Deborah Ruddy, Mina Ryten, Shu Yau, Christopher Bennett, Moira Blyth, Jennifer Campbell, Andrea Coates, Angus Dobbie, Sarah Hewitt, Emma Hobson, Eilidh Jackson, Rosalyn Jewell, Alison Kraus, Katrina Prescott, Eamonn Sheridan, Jenny Thomson, Kirsty Bradshaw, Abhijit Dixit, Jacqueline Eason, Rebecca Haines, Rachel Harrison, Stacey Mutch, Ajoy Sarkar, Claire Searle, Nora Shannon, Abid Sharif, Mohnish Suri, Pradeep Vasudevan, Natalie Canham, Ian Ellis, Lynn Greenhalgh, Emma Howard, Victoria Stinton, Andrew Swale, Astrid Weber, Siddharth Banka, Catherine Breen, Tracy Briggs, Emma Burkitt-Wright, Kate Chandler, Jill Clayton-Smith, Dian Donnai, Sofia Douzgou, Lorraine Gaunt, Elizabeth Jones, Bronwyn Kerr, Claire Langley, Kay Metcalfe, Audrey Smith, Ronnie Wright, David Bourn, John Burn, Richard Fisher, Steve Hellens, Alex Henderson, Tara Montgomery, Miranda Splitt, Volker Straub, Michael Wright, Simon Zwolinski, Zoe Allen, Birgitta Bernhard, Angela Brady, Claire Brooks, Louise Busby, Virginia Clowes, Neeti Ghali, Susan Holder, Rita Ibitoye, Emma Wakeling, Edward Blair, Jenny Carmichael, Deirdre Cilliers, Susan Clasper, Richard Gibbons, Usha Kini, Tracy Lester, Andrea Nemeth, Joanna Poulton, Sue Price, Debbie Shears, Helen Stewart, Andrew Wilkie, Shadi Albaba, Duncan Baker, Meena Balasubramanian, Diana Johnson, Michael Parker, Oliver Quarrell, Alison Stewart, Josh Willoughby, Charlene Crosby, Frances Elmslie, Tessa Homfray, Huilin Jin, Nayana Lahiri, Sahar Mansour, Karen Marks, Meriel McEntagart, Anand Saggar, Kate Tatton-Brown, Rachel Butler, Angus Clarke, Sian Corrin, Andrew Fry, Arveen Kamath, Emma McCann, Hood Mugalaasi, Caroline Pottinger, Annie Procter, Julian Sampson, Francis Sansbury, Vinod Varghese, Diana Baralle, Alison Callaway, Emma J. Cassidy, Stacey Daniels, Andrew Douglas, Nicola Foulds, David Hunt, Mira Kharbanda, Katherine Lachlan, Catherine Mercer, Lucy Side, I. Karen Temple, and Diana Wellesley
Data Availability
Sequence and variant level data and phenotypic data for the DDD study data are available through EGA study ID EGAS00001000775.
RadboudUMC sequence and variant level data cannot be made available through EGA due to the nature of consent for clinical testing. To access the data, please contact christian.gilissen@radboudumc.nl with a request. Data sharing will be dependent on patient consent, diagnostic status of the patient, type of request, and the potential benefit to the patient. GeneDx data cannot be made available through EGA due to the nature of consent for clinical testing. GeneDx-referred patients are consented for aggregate, de-identified research and subject to US HIPAA privacy protection. As such, we are not able to share patient-level BAM or VCF data, which is potentially identifiable without a HIPAA Business Associate Agreement. Access to the de-identified aggregate data used in this analysis is available upon request to GeneDx. GeneDx has contributed deidentified data to this study to improve clinical interpretation of genomic data, in accordance with patient consent and in conformance with the ACMG position statement on genomic data sharing (see Supplementary Note for details). Clinically interpreted variants and associated phenotypes from the DDD study are available through DECIPHER (https://decipher.sanger.ac.uk)
Clinically interpreted variants from RUMC are available from the Dutch national initiative for sharing variant classifications (https://www.vkgl.nl/nl/diagnostiek/vkgl-datashare-database) as well as LOVD (https://databases.lovd.nl/shared/variants), where they are listed with “VKGL-NL_Nijmegen” as the owner
Clinically interpreted variants from GeneDx are deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar). GeneDx’s submitter ID is 26957 (https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/)
Genome Aggregation Database (gnomAD v2.1.1; https://gnomad.broadinstitute.org/)
The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov)
Developmental Disorders Genotype-Phenotype Database (DDG2P; https://www.ebi.ac.uk/gene2phenotype/downloads)
Code Availability
The DeNovoWEST method is available on GitHub (https://github.com/queenjobo/DeNovoWEST) along with code to recreate all figures in the manuscript. DOI: 0.5281/zenodo.3909398. Code to run the Phenopy method is also available on GitHub (https://github.com/GeneDx/phenopy).
References
- 1.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin HC, et al. Quantifying the contribution of recessive coding variation to developmental disorders. Science. 2018;362:1161–1164. doi: 10.1126/science.aar6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samocha KE, et al. Regional missense constraint improves variant deleteriousness prediction. bioRxiv. 2017 doi: 10.1101/148353. 148353. [DOI] [Google Scholar]
- 5.Kosmicki JA, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coe BP, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson JT, et al. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villegas F, et al. Lysosomal Signaling Licenses Embryonic Stem Cell Differentiation via Inactivation of Tfe3. Cell Stem Cell. 2019;24:257–270.e8. doi: 10.1016/j.stem.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Diaz J, Berger S, Leon E. TFE3-associated neurodevelopmental disorder: A distinct recognizable syndrome. Am J Med Genet A. 2020;182:584–590. doi: 10.1002/ajmg.a.61437. [DOI] [PubMed] [Google Scholar]
- 13.Jaganathan K, et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Yilmaz R, et al. A recurrent synonymous KAT6B mutation causes Say-Barber- Biesecker/Young-Simpson syndrome by inducing aberrant splicing. Am J Med Genet A. 2015;167A:3006–3010. doi: 10.1002/ajmg.a.37343. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Pang E, Lin K, Pei Z-M. Improving the measurement of semantic similarity between gene ontology terms and gene products: insights from an edge- and IC-based hybrid method. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066745. e66745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catterall WA, Dib-Hajj S, Meisler MH, Pietrobon D. Inherited neuronal ion channelopathies: new windows on complex neurological diseases. J Neurosci. 2008;28:11768–11777. doi: 10.1523/JNEUROSCI.3901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasser M, Tiber J, Lowery LA. The Role of the Microtubule Cytoskeleton in Neurodevelopmental Disorders. Front Cell Neurosci. 2018;12:165. doi: 10.3389/fncel.2018.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton MJ, et al. Heterozygous mutations affecting the protein kinase domain of cause a syndromic form of developmental delay and intellectual disability. J Med Genet. 2018;55:28–38. doi: 10.1136/jmedgenet-2017-104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martincorena I, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2018;173:1823. doi: 10.1016/j.cell.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi H, Dong C, Chung WK, Wang K, Shen Y. Deep Genetic Connection Between Cancer and Developmental Disorders. Hum Mutat. 2016;37:1042–1050. doi: 10.1002/humu.23040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goriely A, Wilkie AOM. Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am J Hum Genet. 2012;90:175–200. doi: 10.1016/j.ajhg.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 25.Maher GJ, et al. Visualizing the origins of selfish de novo mutations in individual seminiferous tubules of human testes. Proc Natl Acad Sci U S A. 2016;113:2454–2459. doi: 10.1073/pnas.1521325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maher GJ, et al. Selfish mutations dysregulating RAS-MAPK signaling are pervasive in aged human testes. Genome Res. 2018;28:1779–1790. doi: 10.1101/gr.239186.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young LC, et al. SHOC2-MRAS-PP1 complex positively regulates RAF activity and contributes to Noonan syndrome pathogenesis. Proc Natl Acad Sci U S A. 2018;115:E10576–E10585. doi: 10.1073/pnas.1720352115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coe BP, et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet. 2019;51:106–116. doi: 10.1038/s41588-018-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lord J, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet. 2019;393:747–757. doi: 10.1016/S0140-6736(18)31940-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cassa CA, et al. Estimating the selective effects of heterozygous protein-truncating variants from human exome data. Nat Genet. 2017;49:806–810. doi: 10.1038/ng.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satterstrom FK, et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deelen P, et al. Improving the diagnostic yield of exome- sequencing by predicting gene- phenotype associations using large-scale gene expression analysis. Nat Commun. 2019;10:2837. doi: 10.1038/s41467-019-10649-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003671. e1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence and variant level data and phenotypic data for the DDD study data are available through EGA study ID EGAS00001000775.
RadboudUMC sequence and variant level data cannot be made available through EGA due to the nature of consent for clinical testing. To access the data, please contact christian.gilissen@radboudumc.nl with a request. Data sharing will be dependent on patient consent, diagnostic status of the patient, type of request, and the potential benefit to the patient. GeneDx data cannot be made available through EGA due to the nature of consent for clinical testing. GeneDx-referred patients are consented for aggregate, de-identified research and subject to US HIPAA privacy protection. As such, we are not able to share patient-level BAM or VCF data, which is potentially identifiable without a HIPAA Business Associate Agreement. Access to the de-identified aggregate data used in this analysis is available upon request to GeneDx. GeneDx has contributed deidentified data to this study to improve clinical interpretation of genomic data, in accordance with patient consent and in conformance with the ACMG position statement on genomic data sharing (see Supplementary Note for details). Clinically interpreted variants and associated phenotypes from the DDD study are available through DECIPHER (https://decipher.sanger.ac.uk)
Clinically interpreted variants from RUMC are available from the Dutch national initiative for sharing variant classifications (https://www.vkgl.nl/nl/diagnostiek/vkgl-datashare-database) as well as LOVD (https://databases.lovd.nl/shared/variants), where they are listed with “VKGL-NL_Nijmegen” as the owner
Clinically interpreted variants from GeneDx are deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar). GeneDx’s submitter ID is 26957 (https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/)
Genome Aggregation Database (gnomAD v2.1.1; https://gnomad.broadinstitute.org/)
The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov)
Developmental Disorders Genotype-Phenotype Database (DDG2P; https://www.ebi.ac.uk/gene2phenotype/downloads)
The DeNovoWEST method is available on GitHub (https://github.com/queenjobo/DeNovoWEST) along with code to recreate all figures in the manuscript. DOI: 0.5281/zenodo.3909398. Code to run the Phenopy method is also available on GitHub (https://github.com/GeneDx/phenopy).