Abstract
The envelope spike (S) glycoprotein of the severe acute respiratory syndrome associated coronavirus (SARS-CoV) mediates the entry of the virus into target cells. Recent studies point out to a cell entry mechanism of this virus similar to other enveloped viruses, such as HIV-1. As it happens with other viruses peptidic fusion inhibitors, SARS-CoV S protein HR2-derived peptides are potential therapeutic drugs against the virus. It is believed that HR2 peptides block the six-helix bundle formation, a key structure in the viral fusion, by interacting with the HR1 region. It is a matter of discussion if the HIV-1 gp41 HR2-derived peptide T20 (enfuvirtide) could be a possible SARS-CoV inhibitor given the similarities between the two viruses. We tested the possibility of interaction between both T20 (HIV-1 gp41 HR2-derived peptide) and T-1249 with S protein HR1- and HR2-derived peptides. Our biophysical data show a significant interaction between a SARS-CoV HR1-derived peptide and T20. However, the interaction is only moderate (KB = (1.1 ± 0.3) × 105 M−1). This finding shows that the reasoning behind the hypothesis that T20, already approved for clinical application in AIDS treatment, could inhibit the fusion of SARS-CoV with target cells is correct but the effect may not be strong enough for application.
Keywords: T20, Enfuvirtide, T-1249, Fusion inhibitor, HIV-1, SARS-CoV
1. Introduction
The outbreak of severe acute respiratory syndrome (SARS) was caused by a novel coronavirus, the SARS-associated coronavirus (SARS-CoV) [1], [2], [3], [4], [5]. As SARS-CoV can replicate in an animal reservoir, it might be reintroduced in the human population (even as a more dangerous strain), originating a seasonal disease [6]. Considering that there is no specific treatment for this infection, it is necessary to develop antiviral agents to prevent possible future epidemics. Coronavirus are enveloped viruses and their entry into target cells is mediated by the spike (S) envelope glycoprotein [5], [7]. Although SARS-CoV S protein can be classified as a class I viral fusion protein, some features set it apart: for instance, cleavage between the S1 and S2 subunits of the S protein is not a requirement for fusion [5], [7]. The S1 domain (N-terminal region) contains the sequence responsible for the binding to the SARS-CoV cell receptor ACE2 (angiotensin-converting enzyme 2) [8], [9]. The S2 domain (membrane-anchored C-terminal region) mediates membrane fusion, and contains two heptad repeat regions, HR1 and HR2, forming coiled-coil structures [10], [11], [12], [13], [14]. In a number of viruses, including SARS-CoV [10], [11], [12], [13], [14], HR regions have determining importance for viral fusion: these regions form a six-helix bundle structure, consisting of an inner trimeric coiled coil formed by HR1 domains, with HR2 domains surrounding the trimer, packed in antiparallel orientation. This structure brings the viral and cellular membranes into close proximity, leading to membrane fusion. Synthetic peptides derived from the HR were shown to inhibit infection by either retroviruses (e.g., [15], [16], [17], [18]), paramyxoviruses (e.g., [19]) or coronavirus (e.g., [20]). The peptides derived from the HR2 region can inhibit infection by interfering with the six-helix bundle formation. Thus, based on the behavior of other enveloped viruses like HIV-1, recent studies have suggested a cell entry mechanism of SARS-CoV similar to viruses with class I viral fusion proteins [10], [11], [12], [13]. The potential of HR2-derived peptides as inhibitors of SARS-CoV infection was demonstrated [10], [12], [14]. In addition, based on a structural analysis of HIV-1 gp41 and SARS-CoV S protein, Gallaher and Garry proposed that T20 (enfuvirtide) could inhibit SARS-CoV infectivity, although with a limited activity [21]. T20 is a HIV-1 gp41 HR2-derived peptide that blocks entry of HIV-1 into the target cells, which is already in clinical use [22]. T-1249 is a second-generation HIV-1 fusion inhibitor with promising results [23]. In the present study, we have explored the potential of the HIV-1 HR2-derived peptide T20 and the related peptide T-1249 against SARS-CoV, using S protein HR1- and HR2-derived peptides (Table 1 ). The results obtained confirmed the occurrence of an interaction between the HIV-1 peptides and peptides derived from SARS-CoV HR1 as speculated [21]. However, the interaction does not seem to be strong enough to grant therapeutic efficiency, which justifies the limited activity foreseen by Gallaher and Garry [21] and recently confirmed in infectivity assays [24].
Table 1.
Sequences of HIV-1 fusion inhibitor peptides T20 and T-1249, and of the peptides derived from the HR1 and HR2 regions of SARS-CoV spike protein (Pep 1D, Pep 1E and Pep 2B, Pep HR2, respectively)
| Peptide | Protein location | Sequence |
|---|---|---|
| T20 | – | YTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWF |
| T-1249 | – | WQEWEQKITALLEQAQIQQEKNEYELQKLDKWASLWEWF |
| Pep 1D | 900–921 | ENQKQIANQFNKAISQIQESLT |
| Pep 1E | 931–951 | QDVVNQNAQALNTLVKQLSSN |
| Pep 2B | 1157–1178 | SVVNIQKEIDRLNEVAKNLNES |
| Pep HR2 | 1150–1189 | DISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYE |
The corresponding amino acid residues in SARS-CoV S protein of the derived peptides used is in the table.
2. Experimental
2.1. Materials
T20 and T-1249 were a kind gift from Roche (Palo Alto, CA, USA). All the Nα-l-Fmoc-protected amino acids, coupling reagents (DIC) and HOBt were obtained from GL Biochem (Shanghai, China). Fmoc rink MBHA resin (loading 0.44 mmol/g, 1% DVB, 100–200 mesh) was purchased from Hecheng, Co. (Tianjin, China). Other reagents used in peptide synthesis were obtained commercially and were of analytical grade. Unless otherwise stated, all solvents for synthesis were of analytical grade and used without further purification. DMF was redistilled under reduced pressure after dried over 4 Å molecular sieves overnight. 1,8-ANS (8-phenylamino-1-naphthalenesulfonic acid) and di-8-ANEPPS (4-[2-[6-(dioctylamino)-2-naphthalenyl]ethenyl]-1-(3-sulfopropyl)-pyridinium) were purchased from Molecular Probes (Eugene, OR, USA). POPC (1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine) was from Avanti Polar-Lipids (Alabaster, AL, USA). HEPES and NaCl were from Merck (Darmstadt, Germany). 10 mM HEPES buffer pH 7.4, 150 mM NaCl, was used throughout the studies. T20 and T-1249 stock solutions in buffer were diluted to final desired concentrations. Stock solutions of SARS-CoV S protein-derived peptides were prepared in buffer with small amounts of DMSO and diluted to final concentrations. Through the experiments, DMSO concentration in the samples was at most 1.4% (v/v). All peptides solubilization required mild sonication, mainly HR1-derived peptides. The spectrofluorimeter used was a SLM Aminco 8100 with double monochromators and 450 W Xe lamp.
2.2. Methods
2.2.1. Peptide synthesis
The peptides were synthesized by a standard Fmoc/t-Bu protection strategy. After removal of Fmoc of Fmoc-rink MBHA resin by 20% Piperidine/DMF (v:v) treatment for 10 min twice, the resin was thoroughly washed by DMF, Methanol, DCM and DMF, three times each. Fmoc protective amino acid was then coupled onto the resin for 45 min at room temperature using 3-fold excess molar ratio of amino acid and 3-fold excess DIC and HOBt as coupling and additive reagents, respectively. Until Kaiser's test [25] showing to be negative, the Fmoc-deprotection and washing steps were performed again as described above. The next amino acid assembly was repeated. In case of Kaiser's test to be positive or weakly positive, a double coupling step was carried out until Kaiser's test to be negative. Otherwise, the remaining amino group after two-coupling steps was captured by 15% Ac2O/DCM (v:v) for 30 min at room temperature. After all amino acids were successfully assembled onto resin, the last Fmoc group was removed and the resin was rinsed thoroughly with DMF, methanol and DCM. The resin must be completely dried under airflow at room temperature before the peptide was cleaved off resin.
Crude peptide attaching onto 100 mg resin was treated by TFA (4.0 mL), thioanisole (0.2 mL), phenol (0.3 g), deionized water (0.2 mL) and EDT (0.1 mL) for 2 h at room temperature. The ‘peptide + resin’ mixture was filtered and the tube was rinsed with 2–3 mL cleavage mixture. The combined solution was dried under a slow flow of nitrogen gas and then treated with ice-cooled methyl t-butyl ether-petroleum ether (3:1) to precipitate the peptide. The crude peptide was washed twice with ice-cooled methyl t-butyl ether-petroleum ether (3:1) and air dried naturally. The crude peptide was dissolved in 30–60% CH3CN–H2O (v:v) and purified by HPLC (Gilson 322 pump, Gilson UV/vis-152 detector, and Gilson 215 liquid handler) on a semi-preparative Vydac C18 column. The flow rate was 5 mL/min. The solvents were A: 5% CH3CN in H2O, B: 5% H2O in CH3CN. The pH was adjusted with TFA 0.05% (v/v). The detecting wavelength was 214 nm. Depending on the peptide sequences, the gradient was adjusted until the expected HPLC peak was separated very well. Collected fractions were analyzed by our HPLC-MS system on a Vydac C18 analytic column (4.6 μm × 25 cm). When both a purity of peptide over 95% and correct molecular weight were obtained, the peptide was lyophilized and used as a powder for subsequent studies.
2.2.2. SARS-CoV S protein-derived peptides influence on T20 and T-1249 fluorescence
S peptides are not intrinsically fluorescent, except for Pep HR2, which is weakly fluorescent in the operation conditions of our study, due to the Tyr residue. However, the presence of tryptophan residues in T20 and T-1249 enable the use of fluorescence techniques to study these molecules. To study the influence of S peptides on T20 (10 μM) or T-1249 (6.7 μM) fluorescence emission (λ exc = 280 nm; λ em = 350 nm) and conclude on mutual association, these peptides were titrated with successive additions of small amounts of S peptides (or just the solvent for controls). After each addition of peptide the sample was incubated for 10 min before measurement. Fluorescence intensity data was corrected for the dilution effect.
2.2.3. Hydrophobic interactions between T20 or T-1249 and S protein SARS-CoV-derived peptides
The existence of hydrophobic interactions between T20 or T-1249 and S peptides was studied by means of the ANS probe fluorescence (λ exc = 372 nm; λ em = 480 nm). ANS was added (final concentration 26 μM) to a sample of a selected peptide (6.7 μM for T-1249 and 10 μM for all the other peptides), and the other peptide was added in successive aliquots. The inverse situation (i.e., exchange of titrated and added peptide) was also carried out. To account for the possible self-aggregation of the added peptides, the same procedure was repeated in the absence of the titrated peptide, i.e., titration of an aqueous ANS solution only. After each peptide addition, the sample was incubated for 10 min before measurement. DMSO additions, in the same amounts present in peptide solutions, were used to control its effect on ANS fluorescence. Fluorescence intensity data were corrected for the dilution effect.
2.2.4. Membrane partition of SARS-CoV S protein-derived peptides
POPC (in chloroform) and di-8-ANEPPS (from a stock solution in ethanol) were mixed in a round bottom flask, and the solution was dried under a stream of nitrogen. Solvent removal was completed in vacuum, overnight. Large unilamellar vesicles (LUV) were prepared by extrusion techniques [26]. Peptides were added afterwards. Di-8-ANEPPS excitation spectra were obtained with emission at 603 nm [27], [28]. The final concentrations used were 200 μM for lipids, 10 μM for di-8-ANEPPS and 15 μM for all peptides.
2.2.5. Membrane partition of T20 and T-1249 in the presence of SARS-CoV S protein-derived peptides
POPC LUV were prepared by extrusion techniques [26] as before, except that the di-8-ANEPPS probe was not used. Membrane partition studies were performed by successive additions of small volumes of LUV (15 mM) to the peptide samples, with 10 min incubation in between. The peptide samples were composed of T20 or T-1249 alone or in mixtures with equimolar concentration of S peptides (T20 10 μM and T-1249 6.7 μM). The excitation wavelength used was 280 nm and the emission wavelength was 350 nm. Fluorescence intensity data were corrected for the dilution effect.
3. Results
3.1. Pep 1D presence lead to fluorescence variations in T20
S peptides (Table 1) influence on T20 and T-1249 fluorescence emission was used for the evaluation of peptides interactions. Titration of T20 with Pep 1D (Fig. 1A) or Pep 1E (results not shown) leads to a fluorescence emission intensity increase. A similar but not so pronounced result was obtained with T-1249 (Fig. 1B). Fluorescence variation was not detected for any of the others peptide pairs, e.g., T20 or T-1249 with Pep 2B (Fig. 1A and B, respectively). DMSO presence has no effect on T20 and T-1249 fluorescence intensity.
Fig. 1.
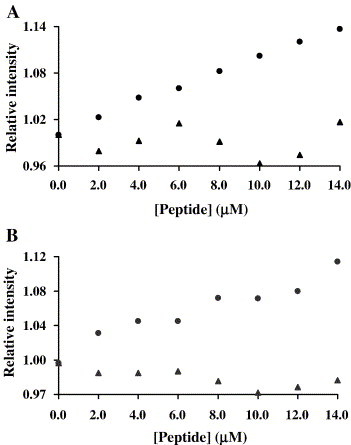
T20 (A) and T-1249 (B) fluorescence intensity variation in the presence of Pep 1D (●) and Pep 2B (▴). Small amounts of S peptides were added to a sample of T20 (10 μM) or T-1249 (6.7 μM).
3.2. Hydrophobic interactions occur between Pep 1D and T20
ANS (an environmentally sensitive probe) is essentially non fluorescent in water, becoming fluorescent in a less polar environment, e.g., when bound to hydrophobic sites in proteins [29], [30]. It was shown before that this probe can be used to study peptide assemblies [30]. Therefore, we used the increase in ANS fluorescence intensity (increased quantum yield) to study the existence of hydrophobic binding between T20 or T-1249 and S peptides because this binding may lead to the formation of hydrophobic grooves where ANS can insert, with a concomitant increase in fluorescence intensity. To evaluate peptides interactions, pair-wise tests were carried out. Small amounts of one peptide were added to the other peptide/ANS sample. For each pair of studied peptides, reverse titration (i.e., titration of the first peptide with the second) was also carried out. To take into account eventual self-aggregation of the titrating peptide, all peptides were used to titrate ANS samples in buffer. The results show that Pep 1D (Fig. 2A), T-1249 (Fig. 2C) and Pep 1E (data not shown) self-aggregate. However, the ANS fluorescence increase observed with the addition of Pep 1D to T20 is larger than the observed with Pep 1D alone (self-aggregation), which is indicative of mutual binding between these peptides—Fig. 2A. The T20/Pep 1E mixture also indicates interaction (data not shown). Although similar, T-1249/Pep 1D results exhibit a larger variability (Fig. 2C). T-1249 does not interact with Pep 1E. Both T20 and T-1249 show no evidence of interaction when the other two S peptides are used (results for the T20/Pep 2B and T-1249/Pep 2B systems are shown in Fig. 2B and D, respectively, as an example). No DMSO effect on ANS fluorescence was detected. Peptides Pep 1D and Pep HR2 interact with each other (data not shown), in agreement with the results obtained by Liu et al. [12]. This result serves as a positive control of the method.
Fig. 2.
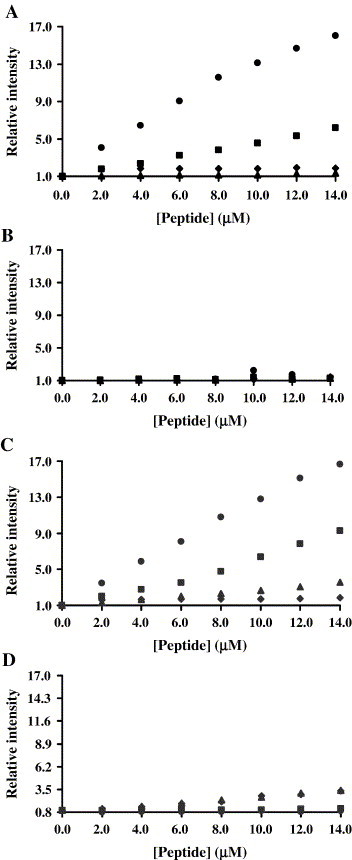
ANS (26 μM) fluorescence intensity for the titration of (A) T20 with Pep 1D (and vice-versa), (B) T20 with Pep 2B (and vice-versa), (C) T-1249 with Pep 1D (and vice-versa) and (D) T-1249 with Pep 2B (and vice-versa). Small amounts of Pep 1D or Pep 2B were added to a T20 or T-1249 sample containing ANS, and vice-versa. T20 or T-1249 added to S peptide (♦) and S peptide added to T20 or T-1249 (●) lead to different results. Small amounts of each peptide (▴—T20 or T-1249; ▪—S peptide, Pep 1D in panel A and C and Pep 2B in panel B and D) were added to a buffer/ANS sample to evaluate self-aggregation.
3.3. S protein-derived peptides do not interact with membranes
Due to the lack of intrinsic fluorescence in S peptides, the interaction studies with model membranes could not be done by a direct method. Thus, di-8-ANEPPS was used as a probe for peptide–membrane interaction. The magnitude of the membrane dipole potential is affected by membrane binding and by the insertion of molecules as such peptides. This change on the potential magnitude may be monitored by means of spectral shifts of the fluorescence indicator di-8-ANEPPS [27], [28]. Fig. 3 shows the fluorescence difference spectra for all S peptides, as well as for T20 and T-1249. Fluorescence difference spectra were obtained by subtracting the excitation spectrum of di-8-ANEPPS-labeled POPC membranes in the absence and presence of the peptides. Before subtraction, the spectra were normalized to the integrated areas so that the difference spectra reflect only spectral shifts. Fluorescence intensity difference spectra for T20 and T-1249 (peptides known to interact with membranes [31], [32]) and S peptides show no interaction of the latter with the membranes. The small DMSO amount present in the samples does not perturb membrane structure [33].
Fig. 3.
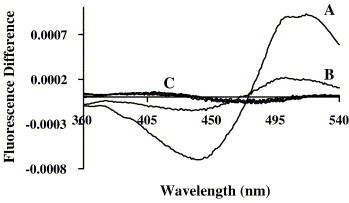
Di-8-ANEPPS-labeled (10 μM) membranes (POPC 200 μM LUV) fluorescence difference spectra (λem = 603 nm). The spectra were obtained by subtracting the excitation spectrum before the addition of peptides from the excitation spectra after the peptides addition (15 μM). Before subtraction, the spectra were normalized to the integrated areas so that the difference spectra reflect only the spectral shifts. (A) T-1249, (B) T20 and (C) S peptides. The latter spectra have no meaningful difference among each other.
3.4. Pep 1D interferes with the membrane partition of T20
T20 and T-1249 membrane partition experiments were done in the presence and absence of S peptides as a tool to enable the calculation of peptide binding constants. A decrease in the partition occurs for T20 in the presence of Pep 1D (Fig. 4A). In the presence of Pep 1E the result is not so striking, although clear. T-1249 results were not conclusive. With Pep 2B and Pep HR2 no difference occurs in T20 and T-1249 partition (Fig. 4B). The small DMSO amounts used do not perturb membranes [33]. In agreement, DMSO had no significant effect on T20 and T-1249 partition to membranes.
Fig. 4.
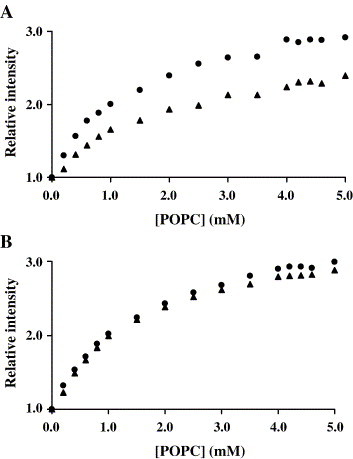
Partition plots of T20 (10 μM) to LUV of POPC. The experiments were done in the absence (●) and in the presence (▴) of equimolar S peptides Pep 1D (A) and Pep 2B (B).
4. Discussion
SARS-CoV membrane fusion mechanism is similar to that of class I viral fusion proteins, where HR regions have a very important role. The HR1 regions form a trimer where the HR2 regions pack onto the grooves in an antiparallel manner to form the six-helix bundle [12]. This structure leads to a close apposition of viral and target cell membranes, favoring the membrane fusion. The SARS-CoV S protein HR2-derived peptides are able to inhibit viral entry. HR2 peptides compete for binding to the HR1 region of the SARS-CoV S protein, blocking the six-helix bundle structure formation, and thus inhibiting fusion [10], [12]. In our studies, we have explored the interaction between T20 (HIV-1 gp41 HR2-derived peptide) or T-1249 and SARS-CoV S protein HR1 (Pep 1D or Pep 1E)- or HR2 (Pep 2B or Pep HR2)-derived peptides, in order to explore a possible SARS-CoV fusion inhibition by the first ones.
T20 fluorescence increases in the presence of Pep 1D or Pep 1E (although with a better signal/noise ratio in the first case), but not in the presence of the other S peptides. The same occurs for T-1249, although the results are not so evident. This increase in the fluorescence quantum yield probably indicates tryptophan residues exposure to a less polar environment, protected from the aqueous environment [34], revealing peptides interaction. Alternative explanations, such as a decrease in Trp fluorescence quenching by other peptide residues, cannot be ruled out. Nevertheless, the increase of ANS fluorescence intensity in the peptide/peptide titrations is an evidence for the formation of hydrophobic pockets. To assure that such increase is not due to the self-aggregation of the titrating peptide, instead of inter-association of different peptides, control experiments were performed in the absence of the titrated peptides. Although self-aggregation occurs for T-1249, Pep 1D and Pep 1E, a T20/Pep 1D inter-association of hydrophobic nature is evident. However, the titration of T20 with Pep 1D does not lead to precisely the same result as the opposite titration. The addition of Pep 1D to T20 lead to an ANS fluorescence increase larger than the increase caused by Pep 1D self-aggregation alone. The addition of T20 to Pep 1D does not result in a similar fluorescence increase. This occurs because T20 is not able to destroy Pep 1D aggregates, which prevent peptides interaction in a larger extent. The T20/Pep 1E and T-1249/Pep 1D systems lead to low signal/noise ratios, preventing straightforward conclusions. Yet, T-1249 and Pep 1E show no evidence for interaction. Both T20 and T-1249 show no interaction with HR2-derived peptides detectable by ANS fluorescence.
Di-8-ANEPPS was used to probe the peptides interaction with membranes. It is known from other studies that both T20 and T-1249 interact with membranes to a reasonable extent [31], [32]. In agreement with the previous results, T-1249 is the peptide that presents the most extensive interaction with membranes. From the comparison between all the fluorescence difference spectra intensity obtained (Fig. 3), it can be concluded that the S peptides have a very low or even null partition to membranes.
T20 and T-1249 membrane partition experiments were done in the absence and presence of S peptides (Fig. 4). T20 partition curves in the absence and in the presence of Pep 1D do not overlap. One possible explanation is that peptides interact in the aqueous medium and this additional equilibrium (interaction) competes with partition, decreasing the molar fraction of the molecules in the lipidic bilayers. The same occurs for T-1249/Pep 1D and for both peptides in the presence of Pep 1E. However, the statistical quality of the results worsens in these cases. In the presence of Pep 2B or Pep HR2, the partition of both T20 and T-1249 is not affected. It is possible to quantify the equilibria balance and rationalize the altered partition plots (Appendix A). This analysis takes into account the partition of T20 and T-1249 to LUV of POPC and the interaction in the aqueous medium with S peptides. Data fitting with Eq. (A1.7) (Appendix A) for T20 in the presence of Pep 1D (Appendix A; Fig. A1.2) leads to a binding coefficient of K B = (1.1 ± 0.3) × 105 M−1 (means ± standard deviation of four replicates). This value is in between the values obtained for very strong and specific binding, such as the binding of the S1 domain of SARS-CoV to ACE2 (∼108 M−1; Ref. [35]) and the values expected for moderately specific association pairs (∼102 M−1; Ref. [36]).
Taken together, the results obtained prove the occurrence of an interaction between T20 and Pep 1D. Taking into account that T20 is an HR2-derived peptide and Pep 1D is an HR1 peptide, our results seem to confirm at a first glance that SARS-CoV could be inhibited by T20, in agreement with the speculative reasoning by others [21]. Nevertheless, the interaction is too moderate to be able to provide therapeutic efficiency, even keeping in mind that SARS-CoV HR2 peptides are less potent fusion inhibitors than the corresponding HR2 peptides from other viruses [10], [12]. For SARS-CoV, the inhibitory peptides concentrations were in the micromolar range, while T20, for instance, has activity in the nanomolar range against HIV-1 entry. Although the inhibitory properties of HR2 peptides could be affected by many different factors, SARS-CoV six-helix bundle lower stability, caused by a weaker interaction between the HR1 and HR2 regions, could be directly correlated with a lower potency of the HR2 peptide in blocking the formation of this structure [10], [12], [37].
In conclusion, our studies indicate a significant interaction between T20 and an S protein HR1-derived peptide (Pep 1D). This interaction could in principle be responsible for the inhibition of SARS-CoV membrane fusion by the most accepted mechanism involving HR2-derived peptides. However, the values found for K B justify the absence of strong anti-SARS activity in the most recent activity studies [24]. Lastly, it is worth mentioning that although a potential use of Pep 1D to block HIV-1 fusion becomes apparent from our work, it is known that HR1-derived peptides usually are less potent fusion inhibitors than HR2-derived peptides [16], [38].
Acknowledgements
This project was partially funded by FCT-MCES (Portugal), including a grant (SFRH/BD/14336/2003) under the program POCTI to A.S.V., and by the Minister of Sciences and Technology, P. R. China (National Key Basic Research Program of China, No. 2003CB514109). T20 and T-1249 were kind gifts from Roche (Palo Alto, CA, USA).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbagen.2005.10.001.
Appendix A. Supplementary data
TX distribution: free in the bulk aqueous media (FW), interacting with PS in the bulk aqueous media (IW) and bound to POPC membranes (L).
Partition plots of T20 to LUV of POPC in the absence (•) and presence (▴) of Pep 1D. Eq. (A1.7) was fitted to the experimental data for T20 in the presence of Pep 1D (solid line) using the calculated Kp for T20 alone. KB is a fitting parameter.
References
- 1.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A., Humphrey C.D., Shieh W., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 4.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C., Chan K.H., Tsang D.N.C., Yung R.W.H., Ng T.K., Yuen K.Y., members of the SARS study group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Peñaranda S., Bankamp B., Maher K., Chen M., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Günther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Überla K., Niedrig M., Pöhlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao X., Dimitrov D.S. The SARS-CoV S glycoprotein. Cell. Mol. Life Sci. 2004;61:2428–2430. doi: 10.1007/s00018-004-4257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosch B.J., Martina B.E.E., van der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J.R., de Groot R., Osterhaus A.D.M.E., Rottier P.J.M. Severe acute respiratory coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingallinella P., Bianchi E., Finotto M., Cantoni G., Eckert D.M., Supekar V.M., Bruckmann C., Carfi A., Pessi A. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8709–8714. doi: 10.1073/pnas.0402753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y., Yan H., Cole D.K., Bell J.I., Rao Z., Tien P., Gao G.F. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 16.Eckert D.M., Kim P.S. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11187–11192. doi: 10.1073/pnas.201392898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilby J.M., Hopkins S., Venetta T.M., DiMassimo B., Cloud G.A., Lee J.Y., Alldredge L., Hunter E., Lambert D., Bolognesi D., Matthews T., Johnson M.R., Nowak M.A., Shaw G.M., Saag M.S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 18.Lu M., Blacklow S.C., Kim P.S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 19.Joshi S.B., Dutch R.E., Lamb R.A. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 20.Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.W.R. Gallaher, R.F. Garry, Model of the pre-insertion region of the spike (S2) fusion glycoprotein of the human SARS coronavirus: implications for antiviral therapeutics, http://www.virology.net/Articles/sars/s2model.html. (Accessed 13 May 2003).
- 22.Kilby J.M., Eron J.J. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 2003;348:2228–2238. doi: 10.1056/NEJMra022812. [DOI] [PubMed] [Google Scholar]
- 23.Eron J.J., Gulick R.M., Bartlett J.A., Merigan T., Arduino R., Kilby J.M., Yangco B., Diers A., Drobnes C., DeMasi R., Greenberg M., Melby T., Raskino C., Rusnak P., Zhang Y., Spence R., Miralles G.D. Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J. Infect. Dis. 2004;189:1075–1083. doi: 10.1086/381707. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto N., Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J., Rabenau H., Doerr H.W., Hunsmann G., Otaka A., Tamamura H., Fujii N., Yamamoto N. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 2004;318:719–725. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G., Lam K.S. One-bead one compound combinatorial library method. In: Fenniri H., editor. Combinatorial Chemistry: Practical Approach. Oxford Univ. Press; Oxford: 2000. pp. 33–49. [Google Scholar]
- 26.Mayer L.D., Hopes M.J., Cullis P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 27.Cladera J., Martin I., O'Shea P. The fusion domain of HIV gp41 interacts specifically with heparan sulfate on the T-lymphocyte cell surface. EMBO J. 2001;20:19–26. doi: 10.1093/emboj/20.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cladera J., O'Shea P. Intramembrane molecular dipoles affect the membrane insertion and folding of a model amphiphilic peptide. Biophys. J. 1998;74:2434–2442. doi: 10.1016/S0006-3495(98)77951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertsch M., Mayburd A.L., Kassner R.J. The identification of hydrophobic sites on the surface of proteins using absorption difference spectroscopy of bromophenol blue. Anal. Biochem. 2003;313:187–195. doi: 10.1016/s0003-2697(02)00590-0. [DOI] [PubMed] [Google Scholar]
- 30.Ganesh S., Jayakumar R. Circular dichroism and Fourier transform infrared spectroscopic studies on self-assembly of tetrapeptide derivative in solution and solvated film. J. Pept. Res. 2003;61:122–128. doi: 10.1034/j.1399-3011.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- 31.Veiga S., Henriques S., Santos N.C., Castanho M. Putative role of membranes in the HIV fusion inhibitor enfuvirtide mode of action at the molecular level. Biochem. J. 2004;377:107–110. doi: 10.1042/BJ20031350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veiga A.S., Santos N.C., Loura L.M.S., Fedorov A., Castanho M.A.R.B. HIV fusion inhibitor peptide T-1249 is able to insert or adsorb to lipidic bilayers. Putative correlation with improved efficiency. J. Am. Chem. Soc. 2004;126:14758–14763. doi: 10.1021/ja0459882. [DOI] [PubMed] [Google Scholar]
- 33.Abdiche Y.N., Myszka D.G. Probing the mechanism of drug/lipid membrane interactions using Biacore. Anal. Biochem. 2004;328:233–243. doi: 10.1016/j.ab.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Santos N.C., Castanho M. Fluorescence spectroscopy methodologies on the study of proteins and peptides. On the 150th anniversary of protein fluorescence. Trends Appl. Spectrosc. 2002;4:113–125. [Google Scholar]
- 35.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.St.C., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAB to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubota Y., Motoda Y., Shigemune Y., Fujisak Y. Fluorescence quenching of 10-methylacridinium chloride by nucleotides. Photochem. Photobiol. 1979;29:1099–1106. [Google Scholar]
- 37.Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfi A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldwin C.E., Sanders R.W., Berkhout B. Inhibiting HIV-1 entry with fusion inhibitors. Curr. Med. Chem. 2003;10:1633–1642. doi: 10.2174/0929867033457124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TX distribution: free in the bulk aqueous media (FW), interacting with PS in the bulk aqueous media (IW) and bound to POPC membranes (L).
Partition plots of T20 to LUV of POPC in the absence (•) and presence (▴) of Pep 1D. Eq. (A1.7) was fitted to the experimental data for T20 in the presence of Pep 1D (solid line) using the calculated Kp for T20 alone. KB is a fitting parameter.


