Abstract
STAT1 belongs to the STAT family of transcription factors, which comprises seven factors: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6. STAT1 is a 91 kDa protein originally identified as the mediator of the cellular response to interferon (IFN) α, and thereafter found to be a major component of the cellular response to IFNγ. STAT1 is, in fact, involved in the response to several cytokines and to growth factors. It is activated by cytokine receptors via kinases of the JAK family. STAT1 becomes phosphorylated and forms a dimer which enters the nucleus and triggers the transcription of its targets. Although not lethal at birth, selective gene deletion of STAT1 in mice leads to rapid death from severe infections, demonstrating its major role in the response to pathogens. Similarly, in humans who do not express STAT1, there is a lack of resistance to pathogens leading to premature death. This indicates a key, non-redundant function of STAT1 in the defence against pathogens. Thus, to successfully infect organisms, bacterial, viral or parasitic pathogens must overcome the activity of STAT1, and almost all the steps of this pathway can be blocked or inhibited by proteins produced in infected cells. Interestingly, some pathogens, like the oncogenic Epstein–Barr virus, have evolved a strategy which uses STAT1 activation.
1. Activation of STAT1
1.1. Molecular structure of STAT1
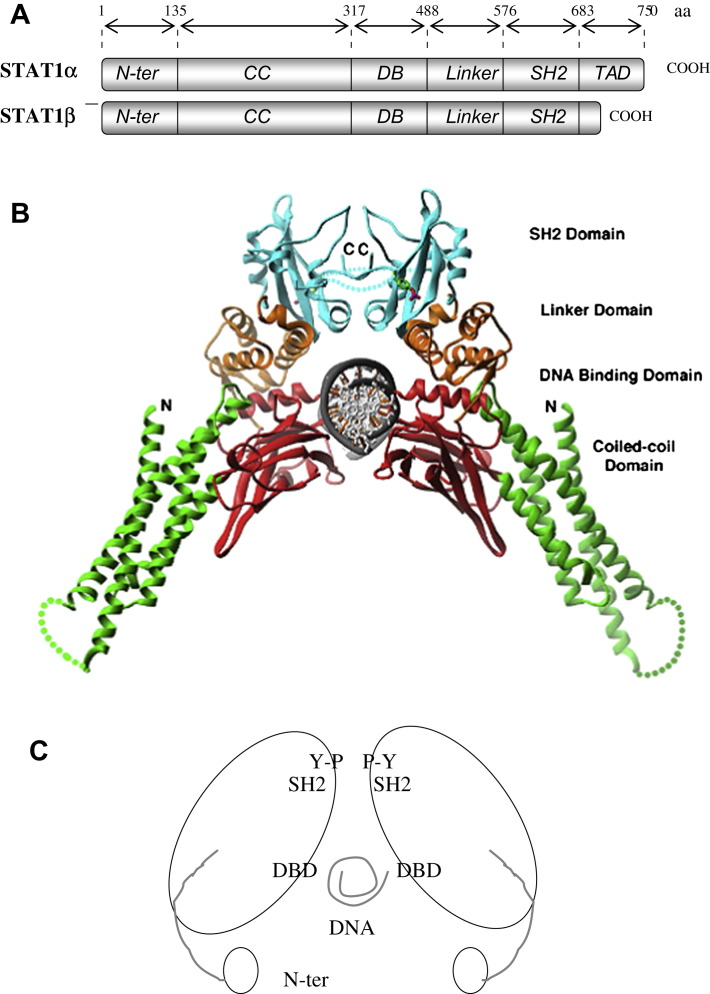
STAT1 was initially identified as an interferon α (IFNα) mediator [1], [2], and thereafter found to be a major component of the cellular response to IFNγ. STAT1 belongs to a family of transcription factors comprising STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6 [3], [4]. The transcript of STAT1 undergoes alternative splicing, resulting in two isoforms: STAT1α (91 kDa) and STAT1β (84 kDa) [5]. The α isoform possesses a complete transactivation domain (TAD) and two major phosphorylated sites: tyrosine 701 and serine 727. The β isoform is shorter and lacks most of the TAD, including serine 727; both isoforms contain an SH2 domain, a DNA-binding domain (DBD) and an N-terminal domain. Diffraction studies show STAT1 crystals dimers forming through interaction of the phosphotyrosine 701 and the SH2 domain (Fig. 1 ).
Fig. 1.
Molecular structure and ribbon model of the STAT1 dimer. A: Schematic molecular organization of STAT1α and STAT1β. B: 3D structure of STAT1 (residues 135–712) (from reference [56]). C. Schematic rendering of the STAT1 dimer in its phosphorylated form showing the SH2 domains interacting with tyrosine 701 (Y–P) the DNA-binding domain (DBD) interacting with DNA and the cup-and-ball-like N-terminal domain (adapted from reference [83]).
1.2. Stimuli that activate STAT1
STAT1 is an essential effector of IFNs. Following activation of the IFNγ receptor, its two subunits, IFNGR1 and IFNGR2 which are isolated in the absence of stimulation [6], [7], [8] become assembled [9], [10]. The two JAK family kinases, JAK1 and JAK2, which are constitutively bound to the inactive chains of the receptor, become activated, resulting in the autophosphorylation of JAK2, which in turn phosphorylates JAK1. The two kinases then phosphorylate the IFNR subunits, forming STAT1 binding sites [11]. STAT1 binds via its SH2 domain [12] and is phosphorylated on tyrosine 701 [13], [14] (Fig. 2 A). The activation of STAT1 following IFNα triggering is somewhat different. The subunit IFNAR2 of the IFNαR forms a complex with TYK2 and STAT2 in the absence of stimulation by IFNα, and the subunit IFNAR1 is associated to JAK1 [15], [16]. Following interaction of IFNα with the two subunits of its receptor [17], [18], JAK1 and TYK2 do not autophosphorylate, but instead phosphorylate one another [19], and subsequently phosphorylate both IFNAR1 and IFNAR2 [20], as well as the tyrosine 690 of STAT2 to which STAT1 binds through its SH2 domain, and STAT1 on tyrosine 701. The phosphorylated STAT1/STAT2 dimer is then released from the IFNAR2 chain (Fig. 2B). STAT1 is also activated in response to several interleukins, including IL2 and IL6 (see Table 1 and references [21], [22], [23], [24]); and in response to growth factors including EGF and PDGF (see Table 1 and reference [21]). Oncostatin M and growth hormone also activate STAT1: this occurs through activation of JAK2 which binds to signaling proteins such as Grb2, Ras or Raf [25], [26], [27]. Other factors such as angiotensin II [28], HGF [29] and TNF [30] activate STAT1. However, they appear to do so without inducing its nuclear translocation nor activating its DNA-binding, suggesting cytoplasmic functions for STAT1.
Fig. 2.
Mechanisms of activation of STAT1 in the cytoplasm following interferon receptor activation. A. Phosphorylation of STAT1 on tyrosine (Y) 701 by JAK1 and JAK2 following IFNγ stimulation. B. Phosphorylation of STAT1 on tyrosine (Y) 701 by JAK1 and TYK2 following stimulation by IFNα.
Table 1.
STAT1 activators.
| IFNs | IFNα, IFNβ et IFNω (omega) | [322] |
| IFNγ | [323] | |
| IFNλ (lambda) | [324] | |
| ILs | IL2, IL3, IL6, IL9–IL12, IL15, IL17, IL22 | [21] |
| IL21 | [22] | |
| IL26 | [23] | |
| IL27 | [24] | |
| FCs | EGF (epidermal growth factor) | [21] |
| VEGF (vascular endothelial growth factor) | ||
| FGF (fibroblaste growth factor) | ||
| HGF (hepatocyte growth factor) | ||
| Hs | GH (growth hormone), Angiotensine, Oncostatine M (OSM) | [21] |
1.3. Phosphorylation of tyrosine 701, serine 727, and of other residues
The phosphorylation on tyrosine 701 that follows the activation of JAK1 and JAK2 by the IFNγR or the activation of JAK1 and TYK2 by IFNαR is necessary for dimerisation [13]. However, if leucine 706 is replaced by a serine, phosphorylation on tyrosine 701 is no longer detectable following treatment of the cells with IFNγ. STAT1 retains nonetheless the capacity to dimerise and to form gamma interferon-activated (GAF) complexes [31], possibly through interaction of the N-terminal domains [32] (Fig. 1, Fig. 3 ).
Fig. 3.
Speculative model for the mechanism of STAT1 activation. Unphosphorylated dimers can form (left side of figure) by interaction of the N-terminal ends (marked N). When phosphorylated, the dimers form by interaction of SH2 domains with phosphotyrosine 701 (P-Y-701). Tetramers can also form by interaction of the N-terminal ends of the phosphorylated dimers, in two different conformations (adapted from reference [59]).
Several different kinases phosphorylate serine 727, including ERK 1/2 following IFNγ stimulation [33], [34], p38α following IFNγ [35], [36], LPS [36], [37], UV [36] and BCR stimulation [38], calmodulin kinase II (CaMKII) following IFNγ [39] or BCR stimulation [38], and PKCδ following IFNα [40] or IFNγ stimulation [41] (see Table 2 ). The mechanisms leading to serine 727 phosphorylation are yet to be elucidated. For instance, while PKCδ can phosphorylate STAT1 on serine 727 in response to stimulation by type II or type I IFNs [40], [41], serine 727 phosphorylation is unchanged in PKCδ-deficient macrophages [42]. In addition, under certain conditions of stimulation such as the addition of IFNγ [43], serine 727 phosphorylation may be entirely dependent on the phosphorylation of tyrosine 701; while under other conditions, such as UV treatment or lipopolysaccharide stimulation [44], [45], it is independent of it, suggesting that different subsets of protein kinases are involved. Interestingly, adenosine, an immunosuppressive compound, has been found to inhibit serine 727 phosphorylation [46].
Table 2.
Stimuli and kinases involved in the phosphorylation of serine 727 of STAT1.
Although on serine 727, phosphorylation is not required for the full activation of STAT1 [47], it seems to be required for certain target genes such as Mx (myxovirus), IRF1 or CBP [42]. Interestingly, mice expressing STAT1 with a S727L mutation were extremely sensitive to bacterial infection and had strongly reduced expression of IFNγ gene targets [48]. The phosphorylation of serine 727 in response to IFNγ is also dependent on the conserved leucine 724 residue [49]. The function of other known phosphorylated sites of STAT1 has not been completely elucidated. For instance, serine 708 phosphorylation following activation of the IκB Kinase ɛ (IKKɛ) by INFα [50] (Table 2). There may be a hierarchy between the phosphorylation sites of STAT1, but probably due to the many effectors involved, it is not currently understood.
1.4. Other mechanisms of activation
Acetylation and methylation are also involved in STAT1 activation. Direct methylation of STAT1 on arginine 31 by methyltransferase PRMT1 (protein R (R for arginine) methyltransferase 1) was suggested [51] but subsequently questionned [52], [53]. Recent work showed that it is the inactivator of STAT1, PIAS1 (protein inhibitor of STAT1-1), which is methylated by PRMT1, leading to increased affinity of STAT1 for its DNA targets [54]. Acetylation of STAT1 on leucine favors tyrosine 701 dephosphorylation (see: [55]).
1.5. Other conformations of STAT1
There are other configurations of the STAT1 dimer which may not depend on phosphorylation. Dimers of STAT1 can form from both phosphorylated and unphosphorylated STAT1 [56], [57]. In the case of unphosphorylated STAT1, the SH2 domains do not participate in dimerisation [57]: they are positioned at opposite ends of the dimer, whose conformation involves the interaction of the N-terminal domains, the coiled-coil domain and the DBD, resulting in an antiparallel conformation [57] (Fig. 3). The role of the N-terminal domain is probably important, although still unclear as its crystal structure is only partially elucidated due to its mobility. It is thought to play a regulatory role by allowing the formation of antiparallel dimers, thereby exposing tyrosine and facilitating its dephosphorylation [58]. In fact, a recent study suggests a regulatory role for the equilibrium between phosphorylated and unphosphorylated STAT1 dimers, this equilibrium is tilted toward parallel dimers by tyrosine phosphorylation [59]. One of the questions that still remains unanswered is the real nature of the functional unit formed by STAT1, including associated proteins.
2. Nucleo-cytoplasmic shuttling of STAT1
STAT1 is activated within the cytoplasm, and exerts its known biological function as a transcription factor in the nucleus. Activated STAT1 is transferred from the cytoplasm to the nucleus, and once released from its targets, returns to the cytoplasm. Several studies have shown that this process is complex and involves a combination of active transfer requiring specialised transfer proteins, and passive transfer, including transfer of non activated STAT1 molecules.
2.1. Nuclear import
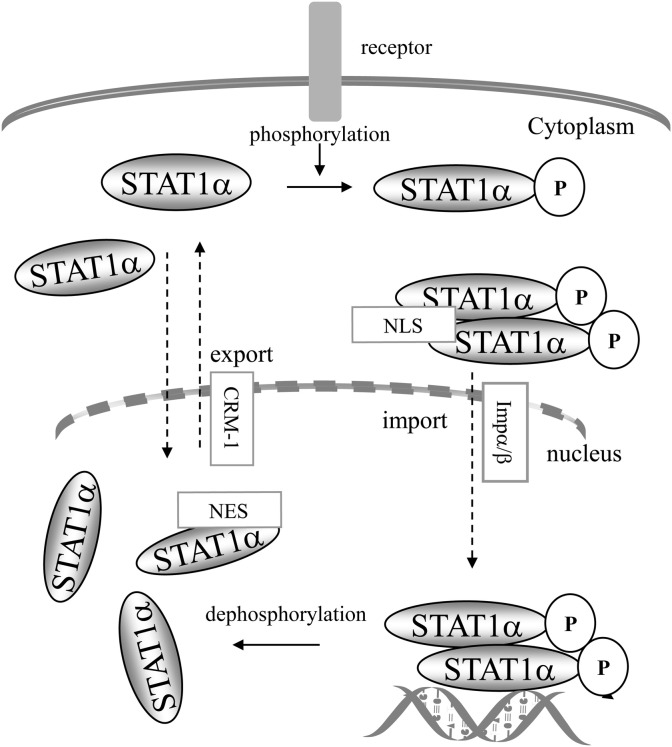
Following IFNα- or IFNγ stimulation, STAT1 becomes nuclear within minutes. It interacts with protein complexes (pore targeting complexes, PTACs) [60], comprising importin α and β (or karyopherin α and β) [61], [62] which interact with the NLS (nuclear localisation sequence) [63]. The NLS of STAT1 is dimer-specific [64], [65], [66], it comprises an arginine/lysine rich motif (R378, K379, K410, K413 and R418) that is located within the DBD of STAT1 [67] in which residue K413 is essential [64]. Residue L407 [65], and the N-terminal domain [68] are also essential, suggesting an important role for spatial conformation in nuclear translocation. Nuclear accumulation of STAT1 requires intact SH2 and tyrosine 701, and its driving force is the binding to DNA which prevents tyrosine dephosphorylation [69], [70]; tyrosine dephosphorylation of STAT1 appears to take place within the nucleus and to be required for STAT1's return to the cytoplasm [71], [72] (Fig. 4 ). However, STAT1 also undergoes a signal-independent constitutive nucleo-cytoplasmic shuttling whose mechanism is still unclear [66], [73]. Interestingly, under certain conditions of stimulation such as angiotensin II [74], TNF-α [30] or HGF [29], phosphorylated STAT1 dimers were found to be unable to enter the nucleus and bind DNA suggesting that some forms of STAT1 may have a stricktly cytoplasmic function. Thus, while it is clear that tyrosine 701 phosphorylation and dimerisation of STAT1 are key processes for the nuclear localisation of STAT1, there are mechanisms that are independent of the formation of dimers, but which are not clearly identified. Moreover, functions of STAT1 within the cytoplasm are probable, as was shown for STAT3, which, in its unphosphorylated form, binds NF-κB and activates a subset of κB-dependent genes [75].
Fig. 4.
Nucleo-cytoplasmic shuttling of STAT1. STAT1 becomes phosphorylated on tyrosine 701 in the cytoplasm and enters the nucleus by interaction of its dimer-specific NLS with importin α/β. Phosphorylated STAT1 interacts with its DNA targets; when released from DNA, STAT1 is dephosphorylated and can return to the cytoplasm involving interaction of the NES with CRM1. There is also a constitutive nucleo-cytoplasmic shuttle of unphosphorylated STAT1. (Adapted from [69]).
2.2. Nuclear export
STAT1 contains an NES (nuclear export sequence) located between residues 392 to 413, within the DBD [65]. This sequence interacts with exportin-1 (or CRM1) an essential component of the export of high molecular weight molecules to the cytoplasm [76], [77]. Nuclear export by exportin-1 requires that it interacts with the component Ran-GTP [78], [79]. The nuclear export of STAT1 requires its dephosphorylation on tyrosine 701, an event which occurs once STAT1 is released from its DNA target [69] (Fig. 4). However, nuclear export of STAT1 appears to be also mediated by other molecules than exportin-1. Comparative analysis of the nuclear shuttling of the α and the β isoforms also revealed interesting differences between both isoforms [80].
2.3. Constitutive nucleo-cytoplasmic shuttling of non activated STAT1
In unstimulated cells, STAT1 is detectable both in the cytoplasm and the nucleus, where it forms unphosphorylated dimers [81] (see Fig. 4). As mentioned above, unphosphorylated dimers have two possible conformations, an antiparallel conformation in which the CC domain of one monomer interacts with the DBD of the other monomer, and a parallel conformation [82], [83]. Nevertheless, active monomeric forms of STAT1 may exist.
3. STAT1 in transcription
3.1. Binding of STAT1 to its gene targets
The recognition sequence of STAT1 is an 8–10 base-pair sequence with the STAT-consensus TTN4–6AA. Two STAT1-specific consensus sequences have been identified, the GAS (Gamma interferon Activated Sequence), and the ISRE (Interferon Stimulated Response Element). They bind STAT1, as shown by EMSA (Electrophoretic Mobility Shift Assay), Chip (Chromatin Immuno-Precipitation) and reporter assays. The GAS sequence consists in a palindromic sequence (see Table 3 ) which interacts mostly with the GAF complex (see above, and Fig. 5 ) [84]. Some promoters, such as those of IFNγ [85] and MIG1 (monokine inducer by gamma 1) [86] contain tandem GAS sequences with which STAT1 homodimers interact via their N-terminal domain, an interaction which apparently stabilises the complex and increases the expression of the target gene [85], [87]. The ISRE sequence is composed of repeats of the motif 5′-TTTC-3′ or its complement 5′-GAAA-3′ (underlined in Table 3) separated by one or two nucleotides [88]. In the case of the promoter of the ISG-15 gene, IRF9 binds the ISRE sequence and interacts with STAT1, which binds the neighbouring half GAS site (highlighted in bold in Table 3), thus stabilizing the complex [89]. Such a combination of consensus motifs is also found in the promoter of the GBP gene, which contains partially overlapping contiguous GAS and ISRE sites (see Table 3) [90]. Both motifs are required for the induction of GBP by IFNα. Their combination may facilitate the induction of GBP by IFNs type I and II [91], [92]. Other ISRE motifs (unusual ISRE) comprise 5′-TTTC-3′ sequences separated by nucleotides, and are also recognised by factors of the IRF family [93], [94], such as the Sp100 gene, a target of IRF1 (see Table 3).
Table 3.
STAT1 binding sequences.
| Sequence | Name | Interactor | Ref. |
|---|---|---|---|
| 5′-TTCNNNT/GAA-3′ | consensus GAS | GAF | [84] |
| 5′-CGTCATTTCCCCGAAATCAG-3′ | IRF1 GAS | STAT1 | |
| AGTTTCNNTTTCNC/T | consensus ISRE | ISGF-3 (STAT1/STAT2/IRF9) | [88] |
| 5′-CTCGGGAAAGGGAAACCGAAACTGAAGCC-3′ | ISG-15 ISRE | IRF9 STAT1 | [325] |
| 5′-AAGTACTTTCAGTTTCATATTACTCTAAATC-3′ | GBP GAS IRSE | [90] | |
| 5′-GGAAAAGAGAAGAGAAAGT-3′ | unusual ISRE | IRF1 | [93], [94] |
Fig. 5.
Transcriptional complexes formed with STAT1 following treatment with IFNα and IFNγ. There are two major complexes: the ISGF-3 complex comprising STAT1, IRF9 and STAT2 which binds the ISRE DNA motif, and the GAF complex comprising a STAT1 homodimer which binds the GAS DNA motif.
Given that the different STATs have very different functions in cells, the similarity between their DNA target sequences [95] is intriguing. For instance, STAT1 and STAT3 share 72% homology in protein sequence and recognise very similar, if not sometimes identical, consensus sequences on their target genes [95]. Yet, STAT1 is mostly an inhibitor of proliferation and promoter of cell death, and STAT3 is mostly involved in cell survival and proliferation. Furthermore, in STAT3-depleted cells, STAT1 was found to loose its ability to induce some targets but not others [96], suggesting a complex coordination of these two STATs, as previously discussed [97]. Identification of the genuine STAT-binding sequences within IFN-treated cells using whole genome analysis such as CHIP (Chromatin Immuno-precipitation) or CHIP–chip (CHIP combined with microarray), although still technically challenging, has started to reveal STAT1's chromosomal targets [98], [99] and suggests that multiple mechanisms direct STAT1 binding to its targets under different activation conditions [98].
3.2. Components of the transcriptional complex
Once in the nucleus, the phosphorylated and dimerised STAT1 needs to interact with other components to induce transcription. The required co-factors include Nmi-1 (N-myc interacting protein 1), CBP/p300 (for CREB (cyclic AMP Response Element) Binding protein) MCM-5 (minichromosome maintenance 5) and BRCA-1 (BReast CAncer susceptibility gene-1) [100]. MCM-5 is a helicase with an ATPase activity involved in DNA replication, which binds the STAT1α isoform and increases its transcriptional activity [101], [102]. CBP/p300 is a histone acetyltransferase involved in chromatin remodeling, which binds both the C-terminal and the N-terminal regions of STAT1 [103]. This interaction is mediated by the phosphorylated serine 727 and the adjacent leucine 724, the latter being required for the binding of the STAT1 complex to RNA polymerase [49]. Interestingly, the CBP/p300 is in limiting amounts in the nucleus and the different transcription factors compete for it. CBP/p300 can form complexes with STAT1 without directly interacting with it, as in the ISGF-3 complex in which CBP/p300 interacts with STAT2, but not STAT1 [104]. Although it binds the same target sequences as the α isoform, the β isoform of STAT1 has not been reported to interact with CBP/p300. If such an interaction did occur, it would involve only the N-terminal domain of STAT1β and might not be functional.
3.3. Transcriptional activity of STAT1
The regulation of gene expression by STAT1 varies with the target and the cellular context. It can be direct or indirect, and it can be an activation or an inhibition. In cellular systems in which STAT1 is activated, the expression of many genes is induced, including CXCL9 (or Mig1) [105], p21waf1/cip1 [106], [107], [108], ifi205 [33], and Hsp70 [109], [110]. STAT1 also stimulates the transcription of genes in cooperation with other transcription factors such as NF-κB [111], as observed for IP10 (induced protein 10) [112] and ICAM1 (intracellular adhesion molecule 1) [113], [114]; STAT1 also cooperates with Sp1 [115] for the induction of several genes including IRF1 [116]. The cooperation of STAT1 with other transcription factors varies with the cellular system. In monocytes STAT1 cooperates with PU.1 in the induction of the FcγR1 (Fc gamma Receptor 1) [117], and in differentiating myeloid cells STAT1 cooperates with IRF1 and PU.1 for the induction of the gene Phox [118]. In these systems, a basal level of STAT1 may be required since its presence in the complex is required for CBP/p300 to associate to the transcriptional complex [119], [120]. STAT1 can also inhibit the transcription of genes such as cyclin A [121], [122], [123], c-myc [124], MMP-9 [125], Bcl2 and BclxL [126]. Even when it is not phosphorylated, STAT1 can induce the constitutive expression of a subset of genes involved in immune regulation [127], in particular LMP2 (low molecular mass polypeptide 2), TAP1 (transporter associated with antigen processing 1) [128], [129] and procaspase 3 [130] (Fig. 6 ). However, STAT1β, which has a truncated TAD, is apparently unable to promote transcription despite its ability to bind the promoter regions of IRF1 [131], LMP2, TAP1 [128] and Fas [35]. It has been suggested that induction of transcription by STAT1β can take place with plasmid-encoded promoters but not with cellular promoters because its lack of TAD impairs chromatin remodeling [101].
Fig. 6.
Components of antigen presentation by CMH1 whose expression is modulated by STAT1α. The figure depicts the antigen presenting machinery; the identified STAT1 targets are highlighted in bold.
4. STAT1 as an inhibitor of infection and of proliferation
STAT1 regulates the immune system, cell differentiation, tumour suppression, cell growth inhibition and apoptosis.
4.1. Stimulation of the immune system
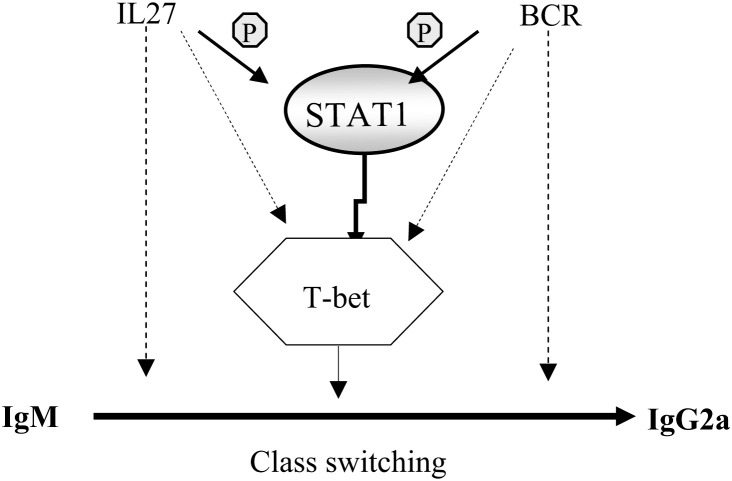
STAT1 plays an essential role in the immune response to viruses [132], [133], [134], bacteria [31], [134], [135], [136] and parasites [137], [138]. STAT1-deficient mice die mainly of viral infection after less than 8 weeks [42], [134], [139]. In humans, high sensitivity to mycobacterial infection has been found to be associated with mutations in STAT1 that interfere with its functions [31], [140], [141], and high sensitivity to viral diseases has been shown to be associated with mutations in the STAT1 gene resulting in the complete absence of the protein [133]. Thus, reduced activity of STAT1 is associated with susceptibility to infectious diseases. STAT1 is involved in all the steps of the processing and the presentation of antigenic peptides by the major histocompatibility complexes (MHC) I and II, including the expression of the proteasome subunits LMP2 [128] and LMP7, TAP1 [129], and indirectly the MHC-I and MHC-II through activation of CIITA (class II transactivator) [142] (see Fig. 6). In mice, STAT1 regulates the expression of immunoglobulins: in its absence, circulating IgE increases [143]. STAT1 is also necessary for class switching, an early step of B cell maturation which is induced by IL27 [24]: IgM-to-IgG2A class switching is abolished in the absence of STAT1[24]. STAT1 also plays an essential role in the production of IgG2A following BCR stimulation [38], [144]. STAT1 directly induces the expression of the protein T-bet [144] which itself activates the class switching of immunoglobulins [145] (Fig. 7 ). There are no studies on the implication of STAT1 in the class switching of immunoglobulins in humans. Recently, the expression of surface IgG was found absent in cells from STAT1-deficient patients [326].
Fig. 7.
Mechanism of the modulation of IgM to IgG2A class switching in mice by STAT1. In mice, the class switching of Ig is under the control of STAT1 through its target T-bet.
4.2. Inhibition of cell growth
IFNα and IFNγ require STAT1 to exert their negative action on proliferation [146]. This is also true of retinoic acid [123]. Interestingly, in systems in which EGF negatively regulates proliferation, the activation of STAT1 is observed, and its expression is required [147], [148]. Active cyclin/CDK complexes are required, and in sufficient amount, for the G1/S and the G2/M transitions to occur normally [149]. Inhibition of cell growth by STAT1 occurs mainly through the regulation of genes involved in cell cycle control: two CDKs (Cyclin Dependent Kinase) inhibitors, p21waf1 and p27kip1, which associate to CDKs and inhibit their kinase activity [150] are induced by STAT1 [123], [148], [151], [152]; and cyclin A [122], [123], and cyclins B, D2, D3 and E [123] are repressed. STAT1 is also responsible for the inhibition of the expression of c-myc [124], [153]. As c-myc represses the expression of p21waf1, this further enhances cell cycle arrest.
4.3. Regulation of cell death
STAT1 induces the expression of procaspases, the latent forms of the caspases which are proteases that transmit the apoptotic pathway in the cytoplasm by sequential cleavage in response to external or internal stimuli. STAT1 was shown to constitutively induce the expression of the procaspases −1, −3 and −11; −1 and −11 are required for the subsequent cleavage of procaspases −3 and −8 in murine lymphocytes [154], [155]. STAT1 also induces the expression of procaspases in response to external stimuli: the expression of procaspase-3 in response to TNF-α requires STAT1 [130], and IFNγ, EGF, 7-ketocholesterol and thrombin have been shown to induce the expression of procaspases in a STAT1-dependent manner but with considerable variation according to the stimuli reaching cells. STAT1 also mediates cell death induced by IL21 in mantle cell lymphoma [156] (see Table 1). Procaspases genes are not the only proapoptotic genes that are induced by STAT1. The Fas gene (CD95/APO-1) is induced in response to IFNγ in colon adenocarcinoma cells [157], [158], in microglial cells [159] and in fibroblasts [158]. In cardiac muscle cells, the ischaemia/reperfusion injury-induced apoptosis is accompanied by activation of STAT1, which induces the expression of the FasL (Fas ligand) Fas and caspase-1 genes [35], [160]. This is inhibited by STAT1 anti-sense RNA. In addition, in this system, the activation of STAT1 leads to the inhibition of the promoters of the anti-apoptotic proteins Bcl2 and Bcl-X [126]. In multiple myeloma cells treated with IFNγ, the expression of the TRAIL gene (TNF-related apoptosis inducing ligand) has been found to increase [161].
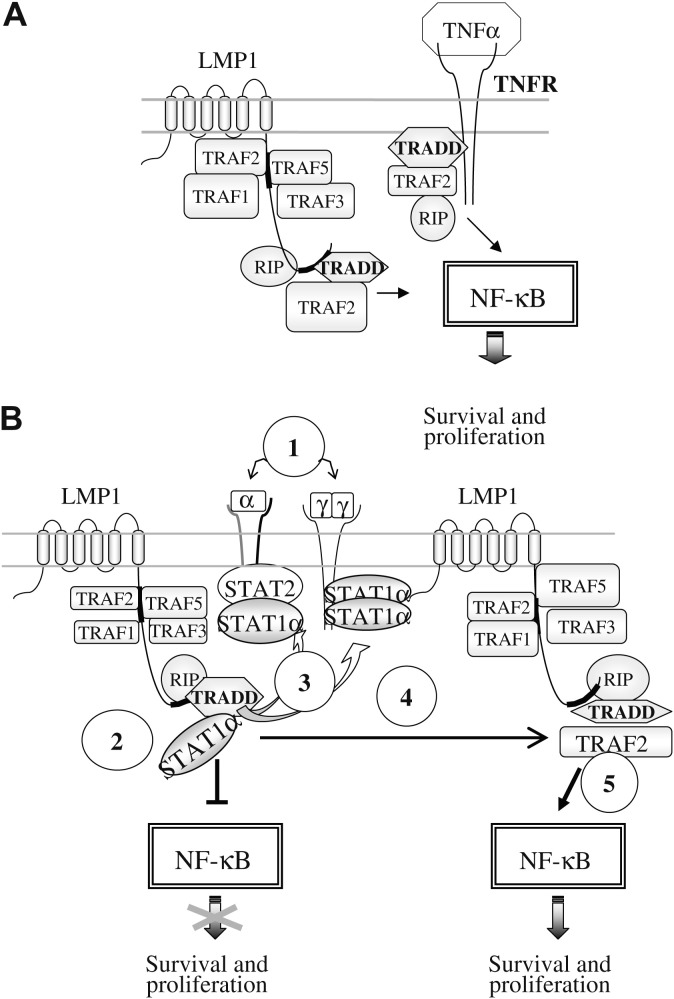
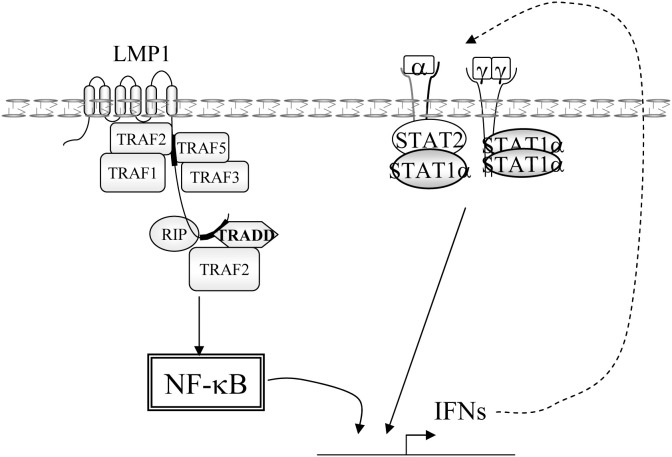
A high level of expression of STAT1 stimulates the TNF-α proapoptotic pathway. STAT1 has been shown to interact with TRADD, thereby inhibiting the activation of NF-κB [162], STAT1 competes with TRAF2, and prevents the formation of the NF-κB activating complex TRADD/TRAF2/RIP [163]. STAT1 operates at two different levels to potentiate TNF-α-induced apoptosis: it inhibits NF-κB signaling [164] and it induces the constitutive expression of procaspase-3, a component which is required for caspase-8-induced apoptosis [130] (see Fig. 11).
Fig. 11.
Actions of STAT1 in EBV-positive lymphoblastoid cells expressing LMP1. A: activation of the NF-κB pathway by LMP1 or the TNFR. B. Complex interaction of STAT1 with the LMP1-activated NF-κB pathway. 1: Induction by NF-κB of IFNα and γ production leading to STAT1 activation. 2: Inhibition of TRADD by STAT1α binding, leading to NF-κB inhibition. 3: Activation of STAT1 by IFNRs following its liberation from TRADD. 4 and 5: activation of the NF-κB pathway by LMP1.
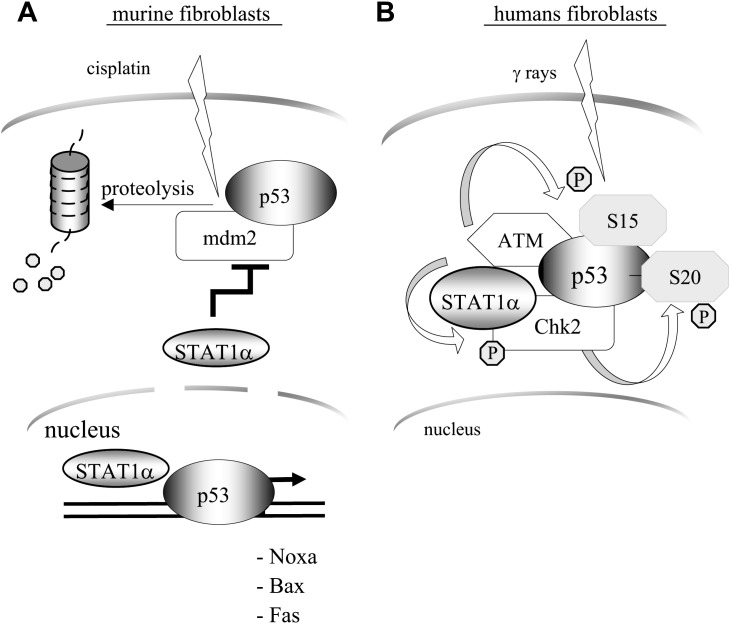
STAT1 also interacts with p53 (Fig. 8 ), a major regulator of apoptosis and cell cycle. The transcription factor p53, present at low basal levels, is induced following hypoxy, nutrient starvation, DNA damage or activation by oncogenes. Activation of p53 involves several post-translational modifications including phosphorylation [165], methylation [166] and acetylation [167]. The phosphorylation of p53 is a key event of its activation and involves some 15 different kinases [168], [169], while ubiquitination by the ubiquitin-ligase Mdm2 (mouse double minute 2) is a negative regulator [170]. The incidence of spontaneous or chemically induced tumours has been found to be higher in STAT1/p53 double knock-out mice than in p53 knock-out mice [171]. In murine embryo fibroblasts (MEF), apoptosis induced by cytotoxic agents such as cisplatin or doxorubicine requires the expression of a functional STAT1. In these cells, STAT1 potentiates p53 by increasing its expression through the repression of the expression of its inhibitor Mdm2; furthermore, STAT1 interacts directly with p53 and increases its transcriptional activity on targets such as Noxa, Bax and Fas [172]. In human fibroblasts, activation of p53 by DNA damage following Xray exposure is regulated by STAT1. In this system, STAT1 regulates the phosphorylation of p53 on serine 15 by ATM (ataxia telengiectasia mutated), and on serine 20 by Chk2 (checkpoint 2). It also facilitates the activation of Chk2 by ATM [173]. In B cells, physical interaction of p53 and STAT1 has also been observed, and it has been found that inhibition of STAT1 protects cells against apoptosis induced by fludarabine [174], a cytotoxic compound used mainly in B cell lymphoma and chronic lymphoid leukemia (CLL) [175], whose efficacy depends on a functional p53 [176]. Fludarabine has also been found to reduce specifically STAT1 protein and mRNA levels in peripheral lymphocytes [177] and lymphoblastoid B cells [178]. However, in the same cells, overexpression of STAT1α sensitised to fludarabine treatment [174], suggesting that STAT1 may act at different antagonistic levels of the pathways triggered by fludarabine. In fact, recent data indicate that treatment of cells with genotoxic agents, such as doxorubicin and to a lesser degree fludarabine, activates STAT1. This activation depends on p53, even if it is transcriptionally inactive [179]. Thus, the function of STAT1 goes beyond the activation of the transcription of proapoptotic genes. By its action on p53 it participates in the selection of which pathway – leading to cell death, or cell cycle arrest – will take place when cells are exposed to different stresses (Fig. 8). Although some of the relevant mechanisms have been clarified, it is still unclear how STAT1 influences the expression of the targets of p53.
Fig. 8.
Modulation of p53 activity by STAT1α. A: STAT1α stabilises p53 by inhibiting Mdm2 expression, and potentiates the transcriptional activity of p53 by forming complexes at the promoter level. B: STAT1α promotes serine 15 and serine 20 phosphorylation of p53.
4.4. Involvement in cell differentiation
A role for STAT1 in cell differentiation has been demonstrated in STAT1 −/− mice. In these mice, excessive osteoblastogenesis is the result of increased activity of the transcriptional factor Runx-2, which is normally sequestered in the cytoplasm by unphosphorylated STAT1 [180], [181]. In human cells, the differentiating action of bryostatin-1 [182] has been found to depend on the expression of STAT1 [183]; and the differentiation of acute promyelocytic leukemia cells induced by retinoic acid was found to require the phosphorylation of STAT1 on serine 727 [184], [185].
4.5. Tumour suppression
STAT1 is a negative regulator of tumorigenesis, angiogenesis and metastasis formation [186]. In STAT1-deficient mice, spontaneous and chemically induced tumours arise more frequently than in wild-type animals, and the anti-tumour activity of IFNα is reduced [187], thus defining STAT1 as a “tumour suppressor” [171]. Indeed, in humans several cancers which resist IFN treatment are associated with a diminished expression of STAT1 [188], [189], [190], and in mammary cancer, the level of activation of STAT1 is linked to a good prognosis, correlating well with the stage of the tumour, the extension to ganglia and the expression of cathepsin D [191]. In Wilms tumours, the phosphorylation of STAT1 on serine 727 is also associated with a good prognosis [192]. In addition, STAT1 inhibits the expression of rho, rac and cdc42, and the activity of the Ras MAPkinase pathway in Ras activated cells [193]. The function of STAT1 as a tumour suppressor is probably linked to its function in the immune system. STAT1 activates directly the expression of the transcription factor CIITA (Class II of MHC transactivating protein), which itself activates the transcription of the MHCII (major histocompatibility complex class II) [194]. In addition, STAT1 is involved in the negative regulation of the MMP-9 which serves as an angiogenic factor [125]. However, although STAT1 is a tumour suppressor, there are indications that it can function positively in tumour growth. One report showed that it can accelerate the development of hematopoietic tumours independently of IFN signaling and in association with upregulation of the MHC class I molecules [195], suggesting a IFN-independent tumorigenic function of STAT1. Another report showed that STAT1 positively regulates some of the enzymes of the glycolysis pathway, thereby linking it to the Marburg effect [196].
5. Pathogens inhibit the STAT1 pathway
5.1. STAT1 is an essential component of the resistance to pathogens
The IFN system is part of a sensing mechanism that detects pathogen invasion and triggers a response which limits the spread of the pathogen. The pathogens are detected by cytoplasmic or endosomal sensors (reviewed in: [197]and [198]) that trigger pathways leading to the activation of IRFs, AP1 and NF-κB. Among the early genes induced by these transcription factors the type I IFNs are key players. They trigger the IFNR/JAK system (see above) which activates the STATs, and particularly STAT1. IFNα and β are potent anti-viral agents, playing a key role in the regulation of the immune system by controlling the proliferation, differentiation, activation and maturation of several cell populations, including: dendritic cells (DC), natural killer cells (NK), Th1 cells, and memory CD8+ lymphocytes. IFNγ is produced mainly by T helper type 1 lymphocytes, but is also produced by many other cell types. Its function is primarily to promote the antimicrobial activity of macrophages. Indeed, the main target of IFNγ is the macrophages in which, in the context of infected cells and through a specific subset of receptors, it principally activates STAT1 (see above). In macrophages, the IFNγ/STAT1 pathway activates microbicidal activity through the induction of NADPH oxidase and iNOS, which are key components of the efficient killing of bacteria, viruses, parasites and fungi (see: [199]). This is highlighted by the efficiency of defence against most pathogens. For instance, there is a 90% inhibition of the replication of Chlamydia in IFNγ-treated cells [200]. Possible mechanisms involve processes that are downstream of the cellular entry of the bacteria [201]. Another example is the intracellular multiplication of Legionella pneumophila in alveolar resident macrophages, which is inhibited by IFNγ-treatment, again indicating the antibacterial effect of IFNγ [202]. The IFN-induced anti-viral state of cells is due in large part to targets of STAT1. Among these, the expression of the protein kinase PKR, a target of STAT1, is induced by IFNs and is subsequently activated by double stranded RNA (dsRNA), an intermediate of RNA viruses' replication. Activated PKR phosphorylates the α subunit of eukaryotic initiation factor 2 resulting in protein synthesis shutdown. The IFN/STAT1 pathway also induces cell cycle arrest through induction of the cyclin kinase inhibitors p21 and p27, and apoptosis through targets of STAT1, or of its early target IRF1. The essential role of STAT1 in the IFNγ pathway is further demonstrated by the high sensitivity of STAT1 −/− mice to infection, including infection by viruses such as the vesicular stomatitis virus (VSV) the mouse cytomegalovirus (MCMV) [134], and by bacteria such as Listeria monocytogenes. Interestingly, there are many different mechanisms by which pathogens inhibit STAT1 (Fig. 9 ).
Fig. 9.
Multiple points of inhibition of the IFN-STAT1 pathway by pathogens. The actions of several pathogens on the different steps of STAT1 activation are illustrated. In the cytoplasm: inhibition of phosphorylation (L. Donovani: Leishmania Donovani); trapping in high molecular weight complexes (hmw) (MV: Measles Virus); induction of degradation (SV: Simian Virus, HPIV: human parainfluenza virus). Inhibition of nuclear transfer (HCMV: human cytomegalovirus, KPNα: karyopherin α). Activation of STAT1β: (L. Mexicana: Leishmania Mexicana, M. Tuberculosis: Mycobacterium Tuberculosis, Pr: proteasome). Within the nucleus: dephosphorylation (HCV: human hepatitis C virus). Inhibition of CBP/p300 binding: (B. mellitensis: Brucella mellitensis). Inhibition of STAT1 following inhibition of the methyltransferase PRMT1 (protein arginine methyltransferase 1) (HBV: human hepatitis B virus, HCV: human hepatitis C virus, meth: methyl group) the putative target of PRMT1, PIAS1 (Protein Inhibitor of STAT1) is indicated. Inhibition of nuclear export.
5.2. Pathogens can inhibit every step of STAT1 activation
Over the course of coevolution, many pathogens, viruses, bacteria or parasites, have become able to efficiently overcome the organism's defences against them. Many of the processes developed by these pathogens are aimed at the IFN-STAT1 pathway, and specifically target steps of the activation process of STAT1, such as its phosphorylation or its nuclear localisation. Pathogens can also induce the dephosphorylation of STAT1 or prevent its methylation in the nucleus, induce the expression of its β isoform, and they also can induce its selective degradation. The mechanisms developed by viruses to overcome the potent effects of the defence system have been well studied: many viral proteins have been identified and several processes of inhibition have been unveiled at least partially (Fig. 9). However, the precise ways in which bacteria and parasites affect the function of STAT1 are much less well-understood. In many instances, although the cellular target has been identified, the molecular process (i.e. the bacterial or parasite proteins) has not.
5.2.1. Degradation of STAT1
Many viruses target STAT1 by inducing its intracellular degradation. Viruses of the Paramyxoviridae family (Mononegavirales order) form a large family of RNA viruses and code for V proteins whose expression greatly reduces the half life of STAT1 and STAT2, thereby inhibiting the JAK-STAT IFN signaling pathway; however, the mechanisms involved for each V protein are different. These viruses are classified into two subfamilies. One is the Paramyxovirinae subfamily, which comprise the Respirovirus genus (formerly Paramyxovirus), including the Sendai virus (SeV) and the human parainfluenza type 3 (HPIV3) virus; the Rubulavirus genus, comprising the simian virus 5 (SV5) the mumps (MuV) and human parainfluenza virus type 2 (HPIV2), and the Morbillivirus genus, comprising the Measles virus. The other is the Pneumovirinae subfamily which consists only of the Pneumovirus genus to which the human respiratory syncytial virus (RSV) belongs. The V proteins from the HPIV2, SV5 and MuV target STAT1 for proteasome-mediated degradation [203], [204], some proteins target STAT1 only, others target both STAT1 and STAT2, some target only STAT2 and some target STAT3 for degradation. Expression of the V proteins by the simian virus 5 (SV5) and HPIV2 by the human parainfluenza virus type 2 (HPIV2) highjacks the polyubiquitinylation pathways of the cell and induces polyubiquitylation of STAT1 and STAT2 resulting in their degradation by the proteasome, an action that is inhibited by proteasome inhibitors such as lactacystine or MG132 [205]. The process driven by the V proteins of HPIV2, SV5 and MuV involves the assembly of cellular Ub-activating enzyme E1, the cellular Ub-conjugating enzyme E2 to STAT1/STAT2 dimers, resulting in the polyubiquitylation of either STAT1 or STAT2 [204], [206], [207]. The Newcastle disease virus (NDV) encodes a V protein which also induces the degradation of STAT1, but the mechanism involved has not been elucidated [208]. The SeV also induces degradation of STAT1 by directing its polyubiquitylation [209].
5.2.2. Inhibition of STAT1 phosphorylation
Many pathogens prevent the phosphorylation of STAT1. The human metapneumovirus (hMPV), a recently discovered Paramyxovirus involved in respiratory tract infections, inhibits IFNα signaling by preventing the phosphorylation and nuclear translocation of STAT1, but the mechanism involved has not been yet elucidated [210]. In the human macrophage U-937 cell line, the parasite Leishmania donovani prevents the phosphorylation of STAT1 in response to IFNγ [137] thereby efficiently impairing the IFNγ-JAK-STAT1 pathway by inhibition of STAT1α binding to the IFNγ response region [211]. In more recent studies performed in the murine J774A.1 and RAW264.7 macrophage cell lines, the different species of Leishmania have been found to affect the IFNγ-JAK-STAT1 pathway differently: the L. donovani, Leishmania major and Leishmania mexicana species all inhibit phosphorylation of STAT1 in response to IFNγ, but L. mexicana, which is apparently a more efficient inhibitor of the IFNγ-JAK-STAT1 pathway, induces the preferential phosphorylation of STAT1β, thereby contributing to the inhibition of STAT1α [212]. However, the mechanism of inhibition of STAT1 by Leishmania may be even more complex as another study found that L. donovani, L. major and L. mexicana trigger the specific degradation of STAT1 by the proteasome; indeed, infection by the parasite was significantly inhibited by the addition of proteasome inhibitors to infected cells [213], raising the question of whether there can be specific targeting of STAT1α and not of STAT1β for the proteasome. The contribution of STAT1 to resistance to L. major appears to be an upregulation of CXCR3, which stimulates the migration of Th1 cells to the infection site [214]. In fact, another species, Leishmania amazonensis, was shown recently to efficiently downregulate the activation pathway of several cytokines, including IFNγ, by inducing decreased phosphorylation of STAT1, STAT2 and STAT3 and also specific degradation of STAT2 [215]-an action that is prevented by proteasome inhibitors. Several pathogens inhibit the IFNγ pathway by interfering with phosphatases; in one study, however, the activation of the SHP-1 phosphatase by L. donovani did not correlate with the inhibition of STAT1, although it contributed to the resistance of the parasite in cultured macrophages. In other words, inhibition of STAT1 by the parasite was independent of the induction of SHP-1 [216]. Another counterintuitive observation is that STAT1 −/− mice are more resistant to visceral leishmaniasis than STAT1 proficient mice [217] suggesting that the parasite needs an efficient IFNγ pathway at some point.
5.2.3. Trapping of STAT1 into high molecular weight complexes
The parainfluenza type 5 virus (HPIV5) sequesters STAT1 [218] as do the Nipah viruses. The Nipah (niV) and Hendra Paramyxovirus-family virus proteins prevent activation and nuclear translocation of STAT1 and STAT2 by trapping them into cytoplasmic high molecular weight complexes [219], [220], [221]. The Nipah virus V protein inactivates STAT1 by forming a complex with STAT2, and its expression into cells results in STAT1 being relocalised to the cytoplasm. The Nipah virus V protein possesses an NES motif which is necessary for the cytoplasmic export of the protein and the cytoplasmic relocation of STAT1; nevertheless, deletion of this NES motif does not abrogate the ability of the protein to block the IFN response, indicating that other domains of the protein play an important role. The P protein of the measles virus directly interacts with STAT1 and prevents its phosphorylation [222]. The Nipah virus P protein, on the other hand, sequesters inactive STAT1 in the nucleus [223].
Although it also belongs to the Paramyxoviridae, the Measles virus (MV) functions differently. Rather than inducing the degradation of STAT1, its V protein interacts with STAT1, STAT2, STAT3 and IRF9, forming high molecular weight complexes that are packaged to cytoplasmic bodies containing an assembly of viral proteins and nucleic acid material of viral origin [224]. Although expression of the V protein of the MV clearly shows that it is a major component of the inhibition of STAT1 in cells, two other viral proteins transcribed from the same P gene are also involved. One of these, the P protein, contributes to immune evasion: its mechanism of action is proposed to be inhibition of the phosphorylation of STAT1, possibly by direct interaction [222]; the P gene of the MV also encodes a C protein: expression of this recombinant protein in cells inhibits IFNα/β and IFNγ signaling, by a mechanism that has not been deciphered [225]. Deletion of regions of the V protein of MV has shown that discrete peptides specifically bind STAT1 and STAT2, preventing phosphorylation by JAK1 [226], [227].
5.2.4. Inhibition of the nuclear translocation of STAT1
The rabies virus is a Rhabdoviridae which belongs to the Mononegavirales order. This neurotrophic single stranded RNA virus replicates in the host's cytoplasm and encodes an RNA polymerase complex consisting of a large protein L and a phosphoprotein P, both of which participate in transcription and replication. A two-hybrid screening system showed that the P protein interacts through its C-terminal domain with the N-terminal domain of STAT1 [228], this results in efficient inhibition of STAT1 nuclear accumulation in response to stimulation by either IFNα or IFNγ. The inhibitory action of protein P does not involve reduced phosphorylation or reduced homodimerisation of STAT1, or heterodimerisation with STAT2 [228], although it interacts much more strongly with phosphorylated STAT1 than with non-phosphorylated STAT1 [229]. Intriguingly, the P protein contains both an NLS and an NES motif [230], and its subcellular location directs that of STAT1, thereby preventing STAT1 nuclear location after IFN stimulation [231]; however, mutant forms of the P protein that do not contain the NES signal appear to be able to inhibit the binding of STAT1 to its DNA target [231]. The nuclear import of proteins can be either facilitated or inhibited by microtubules (MT); the P proteins use MT-facilitated nuclear transfer, but they have the ability to switch to MT-inhibited transfer; in addition, they can impose a switch of STAT1's IFNα-induced nuclear transfer to a MT-inhibited mode, thereby preventing STAT1 nuclear import [232]. The Ebola virus, which causes Ebola hemorrhagic fever with an extremely high mortality (80%), is an efficient inhibitor of IFNα/β signaling. Its protein VP35, when expressed in cells, blocks several components of the anti-viral response, including IRF-3 [233]. A single amino-acid change can reverse this action [234]. Interestingly, a VP24 protein encoded by the virus directly interacts with karyopherins α1, α5 and α6 [235], thereby preventing STAT1's interaction with these karyopherins without modifying its phosphorylation on tyrosine 701 [236], [237], thus indirectly inhibiting STAT1. Another virus which exerts its inhibition of the IFNγ-STAT1 pathway through inactivation of the nuclear transport machinery is severe acute respiratory syndrome virus (SARSV). This virus which induces a severe and frequently fatal acute respiratory syndrome, has the ability to inhibit the IFN response in infected cells. STAT1 has been found to play a role in the resistance of infected animals [238]. Transfection of SARSV viral proteins – the ORF3b, ORF6 and N protein – demonstrated their ability to specifically inhibit the expression of IFN transcriptional targets [239]. The ORF6 protein was found to specifically inhibit the nuclear translocation of STAT1 [239] by tethering karyopherin α2 and β1 to the membrane of the endoplasmic reticulum, thereby disrupting the nuclear transport of STAT1 [240].
5.2.5. Dephosphorylation of STAT1
The vaccinia virus, a Poxviridae family virus, encodes for several proteins which neutralize the IFN host defence system at different levels, including inhibition of PKR, and the release of cytokine homologues which block the IFNα/β and γ Receptors. In addition to this, the virus encodes a protein with dual tyrosine/serine phosphatase activity whose expression is required for virus viability in tissue culture [241], [242]. The dual phosphatase can both prevent STAT1 phosphorylation in infected cells and induce its dephosphorylation, thereby preventing the nuclear translocation of STAT1 and the induction of gene targets [243]. The mosquito-borne Japanese Encephalitis flavivirus also blocks the IFN-induced JAK-STAT pathway: its non-structural protein NS5 induces the dephosphorylation of TYK2 and STAT1, thus preventing STAT1 nuclear translocation and the transcription of its gene targets [244]. However, the phosphatase or phosphatases involved, which are probably tyrosine-phosphatases have not been identified.
The human cytomegalovirus (HCMV), a member of the β-herpesvirus subfamily, is a widespread DNA virus which infects a high percentage of the population. The host's immune system plays a crucial role, and in the defence against the virus, besides TNF-α, IFNγ secreted by T cells can efficiently block HCMV replication in vitro. However, the virus has evolved mechanisms that can counteract the control of infection by inhibiting phosphorylation of tyrosine 701. This process appears to be due to the induction of SHP2, which directly dephosphorylates nuclear STAT1-P-tyr, resulting in its downregulation [245]. Upregulation of a another protein phosphatase, protein phosphatase 2A (PP2A), is involved in the resistance of hepatitis C and B viruses to IFN signaling. The mechanism involves upregulation of PP2A, which inhibits the protein arginine methyltransferase 1 (PRMT1) resulting in reduced STAT1 activity [246]. Other sites of inhibition of the IFN pathway by HCMV include the JAK kinase TYK2 [247], a targeted degradation of STAT2 [248] by the viral 72 kDa protein IE1 which forms physical complexes with STAT1 and STAT2, thereby preventing correct nuclear localisation and association with the promoters of IFN-responsive genes [249].
5.2.6. Inhibition of STAT1 via methylation
The hepatitis B virus (HBV) is able to block IFNα’s action by inhibiting the methylation of STAT1, this results in an increased interaction of STAT1 with PIAS1, thereby protecting the virus against the anti-viral action of the IFN [250].
5.2.7. Inhibition of STAT1 transcriptional activity
The Hepatitis C virus (HCV) efficiently antagonises the anti-viral action of IFN. The molecular mechanism by which this inhibition occurs has not been well characterised. Transfection of full length HCV and subgenomic fragments in the hepatocyte cell line Huh-T7 showed degradation and reduced phosphorylation of STAT1 [251]. Subsequent studies using transfection of the NS5A (non-structural protein5A) of HCV have shown that expression of this protein in Huh7 cells prevents normal activation of STAT1 by inhibiting its phosphorylation on tyrosine 701 and its nuclear translocation [252], [253], an action that was specific to cells of hepatic origin [253]. This discrepancy between observations using transfection of single viral proteins may be due in part to the involvement of other viral proteins. For instance, the transfection of NS3/4A, another non-structural protein of HCV, has been found to inhibit the phosphorylation of STAT1 on serine 727, contributing to the efficient inhibition of the IFN-STAT1 pathway [254]. Differences in published observations may be due in part to the use of different cellular systems; thus, the choice of cellular system may be important when dealing with a hepatotropic virus [253].
Brucella melitensis can develop intracellularly within phagocytes and cause chronic infection, this requires that the IFNγ pathway be silenced: in Brucella-infected macrophages, the IFNγ-induced STAT1-CBP/P300 association, required for a normal response to the cytokine, is disrupted [255].
The parasitic protozoan Toxoplasma gondii is a widespread parasite in human and animal populations, probably in part because it causes asymptomatic infection. This parasite has the ability to simultaneously suppress and trigger innate immune function in the host. Infection by Toxoplasma includes an acute phase in which the parasite disseminates in cells, which is followed by chronic infection in which the parasite is confined within quiescent cysts within tissues (reviewed in [256]). The strong Th1-type immune response raised by Toxoplasma involves an IL-12-driven IFNγ secretion by lymphocytes and the activation of STAT1. In non-professional phagocytic cells that do not express STAT1 there is no anti-Toxoplasma activity [257], and STAT1 −/− mice die of infection although they produce normal levels of IFNγ [258], pointing to a key function of STAT1 in the anti-parasitic function itself. Indeed, IFNγ-inducible genes include genes such as inducible nitric oxyde synthase (iNOS), which is under the control of STAT1 (see: [256]). Part of the parasite's immune evasion may result from its ability to induce the IFNγ-signaling inhibitor SOCS-1 (suppressor of cytokine signaling 1): in murine macrophage cell lines T. gondii infection induced the expression of SOCS-1, resulting in inhibited STAT1 tyrosine phosphorylation, and in SOCS-1 −/− mice, inhibition of the anti-parasitic effect of IFNγ was reduced [259]. However, in a different context, that of human fibroblasts, T. gondii was found to inhibit IFNγ-dependent STAT1 activation without affecting its phosphorylation and nuclear trafficking, by blocking its transcriptional activity on IFNγ-responsive genes, including IRF1 [260]. Interestingly, although the targeting of STAT1 by T. gondii is clearly established, the molecular mechanism of how the parasite's proteins achieve STAT1 inhibition is not known in detail. Among the identified mechanisms, two bring about an increased IL-12 production: the triggering of the Toll Like Receptor by the parasite's surface glycosylphosphatidylinositols (GPI), involving MyD88 and the NF-κB pathway; and the triggering of the chemokine receptor 5 (CCR5) by the parasite cyclophilin C-18 [261]. The blockade in STAT1 signaling, as well as that of NF-κB signaling, has also been attributed to a parasitic heat shock protein (HSP-70) which efficiently attenuates the suppressive action of T. gondii within infected cells (see: [256]), however, the mechanistic details of how STAT1 is blocked are not identified.
5.2.8. Increased expression/phosphorylation of the inhibitory form STAT1β
When infecting cells, the bacterium Mycobacterium tuberculosis induces cell-mediated immunity: infected macrophages secrete IFNα and IFNβ [262], as do dendritic cells (DC) [263]. In the meantime, events downstream from IFN activation are impaired, including a significant reduction in the abundance of the ISGF-3 components STAT1, STAT2 and IRF9 [264]. In addition, IFN-γ-activated human macrophages are unable to restrict the growth of the virulent M. tuberculosis [265], [266] suggesting that the bacterium interferes with the response to IFNγ [267] and efficiently subverts IFN action by acting on events that are downstream of the triggering of IFN receptors. Surprisingly, however, the tyrosine phosphorylation, dimerisation, nuclear transfer and the DNA binding of STAT1 all appear to function normally [264]. Although reduced binding to the CBP/p300 coactivator was noted early on in M. tuberculosis-infected murine macrophages, the transcription of STAT1α-dependent IFNγ targets has been found to be blocked in the absence of any modification of the phosphorylation or stability of STAT1α [268]. As discussed in the above sections, the STAT1α/STAT1β ratio affects the resistance of cells to apoptosis [174] and to viral infection [269]. Interestingly, in one study of M. tuberculosis-infected cells, stabilisation of the mRNA for STAT1β was observed, accompanied by increased expression and phosphorylation of this isoform, suggesting a mechanism for inhibiting the signaling pathway of STAT1 [270].
L. mexicana, as already discussed above, inhibits IFNγ signaling through an IFNR/JAK-independent increased phosphorylation of STAT1β, thereby inhibiting STAT1α, possibly through competition at the level of target gene promoters [212].
6. The paradoxical activation of STAT1 by the oncogenic Epstein–Barr virus
The Epstein–Barr Virus (EBV) has evolved an extremely complex and intertwined interaction with its host's defence system.
6.1. Epstein–Barr virus – transformed cells
The EBV is usually responsible for a mild, often asymptomatic and undetected infection; but this DNA virus is also associated with several malignant diseases, including Burkitt's lymphoma, post-transplant lymphoma, lymphoma associated with HIV infection, Hodgkin's disease, T cell lymphomas and leukemia, epithelial neoplasia such as nasopharynx carcinoma, mammary carcinoma, and gastric carcinoma [271], [272]. Following infection, the EBV persists in a latent form within memory B cells that are CD23- and CD27-positive and CD5- and IgD-negative. There is also a persisting production of virion by the salivary glands of the healthy subject [273], [274]. Among the many genes encoded by the EBV, a limited number have been shown to be essential for latency. Two of these genes code for non-polyadenylated small RNAs, EBER1 and EBER2 (for EBV Encoded RNA), six genes code for nuclear proteins EBNA 1, 2, 3A, 3B, 3C and LP (EBNA for EBV Nuclear Antigen) and three code for membrane proteins LMP1, LMP2A and LMP2B (LMP stands for Latent Membrane Protein) [275], [276]. The expression of these proteins varies with the pathophysiological context. A classification of four viral latencies is used (latencies 0, I, II and III). The EBV has also been shown to transform cells in vitro [277], resulting in lymphoblastoid cell lines (LCL) which grow indefinitely and express the EBV genes corresponding to latency III [278], [279]. It is important to note that LCLs can arise in vivo. They probably originate from a subpopulation of memory B cells which remain positive for EBV and have kept the potential to spontaneously generate LCLs [280], [281], [282] with charateritics similar to the LCLs generated in vitro [276]. Cells from LCLs are similar to lymphoblastic B cells. They are bigger than B lymphocytes, with a larger cytoplasm and numerous short cytoplasmic extensions. Their growth rate is variable, with a doubling time varying from 20 to 48 h. They tend to form aggregates, but do not adhere to the plastic of the culture flask [276]. On their surface they express the B cell markers, CD19 and CD20, and the B cell differentiation markers that are induced by the EBV: CD23, CD38, CD39, CD54, CD58 and CD70 [283], [284], [285]. The LCLs retain the capacity to spontaneously differentiate in so-called plasmacytoid cells. The plasmacytoid cells are similar to plasma cells, they produce high levels of immunoglobulins and have a diminished expression of CD23, CD11a and CD58, and an elevated expression of CD54 and CD38 [286]. Interestingly, these changes also occur during normal differentiation of B lymphocytes to plasma cell. Meanwhile, the expression of the latency proteins EBNA2 and LMP1 diminishes during the differentiation of LCL into plasmacytoid cells [286], [287].
The latency protein LMP1 is a transmembrane protein of 63kDa encoded by the BNLF1 gene of the EBV. It consists of six transmembrane domains (from amino-acid 25 to amino-acid 194). The N-terminus is a short cytoplasmic sequence (1–24) and the C-terminus is a longer cytoplasmic stretch (195–386) which contains two activating regions, CTAR1 (C-terminal activating region 1) and CTAR2 [288] (Fig. 10, Fig. 11 ). LMP1 has been shown to be involved in cell transformation and cell immortalisation [289], [290], [291]. LMP1 forms homo-aggregates through interaction of its cytoplasmic N-terminal ends [292]. Following oligomerisation, LMP1 behaves in a manner similar to an activated TNF receptor (TNFR) [293], [294], [295]. The cytoplasmic regions CTAR1 and CTAR2 allow the association of the signaling molecules: TRAF, (TNFR-associated factor) [296], [297], TRADD (TNFR-1-associated death domain protein) [298], and RIP (receptor-interacting protein) [299]; these proteins activate the kinases p38α, NIK (NF-κB-inducing kinase) and JNK which in turn activate transcription factors including ATF2 (activating transcription factor 2), NF-κB [300], [301], [302], [303] and AP1 [304]. These transcription factors in turn activate the transcription of genes involved in cell growth, such as c-Met, the EGF receptor and cyclin D2; angiogenesis, such as FGF2, VEGF MMP-9 and IL8; and protection against apoptosis such as Bcl2 [305], Blf-1 [306] and A20 [307], [308]. These proteins potentially account for the bulk of the molecular mechanism of cell transformation induced by LMP1. In addition, some of the transcription factors that are activated, such as ATF [309] and AP1 [310], contribute to the maintenance of viral latency by activating the expression of LMP1 itself.
Fig. 10.
Proposed mechanism for the activation of STAT1 by the oncogenic protein LMP1 in Epstein-Barr-transformed lymphoblastoid cell lines. Activation of the oncoprotein LMP1 results in the activation of NF-κB, the induction of the expression of IFNs and their production by cells drives the constitutive activation of STAT1 (adapted from reference [318]).
6.2. Activation of STAT1 by the oncoprotein LMP1
The constitutive phosphorylation of STAT1 on tyrosine 701 and serine 727 has been described in most cells expressing LMP1 such as LCLs or EBV-positive Burkitt lymphoma [178], [311], [312], [313]. However, it is not always observed [314], particularly not in all LCLs [315], and may depend on the cell type. The activation of STAT1 in LMP1-expressing cells was initially thought to result from the binding of JAK3 to CTAR3, identified as a JAK-binding motif in the cytoplasmic region of LMP1 [312]. However, there is no detectable binding of JAK3 to LMP1 in either EBV-infected Burkitt lymphoma or in LCLs [316]. Furthermore, mutation of key amino-acids in either CTAR1, CTAR2 or both, results in the suppression of NF-κB, AP1 and STAT1 activation [317]. By combining inhibition of NF-κB activation and antibody neutralisation of IFNs, we were able to show that activation of STAT1 in LCLs can be accounted for by the constitutive activation of NF-κB by LMP1, resulting in enhanced secretion of IFNα, activating STAT1, which in turn induces IFNγ expression [318]. The direct involvement of LMP1 has further been demonstrated by the observation that STAT1 is phosphorylated on tyrosine 701 in Burkitt cells transfected with an inducible LMP1 [319]. In these experiments, the phosphorylation of STAT1 was detected after 4 h of induction, which is compatible with a secretory loop of IFNs (Fig. 10). However, in some LCLs, phosphorylation of STAT1 was detected on serine 727 but not on tyrosine 701, and DNA-binding capacity was increased, suggesting that perhaps other modifications of STAT1, such as acetylation, may also be involved in its activation by LMP1 [315]. This also points to the importance of the phosphorylation of serine 727 of STAT1, which is due in part to the activation of p38α and JNK by LMP1 through TRAF1 and RIP, but independent of NF-κB, and can be further enhanced by the induction of IFNα and IFNγ secretion, which activate ERK 1/2, CaMKII and PKCδ. STAT1 is probably not the only STAT family member to be activated by LMP1: in the BJAB cell line transfected with inducible LMP1, the phosphorylation of STAT3 is also detected. STAT1 is an inhibitor of cell growth and activator of apoptosis, and it is not clear why in some LCLs it can be constitutively activated. Several explanations can be proposed. Firstly, STAT1α was found to associate to TRADD, thereby inhibiting the TNF-α-induced activation of NF-κB [163]: it follows that IFNγ, by increasing the recrutment of STAT1 monomers to the IFNGR1 receptor, can potentiate the action of TNF-α [320]. Since LMP1 activates NF-κB by mechanisms that are similar to those triggered by the TNFR, it is possible that in LMP1-expressing cells activation of STAT1 results in the potentiation of NF-κB activation. Indeed, the inhibition of STAT1 by overexpression of the β isoform results in a diminished NF-κB capacity to bind the DNA κB sequence. Secondly, the promoter region of LMP1 contains the sequence 5′-TTCctgGAA-3′, which is similar to a classical GAS sequence such as the one present in the IRF1 promoter [314]; however, DNA-binding and reporter gene experiments have shown STAT3 binding to this motif [314]. Nevertheless, the function of STAT1 activation in EBV-transformed cells is not entirely clear. It has recently been shown that LMP2A and LMP2B, whose function is not fully elucidated, induce decreased responsiveness of cells to IFNs by accelerating their surface turnover, thereby reducing STAT1 activation [321]. Although this observation was made in transfected epithelial cells, it indicates that the interaction of the EBV with its host's defence system is complex, and that the survival of EBV-transformed cells in the host must be the result of a subtle equilibrium between the cells' anti-viral defence, including STAT1, and the virus's maintenance proteins.
Ackowlegdments
Part of the research described in this review was supported by: Association pour la Recherche contre le Cancer (ARC) (grant 3133); réseau herpès virus et cancer; Société Française d'Hématologie (SFH); Ligue contre le cancer, and Ministère de la recherche et des technologies (MENRT).
References
- 1.Dale T.C., Imam A.M., Kerr I.M., Stark G.R. Rapid activation by interferon alpha of a latent DNA-binding protein present in the cytoplasm of untreated cells. Proc. Natl. Acad. Sci. U. S. A. 1989;86:1203–1207. doi: 10.1073/pnas.86.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy D.E., Kessler D.S., Pine R., Darnell J.E., Jr. Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 3.Horvath C.M. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 4.Ihle J.N. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 2001;13:211–217. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 5.Schindler C., Fu X.Y., Improta T., Aebersold R., Darnell J.E., Jr. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakatsume M., Igarashi K., Winestock K.D., Garotta G., Larner A.C., Finbloom D.S. The Jak kinases differentially associate with the alpha and beta (accessory factor) chains of the interferon gamma receptor to form a functional receptor unit capable of activating STAT transcription factors. J. Biol. Chem. 1995;270:17528–17534. doi: 10.1074/jbc.270.29.17528. [DOI] [PubMed] [Google Scholar]
- 7.Kotenko S.V., Izotova L.S., Pollack B.P., Mariano T.M., Donnelly R.J., Muthukumaran G., Cook J.R., Garotta G., Silvennoinen O., Ihle J.N. Interaction between the components of the interferon gamma receptor complex. J. Biol. Chem. 1995;270:20915–20921. doi: 10.1074/jbc.270.36.20915. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan D.H., Greenlund A.C., Tanner J.W., Shaw A.S., Schreiber R.D. Identification of an interferon-gamma receptor alpha chain sequence required for JAK-1 binding. J. Biol. Chem. 1996;271:9–12. doi: 10.1074/jbc.271.1.9. [DOI] [PubMed] [Google Scholar]
- 9.Bach E.A., Tanner J.W., Marsters S., Ashkenazi A., Aguet M., Shaw A.S., Schreiber R.D. Ligand-induced assembly and activation of the gamma interferon receptor in intact cells. Mol. Cell Biol. 1996;16:3214–3221. doi: 10.1128/mcb.16.6.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenlund A.C., Schreiber R.D., Goeddel D.V., Pennica D. Interferon-gamma induces receptor dimerization in solution and on cells. J. Biol. Chem. 1993;268:18103–18110. [PubMed] [Google Scholar]
- 11.Greenlund A.C., Morales M.O., Viviano B.L., Yan H., Krolewski J., Schreiber R.D. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity. 1995;2:677–687. doi: 10.1016/1074-7613(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 12.Heim M.H., Kerr I.M., Stark G.R., Darnell J.E., Jr. Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 13.Shuai K., Horvath C.M., Huang L.H., Qureshi S.A., Cowburn D., Darnell J.E., Jr. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 14.Darnell J.E., Jr., Kerr I.M., Stark G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 15.Colamonici O., Yan H., Domanski P., Handa R., Smalley D., Mullersman J., Witte M., Krishnan K., Krolewski J. Direct binding to and tyrosine phosphorylation of the alpha subunit of the type I interferon receptor by p135tyk2 tyrosine kinase. Mol. Cell Biol. 1994;14:8133–8142. doi: 10.1128/mcb.14.12.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colamonici O.R., Uyttendaele H., Domanski P., Yan H., Krolewski J.J. p135tyk2, an interferon-alpha-activated tyrosine kinase, is physically associated with an interferon-alpha receptor. J. Biol. Chem. 1994;269:3518–3522. [PubMed] [Google Scholar]
- 17.Russell-Harde D., Pu H., Betts M., Harkins R.N., Perez H.D., Croze E. Reconstitution of a high affinity binding site for type I interferons. J. Biol. Chem. 1995;270:26033–26036. doi: 10.1074/jbc.270.44.26033. [DOI] [PubMed] [Google Scholar]
- 18.Cohen B., Novick D., Barak S., Rubinstein M. Ligand-induced association of the type I interferon receptor components. Mol. Cell Biol. 1995;15:4208–4214. doi: 10.1128/mcb.15.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauzzi M.C., Velazquez L., McKendry R., Mogensen K.E., Fellous M., Pellegrini S. Interferon-alpha-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J. Biol. Chem. 1996;271:20494–20500. doi: 10.1074/jbc.271.34.20494. [DOI] [PubMed] [Google Scholar]
- 20.Krishnan K., Yan H., Lim J.T., Krolewski J.J. Dimerization of a chimeric CD4-interferon-alpha receptor reconstitutes the signaling events preceding STAT phosphorylation. Oncogene. 1996;13:125–133. [PubMed] [Google Scholar]
- 21.Subramaniam P.S., Torres B.A., Johnson H.M. So many ligands, so few transcription factors: a new paradigm for signaling through the STAT transcription factors. Cytokine. 2001;15:175–187. doi: 10.1006/cyto.2001.0905. [DOI] [PubMed] [Google Scholar]
- 22.Asao H., Okuyama C., Kumaki S., Ishii N., Tsuchiya S., Foster D., Sugamura K. Cutting edge: the common gamma-chain is an indispensable subunit of the IL-21 receptor complex. J. Immunol. 2001;167:1–5. doi: 10.4049/jimmunol.167.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Sheikh F., Baurin V.V., Lewis-Antes A., Shah N.K., Smirnov S.V., Anantha S., Dickensheets H., Dumoutier L., Renauld J.C., Zdanov A., Donnelly R.P., Kotenko S.V. Cutting edge: IL-26 signals through a novel receptor complex composed of IL-20 receptor 1 and IL-10 receptor 2. J. Immunol. 2004;172:2006–2010. doi: 10.4049/jimmunol.172.4.2006. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimoto T., Okada K., Morishima N., Kamiya S., Owaki T., Asakawa M., Iwakura Y., Fukai F., Mizuguchi J. Induction of IgG2a class switching in B cells by IL-27. J. Immunol. 2004;173:2479–2485. doi: 10.4049/jimmunol.173.4.2479. [DOI] [PubMed] [Google Scholar]
- 25.Chauhan D., Kharbanda S.M., Ogata A., Urashima M., Frank D., Malik N., Kufe D.W., Anderson K.C. Oncostatin M induces association of Grb2 with Janus kinase JAK2 in multiple myeloma cells. J. Exp. Med. 1995;182:1801–1806. doi: 10.1084/jem.182.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit L.S., Meyer D.J., Billestrup N., Norstedt G., Schwartz J., Carter-Su C. The role of the growth hormone (GH) receptor and JAK1 and JAK2 kinases in the activation of Stats 1, 3, and 5 by GH. Mol. Endocrinol. 1996;10:519–533. doi: 10.1210/mend.10.5.8732683. [DOI] [PubMed] [Google Scholar]
- 27.Winston L.A., Hunter T. JAK2, Ras, and Raf are required for activation of extracellular signal-regulated kinase/mitogen-activated protein kinase by growth hormone. J. Biol. Chem. 1995;270:30837–30840. doi: 10.1074/jbc.270.52.30837. [DOI] [PubMed] [Google Scholar]
- 28.Marrero M.B., Schieffer B., Paxton W.G., Heerdt L., Berk B.C., Delafontaine P., Bernstein K.E. Direct stimulation of Jak/STAT pathway by the angiotensin II AT1 receptor. Nature. 1995;375:247–250. doi: 10.1038/375247a0. [DOI] [PubMed] [Google Scholar]
- 29.Boccaccio C., Ando M., Tamagnone L., Bardelli A., Michieli P., Battistini C., Comoglio P.M. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 30.Guo D., Dunbar J.D., Yang C.H., Pfeffer L.M., Donner D.B. Induction of Jak/STAT signaling by activation of the type 1 TNF receptor. J. Immunol. 1998;160:2742–2750. [PubMed] [Google Scholar]
- 31.Dupuis S., Dargemont C., Fieschi C., Thomassin N., Rosenzweig S., Harris J., Holland S.M., Schreiber R.D., Casanova J.L. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293:300–303. doi: 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]
- 32.Prabhu A., Coutinho E., Srivastava S. The amino-terminal domain of human signal transducers and activators of transcription 1: overexpression, purification and characterization. J. Biosci. 2005;30:611–618. doi: 10.1007/BF02703561. [DOI] [PubMed] [Google Scholar]
- 33.Gough D.J., Sabapathy K., Ko E.Y., Arthur H.A., Schreiber R.D., Trapani J.A., Clarke C.J., Johnstone R.W. A novel c-Jun-dependent signal transduction pathway necessary for the transcriptional activation of interferon {gamma} response genes. J. Biol. Chem. 2007;282:938–946. doi: 10.1074/jbc.M607674200. [DOI] [PubMed] [Google Scholar]
- 34.David M., Petricoin E., 3rd, Benjamin C., Pine R., Weber M.J., Larner A.C. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 35.Stephanou A., Scarabelli T.M., Brar B.K., Nakanishi Y., Matsumura M., Knight R.A., Latchman D.S. Induction of apoptosis and Fas receptor/Fas ligand expression by ischemia/reperfusion in cardiac myocytes requires serine 727 of the STAT-1 transcription factor but not tyrosine 701. J. Biol. Chem. 2001;276:28340–28347. doi: 10.1074/jbc.M101177200. [DOI] [PubMed] [Google Scholar]
- 36.Kovarik P., Stoiber D., Eyers P.A., Menghini R., Neininger A., Gaestel M., Cohen P., Decker T. Stress-induced phosphorylation of STAT1 at Ser727 requires p38 mitogen-activated protein kinase whereas IFN-gamma uses a different signaling pathway. Proc. Natl. Acad. Sci. U S A. 1999;96:13956–13961. doi: 10.1073/pnas.96.24.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H.S., Lee M.S. Essential role of STAT1 in caspase-independent cell death of activated macrophages through the p38 mitogen-activated protein kinase/STAT1/reactive oxygen species pathway. Mol. Cell Biol. 2005;25:6821–6833. doi: 10.1128/MCB.25.15.6821-6833.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu W., Nair J.S., Malhotra A., Zhang J.J. B cell antigen receptor signaling enhances IFN-gamma-induced Stat1 target gene expression through calcium mobilization and activation of multiple serine kinase pathways. J. Interferon Cytokine Res. 2005;25:113–124. doi: 10.1089/jir.2005.25.113. [DOI] [PubMed] [Google Scholar]
- 39.Nair J.S., DaFonseca C.J., Tjernberg A., Sun W., Darnell J.E., Jr., Chait B.T., Zhang J.J. Requirement of Ca2+ and CaMKII for Stat1 Ser-727 phosphorylation in response to IFN-gamma. Proc. Natl. Acad. Sci. U S A. 2002;99:5971–5976. doi: 10.1073/pnas.052159099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uddin S., Sassano A., Deb D.K., Verma A., Majchrzak B., Rahman A., Malik A.B., Fish E.N., Platanias L.C. Protein kinase C-delta (PKC-delta) is activated by type I interferons and mediates phosphorylation of Stat1 on serine 727. J. Biol. Chem. 2002;277:14408–14416. doi: 10.1074/jbc.M109671200. [DOI] [PubMed] [Google Scholar]
- 41.Deb D.K., Sassano A., Lekmine F., Majchrzak B., Verma A., Kambhampati S., Uddin S., Rahman A., Fish E.N., Platanias L.C. Activation of protein kinase C delta by IFN-gamma. J. Immunol. 2003;171:267–273. doi: 10.4049/jimmunol.171.1.267. [DOI] [PubMed] [Google Scholar]
- 42.Pilz A., Ramsauer K., Heidari H., Leitges M., Kovarik P., Decker T. Phosphorylation of the Stat1 transactivating domain is required for the response to type I interferons. EMBO Rep. 2003;4:368–373. doi: 10.1038/sj.embor.embor802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovarik P., Mangold M., Ramsauer K., Heidari H., Steinborn R., Zotter A., Levy D.E., Muller M., Decker T. Specificity of signaling by STAT1 depends on SH2 and C-terminal domains that regulate Ser727 phosphorylation, differentially affecting specific target gene expression. EMBO J. 2001;20:91–100. doi: 10.1093/emboj/20.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsauer K., Sadzak I., Porras A., Pilz A., Nebreda A.R., Decker T., Kovarik P. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12859–12864. doi: 10.1073/pnas.192264999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu X., Wen Z., Xu L.Z., Darnell J.E., Jr. Stat1 serine phosphorylation occurs independently of tyrosine phosphorylation and requires an activated Jak2 kinase. Mol. Cell Biol. 1997;17:6618–6623. doi: 10.1128/mcb.17.11.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnholt K.E., Kota R.S., Aung H.H., Rutledge J.C. Adenosine blocks IFN-gamma-induced phosphorylation of STAT1 on serine 727 to reduce macrophage activation. J. Immunol. 2009;183:6767–6777. doi: 10.4049/jimmunol.0900331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qureshi S.A., Leung S., Kerr I.M., Stark G.R., Darnell J.E., Jr. Function of Stat2 protein in transcriptional activation by alpha interferon. Mol. Cell Biol. 1996;16:288–293. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varinou L., Ramsauer K., Karaghiosoff M., Kolbe T., Pfeffer K., Muller M., Decker T. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity. 2003;19:793–802. doi: 10.1016/s1074-7613(03)00322-4. [DOI] [PubMed] [Google Scholar]
- 49.Sun W., Xu W., Snyder M., He W., Ho H., Ivashkiv L.B., Zhang J.J. The conserved Leu-724 residue is required for both serine phosphorylation and co-activator recruitment for Stat1-mediated transcription activation in response to interferon-gamma. J. Biol. Chem. 2005;280:41844–41851. doi: 10.1074/jbc.M505797200. [DOI] [PubMed] [Google Scholar]
- 50.Tenoever B.R., Ng S.L., Chua M.A., McWhirter S.M., Garcia-Sastre A., Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 51.Mowen K.A., Tang J., Zhu W., Schurter B.T., Shuai K., Herschman H.R., David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 52.Meissner T., Krause E., Lodige I., Vinkemeier U. Arginine methylation of STAT1: a reassessment. Cell. 2004;119:587–589. doi: 10.1016/j.cell.2004.11.024. (discussion 589–590) [DOI] [PubMed] [Google Scholar]
- 53.Komyod W., Bauer U.M., Heinrich P.C., Haan S., Behrmann I. Are STATS arginine-methylated? J. Biol. Chem. 2005;280:21700–21705. doi: 10.1074/jbc.C400606200. [DOI] [PubMed] [Google Scholar]
- 54.Weber S., Maass F., Schuemann M., Krause E., Suske G., Bauer U.M. PRMT1-mediated arginine methylation of PIAS1 regulates STAT1 signaling. Genes Dev. 2009;23:118–132. doi: 10.1101/gad.489409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Kramer O.H., Heinzel T. Phosphorylation-acetylation switch in the regulation of STAT1 signaling. Mol. Cell Endocrinol. 2009 doi: 10.1016/j.mce.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Chen X., Vinkemeier U., Zhao Y., Jeruzalmi D., Darnell J.E., Jr., Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 57.Mao X., Ren Z., Parker G.N., Sondermann H., Pastorello M.A., Wang W., McMurray J.S., Demeler B., Darnell J.E., Jr., Chen X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 58.Mertens C., Zhong M., Krishnaraj R., Zou W., Chen X., Darnell J.E., Jr. Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 2006;20:3372–3381. doi: 10.1101/gad.1485406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wenta N., Strauss H., Meyer S., Vinkemeier U. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9238–9243. doi: 10.1073/pnas.0802130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sekimoto T., Imamoto N., Nakajima K., Hirano T., Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gorlich D., Henklein P., Laskey R.A., Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 62.Moroianu J., Blobel G., Radu A. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc. Natl. Acad. Sci. U S A. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gorlich D., Kostka S., Kraft R., Dingwall C., Laskey R.A., Hartmann E., Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 64.Fagerlund R., Melen K., Kinnunen L., Julkunen I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J. Biol. Chem. 2002;277:30072–30078. doi: 10.1074/jbc.M202943200. [DOI] [PubMed] [Google Scholar]
- 65.McBride K.M., Banninger G., McDonald C., Reich N.C. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 2002;21:1754–1763. doi: 10.1093/emboj/21.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyer T., Begitt A., Lodige I., van Rossum M., Vinkemeier U. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 2002;21:344–354. doi: 10.1093/emboj/21.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Melen K., Kinnunen L., Julkunen I. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem. 2001;276:16447–16455. doi: 10.1074/jbc.M008821200. [DOI] [PubMed] [Google Scholar]
- 68.Strehlow I., Schindler C. Amino-terminal signal transducer and activator of transcription (STAT) domains regulate nuclear translocation and STAT deactivation. J. Biol. Chem. 1998;273:28049–28056. doi: 10.1074/jbc.273.43.28049. [DOI] [PubMed] [Google Scholar]
- 69.Meyer T., Marg A., Lemke P., Wiesner B., Vinkemeier U. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 2003;17:1992–2005. doi: 10.1101/gad.268003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer T., Vinkemeier U. STAT nuclear translocation: potential for pharmacological intervention. Expert Opin. Ther. Targets. 2007;11:1355–1365. doi: 10.1517/14728222.11.10.1355. [DOI] [PubMed] [Google Scholar]
- 71.Haspel R.L., Darnell J.E., Jr. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10188–10193. doi: 10.1073/pnas.96.18.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.ten Hoeve J., de Jesus Ibarra-Sanchez M., Fu Y., Zhu W., Tremblay M., David M., Shuai K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meyer T., Gavenis K., Vinkemeier U. Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp. Cell Res. 2002;272:45–55. doi: 10.1006/excr.2001.5405. [DOI] [PubMed] [Google Scholar]
- 74.Marrero M.B., Paxton W.G., Schieffer B., Ling B.N., Bernstein K.E. Angiotensin II signalling events mediated by tyrosine phosphorylation. Cell Signal. 1996;8:21–26. doi: 10.1016/0898-6568(95)02016-0. [DOI] [PubMed] [Google Scholar]
- 75.Yang J., Liao X., Agarwal M.K., Barnes L., Auron P.E., Stark G.R. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stade K., Ford C.S., Guthrie C., Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 77.Fornerod M., Ohno M., Yoshida M., Mattaj I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 78.Richards S.A., Carey K.L., Macara I.G. Requirement of guanosine triphosphate-bound ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 79.McBride K.M., McDonald C., Reich N.C. Nuclear export signal located within the DNA-binding domain of the STAT1transcription factor. EMBO J. 2000;19:6196–6206. doi: 10.1093/emboj/19.22.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lodige I., Marg A., Wiesner B., Malecova B., Oelgeschlager T., Vinkemeier U. Nuclear export determines the cytokine sensitivity of STAT transcription factors. J. Biol. Chem. 2005;280:43087–43099. doi: 10.1074/jbc.M509180200. [DOI] [PubMed] [Google Scholar]
- 81.Braunstein J., Brutsaert S., Olson R., Schindler C. STATs dimerize in the absence of phosphorylation. J. Biol. Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 82.Mao X., Chen X. Crystallization and X-ray crystallographic analysis of human STAT1. Acta Crystallogr. Sect. F Struct. Biol. Cryst Commun. 2005;61:666–668. doi: 10.1107/S1744309105017392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhong M., Henriksen M.A., Takeuchi K., Schaefer O., Liu B., ten Hoeve J., Ren Z., Mao X., Chen X., Shuai K., Darnell J.E., Jr. Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc. Natl. Acad. Sci. U S A. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Decker T., Kovarik P., Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 85.Xu X., Sun Y.L., Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 86.Guyer N.B., Severns C.W., Wong P., Feghali C.A., Wright T.M. IFN-gamma induces a p91/Stat1 alpha-related transcription factor with distinct activation and binding properties. J. Immunol. 1995;155:3472–3480. [PubMed] [Google Scholar]
- 87.Vinkemeier U., Cohen S.L., Moarefi I., Chait B.T., Kuriyan J., Darnell J.E., Jr. DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 88.Haque S.J., Williams B.R. Identification and characterization of an interferon (IFN)-stimulated response element-IFN-stimulated gene factor 3-independent signaling pathway for IFN-alpha. J. Biol. Chem. 1994;269:19523–19529. [PubMed] [Google Scholar]
- 89.Bluyssen H.A., Levy D.E. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J. Biol. Chem. 1997;272:4600–4605. doi: 10.1074/jbc.272.7.4600. [DOI] [PubMed] [Google Scholar]
- 90.Lew D.J., Decker T., Strehlow I., Darnell J.E. Overlapping elements in the guanylate-binding protein gene promoter mediate transcriptional induction by alpha and gamma interferons. Mol. Cell Biol. 1991;11:182–191. doi: 10.1128/mcb.11.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Decker T., Lew D.J., Cheng Y.S., Levy D.E., Darnell J.E., Jr. Interactions of alpha- and gamma-interferon in the transcriptional regulation of the gene encoding a guanylate-binding protein. EMBO J. 1989;8:2009–2014. doi: 10.1002/j.1460-2075.1989.tb03608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Decker T., Lew D.J., Darnell J.E., Jr. Two distinct alpha-interferon-dependent signal transduction pathways may contribute to activation of transcription of the guanylate-binding protein gene. Mol. Cell Biol. 1991;11:5147–5153. doi: 10.1128/mcb.11.10.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Escalante C.R., Yie J., Thanos D., Aggarwal A.K. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 94.Kusumoto M., Fujii Y., Tsukuda Y., Ohira T., Kyougoku Y., Taniguchi T., Hakoshima T. Crystallographic characterization of the DNA-binding domain of interferon regulatory dactor-2 complexed with DNA. J. Struct. Biol. 1998;121:363–366. doi: 10.1006/jsbi.1998.3970. [DOI] [PubMed] [Google Scholar]
- 95.Ehret G.B., Reichenbach P., Schindler U., Horvath C.M., Fritz S., Nabholz M., Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 96.Regis G., Icardi L., Conti L., Chiarle R., Piva R., Giovarelli M., Poli V., Novelli F. IL-6, but not IFN-gamma, triggers apoptosis and inhibits in vivo growth of human malignant T cells on STAT3 silencing. Leukemia. 2009 doi: 10.1038/leu.2009.139. [DOI] [PubMed] [Google Scholar]
- 97.Regis G., Pensa S., Boselli D., Novelli F., Poli V. Ups and downs: the STAT1:STAT3 seesaw of Interferon and gp130 receptor signalling. Semin. Cell Dev. Biol. 2008 doi: 10.1016/j.semcdb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 98.Hartman S.E., Bertone P., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhinge A.A., Kim J., Euskirchen G.M., Snyder M., Iyer V.R. Mapping the chromosomal targets of STAT1 by sequence tag analysis of genomic enrichment (STAGE) Genome Res. 2007;17:910–916. doi: 10.1101/gr.5574907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ouchi T., Lee S.W., Ouchi M., Aaronson S.A., Horvath C.M. Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-gamma target genes. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5208–5213. doi: 10.1073/pnas.080469697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J.J., Zhao Y., Chait B.T., Lathem W.W., Ritzi M., Knippers R., Darnell J.E., Jr. Ser727-dependent recruitment of MCM5 by Stat1alpha in IFN-gamma-induced transcriptional activation. EMBO J. 1998;17:6963–6971. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DaFonseca C.J., Shu F., Zhang J.J. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3034–3039. doi: 10.1073/pnas.061487598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang J.J., Vinkemeier U., Gu W., Chakravarti D., Horvath C.M., Darnell J.E., Jr. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bhattacharya S., Eckner R., Grossman S., Oldread E., Arany Z., D'Andrea A., Livingston D.M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 105.Si Q., Zhao M.L., Morgan A.C., Brosnan C.F., Lee S.C. 15-deoxy-Delta12,14-prostaglandin J2 inhibits IFN-inducible protein 10/C × C chemokine ligand 10 expression in human microglia: mechanisms and implications. J. Immunol. 2004;173:3504–3513. doi: 10.4049/jimmunol.173.5.3504. [DOI] [PubMed] [Google Scholar]
- 106.Chen B., He L., Savell V.H., Jenkins J.J., Parham D.M. Inhibition of the interferon-gamma/signal transducers and activators of transcription (STAT) pathway by hypermethylation at a STAT-binding site in the p21WAF1 promoter region. Cancer Res. 2000;60:3290–3298. [PubMed] [Google Scholar]
- 107.Agrawal S., Agarwal M.L., Chatterjee-Kishore M., Stark G.R., Chisolm G.M. Stat1-dependent, p53-independent expression of p21(waf1) modulates oxysterol-induced apoptosis. Mol. Cell Biol. 2002;22:1981–1992. doi: 10.1128/MCB.22.7.1981-1992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bhanoori M., Yellaturu C.R., Ghosh S.K., Hassid A., Jennings L.K., Rao G.N. Thiol alkylation inhibits the mitogenic effects of platelet-derived growth factor and renders it proapoptotic via activation of STATs and p53 and induction of expression of caspase1 and p21(waf1/cip1) Oncogene. 2003;22:117–130. doi: 10.1038/sj.onc.1206065. [DOI] [PubMed] [Google Scholar]
- 109.Stephanou A., Isenberg D.A., Nakajima K., Latchman D.S. Signal transducer and activator of transcription-1 and heat shock factor-1 interact and activate the transcription of the Hsp-70 and Hsp-90beta gene promoters. J. Biol. Chem. 1999;274:1723–1728. doi: 10.1074/jbc.274.3.1723. [DOI] [PubMed] [Google Scholar]
- 110.Chen X.S., Wu N.H., Shen Y.F. Role of STAT1 on the regulation the human hsp90 alpha gene expressionZhongguo Yi Xue Ke Xue Yuan Xue Bao. 2001;23:356–360. [PubMed] [Google Scholar]
- 111.Ohmori Y., Schreiber R.D., Hamilton T.A. Synergy between interferon-gamma and tumor necrosis factor-alpha in transcriptional activation is mediated by cooperation between signal transducer and activator of transcription 1 and nuclear factor kappaB. J. Biol. Chem. 1997;272:14899–14907. doi: 10.1074/jbc.272.23.14899. [DOI] [PubMed] [Google Scholar]
- 112.Majumder S., Zhou L.Z., Chaturvedi P., Babcock G., Aras S., Ransohoff R.M. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J. Immunol. 1998;161:4736–4744. [PubMed] [Google Scholar]
- 113.Jahnke A., Johnson J.P. Synergistic activation of intercellular adhesion molecule 1 (ICAM-1) by TNF-alpha and IFN-gamma is mediated by p65/p50 and p65/c-Rel and interferon-responsive factor Stat1 alpha (p91) that can be activated by both IFN-gamma and IFN-alpha. FEBS Lett. 1994;354:220–226. doi: 10.1016/0014-5793(94)01130-3. [DOI] [PubMed] [Google Scholar]
- 114.Jahnke A., Johnson J.P. Intercellular adhesion molecule 1 (ICAM-1) is synergistically activated by TNF-alpha and IFN-gamma responsive sites. Immunobiology. 1995;193:305–314. doi: 10.1016/s0171-2985(11)80559-0. [DOI] [PubMed] [Google Scholar]
- 115.Look D.C., Pelletier M.R., Tidwell R.M., Roswit W.T., Holtzman M.J. Stat1 depends on transcriptional synergy with Sp1. J. Biol. Chem. 1995;270:30264–30267. doi: 10.1074/jbc.270.51.30264. [DOI] [PubMed] [Google Scholar]
- 116.Book McAlexander M., Yu-Lee L.Y. Sp1 is required for prolactin activation of the interferon regulatory factor-1 gene. Mol. Cell Endocrinol. 2001;184:135–141. doi: 10.1016/s0303-7207(01)00593-7. [DOI] [PubMed] [Google Scholar]
- 117.Aittomaki S., Pesu M., Groner B., Janne O.A., Palvimo J.J., Silvennoinen O. Cooperation among Stat1, glucocorticoid receptor, and PU.1 in transcriptional activation of the high-affinity Fc gamma receptor I in monocytes. J. Immunol. 2000;164:5689–5697. doi: 10.4049/jimmunol.164.11.5689. [DOI] [PubMed] [Google Scholar]
- 118.Kumatori A., Yang D., Suzuki S., Nakamura M. Cooperation of STAT-1 and IRF-1 in interferon-gamma-induced transcription of the gp91(phox) gene. J. Biol. Chem. 2002;277:9103–9111. doi: 10.1074/jbc.M109803200. [DOI] [PubMed] [Google Scholar]
- 119.Aittomaki S., Yang J., Scott E.W., Simon M.C., Silvennoinen O. Distinct functions for signal transducer and activator of transcription 1 and PU.1 in transcriptional activation of Fc gamma receptor I promoter. Blood. 2002;100:1078–1080. doi: 10.1182/blood-2001-12-0236. [DOI] [PubMed] [Google Scholar]
- 120.Aittomaki S., Yang J., Scott E.W., Simon M.C., Silvennoinen O. Molecular basis of Stat1 and PU.1 cooperation in cytokine-induced Fcgamma receptor I promoter activation. Int. Immunol. 2004;16:265–274. doi: 10.1093/intimm/dxh037. [DOI] [PubMed] [Google Scholar]
- 121.Murphy D., Detjen K.M., Welzel M., Wiedenmann B., Rosewicz S. Interferon-alpha delays S-phase progression in human hepatocellular carcinoma cells via inhibition of specific cyclin-dependent kinases. Hepatology. 2001;33:346–356. doi: 10.1053/jhep.2001.21749. [DOI] [PubMed] [Google Scholar]
- 122.Sibinga N.E., Wang H., Perrella M.A., Endege W.O., Patterson C., Yoshizumi M., Haber E., Lee M.E. Interferon-gamma-mediated inhibition of cyclin A gene transcription is independent of individual cis-acting elements in the cyclin A promoter. J. Biol. Chem. 1999;274:12139–12146. doi: 10.1074/jbc.274.17.12139. [DOI] [PubMed] [Google Scholar]
- 123.Dimberg A., Karlberg I., Nilsson K., Oberg F. Ser727/Tyr701-phosphorylated Stat1 is required for the regulation of c-Myc, cyclins, and p27Kip1 associated with ATRA-induced G0/G1 arrest of U-937 cells. Blood. 2003;102:254–261. doi: 10.1182/blood-2002-10-3149. [DOI] [PubMed] [Google Scholar]
- 124.Ramana C.V., Grammatikakis N., Chernov M., Nguyen H., Goh K.C., Williams B.R., Stark G.R. Regulation of c-myc expression by IFN-gamma through Stat1-dependent and -independent pathways. EMBO J. 2000;19:263–272. doi: 10.1093/emboj/19.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sanceau J., Boyd D.D., Seiki M., Bauvois B. Interferons inhibit tumor necrosis factor-alpha-mediated matrix metalloproteinase-9 activation via interferon regulatory factor-1 binding competition with NF-kappa B. J. Biol. Chem. 2002;277:35766–35775. doi: 10.1074/jbc.M202959200. [DOI] [PubMed] [Google Scholar]
- 126.Stephanou A., Brar B.K., Knight R.A., Latchman D.S. Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and Bcl-x promoters. Cell Death Differ. 2000;7:329–330. doi: 10.1038/sj.cdd.4400656. [DOI] [PubMed] [Google Scholar]
- 127.Cheon H., Stark G.R. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chatterjee-Kishore M., Wright K.L., Ting J.P., Stark G.R. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chatterjee-Kishore M., Kishore R., Hicklin D.J., Marincola F.M., Ferrone S. Different requirements for signal transducer and activator of transcription 1alpha and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J. Biol. Chem. 1998;273:16177–16183. doi: 10.1074/jbc.273.26.16177. [DOI] [PubMed] [Google Scholar]
- 130.Kumar A., Commane M., Flickinger T.W., Horvath C.M., Stark G.R. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 131.Zakharova N., Lymar E.S., Yang E., Malik S., Zhang J.J., Roeder R.G., Darnell J.E., Jr. Distinct transcriptional activation functions of STAT1alpha and STAT1beta on DNA and chromatin templates. J. Biol. Chem. 2003;278:43067–43073. doi: 10.1074/jbc.M308166200. [DOI] [PubMed] [Google Scholar]
- 132.Horvath C.M., Darnell J.E., Jr. The antiviral state induced by alpha interferon and gamma interferon requires transcriptionally active Stat1 protein. J. Virol. 1996;70:647–650. doi: 10.1128/jvi.70.1.647-650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., Al Ghonaium A., Tufenkeji H., Frayha H., Al-Gazlan S., Al-Rayes H., Schreiber R.D., Gresser I., Casanova J.L. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 134.Meraz M.A., White J.M., Sheehan K.C., Bach E.A., Rodig S.J., Dighe A.S., Kaplan D.H., Riley J.K., Greenlund A.C., Campbell D., Carver-Moore K., DuBois R.N., Clark R., Aguet M., Schreiber R.D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 135.Nauciel C., Espinasse-Maes F. Role of gamma interferon and tumor necrosis factor alpha in resistance to Salmonella typhimurium infection. Infect. Immun. 1992;60:450–454. doi: 10.1128/iai.60.2.450-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gavrilescu L.C., Butcher B.A., Del Rio L., Taylor G.A., Denkers E.Y. STAT1 is essential for antimicrobial effector function but dispensable for gamma interferon production during Toxoplasma gondii infection. Infect. Immun. 2004;72:1257–1264. doi: 10.1128/IAI.72.3.1257-1264.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nandan D., Reiner N.E. Attenuation of gamma interferon-induced tyrosine phosphorylation in mononuclear phagocytes infected with Leishmania donovani: selective inhibition of signaling through Janus kinases and Stat1. Infect. Immun. 1995;63:4495–4500. doi: 10.1128/iai.63.11.4495-4500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rosas L.E., Keiser T., Pyles R., Durbin J., Satoskar A.R. Development of protective immunity against cutaneous leishmaniasis is dependent on STAT1-mediated IFN signaling pathway. Eur. J. Immunol. 2003;33:1799–1805. doi: 10.1002/eji.200323163. [DOI] [PubMed] [Google Scholar]
- 139.Durbin J.E., Hackenmiller R., Simon M.C., Levy D.E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 140.Chapgier A., Boisson-Dupuis S., Jouanguy E., Vogt G., Feinberg J., Prochnicka-Chalufour A., Casrouge A., Yang K., Soudais C., Fieschi C., Santos O.F., Bustamante J., Picard C., de Beaucoudrey L., Emile J.F., Arkwright P.D., Schreiber R.D., Rolinck-Werninghaus C., Rosen-Wolff A., Magdorf K., Roesler J., Casanova J.L. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2:e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chapgier A., Kong X.F., Boisson-Dupuis S., Jouanguy E., Averbuch D., Feinberg J., Zhang S.Y., Bustamante J., Vogt G., Lejeune J., Mayola E., de Beaucoudrey L., Abel L., Engelhard D., Casanova J.L. A partial form of recessive STAT1 deficiency in humans. J. Clin. Invest. 2009;119:1502–1514. doi: 10.1172/JCI37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Min W., Pober J.S., Johnson D.R. Kinetically coordinated induction of TAP1 and HLA class I by IFN-gamma: the rapid induction of TAP1 by IFN-gamma is mediated by Stat1 alpha. J. Immunol. 1996;156:3174–3183. [PubMed] [Google Scholar]
- 143.Durbin J.E., Fernandez-Sesma A., Lee C.K., Rao T.D., Frey A.B., Moran T.M., Vukmanovic S., Garcia-Sastre A., Levy D.E. Type I IFN modulates innate and specific antiviral immunity. J. Immunol. 2000;164:4220–4228. doi: 10.4049/jimmunol.164.8.4220. [DOI] [PubMed] [Google Scholar]
- 144.Xu W., Zhang J.J. Stat1-dependent synergistic activation of T-bet for IgG2a production during early stage of B cell activation. J. Immunol. 2005;175:7419–7424. doi: 10.4049/jimmunol.175.11.7419. [DOI] [PubMed] [Google Scholar]
- 145.Peng S.L., Szabo S.J., Glimcher L.H. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl. Acad. Sci. U S A. 2002;99:5545–5550. doi: 10.1073/pnas.082114899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bromberg J.F., Horvath C.M., Wen Z., Schreiber R.D., Darnell J.E., Jr. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. U S A. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bromberg J.F., Fan Z., Brown C., Mendelsohn J., Darnell J.E., Jr. Epidermal growth factor-induced growth inhibition requires Stat1 activation. Cell Growth Differ. 1998;9:505–512. [PubMed] [Google Scholar]
- 148.Ohtsubo M., Takayanagi A., Gamou S., Shimizu N. Interruption of NFkappaB-STAT1 signaling mediates EGF-induced cell-cycle arrest. J. Cell Physiol. 2000;184:131–137. doi: 10.1002/(SICI)1097-4652(200007)184:1<131::AID-JCP14>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 149.Sherr C.J., Roberts J.M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 150.Cheng M., Olivier P., Diehl J.A., Fero M., Roussel M.F., Roberts J.M., Sherr C.J. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chin Y.E., Kitagawa M., Su W.C., You Z.H., Iwamoto Y., Fu X.Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 152.Jiang C.K., Flanagan S., Ohtsuki M., Shuai K., Freedberg I.M., Blumenberg M. Disease-activated transcription factor: allergic reactions in human skin cause nuclear translocation of STAT-91 and induce synthesis of keratin K17. Mol. Cell Biol. 1994;14:4759–4769. doi: 10.1128/mcb.14.7.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schlee M., Holzel M., Bernard S., Mailhammer R., Schuhmacher M., Reschke J., Eick D., Marinkovic D., Wirth T., Rosenwald A., Staudt L.M., Eilers M., Baran-Marszak F., Fagard R., Feuillard J., Laux G., Bornkamm G.W. C-myc activation impairs the NF-kappaB and the interferon response: implications for the pathogenesis of Burkitt's lymphoma. Int. J. Cancer. 2007;120:1387–1395. doi: 10.1002/ijc.22372. [DOI] [PubMed] [Google Scholar]
- 154.Lee C.K., Smith E., Gimeno R., Gertner R., Levy D.E. STAT1 affects lymphocyte survival and proliferation partially independent of its role downstream of IFN-gamma. J. Immunol. 2000;164:1286–1292. doi: 10.4049/jimmunol.164.3.1286. [DOI] [PubMed] [Google Scholar]
- 155.Lee C.K., Rao D.T., Gertner R., Gimeno R., Frey A.B., Levy D.E. Distinct requirements for IFNs and STAT1 in NK cell function. J. Immunol. 2000;165:3571–3577. doi: 10.4049/jimmunol.165.7.3571. [DOI] [PubMed] [Google Scholar]
- 156.Gelebart P., Zak Z., Anand M., Dien-Bard J., Amin H.M., Lai R. Interleukin-21 effectively induces apoptosis in mantle cell lymphoma through a STAT1-dependent mechanism. Leukemia. 2009;23:1836–1846. doi: 10.1038/leu.2009.100. [DOI] [PubMed] [Google Scholar]
- 157.Ossina N.K., Cannas A., Powers V.C., Fitzpatrick P.A., Knight J.D., Gilbert J.R., Shekhtman E.M., Tomei L.D., Umansky S.R., Kiefer M.C. Interferon-gamma modulates a p53-independent apoptotic pathway and apoptosis-related gene expression. J. Biol. Chem. 1997;272:16351–16357. doi: 10.1074/jbc.272.26.16351. [DOI] [PubMed] [Google Scholar]
- 158.Xu X., Fu X.Y., Plate J., Chong A.S. IFN-gamma induces cell growth inhibition by Fas-mediated apoptosis: requirement of STAT1 protein for up-regulation of Fas and FasL expression. Cancer Res. 1998;58:2832–2837. [PubMed] [Google Scholar]
- 159.Lee S.J., Zhou T., Choi C., Wang Z., Benveniste E.N. Differential regulation and function of Fas expression on glial cells. J. Immunol. 2000;164:1277–1285. doi: 10.4049/jimmunol.164.3.1277. [DOI] [PubMed] [Google Scholar]
- 160.Stephanou A., Brar B.K., Scarabelli T.M., Jonassen A.K., Yellon D.M., Marber M.S., Knight R.A., Latchman D.S. Ischemia-induced STAT-1 expression and activation play a critical role in cardiomyocyte apoptosis. J. Biol. Chem. 2000;275:10002–10008. doi: 10.1074/jbc.275.14.10002. [DOI] [PubMed] [Google Scholar]
- 161.Miura Y., Tsujioka T., Nishimura Y., Sakaguchi H., Maeda M., Hayashi H., Dong M., Hyodoh F., Yata K., Wada H., Sugihara T., Otsuki T. TRAIL expression up-regulated by interferon-gamma via phosphorylation of STAT1 induces myeloma cell death. Anticancer Res. 2006;26:4115–4124. [PubMed] [Google Scholar]
- 162.Wesemann D.R., Qin H., Kokorina N., Benveniste E.N. TRADD interacts with STAT1-alpha and influences interferon-gamma signaling. Nat. Immunol. 2004;5:199–207. doi: 10.1038/ni1025. [DOI] [PubMed] [Google Scholar]
- 163.Wang Y., Wu T.R., Cai S., Welte T., Chin Y.E. Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol. Cell Biol. 2000;20:4505–4512. doi: 10.1128/mcb.20.13.4505-4512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Suk K., Chang I., Kim Y.H., Kim S., Kim J.Y., Kim H., Lee M.S. Interferon gamma (IFNgamma) and tumor necrosis factor alpha synergism in ME-180 cervical cancer cell apoptosis and necrosis. IFNgamma inhibits cytoprotective NF-kappa B through STAT1/IRF-1 pathways. J. Biol. Chem. 2001;276:13153–13159. doi: 10.1074/jbc.M007646200. [DOI] [PubMed] [Google Scholar]
- 165.Harris S.L., Levine A.J. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 166.Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S., McKinney K., Tempst P., Prives C., Gamblin S.J., Barlev N.A., Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 167.Gu W., Roeder R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 168.Lavin M.F., Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 169.Shieh S.Y., Ikeda M., Taya Y., Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 170.Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 171.Kaplan D.H., Shankaran V., Dighe A.S., Stockert E., Aguet M., Old L.J., Schreiber R.D. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. U S A. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Townsend P.A., Scarabelli T.M., Davidson S.M., Knight R.A., Latchman D.S., Stephanou A. STAT-1 interacts with p53 to enhance DNA damage-induced apoptosis. J. Biol. Chem. 2004;279:5811–5820. doi: 10.1074/jbc.M302637200. [DOI] [PubMed] [Google Scholar]
- 173.Townsend P.A., Cragg M.S., Davidson S.M., McCormick J., Barry S., Lawrence K.M., Knight R.A., Hubank M., Chen P.L., Latchman D.S., Stephanou A. STAT-1 facilitates the ATM activated checkpoint pathway following DNA damage. J. Cell Sci. 2005;118:1629–1639. doi: 10.1242/jcs.01728. [DOI] [PubMed] [Google Scholar]
- 174.Baran-Marszak F., Feuillard J., Najjar I., Le Clorennec C., Bechet J.M., Dusanter-Fourt I., Bornkamm G.W., Raphael M., Fagard R. Differential roles of STAT1alpha and STAT1beta in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood. 2004;104:2475–2483. doi: 10.1182/blood-2003-10-3508. [DOI] [PubMed] [Google Scholar]
- 175.Keating M.J., Kantarjian H., Talpaz M., Redman J., Koller C., Barlogie B., Velasquez W., Plunkett W., Freireich E.J., McCredie K.B. Fludarabine: a new agent with major activity against chronic lymphocytic leukemia. Blood. 1989;74:19–25. [PubMed] [Google Scholar]
- 176.Alvi A.J., Austen B., Weston V.J., Fegan C., MacCallum D., Gianella-Borradori A., Lane D.P., Hubank M., Powell J.E., Wei W., Taylor A.M., Moss P.A., Stankovic T. A novel CDK inhibitor, CYC202 (R-roscovitine), overcomes the defect in p53-dependent apoptosis in B-CLL by down-regulation of genes involved in transcription regulation and survival. Blood. 2005;105:4484–4491. doi: 10.1182/blood-2004-07-2713. [DOI] [PubMed] [Google Scholar]
- 177.Frank D.A., Mahajan S., Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat. Med. 1999;5:444–447. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- 178.Fagard R., Mouas H., Dusanter-Fourt I., Devillers C., Bissieres P., Martin A., Lenoir G., VanTan H., Feuillard J., Raphael M. Resistance to fludarabine-induced apoptosis in Epstein-Barr virus infected B cells. Oncogene. 2002;21:4473–4480. doi: 10.1038/sj.onc.1205554. [DOI] [PubMed] [Google Scholar]
- 179.Youlyouz-Marfak I., Gachard N., Le Clorennec C., Najjar I., Baran-Marszak F., Reminieras L., May E., Bornkamm G.W., Fagard R., Feuillard J. Identification of a novel p53-dependent activation pathway of STAT1 by antitumour genotoxic agents. Cell Death Differ. 2008;15:376–385. doi: 10.1038/sj.cdd.4402270. [DOI] [PubMed] [Google Scholar]
- 180.Kim S., Koga T., Isobe M., Kern B.E., Yokochi T., Chin Y.E., Karsenty G., Taniguchi T., Takayanagi H. Stat1 functions as a cytoplasmic attenuator of Run × 2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17:1979–1991. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Takayanagi H., Kim S., Koga T., Taniguchi T. Stat1-mediated cytoplasmic attenuation in osteoimmunology. J. Cell Biochem. 2005;94:232–240. doi: 10.1002/jcb.20316. [DOI] [PubMed] [Google Scholar]
- 182.Amador M.L., Jimeno J., Paz-Ares L., Cortes-Funes H., Hidalgo M. Progress in the development and acquisition of anticancer agents from marine sources. Ann. Oncol. 2003;14:1607–1615. doi: 10.1093/annonc/mdg443. [DOI] [PubMed] [Google Scholar]
- 183.Battle T.E., Frank D.A. STAT1 mediates differentiation of chronic lymphocytic leukemia cells in response to Bryostatin 1. Blood. 2003;102:3016–3024. doi: 10.1182/blood-2002-09-2972. [DOI] [PubMed] [Google Scholar]
- 184.Pelicano L., Brumpt C., Pitha P.M., Chelbi-Alix M.K. Retinoic acid resistance in NB4 APL cells is associated with lack of interferon alpha synthesis Stat1 and p48 induction. Oncogene. 1999;18:3944–3953. doi: 10.1038/sj.onc.1202802. [DOI] [PubMed] [Google Scholar]
- 185.Dimberg A., Nilsson K., Oberg F. Phosphorylation-deficient Stat1 inhibits retinoic acid-induced differentiation and cell cycle arrest in U-937 monoblasts. Blood. 2000;96:2870–2878. [PubMed] [Google Scholar]
- 186.Huang S., Bucana C.D., Van Arsdall M., Fidler I.J. Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene. 2002;21:2504–2512. doi: 10.1038/sj.onc.1205341. [DOI] [PubMed] [Google Scholar]
- 187.Lesinski G.B., Anghelina M., Zimmerer J., Bakalakos T., Badgwell B., Parihar R., Hu Y., Becknell B., Abood G., Chaudhury A.R., Magro C., Durbin J., Carson W.E., 3rd The antitumor effects of IFN-alpha are abrogated in a STAT1-deficient mouse. J. Clin. Invest. 2003;112:170–180. doi: 10.1172/JCI16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wong L.H., Krauer K.G., Hatzinisiriou I., Estcourt M.J., Hersey P., Tam N.D., Edmondson S., Devenish R.J., Ralph S.J. Interferon-resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48-ISGF3gamma. J. Biol. Chem. 1997;272:28779–28785. doi: 10.1074/jbc.272.45.28779. [DOI] [PubMed] [Google Scholar]
- 189.Wong L.H., Hatzinisiriou I., Devenish R.J., Ralph S.J. IFN-gamma priming up-regulates IFN-stimulated gene factor 3 (ISGF3) components, augmenting responsiveness of IFN-resistant melanoma cells to type I IFNs. J. Immunol. 1998;160:5475–5484. [PubMed] [Google Scholar]
- 190.Dovhey S.E., Ghosh N.S., Wright K.L. Loss of interferon-gamma inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line. Cancer Res. 2000;60:5789–5796. [PubMed] [Google Scholar]
- 191.Widschwendter A., Tonko-Geymayer S., Welte T., Daxenbichler G., Marth C., Doppler W. Prognostic significance of signal transducer and activator of transcription 1 activation in breast cancer. Clin. Cancer Res. 2002;8:3065–3074. [PubMed] [Google Scholar]
- 192.Timofeeva O.A., Plisov S., Evseev A.A., Peng S., Jose-Kampfner M., Lovvorn H.N., Dome J.S., Perantoni A.O. Serine-phosphorylated STAT1 is a prosurvival factor in Wilms' tumor pathogenesis. Oncogene. 2006;25:7555–7564. doi: 10.1038/sj.onc.1209742. [DOI] [PubMed] [Google Scholar]
- 193.Wang S., Koromilas A.E. Stat1 is an inhibitor of Ras-MAPK signaling and Rho small GTPase expression with implications in the transcriptional signature of Ras transformed cells. Cell Cycle. 2009;8:2070–2079. doi: 10.4161/cc.8.13.8891. [DOI] [PubMed] [Google Scholar]
- 194.Muhlethaler-Mottet A., Di Berardino W., Otten L.A., Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 195.Kovacic B., Stoiber D., Moriggl R., Weisz E., Ott R.G., Kreibich R., Levy D.E., Beug H., Freissmuth M., Sexl V. STAT1 acts as a tumor promoter for leukemia development. Cancer Cell. 2006;10:77–87. doi: 10.1016/j.ccr.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 196.Pitroda S.P., Wakim B.T., Sood R.F., Beveridge M.G., Beckett M.A., Macdermed D.M., Weichselbaum R.R., Khodarev N.N. STAT1-dependent expression of energy metabolic pathways links tumour growth and radioresistance to the Warburg effect. BMC Med. 2009;7:68. doi: 10.1186/1741-7015-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Honda K., Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 198.Paun A., Pitha P.M. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schroder K., Hertzog P.J., Ravasi T., Hume D.A. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 200.Rothermel C.D., Rubin B.Y., Murray H.W. Gamma-interferon is the factor in lymphokine that activates human macrophages to inhibit intracellular Chlamydia psittaci replication. J. Immunol. 1983;131:2542–2544. [PubMed] [Google Scholar]
- 201.Rothermel C.D., Byrne G.I., Havell E.A. Effect of interferon on the growth of Chlamydia trachomatis in mouse fibroblasts (L cells) Infect. Immun. 1983;39:362–370. doi: 10.1128/iai.39.1.362-370.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nash T.W., Libby D.M., Horwitz M.A. IFN-gamma-activated human alveolar macrophages inhibit the intracellular multiplication of Legionella pneumophila. J. Immunol. 1988;140:3978–3981. [PubMed] [Google Scholar]
- 203.Didcock L., Young D.F., Goodbourn S., Randall R.E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ulane C.M., Horvath C.M. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304:160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- 205.Yokosawa N., Yokota S., Kubota T., Fujii N. C-terminal region of STAT-1alpha is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J. Virol. 2002;76:12683–12690. doi: 10.1128/JVI.76.24.12683-12690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ulane C.M., Kentsis A., Cruz C.D., Parisien J.P., Schneider K.L., Horvath C.M. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J. Virol. 2005;79:10180–10189. doi: 10.1128/JVI.79.16.10180-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Horvath C.M. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 2004;271:4621–4628. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- 208.Huang Z., Krishnamurthy S., Panda A., Samal S.K. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 2003;77:8676–8685. doi: 10.1128/JVI.77.16.8676-8685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Garcin D., Marq J.B., Strahle L., le Mercier P., Kolakofsky D. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology. 2002;295:256–265. doi: 10.1006/viro.2001.1342. [DOI] [PubMed] [Google Scholar]
- 210.Dinwiddie D.L., Harrod K.S. Human metapneumovirus inhibits IFN-alpha signaling through inhibition of STAT1 phosphorylation. Am. J. Respir. Cell Mol. Biol. 2008;38:661–670. doi: 10.1165/rcmb.2007-0285OC. [DOI] [PubMed] [Google Scholar]
- 211.Ray M., Gam A.A., Boykins R.A., Kenney R.T. Inhibition of interferon-gamma signaling by Leishmania donovani. J. Infect. Dis. 2000;181:1121–1128. doi: 10.1086/315330. [DOI] [PubMed] [Google Scholar]
- 212.Bhardwaj N., Rosas L.E., Lafuse W.P., Satoskar A.R. Leishmania inhibits STAT1-mediated IFN-gamma signaling in macrophages: increased tyrosine phosphorylation of dominant negative STAT1beta by Leishmania mexicana. Int. J. Parasitol. 2005;35:75–82. doi: 10.1016/j.ijpara.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 213.Forget G., Gregory D.J., Olivier M. Proteasome-mediated degradation of STAT1alpha following infection of macrophages with Leishmania donovani. J. Biol. Chem. 2005;280:30542–30549. doi: 10.1074/jbc.M414126200. [DOI] [PubMed] [Google Scholar]
- 214.Barbi J., Snider H.M., Bhardwaj N., Lezama-Davila C.M., Durbin J.E., Satoskar A.R. Signal transducer and activator of transcription 1 in T cells plays an indispensable role in immunity to Leishmania major by mediating Th1 cell homing to the site of infection. FASEB J. 2009;23:3990–3999. doi: 10.1096/fj.09-138057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Xin L., Li K., Soong L. Down-regulation of dendritic cell signaling pathways by Leishmania amazonensis amastigotes. Mol. Immunol. 2008;45:3371–3382. doi: 10.1016/j.molimm.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Forget G., Gregory D.J., Whitcombe L.A., Olivier M. Role of host protein tyrosine phosphatase SHP-1 in Leishmania donovani-induced inhibition of nitric oxide production. Infect. Immun. 2006;74:6272–6279. doi: 10.1128/IAI.00853-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rosas L.E., Snider H.M., Barbi J., Satoskar A.A., Lugo-Villarino G., Keiser T., Papenfuss T., Durbin J.E., Radzioch D., Glimcher L.H., Satoskar A.R. Cutting edge: STAT1 and T-bet play distinct roles in determining outcome of visceral leishmaniasis caused by Leishmania donovani. J. Immunol. 2006;177:22–25. doi: 10.4049/jimmunol.177.1.22. [DOI] [PubMed] [Google Scholar]
- 218.Nishio M., Tsurudome M., Ito M., Kawano M., Komada H., Ito Y. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 2001;75:9165–9176. doi: 10.1128/JVI.75.19.9165-9176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rodriguez J.J., Parisien J.P., Horvath C.M. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 2002;76:11476–11483. doi: 10.1128/JVI.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rodriguez J.J., Wang L.F., Horvath C.M. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J. Virol. 2003;77:11842–11845. doi: 10.1128/JVI.77.21.11842-11845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hagmaier K., Stock N., Goodbourn S., Wang L.F., Randall R. A single amino acid substitution in the V protein of Nipah virus alters its ability to block interferon signalling in cells from different species. J. Gen. Virol. 2006;87:3649–3653. doi: 10.1099/vir.0.82261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Devaux P., von Messling V., Songsungthong W., Springfeld C., Cattaneo R. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology. 2006 doi: 10.1016/j.virol.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 223.Ciancanelli M.J., Volchkova V.A., Shaw M.L., Volchkov V.E., Basler C.F. Nipah virus sequesters inactive STAT1 in the nucleus via a P gene-encoded mechanism. J. Virol. 2009;83:7828–7841. doi: 10.1128/JVI.02610-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Palosaari H., Parisien J.P., Rodriguez J.J., Ulane C.M., Horvath C.M. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 2003;77:7635–7644. doi: 10.1128/JVI.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Shaffer J.A., Bellini W.J., Rota P.A. The C protein of measles virus inhibits the type I interferon response. Virology. 2003;315:389–397. doi: 10.1016/s0042-6822(03)00537-3. [DOI] [PubMed] [Google Scholar]
- 226.Caignard G., Guerbois M., Labernardiere J.L., Jacob Y., Jones L.M., Wild F., Tangy F., Vidalain P.O. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology. 2007;368:351–362. doi: 10.1016/j.virol.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 227.Caignard G., Bourai M., Jacob Y., Tangy F., Vidalain P.O. Inhibition of IFN-alpha/beta signaling by two discrete peptides within measles virus V protein that specifically bind STAT1 and STAT2. Virology. 2009;383:112–120. doi: 10.1016/j.virol.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 228.Vidy A., Chelbi-Alix M., Blondel D. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 2005;79:14411–14420. doi: 10.1128/JVI.79.22.14411-14420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Brzozka K., Finke S., Conzelmann K.K. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 2006;80:2675–2683. doi: 10.1128/JVI.80.6.2675-2683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Chenik M., Chebli K., Blondel D. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J. Virol. 1995;69:707–712. doi: 10.1128/jvi.69.2.707-712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Vidy A., El Bougrini J., Chelbi-Alix M.K., Blondel D. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J. Virol. 2007;81:4255–4263. doi: 10.1128/JVI.01930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Moseley G.W., Lahaye X., Roth D.M., Oksayan S., Filmer R.P., Rowe C.L., Blondel D., Jans D.A. Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. J. Cell Sci. 2009;122:3652–3662. doi: 10.1242/jcs.045542. [DOI] [PubMed] [Google Scholar]
- 233.Hartman A.L., Bird B.H., Towner J.S., Antoniadou Z.A., Zaki S.R., Nichol S.T. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. J. Virol. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hartman A.L., Ling L., Nichol S.T., Hibberd M.L. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J. Virol. 2008;82:5348–5358. doi: 10.1128/JVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mateo M., Reid S.P., Leung L.W., Basler C.F., Volchkov V.E. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signalling. J. Virol. 2009 doi: 10.1128/JVI.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Reid S.P., Leung L.W., Hartman A.L., Martinez O., Shaw M.L., Carbonnelle C., Volchkov V.E., Nichol S.T., Basler C.F. Ebola virus VP24 binds karyopherin {alpha}1 and blocks STAT1 nuclear accumulation. J. Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Reid S.P., Valmas C., Martinez O., Sanchez F.M., Basler C.F. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 2007;81:13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J., Kobinger G.P., Wivel N.A., Crystal R.G., Boyer J., Feldmann H., Voss T.G., Wilson J.M. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 2004;78:11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Liu K., Lemon B., Traktman P. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J. Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Mossman K., Ostergaard H., Upton C., McFadden G. Myxoma virus and Shope fibroma virus encode dual-specificity tyrosine/serine phosphatases which are essential for virus viability. Virology. 1995;206:572–582. doi: 10.1016/s0042-6822(95)80074-3. [DOI] [PubMed] [Google Scholar]
- 243.Najarro P., Traktman P., Lewis J.A. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J. Virol. 2001;75:3185–3196. doi: 10.1128/JVI.75.7.3185-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lin R.J., Chang B.L., Yu H.P., Liao C.L., Lin Y.L. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Baron M., Davignon J.L. Inhibition of IFN-gamma-induced STAT1 tyrosine phosphorylation by human CMV is mediated by SHP2. J. Immunol. 2008;181:5530–5536. doi: 10.4049/jimmunol.181.8.5530. [DOI] [PubMed] [Google Scholar]
- 246.Christen V., Duong F., Bernsmeier C., Sun D., Nassal M., Heim M.H. Inhibition of alpha interferon signaling by hepatitis B virus. J. Virol. 2007;81:159–165. doi: 10.1128/JVI.01292-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Strobl B., Bubic I., Bruns U., Steinborn R., Lajko R., Kolbe T., Karaghiosoff M., Kalinke U., Jonjic S., Muller M. Novel functions of tyrosine kinase 2 in the antiviral defense against murine cytomegalovirus. J. Immunol. 2005;175:4000–4008. doi: 10.4049/jimmunol.175.6.4000. [DOI] [PubMed] [Google Scholar]
- 248.Le V.T., Trilling M., Wilborn M., Hengel H., Zimmermann A. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J. Gen. Virol. 2008;89:2416–2426. doi: 10.1099/vir.0.2008/001669-0. [DOI] [PubMed] [Google Scholar]
- 249.Paulus C., Krauss S., Nevels M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. U S A. 2006;103:3840–3845. doi: 10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Li J., Chen F., Zheng M., Zhu H., Zhao D., Liu W., Chen Z. Inhibition of STAT1 methylation is involved in the resistance of Hepatitis B Virus to Interferon alpha. Antiviral Res. 2009 doi: 10.1016/j.antiviral.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 251.Lin W., Choe W.H., Hiasa Y., Kamegaya Y., Blackard J.T., Schmidt E.V., Chung R.T. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology. 2005;128:1034–1041. doi: 10.1053/j.gastro.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 252.Gong G.Z., Cao J., Jiang Y.F., Zhou Y., Liu B. Hepatitis C virus non-structural 5A abrogates signal transducer and activator of transcription-1 nuclear translocation induced by IFN-alpha through dephosphorylation. World J. Gastroenterol. 2007;13:4080–4084. doi: 10.3748/wjg.v13.i30.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lan K.H., Lan K.L., Lee W.P., Sheu M.L., Chen M.Y., Lee Y.L., Yen S.H., Chang F.Y., Lee S.D. HCV NS5A inhibits interferon-alpha signaling through suppression of STAT1 phosphorylation in hepatocyte-derived cell lines. J. Hepatol. 2007;46:759–767. doi: 10.1016/j.jhep.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 254.Helbig K.J., Yip E., McCartney E.M., Eyre N.S., Beard M.R. A screening method for identifying disruptions in interferon signaling reveals HCV NS3/4a disrupts Stat-1 phosphorylation. Antiviral Res. 2008;77:169–176. doi: 10.1016/j.antiviral.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 255.Bouhet S., Lafont V., Billard E., Gross A., Dornand J. The IFNgamma-induced STAT1-CBP/P300 association, required for a normal response to the cytokine, is disrupted in Brucella-infected macrophages. Microb. Pathog. 2008 doi: 10.1016/j.micpath.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 256.Denkers E.Y. From cells to signaling cascades: manipulation of innate immunity by Toxoplasma gondii. FEMS Immunol. Med. Microbiol. 2003;39:193–203. doi: 10.1016/S0928-8244(03)00279-7. [DOI] [PubMed] [Google Scholar]
- 257.Ceravolo I.P., Chaves A.C., Bonjardim C.A., Sibley D., Romanha A.J., Gazzinelli R.T. Replication of Toxoplasma gondii, but not Trypanosoma cruzi, is regulated in human fibroblasts activated with gamma interferon: requirement of a functional JAK/STAT pathway. Infect. Immun. 1999;67:2233–2240. doi: 10.1128/iai.67.5.2233-2240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lieberman L.A., Banica M., Reiner S.L., Hunter C.A. STAT1 plays a critical role in the regulation of antimicrobial effector mechanisms, but not in the development of Th1-type responses during toxoplasmosis. J. Immunol. 2004;172:457–463. doi: 10.4049/jimmunol.172.1.457. [DOI] [PubMed] [Google Scholar]
- 259.Zimmermann S., Murray P.J., Heeg K., Dalpke A.H. Induction of suppressor of cytokine signaling-1 by Toxoplasma gondii contributes to immune evasion in macrophages by blocking IFN-gamma signaling. J. Immunol. 2006;176:1840–1847. doi: 10.4049/jimmunol.176.3.1840. [DOI] [PubMed] [Google Scholar]
- 260.Kim S.K., Fouts A.E., Boothroyd J.C. Toxoplasma gondii dysregulates IFN-gamma-inducible gene expression in human fibroblasts: insights from a genome-wide transcriptional profiling. J. Immunol. 2007;178:5154–5165. doi: 10.4049/jimmunol.178.8.5154. [DOI] [PubMed] [Google Scholar]
- 261.Aliberti J., Valenzuela J.G., Carruthers V.B., Hieny S., Andersen J., Charest H., Reis e Sousa C., Fairlamb A., Ribeiro J.M., Sher A. Molecular mimicry of a CCR5 binding-domain in the microbial activation of dendritic cells. Nat. Immunol. 2003;4:485–490. doi: 10.1038/ni915. [DOI] [PubMed] [Google Scholar]
- 262.Weiden M., Tanaka N., Qiao Y., Zhao B.Y., Honda Y., Nakata K., Canova A., Levy D.E., Rom W.N., Pine R. Differentiation of monocytes to macrophages switches the Mycobacterium tuberculosis effect on HIV-1 replication from stimulation to inhibition: modulation of interferon response and CCAAT/enhancer binding protein beta expression. J. Immunol. 2000;165:2028–2039. doi: 10.4049/jimmunol.165.4.2028. [DOI] [PubMed] [Google Scholar]
- 263.Remoli M.E., Giacomini E., Lutfalla G., Dondi E., Orefici G., Battistini A., Uze G., Pellegrini S., Coccia E.M. Selective expression of type I IFN genes in human dendritic cells infected with Mycobacterium tuberculosis. J. Immunol. 2002;169:366–374. doi: 10.4049/jimmunol.169.1.366. [DOI] [PubMed] [Google Scholar]
- 264.Prabhakar S., Qiao Y., Hoshino Y., Weiden M., Canova A., Giacomini E., Coccia E., Pine R. Inhibition of response to alpha interferon by Mycobacterium tuberculosis. Infect. Immun. 2003;71:2487–2497. doi: 10.1128/IAI.71.5.2487-2497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kincaid E.Z., Ernst J.D. Mycobacterium tuberculosis exerts gene-selective inhibition of transcriptional responses to IFN-gamma without inhibiting STAT1 function. J. Immunol. 2003;171:2042–2049. doi: 10.4049/jimmunol.171.4.2042. [DOI] [PubMed] [Google Scholar]
- 266.Wang Y., Curry H.M., Zwilling B.S., Lafuse W.P. Mycobacteria inhibition of IFN-gamma induced HLA-DR gene expression by up-regulating histone deacetylation at the promoter region in human THP-1 monocytic cells. J. Immunol. 2005;174:5687–5694. doi: 10.4049/jimmunol.174.9.5687. [DOI] [PubMed] [Google Scholar]
- 267.Nagabhushanam V., Solache A., Ting L.M., Escaron C.J., Zhang J.Y., Ernst J.D. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J. Immunol. 2003;171:4750–4757. doi: 10.4049/jimmunol.171.9.4750. [DOI] [PubMed] [Google Scholar]
- 268.Ting L.M., Kim A.C., Cattamanchi A., Ernst J.D. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J. Immunol. 1999;163:3898–3906. [PubMed] [Google Scholar]
- 269.Ning Q., Berger L., Luo X., Yan W., Gong F., Dennis J., Levy G. STAT1 and STAT3 alpha/beta splice form activation predicts host responses in mouse hepatitis virus type 3 infection. J. Med. Virol. 2003;69:306–312. doi: 10.1002/jmv.10290. [DOI] [PubMed] [Google Scholar]
- 270.Alvarez G.R., Zwilling B.S., Lafuse W.P. Mycobacterium avium inhibition of IFN-gamma signaling in mouse macrophages: Toll-like receptor 2 stimulation increases expression of dominant-negative STAT1 beta by mRNA stabilization. J. Immunol. 2003;171:6766–6773. doi: 10.4049/jimmunol.171.12.6766. [DOI] [PubMed] [Google Scholar]
- 271.Kawa K. Epstein-Barr virus – associated diseases in humans. Int. J. Hematol. 2000;71:108–117. [PubMed] [Google Scholar]
- 272.Gandhi M.K. Epstein-Barr virus-associated lymphomas. Expert Rev. Anti Infect. Ther. 2006;4:77–89. doi: 10.1586/14787210.4.1.77. [DOI] [PubMed] [Google Scholar]
- 273.Babcock G.J., Decker L.L., Volk M., Thorley-Lawson D.A. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 274.Souza T.A., Stollar B.D., Sullivan J.L., Luzuriaga K., Thorley-Lawson D.A. Peripheral B cells latently infected with Epstein-Barr virus display molecular hallmarks of classical antigen-selected memory B cells. Proc. Natl. Acad. Sci. U S A. 2005;102:18093–18098. doi: 10.1073/pnas.0509311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Qu L., Rowe D.T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J. Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Rowe D.T. Epstein-Barr virus immortalization and latency. Front. Biosci. 1999;4:D346–D371. doi: 10.2741/rowe. [DOI] [PubMed] [Google Scholar]
- 277.Pope J.H., Horne M.K., Scott W. Transformation of foetal human keukocytes in vitro by filtrates of a human leukaemic cell line containing herpes-like virus. Int. J. Cancer. 1968;3:857–866. doi: 10.1002/ijc.2910030619. [DOI] [PubMed] [Google Scholar]
- 278.Pope J.H. Establishment of cell lines from Australian leukaemic patients: presence of a herpes-like virus. Aust. J. Exp. Biol. Med. Sci. 1968;46:643–645. doi: 10.1038/icb.1968.171. [DOI] [PubMed] [Google Scholar]
- 279.Sugimoto M., Ide T., Goto M., Furuichi Y. Reconsideration of senescence, immortalization and telomere maintenance of Epstein-Barr virus-transformed human B-lymphoblastoid cell lines. Mech. Ageing Dev. 1999;107:51–60. doi: 10.1016/s0047-6374(98)00131-6. [DOI] [PubMed] [Google Scholar]
- 280.Lewin N., Aman P., Masucci M.G., Klein E., Klein G., Oberg B., Strander H., Henle W., Henle G. Characterization of EBV-carrying B-cell populations in healthy seropositive individuals with regard to density, release of transforming virus and spontaneous outgrowth. Int. J. Cancer. 1987;39:472–476. doi: 10.1002/ijc.2910390411. [DOI] [PubMed] [Google Scholar]
- 281.Yao Q.Y., Rickinson A.B., Epstein M.A. A re-examination of the Epstein-Barr virus carrier state in healthy seropositive individuals. Int. J. Cancer. 1985;35:35–42. doi: 10.1002/ijc.2910350107. [DOI] [PubMed] [Google Scholar]
- 282.Yao Q.Y., Rowe M., Martin B., Young L.S., Rickinson A.B. The Epstein-Barr virus carrier state: dominance of a single growth-transforming isolate in the blood and in the oropharynx of healthy virus carriers. J. Gen. Virol. 1991;72(Pt 7):1579–1590. doi: 10.1099/0022-1317-72-7-1579. [DOI] [PubMed] [Google Scholar]
- 283.Ling N.R., Hardie D., Lowe J., Johnson G.D., Khan M., MacLennan I.C. A phenotypic study of cells from Burkitt lymphoma and EBV-B-lymphoblastoid lines and their relationship to cells in normal lymphoid tissues. Int. J. Cancer. 1989;43:112–118. doi: 10.1002/ijc.2910430122. [DOI] [PubMed] [Google Scholar]
- 284.Rowe M., Rowe D.T., Gregory C.D., Young L.S., Farrell P.J., Rupani H., Rickinson A.B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lacy J., Rudnick H. Transcriptional regulation of the human IgE receptor (Fc epsilon RII/CD23) by EBV. Identification of EBV-responsive regulatory elements in intron 1. J. Immunol. 1992;148:1554–1560. [PubMed] [Google Scholar]
- 286.Rochford R., Hobbs M.V., Garnier J.L., Cooper N.R., Cannon M.J. Plasmacytoid differentiation of Epstein-Barr virus-transformed B cells in vivo is associated with reduced expression of viral latent genes. Proc. Natl. Acad. Sci. U S A. 1993;90:352–356. doi: 10.1073/pnas.90.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wendel-Hansen V., Rosen A., Klein G. EBV-transformed lymphoblastoid cell lines down-regulate EBNA in parallel with secretory differentiation. Int. J. Cancer. 1987;39:404–408. doi: 10.1002/ijc.2910390322. [DOI] [PubMed] [Google Scholar]
- 288.Eliopoulos A.G., Young L.S. LMP1 structure and signal transduction. Semin. Cancer Biol. 2001;11:435–444. doi: 10.1006/scbi.2001.0410. [DOI] [PubMed] [Google Scholar]
- 289.Dawson C.W., Rickinson A.B., Young L.S. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature. 1990;344:777–780. doi: 10.1038/344777a0. [DOI] [PubMed] [Google Scholar]
- 290.Fahraeus R., Rymo L., Rhim J.S., Klein G. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature. 1990;345:447–449. doi: 10.1038/345447a0. [DOI] [PubMed] [Google Scholar]
- 291.Wang D., Liebowitz D., Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 292.Gires O., Zimber-Strobl U., Gonnella R., Ueffing M., Marschall G., Zeidler R., Pich D., Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mosialos G., Birkenbach M., Yalamanchili R., VanArsdale T., Ware C., Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 294.Masy E., Adriaenssens E., Auriault C., Coll J. La protéine oncogène LMP1 du virus d'Epstein-Barr: voies de signalisation et phénotypes associés. Virologie. 2002;6:379–390. [Google Scholar]
- 295.Zhang L., Hong K., Zhang J., Pagano J.S. Multiple signal transducers and activators of transcription are induced by EBV LMP-1. Virology. 2004;323:141–152. doi: 10.1016/j.virol.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 296.Devergne O., Hatzivassiliou E., Izumi K.M., Kaye K.M., Kleijnen M.F., Kieff E., Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-kappaB activation. Mol. Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wu S., Xie P., Welsh K., Li C., Ni C.Z., Zhu X., Reed J.C., Satterthwait A.C., Bishop G.A., Ely K.R. LMP1 protein from the Epstein-Barr virus is a structural CD40 decoy in B lymphocytes for binding to TRAF3. J. Biol. Chem. 2005;280:33620–33626. doi: 10.1074/jbc.M502511200. [DOI] [PubMed] [Google Scholar]
- 298.Izumi K.M., Kieff E.D. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc. Natl. Acad. Sci. U S A. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Izumi K.M., Cahir McFarland E.D., Ting A.T., Riley E.A., Seed B., Kieff E.D. The Epstein-Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-kappaB activation. Mol. Cell Biol. 1999;19:5759–5767. doi: 10.1128/mcb.19.8.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Huen D.S., Henderson S.A., Croom-Carter D., Rowe M. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 301.Luftig M., Prinarakis E., Yasui T., Tsichritzis T., Cahir-McFarland E., Inoue J., Nakano H., Mak T.W., Yeh W.C., Li X., Akira S., Suzuki N., Suzuki S., Mosialos G., Kieff E. Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc. Natl. Acad. Sci. U S A. 2003;100:15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Atkinson P.G., Coope H.J., Rowe M., Ley S.C. Latent membrane protein 1 of Epstein-Barr virus stimulates processing of NF-kappa B2 p100 to p52. J. Biol. Chem. 2003;278:51134–51142. doi: 10.1074/jbc.M304771200. [DOI] [PubMed] [Google Scholar]
- 303.Luftig M., Yasui T., Soni V., Kang M.S., Jacobson N., Cahir-McFarland E., Seed B., Kieff E. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc. Natl. Acad. Sci. U S A. 2004;101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Wan J., Sun L., Mendoza J.W., Chui Y.L., Huang D.P., Chen Z.J., Suzuki N., Suzuki S., Yeh W.C., Akira S., Matsumoto K., Liu Z.G., Wu Z. Elucidation of the c-Jun N-terminal kinase pathway mediated by Estein-Barr virus-encoded latent membrane protein 1. Mol. Cell Biol. 2004;24:192–199. doi: 10.1128/MCB.24.1.192-199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Feuillard J., Schuhmacher M., Kohanna S., Asso-Bonnet M., Ledeur F., Joubert-Caron R., Bissieres P., Polack A., Bornkamm G.W., Raphael M. Inducible loss of NF-kappa B activity is associated with apoptosis and Bcl-2 down-regulation in Epstein-Barr virus-transformed B lymphocytes. Blood. 2000;95:2068–2075. [PubMed] [Google Scholar]
- 306.D'Souza B., Rowe M., Walls D. The bfl-1 gene is transcriptionally upregulated by the Epstein-Barr virus LMP1, and its expression promotes the survival of a Burkitt's lymphoma cell line. J. Virol. 2000;74:6652–6658. doi: 10.1128/jvi.74.14.6652-6658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Laherty C.D., Hu H.M., Opipari A.W., Wang F., Dixit V.M. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappa B. J. Biol. Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 308.Fries K.L., Miller W.E., Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J. Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Sjoblom A., Yang W., Palmqvist L., Jansson A., Rymo L. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J. Virol. 1998;72:1365–1376. doi: 10.1128/jvi.72.2.1365-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Goormachtigh G., Ouk T.S., Mougel A., Tranchand-Bunel D., Masy E., Le Clorennec C., Feuillard J., Bornkamm G.W., Auriault C., Manet E., Fafeur V., Adriaenssens E., Coll J. Autoactivation of the Epstein-Barr virus oncogenic protein LMP1 during type II latency through opposite roles of the NF-kappaB and JNK signaling pathways. J. Virol. 2006;80:7382–7393. doi: 10.1128/JVI.02052-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Weber-Nordt R.M., Egen C., Wehinger J., Ludwig W., Gouilleux-Gruart V., Mertelsmann R., Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 312.Gires O., Kohlhuber F., Kilger E., Baumann M., Kieser A., Kaiser C., Zeidler R., Scheffer B., Ueffing M., Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 1999;18:3064–3073. doi: 10.1093/emboj/18.11.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Baran-Marszak F., Fagard R., Girard B., Camilleri-Broet S., Zeng F., Lenoir G.M., Raphael M., Feuillard J. Gene array identification of Epstein Barr virus-regulated cellular genes in EBV-converted Burkitt lymphoma cell lines. Lab. Invest. 2002;82:1463–1479. doi: 10.1097/01.lab.0000035025.51772.2b. [DOI] [PubMed] [Google Scholar]
- 314.Chen H., Lee J.M., Zong Y., Borowitz M., Ng M.H., Ambinder R.F., Hayward S.D. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J. Virol. 2001;75:2929–2937. doi: 10.1128/JVI.75.6.2929-2937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.McLaren J., Rowe M., Brennan P. Epstein-Barr virus induces a distinct form of DNA-bound STAT1 compared with that found in interferon-stimulated B lymphocytes. J. Gen. Virol. 2007;88:1876–1886. doi: 10.1099/vir.0.82741-0. [DOI] [PubMed] [Google Scholar]
- 316.Higuchi M., Kieff E., Izumi K.M. The Epstein-Barr virus latent membrane protein 1 putative Janus kinase 3 (JAK3) binding domain does not mediate JAK3 association or activation in B-lymphoma or lymphoblastoid cell lines. J. Virol. 2002;76:455–459. doi: 10.1128/JVI.76.1.455-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Brennan P., Floettmann J.E., Mehl A., Jones M., Rowe M. Mechanism of action of a novel latent membrane protein-1 dominant negative. J. Biol. Chem. 2001;276:1195–1203. doi: 10.1074/jbc.M005461200. [DOI] [PubMed] [Google Scholar]
- 318.Najjar I., Baran-Marszak F., Le Clorennec C., Laguillier C., Schischmanoff O., Youlyouz-Marfak I., Schlee M., Bornkamm G.W., Raphael M., Feuillard J., Fagard R. Latent membrane protein 1 regulates STAT1 through NF-kappaB-dependent interferon secretion in Epstein-Barr virus-immortalized B cells. J. Virol. 2005;79:4936–4943. doi: 10.1128/JVI.79.8.4936-4943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Vaysberg M., Lambert S.L., Krams S.M., Martinez O.M. Activation of the JAK/STAT pathway in Epstein Barr virus+-associated posttransplant lymphoproliferative disease: role of interferon-gamma. Am. J. Transplant. 2009;9:2292–2302. doi: 10.1111/j.1600-6143.2009.02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Wesemann D.R., Benveniste E.N. STAT-1 alpha and IFN-gamma as modulators of TNF-alpha signaling in macrophages: regulation and functional implications of the TNF receptor 1:STAT-1 alpha complex. J. Immunol. 2003;171:5313–5319. doi: 10.4049/jimmunol.171.10.5313. [DOI] [PubMed] [Google Scholar]
- 321.Shah K.M., Stewart S.E., Wei W., Woodman C.B., O'Neil J.D., Dawson C.W., Young L.S. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene. 2009;28:3903–3914. doi: 10.1038/onc.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Platanias L.C., Fish E.N. Signaling pathways activated by interferons. Exp. Hematol. 1999;27:1583–1592. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 323.Stark G.R., Kerr I.M., Williams B.R., Silverman R.H., Schreiber R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 324.Donnelly R.P., Sheikh F., Kotenko S.V., Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J. Leukoc. Biol. 2004;76:314–321. doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- 325.Wong L.H., Sim H., Chatterjee-Kishore M., Hatzinisiriou I., Devenish R.J., Stark G., Ralph S.J. Isolation and characterization of a human STAT1 gene regulatory element. Inducibility by interferon (IFN) types I and II and role of IFN regulatory factor-1. J. Biol. Chem. 2002;277:19408–19417. doi: 10.1074/jbc.M111302200. [DOI] [PubMed] [Google Scholar]
- 326.I. Najjar, P.-A. Deglesne, P.O. Schischmanoff, E.E. Fabre, S. Boisson-Dupuis, F. Nimmerjahn, G.W. Bornkamm, I. Dusanter-Fourt, R. Fagard, STAT1-dependent IgG cell-surface expression in a human B cell line derived from a STAT1-deficient patient. J. Leukoc. Biol. 2010 Mar 3. [Epub ahead of print]. [DOI] [PubMed]