Highlights
-
•
Two pheasant coronaviruses (PhCoVs) were isolated in 2017 in China.
-
•
The two PhCoVs were genetically similar to IBV.
-
•
Pathogenicity, replication, and shedding of PhCoV were obvious different when infected chickens and pheasants.
-
•
PhCoVs isolated from different outbreaks may have evolved independently from IBVs by adaption in pheasants.
Keywords: Pheasant coronavirus, Infectious bronchitis virus, S1 gene, Complete genome, Adaption and evolution
Abstract
Two viruses were isolated in 2017 from commercial pheasants with severe clinical signs and mortality in Shandong and Anhui provinces, China, respectively. We examined the pathogenic effects of the viruses in chicken embryos and the size and morphology of the virus particles, performed phylogenetic analysis based on the S1 gene and complete genomic sequences, and examined the antibody responses against infectious bronchitis virus (IBV). The results suggested that the viruses I0623/17 and I0710/17 were avian coronaviruses and were identified as pheasant coronaviruses (PhCoV), with greatest similarity to IBV. Further investigations of the antigenicity, complete genome organization, substitutions in multiple genes, and viral pathogenicity, replication, and shedding in chickens and pheasants showed obvious differences between PhCoV and IBV in terms of antigenicity, and viral pathogenicity, replication, and shedding in chickens and pheasants. The close genetic relationship, but obvious differences between PhCoVs and IBVs suggested the IBVs could be the ancestors of PhCoVs, and that PhCoVs isolated from different outbreaks may have evolved independently from IBVs circulating in the specific region by adaption in pheasants. This hypothesis was supported by analysis of the S1 gene fragments of the two PhCoVs isolated in the current study, as well as PhCoVs isolated in the UK and selected IBV strains. Such analyses indicated different evolution patterns and different tissue tropisms between PhCoVs isolated in different outbreaks. Further studies are needed to confirm this hypothesis by studying the complete genomic sequences of PhCoVs from different outbreaks and the pathogenicity of IBVs in pheasants to compare and clarify the relationships between PhCoVs and IBVs.
1. Introduction
Coronaviruses (CoVs) are enveloped, positive-sense RNA viruses found in a wide range of animals, including mammals and birds. CoVs exhibit marked tropism for epithelial cells in the respiratory, digestive, and urogenital tracts, and are responsible for a diverse spectrum of enteric, hepatic, neurological, and respiratory illnesses of differing severities. CoVs, classified in the subfamily Coronavirinae, family Coronaviridae, order Nidovirales, are now recognized as emerging viruses with a propensity to cross into new host species (Leopardi et al., 2018). CoVs are subdivided into four genera on the basis of genotypic and serological characterization: alpha and beta coronaviruses, which are found mainly in mammals; gamma coronaviruses, detected in birds and marine mammals; and delta coronaviruses found mainly in birds and swine (de Groot, 2012). The 5′ two-thirds of the CoV genome encodes proteins involved in viral RNA synthesis. Most of these proteins are characteristically encoded by the partially-overlapping open reading frames (ORFs), ORF1a and ORF1b, and are translated as the polyproteins pp1a and pp1ab, which are then processed by virus-encoded proteinases into 15 or 16 non-structural proteins (nsps) (Ziebuhr, 2005). The remaining 3′ one-third of the genome encodes virus structural proteins, including the spike (S) glycoprotein, membrane (M) glycoprotein, small envelope (E) protein, and phosphorylated nucleocapsid (N) proteins. The CoV S protein can be divided into an amino-proximal half (S1 subunit or domain) containing the receptor-binding domain, and a carboxyl-proximal half (S2 subunit or domain) containing elements involved in membrane fusion (Lewis et al., 2015). The S protein is an important target for T cell responses and is the major inducer of virus-neutralizing antibodies, which are elicited by epitopes located mostly in the molecule’s S1 domain (Reguera et al., 2012; Satoh et al., 2011). The S1 and S2 domains in some CoVs, e.g. avian CoVs (AvCoVs), are cleaved by a cellular furin-like enzyme (de Haan et al., 2004).
Most viruses in the genus Gammacoronavirus are isolated from birds, including Galliformes (chicken, turkey, quail, guinea fowl, pheasant, and peafowl) (Brown et al., 2016; Cavanagh et al., 2002; Guy, 2008; Liu et al., 2005; Sun et al., 2007; Torres et al., 2017), Anseriformes (duck, goose, teal, swan, and pintail) (Liu et al., 2005; Papineau et al., 2019), Columbiformes (pigeon) (Jonassen et al., 2005), Pelecaniformes (spoonbill and heron), Suliformes (cormorant), Charadriiformes (red knot, oystercatcher, and black-headed gull), and Passeriformes (bulbul) (Chu et al., 2011), although they have also been identified in the beluga whale (Mihindukulasuriya et al., 2008) and in bottlenose dolphins (Woo et al., 2014). Infectious bronchitis virus (IBV) and turkey coronavirus (TCoV) are among the most economically important avian coronaviruses (AvCoVs). IBV was the first recognized CoV in the United States in 1931 (Schalk and Hawn, 1931). IBV is traditionally considered to be a host-specific pathogen in chickens, responsible for respiratory, renal, and genital diseases, with heavy economic consequences worldwide (Cavanagh, 2007). In contrast to alpha and beta coronaviruses, which only occur as one or two different serotypes, IBV has many different serotypes, genotypes, lineages, and variants (Valastro et al., 2016). Furthermore, new IBV genotypes/lineages and variants are continuing to emerge due to its high rate of evolution. This is expressed as an accelerated rate at which viable point mutations, nucleotide insertions, or deletions accumulate in the genome, especially in the S1 gene, associated with a lack of proofreading viral polymerase in IBVs (Cavanagh, 2007), as well as the high rate of virus replication. In addition, evidence suggests that some IBV strains may have arisen by genetic recombination (Cavanagh et al., 1992; Kusters et al., 1989, 1990), or by a combination of mutation and recombination (Jia et al., 1995; Chen et al., 2017; Jiang et al., 2017). TCoV, initially found in the 1970s, is associated with the enteric disease known as transmissible enteritis, coronaviral enteritis of turkeys, or bluecomb (Guy, 2008). TCoV infection was also recently found to be associated with a syndrome comprising several intestinal disorders, usually occurring in turkeys within the first 3 weeks of life and referred to as poult enteritis complex (Barnes et al., 2000). TCoV emergence was proposed to be the result of recombination events involving IBVs and an unidentified CoV, which donated an S gene encoding a protein of low amino acid identity to those of IBVs (Hughes, 2011; Jackwood et al., 2010). These recombinations resulted in a host shift from chickens to turkeys, and in a change in tissue tropism of the virus from the upper respiratory tract to the intestine.
Outbreaks initially associated with CoV infection in pheasants were first reported in the UK in 1980 (Spackman and Cameron, 1983). Affected birds exhibited reduced egg production among breeding hens, accompanied by loss of shell pigmentation and some abnormal shell quality, with later outbreaks of respiratory signs among affected pheasants. However, there was no evidence of kidney damage in the affected birds. Antibodies against IBV were detected by the hemagglutination inhibition (HI) test in 80% of tested pheasants and a CoV was isolated that proved to be identical to the agent responsible for infectious bronchitis (IB) in chickens. Two later outbreaks of nephritis occurred in the UK in 1983 and 1984, resulting in the deaths of 450 out of 1000 8-week-old pheasants and 15 out of 160 adult pheasants, respectively, from which CoVs were isolated using 8–9-day-old embryonating eggs (Lister et al., 1985). However, intranasal instillation of specific-pathogen-free (SPF) chickens with the virus failed to produce any clinical signs of disease. A similar outbreak of nephritis in 1994 resulting in the deaths of over 1000 breeding pheasants out of a total stock of approximately 7000 birds in the UK was considered to be linked to avian IBV (Gough et al., 1996). Sneezing and reduced egg production and hatchability were also reported, but the egg quality was unaffected. Pheasant coronavirus (PhCoV)-associated nephritis and respiratory disease were also subsequently reported in the UK (Pennycott, 2000; Cavanagh et al., 2002). Partial genomic sequence analysis showed that the gene sequences of the pheasant viruses differed from those of IBV to a similar extent as the sequence of one serotype of IBV differs from another (Cavanagh et al., 2002). The above results showed that the CoVs involved in these pheasant infections were closely related to avian IBV; however, the clinical signs and mortality associated with CoV infections in pheasants and the isolated viruses differed from each other and from IBVs. Further studies using molecular techniques are therefore required to determine the complete genomic sequences of CoVs from different outbreaks in pheasants, to compare and fully elucidate their origins and antigenic relationships with strains of IBV.
In this study, we isolated two CoVs from pheasants with nephritis and high mortality for the first time in China. We aimed to determine the full genome sequences of the two isolates and compare them with IBVs, evaluate the pathogenicity of the isolates in pheasants and chickens, and investigate the cross-antigenicity with IBVs, to clarify the origin of PhCoVs and their relationships with strains of IBV.
2. Materials and methods
2.1. Chicken embryos, chickens, and pheasants
White leghorn chickens were hatched from specific pathogen-free (SPF) embryonated eggs (Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences), maintained in Hosfall-type isolators, and provided with food and water ad libitum. Chicken embryonated eggs were used for primary isolation of viruses from the diseased pheasants, viral seed stock preparation and titration, virus-neutralization (VN) tests, re-isolation attempts from oropharyngeal and cloacal swabs, and titration of viruses from selected tissues from chickens and pheasants in the challenge trials. SPF chickens were used for antisera production and challenge trials. One-day-old commercial pheasants (Phasianus colchicus) were purchased from a pheasant-producing company in Shandong province, China, and used in the challenge trials. All experimental procedures and animal care were approved by the Ethical and Animal Welfare Committee of Heilongjiang province, China (License nos. HSY-IACUC-2017-167, -168, -169, and -170).
2.2. Clinical information and sample collection
Several new episodes of serious respiratory diseases occurred in commercial pheasant flocks in China between September 2016 and June 2017. The outbreaks were characterized by clinical signs compatible with IB in chickens, including respiratory signs (all flocks) and kidney lesions (nephritis), leading to economic losses in different geographical locations among the major pheasant-farming areas in Shandong, Anhui, and Henan provinces in China. The birds showed early signs of respiratory disease at around 17 days old, most birds in the flocks showed obvious clinical signs at 20–40 days old, and the clinical signs disappeared at around 55 days old. Mortality occurred at approximately 20 days old and was generally about 30%, but was as high as 50% in some flocks. Gross examination showed severe tracheitis and nephritis. Trachea, kidney, and proventriculus samples from two selected farms in Shandong and Anhui provinces were sent to our laboratory for virus detection in early 2017. The two farms contained 13,500 and 7000 birds, respectively, of which more than 4000 and 2500 died, respectively, during the disease outbreaks.
2.3. Virus detection and isolation
Tracheal and kidney samples from 10 dead birds from the farm in Shandong province, and kidney and proventriculus samples from five dead birds from the farm in Anhui province, respectively, were collected. Samples of each farm were pooled together, ending up with 2 different pools. The samples tested negative for Newcastle disease virus (NDV) (Gohm et al., 2000) and avian influenza virus (AIV) subtypes H5 and H9 by reverse transcription polymerase chain reaction (RT-PCR) (Chaharaein et al., 2009). However, the samples tested positive by RT-PCR using primers initially designed to detect the 3′ end of the IBV N gene (Liu et al., 2009).
PhCoVs can replicate and be isolated in embryonated chicken eggs (Cavanagh et al., 2002). Therefore, each pooled sample was used to inoculate five 9-day-old embryonated eggs (0.2 mL/egg) via the allantoic route. Allantoic fluid was harvested from two inoculated eggs for each pooled sample at 72 h post-inoculation, pooled, and used for virus detection by RT-PCR and direct electron microscopy (Liu and Kong, 2004). The remaining three eggs were incubated at 37 °C, candled daily, and examined for specific lesions at 7 days post-inoculation (dpi). Further blind passages were performed until embryo dwarfing and death were observed at 2–7 dpi.
Two CoVs were isolated from samples from pheasants from Shandong and Anhui provinces, respectively. These PhCoVs were designated γCoV/ph/China/I0623/17 (I0623/17) and γCoV/ph/China/I0710/17 (I0710/17), respectively, according to the standard nomenclature adopted for PhCoV isolates in this study (Ducatez and European Union COST Action FA1207, 2016). The two PhCoVs were isolated and propagated by inoculating and passaging in the allantoic cavity of 9-day-old SPF embryonated chicken eggs (Liu and Kong, 2004). The allantoic fluids from each passage incubated with isolates I0623/17 and I0710/17 were tested for NDV and AIV by hemagglutination (HA) activity. The median titers of isolates I0623/17 and I0710/17 at the final passage (3rd) were 106·6 and 106.2 egg infectious doses (EID50), respectively, determined using the method of Reed and Muench (1938).
2.4. Detection of antibody against IBV in commercial pheasants
Blood samples were collected from six male (A and C) and six layer hen pheasants (B and D) from each of two affected commercial pheasant flocks in Shandong and Anhui provinces, respectively, approximately 2 months after the disappearance of clinical signs. These samples were used for the detection of antibodies against IBV using a commercial enzyme-linked immunosorbent assay (ELISA; IDEXX Corporation, Westbrook, ME, USA) following the manufacturer’s instructions (Liu et al., 2013). Blood samples were also collected from an additional six commercial male (E) and six layer pheasants (F) from another flock without clinical signs, from the same farms as the affected commercial pheasant flocks in Shandong province. None of these birds had received IBV vaccines.
2.5. RNA extraction, RT-PCR amplification, and sequencing of the complete genome
The complete genomic RNAs of isolates I0623/17 and I0710/17 were extracted, from the respective infective allantoic fluids using a viral RNA extraction kit (Takara Bio Inc., Shiga, Japan), according to the manufacturer’s instructions. We initially amplified the complete genomes of the two PhCoV isolates using 14 pairs of partly overlapping primers, originally used to amplify the complete IBV genome (Liu et al., 2013). The sense and antisense primers were also used to amplify the 3′ and 5′ ends of the IBV genome (Liu et al., 2013), together with a PrimeScript™ One-Step RT-PCR Kit ver. 2 (Takara) and a 3′/5′ RACE kit (Takara), according to the manufacturer’s instructions. Two pairs of primers and the sense primer for amplifying the 3′ end of the virus genome were then redesigned to replace the previous corresponding primers, which were unable to amplify the gene fragments of isolates I0623/17 and I0710/17. All primers used in the following procedures are available on request from the authors.
The RT-PCR products were resolved by electrophoresis in 1% agarose gels. The gel-purified RT-PCR products were subjected to direct sequencing or ligated into the PMD-18 T cloning vector (Takara), following the manufacturer’s instructions. Three to five clones per amplicon were sequenced to determine the consensus sequence for any given genomic region. Consensus sequences were assembled from both forward and reverse sequences, edited manually, and corrected on the basis of the different clones of a fragment. The ORFs and deduced proteins were analyzed using Molecular Evolutionary Genetics Analysis 6.0 (Mega 6.0) (Tamura et al., 2013) and BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium), by comparison with the complete genomic sequences of IBV LX4 (GenBank accession numberAY338732) and H120 (FJ888351) strains, and TCoV TCoV-ATCC (EU022526) and TCoV/TX-GL/01 (GQ427174) strains. The sequences reported here have been deposited with the public access GenBank database (www.ncbi.nlm.nih.gov/genbank/). The accession numbers of isolates I0623/17 and I0710/17 are MK423877 and MK423876, respectively.
2.6. S1 gene/fragment analysis
Phylogenetic trees were constructed by aligning sequences using the Clustal W multiple alignment algorithm and the maximum likelihood method with 1000 bootstrap replicates, employing the Kimura 2-parameter and nucleotide substitution model. The S1 gene sequences of 213 reference AvCoV strains were used for phylogenetic analysis, together with those of the two new isolates I0623/17 and I0710/17. The 213 viruses included 199 reference IBV strains reported by Valastro et al. (2016), three IBV GI-28 strains (Chen et al., 2017), three IBV GI-29 strains (Jiang et al., 2018), two IBV GVII-1 strains (Ma et al., 2019), two additional IBV GI-19 strains, ck/CH/LDL/091022 (LDL/091022) (Liu et al., 2013) and ck/CH/LJL/140734 (LJL/140734) (Zhao et al., 2017) which were used the virus-neutralization (VN) test, three TCoVs and one guinea fowl CoV (Brown et al., 2016).
Nine nucleotide sequences of S1 gene fragments of PhCoVs were retrieved from the GenBank database. The sequence fragments were then submitted to a BLAST search in the NCBI database to identify homologous sequences. Six sequences of IBV strains that showed relatively close genetic relationships with the nine PhCoVs were selected for phylogenetic analysis and sequence alignment by comparison with our two isolates, eight GI-19 strains, and three GI-18 strains. Similarly, 16 nucleotide sequences of 3′ UTR fragments of PhCoVs were retrieved from the GenBank database and compared phylogenetically with 44 corresponding sequences from 43 IBV strains and one duck CoV, selected based on the results of phylogenetic analysis of the complete genomes.
2.7. Comparison of complete genomes
The complete genomic sequences of PhCoV isolates I0623/17 and I0710/17 were compared with those of 84 IBV strains and one duck CoV (as an outgroup) from the GenBank database, representing sequences of IBV strains from all continents and each lineage. Phylogenetic trees were constructed as described above. In addition, the complete genomic sequence of isolate I0623/17 was compared with 39 IBV strains and one duck CoV from the GenBank database, which selected on the basis of the results of phylogenetic analysis of the complete genome, using Similarity Plot (SimPlot) version 3.5.1 (http://sray.med.som.jhmi.edu/SCRopftware/simplot/) to analyze possible recombination events. The PhCoV isolate I0710/17 was used as a query virus. The window width and step size were set to 1000 bp and 50 bp, respectively.
2.8. Virus cross-neutralization test
In addition to the two isolates I0623/17 and I0710/17 in this study, three additional IBV strains, LDL/091022, LJL/140734, and H120, were also used in the VN test. LDL/091022 and LJL/140734 were selected because they were genetically related to isolates I0623/17 and I0710/17 based on the phylogenetic trees using both S1 gene/fragment and complete genome sequences. The H120 vaccine was used because it was commonly employed in chicken flocks in China, and was also used for vaccination of some commercial pheasant flocks in China; the H120 vaccine was not used in the affected pheasant flocks in this study. Relatedness values according to the Archetti and Horsfall method (1950) range from 0 for viruses that are antigenically unrelated to 100% for isolates that are identical. This method for assessing the antigenic relatedness of IBV means that viruses of the same serotype may have relatedness values ranging from 50%–100%, whereas viruses of a different serotype typically have relatedness values ranging from 0%–20% (Archetti and Horsfall, 1950).
Sera against the five virus strains were prepared as described previously (Guo et al., 2014). For VN, sera were serially diluted two-fold with sterile phosphate-buffered saline and mixed with 100 EID50 of the virus strains. After incubation for 1 h at 37 °C, virus-serum mixtures were inoculated into the allantoic cavity of SPF chicken embryos and observed for 7 days. Virus replication was determined by the appearance of typical lesions in the inoculated eggs, and by amplification of the IBV N gene from allantoic fluid by RT-PCR (Liu et al., 2009). The tests were performed in the presence of appropriate controls. The end-point titer of each serum sample and the antigenic relatedness values were calculated using the method of Reed and Muench (1938).
In addition, an aliquot of each serum from male and layer hens from each commercial pheasant flock was pooled and tested for PhCoV antibodies by VN, using I0623/17 virus.
2.9. Experimental design
2.9.1. Chicks
A total of 135 1-day-old SPF chicks were divided randomly into nine groups and kept in separate isolation units, with 15 chicks per group (Table 1 ). Chickens in groups 1, 4, and 7 were administered 0.1 ml of 105.0 EID50 of the isolate I0623/17 via the ocular and nasal routes at 1, 30, and 45 day(s) of age, respectively. Chickens in groups 2, 5, and 8 were administered 0.1 ml of 105.0 EID50 of the isolate I0710/17 via the ocular and nasal routes at 1, 30, and 45 day(s) of age, respectively. Chickens in groups 3, 6, and 9 were administered the same amount of sterilized allantoic fluid at the corresponding ages, as negative controls. Five chickens from each inoculated and control group were euthanized at 5 dpi, and trachea, lung, proventriculus, kidney, and cecal tonsil tissues were separated from the surrounding tissue, removed aseptically, and used for virus titration, as described previously (Liu et al., 2009). Trachea and kidney tissue samples were used for the detection of IBV antigen using immunohistochemistry (IHC) with the monoclonal antibody, 6D10 directed against the N protein, as described previously (Han et al., 2013, 2016). The remaining chicks were examined daily for signs of infection for another 25 days after inoculation. Signs, such as eye irritation and/or scratching of the inoculated eye, lethargy, mild coughing, and/or ‘snicking’, were monitored daily for a period of 30 days. Blood samples and oropharyngeal and cloacal swabs were collected from the birds in each group at 4, 8, 12, 16, 20, 24, and 28 dpi. The blood samples were used for antibody detection (ELISA; IDEXX Corporation). The oropharyngeal and cloacal swabs were used for virus recovery in 9-day-old SPF chicken eggs (Liu and Kong, 2004).
Table 1.
Information of experimental design.
| Group | Chicken/Pheasant | Numbera | Ageb | Challenge virus |
|---|---|---|---|---|
| 1 | Chicken | 15 | 1 | I0623/17 |
| 2 | Chicken | 15 | 1 | I0710/17 |
| 3 | Chicken | 15 | 1 | Sterilized allantoic fluid |
| 4 | Chicken | 15 | 30 | I0623/17 |
| 5 | Chicken | 15 | 30 | I0710/17 |
| 6 | Chicken | 15 | 30 | Sterilized allantoic fluid |
| 7 | Chicken | 15 | 45 | I0623/17 |
| 8 | Chicken | 15 | 45 | I0710/17 |
| 9 | Chicken | 15 | 45 | Sterilized allantoic fluid |
| 10 | Pheasant | 15 | 45 | I0710/17 |
| 11 | Pheasant | 15 | 45 | Sterilized allantoic fluid |
A total of 15 birds were included in each group. Five birds from each group were euthanized at 5 dpi, and trachea, lung, proventriculus, kidney, and cecal tonsil tissues were collected and used for virus titration.
The age when the birds were challenged with the virus.
2.9.2. Commercial pheasants
A total of 35 1-day-old commercial pheasants were used in the present study. The birds were housed in isolators under negative pressure 1 week before experimental infection (Table 1). Blood samples were collected from all the birds at 5, 10, 15, 20, 25, 30, 35 and 40 days of age and used for IBV-specific antibody detection (ELISA; IDEXX Corporation). The IBV-specific antibodies had not been detected until 40 days of age. Five birds were selected randomly and euthanized at 40 days, and examined carefully for lesions in each of the tissues and organs. Trachea, lung, kidney, digestive tract, liver, oviduct, spleen, and bursa of Fabricius tissue samples were collected and examined by histopathology to confirm that the birds used in the study were clinically healthy. The samples tested for RT-PCR and PCR were processed differently and NDV (Gohm et al., 2000), IBV (Liu et al., 2009), the AIV subtypes H5, H7, and H9 (Chaharaein et al., 2009) and avian metapneumovirus (Cavanagh et al., 1999) were tested by RT-PCR, and fowl adenovirus (Li et al., 2016), Mycoplasma synoviae (MS), and Mycoplasma galliscepticum (MG) (Moscoso et al., 2004) were tested by PCR using the collected samples. The birds were shown to be free of these pathogen infections (data not shown). Blood samples were collected from the remaining 30 birds. None of them had NDV- or AIV H5-, H7-, or H9-specific HI antibodies at 40 days of age, and none had antibodies against infectious bursal disease virus, avian leukosis virus, reticuloendotheliosis virus, chicken infectious anemia virus, fowl adenovirus, MS, or MG according to ELISAs (IDEXX Corporation). Then, the flock’s health status was observed for another 5 days before the experiments were performed.
Thirty commercial pheasants were divided randomly into two groups of 15 birds per group. Isolate I0710/17 was selected to infect the pheasants in this study because it had the lower rate of virus replication than that of isolate I0623/17 in chickens based on the results of infection to SPF chickens. Birds in group 1 were administered 0.1 ml of 105.0 EID50 of the isolate I0710/17 via the ocular and nasal routes at 45 days of age. Birds in group 2 were administered the same amount of sterilized allantoic fluid at the same age, as a negative control. Five birds from each group were euthanized at 5 dpi, and trachea, lung, proventriculus, kidney, and cecal tonsil tissues were collected and used for virus titration. The titers were calculated to include the total amount of infectious virus recovered from the entire organ. The tracheas and kidneys were also subjected to immunohistochemistry (IHC) for the detection of IBV
Trachea and kidney samples were used to detect IBV antigen using IHC (Han et al., 2013, 2016). The birds were examined daily for signs of infection for another 25 days after inoculation. Signs were monitored daily for a period of 30 days. Blood samples and oropharyngeal and cloacal swabs were collected from the remaining birds in each group at 4, 8, 12, 16, 20, 24, and 28 dpi. The blood samples were used for antibody detection and the oropharyngeal and cloacal swabs were used for virus recovery in 9-day-old SPF chicken eggs (Han et al., 2016), as described above.
2.10. Statistical analysis
Data are expressed as the mean ± standard deviation. Virus titers were analyzed by the Student’s t-test using GraphPad Prism for Windows version 5 (GraphPad Software, La Jolla, CA, USA). Differences were considered significant if the p value was <0.05.
3. Results
3.1. IBV-like coronaviruses were isolated from the diseased commercial pheasants
Tissues from each flock were pooled and inoculated into embryonated SPF chicken eggs, as described previously (Liu and Kong, 2004). No obvious lesions were observed at 7 dpi in the first and second passages. However, stunting and curling of the embryo with urate deposits in the embryonic mesonephros appeared following two additional passages in the allantoic cavity of 9-day-old SPF eggs (Gough et al., 1996). The allantoic fluids from each passage showed no HA activity for chicken red cells. Examination of the concentrated allantoic fluids from the third embryo passage by electron microscopy revealed CoV-like particles (Supplementary Fig. 1).
The allantoic fluids tested positive for IBV by RT-PCR using IBV-specific primers targeting the 3′ end of the N gene, as described previously (Liu et al., 2013). To further identify the virus isolates, we sequenced the PCR products of the two isolates and used the sequences to search the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for similar sequences using the BLASTN program. The two sequences shared the highest nucleotide identities (90%) with two IBV strains, ck/CH/LDL/05II and ck/CH/LDL/05III, isolated in China in 2005 and belonging to the GI-19 lineage (Zhao et al., 2017).
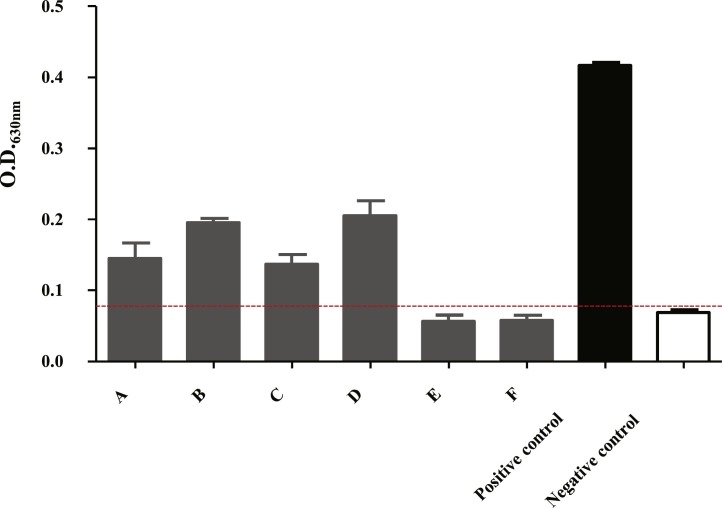
All the affected pheasants collected from four commercial flocks 2 months after the disappearance of clinical signs were positive for the IBV-specific antibodies by ELISA, in contrast to the negative results for serum from pheasants that had not shown clinical signs (Fig. 1 ).
Fig. 1.
Antibody responses of commercial pheasants, using an ELISA kit for detecting IBV antibodies. Blood samples were collected from six male (A and C) and six layer hen pheasants (B and D) from each of two affected commercial pheasant flocks in Shandong and Anhui provinces, respectively. Blood samples were also collected from six additional commercial male (E) and six layer pheasants (F) from other flocks that had not shown any clinical signs. Positive and negative serum controls for IBV are also shown.
Taken together, the embryopathic effect of the virus, the size and morphology of the virus particles, the sequence characteristics of the amplified gene fragments, and the antibody responses against IBV suggested that the agents I0623/17 and I0710/17 were AvCoVs most similar to IBV, and were provisionally designated as PhCoV.
3.2. The two PhCoVs were genetically similar to those of IBVs
The complete genome sequences of the isolates I0623/17 and I0710/17 were obtained by assembly of the fragment sequences. The genome sizes of the two PhCoV isolates were both 27,640 bases. Their genome organizations were typical of AvCoVs, with the gene order (5′ to 3′) of replicase ORF1ab, S, E, M, and N (Fig. 2 A). Both the 5′ and 3′ ends contained short untranslated regions. The replicase ORF1ab in both isolates occupied 19,892 bp of the genomes (Table 2 ). This ORF encodes a number of putative proteins, including nsp3, nsp5, nsp12, nsp13, and other proteins with unknown functions.
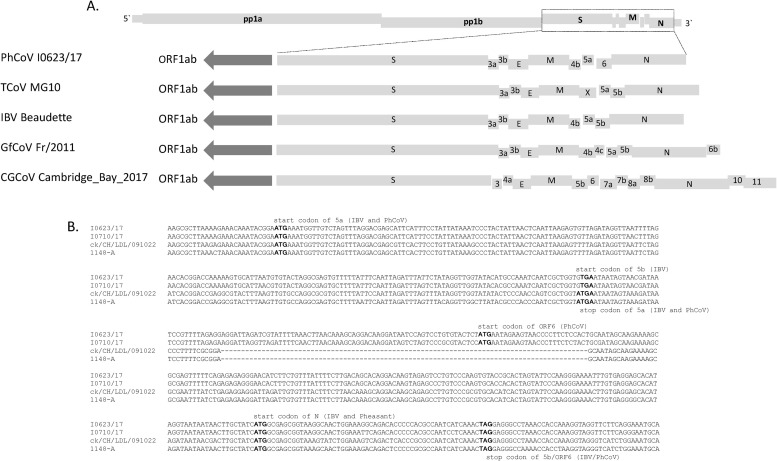
Fig. 2.
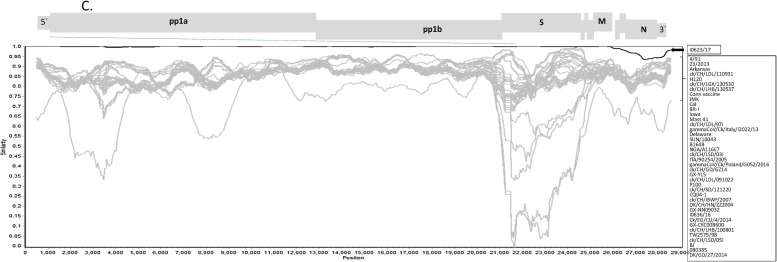
Complete genomic sequence analysis of PhCoV isolate I0623/17 (I0710/17) with those of other AvCoVs. Genomic organization of PhCoV isolate I0623/17 (I0710/17) compared with AvCoVs TCoV MG10, IBV Beaudette, guinea fowl coronavirus (GfCoV) Fr2011, and Canada goose coronavirus (CGCoV) Cambridge_Bay_2017 (A). The names of the main virion genes are given. Genomes of five AvCoVs are drawn to different scales. Alignment of sequences encoding the ORF5 and flanking sequences of PhCoVs I0623/17, I0710/17 and IBV strains LDL/091022and 1148-A (B). A single mutation at the first site of the start codon corresponding to the IBV 5b (ATG→ATT) resulted in absence of 5b in the PhCoV genome. A 90-nt insertion was found in the 5a–ORF6 junction region in the PhCoVs. Analysis was also performed using SimPlot software version 3.5.1 to identify potential recombination breakpoints in the genomes of PhCoV isolates I0623/17 and I0710/17 (C). We used a 1000-bp window with a 50-bp step. Isolate I0710/17 was used as the query strain, and 39 selected IBV strains and one duck CoV were used as reference strains. The names of the main virion genes are given at the top of the SimPlot.
Table 2.
Genome organization and predicted viral proteins encoded by pheasant coronaviruses I0623/17 (I0710/17).
| Gene | Genome position | Size (nucleotide) | Size (amino acid) | Identity to IBV (%) | Most-related lineage/strain | |
|---|---|---|---|---|---|---|
| 5′ UTR | 1-529 | 529 | – | 97.9 | GI-19 | |
| ORF 1a/1ab | NSP2 | 530-2551 | 2022 | 674 | 90.0 | GI-19 |
| NSP3 | 2552-7318 | 4767 | 1588 | 90.6 | GI-22 | |
| NSP4 | 7319-8866 | 1548 | 516 | 92.7 | GI-19 | |
| NSP5 | 8867-9787 | 921 | 307 | 91.1 | GI-22 | |
| NSP6 | 9788-10669 | 882 | 284 | 94.7 | GI-22 | |
| NSP7 | 10670-10918 | 249 | 83 | 98.8 | 1114/14 | |
| NSP8 | 10919-11548 | 630 | 210 | 93.2 | GI-22 | |
| NSP9 | 11549-11881 | 333 | 111 | 92.8 | DK/CH/HN/ZZ2004 | |
| NSP10 | 11882-12316 | 435 | 145 | 94.5 | DK/CH/HN/ZZ2004 | |
| NSP11 | 12317-12385 | 69 | 23 | – | – | |
| NSP12 | 12317-15135 | 2819 | 931 | 94.9 | GI-19 | |
| NSP13 | 15136-16935 | 1800 | 600 | 93.0 | GI-19 | |
| NSP14 | 16936-18498 | 1563 | 521 | 93.9 | GI-22 | |
| NSP15 | 18499-19512 | 1014 | 338 | 93.1 | GI-19 | |
| NSP16 | 19612-20446 | 834 | 278 | 95.2 | GI-19 | |
| S | S1 | 20372-21991 | 1620 | 540 | 92.4 | GI-19 |
| S2 | 21992-23869 | 1878 | 626 | 98.4 | GI-19 | |
| ORF 3 | 3a | 23869-24042 | 174 | 58 | 99.4 | GI-19 |
| 3b | 24042-24236 | 195 | 65 | 100 | GI-9 | |
| 3c (E) | 24217-24546 | 330 | 110 | 95.7 | GI-19 | |
| M | 24518-25195 | 678 | 226 | 95.3 | GI-29 | |
| ORF 4b | 25196-25480 | 285 | 95 | 91.2 | GI-19 | |
| ORF 5a | 25559-25756 | 198 | 66 | 92.4 | GI-19 | |
| ORF 6 | 25849-26091 | 243 | 81 | 87.0 | GI-19 | |
| N | 26034-27260 | 1227 | 409 | 92.5 | GI-19 | |
| 3′ UTR | 27261-27640 | 380 | – | 98.4 | GI-19 | |
The two PhCoV isolates displayed similar genome organizations to IBV strains, and differed only in the number of ORFs downstream of 5a (Fig. 2A). A mutation (ATG→GTG) occurred at a corresponding position to the start codon in our two PhCoV isolates, resulting in the viruses lacking ORF5b. Another ORF, provisionally designated as ORF6, was found between 5a and N in the genomes of the two PhCoVs, which overlapped with N. In addition, a 90-bp insertion sequence was found in the 5a–ORF6 junction region (Fig. 2B). The genomes of the PhCoV isolates were therefore organized in a novel manner: 5′-Gene 1-S-3a-3b-E-M-4b-5a-6-N-3′, which differed from those in IBV and other AvCoVs (Fig. 2A).
In addition, recombination within the genomes of our two PhCoVs was analyzed using SimPlot by comparing them with 40 represented IBVs selected on the basis of the results of phylogenetic analysis. No obvious recombination events were found in the genomes of the PhCoV isolates I0623/17 and I0710/17 (Fig. 2C).
3.3. The PhCoVs differed from IBV to a similar extent as one lineage of IBV differed from another
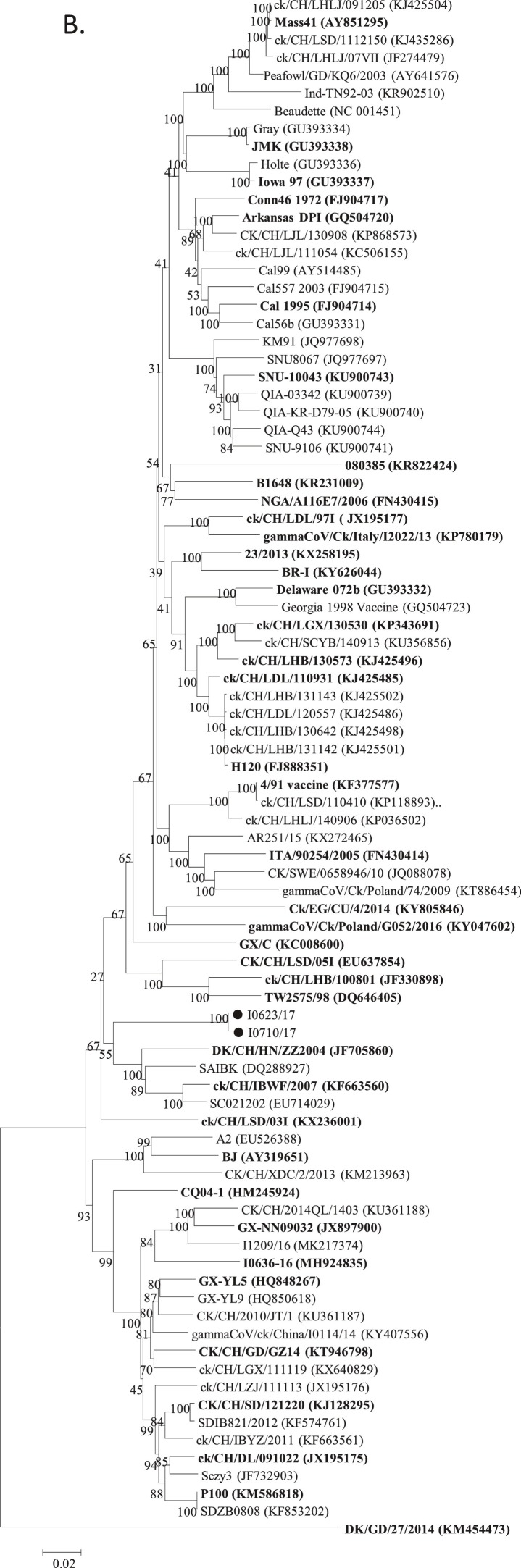
The maximum likelihood trees constructed using the S1 nucleotide sequences of our two PhCoV isolates and 213 reference AvCoV strains are shown in Fig. 3 A. The two PhCoV isolates were clustered together and were not closely related to IBV and other AvCoV strains. Comparatively, the two PhCoV isolates showed a relatively closer relationship with IBV GI-19 lineage strains (<92% and 92% nucleotide and amino acid identities, respectively). According to the complete genomic sequences, the two PhCoV isolates also clustered together and were separate from other IBV strains (Fig. 3B). Isolate I0623/17 (I0710/17) showed the highest sequence identity with the GI-19 lineage (<91%). Trees based on both the S1 and complete genomic sequences, thus, showed that the PhCoVs differed from IBV to a similar extent as one lineage/genotype of IBV differed from another.
Fig. 3.
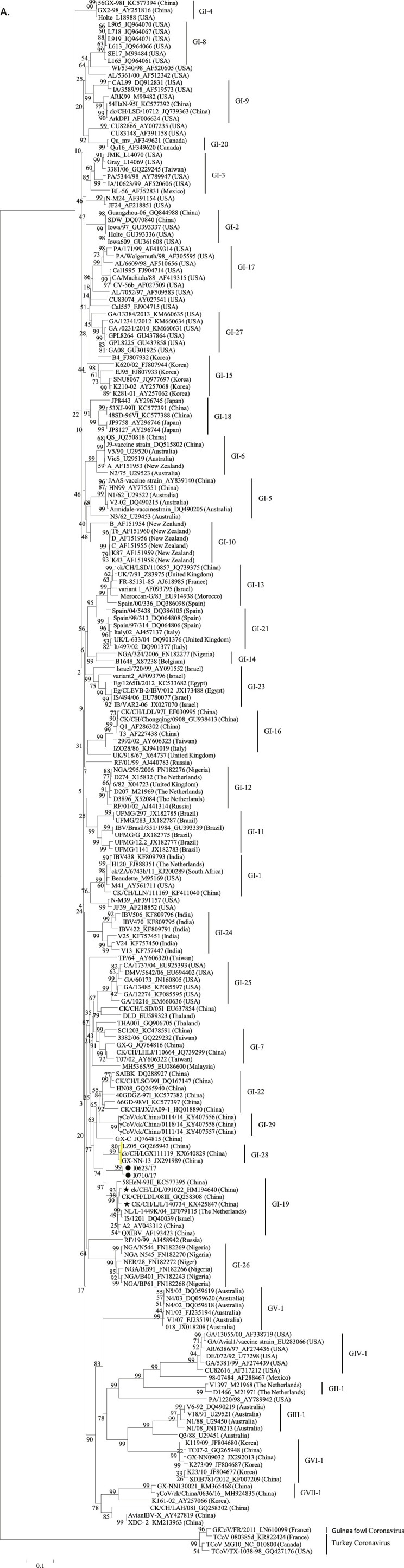
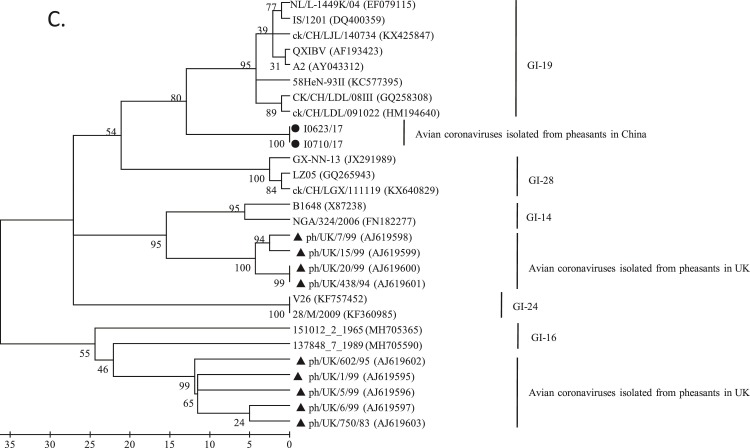
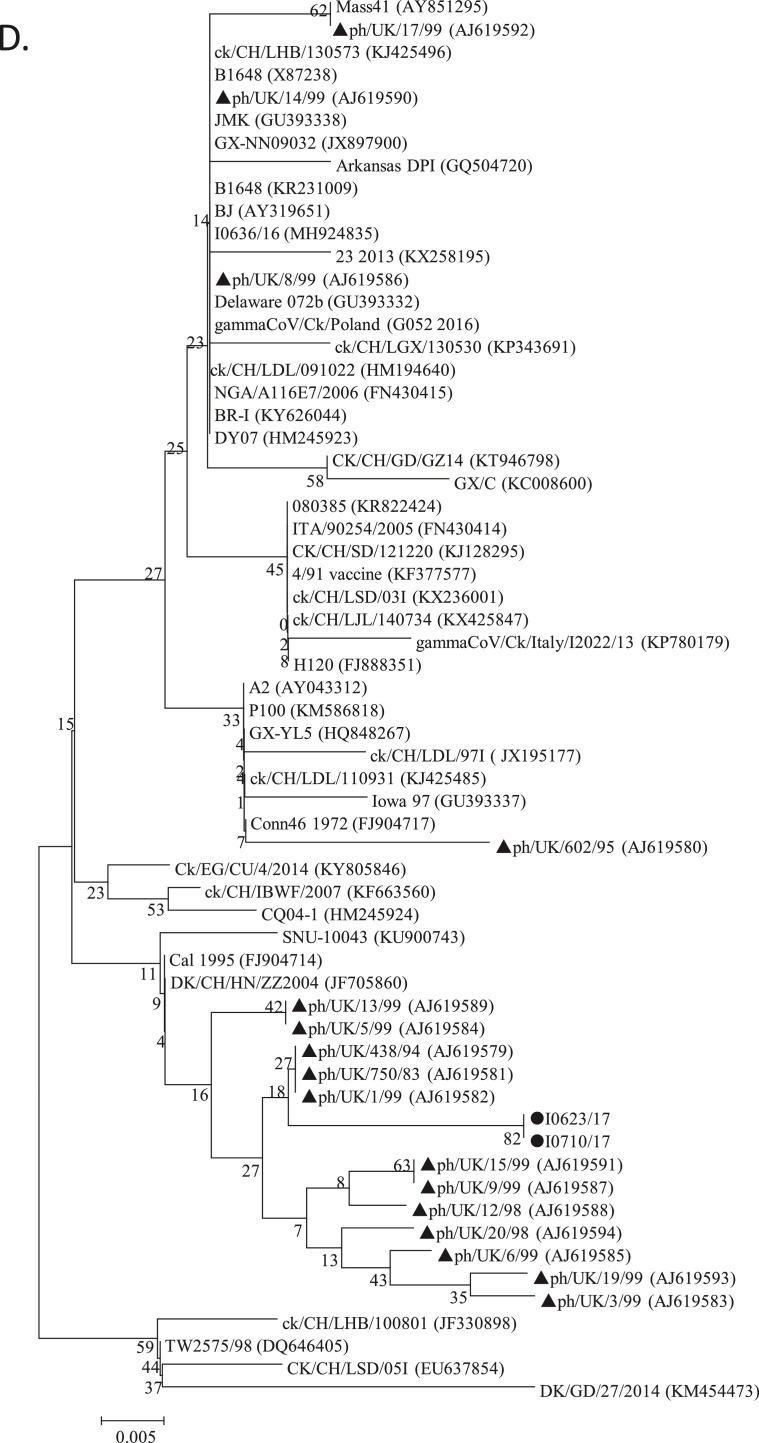
Phylogenetic analysis. Two hundreds and thirteen complete S1 nucleotide sequences of AvCoV reference strains were phylogenetically compared with those of PhCoV isolates I0623/17 and I0710/17 (A). The strains LDL/091022 and LJL/140734 (star) were used in the following VN tests. The complete genomic sequences of our two isolates I0623/17 and I0710/17 were also phylogenetically compared with those of 85 AvCoV reference strains, including 84 IBV strains and one duck CoV strain (B). The 40 sequences used for SimPlot analysis are shown in bold. Phylogenetic trees were constructed using the nucleotide sequences of the S1 gene fragments of PhCoV isolates I0623/17 and I0710/17 (black circle), nine PhCoV strains isolated in the UK (triangle), and 11 selected IBV strains (C). Phylogenetic trees were also conducted using the sequences of 3′ UTR fragments from PhCoV isolates I0623/17 and I0710/17 (black circle), 16 PhCoVs isolated in the UK (triangle), 43 selected IBV strains, and one duck CoV (D). The maximum likelihood method was used to construct the trees with 1000 bootstrap replicates with MEGA4.0 software. GenBank accession numbers are indicated after the names of the viruses PhCoV isolates I0623/17 and I0710/17 (black circle).
Nine sequences of S1 gene fragments from PhCoV strains isolated in the UK were available in GenBank and used for comparison with our two PhCoV isolates and 17 IBV strains isolated in China and Europe. All the PhCoVs were divided into three groups (Fig. 3C). Our two PhCoVs were genetically most closely related to GI-19 lineage viruses, which were believed to originate from China (Liu and Kong, 2004). However, four UK PhCoV strains clustered together and showed a closer relationship with IBV GI-14 lineage strains isolated in Europe and Africa. The remaining five UK PhCoV strains clustered together in a separate group and were most closely related to IBV GI-16 lineage viruses, which were isolated in Europe. Multiple alignments using these sequences confirmed these results and revealed that PhCoVs in each group showed similarities with different IBV lineages (Supplementary Fig. 2), rather than all PhCoVs being most closely related to each other. Interestingly, phylogenetic trees using the 3′ UTR sequences of our two PhCoV isolates, 16 UK PhCoV strains, and 40 IBV strains (selected based on the results of phylogenetic analysis of the complete genomic sequences) showed that most of the PhCoV strains were clustered in a group, but four of which clustered differently (Fig. 3D).
3.4. The PhCoV was antigenically different from the selected IBVs
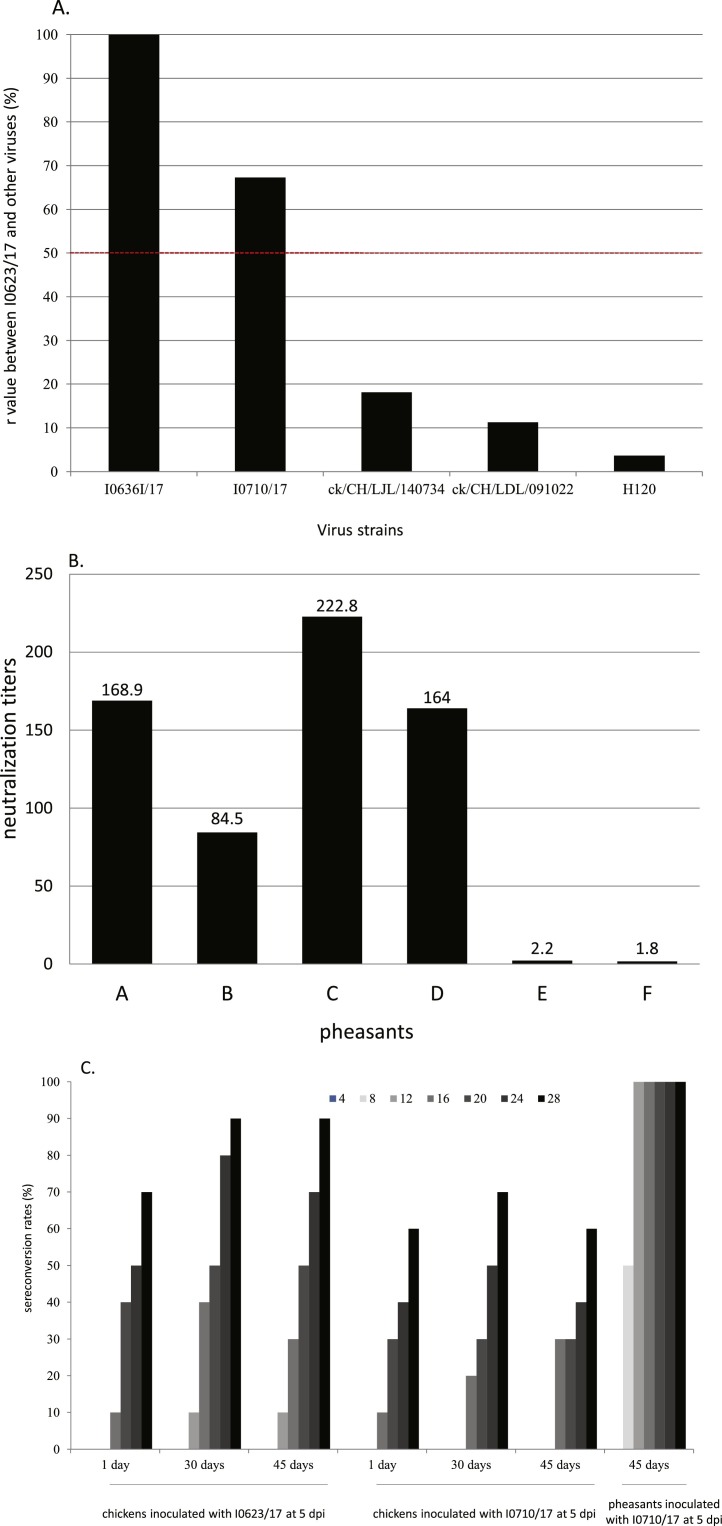
The percentage of antigenic relatedness (r) values determined by VN between the isolate I0623/17 and other viruses, including isolate I0710/17 and three IBV strains, are given in Fig. 4 A. The isolate I0623/17showed a strong antigenic relationship with the isolate I0710/17 on the basis of r value (67.3). In contrast, the r values of the isolate I0623/17 and the IBV strains used in this study were all <18.2%, i.e. below the 50% cutoff value considered to indicate greater antigenic dissimilarity.
Fig. 4.
Neutralization and ELISA tests. The neutralization tests were conducted between PhCoV isolates I0623/17 and I0710/17 and between isolate I0623/17 and selected IBV strains (A). The calculated antigenic relatedness (r) value of PhCoV isolate I0623/17 against I0710/17 and selected IBV strains were shown. The neutralization titers of serum from the commercial pheasants tested by VN for PhCoV antibodies using I0623/17 virus (B). Sera ELISA antibody responses of chickens inoculated with PhCoV isolates I0623/17 and I0710/17 at different ages and pheasants inoculated with PhCoV isolate I0710/17 at 45 days of ages (C).
An aliquot of each serum was pooled and tested by VN for PhCoV antibodies using I0623/17 virus. Neutralization titers ranging from 84.5 (26.4) to 222.8 (27.8) were obtained from pheasant serum after the disappearance of clinical signs (Fig. 4B), indicating previous exposure to the virus. In contrast, the neutralization antibody was negative in serum from pheasants that had not shown any clinical signs.
3.5. The PhCoV was non-pathogenic to SPF chickens but highly pathogenic to commercial pheasants
No clinical signs were observed in chickens inoculated with different PhCoVs at different ages (1, 30, and 45 days), comparable with chickens in the negative control groups. In contrast, the clinical signs were observed in all the pheasants from 2 to 14 dpi with isolate I0710/17 at 45 days of age. The clinical signs, including snicking, watery eyes, nasal discharge, and rales. No gross lesions were observed in the five chickens inoculated with isolate I0710/17 necropsied at 5 dpi, however, gross kidney lesions were observed in three of the five pheasants inoculated with isolate I0710/17. The kidney parenchyma was pale, swollen, and mottled, and the kidney tubules and urethras were distended with uric acid crystals. Three pheasants died at 6 dpi following the clinical signs during the experiment. Gross lesions in the dead birds were also mainly confined to the kidneys. Mild tracheitis was also observed in the dead birds. No clinical signs were observed in the control pheasants.
The antibody responses are illustrated in Fig. 4C. Both chickens and pheasants inoculated with the PhCoV isolates showed negative ELISA antibody results at 4 dpi. No antibody responses were observed in any of the chickens inoculated with PhCoV isolates I0623/17 and I0710/17 at 8 dpi at different ages in ELISA tests; however, 50% of pheasants inoculated with I0710/17 showed seroconversion from 8 dpi. The seroconversion rates were higher in pheasants than in chickens at the corresponding time points. No antibody was detected in the negative control chickens or pheasants.
3.6. The PhCoV had high replication capacity in commercial pheasants
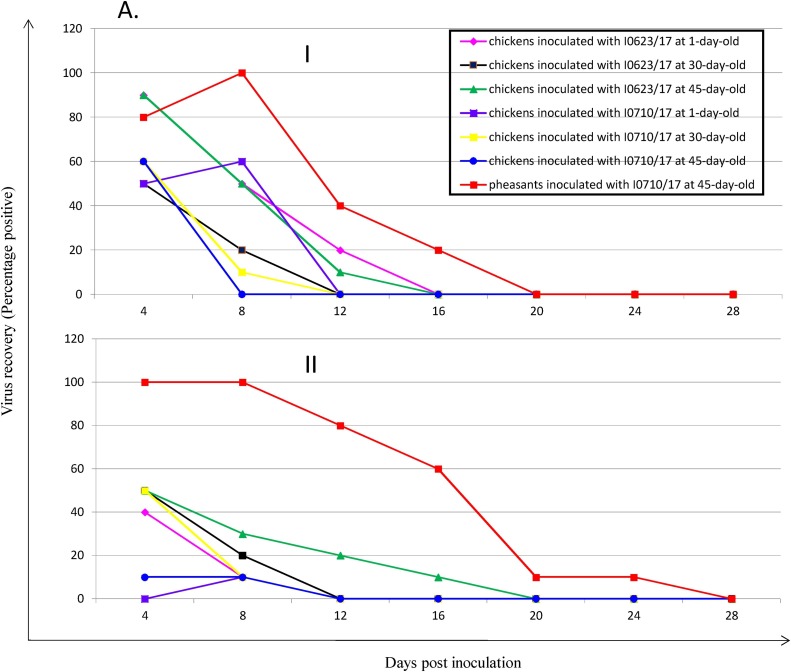
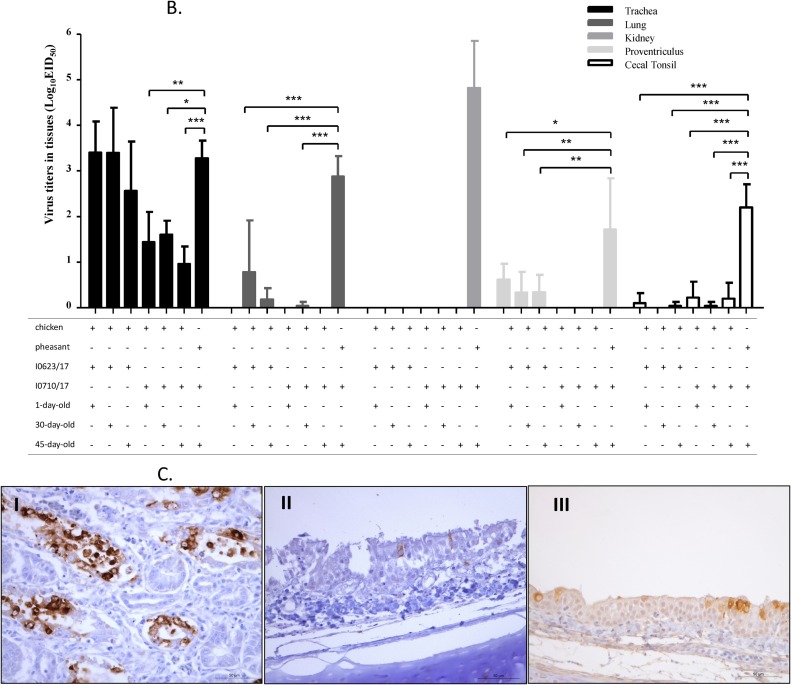
Shedding of the PhCoV isolate in the respiratory and digestive tracts of both chickens and pheasants was determined by virus re-isolation using oropharyngeal and cloacal swabs with 9-day-old SPF chicken eggs (Han et al., 2016). Prolonged virus replication and higher number of birds shedding virus in the respiratory and digestive tracts were observed in pheasants, compared to those of chickens inoculated with PhCoV I0710/17 at different time points (Fig. 5 A). Replication of the challenge viruses was also determined at 5 dpi in the tracheas, kidneys, lungs, proventriculus, and cecal tonsils from chickens and pheasants (Ma et al., 2019) (Fig. 5B). The replication capacity of the PhCoV I0623/17 isolate was detected in the tracheas of chickens and pheasants with comparable titers; however, the titers were somewhat lower in chickens inoculated with I0710/17. Virus titers in the lungs, proventriculus, and cecal tonsils were significantly lower in chickens than in pheasants, with no viruses detected in some chicken tissues. Notably, no virus was detected in the kidneys of chickens challenged with the two PhCoV isolates, in contrast to the high titers in the kidneys of pheasants challenged with I0710/17. In line with this result, viral antigens were detected by IHC in the kidneys of pheasants challenged with I0710/17 (Fig. 5C), but not in the kidneys of chickens inoculated with the two PhCoV isolates. The viral antigens were also detected by IHC in the tracheas of chickens challenged with I0623/17 and pheasants challenged with I0710/17 (Fig. 5C). No virus was detected in any tissues or in oropharyngeal and cloacal swabs from birds in the control group.
Fig. 5.
Infection of chickens with PhCoV isolates I0623/17 and I0710/17 and of pheasants with I0710/17. Virus recovery from oropharyngeal (I) and cloacal (II) swabs from chickens inoculated with PhCoV isolates I0623/17 and I0710/17, and pheasants with PhCoV isolate I0710/17 (A). Virus recovery was performed by inoculating 9-day-old embryonated, specific pathogen-free eggs through the allantoic route with supernatant from the swabs. Replication of our isolates I0623/17 and I0710/17 in the trachea, lung, proventriculus, cecal tonsil, and kidney of chickens at 5 dpi, and replication of I0710/17 in the trachea, lung, proventriculus, cecal tonsil, and kidney of pheasants at 5 dpi (B). Viral titration was performed by inoculating 9-day-old embryonated, specific pathogen-free eggs through the allantoic route. Data are expressed as the mean ± standard deviation. Virus titers were analyzed by Student’s t-test using GraphPad Prism for Windows version 5. Differences were considered significant if the p value was <0.05 (*p < 0.05, **p < 0.01, ***p < 0.001). Detection of the IBV antigen by IHC analysis in the kidney (I) and trachea (II) of pheasants at 5 dpi following PhCoV I0710/17 infection and in the trachea (III) of chickens at 5 dpi following PhCoV I0623/17 infection (C).
4. Discussion
In the current study, we isolated two AvCoVs from commercial pheasants at two respective farms in China in 2017 with disease outbreaks occurring within a 2-month period. The diseased birds showed early respiratory signs, suggesting that they may have been caused by respiratory pathogens, possibly including NDV, AIV subtypes H5 and H9, IBV, or PhCoVs. The embryopathic effects and morphologies of the isolated viruses and the sequence characteristics of the amplified gene fragments suggested that both agents were AvCoVs most similar to IBV. This was confirmed by serological ELISA evidence using IBV as the antigen and VN test using the isolate I0631/17. Since the original description of PhCoV in 1980 (Spackman and Cameron, 1983), the disease caused by PhCoV has become widespread in the UK (Lister et al., 1985; Pennycott, 2000; Cavanagh et al., 2002). Some viruses isolated from pheasants with respiratory disease, reduced egg production, renal problems, and high mortality were believed to be IBV (Spackman &Cameron, 1983; Gough et al., 1996; Lister et al., 1985; Pennycott, 2000), due to similarities with IBV in domestic fowl (Cook et al., 2012). The mortality associated with the outbreaks at pheasant farms in China were comparable to or even higher than those of the outbreak at a large game farm in the UK in 1994, which resulted in the deaths of over 1000 breeding pheasants out of a total stock of approximately 7000 birds (Gough et al., 1996). The clinical signs and gross lesions in dead birds were also similar between the two outbreaks, with upper respiratory signs at the beginning of the outbreaks, and the birds dying rapidly after appearance of the signs of illness. Pale, swollen kidneys and ureters distended with urates were also found in both cases. Some viruses have been successfully isolated and propagated in embryonating domestic fowl eggs (Lister et al., 1985; Gough et al., 1996), as with the two PhCoVs isolated in this study.
Phylogenetic analysis showed that our PhCoVs represented a novel lineage in genotype I of IBV, according to the method of Valastro et al. (2016). The phylogenetic analyses were based on the S1 gene sequences of our PhCoVs and 213 reference AvCoV strains. The nucleotide and putative amino acid compositions of the S1 gene also showed approximately 10% differences between the two current PhCoVs isolates and other AvCoV strains, compared with 13% of nucleotides and 14% of amino acids in the S1 subdomain between IBV lineages, and 30% of nucleotides and 31% (occasionally even 40% or more) of amino acids in the S1 subdomain between IBV serotypes (Valastro et al., 2016). Our PhCoVs thus differed from IBV to a similar extent as one lineage of IBV differed from another. We also found interesting branching of the phylogenetic tree based on the complete genomic sequences of our two PhCoVs and IBVs of different lineages/genotypes. Pairwise comparisons of the complete genomic sequences revealed that our PhCoVs differed by >10%, similar to many IBV lineages and serotypes, further supporting the similarity of the genetic relationship between our two PhCoVs and IBV strains was similar to that between IBV lineages and genotypes. In addition, the robust IBV-specific serum IgG titers in chickens inoculated with PhCoV isolates showed that PhCoV infection stimulated a humoral response in chickens (Raj and Jones, 1997; Aston et al., 2018). However, PhCoVs were antigenically dissimilar to IBVs in the VN tests, suggesting that the antigenic relationship between our two PhCoVs and the IBV strains was similar to those between different IBV genotypes (Gao et al., 2016). Viruses antigenically distinct from the investigated IBVs have also been isolated from pheasants in the UK on several occasions (Lister et al., 1985; Cavanagh et al., 2002).
CoVs typically have a restricted host range and only infect their natural host. As an AvCoV, IBV is considered to be a highly contagious virus that only infects chickens (Gallus gallus) (Cavanagh, 2007); however, IBV has also been reported in other avian species including guinea fowl, partridge, peafowl, and teal, without producing any clinical detectable disease (Cavanagh, 2007; Liu et al., 2005; Sun et al., 2007). Pheasants (P. colchicus) differ from chickens (G. gallus), though both belong to the family Phasianidae, order Galliformes. Although the PhCoVs in this study were very similar to the IBVs, there were some obvious differences. The IBV RNA genome is characterized by the organization 5′-UTR-1a-1b-S-3a-3b-E-M-5a-5b-N-UTR-3′, while the genomes of the PhCoV isolates were organized in a novel 5′-UTR-1a-1b-S-3a-3b-E-M-5a-6-N-3′UTR order, which differed from those of IBV and other AvCoVs. Novel genomic organizations have also been found in other IBVs (Liu et al., 2008a,b; Mardani et al., 2008; Xu et al., 2016) and experiments with engineered mutants suggested that the native order was not functionally essential (Casais et al., 2005; Hodgson et al., 2006). Notably, the current study found differences in the pathogenicity and replication capacity of the PhCoV isolates between chickens and pheasants. Intranasal instillation of 10 3-week-old SPF chickens with 0.2 ml of infectious allantoic fluid containing 103.2 EID50 of PhCoV previously failed to produce any clinical signs of disease (Lister et al., 1985). However, all birds had HI, but precipitating antibodies to IBV M41 strain. In this study, inoculation of chickens via the ocular and nasal routes with 0.1 ml of 105.0 EID50 of the PhCoV isolates I0623/17 and I0710/17 at 1, 30, and 45 days of age, respectively, also failed to produce any clinical signs of disease, while pheasants inoculated with I0710/17 showed obvious clinical signs and 30% mortality, similar to chickens infected with IBV strains (Liu and Kong, 2004). The clinical signs were similar to those in the affected commercial pheasants from which the viruses were isolated, and to those in chickens infected with IBV strains (Cavanagh, 2007). Furthermore, not all the inoculated chickens had ELISA-detected antibodies, compared with seroconversion of all pheasants inoculated with the PhCoV isolate. Strikingly, the viruses could be re-isolated from the tracheas of chickens inoculated with PhCoV isolates I0623/17 and I0710/17, but were absent or present at very low titers in other chicken tissues investigated in this study. In contrast, the PhCoV isolate I0710/17 could be re-isolated from, and had significantly higher viral titers in all pheasant tissues investigated in this study, especially the kidneys. These results were in line with the IHC results, which showed that viral antigen-positive epithelial cells were intensely labeled in the kidneys of pheasants challenged with I0710/17. These results were also consistent with those for viral replication and shedding, which indicated prolonged virus replication and higher number of birds shedding virus in the respiratory and digestive tracts of pheasants compared with chickens inoculated with PhCoVs. These findings support the different biological characteristics of PhCoVs compared with IBVs (Liu et al., 2008a,b).
The close relationship—but with obvious differences between PhCoVs and IBVs—led us to speculate that the PhCoVs may have originated from IBVs. This hypothesis was confirmed, at least in part, by the analyses of S1 fragments from PhCoVs isolated from different outbreaks and selected IBV strains. Instead of being clustered together and showing a close genetic relationship, the PhCoVs isolated from different outbreaks in different years clustered in different groups, with each group showing a relatively close relationship with the IBVs circulating in the regions from which the respective PhCoVs were isolated. We speculated that the IBV strains may have been introduced into and adapted themselves in pheasants, and subsequently evolved independently to different extents resulting in different virulences in pheasants. For example, a previous report on the isolation of a PhCoV reported that the only notable clinical features were respiratory signs, reduced egg production, and shell-quality problems (Spackman and Cameron, 1983). However, the PhCoVs in the current and other studies showed a high affinity to the kidney (Gough et al., 1996; Lister et al., 1985; Pennycott, 2000) and were highly pathogenic in pheasants. The hypothesis that different introductions of the viruses from chickens to pheasants and the independent evolution of the resulting PhCoVs may account for the inconsistent phenotypes of the PhCoVs.
The emergence of CoVs in turkeys was proposed to have resulted from recombination events involving IBVs and an as-yet-unidentified CoV, which donated an S gene encoding a protein with low amino acid identity to IBV (35%) (Jackwood et al., 2010; Brown et al., 2016). This is suspected to have resulted in a host shift from chickens to turkeys, as well as switching the tissue tropism of the virus from the upper respiratory to the intestinal tract. In this study, we detected genome-wide differences, including point mutations, deletions, and insertions, in the ORFs and deduced protein sequences of our PhCoVs, compared with IBVs. The accumulation of substitutions in multiple genes in the PhCoV genome resulted in a host shift from chickens to pheasants, but did not lead to a change in tissue tropism (nephropathogenicity). In addition, we found no recombination events in the genomes of our PhCoVs compared with the IBV strains selected in this study. In contrast, different phylogenetic topologies were observed when the S1 fragments and 3′UTRs of some of the PhCoVs isolated in UK were compared, suggesting that possible recombination events might have occurred. The different evolutionary patterns between our PhCoVs and those isolated in the UK might suggest that PhCoVs isolated in different outbreaks derived differently and have evolved independently.
There is currently no information on the complete genome sequences of PhCoVs isolated in other countries. Further molecular studies are therefore required to determine the complete genomic sequences of PhCoVs from different outbreaks in pheasants, to allow a thorough investigation of the relationships between PhCoVs and IBVs. In addition, further research involving the infection of pheasants with IBVs is also required to ascertain its pathogenicity in pheasants and whether could pheasants be the potential asymptomatic vectors of IBV for possibly spreading the virus.
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2017YFD0500105), the National Natural Science Foundation of China Grant (31872503), the China Agriculture Research System (No. CARS-40-K18), National "Twelfth Five-Year" Plan for Science & Technology Support (2015BAD12B03) and the Provincial Supported Science Foundation of Heilongjiang Province for The National Key Technology R&D Program (GX16B003).The authors declare that they have no competing interests.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2019.108513.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Archetti I., Horsfall F.L.Jr. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exp. Med. 1950;92:441–462. doi: 10.1084/jem.92.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.A., Touzain F., Briand F.X., Gouilh A.M., Courtillon C., Allée C., Lemaitre E., De Boisséson C., Blanchard Y., Eterradossi N. First complete genome sequence of European turkey coronavirus suggests complex recombination history related with US turkey and guinea fowl coronaviruses. J. Gen. Virol. 2016;97:110–120. doi: 10.1099/jgv.0.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Davies M., Cavanagh D., Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005;79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K. Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype. Avian Pathol. 1992;21:401–408. doi: 10.1080/03079459208418858. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Britton P., Naylor C.J. Longitudinal field studies of infectious bronchitis virus and avian pneumovirus in broilers using type-specific polymerase chain reactions. Avian Pathol. 1999;28:593–605. doi: 10.1080/03079459994399. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Welchman Dde.B., Britton P., Gough R.E. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 2002;31:81–93. doi: 10.1080/03079450120106651. [DOI] [PubMed] [Google Scholar]
- Chaharaein B., Omar A.R., Aini I., Yusoff K., Hassan S.S. Detection of H5, H7 and H9 subtypes of avian influenza viruses by multiplex reverse transcription-polymerase chain reaction. Microbiol. Res. 2009;164:174–179. doi: 10.1016/j.micres.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Chen Y., Jiang L., Zhao W., Liu L., Zhao Y., Shao Y., Li H., Han Z., Liu S. Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet. Microbiol. 2017;198:108–115. doi: 10.1016/j.vetmic.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Leung C.Y., Gilbert M., Joyner P.H., Ng E.M., Tse T.M., Guan Y., Peiris J.S., Poon L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011;85:12815–21280. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J.K., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41:239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- de Groot . Family coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy, 9th Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego, CA: 2012. pp. 806–828. [Google Scholar]
- de Haan C.A., Stadler K., Godeke G.J., Bosch B.J., Rottier P.J. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 2004;78:6048–6054. doi: 10.1128/JVI.78.11.6048-6054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez M.F., European Union COST Action FA1207 Recommendations for a standardized avian coronavirus (AvCoV) nomenclature: outcome from discussions within the framework of the European Union COST Action FA1207: "towards control of avian coronaviruses: strategies for vaccination, diagnosis and surveillance". Avian Pathol. 2016;45:602–603. doi: 10.1080/03079457.2016.1211834. [DOI] [PubMed] [Google Scholar]
- Gohm D.S., Thur B., Hofmann M.A. Detection of Newcastle disease virus in organs and faeces of experimentally infected chickens using RT-PCR. Avian Pathol. 2000;29:143–152. doi: 10.1080/03079450094171. [DOI] [PubMed] [Google Scholar]
- Gough R.E., Cox W.J., Winkler C.E., Sharp M.W., Spackman D. Isolation and identification of infectious bronchitis virus from pheasants. Vet. Rec. 1996;138:208–209. doi: 10.1136/vr.138.9.208. [DOI] [PubMed] [Google Scholar]
- Guo H., Liu X., Xu Y., Han Z., Shao Y., Kong X., Liu S. A comparative study of pigeons and chickens experimentally infected with PPMV-1 to determine antigenic relationships between PPMV-1 and NDV strains. Vet. Microbiol. 2014;168:88–97. doi: 10.1016/j.vetmic.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Guy J.S. Isolation and propagation of coronaviruses in embryonated eggs. Methods Mol. Biol. 2008;454:109–117. doi: 10.1007/978-1-59745-181-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Zhang T., Xu Q., Gao M., Chen Y., Wang Q., Zhao Y., Shao Y., Li H., Kong X., Liu S. Altered pathogenicity of a tl/CH/LDT3/03 genotype infectious bronchitis coronavirus due to natural recombination in the 5’- 17kb region of the genome. Virus Res. 2016;213:140–148. doi: 10.1016/j.virusres.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Zhao F., Shao Y., Liu X., Kong X., Song Y., Liu S. Fine level epitope mapping and conservation analysis of two novel linear B-cell epitopes of the avian infectious bronchitis coronavirus nucleocapsid protein. Virus Res. 2013;171:54–64. doi: 10.1016/j.virusres.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L. Recombinational histories of avian infectious bronchitis virus and turkey coronavirus. Arch. Virol. 2011;156:1823–1829. doi: 10.1007/s00705-011-1061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Boynton T.O., Hilt D.A., McKinley E.T., Kissinger J.C., Paterson A.H., Robertson J., Lemke C., McCall A.W., Williams S.M., Jackwood J.W., Byrd L.A. Emergence of a group 3 coronavirus through recombination. Virology. 2010;398:98–108. doi: 10.1016/j.virol.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen C.M., Kofstad T., Larsen I.L., Løvland A., Handeland K., Follestad A., Lillehaug A. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos) J. Gen. Virol. 2005;86:1597–1607. doi: 10.1099/vir.0.80927-0. [DOI] [PubMed] [Google Scholar]
- Jia W., Karaca K., Parrish C.R., Naqi S.A. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhao W., Han Z., Chen Y., Zhao Y., Sun J., Li H., Shao Y., Liu L., Liu S. Genome characterization, antigenicity and pathogenicity of a novel infectious bronchitis virus type isolated from south China. Infect. Genet. Evol. 2017;54:437–446. doi: 10.1016/j.meegid.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters J.G., Jager E.J., Niesters H.G., van der Zeijst B.A. Sequence evidence for RNA recombination in field isolates of avian coronavirus infectious bronchitis virus. Vaccine. 1990;8:605–608. doi: 10.1016/0264-410X(90)90018-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters J.G., Niesters H.G., Lenstra J.A., Horzinek M.C., van der Zeijst B.A. Phylogeny of antigenic variants of avian coronavirus IBV. Virology. 1989;169:217–221. doi: 10.1016/0042-6822(89)90058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopardi S., Holmes E.C., Gastaldelli M., Tassoni L., Priori P., Scaravelli D., Zamperin G., De Benedictis P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018;58:279–289. doi: 10.1016/j.meegid.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C.S., Porter E., Matthews D., Kipar A., Tasker S., Helps C.R., Siddell S.G. Genotyping coronaviruses associated with feline infectious peritonitis. J. Gen.Virol. 2015;96:1358–1368. doi: 10.1099/vir.0.000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang J., Qiu L., Han Z., Liu S. Fowl adenovirus species C serotype 4 is attributed to the emergence of hepatitis-hydropericardium syndrome in chickens in China. Infect. Genet. Evol. 2016;45:230–241. doi: 10.1016/j.meegid.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Lister S.A., Beer J.V., Gough R.E., Holmes R.G., Jones J.M., Orton R.G. Outbreaks of nephritis in pheasants (phasianus colchicus) with a possible coronavirus aetiology. Vet. Rec. 1985;117:612–613. doi: 10.1136/vr.117.23.612. [DOI] [PubMed] [Google Scholar]
- Liu S., Chen J., Chen J., Kong X., Shao Y., Han Z., Feng L., Cai X., Gu S., Liu M. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) J. Gen. Virol. 2005;86:719–725. doi: 10.1099/vir.0.80546-0. [DOI] [PubMed] [Google Scholar]
- Liu S., Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. 2004;33:321–327. doi: 10.1080/0307945042000220697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wang Y., Ma Y., Han Z., Zhang Q., Shao Y., Chen J., Kong X. Identification of a newly isolated avian infectious bronchitis coronavirus variant in China exhibiting affinity for the respiratory tract. Avian Dis. 2008;52:306–314. doi: 10.1637/8110-091307-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang Q., Chen J., Han Z., Shao Y., Kong X., Tong G. Identification of the avian infectious bronchitis coronaviruses with mutations in gene 3. Gene. 2008;412:12–25. doi: 10.1016/j.gene.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang X., Gong L., Yan B., Li C., Han Z., Shao Y., Li H., Kong X. Altered pathogenicity, immunogenicity, tissue tropism and 3’-7kb region sequence of an avian infectious bronchitis coronavirus strain after serial passage in embryos. Vaccine. 2009;27:4630–4640. doi: 10.1016/j.vaccine.2009.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ma H., Xu Q., Sun N., Han Z., Sun C., Guo H., Shao Y., Kong X., Liu S. Characterization of a recombinant coronavirus infectious bronchitis virus with distinct S1 subunits of spike and nucleocapsid genes and a 3’ untranslated region. Vet. Microbiol. 2013;162:429–436. doi: 10.1016/j.vetmic.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Xu L., Ren M., Shen J., Han Z., Sun J., Zhao Y., Liu S. Novel genotype of infectious bronchitis virus isolated in China. Vet. Microbiol. 2019;230:178–186. doi: 10.1016/j.vetmic.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardani K., Noormohammadi A.H., Hooper P., Ignjatovic J., Browning G.F. Infectious bronchitis viruses with a novel genomic organization. J. Virol. 2008;82:2013–2024. doi: 10.1128/JVI.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihindukulasuriya K.A., Wu G., St Leger J., Nordhausen R.W., Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J.Virol. 2008;82:5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso H., Thayer S.G., Hofacre C.L., Kleven S.H. Inactivation, storage, and PCR detection of Mycoplasma on FTA filter paper. Avian Dis. 2004;48:841–850. doi: 10.1637/7215-060104. [DOI] [PubMed] [Google Scholar]
- Papineau A., Berhane Y., Wylie T.N., Wylie K.M., Sharpe S., Lung O. Genome organization of canada goose coronavirus, a novel species identified in a mass die-off of Canada geese. Sci. Rep. 2019;9:5954. doi: 10.1038/s41598-019-42355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennycott T.W. Causes of mortality and culling in adultpheasants. Vet. Rec. 2000;146:273–278. doi: 10.1136/vr.146.10.273. [DOI] [PubMed] [Google Scholar]
- Xu Q., Han Z., Wang Q., Zhang T., Gao M., Zhao Y., Shao Y., Li H., Kong X., Liu S. Emergence of novel nephropathogenic infectious bronchitis viruses currently circulating in Chinese chicken flocks. Avian Pathol. 2016;45:54–65. doi: 10.1080/03079457.2015.1118435. [DOI] [PubMed] [Google Scholar]
- Raj G.D., Jones R.C. Infectious bronchitis virus: immunopathogenesis of infection in the chicken. Avian Pathol. 1997;26:677–706. doi: 10.1080/03079459708419246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Reguera J., Santiago C., Mudgal G., Ordono D., Enjuanes L., Casasnovas J.M. Structural bases of coronavirus attachment to host aminopeptidase N snd its inhibition by neutralizing antibodies. Plos Pathogen. 2012;8 doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J., Furukawa T., Kotake M., Takano T., Motokawa K., Gemma T., Watanabe R., Arai S., Hohdatsu T. Screening and identification of T helper 1 and linear immunodominant antibody-binding epitopes in the spike 2 domain and the nucleocapsid protein of feline infectious peritonitis virus. Vaccine. 2011;29:1791–1800. doi: 10.1016/j.vaccine.2010.12.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk A.F., Hawn M.C. An apparently new respiratory disease of chicks. J. Am. Vet. Med. Assoc. 1931;78:413–422. [Google Scholar]
- Spackman D., Cameron I.R. Isolation of infectious bronchitis virus from pheasants. Vet. Rec. 1983;113:354–355. doi: 10.1136/vr.113.15.354. [DOI] [PubMed] [Google Scholar]
- Sun L., Zhang G.H., Jiang J.W., Fu J.D., Ren T., Cao W.S., Xin C.A., Liao M., Liu W.J. A Massachusetts prototype like coronavirus isolated from wild peafowls is pathogenic to chickens. Virus Res. 2007;130:121–128. doi: 10.1016/j.virusres.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres C.A., Listorti V., Lupini C., Franzo G., Drigo M., Catelli E., Brandão P.E., Cecchinato M. Gamma and Deltacoronaviruses were isolated in quail and pheasants from Northern Italy1. Poult. Sci. 2017;96:717–722. doi: 10.3382/ps/pew332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Tsang A.K., Hui S.W., Fan R.Y., Martelli P., Yuen K.Y. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in Gammacoronavirus. J.Virol. 2014;88:1318–1331. doi: 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao W., Gao M., Xu Q., Xu Y., Zhao Y., Chen Y., Zhang T., Wang Q., Han Z., Li H., Chen L., Liang S., Shao Y., Liu S. Origin and evolution of LX4 genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2017;198:9–16. doi: 10.1016/j.vetmic.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.