Highlights
-
•
Interaction between TGEV N protein and EF1A is found in vitro.
-
•
Interaction between TGEV N protein and EF1A is found in vivo.
-
•
EF1A plays a role in TGEV replication.
-
•
Interaction between TGEV N protein and EF1A was firstly found in this study.
Keywords: EF1A, Transmissible gastroenteritis coronavirus, Nucleocapsid
Abstract
Transmissible gastroenteritis coronavirus (TGEV) is an enteropathogenic coronavirus that causes diarrhea in pigs, which is correlated with high morbidity and mortality in suckling piglets. Using the method of GST pull-down with the nucleocapsid (N), N protein was found to interact with swine testes (ST) cells elongation factor 1-alpha (EF1A), an essential component of the translational machinery with an important role in cells. In vitro and in virus-infected cells interaction was then confirmed by co-precipitation. Knockdown of EF1A impairs N protein proliferation and TGEV replication in host cell. It was demonstrated that EF1A plays a role in TGEV replication. The present study thus provides a protein-related information that should be useful for underlying mechanism of coronavirus replication.
1. Introduction
Coronaviruses (CoVs) includes four genera, alpha-, beta-, gamma-, and deltacoronavirus, which have been clustered in the Coronavirinae subfamily (de Groot et al., 2011, Reguera et al., 2012). Coronaviruses (CoVs) are pleomorphic, enveloped viruses (Perlman and Netland, 2009). Transmissible gastroenteritis virus (TGEV) is a representative CoV in the alphacoronavirus genus; severe acute respiratory syndrome-related coronavirus (SARS-related CoV) is a representative of the betacoronavirus genus; infectious bronchitis virus (IBV) is a representative of the gammacoronavirus genus; and Bulbul-CoV is a representative of the deltacoronavirus genus (de Groot et al., 2011). TGEV is positive RNA viruses, which is a large family of enveloped virus (Masters, 2006). The infection of TGEV causes severe diarrhea in suckling piglets (about 2 weeks old), which results in enormous economic loss in swine-producing areas in the world (Kim and Chae, 2001, Sestak et al., 1996). TGEV genome (28.5 kb) encodes the replicase gene (rep) at the 5′ end and encodes other viral genes at the 3′ end (5′-S-3a-3b-E-M-N-7-3′)(Penzes et al., 2001). TGEV genome encodes four structural proteins: spike (S), membrane (M), minor envelope (E), and nucleocapsid (N).
CoVs N proteins are highly basic with a molecular mass ranging from 40 to 63 kDa, depending on the species and strains. N protein binds to the RNA genome, forming a helical nucleocapsid (Escors et al., 2001, Sturman et al., 1980). N protein has a structural role in coronavirus assembly (Risco et al., 1996) and is a growing evidence for a role in RNA synthesis (Almazan et al., 2004, Baric et al., 1988, Stohlman et al., 1988). Some reports have been studied the response of host cell to TGEV (Ding et al., 2012, Wei et al., 2012). Howerer, there is few report about the interaction of N protein with host cell.
Elongation factor 1-alpha (EF1A) is a major translation factor involved in protein synthesis in mammalian cells. EF1A is an abundant G protein that delivers aminoacyl-tRNA to the elongating ribosome (Carvalho et al., 1984b). EF1A hydrolyzes GTP, dissociates from the aminoacyl-tRNA, and leaves the ribosome (Moldave, 1985). Except a major translation factor, EF1A plays important multifunctional roles in mammalian cells. EF1A Interacts with newly synthesized polypeptides for quality surveillance (Hotokezaka et al., 2002). In ubiquitin-dependent degradation, EF1A interacted with ubiquitinated proteins and is essential for ubiquitin-dependent degradation (Chuang et al., 2005, Gonen et al., 1994). EF1A undergoes several post-translational modifications, mainly phosphorylation and methylation, and plays important role in facilitating apoptosis (Lamberti et al., 2004).
Recently, some reports showed that EF1A interacted with viral proteins. The interaction between EF1A and N protein of SARS-CoV was founded (Zhou et al., 2008). There is no report about whether EF1A interacted with N protein of TGEV. In this study, we demonstrate that EF1A associates with N protein of TGEV and plays a role in virus replication. This study will provide protein-related information for underlying mechanism of coronavirus replication.
2. Materials and methods
2.1. Cells and virus
Swine testes (ST) cells were obtained from ATCC. ST cells were grown in RPMI-1640 medium supplemented with 10% fetal calf serum under standard culture conditions (5% CO2, 37 °C). TGEV infectious strain H (Accession No. FJ755618) and TGEV attenuated strain H (Accession No. EU074218) were propagated on an ST cell monolayer (Wang et al., 2010). Pathogenicity of the TGEV infectious strain H is stronger than TGEV attenuated strain H. However, the attenuated TGEV virus was better to adapt ST cells than infectious TGEV.
2.2. Antibodies
Mouse monoclonal antibody (mAb) to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ab9484) and rabbit polyclonal antibody (pAb) to EF1A (ab140632) were purchased from Abcam. FITC-labeled goat anti-mouse IgG was purchased from Kirkegaard and Perry Laboratories (KPL). TRITC-labeled goat anti-rabbit IgG was purchased from Sigma mAb to N protein of TGEV was prepared in our lab.
2.3. Cell infection
ST cells were infected with TGEV infectious strain H or TGEV attenuated strain H at a multiplicity of infection (MOI) of 1. After adsorption for 1 h, cells were washed and incubated in fresh RPMI-1640 until required post inoculation (hpi).
2.4. Construction of recombinant expression plasmid
N gene of TGEV was amplified with primers F-TGEV-N (5′-CAGGATCCGCCAACCAGGGACAACGT-3′) and R-TGEV-N 5′-CACTCGAGGTTCGTTACCTCATCAATCA-3′) containing Bam HI and Xho I enzyme sites. PCR products were subcloned into a prokaryotic expression pGEX-6p-1 vector (GE Healthcare). Recombinant expression plasmid was designated as pGEX-TGEV-N and confirmed by DNA sequencing.
2.5. GST pull-down assay
GST-N protein was expressed in Escherichia coli BL21 (DE3) under induction of 1 mM isopropyl-β-d-thiogalactopyranoside. GST-N fusion protein was immobilized on beads at 4 °C for 2 h. The lysate of ST cells was prepared using 1 mL RIPA lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) containing a protease inhibitor phenylmethanesulfonyl fluoride (PMSF; 1 mM). After centrifugation at 12,000 × g for 15 min, cell lysate (500 μg) was incubated with the GST-N protein preparation at 4 °C overnight. After washing four times with buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.05% NP-40), the isolated pull-down proteins were then analyzed by 12% PAGE analysis. Expressed GST protein was used as a control.
2.6. Co-immunoprecipitation (Co-IP) assay
The lysate of ST cells infected with TGEV for 24 h was prepared with RIPA lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% deoxycholate) containing a protease inhibitor phenylmethanesulfonyl fluoride (PMSF) (1 mM). After centrifugation at 12,000 × g for 15 min, lysate supernatant was pretreated with protein A/G plus-agarose (Beyotime) for 30 min at 4 °C to eliminate non-specific binding to the agarose beads. The lysate supernatant (500 μg) was incubated with 1 μg of rabbit pAb to EF1A for overnight at 4 °C. Then, 20 μL resuspended Protein A/G PLUS-Agarose was added to this mixture and incubated at 4 °C on a rocker platform for 2 h. After washing four times with lysis buffer, the isolated immunoprecipitated proteins were then analyzed by western blotting using mAb to N protein of TGEV and rabbit pAb to EF1A. The lysate of TGEV mock-infected ST cells was used as a control.
2.7. Western blotting
Equivalent amounts of cell lysates were subjected to 12% PAGE and then transferred to 0.22 μm nitrocellulose membranes (Hybond-C Extra, Amersham Biosciences). After blotting, the membranes were incubated with rabbit pAb to EF1A for 1 h. After washing three times with PBST, the membranes were inoculated with HRP-conjugated goat anti-rabbit IgG (Sigma) at 37 °C for 1 h and visualized using 3,3′,5,5′-tetramethylbenzidine-stabilized substrate (TMB, Amresco).
2.8. Immunofluorescence assay
ST cells inoculated with TGEV were cultured for 24 h. The cells were washed twice with PBS and fixed with paraformaldehyde (4%) for 30 min at 4 °C, and then allowed to air dry. After blotting with 5% skimmed milk powder, the fixed cells were incubated with mAb to TGEV N protein (1:100) and rabbit pAb to EF1A (1:50) for 1 h at 37 °C in a humidified chamber. After washing three times with PBST, the fixed cells were incubated with FITC-labeled goat anti-mouse IgG (1:100, KPL) and TRITC-labeled goat anti-rabbit IgG (1:200, Sigma). The additional nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI, Sigma) was performed as described previously (Jungmann et al., 2001). The triple-stained cells were washed three times with PBST and subsequently examined under a Leica TCS SP5 laser confocal microscopy.
2.9. Transfection of siRNA against EF1A
siRNA against EF1A (GenePharma) was used for transfection. The sequence of the siRNA strands was as follows: 5′-GUGGUAUUACCAUUGACAUTT-3′ (sense) and 5′-AUGUCAAUGGUAAUAACCACTT-3′ (antisense). Transfection with siRNA was performed with X-tremeGENE siRNA reagent (Roche) by following the manufacturer's instructions. ST cells were cultured overnight in six-well tissue culture plates. The siRNA (20 nM) was complexed with X-tremeGENE siRNA reagent by incubating together at room temperature for 30 min. After removing the cell culture supernatant, the complex was added for incubation 36 h.
2.10. Virus titer assay
ST cells were re-plated 1 day before infection in 96 well plates for the 50% infectious dose (TCID50) assays. Treated samples and their paired controls were thawed as described and immediately serially diluted. Cell cultures were then infected for 1 h. After 48 h of incubation, CPE was observed. TCID50 is calculated using the method of Reed and Munch. Virus titer assay were performed three times for each condition and were performed using the Student's t-test.
3. Results
3.1. Expression and purification of TGEV N protein
Full-length TGEV N protein with a GST tag was expressed in E. coli BL21 (DE3) using a T7 polymerase expression system. GST-N protein was successfully expressed and purified in BL21 (DE3) in soluble fractions (Fig. 1 ). Western blot analysis for detection of the GST tag confirmed expression of an ∼70-kDa recombinant GST-N protein (Fig. 1). Purified full-length recombinant GST-N protein was used in subsequent experiments.
Fig. 1.
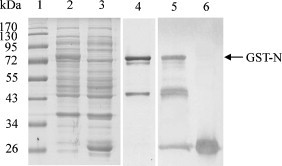
Expression and purification of TGEV GST-N protein. TGEV N protein was expressed in E. coli, and lysates were resolved and purified by 12% PAGE. Proteins were visualized by PhastGel Blue R staining (lanes 1–4) or N protein was detected by western blotting with a GST mAb (lanes 5 and 6). Lane 1, protein molecular weight marker; lane 2, induced culture of E. coli transformed with pGEX-TGEV-N; lanes 4 and 5, recombinant N protein purified by GST agarose; lanes 3 and 6, induced culture of E. coli transformed with pGEX-6p-1.
3.2. EF1A interacting with N protein in vitro
The expressed GST-N protein immobilized on GST-agarose beads was used as a bait to pull down cellular proteins of ST cells that form a complex with N protein. GST protein was used as control to eliminate non-specifically binding proteins. Cellular proteins immobilized on GST-agarose beads in GST pull-down assay were examined with specific antibodies to EF1A (Fig. 2 ). From the GST pull-down results, we can see that the EF1A protein was found in GST-N protein immobilized beads but not in GST protein immobilized beads.
Fig. 2.
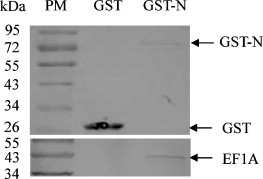
Cellular EF1A interacts with N protein of TGEV in vitro. EF1A binding to GST-N agarose or to GST protein were resolved by Wb. PM, protein marker. GST-N and GST proteins were visualized using mAb to GST; Cellular EF1A of ST cells was visualized using pAb to EF1A.
3.3. Cellular EF1A interacts with N protein of TGEV in virus-infected cells
The immunoprecipitation assay was utilized to elucidate further whether TGEV N protein interacted with cellular EF1A in TGEV-infected ST cells. From the immunoprecipitation results (Fig. 3 ), we can see that the N protein of TGEV was precipitated by the antibody to cellular EF1A in TGEV-infected ST cells but not in mock-infected ST cells. Furthermore, the same results were obtained with TGEV infectious strain or with TGEV attenuated strain (Fig. 3). These results demonstrated that the cellular EF1A interacted with the N protein of TGEV.
Fig. 3.
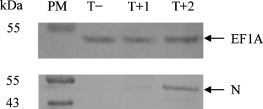
Cellular EF1A interacts with N protein of TGEV in virus-infected cells. N protein of TGEV was precipitated by mAb to cellular EF1A in TGEV-infected ST cells but not in mock-infected ST cells. T+ and T− represent the TGEV infected and uninfected ST cells, respectively. T + 1 represent the TGEV infectious H strain. T + 2 represent the TGEV attenuated H strain.
3.4. Co-localization of EF1A with N protein in TGEV infected cells
The subcellular localization of EF1A was investigated in TGEV-infected ST cells using indirect immunofluorescence confocal microscopy. The results indicated that the subcellular localization of EF1A was distributed in the cytoplasm after TGEV infection (Fig. 4 ). Furthermore, the red fluorescence of the TRITC-labeled goat anti-rabbit IgG binding with cellular EF1A was covered with the green fluorescence of the FITC-labeled goat anti-mouse IgG binding with N protein of TGEV. The evidence indicated that cellular EF1A was co-localized with N protein of TGEV within the ST cells during infection.
Fig. 4.
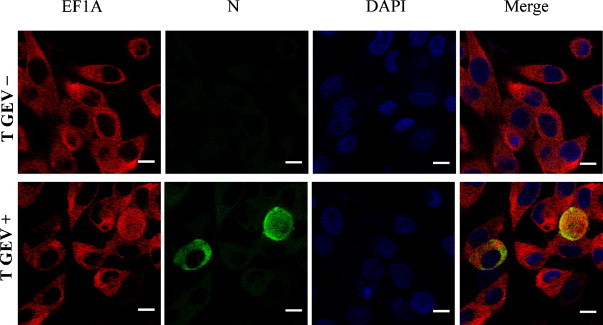
Localization of cellular EF1A and N protein of TGEV. Cells were infected with TGEV. Virus assembly sites were located using antibodies specific for the N protein (green). EF1A was visualized using antibodies specific for EF1A (red). The nucleus was stained with DAPI (blue). The triple-stained cells were observed by Leica TCS SP5 laser confocal microscopy. Bars, 10 μm.
3.5. Knockdown of EF1A impairs TGEV replication in host cell
To further investigate the role of EF1A in TGEV virus replication, EF1A protein of ST cells was inhibited using siRNA. TGEV attenuated virus was used for siRNA analysis. The transfected cells expressed lower protein levels of EF1A when compared with the control siRNA-transfected cells (Fig. 5A and B). ST cells were infected with TGEV for another 8 h or 24 h after transfection with siRNA at an MOI of 1. The viral RNA was measured by quantative real-time RT-PCR and the N protein of TGEV was measured by western blotting. TGEV infection was greatly reduced in the EF1A-knockdown cells, as shown by a reduction in viral N protein expression (Fig. 5A and B). To demonstrate the involvement of EF1A on TGEV replication, we quantified the amounts of cell-associated virus and virus releasing in culture supernatant at 8 h and 24 h after inoculation. Virus titer assay were performed three times for each condition and were performed using the Student's t-test. At 8 h inoculation, the specific numerical TCID50 of supernatant virus in control siRNA group was 103.1/mL and the EF1A siRNA group was 102.3/mL. The specific numerical TCID50 of cell-associated virus in control siRNA group was 103.9/mL and the EF1A siRNA group was 103.2/mL. At 24 h inoculation, the specific numerical TCID50 of supernatant virus in control siRNA group was 104.6/mL and the EF1A siRNA group was 103.7/mL. The specific numerical TCID50 of cell-associated virus in control siRNA group was 104.5/mL and the EF1A siRNA group was 103.6/mL. Fig. 5C shows that knock-down of EF1A resulted in significant reduction of cell-associated virus, which reflected viral replication.
Fig. 5.
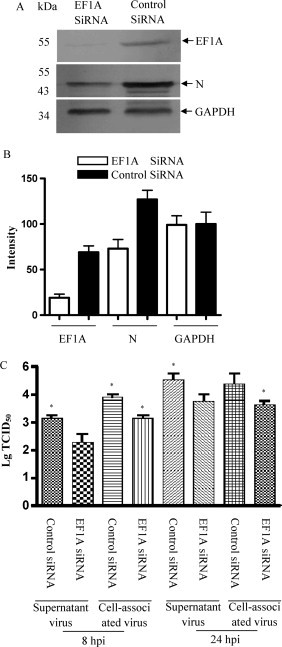
Gene silencing of EF1A reduced TGEV replication in ST cells. EF1A-knockdown cells and negative control knockdown cells were adsorbed with TGEV (MOI = 1) at 37 °C for 1 h. The cells were washed and further incubated with TGEV. The cell lysates were harvested for western blotting with antibodies against EF1A, TGEV N protein and GAPDH, as indicated (A). The averaged densitometric intensity of EF1A and N protein in immunoblot analysis, with GAPDH as a loading control (B). The culture supernatants of cells and the virus-associated cells infected with TGEV for 8 h and 24 h were collected for viral titration (C). The virus titers shown here are the averages and standard deviations of three independent samples. *p < 0.05.
4. Discussion
N protein of CoVs facilitates template switching and is required for efficient transcription (Schelle et al., 2005, Thiel et al., 2003, Zuniga et al., 2010). In addition, N protein displays pleiotropic effect when expressed in host cells, such as induction of apoptosis or cell-cycle arrest (He et al., 2003, Surjit et al., 2004). Some of the functional outcomes that result from N gene expression in host cells are due to direct or indirect interaction between N protein and cellular proteins. Studying the interaction of cellular protein with N protein will provide new information for understanding the mechanism of TGEV infection.
Results from previous study demonstrate that EF1A can anchor mRNA, suggesting that EF1A is involved in sorting and regulating the expression of specific cellular mRNAs (Bassell et al., 1994). EF1 complex is composed of four different subunits, alpha, beta, gamma, and delta (2:1:1:1) in mammalian cells (Carvalho et al., 1984a). In TGEV infected cells, EF1A may interact with N protein of TGEV that bind to TGEV RNA in a similar manner. Instead of an enzymatic activity, EF1A may provide protein–RNA and protein–protein interactions that promote the assembly of TGEV replication complexes. In this study, the results demonstrate that EF1A can interact with N protein of TGEV. It is possible that EF1A is involved in targeting TGEV N onto intracellular membranes that provide a microenvironment for the efficient replication of the viral RNA. From Fig. 3, we can see that the attenuated strain of TGEV N protein appears to be pulled down much more with EF1A than the wild type N protein in. The reason maybe that attenuated TGEV virus was better to adapt ST cells than infectious TGEV (data not shown). We assumed that the interaction of EF1A and N protein maybe play a role in cell culture adaptation.
The intrinsic characteristics of EF1A make it a suitable host protein for RNA virus replication. Results in this study support a role for EF1A in the replication of CoVs. In host cells, EF1A is found in high concentrations, approximately 1% of the total protein in animal cells (Condeelis, 1995) and 5% in plant cells (Browning et al., 1990). For viral replication, the abundance of EF1A would make it unnecessary to compete with cellular processes. CoVs replicate in many hosts (Enjuanes et al., 2006). It is likely that host factors selected for virus replication would be both structurally and functionally conserved across different species. EF1A affords an excellent model system for the further analysis of host protein and virus interactions.
EF1A play an important role in some virus infection. Several viral proteins have been observed to bind to EF1A. The NS5A protein of bovine viral diarrhea virus (BVDV) interacts with EF1A, which may play a role in the replication of BVDV (Johnson et al., 2001). The nucleocapsid protein of SARS-CoV interacted with EF1A and inhibited cell proliferation (Zhou et al., 2008). The RNA-dependent RNA polymerase of Turnip mosaic virus (TuMv) interacts with EF1A in virus-induced vesicles (Thivierge et al., 2008). RNA polymerase of vesicular stomatitis virus (VSV) specifically associates with EF1A (Das et al., 1998). Gag polyprotein of human immunodeficiency virus type 1 (HIV-1) interacts with EF1A requires tRNA, and EF1A may contribute to tRNA incorporation into HIV-1 virions (Cimarelli and Luban, 1999). Studying the mechanism of EF1A and N protein of TGEV will help to understand the pathogenesis of CoVs.
5. Conclusions
In summary, EF1A interaction with N protein of TGEV was found. The efficiency of TGEV replication depends on the presence of EF1A, which may facilitate virus replication. EF1A may promote viral replication by interaction with N protein. The present study thus provides information that should be useful for underlying mechanism of coronavirus replication.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31172350 and 31101823), Heilongjiang Provincial Natural Science Foundation (Grant No. JC201118) and Research Team Program on Scientific and Technological Innovation in Heilongjiang Provincial University (Grant No. 2011TD001).
References
- Almazan F., Galan C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell G.J., Powers C.M., Taneja K.L., Singer R.H. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol. 1994;126:863–876. doi: 10.1083/jcb.126.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning K.S., Humphreys J., Hobbs W., Smith G.B., Ravel J.M. Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J. Biol. Chem. 1990;265:17967–17973. [PubMed] [Google Scholar]
- Carvalho J.F., Carvalho M.D., Merrick W.C. Purification of various forms of elongation factor 1 from rabbit reticulocytes. Arch. Biochem. Biophys. 1984;234:591–602. doi: 10.1016/0003-9861(84)90309-6. [DOI] [PubMed] [Google Scholar]
- Carvalho M.D., Carvalho J.F., Merrick W.C. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch. Biochem. Biophys. 1984;234:603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- Chuang S.M., Chen L., Lambertson D., Anand M., Kinzy T.G., Madura K. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol. Cell. Biol. 2005;25:403–413. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A., Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1999;73:5388–5401. doi: 10.1128/jvi.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem. Sci. 1995;20:169–170. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- Das T., Mathur M., Gupta A.K., Janssen G.M., Banerjee A.K. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1449–1454. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.G., Baric R.S., Enjuanes L., Gorbalenya A.E. Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego: 2011. pp. 774–796. [Google Scholar]
- Ding L., Xu X., Huang Y., Li Z., Zhang K., Chen G., Yu G., Wang Z., Li W., Tong D. Transmissible gastroenteritis virus infection induces apoptosis through FasL- and mitochondria-mediated pathways. Vet. Microbiol. 2012;158:12–22. doi: 10.1016/j.vetmic.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Almazan F., Sola I., Zuniga S. Biochemical aspects of coronavirus replication and virus–host interaction. Annu. Rev. Microbiol. 2006;60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- Escors D., Camafeita E., Ortego J., Laude H., Enjuanes L. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. J. Virol. 2001;75:12228–12240. doi: 10.1128/JVI.75.24.12228-12240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen H., Smith C.E., Siegel N.R., Kahana C., Merrick W.C., Chakraburtty K., Schwartz A.L., Ciechanover A. Protein synthesis elongation factor EF-1 alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7648–7652. doi: 10.1073/pnas.91.16.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Andonov A., Li Y., Bastien N., Cao J., Osiowy C., Dobie F., Cutts T., Ballantine M., Li X. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotokezaka Y., Tobben U., Hotokezaka H., Van Leyen K., Beatrix B., Smith D.H., Nakamura T., Wiedmann M. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J. Biol. Chem. 2002;277:18545–18551. doi: 10.1074/jbc.M201022200. [DOI] [PubMed] [Google Scholar]
- Johnson C.M., Perez D.R., French R., Merrick W.C., Donis R.O. The NS5A protein of bovine viral diarrhoea virus interacts with the alpha subunit of translation elongation factor-1. J. Gen. Virol. 2001;82:2935–2943. doi: 10.1099/0022-1317-82-12-2935. [DOI] [PubMed] [Google Scholar]
- Jungmann A., Nieper H., Muller H. Apoptosis is induced by infectious bursal disease virus replication in productively infected cells as well as in antigen-negative cells in their vicinity. J. Gen. Virol. 2001;82:1107–1115. doi: 10.1099/0022-1317-82-5-1107. [DOI] [PubMed] [Google Scholar]
- Kim B., Chae C. In situ hybridization for the detection of transmissible gastroenteritis virus in pigs and comparison with other methods. Can. J. Vet. Res. 2001;65:33–37. [PMC free article] [PubMed] [Google Scholar]
- Lamberti A., Caraglia M., Longo O., Marra M., Abbruzzese A., Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids. 2004;26:443–448. doi: 10.1007/s00726-004-0088-2. [DOI] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldave K. Eukaryotic protein synthesis. Annu. Rev. Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- Penzes Z., Gonzalez J.M., Calvo E., Izeta A., Smerdou C., Mendez A., Sanchez C.M., Sola I., Almazan F., Enjuanes L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the purdue virus cluster. Virus Genes. 2001;23:105–118. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J., Santiago C., Mudgal G., Ordono D., Enjuanes L., Casasnovas J.M. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Anton I.M., Enjuanes L., Carrascosa J.L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestak K., Lanza I., Park S.K., Weilnau P.A., Saif L.J. Contribution of passive immunity to porcine respiratory coronavirus to protection against transmissible gastroenteritis virus challenge exposure in suckling pigs. Am. J. Vet. Res. 1996;57:664–671. [PubMed] [Google Scholar]
- Stohlman S.A., Baric R.S., Nelson G.N., Soe L.H., Welter L.M., Deans R.J. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J. Virol. 1988;62:4288–4295. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L.S., Holmes K.V., Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Karl N., Schelle B., Disterer P., Klagge I., Siddell S.G. Multigene RNA vector based on coronavirus transcription. J. Virol. 2003;77:9790–9798. doi: 10.1128/JVI.77.18.9790-9798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thivierge K., Cotton S., Dufresne P.J., Mathieu I., Beauchemin C., Ide C., Fortin M.G., Laliberte J.F. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology. 2008;377:216–225. doi: 10.1016/j.virol.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Wang C., Chen J., Shi H., Qiu H., Xue F., Liu C., Zhu Y., Liu S., Almazan F., Enjuanes L., Feng L. Molecular characterization of a Chinese vaccine strain of transmissible gastroenteritis virus: mutations that may contribute to attenuation. Virus Genes. 2010;40:403–409. doi: 10.1007/s11262-010-0467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Burwinkel M., Palissa C., Ephraim E., Schmidt M.F. Antiviral activity of zinc salts against transmissible gastroenteritis virus in vitro. Vet. Microbiol. 2012;160:468–472. doi: 10.1016/j.vetmic.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Liu J., Wang Q., Liu X., Li X., Li P., Ma Q., Cao C. The nucleocapsid protein of severe acute respiratory syndrome coronavirus inhibits cell cytokinesis and proliferation by interacting with translation elongation factor 1 alpha. J. Virol. 2008;82:6962–6971. doi: 10.1128/JVI.00133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Cruz J.L., Sola I., Mateos-Gomez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


