Abstract
Evidence accumulated over the past decade provides support for liquid-liquid phase separation as the mechanism underlying the formation of biomolecular condensates, which include not only “membraneless” organelles such as nucleoli and RNA granules, but additional assemblies involved in transcription, translation and signaling. Understanding the molecular mechanisms of condensate function requires knowledge of the structures of their constituents. Current knowledge suggests that structures formed via multivalent domain-motif interactions remain largely unchanged within condensates. Two different viewpoints exist regarding structures of disordered low-complexity domains within condensates; one argues that low-complexity domains remain largely disordered in condensates and their multivalency is encoded in short motifs called “stickers”, while the other argues that the sequences form cross-β structures resembling amyloid fibrils. We review these viewpoints and highlight outstanding questions that will inform structure-function relationships for biomolecular condensates.
Keywords: liquid-liquid phase separation, demixing, low-complexity domain, multivalent interactions, intrinsically disordered protein region, amyloid-like fibril, super-molecular structure
Graphical Abstract
Introduction
Liquid-liquid phase separation (LLPS) mediates many fundamental biological processes from cell signaling [1] to RNA metabolism [2], stress adaption [3] and transcription [4]. LLPS is rapidly becoming accepted as a key mechanism underlying the formation of biomolecular condensates [5,6], which include typical membraneless organelles such as nucleoli, nuclear speckles and stress granules but also less traditional compartments including heterochromatin [7,8], super-enhancers [4,9], the centrosomes [10], the pre- [11,12] and post-synaptic densities [13] and membrane receptor clusters [1,14,15]. Dysregulation of the (dis-)assembly of biomolecular condensates has been linked to cancer [16,17], neurodegenerative diseases [18,19] and aging [20]. While these observations suggest that phase separation is critically important for function, relatively few examples exist in which the functional requirement for phase separation has been firmly demonstrated. Contributing to this is a lack of understanding of molecular mechanisms mediated by LLPS and our current dearth in knowledge of the molecular structures within liquid dense phases.
LLPS is mediated by multivalent interactions of biomolecules [21–23] and characterized by a density transition and release of solvent [24] above the so-called saturation concentration. This gives rise to two coexisting phases, the dilute and the dense phase. Both phases have a system-specific concentration, which is independent of the total biomolecular concentration in the sample; an increase in the total concentration results in an increase in the volume fraction of the dense phase, at the expense of the volume fraction of the dilute phase, while both dense and dilute concentrations remain constant.
The multivalent interactions that mediate LLPS can be mediated by folded domains that are connected by disordered linkers, or they can be mediated by favorably interacting, so-called “sticky”, residues or motifs within intrinsically disordered regions (IDRs) [25]. Both instantiations of multivalence can be conceptualized by the stickers-and-spacers framework, which has been used to describe the behavior of associative polymers [26] and has been adapted for proteins [27]. In this framework, stickers interact favorably with each other, whereas spacers neither interact with stickers nor with other spacers; their solvation properties, however, are important for determining whether crosslinking of the biomolecules is coupled to a density transition; multivalence without a density transition can lead to the formation of system-spanning networks of interacting molecules, i.e. gels, but does not result in the coexistence of dilute and dense phases [24]. The sticker valence, i.e. the number of interacting motifs within a protein, is anticorrelated with the saturation concentration, i.e. the higher the valence in a given system, the lower is its saturation concentration [21,27]. While many of these concepts directly connect with polymer theories and are useful to uncover general driving forces for phase separation, a complete understanding of the structural features of phase separation-mediating interactions and of the super-molecular structures within dense phases is required to fully appreciate the molecular function of biomolecular condensates. Here we review the current state of knowledge regarding biomolecular structures within condensates. We first review the structural properties of domain-motif interactions and then discuss the structures of intrinsically disordered regions within condensates while addressing the corresponding studies that have provided seemingly divergent yet important insights. We then touch upon the role of RNA structure in condensate formation and close with critical outstanding questions whose answers connect the structural and dynamical features of condensates with their emergent material properties and functions.
Multivalent domain/motif interactions
In many phase-separating proteins that have thus far been characterized, repeats of folded domains in one protein interact with repeats of linear motifs in a binding partner via multivalent interactions (Figure 1). Successive folded domains that are connected by linkers can be present in a single polypeptide chain, as in the classical examples first described by Rosen and colleagues [21]. For example, the protein Nck contains repeats of SH3 domains that interact with multiple proline-rich motifs (PRMs) in the protein N-WASP, and the protein PTB contains multiple RNA-binding domains and interacts with RNA, which is itself a multivalent molecule. The multivalence afforded by multiple folded interaction domains can also be obtained via oligomerization into discrete oligomeric species, as is the case in the nucleolar protein nucleophosmin (NPM1) [28] or by linear polymerization into higher-order structures, as in the case of the ubiquitin ligase adaptor Speckle-type POZ protein (SPOP) [16] and the RNA-binding protein TDP-43 [29]. In all of these examples, irrespective of the architecture that achieves multivalence, the proteins interact with binding partners that are themselves multivalent for binding to the interaction domains. These binding partners typically contain linear motifs and can be proteins or nucleic acids. However, the precise architecture determines the super-molecular structures that make up the dense phase and therefore determines its properties. SPOP oligomers bind multiple DAXX molecules, which results in brush-like structures which phase separate via DAXX-DAXX interactions [16,30]. This is in contrast to networks of SH3 domains that interact with PRMs, wherein single PRM chains can cross-link SH3 domains on different molecules directly (Figure 1).
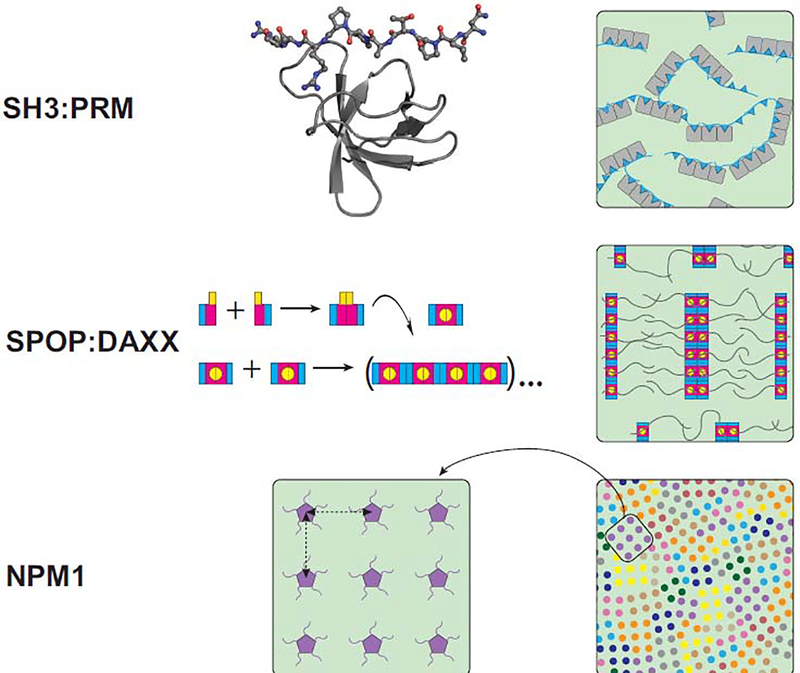
Figure 1. Domain architecture determines super-molecular structure in dense phases.
(Top row) Insight into dense phase structures via high-resolution structures of domain/motif interactions. (Left) A high resolution crystal structure of an SH3 domain bound to a proline-rich motif (PRM) peptide. (PDB: 4WCI) [75]. (Right) A cartoon representation of a dense phase (green background) generated by multivalent SH3:PRM interactions. Three SH3 domains are connected by linkers in a single molecule (gray); a binding partner containing three proline-rich motifs (cyan) interacts with the SH3-protein via multivalent interactions. The two protein types crosslink each other. (Middle row) Multivalency via linear polymerization. (Left) SPOP monomers dimerize and the dimers polymerize into linear structures. The BACK dimerization domain is shown in cyan, the BTB dimerization domain in magenta, and the MATH substrate binding domain in yellow. The curved arrow indicates a 90° rotation of a SPOP dimer onto its side. (Right) SPOP oligomers bind several DAXX molecules via their MATH domains, giving rise to “brush-like” structures. These brushes are crosslinked via DAXX interactions [16,30]. (Bottom row) Local order in a liquid dense phase. The distances between NPM1 pentamers (magenta) repeat (left), giving rise to locally ordered arrays of Npm1 molecules within the dense phase. Npm1 molecules are crosslinked by arginine-rich peptides (not shown). (Right) Over greater distances, the anisotropic arrangement of arrays (each shown in different colors) gives rise to global disorder and liquid-like behavior [28].
The domain-motif interactions in these systems are modular in nature and typically well understood, given that high-resolution structures of individual complexes are often available (Figure 1). The identical structures, strung together by disordered linkers, are also assumed to give rise to the higher-order complexes formed in the multivalent case. It is these higher-order complexes that undergo phase separation due to their reduced solubility compared to individual protein molecules and small complexes. The remaining dilute phase also contains higher-order complexes that coexist with the dense phase [21], and their sizes depend on the effective affinities between the binding partners and the saturation concentration of the system.
The assumption that the structures of building blocks are identical in the dilute and dense phases may be incorrect due to steric constraints imposed by their multivalence, or if the stability of the domains/complexes or their structures are affected by the altered solvation properties of the dense phase. Further, the stability and structure may be influenced by differential partitioning of ions and biomolecules between the light and dense phases. In fact, single molecule FRET experiments demonstrated the presence of distinct NPM1 conformations populating the light versus dense phases. In dilute solution, the A2 tract, a disordered region in NPM1, forms a compact conformation stabilized by electrostatic interactions; in the dense phase the A2 tract is expanded as a consequence of interactions with positively charged nucleolar proteins [28].
Information pertaining to the super-molecular structure of these complexes within dense phases is sparse such that the relative orientations and distances of individual domain/motif complexes in dense phases are largely unknown. The only available information stems from SANS data of the dense phase formed by a truncated form of NPM1 and an arginine-rich, nucleolar-derived peptide [28]. The SANS data shows diffraction peaks, thus implying regularly repeating distances in the dense phase, which can be interpreted as distances between neighboring domains or NPM1 oligomers. This implies a regularly repeating pattern of the NPM1 molecules and at least local order in the dense phase, even if the dense phases are disordered on longer length scales (Figure 1).
It is intuitive to expect that the super-molecular structures within dense phases are dependent on system-specific properties such as linker lengths and sequences. A full description of the structures in the dense phase, the lifetime of individual interactions, and how the dynamics of one building block is correlated with that of its neighbors would open the door to a molecular understanding of the emergent material properties of dense phases. We expect that an integrative approach utilizing solution- [28,31] and solid-state NMR spectroscopy, scattering and single molecule fluorescence techniques [32] together with molecular simulations [33,34] will facilitate our understanding of how behavior at the molecular level correlates with the emergent material properties of condensates. Cryo-EM tomography [3] may play a particularly important role as it will allow bridging between structural biology of condensates in vitro and in cells.
Low complexity domains mediate phase separation
An analysis of the components of membraneless organelles has shown an enrichment of proteins containing intrinsically disordered low complexity domains (LCDs) [21,35,36]. The composition of these sequences is biased and typically enriched with few select amino acids and the chains fail to fold into well-ordered three-dimensional structures. LCDs mediate the phase separation of many proteins and are often necessary and sufficient for phase separation [37–39]. Given these observations, it is likely that LCDs play important roles in the formation of biomolecular condensates and tune their material properties. Among the LCDs used as models for phase separation are Fused in Sarcoma (FUS) and the RNA-binding proteins hnRNPA1, hnRNPA2 and TDP-43. Their LCDs are termed prion-like domains or PrLD due to their compositional similarity to yeast prion proteins; they are enriched in asparagine, glutamine, tyrosine and glycine residues [40]. Two views regarding structures of LCDs within condensates have emerged. One side argues that regions within the LCD fold into a β-sheet structure and that many such structures assemble into amyloid-like fibrils with a cross-β architecture. The opposing view maintains that LCDs remain largely disordered within the dense liquid phase (Figure 2).
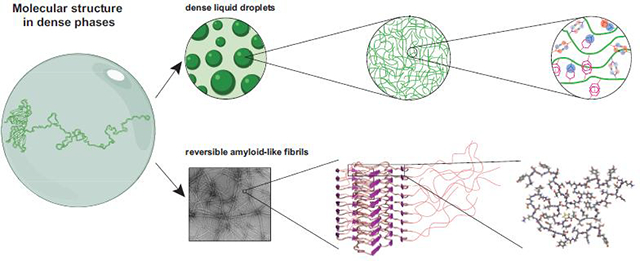
Figure 2. LCDs can form different types of assemblies.
(a) LLPS of intrinsically disordered LCDs. From left to right: (1) A sample with a large dense drop at the bottom of the tube (dark green) overlaid by the dilute phase (light green) is prepared from (2) many small micron-sized droplets (dark green) that fuse. (3) The protein chains remain largely disordered in the dense phase (green mesh). Interactions driving phase separation include hydrophobic, π-π interactions (e.g. between aromatic residues shown in magenta stick representation), cation-π interactions between aromatics (magenta sticks) and positively charged residues (blue sticks), polar interactions (violet sticks), electrostatic interactions between positive (blue sticks) and negative (red sticks) charges, and hydrogen bonding (orange sticks). (b) Hydrogels of LCDs. From left to right: (1) Many low complexity domains such as that of FUS can assemble into hydrogels. (2) The FUS hydrogel is composed of amyloid-like fibrils. (3) Solid-state NMR structure of the core region of FUS that assembles into long filaments with a cross-β architecture. The remaining 77% of the LCD sequence remains disordered, indicated as red lines. (4) The atomic structure of a FUS monomer within a fibril reveals short in-register β-sheets separated by loops. Of note is the absence of all but one hydrophobic residue (yellow). (c) Irreversible amyloid fibrils formed by typical amyloidogenic proteins, not LCDs. (1) Cartoon depiction of irreversible fibrils sedimented in an Eppendorf tube. (2) TEM of α-synuclein fibrils. (3) Structure of the components of the fibrils indicates a long, continuous β-sheet core flanked by disordered regions. (4) Atomic structure of the core of an α-synuclein monomer within a fibril shows an in-register β-sheet stabilized by hydrophobic contacts involving Val, Ile, Ala and Phe residues (yellow). (Some illustrations were made in BioRender (biorender.com).)
Cross-β structure of LCDs in fibrils and hydrogels
A number of reports have shown that LCDs from RNA-binding proteins and similar sequences can generally form hydrogels, which are composed of amyloid-like fibrils [21,41–44]. Characterization of fibrils showed that, despite the presence of cross-β architecture, these structures are distinct from conventional amyloid fibrils such as those of α-synuclein and Aβ [45–50] (Figure 2). Fibril formation of LCDs is often reversible and fibrils dissolve upon dilution, at high temperature or upon treatment with detergents [21,41,51]. Compared to α-synuclein and Aβ, the sequences are enriched in polar amino acids while lacking hydrophobic residues. A combination of solid- and liquid-state NMR spectroscopy were used to elucidate the structure of FUS fibrils; they consist of a β-sheet containing core of 57 residues (23% of the total sequence) within the N-terminal half, while the remainder of the chain (77%) remains disordered [41]. The core region lacks a hydrophobic interface and contains residues that can hydrogen bond to one another.
Recently, short motifs were identified within the LCDs of RNA-binding proteins that can mediate fibrillization and they were termed LARKS (low-complexity aromatic-rich kinked segments) [42,52]. Powder diffraction showed that when isolated as peptides, LARKS formed cross-β structure that was characterized by x-ray crystallography and micro-Electron Diffraction at atomic resolution. The structures are stabilized by aromatic stacking and hydrogen bonds. Kinks within the structures not only allow for close approach of backbones and favorable hydrogen bonding and van der Waals interactions between sheets, but also prevent the sidechains from interdigitating across the β-sheet interface so that less surface area is buried when compared to conventional amyloid fibrils. These structures may thus explain the reversibility of fibrillization and their properties have been rationalized to resemble the transient interactions and high mobility of protein molecules in RNA granules revealed by fast fluorescence recovery after photobleaching (FRAP) [37]. An analysis of the human proteome suggested that LARKS are enriched in hundreds of proteins in biomolecular condensates, the nuclear pore complex and in the extracellular matrix [42], suggesting that the LARK fibril structures may represent relevant modes of interactions in these mesoscale structures in cells. In FUS, LARKS are distributed throughout the sequence and not only in the β-sheet containing core of the fibrils characterized by solid-state NMR. [41,42].
Complementary work has identified additional motifs within the LCD of hnRNPA1 and FUS that form reversible fibrils with cross-β structure; these motifs were termed hnRACS (reversible amyloid cores) [43,44].The core of the fibril consists of Gln residues that form intersheet hydrogen bonds. An FG motif on one side of the core creates a kink that allows the aromatic residue to reach out and engage in a network of interactions, which is required for gelling, i.e. for the branching of fibrils and their arrangement as three-dimensional networks. The reversibility and instability of the fibrils seem to result from stacking of negatively charged Asp residues along the fibril axis; in accordance with this, replacement of Asp with Val abolishes the reversibility of fibrillization and may explain common disease mutations [44,53]. However, the authors observed both reversible and irreversible fibrils that are formed from the same sequence. Future studies should address how reversible fibrils that lack a hydrophobic interface convert over time into irreversible fibrils [44].
A series of reports has presented compelling evidence that features required to form fibril structures are also functionally important inside cells [41,54,55]. Many LCDs of RNA-binding proteins that are associated with stress granules have affinities for FUS hydrogels [21]. Importantly, sequence features that mediate the ability to form fibrils/hydrogels and mediate binding of monomers to hydrogels are also functionally important. This includes the tyrosine residues punctuating the FUS LCD sequence and the methionine residues punctuating the LCD of Pbp1, the yeast ortholog of Ataxin-2. The tyrosine residues in FUS are not only required for hydrogel formation but for transcriptional activity [55] and the methionine residues for redox sensitivity of Pbp1 condensates [54]. This represents a classical biochemical validation of these contacts. Based on these observations, it was postulated that the fibril-containing hydrogel bears similarities to cellular RNA granules. Phosphorylation of residues within the β-sheet core of FUS significantly disrupt hydrogel binding and causes droplet dissolution, further arguing for the importance of the structured core for incorporation into hydrogels and droplets [41]. Further, Li, Liu and colleagues demonstrated that removal of hnRACs from the LCD of hnRNPA1 increases the saturation concentration (csat) while addition of an hnRAC to the sequence reduces csat, suggesting a causative role for the motifs in LLPS [44].
The caveat of studies involving structural characterization of fibrils using solid-state methods is that they do not probe LCDs within the liquid environment of condensed droplets. If these fibrillar structures are in fact present within droplets, the protein fraction in droplets adopting these interactions is often low and, if maturation processes transpire, may grow over time [56]. Conversely, it has been argued that the fibrils offer an opportunity to trap pairwise interactions that are more transient in droplets. However, the interpretation of the fibril structure results are complicated by the fact that these β-sheet structures contain aromatic residues that have themselves been shown to drive LLPS [27,57] possibly without mediating extensive structuring. Additional work is needed to disentangle the importance of sequence features of LARKS and hnRACs vs the requirement of the structural motifs for function.
Whether the formation of fibrillar structures is required for LLPS or a consequence of the process thus remains an open question. Even if the fibrils emerge from liquid dense phases rather than mediate their formation, it is nonetheless crucial to characterize them structurally as they may provide insights into the conversion of liquid-like droplets into solid assemblies which have been implicated in numerous pathologies [37,58–61].
Structures of low complexity domains studied in dense liquid phases
A second set of reports has suggested that LCDs within condensed droplets remain entirely disordered and dynamic [62–65] (Figure 2). These conclusions stem from solution NMR spectroscopy on pure dense phases. In these studies, phase separation is induced in a large protein sample and all droplets are fused into one large drop by centrifugation or gravity.
The narrow chemical shift dispersion in the NMR spectra of these samples argue that the LCD in condensed liquid droplets remains disordered. No evidence for the formation of secondary structure is observed and relaxation experiments that probe the protein dynamics provide evidence for rapid local motions of the backbone and sidechains. Conversely, translational diffusion determined by pulse field gradient measurements is significantly slowed relative to that in the dilute phase [63,65], in line with observations from FRAP on individual microdroplets [38,62]. Comparably, NMR experiments performed on an elastin-like polypeptide showed that molecules within the dense phase exhibit disorder matching that observed in the dilute phase [66].
The slowed diffusion has been attributed to the high viscosity mediated by the high protein concentration within the dense phase (7–40 mM) [62–65] as well as the presence of weak multivalent interactions between the chains [63]. NOESY experiments that can selectively detect intermolecular interactions have detected close contacts between most residue types within the sequence of DDX4 and FUS [63,65]. Electrostatic and cation-π interactions seem to drive phase separation of DDX4 while hydrophobic, π-π and sp2/π interactions and hydrogen bonding likely occur in the condensed phase of FUS. The residue types mediating these interactions may therefore represent stickers in a disordered chain. Extensive NOE networks involving Gln residues were also observed in FUS; mutation of several di-Gln repeats across the sequence results in a reduced propensity for LLPS providing support for the driving force from these contacts for LLPS [65]. All-atom two-chain simulations suggested that hydrogen bonding between these residues may be important. Despite the lack of evidence for secondary structure, these hydrogen bonding patterns could be of a similar variety as those observed in the fibrillar structures, but they would have to be transiently sampled to result in agreement with the NMR results.
In addition to hydrogen boding, other interactions involving the same residues may be present simultaneously, such as sp2/π interactions between the sidechains of Gln residues, between Gln and Tyr residues and with the peptide backbone. Simulations also revealed the presence of hydrophobic and/or π-π interactions between Tyr residues [65]. π-π interactions between nonaromatic residues containing sp2 groups such as Gln, Asn and the backbone may be important for phase separation of LCDs [67]. These conclusions were reached by the analysis of the frequencies of such interactions in ordered structures and the high fraction of π-containing residue types in phase-separating proteins. The dense phase of an elastin-like polypeptide shows evidence for the presence of hydrophobic interactions involving alanine, valine and proline based on intermolecular NOEs [66]. Many types of contacts have been observed, and the biophysical nature of the contacts dominating phase separation is expected to differ by sequence features of the respective LCD [68,69].
Intermolecular PRE experiments, which probe contacts over longer distances, provide further support that interactions are distributed across the sequence of FUS and hnRNPA2 such that all parts of the chain interact with all others, with little preference for contacts between particular regions. Given that PRE effects are present at distances of up to ~35 Å from the spin label, and the density of disordered protein chains within droplets may very well be high enough to approach one another within this range [63–65], these observations leave room for the driving force for phase separation stemming from stickers and spacers arranged along the disordered protein chain.
To date, the presence of ß-sheet structure within purely liquid droplets has not been shown. To test for the presence of ordered structure such as fibrils within the dense phase, Fawzi and coworkers performed NMR studies that probe dynamics across different timescales ranging from sub-microseconds to seconds [65]. None of the methods provided evidence for the presence of ordered structure, even at a low population. Considering the possibility that such a state might be invisible due to dynamics on a timescale that could not be probed by NMR, they turned to Raman spectroscopy but did not see evidence for the presence of fibrils. One would expect that if fibrillar structures were required for LLPS, they would be present at a sufficient abundance to be detected using these methods.
In summary, the currently available experimental data lend themselves to two opposing views with respect to the structural features of contacts within dense LCD phases. They either involve kinked cross-beta structure or interactions between largely disordered motifs. It has been argued that the interactions mediating fibrils could form transiently and exclusively pairwise to give rise to liquid droplets. While evidence argues for the importance of LARK/hnRAC sequence features for function, the same sequence features have also been implicated in the formation of critical contacts in disordered dense phases. Thus, whether LCD fibrils have physiological relevance for explaining liquid condensate structure and what the conformations of interacting motifs are within dense liquids remains to be fully determined.
While biomolecular condensates in cells behave as liquid-like bodies and are in that sense similar to simple in vitro droplets, condensates are complex, multicomponent systems. Different biomolecular condensates have different material properties, on the spectrum from liquid to solid, and these material properties may also evolve over time [5,6,70–72]. Not all proteins from which these properties emerge may sample the same structures and interactions, and they may not be fully captured within single-component systems. Nonetheless, characterizing the vast array of material states encompassing liquids, gels and solids is an important starting point to understanding how biomolecular condensates function in cells. Additionally, studies of LCDs within full-length proteins will be important. Multivalence via modular interaction domains and LCDs often coincide in phase-separating proteins, and the respective modulation of their properties needs to be characterized for a better understanding of the driving forces for phase separation.
RNA structure in biomolecular condensates
RNA is an important constituent in many biomolecular condensates and contributes to phase separation via RNA/RNA and protein/RNA interactions. RNA structure determination is challenging in itself [73], and thus determining the structures of RNA in dense phases and of the super-molecular structures formed presents even larger challenges. Hence, little data currently exists on RNA structure in biomolecular condensates. However, there is intriguing evidence that RNA structure can determine the identity of dense phases. The protein Whi3, a polyQ-rich RNA-binding protein from the filamentous fungus Ashbya gossypii, forms distinct RNA granules with different mRNAs. Careful analysis has revealed that the differences in RNA structures results in the immiscibility of the biomolecular condensates, and that denaturation of the RNAs results in a single type of condensate consisting of Whi3 and both types of RNAs [74]. While the structural characterization of the RNA was lacking atomistic detail and was not performed in the dense phase, the gain in knowledge regarding the molecular origin of specificity has been enormous, promising exciting future insights into the structure and function of RNA in dense phases.
Structural biology meets phase separation – the future
The evidence for the ubiquitous nature of LLPS throughout biology has been mounting quickly over the last few years, as has the realization that dysregulation of phase separation may be causal to diseases or accelerate their progression. These findings demand the attention of the biophysics and structural biology communities, as both can contribute to the quantitation of full phase behaviors, material properties and atomistic and super-molecular structures of dense phases. From the structural perspective, we should be striving to answer the following questions (Figure 3):
Figure 3: (Super-)molecular structural properties that define dense phases.
Clockwise from the top: (1) What are the conformations adopted in phase separation-mediating contacts and what is the rearrangement of building blocks relative to each other (also see Figure 1)? (2) Do the stabilities of folded protein domains differ between the dilute and dense phase? And are the affinities between domains and motifs different between the phases? (3) What is the extent of ordering inside of droplets? (4) What are the lifetimes of individual interactions within the dense phase? (5) Over what distance within the dense phase is the movement of building blocks correlated? (6) How many cross-links form between molecules in the dense phase and what is the resulting mesh size? (7) Do the structures of molecules and their assemblies differ between the bulk dense phase and the phase boundary with the dilute phase?
What are the atomistic details and the biophysical nature of the interactions within LCDs that drive phase separation?
Do the stabilities of domains and affinities of domain/motif interactions differ between dilute and dense phases? And are the structures of modular binding domain/motif interactions identical between dilute and dense phases, or are conformations affected by condensation?
To which extent do motifs/stickers in disordered regions become ordered upon interaction in the dense phase?
What are the lifetimes of the interactions in dense phases?
How is the movement of molecules/building blocks in the dense phase correlated?
How many crosslinks do molecules form in the dense phase and what is the resulting mesh size?
Do the (super-)molecular structures differ between the bulk dense phase and the phase boundary with the dilute phase?
We should aim to tackle these questions in order to understand the molecular origin of material properties. They call for multi-pronged approaches that involve characterization of structure and dynamics on multiple length and time scales. The insight that LLPS plays fundamental roles in cell biology promises an understanding of cell biological processes from a solid biophysical, mechanistic basis. While the progress in our appreciation of the ubiquity of LLPS has been rapid, its characterization, with a few notable exceptions, has remained largely phenomenological. The opportunities for stringent quantitative biophysical and structural characterization are vast, and the potential gain in mechanistic understanding is as well. Such work will undoubtedly benefit from the ever-advancing tools of structural biology and keep the field engaged for years to come.
Highlights.
Liquid-liquid phase separation mediates extensive compartmentalization of cells.
Phase separation is mediated by multivalent interactions.
These encompass domain-motif interactions or interactions between stickers in low-complexity domains.
LCDs can form amyloid-like cross-β structures but stay largely disordered in dense phases.
Dense phase structures may explain the molecular mechanisms of condensate function.
Acknowledgements
T.M. acknowledges funding by NIH grant R01GM112846, St. Jude Children’s Research Hospital, and the American Lebanese Syrian Associated Charities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Case LB, Zhang X, Ditlev JA, Rosen MK: Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 2019, 363:1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H,Lee JH, Jaffrey SR: m(6)A enhances the phase separation potential of mRNA. Nature 2019, 571:424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nuske E, Richter D, Baumeister W, Grill SW, Pappu RV, et al. : Phase separation of a yeast prion protein promotes cellular fitness. Science 2018, 359. [DOI] [PubMed] [Google Scholar]
- 4.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. : Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175:1842–1855 e1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA: Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324:1729–1732. [DOI] [PubMed] [Google Scholar]
- 6.Brangwynne CP, Mitchison TJ, Hyman AA: Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 2011, 108:4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH: Phase separation drives heterochromatin domain formation. Nature 2017, 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ: Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 2017, 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. : Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA: The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 2017, 169:1066–1077 e1010. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Cai Q, Shen Z, Chen X, Zeng M, Du S, Zhang M: RIM and RIM-BP Form Presynaptic Active-Zone-like Condensates via Phase Separation. Mol Cell 2019, 73:971–984 e975. [DOI] [PubMed] [Google Scholar]
- 12.Milovanovic D, Wu Y, Bian X, De Camilli P: A liquid phase of synapsin and lipid vesicles. Science 2018, 361:604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng M, Chen X, Guan D, Xu J, Wu H, Tong P, Zhang M: Reconstituted Postsynaptic Density as a Molecular Platform for Understanding Synapse Formation and Plasticity. Cell 2018, 174:1172–1187 e1116. [DOI] [PubMed] [Google Scholar]
- 14.Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD: Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinkel F, Abraham L, Ko M, Chao J, Bach H, Hui LT, Li H, Zhu M, Ling YM, Rogalski JC, et al. : Phase separation and clustering of an ABC transporter in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2019, 10.1073/pnas.1820683116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR,Lindorff-Larsen K, Salvatella X, et al. : Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol Cell 2018, 72:19–36 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeynaems S, Tompa P, Van Den Bosch L: Phasing in on the cell cycle. Cell Div 2018, 13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nedelsky NB, Taylor JP: Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat Rev Neurol 2019, 15:272–286. [DOI] [PubMed] [Google Scholar]
- 19.Guo L, Fare CM, Shorter J: Therapeutic Dissolution of Aberrant Phases by Nuclear-Import Receptors. Trends Cell Biol 2019, 29:308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alberti S, Hyman AA: Are aberrant phase transitions a driver of cellular aging? Bioessays 2016, 38:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. : Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banani SF, Lee HO, Hyman AA, Rosen MK: Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017, 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentley EP, Frey BB, Deniz AA: Physical Chemistry of Cellular Liquid-Phase Separation. Chemistry 2019, 25:5600–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmon TS, Holehouse AS, Rosen MK, Pappu RV: Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mittag T, Parker R: Multiple Modes of Protein-Protein Interactions Promote RNP Granule Assembly. J Mol Biol 2018, 430:4636–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semenov AN, Rubinstein M: Thermoreversible gelation in solutions of associative polymers. 1. Statics. Macromolecules 1998, 31:1373–1385. [Google Scholar]
- 27.Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. : A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174:688–699 e616.* In this tour de force, the saturation concentration of a large set of RNA-binding proteins and of many sequence variants was determined, leading to the conclusion that interactions between tyrosine and arginine residues are the strongest determinant of the driving force for phase separation in this class of proteins.
- 28.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW: Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 2016, 5.** This singular study reports neutron scattering and single molecule FRET analyses of the dense phase of Nucleophosmin with an arginine-rich peptide, i.e. of a system mediating phase separation via domain/motif interactions. A difference in linker conformations between the dilute and dense phases is revealed, as well as the regularly repeating distances of building blocks in the dense phase.
- 29.Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, et al. : A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J 2018, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmit JD, Bouchard JJ, Martin EW, Mittag T: Protein network structure enables switching between liquid and gel states. bioRxiv 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fromm SA, Kamenz J, Noldeke ER, Neu A, Zocher G, Sprangers R: In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery. Angew Chem Int Ed Engl 2014, 53:7354–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasir I, Onuchic PL, Labra SR, Deniz AA: Single-molecule fluorescence studies of intrinsically disordered proteins and liquid phase separation. Biochim Biophys Acta Proteins Proteom 2019, 1867:980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fei J, Jadaliha M, Harmon TS, Li ITS, Hua B, Hao Q, Holehouse AS, Reyer M, Sun Q, Freier SM,et al. : Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci 2017, 130:4180–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP: Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165:1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. : Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 36.Uversky VN: Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 2017, 44:18–30. [DOI] [PubMed] [Google Scholar]
- 37.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP: Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. : Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 2015, 57:936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP: The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A 2015, 112:7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King OD, Gitler AD, Shorter J: The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 2012, 1462:61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R: Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 2017, 171:615–627 e616.** This manuscript reports the use of solid- and liquid-state NMR spectroscopy to characterize fibrils formed by the FUS low-complexity domain. A 57-residue core is identified, which folds into a β-sheet and partakes in the construction of higher order assemblies displaying cross-β architecture. It is demonstrated that phosphorylation of residues within this core disrupts hydrogel binding and reduces liquid-liquid phase separation.
- 42.Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, Eisenberg DS: Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science 2018, 359:698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo F, Gui X, Zhou H, Gu J, Li Y, Liu X, Zhao M, Li D, Li X, Liu C: Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat Struct Mol Biol 2018, 25:341–346. [DOI] [PubMed] [Google Scholar]
- 44.Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, Gu J, Xie M, Zhao K, Dai B, et al. : Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun 2019, 10:2006.** The manuscript reports that three short motifs within the LCD domain of hnRNPA1, so-called hnRACs, can form reversible amyloid fibrils based on a cross-β architecture. Deletion of the hnRACs results not only in decreased fibrillization but also in a decrease of the driving force for phase separation.
- 45.Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J,Linse S, et al. : Atomic Resolution Structure of Monomorphic Abeta42 Amyloid Fibrils. J Am Chem Soc 2016, 138:9663–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R: Molecular structure of beta-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 2013, 154:1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Bockmann A, Guntert P, Meier BH, Riek R: Atomic-resolution structure of a disease-relevant Abeta(1–42) amyloid fibril. Proc Natl Acad Sci U S A 2016, 113:E4976–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y: Abeta(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol 2015, 22:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schutz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Guntert P, Bockmann A,Glockshuber R, Meier BH: Atomic-resolution three-dimensional structure of amyloid beta fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl 2015, 54:331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM,Kim JK, Barclay AM, Kendall A, et al. : Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nat Struct Mol Biol 2016, 23:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. : Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 2012, 149:768–779. [DOI] [PubMed] [Google Scholar]
- 52.Guenther EL, Cao Q, Trinh H, Lu J, Sawaya MR, Cascio D, Boyer DR, Rodriguez JA, Hughes MP, Eisenberg DS: Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat Struct Mol Biol 2018, 25:463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray DT, Zhou X, Kato M, Xiang S, Tycko R, McKnight SL: Structural characterization of the D290V mutation site in hnRNPA2 low-complexity-domain polymers. Proc Natl Acad Sci U S A 2018, 115:E9782–E9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato M, Yang YS, Sutter BM, Wang Y, McKnight SL, Tu BP: Redox State Controls Phase Separation of the Yeast Ataxin-2 Protein via Reversible Oxidation of Its Methionine-Rich Low-Complexity Domain. Cell 2019, 177:711–721 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL: Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 2013, 155:1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL: The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 2015, 163:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Y, Currie SL, Rosen MK: Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem 2017, 292:19110–19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. : A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 59.Lin Y, Protter DS, Rosen MK, Parker R: Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 2015, 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T, Commins C, et al. : Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J 2018, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, et al. : TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 2017, 95:808–816 e809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burke KA, Janke AM, Rhine CL, Fawzi NL: Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell 2015, 60:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, et al. : Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci U S A 2017, 114:E8194–E8203.** Solution NMR spectroscopy on a large drop demonstrates that the intrinsically disordered region of Ddx4 remains disordered in the dense phase. While translational diffusion and tumbling is slowed significantly, the protein behaves as a monomer and local regions remain rather dynamic. Direct observation of intermolecular interactions between Phe and Arg suggest the importance of π-π and cation-π interactions.
- 64.Ryan VH, Dignon GL, Zerze GH, Chabata CV, Silva R, Conicella AE, Amaya J, Burke KA, Mittal J, Fawzi NL: Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol Cell 2018, 69:465–479 e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murthy AC, Dignon GL, Kan Y, Zerze GH, Parekh SH, Mittal J, Fawzi NL: Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat Struct Mol Biol 2019, 26:637–648.** In this thorough study, the molecular features of the FUS low complexity domain in a large drop are probed by a diverse set of NMR experiments and reveal that the sequence remains disordered in the condensed phase with no indication of any stable structure. NMR experiments combined with simulations and mutagenesis suggest that most residues in the sequence participate in intermolecular interactions, many of which include hydrophobic and/or π-π interactions involving aromatics and Gln, and hydrogen bonding interactions between Gln residues.
- 66.Reichheld SE, Muiznieks LD, Keeley FW, Sharpe S: Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc Natl Acad Sci U S A 2017, 114:E4408–E4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, Forman-Kay JD: Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 2018, 7.* This study uses analyses of high-resolution crystal structures to evaluate the frequency of π-π interactions and to come to the conclusion that phase-separating proteins are enriched in amino acid residues that can mediate such interactions. A phase separation predictor is presented.
- 68.Martin EW, Mittag T: Relationship of Sequence and Phase Separation in Protein Low-Complexity Regions. Biochemistry 2018, 57:2478–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brangwynne CP, Tompa P, Pappu RV: Polymer physics of intracellular phase transitions. Nature Physics 2015, 11:899–904. [Google Scholar]
- 70.Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, Ruer M, Hyman AA, Alberti S: Different Material States of Pub1 Condensates Define Distinct Modes of Stress Adaptation and Recovery. Cell Rep 2018, 23:3327–3339. [DOI] [PubMed] [Google Scholar]
- 71.Boke E, Ruer M, Wuhr M, Coughlin M, Lemaitre R, Gygi SP, Alberti S, Drechsel D, Hyman AA, Mitchison TJ: Amyloid-like Self-Assembly of a Cellular Compartment. Cell 2016, 166:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA: Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 2017, 168:1028–1040 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schlick T, Pyle AM: Opportunities and Challenges in RNA Structural Modeling and Design. Biophys J 2017, 113:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, et al. : mRNA structure determines specificity of a polyQ-driven phase separation. Science 2018, 360:922–927.** This is an important study that shows that RNA structure determines the identity of dense phases formed by the Ashbya gossypii protein Whi3. Whi3 forms distinct dense phases with two different types of RNA, and the condensate properties depend on the structure of the RNA. Denaturation of the RNA results in a single dense phase that contains Whi3 a mixture of both RNAs.
- 75.Rouka E, Simister PC, Janning M, Kumbrink J, Konstantinou T, Muniz JR, Joshi D, O’Reilly N, Volkmer R, Ritter B, et al. : Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3). J Biol Chem 2015, 290:25275–25292. [DOI] [PMC free article] [PubMed] [Google Scholar]