Abstract
Crosstalk between malignant and neighboring cells contributes to tumor growth. In East Asia, infection with the liver fluke is a major risk factor for cholangiocarcinoma (CCA). The liver fluke Opisthorchis viverrini secretes a growth factor termed liver fluke granulin, a homologue of the human progranulin, which contributes significantly to biliary tract fibrosis and morbidity. Here, extracellular vesicle (EV)-mediated transfer of mRNAs from human cholangiocytes to naïve recipient cells was investigated following exposure to liver fluke granulin. To minimize the influence of endogenous progranulin, its cognate gene was inactivated using CRISPR/Cas9-based gene knock-out. Several progranulin-depleted cell lines, termed ΔhuPGRN-H69, were established. These lines exhibited >80% reductions in levels of specific transcript and progranulin, both in gene-edited cells and within EVs released by these cells. Profiles of extracellular vesicle RNAs (evRNA) from ΔhuPGRN-H69 for CCA-associated characteristics revealed a paucity of transcripts for estrogen- and Wnt-signaling pathways, peptidase inhibitors and tyrosine phosphatase related to cellular processes including oncogenic transformation. Several CCA-specific evRNAs including MAPK/AKT pathway members were induced by exposure to liver fluke granulin. By comparison, estrogen, Wnt/PI3K and TGF signaling and other CCA pathway mRNAs were upregulated in wild type H69 cells exposed to liver fluke granulin. Of these, CCA-associated evRNAs modified the CCA microenvironment in naïve cells co-cultured with EVs from ΔhuPGRN-H69 cells exposed to liver fluke granulin, and induced translation of MAPK phosphorylation related-protein in naïve recipient cells in comparison with control recipient cells. Exosome-mediated crosstalk in response to liver fluke granulin promoted a CCA-specific program through MAPK pathway which, in turn, established a CCA-conducive disposition.
Keywords: Opisthorchis viverrini, Liver fluke granulin, Extracellular vesicle, Cellular crosstalk
Introduction
Cholangiocarcinoma (CCA) describes malignancy arising from the biliary epithelium. CCA originates in the cholangiocyte, the specialized epithelial cell that lines the intrahepatic and extrahepatic bile ducts, except for the epithelial cells of the gallbladder. Many CCA are adenocarcinomas [1], [2]. Although the causative agent for many cancers remains obscure including non-liver fluke infection-associated CCA, the principal risk factor in liver fluke-endemic regions is well established: infection with Opisthorchis viverrini and related parasites [3], [4], [5], [6]. Infection with O. viverrini is the principal risk factor for CCA in the Lower Mekong River Basin countries including Thailand, Lao PDR, Vietnam and Cambodia [5], [6]. It has been estimated that 10% of people chronically infected with liver flukes will develop CCA [7]. In regions endemic for opisthorchiasis, the prevalence of CCA can exceed 80 cases per 100,000 residents [8].
Helminth parasites communicate and interact at the host-parasite interface [9]. Communication is facilitated by metabolic products secreted from the tegument and excretory tissues, including via exosomes [10], [11]. The liver fluke O. viverrini releases proteins and other metabolites [12], which influence host cells including cholangiocytes in diverse ways [13], [14], [15], [16]. Whereas the full complement of metabolites released by this parasite remains generally to be investigated for roles of communication and disease, a secreted protein termed liver fluke granulin (Ov-GRN-1) has been the focus of increasing investigation. Liver fluke granulin is a paralogue of human progranulin, which like the progranulin stimulates cell proliferation and wound healing, and which appears to contribute to the pathogenesis of opisthorchiasis [17], [18], [19], [20], [21]. We have exploited this link to more deeply explore the role of liver fluke granulin in pre-malignant lesions of the bile duct by undertaking programmed CRISPR/Cas9 knockout of its cognate gene. Deep sequencing of amplicon libraries from genomic DNA of gene-edited parasites revealed Cas9-catalyzed mutations within the Ov-GRN-1 locus. Gene knockout rapidly depleted levels of Ov-GRN-1 transcripts and depleted cellular levels of liver fluke granulin. Furthermore, experimental infection of hamsters with the gene-edited liver flukes resulted in markedly reduced disease even though gene-edited parasites colonized the biliary tract and developed into the adult developmental stage of the liver fluke. These findings confirmed a role for liver fluke granulin in virulence of the hepatobiliary morbidity characteristic of opisthorchiasis [22].
In the present report, extracellular vesicle-mediated transfer of functional mRNAs to naïve recipient cells from cholangiocytes that had been exposed to liver fluke granulin was investigated. In response, extracellular vesicle-mediated crosstalk promoted CCA-specific translational profile(s) in the recipient cholangiocytes, including via MAPK signaling that, in turn, established a microenvironment supportive of carcinogenesis.
Materials and methods
Cell lines
The cholangiocyte cell line H69 is a SV40-transformed bile duct epithelial cell line derived from a non-cancerous human liver [23]. H69 cells were maintained in H69 complete medium; Ham’s F12 nutrient mixture (25 μg/ml adenine, 5 μg/ml insulin, 1 μg/ml epinephrine, 0.62 μg/ml hydrocortisone, 1.36 μg/ml T3-T, and 10 ng/ml epidermal growth factor (EGF), Dulbecco’s Modified Eagle Medium [DMEM] (Gibco), DMEM/F-12 (Sigma-Aldrich) media containing 10% fetal bovine serum (FBS) and 1× penicillin/streptomycin (pen/strep), as described [21], [24], [25]. H69 cells was maintained in humidified incubator in 5% CO2 in air at 37 °C. H69 cells were Mycoplasma-free as established using the Lookout Mycoplasma PCR detection kit (Sigma-Aldrich) and authenticated using short-tandem repeat profiling (ATCC, Manassas, VA).
Lines of progranulin knockout cholangiocytes
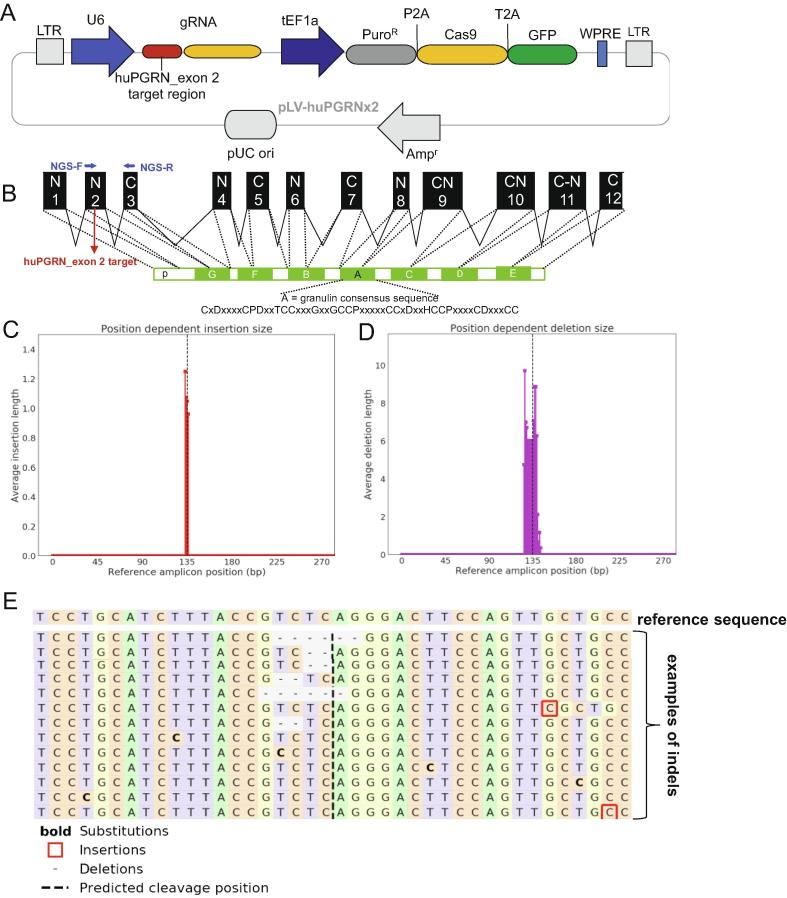
To mutate and disrupt the human progranulin gene, huPGRN, with the aim of minimizing or eliminating endogenous granulin in H69 cells that might obfuscate interpretation of the effect of liver fluke granulin, we employed a pre-designed lentiviral CRISPR/Cas9 vector, ‘All in One CRISPR/Cas9 vector system (Sigma-Aldrich). Our vector encoded a guide RNA targeting human progranulin, huPGRN exon 2, which encodes the granulin-epithelin precursor (GEP); 5′-cctgcaatctttaccgtctc-3′ on chromosome 17: NC_000017.11, region 44,345,086–44-353,106 driven by U6 promoter, and human elongation factor 1α promoter to drive the expression of a fusion protein comprised of the puromycin N-acetyl transferase from Streptomyces alboniger (puromycin resistance marker, PuroR) [26], the Cas9 endonuclease from Streptococcus pyogenes, and green fluorescent protein (GFP), in that order. The gene cargo of the vector is flanked by the long tandem repeat (LTRs) of the HIV-1 lentivirus (Fig. 1A).
Fig. 1.
Programmed CRISPR/Cas9 mutation of the human progranulin gene. Panel A, linear map of a pre-designed lentiviral CRISPR/Cas9 vector containing fused codon-optimized puromycin resistance marker (dark gray bar)-Cas9 (yellow bar) and green fluorescent protein (green bar) driven by the mammalian elongation factor alpha-1 promoter (dark blue arrow); guide RNA (gRNA) targeting human granulin exon 2 (red bar) is expressed from a single vector. The human U6 promoter (blue arrow) drove the gRNA. The vector backbone includes the 5′- and 3′-long terminal repeats (LTR) of the HIV-1 provirus (light gray blocks). Panel B, schematic representation of the partial human granulin gene precursor, huPGRN on chromosome 17: NC_000017.11 regions 44,345,086–44,353,106 (8021 bp) and protein structure. Nucleotide sequence in exon 2 encodes the N-terminus and part of the granulin/epithelin module (GEM) of progranulin; indicating locations of gRNA (4417–4438 nt; red colored-letter) predicted double-stranded break (DSB) (red arrow). Panels C–E, on target INDEL mutations in ΔPGRN-H69 analysis by NGS libraries and CRISPResso bioinformatic platform. Frequency distribution of position-dependent insertions (red bars) (C) and deletion (magenta) (D) the major INDELs; 6 and 2 bp deletions at the programmed CRISPR/Cas9 cleavage site. Other minor mutations including 1 bp insertion (red square) or base substitution (bold) were observed further (>10 bp) predicted cleavage site (E). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Competent Escherichiacoli cells were transformed with the plasmid form of the vector, termed pLV-huPGRNx2, and maintained in LB broth, 100 µg/ml ampicillin. Virions were derived following transfection of human 293T cells producer cells with pLV-huPGRNx2, using FUGENE HD transfection reagent (Promega, Madison, WI) and the MISSION™ lentiviral packaging kit (Sigma-Aldrich), as described [27]. Pooled culture supernatants containing pseudotyped virions were collected at 48–72 h after transfection of 293T cells, clarified by centrifugation at 500×g for 10 min, and passed through a Millipore 0.45 µm pore membrane (Steriflip-GP, Millipore). Virions were concentrated using Lenti-X concentrator (Takara Bio) after which titers were measured by Lenti-X-GoStix Plus (Takara Bio). For programmed knock-out of huPGRN, ∼350,000 H69 cells were exposed to 500 µl of pLV-huPGRNx2 virion (∼5 × 105 infectious units [IFU]/ml) in 2.5 ml complete H69 medium in 6-well plates. One day later, the medium was replaced with medium supplemented with puromycin at 300 ng/ml for the selection and enrichment of cells carrying the proviral form of the gene-editing virus (survival of H69 cells in puromycin ranging from 50 to 400 ng/ml was tested at the outset, aiming to define a concentration to inhibit survival of these cholangiocytes. H69 cells were killed by puromycin within 48 h in at 300 ng/ml puromycin [not shown].) Gene edited cells were maintained in parallel with H69 cells for 72 h, by which point the control H69 cells had died. About 5–10% of cells survived, GFP sorted and entered clonal amplification. Surviving cells were cultured in complete H69 medium supplemented with 300 ng/ml for 20 passages before genotyping. Discrete three biological replicates were undertaken to establish three puromycin-resistant huPGRN knock-out cell lines; b1, b2 and b3 (Fig. 2), which exhibited >70% reductions in levels of huPGRN transcripts and progranulin (Fig. 2A, B). These were termed ΔhuPGRN-H69 cell lines.
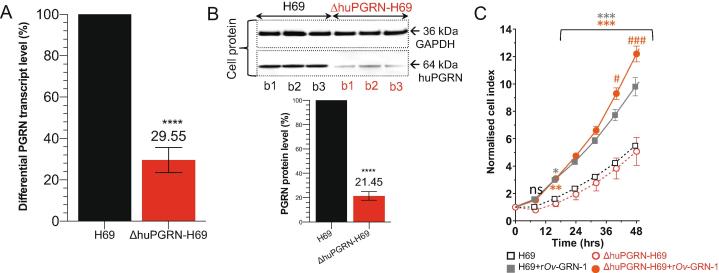
Fig. 2.
Reduction of progranulin transcript and protein expression levels and cell proliferative effects of liver fluke granulin. Panels A and B, reduction of huPGRN transcription levels from ΔhuPGRN-H69 cell; red bar (∼70%) comparing with H69 reference (black bar). The huPGRN differential transcript after normalization with human GAPDH gene; mean ± SD, n = 3 (biological replicates); P < 0.0001 (****), unpaired t-test. Diminished level of huPGRN revealed by WB analysis using anti-PGRN antibody (64 kDa) compared with anti-GAPDH antibody (36 kDa). Progranulin levels in ΔhuPGRN-H69 cells (red bars) was reduced by ∼80%. The GAPDH levels were stable within three biological replicates. These decreased levels of progranulin were significantly different from H69 (black bars); n = 3 (biological replicates); P < 0.0001 (****), unpaired t-test. Panel C, real time monitoring of H69 vs ΔhuGRN-H69 cell proliferation before and after addition of liver fluke granulin. The lower normalized cell index (nCI) of ΔhuGRN-H69 cells (discontinuous red lines) compared with H69 cells (discontinuous gray lines) were monitored over 48 h. The nCI of ΔhuGRN-H69 cells was recovered as in H69 cells nCI value after addition of liver fluke granulin at 100 nM for 24 h, and higher than H69 from 24 to 48 h. The nCI signals are shown as mean ± SD, for ≥3 independent experiments, and assessed using two-way ANOVA with Dunnet’s multiple correction. Comparing 100 nM liver fluke granulin treatments on each cell type to untreated cells (either H69 vs 100 nM liver fluke granulin treated H69 or ΔhuPGRN-H69 vs 100 nM liver fluke granulin -treated ΔhuPGRN-H69): P < 0.05 (*); P < 0.01 (**); P < 0.001 (***). Comparing 100 nM liver fluke granulin-treated H69 to 100 nM liver fluke granulin-treated ΔhuPGRN-H69: P < 0.05 (#); P < 0.001 (###). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Programmed mutation
Following lentiviral transduction and maintenance in 300 ng/ml puromycin, surviving cells were expanded through successive passages. After passage 20, genomic DNAs were extracted from pools of the enriched, puromycin resistant ΔhuPGRN-H69 cells using DNAzol (Molecular Research Center). We amplified the targeted region of exon 2, huPGRN using the NGS primer pair; forward primer 5′-GACAAATGGCCCACAACACT-3′ and reverse primer 5′-GCATAAATGCAGACCTAAGCCC-3′ (Fig. 1B) flanking expected double strand break (DSB) by CRISPR/Cas9 system. The DNA samples were processed for next generation sequencing (NGS) using the Ion Torrent Personal Genome Machine (ThermoFisher), in order to investigate on-target INDELs. The sequencing libraries were prepared from 10 ng DNA using the Ion Torrent Ampliseq kit 2.0-96 LV (ThermoFisher), following the manufacturer’s instructions. The DNA was bar-coded using the Ion Xpress Barcode Adapters kit and quantified by quantitative PCR using the Ion Library TaqMan Quantitation Kit (ThermoFisher) after purification of libraries by Agencourt AMPure XP beads (Beckman). Emulsion PCR was performed using the Ion PGM Hi-Q View OT2 Kit and the Ion OneTouch 2 system (ThermoFisher). Template-positive ion sphere particles (ISPs) were enriched using the Ion Torrent OneTouchES. Enriched ISPs were loaded on a PGM 314R v2 chip and sequenced using the Ion PGM Hi-Q View Sequencing Reagents (ThermoFisher). Raw sequencing data were processed using the Torrent Suite software v5.0 (ThermoFisher), as well as the coverage analysis and variant caller (VC) v5.0 plugins. Processed reads were aligned to the human reference genome (hg38) [28]. The identified variants and the depth of coverage were visually confirmed by the Integrative Genomic viewer (IGV, Broad Institute, MIT, Cambridge, MA). Nine percent of reads were filtered out by post processing. The filtered sequences, average read length 176 bp, were converted to fastq format and analyzed for non-homologous end joining (NHEJ)-mediated mutations using the CRISPResso pipeline [29], [30]. Sequence reads were compared to the reference PCR sequence of the wild type huPGRN gene, GenBank M75161.1 (Fig. 1C–E).
Cellular proliferation
Proliferation of H69 and ΔhuPGRN-H69 cells in response to exposure to 100 nM recombinant liver fluke granulin [17], [21] was quantified using the impedance-based xCELLigence real time cell analysis (RTCA) approach (ACEA Biosciences). The liver fluke granulin peptide (∼10 kDa) was concentrated using Centripep with cut-off 3 kDa (Eppendorf) and resuspended in low salt solution, Opti-MEM. The absorbance at 205 nm and concentration of liver fluke granulin was determined by using a Nanodrop 2000c spectrophotometer (ThermoFisher) [31]. Five thousand cells/well were seeded in 16-well E-plates (ACEA) in H69 complete media. E-plates was inserted in the xCELLigence DP platform at 37 °C, 5% CO2 and changes in impedance reflecting cell adhesion and proliferation record at intervals of 20 min for 24 h. On the following day, the medium was removed and replaced with H69 complete medium supplemented with liver fluke granulin at 100 nM. Cellular proliferation was monitored for 48 h for the wild type H69, liver fluke granulin-treated H69, and ΔhuPGRN-H69 cells with and without liver fluke granulin treatment, and displayed here as change of impedance (Cell Index) after normalizing to the signals for the wild type H69, assigned as the reference cell line (RTCA Software 1.2, ACEA) [32].
Isolation and characterization of extracellular vesicles
The H69 or ΔhuPGRN-H69 cells were treated with 100 nM liver fluke granulin in H69 complete media with 10% exosome depleted-FBS. Forty-eight hours later, were harvested the supernatant from cell culture of H69 with or without, and ΔhuPGRN-H69 with or without liver fluke granulin. The supernatants were collected at 48 h following addition of liver fluke granulin and cellular debris pelleted and removed by centrifugation 2000×g, 15 min. The supernatant was filtered through a 0.22 μm pore size membrane (Millipore, Billerica, MA), mixed with 0.5 volume of tissue culture total exosome isolation reagent (Invitrogen, catalog no. 4478359), and incubated for 16 h at 4 °C. Thereafter, the EV pellet (isolated by centrifugation at 10,000×g, 4 °C, 60 min) was re-suspended in 1× PBS [33]. EVs were investigated in co-culture assays and the EV-associated RNA (evRNA) and protein (evProtein) of the EVs were investigated following extraction of exosomal contents using the Total Exosome RNA and Protein Isolation kit (ThermoFisher).
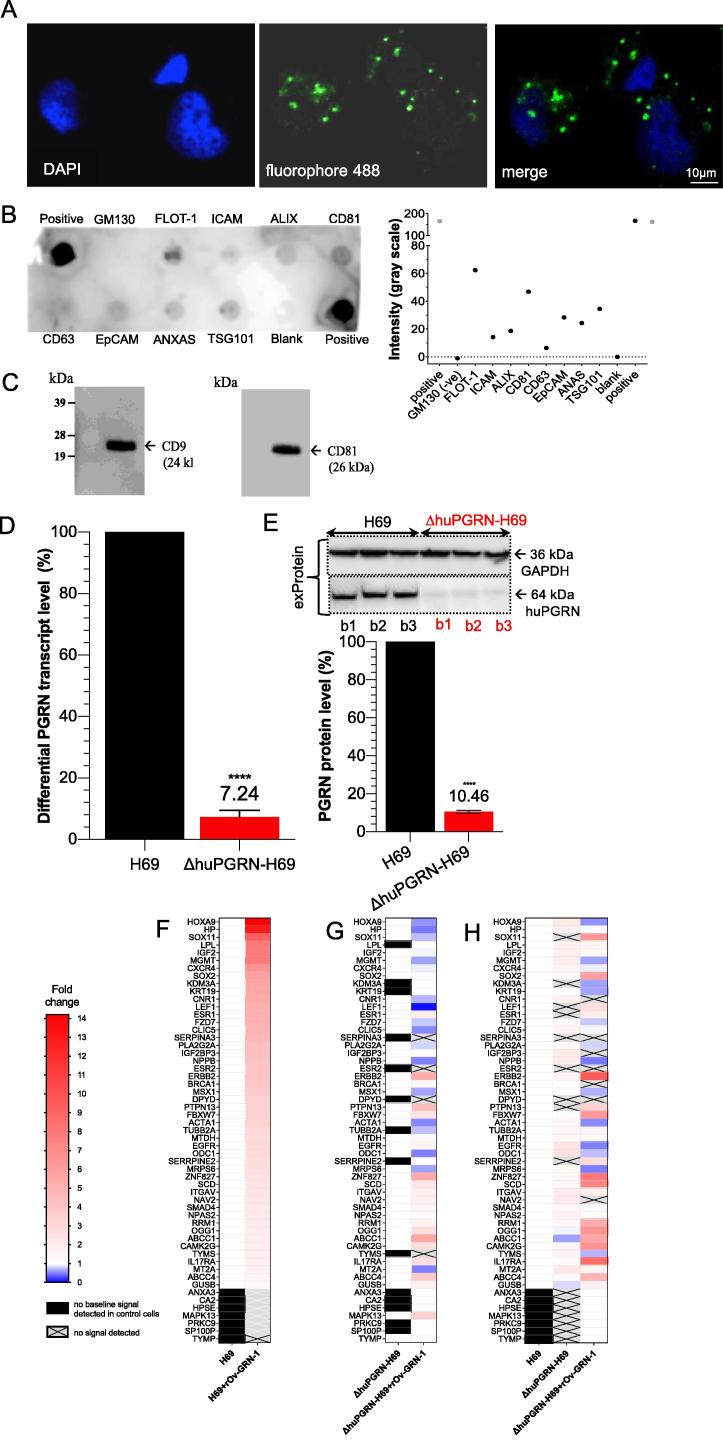
The identification of protein markers on the isolated EVs was undertaken using the commercially available Exo-Check Exosome Antibody Array kit (System Biosciences): the array included 12 pre-printed spots and was used with the cognate antibodies specific for the exosome markers CD63, CD81, ALIX, FLOT1, ICAM1, EpCam, ANXA5 and TSG101. The array included the following controls: a GM130 cis-Golgi marker to monitor cellular contamination, a positive control spot derived from human serum EVs, and a blank spot as a background control. The membrane was developed with SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific) and visualized and quantified on the ChemiDoc Imager (Bio-Rad) [34]. The intensity of EV protein markers was read (gray scale) (ChemiDoc), and subtracted with negative background intensity. The signal intensity for each protein was plotted and compared (Fig. 3B). The evProtein was also confirmed for EV marker expression by western blot analysis against anti-CD9 and CD81 (Abcam). Briefly, 10 µg of the evProtein was separated on gradient (4–12%) SDS-PAGE gel and transferred to nitrocellulose membrane (Bio-Rad). After blocking with 5% skim milk in Tris-buffered saline (TBS)-Tween for 60 min, the membrane was incubated with specific antibody against CD9 or CD81, followed (after washing) by anti-rabbit HRP-linked secondary antibody (DAKO Corporation catalog no. P0448) diluted 1 in 2000. Signals from ECL substrate were detected by chemiluminescence (Amersham Bioscience) and captured and analyzed (ChemiDoc, Bio-Rad) (Fig. 3C).
Fig. 3.
EV-derived H69 cell characterization and PGRN expression. Panel A, the visualization of EVs from H69 cells by in situ hybridization using fluorophore 488-labeled anti-CD81 antibody (green dot, panel A) around DAPI-stained cell nuclei (blue color). The H69-derived EV-like particle sizes ranged from 55 to 80 nm. The protein composition of EVs were determined using the Exosome Antibody Array (SBI System Biosciences). Panel B, dark spots indicate presence of the target protein (upper panel). Absence of a spot for GM130 confirmed the absence of cellular contaminants. The intensity in gray scale was also plotted (lower panel). Panel C, WB analysis for the exosome-specific markers; strongly positive signal for CD9 and CD81 were found. Panels D and E, both the evRNA and evProtein from ΔhuPGRN-H69 showed reduction of ∼90% differential transcript levels [red bar, panel D] after normalization with GAPDH in comparison to H69 [black bar] (red bar, panel D). The levels of progranulin were reduced by ∼90%; unpaired t-test, P < 0.0001 (****), n = 3. Panels F–H, show an induction of CCA-related mRNAs carrying cholangiocyte derived-EVs after exposure to liver fluke granulin. The heat map was plotted based on the differential transcript fold change as determined using the CCA-related gene Array using H69 as the reference from H69 with liver fluke granulin treatment (panel F) and ΔhuPGRN-H69, ΔhuPGRN-with liver fluke granulin treatment (panel H). Similar quantities of evRNAs from each group were probed individually with 88 CCA targets in triplicate; there were slight differences (P < 0.05 by 2way ANOVA gene profiles (55 from 88 transcripts were detected) in ΔhuPGRN-H69 vs H69 (panel A) by absent of DPYD, ESR, KDM3A, LEF1, PRKC9, PTPN13, SERPINA3, SERRPINE2 and SOX11 (panel H). However, most of these genes were recovered after exogenous liver fluke granulin treatment of ΔhuPGRN-H69 (panels G and H). The CCA mRNA profiles after liver fluke granulin treatment of EVs derived from ΔhuPGRN-H69 and H69 were markedly dissimilar following exposure to liver fluke granulin with P < 0.0001, 2way ANOVA (panels F and G). Panel G, shows the CCA-related evRNAs after exposure to liver fluke granulin in ΔhuPGRN-H69 compared with its mRNA profile (P < 0.0001 with two-tailed, paired t-test). The statistical significance was calculated from differential fold changes in transcription. The liver fluke granulin (rOv-GRN-1) treatment group of cells showed 1–15 folds (gradient red bar) level of induction of transcription. The ‘1’ indicates baseline (white bar) level expression, and ‘0’ indicated the absence of expression (gradient blue bar). The transcripts from non- or treated- rOv-GRN-1treatment were not detected (no baseline signal) indicate in black bar and gray-cross bar, respectively. The heat map was plotted from the average transcript level ran in triplicates using Prism v8. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For immunofluorescence staining of EV-like particles from H69 cells were proceeded by culturing the cells in glass slide chamber overnight before fixed at 4 °C in ice-cold methanol for 10 min, washed 3 times in 1× PBS, and permeabilized in 0.1% Triton X-100/PBS for 10 min at room temperature. Nonspecific binding was blocked with 0.5% Tween-20/PBS containing 1% bovine serum albumin (BSA) for 30 min. The primary antibodies against CD81 labeling with fluoroflore-488 was incubated for 60 min at room temperature. The incubated cells were washed 3 times in 1× PBS. Before visualization by confocal microscope, we stained the cell nucleus with DAPI. The EVs were observed and particle sized were measures in H69 cytoplasm (Fig. 3A)
Quantitative real time PCR
Total RNA and evRNA either from H69 or ΔhuPGRN-H69 cells were isolated using RNAzol (Molecular Research Center, Inc.) or total exosome RNA isolation kit (ThermoFisher) following the manufacturer’s instructions. One microgram of RNA was treated for DNase, then used for reverse transcription by an iScript cDNA synthesis kit (Thermo Fisher Scientific). Real time PCR was performed in ABI7300 Real time PCR machine using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). The PCR reaction consisted of 5 µl SsoAdvance SYBR Green PCR master mix, 0.5 µl of 10 µM forward and reverse primers, and 2 µl of 5 times diluted template cDNA in a total volume of 10 µl. The thermal cycle was initiation cycle at 95 °C for 30 s followed by 40 cycles of annealing at 55 °C for 1 min. Samples were analyzed in at least 3 biological replicates (various cell passages) and in typical reactions. The human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was run parallel with human granulin (huPGRN) and used for gene normalization. The differential granulin transcript fold change was calculated by formula 2−ddCt [35]. The specific primers for huPGRN and GAPDH are as follows: PGRN-F: 5′-atgataaccagacctgctgcc-3′, PGRN-R: 5′-aaacacttggtacccctgcg-3′, GAPDH-F; 5′-tgtagttgaggtcaatgaaggg-3′ and GAPDH-F 5′-tgtagttgaggtcaatgaaggg-3′. The fold change was obtained by comparing treated-samples with the untreated control (indicated as a value of 1, 100%). The means and standard deviations of differential transcript expression were calculated by independent Student’s t-tests using Prism version 8 software (GraphPad).
Western blot and densitometry
Protein lysates and protein extracted from EVs (evProtein) from H69 and ΔhuPGRN-H69 cells were prepared using M-PER mammalian protein extraction reagent (ThermoFisher) and exosome protein isolation kit (ThermoFisher), respectively, following the manufacturer’s protocols. Protein concentrations were determined using the Bradford assay [36]. Ten micrograms of cell lysate or 20 µg of evProtein was separated on gradient SDS-polyacrylamide gel (4–12% Bis-Tris, Invitrogen) and transferred to nitrocellulose membrane (Trans-Blot Turbo system, Bio-Rad). Signals were identified with a chemiluminescence substrate (GE Healthcare) following probing with antibodies specific for progranulin (Abcam) or GAPDH (Sigma-Aldrich), and with HRP conjugated-secondary antibodies. Expression levels of huPGRN were imaged using the FluroChem system (Bio-techne) and relative expression levels of progranulin investigated after GAPDH level normalization. Signals were compared using independent Student’s t-tests.
Cholangiocarcinoma (CCA) gene expression panel
A predesigned Cholangiocarcinoma PrimePCR™ Array Pathway Panel (88 targets) (Bio-Rad) was used to investigate the CCA gene expression profile of transcripts in the cargo of EVs released by H69 cells exposed to liver fluke granulin. One microgram of total RNA from EV samples was converted to cDNA (Supermix iScript kit, Bio-Rad). A one in 10 dilution of cDNA was used for qPCRs with a final concentration of 1× SsoAdvanced universal SYBR super mix (Bio-Rad) and 1×PrimePCR assays for the designated target. Reactions were performed (three technical replicates) at 10 μl final volume, using the iQ5 real time PCR system (Bio-Rad) starting with activation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C, 5 s, and annealing/elongation at 60 °C, 30 s. Specificity of target amplification was confirmed by melting-curve analysis. Controls for evaluating reverse transcription, RNA quality, genomic DNA contamination and kit performance were included on the array (Bio-Rad). The reference genes GAPDH, HPRT1 and TBP were assayed for relative gene expression to normalize for variation in quantity of input mRNA. The DNA template served as a positive real-time PCR control for the corresponding gene assay. PrimePCR™ Analysis Software (Bio-Rad) was used for analysis of differential fold changes of target genes. Transcript readings higher than 2-fold change were read as positive signals.
Uptake of PKH26-labeled EVs by cholangiocytes
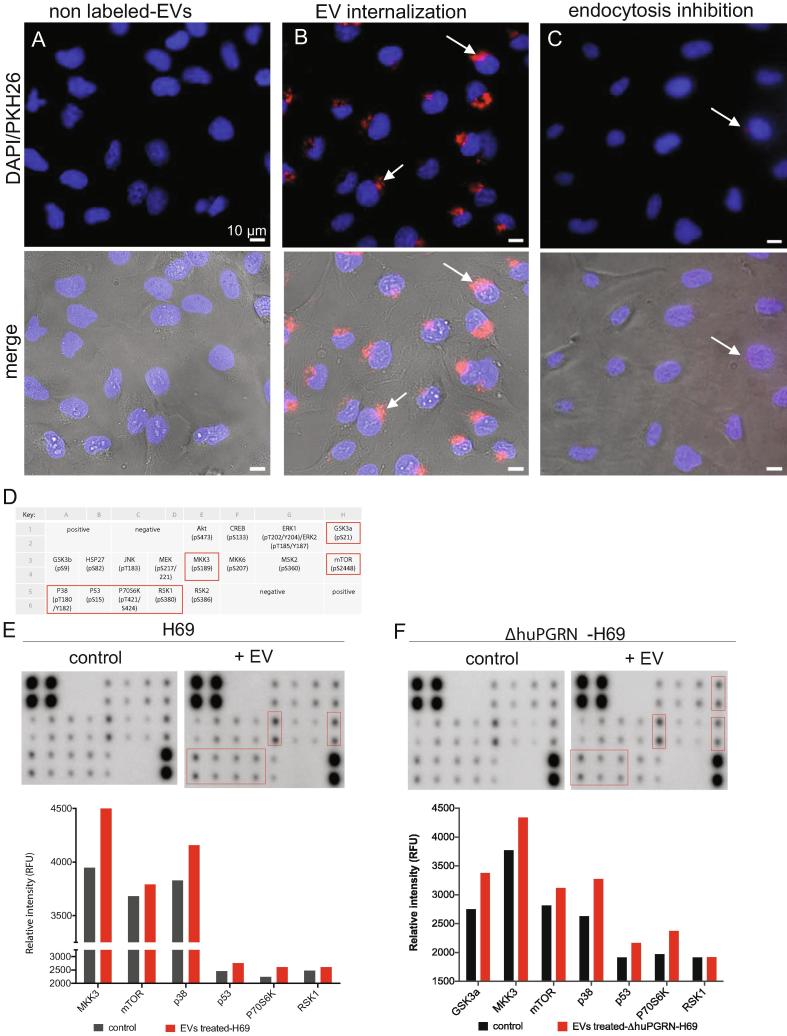
To investigate whether liver fluke granulin-treated H69 communicated CCA-conducive mRNAs to adjacent naïve cells using exosomal cargo, EVs were labeled with PKH26 Fluorescent cell linker kit (Sigma-Aldrich) [37], [38]. In brief, EVs (equally 6.25 μg evProtein) isolated from culture media were re-suspended in one ml of diluent C, to maintain cell viability with maximum dye solubility and staining efficiency [39] and labeled by addition of PKH26 to 2 μM final concentration. The samples were incubated for 5 min with gentle, periodic mixing, after which 2 ml of 1% BSA was added to bind excess dye. The mixture containing PKH26-labeled EVs was diluted into tissue culture exosome isolation reagent (ThermoFisher), as above, after which the EVs were re-suspended in H69 complete medium and co-cultured with H69 or ΔhuPGRN-H69 cells at 80–90% confluence in wells of poly-l-lysine-coated microscope slide chambers (Lab-Tek II). A negative control group was co-cultured with EVs not stained with PKH26. After incubation with EVs for 90 min at 37 °C in 5% CO2, the cells were stained with NucBlue Live Cell Stain ReadyProbes (Invitrogen) and visualized on a Cell Observer SD Spinning Disk Confocal Microscope (Zeiss). A negative control for inhibition of endocytosis was included, which involved addition of Pitstop 2, an inhibitor of clathrin-mediated endocytosis (Abcam) at 30 μM to recipient cells for 15 min [40], before co-culture with labeled-EVs (Fig. 4A–C).
Fig. 4.
Uptake of extracellular vesicles and paracrine transfer of mRNAs from donor to naïve recipient cells. Panels A and B, representative fluorescence micrographs showing uptake of PKH26-labeled EVs into recipient cholangiocytes; blue—nuclei staining with the NucBlu Live Cell Stain ReadyProbe, red—PKH26-labeled EVs. Pitstop 2 blocked clathrin-dependent endocytosis of EVs (C). Merged images shown in lower panel with bright field. Magnification, 40×; scale bar, 10 µm. Arrows indicate internalized EVs in naïve cells. Panels D to F, cholangiocytes were grown to 80% confluence after which they were co-cultured with EVs from liver fluke granulin-activated ΔhuPGRN-H69 cells for 24 h. The cell lysate (500 ng) was used for 17 targets (panel D indicates the key of antibody spot in duplicate) on the human MAPK phosphorylation antibody array; ab211061 (Abcam). The relative density of the spots was quantified by densitometry analysis (panels E and F) (RFU). Red boxes and red bars represent the predominant proteins that significantly higher than control groups (grey bars) namely, GSK3a, MKK3, mTOR, p38, p53, p70S6K and RSK1. The MAPK phosphorylation protein pattern of these protein revealed statistically significant differences in comparison with its control; P < 0.0001, 2way ANOVA. Data represent the average of two individual sets per sample. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Human MAPK phosphorylation array
To investigate whether CCA-associated functional RNAs in EVs were translated by naïve H69 cells (both wild type and the ΔhuPGRN), the cells were incubated with the EVs as described above. For this, 5 × 104 cells were seeded into a well of 24-well plate and maintained overnight in H69 complete medium [41]. Thereafter, EV particles were dispensed into each well and cultured for 24 h. The cells were harvested with cell scraper, washed 3 times with 1× PBS, and thereafter resuspended in lysis buffer provided in the Human MAPK Phosphorylation kit). The protein concentration of the cell lysate was determined by the Bradford method (Bio-Rad) before proceeding to probe the human MAPK phosphorylation array, catalog no. ab211061 (Abcam), following the manufacturer’s instructions. In brief, the protein sample was incubated with each array overnight at 4 °C with gentle orbital shaking. Unbound proteins were removed, and the arrays washed three times with wash buffer. Arrays were incubated with the horseradish peroxidase conjugated anti-rabbit IgG antibody for 120 min at room temperature. Thereafter, the array membranes were washed three times with wash buffer prior to incubation with freshly prepared detection solution for two min at room temperature. Protein signals were visualized using the ChemiDoc system (Bio-Rad). The intensity score of each duplicated array spot was ascertained, and normalized array data were calculated according to the manufacturer’s recommendation. To normalize array data, one array was defined arbitrarily as the reference, to which the other arrays were compared. The following algorithm was used to calculate and determine the signal expression between like analyses as previously described [42], [43]:
X(Ny) = X(y)*P1/P(y), where P1 = mean signal density of positive control spots on reference array; P(y) = mean signal density of positive control spots on array ‘y’; X(y) = mean signal density of spot ‘X’ on array for sample ‘y’; and X(Ny) = normalized signal intensity for spot ‘X’ on Array ‘y’.
We used EVs-derived-ΔhuPGRN-H69 cells (not exposed to liver fluke granulin) as the control to contrast with the in vivo translation of CCA-associated mRNAs in the treatment groups.
Biological and technical replicates, statistics
Biological replicates represented parallel measurements of biologically discrete samples in order to capture random biological variation. Technical replicates were undertaken as well; these represented repeated measurements of the same sample undertaken as independent measurements of the random noise associated with the investigator, equipment or protocol. Responses of knockout versus wild type H69 cell line genotypes including before and after exposure to liver fluke granulin were compared using 2way ANOVA, including Dunnet’s correction for multiple comparisons at successive time points. Thereafter, Tukey's honest significance and Student’s t tests were performed to compare treatment groups. Cell index (CI) assays were performed in triplicate. CI was automatically registered by the RTCA software (ACEA Biosciences Inc., San Diego, CA). Means ± SD indicates plotted using GraphPad Prism 8 (GraphPad Software, San Diego, CA). Values of P < 0.05 were considered to be statistically significant.
Supplementary Fig. 1 provides a schematic overview of experimental design and methods.
Database accessions
Nucleotide sequence reads reported here have been assigned SRA accessions SRR9735031-34 at GenBank, NCBI, and the study has been assigned BioProject ID PRJNA556235 and BioSample accession SAMN12344265.
Results
Progranulin mutation in cholangiocytes by CRISPR knockout
The progranulin gene, huPGRN, in the human H69 cholangiocyte cell line was targeted for CRISPR/Cas9-mediated programmed mutation. The programmed target site encodes the N-terminus and part of the granulin/epithelin module of human PGRN (Fig. 1A, B). Wild type H69 cells were transduced with pLV-huPGRNx2 virions at >5 × 105 infective units (IFU)/ml and, a day later, culture medium supplemented with puromycin at 300 ng/ml added to the virion-transduced cells with the goal of enrichment of transgenic cells exhibiting puromycin resistance [26]. Three daughter cell lines, termed ΔhuPGRN-H69 lines, b1, b2 and b3, exhibited >70% reduction in levels of huPGRN transcripts and of protein as assessed by RT-PCR and western blot (WB), respectively (Fig. 2A, B). Thereafter, discrete amplicon libraries were constructed from each of the three daughter cell lines, from which 139,362, 107,683, and 179,122 sequence reads were obtained. The reads were analyzed using the CRISPResso pipeline to monitor programmed gene mutations and subsequent non-homologous end joining (NHEJ) mediated-repair and insertion-deletion (INDEL) profiles (Fig. 1C, D) [29]. This analysis revealed mutations in 32.3, 41.9 and 44.3% of the alleles in the lines, respectively. Similar INDEL profiles were seen among the three libraries: single nucleotide insertions, 6 and 2 bp deletions, and also other deletions from one to 10 bp in length were common at the position predicted for the programmed double stranded chromosomal break (Fig. 1C, E, dashed line). Fig. 1E presents examples of the mutated alleles. This mutational landscape resembled profiles reported for chromosomal repair of CRISPR/Cas9 double stranded breaks by NHEJ in other mammalian cells [44]. GenBank at NCBI assigned these sequence reads from amplicon- NGS libraries from the mutated PGRN locus of the ΔhuPGRN-H69 lines have been assigned the SRA accessions SRR9735031-34, BioProject ID PRJNA556235, and BioSample accession SAMN12344265.
Proliferation of progranulin knockout cell lines promoted by liver fluke granulin
Prolonged reduction in levels of progranulin were observed for >40 passages in three daughter ΔhuPGRN-H69 lines under puromycin treatment (not shown). By that point, differential huPGRN transcript fold changes were stably reduced to ∼30% (29.8 ± 4.7%) in ΔhuPGRN-H69 cells compared with levels in the parental, wild type cells (Fig. 2A) (t-test, P < 0.0001). When examined by WB, the levels of progranulin in ΔhuPGRN-H69 cells had fallen to 21.5 ± 2.3% of those in the parental H69 line cells, normalized against the human GAPDH reference (Fig. 2B) (t-test, P < 0.0001). A band at the predicted mass of progranulin, 64 kDa, was evident in each of three biological replicates (Fig. 2B, lanes b1-b3), as was the GAPDH signal at 36 kDa.
To assess the consequence of loss of huPGRN following the programmed gene knockout, cellular proliferation and the mitogenic activity of recombinant liver fluke granulin (rOv-GRN-1) were monitored using the xCELLigence approach. This label-free cell-based assay system uses culture plates bearing microelectrodes for non-invasive measurement during cell culture (26). The rate of H69 and ΔhuPGRN cell growth was monitored for 48 h and differences compared every 8 h. Both the wild type and the daughter mutant ΔhuPGRN cholangiocytes proliferated at a similar rate for 48 h, suggesting that the programmed knockout of huPGRN was not detrimental to normal cell growth. Next, the wild type and mutant H69 cell lines were exposed to liver fluke granulin at 100 nM [45]. Upon addition of liver fluke granulin, the rates of proliferation of ΔhuPGRN-H69 and H69 were similar (Fig. 2C, solid lines) over the first 32 h, after which ΔhuPGRN-H69 cells grew significantly quicker than H69s (orange solid circles). Liver fluke granulin induced significantly faster growth from 16 h onwards in both H69 and ΔhuPGRN H69 cells compared to control groups (Fig. 2C, dotted lines) not exposed to liver fluke granulin (two-way ANOVA with Dunnet correction; P < 0.001), confirming earlier findings with H69 and other cells [17], [21].
H69 cholangiocytes released extracellular vesicles
Extracellular vesicle (EV)-like particles recovered from culture supernatants of H69 cells were investigated. Confocal microscopical observation revealed EV-like particles that were probed with anti-CD81 labeled with fluorophore 488 (green) surrounding DAPI-stained nuclei (blue) of H69 cells (Fig. 3A). Dot-blot based biochemical characterization (Exo-Check Array) revealed the following exosomal markers with intensity (gray scale) between 10 and 70: flotillin-1 (FLOT-1), intercellular adhesion molecule 1 (ICAM), ALG-2-interacting protein 1 (ALIX), CD81, epithelial cell adhesion molecule (EpCAM), annexin V (ANXAS) and tumor susceptibility gene 101 (TSG101), while the positive control showed intensity >100 (Fig. 3B). The purified EVs were negative for cis-Golgi matrix protein (GM130) when compared to the blank control sample, indicating the absence of contaminating cellular debris. WB analysis of the EVs confirmed the expression of CD9 and CD81, hallmark surface markers of EVs [46], [47], with chemiluminescent signals on the blot at 24 kDa and 26 kDa, respectively (Fig. 3C). EV particle size distribution ranged from 55 to 80 nm. These findings confirmed the identity as EVs of the particles released from the H69 cells as previous studies [48], [49].
Programmed knockout of huPGRN levels of progranulin in EVs
The programmed mutation induced INDELs at exon 2 of the huPGRN locus lead to reduced levels of progranulin within the protein complement of the EVs. Specifically, we determined the expression of huPGRN extracellular vesicle mRNA (evRNA) and progranulin (evProtein) released by each of three replicate lines of puromycin resistant cells, at passage number 20 in each case. By this point, EVs from ΔhuPGRN-H69 cells included 7.2 ± 2.6% only of the levels of huPGRN transcripts and 10.5 ± 2.0% progranulin compared to EVs from H69 cells, after normalization with GAPDH (Fig. 3D, E) (unpaired t-test, P < 0.0001, both for transcription and protein expression). Expression of progranulin was revealed by WB with an anti-progranulin antibody (target mass, 64 kDa) compared with anti-GAPDH antibody (36 kDa). Levels of the GAPDH, the loading control, were similar both in cell lysates and EV-associated proteins of the parental H69 and daughter ΔhuPGRN cells (Fig. 3E).
CCA-related profiles of transcripts in EVs shed liver fluke granulin-exposed cholangiocytes
To investigate if liver fluke granulin stimulated H69 cells shed EVs containing CCA-related transcripts, the huPGRN gene was mutated by CRISPR/Cas9 knockout (above). As noted, this gene knockout was undertaken in order to minimize levels of endogenous progranulin. The EV-exposed ΔhuPGRN-H69 cells showed similar CCA-related mRNA base line profiles as seen with the CCA-array panel of 88 targets (Bio-Rad) probed with the H69 evRNAs. However, 10 evRNAs were not evident in EVs shed from ΔhuPGRN-H69 cells following gene knockout: dihydropyridine dehydrogenase (DPYD), estrogen receptors (ESR) 1 and 2, lysine-specific demethylase 3A (KDM3A), lymphoid enhancer-binding factor-1 (LEF1), protein tyrosine phosphatase (PRKC9), non-receptor type 13 (PRIN13, a Fas-associated phosphatase), serpin peptidase inhibitor, clade A, member 3 (SERRINA3), serpin peptidase inhibitor, clade E, member 2 (SERRPINE2), and SRY-related HMG-box 11 (SOX11) (Fig. 3H). By contrast, these transcripts were all present in evRNA of H69 cells. DPYD, ESR1, ESR2, KDM3A and SOX11genes participate in pyrimidine catabolism, hormone binding, DNA binding and activation of transcriptions and activate transcription factor, respectively. LEF1encodes a transcription factor that is involved in Wnt signaling. PTPN13 is a member of protein tyrosine phosphatase family, which regulates a variety of cellular processes including oncogenic transformation [50]. The PRINA3 and SERPINA3 genes encode inhibitors of proteases and peptidases. The profiles of evRNA (from three pooled biological replicates) were examined. Following exposure of ΔhuPGRN-H69 to liver fluke granulin, seven transcripts were detected based on the array panel, specifically ESR1, KDM3A, LEF1, PRKC9, PTPN13, SERRPINE2, and SOX11 (Fig. 3F, H, normalized with H69; Fig. 3G, normalized with ΔhuPGRN-H69).
Specific CCA pathways impacted by liver fluke granulin
There were 52 and 43 from 88 CCA-related genes expressed from H69 and ΔhuPGRN-H69 derived-EVs following treatment with liver fluke granulin, respectively. The transcript profiles H69 and ΔhuPGRN-H69 derived-EVs were dissimilar (Fig. 3F, H). For evRNA from H69 cells, elevated levels were induced (4.1- to 14.1-fold changes) for genes encoding the chloride intercellular channel 5 (CLIC5), cannabinoid receptor (CNR1), chemokine (C-X-C-motif) receptor 4 (CXCR4), epidermal growth factor receptor (EGF) family (ERBB2), estrogen pathway (ESR1, ESR2), Frizzled 2-Wnt receptor (FZD7), homeobox A9 (HOXA9), haptoglobin (HP), insulin-like growth factor (IGF), insulin-link growth factor 2 mRNA binding protein 3 (IGF2BP3), zinc finger protein (KDM3A), keratin family (KRT19), transcription factor involved in the Wnt signaling pathway (LEF1), lipoprotein lipase (LPL), MAP kinase family (MAPK13), O6-methylguanine-DNA-methyltransferase (MGMT), natriuretic peptide B (NPPB), phospholipase A2 (PLA2G2A), peptidase inhibitor (SERPINA3), SRY-related HMG-box (SOX 2 and SOX11) family involves in the regulation of embryonic development and in the determination of cell fate (Fig. 3F, H). By comparison, liver fluke granulin activated-ΔhuPGRN-H69 (i.e. without endogenous progranulin) showed elevated levels of transcripts (4.8- to 9.5-fold change) encoding ATP-binding cassette transporter (ABCC1, ABCC4), serine/threonine protein kinase family (CAMK2G), ERBB2 enhancing kinase-mediated activation, F-box protein family (FBXW7), proinflammatory cytokine 17A (IL17A), reductase subunit M1 (RRM1), MAPK13, enzyme responsible for the excision of 8-oxoguanine, enzymes involved in fatty acid biosynthesis (SCD), protein tyrosine phosphatase family involved in cell growth differentiation mitotic cycle and oncogenic transformation (PTPN13), SOX2, SOX1, and zinc finger protein ZNF827 involved in telomere chromosome remodeling. Based on these findings, at least two EV-associated transcripts, ERBB2 and MAPK13 were identified as participants of the MAPK signaling pathway and they exhibited marked fold change (>4) in EVs derived from ΔhuPGRN-H69 cells following activation by liver fluke granulin. These findings concur with a central role hypothesized for the MAPK pathway in liver fluke infection induced-cholangiocarcinogenesis [21], [51].
Human progranulin supplements influence of liver fluke granulin on cholangiocarcinogenesis
Analysis using the CCA PrimePCR Array (Bio-Rad) revealed that 41 of the 55 CCA-related genes induced transcription by two- to four-fold. These 41 genes included actin 1 (ACTA1), breast cancer 1 (BRCA1), epidermal growth factor receptor (EGFR), F-box and WD40 repeat domain containing-7 (FBXW7), MAPK13, mitochondrial ribosomal protein S6 (MRPS6), msh home box 1 (MSX1), metadherin (MTDH), neuron navigator 2 (NAV2), transcription factor (NPAS2), ornithine decarboxylase 1 (ODC1), RRM1, SCD, SERRPINE, TGF-beta signaling family member (SMAD4), tubulin β2α (TUBB2A), ZNF827. Additional genes with elevated levels of transcripts in evRNA (>4-fold change) were noted above. Based on these findings, the endogenous growth factor progranulin appeared to supplement the action of liver fluke granulin in the cholangiocyte in establishing a carcinogenesis-conducive transcriptional profile. This outcome may mimic events during natural infection with O. viverrini where the liver fluke residues within the bile duct, secretes liver fluke granulin that is, in turn, taken up by cholangiocytes [17] and, which contributes to biliary tract inflammation and fibrosis. Cholangiocarcinoma manifests in due course [6], [21], [52].
EVs secreted from liver fluke granulin-activated donor cells transfer CCA-MAPK-related functional mRNAs to naïve cells
To investigate paracrine communication involving transfer of EV-associated mRNAs from donor liver fluke granulin treated-ΔhuPGRN-H69 cells to neighboring (H69 or -ΔhuPGRN-H69 cells), naïve cells were co-cultured with EVs shed by liver fluke granulin-activated ΔhuPGRN-H69. Extracellular vesicles were endocytosed by other cells, as revealed by PKH-26 labelling [53], and delivered their cargo to the recipient cells (Fig. 4A, B). Treatment with Pitstop 2 [40] blocked endocytosis of EVs by H69 (Fig. 4C). H69 cell initiated endocytosis of EVs within 30 min, and endocytosed ∼90% of cell population within 90 min (Fig. 4). CCA-related mRNAs in the cargo of EVs shed by liver fluke granulin-activated ΔhuPGRN-H69 were functional and stimulated changes in gene expression in naïve recipient cholangiocytes. Focusing on MAPK pathway phosphorylation from among these changes in gene expression, we employed a commercial human MAPK phosphorylation array that included 17 kinase phosphorylation sites (Fig. 4D). Following co-culture with the (above) EVs, recipient cells were collected and processed. The levels of seven proteins; GSK3a, MKK3, mTOR, p38, p53, P70S6K and RSK1, were increased significantly in ΔhuPGRN-H69 cells that had been treated with EVs from liver fluke granulin-activated ΔhuPGRN-H69 compared with the control group (Fig. 4D–F) (2way ANOVA, P = 0.0001). Duplicate assays were carried out, each using an identical MAPK phosphorylation array membrane, where each membrane itself included duplicate spots of each target protein in the pathway. The pattern of protein induction was similar in EV-treated H69, except for GSK3a (not significant) (Fig. 4D) (2way ANOVA, P = 0.0001). The indirect amount of each protein was measured with normalized signal intensity (RFU); more of these seven proteins were present in H69 than ΔhuPGRN-H69 cells. For example, MKK3 showed ∼4000 RFU and ∼3700 RFU in the H69 and ΔhuPGRN-H69 control groups, respectively. However, the activation of MAPK phosphorylation by mRNAs from EVs shed by liver fluke granulin-activated ΔhuPGRN-H69 exhibited a similar protein profile. These findings confirmed paracrine signaling among cholangiocytes, which was accomplished by EV-mediated message transfer from liver fluke granulin-exposed cells to naïve cholangiocytes.
Discussion
In East Asia, infection with fish-borne liver flukes is a major risk factor for cholangiocarcinoma (CCA). The liver fluke O. viverrini secretes a growth factor termed liver fluke granulin, Ov-GRN-1. This growth factor is a paralogue of progranulin (PGRN), which is a secreted, cysteine-rich glycoprotein that regulates cell division, survival, motility and migration. Progranulin participates in embryonic development, wound repair, and cancer. Mutations in the human progranulin gene, huPGRN, are associated with a spectrum on neurological disorders [54].
A paralogue from the liver fluke paralogue, liver fluke granulin, stimulates cell proliferation, wound healing, and angiogenesis in vitro and contributes to the pathogenesis of opisthorchiasis in vivo [17], [18], [19], [20], [21], [22]. We have exploited this link infection to explore the role of the secreted liver fluke granulin in pre-malignant lesions by undertaking programmed CRISPR/Cas9 knockout of the Ov-grn-1 gene from the genome of O. viverrini. Infection with liver fluke granulin knockout worms results in markedly reduced disease, which confirmed the key role for this liver fluke granulin in hepatobiliary morbidity during opisthorchiasis [22].
Crosstalk between malignant and neighboring cells contributes to tumor growth [55], [56], [57], [58]. To investigate crosstalk among cholangiocytes during liver fluke infection, here we focused on EV-mediated transfer of mRNAs from cultured cholangiocytes following exposure to liver fluke granulin. To minimize the influence of endogenous progranulin, huPGRN, the cognate gene, was inactivated using CRISPR/Cas9-based gene knock-out. Cholangiocytes of the resulting mutant ΔhuPGRN-H69 lines shown the proliferate rate as same as parental H69 cells (Fig. 2C). Notably, however, cell growth recovered and increased following exposure to liver fluke granulin.
Analysis using a CCA gene array indicated the cooperation of CCA genes with the EVs following exposure to liver fluke granulin, in both the H69 and ΔhuPGRN-H69 cells, highlighting the prominence of signaling pathways in response to intracellular communication. The evRNA expression pattern between H69 cells activated by exposure to liver fluke granulin and, functioning together with human progranulin, induced transcription of CCA-associated mRNAs (41 from 55 genes). By comparison, the evRNA pattern from ΔhuPGRN-H69 (with only minimal levels of progranulin) was substantially different (17 from 55 genes). This finding supported a role for liver fluke granulin during opisthorchiasis-induced carcinogenesis, involving activation of the MAPK pathway [17], [59]. Moreover, functional MAPK evRNAs were transferred to naïve cholangiocytes. Endocytosis of EVs by the H69 and ΔhuPGRN-H69 recipient cells was accomplished within 90 min. Subsequent in vivo translation of MAPK evRNA from EVs shed from liver fluke granulin-activated ΔhuPGRN H69 cells (pooled from 3 biological replicates) was confirmed for GSK3a, MKK3, mTOR, p38, p53, p70S6K and RSK. In overview, EV-mediated crosstalk in response to liver fluke granulin promoted CCA-specific programs including via MAPK signaling that, in turn, may contribute to the CCA-conducive microenvironment.
Cholangiocytes express IL-6 and IL-8 through the TLR4-NF-kB and MAPK signaling pathways [60]. The MAPK pathway is central to cholangiocarcinogenesis activated by Ov-GRN-1 during O. viverrini infection [17], [59]. For CCA development, oncogenic signaling pathways [61] participate at many steps of carcinogenesis [62], [63], [64]. Wnt/β-catenin signaling stimulates the pathobiology associated with CCA [65], with Wnt/β-catenin signaling involved in the inflammation-associated CCA. Suppression of Wnt/β-catenin pathway signaling negatively impacts development of CCA [65]. Apoptosis, growth, cellular intercommunication, and angiogenesis of CCA cells related with receptor tyrosine kinase (RTK) signaling that revealed the multiple kinases associated with PI3K/AKT, Wnt/β-catenin, MAPK, and/or JAK/STAT signaling pathways [66], [67], [68]. Progranulin affects the activation of the MAPK signaling pathway [69], highlighted by the upregulation of huPRGN activity during hepatocellular carcinoma and as the gene target of microRNA-140-3p (miR-140-3p). Overexpression of miR-140-3p disrupts the stimulation in the MAPK signaling pathway by inhibition of expression of progranulin, leading to phosphorylation of ERK and p38. Indeed, in this situation, c-Jun N-terminal kinase is suppressed which, in turn, inhibits migration and invasion by tumor cells [70].
Endogenous progranulin supplemented the action of liver fluke granulin in establishing a carcinogenesis-conducive transcriptional profile. This outcome may mimic events during natural infection with the liver fluke where the parasite residues within the bile duct, secreting liver fluke granulin which, in turn, is taken up by cholangiocytes [17] and which contributes to biliary tract inflammation and fibrosis [6], [21], [52]. Here, transcription levels of the MAPK13 and SOX2 were up regulated following exposure of naïve wild type or the ΔhuPGRN cells to EV-associated RNAs from ΔhuPGRN-H69 cells that had been exposed to liver fluke granulin. By contrast, SOX11 was up regulated in the wild type cells. Perturbation in the MAPK and Wnt signaling that is activated in these cells may reflect the natural history of development of CCA [65] where SOX2 expression is upregulated in the precursor cell and supports self-renewal of the transformed cell [71], [72], [73]. SOX11 plays a role in the tumor cell progression and in maintenance of the protein complex associated with the Wnt and other signaling pathways that mediate angiogenesis [74], [75], [76].
This report has limitations. First, it is currently unclear how to interpret the apparent inconsistency relating to the presence of ERBB2 but not MAPK13 in EVs from both H69 and ΔhuPGRN-H69. However, this might be tractable to investigation by transformation of cholangiocytes with synthetic RNAs encoding these two genes and monitoring MAPK phosphorylation and upregulation of the pathway members. Next, given that this study was undertaken solely in vitro, follow-up studies in the animal models of human opisthorchiasis and infection-induced malignancy should now be planned. Xenografts in immunosuppressed mice may be informative particularly if the positive and negative controls include human CCA cell lines as well as the parental wild type H69 cells are included [77], [78], [79]. In addition, in further support of the hypothesis that liver fluke granulin promotes extracellular vesicle-mediated crosstalk and a cellular microenvironment conducive to CCA, future studies can incorporate three dimensional cell cultures, which have been described with cholangiocytes [80] and/or cholangiocyte organoids [81], [82]. Moreover, studies with tumor registry samples from human cases of liver fluke infection-associated cholangiocarcinoma can be designed to investigate the presence and profiles of EVs and evRNAs from resected tumors [83]. Last, since CCA displays heterogenous morphology and occurs at different sites in the biliary tract [84], [85], investigation of the interaction of liver fluke granulin with discrete hepatic cell types such as hepatocytes and hepatic stellate cells, in addition to cholangiocytes, can likely enhance our understanding of opisthorchiasis-associated bile duct cancer.
Opisthorchiasis appears to induce global epigenetic deregulation in the presence of chronic inflammation, provoked by the mechanical damage inflicted by the feeding and by other activities of the parasite, including secreted factors such as liver fluke granulin, thioredoxin peroxidase and, as shown here, extracellular vesicles that modulate the gene expression profiles [12], [16], [21], [22]. Cholangiocarcinogenesis induced by chronic opisthorchiasis and continuous exposure to liver fluke granulin may act through a RTK signaling, perhaps signaling via ephrin pathway receptor [86], and the interconnections of the MAPK and Wnt/β-catenin pathways. We established a line of the H69 cholangiocyte with markedly diminished levels of endogenous progranulin, using CRISPR/Cas9-mediated programmed gene knockout. The findings presented here with the mutant H69 cell confirmed that EV-mediated cellular communication, induced by exposure of cholangiocytes to liver fluke granulin, modulated conserved signaling pathways in ways reflective of cholangiocarcinogenesis.
Acknowledgements
We thank Drs. Griffin P. Rodgers, NIDDK, National Institutes of Health (NIH) and Yang Liu, Institute of Human Virology, School of Medicine, University of Maryland for support with extracellular vesicle studies and Ion Torrent-based deep sequencing. PA was supported by the Ph.D. program at the Faculty of Medicine, Khon Kaen University and the Thailand Research Fund, Thailand through the Royal Golden Jubilee Ph.D. Program, award number PHD/0111/2557 (PA, TL). We acknowledge support from award R01CA164719 (TL, AL, PJB) from the National Cancer Institute (NCI), NIH, USA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Thailand Research Fund, the NCI or the NIH.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.02.004.
Contributor Information
Paul J. Brindley, Email: pbrindley@gwu.edu.
Wannaporn Ittiprasert, Email: wannaporni@gwu.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Schematic representation of experimental approach and methods, involving the use of intact and gene-edited cholangiocytes to investigate activation by liver fluke granulin and paracrine signaling by extracellular vesicles.
References
- 1.Sirica A.E., Gores G.J., Groopman J.D., Selaru F.M., Strazzabosco M., Wei Wang X., Zhu A.X. Intrahepatic cholangiocarcinoma: continuing challenges and translational advances. Hepatology. 2019;69:1803–1815. doi: 10.1002/hep.30289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan A.S., Dageforde L.A. Cholangiocarcinoma. Surg Clin North Am. 2019;99:315–335. doi: 10.1016/j.suc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Shin H.R., Oh J.K., Masuyer E., Curado M.P., Bouvard V., Fang Y.Y., Wiangnon S., Sripa B., Hong S.T. Epidemiology of cholangiocarcinoma: an update focusing on risk factors. Cancer Sci. 2010;101:579–585. doi: 10.1111/j.1349-7006.2009.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fedorova O.S., Kovshirina Y.V., Kovshirina A.E., Fedotova M.M., Deev I.A., Petrovskiy F.I., Filimonov A.V., Dmitrieva A.I., Kudyakov L.A., Saltykova I.V., Odermatt P., Ogorodova L.M. Opisthorchis felineus infection and cholangiocarcinoma in the Russian Federation: a review of medical statistics. Parasitol Int. 2017;66:365–371. doi: 10.1016/j.parint.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Sripa B., Kaewkes S., Sithithaworn P., Mairiang E., Laha T., Smout M., Pairojkul C., Bhudhisawasdi V., Tesana S., Thinkamrop B., Bethony J.M., Loukas A., Brindley P.J. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007;4 doi: 10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.I.W.G.o.t.E.o.C.R.t. Humans, Biological agents. Volume 100 B. A review of human carcinogens, IARC Monogr Eval Carcinog Risks Hum, 100 (2012) 1–441 [PMC free article] [PubMed]
- 7.Sithithaworn P., Yongvanit P., Duenngai K., Kiatsopit N., Pairojkul C. Roles of liver fluke infection as risk factor for cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2014;21:301–308. doi: 10.1002/jhbp.62. [DOI] [PubMed] [Google Scholar]
- 8.Khan S.A., Tavolari S., Brandi G. Cholangiocarcinoma: epidemiology and risk factors. Liver Int. 2019 doi: 10.1111/liv.14095. [DOI] [PubMed] [Google Scholar]
- 9.Maizels R.M., Smits H.H., McSorley H.J. Modulation of host immunity by helminths: the expanding repertoire of parasite effector molecules. Immunity. 2018;49:801–818. doi: 10.1016/j.immuni.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coakley G., Maizels R.M., Buck A.H. Exosomes and other extracellular vesicles: the new communicators in parasite infections. Trends Parasitol. 2015;31:477–489. doi: 10.1016/j.pt.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcilla A., Trelis M., Cortes A., Sotillo J., Cantalapiedra F., Minguez M.T., Valero M.L., Sanchez del Pino M.M., Munoz-Antoli C., Toledo R., Bernal D. Extracellular vesicles from parasitic helminths contain specific excretory/secretory proteins and are internalized in intestinal host cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulvenna J., Sripa B., Brindley P.J., Gorman J., Jones M.K., Colgrave M.L., Jones A., Nawaratna S., Laha T., Suttiprapa S., Smout M.J., Loukas A. The secreted and surface proteomes of the adult stage of the carcinogenic human liver fluke Opisthorchis viverrini. Proteomics. 2010;10:1063–1078. doi: 10.1002/pmic.200900393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sripa B. Pathobiology of opisthorchiasis: an update. Acta Trop. 2003;88:209–220. doi: 10.1016/j.actatropica.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Chaiyadet S., Smout M., Laha T., Sripa B., Loukas A., Sotillo J. Proteomic characterization of the internalization of Opisthorchis viverrini excretory/secretory products in human cells. Parasitol Int. 2017;66:494–502. doi: 10.1016/j.parint.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaiyadet S., Smout M., Johnson M., Whitchurch C., Turnbull L., Kaewkes S., Sotillo J., Loukas A., Sripa B. Excretory/secretory products of the carcinogenic liver fluke are endocytosed by human cholangiocytes and drive cell proliferation and IL6 production. Int J Parasitol. 2015;45:773–781. doi: 10.1016/j.ijpara.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaiyadet S., Sotillo J., Smout M., Cantacessi C., Jones M.K., Johnson M.S., Turnbull L., Whitchurch C.B., Potriquet J., Laohaviroj M., Mulvenna J., Brindley P.J., Bethony J.M., Laha T., Sripa B., Loukas A. Carcinogenic liver fluke secretes extracellular vesicles that promote cholangiocytes to adopt a tumorigenic phenotype. J Infect Dis. 2015;212:1636–1645. doi: 10.1093/infdis/jiv291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smout M.J., Laha T., Mulvenna J., Sripa B., Suttiprapa S., Jones A., Brindley P.J., Loukas A. A granulin-like growth factor secreted by the carcinogenic liver fluke, Opisthorchis viverrini, promotes proliferation of host cells. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bansal P.S., Smout M.J., Wilson D., Cobos Caceres C., Dastpeyman M., Sotillo J., Seifert J., Brindley P.J., Loukas A., Daly N.L. Development of a potent wound healing agent based on the liver fluke granulin structural fold. J Med Chem. 2017;60:4258–4266. doi: 10.1021/acs.jmedchem.7b00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dastpeyman M., Bansal P.S., Wilson D., Sotillo J., Brindley P.J., Loukas A., Smout M.J., Daly N.L. Structural variants of a liver fluke derived granulin peptide potently stimulate wound healing. J Med Chem. 2018;61:8746–8753. doi: 10.1021/acs.jmedchem.8b00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haugen B., Karinshak S.E., Mann V.H., Popratiloff A., Loukas A., Brindley P.J., Smout M.J. Granulin secreted by the food-borne liver fluke Opisthorchis viverrini promotes angiogenesis in human endothelial cells. Front Med (Lausanne) 2018;5:30. doi: 10.3389/fmed.2018.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smout M.J., Sotillo J., Laha T., Papatpremsiri A., Rinaldi G., Pimenta R.N., Chan L.Y., Johnson M.S., Turnbull L., Whitchurch C.B., Giacomin P.R., Moran C.S., Golledge J., Daly N., Sripa B., Mulvenna J.P., Brindley P.J., Loukas A. Carcinogenic parasite secretes growth factor that accelerates wound healing and potentially promotes neoplasia. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arunsan P., Ittiprasert W., Smout M.J., Cochran C.J., Mann V.H., Chaiyadet S., Karinshak S.E., Sripa B., Young N.D., Sotillo J., Loukas A., Brindley P.J., Laha T. Programmed knockout mutation of liver fluke granulin attenuates virulence of infection-induced hepatobiliary morbidity. Elife. 2019;8 doi: 10.7554/eLife.41463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grubman S.A., Perrone R.D., Lee D.W., Murray S.L., Rogers L.C., Wolkoff L.I., Mulberg A.E., Cherington V., Jefferson D.M. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am J Physiol. 1994;266:G1060–G1070. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- 24.Ninlawan K., O'Hara S.P., Splinter P.L., Yongvanit P., Kaewkes S., Surapaitoon A., LaRusso N.F., Sripa B. Opisthorchis viverrini excretory/secretory products induce toll-like receptor 4 upregulation and production of interleukin 6 and 8 in cholangiocyte. Parasitol Int. 2010;59:616–621. doi: 10.1016/j.parint.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Sa R.O., Zellner M., Grubman M.J. Phylogenetic analysis of segment 10 from African horsesickness virus and cognate genes from other orbiviruses. Virus Res. 1994;33:157–165. doi: 10.1016/0168-1702(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 26.Vara J.A., Portela A., Ortin J., Jimenez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucleic Acids Res. 1986;14:4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ittiprasert W., Mann V.H., Karinshak S.E., Coghlan A., Rinaldi G., Sankaranarayanan G., Chaidee A., Tanno T., Kumkhaek C., Prangtaworn P., Mentink-Kane M.M., Cochran C.J., Driguez P., Holroyd N., Tracey A., Rodpai R., Everts B., Hokke C.H., Hoffmann K.F., Berriman M., Brindley P.J. Programmed genome editing of the omega-1 ribonuclease of the blood fluke, Schistosoma mansoni. Elife. 2019;8 doi: 10.7554/eLife.41337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Telenti A., Pierce L.C., Biggs W.H., di Iulio J., Wong E.H., Fabani M.M., Kirkness E.F., Moustafa A., Shah N., Xie C., Brewerton S.C., Bulsara N., Garner C., Metzker G., Sandoval E., Perkins B.A., Och F.J., Turpaz Y., Venter J.C. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci USA. 2016;113:11901–11906. doi: 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canver M.C., Haeussler M., Bauer D.E., Orkin S.H., Sanjana N.E., Shalem O., Yuan G.C., Zhang F., Concordet J.P., Pinello L. Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments. Nat Protoc. 2018;13:946–986. doi: 10.1038/nprot.2018.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthis N.J., Clore G.M. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013;22:851–858. doi: 10.1002/pro.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ke N., Wang X., Xu X., Abassi Y.A. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol Biol. 2011;740:33–43. doi: 10.1007/978-1-61779-108-6_6. [DOI] [PubMed] [Google Scholar]
- 33.Santos J.C., Lima N.D.S., Sarian L.O., Matheu A., Ribeiro M.L., Derchain S.F.M. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep. 2018;8:829. doi: 10.1038/s41598-018-19339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shenoy G.N., Loyall J., Berenson C.S., Kelleher R.J., Jr., Iyer V., Balu-Iyer S.V., Odunsi K., Bankert R.B. Sialic acid-dependent inhibition of T cells by exosomal ganglioside GD3 in ovarian tumor microenvironments. J Immunol. 2018;201:3750–3758. doi: 10.4049/jimmunol.1801041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.Garnier D., Magnus N., Lee T.H., Bentley V., Meehan B., Milsom C., Montermini L., Kislinger T., Rak J. Cancer cells induced to express mesenchymal phenotype release exosome-like extracellular vesicles carrying tissue factor. J Biol Chem. 2012;287:43565–43572. doi: 10.1074/jbc.M112.401760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puzar Dominkus P., Stenovec M., Sitar S., Lasic E., Zorec R., Plemenitas A., Zagar E., Kreft M., Lenassi M. PKH26 labeling of extracellular vesicles: characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim Biophys Acta Biomembr. 2018;1860:1350–1361. doi: 10.1016/j.bbamem.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Tario J.D., Jr., Humphrey K., Bantly A.D., Muirhead K.A., Moore J.S., Wallace P.K. Optimized staining and proliferation modeling methods for cell division monitoring using cell tracking dyes. J Vis Exp. 2012 doi: 10.3791/4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Kleist L., Stahlschmidt W., Bulut H., Gromova K., Puchkov D., Robertson M.J., MacGregor K.A., Tomilin N., Pechstein A., Chau N., Chircop M., Sakoff J., von Kries J.P., Saenger W., Krausslich H.G., Shupliakov O., Robinson P.J., McCluskey A., Haucke V. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell. 2011;146:471–484. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 41.Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R.L., Martin J.F. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J.C., Lee I.N., Huang C., Wu Y.P., Chung C.Y., Lee M.H., Lin M.H., Yang J.T. Valproic acid-induced amphiregulin secretion confers resistance to temozolomide treatment in human glioma cells. BMC Cancer. 2019;19:756. doi: 10.1186/s12885-019-5843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang T.W., Lin K.F., Lee C.H., Chang H., Lee S.C., Shieh Y.S. The role of thyroid transcription factor-1 and tumor differentiation in resected lung adenocarcinoma. Sci Rep. 2017;7:14222. doi: 10.1038/s41598-017-14651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018;563:646–651. doi: 10.1038/s41586-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smout M.J., Mulvenna J.P., Jones M.K., Loukas A. Expression, refolding and purification of Ov-GRN-1, a granulin-like growth factor from the carcinogenic liver fluke, that causes proliferation of mammalian host cells. Protein Expression Purif. 2011;79:263–270. doi: 10.1016/j.pep.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 46.van Niel G., D'Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 47.Eichenberger R.M., Talukder M.H., Field M.A., Wangchuk P., Giacomin P., Loukas A., Sotillo J. Characterization of Trichuris muris secreted proteins and extracellular vesicles provides new insights into host-parasite communication. J Extracell Vesicles. 2018;7:1428004. doi: 10.1080/20013078.2018.1428004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato K., Meng F., Venter J., Giang T., Glaser S., Alpini G. Author Correction: The role of the secretin/secretin receptor axis in inflammatory cholangiocyte communication via extracellular vesicles. Sci Rep. 2018;8:11238. doi: 10.1038/s41598-018-29609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato K., Meng F., Venter J., Giang T., Glaser S., Alpini G. The role of the secretin/secretin receptor axis in inflammatory cholangiocyte communication via extracellular vesicles. Sci Rep. 2017;7:11183. doi: 10.1038/s41598-017-10694-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Revillion F., Puech C., Rabenoelina F., Chalbos D., Peyrat J.P., Freiss G. Expression of the putative tumor suppressor gene PTPN13/PTPL1 is an independent prognostic marker for overall survival in breast cancer. Int J Cancer. 2009;124:638–643. doi: 10.1002/ijc.23989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jusakul A., Cutcutache I., Yong C.H., Lim J.Q., Huang M.N., Padmanabhan N., Nellore V., Kongpetch S., Ng A.W.T., Ng L.M., Choo S.P., Myint S.S., Thanan R., Nagarajan S., Lim W.K., Ng C.C.Y., Boot A., Liu M., Ong C.K., Rajasegaran V., Lie S., Lim A.S.T., Lim T.H., Tan J., Loh J.L., McPherson J.R., Khuntikeo N., Bhudhisawasdi V., Yongvanit P., Wongkham S., Totoki Y., Nakamura H., Arai Y., Yamasaki S., Chow P.K., Chung A.Y.F., Ooi L., Lim K.H., Dima S., Duda D.G., Popescu I., Broet P., Hsieh S.Y., Yu M.C., Scarpa A., Lai J., Luo D.X., Carvalho A.L., Vettore A.L., Rhee H., Park Y.N., Alexandrov L.B., Gordan R., Rozen S.G., Shibata T., Pairojkul C., Teh B.T., Tan P. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7:1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brindley P.J., Loukas A. Helminth infection-induced malignancy. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu L., Wang J., Zhou X., Xiong Z., Zhao J., Yu R., Huang F., Zhang H., Chen L. Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Sci Rep. 2016;6:32993. doi: 10.1038/srep32993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bateman A., Cheung S.T., Bennett H.P.J. A brief overview of progranulin in health and disease. Methods Mol Biol. 1806;2018:3–15. doi: 10.1007/978-1-4939-8559-3_1. [DOI] [PubMed] [Google Scholar]
- 55.Oliveira A.I., Anjo S.I., Vieira de Castro J., Serra S.C., Salgado A.J., Manadas B., Costa B.M. Crosstalk between glial and glioblastoma cells triggers the “go-or-grow” phenotype of tumor cells. Cell Commun Signal. 2017;15:37. doi: 10.1186/s12964-017-0194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rasanen K., Herlyn M. Paracrine signaling between carcinoma cells and mesenchymal stem cells generates cancer stem cell niche via epithelial-mesenchymal transition. Cancer Discov. 2012;2:775–777. doi: 10.1158/2159-8290.CD-12-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailey J.M., Mohr A.M., Hollingsworth M.A. Sonic hedgehog paracrine signaling regulates metastasis and lymphangiogenesis in pancreatic cancer. Oncogene. 2009;28:3513–3525. doi: 10.1038/onc.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jayatilaka H., Tyle P., Chen J.J., Kwak M., Ju J., Kim H.J., Lee J.S.H., Wu P.H., Gilkes D.M., Fan R., Wirtz D. Synergistic IL-6 and IL-8 paracrine signalling pathway infers a strategy to inhibit tumour cell migration. Nat Commun. 2017;8:15584. doi: 10.1038/ncomms15584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Upontain S., Sereerak P., Laha T., Sripa B., Tangkawatana P., Brindley P.J., Tangkawatana S. Granulin expression in hamsters during Opisthorchis viverrini infection-induced cholangiocarcinogenesis. Asian Pac J Cancer Prev. 2018;19:2437–2445. doi: 10.22034/APJCP.2018.19.9.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yokoyama T., Komori A., Nakamura M., Takii Y., Kamihira T., Shimoda S., Mori T., Fujiwara S., Koyabu M., Taniguchi K., Fujioka H., Migita K., Yatsuhashi H., Ishibashi H. Human intrahepatic biliary epithelial cells function in innate immunity by producing IL-6 and IL-8 via the TLR4-NF-kappaB and -MAPK signaling pathways. Liver Int. 2006;26:467–476. doi: 10.1111/j.1478-3231.2006.01254.x. [DOI] [PubMed] [Google Scholar]
- 61.Kiguchi K., Carbajal S., Chan K., Beltran L., Ruffino L., Shen J., Matsumoto T., Yoshimi N., DiGiovanni J. Constitutive expression of ErbB-2 in gallbladder epithelium results in development of adenocarcinoma. Cancer Res. 2001;61:6971–6976. [PubMed] [Google Scholar]
- 62.Andersen J.B. Molecular pathogenesis of intrahepatic cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2015;22:101–113. doi: 10.1002/jhbp.155. [DOI] [PubMed] [Google Scholar]
- 63.Churi C.R., Shroff R., Wang Y., Rashid A., Kang H.C., Weatherly J., Zuo M., Zinner R., Hong D., Meric-Bernstam F., Janku F., Crane C.H., Mishra L., Vauthey J.N., Wolff R.A., Mills G., Javle M. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ross J.S., Wang K., Gay L., Al-Rohil R., Rand J.V., Jones D.M., Lee H.J., Sheehan C.E., Otto G.A., Palmer G., Yelensky R., Lipson D., Morosini D., Hawryluk M., Catenacci D.V., Miller V.A., Churi C., Ali S., Stephens P.J. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014;19:235–242. doi: 10.1634/theoncologist.2013-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loilome W., Bungkanjana P., Techasen A., Namwat N., Yongvanit P., Puapairoj A., Khuntikeo N., Riggins G.J. Activated macrophages promote Wnt/beta-catenin signaling in cholangiocarcinoma cells. Tumour Biol. 2014;35:5357–5367. doi: 10.1007/s13277-014-1698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dokduang H., Juntana S., Techasen A., Namwat N., Yongvanit P., Khuntikeo N., Riggins G.J., Loilome W. Survey of activated kinase proteins reveals potential targets for cholangiocarcinoma treatment. Tumour Biol. 2013;34:3519–3528. doi: 10.1007/s13277-013-0930-9. [DOI] [PubMed] [Google Scholar]
- 67.Loilome W., Juntana S., Namwat N., Bhudhisawasdi V., Puapairoj A., Sripa B., Miwa M., Saya H., Riggins G.J., Yongvanit P. PRKAR1A is overexpressed and represents a possible therapeutic target in human cholangiocarcinoma. Int J Cancer. 2011;129:34–44. doi: 10.1002/ijc.25646. [DOI] [PubMed] [Google Scholar]
- 68.Yothaisong S., Thanee M., Namwat N., Yongvanit P., Boonmars T., Puapairoj A., Loilome W. Opisthorchis viverrini infection activates the PI3K/ AKT/PTEN and Wnt/beta-catenin signaling pathways in a Cholangiocarcinogenesis model. Asian Pac J Cancer Prev. 2014;15:10463–10468. doi: 10.7314/apjcp.2014.15.23.10463. [DOI] [PubMed] [Google Scholar]
- 69.Chen B.H., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Q.Y., Men C.J., Ding X.W. Upregulation of microRNA-140-3p inhibits epithelial-mesenchymal transition, invasion, and metastasis of hepatocellular carcinoma through inactivation of the MAPK signaling pathway by targeting GRN. J Cell Biochem. 2019 doi: 10.1002/jcb.28750. [DOI] [PubMed] [Google Scholar]
- 71.Engel N.W., Neumann J.E., Ahlfeld J., Wefers A.K., Merk D.J., Ohli J., Schuller U. Canonical Wnt signaling drives tumor-like lesions from Sox2-positive precursors of the murine olfactory epithelium. PLoS One. 2016;11 doi: 10.1371/journal.pone.0166690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schnerch A., Cerdan C., Bhatia M. Distinguishing between mouse and human pluripotent stem cell regulation: the best laid plans of mice and men. Stem Cells. 2010;28:419–430. doi: 10.1002/stem.298. [DOI] [PubMed] [Google Scholar]
- 73.Sui L., Bouwens L., Mfopou J.K. Signaling pathways during maintenance and definitive endoderm differentiation of embryonic stem cells. Int J Dev Biol. 2013;57:1–12. doi: 10.1387/ijdb.120115ls. [DOI] [PubMed] [Google Scholar]
- 74.Brennan D.J., Ek S., Doyle E., Drew T., Foley M., Flannelly G., O'Connor D.P., Gallagher W.M., Kilpinen S., Kallioniemi O.P., Jirstrom K., O'Herlihy C., Borrebaeck C.A. The transcription factor Sox11 is a prognostic factor for improved recurrence-free survival in epithelial ovarian cancer. Eur J Cancer. 2009;45:1510–1517. doi: 10.1016/j.ejca.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 75.Kuci V., Nordstrom L., Conrotto P., Ek S. SOX11 and HIG-2 are cross-regulated and affect growth in mantle cell lymphoma. Leuk Lymphoma. 2016;57:1883–1892. doi: 10.3109/10428194.2015.1121257. [DOI] [PubMed] [Google Scholar]
- 76.Kuci V., Nordstrom L., Jerkeman M., Ek S. Emerging role of SOX11 in mantle cell lymphoma. Blood Lymphat Cancer: Targets Therapy. 2015;2015:35–42. [Google Scholar]
- 77.Obchoei S., Weakley S.M., Wongkham S., Wongkham C., Sawanyawisuth K., Yao Q., Chen C. Cyclophilin A enhances cell proliferation and tumor growth of liver fluke-associated cholangiocarcinoma. Mol Cancer. 2011;10:102. doi: 10.1186/1476-4598-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loeuillard E., Fischbach S.R., Gores G.J., Rizvi S. Animal models of cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 1865;2019:982–992. doi: 10.1016/j.bbadis.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vaeteewoottacharn K., Pairojkul C., Kariya R., Muisuk K., Imtawil K., Chamgramol Y., Bhudhisawasdi V., Khuntikeo N., Pugkhem A., Saeseow O.T., Silsirivanit A., Wongkham C., Wongkham S., Okada S. Establishment of highly transplantable cholangiocarcinoma cell lines from a patient-derived xenograft mouse model. Cells. 2019;8 doi: 10.3390/cells8050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Won J., Cho Y., Lee D., Jeon B.Y., Ju J.W., Chung S., Pak J.H. Clonorchis sinensis excretory-secretory products increase malignant characteristics of cholangiocarcinoma cells in three-dimensional co-culture with biliary ductal plates. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tysoe O.C., Justin A.W., Brevini T., Chen S.E., Mahbubani K.T., Frank A.K., Zedira H., Melum E., Saeb-Parsy K., Markaki A.E., Vallier L., Sampaziotis F. Isolation and propagation of primary human cholangiocyte organoids for the generation of bioengineered biliary tissue. Nat Protoc. 2019;14:1884–1925. doi: 10.1038/s41596-019-0168-0. [DOI] [PubMed] [Google Scholar]
- 82.Shiota J., Zaki N.H.M., Merchant J.L., Samuelson L.C., Razumilava N. Generation of organoids from mouse extrahepatic Bile Ducts. J Vis Exp. 2019 doi: 10.3791/59544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chng K.R., Chan S.H., Ng A.H.Q., Li C., Jusakul A., Bertrand D., Wilm A., Choo S.P., Tan D.M.Y., Lim K.H., Soetinko R., Ong C.K., Duda D.G., Dima S., Popescu I., Wongkham C., Feng Z., Yeoh K.G., Teh B.T., Yongvanit P., Wongkham S., Bhudhisawasdi V., Khuntikeo N., Tan P., Pairojkul C., Ngeow J., Nagarajan N. Tissue microbiome profiling identifies an enrichment of specific enteric bacteria in Opisthorchis viverrini associated cholangiocarcinoma. EBioMedicine. 2016;8:195–202. doi: 10.1016/j.ebiom.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banales J.M., Cardinale V., Carpino G., Marzioni M., Andersen J.B., Invernizzi P., Lind G.E., Folseraas T., Forbes S.J., Fouassier L., Geier A., Calvisi D.F., Mertens J.C., Trauner M., Benedetti A., Maroni L., Vaquero J., Macias R.I., Raggi C., Perugorria M.J., Gaudio E., Boberg K.M., Marin J.J., Alvaro D. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA) Nat Rev Gastroenterol Hepatol. 2016;13:261–280. doi: 10.1038/nrgastro.2016.51. [DOI] [PubMed] [Google Scholar]
- 85.Louis C., Papoutsoglou P., Coulouarn C. Molecular classification of cholangiocarcinoma. Curr Opin Gastroenterol. 2020;36:57–62. doi: 10.1097/MOG.0000000000000611. [DOI] [PubMed] [Google Scholar]
- 86.Neill T., Buraschi S., Goyal A., Sharpe C., Natkanski E., Schaefer L., Morrione A., Iozzo R.V. EphA2 is a functional receptor for the growth factor progranulin. J Cell Biol. 2016;215:687–703. doi: 10.1083/jcb.201603079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of experimental approach and methods, involving the use of intact and gene-edited cholangiocytes to investigate activation by liver fluke granulin and paracrine signaling by extracellular vesicles.