Abstract
Background
Multiple sclerosis (MS) is a complex disease in which the immune system attacks the central nervous system. The molecular mechanisms contributing to the etiology of MS remain poorly understood. Genome-wide association studies (GWAS) of MS have identified a small number of genetic loci significant at the genome level, but they are mainly non-coding variants. Network-assisted analysis may help better interpret the functional roles of the variants with association signals and potential translational medicine application. The Dense Module Searching of GWAS tool (dmGWAS version 2.4) developed in our team is applied to 2 MS GWAS datasets (GeneMSA and IMSGC GWAS) using the human protein interactome as the reference network. A dual evaluation strategy is used to generate results with reproducibility.
Results
Approximately 7500 significant network modules were identified for each independent GWAS dataset, and 20 significant modules were identified from the dual evaluation. The top modules included GRB2, HDAC1, JAK2, MAPK1, and STAT3 as central genes. Top module genes were enriched with functional terms such as “regulation of glial cell differentiation” (adjusted p-value = 2.58 × 10− 3), “T-cell costimulation” (adjusted p-value = 2.11 × 10− 6) and “virus receptor activity” (adjusted p-value = 1.67 × 10− 3). Interestingly, top gene networks included several MS FDA approved drug target genes HDAC1, IL2RA, KEAP1, and RELA,
Conclusions
Our dmGWAS network analyses highlighted several genes (GRB2, HDAC1, IL2RA, JAK2, KEAP1, MAPK1, RELA and STAT3) in top modules that are promising to interpret GWAS signals and link to MS drug targets. The genes enriched with glial cell differentiation are important for understanding neurodegenerative processes in MS and for remyelination therapy investigation. Importantly, our identified genetic signals enriched in T cell costimulation and viral receptor activity supported the viral infection onset hypothesis for MS.
Keywords: GWAS, Multiple sclerosis, dmGWAS, Network module, Gene set enrichment analysis, Drug target
Background
Multiple sclerosis (MS), a chronic and disabling disorder of the central nervous system (CNS), affects nearly 1 million individuals in the United States [1]. MS is characterized by inflammation, demyelination, defective remyelination and neuronal injury that may be extensive, leading to permanent and progressive disability. MS is typically diagnosed between 20 and 40 years of age, and is nearly three times more frequent in women than in men [1, 2]. The cause of MS remains unknown, and it is unclear if its etiology may even vary from patient to patient. MS is thought to occur in genetically predisposed individuals with potential trigger, or at least contribution of environmental factors [3]. One hypothesis involves a yet unknown CNS viral infection triggering T-cell auto-reactivity through a mechanism of molecular mimicry [4]. In light of the high prevalence of the disease and its poorly elucidated mechanism, it is of importance to further study MS pathogenesis.
Studying genetic factors associated with MS may aid in understanding its pathophysiology. The realization that MS risk is significantly higher for family members of affected individuals has prompted a search for genetic predisposing factors [5]. Early genetic studies of MS developed genetic mapping for families affected with MS, which identified human leukocyte antigen (HLA) regions to be associated with MS [5, 6]. Due to limitations of sample size, these early studies were ineffective in validating other genetic risk factors of MS [5]. Pivotally, genome-wide association studies (GWAS), which during recent years have aided in acquiring vast repositories of genetic variations associated with specific phenotypes, were performed in large MS populations [7]. As GWAS were performed for larger MS sample sizes than original genetic mapping studies, several genetic variations, or single nucleotide polymorphisms (SNPs), outside of the HLA region became attributed to MS [8, 9]. Some of these MS GWAS studies are publicly available in the form of GWAS summary statistics datasets, which summarize SNP level association p values in the specific population or subpopulation (i.e., without individual level genotype information to the public) [7]. In this work, two publicly available MS GWAS summary statistics datasets are independently and conjunctively further studied: a MS GWA study performed by the Genetic Multiple Sclerosis Association (GeneMSA) consortium in 2009 and a MS GWA study performed by the International Multiple Sclerosis Genetics Consortium (IMSGC) in 2011 [8, 9]. Both of these GWA studies reported some SNPs with genome-wide significance levels, but these variants are largely in the non-coding regions with unknown function, making biological interpretation difficult.
Although GWAS have come a long way in elucidating genetic variations associated with specific traits, integrating GWAS data with other kinds of datasets can aid in understanding the molecular underpinnings behind these associations [10, 11]. Additionally, differing genotyping platforms used by GWAS and other factors (ethnic background, sample size, statistical tests) can make it difficult to perform meta-analytical studies, but network-based analysis is a promising approach to detect combinatory association signals in network modules. This study aims to apply network approach to find joint association signals at the network modules, and thus, leading to biologically interpretable results [12, 13]. A new version of the dense module searching algorithm (dmGWAS) is used to integrate GWAS signals with a comprehensive human protein-protein interaction (PPI) network so that MS candidate subnetworks can be identified [11]. Uniquely, dense module searching was performed independently on the 2 MS GWAS studies mentioned above. Lastly, this study also aims to jointly examine independent MS GWAS modules by using the dual evaluation function of the dmGWAS package. This evaluation strategy would enhance the results in a reproducible fashion, since the replication rate is often low in GWA studies of complex disease.
Materials and methods
Obtaining gene-level p-values by Pascal
The GeneMSA GWAS and IMSGC GWAS summary statistics data were respectively accessed through the public databases dbGaP and GWAS Catalogue [14, 15]. The human PPI network was downloaded from the public Human Protein Reference Database (HPRD) [16], as demonstrated in [17, 18]. The GeneMSA GWAS data was collected by the Sentrix HumanHap 550-BeadChip genotyping platform, and the IMSGC GWAS data was collected by the Human660-Quad chip genotyping platform [8, 9] (Additional file 1: Table S1).
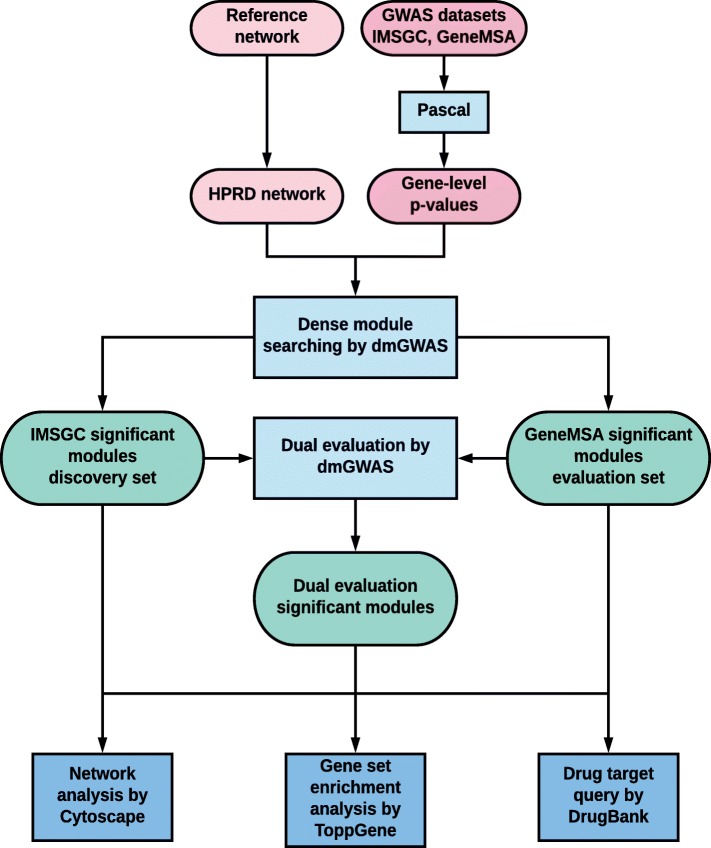
Summary statistics data is presented in the form of SNP-level association p-values. In this study, gene-level p-values (pg) are needed as input for dense module searching by dmGWAS. To obtain pg measures, a pathway scoring algorithm (Pascal) was used, which considers combined SNP-level effects (Fig. 1) [19]. The pg values were obtained with default Pascal settings, which uses the sum-of-chi-squares (SOCS) test and considers the gene region along with 50 kilobase pairs upstream/downstream of the gene region [19]. Pascal pg measurements were obtained independently for each MS GWAS dataset examined. We excluded those genes whose pg was smaller than 10− 12 because they would likely be dominant in the top modules and disrupt our dense module searching process.
Fig. 1.
Pipeline for dmGWAS implementation and gene network assessment. Details are provided in the Materials and methods section
Dense module searching (DMS) and dual evaluation by dmGWAS
Dense module searching was performed with the DMS function of dmGWAS (version 2.4) Linux binary R package that is available at [20]. This function calculates each pg value as a node weight and scores each subgraph based on a quantitative summary of all the weights in each module [11]. This function was executed for each MS GWAS dataset independently. The outcome of DMS is in the form of an RData results file which contains modules with significant z-scores (Zm), where each module is named by its seed gene. Zm measurements by dmGWAS are acquired by
where zi was transferred from pg following the normal distribution function, and k represents the number of genes present within each module [11]. The top modules with the highest Zm values were considered for our independent network analysis and validation.
The dual evaluation function of dmGWAS (dualEval) takes into consideration two module output lists (each from a GWAS dataset in this analysis) from DMS function and performs dense module searching based on the two module lists (Fig. 1) [11]. This was used to assess the overall results of dense module searching from both GeneMSA GWAS and the IMSGC GWAS datasets. Specifically, dual evaluation was performed by using one GWAS dataset (e.g., IMSGC) as the discovery set and the other (e.g., GeneMSA) as the evaluation set, and the consistent results will be reported. Because the IMSGC GWAS had a larger sample size and stronger association signals, we used its DMS modules for the discovery (Additional file 2: Figure S1) and used the DMS modules from GeneMSA GWAS dataset to evaluate IMSGC modules. After implementation of the dualEval function by dmGWAS, significant modules from discovery set were merged.
Network visualization and validation
The software Cytoscape was used for network visualization. Cytoscape is a popular open source tool commonly used for analysis of biomedical networks [21]. Nodes and edges derived from the module results by running dmGWAS version 2.4 were imported into Cytoscape. The NetworkAnalyzer tool, a built-in Cytoscape tool that measures a variety of network parameters, was used to assess the resulting networks [21]. The measure of betweenness and centrality (Betweenness) was used to measure the functional importance of the nodes in top modules. The betweenness value, computed by the Brandes fast algorithm, highlights nodes that join communities of dense subnetworks together [22–24]. Brandes fast algorithm computes betweenness by Freeman’s general formula:
where V is the set of vertices, s and t represent vertices such that σst can denote a path from s to t, and σst(v) can denote the number of shortest paths from s to t that vertex v lies on [23, 25].
Functional enrichment analysis by ToppGene Suite [26] was performed for validation of our resulting MS gene networks. ToppGene (also known as ToppCluster) is a web-based tool that performs enrichment analysis based on annotations of experimentally validated categorical data about human genes [26]. Annotations from the Gene Ontology (GO) database were reported from our analysis [27]. Genes within top 10 modules were considered as the input for each functional enrichment analysis performed on individual GWAS data sets. The complete discovery set of genes from significant dual evaluation modules were considered for the functional enrichment analysis of the dual evaluation results. Calculations of significance level by ToppGene Suite was done by a probability density function [26]. Multiple testing correction was conducted using the Benjamini-Hochberg method for all annotation features, which provided adjusted p-values for enrichment results [28]. A functional enrichment annotation limit of 100 genes was chosen per annotation report, as all gene lists ranged from 26 to 56 genes.
A drug target search within the DrugBank database was performed for further evaluation of the translational potential of the resulting gene networks. DrugBank is a public online database, which contains rich biochemical information about drugs and their gene targets [29]. The DrugBank database was queried for MS FDA approved drugs and their pertaining drug targets. The searched drug targets were then compared to our resulting top 1 % modules and dual evaluation discovery modules. Overlapping drug targets and genes were then reported, along with corresponding pg values.
Results
Multiple sclerosis modules identified by dmGWAS
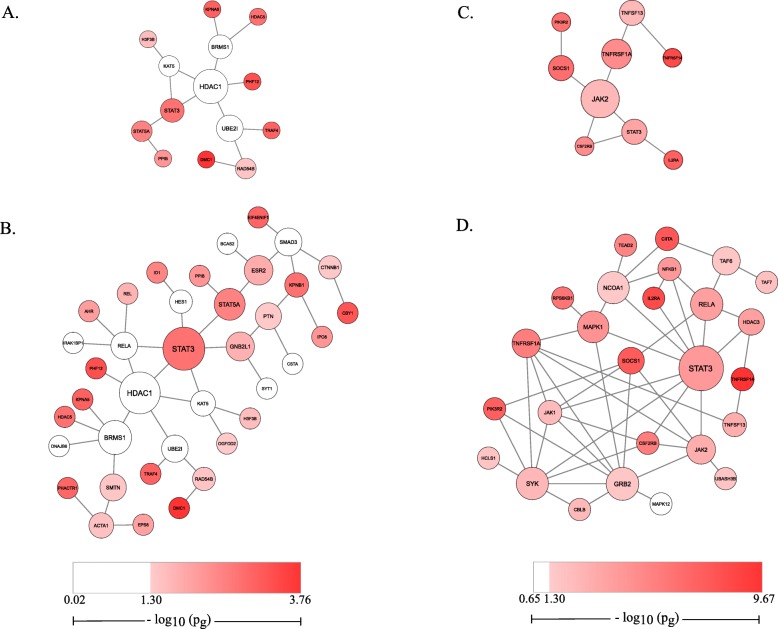
We used dmGWAS tool to search the gene network modules with enriched signals in 2 MS GWAS datasets (Fig. 1). We identified 7458 network modules from GeneMSA GWAS data and 7566 modules from IMSGC GWAS data. We first examined those modules from GeneMSA data. The top module (the highest scored module) included 14 genes and had a normalized Zm score 7.73 and module p-value (pm) of 1.04 × 10− 14. This top module had the central node HDAC1 with a Betweenness measure of 0.76 and a gene-level p-value (pg) of 0.26 (Fig. 2a). The most significant genes within this top module included PHF12 (pg = 4.96 × 10− 4), which directly connected to central node HDAC1, and DMC1 (pg = 4.96 × 10− 4). Note that a network module with enriched association signals does not require each node to have significant pg value; and this is one of the advantages to search a set of genes, each of which has weak or moderate signal, but their interaction contribute to a strong, combinatory signal. Among the 7458 modules identified from the GeneMSA GWAS dataset, the top 10 modules contained 37 genes. All top 10 modules had a normalized Zm greater than 7.66 and a pm less than 2.04 × 10− 14 (values pertaining to 10th top module). Cytoscape visualization of the merged GeneMSA top 10 modules (Fig. 2b) contained the central genes STAT3 (pg = 1.81 × 10− 3, Betweenness = 0.574) and HDAC1 (pg = 0.26, Betweenness = 0.54). Other central genes in GeneMSA top 10 modules included STAT5A (pg = 3.25 × 10− 3, Betweenness = 0.28), ESR2 (pg = 0.02, Betweenness = 0.22) and RELA (pg = 0.08, Betweenness = 0.16). Next, we merged the top 1 % modules (75 modules) to form a network. It contained 144 genes and presented GNB2L1 (pg = 0.02, Betweenness = 0.22), SRC (pg = 0.03, Betweenness = 0.22) and STAT3 (pg = 1.81 × 10− 3, Betweenness = 0.20) as most central (Additional file 3: Figure S2).
Fig. 2.
Top modules associated to GeneMSA GWAS and IMSGC GWAS. The top module of GeneMSA GWAS displays HDAC1 as central (a). The top 10 modules of GeneMSA were merged (b). The top module of IMSGC GWAS shows JAK2 as central (c). The top 10 modules of IMSGC were merged (d). Large nodes represent central genes and small nodes represent less central genes (based on measure of Betweenness). Colors of nodes represent Pascal reported gene-level p-values as specified by legend
The top module from the IMSGC GWAS dataset had a normalized Zm value 11.13 (pm < 2.20 × 10− 16) and contained 9 genes. JAK2 was highly central (Betweenness = 0.75) within this top network with a pg of 1.25 × 10− 3 (Fig. 2c). Highly significant genes within this top module were TNFRSF14 (pg = 2.16 × 10− 10) and IL2RA (pg = 4.37 × 10− 9). Among the 7566 total modules identified from the IMSGC GWAS data, the top 10 modules contained 37 non-redundant genes. All these top 10 modules had Zm values greater than 10.89 (pm < 2.20 × 10− 16). When these top 10 modules were merged and evaluated by Cytoscape, the gene STAT3 (pg = 8.36 × 10− 5, Betweenness = 0.348) appeared central in this network (Fig. 2d). Other central genes in these top 10 modules included GRB2 (pg = 0.02, Betweenness = 0.16), RELA (pg = 1.84 × 10− 4, Betweenness = 0.16), SYK (pg = 3.41 × 10− 3, Betweenness = 0.16) and NCOA1 (pg = 0.03, Betweenness = 0.15). Next, we merged top 1 % modules (76 modules). The merged network included 123 genes and showed two highly central genes: GRB2 (pg = 0.02, Betweenness = 0.31) and EGFR (Pg = 0.07, Betweenness = 0.30) (Additional file 4: Figure S3).
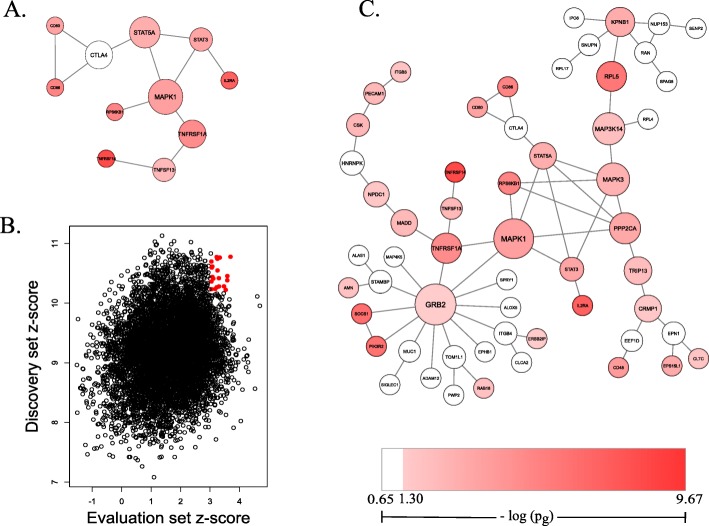
We next reported the dual evaluation results by using IMSGC GWAS as the discovery dataset and GeneMSA GWAS as the evaluation dataset. The top module (Fig. 3a) contained 11 genes and exhibited a normalized Zm value 10.78 (pm < 2.20 × 10− 16). This top module contained MAPK1 (pg = 3.40 × 10− 5) as a central node (Betweenness = 0.60). Two genes, TNFRSF14 (pg = 2.16 × 10− 10) and IL2RA (pg = 4.37 × 10− 9), were highly significant and observed in the dual evaluation top module. In addition, dual evaluation of GeneMSA (evaluation) and IMSGC (discovery) datasets identified 20 significant modules based on discovery and evaluation Zm values (Fig. 3b). All the 20 significant modules from the dual evaluation discovery set contained 56 non-redundant genes (Fig. 3c). The genes GRB2 (pg = 0.02, Betweenness = 0.54) and MAPK1 (pg = 3.40 × 10− 5, Betweenness = 0.51) were the most central genes in the merged network. Other central genes in this dual evaluation gene set included MAPK3 (pg = 5.07 × 10− 4, Betweenness = 0.33), MAP3K14 (pg = 3.78 × 10− 3, Betweenness = 0.31) and PPP2CA (pg = 1.18 × 10− 4, Betweenness = 0.29).
Fig. 3.
Significant modules of dual evaluation regarding IMSGC GWAS as discovery set. The top module of dual evaluation shows MAPK1 as central (a). Modules are regarded as significant based on normalized evaluation and discovery z-scores. Significant modules are represented in red (b). The 20 significant modules were merged to show overall gene network (n = 56) (c). Large nodes represent central genes and small nodes represent less central genes (based on measure of Betweenness). Colors of nodes represent Pascal reported gene-level p-values as specified by legend
Gene set enrichment analysis
We first reported the functional enrichment analysis of gene list (n = 37 genes) from the top 10 modules of GeneMSA GWAS dataset (Additional file 5: Table S2). Interestingly, the most significantly enriched GO Molecular Function terms are all involved in the process of transcription regulation. These transcription regulation functions included “repressing transcription factor binding” (adjusted p-value = 2.79 × 10− 7) as the most enriched GO Molecular Function term, with six contributing genes: CTNNB1, HDAC1, HDAC5, KAT5, RELA, and STAT3. Among the top five enriched GO Biological Process terms, “regulation of glial cell differentiation” was enriched with an adjusted p-value of 2.58 × 10− 3. Contributing genes within the glial cell differentiation process included CTNNB1, HDAC1, HES1 and RELA. Next, we reported the top five GO Cellular Component enriched terms. The second most significant one is “histone deacetylase complex” (adjusted p-value = 3.43 × 10− 4) with four contributing genes: BRMS1, HDAC1, HDAC5 and PHF12. Lastly, the GO Cellular Component “I-kappaB/NF-kappaB complex” (adjusted p-value = 3.53 × 10− 3) was also among the top five enriched for GeneMSA, which included the input genes REL and RELA.
We next performed gene set enrichment analysis of the genes (n = 26 genes) from the top 10 modules from the IMSGC GWAS dataset (Additional file 6: Table S3). Among the enriched GO Molecular Function terms, the top term was “phosphotyrosine residue binding” with an adjusted p-value of 2.74 × 10− 4. This term was identified by the input genes CBLB, GRB2 and MAPK1. Other top enriched GO Molecular Function terms of IMSGC included “protein phosphorylated amino acid binding” (adjusted p-value = 5.46 × 10− 4), “non-membrane spanning protein kinase activity” (adjusted p-value = 1.11 × 10− 3), “growth hormone receptor binding” (adjusted p-value = 1.11 × 10− 3) and “CCR5 chemokine receptor binding” (adjusted p-value = 1.11 × 10− 3). Among the enriched GO Biological Process terms of IMSGC, we found the “regulation of tyrosine phosphorylation of STAT protein” was the most significant (adjusted p-value = 1.58 × 10− 5). Five genes: HCLS1, JAK2, SOCS1, STAT3, and TNFRSF1A contributed to this GO term. Other STAT related processes within the top five IMSGC enriched GO Biological Process were “tyrosine phosphorylation of STAT protein” (adjusted p-value = 1.58 × 10− 5) and “positive regulation of STAT cascade” (adjusted p-value = 1.58 × 10− 5). The top GO Cellular Component term for IMSGC gene list was “I-kappaB/NF-kappaB complex” (adjusted p-value of 1.94 × 10− 3) with the contributing genes NFKB1 and RELA (also observed as significant for the GeneMSA gene set).
Here, we reported the gene set enrichment analysis of the dual evaluation module genes (20 significant modules, 56 non-redundant genes, Table 1). Among the GO Molecular Function terms, the third most significant term is “virus receptor activity” (adjusted p-value of 1.67 × 10− 3), which was supported by four input genes: CD80, CD86, ITGB3 and TNFRSF14. Phosphorylation and kinase related activity was once again observed within top five GO Molecular Function terms enriched for the dual evaluation gene set. These functions included “phosphotyrosine residue binding” (adjusted p-value = 1.67 × 10− 3), “phosphatidylinositol-4,5-bisphosphate 3-kinase activity” (adjusted p-value = 1.67 × 10− 3), “phosphatidylinositol bisphosphate kinase activity” (adjusted p-value = 1.67 × 10− 3), and “protein phosphorylated amino acid binding” (adjusted p-value = 1.67 × 10− 3). The top two enriched GO Biological Process terms in dual evaluation gene set were “T cell costimulation” (adjusted p-value = 2.11 × 10− 6) and “lymphocyte costimulation” (adjusted p-value = 2.11 × 10− 6), which are at times necessary processes in activation of immune response [30, 31]. These costimulation processes both included the same 7 contributing genes: CD80, CD86, CSK, CTLA4, GRB2, MAP3K14 and TNFRSF14. The “JAK-STAT cascade involved in growth hormone signaling” (adjusted p-value = 2.94 × 10− 5) was also present within top five enriched GO Biological Process terms of our dual evaluation. The top GO Cellular Component term enriched for this dual evaluation gene set was the “nuclear pore” with an adjusted p-value of 3.79 × 10− 4.
Table 1.
Gene set enrichment analysis of the modules from dual evaluationa
| GO term | # contributing genes/ term sizeb | Contributing genesc | p-value | adj. p-valued |
|---|---|---|---|---|
| Molecular Function | ||||
| Phosphotyrosine residue binding | 3/18 | GRB2, MAPK1, MAPK3 | 1.916 × 10− 5 | 1.674 × 10− 3 |
| Phosphatidylinositol-4,5-bisphosphate 3-kinase activity | 4/62 | CD80, CD86, GRB2, PIK3R2 | 3.318 × 10− 5 | 1.674 × 10− 3 |
| Virus receptor activity | 4/71 | CD80, CD86, ITGB3, TNFRSF14 | 5.667 × 10−5 | 1.674 × 10−3 |
| Phosphatidylinositol bisphosphate kinase activity | 4/72 | CD80, CD86, GRB2, PIK3R2 | 5.987 × 10−5 | 1.674 × 10−3 |
| Protein phosphorylated amino acid binding | 3/28 | GRB2, MAPK1, MAPK2 | 7.533 × 10−5 | 1.674 × 10−3 |
| Biological Process | ||||
| T cell costimulation | 7/82 | CD80, CD86, CSK, CTLA4, GRB2, MAP3K14, TNFRSF14 | 3.704 × 10−9 | 2.113 × 10−6 |
| Lymphocyte costimulation | 7/83 | CD80, CD86, CSK, CTLA4, GRB2, MAP3K14, TNFRSF14 | 4.037 × 10−9 | 2.113 × 10−6 |
| JAK-STAT cascade involved in growth hormone signaling pathway | 4/15 | MAPK1, MAPK3, STAT3, STAT5A | 8.415 × 10−8 | 2.937 × 10−5 |
| Growth hormone receptor signaling pathway | 4/24 | MAPK1, MAPK3, STAT3, STAT5A | 6.425 × 10−7 | 1.598 × 10− 4 |
| Cellular response to growth hormone stimulus | 4/25 | MAPK1, MAPK3, STAT3, STAT5A | 7.633 × 10−7 | 1.598 × 10−4 |
| Cellular Component | ||||
| Nuclear pore | 5/84 | KPNB1, NUP153, RAN, SENP2, SNUPN | 4.311 × 10−6 | 3.794 × 10−4 |
| Clathrin-coated pit | 4/69 | AMN, CLTC, EPN1, EPS15L1 | 4.666 × 10−5 | 2.053 × 10−3 |
| Endoplasmic reticulum tubular network | 2/12 | KPNB1, RAB18 | 5.296 × 10−4 | 0.0136 |
| Platelet alpha granule membrane | 2/13 | ITGB3, PECAM1 | 6.248 × 10−4 | 0.0136 |
| Cytosolic large ribosomal subunit | 3/67 | RPL4, RPL5, RPL17 | 9.556 × 10−4 | 0.0136 |
aIn this dual evaluation, IMSGC GWAS was the discovery set and GeneMSA was the evaluation set
bContributing genes: the number of genes in the input gene set. Term size: the total number of genes in the corresponding GO term
cContributing genes: those in the input genes that contributed to the enrichment
dAdjusted p-value by the Benjamini-Hochberg method [28]
Search for MS drug targets
MS FDA approved drugs and their drug targets were queried within DrugBank database for further assessment of the genes from our top networks. We found 32 unique MS FDA approved target genes. Among them, we found four drug targets overlapped with our top modules. These four genes are HDAC1, IL2RA, KEAP1, and RELA, which are the targets of 3 MS FDA approved drugs (Table 2). HDAC1, a gene in the histone deacetylase family and the central node in the top module of GeneMSA analysis (Fig. 2a), is a target gene of the MS drug fingolimod [29]. Note that HDAC1 did not have a significant pg value and would be missed in the typical GWAS analysis, but our network analysis could pinpoint this important druggable gene. Another MS drug target gene that overlapped within our modules was IL2RA, a gene which was present within the top module of IMSGC analysis, as well as in the dual evaluation top module. IL2RA is a target gene of the MS drug ocrelizumab. Lastly, the genes RELA and KEAP1 are targets for the MS drug dimethyl fumarate [29]. RELA was present and relatively central within GeneMSA and IMSGC top 10 module gene lists (Fig. 2b, d). KEAP1 was present within top 1 % modules of IMSGC (Additional file 4: Figure S3). Our network analysis indicated its potential to find potential druggable genes from GWAS datasets, which often reported significant variants in the non-coding regions.
Table 2.
MS FDA drug target genes within top modules
| Drug name | Commercial product name | Target gene | Occurrence in top modules | GeneMSA gene-level p-value | IMSGC gene-level p-value |
|---|---|---|---|---|---|
| Fingolimod | Gilenya | HDAC1 | Central in top module of GeneMSA, present in 75/75 top 1 % GeneMSA modules | 0.263 | 0.249 |
| Ocrelizumab | Ocrevus | IL2RA | Present in top modules of IMSGC GWAS and top module of dual evaluation, present in 76/76 top 1 % IMSGC modules | 0.212 | 4.37 × 10−9 |
| Dimethyl fumaratea | Tecfidera | KEAP1 | Present in 1/76 IMSGC top one present modules | 0.172 | 2.22 × 10−6 |
| RELA | Present in 3/75 top 1 % GeneMSA modules and 7/76 IMSGC top 1 % modules | 0.084 | 1.84 × 10−4 |
aDimethyl fumarate has two target genes from module gene list (KEAP1 and RELA)
Discussion
Network approaches can assist the original GWAS analysis to find combinatory signals in a set of genes that are functionally related. This approach will help better interpret the genetic association results toward understanding the molecular mechanisms of the complex disease. In this study, the most significant modules identified by dmGWAS contained central genes that were not addressed in the original GWAS reports. This is mainly due to the genome-wide significance level set in the original GWA studies (e.g. p value < 10− 8 for each SNP). Central genes outside of significance levels may have been revealed as important interactors for their connections with highly associated genes. Four unique MS drug target genes were included in our top modules: HDAC1, IL2RA, KEAP1 and RELA. The drug target genes HDAC1 and RELA in particular were central in top modules and also contributing genes in several notable gene set enrichment results relevant to hypothesized MS mechanisms. Furthermore, GRB2, JAK2, MAPK1 and STAT3 are genes of interest reported based on centrality measures. Common themes relevant to MS observed within our gene set enrichment analysis include immune system pathways, transcription and histone modification pathways, as well as growth hormone signaling pathways. Hypothesis for transcription dysregulation and epigenetic factors in MS were supported. Apart from these common themes, other pertinent enriched functions include the I-kappaB/NF-kappa B complex, glial cell differentiation, virus receptor activity and T-cell and lymphocyte costimulation. These enriched GO terms supported the inflammation and viral infection onset hypotheses for MS, which have not been reported in original GWA studies.
Several genes of interest are FDA-approved MS drug target genes
The genes HDAC1, IL2RA, KEAP1 and RELA present within our top gene networks are of interest, as they are drug-target genes for FDA approved MS disease-modifying therapies. Gene HDAC1, which encodes histone deacetylase 1, was the most central gene in the top module of the GeneMSA GWAS dataset (Fig. 2a) and remained one of the most central genes in merged top 10 modules and merged top 1 % modules (Fig. 2b, Additional file 3: Figure S2). Although HDAC1 is outside of the accepted significance level, it may have been presented as central for being an important interactor with the relatively significant gene PHF12 and other relevant genes. It has been suggested that HDAC1 plays a role in MS pathogenesis. HDAC1 modulates histone acetylation, which plays a role in the epigenetic control of T-cell mediated immunity. Mice with a conditional deletion of HDAC1 have shown to be resistant to experimental autoimmune encephalomyelitis (EAE), a common animal model of MS [32]. Moreover, nuclear export of HDAC1 is associated with impaired mitochondrial function and axonal damage in MS [33]. HDAC1 is a drug-target gene for fingolimod [29, 34] a sphingosine 1-phosphate (S1P) receptor modulator which is thought to mediate its effect in MS by sequestering lymphocytes within secondary lymphoid tissues [35]. The structure of the active metabolite of fingolimod mimics that of S1P, a signaling lipid involved in many cellular functions. It has been shown that fingolimod-P, produced within the nucleus from the phosphorylation of fingolimod, binds and inhibits class I HDAC, including HDAC1, consequently enhancing specific histone acetylation [36]. This effect is carried out independently of S1P receptors.
The gene RELA is also a gene of interest, as it is relatively central in top modules of GeneMSA and IMSGC (Fig. 2b, d), though it was not reported by either GWAS study examined. RELA is a proto-oncogene that encodes for a subunit of NF-kappaB. NF-kappaB is thought to play a central role in MS as it regulates innate and adaptive immunity, and is involved in the activation of astrocytes [37, 38]. Activation of NF-kappaB has been described in MS brain tissue [39]. Transgenic inhibition of astroglial NF-kappaB was shown to improve functional outcome in EAE [40]. RELA has been identified as a drug-target gene of the MS medication dimethyl fumarate. Indeed, monomethyl fumarate, the active metabolite of dimethyl fumarate, was shown to decrease RELA expression in myeloid dendritic cells derived from MS patients [41].
IL2RA and KEAP1 are another two genes of interest within our top gene networks because they are drug target genes in MS. IL2RA is a drug-target gene of ocrelizumad, a recently FDA approved drug for treatment of both relapsing-remitting and primary progressive MS, and was present within our top gene module of IMSGC (Fig. 2c) and the top gene module of our dual evaluation (Fig. 3a). Ocrelizumab is an anti-CD20 monoclonal antibody. Its efficacy in MS is attributed to B-cell depletion. IL2RA encodes for a protein that is a constituent of the IL2 receptor. It was reported by the IMSGC GWAS to be a genetic risk factor of MS. Lastly, although KEAP1 was not reported by either GWAS study, KEAP1 is a drug target gene of the MS drug dimethyl fumarate and was present within one of our top 1 % IMSGC modules. The gene KEAP1 encodes for an adapter protein of the E3 ubiquitin ligase, which are involved in DNA repair and cell cycle controls. Dimethyl fumarate, the drug which targets RELA and KEAP1, is an anti-inflammatory and central nervous system agent [29]. Although it is known that dimethyl fumarate upregulates nuclear-derived 2 (Nrf2) pathway, its relevance to MS is only hypothesized [29].
Additional genes of interest may be promising drug targets
Additional genes of interest were highlighted by their centrality within top gene modules: GRB2, JAK2, MAPK1 and STAT3. Given that the central genes HDAC1 and RELA genes are currently drug targets of FDA approved MS treatment, this suggests other central genes identified may be promising drug targets for future investigation. The protein encoding gene JAK2, which produces a protein that promotes cell proliferation through the JAK/STAT pathway, was central in our top gene module of IMSGC GWAS dataset (Fig. 2c). This gene was not previously reported by either MS GWA study. The gene STAT3, which was central in merged top 10 modules of GeneMSA and IMSGC and encodes for a protein that is a member of the STAT protein family, also plays a key role in the JAK-STAT signaling pathway. STAT3 was previously reported by the IMSGC GWAS and polymorphisms of STAT3 have been considered risk factors of MS, although the mechanism by which this association occurs is not fully understood [42, 43]. Previous work has highlighted the role of the JAK/STAT signaling pathway in the regulation of innate immunity and promotion of oligodendrocyte apoptosis and demyelination [44]. Cytokines and oxidative stress promote inflammation-related gene expression via JAK2 phosphorylation and STAT3 transcription factor activation [45]. Liu et al. demonstrated that JAK2 inhibitors have clinical efficacy in multiple preclinical models of MS by suppressing downstream activation of STAT, particularly STAT3 [46]. Modulation of the JAK/STAT pathway is thought to mediate the beneficial effect of neuroprotective compound linagliptin in cuprizone-induced demyelination [47]. Studies have suggested that JAK/STAT signaling occurs upstream of NF-kappaB activation [48]. Further evidence for the involvement of the JAK/STAT and NF-kappaB pathways in MS pathogenesis was obtained from our gene set enrichment analyses. Regulation of tyrosine phosphorylation of STAT protein, which included JAK2 and STAT3 was also observed within top gene enrichment of IMSGC, along with more STAT regulation pathways. Within the dual evaluation enrichment results, the JAK-STAT cascade involved in growth hormone signaling pathway was observed with the contributing genes of interest MAPK1 and STAT3. The cellular component I-kappaB/NF-kappaB complex was enriched for both the GeneMSA and IMSGC enrichment results and included the contributing drug target gene RELA.
MAPK1, a gene part of the MAP kinase family and an essential part of the growth hormone signaling cascade, was the central gene in the top module of our dual evaluation (Fig. 3a), regarding IMSGC as the discovery set. MAPK1 was previously reported by the IMSGC GWAS, and has been identified as a risk locus for MS [49]. The gene GRB2, a gene that encodes for the growth factor receptor bound protein, was seen as central in the merged significant dual evaluation gene modules (Fig. 3c) and in the top 1 % modules of IMSGC (Additional file 4: Figure S3). This gene was not reported by either GWA study examined. However, GRB2 has also been considered of a biological relevance by another MS network-based report and has been shown to be upregulated in EAE [50, 51]. The genes GRB2, JAK2, MAPK1 and STAT3 may be of consideration for future MS drug target experimentation because of their central presence in our top modules.
Gene set enrichment analysis supports epigenetic hypothesis in MS
Transcription factors have been attributed to subtypes of MS and hypothesized to dysregulate certain genes, thereby contributing to MS onset [52, 53]. The involvement of transcription factor dysregulation is supported by our study, given that our enrichment results showed many transcription factor processes within top five significantly enriched categories. The top enriched GO Molecular Function term in GeneMSA was “repressing transcription factor binding”, which included the genes of interest HDAC1, RELA and STAT3. Other enriched transcription regulation functions reported for GeneMSA include “activating transcription factor binding” and “RNA polymerase II distal enhancer sequence-specific DNA binding”, which both included HDAC1 and RELA. Transcription regulation GO Cellular Component terms enriched for IMSGC were “transcription factor TFTC complex” and “transcription factor TFIID”. Additionally, epigenetic factors are thought to mitigate the environmental factors of MS [34]. This epigenetic factor hypothesis of MS was also supported by our results, as “histone deacetylase complex” was a top enriched GO Molecular Function term and a member of the histone deacetylase family, HDAC1, was highly central in top GeneMSA modules [34]. Another epigenetic enriched function within GeneMSA was the GO Molecular Function term “chromatin DNA binding”, which included the genes of interest HDAC1, RELA and STAT3.
Genes identified for enriched regulation of glial cell differentiation and growth factor signaling
Regulation of glial cell differentiation was observed for the GeneMSA enrichment results. Glial cells include oligodendrocytes, astrocytes, and microglia, which are cells of interest in processes of MS pathogenesis. Oligodendrocytes are cells responsible for myelin sheath synthesis, and microglia and astrocytes are involved in innate immune responses within the central nervous system [54–56]. Myelin restoration is seen as a therapeutic strategy that could potentially aid in reversing neurological disabilities exhibited by people living with MS [57, 58]. As MS remyelinating therapies become more sought after, this suggests that genes involved in the enriched function for regulation of glial cells may be of interest: CTNNB1, HDAC1, HES1 and RELA. Interestingly, epidermal growth factor receptor (EGFR) was shown as central within our top 1 % IMSGC gene modules (Additional file 4: Figure S3), and involvement of growth factors signaling pathways, phosphotyrosine residue binding and protein phosphorylated amino acid binding are supported by our gene set enrichment analysis. EGFR overexpression was found to enhance oligodendrocyte differentiation and axonal myelination, suggesting that EGFR targeting may represent a viable strategy for myelin repair [57].
Viral insult hypothesis for MS etiology supported by virus receptor activity and T cell costimulation
Although the cause of MS is unknown, this complex disease has been attributed to the occurrence of a viral insult at a young age, later causing onset of neurological deficits in genetically predisposed individuals. This hypothesis is regarded as the most probable by some investigators [31]. Functional enrichment analysis of significant dual evaluation gene modules supports this hypothesis as T cell costimulation, lymphocyte costimulation and virus receptor activity were significantly enriched. Costimulation pathways of T cell and B cell activation have been exhibited in pathogen infection, and costimulation pathways of immune response activation have been linked to a viral context [30]. These lymphocyte costimulation pathways, coupled with enrichment of virus receptor activity suggest that the viral hypothesis for MS onset is relevant, and may occur through T cell costimulation. The genes found to be involved in MS virus receptor activity may be of interest for experimental validation of this viral onset hypothesis: CD80, CD86, ITGB3 and TNFRSF14. Finally, the genes involved in T-cell costimulation may also be further investigated to discover if the autoimmune response in MS truly occurs through this immune response activation pathway: CD80, CD86, CSK, CTLA4, GRB2, MAP3K14 and TNFRSF14.
Limitations and future work
There are several limitations in this study. One is data heterogeneity between IMSGC and GeneMSA GWAS datasets. Another limitation of this study is that some genes within our networks may be highly connected with the most significant genes (at individual gene level), and this makes them more likely to be detected due to lack of PPI reference network completeness. On the other hand, the PPI network is still incomplete, and does not include all the protein-coding genes. Thirdly, the highly associated HLA region within MS GWAS datasets is also a limiting factor, as these highly associated regions may mask the association signals of other regions. We will expand our work in future. First, we may include more genetic association data, including both common and rare variants from genome sequencing data, once they are available. Second, we may integrate gene expression and GWAS signal from the same disease (MS) to detect more reliable and causal signals. Tools such as EW-dmGWAS [59] can be applied. Thirdly, functional annotations are available from various resources. We will evaluate these association signals using additional annotations such as epigenetic marks, expression quantitative trait loci (eQTL), tissue-specificity and cell types, among others. Tissue specificity enrichment analysis of genetic variations in association studies has shown interesting results, as demonstrated in our multi-trait GWAS analysis [60]. Finally, further experimental validation of supported and proposed MS mechanisms is needed using cell lines and animal models.
Conclusions
The dmGWAS (version 2.4) tool, previously developed by members of our team, was used to integrate the GeneMSA and IMSGC GWAS with the human interactome to interpret genetic associations. Through a dual evaluation approach, this network-based analysis could effectively yield top modules evaluated by the presence of MS FDA-approved drug target genes: HDAC1, IL2RA, KEAP1 and RELA. Other central genes present within top modules are suggested as potential drug targets for MS: GRB2, JAK2, MAPK1 and STAT3. The independent gene set enrichment results of both GeneMSA and IMSGC top modules supported the hypothesis of epigenetic factors involved in dysregulation of genes in MS. GeneMSA enrichment of top 10 modules presented a pathway involved in regulation of glial cell differentiation, thereby identifying genes that are promising for remyelination therapy investigations. Furthermore, a dual evaluation of both MS GWAS identified genes enriched in virus receptor activity, supporting the viral onset hypothesis for the etiology of MS.
Supplementary information
Additional file 1: Table S1. Publicly available MS GWAS summary statistics data.
Additional file 2: Figure S1. SNP-level and gene-level Manhattan plots comparing individual MS GWAS data.
Additional file 3: Figure S2. Top 1% modules of GeneMSA.
Additional file 4: Figure S3. Top 1% modules of IMSGC.
Additional file 5: Table S2. Gene set enrichment analysis of modules from dual evaluation of GeneMSA.
Additional file 6: Table S3. Gene set enrichment analysis of modules from dual evaluation of IMSGC.
Acknowledgements
We would like to thank the members of Bioinformatics and Systems Medicine Laboratory (BSML) for valuable discussion.
About this supplement
This article has been published as part of BMC Medical Genomics Volume 13 Supplement 5, 2020: The International Conference on Intelligent Biology and Medicine (ICIBM) 2019: Computational methods and application in medical genomics (part 1). The full contents of the supplement are available online at https://bmcmedgenomics.biomedcentral.com/articles/supplements/volume-13-supplement-5.
Authors’ contributions
ZZ and PJ designed the project, AMM, YD, PJ, and ZZ collected the data, AMM and YD analyzed the data, AMM and YD drafted the manuscript, and LAF, ZZ, and PJ edited the manuscript. All authors approved the final manuscript.
Funding
This work was partially supported by National Institutes of Health grants (LM012806, DE027393, DE028103 and DE027711). Publication charges for this article have been funded by R01LM012806. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All the data used in this study are from public sources cited in our reference list. Also, additional files, which may be needed to reproduce the results presented in the manuscript, are made available as additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Astrid M. Manuel, Email: Astrid.M.Manuel@uth.tmc.edu
Yulin Dai, Email: Yulin.Dai@uth.tmc.edu.
Leorah A. Freeman, Email: Leorah.Freeman@uth.tmc.edu
Peilin Jia, Email: peilin.jia@uth.tmc.edu.
Zhongming Zhao, Email: zhongming.zhao@uth.tmc.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12920-020-0674-5.
References
- 1.Wallin MT, Culpepper WJ, Campbell JD, Nelson LM, Langer-Gould A, Marrie RA, Cutter GR, Kaye WE, Wagner L, Tremlett H, Buka SL, Dilokthornsakul P, Topol B, Chen LH, LaRocca NG. The prevalence of MS in the United States: A population-based estimate using health claims data. Neurology. 2019;92:e1–12. doi: 10.1212/WNL.0000000000007035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Confavreux C, Vukusic S. Natural history of multiple sclerosis: a unifying concept. Brain. 2006;129(Pt 3):606–616. doi: 10.1093/brain/awl007. [DOI] [PubMed] [Google Scholar]
- 3.Meltzer EI, Costello FE, Frohman EM, Frohman TC. New ways of “seeing” the mechanistic heterogeneity of multiple sclerosis plaque pathogenesis. J Neuro Ophthalmol. 2018;38:91–100. doi: 10.1097/WNO.0000000000000633. [DOI] [PubMed] [Google Scholar]
- 4.Dendrou CA, Fugger L, Friese MA. Immunopathology of multiple sclerosis. Nat Rev Immunol. 2015;15:545–558. doi: 10.1038/nri3871. [DOI] [PubMed] [Google Scholar]
- 5.Cotsapas C, Mitrovic M. Genome-wide association studies of multiple sclerosis. Clin Transl Immunol. 2018;7:1–9. doi: 10.1002/cti2.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fortunato N. Pathophysiology: the biologic basis for disease in adults and children. AORN J. 2006;10:311. [Google Scholar]
- 7.Pasaniuc B, Price AL. Dissecting the genetics of complex traits using summary association statistics. Nat Rev Genet. 2017;18:117–127. doi: 10.1038/nrg.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, Patsopoulos NA, Moutsianas L, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for Colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia P, Zhao Z. Network-assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum Genet. 2014;133:125–138. doi: 10.1007/s00439-013-1377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Lin CC, Li YY, Zhao Z. Dynamic protein interaction modules in human hepatocellular carcinoma progression. BMC Syst Biol. 2013;7(SUPPL 5):S2. doi: 10.1186/1752-0509-7-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, et al. NCBI’s database of genotypes and phenotypes: DbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, et al. Human Protein Reference Database - 2009 update. Nucleic Acids Res. 2009;37(SUPPL 1):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia P, Zhao Z. VarWalker: personalized mutation network analysis of putative Cancer genes from next-generation sequencing data. PLoS Comput Biol. 2014;10:e1003460. doi: 10.1371/journal.pcbi.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia P, Wang L, Fanous AH, Pato CN, Edwards TL, Zhao Z. Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia. PLoS Comput Biol. 2012;8:e1002587. doi: 10.1371/journal.pcbi.1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. Fast and rigorous computation of gene and Pathway scores from SNP-based summary statistics. PLoS Comput Biol. 2016;12:1–20. doi: 10.1371/journal.pcbi.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.dmGWAS [https://bioinfo.uth.edu/dmGWAS]. Accessed 1 May 2019.
- 21.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinforma. 2014;2014:8.13.1–8.13.24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon J, Blumer A, Lee K. An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics. 2006;22:3106–3108. doi: 10.1093/bioinformatics/btl533. [DOI] [PubMed] [Google Scholar]
- 23.Brandes U. A faster algorithm for betweenness centrality. J Math Sociol. 2001;25:163–177. doi: 10.1080/0022250X.2001.9990249. [DOI] [Google Scholar]
- 24.Jia P, Kao CF, Kuo PH, Zhao Z. A comprehensive network and pathway analysis of candidate genes in major depressive disorder. BMC Syst Biol. 2011;5(SUPPL 3):S12. doi: 10.1186/1752-0509-5-S3-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman LC. Centrality in social networks conceptual clarification. Soc Networks. 1978;1:215–239. doi: 10.1016/0378-8733(78)90021-7. [DOI] [Google Scholar]
- 26.Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: A multiple gene list feature analyzer for comparative enrichment clustering and networkbased dissection of biological systems. Nucleic Acids Res. 2010;38(SUPPL 2):W96–102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gene Ontology Consortium The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:258D–2261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 2018;57:289–300. [Google Scholar]
- 29.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welten SP, Redeker A, Franken KL, Oduro JD, Ossendorp F, Čičin-Šain L, et al. The viral context instructs the redundancy of costimulatory pathways in driving CD8+ T cell expansion. Elife. 2015;4. [DOI] [PMC free article] [PubMed]
- 31.Gilden DH. Infectious causes of multiple sclerosis. Lancet Neurol. 2005;4:195–202. doi: 10.1016/S1474-4422(05)70023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Göschl L, Preglej T, Hamminger P, Bonelli M, Andersen L, Boucheron N, et al. A T cell-specific deletion of HDAC1 protects against experimental autoimmune encephalomyelitis. J Autoimmun. 2018;86:51–61. doi: 10.1016/j.jaut.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Kim JY, Shen S, Dietz K, He Y, Howell O, Reynolds R, et al. HDAC1 nuclear export induced by pathological conditions is essential for the onset of axonal damage. Nat Neurosci. 2010;13:180–189. doi: 10.1038/nn.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koch MW, Metz LM, Kovalchuk O. Epigenetic changes in patients with multiple sclerosis. Nat Rev Neurol. 2013;9:35–43. doi: 10.1038/nrneurol.2012.226. [DOI] [PubMed] [Google Scholar]
- 35.Huwiler A, Zangemeister-Wittke U. The sphingosine 1-phosphate receptor modulator fingolimod as a therapeutic agent: recent findings and new perspectives. Pharmacol Ther. 2018;185:34–49. doi: 10.1016/j.pharmthera.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Hait NC, Wise LE, Allegood JC, O’Brien M, Avni D, Reeves TM, et al. Active, phosphorylated fingolimod inhibits histone deacetylases and facilitates fear extinction memory. Nat Neurosci. 2014;17:971–980. doi: 10.1038/nn.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T, Zhang L, Joo D, Sun S-C. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mc Guire C, Prinz M, Beyaert R, van Loo G. Nuclear factor kappa B (NF-κB) in multiple sclerosis pathology. Trends Mol Med. 2013;19:604–613. doi: 10.1016/j.molmed.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Bonetti B, Stegagno C, Cannella B, Rizzuto N, Moretto G, Raine CS. Activation of NF-κB and c-Jun transcription factors in multiple sclerosis lesions: implications for oligodendrocyte pathology. Am J Pathol. 1999;155:1433–1438. doi: 10.1016/S0002-9440(10)65456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brambilla R, Persaud T, Hu X, Karmally S, Shestopalov VI, Dvoriantchikova G, et al. Transgenic inhibition of Astroglial NF- B improves functional outcome in experimental autoimmune encephalomyelitis by suppressing chronic central nervous system inflammation. J Immunol. 2009;182:2628–2640. doi: 10.4049/jimmunol.0802954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazzola MA, Raheja R, Regev K, Beynon V, von Glehn F, Paul A, et al. Monomethyl fumarate treatment impairs maturation of human myeloid dendritic cells and their ability to activate T cells. Mult Scler J. 2019;25:63–71. doi: 10.1177/1352458517740213. [DOI] [PubMed] [Google Scholar]
- 42.Lill CM, Schjeide BMM, Akkad DA, Blaschke P, Winkelmann A, Gerdes LA, et al. Independent replication of STAT3 association with multiple sclerosis risk in a large German case-control sample. Neurogenetics. 2012;13:83–86. doi: 10.1007/s10048-011-0305-6. [DOI] [PubMed] [Google Scholar]
- 43.Jakkula E, Leppä V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86:285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan Z, Gibson SA, Buckley JA, Qin H, Benveniste EN. Role of the JAK/STAT signaling pathway in regulation of innate immunity in neuroinflammatory diseases. Clin Immunol. 2018;189:4–13. doi: 10.1016/j.clim.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gressens P, Dournaud P, Fafouri A, Bortolotto ZA, Collingridge GL, Doherty A, et al. The role of JAK-STAT signaling within the CNS. Jak-Stat. 2013;2:e22925. doi: 10.4161/jkst.22925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Holdbrooks AT, De Sarno P, Rowse AL, Lora L, Mcfarland BC, et al. Therapeutic efficacy of suppressing the JAK/STAT pathway in multiple models of EAE. J Immunol. 2015;192:59–72. doi: 10.4049/jimmunol.1301513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elbaz EM, Senousy MA, El-Tanbouly DM, Sayed RH. Neuroprotective effect of linagliptin against cuprizone-induced demyelination and behavioural dysfunction in mice: a pivotal role of AMPK/SIRT1 and JAK2/STAT3/NF-κB signalling pathway modulation. Toxicol Appl Pharmacol. 2018;352:153–161. doi: 10.1016/j.taap.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 48.Guzzo C, Mat NFC, Gee K. Interleukin-27 induces a STAT1/3- and NF-κB-dependent proinflammatory cytokine profile in human monocytes. J Biol Chem. 2010;285:24404–24411. doi: 10.1074/jbc.M110.112599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1362. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahurkar S, Moldovan M, Suppiah V, O’Doherty C. Identification of shared genes and pathways: a comparative study of multiple sclerosis susceptibility, severity and response to interferon Beta treatment. PLoS One. 2013;8:e57655. doi: 10.1371/journal.pone.0057655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tseveleki V, Rubio R, Vamvakas SS, White J, Taoufik E, Petit E, et al. Comparative gene expression analysis in mouse models for multiple sclerosis, Alzheimer’s disease and stroke for identifying commonly regulated and disease-specific gene changes. Genomics. 2010;96:82–91. doi: 10.1016/j.ygeno.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parnell GP, Gatt PN, Krupa M, Nickles D, McKay FC, Schibeci SD, et al. The autoimmune disease-associated transcription factors EOMES and TBX21 are dysregulated in multiple sclerosis and define a molecular subtype of disease. Clin Immunol. 2014;151:16–24. doi: 10.1016/j.clim.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Riveros C, Mellor D, Gandhi KS, Mckay FC, Cox MB, Berretta R, et al. A transcription factor map as revealed by a genome- wide gene expression analysis of whole-blood mRNA Transcriptome in multiple sclerosis. PLoS One. 2010;5:e14176. doi: 10.1371/journal.pone.0014176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Needham SR, Roberts SK, Arkhipov A, Mysore VP, Tynan CJ, Zanetti-Domingues LC, et al. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat Commun. 2016;7:13307. doi: 10.1038/ncomms13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreutzberg GW. Microglia: a sensor for pathological events in the CNS. Trends Neurosci. 1996;19:312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- 56.Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat Rev Immunol. 2014;14:463–477. doi: 10.1038/nri3705. [DOI] [PubMed] [Google Scholar]
- 57.Aguirre A, Dupree JL, Mangin JM, Gallo V. A functional role for EGFR signaling in myelination and remyelination. Nat Neurosci. 2007;10:990–1002. doi: 10.1038/nn1938. [DOI] [PubMed] [Google Scholar]
- 58.Plemel JR, Liu WQ, Yong VW. Remyelination therapies: a new direction and challenge in multiple sclerosis. Nat Rev Drug Discov. 2017;16:617–634. doi: 10.1038/nrd.2017.115. [DOI] [PubMed] [Google Scholar]
- 59.Wang Q, Yu H, Zhao Z, Jia P. EW-dmGWAS: edge-weighted dense module search for genome-wide association studies and gene expression profiles. Bioinformatics. 2015;31:2591–2594. doi: 10.1093/bioinformatics/btv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pei G, Dai Y, Zhao Z, Jia P. deTS: tissue-specific enrichment analysis to decode tissue specificity. Bioinformatics. 2019;35:3842–3845. doi: 10.1093/bioinformatics/btz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Publicly available MS GWAS summary statistics data.
Additional file 2: Figure S1. SNP-level and gene-level Manhattan plots comparing individual MS GWAS data.
Additional file 3: Figure S2. Top 1% modules of GeneMSA.
Additional file 4: Figure S3. Top 1% modules of IMSGC.
Additional file 5: Table S2. Gene set enrichment analysis of modules from dual evaluation of GeneMSA.
Additional file 6: Table S3. Gene set enrichment analysis of modules from dual evaluation of IMSGC.
Data Availability Statement
All the data used in this study are from public sources cited in our reference list. Also, additional files, which may be needed to reproduce the results presented in the manuscript, are made available as additional files.