Abstract
The picture beginning to form from genome analyses of viruses, unicellular organisms, and multicellular organisms is that viruses have shared functional modules with cells. A process of coevolution has probably involved exchanges of genetic information between cells and viruses for long evolutionary periods. From this point of view present-day viruses show flexibility in receptor usage and a capacity to alter through mutation their receptor recognition specificity. It is possible that for the complex DNA viruses, due to a likely limited tolerance to generalized high mutation rates, modifications in receptor specificity will be less frequent than for RNA viruses, albeit with similar biological consequences once they occur. It is found that different receptors, or allelic forms of one receptor, may be used with different efficiency and receptor affinities are probably modified by mutation and selection. Receptor abundance and its affinity for a virus may modulate not only the efficiency of infection, but also the capacity of the virus to diffuse toward other sites of the organism. The chapter concludes that receptors may be shared by different, unrelated viruses and that one virus may use several receptors and may expand its receptor specificity in ways that, at present, are largely unpredictable.
I. Introduction
The penetration of animal viruses into cells is the result of an active process of specific interactions with cell surface macromolecules. Differentiated organisms do not express identical sets of surface macromolecules on different cell types, tissues, and organs. This basic feature of organisms results in a compartmentalization of susceptibilities to infection by viruses. As observed repeatedly in nature, different viruses infect distinct target cells in one or a group of host species. This is the expected evolutionary outcome of a population equilibrium between organisms and their parasites that must have been favored by mutual long-term interactions. Although permissivity of virus entry into cells is by no means the only determinant of host cell tropism, it is certainly an important element with implications in viral pathogenesis. Changes in the specificity of virus entry into cells may be of consequence also for viral disease emergence and disease prevention and control.
Early reports (reviewed in McLaren et al., 1959) suggested the presence of receptors for bacteriophage in Escherichia coli cells and for influenza virus, Newcastle disease virus, and enteroviruses in some animal cells. The studies of McLaren, Holland, and their colleagues with poliovirus (Holland 1961, Holland 1959, McLaren 1959) indicated that the productive adsorption of poliovirus was associated with specific antigenic structures possessed only by susceptible primate cells (or subcellular cell debris). We term viral receptor any cell surface macromolecule involved in the recognition of the cell by a virus or in the penetration of a virus into the cell. For simplification, we do not establish a difference between receptor and coreceptor (Young, 2001) because such a distinction is at times ambiguous, and it is not essential to the main aims of the present review. However, we maintain the term coreceptor when referring to some of the molecules that participate in retrovirus entry, as in this case the term is amply employed in the literature. The problems addressed in this review are in the interphase between virus evolution and cell recognition by viruses. Topics include changes in receptor usage, shared use of the same receptors by different viruses, and coevolution of antigenicity and host cell tropism. We then review briefly some of the implications of flexibility in receptor usage for the host range of viruses, for the emergence of viral diseases, and consequences for the use of viruses as vectors for gene delivery. In this regard, we discuss the possible involvement of viruses as agents of lateral gene transfer during the evolution of cellular life on earth as a means to accelerate functional diversification of cells and cell collectivities. In none of theses topics can we (or intend to) be exhaustive. Selected examples are used to unveil the implications of the highly dynamic nature of viral genomes for modifications of receptor specificity, particularly for those viruses with RNA as genetic material. Recent introductions to the mechanisms of virus entry into cells can be found in general virology textbooks (Cann 2001, Flint 2000). Among excellent reviews on viral receptors are those published by Wimmer 1994, Weiss 1995, Miller 1996, Hunter 1997, Evans 1998, Sommerfelt 1999, Berger 1999, Nemerow 2000, Schneider-Schaulies 2000, Skehel 2000, Rossmann 2000, Rossmann 2002, Spear 2000, Speck 2000, Dragic 2001, Young 2001, Eckert 2001, Shukla 2001, Goldsmith 2002, Hogle 2002, Kunz 2002, Bomsel 2003.
Virus evolution has been covered in Domingo 1999, Domingo 2001, Crandall 1999, Flint 2000. A brief review on the evolution of cell tropism of viruses was published by Baranowski et al. (2001). The following sections introduce current concepts of virus evolution and how viral population dynamics affects changes in cell recognition.
II. Basic Concepts of Virus Evolution
A. Types of Interactions between Viruses and Cells
Viruses are highly diverse molecular parasites of cells that have an intracellular phase of replication and an extracellular stage in the form of discrete particles. Despite their remarkable diversity in size, shape, and biological properties, a few definitive features are common to all viruses: (i) they have DNA or RNA (but not both) as genetic material; (ii) their genome encodes a distinctive genetic program, and (iii) the expression of this program, which results in virus replication, is totally dependent on cell structures and cell metabolism.
In their dependence on cells, viruses can produce a wide range of perturbations, such as modifications of specialized (luxury) functions, without an immediate effect on cell survival, or they may cause cell death by apoptosis or necrosis. Viral infections may be inapparent or they may cause acute or chronic disease either directly by affecting cell subpopulations or indirectly by triggering immunopathological responses (Mims 2001, Nathanson 1997). These disparate effects on cells have not been correlated with genome type, virion structure, or receptor usage, presumably because many host functions, as much as viral functions, influence the outcome of an infection.
The genetic complexity of viruses, as reflected in the size of their genome, varies from a few thousand nucleotides in the case of RNA viruses (the genome of bacteriophage MS2 is 3569 nucleotides long, and that of the largest coronaviruses comprises about 32,000 nucleotides) to a broader range in the case of DNA viruses (the double-stranded DNA of hepatitis B virus has about 3200 bp, whereas the large poxviruses, iridoviruses, and herpesviruses have DNA of 130,000 up to 370,000 bp). Genome complexity appears to influence the strategies of the interactions between viruses and cells. Hosts must have evolved defense mechanisms to limit virus replication at the expense of their cells, and in turn, viruses must have evolved mechanisms to counteract cell responses (at least to survive to be an object of interest for a review article). The most complex DNA viruses encode a number of proteins that may or may not have a cellular homologue and that may modulate host defense responses. Classical examples are the glycoprotein C of herpes simplex virus, which blocks complement activation, or proteins E3⧸19K and E1a of adenovirus, which suppress major histocompatibility complex (MHC) class I and class II molecules that are required for the T-cell recognition of infected cells. Other viral proteins are homologues of cytokines, chemokines, or their receptors or may induce or inhibit apoptosis. An increasing number of viral-coded, immunomodulating proteins is being discovered, some with seemingly redundant functions, and others with multiple effects on cells, playing active roles as determinants of virus survival and pathogenesis (reviews in Alcami 2000, Alcami 2003, Seet 2003, Xu 2001). Host-interfering proteins may also be expressed by RNA viruses (HIV-1 nef, RNA silencing suppressor B2 in flock house virus, several interferon antagonists such as NS1 of human influenza virus A, etc.), albeit their number appears to be more limited than for complex DNA viruses. This probably reflects two fundamentally different viral strategies to cope with host defenses: interaction versus evasion.
In contrast to the complex DNA viruses, RNA viruses have condensed essential genetic information for replication in a minimal number of nucleotides. Compressing mechanisms include the presence of overlapping reading frames; ambisense RNA; common leader RNA sequences for the synthesis of subgenomic messenger RNAs; untranslated regions, which include signals for RNA replication and protein synthesis; cis-acting regulatory elements within open reading frames; RNA editing; partial read-through of termination codons, leading to two forms of a protein that differ in a carboxy-terminal extension; leaky ribosome scanning with initiation of protein synthesis occurring at two in-frame AUGs, leading to two forms of a protein that differ by an amino-terminal extension; ribosome hopping, shunting, and bypassing; ribosome frameshifting, resulting in a change of the order of triplet reading to yield a single fused polypeptide from two overlapping open reading frames; and synthesis of a polyprotein, which is then cleaved to produce functional intermediates and fully processed proteins, with processing intermediates and processed proteins having distinct functional roles in interaction with viral RNA, viral proteins, or cellular proteins. In addition, many viral proteins appear to be multifunctional, thereby expanding the role of a single nucleotide stretch in the completion of a virus replication cycle (reviews in Flint 2000, Knipe 2001, Semler 2002). Genome compactness imposes a conflict between the requirement of genetic variation to permit adaptation to changing environments and the need to maintain infectivity in genomes in which virtually every single nucleotide appears to be involved in some structural or functional role. Genetic variation to escape from selective constraints is generally reflected in the survival of subpopulations of genomes that may show little alteration in replication capacity (fitness) or, despite a reduction of fitness, may still replicate to generate new mutant distributions of higher fitness.
B. Genetic Variation and the Dynamics of Viral Populations
The tolerance of viral genomes (or other replicons) to accept mutations decreases with genome complexity (Domingo 2001, Eigen 1988). RNA viruses display mutation rates and frequencies in the range of 10−3 to 10−5 errors per nucleotide copied (Batschelet 1976, Drake 1999), values that imply the continuous generation of dynamic mutant distributions in a replicating RNA virus population. These high mutation rates in RNA genomes would be incompatible with maintenance of the genetic information contained in large viral or cellular DNA genomes (Eigen and Biebricher, 1988). This evolutionary adaptation of mutation rates is mirrored in the biochemical activities of the relevant DNA and RNA polymerases. Cellular DNA polymerases involved in DNA replication generally contain a 3′ → 5′ exonuclease proofreading–repair activity capable of excising misincorporated nucleotides to allow incorporation of the correct complementary nucleotide prior to further elongation of the nascent DNA chain. Such a proofreading–repair activity is absent in viral RNA replicases and reverse transcriptases, as evidenced by both structural and biochemical studies (Menéndez-Arias 2002, Steinhauer 1992). Furthermore, a number of postreplicative repair pathways are active on double-stranded DNA but cannot act on RNA, therefore contributing to a final 105- to 106-fold higher average copying fidelity during cellular DNA replication than during RNA genome replication (Domingo 2001, Drake 1999, Friedberg 1995, Goodman 1998).
Viruses exploit the same mechanisms of genetic variation as cells: mutation, homologous and nonhomologous recombination, and genome segment reassortment in the case of viruses with segmented genomes. Different virus families vary in the extent of utilization of these different variation mechanisms. For example, homologous recombination is very active in positive-strand RNA viruses and retroviruses, but appears to be rare in negative-strand RNA viruses. Because of general high mutation rates, RNA virus populations consist of complex mutant distributions termed viral quasispecies (Domingo 2001, Eigen 1996, Eigen 1988, Eigen 1979, Nowak 1992). Quasispecies was formulated as a general theory of molecular evolution by M. Eigen and colleagues to describe self-organization and adaptation of simple early replicons that could occur at early stages of the development of life (Eigen 1988, Eigen 1979). Although the initial quasispecies theory involved mutant distributions in equilibrium, extensions of the theory to finite replicon populations subjected to environmental changes have been developed (Eigen 2000, Wilke 2001). Therefore, the quasispecies theory was instrumental in understanding the population structure and dynamics of RNA virus populations. Virologists use an extended definition of quasispecies to signify dynamic distributions of nonidentical but closely related mutant and recombinant viral genomes subjected to a continuous process of genetic variation, competition, and selection, and which act as a unit of selection (Domingo, 1999). It must be stressed that because of high mutation rates (Batschelet 1976, Drake 1999), each individual genome in a replicating population, which includes a number of distinctive mutations, has only a fleeting existence because new mutations are arising continuously, even in a single infected cell. Thus, an RNA virus genome population may be defined statistically but it is essentially indeterminate at the level of individual genomes (Domingo 2001, Domingo 1978). Mutant swarms are subjected unavoidably to competition, and those mutant distributions best adapted to replicate in a given environment are those that dominate the population at a given time. Unfit mutant distributions are subjected to negative selection and kept at low (sometimes undetectable) levels in the population. Unfit mutant distributions in one environment may nevertheless be fit in a different environment, and a modulation of frequencies of genome subpopulations is the key to adaptability of RNA viruses. The biological relevance of the quasispecies nature of RNA viruses stems from the fact that mutant spectra may contain genomes with altered biological properties, including modified cell recognition capacity. Relevant parameters are the number of mutations per genome, genome length, and viral population size, as documented with several virus systems both in cell culture and in vivo (Table I ).
TABLE I.
Biological Relevance of Quasispecies Dynamics for RNA Virusesa
| Relevant parameters |
|---|
| 1.Average number of mutations per genome found in a viral population: typically it ranges from 1 to 100 |
| 2.Virus population size: very variable but can reach 109 to 1012 in some infections in vivo |
| 3.Genome length: 3 to 33 kb, with compact genetic information |
| 4.Mutations needed for a phenotypic change: one or few mutations, as documented amply in the text for changes in receptor recognition specificity |
| Examples of phenotypic changes in RNA viruses dependent on one or few mutations |
| 1.Antigenic variation (antibody-, CTL-escape) |
| 2.Virulence |
| 3.Altered pattern of gene expression |
| 4.Resistance to antiviral inhibitors |
| 5.Cell tropism and host range (the topic of this review) |
Based on many published studies reviewed in Domingo 1985, Domingo 1999, Domingo 2001, Crandall 1999, Flint 2000, DeFilippis 2001.
Despite the unlikely occurrence of generalized high mutation rates in complex DNA viruses, hot spots for variation at sequence repeats and extensive diversity among isolates of DNA viruses have been observed (Lua 2002, Smith 1987). Furthermore, small DNA viruses, such as plant geminiviruses or animal parvoviruses, show features of quasispecies dynamics, similar to RNA viruses (Isnard 1998, Lopez-Bueno 2003; reviewed in Domingo et al., 2001).
Diversification of viruses within infected organisms, even when it involves limited genetic change, is of consequence for viral pathogenesis and persistence. This is because one or a few mutations may suffice to change important biological properties of viruses (such as host cell tropism, resistance to antibodies, to cytotoxic T lymphocytes or to inhibitors, among other traits; Table I). Small numbers of mutations in components of mutant spectra are easily attainable by diversifying populations of viruses during acute or chronic infections. Even when a mutation that confers a phenotypic change results in a modest fitness decrease, compensatory mutations can have an opportunity to rescue genomes with normal or nearly normal fitness values (Cassady 2002, Escarmís 1999, Escarmís 2002, Lázaro 2002, Liang 1998, Nijhuis 1999, Wang 1996, Yuan 2000). Viral fitness determined ex vivo may be a relevent indicator of disease progression in vivo (Ball et al., 2003). Within-host variation is the first step in the process of long-term diversification of viruses in successions of transmission events from infected to susceptible hosts. Comparison of consensus nucleotide sequences of independent isolates of the same virus originated from a single source of infection allows a calculation of the rate of evolution. As expected from the complex quasispecies dynamics, rates of evolution for RNA viruses are not constant with time (a “clock” does not operate) and are often in the range of 10−2 to 10−4 substitutions per nucleotide and year. These values can also vary for different genomic segments of the same virus. In sharp contrast, rates of evolution for cellular genes have been estimated in 10−8 to 10−9 substitutions per nucleotide and year. Interestingly, DNA viruses, even those with small genome size, display widely different rates of evolution. As an example, such rates have been estimated in 1.7 × 10−4 substitutions per nucleotide and year for canine parvovirus and in the range of 1 × 10−7 to 3 × 10−8 substitutions per nucleotide and year for some papovaviruses (reviewed in Domingo et al., 2001).
Genomic consensus sequences of independent isolates of a virus allow the establishment of phylogenetic relationships by the application of classical procedures of population genetics (Doolittle 1996, Page 1998, Weiller 1995). Such procedures have confirmed the extensive diversity of extant viruses; it has not been possible to derive a genetic tree that relates the known viruses, not even DNA or RNA viruses separately. This, together with the disparate replication strategies exhibited by viruses, suggests multiple origins for the viruses we study today: viruses are polyphyletic. For viruses that infect distantly related host species, such as the herpesviruses, a parallelism between host phylogeny and virus phylogeny has been observed (McGeoch and Davison, 1999). This suggests a host–virus cospeciation, implying deep evolutionary roots of viruses with their hosts (Gorbalenya 1995, McGeoch 1999) (discussed further in Section V,D).
While we have attained some understanding of the mechanisms involved in the generation of diversity, little is understood of the forces that favor the dominance of some virus types over others in nature. According to the International Committee on Taxonomy of Viruses (van Regenmortel et al., 2000), there are about 3600 virus species recognized and more than 30,000 different viruses, strains, and subtypes. Considering that each “individual” RNA virus circulates as a dynamic quasispecies, the diversity of viral genomes on earth is astonishing. Each replication-competent component of a quasispecies distribution can, in principle, initiate a virus diversification process to generate a spectrum of genotypes and phenotypes. RNA replicons constitute a highly dynamic “RNA world” in a relatively more static [but only relatively! (Bushman, 2002)] DNA-based biosphere of differentiated organisms, as noted two decades ago by Holland and colleagues (1982). The central objective of this review is to examine the effect of virus variation on cell recognition and some of its biological consequences.
III. Nature of Viral Receptors
Viruses recognize target cells by binding to specific receptor molecules at the cell surface. Until the mid-1980s, the only virus receptors that were identified unequivocally were sialic acids for the myxoviruses and paramyxoviruses (Haywood, 1994). Since that time, there has been a deluge of new data about the nature of cell surface molecules participating in viral entry. The molecules that have so far been characterized as viral receptors belong to different families of proteins, carbohydrates, and lipids, often organized as cell surface complexes. Receptors are normally involved in critical cellular functions such as signal transduction, cell adhesion, immune modulation, enzymatic activities, and for some receptors no cellular function has been identified yet (Table II ). It is not clear why some macromolecules and not others act as viral receptors. It has been suggested that one reason is their abundance and availability on the cell surface combined with their capacity to bind and⧸or internalize viruses while triggering initiation of the virus replication cycle. By participating in the first step of virus infection, receptors divert from their usual activity in cellular metabolism and provide yet another example of molecular parasitism, essential in the life cycle of viruses.
TABLE II.
Some Cell Surface Components Proposed to be Involved in Virus Entry into Cells
| Receptor class | Cellular structurea |
|---|---|
| Extracellular matrix components, sugar derivatives and lipids | Galactosylceramide |
| Gangliosides | |
| Glycosaminoglycans (heparan and chondroitin sulfates) | |
| Phospholipids | |
| Sialic acid (N-acetylneuraminic acid, N-acetyl-9-O-acetylneuraminic acid and N-glycolylneuraminic acid | |
| 3-O-sulfated heparan sulfate | |
| Cell adhesion and cell–cell contact proteins | α-Dystroglycan |
| Coxsackievirus-adenovirus receptor (CAR; Ig superfamily) | |
| CD4 (Ig superfamily) | |
| CEACAMs (including Bgp1a, Bgp2, and pregnancy-specific glycoprotein) | |
| Intercellular adhesion molecule type 1 (ICAM-1; Ig superfamily) | |
| Integrins | |
| Junction adhesion molecule (JAM) | |
| Laminin receptor (high affinity) | |
| MHC class I and β2-microglobulin | |
| Neural cell adhesion molecule (NCAM) | |
| Signaling lymphocyte activation molecule (SLAM or CDw150) | |
| Vascular adhesion molecule 1 (VCAM-1) | |
| Chemokine receptors and G-protein-coupled receptors | CC chemokines subfamily: CCR1, CCR2, CCR3, CCR8 |
| CXC chemokines subfamily: CX3CR1, CXCR4 (Fusin), CXCR6 (BONZO) | |
| G-protein-coupled receptor: GPR1, GPR15 (BOB) | |
| Complement control protein superfamily | CD21 (CR2) |
| CD46 | |
| CD55 (DAF) | |
| CD59 | |
| Growth factor receptors | Epidermal growth factor receptor (EGFR) |
| Fibroblast growth factor receptor (FGFR) | |
| Low-affinity nerve growth factor receptor | |
| Low-density lipoprotein receptor-related proteins | Low-density lipoprotein receptor (LDLR) |
| Very low-density lipoprotein receptor (VLDLR) | |
| Tva (receptor for subgroup A avian sarcoma and leukosis virus) | |
| High-density lipoprotein receptor-related proteins | SR-BI (scavenger receptor class B type I) |
| Poliovirus receptor-related proteins | Nectin-1 α (Prr1α, HveC), β (Prr1β, HIgR), and γ (Ig superfamily) |
| Nectin-2 α (Prr2α, HveB) and δ (Prr1δ) (Ig superfamily) | |
| Poliovirus receptor (PVR or CD155; Ig superfamily) | |
| Transporter proteins | Murine cationic amino acid transporter (MCAT-1) |
| Phosphate transporter proteins (Pit1 and Pit2) | |
| RD114⧸simian type D retrovirus receptor (RDR) | |
| Xenotropic and polytropic retrovirus receptor (XPR1) | |
| Tumor necrosis factor receptor-related proteins | HveA (HVEM) |
| CAR1 (receptor for subgroups B and D of avian leukosis virus) | |
| TEF (receptor for subgroup E of avian leukosis virus) | |
| Other proteins | Acetylcholine receptor |
| Aminopeptidase N (APN or CD13; metalloproteinase) | |
| β-Adrenergic receptor | |
| Carboxypeptidase D | |
| Members of the tetraspanin family: CD9, CD81 | |
| Folate receptor-α | |
| Glucose regulated protein 78 (GRP78; member of the heat shock protein 70 family) | |
| Hyaluronidase-2 (HYAL2; tumor suppressor) | |
| Mannose receptor | |
| Transferrin receptor (TfR) | |
| UDP-galactose transporter |
Bgp, biliary glycoprotein; CEACAM, carcinoembryonic antigen-cell adhesion molecule; CR, complement receptor; DAF, decay-accelerating factor; HIgR, herpesvirus immunoglobulin-like receptor; Hve, herpesvirus entry protein; HVEM, herpesvirus entry protein mediator; Ig, immunoglobulin; MHC, major histocompatibility complex; Prr, poliovirus receptor related.
It has not been possible to predict the type of receptors likely to be used by a virus from its phylogenetic position, nor from its biological properties. The diversity of receptors is reciprocated by their unforeseeable exploitation by viruses, independently of their genome structure and replication strategy. The picornaviruses, which share structural features in their capsids, may use as receptors molecules as diverse as integrins, glycoproteins of the immunoglobulin gene superfamily, decay accelerating factor (DAF or CD55), or sialic acid or sialylated proteins or lipids (Table II). A survey of the more complex coronaviruses does not modify the picture: receptors for coronaviruses include aminopeptidase N, the sialic acid N-acetyl-9-O-acetylneuraminic acid, N-glycolylneuraminic acid, and biliary glycoproteins, which belong to the carcinoembryonic antigen family of the immunoglobulin superfamily. Some of these receptors are shared by viruses associated with different pathologies (Table II).
At least three different receptors have been proposed to be involved in the entry of human hepatitis C virus (HCV) into liver cells: CD81, a member of the tetraspanin superfamily of proteins (Pileri et al., 1998), the low-density lipoprotein receptor (LDLR) (Agnello et al., 1999), and the human scavenger receptor class B type I (hSR-BI) (Scarselli et al., 2002) (Table III ). Binding of HCV to CD81 is strain specific, and the binding affinity can be modulated by hypervariable sequences in the envelope protein (Roccasecca et al., 2003). Other hepatitis viruses, despite sharing a specificity for hepatocytes as their main target cells, employ other receptor molecules. The human hepatitis A virus receptor HAVcr-1 is a mucine-like class I integral membrane glycoprotein, and the receptor for duck hepatitis B virus is the C domain of the carboxypeptidase D, gp180 (Urban et al., 2000).
Table III.
Cell Surface Molecules Proposed to be Involved in Virus Entry Processes
| Virusa | Cell surface moleculesb |
|---|---|
| Double-stranded DNA viruses | |
| Adenoviridae | |
| Human adenovirus | CAR (except subgroup B Ad3 and Ad7; and subgroup D Ad8 and Ad37) (Bergelson 1997, Tomko 1997, Roelvink 1998, Roelvink 1999, Bewley 1999, Freimuth 1999); murine CAR (Tomko 1997, Bergelson 1998); integrins αvβ3, αvβ5 (Wickham 1993, Mathias 1998, Chiu 1999), αvβ1 (Li et al., 2001); MHC class I (α2 domain), but not allele HLA-A∗0201 (Hong 1997, Davison 1999); HS-GAG (Dechecchi 2000, Dechecchi 2001) |
| Human adenovirus 8, 19a, 37 | Sialic acid (α2,3-linked) (Arnberg 2000a, Arnberg 2000b, Arnberg 2002) |
| Canine adenovirus 2 | CAR (Soudais et al., 2000) |
| Avian adenovirus CELO | CAR (Tan et al., 2001) |
| Herpesviridae, alphaherpesvirinae | |
| HSV-1⧸HHV-1 | HS-GAG and CS-GAG (WuDunn 1989, Shieh 1992, Banfield 1995); 3-O-sulfated HS (HSV-1 only) (Shukla 1999a, Trybala 2000); HveA⧸HVEM (Montgomery 1996, Whitbeck 1997, Mauri 1998); nectin-1 alpha⧸Prr1α⧸HveC, beta⧸Prr1β⧸HIgR, and gamma (Cocchi 1998, Geraghty 1998, Lopez 2001, Milne 2001); nectin-2 alpha⧸Prr2α⧸HveB and delta⧸Prr1δ (HSV-1 mutant Rid and HSV-2) (Warner 1998, Lopez 2000); murine HveA and HveC (Menotti et al., 2000); porcine HveC (Milne et al., 2001) |
| HSV-2⧸HHV-2 | |
| VZV⧸HHV-3 | HS-GAG (Zhu et al., 1995); manose-6-phosphate-dependent receptor (Zhu et al., 1995) |
| BHV-1 | HS-GAG (Okazaki et al., 1991); nectin-1 alpha⧸Prr1α⧸HveC and beta⧸Prr1β⧸HIgR (Geraghty 1998, Spear 2000); porcine HveC (Milne et al., 2001); murine HveC (Menotti et al., 2000); PVR⧸CD155 (Geraghty et al., 1998) |
| PrV | HS-GAG (Mettenleiter et al., 1990); nectin-1 alpha⧸Prr1α⧸HveC (Geraghty et al., 1998); nectin-2 alpha⧸Prr2α⧸HveB (Warner et al., 1998); murine HveB (Shukla et al., 1999b); porcine HveC (Milne et al., 2001); murine HveC (Menotti et al., 2000); PVR⧸CD155 (Geraghty et al., 1998) |
| Herpesviridae, betaherpesvirinae | |
| HCMV⧸HHV-5 | HS-GAG (Compton et al., 1993); APN⧸CD13 (Soderberg et al., 1993); MHC class I via β2-microglobulin (Grundy et al., 1987) |
| HHV-6 | CD46 (Santoro et al., 1999) |
| HHV-7 | HS-GAG (Secchiero 1997, Skrincosky 2000); CD4 (Lusso et al., 1994) |
| KSHV⧸HHV-8 | HS-GAG (Akula 2001, Birkmann 2001, Wang 2001); integrin α3β1 (Akula et al., 2002) |
| MCMV | MHC class I (H-2Dd and Kb) (Wykes et al., 1993) |
| Herpesviridae, gammaherpesvirinae | |
| EBV⧸HHV-4 | CD21⧸CR2 (Fingeroth 1984, Frade 1985, Nemerow 1985); MHC class II (HLA-DR, -DP, -DQ) (Reisert 1985, Li 1997, Haan 2000, Haan 2000) |
| BHV-4 | HS-GAG (Vanderplasschen et al., 1993) |
| Papovaviridae | |
| Papillomavirus | α6 integrins (Evander 1997, McMillan 1999); HS-GAG (Giroglou et al., 2001) |
| Murine polyomavirus | Sialic acid (N-acetyl neuraminic acid) (Stehle 1994, Stehle 1996); integrin α4β1 (Caruso et al., 2003) |
| SV40 | MHC class I (Atwood 1989, Breau 1992) |
| Poxviridae | |
| Vaccinia virus | HS-GAG and CS-GAG (Chung 1998, Hsiao 1999) |
| Myxoma virus | CCR1, CCR5, CXCR4 (Lalani et al., 1999) |
| Single-stranded DNA viruses | |
| Parvoviridae | |
| AAV2 | HS-GAG (Summerford 1998, Qiu 2000); FGFR 1 (Qing et al., 1999); integrin αcβ5 (Summerford et al., 1999) |
| Human virus B19 | Erytrocyte P antigen (globoside) (Brown 1993, Chipman 1996) |
| Bovine parvovirus | Sialic acid, glycophorin A (Thacker and Johnson, 1998) |
| Canine parvovirus | Sialic acid (Barbis et al., 1992); human, feline and canine TfR (Parker 2001, Hueffer 2003) |
| Feline parvovirus | Sialic acid (Barbis et al., 1992); human and feline TfR (Parker et al., 2001) |
| DNA and RNA reverse transcribing viruses | |
| Hepadnaviridae | |
| HBV | Sialic acid (Komai et al., 1988); asialoglycoprotein receptor (Treichel et al., 1994); endonexin II (Hertogs et al., 1993) |
| Duck hepatitis B virus | Carboxypeptidase D (Breiner 1998, Urban 1998, Tong 1999, Urban 2000) |
| Retroviridae (alpharetrovirus) | |
| ALV-A | Tva (Bates 1993, Connolly 1994) |
| ALV-B, -D | CAR1 (Brojatsch 1996, Adkins 2001) |
| ALV-E | TEF (Adkins et al., 1997) |
| Retroviridae (betaretrovirus) | |
| MMTV | MTVR⧸mouse TfR-1 (Ross et al., 2002) |
| JSRV, ONAV | HYAL2 (Rai 2001, Miller 2003) |
| SRV (receptor group 1) | RDR⧸R-receptor⧸ATB0⧸ASCT2⧸Na(+)-dependent neutral amino acid transporter type 2 (Kewalramani 1992, Koo 1994, Rasko 1999, Tailor 1999b) |
| Retroviridae (gammaretrovirus) | |
| BaEV (receptor group 1) | RDR⧸R-Receptor⧸ATB0⧸ASCT2 ⧸Na(+)-dependent neutral amino acid transporter type 2 (Kewalramani 1992, Koo 1994, Rasko 1999, Tailor 1999b) |
| FeLV-B (receptor group 5) | Pit1⧸Glvr (Takeuchi 1992, Miller 1996); Pit2 ⧸Ram-1⧸Glvr-2 (mutants of FeLV-B only) (Sugai et al., 2001) |
| FeLV-C (receptor group 4) | FLVCR (Tailor et al., 1999c) |
| FeLV-T | Pit1⧸Glvr; FeLIX (Anderson 2000, Lauring 2001) |
| GALV (receptor group 5) | Pit1⧸Glvr (O'Hara 1990, Miller 1994, Miller 1996) |
| MLV-A (receptor group 2) | Pit2 ⧸Ram-1⧸Glvr-2 (Miller 1994, Miller 1994, van Zeijl 1994) |
| MLV-10A1 | Pit1⧸Glvr; Pit2 ⧸Ram-1⧸Glvr-2 (Miller 1994, Miller 1996) |
| MLV-E | MCAT-1⧸REC-1 ⧸ecoR⧸ATRC-1 (Albritton 1989, Wang 1991a) |
| MLV-X (receptor group 3) | XPR1 ⧸X-receptor⧸Rmc-1⧸sxv (Battini 1999, Tailor 1999a, Yang 1999) |
| P-MLV (receptor group 3) | XPR1 ⧸X-receptor⧸Rmc-1⧸sxv (Battini 1999, Tailor 1999a, Yang 1999) |
| RD114 (receptor group 1) | RDR⧸R-receptor⧸ATB0⧸ASCT2 ⧸Na(+)-dependent neutral amino acid transporter type 2 (Kewalramani 1992, Koo 1994, Rasko 1999, Tailor 1999b) |
| REV (receptor group 1) | RDR⧸R-receptor⧸ATB0⧸ASCT2 ⧸Na(+)-dependent neutral amino acid transporter type 2 (Kewalramani 1992, Koo 1994, Rasko 1999, Tailor 1999b) |
| Retroviridae (deltaretrovirus) | |
| BLV (receptor group 6) | BLVRcp1⧸Blvr (Ban et al., 1993) |
| HTLV-I (receptor group 7) | MHC class I; interleukin 2 receptor (Clarke 1983, Lando 1983, Kohtz 1988) |
| Retroviridae (lentiviruses) | |
| FIV-A, -B | CXCR4⧸fusin (Willett 1997, Poeschla 1998, Richardson 1999, Frey 2001); feline homologue of CD9 (Willett et al., 1994) |
| HIV-1 (receptor group 8) | HS-GAG (Patel 1993, Roderiquez 1995, Saphire 2001); CD4 (Dalgleish 1984, Klatzmann 1984, Maddon 1986, McDougal 1986); CXCR4⧸Fusin; CCR5⧸CC-CKR5; CCR2b⧸CC-CKR2b; CCR3⧸CC-CKR3; CCR8⧸Chem-1; BOB⧸GPR15; Bonzo⧸CXCR6⧸STRL33⧸TYMSRT; GPR1; APJ (Alkhatib 1996, Choe 1996, Deng 1996, Doranz 1996, Dragic 1996, Feng 1996, Lapham 1996, Liao 1997, Loetscher 1997, Rucker 1997, Choe 1998, Edinger 1998a, Shimizu 1999); US28 (Pleskoff 1997, Rucker 1997); BLTR (Owman et al., 1998); CD8 (Saha et al., 2001); GalCer (Bhat 1991, Fantini 1993); integrin αvβ3 (Lafrenie et al., 2002) |
| HIV-2 (receptor group 8) | CD4; CXCR4⧸fusin; CCR5⧸CC-CKR5; CCR8⧸Chem-1; GPR1 (Owen 1998, Reeves 1999, Shimizu 1999, Liu 2000); GalCer (Hammache et al., 1998); US28 (Pleskoff et al., 1997) |
| SIV (receptor group 8) | CD4; CCR3⧸CC-CKR3; CCR5⧸CC-CKR5; CCR8⧸Chem-1; BOB⧸GPR15; Bonzo⧸CXCR6⧸STRL33⧸TYMSRT; GPR1; APJ (Alkhatib 1997, Deng 1997, Farzan 1997, Rucker 1997, Choe 1998, Edinger 1998b, Liu 2000) |
| Visna virus | Ovine MHC class II (Dalziel et al., 1991) |
| Double-stranded RNA Viruses | |
| Reoviridae | |
| Reovirus type 1 | JAM (Barton 2001a, Barton 2001b); carbohydrate (unknown nature) (Lerner 1963, Chappell 2000); EGFR (Strong 1993, Tang 1993) |
| Reovirus type 3 | JAM (Barton 2001a, Barton 2001b); sialic acid (Gentsch 1987, Paul 1989, Chappell 2000); type A glycophorin (Paul and Lee, 1987); β-adrenergic receptor (Co 1985b, Choi 1988, Donta 1990) |
| Group A human rotavirus (strain Wa) | Integrins αvβ3 (Guerrero et al., 2000); α2β1 (VLA-2) (Ciarlet et al., 2002) |
| Group A simian rotavirus (strains SA-11 and RRV) | Sialic acid (not RRV variant nar 3) (Yolken 1987, Willoughby 1990, Mendez 1993, Mendez 1999, Ciarlet 1999), gangliosides (Superti 1991, Srnka 1992, Delorme 2001); integrins α2β1 (VLA-2), α4β1 (VLA-4) (Hewish 2000, Zarate 2000, Ciarlet 2002), αvβ3 (Guerrero et al., 2000) |
| Group A porcine rotavirus (strain OSU) | Gangliosides (Rolsma 1994, Rolsma 1998); integrins α2β1 (VLA-2) (Coulson 1997, Ciarlet 2002) |
| Negative-sense, single-stranded RNA viruses | |
| Arenaviridae | |
| LCMV | α-Dystroglycan (Cao 1998, Kunz 2001) |
| Lassa fever virus | α-Dystroglycan (Cao et al., 1998) |
| New World arenavirus (clade C, but not clade A and B viruses) | α-Dystroglycan (Spiropoulou et al., 2002) |
| Bunyaviridae | |
| Hantaviruses | β3 integrins (Gavrilovskaya 1998, Gavrilovskaya 1999) |
| Filoviridae | |
| Ebola virus | FR-α (Chan et al., 2001) |
| Marburg virus | FR-α (Chan et al., 2001); asialoglycoprotein receptor (Becker et al., 1995) |
| Orthomyxoviridae | |
| Influenza A, B | Sialic acid (N-acetylneuraminic acid, α2,3 and α2,6-linked) (Paulson 1979, Weis 1988, Eisen 1997); mannose receptor (Reading et al., 2000) |
| Influenza C | Sialic acid (N-acetyl-9-O-acetylneuraminic acid) (Herrler 1985, Rosenthal 1998) |
| Paramyxoviridae, Paramyxovirinae | |
| Human parainfluenza virus 1, 3 | Sialic acid residues on gangliosides glycolipids (Suzuki et al., 2001) |
| Measles virus | CD46 (Dorig 1993, Naniche 1993, Schneider-Schaulies 1995, Horvat 1996, Buckland 1997, Buchholz 1997, Niewiesk 1997); SLAM (Tatsuo et al., 2000b) |
| Sendai virus | Sialic acid residues on glycophorin and gangliosides GD1a, GT1b, GQ1b (glycolipids) (Markwell 1980, Wu 1980, Markwell 1981, Markwell 1986, Wybenga 1996); asialoglycoprotein receptor (Markwell 1985, Bitzer 1997) |
| NDV | Sialic acid (Crennell et al., 2000) |
| Rinderpest virus | SLAM (Tatsuo et al., 2001) |
| Canine distemper virus | CD9 (Loffler et al., 1997); SLAM (Tatsuo et al., 2001) |
| Paramyxoviridae, pneumovirinae | |
| HRSV | Iduronic acid-containing glycosaminoglycans (heparan sulfate and chondroitin sulfate B) (Feldman 1999, Hallak 2000, Martinez 2000); CX3CR1 (Tripp et al., 2001) |
| Rhabdoviridae | |
| Rabies virus | Acetylcholine receptor (Lentz 1982, Lentz 1984, Hanham 1993, Gastka 1996); NCAM (Thoulouze et al., 1998); low-affinity nerve growth factor receptor p75NTR (Tuffereau 1998, Tuffereau 2001); sialylated gangliosides |
| (Superti et al., 1986); phospholipids (Superti et al., 1984) | |
| VSV | Phosphatidylserine (Schlegel et al., 1983); phosphatidylinositol (Mastromarino et al., 1987) |
| Positive-sense, single-stranded RNA viruses | |
| Arteriviridae | |
| Lactate dehydrogenase virus | Mouse Ia antigens (Inada and Mims, 1984) |
| Coronaviridae | |
| HCoV-229E | Human APN (CD13) (Yeager 1992, Lachance 1998); feline APN (Tresnan et al., 1996) |
| HCoV-OC43 | Sialic acid (N-acetyl-9-O-acetylneuraminic acid) (Vlasak et al., 1988) |
| TGEV | Porcine APN (Delmas et al., 1992); feline APN (Tresnan et al., 1996); canine APN (Benbacer et al., 1997); bovine APN (Benbacer et al., 1997); sialic acid (N-glycolylneuraminic acid) (Schultze 1995, Schultze 1996) |
| BCV | Sialic acid (N-acetyl-9-O-acetylneuraminic acid) (Vlasak 1988, Schultze 1991, Schultze 1992) |
| FIPV | Feline APN (Tresnan et al., 1996); canine APN (Benbacer et al., 1997) |
| CCV | Canine APN (Benbacer et al., 1997); feline APN (Tresnan et al., 1996) |
| MHV | CEACAMs (Dveksler 1991, Williams 1991a, Compton 1992, Yokomori 1992, Dveksler 1993, Chen 1997a), including biliary glycoproteins (Bgp1a, Bgp2) (Nedellec et al., 1994) and pregnancy-specific glycoprotein (Chen et al., 1995) |
| Flaviviridae | |
| Dengue virus | HS-GAG (Chen 1997b, Germi 2002) |
| Yellow fever virus | HS-GAG (Germi et al., 2002) |
| HCV | LDLR protein family (Agnello 1999, Monazahian 1999); CD81 (Pileri 1998, Flint 1999); tamarin CD81 (Allander et al., 2000); human SR-BI (Scarselli et al., 2002) |
| BVDV | LDLR protein family (Agnello et al., 1999) |
| GB virus C⧸hepatitis G virus | LDLR protein family (Agnello et al., 1999) |
| CSFV | HS-GAG (Hulst 2000, Hulst 2001) |
| Picornaviridae (aphthovirus) | |
| FMDV | HS-GAG (Jackson 1996, Sa-Carvalho 1997, Fry 1999, Baranowski 2000); integrins αvβ3 (Fox 1989, Berinstein 1995, Jackson 1997, Neff 1998), αvβ6 (Jackson et al., 2000b), αvβ1 (Jackson et al., 2002), α5β1 (Jackson et al., 2000a) |
| Picornaviridae (cardiovirus) | |
| EMCV | VCAM-1 (Huber, 1994); sialylated glycophorin A (Allaway and Burness, 1986) |
| Picornaviridae (enterovirus) | |
| Coxsackievirus A9 | Integrin αvβ3 (Roivainen 1991, Roivainen 1994, Berinstein 1995, Triantafilou 1999); GRP78 (Triantafilou et al., 2002); MHC class I via β2-microglobulin (Triantafilou et al., 1999) |
| Coxsackievirus B1-6 | CAR (Bergelson 1997, Shafren 1997c, Tomko 1997, Martino 2000, He 2001); murine CAR (Tomko 1997, Bergelson 1998) |
| Coxsackievirus B1, B3, B5 | DAF (Bergelson 1995, Shafren 1995, Martino 1998); integrin αvβ6 (Agrez et al., 1997) |
| Coxsackievirus A21 | DAF (Shafren et al., 1997b); ICAM-1 (Shafren 1997a, Shafren 1997b, Shafren 1997b, Xiao 2001) |
| Echovirus 1, 8 | Integrin α2β1 (VLA-2) (Bergelson 1992, Bergelson 1993, King 1995) |
| Echovirus 9Barty | Integrin αvβ3 (Nelsen-Salz et al., 1999) |
| Echovirus 3, 6, 7, 11–13, 20, 21, 24, 29, 33 | DAF (Bergelson 1994, Ward 1994, Powell 1997); MHC class I via β2-microglobulin (Ward et al., 1998); HS-GAG (Goodfellow et al., 2001); CD59 (Goodfellow et al., 2000) |
| Enterovirus 70 | DAF (Karnauchow et al., 1996) |
| TMEV strains BeAn, DA | Sialic acid (Zhou 1997, Zhou 2000) |
| TMEV strain GDVII | UDP-galactose transporter (or galactose containing glycoprotein) (Hertzler et al., 2001); HS-GAG (Reddi and Lipton, 2002) |
| Poliovirus 1–3 | PVR (Mendelsohn 1989, Racaniello 1996, Belnap 2000, He 2000) |
| Swine vesicular disease virus | CAR (Martino et al., 2000) |
| Picornaviridae (Hepatovirus) | |
| Hepatitis A virus | HAVcr-1 (Kaplan 1996, Feigelstock 1998) |
| Picornaviridae (Rhinovirus) | |
| Major group HRV | ICAM-1 (Greve 1989, Staunton 1989, Tomassini 1989, Olson 1993, Kolatkar 1999) |
| Minor group HRV | VLDLR (Hofer 1994, Marlovits 1998, Hewat 2000); avian homologues of the mammalian LDLR (Gruenberger et al., 1995) |
| HRV 87 | Sialic acid (Uncapher et al., 1991) |
| Picornaviridae (Parechovirus) | |
| Human parechovirus 1⧸echovirus 22 | Integrins αvβ1 and αvβ3 (Triantafilou et al., 2000) |
| Togaviridae | |
| Sindbis virus | High-affinity laminin receptor (Wang et al., 1992); HS-GAG (Byrnes 1998, Klimstra 1998) |
Abbreviations of virus names: AAV2, adeno-associated virus type 2; Ad, adenovirus; ALV, avian leukosis virus; BaEV, baboon endogenous retrovirus; BCV, bovine coronavirus; BHV, bovine herpesvirus; BLV, bovine leukemia virus; BVDV, bovine viral diarrhea virus; CCV canine coronavirus; CSFV classical swine fever virus; EBV, Epstein–Barr virus; EMCV, encephalomyocarditis virus; FeLV, feline leukemia virus; FIPV, feline infectious peritonitis virus; FIV, feline immunodeficiency virus; FMDV, foot-and-mouth disease virus; GALV, gibbon ape leukemia virus; HBV, hepatitis B virus; HCMV, human cytomegalovirus; HCV, hepatitis C virus; HCoV, human coronavirus; HHV, human herpes virus; HIV, human immunodeficiency virus; HRSV, human respiratory syncytial virus; HRV, human rhinovirus; HSV, herpes simplex virus; HTLV, human T-cell leukemia virus; JSRV, Jaagsiekte sheep retrovirus; KSHV, Kaposi's sarcoma-associated herpesvirus; LCMV, lymphocytic choriomeningitis virus; MCMV, murine cytomegalovirus; MHV, mouse hepatitis virus; MLV-A, amphotropic murine leukemia virus; MLV-E, ecotropic murine leukemia virus; MLV-X, xenotropic murine leukemia virus; MMTV, mouse mammary tumor virus; NDV, Newcastle disease virus; ONAV, ovine nasal adenocarcinoma virus; P-MLV, polytropic murine leukemia virus; PrV, pseudorabies virus; RD114, cat endogenous retrovirus; REV, reticuloendotheliosis virus; SIV, simian immunodeficiency virus; SRV, simian retroviruse; SV40, simian virus 40; TEF, name given to the receptor for subgroup E of avian leukosis virus; TGEV, transmissible gastroenteritis virus; TMEV, Theiler's encephalomyelitis virus; VSV, vesicular stomatitis virus; VZV, varicella zoster virus.
Abbreviations of cellular structures: APJ, name given to a ligand-unknown “orphan” seven transmembrane domain receptor of the central nervous system; APN, aminopeptidase N; BLTR, leukotriene B4 receptor; BLVR, bovine leukemia virus receptor; Bgp, biliary glycoprotein; CAR, coxsackievirus-adenovirus receptor; CAR1, name given to the receptor for subgroups B and D avian leukosis virus; CCR, CC-chemokine receptor; CEACAM, carcinoembryonic antigen-cell adhesion molecule; CR, complement receptor; CS-GAG, chondroitin sulfate glycosaminoglycan; CXCR, CXC-chemokine receptor; DAF, decay-accelerating factor; EGFR, epidermal growth factor receptor; FeLIX, FeLV infectivity X-essory protein; FLVCR, feline leukemia virus subgroup C receptor; FGFR, fibroblast growth factor receptor; FR-α, folate receptor-α; GalCer, galactosylceramide; Glvr, gibbon ape leukemia virus receptor; GPR, G protein-coupled receptor; GRP, glucose-regulated protein; HAVcr, hepatitis A virus cellular receptor 1; HIgR, herpesvirus immunoglobulin-like receptor; HS-GAG, heparan sulfate glycosaminoglycan; HLA, human leukocyte antigen; Hve, herpesvirus entry protein; HVEM, herpesvirus entry protein mediator; HYAL, hyaluronidase; ICAM-1, intercellular adhesion molecule type 1; JAM, junction adhesion molecule; LDLR, low-density lipoprotein receptor; MCAT, murine cationic amino acid transporter; MHC, major histocompatibility complex; MTVR, mouse mammary tumor virus receptor; NCAM, neural cell adhesion molecule; Prr, poliovirus receptor related; PVR, poliovirus receptor; SLAM, signaling lymphocyte activation molecule; SR-BI, scavenger receptor class B type I; RDR, name given to the RD114⧸simian type D retrovirus receptor; TfR, transferrin receptor; Tva, name given to the receptor for subgroup A avian sarcoma and leukosis virus; Pit, inorganic phosphate transporter; VCAM, vascular adhesion molecule; VLA, very late antigen; VLDLR, very low-density lipoprotein receptor; XPR1, xenotropic and polytropic retrovirus receptor.
As in the case of viruses with liver tropism, different viruses sharing preferences for other tissues (neural, lymphoid, etc.) do not generally use the same receptor sites. Each class of tissue offers a variety of molecules that can potentially act as virus receptors, and viruses make use of them presumably as the result of ancestral evolutionary processes. As discussed in Section V, analysis of the complete genomic nucleotide sequences of several prokaryotic and eukaryotic organisms, including the first draft of the human genome, has unveiled the presence of several types of mobile genetic elements, suggesting ancestral exchanges of modules among cells, in which viruses likely played an active role. Our present observations on how viruses can penetrate cells and replicate in them are just a snapshot that reflects an instant out of eons of fluid exchanges among cells and autonomous replicons (Bushman, 2002).
The presence on the cell surface of a protein that has been identified as the receptor for a given virus may not be sufficient for a productive viral infection, and there may be multiple mechanisms behind such restrictions: functional domains of the receptor may be blocked in some cellular context, additional proteins (or other cofactors) may be needed, or cells may exhibit impediments for completion of the infection cycle, despite an initial successful interaction with a functional receptor. Mice that are transgenic for the functional form of the poliovirus receptor (PVR or CD155) become susceptible to poliovirus and, upon infection, develop the typical limb paralysis. However, the tissue and organ distribution of the PVR mRNA does not correspond to the sites where virus replicates (Nomoto 1994, Ren 1992, Ren 1990). Likewise, the N-acetylneuraminic acid (sialic acid), which is the receptor for human influenza virus, is common on glycosylated molecules on cell surfaces. As a consequence, influenza viruses can bind to many cell types and yet productive infection occurs generally in the epithelial cells of the respiratory tract when the virus causes disease.
A. A Virus May Use Different Receptor Types
Some viruses apparently use only a single receptor to infect their target cells, whereas others are able to exploit several alternative receptors to initiate their replication in different cell lines or even to enter the same cell type (Table III). Evidence shows that foot-and-mouth disease virus (FMDV) may employ integrins αvβ1, αvβ3, and αvβ6 as receptors. Recognition of these integrins is dependent on an Arg-Gly-Asp (RGD) triplet found in the surface G-H loop of capsid protein VP1, and this triplet is very conserved among natural isolates of the seven FMDV serotypes (Domingo 1990, Domingo 1992, Mateu 1995). However, several studies suggest the use of alternative receptors by FMDV, both in cell culture and in vivo. Upon adaptation of FMDV to cell culture, mutations may occur that result in the acquisition of positively charged amino acids at key residues of the capsid surface that allow FMDV to enter cells via heparan sulfate (HS) and possibly other receptors (Baranowski 1998, Baranowski 2000, Escarmis 1998, Fry 1999, Jackson 1996, Sa-Carvalho 1997, Sevilla 1998, Zhao 2003). Following this seminal observation with FMDV, studies with other viruses have demonstrated that passage in cell culture results in evolutionary changes that allow the viruses to use HS for cell entry. Examples include Sindbis virus adapted to BHK cells (Klimstra et al., 1998), classical swine fever virus propagated in swine kidney cells (Hulst et al., 2000), and variants of human rhinovirus 89 adapted to HEp2 cells (Reischl 2001, Vlasak 2002) (Section II,B).
A well-documented case of use of multiple receptors is that of lentiviruses HIV-1, HIV-2, and simian immunodeficiency virus (SIV). Almost all strains described to date require interactions with two molecules on the cell surface. One of them acts as an attachment receptor, with which viral glycoprotein gp120 interacts. This interaction promotes conformational changes that expose the fusogenic peptide of gp41, which, in turn interacts with the fusion receptor. This is essential for the fusion of viral and cellular membranes and therefore allows the viral capsid to enter the host cell. The group of cellular molecules involved in lentivirus fusion are referred to as coreceptors. It has been suggested that those strains shown to use only the fusion receptor have already undergone, at least partially, the conformational changes normally induced by the attachment receptor (Berger et al., 1999).
The helper T-cell differentiation antigen CD4 is an important attachment receptor for HIV-1, HIV-2, and SIV, but other cell surface molecules, including galactosyl ceramide (GalCer), syndecans, and other glycosaminoglycans, have been found to play a role in the attachment of these viruses (Baba 1988, Ito 1987, Saphire 2001, Zhang 2002). The preferential use of some attachment receptors over others is thought to be influenced by their relative abundance on the cell surface. Thus, on macrophages, CD4 is scarce and HIV attachment takes place mainly through HS proteoglycans, especially syndecans (Saphire et al., 2001). It has been reported that integrin αvβ3 is involved in the infection of macrophages that have differentiated in vitro (Lafrenie et al., 2002); certainly, the significance of this interaction in vivo deserves further study. HIV infection of colon epithelia in vivo is CD4–independent and is believed to take place through GalCer (Bhat 1993, Fantini 1993). Neuronal cells and intestinal epithelia are rarely infected in vivo, but binding of HIV envelope glycoproteins to GalCer contributes to the selection of CCR5-binding variants following vertical transmission (Meng et al., 2002). This GalCer-mediated selection may also contribute to pathogenesis, e.g., in the gastrointestinal disorders associated with HIV infection and in neuronal dysfunction in AIDS dementia. HIV populations that acquired the capacity to bind CD8 and replicate in CD8+ T cells have been recovered from a patient at a late stage of the infection (Saha et al., 2001). A wide variety of molecules from different structural and functional families can act as fusion receptors (or coreceptors). The most important is the chemokine receptor superfamily, for many of its members can bind HIV and⧸or SIV. A related protein is a β-chemokine receptor US28, encoded by human cytomegalovirus (Pleskoff et al., 1997). There are also other coreceptors, such as the leukotriene B4 receptor (BLTR) (which has structural homology with chemokine receptors) (Owman et al., 1998) and an orphan receptor of the central nervous system (APJ). Usually, most HIV strains evolve in vivo from using mainly CCR5 (R5 viruses) to using mainly CXCR4 (X4 viruses), but many other coreceptors can be used simultaneously.
An example in which the functional receptor for a virus is a complex of several cell molecules that the virus uses in a multistep process is found among Reoviridae (Mendez et al., 1999). Rotaviruses are important viral agents of acute gastroenteritis in young children and in many animal species. Rotavirus strains differ in their requirement of sialic acid for initial binding to the cell surface: a minority of animal rotaviruses require the presence of sialic acid residues on the cell surface for efficient binding and infectivity (Arias 2001, Zarate 2000), but most animal and human rotaviruses do not. However, binding to sialic acid is not an essential step for rotavirus infection, as confirmed by the isolation of sialic-independent variants from a sialic-dependent strain (Mendez et al., 1993). The existence of a postattachment cell receptor common to most, if not all, rotavirus strains has been proposed. Integrin αvβ3 blocks the infectivity, although not the binding, of the sialic acid-dependent rhesus rotavirus (RRV), its sialic acid-independent mutant nar3, and the sialic acid-independent human rotavirus strain WA, suggesting that αvβ3 interacts with both neuraminidase-sensitive and neuraminidase-resistant rotaviruses at a postbinding, probably penetration, step (Guerrero et al., 2000). Other integrins, such as α2β1, α4β1, and αxβ2, have also been reported to play a role in rotavirus entry (Ciarlet 2002, Coulson 1997, Hewish 2000, Londrigan 2000).
Cell recognition by herpesviruses offers a classical example of complexity in virus–receptor interactions. The initial contact of herpes simplex virus 1 (HSV-1), the prototype of the Alphaherpesvirinae subfamily, with cells is usually the binding of virus to HS proteoglycans (Shieh 1992, Shukla 2001, WuDunn 1989), mediated by glycoprotein gC and, to a lesser extent, by gB. Then, glycoprotein gD can interact with at least three different classes of molecules that can act as entry mediators for HSV-1. These molecules include HveA (which belongs to the family of tumor necrosis receptor proteins), nectin-1 (HveC) and nectin-2 (HveB) (two members of the immunoglobulin superfamily), and 3-O-sulfated HS (reviewed in Spear 2000, Shukla 2001). The binding of gD to one of these different receptors initiates fusion between the viral envelope and the cell membrane. The related Epstein–Barr virus (EBV), a transforming virus of the gammaherpesvirinae subfamily, displays a marked B-cell lymphotropism associated with the expression of receptor CD21 (or CR2). EBV has been associated with a number of human proliferative diseases, including B-cells lymphomas, a clinical condition encountered in immunocompromised hosts (Krance et al., 1999). However, EBV can replicate in differentiated epithelial cells that do not express CD21, suggesting that other receptors may be involved in EBV infections (reviewed in Schneider-Schaulies, 2000).
Studies with vaccinia virus revealed that two different forms of the same virus can bind to different cellular receptors. Intracellular mature virus and extracellular enveloped virus are two antigenically and structurally distinct infectious virions that bind to unidentified and possibly different cellular receptors (Vanderplasschen and Smith, 1997). Other examples of the use of several receptor types by a virus include adenoviruses, coronaviruses, hepatitis C virus, influenza virus, measles virus, and rabies virus (references and additional examples in Table III and Section IV).
B. A Receptor Type Can Be Used by Several Viruses and Other Microbial Pathogens
Early evidence that viruses can share receptors was obtained in studies on interference in cell binding between different viruses (Lonberg-Holm et al., 1976). Many receptor types are used by viruses of different families, and several examples can be identified by examining Table III (Baranowski et al., 2001). A remarkable example is the shared use of coxsackievirus adenovirus receptor (CAR) by some human and animal adenoviruses and by some picornaviruses of different genera (Bergelson et al., 1997). These viruses belong to immeasurably distant families and are associated with unrelated diseases. Adenoviruses 2 and 5 are agents of respiratory disease in children, whereas coxsackieviruses B1 to B6 may be associated with febrile illness, meningitis, and some types of cardiopathies.
Other examples of shared receptors by viruses include cell surface components belonging to a wide range of receptor classes such as cell adhesion and cell–cell contact proteins (CD4, ICAM-1, integrins, MHC I), chemokine receptors (CXCR4), members of the complement control protein superfamily (CD46, DAF), low-density lipoprotein receptor and poliovirus receptor-related proteins, transporter proteins, and aminopeptidase N, as well as extracellular matrix components and sugar derivatives (references and additional examples in Table III and Section IV) (Baranowski et al., 2001).
Ubiquitous extracellular matrix components such as HS glycosaminoglycans are the preferential class of receptors employed by many viruses (Table III). The abundance of such molecules at the cell surface may facilitate initial contact with the virus, but subsequent interactions with more specific receptors may be needed. For many viral systems, attachment to HS is not an absolute requirement for infection, and cells devoid of HS can be infected, although with a reduced efficiency. Binding to heparin is a phenotypic trait displayed by several viruses propagated in cell culture, presumably because the interaction with HS residues may provide a selective advantage for viruses evolving in cultured cells (Section IV,B). Microbial pathogens that bind to cell surface HS proteoglycans include numerous intracellular (Borrelia burgdorferia, Chlamydia trachomatis, Listeria monocytogenes, Mycobacterium spp., Neisseria gonorrhoeae) and extracellular (Bordetella pertussis, Haemophilus influenzae, Helicobacter pylori, Staphylococcus aureus, Streptococcus spp.) bacteria, fungi, and other cellular parasites (Leishmania spp., Plasmodium spp., Trypanosoma cruzi) (reviewed in Bernfield et al., 1999).
Integrins have several features that make them attractive portals of entry for pathogens (reviewed in Krukonis 1997, Mims 2001, Parkes 2000). Interaction of adenovirus with αv integrins has been reviewed (Nemerow 1999, Nemerow 2000) (Table II). A variety of bacterial pathogens also use integrin receptors to either adhere to or enter into host cells. Yersinia pseudotuberculosis is a gram-negative enteropathogen, which infects cells of the gut wall via binding to β1 integrins (Isberg and Leong, 1990), and Yersinia enterocolitica uses integrins to bind to cells of the intestinal ephitelium. The interaction of Yersinia with host surface integrins induces activation of the cytoskeleton and the rearrangement of actin into pseudopods that engulf the bacteria (Krukonis and Isberg, 1997). A number of microbial pathogens, including B. pertussis, Coxiella burnetti, E. coli, Histoplasma capsulatum, Legionella pneumophila, Leishmania spp., and Rhodococcus equi, make use of integrins αMβ2 and αxβ2, also known as complement receptors (CR3 and CR4), to enter the macrophage and avoid the host microbicidal oxidative burst (Aderem 1999, Capo 1999, Krukonis 1997; see review of cell entry mechanisms by diverse pathogens in Mims et al., 2001).
Another recently described example of a receptor type used by several microbial pathogens is CD81, a putative receptor for HCV (Pileri et al., 1998), which is required on hepatocytes for Plasmodium falciparum and yoelii sporozoite infectivity (Silvie et al., 2003).
C. Virus–Receptor Interactions Revealed by Structural Studies
The specific interactions between a viral protein or glycoprotein and receptor molecules are amenable to structural studies, and such studies are providing essential new information for the understanding of virus–receptor interactions. Enveloped viruses, such as HIV, attach to host cells by means of spike-like membrane glycoproteins, whereas most nonenveloped viruses, such as picornaviruses, attach by means of specialized domains integral to their capsids. Some viruses cannot be assigned to one of the two binding modes. For example, adenoviruses are nonenveloped particles, but have trimeric fibers projecting from the vertices of their icosahedral capsid, which terminate in a globular knob domain responsible for the specific interactions with the cellular receptors (Nemerow, 2000). The interaction of the surface hemagglutinin of human influenza virus with N-acetylneuraminic acid and the conformational alterations associated with the pH-dependent membrane fusion stand as one of the best characterized virus entry processes at both functional and structural levels. The entry of human influenza virus into human cells has been reviewed (Skehel and Wiley, 2000) and is not treated here except as an illustration of some concepts in following sections. Viral receptor-binding sites typically comprise highly conserved amino acid residues (Rossmann et al., 1985), a requirement that appears to guarantee survival of the virus. Early crystallographic studies of picornaviruses indicated that these and other viruses may shield their receptor-binding sites in cavities or surface depressions (“canyons”) that are inaccessible to antibody molecules, and this inaccessibility would confer viruses a selective advantage (Rossmann et al., 1985). Subsequent studies with other viruses have shown that receptor-binding sites may also occur in highly exposed regions of the viral surface (He 2002, Hewat 2000, Verdaguer 1995). It may be predicted that when receptor-binding sites and neutralizing antibody-binding sites overlap at the virus surface, amino acid replacements needed for antibody escape may be deleterious as they affect receptor recognition. However, in line with the dynamics of RNA virus populations (Section II,B), this conflict is just one of the several instances in which negative selection may act to maintain variants at low levels and to rescue fit viruses through compensatory mutations. There is no strict requirement that surface residues involved in receptor recognition be entirely shielded from immune attack. Another proposed mechanism to compensate for possible adverse effects of amino acid replacements regarding cell recognition is provided by the water buffer hypothesis, discussed at the end of this section.
Cryoelectron microscopy (cryo-EM) and image reconstruction methods allow direct visualization of virus–receptor complexes, which are usually too large and unstable to be amenable to analysis by high-resolution X-ray crystallography. In addition, when high-resolution structures of the virus and receptor domains are known, a pseudoatomic model of the virus–receptor complex can be reproduced by docking the atomic structures together using the cryo-EM density map as a guide. Cryo-EM analyses of the major group rhinoviruses 16 and 14 (HRV16, HRV14) in complex with intracellular adhesion molecule-1 (ICAM-1 or CD54) (Kolatkar 1999, Olson 1993) and of the enteroviruses poliovirus type 1 Mahoney (PV1) in complex with poliovirus receptor (PVR or CD155) (Belnap 2000, He 2000, Xing 2000), coxsackievirus A21 (CAV21) in complex with ICAM-1 (Xiao et al., 2001) and coxsackievirus B3 (CBV3) in complex with the coxsackie-adenovirus receptor (CAR) (He et al., 2001) have revealed how various receptors bind differently to structurally similar canyons found on the surface of virus particles (Fig. 1 ) (reviewed in Rossmann et al., 2002).
Fig 1.
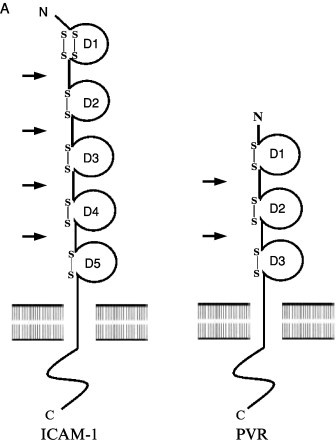
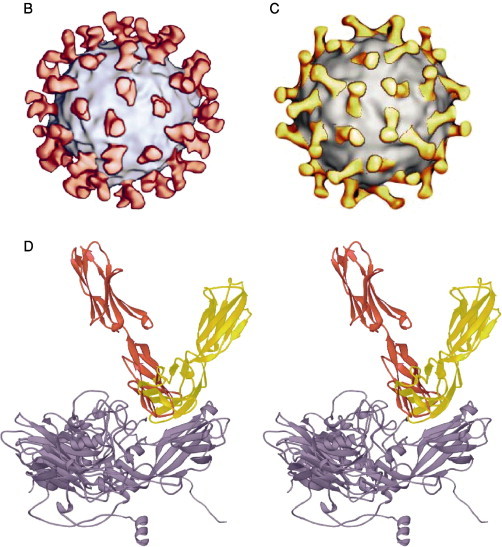
(A) Representation and comparison of the domain structure of ICAM-1 (CD54), the receptor for the major group rhinoviruses, and the poliovirus receptor (PVR or CD155). The immunoglobulin-like domains (labeled D1–D5 or D1–D3) are represented schematically by a circle closed by one or two disulfide bonds. The different Ig domains are linked by a flexible peptide chain. Hinge points are indicated by arrows. (B) Cryo-EM reconstruction showing the complex of HRV16 with its ICAM-1 receptor (from Kolatkar et al., 1999) virus is represented as a gray-scale surface. D1 and D2 domains of ICAM-1 are colored red. (C) Cryo-EM reconstruction of the complex of PV1(M) (gray) with PVR (yellow) (from Xing et al., 2000). (D) ICAM-1 and PVR-binding modes. Stereoview of the ICAM-1 (red) docked onto one icosahedral asymmetric unit of HRV16 (gray) using the cryo-EM map as a guide (PDB accession code 1D3E). The structure of PVR in complex with PV1 (M) (PDB accession code 1DGI) was superimposed for comparison (yellow). ICAM-1 contacts primarily the floor and south wall of the HRV16 canyon. In contrast, PVR overlaps the north and south walls, as well as the floor of the canyon, making additional contacts with the viral surface.
ICAM-1, PVR, and CAR are membrane-anchored glycoproteins that belong to the Immunoglobulin (Ig) superfamily. Their extracellular regions comprise five, three, and two domains, respectively, each with Ig-like folds consisting of a β barrel in which all β strands run parallel or antiparallel to the long axis of the domain (Fig. 1). In the three cases, the amino-terminal domain D1 contains the virus recognition site. This may reflect the steric capacity of the N-terminal Ig domain to penetrate into the picornaviral canyon. All the various receptor molecules utilized by picornaviruses are long, thin, and articulated at hinges between domains, and their properties are consistent with the requirement that the receptor be a molecule able to flex sufficiently to recognize additional sites on the viral surface once the first receptor site has been bound.
The interaction of ICAM-1 with the major group HRVs and with CAV21 and of PVR and CAR with PV1 and CBV3 indicates a common core of partially conserved residues on the canyon of those viruses (He 2001, Rossmann 2000, Xiao 2001). However, the orientation of the receptor molecules is different for each virus–receptor complex (Fig. 1). The orientation of domain D1 is approximately radial in all cases except for poliovirus. The canyon of poliovirus is wider than that of CBVs and HRVs, allowing the tangential binding of PVR into the PV canyon (Belnap 2000, He 2000, Xing 2000). Therefore, the shape and size of the canyon may be important factors that dictate the docking orientation of the receptors. Receptor binding to major group HRVs and enteroviruses is localized within the canyon, at a site adjacent to a hydrophobic pocket within the VP1 β barrel containing an as yet unidentified “pocket factor.” Kinetic analyses of the virus–receptor interaction have shown for both HRVs (Casasnovas and Springer, 1995) and polioviruses (McDermott et al., 2000) that there are two distinct modes of binding whose relative abundance varies with temperature. The binding modes observed in the cryo-EM reconstructions are likely to be the most stable intermediates, although the nature of the interaction may depend on the specific virus–receptor complex.
In contrast to observations with the major group rhinoviruses, cryo-EM reconstructions of the complex between HRV2 and the first three ligand-binding repeats of the very low-density lipoprotein (VLDL) receptor revealed that the receptor for minor group rhinoviruses binds to a star-shaped dome on the five-fold axis and not in the canyon (Hewat et al., 2000). The footprint of V1-3 includes only residues of VP1. Close to the receptor attachment site is the virus-neutralizing immunogenic site A, located within the BC loop of VP1. Because the receptor-binding site of HRV2 is on a protruding domain of the capsid, it is not hidden from immune surveillance. It is remarkable that the receptor-binding site of major group HRVs is very similar to that of enteroviruses, which belong to a different genus, but is essentially different from that of minor group HRVs, which belong to the same genus. Minor group HRVs, which do not bind ICAM-1, are not obviously phylogenetically or structurally distinct from major group HRVs. Furthermore, HRV14, a major group serotype, is more distantly related to another major group serotype, HRV16, than to the minor group representative HRV2. Nevertheless, the residues of HRV2 corresponding to the ICAM-1 footprint on HRV14 or HRV16 lack the charge complementarity observed for major group HRVs (Kolatkar 1999, Verdaguer 2000). In addition, receptors of major group HRVs and enteroviruses both cause virus destabilization, unlike the receptor of minor group HRVs. This is an example of the adaptability to different receptors and the variety of receptor-binding sites exhibited by members of the picornavirus family. A comparison of receptor interactions and entry pathways of different picornaviruses suggests that those receptors that bind into canyons or pits of the capsid induce partial capsid destabilization through displacement of the “pocket factor,” which contributes to capsid stability. This type of receptor-mediated destabilization occurs in poliovirus, some coxsackieviruses, and the major group rhinoviruses, which utilize molecules of the Ig superfamily as receptors. In contrast, those receptor molecules that do not bind into the canyon often do not contribute to virus destabilization and may condition the nature of subsequent steps in the entry process (a destabilizing, acid-mediated step in endosomes, etc.). Examples include the binding of VLDV receptor to the five-fold axis region of the minor group of HRV-2, echovirus 7 binding to DAF and FMDV binding to integrins or other receptor molecules (Table III). It has been suggested that viruses that do not use molecules of the Ig superfamily as receptors may be more variable at receptor recognition sites, facilitating shifts in receptor usage, with consequences for virus pathogenesis (Rossmann et al., 2002).
Other cryo-EM studies have provided indirect evidence about the location of the receptor-binding sites of reoviruses, alphaviruses, adenoviruses, and the picornavirus FMDV. For example, studies on the structure of VP4, the 88-kDa receptor recognition protein of rotavirus, have documented that 60 dimers of VP4 (“spikes”) extend 100 Å above the viral capsid surface in a uniform arrangement that appears to facilitate the binding interaction with cellular receptors (Prasad 1988, Prasad 1990). The distal ends of the spikes are believed to contain the receptor-binding sites, as neutralizing antibodies that bind near the distal ends inhibit viral penetration (Prasad et al., 1990). Cryo-EM and three-dimensional reconstructions have been valuable tools for identifying putative receptor-binding sites on the E2 glycoproteins of two alphaviruses, Sindbis virus and Ross River virus (Smith et al., 1995). This strategy has also facilitated the examination of a highly mobile, Arg-Gly-Asp (RGD)-containing antigenic loop on the adenovirus 2 and 12 particles (Stewart 1997, Chiu 1999) and on the picornavirus FMDV capsid (Hewat 1997, Verdaguer 1999), which harbor integrin-binding sites (Fig. 2 ).
Fig 2.
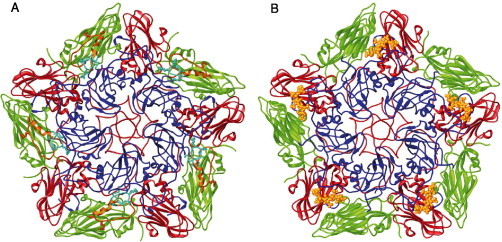
(A) Ribbon representation of a CS8c1 pentamer subunit (VP1, blue; VP2, green; VP3, red). The mobile antigenic G-H loop of VP1 (residues 130–160) is highlighted in yellow in a position corresponding to that found in the complex with the neutralizing antibody SD6 (Hewat et al., 1997) and in cyan for the position determined in the crystallographic structure of the reduced FMDV-O1BFS (Logan et al., 1993). The RGD integrin-binding triplet is depicted as sticks. (B) The structure of FMDV in complex with heparin (Fry et al., 1999). Heparin coordinates for five sugars are shown as yellow ball and sticks.
Detailed understanding of virus–receptor interactions will ultimately require structural analyses at high resolution. Crystallographic studies of interactions between FMDV O1 and HS have shown that the binding site is a shallow depression, of positive electrostatic charge, on the virion surface, contributed by the three external capsid proteins VP1, VP2, and VP3 (Fry et al., 1999) (Fig. 2). The crystal structure revealed that the heparin molecule makes ionic interactions with the viral residues using two sulfates (GlcN-2-2-N-SO3 and GlcN-4-6-O-SO3). Amino acid Arg-56 in VP3 plays a central role in organizing the sulfate groups, making interactions with both. Further polar residues that belong to all three major capsid proteins play a subsidiary role in heparin binding via bridging water molecules. The nonionic interactions observed include van der Waals stacking contact among, His-195 of VP1, the central L-iduronic acid (Idu) ring, and the apolar patch of GlcN-4. The virus surface remained essentially unchanged to accommodate the sugar, suggesting that the shape complementarity may contribute to receptor specificity.
Crystallographic studies of interactions between the HIV gp120 in complex with its receptors (Kwong 1998, Wyatt 1998) and the structurally unrelated adenovirus 12 knob with CAR (Bewley et al., 1999) allowed the identification of key determinants of receptor-binding specificity. These structures revealed that the receptor-binding faces are surface loops exposed to immune selective pressure. Both viruses bury a similar amount of surface area to create an atypical virus–receptor interface because of the presence of a shape mismatch in the surface topography that creates large cavities or channels. The lack of van der Waals interactions between the two protein surfaces may be partially compensated by the presence of solvent molecules filling the cavities. These water molecules would act as a bridge mediating hydrogen bonds between the backbone atoms of the viral protein and the receptor. The viral residues in contact with this water-filled cavity show important sequence variability, whereas surrounding this patch are highly conserved residues, the substitution of which may affect receptor binding. Thus, the observed interfacial cavities may serve a dual purpose as a water buffer between the viral protein and the receptor, and as molecular glue through the establishment of hydrogen bonds between backbone and conserved atoms. The tolerance for variation in the surface of the protein associated with this cavity produces a variational island, which is centrally located between regions required for receptor binding and may help the virus escape from antibodies. By using a noncomplementary interface that traps water molecules, the virus can maintain its receptor specificity while altering its amino acid sequence. This type of interface observed in two very different viruses such as HIV and adenovirus supports the water buffer hypothesis as a new general mechanism by which viruses can complete the first stage of infection successfully. The trapping of water molecules may allow receptor recognition to be less influenced by amino acid substitutions at the relevant sites of the virus surface, minimizing the adverse effects of high mutational pressure (Section II,B) on an essential step in the virus life cycle.
IV. Quasispecies and Shifts in Receptor Usage
A. Minimal Changes in Viral Genomes May Modify Receptor Recognition or Cell Tropism
There are several cases of minimal changes in viral genomes that result in the alteration of receptor specificity or affinity (Baranowski et al., 2001). Early studies with influenza virus identified key residues of virus hemagglutinin (HA) that confer specificity for sialic acid linked to galactose by either an α-2,3 or an α-2,6 linkage. Human H3 influenza viruses bind preferentially to sialic acid linked to galactose by an α-2,6 linkage, whereas avian and equine viruses show a preference for the α-2,3 linkage (Matrosovich 1997, Rogers 1983). Human H1 and H2 viruses, as well as swine H1 viruses, also show preferentially a specificity for Neu5-Acα-2,3Gal. It was established that a single substitution in the 226 position of the HA changed the receptor specificity, which suggests that HAs that differ in the recognition of one sialic acid or another differ in amino acid 226 (Rogers et al., 1983a). Further studies indicated that two mutations in residues 226 and 228 of a human HA allowed replication in ducks. The mutations resulted in a receptor-binding site sequence identical to the known avian influenza virus sequences (Naeve et al., 1984). Other studies on receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates showed that residues 226 and 228 are leucine and serine in human isolates and are glutamine and glycine in avian and equine isolates (Connor et al., 1994), confirming the key role of these two residues in determining the receptor specificity of influenza viruses.
Picornaviruses provide additional examples. Single replacements (Val-160 to Ile in VP1 or Met-62 to Ile in VP4) allow the primate-restricted P1⧸Mahoney strain of poliovirus to paralyze mice (Colston and Racaniello, 1995). The mutation in VP4 may render the virus accessible to a molecule that acts as a virus receptor and which is located on the surface of neurons of the mouse spinal cord. This molecule does not seem to be expressed in the mouse brain (Jia et al., 1999). A mutant of encephalomyocarditis virus termed variant D is usually asymptomatic for rodents, but it can induce diabetes in mice through the destruction of pancreatic β cells. The diabetogenic variant includes amino acid replacement Thr-776 to Ala along the capsid pit, likely affecting receptor interaction and cell tropism (Bae and Yoon, 1993). Important determinants of tropism and pathogenesis of Theiler's murine encephalomyelitis virus (TMEV) have been mapped at various positions on capsid protein VP1, including positions adjacent to the putative virus receptor-binding site. TMEV mutants harboring single amino acid substitutions at these particular VP1 positions displayed altered neurovirulence in susceptible mice and produced only mild symptoms during the acute phase of the infection or were highly attenuated regarding the development of chronic demyelinating disease (Jnaoui 2002, Lin 1998, McCright 1999, Wada 1994). Amino acid residues flanking the RGD integrin-binding motif of FMDV have been shown to influence the selectivity of integrin binding (Jackson 2000a, Jackson 2002).
Research on the Edmonston vaccine strain of measles virus (MV) led to the identification of CD46 (a group of determinants whose function is to protect cells from complement-mediated lysis) as a receptor for this virus. Transgenic mice expressing CD46 may show typical pathogenic manifestations of the virus (Oldstone et al., 1999). Marmosets lacking CD46 were susceptible to several isolates of MV, but not to the Edmonston vaccine strain. Some natural isolates of MV do not enter cells through CD46, instead they use the signaling lymphocyte activation molecule (SLAM or CDW150), a glycoprotein expressed on some types of B and T lymphocytes, as a receptor (Hsu 2001, Tatsuo 2000a, Tatsuo 2000b). Although wild-type MV interacts with SLAM with high affinity, it can also interact with CD46 with low affinity (Masse et al., 2002). A single amino acid replacement in the surface hemagglutinin of the MV envelope determines the ability of the virus to bind CD46 with high affinity (Hsu et al., 1998). Additional receptors may be involved in MV infection (Hashimoto 2002, Oldstone 2002). Despite its relative antigenic stability, it has been estimated that MV mutates at an average rate of 9 × 10−5 substitutions per base copied (Schrag et al., 1999). Therefore, point mutations of the type that led to binding of CD46 should occur frequently during MV replication.
Another example of minimal change in a viral genome that affects receptor recognition, tropism, and pathogenesis is afforded by substitutions in the E2 surface glycoprotein of Sindbis virus. The presence of Arg at position 172 impaired neurovirulence through a decrease of binding to a receptor on neural cells (Tucker and Griffin, 1991); the opposite occurred when position 172 was a Gly. Also, position 55 of E2 is critical for binding to neurons under certain environmental conditions (in media with a similar ionic strength and degree of sulfation than found in interstitial fluid). While Gln-55 increases binding to neurons, His-55 (a substitution that entails a single nucleotide mutation) stabilizes the interaction between Sindbis virus and the surface of neural cells, contributing to a greater neurovirulence (Lee et al., 2002).
Studies on HIV coreceptor usage in cell culture revealed that the presence of either Lys or Arg at position 306, or Lys at position 322 within the variable V3 loop of gp160, led to a switch from using CCR5 to CXCR4 to enter cells (De Jong 1992, Fouchier 1995). In vivo, the change to using the CXCR4 coreceptor was also linked to a number of mutations that implied an acquisition of positive charges and loss of a N-glycosylation site, modifications that clustered between amino acids 190 and 204 og gp160. In other cases, substitutions to nonbasic amino acids at position 440 of gp160 were also linked to the R5 or X4 phenotypes (Hoffman et al., 2002). Furthermore, changes that confer a highly basic character (on loop V3) allow the virus to also use glycosaminoglycans as attachment factors for cell entry, which results in a 10-fold increase in viral production in cell culture (Zhang et al., 2002).
The cellular tropism of another lentivirus, feline immunodeficiency virus (FIV), may be affected by mutations in the V3 region of the surface glycoprotein SU. A change of cell tropism in cell culture was associated with replacement Glu-407 to Lys (Verschoor et al., 1995). Crandell feline kidney (CRFK) cells were transfected with an FIV molecular clone that was unable to infect them. However, high-dose DNA transfection resulted in the recovery of an FIV mutant capable of replicating in these cells. A single point mutation in the SU protein was responsible for this change in viral tropism (Vahlenkamp et al., 1997). In another study, a variant of FIV emerged after the passage of wild-type virus in cell culture. The variant displayed a phenotype markedly different from that of the parental virus, including the capacity to productively infect previously refractory cell lines and the induction of large syncytia. This phenotype could be attributed to the combination of two amino acid substitutions (Gln-224 to Pro, Thr-470 to Pro) in glycoprotein SU and to a premature stop codon that resulted in a truncated transmembrane protein (Lerner and Elder, 2000).
Cases of amino acid replacements affecting cell recognition have also been described in small and complex DNA viruses. Modifications of the host range of parvoviruses have been associated with amino acid substitutions on the viral capsid (Parrish and Truyen, 1999). Binding affinity to the transferrin receptor correlated only partially with the host range displayed by the virus (Hueffer et al., 2003). Remarkable changes in adenovirus (Ad) host cell tropism can be observed upon minimal alterations of virus fiber sequences. Amino acid substitution Glu-240 to Lys in the distal domain of Ad19p fiber conferred binding to human conjunctival cells, whereas the reverse substitution abrogated cell binding when introduced into the phylogenetically distant Ad37 (Huang et al., 1999). The presence of a Lys residue at position 240 of the adenovirus fiber was associated with an outbreak of epidemic keratoconjunctivitis.
Single amino acid substitutions in glycoprotein gD of herpes simplex 1 (HSV-1) can alter receptor preferences (reviewed in Spear et al., 2000). Although wild-type HSV-1 strains can enter cells via HveA (but not via human nectin-2), Rid mutants, which differ from the wild type by an amino acid substitution at position 25 or 27 of gD, can use human nectin-2 (but not human HveA). Studies with the swine alphaherpesvirus pseudorabies virus (PrV) further documented the important potential of herpesviruses to explore alternative mechanisms of cell recognition (Nixdorf 1999, Schmidt 2001, Schmidt 1997). Although glycoprotein gD is critically involved in PrV entry into susceptible cells, the expression of gD is not required for direct viral cell to cell spread and PrV mutants deleted in the gD gene can be propagated in MDBK cells by cocultivating infected and noninfected cells. While infectivity was found to be strictly cell associated in early passages, repeated passaging resulted in the appearance of infectivity in the supernatant, reaching titers as high as 107 PFU⧸ml. Mutations in viral glycoproteins gB and gH were found to correlate with the capacity of PrV to interact with alternative receptors and the development of a gD-independent mode of entry.
This by no means exhaustive list of examples underscores the biological relevance for RNA viruses of quasispecies dynamics regarding alterations in host cell tropism and pathogenic manifestations of viral infections. The weight of single nucleotide substitutions in the phenotype of RNA viruses must be considered in relation to the additional three parameters indicated in Table I (number of mutations per genome, virus population size, and genome length). The fact that a single replacement can have profound biological consequences for an RNA virus was one of the triggers of interest in quasispecies (review of early observations in Domingo et al., 1985). To what extent complex DNA viruses may incorporate mutations at some genomic sites involved in cell recognition, with the flexibility of RNA viruses, is largely unknown. Because of several implications for the biology and uses of DNA viruses (discussed in Section V), this problem certainly deserves further investigation (compare with Section II,B).
B. Changes in Receptor Specificity upon Virus Evolution in Cell Culture and in Vivo
A modification of receptor specificity of FMDV was observed on the long-term passage of FMDV C-S8c1 [a biological clone derived from natural isolate C-Sta Pau Sp⧸70, representative of serotype C FMDV (Sobrino et al., 1983)] in BHK-21 cells. The parental clone C-S8c1 enters BHK-21 cells via an RGD-dependent integrin, as documented by both inhibition of cell entry by synthetic peptides and site-directed mutagenesis of an infectious cDNA clone to modify the RGD (Baranowski et al., 2000). Passage of FMDV C-S8c1 in BHK-21 cells resulted in the dominance of viruses with several amino acid replacements on the capsid surface (Fig. 3 ). These replacements resulted in an expansion of host cell tropism in cell culture, with the acquired capacity to infect a number of primate and human cell lines, a capacity that was absent in the parental C-S8c1 (Baranowski 2000, Ruíz-Jarabo 2003). As expected, some of the amino acid replacements conferred to the multiply passaged virus the capacity to bind heparin. However, these viruses did not need to use HS as a receptor, as mutants deficient in heparin binding (that were selected from the multiply passaged quasispecies) were equally infectious in cell culture (Baranowski et al., 2000). Therefore, a third entry pathway, which is independent of HS and of RGD-dependent integrins, must have been acquired by FMDV C-S8c1 upon repeated passage in BHK-21 cells. The selective changes underlying such cell recognition conversion are not known (Baranowski 2000, Baranowski 2001).
Fig 3.
Flexibility in receptor usage by FMDV. Passage of FMDV in BHK-21 cells resulted in acquisition of amino acid replacements in the capsid, which expanded receptor usage (HS, heparan sulfate; integrin αvβ3; X, an unidentified receptor). Replacements are depicted as Van der Waals spheres in yellow (VP1, blue; VP2 green; VP3, red).
The acquisition of a third entry pathway by FMDV did not abrogate a potential use of RGD-dependent integrins. Indeed, using the mutants from the multiply passaged FMDV C-S8c1 that were deficient in heparin binding, it was documented that RGD containing peptides (but not the same peptides with RGG instead of RGD) inhibited the infection of BHK-21 cells (Baranowski et al., 2000). Therefore, when HS binding was impaired, the virus used the RGD-dependent pathway for cell entry. These results prove that modifications in receptor recognition can be produced readily by modest evolutionary transitions and that a virus can maintain the capacity to penetrate the same cell type via three alternative entry pathways (Baranowski 2000, Baranowski 2001) (Fig. 3).
Some lines of evidence suggest that the use of alternative receptors for FMDV also occurs in vivo. Variants of FMDV harboring replacements within the RGD or at some neighboring positions documented to be critical for integrin binding have been isolated in vivo. One of the relevant experiments involved vaccination of 138 cattle with synthetic peptides representing B-cell and T-cell epitopes of FMDV C3 ARg-85 (Taboga 1997, Tami 2003). The animals were protected only partially as a result of the immune response to the synthetic peptide constructs and, upon challenge with virulent FMDV C3 Arg-85, several of them developed viremia and vesicular lesions. Virus from several lesions included amino acid substitutions within the RGD or at neighboring positions (Fig. 4 ). It is not clear whether such variant viruses would be able to reinitiate infection of cattle and, if they did, whether they would be stable through the completion of infectious cycles in vivo. Passage of FMDV O⧸CHN⧸90 (a type O FMDV used for vaccine production in China) in BHK-21 cells resulted in viruses that displayed altered tropism. Cell culture adaptation of the Cathay prototype foot-and-mouth disease virus from China results in altered tissue culture host range and pathogenic phenotype in pigs (Zhao et al., 2003). This altered virus caused mild disease in swine. Despite this evidence for flexibility in receptor usage, variants of FMDV selected to bind heparan sulfate were attenuated in cattle (Sa-Carvalho et al., 1997). This and other observations suggest that RGD-dependent integrins may be a major class of receptors employed by FMDV in vivo (Neff et al., 1998), although the flexibility of the virus to modify receptor usage in vivo is an open question.
Fig 4.
Selection of FMDV variants in peptide-immunized cattle. In lesions from the immunized animals challenged by a cloned virus, variants with amino acid replacements within or near the RGD integrin receptor-binding triplet were isolated. The position of the RGD in an open turn between a β strand and an helical region of VP1 is indicated in the box (VP1, blue; VP2, green; VP3, red) above the sequence alignment, indicating the amino acid replacements.
Variants of the arenavirus lymphocytic choriomeningitis virus (LCMV) arise during long-term persistent infections in mice (Ahmed et al., 1984; reviewed in Sevilla et al., 2002). Some of the variants had different biological properties, expected from the tissues from which they were isolated. One type of variant predominated in the central nervous system (CNS), whereas another type was dominant in lymphocytes and macrophages. Most CNS isolates caused acute infection when injected intravenously into immunocompetent adult mice. These variants evoked a potent immune response and infection was cleared with the active participation of anti-LCMV CD8+ cytotoxic T lymphocytes (CTLs). In contrast, most LCMV variants from lymphocytes and macrophages evoked a generalized immunosupression in the animals. Interestingly, these critically different phenotypes of LCMV relate to alterations in the affinity for α-dystroglycan, a receptor for LCMV, lassa fever, and clade C new world arenaviruses (Cao 1998, Spiropoulou 2002). A single amino acid substitution in the glycoprotein GP1 of LCMV alters its affinity for α-dystroglycan; high-affinity binding is associated with immunosupression and viral persistence in mice, whereas low-affinity binding results in clearence of infection (Cao 1998, Sevilla 2000, Sevilla 2002, Smelt 2001).
HIV-1 coreceptor usage changes during infection in humans. Most primary isolates belong to the coreceptor specificity R5, but as infection progresses, dualtropic (R5X4) and X4 variants arise and often become dominant (Section III,A). The emergence of X4 HIV-1 strains in infected individuals has been associated with an increase in pathogenic manifestations and development of AIDS. Evolution of coreceptor usage (R5, multitropic, X4) seems to be the rule with HIV-1 subtypes A, B, D, and E during progressive disease. Subtype C apparently does not follow this trend in that the great majority of isolates obtained at different geographic sites are R5 monotropic regardless of the severity of the HIV infection (Fenÿo, 2001). There are variants proven to be able to use other coreceptors as well, such as CCR2b, CCR3, CCR8, BOB, Bonzo, US28, and BLTR (B4 leukotriene receptor), but their relevance for HIV infection in vivo is not clear (Fenÿo, 2001). The host range of HIV-2 in human cells is similar to that of HIV-1, yet differences exist in the use of coreceptors. Primary isolates of HIV-2 use CCR5, but they may simultaneously use a wide variety of other coreceptors, such as CCR1, CCR2b, CCR3, CCR5, and BOB (Table III), despite the main coreceptor being CCR5. Variants that can use CXCR4 become dominant at late stages of the infection (Morner et al., 1999). Multitropic HIV-2 and SIV strains can infect CD4-negative cells (Liu et al., 2000). SIVsm is closer phylogenetically to HIV-2 than to HIV-1 and behaves similarly in several ways. As HIV-2, SIVsm can use a wide variety of different coreceptors, such as CCR3, CCR5, CXCR4, BOB, and Bonzo. They are multitropic, and several isolates can infect CD4-negative cells (this is also true for a small number of HIV-1 strains). Coreceptor usage, in contrast to what it is observed with HIV-1, narrows with time, and as the infection progresses, HIV-2 isolates tend to use only coreceptor CXCR4, and SIV only CCR5 (Vodros et al., 2001).
Feline leukemia virus A (FeLV-A) is the form of FeLV that is transmitted from cat to cat, and for this reason it is called the ecotropic form. It does not cause acute disease and is restricted to grow on feline cells. The receptor for this group of viruses has not been isolated. FeLV-Bs evolve from FeLV-A by recombination with endogenous FeLV-like (enFeLV) envelope sequences, which results in a change to Pit1 receptor specificity. Pit1 is a classic multiple membrane-spanning receptor molecule. Recombinant forms of FeLV-B differ in the amount of envelope surface unit (SU) that is derived from enFeLV, and this may affect whether the virus can also use Pit2 (which has 62% structural identity with Pit1) as a receptor (Anderson 2001, Sommerfelt 1999, Sugai 2001, Tailor 1999b). Viruses that use Pit1 and Pit2 as receptors use the latter with lower efficiency (Boomer et al., 1997). FeLV-C and -T evolve by mutation of FeLV-A. In contrast to group A, groups B, C, and T FeLVs show an expanded host range, which includes infection of human cells (Sommerfelt 1999, Sugai 2001). FeLV-C viruses use a protein called feline leukemia virus subgroup C receptor (FLVCR), and the use of this receptor is associated with amino acid replacements within the 15 to 20 amino acids of the first variable region (V1) of SU. FeLV-C strains differ from FeLV-A in a Lys-to-Arg substitution in V1, but additional substitutions in SU must be involved in the use of FLVCR and the expanded host range (Brojatsch 1992, Sommerfelt 1999). FeLV-T (a T-cell tropic feline leukemia virus) is the first naturally occurring type C retrovirus that cannot infect cells unless both Pit1 and a second coreceptor or entry factor are present. This second receptor component, called FeLIX, is a cellular protein that is closely related to a portion of the FeLV envelope protein. This cellular protein can function either as a transmembrane protein or as a soluble component to facilitate infection (Anderson 2000, Lauring 2001). The most important feature to confer T-cell tropism seems to be an insertion of four to six amino acids in the C-terminal portion of SU, although additional amino acids could also be involved in this phenotype.
Friend virus is a nonneuropathogenic ecotropic murine leukemia virus. Variant PVC-211 isolated after 30 passages of Friend virus in BALB⧸c mice caused a rapidly progressive hind limb paralysis when injected into newborn rats and mice and displayed an expanded tropism in tissue culture. The two mutations responsible for the altered phenotype were Glu-116 to Gly and Glu-129 to Lys in the surface protein of the virus (Kai 1984, Masuda 1992, Masuda 1996a, Masuda 1996b).
C. Nature of the Selective Forces That Drive the Selection of Virus Variants
It is very difficult to identify the selective pressures that act on viruses during an infectious process, not only because of the complexity of host influences involved, but also because selective pressures may vary due to physiological alterations that often accompany the infection. HIV infections, with their erosive effects on the immune system, offer a relevant case. Adaptation of HIV to a changing cellular environment could contribute to the selection of variants with different cell tropism and different pathogenicity (Viscidi, 1999). This can be regarded as a positive selection directed by the immune system, resulting in modifications of host cell tropism of HIV (Crandall, 1999). For example, in the late stages of HIV infection, CD4+ cells are depleted, and variants that use CD8 to enter CD8+ cells arise (Saha et al., 2001). Thus, subpopulations of cells involved in the human immune response, whose abundance may be modified by HIV-1 infection, may in turn become a key selective force for HIV evolution (Crandall, 1999).
The relative availability of chemokine receptor ligands may also be a selective pressure involved in HIV evolution in vivo. Stromal-derived factor 1 (SDF-1), a chemokine that is a CXCR4 ligand, is expressed constitutively by mucosal epithelial cells at sites of HIV transmission and replication (Agace et al., 2000), and this may be one of the factors implicated in the selective transmission of R5 HIV-1 strains. Individuals whose peripheral blood lymphocytes produce high levels of CCR5 ligands (such as RANTES, MIP1α, and MIP1β) are relatively resistant to infection (Garzino-Demo 1998, Paxton 1996, Zagury 1998). Selection by receptor ligands is also supported by experiments using a mouse model in which a modified form of RANTES, a natural ligand for CCR5, selected HIV mutants that used CXCR4 as a coreceptor (Mosier et al., 1999). Also, AMD3100, a bicyclam that is a selective antagonist of CXCR4, led to the complete suppression of X4 variants in cell culture and prevented the switch from the less pathogenic R5 HIV to the more pathogenic X4 strains (Este et al., 1999).
Multiplication in a given type of cell may also be a likely trigger of a tropism alteration. Measles virus isolated on marmoset B-cell lines infected some primate B- and T-cell lines and retained pathogenicity for monkeys. This was not the case for measles virus isolated on Vero cells, as such isolates manifested a different tropism and host range (Tatsuo 2000a, Tatsuo 2000b), suggesting that the type of cell used for measles virus isolation exerted a selective force on the virus. Likewise, the host cell selected an avian retrovirus variant with an expanded host range, consisting of recognition of a receptor on chicken cells and a distinct receptor on quail cells (Taplitz and Coffin, 1997). This evidence must make virologists aware that when they isolate a virus from a biological specimen by passage in an established cell line, a virus with different biological properties (cell tropism or other) than the natural virus may be selected. Variants of LCMV with profoundly different biological properties have been isolated from different organs of infected mice (Section IV,B), emphasizing the impact of quasispecies dynamics during infections in vivo (Sevilla et al., 2002). In most cases summarized in this section, the precise nature of the selective force that acts to perturb mutant distribution is not well understood in molecular terms. In many other cases, even the types of selective constraints that mediate changes in host cell tropism are not obvious. This is perhaps one of the areas of research in evolutionary virology that necessitates efforts because of the multiple implications of variations of host cell tropism for the control of viral disease. Model experiments in cultured cells (that attempt to match the type of cells found in vivo), with the aim of relating variations in the nucleotide sequence (and in the corresponding structure of encoded proteins) with alteration in cell recognition, may provide information on the molecular basis for tropism specificity and tropism changes.
V. Biological Implications of Modifications in Receptor Usage
A. Coevolution of Receptor Usage and Antigenicity
Structural studies have shown in many cases that there is an overlap between the amino acid residues on a viral capsid or envelope that are involved in cell recognition and those that recognize neutralizing antibodies (Section III,C) (Table IV ). The structure of the complex between the G-H loop of VP1 of FMDV with monoclonal antibody 4C4 has clear resemblance with the complex that can be modeled between the virus and its integrin receptor (Fig. 5 ). A change in receptor specificity could involve a modification in binding to antibodies or vice versa. Furthermore, an antigenic domain that coincides with a receptor-binding site may lose constraints for variation once a different receptor that interacts with other capsid residues becomes operational. The latter possibility can be illustrated with results obtained with FMDV.
Table IV.
Examples of Overlap between Antibody and Receptor-Binding Sites in Viruses
| Viral system | Main observations |
|---|---|
| Adenoviridae | |
| Adenovirus | Synthetic peptides representing fiber knob of Ad-3 present cell receptor-binding sites and antigenic epitopes (Liebermann et al., 1998) |
| Coronaviridae | |
| Murine coronavirus | Mab recognized epitopes involved in the binding of virions to cellular receptors (Kubo 1993, Kubo 1994) |
| Flaviviridae | |
| BVDV | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Xue 1993, Minocha 1997) |
| Dengue virus | Amino acid residues critical for mouse neurovirulence are involved in antibody binding (Hiramatsu et al., 1996) |
| Yellow fever virus | Amino acid residues critical for virus neurotropism are involved in antibody binding (Jennings et al., 1994) |
| Hepadnaviridae | |
| Duck hepatitis B virus | Residues critical for virus neutralization are involved in the interaction with cells (Tong 1995, Li 1996, Sunyach 1999) |
| Hepatitis B virus | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Petit 1992, Hertogs 1994, Budkowska 1995) |
| Anti-idiotypic antibodies mimicking cellular structures bind to small hepatitis B surface antigen (Neurath et al., 1986) | |
| A synthetic peptide analogue is recognized by both cell receptors and anti-HBV antibodies (Neurath et al., 1986) | |
| Herpesviridae | |
| BHV-1 | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Thaker 1994, Varthakavi 1996) |
| HCMV | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Keay 1989, Keay 1991) |
| HSV | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Huang and Campadelli-Fiume, 1996) |
| Overlap between major neutralizing antigenic site and a receptor-binding domain of gD (Whitbeck et al., 1999) | |
| Orthomyxoviridae | |
| Influenza virus | Amino acid residues within the sialic acid-binding pocket of virus hemagglutinin are accessible to neutralizing antibodies (Stewart and Nemerow, 1997) |
| Antigenic and hemagglutinin variants selected upon egg adaptation (Robertson et al., 1987) | |
| Low-affinity neutralizing antibody response selected for receptor-binding variants of influenza virus HA (Laeeq et al., 1997) | |
| Amino acid changes at residues involved in antibody binding can modulate the hemagglutinating activity of influenza C virus (Matsuzaki et al., 1992) | |
| Passage of influenza C virus in HMV-II cells resulted in selection of antigenically distinct variants, which have an advantage in binding to the cell surface receptors (Umetsu et al., 1992) | |
| Hemagglutinin variants displayed increased resistance to neutralization (Nohinek et al., 1985) | |
| The receptor-binding specificity of the hemagglutinin can markedly influence the antigenic analysis obtained with monoclonal antibodies in HI tests (Yamada et al., 1984) | |
| Picornaviridae | |
| FMDV | Overlap of integrin- and antibody-binding sites (Verdaguer et al., 1995) |
| Monoclonal antibodies selected variants with altered integrin recognition (Martinez 1997, Baranowski 2000, Ruíz-Jarabo 2003) | |
| Adaptation to cell culture may result in antigenic variation (Curry 1996, Sa-Carvalho 1997, Baranowski 2000) | |
| Some amino acid residues involved in heparin-binding map at antigenic sites (Sa-Carvalho 1997, Fry 1999, Baranowski 2000) | |
| Antigenic variants with altered receptor specificity can be selected in vivo (Taboga et al., 1997; Tami et al., 2003) | |
| Poliovirus | Receptor recognition influenced by residues of antigenic sites (Murray 1988, Harber 1995) |
| The exposed BC loop of capsid protein VP1 plays a critical role in receptor interactions in the mouse central nervous system (Yeates et al., 1991) | |
| HRV | Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon (Smith et al., 1996) |
| The footprint of very low density lipoprotein receptor on HRV-2 surface covers two exposed loops of capsid protein VP1 (BC- and HI-loops) (Hewat et al., 2000) | |
| TMEV | Neutralization epitopes map close to the putative receptor binding region (Sato et al., 1996) |
| Mutations associated with adaptation to some culture cells map in antigenic sites (Jnaoui and Michiels, 1998) | |
| Reoviridae | |
| Bluetongue virus | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Xu et al., 1997) |
| Reovirus | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Co 1985a, Gaulton 1985, Williams 1988, Williams 1989, Williams 1991b) |
| Rhabdoviridae | |
| Rabies virus | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Hanham et al., 1993) |
| Amino acid residues critical for virus neurotropism are involved in antibody binding (Coulon et al., 1998) | |
| Togaviridae | |
| Sindbis virus | Anti-idiotypic antibodies mimicking viral antigens bind to cellular receptors (Ubol 1991, Wang 1991b, Strauss 1994) |
| Several antibodies bind to regions of the virions implicated in cell-receptor recognition (Smith et al., 1995) | |
| Ross River virus | Several antibodies bind to regions of the virion implicated in cell-receptor recognition (Smith et al., 1995) |
Fig 5.
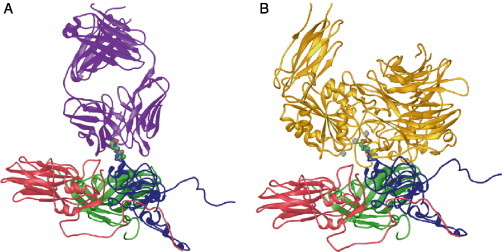
(A) Cryoelectron microscopy structure of the complex between FMDV and the Fab fragment from MAb 4C4 bound to the VP1 G-H loop of the virus (Verdaguer et al., 1999). One protomer subunit of FMDV is shown as a ribbon diagram (VP1, blue; VP2, green; VP3, red) and the Fab is in violet. The flexible G-H loop of FMDV is located in an extended orientation with the RGD motif (depicted as a Van de Waals spheres) occupying a fully exposed position. The RGD triplet, in this complex, shows a similar conformation to that found for the same triplet when bound to the integrin αvβ3 (Xiong et al., 2002). A least-squares superimposition of the main chain atoms from RGD residues in the FMDV loop with the equivalent residues in the integrin RGD ligand gives an average rms deviation of only 0.32 Å. The transformation necessary to superimpose the FMDV loop to the integrin ligand can also be used to superimpose the integrin αvβ3 onto the viral capsid to obtain an approximate docking model for the FMDV–αvβ3 complex. (B) Ribbon drawing of a FMDV protomer together with the docked αvβ3 receptor (yellow). For clarity, only the β propeller of subunit α and the βA and hybrid domains of the subunid β are represented. The docking model suggests that the αvβ3 receptor binds the FMDV G-H loop in an exposed position similar to that found when the loop is recognized by neutralizing antibodies.
The capacity of FMDV to use multiple alternative receptors for entry even into the same cell type (Section IV,C) confers to this virus the potential to modulate receptor usage in response to selective constraints (Baranowski et al., 2000), which has considerable implications for the evolution of virus antigenicity. Because the RGD triplet located at the surface G-H loop of capsid protein VP1 is a key part of several epitopes recognized by neutralizing antibodies (Mateu 1995, Verdaguer 1995), dispensability of the RGD integrin-binding motif for cell entry expanded the repertoire of antigenic variants of FMDV greatly and prompted the isolation of viable antibody-escape mutants with profoundly altered antigenicity that contained mutations at the RGD triplet (Martinez 1997, Ruiz-Jarabo 1999). Mutants with RED, RGG, and even GGG instead of RGD were isolated. Viable viruses were obtained when the relevant mutations were engineered in an infectious clone with the sequence context of the capsid of the multiply passaged C-S8cl; RNA transcripts did not give rise to variable virus when the same mutations were engineered in the sequence context of the parental clone C-S8cl (Baranowski et al., 2000). Studies with FMDV containing RGG showed impaired reactivity with monoclonal antibodies specific for the relevant antigenic sites and with polyclonal antibodies raised in swine and guinea pigs using the wild-type virus as immunogen (Ruiz-Jarabo et al., 1999).
The genomic changes that can endow FMDV with the capacity to use alternative mechanisms of cell recognition (Section V,A) are minimal, and viruses with alterations in the RGD and with unusual receptor-binding specificities are likely to be present in the mutant spectrum of FMDV replicating in the animal host (Taboga et al., 1997) (described in Section IVB). The antigenic alterations produced by replacements at or around the RGD suggest that the emergence of these particular FMDV mutants in vivo was the result of selection of antigenic variants that escaped neutralization by anti-FMDV antibodies in peptide-vaccinated cattle (Fig. 4). A study analyzing the genetic changes selected during the adaptation of FMDV to guinea pig documented the progressive dominance of an unusual amino acid replacement (Leu-147 to Pro) affecting the antigenic structure of the G-H loop of capsid protein VP1 in the course of adaptation of FMDV to this new host (Núñez et al., 2001). Construction of infectious cDNA clones of FMDV confirmed that this Leu-147 was essential for virus interaction with integrin receptor molecules expressed in BHK cells and various other cell lines used commonly to propagate FMDV and that mutants with Pro-147 do not form plaques on BHK-21 cells (Núñez et al., 2001). The isolation of FMDV mutants displaying altered cell tropism in association with antigenic changes illustrates the important adaptive potential of FMDV and the capacity of this virus to explore new antigenic⧸receptor recognition structures upon replication in its hosts.
A mutant of rabies virus harboring changes Lys-330 to Asn and Arg-333 to Met at antigenic site III of its surface glycoprotein manifested a modification in both cell tropism and antigenicity (Coulon et al., 1998). This double mutant, selected by the successive use of two neutralizing antiglycoprotein monoclonal antibodies, was not pathogenic for adult mice and could not penetrate the nervous system either by the motor or by the sensory routes. In vitro experiments showed that the double mutant was able to infect BHK cells, neuroblastoma cells, and freshly prepared embryonic motoneurons, albeit with a lower efficiency than the parental strain CVS. Upon further incubation at 37 °C, the motoneurons became resistant to infection by the mutant while remaining permissive to infection by the CVS strain (Coulon et al., 1998). Thus, rabies virus can use different types of receptors: a molecule that is expressed ubiquitously at the surface of continuous cell lines and that is recognized by both CVS and the double mutant and a neuron-specific molecule that is not recognized by the double mutant.
One of the first lines of evidence of coevolution of receptor usage and antigenicity in viruses was documented with human influenza type A with single amino acid replacements in the hemagglutinin, which modified receptor specificity or receptor-binding affinity (Robertson et al., 1987; reviewed in Skehel and Wiley, 2000). Additional cases of an overlap of receptor-binding and antigenic sites are given in Table IV.
B. Use of Soluble Receptor Analogs and Receptor Ligands, and Selection of Resistant Viruses
Ever since viral receptors were discovered, antiviral therapies have been designed based on the administration of soluble receptors and receptor ligands. The discovery that chemokine receptors act as cofactors essential for HIV entry into target cells identified new targets for antiretroviral therapy. Viral entry can be inhibited in vitro by the natural ligands for CXCR4, the CXC chemokine SDF-1, and CCR5, the CC chemokines RANTES, MIP-1α and MIP-1β. Several peptidic compounds have been identified as CXCR4 antagonists that display anti-HIV activity, and the HIV-1 tat protein has been described as a “natural” CXCR4 antagonist with anti-HIV-1 activity. The bicyclam derivatives are among the most potent and specific CXCR4 antagonists described and they block X4 HIV replication very efficiently (De Clercq and Schols, 2001). Mutants resistant to CCR5 antagonists have been isolated. This resistance is not mediated by a change in coreceptor usage, but rather by the ability to use CCR5 despite the presence of the inhibitor (Trkola et al., 2002). However, much remains to be learned about the possible physiological impact of the administration of receptor analogs and receptor ligands. The multitude of different receptors involved in HIV entry into cultured cells and their important physiological roles in vivo, together with their implications in distinct pathogenic processes, have added further complexity to this field of research (Heveker, 2001).
One of the most obvious potential difficulties with the use of receptor analogs as an antiviral therapy is the selection of soluble receptor-resistant mutants from the quasispecies swarms. This was illustrated with the avian sarcoma-leukosis virus subgroup A (ASLV-A), in which virus with a six amino acid deletion in the heterogeneous region 1 of the surface protein (SU) was selected by a soluble receptor in cell culture (Holmen and Federspiel, 2000). In some other instances, soluble receptors were capable of inducing conformational changes in viral envelopes, and nonsusceptible cells became susceptible to viral infection. In the case of ASLV-A, soluble Tva could activate SU and convert it to a fusogenic conformation-competent form capable of mediating the fusion of viral and cellular membranes, even when the membranes did not harbor viral receptors (Damico and Bates, 2000). This has also been observed with murine leukemia viruses (Lavillette et al., 2000), with mouse hepatitis virus (Taguchi and Matsuyama, 2002), and with HIV-1 in that CD4 on the surface of a cell could be used by the virus to fuse to neighboring cells, provided the latter had a coreceptor (even in the absence of CD4) (Speck et al., 1999).
There is no solid reason to assume that the selection of viruses resistant to receptor analogs or to receptor ligands should be substantially more difficult than the selection of antibody- or inhibitor-resistant mutants in the course of viral replication, selections that have been documented profusely not only for HIV but for many other RNA viruses (Table I) (reviewed in Domingo et al., 2001). From the mechanisms exploited by viruses to escape antibodies, CTLs, or inhibitors and from the molecular basis of fitness gain, a number of pathways for the selection of mutants resistant to soluble receptors (or analogs) are plausible. Viral mutants could be selected that display an increased affinity for the receptor attached to the cell surface, and not for its soluble form (or analog). A completely independent pathway could be the selection of mutants able to use an unrelated receptor, as this has already been described even for viruses not subjected to the presence of receptor analogs (Section V,A). Yet another possibility is the selection of mutants capable of binding both the cellular receptor and the authentic receptor on the cell, with a slight affinity bias to permit cell entry, among other conceivable variations of these mechanisms.
If an antiviral therapy based on soluble receptors, receptor analogs or receptor ligands is to be pursued, it may be necessary—in line with antiviral strategies based on combination therapy—to administer simultaneously several types of soluble analogs when potential alternative receptors for the virus have been identified. An obvious problem is the possible physiological impact of such a combination of receptors or their analogs. These difficulties should also be evaluated carefully if strategies employing combinations of inhibitors, receptor analogs, and immunotherapy are considered. Viruses have learned to confront many selective constraints over extended periods of coevolution with their hosts (I, V) and it is not known whether artificial constraints may be found that would suppress viral replication in a way as to prevent selection of escape mutants.
C. Emergence and Reemergence of Viral Diseases
The emergence and reemergence of infectious diseases are increasing concerns for human and animal health and for a number of economically significant activities related to agriculture and farming. More than 40 emergent and reemergent human viral diseases have been described over the last two decades, and the adaptive potential of viruses, including changes in host range, undoubtedly has played a very relevant role (reviewed in Brault 2002, Leitmeyer 1999; specific recent examples in Morse 1993, Morse 1994). However, a complex set of interconnected influences are involved in viral disease emergence. These include environmental and climatic changes, socioeconomic and political factors, agricultural practices, and technological developments. The recent reemergence of the foot-and-mouth disease in Europe illustrates how a number of rather unpredictable and complex factors can produce a devastating animal disease epizootic (Samuel 2001, Sobrino 2001). Additional examples have been documented and discussed (Domingo 2001, Mahy 1997, Morse 1993, Morse 1994, Murphy 1994). These multiple influences have, as a net result, an alteration of viral traffic and of the demography of viral vectors and susceptible hosts (Morse, 1994), with the many implications of alterations of viral ecology (Hurst, 2000).
A majority of the emergent and reemergent viral diseases are associated with RNA viruses, and frequently those displaying high recombination rates. Examples are the expansion of dengue fever and dengue hemorrhagic fever (Gubler, 1998), recent outbreaks of poliomyelitis (Kew et al., 2002), and the appearance of severe acute respiratory syndrome (SARS). The emergence of HIV stands as the most significant introduction of a retrovirus into the human population, likely from several simian virus ancestors, and the virus is evolving continuously through mutation and recombination (Crandall, 1999). Poliovirus variation has been signaled as a major problem for the successful eradication of poliovirus (Nomoto and Arita, 2002). Furthermore, poliovirus and the C-cluster coxsackie A viruses are grouped together based on phylogenetic analyses. It has been suggested that if polioviruses were eradicated, some coxsackie A viruses may have an opportunity to modify their receptor specificity from ICAM-1 to CD155 (Table II) and evolve toward a polio-like virus (Nomoto and Arita, 2002).
Avian and equine hemagglutinins bind preferentially to N-acetyl sialic acid linked to galactose by α-2,3 linkages (Neu5-Acα-2,3Gal), whereas most human HAs bind preferentially to Neu5-Acα-2,6Gal. Changes in binding sites have been reported to cause host range switches among different hosts (Aytay 1991, Matrosovich 1997, Matrosovich 1999, Matrosovich 2000, Naeve 1984, Rogers 1983a, Rogers 1983b). The receptor-binding specificity of the avian influenza hemagglutinin was altered early after the transmission to humans and pigs (Matrosovich et al., 2000), constituting a case of positive selection by the recipient host.
Modifications in host cell tropism need not be associated with a modification in receptor specificity. As discussed in Section V A, adaptation of a FMDV clone to the guinea pig resulted in the dominance of FMDV mutants with replacement Leu-147 to Pro in the G-H loop of VP1, which altered integrin recognition by the virus. However, the critical amino acid replacement for FMDV to cause disease in guinea pig was located in nonstructural protein 3A (Núñez et al., 2001). It is not known what is the actual contribution of Pro-147 in VP1 to the pathogenesis of FMDV in guinea pigs, but this model study with FMDV illustrates yet another possibility in a cascade of adaptive events: a virus may emerge as pathogenic in a new species without a strict requirement for a shift in receptor recognition, but in the course of replication in the new host, additional mutations that entail modifications in receptor recognition become permissible and perhaps favored (Núñez et al., 2001).
D. Gene Flow and Gene Therapy: Role of Viruses
The growing list of complete genomic nucleotide sequences of many cellular organisms and viral genomes suggests that all life forms share some basic functional motifs (Mount, 2001). No viral functions that depart in any outstanding way from similar cellular functions have been identified. This includes similarities between viral and cellular proteins involved in genome replication, proteolytic activities, and general patterns of genome organization and expression. These important advances in molecular genetics suggest that the exchange of modules, together with mutation, has contributed to the coadaptation of cells and autonomous replicons over long evolutionary periods (DeFilippis 2001, Domingo 2003, Domingo 1999, Gorbalenya 1995, Holland 1998). In cases in which viruses infect multiple host species, convergent phylogenies of viruses with their host can sometimes be seen (Gibbs 1999, McGeoch 1999). Some of the features of variation of cell tropism discussed in A, A may not be foreign to differentiated organisms. The insertion of two amino acids into ectodysplasin—a member of the tumor necrosis-binding family—resulted in a change of its cellular receptor specificity, and the differential expression of the two forms of ectodysplasin plays a role in epidermal morphogenesis (Yan et al., 2000). Not only functional modules but also biological strategies are shared among viruses and cells.
In the case of viruses, the capacity to use alternative receptors and to change receptor specificities by modest genetic variation (involving short genetic distances) may represent an adaptation of viruses to cope with increasingly differentiated organisms. The absolute requirement of a cellular environment for virus replication (one of the definitive features of viruses; Section II,A) implies a permanent coexistence of cellular and viral genomes. The uptake of cellular genes by viruses has been documented in transducing bacterial viruses, RNA and DNA tumor viruses and, as remarkable cases, in cytopathic variants of the flavivirus bovine viral diarrhea virus, and defective viruses that have acquired host sequences by nonhomologous recombination events (examples reviewed in Domingo 1999, Bushman 2002, Domingo 2003).
The elucidation and interpretation of the complete genomic sequences of an increasing number of prokaryotic and eukaryotic organisms indicate that a substantial part of many of these genomes consist of mobile genetic elements or their relics (reviewed in McClure 1999, Mount 2001, Bushman 2002). It has been estimated that this part of the human genome is 40% of the total! The process by which cells from one organism have captured DNA fragments from another organism is known as horizontal gene transfer. This transfer appears to be particularly frequent in prokaryotes, but it does also occur in eukaryotes. In addition to virus infection, lateral gene transfers are mediated by conjugative plasmids and several classes of mobile elements. Transfers are affected by viral infection (transduction), conjugation, and transformation. The transposition of sequences may occur within the DNA of the same cell.
Virus receptor specificity and its variations (as discussed in previous sections) may facilitate the access of cellular genes to specific cell types in tissues and organs. The human body comprises 50 trillion cells organized in hundreds of organs, tissues, and classes of cell subpopulations. Viruses may spread differentially within organs. As an example, the dissemination of neurotropic viruses in the brain is used increasingly to define connections among subsets of neurons (Card, 2001). To complete its incorporation into a new host cell, the transferred DNA must integrate into the recipient chromosome by some type of molecular recombination event. In this respect, integrative bacteriophages and retroviruses provide the adequate mechanisms as an integral part of their life cycles (Coffin et al., 1997). The retroviral reverse transcriptases must jump strands to complete the synthesis of cDNA, and this property may favor the capture of cellular sequences for transfer to other cells. The mechanisms that mediate the integration of lysogenic bacteriophage DNA, retroviral cDNAs, and a class of transposons termed non-LTR retrotransposons are different, as are the proteins involved (reviewed in Ptashne 1992, Bushman 2002, Coffin 1997). Nonidentical requirements for integration may have broadened the possibilities of DNA flow among cells in the course of evolution. About 8% of the human genome appears to have been derived from retroviral-like sequences. Retroviral-mediated gene transfer can have deep phenotypic consequences for cells, such as a variety of disease manifestations in differentiated organisms (McClure, 1999), including alterations of cell growth control, and in some cases of infertility in humans (Sun et al., 2000). Estimates using mice suggest that 40% of DNA insertion events may have phenotypic consequences for the animal (reviewed in Bushman, 2002).
This brief account of the extensive evidence of lateral gene transfers in the cellular world underscores the profound evolutionary implications of the rapid changes in receptor specificities that viruses can undergo. Indeed, many of the viruses referred to in previous sections may infect multiple host species, and through genetic change they may acquire the potential to infect other host species, therefore permitting gene transfers among distant host species and, perhaps occasionally, also among taxa. Many lines of evidence (too numerous to be reviewed here) have clearly documented DNA transfers among distant domains of life (bacteria to plants, bacteria to animal cells, etc.) (reviewed in Bushman, 2002). It is not known how many DNA transfer events are initiated and never completed, and of those in which the transfer is completed, how many lead to viable genotypes. Perhaps the sorting of viable recombinant genomes is parallel on long evolutionary times to the sorting of viable RNA genomes subjected to mutational pressure that occurs over infinitely short evolutionary time scales (Section II,B). In both cases, negative selection may eliminate many nascent mutant and recombinant forms to leave a surviving minority endowed with a minimally required relative fitness value. Genetic disease in differentiated organisms may be viewed as an unavoidable price to pay to ensure a source of genome fluidity for long-term survival.
Gene therapy is a growing field of research in molecular medicine, whose aim is the supply of a functional gene into human cells for the treatment of inherited or acquired genetic disorders (Lemoine, 2000). The application of DNA recombinant techniques, together with an understanding of the life cycle of viruses, has permitted the development of virus vectors for the transport of foreign genes into target cells. This technology takes advantage of one of the natural tendencies of viruses to act occasionally as agents of horizontal gene transfers, although for therapeutic purposes, viruses must be modified carefully to act as safe and efficient vectors. Viruses that have been used as gene vectors include adenoviruses, parvoviruses (adeno-associated viruses), herpesviruses, retroviruses (simple and complex lentiviruses such as HIV), SV40, and some RNA viruses such as Sindbis virus (reviewed in Pfeifer and Verma, 2001). Viral vectors are engineered to have the foreign “transgene” replace deleted viral genes, while maintaining the cis-acting elements to form virions and to integrate the transgene. Packaging cells or plasmids provide in trans those proteins required for particle formation. A number of safety features (that necessitate different approaches for the different vector types) must be introduced to avoid the spread of replication-competent viruses in the organism, cytotoxic effects, or undesired gene damage due to unrestricted and random integration of transgene into human DNA. Important difficulties encountered in viral-mediated gene therapy are the immune response (antibody or cellular responses evoked by the vector or preexistent in the human recipient), vector stability, dependence of the transduction efficiency on the metabolic state, and cell cycle phase of the target cells, among others. From the point of view of targeting the desired target cells, the receptor specificity of the viral vectors is an essential element of vector safety and efficacy, and many of the considerations discussed in previous sections concerning variations in receptor specificity are relevant to the design of viral vectors, both regarding the basal amplitude of host range afforded by each vector type and the possibilities of engineered alterations in the vector surface proteins to attain a more defined cellular specificity. Vectors based on adeno-associated virus type 2 or retroviral pseudotypes containing the surface glycoprotein of vesicular stomatitis virus display a broad host cell tropism. In contrast, vectors based on murine leukemia virus or HIV-1 often show restricted tropism, imposed by the receptors recognized by their surface envelope proteins (receptor types utilized by some viruses used for vector construction are listed in Table III). For some vectors, the host range can be expanded by replacing surface proteins (or subdomains) by heterologous counterparts, such as heterologous envelope proteins of retroviral vectors (Pfeifer and Verma, 2001).
VI. Conclusions and Overview
The picture beginning to form from genome analyses of viruses, unicellular organisms, and multicellular organisms is that viruses have shared functional modules with cells. A process of coevolution has probably involved exchanges of genetic information between cells and viruses for long evolutionary periods. From this point of view, it is perhaps not surprising that present-day viruses show flexibility in receptor usage and a capacity to alter through mutation their receptor recognition specificity. Shifts in receptor usage have been documented for a variety of DNA and RNA viruses, and the list is increasing continuously. This is just one facet of the fluidity exhibited by cells and viruses regarding exchanges and variation of their genetic material. This is particularly spectacular for RNA viruses because they cannot replicate without producing mutants. The mutation rates, mutation frequencies, and population dynamics of DNA viruses during short-term evolution have been investigated far less than those of RNA viruses. For this reason, it is not possible to anticipate at present whether some DNA viruses may share with RNA viruses a rapid adaptation to using new receptors. It is possible that for the complex DNA viruses, due to a likely limited tolerance to generalized high mutation rates, modifications in receptor specificity will be less frequent than for RNA viruses, albeit with similar biological consequences once they occur.
RNA viruses consist essentially of pools of mutants, and minority components of such pools may rise to dominance under appropriate environmental conditions. Just by exploring constellations of modest numbers of amino acid substitutions in their capsid or envelopes, viruses can extend the range of receptors they can utilize or coutilize to enter the same cell type, a means for the virus to secure its survival. Different receptors, or allelic forms of one receptor, may be used with different efficiency, and receptor affinities are probably modified by mutation and selection. Receptor abundance and its affinity for a virus may modulate not only the efficiency of infection, but also the capacity of the virus to diffuse toward other sites of the organism.
The acquired ability of an RNA virus to enter a new cell type may have yet another consequence for the biological behavior of the virus progeny due to the new intracellular environment encountered by the virus in which the subsets of genomes undergoing positive or negative selection may be different than those that underwent the same processes in the previous host cell type. Therefore, a change in receptor specificity represents biological novelty not only in that a new cell type is infected, but also because the new environment may alter the mutant repertoire participating in subsequent rounds of infection. The application of microarray-based gene expression screening methods suggests considerable variation in expression patterns not only between different tissues and organs but also, on occasion, among cells of the same tissue. Thus, there is a great potential for environmental heterogeneity to modulate the composition of quasispecies swarms.
The main conclusion of this review is that receptors may be shared by different, unrelated viruses and that one virus may use several receptors and may expand its receptor specificity in ways that, at present, are largely unpredictable. We have suggested that this may have consequences for viral pathogenesis, coevolution of cell receptor specificity and antigenicity, a number of possible medical applications of receptor analogs and receptor ligands, the host range of viruses, the emergence of viral disease, and the application of viral vectors for viral therapy. Most of the studies on which we have based our arguments have been carried out with a single virus isolate, clone, or reference strain. Findings may be dependent on a particular sequence context of the viral genome in view of the compactness of information in viral genomes and the connections among genomic regions. It would not be surprising if similar studies with other representatives of the same virus genus yielded different results in the detail of the specific mutations associated with a phenotypic change regarding receptor recognition. However, we would also expect that the results would still convey the same general concepts and conclusions, as they derive from the phenotypic flexibility resulting from high mutation rates and quasispecies dynamics.
Acknowledgements
Work at the CBMSO was supported by Grants PM97-0060-C02-01, BMC 2001-1823-C02-01, and Fundación Ramón Areces. Work at CISA-INIA was supported by institutional grants from INIA and a contract “Ramón y Cajal” to E.B. Work at IBM was supported by Grant BIO99-0865. We acknowledge J. Bella and M. Rossmann for supplying the reconstruction shown in Fig. 1B and H. Cheng and J. M. Casasnovas for the reconstruction shown in Fig. 1C.
References
References
- Aderem A, Underhill D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- Adkins H.B, Blacklow S.C, Young J.A. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J. Virol. 2001;75(8):3520–3526. doi: 10.1128/JVI.75.8.3520-3526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins H.B, Brojatsch J, Naughton J, Rolls M.M, Pesola J.M, Young J.A. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc. Natl. Acad. Sci. USA. 1997;94(21):11617–11622. doi: 10.1073/pnas.94.21.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agace W.W, Amara A, Roberts A.I, Pablos J.L, Thelen S, Uguccioni M, Li X.Y, Marsal J, Arenzana-Seisdedos F, Delaunay T, Ebert E.C, Moser B, Parker C.M. Constitutive expression of stromal derived factor-1 by mucosal epithelia and its role in HIV transmission and propagation. Curr. Biol. 2000;10(6):325–328. doi: 10.1016/s0960-9822(00)00380-8. [DOI] [PubMed] [Google Scholar]
- Agnello V, Abel G, Elfahal M, Knight G.B, Zhang Q.X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA. 1999;96(22):12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrez M.V, Shafren D.R, Gu X, Cox K, Sheppard D, Barry R.D. Integrin alpha v beta 6 enhances coxsackievirus B1 lytic infection of human colon cancer cells. Virology. 1997;239(1):71–77. doi: 10.1006/viro.1997.8831. [DOI] [PubMed] [Google Scholar]
- Ahmed R, Salmi A, Butler L.D, Chiller J.M, Oldstone M.B. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice: Role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 1984;160(2):521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akula S.M, Pramod N.P, Wang F.Z, Chandran B. Integrin alpha3beta1 (CD 49c⧸29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV⧸HHV-8) entry into the target cells. Cell. 2002;108(3):407–419. doi: 10.1016/s0092-8674(02)00628-1. [DOI] [PubMed] [Google Scholar]
- Akula S.M, Wang F.Z, Vieira J, Chandran B. Human herpesvirus 8 interaction with target cells involves heparan sulfate. Virology. 2001;282(2):245–255. doi: 10.1006/viro.2000.0851. [DOI] [PubMed] [Google Scholar]
- Albritton L.M, Tseng L, Scadden D, Cunningham J.M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57(4):659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- Alcamí A. Viral mimicry of cytokines, chemokines, and their receptors. Nature Reviews, Immunology. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- Alcamí A, Koszinowski U.H. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8(9):410–418. doi: 10.1016/S0966-842X(00)01830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatib G, Combadiere C, Broder C.C, Feng Y, Kennedy P.E, Murphy P.M, Berger E.A. CC CKR5: A RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272(5270):1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Alkhatib G, Liao F, Berger E.A, Farber J.M, Peden K.W. A new SIV co-receptor, STRL33. Nature. 1997;388(6639):238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- Allander T, Forms X, Emerson S.U, Purcell R.H, Bukh J. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology. 2000;277(2):358–367. doi: 10.1006/viro.2000.0617. [DOI] [PubMed] [Google Scholar]
- Allaway G.P, Burness A.T. Site of attachment of encephalomyocarditis virus on human erythrocytes. J. Virol. 1986;59(3):768–770. doi: 10.1128/jvi.59.3.768-770.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.M, Lauring A.S, Burns C.C, Overbaugh J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science. 2000;287(5459):1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- Anderson M.M, Lauring A.S, Robertson S, Dirks C, Overbaugh J. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J. Virol. 2001;75(22):10563–10572. doi: 10.1128/JVI.75.22.10563-10572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C.F, Guerrero C.A, Mendez E, Zarate S, Isa P, Espinosa R, Romero P, Lopez S. Early events of rotavirus infection: the search for the receptor(s) Novartis Found. Symp. 2001;238:47–60. doi: 10.1002/0470846534.ch4. [DOI] [PubMed] [Google Scholar]
- Arnberg N, Edlund K, Kidd A.H, Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 2000;74(1):42–48. [PMC free article] [PubMed] [Google Scholar]
- Arnberg N, Kidd A.H, Edlund K, Olfat F, Wadell G. Initial interactions of subgenus D adenoviruses with A549 cellular receptors: Sialic acid versus alpha(v) integrins. J. Virol. 2000;74(16):7691–7693. doi: 10.1128/jvi.74.16.7691-7693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnberg N, Pring-Akerblom P, Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor on Chang C cells. J. Virol. 2002;76(17):8834–8841. doi: 10.1128/JVI.76.17.8834-8841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood W.J, Norkin L.C. Class I major histocompatibility proteins as cell surface receptors for simian virus 40. J. Virol. 1989;63(10):4474–4477. doi: 10.1128/jvi.63.10.4474-4477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aytay S, Schulze I.T. Single amino acid substitutions in the hemagglutinin can alter the host range and receptor binding properties of H1 strains of influenza A virus. J. Virol. 1991;65(6):3022–3028. doi: 10.1128/jvi.65.6.3022-3028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Snoeck R, Pauwels R, de Clercq E. Sulfated polysaccharides are potent and selective inhibitors of various enveloped viruses, including herpes simplex virus, cytomegalovirus, vesicular stomatitis virus, and human immunodeficiency virus. Antimicrob Agents Chemother. 1988;32(11):1742–1745. doi: 10.1128/aac.32.11.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y.S, Yoon J.W. Determination of diabetogenicity attributable to a single amino acid, Ala776, on the polyprotein of encephalomyocarditis virus. Diabetes. 1993;42(3):435–443. doi: 10.2337/diab.42.3.435. [DOI] [PubMed] [Google Scholar]
- Ball S.C, Abraha A, Collins K.R, Marozsan A.J, Baird H, Quinones-Mateu M.E, Penn-Nicholson A, Murray M, Richard N, Lobritz M, Zimmerman P.A, Kawamura T, Blauvelt A, Arts E.J. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 2003;77:1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban J, Portetelle D, Altaner C, Horion B, Milan D, Krchnak V, Burny A, Kettmann R. Isolation and characterization of a 2.3-kilobase-pair cDNA fragment encoding the binding domain of the bovine leukemia virus cell receptor. J. Virol. 1993;67(2):1050–1057. doi: 10.1128/jvi.67.2.1050-1057.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield B.W, Leduc Y, Esford L, Visalli R.J, Brandt C.R, Tufaro F. Evidence for an interaction of herpes simplex virus with chondroitin sulfate proteoglycans during infection. Virology. 1995;208(2):531–539. doi: 10.1006/viro.1995.1184. [DOI] [PubMed] [Google Scholar]
- Baranowski E, Ruiz-Jarabo C.M, Domingo E. Evolution of cell recognition by viruses. Science. 2001;292(5519):1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- Baranowski E, Ruiz–Jarabo C.M, Sevilla N, Andreu D, Beck E, Domingo E. Cell recognition by foot-and-mouth disease virus that lacks the RGD integrin-binding motif: flexibility in aphthovirus receptor usage. J. Virol. 2000;74(4):1641–1647. doi: 10.1128/jvi.74.4.1641-1647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowski E, Sevilla N, Verdaguer N, Ruíz-Jarabo C.M, Beck E, Domingo E. Multiple virulence determinants of foot-and-mouth disease virus in cell culture. J. Virol. 1998;72(8):6362–6372. doi: 10.1128/jvi.72.8.6362-6372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbis D.P, Chang S.F, Parrish C.R. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology. 1992;191(1):301–308. doi: 10.1016/0042-6822(92)90192-r. [DOI] [PubMed] [Google Scholar]
- Barton E.S, Chappell J.D, Connolly J.L, Forrest J.C, Dermody T.S. Reovirus receptors and apoptosis. Virology. 2001;290(2):173–180. doi: 10.1006/viro.2001.1160. [DOI] [PubMed] [Google Scholar]
- Barton E.S, Forrest J.C, Connolly J.L, Chappell J.D, Liu Y, Schnell F.J, Nusrat A, Parkos C.A, Dermody T.S. Junction adhesion molecule is a receptor for reovirus. Cell. 2001;104(3):441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- Bates P, Young J.A, Varmus H.E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74(6):1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- Batschelet E, Domingo E, Weissmann C. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene. 1976;1(1):27–32. doi: 10.1016/0378-1119(76)90004-4. [DOI] [PubMed] [Google Scholar]
- Battini J.L, Rasko J.E, Miller A.D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: Possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA. 1999;96(4):1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Spiess M, Klenk H.D. The asialoglycoprotein receptor is a potential liver-specific receptor for Marburg virus. J. Gen. Virol. 1995;76(Pt 2):393–399. doi: 10.1099/0022-1317-76-2-393. [DOI] [PubMed] [Google Scholar]
- Belnap D.M, McDermott B.M., Jr, Filman D.J, Cheng N, Trus B.L, Zuccola H.J, Racaniello V.R, Hogle J.M, Steven A.C. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA. 2000;97(1):73–78. doi: 10.1073/pnas.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbacer L, Kut E, Besnardeau L, Laude H, Delmas B. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 1997;71(1):734–737. doi: 10.1128/jvi.71.1.734-737.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J.M, Chan M, Solomon K.R, St. John N.F, Lin H, Finberg R.W. Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc. Natl. Acad. Sci. USA. 1994;91(13):6245–6249. doi: 10.1073/pnas.91.13.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J.M, Cunningham J.A, Droguett G, Kurt-Jones E.A, Krithivas A, Hong J.S, Horwitz M.S, Crowell R.L, Finberg R.W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275(5304):1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- Bergelson J.M, Krithivas A, Celi L, Droguett G, Horwitz M.S, Wickham T, Crowell R.L, Finberg R.W. The murine CAR homolog is a receptor for coxsackie B viruses and adenoviruses. J. Virol. 1998;72(1):415–419. doi: 10.1128/jvi.72.1.415-419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J.M, Mohanty J.G, Crowell R.L, St John N.F, Lublin D.M, Finberg R.W. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55) J. Virol. 1995;69(3):1903–1906. doi: 10.1128/jvi.69.3.1903-1906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J.M, Shepley M.P, Chan B.M, Hemler M.E, Finberg R.W. Identification of the integrin VLA-2 as a receptor for echovirus 1. Science. 1992;255(5052):1718–1720. doi: 10.1126/science.1553561. [DOI] [PubMed] [Google Scholar]
- Bergelson J.M, St John N, Kawaguchi S, Chan M, Stubdal H, Modlin J, Finberg R.W. Infection by echoviruses 1 and 8 depends on the alpha 2 subunit of human VLA-2. J. Virol. 1993;67(11):6847–6852. doi: 10.1128/jvi.67.11.6847-6852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger E.A, Murphy P.M, Farber J.M. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Berinstein A, Roivainen M, Hovi T, Mason P.W, Baxt B. Antibodies to the vitronectin receptor (integrin alpha V beta 3) inhibit binding and infection of foot-and-mouth disease virus to cultured cells. J. Virol. 1995;69(4):2664–2666. doi: 10.1128/jvi.69.4.2664-2666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfield M, Götte M, Woo Park P, Reizes O, Fitzgerald M.L, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Bewley M.C, Springer K, Zhang Y.B, Freimuth P, Flanagan J.M. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286(5444):1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- Bhat S, Mettus R.V, Reddy E.P, Ugen K.E, Srikanthan V, Williams W.V, Weiner D.B. The galactosyl ceramide⧸sulfatide receptor binding region of HIV-1 gp120 maps to amino acids 206–275. AIDS Res. Hum. Retrovir. 1993;9(2):175–181. doi: 10.1089/aid.1993.9.175. [DOI] [PubMed] [Google Scholar]
- Bhat S, Spitalnik S.L, Gonzalez-Scarano F, Silberberg D.H. Galactosyl ceramide or a derivative is an essential component of the neural receptor for human immunodeficiency virus type 1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA. 1991;88(16):7131–7134. doi: 10.1073/pnas.88.16.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkmann A, Mahr K, Ensser A, Yaguboglu S, Titgemeyer F, Fleckenstein B, Neipel F. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 2001;75(23):11583–11593. doi: 10.1128/JVI.75.23.11583-11593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzer M, Lauer U, Baumann C, Spiegel M, Gregor M, Neubert W.J. Sendai virus efficiently infects cells via the asialoglycoprotein receptor and requires the presence of cleaved F0 precursor proteins for this alternative route of cell entry. J. Virol. 1997;71(7):5481–5486. doi: 10.1128/jvi.71.7.5481-5486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsel M, Alfsen A. Entry of viruses through the epithelial barrier: pathogenic trickery. Nature Rev. Mol. Cell. Biol. 2003;4(1):57–68. doi: 10.1038/nrm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomer S, Eiden M, Burns C.C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J. Virol. 1997;71(11):8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A.C, Powers A.M, Holmes E.C, Woelk C.H, Weaver S.C. Positively charged amino acid substitutions in the e2 envelope glycoprotein are associated with the emergence of venezuelan equine encephalitis virus. J. Virol. 2002;76(4):1718–1730. doi: 10.1128/JVI.76.4.1718-1730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breau W.C, Atwood W.J, Norkin L.C. Class I major histocompatibility proteins are an essential component of the simian virus 40 receptor. J. Virol. 1992;66(4):2037–2045. doi: 10.1128/jvi.66.4.2037-2045.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiner K.M, Urban S, Schaller H. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J. Virol. 1998;72(10):8098–8104. doi: 10.1128/jvi.72.10.8098-8104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brojatsch J, Kristal B.S, Viglianti G.A, Khiroya R, Hoover E.A, Mullins J.I. Feline leukemia virus subgroup C phenotype evolves through distinct alterations near the N terminus of the envelope surface glycoprotein. Proc. Natl. Acad. Sci. USA. 1992;89(18):8457–8461. doi: 10.1073/pnas.89.18.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brojatsch J, Naughton J, Rolls M.M, Zingler K, Young J.A. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87(5):845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- Brown K.E, Anderson S.M, Young N.S. Erythrocyte P antigen: Cellular receptor for B19 parvovirus. Science. 1993;262(5130):114–117. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- Buckland R, Wild T.F. Is CD46 the cellular receptor for measles virus? Virus Res. 1997;48(1):1–9. doi: 10.1016/s0168-1702(96)01421-9. [DOI] [PubMed] [Google Scholar]
- Buchholz C.J, Koller D, Devaux P, Mumenthaler C, Schneider-Schaulies J, Braun W, Gerlier D, Cattaneo R. Mapping of the primary binding site of measles virus to its receptor CD46. J. Biol. Chem. 1997;272(35):22072–22079. doi: 10.1074/jbc.272.35.22072. [DOI] [PubMed] [Google Scholar]
- Budkowska A, Bedossa P, Groh F, Louise A, Pillot J. Fibronectin of human liver sinusoids binds hepatitis B virus: Identification by an anti-idiotypic antibody bearing the internal image of the pre-S2 domain. J. Virol. 1995;69(2):840–848. doi: 10.1128/jvi.69.2.840-848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. “Lateral DNA transfer. Mechanisms and Consequences.”. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2002. [Google Scholar]
- Byrnes A.P, Griffin D.E. Binding of Sindbis virus to cell surface heparan sulfate. J. Virol. 1998;72(9):7349–7356. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann A.J. “Principles of Molecular Virology.”. Academic Press; San Diego: 2001. [Google Scholar]
- Cao W, Henry M.D, Borrow P, Yamada H, Elder J.H, Ravkov E.V, Nichol S.T, Compans R.W, Campbell K.P, Oldstone M.B. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282(5396):2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- Capo C, Lindberg F.P, Meconi S, Zaffran Y, Tardei G, Brown E.J, Raoult D, Mege J.L. Subversion of monocyte functions by coxiella burnetti: Impairment of the cross-talk between alphavbeta3 integrin and CR3. J. Immunol. 1999;163(11):6078–6085. [PubMed] [Google Scholar]
- Card J.P. Pseudorabies virus neuroinvasiveness: A window into the functional organization of the brain. Adv. Virus. Res. 2001;56:39–71. doi: 10.1016/s0065-3527(01)56004-2. [DOI] [PubMed] [Google Scholar]
- Casasnovas J.M, Springer T.A. Kinetics and thermodynamics of virus binding to receptor: Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J. Biol. Chem. 1995;270(22):13216–13224. doi: 10.1074/jbc.270.22.13216. [DOI] [PubMed] [Google Scholar]
- Cassady K.A, Gross M, Gillespie G.Y, Roizman B. Second-site mutation outside of the U(S)10–12 domain of Deltagamma(1)34.5 herpes simplex virus 1 recombinant blocks the shutoff of protein synthesis induced by activated protein kinase R and partially restores neurovirulence. J. Virol. 2002;76(3):942–949. doi: 10.1128/JVI.76.3.942-949.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.Y, Empig C.J, Welte F.J, Speck R.F, Schmaljohn A, Kreisberg J.F, Goldsmith M.A. Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell. 2001;106(1):117–126. doi: 10.1016/s0092-8674(01)00418-4. [DOI] [PubMed] [Google Scholar]
- Chappell J.D, Duong J.L, Wright B.W, Dermody T.S. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 2000;74(18):8472–8479. doi: 10.1128/jvi.74.18.8472-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.S, Asanaka M, Chen F.S, Shively J.E, Lai M.M. Human carcinoembryonic antigen and biliary glycoprotein can serve as mouse hepatitis virus receptors. J. Virol. 1997;71(2):1688–1691. doi: 10.1128/jvi.71.2.1688-1691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.S, Asanaka M, Yokomori K, Wang F, Hwang S.B, Li H.P, Lai M.M. A pregnancy-specific glycoprotein is expressed in the brain and serves as a receptor for mouse hepatitis virus. Proc. Natl. Acad. Sci. USA. 1995;92(26):12095–12099. doi: 10.1073/pnas.92.26.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Maguire T, Hileman R.E, Fromm J.R, Esko J.D, Linhardt R.J, Marks R.M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nature Med. 1997;3(8):866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- Chipman P.R, Agbandje-McKenna M, Kajigaya S, Brown K.E, Young N.S, Baker T.S, Rossmann M.G. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc. Natl. Acad. Sci. USA. 1996;93(15):7502–7506. doi: 10.1073/pnas.93.15.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.Y, Mathias P, Nemerow G.R, Stewart P.L. Structure of adenovirus complexed with its internalization receptor, alphavbeta5 integrin. J. Virol. 1999;73(8):6759–6768. doi: 10.1128/jvi.73.8.6759-6768.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M.E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J. Virol. 1998;72(7):6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P.D, Wu L, Mackay C.R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Choi A.H, Lee P.W. Does the beta-adrenergic receptor function as a reovirus receptor? Virology. 1988;163(1):191–197. doi: 10.1016/0042-6822(88)90246-2. [DOI] [PubMed] [Google Scholar]
- Chung C.S, Hsiao J.C, Chang Y.S, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 1998;72(2):1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarlet M, Crawford S.E, Cheng E, Blutt S.E, Rice D.A, Bergelson J.M, Estes M.K. VLA-2 (alpha2beta1) integrin promotes rotavirus entry into cells but is not necessary for rotavirus attachment. J. Virol. 2002;76(3):1109–1123. doi: 10.1128/JVI.76.3.1109-1123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarlet M, Estes M.K. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 1999;80(Pt 4):943–948. doi: 10.1099/0022-1317-80-4-943. [DOI] [PubMed] [Google Scholar]
- Clarke M.F, Gelmann E.P, Reitz M.S., Jr. Homology of human T-cell leukaemia virus envelope gene with class I HLA gene. Nature. 1983;305(5929):60–62. doi: 10.1038/305060a0. [DOI] [PubMed] [Google Scholar]
- Co M.S, Gaulton G.N, Fields B.N, Greene M.I. Isolation and biochemical characterization of the mammalian reovirus type 3 cell-surface receptor. Proc. Natl. Acad. Sci USA. 1985;82(5):1494–1498. doi: 10.1073/pnas.82.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co M.S, Gaulton G.N, Tominaga A, Homcy C.J, Fields B.N, Greene M.I. Structural similarities between the mammalian beta-adrenergic and reovirus type 3 receptors. Proc. Natl. Acad. Sci. USA. 1985;82(16):5315–5318. doi: 10.1073/pnas.82.16.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 1998;72(12):9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J.M, Hughes S.H, Varmus H.E. “Retroviruses.”. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- Colston E.M, Racaniello V.R. Poliovirus variants selected on mutant receptor-expressing cells identify capsid residues that expand receptor recognition. J. Virol. 1995;69(8):4823–4829. doi: 10.1128/jvi.69.8.4823-4829.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton S.R, Stephensen C.B, Snyder S.W, Weismiller D.G, Holmes K.V. Coronavirus species specificity: Murine coronavirus binds to a mouse-specific epitope on its carcinoembryonic antigen-related receptor glycoprotein. J. Virol. 1992;66(12):7420–7428. doi: 10.1128/jvi.66.12.7420-7428.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton T, Nowlin D.M, Cooper N.R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193(2):834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- Connolly L, Zingler K, Young J.A. A soluble form of a receptor for subgroup A avian leukosis and sarcoma viruses (ALSV-A) blocks infection and binds directly to ALSV-A. J. Virol. 1994;68(4):2760–2764. doi: 10.1128/jvi.68.4.2760-2764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor R.J, Kawaoka Y, Webster R.G, Paulson J.C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205(1):17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- Coulon P, Ternaux J.P, Flamand A, Tuffereau C. An avirulent mutant of rabies virus is unable to infect motoneurons in vivo and in vitro. J. Virol. 1998;72(1):273–278. doi: 10.1128/jvi.72.1.273-278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson B.S, Londrigan S.L, Lee D.J. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc. Natl. Acad. Sci. USA. 1997;94(10):5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall K.A, editor. Johns Hopkins Univ. Press; Baltimore: 1999. (“The Evolution of HIV.”). [Google Scholar]
- Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nature Struct. Biol. 2000;7(11):1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- Curry S, Fry E, Blakemore W, Abu-Ghazaleh R, Jackson T, King A, Lea S, Newman J, Rowlands D, Stuart D. Perturbations in the surface structure of A22 Iraq foot-and-mouth disease virus accompanying coupled changes in host cell specificity and antigenicity. Structure. 1996;4(2):135–145. doi: 10.1016/s0969-2126(96)00017-2. [DOI] [PubMed] [Google Scholar]
- Dalgleish A.G, Beverley P.C, Clapham P.R, Crawford D.H, Greaves M.F, Weiss R.A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312(5996):763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Dalziel R.G, Hopkins J, Watt N.J, Dutia B.M, Clarke H.A, McConnell I. Identification of a putative cellular receptor for the lentivirus visna virus. J. Gen. Virol. 1991;72(Pt 8):1905–1911. doi: 10.1099/0022-1317-72-8-1905. [DOI] [PubMed] [Google Scholar]
- Damico R, Bates P. Soluble receptor-induced retroviral infection of receptor-deficient cells. J. Virol. 2000;74(14):6469–6475. doi: 10.1128/jvi.74.14.6469-6475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison E, Kirby I, Elliott T, Santis G. The human HLA-A∗0201 allele, expressed in hamster cells, is not a high-affinity receptor for adenovirus type 5 fiber. J. Virol. 1999;73(5):4513–4517. doi: 10.1128/jvi.73.5.4513-4517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechecchi M.C, Melotti P, Bonizzato A, Santacatterina M, Chilosi M, Cabrini G. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J. Virol. 2001;75(18):8772–8780. doi: 10.1128/JVI.75.18.8772-8780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechecchi M.C, Tamanini A, Bonizzato A, Cabrini G. Heparan sulfate glycosaminoglycans are involved in adenovirus type 5 and 2-host cell interactions. Virology. 2000;268(2):382–390. doi: 10.1006/viro.1999.0171. [DOI] [PubMed] [Google Scholar]
- De Clercq E, Schols D. Inhibition of HIV infection by CXCR4 and CCR5 chemokine receptor antagonists. Antivir. Chem. Chemother. 2001;12(Suppl. 1):19–31. [PubMed] [Google Scholar]
- DeFilippis V.R, Villarreal L.P. Virus evolution. In: Knipes D.M, Howley P.M, Griffin D.E, Lamb R.A, Martin M.A, Roizman B, Straus S.E, editors. 4th ed. Lippincott-Raven; Philadelphia: 2001. pp. 353–370. (“Fields Virology”). [Google Scholar]
- De Jong J.J, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: Analysis by single amino acid substitution. J. Virol. 1992;66(11):6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B, Gelfi J, L'Haridon R, Vogel L.K, Sjostrom H, Noren O, Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357(6377):417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme C, Brussow H, Sidoti J, Roche N, Karlsson K.A, Neeser J.R, Teneberg S. Glycosphingolipid binding specificities of rotavirus: Identification of a sialic acid-binding epitope. J. Virol. 2001;75(5):2276–2287. doi: 10.1128/JVI.75.5.2276-2287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R.E, Hill C.M, Davis C.B, Peiper S.C, Schall T.J, Littman D.R, Landau N.R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381(6584):661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Deng H.K, Unutmaz D, KewalRamani V.N, Littman D.R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388(6639):296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- Domingo E. Quasispecies. In: Granoff A, Webster R.G, editors. “Encyclopedia of Virology”. Academic Press; London: 1999. pp. 1431–1436. [Google Scholar]
- Domingo E. Complexities of virus–cell interactions. Current Opinions in Microbiology. 2003;6:383–385. doi: 10.1016/s1369-5274(03)00094-8. [DOI] [PubMed] [Google Scholar]
- Domingo E, Biebricher C, Eigen M, Holland J.J. “Quasispecies and RNA Virus Evolution: Principles and Consequences.”. Landes Bioscience; Austin: 2001. [Google Scholar]
- Domingo E, Escarmis C, Martinez M.A, Martinez-Salas E, Mateu M.G. Foot-and-mouth disease virus populations are quasispecies. Curr. Top. Microbiol. Immunol. 1992;176:33–47. doi: 10.1007/978-3-642-77011-1_3. [DOI] [PubMed] [Google Scholar]
- Domingo E, Martinez-Salas E, Sobrino F, de la Torre J.C, Portela A, Ortin J, Lopez-Galindez C, Pérez-Breña P, Villanueva N, Nájera R. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: Biological relevance. Gene. 1985;40(1):1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- Domingo E, Mateu M.G, Martinez M.A, Dopazo J, Moya A, Sobrino F. Genetic variability and antigenic diversity of foot-and-mouth disease virus. In: Kurkstak E, Marusyk R.G, Murphy S.A, Van-Regenmortel M.H.V, editors. Vol. 2. Plenum; New York: 1990. pp. 233–266. (“Applied Virology Research”). [Google Scholar]
- Domingo E, Sabo D, Taniguchi T, Weissmann C. Nucleotide sequence heterogeneity of an RNA phage population. Cell. 1978;13(4):735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- Domingo E, Webster R.G, Holland J.J, editors. Academic Press; San Diego: 1999. (“Origin and Evolution of Viruses.”). [Google Scholar]
- Donta S.T, Shanley J.D. Reovirus type 3 binds to antagonist domains of the beta-adrenergic receptor. J. Virol. 1990;64(2):639–641. doi: 10.1128/jvi.64.2.639-641.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R.F, editor. Computer methods for macromolecular sequence analysis. Vol. 266. Academic Press; San Diego: 1996. (“Methods in Enzymology”). [Google Scholar]
- Doranz B.J, Rucker J, Yi Y, Smyth R.J, Samson M, Peiper S.C, Parmentier M, Collman R.G, Doms R.W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85(7):1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- Dorig R.E, Marcil A, Chopra A, Richardson C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75(2):295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- Dragic T. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 2001;82(Pt 8):1807–1814. doi: 10.1099/0022-1317-82-8-1807. [DOI] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway G.P, Martin S.R, Huang Y, Nagashima K.A, Cayanan C, Maddon P.J, Koup R.A, Moore J.P, Paxton W.A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- Drake J.W, Holland J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dveksler G.S, Dieffenbach C.W, Cardellichio C.B, McCuaig K, Pensiero M.N, Jiang G.S, Beauchemin N, Holmes K.V. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 1993;67(1):1–8. doi: 10.1128/jvi.67.1.1-8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dveksler G.S, Pensiero M.N, Cardellichio C.B, Williams R.K, Jiang G.S, Holmes K.V, Dieffenbach C.W. Cloning of the mouse hepatitis virus (MHV) receptor: Expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 1991;65(12):6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D.M, Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Edinger A.L, Hoffman T.L, Sharron M, Lee B, O'Dowd B, Doms R.W. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology. 1998;249(2):367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- Edinger A.L, Hoffman T.L, Sharron M, Lee B, Yi Y, Choe W, Kolson D.L, Mitrovic B, Zhou Y, Faulds D, Collman R.G, Hesselgesser J, Horuk R, Doms R.W. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 1998;72(10):7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M. On the nature of virus quasispecies. Trends. Microbiol. 1996;4(6):216–218. doi: 10.1016/0966-842X(96)20011-3. [DOI] [PubMed] [Google Scholar]
- Eigen M. Natural selection: A phase transition? Biophys. Chem. 2000;85:101–123. doi: 10.1016/s0301-4622(00)00122-8. [DOI] [PubMed] [Google Scholar]
- Eigen M, Biebricher C.K. Sequence space and quasispecies distribution. In: Domingo E, Ahlquist P, Holland J.J, editors. Vol. 3. CRC Press; Boca Raton, FL: 1988. pp. 211–245. (“RNA Genetics”). [Google Scholar]
- Eigen M, Schuster P. “The Hypercycle: A Principle of Natural Self-Organization.”. Springer; Berlin: 1979. [DOI] [PubMed] [Google Scholar]
- Eisen M.B, Sabesan S, Skehel J.J, Wiley D.C. Binding of the influenza A virus to cell-surface receptors: Structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology. 1997;232(1):19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- Escarmís C, Carrillo E.C, Ferrer M, Arriaza J.F, Lopez N, Tami C, Verdaguer N, Domingo E, Franze-Fernández M.T. Rapid selection in modified BHK-21 cells of a foot-and-mouth disease virus variant showing alterations in cell tropism. J. Virol. 1998;72(12):10171–10179. doi: 10.1128/jvi.72.12.10171-10179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escarmís C, Dávila M, Domingo E. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller's ratchet. J. Mol. Biol. 1999;285:495–505. doi: 10.1006/jmbi.1998.2366. [DOI] [PubMed] [Google Scholar]
- Escarmís C, Gómez-Mariano G, Dávila M, Lázaro E, Domingo E. Resistance to extinction of low fitness virus subjected to plaque-to-plaque transfers: Diversification by mutation clustering. J. Mol. Biol. 2002;315(4):647–661. doi: 10.1006/jmbi.2001.5259. [DOI] [PubMed] [Google Scholar]
- Este J.A, Cabrera C, Blanco J, Gutierrez A, Bridger G, Henson G, Clotet B, Schols D, De Clercq E. Shift of clinical human immunodeficiency virus type 1 isolates from X4 to R5 and prevention of emergence of the syncytium-inducing phenotype by blockade of CXCR4. J. Virol. 1999;73(7):5577–5585. doi: 10.1128/jvi.73.7.5577-5585.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evander M, Frazer I.H, Payne E, Qi Y.M, Hengst K, McMillan N.A. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J. Virol. 1997;71(3):2449–2456. doi: 10.1128/jvi.71.3.2449-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.J, Almond J.W. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 1998;6(5):198–202. doi: 10.1016/s0966-842x(98)01263-3. [DOI] [PubMed] [Google Scholar]
- Fantini J, Cook D.G, Nathanson N, Spitalnik S.L, Gonzalez-Scarano F. Infection of colonic epithelial cell lines by type 1 human immunodeficiency virus is associated with cell surface expression of galactosylceramide, a potential alternative gp120 receptor. Proc. Natl. Acad. Sci. USA. 1993;90(7):2700–2704. doi: 10.1073/pnas.90.7.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 1997;186(3):405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigelstock D, Thompson P, Mattoo P, Zhang Y, Kaplan G.G. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J. Virol. 1998;72(8):6621–6628. doi: 10.1128/jvi.72.8.6621-6628.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman S.A, Hendry R.M, Beeler J.A. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 1999;73(8):6610–6617. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Broder C.C, Kennedy P.E, Berger E.A. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272(5263):872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Fenÿo E.M. The role of virus biological phenotype in human immunodeficiency virus pathogenesis. AIDS Rev. 2001;3:157–168. [Google Scholar]
- Fingeroth J.D, Weis J.J, Tedder T.F, Strominger J.L, Biro P.A, Fearon D.T. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. USA. 1984;81(14):4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint M, Maidens C, Loomis-Price L.D, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating J.A. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 1999;73(8):6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint S.J, Enquist L.W, Krug R.M, Racaniello V.R, Skalka A.M. “Virology, Molecular Biology, Pathogenesis and Control.”. ASM Press; Washington, DC: 2000. [Google Scholar]
- Fouchier R.A, Brouwer M, Broersen S.M, Schuitemaker H. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J. Clin. Microbiol. 1995;33(4):906–911. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G, Parry N.R, Barnett P.V, McGinn B, Rowlands D.J, Brown F. The cell attachment site on foot-and-mouth disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid) J. Gen. Virol. 1989;70(Pt 3):625–637. doi: 10.1099/0022-1317-70-3-625. [DOI] [PubMed] [Google Scholar]
- Frade R, Barel M, Ehlin-Henriksson B, Klein G. gp140, the C3d receptor of human B lymphocytes, is also the Epstein-Barr virus receptor. Proc. Natl. Acad. Sci. USA. 1985;82(5):1490–1493. doi: 10.1073/pnas.82.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimuth P, Springer K, Berard C, Hainfeld J, Bewley M, Flanagan J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J. Virol. 1999;73(2):1392–1398. doi: 10.1128/jvi.73.2.1392-1398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S.C, Hoover E.A, Mullins J.I. Feline immunodeficiency virus cell entry. J. Virol. 2001;75(11):5433–5440. doi: 10.1128/JVI.75.11.5433-5440.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C, Walker G.C, Siede W. “DNA Repair and Mutagenesis.”. American Society for Microbiology; Washington, DC: 1995. [Google Scholar]
- Fry E.E, Lea S.M, Jackson T, Newman J.W, Ellard F.M, Blakemore W.E, Abu-Ghazaleh R, Samuel A, King A.M, Stuart D.I. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 1999;18(3):543–554. doi: 10.1093/emboj/18.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzino-Demo A, DeVico A.L, Cocchi F, Gallo R.C. Beta-chemokines and protection from HIV type 1 disease. AIDS Res. Hum. Retrovir. 1998;14(Suppl. 2):S177–S184. [PubMed] [Google Scholar]
- Gastka M, Horvath J, Lentz T.L. Rabies virus binding to the nicotinic acetylcholine receptor alpha subunit demonstrated by virus overlay protein binding assay. J. Gen. Virol. 1996;77(Pt 10):2437–2440. doi: 10.1099/0022-1317-77-10-2437. [DOI] [PubMed] [Google Scholar]
- Gaulton G, Co M.S, Greene M.I. Anti-idiotypic antibody identifies the cellular receptor of reovirus type 3. J. Cell Biochem. 1985;28(1):69–78. doi: 10.1002/jcb.240280110. [DOI] [PubMed] [Google Scholar]
- Gavrilovskaya I.N, Brown E.J, Ginsberg M.H, Mackow E.R. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by beta3 integrins. J. Virol. 1999;73(5):3951–3959. doi: 10.1128/jvi.73.5.3951-3959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilovskaya I.N, Shepley M, Shaw R, Ginsberg M.H, Mackow E.R. beta3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA. 1998;95(12):7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch J.R, Pacitti A.F. Differential interaction of reovirus type 3 with sialylated receptor components on animal cells. Virology. 1987;161(1):245–248. doi: 10.1016/0042-6822(87)90192-9. [DOI] [PubMed] [Google Scholar]
- Geraghty R.J, Krummenacher C, Cohen G.H, Eisenberg R.J, Spear P.G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280(5369):1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- Germi R, Crance J.M, Garin D, Guimet J, Lortat-Jacob H, Ruigrok R.W, Zarski J.P, Drouet E. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology. 2002;292(1):162–168. doi: 10.1006/viro.2001.1232. [DOI] [PubMed] [Google Scholar]
- Gibbs M.J, Weiller G.F. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc. Natl. Acad. Sci. USA. 1999;96(14):8022–8027. doi: 10.1073/pnas.96.14.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroglou T, Florin L, Schafer F, Streeck R.E, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 2001;75(3):1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith M.A, Doms R.W. HIV entry: Are all receptors created equal? Nature Immunol. 2002;3(8):709–710. doi: 10.1038/ni819. [DOI] [PubMed] [Google Scholar]
- Goodfellow I.G, Powell R.M, Ward T, Spiller O.B, Almond J.W, Evans D.J. Echovirus infection of rhabdomyosarcoma cells is inhibited by antiserum to the complement control protein CD59. J. Gen. Virol. 2000;81(Pt 5):1393–1401. doi: 10.1099/0022-1317-81-5-1393. [DOI] [PubMed] [Google Scholar]
- Goodfellow I.G, Sioofy A.B, Powell R.M, Evans D.J. Echoviruses bind heparan sulfate at the cell surface. J. Virol. 2001;75(10):4918–4921. doi: 10.1128/JVI.75.10.4918-4921.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M.F, Fygenson K.D. DNA polymerase fidelity: From genetics toward a biochemical understanding. Genetics. 1998;148(4):1475–1482. doi: 10.1093/genetics/148.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E. Origin of RNA viral genomes: Approaching the problem by comparative sequence analysis. In: Gibbs A.J, Calisher C.H, Garcia-Arenal F, editors. “Molecular Basis of Virus Evolution”. Cambridge Univ. Press; Cambridge: 1995. pp. 49–66. [Google Scholar]
- Greve J.M, Davis G, Meyer A.M, Forte C.P, Yost S.C, Marlor C.W, Kamarck M.E, McClelland A. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56(5):839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- Gruenberger M, Wandl R, Nimpf J, Hiesberger T, Schneider W.J, Kuechler E, Blaas D. Avian homologs of the mammalian low-density lipoprotein receptor family bind minor receptor group human rhinovirus. J. Virol. 1995;69(11):7244–7247. doi: 10.1128/jvi.69.11.7244-7247.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy J.E, McKeating J.A, Ward P.J, Sanderson A.R, Griffiths P.D. Beta 2 microglobulin enhances the infectivity of cytomegalovirus and when bound to the virus enables class I HLA molecules to be used as a virus receptor. J. Gen. Virol. 1987;68(Pt 3):793–803. doi: 10.1099/0022-1317-68-3-793. [DOI] [PubMed] [Google Scholar]
- Gubler D.J. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998;11(3):480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C.A, Mendez E, Zarate S, Isa P, Lopez S, Arias C.F. Integrin alpha(v)beta(3) mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA. 2000;97(26):14644–14649. doi: 10.1073/pnas.250299897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan K.M, Kwok W.W, Longnecker R, Speck P. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 2000;74(5):2451–2454. doi: 10.1128/jvi.74.5.2451-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan K.M, Longnecker R. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc. Natl. Acad. Sci. USA. 2000;97(16):9252–9257. doi: 10.1073/pnas.160171697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallak L.K, Collins P.L, Knudson W, Peeples M.E. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology. 2000;271(2):264–275. doi: 10.1006/viro.2000.0293. [DOI] [PubMed] [Google Scholar]
- Hammache D, Pieroni G, Yahi N, Delezay O, Koch N, Lafont H, Tamalet C, Fantini J. Specific interaction of HIV-1 and HIV-2 surface envelope glycoproteins with monolayers of galactosylceramide and ganglioside GM3. J. Biol. Chem. 1998;273(14):7967–7971. doi: 10.1074/jbc.273.14.7967. [DOI] [PubMed] [Google Scholar]
- Hanham C.A, Zhao F, Tignor G.H. Evidence from the anti-idiotypic network that the acetylcholine receptor is a rabies virus receptor. J. Virol. 1993;67(1):530–542. doi: 10.1128/jvi.67.1.530-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harber J, Bernhardt G, Lu H.H, Sgro J.Y, Wimmer E. Canyon rim residues, including antigenic determinants, modulate serotype-specific binding of polioviruses to mutants of the poliovirus receptor. Virology. 1995;214(2):559–570. doi: 10.1006/viro.1995.0067. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Ono N, Tatsuo H, Minagawa H, Takeda M, Takeuchi K, Yanagi Y. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 2002;76(13):6743–6749. doi: 10.1128/JVI.76.13.6743-6749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood A.M. Virus receptors: Binding, adhesion strengthening, and changes in viral structure. J. Virol. 1994;68(1):1–5. doi: 10.1128/jvi.68.1.1-5.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Bowman V.D, Mueller S, Bator C.M, Bella J, Peng X, Baker T.S, Wimmer E, Kuhn R.J, Rossmann M.G. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA. 2000;97(1):79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chipman P.R, Howitt J, Bator C.M, Whitt M.A, Baker T.S, Kuhn R.J, Anderson C.W, Freimuth P, Rossmann M.G. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nature Struct. Biol. 2001;8(10):874–878. doi: 10.1038/nsb1001-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Lin F, Chipman P.R, Bator C.M, Baker T.S, Shoham M, Kuhn R, Medof M.E, Rossmann M.G. Structure of decay-accelerating factor bound to echovirus 7: A virus receptor complex. Proc. Natl. Acad. Sci. USA. 2002;99(16):10325–10329. doi: 10.1073/pnas.152161599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrler G, Rott R, Klenk H.D, Muller H.P, Shukla A.K, Schauer R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. EMBO J. 1985;4(6):1503–1506. doi: 10.1002/j.1460-2075.1985.tb03809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertogs K, Depla E, Crabbe T, De Bruin W, Leenders W, Moshage H, Yap S.H. Spontaneous development of anti-hepatitis B virus envelope (anti-idiotypic) antibodies in animals immunized with human liver endonexin II or with the F(ab′)2 fragment of anti-human liver endonexin II immunoglobulin G: Evidence for a receptor-ligand-like relationship between small hepatitis B surface antigen and endonexin II. J. Virol. 1994;68(3):1516–1521. doi: 10.1128/jvi.68.3.1516-1521.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertogs K, Leenders W.P, Depla E, De Bruin W.C, Meheus L, Raymackers J, Moshage H, Yap S.H. Endonexin II, present on human liver plasma membranes, is a specific binding protein of small hepatitis B virus (HBV) envelope protein. Virology. 1993;197(2):549–557. doi: 10.1006/viro.1993.1628. [DOI] [PubMed] [Google Scholar]
- Hertzler S, Kallio P, Lipton H.L. UDP-galactose transporter is required for Theiler's virus entry into mammalian cells. Virology. 2001;286(2):336–344. doi: 10.1006/viro.2001.0981. [DOI] [PubMed] [Google Scholar]
- Heveker N. Chemokine receptors as anti-retroviral targets. Curr. Drug Targets. 2001;2(1):21–39. doi: 10.2174/1389450013348849. [DOI] [PubMed] [Google Scholar]
- Hewat E.A, Neumann E, Conway J.F, Moser R, Ronacher B, Marlovits T.C, Blaas D. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: A structural view. EMBO J. 2000;19(23):6317–6325. doi: 10.1093/emboj/19.23.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewat E.A, Verdaguer N, Fita I, Blakemore W, Brookes S, King A, Newman J, Domingo E, Mateu M.G, Stuart D.I. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: Positioning of a highly mobile antigenic loop. EMBO J. 1997;16(7):1492–1500. doi: 10.1093/emboj/16.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish M.J, Takada Y, Coulson B.S. Integrins alpha2beta1 and alpha4beta1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 2000;74(1):228–236. doi: 10.1128/jvi.74.1.228-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu K, Tadano M, Men R, Lai C.J. Mutational analysis of a neutralization epitope on the dengue type 2 virus (DEN2) envelope protein: Monoclonal antibody resistant DEN2⧸DEN4 chimeras exhibit reduced mouse neurovirulence. Virology. 1996;224(2):437–445. doi: 10.1006/viro.1996.0550. [DOI] [PubMed] [Google Scholar]
- Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blass D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. USA. 1994;91(5):1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N.G, Seillier-Moiseiwitsch F, Ahn J, Walker J.M, Swanstrom R. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 2002;76(8):3852–3864. doi: 10.1128/JVI.76.8.3852-3864.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle J.M. Poliovirus cell entry: Common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmen S.L, Federspiel M.J. Selection of a subgroup A avian leukosis virus [ALV(A)] envelope resistant to soluble ALV(A) surface glycoprotein. Virology. 2000;273(2):364–373. doi: 10.1006/viro.2000.0424. [DOI] [PubMed] [Google Scholar]
- Holland J, Domingo E. Origin and evolution of viruses. Virus Genes. 1998;16(1):13–21. doi: 10.1023/a:1007989407305. [DOI] [PubMed] [Google Scholar]
- Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Holland J.J. Receptor affinities as major determinants of enterovirus tissue tropism in humans. Virology. 1961;15:312–326. doi: 10.1016/0042-6822(61)90363-4. [DOI] [PubMed] [Google Scholar]
- Holland J.J, McLaren L.C. The mammalian cell-virus relationship. II. Adsorption, reception, and eclipse by HeLa cells. J. Exp. Med. 1959;109:487–504. doi: 10.1084/jem.109.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.S, Karayan L, Tournier J, Curiel D.T, Boulanger P.A. Adenovirus type 5 fiber knob binds to MHC class I alpha2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1997;16(9):2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvat B, Rivailler P, Varior-Krishnan G, Cardoso A, Gerlier D, Rarourdin-Combe C. Transgenic mice expressing human measles virus (MV) receptor CD46 provide cells exhibiting different permissivities to MV infections. J. Virol. 1996;70(10):6673–6681. doi: 10.1128/jvi.70.10.6673-6681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao J.C, Chung C.S, Chang W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 1999;73(10):8750–8761. doi: 10.1128/jvi.73.10.8750-8761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu E.C, Iorio C, Sarangi F, Khine A.A, Richardson C.D. CDw150 (SLAM) is a receptor for a lymphotropic strain of measles virus and may account for the immunosuppressive properties of this virus. Virology. 2001;279(1):9–21. doi: 10.1006/viro.2000.0711. [DOI] [PubMed] [Google Scholar]
- Hsu E.C, Sarangi F, Iorio C, Sidhu M.S, Udem S.A, Dillehay D.L, Xu W, Rota P.A, Bellini W.J, Richardson C.D. A single amino acid change in the hemagglutinin protein of measles virus determines its ability to bind CD46 and reveals another receptor on marmoset B cells. J. Virol. 1998;72(4):2905–2916. doi: 10.1128/jvi.72.4.2905-2916.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Reddy V, Dasgupta N, Nemerow G.R. A single amino acid in the adenovirus type 37 fiber confers binding to human conjunctival cells. J. Virol. 1999;73(4):2798–2802. doi: 10.1128/jvi.73.4.2798-2802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Campadelli-Fiume G. Anti-idiotypic antibodies mimicking glycoprotein D of herpes simplex virus identify a cellular protein required for virus spread from cell to cell and virus-induced polykaryocytosis. Proc. Natl. Acad. Sci. USA. 1996;93(5):1836–1840. doi: 10.1073/pnas.93.5.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S.A. VCAM-1 is a receptor for encephalomyocarditis virus on murine vascular endothelial cells. J. Virol. 1994;68(6):3453–3458. doi: 10.1128/jvi.68.6.3453-3458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueffer K, Parker J.S, Weichert W.S, Geisel R.E, Sgro J.Y, Parrish C.R. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 2003;77(3):1718–1726. doi: 10.1128/JVI.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulst M.M, van Gennip H.G, Moormann R.J. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein E(rns) J. Virol. 2000;74(20):9553–9561. doi: 10.1128/jvi.74.20.9553-9561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulst M.M, van Gennip H.G, Vlot A.C, Schooten E, de Smit A.J, Moormann R.J. Interaction of classical swine fever virus with membrane-associated heparan sulfate: role for virus replication in vivo and virulence. J. Virol. 2001;75(20):9585–9595. doi: 10.1128/JVI.75.20.9585-9595.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter E. Viral entry and receptors. In: Coffin J.M, Hughes S.H, Varmus H.E, editors. “Retroviruses”. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 71–119. [Google Scholar]
- Hurst C.J, editor. Academic Press; San Diego: 2000. (“Viral Ecology”). [Google Scholar]
- Inada T, Mims C.A. Mouse Ia antigens are receptors for lactate dehydrogenase virus. Nature. 1984;309(5963):59–61. doi: 10.1038/309059a0. [DOI] [PubMed] [Google Scholar]
- Isberg R.R, Leong J.M. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell. 1990;60(5):861–871. doi: 10.1016/0092-8674(90)90099-z. [DOI] [PubMed] [Google Scholar]
- Isnard M, Granier M, Frutos R, Reynaud B, Peterschmitt M. Quasispecies nature of three maize streak virus isolates obtained through different modes of selection from a population used to assess response to infection of maize cultivars. J. Gen. Virol. 1998;79:3091–3099. doi: 10.1099/0022-1317-79-12-3091. [DOI] [PubMed] [Google Scholar]
- Ito M, Baba M, Sato A, Pauwels R, De Clercq E, Shigeta S. Inhibitory effect of dextran sulfate and heparin on the replication of human immunodeficiency virus (HIV) in vitro. Antiviral Res. 1987;7(6):361–367. doi: 10.1016/0166-3542(87)90018-0. [DOI] [PubMed] [Google Scholar]
- Jackson T, Blakemore W, Newman J.W, Knowles N.J, Mould A.P, Humphries M.J, King A.M. Foot-and-mouth disease virus is a ligand for the high-affinity binding conformation of integrin alpha5beta1: Influence of the leucine residue within the RGDL motif on selectivity of integrin binding. J. Gen. Virol. 2000;81(Pt5):1383–1391. doi: 10.1099/0022-1317-81-5-1383. [DOI] [PubMed] [Google Scholar]
- Jackson T, Ellard F.M, Ghazaleh R.A, Brookes S.M, Blakemore W.E, Corteyn A.H, Stuart D.I, Newman J.W, King A.M. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 1996;70(8):5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T, Mould A.P, Sheppard D, King A.M. Integrin alphavbetal is a receptor for foot-and-mouth disease virus. J. Virol. 2002;76(3):935–941. doi: 10.1128/JVI.76.3.935-941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T, Sharma A, Ghazaleh R.A, Blakemore W.E, Ellard F.M, Simmons D.L, Newman J.W, Stuart D.I, King A.M. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth disease viruses to the purified integrin alpha(v)beta3 in vitro. J. Virol. 1997;71(11):8357–8361. doi: 10.1128/jvi.71.11.8357-8361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T, Sheppard D, Denyer M, Blakemore W, King A.M. The epithelial integrin alphavbeta6 is a receptor for foot-and-mouth disease virus. J. Virol. 2000;74(11):4949–4956. doi: 10.1128/jvi.74.11.4949-4956.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings A.D, Gibson C.A, Miller B.R, Mathews J.H, Mitchell C.J, Roehrig J.T, Wood D.J, Taffs F, Sil B.K, Whitby S.N. Analysis of a yellow fever virus isolated from a fatal case of vaccine-associated human encephalitis. J. Infect. Dis. 1994;169(3):512–518. doi: 10.1093/infdis/169.3.512. [DOI] [PubMed] [Google Scholar]
- Jia Q, Ohka S, Iwasaki K, Tohyama K, Nomoto A. Isolation and molecular characterization of a poliovirus type 1 mutant that replicates in the spinal cords of mice. J. Virol. 1999;73(7):6041–6047. doi: 10.1128/jvi.73.7.6041-6047.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jnaoui K, Michiels T. Adaptation of Theiler's virus to L929 cells: mutations in the putative receptor binding site on the capsid map to neutralization sites and modulate viral persistence. Virology. 1998;244(2):397–404. doi: 10.1006/viro.1998.9134. [DOI] [PubMed] [Google Scholar]
- Jnaoui K, Minet M, Michiels T. Mutations that affect the tropism of DA and GDVII strains of Theiler's virus in vitro influence sialic acid binding and pathogenicity. J. Virol. 2002;76(16):8138–8147. doi: 10.1128/JVI.76.16.8138-8147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai K, Furuta T. Isolation of paralysis-inducing murine leukemia viruses from Friend virus passaged in rats. J. Virol. 1984;50(3):970–973. doi: 10.1128/jvi.50.3.970-973.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan G, Totsuka A, Thompson P, Akatsuka T, Moritsugu Y, Feinstone S.M. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 1996;15(16):4282–4296. [PMC free article] [PubMed] [Google Scholar]
- Karnauchow T.M, Tolson D.L, Harrison B.A, Altman E, Lublin D.M, Dimock K. The HeLa cell receptor for enterovirus 70 is decay-accelerating factor (CD55) J. Virol. 1996;70(8):5143–5152. doi: 10.1128/jvi.70.8.5143-5152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keay S, Baldwin B. Anti-idiotype antibodies that mimic gp86 of human cytomegalovirus inhibit viral fusion but not attachment. J. Virol. 1991;65(9):5124–5128. doi: 10.1128/jvi.65.9.5124-5128.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keay S, Merigan T.C, Rasmussen L. Identification of cell surface receptors for the 86-kilodalton glycoprotein of human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 1989;86(24):10100–10103. doi: 10.1073/pnas.86.24.10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew O, Morris-Glasgow V, Landaverde M, Burns C, Shaw J, Garib Z, Andre J, Blackman E, Freeman C.J, Jorba J, Sutter R, Tambini G, Venczel L, Pedreira C, Laender F, Shimizu H, Yoneyama T, Miyamura T, van Der Avoort H, Oberste M.S, Kilpatrick D, Cochi S, Pallansch M, de Quadros C. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science. 2002;296(5566):356–359. doi: 10.1126/science.1068284. [DOI] [PubMed] [Google Scholar]
- Kewalramani V.N, Panganiban A.T, Emerman M. Spleen necrosis virus, an avian immunosuppressive retrovirus, shares a receptor with the type D simian retroviruses. J. Virol. 1992;66(5):3026–3031. doi: 10.1128/jvi.66.5.3026-3031.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.L, Cunningham J.A, Finberg R.W, Bergelson J.M. Echovirus 1 interaction with the isolated VLA-2 I domain. J. Virol. 1995;69(5):3237–3239. doi: 10.1128/jvi.69.5.3237-3239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J.C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312(5996):767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- Klimstra W.B, Ryman K.D, Johnston R.E. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 1998;72(9):7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe D.M, Howley P.M, editors. Lippincott Williams and Wilkins; Philadelphia: 2001. (“Fields Virology.”). [Google Scholar]
- Kohtz D.S, Altman A, Kohtz J.D, Puszkin S. Immunological and structural homology between human T-cell leukemia virus type I envelope glycoprotein and a region of human interleukin-2 implicated in binding the beta receptor. J. Virol. 1988;62(2):659–662. doi: 10.1128/jvi.62.2.659-662.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolatkar P.R, Bella J, Olson N.H, Bator C.M, Baker T.S, Rossmann M.G. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18(22):6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komai K, Kaplan M, Peeples M.E. The Vero cell receptor for the hepatitis B virus small S protein is a sialoglycoprotein. Virology. 1988;163(2):629–634. doi: 10.1016/0042-6822(88)90306-6. [DOI] [PubMed] [Google Scholar]
- Koo H.M, Parthasarathi S, Ron Y, Dougherty J.P. Pseudotyped REV⧸SRV retroviruses reveal restrictions to infection and host range within members of the same receptor interference group. Virology. 1994;205(1):345–351. doi: 10.1006/viro.1994.1651. [DOI] [PubMed] [Google Scholar]
- Krance R.A, Heslop H.E, Brenner M.K. Transplantation and virus infections. In: Granoff A, Webster R.G, editors. Vol. 3. Academic Press; San Diego: 1999. pp. 1823–1833. (“Encyclopedia of Virology”). [Google Scholar]
- Krukonis E.S, Isberg R.R. Microbial pathogens and integrin interactions. In: Eble J.A, Kühn K, editors. “Integrin-Ligand Interaction”. Landes Bioscience; Austin: 1997. pp. 175–197. [Google Scholar]
- Kubo H, Takase-Yoden S, Taguchi F. Neutralization and fusion inhibition activities of monoclonal antibodies specific for the S1 subunit of the spike protein of neurovirulent murine coronavirus JHMV c1-2 variant. J. Gen. Virol. 1993;74(Pt 7):1421–1425. doi: 10.1099/0022-1317-74-7-1421. [DOI] [PubMed] [Google Scholar]
- Kubo H, Yamada Y.K, Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68(9):5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz S, Borrow P, Oldstone M.B. Receptor structure, binding, and cell entry of arenaviruses. Curr. Top. Microbiol. Immunol. 2002;262:111–137. doi: 10.1007/978-3-642-56029-3_5. [DOI] [PubMed] [Google Scholar]
- Kunz S, Sevilla N, McGavern D.B, Campbell K.P, Oldstone M.B. Molecular analysis of the interaction of LCMV with its cellular receptor [alpha]-dystroglycan. J. Cell. Biol. 2001;155(2):301–310. doi: 10.1083/jcb.200104103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D, Wyatt R, Robinson J, Sweet R.W, Sodroski J, Hendrickson W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance C, Arbour N, Cashman N.R, Talbot P.J. Involvement of aminopeptidase N (CD13) in infection of human neural cells by human coronavirus 229E. J. Virol. 1998;72(8):6511–6519. doi: 10.1128/jvi.72.8.6511-6519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeeq S, Smith C.A, Wagner S.D, Thomas D.B. Preferential selection of receptor-binding variants of influenza virus hemagglutinin by the neutralizing antibody repertoire of transgenic mice expressing a human immunoglobulin mu minigene. J. Virol. 1997;71(4):2600–2605. doi: 10.1128/jvi.71.4.2600-2605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafrenie R.M, Lee S.F, Hewlett I.K, Yamada K.M, Dhawan S. Involvement of integrin alphavbeta3 in the pathogenesis of human immunodeficiency virus type 1 infection in monocytes. Virology. 2002;297(1):31–38. doi: 10.1006/viro.2002.1399. [DOI] [PubMed] [Google Scholar]
- Lalani A.S, Masters J, Zeng W, Barrett J, Pannu R, Everett H, Arendt C.W, McFadden G. Use of chemokine receptors by poxviruses. Science. 1999;286(5446):1968–1971. doi: 10.1126/science.286.5446.1968. [DOI] [PubMed] [Google Scholar]
- Lando Z, Sarin P, Megson M, Greene W.C, Waldman T.A, Gallo R.C, Broder S. Association of human T-cell leukaemia⧸lymphoma virus with the Tac antigen marker for the human T-cell growth factor receptor. Nature. 1983;305(5936):733–736. doi: 10.1038/305733a0. [DOI] [PubMed] [Google Scholar]
- Lapham C.K, Ouyang J, Chandrasekhar B, Nguyen N.Y, Dimitrov D.S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274(5287):602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- Lauring A.S, Anderson M.M, Overbaugh J. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: Implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 2001;75(19):8888–8898. doi: 10.1128/JVI.75.19.8888-8898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavillette D, Ruggieri A, Russell S.J, Cosset F.L. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 2000;74(1):295–304. doi: 10.1128/jvi.74.1.295-304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro E, Escarmis C, Domingo E, Manrubia S.C. Modeling viral genome fitness evolution associated with serial bottleneck events: Evidence of stationary states of fitness. J. Virol. 2002;76:8675–8681. doi: 10.1128/JVI.76.17.8675-8681.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Knight R, Smit J.M, Wilschut J, Griffin D.E. A single mutation in the e2 glycoprotein important for neurovirulence influences binding of sindbis virus to neuroblastoma cells. J. Virol. 2002;76(12):6302–6310. doi: 10.1128/JVI.76.12.6302-6310.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitmeyer K.C, Vaughn D.W, Watts D.M, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 1999;73(6):4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine N.R, editor. Springer-Verlag; Berliin: 2000. (“Understanding Gene Therapy.”). [Google Scholar]
- Lentz T.L, Burrage T.G, Smith A.L, Crick J, Tignor G.H. Is the acetylcholine receptor a rabies virus receptor? Science. 1982;215(4529):182–184. doi: 10.1126/science.7053569. [DOI] [PubMed] [Google Scholar]
- Lentz T.L, Wilson P.T, Hawrot E, Speicher D.W. Amino acid sequence similarity between rabies virus glycoprotein and snake venom curaremimetic neurotoxins. Science. 1984;226(4676):847–848. doi: 10.1126/science.6494916. [DOI] [PubMed] [Google Scholar]
- Lerner A.M, Cherry J.D, Finland M. Haemagglutination with reoviruses. Virology. 1963;19:58–65. doi: 10.1016/0042-6822(63)90024-2. [DOI] [PubMed] [Google Scholar]
- Lerner D.L, Elder J.H. Expanded host cell tropism and cytopathic properties of feline immunodeficiency virus strain PPR subsequent to passage through interleukin-2-independent T cells. J. Virol. 2000;74(4):1854–1863. doi: 10.1128/jvi.74.4.1854-1863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Brown S.L, Stupack D.G, Puente X.S, Cheresh D.A, Nemerow G.R. Integrin alpha(v)beta1 is an adenovirus coreceptor. J. Virol. 2001;75(11):5405–5409. doi: 10.1128/JVI.75.11.5405-5409.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.S, Tong S.P, Wands J.R. Characterization of a 120-kilodalton pre-S-binding protein as a candidate duck hepatitis B virus receptor. J. Virol. 1996;70(9):6029–6035. doi: 10.1128/jvi.70.9.6029-6035.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Spriggs M.K, Kovats S, Turk S.M, Comeau M.R, Nepom B, Hutt-Fletcher L.M. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 1997;71(6):4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Rong L, Laughrea M, Kleiman L, Wainberg M.A. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J. Virol. 1998;72(8):6629–6636. doi: 10.1128/jvi.72.8.6629-6636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F, Alkhatib G, Peden K.W, Sharma G, Berger E.A, Farber J.M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J. Exp. Med. 1997;185(11):2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann H, Mentel R, Bauer U, Pring-Akerblom P, Dolling R, Modrow S, Seidel W. Receptor binding sites and antigenic epitopes on the fiber knob of human adenovirus serotype 3. J. Virol. 1998;72(11):9121–9130. doi: 10.1128/jvi.72.11.9121-9130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Sato S, Patick A.K, Pease L.R, Roos R.P, Rodriguez M. Molecular characterization of a nondemyelinating variant of Daniel's strain of Theiler's virus isolated from a persistently infected glioma cell line. J. Virol. 1998;72(2):1262–1269. doi: 10.1128/jvi.72.2.1262-1269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.Y, Soda Y, Shimizu N, Haraguchi Y, Jinno A, Takeuchi Y, Hoshino H. CD4-dependent and CD4-independent utilization of coreceptors by human immunodeficiency viruses type 2 and simian immunodeficiency viruses. Virology. 2000;278(1):276–288. doi: 10.1006/viro.2000.0623. [DOI] [PubMed] [Google Scholar]
- Loetscher M, Amara A, Oberlin E, Brass N, Legler D, Loetscher P, D'Apuzzo M, Meese E, Rousset D, Virelizier J.L, Baggiolini M, Arenzana-Seisdedos F, Moser B. TYMSTR, a putative chemokine receptor selectively expressed in activated T cells, exhibits HIV-1 coreceptor function. Curr. Biol. 1997;7(9):652–660. doi: 10.1016/s0960-9822(06)00292-2. [DOI] [PubMed] [Google Scholar]
- Loffler S, Lottspeich F, Lanza F, Azorsa D.O, ter Meulen V, Schneider-Schaulies J. CD9, a tetraspan transmembrane protein, renders cells susceptible to canine distemper virus. J. Virol. 1997;71(1):42–49. doi: 10.1128/jvi.71.1.42-49.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan D, Abu-Ghazaleh R, Blakemore W, Curry S, Jackson T, King A, Lea S, Lewis R, Newman J, Parry N. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature. 1993;362(6420):566–568. doi: 10.1038/362566a0. [DOI] [PubMed] [Google Scholar]
- Lonberg-Holm K, Crowell R.L, Philipson L. Unrelated animal viruses share receptors. Nature. 1976;259(5545):679–681. doi: 10.1038/259679a0. [DOI] [PubMed] [Google Scholar]
- Londrigan S.L, Hewish M.J, Thomson M.J, Sanders G.M, Mustafa H, Coulson B.S. Growth of rotaviruses in continuous human and monkey cell lines that vary in their expression of integrins. J. Gen. Virol. 2000;81(Pt 9):2203–2213. doi: 10.1099/0022-1317-81-9-2203. [DOI] [PubMed] [Google Scholar]
- Lopez M, Cocchi F, Avitabile E, Leclerc A, Adelaide J, Campadelli-Fiume G, Dubreuil P. Novel, soluble isoform of the herpes simplex virus (HSV) receptor nectin1 (or PRR1-HIgR-HveC) modulates positively and negatively susceptibility to HSV infection. J. Virol. 2001;75(12):5684–5695. doi: 10.1128/JVI.75.12.5684-5691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M, Cocchi F, Menotti L, Avitabile E, Dubreuil P, Campadelli-Fiume G. Nectin2alpha (PRR2alpha or HveB) and nectin2delta are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 2000;74(3):1267–1274. doi: 10.1128/jvi.74.3.1267-1274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bueno A, Mateu M.G, Almendral J.M. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J. Virol. 2003;77:2701–2708. doi: 10.1128/JVI.77.4.2701-2708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lua L.H, Pedrini M.R, Reid S, Robertson A, Tribe D.E. Phenotypic and genotypic analysis of Helicoverpa armigera nucleopolyhedrovirus serially passaged in cell culture. J. Gen. Virol. 2002;83(Pt 4):945–955. doi: 10.1099/0022-1317-83-4-945. [DOI] [PubMed] [Google Scholar]
- Lusso P, Secchiero P, Crowley R.W, Garzino-Demo A, Berneman Z.N, Gallo R.C. CD4 is a critical component of the receptor for human herpesvirus 7: Interference with human immunodeficiency virus. Proc. Natl. Acad. Sci. USA. 1994;91(9):3872–3876. doi: 10.1073/pnas.91.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddon P.J, Dalgleish A.G, McDougal J.S, Clapham P.R, Weiss R.A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47(3):333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- Mahy B.W. Human viral infections: An expanding frontier. Antiviral Res. 1997;36(2):75–80. doi: 10.1016/s0166-3542(97)00042-9. [DOI] [PubMed] [Google Scholar]
- Markwell M.A, Moss J, Hom B.E, Fishman P.H, Svennerholm L. Expression of gangliosides as receptors at the cell surface controls infection of NCTC 2071 cells by Sendai virus. Virology. 1986;155(2):356–364. doi: 10.1016/0042-6822(86)90199-6. [DOI] [PubMed] [Google Scholar]
- Markwell M.A, Paulson J.C. Sendai virus utilizes specific sialyloligosaccharides as host cell receptor determinants. Proc. Natl. Acad. Sci. USA. 1980;77(10):5693–5697. doi: 10.1073/pnas.77.10.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell M.A, Portner A, Schwartz A.L. An alternative route of infection for viruses: Entry by means of the asialoglycoprotein receptor of a Sendai virus mutant lacking its attachment protein. Proc. Natl. Acad. Sci USA. 1985;82(4):978–982. doi: 10.1073/pnas.82.4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell M.A, Svennerholm L, Paulson J.C. Specific gangliosides function as host cell receptors for Sendai virus. Proc. Natl. Acad. Sci. USA. 1981;78(9):5406–5410. doi: 10.1073/pnas.78.9.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits T.C, Zechmeister T, Gruenberger M, Ronacher B, Schwihla H, Blaas D. Recombinant soluble low density lipoprotein receptor fragment inhibits minor group rhinovirus infection in vitro. FASEB J. 1998;12(9):695–703. doi: 10.1096/fasebj.12.9.695. [DOI] [PubMed] [Google Scholar]
- Martinez I, Melero J.A. Binding of human respiratory syncytial virus to cells: Implication of sulfated cell surface proteoglycans. J. Gen. Virol. 2000;81(Pt 11):2715–2722. doi: 10.1099/0022-1317-81-11-2715. [DOI] [PubMed] [Google Scholar]
- Martinez M.A, Verdaguer N, Mateu M.G, Domingo E. Evolution subverting essentiality: Dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc. Natl. Acad. Sci. USA. 1997;94(13):6798–6802. doi: 10.1073/pnas.94.13.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino T.A, Petric M, Brown M, Aitken K, Gauntt C.J, Richardson C.D, Chow L.H, Liu P.P. Cardiovirulent coxsackieviruses and the decay-accelerating factor (CD55) receptor. Virology. 1998;244(2):302–314. doi: 10.1006/viro.1998.9122. [DOI] [PubMed] [Google Scholar]
- Martino T.A, Petric M, Weingartl H, Bergelson J.M, Opavsky M.A, Richardson C.D, Modlin J.F, Finberg R.W, Kain K.C, Willis N, Gauntt C.J, Liu P.P. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B and by swine vesicular disease virus. Virology. 2000;271(1):99–108. doi: 10.1006/viro.2000.0324. [DOI] [PubMed] [Google Scholar]
- Masse N, Barrett T, Muller C.P, Wild T.F, Buckland R. Identification of a second major site for CD46 binding in the hemagglutinin protein from a laboratory strain of measles virus (MV): Potential consequences for wild-type MV infection. J. Virol. 2002;76(24):13034–13038. doi: 10.1128/JVI.76.24.13034-13038.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastromarino P, Conti C, Goldoni P, Hauttecoeur B, Orsi N. Characterization of membrane components of the erythrocyte involved in vesicular stomatitis virus attachment and fusion at acidic pH. J. Gen. Virol. 1987;68(Pt 9):2359–2369. doi: 10.1099/0022-1317-68-9-2359. [DOI] [PubMed] [Google Scholar]
- Masuda M, Hanson C.A, Alvord W.G, Hoffman P.M, Ruscetti S.K. Effects of subtle changes in the SU protein of ecotropic murine leukemia virus on its brain capillary endothelial cell tropism and interference properties. Virology. 1996;215(2):142–151. doi: 10.1006/viro.1996.0017. [DOI] [PubMed] [Google Scholar]
- Masuda M, Hanson C.A, Hoffman P.M, Ruscetti S.K. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J. Virol. 1996;70(12):8534–8539. doi: 10.1128/jvi.70.12.8534-8539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Remington M.P, Hoffman P.M, Ruscetti S.K. Molecular characterization of a neuropathogenic and nonerythroleukemogenic variant of Friend murine leukemia virus PVC-211. J. Virol. 1992;66(5):2798–2806. doi: 10.1128/jvi.66.5.2798-2806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu M.G. Antibody recognition of picornaviruses and escape from neutralization: A structural view. Virus Res. 1995;38(1):1–24. doi: 10.1016/0168-1702(95)00048-u. [DOI] [PubMed] [Google Scholar]
- Mathias P, Galleno M, Nemerow G.R. Interactions of soluble recombinant integrin alphav beta5 with human adenoviruses. J. Virol. 1998;72(11):8669–8675. doi: 10.1128/jvi.72.11.8669-8675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci M.R, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 2000;74(18):8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 1999;73(2):1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M.N, Gambaryan A.S, Teneberg S, Piskarev V.E, Yamnikova S.S, Lvov D.K, Robertson J.S, Karlsson K.A. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233(1):224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Sugawara K, Adachi K, Hongo S, Nishimura H, Kitame F, Nakamura K. Location of neutralizing epitopes on the hemagglutinin-esterase protein of influenza C virus. Virology. 1992;189(1):79–87. doi: 10.1016/0042-6822(92)90683-g. [DOI] [PubMed] [Google Scholar]
- Mauri D.N, Ebner R, Montgomery R.I, Kochel K.D, Cheung T.C, Yu G.L, Ruben S, Murphy M, Eisenberg R.J, Cohen G.H, Spear P.G, Ware C.F. LIGHT, a new member of the TNF superfamily, and lymphotoxin alpha are ligands for herpesvirus entry mediator. Immunity. 1998;8(1):21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- McClure M.A. The retroid agents: Disease, function and evolution. In: Domingo E, Webster R.G, Holland J.J, editors. “Origin and Evolution of Viruses”. Academic Press; San Diego: 1999. pp. 163–195. [Google Scholar]
- McCright I.J, Tsunoda I, Whitby F.G, Fujinami R.S. Theiler's viruses with mutations in loop I of VP1 lead to altered tropism and pathogenesis. J. Virol. 1999;73(4):2814–2824. doi: 10.1128/jvi.73.4.2814-2824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott B.M, Jr., Rux A.H, Eisenberg R.J, Cohen G.H, Racaniello V.R. Two distinct binding affinities of poliovirus for its cellular receptor. J. Biol. Chem. 2000;275(30):23089–23096. doi: 10.1074/jbc.M002146200. [DOI] [PubMed] [Google Scholar]
- McDougal J.S, Kennedy M.S, Sligh J.M, Cort S.P, Mawle A, Nicholson J.K. Binding of HTLV-III⧸LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science. 1986;231(4736):382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- McGeoch D.J, Davison A.J. The molecular evolutionary history of the herpesviruses. In: Domingo E, Webster R.G, Holland J.J, editors. “Origin and Evolution of Viruses”. Academic Press; San Diego: 1999. pp. 441–465. [Google Scholar]
- McLaren L.C, Holland J.J, Syverton J.T. The mammalian cell-virus relationship. I. Attachment of poliovirus to cultivated cells of primate and nonprimate origin. J. Exp. Med. 1959;109:475–485. doi: 10.1084/jem.109.5.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan N.A, Payne E, Frazer I.H, Evander M. Expression of the alpha6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virology. 1999;261(2):271–279. doi: 10.1006/viro.1999.9825. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C.L, Wimmer E, Racaniello V.R. Cellular receptor for poliovirus: Molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56(5):855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- Mendez E, Arias C.F, Lopez S. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J. Virol. 1993;67(9):5253–5259. doi: 10.1128/jvi.67.9.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez E, Lopez S, Cuadras M.A, Romero P, Arias C.F. Entry of rotaviruses is a multistep process. Virology. 1999;263(2):450–459. doi: 10.1006/viro.1999.9976. [DOI] [PubMed] [Google Scholar]
- Menéndez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Progr. Nucleic Acid Res. Mol. Biol. 2002;71:91–147. doi: 10.1016/s0079-6603(02)71042-8. [DOI] [PubMed] [Google Scholar]
- Meng G, Wei X, Wu X, Sellers M.T, Decker J.M, Moldoveanu Z, Orenstein J.M, Graham M.F, Kappes J.C, Mestecky J, Shaw G.M, Smith P.D. Primary intestinal epithelial cells selectively transfer R5 HIV-1 to CCR5+ cells. Nature Med. 2002;8(2):150–156. doi: 10.1038/nm0202-150. [DOI] [PubMed] [Google Scholar]
- Menotti L, Lopez M, Avitabile E, Stefan A, Cocchi F, Adelaide J, Lecocq E, Dubreuil P, Campadelli-Fiume G. The murine homolog of human Nectin 1 delta serves as a species nonspecific mediator for entry of human and animal alpha herpesviruses in a pathway independent of a detectable binding to gD. Proc. Natl. Acad. Sci. USA. 2000;97(9):4867–4872. doi: 10.1073/pnas.97.9.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T.C, Zsak L, Zuckermann F, Sugg N, Kern H, Ben-Porat T. Interaction of glycoprotein gIII with a cellular heparinlike substance mediates adsorption of pseudorabies virus. J. Virol. 1990;64(1):278–286. doi: 10.1128/jvi.64.1.278-286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.D. Cell-surface receptors for retroviruses and implications for gene transfer. Proc. Natl. Acad. Sci. USA. 1996;93(21):11407–11413. doi: 10.1073/pnas.93.21.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.D. Identification of Hya12 as the cell-surface receptor for jaagsiekte sheep retrovirus and ovine nasal adenocarcinoma virus. Curr Top Microbiol Immunol. 2003;275:179–199. doi: 10.1007/978-3-642-55638-8_7. [DOI] [PubMed] [Google Scholar]
- Miller D.G, Edwards R.H, Miller A.D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA. 1994;91(1):78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.G, Miller A.D. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 1994;68(12):8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne R.S, Connolly S.A, Krummenacher C, Eisenberg R.J, Cohen G.H. Porcine HveC, a member of the highly conserved HveC⧸nectin 1 family, is a functional alphaherpesvirus receptor. Virology. 2001;281(2):315–328. doi: 10.1006/viro.2000.0798. [DOI] [PubMed] [Google Scholar]
- Mims C, Nash A, Stephen J. “Mims' Pathogenesis of Infectious Disease.”. Academic Press; San Diego: 2001. [Google Scholar]
- Minocha H.C, Xue W, Reddy J.R. A 50 kDa membrane protein from bovine kidney cells is a putative receptor for bovine viral diarrhea virus (BVDV) Adv. Exp. Med. Biol. 1997;412:145–148. doi: 10.1007/978-1-4899-1828-4_22. [DOI] [PubMed] [Google Scholar]
- Monazahian M, Bohme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 1999;57(3):223–229. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Montgomery R.I, Warner M.S, Lum B.J, Spear P.G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF⧸NGF receptor family. Cell. 1996;87(3):427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- Morner A, Bjorndal A, Albert J, Kewalramani V.N, Littman D.R, Inoue R, Thorstensson R, Fenyo E.M, Bjorling E. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol. 1999;73(3):2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S.S, editor. Oxford Univ. Press; Oxford: 1993. (“Emerging Viruses.”). [Google Scholar]
- Morse S.S, editor. Raven Press; New York: 1994. (“The Evolutionary Biology of Viruses.”). [Google Scholar]
- Mosier D.E, Picchio G.R, Gulizia R.J, Sabbe R, Poignard P, Picard L, Offord R.E, Thompson D.A, Wilken J. Highly potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J. Virol. 1999;73(5):3544–3550. doi: 10.1128/jvi.73.5.3544-3550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D.W. “Bioinformatics. Sequence and Genome Analysis.”. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Murphy F.A, Nathanson N. The emergence of new viral diseases: An overview. Semin. Virol. 1994;5:87–102. [Google Scholar]
- Murray M.G, Bradley J, Yang X.F, Wimmer E, Moss E.G, Racamello V.R. Poliovirus host range is determined by a short amino acid sequence in neutralization antigenic site I. Science. 1988;241(4862):213–215. doi: 10.1126/science.2838906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeve C.W, Hinshaw V.S, Webster R.G. Mutations in the hemagglutinin receptor-binding site can change the biological properties of an influenza virus. J. Virol. 1984;51(2):567–569. doi: 10.1128/jvi.51.2.567-569.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naniche D, Varior-Krishnan G, Cervoni F, Wild T.F, Rossi B, Rabourdin-Combe C, Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 1993;67(10):6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson N, Ahmed R, Gonzalez-Scarano F, Griffin D.E, Holmes K.V, Murphy F.A, Robinson H.L, editors. Lippincott-Raven; Philadelphia: 1997. (“Viral Pathogenesis.”). [Google Scholar]
- Nedellec P, Dveksler G.S, Daniels E, Turbide C, Chow B, Basile A.A, Holmes K.V, Beauchemin N. Bgp2, a new member of the carcinoembryonic antigen-related gene family, encodes an alternative receptor for mouse hepatitis viruses. J. Virol. 1994;68(7):4525–4537. doi: 10.1128/jvi.68.7.4525-4537.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff S, Sa-Carvalho D, Rieder E, Mason P.W, Blystone S.D, Brown E.J, Baxt B. Foot-and-mouth disease virus virulent for cattle utilizes the integrin alpha(v)beta3 as its receptor. J. Virol. 1998;72(5):3587–3594. doi: 10.1128/jvi.72.5.3587-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen-Salz B, Eggers H.J, Zimmermann H. Integrin alpha(v)beta3 (vitronectin receptor) is a candidate receptor for the virulent echovirus 9 strain Barty. J. Gen. Virol. 1999;80(Pt 9):2311–2313. doi: 10.1099/0022-1317-80-9-2311. [DOI] [PubMed] [Google Scholar]
- Nemerow G.R. Cell receptors involved in adenovirus entry. Virology. 2000;274(1):1–4. doi: 10.1006/viro.2000.0468. [DOI] [PubMed] [Google Scholar]
- Nemerow G.R, Stewart P.L. Role of alpha(v) integrins in adenovirus cell entry and gene delivery. Microbiol. Mol. Biol. Rev. 1999;63(3):725–734. doi: 10.1128/mmbr.63.3.725-734.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemerow G.R, Wolfert R, McNaughton M.E, Cooper N.R. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2) J. Virol. 1985;55(2):347–351. doi: 10.1128/jvi.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath A.R, Kent S.B, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46(3):429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- Niewiesk S, Schneider-Schaulies J, Ohnimus H, Jassoy C, Schneider-Schaulies S, Diamond L, Logan J.S, ter Meulen V. CD46 expression does not overcome the intracellular block of measles virus replication in transgenic rats. J. Virol. 1997;71(10):7969–7973. doi: 10.1128/jvi.71.10.7969-7973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhuis M, Schuurman R, de Jong D, Erickson J, Gustchina E, Albert J, Schipper P, Gulnik S, Boucher C.A. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. Aids. 1999;13(17):2349–2359. doi: 10.1097/00002030-199912030-00006. [DOI] [PubMed] [Google Scholar]
- Nixdorf R, Schmidt J, Karger A, Mettenleiter T.C. Infection of Chinese hamster ovary cells by pseudorabies virus. J. Virol. 1999;73(10):8019–8026. doi: 10.1128/jvi.73.10.8019-8026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohinek B, Gerhard W, Schulze I.T. Characterization of host cell binding variants of influenza virus by monoclonal antibodies. Virology. 1985;143(2):651–656. doi: 10.1016/0042-6822(85)90407-6. [DOI] [PubMed] [Google Scholar]
- Nomoto A, Arita I. Eradication of poliomyelitis. Nature Immunol. 2002;3(3):205–208. doi: 10.1038/ni0302-205. [DOI] [PubMed] [Google Scholar]
- Nomoto A, Koike S, Aoki J. Tissue tropism and species specificity of poliovirus infection. Trends Microbiol. 1994;2(2):47–51. doi: 10.1016/0966-842x(94)90125-2. [DOI] [PubMed] [Google Scholar]
- Nowak M.A. What is a quasispecies? Trends Ecol. Evol. 1992;4:118–121. doi: 10.1016/0169-5347(92)90145-2. [DOI] [PubMed] [Google Scholar]
- Núñez J.I, Baranowski E, Molina N, Ruiz-Jarabo C.M, Sánchez C, Domingo E, Sobrino F. A single amino acid substitution in nonstructural protein 3A can mediate adaptation of foot-and-mouth disease virus to the guinea pig. J. Virol. 2001;75(8):3977–3983. doi: 10.1128/JVI.75.8.3977-3983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara B, Johann S.V, Klinger H.P, Blair D.G, Rubinson H, Dunn K.J, Sass P, Vitek S.M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1(3):119–127. [PubMed] [Google Scholar]
- Okazaki K, Matsuzaki T, Sugahara Y, Okada J, Hasebe M, Iwamura Y, Ohnishi M, Kanno T, Shimizu M, Honda E. BHV-1 adsorption is mediated by the interaction of glycoprotein gIII with heparinlike moiety on the cell surface. Virology. 1991;181(2):666–670. doi: 10.1016/0042-6822(91)90900-v. [DOI] [PubMed] [Google Scholar]
- Oldstone M.B, Homann D, Lewicki H, Stevenson D. One, two, or three step: Measles virus receptor dance. Virology. 2002;299(2):162–163. doi: 10.1006/viro.2002.1507. [DOI] [PubMed] [Google Scholar]
- Oldstone M.B, Lewicki H, Thomas D, Tishon A, Dales S, Patterson J, Manchester M, Homann D, Naniche D, Holz A. Measles virus infection in a transgenic model: Virus-induced immunosuppression and central nervous system disease. Cell. 1999;98(5):629–640. doi: 10.1016/s0092-8674(00)80050-1. [DOI] [PubMed] [Google Scholar]
- Olson N.H, Kolatkar P.R, Oliveira M.A, Cheng R.H, Greve J.M, McClelland A, Baker T.S, Rossmann M.G. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA. 1993;90(2):507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen S.M, Ellenberger D, Rayfield M, Wiktor S, Michel P, Grieco M.H, Gao F, Hahn B.H, Lal R.B. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J. Virol. 1998;72(7):5425–5432. doi: 10.1128/jvi.72.7.5425-5432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owman C, Garzino-Demo A, Cocchi F, Popovic M, Sabirsh A, Gallo R.C. The leukotriene B4 receptor functions as a novel type of coreceptor mediating entry of primary HIV-1 isolates into CD4-positive cells. Proc. Natl. Acad. Sci. USA. 1998;95(16):9530–9534. doi: 10.1073/pnas.95.16.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.D.M, Holmes E.C. “Molecular Evolution: A Phylogenetic Approach.”. Blackwell; Science, Oxford: 1998. [Google Scholar]
- Parker J.S, Murphy W.J, Wang D, O'Brien S.J, Parrish C.R. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 2001;75(8):3896–3902. doi: 10.1128/JVI.75.8.3896-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes R.J, Hart S.L. Adhesion molecules and gene transfer. Adv. Drug Deliv. Rev. 2000;44(2–3):135–152. doi: 10.1016/s0169-409x(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Parrish C.R, Truyen U. Parvovirus variation and evolution, In: Domingo E, Webster R.G, Holland J.J, editors. “Origin and Evolution of Viruses”. Academic Press; San Diego: 1999. pp. 421–439. [Google Scholar]
- Patel M, Yanagishita M, Roderiquez G, Bou-Habib D.C, Oravecz T, Hascall V.C, Norcross M.A. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res. Hum. Retrovir. 1993;9(2):167–174. doi: 10.1089/aid.1993.9.167. [DOI] [PubMed] [Google Scholar]
- Paul R.W, Choi A.H, Lee P.W. The alpha-anomeric form of sialic acid is the minimal receptor determinant recognized by reovirus. Virology. 1989;172(1):382–385. doi: 10.1016/0042-6822(89)90146-3. [DOI] [PubMed] [Google Scholar]
- Paul R.W, Lee P.W. Glycophorin is the reovirus receptor on human erythrocytes. Virology. 1987;159(1):94–101. doi: 10.1016/0042-6822(87)90351-5. [DOI] [PubMed] [Google Scholar]
- Paulson J.C, Sadler J.E, Hill R.L. Restoration of specific myxovirus receptors to asialoerythrocytes by incorporation of sialic acid with pure sialyltransferases. J. Biol. Chem. 1979;254(6):2120–2124. [PubMed] [Google Scholar]
- Paxton W.A, Martin S.R, Tse D, O'Brien T.R, Skurnick J, VanDevanter N.L, Padian N, Braun J.F, Kotler D.P, Wolinsky S.M, Koup R.A. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nature Med. 1996;2(4):412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- Petit M.A, Capel F, Dubanchet S, Mabit H. PreS1-specific binding proteins as potential receptors for hepatitis B virus in human hepatocytes. Virology. 1992;187(1):211–222. doi: 10.1016/0042-6822(92)90309-d. [DOI] [PubMed] [Google Scholar]
- Pfeifer A, Verma I.M. Virus vectors and their applications. In: Knipe D.M, Howley P.M, editors. “Fields Virology”. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 469–491. [Google Scholar]
- Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A.J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282(5390):938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276(5320):1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- Poeschla E, Gilbert J, Li X, Huang S, Ho A, Wong-Staal F. Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. J. Virol. 1998;72(8):6527–6536. doi: 10.1128/jvi.72.8.6527-6536.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell R.M, Ward T, Evans D.J, Almond J.W. Interaction between echovirus 7 and its receptor, decay-accelerating factor (CD55): Evidence for a secondary cellular factor in A-particle formation. J. Virol. 1997;71(12):9306–9312. doi: 10.1128/jvi.71.12.9306-9312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B.V, Burns J.W, Marietta E, Estes M.K, Chiu W. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature. 1990;343(6257):476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- Prasad B.V, Wang G.J, Clerx J.P, Chiu W. Three-dimensional structure of rotavirus. J. Mol. Biol. 1988;199(2):269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. “A Genetic Swith: Phage λ and Higher Organisms.”. Cell Press and Blackwell Scientific; Cambridge, MA: 1992. [Google Scholar]
- Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nature Med. 1999;5(1):71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- Qiu J, Handa A, Kirby M, Brown K.E. The interaction of heparin sulfate and adeno-associated virus 2. Virology. 2000;269(1):137–147. doi: 10.1006/viro.2000.0205. [DOI] [PubMed] [Google Scholar]
- Racaniello V.R. Early events in poliovirus infection: Virus-receptor interactions. Proc. Natl. Acad. Sci. USA. 1996;93(21):11378–11381. doi: 10.1073/pnas.93.21.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai S.K, Duh F.M, Vigdorovich V, Danilkovitch-Miagkova A, Lerman M.I, Miller A.D. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA. 2001;98(8):4443–4448. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko J.E, Battini J.L, Gottschalk R.J, Mazo I, Miller A.D. The RD114⧸simian type D retrovirus receptor is a neutral amino acid transporter. Proc. Natl. Acad. Sci. USA. 1999;96(5):2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading P.C, Miller J.L, Anders E.M. Involvement of the mannose receptor in infection of macrophages by influenza virus. J. Virol. 2000;74(11):5190–5197. doi: 10.1128/jvi.74.11.5190-5197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi H.V, Lipton H.L. Heparan sulfate mediates infection of high-neurovirulence Theiler's viruses. J. Virol. 2002;76(16):8400–8407. doi: 10.1128/JVI.76.16.8400-8407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves J.D, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira J.M, Moniz-Pereira J, Clapham P.R. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: Comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J. Virol. 1999;73(9):7795–7804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl A, Reithmayer M, Winsauer G, Moser R, Gosler I, Blaas D. Viral evolution toward change in receptor usage: Adaptation of a major group human rhinovirus to grow in ICAM-1-negative cells. J. Virol. 2001;75(19):9312–9319. doi: 10.1128/JVI.75.19.9312-9319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert P.S, Spiro R.C, Townsend P.L, Stanford S.A, Sairenji T, Humphreys R.E. Functional association of class II antigens with cell surface binding of Epstein-Barr virus. J. Immunol. 1985;134(6):3776–3780. [PubMed] [Google Scholar]
- Ren R, Racaniello V.R. Human poliovirus receptor gene expression and poliovirus tissue tropism in transgenic mice. J. Virol. 1992;66(1):296–304. doi: 10.1128/jvi.66.1.296-304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R.B, Costantini F, Gorgacz E.J, Lee J.J, Racaniello V.R. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell. 1990;63(2):353–362. doi: 10.1016/0092-8674(90)90168-e. [DOI] [PubMed] [Google Scholar]
- Richardson J, Pancino G, Merat R, Leste-Lasserre T, Moraillon A, Schneider-Mergener J, Alizon M, Sonigo P, Heveker N. Shared usage of the chemokine receptor CXCR4 by primary and laboratory-adapted strains of feline immunodeficiency virus. J. Virol. 1999;73(5):3661–3671. doi: 10.1128/jvi.73.5.3661-3671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J.S, Bootman J.S, Newman R, Oxford J.S, Daniels R.S, Webster R.G, Schild G.C. Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology. 1987;160(1):31–37. doi: 10.1016/0042-6822(87)90040-7. [DOI] [PubMed] [Google Scholar]
- Roccasecca R, Ansuini H, Vitelli A, Meola A, Scarselli E, Acali S, Pezzanera M, Ercole B.B, McKeating J, Yagnik A, Lahm A, Tramontano A, Cortese R, Nicosia A. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 2003;77(3):1856–1867. doi: 10.1128/JVI.77.3.1856-1867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderiquez G, Oravecz T, Yanagishita M, Bou-Habib D.C, Mostowski H, Norcross M.A. Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J. Virol. 1995;69(4):2233–2239. doi: 10.1128/jvi.69.4.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelvink P.W, Lizonova A, Lee J.G, Li Y, Bergelson J.M, Finberg R.W, Brough D.E, Kovesdi I, Wickham T.J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 1998;72(10):7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelvink P.W, Mi Lee G, Einfeld D.A, Kovesdi I, Wickham T.J. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science. 1999;286(5444):1568–1571. doi: 10.1126/science.286.5444.1568. [DOI] [PubMed] [Google Scholar]
- Rogers G.N, Paulson J.C. Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127(2):361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Rogers G.N, Paulson J.C, Daniels R.S, Skehel J.J, Wilson I.A, Wiley D.C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- Rogers G.N, Pritchett T.J, Lane J.L, Paulson J.C. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology. 1983;131(2):394–408. doi: 10.1016/0042-6822(83)90507-x. [DOI] [PubMed] [Google Scholar]
- Roivainen M, Hyypia T, Piirainen L, Kalkkinen N, Stanway G, Hovi T. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J. Virol. 1991;65(9):4735–4740. doi: 10.1128/jvi.65.9.4735-4740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roivainen M, Piirainen L, Hovi T, Virtanen I, Riikonen T, Heino J, Hyypia T. Entry of coxsackievirus A9 into host cells: Specific interactions with alpha v beta 3 integrin, the vitronectin receptor. Virology. 1994;203(2):357–365. doi: 10.1006/viro.1994.1494. [DOI] [PubMed] [Google Scholar]
- Rolsma M.D, Gelberg H.B, Kuhlenschmidt M.S. Assay for evaluation of rotavirus-cell interactions: Identification of an enterocyte ganglioside fraction that mediates group A porcine rotavirus recognition. J. Virol. 1994;68(1):258–268. doi: 10.1128/jvi.68.1.258-268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolsma M.D, Kuhlenschmidt T.B, Gelberg H.B, Kuhlenschmidt M.S. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 1998;72(11):9079–9091. doi: 10.1128/jvi.72.11.9079-9091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal P.B, Zhang X, Formanowski F, Fitz W, Wong C.H, Meier-Ewert H, Skehel J.J, Wiley D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396(6706):92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S.R, Schofield J.J, Farr C.J, Bucan M. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA. 2002;99(19):12386–12390. doi: 10.1073/pnas.192360099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M.G, Arnold E, Erickson J.W, Frankenberger E.A, Griffith J.P, Hecht H.J, Johnson J.E, Kamer G, Luo M, Mosser A.G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317(6033):145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G, Bella J, Kolatkar P.R, He Y, Wimmer E, Kuhn R.J, Baker T.S. Cell recognition and entry by rhino- and enteroviruses. Virology. 2000;269(2):239–247. doi: 10.1006/viro.2000.0258. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G, He Y, Kuhn J. Picornavirus–receptor interactions. Trends Microbiol. 2002;10:324–331. doi: 10.1016/s0966-842x(02)02383-1. [DOI] [PubMed] [Google Scholar]
- Rucker J, Edinger A.L, Sharron M, Samson M, Lee B, Berson J.F, Yi Y, Margulies B, Collman R.G, Doranz B.J, Parmentier M, Doms R.W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J. Virol. 1997;71(12):8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruíz-Jarabo C.M, Pariente N, Baranowski E, Domingo E. Manuscript in preparation. 2003. [Google Scholar]
- Ruiz-Jarabo C.M, Sevilla N, Davila M, Gomez-Mariano G, Baranowski E, Domingo E. Antigenic properties and population stability of a foot-and-mouth disease virus with an altered Arg-Gly-Asp receptor-recognition motif. J. Gen. Virol. 1999;80(Pt 8):1899–1909. doi: 10.1099/0022-1317-80-8-1899. [DOI] [PubMed] [Google Scholar]
- Sa-Carvalho D, Rieder E, Baxt B, Rodarte R, Tanuri A, Mason P.W. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J. Virol. 1997;71(7):5115–5123. doi: 10.1128/jvi.71.7.5115-5123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Zhang J, Gupta A, Dave R, Yimen M, Zerhouni B. Isolation of primary HIV-1 that target CD8+ T lymphocytes using CD8 as a receptor. Nature Med. 2001;7(1):65–72. doi: 10.1038/83365. [DOI] [PubMed] [Google Scholar]
- Samuel A.R, Knowles N.J. Foot-and-mouth disease virus: Cause of the recent crisis for the UK livestock industry. Trends Genet. 2001;17(8):421–424. doi: 10.1016/s0168-9525(01)02374-5. [DOI] [PubMed] [Google Scholar]
- Santoro F, Kennedy P.E, Locatelli G, Malnati M.S, Berger E.A, Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell. 1999;99(7):817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- Saphire A.C, Bobardt M.D, Zhang Z, David G, Gallay P.A. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J. Virol. 2001;75(19):9187–9200. doi: 10.1128/JVI.75.19.9187-9200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Zhang L, Kim J, Jakob J, Grant R.A, Wollmann R, Roos R.P. A neutralization site of DA strain of Theiler's murine encephalomyelitis virus important for disease phenotype. Virology. 1996;226(2):327–337. doi: 10.1006/viro.1996.0660. [DOI] [PubMed] [Google Scholar]
- Scarselli E, Ansuini H, Cerino R, Roccasecca R.M, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. Embo. J. 2002;21(19):5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel R, Tralka T.S, Willingham M.C, Pastan I. Inhibition of VSV binding and infectivity by phosphatidylserine: Is phosphatidylserine a VSV-binding site? Cell. 1983;32(2):639–646. doi: 10.1016/0092-8674(83)90483-x. [DOI] [PubMed] [Google Scholar]
- Schmidt J, Gerdts V, Beyer J, Klupp B.G, Mettenleiter T.C. Glycoprotein D-independent infectivity of pseudorabies virus results in an alteration of in vivo host range and correlates with mutations in glycoproteins B and H. J. Virol. 2001;75(21):10054–10064. doi: 10.1128/JVI.75.21.10054-10064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J, Klupp B.G, Karger A, Mettenleiter T.C. Adaptability in herpesviruses: Glycoprotein D-independent infectivity of pseudorabies virus. J. Virol. 1997;71(1):17–24. doi: 10.1128/jvi.71.1.17-24.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Schaulies J. Cellular receptors for viruses: Links to tropism and pathogenesis. J. Gen. Virol. 2000;81(Pt 6):1413–1429. doi: 10.1099/0022-1317-81-6-1413. [DOI] [PubMed] [Google Scholar]
- Schneider-Schaulies J, Dunster L.M, Schwartz-Albiez R, Krohne G, ter Meulen V. Physical association of moesin and CD46 as a receptor complex for measles virus. J. Virol. 1995;69(4):2248–2256. doi: 10.1128/jvi.69.4.2248-2256.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag S.J, Rota P.A, Bellini W.J. Spontaneous mutation rate of measles virus: Direct estimation based on mutations conferring monoclonal antibody resistance. J. Virol. 1999;73(1):51–54. doi: 10.1128/jvi.73.1.51-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B, Enjuanes L, Herrler G. Analysis of the sialic acid-binding activity of the transmissible gastroenteritis virus. Adv. Exp. Med. Biol. 1995;380:367–370. doi: 10.1007/978-1-4615-1899-0_59. [DOI] [PubMed] [Google Scholar]
- Schultze B, Gross H.J, Brossmer R, Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J. Virol. 1991;65(11):6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B, Herrler G. Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J. Gen. Virol. 1992;73(Pt 4):901–906. doi: 10.1099/0022-1317-73-4-901. [DOI] [PubMed] [Google Scholar]
- Schultze B, Krempl C, Ballesteros M.L, Shaw L, Schauer R, Enjuanes L, Herrler G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 1996;70(8):5634–5637. doi: 10.1128/jvi.70.8.5634-5637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchiero P, Sun D, De Vico A.L, Crowley R.W, Reitz M.S, Jr., Zauli G, Lusso P, Gallo R.C. Role of the extracellular domain of human herpesvirus 7 glycoprotein B in virus binding to cell surface heparan sulfate proteoglycans. J. Virol. 1997;71(6):4571–4580. doi: 10.1128/jvi.71.6.4571-4580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet B.T, Johnston J.B, Brunetti C.R, Barrett J.W, Everett H, Cameron C, Sypula J, Nazarian S.H, Lucas A, McFadden G. Poxviruses and immune evasion. Annu. Rev. Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- Semler B, Wimmer E, editors. ASM Press; 2002. (“Molecular Biology of Picornaviruses.”). [Google Scholar]
- Sevilla N, Domingo E, de la Torre J.C. Contribution of LCMV towards deciphering biology of quasispecies in vivo. Curr. Top. Microbiol. Immunol. 2002;263:197–220. doi: 10.1007/978-3-642-56055-2_10. [DOI] [PubMed] [Google Scholar]
- Sevilla N, Kunz S, Holz A, Lewicki H, Homann D, Yamada H, Campbell K.P, de La Torre J.C, Oldstone M.B. Immunosuppression and resultant viral persistence by specific viral targeting of dendritic cells. J. Exp. Med. 2000;192(9):1249–1260. doi: 10.1084/jem.192.9.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla N, Ruiz-Jarabo C.M, Gómez-Mariano G, Baranowski E, Domingo E. An RNA virus can adapt to the multiplicity of infection. J. Gen. Virol. 1998;79:2971–2980. doi: 10.1099/0022-1317-79-12-2971. [DOI] [PubMed] [Google Scholar]
- Shafren D.R, Bates R.C, Agrez M.V, Herd R.L, Burns G.F, Barry R.D. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J. Virol. 1995;69(6):3873–3877. doi: 10.1128/jvi.69.6.3873-3877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafren D.R, Dorahy D.J, Greive S.J, Burns G.F, Barry R.D. Mouse cells expressing human intercellular adhesion molecule-1 are susceptible to infection by coxsackievirus A21. J. Virol. 1997;71(1):785–789. doi: 10.1128/jvi.71.1.785-789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafren D.R, Dorahy D.J, Ingham R.A, Burns G.F, Barry R.D. Coxsackievirus A21 binds to decay-accelerating factor but requires intercellular adhesion molecule 1 for cell entry. J. Virol. 1997;71(6):4736–4743. doi: 10.1128/jvi.71.6.4736-4743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafren D.R, Williams D.T, Barry R.D. A decay-accelerating factor-binding strain of coxsackievirus B3 requires the coxsackievirus-adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J. Virol. 1997;71(12):9844–9848. doi: 10.1128/jvi.71.12.9844-9848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh M.T, WuDunn D, Montgomery R.I, Esko J.D, Spear P.G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell Biol. 1992;116(5):1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Soda Y, Kanbe K, Liu H.Y, Jinno A, Kitamura T, Hoshino H. An orphan G protein-coupled receptor, GPR1, acts as a coreceptor to allow replication of human immunodeficiency virus types 1 and 2 in brain-derived cells. J. Virol. 1999;73(6):5231–5239. doi: 10.1128/jvi.73.6.5231-5239.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D, Liu J, Blaiklock P, Shworak N.W, Bai X, Esko J.D, Cohen G.H, Eisenberg R.J, Rosenberg R.D, Spear P.G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99(1):13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- Shukla D, Rowe C.L, Dong Y, Racaniello V.R, Spear P.G. The murine homolog (Mph) of human herpesvirus entry protein B (HveB) mediates entry of pseudorabies virus but not herpes simplex virus types 1 and 2. J. Virol. 1999;73(5):4493–4497. doi: 10.1128/jvi.73.5.4493-4497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D, Spear P.G. Herpesviruses and heparan sulfate: An intimate relationship in aid of viral entry. J. Clin. Invest. 2001;108(4):503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvie O, Rubinstein E, Franetich J.F, Prenant M, Belnoue E, Renia L, Hannoun L, Eling W, Levy S, Boucheix C, Mazier D. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nature Med. 2003;9(1):93–96. doi: 10.1038/nm808. [DOI] [PubMed] [Google Scholar]
- Skehel J.J, Wiley D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Skrincosky D, Hocknell P, Whetter L, Secchiero P, Chandran B, Dewhurst S. Identification and analysis of a novel heparin-binding glycoprotein encoded by human herpesvirus 7. J. Virol. 2000;74(10):4530–4540. doi: 10.1128/jvi.74.10.4530-4540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smelt S.C, Borrow P, Kunz S, Cao W, Tishon A, Lewicki H, Campbell K.P, Oldstone M.B. Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor alpha-dystroglycan correlate with viral tropism and disease kinetics. J. Virol. 2001;75(1):448–457. doi: 10.1128/JVI.75.1.448-457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.B, Inglis S.C. The mutation rate and variability of eukaryotic viruses: An analytical review. J. Gen. Virol. 1987;68(Pt 11):2729–2740. doi: 10.1099/0022-1317-68-11-2729. [DOI] [PubMed] [Google Scholar]
- Smith T.J, Chase E.S, Schmidt T.J, Olson N.H, Baker T.S. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature. 1996;383(6598):350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.J, Cheng R.H, Olson N.H, Peterson P, Chase E, Kuhn R.J, Baker T.S. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA. 1995;92(23):10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrino F, Dávila M, Ortín J, Domingo E. Multiple genetic variants arise in the course of replication of foot-and-mouth disease virus in cell culture. Virology. 1983;128:310–318. doi: 10.1016/0042-6822(83)90258-1. [DOI] [PubMed] [Google Scholar]
- Sobrino F, Domingo E. Foot-and-mouth disease in Europe. EMBO Rep. 2001;2(6):459–461. doi: 10.1093/embo-reports/kve122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg C, Giugni T.D, Zaia J.A, Larsson S, Wahlberg J.M, Moller E. CD13 (human aminopeptidase N) mediates human cytomegalovirus infection. J. Virol. 1993;67(11):6576–6585. doi: 10.1128/jvi.67.11.6576-6585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerfelt M.A. Retrovirus receptors. J. Gen. Virol. 1999;80(Pt 12):3049–3064. doi: 10.1099/0022-1317-80-12-3049. [DOI] [PubMed] [Google Scholar]
- Soudais C, Boutin S, Hong S.S, Chillon M, Danos O, Bergelson J.M, Boulanger P, Kremer E.J. Canine adenovirus type 2 attachment and internalization: coxsackievirus-adenovirus receptor, alternative receptors, and an RGD-independent pathway. J. Virol. 2000;74(22):10639–10649. doi: 10.1128/jvi.74.22.10639-10649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear P.G, Eisenberg R.J, Cohen G.H. Three classes of cell surface receptors for alphaherpesvirus entry. Virology. 2000;275(1):1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- Speck P, Haan K.M, Longnecker R. Epstein-Barr virus entry into cells. Virology. 2000;277:1–5. doi: 10.1006/viro.2000.0624. [DOI] [PubMed] [Google Scholar]
- Speck R.F, Esser U, Penn M.L, Eckstein D.A, Pulliam L, Chan S.Y, Goldsmith M.A. A trans-receptor mechanism for infection of CD4-negative cells by human immunodeficiency virus type 1. Curr. Biol. 1999;9(10):547–550. doi: 10.1016/s0960-9822(99)80241-3. [DOI] [PubMed] [Google Scholar]
- Spiropoulou C.F, Kunz S, Rollin P.E, Campbell K.P, Oldstone M.B. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J. Virol. 2002;76(10):5140–5146. doi: 10.1128/JVI.76.10.5140-5146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srnka C.A, Tiemeyer M, Gilbert J.H, Moreland M, Schweingruber H, de Lappe B.W, James P.G, Gant T, Willoughby R.E, Yolken R.H. Cell surface ligands for rotavirus: Mouse intestinal glycolipids and synthetic carbohydrate analogs. Virology. 1992;190(2):794–805. doi: 10.1016/0042-6822(92)90917-e. [DOI] [PubMed] [Google Scholar]
- Staunton D.E, Merluzzi V.J, Rothlein R, Barton R, Marlin S.D, Springer T.A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56(5):849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- Stehle T, Harrison S.C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4(2):183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- Stehle T, Yan Y, Benjamin T.L, Harrison S.C. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369(6476):160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- Steinhauer D.A, Domingo E, Holland J.J. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122(2):281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- Stewart P.L, Chiu C.Y, Huang S, Muir T, Zhao Y, Chait B, Mathias P, Nemerow G.R. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 1997;16(6):1189–1198. doi: 10.1093/emboj/16.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P.L, Nemerow G.R. Recent structural solutions for antibody neutralization of viruses. Trends Microbiol. 1997;5(6):229–233. doi: 10.1016/S0966-842X(97)01049-4. [DOI] [PubMed] [Google Scholar]
- Strauss J.H, Wang K.S, Schmaljohn A.L, Kuhn R.J, Strauss E.G. Host-cell receptors for Sindbis virus. Arch. Virol. Suppl. 1994;9:473–484. doi: 10.1007/978-3-7091-9326-6_46. [DOI] [PubMed] [Google Scholar]
- Strong J.E, Tang D, Lee P.W. Evidence that the epidermal growth factor receptor on host cells confers reovirus infection efficiency. Virology. 1993;197(1):405–411. doi: 10.1006/viro.1993.1602. [DOI] [PubMed] [Google Scholar]
- Sugai J, Eiden M, Anderson M.M, Van Hoeven N, Meiering C.D, Overbaugh J. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or Pit2. J. Virol. 2001;75(15):6841–6849. doi: 10.1128/JVI.75.15.6841-6849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerford C, Bartlett J.S, Samulski R.J. AlphaVbeta5 integrin: A co-receptor for adeno-associated virus type 2 infection. Nature Med. 1999;5(1):78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- Summerford C, Samulski R.J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 1998;72(2):1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Page D.C. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum. Mol. Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- Sunyach C, Rollier C, Robaczewska M, Borel C, Barraud L, Kay A, Trepo C, Will H, Cova L. Residues critical for duck hepatitis B virus neutralization are involved in host cell interaction. J. Virol. 1999;73(4):2569–2575. doi: 10.1093/gao/9781884446054.article.t048360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Superti F, Donelli G. Gangliosides as binding sites in SA-11 rotavirus infection of LLC-MK2 cells. J. Gen. Virol. 1991;72(Pt 10):2467–2474. doi: 10.1099/0022-1317-72-10-2467. [DOI] [PubMed] [Google Scholar]
- Superti F, Hauttecoeur B, Morelec M.J, Goldoni P, Bizzini B, Tsiang H. Involvement of gangliosides in rabies virus infection. J. Gen. Virol. 1986;67(Pt 1):47–56. doi: 10.1099/0022-1317-67-1-47. [DOI] [PubMed] [Google Scholar]
- Superti F, Seganti L, Tsiang H, Orsi N. Role of phospholipids in rhabdovirus attachment to CER cells. Arch. Virol. 1984;81(3–4):321–328. doi: 10.1007/BF01310002. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Portner A, Scroggs R.A, Uchikawa M, Koyama N, Matsuo K, Suzuki Y, Takimoto T. Receptor specificities of human respiroviruses. J. Virol. 2001;75(10):4604–4613. doi: 10.1128/JVI.75.10.4604-4613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboga O, Tami C, Carrillo E, Nunez J.I, Rodriguez A, Saiz J.C, Blanco E, Valero M.L, Roig X, Camarero J.A, Andreu D, Mateu M.G, Giralt E, Domingo E, Sobrino F, Palma E.L. A large-scale evaluation of peptide vaccines against foot-and-mouth disease: Lack of solid protection in cattle and isolation of escape mutants. J. Virol. 1997;71(4):2606–2614. doi: 10.1128/jvi.71.4.2606-2614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F, Matsuyama S. Soluble receptor potentiates receptor-independent infection by murine coronavirus. J. Virol. 2002;76(3):950–958. doi: 10.1128/JVI.76.3.950-958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor C.S, Nouri A, Lee C.G, Kozak C, Kabat D. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. USA. 1999;96(3):927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor C.S, Nouri A, Zhao Y, Takeuchi Y, Kabat D. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J. Virol. 1999;73(5):4470–4474. doi: 10.1128/jvi.73.5.4470-4474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor C.S, Willett B.J, Kabat D. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J. Virol. 1999;73(8):6500–6505. doi: 10.1128/jvi.73.8.6500-6505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Vile R.G, Simpson G, O'Hara B, Collins M.K, Weiss R.A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 1992;66(2):1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tami C, Taboga O, Berinstein A, Nuñez J.I, Palma E.L, Domingo E, Sobrino F, Carrillo E. Evidence of the coevolution of antigenicity and host cell tropism of foot-and-mouth disease virus in vivo. J. Virol. 2003;77(2):1219–1226. doi: 10.1128/JVI.77.2.1219-1226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P.K, Michou A.I, Bergelson J.M, Cotten M. Defining CAR as a cellular receptor for the avian adenovirus CELO using a genetic analysis of the two viral fibre proteins. J. Gen. Virol. 2001;82(Pt 6):1465–1472. doi: 10.1099/0022-1317-82-6-1465. [DOI] [PubMed] [Google Scholar]
- Tang D, Strong J.E, Lee P.W. Recognition of the epidermal growth factor receptor by reovirus. Virology. 1993;197(1):412–414. doi: 10.1006/viro.1993.1603. [DOI] [PubMed] [Google Scholar]
- Taplitz R.A, Coffin J.M. Selection of an avian retrovirus mutant with extended receptor usage. J. Virol. 1997;71(10):7814–7819. doi: 10.1128/jvi.71.10.7814-7819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuo H, Okuma K, Tanaka K, Ono N, Minagawa H, Takade A, Matsuura Y, Yanagi Y. Virus entry is a major determinant of cell tropism of Edmonston and wild-type strains of measles virus as revealed by vesicular stomatitis virus pseudotypes bearing their envelope proteins. J. Virol. 2000;74(9):4139–4145. doi: 10.1128/jvi.74.9.4139-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406(6798):893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- Tatsuo H, Ono N, Yanagi Y. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J. Virol. 2001;75(13):5842–5850. doi: 10.1128/JVI.75.13.5842-5850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker T.C, Johnson F.B. Binding of bovine parvovirus to erythrocyte membrane sialylglycoproteins. J. Gen. Virol. 1998;79(Pt 9):2163–2169. doi: 10.1099/0022-1317-79-9-2163. [DOI] [PubMed] [Google Scholar]
- Thaker S.R, Stine D.L, Zamb T.J, Srikumaran S. Identification of a putative cellular receptor for bovine herpesvirus 1. J. Gen. Virol. 1994;75(Pt 9):2303–2309. doi: 10.1099/0022-1317-75-9-2303. [DOI] [PubMed] [Google Scholar]
- Thoulouze M.I, Lafage M, Schachner M, Hartmann U, Cremer H, Lafon M. The neural cell adhesion molecule is a receptor for rabies virus. J. Virol. 1998;72(9):7181–7190. doi: 10.1128/jvi.72.9.7181-7190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassini J.E, Graham D, DeWitt C.M, Lineberger D.W, Rodkey J.A, Colonno R.J. cDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc. Natl. Acad. Sci. USA. 1989;86(13):4907–4911. doi: 10.1073/pnas.86.13.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko R.P, Xu R, Philipson L. HCAR and MCAR: The human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA. 1997;94(7):3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li J, Wands J.R. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J. Virol. 1995;69(11):7106–7117. doi: 10.1128/jvi.69.11.7106-7112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li J, Wands J.R. Carboxypeptidase D is an avian hepatitis B virus receptor. J. Virol. 1999;73(10):8696–8702. doi: 10.1128/jvi.73.10.8696-8702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treichel U, Meyer zum Buschenfelde K.H, Stockert R.J, Poralla T, Gerken G. The asialoglycoprotein receptor mediates hepatic binding and uptake of natural hepatitis B virus particles derived from viraemic carriers. J. Gen. Virol. 1994;75(Pt 11):3021–3029. doi: 10.1099/0022-1317-75-11-3021. [DOI] [PubMed] [Google Scholar]
- Tresnan D.B, Levis R, Holmes K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70(12):8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K, Fradelizi D, Wilson K, Triantafilou M. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J. Virol. 2002;76(2):633–643. doi: 10.1128/JVI.76.2.633-643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K, Triantafilou M, Takada Y, Fernandez N. Human parechovirus 1 utilizes integrins alphavbeta3 and alphavbeta1 as receptors. J. Virol. 2000;74(13):5856–5862. doi: 10.1128/jvi.74.13.5856-5862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou M, Triantafilou K, Wilson K.M, Takada Y, Fernandez N, Stanway G. Involvement of beta2-microglobulin and integrin alphavbeta3 molecules in the coxsackievirus A9 infectious cycle. J. Gen. Virol. 1999;80(Pt 10):2591–2600. doi: 10.1099/0022-1317-80-10-2591. [DOI] [PubMed] [Google Scholar]
- Tripp R.A, Jones L.P, Haynes L.M, Zheng H, Murphy P.M, Anderson L.J. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nature Immunol. 2001;2(8):732–738. doi: 10.1038/90675. [DOI] [PubMed] [Google Scholar]
- Trkola A, Kuhmann S.E, Strizki J.M, Maxwell E, Ketas T, Morgan T, Pugach P, Xu S, Wojcik L, Tagat J, Palani A, Shapiro S, Clader J.W, McCombie S, Reyes G.R, Baroudy B.M, Moore J.P. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA. 2002;99(1):395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybala E, Liljeqvist J.A, Svennerholm B, Bergstrom T. Herpes simplex virus types 1 and 2 differ in their interaction with heparan sulfate. J. Virol. 2000;74(19):9106–9114. doi: 10.1128/jvi.74.19.9106-9114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P.C, Griffin D.E. Mechanism of altered Sindbis virus neurovirulence associated with a single-amino-acid change in the E2 glycoprotein. J. Virol. 1991;65(3):1551–1557. doi: 10.1128/jvi.65.3.1551-1557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffereau C, Benejean J, Blondel D, Kieffer B, Flamand A. Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 1998;17(24):7250–7259. doi: 10.1093/emboj/17.24.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffereau C, Desmezieres E, Benejean J, Jallet C, Flamand A, Tordo N, Perrin P. Interaction of lyssaviruses with the low-affinity nerve-growth factor receptor p75NTR. J. Gen. Virol. 2001;82(Pt 12):2861–2867. doi: 10.1099/0022-1317-82-12-2861. [DOI] [PubMed] [Google Scholar]
- Ubol S, Griffin D.E. Identification of a putative alphavirus receptor on mouse neural cells. J. Virol. 1991;65(12):6913–6921. doi: 10.1128/jvi.65.12.6913-6921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umetsu Y, Sugawara K, Nishimura H, Hongo S, Matsuzaki M, Kitame F, Nakamura K. Selection of antigenically distinct variants of influenza C viruses by the host cell. Virology. 1992;189(2):740–744. doi: 10.1016/0042-6822(92)90597-i. [DOI] [PubMed] [Google Scholar]
- Uncapher C.R, DeWitt C.M, Colonno R.J. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180(2):814–817. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]
- Urban S, Breiner K.M, Fehler F, Klingmuller U, Schaller H. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J. Virol. 1998;72(10):8089–8097. doi: 10.1128/jvi.72.10.8089-8097.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Schwarz C, Marx U.C, Zentgraf H, Schaller H, Multhaup G. Receptor recognition by a hepatitis B virus reveals a novel mode of high affinity virus-receptor interaction. EMBO. J. 2000;19(6):1217–1227. doi: 10.1093/emboj/19.6.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahlenkamp T.W, Verschoor E.J, Schuurman N.N, van Vliet A.L, Horzinek M.C, Egberink H.F, de Ronde A. A single amino acid substitution in the transmembrane envelope glycoprotein of feline immunodeficiency virus alters cellular tropism. J. Virol. 1997;71(9):7132–7135. doi: 10.1128/jvi.71.9.7132-7135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderplasschen A, Bublot M, Dubuisson J, Pastoret P.P, Thiry E. Attachment of the gammaherpesvirus bovine herpesvirus 4 is mediated by the interaction of gp8 glycoprotein with heparinlike moieties on the cell surface. Virology. 1993;196(1):232–240. doi: 10.1006/viro.1993.1471. [DOI] [PubMed] [Google Scholar]
- Vanderplasschen A, Smith G.L. A novel virus binding assay using confocal microscopy: Demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J. Virol. 1997;71(5):4032–4041. doi: 10.1128/jvi.71.5.4032-4041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Regenmortel M.H.V, Fauquet C.M, Bishop D.H, Carstens E.B, Estes M.K, Lemon S.M, Maniloff J, Mayo M.A, Mc Geoch D.J, Pringle C.R, Wickner R.B. “Virus Taxonomy.”. Academic Press; San Diego: 2000. [Google Scholar]
- van Zeijl M, Johann S.V, Closs E, Cunningham J, Eddy R, Shows T.B, O'Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA. 1994;91(3):1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varthakavi V, Minocha H.C. Identification of a 56 kDa putative bovine herpesvirus 1 cellular receptor by anti-idiotype antibodies. J. Gen. Virol. 1996;77(Pt 8):1875–1882. doi: 10.1099/0022-1317-77-8-1875. [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Blaas D, Fita I. Structure of human rhinovirus serotype 2 (HRV2) J. Mol. Biol. 2000;300(5):1179–1194. doi: 10.1006/jmbi.2000.3943. [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Mateu M.G, Andreu D, Giralt E, Domingo E, Fita I. Structure of the major antigenic loop of foot-and-mouth disease virus complexed with a neutralizing antibody: Direct involvement of the Arg-Gly-Asp motif in the interaction. EMBO J. 1995;14(8):1690–1696. doi: 10.1002/j.1460-2075.1995.tb07158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer N, Schoehn G, Ochoa W.F, Fita I, Brookes S, King A, Domingo E, Mateu M.G, Stuart D, Hewat E.A. Flexibility of the major antigenic loop of foot-and-mouth disease virus bound to a Fab fragment of a neutralising antibody: Structure and neutralisation. Virology. 1999;255(2):260–268. doi: 10.1006/viro.1998.9554. [DOI] [PubMed] [Google Scholar]
- Verschoor E.J, Boven L.A, Blaak H, van Vliet A.L, Horzinek M.C, de Ronde A. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J. Virol. 1995;69(8):4752–4757. doi: 10.1128/jvi.69.8.4752-4757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscidi R.P. HIV evolution and disease progression via longitudinal studies. In: Crandall K.A, editor. “The Evolution of HIV”. Johns Hopkins University Press; Baltimore: 1999. pp. 346–389. [Google Scholar]
- Vlasak M, Goesler I, Blaas D. “EUROPIC America 2002,”. 2002. HEp2 cell adapted HRV89 variants use heparan sulfate proteoglycan for cell entry; p. C23. [Google Scholar]
- Vlasak R, Luytjes W, Spaan W, Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA. 1988;85(12):4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodros D, Thorstensson R, Biberfeld G, Schols D, De Clercq E, Fenyo E.M. Coreceptor usage of sequential isolates from cynomolgus monkeys experimentally infected with simian immunodeficiency virus (SIVsm) Virology. 2001;291(1):12–21. doi: 10.1006/viro.2001.1164. [DOI] [PubMed] [Google Scholar]
- Wada Y, Pierce M.L, Fujinami R.S. Importance of amino acid 101 within capsid protein VP1 for modulation of Theiler's virus-induced disease. J. Virol. 1994;68(2):1219–1223. doi: 10.1128/jvi.68.2.1219-1223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.Z, Akula S.M, Pramod N.P, Zeng L, Chandran B. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J. Virol. 2001;75(16):7517–7527. doi: 10.1128/JVI.75.16.7517-7527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kavanaugh M.P, North R.A, Kabat D. Cell-surface receptor for ecotropic murine retroviruses is a basic amino-acid transporter. Nature. 1991;352(6337):729–731. doi: 10.1038/352729a0. [DOI] [PubMed] [Google Scholar]
- Wang K.S, Kuhn R.J, Strauss E.G, Ou S, Strauss J.H. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J. Virol. 1992;66(8):4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.S, Schmaljohn A.L, Kuhn R.J, Strauss J.H. Antiidiotypic antibodies as probes for the Sindbis virus receptor. Virology. 1991;181(2):694–702. doi: 10.1016/0042-6822(91)90903-o. [DOI] [PubMed] [Google Scholar]
- Wang W.K, Essex M, Lee T.H. Single amino acid substitution in constant region 1 or 4 of gp120 causes the phenotype of a human immunodeficiency virus type 1 variant with mutations in hypervariable regions 1 and 2 to revert. J. Virol. 1996;70(1):607–611. doi: 10.1128/jvi.70.1.607-611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T, Pipkin P.A, Clarkson N.A, Stone D.M, Minor P.D, Almond J.W. Decay-accelerating factor CD55 is identified as the receptor for echovirus 7 using CELICS, a rapid immuno-focal cloning method. EMBO J. 1994;13(21):5070–5074. doi: 10.1002/j.1460-2075.1994.tb06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T, Powell R.M, Pipkin P.A, Evans D.J, Minor P.D, Almond J.W. Role for beta2-microglobulin in echovirus infection of rhabdomyosarcoma cells. J. Virol. 1998;72(7):5360–5365. doi: 10.1128/jvi.72.7.5360-5365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner M.S, Geraghty R.J, Martinez W.M, Montgomery R.I, Whitbeck J.C, Xu R, Eisenberg R.J, Cohen G.H, Spear P.G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246(1):179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- Weiller G.F, McClure M.A, Gibbs A.J. Molecular phylogenetic analysis. In: Gibbs A.J, Calisher C.H, Garcia-Arenal F, editors. “Molecular Basis of Virus Evolution”. Cambridge Univ. Press; Cambridge: 1995. pp. 553–585. [Google Scholar]
- Weis W, Brown J.H, Cusack S, Paulson J.C, Skehel J.J, Wiley D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333(6172):426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Weiss R.A, Tailor C.S. Retrovirus receptors. Cell. 1995;82(4):531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Whitbeck J.C, Muggeridge M.I, Rux A.H, Hou W, Krummenacher C, Lou H, van Geelen A, Eisenberg R.J, Cohen G.H. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J. Virol. 1999;73(12):9879–9890. doi: 10.1128/jvi.73.12.9879-9890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitbeck J.C, Peng C, Lou H, Xu R, Willis S.H, Ponce de Leon M, Peng T, Nicola A.V, Montgomery R.I, Warner M.S, Soulika A.M, Spruce L.A, Moore W.T, Lambris J.D, Spear P.G, Cohen G.H, Eisenberg R.J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 1997;71(8):6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham T.J, Mathias P, Cheresh D.A, Nemerow G.R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73(2):309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- Wilke C.O, Ronnewinkel C, Martinez T. Dynamic fitness landscape in molecular evolution. Physics Rep. 2001;349:395–446. [Google Scholar]
- Willett B.J, Hosie M.J, Jarrett O, Neil J.C. Identification of a putative cellular receptor for feline immunodeficiency virus as the feline homologue of CD9. Immunology. 1994;81(2):228–233. [PMC free article] [PubMed] [Google Scholar]
- Willett B.J, Picard L, Hosie M.J, Turner J.D, Adema K, Clapham P.R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J. Virol. 1997;71(9):6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.K, Jiang G.S, Holmes K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA. 1991;88(13):5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W.V, Guy H.R, Rubin D.H, Robey F, Myers J.N, Kieber-Emmons T, Weiner D.B, Greene M.I. Sequences of the cell-attachment sites of reovirus type 3 and its anti-idiotypic⧸antireceptor antibody: Modeling of their three-dimensional structures. Proc. Natl. Acad. Sci. USA. 1988;85(17):6488–6492. doi: 10.1073/pnas.85.17.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W.V, Kieber-Emmons T, Weiner D.B, Rubin D.H, Greene M.I. Contact residues and predicted structure of the reovirus type 3-receptor interaction. J. Biol. Chem. 1991;266(14):9241–9250. [PubMed] [Google Scholar]
- Williams W.V, Weiner D.B, Greene M.I. Development and use of antireceptor antibodies to study interaction of mammalian reovirus type 3 with its cell surface receptor. Methods Enzymol. 1989;178:321–341. doi: 10.1016/0076-6879(89)78024-1. [DOI] [PubMed] [Google Scholar]
- Willoughby R.E, Yolken R.H. SA11 rotavirus is specifically inhibited by an acetylated sialic acid. J. Infect. Dis. 1990;161(1):116–119. doi: 10.1093/infdis/161.1.116. [DOI] [PubMed] [Google Scholar]
- Wimmer E, editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. (“Cellular Receptors for Animal Viruses.”). [Google Scholar]
- Wu P.S, Ledeen R.W, Udem S, Isaacson Y.A. Nature of the Sendai virus receptor: glycoprotein versus ganglioside. J. Virol. 1980;33(1):304–310. doi: 10.1128/jvi.33.1.304-310.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WuDunn D, Spear P.G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 1989;63(1):52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Kwong P.D, Desjardins E, Sweet R.W, Robinson J, Hendrickson W.A, Sodroski J.G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wybenga L.E, Epand R.F, Nir S, Chu J.W, Sharom F.J, Flanagan T.D, Epand R.M. Glycophorin as a receptor for Sendai virus. Biochemistry. 1996;35(29):9513–9518. doi: 10.1021/bi9606152. [DOI] [PubMed] [Google Scholar]
- Wykes M.N, Shellam G.R, McCluskey J, Kast W.M, Dallas P.B, Price P. Murine cytomegalovirus interacts with major histocompatibility complex class I molecules to establish cellular infection. J. Virol. 1993;67(7):4182–4189. doi: 10.1128/jvi.67.7.4182-4189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Bator C.M, Bowman V.D, Rieder E, He Y, Hebert B, Bella J, Baker T.S, Wimmer E, Kuhn R.J, Rossmann M.G. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J. Virol. 2001;75(5):2444–2451. doi: 10.1128/JVI.75.5.2444-2451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Tjarnlund K, Lindqvist B, Kaplan G.G, Feigelstock D, Cheng R.H, Casasnovas J.M. Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J. 2000;19(6):1207–1216. doi: 10.1093/emboj/19.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.P, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman S.L, Arnaout M.A. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296(5565):151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Xu G, Wilson W, Mecham J, Murphy K, Zhou E.M, Tabachnick W. VP7: An attachment protein of bluetongue virus for cellular receptors in Culicoides variipennis. J. Gen. Virol. 1997;78(Pt 7):1617–1623. doi: 10.1099/0022-1317-78-7-1617. [DOI] [PubMed] [Google Scholar]
- Xu X.N, Screaton G.R, McMichael A.J. Virus infections: Escape, resistance, and counterattack. Immunity. 2001;15(6):867–870. doi: 10.1016/s1074-7613(01)00255-2. [DOI] [PubMed] [Google Scholar]
- Xue W, Minocha H.C. Identification of the cell surface receptor for bovine viral diarrhoea virus by using anti-idiotypic antibodies. J. Gen. Virol. 1993;74(Pt 1):73–79. doi: 10.1099/0022-1317-74-1-73. [DOI] [PubMed] [Google Scholar]
- Yamada A, Brown L.E, Webster R.G. Characterization of H2 influenza virus hemagglutinin with monoclonal antibodies: Influence of receptor specificity. Virology. 1984;138(2):276–286. doi: 10.1016/0042-6822(84)90351-9. [DOI] [PubMed] [Google Scholar]
- Yan M, Wang L.C, Hymowitz S.G, Schilbach S, Lee J, Goddard A, de Vos A.M, Gao W.Q, Dixit V.M. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science. 2000;290(5491):523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- Yang Y.L, Guo L, Xu S, Holland C.A, Kitamura T, Hunter K, Cunningham J.M. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nature Genet. 1999;21(2):216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]
- Yeager C.L, Ashmun R.A, Williams R.K, Cardellichio C.B, Shapiro L.H, Look A.T, Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357(6377):420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates T.O, Jacobson D.H, Martin A, Wychowski C, Girard M, Filman D.J, Hogle J.M. Three-dimensional structure of a mouse-adapted type 2⧸type 1 poliovirus chimera. EMBO J. 1991;10(9):2331–2341. doi: 10.1002/j.1460-2075.1991.tb07772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K, Lai M.M. Mouse hepatitis virus utilizes two carcinoembryonic antigens as alternative receptors. J. Virol. 1992;66(10):6194–6199. doi: 10.1128/jvi.66.10.6194-6199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolken R.H, Willoughby R, Wee S.B, Miskuff R, Vonderfecht S. Sialic acid glycoproteins inhibit in vitro and in vivo replication of rotaviruses. J. Clin. Invest. 1987;79(1):148–154. doi: 10.1172/JCI112775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.A.T. Virus entry and uncoating. In: Knipe D.M, Howley P.M, editors. “Fields Virology”. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 87–103. [Google Scholar]
- Yuan T.T, Shih C. A frequent, naturally occurring mutation (P130T) of human hepatitis B virus core antigen is compensatory for immature secretion phenotype of another frequent variant (197L) J. Virol. 2000;74(10):4929–4932. doi: 10.1128/jvi.74.10.4929-4932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagury D, Lachgar A, Chams V, Fall L.S, Bernard J, Zagury J.F, Bizzini B, Gringeri A, Santagostino E, Rappaport J, Feldman M, O'Brien S.J, Burny A, Gallo R.C. C-C chemokines, pivotal in protection against HIV type 1 infection. Proc. Natl. Acad. Sci. USA. 1998;95(7):3857–3861. doi: 10.1073/pnas.95.7.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate S, Espinosa R, Romero P, Guerrero C.A, Arias C.F, Lopez S. Integrin alpha2beta1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology. 2000;278(1):50–54. doi: 10.1006/viro.2000.0660. [DOI] [PubMed] [Google Scholar]
- Zhang Y.J, Hatziioannou T, Zang T, Braaten D, Luban J, Goff S.P, Bieniasz P.D. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J. Virol. 2002;76(12):6332–6343. doi: 10.1128/JVI.76.12.6332-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Pacheco J.M, Mason P.W. Evaluation of genetically engineered derivatives of a Chinese strain of foot-and-mouth disease virus reveals a novel cell-binding site which functions in cell culture and in animals. J. Virol. 2003;77(5):3269–3280. doi: 10.1128/JVI.77.5.3269-3280.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lin X, Green T.J, Lipton H.L, Luo M. Role of sialyloligosaccharide binding in Theiler's virus persistence. J. Virol. 1997;71(12):9701–9712. doi: 10.1128/jvi.71.12.9701-9712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Luo Y, Wu Y, Tsao J, Luo M. Sialylation of the host receptor may modulate entry of demyelinating persistent Theiler's virus. J. Virol. 2000;74(3):1477–1485. doi: 10.1128/jvi.74.3.1477-1485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Gershon M.D, Ambron R, Gabel C, Gershon A.A. Infection of cells by varicella zoster virus: Inhibition of viral entry by mannose 6-phosphate and heparin. Proc. Natl. Acad. Sci. USA. 1995;92(8):3546–3550. doi: 10.1073/pnas.92.8.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmern D. Evolution of RNA viruses. In: Domingo E, Holland J.J, Ahlquist P, editors. Vol. 2. CRC Press; Boca Raton, FL: 1988. pp. 211–240. (“RNA Genetics”). [Google Scholar]


