Abstract
The proposed study evaluates the efficacy of knowledge transfer gained through an ensemble of modality-specific deep learning models toward improving the state-of-the-art in Tuberculosis (TB) detection. A custom convolutional neural network (CNN) and selected popular pretrained CNNs are trained to learn modality-specific features from large-scale publicly available chest x-ray (CXR) collections including (i) RSNA dataset (normal = 8851, abnormal = 17833), (ii) Pediatric pneumonia dataset (normal = 1583, abnormal = 4273), and (iii) Indiana dataset (normal = 1726, abnormal = 2378). The knowledge acquired through modality-specific learning is transferred and fine-tuned for TB detection on the publicly available Shenzhen CXR collection (normal = 326, abnormal =336). The predictions of the best performing models are combined using different ensemble methods to demonstrate improved performance over any individual constituent model in classifying TB-infected and normal CXRs. The models are evaluated through cross-validation (n = 5) at the patient-level with an aim to prevent overfitting, improve robustness and generalization. It is observed that a stacked ensemble of the top-3 retrained models demonstrates promising performance (accuracy: 0.941; 95% confidence interval (CI): [0.899, 0.985], area under the curve (AUC): 0.995; 95% CI: [0.945, 1.00]). One-way ANOVA analyses show there are no statistically significant differences in accuracy (P = .759) and AUC (P = .831) among the ensemble methods. Knowledge transferred through modality-specific learning of relevant features helped improve the classification. The ensemble model resulted in reduced prediction variance and sensitivity to training data fluctuations. Results from their combined use are superior to the state-of-the-art.
Keywords: Classification, confidence interval, convolutional neural network, deep learning, ensemble, knowledge transfer, modality-specific learning, tuberculosis
I. Introduction
Data-driven deep learning (DL) algorithms such as convolutional neural networks (CNNs) self-discover hierarchical feature representations from raw data pixels and perform end-to-end feature extraction and classification with minimal expert intervention. These models are shown to achieve state-of-the-art performance in visual recognition tasks [1]. State-of-the-art, computer-aided diagnostic tools (CADx) applied to chest X-ray (CXR) analysis make use of CNNs to support expert radiologist decisions by analyzing the CXRs for the existence of typical disease manifestations and localizing the suspicious regions for interpretation [2]. Unlike rule-based feature descriptors [3][4], CNNs have demonstrated superior results in medical visual recognition tasks, such as detecting parasitized cells in thin-blood smear images [5], cardiomegaly [6], and Tuberculosis (TB) manifestations in CXRs [7].
TB is a dreadful infectious disease caused by Mycobacterium tuberculosis. According to the 2019 World Health Organization (WHO) report, TB remains the top infectious killer across the world, with 10 million people falling ill with the disease in 2018 [8]. People from the Asian and African sub-continents accounted for more than 60% of those suffering from the infection. CXRs are the most common imaging modality used to diagnose conditions affecting the chest and its contents [9] and are particularly useful in establishing a possible diagnosis of TB.
The study of the literature reveals that researchers are working with CXR collections toward improving the performance of automated TB screening. The authors of [9] extracted the lung region of interest (ROI) using a graph-cut segmentation approach and computed texture and shape feature descriptors including histogram of oriented gradients (HOG), local binary patterns (LBP), Hu moments, and Tamura texture descriptors using the publicly available Shenzhen CXR dataset [3] to classify them into normal and abnormal classes. Different classifiers including multilayer perceptron (MLP), support vector machine (SVM), decision trees, and logistic regression were evaluated. The authors reported superior performance with the linear SVM classifier that obtained an area under the curve (AUC) of 0.90 and an accuracy of 0.84. The authors of [10] designed a CADx system using deep CNNs toward automating TB screening. They used custom and pretrained CNNs and trained them on a large-scale private CXR collection. The trained models were used to classify the radiographic images in the Shenzhen CXR dataset. It was observed that the pretrained CNNs delivered a superior performance with an accuracy of 0.837 and AUC of 0.926, as compared to randomly initialized models that gave an accuracy of 0.77 and an AUC of 0.82.
The promising performance of CNNs is accompanied by the availability of huge amounts of annotated data. Under conditions of limited data availability, the models are pretrained on a large-scale collection of natural, stock-photographic images such as ImageNet [1]. This is called transfer learning (TL) where the learned feature representations are transferred and fine-tuned for a similar task.
It has been asserted that visual characteristics of medical images, such as shape, color, texture, spatial dimension, resolution, appearance, and their combinations, tend to be different from those in natural images [11]. For instance, unlike natural images, CXRs exhibit high inter-class similarity and low intra-class variance. Further, some popular disease-specific datasets, such as the Shenzhen TB CXR dataset, are often too small for the conventional TL to be reliable. Small sets result in the models overfitting to the training samples and consequently generalizing poorly to the unseen data. It is believed that improved generalization in the transferred knowledge is possible with the use of pretrained model architectures combined with modality-specific features to improve performance on similar tasks, hereafter referred to as modality-specific learning. Then, transferring knowledge to the specific tasks which may suffer from small sets is expected to allow better adaptation of the models as compared to conventional TL strategy. It is sensible to mention that the current literature leaves much room for progress in studying the efficacy of these strategies.
CNNs learn through error backpropagation and stochastic optimization to minimize the cross-entropic loss and categorize the images to their respective classes. However, these models are highly sensitive to the training data fluctuations. This results in modeling random noise and overfitting during model training, leading to high prediction variance and limited performance. The variance of these models could be reduced by combining the predictions of multiple, diverse CNNs that are accurate in different regions in the feature space and make different errors. The process is called ensemble learning and is expected to deliver promising predictions as compared to any individual constituent learning algorithm [12][13][14][15][16][17]. There are several approaches to constructing model ensembles, such as majority voting, simple averaging, weighted averaging, stacking, and blending. These methods are shown to minimize model variance and enhance learning. The authors of [18] evaluated three different proposals including CNN based feature extraction, bag of words (BOW) generation and multiple instance learning, and model ensembles toward classifying the radiographic images in the Shenzhen CXR dataset. For ensemble learning, the pretrained CNNs including VGGNet [19], ResNet [20], and GoogLeNet [21] were used to extract features to be fed into an SVM classifier and the final predictions were averaged. It was observed that, in terms of accuracy, multiple instance learning demonstrated superior performance. In terms of AUC, model ensembles attained similar performance as in [10], with an AUC of 0.926. The authors of [7] used four de-identified CXR datasets including the publicly available Shenzhen and Montgomery CXR collections, and those collected from Thomas Jefferson University Hospital, Philadelphia, and the Belarus TB Portal and evaluated untrained and pretrained CNN models including AlexNet [1] and GoogLeNet toward detecting pulmonary TB. The authors observed that the averaging ensemble of the pretrained CNN models demonstrated superior performance with an AUC of 0.99, as compared to the untrained models. The authors of [22] trained different pretrained CNN models including AlexNet, VGGNet, and ResNet and created a model ensemble by averaging their predictions toward detecting cardiomegaly in CXRs. It is observed that the model ensemble classified cardiomegaly with an accuracy of 92% as compared to rule-based feature descriptors that attained 76.5%. The combination of DL and ensemble learning is shown to efficiently handle visual recognition tasks and improve predictions through their inherent characteristics of constructing complex, non-linear decision-making functions.
In this study, we propose an ensemble of modality-specific DL models toward TB detection using the Shenzhen CXR dataset and demonstrate improved performance. The customized CNN and pretrained models are trained on a large-scale CXR collection to learn modality-specific features. The retrained models are repurposed to classify TB-infected and normal CXRs. We propose the advantages of combining model predictions through different ensemble methods, such as majority voting, simple averaging, weighted averaging, and stacking, to reduce prediction variance, training data sensitivity, and improve predictions than any individual constituent model. The combined use of modality-specific knowledge transfer and ensemble learning is expected to demonstrate improved generalization and be applied to an extensive range of visual recognition tasks.
II. Materials And Methods
A. Data Collection And Preprocessing
The following publicly available CXR datasets are used in this retrospective study:
Pediatric pneumonia dataset [23]: The dataset includes anterior-posterior (AP) CXRs of children from 1 to 5 years of age, collected from Guangzhou Women and Children’s Medical Center in Guangzhou, China. The imaging has been performed as part of routine clinical care with the approval of the institutional review board (IRB). The study has been conducted in compliance with the United States Health Insurance Portability and Accountability Act (HIPAA). The collection includes 1,583 normal CXRs and 4,273 radiographs infected with bacterial and viral pneumonia. The dataset is curated by expert radiologists and screened to remove low-quality, unreadable radiographs.
Radiological Society of North America (RSNA) pneumonia dataset [24]: The dataset is hosted by the radiologists from RSNA and Society of Thoracic Radiology (STR) for the Kaggle pneumonia detection challenge toward predicting pneumonia in a collection of AP and posterior-anterior (PA) frontal CXRs. It includes a total of 17833 abnormal and 8851 normal radiographs in DICOM format with a spatial resolution of 1024×1024 pixel dimensions and 8-bit depth. The authors didn’t obtain IRB approval since the examinations were part of the publicly available NIH CXR dataset [25].
Indiana dataset [26]: The dataset includes 2,378 abnormal and 1726 normal, PA chest radiographs, collected from hospitals affiliated with the Indiana University School of Medicine, and archived at the National Library of Medicine (NLM) (OHSRP# 5357). The images and reports were automatically de-identified and manually verified. The collection is made publicly available through the OpenI® search engine developed by NLM.
Shenzhen dataset [3]: The dataset includes 336 TB-infected and 326 normal CXRs (both AP and PA) collected from the outpatient clinics of Shenzhen No.3 People’s Hospital, China. The images were de-identified by the data providers and are exempted from IRB review at their institutions. The data was exempted from IRB review (OHSRP# 5357) by the NIH Office of Human Research Protection Programs. Radiologist readings are made available to be considered as ground-truth.
We collected the data from RSNA pneumonia, pediatric pneumonia, and Indiana datasets and divided them at the patient-level into training (80.0%) and test (20.0%) sets. We randomly allocated 10% of the training for validation. The performance of the retrained predictive models is cross-validated using Shenzhen TB CXR collection at the patient-level to provide a more realistic performance evaluation as the test images represent truly unseen information for the training process, with no clues about the disease manifestations or other artifacts leaking into the training data with an aim to improve model robustness and generalization.
Prior to model training, the following preprocessing steps are applied in common to the CXR datasets used in this study: (a) median-filtering with a 3×3 window for edge preservation and noise removal; (b) resizing to 224×224 pixel resolution to reduce computational complexity and memory requirements; (c) rescaling to restrict the pixels in the range [0 1]; and (d) normalization and standardization through mean subtraction and division by standard deviation to ensure similar distribution range for the extracted features.
B. Models And Computational Resources
The performance of the following CNNs are evaluated toward the task of detecting TB in CXRs: (a) customized CNN; (b) VGG-16; (c) Inception-V3 [21]; (d) InceptionResNet-V2 [21]; (e) Xception [27]; and (f) DenseNet-121 [28]. The pretrained models are selected based on several aspects: We observed their performance on the ImageNet validation dataset. Considering the top-1 and top-5 accuracy, the pretrained models used in this study are found to deliver promising performance as compared to other models. The authors of [29] evaluated several DL models including ResNet-152, DenseNet-121, Inception-V4, and SEResNeXt-101 toward CXR lung disease classification. In the process, it was observed that DenseNet-121 produced the best results. In another study [30], the authors used the DenseNet-121 model to train on the NIH CXR dataset and achieved state-of-the-art results.
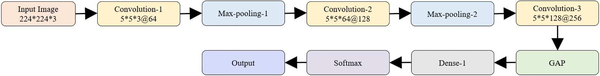
We designed and evaluated the performance of a baseline, custom, sequential CNN model toward the current task. Fig. 1 shows the architecture of the customized CNN used in this study.
Fig. 1.
Architecture of the customized CNN.
Each CNN block has a batch normalization layer, followed by separable convolution, non-linear activation, and dropout layers. We performed zero paddings at the convolutional layers to ensure that the spatial output dimensions match that of the original input. We initialized the number of convolutional filters to 64 and increased the number by a factor of two, every time a max-pooling layer is added. This is done to ensure the amount of computation roughly remains the same across all the separable convolutional layers. We used 5×5 kernels uniformly across the convolutional layers. Batch normalization is performed to normalize the output of the previous activation layers in an attempt to reduce overfitting and improve generalization. Separable convolutional dropouts offer regularization by reducing the sensitivity of the model to training data fluctuations [27]. A global average pooling (GAP) layer is added to the deepest separable convolutional layer to reduce feature dimensionality by spatially averaging the feature maps. The output of the GAP layer is fed to the first dense, fully-connected layer, followed by a dropout and final dense layer to predict on the current task. The customized model is trained to learn and minimize the cross-entropic loss toward classifying the CXRs into their respective categories.
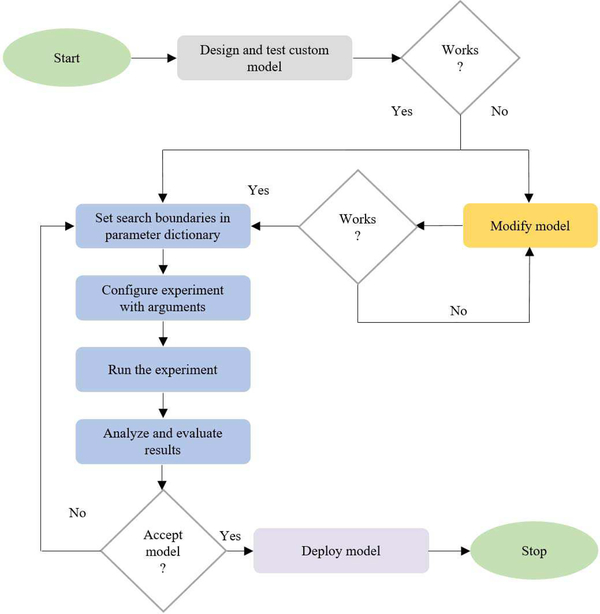
The customized CNN is optimized for its parameters and hyperparameters including (a) hidden neurons in the first dense layer, (b) separable-convolutional dropout, (c) dense layer dropout, (d) optimizer function, and (e) non-linear activation using Talos optimization tool [31]. Fig. 2 shows the process flow diagram toward optimizing the custom model hyper-parameters. The pretrained models are instantiated with the ImageNet-trained weights.
Fig. 2.
Process flow diagram toward the automated optimization of custom CNN hyperparameters using the Talos optimization algorithm.
The models are truncated at their deepest convolutional layer and added with a GAP and dense layer. The models are fine-tuned with smaller weight updates through stochastic gradient descent optimization to minimize the categorical cross-entropic loss toward the current task.
C. Modality Specific Learning
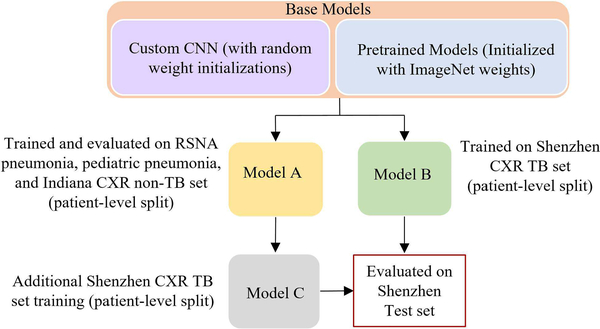
We propose a modality-specific learning strategy to improve generalization in the transferred knowledge and prediction performance by using pretrained model architectures combined with modality-specific features. The customized CNN and pretrained models are trained on a large-scale CXR collection to learn modality-specific features. The retrained models are fine-tuned to classify TB-infected and normal CXRs. Fig. 3 shows the process flow diagram for the proposed strategy. The overall process is described herewith:
Fig. 3.
Modality-specific knowledge transfer showing the base and retrained models along with the patient-level train/test split for each model.
Model A: The custom and pretrained models, otherwisecalled the base models, are trained on a collection of datasets including RSNA pneumonia, pediatric pneumonia, and Indiana collections to learn the CXR modality-specific features and classify them into abnormal and normal categories. Callbacks and model checkpoints are used to investigate the performance of the models after each epoch. The models are evaluated for 100 epochs or until the performance plateau. The learning rate is reduced whenever the validation accuracy ceased to improve. The retrained models with the best test classification accuracy are stored for further evaluation.
Model B: The base models are trained and evaluatedwith the Shenzhen TB CXR collection, to categorize into TB-infected and normal classes. Due to limited data availability, the models are evaluated through five-fold cross-validation with an aim to prevent overfitting and improve robustness and generalization. The retrained base models with the best model weights, giving the highest test classification accuracy for each cross-validated fold are stored for further evaluation.
Model C: Retrained models from Model A with CXRmodality-specific knowledge are fine-tuned on Shenzhen TB CXR collection to categorize into TB-infected and normal classes. Embedding modality-specific knowledge is expected to improve model adaption to the target task. The retrained models showing the best performance for each cross-validated fold are stored for further evaluation. With modality-specific knowledge transfer, Model C is expected to demonstrate improved TB detection performance as compared to Model B.
D. Ensemble Learning
Ensemble learning helps to reduce variance and improve generalization by combining the predictions of multiple models and obtain promising predictions than any individual, constituent model.
In this study, the predictions of the models from Model C are combined through majority voting, simple averaging, weighted averaging, and stacking to classify the CXRs into TB-infected and normal classes. In majority voting, the predictions of multiple models are considered as votes. Final predictions are made based on these votes obtained from the majority of the models. Simple averaging averages the prediction probabilities from multiple models to arrive at the final predictions. Weighted averaging is an extension of simple averaging in which the models are assigned different weights based on their importance in making the predictions.
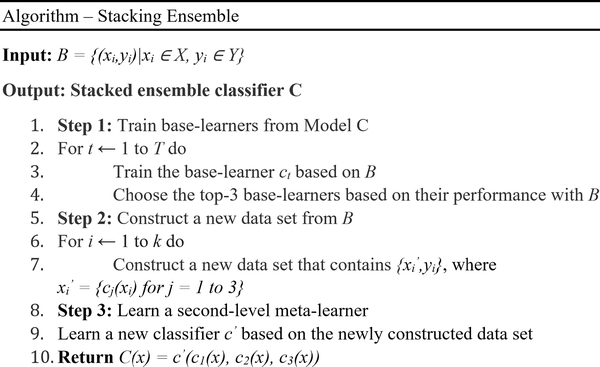
Stacking or stacked generalization is an ensemble method where a meta-learner learns how to best combine the predictions from individual models (base-learners) [32]. A stacking ensemble has two levels: (a) Level-0 includes the training data input and base-learners, and (b) Level-1 takes the predictions of base-learners as input and a meta-learner learns to optimally combine the predictions of base-learners. In this study, we used a neural network-based meta-learner to learn from the predictions of the top-performing models from Model C. The layers in the base-learners are marked as not trainable so the weights are not updated when the stacking ensemble is trained. The outputs of the base-learners are concatenated. A hidden layer is defined to interpret these predictions to the meta-learner and an output layer to arrive at probabilistic predictions. Fig. 4 shows the algorithm for training the stacking ensemble proposed in this study.
Fig. 4.
Stacking ensemble approach.
Unlike other ensemble methods, stacking uses the predictions of the base-learners as a context and conditionally decides to differentially weigh these predictions to deliver better performance than any individual, constituent model. The benefit of this approach is that the outputs of the base-learners are fed directly to the meta-learner and the stacking ensemble is treated as a single model where the base-learners are embedded in a larger multi-headed neural network.
The models in modality-specific knowledge transfer and ensemble pipeline are evaluated in terms of the following performance metrics: (a) accuracy; (b) AUC; (c) sensitivity; (d) specificity; (e) F-score; and (f) Matthews Correlation Coefficient (MCC). The models are trained and evaluated on a Windows system with Xeon CPU, 32GB RAM, NVIDIA 1080Ti GPU and CUDA/CUDNN for GPU acceleration. The models are configured in Python using Keras API with a Tensorflow backend.
E. Statistical Analysis
DL models are statistical and probabilistic in nature that captures data patterns through the use of computational methods. It is highly probable that observations that involve drawing samples from a population demonstrate an effect that would have occurred due to sampling errors. However, if the observed effect demonstrates P < 0.05 (95% confidence interval (CI)), a conclusion is made that the observed effect reflects the characteristics of the entire population. Tests for statistical significance help to measure whether the differences between the studied groups are significant or occurred by chance.
In this study, statistical analyses are performed to analyze for the existence of a statistically significant difference in the mean values of the performance metrics achieved with different ensemble methods. One-way analysis of variance (ANOVA) is performed to determine the existence of these statistically significant performance differences. However, to perform this analysis, the data should satisfy the following assumptions: (a) normal distribution; (b) homogenous variance; (c) absence of significant outliers; and (d) independence of observations [33]. Shapiro-Wilk normality analysis [34] is performed to investigate for data normality and Levene’s analysis [35], to check for homogeneous variances. The data is analyzed for the presence of outliers and the independence of observations. The null hypothesis (H0) that all ensemble methods demonstrate similar performance is accepted if no statistically significant difference is observed in the mean value of the performance metrics for the different ensemble methods under study. The alternate hypothesis (H1) is accepted and H0 is rejected if a statistically significant performance difference (P < 0.05) is found to exist.
One-way ANOVA is an omnibus test and needs a post-hoc study to identify the specific ensemble methods demonstrating this statistically significant performance differences. In this study, a Tukey post-hoc test [36] is performed to identify the ensemble methods demonstrating these statistically significant performance differences. We used the IBM SPSS [37] package to perform statistical analyses.
III. Results
The optimal hyperparameter values obtained with the Talos optimization tool for the customized CNN are as follows: (a) hidden neurons in the first dense layer (256); (b) separable-convolutional dropout (0.25); (c) dense layer dropout (0.5); (d) optimizer function (Adam); and (e) non-linear activation (ReLU).
The performance of the customized CNN and pretrained models in Model A toward classifying abnormal and normal CXRs are evaluated and the obtained results are shown in Table I. This is the first step in the modality-specific knowledge transfer pipeline where the customized CNN and pretrained models are trained to learn the CXR modality-specific features across the normal and abnormal categories.
Table I.
PERFORMANCE METRICS ACHIEVED WITH MODELS IN MODEL A
| Models | Acc. | AUC | Sens. | Spec. | FI | MCC |
|---|---|---|---|---|---|---|
| Custom | 0.861 | 0.940 | 0.869 | 0.845 | 0.893 | 0.697 |
| VGG-16 | 0.896 | 0.960 | 0.922 | 0.841 | 0.922 | 0.764 |
| Inception-V3 | 0.896 | 0.960 | 0.909 | 0.869 | 0.921 | 0.769 |
| InceptionResNet-V2 | 0.896 | 0.960 | 0.919 | 0.850 | 0.922 | 0.766 |
| Xception | 0.887 | 0.959 | 0.888 | 0.886 | 0.913 | 0.755 |
| DenseNet-121 | 0.897 | 0.962 | 0.926 | 0.837 | 0.930 | 0.766 |
Acc. = Accuracy; Sens. = Sensitivity; Spec. = Specificity; FI = F-Score.
Accuracy demonstrates the model’s ability to correctly classify positive and negative cases. Specificity gives a measure of the models’ ability to correctly identify negative cases. Sensitivity (recall) demonstrates the ability to correctly identify positive cases. A measure of F-score gives the harmonic average of recall and precision, and MCC, the degree of agreement between the predictions and ground-truth values. It is observed that the DenseNet-121 showed better performance in terms of accuracy (0.897), AUC (0.962), and sensitivity (0.926). The Xception model gave higher values for specificity (0.887). However, considering the balance between precision and recall, as demonstrated by the F-score, the DenseNet-121 demonstrated superior performance in classifying the abnormal and normal CXRs.
The performance of the customized and pretrained models in Model B, cross-validated with the Shenzhen TB CXR dataset, toward classifying TB-infected and normal CXRs are evaluated and the results are shown in Table II. It is observed that DenseNet-121 demonstrated better performance for metrics including accuracy (0.899), AUC (0.948), specificity (0.933), F-score (0.897), and MCC (0.801). The Inception-V3 model showed higher values for sensitivity (0.908).
Table II.
PERFORMANCE METRICS ACHIEVED WITH MODELS IN MODEL B
| Models | Acc. | AUC | Sens. | Spec. | FI | MCC |
|---|---|---|---|---|---|---|
| Custom | 0.783 | 0.830 | 0.759 | 0.807 | 0.780 | 0.570 |
| VGG-16 | 0.887 | 0.934 | 0.872 | 0.902 | 0.887 | 0.778 |
| Inception-V3 | 0.885 | 0.942 | 0.908 | 0.862 | 0.890 | 0.773 |
| InceptionResNet-V2 | 0.885 | 0.936 | 0.887 | 0.884 | 0.887 | 0.772 |
| Xception | 0.879 | 0.930 | 0.872 | 0.887 | 0.880 | 0.760 |
| DenseNet-121 | 0.899 | 0.948 | 0.866 | 0.933 | 0.897 | 0.801 |
The retrained custom and pretrained models in Model A are fine-tuned and cross-validated with the Shenzhen TB CXR collection to obtain the models in Model C to classify TB-infected and normal CXRs and the results are shown in Table III. The notable results are as follows: (a) the performance of the models in Model C is promising compared to that of Model B models. This may be because the CXR modality-specific features learned from a large-scale data collection resulted in a generalized transfer of knowledge, suitable to be repurposed for the task of TB detection; (b) the standard deviation of the evaluated metrics for the Model C models are significantly lower than that of Model B. This may be because of the improved generalization, reduced bias, and overfitting, resulted from the modality-specific knowledge transfer toward the current task. It is observed that Inception-V3 demonstrated better performance for the metrics including accuracy (0.940), AUC (0.974), sensitivity (0.938), F-score (0.941), and MCC (0.880). The VGG-16 model demonstrated higher values for specificity (0.963). However, considering the usage as a screening tool, the sensitivity metrics carry high prominence. Also, considering the F-score that demonstrates the balance between precision and recall, the Inception-V3 model showed superior performance. These results indicated that modality-specific learning improved the models’ robustness, generalization, and reduced bias and overfitting toward giving promising results in classifying TB-infected and normal CXRs.
Table III.
PERFORMANCE METRICS ACHIEVED WITH MODELS IN MODEL C
| Models | Acc. | AUC | Sens. | Spec. | FI | MCC |
|---|---|---|---|---|---|---|
| Custom | 0.872 | 0.920 | 0.866 | 0.878 | 0.872 | 0.748 |
| VGG-16 | 0.923 | 0.964 | 0.884 | 0.963 | 0.921 | 0.850 |
| Inception-V3 | 0.940 | 0.974 | 0.938 | 0.942 | 0.941 | 0.880 |
| InceptiohResNet-V2 | 0.925 | 0.968 | 0.905 | 0.945 | 0.924 | 0.852 |
| Xception | 0.891 | 0.944 | 0.875 | 0.908 | 0.891 | 0.786 |
| DenseNet-121 | 0.928 | 0.960 | 0.920 | 0.936 | 0.928 | 0.856 |
We evaluated the cross-validated performance of multiple ensemble methods, including majority voting, simple averaging, weighted averaging, and stacking, using the top-3 performing models in Model C, including InceptionResNet-V2, Inception-V3, and DenseNet-121 toward improving the performance of classifying TB-infected and normal CXRs in the Shenzhen CXR dataset. Table IV shows the results obtained with the different ensemble methods toward the current task.
Table IV.
PERFORMANCE METRICS ACHIEVED WITH THE ENSEMBLE OF TOP-3 MODELS IN MODEL C (INCEPTIONRESNET-V2, INCEPTION-V3, AND DENSENET-121)
| Ensemble method | Acc. | AUC | Sens. | Spec. | FI | MCC |
|---|---|---|---|---|---|---|
| Majority Voting | 0.925 | - | 0.923 | 0.927 | 0.926 | 0.852 |
| Simple Averaging |
0.931 | 0.970 | 0.920 | 0.942 | 0.931 | 0.863 |
| Weighted Averaging |
0.934 | 0.975 | 0.923 | 0.945 | 0.934 | 0.868 |
| Stacking | 0.941 | 0.995 | 0.926 | 0.957 | 0.941 | 0.884 |
Bold text indicates thè performance measures of the best-performing ensemble.
For weighted averaging, we empirically observed that the use of weights (InceptionResNet-V2 (0.25), Inception-V3 (0.5), and DenseNet-121 (0.25)) gave the best results. The notable results are as follows: (a) stacking ensemble demonstrated better performance in terms of all performance metrics (accuracy (0.941), AUC (0.995), sensitivity (0.926), specificity (0.957), F-Score (0.941), and MCC (0.884)); and (b) the performance of the stacking ensemble appeared promising because the meta-learner learned to correct the predictions of the individual base-learners by differentially weighing their predictions to deliver optimal predictions than any individual constituent model. The results demonstrated that the classification task is benefited by the combination of modality-specific knowledge transfer and ensemble learning to deliver superior performance.
The performance of the stacking ensemble appears visually significant. However, the test for statistical significance helps to ensure whether the observed difference in performance reflects the population characteristics. These tests measure whether the differences between the studied ensemble methods are statistically significant in the 95% CI. The tests for data normality and homogeneity of variances using Shapiro-Wilk and Levene’s analysis respectively demonstrated P > 0.05 to signify that the assumptions of data normality and homogeneity of variances hold good. Thus, we performed a one-way ANOVA analysis to investigate the existence of a statistically significant difference in the mean values of the performance metrics for the different ensemble methods under study. For the accuracy metric, it is observed that no statistically significant difference exists between the different ensemble methods (P = .759). Similar characteristics are observed for AUC (P = .831), sensitivity (P = .997), specificity (P = .701), F-score (P = .788), and MCC (P = .756). These results signify that there exists no statistically significant difference in performance between the different ensemble methods toward classifying the TB-infected and normal CXRs in the Shenzhen CXR dataset under study.
The performance of the stacking ensemble in classifying TB-infected and normal CXRs is compared to that of the state-of-the-art literature as shown in Table V.
Table V.
COMPARING THE RESULTS WITH THE STATE-OF-THE-ART LITERATURE
| Parameters | Jaeger et al. (8) | Hwang et al. (9) | Lopes et al. (12) | Proposed method |
|---|---|---|---|---|
| Accuracy | 0.840 | 0.837 | 0.847 | 0.941 [0.899 0.985] |
| AUC | 0.900 | 0.926 | 0.926 | 0.990 [0.945 1.00] |
| Sensitivity | - | - | - | 0.926 [0.850 1.00] |
| Specificity | - | - | - | 0.957 [0.883 1.00] |
| F-Score | - | - | - | 0.941 [0.898 0.985] |
| MCC | - | - | - | 0.884 [0.802 0.9671 |
It is observed that the proposed ensemble outperformed the state-of-the-art in all performance metrics.
IV. Discussion
The customized CNN used in this study converges to a promising solution due to (a) hyperparameter optimization, (b) implicit regularization with batch normalization, and (c) reduced bias, improved generalization through use of separable-convolutional and dense layer dropouts. The use of depth-wise separable convolutions ensured a reduction in the trainable parameters, offering the benefit of reduced computational overhead and memory requirements. The models are evaluated through cross-validation studies to present a realistic and generalized performance measure. Modality-specific knowledge transfer helped to embed CXR modality-specific knowledge into the predictive models that resulted in a generalized knowledge transfer, appropriate to be fine-tuned for the task of TB detection. It is observed that the pretrained CNN models retrained on the large-scale CXR collection found superior solutions in the feature space as compared to the custom model with random weight initializations. Ensemble learning reduced models’ prediction variance and sensitivity to training data fluctuations by combining the predictions and deliver optimal performance. In the process, the performance of the stacking ensemble demonstrated superior performance by differentially weighing the predictions to deliver superior performance than any individual, constituent model.
The performance of the ensemble methods is analyzed for the existence of a statistically significant difference to ensure the observed performance difference reflects the characteristics of the entire population. It is observed that there existed no statistically significant performance difference between the ensemble methods. The stacked modality-specific model ensemble significantly outperformed the state-of-the-art in terms of accuracy and AUC. The values for the other performance metrics are not reported in the literature.
This preliminary study, however, has some limitations. The proposed combination of modality-specific knowledge transfer and ensemble learning pipeline is evaluated with the Shenzhen TB CXR collection with small sample size. Future work would include evaluating the efficacy with a larger CXR collection. There are several ensemble methods, each with its own advantages/disadvantages, the method to use depends on the problem under study. CNNs are perceived as black-boxes due to lack of interpretability and their predictions need explanations. Visualization studies need to be performed with model ensembles to give an explanation of the predictions since a poorly understood model behavior could adversely impact medical decision-making. Ensemble methods are computationally expensive, adding training time and memory constraints to the problem. It may not be practicable to implement model ensembles, however, with the advent of low-cost GPU solutions and cloud technology, model ensembles could become practically feasible for real-time applications. Future research could include transferring the knowledge of model ensembles into small, portable models.
We observe that knowledge transfer imposed using modality-specific medical images (large-scale CXR collection) for enhancing pretrained models aided them in improving decision-making. They learned features that are relevant to detect TB manifestations. The predictions of these models are combined through ensemble learning that reduced prediction variance and sensitivity to training data fluctuations. The combined use of modality-specific knowledge transfer and ensemble learning demonstrated superior results as compared to the state-of-the-art and led to reduced overfitting and improved generalization. Since the proposed methodology is not problem-specific it could be used to develop clinically valuable solutions and enable the application to a broad range of visual recognition tasks.
Biography

SIVARAMAKRISHNAN RAJARAMAN received his Ph.D. in Information and Communication Engineering from Anna University, India. He is involved in projects that aim to apply computational sciences and engineering techniques toward advancing life science applications. These projects involve the use of medical images for aiding healthcare professionals in low-cost decision-making at the point of care screening/diagnostics. He is a versatile researcher with expertise in machine learning, data science, biomedical image analysis/understanding, and computer vision. He is a Member of the International Society of Photonics and Optics, Institute of Electrical and Electronics Engineers (IEEE) and the IEEE Engineering in Medicine and Biology Society.

SAMEER K. ANTANI received the B.S. and M.S. degrees in aerospace engineering from the University of Virginia, Charlottesville, in 2001 and the Ph.D. degree in mechanical engineering from Drexel University, Philadelphia, PA, in 2008. He is a versatile lead researcher advancing the role of computational sciences and automated decision making in biomedical research, education, and clinical care. His research interests include topics in medical imaging and informatics, machine learning, data science, artificial intelligence, and global health. He applies his expertise in machine learning, biomedical image informatics, automatic medical image interpretation, data science, information retrieval, computer vision, and related topics in computer science and engineering technology His primary R&D areas include cervical cancer, HIV/TB, and visual information retrieval, among others.
He is a Senior Member of the International Society of Photonics and Optics, Institute of Electrical and Electronics Engineers (IEEE) and the IEEE Computer Society.
References
- [1].Krizhevsky A, Sutskever I, Hinton GE, “Imagenet classification with deep convolutional neural networks”, Proc. Int. Conf. Neural Inf. Process. Syst., pp. 1–9, 2012. [Google Scholar]
- [2].Rajaraman S et al. , “Visualization and Interpretation of Convolutional Neural Network Predictions in Detecting Pneumonia in Pediatric Chest Radiographs”, Appl Sci, vol. 8, no. 10, 1715, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jaeger S et al. , “Two public chest X-ray datasets for computer-aided screening of pulmonary diseases”, Quant Imaging Med Surg, vol. 4, no. 6, pp. 475–477, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Candemir S et al. , “Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration”, IEEE Trans. Med. Imag, vol. 33, no. 2, pp. 577–590, 2014. [DOI] [PubMed] [Google Scholar]
- [5].Rajaraman S et al. , “Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images”, PeerJ, 6:e4568, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Candemir S et al. , “Deep learning for grading cardiomegaly severity in chest x-rays: An investigation”, Proc. IEEE Life Sci. Conf., pp. 109–113, October 2018. [Google Scholar]
- [7].Lakhani P, Sundaram B, “Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks”, Radiology, vol. 284, no. 2, pp. 574–582, 2017. [DOI] [PubMed] [Google Scholar]
- [8].World Health Organization (WHO) Global tuberculosis report, Oct. 2019. Accessed on: Oct. 20, 2019 [Online]. Available: https://www.who.int/tb/publications/global_report/en/.
- [9].Jaeger S et al. , “Automatic tuberculosis screening using chest radiographs”, IEEE Trans. Med. Imag, vol. 33, no. 2, pp. 233–245, 2014. [DOI] [PubMed] [Google Scholar]
- [10].Hwang S et al. , “A novel approach for tuberculosis screening based on deep convolutional neural networks”, Proc. SPIE Medical Imaging: Computer-Aided Diagnosis, 97852W, 2016. [Google Scholar]
- [11].Suzuki K, “Overview of deep learning in medical imaging”, Radiol Phys Technol, vol. 10, no. 3, pp. 257–273, 2017. [DOI] [PubMed] [Google Scholar]
- [12].Dietterich TG, “Ensemble Methods in Machine Learning. In: Multiple Classifier Systems”, Lecture Notes in Computer Science: Springer, Berlin, Heidelberg, vol. 1857, pp. 1–15, 2000. [Google Scholar]
- [13].Nanni L, Ghidoni S, Brahnam S, “Ensemble of convolutional neural networks for bioimage classification”, Applied Computing and Informatics, Article in Press, [Online] Available: https://www.sciencedirect.com/science/article/pii/S2210832718301388.
- [14].Nanni L et al. , General Purpose (GenP) Bioimage Ensemble of Handcrafted and Learned Features with Data Augmentation, 2019, [Online] Available: https://arxiv.org/abs/1904.08084.
- [15].Zhang W et al. , “SFPEL-LPI: Sequence-based feature projection ensemble learning for predicting LncRNA-protein interactions’, PLoS Comput. Biol, vol. 14, no. 12, e1006616, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tang G et al. , “Sequence-based bacterial small RNAs prediction using ensemble learning strategies”, BMC Bioinformatics, vol. 19, no. 20, 503, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang W et al. , “SFLLN: A sparse feature learning ensemble method with linear neighborhood regularization for predicting drug–drug interactions”, Inf. Sci, vol. 497, pp. 189–201, 2019. [Google Scholar]
- [18].Lopes UK, Valiati JF, “Pre-trained convolutional neural networks as feature extractors for tuberculosis detection”, Comput Biol Med, vol. 89, pp. 135–143, 2017. [DOI] [PubMed] [Google Scholar]
- [19].Simonyan K, Zisserman A, Very deep convolutional networks for large-scale image recognition, 2014, [Online] Available: https://arxiv.org/abs/arXiv:1409.1556.
- [20].He K, Zhang X, Ren S, Sun J, “Deep residual learning for image recognition”, Proc. IEEE Conf. Comput. Vis. Pattern Recognit., pp. 770–778, Jun. 2016. [Google Scholar]
- [21].Szegedy C et al. , “Going deeper with convolutions”, Proc. IEEE Conf. Comput. Vis. Pattern Recognit, pp. 1–9, Jun. 2015. [Google Scholar]
- [22].Islam MT et al. , Abnormality Detection and Localization in Chest X-Rays using Deep Convolutional Neural Networks, 2017, [Online] Available: https://arxiv.org/abs/1705.09850.
- [23].Kermany DS et al. , “Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning”, Cell, vol. 172, no. 5, pp. 1122–1131, 2018. [DOI] [PubMed] [Google Scholar]
- [24].Shih G et al. , “Augmenting the National Institutes of Health Chest Radiograph Dataset with Expert Annotations of Possible Pneumonia”, Radiol Artif Intell, vol. 1, no. 1, pp. 1–5, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang X et al. , “ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases”, Proc. IEEE Conf. Comput. Vis. Pattern Recognit., pp. 3462–3471, July 2017. [Google Scholar]
- [26].Demner-Fushman D et al. , “Preparing a collection of radiology examinations for distribution and retrieval”, J Am Med Inform Assoc, vol. 23, no. 2, pp. 304–310, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chollet F, Xception: Deep Learning with Depthwise Separable Convolutions, 2018, [Online] Available: https://arxiv.org/abs/1610.02357.
- [28].Huang G, Liu Z, van der Maaten L, Weinberger KQ, “Densely connected convolutional networks”, Proc. IEEE Conf. Comput. Vis. Pattern Recognit, vol. 1, no. 2, pp. 4700–4708, Jul. 2017. [Google Scholar]
- [29].Jeremy I et al. , CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison, 2019, [Online] Available: https://arxiv.org/abs/1901.07031.
- [30].Rajpurkar P et al. , CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning, 2018, [Online] Available: https://arxiv.org/abs/1711.05225.
- [31].Autonomio Talos [Computer software], Mar. 20, 2019. Accessed on: Apr. 3, 2019 [Online]. Available: https://autonomio.github.io/docs_talos/#introduction.
- [32].Wolpert DH, “Stacked generalization”, Neural Networks, vol. 5, no. 2, pp. 241–259, 1992. [Google Scholar]
- [33].Kim TK, “Understanding one-way anova using conceptual figures”, Korean J Anesthesiol, vol. 70, no. 1, pp. 22–26, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yap BW, Sim CH, “Comparisons of various types of normality tests”, J Stat Comput Simul, vol. 81, no. 12, pp. 2141–2155, 2011. [Google Scholar]
- [35].Kim YJ, Cribbie RA, “ANOVA and the variance homogeneity assumption: Exploring a better gatekeeper”, Br J Math Stat Psychol, vol. 71, no. 1, pp. 1–12, 2018. [DOI] [PubMed] [Google Scholar]
- [36].Opitz D, Maclin R, “Popular Ensemble Methods: An Empirical Study”, J Artif Intell Res, vol. 11, pp. 169–198, 1999. [Google Scholar]
- [37].IBM SPSS Statistics 25, Apr. 2019. Accessed on: May. 15, 2019 [Online]. Available: http://www-01.ibm.com/support/docview.wss?uid=swg24043678