Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is an emerging zoonotic pathogen with a broad host range. The extent of MERS-CoV in nature can be traced to its adaptable cell entry steps. The virus can bind host-cell carbohydrates as well as proteinaceous receptors. Following receptor interaction, the virus can utilize diverse host proteases for cleavage activation of virus-host cell membrane fusion and subsequent genome delivery. The fusion and genome delivery steps can be completed at variable times and places, either at or near cell surfaces or deep within endosomes. Investigators focusing on the CoVs have developed several methodologies that effectively distinguish these different cell entry pathways. Here we describe these methods, highlighting virus-cell entry factors, entry inhibitors, and viral determinants that specify the cell entry routes. While the specific methods described herein were utilized to reveal MERS-CoV entry pathways, they are equally suited for other CoVs, as well as other protease-dependent viral species.
Key words: Middle East respiratory syndrome (MERS), Coronavirus (CoV), Protease, Pseudovirus, Spike (S), Viral entry, Endosome, Virus concentration, Virus purification, Protease inhibitor, HR2 peptide, IFITM3, TMPRSS2, Transfection
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) is endemic in bats, and also in dromedary camels, and can be transmitted zoonotically from camels to humans [1–6]. The virus was discovered in humans in 2012, and since then there has been over 2,000 laboratory-confirmed cases worldwide, with 35% of infected humans suffering fatal outcomes [7–9]. Although MERS-CoV zoonotic and human-to-human transmission rates have declined due to general awareness and improved hospital practices, there are continued possibilities for epidemics, and there is a need for preventive vaccines and therapeutic antivirals. Mechanistic insights into human MERS-CoV entry will promote vaccine and antiviral drug developments.
MERS-CoV, like all other coronaviruses, exists as enveloped extracellular particles with protruding spike (S) proteins. Infection is initiated through viral S protein binding to host cell receptors. Subsequent proteolytic cleavage of cell-bound S proteins triggers S protein-mediated coalescence (fusion) of viral and cell membranes. The triggering process involves a series of currently obscure S protein conformational changes, from “ pre-fusion” metastable states to “ extended fusion intermediates” to “ post-fusion” collapsed states that exist after virus and cell membranes have coalesced and viral mRNA genomes have dispensed into the host cell cytoplasm [10, 11]. These events depend on cellular proteases, and as one may expect, the availability of particular proteases immediately following receptor engagement is a rate-limiting step in CoV entry.
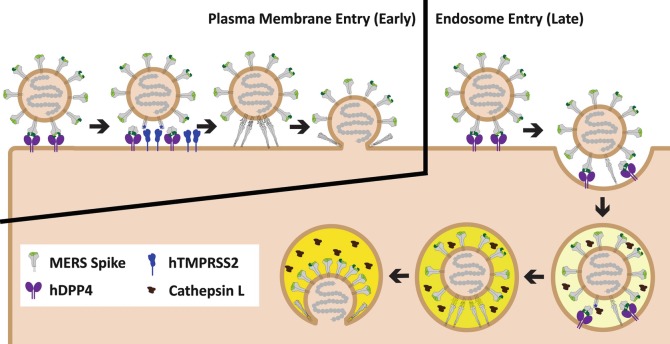
Cellular proteases accumulate in distinct subcellular locations on the endocytic CoV entry pathway: serine proteases such as trypsin and elastase are extracellular; the type II transmembrane serine proteases (TTSPs) are anchored into plasma membranes; and the cysteine-type cathepsin proteases are enriched in endosomes [12, 13]. For several CoV infections, not all of these proteases are required; however, it is possible that each has distinct potential to activate fusion such that a productive infection ensues. There is evidence that particular CoVs have “preferred” in vivo entry routes (e.g., MERS-CoV and 229E-CoV prefer plasma membrane entry, while some MHV-CoVs prefer endosome entry [14–19], (see Fig. 1). Knowledge of these preferred routes, and their relation to virus-induced disease, is necessary to identify virus variants that might have high transmissibility and disease potential, and to recognize the host factors that might be targeted therapeutically such that infections are suppressed at the cell entry stage.
Fig. 1.
MERS-CoV enters host either at or near the plasma membrane or in the endosomes. The MERS-CoV spike (S) proteins (gray) engage human DiPeptidyl Peptidase 4 ( hDPP4, purple) via their receptor-binding domains (green). Receptor engagement exposes protease cleavage sites (blue stars) on S proteins. If cell surface proteases such as hTMPRSS2 (blue) are present, S proteins are cleaved and viral fusion occurs at or near the plasma membrane. If hTMPRSS2 or similar cell-surface proteases are not present, then MERS-CoV is endocytosed, and can be triggered by endosomal proteases such as cathepsin L (brown) to complete viral entry
Here we provide protocols to dissect CoV entry pathways. These include procedures for pseudovirus production, particle purification and concentration, as well as specific assays to differentiate CoV entry pathways. While the protocols are set for characterizing MERS-CoV entry, they can be readily adjusted to evaluate other CoV and other protease-dependent virus entry events.
Materials
Particle Production
150 mm Tissue culture dishes.
HEK-293T cells.
293T cell media: Dulbecco’s Modified Eagle Media (DMEM) with l-glut, 4.5 g/l glucose and 100 mM sodium pyruvate, additional supplements include 10% fetal bovine serum, 10 mM HEPES, 0.1 mM nonessential amino acids, 100 U/ml penicillin G, and 100 μg/ml streptomycin.
Transfection media: DMEM with l-glut, 4.5 g/l glucose and 100 mM sodium pyruvate, and 10% fetal bovine serum.
Serum-free media: DMEM with L-glut, 4.5 g/l glucose and 100 mM sodium pyruvate, additional supplements include 10 mM HEPES, 0.1 mM nonessential amino acids, 100 U/ml penicillin G, and 100 μg/ml streptomycin.
Polyethylenimine (PEI) at 1 mg/ml dissolved in ddH2O.
OptiMEM reduced serum medium.
Expression plasmids for MERS-CoV-spike.
Expression plasmid for HIV core-Fluc (pNL4.3HIVluc).
Transducing particle: VSVΔG-Fluc pseudotyped with Junin virus (JUNV) GP.
Particle Purification and Concentration
Centrifuge: Eppendorf 5810 or equivalent.
Ultracentrifuge: Beckman Coulter’s or equivalent. SW28 swinging-bucket rotor, buckets, and Ultra-Clear tubes.
Falcon 15 and 50 ml conical centrifuge tubes.
Sucrose solution: 20% sucrose (w/v) in serum-free media.
Characterizing Viral Entry Pathways
Falcon 6-well and 96-well cell culture plates.
5x Cell Culture Lysis Reagent (CCLR): 125 mM Tris–HCl pH 7.8, 10 mM DTT , 10 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 50% glycerol, 5% Triton X-100.
Firefly luciferase substrate: 1 mM D-luciferin, 3 mM ATP, 15 mM MgSO4·H2O, 30 mM HEPES [pH 7.8].
Protease inhibitor cocktail: 200 μM Camostat, 20 μM proprotein convertase inhibitor, 20 μM E64D in serum-free media.
Vehicle control: DMSO in serum-free media at equivalent levels to the protease inhibitor cocktail.
CoV fusion antagonists: CoV species-matching HR2 peptides.
Expression plasmids for: hTMPRSS2, hCD9, hIFITM3.
Methods
Carry out all incubations at 37 °C with 5% CO2 unless otherwise specified.
VSV-Based Pseudovirus Production (seeNote 1)
Plate enough 293T cells (5 × 106) in 20 ml into a 15 cm dish to reach 80% confluency on the next day.
On the following day, make transfection mixture by adding 20 μg of MERS-CoV-spike plasmid (see Note 2) and 110 μl of PEI into 2 ml of OptiMEM. Incubate the mixture in the dark for 15 min at room temperature.
Replace existing media with 20 ml of transfection media (pre-warmed to 37 °C, see Note 3). Add transfection mixture dropwise onto the cells. Incubate the cells for 6–8 h (see Note 4).
Replace transfection media with 20 ml of 293T cell media and incubate overnight.
Dilute 100× transducing particle ( VSVΔG-Fluc pseudotyped with Junin virus (JUNV) GP, see Note 5) into 15 ml of pre-warmed serum-free media, which is then used to replace existing media on the transfected cells. Incubate cells for 2 h.
Remove supernatant, rinse cells with 10 ml of pre-warmed serum-free media three times, then add back 13 ml of pre-warmed 293T cell media. Incubate cells overnight.
Collect supernatant (first collection) with a 15 ml Falcon tube, add back 13 ml of pre-warmed 293T cell media, and incubate cells overnight (see Note 6).
Spin supernatant at 300 × g for 10 min at 4 °C.
Transfer supernatant into a fresh tube and spin at 3000 × g for 10 min at 4 °C. Discard pellet.
Transfer supernatant into a fresh tube and freeze it at −80 °C.
On the following day, repeat steps 7–10 (second collection).
On the final day, collect supernatant (third collection), discard cells, repeat steps 8–10.
HIV-Based Pseudovirus Production
Plate enough 293T cells (5 × 106) in 20 ml into a 15 cm dish to reach 80% confluency on the next day.
On the following day, make transfection mixture by adding 10 μg of MERS-CoV-spike plasmid, 10 μg of HIV core-Fluc-expressing plasmid, and 110 μl of PEI into 2 ml of OptiMEM. Incubate the mixture in the dark for 15 min at room temperature.
Replace existing media with 20 ml of transfection media (pre-warmed to 37 °C). Add transfection mixture dropwise onto the cells. Incubate the cells for 6–8 h.
Replace transfection media with 20 ml of 293T cell media and incubate overnight.
Remove supernatant, and add back 13 ml of pre-warmed 293T cell media. Incubate cells overnight.
Collect supernatant (first collection) with a 15 ml Falcon tube, add back 13 ml of pre-warmed 293T cell media, and incubate cells overnight.
Spin supernatant at 300 × g for 10 min at 4 °C.
Transfer supernatant into a fresh tube and spin at 3000 × g for 10 min at 4 °C. Discard pellet.
Transfer supernatant into a fresh tube and freeze it at −80 °C.
On the following day, repeat steps 6–9 (second collection).
On the final day, collect supernatant (third collection), discard cells, repeat steps 7–9.
Particle Purification and Concentration
We noted that pseudoviruses lose their transduction capabilities (up to 90%!) upon exposure to the high g-forces (~100,000 × g) commonly used in traditional viral concentration methods. Therefore, we adopted a low-speed viral concentration and purification protocol that achieves viral concentration without compromising viral transduction capabilities.
Thaw and pool collected pseudovirus-containing supernatants (see Subheadings 3.1 or 3.2).
Transfer 32 ml of the pooled supernatant into a SW28 Ultra-Clear tube.
Use a 3 ml syringe with needle to add a cushion of 3 ml of 20% sucrose to the bottom of the tube. Eject from syringe slowly to avoid sucrose mixing with the sample. After the placement of the cushion, gently add the remaining sample (~3 ml) into the tube. If there is still space, add serum-free media to the brim of the Ultra-Clear tube.
Load the Ultra-Clear tube into a SW28 bucket, and spin at 6500 rpm (5591 × g) for 18 h at 4 °C.
After the spin, carefully take out the Ultra-Clear tube, and remove all the supernatant without disturbing the pellet at the bottom center (may be invisible). Quickly add back ~350 ml of serum-free media, and gently resuspend the pellet with a 1 ml pipette (see Note 8). Aliquot and store the fully resuspended sample (now 100× and purified) at −80 °C for future use.
Characterizing CoV Entry Pathways
Subsequent to receptor engagement, CoV spikes require proteolytic cleavage to trigger membrane fusion. Two classes of cellular proteases have been identified to trigger CoV fusions: Serine proteases that are either secreted or expressed on the plasma membrane [14, 17], and cysteine proteases that resides in the endosome [18, 19]. Therefore, the utilization of specific proteases by a given CoV also dictates its site of entry. We utilize several assays to differentiate the preferred site of entry for wild-type (WT) and mutant MERS spikes, the efficacies of entry inhibitors, and the identification of pro- or antiviral host factors.
Characterizing CoV Entry Kinetics Using Protease Inhibitor Cocktails (seeNote 9)
The CoV site of entry is correlated with entry kinetics, with viral entry at the plasma membrane being “early,” and entry through endosomes “late,” in relation to the different virus trafficking times prior to membrane fusion [15–17]. Together with the knowledge that CoV entry requires proteolysis, we utilize a time-course assay to characterize CoV entry kinetics, where protease inhibitors are added at various time points to arrest future entry, allowing readouts for infection within short inoculation time windows.
Plate sufficient permissive cells into a 96-well plate, 60 μl per well, to reach 95% confluency on the next day.
On the following day, add 40 μl of MERS spike-bearing viral particles (Subheadings 3.1 or 3.2) onto cells. Incubate the plate at 4 °C for 1 h to allow viral binding.
After 1 h, remove unbound particles by aspirating the supernatant (see Note 10). Add back 50 μl of fresh media. Incubate the plate at 37 °C.
At various time intervals (0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 60 min, and so on, see Note 11), add 50 μl of protease inhibitor cocktail to the cells. At time interval of 0 min, also have a condition where 50 μl of vehicle control (see Note 12) is added instead. Leave the drugs/vehicle on cells and incubate at 37 °C overnight (~18 h post viral inoculation).
Remove media, add 50 μl of 1×CCLR (5x CCLR diluted in ddH2O) to lyse cells. Freeze the plate at −80 °C for 30 min (see Note 13). Thaw the plate and transfer 20 μl to a white reading plate to analyze firefly luciferase activity. Normalize enzyme activity from all conditions to the vehicle control, which is set to “100%”. Plot data as “% viral entry.”
Characterizing Entry Routes Using Specific Protease Inhibitors
The entry assay (Subheading 3.4.1) can sensitively distinguish viruses with accelerated or delayed kinetics, but it is frequently time and reagent consuming. The assay can be simplified by an alternative where titration of a particular inhibitor is applied for a short period of time.
Plate sufficient permissive cells into a 96-well plate, 60 μl per well, to reach 95% confluency on the next day.
On the following day, at −1 h, remove media and add 60 μl of protease inhibitor or vehicle-containing serum-free media (1–1000 μM camostat to inhibit plasma membrane protease TMPRSS2, or 1–1000 μM E64D to inhibit endosomal cathepsins). Incubate cells for 1 h at 37 °C.
Add 40 μl of MERS spike-bearing viral particles (Subheading 3.1 or 3.2) onto cells. Incubate the plate at 37 °C for 2 h to allow viral entry.
After 2 h, remove unbound particles by aspirating the supernatant. Rinse twice with 100 μl PBS. Add back 50 μl of fresh media. Incubate the plate at 37 °C overnight (~18 h post viral inoculation).
Remove media, add 50 μl of 1×CCLR to lyse cells. Freeze the plate at −80 °C for 30 min. Thaw the plate and transfer 20 μl to a white reading plate to analyze firefly luciferase activity. Normalize enzyme activity from all conditions to the vehicle control, which is set to “100%.” Plot data as “% viral entry.”
Characterizing Entry Routes Using Spike Fusion Antagonists
The protocol described in Subheading 3.4.2 is flexible and can be tailored to evaluate different viral inhibitors. These include fusion inhibitors. CoV spike proteins facilitate membrane fusion by transiting from “ extended fusion intermediates” to “ post-fusion” conformations. This transition requires interactions between antiparallel helices termed heptad repeat region 1 (HR1) and 2 (HR2) [20, 21], and can be arrested by exogenous HR2 peptides, which bind to the fusion intermediates. Typically, HR2 peptides do not enter endosomes and therefore only arrest viruses that transition into intermediate conformations extracellularly, i.e., at target cell plasma membranes. However, lipid-conjugated HR2 peptides can bind plasma membranes and endocytose, accumulating in endosomes such that they will arrest intracellular virus-cell membrane fusion. Using a modified Subheading 3.4.2, we tested the efficacy of native vs. lipid-conjugated HR2 on blocking CoV endosomal entry [22].
Plate sufficient permissive cells into a 96-well plate, 60 μl per well, to reach 95% confluency on the next day.
On the following day, at −1 h, remove media and add 60 μl of native vs. lipid-conjugated HR2 peptides at 0.01–1 μM, or vehicle control. Incubate cells for 1 h at 37 °C.
Add 40 μl of MERS spike-bearing viral particles (Subheadings 3.1 or 3.2) onto cells. Incubate the plate at 37 °C for 1 h to allow viral entry.
After 1 h, remove unbound particles by aspirating the supernatant. Rinse twice with 100 μl PBS. Add back 50 μl of fresh media. Incubate the plate at 37 °C overnight (~18 h post viral inoculation).
Remove media, add 50 μl of 1x CCLR to lyse cells. Freeze the plate at −80 °C for 30 min. Thaw the plate and transfer 20 μl to a white reading plate to analyze firefly luciferase activity. Normalize enzyme activity from all conditions to the vehicle control, which is set to “100%.” Plot data as “% viral entry.”
Identifying Host Factors Participating in MERS-CoV Entry
With the ability to differentiate between MERS-CoV entry at the plasma membrane and within endosomes, we and others have identified several host factors that affect viral routes of entry [14, 15, 17, 23, 24]. These include but are not limited to: (1) transmembrane protease serine subtype 2 (hTMPRSS2), which facilitates MERS-CoV entry at the plasma membrane. (2) tetraspanin hCD9, which ferries the MERS-CoV receptor hDPP4 into close proximity with hTMPRSS2 to potentiate MERS-CoV entry at the plasma membrane. (3) interferon-induced transmembrane protein 3 (hIFITM3), which blocks CoV endosomal entry.
Plate sufficient permissive cells into a 6-well plate, 2 ml per well, to reach 85% confluency on the next day.
- On the following day, transfect cells with expression plasmids for vector control, hTMPRSS2, hCD9, or hIFITM3 (see Note 14):
- Make transfection mixture by adding 1 μg of plasmid and 12 μl of PEI into 200 μl of OptiMEM. Incubate the mixture in the dark for 15 min at room temperature.
- Replace existing media with 2 ml of transfection media (pre-warmed to 37 °C). Add transfection mixture dropwise onto the cells. Incubate the cells for 6–8 h.
- Replace transfection media with 2 ml of fresh media and incubate overnight.
On the next day, plate sufficient transfected cells into a 96-well plate, 60 μl per well, to reach 95% confluency on the next day.
Add 40 μl of MERS spike-bearing viral particles (Subheadings 3.1 or 3.2) onto cells. Incubate the plate at 4 °C for 1 h to allow viral binding.
After 1 h, remove unbound particles by aspirating the supernatant. Add back 50 μl of fresh media. Incubate the plate at 37 °C.
At various time intervals (0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 60 min, and so on), add 50 μl of protease inhibitor cocktail to the cells. At time interval of 0 min, also have a condition where 50 μl of vehicle control is added instead. Leave the drugs/vehicle on cells and incubate at 37 °C overnight (~18 h post viral inoculation).
Remove media, add 50 μl of 1×CCLR (5×CCLR diluted in ddH2O) to lyse cells. Freeze the plate at −80 °C for 30 min. Thaw the plate and transfer 20 μl to a white reading plate to analyze firefly luciferase activity. Normalize enzyme activity from all conditions to the vehicle control, which is set to “100%.” Plot data as “% viral entry.”
Notes
The current protocol describes particle production in 15-cm diameter plates. This protocol can be scaled up or down as long as the ratio of all components remains constant. However, for consistency and reproducibility we do not recommend producing particles in container sizes smaller than 5-cm diameter.
When using PEI as a transfection reagent, use 1 μg of DNA per million cells.
293T cells can detach from the plate, so exert care when replacing media. Always add back media after aspiration as soon as possible to prevent cell-drying. When adding media, liquid should land on the side wall of the plate, not directly onto the cells. Always pre-warm media.
The DNA:PEI ratio (1:~6) is optimized specifically for a transfection period of 6–8 h. If transfection is allowed to go overnight, lower the DNA:PEI ratio to 1:3.
JUNV GP-vsvΔG pseudovirus was chosen for its short half-life, which reduces the amount of inoculum JUNV GP pseudovirus contaminating the desired CoV-spike- vsvΔG.
This protocol maximizes pseudovirus production by taking three consecutive harvests from the producer cells (24–48, 48–72, and 72–96 h post-transfection). Each harvest period brings similar pseudovirus yields.
At the indicated spin speed, the k-factor is 4556. Since the sedimentation coefficients of VSV [25] or HIV [26] pseudovirus are around 500, a 9-h spin is sufficient. The actual spin time is doubled to insure that the pseudoviruses are pelleted through the more viscous 20% sucrose cushion.
When resuspending pellet, set 1 ml pipette to 200 μl and pipette up and down gently to avoid bubbles. Make sure to rinse the entire bottom of the tube to maximize collection.
The current protocol describes viral entry into cells seeded in 96-well plates. This protocol can be scaled up as long as the ratio of all components remains constant.
After removing media via aspiration, return fresh media to cells as soon as possible to prevent cell drying. Aspirate and return media from a maximum of 24 wells at a time.
CoV entry kinetics are unique to each virus and host cell combination. In LET-1 cells, MERS-CoV entry completes in around 1 h. In HeLa cells, 229E-CoV entry takes 4+ hours to complete. Pilot experiments with larger time intervals (30–60 min) are recommended.
The vehicle control for protease inhibitors is DMSO, which is cytotoxic at high concentrations. Pilot experiments are recommended to identify nontoxic DMSO concentrations.
Samples can be kept at −80 °C for up to a month before reading Fluc. To further prevent signal loss, protease inhibitor cocktail can be added to 1x CCLR before use.
For cell types that are killed by PEI:DNA transfection, lipid-based transfection vectors such as lipofectamine can be used instead. For cell types that are resistant to transfection, retro- or lenti-viral-based or adenovirus-based transduction systems can be used to introduce genes of interest [15].
Acknowledgment
This work is supported by NIAID grant AI060699 and by the Loyola University Chicago Center for Translational Research and Education.
Contributor Information
Rahul Vijay, Email: rahul-vijay@uiowa.edu.
Tom Gallagher, Email: tgallag@luc.edu.
References
- 1.Middle East respiratory syndrome coronavirus (MERS-CoV). https://www.who.int/en/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov). Accessed 4 Mar 2019
- 2.Sabir JSM, Lam TTY, Ahmed MMM, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 3.Reusken CBEM, Haagmans BL, Müller MA, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haagmans BL, Al Dhahiry SHS, Reusken CBEM, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memish ZA, Mishra N, Olival KJ, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilardi K, Byarugaba DK, Baric RS, et al. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. MBio. 2017;8:1–13. doi: 10.1128/mbio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(2019)WHO | Middle East respiratory syndrome coronavirus (MERS-CoV). In: WHO. https://www.who.int/emergencies/mers-cov/en/. Accessed 4 Mar 2019
- 8.Fouchier RAM, van Boheemen S, Osterhaus ADME, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- 9.Hijawi B, Abdallat M, Sayaydeh A, et al. Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. East Mediterr Health J. 2013;19(Suppl 1):S12–S18. doi: 10.26719/2013.19.supp1.S12. [DOI] [PubMed] [Google Scholar]
- 10.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulswit RJG, de Haan CAM, Bosch B-J. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugge TH, Antalis TM, Wu Q. Type II Transmembraneserine proteases. J Biol Chem. 2009;284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J, Li K, Barlan A, et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Earnest JT, Hantak MP, Li K, et al. Thetetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog. 2017;13:e1006546. doi: 10.1371/journal.ppat.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li K, Wohlford-Lenane CL, Channappanavar R, et al. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirato K, Kanou K, Kawase M, Matsuyama S (2017) Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J Virol 91. 10.1128/JVI.01387-16 [DOI] [PMC free article] [PubMed]
- 18.Qiu Z, Hingley ST, Simmons G, et al. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J Virol. 2006;80:5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama S, Taguchi F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J Virol. 2009;83:11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch BJ, Martina BEE, Van Der Zee R, et al. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci U S A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosch BJ, Rossen JWA, Bartelink W, et al. Coronavirus escape from heptad repeat 2 (HR2)-derived peptide entry inhibition as a result of mutations in the HR1 domain of the spike fusion protein. J Virol. 2008;82:2580–2585. doi: 10.1128/JVI.02287-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JE, Gallagher T. Lipidation increases antiviral activities of coronavirus fusion-inhibiting peptides. Virology. 2017;511:9–18. doi: 10.1016/j.virol.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrensch F, Winkler M, Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6:3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Sehgal M, Hou Z, et al (2017) Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J Virol 92. 10.1128/JVI.01535-17 [DOI] [PMC free article] [PubMed]
- 25.Bradish C. J., Brooksby J. B., Dillon J. F. Biophysical Studies of the Virus System of Vesicular Stomatitis. Journal of General Microbiology. 1956;14(2):290–314. doi: 10.1099/00221287-14-2-290. [DOI] [PubMed] [Google Scholar]
- 26.Vogt V (1997) Retroviral Virions and Genomes. Cold Spring Harbor Laboratory Press [PubMed]