Abstract
Novel treatments for glioblastoma (GBM) are urgently needed, particularly those which can simultaneously target GBM cells’ ability to grow and migrate. Herein, we describe a synthetic, bioreducible, biodegradable polymer that can package and deliver hundreds of siRNA molecules into a single nanoparticle, facilitating combination therapy against multiple GBM-promoting targets. We demonstrate that siRNA delivery with these polymeric nanoparticles is cancer-selective, thereby avoiding potential side effects in healthy cells. We show that we can deliver siRNAs targeting several anti-GBM genes (Robol, YAP1, NKCC1, EGFR, and survivin) simultaneously and within the same nanoparticles. Robol (roundabout homolog 1) siRNA delivery by biodegradable particles was found to trigger GBM cell death, as did non-viral delivery of NKCC1, EGFR, and survivin siRNA. Most importantly, combining several anti-GBM siRNAs into a nanoparticle formulation leads to high GBM cell death, reduces GBM migration in vitro, and reduces tumor burden over time following intratumoral administration. We show that certain genes, like survivin and EGFR, are important for GBM survival, while NKCC1, is more crucial for cancer cell migration. This represents a powerful platform technology with the potential to serve as a multimodal therapeutic for cancer.
Keywords: nanoparticle, siRNA, combination therapy, cancer therapy, gene therapy
Graphical Abstract
Bioreducible polymeric nanoparticles encapsulating siRNA combinations are successfully delivered to human glioblastoma cells in vitro and in vivo. The delivered siRNA molecules, including Survivin, EGFR, NKCC1, YAP1, and Robo1, result in tumor cell death, reduced tumor migration in vitro, and reduced tumor burden over time in vivo while avoiding major effects on healthy brain cells.
1. Introduction
Glioblastoma (GBM) is the most malignant primary human brain cancer, and improved treatments for this disease are needed.[1–7] Increasing evidence has shown that a subset of GBM cells have stem-like properties and the ability to initiate tumors.[8, 9] These cells, being resistant to conventional treatments, can migrate away from the primary tumor mass and form new tumors, which is thought to be responsible, in part, for the nearly inevitable recurrence of GBM despite aggressive treatment.[10, 11] A novel treatment modality with potential for clinical success is the use of RNA interference (RNAi), a natural cellular process that suppresses gene expression with high sequence specificity following introduction of short interfering RNA (siRNA) into cells.[12] By knocking down the expression of genes that promote cancer cell survival, migration, and tumorigenicity, siRNA can be a powerful tool for treating GBM.
However, siRNA must reach the cytoplasm of cells to be effective, and delivering these oligonucleotides into cells safely, efficiently, and in combination is challenging. Viruses have historically been used as very efficient nucleic acid delivery vectors, but the toxicity and immunogenicity associated with viruses hinders their clinical use. Also, viral vectors are generally limited to the delivery of only one siRNA sequence per virus particle due to size constraints on the nucleic acid cargo.[13] In previous work, we have developed a synthetic bioreducible poly(beta-amino ester) (PBAE) that is highly effective at delivering siRNA to GBM cells.[14] Because this PBAE nanoparticle is degradable by hydrolysis as well as by bioreduction in the relatively reducing cytoplasmic environment, it exhibits low non-specific toxicity. Importantly, the cationic PBAE forms electrostatic complexes with negatively charged cargo like siRNA spontaneously in aqueous conditions. We have also found that each nanocomplex contains many copies of siRNA, thus allowing multiple siRNA molecules, including multiple different siRNA sequences, to be delivered simultaneously in the same particle targeting various genes.[15]
siRNA is promising as a therapeutic to treat brain cancer for several reasons: i) it can hit targets that are seen as “undruggable” by conventional medicinal approaches; ii) it has the potential of overcoming drug resistances by affecting multiple disease-causing biochemical pathways in parallel; iii) it does not necessarily require reformulation of a drug delivery particle when the type of siRNA cargo changes; and, unlike DNA, iv) it does not risk introducing inheritable genetic changes. Protein targets of particular interest to us include Roundabout homolog 1 (Robol), a protein identified as key to normal and cancer cell migration;[16–18] yes-associated protein 1 (YAP1), which promotes growth of GBM cells;[19, 20] sodium-potassium-chloride cotransporter (NKCC1), an ion transporter that affects cancer cell migration;[21, 22] survivin, an anti-apoptotic gene whose expression in GBM correlates with proliferation[23] and with poor prognosis;[24] and endothelial growth factor receptor (EGFR), an oncogene whose aberrant expression is among the most common mutations in GBM.[25–27] Other reports of knockdown of Robo1,[18] YAP1,[28] NKCC1,[21, 29] survivin,[30] or EGFR[31] often focus on knockdown of a single gene to isolate cellular mechanisms of GBM survival, growth, and invasion. Of the studies that have used RNAi against one of these targets as a therapeutic strategy, to our knowledge, this is the first report of a delivery system that causes knockdown of all five of these targets at once for GBM therapy.
In this study, we design and use bioreducible PBAE nanoparticles to deliver siRNA to primary human GBM cells in vitro and in vivo and show both effective gene knockdown and also the functional effect of reducing cancer cell viability. For the first time, the potential to deliver five separate siRNAs within a single type of PBAE nanoparticle has been validated, opening up new avenues for combination cancer therapy. We show the efficacy of this treatment strategy in vivo by using the PBAE to intratumorally administer a combination of siRNAs to an orthotopic brain tumor, causing a reduction in the tumor burden compared to the control. This strategy may serve as a widely applicable platform to treat many different types of cancer that are otherwise refractory to treatment.
2. Results and Discussion
2.1. Characterization of R646/siRNA nanoparticles
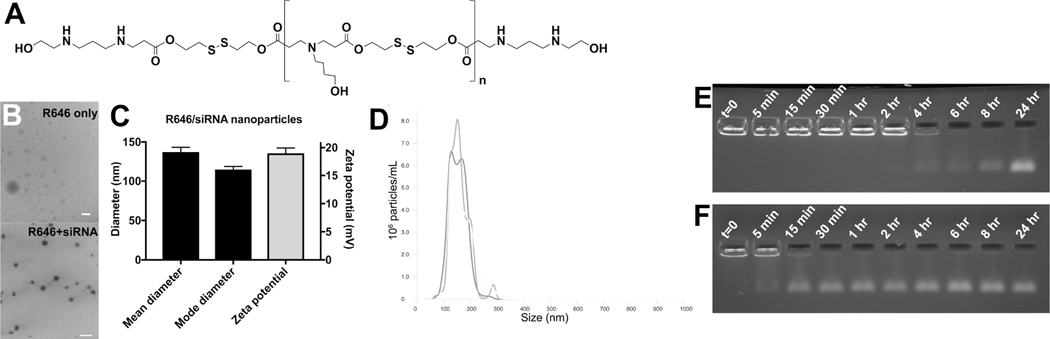
Bioreducible PBAEs were synthesized via polymerization of monomer bis(2-hydroxyethyl) disulfide (BR6) with monomer 4-amino-1-butanol (S4), followed by polymer end-capping with 2-((3-aminopropyl)amino)ethanol (E6), to yield polymer BR6-S4-E6 (R646), based on the bioreducible linear BR6-S4 polymer that we found promising for oligonucleotide delivery.[32] R646 (Figure 1A) spontaneously self-assembled into nanoparticles when mixed with siRNA via electrostatic interactions. Nanoparticle tracking analysis (NTA) showed that the mean and mode hydrodynamic diameter of the nanoparticles in PBS buffer was 137 ± 6 nm and 115 ± 4 nm, respectively (mean ± standard deviation of n=3 particle batches), indicating that the particles are within the ~100 nm size range conducive to cellular uptake via endosomal engulfment.[33] This measurement was verified by transmission electron microscopy (TEM) (Figure 1B), with the dry diameter of the nanoparticles being slightly smaller than the hydrodynamic diameter measured by NTA. Without siRNA being present, the homogenous spherical polyplex nanoparticles did not form (Figure 1B). The zeta potential of the R646 siRNA nanoparticles was positive, measured at 18±1 mV (mean ± standard deviation of n=3 particle batches) (Figure 1C). This is expected due to the cationic nature of R646, as is typical of PBAE-based nanoparticles, and may aid in facilitating particle uptake by cells. Particle size distribution of the self-assembled R646 siRNA NPs was found to be approximately monodisperse (Figure 1D).
Figure 1. Physicochemical characterization of R646/siRNA nanoparticles.
siRNA-containing nanoparticles (NPs) based on (A) polymer structure R646 are of (B) Spherical morphology, (C) Small ~100 nm size, and have positive zeta potential. (D) Particle size distribution of the self-assembled R646 siRNA NPs is approximately monodisperse. Gel retention assay results of siRNA release from NPs over time show (E) Persistence of the NPs over longer times in aCSF, mimicking the extracellular space, compared to (F) Faster release of siRNA from the NPs in conditions mimicking the cytosol with 5 mM GSH. Scale bars = 200 nm.
siRNA is hypothesized to be released upon degradation of the PBAE. Because R646 contains both ester and disulfide bonds, its degradation can be caused by either hydrolysis in aqueous conditions or reduction in reducing environments. A gel retardation assay was used to assess the rate of siRNA release from particles after incubation in an aqueous, physiological salt solution with or without the reducing agent glutathione (GSH). siRNA still bound to the PBAE would be unable to move through an agarose gel, whereas a distinct band in the gel indicates siRNA release from the polymer. As can be seen in Figure 1E, R646/siRNA particles incubated at physiological temperature in artificial cerebrospinal fluid (aCSF) did not begin to release siRNA until 4 hr, and release was not complete until >8 hr. On the other hand, R646/siRNA incubated in PBS with 5 mM GSH showed some siRNA release almost immediately, and release appeared to be complete after approximately 30 min (Figure 1F).
Because all of the siRNA is complexed with the R646 polymer upon nanoparticle formation (Figure 1E,F), by measuring the concentration of nanoparticles in the suspension, it is possible to calculate the average number of siRNA molecules per particle using NTA. [34] As 720 nM RNA was encapsulated by the particles and the average concentration of particles was 2.9±0.2 ×1011 particles/mL, each particle contained approximately 1520 ± 90 siRNA molecules.
2.2. siRNA delivery to human GBM and healthy brain cells
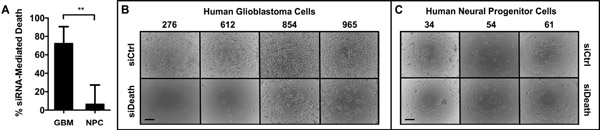
To verify that R646 could deliver siRNA into cells and enable gene knockdown, we formed nanoparticles with R646 and AllStars Human Cell Death Control siRNA (siDeath), a blend of siRNA oligos that kill human cells upon successful intracellular delivery. We used a scrambled-sequence control (scRNA) as our negative control RNA. Cells were treated with either R646/siDeath or R646/scRNA nanoparticles, and transfection efficacy was measured as the difference in cell death between the two groups. Primary human GBM cells obtained intraoperatively from four separate patient samples served as our GBM cell model, and healthy neural progenitor cells (NPCs), obtained from three separate primary human tissue samples served as our non-cancer cell model.[35–38] As shown in Figure 2A–B, GBM cells treated with R646/siDeath showed significantly lower viability after five days than GBM cells treated with the R646/scRNA control. Despite heterogeneity among the GBM tissue samples, the response we observed was consistent, with 72 ± 9% of GBM cells killed by siRNA-induced gene knockdown.
Figure 2.
Bioreducible PBAE nanoparticles selectively transfect primary human GBM cells versus primary human neural progenitor cells (NPCs) (A) siRNA-mediated death 5 days following transfection using a death positive control siRNA shows significantly more cell killing in the (B) Four GBM cell samples tested versus (C) Three NPC cell samples tested with low non-specific toxicity due to the polymer. Scale bars = 100 μm.
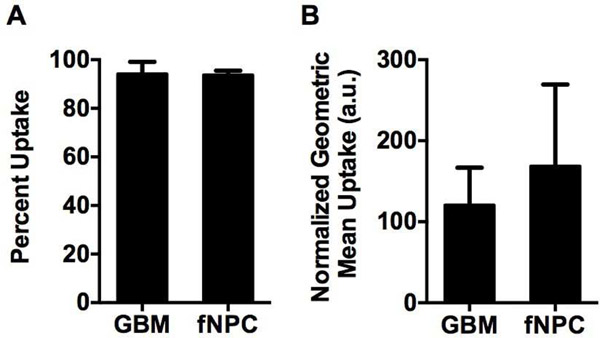
Importantly, there was significantly (p<0.01) less siRNA-induced cell death seen in the transfected NPCs (Figure 2A, C). Interestingly, this difference between GBM and healthy cells does not appear to be due to differences in the overall nanoparticle uptake efficiency. Figure 3 shows that 94 ± 2% of GBM cells and 94 ± 1% of NPCs internalized particles (Figure 3A). There was also no statistically significant difference in the amount of particles taken up per cell, measured as the mean fluorescence intensity per cell (120 ± 20 vs. 170 ± 60 normalized fluorescence units in GBM cells and NPCs, respectively) (Figure 3B). While this does not account for potentially different routes of particle uptake between the cell types,[39] it indicates that total nanoparticle uptake by cells is not the limiting factor to successful transfection in this system.
Figure 3.
Nanoparticle uptake is not statistically different between primary human GBM cells and NPCs. (A) The percent of cells positive for fluorophore-labeled siRNA is not statistically different between GBM and NPC samples. (B) The geometric mean fluorescence of fluorophore-labeled siRNA within each cell, a measure of nanoparticles per cell, is not statistically different between the GBM and NPC samples.
2.3. Knockdown of therapeutically relevant genes
Moving forward, we completed all in vitro experiments with patient-derived primary human GBM cell line 612, as we found this cell line to have the highest transfection efficacy in our previous experiment (Figure 2). Additionally, GBM line 612 has been shown to be highly migratory, a trait which has been shown to negatively correlate with positive patient outcomes.[21, 27, 40, 41] Cells were transfected with siRNA oligos targeting Robo1, YAP1, NKCC1, survivin, and EGFR. Our first goal was to determine the timeline of gene knockdown. We separately targeted Robo1, YAP1, and NKCC1, using scRNA as a negative control, and harvested cells at days 3 – 7 post-transfection for analysis via Western blotting (Supplementary Figure S1). For all targeted genes, we saw a decrease in protein expression vs. scRNA over all days tested, and for all further Western blotting analyses, cells were harvested at day 3 post-transfection. For all 5 gene targets, we then needed to determine which siRNA oligo would lead to the strongest decrease in protein expression. Each siRNA oligo targets only a region of a gene’s mRNA transcript, and mutations and splice variants can lead to oligos being ineffective in some cell types. Using scRNA as a negative control and a blend of three siRNA oligo variants as a positive control, we were able to determine which particular oligo was most effective, and then to use only this one for all future experiments (Supplementary Figure S2, Supplementary Table S1).
We completed a dose-response test for siRNAs individually, targeting either Robol, YAP1, or NKCC1 and analyzed the reduction in protein expression via Western blotting (Supplementary Figure S3). In this experiment, the total siRNA dose was maintained at 120 nM, but the percentage containing targeting siRNA was varied, using scRNA to fill the remaining dose. For all gene targets, we found protein expression reduction when 10% of the dose contained targeting siRNA to be equal to the reduction when 100% of the dose was targeting siRNA. We also found some reduction in protein expression when 5% and even 1% of the dose was targeting siRNA, versus 0% (scRNA only). This indicated that 90% of our deliverable siRNA was in biological excess for gene knockdown, and led us to test the hypothesis that we could knockdown more than one gene simultaneously.
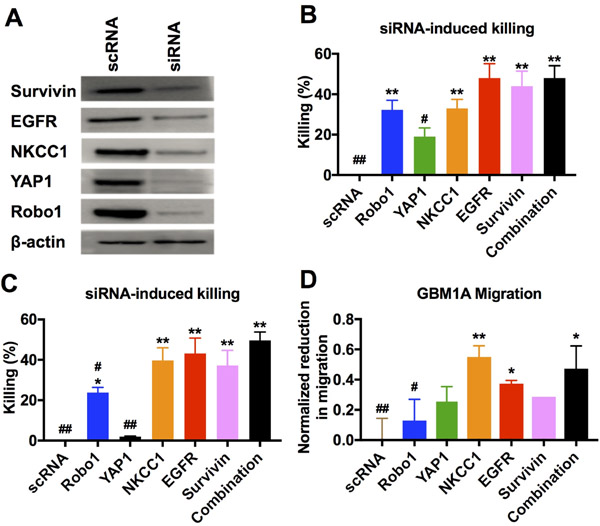
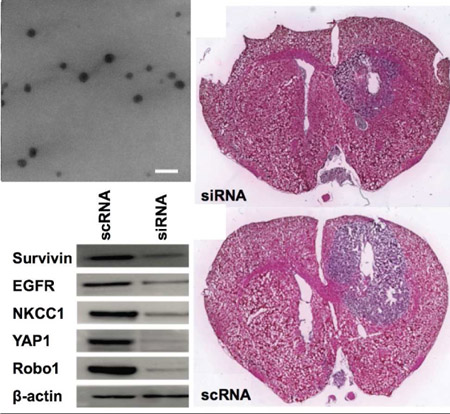
For functional analysis of GBM behavior both in vitro and in vivo, and to demonstrate robustness of the nanoparticles, we performed further experiments using patient-derived primary human GBM cell line GBM1A. GBM1A, which is capable of forming measurable mass-effect tumors when injected orthotopically, mimics what is seen in the clinic and is complimentary to GBM line 612. GBM1A cells were transfected with a mixture of five siRNA oligos targeting Robo1, YAP1, NKCC1, survivin, and EGFR. Western blotting showed simultaneous knockdown of all five genes in transfected cells compared to cells treated with the scRNA control (Figure 4A). In addition, transfection with each siRNA sequence separately as well as the mixture of all five siRNA sequences caused a decrease in the viability of GBM1A cells compared to the scRNA-treated controls (Figure 4B). It should be noted that the cells transfected with all five siRNAs received a 120 nM total siRNA dose, or 24 nM of each siRNA, and the cells transfected with only a single siRNA sequence received the relatively higher dose of 120 nM of only that single siRNA. The effect of the combination therapy was even more pronounced when comparing 24 nM dosage of each individual siRNA to 24 nM of all siRNAs in combination (Figure 4C).
Figure 4.
(A) Primary human GBM cells GBM1A transfected with a single formulation of bioreducible R646 nanoparticles containing a mixture of five siRNA sequences showed simultaneous knockdown of all five targeted genes. (B) Cells transfected with 5x doses of each individual siRNA sequence showed reduced viability compared to control cells as did the siRNA mixture combination condition. (C) Reducing the functional siRNA dose while keeping the total siRNA amount the same by mixing in control scRNA results in an even more pronounced effect of the combination therapy. (D) Cells transfected with certain single siRNAs showed reduced migration capacity compared to control cells. Cells transfected with the mixture condition of all five siRNA sequences in combination showed optimal cell killing with a strong reduction in migration capacity. For B-D, a one-way ANOVA with post-hoc Dunnett’s test was used to determine statistically significant differences from the scRNA control (*p<0.05; **p<0.01) or the combination treatment (#p<0.05, ##p<0.01). A Bonferroni correction was used for the separate comparisons.
The viability of the cell line tested here, GBM1A, was sensitive to the knockdown of certain genes, particularly EGFR and survivin, while knockdown of some, like YAP1, had only a modest effect on GBM1A viability; interestingly, however, all five sequences in combination (24 nM each) had a statistically similar (p>0.05) effect on GBM1A cells to that of a 5-fold higher dose (120 nM) of EGFR or survivin knockdown alone. Similarly, certain genes, particularly NKCC1 and EGFR, played in important role in GBM1A migration while others, like Robo1 and survivin, had little or no effect. Transfection with a mixture of all five genes resulted in decreased migration statistically similar to that seen with NKCC1, the siRNA that had the greatest effect on migration individually (Figure 4D). This highlights the benefit of our combination delivery system, in that we are able to target multiple pathways of GBM malignancy simultaneously and without a loss in efficacy of each individual siRNA. The independence of each siRNA’s efficacy could be useful in tailoring siRNA combinations to target specific tumor subtypes with different behaviors or gene expression profiles.
2.4. In vivo efficacy of siRNA delivered in combination via PBAEs
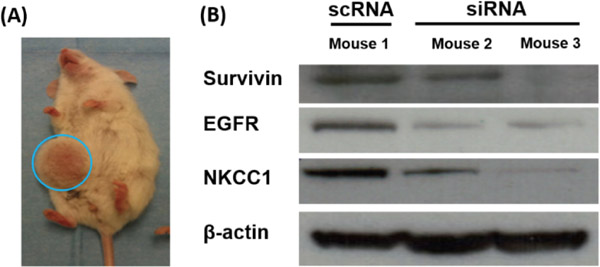
In addition to verifying that in vitro delivery of siRNA combinations could cause human glioma cell death, it was important to evaluate if this strategy could also be effective in vivo. We first used a flank tumor model for ease of tumor access for protein analysis. As shown in Figure 5, co-delivery of siRNAs against survivin, EGFR, and NKCC1 in combination resulted in simultaneous in vivo knockdown of all three proteins, measured by Western blotting, demonstrating that these nanoparticles can successfully transfect tumor cells in vivo.
Figure 5.
Simultaneous knockdown of three genes following in vivo transfection of primary human GBM in a subcutaneous tumor model with a single formulation of bioreducible PBAE nanoparticles.
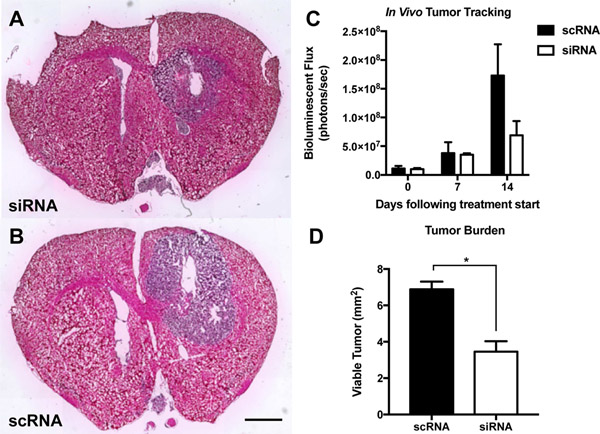
In addition to the subcutaneous model, we also established a more clinically relevant human orthotopic GBM1A tumor model in mice and intratumorally injected nanoparticles to co-deliver all five functional siRNA sequences (siRobo, siYAP1, siNKCC1, siEGFR, and siSurvivin) simultaneously (Figure 6). In previous work, we have demonstrated that intratumoral injection of R646 RNA nanoparticles into orthotopic brain tumors results in nanoparticle penetration throughout the tumor tissue.[42] In this work, our goal was to change the malignant, proliferative phenotype of these cells to lead to either tumor death or slowed tumor growth. Using an In Vivo Imaging System (IVIS) and systemically administered luciferin, we tracked the size of the luciferase-positive tumor over time and also performed histology on excised brains 14 days after the start of treatment to measure differences in the tumor burden among the groups. Tumors treated with the five siRNA sequences were less luminescent than those treated with control scRNA (7 ± 3 ×107 photons/sec flux vs. 17 ± 5 ×107 photons/sec, respectively, Figure 6C). Image analysis of histological sections also showed significantly (p<0.05) lower tumor burden in combination siRNA-treated mice (3.5 ± 0.6 mm2) than in scRNA-treated mice (6.9 ± 0.4 mm2) (Figure 6A–B, D).
Figure 6.
PBAE nanoparticle co-delivery of five siRNAs reduced tumor growth in an orthotopic model of human GBM. Hemotoxylin- and eosin-stained histological slides of siRNA treated tumors (A) show reduced tumor burden versus (B) scRNA treated tumors. (C) IVIS tracking of luciferase-positive GBM tumors shows inhibited tumor growth in siRNA-treated tumors. (D) Quantification of siRNA-mediated reduction in tumor burden (n = 3 per group). Scale bar = 2 mm.
2.5. Discussion
This study demonstrates the ability of bioreducible PBAE-based nanoparticles to deliver functional siRNA to GBM cells both in vitro and in vivo, resulting in tumor cell death or slowed growth while avoiding major effects on healthy brain cells. The bioreducible, disulfide-containing PBAE R646 is of particular interest as an siRNA delivery agent, as it is achieves siRNA release specifically in the cytosol of cells, which is roughly three orders of magnitude more reducing than the extracellular space.[43] This targeted release is thought to improve siRNA delivery efficacy while also preventing any unwanted toxicity from the polymer by promoting its quick degradation inside the cell.
The specificity for human primary brain cancer cells over human primary healthy neural cells highlights one of the advantages of PBAE nanoparticles and could greatly improve nanotherapeutics for cancer by minimizing adverse side effects to off-target cells. This is in agreement with recent results showing that PBAE/nucleic acid nanoparticles typically exhibit high specificity for cancer cells over healthy cells[15, 44, 45] and makes this technology attractive for in vivo cancer treatment.
Polymer-based synthetic delivery systems are also advantageous versus traditional nucleic acid delivery vectors like viruses, which can generally carry only one nucleic acid sequence per particle. As we showed here, approximately 1500 siRNA molecules are estimated to be packaged into a single PBAE/siRNA nanoparticle, and thus, many copies of different types of siRNA can be complexed together within a single particle. This opens the possibility of combination siRNA therapy, which may be crucial for effective cancer treatment, and overcoming tumor resistances, allowing this technology to target multiple different pathways relevant to tumor cell proliferation and malignancy. Even within a single patient, tumors often comprise a highly heterogeneous population of malignant cells with a range of different mutations. Thus, knocking down multiple genes or pathways at the same time can help ensure that few or no GBM cells would evade the therapeutic effect of the nanoparticles.
Multimodal agent delivery is a particularly attractive approach to treat diseases like GBM, which have heterogeneous characteristics between patients and even within the same tumor. [46] Our results showing codelivery of siRNAs targeting GBM-promoting genes such as YAP1, NKCC1, survivin, Robol and EGFR suggest that while some sequences are effective in causing cell death and others decrease tumor cell migration, all five sequences in combination are effective at disrupting multiple pathways in parallel. This is particularly relevant to target the “go or grow” aspect of cancer that describes how the proliferation and migration behaviors of cancer cells are inversely proportional, and when one of the two is targeted individually, the other one is enhanced. [47] This also broadens the potential utility of combination siRNA delivery, as different patients’ tumors may have different sensitivities to knockdown of specific genes and drug resistances. In this study, we chose five gene targets known to promote GBM proliferation and migration to show that combination delivery enables multi-gene knockdown and yields simulataneous reduction in both of these behaviors. Our results suggest that this codelivery system could work with theoretically any combination of siRNAs. A combination of multiple siRNA sequences can therefore be delivered to each patient to hit multiple targets simultaneously without significantly lessening the effect of each sequence and working effectively in a wide array of heterogeneous tumors. This would allow for the delivery of patient-specific siRNA combinations that are tuned to the gene expression profile of individual GBMs. As GBM is often heterogeneous even within single tumors, this also would allow us to use a single formulation to deliver siRNAs that would target aberrant behavior within differing tumor subpopulations. Additionally, while 120 nM of siRNA was delivered under the optimized transfection conditions, our dose-response experiment showed that up to 90% of the total siRNA could be replaced with scrambled control scRNA without affecting the extent of protein knockdown (Supplementary Figure S3), suggesting that potentially 10 siRNA sequences can be mixed and formulated into the same particles without compromising the effect of any individual sequence.
Combination siRNA delivery via bioreducible PBAE nanoparticles is a promising strategy for the treatment of glioblastoma. This advanced therapeutic strategy of delivering YAP1, NKCC1, survivin, Robol, and EGFR siRNA together for intracellular delivery is complementary to recent advancements in the field. For example, delivery of lipopolymeric nanoparticles have recently been shown encapsulating siRNAs against the four transcription factors SOX2, OLIG2, SALL2, and POU3F2 to stop the growth of brain tumor-initiating cells.[48] In contrast, we demonstrate the potential of bioreducible non-lipid nanoparticles that may be safer due to their quick environmentally-triggered biodegradation and their less hydrophobic character. Our work also demonstrates the potential of targeting non-canonical targets with non-viral combination nanoparticles. Our combination approach may also be able to be extended to polymeric nanoparticles that contain DNA[45] and other RNAs in addition to siRNA[42] in order to have a multimodal cancer pathway reprogramming effect.
3. Conclusions
This study describes an advanced therapeutic platform the delivery of combination siRNA molecules intracellularly to the cytosol of tumor cells. Bioreducible PBAE-based nanoparticles are used to deliver five anti-GBM genes (Robol, YAP1, NKCC1, EGFR, and survivin) in combination. Although the dose of each siRNA was relatively low (24 nM), significant functional effects were observed in human glioblastoma cells including inhibition of cancer cell growth and migration. The nanomedicine-induced knockdown was found to be specific to GBM while sparing healthy cells. In animal models, tumor burden is reduced over time by treatment with the bioreducible polymeric nanoparticles encapsulating siRNA. This multimodal nucleic-acid based therapy is promising for the treatment of glioblastoma and other solid tumors.
4. Experimental Section
Materials
Chemicals used to synthesize the bioreducible PBAE monomer BR6 were purchased from Sigma-Aldrich (St. Louis, MO) and used as received. All other PBAE monomers were purchased from Alfa Aesar (Ward Hill, MA). AllStars Human Cell Death siRNA (siDeath) and a scrambled negative control siRNA (scRNA) were purchased from Qiagen; all other siRNA oligos were purchased from Origene Technologies (Rockville, MD). Antibodies against Yes-associated protein 1 (YAP1), sodium-potassium-chloride cotransporter 1 (NKCC1), survivin, roundabout homolog 1 (Robol), endothelial growth factor receptor (EGFR), and β-actin for use in Western blotting were purchased from the vendors listed in Supplementary Table S2. Cannulas used to implant orthotopic tumors and administer nanoparticles were purchased from Plastics One, Inc. (Roanoke, VA).
Bioreducible polymer synthesis
Monomer BR6 was synthesized as previously described.[14] Briefly, bis(2-hydroxyethyl) disulfide was acrylated with acryloyl chloride under anhydrous conditions in the presence of triethylamine (TEA). The TEA HCl precipitate was removed via filtration, and other impurities were removed by washing with Na2CO3 and then with water. The final product was purified from organic solvents using rotary evaporation. Synthesis of the bioreducible polymer, referred to hereafter as R646, was carried out in a method similar to that described previously.[15] Briefly, the BR6 backbone monomer was mixed with the side chain monomer 4-amino-1-butanol (S4) at a 1.01:1 molar ratio of BR6 to S4 in anhydrous tetrahydrofuran (THF). The monomer concentration at the start of the reaction was 500 mg/mL. The polymerization reaction was carried out for 24 hr while stirring at 60°C. Then, a ten-fold molar excess of the end-cap 2-((3-aminopropyl)amino)ethanol (E6) was added to the mixture and allowed to react with stirring at room temperature for one hour. This end-capped polymer was twice precipitated and centrifuged with diethyl ether to remove unreacted monomers. Residual ether was removed by evaporation under vacuum for 48 hr. Finally, the polymer R646 was dissolved in anhydrous DMSO at 100 mg/mL stored with desiccant at −20°C. The structure of R646 is shown in Figure 1 along with the physicochemical characterization of R646/siRNA nanoparticles. The structure of R646 was previously characterized via H1-NMR, as well as the polymer molecular weight and polydispersity via gel permeation chromatography (MW=3,978 Da and PDI=1.349).[42]
PBAE/siRNA nanoparticle characterization
All physicochemical characterization of PBAE/siRNA nanoparticles was carried out using scRNA. For sizing and zeta potential measurements, scRNA was diluted to 1.44 μM in 25 mM sodium acetate buffer (pH 5, NaAc), and R646 polymer was diluted separately in NaAc to 3.24 mg/mL. The R646 and scRNA solutions were mixed 1:1, vortexed briefly, and allowed to form nanoparticles over 10 min. The nanoparticle suspension was then diluted in 1×PBS to an appropriate concentration range for hydrodynamic size measurement by nanoparticle tracking analysis (NTA, NanoSight NS300, Malvern Instruments, Worcestershire, UK) or zeta potential measurement (Zetasizer Nano, Malvern Instruments).
siRNA release from the PBAE/siRNA complexes was measured using a gel retention assay. Particles were made at higher concentration and then diluted 10-fold in either artificial cerebral spinal fluid (aCSF) or 1×PBS with 5 mM glutathione (PBS/GSH) to simulate the extracellular and intracellular environments, respectively. The diluted particles were incubated at 37°C. At predetermined time points, some of the diluted particles were removed, added to sucrose as a lyoprotectant (final concentration 30 mg/mL sucrose), and lyophilized for 48 hr. After all time points were collected, samples were rehydrated simultaneously and analyzed by gel electrophoresis using a 1% agarose gel (Ultrapure™ agarose, Life Technologies) at 100 V. To minimize any binding interference, 30% glycerol without any other dyes was used as a loading buffer. The siRNA was visualized by ethidium bromide staining.
The morphology of nanoparticles was also visualized by transmission electron microscopy (TEM). Nanoparticles were formed as described above at final concentrations of 800 nM scRNA and 1.8 mg/mL R646 in NaAc. A 30 μL solution of nanoparticles were added to 400-square mesh carbon coated TEM grids and allowed to coat grids for 20 min. Grids were rinsed with ultra pure water, allowed to dry fully, and imaged with a Philips CM120 TEM.
Cell Culture
Primary fetal neural progenitor cells (NPCs) were utilized as a healthy brain tissue control in order to assess the cell type specificity of siRNA delivery via R646.[16, 17, 49] NPCs are obtained as described previously following procedures approved by the Johns Hopkins University Institutional Review Board.[50] NPCs 34, 54, and 61, with each number indicating different tissue samples, were used to provide three separate cell source samples. NPCs were grown in a 2:1 mixture of high glucose DMEM to Ham’s F-12 nutrient mixture (Gibco®) supplemented with 1× B-27® Serum-Free Supplement, 1% Antibiotic-Antimycotic (Invitrogen), 10 ng/mL basic fibroblast growth factor (bFGF, Roche Applied Science), 10 ng/mL epidermal growth factor (EGF, Sigma), 5 ng/mL leukemia inhibitory factor (LIF, Millipore), and 2.5 μg/mL heparin (Sigma).
Primary human GBM cell samples 276, 612, 854, and 965, each number indicating samples harvested from a different patient tumor, were isolated from intraoperative samples by the Quiñones laboratory.[50] GBM cells were grown in in 1:1 DMEM/F-12 (Gibco®) supplemented with 1% Antibiotic-Antimycotic (Invitrogen), 1× B-27® Serum-Free Supplement, and 20 ng/mL each of bFGF and EGF for all experiments. Tissue culture plates were coated with 5 μg/mL laminin (Sigma) to allow GBM and NPC cells to adhere for all experiments. Primary human neurospheres (GBM1A, used for orthotopic tumors) were grown in suspension culture using the same medium as that used for other GBM cells, and GBM1A cells were only plated as adherent culture on laminin for in vitro experiments. For all in vitro experiments, cells were plated at a density of 4.7 × 104 cells/cm2 and allowed to adhere overnight.
In vitro siRNA delivery
For all transfections, polymer R646 and siRNA were separately diluted in NaAc, mixed in a 1:1 v/v ratio, and allowed to self-assemble into nanoparticles for 10 min before being added to cells. Nanoparticles were incubated with cells for 2 h at 37°C, and then the media and remaining nanoparticles were removed and replaced with fresh media. For all transfections, the total concentrations of R646 and siRNA were 270 μg/mL and 120 nM, respectively. For experiments in which more than one siRNA sequence was used in a nanoparticle formulation, siRNA oligos were blended in 25 mM NaAc before being mixed with the polymer. For functional siRNA oligos, the best time point for assessing knockdown was measured via Western blotting (Supplementary Figure S1), and the most effective isoform was selected by comparing the protein knockdown using Western blotting (Supplementary Figure S2); the sequences of siRNA oligos are listed in Supplementary Table S1.
For cellular uptake experiments, scrambled control RNA (scRNA) was first labeled with a Cy5 fluorophore using the MirusBio Label IT® Nucleic Acid labeling kit according to the manufacturer’s instructions and then mixed with unlabeled scRNA (1:4 labeled to unlabeled scRNA). Nanoparticles on the external surface of cells were removed by washing with 50 μg/mL heparin sulfate prior to flow cytometry. The cells were then detached from the plate using Accutase® (Life Technologies) and analyzed using a high-throughput BD Accuri™ C6 flow cytometer equipped with a Hypercyt autosampler. The percentage of cells that had taken up particles was calculated using the Cy5 signal from the FL4 detector (excitation: 640 nm, emission: 675/25 nm). The data were analyzed using FlowJo 7 software.
Quantification of siRNA-induced cell killing in vitro
For initial experiments, cells were transfected with either siDeath or scRNA. Five days later, cells were stained with 5 μg/mL propidium iodide (PI), fixed with 10% formalin, and stained with 750 nM 4’,6-diamidino-2-phenylindole (DAPI). Microscopic images were captured at 5X magnification using a Zeiss Axio observer A1 microscope with a Zeiss Axiocam MRm camera and AxioVision software. Live and dead cells were quantified using ImageJ v1.47 software, and dead cells were subtracted from the live cell count to yield the total cell count for each well. To measure the effect of the functional siRNAs, cells were transfected with scRNA, siYAPl, siNKCCl, siRobol, siEGFR, siSurvivin, or a combination of all five functional sequences. For all groups except the scRNA control, cells were administered 120 nM total functional siRNA. Cell death was measured and quantified as above.
In vitro cell migration after siRNA transfection
GBM1A cells were seeded in laminin-coated 24-well plates (6×104 cells per well in 600 (μL medium). After adhering overnight, they were transfected with 120 nM functional siRNA (single oligo or mixture of all five oligos) or 120 nM control scRNA. Three days after transfection, cells were collected using Accutase, counted, and seeded onto a CIM-Plate 16 (ACEA BioSciences, San Diego, CA) according to the manufacturer’s instructions. Complete GBM1A culture medium supplemented with 10% FBS was used as the chemoattractant. The migration of GBM1A cells across the CIM-Plate membrane was measured by the increase in impedance using an xCELLigence RTCA DP (ACEA BioSciences).
Subcutaneous human GBM model and nanoparticle administration in vivo
All animal experiments were conducted following the guidelines and approval of the Johns Hopkins University Animal Care and Use Committee. Four-week-old athymic nu/nu mice were anesthetized by isofluorane inhalation. Two million human GBM 612 cells were injected subcutaneously into the flank of the mice to create tumors. After one month, when tumors had reached approximately 1 cm3, nanoparticles containing 27 μg siRNA were injected directly into the tumor. Gene knockdown was assessed via Western blotting as described below.
Protein expression analysis
Except where noted otherwise, cells were harvested three days after in vitro transfection experiments. Protein lysates were isolated using 5.2 μL/cm2 radioimmunoprecipitation assay (RIPA) buffer in tissue culture flasks. Cells were dissociated via cell scraping, lysates were kept on ice for 30 min with vortexing at 5 min intervals. Cell debris was removed by centrifugation at 19.2 × 103 RCF, and the supernatant was collected and stored at −80°C.
To measure in vivo protein expression, animals with subcutaneous tumors were treated with nanoparticles and then sacrificed after two days, and the tumors were cut from the surrounding tissue. The tumor tissue was homogenized in RIPA buffer and the debris removed via centrifugation as described above.
All protein electrophoresis was carried out using 4–12% NuPAGE Novex Bis-Tris gels (Thermo Fisher Scientific) in 10% 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (Thermo Fisher Scientific) at 135 V for 90 min. Proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane using a Pierce G2 Fast Blotter (Thermo Fisher Scientific) according manufacturer’s instructions. The membranes were blocked for 1 hr with blocking buffer, 5% milk in 1× Tris-buffered saline with Tween® 20 (TBST) at room temperature, and then incubated with primary antibodies overnight at 4°C in blocking buffer. After washing with TBST, membranes were incubated with secondary antibodies for 1 hr at room temperature in blocking buffer. The antibody dilutions and specifications are listed in Table S2.
Orthotopic human GBM model and nanoparticle administration
All animal experiments were conducted following the guidelines and approval of the Johns Hopkins University Animal Care and Use Committee. Experiments were conducted using 4–8 week-old athymic nu/nu mice. Transcranial cannulas (Plastics One Inc., Roanoke, VA) were implanted stereotactically into the right striatum of the brain. (X: 1.5 mm, Y: 1.34 mm, Z: −3.4 mm from bregma). A cyanoacrylate glue was used to attach the cannula to the skull, the surrounding skin was sutured closed. Ketamine/xylazine was used as an anesthetic.
Animals were inoculated with 500,000 luciferase-positive GBM1A cells using an internal cannula inserted within the transcranial cannula. A cell suspension (4 μL) was injected at a rate of 1 μL/min.
Magnetic resonance imaging (MRI) acquisition and Tumor Size:
Animals were monitored for tumor growth via small animal Bruker 9.4T BiosSpin magnetic resonance imaging system with basic repetition time (TR) of 16 milliseconds and echo time (TE) of 3000 milliseconds. Rapid acquisition with refocused echoes (RARE) were taken with 3 averages; 128×128 voxels; 0.25 mm x 0.25 mm in plane resolution and 1.5 mm slice thickness.
After 14 days, tumors approximately 1 mm in diameter were visible via MRI using T2-weighted images and confirmed with hematoxylin and eosin staining for an n=4 mice. Tumor volumetric analysis was based on calculating the volume of a sphere in mm3 taken from the axial and coronal sections of MRI images. Corresponding H&E staining of the 14-day time-point confirmed radius of tumor.
Intratumoral treatment with nanoparticles began and was repeated twice weekly for two weeks. For each treatment, 4 μL of lyophilized nanoparticles, prepared as previously described[51] using 30 mg/mL sucrose as a lyoprotectant, were injected at 1 μL/min through the cannulas. The total dose of siRNA per mouse was 0.6 μg.
Analysis of tumor growth
Luciferase-positive GBM tumors were imaged using an In Vivo Imaging System (IVIS) Spectrum (Caliper Life Sciences) and analyzed using Living Image® software (PerkinElmer). Animals were injected intraperitoneally with 4.5 mg D-luciferin potassium salt (GoldBio) in 1× PBS 10 min before imaging. Mice (n=5 at experiment start date) were imaged 14, 17, and 21 days after tumor inoculation.
Analysis of brain tissue and tumor size
Four weeks after tumor inoculation, animals were anesthetized using ketamine/xylazine and perfused with 10% formalin. The skulls were removed and brains stored in 10% formalin overnight. Brains were immersed in 30% sucrose for two days and then in Optimal Cutting Temperature (OCT, Fisher Scientific) overnight. The brains in OCT were frozen and sectioned with a Leica CM1905 cryostat (14-μm sections). Six sections from each animal (n=3 mice per group) were stained with hematoxylin and eosin (H&E), and the slides were imaged with a Zeiss Axio observer A1 microscope with a Zeiss Axiocam MRm camera at 2.5× magnification. The tumor area in each H&E-stained section was measured via computer-assisted morphometric quantification using AxioVision 4.8 software.
Statistical Analysis
Unless otherwise indicated, the data are presented as the mean ± standard error of the mean. In vitro siRNA nanoparticle uptake and transfection efficacy in GBM versus NPC cells and differences in tumor burden between groups were analyzed using a two-tailed t-test. One-way ANOVA with Dunnett’s post-tests were used to evaluate multiple comparisons to a control group and a Bonferroni correction was used when comparing a greater number of groups. p-values < 0.05 were considered statistically significant in all cases.
Supplementary Material
Acknowledgements
The authors would like to thank Montserrat Lara-Velazquez, Paola Suarez-Meade, Carla Vazquez-Ramos, and Emily Lavell, for assistance with animal surgery and histological processing, Rachel Sarabia-Estrada for assistance with animal surgery and the use of surgical equipment, and Barbara Kim for assistance with transfections and Western blotting. The authors thank the NIH (R01EB016721, R01CA195503, and R01CA228133) for support of this work. AQH is also supported by the Mayo Clinician Investigator Award, Mayo Professorship, and the State of Florida. HGC is also supported by the NCI (5R21CA199295). JJG thanks the Bloomberg~Kimmel Institute for Cancer Immunotherapy for support. KLK thanks the National Cancer Institute (NIH F31CA196163), and the ARCS Foundation for fellowship support. JK thanks Samsung for scholarship support.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Data Availability
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
Supplementary Data
Supplementary Data is available from the Wiley Online Library or from the author.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Chaichana KL, Zadnik P, Weingart JD, Olivi A, Gallia GL, Blakeley J, Lim M, Brem H, Quinones-Hinojosa A, Multiple resections for patients with glioblastoma: prolonging survival, J Neurosurg 118(4) (2013) 812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McGirt MJ, Chaichana KL, Gathinji M, Attenello FJ, Than K, Olivi A, Weingart JD, Brem H, Quinones-Hinojosa AR, Independent association of extent of resection with survival in patients with malignant brain astrocytoma, Journal of Neurosurgery 110(1) (2009) 156–62. [DOI] [PubMed] [Google Scholar]
- [3].Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma, New England Journal of Medicine 352(10) (2005) 987–996. [DOI] [PubMed] [Google Scholar]
- [4].Chaichana KL, Jusue-Torres I, Navarro-Ramirez R, Raza SM, Pascual-Gallego M, Ibrahim A, Hernandez-Hermann M, Gomez L, Ye X, Weingart JD, Establishing percent resection and residual volume thresholds affecting survival and recurrence for patients with newly diagnosed intracranial glioblastoma, Neuro-oncology 16(1) (2013) 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lara-Velazquez M, Al-Kharboosh R, Jeanneret S, Vazquez-Ramos C, Mahato D, Tavanaiepour D, Rahmathulla G, Quinones-Hinojosa A, Advances in brain tumor surgery for glioblastoma in adults, Brain sciences 7(12) (2017) 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, Xia S, Trageser D, Guerrero-Cázares H, Eberhart CG, c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype, Proceedings of the National Academy of Sciences 108(24) (2011) 9951–9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin C, Guerrero-Cázares H, Quinones-Hinojosa A, Laterra J, Xia S, Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition, Oncogene 30(31) (2011) 3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Auvergne R, Wu C, Connell A, Au S, Cornwell A, Osipovitch M, Benraiss A, Dangelmajer S, Guerrero-Cazares H, Quinones-Hinojosa A, Goldman SA, PAR1 inhibition suppresses the self-renewal and growth of A2B5-defined glioma progenitor cells and their derived gliomas in vivo, Oncogene 35(29) (2016) 3817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tilghman J, Wu H, Sang Y, Shi X, Guerrero-Cazares H, Quinones-Hinojosa A, Eberhart CG, Laterra J, Ying M, HMMR maintains the stemness and tumorigenicity of glioblastoma stem-like cells, Cancer Res 74(11) (2014) 3168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Woolard K, Fine HA, Glioma Stem Cells: Better Flat Than Round, Cell Stem Cell 4(6) (2009) 466–467. [DOI] [PubMed] [Google Scholar]
- [11].Zaidi H, DiMeco A,F, Quinones-Hinojosa A, Brain Tumor Stem Cells, Youman’s Neurological Surgery 2009. [Google Scholar]
- [12].Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC, Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans, Nature 391(6669) (1998) 806–811. [DOI] [PubMed] [Google Scholar]
- [13].Thomas CE, Ehrhardt A, Kay MA, Progress and problems with the use of viral vectors for gene therapy, Nature Reviews Genetics 4(5) (2003) 346–358. [DOI] [PubMed] [Google Scholar]
- [14].Kozielski KL, Tzeng SY, Green JJ, A bioreducible linear poly(beta-amino ester) for siRNA delivery, Chem Commun 49(46) (2013) 5319–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kozielski KL, Tzeng SY, De Mendoza BAH, Green JJ, Bioreducible Cationic Polymer-Based Nanoparticles for Efficient and Environmentally Triggered Cytoplasmic siRNA Delivery to Primary Human Brain Cancer Cells, ACS Nano 8(4) (2014) 3232–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Guerrero - Cazares H, Lavell E, Chen L, Schiapparelli P, Lara - Velazquez M, Capilla - Gonzalez V, Clements AC, Drummond G, Noiman L, Thaler K, Brief report: robo1 regulates the migration of human subventricular zone neural progenitor cells during development, Stem cells 35(7) (2017) 1860–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang Y-J, Schiapparelli P, Kozielski K, Green J, Lavell E, Guerrero-Cazares H, Quinones-Hinojosa A, Searson P, Electrophoresis of cell membrane heparan sulfate regulates galvanotaxis in glial cells, J Cell Sci (2017) jcs. 203752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mertsch S, Schmitz N, Jeibmann A, Geng JG, Paulus W, Senner V, Slit2 involvement in glioma cell migration is mediated by Robo1 receptor, J Neuro-Oncol 87(1) (2008) 1–7. [DOI] [PubMed] [Google Scholar]
- [19].Orr BA, Bai HB, Odia Y, Jain D, Anders RA, Eberhart CG, Yes-Associated Protein 1 Is Widely Expressed in Human Brain Tumors and Promotes Glioblastoma Growth, J Neuropath Exp Neur 70(7) (2011) 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shah SR, David JM, Tippens ND, Mohyeldin A, Martinez-Gutierrez JC, Ganaha S, Schiapparelli P, Hamilton DH, Palena C, Levchenko A, Brachyury-YAP regulatory axis drives stemness and growth in cancer, Cell reports 21(2) (2017) 495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schiapparelli P, Guerrero-Cazares H, Magana-Maldonado R, Hamilla SM, Ganaha S, Goulin Lippi Fernandes E, Huang CH, Aranda-Espinoza H, Devreotes P, Quinones-Hinojosa A, NKCC1 Regulates Migration Ability of Glioblastoma Cells by Modulation of Actin Dynamics and Interacting with Cofilin, EBioMedicine 21 (2017) 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Garzon-Muvdi T, Schiapparelli P, ap Rhys C, Guerrero-Cazares H, Smith C, Kim DH, Kone L, Farber H, Lee DY, An SS, Levchenko A, Quinones-Hinojosa A, Regulation of Brain Tumor Dispersal by NKCC1 Through a Novel Role in Focal Adhesion Regulation, Plos Biol 10(5) (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mellai M, Caldera V, Patrucco A, Annovazzi L, Schiffer D, Survivin expression in glioblastomas correlates with proliferation, but not with apoptosis, Anticancer research 28(1A) (2008) 109–18. [PubMed] [Google Scholar]
- [24].Chakravarti A, Noll E, Black PM, Finkelstein DF, Finkelstein DM, Dyson NJ, Loeffler JS, Quantitatively determined survivin expression levels are of prognostic value in human gliomas, Journal of clinical oncology : official journal of the American Society of Clinical Oncology 20(4) (2002) 1063–8. [DOI] [PubMed] [Google Scholar]
- [25].Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, Bronson RT, Chen JW, Weissleder R, Housman DE, Charest A, Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis, Proceedings of the National Academy of Sciences of the United States of America 106(8) (2009) 2712–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Galvez-Contreras AY, Quiñones-Hinojosa A, Gonzalez-Perez O, The role of EGFR and ErbB family related proteins in the oligodendrocyte specification in germinal niches of the adult mammalian brain, Frontiers in cellular neuroscience 7 (2013) 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kondapalli KC, Llongueras JP, Capilla-González V, Prasad H, Hack A, Smith C, Guerrero-Cázares H, Quiñones-Hinojosa A, Rao R, A leak pathway for luminal protons in endosomes drives oncogenic signalling in glioblastoma, Nature communications 6 (2015) 6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Orr BA, Bai H, Odia Y, Jain D, Anders RA, Eberhart CG, Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth, J Neuropathol Exp Neurol 70(7) (2011) 568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ma H, Li T, Tao Z, Hai L, Tong L, Yi L, Abeysekera IR, Liu P, Xie Y, Li J, Yuan F, Zhang C, Yang Y, Ming H, Yu S, Yang X, NKCC1 promotes EMT-like process in GBM via RhoA and Racl signaling pathways, Journal of cellular physiology (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hendruschk S, Wiedemuth R, Aigner A, Topfer K, Cartellieri M, Martin D, Kirsch M, Ikonomidou C, Schackert G, Temme A, RNA interference targeting survivin exerts antitumoral effects in vitro and in established glioma xenografts in vivo, Neuro Oncol 13(10) (2011) 1074–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Padfield E, Ellis HP, Kurian KM, Current Therapeutic Advances Targeting EGFR and EGFRvIII in Glioblastoma, Frontiers in oncology 5 (2015) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kozielski KL, Tzeng SY, Green JJ, A bioreducible linear poly(beta-amino ester) for siRNA delivery, Chem. Commun. (Camb.) 49(46) (2013) 5319–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].He C, Hu Y, Yin L, Tang C, Yin C, Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles, Biomaterials 31(13) (2010) 3657–3666. [DOI] [PubMed] [Google Scholar]
- [34].Bhise NS, Shmueli RB, Gonzalez J, Green JJ, A Novel Assay for Quantifying the Number of Plasmids Encapsulated by Polymer Nanoparticles (vol 8, pg 367, 2012), Small 8(8) (2012) 1129–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Auvergne R, Wu C, Connell A, Au S, Cornwell A, Osipovitch M, Benraiss A, Dangelmajer S, Guerrero-Cázares H, Quinones-Hinojosa A, PAR1 inhibition suppresses the selfrenewal and growth of A2B5-defined glioma progenitor cells and their derived gliomas in vivo, Oncogene 35(29) (2016) 3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Auvergne RM, Sim FJ, Wang S, Chandler-Militello D, Burch J, Al Fanek Y, Davis D, Benraiss A, Walter K, Achanta P, Transcriptional differences between normal and glioma-derived glial progenitor cells identify a core set of dysregulated genes, Cell reports 3(6) (2013) 2127–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chaichana KL, Guerrero-Cazares H, Capilla-Gonzalez V, Zamora-Berridi G, Achanta P, Gonzalez-Perez O, Jallo GI, Garcia-Verdugo JM, Quiñones-Hinojosa A, Intra-operatively obtained human tissue: protocols and techniques for the study of neural stem cells, Journal of neuroscience methods 180(1) (2009) 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guerrero-Cázares H, Chaichana KL, Quiñones-Hinojosa A, Neurosphere culture and human organotypic model to evaluate brain tumor stem cells, Cancer Stem Cells, Springer2009, pp. 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kim J, Sunshine JC, Green JJ, Differential polymer structure tunes mechanism of cellular uptake and transfection routes of poly(beta-amino ester) polyplexes in human breast cancer cells, Bioconjugate chemistry 25(1) (2014) 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Abbadi S, Rodarte JJ, Abutaleb A, Lavell E, Smith CL, Ruff W, Schiller J, Olivi A, Levchenko A, Guerrero-Cazares H, Glucose-6-phosphatase is a key metabolic regulator of glioblastoma invasion, Molecular Cancer Research (2014) molcanres. 0106.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Smith CL, Kilic O, Schiapparelli P, Guerrero-Cázares H, Kim D-H, Sedora-Roman NI, Gupta S, O’Donnell T, Chaichana KL, Rodriguez FJ, Migration phenotype of brain-cancer cells predicts patient outcomes, Cell reports 15(12) (2016) 2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lopez-Bertoni H, Kozielski KL, Rui Y, Lal B, Vaughan H, Wilson DR, Mihelson N, Eberhart CG, Laterra J, Green JJ, Bioreducible Polymeric Nanoparticles Containing Multiplexed Cancer Stem Cell Regulating miRNAs Inhibit Glioblastoma Growth and Prolong Survival, Nano Lett 18(7) (2018) 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Griffith OW, Biologic and pharmacologic regulation of mammalian glutathione synthesis, Free Radical Biology and Medicine 27(9) (1999) 922–935. [DOI] [PubMed] [Google Scholar]
- [44].Tzeng SY, Higgins LJ, Pomper MG, Green JJ, Biomaterial-mediated cancer-specific DNA delivery to liver cell cultures using synthetic poly(beta-amino ester)s, Journal of biomedical materials research. Part A 101(7) (2013) 1837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Guerrero-Cazares H, Tzeng SY, Young NP, Abutaleb AO, Quinones-Hinojosa A, Green JJ, Biodegradable Polymeric Nanoparticles Show High Efficacy and Specificity at DNA Delivery to Human Glioblastoma in Vitro and in Vivo, Acs Nano 8(5) (2014) 5141–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Soeda A, Hara A, Kunisada T, Yoshimura S, Iwama T, Park DM, The Evidence of Glioblastoma Heterogeneity, Sci Rep-Uk 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Garay T, Juhasz E, Molnar E, Eisenbauer M, Czirok A, Dekan B, Laszlo V, Hoda MA, Dome B, Timar J, Klepetko W, Berger W, Hegedus B, Cell migration or cytokinesis and proliferation?--revisiting the “go or grow” hypothesis in cancer cells in vitro, Exp Cell Res 319(20) (2013) 3094–103. [DOI] [PubMed] [Google Scholar]
- [48].Yu D, Khan OF, Suva ML, Dong B, Panek WK, Xiao T, Wu M, Han Y, Ahmed AU, Balyasnikova IV, Zhang HF, Sun C, Langer R, Anderson DG, Lesniak MS, Multiplexed RNAi therapy against brain tumor-initiating cells via lipopolymeric nanoparticle infusion delays glioblastoma progression, Proceedings of the National Academy of Sciences of the United States of America 114(30) (2017) E6147–E6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhu M, Feng Y, Dangelmajer S, Guerrero-Cázares H, Chaichana KL, Smith CL, Levchenko A, Lei T, Quinones-Hinojosa A, Human cerebrospinal fluid regulates proliferation and migration of stem cells through insulin-like growth factor-1, Stem cells and development 24(2) (2014) 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ravin R, Blank PS, Steinkamp A, Rappaport SM, Ravin N, Bezrukov L, Guerrero-Cazares H, Quinones-Hinojosa A, Bezrukov SM, Zimmerberg J, Shear forces during blast, not abrupt changes in pressure alone, generate calcium activity in human brain cells, PloS one 7(6) (2012) e39421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tzeng SY, Guerrero-Cázares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ, Non-viral gene delivery nanoparticles based on poly(beta-amino esters) for treatment of glioblastoma, Biomaterials 32(23) (2011) 5402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.