Using proteomic and RT-qPCR approaches, we investigated how Pseudomonas aeruginosa adapts the level of expression of its various iron-uptake pathways in response to different growth conditions (planktonic growth in different media and an epithelial cells infection assay) in the presence and absence of four different exosiderophores. The results show a complex phenotypic plasticity in the expression of the various iron-uptake pathways indicating a high potential of adaptation of P. aeruginosa to a large variety of biotopes.
Keywords: Label-free quantification, mass spectrometry, microbiology, molecular biology, pathogens, iron homeostasis, phenotypic plasticity, Pseudomonas, siderophore, TonB transporters
Graphical Abstract
Highlights
P. aeruginosa grown with exosiderophores and analyzed by proteomic and RT-qPCR.
Catechol-type exosiderophores strongly induce the expression of their transporters.
Repression of the endogenous iron uptake pathways.
Complex phenotypic plasticity in the expression of the various iron-uptake pathways.
Abstract
Bacteria secrete siderophores to access iron, a key nutrient poorly bioavailable and the source of strong competition between microorganisms in most biotopes. Many bacteria also use siderophores produced by other microorganisms (exosiderophores) in a piracy strategy. Pseudomonas aeruginosa, an opportunistic pathogen, produces two siderophores, pyoverdine and pyochelin, and is also able to use a panel of exosiderophores. We first investigated expression of the various iron-uptake pathways of P. aeruginosa in three different growth media using proteomic and RT-qPCR approaches and observed three different phenotypic patterns, indicating complex phenotypic plasticity in the expression of the various iron-uptake pathways. We then investigated the phenotypic plasticity of iron-uptake pathway expression in the presence of various exosiderophores (present individually or as a mixture) under planktonic growth conditions, as well as in an epithelial cell infection assay. In all growth conditions tested, catechol-type exosiderophores were clearly more efficient in inducing the expression of their corresponding transporters than the others, showing that bacteria opt for the use of catechol siderophores to access iron when they are present in the environment. In parallel, expression of the proteins of the pyochelin pathway was significantly repressed under most conditions tested, as well as that of proteins of the pyoverdine pathway, but to a lesser extent. There was no effect on the expression of the heme and ferrous uptake pathways. Overall, these data provide precise insights on how P. aeruginosa adjusts the expression of its various iron-uptake pathways (phenotypic plasticity and switching) to match varying levels of iron and competition.
Natural biotopes host diverse microbial species, all in competition for space and resources. Iron is a key nutrient that is indispensable for microorganism survival and growth because it is involved in many essential biological processes (cofactors of enzymes and redox proteins). Indeed, iron is often the source of competition between species in the same biotope and is a growth-limiting factor because it is poorly soluble in aqueous solutions under aerobic conditions and at neutral pH and consequently, insufficiently bioavailable. Such competition for iron also exists in vertebrate microbiota between beneficial, commensal, and pathogenic bacteria. In addition, competition for iron occurs as well between pathogens and hosts during infection: vertebrate immune systems have developed nutrient immunity defense mechanisms to protect them against invading pathogens by sequestering iron and consequently starving pathogens (1, 2).
In such a context of competition for iron, bacteria have evolved several strategies to efficiently access it, the most common being the production and release of siderophores (small iron-chelating compounds) into their environment (3). Siderophores have a molecular weights ranging from 200 to 2000 Da and are produced by bacteria under iron-limiting conditions. They are excreted into the bacterial environment where they very efficiently scavenge iron before being taken up by highly selective outer membrane TonB-dependent transporters (TBDT)1 (4, 5). Bacterial species generally produce one or several siderophores and express a specific corresponding TBDT for the recapture of the siderophore once they are loaded with iron (6, 7). In addition, most bacteria are often able to also use exosiderophores (chelators produced by other bacteria) because they are able to express the corresponding specific TBDTs, allowing the capture and import of the ferri-exosiderophores (6, 7). TBDTs are characterized by very high siderophore binding selectivity and there is a strong correlation between the amount of various siderophores and exosiderophores that can be used by a bacterium and the number of genes encoding TBDTs in its genome (6).
Siderophore-mediated interspecies competition for iron exists at least at two major levels. As already mentioned above, many bacterial species can use exosiderophores, stealing those of others and economizing on their own siderophore production (8–17). In contrast, bacteria can produce several siderophores and steal iron away from competitors unable to express compatible TBDTs and/or competitive siderophores (18–25). In the second case, the iron-chelating affinities of the various siderophores produced mediate the competition: the bacteria producing the siderophore with the highest affinity for iron will have the highest chance to scavenge iron and access it.
P. aeruginosa, an opportunist human pathogen (WHO top-priority bacteria because of its intrinsic multi-resistance properties), can express at least 15 different iron-uptake pathways: (1) one ferrous (Fe2+) iron-uptake pathway, (2) three heme-acquisition pathways, (3) ferric (Fe3+) iron-uptake pathways by the two main siderophores, pyoverdine (PVD) and pyochelin (PCH), produced by the pathogen, and (4) at least 10 different “siderophore piracy” strategies to uptake Fe3+ using exosiderophores (7). Such a piracy strategy is possible because of the ability of P. aeruginosa to express various iron uptake pathways with specific TBDTs (26). TBDTs are generally expressed at very low levels and pathogens only induce the expression of the most efficient pathway(s) for iron acquisition, depending on the environment (13, 27, 28). P. aeruginosa detects the presence of exosiderophores in its environment using sigma and anti-sigma factors, two component systems, and a transcriptional regulator of the AraC family (13, 27–31). This transcriptional regulators activate in the presence of ferri-exosiderophore the transcription of the exosiderophore-corresponding TBDT as well as the proteins needed to get release of iron from the chelator once in the bacteria (28, 31–34).
Currently, little is known concerning siderophore piracy and competition in biotic interactions and the bacterial phenotypic adaptations involved. For example, how bacteria select, regulate, and adapt the expression levels of their various iron-uptake pathways in response to external environment stimuli is largely unknown. As the expression of these various iron uptake pathways is costly, there must be regulating networks that allow the bacteria to rapidly adapt to environmental changes and express the optimal iron uptake pathway(s). The various iron-uptake pathways present in bacterial genomes are probably solely expressed when required and their level of expression adjusted to the level of competition. Thus, the phenotypic switches concerning the expression of bacterial iron-uptake pathways may be very diverse, depending on bacterial growth conditions and environmental stimuli.
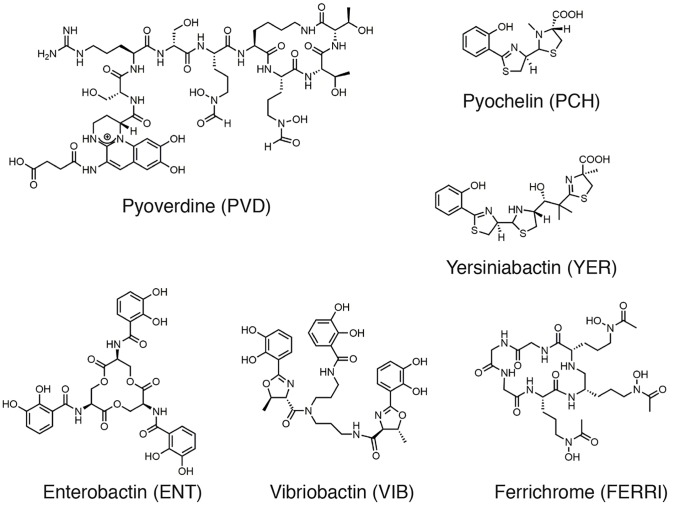
Here, we investigated such phenotypic plasticity in P. aeruginosa using proteomic and RT-qPCR approaches. We deciphered how this pathogen adapts the level of expression of its various iron-uptake pathways in response to different growth conditions (planktonic growth in different media and an epithelial cells infection assay) in the presence and absence of four different exosiderophores (Fig. 1): enterobactin (ENT), a catechol siderophore produced by E. coli (35); vibriobactin (VIB), another catechol siderophore produced by Vibrio cholera (36); ferrichrome (FERRI), a hydroxamate siderophore produced by fungi of the Aspergillus, Ustilago and Penicillium genera (37); and yersiniabactin (YER), produced by Yersinia pestis (38). Purified siderophores were used, rather than bacterial co-cultures, to evaluate only the impact of the exosiderophores (and no other metabolites) on the plasticity of the phenotypic switching. In the absence of competition with other siderophores, these exosiderophores were all able to induce the expression of their corresponding uptake pathways in P. aeruginosa and 55Fe uptake assays showed that all, except YER, have the capacity to transport iron into P. aeruginosa cells with similar uptake rates. In the presence of a mixture of siderophores in the bacterial environment, the affinity of the various siderophores for iron plays a key role and catechol siderophores (very strong iron chelators among the siderophores) become the major players in this competition by scavenging iron from other siderophores. Thus, catechol siderophores are the most efficient in inducing the expression of their corresponding TBDTs in the presence of other siderophores under iron-restricted growth conditions, as well as in more complex systems, such as an epithelial cell infection assay. This phenotype goes hand in hand with significant repression of the proteins of the PCH pathway and, in some conditions, also that of the proteins of the PVD pathway. However, there was no effect on the level of expression of heme, ferrous and ferri-citrate uptake pathways.
Fig. 1.
Chemical structures of siderophores. Affinities of the siderophores for iron: ENT, 1042 m−1 (35), PVD, 1032 m−1 (56), FERRI, 1029 m−1 (57) and PCH, 1018 m−2 (58). PVD, ENT, FERRI, VIB and YER chelate iron with a stoichiometry of 1 :1, and PCH with a stoichiometry of 2 : 1 (Sid : metal).
EXPERIMENTAL PROCEDURES
Chemicals
The pyochelin (PCH) was synthesized and purified as previously described (39, 40). Pyoverdine (PVD) was purified from bacterial culture supernatants as previously described (41). Enterobactin (ENT) and ferrichrome (FERRI) were purchase from Sigma-Aldrich, vibriobactin (VIB) and yersiniabactin (YER) from EMC Microcollections. The protonophore CCCP (carbonyl cyanide m-chlorophenylhydrazone) was purchased from Sigma-Aldrich. 55FeCl3 was obtained from Perkin Elmer Life and Analytical Sciences (Billerica, MA). RPMI was purchased from Thermo-Fisher.
Bacterial Strains, Plasmids, and Growth Conditions
The P. aeruginosa strains used in this study are listed in supplemental Table S1 in supplemental materials. P. aeruginosa strains were first grown overnight at 30 °C in LB broth and were then washed, resuspended and cultured overnight at 30 °C in iron-deficient CAA medium (casamino acid medium, composition: 5 g l−1 low-iron CAA (Difco), 1.46 g l−1 K2HPO4 3H2O, 0.25 g l−1 MgSO4 7H2O) or RPMI.
Plasmid and Strain Construction
Enzymes were obtained from ThermoFisher Scientific. Escherichia coli strains TOP10 (Invitrogen) was used as the host strain for the plasmids. The DNA fragments from P. aeruginosa used for cloning were amplified from the genomic DNA of strain PAO1 with Phusion High-Fidelity DNA polymerase (ThermoFisher Scientific). Primers are listed in supplemental Table S4. For construction of the pEXG2 ΔfoxA and pEXG2 ΔfiuA plasmids, a 1400 bp insert containing the 700 bp flanking sequences of the gene was amplified by PCR, phosphorylated using T4 polynucleotide kinase and cloned into a PCR amplified linear pEXG2 vector using blunt end ligation with T4 DNA ligase according to manufacturer's instructions. Mutations in the chromosomal genome of P. aeruginosa were generated as previously described, by sequentially introducing both pEXG2 vectors using triparental mating into P. aeruginosa PAO1 ΔpvdFΔpchA and selecting for deletion mutants before verification by PCR and sequencing (33).
Cell Culture
A549 (ATCC® CCL-185™) human pulmonary epithelial cells were routinely cultivated at 37 °C, 5% CO2 in RPMI 1640 medium (Gibco) supplemented with 10% v/v FBS (Gibco) and passaged every 3 to 4 days. The cells used for the infection assays were passaged at least once after thawing and consequently were used between passage 15 to 30.
Cell Viability Assay
A549 cell viability was estimated using the Real Time-Glo™ MT cell viability assay kit (Promega™). A549 cells were seeded at a density of 1 × 104 cell per well in cell culture-treated 96-wall plates (Thermo Scientific) and left to incubate overnight at 37 °C with 5% CO2. A saturated P. aeruginosa PAO1 LB culture was diluted to an OD600 nm of 0.1 in LB and incubated for 2 h 30 at 30 °C, until it reached exponential phase of growth. The growth medium in each well of the 96-well plate was removed and A549 cells were washed with 1× PBS before addition of 100 μl of fresh pre-warmed RPMI medium containing siderophores and/or 5 × 105 bacteria (multiplicity of infection of 50). In addition, 100 μl pre-warmed RPMI medium containing both reagents from the viability kit diluted at a 1:1000 ratio were also added to each well. Luminescence data was acquired by a TECAN Infinite M200 Pro plate reader over 10 h, with the temperature set to 37 °C.
Iron Uptake
55FeCl3 was obtained from Perkin Elmer Life and Analytical Sciences (Billerica, MA), in solution, at a concentration of 71.1 mm, with a specific activity of 10.18 Ci/g. Siderophore-55Fe complexes were prepared at 55Fe concentrations of 50 μm, with a siderophore/iron (mol:mol) ratio of 20:1 for PVD, ENT, FERRI, and YER and 40:1 for PCH because of its 2:1 iron chelating stoichiometry. Bacteria were grown in CAA medium in the presence of 10 μm of the different exosiderophores in order to express the corresponding iron uptake pathways. The bacteria were then washed with 50 mm Tris-HCl pH 8.0, to eliminate the siderophores used to induce transporters expression, and diluted to an OD600 nm of 1. Bacteria were incubated in the presence of 500 nm chelator-55Fe. After 30 min incubated, bacteria were harvested by centrifugation and the radioactivity in the pellet monitored. The experiments were repeated with cells pretreated with 200 μm CCCP. This compound inhibits the protonmotive force across the bacterial cell membrane, thereby inhibiting TonB-dependent iron uptake (42). CCCP was therefore used to evaluate the radioactivity associated to the pellet because of 55Fe precipitation or binding of siderophore-55Fe to the bacterial cell surface. By subtracting the data obtained in the presence of CCCP from the data in the absence of CCCP, the radioactivity due only to 55Fe uptake was obtained.
When two siderophores were used in competition in the same iron uptake assay, both siderophores were incubated at 10 μm for PVD, ENT, and FERRI and 40 μm in the case of PCH in the presence of 500 nm 55Fe. The uptake assay was then carried out as described above using ΔpvdFΔpchAΔpfeA or ΔpvdFΔpchAΔfiuAΔfoxA cells.
Iron Scavenging from PVD-Fe and CAS-Fe Complexes
All siderophores were prepared in solution at 10 mm in DMSO (ENT), ethanol (YER), methanol (VIB and PCH) or water (FERRI and PVD). PVD-Fe, in solution at 10 μm in 100 μl of 100 mm HEPES buffer pH 7.4, was incubated at 25 °C in the presence of increasing concentrations of siderophores (ENT, FERRI, VIB, YER, and PCH) for 48 h. The fluorescence at 447 nm corresponding to apo PVD (PVD-Fe being not fluorescent) was monitored (excitation at 400 nm) for each adding in a TECAN Infinite M200 plate reader. When the kinetic of iron scavenging from PVD-Fe was followed, PVD-Fe at 10 μm in 100 μl of HEPES buffer was incubated at 25 °C in the presence of 100 μm siderophores (ENT, FERRI, VIB, YER, and PCH). The fluorescence was monitored in function of time during 1 h in a TECAN Infinite M200 plate reader.
For the experiment using CAS (Chrome Azurol S, purchased from Sigma), the CAS-shuttle (CAS-Fe) solution was prepared according to Schwyn & Neilands 1987 (43): 9 mg of CAS were dissolved in 7.5 ml of water before addition of 1.5 ml of a 1 mm iron(III) chloride solution. Twenty-two milligrams of hexadecyltrimethylammonium bromide were dissolved in 50 ml of water and the CAS-Fe solution was added dropwise. The solution was then buffered to pH 5.6 by addition of 4.307 g of piperazine and 6.25 ml of concentrated hydrochloric acid before adjusting the volume to 100 ml with water. Finally, sulfosalicylic acid was added at a concentration of 4 mm. To carry out the iron scavenging experiment, 100 μl of the CAS-shuttle reagent at 7.5 μm was incubated at room temperature in the presence of increasing concentrations of siderophores (ENT, FERRI, VIB, YER, and PCH) for 2 h. The absorbance at 630 nm was monitored for each concentration of siderophore added in a TECAN Infinite M200 plate reader.
Quantitative Real-time PCR on Bacteria Grown in Liquid Culture
Specific gene expression was measured by reverse transcription-quantitative PCR (RT-qPCR), as previously described (32, 44). Briefly, overnight cultures of P. aeruginosa PAO1 grown in CAA medium were pelleted, resuspended and diluted in fresh medium to obtain an OD600 nm of 0.1 units. The cells were then incubated in the presence or absence of 10 μm ENT, VIB, FERRI or YER, with vigorous shaking, at 30 °C for 8 h (CAA medium). An aliquot of 2.5 × 108 cells from this culture was added to two volumes of RNAprotect Bacteria Reagent (Qiagen). Total RNA was extracted with a RNeasy Mini kit (Qiagen), treated with DNase (RNase-Free DNase Set, Qiagen) and purified with an RNeasy Mini Elute cleanup kit (Qiagen). We then reverse-transcribed 1 μg of total RNA with a High-Capacity RNA-to-cDNA Kit, in accordance with the manufacturer's instructions (Applied Biosystems). The amounts of specific cDNAs were assessed in a StepOne Plus instrument (Applied Biosystems) with Power Sybr Green PCR Master Mix (Applied Biosystems) and the appropriate primers (supplemental Table S1), with the uvrD mRNA used as an internal control. The transcript levels for a given gene in each strain were normalized with respect to those for uvrD and are expressed as a base two logarithm of the ratio (fold-change) relative to the reference conditions.
Quantitative Real-time PCR on Bacteria Incubated in the Presence of Epithelial Cells
A549 cells were seeded at a density of 1 × 106 cell per plate in 10 cm tissue culture dishes (Corning), 1 day before infection with P. aeruginosa cells. P. aeruginosa PAO1 was grown overnight in LB medium at 30 °C under vigorous shaking, before being diluted to an OD600 nm of 0.1 and incubated for 2 h 30 min, until the OD600 nm of the suspension reached 0.4 to 0.6 indicating that bacteria are in exponential phase of growth. A549 cells were infected with a volume of this P. aeruginosa suspension equivalent to 50 × 106 CFU (multiplicity of infection of 50). Siderophores were added to the A549 cells at the same moment as P. aeruginosa cells. The infection was allowed to carry on for 3 h at 37 °C, 5% CO2. Afterward, the plates were washed with cold PBS 1× buffer and the A549 cells and bacteria still adhering to the bottom of the plate harvested with a cell scraper. A549 cell and bacteria were then harvested by centrifugation and the dry pellet was re-suspended in one volume of PBS 1× buffer and two volumes of RNAprotect Bacteria Reagent (Qiagen) before centrifugation and storage at −80 °C overnight. On the following day, lysis of the sample was carried out using Tris-EDTA pH 8 buffer containing 15 mg/ml lyzozyme. Lysates were homogenized using the QIAshredder kit (Qiagen) and total RNA extracted used RNeasy Mini kit (Qiagen). Genomic DNA digestion was then performed with DNase (RNase-Free DNase Set, Qiagen) and purified with a RNeasy Mini Elute cleanup kit (Qiagen). The same method was then used as for the quantitative real-time PCR analysis of transcripts in bacteria grown in liquid culture, the only difference being the quantification of GAPDH as a control in order to assess for homogeneity of the amount of A549 cell RNA present in each sample.
Proteomics Analysis on Bacteria Grown in Liquid Culture
PAO1 strain grown in CAA medium was diluted to have an OD600 nm of 0.1 units. Cells were then incubated with or without 10 μm FERRI, YER or VIB at 30 °C for 8 h. A sample of 5 × 108 cells from each culture were used for proteomic analysis. Each sample was prepared in biological triplicate for each cell culture condition. Cell pellets were resuspended in 200 μl of lysis buffer (50 mm Tris-HCl pH 7.6, 50 mm NaCl and 1% Triton) and sonication was carried out to perform efficient cell lysis (5 cycles of 30 s pulse of sonication at 4 °C followed by 5 min of incubation on ice). Protein concentrations were determined by Bradford assay using bovine serum albumin as standard. Proteins were further precipitated overnight with glacial 0.1 m ammonium acetate in 100% methanol (5 volumes, −20 °C). After centrifugation at 12,000 × g and 4 °C during 15 min, the resulting pellets were washed twice with 0.1 m ammonium acetate in 80% methanol and further dried under vacuum (SpeedVac concentrator). Pellets were resuspended in 100 μl of 50 mm ammonium bicarbonate and submitted to reduction (5 mm Dithiothreitol, 95 °C, 10 min) and alkylation (10 mm Iodoacetamide, room temperature, 20 min). Proteins were finally digested overnight with 150 ng of sequencing-grade trypsin (Promega). The proteomic data sets were obtained by the injection of 750 ng of each peptidic mixture on a Q-Exactive Plus mass spectrometer coupled to an EASY-nanoLC-1000 (Thermo-Fisher Scientific) as described previously (45).
The raw data obtained were converted into .mgf files with Proteome Discoverer Daemon software (Thermo-Fisher Scientific, script Export-to-mgf, version 2.2). For both differential proteomic analyses, data were searched against the Pseudomonas aeruginosa UniprotKB sub-database with a decoy strategy (UniprotKB release 2016_12, taxon 208964, Pseudomonas aeruginosa strain PAO1, 5564 forward protein sequences). Peptides and proteins were identified with Mascot algorithm (version 2.5.1, Matrix Science, London, UK). The following parameters were used: (1) Trypsin/P was selected as enzyme, (2) two missed cleavages were allowed, (3) methionine oxidation and acetylation of protein N-term were set as variable modifications and carbamidomethylation of cysteine as fixed modification, (4) mass tolerance for precursor ions was set at 10 ppm, and at 0.02 Da for fragment ions. Mascot data were further imported into Proline v1.4 software (http://proline.profiproteomics.fr/) (46). Proteins were validated on Mascot rank equal to 1, and 1% FDR on both peptide spectrum matches (PSM score) and protein sets (Protein Set score). The total number of MS/MS fragmentation spectra was used to quantify each protein from at least three independent biological replicates: this BasicSC value calculated by Proline includes all PSMs of all peptides, including the modified peptides (3 fixed and variable modifications) and the peptides shared by different protein sets. A Specific (without shared peptides) and a Weighted Spectral Count value is also available for each protein in the supplemental data files supplemental Data S1 and supplemental Data S2. After a column-wise normalization of the data matrix, the BasicSC spectral count values were submitted to a negative-binomial test using an edgeR GLM regression through R (R v3.2.5). The statistical test was based on the published msmsTests R package available in Bioconductor to process label-free LC-MS/MS data by spectral counts (47). For each identified protein, an adjusted p value (adjp) corrected by Benjamini-Hochberg was calculated, as well as a protein fold-change (FC).
Label-free Proteomic Analysis on Bacteria Incubated in the Presence of Epithelial Cells
A549 cells were infected as described above for the RT-qPCR analyses. The dry pellet of A549 cells and bacteria (five independent biological replicates) were resuspended in Laemmli buffer before undergoing lysis by sonication for 1 min total in short bursts of 5 s (Branson Digital Sonifier). Proteins in lysate were titrated using the colorimetric DC Protein Assay (Bio-Rad) and 12.5 μg of protein were cast into a 7.5% acrylamide tube-gel as described (49). After fixation, the gels were cut into pieces and washed four times. The dehydrated gel pieces were covered with sequencing-grade trypsin (Promega, Fitchburg) with a 1:25 enzyme/protein ratio for 14 h at 37 °C. The peptides were extracted using 80% ACN and 0.1% HCOOH for 90 min at room temperature. After evaporation, the peptides were suspended in 50 μl of 2% ACN in 0.1% HCOOH, and containing 11 iRT-peptides (RT-Kit; Biognosys, Schlieren, Switzerland) to monitor the retention time reproducibility along successive analyses (mean CV = 6% between analyses).
NanoLC-MS/MS analyses (750 ng injected) were performed on a nanoACQUITY Ultra-Performance-LC systems hyphenated to a Q-Exactive Plus mass spectrometer as previously described (49), except an elution gradient from 1 to 35% ACN in 0.1% HCOOH over 120 min, then 35% to 90% ACN in 0.1% HCOOH over 1 min, and a flow rate of 450 nL/min.
The raw data obtained were converted into “.mfg” files with MSConvert software (ProteomeWizard, version 3.0.6090). Peaks were assigned with Mascot (Matrix Science, version 2.6.2) against an in-house database containing human entries from SwissProt database (17 March 2017, 20194 entries) and Pseudomonas aeruginosa PAO1 entries from UniProtKB database (17 March 2017, 5677 entries). Common contaminant proteins such human keratins and trypsin were added to the database and concatenated with the reverse copies of all sequences. Trypsin was selected as enzyme; one missed cleavage was allowed. Methionine oxidation was set as variable modification and carbamidomethylation of cysteine as fixed modification. Mass tolerance for precursor ions was set at 5 ppm, and at 0.07 Da for fragment ions. Mascot .dat results files were loaded into Proline software (Proline Studio Release, version 2.0) (46). PSM were validated on pretty rank equal to 1, and 1% FDR on both PSM (adjusted e-value) and protein sets (Protein Set score) levels. For quantification purpose, the “.raw” files were converted into “.mzDB” files with MS Angel software (version 1.6.2). XIC quantification was performed using 5 ppm as m/z tolerance for the peptide abundance extraction. Loess smoothing was performed for the LC-MS runs alignments. Cross assignments of peptide ions were performed using 5 ppm as m/z tolerance and 60 s as retention time tolerance. Only proteins identified with at least one unique peptide were considered and only specific peptides were kept for the sum of protein abundances. The contaminants were excluded from the protein list prior to statistical analysis.
Experimental Design and Statistical Rationale
The type and the number of replicates was determined from a preliminary study conducted on A549 cells infected by P. aeruginosa PAO1. We observed identical reproducibility (67%) in the identified Pseudomonas proteins between three independent cultures, or three technical replicates (injection replicates). Thus, we privileged biological replicates for the label free study in the absence or the presence of ENT, VIB, FERRI, or YER. Statistical analysis was performed using the ProStaR software (version 1.16.6) (50). Proteins identified in at least 4 replicates per condition were considered, normalization was performed using median quantile centering mode for all conditions. Imputation was performed at the protein level SLSA mode for partial observed values per condition and DetQuantile (2.5%, factor 1) mode for values missing in entire condition. Welch's t test was used to identify statistically differentially expressed proteins between two conditions. For each comparison, the p value calibration was performed using the Benjamini-Hochberg procedure. Proteins with p value below 0.001 were considered as differentially expressed, corresponding to a FDR between 1–5%. Following values were obtained: 2.34% FDR for Control versus ENT, 1.97% FDR for Control versus VIB, 4.24% FDR for Control versus FERRI and 1.96% FDR for Control versus YER.
PVD Titration
PVD present in growth media of bacterial cultures was titrated spectrophotometrically. Bacteria were grown in the same conditions as for the RT-qPCR and proteomic analysis before being separated from growth media by centrifugation. Supernatants were sterile filtered and OD400 nm was measured in a TECAN Infinite M200 multi-plate reader. The PVD concentration (n = 3) in culture media was inferred using a standard curve generated with purified apo-PVD.
RESULTS
Ability of Various Exosiderophores to Transport Iron Into P. aeruginosa Cells
We first investigated the ability of the various exosiderophores used in this study to import iron into P. aeruginosa cells in the absence of any competition with other siderophores (endogenous or exosiderophores) using a PVD- and PCH-deficient P. aeruginosa strain (ΔpvdFΔpchA, Table I supplemental material). The strain was grown under iron-restricted conditions (CAA medium; iron concentration: 20 nm (52)) in the presence of one of the exosiderophores to induce the expression of the proteins of the corresponding exosiderophore-dependent iron-uptake pathway(s) (13). Bacteria were then incubated with the exosiderophore loaded with 55Fe and iron incorporation into the bacteria monitored (Fig. 2A). For Sid-55Fe complexes preparation, one equivalent of ENT, PVD, FERRI and YER were mixed with one of 55Fe, and for PCH, two equivalent of siderophores with one of 55Fe (these two siderophores chelating iron with a 2 :1 stoichiometry). All exosiderophores, except YER, showed the same magnitude of 55Fe-uptake capacity: ∼94 pmol 55Fe/ml/OD600 nm for PVD, slightly lower for FERRI (80 pmol 55Fe/ml/OD600 nm), and ∼55 pmol 55Fe/ml/OD600 nm for the other siderophores, including PCH. We observed no significant 55Fe uptake for YER, suggesting that P. aeruginosa cannot apparently access iron via this siderophore. Under the experimental conditions used here, 55Fe uptake rates are mostly a function of the iron uptake turnover of the various bacterial transporters involved and their expression levels.
Table I. TBDT expressed in P. aeruginosa cells grown in LB, CAA, or RPMI media. Proteomic analyses were performed on P. aeruginosa PAO1 cells grown in LB, CAA, or RPMI media. The gene list on the left corresponds to the TBDT genes present in the P. aeruginosa genome. TBDTs detected in the media by proteomic analysis are shown in yellow and those not detected are shown in grey. For the complete list of genes coding for TBDTs in P. aeruginosa PAO1 genome see supplemental Table S2 in supplemental materials.
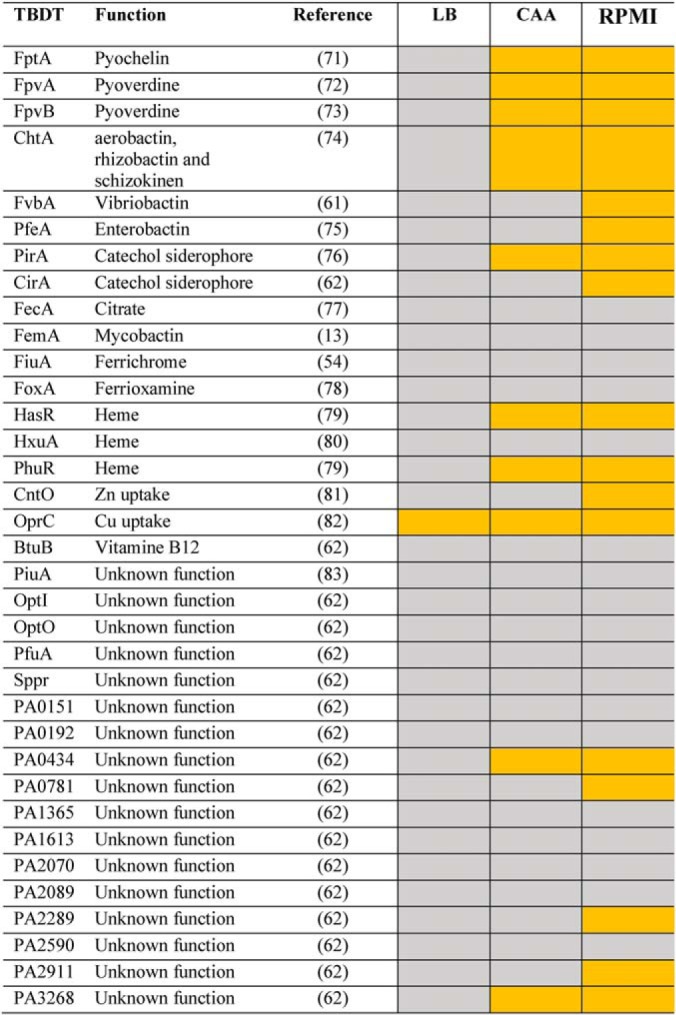
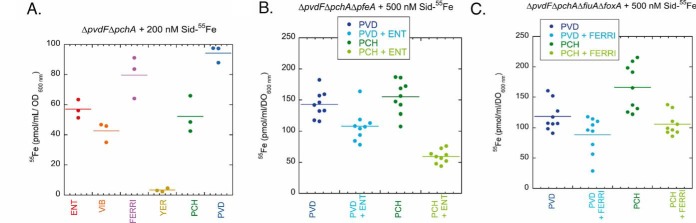
Fig. 2.
A, 55Fe uptake by ΔpvdFΔpchA cells in the presence of various exosiderophores. ΔpvdFΔpchA cells were grown in CAA medium supplemented with one exosiderophore to induce the expression of its corresponding transporters (13). Bacteria were then washed with 50 mm Tris-HCl (pH 8.0) and transport assays initiated by adding the corresponding exosiderophore loaded with 55Fe to a concentration of 200 nm, as described in Materials and Methods. After 30 min incubation, samples were centrifuged, and the radioactivity retained in the cells monitored. Uptake experiments were also carried out for each exosiderophore in the presence of the protonophore CCCP to evaluate the radioactivity because of binding of 55Fe-exosiderophore complexes to the bacterial cell surface or to precipitation. These values were subtracted from those obtained in the absence of CCCP to consider only the radioactivity because of 55Fe uptake. The results are expressed as pmol of 55Fe transported per ml/OD600 nm. The data represent 3 independent experiments. B, 55Fe uptake by PVD or PCH in ΔpvdFΔpchAΔpfeA cells in the presence of ENT acting as a competitor for iron chelation. 55Fe uptake assays were carried out with strain ΔpvdFΔpchAΔpfeA, which is unable to transport 55Fe-ENT. Siderophore complexes were prepared by either mixing 500 nm 55Fe with 10 μm ENT (red dots), 10 μm PVD (dark blue dots), 10 μm PVD and 10 μm ENT (light blue dots), 20 μm PCH (dark green dots), or 20 μm PCH and 10 μm ENT (light green dots). ΔpvdFΔpchAΔpfeA cells were incubated for 30 min in the presence of the various siderophore-55Fe mixtures at 500 nm. Bacteria were then harvested by centrifugation and the radioactivity counted. As in panel A, each uptake assay was repeated in the presence of 200 μm CCCP to specifically evaluate the radioactivity because of 55Fe uptake from that because of binding of the siderophore-55Fe complexes to the cell surface and these values were subtracted from those obtained in the absence of CCCP. The data represent 3 independent experiments with 3 technical replicates. C, 55Fe uptake by PVD or PCH in ΔpvdFΔpchAΔfiuAΔfoxA cells in the presence of FERRI acting as a competitor for iron chelation. 55Fe uptake assays were carried out with the ΔpvdFΔpchAΔfiuAΔfoxA strain, which is unable to efficiently transport 55Fe-FERRI. Siderophore complexes were prepared by either mixing 500 nm 55Fe with 10 μm FERRI (red dots), 10 μm PVD (dark blue dots), 10 μm PVD and 10 μm FERRI (light blue dots), 20 μm PCH (dark green dots), or 20 μm PCH and 10 μm FERRI (light green dots). The uptake assays were carried out as in graph B, with or without 200 μm CCCP. The data represent 3 independent experiments with 3 technical replicates.
Under experimental conditions in which several siderophores compete for Fe, the 55Fe-uptake rates presented in Fig. 2A may vary highly, as they will more strongly depend on the affinity of each siderophore for iron and its ability to efficiently scavenge iron from other siderophores. We investigated this issue by simultaneously incubating one or two siderophores in the presence of 55Fe: 500 nm 55Fe with 20 μm PCH or 10 μm PVD (iron chelation stoichiometry of 2 and 1 for PCH and PVD, respectively), with or without either 10 μm ENT or FERRI as a competitor for 55Fe chelation (Fig. 2B and 2C). These siderophore-iron mixtures were added to ΔpvdFΔpchAΔpfeA or ΔpvdFΔpchAΔfiuAΔfoxA cells (supplemental Table S1) grown under iron-restricted conditions. Both strains are unable to produce PVD or PCH and ΔpvdFΔpchAΔpfeA and ΔpvdFΔpchAΔfiuAΔfoxA are also unable to import ENT-55Fe and FERRI-55Fe, respectively (53, 54). Thus, ENT and FERRI only acted as competitors for iron chelation and were unable to carry out 55Fe uptake into the bacteria; only 55Fe uptake by PVD or PCH was monitored. In the presence of ENT, the uptake of 55Fe into ΔpvdFΔpchAΔpfeA cells by PCH was more affected (63% inhibition) than uptake by PVD (23% inhibition) (Fig. 2B). We obtained similar results in the presence of FERRI as competitor: 63% inhibition for 55Fe uptake by PCH and 25% by PVD (Fig. 2C). The presence of ENT and FERRI more strongly affected the ability of PCH to transport iron than PVD, consistent with the affinities of these two siderophores for iron: 1049 m−1, 1032 m−1, 1029 m−1, and 1018 m−2 for ENT (55), PVD (56), FERRI (57), and PCH (58), respectively.
Overall, these data show that P. aeruginosa is able to access iron using various siderophores, but the efficiency of iron uptake of each chelator depends on their ability to scavenge iron.
Catechol Siderophores Are the Most Powerful Iron Chelators
The various siderophores used in this study do not all have the same denticity and therefore chelate ferric iron with different affinities and stoichiometries. For example, ENT is a very strong iron chelator (Ka of 1049 m−1), whereas PCH has a lower affinity (Ka of 1018 m−2) (55, 58). Thus, when these siderophores are in the bacterial environment in the presence of other siderophores, they will non-equivalently compete for iron. Here, we compared the ability of various siderophores to scavenge iron by monitoring their ability to dissociate PVD-Fe and CAS-Fe complexes (Fig. 3A–3C).
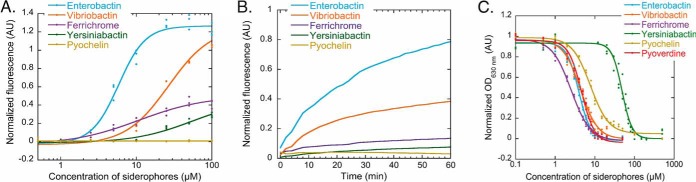
Fig. 3.
A, Ability of various siderophores to steal iron from PVD-Fe complexes. For these titration assays, PVD-Fe at 10 μm in 100 mm HEPES pH 7.4 was incubated with various concentrations of ENT, VIB, FERRI, YER and PCH (until equilibrium was reached, 48 h) as described in Materials and Methods. Apo PVD is fluorescent and PVD-Fe is not. Apo PVD formation was thus followed by monitoring its fluorescent at 447 nm (excitation at 400 nm). The data were normalized using (FMEASURED - FPVD-Fe)/(FPVD - FPVD-Fe), FMEASURED for the fluorescence measured for each experimental condition, FPVD-Fe for the fluorescence of 10 μm PVD-Fe, and FPVD for the fluorescence of 10 μm PVD. B, Kinetics of PVD-Fe dissociation in the presence of various siderophores. PVD-Fe (10 μm in 100 mm HEPES buffer pH 7.4) was incubated with 100 μm ENT, VIB, FERRI, YER, or PCH as described in Materials and Methods. The kinetics of apo PVD formation were followed by monitoring the fluorescence emission at 447 nm (excitation at 400 nm). C, Ability of various siderophores to steal iron from the CAS-Fe complex. CAS-Fe at 7.5 μm was incubated with various concentrations of ENT, VIB, FERRI, YER, PCH, or PVD as described in Materials and Methods. CAS-Fe dissociation was followed by monitoring the absorbance at 630 nm. The data were normalized using (AMEASURED - AapoCAS)/(AapoCAS - ACAS-Fe), AMEASURED for the absorbance measured for each experimental condition, AapoCAS for the absorbance of CAS without iron and ACAS-Fe for the absorbance of CAS loaded with iron.
PVD, the major siderophore produced by P. aeruginosa PAO1, has unique fluorescent properties. It is fluorescent in its apo form (emission of fluorescence at 447 nm when excited at 400 nm) and non-fluorescent when complexed with iron and can be considered to be a strong iron chelator with an affinity for iron of 1032 m−1 (iron chelation stoichiometry of 1) (56, 59). Incubation of 10 μm of PVD-Fe (non-fluorescent) for 48 h in the presence of increasing concentrations of apo siderophores resulted in complete removal of iron by only the catechol-type siderophores ENT and VIB at concentrations of ∼70 μm (Fig. 3A). We did not observe complete removal of iron from PVD by FERRI and YER at the concentrations tested (0.5 μm to 1 mm). Surprisingly PCH was unable to scavenge iron from PVD at the concentrations tested.
The kinetics of PVD-Fe dissociation were also monitored by incubating 10 μm of PVD-Fe in the presence of 100 μm siderophores (Fig. 3B). The kinetics were very different, depending on the siderophores and showed the following sequence for the ability of these siderophores to scavenge iron from PVD: ENT>VIB>FERRI>YER>PCH. Catechol-type siderophores clearly compete more efficiently for iron than the other siderophores (hydroxamates or siderophores such as PCH and YER) consistent with the affinities of these chelators for iron.
We also carried out a competition experiment using CAS-Fe (Chrome azurol S) (Fig. 3C). In this test, developed by Schwyn and Neiland, the ternary complex chrome azurol S-Fe3+-hexadecyltrimethylammonium bromide, with a typical absorbance at 630 nm, serves as an indicator (43). Its color changes from blue to orange when a strong chelator removes the iron from the dye. All siderophores were able to scavenge iron from CAS with equivalent efficiency, except PCH and YER, for which slightly higher concentrations were needed. CAS has a lower affinity for iron than PVD, which explains why PCH can compete for iron with this chelator.
In conclusion, PVD-Fe dissociation assays in the presence of siderophores showed that catechol chelators are more efficient than the others in chelating iron and in competition for iron with PVD, as expected (Fig. 3A–3C). Consequently, the presence of any catechol siderophore in the environment of P. aeruginosa may affect the ability of PVD and, above all, PCH to supply the bacteria with iron.
Adjustment of the Expression of the Proteins of the Various Iron Uptake Pathways When Grown in Iron-restricted Versus Iron-rich Medias
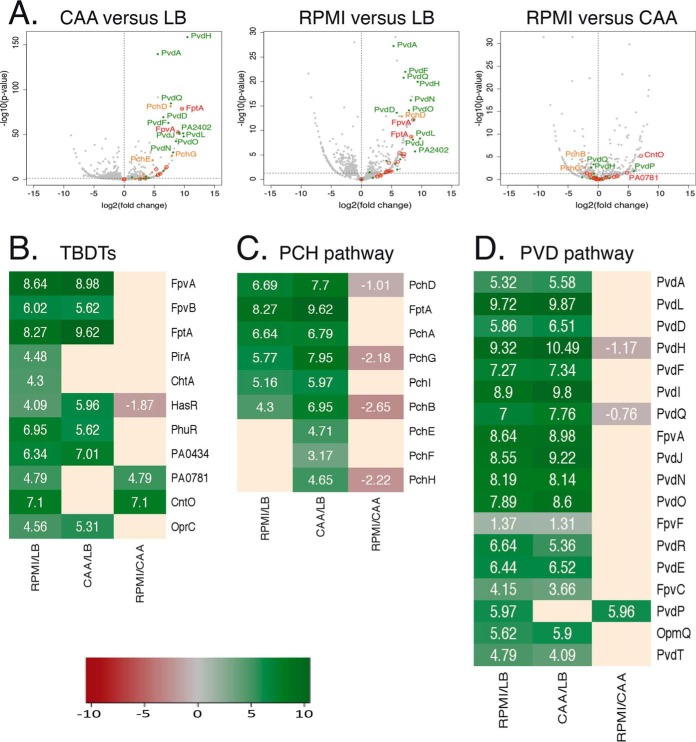
LB medium contains 4.3 μm iron and consequently the proteins of the different iron uptake pathways are poorly expressed and no siderophores are produced (52). Proteomic analysis of P. aeruginosa cells grown in LB detected only peptides corresponding to one TBDT, OprC, involved in Cu acquisition (Table I), indicating that OprC is expressed by the bacteria in LB. This result does not exclude that some of the other TBDTs were also expressed but at low levels. When grown in CAA (an iron-limited medium, 20 nm iron (52)), we detected peptides corresponding to several TBDTs (Table I): FptA, FpvA, and FpvB, corresponding to the siderophores PCH and PVD, HasR and PhuR, involved in heme acquisition, PirA, involved in iron acquisition via catechol-type siderophores, ChtA (aerobactin transporter), two TBDTs of unknown function (PA0434 and PA3268) and as in LB OprC. Analysis of the differences in protein expression between P. aeruginosa cells grown in CAA medium versus those grown in LB medium confirmed expression of the proteins of the PVD and PCH iron-uptake pathways (Fig. 4A–4D) and that of the TBDTs PhuR, HasR, OprC, and PA0434.
Fig. 4.
Analysis of changes in the expression of proteins involved in iron-uptake pathways in P. aeruginosa cells grown in LB, CAA, or RPMI media. A, Proteomic analyses were performed on P. aeruginosa PAO1 cells grown over night in LB, CAA, or RPMI media. Average values measured in CAA or RPMI were plotted against average values measured in LB (two first panels) or CAA (third panels). Average values represent the average of the relative intensity of each protein, normalized against all proteins detected by shotgun analysis (n = 3). In red, proteins corresponding to TBDTs; in green, proteins other than FpvA of the PVD pathway; and in orange, proteins other than FptA of the PCH pathway. B–D, Heat maps of various TBDTs (B) and proteins involved in the PCH (C) and PVD (D) pathways: the darker the shade of green, the higher the expression of the protein is induced; the darker the shade of red, the stronger the expression of protein is repressed. Only the proteins for which a change in the level of expression was observed are shown. Supplemental Data S1 shows the detailed results of protein identification and quantitation.
When grown in RPMI medium (used in the epithelial cell infection described below), all the TBDTs already detected in bacteria grown in CAA medium appeared to be expressed as well (Table I). In addition, we also detected CirA, FvbA, and PfeA (three TBDTs involved in iron acquisition by catechol-type siderophores), CntO (involved in Zn acquisition), PA0781, PA2289, and PA2911 (three TBDTs of unknown function), suggesting that these transporters are also expressed under these growth conditions. Analysis of differences in protein expression between P. aeruginosa cells grown in RPMI medium versus those grown in LB medium (Fig. 4A–4D) confirmed the expression of proteins of both PVD and PCH pathways, heme-uptake pathways, PirA, ChtA, OprC, CntO, PA0434, and PA0781. Analysis of differences in protein expression between P. aeruginosa cells grown in RPMI medium versus those grown in CAA medium showed no change in the expression levels of most of the proteins mentioned above, except HasR (which appears to be slightly less expressed in cells grown in RPMI than in those grown in the CAA medium) and CntO and PA0781 (higher induction). The iron concentration in RPMI medium could not be measured by ICP-AES, because it is under the detection level of the machine and thus lower than that in CAA medium.
In conclusion, we observed three different phenotypic patterns for the expression of the TBDTs present in the genome of P. aeruginosa when the cells were grown in three different media, LB, CAA, and RPMI, suggesting high phenotypic plasticity.
The Presence of an Exosiderophore Induces the Transcription and Expression of Its Corresponding TBDT(s) in CAA Medium
It is well known that bacteria can sense the presence of exosiderophores in their environment via sigma factors, two component systems, or AraC regulators (13, 27–31). Here, P. aeruginosa PAO1 was grown in CAA medium with or without 10 μm of one of the exosiderophores and a proteomic approach was used to identify the proteins, especially TBDTs having their expression induced or repressed (Fig. 5A). In a second step, RT-qPCR was used to confirm the proteomic data (Fig. 5B). For the RT-qPCR experiments we focused only on TBDT genes, because an induction of the transcription and expression of the outer membrane transporter is usually associated as well with an induction of the transcription and expression of the inner membrane transporters or proteins needed to get iron release from the siderophore (like the esterase PfeE in the ENT pathway (33) or the inner membrane transporter FptX in the PCH pathway (60)).
Fig. 5.
Analysis of the changes in the expression (panel A) and the transcription (panels B, C, and D) of genes involved in iron-uptake pathways in P. aeruginosa cells grown under iron-limited conditions (CAA medium) in the absence or presence of exosiderophores. A, Proteomic analyses were performed on P. aeruginosa PAO1 cells grown over night in CAA supplemented, or not, with 10 μm FERRI, VIB, or YER. Average values measured in CAA in the absence of any supplementation with siderophores were plotted against average values measured in CAA supplemented with either 10 μm FERRI, VIB, or YER. Median values represent the median of the relative intensity of each protein, normalized against all proteins detected by shotgun analysis (n = 3). Supplemental Data S2 shows the detailed results of protein identification and quantitation. B, fptA and fpvA encode the TBDTs of PCH and PVD, respectively, pfeA of ENT, fvbA of VIB, femA of mycobactin and carboxymycobatin, fiuA of FERRI, foxA of ferrioxamine B and PA0434 of a hypothetical TBDT. RT-qPCR was performed on P. aeruginosa PAO1 cells grown in CAA medium during 8 h as described in Materials and Methods, with or without 10 μm ENT, FERRI, VIB, or YER. The data were normalized relative to the reference gene uvrD and are representative of three independent experiments performed in triplicate (n = 3). Results are given as the ratio between the values obtained in the presence of siderophores over those obtained in their absence. C, RT-qPCR analyses were performed on P. aeruginosa PAO1 cells grown in CAA during 8 h as described in Materials and Methods and supplemented, or not, with a mixture of 2 μm ENT, 2 μm FERRI, 2 μm VIB, and 2 μm YER (total of 8 μm siderophores). The experiment was repeated with a mixture of 10 μm of each siderophore (total of 40 μm siderophores). As for panel B, the data were normalized relative to the reference gene uvrD and are representative of three independent experiments performed in triplicate (n = 3). Results are given as the ratio between the values obtained in the presence of the siderophores over those obtained in their absence. The data represent 3 independent experiments with 3 technical replicates. D, RT-qPCR was performed on P. aeruginosa PAO1 grown in CAA medium, with or without 2 μm ENT, 2 μm FERRI, 2 μm VIB, and 2 μm YER (total concentration of siderophores 8 μm) for 3 and 8 h. As for panel B, the data were normalized relative to the reference gene uvrD and are representative of three independent experiments performed in triplicate (n = 3). Results are given as the ratio between the values obtained in the presence of the siderophores over those obtained in their absence. The data represent 3 independent experiments with 3 technical replicates.
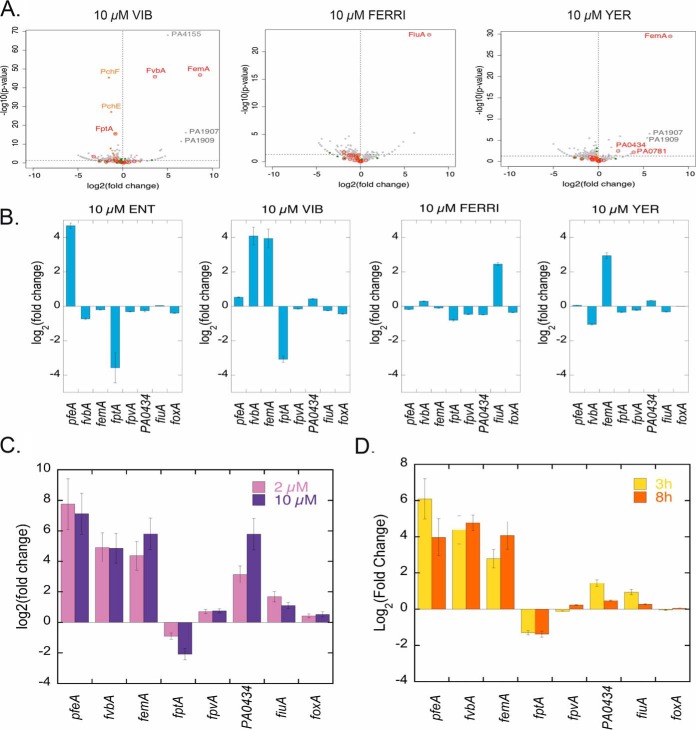
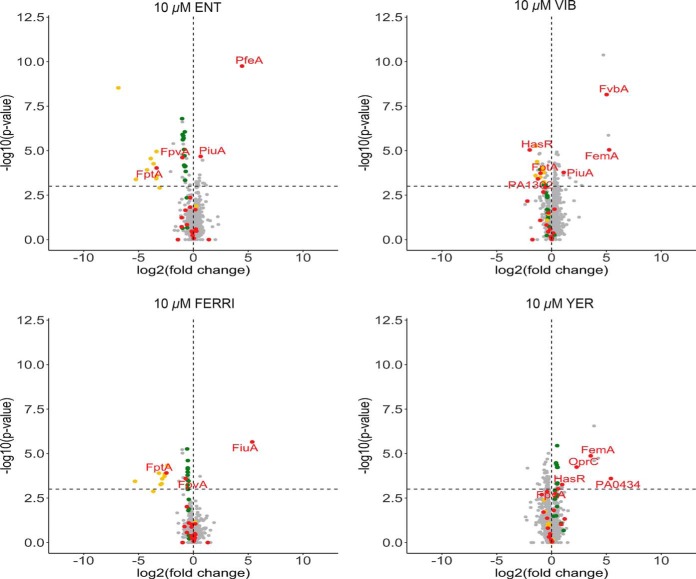
Previous proteomic data from our group showed that 10 μm of ENT in the growth medium of P. aeruginosa induces the expression of only one TBDT, PfeA, the specific TBDT of ENT and PfeE the enzyme hydrolyzing Ferri-ENT in the bacterial periplasm to get iron release (32, 33). This was confirmed here by RT-qPCR, which showed a 26-fold (log2(26) = 4.68, Fig. 5B) increase in transcription of pfeA. The presence of VIB, the other catechol siderophore used in this study, induced transcription and expression of two TBDTs, fvbA and femA (Fig. 5A and 5B), with a lower level of induction of the transcription than that observed with ENT for pfeA (17 and 15 fold change for fvbA and femA, respectively). According to the literature and gene annotation, fvbA is the specific TBDT of VIB, and femA of mycobactins and carboxymycobactins (a chemically diverse family of siderophores produced by Mycobacterium that chelates iron through a hydroxyphenyloxazolin moiety and either hydroxamate or carboxylate functions) (13, 61). The expression of femA can be explained by the presence of certain common chemical motifs in both VIB and mycobactin (or carboxymycobactin). Proteomic data for the growth conditions in the presence of VIB also showed an induction of the expression of gene PA4155, a gene located next to fvbA on P. aeruginosa PAO1 genome and encoding for an oxidoreductase according the genome annotation (62) and of genes PA1907 and PA1909, located next to femA (PA1910). PA1907 and PA1909 may have an esterase and reductase activity according to genome annotation (62) and could be involved in the mechanism of iron release from VIB (nothing being known yet about the proteins and molecular mechanisms involved in iron release from VIB in P. aeruginosa). Surprisingly, the presence of FERRI induced only the transcription and expression of fiuA and not foxA, even though foxA has been shown to be capable of importing FERRI into the bacteria in the absence of fiuA (54). It should be noted that the fold change in transcription observed for fiuA in the presence of FERRI (5.5 fold change -log2(5.5) = 2.46, Fig. 5B) was quite low relative to that of pfeA in the presence of ENT (26 fold change -log2(26) = 4.68-). Finally, and surprisingly, YER, which is not a catechol siderophore and which was not able to transport 55Fe into P. aeruginosa cells, also induced the transcription of femA, with a 7.8-fold change in transcription, and not significantly that of any other TBDT. Again, certain common chemical motifs present in mycobactins and carboxymycobactins can also be found in YER and may explain such induction of femA transcription. Comparison of the fold changes in transcription of these TBDTs in the presence of different exosiderophores suggest that catechol siderophores (ENT and VIB) are slightly more efficient than FERRI and YER in inducing the transcription of their corresponding TBDTs (Fig. 5B). The induction of femA expression by the presence of VIB and YER suggests that this transporter could have a broader specificity for siderophores than previously reported.
The proteomic and qRT-PCR studies also showed that two catechol siderophores ENT and VIB, but not FERRI or YER, repressed the expression of genes of the PCH pathway (Fig. 5A) and, among others, fptA (Fig. 5B), the TBDT of the endogenous siderophore PCH. None of the exosiderophores tested influenced expression of the genes of the PVD pathway under the growth conditions tested here, suggesting that PVD is produced in sufficient amounts to remain competitive for iron chelation with ENT. Indeed 54.2 ± 1.1 μm of PVD was produced in the presence of 10 μm ENT, which involves the presence of both PVD-Fe and ENT-Fe complexes, both able to induce the transcription of their corresponding TBDTs. In addition, we observed no effect on the transcription of genes of the heme and ferrous uptake pathways. The expression of other TBDTs (CntO, OprC, PA0434, PA0781, PirA and PA4675), for which their expression was induced in CAA medium (Fig. 4B), was not affected by the presence of the exosiderophores tested.
In conclusion, (1) all exosiderophores were able to induce the transcription and expression of specific TBDTs and (2) in parallel, only the catechol siderophores repressed the transcription and expression of fptA (PCH TBDT), as well as all genes of the PCH pathway, with (iii) no effect on the expression of the genes of the PVD pathway, the heme and ferrous uptake pathways, or those for any other TBDTs.
Adjustment of the Expression of the Various Iron-uptake Pathways When Grown in the Presence of a Mixture of Four Different Exosiderophores in CAA Medium
P. aeruginosa PAO1 cells were grown in the presence of a mixture of all four exosiderophores at two different concentrations (2 and 10 μm of each siderophore, resulting in a total concentration of siderophores of 8 and 40 μm) and the transcription of the TBDTs for which transcription was even slightly affected in Fig. 5B was analyzed by RT-qPCR (Fig. 5C). The experiment was also repeated with 2 μm of each siderophore (Fig. 5D) at two different times of culture: 8 h (as for the proteomic analysis presented in Fig. 5A) and 3 h.
We observed strong induction of the transcription of the three TBDTs pfeA, fvbA (both involved in iron acquisition via catechol siderophores), and femA, of similar intensity for fvbA and femA and of slightly stronger intensity for pfeA (Fig. 5C), with both 2 and 10 μm of each exosiderophore. On the contrary, the induction of transcription of fiuA was low, again showing that hydroxamate siderophores, such as FERRI, have difficulty scavenging iron in the presence of catechol-type siderophores and afterward induce the expression of their corresponding TBDT. There were no significant differences in the transcription of these three TBDTs between 3 and 8 h of culture.
Unexpectedly, the transcription of PA0434 was significantly more induced by this mixture of siderophores, especially in the presence of 10 μm of each siderophore, than by the siderophores alone (very small induction of transcription with VIB and YER alone in Fig. 5B). This induction was stronger at the beginning of the culture (3 h, Fig. 5D) than after 8 h. No information is available in the literature concerning the molecules that could be transported by this TBDT. However, there does not appear to be a Fur box in front of this gene and thus this TBDT may be involved in the uptake of a molecule other than a siderophore.
Finally, we observed no repression of fpvA transcription with this siderophore mixture, only repression of fptA, indicating again that PCH is not sufficiently competitive for iron chelation relative to PVD and the catechol siderophores. Repression of fptA was stronger when 10 μm of each siderophore was present (total of 40 μm siderophores) than when 2 μm was used, as expected, and did not change in intensity between 3 and 8 h of culture.
In conclusion, P. aeruginosa cells adapted the expression of their iron-uptake pathways in the presence of our mixture of four siderophores, including two catechol siderophores, by (1) increasing the transcription and expression of the catechol siderophore-dependent iron uptake pathways, (2) repressing the expression of the PCH pathway (no effect on the PVD pathway), and (3) inducing the transcription of PA0434, already induced in CAA (versus growth in LB).
Adjustment of the Expression of the Proteins of the Various Iron-uptake Pathways in the Presence of Exosiderophores in the Epithelial Cell Infection Assay
We also investigated whether the ability to detect the presence of exosiderophores also operates in the epithelial cell infection assay. Epithelial A549 cells were infected with P. aeruginosa PAO1 cells grown over night in LB. The bacteria/epithelial cell ratio used resulted in 40 to 50% viability of the cells 10 h after infection (supplemental Fig. S1). Bacteria associated with the epithelial cells were harvested after 3 h of infection, when apoptosis of the epithelial cells started to be observed. The sample preparation that we developed and the proteomic workflows used in this infection assay were different from those used for bacteria growing under planktonic growth conditions and allowed us to identify 1300 proteins from P. aeruginosa in the presence of a large amount of protein from the epithelial cells. Because of the proteomic workflows used, we were unable to carry out a differential analyses to compare the expression of proteins between bacteria incubated in RPMI alone and those incubated with A549 cells. However, we monitored the change in transcription of several genes in P. aeruginosa cells incubated for 3 h in RPMI in the absence or presence of epithelial cells by RT-qPCR (Fig. 6). Phenotypic changes in the P. aeruginosa cells were already observable after 3 h of infection. The presence of epithelial cells induced weakly transcription of fpvA, hasR, and phuR genes (fold changes of 1.6, 2.2, and 2.5) as well as of aprA and toxA (fold changes of 3 and 5.4), respectively, two genes coding for virulence factors. These data indicate that bacteria detect the presence of epithelial cells and adjust the expression of their genes.
Fig. 6.
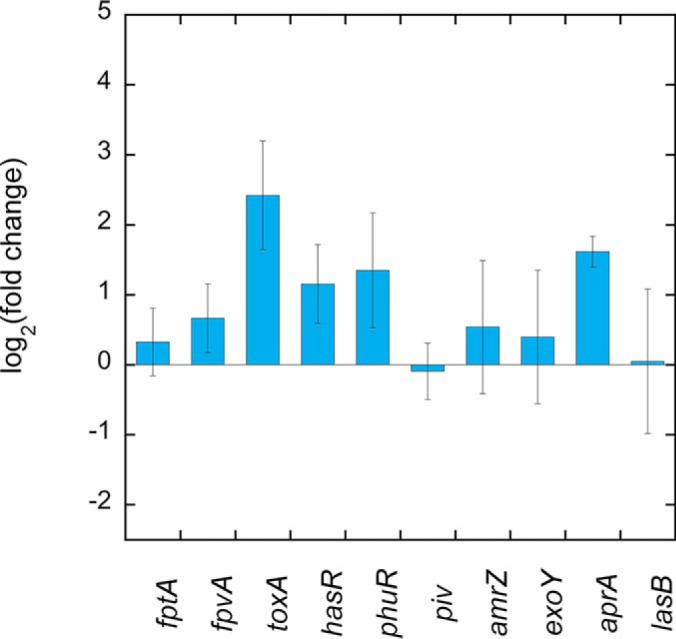
Analysis of changes in the transcription of genes involved in iron-uptake pathways and coding for virulence factors in P. aeruginosa cells in RPMI medium for 3 h in the absence or presence of A549 epithelial cells. The genes piv, amrZ, aprA, toxA, and exoY code for virulence factors (62).The data were normalized relative to the reference gene uvrD and are representative of three independent experiments. Results are given as the ratio between the values obtained in the presence of the epithelial cells over those obtained in their absence after 3 h of incubation in RPMI medium. The data represent 3 independent experiments with 3 technical replicates.
In addition, we also used RT-qPCR to compare (supplemental Fig. S2) the transcription levels of TBDTs involved in iron acquisition in bacteria grown over night in LB and then incubated for 3 h with A549 cells in RPMI versus bacteria grown in the conditions used in Fig. 4, i.e. overnight in RPMI and afterward 8 h again in RPMI. There was less transcription of all TBDTs tested except fptA under the infection conditions (for example fold change of 0.5 and 0.13 for pfeA and PA0434 respectively) compared with the planktonic growth conditions used in Fig. 4. The iron-uptake pathways of bacteria (except PCH uptake pathway) were apparently less expressed after 3 h of incubation in RPMI containing epithelial cells relative to bacteria grown in RPMI. Bacteria, which were grown in LB before use in the epithelial cell infection assay were not yet as iron-starved after 3 h in RPMI medium as bacteria cultivated in RPMI (overnight in RPMI and re-cultured for 8 h in RPMI before RT-qPCR analysis).
The infection assay was then carried out, with or without 10 μm ENT, VIB, FERRI, or YER. The siderophores were added to the infection assay at the same moment as the bacteria (grown in LB without any siderophore). Thus, the bacteria were in contact with the exosiderophores only during the duration of infection. After 3 h of infection, cells (A549 and bacteria) were harvested and protein expression analyzed by proteomic analysis and the transcription of a few selected genes investigated by RT-qPCR (Fig. 7 and in Fig. 8A the histograms with blue bars). The same experiment was repeated in the absence of A549 cells and analyzed only by RT-qPCR (Fig. 8A, histograms with green bars).
Fig. 7.
Analysis of changes in the expression of proteins involved in iron uptake pathways in P. aeruginosa cells during A549 epithelial cell infection, in the absence and presence of exosiderophores. Proteomic analyses were performed on P. aeruginosa PAO1 cells after 3 h of incubation with A549 epithelial cells in RPMI medium, with or without 10 μm FERRI, VIB, or YER. Median values measured in the infection assay in the absence of any supplementation with siderophores were plotted against median values measured in the infection assay supplemented with either 10 μm FERRI, VIB, or YER. Median values represent the median of the relative intensity of each protein, normalized against all proteins detected by shotgun analysis (n = 5). Supplemental Data S3 shows the detailed results of protein identification and quantitation.
Fig. 8.
Analysis of changes in the transcription of genes involved in iron uptake pathways in P. aeruginosa cells during A549 epithelial cell infection, in the absence and presence of exosiderophores. A, Transcription analyses. Histograms with green bars: RT-qPCR was performed on P. aeruginosa PAO1 cells after 3 h of incubation in RPMI without A549 epithelial cells and with or without 10 μm ENT, FERRI, VIB, or YER. Histograms with blue bars: RT-qPCR was performed on P. aeruginosa PAO1 cells after 3 h of incubation with A549 epithelial cells in RPMI medium, with or without 10 μm ENT, FERRI, VIB, or YER. The data were normalized relative to the reference gene uvrD and are representative of three independent experiments performed in triplicate (n = 3). For all eight histograms, the results are given as the ratio between the values obtained in the presence of siderophores over those obtained in their absence. B, RT-qPCR analyses were performed on P. aeruginosa PAO1 cells after 3 h of incubation with A549 epithelial cells in RPMI medium, supplemented or not with a mixture of 2 μm ENT, 2 μm FERRI, 2 μm VIB, and 2 μm YER (total of 8 μm siderophores). The experiment was repeated with a mixture of 10 μm of each siderophore (total of 40 μm siderophores). The data are represented as an histogram and a heat map. As for panel A, the data were normalized relative to the reference gene uvrD and are representative of three independent experiments performed in triplicate (n = 3). Results are given as the ratio between the values obtained in the presence of siderophores over those obtained in their absence. For both A and B, the data represent 3 independent experiments with 3 technical replicates.
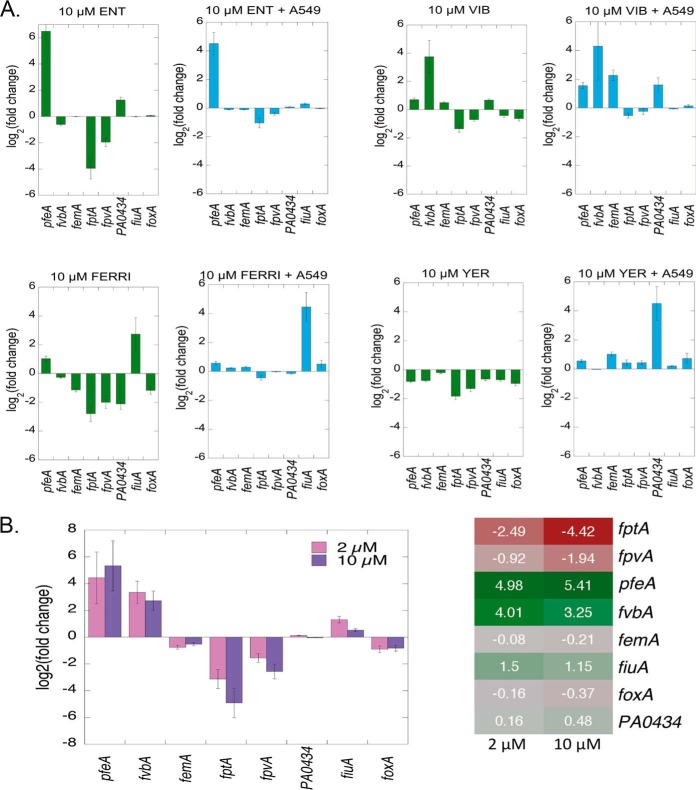
All iron-uptake pathways of P. aeruginosa in the infection assay were expressed at a much lower level than in bacteria grown overnight in RPMI medium, because the bacteria were less iron-starved (Fig. 6). Despite this context, all four exosiderophores induced the transcription and expression of their specific TBDTs during the 3 h infection period. The presence of ENT efficiently induced the specific transcription and expression of pfeA and FERRI of fiuA (Fig. 7 and 8A) under both conditions, with and without A549 cells, showing that the ability of exosiderophores to induce the expression of their corresponding TBDT also functions in complex systems, such as this epithelial cell infection assay. VIB induced the transcription of four transporters, fvbA, pfeA, femA, and PA0434, in RPMI in the presence of A549 cells (Fig. 7 and 8A), whereas only fvbA transcription was clearly activated in the absence of A549 cells. These two phenotypes are different from that observed with CAA medium: induction of the transcription of both fvbA and femA TBDTs with equivalent efficiencies. For YER, a siderophore unable to transport iron into P. aeruginosa cells, the results were also completely different from those observed in CAA medium. There was strong induction of transcription and expression of PA0434 in the epithelial infection assay (Fig. 7 and 8A), whereas femA expression was induced in CAA medium and no TBDT was induced in RPMI alone.
Another important observation was the absence of additional repression of PCH, PVD, or any other iron-uptake pathways in the presence of A549 cells and one of the exosiderophores, whereas we observed repression of transcription of the PCH and PVD iron-uptake pathways in the same experiment in the absence of A549 cells (fold change between 0.06 (log2(0.06) = −4) and 0.39 (log2(0.39) = −1.4) for fptA, and between 0.25 (log2(0.25) = −2) and 0.61 (log2(0.61) = −0.7) fpvA transcription; Fig. 8A). PCH and PVD appear to have more access to iron (and consequently induce the transcription of their corresponding uptake pathways) in the presence of A549 cells than in their absence, probably because the A549 cells serve as an iron source, with this nutrient being released during epithelial cell lysis. Therefore, bacteria are in a less iron-restricted environment in the presence of A549 cells, but one which still allows activation of the transcription and expression of iron-uptake pathways dedicated to exosiderophores (when these siderophores are present in the growth media). In the absence of A549 cells, the addition of an exosiderophore makes iron less accessible for PVD and PCH, resulting in additional repression of the transcription of these pathways.
We also investigated the effect of the mixture of 2 or 10 μm of the four siderophores, ENT, VIB, FERRI, and YER, on the transcription of P. aeruginosa iron-uptake pathways when the bacteria infected A549 cells (Fig. 8B). The mixture of the four exosiderophores (1) again strongly induced the transcription of pfeA and fvbA, as in CAA, (2) not that of fiuA or foxA, (3) nor that of femA or PA0434, and finally (iv) fptA and, surprisingly, also fpvA transcription were significantly repressed. The presence of high concentrations of exosiderophores makes iron poorly accessible to PCH and even PVD, despite the presence of epithelial cells as an iron source.
In conclusion, our results show high phenotypic plasticity for the transcription and expression of iron-uptake pathways. P. aeruginosa cells adapt and modulate the expression of their iron-uptake pathways as a function of environmental stimuli, which can be highly diverse. The presence of exosiderophores induces the transcription and expression of their corresponding TBDTs, even in an infection assay, in which bacteria are poorly iron starved. Moreover PA0434, a TBDT of unknown function, likely plays a significant role in P. aeruginosa growth and metal homeostasis, as its transcription and expression was activated in RPMI medium.
DISCUSSION
Most bacteria do not live in planktonic conditions, but rather in biofilms and are members of large communities called microbiota. Microbiota play diverse and key roles, both across the planet (in the carbon and nitrogen cycles and across all ecosystems) and in humans (involved in many diseases, such as obesity and inflammatory or autoimmune diseases) (63, 64). Such microbiota display very wide bacterial species diversity with, for example, more than 1,000 bacterial species making up the human microbiome. In such communities, iron is at the origin of strong competition and the ability to produce siderophores plays a fundamental role in the capacity of bacteria to access this nutrient and survive. For example, siderophore concentrations in soil range from tens of micromoles to a few millimoles per liter, indicating that bacteria are generally in the presence of siderophores, probably essentially those produced by other microorganisms (65).
Bacterial genome sequencing has shown that most bacteria can produce siderophores and have genes within their genomes that encode transporters involved in the import of ferri-siderophore complexes. In addition, the ability to hack siderophores produced by other bacteria is not restricted to P. aeruginosa but is frequently encountered among bacterial species. The presence of many potential iron uptake pathways in the genome does not mean that they are all expressed simultaneously. In contrast to the large amount of genomic data generated by bacterial genome sequencing, almost no data are available concerning the phenotypic plasticity linked to the expression of iron-uptake pathways. As already mentioned, P. aeruginosa is able to express (1) one ferrous (Fe2+) iron-uptake pathway, (2) three heme-acquisition pathways, (3) ferric (Fe3+) iron-uptake pathways by the two main siderophores PVD and PCH, produced by the pathogen, and (4) at least 10 different “siderophore piracy” strategies to take up Fe3+ using exosiderophores (7). The presence of such a large panel of iron-acquisition pathways in the genome involves even a larger panel of possible phenotypes, with various combinations of expression of these iron-uptake pathways, depending on the environmental stimuli.
Growth of P. aeruginosa in three different media (LB, CAA, and RPMI) clearly showed three different phenotypic patterns for the expression of the various iron-uptake pathways present in the genome. None of these iron uptake pathways were apparently expressed in LB (iron-rich medium) (Table I). Growth in CAA resulted in the induction of the expression of the PVD and PCH pathways, as well as the heme-uptake pathways and two TBDT transporters of unknown function relative to the cultures in LB (Table I and Fig. 4). Growth in RPMI medium resulted in a more complex phenotype in terms of iron-uptake pathway expression than for bacteria grown in CAA: the same iron-uptake pathways as in CAA were expressed along with the induction of the expression of three other TBDTs (PirA, ChtA, PA0781) potentially involved in iron acquisition (Table I and Fig. 4). These data also demonstrate that bacteria do not express just one iron uptake pathway but a combination of a few, probably those with the highest chance to scavenge iron from the growth environment. Bacteria first grown in LB and then incubated for 3 h in RPMI in the presence of A549 cells showed lower transcription of all TBDTs tested relative to bacteria grown overnight in RPMI (supplemental Fig. S2), as P. aeruginosa was not yet iron starved after 3 h of incubation in RPMI. However, the presence of epithelial cells slightly induced more significantly the transcription of several genes coding for virulence factors (Fig. 6), indicating that the bacteria sensed the presence of the epithelial cells. The results also show that the media, the chemical environment (iron concentrations), more strongly affects the transcription and expression levels of the various iron-uptake pathways than the presence of epithelial cells.
The presence of exosiderophores (ENT, VIB, FERRI, or YER) induced the expression of specific TBDTs, regardless of the growth conditions tested (Figs. 5, 7, and 8), as well as other proteins involved in iron release from the siderophores (for example the esterase PfeE in the presence of ENT). In each case, the bacteria sensed the presence of the chelators in their environment and adapted their phenotype to access iron via the exosiderophore present. Consequently, the more iron-uptake pathways dedicated to the uptake of ferric forms of exosiderophores are present in the genome of a bacterium, the higher are the number of possible combinations of their expression, allowing greater potential adaptation of the bacteria to diverse environments and microbiota in which many different siderophores are present.
When P. aeruginosa was in the presence of a mixture of the four siderophores, the level of induction of expression of the corresponding TBDTs was strongly mediated by the affinity of the siderophores for ferric iron, resulting in the catechol-type siderophores having the strongest capacity to induce their corresponding uptake pathways (PfeA and FvbA) (Figs. 4, 7 and 8). PCH and hydroxamate siderophores, such as FERRI, had difficulty scavenging iron in the presence of tris-catechol-type siderophores and subsequently inducing the expression of their corresponding TBDTs. The epithelial cell infection assay also showed that the presence of eukaryotic cells had no inhibitory effect on the ability of exosiderophores to induce the transcription of their corresponding uptake pathways (Fig. 7 and 8). The key element in siderophore-mediated bacterial interspecies competition for iron clearly lies in the ability of the various siderophores to scavenge iron from other chelating compounds in the bacterial environment, which directly affects the level of expression of the proteins of the various iron-uptake pathways present in the bacterial genome. Bacteria that can produce and/or use catechol siderophores have an environmental advantage over other bacteria.
Under most of the conditions tested here, catechol siderophores significantly repressed the transcription and expression of the proteins of the PCH pathway as well as that of the PVD pathway, but to a lesser extent. This can be explained by the higher affinity of PVD than PCH for iron (1032 m−1 and 1018 m−2 for PVD (56) and PCH (58)). Previously, Dumas et al. have shown that PCH is first produced and is replaced by PVD when the concentration of iron decreases (66) because a siderophore with a higher affinity for iron is needed to scavenge this metal from strongly iron-restricted environments. Consistent with the work of Dumas, we have previously shown that P. aeruginosa grown in CAA medium produces higher amounts of PVD than PCH (52), because this medium is highly iron restricted. Here, in CAA medium and in the presence of exosiderophores, PVD was still produced and in sufficiently high amounts (54.2 ± 1.1 μm in the presence of 10 μm ENT) to remain competitive with ENT and VIB for iron chelation and induce the expression of the proteins of the PVD pathway. Thus, these data show that in the presence of catechol siderophores (siderophores with a strong affinity for iron 1049 m−1 for ENT (55)), P. aeruginosa cells switch off first PCH production (this siderophore being no more able to chelate the metal and activate its auto-regulating loop). Moreover, the ability of bacteria to produce large amounts of a given siderophore (up to 150 μm in the case of PVD in the absence of any exosiderophore (52)) is also an advantage for the expression of a pathway. We observed no repression of the transcription of the ferrous and heme-uptake pathways in the presence of exosiderophores under any of the growth conditions tested.
There was also significant induction of femA and PA0434 gene expression under several growth conditions tested, indicating that femA may have broader siderophore specificity than previously reported and that both transporters must play a critical role in iron homeostasis under certain growth conditions. Moreover, a siderophore such as YER, which is unable to transport iron into P. aeruginosa cells but sequesters iron in the bacterial environment, also clearly affects the expression of various iron-uptake pathways and induces phenotypic switching.
Overall, the results show very high and complex phenotypic plasticity for the expression of the iron-uptake pathways present in the P. aeruginosa genome. This involves very fine modulation for the adaptation of the bacteria to different environments.
55Fe uptake assays (Fig. 2) have shown that the siderophores tested (except YER) have very similar uptake rates, except when another siderophore enters into competition for iron, and are consistent with those previously reported (54, 67, 68). Bacteria express a much higher level of TBDTs at their outer membrane than are needed for iron uptake; only ∼10% of the TBDT of a siderophore is involved in iron uptake (42). This is not surprising, as the ratio between the amount of a given TBDT present in the outer membrane and the inner membrane TonB protein (TonB being the protein which transfers the energy from the inner membrane to the TBDT for uptake of ferri-siderophore complexes) has been estimated to at least 10 (69). Bacteria probably express more TBDT than needed to maintain a certain concentration of siderophore-Fe complexes bound at the bacterial surface, a strategy to sequester iron with a usable siderophore at the bacterial surface. Consequently, induction of the expression of a given siderophore-dependent iron-uptake pathway allows uptake of the corresponding siderophore-Fe complex by the bacteria, as well as storage of a certain amount of ferri-siderophore complexes bound to their corresponding TBDT at the bacterial surface awaiting TonB activation.
In conclusion, the data provide new and clear insights into how bacteria adapt the expression of the iron-uptake pathways present in their genome according to their growth environment, with the simultaneous expression of several iron-uptake pathways. These phenotypic patterns show amazing plasticity and can be finely modulated and adjusted by environmental stimuli, especially the iron concentration and the presence of exosiderophores. Among siderophores, tris-catechol siderophores are clearly the most powerful iron chelators, with a strong capacity to induce the expression of their corresponding iron-uptake pathways in bacteria, even in bacteria that are not completely iron starved, and consequently must play dominant roles in the competition for iron in microbiota. The repression of the transcription and expression of the endogenous siderophore-dependent iron uptake pathways is almost always observed in the presence of tris-catechol siderophores, indicating that the bacteria use the tris-catechol siderophores to access iron, rather than their own siderophores. Such high phenotypic plasticity concerning the various iron-uptake pathways present in P. aeruginosa genome indicates a high potential of adaptation to a large variety of biotopes.
An increasing amount of data show that vectorization of antibiotics by siderophores may be an attractive strategy in the context of antibiotic resistance, providing the possibility to efficiently transport any potential antibiotic unable to cross the bacterial envelop into bacteria (70). The high phenotypic plasticity of P. aeruginosa and probably that of other bacteria may initially appear to be a handicap for the development of such a strategy. However, the strong capacity of tris-catechol siderophores to induce the transcription of their corresponding TBDTs in various growth media, and even in the presence of other siderophores, is a property that should be considered and explored in siderophore-dependent Trojan horse strategies.
DATA AVAILABILITY
The MS data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD011950 (48) and PXD015638 (51).
Supplementary Material
Footnotes
* This work was partially funded by RF20160501642 from the associations Vaincre la Mucoviscidose and Gregory Lemarchal. In addition, these results were generated as part of the work of the Translocation Consortium (www.translocation.com), supported by the Innovative Medicines Joint Undertaking under Grant Agreement no. 115525, through financial contributions from the European Union's Seventh Framework Program (FP7/2007–2013) and contributions in kind from EFPIA companies. The mass spectrometry instrumentation at the IBMC was funded by the University of Strasbourg, IdEx “Equipement mi-lourd” 2015. The equipment at the IPHC was partly funded by the French Proteomics Infrastructure (ProFI; ANR-10-INSB-08-03). The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- TBDT
- tonB-dependent transporters
- ACN
- acetonitrile
- CAA
- casamino acid medium
- CAS
- chrome azurol S
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazine
- CV
- coefficient of variation
- Da
- dalton
- E. coli
- Escherichia coli
- EDTA
- ethylenediaminetetraacetic acid
- ENT
- enterobactin
- FERRI
- ferrichrome
- HEPES
- (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- LB
- luria-bertani
- LC-MS
- liquid chromatography-mass spectrometry
- MS
- mass spectrometry/spectroscopy
- MS/MS
- tandem mass spectrometry/spectroscopy
- NanoLC-MS
- nano liquid chromatography-mass spectrometry
- OD
- optical density
- PBS
- phosphate buffered saline
- PCH
- pyochelin
- PCR
- polymerase chain reaction
- P. aeruginosa
- Pseudomonas aeruginosa
- PSM
- Peptide spectrum matches
- PVD
- pyoverdine
- RPMI
- roswell park memorial institute medium
- RT-qPCR
- reverse transcriptase polymerase chain reaction
- SID
- siderophore
- SLSA
- structured least square adaptative
- VIB
- vibriobactin
- YER
- yersiniabactin.
REFERENCES
- 1. Núñez G., Sakamoto K., and Soares M. P. (2018) Innate nutritional immunity. J. Immunol. 201, 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Begg S. L. (2019) The role of metal ions in the virulence and viability of bacterial pathogens. Biochem. Soc. Trans. 47, 77–87 [DOI] [PubMed] [Google Scholar]
- 3. Hider R. C., and Kong X. (2011) Chemistry and biology of siderophores. Nat. Prod. Reports 27, 637–657 [DOI] [PubMed] [Google Scholar]
- 4. Schalk I. J. (2013) Innovation and originality in the strategies developed by bacteria to get access to iron. Chembiochem 14, 293–294 [DOI] [PubMed] [Google Scholar]
- 5. Schalk I. J., and Guillon L. (2013) Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 44, 1267–1277 [DOI] [PubMed] [Google Scholar]
- 6. Schalk I. J., Mislin G. L. A., and Brillet K. (2012) Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Curr. Top. Membr. 69, 37–66 [DOI] [PubMed] [Google Scholar]
- 7. Cornelis P., and Dingemans J. (2013) Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infection Microbiol. 3, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luckey M., Pollack J. R., Wayne R., Ames B. N., and Neilands J. B. (1972) Iron uptake in Salmonella typhimurium: utilization of exogenous siderochromes as iron carriers. J. Bacteriol. 111, 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schubert S., Fischer D., and Heesemann J. (1999) Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J. Bacteriol. 181, 6387–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Griffin A. S., West S. A., and Buckling A. (2004) Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 11. West S. A., Griffin A. S., Gardner A., and Diggle S. P. (2006) Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607 [DOI] [PubMed] [Google Scholar]
- 12. Ross-Gillespie A., Gardner A., West S. A., and Griffin A. S. (2007) Frequency dependence and cooperation: theory and a test with bacteria. Am. Nat. 170, 331–342 [DOI] [PubMed] [Google Scholar]
- 13. Llamas M. A., Mooij M. J., Sparrius M., Vandenbroucke-Grauls C. M., Ratledge C., and Bitter W. (2008) Characterization of five novel Pseudomonas aeruginosa cell-surface signalling systems. Mol. Microbiol. 67, 458–472 [DOI] [PubMed] [Google Scholar]
- 14. Kümmerli R., Griffin A. S., West S. A., Buckling A., and Harrison F. (2009) Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc. Biol. Sci. 276, 3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. D'Onofrio A., Crawford J. M., Stewart E. J., Witt K., Gavrish E., Epstein S., Clardy J., and Lewis K. (2010) Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 17, 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scholz R. L., and Greenberg E. P. (2015) Sociality in Escherichia coli: enterochelin is a private good at low cell density and can be shared at high cell density. J. Bacteriol. 197, 2122–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kümmerli R., Santorelli L. A., Granato E., Dumas Z., Dobay A., Griffin A. S., and West S. A. (2015) Co-evolutionary dynamics between public good producers and cheats in the bacterium Pseudomonas aeruginosa. J. Evol. Biol. 28, 64. [DOI] [PubMed] [Google Scholar]
- 18. Weaver V. B., and Kolter R. (2004) Burkholderia spp. alter Pseudomonas aeruginosa physiology through iron sequestration. J. Bacteriol. 186, 2376–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Joshi F., Archana G., and Desai A. (2006) Siderophore cross-utilization amongst rhizospheric bacteria and the role of their differential affinities for Fe3+ on growth stimulation under iron-limited conditions. Curr. Microbiol. 53, 141–147 [DOI] [PubMed] [Google Scholar]
- 20. Harrison F., Paul J., Massey R. C., and Buckling A. (2008) Interspecific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa. ISME J 2, 49–55 [DOI] [PubMed] [Google Scholar]
- 21. Butaitė E., Baumgartner M., Wyder S., and Kümmerli R. (2017) Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat. Commun. 8, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leinweber A., Fredrik Inglis R., and Kümmerli R. (2017) Cheating fosters species co-existence in well-mixed bacterial communities. ISME. J. 11, 1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niehus R., Picot A., Oliveira N. M., Mitri S., and Foster K. R. (2017) The evolution of siderophore production as a competitive trait. Evolution 71, 1443–1455 [DOI] [PubMed] [Google Scholar]
- 24. Schiessl K. T., Janssen E. M.-L., Kraemer S. M., McNeill K., and Ackermann M. (2017) Magnitude and mechanism of siderophore-mediated competition at low iron solubility in the Pseudomonas aeruginosa Pyochelin System. Front. Microbiol. 8, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leinweber A., Weigert M., and Kümmerli R. (2018) The bacterium Pseudomonas aeruginosa senses and gradually responds to inter-specific competition for iron. Evolution 72, 1515–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cornelis P., and Bodilis J. (2009) A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ. Microbiol. Reports 1, 256–262 [DOI] [PubMed] [Google Scholar]
- 27. Visca P., Leoni L., Wilson M. J., and Lamont I. L. (2002) Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45, 1177–1190 [DOI] [PubMed] [Google Scholar]
- 28. Llamas M. A., Imperi F., Visca P., and Lamont I. L. (2014) Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol. Rev. 38, 569–597 [DOI] [PubMed] [Google Scholar]
- 29. Dean C. R., and Poole K. (1993) Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol. Microbiol. 8, 1095–1103 [DOI] [PubMed] [Google Scholar]
- 30. Heinrichs D. E., and Poole K. (1996) PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178, 2586–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Michel L., Gonzalez N., Jagdeep S., Nguyen-Ngoc T., and Reimmann C. (2005) PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol. Microbiol. 58, 495–509 [DOI] [PubMed] [Google Scholar]
- 32. Gasser V., Baco E., Cunrath O., August P. S., Perraud Q., Zill N., Schleberger C., Schmidt A., Paulen A., Bumann D., Mislin G. L. A., and Schalk I. J. (2016) Catechol siderophores repress the pyochelin pathway and activate the enterobactin pathway in Pseudomonas aeruginosa: an opportunity for siderophore-antibiotic conjugates development. Environ. Microbiol. 18, 819–832 [DOI] [PubMed] [Google Scholar]
- 33. Perraud Q., Moynié L., Gasser V., Munier M., Godet J., Hoegy F., Mély Y., Mislin G. L. A., Naismith J. H., and Schalk I. J. (2018) A key role for the periplasmic PfeE esterase in iron acquisition via the siderophore enterobactin in Pseudomonas aeruginosa. ACS Chem. Biol. 13, 2603–2614 [DOI] [PubMed] [Google Scholar]
- 34. Michel L., Bachelard A., and Reimmann C. (2007) Ferripyochelin uptake genes are involved in pyochelin-mediated signalling in Pseudomonas aeruginosa. Microbiology 153, 1508–1518 [DOI] [PubMed] [Google Scholar]
- 35. Raymond K. N., Dertz E. A., and Kim S. S. (2003) Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wyckoff E. E., Mey A. R., and Payne S. M. (2007) Iron acquisition in Vibrio cholerae. Biometals 20, 405–416 [DOI] [PubMed] [Google Scholar]
- 37. Leong S. A., and Winkelmann G. (1998) Molecular biology of iron transport in fungi. Metal Ions Biol. Syst. 35, 147–186 [PubMed] [Google Scholar]
- 38. Perry R. D., and Fetherston J. D. (2011) Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infection 13, 808–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Youard Z. A., Mislin G. L., Majcherczyk P. A., Schalk I. J., and Reimmann C. (2007) Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J. Biol. Chem. 282, 35546–35553 [DOI] [PubMed] [Google Scholar]
- 40. Zamri A., and Abdallah M. A. (2000) An improved stereocontrolled synthesis of pyochelin, a siderophore of Pseudomonas aeruginosa and Burkholderia cepacia. Tetrahedron 56, 249–256 [Google Scholar]
- 41. Demange P., Wendenbaum S., Linget C., Mertz C., Cung M. T., and Dell A., Abdallah M. A. (1990) Bacterial siderophores: structure and NMR assigment of pyoverdins PaA, siderophores of Pseudomonas aeruginosa ATCC 15692. Biol. Metals 3, 155–170 [Google Scholar]
- 42. Clément E., Mesini P. J., Pattus F., Abdallah M. A., and Schalk I. J. (2004) The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry 43, 7954–7965 [DOI] [PubMed] [Google Scholar]
- 43. Schwyn B., and Neilands J. B. (1987) Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56 [DOI] [PubMed] [Google Scholar]
- 44. Gross H., and Loper J. E. (2009) Genomics of secondary metabolite production by Pseudomonas spp. Nat. Product Reports 26, 1408–1446 [DOI] [PubMed] [Google Scholar]
- 45. Waltz F., Nguyen T.-T., Arrivé M., Bochler A., Chicher J., Hammann P., Kuhn L., Quadrado M., Mireau H., Hashem Y., and Giegé P. (2019) Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 5, 106–117 [DOI] [PubMed] [Google Scholar]
- 46. Carapito C., Lane L., Benama M., Opsomer A., Mouton-Barbosa E., Garrigues L., Gonzalez de Peredo A., Burel A., Bruley C., Gateau A., Bouyssié D., Jaquinod M., Cianferani S., Burlet-Schiltz O., Van Dorsselaer A., Garin J., and Vandenbrouck Y. (2015) Computational and mass-spectrometry-based workflow for the discovery and validation of missing human proteins: application to chromosomes 2 and 14. J. Proteome Res. 14, 3621–3634 [DOI] [PubMed] [Google Scholar]
- 47. Gregori J., Sanchez A., and Villanueva J. (2019) msmsTests: LC-MS/MS Differential Expression Tests (Bioconductor version: Release (3.9)) [Google Scholar]
- 48. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.-W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muller L., Fornecker L., Van Dorsselaer A., Cianférani S., and Carapito C. (2016) Benchmarking sample preparation/digestion protocols reveals tube-gel being a fast and repeatable method for quantitative proteomics. Proteomics 16, 2953–2961 [DOI] [PubMed] [Google Scholar]
- 50. Wieczorek S., Combes F., Lazar C., Giai Gianetto Q., Gatto L., Dorffer A., Hesse A.-M., Couté Y., Ferro M., Bruley C., and Burger T. (2017) DAPAR & ProStaR: software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics 33, 135–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P.-A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H.-J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cunrath O., Geoffroy V. A., and Schalk I. J. (2016) Metallome of Pseudomonas aeruginosa: a role for siderophores. Environ. Microbiol. 18, 3258–3267 [DOI] [PubMed] [Google Scholar]
- 53. Poole K., Young L., and Neshat S. (1990) Enterobactin-mediated iron transport in Pseudomonas aeruginosa. J. Bacteriol. 172, 6991–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hannauer M., Barda Y., Mislin G. L., Shanzer A., and Schalk I. J. (2010) The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechansim with acylation of the siderophore and a recycling of the modified desferrichrome. J. Bacteriol. 192, 1212–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loomis L., and Raymond K. N. (1991) Solution equilibria of enterobactin complexes. Inorg. Chem. 30, 906–911 [Google Scholar]
- 56. Albrecht-Gary A. M., Blanc S., Rochel N., Ocacktan A. Z., and Abdallah M. A. (1994) Bacterial iron transport: coordination properties of pyoverdin PaA, a peptidic siderophore of Pseudomonas aeruginosa. Inorg. Chem. 33, 6391–6402 [Google Scholar]
- 57. Anderegg G., L'Eplattenier F., and Schwarzenbach G. (1963) Hydroxamatkomplexe III. Eisen(III)-Austausch zwischen Sideraminen und Komplexonen. Diskussion der Bildungskonstanten der Hydroxamatkomplexe. Helvetica Chimica. Acta 46, 1409–1422 [Google Scholar]
- 58. Brandel J., Humbert N., Elhabiri M., Schalk I. J., Mislin G. L. A., and Albrecht-Garry A.-M. (2012) Pyochelin, a siderophore of Pseudomonas aeruginosa: Physicochemical characterization of the iron(III), copper(II) and zinc(II) complexes. Dalton Trans. 41, 2820–2834 [DOI] [PubMed] [Google Scholar]
- 59. Folschweiller N., Gallay J., Vincent M., Abdallah M. A., Pattus F., and Schalk I. J. (2002) The interaction between pyoverdin and its outer membrane receptor in Pseudomonas aeruginosa leads to different conformers: a time-resolved fluorescence study. Biochemistry 41, 14591–14601 [DOI] [PubMed] [Google Scholar]
- 60. Cunrath O., Gasser V., Hoegy F., Reimmann C., Guillon L., and Schalk I. J. (2015) A cell biological view of the siderophore pyochelin iron uptake pathway in Pseudomonas aeruginosa. Environ. Microbiol. 17, 171–185 [DOI] [PubMed] [Google Scholar]
- 61. Elias S., Degtyar E., and Banin E. (2011) FvbA is required for vibriobactin utilization in Pseudomonas aeruginosa. Microbiology 157, 2172–2180 [DOI] [PubMed] [Google Scholar]
- 62. Winsor G. L., Griffiths E. J., Lo R., Dhillon B. K., Shay J. A., and Brinkman F. S. L. (2016) Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Esteve E., Ricart W., and Fernández-Real J.-M. (2011) Gut microbiota interactions with obesity, insulin resistance and type 2 diabetes: did gut microbiote co-evolve with insulin resistance? Curr. Opin. Clin. Nutr. Metab. Care 14, 483–490 [DOI] [PubMed] [Google Scholar]
- 64. Moran M. A. (2015) The global ocean microbiome. Science 350, aac8455. [DOI] [PubMed] [Google Scholar]
- 65. Hersman L., Lloyd T., and Sposito G. (1995) Siderophore-promoted dissolution of hematite. Geochim. Cosmochim. Acta 59, 3327–3330 [Google Scholar]
- 66. Dumas Z., Ross-Gillespie A., and Kümmerli R. (2013) Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc. Biol. Sci. 280, 20131055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chakraborty R., Lemke E. A., Cao Z., Klebba P. E., and van der Helm D. (2003) Identification and mutational studies of conserved amino acids in the outer membrane receptor protein, FepA, which affect transport but not binding of ferric-enterobactin in Escherichia coli. Biometals. 16, 507–518 [DOI] [PubMed] [Google Scholar]
- 68. Hoegy F., Lee X., Noël S., Mislin G. L., Rognan D., Reimmann C., and Schalk I. J. (2009) Stereospecificity of the siderophore pyochelin outer membrane transporters in fluorescent Pseudomonads. J. Biol. Chem. 284, 14949–14957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Higgs P. I., Larsen R. A., and Postle K. (2002) Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44, 271–281 [DOI] [PubMed] [Google Scholar]
- 70. Mislin G. L. A., and Schalk I. J. (2014) Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics 6, 408–420 [DOI] [PubMed] [Google Scholar]
- 71. Ankenbauer R. G., and Quan H. N. (1994) FptA, the Fe(III)-pyochelin receptor of Pseudomonas aeruginosa: a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J. Bacteriol. 176, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Poole K., Neshat S., Krebes K., and Heinrichs D. E. (1993) Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J. Bacteriol. 175, 4597–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ghysels B., Dieu B. T., Beatson S. A., Pirnay J. P., Ochsner U. A., Vasil M. L., and Cornelis P. (2004) FpvB, an alternative type I ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology 150, 1671–1680 [DOI] [PubMed] [Google Scholar]
- 74. Cuiv P. O., Clarke P., and O'Connell M. (2006) Identification and characterization of an iron-regulated gene, chtA, required for the utilization of the xenosiderophores aerobactin, rhizobactin 1021 and schizokinen by Pseudomonas aeruginosa. Microbiology 152, 945–954 [DOI] [PubMed] [Google Scholar]
- 75. Dean C. R., and Poole K. (1993) Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J. Bacteriol. 175, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ghysels B., Ochsner U., Mollman U., Heinisch L., Vasil M., Cornelis P., and Matthijs S. (2005) The Pseudomonas aeruginosa pirA gene encodes a second receptor for ferrienterobactin and synthetic catecholate analogues. FEMS Microbiol. Lett. 246, 167–174 [DOI] [PubMed] [Google Scholar]
- 77. Marshall B., Stintzi A., Gilmour C., Meyer J.-M., and Poole K. (2009) Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology 155, 305–315, [DOI] [PubMed] [Google Scholar]
- 78. Llamas M. A., Sparrius M., Kloet R., Jimenez C. R., Vandenbroucke-Grauls C., and Bitter W. (2006) The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 188, 1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ochsner U. A., Vasil M. L., Alsabbagh E., Parvatiyar K., and Hassett D. J. (2000) Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182, 4533–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Otero-Asman J. R., García-García A. I., Civantos C., Quesada J. M., and Llamas M. A. (2019) Pseudomonas aeruginosa possesses three distinct systems for sensing and using the host molecule haem. Environ. Microbiol. 21, 4629–4647 [DOI] [PubMed] [Google Scholar]
- 81. Lhospice S., Gomez N. O., Ouerdane L., Brutesco C., Ghssein G., Hajjar C., Liratni A., Wang S., Richaud P., Bleves S., Ball G., Borezée-Durant E., Lobinski R., Pignol D., Arnoux P., and Voulhoux R. (2017) Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep. 7, 17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yoneyama H., and Nakae T. (1996) Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 142, 2137–2144 [DOI] [PubMed] [Google Scholar]
- 83. Moynié L., Luscher A., Rolo D., Pletzer D., Tortajada A., Weingart H., Braun Y., Page M. G. P., Naismith J. H., and Köhler T. (2017) Structure and function of the PiuA and PirA siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother. 61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD011950 (48) and PXD015638 (51).