Abstract
The relevance of protein tyrosine phosphatase (PTP) to host–pathogen interaction is highlighted in mammalian studies, whereas less is known in insects. Here we presented the categorization of the PTP complement of silkworm and characterized their homologous relationship with human and fruit fly PTPs. Among the 36 PTP genes, ptp-h, which was proposed to be the origin of baculovirus ptp belongs to atypical VH1-like dual-specific PTP subset and encodes a catalytic active protein. The maximum expression level of Bmptp-h was at 5th instar and in fat body. Bombyx mori nucleopolyhedrovirus (BmNPV) infection potently induced its expression in silkworm larvae and in BmE cells. Knock-down of Bmptp-h by RNA interference significantly inhibited viral replication, and over-expression enhanced viral replication as determined by viral DNA abundance and BmNPV-GFP positive cells. These results suggest that BmPTP-h might be one of the host factors that is beneficial to baculovirus infection by promoting viral replication.
Keywords: Protein tyrosine phosphatase, Bombyx mori, BmPTP-h, Nucleopolyhedrovirus, Viral replication
Highlights
-
•
36 PTP genes were identified and classified in silkworm genome.
-
•
BmNPV infection potently induced the expression of Bmptp-h.
-
•
BmPTP-h has catalytic activity.
-
•
Knock-down of Bmptp-h by RNAi significantly inhibited baculovirus replication.
-
•
Over-expression of Bmptp-h enhanced baculovirus replication.
1. Introduction
Reversible protein tyrosine phosphorylation provides a potent and versatile mechanism for regulating various physiological processes in eukaryotic cells. Protein tyrosine kinases (PTKs), which catalyze the transfer of phosphoryl group to tyrosine residues, and protein tyrosine phosphatases (PTPs), which remove phosphate moiety from phosphotyrosine residues, work together to modulate signaling pathways. In some cases, PTPs play a dominate role in shaping the spatio-temporal dynamics of protein tyrosine phosphorylation network, making it a pivotal target for functional manipulation (Mustelin et al., 2005). Although the study of PTPs lagged behind of PTKs for historical reasons and technical difficulties, a large number of PTPs have been discovered in recent years, paralleling PTKs in sequence diversity and functional complexity (Alonso et al., 2004). Despite a minority reported to be Asp-based PTPs, all others possess at least one signature motif, [I/V]HCX5R[S/T] and share a Cys-based catalytic mechanism. Some PTPs serve as pTyr-specific enzymes, others can also dephosphorylate Ser/Thr residues, thus they are operationally defined as dual specificity phosphatase (DUSP). And the ability of removing phosphate group from phospholipid or RNA is also reported in some DUSPs (Tonks, 2013).
Intriguingly, numerous host–virus interactions are based on the interplay of PTKs and PTPs as revealed by studies on vertebrates and invertebrates (Devasthanam, 2014, Kingsolver et al., 2013). Mounting a potent immune response to invading virus relies on proper phosphorylation or dephosphorylation of signaling molecules in host cells. Reciprocally, virus develops strategies to utilize or disrupt this regulatory mechanism for survival and transmission. For instance, in vitro studies using mammalian cell lines demonstrated that DUSP1, which dephosphorylates MAPKs, facilitates the infection of hepatitis C virus (HCV) and coronavirus infectious bronchitis virus (IBV) (Choi et al., 2015, Liao et al., 2011). And congenital human cytomegalovirus (HCMV) can take advantage of CDC25, a cell cycle regulator to promote viral replication (Fu et al., 2015). However, due to the lack of systematic analysis of insect PTPs and distinct antiviral mechanism that insect may take, few surveys reported the relevance of insect PTPs to antiviral immune response.
Some virus also encode PTPs, which have been hypothesized or demonstrated to function as virulence factors that impair host immune response or other functions (Heneberg, 2012, Peters et al., 2002, Provost et al., 2004). The best-known example in insect is the expression of several bracovirus-encoded PTPs in lepidopteran hosts promoted the apoptosis of granulocytes and inhibited spreading and phagocytosis of plasmatocytes, therefore facilitating parasitoid development (Ibrahim et al., 2007, Ibrahim and Kim, 2008, Pruijssers and Strand, 2007). Interestingly, phylogenic analysis showed that those PTP genes were acquired from braconid genome and experienced complex evolutionary change (Espagne et al., 2004, Serbielle et al., 2012). Recent studies on another subset of insect virus identified a nucleopolyhedrovirus (NPV)-encoded PTP, which was presumably acquired from a lepidopteran host by horizontal gene transfer (HGT) as an essential factor to induce an enhanced locomotory activity (ELA) in host caterpillars (Kamita et al., 2005, van Houte et al., 2012). And NPV ptp deletion mutant lost its ability to manipulate its host behavior. Surprisingly, later study found BmNPV PTP functions as a virus-associated structural protein rather than as an enzyme in regard to the induction of ELA, although it still maintains the catalytic activity (Katsuma et al., 2012). Moreover, BmNPV ptp deletion mutant had reduced progeny production in silkworm larvae and showed a delay in late gene expression in cell lines. An earlier study identified a BmNPV ptp homolog encoded by Autographa californica multicapsid NPV (AcMNPV) as a dual phosphatase containing RNA 5’-triphosphatase activity which is implicated in processing of viral late mRNAs (Takagi et al., 1998). In contrast to BmNPV ptp mutant, AcMNPV ptp mutant seems to have no defect in replication in insect larvae, but partially defect in occluded virus production in a cell-specific manner (Li and Miller, 1995).
Given that HGT between host and pathogen has been proposed to be an important mechanism to increase pathogen survival and propagation (de la Casa-Esperon, 2012), it is interesting to trace back the function of the host copies to better understand the interaction between the host and pathogen. In the present study, we first analyzed BmPTP-h based on its functional categorization, then we investigated the immune-regulatory role it would play under the challenge of BmNPV in vitro and in vivo. We found that it increased expression level after viral infection and benefited viral replication. Our result provides complementary insights into the molecular machinery exploited by insect virus which could be pursued for antiviral strategy.
2. Experimental procedures
2.1. PTP identification and classification
Based on the annotated information of Bombyx mori comprehensive gene sets (http://sgp.dna.affrc.go.jp/ComprehensiveGeneSet/), sequence of protein predicted to possess tyrosine phosphatase domain was extracted and analyzed by online software SMART (http://smart.embl-heidelberg.de/). Protein that contains the PTP signature motif (CX5R) was then used as the query in BLASTP search against NCBI sequence repository to identify their paralogs. B. mori PTPs were classified into subfamilies according to their orthologous PTPs in human and fruit fly (Alonso et al., 2004, Hatzihristidis et al., 2015).
2.2. Insect, cells and virus
Silkworm larvae (DaZao P50 strain) were reared on fresh mulberry leaves at 25 °C and relative humidity of 80%. BmE cells were maintained at 27 °C in Grace medium (Gibco, UK) supplemented with 10% FBS (Hyclone, USA). Wide-type BmNPV (Guangdong strain, China) and recombinant BmNPV-GFP (courtesy of Dr. Xiaofeng Wu, Zhejiang University, China) were propagated in BmE cells and silkworm larvae following the standard procedure. Viral titer were determined by plaque assay in BmE cells (Katsuma et al., 1999).
2.3. Bmptp-h over-expression and RNAi
BmPTP-h-coding sequence (NCBI Gene ID: 692515) was cloned into the pSL1180 expression vector with a Flag tag at the N-terminus using primer set 1 in Table 1 dsRNA to Bmptp-h or red fluorescent protein gene (RFP) (dsBmPTP-h or dsRFP) were in vitro synthesized using T7 RiboMAX Large Scale RNA Production System (Promega, USA) with the primer sets 2 and 4. Plasmids or dsRNA were transfected into BmE cells using X-treme GENE transfection reagent (Roche, Switzerland) following the manufacturer's instruction. dsRNA was injected into silkworm larvae (20 μg/larva) at second day of fifth instar through the second last stoma in abdomen with a fine needle.
Table 1.
Sequence of primers used in this work.
| Gene | Primer set no. | Sequence of primers (5′-3′) |
|---|---|---|
| Bmptp-h | 1 | F: CGGGATCCATGGATTACAAGGATGACGACGATAAGCCTAAACTTCCCGATAGATGG R: ATTTGCGGCCGCTTTAACGATACCTTCTTCTAGTTGTTTCAG |
| 2 | F: TAATACGACTCACTATAGGGAGAGACTCGGTGCAGTCATAGAT R: TAATACGACTCACTATAGGGAGACTGCTGTTAGTCCGCTCTT |
|
| 3 | F: ACAATGCTTGCGGACGGGTAAT R: CCTCTAGAAGCGCCGGAATGTC |
|
| RFP | 4 | F: TAATACGACTCACTATAGGG GTACGGCTCCAAGGTGTACG R: TAATACGACTCACTATAGGGGGTGTAGTCCTCGTTGTGGG |
| gp64 | 5 | F: CCATCGTGGAGACGGACTA R: CTCGCACTGCTGCCTGA |
| sw22934 | 6 | F: TTCGTACTGGCTCTTCTCGT R: CAAAGTTGATAGCAATTCCCT |
| BmGAPDH | 7 | F: CATTCCGCGTCCCTGTTGCTAAT R: GCTGCCTCCTTGACCTTTTGC |
F: forward, R: reverse, Flag tag sequence is underlined.
2.4. Viral infection
For infection of cells, 5000 virions were incubated with 1×106 BmE cells 48 h after transfection. For infection of silkworm larvae, 10 000 virions were injected into silkworm larvae at Day 3 of 5th instar. At different time points after infection as indicated in graph, the expression level of Bmptp-h or BmNPV gp64 was subjected to quantitative PCR analysis.
2.5. Quantitative PCR (qPCR) analysis
Total RNA was extracted from cells or tissue samples using Total RNA Kit (Omega, USA) at different time points post viral infection and qPCR was performed using SYBR Premix Ex Taq II (TaKaRa, Japan) on a StepOne Plus Real-Time PCR System (Applied Biosystems, USA) with a program consisting of an initial denaturing step of 30 s at 95 °C and 40 amplification cycles consisting of 5 s at 95 °C followed by 30 s at 60 °C. Bmptp-h and sw22934 were amplified with primer sets 3 and 6 accordingly. For analysis of BmNPV replication, total DNA was extracted from viral infected BmE cells using the Universal Genomic DNA Extraction Kit (Takara) as described. BmNPV gp64 and BmGAPDH were amplified with primer sets 5 and 7 respectively. The expression level of Bmptp-h or BmNPV gp64 genes were normalized to the control sw22934 or BmGAPDH. The relative viral abundance was determined as described previously (Jiang et al., 2012).
2.6. Western blotting
Cells were solubilized in lysis buffer (100 mM NaCl, 50 mM Tris–HCl, 0.1% SDS, 1% NP-40 pH 7.5) containing protease inhibitor cocktail (Roche). After centrifugation at 17 000 g for 15 min at 4 °C, the supernatant was collected on ice for western blot analysis. Concentration of cell lysate was determined by BCA assay. A total of 20 μg protein per sample was resolved on 12% SDS-PAGE. After transfer to PVDF membrane (GE Health Care, USA), samples were immuno-blotted with anti-FLAG mAb or anti-Tubulin mAb (Sigma, USA) following the standard procedure.
2.7. Tyrosine phosphatase assay
1×106 BmE cells transfected with BmPTP-h expression vector were solubilized in 200 μl lysis buffer, then pre-cleared and incubated with casein (0.2 mM) for 10 min at 30 °C. The reaction was terminated by addition of TCA buffer. After centrifugation at 17 000 g for 10 min, 200 μl supernatant was mixed with 20 μl molydbate dye and liberated phosphate was visualized at 750 nm using a microplate reader (Promega).
2.8. Statistical analysis
Data were presented as the mean ± S.D. (n = 3). Statistical significant differences were determined by Student's t-test (for comparison of two means), or one-way analysis of variance (ANOVA) for multiple comparison test (*p < 0.05, **p < 0.01).
3. Results
3.1. Identification and classification of BmPTPs
Table 2 summarizes the 36 putative Cys-dependent tyrosine phosphatases identified in silkworm. All of them have full-length cDNA or matching EST sequence which indicates that they are not pseudogenes, except three genes (NCBI Gene ID: 101747207, 101735618, 101737822) which were predicted based on genome information. These tyrosine phosphatases were classified into four subfamilies based on their phosphatase catalytic domain sequences, including classical pTyr-specific PTPs, VH1-like dual-specific PTPs (DUSPs), low molecular weight PTP (LMWP) and CDC25. The classical PTPs were then divided in two subgroups, receptor- or non-receptor type PTPs based on whether they contained transmembrane domain or not. And DUSPs were annotated as MKPs (mitogen-activated protein kinases phosphatase), PTEN (phosphatase and tensin homologue deleted on chromosome 10), PRL (phosphatase of regenerating liver), CDC14, Myotubularins and atypical DUSPs based on their human and fruitfly orthologs, respectively. There are 14 classical pTyr-specific PTPs, 20 DUSPs, 1 LMWP and 1 CDC25 (and 1 Asp-dependent tyrosine phosphatase, EYA, NCBI GI: 827538455). Compared with the 43 Cys-dependent tyrosine phosphatases identified in Drosophila melanogaster, silkworm PTP gene set is smaller. Particularly, silkworm has less members in receptor-type, Myotubularin and LMWP subfamilies. Although no clear orthology relationship can be ascribed to some silkworm atypical DUSPs, they bear considerable similarity to human and Drosophila DUSPs, especially in PTP domains. Most of silkworm tyrosine phosphatases contain the conserved signature motif sequence [I/V]HCX5R[S/T], a few display variations at the first two amino acid residues and one has Gln instead of Ser/Thr at last position which is predicted to be catalytic inactive. Whereas 79 out of the 107 human PTPs are multidomain proteins, less than a half of silkworm PTPs harbor at least one characteristic domain, which may contribute to the specific interaction with regulatory or targeting molecules.
Table 2.
Categories of silkworm PTPs.
| PTP subfamily/Gene ID | Signature motif | Characteristic domain | Human ortholog | Fruitfly ortholog | Sequence source | |
|---|---|---|---|---|---|---|
| Classical pTyr-specific PTPs | ||||||
| Receptor | 101746939 | VHCSAGVGRT VHCQTGCERS |
IG, FN3 | RPTPα | PTP69D | EST,CDS |
| 101736187 | VHCSAGVGRS | FN3 | RPTPβ | PTP10D | FL cDNA | |
| 101737535 | VTCASGAGRS | PTPRγ | CG42327 | FL cDNA | ||
| 101738718 | VHCSDGAGRS | RPTPN2 | RPTPN2 | IA-2 | FL cDNA | |
| 101736712 | IHCSAGIGRT | PTPRR | PTP-ER | FL cDNA | ||
| 101747207 | VHCSAGVGRT | PTPRS | PTP99A | CDS | ||
| Non-receptor | 100579140 | VHCSAGIGRS | PTP1B | PTP61F | FL cDNA | |
| 101737573 | VHCSAGVVRT | ERM,FERM_C,PDZ | PTPN4 | PTPmeg | EST,CDS | |
| 101744807 | VHCSAGIGRT | CRAL_TRIO_N,Sec14 | PTPN9 | l(1)G0232 | FL cDNA | |
| 101735618 | YCCGEGAGRS | ERM,FERM_C | PTPN14 | Pez | CDS | |
| 101736631 | IQCGAGAGRS | BRO1-like | PTPN23 | MOP | EST,CDS | |
| 101736863 | VHCSAGVGRT | SH2 | SHP2 | Csw | EST,CDS | |
| 101737822 | VHCSAGIGRT | SH2 | SHP2 | Csw | CDS | |
| 101735492 | VHCNDGGGRS VHCNDGGGRS | RPTPκ | PTP36E | EST,CDS | ||
| VH1-like dual-specificity PTPs | ||||||
| MKP | 101744546 | VHCHFGVSRS | MKP-4 | MKP-4 | FL cDNA | |
| 101740325 | VHCVAGVSRS | DUSP7 | MKP-3 | EST,CDS | ||
| PTEN | 101745952 | VHCKAGKGRT | PTEN_C2 | PTEN | PTEN | EST,CDS |
| PRL | 101743511 | VHCVAGLGRA | PRL-1 | PRL-1 | FL cDNA | |
| Slingshot | 101737293 | VHCKMGISRS | SSH-N, DEK_C | SSH1 | SSH | FL cDNA |
| CDC14 | 101740096 | VHCKAGLGRT | CDC14A | CDC14 | EST,CDS | |
| 101744892 | IHCHAGLGRT | PTP9Q22 | CDC14 | EST,CDS | ||
| Myotubularin | 101740354 | VHCSDGWDRT | FYVE | MTMR3 | CG3632 | EST,CDS |
| 101736509 | VHCSDGWDRT | FYVE | MTMR6 | CG3530 | FL cDNA | |
| 101739438 | FLCDTDQERQ | MTMR9a | CG5026 | EST,CDS | ||
| 101736147 | VHCSDGWDRT | GRAM | MTMR2 | MTM | FL cDNA | |
| Atypical | 692515 | VHCTHGLNRT | DUSP11 | CG13197 | FL cDNA | |
| 101738891 | VHCYFGVSRS | DUSP12 | MKP-4 | FL cDNA | ||
| 101738231 | VHCMIGVSRS | DUSP13 | CG7378 | FL cDNA | ||
| 101743164 | VHCVAGVSRS | DUSP14 | CG15528 | EST,CDS | ||
| 101735761 | IHCLAGMSRS | DUSP22 | CG10089 | FL cDNA | ||
| 101744063 | IHCRQGRSRS | DUSP23b | CDC14b | FL cDNA | ||
| 101742480 | VHCRHGRGRT | DUSP23b | CDC14b | EST,CDS | ||
| 101737334 | VHCKAGRTRS | PTPMT1 | Plip | FL cDNA | ||
| 101740551 | VHCTHGFNRT | NADH_4Fe–4S | mRNA-cap | mRNA-cap | FL cDNA | |
| Low molecular weight PTP | ||||||
| 692857 | FICLGNICRS | LMWP | Primo-1 | FL cDNA | ||
| CDC25 | ||||||
| 100216491 | FHCEFSLERG | RHOD | CDC25C | Stg | EST,CDS | |
Information of BmPTP-h is highlighted in bold.
Human MTMR9 has been proved to be a catalytic inactive phosphatase.
No clear orthology relationship could be ascribed to two silkworm atypical PTPs (101744063, 101742480).
Bmptp-h (NCBI Gene ID: 692515), which was proposed to be the origin of BmNPV ptp, encodes an atypical VH1-like DUSP. It is homologous to Drosophila CG13197 and distantly related to human DUSP11, and the latter binds splicing complex and has been implicated in inflammatory bowel disease (Hasler et al., 2011, Yuan et al., 1998). BmPTP-h may also have 5’-triphosphatase activity using RNA as a substrate like DUSP11 (Deshpande et al., 1999). Clear function of these molecules is unknown.
3.2. Expression profile of Bmptp-h during the development and in different tissues
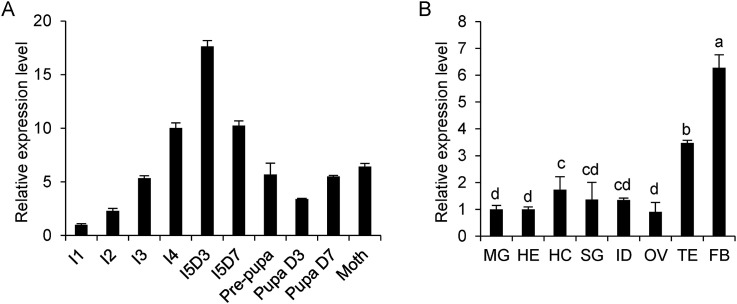
To evaluate the physiological functions of BmPTP-h, we first analyzed its expression profiles at different developmental stages or in different tissues by qPCR. We found the expression level of Bmptp-h increased gradually at larval stage, and it reached the highest level on the 3rd day of fifth instar (I5D3), which was more than 15-times higher than in 1st instar (I1), then decreased afterwards until the end of pupal stage (Fig. 1 A). Next, we examined Bmptp-h transcriptional levels in different tissues dissected from I5D3 larvae (Fig. 1B). A relative high level of Bmptp-h was detected in the fat body, followed by testes.
Fig. 1.
Expression profile of Bmptp-h during the development and in different tissues. (A) Expression profile at different developmental stages. (B) Expression profile in different tissues of larvae at third day of fifth instar. The Bmptp-h mRNA level measured by qPCR was normalized to the internal control (sw22934) and represented as mean ± S.D. (n = 3). The letters marked at the top of columns indicate statistical significance of means detected by ANOVA analysis. Means with the same letter are not significantly different from each other. I, instar; D, day; FB, fat body; HE, head; HC, haemocyte; MG, midgut; SG, silk gland; ID, imaginal disc; OV, ovary; TE, testis.
3.3. Expression profile of Bmptp-h under NPV infection
Since previous studies have shown BmNPV PTP is important for viral productive infection in silkworm larvae and cell line, and BmPTP-h shares 48.2% identity (80.7% similarity) with BmNPV PTP on amino acid sequence (Takagi et al., 1998), we speculated that BmPTP-h also plays a role in regulating interaction between virus and host. For that reason, we injected NPV virions into I5D3 larvae and investigated the expression profile of Bmptp-h during infection. Although the expression level of Bmptp-h changed over time during the progression, viral infection induced higher level of Bmptp-h transcription compared to the uninfected larvae at the same time point (Fig. 2 A). The expression level of Bmptp-h in uninfected I5D7 larvae (96 h) was lower than that at I5D3 (0 h), which is in accordance with the result shown in Fig. 1A. However, in viral-infected larvae Bmptp-h transcript level was much higher at 96 h. We also found expression of Bmptp-h was potently induced by BmNPV infection in BmE cells after 48 h compared with uninfected cells and increased sharply afterwards (Fig. 2B).
Fig. 2.
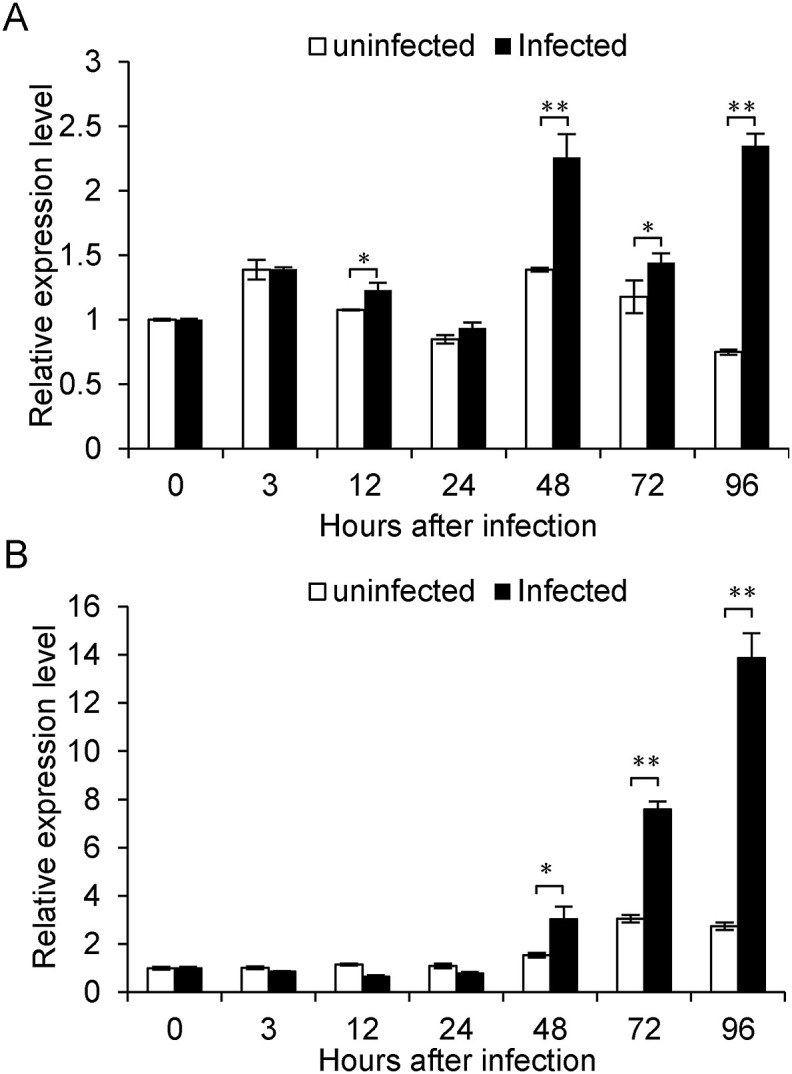
Expression profile of Bmptp-h in silkworm larvae and BmE cells under BmNPV infection. (A) Expression of Bmptp-h in silkworm larvae infected with BmNPV. (B) Expression of Bmptp-h in BmE cells infected with BmNPV. The Bmptp-h mRNA level measured by qPCR was normalized to the internal control (sw22934) and annotated as fold increase (±S.D., n = 3) over the normalized value at 0 min. Statistical significance between the expression level of Bmptp-h in uninfected and BmNPV-infected larvae or BmE cells at the same time point was assessed using Student's t-test. *p < 0.05, **p < 0.01.
3.4. Effects of Bmptp-h knock-down and over-expression to NPV infection in BmE cells
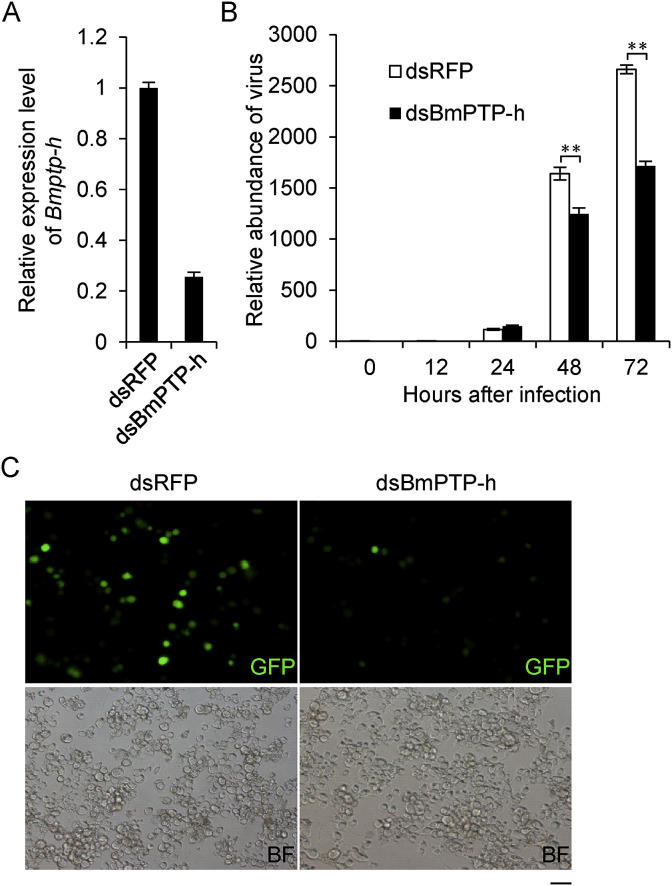
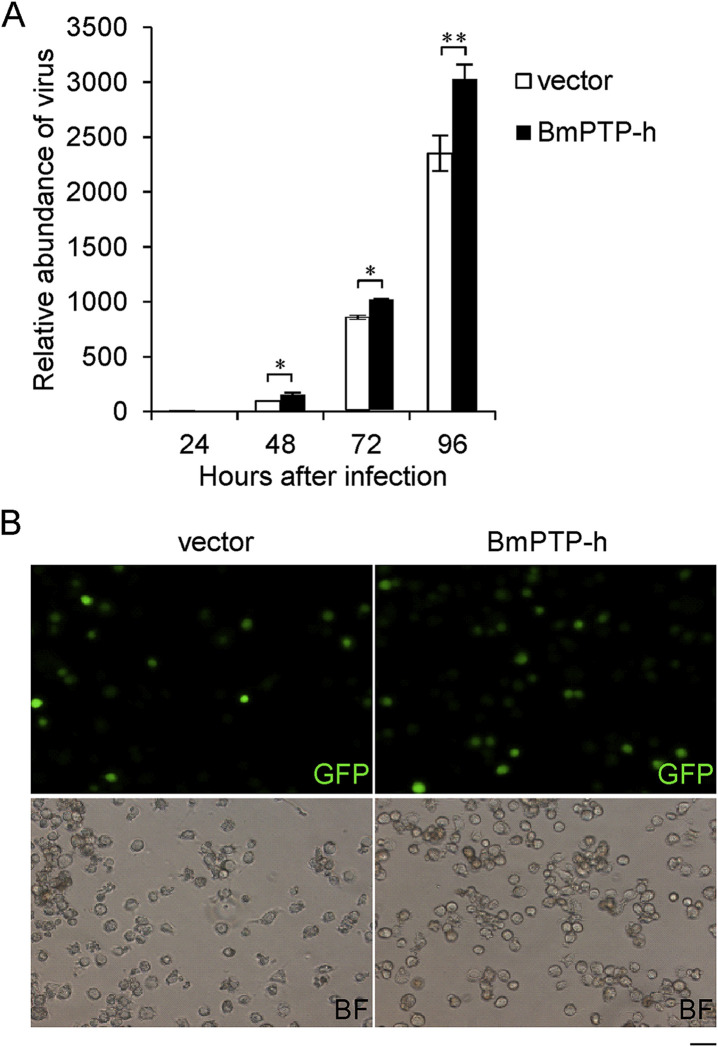
To test the tyrosine phosphatase activity of BmPTP-h, we expressed a Flag-tagged BmPTP-h in BmE cells (Fig. 3 A), and measured the enzymatic level of cell lysates (Fig. 3B). Cells expressing Flag-BmPTP-h showed a significant increase in the overall PTP activity level compared to the control, indicating that BmPTP-h is catalytic active and able to regulate the cellular functions via tyrosine dephosphorylation. To investigate whether BmPTP-h is involved in anti-viral response, we performed dsRNA-mediated RNAi to deplete the expression of Bmptp-h in BmE cells (Fig. 4 A). About 80% decrease in transcript level was obtained by dsBmPTP-h compared with dsRFP. 48 h after dsRNA transfection, we challenged cells with BmNPV or BmNPV-GFP, and analyzed the viral abundance by qPCR or fluorescence microscopy (Fig. 4B and C). Replication of viral DNA or the amount of GFP-positive cells was remarkably reduced when Bmptp-h was depleted. We then over-expressed Bmptp-h in BmE cells and measured the susceptibility of these cells to BmNPV or BmNPV-GFP infection (Fig. 5 ). Viral replication or GFP-positive cells significantly increased in BmPTP-h-overexpressing cells than in control. These results suggest that BmPTP-h promotes viral productive infection in cells.
Fig. 3.
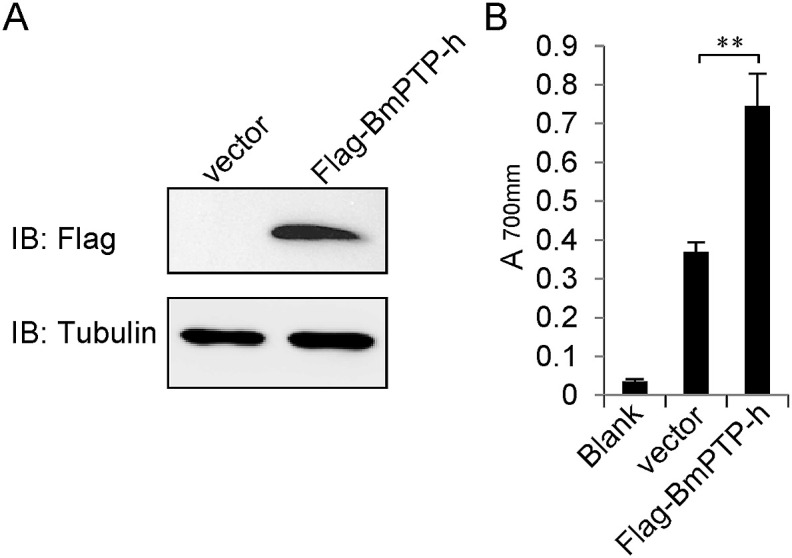
Phosphatase activity of BmPTP-h. (A) Lysates of BmE cells transfected with pSL1180 empty vector or Flag-BmPTP-h expressing plasmid were subjected to immunoblotting with anti-Flag antibody. (B) Cell lysates were precleared of free phosphate and incubated with casein, then mixed with molydbate dye and visualized at 750 nm. The results are given as the mean ± S.D. (n = 3). Statistical significance was assessed using Student's t-test. *p < 0.05, **p < 0.01.
Fig. 4.
Knock-down of Bmptp-h reduces BmNPV replication in BmE cells. (A) BmE cells were transfected with dsRFP or dsBmPTP-h, and knock-down efficiency of Bmptp-h was measured by qPCR analysis. (B) 48 h after dsRNA transfection, cells were infected with BmNPV. Relative abundance of virus determined by BmNPV gp64 expression level was measured by qPCR analysis. (C) Fluorescence (GFP) and bright-field (BF) images of dsRFP- or dsBmPTP-h-treated cells at 72 h post infection of BmNPV-GFP. The mRNA level of target genes was normalized to the internal control (sw22934 or BmGAPDH) and represented as mean ± S.D. (n = 3). Statistical significance was assessed using Student's t-test. *p < 0.05. Scale: 50 μm.
Fig. 5.
Over-expression of Bmptp-h enhances BmNPV replication in BmE cells. (A) BmE cells were transfected with vector or Flag-BmPTP-h expressing plasmid 48 h before BmNPV infection. Relative abundance of virus determined by BmNPV gp64 expression level was measured by qPCR analysis. (B) Fluorescence (GFP) and bright-field (BF) images of vector- or BmPTP-h-transfected cells at 72 h post infection of BmNPV-GFP. The mRNA level of target genes was normalized to the internal control (sw22934 or BmGAPDH) and represented as mean ± S.D. (n = 3). Statistical significance was assessed using Student's t-test. *p < 0.05, **p < 0.01. Scale: 50 μm.
3.5. Effects of Bmptp-h knock-down to NPV infection in vivo
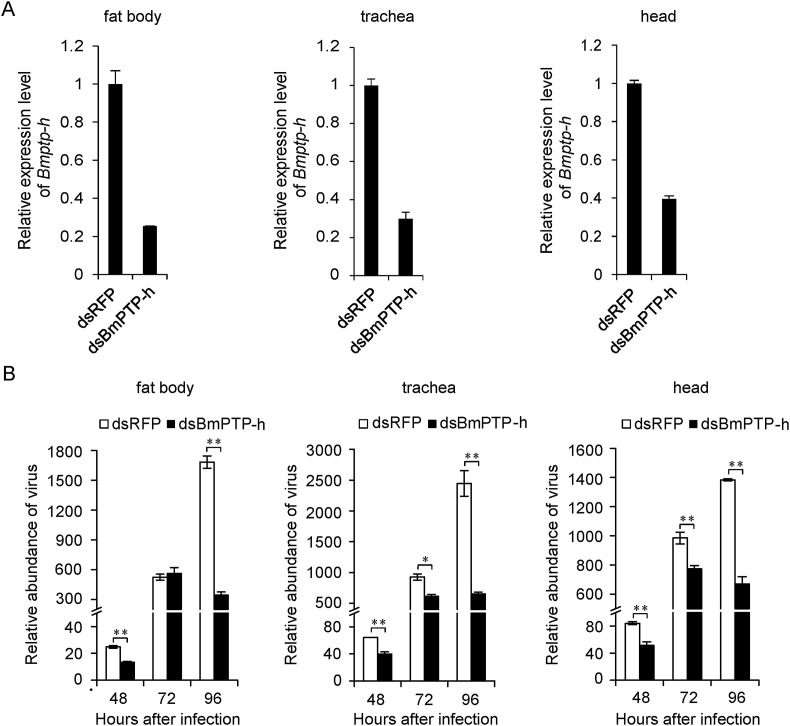
In order to investigate whether depletion of Bmptp-h has any effect to viral infection in vivo, we injected dsRNA into silkworm larvae 12 h before BmNPV infection and measured viral abundance. Since RNAi efficiency might not be the same in different tissues, we dissected several tissues and examined the expression level of Bmptp-h in them separately (Fig. 6 A). With the exception of midgut (data not shown), the relative expression levels of Bmptp-h decreased to 25%–40% after injection of dsRNA targeting Bmptp-h (dsBmPTP-h) compared to the control dsRFP in fat body, trachea and head. Furthermore, in those tissues viral abundance was all significantly decreased (Fig. 6B), and the reduction was most dramatic at 96 h post infection, suggesting that Bmptp-h silencing might have adverse effect to viral replication in vivo.
Fig. 6.
Knock-down of Bmptp-h reduced BmNPV replication in vivo. (A) dsRFP or dsBmPTP-h was injected into silkworm larvae at second day of fifth instar and knock-down efficiency of Bmptp-h was measured in different tissues. (B) 24 h after dsRNA treatment, silkworm larvae were injected with BmNPV. Relative abundance of virus post infection determined by BmNPV gp64 expression level was measured in corresponding tissues. The mRNA level of target genes was normalized to the internal control (sw22934 or BmGAPDH) and represented as mean ± S.D. (n = 3). Statistical significance was assessed using Student's t-test. *p < 0.05, **p < 0.01.
4. Discussion
In this study, we found BmNPV infection induced the expression of BmPTP-h, a dual-specific phosphatase in silkworm larvae and BmE cells. Partial depletion of Bmptp-h by dsRNA inhibited BmNPV replication, and over-expression of Bmptp-h resulted in enhanced viral replication. The results suggest that BmPTP-h might be one of the host factors that are beneficial to the replication of BmNPV. Although RNA interference is considered to be the major antiviral mechanism in insect, induction of immune signaling pathways, including Toll, IMD and JAK-STAT pathways have been observed upon viral infection (Merkling and van Rij, 2013, Sabin et al., 2010, Zambon et al., 2005). Since the signal transduction largely relies on protein phosphorylation, as a dual-specific phosphatase, BmPTP might be exploited by virus to disrupt the signaling cascades through targeting the phosphorylated molecules, resulting in immunosuppression. Furthermore, since double mutation of NPV lef-4 and NPV ptp, which are the only two viral genes encoding proteins with RNA triphosphatase activity did not alter viral RNA processing, it was proposed that host mRNA-capping enzymes might be involved in viral RNA maturation (Li and Guarino, 2008). Considering the RNA triphosphatase activity that BmPTP-h possibly possesses, it could also be hijacked by virus for efficient viral RNA synthesis.
Identification of host factors important for viral infection, especially for viral replication have been carried out using Drosophila and human cells by genome-wide RNAi screening (Hao et al., 2008, Konig et al., 2010). Those studies reported that molecules involved in kinase-regulated signaling, ubiquitination and phosphatase activity are enriched in host–pathogen interaction network. However, the mechanism on how virus induces the expression of those “benefit” factors in host is unclear. One hypothesis is that production of viral proteins down-regulates the repressors of those factors in host, such as microRNA, resulting in increased expression of molecules favoring viral replication (Fu et al., 2015). A few studies suggested that baculovirus infection leads to an overall suppression of host cellular microRNA (Mehrabadi et al., 2013, Singh et al., 2012), whether there is a microRNA targeting Bmptp-h needs further identification.
It is interesting to note that besides BmNPV ptp, several other baculoviral auxiliary genes are also acquired from host insect genome via horizontal gene transfer (HGT), such as ecdysteroid UDP-glucosyltransferase (egt) and fibroblast growth factor (vfgf) (Katsuma et al., 2008). Although they are dispensable for virus production in cell line, they are reported to be involved in controlling host physiology. No direct evidence is available showing that BmPTP-h is also playing roles in regulating silkworm wandering behavior, however our study demonstrated that it promotes viral replication, facilitating rapid spread throughout the whole body of the silkworm to overwhelm the immune system. In contrast to this “collaboration” strategy employed by baculovirus in the case of BmPTP-h, HGT of host genes usually allows pathogen to compete with hosts for the molecules necessary to maintain physiological integrity or dampen host response by acting as dysfunctional mimics (Liu et al., 1997, Zhao et al., 2014). Kamita et al. showed inserting Bmptp-h into the BmNPV ptp-deleted virus rescued the inability to induce ELA, but to a much lesser extent than WT virus, suggesting that the modification on BmNVP ptp sequence during evolution may have provided selective advantages and drive their fixation in the viral genome to counteract the host defense.
In conclusion, we did a whole genome survey for silkworm PTP gene sets and characterized their homologous relationship with human and fruit fly PTPs. We also found BmPTP-h acts as a host factor facilitating baculovirus infection via promoting its DNA replication. Further understanding of the pathways, in which BmPTP-h participates will provide new insight into the host–virus interactions that control the viral replication.
Acknowledgments
This research was supported by grants of “National Basic Research Program of China” (No. 2012CB114600) and “Natural Science Foundation of China” (No. 31201854).
References
- Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Choi J.E., Kwon J.H., Kim J.H., Hur W., Sung P.S., Choi S.W., Yoon S.K. Suppression of dual specificity phosphatase I expression inhibits hepatitis C virus replication. PLoS One. 2015;10:e0119172. doi: 10.1371/journal.pone.0119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Casa-Esperon E. Horizontal transfer and the evolution of host-pathogen interactions. Int. J. Evol. Biol. 2012;2012:679045. doi: 10.1155/2012/679045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande T., Takagi T., Hao L., Buratowski S., Charbonneau H. Human PIR1 of the protein-tyrosine phosphatase superfamily has RNA 5'-triphosphatase and diphosphatase activities. J. Biol. Chem. 1999;274:16590–16594. doi: 10.1074/jbc.274.23.16590. [DOI] [PubMed] [Google Scholar]
- Devasthanam A.S. Mechanisms underlying the inhibition of interferon signaling by viruses. Virulence. 2014;5:270–277. doi: 10.4161/viru.27902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E., Dupuy C., Huguet E., Cattolico L., Provost B., Martins N., Poirie M., Periquet G., Drezen J.M. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science. 2004;306:286–289. doi: 10.1126/science.1103066. [DOI] [PubMed] [Google Scholar]
- Fu Y.R., Liu X.J., Li X.J., Shen Z.Z., Yang B., Wu C.C., Li J.F., Miao L.F., Ye H.Q., Qiao G.H. MicroRNA miR-21 attenuates human cytomegalovirus replication in neural cells by targeting Cdc25a. J. Virol. 2015;89:1070–1082. doi: 10.1128/JVI.01740-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L., Sakurai A., Watanabe T., Sorensen E., Nidom C.A., Newton M.A., Ahlquist P., Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler R., Kerick M., Mah N., Hultschig C., Richter G., Bretz F., Sina C., Lehrach H., Nietfeld W., Schreiber S. Alterations of pre-mRNA splicing in human inflammatory bowel disease. Eur. J. Cell. Biol. 2011;90:603–611. doi: 10.1016/j.ejcb.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Hatzihristidis T., Desai N., Hutchins A.P., Meng T.C., Tremblay M.L., Miranda-Saavedra D. A Drosophila-centric view of protein tyrosine phosphatases. FEBS Lett. 2015;589:951–966. doi: 10.1016/j.febslet.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Heneberg P. Finding the smoking gun: protein tyrosine phosphatases as tools and targets of unicellular microorganisms and viruses. Curr. Med. Chem. 2012;19:1530–1566. doi: 10.2174/092986712799828274. [DOI] [PubMed] [Google Scholar]
- Ibrahim A.M., Choi J.Y., Je Y.H., Kim Y. Protein tyrosine phosphatases encoded in Cotesia plutellae bracovirus: sequence analysis, expression profile, and a possible biological role in host immunosuppression. Dev. Comp. Immunol. 2007;31:978–990. doi: 10.1016/j.dci.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Ibrahim A.M., Kim Y. Transient expression of protein tyrosine phosphatases encoded in Cotesia plutellae bracovirus inhibits insect cellular immune responses. Naturwissenschaften. 2008;95:25–32. doi: 10.1007/s00114-007-0290-7. [DOI] [PubMed] [Google Scholar]
- Jiang L., Wang G., Cheng T., Yang Q., Jin S., Lu G., Wu F., Xiao Y., Xu H., Xia Q. Resistance to Bombyx mori nucleopolyhedrovirus via overexpression of an endogenous antiviral gene in transgenic silkworms. Arch. Virol. 2012;157:1323–1328. doi: 10.1007/s00705-012-1309-8. [DOI] [PubMed] [Google Scholar]
- Kamita S.G., Nagasaka K., Chua J.W., Shimada T., Mita K., Kobayashi M., Maeda S., Hammock B.D. A baculovirus-encoded protein tyrosine phosphatase gene induces enhanced locomotory activity in a lepidopteran host. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2584–2589. doi: 10.1073/pnas.0409457102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuma S., Kawaoka S., Mita K., Shimada T. Genome-wide survey for baculoviral host homologs using the Bombyx genome sequence. Insect Biochem. Mol. Biol. 2008;38:1080–1086. doi: 10.1016/j.ibmb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Katsuma S., Koyano Y., Kang W., Kokusho R., Kamita S.G., Shimada T. The baculovirus uses a captured host phosphatase to induce enhanced locomotory activity in host caterpillars. PLoS Pathog. 2012;8:e1002644. doi: 10.1371/journal.ppat.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuma S., Noguchi Y., Zhou C.L., Kobayashi M., Maeda S. Characterization of the 25K FP gene of the baculovirus Bombyx mori nucleopolyhedrovirus: implications for post-mortem host degradation. J. Gen. Virol. 1999;80:783–791. doi: 10.1099/0022-1317-80-3-783. [DOI] [PubMed] [Google Scholar]
- Kingsolver M.B., Huang Z., Hardy R.W. Insect antiviral innate immunity: pathways, effectors, and connections. J. Mol. Biol. 2013;425:4921–4936. doi: 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R., Stertz S., Zhou Y., Inoue A., Hoffmann H.H., Bhattacharyya S., Alamares J.G., Tscherne D.M., Ortigoza M.B., Liang Y. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Guarino L.A. Roles of LEF-4 and PTP/BVP RNA triphosphatases in processing of baculovirus late mRNAs. J. Virol. 2008;82:5573–5583. doi: 10.1128/JVI.00058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Miller L.K. Properties of a baculovirus mutant defective in the protein phosphatase gene. J. Virol. 1995;69:4533–4537. doi: 10.1128/jvi.69.7.4533-4537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Wang X., Huang M., Tam J.P., Liu D.X. Regulation of the p38 mitogen-activated protein kinase and dual-specificity phosphatase 1 feedback loop modulates the induction of interleukin 6 and 8 in cells infected with coronavirus infectious bronchitis virus. Virology. 2011;420:106–116. doi: 10.1016/j.virol.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., de Waal Malefyt R., Briere F., Parham C., Bridon J.M., Banchereau J., Moore K.W., Xu J. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J. Immunol. 1997;158:604–613. [PubMed] [Google Scholar]
- Mehrabadi M., Hussain M., Asgari S. MicroRNAome of Spodoptera frugiperda cells (Sf9) and its alteration following baculovirus infection. J. Gen. Virol. 2013;94:1385–1397. doi: 10.1099/vir.0.051060-0. [DOI] [PubMed] [Google Scholar]
- Merkling S.H., van Rij R.P. Beyond RNAi: antiviral defense strategies in Drosophila and mosquito. J. Insect Physiol. 2013;59:159–170. doi: 10.1016/j.jinsphys.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Mustelin T., Vang T., Bottini N. Protein tyrosine phosphatases and the immune response. Nat. Rev. Immunol. 2005;5:43–57. doi: 10.1038/nri1530. [DOI] [PubMed] [Google Scholar]
- Peters M.A., Jackson D.C., Crabb B.S., Browning G.F. Chicken anemia virus VP2 is a novel dual specificity protein phosphatase. J. Biol. Chem. 2002;277:39566–39573. doi: 10.1074/jbc.M201752200. [DOI] [PubMed] [Google Scholar]
- Provost B., Varricchio P., Arana E., Espagne E., Falabella P., Huguet E., La Scaleia R., Cattolico L., Poirie M., Malva C. Bracoviruses contain a large multigene family coding for protein tyrosine phosphatases. J. Virol. 2004;78:13090–13103. doi: 10.1128/JVI.78.23.13090-13103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A.J., Strand M.R. PTP-H2 and PTP-H3 from Microplitis demolitor Bracovirus localize to focal adhesions and are antiphagocytic in insect immune cells. J. Virol. 2007;81:1209–1219. doi: 10.1128/JVI.02189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin L.R., Hanna S.L., Cherry S. Innate antiviral immunity in Drosophila. Curr. Opin. Immunol. 2010;22:4–9. doi: 10.1016/j.coi.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbielle C., Dupas S., Perdereau E., Hericourt F., Dupuy C., Huguet E., Drezen J.M. Evolutionary mechanisms driving the evolution of a large polydnavirus gene family coding for protein tyrosine phosphatases. BMC Evol. Biol. 2012;12:253. doi: 10.1186/1471-2148-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C.P., Singh J., Nagaraju J. A baculovirus-encoded MicroRNA (miRNA) suppresses its host miRNA biogenesis by regulating the exportin-5 cofactor Ran. J. Virol. 2012;86:7867–7879. doi: 10.1128/JVI.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T., Taylor G.S., Kusakabe T., Charbonneau H., Buratowski S. A protein tyrosine phosphatase-like protein from baculovirus has RNA 5'-triphosphatase and diphosphatase activities. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9808–9812. doi: 10.1073/pnas.95.17.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks N.K. Protein tyrosine phosphatases–from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte S., Ros V.I., Mastenbroek T.G., Vendrig N.J., Hoover K., Spitzen J., van Oers M.M. Protein tyrosine phosphatase-induced hyperactivity is a conserved strategy of a subset of baculoviruses to manipulate lepidopteran host behavior. PLoS One. 2012;7:e46933. doi: 10.1371/journal.pone.0046933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Li D.M., Sun H. PIR1, a novel phosphatase that exhibits high affinity to RNA · ribonucleoprotein complexes. J. Biol. Chem. 1998;273:20347–20353. doi: 10.1074/jbc.273.32.20347. [DOI] [PubMed] [Google Scholar]
- Zambon R.A., Nandakumar M., Vakharia V.N., Wu L.P. The toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Xu C., Lu H.L., Chen X., St Leger R.J., Fang W. Host-to-pathogen gene transfer facilitated infection of insects by a pathogenic fungus. PLoS Pathog. 2014;10:e1004009. doi: 10.1371/journal.ppat.1004009. [DOI] [PMC free article] [PubMed] [Google Scholar]