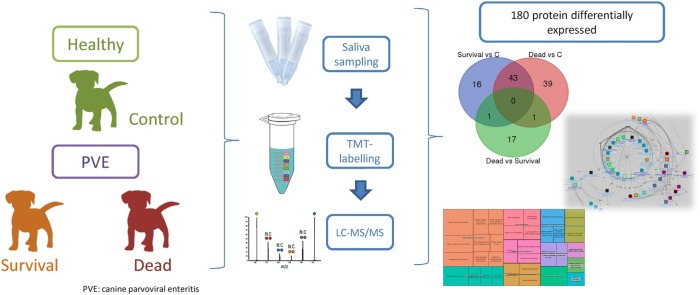
Graphical abstract
Keywords: Bioinformatics, Biomarker, Canine parvovirosis, Dog, Label-based proteomics, Saliva
Highlights
-
•
Saliva proteome showed 190 differentially expressed proteins in dogs with CPV.
-
•
Saliva could be a potential source of biomarkers of diagnosis in CPV.
-
•
Catalytic activity and binding are modulated in saliva proteome of dogs with CPV.
Abstract
The present study evaluated the changes in salivary proteome in parvoviral enteritis (PVE) in dogs through a high-throughput quantitative proteomic analysis. Saliva samples from healthy dogs and dogs with severe parvovirosis that survived or perished due to the disease were analysed and compared by Tandem Mass Tags (TMT) analysis. Proteomic analysis quantified 1516 peptides, and 287 (corresponding to 190 proteins) showed significantly different abundances between studied groups. Ten proteins were observed to change significantly between dogs that survived or perished due to PVE.
Bioinformatics’ analysis revealed that saliva reflects the involvement of different pathways in PVE such as catalytic activity and binding, and indicates antimicrobial humoral response as a pathway with a major role in the development of the disease. These results indicate that saliva proteins reflect physiopathological changes that occur in PVE and could be a potential source of biomarkers for this disease.
1. Introduction
Parvoviruses are small icosahedral viruses that infect many animal species including humans worldwide. Canine parvovirus type-2 is the causal agent of canine parvoviral enteritis (PVE), a severe disease that causes nearby 100% morbidity and up to 10–90% mortality in unvaccinated adults and puppies, respectively [1]. The clinical presentation of PVE ranges from mild to severe, based on clinical signs and physical examination [2,3]. The most common clinical signs in puppies include acute severe vomiting and bloody diarrhoea, fever, dehydration and lethargy [4]. Complications such as cardiac damage, which may result in death or permanent myocardial damage [5], and systemic inflammatory response syndrome (SIRS) prior to sepsis have been described [6].
In dogs, saliva has proven to be an useful biofluid for the diagnostic of several conditions such as leishmaniosis [7] and helicobacter infection [8], as well as for evaluation of stress [9]. The use of non-invasive specimens such as saliva provides several advantages compared to invasive methods, being safer for the personal and the patient, easier to collect, pain-free and causes reduced sampling stress [10]. Saliva can have potential advantages versus others non-invasive samples such as faeces, which usually need a laborious pre-treatment prior to its analysis. In particular, in the case of PVE, it has been described a poor sensitivity of faeces when used in rapid antigen ELISA test [11]. For this, it is envisioned that saliva can play an increasingly important role in the early diagnosis and monitoring of diseases [12].
However, to the author’s best knowledge, there are no reported studies about changes in saliva composition in PVE. The knowledge of the changes that occur in saliva in dogs with parvovirus could help to detect possible biomarkers in this fluid for PVE disease.
Proteomics sciences focus on the study of the proteome content of cells, organs and organisms [13]. Proteomic techniques have become widely used with the aim of discovering disease-related biomarkers in diverse biological fluids that could be used in routine practice for early diagnosis, prognosis or treatment monitoring [14,15]. Over the last several years, novel technologies such as the use of isobaric tags have greatly increase the sensitivity of the proteomic analysis [[16], [17], [18]]. Tandem Mass Tags (TMT) allow for relative simultaneous quantification of differentially labelled peptides [19] since each sample is marked and distinguished by differences on their reporter ion masses [20,21]. However, there is still a scarcity of reports using TMT technology in saliva samples, and no reports have been found describing the use of TMT to detected possible biomarkers of diagnosis and prognosis in PVE.
The objective of the present study is to evaluate possible changes in saliva proteins associated to canine parvovirus infection. This would help to clarify if saliva components can reflect the physiopathological changes associated to the disease and to evaluate the possibility of identifying potential biomarkers of PVE in saliva samples. For this, saliva samples were collected from healthy dogs (control group) and from dogs with PVE that survived or died because the infection. Saliva samples were analysed by quantitative high-throughput TMT-based proteomic approach enabling the identification of significantly varying proteins among different groups. Subsequently, the differentially expressed proteins were used as a starting point for creating protein interaction networks.
2. Materials and methods
2.1. Animals
A total of 11 client-owned dogs that were presented to the Veterinary Teaching Hospital, Small Animal Clinics, Bursa Uludag University, Bursa, Turkey, of different breeds were involved in this study (Table 1 ). Parvovirosis infection was diagnosed by compatible clinical (acute bloody diarrhoea, vomiting, anorexia, dehydration etc.) and haematological signs (leukopenia, neutropenia and lymphopenia) in combination with the positive test results of the commercially available faecal diagnostic test (Antigen Rapid PVE kit, Animal Genetics, Inc., Suwon, Korea). Selected clinical (body temperature and heart and respiratory rates) and haematological (leukogram, eritrogram and thrombogram) findings were used to assess the general health status of the animal. Dogs were excluded if they were co-infected with other viral (canine distemper or coronavirus), parasite (coccidiosis, giardiasis, ascaridiosis etc.), or vector borne diseases by use of faecal screening test, fecal microscopic examinations, or speed test for ehrlichiosis, anaplasmosis, lyme, and dirofilariasis (Anigen Rapid CaniV-4, Bionote, Korea), respectively.
Table 1.
Main exploratory and haematological findings in healthy and dogs with PVE.
| ID | Breed | Gender / age (months) | T °C |
P /min | R /min | WBC / N / L X 103/μL |
Hct % | Total protein gr/dL | Group |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Mixed | M / 2 | 39.2 | 120 | 22 | 11.5 / 8.7 / 2.1 | 27.6 | ND | Healthy |
| 2 | Mixed | M / 2 | 39.3 | 110 | 24 | 10.7 / 6.8 / 3.2 | 36.1 | ND | Healthy |
| 3 | Mixed | F / 2 | 38.9 | 112 | 18 | 12.9 / 7.8 / 4.2 | 32.8 | 5.3 | Healthy |
| 4 | Mixed | M / 5 | 38.7 | 124 | 16 | 13.5 /10.2/ 2.0 | 40.6 | 451 | Healthy |
| 5 | Mixed | M / 10 | 38.5 | 114 | 16 | 15.0/ 12.8/ 1.5 | 48.8 | 6.9 | Healthy |
| 6 | Rottweiler | F / 5 | 39.8 | 128 | 44 | 2.1 / 0.6 / 1.4 | 31.8 | 4 | Survival |
| 7 | Cocker | M / 3 | 38.3 | 200 | 44 | 2.5 / 1.0 / 1.3 | 39.1 | 4.7 | Survival |
| 8 | Mixed | F / 3 | 37.5 | 148 | 20 | 1.6 / 0.6 / 0.8 | 29.7 | 4.1 | Survival |
| 9 | Anatolian sheepdog | M / 5 | 39.8 | 120 | 44 | 0.3 / 0.1 / 0.1 | 34.2 | 3.7 | Not-survival |
| 10 | Rottweiler | M / 7 | 40.7 | 108 | 28 | 1.2 / 0.3 / 0.8 | 43.2 | 5.6 | Not-survival |
| 11 | Cocker | F / 8 | 37.0 | 108 | 32 | 0.3 / 0.0 / 0.2 | 37.5 | 3.3 | Not-survival |
M: male, F: female, ND: not determined, P: pulsation, R: respiration, Hct: Hematocrite, WBC: white blood cell count, N: Neutrophil count, L: Lymphocyte count.
The dogs were divided into three groups according to their clinical condition and development of the disease. A group with 5 healthy dogs was considered as control group. Virus is shed for a few days before the onset of clinical signs. Therefore, even if all dogs in control group were considered healthy based on the clinical, haematological and serum biochemistry findings (comprehensive profile, VetScan, Abaxis), faecal screening test for CPV-Ag were applied. Dogs with negative test result were enrolled to study. All dogs in test group were treated as described previously [22]. Briefly, fluid replacement therapy (dextrose 5% with a balanced crystalloid solution - lactated ringer, and hydroxyethyl starch), anti-emetics (metoclopramid 0.2-0.4 mg/kg, BID, subcutaneously, and ranitidin 2–4 mg/kg, BID, intramusculary), parenteral antibiotics (ampicillin 30 mg/kg, BID + amikasin 5–10 mg/kg, BID + metronidazol 25 mg/kg, BID), immune stimulator (imunex), probiotics and vitamins (E and C) were used. Dogs with clinical diagnosis of severe parvovirosis were divided into two groups according to the progress of the disease: 3 dogs that survived (survival group) (age 3.67 ± 0.94 months, 1 male and 2 female) and 3 that died because of the disease (non-surviving group) (age 6.67 ± 1.25 months, 2 males and 1 female). Therefore, dogs were classified according to the severity of their clinical and haematological signs (Table 1). There was a similarity of presented clinical signs and its duration (2–4 days) in dogs within test group. Despite the treatment, three dogs died within 5 days (one on day 3, one on day 4 and other on day 5).
All the procedures were approved by the Local Ethical Committee of the University of Uludag.
2.2. Saliva sampling
At least 0.5 ml of unstimulated saliva specimens were collected from each patient by placing small cotton swabs around the mouth. When the cotton swabs were thoroughly moist, they were placed in collection devices (Salivette saliva collection tube / V-Bottom, Sarstedt, Aktiengesellschaft & Co, Nümbrecht, Germany), centrifuged (3000 x g for 10 min, 4 °C), and the supernatant was stored at −80 °C until analysis [23]. Due to dehydration and subsequent dry mouth, for some animals the sampling protocol was performed twice in order to obtain at least 0.5 ml of unstimulated saliva.
2.3. Proteomic study of saliva samples and LC–MS/MS analysis
From each sample, 35 μg of acetone-precipitated proteins were subjected to reduction, alkylation, digestion and labelled using 6-plex Tandem Mass Tag reagents according to manufacturer instructions (Thermo Scientific) with some modification, as described previously [24]. In short, 35 μg of sample and internal standards were reduced with 200 mM DTT (Sigma-Aldrich), alkylated with 375 mM iodoacetamide (Sigma- Aldrich) and precipitated with ice-cold acetone (VWR, Pennsylvania, USA) overnight. Samples were then centrifuged and acetone was decanted. Pellets were resuspended with 50 μl of 100 mM TEAB buffer and digested with trypsin (Promega) overnight at 37 °C (2.5 μg of trypsin per 100 μg of protein). TMT label reagents were equilibrated to room temperature, resuspended in anhydrous acetonitrile LC–MS grade (Thermo Scientific) and added to each sample. Labelling reaction was incubated for 1 h at room temperature and then quenched by adding 5% hydroxylamine (Thermo Scientific) for 15 min. Samples were then combined at equal amounts and 6 μg of each mixed sample set was placed in a well of a microplate, vacuum-dried and stored at - 20 °C before further LC–MS/MS analysis. The LC − MS/MS analysis was performed on Dionex Ultimate 3000 RSLS nano flow system (Dionex, Camberley, UK) and Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) as described elsewhere [25].
2.4. Statistical analysis
In order to compare the abundances of peptides detected in proteomic analysis between 2 groups in 3 comparisons (Control versus Survival, Control versus Non-surviving and Survival versus Non-surviving), data were normalized by logarithmic transformation and Student’s t-test was used (one-tailed, unpaired) were performed. In all cases, values of P < 0.05 were considered to be significant. Fold changes (FC) have been calculated as follow FC = log2 (Group 1 / Group 2). Statistics were performed with RStudio (v1.0.143) (R Studio Team. RStudio [26] “RStudio Team. RStudio: Integrated Development Environment for R [Internet]. Boston, MA: RStudio, Inc.; 2015. Available from: http://www.rstudio.com/,” 2015).
2.5. GO pathways
The data obtained in the proteomic study were used for the Gene Ontology (GO) analysis. Canine genes encoding proteins differentially expressed were converted to their human orthologs using the Ensembl orthologs database and its BioMart tool for data mining (www.ensembl.org). Obtained genes were used to determine the GO terms over-represented in two conditions (Survival and Non-surviving), by the utilization of the Cytoscape (v3.6.1) plug-in ClueGO (v2.5.0) [27,28] on the Homo sapiens GO-biological process (22/01/2018). GO terms over-represented for each group were submitted to analysis by REVIGO (allowed similarity = 0.7, SimRel) to remove redundant GO terms and groups related GO terms based on their functional description [29]. Finally, pathways interactomes were designed in Cytoscape using the radial layout.
3. Results
3.1. High resolution quantitative proteomic analysis
High-resolution quantitative proteomic analysis allowed the identification of 1516 canine peptides from the eleven non-depleted canine saliva samples. For the comparison Control versus Survival, 148 peptides were differentially expressed, corresponding to 90 unique proteins. For the comparison Control versus Non-surviving, 125 peptides were differentially expressed, corresponding to 90 unique proteins. For the comparison Survival versus Non-surviving, 14 peptides were differentially expressed, corresponding to 10 unique proteins. Therefore a total of 190 proteins were identified that differed between groups.
Of the 90 differentially expressed proteins between survival and control groups, the proteins most down-regulated in the survival group were cathelicidin antimicrobial peptide (CAMP), rho-GDP dissociation inhibitor beta (ARHGDIB), apolipoprotein A-1 (APO-A1), neutrophil elastase (ELANE), matrix metalloproteinase-9 (MMP9), EF-hand domain containing protein D2 (EFHD2), CD177 antigen (CD177), plastin-2 (LCP1), retinol binding protein 4 (RBP4), and maltase-glucoamylase intestinal (MGAM). Epididymal – specific lipocalin-9 (LCN9), BPI fold-containing family B member 2 (BPIFB2), lymphocyte antigen 6D (LY6D), desmoglein (-1 and -3, DSG1 and DSG3, respectively), alpha-2-macrogloulin-like protein 1 (A2ML1), democollin-2 (DSC2), leucine-rich-alpa-2-glycoprotein (LRG1), polymeric immunoglobulin receptor (PIGR), and l-amino acid oxidase (IL4I1) were the proteins most up-regulated in the survival group. Of these, CAMP and ARHGDIB were the proteins most down-regulated (-2.29 and -2.26-fold lower expression, respectively), while LCN9 and BPIFB2 were the most up-regulated (2.28 and 1.62-fold higher) in the survival group compared to the control group.
When non-surviving and control groups were compared, 90 proteins showed differences in abundance. The proteins most down-regulated were ARHGDIB, CAMP, APOA1, hemoglobin subunit beta (HBB), RBP4, CD177, MMP9, vitamin D binding protein (GC) and ELANE. On the other hand, immunoglobulin heavy chain (IGH), LCN9, clusterin (CLU), trefoil factor 1 (TFF1), LY6D, olfactomedin 4 (OLFM4), LRG1, glyoxalase I (GLO1), BPIFB2, and DSG3 were the proteins most up-regulated.
Finally, when the two groups of dogs with parvovirosis were compared, 10 proteins showed different abundances. Ezrin, allergen Dog 1, lactoperoxidase, l-lactate dehydrogenase A, cystatin-M and macrophage-capping protein were higher in survival group, whereas alpha-actinin-1, peptidyl-prolyl cis-trans isomerase A, ribosyldihydronicotinamide dehydrogenase and lactoylglutathione lyase were upregulated in the dogs that died due to the disease.
3.2. Bioinformatics
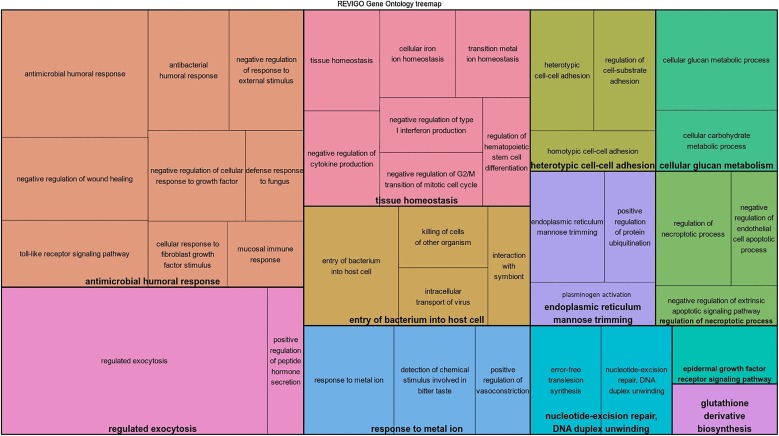
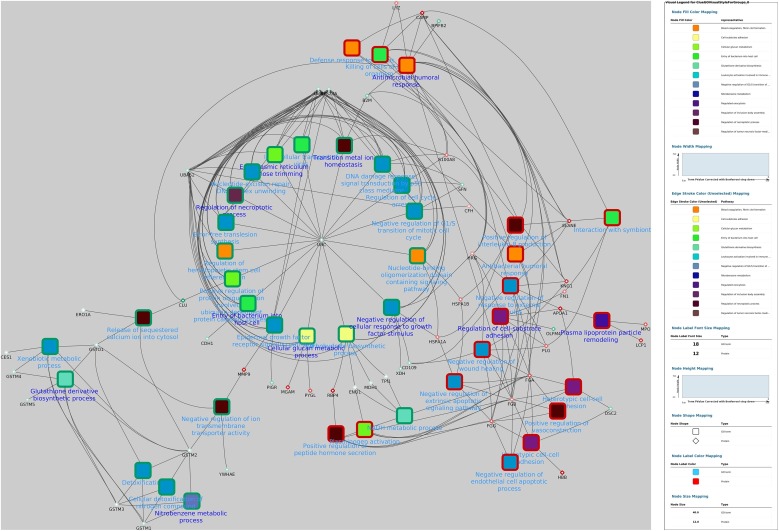
The GO analysis performed using Cytoscape plug-in ClueGO identified 34 GO terms between survival and controls, which were filtered for redundancies and then grouped into 12 main groups using REVIGO. The functional groups included antimicrobial humoral response, entry of bacterium into host cell, tissue homeostasis, heterotypic cell-cell adhesion and endoplasmic reticulum mannose trimming (Fig. 1 ).
Fig. 1.
Over-represented GO terms in dogs that survived canine parvovirosis, grouped by REVIGO, based on their description. Leading GO terms (N = 12) for each group was defined as the highest significance inside the group.
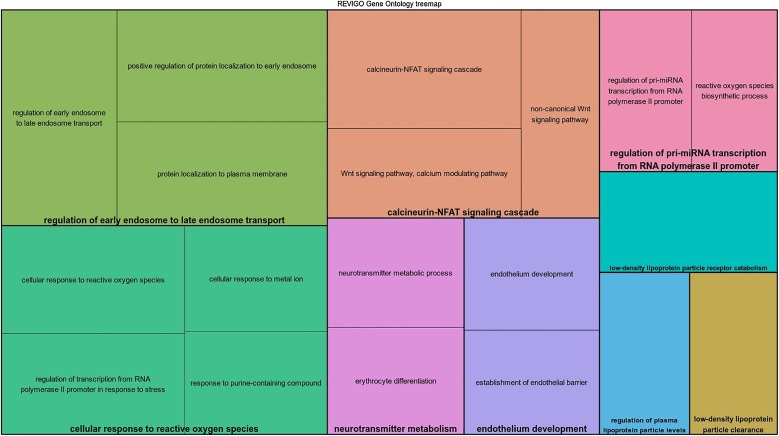
When comparing non-surviving and control groups, 42 GO terms were identified, which were grouped into 11 main functional groups including negative regulation of cellular response to growth factor stimulus, antimicrobial humoral response, transition metal ion homeostasis, endoplasmic reticulum mannose trimming and glutathione derivate biosynthesis (Fig. 2 ).
Fig. 2.
Over-represented GO terms in dogs that dead due to canine parvovirosis, grouped by REVIGO, based on their description. Leading GO terms (N = 12) for each group was defined as the highest significance inside the group.
Comparison of survival versus non-surviving groups did not identify enough proteins significantly expressed to generate a reliable GO analysis using only experimental results.
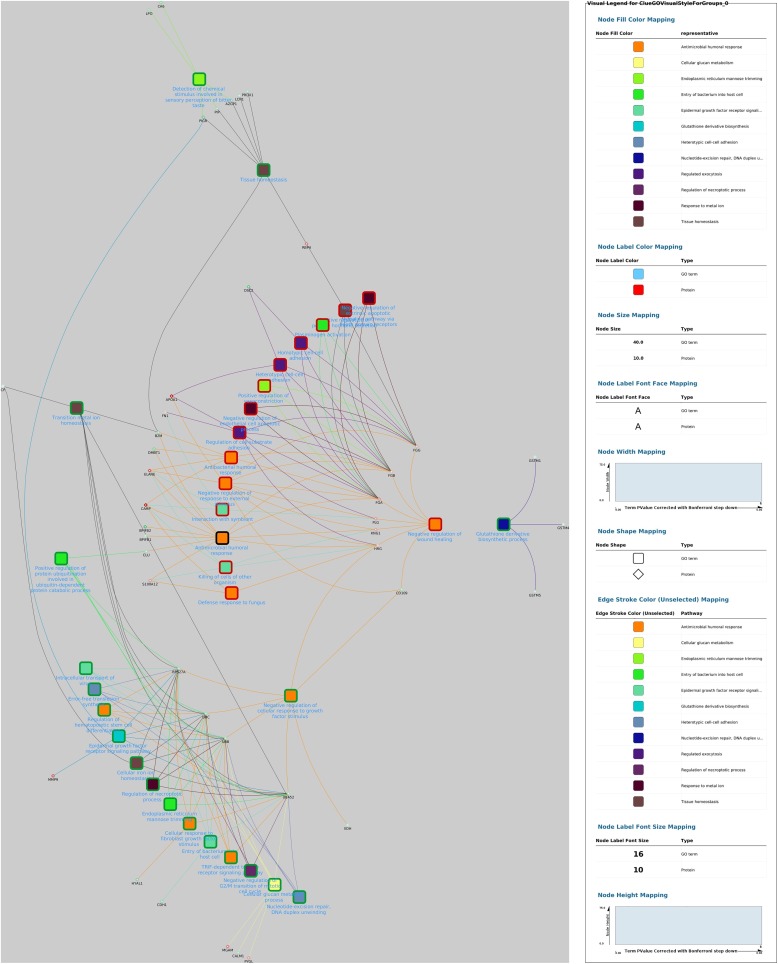
Finally, GO terms most represented in this study after being filtered by REVIGO and depicted by Cytoscape are shown in Fig. 3, Fig. 4, Fig. 5, Fig. 6 . When the network from control and survival group was analysed, a negative regulation of wound healing (related with the down-regulation of fibrinogen FGA, FGB and FGG genes) and negative regulation of cellular response to growth factor stimulus (related to the up-regulation of the ubiquitin complex composed by RPS27 A, UBC, UBB and UBA52 genes) nodes demonstrated central roles in the development of the disease since they are involved in many pathways.
Fig. 3.
Over-represented GO terms generated by the comparison of dogs that survived and dogs that died due to parvovirosis, grouped by REVIGO, based on their description. Leading GO term (N = 9) for each group was defined as the highest significance inside the group.
Fig. 4.
In silico inferred interactome network of identified GO terms over-represented in canine parvovirosis (survival versus control groups). Differentially expressed proteins interacting with at least 1 term were added. Radial layout was applied and the GO group leader terms are in dark blue text. Nodes colours represent the group of GO terms (determined by ReviGO) and border are represented in green and red for over-expressed and lower-expressed proteins, respectively, when compared to control group.
Fig. 5.
In silico inferred interactome network of identified GO terms over-represented in canine parvovirosis (dead versus control groups). Differentially expressed proteins interacting with at least 1 term were added. Radial layout was applied and the GO group leader terms are in dark blue text. Nodes colours represent the group of GO terms (determined by ReviGO) and border are represented in green and red for over-expressed and lower-expressed proteins, respectively, when compared to control group.
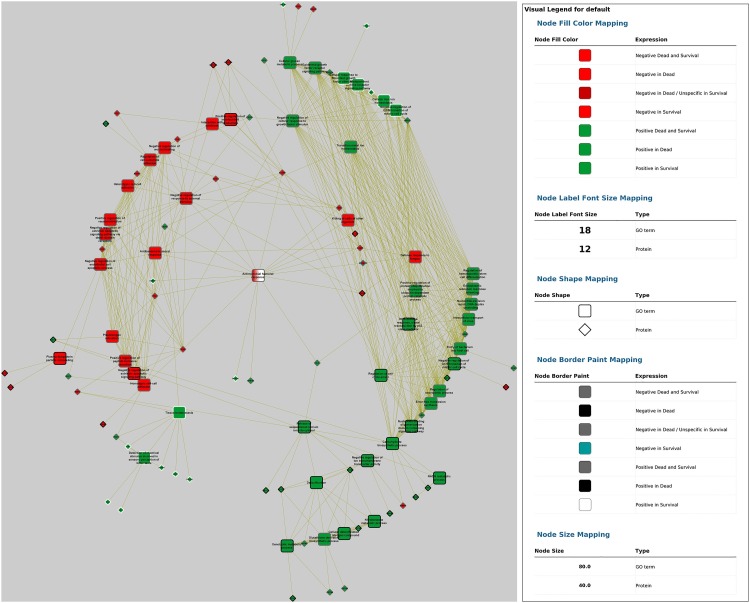
Fig. 6.
In silico inferred interactome network of identified GO terms over-represented in canine parvovirosis (survival and dead versus control groups). Differentially expressed proteins interacting with at least 1 term were added. Radial layout was applied. Nodes fill colour figure protein and GO terms regulation (red for negatively and green for positively regulated proteins) and node border represent the presence of the term in dead only (black), survival only (white) or in both groups (grey).
The network obtained from comparison of non-surviving and control groups suggested that the over-expressed UBC gene (which expresses ubiquitin C protein) appears to have a central role by affecting or being affected by numerous pathways identified as related with non-surviving group in parvovirosis. Interestingly, this ubiquitin C protein is related only with GO terms over-expressed in non-surviving group. This indicate that, in the present study, all cellular pathways which use ubiquitin form a pertinent group, which is positively correlated with the non-survival status. Similar to the previously mentioned network, fibrinogen expression is down-regulated in saliva of dogs with parvovirosis.
When merging fold changes data and over-represented pathways in both non-surviving and survival groups, the obtained network highlights that antimicrobial humoral response was shown as central term. This node is the only one which is over-represented in both parvovirus groups (survival and non-surviving) but with different expression when compared to controls: it was lower-expressed in the non-surviving group, but was not differentially expressed in survival group as compared to controls. Antibacterial humoral response, negative regulation of response to external stimulus, and killing of cells of other organisms are the three down-regulated nodes which are directly linked with antimicrobial humoral response. The network also showed 14 proteins surrounding this central node, 9 of which were down-regulated (ELANE, kininogen 1 [KNG-1], FGA, FGB, S100 calcium binding protein A8 [S100A8], S100 calcium binding protein A12 [S100A12], CAMP, lysozyme [LYZ] and HRG), and 5 up-regulated (beta-2 microglobulin [B2M], deleted in malignant brain tumours 1 [DMBT1], BPI fold-containing family B member 1 [BPIFB1], BPIFB2, and CLU) when compared to controls.
4. Discussion
This paper was focused in the identification of the differentially expressed proteins in saliva and in describing the biological roles which may be affected due to PVE. The present study identified for the first time 190 differentially expressed proteins in saliva from healthy dogs and dogs with severe parvovirosis using TMT technology, 10 of which were differentially expressed between animals that survived and perish due to the disease. These differences in proteins indicate that saliva can reflect several physiological pathways in canine parvovirosis.
Overall changes in protein expression in saliva from dogs with parvovirosis obtained in this study by high-resolution quantitative proteomic analysis suggested alterations in coagulation and inflammation systems, which are closely related pathways since the activation of one mechanism may lead to the activation of the other [30]. Coagulation products such as thrombin can promote inflammatory responses, while inflammation is known to suppresses anticoagulation mechanisms [31]. The immune system provides protection against infection; however, its activation needs to be tightly regulated in order to prevent excessive inflammation and subsequent damage to the host [32].
From all the proteins that change in saliva of dogs with parvovirosis, two that are most down-regulated and the two proteins that increased the most in the canine patients are worthy of specific mention. The two most down-regulated proteins in the dogs with parvovirosis that survived, cathelicidin antimicrobial peptide (CAMP) and Rho-GDP dissociation inhibitor beta (ARHGDIB), have protective role against infections and are involved in the inflammatory response. Cathelicidins are a group of short cationic antimicrobial peptides with an important role in the protection against infections [[33], [34], [35]]. The main source of CAMPs are the specific granules of neutrophils, which are released upon neutrophil activation [36,37], although they can be produced by other cells such as epithelial cells at mucosal surfaces [38]. CAMPs are known to regulate the activation of a wide variety of toll-like receptors, which plays important roles in the modulation of the inflammatory response during infections [39,40]. In addition, antimicrobial activity has been described in CAMPs, and the loss of cathelidicin expression has been reported to increase the susceptibility to Escherichia coli infection in mice [33,34]. Rho-GDP dissociation inhibitor beta (ARHGDIB) is primarily expressed in cells of a hematopoietic lineage and belongs to the RhoGDI family, which are known to regulate Rho-GTPases [41]. ARHGDIB showed an ability to decrease human immunodeficiency virus type 1 (HIV-1) replication in cell lines and can be used as a biomarker of HIV-1 infection [42]. ARHGDIB expression has been also proposed as suppressor of metastasis and predictor of prognosis in human patients with bladder cancer, in which ARHGDIB positive tumours are related with longer disease-free and disease-specific survival rates [43].
On the other hand, the two most up-regulated proteins in dogs with parvovirosis that survived, were lipocalin-9 (LCN9) and BPI fold-containing family B member 2 (BPiFB2). Lipocalins are a family of proteins that usually carry lipids or other hydrophobic or amphiphilic compounds accommodated in a cavity within their conformation, including steroids, hormones and metabolites such as vitamins and cofactors [44]. Furthermore, lipocalins have been reported to be modulators of cell growth and metabolism, and regulators of the immune response [45,46]. Although little is known about LCN9, a related protein, neutrophil gelatinase-associated lipocalin, NGAL, has been associated with bacterial infection in humans [[47], [48], [49]]. BPiFB2 belongs to a family of proteins considered as a key component of the innate immune response against Gram-negative bacteria [50], but the physiological functions of the BPIFB family still remain largely unknown [51]. Despite this, a previous study on BPIFB2 has reported the involvement of BPIFB proteins regulating enterovirus infection, with BPIFB6 assisting the enterovirus replication by controlling secretory pathway trafficking and Golgi complex morphology [51].
In addition to these 4 proteins, 86 other proteins were significantly differentially expressed in saliva of parvovirus infected dogs compared to healthy dogs. By analysing the over-represented cellular pathways defined by those proteins by gene ontology analyses, it could be observed that they are involved in other physiopathological pathways such as catalytic activity, binding, transporter activity, antioxidant activity, structural molecule activity, receptor activity and translation regulator activity. Since more than 85% of the genes differentially expressed belong to the proteins related with catalytic activity and binding, the discussion will focus on these and related proteins.
In regard to the proteins related to catalytic activity, some were down-regulated, including, apolipoprotein A-1 (APO-A1), neutrophil esterase (ELANE), complement C3 (C3) and histidine-rich glycoprotein (HRG). APO-A1 is the major protein component of high density lipoproteins in human and other mammals [52] and the decreased levels APO-A1 observed in the present study are in concordance with other responses described in different canine diseases such as leishmaniosis [53] or canine idiopathic dilated cardiomyopathy [25], suggesting that APO-A1 could be a feasible biomarker in dogs. Lower expression of neutrophil esterase was observed in both groups of dogs with PVE when compared to control group. Neutrophil esterase is a serine proteinase with important antimicrobial effects that are released by neutrophils and enhances inflammatory responses. Genetic mutations in its gene (ELANE) has been identified in forms of hereditary neutropenia in humans and dogs [54]. There is a case reported of a mutation in ELANE gene as the cause of chronic parvovirosis in a woman [55]. The activation of complement C3 supports local inflammatory responses against pathogens and initiates the humoral immune response, playing a central role in the activation of the complement system [56]. Increased C3 expression has also been reported in dogs with canine babesiosis [57]. In our study, different hypotheses could explain the down-regulation of complement C3 in saliva of dog with PVE such as a reduction due to cleavage of the protein in order to activate complement cascade, or as a consequence of an immunodeficiency [58]. Histidine-rich glycoprotein is a relatively abundant plasma protein with a wide range of targets and functions, such as regulating clotting and fibrinolysis, and angiogenesis [59,60]. HRG roles in anti-inflammatory effects and pathogen control have been proposed since, after experimental Streptococcus pyogenes infection, HRG-deficient mice showed higher bacterial burden and pro-inflammatory cytokines than wild-type mice [61].
Thirty-four of the proteins differentially expressed in saliva between controls and dogs with parvovirosis have binding activity, including many of the proteins described above such as APO-A1, A2M, C3 and LCN9. Other important proteins differentially expressed in saliva between healthy and dogs with parvovirus with binding activity are desmoglein-1 and -3 (DSG1 and DSG3, with 1.39 and 1.43- fold higher in dogs with parvovirus, respectively), and clusterins (CLU, 0.71-fold higher). Desmogleins are a family of calcium-binding transmembrane glycoproteins which play an important role in the desmosomes transmembrane component [62]. In humans, salivary levels of DSG1 and DSG3 are correlated with serum anti DSG titres, proposing salivary DSG as suitable markers of diagnosis of Pemphigus vulgaris [63]. Clusterins are secretory glycoproteins that are involved in multiple physiopathological processes, including lipid metabolism, tissue remodelling, reproduction, transport and cell apoptosis [64]. These proteins have been classically considered as markers of cell death [65]; however, protective roles such as anti-apoptotic or prevention of complement attacking complex functions of clusterins have been suggested [66,67].
The bioinformatics analysis of the in silico inferred proteins network allowed the generation of more information from the experimental results, including data on intracellular pathways. This revealed the impact on processes such as an inhibition of humoral and cellular responses, and a positive effect in the entry of bacterium into host cell. These results are in concordance with the pathogenicity of PVE in which, after the ingestion, the virus replicates in the lymphoid tissue in the dog’s throat and then spreads to the bloodstream infecting rapidly dividing cells, resulting in depletion of lymphocytes in lymph nodes and necrosis of the intestinal crypts [68]. These mucosal intestinal lesions may give rise to secondary bacterial invasions from the intestines, leading to systemic inflammatory response syndrome (SIRS), sepsis and endotoxemia [[69], [70], [71], [72]].
Bioinformatics allowed several pathways and proteins to be highlighted which could be useful in the diagnosis of PVE with saliva samples. For example, the network created by comparison of survival and control groups revealed that fibrinogen and ubiquitin complexes could play important roles in the development of the disease. The down-regulation of fibrinogen observed in the present study is in contrast with other reports that showed increased levels of fibrinogen in dogs with parvovirosis [73], leishmaniosis [74] or babesiosis [75]; although dogs with uncomplicated babesiosis showed higher levels of fibrinogen that those with SIRS or multiple organ dysfunction syndrome (MODS) [75]. This could be explained by the fact that, although fibrinogen is considered as a positive acute phase protein in dogs, in disseminated intravascular coagulation (DIC) and hyperfibrinolysis the concentration of fibrinogen falls due to fibrinogen cleavage to form soluble fibrin monomers [76,77]. Progressive decrease of fibrinogen levels was also reported in dogs during acute haemorrhage [78], as occurs in parvovirosis. On the other hand, ubiquitins have been shown to be involved in numerous intracellular processes such as cell cycle control, apoptosis, DNA repair, regulation of transcription, stress responses, and targeting cellular proteins for degradation by the 26S proteasome [79]. As described in previous reports, viruses often make use of the ubiquitin conjugation to target for degradation of cellular proteins that might otherwise affect the replication of the virus [80].
One of the limitations of the present study may be the sample size, which could have limited the potential to identify proteins that are reproducibly modulated in concentration in dogs with PVE due to the biological inter-individual variations. Therefore this could be considered as a pilot study, and further validation studies with a larger number of dogs should be made to confirm these findings. The changes in protein expression and the subsequent bioinformatics analysis described in the present study were based on the analysis of saliva and, therefore, further studies are desirable to discern if they reflect systemic alterations. Nevertheless, despite these limitations, our results indicate that PVE can produce changes in saliva proteome that can reflect an inflammatory response and changes in coagulation system and immune response – among other biological processes.
This paper was focused on the identification of the differentially expressed proteins in saliva and in the description of the biological roles of the proteins affected due to the disease. Further large-scale clinical studies with larger populations would be desirable to elucidate if the proteins identified in this report can be of help in the earlier detection, predicting the outcome, or giving information to the clinician to adjust the treatment. If these studies are successful, these proteins can be used for developing rapid diagnostic tests such as lateral flow kits in saliva for clinical practice.
5. Conclusions
TMT-based proteomic analysis allowed the identification of 190 proteins with different abundances between healthy dogs and dogs with PVE, most of them not previously reported in canine parvovirosis. These proteins are involved in various physiopathological processes such as coagulation, inflammation and defence mechanisms, and could be considered as potentially suitable biomarkers of the disease. This study demonstrated that saliva proteome is a suitable biofluid for both study and diagnostic of canine parvovirosis.
Disclosure statement
The authors report no other conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Acknowledgements
LFM was granted with predoctoral contract ‘FPU’ of University of Murcia, Spain. AT has a post-doctoral fellowshisp “Juan de la Cierva Incorporación” and DE has a post-doctoral fellowshisp “Juan de la Cierva Formación” supported by the “Ministerio de Economía y Competitividad”, Spain. This work was supported by a grant from the Program for Research Groups of Excellence of the Seneca Foundation, Murcia, Spain (grant 19894/GERM/15) and European Commission ERA chair FP7 grant (VetMedZg #621394).
References
- 1.Nandi S., Kumar M. Canine parvovirus: current perspective. Indian J. Virol. 2010;21(1):31–44. doi: 10.1007/s13337-010-0007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matijatko V., Mrljak V., Kiš I., Kučer N., Foršek J., Živičnjak T., Romić Ž., Šimec Z., Ceron J.J. Evidence of an acute phase response in dogs naturally infected with Babesia canis. Vet. Parasitol. 2007;144:242–250. doi: 10.1016/j.vetpar.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Panda D., Patra R.C., Nandi S., Swarup D. Oxidative stress indices in gastroenteritis in dogs with canine parvoviral infection. Res. Vet. Sci. 2009;86:36–42. doi: 10.1016/j.rvsc.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kocaturk M., Tvarijonaviciute A., Martinez-Subiela S., Tecles F., Eralp O., Yilmaz Z., Ceron J.J. Inflammatory and oxidative biomarkers of disease severity in dogs with parvoviral enteritis. J. Small Anim. Pract. 2015;56:119–124. doi: 10.1111/jsap.12250. [DOI] [PubMed] [Google Scholar]
- 5.Ford J., McEndaffer L., Renshaw R., Molesan A., Kelly K. Parvovirus infection is associated with myocarditis and myocardial fibrosis in young dogs. Vet. Pathol. 2017;54:964–971. doi: 10.1177/0300985817725387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohr A.J., Leisewitz A.L., Jacobson L.S., Steiner J.M., Ruaux C.G., Williams Da. Effect of early enteral nutrition on intestinal permeability, intestinal protein loss, and outcome in dogs with severe parvoviral enteritis. J. Vet. Intern. Med. 2003;17:791–798. doi: 10.1111/j.1939-1676.2003.tb02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantos-Barreda A., Escribano D., Bernal L.J., Cerón J.J., Martínez-Subiela S. Quantification of anti-Leishmania antibodies in saliva of dogs. Vet. Parasitol. 2017;242:54–58. doi: 10.1016/j.vetpar.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Jankowski M., Spużak J., Kubiak K., Glińska-Suchocka K., Biernat M. An evaluation of the usefulness of invasive and non-invasive methods used to diagnose Helicobacter spp. Infections in dogs. Pol. J. Vet. Sci. 2017;20:491–499. doi: 10.1515/pjvs-2017-0059. [DOI] [PubMed] [Google Scholar]
- 9.Di Nardo F., Anfossi L., Ozella L., Saccani A., Giovannoli C., Spano G., Baggiani C. Validation of a qualitative immunochromatographic test for the noninvasive assessment of stress in dogs. J. Chromatogr. B. 2016;1028:192–198. doi: 10.1016/j.jchromb.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Lindsay A., Costello J.T. Realising the potential of urine and saliva as diagnostic tools in sport and exercise medicine. Sport. Med. 2016;47:1–21. doi: 10.1007/s40279-016-0558-1. [DOI] [PubMed] [Google Scholar]
- 11.Desario C., Decaro N., Campolo M., Cavalli A., Cirone F., Elia G., Martella V., Lorusso E., Camero M., Buonavoglia C. Canine parvovirus infection: which diagnostic test for virus? J. Virol. Methods. 2005;126:179–185. doi: 10.1016/j.jviromet.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Tabak L.A. A revolution in biomedical assessment: the development of salivary diagnostics. J. Dent. Educ. 2001;65(12):1335–1339. [PubMed] [Google Scholar]
- 13.Guillemin N., Horvatić A., Kuleš J., Galan A., Mrljak V., Bhide M. Omics approaches to probe markers of disease resistance in animal sciences. Mol. Biosyst. 2016;12:2036–2046. doi: 10.1039/C6MB00220J. [DOI] [PubMed] [Google Scholar]
- 14.Li H., Li C., Wu H., Zhang T., Wang J., Wang S., Chang J. Identification of Apo-A1 as a biomarker for early diagnosis of bladder transitional cell carcinoma. Proteome Sci. 2011;9(21) doi: 10.1186/1477-5956-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz-Romero C., Blanco F.J. Proteomics role in the search for improved diagnosis, prognosis and treatment of osteoarthritis. Osteoarthr. Cartil. 2010;18:500–509. doi: 10.1016/j.joca.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Baeumlisberger D., Arrey T.N., Rietschel B., Rohmer M., Papasotiriou D.G., Mueller B., Beckhaus T., Karas M. Labeling elastase digests with TMT: informational gain by identification of poorly detectable peptides with MALDI-TOF/TOF mass spectrometry. Proteomics. 2010;10:3905–3909. doi: 10.1002/pmic.201000288. [DOI] [PubMed] [Google Scholar]
- 17.Dayon L., Turck N., García-Berrocoso T., Walter N., Burkhard P.R., Vilalta A., Sahuquillo J., Montaner J., Sanchez J.-C. Brain extracellular fluid protein changes in acute stroke patients. J. Proteome Res. 2011;10:1043–1051. doi: 10.1021/pr101123t. [DOI] [PubMed] [Google Scholar]
- 18.Giron P., Dayon L., Turck N., Hoogland C., Sanchez J.-C. Quantitative analysis of human cerebrospinal fluid proteins using a combination of cysteine tagging and amine-reactive isobaric labeling. J. Proteome Res. 2011;10:249–258. doi: 10.1021/pr100535f. [DOI] [PubMed] [Google Scholar]
- 19.Ross P.L., Huang Y.N., Marchese J.N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D.J. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Baldini C., Giusti L., Ciregia F., Da Valle Y., Giacomelli C., Donadio E., Ferro F., Galimberti S., Donati V., Bazzichi L., Bombardieri S., Lucacchini A. Correspondence between salivary proteomic pattern and clinical course in primary Sjögren syndrome and non-Hodgkin’s lymphoma: a case report. J. Transl. Med. 2011;9(188) doi: 10.1186/1479-5876-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beckett P. The basics of 2D DIGE. Methods Mol. Biol. 2012;854:9–19. doi: 10.1007/978-1-61779-573-2_2. [DOI] [PubMed] [Google Scholar]
- 22.Yilmaz Z., Senturk S. Characterisation of lipid profiles in dogs with parvoviral enteritis. J. Small Anim. Pract. 2007;48:643–650. doi: 10.1111/j.1748-5827.2007.00391.x. [DOI] [PubMed] [Google Scholar]
- 23.Contreras-Aguilar M.D., Tecles F., Martínez-Subiela S., Escribano D., Bernal L.J., Cerón J.J. Detection and measurement of alpha-amylase in canine saliva and changes after an experimentally induced sympathetic activation. BMC Vet. Res. 2017;13:266. doi: 10.1186/s12917-017-1191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-Subiela S., Ceron J.J., Yilmaz Z., Martinez-Subiela S., Horvatic A., Escribano D., Pardo-Marin L., Kocaturk M., Mrljak V., Burchmore R., Ceron J.J., Yilmaz Z. Identification of novel biomarkers for treatment monitoring in canine leishmaniosis by high-resolution quantitative proteomic analysis. Vet. Immunol. Immunopathol. 2017;191:60–67. doi: 10.1016/j.cimid.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Bilić P., Guillemin N., Kovačević A., Beer Ljubić B., Jović I., Galan A., Eckersall P.D., Burchmore R., Mrljak V. Serum proteome profiling in canine idiopathic dilated cardiomyopathy using TMT-based quantitative proteomics approach. J. Proteomics. 2018;179:110–121. doi: 10.1016/j.jprot.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 26.RStudio Team. RStudio . RStudio, Inc.; Boston, MA: 2015. Integrated Development Environment for R.http://www.rstudio.com/ 2015. [Internet].Available from: [Google Scholar]
- 27.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Supek F., Bošnjak M., Škunca N., Šmuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Hinsbergh V.W.M. Endothelium - Role in regulation of coagulation and inflammation. Semin. Immunopathol. 2012;34(1):93–106. doi: 10.1007/s00281-011-0285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esmon C.T., Fukudome K., Mather T., Bode W., Regan L.M., Stearns-Kurosawa D.J., Kurosawa S. Inflammation, sepsis, and coagulation. Haematologica. 1999;84:254–259. https://doi.org/10189392. [PubMed] [Google Scholar]
- 32.Coorens M., Schneider V.A.F., de Groot A.M., van Dijk A., Meijerink M., Wells J.M., Scheenstra M.R., Veldhuizen E.J.A., Haagsman H.P. Cathelicidins inhibit Escherichia coli-Induced TLR2 and TLR4 activation in a viability-dependent manner. J. Immunol. 2017;199:1418–1428. doi: 10.4049/jimmunol.1602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chromek M., Arvidsson I., Karpman D. The antimicrobial peptide cathelicidin protects mice from Escherichia coli O157:H7-Mediated disease. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chromek M., Slamová Z., Bergman P., Kovács L., Podracká L., Ehrén I., Hökfelt T., Gudmundsson G.H., Gallo R.L., Agerberth B., Brauner A. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- 35.Nizet V., Ohtake T., Lauth X., Trowbridge J., Rudisill J., Dorschner R.A., Pestonjamasp V., Piraino J., Huttner K., Gallo R.L. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 36.Jann N.J., Schmaler M., Kristian S.A., Radek K.A., Gallo R.L., Nizet V., Peschel A., Landmann R. Neutrophil antimicrobial defense against Staphylococcus aureus is mediated by phagolysosomal but not extracellular trap-associated cathelicidin. J. Leukoc. Biol. 2009;86:1159–1169. doi: 10.1189/jlb.0209053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dijk A., Tersteeg-Zijderveld M.H.G., Tjeerdsma-van Bokhoven J.L.M., Jansman A.J.M., Veldhuizen E.J.A., Haagsman H.P. Chicken heterophils are recruited to the site of Salmonella infection and release antibacterial mature Cathelicidin-2 upon stimulation with LPS. Mol. Immunol. 2009;46:1517–1526. doi: 10.1016/j.molimm.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 38.Schauber J., Svanholm C., Term??n S., Iffland K., Menzel T., Scheppach W., Melcher R., Agerberth B., L??hrs H., Gudmundsson G.H. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52:735–741. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott M.G., Davidson D.J., Gold M.R., Bowdish D., Hancock R.E.W. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J. Immunol. 2002;169:3883–3891. doi: 10.4049/jimmunol.169.7.3883. [DOI] [PubMed] [Google Scholar]
- 40.Van Dijk A., Van Eldik M., Veldhuizen E.J.A., Tjeerdsma-Van Bokhoven H.L.M., De Zoete M.R., Bikker F.J., Haagsman H.P. Immunomodulatory and anti-inflammatory activities of chicken cathelicidin-2 derived peptides. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Togawa A., Miyoshi J., Ishizaki H., Tanaka M., Takakura A., Nishioka H., Yoshida H., Doi T., Mizoguchi A., Matsuura N., Niho Y., Nishimune Y., Nishikawa S.I., Takai Y. Progressive impairment of kidneys and reproductive organs in mice lacking Rho GDIα. Oncogene. 1999;18:5373–5380. doi: 10.1038/sj.onc.1202921. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe T., Urano E., Miyauchi K., Ichikawa R., Hamatake M., Misawa N., Sato K., Ebina H., Koyanagi Y., Komano J. The hematopoietic cell-specific rho GTPase inhibitor ARHGDIB/D4GDI limits HIV type 1 replication. AIDS Res. Hum. Retroviruses. 2012;28:913–922. doi: 10.1089/aid.2011.0180. [DOI] [PubMed] [Google Scholar]
- 43.Theodorescu D., Sapinoso L.M., Conaway M.R., Oxford G., Hampton G.M., Frierson H.F. Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin. Cancer Res. 2004;10:3800–3806. doi: 10.1158/1078-0432.CCR-03-0653. [DOI] [PubMed] [Google Scholar]
- 44.Jensen-Jarolim E., Pacios L.F., Bianchini R., Hofstetter G., Roth-Walter F. Structural similarities of human and mammalian lipocalins, and their function in innate immunity and allergy. Allergy. 2016;71:286–294. doi: 10.1111/all.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Åkerstrom B., Flower D.R., Salier J.P. Lipocalins: unity in diversity. Biochim. Biophys. Acta - Protein Struct. Mol. Enzymol. 2000;1482(1–2):1–8. doi: 10.1016/S0167-4838(00)00137-0. [DOI] [PubMed] [Google Scholar]
- 46.Lögdberg L., Wester L. Immunocalins: a lipocalin subfamily that modulates immune and inflammatory responses. Biochim. Biophys. Acta - Protein Struct. Mol. Enzymol. 2000;1482(1–2):284–297. doi: 10.1016/S0167-4838(00)00164-3. [DOI] [PubMed] [Google Scholar]
- 47.Fjaertoft G., Foucard T., Xu S., Venge P. Human neutrophil lipocalin (HNL) as a diagnostic tool in children with acute infections: a study of the kinetics. Acta Paediatr. 2005;94:661–666. doi: 10.1080/08035250510031610. [DOI] [PubMed] [Google Scholar]
- 48.Mårtensson J., Bell M., Oldner A., Xu S., Venge P., Martling C.R. Neutrophil gelatinase-associated lipocalin in adult septic patients with and without acute kidney injury. Intensive Care Med. 2010;36:1333–1340. doi: 10.1007/s00134-010-1887-4. [DOI] [PubMed] [Google Scholar]
- 49.Parravicini E., Nemerofsky S.L., Michelson K.A., Huynh T.K., Sise M.E., Bateman D.A., Lorenz J.M., Barasch J.M. Urinary neutrophil gelatinase-associated lipocalin is a promising biomarker for late onset culture-positive sepsis in very low birth weight infants. Pediatr. Res. 2010;67:636–640. doi: 10.1203/PDR.0b013e3181da75c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bingle L., Barnes F.A., Lunn H., Musa M., Webster S., Douglas C.W.I., Cross S.S., High A.S., Bingle C.D. Characterisation and expression of SPLUNC2, the human orthologue of rodent parotid secretory protein. Histochem. Cell Biol. 2009;132:339–349. doi: 10.1007/s00418-009-0610-4. [DOI] [PubMed] [Google Scholar]
- 51.Morosky S., Lennemann N.J., Coyne C.B. BPIFB6 Regulates Secretory Pathway Trafficking and Enterovirus Replication. J. Virol. 2016;90:5098–5107. doi: 10.1128/JVI.00170-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y., DiDonato J.A., Levison B.S., Schmitt D., Li L., Wu Y., Buffa J., Kim T., Gerstenecker G.S., Gu X., Kadiyala C.S., Wang Z., Culley M.K., Hazen J.E., Didonato A.J., Fu X., Berisha S.Z., Peng D., Nguyen T.T., Liang S., Chuang C.-C., Cho L., Plow E.F., Fox P.L., Gogonea V., Tang W.H.W., Parks J.S., Fisher E.A., Smith J.D., Hazen S.L. An abundant dysfunctional apolipoprotein A1 in human atheroma. Nat. Med. 2014;20:193–203. doi: 10.1038/nm.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Escribano D., Tvarijonaviciute A., Kocaturk M., Cerón J.J., Pardo-Marín L., Torrecillas A., Yilmaz Z., Martínez-Subiela S. Serum apolipoprotein-A1 as a possible biomarker for monitoring treatment of canine leishmaniosis. Comp. Immunol. Microbiol. Infect. Dis. 2016;49:82–87. doi: 10.1016/j.cimid.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Horwitz M., Benson K.F., Duan Z., Li F.-Q., Person R.E. Hereditary neutropenia: dogs explain human neutrophil elastase mutations. Trends Mol. Med. 2004;10:163–170. doi: 10.1016/j.molmed.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Thanarajasingam U., Jensen M.A., Dorschner J.M., Wampler Muskardin T., Ghodke-Puranik Y., Purmalek M., Eliopoulos E., Zervou M.I., Goulielmos G.N., Howard M., Kaplan M.J., Niewold T.B. Brief report: a novel ELANE mutation associated with inflammatory arthritis, defective NETosis, and recurrent parvovirus infection. Arthritis Rheumatol. (Hoboken, N.J.) 2017;69:2396–2401. doi: 10.1002/art.40314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chee C.S., Chang K.M., Loke M.F., Angela Loo V.P., Subrayan V. Association of potential salivary biomarkers with diabetic retinopathy and its severity in type-2 diabetes mellitus: a proteomic analysis by mass spectrometry. PeerJ. 2016;4:e2022. doi: 10.7717/peerj.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuleš J., Mrljak V., Barić Rafaj R., Selanec J., Burchmore R., Eckersall P.D. Identification of serum biomarkers in dogs naturally infected with Babesia canis canis using a proteomic approach. BMC Vet. Res. 2014;10:111. doi: 10.1186/1746-6148-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Neil K.M., Ochs H.D., Heller S.R., Cork L.C., Morris J.M., Winkelstein J.A. Role of C3 in humoral immunity. Defective antibody production in C3-deficient dogs. J. Immunol. 1988;140:1939–1945. [PubMed] [Google Scholar]
- 59.Lee C., Bongcam-Rudloff E., Sollner E., Jahnen-Dechent W., Claesson-Welsh L. Type 3 cystatins; fetuins, kininogen and histidine-rich glycoprotein. Front Biosci (Landmark Ed). 2009;1:2911–2922. doi: 10.1111/febs.12167. [DOI] [PubMed] [Google Scholar]
- 60.Poon I.K.H., Patel K.K., Davis D.S., Parish C.R., Hulett M.D. Histidine-rich glycoprotein: the Swiss Army knife of mammalian plasma. Blood. 2011;117(7):2093–2101. doi: 10.1182/blood-2010-09-303842. [DOI] [PubMed] [Google Scholar]
- 61.Shannon O., Rydengård V., Schmidtchen A., Mörgelin M., Alm P., Sørensen O.E., Björck L. Histidine-rich glycoprotein promotes bacterial entrapment in clots and decreases mortality in a mouse model of sepsis. Blood. 2010;116:2365–2372. doi: 10.1182/blood-2010-02-271858. [DOI] [PubMed] [Google Scholar]
- 62.Ellebrecht, C.T., Payne, A.S., n.d. Plakophilins, desmogleins, and pemphigus: the tail wagging the dog Desmosomes and disease. 10.1038/jid.2013.491. [DOI] [PMC free article] [PubMed]
- 63.Hallaji Z., Mortazavi H., Lajevardi V., Tamizifar B., Amirzargar A., Daneshpazhooh M., Chams-Davatchi C. Serum and salivary desmoglein 1 and 3 enzyme-linked immunosorbent assay in pemphigus vulgaris: correlation with phenotype and severity. J. Eur. Acad. Dermatol. Venereol. 2010;24:275–280. doi: 10.1111/j.1468-3083.2009.03408.x. [DOI] [PubMed] [Google Scholar]
- 64.Rosenberg M.E., Silkensen J. Clusterin: Physiologic and pathophysiologic considerations. Int. J. Biochem. Cell Biol. 1995;27(7):633–645. doi: 10.1016/1357-2725(95)00027-M. [DOI] [PubMed] [Google Scholar]
- 65.Wong, P., Borst, D.E., Farber, D., Danciger, J.S., Tenniswood, M., Chader, G.J., van Veen, T., n.d. Increased TRPM-2/clusterin mRNA levels during the time of retinal degeneration in mouse models of retinitis pigmentosa. Biochem. Cell Biol. 72, 439–446. [DOI] [PubMed]
- 66.Martynova E.V., Maksudova A.N., Shakirova V.G., Abdulkhakov S.R., Khaertynova I.M., Anokhin V.A., Ivanova V.V., Abiola I.M., Garanina E.E., Tazetdinova L.G., Valiullina A.H., Khaiboullina S.F. Urinary clusterin is upregulated in Nephropathia Epidemica. Dis. Markers. 2018;2018:1–7. doi: 10.1155/2018/8658507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang F., Kumano M., Beraldi E., Fazli L., Du C., Moore S., Sorensen P., Zoubeidi A., Gleave M.E. Clusterin facilitates stress-induced lipidation of LC3 and autophagosome biogenesis to enhance cancer cell survival. Nat. Commun. 2014;5 doi: 10.1038/ncomms6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kocaturk M., Martinez S., Eralp O., Tvarijonaviciute A., Ceron J., Yilmaz Z. Prognostic value of serum acute-phase proteins in dogs with parvoviral enteritis. J. Small Anim. Pract. 2010;51:478–483. doi: 10.1111/j.1748-5827.2010.00965.x. [DOI] [PubMed] [Google Scholar]
- 69.Macintire D.K., Smith-Carr S. Canine parvovirus. Part II. Clinical signs, diagnosis, and treatment. Compend. Contin. Vet. Educ. Pract. 1997;19:291–302. [Google Scholar]
- 70.Mantione N.L., Otto C.M. Characterization of the use of antiemetic agents in dogs with parvoviral enteritis treated at a veterinary teaching hospital: 77 cases (1997-2000) J. Am. Vet. Med. Assoc. 2005;227:1787–1793. doi: 10.2460/javma.2005.227.1787. [DOI] [PubMed] [Google Scholar]
- 71.Turk J., Fales W., Miller M., Pace L., Fischer J., Johnson G., Kreeger J., Turnquist S., Pittman L., Rottinghaus A. Enteric Clostridium perfringens infection associated with parvoviral enteritis in dogs: 74 cases (1987-1990) J. Am. Vet. Med. Assoc. 1992;200:991–994. [PubMed] [Google Scholar]
- 72.Turk J., Miller M., Brown T., Fales W., Fischer J., Gosser H., Nelson S., Shaw D., Solorzano R. Coliform septicemia and pulmonary disease associated with canine parvoviral enteritis: 88 cases (1987-1988) J. Am. Vet. Med. Assoc. 1990;196:771–773. [PubMed] [Google Scholar]
- 73.Otto C.M., Rieser T.M., Brooks M.B., Russell M.W. Evidence of hypercoagulability in dogs with parvoviral enteritis. J. Am. Vet. Med. Assoc. 2000;217:1500–1504. doi: 10.2460/javma.2000.217.1500. [DOI] [PubMed] [Google Scholar]
- 74.Britti D., Gaspari M., Massimini G., Casalinuovo F., Morittu V.M., Cuda G. Veterinary Research Communications. 2010. Proteomic analysis in canine leishmaniasis; pp. 91–96. [DOI] [PubMed] [Google Scholar]
- 75.Kuleš J., de Torre-Minguela C., Barić Rafaj R., Gotić J., Nižić P., Ceron J.J., Mrljak V. Plasma biomarkers of SIRS and MODS associated with canine babesiosis. Res. Vet. Sci. 2016;105:222–228. doi: 10.1016/J.RVSC.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 76.Carretón E., Morchón R., Simón F., Juste M.C., Méndez J.C., Montoya-Alonso J.A. Cardiopulmonary and inflammatory biomarkers in the assessment of the severity of canine dirofilariosis. Vet. Parasitol. 2014;206:43–47. doi: 10.1016/j.vetpar.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 77.Cerón J.J., Eckersall P.D., Martínez-Subiela S. Acute phase proteins in dogs and cats: current knowledge and future perspectives. Vet. Clin. Pathol. 2005;34(2):85–99. doi: 10.1111/j.1939-165X.2005.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 78.Lynch A.M., DeLaforcade A.M., Meola D., Shih A., Bandt C., Guerrero N.H., Riccó C. Assessment of hemostatic changes in a model of acute hemorrhage in dogs. J. Vet. Emerg. Crit. Care San Antonio (San Antonio) 2016;26:333–343. doi: 10.1111/vec.12457. [DOI] [PubMed] [Google Scholar]
- 79.Johnston S.C., Larsen C.N., Cook W.J., Wilkinson K.D., Hill C.P. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 angstrom resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Randow F., Lehner P.J. Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 2009;11(5):527–534. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]