Abstract
Although recent research has revealed high genomic complexity in the earliest-splitting animals and their ancestors, the macroevolutionary trends orchestrating gene repertoire evolution throughout the animal phyla remain poorly understood. We used a phylogenomics approach to interrogate genome evolution across all animal phyla. Our analysis uncovered a bimodal distribution of recruitment of orthologous genes, with most genes gained very ‘early’ (i.e. at deep nodes) or very ‘late’, representing lineage-specific acquisitions. The emergence of animals was characterized by both a high gene birth and duplication ratio. Deuterostomes, ecdysozoans and xenacoelomorphans were characterized by no gene gain but rampant differential gene loss. Genes considered as animal hallmarks, such as Notch/Delta, were convergently duplicated in all phyla and at different evolutionary depths. Genes duplicated in all nodes from Metazoa to phylum-specific levels were enriched in functions related to the neural system, suggesting that this system has been continuously and independently reshaped throughout evolution across animals. Our results support that animal genomes evolved by unparalleled gene duplication followed by differential gene loss, and provide an atlas of gene repertoire evolution throughout the Animal Tree of Life to navigate how, when and how often each gene in each genome was gained, duplicated or lost.
The Animal Tree of Life includes 36 main phyla that entail an impressive diversity in terms of morphology, physiology and life-styles. This diversity is the result of radical morphological or physiological innovations such as the development of gonads, muscles or brains. Although the lineages in which these innovations appear can be identified with confidence, we know very little about the genomic changes underlying their appearance. Recent studies based on sequence comparison and clustering of up to 14 animal phyla 1–3 have shown that thousands of homologous gene groups could be detected at the level of Opisthokonta, Holozoa, Metazoa, Eumetazoa or Bilateria, among other clades, supporting a shared ancestry of many of the genes present in extant animals and describing a minimal genome content for the hypothetical Last Common Ancestor (LCA) of each of these lineages. However, besides failing to cover more than half of the metazoan diversity, none of these studies used a phylogenetic approach, and thus provide limited resolution of the reconstructed gene evolutionary histories, hampering our understanding of the evolutionary dynamics governing gene repertoire evolution. Herein, we undertook a phylogenomic approach to gene family evolution across the Animal Tree of Life including representatives of all animal phyla. Our methodological approach, centered around the use of gene phylogenies to infer pairwise orthology and paralogy relationships for all genes in all genomes, is robust to the effect of horizontal gene transfer, gene loss, genome completeness and variable genome size, factors that strongly affect analyses based on gene family inference. Since we inferred these orthology/paralogy relationships and their evolutionary dynamics for each gene in one species at a time, our results are independent for each lineage, thus providing a robust backbone of global evolutionary patterns of gene evolution across metazoans. Moreover, since orthologous genes have significantly more similar functions than paralogous genes 45, our approach allowed us to shed light on the evolution of function across different phyla, even if putatively.
Results and Discussion
A phylogenomic approach to gene repertoire evolution across the Metazoa Tree of Life reveals unbalanced distributions of gene gains, losses and duplications
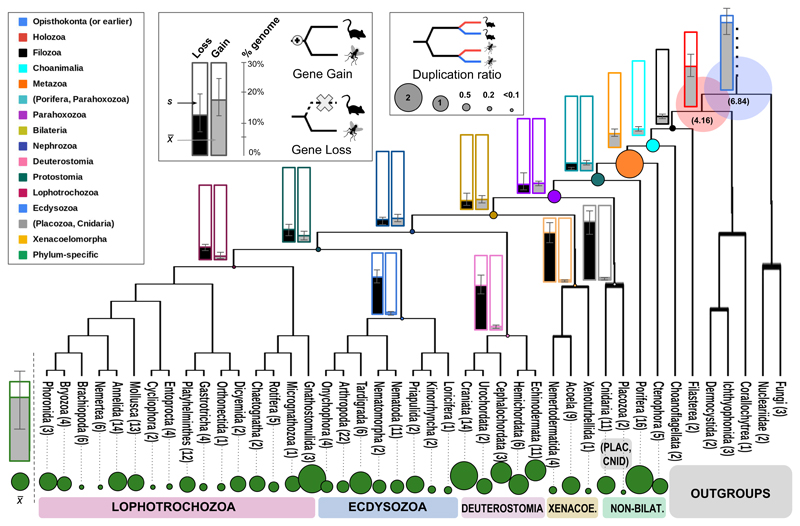
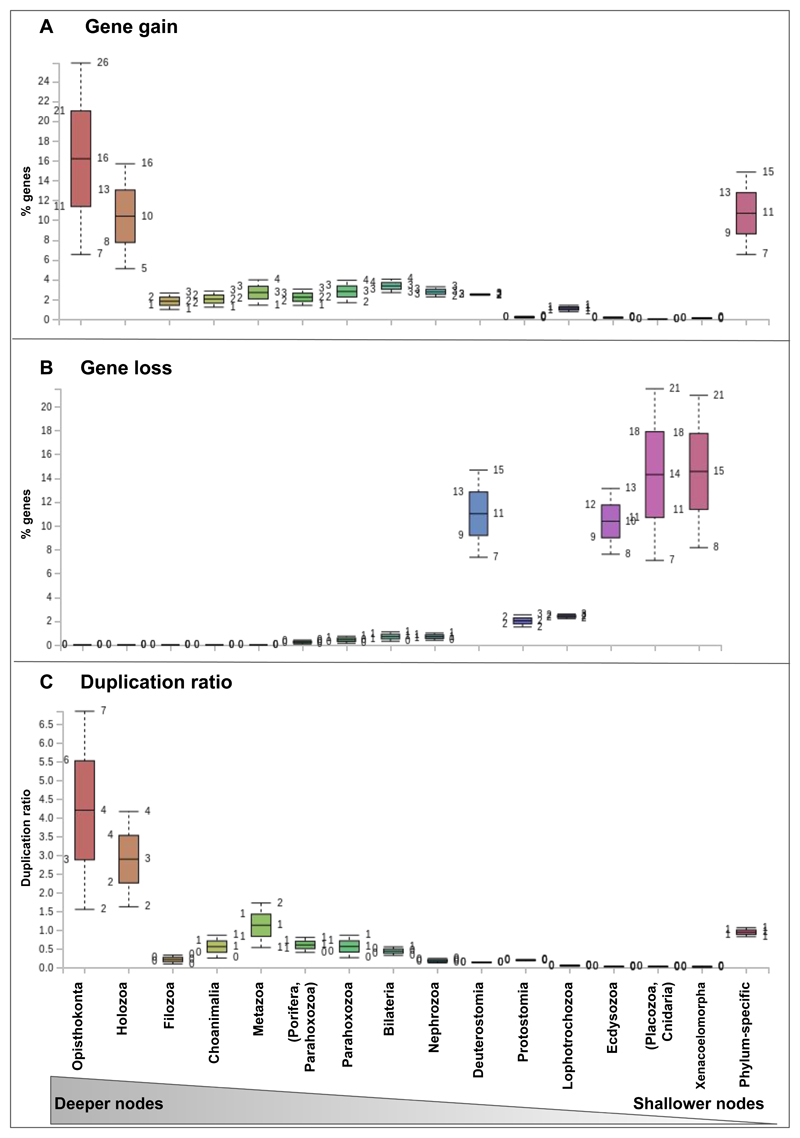
We used a phylogenetic approach to reconstruct evolutionary histories for each gene in each genome and infer pairwise orthology/paralogy relationships in a data set composed of 231 genomes and transcriptomes. For this, we rooted the gene trees with the topology depicted in Fig. 1a, based on our current knowledge on animal phylogeny 6–8. Results were not significantly different when rooting with alternative topologies (e.g., considering the ‘Porifera-sister’ hypothesis; p>0.05). Note that the orthology inference algorithm we used is robust to a non-resolved phylogeny - see Methods, nevertheless our results are based on a species tree assumption and may vary upon future reshaping of the animal tree of life. Remarkably, a consistent pattern of ortholog gain/loss was independently recovered in all lineages (Fig. 1b,c), which consisted of strikingly unbalanced distributions of gene gain, loss and duplication (Fig. 2a,b, see Glossary). Gene gain patterns displayed a bimodal distribution. Approximately one quarter of the genes in each genome was gained or already present at the node Opisthokonta or gained at Holozoa (Fig. 1b, see Suppl. Mat. S3, S4.). The following nodes from Filozoa to Nephrozoa exhibited much lower values of ortholog gain, up to 10%, a value that decreased further towards shallower nodes (Fig. 1b). Around one third of each genome comprised lineage-specific genes, i.e. genes that had no homologs in the rest of the taxa (Fig. 1b), reflecting that lineage-specific gene gain plays a crucial role during animal genome evolution. Notably, Xenacoelomorpha, Deuterostomia and Ecdysozoa were defined by high values of ortholog gene loss, as compared to all earlier-splitting lineages, whereas virtually no ortholog gene gain was observed in any of the phyla comprised in these clades (less than 2%; Fig. 1c).
Figure 1. Gene gain, loss and duplication ratios show a similar pattern across animal phyla.
Phylogenetic hypothesis of animal phyla interrelationships. Number of taxa per phyla corresponding to dataset 1 is indicated in each case. Nodes of interest are highlighted in different colors. Gene gain, loss (histograms in main branches) and duplication ratios (coloured circles) as inferred for each phyla in dataset 2 are shown. Colors correspond to nodes as defined in the legend. Mean (x) and standard deviation (S) values of gene gain and loss are indicated in each histogram. Size of circles representing duplication ratios are proportional to the size shown in the corresponding legend, excepting for Opisthokonta and Holozoa, where values are shown in brackets.
Figure 2. Gene gain and duplication ratios are high at deeper nodes, and gene loss at shallower ones.
Box plot graphs of gene gain (a), loss (b) and duplication ratios (c) per node as inferred from dataset 1. Mean and standard deviation values are shown for each node. Colors indicatenodes as shown in the legend.
High gene duplication at the origin of Metazoa
We next calculated genome-wide average ratios of gene duplication for each genome in each tree node (Fig. 1d, 2c). Our results recovered high duplication ratios at Opisthokonta and Holozoa, followed by a low ratio at Choanimalia that increased again at Metazoa. The following nodes were characterized by progressively descending ratios towards the tips of the tree, with those at Parahoxozoa and Bilateria being twice than ratios at shallower nodes (Xenacoelomorpha, Deuterostomia, Protostomia, Lophotrochozoa, Ecdysozoa). Finally, lineage-specific duplication ratios were again high, consistent with widespread lineage-specific expansions for all taxa. These results show that both ancient duplication events and a high rate of retention of duplicate genes have contributed to an abundance of duplicate genes in animal genomes. Altogether our results support a scenario of high gene birth rate in the branch leading to metazoans that decreased progressively as animals diversified into clades (Fig. 2a). Interestingly, some of the most highly duplicated gene families included hallmark genes of animal genomes such as Notch/Delta ligands (Fig. 3a, see also Suppl. Mat.), together with transposon-related gene families such as Pao retrotransposon peptidase and proteins involved in neuropeptide reception (somatostatin/galanin receptors). Notably, these gene families were highly duplicated in metazoans but did not have virtually any homologs in the outgroups (and if they had, both their taxon representation and duplication ratio were very low; see Suppl. Mat.). In contrast, other highly-duplicated gene families were present both in animals and most outgroups, but duplication ratios were only high in metazoans: such is the case of tyrosine kinase receptors, transmembrane proteases and, remarkably, dynein, a family of cytoskeletal motor proteins that move along microtubules in cells and that drives the beat of eukaryotic cilia and flagella.
Figure 3. Genes related to the neural system are amongst the most highly duplicated.
a. List of top 10 most duplicated gene families as defined by OrthoFinder2. Group of taxa in which these families were inferred (metazoa or outgroups), mean and maximum number of duplications across animals and putative functions are indicated. b. Treemap representation of GO enrichment analysis of duplicated genes related to neural system (light blue) in all nodes in Ctenophora, Porifera, Placozoa and Craniata (see also Suppl. Mat.). Other colors represent other functions as shown in the legend. Each main square represents hierarchical GO term relationships. Square size is proportional to the p value for each GO term as found in the enrichment analyses. Scales are irrelevant.
Orthology-informed putative functions of duplicated genes
To further understand animal gene repertoire evolution at a functional level, we performed an evolutionary-informed interrogation of the putative function of gene duplications, gains and losses in our data set. The fraction of annotated genes ranged from 50-60% in some phyla (e.g., Tardigrada, Rotifera) to 90-100% in others (e.g., most deuterostomes), indicating a ‘hidden biology’ effect of non-annotated genes that is biasing our understanding on gene function evolution. This limitation notwithstanding, we need to focus our discussion on the annotated subset of genes. To attenuate this bias, we were careful to limit functional gene ontology (GO) term propagation to orthologous, not just homologous, genes, and to focus on high levels of the GO hierarchy, where function is more likely to be conserved 45. In all phyla, orthologs gained or already present at Opisthokonta and Holozoa were significantly enriched in complex functions such as DNA and protein modification, signal transduction, cell division and embryogenesis and developmental structures such as muscle, mesoderm or neural system formation, these patterns being highly similar across phyla. In Choanimalia, enriched functions among gained genes were related to cilia assembly and movement and chemotaxis (Fig. 3b, Suppl. Mat.). In orthologs gained at Metazoa to Nephrozoa, functions related to synapsis establishment and nervous system development were enriched and prevalent in virtually all genomes, together with various functions related to morphogenesis and reproduction. Virtually no gene gain (and therefore no enrichment) was detected in shallower nodes. These results suggest that orthologs involved in such complex developmental features were gained very ‘early’ in evolution, and supports that co-option of existing genes to novel functions was potentially a critical factor generating the increasingly-complex morphological variation observed across animal phyla.
For duplications, GO enrichment analyses in most nodes revealed a strong correlation with enriched functions also enriched among acquired genes (Suppl. Mat. S6, S7, S9). In contrast, while no functional enrichment was detected in the few genes gained at relatively shallower levels due to their low number (e.g., Deuterostomia, Protostomia, Xenacoelomorpha, etc.), genes duplicated at these nodes were strongly enriched in functions related to neural system, such as synapsis, neurotransmission or neural development (Fig. 3b). From the recurrent enrichment of related terms across the metazoan tree, we conclude that the neural system has been continuously shaped during metazoan evolution, being refined independently in all animal phyla - even in those without a complex nervous system (as defined by morphology in other phyla) such as Porifera and Placozoa. This result was further validated with the finding of a high level of duplications in orthogroups from the metazoan core gene repertoire (see below), in which genes involved in KEGG pathways related to neural system development and signal transduction were more highly duplicated relative to other pathways (Fig. 5a, c and d; see also Suppl. Mat. S9). We hypothesize that ancient gene duplicates related to neural activity may have been co-opted in a convergent manner to generate a growing neural complexity in animal phyla, while subsequent lineage-specific duplications potentially enabled an expanding structural plasticity, neuronal morphology and connectivity.
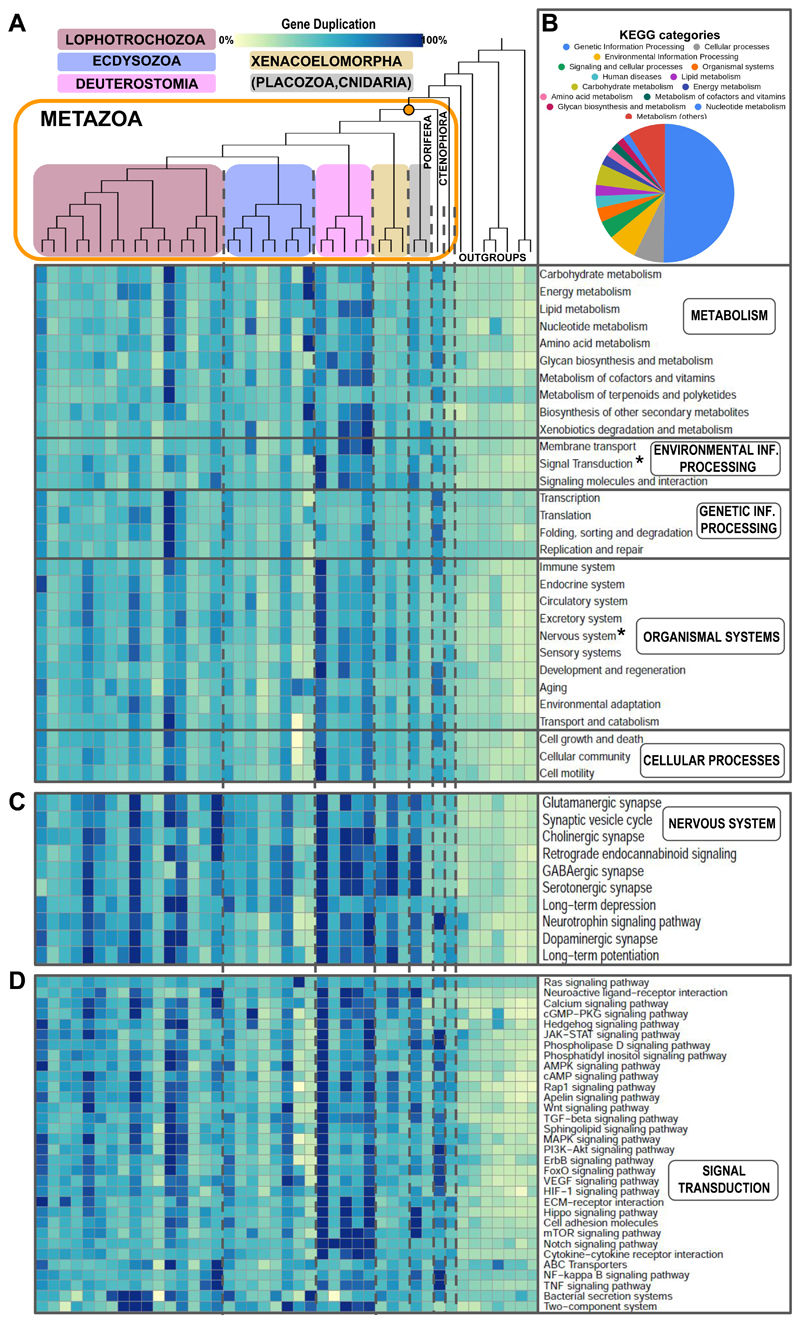
Figure 5. The core gene repertoire of metazoans includes genes from a plethora of KEGG pathways that have undergone different degrees of duplication.
a. Heatmap of pathway conservation and duplication level in the core gene repertoire of metazoans. The reference pathways were selected from the KEGG pathways collection. The color in each cell depicts the relative number of duplicates within each KEGG category found in each phyla as inferred for dataset 2 (for total number of duplicates and specific KEGG pathway annotation per orthogroup see Suppl. Mat. S9). Tree topology and colors for each clade are as in Fig. 1. Asterisks indicate specific pathways expanded in 5b and 5c. b. Percentage of each KEGG category annotated in all orthogroups from the metazoan core gene repertoire. c,d. Heatmap of KEGG nervous system (5c) and signal transduction (5d) pathway conservation and duplication level in the core repertoire of metazoans. The color in each cell is calculated as in 5a.
Rampant gene loss and differential retention of paralogs across animal lineages
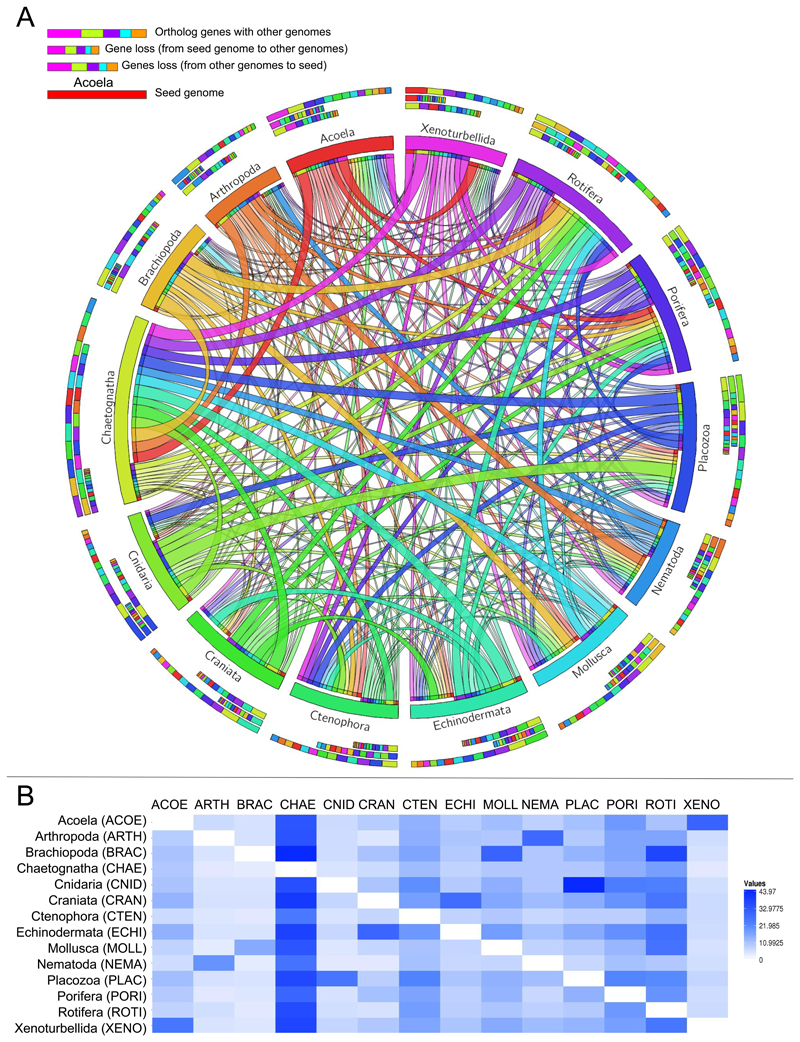
On the other hand, we detected virtually no enrichment among genes lost at any node in any phyla, suggesting that gene loss and differential retention of paralogs reshaped the whole biology of the organisms at all evolutionary depths. When exploring absolute gene loss pairwise between taxa (i.e., total number of genes lost in one taxa compared to the other, without taking into account in which node they were lost), we observed that values in all comparisons were remarkably high (Fig. 4). Particularly high values correlated with some recalcitrant positions in the Animal Tree of Life, such as Xenoturbellida (i.e., Xenoturbellida has lost a high percentage of orthologs in each pairwise inference with the remaining genomes independently) and Placozoa and Cnidaria (with Cnidaria having lost 44% of all its orthologs relative to Placozoa, and Placozoa 26% to Cnidaria). Our results thus may indicate a pervasive effect of extensive hidden paralogy hampering the phylogenomic reconstruction of deep animal relationships. The relevance of gene loss to metazoan evolution has already been anticipated. For instance, homology searches and clustering approaches9 defined a core bilaterian gene repertoire shared between Lophotrochozoa and Deuterostomia that had been apparently lost in Ecdysozoa and Platyzoa (rotifers, flatworms and their kin), and which is putatively involved in the control of homoeostasis and multicellularity. This study also observed that the number of gene families shared within Ecdysozoa and within Platyzoa was much lower than within Lophotrochozoa or Deuterostomia, pointing to extensive differential gene loss. Nevertheless, a phylogenomic-center approach based on orthology/paralogy inference had never been explored to date. In addition to placing these findings in a broader evolutionary context, our results further validate the role of differential gene loss following gene duplication as a prevailing force shaping metazoan gene repertoire evolution.
Figure 4. Pairwise gene loss is pervasive across phyla.
a. Chord diagram representation of the dynamics of gene loss observed between each pair of taxa measured as the percentage of genes in each genome that were lost, as inferred for dataset 3 (phylome approach). Each seed phyla is represented by one color. Thicker chords between two given phyla represent higher percentages of gene loss. Directionality of chords is defined by each color (i.e., if a chord and a phylum share a color, the percentage of gene loss is calculated based on the genome size of that phylum). b. Percentage values of gene loss as inferred for dataset 3. Values are polarized with the genome size of each seed genome shown in the left-most column.
The role of gene duplication in the evolution of the metazoan core gene repertoire
We explored pathway conservation in the metazoan core gene repertoire (defined as the orthogroups gained at the branch leading to metazoa or more basal branches, present in at least 80% of the phyla, and in a minimum of 50% of phyla within Xenacoelomorpha, Deuterostomia, Lophotrochozoa and Ecdysozoa). This core repertoire consisted of 3,196 orthogroups, from which 2,479 yielded KEGG annotations (Fig. 5, Suppl. Mat. S9). Overall, half of the orthogroups in the metazoan core gene repertoire with KEGG annotations in the main KEGG categories represented (clustered in the overarching categories of metabolism, environmental information processing, genetic information processing, organismal systems and cellular processes) corresponded to genetic information processing, including transcription, translation, DNA replication and repair and folding, sorting and degradation (Fig. 5a). Duplication levels of genes in these categories for some animal phyla were similar to the ones in the outgroups. In contrast, the remaining categories showed a higher number of duplications in metazoans than in their unicellular outgroups, reinforcing the role of gene duplication and not merely presence or absence of genes, as a main driver of genome evolution across metazoans. Remarkably, genes involved in pathways related to development of organismal systems (i.e., excretory, nervous, circulatory and excretory systems, among others) showed higher levels of duplication in metazoans, underpinning again the link between gene duplication and morphological complexity. From all animal phyla, deuterostomes (and, within deuterostomes, Craniata in particular) exhibited the highest levels of gene duplication, pointing again to co-option of gene duplicates as a driving force increasing morphological complexity. In contrast, Porifera also exhibited high levels of gene duplication in multiple KEGG pathways, which indicates that probably regulatory complexity or other evolutionary processes are required for such an increase in morphological disparity. Finally, pathways that included genes with particularly high levels of duplication in metazoans were related to neural system development and signal transduction (Fig. 5c and d), indicating again - as shown above in the enrichment analyses - that the evolution of the neural systems involves ancient genes and seems to be strongly influenced by gene duplication across metazoans.
Conclusions
Altogether, our results provide support for a consistent pattern of gene repertoire evolution across the Animal Tree of Life, characterized by a bimodal distribution of gene gain and duplications with peaks at both the deepest and phylum-level nodes, and rampant gene loss in the branches leading to the most diverse clades. We show that waves of acquisition and loss of orthologous genes are coupled to different levels of gene duplication, with high gene gain rates associated to high duplication ratios and high levels of orthologous gene loss preferentially occurring at nodes where duplication ratios are low. These results underscore the key role of gene duplications but also gene loss in animal genome evolution. In other words, it is not the presence of key innovations what explains animal origins, but rather their complex evolutionary dynamics, providing an arena that potentially fueled an increase in cell type and tissue complexity throughout the animal kingdom. Our results therefore challenge the idea that the gene repertoire underlying the vast morphological disparity displayed by animals has become more complex through time (e.g.,10)12. Losses are compensated by gene gains and duplications in each lineage independently, and thus the size of the gene repertoire is not necessarily altered. Consequently, increase in morphological complexity must result from other evolutionary phenomena, such as co-option following gene duplication, integration of genes in functional modules or through the potential implication of non-coding sequences leading to an increase of genomic regulatory complexity (e.g., through the contribution of transposable elements in cis-regulatory elements13,14). All in all, our results point towards a highly complex evolutionary history of each individual gene and lineage. The correlation between morphological and genetic complexity across the Animal Tree of Life - epitomised by the early gain and continuous duplication of genes involved in the evolutionary development of the nervous system - is therefore strongly challenged. We emphasize that phylogenomic-centered studies are consequently most needed to further understand gene repertoire evolution in nonmodel organisms.
Methods
Taxon sampling
We included in our study 216 genomes and transcriptomes representing all animal phyla, with each phylum being represented by a number of taxa ranging from 1 to 22 (Suppl. Mat. S1). Special attention was paid to maximize lineage representation within each phylum by including representatives of the main clades below the level of phylum whenever possible. Furthermore, we included 15 outgroup species, comprising representatives of the clades Fungi, Nucleariidae, Ichthyophonida, Dermocystida, Corallochytrea, Filasterea and Choanoflagellata (Suppl. Mat. S1). Our dataset comprises a total of 231 genomes and transcriptomes considering ingroup and outgroup species.
Glossary and definitions
Gene gain: the branch leading to a clade were a gene was gained correspond to the branch leading to the last common ancestor (LCA) of all the orthologs of that gene (i.e. homologous genes derived from speciation event) present in our dataset. Thus, by definition that gene lacks orthologs outside the clade, and it has orthologs in at least another lineage within the clade. Note that the presence of subsequent gene duplications and gene losses of that gene family within the clade are possible.
Gene loss: we consider that a gene was lost in the branch leading to a given clade when that gene has no homologs within that clade, but that homologs of that gene are present in the closest sister lineage of that clade. Note that gene losses must be necessarily be evaluated from the perspective of genes present in other clades.
Duplication ratio: the total number of gene duplications mapped to a given branch in the species tree divided by the total number of genes.
Lineage-specific gene: A gene that does not have any detectable homologous genes outside its lineage, regardless of the taxonomic level of that lineage (e.g., phylum-specific refers to genes with no detectable homologous genes outside that phylum). Note that it is consequently conditioned to the taxon sampling of each study.
Inference of orthogroups and pairwise orthology relationships
Most comparative genomic studies on metazoa genome evolution so far have focused on gene family inference, i.e., the inference of homologous genes clustered in orthogroups (i.e., the set of genes from multiple species descended from a single gene in the last common ancestor of a set of species) (e.g., 1,2). While this approach can definitely be most helpful in understanding gross patterns of change in the gene repertoire (for instance, presence/absence of a gene family in a clade), it fails to provide a detailed scenario of how homologous genes are related, particularly if they arose through a speciation event (i.e., they are orthologous) or a duplication event (i.e., they are paralogous)15. For instance, this second approach would allow to understand not only the presence/absence of orthogroups in lineages, but also how (speciation or duplication), when (positioning the event in a node) and how often a certain gene has been gained, duplicated or lost in the context of a certain phylogeny. Furthermore, gene family inference is strongly biased by the methodological approach used, such as the inflation value in the MCL clustering step that will infer a higher number of families as we increase its value 16.
To overcome this methodological limitation, and with the goal of generating a compelling atlas of gene repertoire evolution across the Metazoa Tree of Life that allows us to understand how, when and how often each gene in each taxon was gained, duplicated or lost, we designed a two-step orthology inference protocol. First, we inferred orthogroups for the whole dataset (dataset 1 hereafter, n=231) with OrthoFinder v. 2.3.1 17. Since higher inflation values during MCL clustering can result in higher numbers of inferred gene families (i.e., an orthogroup that has undergone a high number of duplications will be split in several smaller orthogroups), we chose an inflation value of 1.5 as a conservative approach, as discussed in other studies (e.g., 2). The inference of orthogroups allowed us to compare our results with previous studies and, combined with the pairwise orthology inference as described below, to characterize which gene families had undergone higher rates of duplications, expansions or losses.
Second, after inferring orthogroups we inferred pairwise orthology relationships for each gene in each genome/transcriptome. Since orthogroups in this whole dataset (dataset 1) contained up to 24,178 sequences, we inferred gene trees with DendroBLAST 19 as implemented in the OrthoFinder pipeline to overcome computational constraints. A species-overlap algorithm, as implemented in ETE v3 20 was used to infer orthology and paralogy relationships from the phylogenetic trees reconstructed in the phylome. The algorithm traverses the tree and calls speciation or duplication events at internal nodes based on the presence of common species at both daughter partitions defined by the node. This algorithm is robust to a high degree of topological diversity observed in the individual gene trees; in contrast to any algorithm based on reconciliation with a specific species tree (that would inevitably infer false duplication events in the trees showing topologies that depart from the canonical species tree), the species-overlap algorithm does not require a fully resolved species phylogeny and a reconciliation phase 21. Gene gains and losses were calculated on this basis. Duplication ratios per node were calculated by dividing the number of duplications observed in each node by the total number of gene trees containing that node: theoretically, a value of 0 would indicate no duplication, a value of 1 an average of one duplication per gene in the genome, and >1 multiple duplications per gene and node. We used the species tree depicted in Fig. 1a, based on our current knowledge on animal phylogeny 6–8,22. Despite the robustness of the species-overlap algorithm, and due to the ongoing debate about the phylogenetic position of Porifera and Ctenophora, we repeated the approach constraining the tree topology to show the ‘Porifera-sister’ hypothesis. Since the results were virtually the same in all nodes under both topology constraints, we report only the results considering the ‘Ctenophora-sister’ hypothesis (see also Suppl. Mat.).
We detected expanded protein families with ETE v3 20. Only those nodes with more than 5 sequences per taxon were considered as expansions for that specific taxon. Overlapping expansions (i.e., partial gene trees with terminals in common) were fused when they shared more than 20% of their members.
With the goal of testing the robustness of this approach - particularly the effect of the inclusion of transcriptomes of potentially lower quality- we selected a subset of the best genome/transcriptome for each phylum (one per phylum) and seven outgroups (dataset 2 hereafter, n=43) and repeated the same analytical pipeline. Furthermore, and with the goal of testing our results now in the light of a much more accurate individual gene tree inference analytical procedure, we built a third dataset (dataset 3 hereafter, n=18) composed only of 15 genomes and 3 high quality transcriptomes of taxa from key lineages where sequenced genomes are not available (Xenoturbellida, Acoela and Chaetognatha, BUSCO completeness > 90%, only longest isoform). We constructed the phylome (i.e., the complete collection of individual gene trees for each genome) for each taxon using the PhylomeDB pipeline 23, which resulted in one of the most accurate orthology inference pipeline after standardized benchmarking using sets of reference gene trees (see Fig. 3 in 24), therefore minimizing the error due to inaccurate inference of individual gene trees. In brief, for each protein encoded in the first genome (referred to as seed hereafter), we performed a BLAST search against the custom proteome database built from all the 18 genomes and transcriptomes, that included a total of 397,671 proteins. Results were filtered using an e-value threshold of 1e-05 and a minimum overlapping region of 0.5 (i.e., minimum of 50% overlap between the query and the hit sequence). Multiple sequence alignments were reconstructed in both sequence orientations using three different programs: MUSCLE v3.8 25, MAFFT v6.712b 26, and Kalign 27. The resulting six alignments were then combined using M-COFFEE 28. A trimming step based on non-consistent regions in the consensus alignment was performed using trimAl v1.3 29(consistency-score cutoff 0.1667, gap-score cutoff 0.9). The best-fitting model was selected by reconstructing neighbor joining trees as implemented in BioNJ 30using seven different models (JTT, LG, WAG, Blosum62, MtREV, VT and Dayhoff). The best model in terms of likelihood as selected by the Akaike Information Criterion (AIC) was selected for tree reconstruction. Trees were reconstructed using PhyML 31. Four rate categories were used and invariant positions were inferred from the data. Branch support was computed using an aLRT (approximate likelihood ratio test) based on a chi-square distribution. The same procedure was repeated with the remaining 17 genomes and transcriptomes (i.e., all genomes and transcriptomes were treated as seed, one at a time for each phylome reconstruction). Resulting trees and alignments -a total of 260,000 considering all phylomes- are can be visualized in PhylomeDB 4.0 23,31 (http://phylomedb.org).
Similarity searches are not infallible. Any method holds a different compromise between different sources of errors and entails a balance between false positives and negatives (e.g., 32). The phylomeDB approach is not based on an all-by-all BLAST but it searches for homologues (and then infers orthology/paralogy relationships) only for one proteome at a time, the so-called seed. For instance, in our study we analyzed 18 different phylomes, meaning that we used each taxon as a seed in an independent phylome analysis. This has an important implication: since we are using the same species tree to polarize our results, the values of gene gain and loss for each node are informed by completely different homology searches (i.e., one per taxon). Therefore, this approach supports the high robustness of our results since the values of gene gain, duplication and loss are strongly similar for each node regardless of the seed taxon, as shown in Figs. 1 and 2 (see also Suppl. Mat. S3). This stands even if we compare the percentage of gene gain and loss per node inferred for fast-evolving lineages (such as nematodes, see phylome of Caenorhabditis elegans) with slow-evolving ones, such as Drosophila melanogaster. Moreover, in the case of duplications (including expansions), the phylomeDB pipeline applies a clustering approach that clusters together individual gene trees that share a minimum of 50% of the genes. This means that if a BLAST search fails to recover homologs of a given sequence based on length or sequence similarity, which could result in splitting a single gene family into two or more if we were using a program based on gene family inference, it will be corrected through this step. The OrthoFinder2 approach is likewise based in BLAST reciprocal best hits, the main difference with phylomeDB being that the former is based in an all-by-all approach (i.e., inference of gene families through a single BLAST search). From the orthology inference programs based on the inference of gene families, OrthoFinder2 has been shown to be among the best ones, as discussed in 17. We favoured OrthoFinder over the commonly-used software OrthoMCL 15 because despite using a similar inference pipeline (consisting of a BLAST search followed by MCL clustering), it has been shown that with the OrthoMCL algorithm short sequences suffer from low recall rate and long sequences suffer from low precision (that is, many short sequences fail to be assigned to an orthogroup while many long sequences are assigned to the incorrect orthogroup) 13.
Benchmarking studies also support the robustness of our chosen methodologies. PhylomeDB scored amongst the best methods to correctly retrieve highly-curated individual gene trees and their orthology/paralogy relationships in a thorough benchmarking study 24, with very high rates of precision (i.e., positive predicted value) and recall (i.e., true positive rate)(see Fig. 3). Likewise, the score of OrthoFinder2 was amongst the highest as benchmarked in 16,17(see Fig. 2 in the later).
While alternative methodologies to BLAST based on Smith-Waterman alignments and profile Hidden markov model searches have been proven to perform similarly to BLAST searches for phylostratigraphic analysis 32, methods based in the later continue to be one of the most adequate approaches consequently due to a good trade-off between computational time and error rate. Two parameters have been shown to particularly affect BLAST searches: evolutionary rate and length of sequence. For instance, it has been shown that higher error rates in relative age estimation are associated with higher evolutionary rates and shorter sequence lengths 33. In order to further test if our results were robust to these two factors, we selected four of the phylomes representing three of the main lineages in the Metazoa Tree of Life differing in their genome size (measured as number of proteins in their proteome) and overall evolutionary rate: Craniata as a representative of Deuterostomia, Mollusca as a representative of Lophotrochozoa, Arthropoda as a representative of a slow-evolving Ecdysozoa, and Nematoda as a representative of a fast-evolving Ecdysozoa. We took all multiple sequence alignment (MSAs) of proteins where homologs were detected following the phylomeDB pipeline and calculated two parameters. First, we calculated the average identity score as a proxy of evolutionary rate. The identity score for each possible pair of sequences in the alignment is the number of identical residues aligned between these two sequenced divided by the length of the longer sequence. We have calculated the average identity score (average IdSc hereafter) for each MSA with trimAL v1.2.129, flag -sident. The identity score ranges from 0 (highly dissimilar sequences) to 1 (highly similar sequences). Second, we calculated the average protein length in each MSA. All phylomes showed a highly similar distribution pattern (Suppl. Mat. 8). In order to test if our estimates of gene gain and loss per node were affected by these two parameters, for each phylome we divided all MSAs in four quartiles based on average IdSc and length of MSA. Next, for each quartile in each phylome we re-analysed orthology/paralogy inference and gene gain, duplication and loss patterns, totalling 32 new analyses for each phylome. We statistically compared the values of gene gain, duplication and loss between them and between the quartiles and the full set of MSA for each phylome (i.e., the original analysis) by means of one-way ANOVA. Especifically, we compared if the values of gene gain, duplication and loss were different between the five treatments (the full phylome and the four quartiles of each phylome) for both the average identity score and length of MSA. None of the ANOVA analyses showed significant differences between any of the treatments (p-value > 0.05; Suppl. Mat. 8). Therefore, we can conclude that after accounting for error and biases due to evolutionary rate and length of sequence, our results remained largely unaltered, which attest to the high robustness of our approach.
GO term annotation and enrichment analysis
To assign Gene Ontology (GO) terms to all genomes and transcriptomes in datasets 1 and 2, GO terms based on orthology relationship were propagated with eggNOG-mapper 34. For that, we selected the eukaryotic eggNOG database (euNOG 35) and prioritised coverage (i.e., GO terms were propagated if any type of orthologs to a gene in a genome were detected: one-to-one, one-to-many, many-to-one or many-to-many). Compared with InterProScan 36, eggNOG-mapper has been shown to achieve similar proteome coverage and precision while predicting on average more terms per protein and increasing the rate of experimentally validated terms recovered per protein by 35%. In addition, eggNOG-mapper predictions scored within the top-5 methods in the three GfatiGOO categories using the CAFA2 NK-partial benchmark 34. Moreover, we annotated the orthogroups inferred for datasets 1 and 2 to be able to link particular genes to gene families and characterize them functionally.
For dataset 3, GO terms were propagated based on orthology relationship considering the genomes in the PhylomeDB database. This database includes both manually- and automatically- curated genome annotations, therefore providing more accurate information about gene function than other databases. All orthologs were selected to propagate GO terms, thus prioritizing coverage as in the annotation with eggNOG-mapper. With the goal of understanding the degree of functional annotation in each genome or transcriptome, we calculated the percentage of genes annotated. In addition, we clustered together all genomes/transcriptomes per phyla and calculated the mean percentage of annotation to enable their comparison.
We next checked functional enrichment of genes gained at different nodes with FatiGO 37. For that, for each taxon we parsed the genes inferred as gained in each node and tested enrichment against the remaining genes in the genome of that taxon. This analysis was done independently for all taxa. Whenever enrichment was detected, sets of enriched GO terms were summarized and visualized in REVIGO38. The same pipeline was repeated with the gene expansions in each genome in our core data set to further understand GO terms enrichment in each phyla independently compared to the background (i.e., the sum of the non-expanded genes in each of the scrutinized genomes), and with genes lost in each node per taxon as well.
Metazoan core gene repertoire inference and annotation
To infer the metazoan core gene repertoire, we selected the orthogroups as inferred in OrthoFinder2 that (i) were present in at least 80% of all phyla, and (ii) were represented in at least 50% of the phyla of each main lineage (Xenacoelomorpha, Deuterostomia, Lophotrochozoa and Ecdysozoa). To further understand the pathway conservation of this core, KEGG pathway annotation and mapping was performed with KEGG Mapper and BlastKOALA 39,40. KEGG pathway annotation for 30 selected general categories was combined with the total number of duplication values (i.e., not referred to duplications in any specific node) for each annotated orthogroup in each category and visualized through heatmaps with the pheatmap R package.
Supplementary Material
Acknowledgements
We are really thankful to Marina Marcet-Houben and Irene Julca for multiple discussions that contributed to greatly improve this study.
Funding: RF was funded by a Juan de la Cierva-Incorporación Fellowship (Government of Spain) and a Marie Skłodowska-Curie Fellowship (747607). TG acknowledges support from the Spanish Ministry of Economy, Industry, and Competitiveness (MEIC) for the EMBL partnership, and grants ‘Centro de Excelencia Severo Ochoa 2013-2017’ SEV-2012-0208, and BFU2015-67107 cofunded by European Regional Development Fund (ERDF); from the CERCA Programme / Generalitat de Catalunya; from the Catalan Research Agency (AGAUR) SGR857, and grant from the European Union’s Horizon 2020 research and innovation programme under the grant agreement ERC-2016-724173 the Marie Sklodowska-Curie grant agreement No H2020-MSCA-ITN-2014-642095.
Footnotes
Author contributions: RF and TG developed the overall conceptual approach and analysis. RF compiled and analysed the data. RF and TG wrote the manuscript. TG supervised the study.
Competing interests: the authors declare no competing interests.
Data Availability
All data, code and supplemental information are available in the manuscript, the supplementary materials and the Harvard dataverse repository https://doi.org/10.7910/DVN/ZKDAE2. Accession numbers of all taxa included in each analysis is indicated in Suppl. Mat. S1. All phylomes can be accessed at PhylomeDB 4.0 under phylome numbers 431, 462, 747, 778, 782, 812, 819, 824, 875, 888, 937, 950 and 953 (i.e. http://www.phylomedb.org/phylome_431 for direct access).
References
- 1.Paps J, Holland PWH. Reconstruction of the ancestral metazoan genome reveals an increase in genomic novelty. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04136-5. 1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richter DJ, Fozouni P, Eisen MB, King N. Gene family innovation, conservation and loss on the animal stem lineage. eLife. 2018;7 doi: 10.7554/eLife.34226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domazet-Loso T, Brajković J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23:533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology. Nat Rev Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurent JM, et al. Humanization of yeast genes with multiple human orthologs reveals principles of functional divergence between paralogs. doi: 10.1101/668335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon JT, et al. Xenacoelomorpha is the sister group to Nephrozoa. Nature. 2016;530:89–93. doi: 10.1038/nature16520. [DOI] [PubMed] [Google Scholar]
- 7.Laumer CE, et al. Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias. Elife. 2018;7 doi: 10.7554/eLife.36278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen X-X, Hittinger CT, Rokas A. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol. 2017;1:126. doi: 10.1038/s41559-017-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Y-J, et al. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat Ecol Evol. 2018;2:141–151. doi: 10.1038/s41559-017-0389-y. [DOI] [PubMed] [Google Scholar]
- 10.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 11.Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. 2016;17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 12.Shen X-X, et al. Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell. 2018;175:1533–1545.e20. doi: 10.1016/j.cell.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebé-Pedrós A, et al. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat Ecol Evol. 2018;2:1176–1188. doi: 10.1038/s41559-018-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabaldón T. Large-scale assignment of orthology: back to phylogenetics? Genome Biol. 2008;9:235. doi: 10.1186/gb-2008-9-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emms DM, Kelly S. OrthoFinder2: fast and accurate phylogenomic orthology analysis from gene sequences. 2018 doi: 10.1101/466201. [DOI] [Google Scholar]
- 18.Li L. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Research. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly S, Maini PK. DendroBLAST: approximate phylogenetic trees in the absence of multiple sequence alignments. PLoS One. 2013;8:e58537. doi: 10.1371/journal.pone.0058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huerta-Cepas J, Dopazo J, Gabaldón T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldón T. The human phylome. Genome Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-6-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marlétaz F, Katja TC, Goto T, Satoh N, Rokhsar DS. A New Spiralian Phylogeny Places the Enigmatic Arrow Worms among Gnathiferans. Current Biology. 2019;29:312–318.e3. doi: 10.1016/j.cub.2018.11.042. [DOI] [PubMed] [Google Scholar]
- 23.Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014;42:D897–902. doi: 10.1093/nar/gkt1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altenhoff AM, et al. Standardized benchmarking in the quest for orthologs. Nat Methods. 2016;13:425–430. doi: 10.1038/nmeth.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Standley DM. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- 27.Lassmann T, Sonnhammer ELL. Kalign, Kalignvu and Mumsa: web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–9. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace IM, O’Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 31.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 32.Moyers BA, Zhang J. Toward Reducing Phylostratigraphic Errors and Biases. Genome Biol Evol. 2018;10:2037–2048. doi: 10.1093/gbe/evy161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moyers B, Zhang J. Phylostratigraphic Bias Creates Spurious Patterns of Genome Evolution. Molecular Biology and Evolution. 2016;33:3031–3031. doi: 10.1093/molbev/msw202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huerta-Cepas J, et al. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Molecular Biology and Evolution. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huerta-Cepas J, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Research. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 38.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M. Enzyme Annotation and Metabolic Reconstruction Using KEGG. Methods Mol Biol. 2017;1611:135–145. doi: 10.1007/978-1-4939-7015-5_11. [DOI] [PubMed] [Google Scholar]
- 40.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.