Abstract
Artificial Intelligence and Nanotechnology are two fields that have been instrumental in realizing the goal of Precision Medicine – tailoring the best treatment for each cancer patient. Recent conversion between these two fields is enabling better patient data acquisition and improved design of nano-materials for precision cancer medicine. Diagnostic nanomaterials are used to assemble a patient-specific disease profile, which is then leveraged, through a set of therapeutic nanotechnologies, to improve the treatment outcome. However, high intra-tumor and inter-patient heterogeneities make the rational design of diagnostic and therapeutic platforms, and analysis of their output, extremely difficult. Integration of AI approaches can bridge this gap, using pattern analysis and classification algorithms for improved diagnostic and therapeutic accuracy. Nanomedicine design also benefits from the application of AI, by optimizing material properties according to predicted interactions with the target drug, biological fluids, immune system, vasculature and cell membranes, all affecting therapeutic efficacy.
Here, fundamental concepts in AI are described and the contributions and promise of nanotechnology coupled with AI to the future of precision cancer medicine is reviewed.
Keywords: nanotechnology, artificial intelligence, big-data, cancer, precision medicine
1. Introduction
Every patient is unique. Alongside our apparent differences, such as age, gender, height, eye color and blood type, we also have unique molecular signatures. This leads to different phenotypic changes and varied drug-responses among patients.[1] Diversity among patients is especially apparent in different types of cancer, which are affected by accumulation of driver mutations leading to intra-tumor and inter-patient heterogeneities, complicating diagnosis and treatment.[2] Precision medicine aims to tailor a specific treatment regime to each patient by accounting for multiple genetic and epigenetic characteristics.[3]
Nanomaterials have contributed to the evolution of precision medicine, throughout all of the medical stages. New omics collection technologies, such as single-molecule nanopore sequencing, enable fast and sensitive single-molecule detection along with longer sequence read length, thus maintaining genetic context.[4] Diagnostic assays based on nanosensors allow biomarker detection in femtomolar concentrations as well as scanning for multiple disease biomarkers simultaneously in liquid biopsies (blood, urine, saliva) and in cell cultures.[5] Nanomedicine-based cancer treatments have been evolving over the past decades, from a population wide treatment approach, aimed primarily at improving efficacy and reducing side effects, to targeted systems that report about drug activity inside the patient’s body.
Advances in nanomedicine fabrication techniques coupled with increased understanding of cancer biology, promoted the rational design of targeted therapy approaches utilizing endogenous and external stimuli for improved drug delivery. These advancements also supported the development of theranostic nanomedicines that combine a drug and an imaging agent to further analyze the treatment efficacy inside the patient’s body.[6] Nevertheless, current nanosensors and targeted nanomedicine have had limited success in clinical translation in the field of cancer.[7] Artificial intelligence (AI, see Box 1) is a branch of computer science that relates to machines that perform tasks that require “human intelligence”. Machine learning (ML, see Box 1), an area in AI, is an approach that trains an algorithm using large datasets of previous examples. It is applied in order to, inter alia, find patterns and classify data or find an optimal solution to a presented problem. Machine learning and AI in general have been used in different fields of medicine including medical imaging and analysis of gene expression patterns.[8] In nanoinformatics, AI and other computational methods are applied for nanomaterial design and implementation.[9]
Box 1. Basic Terms in Artificial Intelligence, Machine learning and Computational Models.
Artificial intelligence (AI) –
The ability of machines to execute tasks that require “human intelligence”, such as problem solving and learning. For example, an algorithm that learns to distinguish between healthy and diseased individuals.
Machine learning (ML) –
An area in AI based on construction of algorithms supplied with large datasets that are used as an input for training and improving the algorithm's output results. ML is used for numerous applications including decision making, classification and pattern recognition problems.
Supervised and Unsupervised Learning –
Supervised learning is a machine learning task in which the training data is already labeled with the required output and the algorithm modifies its variables in order to optimize the obtained results from the data as requested by the user. In unsupervised learning, the data is classified without prior labeling and categorization according to patterns discovered by the algorithm.
Artificial Neural Networks (ANN) –
An ANN is a framework of connected layers of nodes that can be used for implementing machine learning algorithms. It is composed of an input layer, an output layer, and usually also contains hidden layers. A node can receive input from multiple connections from the preceding layer, each assigned with a specific weight that is considered when calculating the node's output. The network is trained to optimize the weights of each node-to-node connection for achieving increased accuracy of the output.
Support Vector Machine (SVM) –
SVM is a machine learning algorithm that is trained to classify data by constructing an n-dimensional space according to the input features and optimizing the separation of different groups across this space.
Decision Tree Learning –
A decision tree is a method for data classification and regression that is based on constructing a tree-like structure that performs sequential tests on selected features.
Random Forest Classifier –
A Random Forest Classifier is built from a combination of decision trees. The randomness of this approach is due to a selection of the feature for testing from a random subset of the total features during the tree construction process, which leads to variant decision trees that comprise the forest.
Principal Component Analysis (PCA) –
PCA is a method that receives a dataset of examples with multiple features and uses linear combinations in order to generate a smaller number of new features called principal components (PCs). These PCs are arranged from top to bottom according to their variance, and therefore by using the top PCs the data can be classified and presented in lower dimensionality.
Feature Selection –
Feature selection is used in order to reduce the complexity of the problem by detecting the important features that contribute most to the results.
Levenberg–Marquardt (LM) Algorithm –
The LM algorithm is a fitting algorithm that is used for non-linear problems in an iterative process in order to minimize errors. It can be used in the training process of machine learning models.
Metropolis Monte Carlo Algorithm –
The Metropolis Monte Carlo Algorithm is used for randomly generating a set of configurations for an investigated system and calculate their probability distribution.
Iterative Stochastic Elimination (ISE) –
ISE is an algorithm for complex problem solving that uses stepwise variable scoring and rejection of the variables that lead to the worst results after each step, thus simplifying the problem.
The ability of AI algorithms to process large datasets and recognize complex patterns can be exploited for improved design of nanotechnologies for diagnostics and treatment. Prediction of nanoparticle interactions with the target drug, biological media and cell membranes, in addition to drug encapsulation efficiency and release kinetics can help optimize nanomedicine formulations.[10] Moreover, pattern recognition and classification algorithms can be used in order to differentiate between healthy and diseased patients and predict drug efficacy in patients.[11, 12] These analysis capabilities are especially essential in the case of cancer, due to its high complexity.
In this review we discuss the implementation of nanomaterials to the development of omics, diagnostics and treatment technologies for precision cancer medicine, emphasizing the contribution of AI for precision medicine data analysis and nanomedicine design.
2. Nanomaterials and AI in Precision Diagnostics
2.1. Expanding the Omics Toolkit
An essential requirement for successfully practicing precision medicine is assembling a molecular profile for each patient. This includes disease-relevant biomarkers that will provide a roadmap for a personalized treatment plan. A disease profile based on omics data usually comprises: genomic, epigenomic, transcriptomic, proteomic, metabolomic and microbiomic data, all compiled into a distinct molecular signature of the patient.[13] The recognition process of relevant disease biomarkers and their distribution and variance among different patients has evolved greatly with the development of big data analysis of population-wide omics. For example, classification of healthy individuals and patients with localized and metastatic tumors was performed by generating an RNA-based molecular signature that distinguished between the populations.[14] The molecular signature was obtained using RNA sequencing of tumor educated platelets (TEPs, blood platelets with altered RNA profile affected by platelet-tumor interactions) and provided accurate localization of the primary tumor in more than 70% of the cases. Rapid, accurate and affordable data collection tools are essential for collecting large amounts of data from a diverse population of patients and efficiently recognizing new biomarkers.
Nanotechnology improves the speed and precision of sequencing technologies used for omics data collection. In particular, third-generation sequencing methods, such as single-molecule real-time (SMRT) sequencing and nanopore sequencing, allow direct analysis of single DNA molecules without a need to amplify the template, thereby minimizing reading errors.[15] The SMRT system, is based on 60-100 nm cavities prepared using electron beam lithography on a thin aluminum 100-nm sheet deposited on a silica substrate.[16] These sub-wavelength cavities, each containing a single DNA polymerase, are then utilized as a confined observation volume for optical monitoring of the addition of fluorescent nucleotides to a complement strand of the target DNA.[17] In addition to obtaining real time data, another major advantage of this technology is its ability to sequence long reads, maintaining the genetic context and overcoming challenges in sequencing repetitive genetic elements.[18] Moreover, identification methods of DNA methylations and lesions based on SMRT sequencing provide epigenomic data for obtaining additional biomarkers for cancer and other malignancies.[19]
Nanopores present a different single-molecule method for DNA and RNA sequencing based on measuring the changes in ionic current as a DNA strand is translocated across a lipid membrane.[20] An important advantage of nanopore sequencing is that it does not require nucleotide labeling and extensive sample preparation, in addition to its long read length capabilities. Furthermore, recent portable and simple hand-held platforms expanded the use of nanopore sequencing to remote locations and enabled rapid identification of biological targets such as the Ebola virus.[21]
The first nanopore technologies were based on protein nanopores from bacterial origin, such as α-hemolysin, that enabled translocation of single-stranded DNA under an applied voltage and detection of current changes as the nucleic acid passed through the nanopore constriction.[22] Yet, achieving single nucleotide resolution with protein-based nanopore sequencing is challenging since multiple nucleotides are localized in the nanopore at a given time point, all contributing to the measured ionic current. Solid-state nanopores present a possible alternative to protein-based nanopores. These nanopores, made of a variety of materials, including SiO2, graphene, Boron Nitride and MoS2, demonstrate higher stability and controlled pore size.[23] Ion beam sculpting, electron-beam lithography and chemical vapor deposition have all been used for solid-state nanopore fabrication, achieving diverse nanopore geometries and controlled diameters (as small as 1 nm) that expand the range of applications from DNA sequencing to protein and DNA-protein complex sensing.[24] Nevertheless, single nucleotide resolution in solid-state nanopores still remains a challenge. Even in membranes with single-atom thickness (such as graphene) that contain a single nucleotide in the pore, the applied electrical field-effect extends also to nucleotides that are located near the nanopore surface affecting the measured current.[25]
Therefore, AI is applied in the translation process of the nanopore raw signal to a nucleotide sequence. Artificial Neural Networks (ANNs, see Box 1 and Figure 1) are a set of algorithms often used in nanopore sequencing. In general, an ANN is composed of layers of connected nodes. Using training data with known output (known oligonucleotide sequences in the case of nanopores), the weight of each connection is altered according to its effect on the output in order to obtain optimal results. In current nanopore sequencing algorithms the accuracy rate reaches 90%, and therefore further post-sequencing computational analysis is required for read correction. These additional algorithms generate consensus sequences from multiple reads and exploit them to increase the sequencing accuracy level to 97% and higher, depending on the DNA coverage level.[26]
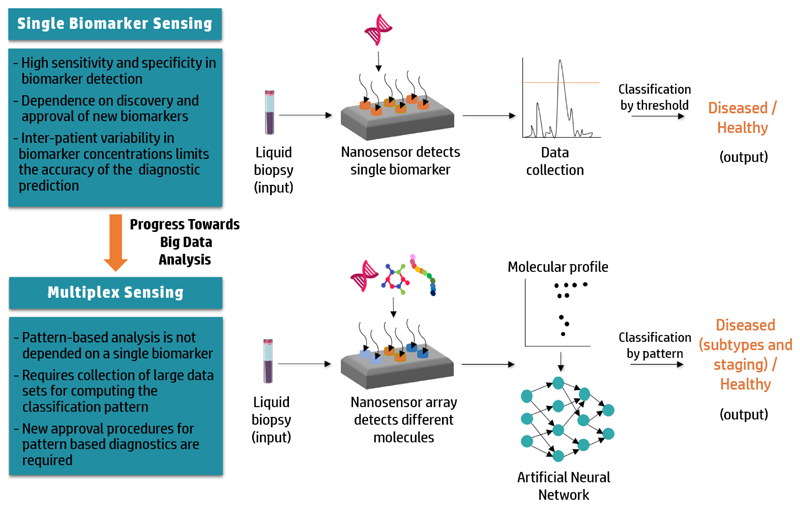
Figure 1.
Advancing from single biomarker sensing to multiplex sensing. Diagnostic screening of patient-derived liquid biopsies with single biomarkers sensors demonstrates high sensitivity and specificity, but is limited by inter-patient heterogeneity in biomarker expression and the low number of approved single biomarkers. Integration of AI in the data analysis of multiplex nanosensors that can detect a number of target molecules enables identification of disease-specific biomarker patterns. These patterns can be used for patient screening, overcoming the variability in biomarker expression.
Quantum sequencing aims to overcome the single nucleotide resolution challenge by integrating nanoscale electrodes perpendicularly to a nanopore, and measuring the electron tunneling at a single nucleotide resolution.[27] Each nucleotide has a unique electronic signature that can be measured by the changes in the electron tunneling current between these electrodes as the nucleotide passes through it.[28] However, the scalability of the solid-state nanopores fabrication techniques with integrated electrodes still needs to be improved in order to fulfill the promise of this approach. Quantum sequencing also exploits machine learning algorithms for raw signal interpretation. For example, first steps in amino acid identification and peptide sequencing were performed with quantum sequencing, implementing a support vector machine (SVM) algorithm (see Box 1) for recognizing the amino acid sequence.[29] The SVM algorithm is trained to classify signals of known amino acids according to the signal features, and use this training to classify new signals of unknown amino acid sequences.
Combining machine learning algorithms with feature selection (see Box 1) can be useful for detecting disease specific signatures in omics data.[30] Feature selection aims to recognize relevant features in a selected target from a larger pool of features. For example, a classification algorithm analyzed nearly 10,000 genes from two-hundred prostate cancer patients, recognizing 50 of them to be related to metastatic prostate cancer.[31] The selected features can then be utilized as biomarker signature criteria in a machine learning algorithm for patient classification and diagnostics. Moreover, current efforts in combining omics data from multiple sources (transcriptomics, proteomics, metabolomics) in order to create integrated signatures, will further improve the accuracy of biomarker signatures and patient’ diagnoses.[32]
2.2. Nano-sensors compose the patient's biomarker profile
Nanosensors are designed to detect a target analyte through measureable optical, electrical, magnetic or mechanical signals (Figure 1). A variety of nanomaterials for sensing applications, including gold nanoparticles, quantum dots, carbon nanotubes and nanowires, have been studied.[33] We focus here on the application of these sensors for composing a diagnostic profile for cancer patients in order to achieve early disease detection, personalize the patient's treatment plan and monitor the disease progression.
The intrinsic spectral, mechanical and electrochemical properties of nanosensors enable an increased signal-to-noise ratio output, making them preferable for cancer biomarker sensing. Effective nanosensors must be able to detect extremely low concentrations of the molecular signature in an environment of other interfering biological components. For example, the serum levels of prostate specific antigen (PSA), one of the few FDA-approved biomarkers, are elevated in prostate cancer patients from 0.5-2 ng/ml in healthy individuals to above 4 ng/ml, and need to be detected in the serum environment that contains the abundant human serum albumin in a concentration range of approximately 35-50 g/L, a thirty-million times greater concentration.[34] In vitro diagnostic platforms are rationally-designed to identify biological biomarkers (DNA, RNA, proteins and metabolites) in liquid biopsies taken from blood, urine, sweat, saliva and breath by conjugating a ligand that can bind the target molecule to a designed nanomaterial.[35, 36] The high sensitivity of these platforms, reaching femtomolar concentrations, is essential for early detection of cancer biomarkers that exist at early stages of the disease.[37] For example, a streptavidin-conjugated quantum-dot nanosensor, enabled distinguishing between the wild type KRAS gene and a cancerous KRAS with a single point mutation point by generating a fluorescent signal.[38] The sensor is comprised of DNA strands that emit a distinct fluorescence resonance energy transfer (FRET) signal upon hybridization with the mutated gene. Such sensors can be modified to detect almost any mutant DNA or RNA. In another study, a hybrid mechanical and opto-plasmonic nanosensor based on a sandwich assay was used for serum-based protein detection, including the detection of the FDA-approved biomarker carcinoembryonic antigen (CEA) that is used as a diagnostic marker of colorectal cancer.[35] This sensor captures the required protein using two antibodies for two different epitopes on the target. The first antibody is anchored to the surface of the sensor and the second is conjugated to gold nanoparticles that supply the plasmonic output which is subsequently analyzed for protein quantification with a 1x10-16 g/ml sensitivity.
Improving the sensitivity and portability of these sensors will makes them attractive point-of-care (POC) devices for diagnosing and monitoring cancer patients. Clinically-approved devices are already used for isolating circulating tumor cells (CTCs) from blood using magnetic beads functionalized with antibodies against the epithelial cell adhesion molecule (EpCAM).[39] Yet large-scale fabrication issues, slow capture rate of the target molecules and lack of defined validation and analytical testing hinder the translation of point-of-care nanosensors to the clinic.[40] Moreover, the current number of clinically approved biomarkers must be expanded, as the specificity of cancer prognosis by a single biomarker is not sufficient.[41] For example, CA125, a clinically-approved protein biomarker for ovarian cancer is elevated in only 50-60% of the patients with stage I disease, rendering the classification of additional biomarkers to improve false positive detections.[42]
Multiplex sensing of a number of biomarkers for constructing a disease signature based on computational analysis can increase the accuracy of diagnosis. For example, the clinically approved OVA1 test uses five biomarkers for assessing the likelihood of a diagnosed ovarian adnexal mass to be malignant.[43] This test uses a designated algorithm which calculates an integrated score on a scale of 0.0 to 10.0 based on five separate immunoassays performed for each of the biomarkers to indicate the risk for malignancy. Multiplex nanosensor arrays can detect a number of target biomarkers in a single test, saving time and providing with high sensitivity and selectivity. Electrical detection of three prostate cancer biomarkers in non-diluted serum samples was performed using silicon-nanowires synthesized by chemical vapor deposition.[44] The nanowires were deposited on a microfluidic chip, and controlled clusters of nanowires were each functionalized with a specific antibody to one of the target biomarkers. The changes in conductance due to protein-antibody binding were monitored and translated into biomarker concentrations. Similar multiplex sensing was also performed in quantum-dot-based fluorescent sensors and gold nanorod-based localized surface plasmon resonance (LSPR) sensors, the latter enabling detection of 6 cytokine biomarkers in approximately 40 minutes.[45] The development of multiplex nanosensors is promoting the transition from single biomarker detection to multiple biomarker signatures.[46] Combining multiple variables for the calculated diagnostic output reduces the error rates and can be used not only for cancer detection, but also for cancer staging.[47]
“Electronic nose” nanosensors are highly adequate for multiplex analysis of a large number of analytes. The electronic nose was designed to mimic the olfactory system and utilizes an array of selected sensors that can detect and quantify complex mixtures of volatile organic compounds (VOCs) in the gaseous phase.[48] Different types of electronic nose nanosensors detected and analyzed specific VOC patterns in the breath of cancer patients.[11, 49] One of the main advantages of electronic nose nanosensors is that they do not depend on specific aptamers or antibodies for recognition of a specific entity. Instead, each one of the detected VOCs generates a unique pattern when reacting with the different sensors in the nanosensor array, making them suitable for a wide range of targets. For example, a nanosensor array based on multiple 5-nm gold nanoparticles functionalized with different ligands, was able to distinguish between healthy and diseased breath samples in cancer patients diagnosed with breast, lung, colorectal and prostate cancer.[50]
Computational analysis plays an important role in realizing the shift from rationally-designed sensors targeting specific biomarkers to pattern-based nanosensors, which require advanced methods for clustering and classification of large datasets.[51] Principal component analysis (PCA) (see Box 1) is a commonly used method for electronic nose data analysis that uses linear combinations of the input data features to create a smaller number of new variables that exhibit high variance. These are then used to classify the data to distinct groups.[52] One shortage of this method is that it only relates to linear combination and therefore non-linear relations between the variables will not be preserved. AI methods, such as Neural Network models, allow to maintain non-linear correlations between the variables and increase the accuracy of data classification in electronic-nose applications.[53] For example, Kermani, et al. used a Levenberg–Marquardt (LM) algorithm (see Box 1), which is used to solve problems with non-linear correlations, as a training method of a Neural Network for fragrance classification.[54] The implementation of nanosensors arrays for biomarker-free, quick and accurate cancer diagnostics and staging seems promising, yet one issue that needs to be addressed is the requirement for large data collection from a wide variety of populations. This is especially essential for pattern recognition in nanosensor arrays that require the clustering algorithm to take into account the inherent variabilities between populations.
In addition to the efforts performed in developing efficient nanosensors for markers in liquid biopsies, analysis of the tumor tissue remains imperative for the diagnostic process. Therefore, many of the sensing technologies are being modified for quick detection of biomarkers in cell culture. In case the target analyte is found inside the cell, a method for intracellular penetration is required. For example, plasmonic gold nanoparticles functionalized with a fusogenic transduction peptide and a targeting antibody were able to the access the cytoplasm of NIH3T3 fibroblasts and bind to their target protein.[55] For detection of oligonucleotides, fluorescent nanoprobes known as molecular beacons, can detect specific sequences with high accuracy.[56] These probes are based on a DNA hairpin conjugated to a fluorescent entity and a quencher that are released upon binding to the target sequence, exposing the fluorescent signal.[57] Moreover, nanosensors that are based on pattern recognition, rather than biomarker targeting, were developed for cell culture analysis as well. Electronic noses that sense the headspace of cell cultures and arrays of fluorescently-labeled nanoparticles with the ability to specifically identify cancer stem cells in a tumor-derived cell culture are examples of these kind of sensors.[58]
3. Using Nanotechnology to Predict Personalized Drug Potency
Despite advances in the analysis of personalized omics data, it is still difficult to predict a patient's response to medication based on omics alone. This difficulty is specifically apparent in the field of cancer, where predicting the tumor's response must account for tumor/metastasis heterogeneity and the development of resistance over time.[59] Testing a candidate drug inside the patient's body to gauge its response can be used to optimize the treatment course.
Nanotechnology offers the means to perform such in situ diagnostics. Barcoded liposomes, each encapsulating a drug and a specific DNA barcode, can be used in order to predict the drugs' potency in the tumor of a specific patient.[60] For example, liposomes containing four potential drugs (gemcitabine, cisplatin, doxorubicin, caffeine), each marked with a unique DNA barcode, were injected to murine breast cancer models in doses lower than 0.1% of the therapeutic drug dose. After 24 hours, a biopsy was taken from the tumor and the barcode distribution between live and dead tumor cells was analyzed. Barcodes matching the drugs with high potency were found in higher concentrations in dead tumor cells and in low levels in live cells. Currently, the data analysis methods used in this approach are based on direct counts of the barcodes and their delivered percentage in live and dead cells. Integration of AI algorithms in the data analysis process can extend its applicability and allow recognition of combinatorial treatment effects, taking into account complex patterns of barcode distribution.
Moreover, the use of computational methods for in silico drug screening as a preceding step can further improve the results of the in situ nano-based screening (Figure 2).[61] For example, a study performed on tumor samples from 48 individuals identified mutational cancer drivers and classified them according to their mode of action, using a random forest classifier learning-based algorithm (see Box 1).[12, 62] This algorithm is based on a combination of decision trees that test selected features after training the algorithm on previous data. Then, a dataset of available drugs which are able to target these cancer drivers was constructed based on drug-target interactions, followed by automatic fitting of therapeutic agents to the mutational landscape of each patient. Despite the limitations in this method, including possible inaccuracies due to errors in mutation classification, disregard of the combinatorial effect of drugs and intra-tumor heterogeneity, this computational approach offers a unique point of view on possible therapeutic strategies.
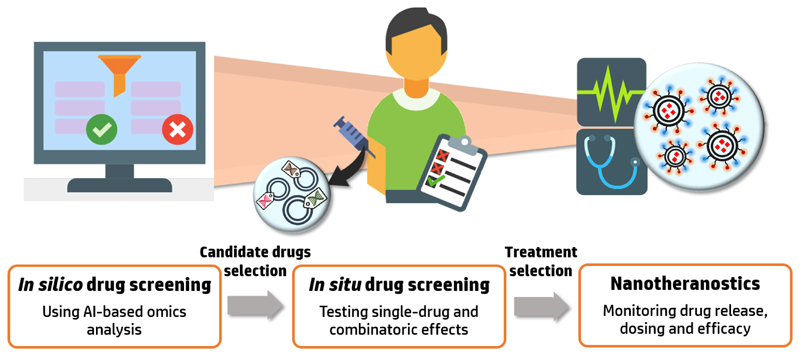
Figure 2.
Exploiting AI and nanomedicine for tailoring a patient-specific treatment regime. Initial drug screening with computational methods based on the patient's specific omics profile will provide a list of drugs with therapeutic potential. These drugs can then be tested in situ with nanoparticle-based technologies in order to select the optimal treatment regime. Applying nanotheranostic methods combining the nanomedicine with an imaging agent will allow to tune the treatment protocol by monitoring the drug's pharmacokinetics and release in the target site.
4. Computation in Nanotherapeutics - Targeting and Personalized Dosing
4.1. Designing Nanoparticles for improved targeting
One of the first ideas that come to mind when discussing precision cancer therapeutics is using targeted drugs that will recognize and activate only in the disease target site, not harming healthy tissues. This concept was first introduced by Paul Erlich in the early 1900's as the “Magic Bullet” theory and has been widely discussed since.[63] Nanomedicines provide a tool for personalized targeting by coating the surface of drug-loaded nanoparticles with specific ligands, such as antibodies, membrane-bound receptor ligands, and other cellular markers that enable to selectively bind to the target cells once the particles are in the vicinity of the target tissue.[64] A parallel approach suggests using synthetic drug delivery systems with biological properties – namely, biomimetic Trojan systems that have the outer appearance of a natural cell or extra-cellular vesicle, that are loaded with drugs.[65] These systems can take advantage of the natural properties of the mimicked cells and reduce undesired reactions of nanoparticles in the plasma such as uptake by the reticuloendothelial system (RES). For example, nanoparticles derived from the membrane of mesenchymal stem cells (MSCs) loaded with a plasmid DNA encoding for the cancer inhibitory protein hemopexin-like domain (PEX), retained the surface characteristics of the MSCs and displayed tumor-targeting abilities in murine models of lung and prostate cancers.[66]
External stimuli can also be used to drive nanoparticles towards each patient’s disease site or trigger their activation in a selected location.[67] Heat, magnetic fields, ultrasonic irradiation and light have been used to trigger drug release from nanoparticles at the disease site.[68] Endogenous patient biomarkers expressed in the diseased tissue are utilized as cues for triggering drug release onsite as well. The acidic environment in solid tumors has been used to reduce the inter-molecular bond strengths in nanomedicines, thereby allowing loaded drugs to diffuse rapidly out of the particle at the target site.[69] Similarly, the altered expression of extracellular enzymes has been exploited to cleave molecular bonds in nanoparticles in order to release encapsulated drugs.[70] Yet the implementation of targeted nanomedicine in the clinic hasn't come of age so far, with few formulations currently in clinical studies.[71] A number of recently published reviews have discussed in detail many of the challenges that have to be overcome in order to successfully translate targeting to the clinic.[72] We want to highlight two important aspects that are highly relevant for the success of this approach for precision medicine. The first aspect is the significance of accurate biomarker profiling of the patient's tumor, based on omics and nanosensor-based techniques, before administrating a targeted drug. This is a critical issue in order to find the right population of patients that fit the targeted drug, and was evident from the clinical study stages of different targeted therapies. For example, the efficacy of cetuximab and panitumumab, monoclonal antibodies targeted to the epidermal growth factor receptor (EGFR), was shown to be associated not only with EGFR expression levels, but also to KRAS-associated resistance, EGFR ligands mRNA levels and EGFR downstream pathway genes.[73] Omics-based classification of the responsive and irresponsive patients to the drug, followed by nanosensor-based patient pre-screening will increase the response rate and improve patient selection. This again highlights the potential that biomarker signature profiles generated with AI-based algorithms hold.
Integrating computational modeling into the nanoparticle design stage is another important aspect in increasing the success rate of targeted therapies. It is now clear that simply tagging drug-loaded nanoparticles with a targeting moiety does not promise successful delivery and release at the diseased site. The effect of the nanomedicine properties on the interactions with plasma, the vasculature endothelium and cellular membranes is not easily rationalized and can be significantly improved with computational methods. For example, an optimal antibody surface coverage of nanoparticles for specific vascular endothelium binding was computed in a computational model that calculated the binding free energy of nanoparticles using a Metropolis Monte Carlo algorithm (see Box 1).[74] The process initiates from a randomly selected state of the nanoparticle and ligand system, and by random changes of the defined variables it generates a probability distribution of the system's configurations. The effect of the nanoparticle's antibody surface coverage on the binding energy of the system was obtained by changing the former parameter in the computational model and simulating the changes in the system's states. The obtained results, displaying optimal targeting at a coverage ratio of above 100 antibodies per nanoparticle, were in agreement with experimental data obtained from cell culture and in vivo murine models performed in that study.
Moreover, computational models are imperative for predicting the ability of nanoparticles to cross barriers on their way to the target organ. Several model types have been used for predicting the ability of nanoparticles to permeate across the blood-brain barrier (BBB) and their potential toxicity.[75] These models can be implemented for improving the formulations of brain targeting nanoparticles. Yet the construction of these models is exceptionally difficult due to the high complexity of the permeation process; therefore, it requires large computation capabilities and understanding of the principles governing the biological and physical mechanisms. Recently, a machine learning approach for blood-brain permeability prediction was presented based on the drugs' indications, side effects and chemical properties.[76]
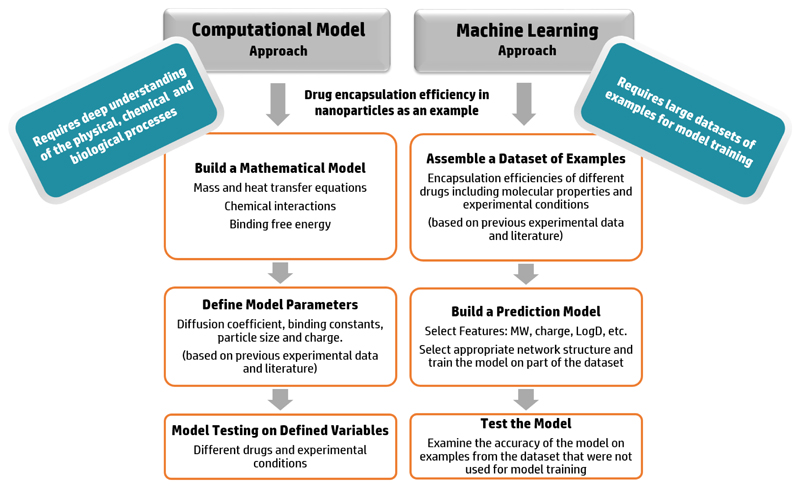
It is important to note that the design of machine learning methods requires different types of data than traditional computational models (Figure 3). In the latter, extensive physical, chemical and biological knowledge is necessary for constructing a computational model that accounts for the governing mechanisms in the process. However, in machine learning methods prior understanding of the process is not a prerequisite; these methods utilize large datasets of experimental results related to the investigated subject and detect correlations in the data which are translated into a prediction model. Nevertheless, understanding of the data can provide additional insights when choosing the machine learning model and algorithms in order to achieve maximal accuracy. The type of the data (text, numbers, images, etc.), mathematical behavior of the involved mechanisms and dependent relationships between data features are examples of variables that can be taken into consideration when designing a machine learning model. For example, DNA sequences were converted to 2-dimensional images and used in Neural Networks optimized for image analysis to predict the DNA chromatin structure.[77] Every DNA sequence was divided into overlapping 4-codon oligonucleotides that were converted into a pixel in the image. The conversion of the data from text to image facilitated the use of machine learning models optimal for image processing and improved the obtained prediction accuracy.
Figure 3.
Differences between computational models and machine learning algorithms: prediction of drug encapsulation as an example. Computational models depend on a devised mathematical model for simulation of the physio-chemical process and therefore prior physical, chemical and biological knowledge of the mechanisms is essential. Machine learning algorithms on the other hand, are based on training on large datasets of previous examples and detecting key features and correlations in the data for increasing the prediction accuracy.
Other computational methods considered the optimal shape and size for nanoparticle binding and drug release.[78] For example, an Artificial Neural Network (ANN) was used for predicting the size and initial burst rates of poly(lactic-co-glycolic acid) (PLGA) nanoparticles.[79] The ANN received an input of the PLGA molecular weight and concentration, in addition to the poly(vinyl alcohol) (PVA) molecular weight and the sonication rate which were used in the preparation process, and returned the predicted nanoparticle size and initial burst rate. Although the algorithm provided results with less than 5% error rate when tested on the provided data in the study, a major challenge in this computational approach is the incorporation of a large training set that is required for achieving increased accuracy. Harnessing high throughput screening methods for obtaining large datasets in a relatively short amount of time can help address the data acquisition challenge.[80] Yamankurt, et al. developed a high throughput screening protocol to test the activity of almost 1000 nano-formulations of spherical nucleic acids (SNAs) and used the obtained results to train a machine learning algorithm to predict the activity of new SNA formulations.[81] They furtherer showed that reducing the size of the dataset to approximately 150 examples of formulations provided similar accuracy rates for activity prediction.
4.2. Nanomaterials and AI for correlating drug dosing and therapeutic efficacy
Nanomaterials can provide controlled drug dosing by fitting the drug release rate to the specific pharmacokinetic and pharmacodynamics profiles of the patient. In a similar manner to the targeting approach, external stimuli are exploited for this purpose. Porous scaffolds that can release the target agent on-demand present an attractive approach for controlled release and dosing. For example, a macroporous ferrogel based on alginate patterned with Fe3O4 nanoparticles displayed reversible deformation under an external magnetic field enabling step-wise release of different moieties ranging from low molecular weight drug Mitoxantrone (type II topoisomerase inhibitor), to plasmid DNA and even whole cells.[82] Electric field and ultrasound were also used in in vitro and in vivo animal models for achieving pulsatile release in various nanomaterials including electro-sensitive polymeric nanoporous membranes and mesoporous silica nanoparticles.[83] However, in order to improve personalized dosing technologies, the pulsatile release systems must be coupled to real-time sensing technologies that will monitor the drug levels in the plasma or in the target site in a similar manner to the operation of insulin pumps. This will introduce challenges in fabricating nanosensing technologies that are also stable for prolonged time periods.
Dosing control is not always sufficient in order to personalize the treatment, as patients with different pharmacogenomic profiles respond differently to varying drug doses. In these cases, AI can be exploited in order to correlate between drug dosing and the therapeutic outcome. For example, Artificial Neural Networks were developed for constructing tailored radiotherapy treatment plans for cancer patients according to the radiation's physical specifications, the treatment goal and the patients' physiological and anatomical parameters.[84] These methods are currently being adapted to predict drug-response relationships for chemical drugs as well, based on drug properties, physiological measurements and gene expression profiles.[85] Other AI models link directly between the patient's condition and a suggested treatment to predict the treatment efficacy without considering patient-specific dosing. For example, a pharmacogenetic predictor was used to predetermine the response of breast cancer patients to a combination treatment of Paclitaxel, Fluorouracil, Doxorubicin and Cyclophosphamide. For that purpose, a classification model was designed based on training data obtained from gene expression profiles of 82 breast cancer patients treated with the aforementioned drug combination. The AI model successfully predicted pathologic complete response in 92% of the 51 patients that were analyzed.[86] Implementing these dosing and treatment efficacy predictors to nanomedicine will help improve their performance in clinical settings.
5. Nanotechnology Facilitates Personalized Gene Therapy
Gene therapy is an ultimate example of precision medicine, as patient-specific mutations are corrected, removed or inhibited. Nano-carriers are vital for successful delivery of oligonucleotides for gene therapy, enabling their activity at the target site. Gene silencing with RNA interference (RNAi) was first demonstrated by Fire and Mello, and has been used since to target complimentary mRNA molecules in cells and lead to their degradation.[87] This mechanism was exploited for precision treatment in various diseases, including cancer. Silencing of oncogenes, proteases that mediate cell invasion and metastasis, genes associated with drug resistance, and angiogenic factors was shown to have positive therapeutic effects in different types of cancer.[88]
However, in order to implement RNAi as a precision treatment in the clinic, efficient delivery vehicles are essential.[89] These delivery vehicles must be able to avoid phagocytic uptake in the blood stream, successfully transport through the endothelial barrier and plasma membrane, escape the endosome and release the small interfering RNA (siRNA) in the cytoplasm.[90] In addition, using delivery vehicles that will direct the payload towards the target site can reduce innate immune responses and off-target effects.[91] Many different types of nanoparticles have been tested in animal models for siRNA delivery. Lipid nanoparticles, poly(ethylene imine) polymer nanoparticles, self-assembled nucleic acid nanoparticles and polysaccharide-based nanoparticles composed of chitosan or cyclodextrin were all shown to deliver siRNA in vivo.[92] Recently, an enormous step forward was taken with the clinical approval of the first siRNA-based nanomedicine – Patisiran, a lipid nanoparticle encapsulating siRNA for treating hereditary transthyretin amyloidosis.[93] Nevertheless, the use of these delivery tools in cancer remains a challenge. Delivery issues including inter-patient variability in tumor vasculature and high interstitial fluid pressure in the tumor environment need to be overcome in order to translate this technology to clinical use in cancer. A number of clinic trials are currently running with siRNA formulations against different targets in several cancer types, attempt to make this step forward.[94]
The use of nanotechnology for gene therapy delivery is not limited to RNAi alone. Delivery of mRNA vaccines and mRNAs for immune-oncology is currently tested in clinical trials for a number of malignant diseases including metastatic melanoma, pancreatic and colorectal cancers.[95] For example, lipid nanoparticles encapsulating OX40L mRNA that amplifies the immune-stimulatory response of DCs are currently in phase I trials for metastatic solid tumors and lymphoma.[96] Moreover, the discovery and development of the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 technology provides another tool for genome editing which requires efficient delivery tools and can be utilized for cancer therapy.[97] Liu, et al. attempted to create allogenic chimeric antigen receptor (CAR) T-cells by knocking out T-cell genes known to induce immunogenic graft-vs-host reactions.[98] Although these cells showed comparable activities to unmodified CAR T-cells, their immunogenicity needs to be further investigated.
Computational methods are introduced in several stages in the engineering process of gene therapy systems. Numerous AI-based models, including Neural Networks, SVMs and decision trees, were used for classification of effective and ineffective sequences for RNAi in order to recognize key features in their design.[99] However, these models only refer to the efficiency of the siRNA sequence itself, and don't take into account the delivery method. The use of predictive algorithms for delivery of oligonucleotides is not as frequent. Instead, laborious experimental scanning of chemical libraries were performed in several studies for testing parameters in the design of carrier systems.[100] Nevertheless, this data can be exploited for training machine learning algorithms and help uncover vital design parameters which might have been overlooked in current designs. Moreover, specific modeling of membrane-nanoparticle interactions can provide insights on the uptake mechanism of the particle and its inter-cellular pathway, as well as the effects of the nanoparticle properties on these processes.[101] By taking these considerations into account and altering the nanoparticle's properties, the transfection efficacy of the nanoparticles can be further increased.
6. Deciding when to use Nanomedicine
Despite the numerous advantages of nanomedicine-based therapies, nanomedicine are not suitable for every patient and malignancy. The most widely discussed example for this issue is the great variability in the enhanced permeability and retention (EPR) effect between different patients and cancer types.[102, 103] While most clinically approved nanomedicine to date rely on the EPR effect for effective delivery to the target site, its heterogeneity among patients due to different clinical and physiological conditions, such as varied vascular architecture, leads to different drug response among patients.[103] Similar variability exists in targeting moieties' expression in the case of ligand-targeted nanoparticles.[104] Therefore, prescreening the patient's compliance to the specific nanomedicine is crucial before deciding on this type of treatment.[105] Nanoparticle-based imaging has been widely used in pre-clinical studies and was also tested in several clinical studies.[106] The intrinsic properties of nanomaterials make them suitable for various imaging modalities. superparamagnetic iron oxide nanoparticles (SPIONs) for example, are clinically approved imaging contrast agents exhibiting improved contrast in MRI imaging due to their unique magnetic properties.[107] Bourdeau, et al. used protein-based gas nano-vesicles as ultrasound and MRI contrast agents in animal models.[108] “C-dots”, dye-labeled mesoporous silica nanoparticles functionalized with a targeting peptide, are currently under clinical trials for image-guided intra-operative mapping of lymph node metastasis in head and neck, breast and colorectal cancer.[109] Nevertheless, only a few nanoparticle-based imaging agents have been clinically approved. One reason for their limited use is their unique pharmacokinetic profile that does not always fit the requirements of an imaging agent. Nanoparticles often display long circulation times, limited tissue penetration which is highly dependent on the tumor vascularization, and are mostly taken up by the mononuclear phagocytic system. Therefore, in certain tumor types these agents do not achieve the desirable rapid accumulation in the target site in comparison to low-molecular-weight imaging agents.[110] Concerns about toxicity and compatibility of certain nanomaterials, as well as high development costs pose additional difficulties for translating these imaging technologies to the clinic.
Nanotheranostics offer an alternative solution for pre-screening nanomedicine suitability in different patients using nanomedicines that contain both a drug and an imaging agent in one nanoparticle.[111] These nanoparticles can then be used in order to study the nanomedicine's pharmacokinetics, targeting efficacy and drug release profile.[112] For example, a radiolabeled cyclodextrin-based nanoparticle encapsulating the topoisomerase inhibitor camptothecin, was used for pharmacokinetic studies with PET imaging in mice neuroblastoma models.[113] Nanotheranostics were also used for increasing the accuracy of nanoparticle-based photothermal and radiation therapies, by localizing the nanoparticles in the tumor area prior to the treatment in animal models.[114] For example, PEGylated WS2 nanosheets displaying high absorbance of NIR irradiation for photothermal therapy were detected with CT imaging prior to their photo-activation in a murine breast cancer model.[115] These nanosheets displayed good biocompatibility and effectively photo-ablated the tumors both in intra-tumoral and intravenous administration.
Yet although nanotheranosic formulations present an attractive approach, current theranostic clinical studies are mostly based on direct labeling of drugs or targeting ligands with imaging moieties (mostly for cancer radiation therapy) without using nanoparticles.[116] The high complexity of designing, testing and clinically approving a nanomedicine, even before adding an imaging agent, may withhold attempts to add an imaging agent for theranostic purposes. Therefore, simple techniques that will allow to efficiently label an approved nanomedicine after the fabrication process will enable more frequent use of nanomedicine pre-screening in patients. For example, remote loading of 52Mn-DOTA to liposomes enabled their radiolabeling for PET imaging without effecting their encapsulated materials or their external surface properties.[117] The utilization of AI in nanotheranostic formulation can provide essential insights (Figure 4). In case the imaging agent and the drug need to be loaded into the particle, predictive machine-learning algorithms can be applied to predict their encapsulation efficiency.[118] For example, a quantitative structure-property relationship (QSPR) model was used to predict the ability of the molecules to be remotely loaded into liposomes with over 90% accuracy according to their chemical structure and the conditions of the encapsulation process.[119] This model was implemented with several different algorithms, including SVM, decision trees and Iterative Stochastic Elimination (ISE, see Box 1). A similar method was used to predict the cytotoxicity of metal oxide nanoparticles.[120] Extrapolating this method to other types of nanoparticles, will allow to assess the effect of surface labeling of nanoparticles for imaging purposes on their biocompatibility. The contribution of AI to image analysis must also be recognized when discussing medical imaging.[121] Machine learning algorithms for tumor detection, characterization and monitoring are constantly improved in their accuracy and reproducibility, aiming to save time and improve the diagnostic abilities of medical teams. Implementing these algorithms in nanotheranostic imaging could provide further insights on the particles bio-distribution profiles and therapeutic efficacies.
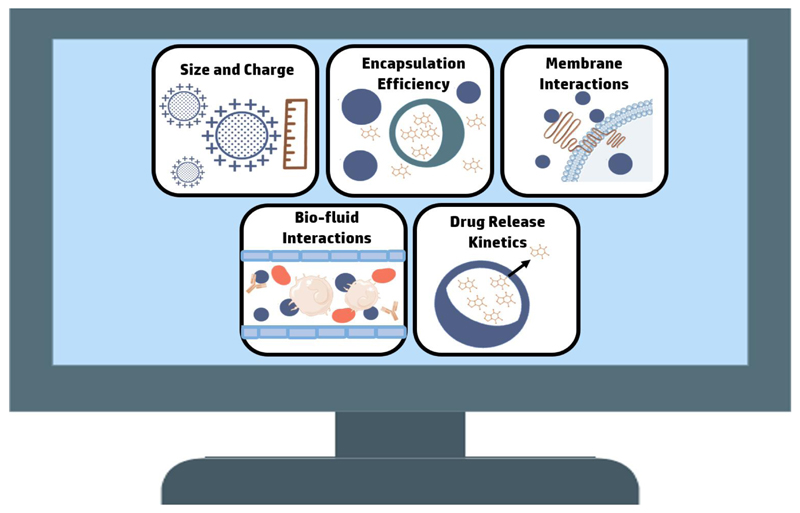
Figure 4.
Computational methods contribute to various aspects of nanoparticle design. Current machine-learning algorithms and computational models provide tools for predicting the nanoparticles' size and charge, drug encapsulation efficiency, interactions with biological membranes, biological fluids and drug release kinetics.
A complimentary approach to patient pre-screening before selecting nanomedicine-based treatments, is based on actively improving the patients' nanomedicine compatibility. This can be performed by reducing the tumor stromal barrier in order to achieve better nanoparticle localization and penetration. For example, conjugation of recombinant human hyaluronidase to PLGA-PEG nanoparticles increased their penetration into the tumor in tumor-bearing mice by eliminating hyaluronic acid in the tumor microenvironment.[122] This technology is currently under clinical studies in combination with Nab-Paclitaxel and other therapeutic agents in pancreatic cancer patients.[123] Von Maltzahn, et al, presented another approach for enhanced particle accumulation with a system based on two types of nanoparticles that can enhance nanoparticle localization in murine tumors by triggering endogenous biological processes in the cancer tissue.[124] First, gold nanorods were injected and localized to the tumor utilizing the EPR effect. Photothermal heating of these nanorods initiated extravascular coagulation and fibrin deposition specifically in the tumor area. This in turn led to amplified tumor localization of a second wave of injected nanoparticles functionalized with fibrin binding peptides that were recruited to the target site by the coagulation signal of the first wave of nanoparticles.
7. Future Outlook
The development and implementation of precision medicine has begun revolutionizing the diagnosis and care of cancer patients. Cancer multi-gene panel scans adapted for different cancer types are available in clinics worldwide, aimed to assist in treatment selection. Yet, further improvements and considerations must be addressed in order to fulfill the potential of precision medicine.
Patient classification is an important issue that must be improved in order to optimally personalize the treatment regime. To date, patient classification is based on differentiating healthy vs. diseased individuals, with several technologies that enable cancer staging and differentiating between cancer types.[47, 50] Implementation of unsupervised learning methods (see Box 1) that will cluster patient groups without predefined categorization, can assist in detection of new features that are important to take into account when tailoring a treatment regime for a patient. For example, results from pharmacogenomic studies indicate that gender is an important factor to consider for drug administration, since it influences the therapeutic response, adverse effects and pharmacokinetics, partially due to differences in drug metabolizing enzymes expression patterns and effects of sex hormones.[125] A recent study emphasized the importance of this issue specifically for nanomedicine, demonstrating differences in nanoparticle uptake between male and female amniotic stem cells.[126]
Patient follow-up after completing the treatment is another important issue that is not discussed as often in comparison to the diagnostic and treatment stages. Current protocols of long-term patient follow-up vary for different cancer types, treatment stages and treatment regimens, and raise controversies among medical teams.[127] Major effectors on the success of long-term follow-up are the sensitivity and accuracy of the performed tests, as well as their cost, time-consumption, complexity and patient-compliance. Many of the sensing technologies that are developed for diagnostic purposes could be adjusted to fit follow-up purposes. For example, successful differentiation between pre- and post-surgery states of lung cancer patients using a VOC nanosensor array, initiated a clinical study for its use as a shot-term follow-up tool.[128]
Portable and easy-to-operate nanosensors can improve the implementation of follow-up protocols for cancer patients. Advancements in the fabrication of flexible and self-healing nanomaterials should pave the way towards electronic skin nanosensors that will enable continuous monitoring of selected biomarkers through sweat, saliva and non-invasive blood analysis.[129] In a recently published study by Gao, et al., a flexible sensing array for detection of sweat metabolites and electrolytes was designed and connected to a mobile app via Bluetooth.[130] This on-body nanosensor was used for real time monitoring of subject's sweat profile under different physiological conditions. In addition, combination of microfluidic techniques in point-of-care devices and smartphone-integrated platforms can also prove essential for simplifying patient-operated devices that will allow more frequent follow up without increasing the burden on medical teams.[131]
Computational methods are essential in the implementation of precision medicine, beginning from molecular data collection, to the diagnostic and treatment stages. AI algorithms for patient classification, screening patients' drug suitability and for optimizing nanomedicine properties demonstrated their value for precision medicine in numerous studies. However, in order to reach clinical implementation of these algorithms, several challenges must be addressed. One of the most important issues for achieving high accuracy in these computational methods is obtaining large datasets that will be used for training the algorithms. Therefore, data standardization as well as data collection from heterogenic patient populations are crucial for the clinical success of these models. Moreover, stronger connection between the experts in the fields of nanomaterials, medicine and computer science and implementation of computation in all stages of academic and industrial research will help to optimize their performance and clinical relevance.
The design of specific computational models for different patients is another emerging approach for improving the accuracy of precision medicine. Bordbar, et al. took initial steps in this direction and designed personalized computational models for cellular metabolism kinetics that provided insights on the patients' personal pharmacodynamics.[132] These models were based on several measurements of metabolite concentrations in erythrocytes and in the plasma extracted from each subject. These measurements were used to calculate the metabolite baseline levels and a single rate constant that represented a combination of the kinetic constants for the metabolic network. Simulations of the response of each of the models to ribavirin, a drug used for treatment in Hepatitis C patients, revealed drug tolerability issues in some of the individuals. Translating this approach for cancer patients can prove beneficial for metabolomics-based diagnosis and drug pre-screening.
A frequently asked question when discussing precision medicine treatment's is: can we personalize a nanomedicine for every patient? Although present techniques enable versatile conjugation of any desired antibody to nanoparticles, as well as variability in the choice of cargo (e.g., a specific siRNA sequence that matches the chosen gene for inhibition), many further obstacles remain in order to make this approach realizable.[133] Besides the complicated clinical approval process for personalized nanomedicine, limitations of current fabrication techniques and the high costs of nanomedicine development must be addressed. More accurate use of existing drugs by utilizing precision diagnostic platforms and personalized drug-tailoring techniques can significantly improve treatment outcome by constructing a combinatorial treatment protocol for each patient targeting several pathways simultaneously. This will enable to overcome drug resistance and improve therapeutic efficacy. AI and other computational models will play an important role in the development, design and implementation of these nanotechnologies.
Acknowledgments
This work was supported by ERC-STG-2015-680242.
The authors also acknowledge the support of the Technion Integrated Cancer Center (TICC), the Russell Berrie Nanotechnology Institute, the Lorry I. Lokey Interdisciplinary Center for Life Sciences & Engineering, The Israel Ministry of Economy for a Kamin Grant (52752); the Israel Ministry of Science Technology and Space – Office of the Chief Scientist (3-11878); the Israel Science Foundation (1778/13, 1421/17); the Israel Cancer Association (2015-0116); the German-Israeli Foundation for Scientific Research and Development for a GIF Young grant (I-2328-1139.10/2012); the European Union FP-7 IRG Program for a Career Integration Grant (908049); the Phospholipid Research Center Grant; a Mallat Family Foundation Grant; The Unger Family Fund; A. Schroeder acknowledges Alon and Taub Fellowships. O. Adir acknowledges the Sherman and Gutwirth fellowships. M. Poley acknowledges the Shulamit Aloni fellowship (Ministry of Science, Technology and Space, Israel). G. Chen acknowledges the Sherman Fellowship. N. Krinsky acknowledges the “Baroness Ariane de Rothschild Women Doctoral Program” from the Rothschild Caesarea Foundation.
Biographies
Avi Schroeder is an Associate Professor of Chemical Engineering at the Technion – Israel Institute of Technology where he heads the Laboratory for Targeted Drug Delivery and Personalized Medicine Technologies (https://www.schroederlab.com/). His research focuses on the use of nanotechnology for precision cancer medicine.
Omer Adir is a Ph.D. candidate in Prof. Schroeder's Laboratory for Targeted Drug Delivery and Personalized Medicine Technologies, Technion – Israel Institute of Technology. He earned his B.Sc. degree in biochemical engineering from the Technion – Israel Institute of Technology. His research focuses on development of synthetic cells for therapeutic purposes.
Contributor Information
Omer Adir, Department of Chemical Engineering, Technion – Israel Institute of Technology, Haifa 3200003, Israel; The Norman Seiden Multidisciplinary Program for Nanoscience and Nanotechnology, Technion – Israel Institute of Technology, Haifa 32000, Israel.
Sahar Froim, Department of Physical Electronics, School of Electrical Engineering, Fleischman Faculty of Engineering, Tel Aviv University, Tel Aviv 69978, Israel.
Twan Lammers, Institute for Experimental Molecular Imaging, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Dr. Avi Schroeder, Department of Chemical Engineering, Technion – Israel Institute of Technology, Haifa 3200003, Israel.
References
- [1].Schork NJ. Nature. 2015;520:609. doi: 10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- [2].Longo DL. N Engl J Med. 2012;366:956. doi: 10.1056/NEJMe1200656. [DOI] [PubMed] [Google Scholar]; McGranahan N, Swanton C. Cell. 2017;168:613. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- [3].Collins FS, Varmus H. N Engl J Med. 2015;372:793. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gupta PK. Trends in biotechnology. 2008;26:602. doi: 10.1016/j.tibtech.2008.07.003. [DOI] [PubMed] [Google Scholar]
- [5].Xia Z, Xing Y, So M-K, Koh AL, Sinclair R, Rao J. Analytical chemistry. 2008;80:8649. doi: 10.1021/ac801562f. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hahm J-i, Lieber CM. Nano letters. 2004;4:51. [Google Scholar]
- [6].Wang S, Huang P, Chen X. Acs Nano. 2016;10:2991. doi: 10.1021/acsnano.6b00870. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sumer B, Gao J. :2008. [Google Scholar]
- [7].Min Y, Caster JM, Eblan MJ, Wang AZ. Chemical reviews. 2015;115:11147. doi: 10.1021/acs.chemrev.5b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wernick MN, Yang Y, Brankov JG, Yourganov G, Strother SC. IEEE signal processing magazine. 2010;27:25. doi: 10.1109/MSP.2010.936730. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, Ferrero E, Agapow P-M, Zietz M, Hoffman MM. Journal of The Royal Society Interface. 2018;15 doi: 10.1098/rsif.2017.0387. 20170387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maojo V, Fritts M, de la Iglesia D, Cachau RE, Garcia-Remesal M, Mitchell JA, Kulikowski C. International journal of nanomedicine. 2012;7:3867. doi: 10.2147/IJN.S24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shamay Y, Shah J, Işik M, Mizrachi A, Leibold J, Tschaharganeh DF, Roxbury D, Budhathoki-Uprety J, Nawaly K, Sugarman JL, Baut E, et al. Nature Materials. 2018 doi: 10.1038/s41563-017-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ding H-m, Tian W-d, Ma Y-q. Acs Nano. 2012;6:1230. doi: 10.1021/nn2038862. [DOI] [PubMed] [Google Scholar]
- [11].Altomare D, Di Lena M, Porcelli F, Trizio L, Travaglio E, Tutino M, Dragonieri S, Memeo V, De Gennaro G. British journal of surgery. 2013;100:144. doi: 10.1002/bjs.8942. [DOI] [PubMed] [Google Scholar]
- [12].Rubio-Perez C, Tamborero D, Schroeder MP, Antolín AA, Deu-Pons J, Perez-Llamas C, Mestres J, Gonzalez-Perez A, Lopez-Bigas N. Cancer cell. 2015;27:382. doi: 10.1016/j.ccell.2015.02.007. [DOI] [PubMed] [Google Scholar]
- [13].Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE. Cell. 2012;148:1293. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Von Hoff DD, Stephenson JJ, Jr, Rosen P, Loesch DM, Borad MJ, Anthony S, Jameson G, Brown S, Cantafio N, Richards DA, Fitch TR, et al. J Clin Oncol. 2010;28:4877. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- [14].Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, Schellen P, Verschueren H, Post E, Koster J. Cancer cell. 2015;28:666. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Korlach J, Bjornson KP, Chaudhuri BP, Cicero RL, Flusberg BA, Gray JJ, Holden D, Saxena R, Wegener J, Turner SW. Methods in enzymology. Vol. 472. Elsevier; 2010. p. 431. [DOI] [PubMed] [Google Scholar]
- [16].Foquet M, Samiee KT, Kong X, Chauduri BP, Lundquist PM, Turner SW, Freudenthal J, Roitman DB. Journal of Applied Physics. 2008;103 034301. [Google Scholar]
- [17].Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, et al. Science. 2009;323:133. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- [18].Beatson SA, Walker MJ. Science. 2014;345:1454. doi: 10.1126/science.1260471. [DOI] [PubMed] [Google Scholar]
- [19].Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Nature methods. 2010;7:461. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]; Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Proceedings of the National Academy of Sciences. 1998;95:11891. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lofton-Day C, Model F, DeVos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M. Clinical chemistry. 2008;54:414. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- [20].Schneider GF, Dekker C. Nature biotechnology. 2012;30:326. doi: 10.1038/nbt.2181. [DOI] [PubMed] [Google Scholar]; Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A. Nature methods. 2018;15:201. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- [21].Jain M, Olsen HE, Paten B, Akeson M. Genome biology. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Quick J, Loman NJ, Duraffour S, Simpson JT, Ettore S, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A, et al. Nature. 2016;530:228. [Google Scholar]
- [22].Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proceedings of the National Academy of Sciences. 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]; Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. Nature biotechnology. 2012;30:349. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]; Carter J-M, Hussain S. Wellcome open research. 2017;2 doi: 10.12688/wellcomeopenres.11246.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Farimani AB, Min K, Aluru NR. ACS nano. 2014;8:7914. doi: 10.1021/nn5029295. [DOI] [PubMed] [Google Scholar]; Liu S, Lu B, Zhao Q, Li J, Gao T, Chen Y, Zhang Y, Liu Z, Fan Z, Yang F. Advanced materials. 2013;25:4549. doi: 10.1002/adma.201301336. [DOI] [PubMed] [Google Scholar]
- [24].Dekker C. Nature nanotechnology. 2007;2:209. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]; Han A, Creus M, Schürmann G, Linder V, Ward TR, de Rooij NF, Staufer U. Analytical chemistry. 2008;80:4651. doi: 10.1021/ac7025207. [DOI] [PubMed] [Google Scholar]; Yusko EC, Bruhn BR, Eggenberger OM, Houghtaling J, Rollings RC, Walsh NC, Nandivada S, Pindrus M, Hall AR, Sept D. Nature nanotechnology. 2017;12:360. doi: 10.1038/nnano.2016.267. [DOI] [PubMed] [Google Scholar]
- [25].Lindsay S. Nature nanotechnology. 2016;11:109. doi: 10.1038/nnano.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Goodwin S, Gurtowski J, Ethe-Sayers S, Deshpande P, Schatz MC, McCombie WR. Genome research. 2015 doi: 10.1101/gr.191395.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Di Ventra M, Taniguchi M. Nature nanotechnology. 2016;11:117. doi: 10.1038/nnano.2015.320. [DOI] [PubMed] [Google Scholar]
- [28].Huang S, He J, Chang S, Zhang P, Liang F, Li S, Tuchband M, Fuhrmann A, Ros R, Lindsay S. Nature nanotechnology. 2010;5:868. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhao Y, Ashcroft B, Zhang P, Liu H, Sen S, Song W, Im J, Gyarfas B, Manna S, Biswas S. Nature nanotechnology. 2014;9:466. doi: 10.1038/nnano.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Computational and structural biotechnology journal. 2015;13:8. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tan J, Ung M, Cheng C, Greene CS. Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders. Pacific Symposium on Biocomputing Co-Chairs; 2014. [PMC free article] [PubMed] [Google Scholar]
- [31].Ren X, Wang Y, Chen L, Zhang X-S, Jin Q. Nucleic acids research. 2012;41:e53. doi: 10.1093/nar/gks1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim M, Rai N, Zorraquino V, Tagkopoulos I. Nature communications. 2016;7 doi: 10.1038/ncomms13090. 13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen A, Chatterjee S. Chemical Society Reviews. 2013;42:5425. doi: 10.1039/c3cs35518g. [DOI] [PubMed] [Google Scholar]; Kneipp J, Kneipp H, Wittig B, Kneipp K. Nanomedicine: Nanotechnology, Biology and Medicine. 2010;6:214. doi: 10.1016/j.nano.2009.07.009. [DOI] [PubMed] [Google Scholar]; Swierczewska M, Liu G, Lee S, Chen X. Chemical Society Reviews. 2012;41:2641. doi: 10.1039/c1cs15238f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang RE, Tian L, Chang Y-H. Journal of pharmaceutical and biomedical analysis. 2012;63:165. doi: 10.1016/j.jpba.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kosaka PM, Pini V, Ruz JJ, Da Silva R, González M, Ramos D, Calleja M, Tamayo J. Nature nanotechnology. 2014;9:1047. doi: 10.1038/nnano.2014.250. [DOI] [PubMed] [Google Scholar]
- [36].Warren AD, Kwong GA, Wood DK, Lin KY, Bhatia SN. Proceedings of the National Academy of Sciences. 2014;111:3671. doi: 10.1073/pnas.1314651111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gau V, Wong D. Annals of the New York Academy of Sciences. 2007;1098:401. doi: 10.1196/annals.1384.005. [DOI] [PubMed] [Google Scholar]; Konvalina G, Haick H. Accounts of chemical research. 2013;47:66. doi: 10.1021/ar400070m. [DOI] [PubMed] [Google Scholar]
- [37].Acimovic SS, Ortega MA, Sanz V, Berthelot J, Garcia-Cordero JL, Renger J, Maerkl SJ, Kreuzer MP, Quidant R. Nano letters. 2014;14:2636. doi: 10.1021/nl500574n. [DOI] [PubMed] [Google Scholar]
- [38].Zhang C-Y, Yeh H-C, Kuroki MT, Wang T-H. Nature materials. 2005;4:826. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- [39].Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW. New England Journal of Medicine. 2004;351:781. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]; Green BJ, Saberi Safaei T, Mepham A, Labib M, Mohamadi RM, Kelley SO. Angewandte Chemie International Edition. 2016;55:1252. doi: 10.1002/anie.201505100. [DOI] [PubMed] [Google Scholar]
- [40].Nair P, Alam M. Applied physics letters. 2006;88 233120. [Google Scholar]; Zhang Z, Chan DW. Cancer Epidemiology and Prevention Biomarkers. 2010;19:2995. doi: 10.1158/1055-9965.EPI-10-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kirwan A, Utratna M, O’Dwyer ME, Joshi L, Kilcoyne M. BioMed research international. 2015;2015 doi: 10.1155/2015/490531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rosen DG, Wang L, Atkinson JN, Yu Y, Lu KH, Diamandis EP, Hellstrom I, Mok SC, Liu J, Bast RC., Jr Gynecologic oncology. 2005;99:267. doi: 10.1016/j.ygyno.2005.06.040. [DOI] [PubMed] [Google Scholar]
- [43].Bristow RE, Smith A, Zhang Z, Chan DW, Crutcher G, Fung ET, Munroe DG. Gynecologic oncology. 2013;128:252. doi: 10.1016/j.ygyno.2012.11.022. [DOI] [PubMed] [Google Scholar]
- [44].Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat Biotechnol. 2005;23:1294. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- [45].Hu M, Yan J, He Y, Lu H, Weng L, Song S, Fan C, Wang L. ACS nano. 2009;4:488. doi: 10.1021/nn901404h. [DOI] [PubMed] [Google Scholar]; Chen P, Chung MT, McHugh W, Nidetz R, Li Y, Fu J, Cornell TT, Shanley TP, Kurabayashi K. ACS nano. 2015;9:4173. doi: 10.1021/acsnano.5b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Borrebaeck CA. Nature Reviews Cancer. 2017;17:199. doi: 10.1038/nrc.2016.153. [DOI] [PubMed] [Google Scholar]
- [47].Hizir MS, Balcioglu M, Rana M, Robertson NM, Yigit MV. ACS applied materials & interfaces. 2014;6:14772. doi: 10.1021/am504190a. [DOI] [PubMed] [Google Scholar]
- [48].Wilson A, Baietto M. Sensors. 2009;9:5099. doi: 10.3390/s90705099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Phillips M, Gleeson K, Hughes JMB, Greenberg J, Cataneo RN, Baker L, McVay WP. The Lancet. 1999;353:1930. doi: 10.1016/S0140-6736(98)07552-7. [DOI] [PubMed] [Google Scholar]
- [50].Peng G, Hakim M, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, Tisch U, Haick H. British journal of cancer. 2010;103:542. doi: 10.1038/sj.bjc.6605810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Scott SM, James D, Ali Z. Microchimica Acta. 2006;156:183. [Google Scholar]
- [52].Peng G, Tisch U, Adams O, Hakim M, Shehada N, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, Haick H. Nature nanotechnology. 2009;4:669. doi: 10.1038/nnano.2009.235. [DOI] [PubMed] [Google Scholar]; Dragonieri S, Annema JT, Schot R, van der Schee MP, Spanevello A, Carratú P, Resta O, Rabe KF, Sterk PJ. Lung cancer. 2009;64:166. doi: 10.1016/j.lungcan.2008.08.008. [DOI] [PubMed] [Google Scholar]
- [53].Wang B, Cancilla JC, Torrecilla JS, Haick H. Nano letters. 2014;14:933. doi: 10.1021/nl404335p. [DOI] [PubMed] [Google Scholar]
- [54].Kermani BG, Schiffman SS, Nagle HT. Sensors and Actuators B: Chemical. 2005;110:13. [Google Scholar]
- [55].Kumar S, Harrison N, Richards-Kortum R, Sokolov K. Nano letters. 2007;7:1338. doi: 10.1021/nl070365i. [DOI] [PubMed] [Google Scholar]
- [56].Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Y, Medley CD, Cao Z, Li J. Angewandte Chemie International Edition. 2009;48:856. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Nucleic acids research. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Geng Y, Goel HL, Le NB, Yoshii T, Mout R, Tonga GY, Amante JJ, Mercurio AM, Rotello VM. Nanomedicine: Nanotechnology, Biology and Medicine. 2018;14:1931. doi: 10.1016/j.nano.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gendron KB, Hockstein NG, Thaler ER, Vachani A, Hanson CW. Otolaryngology--Head and Neck Surgery. 2007;137:269. doi: 10.1016/j.otohns.2007.02.005. [DOI] [PubMed] [Google Scholar]
- [59].Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P. New England journal of medicine. 2012;366:883. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dahlman JE, Kauffman KJ, Xing Y, Shaw TE, Mir FF, Dlott CC, Langer R, Anderson DG, Wang ET. Proceedings of the National Academy of Sciences. 2017 doi: 10.1073/pnas.1620874114. 201620874. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yaari Z, Da Silva D, Zinger A, Goldman E, Kajal A, Tshuva R, Barak E, Dahan N, Hershkovitz D, Goldfeder M. Nature communications. 2016;7 doi: 10.1038/ncomms13325. 13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Majumder B, Baraneedharan U, Thiyagarajan S, Radhakrishnan P, Narasimhan H, Dhandapani M, Brijwani N, Pinto DD, Prasath A, Shanthappa BU. Nature communications. 2015;6 doi: 10.1038/ncomms7169. 6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Schroeder MP, Rubio-Perez C, Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. Bioinformatics. 2014;30:i549. doi: 10.1093/bioinformatics/btu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Strebhardt K, Ullrich A. Nature Reviews Cancer. 2008;8:473. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- [64].El-Sayed IH, Huang X, El-Sayed MA. Cancer letters. 2006;239:129. doi: 10.1016/j.canlet.2005.07.035. [DOI] [PubMed] [Google Scholar]; Lu Y, Low PS. Advanced drug delivery reviews. 2012;64:342. doi: 10.1016/s0169-409x(02)00042-x. [DOI] [PubMed] [Google Scholar]
- [65].Molinaro R, Evangelopoulos M, Hoffman JR, Corbo C, Taraballi F, Martinez JO, Hartman KA, Cosco D, Costa G, Romeo I. Advanced Materials. 2018;30 doi: 10.1002/adma.201702749. 1702749. [DOI] [PubMed] [Google Scholar]; Parodi A, Molinaro R, Sushnitha M, Evangelopoulos M, Martinez JO, Arrighetti N, Corbo C, Tasciotti E. Biomaterials. 2017;147:155. doi: 10.1016/j.biomaterials.2017.09.020. [DOI] [PubMed] [Google Scholar]
- [66].Kaneti L, Bronshtein T, Malkah Dayan N, Kovregina I, Letko Khait N, Lupu-Haber Y, Fliman M, Schoen BW, Kaneti G, Machluf M. Nano letters. 2016;16:1574. doi: 10.1021/acs.nanolett.5b04237. [DOI] [PubMed] [Google Scholar]
- [67].Mura S, Nicolas J, Couvreur P. Nature materials. 2013;12:991. doi: 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- [68].Grass RN, Athanassiou EK, Stark WJ. Angewandte Chemie International Edition. 2007;46:4909. doi: 10.1002/anie.200700613. [DOI] [PubMed] [Google Scholar]; Schroeder A, Honen R, Turjeman K, Gabizon A, Kost J, Barenholz Y. Journal of controlled release. 2009;137:63. doi: 10.1016/j.jconrel.2009.03.007. [DOI] [PubMed] [Google Scholar]; Yang X, Liu X, Liu Z, Pu F, Ren J, Qu X. Advanced materials. 2012;24:2890. doi: 10.1002/adma.201104797. [DOI] [PubMed] [Google Scholar]; Ta T, Porter TM. Journal of controlled release. 2013;169:112. doi: 10.1016/j.jconrel.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Gao W, Chan JM, Farokhzad OC. Molecular pharmaceutics. 2010;7:1913. doi: 10.1021/mp100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].De La Rica R, Aili D, Stevens MM. Advanced drug delivery reviews. 2012;64:967. doi: 10.1016/j.addr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- [71].Laeubli H. Available from: https://clinicaltrials.gov/ct2/show/NCT03603379?term=Anti-EGFR+immunoliposomes&rank=3. [Google Scholar]; Shi J, Kantoff PW, Wooster R, Farokhzad OC. Nature Reviews Cancer. 2017;17:20. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Brannon-Peppas L, Blanchette JO. Advanced drug delivery reviews. 2012;64:206. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]; Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Nature communications. 2018;9:1410. doi: 10.1038/s41467-018-03705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Meriggi F, Di Biasi B, Abeni C, Zaniboni A. Current drug targets. 2009;10:1033. doi: 10.2174/138945009789577891. [DOI] [PubMed] [Google Scholar]
- [74].Liu J, Weller GE, Zern B, Ayyaswamy PS, Eckmann DM, Muzykantov VR, Radhakrishnan R. Proceedings of the National Academy of Sciences. 2010;107:16530. doi: 10.1073/pnas.1006611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shityakov S, Roewer N, Broscheit J-A, Förster C. Computational Toxicology. 2017;2:20. doi: 10.1016/j.compbiolchem.2017.06.004. [DOI] [PubMed] [Google Scholar]
- [76].Gao Z, Chen Y, Cai X, Xu R. Bioinformatics. 2016;33:901. doi: 10.1093/bioinformatics/btw713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yin B, Balvert M, Zambrano D, Schönhuth A, Bohte S. arXiv preprint. 2018 arXiv:1806.04931. [Google Scholar]
- [78].Shah S, Liu Y, Hu W, Gao J. Journal of nanoscience and nanotechnology. 2011;11:919. doi: 10.1166/jnn.2011.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Baghaei B, Saeb MR, Jafari SH, Khonakdar HA, Rezaee B, Goodarzi V, Mohammadi Y. Journal of Applied Polymer Science. 2017;134:45145. [Google Scholar]
- [80].Anderson DG, Lynn DM, Langer R. Angewandte Chemie International Edition. 2003;42:3153. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- [81].Yamankurt G, Berns EJ, Xue A, Lee A, Bagheri N, Mrksich M, Mirkin CA. Nature Biomedical Engineering. 2019;1 doi: 10.1038/s41551-019-0351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhao X, Kim J, Cezar CA, Huebsch N, Lee K, Bouhadir K, Mooney DJ. Proceedings of the National Academy of Sciences. 2011;108:67. doi: 10.1073/pnas.1007862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jeon G, Yang SY, Byun J, Kim JK. Nano letters. 2011;11:1284. doi: 10.1021/nl104329y. [DOI] [PubMed] [Google Scholar]; Paris JL, Cabañas MV, Manzano M, Vallet-Regí M. ACS nano. 2015;9:11023. doi: 10.1021/acsnano.5b04378. [DOI] [PubMed] [Google Scholar]; Ge J, Neofytou E, Cahill TJ, III, Beygui RE, Zare RN. ACS nano. 2011;6:227. doi: 10.1021/nn203430m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Valdes G, Simone CB, II, Chen J, Lin A, Yom SS, Pattison AJ, Carpenter CM, Solberg TD. Radiotherapy and Oncology. 2017;125:392. doi: 10.1016/j.radonc.2017.10.014. [DOI] [PubMed] [Google Scholar]
- [85].Linden A, Yarnold PR, Nallamothu BK. Journal of Evaluation in Clinical Practice. 2016;22:860. doi: 10.1111/jep.12573. [DOI] [PubMed] [Google Scholar]; Menden MP, Iorio F, Garnett M, McDermott U, Benes CH, Ballester PJ, Saez-Rodriguez J. PLoS one. 2013;8:e61318. doi: 10.1371/journal.pone.0061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ. Journal of clinical oncology. 2006;24:4236. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- [87].Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. nature. 2001;411:494. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]; Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- [88].Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M. Cancer discovery. 2013 doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- [89].Whitehead KA, Langer R, Anderson DG. Nature reviews Drug discovery. 2009;8:129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]; Daka A, Peer D. Advanced drug delivery reviews. 2012;64:1508. doi: 10.1016/j.addr.2012.08.014. [DOI] [PubMed] [Google Scholar]
- [90].Kanasty R, Dorkin JR, Vegas A, Anderson D. Nature materials. 2013;12:967. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- [91].Whitehead KA, Dahlman JE, Langer RS, Anderson DG. Annual review of chemical and biomolecular engineering. 2011;2:77. doi: 10.1146/annurev-chembioeng-061010-114133. [DOI] [PubMed] [Google Scholar]
- [92].Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E. Nature biotechnology. 2010;28:172. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]; Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel J, Ribas A. nature. 2010;464:1067. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M. Nature nanotechnology. 2012;7:389. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E. New England Journal of Medicine. 2013;369:819. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- [94].Kim HJ, Kim A, Miyata K, Kataoka K. Advanced drug delivery reviews. 2016;104:61. doi: 10.1016/j.addr.2016.06.011. [DOI] [PubMed] [Google Scholar]; Coleman R. Retrieved from: https://clinicaltrials.gov/ct2/show/NCT01591356?term=siRNA&cond=Cancer&rank=2.; O'Reilly EM, Golan T. Retrieved from: https://clinicaltrials.gov/ct2/show/NCT01676259?term=siRNA&cond=Cancer&rank=13.
- [95].Rosenberg SA. Retrieved from: https://clinicaltrials.gov/ct2/show/record/NCT03480152?term=mRNA&cond=cancer.; Zhan X. Retrieved from: https://clinicaltrials.gov/ct2/show/NCT03468244?term=mRNA&cond=cancer&rank=50.; Kulkarni JA, Cullis PR, Van Der Meel R. nucleic acid therapeutics. 2018;28:146. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- [96].Retrieved from: https://clinicaltrials.gov/ct2/show/NCT03323398?term=mRNA&cond=cancer&rank=13
- [97].Hsu PD, Lander ES, Zhang F. Cell. 2014;157:1262. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yi L, Li J. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2016;1866:197. doi: 10.1016/j.bbcan.2016.09.002. [DOI] [PubMed] [Google Scholar]
- [98].Liu X, Zhang Y, Cheng C, Cheng AW, Zhang X, Li N, Xia C, Wei X, Liu X, Wang H. Cell research. 2017;27:154. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Peek AS. BMC bioinformatics. 2007;8:182. doi: 10.1186/1471-2105-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ge G, Wong GW, Luo B. Biochemical and biophysical research communications. 2005;336:723. doi: 10.1016/j.bbrc.2005.08.147. [DOI] [PubMed] [Google Scholar]; Jagla B, Aulner N, Kelly PD, Song D, Volchuk A, Zatorski A, Shum D, Mayer T, De Angelis DA, Ouerfelli O. Rna. 2005;11:864. doi: 10.1261/rna.7275905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Alabi CA, Love KT, Sahay G, Yin H, Luly KM, Langer R, Anderson DG. Proceedings of the National Academy of Sciences. 2013;110:12881. doi: 10.1073/pnas.1306529110. [DOI] [PMC free article] [PubMed] [Google Scholar]; Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R. Nature biotechnology. 2008;26:561. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ding Hm, Ma Yq. Small. 2015;11:1055. doi: 10.1002/smll.201401943. [DOI] [PubMed] [Google Scholar]
- [102].Torchilin V. Advanced drug delivery reviews. 2011;63:131. doi: 10.1016/j.addr.2010.03.011. [DOI] [PubMed] [Google Scholar]; Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. AACR. 2013 doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Maeda H. Advanced drug delivery reviews. 2015;91:3. doi: 10.1016/j.addr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- [104].Bae YH, Park K. Journal of Controlled Release. 2011;153:198. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Miller MA, Gadde S, Pfirschke C, Engblom C, Sprachman MM, Kohler RH, Yang KS, Laughney AM, Wojtkiewicz G, Kamaly N. Science translational medicine. 2015;7:314ra183. doi: 10.1126/scitranslmed.aac6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Harrington KJ, Mohammadtaghi S, Uster PS, Glass D, Peters AM, Vile RG, Stewart JSW. Clinical Cancer Research. 2001;7:243. [PubMed] [Google Scholar]
- [107].Thorek DL, Chen AK, Czupryna J, Tsourkas A. Annals of biomedical engineering. 2006;34:23. doi: 10.1007/s10439-005-9002-7. [DOI] [PubMed] [Google Scholar]
- [108].Bourdeau RW, Lee-Gosselin A, Lakshmanan A, Farhadi A, Kumar SR, Nety SP, Shapiro MG. Nature. 2018;553:86. doi: 10.1038/nature25021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Stambuk H. Retrieved from: https://clinicaltrials.gov/ct2/show/record/NCT02106598?term=targeted%2C+nanoparticle&rank=3.
- [110].Kiessling F, Mertens ME, Grimm J, Lammers T. Radiology. 2014;273:10. doi: 10.1148/radiol.14131520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Theek B, Rizzo LY, Ehling J, Kiessling F, Lammers T. Clinical and translational imaging. 2014;2:67. doi: 10.1007/s40336-014-0051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kelkar SS, Reineke TM. Bioconjugate chemistry. 2011;22:1879. doi: 10.1021/bc200151q. [DOI] [PubMed] [Google Scholar]
- [112].Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Proceedings of the National Academy of Sciences. 2007;104:15549. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Schluep T, Hwang J, Hildebrandt IJ, Czernin J, Choi CHJ, Alabi CA, Mack BC, Davis ME. Proceedings of the National Academy of Sciences. 2009;106:11394. doi: 10.1073/pnas.0905487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sancey L, Lux F, Kotb S, Roux S, Dufort S, Bianchi A, Cremillieux Y, Fries P, Coll J-L, Rodriguez-Lafrasse C. The British journal of radiology. 2014;87 doi: 10.1259/bjr.20140134. 20140134. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ke H, Wang J, Dai Z, Jin Y, Qu E, Xing Z, Guo C, Yue X, Liu J. Angewandte Chemie International Edition. 2011;50:3017. doi: 10.1002/anie.201008286. [DOI] [PubMed] [Google Scholar]
- [115].Cheng L, Liu J, Gu X, Gong H, Shi X, Liu T, Wang C, Wang X, Liu G, Xing H. Advanced materials. 2014;26:1886. doi: 10.1002/adma.201304497. [DOI] [PubMed] [Google Scholar]
- [116].Hofman M, MacCallum P. Retrieved from: https://clinicaltrials.gov/ct2/show/NCT03392428?term=theranostic&draw=2&rank=6 [Google Scholar]; Erba PA, Ospedaliero A. Retrieved from: https://clinicaltrials.gov/ct2/show/NCT03246659?term=theranostic&draw=2&rank=11 [Google Scholar]; Bushnell D. Retrieved from: https://clinicaltrials.gov/ct2/show/NCT03044977?term=theranostic&draw=2&rank=27 [Google Scholar]; Galgano S. Retrieved from: https://clinicaltrials.gov/ct2/show/NCT03648073?term=theranostic&draw=2&rank=18 [Google Scholar]
- [117].Jensen AI, Severin GW, Hansen AE, Fliedner FP, Eliasen R, Parhamifar L, Kjær A, Andresen TL, Henriksen JR. Journal of Controlled Release. 2018;269:100. doi: 10.1016/j.jconrel.2017.11.006. [DOI] [PubMed] [Google Scholar]
- [118].Hathout RM, Metwally AA. European Journal of Pharmaceutics and Biopharmaceutics. 2016;108:262. doi: 10.1016/j.ejpb.2016.07.019. [DOI] [PubMed] [Google Scholar]
- [119].Cern A, Barenholz Y, Tropsha A, Goldblum A. Journal of controlled release. 2014;173:125. doi: 10.1016/j.jconrel.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Puzyn T, Rasulev B, Gajewicz A, Hu X, Dasari TP, Michalkova A, Hwang H-M, Toropov A, Leszczynska D, Leszczynski J. Nature nanotechnology. 2011;6:175. doi: 10.1038/nnano.2011.10. [DOI] [PubMed] [Google Scholar]
- [121].Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJ. Nature Reviews Cancer. 2018;1 doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Zhou H, Fan Z, Deng J, Lemons PK, Arhontoulis DC, Bowne WB, Cheng H. Nano letters. 2016;16:3268. doi: 10.1021/acs.nanolett.6b00820. [DOI] [PubMed] [Google Scholar]
- [123].Chondros D. Retrieved from: https://clinicaltrials.gov/ct2/show/record/NCT01839487?term=PEGPH20.
- [124].Von Maltzahn G, Park J-H, Lin KY, Singh N, Schwöppe C, Mesters R, Berdel WE, Ruoslahti E, Sailor MJ, Bhatia SN. Nature materials. 2011;10:545. doi: 10.1038/nmat3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Franconi F, Campesi I. Brit J Pharmacol. 2014;171:580. doi: 10.1111/bph.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Serpooshan V, Sheibani S, Pushparaj P, Wojcik M, Jang AY, Santoso MR, Jang JH, Huang H, Safavi-Sohi R, Haghjoo N, Nejadnik H, et al. ACS Nano. 2018;12:2253. doi: 10.1021/acsnano.7b06212. [DOI] [PubMed] [Google Scholar]
- [127].Jeffery M, Hickey BE, Hider PN, See AM. Cochrane Database of Systematic Reviews. 2016 [Google Scholar]
- [128].Broza YY, Kremer R, Tisch U, Gevorkyan A, Shiban A, Best LA, Haick H. Nanomedicine: Nanotechnology, Biology and Medicine. 2013;9:15. doi: 10.1016/j.nano.2012.07.009. [DOI] [PubMed] [Google Scholar]
- [129].Jin H, Huynh T-P, Haick H. Nano letters. 2016;16:4194. doi: 10.1021/acs.nanolett.6b01066. [DOI] [PubMed] [Google Scholar]
- [130].Gao W, Emaminejad S, Nyein HYY, Challa S, Chen K, Peck A, Fahad HM, Ota H, Shiraki H, Kiriya D. Nature. 2016;529:509. doi: 10.1038/nature16521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Gaster RS, Hall DA, Wang SX. Lab Chip. 2011;11:950. doi: 10.1039/c0lc00534g. [DOI] [PubMed] [Google Scholar]
- [132].Bordbar A, McCloskey D, Zielinski DC, Sonnenschein N, Jamshidi N, Palsson BO. Cell systems. 2015;1:283. doi: 10.1016/j.cels.2015.10.003. [DOI] [PubMed] [Google Scholar]
- [133].Kedmi R, Veiga N, Ramishetti S, Goldsmith M, Rosenblum D, Dammes N, Hazan-Halevy I, Nahary L, Leviatan-Ben-Arye S, Harlev M. Nature nanotechnology. 2018;13:214. doi: 10.1038/s41565-017-0043-5. [DOI] [PubMed] [Google Scholar]