Abstract
Purpose:
Although KRAS represents the most commonly mutated oncogene, it has long been considered an “undruggable” target. Novel covalent inhibitors selective for the KRASG12C mutation offer the unprecedented opportunity to target KRAS directly. However, prior efforts to target the RAS-MAPK pathway have been hampered by adaptive feedback, which drives pathway reactivation and resistance.
Experimental Design:
A panel of KRASG12C cell lines were treated with the KRASG12C inhibitors ARS-1620 and AMG 510 to assess effects on signaling and viability. Isoform-specific pulldown of activated GTP-bound RAS was performed to evaluate effects on the activity of specific RAS isoforms over time following treatment. RTK inhibitors, SHP2 inhibitors, and MEK/ERK inhibitors were assessed in combination with KRASG12C inhibitors in vitro and in vivo as potential strategies to overcome resistance and enhance efficacy.
Results:
We observed rapid adaptive RAS pathway feedback reactivation following KRASG12C inhibition in the majority of KRASG12C models, driven by RTK-mediated activation of wild type RAS, which cannot be inhibited by G12C-specific inhibitors. Importantly, multiple RTKs can mediate feedback, with no single RTK appearing critical across all KRASG12C models. However, co-inhibition of SHP2, which mediates signaling from multiple RTKs to RAS, abrogated feedback reactivation more universally, and combined KRASG12C/SHP2 inhibition drove sustained RAS pathway suppression and improved efficacy in vitro and in vivo.
Conclusions:
These data identify feedback reactivation of wild type RAS as a key mechanism of adaptive resistance to KRASG12C inhibitors and highlight the potential importance of vertical inhibition strategies to enhance the clinical efficacy of KRASG12C inhibitors.
Keywords: KRAS, adaptive resistance, SHP2, KRASG12C
STATEMENT OF TRANSLATIONAL RELEVANCE
KRAS is the most commonly mutated oncogene in human cancer, and new mutant-specific inhibitors of KRAS, such as covalent inhibitors of KRASG12C, offer the unprecedented opportunity to target mutant KRAS directly. However, prior efforts targeting the RAS-MAPK pathway have been constrained by adaptive feedback reactivation of pathway signaling. We describe how adaptive feedback through multiple RTKs can drive resistance to KRASG12C inhibition through compensatory activation of wild type RAS isoforms, which cannot be inhibited by G12C-specific inhibitors. Our data suggest that vertical pathway inhibition strategies, and in particular combinations of KRASG12C inhibitors with SHP2 inhibitors—which can interrupt feedback from multiple RTKs—may be critical to abrogate feedback reactivation of the RAS pathway following KRASG12C inhibition and may represent a promising therapeutic approach for KRASG12C cancers.
INTRODUCTION
RAS is the most frequently mutated oncogene in cancer, with KRAS mutations being the most predominant of the three RAS isoforms (HRAS, NRAS and KRAS) (1). In its wild type form, RAS cycles between the GDP-bound inactive state and GTP-bound active state, and when mutated at the most common G12, G13, and Q61 loci, KRAS is in a constitutively active GTP-bound state. Mutant RAS has long been considered an undruggable target, and thus most therapeutic strategies have focused on targeting downstream effector pathways such as the ERK MAPK cascade (2). However, there has been limited clinical success in targeting downstream effectors, and other approaches of targeting RAS function have been met with limited success (2).
Recently, covalent inhibitors targeting a specific KRAS mutation—Glycine 12 to cysteine (G12C)—have been developed that show encouraging preclinical efficacy in KRASG12C tumor models (3–5). These inhibitors undergo an irreversible reaction with the mutant cysteine present only in G12C mutant KRAS, making them highly selective for KRASG12C versus wild type KRAS or other RAS isoforms. The inhibitors function by locking KRASG12C in an inactive GDP bound state, exploiting the unique property of KRASG12C to cycle between the GDP- and GTP-bound states (6,7). The KRASG12C mutation represents 11% of all KRAS mutations (COSMIC v89)(1,8), but is the most common RAS mutation in lung cancer and also occurs in many other types of cancer, such as colon and pancreatic cancers. Two KRASG12C inhibitors have entered clinical trials: AMG510 ( NCT03600883) and MRTX1257 ( NCT03785249). As the first such agents capable of inhibiting mutant KRAS directly, this class of agents offers an unprecedented therapeutic opportunity to target this critical oncogene.
However, previous efforts to target the RAS-RAF-MEK pathway have been hindered by adaptive feedback reactivation of pathway signaling as a major mode of therapeutic resistance. For example, BRAF inhibition in BRAFV600 mutant cancers leads to loss of negative feedback signals regulated by the MAPK pathway, leading to receptor tyrosine kinase (RTK)-mediated reactivation of MAPK signaling through wild type RAS and RAF, particularly in specific tumor types, such as colorectal cancer (9–12). Similarly, in KRAS mutant cancers, MEK inhibitor treatment leads to adaptive feedback activation of RAS signaling, often through EGFR or other human epidermal growth factor receptor (HER) family members or FGFR, limiting efficacy (13,14). Indeed, early preclinical studies with KRASG12C inhibitors, such as ARS-1620, have suggested a potential role for adaptive feedback as a mechanism of resistance (7) (15). Early clinical data with the KRASG12C inhibitor AMG510 was recently reported, showing an initial overall response rate of 24% (13/55) in patients with KRASG12C mutant cancers, raising the possibility that overcoming adaptive resistance could be a vital next step in improving efficacy (16). Therefore, understanding the potential mechanisms driving adaptive feedback resistance to KRAS inhibition may be critical to the development of future therapeutic strategies.
Here, we evaluate the adaptive feedback response to KRASG12C inhibition and find evidence of rapid RAS pathway reactivation in the majority of KRASG12C models. We observe that feedback reactivation is driven in large part by RTK-mediated activation of wild type RAS (NRAS and HRAS), which is not inhibited by G12C-specific inhibitors. Importantly we find that this process is mediated by multiple RTKs, and that no single RTK dominates across all KRASG12C models, suggesting that a strategy co-targeting a single RTK to block adaptive resistance may be ineffective. However, we observe that co-targeting of SHP2 (17,18), a key phosphatase that mediates signaling from multiple RTKs to RAS, is able to abrogate feedback reactivation of RAS signaling following KRASG12C inhibition across all models, and that the combination of KRASG12C and SHP2 inhibitors leads to sustained RAS pathway suppression and improved efficacy in vitro and in vivo. These data suggest that vertical inhibition strategies to block adaptive RAS pathway reactivation may be key to enhance the efficacy of KRASG12C inhibitors.
MATERIALS AND METHODS
Cell lines and inhibitors
Cell lines were obtained from ATCC or the Center for Molecular Therapeutics at the MGH Cancer Center (Boston, MA), which routinely performs cell line authentication testing by SNP and short tandem repeat analysis, and maintained in DMEM/F12 supplemented with 10% fetal calf serum, and were not cultured longer than 6 months after receipt from cell banks. ARS-1620, SHP099, RMC-4550 erlotinib, afatinib, crizotinib, and BGJ398 used for in vitro studies were purchased from Selleckchem and AMG 510 was purchased from MedChemExpress. ARS-1620 used in in vivo studies was purchased from MedchemExpress and SHP099, afatinib, and BGJ398 used in in vivo studies were purchased from Selleckchem.
Inhibitor treatment assays
Short term sensitivity to ARS-1620 was determined by CellTiter-Glo (Promega). Briefly, cell lines were seeded at 1–2×103 cells/well of a 96-well plate and 24 h after seeding, a serial dilution of ARS-1620 was added to cells After 72 h of inhibitor treatment, plates were developed with CellTiter-Glo and luminescence read on a plate reader (Molecular Devices). For long-term viability assays, cells were plated at low density 2×102-3×103 in 6-well plates and treated with ARS-1620 (1 µM) alone or in combination with SHP099 (10 µM), erlotinib, afatinib, crizotinib or BGJ398 (all 1µM) for 10–14 days with drug refreshed every 2–3 days. Assays were fixed and stained with a crystal violet solution (4% formaldehyde) and plates were scanned using a photo scanner and cell growth was quantified using ImageJ software.
Inhibitor treatment and Western analyses
Cell lines were treated with ARS-1620, SHP099, or a combination for 4, 24, 48 or 72 h before samples were collected in NP40 lysis buffer. Whole cell lysates were resolved on 4–12% Bis-Tris gels (Thermo Fisher) and western blotting was performed using antibodies against phospho-CRAF (S338), phospho-MEK (S217/221), phospho-ERK (T202/Y204), MYC, phospho-AKT (S473) (Cell Signaling Technologies), phospho-p90 RSK (Abcam), and GAPDH (Millipore). Densitometry analysis was performed using ImageJ software.
RAS-GTP pulldown
After indicated inhibitor treatment, RAS activity was assessed by GST-RAF-RBD pulldown (Cell Signaling Technologies), followed by immunoblotting with pan-RAS or RAS isoform–specific antibodies. Pulldown samples and whole-cell lysates were resolved on 4–12% Bis-Tris Gels and western blotting was performed using antibodies against KRAS (Sigma), NRAS (Santa Cruz), HRAS (Proteintech), and pan-RAS (Cell signaling). Densitometry analysis was performed using ImageJ software.
Quantitative PCR
Total RNA was isolated using an RNeasy kit (Qiagen) and reverse transcription was performed using the qScript cDNA SuperMIx (Quantabio ) Real time quantitative Taqman PCR was performed on the Light Cycler 480 (Roche) with FAM/MGB labeled probes against DUSP6 (Hs04329643_s1, Thermo Fisher) and endogenous control VIC/MGB labeled β-actin (Thermo Fisher).
Xenograft studies
SW837 and MIA PaCa-2 cell lines (5×106) were injected into 6–8 week old female athymic nude mice (Charles River Laboratories. Treatment of ARS-1620 (200mg/kg), afatinib (12.5 mg/kg), BGJ398 (20 mg/kg) and SHP099 (75mg/kg) by daily oral gauvage was initiated when tumor size reached 100–200 mm3 and tumor size was assessed by caliper measurements for 21–25 days. All animal studies were performed through the Institutional Animal Care and Use Committee (IACUC).
Graphical analysis
Data were analyzed using GraphPad Prism 6 software and curve fit and GI50 values were generated as indicated in the Figure Legends.
RESULTS
Feedback reactivation of RAS signaling following KRASG12C inhibition
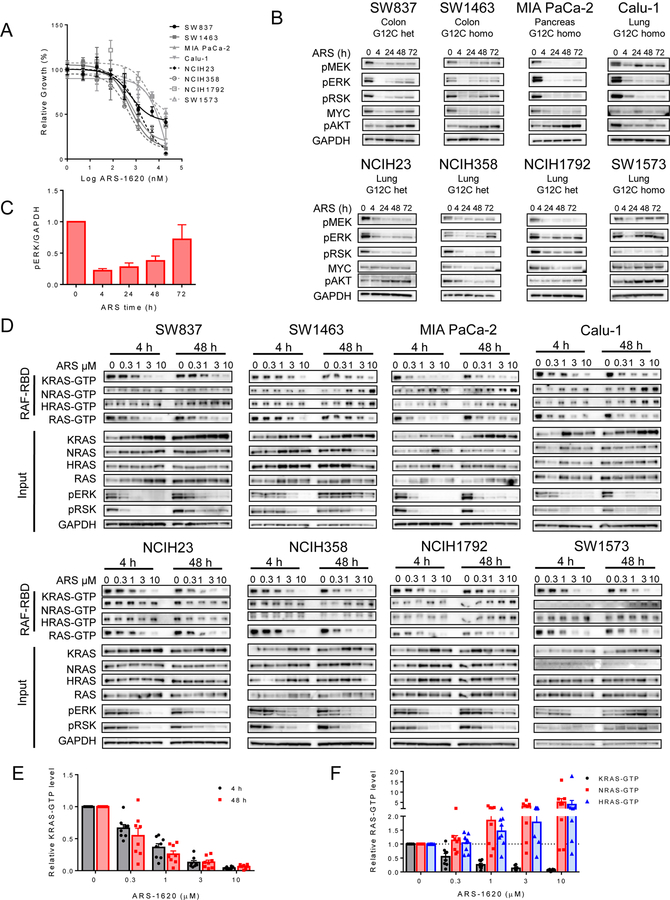
To model potential mechanisms constraining the efficacy of KRASG12C inhibition, we evaluated the effects of the KRASG12C inhibitor ARS-1620 across a panel of eight KRASG12C-mutant cell lines (Fig. 1A). To explore the potential role of adaptive feedback in the setting of KRASG12C inhibition, we assessed the effects of ARS-1620 in each model over time from 4 hours to 72 hours. In all cell lines, ARS-1620 suppressed MAPK pathway signaling, a key downstream effector pathway of KRAS, as measured by inhibition of phospho-MEK, phospho-ERK, and phospho-RSK at 4 hours (Figs. 1B–C). In addition, suppression of total MYC protein levels, which are highly regulated by RAS and ERK activity, was observed (19,20). However, by 24–48 hours, RAS-MAPK pathway signaling began to rebound from the nadir of pathway suppression, leading to pathway reactivation and incomplete suppression by 72 hours (Figs. 1B–C). A similar rebound in RAS-MAPK signaling was also observed with the more recently available clinical KRASG12C inhibitor AMG 510 (Supplementary Fig. S1) Importantly, while some variability in the degree of pathway reactivation was observed across models, with some such as the Calu-1 cell exhibiting little to no rebound, RAS-MAPK pathway activity, as assessed by phospho-ERK levels, rebounded on average to ~75% of baseline levels by just 72 hours. The rapid and consistent reactivation of signaling observed following KRASG12C inhibition suggests that adaptive feedback may have the potential to limit efficacy for this class of inhibitors.
Figure 1. Feedback reactivation of RAS signaling occus following KRASG12C inhibition.
(A) Cell lines were treated for 72 h with a dose titration of ARS-1620 and viability was measured by CellTiter-Glo. (B) KRAS-G12C mutant cell lines were treated with ARS-1620 (1 µM) for 0, 4, 24, 48, and 72 h. Blot analysis was performed for phospho- (p)MEK, pERK, pRSK, pAKT, and total MYC with GAPDH as a loading control. (C) Densitometry of phospho-ERK normalized to GAPDH for blots in, results represent an average of phospho-ERK across all 8 cell lines (A).(D) Cell lines were treated with a dose titration of 0.3–10 µM ARS-1620 for 4 or 48 h and lysates were subject to a RAF-RBD pulldown and blot analysis of KRAS, NRAS, HRAS and total RAS as well as pERK, pRSK and GAPDH for input samples. (E) Densitometry analysis of 4 and 48 h KRAS-GTP levels normalized to input KRAS and GAPDH loading control in D (F) Densitometry analysis of 48 h KRAS-GTP, NRAS-GTP, and HRAS-GTP levels normalized to input RAS and GAPDH loading control of blots in (D). Densitometry results in (E) and (F) represent an average across all 8 cell lines.
To better understand the mechanism of feedback reactivation in the setting of KRASG12C inhibition, we assessed the effects of KRASG12C inhibition with ARS-1620 across a range of concentrations from 0.3 to 10 µM at 4 and 48 hours on levels of active GTP-bound KRAS and wild type RAS (NRAS, HRAS), as well as downstream signaling (Fig 1D). ARS-1620 effectively suppressed KRAS-GTP levels as measured by a RAF-RAS binding domain (RBD) pulldown assay in all cell lines in a dose-dependent manner, consistent with suppression of KRASG12C activity (Figs. 1D–E). However, consistent with our data from Figs. 1B–C, evidence of a rebound in downstream pathway activation (phospho-ERK, phospho-RSK) was observed by 48 hours, relative to 4 hours. Importantly, this rebound reactivation was observed even at the highest concentrations of ARS-1620, suggesting that pathway rebound is not due simply to inadequate levels of inhibitor. Importantly, in most cell lines, increases in the levels of the active GTP-bound forms of the wild type RAS isoforms (NRAS-GTP, HRAS-GTP) were noted by 48 hours in a dose dependent manner (although levels of HRAS and NRAS expression were quite low in some cell models) indicating that induction of wild type RAS activity may play a key role in feedback reactivation, particularly since no clear rebound in KRAS-GTP levels was noted between 4 and 48 h of treatment at any inhibitor concentration (Figs. 1E–F). In fact, on average we observed a 4 to 5-fold increase in NRAS-GTP and HRAS-GTP levels by 48 hours following treatment with the highest concentrations of ARS-1620 across all models. Similarly, we also observed a strong induction of both NRAS-GTP and HRAS-GTP levels and rebound in downstream MAPK signaling (phospho-ERK, phospho-RSK) upon treatment with AMG 510 (Supplementary Fig. S1). Taken together, these data suggest that adaptive feedback reactivation of pathway signaling following KRASG12C inhibition corresponds with a rebound in RAS activity, in particular an increase in wild-type RAS activity, although the exact mechanism of RAS reactivation may differ across KRASG12C cancers. Importantly, these data suggest that blocking adaptive feedback signals driving pathway may be necessary to enhance the efficacy of KRASG12C inhibitors.
Adaptive resistance to KRASG12C inhibition is driven by multiple RTKs
To identify potential strategies to inhibit feedback reactivation following KRASG12C inhibition, we attempted to define the specific mechanisms driving adaptive pathway rebound. Prior work has demonstrated that adaptive feedback reactivation of RAS-MAPK signaling can be driven by RTKs in response to RAF inhibition and MEK inhibition in BRAF- and KRAS-mutant cancers (11,13,21,22) (21–23). To assess the role of RTKs in feedback reactivation in response to KRASG12C inhibition, we analyzed the levels of RTK activation at baseline and after 48 hours of ARS-1620 treatment by a phospho-RTK array (Supplementary Fig. S2). We observed high basal levels of several phosphorylated RTKs, including phospho-EGFR in most cell lines. Additionally, following 48 hours of treatment with ARS-1620, we observed an adaptive increase in the phosphorylation levels of multiple RTKs, including EGFR, HER2, FGFR, and c-MET, but with a highly heterogeneous pattern across cell line models, suggesting that multiple RTKs may play a role in adaptive feedback to KRASG12C inhibition. Previous studies have suggested that key RTKs may play dominant roles in driving adaptive feedback to inhibitors of the MAPK pathway, including EGFR in response to BRAF inhibition in BRAFV600E colorectal cancer and EGFR/HER family kinases or FGFR in response to MEK inhibitors in KRAS-mutant lung cancers (13,14) (21,24). However, the potential for multiple RTKs to contribute to adaptive resistance in each of the paradigms above may limit efforts to block adaptive feedback by targeting a single RTK. Similarly, the variable induction of multiple RTKs across different KRASG12C models following ARS-1620 treatment and different levels of phospho-RTKs at baseline suggests that distinct and/or multiple RTKs may drive adaptive feedback across different KRASG12C cancers, and that strategies targeting a single RTK may not be universally effective. In this scenario, the RTK associated phosphatase SHP2 may represent a common node through which to inhibit feedback reactivation of RAS signaling by multiple RTKs that may preserve efficacy across heterogeneous KRASG12C cancers.
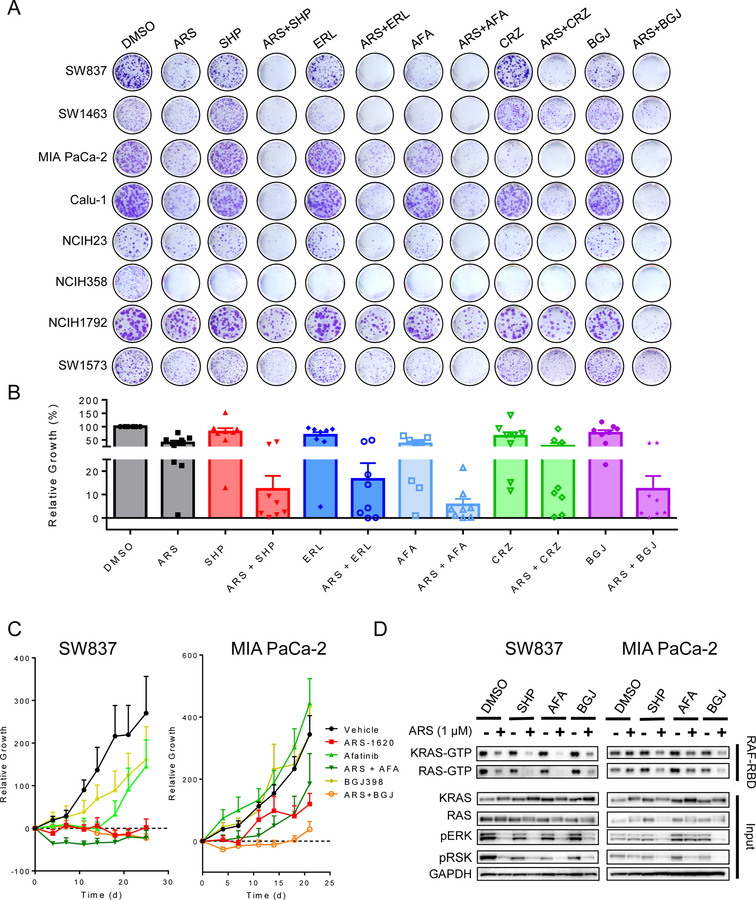
Thus, to assess whether vertical inhibition of the RAS-MAPK pathway could block feedback reactivation and enhance the efficacy of KRASG12C inhibition, we evaluated whether co-inhibition of individual RTKs or SHP2 could augment the efficacy of ARS-1620 in suppressing cell viability (Figs. 2A, B; Supplementary Fig. S3). Interestingly, in each cell line, inhibition of a specific RTK in combination with ARS-1620 led to a marked decrease in cell viability relative to ARS-1620 alone. However, KRASG12C cell lines demonstrated a heterogeneous response to RTK inhibition, with select inhibitors enhancing anti-tumor effect in combination, relative to ARS-1620 alone. Overall, the pan-HER inhibitor afatinib and the FGFR inhibitor BGJ398 displayed the greatest and most consistent cooperativity with ARS-1620, though neither combination was universally effective in all models. For example, SW1463 showed primary dependence on EGFR/HER family signaling (erlotinib, afatinib), as compared to FGFR signaling (BGJ398), whereas MIA Paca-2 appeared to be more dependent on FGFR signaling and showed less dependence on EGFR/HER family signaling. Notably, while some cell lines showed heightened dependence on a single RTK, SHP2 inhibition was effective in suppressing adaptive resistance to KRASG12C inhibition across all cell lines. These results support that multiple RTKs can contribute to adaptive feedback resistance to KRASG12C inhibition, and that combined inhibition of RTKs or SHP2 and KRASG12C could represent a more effective therapeutic strategy to overcome resistance and improve efficacy in KRAS G12C-mutant tumors. Furthermore, as strong feedback reactivation of MAPK signaling downstream of RAS is observed following KRASG12C inhibition, we also assessed potential combinations of KRASG12C inhibition with MEK and ERK inhibitors, which also displayed potential cooperativity across the panel of KRASG12C models, though strong induction of RAS activity was observed with these combinations (Supplementary Fig. S5). Together, these data support the potential of vertical pathway inhibition strategies to overcome adaptive resistance to KRASG12C inhibition.
Figure 2. KRASG12C inhibitors are subject to adaptive feedback through activation of RTKs.
(A) Cell lines were treated with ARS-1620 (1 µM), SHP099 (10 µM), erlotinib (1 µM), afatinib (1 µM), crizotinib (1 µM), or BGJ398 (1 µM) or a combination for 10–14 days and then stained with crystal violet. (B) Quantification of assays in A. (C) SW837 and MIA PaCa-2 xenografts treated daily with ARS-1620 (200 mg/kg), afatinib (12.5 mg/kg), BGJ398 (20 mg/kg) or combination for 25 and 21 days respectively. (D) SW837 and MIA PaCa-2 cell lines were treated ARS-1620 (1 µM), SHP099 (10 µM), afatinib (1 µM), or BGJ398 (1 µM) or a combination and lysates were subject to a RAF-RBD pulldown and blot analysis of KRAS-GTP and total RAS-GTP as well as pERK, pRSK and GAPDH for input samples.
To test whether individual RTK inhibition in combination with KRASG12C inhibition might represent a promising therapeutic strategy, we investigated the in vivo efficacy of KRASG12C inhibition in combination with the RTK inhibitors that demonstrated the greatest cooperativity with ARS-1620 in vitro: the pan-HER inhibitor afatinib and the FGFR inhibitor BGJ398. ARS-1620 demonstrated modest efficacy as a single agent in both the SW837 and MIA PaCa-2 xenograft models (Fig. 2C; Supplementary Fig. S4). Interestingly, while neither reached statistical significance, a clear trend toward enhanced efficacy relative to ARS-1620 alone was observed with a specific RTK combination in each model. However, the predominant RTK dependency of each model differed, with the afatinib combination appearing to produce the greatest effect in SW837 xenografts (though not significant, p=0.37 vs ARS-1620 alone), in contrast with MIA PaCa-2 xenografts, in which the BGJ398 combination appeared to produce the greatest effect (though not significant, p=0.13 vs ARS-1620 alone), similar to their in vitro profiles (Fig. 2A). Consistent with these observations, we found that adaptive feedback reactivation in the setting of KRASG12C inhibition exhibited a similarly variable RTK dependence in each model for maintaining both KRAS-GTP and total RAS-GTP levels (Fig. 2D), with the SW837 model more dependent on HER signaling, and MIA PaCa-2 more dependent on FGFR signaling. However, we found that SHP2 inhibition could enhance suppression of active RAS in combination with ARS-1620 across both models, supporting the potential for SHP2 as a common downstream node through which to inhibit signaling from multiple RTKs. Taken together, these data support the potential for vertical inhibition strategies to suppress adaptive feedback resistance to KRASG12C inhibition, but also suggest that co-inhibition of individual RTKs may not be broadly effective across KRASG12C cancers.
SHP2 inhibition reduces feedback reactivation in response to KRASG12C inhibitors
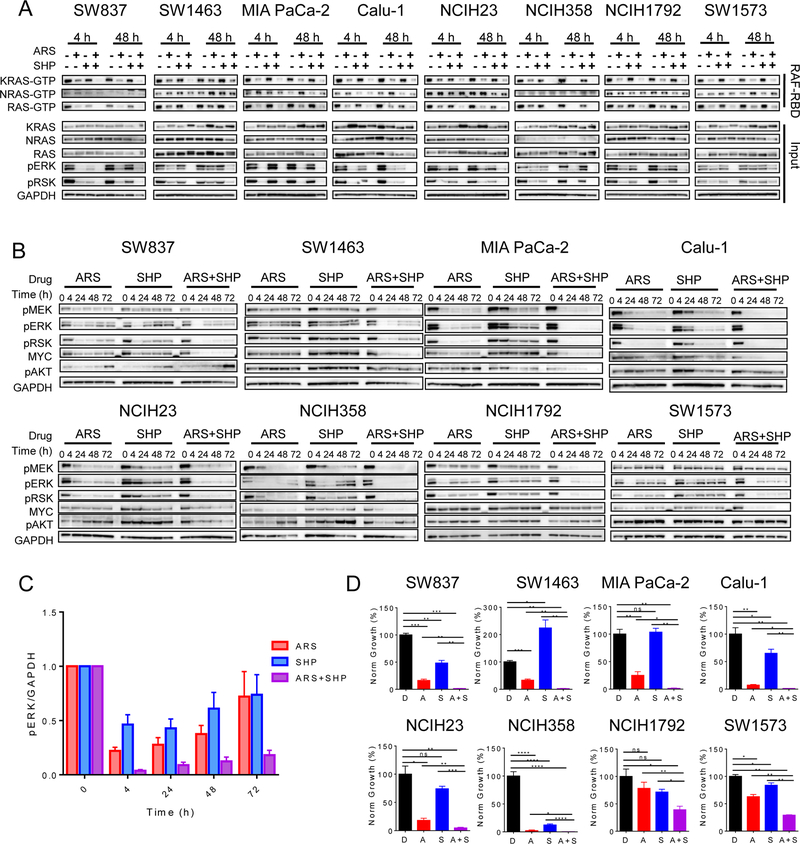
While our data suggest that SHP2 inhibitors could be a promising combination partner for KRASG12C inhibitors, SHP2 inhibitors as single agents were recently proposed as a potential therapeutic strategy for KRASG12C cancers (25). SHP2 acts to dephosphorylate and activate multiple nodes of RAS signaling downstream of RTKs, enhancing signaling through the RAS-RAF-MEK-ERK cascade (25–28). Thus, inhibition of SHP2 has the potential to block the ability of multiple RTKs to activate RAS. Because KRASG12C actively cycles between its GDP-bound inactive state and its GTP-bound active state, KRASG12C may be uniquely dependent on some level of basal upstream activation from RTKs to maintain activity. Indeed, SHP2 inhibition alone was shown to have anti-tumor effects in vitro and in vivo (25). However, we found that the SHP2 inhibitor SHP099 led to incomplete suppression of KRAS-GTP levels as a single agent (Fig. 3A).
Figure 3. SHP2 inhibition enhances the efficacy of KRASG12C inhibition.
(A) Cell lines were treated with ARS-1620 (1 uM), SHP099 (10 uM), or a combination for 4 or 48 h and lysates were subject to a RAF-RBD pulldown and blot analysis of KRAS, NRAS, HRAS and total RAS as well as pERK, pRSK and GAPDH for input samples. (B) KRASG12C mutant cell lines were treated with ARS-1620 (1 µM), SHP099 (10 µM) or a combination for 0, 4, 24, 48, and 72 h. Blot analysis was performed for pMEK, pERK, pRSK, pAKT, total MYC with GAPDH as a loading control. (C) Densitometry analysis of pERK normalized to GAPDH was performed for all cell lines (D) Quantification of crystal violet stain of cell lines treated with ARS-1620 (1 µM), SHP099 (10 µM), or a combination for 10–14 days, statistical significance was evaluated by Student’s t-test, where *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001.
However, in all cell lines, concurrent ARS-1620 and SHP099 treatment led to more complete suppression of both KRAS-GTP and total RAS-GTP levels when compared to either agent alone at 48 hours (Fig. 3A). SHP099 also reduced NRAS-GTP levels in several cell lines and abrogated the adaptive increase in activated NRAS-GTP levels by 48 hours induced by ARS-1620 in some models. The reduction in KRAS-GTP and RAS-GTP levels translated to further suppression of the MAPK pathway downstream of RAS by 48 hours. Thus, our results suggest that SHP2 inhibition can mitigate the induction of wild type RAS activity that drives adaptive feedback reactivation of RAS and MAPK pathway signaling in response to KRASG12C inhibition.
Combined inhibition of KRASG12C and SHP2 with ARS-1620 and SHP099 also led to more complete suppression of the MAPK pathway over time in all models, exhibiting a strong reduction in feedback reactivation of the pathway (Fig. 3B–C; Supplementary Fig. S6). A similar pattern of more complete suppression of the MAPK pathway was also observed with ARS-1620 combined with the SHP2 inhibitor RMC-4550 (Supplementary Fig. S5) and AMG 510 combined with either SHP099 or RMC-4550 (Supplementary Fig. S1). Interestingly, the effects of KRASG12C or SHP2 inhibition on PI3K/AKT signaling, as measured by phospho-AKT, was highly variable across cell lines, with a reduction noted in some cell lines (SW1463, MIA PaCa-2, NCIH23, NCIH358) and unaffected or increased levels noted in other cell lines, which is consistent with prior studies suggesting that PI3K is not universally tied to RAS activity (15,29). Combined KRASG12C and SHP2 inhibition more effectively suppressed tumor cell viability at 72 hours, leading to GI50 shifts of ~5 to 30-fold in six of eight KRASG12C models (Supplementary Fig. S7). Interestingly, the effects of combined KRASG12C and SHP2 inhibition on cell viability were more pronounced in long-term viability assays. While some models (NCIH358, Calu-1) demonstrated marked suppression of viability with ARS-1620 alone, combined KRASG12C and SHP2 inhibition led to a statistically significant reduction in cell number relative to either agent alone in all KRASG12C models, while having no effect in the KRASG12D Ls174T model (Fig. 3D, Supplementary Fig. S7, S8).
Combined KRASG12C and SHP2 inhibition drives tumor regressions in vivo
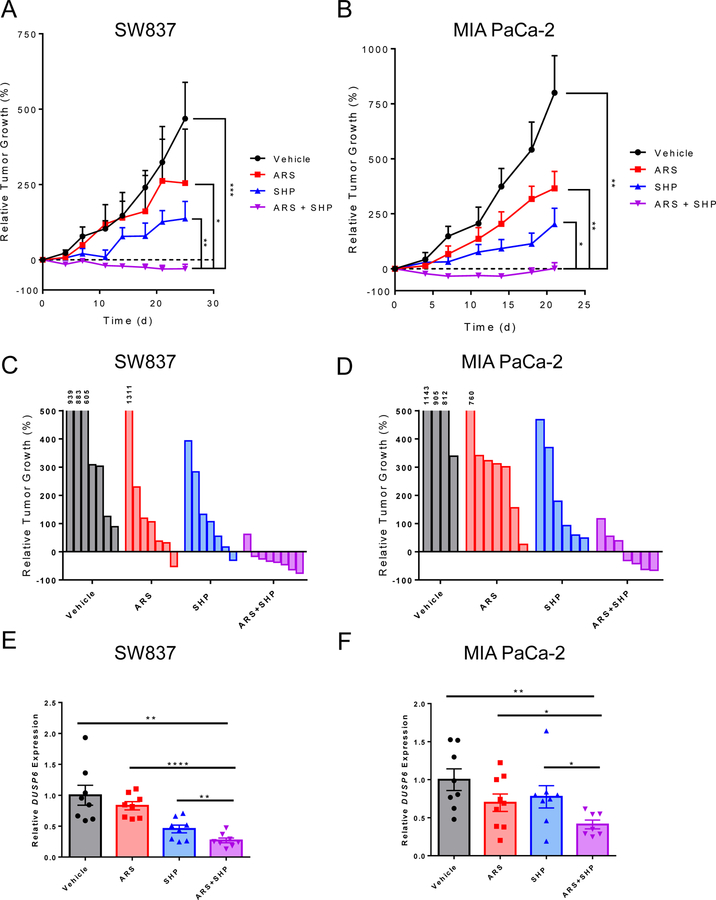
To better assess combined KRASG12C and SHP2 inhibition as a potential therapeutic strategy for KRASG12C cancers, we investigated the efficacy of combined KRASG12C and SHP2 inhibition in vivo. While a reduction in tumor growth relative to vehicle control was observed in both SW837 and MIA PaCa-2 xenograft models with single agent ARS-1620 or SHP099, the combination of ARS-1620 and SHP099 showed a statistically significant increase in anti-tumor efficacy compared to each agent alone and led to tumor regressions in the majority of tumors in each model (Figs. 4A, B; Supplemental Fig. S9). The efficacy of SHP099 monotherapy in vivo was more striking than seen in our in vitro experiments, supporting previous findings that SHP2 perhaps plays a more important role in KRASG12C-driven tumor growth in vivo (27,28). Since our in vitro data suggest that SHP2 inhibition can abrogate adaptive feedback reactivation of RAS pathway signaling following KRASG12C inhibition, we evaluated the degree of RAS-MAPK pathway inhibition in tumors after 4 days of each treatment by assessing transcript levels of ERK transcriptional targets (30,31). Consistent with the feedback reactivation observed in vitro, we observed modest and incomplete suppression of ERK-dependent transcripts, such as DUSP6 after 4 days of treatment with either ARS-1620 or SHP099 alone (Figs. 4E,F and Supplementary Fig. S9). However, combined treatment with ARS-1620 and SHP099 maintained a greater decrease after 4 days of treatment compared to each inhibitor alone. These results support that combined KRASG12C and SHP2 inhibition may overcome adaptive feedback resistance and may represent a promising therapeutic strategy for future clinical trials in KRASG12C mutant cancers.
Figure 4. SHP2 inhibition enhances the efficacy of KRASG12C inhibition in vivo.
(A) SW837 and (B) MIA PaCa-2 xenografts treated daily with ARS-1620 (200 mg/kg), SHP099 (75 mg/kg), or both for 25 and 21 days respectively; statistical significance was evaluated by Mann-Whitney test where *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001. Waterfall plots of (C) SW837 and (D) MIA PaCa-2 tumors endpoint tumors from (A) and (B) respectively (E) SW837 and (F) MIA PaCa-2 tumors treated with ARS-1620, SHP099, or both for 4 days and Taqman quantitative PCR was performed to monitor changes in DUSP6 gene transcription and β-actin was used as an endogenous control, statistical significance was evaluated by Student’s t-test, where *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001.
DISCUSSION
KRASG12C inhibitors represent the first potential opportunity to target mutant KRAS directly in patients. However, adaptive feedback resistance has been a key limitation in prior clinical efforts to target the RAS-MAPK pathway. For example, RTK-driven feedback reactivation of RAS-MAPK signaling has been identified as a key driver of resistance in BRAFV600E cancers treated with BRAF inhibitors (colorectal cancer in particular) and in KRAS-mutant cancers treated with MEK inhibitors (11,12,32). Thus, this prior experience predicts that adaptive resistance may pose a key obstacle for KRASG12C inhibitors, as well, and that strategies to overcome this mechanism may be key to improving clinical efficacy. In line with this hypothesis, we find consistent evidence of rapid feedback reactivation of RAS pathway signaling in KRASG12C cancer models following treatment with KRASG12C inhibitors. We observe that feedback reactivation is driven by multiple RTKs and that the pattern of RTK-dependence is highly heterogeneous across KRASG12C models. RTK-driven feedback may contribute to RAS reactivation through two mechanisms. First, as described by Lito et al, increased RTK activity may lead to increased cycling of KRASG12C to its active GTP-bound form that hinders the binding of most KRASG12C inhibitors, which bind specifically to the inactive GDP-bound form (7). Second, in this study we see clear evidence of RTK-driven induction of wild type RAS (NRAS or HRAS) activity following KRASG12C inhibitor treatment, which can reactivate signaling in a KRASG12C-independent manner. Importantly, while GTP-bound state-selective KRASG12C inhibitors have been proposed as one possible solution to the first mechanism (increased RTK-driven cycling of KRASG12C to its active GTP-bound state), this mechanism of action is unlikely to be effective against the second mechanism of adaptive feedback (RTK-driven activation of wild type RAS) (33). Thus, given this novel potential role for wild type RAS, our data suggest that interrupting the adaptive feedback loop following KRASG12C inhibition may be critical to overcoming adaptive resistance.
Targeting a dominant RTK driving adaptive feedback reactivation is an attractive strategy to suppress adaptive resistance, as it intercepts the critical feedback loop at its most upstream point. Indeed, in BRAFV600 colorectal cancer, targeting EGFR—the RTK thought to be the primary driver of feedback reactivation from preclinical studies—in combination with BRAF inhibition, has led to an improvement in clinical efficacy (34–39). However, preclinical and clinical data have demonstrated that other RTKs can drive feedback in an EGFR-independent manner, and that the combination of BRAF and EGFR only suppresses signaling in a subset of patients (34). Similarly, we note that multiple RTKs appear to be involved in feedback reactivation following KRASG12C inhibition and that the pattern of RTK-dependence is highly variable between KRASG12C mutant cancers. Interestingly, our data do indicate that some RTK inhibitors—in particular the pan-HER inhibitor afatinib and the FGFR inhibitor BGJ398—are effective in combination with KRASG12C inhibitors in many models, suggesting that pan-HER or FGFR co-targeting could be promising strategies. This finding is consistent with prior studies identifying HER family and FGFR signaling as key mediators of adaptive feedback to RAS-MAPK pathway inhibitors (10–14,21,22) and recent functional genomic screens(40). However, for each of these inhibitors, there are models in which pathway feedback reactivation appears independent of each specific RTK—i.e. MIA PaCa2 for afatinib, and SW1463 for BGJ398 (Fig. 2A)—and RTK combinations showed differential effects across in vivo models (Fig. 2C), suggesting that co-targeting a single RTK is unlikely to be universally effective.
Conversely, targeting SHP2 provides the potential opportunity to intercept a common signaling node linking multiple RTKs to RAS signaling. Indeed, inhibition of SHP2 was recently shown to overcome RTK-driven adaptive feedback mechanisms to MEK inhibition, and enhance the efficacy of MEK inhibitor induced growth suppression in vitro and in vivo in RAS-driven cancers (27,41–44). Furthermore, SHP2 inhibition was also demonstrated to have single-agent efficacy in preclinical KRASG12C models by reducing cycling to the active GTP-bound state, offering an additional advantage as a potential combination partner (25). Our data support SHP2 as a promising combination partner for KRASG12C inhibitors, capable of abrogating adaptive feedback reactivation from multiple RTKs to maintain RAS pathway suppression and enhance efficacy in vitro and in vivo. Although some models showed the greatest sensitivity to KRASG12C inhibition in combination with a specific RTK inhibitor, combined KRASG12C and SHP2 inhibition suppressed pathway signaling and enhanced efficacy in all models, supporting its potential as a more universal approach. Moreover, we observed that SHP2 inhibition could consistently block the adaptive increase in wild type RAS (HRAS, NRAS) activation observed after KRASG12C inhibition in many models (Fig. 3A). While the results of initial clinical trials will demonstrate whether SHP2 inhibitors will have a viable therapeutic index in patients, these data suggest that SHP2 inhibitors, either alone or perhaps in combination with specific RTK inhibitors or downstream inhibitors of the RAS-MAPK pathway, could be promising therapeutic strategies for future clinical trials.
The initial reports of single-agent activity of KRASG12C inhibitors in patients with KRASG12C cancers are promising and support that mutant RAS represents a valid clinical target. However, not all patients respond to therapy, and the durability of benefit may be limited, suggesting that strategies to target key resistance mechanisms may improve clinical efficacy. In 55 patients with KRASG12C cancers treated with the KRASG12C inhibitor AMG-510, a 24% overall response rate was reported, with 11 responses observed in 23 non-small cell lung cancer (NSCLC) patients, but only 1 response seen in 26 colorectal cancer patients(16). Interestingly, this pattern of tumor type-specific difference in response is highly consistent with prior examples of adaptive resistance. For instance, BRAF inhibitors led to a monotherapy response rate of >50% in BRAFV600E melanoma and ~30% in BRAFV600E NSCLC, but only ~5% in BRAFV600E colorectal cancer, likely due to the variable robustness of adaptive feedback networks in each tumor type (45–49). Still, in all cases, combination therapies to suppress adaptive feedback proved key to optimizing the frequency and durability of response in each tumor type (34,50,51). Similarly, our data suggest that therapeutic combinations will be key to overcoming adaptive resistance and enhancing clinical benefit of KRASG12C inhibitors. Initially, combinations may need to focus on improving suppression of RAS signaling by combining KRASG12C inhibitors with agents targeting adaptive feedback—such as RTK inhibitors, SHP2 inhibitors, or perhaps downstream pathway inhibitors (i.e. MEK or ERK)—and determining the most effective clinical strategy may be a critical first step. However, additional combinations exploring potential synthetic lethal interactions with other pathways may also be key to maximizing efficacy. For example, a study by Misale et al, suggested that targeting the PI3K/AKT pathway, which is often activated independently of RAS, may improve the efficacy of KRASG12C inhibitors and a study by Molina-Arcas et al suggested targeting IGFR and mTOR (15,52). Taken together, our data suggest that adaptive feedback is likely to play a key role in resistance to KRASG12C inhibitors, and that vertical inhibition of the RAS pathway, such as combined SHP2 and KRASG12C inhibition, may represent a promising strategy for evaluation in future clinical trials.
Supplementary Material
Acknowledgments
Grant Support:
R.B.C. is supported by NIH/NCI Gastrointestinal Cancer SPORE P50 CA127003, R01CA208437, U54CA224068, and a Stand Up To Cancer Colorectal Dream Team Translational Research Grant (SU2C-AACR-DT22–17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C.
Disclosure of Potential Conflicts of Interest:
R.B.C. is a consultant/advisory board member for Amgen, Array Biopharma, Astex Pharmaceuticals, Avidity Biosciences, BMS, C4 Therapeutics, Chugai, Fog Pharma, Fount Therapeutics, Genentech, LOXO, Merrimack, N-of-one, Novartis, nRichDx, Revolution Medicines, Roche, Roivant, Spectrum Pharmaceuticals, Shionogi, Symphogen, Taiho, and Warp Drive Bio; holds equity in Avidity Biosciences, C4 Therapeutics, Fount Therapeutics, nRichDx, and Revolution Medicines; and has received research funding from Asana and AstraZeneca.
REFERENCES
- 1.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. Journal of cell science 2016;129(7):1287–92 doi 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nature reviews Clinical oncology 2018;15(11):709–20 doi 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 3.Patricelli MP, Janes MR, Li L-S, Hansen R, Peters U, Kessler LV, et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer discovery 2016;6(3):316–29 doi 10.1158/2159-8290.cd-15-1105. [DOI] [PubMed] [Google Scholar]
- 4.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018;172(3):578–89.e17 doi 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Fell JB, Fischer JP, Baer BR, Ballard J, Blake JF, Bouhana K, et al. Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity. ACS medicinal chemistry letters 2018;9(12):1230–4 doi 10.1021/acsmedchemlett.8b00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostrem JM, Shokat KM. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nature reviews Drug discovery 2016;15(11):771–85 doi 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 7.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science (New York, NY) 2016;351(6273):604–8 doi 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47(D1):D941–d7 doi 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paraiso KH, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. British journal of cancer 2010;102(12):1724–30 doi 10.1038/sj.bjc.6605714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montero-Conde C, Ruiz-Llorente S, Dominguez JM, Knauf JA, Viale A, Sherman EJ, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer discovery 2013;3(5):520–33 doi 10.1158/2159-8290.cd-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery 2012;2(3):227–35 doi 10.1158/2159-8290.cd-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483(7387):100–3 doi 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 13.Manchado E, Weissmueller S, Morris JPt, Chen CC, Wullenkord R, Lujambio A, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016;534(7609):647–51 doi 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, et al. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer discovery 2016;6(7):754–69 doi 10.1158/2159-8290.cd-15-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misale S, Fatherree JP, Cortez E, Li C, Bilton S, Timonina D, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research 2019;25(2):796–807 doi 10.1158/1078-0432.ccr-18-0368. [DOI] [PubMed] [Google Scholar]
- 16.Govindan R, Fakih MG, Price TJ, Falchook GS, Desai J, Kuo JC, et al. 446PDPhase I study of AMG 510, a novel molecule targeting KRAS G12C mutant solid tumours. Annals of Oncology 2019;30(Supplement_5) doi 10.1093/annonc/mdz244.008. [DOI] [Google Scholar]
- 17.Agazie YM, Hayman MJ. Molecular Mechanism for a Role of SHP2 in Epidermal Growth Factor Receptor Signaling. Molecular and cellular biology 2003;23(21):7875–86 doi 10.1128/mcb.23.21.7875-7886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dance M, Montagner A, Salles JP, Yart A, Raynal P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cellular signalling 2008;20(3):453–9 doi 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes & development 2000;14(19):2501–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer cell 2016;29(1):75–89 doi 10.1016/j.ccell.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, et al. Intrinsic Resistance to MEK Inhibition in KRAS Mutant Lung and Colon Cancer through Transcriptional Induction of ERBB3. Cell reports 2014;7(1):86–93 doi 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 22.Johnson GL, Stuhlmiller TJ, Angus SP, Zawistowski JS, Graves LM. Molecular pathways: adaptive kinome reprogramming in response to targeted inhibition of the BRAF-MEK-ERK pathway in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2014;20(10):2516–22 doi 10.1158/1078-0432.ccr-13-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 2012;149(2):307–21 doi 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moll HP, Pranz K, Musteanu M, Grabner B, Hruschka N, Mohrherr J, et al. Afatinib restrains K-RAS–driven lung tumorigenesis. Science translational medicine 2018;10(446):eaao2301 doi 10.1126/scitranslmed.aao2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nature cell biology 2018;20(9):1064–73 doi 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankson R, Yu ZH, Bai Y, Li Q, Zhang RY, Zhang ZY. Therapeutic Targeting of Oncogenic Tyrosine Phosphatases. Cancer research 2017;77(21):5701–5 doi 10.1158/0008-5472.can-17-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nature medicine 2018;24(7):961–7 doi 10.1038/s41591-018-0023-9. [DOI] [PubMed] [Google Scholar]
- 28.Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nature medicine 2018;24(7):954–60 doi 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 29.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. The Journal of clinical investigation 2011;121(11):4311–21 doi 10.1172/jci57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kidger AM, Keyse SM. The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs). Semin Cell Dev Biol 2016;50:125–32 doi 10.1016/j.semcdb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing J, Greshock J, Holbrook JD, Gilmartin A, Zhang X, McNeil E, et al. Comprehensive Predictive Biomarker Analysis for MEK Inhibitor GSK1120212. Molecular cancer therapeutics 2012;11(3):720–9 doi 10.1158/1535-7163.mct-11-0505. [DOI] [PubMed] [Google Scholar]
- 32.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014;508(7494):118–22 doi 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 33.Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer discovery 2013;3(1):112–23 doi 10.1158/2159-8290.cd-12-0231. [DOI] [PubMed] [Google Scholar]
- 34.Corcoran RB, Andre T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, et al. Combined BRAF, EGFR, and MEK Inhibition in Patients with BRAFV600E-Mutant Colorectal Cancer. Cancer discovery 2018. doi 10.1158/2159-8290.cd-17-1226. [DOI] [PMC free article] [PubMed]
- 35.van Geel R, Tabernero J, Elez E, Bendell JC, Spreafico A, Schuler M, et al. A Phase Ib Dose-Escalation Study of Encorafenib and Cetuximab with or without Alpelisib in Metastatic BRAF-Mutant Colorectal Cancer. Cancer discovery 2017;7(6):610–9 doi 10.1158/2159-8290.cd-16-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaeger R, Cercek A, O’Reilly EM, Reidy DL, Kemeny N, Wolinsky T, et al. Pilot trial of combined BRAF and EGFR inhibition in BRAF-mutant metastatic colorectal cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21(6):1313–20 doi 10.1158/1078-0432.ccr-14-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopetz S, McDonough SL, Lenz H-J, Magliocco AM, Atreya CE, Diaz LA, et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG S1406). Journal of Clinical Oncology 2017;35(15_suppl):3505– doi 10.1200/JCO.2017.35.15_suppl.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cutsem EV, Huijberts S, Grothey A, Yaeger R, Cuyle P-J, Elez E, et al. Binimetinib, Encorafenib, and Cetuximab Triplet Therapy for Patients With BRAF V600E–Mutant Metastatic Colorectal Cancer: Safety Lead-In Results From the Phase III BEACON Colorectal Cancer Study. Journal of Clinical Oncology 2019;37(17):1460–9 doi 10.1200/jco.18.02459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373(8):726–36 doi 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lou K, Steri V, Ge AY, Hwang YC, Yogodzinski CH, Shkedi AR, et al. KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Science signaling 2019;12(583) doi 10.1126/scisignal.aaw9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fedele C, Ran H, Diskin B, Wei W, Jen J, Geer MJ, et al. SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models. Cancer discovery 2018;8(10):1237–49 doi 10.1158/2159-8290.cd-18-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed TA, Adamopoulos C, Karoulia Z, Wu X, Sachidanandam R, Aaronson SA, et al. SHP2 Drives Adaptive Resistance to ERK Signaling Inhibition in Molecularly Defined Subsets of ERK-Dependent Tumors. Cell reports 2019;26(1):65–78.e5 doi 10.1016/j.celrep.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu H, Liu C, Velazquez R, Wang H, Dunkl LM, Kazic-Legueux M, et al. SHP2 inhibition overcomes RTK-mediated pathway re-activation in KRAS mutant tumors treated with MEK inhibitors. Molecular cancer therapeutics 2019. doi 10.1158/1535-7163.mct-18-0852. [DOI] [PubMed]
- 44.Wong GS, Zhou J, Liu JB, Wu Z, Xu X, Li T, et al. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nature medicine 2018;24(7):968–77 doi 10.1038/s41591-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. The New England journal of medicine 2012;366(8):707–14 doi 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Planchard D, Kim TM, Mazieres J, Quoix E, Riely G, Barlesi F, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. The Lancet Oncology 2016;17(5):642–50 doi 10.1016/s1470-2045(16)00077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kopetz S, Desai J, Chan E, Hecht JR, O’Dwyer PJ, Maru D, et al. Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2015;33(34):4032–8 doi 10.1200/jco.2015.63.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay J-Y, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. The New England journal of medicine 2015;373(8):726–36 doi 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karoulia Z, Gavathiotis E, Poulikakos PI. New perspectives for targeting RAF kinase in human cancer. Nature reviews Cancer 2017;17(11):676–91 doi 10.1038/nrc.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367(18):1694–703 doi 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Planchard D, Besse B, Groen HJM, Souquet P-J, Quoix E, Baik CS, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. The Lancet Oncology 2016;17(7):984–93 doi 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molina-Arcas M, Moore C, Rana S, van Maldegem F, Mugarza E, Romero-Clavijo P, et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Science translational medicine 2019;11(510) doi 10.1126/scitranslmed.aaw7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.