Graphical abstract
Keywords: Ras dimerization, Nanoclusters, KRAS, C-Raf, RBD, CRD, MAPK pathway, Anionic bilayer, Molecular dynamics
Abstract
Proximally located in the membrane, oncogenic Ras dimers (or nanoclusters) can recruit and promote Raf dimerization and MAPK (Raf/MEK/ERK) signaling. Among Ras isoforms, KRas4B is the most frequently mutated. Recent data on the binary KRas4B–Raf-1 complex suggested that Raf-1 CRD not only executes membrane anchorage, but also supports the high-affinity interaction of Raf-1 RBD with KRas4B catalytic domain. For a detailed mechanistic picture of Raf activation at the membrane, we employ explicit MD simulations of the quaternary KRas4B–Raf-1 complex. The complex contains two active GTP-bound KRas4B proteins forming a dimer through the allosteric lobe interface and two tandem RBD-CRD segments of Raf-1 interacting with the effector lobes at both ends of the KRas4B dimer. We show that Raf-1 RBD-CRD supports stable KRas4B dimer at preferred interface and orientation at the membrane, thereby cooperatively enhancing the affinity of the KRas4B–Raf-1 interaction. We propose that a Ras dimer at the membrane can increase the population of proximal Raf kinase domains, promoting kinase domain dimerization in the cytoplasm. Collectively, the dynamic Ras–Raf assembly promotes Raf activation not by allostery; instead, Ras activates Raf by shifting its ensemble toward kinase domain-accessible states through enhanced affinity at the membrane.
1. Introduction
Membrane-anchored Ras controls cell survival and proliferation. It activates Raf and stimulates the mitogen-activated protein kinase (MAPK, Raf/MEK/ERK) signalling pathways [1], [2], [3]. Ras proteins also activate other effectors, such as phosphatidylinositide-3-kinase (PI3K), Ras association domain family 5 (RASFF5), and Ras-Ral guanine nucleotide dissociation stimulator (RalGDS) [4], [5], [6], [7], [8], [9]. All events occur at the membrane when the C-terminal tail of the hypervariable region (HVR) with the post-translational modifications (PTMs) anchors to it [10], [11] (Fig. 1A). The PTMs involve methylation and hydrophobic prenyl modifications including farnesylation and palmitoylation. Ras isoforms HRas, NRas, and KRas (with two splice variants KRas4A and KRas4B) contain a farnesylated/methylated cysteine at the C-terminus, but the palmitoyl modifications at other cysteine residues in the HVR differ [12], [13]. Membrane anchorage is necessary for formation of Ras nanoclusters, which are required for Raf’s kinase domain dimerization and activation [14], [15], [16]. Ras side-to-side dimers (and higher oligomers) in the nanoclusters effectively promote Raf dimerization [17], [18]. Blocking Ras dimerization and nanoclustering abolishes MAPK signalling, albeit not the PI3K/Akt/mTOR pathway [16], [19].
Fig. 1.
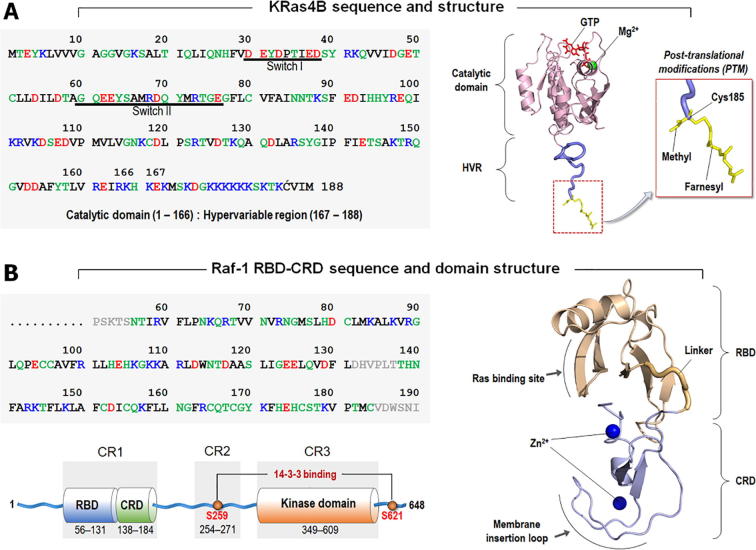
Sequences and structures. (A) KRas4B sequence and the modeled structure with the post-translational modifications including farnesylation and methylation at the C-terminal Cys185. (B) Raf-1 RBD-CRD sequence and the domain structure. All Raf kinases share three conserved regions; conserved region 1 involves the tandem RBD-CRD segment, conserved region 2 contains the Ser/Thr-rich region at the flexible linker, and conserved region 3 is the kinase domain. In the sequence, hydrophobic, polar/glycine, positively charged, and negatively charged residues are colored black, green, blue, and red, respectively. In Raf-1 RBD-CRD sequence, gray denotes the unstructured loop region. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Nuclear magnetic resonance (NMR) and computational studies have recently demonstrated that KRas4B can form a dimer with two distinct dimeric interfaces, symmetrically facing each other at the allosteric and effector lobes of the catalytic domain [17], [20]. Since they are located at opposite surfaces of the catalytic domain, dimer-to-dimer or multimeric combinations of Ras molecules are possible in the nanocluster, pointing to nanoclustering as a dynamic molecular assembly at the membrane [21]. When Raf is recruited to the membrane, its Ras binding domain (RBD) only targets the effector binding site of the Ras catalytic domain. Measurements of affinity between Ras and Raf’s RBD in solution show that it is high, in the low nanomolar range [22]. Raf RBD is expected to easily compete with Ras molecules assembled through the effector lobe dimer interface. Thus, when two Raf kinases interact with two adjacent Ras proteins, only the allosteric lobe dimer interface is available for the Ras side-to-side dimeric interaction. Recent atomistic models of KRas4B dimer at the anionic membrane provided detailed information of the dimeric interface. They indicated that in this environment KRas4B forms a dimer with the helical interface involving α3 and α4 helices, but the population of a dimer with the α4 and α5 helical interface is low [17].
Raf kinase consists of the N-terminal tail, RBD, cysteine-rich domain (CRD), Ser/Thr-rich flexible linker, kinase domain, and the C-terminal tail [23] (Fig. 1B). All Raf kinases share three conserved regions; conserved region 1 contains the RBD-CRD segment, conserved region 2 involves the Ser/Thr-rich segment, and conserved region 3 is the kinase domain. Raf dimerization occurs at conserved region 3, while conserved region 1 is responsible for Ras binding and membrane anchorage [24], [25]. Using molecular dynamics (MD) simulations of farnesylated/methylated KRas4B-GTP in complex with tandem Raf-1 RBD-CRD at the anionic membrane, we identified key basic CRD residues that are responsible for Raf-1 membrane attachment [26], consistent with earlier experimental observations [27], [28]. We observed that Raf-1 CRD uses an insertion loop comprising positively charged and hydrophobic residues to engage in membrane attachment. These intrinsic features suggest that it serves as a membrane binding segment [26], [29], [30]. Acting as an anchor point in the membrane, the role of Raf-1 CRD resembles that of KRas4B HVR. For the binary KRas4B–Raf-1 complex attached to the membrane, two anchor points, one from the KRas4B HVR and the other from the Raf-1 CRD, can restrict the fluctuations (including mobility [21]) of the catalytic domain of KRas4B, increasing the affinity of KRas4B to the RBD of Raf-1, which is expected to be lower than that measured in solution [22].
Our previous model of the KRas4B dimer at the membrane was constructed in the absence of Raf [17]. Subsequent studies of the Ras–Raf interaction at the membrane were only conducted for the binary KRas4B–Raf-1 complex [26]. To obtain a complete mechanistic picture of the Ras–Raf interaction, we model the farnesylated/methylated KRas4B-GTP dimer interacting with two Raf-1 conserved region 1 segments at both sides of the KRas4B dimer. In the quaternary KRas4B–Raf-1 complex, the KRas4B dimer is aligned through the allosteric lobe interface, and the covalently connected tandem Raf-1 RBD-CRD structures interact with KRas4B catalytic domain and the membrane. Our comprehensive explicit solvent simulations and detailed analyses reveal that the tethered KRas4B–Raf-1 organization increases the residence times of KRas4B orientation with the α3 and α4 helical interface. However, it loses its integrity at the α4 and α5 helical interface, since the α4-α5 dimer orientation hampers Raf-1 CRD contact with the membrane. With the separation into two binary complexes, Raf-1 CRD can anchor to the membrane, stimulating a reorientation of KRas4B catalytic domain toward a dimerization- and membrane-interaction-favored state. We suggest that Raf-1 conserved region 1 actions enrich the population of the KRas4B active signalling complex. Concomitantly, Ras dimerization further increases the affinity of KRas4B interaction with Raf-1 RBD-CRD. In oncogenic Ras nanoclusters, Ras binding to Raf’s RBD recruits Raf to the plasma membrane [22], [31]. CRD’s anchorage to the membrane reduces the Ras–RBD fluctuations. This enhanced stability at the membrane promotes Raf-1 kinase domain dimerization in the cytoplasm, thus MAPK signalling. Because the affinity of the Raf kinase domain interaction with the RBD-CRD segment in the autoinhibited state is low, the cooperatively increased Ras–Raf-1 affinity intensifies the shift of the Raf-1 population toward relieving Raf’s autoinhibition [23]. Taken together, population shift, rather than allostery via the long linker between the RBD-CRD and the kinase domain, is the key in Raf’s activation by Ras [32].
2. Methods
2.1. Preparing the quaternary KRas4B–Raf-1 complex interacting with the anionic membrane
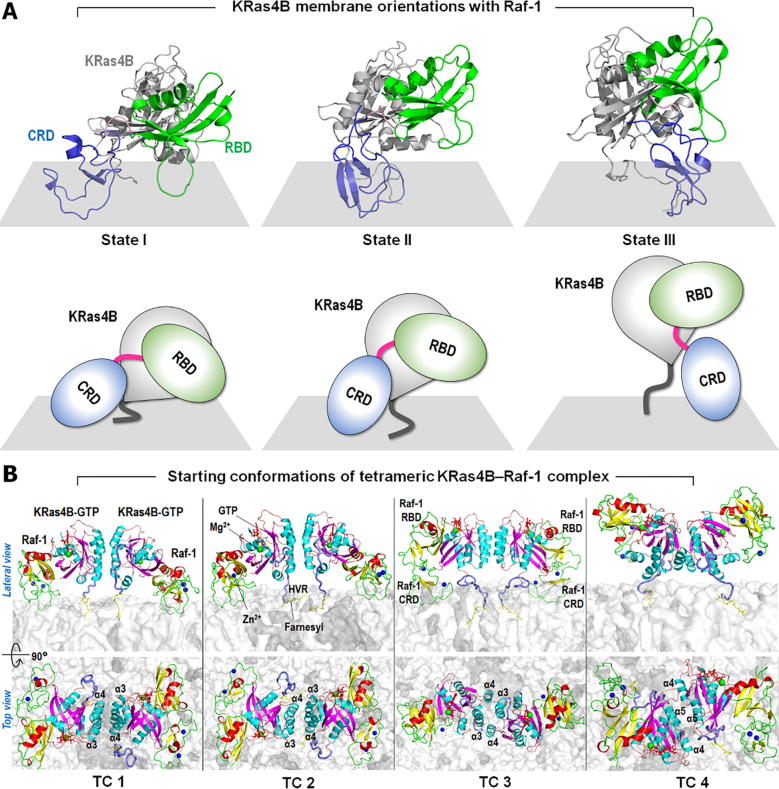
To generate the initial configurations of quaternary KRas4B–Raf-1 complex, we adopted three different KRas4B membrane orientations, states I, II, and III from previous studies [26] (Fig. 2A). In our previous studies, the crystal structure of the catalytic domain of KRas4B (PDB ID: 3GFT) was used to model the full-length KRas4B protein (Fig. 1A). For Raf-1, the solution structure of Raf-1 CRD (PDB ID: 1FAR) and the crystal structure Raf-1 RBD (PDB ID: 4G0N) were used to model the tandem RBD-CRD segment (Fig. 1B). As a building block towards construction of a tetrameric assembly, we extracted three binary complexes of KRas4B interacting with Raf-1’s RBD-CRD from previous simulations. These binary complexes depict the final conformations of membrane anchored KRas4B–RBD-CRD from three independent trajectories, representing three different KRas4B membrane orientations states as described in Fig. 2A. Since the Raf-1 RBD-CRD interaction with KRas4B occurs at the effector lobe of the catalytic domain, the allosteric lobe opposite to the Raf-1 binding site is only available for KRas4B dimerization. Thus, we performed a 2-fold rotational symmetry operation for the binary complex with respect to the allosteric lobe of KRas4B one at a time, obtaining three different quaternary configurations. Each quaternary complex contains two identical binary complexes symmetrically assembled through the allosteric lobe dimer interface of KRas4B. The initial construction reveals a linear shape of the quaternary complex that contains two farnesylated/methylated GTP-bound KRas4B in the central region, forming a dimer with the symmetric α3-α4/α3-α4 helical alignment. At both ends of the KRas4B dimer, two Raf-1 conserved region 1 with covalently connected tandem RBD-CRD structure are placed in the effector lobes of the KRas4B dimer. To test stability of the helical interfaces of KRas4B dimer (Fig. S1), the fourth quaternary complex was generated by the superimposition of two identical binary complexes into the symmetric α4-α5/α4-α5 helical alignment of KRas4B dimer. We employed the binary complex with the KRas4B membrane orientation in state III in the fourth quaternary construction. In the initial construction, the positions of RBD and CRD are highly restricted due to the α4-α5/α4-α5 helix alignment of KRas4B, resulting in none of the Raf-1 RBD-CRD models satisfying the CRD–membrane interaction. A total of 4 different initial configurations were prepared for explicit MD simulations with anionic lipid bilayer containing 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS) in 4:1, molar ratio.
Fig. 2.
Initial configurations of the KRas4B–Raf-1 systems at the membrane. (A) Three initial configurations of the binary KRas4B–Raf-1 complex with three different KRas4B membrane orientational states I, II, and III. Molecular topologies for each binary system are shown below. (B) Snapshots representing the starting points for four different initial TCs (1–4) of quaternary KRas4B–Raf-1 complex at the anionic bilayer composed of DOPC:DOPS (molar ratio 4:1). TCs 1–3 were constructed by symmetric α3-α4/α3-α4 helical alignment of each binary complex with the orientational states I, II, and III, respectively. TC 4 was obtained by symmetric α4-α5/α4-α5 helical alignment. In the bilayer structure, DOPC and DOPS are shown as white and grey surfaces, respectively.
2.2. Atomistic molecular dynamics simulations
We performed MD simulations using the updated CHARMM [33] all-atom additive force (version 36) [34] for constructing the set of starting points and relaxing the systems to a production-ready stage. Our simulations closely followed the same protocol as in our previous works [5], [7], [10], [17], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47]. The membrane simulations were performed with the anionic lipid bilayer generated by the bilayer-building protocol involving the interactions of pseudospheres through the vdW (van der Waals) force field [48], [49]. A unit cell containing a total of 500 lipids (400 DOPC and 100 DOPS) constitutes the bilayer with TIP3P waters, added at both sides with lipid/water ratio of ~1/140. The cross-sectional areas per lipid for DOPC and DOPS are 72.4 Å2 and 65.3 Å2, respectively [50]. With a choice for the number of lipid molecules, the optimal value of lateral cell dimension of 133.2 Å × 133.2 Å for the unit cell can be determined. To electrically neutralize the system, 15 Mg2+ and 46 Na+ were inserted. To obtain a physiological salt concentration near 100 mM, additional 45 Na+ and 45 Cl- were added. The initial placement of the ions is at the bulk water regions at both sides of the bilayer, far from the protein complex and the bilayer surfaces. The bilayer system containing a quaternary KRas4B–Raf-1 complex, lipids, salts, and water has almost 283,000 atoms.
A series of minimization cycles were performed for the solvents including lipids around the harmonically restrained protein complex. The preequilibrium simulations for 5 ns were performed on each quaternary system with the restrained backbones of KRas4B and Raf-1 RBD-CRD until the solvent reached 310 K. The harmonic restraints on the backbones of KRas4B–Raf-1 complex were gradually removed through dynamic cycles with the particle mesh Ewald electrostatics calculation [51], completing the final preequilibrium stage. A total of 4.0 μs simulations were performed for the 4 systems, each with 1 μs, and additional simulations for selected systems were also performed to check reproducibility. In the production runs, the Langevin temperature control maintained the constant temperature at 310 K [52], and the Nosé-Hoover Langevin piston pressure control sustained the pressure at 1 atm. The SHAKE algorithm was applied to constrain the motion of bonds involving hydrogen atoms [53]. Two Zn2+ in each CRD were coordinated throughout the simulations by employing the collective variable-based calculations (Colvars) [54] in the NAMD code [55]. The production runs were performed with the NAMD parallel-computing code [55] on a Biowulf cluster at the National Institutes of Health (Bethesda, MD). In the analysis, the first 100 ns trajectories were removed, and thus averages were taken afterward.
3. Results
3.1. Raf-1 supports KRas4B dimerization through the α3 and α4 helical interface
In our previous simulations [26], we provided highly populated membrane orientations of KRas4B-GTP in complex with tandem RBD-CRD of Raf-1. To model the quaternary KRas4B–Raf-1 complex, we extracted three dimeric configurations from previous studies [26]. These configurations represent three different KRas4B membrane orientations in states I, II, and III (Fig. 2A). To construct tetrameric configurations (TCs), the binary KRas4B–Raf-1 complex in each dimeric configuration and its copy were symmetrically aligned with respect to the allosteric lobe dimer interface of KRas4B [17], [20] (Fig. S1). Three TCs 1, 2, and 3 with the KRas4B orientation in the states I, II, and III, respectively, contain the symmetric α3-α4/α3-α4 helical alignment at the KRas4B dimer interface (Fig. 2B). It was observed earlier that KRas4B dimer favors the α3 and α4 helical interface [17]. To corroborate this helical interface as a major KRas4B dimeric interface, an additional quaternary complex was constructed by the symmetric α4-α5/α4-α5 helical alignment at the KRas4B dimer interface (TC 4).
To validate the tetramer KRas4B–Raf-1 conformation in the lipid environment, we performed MD simulations on the quaternary complex at an anionic lipid bilayer composed of DOPC and DOPS lipids in 4:1, molar ratio. During the simulation, we observed that TC 1 preserved the symmetric α3-α4/α3-α4 helical alignment (Fig. 3A), while both TCs 2 and 3 slightly altered their dimer-to-dimer interfaces, shifting to an asymmetric α3-α4/α3 interface (Fig. 3B, C). Superimpositions of the final conformation onto the starting point clearly denote the differences in the KRas4B dimer interfaces (Fig. S2). For TC 4, the quaternary conformation was completely dissociated, reducing into two separated binary complexes (Fig. 3D). The dissociation occurred immediately at the early stage of the simulation (Fig. 3E), suggesting that Raf-1 predominantly supports the α3 and α4 helical interface. To quantify the dimeric interface, we calculated the interface area by using the Proteins Interfaces Structures and Assemblies (PISA) program [56]. For the Ras-Ras interface area, we obtained ~380 Å2, ~370 Å2, and ~330 Å2 for TCs 1, 2, and 3, respectively. These values are very close to the crystal interface area in the range of ~350–450 Å2 for Ras dimers involving the α3 and α4 helices (PDB IDs: 4EFL, 4EFM, 3KKN). For TC 4, no interface area was obtained due to dissociation. In the Ras-Raf interaction, the interface area was in the range of ~700–810 Å2, which is slightly larger than the crystal interface area of 614 Å2 for the Ras-RBD assembly (PDB ID: 4G0N). The larger values indicate that CRD partially contributes to the interface area.
Fig. 3.
Simulated configurations of the KRas4B–Raf-1 systems at the membrane. (A-D) Snapshots representing the final structures for four different TCs (1–4) of quaternary KRas4B–Raf-1 complex at the anionic bilayer composed of DOPC:DOPS (molar ratio 4:1). Boxes highlight the KRas4B allosteric lobe dimer interface. (E) Shown is the time series of the center of mass distance between two KRas4B molecules as a function of time for TCs 1 – 4.
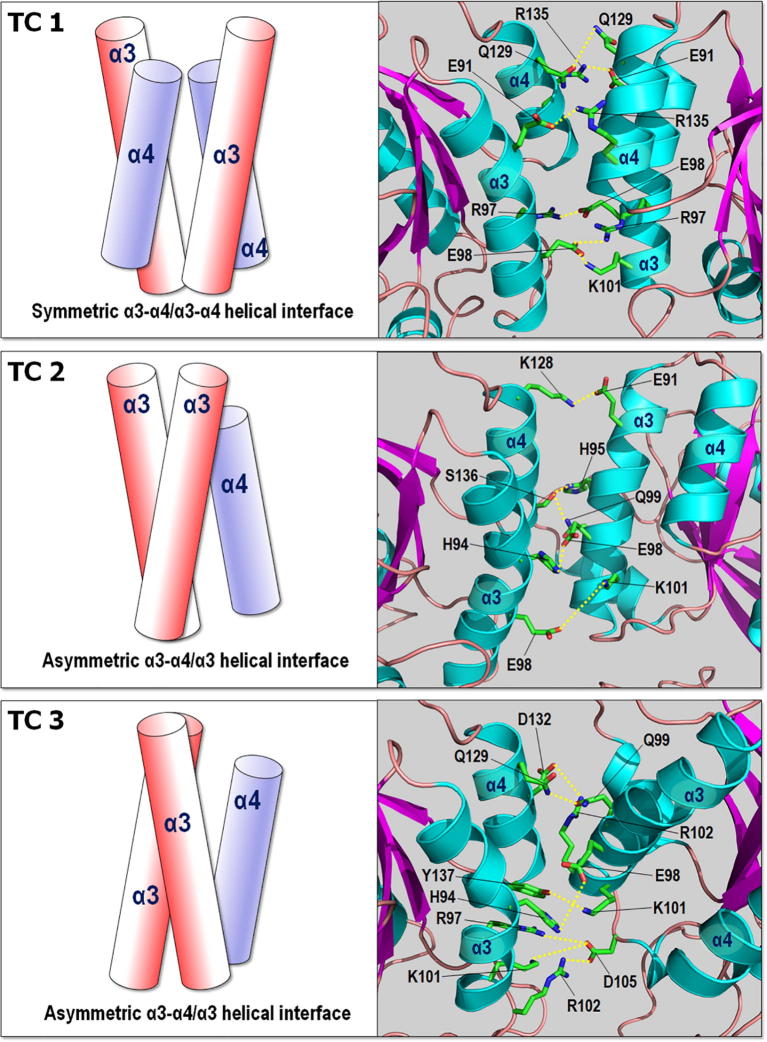
The asymmetry in the dimeric interface was also observed for the membrane-attached KRas4B dimer in the absence of Raf [17]. The KRas4B dimeric interface is stabilized by several salt bridge and hydrophilic interactions between residues at the α3 and α4 helices (Fig. 4). The most common residues involved in the interaction are His94, Arg97, Glu98, and Lys101 at the α3 helix. The residues, Lys128, Gln129, Asp132, Arg135, and Ser136 at the α4 helix also contribute to the interaction. Note that both TCs 2 and 3 exhibit the asymmetric helical alignment, but the helices cross each other with different orientations. This is mainly due to the different conformations of Raf-1 RBD-CRD (Fig. 2A) that support the active KRas4B membrane orientation [26]. In our previous studies of KRas4B dimer with the same α3 and α4 helical interface, we obtained the interaction energy of ~−470 kcal/mol between the catalytic domains in the absence of Raf-1 [17]. Here, we obtained similar values of the interaction energies, −480 ± 104 kcal/mol, −432 ± 108 kcal/mol, and −412 ± 110 kcal/mol for TCs 1, 2, and 3, respectively. These similar values of the interaction energies between Ras proteins reflect that the residues involved in the interaction are highly conserved at the same α3 and α4 helical interface.
Fig. 4.
Topological diagrams representing the allosteric helices, α3 and α4 (left panels), and snapshots highlighting the allosteric lobe dimer interface (right panels) for TCs 1, 2, and 3. No dimer interface was observed for TC 4 due to separation. Residues involving in the salt bridge and hydrophilic interactions are marked.
3.2. Raf-1 promotes active KRas4B membrane orientation
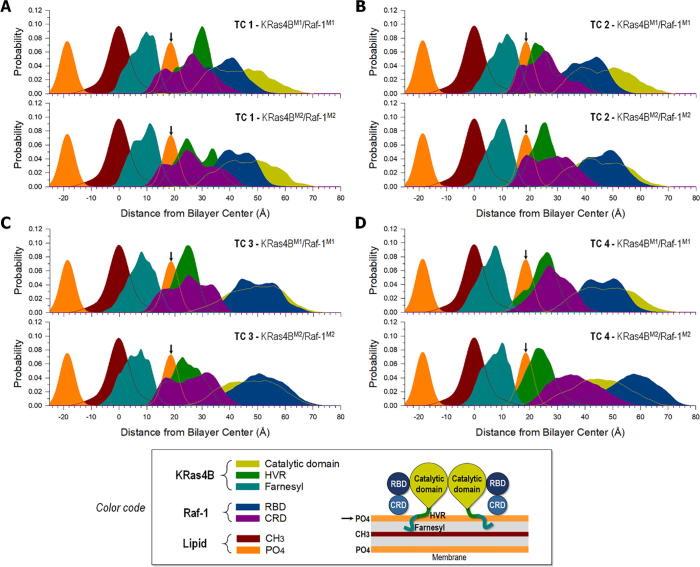
For membrane-attached Ras, the accessibility of the effector binding site is crucial for function. In the absence of effectors, an active KRas4B exhibits multiple orientations resulting in large fluctuations of the catalytic domain at the membrane [10]. Effectors can restrain the fluctuations when attached to Ras binding site, restricting the orientation and location of the catalytic domain on the membrane surface. To observe how the quaternary association is influenced by membrane localization of the KRas4B–Raf-1 complex, we calculated the average positions of each protein domain and lipid group over the simulation trajectories (Fig. 5). Position probability distribution functions for phosphate (PO4) and the terminal methyl (CH3) groups of DOPC and DOPS lipids, for RBD and CRD of Raf-1, and for the catalytic domain, HVR, and farnesyl of KRas4B were calculated as a function of distance from the bilayer center. The peaks in the distribution curves reflect the highly populated locations of each component. The symmetric distributions of the PO4 group at both leaflets at d = ~ ±20 Å (where d is the distance from the bilayer center) constitute the lipid bilayer, and a CH3 peak between them denotes the bilayer center at d = 0. For convenience, since the protein complex is located at the one side of the bilayer, we set the bilayer surface at d = ~20 Å to z = 0. For all configurations, high distribution peaks below the bilayer surface indicate that the farnesyl is located at z = ~−10 Å, where the arrow in the figure indicates the bilayer surface at z = 0. All farnesyls stably anchor to the interior of the bilayer. For the quaternary complexes (TCs 1, 2, and 3), the catalytic domain of KRas4B and Raf-1 RBD are elevated from the bilayer surface as TC 1 → TC 3 (Table S1). In contrast, the HVR locates slightly toward the bilayer surface as TC 1 → TC 3, but the location of Raf-1 CRD is rather similar. The RBD is located at a position below the catalytic domain in TCs 1 and 2, but it is located at the same position as the catalytic domain in TC 3. For TC 4, the distributions of the first monomeric units, KRas4BM1 and Raf-1M1, are similar to those in TC 3, but this is not the case for the second monomeric units. In the KRas4B dimer with the α4 and α5 helical interface, Raf-1 RBD needs to be positioned above the catalytic domain due to the effector binding site of KRas4B facing opposite to the bilayer surface (Fig. 2B). Thus, the α4-α5/α4-α5 helix alignment disfavors CRD effectively contacting the lipid bilayer. However, after separation into two binary complexes, we observed that the first binary complex quickly adjusted its orientation and attached the CRD to the bilayer at t ~ 150 ns (Fig. S3). Although the second binary complex exhibited large fluctuations, it also attached the CRD to the bilayer later.
Fig. 5.
Probability distribution across the lipid bilayer. Probability distribution functions across the lipid bilayer for different component groups of lipids (CH3, methyl group, red; PO4, phosphate group, orange), different domains of KRas4B (catalytic domain, yellow; HVR, green; farnesyl, blue), and different domains of Raf-1 (RBD, dark blue; CRD, purple) as a function of distance from the bilayer center, d, for four different TCs (1–4) of quaternary KRas4B–Raf-1 complex at the anionic bilayer composed of DOPC:DOPS (molar ratio 4:1). The first (KRas4BM1/Raf-1M1) and the second (KRas4BM2/Raf-1M2) monomeric units are separately shown in each TC. The arrows at d = ~ 20 Å denote the bilayer surface at z = 0. Color code denotes different colors used for the domains of KRas4B, Raf-1, and lipid groups as in the schematic diagram of the quaternary complex. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Raf-1 RBD’s position at the bilayer with respect to the catalytic domain is highly correlated with the variation in the catalytic domain orientation at the membrane. To quantify the orientation, we generated two vectors connecting the atom pairs in GTP, PA → O3A () and C6 → O6 (), and then measured the angle between each vector and the bilayer normal. The ensembles of the KRas4B orientation were sampled from the population distribution of the angles of the two vectors. In our previous studies [26], we applied this protocol to the binary KRas4B–Raf-1 complex and defined the KRas4B active-state orientation when the angles were populated in the ranges of 60° < θPO < 90° and 40° < θCO < 100°. For the quaternary complex, we observed that except TC 4, KRas4B catalytic domain roughly retains the active-state orientation, although its orientation drifted slightly from the initial setting (Fig. S4). Each monomer in the KRas4B dimer evolves into different orientational states even if the monomers start from the same orientation. TC 1 highly populates the KRas4B orientation in states II and III, converging from state I. TC 2 also populates the KRas4B orientation in states II and III, which are near its initial orientation. However, TC 3 appears to yield the KRas4B orientation in states V with (θPO, θCO) ≈ (90°, 50°) and VI with (θPO, θCO) ≈ (120°, 40°), which are less populated states for the binary complex, even though the initial configuration was assigned to the highly populated active-state orientation in state III [26]. In TC 4, the first binary complex approaches the KRas4B orientation to (θPO, θCO) ≈ (70°, 60°) after separation, which is the highly populated state III for the binary complex, while the second binary complex lags toward the active-state orientation. It can be seen that the quaternary complex shifts the population of KRas4B orientation from monomer-specific to dimer-specific catalytic domain orientation. This prompts us to modify the definition of the landscape for highly populated catalytic domain orientation for the KRas4B monomer in the binary complex [26].
To better quantify dimer-specific KRas4B orientations, we considered the angles of two vectors connecting the atom pairs, Leu79 → Tyr96 () and Leu79 → Val8 (), in the catalytic domain of KRas4B. The selected atoms are highly stable in the protein core, and the vectors are almost perpendicular to each other. The population distribution of the angles of these two vectors with respect to the membrane normal can provide information of the Ras membrane orientation and, at the same time, the direction of the catalytic domain inclination (Fig. 6A). In the figure, the red circle measures the degree of the catalytic domain inclination. For example, if the orientational distribution map is located outside the red circle, Ras can be regarded as occluded. We note that the limit of the red circle was roughly assigned, and that it does not quantitatively represent the measure of Ras membrane occlusion. It is based on the observation that the catalytic domain begins to contact with the membrane surface when the map is located outside the red circle. The radial direction of blue dots characterizes the direction of the catalytic domain inclination. For instance, if the distribution map is located at the 4th quadrant, Ras’ β2 strand (or Switch I) faces toward the bilayer surface. We observed that TCs 1, 2, and 3 populate the distribution map of KRas4B orientation within the red circle, suggesting that the catalytic domains represent the active-state orientation (Fig. 6B-D). For KRas4B catalytic domain, any orientational state within the red circle can facilitate both Raf binding at the effector lobe and dimerization with another Ras at the allosteric lobe. That is, Raf binding and Ras dimerization restrict the distribution map of the catalytic domain orientation to be populated within the red circle. Our simulations showed that these quaternary complexes were highly stable with confined KRas4B membrane orientation, supported by the Raf-1 interactions with both KRas4B and membrane. For TC 4, KRas4BM1 in the first binary complex exhibits the active-state orientation just after the separation (Fig. 6E), suggesting that Raf-1 promotes active KRas4B membrane orientation. However, KRas4BM2 in the second binary complex slowly recovers the active-state orientation from the occluded orientation after the separation.
Fig. 6.
KRas4B membrane orientation. (A) A reference plot describing two-dimensional probability distributions of orientation angles for two vectors, Leu79 → Tyr96 () and Leu79 → Val8 () in the core of KRas4B with respect to the bilayer normal (out of the page). Each colored map corresponds to each Ras catalytic domain’s membrane orientation in the quaternary complex. Population map inside the red circle represents the active-state orientation of KRas4B. Occlusion can be observed with the distribution map outside the red circle. The blue dots in the blue circle denote the direction of catalytic domain occlusion (see details in the main text). (B-E) The sampled distributions from the simulation trajectory for four different TCs (1–4) of quaternary KRas4B–Raf-1 complex at the anionic bilayer composed of DOPC:DOPS (molar ratio 4:1). KRas4B dimers populate the distribution maps within the red circle. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Membrane interaction of Raf-1: both RBD and CRD involve in the interaction with the membrane
To quantify how Raf-1 effectively localizes on the membrane and supports the KRas4B orientation in the quaternary complex, we measured probability distribution functions of membrane contacts for Raf-1 residues. As expected, the quaternary complexes, TCs 1, 2, and 3, show high contact probability for the CRD residues (Fig. 7), indicating that CRD acts as a membrane binding domain of Raf-1. Interestingly, although RBDs in TC 3 hesitate to interact with the membrane, RBD residues nearby Lys106 in TCs 1 and 2 yield high contact probability, indicating that Raf-1 RBD involves in the interaction with the membrane. For TC 4 with two separated binary complexes, the Raf-1M1 RBD shows high membrane contact probability, suggesting that RBD anchoring to the membrane can promote membrane attachment of CRD. We observed that the RBD residue Lys106 first touches the membrane at t ~ 110 ns and CRD residue Lys148 follows at t ~ 350 ns (Fig. S5). Once the CRD establishes membrane attachment, the RBD–membrane interaction becomes sporadic, depending on KRas4B catalytic domain orientation at the membrane.
Fig. 7.
Lipid contact probability. The probability of lipid contacts for the residues of Raf-1 for four different TCs (1–4) of quaternary KRas4B–Raf-1 complex at the anionic bilayer composed of DOPC:DOPS (molar ratio 4:1). The first (Raf-1M1) and the second (Raf-1M2) monomeric unit of Raf-1 are marked in each TC.
Raf-1 RBD contains twelve positively charged residues; seven basic residues (Arg59, Lys65, Arg67, Arg73, Lys84, Lys87, and Arg89) are located near the Ras binding interface in the N-lobe and five basic residues (Arg100, Lys106, Lys108, Lys109, and Arg111) are found at a loop in the C-lobe. Among the C-lobe basic residues, three lysine residues Lys106, Lys108, and Lys109 are involved in the interaction with the anionic lipid bilayer (Fig. S6). The similar profiles in the membrane of the deviation of these basic residues among all RBDs in TCs suggest that membrane localization and orientation of Raf-1 RBD in complex with KRas4B are highly conserved. For example, Lys106 has the lowest deviation from the bilayer surface for all RBD conformations. In our previous simulations [26], for Raf-1 CRD we defined the membrane insertion loop at 144KTFLKLAFCDICQKFLLN161 and discovered that three key basic residues, Lys144, Lys148, and Lys157 are responsible for CRD-membrane binding. Lys148 has a high probability of inserting into the membrane. Surface scanning mutagenesis showed that mutations in the insertion loop, K144A/R164A or K144A/L160A inhibits Raf-1 activation but did not interfere significantly with Ras binding [57]. This indicates that the insertion loop is crucial for Raf activation targeting membrane attachment. Here, for Raf-1 RBD we designate the region with 101LLHEHKGKKA110 as a “membrane contact loop” and suggest that three key basic residues, Lys106, Lys108, and Lys109 lead to RBD-membrane contacts (Fig. 8). Unlike the CRD’s membrane insertion loop, which contains the hydrophobic residues next to Lys148, the RBD’s membrane contact loop does not have hydrophobic residues next to the key basic residue Lys106. This suggests that the membrane contact loop has an auxiliary role in the Raf-1 membrane interaction, while the membrane insertion loop has an intrinsic role in the membrane attachment.
Fig. 8.
Membrane interaction of Raf-1. Snapshots of membrane interacting Raf-1 (left panel) and its topology diagram (right panel) for (A) the first Raf-1 (Raf-1M1) molecule in TC 1 and also (B) the first Raf-1 (Raf-1M1) molecule in TC 3. The key basic residues, Lys106, Lys108, and Lys109 in the membrane contact loop of RBD, and Lys144, Lys148, and Lys157 in the membrane insertion loop of CRD are highlighted in the topology diagrams.
4. Discussion
Here, we provide a mechanistic picture detailing how two Raf molecules attach to the membrane and interact with Ras dimer, and how this can activate Raf using explicit MD simulations. In the quaternary KRas4B–Raf-1 complex at the anionic bilayer, GTP-bound KRas4B proteins form a dimer through the allosteric lobe interface, and two tandem Raf-1 RBD-CRD bind to the exposed effector lobes at both ends of the dimer. Previous models only provided the binary KRas4B–Raf-1 complex at the membrane [26], [29]. However, to activate Raf, two Raf molecules are required for kinase domain dimerization, and proximal, nanoclustered or dimeric, Ras molecules are needed to accomplish this aim. Physiological Raf activation can be via kinase domain homodimerization or heterodimerization with other Raf isoforms with the kinase domains catalysing cis autophosphorylation of each activation loop [58], [59], [60], [61]. However, autoinhibited, full-length Raf may not achieve a sufficiently high local concentration to accomplish dimerization in the cytoplasm. Raf interaction with Ras molecules organized in nanoclusters [14], [62], coupled with the strong Ras–RBD interaction and further enhanced by CRD, significantly increases Raf’s effective concentration at the membrane, essentially scaling down Raf’s distribution from 3-dimentional to 2-dimentional organization.
It was reported that the lateral diffusion of Ras in the plasma membrane is as fast as lipid probes and significantly faster than a typical membrane protein [63], [64]. In a nanocluster, Ras molecules congregate in specific membrane microdomains with favored lipid composition [11], [65]. A major driving force gathering the Ras molecules is the interactions between their catalytic domains, with the prenylated HVR engaging in the membrane association. Membrane-unbound Ras catalytic domain can obtain fast lateral diffusion and facilitate nanocluster formation. In contrast, highly occluded Ras with membrane-bound catalytic domain dampens its lateral mobility, restricting nanocluster formation. Ras nanocluster is the active signalling platform for the MAPK pathway, and GTP-bound Ras molecules with membrane-unbound catalytic domain are likely to exist in the cluster. Monomeric Ras with membrane-unbound catalytic domain exhibits high fluctuations unless lipid interactions secure the catalytic domain at the membrane surface [10]. When gathered, the continuous network of the catalytic domain interactions decreases the fluctuations. Ras catalytic domain interactions are transient with low affinity [17], [20], suggesting that the Ras nanocluster is a dynamic, lateral assembly of Ras molecules in the membrane. In the nanocluster, Ras can associate through the allosteric and effector lobe interfaces. The allosteric lobe dimer interface involves α3, α4, and α5 helices, while the effector lobe dimer interface contains a shifted β-sheet extension with relatively higher affinity [20].
KRas4B forms a dimer in a GTP-dependent manner [17], [20], and assembles into higher order nanoclusters [15], which can contain 6 to 8 Ras proteins [66], [67]. Thus, a KRas4B nanocluster may be an array of multiple combinations of molecular interactions through two distinct dimeric interfaces, or consist of spatially adjacent, albeit loose monomers. When Raf-1 is recruited to the membrane, its RBD targets the effector lobe. Two Raf-1 RBDs can bind to the exposed effector lobes of an allosteric lobe interface-mediated KRas4B dimer. The reduced fluctuations secure the weakly aligned KRas4B allosteric lobe dimer interface, enhancing the KRas4B–Raf-1 interaction at the membrane. Our simulations provide atomistic description of this cooperative mechanism and the favored mode of attachment of the quaternary assembly to the membrane. The simulations show that the populated asymmetric membrane-bound allosteric lobe KRas4B dimer interfaces are mainly through the α3 and α4 helices. Helical interface asymmetry results in a bent tetrameric conformation. This delineates the shape of the nanocluster as less likely linear, and more probably curved or circular-like, accommodating, or promoting, local membrane curvature [68]. An additional anchor point provided by Raf-1 RBD’s membrane contact loop further secures Raf-1’s membrane attachment. The quaternary complex is highly stable at the membrane with a total of six anchor points; two by HVRs, two by CRDs, and two by RBDs. However, unlike the Raf-1 CRD’s membrane insertion loop containing both key basic residues and hydrophobic residues [26], [27], [28], the membrane contact loop of RBD lacks hydrophobic residues, suggesting its auxiliary role in the Raf-1 membrane anchorage.
Does Ras dimerization promote Raf dimerization or vice versa as has sometimes been hypothesized? Our studies suggest that KRas4B dimer may promote Raf-1 kinase domain dimerization, possibly yielding a proximity of two Raf kinase domains in the cytoplasm, by cooperatively amplifying the affinity of the Ras–Raf-1 RBD-CRD interaction. The enhanced affinity acts to shift the Raf-1 ensemble thereby relieve its autoinhibition toward a kinase domain-accessible state. We propose that Raf-1 RBD-CRD binds to the effector lobes of the Ras dimer with high affinity. This binding reduces the fluctuations of the Ras dimer in the membrane, which further promotes Ras–Raf-1 affinity to effectively accomplish the population shift. Reduced fluctuations in the membrane of spatially proximal Raf-1 binding to Ras monomers can similarly cooperatively accomplish this role. The reduced fluctuations of the dimer (or of spatially proximal Ras monomers) at the bilayer cooperatively enhances it. The quaternary assembly promotes Raf activation by shifting its equilibrium to the kinase domain accessible state [32] thereby enhancing active Ras signalling.
CRediT authorship contribution statement
Hyunbum Jang: Conceptualization, Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Project administration. Mingzhen Zhang: Validation, Resources, Data curation, Writing - review & editing. Ruth Nussinov: Conceptualization, Writing - review & editing, Visualization, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. All simulations had been performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov/).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.03.018.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.McCubrey J.A., Steelman L.S., Chappell W.H., Abrams S.L., Wong E.W., Chang F. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant K.L., Mancias J.D., Kimmelman A.C., Der C.J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu S., Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R. Ras conformational ensembles, allostery, and signaling. Chem Rev. 2016;116:6607–6665. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 4.Castellano E., Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011;2:261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M., Jang H., Nussinov R. The structural basis for Ras activation of PI3Kα lipid kinase. Phys Chem Chem Phys. 2019;21:12021–12028. doi: 10.1039/c9cp00101h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao T.J., Tsai C.J., Jang H., Fushman D., Nussinov R. RASSF5: an MST activator and tumor suppressor in vivo but opposite in vitro. Curr Opin Struct Biol. 2016;41:217–224. doi: 10.1016/j.sbi.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Liao T.J., Jang H., Tsai C.J., Fushman D., Nussinov R. The dynamic mechanism of RASSF5 and MST kinase activation by Ras. Phys Chem Chem Phys. 2017;19:6470–6480. doi: 10.1039/c6cp08596b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neel N.F., Martin T.D., Stratford J.K., Zand T.P., Reiner D.J., Der C.J. The RalGEF-Ral effector signaling network: the road less traveled for anti-Ras drug discovery. Genes Cancer. 2011;2:275–287. doi: 10.1177/1947601911407329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nussinov R., Tsai C.J., Jang H. Ras assemblies and signaling at the membrane. Curr Opin Struct Biol. 2020;62:140–148. doi: 10.1016/j.sbi.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Jang H., Banerjee A., Chavan T.S., Lu S., Zhang J., Gaponenko V. The higher level of complexity of K-Ras4B activation at the membrane. FASEB J. 2016;30:1643–1655. doi: 10.1096/fj.15-279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee A., Jang H., Nussinov R., Gaponenko V. The disordered hypervariable region and the folded catalytic domain of oncogenic K-Ras4B partner in phospholipid binding. Curr Opin Struct Biol. 2016;36:10–17. doi: 10.1016/j.sbi.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nussinov R., Tsai C.J., Jang H. Oncogenic Ras isoforms signaling specificity at the membrane. Cancer Res. 2018;78:593–602. doi: 10.1158/0008-5472.CAN-17-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pantar T. The current understanding of KRAS protein structure and dynamics. Comput Struct Biotechnol J. 2020;18:189–198. doi: 10.1016/j.csbj.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y., Hancock J.F. Ras nanoclusters: versatile lipid-based signaling platforms. Biochim Biophys Acta. 2015;1853:841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Nan X., Tamguney T.M., Collisson E.A., Lin L.J., Pitt C., Galeas J. Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway. Proc Natl Acad Sci U S A. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nussinov R., Tsai C.J., Jang H. Is Nanoclustering essential for all oncogenic KRas pathways? Can it explain why wild-type KRas can inhibit its oncogenic variant? Semin Cancer Biol. 2019;54:114–120. doi: 10.1016/j.semcancer.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R. Membrane-associated Ras dimers are isoform-specific: K-Ras dimers differ from H-Ras dimers. Biochem J. 2016;473:1719–1732. doi: 10.1042/BCJ20160031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M., Peters A., Huang T., Nan X. Ras dimer formation as a new signaling mechanism and potential cancer therapeutic target. Mini Rev Med Chem. 2016;16:391–403. doi: 10.2174/1389557515666151001152212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer-Smith R., Koide A., Zhou Y., Eguchi R.R., Sha F., Gajwani P. Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol. 2017;13:62–68. doi: 10.1038/nchembio.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muratcioglu S., Chavan T.S., Freed B.C., Jang H., Khavrutskii L., Freed R.N. GTP-dependent K-Ras dimerization. Structure. 2015;23:1325–1335. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nussinov R., Tsai C.J., Jang H. Oncogenic KRas mobility in the membrane and signaling response. Semin Cancer Biol. 2019;54:109–113. doi: 10.1016/j.semcancer.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann C., Martin G.A., Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J Biol Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 23.Nussinov R., Zhang M., Tsai C.J., Liao T.J., Fushman D., Jang H. Autoinhibition in Ras effectors Raf, PI3Kα, and RASSF5: a comprehensive review underscoring the challenges in pharmacological intervention. Biophys Rev. 2018;10:1263–1282. doi: 10.1007/s12551-018-0461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavoie H., Thevakumaran N., Gavory G., Li J.J., Padeganeh A., Guiral S. Inhibitors that stabilize a closed RAF kinase domain conformation induce dimerization. Nat Chem Biol. 2013;9:428–436. doi: 10.1038/nchembio.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cutler R.E., Jr., Stephens R.M., Saracino M.R., Morrison D.K. Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci U S A. 1998;95:9214–9219. doi: 10.1073/pnas.95.16.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S., Jang H., Zhang J., Nussinov R. Raf-1 cysteine-rich domain increases the affinity of K-Ras/Raf at the membrane, promoting MAPK signaling. Structure. 2018;26(513–525) doi: 10.1016/j.str.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Improta-Brears T., Ghosh S., Bell R.M. Mutational analysis of Raf-1 cysteine rich domain: requirement for a cluster of basic aminoacids for interaction with phosphatidylserine. Mol Cell Biochem. 1999;198:171–178. doi: 10.1023/a:1006981411691. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh S., Xie W.Q., Quest A.F., Mabrouk G.M., Strum J.C., Bell R.M. The cysteine-rich region of raf-1 kinase contains zinc, translocates to liposomes, and is adjacent to a segment that binds GTP-ras. J Biol Chem. 1994;269:10000–10007. [PubMed] [Google Scholar]
- 29.Li Z.L., Prakash P., Buck M. A “Tug of War” maintains a dynamic protein-membrane complex: molecular dynamics simulations of C-Raf RBD-CRD bound to K-Ras4B at an anionic membrane. ACS Cent Sci. 2018;4:298–305. doi: 10.1021/acscentsci.7b00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Travers T., Lopez C.A., Van Q.N., Neale C., Tonelli M., Stephen A.G. Molecular recognition of RAS/RAF complex at the membrane: role of RAF cysteine-rich domain. Sci Rep. 2018;8:8461. doi: 10.1038/s41598-018-26832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chong H., Guan K.L. Regulation of Raf through phosphorylation and N terminus-C terminus interaction. J Biol Chem. 2003;278:36269–36276. doi: 10.1074/jbc.M212803200. [DOI] [PubMed] [Google Scholar]
- 32.Nussinov R., Tsai C.J., Jang H. Does Ras activate Raf and PI3K allosterically? Front Oncol. 2019;9:1231. doi: 10.3389/fonc.2019.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks B.R., Brooks C.L., 3rd, Mackerell A.D., Jr., Nilsson L., Petrella R.J., Roux B. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klauda J.B., Venable R.M., Freites J.A., O'Connor J.W., Tobias D.J., Mondragon-Ramirez C. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chavan T.S., Jang H., Khavrutskii L., Abraham S.J., Banerjee A., Freed B.C. High-affinity interaction of the K-Ras4B hypervariable region with the Ras active site. Biophys J. 2015;109:2602–2613. doi: 10.1016/j.bpj.2015.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang H., Abraham S.J., Chavan T.S., Hitchinson B., Khavrutskii L., Tarasova N.I. Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J Biol Chem. 2015;290:9465–9477. doi: 10.1074/jbc.M114.620724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang H., Banerjee A., Chavan T., Gaponenko V., Nussinov R. Flexible-body motions of calmodulin and the farnesylated hypervariable region yield a high-affinity interaction enabling K-Ras4B membrane extraction. J Biol Chem. 2017;292:12544–12559. doi: 10.1074/jbc.M117.785063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao T.J., Jang H., Fushman D., Nussinov R. Allosteric KRas4B Can modulate SOS1 fast and slow Ras activation cycles. Biophys J. 2018;115:629–641. doi: 10.1016/j.bpj.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu S., Banerjee A., Jang H., Zhang J., Gaponenko V., Nussinov R. GTP binding and oncogenic mutations may attenuate hypervariable region (HVR)-catalytic domain interactions in small GTPase K-Ras4B, exposing the effector binding site. J Biol Chem. 2015;290:28887–28900. doi: 10.1074/jbc.M115.664755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu S., Jang H., Nussinov R., Zhang J. The structural basis of oncogenic mutations G12, G13 and Q61 in small GTPase K-Ras4B. Sci Rep. 2016;6:21949. doi: 10.1038/srep21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muratcioglu S., Jang H., Gursoy A., Keskin O., Nussinov R. PDEδ binding to Ras isoforms provides a route to proper membrane localization. J Phys Chem B. 2017;121:5917–5927. doi: 10.1021/acs.jpcb.7b03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozdemir E.S., Jang H., Gursoy A., Keskin O., Li Z., Sacks D.B. Unraveling the molecular mechanism of interactions of the Rho GTPases Cdc42 and Rac1 with the scaffolding protein IQGAP2. J Biol Chem. 2018;293:3685–3699. doi: 10.1074/jbc.RA117.001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozdemir E.S., Jang H., Gursoy A., Keskin O., Nussinov R. Arl2-mediated allosteric release of farnesylated KRas4B from shuttling factor PDEδ. J Phys Chem B. 2018;122:7503–7513. doi: 10.1021/acs.jpcb.8b04347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakrabarti M., Jang H., Nussinov R. Comparison of the conformations of KRAS isoforms, K-Ras4A and K-Ras4B, points to similarities and significant differences. J Phys Chem B. 2016;120:667–679. doi: 10.1021/acs.jpcb.5b11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M., Jang H., Gaponenko V., Nussinov R. Phosphorylated calmodulin promotes PI3K activation by binding to the SH2 domains. Biophys J. 2017;113:1956–1967. doi: 10.1016/j.bpj.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M., Jang H., Nussinov R. The mechanism of PI3Kα activation at the atomic level. Chem Sci. 2019;10:3671–3680. doi: 10.1039/c8sc04498h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jang H., Banerjee A., Marcus K., Makowski L., Mattos C., Gaponenko V. The structural basis of the farnesylated and methylated KRas4B interaction with calmodulin. Structure. 2019;27:1647–1659. doi: 10.1016/j.str.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woolf T.B., Roux B. Molecular dynamics simulation of the gramicidin channel in a phospholipid bilayer. Proc Natl Acad Sci U S A. 1994;91:11631–11635. doi: 10.1073/pnas.91.24.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woolf T.B., Roux B. Structure, energetics, and dynamics of lipid-protein interactions: a molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 1996;24:92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 50.Kucerka N., Tristram-Nagle S., Nagle J.F. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 51.Darden T, York D, Pedersen L: Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem Phys 1993, 98:10089–10092.
- 52.Wu X.W., Brooks B.R. Self-guided Langevin dynamics simulation method. Chem Phys Lett. 2003;381:512–518. [Google Scholar]
- 53.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 54.Fiorin G., Klein M.L., Henin J. Using collective variables to drive molecular dynamics simulations. Mol Phys. 2013;111:3345–3362. [Google Scholar]
- 55.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 57.Daub M., Jockel J., Quack T., Weber C.K., Schmitz F., Rapp U.R. The RafC1 cysteine-rich domain contains multiple distinct regulatory epitopes which control Ras-dependent Raf activation. Mol Cell Biol. 1998;18:6698–6710. doi: 10.1128/mcb.18.11.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohler M., Brummer T. B-Raf activation loop phosphorylation revisited. Cell Cycle. 2016;15:1171–1173. doi: 10.1080/15384101.2016.1159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang B.H., Guan K.L. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J. 2000;19:5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu J., Stites E.C., Yu H., Germino E.A., Meharena H.S., Stork P.J.S. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell. 2013;154:1036–1046. doi: 10.1016/j.cell.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw A.S., Kornev A.P., Hu J., Ahuja L.G., Taylor S.S. Kinases and pseudokinases: lessons from RAF. Mol Cell Biol. 2014;34:1538–1546. doi: 10.1128/MCB.00057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Y., Prakash P., Liang H., Cho K.J., Gorfe A.A., Hancock J.F. Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell. 2017;168(239–251) doi: 10.1016/j.cell.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kenworthy A.K., Nichols B.J., Remmert C.L., Hendrix G.M., Kumar M., Zimmerberg J. Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niv H., Gutman O., Kloog Y., Henis Y.I. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J Cell Biol. 2002;157:865–872. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murakoshi H., Iino R., Kobayashi T., Fujiwara T., Ohshima C., Yoshimura A. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci U S A. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plowman S.J., Muncke C., Parton R.G., Hancock J.F. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci U S A. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abankwa D., Gorfe A.A., Hancock J.F. Ras nanoclusters: molecular structure and assembly. Semin Cell Dev Biol. 2007;18:599–607. doi: 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee Y, Phelps C, Huang T, et al. High-throughput, single-particle tracking reveals nested membrane domains that dictate KRas(G12D) diffusion and trafficking. Elife 2019, 8. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.