Abstract
Lower respiratory infections remain one of the top global causes of death and the emergence of new diseases continues to be a concern. In the first two decades of the 21st century, we have born witness to the emergence of newly recognized coronaviruses that have rapidly spread around the globe, including severe acute respiratory syndrome virus (SARS) and Middle Eastern respiratory syndrome virus (MERS). We have also experienced the emergence of a novel H1N1 pandemic influenza strain in 2009 that caused substantial morbidity and mortality around the world and has transitioned into a seasonal strain. Although we perhaps most frequently think of viruses when discussing emerging respiratory infections, bacteria have not been left out of the mix, as we have witnessed an increase in the number of infections from Legionella spp. since the organisms’ initial discovery in 1976. Here, we explore the basic epidemiology, clinical presentation, histopathology, and clinical laboratory diagnosis of these four pathogens and emphasize themes in humans’ evolving relationship with our natural environment that have contributed to the infectious burden. Histology alone is rarely diagnostic for these infections, but has been crucial to bettering our understanding of these diseases. Together, we rely on the diagnostic acumen of pathologists to identify the clinicopathologic features that raise the suspicion of these diseases and lead to the early control of the spread in our populations.
Keywords: SARS, MERS, Influenza, Legionella, Pathology
Emerging respiratory infections have captured the public's and scientific/medical communities’ fascination and concern since the time of the pneumonic plague and have manifested more recently in popular movies depicting airborne virus outbreaks and debate about the potential for respiratory spread of the Ebola virus. Common themes in emerging infections include zoonotic hosts and changes in human behavior, including increased international travel and/or modification of our physical environment. While several emerging agents may exhibit respiratory involvement, herein we focus on emerging pathogens that primarily involve the respiratory system and focus on 4 representative agents that demonstrate a range of features of emerging diseases: SARS, MERS, 2009 pandemic influenza, and Legionella spp.
SARS
Etiology, epidemiology, and clinical presentation
Severe acute respiratory virus (SARS) is a rapidly fatal pulmonary infection caused by the SARS coronavirus (SARS-CoV). The first documented outbreak occurred primarily in China from 2002–2004 with regional outbreaks in North America and Europe attributed to human-to-human transmission from infected international travelers. Genetic characterization suggests that the introduction into the human population occurred from civets or other mammals found in live-animal markets of China.1 A total of 8096 cases from 29 countries, including 774 fatalities were identified during the course of the outbreak (case fatality rate: 9.6%).2 A robust international response costing an estimated 40 billion dollars helped contain the outbreak by 2004, with no new cases reported since then.2 In November 2017, a colony of horseshoe bats in southwestern China harboring a genetically similar coronavirus was identified and raised the possibility that the virus originated in bats before spreading to the aforementioned animal markets.3
SARS-CoV is transmitted between humans through respiratory droplets and close interactions with some individuals acting as so-called super spreaders.4 Infected patients generally present with symptoms 2–12 days after infection. Epidemiological studies during the outbreak identified a disproportionate number of deaths occurring in the elderly and those with immunosuppression. Severe disease was limited in those 12 years of age and younger.5 Symptoms are generally non-specific with fever, myalgia, and malaise among the most common. Laboratory derangements include lymphopenia, thrombocytopenia, elevated C-reactive protein, and elevated lactate dehydrogenase (LDH). Prognostic studies suggest an increased risk for serious outcomes with increased age, elevated pulse, and LDH levels.6
Histopathologic features
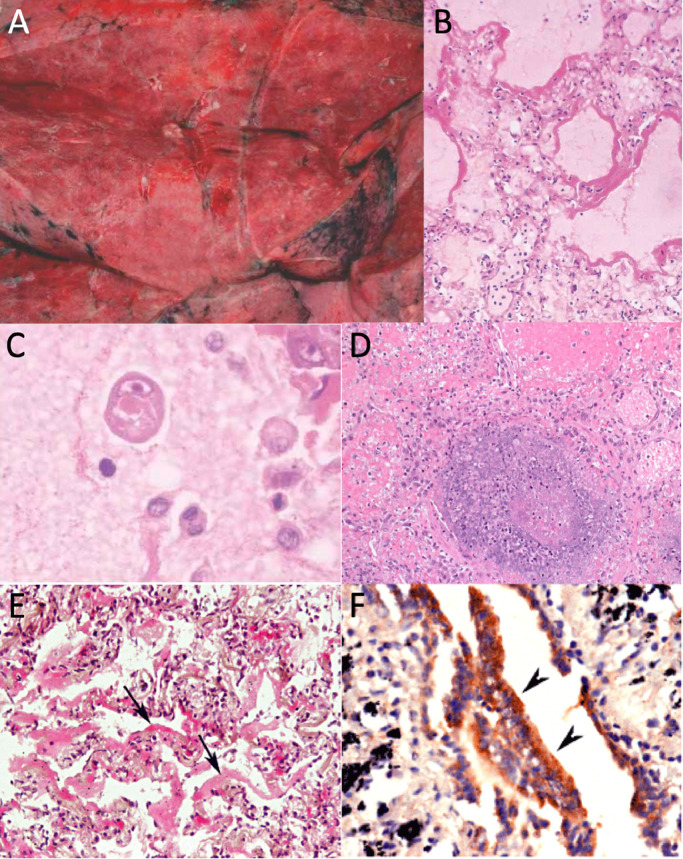
The available histopathologic data for SARS-CoV has been primarily ascertained from autopsy cases. In reported cases of fatal SARS, gross evaluation of the organs found edematous, heavy lungs weighing up to 2100 g with multiple areas of congestion (Fig. 1 A). Cut sections demonstrated irregular, patchy areas of consolidation suspicious for pneumonia. Inspection of the upper respiratory tract demonstrated mucopurulent material.7, 8
Fig. 1.
Emerging and reemerging viral respiratory infections, examples: A) SARS gross pathology,9 B) SARS hyaline membranes,9 C) SARS intracytoplasmic inclusions,9 D) MERS pulmonary necrosis,30 E) H1N1pdm09 hyaline membranes,52 and F) H1N1pdm09 antigens in bronchiolar epithelial cells.52 All panel images ©John Wiley and Sons as cited.
The histological presentation of pulmonary SARS infections is non-specific and dependent on the time since symptom onset. Early phase disease (<11 days) is most commonly associated with acute diffuse alveolar damage. Later phase disease demonstrates a combination of diffuse alveolar damage (Fig. 1B) and acute fibrinous and organizing pneumonia. In addition, features of squamous metaplasia, multinuclear giant cell formation, and intravascular microthrombi may be apparent. Intracytoplasmic viral inclusions have also been described in alveolar epithelial cells (Fig. 1C).9 Autopsy cases from the SARS outbreak have not revealed a correlation between the pattern of damage and a clinically significant outcome. Furthermore, no pathognomic autopsy features have been identified.7, 10 As the majority of patient specimens were taken from autopsy, there is little documentation of the pathologic presentation of SARS-CoV in living patients. One report documents a single open lung biopsy that was taken approximately one week after symptom onset in a patient subsequently diagnosed with SARS. Pathologic findings included a mild increase in interstitial lymphocytes and a moderate increase of alveolar macrophages with hyaline membrane formation. There were occasional pneumocytes demonstrating viral cytopathic-like changes including cytomegaly with nuclear enlargement and prominent nucleoli.7
In situ hybridization and immunohistochemical stains have been used to localize viral elements within tissue samples—potentially useful diagnostically and to study viral tropism. Studies have identified SARS-CoV in the lungs, intestinal enterocytes, and splenic white pulp.8 Within the lungs, immunofluorescent in situ hybridization studies have colocalized viral RNA with cellular cytokeratins, suggesting the infected cells are pneumocytes.11 SARS-CoV may occasionally be identified within the pulmonary macrophages. Ultrastructural studies similarly demonstrate SARS-CoV virions and nucleocapsid inclusions within pneumocytes.12
Clinical laboratory diagnostics
Due to the non-specific nature of SARS-CoV infection and rapid spread, easily implementable and accurate diagnostics were a necessary component of disease control. Tests to detect viral RNA by RT-PCR were rapidly developed following the initial SARS outbreak. The sensitivity of molecular detection is affected by the sample collection site and time since infection. In general, blood is most often the first body fluid in which viral RNA is detected. Over 70% of patients will be positive 3 days after symptom onset with peak values at days 5–6.13 In comparison, viral RNA is detected in only 30–40% of respiratory tract samples during the first four days and peaks at day 10. If nasopharyngeal swabs are used diagnostically, serial testing should be considered, as a minority of infected patients are positive at the time of presentation.14
Viral RNA has also been detected in up to 90% of stool samples 15–17 days post-symptom onset in serologically-confirmed SARS patients.14 For these reasons, a multimodal approach sampling different body fluids at various time points early in infection may be helpful for disease identification when there is a high pre-test probability. The clinical sensitivity of RT-PCR tests may be affected by the region of the virus targeted: The initial SARS PCR assays were directed towards the polymerase gene, while subsequent in vitro data demonstrated a 100-fold increase in sensitivity by targeting the nucleocapsid gene.15
Serologic tests using ELISA and indirect immunofluorescence have been used to monitor convalescent patients and perform serosurveys. The majority of infected patients will seroconvert: IgM values peak between weeks 2–3 after onset of symptoms while remaining detectable up to 12 weeks post-infection; IgG values reach peak titers more slowly and may persist indefinetly.16, 17 Serological studies have used a range of reagents and methods from indirect immunofluorescence to ELISAs with purified virus or recombinant nucleocapsid protein.18
MERS
Etiology, epidemiology and clinical presentation
Middle East Respiratory Syndrome (MERS) was first identified during September 2012 in a Saudi Arabian patient with respiratory failure.19 In comparison to the rapid spread and subsequent quiescence of SARS-CoV, MERS-CoV has continued to circulate and produce sporadic outbreaks both within the Arabian Peninsula and in countries where infected patients have traveled. There have been 2266 confirmed cases of MERS and 804 fatalities (case fatality: 35.5%).20 Significant molecular and serological data point to dromedary camels as the source of transmission into human populations.21 Whether camels are an intermediate host or reservoir is still under research. Studies have found fragments of MERS-CoV genomic material identical to human isolates in bat populations.22 While transmission from camels may introduce MERS-CoV into human populations, the majority of reported cases have resulted from human-to-human nosocomial transmission. In one outbreak, a single ill patient admitted to a Korean hospital led to 186 infections including 36 fatal cases.23 Person-to-person spread has also been documented within households, where patient respiratory secretions and close proximity carry the highest risk of tranmission.24 For these reasons, patients with symptoms and epidemiological features suggestive of MERS should be rapidly quarantined and tested for viral infection.
The clinical presentation varies from asymptomatic infection to severe disease. In general, older patients and those with chronic comorbid conditions (diabetes, heart disease) are at greatest risk for progression to respiratory failure.23 Despite the high mortality associated with symptomatic cases, studies have revealed that approximately 25% of patients are asymptomatic when infected with MERS-COV.25 Clinical disease in children is uncommon.23 A seroepidemiological study of over 10,000 patient samples from Saudi Arabia found positive antibodies in 0.15% of patients. Those with camel-exposure had a higher probability of positive serology and may serve as an asymptomatic or subclinical transmission source.26 Clinical symptoms are non-specific and include runny nose, sore throat, and myalgias. The incubation period ranges from 2 to 14 days.23 Symptoms appear before the patient reaches a detectable viremia. In addition to respiratory symptoms, gastrointestinal distress and neurological sequelae have been documented.23 The most common laboratory derangements observed are leukopenia, thrombocytopenia, and anemia.
Histopathologic features
The pathological findings associated with human MERS-CoV infection have been limited due to the rare number of autopsies performed. In a case reported by Ng et al. a man died 8 days after admission due to respiratory failure and shock.27, 28 Studies included histology, immunohistochemistry with multiple MERS-CoV antibodies, and ultrastructural evaluation. The primary finding at autopsy was viral-mediated lung damage with features of diffuse alveolar damage. Infected cell types included pneumocytes, multinucleated epithelial cells, and bronchial submucosal glands. These infected cells expressed DPP-4 surface antigen which serves as the cell receptor of MERS-CoV. Samples of other DPP-4 positive tissues including lymphoreticular system, kidneys, gastrointestinal tract, and brain were negative for virus by multiple methods.29 In a second documented autopsy, the patient was a 33-year-old man receiving chemotherapy for T-cell lymphoma who received an antemortem diagnosis of MERS-CoV infection by sputum PCR; cause of death was attributed to respiratory and renal failure. The autopsy revealed a necrotizing pneumonia with diffuse alveolar damage (Fig. 1D), but was partly limited by possible confounding effects of therapy and tissue samples limited to needle core biopsies. Viral inclusions were identified by electron microscopy not only in the respiratory epithelium but also the epithelial cells of the renal proximal tubules.30 Also reported in the literature is a case of a man who 8 weeks following infection with MERS-CoV underwent renal biopsy. Histology demonstrated acute tubular sclerosis with proteinaceous cast formation, acute tubulointerstitial nephritis, and no glomerulosclerosis. In situ hybridization and electron microscopy of renal tissue was negative for viral components.31
Despite the paucity of autopsy cases, multiple studies have examined the pathogenesis of MERS-CoV in human tissue ex vivo and in animal models.23 , 32 In a study comparing the replication of camel-isolated to human-isolated MERS-CoV strains, researchers found similar replication kinetics and cellular tropism. All strains infected non-ciliated bronchial epithelium and alveolar epithelial cells including type II pneumocytes. No infection of the pulmonary macrophages was identified.33 Studies in kidney explants demonstrated infection of multiple cell types including renal tubular cells, vascular endothelial cells, and podocytes. 34 Experimental infection of small intestine explants with MERS-CoV demonstrated infection limited to the surface enterocytes and formation of syncytial cells. In keeping with these observations, infected patients have been found to shed virus in their urine and stool.35 Mouse models expressing the human DPP-4 receptor were rapidly developed after the discovery of the virus. These models have been used to further understand the inflammatory patterns and viral localization, which identified evidence of the virus in the lungs, brain, heart, spleen, and intestines.36 A Rhesus macaque model demonstrated a similar disease phenotype as seen in humans, including pulmonary consolidation, edematous lung lesions, and pneumonia. Also frequently observed were multinucleated giant cells and acute diffuse alveolar damage with hyaline membrane formation. In one study, infectious virus was identified in the lung, but not in the upper respiratory track, blood, or other solid organs.37, 38, 39
Clinical laboratory diagnostics
During transmission events, RT-PCR has served as the primary clinical laboratory diagnostic test. Important to the success of these tests is an understanding of the viral kinetics and tissue tropism found in MERS-CoV cases. Several studies have documented that lower respiratory tract samples provide the highest viral loads. The virus may also be detected in upper respiratory swabs, whole blood or serum, feces, and urine.35 Despite 10 to 100 times lower virus levels, samples from the upper respiratory tract and blood can provide additional diagnostic utility by providing a convenient sample type. Blood is only positive in one-half to one-third of cases, but detectable viremia at the time of diagnosis has been associated with an increased requirement for mechanical ventilation and patient death. 40 The limited viremia rate is in contrast to SARS-CoV, where RT-PCR on blood can be helpful for initial diagnosis and is often the first positive site identified. For MERS-CoV, testing of upper and lower respiratory samples along with patient blood may help maximize the sensitivity while also stratifying risk. Two RT-PCR testing strategies have been granted emergency use authorization by the FDA: both target a region upstream of the envelope gene; while one additionally targets a region of the ORF1a gene, the other targets two regions within the nucleocapsid gene.41, 42
Serology in MERS-CoV infection follows similar kinetics to that of SARS-CoV. The majority of patients develop detectable levels of IgM and IgG antibodies 2–3 weeks following symptom onset. In most cases, the detection of IgM does not add diagnostic value compared to IgG.35 If serologic testing is used to diagnose recent infection, a neutralization assay and 4-fold increase in titer after 14 days should be used to confirm a specific immune response.43 Antibody responses may be affected by the disease severity. Several studies have demonstrated PCR-positive patients with mild disease often do not produce detectable levels of antibodies, particularly when followed past the acute phase of the disease. 44
Pandemic influenza 2009
Etiology, epidemiology, and clinical presentation
In contrast to the emergence of MERS and SARS, the 2009 H1N1 influenza pandemic was a re-emergence of a pathogen most commonly responsible for seasonal infections, but with demonstrated pandemic potential. The influenza genome contains 8 segments and is capable of both antigenic drift and shift, where drift represents small nucleotide mutations and shift represents exchange of genome segments. These larger shifts are central to the pandemic potential of the virus as it can result in novel antigens not previously seen in the human population. The virus (H1N1pdm09) first emerged in Mexico during the spring of 2009.45 The virus contains gene segments from human, porcine, and avian influenza strains.46 The WHO documented 17,483 laboratory-confirmed influenza fatalities from over 213 countries during the 2009 outbreak, although this is likely a gross under-representation as the CDC estimated 12,469 influenza fatalities in the US alone.47 Estimates of the global mortality resulting from influenza pandemics has decreased from 1–3% in 1918 to 0.001–0.007% during 2009.47 A significant contributing factor to the mortality difference is thought to be due to expanded medical interventions, including improved treatment of acute respiratory distress and secondary bacterial pneumonias. Since its introduction in human populations, the 2009 strain has become a commonly circulating strain that is now incorporated into the seasonal vaccine.
The full clinical spectrum of pandemic influenza infection differs little from seasonal influenza. However, during the 2009 pandemic, patients infected with the pandemic strain were younger, less likely to have underlying conditions, more frequently required ventilatory support or ICU care, and had extrapulmonary complications.48, 49 Younger patients also had higher rates of adverse outcomes as compared to seasonal cases.48 The majority of infections ranged from subclinical to significant malaise with fever, myalgias, and rhinorrhea. In those cases progressing to severe disease, acute onset hypoxemia and acute respiratory distress were frequent. In addition, patients with severe influenza requiring hospitalization were at increased risk of developing secondary bacterial pneumonia, acute kidney injury, and requiring mechanical ventilation.50
Histopathologic features
Prior to 2009, the most recent pandemic influenza outbreak occurred in 1968, before the broad adoption of immunohistochemical and molecular techniques for characterizing infectious diseases. During the 2009 pandemic healthcare researchers expanded their study of viral receptor binding, replication within the lower respiratory system, and molecular characterization of secondary bacterial pneumonia. Autopsy case series from multiple countries were published and used a variety of techniques including ultrastructural, immunohistochemical, in situ hybridization, and molecular methods to varying degrees to gain insight into the pathology of pandemic influenza infections. The majority of reports came from public health laboratories and varied from single case reports to cohorts of up to 100 fatal cases.51, 52, 53
Gross organ findings from H1N1pdm09 infections demonstrated edematous and heavy lungs.54 Cut sections frequently revealed thrombosis of small pulmonary vessels and areas of consolidation suggestive of bacterial pneumonia.55 Other gross organ findings included cerebral edema, hepatic congestion, and hemorrhagic necrosis of the adrenal glands.54, 56 Underlying conditions including obesity, cardiovascular disease, asthma, and pregnancy were common in fatal cases.51
Microscopically, acute respiratory distress manifested in the lungs as diffuse alveolar damage with intra-alveolar hyaline membranes (Fig. 1E) in various degrees of organization and thrombotic pulmonary vessels. Additional histologic findings included bronchoalveolar pneumonia, interstitial septal thickening, and type II pneumocyte hyperplasia. Less frequently observed was fibrosis and squamous metaplasia.51, 52, 53, 54 , 56, 57 Within the upper respiratory tract, tracheitis and bronchitis ranged from focal to severe. The epithelium was frequently ulcerated and hemorrhagic and submucosal glands were commonly inflammed.51, 53, 57 Immunohistochemical stains and electronic microscopy indicated viral localization in the epithelium (Fig. 1F), submucosal glands, pneumocytes, alveolar macrophages, and rarely in endothelial cells, but not in non-respiratory tissues.51
Extrapulmonary manifestations of influenza were observed in the 2009 pandemic, especially neurologic and cardiac dysfunction, as has been described in seasonal cases.48, 54, 58 In a case of fatal 2009 pandemic influenza with encephalitis, examination of the brain revealed global edema, tonsillar herniation, punctate hemorrhages in the medulla and amyloid precursor protein deposition consistent with a pattern of ischemic injury.54 The same series from New Mexico reported a case of fulminant pediatric myocarditis.54 Similarly, myocarditis has been described in fatal pediatric seasonal cases.59 When documented, hepatic and renal changes were generally non-specific.
Animal models of 2009 pandemic influenza indicate better replication within the lower respiratory tract and higher titer virus shedding as compared to seasonal influenza.60 Histopathologic findings in these models mimic that of human disease and display features of necrotizing bronchiolitis, alveolar exudates, and antigen positive pneumocytes. In non-human primates infected with pandemic influenza, more severe acute cytopathic effects within the bronchioles and alveoli were observed as comparted to a seasonal strain. Within the lower respiratory tract, pandemic influenza demonstrated a similar increase in virulence with viral titers peaking at higher levels and persisting for longer duration.61, 62
In addition to the histopathological effects caused by viral infection, secondary bacterial infections were also common during the 2009 influenza pandemic. Studies estimated anywhere from one-quarter to one-half of patients with fatal influenza infections had a co-existing bacterial pneumonia.51, 57 In cases with histologic evidence of acute pneumonia, morphologic confirmation of bacterial forms by hematoxylin and eosin or special stains was possible in over 75% of cases.51 The histologic features most often observed in conjunction with bacterial pneumonia were epithelial necrosis and neutrophilic inflammation. Finding of bacterial pneumonia were more prominent in later stages of influenza infection.63 In cases with bronchopneumonia identified on histology, post-mortem culture or PCR identified bacteria in 50% of 2009 pandemic influenza the cases. The most common organisms identified in these patients were Streptococcus pneumoniae and Streptococcus pyogenes. Other less common pathogens included Streptococcus mitis, methicillin-resistant Staphylococcus aureus, and Pseudomonas aeruginosa.51, 53, 57
Clinical laboratory diagnostics
In patients with suspected influenza infection, diagnostic methods include viral culture, rapid antigen tests, and molecular assays. The majority of clinical diagnostic labs have shifted to using rapid molecular tests, which may be performed as a laboratory-developed real-time reverse transcription PCR (rRT-PCR) panel or cartridge-based assay.64, 65 These methods demonstrate excellent analytic parameters and have a turn-around time on the order of minutes to hours. However, the shortcomings of these rapid molecular tests were made evident during the 2009 pandemic. As the primer and probe sets were designed around recent seasonal strains of influenza A and B, the novel genetic elements of the 2009 pandemic strain significantly reduced the sensitivity and specificity of these tests.66 Fortunately, molecular assays were rapidly adapted to detect the 2009 pandemic strain. Rapid antigen tests, already less sensitive than molecular testing, were noted to be particularly insensitive to the 2009 pandemic strain.66 Clinical laboratories should increase their suspicion of negative results during a novel pathogen outbreak and work closely with state public health laboratories to validate new strain testing.
With regards to testing patients for influenza-like illnesses, nasal/nasopharyngeal swabs or aspirates are the preferred, and most often FDA-cleared, specimens. These specimens can also be obtained post-mortem for autopsy investigation and epidemiological surveillance. The majority fatal cases during the 2009 pandemic had positive nasopharyngeal or tracheal swabs by rRT-PCR and greater than 50% of cases had viral nucleic acid present in formalin fixed, paraffin embedded (FFPE) tissues.51 H1N1pdm09 has been shown to be detectable in stool, blood, and urine, although these samples were not closely examined during pandemic autopsy series.67, 68
Legionella spp.
Etiology, epidemiology, and clinical presentation
Legionella pneumophila was initially recognized as the agent of atypical pneumonia associated with the namesake Legionnaire's convention in Philadelphia in 1976. Legionella species are opportunistic pathogens comprising a diverse genus with over 50 species and over 70 serogroups. Roughly one-half have been described associated with human disease.69 L. pneumophila serogroup 1 is most commonly associated with human infection, although other serogroups and species together cause a substantial disease burden.70, 71 Legionella can also cause non-respiratory diseases such as Pontiac fever as well as rare wound and other infections. The organism is a facultative intracellular Gram-negative rod that is common in the environment and naturally infects several species of free-living amoeba in a process that parallels infection of the mammalian macrophage. Despite highly evolved virulence traits the disease typically occurs in immunocompromised patients and those with respiratory compromise.72 Host amoeba include both soil and water-dwelling species; most human infections in the western hemisphere with known sources are attributable to inhalation of water from human-engineered water systems. The organism, likely in concert with its host amoeba, colonize fresh water systems including air conditioners, heater-coolers, potable water systems, etc. and form tenacious biofilms that are extremely challenging to clear. Showers, fountains, artificial waterfalls, and other similar sources that generate aerosols or droplets are a particular risk. Infection is both nosocomial and community-acquired. Only one case of possible human-to-human transmission has been documented and transmission from natural water sources is not widely recognized.73 The disease is thought to cause ∼2–9% of community-acquired pneumonias.72 Legionella is a nationally reportable infection and has demonstrated a 5.5-fold increase in cases from 2000–2017 to an approximate 7500 reported cases in 2017, although that figure may represent as few as 5% of actual cases.72, 74 The increase in cases is not entirely understood, but may represent increased reporting, changing environmental factors, or increased host susceptibility.74 Infections from species other than L. pneumophila can cause disease in a high portion of patients in some areas, but these species are often under-appreciated because the most common diagnostic test only detects L. pneumophila serogroup 1. Many reports indicate Legionella micdadei is the second most common species to cause human disease.70, 75
Most patients are symptomatic within 2–10 days, however, incubation periods of up to 26 days have been described.76 Complicating the diagnosis, especially early in infection, is a relative paucity of respiratory symptoms. Cough, when present, is frequently non-productive. Fever, gastrointestinal symptoms, bradycardia, altered mental status, leukocytosis, hyponatremia, and elevated transaminases are common and may present before respiratory symptoms.77 The presence of these extrapulmonary findings should raise the clinical suspicion for the disease compared to the more common bacterial pneumonias. Radiologic findings are non-specific and can include progressive patchy infiltrates, consolidation, and pleural effusions. The disease can also present with cavitary lesion(s), especially L. micdadei: In one series of 27 patients, 20% had cavitary disease.78 Case fatality rates are highly variable from ∼5–40%. Most fatalities are attributed to L. pneumophila, however, death from L. micdadei is also common despite demonstrated lower virulence in in vitro models.75, 79 Treatment is with levofloxacin or azithromycin; empiric treatment with beta-lactams can delay time to effective therapy, as they are considered ineffective.
Histopathologic features
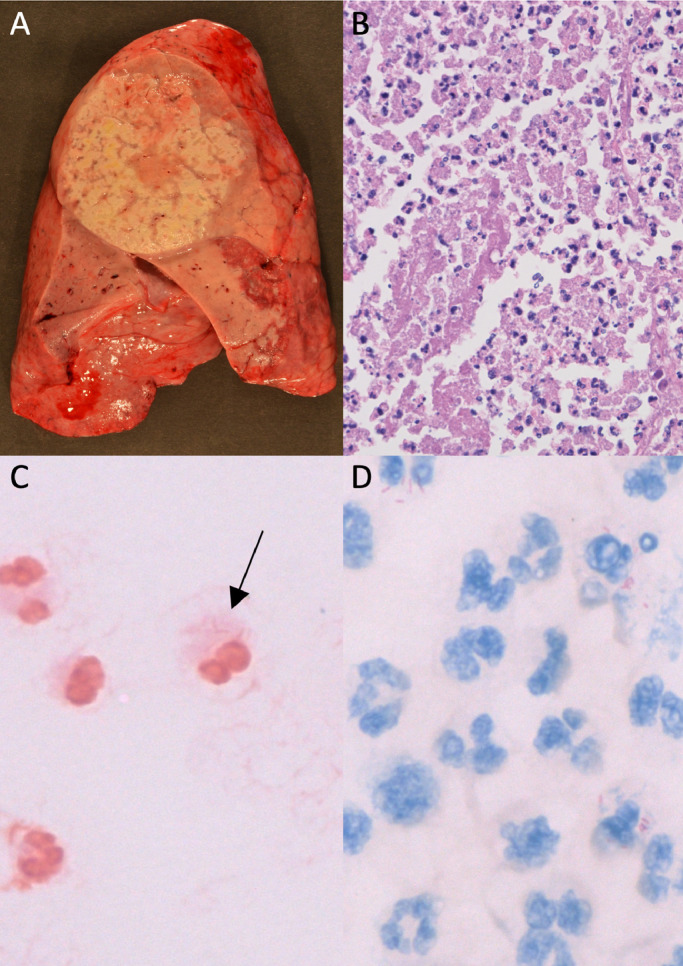
Similar to the emerging viral respiratory infections and the clinical and radiologic features of Legionella species infection, there are no pathognomonic histopathologic features diagnostic of Legionella Infections. However, in the appropriate clinicopathologic context, the presence of several features should raise the question of Legionella in the differential diagnosis and the appropriate clinical laboratory testing should be considered. Gross findings include congested firm and distended lungs, weighing an average of ∼2000–2500 g; disease presents unilaterally or bilaterally and most commonly as lobular or confluent lobular pneumonia.80, 81 Cavitations, nodules, abscesses, and empyema may be present, particularly with L. micdadei (Fig. 2 A).81 Microscopic findings include marked fibrinopurulent exudate with a mixed necrotic infiltrate of neutrophils and macrophages that completely fills the alveolar spaces (Fig. 2B).80 Compared to other acute bacterial pneumonias, there is a relative abundance of macrophages.80 The organism stains weakly or not at all by Gram or Brown and Hopps (Fig. 2D). The organism is most evident on silver stains, including Warthin-Starry, Steiner, or Dieterle. The organism can be seen intracellularly within macrophages or free in the necrotic milieu. L. micdadei, but not other species, stain using both microbiologic and histopathologic modified acid-fast stains (Fite) (Fig. 2D), which can lead to diagnostic mis-adventures.81, 82 In addition to Mycobacteria and several parasites, a handful of Gram-positive organisms including Nocardia, Rhodococcus, Tsukamurella, Gordonia, and Dietzia can also stain by modified acid-fast stains. In contrast to L. micdadei, all of these organisms should stain Gram positive. In addition, Nocardia appears as a branching Gram positive rods, helping differentiate it from L. micdadei. Independently, positive staining with Fite can be particularly helpful when culture was not performed and molecular studies fail to amplify nucleic acid. Immunohistochemical stains are not widely available. Electron microscopy was critical in the early understanding of the infection and organism's life cycle, but does not play a role in routine diagnosis. The histopathologic infectious differential includes influenzae and non-necrotizing hemorrhagic bacterial pneumonias, including Klebsiella pneumoniae, Haemophilis influenzae, and S. pneumoniae.
Fig. 2.
Legionella micdadei: A) Gross pathology, B) hematoxylin and eosin stain, C) faint Gram staining characteristic of Legionella species, and D) modified acid-fast staining characteristic of L. micdadei, but not other Legionella species. Prosector acknowledgement: Drs. Kelly Smith and Gregory Cheeney.
Clinical laboratory diagnostics
It is crucial for the anatomic pathologist to understand the different clinical laboratory diagnostics for Legionella species given the array of available tests and their limitations. For example, the diligent autopsy pathologist who reviews a patient chart and notes a negative lower respiratory culture and urine antigen test may be misled away from a diagnosis of Legionella infection even though the etiology could still be a Legionella sp. not detected by these methods. The gold standard clinical laboratory diagnostic is culture. Legionella spp. are relatively slow growing in vitro, typically taking 3–4 days to form colonies, and do not grow on routine clinical solid media. Agar plates supplemented with activated charcoal are required for growth (i.e. charcoal-buffered yeast extract; BCYE). Because of these limitations and cumbersome nature of these cultures, many clinical microbiology laboratories do not perform cultures for the organism and even when they do, these special media may not be included in all routine lower respiratory cultures. More widely available and used is the urine antigen test, which is available as an FDA-cleared simple lateral flow assay with high sensitivity and specificity that can be performed within ∼15 min. The critical caveat is that the assay only detects the excreted antigens of L. pneumophila serogroup 1. Additional caveats include a small percent of patients that do not excrete the antigen and frequently the antigen can be detected for several weeks after infection, which complicates interpretation of the time-course of infection.69, 83 Studies vary greatly in the local species and serogroup distributions; likely components of actual geographic distribution of the organism and reporting bias. In one study, half of all sporadic cases over a 15 year period were due to species other than L. pneumophila serogroup 1.75 Culture has the additional benefit of supporting epidemiologic investigations and infection control. It yields a bacterial isolate that can be compared to environmental isolates from hospital water sources or other human-engineered systems in question by strain typing assays. This has historically been performed by pulse field gel electrophoresis or sequence-based typing of a limited number of alleles, but is contemporarily performed with a higher degree of resolution by whole genome sequencing (WGS).84 WGS is most commonly performed by public health laboratories for outbreak investigations, but is also available in at least one CLIA laboratory to support rapid decision making by hospital infection control experts.
Molecular diagnostics for the detection of L. pneumophila on direct patient specimens are also available in several clinical laboratories. Methodologies and target sequences vary, but frequently employ RT-PCR with or without amplicon sequencing. Legionella genus-specific regions of the 16S rRNA gene, 5S rRNA, and mip, are often targeted.85 Assays vary as to whether they detect all or most Legionella species, L. pneumophila only, or a limited number of species. Although these molecular assays are not as rapid as the urine antigen test and are more expensive, they are highly sensitive, can be performed faster than culture, and have the distinct advantage of detecting strains other than L. pneumophila serogroup 1. Legionella-specific PCR assays are available for FFPE samples, although in a limited number of public health and CLIA reference laboratories. The availability of these assays for FFPE can be particularly useful for the autopsy pathologist when Legionella was not suspected antemortem. Broad-range 16S rRNA also may detect the organism, however, frequently at a reduced sensitivity compared to the species-specific assays. Serology and immunofluorescence have historical roles, but have a limited role in contemporary clinical diagnosis. The organism stains poorly by safranin Gram counter stain. L. micdadei is modified acid-fast stain positive (mAFB), as discussed above.
Conclusions
Pathologists are an essential component of monitoring and identifying the emergence of new pathogens. Classic populations of concern include those who are immunocompromised and those at the interface of animal-human boundaries. Even when an infectious agent is well known, a lack of herd immunity from antigenic shift or waning immunity from vaccine-preventable diseases or voluntary abstinence can contribute to rapid spread of a disease within the human population. Particularly for respiratory infections, international travel presents an ever-increasing risk: The International Air Transport Association reported that in 2017, there was a record 4.1 billion passenger trips.86
The role of histopathology during outbreaks and pandemics helps to recognize the extent of respiratory involvement as well as extent of other tissues affected. Although many of the different infectious agents cause similar histopathologic features, some features can help narrow sufficiently the differential diagnosis to successfully harness more specific molecular methods. On the other hand, many molecular laboratory assays target only specific pathogens and the assays require intact nucleic acid in the specimen. Cultures cast a broad net, but may also require specific growth conditions to isolate microorganisms. Knowing the broad array of different diagnostic methods is paramount in identifying emerging and reemerging pathogens.
References
- 1.Guan Y., Zheng B.J., He Y.Q., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 2.CDC. Disease of the Week week - SARS. Centers for Disease Control and Prevention. http://www.cdc.gov.offcampus.lib.washington.edu/dotw/sars/index.html. Published March 3, 2016. Accessed 15 January 2019.
- 3.Hu B., Zeng L.-P., Yang X.-L., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13(11) doi: 10.1371/journal.ppat.1006698. Drosten C., ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen S.J., Chang H.-L., Cheung T.Y.-Y., et al. Transmission of the severe acute respiratory syndrome on aircraft. N Engl J Med. 2003;349(25):2416–2422. doi: 10.1056/NEJMoa031349. [DOI] [PubMed] [Google Scholar]
- 5.Leung C., Kwan Y., Ko P., et al. Severe acute respiratory syndrome among children. Pediatrics. 2004;113(6):e535–e543. doi: 10.1542/peds.113.6.e535. [DOI] [PubMed] [Google Scholar]
- 6.Chan J.C.K., Tsui E.L.H., Wong V.C.W., Group* THASC Prognostication in severe acute respiratory syndrome: a retrospective time-course analysis of 1312 laboratory-confirmed patients in Hong Kong. Respirology. 2007;12(4):531–542. doi: 10.1111/j.1440-1843.2007.01102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholls J.M., Poon L.L.M., Lee K.C., et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu J., Gong E., Zhang B., et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202(3):415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Y., Wang H., Shen H., et al. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol. 2003;200(3):282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franks T.J., Chong P.Y., Chui P., et al. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum Pathol. 2003;34(8):743–748. doi: 10.1016/S0046-8177(03)00367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.To K.F., Tong J.H.M., Chan P.K.S., et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in‐situ hybridization study of fatal cases. J Pathol. 2004;202(2):157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shieh W.-J., Hsiao C.-H., Paddock C.D., et al. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum Pathol. 2005;36(3):303–309. doi: 10.1016/j.humpath.2004.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant P.R., Garson J.A., Tedder R.S., Chan P.K.S., Tam J.S., Sung J.J.Y. Detection of SARS coronavirus in plasma by real-time RT-PCR. N Engl J Med. 2003;349(25):2468–2469. doi: 10.1056/NEJM200312183492522. [DOI] [PubMed] [Google Scholar]
- 14.Chan K.H., Poon L.L.L.M., Cheng V.C.C., et al. Detection of SARS coronavirus in patients with suspected SARS. Emerg Infect Dis. 2004;10(2):294–299. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuiken T., Fouchier R.A.M., Schutten M., et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349(5):508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 17.Peiris J.S.M., Chu C.M., Cheng V.C.C., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo P.C.Y., Lau S.K.P., Wong B.H.L., et al. Longitudinal Profile of Immunoglobulin G (IgG), IgM, and IgA Antibodies against the Severe Acute Respiratory Syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin Diagn Lab Immunol. 2004;11(4):665–668. doi: 10.1128/CDLI.11.4.665-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 20.WHO | Middle East respiratory syndrome coronavirus (MERS-CoV) 2019. WHO. [Google Scholar]; http://www.who.int/emergencies/mers-cov/en/. Accessed 16 January 2019.
- 21.Azhar E.I., El-Kafrawy S.A., Farraj S.A., et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 22.Memish Z.A., Mishra N., Olival K.J., et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19(11):1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arabi Y.M., Balkhy H.H., Hayden F.G., et al. Middle East respiratory syndrome. N Engl J Med. 2017;376(6):584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arwady M.A., Alraddadi B., Basler C., et al. Middle East respiratory syndrome coronavirus transmission in extended family, Saudi Arabia, 2014. Emerg Infect Dis. 2016;22(8):1395–1402. doi: 10.3201/eid2208.152015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oboho I.K., Tomczyk S.M., Al-Asmari A.M., et al. 2014 MERS-CoV outbreak in Jeddah–a link to health care facilities. N Engl J Med. 2015;372(9):846–854. doi: 10.1056/NEJMoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller M.A., Meyer B., Corman V.M., et al. Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infect Dis. 2015;15(5):559–564. doi: 10.1016/S1473-3099(15)70090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker D.H. Value of autopsy emphasized in the case report of a single patient with Middle East respiratory syndrome. Am J Pathol. 2016;186(3):507–510. doi: 10.1016/j.ajpath.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng D.L., Hosani F., Keating M.K., et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol. 2016;186(3):652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng D.L., Al Hosani F., Keating M.K., et al. Clinicopathologic, Immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol. 2016;186(3):652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alsaad K.O., Hajeer A.H., Al Balwi M., et al. Histopathology of Middle East respiratory syndrome coronovirus (MERS‐CoV) infection – clinicopathological and ultrastructural study. Histopathology. 2018;72(3):516–524. doi: 10.1111/his.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cha R., Yang S.H., Moon K.C., et al. A case report of a Middle East respiratory syndrome survivor with kidney biopsy results. J Korean Med Sci. 2016;31(4):635–640. doi: 10.3346/jkms.2016.31.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J., Li C., Zhao G., et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv. 2017;3(11):eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan R.W.Y., Hemida M.G., Kayali G., et al. Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study. Lancet Respir Med. 2014;2(10):813–822. doi: 10.1016/S2213-2600(14)70158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeung M.-L., Yao Y., Jia L., et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol. 2016;1(3):16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corman V.M., Albarrak A.M., Omrani A.S., et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis Off Publ Infect Dis Soc Am. 2016;62(4):477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li K., Wohlford-Lenane C.L., Channappanavar R., et al. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci. 2017;114(15):E3119–E3128. doi: 10.1073/PNAS.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Wit E., Rasmussen A.L., Falzarano D., et al. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci. 2013;110(41):16598–16603. doi: 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baseler L.J., Falzarano D., Scott D.P., et al. An acute immune response to Middle East respiratory syndrome coronavirus replication contributes to viral pathogenicity. Am J Pathol. 2016;186(3):630–638. doi: 10.1016/j.ajpath.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao Y., Bao L., Deng W., et al. An animal model of MERS Produced by infection of rhesus macaques With MERS coronavirus. J Infect Dis. 2014;209(2):236–242. doi: 10.1093/infdis/jit590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S.Y., Park S.J., Cho S.Y., et al. Viral RNA in blood as indicator of severe outcome in Middle East respiratory syndrome coronavirus infection. Emerg Infect Dis. 2016;22(10):1813–1816. doi: 10.3201/eid2210.160218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corman V.M., Ölschläger S., Wendtner C.-M., Drexler J.F., Hess M., Drosten C. Performance and clinical validation of the RealStar® MERS-CoV Kit for detection of Middle East respiratory syndrome coronavirus RNA. J Clin Virol. 2014;60(2):168–171. doi: 10.1016/j.jcv.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu X., Whitaker B., Sakthivel S.K.K., et al. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014;52(1):67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Abdallat M.M., Payne D.C., Alqasrawi S., et al. Hospital-associated outbreak of Middle East respiratory syndrome coronavirus: a serologic, epidemiologic, and clinical description. Clin Infect Dis. 2014;59(9):1225–1233. doi: 10.1093/cid/ciu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choe P.G., Perera R.A.P.M., Park W.B., et al. MERS-CoV antibody responses 1 year after symptom onset, South Korea, 2015. Emerg Infect Dis. 2017;23(7):1079–1084. doi: 10.3201/eid2307.170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Writing committee of the WHO consultation on clinical aspects of pandemic (H1N1) 2009 influenza. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N Engl J Med. 2010;362(18):1708–1719. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- 46.Garten R.J., Davis C.T., Russell C.A., et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Past Pandemics | Pandemic Influenza (Flu) | CDC. http://www.cdc.gov/flu/pandemic-resources/basics/past-pandemics.html. Published August 10, 2018. Accessed 18 January 2019.
- 48.Lee N., Chan P.K.S., Lui G.C.Y., et al. Complications and outcomes of pandemic 2009 influenza A (H1N1) virus infection in hospitalized adults: how do they differ from those in seasonal influenza. J Infect Dis. 2011;203(12):1739–1747. doi: 10.1093/infdis/jir187. [DOI] [PubMed] [Google Scholar]
- 49.Cheng V.C.C., To K.K.W., Tse H., Hung I.F.N., Yuen K.-Y. Two years after pandemic influenza A/2009/H1N1: what have we learned? Clin Microbiol Rev. 2012;25(2):223–263. doi: 10.1128/CMR.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar A., Zarychanski R., Pinto R., et al. Critically ill patients with 2009 influenza A(H1N1) infection in Canada. JAMA. 2009;302(17):1872. doi: 10.1001/jama.2009.1496. [DOI] [PubMed] [Google Scholar]
- 51.Shieh W.-J., Blau D.M., Denison A.M., et al. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol. 2010;177(1):166–175. doi: 10.2353/AJPATH.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shelke V.N., Kolhapure R.M., Kadam D., et al. Pathologic study of pandemic influenza A (H1N1) 2009 cases from India. Pathol Int. 2012;62(1):36–42. doi: 10.1111/j.1440-1827.2011.02751.x. [DOI] [PubMed] [Google Scholar]
- 53.Nakajima N., Sato Y., Katano H., et al. Histopathological and immunohistochemical findings of 20 autopsy cases with 2009 H1N1 virus infection. Mod Pathol. 2012;25(1):1–13. doi: 10.1038/modpathol.2011.125. [DOI] [PubMed] [Google Scholar]
- 54.Brooks E.G., Bryce C.H., Avery C., Smelser C., Thompson D., Nolte K.B. 2009 H1N1 fatalities: the new mexico experience. J Forensic Sci. 2012;57(6):1512–1518. doi: 10.1111/j.1556-4029.2012.02163.x. [DOI] [PubMed] [Google Scholar]
- 55.Mauad T., Hajjar L.A., Callegari G.D., et al. Lung pathology in fatal novel human influenza A (H1N1) infection. Am J Respir Crit Care Med. 2010;181(1):72–79. doi: 10.1164/rccm.200909-1420OC. [DOI] [PubMed] [Google Scholar]
- 56.Soto-Abraham M.V., Soriano-Rosas J., Díaz-Quiñónez A., et al. Pathological changes associated with the 2009 H1N1 virus. N Engl J Med. 2009;361(20):2001–2003. doi: 10.1056/NEJMc0907171. [DOI] [PubMed] [Google Scholar]
- 57.Gill J.R., Sheng Z.-M., Ely S.F., et al. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med. 2010;134:235–243. doi: 10.5858/134.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sellers S.A., Hagan R.S., Hayden F.G., Fischer W.A. The hidden burden of influenza: a review of the extra-pulmonary complications of influenza infection. Influenza Other Respir Viruses. 2017;11(5):372–393. doi: 10.1111/irv.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guarner J., Paddock C.D., Shieh W., et al. Histopathologic and Immunohistochemical features of fatal influenza virus infection in children during the 2003–2004 season. Clin Infect Dis. 2006;43(2):132–140. doi: 10.1086/505122. [DOI] [PubMed] [Google Scholar]
- 60.Munster V.J., de Wit E., van den Brand J.M.A., et al. Pathogenesis and transmission of swine-origin 2009 A (H1N1) influenza virus in ferrets. Science. 2009;325(5939):481–483. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh Y., Shinya K., Kiso M., et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460(7258):1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Safronetz D., Rockx B., Feldmann F., et al. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J Virol. 2011;85(3):1214–1223. doi: 10.1128/JVI.01848-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taubenberger J.K., Morens D.M. The pathology of influenza virus infections. Annu Rev Pathol Mech Dis. 2008;3(1):499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merckx J., Wali R., Schiller I., et al. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann Intern Med. 2017;167(6):394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]
- 65.Chartrand C., Leeflang M.M.G., Minion J., Brewer T., Pai M. Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann Intern Med. 2012;156(7):500–511. doi: 10.7326/0003-4819-156-7-201204030-00403. [DOI] [PubMed] [Google Scholar]
- 66.Kumar S., Henrickson K.J. Update on influenza diagnostics: lessons from the novel H1N1 influenza A pandemic. Clin Microbiol Rev. 2012;25(2):344–361. doi: 10.1128/CMR.05016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee N. Pathogenesis of pandemic H1N1 in humans. In: Presented at the XII in - ternational symposium on respiratory viral infections, Taipei, Taiwan.
- 68.To K.K.W., Hung I.F.N., Li I.W.S., et al. Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin Infect Dis. 2010;50(6):850–859. doi: 10.1086/650581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pierre D.M., Baron J., Yu V.L., Stout J.E. Diagnostic testing for Legionnaires’ disease. Ann Clin Microbiol Antimicrob. 2017:16. doi: 10.1186/s12941-017-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muder R.R., Victor L.Y. Infection due to Legionella species other than L. pneumophila. Clin Infect Dis. 2002;35(8):990–998. doi: 10.1086/342884. [DOI] [PubMed] [Google Scholar]
- 71.Sivagnanam S., Podczervinski S., Butler-Wu S.M., et al. Legionnaires’ disease in transplant recipients: a 15-year retrospective study in a tertiary referral center. Transpl Infect Dis Off J Transplant Soc. 2017;19(5) doi: 10.1111/tid.12745. [DOI] [PubMed] [Google Scholar]
- 72.Cunha B.A., Burillo A., Bouza E. Legionnaires’ disease. The Lancet. 2016;387(10016):376–385. doi: 10.1016/S0140-6736(15)60078-2. [DOI] [PubMed] [Google Scholar]
- 73.Probable Person-to-Person Transmission of Legionnaires’ Disease | NEJM. https://www-nejm-org.offcampus.lib.washington.edu/doi/full/10.1056/NEJMc1505356?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dpubmed. Accessed 17 January 2019. [DOI] [PubMed]
- 74.Legionnaires Disease History and Patterns | Legionella | CDC. http://www.cdc.gov/legionella/about/history.html. Published November 26, 2018. Accessed 17 January 2019.
- 75.Sivagnanam S., Podczervinski S., Butler‐Wu S.M., et al. Legionnaires’ disease in transplant recipients: a 15-year retrospective study in a tertiary referral center. Transpl Infect Dis. 2017;19(5):e12745. doi: 10.1111/tid.12745. [DOI] [PubMed] [Google Scholar]
- 76.Bargellini A., Marchesi I., Marchegiano P., et al. A culture-proven case of community-acquired legionella pneumonia apparently classified as nosocomial: diagnostic and public health implications. Case Rep Med. 2013;2013 doi: 10.1155/2013/303712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cunha B.A., Cunha C.B. Legionnaire's disease: a clinical diagnostic approach. Infect Dis Clin North Am. 2017;31(1):81–93. doi: 10.1016/j.idc.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 78.Rudin J.E., Wing E.J. A comparative study of Legionella micdadei and other nosocomial acquired pneumonia. Chest. 1984;86(5):675–680. doi: 10.1378/chest.86.5.675. [DOI] [PubMed] [Google Scholar]
- 79.Joshi A.D., Swanson M.S. Comparative analysis of Legionella pneumophila and Legionella micdadei virulence traits. Infect Immun. 1999;67(8):4134–4142. doi: 10.1128/iai.67.8.4134-4142.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blackmon J.A., Chandler F.W., Cherry W.B., et al. Legionellosis. Am J Pathol. 1981;103(3):429–465. [PMC free article] [PubMed] [Google Scholar]
- 81.Winn W.C., Myerowitz R.L. The pathology of the legionella pneumonias: a review of 74 cases and the literature. Hum Pathol. 1981;12(5):401–422. doi: 10.1016/S0046-8177(81)80021-4. [DOI] [PubMed] [Google Scholar]
- 82.Waldron P.R., Martin B.A., Ho D.Y. Mistaken identity: Legionella micdadei appearing as acid fast bacilli on lung biopsy of a hematopoietic stem cell transplant patient. Transpl Infect Dis Off J Transplant Soc. 2015;17(1):89–93. doi: 10.1111/tid.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kohler R.B., Winn W.C., Wheat L.J. Onset and duration of urinary antigen excretion in Legionnaires disease. J Clin Microbiol. 1984;20(4):605–607. doi: 10.1128/jcm.20.4.605-607.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salipante S.J., SenGupta D.J., Cummings L.A., Land T.A., Hoogestraat D.R., Cookson B.T. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol. 2015;53(4):1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Avni T., Bieber A., Green H., Steinmetz T., Leibovici L., Paul M. Diagnostic accuracy of PCR alone and compared to urinary antigen testing for detection of Legionella spp.: a systematic review. J Clin Microbiol. 2016;54(2):401–411. doi: 10.1128/JCM.02675-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.IATA. Traveler Numbers Reach New Heights. https://www.iata.org/pressroom/pr/pages/2018-09-06-01.aspx. Accessed 18 January 2019.