Abstract
During RNA viruses's replication, double-stranded RNA (dsRNA) is normally produced and induce host innate immune response. Most of gene activation due cytokine mediated but which are due to methylation mediated is still unknown. In the study, DNA methylome was integrated with our previous transcriptome data to investigate the differentially methylated regions and genes using MeDIP-chip technology. We found that the transcriptional expressions of 15, 37 and 18 genes were negatively related with their promoter DNA methylation levels in the cells treated by PolyI:C, Aza-CdR, as well as PolyI:C plus Aza-CdR, respectively, compared with the untreated cells. GO analysis revealed hypo-methylated genes (BNIP3L and CDK9) and a hyper-methylated gene (ZC3HAV1) involved in the host response to viral replication. Our results suggest that these novel genes targeted by DNA methylation can be potential markers relevant to virus replication and host innate immune response to set up a medical model of infectious diseases.
Abbreviations: PK15, porcine kidney epithelial cell line; dsRNA, double-stranded RNA; PolyI:C, a synthetic viral-like dsRNA analogplolyinosinic: polycyticdylic acid; Aza-CdR, DNA methyltransferases inhibitor 5-aza-2′-deoxycytidine; P, PK15 cells extracellular treatment with PolyI:C; A, PK15 cells treated with Aza-CdR; P+A, PK15 cells treated with PolyI:C followed by the addition of Aza-CdR; C, control (untreated PK15 cells); DEPs, differentially enrichment peaks; DMRs, differentially methylated regions; HCP, high-CpG-containing promoters; ICP, inter mediate-CpG-containing promoters; LCP, low-CpG-containing promoters; BPs, biological processes; DEGs, differentially expressed genes
Keywords: DNA methylome, Transcriptome, Methylation targeted genes, Viral replication, PolyI:C, Aza-CdR
1. Introduction
Health is a fundamental issue in modern swine industry. Nowadays, porcine reproductive and respiratory syndrome virus (PRRSV), class swine fever virus (CSFV) and many other RNA viruses are threatening the porcine health worldwide. During the replication of the viruses in swine cells, double-stranded RNA (dsRNA) is normally produced by most RNA viruses. The majority diseases of economic importance in swine industry such as Porcine reproductive and respiratory syndrome (PRRS), Classical swine fever (CSF), Swine vesicular disease (SVD), Foot and mouth disease (FMD), Transmissible gastroenteritis of pigs (TGE) and Influenza are caused by RNA viruses. The dsRNA intermediates during virus replication induce host innate immune response. During innate immune response, host toll-like receptor 3 (TLR3) recognizes these genomic viral dsRNA generated during viral replication [1]. The interaction of dsRNA with TLR3 is a potent inducer of type I IFN cascade genes for antiviral action.
Synthetic polyinosinic:polycyticdylic acid (PolyI:C) is one kind of mimics of virus dsRNA. Our earlier work discovered that compared with the untreated porcine kidney epithelial cell lines (PK15), 76 significantly differentially expressed genes (DEGs) were detected in PolyI:C treated cells (P < 0.05) [2]. Out of these DEGs, eighteen are involved in epigenetic modifications or encode DNA methylation enzymes, such as DNA methylation-related gene DNMT3A (DNA methyltransferase 3A), MGMT (O-6-methylguanine-DNA methyltransferase) and etc. [2]. Galli et al. demonstrated that PolyI:C-mediated activation of TLR3 induces miRNAs targeting DNA methyltransferases, leading to demethylation and re-expression of the oncosuppressor retinoic acid receptor beta (RARß) [3]. DNA methylation is one kind of key factors in epigenetic modifications [2,4]. Previous studies proved that hyper-methylated promoter and consequently decreased expression of immune related genes can be induced by virus challenge [2,5,6]. However, the DNA methylation profiles and how they regulate target genes expressions in dsRNA infected porcine cells remain largely unknown.
Cumulative evidence shows DNA methylation in relation to viral infection. It is well known that indigenous Chinese Tongcheng pigs reportedly showed strong resistance to PRRSV infection. The integrative analysis of mRNA and miRNA expression revealed that down-regulated methylation-related genes (DNMT1 and DNMT3b) were targeted by five up-regulated differentially expressed miRNAs. The results lay a strong foundation for developing novel therapies to control PRRS in pigs [7]. These data suggest that the methylation variation of vital genes may be beneficial for the host to fight against RNA virus infection.
Methylated DNA immunoprecipitation microarray (MeDIP-chip) is an effective technique to map DNA methylome for different kinds of cells [4,8]. It is also efficient to well understand the regulatory effects of DNA methylation on genes expressions on genome-wide [9,10]. In the present study, MeDIP-chip and transcriptome assays were conducted and combinative analyzed to draw the DNA methylome profiles and to identify the targeted gene of the PK15 cells upon the extracellular treatment of PolyI:C (P), Aza-CdR (A, 5-aza-2′-deoxycytidine, an inhibitor of DNA methyltransferase [[11], [12], [13]]) alone, or both (P + A). Further, by comparing each treatment to the untreated control cells (C), the differentially enriched methylation peaks (DEPs) and DNA methylated markers were detected and then confirmed in PK15 cells. Our results presented a comprehensive map of DNA methylome in dsRNA and/or Aza-CdR treated porcine cells and also revealed candidate-resistance genes relevant to virus replication.
2. Results
2.1. DNA methylome patterns of PK15 cells response to virus mimics extracellular treatment
To decipher DNA methylation profiles of the porcine PK15 cells response to the extracellular treatment of virus mimics dsRNA (PolyI:C), we detected and analyzed whole genome DNA methylation levels of PK15 cells treated by PolyI:C (P), DNMTs inhibitor (Aza-CdR, A), and both (P + A) by comparing to the untreated control cells (C). The optimization of dose- and time-series experiments of PolyI:C, Aza-CdR, or both treatment in the cells were conducted referring to the previous study [2].
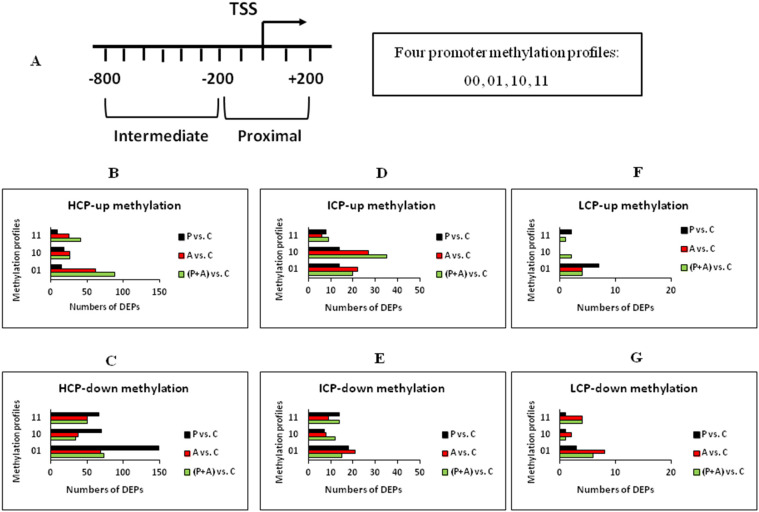
Gene's promoter is the main target of DNA methylation modification, which is the key switch of turning on or off gene expressions. Based on the CpG density, gene promoter was divided into three molds: High-, Intermediate- and Low-CpG-containing promoter (abbreviated as HCP, ICP and LCP, respectively). To clearly map the patterns of DNA methylation on promoters, each promoter was divided into two regions relative to the transcriptional start site (TSS) of a gene: proximal (−200 to +200 bp) and intermediate (−800 to −200 bp) (Fig. 1 ). Each region was then identified as methylated (represented as ‘1’) or unmethylated (‘0’) based on the enrichment peaks of DNA methylation. Thus different promoters were classified into four types: ‘00’, ‘11’, ‘10’ and ‘01’ were denoted as extensively unmethylated (two regions are both non-peak-finding), fully methylated (two regions are both peak-finding), intermediate methylated (the region of −800 to −200 bp is peak-finding) and proximally methylated (the region of −200 to +200 bp is peak-finding), respectively (Fig. 1A, Fig. S1A).
Fig. 1.
The distribution of differential enrichment DNA methylation peaks in PK15 cells induced by PolyI:C, Aza-CdR, or both compared with controls. (A) Diagram showing the two promoter regions relative to the TSS: Proximal (−200 to +200 bp), Intermediate (−800 to −200 bp). Each region is defined as methylated (‘1’) or unmethylated (‘0’) if DEPs located in the region relative to the TSS. ‘00’: extensively unmethylated, ‘01’: proximally methylated, ‘10’: intermediate methylated, ‘11’: fully methylated. (B) Number of up-methylation peaks in HCPs for each comparison and for each methylation profiles. (C) Number of down-methylation peaks in HCPs for each comparison and for each methylation profiles. (D, E, F, G) The same as B or C in ICPs and LCPs.
The MeDIP-chip data were filtered by screening differential enrichment peaks (DEPs) overlapping the promoter regions. When DEP was identified in PolyI:C treated group not in control group, we defined it as up-methylated DEP in P vs. C. In contrast, if DEP was detected in control group not in PolyI:C treated group, it was defined as down-methylated DEP in P vs. C. The number of DEPs of promoters with up- or down-methylation within each comparison was shown in Fig. 1. There were no significant differences (P > 0.05) between the counts of up- and down-methylation ICP (see Fig. 1D, E), while HCP (see Fig. 1B, C) and LCP (see Fig. 1F, G) showed heterogeneous methylation patterns. Most HCPs in PK15 cells extracellular treatment by PolyI:C alone were hypomethylated (‘01’ profile, see Fig. 1C, black bar) compared with those in untreated PK15 cells, account for 52% of the three kinds of down-regulated methylation profiles (‘01’, ‘10’ and ‘11’were of 149, 70 and 67 DEPs, respectively). LCPs were hyperrmethylated (‘01’ profile, see Fig. 1F, black bar) in P vs. C, account for 78% of three up-regulated methylation profiles (‘01’, ‘10’ and ‘11’were of 7, 0 and 2 DEPs, respectively). The data indicated that differential methylation of promoters in PolyI:C and Aza-CdR treated cells is enriched in specific regions relative to TSS and dependent on overall CpG density. Other differential comparisons also provided some relevant information, including (P + A) vs. P and (P + A) vs. A. (Fig. S1B).
2.2. Genome-wide transcription associated with promoter DNA methylation level
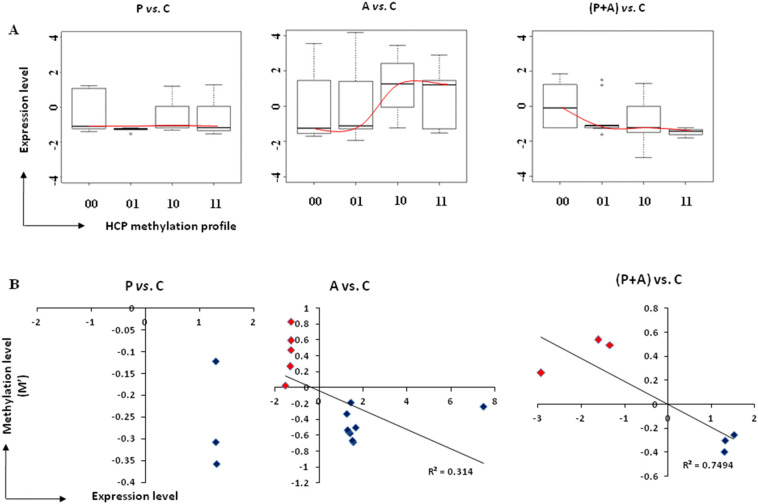
In order to access the target site of promoter methylation relevant to transcription repression, gene expression was evaluated with transcriptional microarray. Fig. 2A showed the expression levels of the genes with different metyhylation profiles (‘00’, ‘01’, ‘10’ and ‘11’) in HCP. The results indicated that the location of methylation peaks to the TSS in HCPs were significantly negative correlated with genes expression in (P + A) vs. C, while the relationship was not detected in the comparison of P vs. C (see Fig. 2A, P < 0.05). In the comparison of A vs. C, HCPs with proximal methylation profile (‘01’) seems to be less transcriptionally active than intermediate and fully methylation profiles (‘10’ and ‘11’) (Fig. 2A, P < 0.01). This finding suggest that hypermethylation on genes promoter is associated with decreases in cellular transcript levels and Aza-CdR has genome-wide effect on viral mimic dsRNA in vitro.
Fig. 2.
Integrated analysis of promoter methylation and transcriptional expression in HCPs for P vs. C, A vs. C and (P + A) vs. C. (A) Gene expression in HCPs for three comparisons under the control of promoters with each methylation profile (‘00’, ‘01’, ‘10’, ‘11’), red curve indicate trend for the relationship between methylation profiles and transcriptional expression. DEGs were screened from microarray, P < 0.05. Up line and down line in each boxplot represents gene expression levels from the 1/4-3/4, the middle line represents the average expression level. (B) Regression analysis of gene expression level (|Fold change| > 1.2, P < 0.05) and DNA methylation degree of gene promoter. Blue scatter plots indicate hypo-methylation-activated genes, red scatter plots indicate hyper-methylation-silenced genes. R represents correlation coefficient. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We further analyzed DNA methylation degree with the transcription expression level to evaluate the possibility of the quantitative relationship for each comparison. M' value represents methylation degree for the comparison which is the differential average log2 (MeDIP/Input) ratio of three parallel samples between experiment group and control. All comparisons in HCPs showed a clear negative correlation between DNA methylation level and gene expression level in the second quadrant and the fourth quadrant (see Fig. 2B, |Fold change| > 1.2, P < 0.05). Of 15 differentially expressed genes (DEGs) in the comparisons of P vs. C, 3 genes expression were up-regulated driven by lower methylation (accounting for 20% of 15 genes, blue scatter plots in Fig. 2B). Of 37 DEGs in the comparisons of A vs. C, 9 were up-regulated expression driven by lower methylation (24%, blue scatter plots in Fig. 2B), 5 were down-regulation expression driven by higher methylation (14%, red scatter plots in Fig. 2B). Especially, of 18 DEGs in the comparisons of (P + A) vs. C, 3 were up- and 3 were down-regulated expression driven by lower and higher methylation (each account for 17%, blue and red scatter plots in Fig. 2B), respectively. The details of the differentially expressed genes (DEGs) regulated by promoter methylation were showed in Table S1. We found that around 20% of differentially methylated genes (three DEGs were PPARG, TNKS2 and ERV-PK15) in PolyI:C extracellular treated PK15 cells were negatively regulated by promoter methylation at transcriptional level. The data also indicated that around 38% of DEGs in A vs. C (14 DEGs were showed in Table S1) and 34% of DEGs in (P + A) vs. C (6 DEGs were showed in Table S1) were negatively regulated by promoter methylation at transcriptional level. Surprisingly, remaining 80% DEGs hypomethylated and decreased in gene expression were observed in P vs. C (Fig. S2, blue scatters in the third quadrant), suggesting active demethylation rather than only passive demethylation, as might be expected in dsRNA treated porcine cells.
2.3. Functional pathways of the differentially methylated porcine genes involved in the stimulation of PolyI:C, Aza-CdR and both
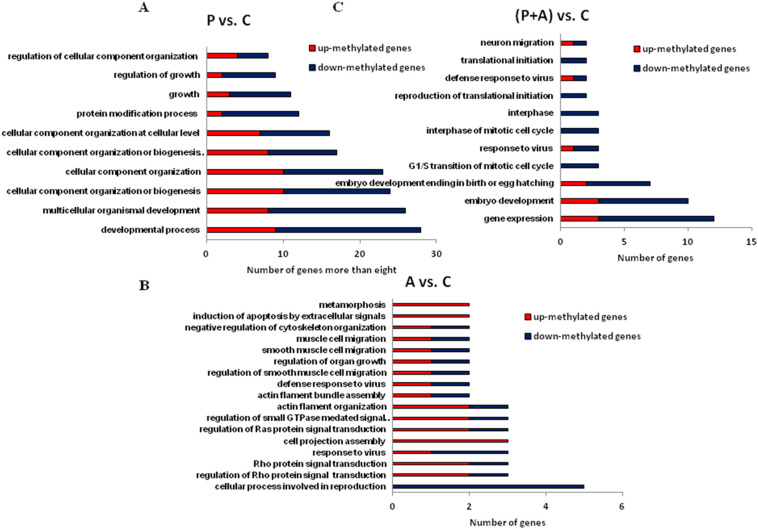
To annotate the functions of the differentially methylated genes affected by the stimulation of PolyI:C, Aza-CdR, and both, Gene Ontology (GO) were conducted to capture the relevant biological processes (see Fig. 3 , Table S2). A total of 58, 75 and 82 genes with different methylation modification were identified in the comparisons of P vs. C, A vs. C and (P + A) vs. C, respectively (see Table S3). Using UCSC and RefSeq database, these genes were found involved in different biological function. In ten down-methylated biological processes (BPs, numbers of gene more than eight in each BP), the critical five in P vs. C include regulation of growth, protein modification process, developmental process, cellular component organization, multicellular organismal development. In (P + A) vs. C, the top five of nine down-methylated BPs were gene expression, embryo development, G1/S transition of mitotic cell cycle, response to virus, reproduction of translational initiation. In A vs. C, just 2/17 BPs were down-methylation, containing cellular process involved in reproduction and response to virus. Up-methylated genes mainly enriched in induction of apoptosis by extracellular signals, metamorphosis and cell projection assembly and regulation of Rho protein signal transduction (see Fig. 3, Table S2 and S4, red bars: up-methylated genes, blue bars: down-methylated genes). These data indicated that the effects of Aza-CdR on gene expression in PK15 cells were largely independent of cell growth and cellular morphology. The common significant GO terms (P < 0.05) in the comparisons of A vs. C and (P + A) vs. C were “response to virus” (GO ID: 0009615) and “defense response to virus” (GO ID: 0051607), which imply that Aza-CdR has a favorable defense effects on the viral-intermediate dsRNA extracellular treatment. Here, Aza-CdR is expected to inactivate the effect of PolyI:C. The methyl transferase inhibitor Aza-CdR was used to inhibit specific methylation due to Poly I:C.
Fig. 3.
The differential methylation genes clustered into the functional gene ontology in the comparison of P vs. C (A), A vs. C (B), (P + A) vs. C (C). Red bars indicate up-methylated gene numbers and blue bars indicate down-methylated gene numbers. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
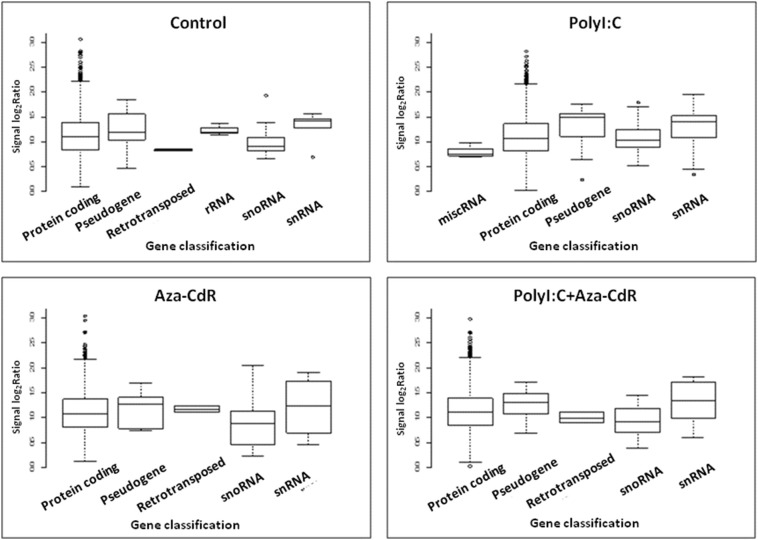
Porcine transcriptional microarray included 44,034 transcripts, which was functionally classified into protein-coding genes, miRNA, rRNA, tRNA and etc. Different gene types with various methylation patterns showed different methylation characteristics. Comparing the control to each treatment, the DNA methylation levels of the genes encoding porcine proteins, miRNA, miscRNA, pseudogenes, retrotransposon, rRNA, snoRNA and snRNA were analyzed (see Fig. 4 ). We found that the promoter regions of pseudogenes showed hypermethylation causing transcriptional repression. The DNA methylation level of retrotransposon genes was hypomethylated in the control cells (see Fig. 4, control), nevertheless after treated with PolyI:C followed by the addition of Aza-CdR, the retrotransposon was up-methylated (see Fig. 4, PolyI:C + Aza-CdR) in PK15 cells. Human endogenous retroviruses (HERV) represents a potential marker or mediator of environmental exposures (e.g., virus infection) in the development of chronic complex diseases. ERVWE1 is so far the only verified HERV proviral locus that has retained a long env ORF. Hypermethylation of ERVWE1 observed in non-placental cells has been linked with transcriptional repression [14]. Here, we found the up-methylation induced by Aza-CdR may repress the activation of retrotransposon in PolyI:C treated PK15 cells.
Fig. 4.
Average methylation level of different gene categories in the porcine kidney epithelial cells' genome. Boxplots of the methylation level distribution of each gene category. Gene methylation levels from the 1/4-3/4 were used and the middle line represents the average methylation level.
2.4. Functional methylation markers and targeted genes triggered by virus mimics
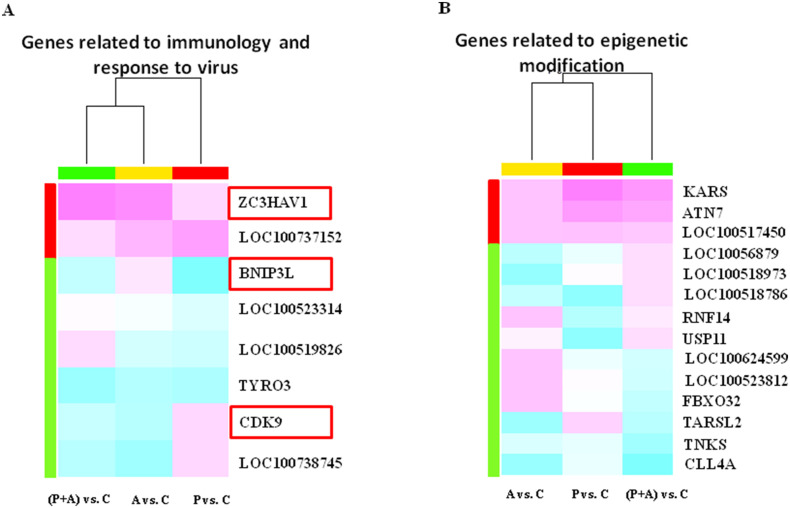
We further clustered these differentially methylated genes into two classifications involved in response to virus and epigenetic modifications (Fig. 5 ). The genes screened in PolyI:C plus Aza-CdR treated cells are related to immunology and response to virus that first clustered with those of Aza-CdR (Fig. 5A X-axis, green and yellow colors), while genes related to epigenetic modification were foremost merging in P vs. C and A vs. C (Fig. 5B X-axis, yellow and red colors). The effect of PolyI:C plus Aza-CdR on immune response more closely resembled the effects of Aza-CdR, inducing the epigenetic effect similar with PolyI:C. As the red box shown in Fig. 5A 3 out of 8 genes in immune-related pathway were related with response to virus, including cyclin-dependent kinase 9 (CDK9), BCL2/adenovirus E1B interacting protein 3-like (BNIP3L) and zinc finger CCCH-type of antiviral 1 (ZC3HAV1). CDK9 participated in the GO term of cellular response to cytokine stimulus (GO ID: 0071345), regulation of DNA repair (GO ID: 0006282) and regulation of histone modification (GO ID: 0031056). The promoter methylation of BNIP3L and CDK9 genes significantly decreased following dsRNA mimics and Aza-CdR treatment. The promoter methylation of ZC3HAV1 gene significantly increased in (P + A) vs. C. The remaining five genes (LOC100519826, LOC100523314, LOC100737152, LOC100738745, TYRO3) were participated in GO term of inflammatory response. Meanwhile, 14 out of all common genes (KARS, ATN7, ARF14 etc.) were found involved in the epigenetic modifications (see Fig. 5B). Table 2 and S5 showed the detailed differential methylation peaks information in the related gene promoters in the comparison of (P + A) vs. C and A vs. C.
Fig. 5.
Heat-map of selected genes related to immunology, response to virus and epigenetic modification. DNA methylation levels are depicted by a pseudocolor scale as indicated from pink (hyper-methylation) to blue (hypo-methylation). Red, yellow, green colors on the X-axis mean the M' value of each gene in comparison of P vs. C, A vs. C, (P + A) vs. C. Red or green colors on the Y-axis mean average hyper-/hypo-methylated level in three comparisons of each genes. (A) Genes related to immunology and response to virus. Red box indicate the genes related to defense response to the virus. (B) Genes related to epigenetic modification. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Information of differential methylated genes.
| Gene symble | Description | CpG location | Strand | Position | Type | Gene name |
|---|---|---|---|---|---|---|
| CDK9 | Protein coding | Chr1: 282627136282,627,682 | + | Promoter | HCP | cyclin-dependent kinase 9 |
| BNIP3L | Protein coding | Chr14: 10460236–10,460,615 | + | Promoter | HCP | BCL2/adenovirus E1B interacting protein 3-like |
| ZC3HAV1 | Protein coding | Chr18: 8723234–8,723,868 | + | Promoter | HCP | zinc finger CCCH-type, antiviral 1 |
| TYRO3 | Protein coding | Chr1: 136238734–136,239,488 | + | Promoter | HCP | TYRO3 protein tyrosine kinase 3 |
| FBXO32 | Protein coding | Chr4: 15473215–15,473,744 | + | Promoter | HCP | F-box protein 32 |
| TARSL2 | Protein coding | Chr1:281862813–291,917,305 | + | Promoter | HCP | threonyl-tRNA synthetase-like 2 |
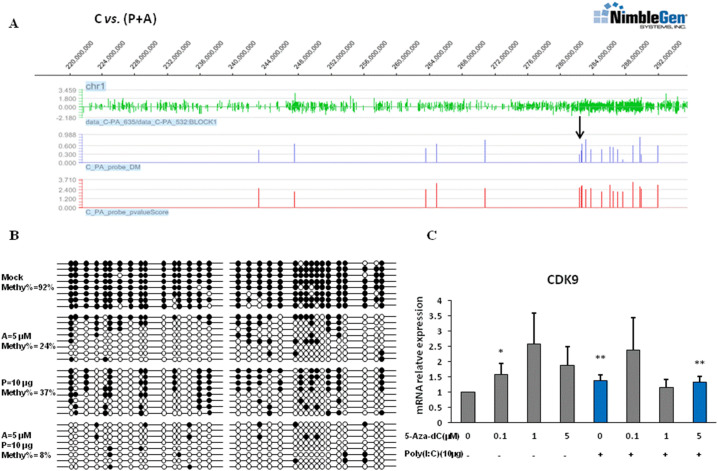
To validate the influence of promoter differential methylation regions (p-DMRs) on the expression of the functional genes, we picked three genes of CDK9, BNIP3L and ZC3HAV1 to dissect the methylation markers (see Fig. 6 and Fig. S3) via bisulfite-PCR and sequencing. As regards to BNIP3L and ZC3HAV1 genes, the DEPs on their promoter regions were shown in Fig. S3A and S3C (black arrows, P-value score > 2) and Table 2. The significant up- and down-regulated gene expression level were also validated (Fig. S3B and S3D, blue bars, P < 0.05). As for CDK9 gene, we observed that the differential methylation region in its promoter (Chr 1: 282627136–282,627,682, black arrow in Fig. 6A P-value score > 2) was enriched in the control cells (92%) compared to the cells treated with Aza-CdR (24%), PolyI:C (37%), and PolyI:C plus Aza-CdR (8%) (see Fig. 6B).
Fig. 6.
DNA methylation and expression variation of porcine CDK9 gene induced by PolyI:C, Aza-CdR or both. (A) Graphical representation of differential enrichment peaks by methylated DNA immunoprecipitation chip (Signalmap software, NimbleGen). The panels show the DNA hyper-methylation profile at CDK9 promoter in controls compared with PK15 cells treated with PolyI:C plus Aza-CdR. First panel: green peaks represent probe-level log2 (MeDIP/Input) ratio. Second panel: blue bars represent different M' value. Third panel: red bars represent P-value score cutoff 2. The differential methylated CpG islands are indicated by black arrow in C vs. (P + A). Chromosomal location is indicated at the top of the diagram. (B) Bisulfite-sequencing validation of hypermethylated and hypomethylated CDK9 promoter detected by MeDIP-chip. The black circle represents the methylated CG site, while the white circle represents the unmethylated site. (C) mRNA expression level of CDK9 based on the dose-response assay of PolyI: C, Aza-CdR, or both. Significant levels were determined using a t-test by comparing the expression levels of CDK9 between each treatment to the non-treated control. Blue bars represent mRNA relative expression level in PolyI:C alone treated cells and P + A treated cells, respectively. ⁎P < 0.05, ⁎⁎P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To determine whether PolyI:C and demethylation agent (Aza-CdR) initiate CDK9 transcription by de-methylation in PK15 cells, we further examined the transcriptional expression. We found that the expression of CDK9 gene was significantly promoted in the cells treated by PolyI:C (10 μg/mL) alone or by PolyI:C (10 μg/mL) plus Aza-CdR (5 μM) (see Fig. 6C, blue bars, P < 0.01). However, the treatment of 5 μM Aza-CdR alone did not significantly influence the expression of CDK9 compared to the untreated cells (see Fig. 6CP > 0.05). These data revealed the anti-viral resistance of CDK9 gene in PK15 cells by means of DNA methylation regulation.
3. Discussion
DNA methylation regulation is of critical significance to gain insight into the interaction between antiviral innate immunity and virus replication. This study provides the first DNA methylome and its regulation effects on genes expression in porcine PK15 cell lines post extracellular treatment of mimics virus (PolyI:C), DNA methyltransferases inhibitor (Aza-CdR), and both (PolyI:C plus Aza-CdR). The targeted genes regulated by DNA methylation and the vital pathways involved in dsRNA treatment were identified in the PK15 cell model.
Host's innate immune response in mammalian cells was dysregulated by viral-like dsRNA treatment and DNA methyltransferase inhibitor (Aza-CdR) treatment [2,[15], [16], [17], [18]]. The main reason was declared that epigenetic modification alteration is a pivotal step in the initiation and progression of most oncogenic viruses induced cancers [19]. Differentially methylated genes involved in response to virus, protein modification process and gene expression were discovered in the current study. Two significant GO terms, response to virus and defense response to virus, were detected in the comparisons of A vs. C and (P + A) vs. C. The results suggest that the regression effect of Aza-CdR on the stimulant of PolyI:C may be regulated by DNA methylation in the porcine kidney epithelial cells.
The DNA methylation as an indirect effects on gene expression was observed remarkably consistent across dosage and duration of both drugs exposure, and were largely independent of cells growth and morphology [[4], [5], [6],19]. The results showed remaining of 62–80% DEGs in PolyI:C and/or Aza-CdR treated cells are not negatively regulated by methylation, which is consistent with the previous data that the effects of Aza-CdR on gene expression more closely resembled chromatin decondensation than to DNMTs inactivation [11]. The indirect effects on gene expression we observed were also remarkably consistent across dosage and duration of both drugs exposure, and were largely independent of cells growth and morphology. In addition, the lower concentration of Aza-CdR (200–300 nM) was not generally used to modulate gene expression, may explain the small number of genes up-regulated after Aza-CdR exposure [20].
The approach taken here is an effective way to identify methylation-silenced genes and hypomethylated genes [21]. By means of the treatment of virus mimics and chemical genomic screening for epigenetic modification, MeDIP-chip and transcriptional microarray are cost-effective methods to identify methylation targeted regions and genes [22,23]. Combined methylation alteration regions with gene expression information can dissect the functional-related gene involved in virus replication and anti viral responses. Promoter methylation variation was observed in CDK9 gene which is enriched in the GO term of response to virus, as well as in ZC3HAV1 and BNIP3L genes that are involved in defense response to virus in P vs. C and (P + A) vs. C. Of which, CDK9 gene encodes cyclin-dependent kinase 9 and is a positive transcriptional elongation factor for the virus-encoded Tat protein activity [24,25]. Cyclin T1/CDK9 interacts with influenza A virus polymerase to inhibit virus transcription and replication [26]. Its character of down-methylation while up-expression in PolyI:C or PolyI:C plus Aza-CdR treated cells could serve as biomarker for virus replication in the swine cells. BNIP3L gene (BCL2/adenovirus E1B 19kD-interacting protein 3-like) can function as virus-induced tumor repressor, which encodes a homologue of NIP3, a protein that interacts with adenovirus E1B19kD protein and host BCL2 protein [27,28]. The mRNA of the pro-apoptotic gene BNIP3L was significantly decreased following murine coronavirus (a signal strand RNA) infection, evades host antiviral defense and promotes cell survival [29]. Its down-methylation while very significant up-expression in PolyI:C plus Aza-CdR treated cells (Fig. S3A and Table S5) means BNIP3L could be a powerful candidate gene of anti virus infection. ZC3HAV1 is an interferon-inducible genes and encodes zinc-finger antiviral protein (ZAP) [30]. Through binding to the viral RNA and recruiting the processing exosome, human ZAP causes the degradation of viral RNA [31]. Although the expression of ZC3HAV1 was up-regulated by PolyI:C or Aza-CdR alone (Fig. S3D), its up-methylation but down-expression in PolyI:C plus Aza-CdR treated cells (blue bar in Fig. S3D) suggest that the anti viral function of ZAP in PK15 cells can be induced by dsRNA or methyltransferase inhibitor but cannot be improved by both.
In addition, down-methylation alteration in tRNA methyltransferase gene (TRMT6) (Table S4) was only observed in (P + A) vs. C, which is consistent with the previous study that TRMT6 plays a key role in translational initiation and RNA modification [32,33]. TRMT6 is part of a two component methyltransferase in yeast, which is involved in the post-translational modification and produces the modified nucleoside 1-methyladenosine in tRNAs [34]. In human, modified nucleoside 1-methyladenosine causes initiator methionine stability and may be involved in the replication of human immunodeficiency virus type 1 [35,36]. The up-expression of tRNA methyltransferase may cause hypermethylation of tRNA. RNA virus usually encode proteins to encounter the dsRNA-induced cellular defense. Cell nucleolin is a central hub for the replication of pathogenic RNA viruses. Previous study suggests that non-structural 1 protein (NS1) of influenza A (H3N2) could induce nucleolar stress based on epigenetic alteration of rRNA gene promoter via interaction with nucleolin [37]. In A vs. C comparison, promoter methylation of LOC100511875 has been linked with maturation of 5.8S rRNA and defense response to virus. Moreover, porcine retroviral oncogene MYB was down-methylated and activated in dsRNA treated PK15 cells. MYB encodes nuclear DNA-binding proteins and is involved in G1/S transition of mitotic cell cycle [36]. The activation of oncogene MYB in PolyI:C treated cells means it could be a biomarker of dsRNA treatment. Retroviruses can selectively trigger an array of innate immune responses through various pattern recognition receptors (PRRs), ZC3HAV1 was decreased two to eight folds that examined whether avian leukosis virus induces or inhibits innate immune host responses [38], the results resembled to our study.
Researchers suggested that DNA hypomethylation is involved in porcine endogenous retrovirus (PERV) activation [39]. PK15 cell lines have been shown to be positive for PERV. Matouskováatt. al found that 5′long terminal repeats (LTRs) of PERV amplified from PK15 contained 2.7% methylated CpG and no specific patterns of CpG methylation were observed in PERV 5′LTR sequences [39]. These results suggest minor methylation variation in PERV 5′LTR in PK15. Nevertheless, PK15 is possible contaminated with non-pathogenic porcine circovirus type 1 (PCV1) [40]. These potential viral sources could alter cellular promoter methylation/demethylation response against PolyI:C extracellular treatment. Therefore, to eliminate the possible influence of PERV and PCV1 contamination of the PK15, we designed untreated cells to as the controls. In addition, further study is warranted to repeat dsRNA mimic experiment and RNA virus infection study with different porcine cell types.
A reverse correlation between promoter methylation and transcription expression of target genes has been well established [41,42]. The methylation level on genes promoter regions was more important than that on gene body [21]. Koga et al. reported a stronger correlation between DNA methylation and gene repression in high-CpG containing promoters (HCPs) compared with intermediate-CpG (ICPs) and low-CpG containing promoters (LCPs) [4]. This is consistent with our findings that the methylation levels in proximal regions of the TSS in HCPs (Fig. 2A, red curve, P < 0.05) and ICP (Fig. S4, red curve) were strongly associated with transcriptional expression levels in (P + A) vs. C, while no correlation in LCP (Fig. S4).
Poly I:C, the dsRNA mimic regularly used as an effective tool to study RNA virus replication in different mammalian cells as an intermediate. PolyI:C is at least 3 kb in length. By producing PolyI:C of various lengths, it was determined that short segments of the polymer (~300 bp) are potent ligands for RIG-I (retinoic acid inducible gene I), longer segments of PolyI:C preferentially activated MDA-5 (melanoma differentiation-associated gene 5) and usually induces TLR3 pathway [43,44]. Our microarray results interpreted Toll-like receptor/RIG-1-like receptor signaling pathway responsible for inducing IFN was enriched in porcine kidney cells treated with PolyI:C (272-4242 bp) plus Aza-CdR at 10 h [2]. PolyI:C challenge can elicit certain gene expression changes relevant to TLR3 pathway similar to acute viral infection in PBMCs cells at 3 h treatment [16]. Thus, TLR3 receptor can be specially expressed in immune cells and non-immune cells, e.g. kidney epithelial cells [1]. The experiment was focusing on the extracellular PolyI:C stimulation rather than transfected PolyI:C. With regard to dsRNAs, TLR3, RIG-1 and MDA-5 pathways were different each other, thus there could be difference between the treatment of extracellular PolyI:C and transfected PolyI:C [45,46]. DNA methylation mechanism underlying the discrimination between extracellular delivery of PolyI:C treatment for different times remains to be elucidated.
In summary, many contagious viruses are RNA virus which causes seasonal pandemics and imposes great economic losses on livestock industry and harmful to human health. To identify DNA methylation regulated transcriptome variations and potential methylation marks underlying the interaction of virus infection and host, the study first presents the integrative analysis of DNA promoter methylome and transcriptome in mimic virus dsRNA and DNA methyltransferases inhibitor extracellular treated porcine kidney cells (PK15). The results revealed several key promoter methylation marks and three powerful candidate methylated genes (CDK9, BNIP3L and ZC3HAV1) can defense virus replication in porcine kidney epithelial cells and warrant to do further functional studies involving against RNA virus infections. This study provides new insights into the epigenetic regulatory mechanisms of genes expression in virus-like dsRNA treated cells.
4. Materials and methods
All protocols were reviewed and approved by the Institutional Animal Care and Use Committee at China Agricultural University in Beijing, China (permit number: DK996).
4.1. Cell culture and treatments
Porcine kidney cell line (PK15) was bought from the China Center for Type Culture Collection (CCTCC, Wuhan, China). The PK15 cells were grown in Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) containing 10% heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT, SH30084.03, which were free of foot-and-mouth disease virus and bovine spongiform encephalopathy virus contamination), 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen) at 37 °C with 5% CO2. After the cells were serum-starved for 2 h, we designed three treatments: (1) P: PolyI:C (10 μg/mL; polyinosinic: polycytidylic acid; Sigma-Aldrich, Shanghai Trading Co., Ltd) was added to the culture medium and treated for 10 h. (2) A: Four hours later, different concentrations of Aza-CdR (0.1, 1, 5 μM; Sigma-Aldrich, St. Louis, MO) were added and treated for 6 h. (3) P + A: with 10 μg/mL PolyI:C treated the PK15 cell for 4 h, then added 5 μM/L Aza-CdR and treated for 6 h [2]. The three group cells and the untreated control cells (C) were harvested and applied to the subsequent study.
4.2. Extraction and bisulfite treatment of genomic DNA
Genomic DNA was extracted from each treatment of the PK15 cells using the Wizard Genomic DNA Purification kit (Promega, US). DNA concentration and quality were measured with the NanoDropTM ND-2000c Spectrophotometer (Thermo Scientific, Inc., US). Sodium bisulfite conversion of 1 μg genomic DNA for each sample was conducted using the EZ DNA Methylation Golden kit according to the manufacturer instructions (ZYMO Research, California, US). Bisulfite-converted DNA was eluted in 20 μL elution buffer (ZYMO Research).
4.3. DNA methylation profiling by MeDIP-chip
A standard methyl-DNA immunoprecipitation microarray (MeDIP-chip) assay was performed as described previously [9]. Five μg of genomic DNA of each sample was sonicated to produce random fragments in size of 400-500 bp. The sheared PK15 genomic DNA for each treatment was immunoprecipitated with an antibody which specifically recognizes 5-methylcytosine, followed by hybridization to NimbleGen array (MeDIP-chip), which probes porcine 385 K CpG islands plus ensemble promoters. Briefly, after denaturation at 94 °C for 10 min, immunoprecipitation was performed using 5 μg monoclonal antibody against 5-methylcytidine (1 μg/μL, Diagenode) in a final volume of 500 μL IP buffer (0.5% NP 40; 1.1% Triton X-100; 1.5 mM EDTA; 50 mM Tris-HCl; 150 mM NaCl) at 4 °C for overnight. Add 5 μg mouse IgG to mock IP sample tube. Add 50 μL of magnetic beads (Bangs laboratories. Inc) coupled anti-mouse IgG and mix for 2 h at 4 °C by end-over-end rotation. After the incubation, immunoprecipitated complexes were collected with pellet beads by Magnetic Separation Rack for 2 min at 4 °C, washed with buffer for 6 times, eluted with 200 μL elution buffer, and purified by phenol:chloroform extraction and isopropanol precipitation. Immunoprecipitated methylated DNA was labeled with Cy5 fluorophere and the input genomic DNA was labeled with Cy3 fluorophere. The labeled DNA were hybridized to custom NimbleGen CpG island plus Ensemble promoter array (Roche, Germany), which covered all known CpG islands annotated by UCSC and all well-characterized RefSeq promoter regions (from −800 bp to +200 bp transcription start sites, TSS) totally covered by 385 K probes. A total of twelve two-colour arrays were then scanned with GenePix 4000 Binstrument (NimbleGen).
4.4. Data normalization and differential methylation enrichment peak-finding
Raw data were subjected median-centering, quantile normalization, and linear smoothing by Bioconductor packages. After normalization, a normalized log2 (MeDIP/Input) ratio data was created for each sample. A customized peak-finding algorithm provided by NimbleGen was applied to analyze methylation data from MeDIP-chip (Roche-NimbleGen) as previously described. The algorithm was used to perform the modified Kolmogorov-Smirnov test on several adjacent probes using sliding windows to predict enriched regions across the array. We filtered the differential methylation peaks according to two principles suggested by NimbleGen: (1) At least one group log2 (MeDIP/Input) ratio > 0.3; (2) At least in one peak, the coefficient variability (CV) of the half probes ≤ 0.8 between two groups. Differential probe-level log2 (MeDIP/Input) ratio between each treatment and untreated cells was applied to the MeDIP hybridization to determine differential methylation region (DMR).
Furthermore, differential enrichment peaks (DEPs) analysis for each comparison using M' method was completed and M' = Average (log2 MeDIPE/InputE)—Average (log2 MeDIPC/InputC). Then the identified DEPs were mapped to genomic features: transcripts and CpG Islands. The mapping data of DEP in promoter region for each comparison was saved in ‘Islands. The mapping “DEP data” was input into Signal Map software (version 8.0.0.0, Roche-NimbleGen) to observe and evaluate the differential methylation peaks between two groups.
Peak-finding was used to determine the differential methylated regions. NimbleScan detects peaks by searching for at least 2 probes above a P-value minimum cutoff (−log10) of 2 (P < 0.01).
4.5. Hot start PCR and bisulfite cloning sequencing
PCR primers of the porcine CDK9 (NCBI: Gene ID_100307051) promoter used for bisulphite DNA methylation detection were designed with Oligo 6.0 software (see Table 1 ). Hot start PCR was carried out in 25 μL solution including 15 to 20 ng bisulfite-treated DNA, 12.5 μL Hot start PCR premix (ZYMO Research), 0.5 μM forward primer, and 0.5 μM reverse primer. PCR cycling conditions were 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s, 50 to 60 °C for 45 s, and 72 °C for 45 s, and a final incubation at 72 °C for 10 min. The PCR products were checked using 2% agarose gels with ethidium bromide. The PCR products were cloned into the pGM-T vector and sequenced using an ABI 377 automated sequencer. Eight clones were sequenced for each sample.
Table 1.
Primers of bisulfite sequencing PCR and RT-PCR.
| Gene | Primer name | Sequence | Product Type | Product Size | Annealing Temperature |
|---|---|---|---|---|---|
| CDK9 | Forward | 5′-TTGAAGTTGTTGGTATGTGTG-3′ | DNA | 221 bp | 60 °C |
| Reverse | 5′-CTCCTCCCTCCCACCCTT-3′ | (Bisulfite PCR) | |||
| CDK9 | Forward | 5′-CGGAGATCAAGCGGGTCAT-3′ | cDNA | 109 bp | 60 °C |
| Reverse | 5′-GGTGATGAGCACGTTGGC-3′ | (real-time qPCR) | |||
| BNIP3L | Forward | 5′-CAACAACTGTGAGGAAGGTG-3′ | cDNA | 150 bp | 60 °C |
| Reverse | 5′-ATAGAGGATGAGGAAGGTACA-3′ | (real-time qPCR) | |||
| ZC3HAV1 | Forward | 5′-GAAGATCCAGAACACGAAGC-3′ | cDNA | 120 bp | 60 °C |
| Reverse | 5′-GAGGATTCTTTCAACTTTCAC-3′ | (real-time qPCR) | |||
| GAPDH | Forward | 5′-ACTCACTCTTCTACCTTTGATGCT-3′ | cDNA | 100 bp | 60 °C |
| Reverse | 5′-TGTTGCTGTAGCCAAATTCA-3′ | (real-time qPCR) |
4.6. Hybridization of cDNA microarray and integration analysis
The detailed procedures of cDNA microarray were as described in our previous work [2]. Briefly, total RNA was extracted from PK15 cells using the Trizol kit (Invitrogen, Carlsbad, CA). Contaminated DNA was cleared by DNAse (Qiagen). Twelve samples in the hybridization slides were scanned using an Agilent Microarray Scanner (Agilent). The results regarding differential expressed genes (DEGs) were considered significant for a fold change (|FC|) > 1.2 and P < 0.05. The gene ontology (GO) analyses of the DEGs were assessed using Fisher's exact test based on the two public databases: National Center of Biotechnology Information (NCBI) Entrez, Gene Ontology (http://www.geneontology.org). The P-value < 0.05 denotes the significance of GO terms enrichment in the differential enrichment genes.
The integration analysis of the transcriptome and methylome were conducted by R language tools based on the data presented. We used P ≤ 0.01 and |log2 ratio| ≥ 0.3 as the threshold to assess the significance of differentially methylated genes which matched differentially expressed genes (|FC|) > 1.2 and P < 0.05), then the common genes were evaluate the differential methylation peaks (DEPs) between the comparison and control (P vs. C, A vs. C, (P + A) vs. C).
4.7. Real-time qRT-PCR validation and statistical analysis
Real-time quantitative RT-PCR (qRT-PCR) reactions were performed in 20 μL solution with a LightCycler480 SYBR Green I Master (Roche) according to the manufacturer instructions. Each reaction was performed in triplicate. The mRNA expression level of the validated genes (CDK9, BNIP3L and ZC3HAV1) was normalized against the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) in the corresponding samples. The PCR reactions were cycled 40 times after the initial denaturation (95 °C, 5 min) with the following parameters: denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 35 s. The primers are shown in Table 1.
A 2(−△△Ct) method was used to estimate the relative expression level of the genes. Student's t-test was used to analyze the differences of the genes expression levels between the treatments.
Acknowledgements
The authors thank Dr. Tahir Usman from Pakistan for his careful proof reading of our manuscript. This work was supported by the Natural Science Foundation of Inner Mongolia (2016BS0305), the National Natural Science Foundation of China (31572361), the Innovation Research Team for Modern Agricultural Industry and Technology in Beijing City (25019126), the Program of New Breed Development via Transgenic Technology (2013ZX080011-006), the Twelfth Five-Year Plan for National Science and Technology Projects in Rural Areas (2011BAD28B01).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2018.09.020.
Contributor Information
Chuduan Wang, Email: cdwang@cau.edu.cn.
Ying Yu, Email: yuying@cau.edu.cn.
Appendix A. Supplementary data
Fig. S1A: ‘00’ methylation profile. ‘00’ profile means unmethylated probes in the CpG islands of gene promoter. The average number of unmethylated probes in HCP in three comparisons of P vs. C, A vs. C and (P + A) vs. C reached 786, more than that in ICP (212) and LCP (39).; Fig. S1B The three types of distribution of differential enrichment DNA methylation peaks in (P + A) vs. P (purple bar) and (P + A) vs. A (blue bar).; Fig. S2: DEGs (P < .05) in HCP of MeDIP-chip integrated with microarray. 15, 37, 18 DEGs in P vs. C, A vs. C, (P + A) vs. C.; Fig. S3: Relationship between DNA methylation and expression of porcine BNIP3L and ZC3HAV1 gene. (A) The DEPs of BNIP3L promoter in controls compared with PK15 cells treated with PolyI:C plus Aza-CdR. Black arrow represents the differential hyper-methylated CpG islands in C vs. (P + A). (B) Significant expression levels were determined in (P + A) cells compared with the non-treated control cells (blue bars), *P < 0.05. (C) The DEPs of ZC3HAV1 promoter in PK15 cells treated with PolyI:C plus Aza-CdR compared with controls. Black arrow represents the differential hyper-methylated CpG islands in (P + A) vs. C. First panel: green peaks represent probe-level log2 (MeDIP/Input) ratio. Second panel: blue bars represent different M’ value. Third panel: red bars represent P-value score of a minimum cutoff (−log10) of 2. Numbers on the top of panel represent DMRs location. (D) Transcriptional expression of of ZC3HAV1 gene in PK15 cells treated by PolyI:C, Aza-CdR, or both. Significant expression levels were determined in the cells treated by (P + A) (blue bars) compared with the non-treated control cells. *P < 0.05.; Fig. S4: Integrated analysis of promoter methylation and transcriptional expression in HCPs (A), ICPs (B), LCPs (C) for P vs. C, A vs. C and (P + A) vs. C. Gene expression for three comparisons under the control of promoters with each methylation profile (‘01’, ‘10’, ‘11’). DEGs were screened from microarray. In each boxplot, up line and down line represents gene expression levels from the 1/4–3/4 and the middle line represents the average expression level. Red curve indicate trend for the relationship between methylation profiles and transcriptional expression.
Supplementary tables
References
- 1.Anders H.J. Innate pathogen recognition in the kidney: toll-like receptors, NOD-like receptors, and RIG-like helicases. Kidney Int. 2007;72:1051–1056. doi: 10.1038/sj.ki.5002436. [DOI] [PubMed] [Google Scholar]
- 2.Wang X.S., Ao H., Zhai L.W., Bai L.J., He W.Y., Yu Y., Wang C.D. Genome-wide effects of DNA methyltransferases inhibitor on gene expression in double-stranded RNA transfected porcine PK15 cells. Genomics. 2014;103:371–379. doi: 10.1016/j.ygeno.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Galli R., Panoe A., Fabbri M., Zanesi N., Calore F., Cascione L., Acunzo M., Stoppacciaro A., Tubaro A., Lovat F., Gasparini P., Fadda P., Alder H., Volinia S., Filippini A., Ziparo E., Riccioli A., Croce C.M. Toll-like receptor 3 (TLR3) activation induces microRNA-dependent reexpression of functional RARβ and tumor regression. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9812–9817. doi: 10.1073/pnas.1304610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koga Y., Pelizzola M., Cheng E., Krauthammer M., Sznol M., Ariyan S., Narayan D., Molinaro A.M., Halaban R., Weissman S.M. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res. 2009;19:1462–1470. doi: 10.1101/gr.091447.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amara K., Trimeche M., Ziadi S., Laatiri A., Hachana M., Sriha B., Mokni M., Korbi S. Presence of simian virus 40 DNA sequencesin diffuse large B-cell lymphomas in Tunisia correlates with aberrant promoter hypermethylation of multiple tumor suppressor genes. Int. J. Cancer. 2007;121:2693–2702. doi: 10.1002/ijc.23038. [DOI] [PubMed] [Google Scholar]
- 6.Luo J., Yu Y., Chang S., Tian F., Zhang H., Song J.Z. DNA methylation fluctuation induced by virus infection differs between MD-resistant and -susceptible chickens. Front. Genet. 2012;3:20. doi: 10.3389/fgene.2012.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X., Michal J.J., Jiang Z., Liu B. MicroRNA expression profiling in alveolar macrophages of indigenous Chinese Tongcheng pigs infected with PRRSV in vivo. J. Appl. Genet. 2017;58:539–544. doi: 10.1007/s13353-017-0410-9. [DOI] [PubMed] [Google Scholar]
- 8.Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D. Chromosome wide and promoter-specific analyses identify sites of differential DNA methylationin normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 9.Jacinto F.V., Ballestar E., Esteller M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. BioTechniques. 2008;44:35–43. doi: 10.2144/000112708. [DOI] [PubMed] [Google Scholar]
- 10.Pälmke N., Santacruz D., Walter J. Comprehensive analysis of DNA methylation in mammalian tissues using MeDIP-chip. Methods. 2011;53:175–184. doi: 10.1016/j.ymeth.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Gius D., Cui H., Bradbur C.M., Cook J., Smart D.K., Zhao S., Young L., Brandenburg S.A., Hu Y., Bisht K.S., Ho A.S., Mattson D., Sun L., Munson P.J., Chuang E.Y., Mitchell J.B., Feinberg A.P. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell. 2004;6:361–371. doi: 10.1016/j.ccr.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Patel K., Dickson J., Din S., Macleod K., Jodrell D., Ramsahoye B. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karpf A.R., Lasek A.W., Ririe T.O., Hanks A.N. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. Mol. Pharmacol. 2004;65:18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- 14.Li F., Karlsson H. Expression and regulation of human endogenous retrovirus W elements. APMIS. 2016;124:52–66. doi: 10.1111/apm.12478. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Vargas H., Lambert M.P., Le Calvez-Kelm F., Gouysse G., McKay-Chopin S., Tavtigian S.V., Scoazec J.Y., Herceg Z. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C.C., Duffy K.E., San Mateo L.R., Amegadzie B.Y., Sarisky R.T., Mbow M.L. A pathway analysis of polyI:C-induced global gene expression change in human peripheral blood mononuclear cells. Physiol. Genomics. 2006;26:125–133. doi: 10.1152/physiolgenomics.00002.2006. [DOI] [PubMed] [Google Scholar]
- 17.Flori L., Rogel-Gaillard C., Cochet M., Lemonnier G., Hugot K., Chardon P., Robin S., Lefin S.F. Transcriptomic analysis of the dialogue between pseudorabies virus and porcine epithelial cells during infection. BMC Genomics. 2008;9:123. doi: 10.1186/1471-2164-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X., Cheng G., Yan W.Y., Liu M., He Y., Zheng Z. Characterization and virus-induced expression profiles of the porcine interferon- helial cells durin. J. Interf. Cytok. Res. 2009;29:687–693. doi: 10.1089/jir.2008.0060. [DOI] [PubMed] [Google Scholar]
- 19.Flanagan J.M. Host epigenetic modifications by oncogenic viruses. Br. J. Cancer. 2007;96:183–188. doi: 10.1038/sj.bjc.6603516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki H., Gabrielson E., Chen W., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita S., Hosoya K., Gyobu K., Takeshima H., Ushijima T. Development of a novel output value for quantitative assessment in methylated DNA immunoprecipitation-CpG island microarray analysis. DNA Res. 2009;16:275–286. doi: 10.1093/dnares/dsp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashit S., Tsujino Y., Moriguchi K., Tatematsu M., Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2′-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattori N., Okochi-Takad E., Kikuyam M., Wakabayash M., Yamashita S., Ushijima T. Methylation silencing of angiopoietin-like 4 in rat and human mammary carcinomas. Cancer Sci. 2011;102:1337–1343. doi: 10.1111/j.1349-7006.2011.01955.x. [DOI] [PubMed] [Google Scholar]
- 24.Asamitsu K., Omagari K., Okuda T., Hibi Y., Okamoto T. Quantification of the HIV transcriptional activator complex in live cells by image-based protein-protein interaction analysis. Genes Cells. 2016;21:706–716. doi: 10.1111/gtc.12375. [DOI] [PubMed] [Google Scholar]
- 25.Li D.Y., Sun Y., Peng F., Kang Z.Z., Wang J.N., Hua L.L., Zhang Y., Zhang D.L. Effect of H3N2 and H1N1 influenza A virus infection on expression of porcine CDK9 gene. J. Northwest Univ. 2013;41:14–18. [Google Scholar]
- 26.Zhang J.J., Li G., Ye X. Cyclin T1/CDK9 interacts with influenza A virus polymerase and facilitates its association with cellular RNA polymerase II. J. Virol. 2010;84:12619–12627. doi: 10.1128/JVI.01696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsushima M., Fujiwara T., Takahashi E., Minaguchi T., Eguchi Y., Tsujimoto Y., Suzumori K., Nakamura Y. Isolation, mapping, and functional analysis of a novel human cDNA (BNIP3L) encoding a protein homologous to human NIP3. Genes Chromosome Canc. 1998;21:230–235. [PubMed] [Google Scholar]
- 28.Fei P., Wang W., Kim S., Wang S., Burns T.F., Sax J.K., Buzzai M., Dicker D.T., McKenna W.G., Bernhard E.J., El-Deiry W.S. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Cai Y.Y., Liu Y., Yu D.D., Zhang X.M. Down-regulation of transcription of the proapoptotic gene BNip3 in cultured astrocytes by murine coronavirus infection. Virology. 2003;316:104–115. doi: 10.1016/j.virol.2003.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cagliani R., Guerini F.R., Fumagalli M., Riva S., Agliardi C., Galimberti D., Pozzoli U., Goris A., Dubois B., Fenoglio C., Forni D., Sanna S., Zara I., Pitzalis M., Zoledziewska M., Cucca F., Marini F., Comi G.P., Scarpini E., Bresolin N., Clerici M., Sironi M. A trans-specific polymorphism in ZC3HAV1 is maintained by long-standing balancing selection and may confer susceptibility to multiple sclerosis. Mol. Biol. Evol. 2012;29:1599–1613. doi: 10.1093/molbev/mss002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo X., Ma J., Sun J., Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. U. S. A. 2007;104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu X., Hother C., Ralfkiær U.M., Søgaard A., Lu Q., Workman C.T., Liang G., Jones P.A., Grønbæk K. Equitoxic doses of 5-azacytidine and 5-aza-2′deoxycytidine induce diverse immediate and overlapping heritable changes in the transcriptome. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer M., Hagemann S., Hanna K., Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 34.Momparler R.L., Siegel S., Avila F., Lee T., Karon M. Effect of tRNA from 5-azacytidine-treated hamster fibrosarcoma cells on protein synthesis in vitro in a cell-free system. Biochem. Pharmacol. 1976;25:389–392. doi: 10.1016/0006-2952(76)90338-5. [DOI] [PubMed] [Google Scholar]
- 35.Jackson-Grusby L., Beard C., Possemato R., Tudor M., Fambrough D., Csankovszki G., Dausman J., Lee P., Wilson C., Lander E., Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 36.Biedenkapp H., Borgmeyer U., Sippel A.E., Klempnauer K.H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- 37.Yan Y.X., Du Y.M., Wang G.F., Li K.S. Non-structural protein 1 of H3N2 influenza A virus induces nucleolar stress via interaction with nucleolin. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-18087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng M., Dai M., Xie T.T., Li Z.H., Shi M.Q., Zhang X.Q. Innate immune responses in ALV-J infected chicks and chickens with hemangioma in vivo. Front. Microbiol. 2016;7:786. doi: 10.3389/fmicb.2016.00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matousková M., Vesely P., Daniel P., Mattiuzzo G., Hector R.D., Scobie L., Takeuchi Y., Hejnar J. Role of DNA methylation in expression and transmission of porcine endogenous retroviruses. J. Virol. 2013;87:12110–12120. doi: 10.1128/JVI.03262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung A.K. Identification of the essential and non-essential transcription units for protein synthesis, DNA replication and infectious virus production of Porcine circovirus type 1. Arch. Virol. 2004;149:975–978. doi: 10.1007/s00705-003-0249-8. [DOI] [PubMed] [Google Scholar]
- 41.Bird A.P., Wolffe A.P. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 42.Bock C. Analysing and interpreting DNA methylation data. Nat. Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 43.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J.J., Guo Y.M., Hirokawa M., Iwamoto K., Ubukawa K., Michishita Y., Fujishima N., Tagawa H., Takahashi N., Xiao W., Yamashita J., Ohteki T., Sawada K. A synthetic double-stranded RNA, polyI:C, induces a rapid apoptosis of human CD34+ cells. Exp. Hematol. 2012;40:330–341. doi: 10.1016/j.exphem.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Palchetti S., Starace D., De Cesaris P., Filippini A., Ziparo E., Riccioli A. Transfected poly(I:C) activates different dsRNA receptors, leading to apoptosis or immunoadjuvant response in androgen-independent prostate cancer cells. J. Biol. Chem. 2015;290:5470–5483. doi: 10.1074/jbc.M114.601625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frank-Bertoncelj M., Pisetsky D.S., Kolling C., Michel B.A., Gay R.E., Jüngel A., Gay S. TLR3 ligand poly(I:C) exerts distinct actions in Synovial Fibroblasts when delivered by extracellular vesicles. Front. Immunol. 2018;9:28. doi: 10.3389/fimmu.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1A: ‘00’ methylation profile. ‘00’ profile means unmethylated probes in the CpG islands of gene promoter. The average number of unmethylated probes in HCP in three comparisons of P vs. C, A vs. C and (P + A) vs. C reached 786, more than that in ICP (212) and LCP (39).; Fig. S1B The three types of distribution of differential enrichment DNA methylation peaks in (P + A) vs. P (purple bar) and (P + A) vs. A (blue bar).; Fig. S2: DEGs (P < .05) in HCP of MeDIP-chip integrated with microarray. 15, 37, 18 DEGs in P vs. C, A vs. C, (P + A) vs. C.; Fig. S3: Relationship between DNA methylation and expression of porcine BNIP3L and ZC3HAV1 gene. (A) The DEPs of BNIP3L promoter in controls compared with PK15 cells treated with PolyI:C plus Aza-CdR. Black arrow represents the differential hyper-methylated CpG islands in C vs. (P + A). (B) Significant expression levels were determined in (P + A) cells compared with the non-treated control cells (blue bars), *P < 0.05. (C) The DEPs of ZC3HAV1 promoter in PK15 cells treated with PolyI:C plus Aza-CdR compared with controls. Black arrow represents the differential hyper-methylated CpG islands in (P + A) vs. C. First panel: green peaks represent probe-level log2 (MeDIP/Input) ratio. Second panel: blue bars represent different M’ value. Third panel: red bars represent P-value score of a minimum cutoff (−log10) of 2. Numbers on the top of panel represent DMRs location. (D) Transcriptional expression of of ZC3HAV1 gene in PK15 cells treated by PolyI:C, Aza-CdR, or both. Significant expression levels were determined in the cells treated by (P + A) (blue bars) compared with the non-treated control cells. *P < 0.05.; Fig. S4: Integrated analysis of promoter methylation and transcriptional expression in HCPs (A), ICPs (B), LCPs (C) for P vs. C, A vs. C and (P + A) vs. C. Gene expression for three comparisons under the control of promoters with each methylation profile (‘01’, ‘10’, ‘11’). DEGs were screened from microarray. In each boxplot, up line and down line represents gene expression levels from the 1/4–3/4 and the middle line represents the average expression level. Red curve indicate trend for the relationship between methylation profiles and transcriptional expression.
Supplementary tables