Highlights
-
•
–1PRF occurs when ribosomes move over a slippery sequence.
-
•
A frameshifting pseudoknot/stem-loop element stalls ribosomes in a metastable state.
-
•
–1PRF may contribute to the quality-control machinery in eukaryotes.
-
•
Trans-acting factors (proteins, miRNAs, or antibiotics) can modulate –1PRF.
Keywords: gene expression, translation, protein synthesis, decoding, ribosome, mRNA reading frame maintenance
Abstract
Programmed −1 ribosomal frameshifting (−1PRF) is an mRNA recoding event commonly utilized by viruses and bacteria to increase the information content of their genomes. Recent results have implicated −1PRF in quality control of mRNA and DNA stability in eukaryotes. Biophysical experiments demonstrated that the ribosome changes the reading frame while attempting to move over a slippery sequence of the mRNA – when a roadblock formed by a folded downstream segment in the mRNA stalls the ribosome in a metastable conformational state. The efficiency of −1PRF is modulated not only by cis-regulatory elements in the mRNA but also by trans-acting factors such as proteins, miRNAs, and antibiotics. These recent results suggest a molecular mechanism and new important cellular roles for −1PRF.
Accurate decoding versus programmed recoding
Ribosomes (see Glossary) are cellular factories that produce proteins in all cells using the nucleotide sequence of mRNAs as a blueprint. Nucleotide triplets of an mRNA – the codons – are translated into an amino acid sequence of a protein. The selection of the translation start and the reading frame on an mRNA is tightly controlled during the initiation phase of protein synthesis. The subsequent elongation phase entails repeated cycles of codon decoding by aminoacyl-tRNA, peptide bond formation, and tRNA–mRNA translocation. Elongation cycles continue until the ribosome reaches a stop codon, on which translation is terminated. It is intuitively clear that translation of an mRNA sequence into a protein must be colinear and highly accurate. Errors can lead to the formation of toxic or misfolded proteins, increase the energetic cost of translation, and cause additional load on the cellular clean-up and quality-control machineries [1]. To avoid this burden, cells have evolved sophisticated control mechanisms that ensure the fidelity of decoding and reading frame maintenance. However, in special cases, the ribosomes, guided by signals encoded in the mRNA, abandon the principle of mRNA–protein colinearity and read the message in an alternative way, which results in mRNA recoding (Box 1 ) 2, 3, 4. Programmed frameshifting is a recoding event that can occur in the + or − direction relative to the normal 0-frame mRNA translation by shifting the ribosome by one or two nucleotides, thereby producing two (or even three) different proteins from one mRNA. In contrast to spontaneous frameshifting, which is infrequent, the efficiency of programmed frameshifting may be as high as 80%, although in many cases it is only a few percent [5]. Programmed frameshifting increases the coding potential of the genome and is often used to expand the variability of cellular proteomes, adapt to changing environments, or ensure a defined stoichiometry of protein products. The mechanisms of +1 and −1 frameshifting appear to be different, and particularly the mechanism of −1 programmed ribosome frameshifting (−1PRF), its abundance, and physiological significance have remained unclear for a long time. The advances of biophysical techniques and reconstitution of highly-purified translation systems has recently provided new insight into how and when ribosomes slip into the −1 reading frame. New examples of −1PRF in eukaryotes and the identification of previously unknown trans-acting elements show how cells can regulate gene expression through frameshifting. This review is intended to summarize these recent breakthroughs in understanding the mechanism and biological importance of −1PRF.
Box 1. Translational recoding events.
Protein synthesis is usually very accurate. If nevertheless an error occurs it can lead to insertion of a wrong amino acid into a protein at a particular position due to incorrect decoding of a sense codon (a missense error) or a stop codon (stop-codon read-through). Alternatively, the ribosome may spontaneously change the reading frame on the mRNA, resulting in a completely different protein sequence (frameshifting). Spontaneous missense, nonsense, or frameshifting errors occur at frequencies of 10−7 to 10−3 per codon 57, 58, 59, 60.
In contrast to spontaneous errors during elongation, recoding, or reprogrammed genetic decoding, is a process that alters the reading of individual codons (usually stop codons), alleviates the colinearity of mRNA and protein, or changes the reading frame of an mRNA. Recoding events are usually ‘programmed’ by stimulatory elements in the mRNA. The efficiency of recoding can reach up to 80% of normal translation 61, 62, 63. Main types of recoding are read-through, ribosomal frameshifting, and bypassing (Figure I ) 3, 4. A stop codon can be interpreted as a sense codon for non-standard amino acids, such as selenocysteine or pyrrolysine (read-through). The ribosome can skip a part of the mRNA producing a single protein from a discontinuous reading frame (bypassing); examples of bypassing are in gene60 of bacteriophage T4 and in the mitochondrial genome of the yeast Magnusiomycetes 62, 64, 65. Slippage of the ribosome in + or − direction changes the reading frame on the mRNA. The best known examples of +1 frameshifting are the RF2 gene in bacteria [66] and the ornithine decarboxylase gene [67] in eukaryotes; examples of −1PRF are summarized in Table 1.
Figure I.
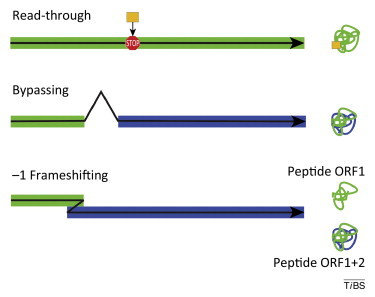
Three major types of recoding events. Read-through entails the insertion of an unusual amino acid (gold) at the position of a stop codon. Bypassing connects two parts of a discontinuous reading frame (green and blue linear segments) resulting in a single protein (green-blue). Frameshifting may be used to regulate the synthesis of a single protein or to produce two proteins (peptide ORF1 and peptide ORF1+2) from a single mRNA with two overlapping open reading frames (ORFS; green and blue segments). The black arrows indicate the direction of ribosome movement.
Prevalence of ribosomal frameshifting
Although the large majority of reported frameshifting sequences have so far been found in viral genomes, programmed frameshifting exists in all branches of life from bacteria to higher eukaryotes. The important role of −1PRF in viruses and bacteria is well documented [6]. In bacteria, −1PRF is required to produce the γ subunit of DNA polymerase III and is particularly abundant in bacterial transposable elements, which use −1PRF to generate their transposase 2, 7. −1PRF is also found in several families of eukaryotic viruses, where it is often used for the expression of viral replicases [8]. In retroviruses such as HIV, −1PRF is necessary to produce the DNA polymerase Pol and defines the ratio of Gag to Pol proteins. Because this ratio should not exceed a particular threshold value to maintain efficient virus assembly, genome packaging and maturation [9], changing −1PRF efficiency can be detrimental to the virus 10, 11. In comparison to viruses and bacterial mobile elements, only a few examples of −1PRF have been described in higher eukaryotes, although computational analysis have predicted that ∼10% of cellular mRNAs may be controlled by −1PRF [12]. Recent evidence suggests that −1PRF may have unexpected roles in regulating not only the production of particular proteins but also in regulating mRNA and DNA stability (Table 1 ). Dinman and colleagues demonstrated operational −1PRF signals in four mRNAs encoding proteins crucial for yeast telomerase maintenance [13], and reported an example of eukaryotic −1 PRF in ccr5 and six other genes for cytokine receptors [14]. Overexpression of the telomerase gene Est2 through inhibition of frameshifting leads to formation of shorter telomeres, suggesting the importance of −1PRF for the maintenance of eukaryotic genomes [13]. Translation in the −1 frame of ccr5 leads to a premature stop, which targets the mRNA for degradation by nonsense-mediated decay (NMD) [14]. This appears to be a general phenomenon because, for the large majority of predicted genes, −1PRF leads to premature termination less than 30 codons beyond the frameshifting site [15]. Thus, in eukaryotic genomes −1PRF can be used to regulate gene expression. Another notable example is the embryonal carcinoma differentiation regulated gene (edr), where −1PRF regulates the synthesis of two distinct polypeptides [16]. Edr is a single-copy gene in mice and humans. Its expression is regulated in a spatiotemporal manner during embryogenesis, suggesting a role of −1PRF in development [17]. Global analysis of dual decoding potential in human cells revealed several new examples of alternative frame reading in ribosome profiling data [18]. By analogy, one can expect more examples of −1PRF emerging with advances in high-throughput analysis of cellular proteomes. Together, these findings indicate that −1PRF may be more abundant and more important in eukaryotic cells than previously thought.
Table 1.
Examples of –1PRF
| Gene (protein function) | Systema | Slippery site | Stimulator | Refs |
|---|---|---|---|---|
| Insertion sequences of IS1 IS3 family | Bacteria | Various | Stem loop, SD like element | [7] |
| dnaX (polymerase III subunits) | Bacteria (Escherichia coli and relatives) | AAAAAAG | Stem loop, SD like element | [61] |
| cdd (cytidine deaminase) | Bacteria | ACGAAAG | SD like element (GGAGG) | [70] |
| ermCL | Bacteria (Erm resistant strains) | AAAAAA | Putative stem loop | [49] |
| orf 1a/1b | Coronavirus (IBV, MHV, SARS Co-V, HCV) | UUUAAAC | Pseudoknot | 71, 72, 73, 74 |
| gag/pol (polymerase) | Lentivirus (VMV, FIV) | GGGAAAC | Pseudoknot | 75, 76 |
| gag/pol (polymerase) | Lentivirus (HIV, SIV) | UUUUUUA | Stem loop | 77, 78 |
| orf2/3 (p2/p3) | Polerovirus (BWYV, ScYLV, PEMV) | GGGAAAC | Pseudoknot | 79, 80 |
| orf1/2 | Totivirus (ScV-L-A) | GGGUUUA | Stem loop | [81] |
| orf 1a/1b | Astrovirus (Hast-V1) | AAAAAAC | Stem loop | [82] |
| orf 1a/1b | Arterivirus (BEV, EAV) | U/GUUAAAC | Pseudoknot | 83, 84 |
| orf 1a/1b (replicase) | Arterivirus (PRRSV) | GGUUUUU | CCCANCUCC | [31] |
| Peg10/Edr (embryonic regulator) | Higher eukaryotes | GGGAAAC | Pseudoknot | [16] |
| Ma3 (paraneoplastic antigen) | Higher eukaryotes | GGGAAAC | Pseudoknot | [85] |
| ccr5 (HIV cytokine receptor) | Higher eukaryotes | UUUAAAA | Pseudoknot | [14] |
| ILR (interleukin receptor subunits) | Higher eukaryotes | Various | Putative pseudoknot | [14] |
BEV, bovine enterovirus; BWYV, beet western yellows virus; EAV, equine arteritis virus; FIV, feline immunodeficiency virus; Hast-V1, human astrovirus type 1; HCV, human coronavirus; IBV, avian infectious bronchitis virus; MHV, mouse hepatitis virus; PEMV, pea enation mosaic virus; PRRSV, porcine reproductive and respiratory syndrome virus; SARS Co-V, severe acute respiratory syndrome coronavirus; ScV-L-A, Saccharomyces cerevisiae virus L-A; ScYLV, sugarcane yellow leaf virus; SIV, simian immunodeficiency virus; VMV, visna-maedi retrovirus.
When do ribosomes slip?
−1PRF is promoted by several stimulatory elements embedded in the mRNA sequence (Figure 1 ) 5, 19. The crucial regulatory element is the so-called slippery sequence, which is usually a heptameric sequence with the pattern X XXY YYZ (the underlined sequence denotes the 0-frame codons). The slippery sequence ensures correct base-pairing between the mRNA codon and the tRNA anticodon before and after the slippage, provided that the 0-frame and −1-frame codons can be decoded by the same tRNA through wobble interactions at the third codon position [XXY (0-frame) vs XXX (−1-frame), or YYZ (0-frame) vs YYY (−1-frame)]. Another stimulatory element is an mRNA secondary structure element, usually a pseudoknot or a stem-loop located 5–8 nt downstream of the slippery sequence. Interestingly, antisense oligonucleotides or guanine-rich sequences that can fold into four-stranded structures of stacked guanine-tetrads, so-called G-quadruplexes, can also stimulate −1PRF, reaching efficiencies up to 40% 20, 21. In addition, in some cases a Shine–Dalgarno-like element 11–14 nt upstream of the first slippery codon [22] or even long-distance base-pairing appear to stimulate frameshifting [23].
Figure 1.
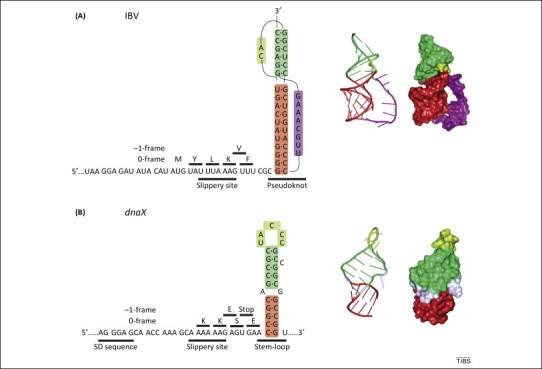
Frameshifting stimulatory signals on mRNA. (A) Left panel, schematic of the modified frameshifting sequence of the avian infectious bronchitis virus (IBV) 1a/1b mRNA [27]. The original IBV slippery sequence (U UUA AAC) was changed to ensure maximum frameshifting efficiency in Escherichia coli[37]. The amino acids incorporated upon translation in the 0- or −1-frame are indicated above the mRNA sequence. Middle and right panels, structure and surface representation of the frameshifting pseudoknot. (B) Left panel, schematic of the programmed −1 ribosomal frameshifting (−1PRF) regulatory elements in dnaX from E. coli. In addition to the slippery sequence and stimulatory stem-loop, frameshifting on dnaX is modulated by a Shine–Dalgarno (SD)-like sequence upstream of the slippery site [22]. Middle and right panels, structure and surface representation of the frameshifting stem-loop. Images were prepared using PyMOL (http://www.pymol.org/).
To understand the mechanism of −1PRF, it is first essential to identify the precise timing of slippage and the position of the frameshifting ribosome on the slippery sequence. One straightforward way to determine the timing of slippage is to analyze the kinetics of translation at each codon before, over, and after the slippery sequence (through a ‘codon walk’) to find the step which is different for ribosomes that undergo −1PRF compared to those continuing unperturbed translation in the 0-frame. Previous reports suggested that a ribosome, when it encounters a frameshifting pseudoknot or a stem-loop, pauses or even drops off the mRNA track 24, 25, 26. Such losses in translation rate and efficiency can be reliably monitored by rapid kinetic analysis that monitors the chemical reactions during translation. For example, the stepwise insertion of amino acids into the nascent peptide can be measured using the quench-flow technique (Figure 2 ) [27]. The outcome of −1PRF can be also monitored by mass spectrometry or peptide analysis 28, 29, 30, 31, 32, 33, which may provide information about the slippage pathway. Alternatively, the timing of −1PRF can be identified by following single ribosomes while they translate the mRNA using various techniques, such as optical tweezers [28] or single-molecule fluorescence resonance energy transfer (FRET) 34, 35, 36. In the latter case, fluorescence labels must be attached to elements of the ribosome or to tRNAs. Translation is then monitored using changes in FRET efficiency between two reporters, or fluorescence changes of single reporters 35, 36. Fluorescence and FRET approaches were also used in ensemble kinetic experiments using the stopped-flow technique [27].
Figure 2.
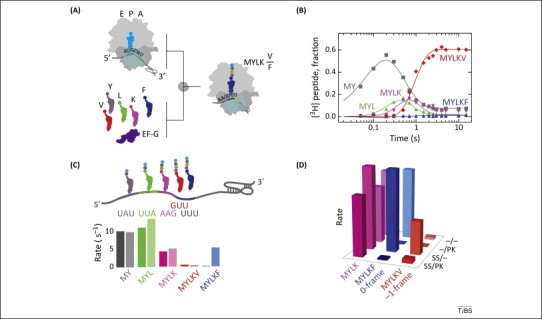
Codon walk over the frameshifting sequence of avian infectious bronchitis virus (IBV). (A) Schematic of the experiments. Purified translation components are rapidly mixed in a quench-flow device. After the desired time the reactions are stopped by rapid mixing with a strong base (quencher) and the peptide products are analyzed by HPLC, measuring [3H]Met radioactivity. (B) Example of a time-course of peptide synthesis on a frameshifting mRNA. Monitored peptides are fMetTyr (MY), fMetTyrLeu (MYL), fMetTyrLeuLys (MYLK), fMetTyrLeuLysPhe (MYLKF), and fMetTyrLeuLysVal (MYLKV). The time-courses are used to calculate the rates and efficiencies of insertion into peptide of each amino acid in 0- and −1-frame. (C) Comparison of the amino acid incorporation rates (s−1) for frameshifting (dark colors) and non-frameshifting (light colors) mRNAs. The frameshifting mRNA contained both a slippery sequence and pseudoknot, the non-frameshifting mRNA had no stimulatory elements for programmed −1 ribosomal frameshifting (−1PRF). (D) Contributions of the regulatory elements in the mRNA. SS and PK indicate the presence of a slippery site or a pseudoknot, respectively, in the mRNA [27].
A codon walk over the modified frameshifting sequence of avian infectious bronchitis virus (IBV) 1a/1b (Figure 1A) identified the step at which frameshifting occurs (Figure 2). IBV 1a/1b is a model system that – after some adjustments in the frameshifting signals – works well in eukaryotic and bacterial cells 27, 37. The frameshifting site of the IBV 1a/1b construct optimized for E. coli contains a slippery sequence U UUA AAG followed by a pseudoknot 6 nt downstream of the slippery sequence. When translation in the 0-frame is not perturbed by −1PRF signals, the ribosome rapidly moves over the mRNA incorporating Tyr, Leu, Lys, and Phe (the 0-frame amino acids) at rates of 4–15 s−1 [27]. Tyr, Leu, and Lys are also rapidly incorporated when the −1PRF stimulatory signals are present (Figure 2C). However, Phe insertion is delayed (insertion into peptide is 0.1 s−1, 40-fold slower than during normal translation) and inefficient (on only 25% of the ribosomes). Instead, the −1-frame amino acid Val is preferentially incorporated (on 75% of the ribosomes), albeit also at a low rate (0.3 s−1). Notably, Phe and Val tRNAs do not compete with one another for binding to the ribosome, indicating that the partitioning of the ribosomes between the 0- and −1-frames occurs after Lys insertion, but before the next codon is decoded. These data indicate that commitment to the alternative frame occurs sometime after Lys insertion, but before the next decoding step. Thus, −1PRF must take place during the movement of the ribosome over the slippery sequence, when peptidyl-tRNALys is translocated from the A to the P site of the ribosome, while tRNALeu moves from the P to the E site [27]. Notably, as the ribosome sets out to move over the slippery codons, the pseudoknot/stem-loop is located 12 nt from position 1 of the P-site codon, very close to the entry to the mRNA entry channel of the ribosome [38]. Translocation over the slippery sequence would require that the first two base-pairs of the pseudoknot/stem-loop structure open up to enter the channel. Thus, to continue translation, the ribosome must unwind the double strand at the base of the pseudoknot.
While −1PRF upon IBV 1a/1b translation occurs at a single defined site, the situation is more complex in a different model system, the bacterial dnaX gene, that is capable of dual coding. The frameshifting sequence of dnaX has the slippery sequence A AAA AAG, an mRNA stem-loop structure as a stalling element located 6 nt downstream, and a Shine–Dalgarno-like sequence upstream of the slippery sequence (Figure 1B). Mass spectrometry of translated products showed that ribosomes can slip not only by −1 but also by −4 or +2 nt, suggesting the existence of alternative frameshifting sites [28]. Similarly to IBV 1a/1b, slippage on dnaX occurs during tRNA translocation 28, 35, 36. Earlier work suggested that −PRF may occur by different pathways [29] and, accordingly, Chen et al. [35] reported that tRNA accommodation contributed to frameshifting in addition to translocation. By contrast, Yan et al. [28] did not find any evidence for −1PRF pathways via schemes other that translocation; such alternative pathways were rigorously ruled out for IBV 1a/1b [27]. The timing of −1PRF on dnaX may depend on the position and strength of the Shine–Dalgarno-like sequence and the exact location of the stem-loop base. A codon walk over the dnaX sequence, and over other examples of frameshifting sequences, should clarify how the regulatory elements govern the timing and mechanism of −1PRF.
What exactly happens during slippage?
Recent results from ensemble kinetics and smFRET suggest a picture of what takes place on the ribosome during frameshifting (Figure 3 ). Frameshifting signals have only a small effect on the conformational dynamics of the ribosome before tRNA-mRNA translocation [in the pre-translocation (PRE) state, Box 2 ] [36]. The rate of EF-G binding to the PRE state is somewhat reduced by the presence of the pseudoknot [27]. However, these effects are mild and do not limit the rate of subsequent steps. In addition, the rate of tRNA movement after EF-G recruitment is not affected by frameshifting 27, 35, 36. Notably, at this stage the tRNA anticodons move together with the head of the 30S subunit [27]. However, from this point on translocation on frameshifting ribosomes is perturbed. The tRNA bound on the first slippery codon (XXY) is retained in the E site longer than normally. EF-G apparently must undertake multiple binding attempts and remains bound to the ribosome for a longer time before translocation is completed 27, 35, 36. The ribosome is stalled in a long-lived rotated (or even hyper-rotated) state 35, 39, and backward movement of the 30S subunit is impeded [27]. Thus, during −1PRF ribosomes are trapped in a metastable state in which both backward movement of the 30S head and reverse rotation of the 30S subunit are impeded, most probably by the pseudoknot/stem-loop blocking the entrance to the mRNA tunnel. Although both dissociation of the E-site tRNA and backward rotation of the 30S subunit are slow, the E-site tRNA is released before the ribosome rotates backwards [35]. This would suggest that there is a time-window when only one tRNA is bound to the ribosome. Furthermore, such a metastable ribosome conformation may also destabilize the tRNA in the P site [40], which may increase the chance of slippage. The ribosomes can also undergo back-and-forth translocation excursions exploring alternative reading frames [28]
Figure 3.
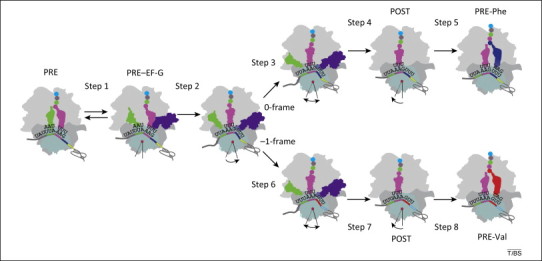
Kinetic model of programmed −1 ribosomal frameshifting (−1PRF). Frameshifting occurs during translocation of the two tRNAs bound to the slippery sequence (tRNALeu in the P site and MYLK-tRNALys in the A site). Recruitment of EF-G (step 1) to the pre-translocation (PRE) complex facilitates rapid tRNA movement (step 2) into a state where translocation on the 50S subunit is completed; however, the following steps are inhibited by the presence of the pseudoknot. tRNALeu moves on the 50S subunit while the distance to the 30S subunit is not changed (steps 3 and 6 in 0-frame and −1-frame, respectively). Then, tRNALeu and the 30S subunit move apart (steps 4 and 7). Steps 3 and 4 are particularly slow for the tRNA that remains in 0-frame, which limits the decoding rate in the 0-frame by Phe-tRNAPhe (step 5). By contrast, tRNALeu movement on those ribosomes that shift to the −1-frame is faster (step 6), followed by dissociation of tRNALeu from the 30S subunit, 30S head rotation, dissociation of EF-G (step 7), and binding of Val-tRNAVal (step 8). Reproduced, with permission, from [27].
Box 2. Translocation.
Movement of the ribosome along the mRNA is a complex, multiphasic process which is catalyzed by the translation elongation factor G (EF-G) in bacteria (Figure I ) (for recent review, see [68] and references therein). Before translocation, deacylated tRNA and peptidyl-tRNA reside in the P and A sites of the ribosome, respectively, with their anticodons attached to the mRNA codons on the 30S ribosomal subunit (the pre-translocation or PRE complex). On the 50S subunit, the 3′ ends of the tRNAs can spontaneously move from their classical (C) positions in the P and A sites towards E and P sites, respectively, entering the so-called hybrid (H) states. Concomitantly, ribosomal subunits rotate relative to one another from the classical non-rotated (N) to the rotated (R) state. Recruitment of EF-G and GTP hydrolysis promote rapid translocation of the tRNAs on both ribosomal subunits. At this point in time, the head of the 30S subunit moves together with the tRNAs and mRNA. Then, the E-site tRNA and EF-G become free to spontaneously dissociate from the ribosome, the head of the 30S subunit moves backwards, and the 30S subunit as a whole rotates relative to the 50S subunit into the N state, resulting in a post-translocation state (POST). These steps form the conformational landscape of translocation and couple the energy of Brownian motion and GTP hydrolysis of EF-G to the directed movement of the ribosome along the mRNA.
Figure I.
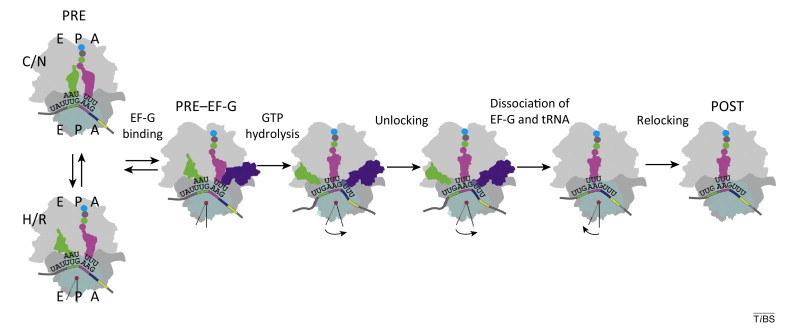
Schematic of translocation. Figure modified from [27].
Although translocation is obviously a complex process, there are well-established methods to follow the elemental steps by ensemble kinetics and single-molecule techniques. One reliable diagnostic test to determine whether the peptidyl-tRNA has been translocated to the P site is the ability of peptidyl-tRNA to react with the antibiotic puromycin that replaces aminoacyl-tRNA as the A-site substrate: if peptidyl-tRNA rapidly reacts with puromycin it is in the authentic P site on the 50S subunit. To monitor the kinetic steps of translocation a variety of fluorescence labels attached to mRNA, tRNA, or the ribosome have been utilized, for example to measure FRET between two ribosome-bound tRNA; between the P/E site tRNA and ribosomal proteins L1 or S13; or the A/P site tRNA and the ribosomal protein L11 27, 34, 35, 36. Rotation is monitored using fluorescence reporters attached to the 30S and 50S subunits [69]. Finally, EF-G binding and dissociation can be followed using labels in EF-G on one side and ribosomal proteins L11 or L12 on the other side 27, 34.
There are two ways to resolve the metastable stalled state: either the ribosome waits for spontaneous mRNA unwinding, or it slips into the −1 direction (Figure 3). The latter scenario may be advantageous because it moves the base of the pseudoknot/stem-loop to the entrance of the mRNA tunnel where the helicase center of the ribosome can actively unwind the mRNA secondary structure 38, 41, allowing more rapid progression of the ribosomes. As a result of kinetic partitioning, a small fraction of ribosomes continues translation in the 0-frame, while the majority switches into the −1-frame [27].
The role of cis-acting elements
Deciphering the molecular mechanism of −1PRF provides novel insights into the roles of the regulatory elements in the mRNA. In general, the presence of the pseudoknot/stem-loop structure stalls the ribosome by impeding translocation 27, 34, 35, 36 (Figure 2D). Optical-trap experiments have shown that folded elements in the mRNA decrease elongation rates and that the mRNA structure stability correlates with elongation velocity [42]. The pseudoknot/stem-loop may act as a roadblock for rapid translation. Because the size of the double-helical stem region of the pseudoknot or a hairpin (21 Å) is larger than the average diameter of the mRNA tunnel (15 Å) [43], the mRNA must unwind before entering the tunnel. The bacterial ribosome has an intrinsic helicase activity attributed to the 30S subunit proteins S3, S4, and S5 [38]. This mechanism may work in both pro- and eukaryotic systems because the helicase residues in S3 and S5 are evolutionarily conserved. The ribosome may exploit charged residues in proteins S3, S4, and S5 to unwind the mRNA either actively, acting as a processivity clamp, or passively, by stabilizing the open form of the mRNA at the entrance of the mRNA tunnel [41]. For complex RNA structures such as a pseudoknot, its overall thermodynamic stability does not directly correlate with the efficiency of frameshifting; instead, the stability of the double strand at the mRNA entrance channel of the ribosome at the base of the pseudoknot is important [44]. In addition, the downstream RNA secondary structure may dynamically switch between different conformations [45], explaining why structural plasticity is important for −1PRF [46]. Interestingly, ribosome stalling at an mRNA roadblock inhibits dissociation of the E-site tRNA and EF-G even in the absence of frameshifting 27, 34, suggesting a global effect of an mRNA roadblock on ribosome dynamics.
The slippery sequence is essential for −1PRF to occur when the ribosome is stalled at the pseudoknot/stem-loop element. In addition, kinetic data suggest that slippery sequences may cause a significant percentage of ribosomes to change the reading frame even in the absence of a roadblock on the mRNA (Figure 2D) [27]. Such slippage would provide an additional, yet unexplored potential for dual coding in genes. Computational analysis of the yeast genome identified 43 841 potential slippery sites, compared to the 17 109 expected assuming a random occurrence of such sequences [47]. This suggests that slippery sites are over-represented in the yeast genome, which may reflect an evolutionary bias in favor of such sequences; however, it is not known whether and to what extent this potential for dual coding is utilized.
Trans-acting factors regulating frameshifting
Stimulation or inhibition of frameshifting by cellular factors is another emerging area of particular interest. One group of potential −1PRF regulators acting in trans are antibiotics [48]. Ketolide antibiotics such as erythromycin (Erm) promote frameshifting at the ermC mRNA leader segment (ermCL). Expression of ermCL regulates the expression of the downstream ermC resistance gene, allowing the translating ribosome to enter the intergenic spacer and alter the mRNA structure in a way that unmasks the start codon of ermC and leads to the translation of the main ermC open reading frame (ORF) [49]. The slippage in ermCL occurs on a run of adenine residues, AAA AAA, at the last two sense codons of ermCL. Such a slippery sequence would allow tRNALys (anticodon UUU) to re-pair to the alternative Lys codon in the −1-frame, similarly to what is observed in the IBV 1a/1b system. How exactly ketolides, which bind in the peptide exit channel of the 50S subunit, promote frameshifting is not known. One possibility is that stalling of translation by a roadblock in the exit tunnel has a similar effect on the slippage as a roadblock in the mRNA. A similar regulatory principle may be utilized in other bacterial and eukaryotic genes that are activated in response to different environmental conditions 49, 50.
Another group of trans-regulators of −1PRF are proteins. For example, nsp1β, the replicase subunit of the porcine reproductive and respiratory syndrome virus (PRRSV), can activate −1 and −2 ribosomal frameshifting, resulting in the synthesis of three different proteins from a single mRNA [31]. A putative RNA-binding domain in the nsp1β protein is likely to bind to a conserved CCCANCUCC motif located 11 nt downstream of the slippery sequence. The complex (alone or with other cellular components) may provide a roadblock analogous to a pseudoknot/stem-loop structure, favoring ribosome rearrangements that may lead to −1PRF. Notably, nsp1β appears to interact with rpS14, a ribosomal protein located close to the ribosomal helicase site [51]. It is tempting to speculate that nsp1β may affect the helicase activity, thereby modulating −1PRF.
A special group of trans-acting factors comprises proteins or RNAs that stabilize or destabilize the stimulatory mRNA element. For example, annexin A2 (ANXA2) binds to the IBV pseudoknot; overexpression of ANXA2 reduces −1PRF, whereas knockdown of the protein stimulates −1PRF [52]. Because ANXA2 has been implicated in diverse cellular functions, including exocytosis, DNA synthesis and cell proliferation, the protein may contribute to the host defense in virus-infected cells by inhibiting frameshifting. Micro-RNAs (miRNAs) can also modulate −1PRF by binding to the pseudoknot. The efficiency of −1PRF on ccr5 mRNA, coding for the cytokine receptor used by HIV, appears to be modulated by at least two different miRNAs [14]. Binding of miRNAs stabilizes the frameshifting pseudoknot, thereby leading to increased −1PRF. miRNA-regulated −1PRF has also been demonstrated in mRNAs encoding six other cytokine receptors. Finally, a more complex example of regulation by a protein (or a group of proteins) is provided by the viral Tat protein, a regulatory protein that upregulates the efficiency of HIV transcription. Tat modulates −1PRF by binding to the TAR region in the 5′ untranslated region (UTR) on the HIV mRNA or by some indirect effect related to disruption of Tat–TAR interaction [53]. Destabilization of the Tat–TAR interactions in the mRNA 5′ UTR would facilitate the ribosome access to the start codon of gag/pol mRNA and accelerate ribosome loading. As a result, the distance between the ribosomes translating HIV mRNA would not provide enough time for the frameshifting stem-loop structure to refold between the passages of consecutive ribosomes [54]. Furthermore, Tat also regulates translation by favoring cap- over internal ribosome entry site (IRES)-dependent translation initiation. Because IRES-dependent translation makes much less −1PRF than does cap-dependent translation, the switch from cap- to IRES-mediated initiation alters the overall amount of the −1-frame Pol product [53]. Thus, translation efficiency per se can modulate −1PRF 25, 55. Along the same lines, changes in global translation rates may account for the stimulation of −1PRF upon downregulation of eukaryotic translation release factor levels [56]. Together, these results highlight a complex network of trans-acting factors that can modulate −1PRF.
Concluding remarks
Advances in understanding −1PRF provide new insights into the mechanism of recoding, highlight novel principles for the regulation of gene expression, and suggest new strategies for drug discovery (Box 3 ). It remains to be seen whether slippage during translocation, as observed for the IBV frameshift, is a universal mechanism or whether there are cases where alternative pathways are utilized. The role of the ribosomal helicase activity in resuming translation after −1PRF is one of the unresolved issues. As we start to rationalize the effect of the cis-acting regulatory elements of the mRNA – the slippery sequence and secondary structure elements – it will be interesting to see how slippery sequences can contribute to the diversity of the cellular proteome at changing environmental conditions, alone or in combination with trans-acting factors.
Box 3. Outstanding questions.
-
•
What is the true frequency of frameshifting in cells under changing environmental conditions, different developmental and differentiation stages, in various tissues, and in health and disease?
-
•
How universal is the timing of −1PRF during translocation over the slippery codons?
-
•
What is the role of the ribosomal helicase activity, and what is the effect of conformational fluctuations of the RNA structure downstream of the slippage site?
-
•
What is the potential of slippery sequences in the genome to generate alternative proteins?
-
•
What is the link between the cellular quality-control networks and frameshifting, particularly in eukaryotic cells?
-
•
How can one use modulation of frameshifting to combat diseases?
The new insights into the mechanism also provide the basis for understanding of how proteins, RNAs, and antibiotics modulate −1PRF. The emerging hallmark of −1PRF is stalling of the ribosome over the slippery sequence in a metastable state that favors slippage. Additional stabilization or destabilization of the roadblock for translation can modulate the partitioning between 0- and −1-frames; further examples of regulatory factors will provide a more detailed picture of how exactly this can work. It also becomes increasingly clear that, in addition to ensuring the double (or even triple) coding capacity of some genes, −1PRF serves as a regulatory checkpoint which links translation to the cellular quality-control machinery. Future work will show how these control modules are coupled, in particular in tissue- or disease-specific gene expression. Finally, −1PRF, with its idiosyncratic regulatory mRNA elements, may be used as a target for the development of new therapeutic strategies to treat diseases caused by altered gene expression, combat viral infection, and modulate host immune responses.
Acknowledgments
We thank Wolfgang Wintermeyer, Max Planck Institute for Biophysical Chemistry, for critical reading the manuscript. The work in our laboratory is supported by the Max Planck Society and grants from the Deutsche Forschungsgemeinschaft.
Glossary
- Avian infectious bronchitis virus (IBV)
a coronavirus. The IBV 1a/1b sequence encodes two proteins: a shorter protein 1a and a longer 1a/1b polypeptide which is synthesized due to −1PRF. IBV is one of the best-studied examples of −1PRF. The efficiency of −1PRF is very high: 30–70%. The IBV frameshifting sequence is also operational in mammalian, plant, yeast and bacterial cells, suggesting that the principles of −1PRF are universal.
- Cis-acting elements
the elements in the mRNA that modulate −1PRF.
- Colinearity
denotes the linear correspondence between the mature mRNA and its protein product. Colinearity is ensured by the ribosomes, because they usually translate codons one after another in a very accurate manner.
- dnaX
an example of dual coding in a chromosomal gene in bacteria. The dnaX gene encodes two products, the τ and γ subunits of DNA polymerase III. τ is the longer protein; the shorter γ polypeptide is synthesized as a result of −1PRF.
- Elongation factor G (EF-G)
the translocase in bacteria that promotes the movement of the ribosome over the mRNA at the cost of GTP hydrolysis. The homologous eukaryotic translocase is eEF2.
- Fluorescence resonance energy transfer (FRET)
energy transfer can occur between two fluorescent reporter groups where the donor in the excited state transfers energy to an acceptor, provided that the two are close to one another (<10 nm). FRET efficiency is inversely proportional to the sixth power of the distance between donor and acceptor, making FRET suitable for measuring small changes in distance. FRET can be utilized to measure the thermodynamics, kinetics, and dynamics of interactions between interacting molecules or within macromolecules. smFRET, single-molecule FRET, is a technique to monitor FRET changes of individual molecules or molecular assemblies. smFRET has been extensively used to study tRNA–mRNA translocation on the ribosome and, recently, −1PRF.
- Frameshift (FS) efficiency
a measure for the preference for translation in the −1-frame compared to overall translation, calculated as FS% = −1-frame product/(−1-frame + 0-frame products) × 100%.
- Optical tweezers
a single-molecule technique for measuring the force required to unwind an mRNA secondary structure (or any other process that generates force). An experiment is performed by focusing a laser beam on a bead attached to the macromolecule under study such that small movements from the center of the laser beam generate a counteracting force. In the experimental setup to measure unwinding forces with optical tweezers, the ends of the RNA molecules are flanked by DNA handles that are attached to beads. While one bead is trapped by the laser beam, the second is pulled by a micropipette or another laser beam to generate the tension force. The force is proportional to the displacement of the object from the center of the optical trap.
- Pseudoknot
a complex RNA structure that is formed by base-pairing between the loop of a stem-loop structure in an RNA molecule and a single-stranded region located upstream or downstream of the stem-loop structure. Pseudoknots have been implicated in gene expression and viral genome replication.
- Quench flow
a rapid-kinetics technique which allows monitoring the kinetics of reactions in the ms to s time-range. After rapid mixing of two ligands, the reaction is allowed to proceed for a desired time and then stopped by the addition of a quencher. In the present context the technique was used for performing a codon walk on the mRNA.
- Rapid kinetics
an approach to monitor reactions in the μs to s time-range and to determine the rates of rapid reactions. The term is usually applied to measurements on ensembles of molecules.
- Ribosomes
large cellular organelles (about 2.5 MDa in prokaryotes) that synthesize proteins. They are ribonucleoprotein complexes composed of two subunits, the large (50S) and the small (30S).
- 30S subunit
the small subunit of bacterial ribosomes that is responsible for mRNA recruitment and decoding of mRNA codons. The number indicates the sedimentation coefficient of the particle (in Svedberg units).
- 50S subunit
large subunit of bacterial ribosomes; responsible for peptide bond formation and provides the exit tunnel for the emerging peptide.
- Stopped flow
a rapid kinetics technique in which, following rapid mixing of the reactants, the reaction progress is monitored by changes of an optical parameter, such as fluorescence, FRET, light scattering, or absorption. The technique has been useful in dissecting different steps of the translation cycle and gaining kinetic information about individual steps. Stopped flow has been used to solve the mechanism of −1PRF by measuring changes in fluorescence of tRNA, FRET between tRNA and the 30S ribosome head, as well as EF-G binding and dissociation.
- Trans-acting elements
modulators of −1PRF that do not reside in the mRNA itself.
References
- 1.Drummond D.A., Wilke C.O. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranov P.V. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Baranov P.V. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 4.Gesteland R.F., Atkins J.F. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 5.Fayet O., Prere M.F. Programmed ribosomal-1 frameshifting as a tradition: the bacterial transposable elements of the IS3 family. In: Atkins J.F., Gesteland R.F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; 2010. pp. 259–280. [Google Scholar]
- 6.Atkins J.F., Gesteland R.F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; 2010. [Google Scholar]
- 7.Chandler M., Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 8.Brierley I., Dos Ramos F.J. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brakier-Gingras L. Targeting frameshifting in the human immunodeficiency virus. Expert Opin. Ther. Targets. 2012;16:249–258. doi: 10.1517/14728222.2012.665879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulude D. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology. 2006;345:127–136. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 11.Hung M. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 1998;72:4819–4824. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belew A.T. PRFdb: a database of computationally predicted eukaryotic programmed −1 ribosomal frameshift signals. BMC Genomics. 2008;9:339. doi: 10.1186/1471-2164-9-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Advani V.M. Yeast telomere maintenance is globally controlled by programmed ribosomal frameshifting and the nonsense-mediated mRNA decay pathway. Translation. 2013;1:e24418. doi: 10.4161/trla.24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belew A.T. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature. 2014;512:265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belew A.T., Dinman J.D. Cell cycle control (and more) by programmed −1 ribosomal frameshifting: implications for disease and therapeutics. Cell Cycle. 2015;14:172–178. doi: 10.4161/15384101.2014.989123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manktelow E. Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–1563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigemoto K. Identification and characterisation of a developmentally regulated mammalian gene that utilises −1 programmed ribosomal frameshifting. Nucleic Acids Res. 2001;29:4079–4088. doi: 10.1093/nar/29.19.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel A.M. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012;22:2219–2229. doi: 10.1101/gr.133249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brierley I. Pseudoknot-dependent programmed −1 ribosomal frameshifting: structures, mechanisms and models. In: Atkins J.F., Gesteland R.F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; 2010. pp. 149–174. [Google Scholar]
- 20.Howard M.T. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA. 2004;10:1653–1661. doi: 10.1261/rna.7810204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu C.H. Stimulation of ribosomal frameshifting by RNA G-quadruplex structures. Nucleic Acids Res. 2014;42:1887–1892. doi: 10.1093/nar/gkt1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen B. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller W.A., Giedroc D.P. Ribosomal frameshifting in decoding plant viral RNAs. In: Atkins J.F., Gesteland R.F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; 2010. pp. 193–220. [Google Scholar]
- 24.Somogyi P. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopinski J.D. Kinetics of ribosomal pausing during programmed −1 translational frameshifting. Mol. Cell. Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu C. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caliskan N. Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014;157:1619–1631. doi: 10.1016/j.cell.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan S. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao P.Y. The many paths to frameshifting: kinetic modelling and analysis of the effects of different elongation steps on programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2011;39:300–312. doi: 10.1093/nar/gkq761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yelverton E. The function of a ribosomal frameshifting signal from human immunodeficiency virus-1 in Escherichia coli. Mol. Microbiol. 1994;11:303–313. doi: 10.1111/j.1365-2958.1994.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Y. Efficient −2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dayhuff T.J. Characterization of ribosomal frameshift events by protein sequence analysis. J. Biol. Chem. 1986;261:7491–7500. [PubMed] [Google Scholar]
- 33.Jiang H. Orsay virus utilizes ribosomal frameshifting to express a novel protein that is incorporated into virions. Virology. 2014;450–451:213–221. doi: 10.1016/j.virol.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 2013;20:582–588. doi: 10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J. Dynamic pathways of −1 translational frameshifting. Nature. 2014;512:328–332. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H.K. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5538–5543. doi: 10.1073/pnas.1403457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brierley I. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J. Mol. Biol. 1997;270:360–373. doi: 10.1006/jmbi.1997.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takyar S. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 39.Qin P. Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep. 2014;15:185–190. doi: 10.1002/embr.201337762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fredrick K., Noller H.F. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol. Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 41.Qu X. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen J.D. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yusupova G. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 44.Mouzakis K.D. HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome. Nucleic Acids Res. 2013;41:1901–1913. doi: 10.1093/nar/gks1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houck-Loomis B. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature. 2011;480:561–564. doi: 10.1038/nature10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie D.B. Programmed −1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16167–16172. doi: 10.1073/pnas.1204114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theis C. KnotInFrame: prediction of −1 ribosomal frameshift events. Nucleic Acids Res. 2008;36:6013–6020. doi: 10.1093/nar/gkn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harger J.W. An ‘integrated model’ of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- 49.Gupta P. Regulation of gene expression by macrolide-induced ribosomal frameshifting. Mol. Cell. 2013;52:629–642. doi: 10.1016/j.molcel.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brierley I. Macrolide-induced ribosomal frameshifting: a new route to antibiotic resistance. Mol. Cell. 2013;52:613–615. doi: 10.1016/j.molcel.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 51.Beura L.K. Cellular poly(C) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1β and support viral replication. J. Virol. 2011;85:12939–12949. doi: 10.1128/JVI.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwak H. Annexin A2 binds RNA and reduces the frameshifting efficiency of infectious bronchitis virus. PLoS ONE. 2011;6:e24067. doi: 10.1371/journal.pone.0024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charbonneau J. The 5′ UTR of HIV-1 full-length mRNA and the Tat viral protein modulate the programmed −1 ribosomal frameshift that generates HIV-1 enzymes. RNA. 2012;18:519–529. doi: 10.1261/rna.030346.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gendron K. The presence of the TAR RNA structure alters the programmed −1 ribosomal frameshift efficiency of the human immunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res. 2008;36:30–40. doi: 10.1093/nar/gkm906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi Y. Identification of a cellular factor that modulates HIV-1 programmed ribosomal frameshifting. J. Biol. Chem. 2010;285:19776–19784. doi: 10.1074/jbc.M109.085621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park H.J. Increased −1 ribosomal frameshifting efficiency by yeast prion-like phenotype [PSI+] FEBS Lett. 2009;583:665–669. doi: 10.1016/j.febslet.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 57.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manickam N. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014;20:9–15. doi: 10.1261/rna.039792.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kramer E.B., Farabaugh P.J. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kurland C.G. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 61.Tsuchihashi Z., Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W.M. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988;239:1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- 63.Loughran G. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014;42:8928–8938. doi: 10.1093/nar/gku608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lang B.F. Massive programmed translational jumping in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5926–5931. doi: 10.1073/pnas.1322190111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samatova E. High-efficiency translational bypassing of non-coding nucleotides specified by mRNA structure and nascent peptide. Nat. Commun. 2014;5:4459. doi: 10.1038/ncomms5459. [DOI] [PubMed] [Google Scholar]
- 66.Mikuni O. Sequence and functional analysis of mutations in the gene encoding peptide-chain-release factor 2 of Escherichia coli. Biochimie. 1991;73:1509–1516. doi: 10.1016/0300-9084(91)90185-4. [DOI] [PubMed] [Google Scholar]
- 67.Matsufuji S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holtkamp W. Synchronous tRNA movements during translocation on the ribosome are orchestrated by elongation factor G and GTP hydrolysis. Bioessays. 2014;36:908–918. doi: 10.1002/bies.201400076. [DOI] [PubMed] [Google Scholar]
- 69.Ermolenko D.N. Observation of intersubunit movement of the ribosome in solution using FRET. J. Mol. Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 70.Mejlhede N. Ribosomal -1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J. Bacteriol. 1999;181:2930–2937. doi: 10.1128/jb.181.9.2930-2937.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brierley I. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bredenbeek P.J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Herold J., Siddell S.G. An ‘elaborated’ pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 1993;21:5838–5842. doi: 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su M.C. An atypical RNA pseudoknot stimulator and an upstream attenuation signal for -1 ribosomal frameshifting of SARS coronavirus. Nucleic Acids Res. 2005;33:4265–4275. doi: 10.1093/nar/gki731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pennell S. The stimulatory RNA of the Visna-Maedi retrovirus ribosomal frameshifting signal is an unusual pseudoknot with an interstem element. RNA. 2008;14:1366–1377. doi: 10.1261/rna.1042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morikawa S., Bishop D.H. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marcheschi R.J. Programmed ribosomal frameshifting in SIV is induced by a highly structured RNA stem-loop. J. Mol. Biol. 2007;373:652–663. doi: 10.1016/j.jmb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacks T. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 79.Cornish P.V. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated -1 ribosomal frameshifting. Proc. Natl. Acad. Sci U.S.A. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nixon P.L. Thermodynamic analysis of conserved loop-stem interactions in P1-P2 frameshifting RNA pseudoknots from plant Luteoviridae. Biochemistry. 2002;41:10665–10674. doi: 10.1021/bi025843c. [DOI] [PubMed] [Google Scholar]
- 81.Dinman J.D., Wickner R.B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marczinke B. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J. Virol. 1994;68:5588–5595. doi: 10.1128/jvi.68.9.5588-5595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.den Boon J.A. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Snijder E.J. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990;18:4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wills N.M. A functional -1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J. Biol. Chem. 2006;281:7082–7088. doi: 10.1074/jbc.M511629200. [DOI] [PubMed] [Google Scholar]


