Abstract
The ribosomal frameshifting signal present in the genomic RNA of the coronavirus infectious bronchitis virus (IBV) contains a classic hairpin-type RNA pseudoknot that is believed to possess coaxially stacked stems of 11 bp (stem 1) and 6 bp (stem 2). We investigated the influence of stem 1 length on the frameshift process by measuring the frameshift efficiency in vitro of a series of IBV-based pseudoknots whose stem 1 length was varied from 4 to 13 bp in single base-pair increments. Efficient frameshifting depended upon the presence of a minimum of 11 bp; pseudoknots with a shorter stem 1 were either non-functional or had reduced frameshift efficiency, despite the fact that a number of them had a stem 1 with a predicted stability equal to or greater than that of the wild-type IBV pseudoknot. An upper limit for stem 1 length was not determined, but pseudoknots containing a 12 or 13 bp stem 1 were fully functional. Structure probing analysis was carried out on RNAs containing either a ten or 11 bp stem 1; these experiments confirmed that both RNAs formed pseudoknots and appeared to be indistinguishable in conformation. Thus the difference in frameshifting efficiency seen with the two structures was not simply due to an inability of the 10 bp stem 1 construct to fold into a pseudoknot. In an attempt to identify other parameters which could account for the poor functionality of the shorter stem 1-containing pseudoknots, we investigated, in the context of the 10 bp stem 1 construct, the influence on frameshifting of altering the slippery sequence-pseudoknot spacing distance, loop 2 length, and the number of G residues at the bottom of the 5′-arm of stem 1. For each parameter, it was possible to find a condition where a modest stimulation of frameshifting was observable (about twofold, from seven to a maximal 17 %), but we were unable to find a situation where frameshifting approached the levels seen with 11 bp stem 1 constructs (48-57 %). Furthermore, in the next smaller construct (9 bp stem 1), changing the bottom four base-pairs to G·C (the optimal base composition) only stimulated frameshifting from 3 to 6 %, an efficiency about tenfold lower than seen with the 11 bp construct. Thus stem 1 length is a major factor in determining the functionality of this class of pseudoknot and this has implications for models of the frameshift process.
Keywords: RNA pseudoknot, ribosomal frameshifting, RNA structure probing, coronavirus, retrovirus
Abbreviations: ORF, open reading frame
Introduction
The process of −1 ribosomal frameshifting on viral RNAs requires the presence of two cis-acting mRNA signals; a slippery sequence, where the ribosome changes reading frame, and a stimulatory RNA structure located immediately downstream (reviewed by Brierley 1995, Farabaugh 1996). In most cases, the stimulatory element takes the form of an RNA pseudoknot Brierley et al 1989, ten Dam et al 1990, which is thought to function by modulation of the ribosomal elongation cycle (Dinman et al., 1997), perhaps during a ribosomal pause Tu et al 1992, Somogyi et al 1993. The role of pseudoknots in the frameshift process was first investigated using a signal derived from the genome of the avian coronavirus infectious bronchitis virus (IBV; Brierley et al 1989, Brierley et al 1991). The IBV signal (see Figure 1), which is present at the overlap of the 1a and 1b open reading frames (ORFs) of the genomic RNA, is highly efficient and instructs about 30 % of ribosomes to change frame at the overlap region (Brierley et al., 1987). Computer RNA folding programs (ten Dam et al., 1990) suggest that the IBV pseudoknot is of the hairpin type (ten Dam et al., 1992), possessing coaxially stacked stems of 11 bp (stem 1) and 6 bp (stem 2) connected by single-stranded loops of 1 nt (loop 1) and 32 nt (loop 2). In earlier experiments, variants of the IBV pseudoknot containing complementary and compensatory changes within the stems and various loop mutations were tested for their ability to promote frameshifting in a rabbit reticulocyte lysate in vitro translation system (RRL; Brierley et al., 1991). The effects on frameshifting of these mutations were consistent with the predicted organisation of the IBV pseudoknot. Only those loop mutations which reduced loop length to below the functional minimum (as predicted from measurements of inter-phosphate distances using model RNA A-type helices; Pleij et al., 1985) had a detrimental effect on frameshifting and in general, the destabilisation of a single base-pair in either stem was sufficient to reduce or abolish frameshifting. Additionally, no primary sequence determinants in the stems or loops appeared to be required; as long as the predicted overall structure was maintained, frameshifting occurred efficiently.
Figure 1.
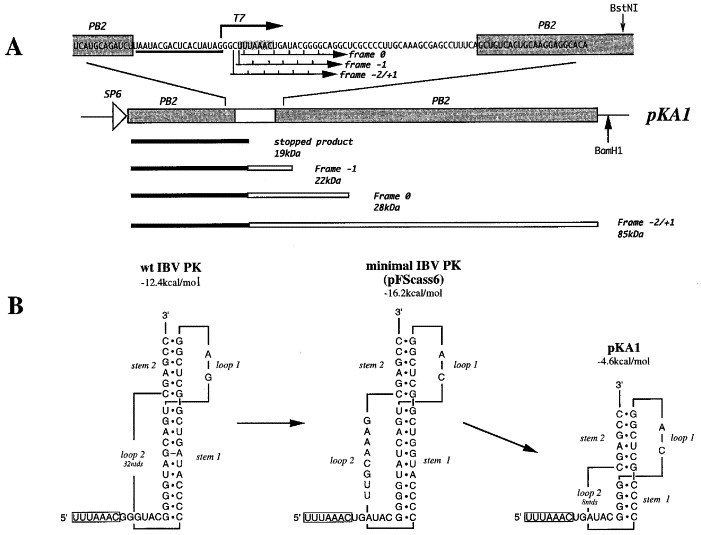
The frameshift reporter construct pKA1. (a) Plasmid pKA1 was derived from pFScass 6 (Brierley et al., 1992) by site-directed mutagenesis (see Materials and Methods). pKA1 contains a truncated version of the minimal IBV pseudoknot (white box) cloned into a reporter gene, the influenza PB2 gene (shaded boxes). Linearisation of the plasmid with BamH1 and in vitro transcription using SP6 RNA polymerase yields an mRNA (2.4kb) that, when translated in RRL, is predicted to produce a 19 kDa non-frameshift product corresponding to ribosomes that terminate at the UGA termination codon (located immediately downstream of the slippery sequence UUUAAAC, shaded), and a 22 kDa −1 frameshift product. The 0-frame and −2/+1 frames are also open (to some extent) in this construct. Ribosomes which enter these frames produce 28 kDa and 85 kDa products respectively. A bacteriophage T7 promoter is present just upstream of the frameshift region; this promoter is employed to generate short, pseudoknot-containing transcripts from BstNI-digested templates for secondary structure analysis. (b) The wild-type (wt) IBV, the minimal IBV and the pKA1 pseudoknots (PK). The minimal IBV frameshift signal present in pFScass 6 is based on the wild-type IBV frameshift signal and is fully functional in frameshifting (Brierley et al., 1992). It differs from the wild-type in a number of ways; a termination codon (UGA) is present immediately downstream of the slippery sequence (UUUAAAC, boxed) to terminate zero frame ribosomes, loop 2 of the pseudoknot contains 8 rather than 32 nt, the G·A mismatched pair in stem 1 of the wild-type pseudoknot is replaced by a U·G pair, the G nucleotide of loop 1 is replaced by a C nucleotide and, finally, the minimal pseudoknot has no stop codons. Plasmid pKA1 was derived from pFScass 6 by deletion mutagenesis (see Materials and Methods). The type and number of bases in the loops and stem 2 remained unaltered. The predicted stability of stem 1 of each construct is shown (calculated according to the rules by Turner et al., 1988, using a loop length of 8 nt; see the text).
The structure of the IBV pseudoknot is representative of those pseudoknots present at the frameshift sites of the corona-, toro- and arteriviruses that possess a long stem 1 of 11-12 bp and usually, a long loop 2 (30-164 nt). At viral frameshift sites that contain pseudoknots, a second main class is distinguishable, members of which are predicted to contain stems of 5 to 7 bp and shorter loops ten Dam et al 1990, Brierley 1995. Much of the recent research on pseudoknot structure and function has concentrated on this class of pseudoknot, and has been performed on wild-type and mutant variants of the pseudoknots present at the frameshift sites of simian retrovirus 1 gag-pro (SRV-1; ten Dam et al 1994, ten Dam et al 1995, Du et al 1997, Sung and Kang 1998) and the retrovirus mouse mammary tumour virus (MMTV) gag-pro Chen et al 1995, Chen et al 1996, Shen and Tinoco 1995, Kang et al 1996, Kang and Tinoco 1997. Of particular relevance are the NMR studies of MMTV-derived functional and non-functional pseudoknots Shen and Tinoco 1995, Kang et al 1996, Chen et al 1996, Kang and Tinoco 1997, which have provided a possible insight into pseudoknot function. The solution structure of a 34 nt RNA pseudoknot VPK, a modified but functional version of the MMTV pseudoknot, has revealed that the two pseudoknot stems are not coaxially stacked, since an unpaired, intercalated A residue is present at the junction between the two stems which introduces a pronounced bend. This bent conformation appears to be essential for efficient frameshifting and may reflect a requirement for an interaction between a component of the translation apparatus and a specific pseudoknot conformation Chen et al 1996, Kang and Tinoco 1997.
Despite these advances, our understanding of the molecular basis of the frameshift process is rudimentary. Nevertheless, the occurrence of two apparently distinct classes of RNA pseudoknot provides the opportunity to gain mechanistic insights from a functional comparison of the two. Unfortunately, our knowledge of the conformation of pseudoknots of the IBV class is much more limited. To date, neither NMR analyses nor secondary structure mapping with chemical and enzymatic probes have been carried out. However, a notable feature of these pseudoknots is that they are predicted to possess a stem 1 with a length equal to or greater than the predicted number of base-pairs in one turn of an A-form RNA helix (11 bp; Arnott et al., 1973), raising the possibility that frameshifting at such pseudoknots may require the maintenance of a defined stem 1 length. Here, we address this issue by measuring the frameshift efficiency in RRL of a series of IBV-related pseudoknots whose stem 1 length was varied from 4 to 13 bp in single base-pair increments. We found that a stem 1 length of 11 bp or more was required for efficient frameshifting, and that shorter pseudoknots had greatly reduced or did not stimulate frameshifting. The evidence from additional mutagenesis experiments is consistent with the view that the defect in many of the shorter pseudoknots is simply one of stem 1 length rather than, for example, inappropriate stem 1 stability, loop length or sequence composition. Furthermore, direct RNA structure probing demonstrated that a 10 bp stem 1- containing construct with greatly reduced frameshifting was conformationally indistinct from a fully functional 11 bp stem-1 containing construct and that both RNAs formed pseudoknots. The functional requirement for a minimal stem 1 length in pseudoknots of the IBV class has implications for models of the frameshift process.
Results
IBV-pseudoknot induced −1 frameshifting requires a minimum stem 1 length
To investigate the influence of IBV pseudoknot stem 1 length on frameshift efficiency, we made a series of constructs (the KA series of plasmids) with different stem 1 lengths containing only G·C (or C·G) base-pairs. We began with a frameshift reporter construct, pFScass 6 (Brierley et al., 1992), which contains a fully functional, trimmed version of the IBV frameshift signal (the “minimal” IBV pseudoknot; see the legend to Figure 1) cloned into the influenza A/PR8/34 PB2 gene (Young et al., 1983) at a unique BglII site (Figure 1). The position of the pseudoknot in this construct is just upstream of a region of the PB2 gene where significant lengths of ORF are present in all three frames. A frameshift into any one of these frames generates a product with a characteristic size (22, 28 or 85 kDa). This property was crucial for the analysis described here, since we wished to introduce single or pairs of nucleotides into the pseudoknot, which would change the exit phase of −1 frameshifted ribosomes. The founder member of the KA series of plasmids, pKA1 (Figure 1), was derived from pFScass 6 by deletion mutagenesis (see Materials and Methods) such that the pseudoknot encoded by this plasmid would have only four G·C base-pairs in stem 1. The type and number of bases in the loops and stem 2 remained unaltered. The length of stem 1 of pKA1 was then increased sequentially in single G·C (or C·G) base-pair steps from 4 to 12 bp and frameshifting measured by in vitro translation in the RRL of SP6-derived transcripts from BamHI-digested plasmids. The base composition of the stem 1 length variants, the predicted stability of each stem 1 region, and the predicted size of the frameshift products are shown in Figure 2. In vitro translations of the various pKA-derived mRNAs are also shown, along with the calculated frameshift efficiencies. As the energetics of pseudoknot formation is not fully understood, the stem 1 stability predictions, calculated according to the rules of Turner et al. (1988), were performed on the basis that stem 1 formed independently and had a loop of 8 nt. For pKA-derived plasmids, ribosomes which terminate without frameshifting synthesise a 19 kDa non-frameshifted species. Ribosomes which undergo −1 frameshifting at the slippery sequence continue translation to produce a −1 frameshift product whose size is 22, 28 or 85 kDa depending upon the exit reading frame, which is determined by the number of nucleotides introduced into stem 1. From Figure 2(b) it is clear that only certain pseudoknots are functional in frameshifting. Those with a stem 1 length of 4 to 8 bp were unable to stimulate frameshifting above the background noise (<2 %) seen in RRL with pFScass-based constructs (the background polypeptides arise as a result of low-level abberant initiation events; see Brierley et al., 1992). Frameshifting at pseudoknots with stem 1 lengths of 9 and 10 bp was detectable, but occured at a low level (3 % and 7 % respectively). However, when stem 1 length was increased to 11 bp, frameshifting occured very efficiently (pKA18; 48 %). It was apparent that the lack of function of the shorter constructs was unrelated to the predicted overall stability of stem 1, since several of them (pKA7, 9, 11, 13) had stabilities equal to or greater than those predicted for stem 1 of either the wild-type (−12.4 kcal/mol) or the minimal (−16.2 kcal/mol) IBV pseudoknots. To test whether longer stem 1 constructs were also capable of promoting high efficiency frameshifting, we prepared a 12 bp stem 1 construct (pKA26) using an oligonucleotide cloning strategy (see Materials and Methods), since we were unable to add an extra stem 1 base-pair to pKA18 by mutagenesis, probably due to extensive regions of self-complementarity in the G+C-rich mutagenic oligonucleotides. In this construct, frameshifting was at a high level (40 %) indicating that a 12 bp stem 1 is capable of stimulating high efficiency frameshifting. To corroborate these observations, we prepared another series of pseudoknots (the pSL series; see Figure 3) with a stem 1 length of 10, 12 or 13 bp. Like the pKA series, these constructs were based on the minimal IBV pseudoknot (pFScass 6, Figure 1), but base-pairs were removed or inserted near the top of stem 1 and the A·U content of stem 1 of each construct was much greater than the equivalent constructs of the pKA series. The results obtained from in vitro translation of these mRNAs closely paralleled those obtained with the G+C-rich stem 1 constructs of the pKA series. Only low level frameshifting was seen with the 10 bp stem 1 construct pSL3 (7 %), but the 12 and 13 bp variants (pSL4, pSL7) stimulated frameshifting efficiently (55 and 47 %, respectively). In the pSL7 translations, a small quantity of 85 kDa product was unexpectedly observed (in this construct, the −1 frameshift generates a 28 kDa protein) which corresponds to translation by ribosomes in the zero reading frame. This product may arise following suppression (at about 1 % efficiency) of the UGA termination codon located immediately downstream of the slippery sequence; pseudoknot-induced termination codon suppression has been described in the production of the Gag-Pol polyprotein of the type C retroviruses Wills et al 1991, Feng et al 1992. We also measured frameshifting at the intermediate constructs (pSL1, 2, 5 and 6) with an unpaired base near the top of the stem. For those constructs with a stem 1 containing 12 bp residues (pSL5 and 6), high efficiency frameshifting was observed (48 %). The two constructs with fewer base-paired residues (pSL1, 10 bp; pSL2, 11 bp) showed inefficient frameshifting (10 % and 7 %, respectively). From these studies, it seems that efficient frameshifting, at least of the IBV-type, requires a pseudoknot with a minimal stem 1 length of 11 bp. However it was necessary to rule out several non-trivial alternative explanations for the lack of functionality of the shorter pseudoknots. These included the possible influence of slippery sequence-pseudoknot spacing distance, the length of loop 2 and the base composition at the start of stem 1.
Figure 2.
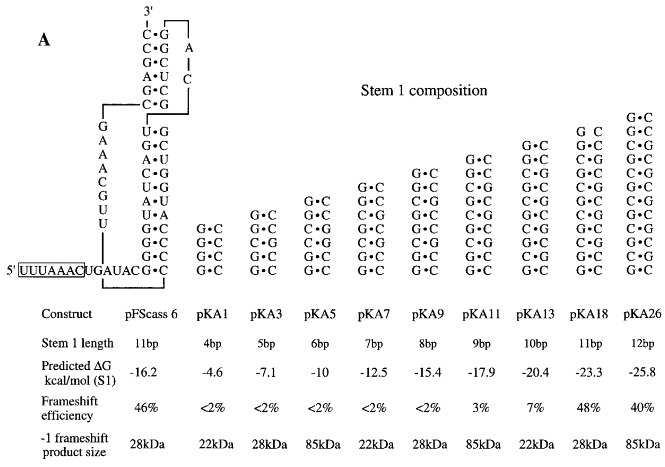
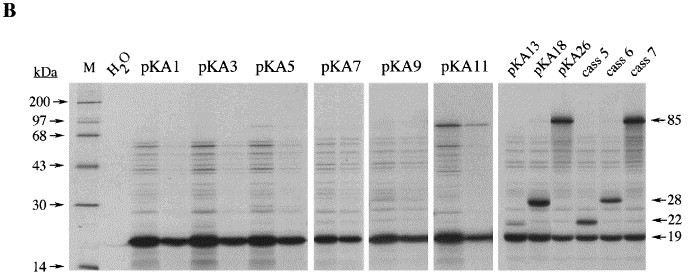
Influence of stem 1 length on −1 ribosomal frameshifting (part I). (a) The stem 1 base composition of several members of the pKA series. Starting with pKA1, stem 1 length was increased sequentially by single G·C (or C·G) pairs up to 12 bp (pKA26). The predicted stability of stem 1 of each construct is indicated. Also shown is the size of the −1 frameshift product expected for each frameshift construct and the efficiency of frameshifting seen in RRL. All constructs possess the same loop 1, loop 2 and stem 2 sequences as in pFScass 6. (b) RRL translation products synthesised in response to mRNAs derived from BamHI-digested pKA1 and mutant derivatives. Products were labelled with [35S]methionine, separated on a SDS-15 % polyacrylamide gel and detected by autoradiography. The frameshifted (22, 28 or 85 kDa) and non-frameshifted (19 kDa) species are marked with arrows. For pKA1, 3, 5, 7, 9 and 11, mRNAs were translated at two concentrations (25 μg/ml and 10 μg/ml). The other RNAs were at 20 μg/ml. RNAs derived from pFScass 5, 6 and 7 were translated to provide size markers for the three potential −1 frameshift exit frames. Plasmids pFScass 5 and 7 differ from pFScass 6 only by the addition of one (pFScass 5) or two (pFScass 6) C nucleotides downstream of the pseudoknot-encoding region. Track M was loaded with [14C]high molecular mass markers (Amersham). A control translation programmed with water was also included. The frameshift efficiencies quoted are the average of three independent measurements which varied by less than 10 %, i.e. a measurement of 40 % frameshift efficiency was between 36 % and 44 %.
Figure 3.
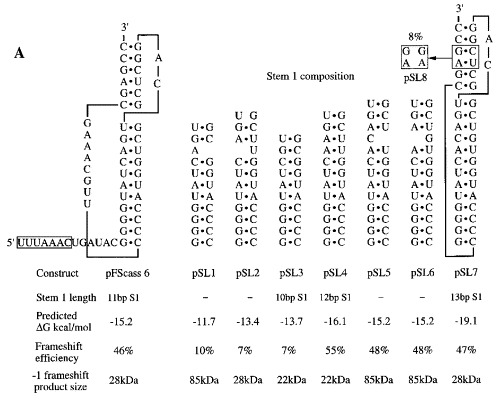
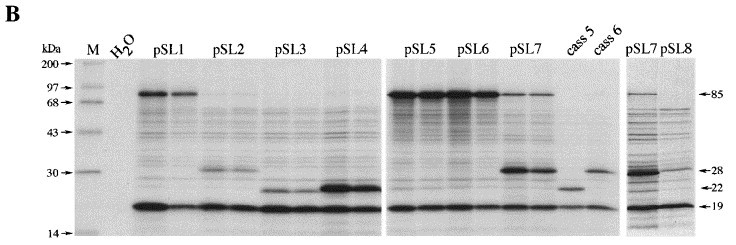
Influence of stem 1 length on −1 ribosomal frameshifting (part II). (a) The stem 1 base composition of members of the pSL series. Mutations were introduced into stem 1 of constructs pFScass 5, 6 or 7 such that stem 1 length was increased or decreased in single base-pair steps. The predicted stability of stem 1 of each construct is indicated. Also shown is the size of the −1 frameshift product specified by each frameshift signal and the efficiency of frameshifting seen in RRL. All constructs possess the same loop 1, loop 2 and stem 2 sequences as pFScass 6 except plasmid pSL8, a variant of pSL7 with a destabilised stem 2. (b) RRL translation products synthesised in response to mRNAs derived from BamHI-digested pSL1 and mutant derivatives. Products were labelled with [35S]methionine, separated on a SDS-15 % polyacrylamide gel and detected by autoradiography. The frameshifted (22, 28 or 85 kDa) and non-frameshifted (19 kDa) species are marked with arrows. For the pSL series, mRNAs were translated at two concentrations (25 μg/ml and 12.5 μg/ml). Track M was loaded with [14C] high molecular mass markers (Amersham) and a control translation programmed with water was also included.
In earlier investigations, the optimal spacing distance between the IBV slippery sequence and pseudoknot was found to be 6 nt; addition or deletion of one, two or three nucleotides to the spacer reduced frameshifting by two-, four- and tenfold, respectively Brierley et al 1989, Brierley et al 1992. Hence it was possible that the non-functional pseudoknots of the KA series were simply inappropriately spaced. Given the large number of constructs, it was impractical to alter the spacing distance of all pseudoknots, so we concentrated on spacing variants of pKA13, which, with a 10 bp stem 1, was the closest relative of the efficient structures of pKA18 and 26. We inserted or deleted one, two or three nucleotides from the spacer region of pKA13 and tested for frameshifting. As can be seen in Figure 4, the frameshift efficiency varied depending upon the spacer distance tested (3-9 nt), with an optimum at 8 nt. At this optimal distance, the frameshifting efficiency was 12 %, about twofold higher than that seen with a 6 nt spacer, but still considerably lower than that seen with the 11 bp stem 1 construct, indicating that the pseudoknot is defective in some other way.
Figure 4.
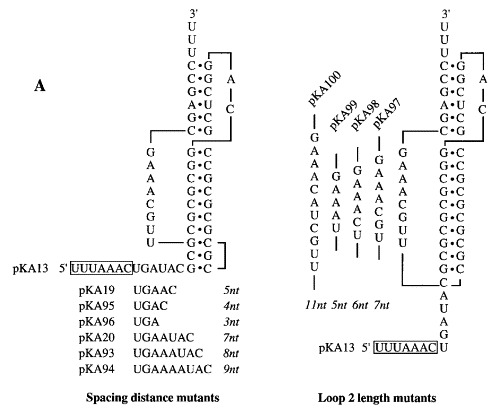
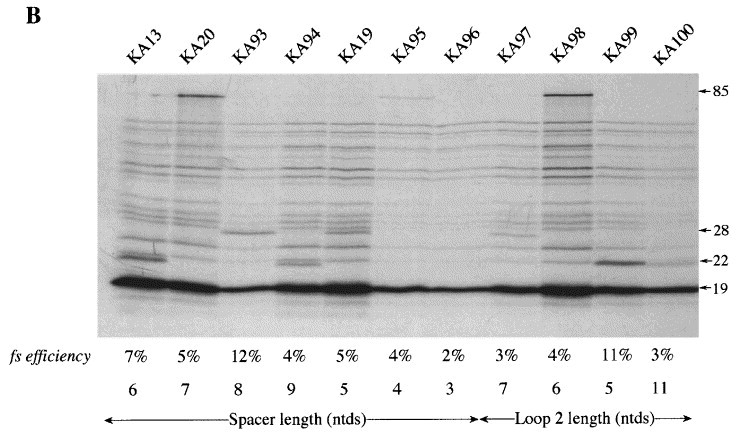
Modulation of the length of loop 2 or spacer region of pKA13. (a) The length of the spacer region or loop 2 in each construct is shown in italics. (b) RRL translation products synthesised in response to mRNAs derived from BamHI-digested pKA13 and mutant derivatives. Products were labelled with [35S]methionine, separated on a SDS-15 % polyacrylamide gel and detected by autoradiography. The frameshifted (22, 28 or 85 kDa) and non-frameshifted (19 kDa) species are marked with arrows.
The folding of an RNA pseudoknot requires that loop 2 span the helix of stem 1. As we had altered the length of stem 1 in the pKA series, it was possible that the consequent change in ratio of loop length to stem 1 helix length may influence frameshifting. To investigate this we increased (by 3 nt) or decreased (by 1, 2 or 3 nt) the length of loop 2 in the context of the 10 bp stem 1-containing construct pKA13 and tested for frameshifting (Figure 4). We found that most loop 2 changes reduced frameshifting to a value of between 2 and 4 %. A modest increase (from 7 % to 11 %) was seen with a loop 2 length of 5 nt. Thus the reduced functionality of the 10 bp stem 1 pseudoknot is not a consequence of an inappropriate loop 2 length.
The final possibility we considered was that the base-pairs present at the bottom of stem 1 in pKA13 were inappropriate. A large number of the pseudoknots involved in −1 ribosomal frameshifting have three or four G·C base-pairs at the start of stem 1, and there is a preference for G bases in the 5′-arm of the stem (ten Dam et al., 1990). Indeed, a variant of the SRV-1 gag-pro pseudoknot with a G·C to C·G change at the third base-pair of stem 1 (the 5′-arm of the stem having the sequence 5′ GGCGCC 3′ rather than 5′ GGGGCC 3′) showed only 50 % of the wild-type frameshift efficiency in RRL (ten Dam et al., 1995). During construction of the pKA series, base-pairs were added as G·C or C·G pairs in such a way that the predicted stability of stem 1 increased in a reasonably linear fashion as the length increased. A consequence of this was that the composition of the bottom of stem 1 varied. The general importance of base-pairs at the start of stem 1 was tested by replacing the four base-pairs at the start of stem 1 of the pKA18 pseudoknot with other combinations including A·U pairs (Figure 5). We found that the inclusion of A·U pairs reduced frameshifting, particularly when two A·U pairs were present (pKA40; 7 %) or a single A·U pair was included near the bottom of stem 1 (pKA41, 18 %). Replacing G·C by A·U at the fourth pair in the stem also gave a measurable reduction in frameshifting efficiency (pKA49, 35 %). In contrast, a mutant derivative that contained four G·C pairs with the G bases all in the 5′-arm showed the greatest frameshifting efficiency (pKA50; 57 %). To test whether the bases at the start of stem 1 of pKA13 (5′ GCGC 3′) were thus inappropriate for efficient frameshifting, we changed the bottom four pairs to those present in the functional pKA18 (5′ GGCG 3′). In the resultant construct pKA27 (see Figure 5), frameshifting did increase (from 7 % to 17 %), although it was still at a threefold lower level than that seen in pKA18 (48 %). Nevertheless, this partial restoration of function raised the possibility that other G-rich stretches may be even more effective. For this reason we changed the base-pairs at the bottom of stem 1 of the poorly functional 9 bp stem 1-containing construct pKA11 (3 %; 5′ GGCG 3′) to the optimal sequence 5′ GGGG 3′. In the resulting constuct (pKA130, Figure 5), frameshifting was increased (to 6 %) but remained at a very low level in comparison to the 11 bp stem 1-containing constructs. These results are consistent with the view that the defect in many of the shorter pseudoknots is primarily one of stem 1 length rather than inappropriate stem 1 stability, loop length or sequence composition at the base of the stem.
Figure 5.
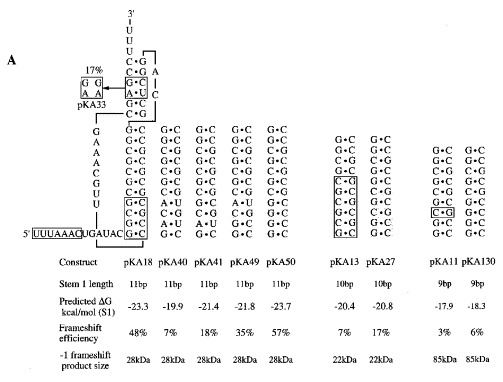
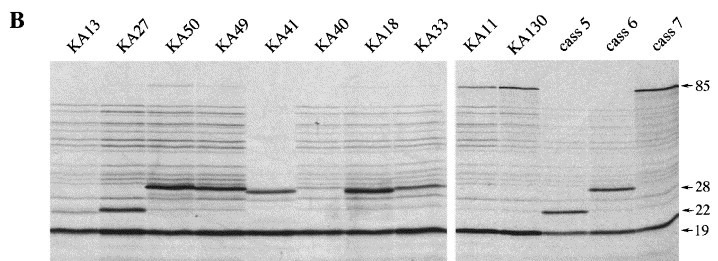
The nucleotide composition at the bottom of stem 1 influences frameshifting. (a) Base-pairs at the bottom of stem 1 of pseudoknots of pKA11, 13 and 18 were changed using site-directed mutagenesis. The parental sequences present in stem 1 are boxed and the variant sequences are shaded. Also shown is pKA33, a variant of pKA18 with a destabilised stem 2. (b) RRL translation products synthesised in response to mRNAs derived from BamHI-digested pKA11, 13, 18 and mutant derivatives. Products were labelled with [35S]methionine, separated on a SDS-15 % polyacrylamide gel and detected by autoradiography. The frameshifted (22, 28 or 85 kDa) and non-frameshifted (19 kDa) species are marked with arrows.
RNA structure mapping of the pseudoknots of constructs pKA13 and pKA18
Throughout the preceding section, we referred to the various RNA structures as non-functional or functional pseudoknots. Clearly it was important to confirm that the relevant RNAs indeed formed pseudoknots. As a first step towards this, we introduced a destabilizing mutation into stem 2 of the functional constructs pKA18 and pSL7 (11 and 13 bp stem 1, respectively) and tested for frameshifting (Figure 5, pKA33, 17 %; Figure 3, pSL8, 8 %). In each case, destabilizing stem 2 reduced frameshifting, supporting the idea that these RNAs are pseudoknotted. Such a strategy was not practical for the non-functional (or barely functional) constructs, so we determined the secondary structure of a poorly functional construct (pKA13, 7 %) by chemical and enzymatic structure probing using an end-labelling procedure van Belkum et al 1988, Wyatt et al 1990, Polson and Bass 1994 in comparison with the functional pKA18 (48 %). Plasmids of the pKA series have an internal T7 promoter just upstream of the frameshift signal, allowing run-off transcripts to be prepared following linearisation with BstN1 (Figure 1). Transcripts of 130 and 132 nt were prepared from pKA13 and pKA18, respectively, end-labelled with [γ-33P]ATP, gel-purified and subjected to limited chemical and enzymatic digestion prior to analysis on denaturing 10 % or 15 % polyacrylamide gels. In these experiments, the Mg2+ level was kept at 2 mM (except for imidazole probing, where 10 mM Mg2+ was used) which is the approximate concentration of this ion in RRL (Jackson & Hunt, 1983), and sites of cleavage were scored only from those reactions where 80-90 % of the full-length RNA remained intact. A representative selection of the individual biochemical analyses and a diagrammatic summary are shown in Figure 6. The bases are numbered from the start of the transcript. The single strand-specific enzymatic probes employed were RNase T1, which cleaves 3′ of unpaired G residues, and RNase U2, which cleaves 3′ of single-stranded A or G, with a preference for A. To probe double-stranded regions, RNase V1 was employed, which cleaves in helical regions. RNase V1 is not base-specific but cleaves RNA that is in helical conformation, whether base-paired (a minimum of 4-6 bp are required) or single-stranded and stacked. We also used the single strand-specific chemical probes imidazole (Vlassov et al., 1995) and lead acetate Krzyzosiak et al 1988, Kolchanov et al 1996.
Figure 6.
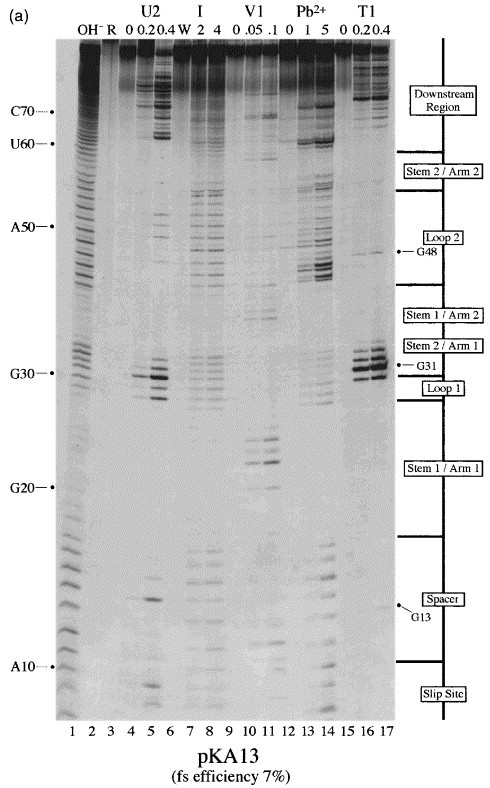
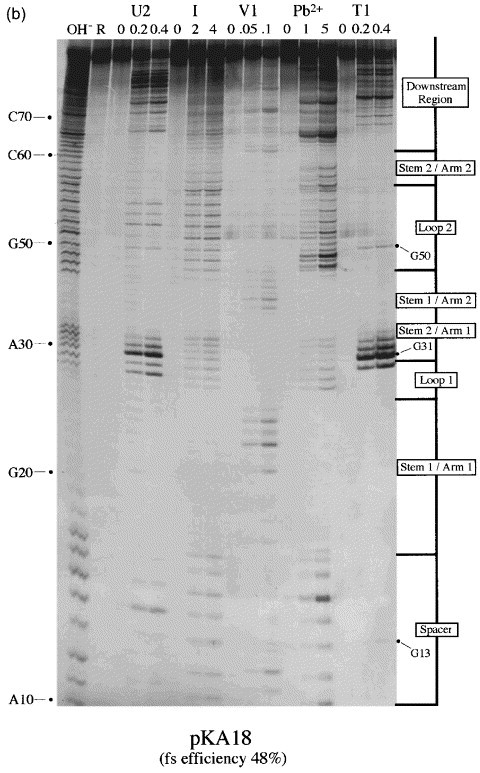
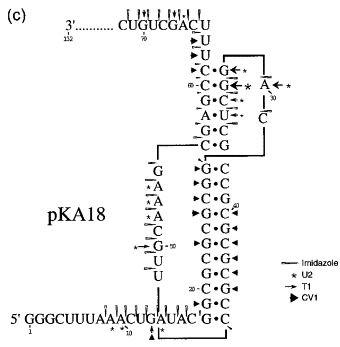
Structure probing of the pKA13 and 18 frameshift signals. RNA derived by T7 transcription of (a) pKA13/BstN1 or (b) pKA18/BstN1 was 5′ end-labelled with [γ-33P]ATP and subjected to limited RNase or chemical cleavage using structure-specific probes. Sites of cleavage were identified by comparison with a ladder of bands created by limited alkaline hydrolysis of the RNA (OH−) and the position of known RNase U2 and T1 cuts, determined empirically. Products were analysed on 15 % acrylamide/7 M urea gels. Data was also collected from 10 % gels (gels not shown). Enzymatic structure probing was with RNases U2, V1 and T1. Uniquely cleaved nucleotides were identified by their absence in untreated control lanes (0). The number of units of enzyme added to each reaction is indicated. Chemical structure probing was with imidazole (I, hours) or lead acetate (Pb2+; mM concentration in reaction). The water lane (W) in the imidazole panel represents RNA which was dissolved in water, incubated for four hours and processed in parallel with the imidazole-treated samples. R represents an aliquot of the purified RNA loaded directly onto the gel without incubation in a reaction buffer. (c) Summary of the KA18 probing results. The sensitivity of bases in the KA18 frameshift region to the various probes is shown. The size of the symbols is approximately proportional to the intensity of cleavage at that site. The lead acetate probing data, which was was similar to that seen with imidazole, is omitted for clarity. The KA13 probing results are not summarised since they were essentially indistinguishable from those of KA18.
It is clear from the structure-probing data that both constructs are pseudoknots and indeed, they appear to be almost identical at the resolution of this methodology. Structure probing with either imidazole or lead (lanes 7-8, 13-14) generated a very similar pattern of bands clearly highlighting the major regions of single and double-stranded RNA. As predicted, the RNA folds into a structure with two stem regions joined by single-stranded loops. Both pseudoknot stems show resistance to chemical cleavage, especially the G+C-rich stem 1. Pseudoknot stem 1 appears to be remarkably stable; there was even some partial resistance to boiling in an alkaline solution as seen in the hydroxyl ladder marker lane (lane 1). The stability of stem 1 is also apparent in that the bands corresponding to (approximately) nucleotides C35 to C45 are heavily compressed into a smaller than expected region of the gel. This compression was not greatly relieved by including formamide (to 20 % (v/v)) in the denaturing gels. RNase V1 cuts were especially evident in stem 1 and were also seen at the top of the 3′-arm of stem 2 (lanes 10-11). In most cases, the single strand-specific enzymes RNase T1 and U2 (lanes 4-5, 16-17) reacted as expected, with those slip-site, spacer, loop or downstream region residues predicted to be single-stranded cleaving as such. For example in pKA18, RNases T1 and U2 cleaved G and A residues, respectively, in loop 2, but not within stems. Perhaps the only unexpected feature which emerged was the highly reactive nature of the residues at the top of the 5′-arm of stem 2 to enzymatic probes (G30, G31 and C32 in pKA13; G31, G32 and C33 in pKA18). These residues were predicted to be in double-stranded conformation, but were clearly recognised by single strand-specific reagents. The cleavage of C residues by RNases T1 and U2 was unexpected, but may be a reflection of the highly reactive nature of the adjacent residues. Most of the first arm of stem 2 was also cleaved by the chemical probes imidazole and lead acetate. On the opposite arm of stem 2, the complementary bases appeared to be much less reactive to single-stranded probes. Base G55 of pKA13 (or G57 of pKA18) was resistant to RNase T1 cleavage, despite the reactivity of its partner discussed above. Furthermore, this arm of stem 2 was largely resistant to imidazole or lead cleavage. Only the the last nucleotide of stem 2 of pKA13 (C59) or pKA18 (C61) showed sensitivity to imidazole, and such cleavage is often seen at the ends of helices. These observations are consistent with the formation of stem 2, but a determination of its precise conformation will require further study.
Discussion
Here, we demonstrate that the promotion of efficient −1 ribosomal frameshifting by pseudoknots of the IBV class requires a minimum stem 1 length of 11 bp. Furthermore, our results are consistent with the view that the defect in many of the shorter pseudoknots is simply a stem 1 length deficiency rather than an inappropriate stem 1 stability, loop length or sequence composition. How do these observations fit with current models of the frameshift process? Several models have been postulated to account for the role of RNA pseudoknots (and hairpin-loops) at frameshift sites (see Farabaugh et al., 1996). The basic hypothesis is that interaction of the elongating ribosome with the pseudoknot leads to ribosome pausing, increasing the opportunity that the ribosome-bound tRNAs will slip into the −1 reading frame at the slippery sequence (Jacks et al., 1988). Pausing could be induced for example, by interaction of the pseudoknot with a component of the translation apparatus (protein or RNA) or following the failure of a ribosome-associated helicase to unwind the pseudoknot effectively. However, we do not know whether the pause itself is required for frameshifting or is simply a consequence of the kinds of interactions described above. Our mutagenic analysis of the base-pairs at the bottom of stem 1 of pseudoknot KA18 (11 bp stem 1) is consistent with a requirement for an initial pause upon encounter of the pseudoknot by the ribosome. Although only a limited number of pseudoknots were assayed (five constructs), there was a direct correlation between the number of G residues (as part of a G·C base-pair) present in the 5′-arm at the bottom of stem 1 and the efficiency of frameshifting observed, with an optimum at four G bases. The A·U pairs were in fact inhibitory in this region. The overall stability of stem 1, however, seemed to be less important as long as the stem was sufficiently long. Several of the shorter pseudoknots tested had stem 1 structures that were G-rich at the base, and predicted to be at least as stable overall as stem 1 of the wild-type IBV pseudoknot (e.g. pKA7, 9, 11; pSL3), yet were non-functional. This length requirement may exist for a number of reasons. In support of models in which a pseudoknot-ribosome interaction occurs, the minimum length requirement may allow an important pseudoknot feature to be placed at a precise location in the ribosome. Alternatively, the requirement for a long stem 1 may have consequences for unwinding of the pseudoknot; it may be significant that the minimum stem 1 length corresponds to one turn of an RNA helix (Arnott et al., 1973). Computer-assisted RNA folding analysis of other frameshifter pseudoknots of the IBV class indicate that stem 1 lengths of 11-12 bp are present in the natural frameshift signals (Brierley, 1995). Sequencing studies of the frameshift region of transmissible gastroenteritis virus (TGEV; Eleouet et al., 1995) suggested a 14 bp stem 1-containing pseudoknot located 3 nt downstream of the slippery sequence, but it is more likely that the spacing distance is in fact 6 nt and stem 1 only 11 bp. Although we did not determine an upper limit for stem 1 length in this study, longer pseudoknots with up to 13 bp in stem 1 remained functional and tolerance to an unpaired residue near the top of the stem was also seen in two of the longer stem 1-containing constructs. This may also have implications in models of pseudoknot-ribosome interactions, since the ribosome would presumably encounter various features of the pseudoknot differently as the length of stem 1 increased and the stem 1 helix turned.
Here, we did not address whether a minimal stem 1 stability is required for IBV frameshifting. Previously, the often inhibitory effects of introducing stem 1 mutations in pseudoknots of the IBV class have been attributed to a straightforward destabilisation of the stem, but some of these mutations may in fact have acted by impairing a functionality normally provided by a long stem. It would be of interest to construct an A+U-rich 11 bp stem 1-containing pseudoknot with a predicted stability less than the wild-type IBV pseudoknot. However, given the requirement for a G+C-rich stem base, a mismatch would be required in the middle of the stem in order to achieve the necessary reduction in stability which would cloud interpretation of the experiment. A related experiment has been carried out using the stem-loop stimulated frameshift signal of the human immunodeficiency virus (HIV-1). Variants of the HIV-1 stem-loop were prepared in which the length of the stem was unchanged (11 bp) but the predicted stability varied by inclusion of A·U pairs and frameshifting measured in vivo (Bidou et al., 1997). As overall stem stability increased, a concomitant increase in frameshift efficiency was found up to a certain point when a plateau was reached. A comparison of the HIV-1 experiments with those presented here for IBV is difficult given the different stimulatory RNA signals and translational environments involved. However, with these restrictions in mind, the pattern seen with HIV-1 does not appear to be reflected in IBV pseudoknot-induced frameshifting. A number of IBV-based pseudoknots with stem 1 regions of identical length yet different predicted overall stabilities were found to promote similar frameshift efficiencies; for example, the 11 bp stem 1 constructs pFScass 6 (46 % and −16.2 kcal/mol) and pKA18 (48 % and −23.3 kcal/mol) or the 12 bp stem 1 constructs pSL4; (55 % and −16.1 kcal/mol) and pKA26 (40 % and −25.8 kcal/mol).
An important question to address is whether IBV pseudoknot-induced frameshifting requires a kinked pseudoknot conformation as seen with MMTV. On the surface, the two structures are expected to be somewhat dissimilar in that the IBV pseudoknot has a requirement for a much longer stem 1 and is predicted to have coaxially stacked stems. It has been noted that the base-pair at the top of stem 1 of the wild-type IBV pseudoknot is a relatively weak U·G pair, and if this pair did not form, an unpaired G residue would be present at the stem junction that could lead to a kinked structure (Chen et al., 1996). However, an IBV pseudoknot mutant has been constructed that contains a more stable C·G pair at the top of stem 1 (pFS8.40; Brierley et al., 1991). This construct was found to stimulate frameshifting even more than the wild-type structure, arguing against a role for an unpaired base at the top of stem 1. Furthermore, introduction of an A triplet into loop 1 had no effect on frameshifting (pFS8.23; Brierley et al., 1991), whereas if the pseudoknot was kinked, lengthening loop 1 would be expected to reduce kinking and hence lower frameshifting. Also, several of the functional pseudoknots tested in the present study are predicted to possess stable G·C pairs at the top of stem 1. Thus the evidence to date favours the idea that the two types of pseudoknot are distinct.
Nevertheless, there may be similarities between the two in the stem 2 region. Structure probing of the ten and 11 bp stem 1-containing pseudoknots described here revealed a region in the 5′-arm of stem 2 that was hypersensitive to enzymatic probes. This hypersensitivity was not seen on RNase T1 probing of the kinked VPK pseudoknot (Chen et al., 1995) which was probed at higher salt and magnesium concentrations (100 mM KCl, 10 mM MgCl2 compared with 50 mM Na cacodylate, 2 mM MgCl2). Hence the reactivity could be interpreted as arising from periodic “breathing” of stem 2 of the IBV-derived pseudoknot under the more destablilising ionic conditions. However, the extent of the reactivity at this site compared with other single-stranded G residues in the pseudoknot, and the unreactivity of G residues on the opposite arm of stem 2 argues against breathing of stem. It may be that this region of stem 2 adopts an unusual conformation. The same region of stem 2 of VPK has been shown to react with the conformation-sensitive probe NiCR ([2, 12-dimethyl-3, 7, 11, 17-tetraazabicyclo (11.3.1) heptadeca-1(17), 2, 11, 13, 15-pantaene]-nickel (II) perchlorate; Chen et al., 1995) which cleaves at the most solvent-accessible G residues. Thus the sensitivity of the equivalent region of IBV-based pseudoknots to enzymatic probes may be explicable in terms of a common structural feature shared by the two pseudoknots. In recent experiments, we have found that the introduction of an unpaired A residue at the junction between the two stems of pKA5 (stem 1 of six nucleotides; see Figure 2) can, under certain circumstances, allow a functional, presumably kinked pseudoknot to be prepared (Liphardt et al., 1999). Structure mapping of this construct with RNase T1 revealed a similar stem 2 hypersensitivity as we saw with pKA13 and 18, supporting the idea that this region of the two classes of pseudoknot has a similar conformation. The structural studies needed to confirm these hypotheses are underway.
Materials and methods
Site-specific mutagenesis
Site-directed mutagenesis was carried out by a procedure by Kunkel (1985) as described (Brierley et al., 1989). Mutants were identified by dideoxy sequencing of single-stranded templates (Sanger et al., 1977). Sequencing through G+C-rich regions was facilitated by including formamide (to 20 % (v/v)) in the sequencing gel and replacing dGTP with deaza-GTP in the sequencing mixes.
Construction of plasmids
The starting plasmid for these studies, pFScass 6 (Brierley et al., 1992) is a frameshift reporter construct which contains the minimal IBV frameshift region cloned into the influenza A/PR8/34 PB2 gene (Young et al., 1983) at a unique BglII site (position 456), a site at which significant lengths of open reading frame are present in each of the three frames downstream (Figure 1). Plasmid KA1 (Figure 1) was derived from pFScass 6 by deletion of two blocks of 7 bp (TATCAGT and GCTGGTA) from the pseudoknot-encoding region in separate mutagenesis reactions, such that the pseudoknot encoded by this plasmid would have only four G·C base-pairs in stem 1. Subsequent plasmids in the pKA series that contained 11 bp or fewer in stem 1 were prepared by site-directed mutagenesis of the next smaller pKA construct. For example, pKA5, which possesses a 6 bp stem 1 was prepared from pKA3, containing a 5 bp stem 1, by two rounds of site-directed mutagenesis to introduce an additional G·C pair. We were unable to make a 12 bp stem 1 construct by mutagenesis, probably due to extensive regions of self-complementarity in the G+C-rich mutagenic oligonucleotides. For this reason, we adopted an oligonucleotide cloning strategy. Plasmid pFScass 6 was mutated to introduce a unique MluI site immediately downstream of the pseudoknot-encoding region to generate pFScass9. This plasmid was digested with BglII and MluI (which removes the minimal IBV frameshift region) and ligated with four pairs of kinased, annealed oligonucleotides to produce pKA22, which contains a 12 bp stem 1 pseudoknot-encoding region. The six extra base-pairs (ACGCGT) introduced into pKA22 (at a position ten bases downstream of the 3′-end of the pseudoknot) by this cloning strategy were deleted in plasmid pKA26 to ensure the same 3′-end context. Optimal ligation conditions for efficient cloning of the oligonucleotides into the vector were with 5 ng of each of the eight oligonucleotides (average length 25 nt), 30 ng vector DNA and 1 unit T4 DNA ligase (Bethesda Research Laboratories) for 12 hours at 14°C. A second series of stem 1 length variants of the minimal IBV pseudoknot (the pSL series) were prepared by introduction or deletion of base-pairs using site-directed mutagenesis. The starting plasmids for this series were pFScass 5 and pFScass 7, which differ from pFScass 6 only in the possession of one (pFScass 5) or two (pFScass 7) additional C residues a few nucleotides downstream of the pseudoknot-forming region (Brierley et al., 1992).
In vitro transcription and translation
Plasmids for in vitro transcription were prepared as described (Brierley et al., 1989). In vitro transcription reactions employing the bacteriophage SP6 RNA polymerase were carried out essentially as described by Melton et al. (1984) and included the synthetic cap structure 7meGpppG (New England Biolabs) to generate capped mRNA. Product RNA was recovered by a single extraction with phenol/chloroform/isoamyl alcohol (49:49:2) followed by precipitation in ethanol in the presence of 2 M ammonium acetate. The RNA pellet was dissolved in water, and the remaining unincorporated nucleotide triphosphates removed by Sephadex G-50 chromatography. RNA was recovered by precipitation in ethanol, dissolved in water and checked for integrity by electrophoresis on 1.5 % (w/v) agarose gels containing 0.1 % (w/v) sodium dodecyl sulphate. In ribosomal frameshift assays, serial dilutions of purified mRNAs were translated in RRL as described (Brierley et al., 1989). Translation products were analysed on SDS-15 % (w/v) polyacrylamide gels according to standard procedures (Hames, 1981). The relative abundance of non-frameshifted or frameshifted products on the gels was determined by direct measurement of [35S]methionine incorporation using a Packard Instant Imager 2024. Frameshift efficiencies were calculated from those dilutions of RNA where translation was highly processive (RNA concentrations of 10 μg to 25 μg RNA/ml of reticulocyte lysate). The frameshift efficiencies quoted are the average of at least three independent measurements which varied by less than 10 %, i.e. a measurement of 40 % frameshift efficiency was between 36 % and 44 %. The calculations of frameshift efficiency take into account the differential methionine content of the various products (19 kDa, 11; 22 kDa and 28 kDa, 12; 85 kDa, 35).
RNA structure probing
RNAs for secondary structure probing were prepared by in vitro transcription of BstNI-cut plasmid templates using bacteriophage T7 RNA polymerase. Transcription reactions were on a 200 μl scale and contained 20 μg plasmid DNA, 2.5 mM of each rNTP and 500 units of T7 RNA polymerase (New England Biolabs) in a buffer containing 40 mM Tris (pH 8), 15 mM MgCl2, 5 mM DTT. After three hours at 37°C, 100 units of DNase I was added and the incubation continued for a further 30 minutes. Nucleic acids were harvested by extraction with phenol/chloroform (1:1) and precipitation in ethanol. DNA fragments were removed by Sephadex G-50 chromatography and the RNA transcripts concentrated by ethanol precipitation. The RNA was quantified by spectrophotometry and its integrity checked by electrophoresis on a 2 % (w/v) agarose gel containing 0.1 % sodium dodecyl sulphate. Transcripts (10 μg) were 5′ end-labelled with [γ-33P]ATP using a standard dephosphorylation-rephosphorylation strategy (ten Dam et al., 1995), purified from 10 % acrylamide-urea gels and dissolved in water. The structure probing experiments followed the general principles outlined by others van Belkum et al 1988, Wyatt et al 1990, Polson and Bass 1994. All reactions contained 10-50,000 cpm 5′ end-labelled RNA transcript and 10 μg baker’s yeast carrier tRNA. RNAse probing reactions were carried out in 50 μl reaction volumes. Probing with RNase T1 (Amersham) was on ice for 20 minutes in 50 mM sodium cacodylate (pH 7), 2 mM MgCl2 and 0-1 units T1; RNase V1 (Pharmacia) at 25°C for 20 minutes in 10 mM Tris (pH 8), 2 mM MgCl2, 0.1 M KCl and 0-0.35 units V1; RNase U2 (USB) on ice for 20 minutes in 20 mM sodium acetate (pH 4.8), 2 mM MgCl2, 100 mM KCl and 0-0.2 units U2. Enzymatic reactions were stopped by the addition of 150 μl ethanol and the RNA recovered by centrifugation. RNAs were prepared for analysis on 10 or 15 % polyacrylamide-7 M urea sequencing-type gels by dissolution in water, mixing with an equal volume of formamide gel loading buffer (95 % (v/v) formamide, 10 mM EDTA, 0.1 % bromophenol blue, 0.1 % xylene cyanol) and heating to 80°C for three minutes. Chemical probing experiments were performed with lead acetate and imidizole in 10 μl reaction volumes. Lead probing was at 25°C for five minutes in 20 mM Hepes-NaOH (pH 7.5), 5 mM Mg acetate, 50 mM K acetate and 1-5 mM Pb acetate. Reactions were stopped by addition of EDTA to 33 mM, the RNA recovered by precipitation in ethanol, redissolved in water and prepared for gel loading as above. For imidizole probing, the end-labelled RNA was mixed with 10 μg carrier tRNA, dried in a desiccator and redissolved in 10 μl of 2 M imidizole (pH 7) containing 40 mM NaCl and 10 mM MgCl2. After incubation at 37°C for one to four hours, the reaction was stopped by addition of 10 μl of a fresh solution of 2 % (w/v) lithium perchlorate in acetone. The RNA was recovered by centrifugation, washed with acetone, dried, dissolved in water and prepared for gel loading as above. All structure probing gels included an alkaline hydrolysis ladder of the relevant RNA as a size marker, prepared by dissolving the dried pellet from 3 μl of end-labelled RNA and 10 μg carrier tRNA in 3 μl of 22.5 mM NaHCO3, 2.5 mM Na2CO3 and boiling for one to 2.5 minutes. After the addition of an equal volume of formamide gel loading buffer, the sample was heated to 80°C for three minutes and loaded immediately onto the gel. The structure probing was carried out on at least three independent preparations of end-labelled RNA.
Acknowledgements
This work was supported by the Medical Research Council, UK and the Biotechnology and Biological Sciences Research Council, UK. We thank Dr Paul Digard for critical reading of the manuscript.
Footnotes
Edited by D. E. Draper
References
- Arnott S., Hukins D.W., Dover S.D., Fuller W., Hodgson A.R. Structures of synthetic polynucleotides in the A-RNA and A′-RNA conformations: X-ray diffraction analyses of the molecular conformations of polyadenylic acid-polyuridylic acid and polyinosinic acid-polycytidylic acid. J. Mol. Biol. 1973;81:107–122. doi: 10.1016/0022-2836(73)90183-6. [DOI] [PubMed] [Google Scholar]
- Bidou L., Stahl G., Grima B., Liu H., Cassan M., Rousset J-P. In vivo HIV-1 frameshift efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA. 1997;10:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- Brierley I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E.G., Binns M.M., Bilimoria B., Blok V.C., Brown T.D.K., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterisation of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N.J., Jenner A.J., Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chamorro M., Lee S.I., Shen L.X., Hines J.V., Tinoco I., Jr, Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.Y., Kang H.S., Shen L.X., Chamorro M., Varmus H.E., Tinoco I. A characteristic bent conformation of RNA pseudoknots promotes −1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- Dinman J.D., Ruiz-Echevarria M.J., Czaplinski K., Peltz S.W. Peptidyl-transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficienciesm: development of model systems. Proc. Natl Acad. Sci. USA. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.H., Holland J.A., Hansen M.R., Giedroc D.P., Hoffman D.W. Base-pairings within the RNA pseudoknot associated with the simian retrovirus-1 gag-pro frameshift site. J. Mol. Biol. 1997;270:464–470. doi: 10.1006/jmbi.1997.1127. [DOI] [PubMed] [Google Scholar]
- Eleouet J.F., Rasschaert D., Lambert P., Levy L., Vende P., Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microbial Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y-X., Yuan H., Rein A., Levin J.G. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J. Virol. 1992;66:5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames B.D. An introduction to polyacrylamide gel electrophoresis. In: Hames B.D., Rickwood D., editors. IRL Press; Oxford: 1981. pp. 1–91. (Gel Electrophoresis of Proteins-A Practical Approach). [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Kang H.S., Tinoco I. A mutant RNA pseudoknot that promotes ribosomal frameshifting in mouse mammary tumor virus. Nucl. Acids Res. 1997;25:1943–1949. doi: 10.1093/nar/25.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.S., Hines J.V., Tinoco I. Conformation of a non-frameshifting RNA pseudoknot from mouse mammary tumor virus. J. Mol. Biol. 1996;259:135–147. doi: 10.1006/jmbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- Kolchanov N.A., Titov I.I., Vlassova I.E., Vlassov V.V. Chemical and computer probing of RNA structure. Prog. Nucl. Acid Res. Mol. Biol. 1996;53:131–196. doi: 10.1016/S0079-6603(08)60144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzosiak W.J., Marciniec T., Wiewiorowski M., Romby P., Ebel J.-P., Giege R. Characterisation of the lead (II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNA Phe. Biochemistry. 1988;27:5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J., Napthine S., Kontos H., Brierley I. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D.A., Krieg P.A., Robagliati M.R., Maniatis T., Zinn K., Green M.R. Efficient in vitro synthesis of biologically active RNA and RNA hybridisation probes from plasmids containing a bacteriophage SP6 promoter. Nucl. Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C.W.A., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucl. Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson A.G., Bass B.L. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.X., Tinoco I. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Somogyi P., Jenner A.J., Brierley I., Inglis S.C. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D., Kang H. Mutational analysis of the RNA pseudoknot involved in efficient ribosomal frameshifting in simian retrovirus 1. Nucl. Acids Res. 1998;26:1369–1372. doi: 10.1093/nar/26.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E.B., Pleij C.W.A., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E., Pleij K., Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- ten Dam E., Brierley I., Inglis S.C., Pleij C. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucl. Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E., Verlaan P., Pleij C. Analysis of the role of the pseudoknot component in the SRV-1 gag-pro ribosomal frameshift signal: loop lengths and stability of the stem regions. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- Tu C., Tzeng T-H., Bruenn J.A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl Acad. Sci. USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.H., Sugimoto N., Freier S.M. RNA structure prediction. Annu. Rev. Biophys. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- van Belkum A., Verlaan P., Bing KunJ., Pleij C., Bosch L. Temperature-dependent chemical and enzymatic probing of the tRNA-like structure of TYMV RNA. Nucl. Acids Res. 1988;16:1931–1950. doi: 10.1093/nar/16.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov V.V., Zuber G., Felden B., Behr J.-P., Giege R. Cleavage of tRNA with imidazole and spermine imidazole constructs: a new approach for probing RNA structure. Nucl. Acids Res. 1995;23:3161–3167. doi: 10.1093/nar/23.16.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills N.M., Gesteland R.F., Atkins J.F. Evidence that a downstream pseudoknot is required for translational readthrough of the Moloney murine leukaemia virusgag stop codon. Proc. Natl Acad. Sci. USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J.R., Puglisi J.D., Tinoco I. RNA pseudoknots: stability and loop size requirements. J. Mol. Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- Young J.F., Desselberger U., Graves P., Palese P., Shatzman A., Rosenberg M. Cloning and expression of influenza virus genes. In: Laver W.G., editor. Elsevier Science; Amsterdam: 1983. pp. 129–138. (The Origin of Pandemic Influenza Viruses). [Google Scholar]


