Abstract
Programmed −1 ribosomal frameshifting has become the subject of increasing interest over the last several years, due in part to the ubiquitous nature of this translational recoding mechanism in pathogenic animal and plant viruses. All cis-acting frameshift signals encoded in mRNAs are minimally composed of two functional elements: a heptanucleotide “slippery sequence” conforming to the general form X XXY YYZ, followed by an RNA structural element, usually an H-type RNA pseudoknot, positioned an optimal number of nucleotides (5 to 9) downstream. The slippery sequence itself promotes a low level (≈1 %) of frameshifting; however, downstream pseudoknots stimulate this process significantly, in some cases up to 30 to 50 %. Although the precise molecular mechanism of stimulation of frameshifting remains poorly understood, significant advances have been made in our knowledge of the three-dimensional structures, thermodynamics of folding, and functional determinants of stimulatory RNA pseudoknots derived from the study of several well-characterized frameshift signals. These studies are summarized here and provide new insights into the structural requirements and mechanism of programmed −1 ribosomal frameshifting.
Keywords: RNA pseudoknot, ribosomal frameshifting, recoding, RNA structure, RNA thermodynamics
Abbreviations: MMTV, mouse mammery tumor virus; FIV, feline immunodeficiency virus
Introduction to RNA pseudoknots
As anticipated by the crystallographic structure of transfer RNA nearly thirty years ago (Robertus et al., 1974), it is now widely recognized that all classes of biological RNA molecules fold into complex three-dimensional shapes and structures in order to carry out their diverse biological functions Doudna and Cate 1997, Hermann and Patel 1999. Pseudoknotted RNA structures are among the simplest of RNA folding motifs. An RNA pseudoknot is minimally comprised of two helical structures connected by two single-stranded loops, thereby providing a simple way in which a single-strand of RNA can fold back on itself (Figure 1(a)). As such, RNA pseudoknots are widely recognized to play diverse fundamental roles in structurally organizing complex RNAs, in the assembly of ribonucleoprotein complexes, and in translational regulation and recoding when present within messenger RNAs (for a review, see Gesteland & Atkins, 1996).
Figure 1.
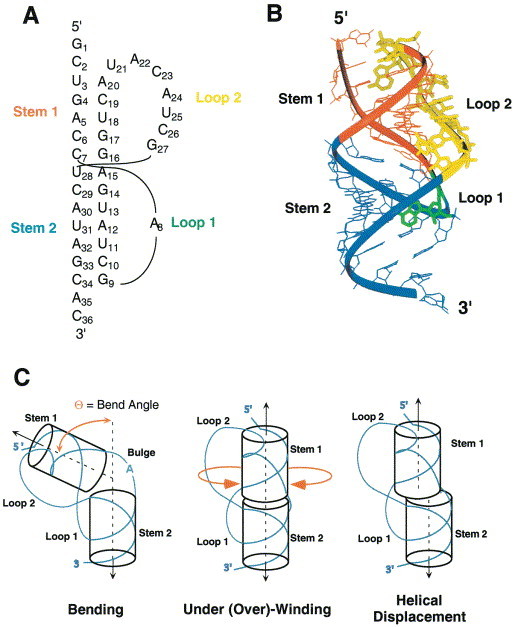
(a) Secondary structural representation of an H-type pseudoknot. The sequence shown corresponds to the autoregulatory gene 32 mRNA pseudoknot from bacteriophage T2 (Du et al., 1996). (b) Tertiary structural representation of the simple pseudoknotted RNA from the T2 gene 32 mRNA showing that the single-stranded loops L1 (green) and L2 (yellow) will cross the major and minor grooves of stems S2 (blue) and S1 (red), respectively, on the same face of the molecule provided the junction region conforms to near-normal A-form geometry (PDB code 2TPK). (c) Schematic representation of the structural diversity of the helical junction of simple H-type RNA pseudoknots. Note that various circular permutations of this motif can theoretically be formed, e.g. by breaking the chain in loop L1 or L2 and connecting the 5′ and 3′ ends shown with an extended loop. The standard H-type folding topology shown is nearly exclusively observed in natural RNA sequence contexts.
An RNA pseudoknot was first recognized as a novel RNA folding motif by Rietveld, Pleij and coworkers in transfer RNA-like structures found at the 3′ end of turnip yellow mosaic virus (TYMV) genomic RNA Rietveld et al 1982, Rietveld et al 1983, the NMR solution structure of which was recently solved (Kolk et al., 1998). Similar pseudoknots have also been found in related positive-strand plant RNA viruses, including brome mosaic virus and tobacco mosaic virus (Hall, 1979). These tRNA-like structures are substrates for specific aminoacyl-tRNA synthetases, and must be aminoacylated for viral replication and propagation in plant cells (Dreher et al., 1996). Pseudoknotted RNA structures have since been identified in virtually all types of naturally occurring RNAs, including ribosomal RNAs (Powers & Noller, 1991), messenger RNAs McPheeters et al 1988, Jacks et al 1987, Jacks et al 1988a, Jacks et al 1988b, Wills et al 1991, Feng et al 1992, transfer-messenger RNA (tmRNA) (Nameki et al., 1999), catalytic and self-splicing RNAs Hass et al 1994, Jabri et al 1997, Ferre-D’Amare et al 1998, RNA components of ribonucleoprotein complexes, e.g. telomerase (Gilley & Blackburn, 1999), and in viral genomic RNAs (for a review, see ten Dam et al., 1992).
An RNA pseudoknot is a structural element of RNA which forms when nucleotides within one of the four types of single-stranded loops in a secondary structure (hairpin loop, bulge loop, interior loop and bifurcation loop) base-pair with nucleotides outside that loop (for reviews, see Pleij and Bosch 1989, Schimmel 1989, Draper 1990, Pleij 1990, Pleij 1994, Westhof et al 1996, ten Dam et al 1992). Pseudoknots can possess many distinct folding topologies; however, the majority of pseudoknots that have been described to date, including the paradigm plant viral RNAs, are of the so-called H-type (Hairpin) topology in which nucleotides from a hairpin loop base-pair with single-stranded regions outside of the hairpin. This topology represents the simplest way to form a pseudoknotted structure, featuring two stem regions (S1 and S2) and two connecting loops (designated L1 and L2) (Figure 1(a)). The two helical stems of S1 and S2 base-pairs combine to form a quasi-continuous RNA double helical structure of S1 + S2 base-pairs (Pleij et al., 1985) (Figure 1(b)), containing one continuous and one discontinuous complementary strand. The non-equivalent single-stranded loops (L1 and L2) cross the major and minor grooves of stem S2 and stem S1, respectively (Figure 1(b)).
Even within this simple folded motif, a striking range of structural diversity can be accommodated. Much of this structural diversity derived from differences in the helix-helix junction, defined by the extent to which the two stems are coaxially stacked on one another, the degree of under- or over-rotation at the base-pair step at the helical junction relative to A-form helical geometry, the presence or absence of intervening nucleotides on the continuous strand, and the interhelical angle between the S1 and S2 stems (Figure 1(c)). As is readily apparent upon inspection of simple cylinder model representations of pseudoknots (Figure 1(c)), even small differences in the structure of the helical junction will significantly affect the global conformation of the molecule, how each of the two loops are arranged on the surface of the molecule, how these loops interact with the helical stems, as well as define where the single-stranded RNA enters and exits this motif.
In addition to structural roles in the folding of complex RNAs, RNA pseudoknots also play a variety of regulatory roles in protein synthesis. In translation initiation, a pseudoknot is sometimes positioned in the non-protein coding leader sequence or overlapping the ribosome binding site and/or initiation codon of the messenger RNAs (cf. Tang and Draper 1989, McPheeters et al 1988, Gluick and Draper 1994, Ehresmann et al 1995). Here, they modulate the specific binding of proteins to their cognate mRNAs, thereby regulating or autoregulating the expression of the downstream gene. RNA pseudoknots can also be present within the coding regions of mRNAs, where they stimulate programmed −1 ribosomal frameshifting, an essential mechanism employed by viruses, DNA insertion sequences, yeast, and bacteria to tightly regulate the relative expression levels of protein products present in two overlapping translational reading frames (for reviews, see Gesteland and Atkins 1996, Farabaugh 1996). In addition, pseudoknots have been shown to stimulate translational readthrough by amber codon suppression (Wills et al., 1991). This review focuses on the structural and functional diversity of these RNA pseudoknots which affect translation.
Translational frameshifting and ribosome reprogramming
In the vast majority of retroviral mRNAs, overlapping ORFs encode the gag gene (for viral structural proteins), the pro gene (which encodes the viral protease), and the pol gene (pol encodes the replicative enzymes of the virus including integrase and reverse transcriptase) or a pro/pol gene depending on the virus. Since translational initiation signals which could be used to synthesize the Pol and/or Pro proteins are not present in the genomic mRNA, these retroviruses employ −1 ribosomal frameshifting to create Gag-Pol or Gag-Pro-Pol fusion proteins from a single gag-pol (or gag-pro-pol) translational unit (Figure 2(a)) (Jacks et al., 1987). At some intrinsic frequency (1 to 50 %), the translating ribosome shifts into the −1 reading frame at a heptanucleotide sequence conforming to the general sequence X XXY YYZ (termed the slippery sequence) in the gag-pol (or gag-pro) overlapping region (for reviews, see Jacks 1990, Atkins et al 1990, Hatfield and Oroszlan 1990). The simultaneous-slippage model for −1 ribosomal frameshifting, first proposed by Jacks et al. (1988a) in their studies of the Rous sarcoma virus gag-pro region, is consistent with many experiments and explains the nucleotide sequence determinants of the slippery sequence at the frameshift site (Figure 2(b)). The model proposes that each of the two ribosome-bound tRNAs (the peptidyl- and aminoacyl-tRNAs bound in the P and A sites, respectively) slip backwards in the 5′ direction simultaneously from their initial positions in the zero frame (XXY YYZ) to the −1 frame (XXX YYY) and that frameshifting occurs only when each tRNA maintains at least two codon-anticodon base-pairs with the mRNA in the −1 shifted frame. A variety of slippery sequences are utilized by viruses with some intrinsically more efficient than others (Brierley et al., 1992), but G- and C-rich codons are not functional in the YYY position, due perhaps to the fact that these tRNA-mRNA contacts would be too stable to break during tRNA slippage. Replication competence, viral propagation, and viability of HIV-1 and the yeast double-stranded RNA virus L-A absolutely require that the pol gene be translated at a precise ratio relative to the gag gene Dinman 1995, Hung et al 1998. This observation, in turn, has motivated attempts to find small molecules which can modulate the efficiency of the frameshifting process and therefore act as antiviral agents Hung et al 1998, Dinman et al 1998, as well as identify endogenous protein factors which modulate frameshifting efficiency in vivo (Cui et al., 1998). In the retroviruses which do not utilize frameshifting to express the pol gene products, e.g. type-C retroviruses such as Moloney murine leukemia virus (MuLV), the elongating ribosome “reads through” an in-frame amber stop codon separating gag and pol genes some fraction of the time (≈10 %) to express the Gag-Pol fusion protein required for assembly and replication Wills et al 1991, Feng et al 1992.
Figure 2.
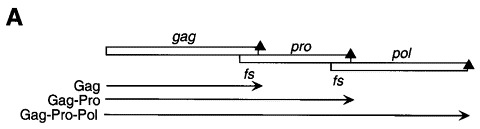
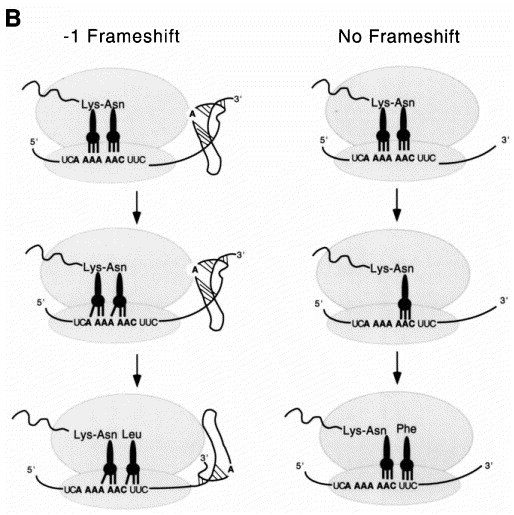
(a) Production of Gag, Gag-Pro, and Gag-Pro-Pol fusion proteins from a single mRNA by way of two isolated −1 ribosomal frameshifting events. (b) Simultaneous slippage model (Jacks et al., 1988a) for translational regulation of ribosomal frameshifting. Adapted from Gesteland & Atkins (1996).
In most instances where translational read-through or ribosomal frameshifting is suspected or confirmed to occur, the RNA sequence 5 to 9 nt downstream from the gag termination codon or the 3′ end of the slippery sequence is known or postulated to harbor an H-type pseudoknotted RNA structure (Figure 2(b)). The existence of these pseudoknots and their essential involvement in ribosomal frameshifting or read-through events have been confirmed by mutagenesis studies in a number of viruses, including the coronavirus avian infectious bronchitis virus (IBV) Brierley et al 1989, Brierley et al 1991; animal retroviruses including RSV Jacks et al 1988a, Jacks et al 1988b, mouse mammary tumor virus (MMTV) (Chamorro et al., 1992), feline immunodeficiency virus (FIV) (Morikawa & Bishop, 1992), simian retrovirus type-1 (SRV-1) (ten Dam et al., 1994), and MuLV Wills et al 1991, Feng et al 1992; double-stranded RNA viruses of Saccharomyces cerevisiae, L-A and L-1 Dinman et al 1991, Tzeng et al 1992; and in plant luteoviruses and enamoviruses, including beet western yellows virus (Garcia et al., 1993; Miller et al., 1996; Kim et al., 1999). A prominent exception to this general rule is in HIV-1, where a simple 3′ hairpin appears to stimulate -1 frameshifting at the gag-pol junction (Parkin et al., 1992), but only 3 to 5-fold above that obtained for this rather efficient slippery sequence (U UUU UUA) alone Jacks et al 1988b, Bidou et al 1997. In addition, other more complex folded structures or multiple molecular determinants are known or predicted to function as modest stimulators of −1 frameshifting in other natural contexts (cf. Rettberg et al., 1999).
How do downstream pseudoknots stimulate ribosome recoding?
The precise mechanism by which downstream RNA pseudoknots stimulate frameshifting or amber stop codon suppression is unknown. Recent structural studies of intact ribosome-tRNA-mRNA complexes as well as individual ribosomal subunits, coupled with extensive biochemical data, serve to provide some structural insight here Clemons et al 1999, Ban et al 1999, Cate et al 1999. Biochemical and modeling experiments suggest that docking the bipartite frameshifting signal of the gag-pro junction of MMTV into the ribosome can be accommodated in such a way that the folded pseudoknot is not necessarily engulfed or unfolded by the ribosome while maintaining base-pairing of the mRNA slippery sequence to the A- and P-site RNAs in the zero or −1 frames. Instead, the folded pseudoknot may be able to come into very close proximity to the ribosome itself, while maintaining base-pairing of the slippery sequence to the transfer RNAs in the peptidyl transferase center (Figure 3). This simple modeling exercise provides an immediate explanation as to why decreasing or increasing the spacer length between the slippery sequence (or amber codon) and the pseudoknot usually greatly decreases recoding efficiency. In either case, the slippery sequence would no longer be optimally positioned in the active site of the ribosome, and the way in which the ribosome “interacts” with the pseudoknot would be altered. The importance of nucleotide sequence in this connecting region has only been tested extensively in the MuLV read-through site, where the identities of some nucleotides were found to be more important than others (Wills et al., 1994). In addition, the topological constraints of the classic pseudoknot fold, in which the 5′ (closest to an approaching ribosome) and 3′ structural boundaries of the pseudoknot are on opposite ends of the molecule, when compared to a standard hairpin, must also be important Draper 1990, ten Dam et al 1992. In fact, replacement of the pseudoknot in the IBV frameshifting site with an RNA hairpin conforming to the S1 + S2 base-pairs of the pseudoknot greatly diminishes frameshifting efficiency (Somogyi et al., 1993).
Figure 3.
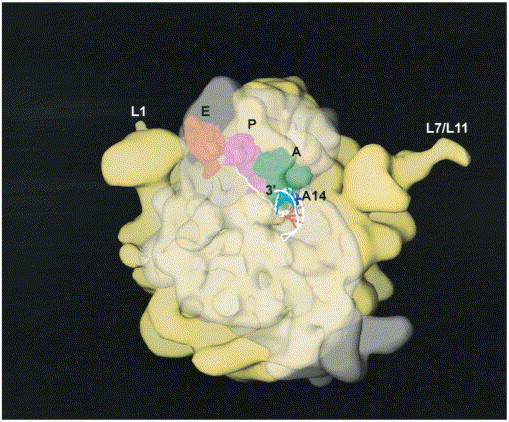
Model of the MMTV gag-pro mRNA frameshifting signal docked into the low resolution structure of the E. coli ribosome derived from the 25 Å cryo-electron microscopy electron density map (Frank et al., 1995a). The large 50 S ribosomal subunit (yellow) is shown to the rear of this view with the L1 stalk and the L7/L12 regions labeled. The small 30 S subunit is shown to the front, but made transparent so as not to obscure the aminoacyl (A), peptidyl (P) and exit (E) tRNAs, as well as the proposed path of the mRNA (Frank et al., 1995a). The A, P, and E-site tRNAs were positioned as described in the 7.8 Å resolution crystallographic structure of the T. thermophilus ribosome (Cate et al., 1999) (PDB code 486D) by manually docking them into the structure of the E. coli ribosome. The nucleotides of the slippery sequence are positioned opposite the A- and P-site tRNA anticodon triplets in the zero reading frame as described in the Cate et al. (1999) structure. The seven nucleotides between the 3′ base of the slippery sequence and the 5′ nucleotide of stem S1 are drawn in an extended conformation. The structure suggests that the pseudoknot need not be drawn into the ribosome and/or become largely unfolded when the slippery sequence is positioned in the active site of the ribosome. The structure further reveals how the 5′-side of loop L2 (as well as the base of stem S1) may well be first to physically interact with the translating ribosome. Adapted from Frank et al. (1995b).
When an elongating ribosome encounters a pseudoknot, it has been shown to pause or stall over the slippery sequence, implying that ribosomal pausing is a necessary condition for −1 frameshifting Tu et al 1992, Somogyi et al 1993. Consistent with this, some peptidyl transferase inhibitors have been shown to specifically alter the efficiencies of −1 frameshifting (Dinman et al., 1997). At least one of these (sparsomycin) would be expected to increase the amount of time the ribosome remains stalled over the slippery sequence and has been shown to strongly increase frameshifting efficiency (Dinman et al., 1997). However, the degree of frameshifting may only be partially correlated with the extent of ribosomal pausing on the slippery sequence since RNA stem-loops also induce ribosomes to pause, albeit at reduced levels (Somogyi et al., 1993), suggesting that ribosomal pausing is necessary but not sufficient to promote efficient frameshifting.
Any model for a mechanism of frameshifting must also account for the fact that the intrinsic efficiency of frameshifting varies dramatically from one natural site to another, from as low as 1 to 4 % in BWYV and other plant virus frameshift sites, to as much as 50 % in some instances, e.g. Escherichia coli dnaX (Larsen et al., 1997). A further complication is that absolute determinations of frameshifting efficiency for the same site have been shown to differ between in vitro translation systems versus in vivo Garcia et al 1993, Brierley 1995, Grentzmann et al 1998, with in vivo efficiencies in eukaryotic cells generally lower (cf. Kim et al., 1999). This makes comparisons of functional data from laboratory to laboratory and from site to site difficult. Nonetheless, most retroviral and coronoviral frameshift sites are characterized by frameshifting efficiencies which typically range from ≈15 to 40 %. In all cases, the presence of the slippery heptanucleotide sequence alone stimulates frameshifting, often up to 1 % or so in some cases, considerably greater than the extremely low level of error frameshifting on random sequence mRNA which is on the order of 0.01 % (Kurland, 1992). Thus, while some pseudoknots are intrinsically able to stimulate higher levels of frameshifting than others, all represent only a subtle modulation of a process which occurs to a significant extent in the absence of a downstream stimulatory element.
Another point to consider is that recoding pseudoknots must ultimately be unfolded by the ribosome following recoding, since they are subsequently decoded by the ribosome. Therefore, a subtle modulation of the structure, global or local stability, or dynamics of the pseudoknot may be all that is required to endow a particular structure with efficient function, to the exclusion of other similar conformations. If this is the case, it will be challenging to unravel the molecular determinants of frameshifting and/or read-through from structural studies alone. This review will summarize the significant progress made in the last several years in determining the structure, stability, dynamics and functional efficacy of wild-type and variant mRNA sites and how these findings might be combined to develop a mechanistic framework for understanding ribosomal frameshifting. For earlier more extensive reviews of ribosomal frameshifting and translational recoding, the reader is referred to Brierley 1995, Gesteland and Atkins 1996, Farabaugh 1996.
Structural studies of frameshifting mRNA pseudoknots
Based on the solution structure of the phage T2 gene 32 autoregulatory mRNA pseudoknot (Figure 1(a) and (b)), Du et al. (1996) hypothesized that the structures of many frameshifting mRNAs would contain certain features present in the T2 pseudoknot. Specifically predicted were the presence of a stem S2 of six or seven base-pairs crossed by a short single-nucleotide loop L1, with more variability in the lengths of stem S1 and loop L2. Consistent with this hypothesis, it was subsequently shown that a stem S2 of six base-pairs, but not five or eight base-pairs, was fully compatible with a single-nucleotide L1 pseudoknotted conformation (Du & Hoffman, 1997), as originally predicted by Pleij et al. (1985). If one allows for an expansion of loop L1 from one to two nucleotides within this general motif (Theimer et al., 1998), this hypothesis is consistent with structural analyses of a number of frameshifting Brierley et al 1991, Chamorro et al 1992, ten Dam et al 1995 and read-through Alam et al 1999a, Alam et al 1999b RNA pseudoknots, and suggests that significant structural and/or functional features are encoded in this motif. Interestingly, a high affinity biotin-binding RNA aptamer was recently shown to be modeled as an H-type RNA pseudoknot with precisely these structural features (Wilson et al., 1998).
High resolution structures derived from NMR spectroscopy and X-ray crystallography, as well as detailed structure probing experiments using structure- and sequence-specific ribonucleases and chemical probing, have provided insight into structural features associated with functional versus non-functional frameshifting RNAs. These fall into what appear to be, at least currently, four structural classes defined by: (1) MMTV gag-pro pseudoknot; (2) the SRV-1 gag-pro pseudoknot; (3) the pseudoknot derived from the 1a-1b junction in the coronavirus avian infectious bronchitis virus (IBV), and (4) the pseudoknot derived from the P1–P2 junction from a plant luteovirus, beet western yellows virus (BWYV). Recent structural and functional studies of these four frameshift signals are discussed below.
MMTV
The MMTV gag-pro frameshifting pseudoknot has a five base-pair stem S1 with a relatively short loop L2 (eight nucleotides) and is characterized by an unpaired adenosine wedged between the two helical stems. Stem S2 has six base-pairs and is crossed by a two-nucleotide loop L2 (Figure 4). The MMTV pseudoknot and structural variants have been subjected to mutational analysis and structure probing Chamorro et al 1992, Chen et al 1995 and the solution structure of MMTV-vpk has been solved by NMR spectroscopy (Shen & Tinoco, 1995) (Figure 4). Although a refined NMR solution structure of the MMTV pseudoknot contains additional loop-to-stem constraints for loop L1 due to the presence of a coordinated Co(NH3)6 3+ ion, the precise conformation of the nucleotides in loop L1 remain undefined, as do nucleotides in loop L2 (Gonzalez & Tinoco, 1999). The motional dynamics of nucleotides in loop L2 are distinct from the rest of the molecule, and might be characterized by rapid internal motion. The unpaired adenosine at the helical junction (A14) appears to induce a pronounced ≈60 ° bend angle between helices; in RNAs in which this adenosine is deleted (Figure 4), the helical stems are reported to be coaxially stacked as determined by NMR spectroscopy. Since standard NMR spectroscopy is not a particularly good method to precisely define the global conformation of a extended RNA helical molecule Dallas and Moore 1997, Holland et al 1999, the MMTV structure is likely compatible with a range of interhelical angles. Nonetheless, mutational studies in this system suggest that frameshifting and non-frameshifting variants appear to differ primarily by the presence or absence of the wedged adenosine Chamorro et al 1992, Chen et al 1996, Kang et al 1996, Kang and Tinoco 1997 consistent with the hypothesis that frameshifting efficiency at the MMTV gag-pro site derives at least in part, from a specific bent conformation of the pseudoknot.
Figure 4.
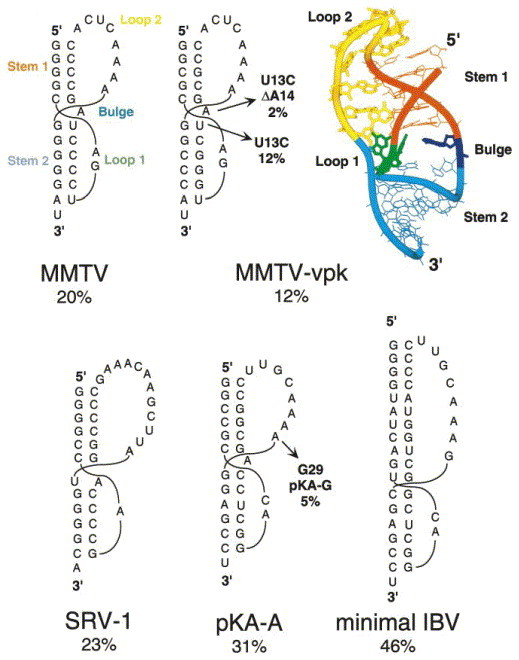
Secondary structural representations and reported in vitro frameshifting efficiencies of the MMTV and MMTV-vpk gag-pro pseudoknots Chamorro et al 1992, Chen et al 1995, the SRV-1 gag-pro pseudoknot ten Dam et al 1994, Du et al 1997, and the minimal wild-type IBV (Liphardt et al., 1999) and chimeric MMTV-IBV (pKA-A) (Napthine et al., 1999) pseudoknots. The solution structure of the MMTV-vpk pseudoknot is also shown (PDB code 1RNK) (Shen & Tinoco, 1995).
SRV-1
The pseudoknot from the gag-pro junction of simian retrovirus type-1 (SRV-1) (Figure 4) was among the first targets of extensive mutational studies designed to define functional requirements regarding stem and loop composition and length in a retroviral mRNA pseudoknot (ten Dam et al., 1995). It was shown that the identity and number of nucleotides in the loops were largely unimportant, provided that a minimum number of nucleotides were present so that the basic structure of the pseudoknot was maintained (Pleij et al., 1985). For example, the single adenosine in loop L1 could be replaced with any other nucleotide or the loop enlarged by one nucleotide with little discernable effect on frameshifting efficacy (ten Dam et al., 1995). Interestingly, within the context of the native stem S1, ten Dam et al. (1995) identified a rough correlation between the stability of stem S2 and frameshifting efficiency. A similar correlation was not found when compensatory mutations were introduced into the SRV-1 stem S1, although the interpretation of these experiments is complicated by the fact that a guanosine-rich 5′ arm of stem S1 appears to be important for function (cf. Napthine et al 1999, ten Dam et al 1995). These results lead the authors to propose that a certain threshold stability of the proximal stem S1 was required to fix the ribosome positioned at or over the slippery sequence, and provided this was maintained, the approaching ribosome might then “sense” the stability of the distal stem S2.
Given the proposal that a specific, bent conformation induced by an unpaired junction nucleotide is required for efficient stimulation of frameshifting by the MMTV gag-pro pseudoknot (Chen et al., 1996), it was of interest to determine the extent to which this structural motif characterized other gag-pro pseudoknots evolutionarily related to MMTV. The frameshifting pseudoknots from the gag-pro junction in SRV-1 and the gag-pro junction in feline immunodeficiency virus (FIV) each have the potential to form either an A-U base-pair or an unpaired adenosine at the helical junction Chen et al 1996, Morikawa and Bishop 1992, i.e. the nucleotide sequence suggests that there is structural ambiguity at the junction (Figure 4). NMR evidence suggests that the closing A-U base-pair at the helical junction in SRV-1 is formed (Du et al., 1997) but is characterized by a significantly faster base-pair opening frequency than are other stem base-pairs. Subsequent mutational analyses suggested that the identity of these two nucleotides and their ability to base-pair have no affect on frameshifting efficiency in SRV-1 or FIV Chen et al 1996, Sung and Kang 1998. In the case of the SRV-1 gag-pro pseudoknot and its variants, the structure of the helical junction may well be dynamic and perhaps not easily defined in terms of a single conformation at equilibrium.
IBV
The IBV 1a-1b pseudoknot is representative of frameshift-stimulating pseudoknots derived from coronaviruses, toroviruses and arteriviruses (Brierley, 1995). Unlike the retroviral gag-pro pseudoknots, these pseudoknots are characterized by a long stem S1 of 11–12 base-pairs coupled with a long loop L2 (Figure 4). Like the MMTV site (Du et al., 1996), they are associated with a six base-pair stem S2 with two nucleotides in loop L1. The native IBV pseudoknot as well as one with a minimal length loop 2 are both highly efficient frameshift enhancers when placed downstream of the efficient slippery sequence U UUA AAC, with recoding efficiencies of 40–55 % not uncommon with these structures. Early mutational studies suggested that there were no primary nucleotide sequence determinants in the stems and loops as long as the predicted structure was maintained Brierley et al 1989, Brierley et al 1991, i.e. the connecting loops were long enough to span the helical stems without disrupting the structure (Pleij et al., 1985).
Napthine et al. (1999) recently systematically investigated the influence of the length of stem S1 on functional activity since this seemed to be the primary structural difference between the IBV and retroviral gag-pro mRNA molecules. The remarkable finding from these studies was that a minimal stem S1 length of 11 to 12 base-pairs was found to be absolutely required for high-level frameshifting in vitro (48 %); a stem S1 length of ten base-pairs was only poorly effective (7 %), and shorter stem S1 constructs even less so. Increasing the length of loop L2 in the context of a ten base-pair stem S1 construct did not increase the efficiency of frameshifting, as would be expected if the reduced activity of the ten base-pair S1 RNA was not a trivial consequence of a loop L2 which was too short to span stem S1. Why an 11 or 12 base-pair stem is required in this context is unknown, although it may be significant that this corresponds to essentially one turn of A-form helix. This hints at a topological requirement for frameshifting, rather than one based on simple stability and structural determinants. Notably, pseudoknots containing the same number of base-pairs in stem S1 (11 base-pairs), but predicted to differ by over 8 kcal mol−1 in ΔG°37 due to differences in base-pair composition, were found to give rise to uniformly high levels of frameshifting (⩾45 %) characteristic of this system (Napthine et al., 1999). Systematic studies on the impact that changes in the predicted stability of stem S2 and frameshifting efficiency, like those reported for the SRV-1 gag-pro pseudoknot ten Dam et al 1994, ten Dam et al 1995 have not yet been reported for IBV.
Although the precise conformation of the S1–S2 helical junction region in the IBV pseudoknot is unknown, mutagenesis experiments suggest that both putative junction base-pairs are indeed paired, with the G·U to G·C base-pair substitution at the top of stem S2 (Figure 4) strongly stimulatory for frameshifting. This suggests that the junction is coaxially stacked like that in the T2 molecule (Figure 1(b)) and is structurally distinct from the helical junction in MMTV. In addition, the base-pair composition at the top of stem S1, closest to the slippery site, appeared to influence frameshifting efficiency, with a run of four guanines on the 5′ strand optimal for frameshifting; a similar requirement has been observed in SRV-1 (ten Dam et al., 1995). Finally, from structure probing experiments, there was some evidence of structural complexity or heterogeneity at the base of stem S2, near the 3′ side of loop L1, where there is predicted to be a sharp turn in the polynucleotide backbone.
Liphardt et al. (1999) propose that the MMTV gag-pro frameshifting pseudoknot may be largely representative of frameshift enhancers which maintain a shorter stem S1 and loop L2, coupled with similar stem S2 and loop L1 features. In a clever series of experiments, Liphardt et al. (1999) started with an inactive IBV-like pseudoknot with six base-pairs in each stem and a loop L2 of 8 nucleotides and asked what minimal structural features were required to confer functional activity on this inactive pseudoknot. It was found that an intercalated adenosine derived from the continuous strand had to be present at the junction of the two helical stems, like that found in the MMTV site, and that the 3′ nucleotide in loop L2 closest to the junction had to be an adenosine base (A29). These structural features are embodied in the pKA-A pseudoknot shown in Figure 4. Furthermore, compensatory base-pair substitutions in two base-pairs near the helical junction in stem S1 (C4-G18 and G5-C17) revealed that the requirement for A29 was manifest only when these two base-pairs were maintained; i.e. flipped G4-C18 and C5-G17 base-pair substitutions led to a fourfold loss in frameshifting efficiency and bypassed the requirement for A29 in loop L2. This was taken as functional evidence for a key loop L2-stem S1 interaction, much like that observed in the NMR structure of the TYMV pseudoknot (Kolk et al., 1998). Surprisingly, the functional requirement for this interaction could be largely bypassed if the length of loop L2 was significantly increased (to 13 nt). Consistent with these findings, the SRV-1 gag-pro pseudoknot, which has a longer loop L2 than MMTV (11 or 12 versus eight nucleotides) and a uridine-rich rather than adenosine-rich 3′ end of loop L2 (Figure 4), does not show this functional requirement.
High resolution structural studies of pKA-A will be necessary to provide additional insight into this functional requirement. However, ribonuclease and chemical structure probing experiments reveal that pKA-A and pKA-G RNAs (in which A29 is replaced by G29 with a concomitant reduction in frameshift efficiency by ≈sixfold) are essentially indistinguishable using this methodology (Liphardt et al., 1999). This suggests that the structural differences, if they are present, must be subtle indeed. In even the MMTV gag-pro pseudoknot, the precise nature of these loop L2-stem S1 interactions have thus far eluded efforts to unambiguously identify them (Gonzales & Tinoco, 1999).
BWYV
An H-type pseudoknot in BWYV and related plant luteoviruses regulates the expression of an RNA-dependent RNA polymerase encoded by viral gene P2 expressed as a P1–P2 fusion protein by promoting −1 frameshifting within the P1 gene (Garcia et al., 1993), in an analogous fashion to gag-pol retroviral frameshifting. The combination of the G GGA AAC slippery sequence and the pseudoknot found six nucleotides downstream is sufficient to induce ribosomes to simultaneously slip into the −1 frame about 1 to 4 % of the time Garcia et al 1993, Kim et al 1999. The 1.6 Å crystal structure of a 28 nucleotide RNA encompassing the proposed minimal structure of the pseudoknot reveals some remarkable structural features (Figure 5) (Su et al., 1999). As found in the MMTV gag-pro pseudoknot, the helical stems are not coaxially stacked. However, in contrast to MMTV, this is largely due to a 48 ° rotation between the top base of stem S2 (A25) and G7 in stem S1 which results in a ≈5 Å helical displacement of the two stems relative to one another, with U13, the potential base-pairing partner of A25 at the helical junction, extruded from the helix (Figure 5(b)). The base-pairs in stem S2 are considerably distorted from A-form helical geometry, perhaps to accommodate the short loop L1 in this context (Pleij et al., 1985) and the interhelical angle is ≈25 °. There is a clear base-triple (or base quadruple) interaction between C8 from loop L1 and the G12–G26 base-pair (A25 may also be involved in further stabilizing this unusual loop-stem interaction) (Figure 5). Another remarkable feature of the structure is a novel adenosine-rich RNA triplex, in which loop L2 snakes down the minor groove of stem S1, forming a series of non-canonical hydrogen bonding interactions involving the 2′-OH, N1, N3 and N7 of A20, as well as N1 and N6 of A24 (Su et al., 1999). The sequence of loop L2 in related plant luteoviral pseudoknots is strongly conserved with adenosines at the 3′ end of the loop (Miller et al., 1995).
Figure 5.
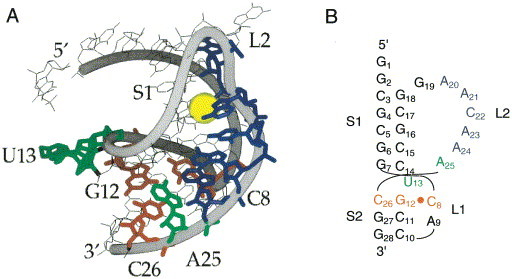
(a) The structure of the 28-nucleotide BWYV pseudoknot as determined by X-ray crystallography (Su et al., 1999) (PDB code 437D). The C8·G12-C26 base-triple interaction is shown in red and the loop 2 nucleotides that make crystal contacts with stem 1 are in blue. Also shown are U13 and A25 which would be predicted to form a base-pair in stem S2 (green) and a Na+ ion bound between loop L2 and stem S1 nucleotides (yellow). (b) Secondary structural rendering of the BWYV pseudoknot with nucleotides colored as in (a).
This complex structure makes the prediction that the minor groove triplex as well as the proposed loop L1-stem S2 base-triple interaction would be important for maintaining frameshifting activity. An extensive mutational analysis of the BWYV pseudoknot reveals that this is largely the case, with a few unanticipated results (Kim et al., 1999). For example, the identity of loop L1 C8 and much of the minor groove triplex was found to be essential for maintaining high levels of frameshifting activity. The effects of these and other mutations which diminished frameshifting efficiency were generally readily rationalized on the basis of a lower predicted stability of the mutant pseudoknots. There was, however, one prominent exception. A series of mutants which contained a single nucleotide insertion at the 5′ end of loop L2, the region which connects stem S1 and the loop L2 nucleotides, increased frameshifting efficiencies by up to 3-fold of the wild-type level (Kim et al., 1999). Why this is the case is unknown, but may derive from an increased stability, through the reduction of the entropic loop closing penalty (Gultyaev et al., 1999), or through favorable interactions with the ribosome (cf. Figure 3). In any case, these findings, which document the functional importance of the identity of loop nucleotides in the BWYV pseudoknot, contrast sharply with previous mutational studies on SRV-1 and IBV where loop L2 sequence substitutions were found to have no significant functional affect Brierley et al 1991, ten Dam et al 1994.
Determinants of the thermodynamic stability of frameshifting pseudoknots
Considerable effort has gone into the study of the folding and unfolding of pseudoknots at equilibrium. These studies are required to determine the contributions of specific structural features to the stability of frameshifting pseudoknots, as well as the degree to which functionally debilitating mutations alter the stability of the pseudoknot relative to the partially folded conformers. Such studies allow meaningful correlations to be drawn between the structure, stability and function of pseudoknot structural domains. In addition, study of equilibrium unfolding pathways provides insight into the highly populated partially folded intermediates which may be encountered by the ribosome, as well as defining the corresponding free energy separation between folded and partially folded states.
Due to the nature of the connectivity of the polynucleotide chain in a pseudoknot, equilibrium unfolding experiments also provide a sensitive reporter on the energetic coupling between distal regions of the molecule, which must be communicated by the connecting single-stranded loops and through the helical junction. This coupling can be strong when the pseudoknot contains a tightly stacked helical junction that may increase the apparent cooperativity of molecular unfolding Draper 1996, Gluick and Draper 1994. In contrast, the coupling of helical unfolding events is anticipated to be weak if the helical junction is not strongly stacked or other noncanonical loop-stem stabilizing interactions are absent in the folded molecule.
Equilibrium unfolding pathways for pseudoknotted RNAs contain multiple unfolding steps
Unlike many single-domain globular proteins, the unfolding of a simple RNA is well-modeled as a series of sequential two-state unfolding steps, where each ith unfolding transition is characterized by an unfolding enthalpy (ΔH i), melting temperature (t m), and via the van’t Hoff relationship, an equilibrium constant K i (Figure 6(a)). Two important pieces of information result from such an analysis: (1) The equilibrium unfolding pathway, which provides a direct indication of the nature of the most stable partially folded intermediates which the translating ribosome might encounter (cf. Figure 6(b)), and (2) The free energy landscape of unfolding and how nucleotide substitutions might alter this landscape. As expected from microscopic reversibility, the equilibrium unfolding pathway will mirror the equilibrium folding pathway, although this has not always been rigorously established.
Figure 6.
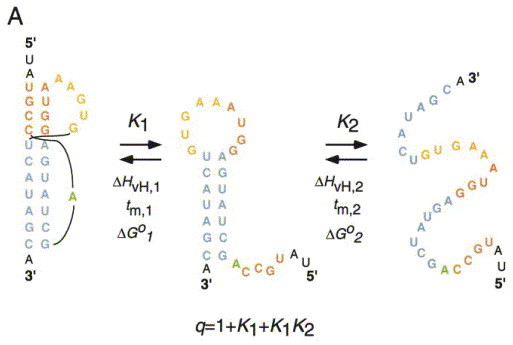
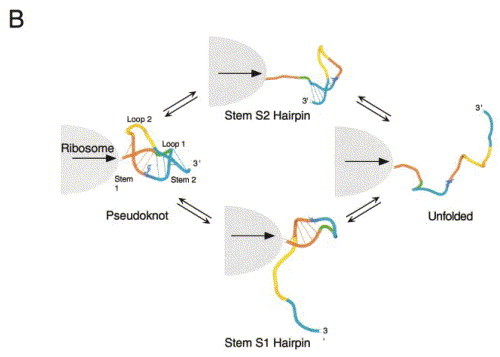
(a) The equilibrium unfolding pathway of the bacteriophage T4 gene 32 autoregulatory mRNA pseudoknot. q is the partition function for this coupled equilibrium with the folded pseudoknot as the reference state. Derived from Theimer et al. (1998). (b) Cartoon representations of two possible unfolding pathways which might occur at the ribosome.
Findings from such an analysis carried out on several pseudoknotted RNA molecules are briefly discussed below.
Natural sequence H-type pseudoknots fold in the absence of divalent cations
Early studies carried out on pseudoknotted RNAs with relatively short helical stems appeared to require a minimal concentration of divalent cation (usually Mg2+) to fold (Puglisi et al., 1988). It is now known that this is not a general feature of frameshifting and autoregulatory pseudoknots which adopt fully folded pseudoknotted conformations in the presence of low concentrations of monovalent salt Gluick and Draper 1994, Gluick et al 1997, Theimer et al 1998, Nixon and Giedroc 1998, Theimer and Giedroc 1999. The presence of high concentrations of monovalent ions or more modest concentrations of divalent and trivalent ions do strongly stabilize these molecules against thermal denaturation without apparently inducing much in the way of thermodynamically detectable tertiary structural or loop-stem interactions. This contrasts sharply with folding studies carried out on more complex RNAs Pan and Sosnick 1997, Laing and Draper 1994, Szewczak et al 1998.
Delocalized or weakly localized mono- or multivalent ions appear to play a major role in stabilizing RNA pseudoknots
RNA is a polyanion at neutral pH, and as such, is associated with a considerable number of positively charged counterions in solution. Metal binding sites in RNAs can be conceptually divided into two general classes (Laing et al., 1994): (1) highly specific complexes of a defined coordination structure and geometry employing both inner sphere and outer sphere coordination bonds; and (2) non-specific complexes which form a delocalized cloud of fully hydrated, mobile ions. However, as discussed below, it may be more appropriate to think of these two classes of ion binding sites as extrema of a continuum of sites and affinities.
Structural and thermodynamic studies with pseudoknotted RNAs are consistent with the idea that Mg2+ and other divalent ions stabilize the pseudoknot by associating as weakly or partially localized ions in regions of the molecule characterized by modestly higher affinities relative to those typically present in an RNA duplex or hairpin structure Gluick et al 1997, Theimer and Giedroc 1999, Nixon and Giedroc 1998, Nixon et al 1999. As a result, the pseudoknotted conformation sequesters slightly more counterions per phosphate at lower metal ion concentrations relative to the partially folded forms of the molecule. This sequestration likely arises from patches of relatively larger negative electrostatic surface potential; recent success in calculating such surfaces has been obtained through numerical solutions of the nonlinear Poisson-Boltzmann equation (Chin et al., 1999). In RNA pseudoknots, these regions might be present at or near the helix-helix junction, where the two loops of the pseudoknot exit and rejoin the duplex regions of the molecule. While these site(s) are of intermediate affinity and bind only weakly localized ions, it is sometimes possible to structurally define these sites in solution using trivalent Co(NH3)6 3+ ions and NMR spectroscopy. Gonzales & Tinoco (1999) successfully identified a binding site for Co(NH3)6 3+ in the major groove, near the base of stem S2, in an ion binding pocket formed by the two nucleotide loop L1 and the major groove of stem S2 of the MMTV-vpk pseudoknot (cf. Figure 4); this “site” was predicted by Brownian motion molecular dynamics simulations (Hermann & Westhof, 1998). Interestingly, a Co(NH3)6 3+ ion(s) also appears localized near the loop L1-stem S2 region in the T2 and T4 gene 32 mRNA pseudoknots as well (Nixon et al., 1999).
The stability of many H-type pseudoknots derives almost exclusively from formation of their secondary structure
Base stacking and hydrogen bonding are the primary determinants of the stability and specificity of folding in simple RNA structures (Turner et al., 1988); not surprisingly, the enthalpy of denaturation of secondary structure dominates the enthalpy term in pseudoknot unfolding (Table 1). With one striking exception (the BWYV pseudoknot, discussed below), this occurs to such a degree that if other pseudoknot loop-stem interactions do form in aqueous solution, they appear to contribute minimally to the net enthalpy term of the stability. The simple prediction from these findings is that in many cases, pseudoknot loops would function only as connecting linkers, substitution of which would be energetically silent. Although the data are limited Theimer et al 1998, Theimer and Giedroc 1999 this is often found not to be the case, revealing that substitutions of loop nucleotides perturb the energetics of loop closure in pseudoknots in a way that is not fully understood. The molecular origin of these perturbations are not known and may have to do with subtle differences in the energetics of the unfolded state, but are often quite significant and can even dictate whether or not the pseudoknotted conformation will be the major conformer at equilibrium at 37 °C Theimer et al 1998, Theimer and Giedroc 1999.
Table 1.
Experimental van’t Hoff (ΣΔHvH) and calorimetric (ΔHcal) enthalpies of unfolding of various RNA pseudoknots compared to predicted enthalpies (ΔHpred) derived from secondary structure unfolding alone (cf. Turner et al., 1988)
| RNA | Equilibrium unfolding pathway | # Transitions | ΣΔHvHa (kcal mol−1) | ΔHcal (kcal mol−1) | ΔHpredb (kcal mol−1) |
|---|---|---|---|---|---|
| T4 gene 32 mRNAc | F ↔ S2 hp ↔ U | 2 | 103 | 98 | 107 |
| T2 gene 32mRNAd | F ↔ J ↔ S1 hp ↔ U | 3 | 119 | 116 | 122 |
| mIAP gag-proe | F ↔ S1/J ↔ I ↔ S1 hp ↔ U | 3f | 141 | 137 | 146 |
| MMTV-vpk gag-prog | F ↔ hp ↔ U | 2 | 105 | 117 | 113 |
| Mo-MLV gag-proh | F ↔ S2 hp ↔ U | 2 | 145 | 165 | 163 |
| BWYVi | F ↔ PK ↔ S1 hp ↔ U | 3 | 115 | 121 | 83 |
Solution Conditions: [Mg2+]⩾2.0 mM; or 1.0 M Na+.
From UV optical melting profiles.
Includes contributions from base stacking within the helical stems, coaxial stacking of the two helices, and stacking of the 3′ single-stranded nucleotides on each helical stem.
Not including contribution from the I ↔ S1 hp transition which reflects the unfolding of structure not present in the pseudoknotted conformation (F).
Gluick et al. (1997). van’t Hoff anlysis of the UV optical melting profiles underestimates the true ΔHcal.
pH = 6.0, 0.5 M K+(Nixon & Giedroc, 2000).
The BWYV pseudoknot is unique in that significant 3° structural stabilization is derived from loop-stem interactions
Equilibrium unfolding studies of wild-type and sequence variants of the BYWV pseudoknot reveal that this RNA exhibits some remarkable stability determinants not previously observed in other pseudoknotted RNAs (Nixon & Giedroc, 2000). The BYWV pseudoknot is characterized a strongly pH-dependent tertiary structural folding, which contributes at pH 6.0 ≈30 kcal mol−1 in unfolding enthalpy (Table 1) and nearly 4 kcal mol−1 in stability beyond that which can be attributed to secondary structure alone (Nixon & Giedroc, 2000). Characterization of wild-type and mutant RNAs as a function of pH suggests that protonation of N3 of C8 in loop L1, to form a third hydrogen bond to the G12-C26 base-pair of stem S2, is solely responsible for the pH-dependence (cf. Figure 5). Disruption of the base-triple interaction by nucleotide substitution reduces the stability of the tertiary structure to that of the wild-type RNA at pH 8, provided a native loop L2 is present. Loop-stem interactions are absolutely required to stabilize the pseudoknotted conformation in the BWYV pseudoknot; this is very likely a consequence of the low intrinsic stability of stem S2, which does not benefit from strong helix-helix stacking with stem S1. Functional studies reveal that the native C8+·G12-C26 base-triple interaction is absolutely required to stimulate frameshifting (Kim et al., 1999).
The helical junction in autoregulatory and retroviral frameshifting pseudoknots is the weak point of these molecules thermodynamically
From work on model systems, it was anticipated that coaxial stacking of the two helices in simple RNA pseudoknots might provide a major stability determinant for these molecules. In a model duplex system which recapitulates the primary sequence determinants of helix-helix stacking in RNA, it was found that the free energy increment for base stacking at a coaxial helix junction consisting of one continuous and one non-continuous RNA strand like that found in a pseudoknot was more stabilizing (by ≈1 kcal mol−1 at 37 °C) than was the same base stack in a continuous helix Walter et al 1994, Walter and Turner 1994. It was therefore surprising that studies of wild-type and variant T4 gene 32 mRNA pseudoknots revealed that compensatory base-pairing mutations introduced into the helical junction were not as destabilizing or stabilizing as predicted on the basis of these model studies (Theimer et al., 1998). In fact, unfolding of the wild-type T2 gene 32 pseudoknot appears to initiate at the junction stack itself, identifying this region as the weakest point in the molecule (Nixon & Giedroc, 1998). NMR structural studies provide a possible explanation for this (Holland et al., 1999). They reveal that the base-pair step at the helical junction at low salt is over-rotated by ≈18 ° in order to remove close approach of phosphate groups in this region; this reduces base-pair stacking at the junction, which would substantially weaken the interface. Consistent with this, recent solvent exchange studies of the T2 pseudoknot reveal that the A15-U28 base-pair (cf. Figure 1) is characterized by a substantially increased base-pair opening frequency relative to other base-pairs of the molecule (J. Lillemoen & D. Hoffman, unpublished results); the same was found for the SRV-1 gag-pro pseudoknot (Du et al., 1997). A weakly base-paired interhelical junction is also consistent with studies which show that the global stability of the helical junction region can be strongly modulated by the nature and number of nucleotides in each of the two loops Theimer et al 1998, Theimer and Giedroc 1999. In addition, in at least one case, the presence of a 3′ dangling single-stranded nucleotide stacked at the base of stem S2 can strongly stabilize the junction region through non-nearest neighbor effects apparently transmitted through the single-nucleotide loop L1 in the T4 gene 32 mRNA pseudoknot (Theimer et al., 1998).
In frameshifting mRNA pseudoknots, the presence of a weakened or non-canonical helical junction is also the rule rather than the exception. For example, a recent investigation of the folding of MMTV gag-pro pseudoknots reveal that the intercalated adenosine contributes 0.7 to 1.4 kcal mol−1 (37 °C) to global stability, depending on the salt concentration (Theimer & Giedroc, 2000). This free energy increment, while significant, is comparable to expectations for a single-stranded adenosine stack on a helical terminus but less than that which would be expected for a strongly coaxially stacked junction. A thermodynamic analysis of the folding of the closely evolutionarily related gag-pro frameshifting pseudoknot from mouse intracisternal A-type particles (mIAP) reveals that the deletion of the analogous adenosine is also destabilizing by ≈0.5 to 1 kcal mol−1 (37 °C) (Theimer & Giedroc, 1999). Functional studies reveal that this deletion reduces frameshifting efficiencies by ten-fold, analogous to previous studies of MMTV gag-pro and MMTV-IBV chimeric pseudoknots (E. Sulistijo, C. Theimer & D. Giedroc, unpublished observations).
Pseudoknot unfolding intermediates and ribosomal frameshifting
All things being equal, an RNA hairpin positioned downstream from the same slippery sequence in exactly the same place as a pseudoknot has been shown not to stimulate high levels of frameshifting. Further, in at least one case, the hairpin has been shown to reduce the degree of ribosomal pausing relative to a pseudoknot containing the same number of base-pairs (by about fivefold) (Somogyi et al., 1993). Thus, as previously suggested by Dinman et al., (1997), it seems reasonable to propose that increasing the amount of time the ribosome remains stalled over the slippery sequence will increase the likelihood that the ribosome will shift backward, irrespective of the actual mechanism. If one further accepts the idea that any unique stabilizing, structural and/or topological features of the pseudoknot are lost upon unfolding to a hairpin intermediate, and that partial unfolding is rate-limiting for frameshifting, it follows that the rate at which the pseudoknot is unfolded to the hairpin intermediate might be tightly tied to the efficiency of recoding.
The extent to which equilibrium unfolding intermediates observed with isolated pseudoknotted RNA fragments are populated during unfolding of the mRNA frameshift signal by the ribosome is obviously not known. However, in order for the ribosome to pass through this structure, at least two mutually exclusive, partially folded pseudoknot folding intermediates could potentially be transiently formed. These are the stem S1 and stem S2 hairpins (Figure 6(b)). Formation of the stem S1 hairpin intermediate requires that the distal stem S2 is denatured first. This might occur through a physical interaction of the paused ribosome with the proximal stem S1 and loop L2, which destabilizes the helical junction and the distal stem S2, resulting in S2 helix unwinding. This in turn removes the topological constraint imposed by the pseudoknot, leaving a partially folded S1 hairpin, which following frameshifting, is denatured and decoded in the normal fashion by the ribosome. This unfolding model predicts that the S1 stem is of a stability appropriate to function as a physical block or a pause determinant (Somogyi et al., 1993) and could occur with or without partial unwinding of stem S1.
This unfolding model sequence characterizes the mIAP gag-pro and BWYV frameshifting pseudoknots, and perhaps the MMTV gag-pro mRNA as well Theimer and Giedroc 1999, Theimer and Giedroc 2000, Nixon and Giedroc 2000. It makes the prediction that frameshifting efficiency will be tightly correlated with the stability of the weakest part of the molecule, e.g. stem S2 in the SRV-1 gag-pro site (ten Dam et al., 1995) or the helical junction in other retroviral frameshifters. It also predicts that there would be no correlation with the stability of stem S1, as already determined in several systems Napthine et al 1999, ten Dam et al 1995. Furthermore, this scenario readily accommodates findings which reveal that, at least in some cases, the pseudoknot loops function as strong thermodynamic coupling agents: mutations in loop L2 of mIAP gag-pro induce a long-range destabilization of stem S2, or more generally the weakest structural determinant in the molecule (Theimer & Giedroc, 1999). This recoding model may well characterize frameshifting pseudoknots that contain a relatively short stem S1, e.g. the luteoviral and retroviral gag-pro pseudoknots, and predicts that destabilization of the helical junction region may have a large impact on frameshifting efficiencies, as has been observed Liphardt et al 1999, Kim et al 1999.
The alternative unfolding pathway, in which a stem S2 hairpin intermediate would be populated at the ribosome, requires that stem S1 is melted first. This might well be operative in frameshifting pseudoknots which are characterized by a distal stem S2 and loop L2 that are not strongly thermodynamically or physically coupled to the S1 stem. Moving the distal stem S2 far from the ribosome would satisfy either criterion, within limits. This could be accomplished by lengthening stem S1 to a full turn of helix like that which appears to be critical for maintaining high efficiency frameshifting in IBV (Napthine et al., 1999). Furthermore, weak coupling of the distal stem S2 and loop L2 to the rest of the pseudoknot could potentially accommodate considerable structural complexity within loop L2 with essentially no effect on frameshifting efficiency, as has been found in IBV and related coronoviruses (Brierley, 1995). Such complexity is not accommodated in retroviral and plant viral frameshifting signals (ten Dam et al., 1995).
Mechanism of stimulation of frameshifting by RNA pseudoknots
How do the downstream pseudoknots stimulate frameshifting? Below we present three simple models, each of which implicates a critical role for the stalling of the elongating ribosome over the slippery sequence upon encountering the pseudoknot. These models are not mutually exclusive, but merely provide a physicochemical view on what features of the structure and stability of the pseudoknot, relative to a simple hairpin, may induce the ribosome to pause, and thus increase the likelihood of shifting reading frames. Other more complex functions of the pseudoknot are of course possible, including allosteric modulation of the peptidyl transferase activity, or the rate of aminoacyl-tRNA·GTP·EF-Tu loading, GTP hydrolysis or release of GDP·EF-Tu, the latter of which is normally rate-limiting (Pape et al., 1998).
Model 1
The structure of the pseudoknot plays a primary role in stimulating frameshifting. In this model, the downstream pseudoknot might function as a binding site for a ribosomal or ribosome-associated protein which directly communicates to the ribosome to stall, or simply lowers the free energy of folding the pseudoknot so that it becomes more difficult to denature (cf. Figure 7(a)). This scenario seems unlikely since the existing structural and functional data suggest that there are two or more clearly distinct structural classes of frameshifting mRNAs, which can be easily distinguished primarily on the basis of the length of stem S1 (Liphardt et al. 1999). In addition, ten Dam et al. (1994) showed that if an exogenous binding factor existed for the SRV-1 gag-pro pseudoknot, the frameshifting site on the mRNA could not be competed out with a large molar excess of short RNAs which contain the SRV-1 pseudoknot, as might be expected by this model.
Figure 7.
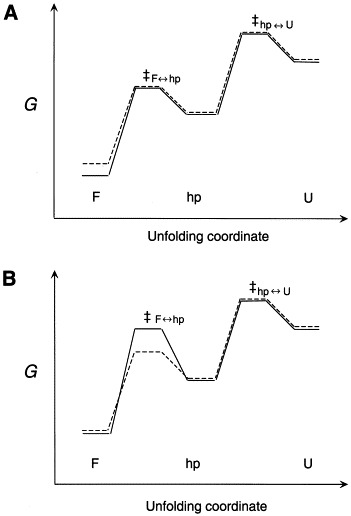
Hypothetical reaction coordinate diagrams which illustrate thermodynamic (a) versus primarily kinetic (b) control of partial unfolding of the pseudoknot. F, folded; hp, hairpin intermediate; U, unfolded RNAs schematized for a wild-type, functional pseudoknot (—) and a weakly destabilized mutant pseudoknot (– – –). In (a), the transition state free energies are identical; thus ΔΔGF↔hp‡ is totally determined by ΔΔGF↔hp or the ground-state structure. In (b), a small ΔΔGF↔hp is shown accompanied by a significant difference in GF↔hp‡ for each of two RNAs such that ΔΔGF↔hp‡ are different. In both cases, a more positive ΔGF↔hp‡ for the functional pseudoknot would slow the rate of interconversion between the F and partially unfolded forms, potentially increasing the efficiency of frameshifting (see the text for details).
Model 2
The stability increment (intrinsic free energy difference) between the folded and partially folded states of the pseudoknot plays a primary role in stimulating frameshifting. This model is easiest to understand from the standpoint of pseudoknot unfolding equilibria (Figure 6). The pseudoknotted conformation is in equilibrium with partially folded stem-loop structures; at a minimum, this conformation must be more stable than the component hairpin structures in order to dominate the population distribution under frameshifting conditions. Thus, beyond this threshold stability, increasing the free energy difference between folded and partially folded structures even slightly might then be expected to further enhance frameshifting efficiency by effectively increasing the kinetic barrier (slowing the rate) to formation of the same partially folded hairpin structure, provided of course transition state energies were of the same energy (Figure 7(a)).
Although the available data are not extensive, most functional and thermodynamic data in the IBV and MMTV systems, respectively, suggest that this free energy increment will not be strongly correlated with frameshifting. For example, in the MMTV gag-pro site, the MMTV-vpk variant and the mutant U13C RNAs (Figure 4) differ in their stabilities by approximately 2 kcal mol−1 (Theimer & Giedroc, 2000); however, Chen et al. (1995) report identical frameshifting efficiencies for these two molecules. This is inconsistent with simple expectations of this model. This contrasts with the situation in the HIV-1 stem-loop stimulatory element, in which a correlation was found between global stability of the hairpin and frameshifting efficiency, within certain boundaries (Bidou et al., 1997).
Model 3
Differential transition state energy barriers dictated by small differences in local structure, stability or dynamics are a primary determinant of frameshifting efficiency. There is virtually no support for or against this model, since experiments along these lines have yet to be carried out. However, pseudoknots differing in the presence of even a small number of loop-stem interactions at the helical junction versus a helical junction with significant flexibility might be expected to subtly differ in their rates of unfolding, in the absence of large, measurable differences in free energies of the folded versus the partially folded intermediates. In this scenario, the efficiency of frameshifting would be kinetically, rather than thermodynamically, controlled. A reaction coordinate diagram which illustrates this idea is shown in Figure 7(b).
For example, increasing the activation energy barrier by only 1.0 kcal mol−1 at 37 °C would result in a decrease in the rate of unfolding by as much as fivefold; if the rate of unfolding is directly tied to frameshifting efficiency, this would lead to frameshifting efficiencies which differ by as much of fivefold, a large and easily measurable effect. Indeed, this is nearly the extent to which the IBV-MMTV chimeric pKA-A and pKA-G pseudoknots differ in their frameshifting efficiencies (cf. Figure 4) (Liphardt et al., 1999). The disruption of a favorable interaction involving A29 in pKA-A by substitution with G29 in pKA-G might make the helical junction more dynamically mobile, thereby lowering the transition state energy barrier for stem-loop unfolding.
Future studies
Many questions remain regarding the details of the mechanism by which RNA pseudoknots stimulate ribosomal recoding events. Clearly, additional high resolution structural studies of evolutionary diverse frameshift-stimulating pseudoknots, such as the minimal IBV sequence, would lend evidence for or against the hypothesis that clearly dissimilar structural motifs are capable of stimulating high levels of frameshifting. Structural studies should be complemented by systematic thermodynamic and kinetic analyses, to probe for correlations between stability, unfolding pathway and functional activity. Detailed studies of the base and backbone-specific motional dynamics of the helical junction regions in similar molecules using NMR approaches in partially oriented samples (Hansen et al., 1998), heteronuclear NMR relaxation experiments (Akke et al., 1997) and steady-state and time-resolved fluorescence techniques (Eis & Millar, 1993) are likely to shed significant molecular insight into the nature of the helix-helix junction, as well as the structure and dynamics of the globular conformation of the molecule, and how this might differ between functional and weakly functional pseudoknots.
In another potentially informative line of investigation, the rates of interconversion between fully and partially folded pseudoknots could be investigated, in solution (cf. Wyatt et al., 1990) and possibly at the ribosome. These interconversion rates could be compared with the rate of ribosome movement as the pseudoknotted region is being translated, as well as the kinetics of unwinding of pseudoknot stem S1 or stem S2. Experiments designed to probe the nature of pseudoknot-ribosome interactions at nucleotide resolution, and how these might change during the recoding process, are also of interest, even more so now that the high resolution structure of the ribosome is forthcoming Clemons et al 1999, Ban et al 1999, Cate et al 1999. In addition, the future identification and characterization of an 80S ribosome-associated RNA helicase(s), and how the rate of helix unwinding and substrate specificity differs for pseudoknotted versus hairpin-like RNA substrates is a particularly critical area for future investigation. Further biochemical characterization of maintenance-of-frame (mof) mutants already isolated from Saccharomyces cerevisiae which modulate the degree of frameshifting in ScV/L-A will also surely be informative with regard to cellular factors might be influencing this process (Dinman & Wickner, 1994). Indeed, some of these have mapped to mutations in the yeast homologue of EF-Tu (Dinman & Kinzy, 1997).
All of these questions represent fundamental issues in RNA function, structure, stability and dynamics that are of high interest in and of themselves, but also represent approaches which might help resolve outstanding mechanistic issues regarding the fascinating process of ribosomal recoding.
Acknowledgements
Work in the author’s laboratory on RNA pseudoknots is supported by a grant from the NIH to D. P. G. and David W. Hoffman (AI-40187). We thank our colleagues Drs David Hoffman, Tao Pan, Victoria DeRose and Art Johnson for helpful comments on the manuscript. We would also like to express our appreciation to Dr Joachim Frank for the cryo-EM electron density map of the E. coli ribosome which was used in the modeling experiments.
Footnotes
Edited by D. E. Draper
References
- Akke M, Fiala R, Jiang F, Patel D, Palmer A.G.I. Base dynamics in a UUCG tetraloop RNA hairpin characterized by 15N spin relaxation: Correlations with structure and stability. RNA. 1997;3:702–709. [PMC free article] [PubMed] [Google Scholar]
- Alam S.L, Atkins J.F, Gesteland R.F. Programmed ribosomal frameshifting: much ado about knotting! Proc. Natl Acad. Sci. USA. 1999;96:14177–14179. doi: 10.1073/pnas.96.25.14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.L, Wills N.M, Ingram J.A, Atkins J.F, Gesteland R.F. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J. Mol. Biol. 1999;288:837–852. doi: 10.1006/jmbi.1999.2713. [DOI] [PubMed] [Google Scholar]
- Atkins J.F, Weiss R.B, Gesteland R.F. Ribosome gymnastics: degree of difficulty 9.5, style 10.0. Cell. 1990;62:413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Capel M, Moore P.B, Steitz T.A. Placement of protein and RNA structures into a 5 Å-resolution map of the 50 S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- Bidou L, Stahl G, Grima B, Liu H, Cassan M, Rousset J.-P. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA. 1997;3:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- Brierley I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Brierley I, Digard P, Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Rolley N.J, Jenner A.J, Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Jenner A.J, Inglis S.C. Mutational analysis of the “slippery-sequence” component of the coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H, Yusupov M.M, Yusupova G.Z, Earnest T.H, Noller H.F. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- Chamorro M, Parkin N, Varmus H.E. An RNA pseudoknot required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl Acad. Sci. USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chamorro M, Lee S.I, Shen L.X, Hines J.V, Tinoco I, Jr, Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kang H, Shen L.X, Chamorro M, Varmus H.E, Tinoco I., Jr A characteristic bent conformation of RNA pseudoknots promotes −1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- Chin K, Sharp K.A, Honig B, Pyle A.M. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nature Struct. Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- Clemons W.M, Jr, May J.L.C, Wimberley B.T, McCutcheon J.P, Capel M.S, Ramakrishnan V. Structure of a bacterial 30 S ribosomal subunit at 5.5 Å resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- Cui Y, Dinman J.D, Kinzy T.G, Peltz S.W. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol. Cell. Biol. 1998;18:1506–1516. doi: 10.1128/mcb.18.3.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas A, Moore P.B. The loop E-loop D region of Escherichia coli: 5 S rRNA: the solution structure reveals an unusual loop that may be important for binding ribosomal proteins. Structure. 1997;5:1639–1653. doi: 10.1016/s0969-2126(97)00311-0. [DOI] [PubMed] [Google Scholar]
- Dinman J.D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D, Kinzy T.G. Translational misreading: mutations in translation elongation factor 1-alpha differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D, Wickner R.B. Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered −1 frameshifting efficiencies. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D, Icho T, Wickner R.B. A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl Acad. Sci. USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D, Ruiz-Echevarria M.J, Czaplinski K, Peltz S.W. Peptidyl-transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: development of model systems. Proc. Natl Acad. Sci. USA. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D, Ruiz-Echevarria M.J, Peltz S.W. Translating old drugs into new treatments: Ribosomal frameshifting as a target for antiviral drugs. Trends Biotechnol. 1998;16:190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A, Cate J.H. RNA structure: crystal clear? Curr. Opin. Stru. Biol. 1997;7:310–316. doi: 10.1016/s0959-440x(97)80045-0. [DOI] [PubMed] [Google Scholar]
- Draper D.E. Pseudoknots and control of protein synthesis. Curr. Opin. Cell Biol. 1990;2:1099–1103. doi: 10.1016/0955-0674(90)90162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. Strategies for RNA folding. TIBS. 1996;21:145–149. [PubMed] [Google Scholar]
- Dreher T.W, Tsai C, Skuzeski J.M. Aminoacylation identity switch of turnip yellow mosaic virus RNA from valine to methionine results in an infectious virus. Proc. Natl Acad. Sci. USA. 1996;93:12212–12216. doi: 10.1073/pnas.93.22.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Hoffman D.W. An NMR and mutational study of the pseudoknot within the gene 32 mRNA of bacteriophage T2: insights into a family of structurally related RNA pseudoknots. Nucl. Acids Res. 1997;25:1130–1135. doi: 10.1093/nar/25.6.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Giedroc D.P, Hoffman D.W. Structure of the autoregulatory pseudoknot within the gene 32 messenger RNA of bacteriophages T2 and T6: a model for a possible family of structurally related RNA pseudoknots. Biochemistry. 1996;35:4187–4198. doi: 10.1021/bi9527350. [DOI] [PubMed] [Google Scholar]
- Du Z, Holland J.A, Hansen M.R, Giedroc D.P, Hoffman D.W. Base-pairings within the RNA pseudoknot associated with the simian retrovirus-1 gag-pro frameshift site. J. Mol. Biol. 1997;270:464–470. doi: 10.1006/jmbi.1997.1127. [DOI] [PubMed] [Google Scholar]
- Ehresmann C, Philippe C, Westhof E, Benard L, Portier C, Ehresmann B. A pseudoknot is required for efficient translational initiation and regulation of the Escherichia coli rpsO gene coding for ribosomal protein S15. Biochem. Cell Biol. 1995;73:1131–1140. doi: 10.1139/o95-122. [DOI] [PubMed] [Google Scholar]
- Eis P.S, Millar D.P. Conformational distributions of a four-way DNA junction revealed by time-resolved fluorescence resonance energy transfer. Biochemistry. 1993;32:13852–13860. doi: 10.1021/bi00213a014. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.-X, Yuan H, Rein A, Levin J.G. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J. Virol. 1992;66:5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D’Amaré A.R, Zhou K, Doudna J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata R.K, Agrawal R.K. A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature. 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- Frank J, Verschoor A, Li Y, Zhu J, Lata R.K, Radermacher M, Penczek P, Grassucci R, Agrawal R.K, Srivastave S. A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem. Cell Biol. 1995;73:757–765. doi: 10.1139/o95-084. [DOI] [PubMed] [Google Scholar]
- Garcia A, van Duin J, Pleij C.W.A. Differential response to frameshift signals in eukaryotic and prokaryotic translational systems. Nucl. Acids Res. 1993;21:401–406. doi: 10.1093/nar/21.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R.F, Atkins J.F. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- Gilley D, Blackburn E.H. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl Acad. Sci. USA. 1999;96:6621–6625. doi: 10.1073/pnas.96.12.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluick T.C, Draper D.E. Thermodynamics of folding a pseudoknotted mRNA fragment. J. Mol. Biol. 1994;241:246–262. doi: 10.1006/jmbi.1994.1493. [DOI] [PubMed] [Google Scholar]
- Gluick T.C, Wills N.M, Gesteland R.F, Draper D.E. Folding of an mRNA pseudoknot required for stop codon readthrough: effects on mono- and divalent ions on stability. Biochemistry. 1997;36:16173–16186. doi: 10.1021/bi971362v. [DOI] [PubMed] [Google Scholar]
- Gonzales R.L, Jr, Tinoco I., Jr Solution structure and thermodynamics of a divalent metal ion binding site in an RNA pseudoknot. J. Mol. Biol. 1999;289:1267–1282. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- Grentzmann G, Ingram J.A, Kelly P.J, Gesteland R.F, Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Gultyaev A.P, van Batenberg F.H.D, Pleij C.W.A. An approximation of loop free energy values of RNA H-pseudoknots. RNA. 1999;5:609–617. doi: 10.1017/s135583829998189x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass E.S, Brown J.W, Pitulle C, Pace N.R. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl Acad. Sci. USA. 1994;91:2527–2531. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.C. Transfer RNA-like structures in viral genomes. Internatl. Rev. Cytol. 1979;60:1–26. doi: 10.1016/s0074-7696(08)61257-7. [DOI] [PubMed] [Google Scholar]
- Hansen M.R, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nature Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- Hatfield D, Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. TIBS. 1990;15:186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T, Patel D.J. Stitching together RNA tertiary architectures. J. Mol. Biol. 1999;294:829–849. doi: 10.1006/jmbi.1999.3312. [DOI] [PubMed] [Google Scholar]
- Hermann T, Westhof E. Exploration of metal ion binding sites in RNA folds by Brownian-dynamics simulations. Structure. 1998;6:1303–1314. doi: 10.1016/s0969-2126(98)00130-0. [DOI] [PubMed] [Google Scholar]
- Holland J.A, Hansen M.R, Du Z, Hoffman D.W. An examination of coaxial stacking of helical stems in a pseudoknot motif: the gene 32 messenger RNA pseudoknot of bacteriophage T2. RNA. 1999;5:257–271. doi: 10.1017/s1355838299981360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung M, Patel P, Davis S, Green S.R. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 1998;72:4819–4824. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri E, Aigner S, Cech T.R. Kinetic and secondary structure analysis of Naegleria andersoni GIR1, a group I ribozyme whose putative biological function is site-specific hydrolysis. Biochemistry. 1997;36:16345–16354. doi: 10.1021/bi9718595. [DOI] [PubMed] [Google Scholar]
- Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr. Top. Microbiol. Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- Jacks T, Townsley K, Varmus H.E, Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc. Natl Acad. Sci. USA. 1987;84:4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Madhani H.D, Masiarz F.R, Varmus H.E. Signals for ribosomal frameshifting in the Rous Sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Power M.D, Masiarz F.R, Lucius P.A, Barr P.J, Varmus H.E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Kang H, Tinoco I., Jr A mutant RNA pseudoknot that promotes ribosomal frameshifting in mouse mammary tumor virus. Nucl. Acids Res. 1997;25:1943–1949. doi: 10.1093/nar/25.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Hines J.V, Tinoco I., Jr Conformation of a non-frameshifting RNA pseudoknot from mouse mammary tumor virus. J. Mol. Biol. 1996;259:135–147. doi: 10.1006/jmbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- Kim Y.-G, Su L, Maas S, O’Neill A, Rich A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl Acad. Sci. USA. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk M.H, van der Graaf M, Wijmenga S.S, Pleij C.W.A, Heus H.A, Hilbers C.W. NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science. 1998;280:434–438. doi: 10.1126/science.280.5362.434. [DOI] [PubMed] [Google Scholar]
- Kurland C.G. Translational accuracy and the fitness of bacteria. Annu. Rev. Genetics. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- Laing L.G, Draper D.E. Thermodynamics of RNA folding in a conserved ribosomal RNA domain. J. Mol. Biol. 1994;237:560–576. doi: 10.1006/jmbi.1994.1255. [DOI] [PubMed] [Google Scholar]
- Laing L.G, Gluick T.C, Draper D.E. Stabilization of RNA structure by Mg ions, specific and nonspecific effects. J. Mol. Biol. 1994;237:577–587. doi: 10.1006/jmbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- Larsen B, Gesteland R.F, Atkins J.F. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50 % J. Mol. Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J, Napthine S, Kontos H, Brierley I. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters D.S, Stormo G.D, Gold L. Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J. Mol. Biol. 1988;201:517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- Miller W.A, Dinesh-Kumar S.P, Paul C.P. Luteovirus gene expression. Crit. Rev. Plant Sci. 1995;14:179–211. [Google Scholar]
- Morikawa S, Bishop D.H.L. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:387–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameki N, Felden B, Atkins J.F, Gesteland R.F, Himeno H, Muto A. Functional and structural analysis of a pseudoknot upstream of the Tag-encoded sequence in E. coli tmRNA. J. Mol. Biol. 1999;286:733–744. doi: 10.1006/jmbi.1998.2487. [DOI] [PubMed] [Google Scholar]
- Napthine S, Liphardt J, Bloys A, Routledge S, Brierley I. The role of RNA pseudoknot stem 1 length in the promotion of efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P.L, Giedroc D.P. Equilibrium unfolding (folding) pathway of a model H-type pseudoknotted RNA: the role of magnesium ions in stability. Biochemistry. 1998;37:16116–16129. doi: 10.1021/bi981726z. [DOI] [PubMed] [Google Scholar]
- Nixon P.L, Giedroc D.P. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- Nixon P.L, Theimer C.A, Giedroc D.P. Thermodynamics of stabilization of RNA pseudoknots by cobalt(III) hexaammine. Biopolymers. 1999;50:443–458. doi: 10.1002/(SICI)1097-0282(19991005)50:4<443::AID-BIP9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Pan T, Sosnick T.R. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nature Struct. Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M.V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N.T, Chamorro M, Varmus H.E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C.W.A. Pseudoknots: a new motif in the RNA game. TIBS. 1990;15:143–147. doi: 10.1016/0968-0004(90)90214-v. [DOI] [PubMed] [Google Scholar]
- Pleij C.W.A. RNA pseudoknots. Curr. Opin. in Stru. Biol. 1994;4:337–344. [Google Scholar]
- Pleij C.W.A, Rietveld K, Bosch L. A new principle of RNA folding based on pseudoknotting. Nucl. Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C.W.A, Bosch L. RNA pseudoknots: structure, detection, and prediction. Methods Enzymol. 1989;180:289–303. doi: 10.1016/0076-6879(89)80107-7. [DOI] [PubMed] [Google Scholar]
- Powers T, Noller H.F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J.D, Wyatt J.R, Tinoco I., Jr A pseudoknotted RNA oligonucleotide. Nature. 1988;331:283–286. doi: 10.1038/331283a0. [DOI] [PubMed] [Google Scholar]
- Retberg C.C, Prere M.F, Gesteland R.F, Atkins J.F, Fayet O. A three-way junction and constituent stem-loops as the stimulator for programmed: −1 frameshifting in bacterial insertion sequence I5911. J. Mol. Biol. 1999;286:1365–1378. doi: 10.1006/jmbi.1999.2546. [DOI] [PubMed] [Google Scholar]
- Rietveld K, Van Poelgeest R, Pleij C.W, Van Boom J.H, Bosch L. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucl. Acids Res. 1982;10:1929–1946. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K, Pleij C.W.A, Bosch L. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 1983;2:1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus J.D, Ladner J.E, Finch J.T, Rhodes D, Brown R.S, Clark B.F.C, Klug A. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Szewczak A.A, Podell E.R, Bevilacqua P.C, Cech T.R. Thermodynamic stability of the P4-P6 domain RNA tertiary structure measured by temperature gradient gel electrophoresis. Biochemistry. 1998;37:11162–11170. doi: 10.1021/bi980633e. [DOI] [PubMed] [Google Scholar]
- Schimmel P. RNA pseudoknots that interact with the components of the translation apparatus. Cell. 1989;58:9–12. doi: 10.1016/0092-8674(89)90395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.X, Tinoco I., Jr The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Jenner A.J, Brierley I, Inglis S.C. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Chen L, Egli M, Berger J.M, Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nature Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D, Kang H. Mutational analysis of the RNA pseudoknot involved in efficient ribosomal frameshifting in simian retrovirus-1. Nucl. Acids Res. 1998;26:1369–1372. doi: 10.1093/nar/26.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.K, Draper D.E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989;57:531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- ten Dam E.B, Pleij K, Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- ten Dam E, Brierley I, Inglis S, Pleij C. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucl. Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E, Verlaan P.W.G, Pleij C.W.A. Analysis of the role of the pseudoknot component in the SRV-1 gag-pro ribosomal frameshift signal: loop lengths and stability of the stem regions. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A, Giedroc D.P. Equilibrium unfolding pathway of an H-type RNA pseudoknot which promotes programmed −1 ribosomal frameshifting. J. Mol. Biol. 1999;289:1283–1299. doi: 10.1006/jmbi.1999.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A, Giedroc D.P. Contribution of the intercalated adenosine at the helical junction to the stability of the gag-pro frameshifting pseudoknot from mouse mammary tumor virus. RNA. 2000;6:409–442. doi: 10.1017/s1355838200992057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A, Wang Y, Hoffman D.W, Krisch H.M, Giedroc D.P. Non-nearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J. Mol. Biol. 1998;279:545–564. doi: 10.1006/jmbi.1998.1812. [DOI] [PubMed] [Google Scholar]
- Tu C.-L, Tzeng T.-H, Bruenn J.A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl Acad. Sci. USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.H, Sugimoto N, Freier S.M. RNA structure prediction. Annu. Rev. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Tzeng T.-H, Tu C.-L, Bruenn J.A. Ribosomal frameshifting requires a pseudoknot in the Saccharomyces cerevisiae double-stranded RNA virus. J. Virol. 1992;66:999–1006. doi: 10.1128/jvi.66.2.999-1006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A.E, Turner D.H. Sequence dependence of stability for coaxial stacking of RNA helixes with Watson-Crick base-paired interfaces. Biochemistry. 1994;33:12715–12719. doi: 10.1021/bi00208a024. [DOI] [PubMed] [Google Scholar]
- Walter A.E, Turner D.H, Kim J, Lyttle M.H, Muller P, Mathews D.H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl Acad. Sci. USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Masiquida B, Jaeger L. RNA tectonics: toward RNA design. Folding Design. 1996;1:78–88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- Wills N.M, Gesteland R.F, Atkins J.F. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl Acad. Sci. USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills N.M, Gesteland R.F, Atkins J.F. Pseudoknot dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 1994;17:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Nix J, Szostak J. Functional requirements for specific ligand recognition by a biotin-binding RNA pseudoknot. Biochemistry. 1998;37:14410–14419. doi: 10.1021/bi981371j. [DOI] [PubMed] [Google Scholar]
- Wyatt J.R, Puglisi J.D, Tinoco I., Jr RNA pseudoknots: stability and loop size requirements. J. Mol. Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]


