Abstract
The equilibrium unfolding pathway of a 41-nucleotide frameshifting RNA pseudoknot from the gag-pro junction of mouse intracisternal A-type particles (mIAP), an endogenous retrovirus, has been determined through analysis of dual optical wavelength, equilibrium thermal melting profiles and differential scanning calorimetry. The mIAP pseudoknot is an H-type pseudoknot proposed to have structural features in common with the gag-pro frameshifting pseudoknots from simian retrovirus-1 (SRV-1) and mouse mammary tumor virus (MMTV). In particular, the mIAP pseudoknot is proposed to contain an unpaired adenosine base at the junction of the two helical stems (A15), as well as one in the middle of stem 2 (A35). A mutational analysis of stem 1 hairpins and compensatory base-pair substitutions incorporated into helical stem 2 was used to assign optical melting transitions to molecular unfolding events. The optical melting profile of the wild-type RNA is most simply described by four sequential two-state unfolding transitions. Stem 2 melts first in two closely coupled low-enthalpy transitions at low tm in which the stem 3′ to A35, unfolds first, followed by unfolding of the remainder of the helical stem. The third unfolding transition is associated with some type of stacking interactions in the stem 1 hairpin loop not present in the pseudoknot. The fourth transition is assigned to unfolding of stem 1. In all RNAs nvestigated, ΔHvH ≈ΔHcal, suggesting that ΔCp for unfolding is small. A35 has the thermodynamic properties expected for an extrahelical, unpaired nucleotide. Deletion of A15 destabilizes the stem 2 unfolding transition in the context of both the wild-type and ΔA35 mutant RNAs only slightly, by ΔΔG°≈1 kcal mol−1 (at 37 °C). The ΔA15 RNA is considerably more susceptible to thermal denaturation in the presence of moderate urea concentrations than is the wild-type RNA, further evidence of a detectable global destabilization of the molecule. Interestingly, substitution of the nine loop 2 nucleotides with uridine residues induces a more pronounced destabilization of the molecule (ΔΔG°≈2.0 kcal mol−1), a long-range, non-nearest neighbor effect. These findings provide the thermodynamic basis with which to further refine the relationship between efficient ribosomal frameshifting and pseudoknot structure and stability.
Keywords: RNA pseudoknot, RNA folding, thermodynamics, nucleic acid stability, frameshifting
Abbreviations: UTR, untranslated region; IRE, iron-responsive element; IBV, infectious bronchitis virus; ScV, Saccharomyces cerevisiae virus; mIAP, mouse intracisternal A-type particle; RSV, Rous sarcoma virus; MMTV, mouse mammary tumor virus; SRV-1, simian retrovirus 1; MuLV, Moloney murine leukemia virus; Mops, 3-N-morpholino]propanesulfonic acid; PAGE, polyacrylamide gel electrophoresis; DEPC, diethylpyrocarbonate
Introduction
Our understanding of the role of RNA in biological systems has continued to expand as more catalytic RNAs, RNA structural motifs, and non-canonical interactions have been discovered and characterized (for a review, see Conn & Draper, 1998). It has become clear that mRNAs contain secondary and tertiary structures both in the coding and non-coding regions and that these interactions are critical to regulation of translation in both prokaryotic and eukaryotic systems. mRNA structures can regulate translation during initiation, elongation, and termination or by affecting the rate of mRNA degradation. Regulation of translation initiation has been observed when proteins interact with 5′ untranslated region (UTR) mRNA structures: for example, a hairpin in the case of the iron-responsive element (IRE) on the ferritin mRNA and an RNA pseudoknot in the case of the Escherichia coliα operon (Tang & Draper, 1990). Hairpins and pseudoknots have also been implicated in ribosomal frameshifting and hopping during elongation, stop codon readthrough at termination and regulation of mRNA degradation (for a review, seeGesteland & Atkins, 1996).
RNA pseudoknots are simple tertiary structures composed of single-stranded loop regions and helical stems. The H-type or hairpin-type pseudoknot consists of a hairpin that forms the 5′ helical stem of the pseudoknot and the first loop of the pseudoknot. This is followed by a single-stranded region which forms the second loop of the pseudoknot and a region complementary to part of the hairpin loop sequence, forming the second helical stem of the pseudoknot (Figure 1). The lengths of the loops and stems vary, with certain minimal loop lengths required to span specific helical stem lengths Pleij et al 1985, Wyatt et al 1990. The helical stems can be coaxially stacked Du and Hoffman 1997, Holland et al 1999 or bent Shen and Tinoco 1995, Su et al 1999 with respect to one another.
Figure 1.
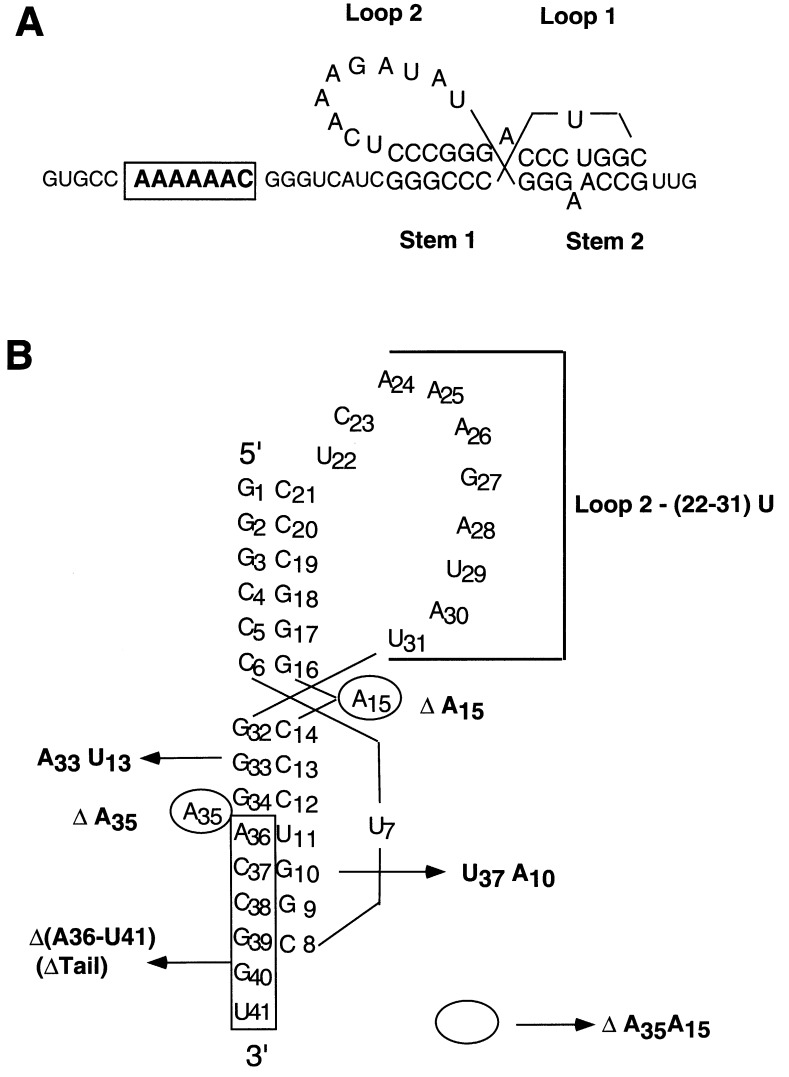
(a) The mIAP gag-pro frameshift site (Fehrmann et al., 1997). The slip site heptamer is enclosed in a box and the pseudoknot structural elements are annotated. The structure of this pseudoknot has yet to be determined. The adenosine at the junction (A15), shown unpaired in this model, may be paired with the final uridine in loop 2 (U31) and it is not clear which of the two adenosine bases (A35 and A36) in stem 2 is paired with U11. (b) Mutations incorporated in the mIAP pseudoknot sequence. The double mutant, ΔA35A15 RNA, is indicated by the open circles.
Ribosomal frameshifting occurs when a translating ribosome changes reading frame at a specific site in an mRNA causing the synthesis of an altered protein product. Frameshifting can occur in either direction to the −1, −2, or +1 frames Matsufuji et al 1996, Farabaugh 1996. The −1 ribosomal frameshifting is a common mechanism used by RNA viruses to regulate the synthesis level of certain proteins and to ensure packaging of pol and pro gene products by expressing them as fusion proteins with the gag gene capsid protein. The frequency at which frameshifting occurs is variable, from 1-5 % in some plant viruses up to 50 % in the dnaX frameshift site of E. coli (Larsen et al., 1997) with typical retroviral efficiencies between 15 and 30 % Brierley 1995. The −1 ribosomal frameshift site is typically composed of a heptameric nucleotide sequence X XXY YYZ in the zero coding frame. The identities of the nucleotides vary in different sites and between prokaryotes and eukaryotes. The primary sequence requirement appears to be that at least two of the three mRNA codon-tRNA anticodon base-pairing interactions can be maintained in the new frame (XXX YYY Z).
The simultaneous slippage model was proposed to describe the mechanism of −1 ribosomal frameshifting from studies of the gag-pol junction frameshift site in Rous sarcoma virus (RSV) (Jacks et al., 1988). In this model, both the A and P sites of the ribosome contain tRNAs and the frameshift occurs when both the A and P-site tRNAs slip into the −1 frame on the slip site heptamer. Sites with high frameshifting efficiencies typically have mRNA structures required for maintaining high efficiency located three to nine nucleotides downstream of the slippery heptamer. In many, though not all cases, the structural element downstream of the slippery site is proposed to form an H-type RNA pseudoknot. One explanation is that these RNA structures simply slow or stall the ribosome temporarily, increasing the amount of time the ribosome spends on the slippery sequence, and thereby increasing the probability of a frameshifting event.
Ribosomal pausing mediated by RNA structure has been observed for the pseudoknots in the Saccharomyces cerevisiae virus (ScV) at the cap-pol junction (Tu et al., 1992) and also in the coronavirus infectious bronchitis virus (IBV) at the 1a-1b junction (Somogyi et al., 1993). In ScV, ribosomal pausing was assayed using “heelprinting” to locate regions of the mRNA protected from nuclease digestion in the presence of translating ribosomes. Ribosomal pausing was monitored at the IBV frameshift site by examining the size and distribution of incomplete polypeptide intermediates generated from initiation-synchronized in vitro translation over time. Pausing was shown to correlate well with frameshift efficiency but both studies observed structures with normal pausing but no detectable frameshifting. This suggests that ribosomal pausing is necessary, but not sufficient, for frameshifting. In addition, thermodynamic stability of the RNA structure alone apparently cannot account for frameshifting efficiency at the ScV and IBV frameshift sites, although this point has not been thoroughly investigated.
Structurally, the most well-characterized frameshifting pseudoknot occurs at the gag-pro frameshift site in mouse mammary tumor virus (MMTV). This pseudoknot has been subjected to mutational analysis and structure probing Chamorro et al 1992, Chen et al 1995 and the solution structure has been solved by NMR (Shen & Tinoco, 1995). In addition, the structures of frameshifting and non-frameshifting variants have also been determined Chen et al 1996, Kang et al 1996, Kang and Tinoco 1997. Frameshifting efficiency in these MMTV pseudoknot derivatives was found to correlate with a specifically bent conformation, where the two helical stems were prevented from coaxially stacking by a single unpaired adenosine at the helical junction. An extensive mutational study was also performed on the pseudoknot at the gag-pro frameshift site in simian retrovirus 1 (SRV-1) (ten Dam et al., 1995). The frameshifting efficiency of base-pair substitutions in the second stem of the pseudoknot correlated qualitatively with the predicted thermodynamic stability of that stem. NMR evidence suggests that the closing A-U base-pair in stem 2 at the helical junction is formed (Du et al., 1997) while mutational analysis suggests that the identity of the two nucleotides and their ability to base-pair have no affect on frameshifting efficiency Chen et al 1996, Sung and Kang 1998, prompting these authors to hypothesize that the SRV-1 pseudoknot is also bent, like in MMTV.
These studies reveal that a detailed investigation of a number of closely related frameshifting pseudoknots will be required to map the general determinants of frameshifting efficiency. Here, we have examined the thermodynamic stability of the pseudoknot which promotes efficient −1 frameshifting from the gag-pro frameshift site of mouse intracisternal A-type particles (mIAP) (Fehrmann et al., 1997). The proposed pseudoknot shares some similarities with the SRV-1 and MMTV pseudoknots, including potential structural ambiguity at the junction of the two helical stems in which A15 may or may not base-pair with U31 in loop 2 (Figure 1(a)). mIAP is an endogenous mouse retrovirus which phylogenetic analysis of the nucleic acid and amino acid sequences of the pol gene reveals is a member of a closely related subclass of retroviruses that includes both SRV-1 and MMTV (Li et al., 1995). Study of mIAP will therefore allow us to examine the general structural and stability features of this group of retroviral frameshifting pseudoknots.
Results
In order to define the thermodynamics of unfolding for the mIAP frameshifting pseudoknot, the compensatory base-pair substitution and deletion mutants shown in Figure 1(b)were prepared and subjected to thermal denaturation. Unless otherwise indicated, all thermal melts were collected in 50 mM monovalent K+ and no added divalent cation, so that the complete melting profile could be obtained in the accessible temperature range (5-120 °C).
Optical melting profiles of the wild-type mIAP pseudoknot and stem 1 hairpins
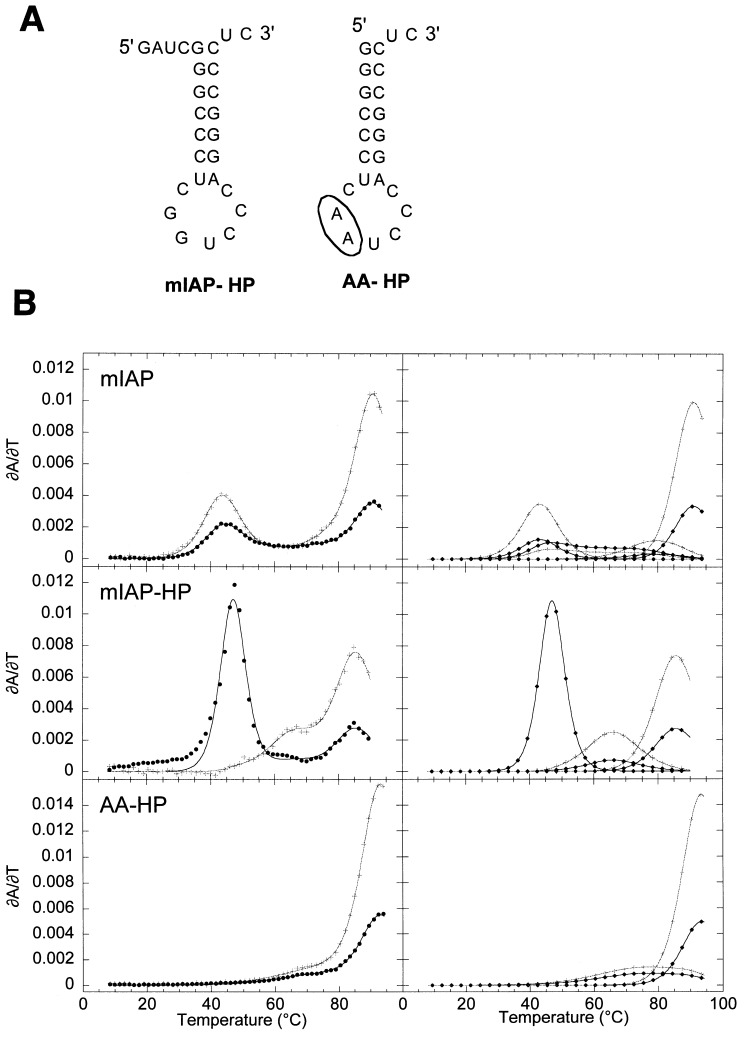
The optical melting profiles acquired at 260 nm and 280 nm (plotted as ∂A/∂T versus T) (Banerjee et al., 1993) for the wild-type 41-nucleotide mIAP pseudoknot and two stem 1 hairpins in the absence of exogenous Mg2+ are shown in Figure 2(b) with the thermodynamic parameters compiled in Table 1. Unless otherwise noted, the experimental data are presented in the left-hand side of each panel and are superimposed with a continuous curve which describes an optimized non-linear least-squares fit to an unfolding model consisting of a set of sequential, interacting, two-state transitions. The individual transition fits are shown on the right-hand side of the Figure for clarity. Initially, melts were collected on a wild-type mIAP pseudoknot and mIAP-HP with a short single-stranded 5′ tail. The 5′ single-stranded tail was omitted from the mIAP wild-type pseudoknot RNA and subsequent mutant RNAs when it was observed that it did not significantly affect the stability of the pseudoknot as determined from optical melting profiles (data not shown).
Figure 2.
(a) The sequences of the mIAP pseudoknot stem 1 hairpin RNAs. (b) Melting profiles of the wild-type mIAP pseudoknot and two stem 1 hairpins at 50 mM KCl in the absence of exogenously added Mg2+. On the left, the composite fits are superimposed on the experimental data. On the right, the individual transitions of the composite fit are shown for clarity. Black (•) and gray-scale (+) correspond to ∂A/∂T monitored at 260 nm and 280 nm, respectively.
Table 1.
Summary of the thermodynamic parameters obtained for the unfolding of stem assignment mutants at 50 mM K+, pH 7.0
| Transition 1 |
Transition 2 |
Transition 3 |
Transition 4 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNA | ΔHvHT | ΔHvH | tm | 260/280a | ΔHvH | tm | 260/280 | ΔHvH | tm | 260/280 | ΔHvH | tm | 260/280 |
| mIAP | 181 | 47.7 | 43.9 | 0.36 | 27.7 | 47.0 | 1.29 | 38.6 | 70.0 | 0.98 | 66.9 | 92.4 | 0.36 |
| mIAP-HP | 99 | 76.3 | 47.0 | (260)b | 42.2 | 66.2 | 0.26 | 57.1 | 85.8 | 0.36 | |||
| AA-HP | 86 | 24.9 | 76.2 | 0.65 | 61.6 | 93.0 | 0.33 | ||||||
| A33 U13 | 182 | 36.8 | 32.4 | 0.84 | 16.0 | 38.8 | 0.8 | 44.7 | 78.5 | (280)b | 62.0 | 86.7 | 0.30 |
| U37 A10 | 168 | 41.8 | 36.7 | 1.35 | 20.5 | 52.1 | 1.73 | 40.4 | 76.0 | 0.53 | 65.0 | 91.0 | 0.31 |
| Δ tailc | 126 | 27.6 | 38.4 | 1.44 | 36.0 | 69.5 | 0.74 | 62.1 | 91.7 | 0.37 | |||
| 5 mM Mg2+ | 37.9 | 68.2 | 0.95 | 46.5 | 86.3 | 0.70 | - | - | - | ||||
Enthalpy is in kcal mol−1 and tm is in °C. ΔHvHT is the total van’t Hoff enthalpy summed over all four transitions.
The 260/280 ratio represents (∂A260/∂T)/(∂A280/∂T), the ratio of the hyperchromic effect at 260 and 280 nm.
260/280 ratios in parentheses indicate that hyperchromicity was observed only at the wavelength in parentheses.
The Δ tail data are shown at 0 and 5 mM Mg2+ as the enthalpy of the first transition increased with Mg2+ concentration. At 5 mM Mg2+, the final transition is off-scale and cannot be measured optically.
The wild-type mIAP pseudoknot optical melting profile is shown in Figure 2(b). The profile exhibits hyperchromicity in two transitions, a broad low intensity transition between 30 and 50 °C and a sharper, higher intensity transition above 80 °C, both with significant hyperchromicity at 280 nm as expected for the high G-C content of the two helical stems (Fresco et al., 1963). Even though initial inspection gives the appearance of only two transitions, it was necessary to fit the data to four sequential two-state transitions in order to eliminate the periodic deviation in the residuals observed when fitting to fewer transitions. Turner rules compiled at 1 M NaCl predict an enthalpy of 68 and 77 kcal mol−1 for the melting of stems 1 and 2, respectively, provided seven base-pairs are formed in stem 2 and the 3′ single-stranded nucleotide stacks against the stem Turner et al 1988, He et al 1991, Xia et al 1998. The predicted stabilization due to stacked Watson-Crick base-pairs was calculated using the modified individual nearest-neighbor model (INN-HB) parameters which account for the difference in stabilization between internal and closing A-U base-pairs (Xia et al., 1998). The thermodynamic parameters for hairpin and bulge loops reported byTurner et al. (1988) were used. In good agreement with these predictions, the first two transitions have a combined ΔH=75 kcal mol−1 and the final transition has an ΔH=66.7 kcal mol−1, suggesting that stem 2 unfolds first in two steps with stem 1 unfolding at much higher temperature.
Two stem 1 hairpin RNAs were constructed to unambiguously define the optical signature of stem 1 unfolding:these are mIAP-HP RNA, which consists of the first 23 nucleotides of the mIAP pseudoknot sequence, and AA-HP RNA, a variant of the mIAP-HP RNA in which the hairpin loop nucleotides 9 and 10 were converted from guanosine to adenosine as shown in Figure 2(a). Unexpectedly, the optical melting profile of the mIAP-HP shows three distinct transitions. The first transition is unusual in that it has significant hyperchromicity (∂A/∂T) only at 260 nm with a very high enthalpy (ΔH=76 kcal mol−1) at t m =47 °C. The second transition has an ΔH=42 kcal mol−1 and an (∂A 260/∂T)/(∂A 280/∂T) value (A 260/A 280 ratio) of ≈0.26, while the third transition is characterized by ΔH=57 kcal mol−1 and A 260/A 280 ≈0.36. The melting profile was superimposable in both the folding and unfolding directions (data not shown). The combined enthalpy of these transitions is 175 kcal mol−1, much larger than would be predicted from the nearest-neighbor model for the stem 1 hairpin (64 kcal mol−1). The molecular origin of the first unfolding transition is unknown; however, it is observed calorimetrically as well with essentially identical van’t Hoff thermodynamic parameters (data not shown). The second transition shows a concentration dependence over the 2-55 μM range consistent with predictions for formation of some kind of intermolecular structure. The high enthalpy and t m and low A 260/A 280 ratio of the third optical transition are consistent with predictions for the unfolding of a stem consisting of six G-C base-pairs.
In an attempt to simplify the hairpin melting profile, a second stem 1 hairpin (AA-HP) was constructed with the 5′ single-stranded tail eliminated and two G→A substitutions in the hairpin loop to potentially disrupt any loop structure. As shown in Figure 2(b), the melting profile is simplified with the complete disappearance of the first mIAP-HP transition and a significant reduction in the intensity of the second unfolding transition. The optical signature of the final transition was identical in the two hairpin RNAs and in the wild-type mIAP pseudoknot and was therefore assigned to the unfolding of stem 1. We interpret the penultimate transition as some type of unstacking in the loop, the intensity of which is strongly affected by the identity of the loop purines.
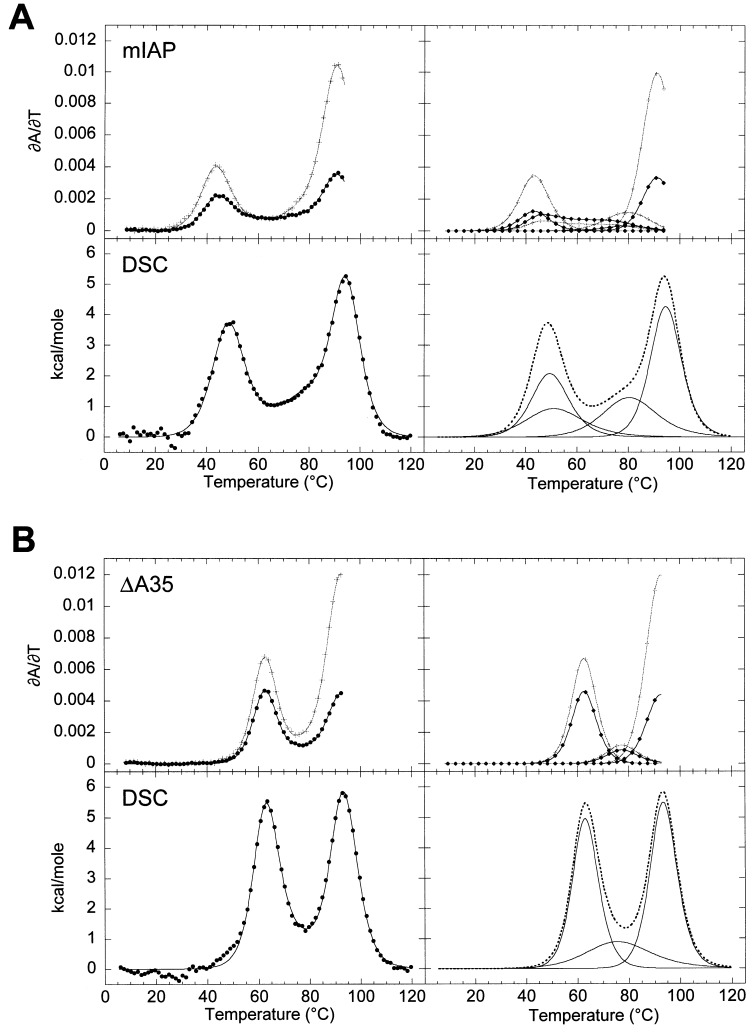
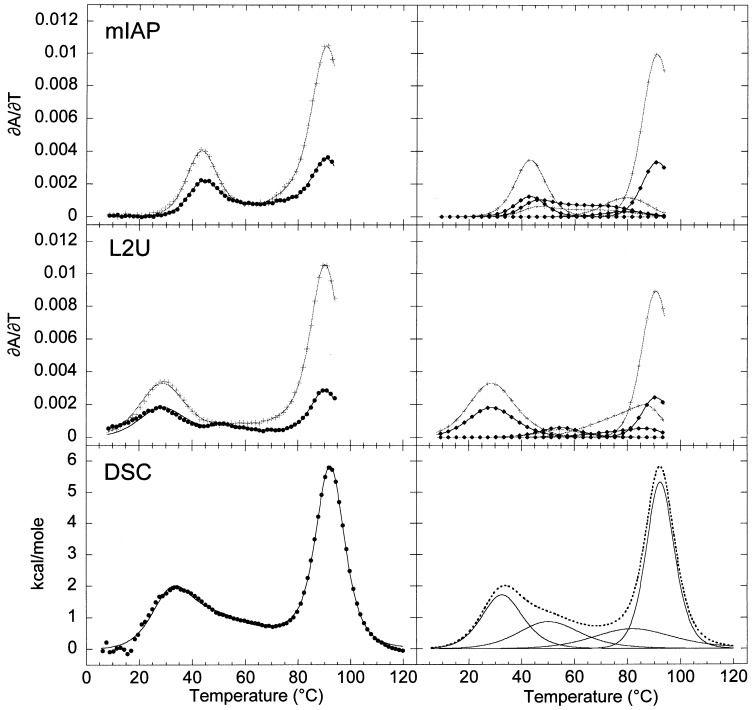
Differential scanning calorimetry (DSC) was performed on the wild-type mIAP pseudoknot and compared to that obtained for a nucleotide deletion mutant, termed ΔA35 RNA, in which a stem 2 unpaired adenosine is deleted and all seven possible stem 2 base-pairs are predicted to form (Figure 1(b)). Comparisons of the optical and calorimetric melting profiles for these two RNAs are shown in Figure 3 with the fitted thermodynamic parameters from the optical and calorimetric data presented in Table 2. The calorimetric and optical melting profiles were fit independently and returned internally consistent thermodynamic parameters for the individual unfolding transitions, with the caveat that the enthalpies and t m values of the final transitions are more accurately determined from calorimetry, since the optical data could only be collected to 95 °C.
Figure 3.
Optical and calorimetric melting profiles for (a) the mIAP pseudoknot RNA and (b) the deletion mutant, ΔA35 RNA. The composite fit is superimposed on the calorimetry data ( •) on the left, with the individual transitions shown for clarity on the right with the composite fit shown with a broken line. Optical data are represented as described previously with black ( •) and gray-scale (+) corresponding to ∂A/∂T monitored at 260 nm and 280 nm, respectively.
Table 2.
Summary of the thermodynamic parameters obtained for the unfolding of bulged adenosine and loop 2 mutants at 50 mM K+, pH 7.0
| Transition 1 |
Transition 2 |
Transition 3 |
Transition 4 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNA | ΔHcala | ΔHvHT | ΔHvH | tm | 260/280b | ΔHvH | tm | 260/280 | ΔHvH | tm | 260/280 | ΔHvH | tm | 260/280 | |
| mIAP | UV | 181 | 47.7 | 43.9 | 0.36 | 27.7 | 47.0 | 1.29 | 38.6 | 70.0 | 0.98 | 66.9 | 92.4 | 0.36 | |
| cal | 169 | 173 | 40.5 | 49.1 | 27.7 | 51.0 | 36.0 | 80.2 | 68.6 | 94.3 | |||||
| Δ A15 | UV | 168 | 46.2 | 43.5 | 0.24 | 19.9 | 45.8 | 1.15 | 40.0 | 81.8 | 0.40 | 62.3 | 92.9 | 0.32 | |
| cal | 159 | 165 | 38.2 | 44.2 | 30.2 | 48.2 | 27.8 | 71.9 | 68.3 | 93.1 | |||||
| ΔA35 | UV | 161 | 66.6 | 62.7 | 0.67 | 30.0 | 76.7 | 0.74 | 64.6 | 92.7 | 0.35 | ||||
| cal | 172 | 169 | 64.7 | 63.1 | 30.9 | 77.8 | 73.4 | 93.3 | |||||||
| ΔA35A15 | UV | 146 | 57.3 | 58.8 | 0.55 | 30.0 | 71.5 | 0.72 | 58.8 | 90.6 | 0.31 | ||||
| cal | 140 | 138 | 52.8 | 61.5 | 23.2 | 74.7 | 62.3 | 91.6 | |||||||
| L2U | UV | 162 | 30.8 | 29.0 | 0.55 | 40.0 | 55.0 | 1.27 | 26.9 | 82.5 | 0.28 | 64.5 | 89.6 | 0.27 | |
| cal | 160 | 160 | 36.1 | 32.8 | 28.3 | 52.7 | 32.0 | 88.4 | 63.3 | 91.6 | |||||
Enthalpy is reported in kcal mol−1 and tm is reported in °C. For each RNA, thermodynamic parameters determined from optical melting (UV) are presented in the first row and from calorimetry (cal) in the second. ΔHvHT is the total van’t Hoff enthalpy summed over the four transitions.
Hcal is a model-independent enthalpy derived from the differential scanning calorimetry data.
The 260/280 ratio represents (∂A260/∂T)/(∂A280/∂T), the ratio of the hyperchromic effect at 260 and 280 nm.
The optical and calorimetric melting profiles of the ΔA35 RNA (shown in Figure 3(b)) were fit to three sequential transitions. The second and third transitions were observed to be consistent with the hairpin loop unstacking and stem 1 unfolding transitions previously assigned in the wild-type pseudoknot. The first unfolding transition at 63 °C is assigned to unfolding of stem 2. Comparing the wild-type mIAP and ΔA35 RNAs, the transitions observed at lower t m differ significantly in hyperchromic intensity. However, the first ΔA35 RNA transition and the combined first and second mIAP RNA transitions return the same total calorimetric enthalpy as determined by the area under the curve. The calorimetric enthalpy observed for these two molecules confirms that stem 2 unfolding in the wild-type mIAP pseudoknot occurs in two tightly coupled transitions with base stacking interactions similar to that observed in the ΔA35 RNA. More importantly, the van’t Hoff. Enthalpics determined for each transition from calorimetry and from optical melts are in excellent agreement with one another and, whem summed, agree well with the osberved total calorimetric enthalpy. This suggests that ΔC p for the overall unfolding process is small. As discussed below, the impact of this A35 deletion on the global stability of the molecule is essentially as predicted for an extrahelical, unpaired nucleotide.
Optical melting profiles of the stem assignment mutant RNAs
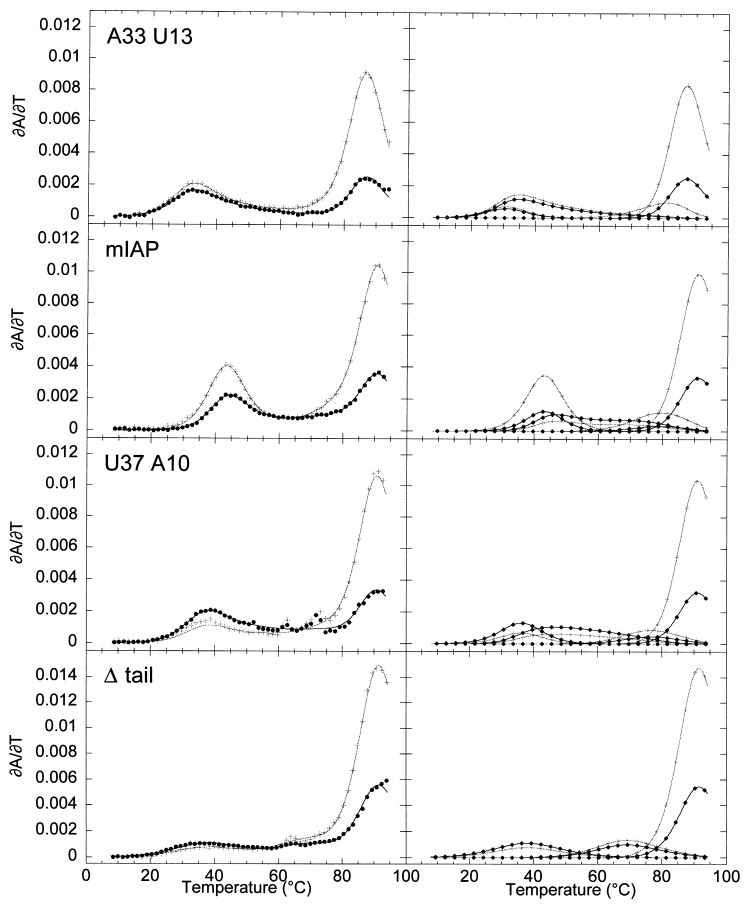
In order to deconvolute the two stem 2 unfolding transitions of the mIAP pseudoknot and assign each to the unfolding of one of the two substems of stem 2 (Figure 1(b)), an RNA in which the 3′ five nucleotides (C37 to U41) were deleted, Δ tail, was prepared. In addition, two stem 2 base-pair substitution RNAs were constructed as well (Figure 1(b)). The stem 2 variants were A33 U13 RNA, which substitutes an A-U base-pair for a G-C base-pair above the unpaired adenosine base, and U37 A10 RNA, which substitutes an A-U base-pair for a G-C base-pair below the unpaired adenosine base. The optical melting profiles acquired at 260 nm and 280 nm for the stem 2 variants and the wild-type mIAP pseudoknot in the absence of exogenously added Mg2+ are shown in Figure 4 with the thermodynamic parameters compiled in Table 1. As expected, in all three cases the compensatory stem 2 mutations primarily affect the low temperature transitions with the hairpin loop and stem 1 unfolding transitions essentially as observed in the wild-type pseudoknot.
Figure 4.
Melting profiles of the wild-type mIAP pseudoknot and stem assignment mutant RNAs at 50 mM KCl in the absence of exogenously added Mg2+. On the left, the composite fits are superimposed on the experimental data. On the right, the individual transitions of the composite fit are shown for clarity. Black ( •) and gray-scale (+) correspond to ∂A/∂T monitored at 260 and 280 nm, respectively.
The 3′ base-pair substitution mutant RNA, U37 A10, has a significantly different melting profile relative to the wild-type pseudoknot in the stem 2 transition region. The enthalpies of both transitions are decreased by 6-8 kcal mol−1. The first transition in U37 A10 occurs at a lower t m and has an inverted 260/280 ratio compared to the wild-type RNA as well as a significantly lower intensity. The second transition occurs at a higher t m than wild-type RNA also with a slightly increased 260/280 ratio. Turner rules predict an enthalpy of 75 kcal mol−1 for the substituted stem 2, 12 kcal mol−1 more than is observed experimentally for the sum of the first two transitions Turner et al 1988, Xia et al 1998. This mutation therefore destabilizes the first transition both enthalpically and entropically, suggesting that the first transition corresponds to unfolding of the substem 3′ to the unpaired adenosine base in stem 2. The entropic destabilization of the second transition is likely due to the tight coupling of the first two transitions on the temperature coordinate, since it reports on the unfolding of the same secondary structural unit.
Like the U37 A10 RNA, unfolding of the A33 U13 RNA shows significant perturbation of both stem 2 transitions. Although modified Turner rules predict an enthalpy of 73 kcal mol−1 for the melting of stem 2 (Xia et al., 1998), only 53 kcal mol−1 are recovered experimentally under these conditions. Both transitions show a decrease in van’t Hoff enthalpy by ≈10 kcal mol−1 and a decrease in t m by 10 deg. C with respect to the wild-type RNA. The resolved A 260/A 280 ratios of both transitions are significantly affected, increasing in the first transition and decreasing in the second. One explanation for the effect this mutation has on both transitions is that the lower half of stem 2 is stabilized by stacking interactions with the upper half as exhibited by the tight coupling of the two transitions on the temperature coordinate. Destabilization of the upper half of the stem as measured in the second optical unfolding transition would cause an observed destabilization of transition 1 and the lower half of the stem due to the cooperativity of unfolding the helical stem.
The 3′ tail deleted molecule, Δ tail RNA, is predicted to have a single transition at low t m and should model the unfolding of the upper substem of stem 2 as the base-pairs derived from the lower half of stem 2 cannot form. The enthalpy and A 260/A 280 ratio are consistent with those observed for the second pseudoknot unfolding transition although the t m is lower. The predicted enthalpy for three G-C base-pairs, an A-U base-pair, and a stacked 3′ adenosine base is 40 kcal mol−1. The enthalpy of the first transition in this molecule increases from 28 to 38 kcal mol−1 when ⩾1 mM Mg2+ is added, in accord with predictions. The unfolding enthalpies of the wild-type pseudoknot transitions are not Mg2+-dependent up to 1 mM Mg2+(data not shown).
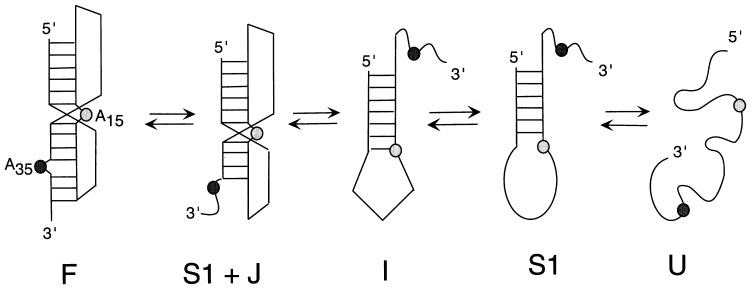
Our detailed investigation of the stem-assignment mutant and stem 1 hairpin RNAs make it possible to assign a temperature-induced equilibrium unfolding pathway for the mIAP gag-pro frameshifting pseudoknot as shown in Figure 5. The unfolding proceeds through four optically and calorimetrically defined steps. The fully folded pseudoknot (F) unfolds initially at the four 3′ base-pairs of stem 2 below the unpaired adenosine (S1+J), followed by the rest of the helical stem 2 to form an intermediate (I), which consists of the stem 1 hairpin and some structure in the hairpin loop. The hairpin loop structure is then unfolded to give the stem 1 hairpin (S1) and finally the fully unfolded form (U):F ⇔ S1+J ⇔ I ⇔ S1 ⇔ U The stability of the hairpin loop structure is not expected to contribute to the overall pseudoknot stability as the loop structure must be disrupted in order to form the stem 2 base-pairing in the pseudoknot.
Figure 5.
The proposed equilibrium unfolding pathway of the mIAP pseudoknot based on a multiple, interacting, sequential two-state transition analysis of the optical and calorimetric unfolding data.
Mg2+-dependence of the thermodynamics of unfolding of the mIAP pseudoknot
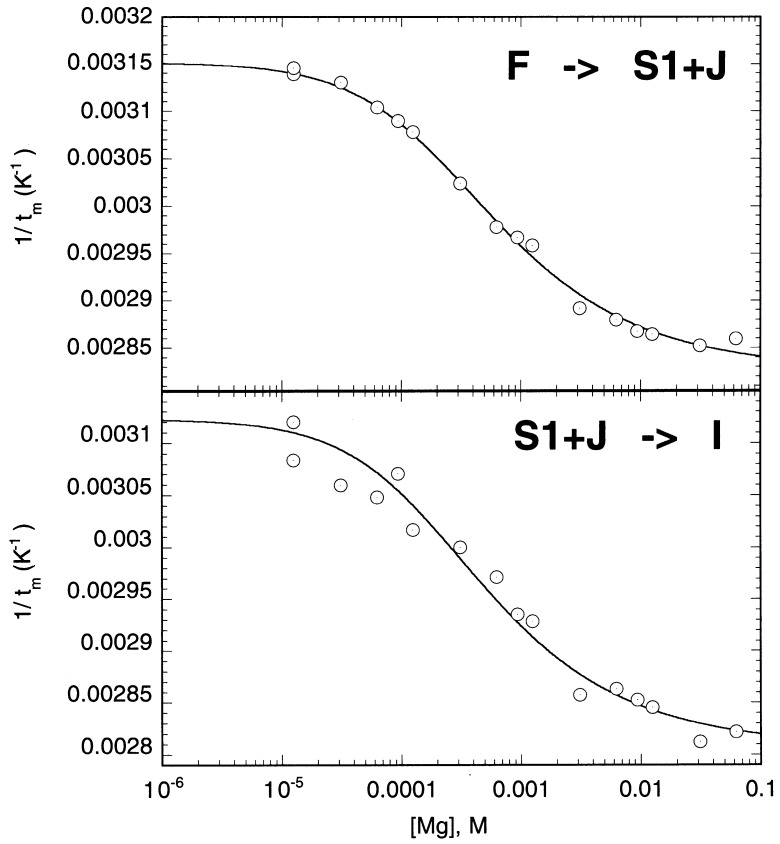
All of the above unfolding experiments were carried out in 50 mM KCl (pH 7.0) in the absence of added divalent cation. Under these conditions, all of the RNA molecules studied here appear fully folded, since the total recovered unfolding enthalpy is comparable to nearest-neighbor predictions and does not change greatly with added Mg2+ (Table 2). To further probe the stabilization of the wild-type mIAP pseudoknot by Mg2+, optical melts were collected over a wide range of Mg2+ concentrations (50 μM-50 mM) in this background of 50 mM KCl. The data are shown in Figure 6 for the F→S1+J and S1+J→I transitions in the format of a 1/t m versus log[Mg2+] plot. Given the non-linear dependence of 1/t m on log[Mg2+] for these two unfolding transitions, the data were fit to a non-specific binding model for divalent ions in which the folded and unfolded forms of the RNA participating in the transition have non-negligible affinities for Mg2+ Laing et al 1994, Nixon and Giedroc 1998, Gluick et al 1997. The number of phosphate groups participating in these two unfolding steps was fixed at 8 and 6, respectively. The magnesium binding affinities for the folded (K f) and unfolded (K u) forms were found to be 3500(±200) M−1 and 500(±40) M−1, respectively, for the first transition and 3500(±600) M−1 and 700(±150) M−1 for the second transition. The non-specific model fits the data extremely well with a total of ≈2.3 Mg2+ released upon unfolding of the pseudoknot to the partially folded stem 1 hairpin intermediate at 0.4 mM Mg2+. For comparison, the unfolding of a seven base-pair hairpin releases ≈1 Mg2+ at slightly higher monovalent salt (0.10 M K+), suggesting that the fully and partially folded pseudoknot sequesters slightly more Mg2+ than do similarly sized RNA hairpins (Nixon et al., 1999).
Figure 6.
Effect of [Mg2+] on the tm for the first two unfolding transitions in the wild-type mIAP pseudoknot. The dependence of 1/tm on [Mg2+] for the first transition (F→S1+J) is shown at the top and for the second transition (S1+J→I) at the bottom (seeFigure 5). The continuous line represents the fit of the experimental data to a non-specific binding model (Nixon & Giedroc, 1998).
Optical and calorimetric melting of unpaired adenosine deletion mutant RNAs
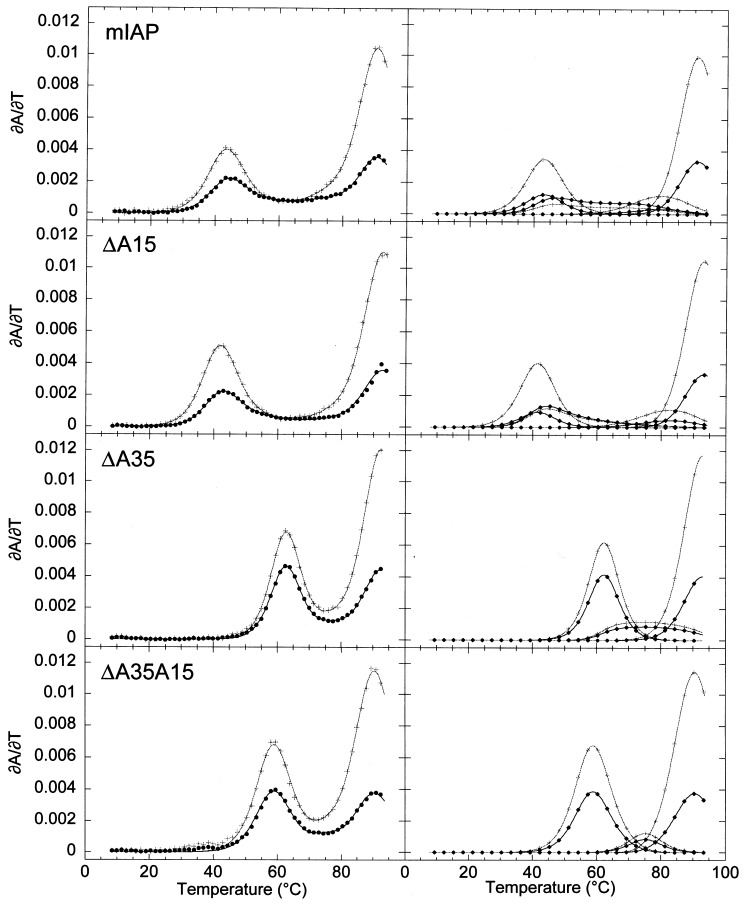
Three unpaired adenosine deletion molecules were characterized to determine the extent to which these nucleotides contribute to the thermodynamic stability of the mIAP frameshifting pseudoknot. These mutations are ΔA15 RNA, where the putative unpaired adenosine base at the junction has been removed, Delta;A35 RNA, where the extrahelical adenosine base in the discontinuous seven base-pair stem 2 has been removed, and the double-mutant, ΔA35A15 RNA, in which both adenosine bases are removed. Optical melting profiles for these RNAs are shown in Figure 7 with optical and calorimetric data compiled in Table 2.
Figure 7.
Melting profiles of the wild-type mIAP pseudoknot and unpaired adenosine deletion mutant RNAs at 50 mM KCl in the absence of exogenously added Mg2+. On the left, the composite fits are superimposed on the experimental data. On the right, the individual transitions of the composite fit are shown for clarity. Black (•) and gray-scale (+) correspond to ∂A/∂T monitored at 260 nm and 280 nm, respectively.
The ΔA15 mutation occurs at the junction of the two stems and it was therefore not possible to predict the affect that this mutation would have on the stability of the pseudoknot structure. The mutation could be stabilizing if the deletion allows coaxial stacking across the junction otherwise not present, neutral if A15 has no affect on pseudoknot stability, or destabilizing if A15 is involved in stacking of the helical stems, tertiary interactions, or maintenance of the correct junction conformation through an A15-U31 base-pair or other interactions. This is a particularly interesting mutant, since the analogous adenosine residue in the MMTV gag-pro pseudoknot is wedged between the two pseudoknot stems leading to a bent structure (Shen & Tinoco, 1995).
The melting profile of the ΔA15 RNA is very similar to that of the wild-type pseudoknot. The optical profile shows a small reduction in the enthalpy and t m of both stem 2 transitions. The calorimetry data also show some destabilization of the first two transitions reflected in both t m and enthalpy, with a total 10 kcal mol−1 decrease in the calorimetric enthalpy of the ΔA15 RNA (Table 2) and a free energy difference ΔΔG°(37 °C)≅1 kcal mol−1 (Table 3) compared to the wild-type RNA. This suggests that A15 plays a small role in stabilization of the pseudoknotted structure. In addition, investigation of the Mg2+-dependence of the unfolding of the ΔA15 RNA reveals that the binding affinities for Mg2+ determined for the ΔA15 RNA are virtually identical with those observed for the wild-type mIAP RNA, with the same net release of Mg2+ upon unfolding of stem 2 (data not shown).
Table 3.
Calculated and predicted thermodynamic parameters for the unfolding of the unpaired adenosine and loop 2 mutants at 50 mM K+, pH 7.0
| RNA | Stem | ΔHstema | ΔHpredb | ΔG° | ΔG°predc | ΔΔG°wt-mutant |
|---|---|---|---|---|---|---|
| mIAP | 2 | 75.4 (68.2) | 77.3 | 1.9 (2.7) | 5.3 | - |
| 1 | 66.9 (68.6) | 68.4 | 10.1 (10.7) | 12.3 | - | |
| Δ A15 | 2 | 66.1 (68.4) | 77.3 | 1.5 (1.9) | 5.3 | 0.4 (0.8) |
| 1 | 62.3 (68.3) | 68.4 | 9.5 (10.5) | 12.3 | 0.6 (0.2) | |
| Δ A35 | 2 | 66.6 (64.7) | 81.0 | 5.1 (5.0) | 9.0 | −3.2(-2.3) |
| 1 | 64.6 (73.4) | 68.4 | 9.8 (11.3) | 12.3 | 0.3 (−0.6) | |
| Δ A35A15 | 2 | 57.3 (52.8) | 81.0 | 3.8 (3.9) | 9.0 | −1.9 (−1.2) |
| 1 | 58.8 (62.3) | 68.4 | 8.7 (9.3) | 12.3 | 1.4 (1.4) | |
| L2U | 2 | 70.8 (64.4) | 77.3 | 1.4 (0.9) | 5.3 | 0.5 (1.8) |
| 1 | 64.5 (63.3) | 68.4 | 9.4 (9.5) | 12.3 | 0.7 (1.2) | |
| AA-HP | 1 | 61.6 | 68.4 | 9.4 | 12.3 | 0.6 |
Enthalpy and free energy are reported in kcal mol−1 for the sum of unfolding transitions 1, 2 and 4. ΔG° values are calculated at 37 °C. Parameters determined from calorimetry data are shown in parentheses.
The two stem 2 transition enthalpies are summed for mIAP, ΔA15, and L2U.
Does not include a contribution from the helical junction (ΔH=10.6 kcal mol−1) but does include a contribution from the 3′ stack.
Calculated at 1.0 M Na+. Incorporates a fixed value of 9 kcal mol−1 for the pseudoknot loop closing entropy estimated from the unfolding of H-type pseudoknots previously studied Theimer et al 1998, Nixon and Giedroc 1998. Previous studies show that the free energy calculated for a simple hairpin at 100 mM KCl is 2-4 kcal mol−1 smaller than that observed at high Mg2+ (equivalent stabilization to high monovalent concentrations) (Theimer et al., 1998); thus, the experimental ΔG° values are in good agreement with predicted ΔG° values.
The ΔA35 deletion RNA was predicted to allow stacking of both substems of stem 2, which could then potentially unfold in one cooperative unit exhibiting a single optical transition with a predicted enthalpy of 81 kcal mol−1 in 1 M NaCl. Both calorimetry and optical melting data reveal that this transition melts in a single cooperative transition at 63 °C with an enthalpy of 65-70 kcal mol−1 measured in 50 mM KCl (pH 7.0), qualitatively in accordance with predictions.
As expected, the double deletion mutant RNA, ΔA35A15 RNA, shows a single stem 2 transition, similar to the single mutant ΔA35 RNA. Interestingly, both stem 2 and stem 1 are destabilized enthalpically and in t m compared to the ΔA35 deletion alone. In particular, stem 2 in the double mutant appears enthalpically destabilized by ≈12 kcal mol−1 with the total calorimetric enthalpy for the pseudoknot transitions 23 kcal mol−1 less than observed for the ΔA35 RNA. This suggests that the junction of the two stems causes a somewhat larger enthalpic destabilization of stem 2 in the context of the ΔA35 mutation than that observed in the wild-type pseudoknot; however, in free energy, the overall effect is comparable in magnitude to the ΔA15 mutation.
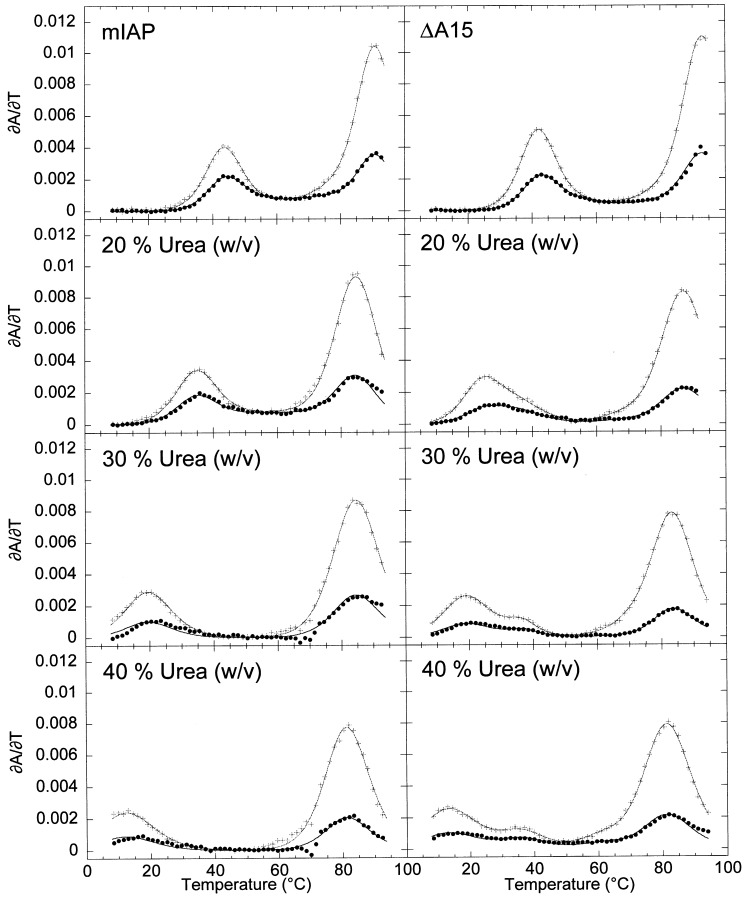
Optical melting profiles were collected in the presence of urea to more completely determine the thermodynamic parameters for unfolding for, in particular, the high temperature transitions. Urea was used to decrease the t m of the transitions in a pseudoknot involved in stop-codon readthrough at the gag-pol junction in the Moloney murine leukemia virus mRNA (MuLV) (Gluick et al., 1997). In that study, a linear and shallow dependence of the t m on the urea concentration was observed, while the van’t Hoff enthalpies were found to decrease by only a small extent. When the wild-type and ΔA15 pseudoknots were thermally denatured in the presence of increasing concentrations of urea, significant differences in their melting profiles were observed as shown in Figure 8. In the wild-type pseudoknot, the two low temperature transitions remain tightly coupled and appear to move together to lower t m, although urea-induced destabilization of stem 2 is considerably more pronounced than that of stem 1. In the ΔA15 RNA, both stem 2 unfolding transitions move to lower t m at low urea concentrations; at greater than 20 % (w/v) urea, the first transition continues to move to lower t m, while the second transition appears to stop, allowing clear separation of the two stem 2 transitions on the temperature coordinate. Although the molecular origins of these effects of urea are unknown, these data provide additional evidence that the helical junction regions of these two pseudoknots may well be different, with the ΔA15 deletion modestly destabilizing at low urea and ultimately stabilizing in high urea.
Figure 8.
Urea dependence of the optical melting profiles for the mIAP pseudoknot and he deletion mutant ΔA15 RNAs in 50 mM KCl. (a) Wild-type mIAP in the left column and (b) ΔA15 in the right column at 0, 20, 40, and 50 % (w/v) urea. Black ( •) and gray-scale (+) correspond to ∂A/∂T monitored at 260 and 280 nm, respectively with the composite fits superimposed on the experimental data.
Optical melting profile of a loop 2 substitution mutant
Finally, a loop 2 substitution mutant RNA, L2U, was constructed to determine if the sequence of loop 2 contributes in any significant way to pseudoknot stability. The nine nucleotide loop sequence was changed to uridine bases as outlined in Figure 1(b), which would be expected to eliminate any significant helix-single-strand tertiary contacts (Wyatt et al., 1990). The optical and calorimetric profiles for L2U are shown compared to the wild-type pseudoknot in Figure 9 with the thermodynamic data compiled in Table 2, Table 3. This substitution has a dramatic impact on the appearance of the melting profile of the molecule. Analysis of the optical melting profile shows that the F→S1+J transition is significantly destabilized with the van’t Hoff enthalpy and t m decreased by 17 kcal mol−1 and 15 deg. C, respectively, while the ΔH vH and the t m of the S1+J→I transition appear to increase by 12 kcal mol−1 and 8 deg. C, respectively. The calorimetric data show trends that mirror the optical data and emphasize the strong, largely entropically derived destabilization of the 3′ substem of stem 2 (compare Figure 3, Figure 9). This is the only mutant RNA for which the F→S1+J and S1+J→I unfolding steps are not tightly coupled on the temperature coordinate at 0 mM Mg2+. The simplest interpretation of the effects of the L2U substitution is that the entropic penalty for pseudoknot loop closure is significantly larger in this mutant relative to the wild-type pseudoknot.
Figure 9.
Melting profiles of the wild-type mIAP pseudoknot compared to the loop 2 substitution mutant, L2U RNA. On the left, the composite fits are superimposed on the experimental data. On the right, the individual transitions of the composite fit are shown for clarity. Black ( •) and gray-scale (+) correspond to ∂A/∂T monitored at 260 and 280 nm, respectively. The composite fit is superimposed on the L2U calorimetry data ( •) on the left, with the individual transitions shown for clarity on the right with the composite fit shown with a broken line.
Discussion
The equilibrium unfolding pathway of the mIAP pseudoknot proceeds in four discreet steps
The temperature-induced equilibrium unfolding pathway for the mIAP gag-pro frameshifting pseudoknot proceeds through four optically and calorimetrically defined steps. The fully folded pseudoknot (F) unfolds initially at the four 3′ base-pairs of stem 2 below the unpaired adenosine to form a non-native pseudoknot intermediate (S1+J). This is followed by the unfolding of the remainder of the helical stem 2 to form an intermediate (I) which consists of the stem 1 hairpin with some as yet unknown structure in the hairpin loop. The hairpin loop structure is then unfolded to give the stem 1 hairpin (S1), which is denatured to the fully unfolded form (U):F ⇔ S1+J ⇔ I ⇔ S1 ⇔ U Interestingly, the two RNAs in which A35 has been deleted follow this pathway, in spite of the fact that the seven base-pair 3′ helical stem 2 of this pseudoknot is predicted to be more stable than the six base-pair 5′ helical stem 1. This general unfolding behavior was also observed for the gene 32 autoregulatory H-type pseudoknot from bacteriophage T2 (Nixon & Giedroc, 1998) in which the 3′ helical stem, predicted to be more stable thermodynamically, unfolds first in two discernable transitions, followed by the 5′ helical stem. The mIAP pseudoknot therefore appears to unfold in an obligate pathway in which the 3′ stem 2 is considerably less stable in the context of the pseudoknot than is stem 1.
The role of Mg2+ in the stability of the mIAP pseudoknot
The observed and predicted enthalpies for the two stems in mIAP indicate that the observed van’t Hoff enthalpy (in the absence of Mg2+) is sufficient to account for all or nearly all of the predicted secondary structure in the pseudoknot (Table 2). Tertiary structure in RNA is folded after and unfolded before secondary structure (Draper, 1996). If Mg2+ induces tertiary structure in the pseudoknot, the unfolding of the tertiary structure would be expected to occur before, or concomitant with, the unfolding of the secondary structure. The van’t Hoff enthalpy observed for the two stem 2 transitions in the wild-type mIAP does not increase with increasing [Mg2+]⩾1.0 mM and no new unfolding transitions are observed, suggesting that Mg2+ does not induce the additional folding or energetically significant loop-stem interactions found in more complex RNAs (Draper, 1996). Thermodynamic analysis of the unfolding of other simple H-type RNA pseudoknots suggests that this may be a general property of these molecules Gluick et al 1997, Nixon and Giedroc 1998, Theimer et al 1998. Consistent with this, the studies reported here suggest that the unfolding of these RNAs does not seem to be accompanied by a large change in the total heat capacity of the system, given the excellent agreement between the van’t Hoff enthalpies determined from optical and calorimetric experiments (Table 2).
Deletion of the unpaired adenosine base at the helical junction is only slightly destabilizing
The calculated and predicted thermodynamic parameters for the wild-type mIAP and selected mutants are summarized in Table 3. The mIAP pseudoknot shares some similarities with the SRV-1 and MMTV pseudoknots including structural ambiguity at the junction of the two helical stems. Deletion of the junction adenosine base in the mIAP pseudoknot, A15, is only slightly destabilizing with ΔΔG°(37 °C)≈1 kcal mol−1. This destabilization is smaller than would be predicted for the loss of an A-U base-pair and coaxial stacking on stem 1. The close approach of backbone phosphate groups at the helical junction in RNA pseudoknots sometimes creates a region high in negative electrostatic potential which has been suggested as a possible location for chelation of divalent metal ions with higher affinity than duplex RNA Gluick et al 1997, Puglisi et al 1990, Nixon and Giedroc 1998. Therefore, mutations at the helical junction might destabilize an RNA pseudoknot by significantly altering higher affinity divalent binding sites. The wild-type mIAP and ΔA15 pseudoknots were studied over a wide range of Mg2+ concentrations (50 μM-50 mM), and the dependence of t m on [Mg2+] for the F→S1+J and S1+J→I transitions fit to a non-specific binding model for divalent ions (Figure 6). The magnitude of K f andK u, which represent the average affinities of the RNAs for Mg2+, were found to be reasonable for non-specific binding in a background of 50 mM KCl compared to the magnitudes of K f and K u values determined for the pseudoknot from bacteriophage T2 gene32 mRNA at 100 mM KCl Nixon and Giedroc 1998, Nixon et al 1999. More importantly, the Mg2+ binding affinities were found to be virtually identical for these two RNAs, suggesting that it is unlikely that the A15 deletion causes the loss of a specific ion binding site or results in differential accumulation of ions in the pseudoknot. The uncoupling of the two stem 2 transitions in the ΔA15 molecule in the presence of urea (Figure 8), with the region of stem 2 closest to the junction becoming stabilized toward urea denaturation compared to the mIAP pseudoknot, however, suggests that the helical junction in the F and S1+J conformations of the ΔA15 pseudoknot may be better stacked than in the wild-type mIAP pseudoknot.
Preliminary 2D NMR experiments suggest the presence of only one A-U base-pair in the mIAP pseudoknot (C.A.T., D. Hoffman & D.P.G., unpublished observations). In combination with the small destabilization observed upon deletion of A15, these data suggest that A15 is unpaired or not strongly paired. Structure probing experiments indicate that the unpaired adenosine base in stem 2, A35, and the adenosine bases in loop 2 are substantially more susceptible to DEPC modification than is A15 in the wild-type mIAP pseudoknot (T. Young & C.A.T., unpublished results). The reduced susceptibility of A15 to both chemical modification and enzymatic cleavage may suggest that some or all of the A15 nucleotide is not totally exposed to solvent; additional structural insight will have to await further structural studies. Analogous thermodynamic studies of the structurally characterized MMTV-vpk pseudoknot (Shen & Tinoco, 1995) suggest that the unpaired adenosine base at the junction does not contribute exceptional stability to the pseudoknot and in fact provides stabilization equivalent to that predicted for a 3′ single-stranded nucleotide stacked on the junction base-pair of stem 2 (unpublished results).
The thermodynamic effects of deletion of A15 and A35 are very nearly additive
Three RNAs, ΔA15, ΔA35, and ΔA35A15, were prepared to examine the contribution that the unpaired adenosine bases make to the thermodynamic stability of the mIAP frameshifting pseudoknot. Deletion of A35 is predicted to be stabilizing, allowing the formation of an uninterrupted stem 2, which would unfold in a single cooperative transition. A15 and A35 were both deleted in the ΔA35A15 RNA, to determine if the A15 deletion, which is only modestly destabilizing in the context of the wild-type pseudoknot, has the same effect in the context of a continuous stem 2. The calculated and predicted thermodynamic parameters for the ΔA35 and ΔA35A15 RNAs are summarized in Table 3. The ΔA35 RNA stabilizes the mIAP pseudoknot by 3.2 kcal mol−1, exactly as expected for the elimination of the entropic destabilization predicted for a single nucleotide bulge in a helical stem (Turner et al., 1988). Although the molecular origin may well be different, deletion of A15 in the context of the ΔA35 deletion reveals that the free energy of unfolding of stem 2 is also ≈1 kcal mol−1 smaller than ΔA35 RNA, coupled with a small perturbation in the stability of stem 1. These results suggest that the thermodynamic impact of these two deletions are very nearly additive, suggesting that it is unlikely that the unpaired adenosine bases cooperate in some way to globally stabilize the molecule.
Substitution of the loop 2 sequence with uridine bases destabilizes the mIAP pseudoknot
In pseudoknots that promote frameshifting and stop codon readthrough, the sequence and length of loop 2 has been shown in some cases to be functionally significant. For example, sequences in loop 2 seem to be important for stop codon readthrough at the UAG terminator between gag and pol in Moloney murine leukemia virus (MuLV) Wills et al 1991, Wills et al 1994. On the other hand, the entire loop 2 sequence in the gag-pro frameshift site in SRV-1 can be changed with essentially no effect on frameshifting, and reducing the length of loop 2 actually increases frameshifting activity (ten Dam et al., 1995). The effect of loop 2 sequence and length on the stability of the pseudoknot cannot be predicted from known thermodynamic parameters although the minimum loop length requirements have been experimentally examined Pleij et al 1985, Wyatt et al 1990. The substitution of the loop 2 sequence with uridine bases in the mIAP pseudoknot uncouples the F→S1+J and S1+J→I unfolding events, destabilizing the 3′ substem and stabilizing the junction substem. The combined enthalpy of the first two stem 2 transitions is close to that predicted for the entire helical stem, and the overall destabilization compared to the mIAP pseudoknot ΔΔG° (37 °C) is 1.2 kcal mol−1 from optical melts and 3.0 kcal mol−1 determined calorimetrically. Interestingly, the L2U mutant melting profile bears a striking resemblance to the effect of 20-25 % urea on the melting profile of the ΔA15 RNA pseudoknot (Figure 8). This suggests that the origin of this perturbation of the stability of the molecule by uridine substitution of loop 2 may be similar to that induced by low concentrations of urea. The mechanism by which urea destabilizes RNA is not known, but hydrogen bonding to RNA functional groups or a change in solvent ordering are two possibilities. In any case, both effects are likely transmitted through a selectively weakened junction region. These findings suggest that caution must be exercised in ascribing changes in frameshifting efficiency to nucleotide sequence effects, due to the possibility of a change in the global stability of the molecule.
Conclusions
Here, we have determined the equilibrium unfolding pathway and quantified the thermodynamic effects of deletions of unpaired adenosine nucleotides and loop 2 sequence substitution on the stability of an H-type RNA pseudoknot involved in programmed ribosomal frameshifting using thermal denaturation monitored by UV hyperchromicity and differential scanning calorimetry. We have determined that (1) the unfolding pathway proceeds in four definable steps, (2) the unfolding thermodynamics suggest that ΔC°p is small in this system, (3) stabilization by Mg2+ can be fully accounted for by the impact of delocalized, non-specifically bound ions, (4) the unpaired adenosine base at the helical junction A15 is only modestly stabilizing, and (5) substitution of the loop 2 nucleotide sequence with uridine bases is destabilizing in the context of the mIAP pseudoknot. Experiments are in progress in this laboratory to characterize the structures and frameshifting activity of these molecules. These experiments will allow us to systematically explore the relationships between thermodynamic stability, frameshifting activity, structure, and dynamics and help us to clarify the role of downstream pseudoknots on programmed ribosomal frameshifting.
Materials and methods
RNA synthesis and purification
RNAs with the sequences shown in Figure 1(b)were prepared by in vitro transcription using T7 RNA polymerase and partially double-stranded templates as described Qiu et al 1996, Du et al 1996. Crude RNAs were purified by denaturing PAGE, visualized by UV shadowing and electroeluted using an S&S electroeluter. The recovered RNA was loaded onto a C18-cartridge (Alltech), eluted with a 50 % (v/v) methanol wash, taken to dryness and subjected to exhaustive dialysis against 10 mM Mops (pH 7.0), 50 mM KCl. The first change of dialysis buffer contained 10 mM EDTA, with the subsequent (two or three) changes containing no EDTA. RNAs were stored at −20 °C. Samples were prepared for thermal denaturation by diluting into final dialysis buffer, heating at 90 °C for five minutes, slow cooling to room temperature, and then adding the appropriate concentration of MgSO4. This procedure was found to be most effective at removing RNA dimers formed at the stock solution concentration. Melt samples which contained urea were prepared by diluting into final dialysis buffer, adding the appropriate concentration of urea, heating at 65 °C for 20 minutes, and then slow cooling to room temperature. Dimer formation was substantially reduced in the presence of urea.
Thermal denaturation experiments
All RNA melts were collected on a temperature controller equipped Cary 1 spectrophotometer operating in double beam mode. All melts were collected in 10 mM Mops (pH 7.0), 50 mM KCl with the indicated concentration of MgSO4 or urea added. Capped cells were used and percentage transmittance was recorded as a function of temperature at both 260 nm and 280 nm simultaneously. Samples were loaded into room temperature cuvettes and the cells allowed to equilibrate at 5 °C for 30 minutes. The temperature controller was ramped at a rate of 0.3 deg. C/minute from 5-100 °C. Data points were collected every 0.3-0.4 deg. C as determined by a temperature probe inserted into a cuvette containing final dialysis buffer.
Analysis of UV thermal melting profiles
Cary 1 report files were converted from percentage transmittance to absorbance and subjected to piece-wise linear interpolation. The interpolated sets of data were then smoothed over a 4 deg. C window and the melting profile was generated by taking the first derivative of absorbance with respect to temperature (∂A/∂T) and plotted as a function of temperature. Melting profiles were subjected to non-linear least-squares parameter estimation of t m, i, ΔH i, and A i, for each i th sequential unfolding transition via application of an unfolding model involving interacting sequential two-state unfolding transitions (Laing & Draper, 1994) using the t -melt program running on an SGI O2 Unix workstation as described (Theimer et al., 1998). The van’t Hoff analysis of the UV melting data assumes that ΔC p is zero, which is a reasonable approximation given the observed agreement between the UV and calorimetric melting data.
In the t-melt algorithm, each optical melting transition is assumed to be two-state and is characterized by an equilibrium constant, K i, that is a function of the enthalpy of the transition, ΔH i, and the melting temperature for that transition, t m. All species in the solution at temperature T are described by the partition functionq:q=1+∑j=1NTRANSπi=1jKi where NTRANS is the total number of unfolding transitions. We define z n as the sum of the terms in which an individual transition, n, participates. The absorbance at a given temperature is therefore the sum over all transitions of the contribution of each transition, z n, multiplied by its intrinsic amplitude,A n, divided by the partition function for the system as a whole,q:signal=∑n=1NTRANSAnznq=∑n=1NTRANSAn∑j=nNTRANSπi=1jKi1+∑j=1NTRANSπi=1jKi
The derivative with respect to temperature of the absorbance signal at 260 and 280 nm (∂A/∂T) is calculated and the ∂A/∂T at each wavelength is simultaneously subjected to least-squares parameterization of t m, ΔH, and amplitude at 260 nm and 280 nm:∂A∂T=∑n=1NTRANSAn∑j=nNTRANS∑m=1jπi=1m-1Kiπi=m+1jKiHmKmqT2R−AnZn∑j=nNTRANS∑m=1jπi=1m-1Kiπi=m+1jHmKmq2T2R
Calorimetry experiments
All calorimetry scans were collected on a Microcal VP-DSC. RNA concentrations in the sample cell varied between 15 and 50 μM in a 0.5 ml cell. Samples prepared for calorimetry were subjected to exhaustive dialysis against 10 mM Mops (pH 7.0), 50 mM KCl and the final dialysis buffer was used for dilutions and references. Samples were diluted into final dialysis buffer, heated at 90 °C for five minutes, slow cooled to room temperature, and degassed. Samples were then loaded into room temperature cells, cooled to 1 °C, and allowed to equilibrate for 30 minutes. The temperature was ramped at a rate of 1 deg. C/minute from 1-120 °C. Baseline scans were run under identical conditions using final dialysis buffer in both the sample and reference cells.
Analysis of differential scanning calorimetry data
VP-DSC data files were analyzed using the program Origin from Microcal. The collected buffer baseline scan was subtracted from the RNA sample scan and the data corrected for the concentration of RNA in the cell. Data were then subjected to a linear baseline correction, and the calorimetric enthalpy, H cal, calculated from the area under the baseline-corrected curve. van’t Hoff enthalpies and t m values were determined utilizing a sequential two-state unfolding model fit and the fit minimized to represent the experimental data and also minimize the difference between the sum of the van’t Hoff enthalpies and the total calorimetric enthalpy, H cal Multiple baseline correction routines were applied to ensure that subtle variations in baselining the data did not have large effects on the observed parameters.
The sequential unfolding model used for fitting the calorimetry data is identical with that used for the optical data with the equilibrium constants, K i, the partition function, q, and the sum of the terms in which an individual transition participates, z n, described in the same way. Instead of an absorbance change, the signal observed is a change in the total molar heat content of the system, which is a function of the molar enthalpy, ΔH n, which changes for each individual transition,n. This function can be expressed as the sum over all transitions of the contribution of each transition, z n, multiplied by its intrinsic molar enthalpy, ΔH n, divided by the partition function for the system as a whole, q:H=∑n=1NTRANSΔHnZnq=∑n=1NTRANSΔHn∑j=nNTRANSπi=1jKi1+∑j=1NTRANSπi=1jKi
This equation can then be differentiated with respect to temperature to obtain the system heat capacity, C p, which gives an equation identical in form with the ∂A/∂T equation for fitting the derivative of the optical data in which intrinsic molar enthalpy, ΔH n, is substituted for the intrinsic amplitude parameter, A n.
Acknowledgements
This work was supported by NIH grant AI40187 to D.P.G. and D. W. Hoffman. The Microcal VP-DSC was supported, in part, by grants from the Texas Agricultural Experiment Station and NIH grant GM42569 to D.P.G. We thank Dr Jon A. Christopher for the program t -melt (Theimer et al., 1998) and we also thank Mr Brian Cannon for highly purified T7 RNA polymerase. Mr Cannon was supported by a Howard Hughes Medical Institute Undergraduate Biological Sciences Education Program Grant (7119571195-539101).
Footnotes
Edited by D. E. Draper
References
- Banerjee A.R., Jaeger J.A., Turner D.H. Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry. 1993;32:153–163. doi: 10.1021/bi00052a021. [DOI] [PubMed] [Google Scholar]
- Brierley I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H.E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl Acad. Sci. USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chamorro M., Lee S.I., Shen L.X., Hines J.V., Tinoco I., Jr, Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Kang H., Shen L.X., Chamorro M., Varmus H.E., Tinoco I., Jr A characteristic bent conformation of RNA pseudoknots promotes −1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- Conn G.L., Draper D.E. RNA structure. Curr. Opin. Struct. Biol. 1998;8:278–285. doi: 10.1016/s0959-440x(98)80059-6. [DOI] [PubMed] [Google Scholar]
- Draper D.E. Strategies for RNA folding. Trends Biochem. Sci. 1996;21:145–149. [PubMed] [Google Scholar]
- Du Z., Hoffman D.W. An NMR and mutational study of the pseudoknot within the gene 32 mRNA of bacteriophage T2: insights into a family of structurally related RNA pseudoknots. Nucl. Acids Res. 1997;25:1120–1135. doi: 10.1093/nar/25.6.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Giedroc D.P., Hoffman D.W. Structure of the autoregulatory pseudoknot within the gene 32 messenger RNA of bacteriophages T2 and T6: a model for a possible family of structurally related RNA pseudoknots. Biochemistry. 1996;35:4187–4198. doi: 10.1021/bi9527350. [DOI] [PubMed] [Google Scholar]
- Du Z., Holland J.A., Hansen M.R., Giedroc D.P., Hoffman D.W. Base pairings within the RNA pseudoknot associated with the Simian retrovirus-1 gag-pro frameshift site. J. Mol. Biol. 1997;270:464–470. doi: 10.1006/jmbi.1997.1127. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann F., Welker R., Kräusslich H.-G. Intracisternal A-type particles express their proteinase in a separate reading frame by translational frameshifting, similar to D-type retroviruses. Virology. 1997;235:352–359. doi: 10.1006/viro.1997.8708. [DOI] [PubMed] [Google Scholar]
- Fresco J.R., Klotz L.C., Richards E.G. A new spectroscopic approach to the determination of helical secondary structure in ribonucleic acids. Cold Spring Harbor Symp. Quant. Biol. 1963;28:83–90. [Google Scholar]
- Gesteland R.F., Atkins J.F. RECODING: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- Gluick T.C., Wills N.M., Gesteland R.F., Draper D.E. Folding of an mRNA pseudoknot required for stop codon readthrough: effects of mono- and divalent ions on stability. Biochemistry. 1997;36:16173–16186. doi: 10.1021/bi971362v. [DOI] [PubMed] [Google Scholar]
- He L., Kierzek R., SantaLucia J., Walter A.E., Turner D.H. Nearest-neighbor parameters for GïU mismatches: 5′GU3′/3′UG5′ is destabilizing in the contexts CGUG/GUGC, UGUA/AUGU, and AGUU/UUGA but stabilizing in GGUC/CUGG. Biochemistry. 1991;30:11124–11132. doi: 10.1021/bi00110a015. [DOI] [PubMed] [Google Scholar]
- Holland J.A., Hansen M.R., Du Z., Hoffman D.W. An examination of coaxial stacking of helical stems in a pseudoknot motif: the gene 32 messenger RNA pseudoknot of bacteriophage T2. RNA. 1999;5:257–271. doi: 10.1017/s1355838299981360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Tinoco I., Jr A mutant RNA pseudoknot that promotes ribosomal frameshifting in mouse mammary tumor virus. Nucl. Acids Res. 1997;25:1943–1949. doi: 10.1093/nar/25.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Hines J.V., Tinoco I., Jr Conformation of a non-frameshifting RNA pseudoknot from mouse mammary tumor virus. J. Mol. Biol. 1996;259:135–147. doi: 10.1006/jmbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- Laing L.G., Draper D.E. Thermodynamics of RNA folding in a conserved ribosomal RNA domain. J. Mol. Biol. 1994;237:560–576. doi: 10.1006/jmbi.1994.1255. [DOI] [PubMed] [Google Scholar]
- Laing L.G., Gluick T.C., Draper D.E. Stabilization of RNA structure by Mg ions. Specific and non-specific effects. J. Mol. Biol. 1994;237:577–587. doi: 10.1006/jmbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- Larsen B., Gesteland R.F., Atkins J.F. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshiftting: programmed efficiency of 50 % J. Mol. Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.D., Bronson D.L., Lemke T.D., Faras A.J. Phylogenetic analyses of 55 retroelements on the basis of the nucleotide and product amino acid sequences of thepol gene. Mol. Biol. Evol. 1995;12:657–670. doi: 10.1093/oxfordjournals.molbev.a040231. [DOI] [PubMed] [Google Scholar]
- Matsufuji S., Matsufuji T., Wills N.M., Gesteland R.F., Atkins J.F. Reading two bases twice: mammalian antizyme frameshifting in yeast. EMBO J. 1996;15:1360–1370. [PMC free article] [PubMed] [Google Scholar]
- Nixon P.L., Giedroc D.P. Equilibrium unfolding (folding) pathway of a model H-type pseudoknotted RNA: the role of magnesium ions in stability. Biochemistry. 1998;37:16116–16129. doi: 10.1021/bi981726z. [DOI] [PubMed] [Google Scholar]
- Nixon P.L., Theimer C.A., Giedroc D.P. Thermodynamics of stabilization of RNA pseudoknots by cobalt(III) hexammine. Biopolymers. 1999 doi: 10.1002/(SICI)1097-0282(19991005)50:4<443::AID-BIP9>3.0.CO;2-C. in the press. [DOI] [PubMed] [Google Scholar]
- Pleij C.W.A., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucl. Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J.D., Wyatt J.R., Tinoco I., Jr Conformation of an RNA pseudoknot. J. Mol. Biol. 1990;214:437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Kaluarachchi K., Du Z., Hoffman D.W., Giedroc D.P. Thermodynamics of folding of the RNA pseudoknot of the T4 gene 32 autoregulatory messenger RNA. Biochemistry. 1996;35:4176–4186. doi: 10.1021/bi9527348. [DOI] [PubMed] [Google Scholar]
- Shen L.X., Tinoco I., Jr The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Somogyi P., Jenner A.J., Brierley I., Inglis S.C. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Chen L., Egli M., Berger J.M., Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nature Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D., Kang H. Mutational analysis of the RNA pseudoknot involved in efficient ribosomal frameshifting in simian retrovirus-1. Nucl. Acids Res. 1998;26:1369–1372. doi: 10.1093/nar/26.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.K., Draper D.E. Evidence for allosteric coupling between the ribosome and repressor binding sites of a translationally regulated mRNA. Biochemistry. 1990;29:4434–4439. doi: 10.1021/bi00470a025. [DOI] [PubMed] [Google Scholar]
- ten Dam E.B., Verlaan P.W.G., Pleij C.W.A. Analysis of the role of the pseudoknot component in the SRV-1 gag-pro ribosomal frameshift signal: loop lengths and stability of the stem regions. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A., Wang Y., Hoffman D.W., Krisch H.M., Giedroc D.P. Non-nearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J. Mol. Biol. 1998;279 doi: 10.1006/jmbi.1998.1812. 5454–564. [DOI] [PubMed] [Google Scholar]
- Tu C., Tzeng T.-H., Bruenn J.A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl Acad. Sci. USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.H., Sugimoto N., Freier S.M. RNA structure prediction. Annu. Rev. Biophys. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Wills N.M., Gesteland R.F., Atkins J.A. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virusgag stop codon. Proc. Natl Acad. Sci. USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills N.M., Gesteland R.F., Atkins J.A. Pseudoknot-dependent readthrough of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J.R., Puglisi J.D., Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J. Mol. Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- Xia T., SantaLucia J., Jr, Burkard M.E., Kierzek R., Shroeder S.J., Jiao X., Cox C., Turner D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base-pairs. Biochemistry. 1998;17:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]