Abstract
The Ebola virus membrane-associated matrix protein VP40 is thought to be crucial for assembly and budding of virus particles. Here we present the crystal structure of a disk-shaped octameric form of VP40 formed by four antiparallel homodimers of the N-terminal domain. The octamer binds an RNA triribonucleotide containing the sequence 5′-U-G-A-3′ through its inner pore surface, and its oligomerization and RNA binding properties are facilitated by two conformational changes when compared to monomeric VP40. The selective RNA interaction stabilizes the ring structure and confers in vitro SDS resistance to octameric VP40. SDS-resistant octameric VP40 is also found in Ebola virus-infected cells, which suggests that VP40 has an additional function in the life cycle of the virus besides promoting virus assembly and budding off the plasma membrane.
Keywords: filovirus, Ebola virus, matrix protein VP40, assembly, budding, X-ray structure
Introduction
Filoviruses belong to the order of negative-stranded nonsegmented RNA viruses and are classified into the two genera, Ebola and Marburg viruses. They are the causative agents of a severe, mostly lethal, haemorraghic fever in humans, with occasional epidemic outbreaks of the disease in regions of central Africa [1]. Filoviruses are enveloped by a lipid bilayer, which anchors the glycoprotein GP to the surface of viral particles. GP undergoes a number of posttranslational modifications, which have been associated with virus entry and possibly pathogenicity 2, 3. The matrix protein VP40 forms a layer underneath the viral membrane and provides a link to the ribonucleoprotein complex (RNP) composed of the viral RNA packaged by the nucleoprotein (NP) as well as the associated proteins VP35, VP30, and polymerase L 4, 5, 6, 7. The mature viral particles have a filamentous shape, which varies in length 4, 8.
Structural analysis of VP40 shows a monomeric metastable conformation consisting of two structurally related β sandwich domains that are loosely associated with each other [9]. Trypsin cleavage within the C-terminal domain and destabilization of the monomer by urea treatment, mutagenesis, and binding to synthetic liposomes leads to the formation of ring-like structures in vitro, a process that has been suggested to play a role in assembly and budding 10, 11. The change in the oligomeric state is arbitrated by the N-terminal domain [9], while the C-terminal domain is required for membrane association in vitro [10] and in vivo [12].
Filovirus assembly and budding takes place at the plasma membrane, which requires lipid raft microdomains 4, 13. Expression of Ebola virus VP40 was shown to be sufficient to induce the release of virus-like particles from mammalian cells 12, 14, whereas coexpression of GP improved the efficiency of particle production dramatically 13, 15. Furthermore, VP40 contains short sequence motifs (PPXY and PTAP) at its N terminus whose presence has been implicated in virus particle release by interacting with cellular factors 12, 16, 17, 18.
VP40 also associates with RNP structures in vivo [19] and it has been reported to localize to late endosomal membranes [20]. Matrix protein/RNP interactions have been reported for a number of enveloped viruses 21, 22, 23, 24. This process may generally involve interactions with viral RNA (vRNA), such as the Corona virus M/mRNA1 recognition, which seems to be crucial for the M-NP association [25]. Likewise, influenza virus M1 and vRNA were reported to promote the assembly of helical NP structures, leading to the translocation of RNP from the nucleus to the cytoplasm [26]. In addition, other studies implicate matrix proteins in the interference with the cellular RNA metabolism 27, 28.
Here we present the X-ray structure of octameric VP40 assembled through its N-terminal domain (NTD). Oligomerization is accompanied by structural rearrangements, when compared to the full-length monomeric conformation of VP40. Notably, we find that a short triribonucleotide (5′-U-G-A-3′), derived from the expression host, is specifically bound at the inner pore dimer-dimer interface. This, together with the detection of a small amount of octameric SDS-resistant full-length VP40 in Ebola virus-infected cells, suggests that VP40 is not only active in bilayer interaction during the assembly process, but also plays an important role either in viral or host cell RNA metabolism during its replication cycle.
Results
Structure Determination
Both VP40(31–212) and VP40(31–194) form ring-like oligomeric structures in solution and were crystallized in space groups P6222 and P422, respectively. The P6222 crystals contained the full-length construct VP40(31–212), while the P422 crystals were only obtained upon proteolysis at residue 54 in the crystallization drop, as confirmed by SDS-PAGE analysis and N-terminal sequencing of dissolved crystals. The structure of the P422 crystal form, thus containing VP40(55–194), was solved by a combination of single isomorphous replacement (SIR) and multiwavelength anomalous diffraction (MAD) methods. The final model contains residues 69 to 192 and a short stretch of very well-defined single-stranded RNA, containing the sequence 5′-U-G-A-3′ (phasing and refinement statistics are presented in Table 1).
Table 1.
Crystallographic Statistics
| A. Data Collection |
||||||
|---|---|---|---|---|---|---|
| Data Set | VP40(55–194) (native) | VP40(55–194) (Pt CuKα) | VP40(55–194) (Pt peak) | VP40(55–194) (Pt remote) | VP40(55–194) (Pt peak and remote merged) | VP40(31–212) (native) |
| Space group | P422 | P422 | P422 | P422 | P422 | P6222 |
| Cell constantsa (Å) | 80.51, 46.77 | 80.39, 46.92 | 80.57, 46.96 | 80.57, 46.96 | 80.57, 46.96 | 79.61, 239.18 |
| Wavelength (Å) | 0.933 | 1.542 | 1.072 | 1.069 | 1.072 + 1.069 | 0.979 |
| X-ray source/beamline | ID14-EH1 | rotating anode | BM-14 | BM-14 | BM-14 | ID14-EH4 |
| Max. resolution (Å) | 1.60 | 2.30 | 1.63 | 1.63 | 1.63 | 2.60 |
| Rmergeb | 6.5 (34.1)c | 6.9 (25.3) | 4.6 (23.8) | 4.4 (23.9) | 6.5 (28.7) | 8.5 (40.6) |
| Completeness (%) | 95.9 (78.2)c | 99.6 (99.2) | 99.9 (99.7) | 99.9 (99.6) | 99.9 (99.8) | 97.8 (86.0) |
| No. reflections | 167,556 | 64,896 | 95,075 | 94,570 | 189,216 | 63,464 |
| No. unique reflections | 19,961 | 7,179 | 19,721 | 19,726 | 19,736 | 14,356 |
| <I/σ(I)> | 7.1 (1.6)c | 10.1 (2.9) | 10.6 (2.4) | 11.0 (2.5) | 7.4 (2.2) | 4.5 (1.8) |
| Average multiplicity | 8.4 (2.9)c | 9.0 (8.5) | 4.8 (4.3) | 4.8 (4.4) | 9.6 (8.6) | 4.4 (3.6) |
| Wilson B factor (Å2) | 14.2 | 27.0 | 16.3 | 16.6 | 16.3 | 67.2 |
| B. MAD Phasing (VP40(55–194)) | ||||||
| Phasing Powerd |
Figure of Merite |
|||||
| ano | 2.3 | acentric | 0.68 | |||
| iso |
3.9 |
centric |
0.66 |
|||
| C. Final Refinement Statistics | ||||||
| Structure | VP40(55–194) | VP40(31–212) | ||||
| Resolution range used for refinement (Å) | 40.0–1.60 | 59.8–2.60 | ||||
| No. of reflections in working set/test set | 18,914/1,026 | 13,004/995 | ||||
| Rcrystf | 0.164 | 0.305 | ||||
| Rfreef | 0.182 | 0.329 | ||||
| Bond lengths rmsd (Å) | 0.008 | 0.009 | ||||
| Bond angles rmsd (°) | 1.18 | 1.42 | ||||
| Bonded B's (main chain/side chain) rmsd (Å2) | 0.58/1.69 | 0.60/1.23 | ||||
| Average B (Å2) | 11.1 | 39.7 | ||||
| Protein residues in final model | 124 (out of 140) | 245 (out of 363) | ||||
| No. of nonhydrogen atoms used in refinement | 1195 | 2153 | ||||
| RNA residues | 3 | 6 | ||||
| Solvent molecules | 158 | 113 | ||||
| Ions/cations |
– |
2 (Cl−) |
||||
Values for a = b and for c.
Rmerge = [ΣhΣI|Ii(h) − <I(h>)|/ΣhΣIIi(h)] × 100, where Ii(h) is the ith measurement and <I(h)> is the weighted mean of all measurements of reflec-tion h.
Numbers in parentheses are for last shell.
Phasing power = <|FH|>/E, where <|FH|> is the rms structure factor amplitude for the heavy atom and E is the estimated lack-of-closure error.
Figure of merit = <|∑P(α)eiα/∑|P(α)|>, where α is the phase and P(α) is the phase probability distribution.
Rcryst and Rfree = ∑h||F(h)obs|− k|F(h)calc||/∑h|F(h)obs| for reflections in the working and test sets (>500), respectively.
The P6222 crystal form [VP40(31–212)], which contains two molecules per asymmetric unit, was subsequently solved by Patterson search techniques. The chain traces in both crystal forms are well superimposable, with only slight rms deviations for the Cα atoms of both molecules (0.37 Å for molecule A and 0.42 Å for molecule B). Accordingly, only the structure as present in the tetragonal crystals will be discussed. The P6222 model contains residues 69 to 190 (or 191 in molecule B) and two single-stranded triribonucleotides as found in the P422 structure. Residues 31 to 68 and 189(190) to 212 were disordered in the crystal and could not be assigned (for refinement statistics, see Table 1).
General Architecture
Both NTD protein variants analyzed here form toroidal octameric rings (assembled by crystallographic symmetry) with a wide central pore (Figure 1) . The ring structures are composed of four dimeric antiparallel β sandwich domains (Figure 1A). Each domain contains six antiparallel β strands forming two sheets and three α helices that pack laterally against the β structure. The ring structure has an outer diameter of ∼84 Å and an inner diameter of ∼17 Å (Figure 1B); the width of the particle is ∼42 Å (Figures 1C). In each monomer, the C-terminal ends (residues 184 to 194) reach over to the neighboring monomer, which contacts them in an extended conformation. This arrangement leads to the positioning of the C-terminal ends at the outer side of the ring, pointing away from it, almost at the same horizontal level and only 15 Å apart (Figure 1C). This suggests that the C-terminal domains, which were not present in the construct, could be positioned at the side, above or below the ring. The structure lacks ∼70 residues at the N terminus and its N-terminal end is positioned at the outside of the ring, close to the C-terminal end (Pro 185) (Figure 1B). Each monomer binds a short stretch of RNA at the dimer-dimer interface, characterized by excellent electron density for a triribonucleotide (Figure 1D).
Figure 1.
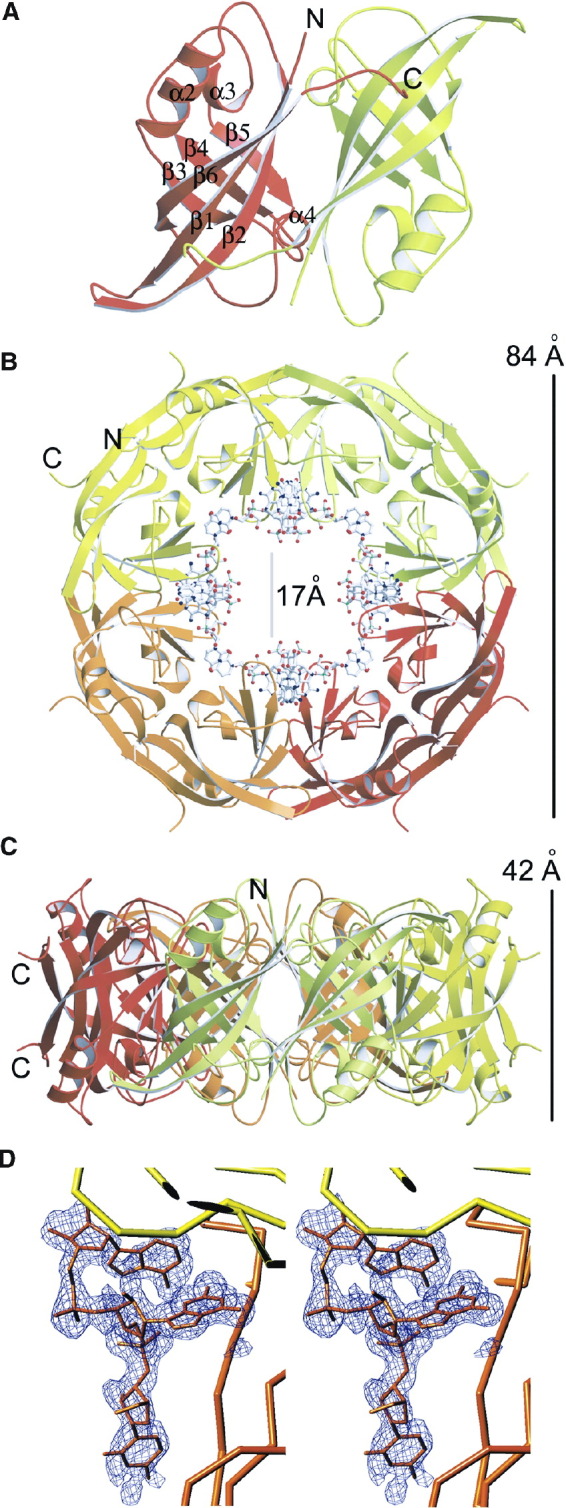
The N-Terminal Domain of Ebola Virus VP40 Forms Octameric Ring-like Structures Mediated by Specific ssRNA Binding
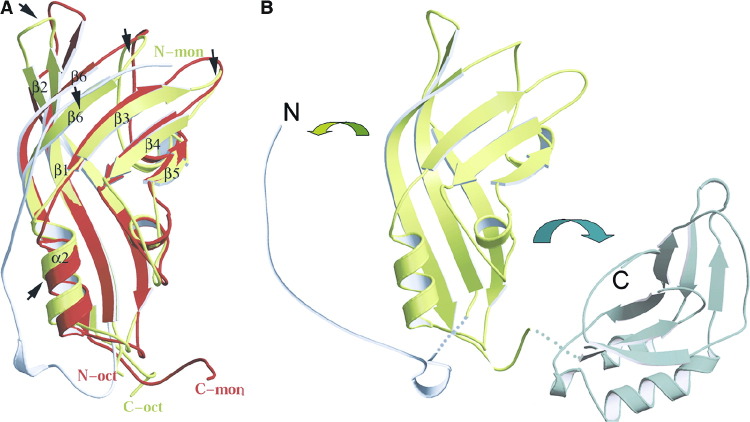
(A) Ribbon drawing of the antiparallel dimer formed by the N-terminal domain of VP40. The two monomers are shown in different colors and the secondary structure elements are labeled [9]. Note that the C-terminal end (residues 189 to 194) interacts in an extended conformation with the outer β sheet of its neighbor.
(B and C) Ribbon drawing of the ring structure of VP40. (B) View from the top, and (C) view from the side. The RNA molecule, bound at the dimer-dimer interface, is shown as an all-atom model. Each dimer (such as the one shown in A) is drawn in a different color and the dimensions as well as the positions of the N and C termini are indicated (the RNA has been omitted in C).
(D) Stereo view of a σA-weighted electron density omit map (mFobs − DFcalc) contoured at 2σ showing the superimposed final model for the triribonucleotide with the sequence U-G-A as an all-atom model and with a Cα tracing of the adjacent NTD molecule.
Molecular Interface within the Antiparallel Dimer
The interface of the antiparallel dimer occupies 1250 Å2. The dimer is stabilized by salt bridges from Glu160A (molecule A) to Arg148B (molecule B) and Arg151B, respectively; further polar interactions are found between Trp95A and Gln184B (Figure 2A) . The effect of the interactions is 2-fold due to the antiparallel nature of the dimer. The stability of the dimer is further enhanced by hydrophobic core interactions (residues Trp95, Pro97, Phe161, Ile74, and Ile82 from molecules A and B) (Figure 2B) as well as residues 189 to 194 (A and B), which interact in an extended conformation on the outside of the ring with a neighboring molecule (Figure 1A). All polar residues except Glu160, which is substituted by Asn, and all of the hydrophobic residues involved in the stabilization of the interface are conserved between VP40 sequences from Ebola and Marburg viruses [9]. The interface of the antiparallel dimer coincides with the interface occupied by the N- and C-terminal domains in the closed monomeric conformation, and residues covered by both interfaces are approximately the same [9]. This indicates that the C-terminal domain has to move away to allow the formation of the NTD dimer as suggested [11].
Figure 2.
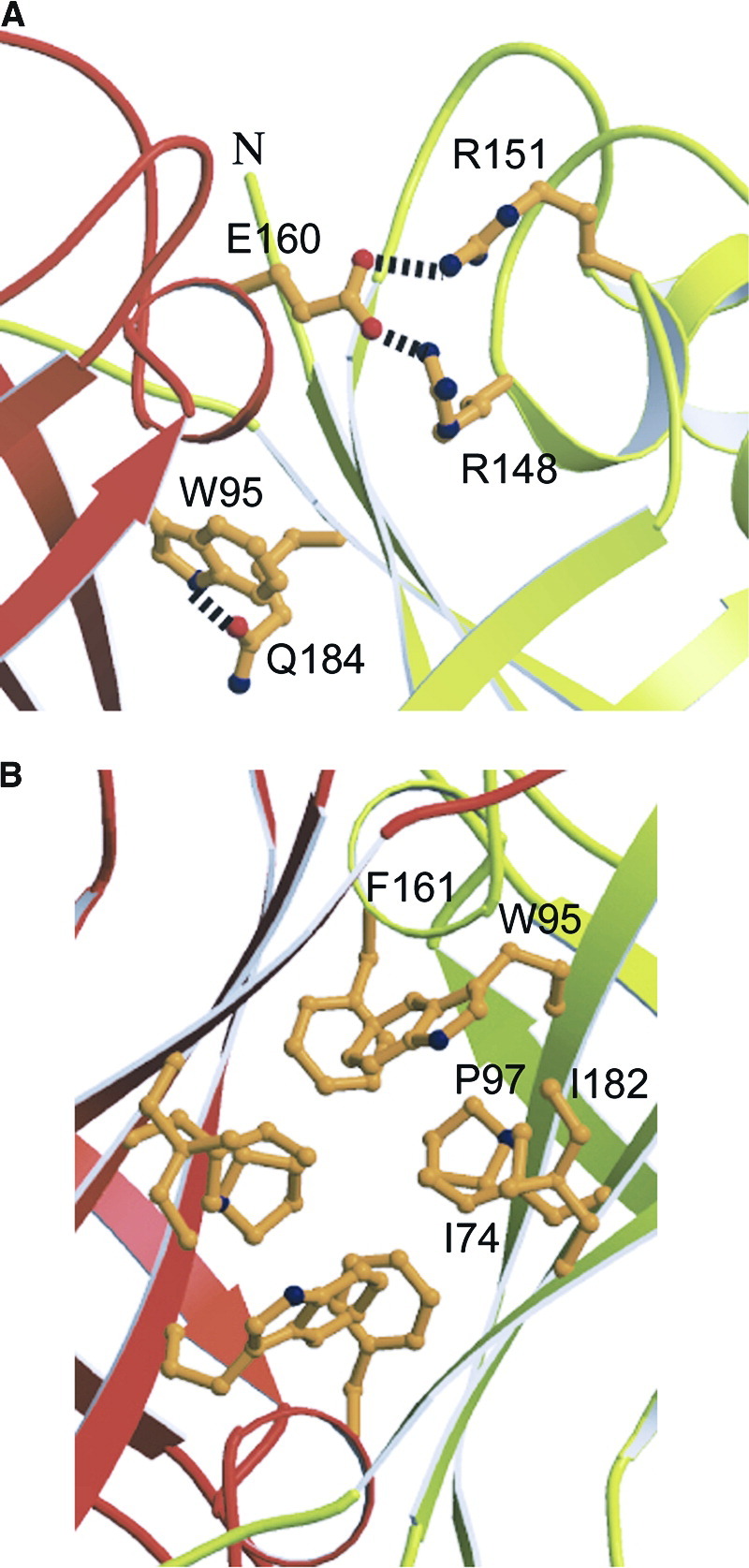
Close-Up View of the Molecular Interactions at the Interface of the Antiparallel Dimer
The view is along the two-fold axis similar to Figure 1A (rotated by 180°). Panel (A) shows hydrophilic interactions and panel (B) focuses on the hydrophobic core in the center of the two-fold axis. Secondary structure elements are labeled accordingly. Note that most of the residues involved in the specific contacts are conserved between Ebola and Marburg virus VP40 (see Figure 3 in [9]).
Molecular Interface between Two Dimers
The dimer-dimer interactions bury a surface of 990 Å2 (involving molecules A and C). This is dominated by hydrophobic interactions complemented with polar main chain contacts, including hydrogen bonds between the amide of Gly141A and the carbonyl of Tyr171C, and the oxygen of Thr173A and the amide of Gly139C. In addition, residues from the loop structure connecting β strand 4 and α helix 3 are sandwiched between two loop structures from a neighboring molecule, namely one connecting β strands 1 and 2 and the other one bridging α helix 4 with β strand 6. This creates a weak hydrophobic core on both corresponding ends of the β sandwich structures. The formation of the dimer-dimer interface also generates the binding pocket for the specific recognition of a single-stranded ribonucleotide segment, which dominates the interface (Figure 3A) .
Figure 3.
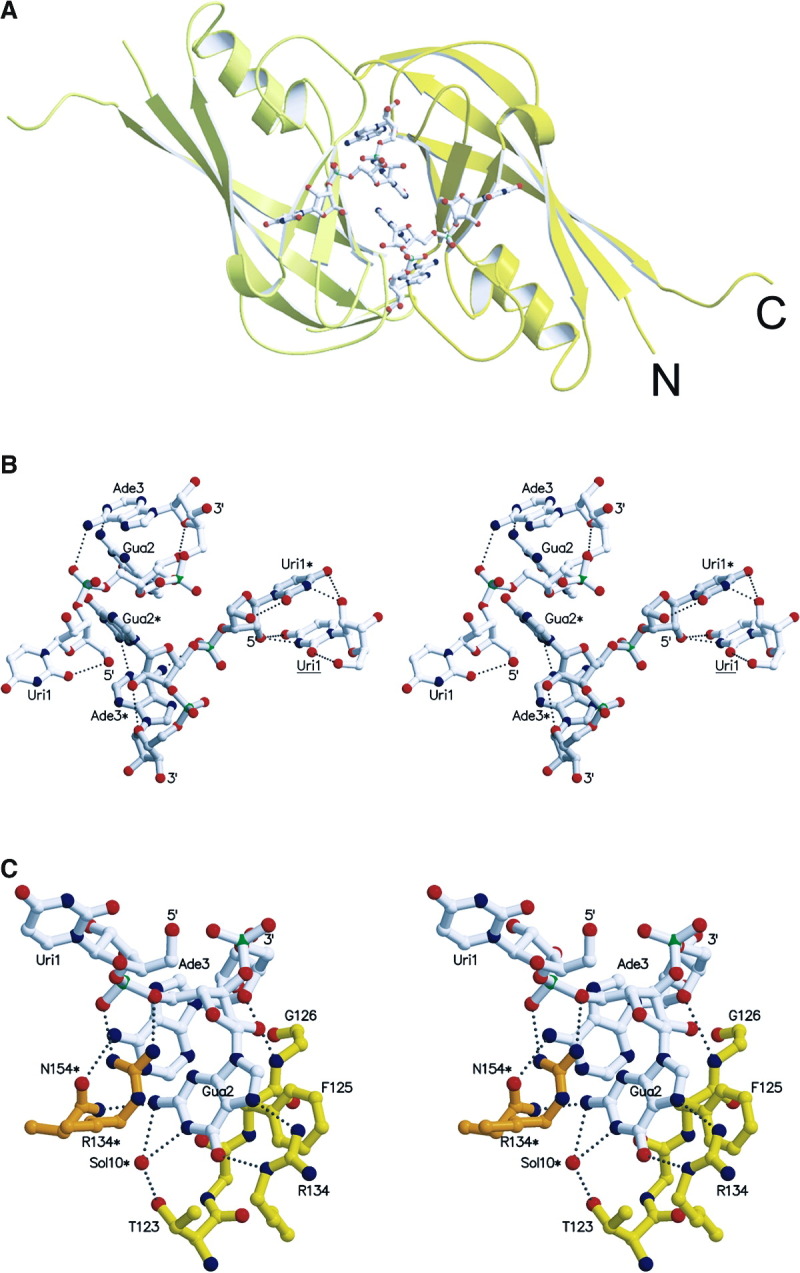
RNA Conformation and RNA Protein Interactions
(A) Two RNA molecules bind at the molecular interface generated by the dimer-dimer formation. The two protomers are shown in yellow and light yellow and the triribonucleotide is drawn as an all-atom model.
(B) Stereo view of two complete symmetry-related triribonucleotides containing the sequence 5′-U-G-A-3′, bound at the dimer-dimer interface. The two molecules are in white and in light gray with labels containing one asterisk. In addition, the right panel includes also a uridine (label underlined) from a neighboring RNA molecule, interacting specifically with Uri1* (for clarity, only Uri1 of the triribonucleotide is shown). The hydrogen bonding distances within the triribonucleotides are indicated as dashed lines. Note the parallel base pair stacking by the central guanine (Gua2 and Gua2*) as well as the parallel orientation of two symmetry-related uracil bases (Uri1* and Uri1). Both riboses from the uridine and the guanine are in the C2′-endo conformation while the guanine ribose is in C3′-endo [29].
(C) Close-up stereo view of protein-RNA interactions, shown for a single RNA molecule; van der Waals contacts are not shown with the exception of the parallel base pair stacking of F125. Note that most of the interactions are contributed by the guanosine phoshate. Hydrogen bonding distances are indicated by dashed lines and contributions of symmetry-related residues are marked with an asterisk (N154*).
VP40 Binds RNA Specifically
The RNA oligonucleotide is composed of a 5′-phosphate-depleted uridine residue (Uri1R, bound to molecule A), followed by a guanosine phosphate (Gua2R) and an adenosine phosphate (Ade3R) at the 3′ end. Both termini are pointing into the interior of the pore, being accessible for bulk solvent and, putatively, for 3′- and/or 5′-elongated RNA molecules Figure 1, Figure 3. The high quality of the final electron density map, including the observed interactions, permitted unambiguous identification of this Uri-Gua-Ade nucleotide (Figure 1D). Uri1R displays its sugar puckering as C2′-endo [29] (Table 2) , which is stabilized by a number of polar interactions between symmetry-related Uri1R residues. The base moiety is positioned in syn conformation, fixed by an internal hydrogen bond (Uri1R O5′-O2, 3.3 Å). This positions the aromatic base ring perfectly parallel to that of a symmetry-equivalent Uri1R* residue (3.9 Å between planes), resulting in a very strong π interaction (Figure 3B).
Table 2.
Conformational Parameters of the Bound Triribonucleotide
| Residue Number | Residue Name | Base Position Relative to Sugar | Value of χ | Orientation around C4′–C5′ | Sugar Puckering | |
|---|---|---|---|---|---|---|
| 5′ | ||||||
| 1R | Uri | syn | 52.8° | gauche(+) | C2′-endo | |
| 2R | Gua | syn | 67.6° | gauche(−) | C2′-endo | |
| 3R | Ade | anti | −102.2° | gauche(+) | C3′-endo | |
| 3′ |
Nucleoside Uri1R is connected with Gua2R by a phosphate-linker in extended conformation, and no interactions are observed between these two residues. Gua2R is also puckered as purely C2′-endo, with its O2′ atom stabilized by an interaction with the protein moiety (see below). The base, equally in syn conformation, is accommodated in a pocket in the inner pore surface and at the interface between protein protomers (see below). This pocket is formed by protein residues and by the perfectly parallel base plane of a symmetry-related Gua2R* residue.
The sugar-phosphate backbone folds back after Gua2R to reach Ade3R, so that both the guanine and adenine bases come to interact with each other, being approximately perpendicular (Gua2R N2-Ade3R N1, 2.9 Å). A further interaction of the adenine is observed with the preceding phosphate group (Ade3R N6-Gua2R O2P, 3.3 Å). The sugar puckering of this 3′-terminal residue is C3′-endo, enabling another interaction with the sugar of the preceding residue (Gua2R O2′-Ade3R O4′, 3.4 Å), and the base is positioned in anti (Figure 3B and Table 2).
RNA-Protein Interactions
Inside of the central cavity of the ring, the dimer-dimer interface is stabilized by the interaction with the ssRNA segment (Figure 3A). The main specificity of the observed trinucleotide binding resides in the central guanosine phosphate. The guanine base is placed in a deep pocket located at the interface between two protein protomers related by crystallographic symmetry (residues from the symmetry-related polypeptide are characterized by an asterisk) and interacts mainly with Arg134 (Gua2R O6-Arg134 Nϵ, 2.9 Å; Gua2R N7-Arg134 Nη2, 2.9 Å) and Phe125 (parallel ring stacking, 3.4 Å between aromatic planes) of one protein chain. The backbone phosphate group of Gua2R is linked via a double-headed salt bridge with Arg134* (Gua2R O2P-Arg134 Nη1, 2.9 Å; Gua2R O5′-Arg134 Nη2, 3.0 Å), while the sugar moiety contacts the protein main chain (Gua2R O2′-Gly126 N, 3.1 Å) (Figure 3C)
Fewer contacts are observed between the flanking residues of the oligonucelotide and the protein. Nucleoside Uri1R is not involved in any polar interactions with the protein octamer, and only hydrophobic forces are established by the aromatic base to the side chains of Leu132* and Leu158*. The 3′-terminal nucleotide Ade3R is placed in a shallow cavity lined out by the protein main chain (His123-Gly126) and the side chains of Tyr171, Asn154*, and Ile152*. It interacts directly via its base with the protein through one hydrogen bond (Ade3R N1-Asn154* Nδ2, 3.0 Å) and via its sugar moiety through a second one (Ade3R O2′-Gly126 O, 3.0 Å) (Figure 3C).
The observed structural features and the fact that the protein oligomer was capable of sequestering a triribonucleotide of a particular sequence from the expression host E. coli strongly suggest that the octameric VP40 N-terminal domain displays a marked selectivity for the specified sequence, in particular for a central guanosine phosphate. Furthermore, the implication of the sugar O2′ atoms in a number of key interactions (both intra- and intermolecular) further permit to discard the potential binding of a trideoxyribonucleotide with an equivalent sequence. Therefore, the observed binding must be considered both RNA- and sequence-specific. In addition, all polar residues involved in the RNA interaction are strictly conserved between Ebola and Marburg VP40 sequences and some van der Waals contacts are substituted by conservative changes.
Structural Rearrangements of VP40 upon Oligomerization
The NTD of the monomeric closed conformation [9] and the protomer within the oligomeric structure can be overlaid with an rms deviation (matching the Cα atoms of residues 72 to 189) of 2.8 Å. Most of the differences observed lie in the loop regions and the positions of α helix 2 and β strand 6. In particular, α helix 2 moves 3.3 Å (at Cα position 117) and β strand 6 4.3 Å (at Cα position 173), resulting in major differences within this region (Figure 4A) .
Figure 4.
Comparison of the NTDs Derived from the Closed Monomeric Conformation and from the Ring Structure Unveils Major Conformational Changes
(A) Superposition of Cα atoms 71 to 191 results in an rms deviation of 2.8 Å. Sites of major conformational movements are indicated with arrows. The NTD from the closed monomeric conformation is shown in red and the one from the octamer structure in yellow.
(B) Schematic overview of the two major conformational changes in VP40. An N-terminal loop (gray) and the C-terminal domain (gray) from the closed VP40 conformation must change their conformation to achieve octamerization. This is indicated by the ribbon drawing of the three regions involved; the potential movement of the two domains with respect to the N-terminal domain is highlighted by arrows.
Therefore, two conformational changes are observed to facilitate the transition from the monomer to the octamer. First, N-terminal residues 31 to 70, which pack in an extended conformation together with a 310-helix (helix 1) against the core of the β sandwich structure in the monomeric conformation, have to unfold and be expelled from the shallow cleft created by β strands 3 and 6 to allow the formation of the dimer-dimer interface with its RNA binding pocket (Figure 4B). The N-terminal chain runs between α helix 2 and β strand 6 and both structural features move substantially in the oligomeric conformation (Figure 4A). In addition, the original chain direction from the monomeric form would also position the N terminus at the inside of the ring structure, which may not be large enough to accommodate eight additional polypeptide chains.
The second conformational change includes the movement of the C-terminal domain out of its position to facilitate the formation of the antiparallel dimer, which occupies an almost identical interface as seen between the N- and C-terminal domains of the closed monomeric conformation (Figure 3, Figure 4; see also Figure 4 in [9]). The linker between both domains could be as long as ∼30 Å, as it contains 11 residues up to the first β strand (β strand 7) of the C-terminal domain [9] and may therefore position the C-terminal domain in a random position with respect to the NTD.
Detection of Oligomeric VP40 In Vivo
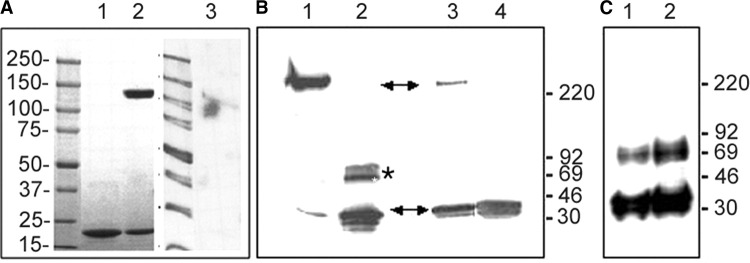
Part of the NTD construct VP40(31–212), which was used for crystallization, reveals SDS resistance when separated under nonboiling conditions on SDS-PAGE. A high molecular weight form is detected that migrates close to the 150 kDa marker protein (Figure 5A , lane 2). The same oligomeric form is observed when crystals belonging to space group P6222 are dissolved and separated under nonboiling conditions (Figure 5A, lane 3). This indicates that the SDS resistance is an intrinsic feature of octameric ring-like VP40 in complex with RNA. In addition, monomeric VP40 adopts the same oligomeric SDS-resistant structure when incubated with 2 M urea in the presence of E. coli nucleic acids. The formation of ring-like structures was also confirmed by electron microscopy (data not shown). The oligomeric form of SDS-resistant VP40 in complex with nucleic acids thus migrates above the 220 kDa marker protein (Figure 5B; lane 1), while monomers and dimers are detected under boiling conditions (Figure 5, lane 2).
Figure 5.
SDS Resistance of Oligomeric Ring-like VP40 in Complex with RNA
(A) SDS resistance of VP40(31–212). Lane 1, boiled sample; lane 2, nonboiled sample; lane 3, nonboiled sample of crystals of VP40(31–212) in space group P6222.
(B) SDS resistance of full-length VP40. Lane 1, full-length recombinant VP40 destabilized in the presence of E. coli nucleic acids and separated on SDS-PAGE; lane 1, nonboiled; lane 2, boiled. Lane 3, detection of full-length VP40 in Ebola virus infected cells under nonboiling conditions and boiled (lane 4). Note that the high molecular weight form in lane 3 corresponds to the size of the oligomeric full-length VP40 generated in vitro (lane 1). The monomeric form and the proposed octameric form are both indicated by arrows and a band corresponding to a dimer (lane 2) is indicated by an asterisk.
(C) SDS resistance analysis of VP40 present in purified Ebola virus particles. Samples were separated under boiled (lane 1) and nonboiled conditions (lane 2).
Samples were separated on gradient SDS-PAGE and positions of marker proteins are indicated. Bands in (A) were detected by Coomassie blue staining and in (B) and (C) by Western blot.
Analysis of VP40 SDS resistance in cells infected with Ebola virus reveals that a fraction of the total protein also shows SDS resistance, specified by the high molecular weight band that migrates at the same position as SDS-resistant recombinant VP40 (Figure 5B, lanes 3 and 1). In addition, no dimeric form of VP40 can be detected in infected cells under either separation condition. This strongly indicates, together with the RNA binding properties of full-length oligomeric VP40 in vitro, that octameric VP40 bound to a specific RNA sequence plays a role in the virus life cycle. We then analyzed purified Ebola virus particles to determine whether octameric VP40 in complex with RNA is part of the virus particle. Separation of samples under boiled and nonboiled conditions produced mostly monomers and a band corresponding to dimers of VP40. However, no high molecular weight band corresponding to the SDS-resistant form detected in virus-infected cells could be detected, indicating that octameric VP40 might not be abundantly present in virus particles (Figure 5C).
Discussion
Viral matrix proteins from negative-strand RNA viruses participate in the assembly of lipid-enveloped viruses by providing a link between the surrounding membrane and the nucleocapsid structure. The unraveling of the crystal structure of octameric VP40 in complex with RNA may suggest yet another still uncharacterized function for the matrix protein. The structure shows that VP40 has to undergo two major conformational changes in order to allow the observed oligomerization. This involves the movement of the C-terminal domain, which had been suggested earlier [11], as well as the displacement of the N-terminal region (residues 31 to 70), which then generates the binding pocket for the specific recognition of the ssRNA motif U-G-A. Indeed, the octamer structure shows that the interface of the dimeric subunit is similar to the interface occupied by the N- and C-terminal domains in the closed conformation.
The binding of the ssRNA is typical for protein-RNA interactions with the characteristic parallel stacking of bases and aromatic residues, such as Phe125. As reported for many protein/RNA complexes, adenine is coordinated by recognition of its N1 and N6 groups and the recognition of guanine involves arginines, a role that is fulfilled by Arg134 [30]. Phe125 and Arg134 are the most important residues in the interaction and both are positioned next to each other and exposed close to the interface of the N- and C-terminal domains in the monomeric conformation (see Figure 5C in [9]), which might implicate them in the transition process. Uracil at the 5′ end is less well fixed by protein-RNA interactions; however, it is still unambiguously defined by electron density and well coordinated by stacking against hydrophobic residues Leu132 and Leu158 as well as a symmetry-related uracil group, including polar interactions between neighboring uridines. Interestingly, all polar residues involved in the protein-RNA interaction are strictly conserved and all hydrophobic residues are either conserved, such as Phe125, or substituted by conservative changes among all available sequences from Ebola virus subtypes (Zaire, Reston) and Marburg virus strains (Musoke and Popp). We propose that this conservation is significant (34% identity between Ebola [Zaire] and Marburg [Popp] VP40 NTDs), and it indicates that Marburg virus VP40 may also interact specifically with nucleic acids.
A number of RNA binding proteins form oligomers that assemble into ring-like structures with specific RNA binding properties on the outside of a ring framework, as in case of octameric rotavirus NSP2 [31] and TRAP (trp RNA binding attenuation protein) [32]. In addition, RNA recognition can also occur on the inside of the ring, as in the case of the Sm protein family, which assembles into hexamers or heptamers and functions in the biogenesis and stability of mRNA 33, 34 and TB-RBP, which forms octamers and controls the translation of stored mRNAs in testis and brain [35]. The general function of binding multiple RNA copies could be as simple as concentrating multiple copies at specific sites. In case of VP40, the binding of the RNA seems to follow an induced fit pathway, as suggested for general RNA-protein recognition [36], which evolved to accommodate multiple functions within a single polypeptide chain by creating a symmetric homo-oligomer. The C-terminal domains, which are not present in the structure, could be still involved in membrane association and might therefore localize specific action to membrane compartments as described for positive-strand RNA virus replication strategies [37].
The RNA ligand greatly stabilizes the dimer-dimer interface, which is similar to the RNA-induced dimerization of rotavirus NSP3 [38]. VP40(31–212) shows SDS resistance when separated under nonboiling conditions on SDS-PAGE, which can be attributed to the presence of nucleic acids in the sample 17, 56. Only SDS-resistant high molecular weight VP40 is present in the hexagonal crystal form, which indicates preferential crystallization of this form. Furthermore, we show that full-length VP40 can be transformed into a SDS-resistant form in the presence of E. coli nucleic acids, assembling into ring-like structures, as determined by electron microscopy. Interestingly, the same SDS-resistant high molecular weight form is found in cells infected with Ebola virus. Although the amount present in these cells is low, it clearly indicates that this oligomeric form of VP40, which depends on RNA binding, is present in infected cells and plays a specific role in the viral life cycle. In contrast, the high molecular weight SDS-resistant form is not present in virus particles, indicating a role distinct from the direct assembly of particles. We find, however, a SDS-resistant dimeric form of VP40, which could be part of higher-order oligomers or hexamers as suggested 10, 11. Notably, the SDS-resistant dimeric form is not detected in infected cells.
VP40 had been previously shown to form ring-like structures in solution, which had been suggested to contain antiparallel dimers forming trimers 10, 11. Further analysis of our previous data revealed that the original preparations contained both a mixture of hexamers and octamers due to the presence of nucleic acids in the sample [56].
The function of octameric VP40 in the virus life cycle is still elusive. It might have a role in RNP formation as described for a number of matrix proteins 21, 22, 23, 24, 25, 26, although recent evidence would not directly support such a role [7]. On the other hand, it might be in agreement with the detection of viral RNPs associated with VP40 in inclusions of Marburg virus-infected cells [20]. Other potential interactions besides those with the viral genome may involve filovirus mRNAs containing the triribonucleotide sequence [39], and such complexes may function in transcription or translation control. Lastly, we cannot exclude the possibility that the bound (cellular or viral) RNA acts solely as an adaptor in order to generate a conformation of VP40 suitable to interact with an unknown regulatory protein, active in the virus life cycle.
Biological Implications
Viral matrix proteins play an important role in the assembly and budding processes of enveloped viruses. They are positioned underneath the viral membrane and ensure the integrity of mature viral particles. VP40 is a monomer in solution and contains two domains [9]. The N-terminal domain is involved in homo-oligomerization [10] and interaction with cellular budding factors 16, 17, 18, while the C-terminal domain is instrumental for membrane association 10, 12.
The structure of octameric VP40 binds sequence-specific ssRNA and represents the first crystal structure of a matrix protein from an enveloped virus in complex with ssRNA. As the limiting size of a viral genome to be packaged into virus particles leads to evolutionary pressure to increase the coding content of the genome without changing its size, several conformations of VP40 will therefore allow the matrix protein to exert multiple tasks. We provide evidence that the RNA bound form of octameric VP40 is not an abundant component of the assembled particle, as its SDS-resistant form is only found in infected cells and not in mature viral particles. It is therefore most likely involved in a yet unknown regulatory step during the life cycle of the virus. Our data also suggest that Marburg viruses employ a similar strategy, as all residues involved in RNA interaction are conserved between Ebola and Marburg virus strains. The octamer structure of VP40 now provides the framework for a precise functional analysis. In addition, the presence of the octameric form of VP40 in infected cells together with the well-defined RNA binding pocket may render VP40 a new target for antiviral drug development.
Experimental Procedures
Expression and Crystallization
VP40(31–212) was expressed and purified as described [10], and a C-terminally truncated form of VP40, VP40(31–194), was cloned, expressed, and purified as described for VP40(31–212). For crystallization, 1 μl of VP40(31–212) (10 mg/ml) and 1 μl of well solution (100 mM HEPES [pH 7.5], 1.5 M NH4H2PO4, 15% MPD), and 1 μl of VP40(31–194) (13 mg/ml) and 1 μl of well solution (100 mM Na-acetate [pH 4.6], 35% MPD, 4% glycerol) were mixed and equilibrated against the well buffer. VP40(31–194) was nonspecifically proteolyzed in the crystallization drop, and the N terminus of a crystal was determined to start at position 55 by chemical sequencing [VP40(55–194)]. For cryo-protection, VP40(31–212) crystals were briefly transferred to well solution containing 5% glycerol, and VP40(31–194) crystals were transferred into well solution containing 15% glycerol before flash cooling in a stream of liquid nitrogen.
VP40(55–194): Diffraction Data Collection and Processing
VP40(55–194) crystallizes in space group P422 with unit cell dimensions of a = b = 80.51 Å and c = 46.77 Å, with one molecule per asymmetric unit and a Matthews-parameter VM = 2.5 Å3/Da (50% solvent; [40]). One native data set was collected at beamline ID14-EH1 (ESRF, Grenoble). A heavy-ion derivative was obtained by soaking a crystal during 6 hr in the cryo-protecting buffer containing 200 μM potassium tetra-chloroplatinate(II), and one data set was collected on a rotating anode source. A multiple-anomalous diffraction (MAD) experiment at the platinum LIII absorption edge was performed at beamline BM-14 (ESRF, Grenoble). Therefore, two data sets, at λ = 1.069 Å (absorption peak) and λ = 0.979 Å (hard remote), were collected from the same crystal, with data extending to 1.63 Å (Table 1). All diffraction data were indexed and integrated with DENZO [41], scaled, merged, and reduced with SCALA within the CCP4 package [42].
VP40(55–194): Structure Solution, Model Building, and Refinement
A single platinum site was located in an anomalous difference-Patterson map (bound to Met89 Sδ). Phases up to 1.63 Å resolution were calculated from this single refined site with the program SHARP [43], from the phases provided by the two-wavelength MAD (synchrotron data) and the SIRAS (rotating-anode data) experiments (Table 1). A posterior density modification step (solvent flipping) with SOLOMON [42] rendered an electron density map of excellent quality that permitted density improvement and automated chain tracing using wARP/ARP [44]. Subsequent final corrections of the model, the introduction of solvent molecules where appropriate if present in σA-weighted 2mF obs − DF calc and mF obs − DF calc maps, and identification and building of the triribonucleotide were performed with the programs O [45] and TURBO-FRODO [46]. Positional and temperature-factor crystallographic refinement was performed with the program CNS v. 1.0 [47]. As a last step, REFMAC5 was used with TLS refinement [48] to better account for the anisotropic thermal displacements. The final refined model consists of protein residues 69 to 192 (molecule A), ribonucleotides 1 to 3 (chain R), and 158 solvent molecules. It has been refined employing native data to 1.6 Å resolution, with a final crystallographic R value of 16.4% and an Rfree value of 18.2%. The model has good stereochemistry (Table 1) and all residues lie within the allowed regions of a Ramachandran plot, as checked with PROCHECK [49].
VP40(31–212): Data Collection, Structure Solution, and Refinement
VP40(31–212) crystallizes in space group P6222 with unit cell dimensions a = b = 79.61 Å and c = 239.18 Å, and two monomers per asymmetric unit (VM = 2.7 Å3/Da; 55% solvent contents). A native data set to 2.6 Å resolution was collected at beamline ID14-EH4 at the European Synchrotron Radiation Facility (ESRF, Grenoble) and processed as described (see Table 1). The structure was solved by Patterson-search techniques using the superimposed coordinates of the VP40(55–194) structure and those of the NTD as present in the full-length molecule [9]. The rotation function could only be solved employing the program BEAST [50]. Translation functions were then computed with AMoRe [51], resulting in a correlation coefficient (CC) of 47.0% and an R value of 47.9%. This calculation corroborated P6222 as the correct space group. An electron density map calculated with the obtained model phases revealed extra density in the central cavity of the octameric protein ring, confirming the presence of the triribonucleotide (not used during the searches nor for phasing) and thus the correctness of the solution. The structure was improved in several cycles of manual model building and crystallographic refinement as mentioned above. The final model includes protein residues 69 to 190 (molecule A), residues 69 to 191 (molecule B), two triribonucleotides of sequence 5′-U-G-A-3′ (chains R and S), and 113 water molecules. Two chloride anions at the interface between octamers were tentatively assigned based on very strong electron density peaks that could not be explained by solvent molecules. Residues 31 to 68 and 191/192 to 212 in each protomer are disordered (33% of the total). Therefore, the structure could not be refined beyond a crystallographic R value of 30.6% (Rfree value of 32.9%; see Table 1). The model has good stereochemistry (Table 1) and all residues lie in allowed regions of the Ramachandran plot, with just one exception. The final model displays high temperature factors for the ribonucleotide moieties. One of the two triribonucleotides clearly displays electron density for a phosphate group on the 5′ terminus, indicating that ssRNA extends in a flexible way in that direction.
Structure Analysis
Figures were generated using the programs MOLSCRIPT [52], RASTER 3D [53], and TURBO-FRODO [46]. Secondary structure elements were assigned with DSSP [54], and the superposition of Cα traces was performed with the program LSQKAB [42]. The atomic coordinates have been deposited with the RCSB Protein Data Bank under access codes 1h2c and 1h2d.
SDS Resistance Analysis of Recombinant VP40
VP40(31–212) was separated on SDS-PAGE gradient gels with and without prior boiling of the samples in standard sample buffer containing 0.5% SDS (w/v). Hexagonal crystals of VP40(31–212) were washed extensively in the crystallization buffer and then dissolved in SDS loading buffer and separated under nonboiling conditions. Full-length monomeric VP40 was purified as described [10] and then destabilized in 2 M urea in the presence of an excess of E. coli nucleic acids (1:1; w/w).
VP40 Analysis in Ebola Virus-Infected Cells
Ebola virus-Zaire strain Mayinga was grown, passaged, and purified as described [55]. Ebola virus-infected cells were washed 24 hr postinfection and lysed for 5 min at 4°C in PBS containing 1% Triton X-100. Lysates were centrifuged for 2 min at 9200 rpm and supernatants were mixed with sample buffer resulting in a final concentration of 0.5% SDS. Samples were divided and one part was boiled for 5 min while the other part left at RT. As a positive control, recombinant octameric VP40 was used and treated under the same conditions as described above. Samples were loaded onto an SDS-PAGE and blotted onto PVDF membranes that were incubated with a mouse monoclonal anti-VP40 antibody (dilution 1:100), followed by a goat anti-mouse IgG (dilution 1:40,000) as secondary antibody.
VP40 Analysis in Purified Ebola Viruses
SDS resistance of virion-associated VP40 was tested using 20 μl of purified virions (∼2 × 108 particles) that were incubated with PBS/1% Triton X-100 for 5 min at 4°C and further treated as described above.
Acknowledgements
We thank all members of the ESRF/EMBL Joint Structural Biology Group for support at the ESRF beamlines and S. Scianimanico for excellent technical assistance. W.W. acknowledges the financial support from EMBL and F.X.G.-R by grants BIO2000-1659 (Ministerio de Ciencia y Tecnología, Spain) and SGR2001-346 (Generalitat de Catalunya, Catalunya) and the Visiting Scientist status at EMBL Outstation Grenoble. S.B. acknowledges the financial support by the Deutsche Forschungsgemeinschaft (SFB 535, TP B9, and SFB 286, TP A6).
Published: April 1, 2003
References
- 1.Bruce J., Brysiewicz P. Ebola fever: the African emergency. Int. J. Trauma Nurs. 2002;8:36–41. doi: 10.1067/mtn.2002.123083. [DOI] [PubMed] [Google Scholar]
- 2.Volchkov V.E., Volchkova V.A., Mühlberger E., Kolesnikova L.V., Weik M., Dolnik O., Klenk H.-D. Recovery of infectious Ebola virus from cDNA: RNA editing of the GP gene and viral cytotoxicity. Science. 2001;291:1965–1969. doi: 10.1126/science.1057269. [DOI] [PubMed] [Google Scholar]
- 3.Feldmann H., Volchkov V.E., Volchkova V.A., Stroher U., Klenk H.D. Biosynthesis and role of filoviral glycoproteins. J. Gen. Virol. 2001;82:2839–2848. doi: 10.1099/0022-1317-82-12-2839. [DOI] [PubMed] [Google Scholar]
- 4.Geisbert T.W., Jahrling P.B. Differentiation of filoviruses by electron microscopy. Virus Res. 1995;39:129–150. doi: 10.1016/0168-1702(95)00080-1. [DOI] [PubMed] [Google Scholar]
- 5.Becker S., Huppertz S., Klenk H.D., Feldmann H. The nucleoprotein of Marburg virus is phosphorylated. J. Gen. Virol. 1994;75:809–818. doi: 10.1099/0022-1317-75-4-809. [DOI] [PubMed] [Google Scholar]
- 6.Becker S., Rinne C., Hofsass U., Klenk H.D., Mühlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology. 1998;249:406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y., Xu L., Sun Y., Nabel G.J. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- 8.Ellis D.S. In: Animal Virus Structure. Nermut M.V., Steven A.C., editors. Elsevier; Amsterdam: 1987. The filoviridae. 313–321.pp. [Google Scholar]
- 9.Dessen A., Volchkov V., Dolnik O., Klenk H.D., Weissenhorn W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000;19:4228–4236. doi: 10.1093/emboj/19.16.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruigrok R.W., Schoehn G., Dessen A., Forest E., Volchkov V., Dolnik O., Klenk H.D., Weissenhorn W. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 2000;300:103–112. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- 11.Scianimanico S., Schoehn G., Timmins J., Ruigrok R.H., Klenk H.D., Weissenhorn W. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 2000;19:6732–6741. doi: 10.1093/emboj/19.24.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timmins J., Scianimanico S., Schoehn G., Weissenhorn W. Vesicular release of Ebola virus matrix protein VP40. Virology. 2001;283:1–6. doi: 10.1006/viro.2001.0860. [DOI] [PubMed] [Google Scholar]
- 13.Bavari S., Bosio C.M., Wiegand E., Ruthel G., Will A.B., Geisbert T.W., Hevey M., Schmaljohn C., Schmaljohn A., Aman M.J. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasenosky L.D., Neumann G., Lukashevich I., Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 2001;75:5205–5214. doi: 10.1128/JVI.75.11.5205-5214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noda T., Sagara H., Suzuki E., Takada A., Kida H., Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty R.N., Brown M.E., Wang G., Huibregtse J., Hayes F.P. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timmins J., Schoehn G., Ricard-Blum S., Scianimanico S., Vernet T., Ruigrok R.W.H., Weissenhorn W. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 2003;326:493–502. doi: 10.1016/s0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- 18.Martín-Serrano J., Zang T., Bieniasz P.D. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 19.Kolesnikova L., Mühlberger E., Ryabchikova E., Becker S. Ultrastructural organization of recombinant Marburg virus nucleoprotein: comparison with Marburg virus inclusions. J. Virol. 2000;74:3899–3904. doi: 10.1128/jvi.74.8.3899-3904.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolesnikova L., Bugany H., Klenk H.D., Becker S. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J. Virol. 2002;76:1825–1838. doi: 10.1128/JVI.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaptur P.E., Rhodes R.B., Lyles D.S. Sequences of the vesicular stomatitis virus matrix protein involved in binding to nucleocapsids. J. Virol. 1991;65:1057–1065. doi: 10.1128/jvi.65.3.1057-1065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coronel E.C., Takimoto T., Murti K.G., Varich N., Portner A. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 2001;75:1117–1123. doi: 10.1128/JVI.75.3.1117-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stricker R., Mottet G., Roux L. The Sendai virus matrix protein appears to be recruited in the cytoplasm by the viral nucleocapsid to function in viral assembly and budding. J. Gen. Virol. 1994;75:1031–1042. doi: 10.1099/0022-1317-75-5-1031. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt A.P., Leser G.P., Waning D.L., Lamb R.A. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 2002;76:3952–3964. doi: 10.1128/JVI.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X., Liu T., Muller J., Levandowski R.A., Ye Z. Effect of influenza virus matrix protein and viral RNA on ribonucleoprotein formation and nuclear export. Virology. 2001;287:405–416. doi: 10.1006/viro.2001.1067. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed M., Lyles D.S. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J. Virol. 1998;72:8413–8419. doi: 10.1128/jvi.72.10.8413-8419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Kobbe C., van Deursen J.M., Rodrigues J.P., Sitterlin D., Bachi A., Wu X., Wilm M., Carmo-Fonseca M., Izaurralde E. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 29.Sänger W. Principles of nucleic acid structure. Springer Verlag; New York: 1984. [Google Scholar]
- 30.Allers J., Shamoo Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 31.Jayaram H., Taraporewala Z., Patton J.T., Prasad B.V. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature. 2002;417:311–315. doi: 10.1038/417311a. [DOI] [PubMed] [Google Scholar]
- 32.Antson A.A., Dodson E.J., Dodson G., Greaves R.B., Chen X., Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 33.Hermann H., Fabrizio P., Raker V.A., Foulaki K., Hornig H., Brahms H., Lührmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Törö I., Thore S., Mayer C., Basquin J., Seraphin B., Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pascal J.M., Hart P.J., Hecht N.B., Robertus J.D. Crystal structure of TB-RBP, a novel RNA-binding and regulating protein. J. Mol. Biol. 2002;319:1049–1057. doi: 10.1016/S0022-2836(02)00364-9. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Canadillas J.M., Varani G. Recent advances in RNA-protein recognition. Curr. Opin. Struct. Biol. 2001;11:53–58. doi: 10.1016/s0959-440x(00)00164-0. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell. 2002;93:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 38.Deo R.C., Groft C.M., Rajashankar K.R., Burley S.K. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell. 2002;108:71–81. doi: 10.1016/s0092-8674(01)00632-8. [DOI] [PubMed] [Google Scholar]
- 39.Mühlberger E., Trommer S., Funke C., Volchkov V., Klenk H.D., Becker S. Termini of all mRNA species of Marburg virus: sequence and secondary structure. Virology. 1996;223:376–380. doi: 10.1006/viro.1996.0490. [DOI] [PubMed] [Google Scholar]
- 40.Matthews B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z., Minor W. Processing of x-ray data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.CCP4 The CCP4 (Collaborative Computational Project 4) suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.de la Fortelle E., Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 44.Perrakis A., Morris R., Lamzin V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 45.Jones T.A., Kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 46.Roussel, A., and Cambilleau, C. (1989). Turbo-Frodo. In Silicon Graphics Geometry Partners Directory (Mountain View, CA: Silicon Graphics), pp. 77–79.
- 47.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Winn M.D., Isupov M.N., Murshudov G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–290. [Google Scholar]
- 50.Read R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 51.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 52.Kraulis P. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:924–950. [Google Scholar]
- 53.Merritt E.A., Bacon D.J. Raster3D photorealistic graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 54.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 55.Becker S., Feldmann H., Will C., Slenczka W. Evidence for occurrence of filovirus antibodies in humans and imported monkeys: do subclinical filovirus infections occur worldwide? Med. Microbiol. Immunol. (Berl.) 1992;181:43–55. doi: 10.1007/BF00193395. [DOI] [PubMed] [Google Scholar]
- 56.Timmins J., Schoehn G., Kohlhaas C., Klenk H.-D., Ruigrok R.W.H., Weissenhorn W. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology. 2003;in press doi: 10.1016/s0042-6822(03)00260-5. [DOI] [PubMed] [Google Scholar]