Abstract
The initial steps of the Human Immunodeficiency Virus (HIV) replication cycle play a crucial role that arbitrates viral tropism and infection efficiency. Before the release of its genome into the host cell cytoplasm, viruses operate a complex sequence of events that take place at the plasma membrane of the target cell. The first step is the binding of the HIV protein envelope (Env) to the cellular receptor CD4. This triggers conformational changes of the gp120 viral protein that allow its interaction with a co-receptor that can be either CCR5 or CXCR4, defining the tropism of the virus entering the cell. This sequential interaction finally drives the fusion of the viral and host cell membrane or to the endocytosis of the viruses. Here, we discuss how the membrane composition and organization of both the virus and the target cell can affect these steps and thus influence the capability of the viruses to infect cells.
Keywords: HIV, Lipids, Membrane dynamics
Highlights
-
•
An overview of lipid role in HIV infection is proposed.
-
•
We discuss the influence of lipid composition on HIV early steps of infection.
-
•
We discuss the role of membrane organization an dynamics in HIV entry.
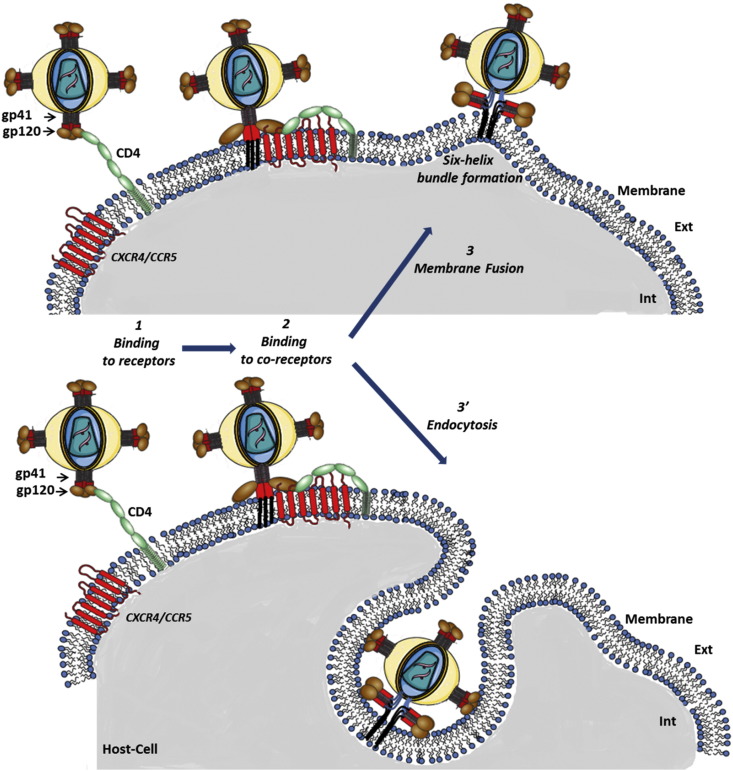
1. The entry of HIV virus in the target cell follows a complex sketch
The entry of the virus into the cell can be decomposed in three main successive steps: 1/the interactions of the viruses with their specific receptors CD4, 2/the interactions of the viruses with co-receptors (CCR5 or CXCR4) and 3 or 3′/either the fusion of the membrane of the virus with the one of the infected cell or the endocytosis of the virus (Fig. 1 ). The efficiency of each of these steps is influenced by the membrane protein and lipid composition and by their lateral organization and dynamics at the surface of the target cell.
Fig. 1.
Schematic view of HIV-1 early steps of infection. The upper part represents the HIV-1 virus envelope. The bottom part corresponds to the infected immune cell (T-lymphocyte, macrophage or dendritic cell). 1- Binding of CD4 to gp120 leads to exposure of a co-receptor binding site in gp120. 2- Binding of gp120 to co-receptor: depending on virus tropism co-receptor can be either CCR5 (R5 tropism) or CXCR4 (X4 tropism). Co-receptor binding triggers new conformational changes of gp120, exposing gp41 protein that inserts into the membrane of host cell. 3- Fusion: gp41 protein bends back on itself, forming a six helix bundle that bring closer the viral and target cell membranes leading to their fusion. 3'- Endocytosis: an alternative pathway to fusion is the entry of virus into its cell host via endocytosis. Virus particle then can be degraded or they can fuse with vesicular membrane, releasing virus content into the cytoplasm of the cell.
1.1. Binding to CD4 receptors
It has been shown that HIV attachment to immune cells first occurs via non-specific adhesion molecules that restrict the virus envelope in vicinity of more specific receptors [1], [2]. CD4 is the principal and mandatory receptor. Upon binding to CD4 (step1), the viral gp120 protein undergoes a conformational change allowing additional interactions with a co-receptor (step 2). The main parameter that can affect the efficiency of this first step is the density of CD4 receptors that are present at the surface of the target cell [3], [4], [5]. This amount varies a lot from cell to cell type – in macrophages CD4 expression levels are 10–20 fold lower than what they are in CD4+ T cells – but also depends on the activation state of immune cells [6], [7]. For these reasons, there is a huge variability of infection efficacy depending both on the virus strain and the cell type which is infected, rendering difficult the interpretation and the comparisons of experiments carried out in different studies.
Beyond the expression level of CD4, a second factor that has to be taken into account is the lateral organization of the proteins. It is now well known that the lateral organization of proteins in the plasma membrane of the cells is heterogeneous [8], [9] and might depend on lipid composition [10]. In this context, many laboratories have observed that CD4 proteins are not randomly distributed at the surface of the plasma membrane of cells but rather confined into domains [11], [12], [13]. Such a clustering may promote HIV binding since viruses could engage several interactions with multiple CD4 confined in a small area.
CD4 has been shown to be partially located into lipid rafts [14]. Lipid rafts microdomains result from the preferential association between sphingolipids, saturated lipids, sterols and specific proteins in the external leaflet of the plasma membrane. These microdomains are found in a liquid-ordered state (Lo) that renders them resistant to solubilization by detergents at low temperature [15], [16], [17]. There is a growing amount of evidence that the presence of cholesterol in the target cell is essential for the infection by many viruses. Ebola virus [18], herpes simplex [19], [20] or murine coronavirus [21] all require cholesterol in the target cell membrane. A long controversy started at the beginning of the millennium to know whether HIV preferentially binds to CD4 proteins that are confined into lipid rafts or not. The putative role of lipid rafts first came from the observation that cholesterol depletion impaired HIV infection [22], [23], [24]. Similar results have also been obtained by using glucosylceramide synthase inhibitors that blocks glycosphingolipid biosynthesis [25]. Nevertheless, these approaches present two drawbacks. First, these treatments are very stressful for the cells and one cannot be sure that the observed effects are the result of the sole decrease in cholesterol content. Second, it does not reveal at which step lipid rafts might be involved in the infection process (receptor or co-receptor binding, fusion…). To go further in the comprehension of the role of lipid rafts in HIV infection, detergent extractions and optical microscopy studies have been used and produced contradictory results. As an example, one study showed that a CD4 mutant excluded from lipid raft domains membranes did not allow efficient HIV entry [26], whereas a similar study, using alternative mutants that also prevent CD4 association with lipid rafts, established that CD4 excluded from lipid rafts do support HIV entry [14]. These two studies have been carried out on different cell types (HEK and T cells, respectively). These cell lines very likely have different lipid composition and organization. This might explain the discrepancies in the results that have been described above. This also enlightens the crucial role of membrane composition and organization on the mechanisms of infection process. Apparent contradictions also arise from works published by the same authors that successively showed that HIV uses lipid raft-co-localized CD4 for productive entry into cells [24] and that CD4 receptor localized to non-raft membrane microdomains supports HIV-1 entry [27]. One hypothesis, that renders these puzzling observations compatible, is to take into consideration that these studies are probing different steps of the infection process (i.e. binding to CD4 receptor, binding to co-receptor or fusion). Our proposal is that the lipid raft localization of CD4 is not required for virus binding while the following steps are dependent on the presence of lipid rafts. This hypothesis is reinforced by the observation that CD4 lateral mobility affects HIV-1 envelope glycoprotein mediated fusion [28]. In other words, once viruses are bound to the cell surface they might migrate to lipid raft domains where the infection can be completed. Furthermore, this model is in agreement with the observation that murine leukemia virus “surfing” at the surface of infected cells precedes entry [29]. This shows that the different steps of HIV infection do not occur at the same place rendering necessary the development of dynamic approaches in order to study the time-course of infection process.
1.2. Binding to co-receptors
The major co-receptors of HIV-1 are the chemokine receptors CCR5 and CXCR4. The use of the co-receptors defines the viral tropism of the strain that will infect the cell. Viruses using CCR5 as a co-receptor are called R5, those using CXCR4 are named X4 while those that use both are termed R5X4 [30], [31]. R5 viruses prevail during the chronic phase of the infection while the emergence, after several years and for half the patients, of variants capable of using CXCR4 is associated to the rapid progress towards a pathologic state [32]. The mechanisms by which X4 viruses emerge and precipitate disease progression are still unknown. Different studies have proposed that the infection process requires the attachment of several gp120 envelope trimers to multiple CD4 and multiple co-receptors [33], [34], [35], [36]. Considering the weakness of CD4-gp120 interaction [2], the viruses should have a very short time (less than 0.2 s) to encounter their co-receptors once bound to CD4 [37], [38]. Virus entry might thus not only depend on the local membrane density of CD4 but also on that of chemokine receptors. This local density is expected to be favored by the sequestration of CCR5 and/or CXCR4 with CD4 into delimited membrane domains. This is further supported by high-resolution electron microscopy experiments showing that, CCR5, CXCR4 and CD4 are clustered on microvilli of human macrophages and T cells [13].
1.2.1. CCR5 co-receptor
Co-immunoprecipitation studies have shown a constitutive association between CD4 and CCR5 at the surface of T cells, macrophages and monocytes [39]. Similar studies performed on HEK cells showed the opposite [40]. A constitutive interaction has later been confirmed by FRET experiments that also revealed that this interaction takes place outside of lipid raft domains and is reinforced by the viral gp120 protein [41]. Another clue of CD4-CCR5 interaction came from the study of the membrane dynamics of these receptors. It has first been observed that CD4 and CCR5 presented different mobility [42]. More interestingly, we have shown that the presence of CD4 strongly affects the dynamics of CCR5. Indeed, when CCR5 was expressed alone in HEK 293T cells only one diffusing population confined into large domains was detected. This behavior was markedly modified in the presence of CD4 since two diffusing populations of CCR5 were then observed, leading to the conclusion that the interaction of one CD4 with several CCR5 leads to their confinement into smaller membrane domains than in the absence of CD4 [11]. These confinement zones that concentrate both CD4 and CCR5 might constitute platforms that would facilitate the binding (and/or consecutive fusion) of HIV to its target cell.
1.2.2. CXCR4 co-receptor
Because of the prevalence of R5 virus strain in primo-infections, most of studies concern CCR5 receptor and, comparatively, only few studies have been carried out on CXCR4 protein organization within the membrane of immune cells while many works concerns clinical investigations [43]. As for CCR5, a co-immunoprecipitation study has shown a constitutive association between CD4 and CXCR4 [40]. However, this study has been performed in HEK 293T cells transiently expressing CD4 with CXCR4 or CCR5 that gave contradictory results regarding CCR5 [39] as mentioned in the above CCR5 co-receptor chapter. More recent publications, respectively based on the use of cyclodextrin and ceramide modulators, have also shown that CXCR4 proteins are partially localized into lipid rafts domains and that this localization into lipid rafts is required for efficient infection [44], [45].
Even though literature can be contradictory in some points, it seems to be clear that receptor and co-receptors of HIV virus are confined into domains that could act as ports of entry for HIV viruses. This is for example illustrated by the fact that statins inhibit both R5 and X4 virus infection by down-regulating the activity of Rho GTPases which is expected to impede the clustering of host lipid raft–associated receptors [48]. The existence of different and independent confinement zones might also give an explanation for R5 prevalence at the expense of X4. For instance one can hypothesize that CD4 is preferentially confined with CCR5 into microdomains different from those containingCXCR4 [22]. This hypothesis is in agreement with the observation that reducing the expression of CD4 decreases the susceptibility of human cells to infection by X4 viruses [46]. Furthermore, it has been shown that lowering CCR5 expression level with molecules modulating cholesterol content (lovastatin, mevastatin and simvastatin), favors the infection by X4 viruses [47].
Taken together these results raise the question of the lateral distribution and the stoichiometry of the different receptors at the surface of the target cell. Recently, Johnston and collaborators have developed affinofile cells in which the expression level of both CD4 and CCR5 can be independently modulated [49]. Several strains stably expressing various amounts of CXCR4 have been generated; this allows obtaining any ratio of CD4, CCR5 and CXCR4. This tool provides a powerful method to characterize viral entry efficiency as a function of CD4 and CCR5 expression levels [50]. These affinofile cell lines also permitted to demonstrate that several CCR5 conformations (distinguished thanks to specific monoclonal antibodies), exhibiting different localization and G-protein association, might have implications in selective targeting of HIV-1 [51]. This system is also used in our lab to check whether the expression level of receptors and co-receptors can impact the dynamic of each other and the infection efficiency of various virus strains.
1.3. Fusion
Receptor binding and subsequent association with the co-receptor, lead to conformational changes of the viral trimeric gp120 protein. This induces the exposure of the hydrophobic gp41 peptide which inserts into the host cell membrane. This brings closer the viral and host membranes. The gp41 protein then bends back on itself, forming a six helix bundle [52], [53]. At this stage, the viral membrane and the target cell membrane have bypassed the repulsion forces and are close enough to spontaneously form the fusion pore [54], [55].
The role of lipids in the modulation of this step has been extensively studied and the implication of lipid rafts is now admitted by most of researchers. It has first been shown on large unilamellar vesicles (LUV) that sphingomyelin and cholesterol promote gp41 surface aggregation and membrane restructuring [56]. Such an aggregation is thought to be important to reach a stoichiometry allowing the fusion to occur. Indeed it has been proposed that at least two envelop proteins have to bind CD4 and a co-receptor and that two fusion proteins of each trimer have to insert into the target cell to promote viral entry [57]. The gp41 protein presents an extracellular domain that contains the fusion peptide whose structure strongly depends on its lipid environment. Some authors have proposed that a small amino acid sequence (LWYIK), within the gp41 protein, constitute a cholesterol-binding domain [58] which is important for HIV infectivity [59]. This sequence presents homology with the Cholesterol Recognition Amino-acid Consensus (CRAC) motifs described by Epand [60]. Further works realized on model membranes confirmed that the ectodomain membrane proximal region of gp41 was able to bind to cholesterol membrane domains [61]. The binding to such a cholesterol-rich (>30 mol%) zone promotes β-sheet secondary structuration of the fusion peptide that deeply embeds into the host membrane. This deep insertion has been shown to favor fusogenicity [62], [63], [64]. Beyond these effects on insertion and structuration of the gp41 virus protein, cholesterol might play a role in the membrane fusion itself. The fusion involves the coalescence of viral and hosts cell membranes, accompanied by the mixing of their cytoplasm. This implies that lipids transiently adopt a non-lamellar organization as described by Chernomordik and Kozlov [65]. Such an organization, named “hemifusion stalk”, can be stabilized by inverted cone-shaped and/or by cone-shaped lipids, depending on their outer or inner leaflet location. From this point of view, it is interesting to note that HIV membrane is enriched in phopshoinositides [66] that can induce a positive curvature that favor the initiation of fusion while cholesterol might mediate membrane curvature of host cell during fusion [67]. It has to be noticed that this effect of cholesterol is not specific to HIV fusion process and has been observed in other processes such as nuclear membrane formation [68].
Other authors have shown that sphingomyelin plays a role in HIV infection. A study based on surface aggregation assays of rhodamine-labeled gp41 subunit demonstrated that sphingomyelin and cholesterol promoted gp41-aggregation [56]. The effect of sphingomyelinase has also been studied by extraction of rafts, virus binding assays and measurements of the dynamics of HIV receptors. The authors observed that sphingomyelinase treatments decreased CD4 lateral mobility and inhibited HIV fusion with the membrane of the target cell. They propose that these effects are due to the clustering of CD4 molecules that prevents co-receptor engagement and HIV fusion [69].
It has to be noticed that the fusion of the viral membrane and the plasma membrane of the target cell does not necessarily conduce to productive infection since the viral content can be degraded inside the target cell. Furthermore, lipid mixing of viral and cell membranes have been observed without release of the viral content inside the target cell [70], [71]. This incomplete fusion is an argument in favor of an endocytotic pathway discussed in the following section.
1.4. Endocytosis
Although the mechanism of HIV fusion is now well documented (see part 3 of this manuscript), several lines of evidence suggest that HIV entry into their cell host can also occur via endocytosis [55], [72], [73], [74], [75], [76]. This process has moreover been proposed to be predominant in HIV internalization into macrophages, endothelial cells or epithelial cells [31], [77], [78], [79], [80] and has also been shown to depend on the cell activation status [81]. This mini-review is focused on the role specifically played by lipids in this process, we invite the reader to refer to the recent review by Melikian for a more general discussion of endocytosis versus fusion in HIV entry [71].
Kinetic measurement of fusion performed at different temperatures and the use of specific inhibitors pointed out that HIV viruses enter the cells via endocytosis followed by fusion with endosomes [70]. This work also showed that inhibiting endocytosis allowed lipid mixing of viral and cell membrane without conducing to a complete fusion. Interestingly, the obtained results were independent of virus tropism but kinetics appeared to depend on the target cell type. This might be due to differences in endosome trafficking, but it is tempting to propose that these variations might be due to differences in lipid composition of these cell lines. As an example, in similar experiments, Markosyan and collaborators have proposed that membrane tension might differ between cell types and affect pore formation and growth [82].
A study, based on cholesterol disruption of macrophage membranes, demonstrate that HIV entry into macrophages is dependent on lipid rafts [83]. This has later been confirmed by a study showing that endocytosis of HIV involves rafts [84]. The originality of this work comes from the fact it does not use pharmacological agents that may have non-specific effects. Thanks to genetically modified macrophages allowing manipulating the sub-cellular distribution of CD4, these authors have that the entry of HIV into macrophages is dependent on endocytosis through lipid raft containing CD4 [84].
Only few works have been carried out to specifically study the role of lipids in HIV endocytosis and it will be interesting to go further in this field.
2. Concluding remarks
One difficulty to understand HIV infection mechanism comes from the fact that different entry pathways exist and the importance of each being still a subject of controversy. Since most of HIV particles are degraded by the cells and only a small fraction of viruses establishes infection, the identification of productive entry pathway remains difficult.
However, whatever the mode of entry of viruses, many observations suggest that membrane composition and organization play important roles. Cholesterol, sphingomyelin and lipid raft domains seem to be of first importance not only in the infection process but also in virus prevalence [47]. Glycosphingolipids and gangliosides have also been reported to facilitate HIV infection such as galactosylceramides [85] or GM1 and GM3 [86], [87]. These lipids might serve as HIV binding sites or regulate dynamic and clustering of CD4, CCR5 and CXCR4. This information has been recently reinforced by the fact that peptides conjugated to lipid comprising sphingomyelin backbones impair early and late HIV-1 membrane fusion [88].
In this context, the development of new original and non-invasive techniques should allow to discriminate the entry pathway of viruses and to decipher the molecular mechanisms involved in the infection process. Among them, the Single Particle Tracking technique is promising. It has been used to study the dynamics and the organization of HIV receptors and co-receptors, revealing the existence of subpopulation of CD4 with different behaviors in T-lymphocytes [12]. Tracking entire viruses consists in labeling viruses with fluorescent probes and recording their movements with a videomicroscope. This might be useful to study the time-course of infection process. This approach gives information regarding the percentage of aborted binding (i.e. virus that bind to CD4 without encountering co-receptors and finally unbind). It also permits to measure the time-lapse from binding to cell surface until virus and target cell membranes fusion. These two parameters give real time insights onto the efficiency of the infection process. This method will be useful to verify previously obtained data on the behavior of single proteins in a more physiological and relevant context. Such an approach has already been successfully used with influenza viruses [89], [90] and might be extended to inactivated HIV viruses that keep their binding and fusion properties [91], [92]. Combined with the use of drugs to modulate lipid membrane composition these techniques will permit to understand the mechanisms involved in infection and more specifically the role played by lipids in this process.
Contributor Information
Fabrice Dumas, Email: fabricedumas@yahoo.fr, fabrice.dumas@ipbs.fr.
Laurence Salomé, Email: laurence.salome@ipbs.fr.
References
- 1.Saphire A.C., Bobardt M.D., Zhang Z., David G., Gallay P.A. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J. Virol. 2001;75:9187–9200. doi: 10.1128/JVI.75.19.9187-9200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ugolini S., Mondor I., Sattentau Q.J. HIV-1 attachment: another look. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- 3.Kabat D., Kozak S.L., Wehrly K., Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J. Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak S.L. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J. Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platt E.J., Wehrly K., Kuhmann S.E., Chesebro B., Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazazi F., Mathijs J.M., Foley P., Cunningham A.L. Variations in CD4 expression by human monocytes and macrophages and their relationships to infection with the human immunodeficiency virus. J. Gen. Virol. 1989;70(Pt 10):2661–2672. doi: 10.1099/0022-1317-70-10-2661. [DOI] [PubMed] [Google Scholar]
- 7.Mariani S.A., Vicenzi E., Poli G. Asymmetric HIV-1 co-receptor use and replication in CD4(+) T lymphocytes. J. Transl. Med. 2011;9(Suppl. 1):S8. doi: 10.1186/1479-5876-9-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagatolli L.A., Mouritsen O.G. Is the fluid mosaic (and the accompanying raft hypothesis) a suitable model to describe fundamental features of biological membranes? What may be missing? Front. Plant Sci. 2013;4:457. doi: 10.3389/fpls.2013.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelman D.M. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 10.Dumas F., Lebrun M.C., Tocanne J.F. Is the protein/lipid hydrophobic matching principle relevant to membrane organization and functions? FEBS Lett. 1999;458:271–277. doi: 10.1016/s0014-5793(99)01148-5. [DOI] [PubMed] [Google Scholar]
- 11.Baker A.M. CD4 interacts constitutively with multiple CCR5 at the plasma membrane of living cells. A fluorescence recovery after photobleaching at variable radii approach. J. Biol. Chem. 2007;282:35163–35168. doi: 10.1074/jbc.M705617200. [DOI] [PubMed] [Google Scholar]
- 12.Mascalchi P., Lamort A.S., Salomé L., Dumas F. Single particle tracking reveals two distinct environments for CD4 receptors at the surface of living T lymphocytes. Biochem. Biophys. Res. Commun. 2012;417:409–413. doi: 10.1016/j.bbrc.2011.11.129. [DOI] [PubMed] [Google Scholar]
- 13.Singer I.I. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells. J. Virol. 2001;75:3779–3790. doi: 10.1128/JVI.75.8.3779-3790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Percherancier Y. HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains. J. Biol. Chem. 2003;278:3153–3161. doi: 10.1074/jbc.M207371200. [DOI] [PubMed] [Google Scholar]
- 15.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 16.Pike L.J. Rafts defined: a report on the keystone symposium on lipid rafts and cell function. J. Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 18.Freitas M.S. Measuring the strength of interaction between the ebola fusion peptide and lipid rafts: implications for membrane fusion and virus infection. PLoS One. 2011;6:e15756. doi: 10.1371/journal.pone.0015756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bender F.C. Specific association of glycoprotein B with lipid rafts during herpes simplex virus entry. J. Virol. 2003;77:9542–9552. doi: 10.1128/JVI.77.17.9542-9552.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gianni T., Gatta V., Campadelli-Fiume G. {alpha}V{beta}3-integrin routes herpes simplex virus to an entry pathway dependent on cholesterol-rich lipid rafts and dynamin2. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22260–22265. doi: 10.1073/pnas.1014923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi K.S., Aizaki H., Lai M.M.C. Murine coronavirus requires lipid rafts for virus entry and cell-cell fusion but not for virus release. J. Virol. 2005;79:9862–9871. doi: 10.1128/JVI.79.15.9862-9871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak S.L., Heard J.M., Kabat D. Segregation of CD4 and CXCR4 into distinct lipid microdomains in T lymphocytes suggests a mechanism for membrane destabilization by human immunodeficiency virus. J. Virol. 2002;76:1802–1815. doi: 10.1128/JVI.76.4.1802-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Z., Cimakasky L.M., Hampton R., Nguyen D.H., Hildreth J.E. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retroviruses. 2001;17:1009–1019. doi: 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]
- 24.Popik W., Alce T.M., Au W.C. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells. J. Virol. 2002;76:4709–4722. doi: 10.1128/JVI.76.10.4709-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hug P. Glycosphingolipids promote entry of a broad range of human immunodeficiency virus type 1 isolates into cell lines expressing CD4, CXCR4, and/or CCR5. J. Virol. 2000;74:6377–6385. doi: 10.1128/jvi.74.14.6377-6385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del Real G. Blocking of HIV-1 infection by targeting CD4 to nonraft membrane domains. J. Exp. Med. 2002;196:293–301. doi: 10.1084/jem.20020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popik W., Alce T.M. CD4 receptor localized to non-raft membrane microdomains supports HIV-1 entry identification of a novel raft localization marker in CD4. J. Biol. Chem. 2004;279:704–712. doi: 10.1074/jbc.M306380200. [DOI] [PubMed] [Google Scholar]
- 28.Rawat S.S. Restricted lateral mobility of plasma membrane CD4 impairs HIV-1 envelope glycoprotein mediated fusion. Mol. Membr. Biol. 2008;25:83–94. doi: 10.1080/09687680701613713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehmann M.J., Sherer N.M., Marks C.B., Pypaert M., Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J. Cell. Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger E.A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 31.Lu Z. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verhofstede C., Nijhuis M., Vandekerckhove L. Correlation of coreceptor usage and disease progression. Curr. Opin. HIV AIDS. 2012;7:432–439. doi: 10.1097/COH.0b013e328356f6f2. [DOI] [PubMed] [Google Scholar]
- 33.Kuhmann S.E., Platt E.J., Kozak S.L., Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Layne S.P., Merges M.J., Dembo M., Spouge J.L., Nara P.L. HIV requires multiple gp120 molecules for CD4-mediated infection. Nature. 1990;346:277–279. doi: 10.1038/346277a0. [DOI] [PubMed] [Google Scholar]
- 35.Mulampaka S.N., Dixit N.M. Estimating the threshold surface density of Gp120-CCR5 complexes necessary for HIV-1 envelope-mediated cell-cell fusion. PLoS One. 2011;6:e19941. doi: 10.1371/journal.pone.0019941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sougrat R. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 2007;3:e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang M.I., Panorchan P., Dobrowsky T.M., Tseng Y., Wirtz D. Single-Molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells. J. Virol. 2005;79:14748–14755. doi: 10.1128/JVI.79.23.14748-14755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobrowsky T.M., Zhou Y., Sun S.X., Siliciano R.F., Wirtz D. Monitoring early fusion dynamics of human immunodeficiency virus type 1 at single-molecule resolution. J. Virol. 2008;82:7022–7033. doi: 10.1128/JVI.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao X. Constitutive cell surface association between CD4 and CCR5. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basmaciogullari S., Pacheco B., Bour S., Sodroski J. Specific interaction of CXCR4 with CD4 and CD8alpha: functional analysis of the CD4/CXCR4 interaction in the context of HIV-1 envelope glycoprotein-mediated membrane fusion. Virology. 2006;353:52–67. doi: 10.1016/j.virol.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 41.Gaibelet G. CD4 and CCR5 constitutively interact at the plasma membrane of living cells: a confocal fluorescence resonance energy transfer-based approach. J. Biol. Chem. 2006;281:37921–37929. doi: 10.1074/jbc.M607103200. [DOI] [PubMed] [Google Scholar]
- 42.Steffens C.M., Hope T.J. Mobility of the human immunodeficiency virus (HIV) receptor CD4 and coreceptor CCR5 in living cells: implications for HIV fusion and entry events. J. Virol. 2004;78:9573–9578. doi: 10.1128/JVI.78.17.9573-9578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vicenzi E., Liò P., Poli G. The puzzling role of CXCR4 in human immunodeficiency virus infection. Theranostics. 2013;3:18–25. doi: 10.7150/thno.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamiyama H. CXCR4-tropic, but not CCR5-tropic, human immunodeficiency virus infection is inhibited by the lipid raft-associated factors, acyclic retinoid analogs, and cholera toxin B subunit. AIDS Res. Hum. Retroviruses. 2013;29:279–288. doi: 10.1089/aid.2012.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamiyama H. Raft localization of CXCR4 is primarily required for X4-tropic human immunodeficiency virus type 1 infection. Virology. 2009;386:23–31. doi: 10.1016/j.virol.2008.12.033. [DOI] [PubMed] [Google Scholar]
- 46.Lee S. Coreceptor competition for association with CD4 may change the susceptibility of human cells to infection with T-tropic and macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 2000;74:5016–5023. doi: 10.1128/jvi.74.11.5016-5023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabatov A.A., Pollakis G., Linnemann T., Paxton W.A., de Baar M.P. Statins Disrupt CCR5 and RANTES expression levels in CD4+ T lymphocytes in vitro and preferentially decrease infection of R5 versus X4 HIV-1. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Real G. Statins inhibit HIV-1 infection by down-regulating rho activity. J. Exp. Med. 2004;200:541–547. doi: 10.1084/jem.20040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston S.H. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J. Virol. 2009;83:11016–11026. doi: 10.1128/JVI.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chikere K., Chou T., Gorry P.R., Lee B. Affinofile profiling: how efficiency of CD4/CCR5 usage impacts the biological and pathogenic phenotype of HIV. Virology. 2013;435:81–91. doi: 10.1016/j.virol.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flegler A.J., Cianci G.C., Hope T.J. CCR5 conformations are dynamic and modulated by localization, trafficking and G Protein association. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 53.Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 54.Blumenthal R., Durell S., Viard M. HIV entry and envelope glycoprotein-mediated fusion. J. Biol. Chem. 2012;287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melikyan G.B. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sáez-Cirión A. Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J. Biol. Chem. 2002;277:21776–21785. doi: 10.1074/jbc.M202255200. [DOI] [PubMed] [Google Scholar]
- 57.Magnus C., Regoes R.R. Analysis of the subunit stoichiometries in viral entry. PLoS One. 2012;7:e33441. doi: 10.1371/journal.pone.0033441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincent N., Genin C., Malvoisin E. Identification of a conserved domain of the HIV-1 transmembrane protein gp41 which interacts with cholesteryl groups. Biochim. Biophys. Acta. 2002;1567:157–164. doi: 10.1016/s0005-2736(02)00611-9. [DOI] [PubMed] [Google Scholar]
- 59.Salzwedel K., West J.T., Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for env-mediated fusion and virus infectivity. J. Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Epand R.M. Cholesterol and the interaction of proteins with membrane domains. Prog. Lipid Res. 2006;45:279–294. doi: 10.1016/j.plipres.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 61.Veiga A.S., Castanho M.A.R.B. The influence of cholesterol on the interaction of HIV gp41 membrane proximal region-derived peptides with lipid bilayers. FEBS J. 2007;274:5096–5104. doi: 10.1111/j.1742-4658.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- 62.Lai A.L., Freed J.H. HIV gp41 fusion peptide increases membrane ordering in a cholesterol-dependent fashion. Biophys. J. 2014;106:172–181. doi: 10.1016/j.bpj.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai A.L., Moorthy A.E., Li Y., Tamm L.K. Fusion activity of HIV gp41 fusion domain is related to its secondary structure and depth of membrane insertion in a cholesterol-dependent fashion. J. Mol. Biol. 2012;418:3–15. doi: 10.1016/j.jmb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiang W., Sun Y., Weliky D.P. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan R. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ivankin A., Kuzmenko I., Gidalevitz D. Cholesterol mediates membrane curvature during fusion events. Phys. Rev. Lett. 2012;108:238103. doi: 10.1103/PhysRevLett.108.238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dumas F. Spatial regulation of membrane fusion controlled by modification of phosphoinositides. PLoS One. 2010;5:e12208. doi: 10.1371/journal.pone.0012208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finnegan C.M. Sphingomyelinase restricts the lateral diffusion of CD4 and inhibits human immunodeficiency virus fusion. J. Virol. 2007;81:45294–45304. doi: 10.1128/JVI.02553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De la Vega M. Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology. 2011;8:99. doi: 10.1186/1742-4690-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Melikyan G.B. HIV entry: a game of hide-and-fuse? Curr. Opin. Virol. 2014;4:1–7. doi: 10.1016/j.coviro.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fackler O.T., Peterlin B.M. Endocytic entry of HIV-1. Curr. Biol. 2000;10:1005–1008. doi: 10.1016/s0960-9822(00)00654-0. [DOI] [PubMed] [Google Scholar]
- 73.Kubo Y., Hayashi H., Matsuyama T., Sato H., Yamamoto N. Retrovirus entry by endocytosis and cathepsin proteases. Adv. Virol. 2012:e640894. doi: 10.1155/2012/640894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G.B. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Permanyer M., Ballana E., Esté J.A. Endocytosis of HIV: anything goes. Trends Microbiol. 2010;18:543–551. doi: 10.1016/j.tim.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 76.Wilen C.B., Tilton J.C., Doms R.W. HIV: cell binding and entry. Cold Spring Harb. Perspect. Med. 2012;2:a006866. doi: 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carter G.C., Bernstone L., Baskaran D., James W. HIV-1 infects macrophages by exploiting an endocytic route dependent on dynamin, Rac1 and Pak1. Virology. 2011;409:234–250. doi: 10.1016/j.virol.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 78.Gobeil L.A., Lodge R., Tremblay M.J. Macropinocytosis-like HIV-1 internalization in macrophages is CCR5 dependent and leads to efficient but delayed degradation in endosomal compartments. J. Virol. 2013;87:735–745. doi: 10.1128/JVI.01802-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bomsel M. Transcytosis of infectious human immunodeficiency virus across a tight human epithelial cell line barrier. Nat. Med. 1997;3:42–47. doi: 10.1038/nm0197-42. [DOI] [PubMed] [Google Scholar]
- 80.Maréchal V. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J. Virol. 2001;75:11166–11177. doi: 10.1128/JVI.75.22.11166-11177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gobeil L.A., Lodge R., Tremblay M.J. Differential HIV-1 endocytosis and susceptibility to virus infection in human macrophages correlate with cell activation status. J. Virol. 2012;86:10399–10407. doi: 10.1128/JVI.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Markosyan R.M., Cohen F.S., Melikyan G.B. Time-resolved imaging of HIV-1 env-mediated lipid and content mixing between a single virion and cell membrane. Mol. Biol. Cell. 2005;16:5502–5513. doi: 10.1091/mbc.E05-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carter G.C. HIV entry in macrophages is dependent on intact lipid rafts. Virology. 2009;386:192–202. doi: 10.1016/j.virol.2008.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Wilgenburg B., Moore M.D., James W.S., Cowley S.A. The productive entry pathway of HIV-1 in macrophages is dependent on endocytosis through lipid rafts containing CD4. PLoS One. 2014;9:e86071. doi: 10.1371/journal.pone.0086071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhat S. The galactosyl ceramide/sulfatide receptor binding region of HIV-1 gp120 maps to amino acids 206–275. AIDS Res. Hum. Retroviruses. 1993;9:175–181. doi: 10.1089/aid.1993.9.175. [DOI] [PubMed] [Google Scholar]
- 86.Rawat S.S., Johnson B.T., Puri A. Sphingolipids: modulators of HIV-1 infection and pathogenesis. Biosci. Rep. 2005;25:329–343. doi: 10.1007/s10540-005-2894-5. [DOI] [PubMed] [Google Scholar]
- 87.Hammache D. Specific interaction of HIV-1 and HIV-2 surface envelope glycoproteins with monolayers of galactosylceramide and ganglioside GM3. J. Biol. Chem. 1998;273:7967–7971. doi: 10.1074/jbc.273.14.7967. [DOI] [PubMed] [Google Scholar]
- 88.Klug Y.A. Early and late HIV-1 membrane fusion events are impaired by sphinganine lipidated peptides that target the fusion site. Biochem. J. 2014;461:213–222. doi: 10.1042/BJ20140189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lakadamyali M., Rust M.J., Zhuang X. Endocytosis of influenza viruses. Microbes Infect. 2004;6:929–936. doi: 10.1016/j.micinf.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lakadamyali M., Rust M.J., Babcock H.P., Zhuang X. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morcock D.R. Elimination of retroviral infectivity by N-ethylmaleimide with preservation of functional envelope glycoproteins. J. Virol. 2005;79:1533–1542. doi: 10.1128/JVI.79.3.1533-1542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rossio J.L. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 1998;72:7992–8001. doi: 10.1128/jvi.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]