Abstract
5′ and 3′ UTR sequences on the coronavirus genome are known to carry cis-acting elements for DI RNA replication and presumably also virus genome replication. 5′ UTR-adjacent coding sequences are also thought to harbor cis-acting elements. Here we have determined the 5′ UTR and adjacent 289-nt sequences, and 3′ UTR sequences, for six group 2 coronaviruses and have compared them to each other and to three previously reported group 2 members. Extensive regions of highly similar UTR sequences were found but small regions of divergence were also found indicating group 2 coronaviruses could be subdivided into those that are bovine coronavirus (BCoV)-like (BCoV, human respiratory coronavirus-OC43, human enteric coronavirus, porcine hemagglutinating encephalomyelitis virus, and equine coronavirus) and those that are murine hepatitis virus (MHV)-like (A59, 2, and JHM strains of MHV, puffinosis virus, and rat sialodacryoadenitis virus). The 3′ UTRs of BCoV and MHV have been previously shown to be interchangeable. Here, a reporter-containing BCoV DI RNA was shown to be replicated by all five BCoV-like helper viruses and by MHV-H2 (a human cell-adapted MHV strain), a representative of the MHV-like subgroup, demonstrating group 2 common 5′ and 3′ replication signaling elements. BCoV DI RNA, furthermore, acquired the leader of HCoV-OC43 by leader switching, demonstrating for the first time in vivo recombination between animal and human coronavirus molecules. These results indicate that common replication signaling elements exist among group 2 coronaviruses despite a two-cluster pattern within the group and imply there could exist a high potential for recombination among group members.
Keywords: Group 2 coronaviruses, RNA replication signals, RdRp promoter, DI RNA, Leader switching, Recombination between human and animal coronaviruses
Introduction
The coronavirus genus in the family Coronaviridae has been divided into three groups, initially on the basis of serologic relatedness but more recently on the basis of genome sequence similarities Cavanagh et al., 1997, Lai and Cavanagh, 1997, Siddell, 1995. The recently described human severe acute respiratory syndrome (SARS) coronavirus appears not to fall precisely into any of these groups Marra, et al., 2003, Rota, et al., 2003. Members of group 1 share identity with the porcine transmissible gastroenteritis virus (TGEV) and [officially, as of 2000 (Enjuanes et al., 2000)] include TGEV [and strain porcine respiratory coronavirus (PRCoV)], canine coronavirus (CCoV), feline coronavirus (FECoV) [and strain feline infectious peritonitis virus (FIPV)], human respiratory coronavirus-229E (HCoV-229E), and porcine epidemic diarrhea virus (PEDV). Members of group 2 share identity with the mouse hepatitis coronavirus (MHV) and include bovine coronavirus (BCoV), human respiratory coronavirus-OC43 (HCoV-OC43), porcine hemagglutinating encephalomyelitis coronavirus (HEV), rat coronavirus (RtCoV) [and strain sialodacryoadenitis coronavirus (SDAV)] (Enjuanes et al., 2000). Members of group 3 share identity with the avian infectious bronchitis virus (IBV) and include IBV and turkey enteric coronavirus (TCoV) Enjuanes et al., 2000, Guy, 2000. Coronaviruses likely to become officially classified as group 2 members on the basis of antigenic relatedness and genome sequence similarities include human enteric coronavirus-4408 (HECoV-4408) (Zhang et al., 1994), puffinosis virus (PV) (Klausegger et al., 1999), and equine enteric coronavirus (ECoV) (Guy et al., 2000).
Recombination among coronaviruses is an attribute of the genus and is thought to contribute to the emergence of new pathotypes Lai, 1992, Lai and Cavanagh, 1997. Curiously, recombination so far has been noted only between species of the same group, for example, between the feline and canine enteric coronaviruses (Herrewegh et al., 1998), between strains of MHV (Keck et al., 1988), and between strains of IBV Jia et al., 1995, Kottier et al., 1995, although in principle intergroup recombination is possible based on reverse genetics experimentation in which an MHV chimera was produced with the ectodomain portion of the FeCoV spike protein gene (Kuo et al., 2000). Inasmuch as natural recombination events among coronaviruses probably result from a polymerase jumping mechanism during coinfection Brian and Spaan, 1997, Lai, 1992, Lai and Cavanagh, 1997, signals read by the RNA polymerase complex to initiate and carry out genome replication are likely to contribute to the recombination process. It would therefore be expected that viruses sharing signals for RNA polymerase recognition would be more likely to undergo recombination than those that do not. Characterization of common signals for RNA synthesis, therefore, may aid in predicting which viruses are likely to recombine during a natural coinfection.
To date, common 3′-proximal replication signaling elements have been noted between MHV and BCoV, both group 2 members for which DI RNAs have been studied. In an MHV-A59-derived DI RNA, which comprised only the virus genomic termini, and in the MHV-A59 genome, it has been shown that the entire 3′ UTR of BCoV is able to replace the equivalent region without loss of DI RNA replicability or virus viability Hsue and Masters, 1997, Hsue et al., 2000. Likewise, in a BCoV DI RNA that comprised only the virus genomic termini (Fig. 1; Chang et al., 1994) it has been shown that the MHV-A59 3′ UTR was able to replace the BCoV 3′ UTR with no detectable loss of replicating ability (S. Ku, G.D. Williams, and D.A. Brian, unpublished data). The partially characterized 3′ UTR-mapping cis-acting elements in BCoV and MHV include a 5′-proximal bulged stem-loop Hsue and Masters, 1997, Hsue et al., 2000, a 5′-proximal hairpin-type pseudoknot (Williams et al., 1999), a centrally located stem-loop (Liu et al., 2001), and a 3′-terminal poly(A) tail (Spagnolo and Hogue, 2000). What roles the higher order structures play are not clear since the 3′-terminal 55-nts and poly(A) tail are thought to be the minimal sequence requirements for minus-strand synthesis (Lin et al., 1994). Common 5′-proximal replication signals between MHV and BCoV have not yet been described. For both MHV and BCoV DI RNAs it has been shown that the 5′-terminal sequence is required for DI RNA replication Chang et al., 1994, Kim et al., 1993 and in BCoV DI RNA at least two stem-loops identified as stem-loops III and IV function as higher order cis-acting signaling elements (Raman et al., 2003; S. Raman and D. Brian, unpublished data). A higher order cis-acting structure mapping within the first 290 nts of ORF1 has also been found (C. Brown, K. Nixon, S. Senanayake, and D. Brian, unpublished data). Might these elements be common between MHV and BCoV as well? To what extent might 5′- and 3′-proximal replication signals be shared among other group 2 members?
Fig. 1.
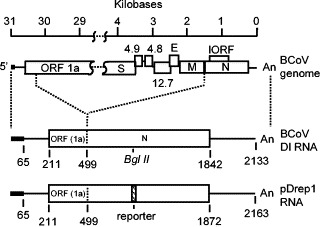
Structure of the bovine coronavirus DI RNA and pDrep1 as they relate to the parent bovine coronavirus genome. Note that 289 nts of ORF 1a and the entire N ORF are part of the DI RNA. The reporter is a 30-nt sequence derived from the TGEV N gene that was cloned in-frame into the BglII site of the BCoV N gene (Chang et al., 1994).
Here we examine the 5′ UTR and adjacent 289-nt sequences of four established and two new putative group 2 coronavirus members, the 3′ UTR sequences of four established and one new putative member, and compare them with previously reported sequences of three other group 2 coronaviruses. Our results suggest that the 5′ and 3′ UTR, and 5′ UTR-adjacent sequence patterns, may be subgrouped into those that are BCoV-like and those that are MHV-like. This bifurcated pattern is consistent with a proposed subgrouping of group 2 coronaviruses on the basis of activities of the hemagglutinin-esterase proteins (Wurzer et al., 2002). The diverged UTR patterns, therefore, raised the question as to whether the two subgroups of group 2 coronaviruses might differ with respect to their RNA replication signals as measured by replication assays. To begin to answer this, we tested replication in trans of a BCoV DI RNA with various group 2 helper viruses and learned that replication was supported by all five viruses within the BCoV-like subgroup, and by MHV-H2, the only virus tested within the MHV-like subgroup. It was also found that the BCoV DI RNA acquired the leader sequence of HCoV-OC43 helper virus, thereby demonstrating for the first time an in vivo (in this case, in cell-culture) recombination event between molecules of an animal and human coronavirus. These results indicate that the signals for recognition and engagement by the RNA replication machinery are probably common among group 2 coronaviruses, regardless of subgroup classification, and indicate a potential for recombination among members of the group should mixed infections occur.
Results
Nucleotide sequence analyses of the 5′ UTR and adjacent 289 nts, and 3′ UTR, of the group 2 coronaviruses reveal a two cluster pattern
Structural elements acting as cis-acting replication signals in genomic and DI RNAs have been found in the 5′ UTR and adjacent 289 nts, and in the 3′ UTR, of MHV and BCoV (reviewed under Introduction above). To examine potentially conserved features in these regions among the group 2 coronaviruses, the 5′ UTR sequences, 5′ UTR-adjacent 289 nts, and 3′ UTR sequences of HCoV-OC43, HECoV-4408, HEV-TN11, ECoV-NC99, and SDAV were determined and compared with the published sequences for BCoV-Mebus, MHV-A59, MHV-2, and MHV-JHM (see GenBank Accession Nos. described below and Fig. 2, Fig. 3). For PV, the 3′ UTR was also compared (Fig. 3). Upon alignment it became apparent by inspection that there were several regions, up to 18 nts in length in the 5′ UTR and adjacent 289 nts and up to 29 nts in the 3′ UTR, that were identical among all the viruses examined (identified by shading in Fig. 2, Fig. 3). It was likewise apparent that there were several smaller regions in which sequences diverged considerably, yielding patterns that would suggest the viruses could be grouped into those that are either BCoV-like or MHV-like. The existence of a two-cluster pattern was confirmed in separate phylogenetic analyses of the 5′ and 3′ UTRs and of the amino acid sequences in the partial ORF adjacent to the 5′UTR (Figs. 4A–C).
Fig. 2.
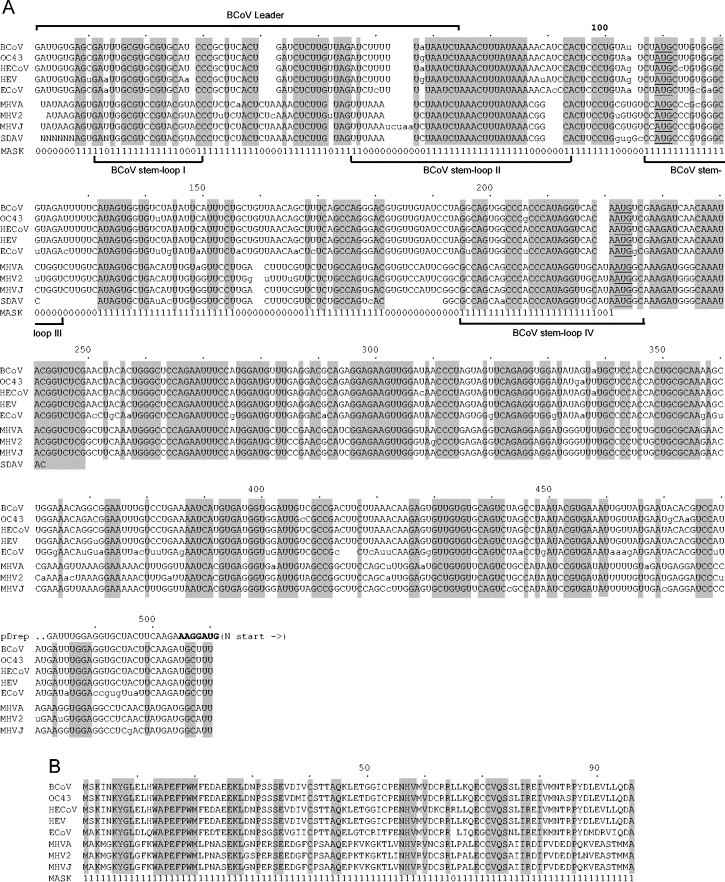
Aligned 5′-terminal sequences of group II coronaviruses. (A) Aligned 5′ UTR and adjacent 289-nt (partial ORF 1a) sequences. The alignment was constructed to minimize inserted gaps; only unambiguously aligned nucleotide positions in the UTR (identified by 1s in the MASK) were used to calculate the phylogenetic tree (shown in Fig. 4). Identical regions among all nine group 2 coronaviruses are noted by shading. Note that the SDAV sequence is known for only 21 nts into ORF 1a, so sequences of only eight viruses are shaded beyond this point. Positions of the BCoV leader sequence (65 nts) and of BCoV stem loops I, II, III and IV, of which stem loops III and IV are phylogenetically conserved among group 2 coronaviruses Chang et al., 1996, Raman et al., 2003; Raman et al., unpublished data), are noted. Numbers above refer to positions in the sequence alignment. Lowercase letters identify nonconforming nucleotides within the respective BCoV-like or MHV-like subgroups. Part of the pDrep1 5′-terminal sequence is shown to indicate its point of divergence from BCoV genome sequence. (B) Aligned amino acid sequences within the partial ORF 1a. Only unambiguously aligned amino acid positions (identified by 1s in the MASK) were used to calculate the phylogenetic tree (shown in Fig. 4). Numbers above refer to positions in the sequence alignment. Abbreviations: BCoV, bovine coronavirus-Mebus; OC43, human coronavirus-OC43; HECoV-OC43, human enteric coronavirus-4408; HEV, porcine hemagglutinating encephalomyelitis virus-TN11; ECoV, equine coronavirus-NC99; MHVA, mouse hepatitis virus-A59; MHV2, mouse hepatitis virus-2; MHVJ, mouse hepatitis virus-JHM; SADV, rat sialodacryoadenitis virus.
Fig. 3.
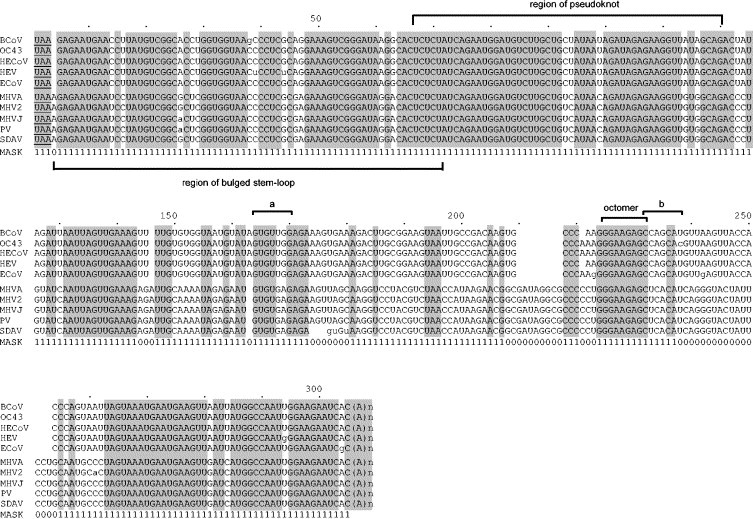
Aligned 3′ UTR sequences of group II coronaviruses. The alignment was constructed to minimize inserted gaps; only unambiguously aligned nucleotide positions (identified by 1s in the MASK) were used to calculate the phylogenetic tree (shown in Fig. 4). Shaded areas identify sequences identical among all 10 viruses. Numbers above refer to the sequence alignment. The underlined UAA stop codon of the N ORF begins the 3′ UTR and a poly (A) tail, identified as (A)n, terminates the sequence. Sequences making up a cis-acting bulged stem-loop (Hsue and Masters, 1997), a pseudoknot (Williams et al., 1999), a long-range stem interaction (identified as a and b) (Liu et al., 2001), and a conserved GGAAGAGC octomer, are bracketed. Lowercase letters indicate nonconforming nucleotides within the respective BCoV-like or MHV-like subgroups. Abbreviations are as described in Fig. 2, with the addition of PV, puffinosis virus.
Fig. 4.
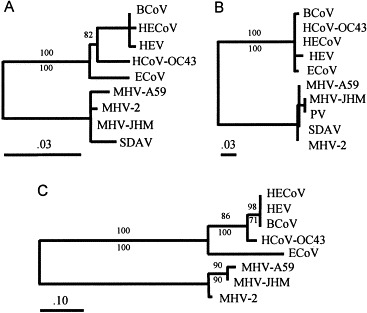
Phylogenetic relationships inferred from coronavirus sequences: (A) 5′ UTR (145 nucleotide positions), (B) 3′ UTR (260 nucleotide positions), and (C) partial 5′ ORF (95 amino acid positions). The phylogentic frameworks in A and B were inferred from nucleic acid sequences by maximum likelihood (-ln L = 404.60992 and 539.07673 for A and B, respectively); the framework in C was inferred from translated amino acid sequences by a distance method. Paired numbers represent bootstrap proportions calculated by distance (top numbers) and parsimony (bottom numbers) methods. Scale bar units indicate nucleotide (A and B) or amino acid (C) substitutions per sequence position. Abbreviations are as described in Fig. 1, Fig. 2.
Helper virus from either cluster can support the replication of BCoV DI RNA in HRT cells
It has been previously shown that the 3′ UTRs of MHV and BCoV are functionally interchangeable in the context of the MHV genome Hsue and Masters, 1997, Hsue et al., 2000 and BCoV DI RNA (S. Ku, G.D. Williams, and D.A. Brian, unpublished data), indicating the presence of common 3′-proximal replication signals. Common 5′ proximal RNA replication signals have not been similarly sought. To test for the presence of common replication signals, the accumulation of reporter-containing BCoV DI RNA (transcripts of pDrep1; Fig. 1) was tested with five of the BCoV-like and one of the MHV-like group 2 helper viruses described (Figs. 5A–C). For this, HRT-18 cells were mock infected or infected with BCoV-Mebus, HCoV-OC43, ECoV-NC99, HECoV-4408, HEV-TN, or MHV-H2, transfected 1 h later with pDrep1 RNA, and total RNA from cells was extracted at 1, 24, and 48 h posttransfection, and at 24 h after infection with passage 1 virus (and in some cases passage 2 virus) for measurement of DI RNA accumulation. Accumulation was measured by Northern blotting using a pDrep1 reporter-specific radiolabeled probe. In the absence of helper virus, transfected DI RNA has a measured half-life of less than 4 h and no accumulation is observed at 24 h posttransfection Chang et al., 1994, Chang and Brian, 1996, Senanayake and Brian, 1999; data not shown). The results for the BCoV-like viruses shown in Figs. 5A and C illustrate a net accumulation of progeny DI RNA molecules by 24 and 48 h posttransfection, and by 24 h after virus passaging (Fig. 5A, lanes 1–4), establishing that common replication signals are recognized by the BCoV-like coronaviruses tested. It should be noted that whereas helper virus replication rates for the viruses other than BCoV were less than that for BCoV at 24 hpi through VP1 [ranging from 1 to 50%, as measured by reprobing the stripped blot for viral mRNAs with a universally sensitive 3′ UTR-detecting probe (3′UTRPSEU, Table 1)] (Figs. 5A, B, and D), BCoV DI RNA replication measured at the same time points was relatively higher (ranging from 5 to 100%) (Figs. 5A–C). While the quantitative measurements made here are not precise enough to establish the relative efficiencies of DI RNA replication by the various helper viruses, or the relative inhibitory effects on virus replication by the DI RNA, they do establish a helper virus function for DI RNA replication in each case.
Fig. 5.
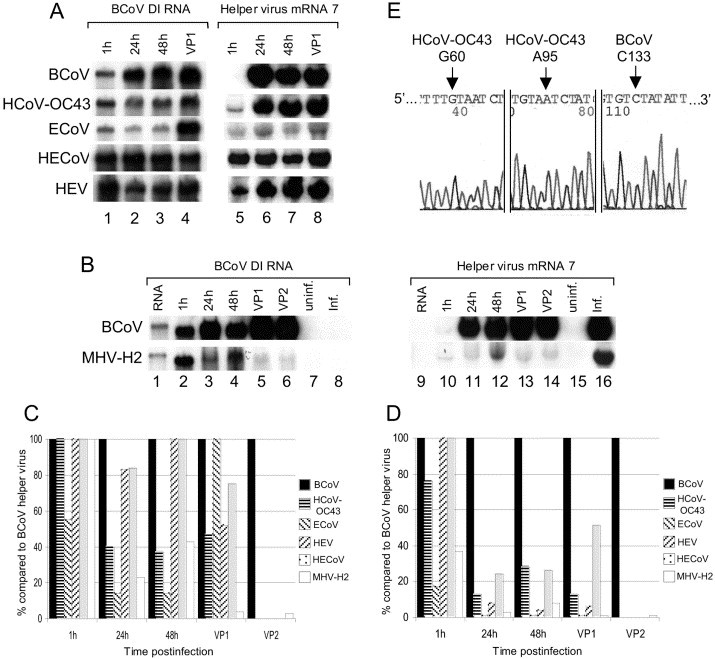
Replication of BCoV DI RNA by the helper virus functions of group 2 coronaviruses. (A) Northern blot analyses showing replication (accumulation) of reporter-containing BCoV DI RNA in the presence of BCoV, HCoV-OC43, ECoV, HECoV, and HEV, and accumulation of mRNA 7 as an indicator of helper virus replication. RNA was extracted at the indicated times posttransfection, or at 24 h after virus passaging (VP1). Lanes 1–4, blots were probed with a DI RNA reporter-specific radiolabled probe. Lanes 5–8, blots used in lanes 1–4 were probed with a 3′ UTR-specific probe that recognizes an identical sequence in all the helper viruses. (B) Northern blot analyses showing replication (accumulation) of reporter-containing BCoV DI RNA in the presence of BCoV and MHV-H2 helper viruses, and accumulation of mRNA 7 as an indicator of helper virus replication. RNA was extracted at the indicated times postinfection, or at 24 h after virus passaging (VP1 and VP2). Lanes 1–8, blots were probed to detect DI RNA only as were lanes 1–4 in (A). Lanes 9–16, blots used in lanes 1–8 were stripped and probed to detect viral mRNAs as were lanes 5–8 in (A). Lanes 1 and 9, pDrep1 marker transcript. RNA from mock-infected (uninf) cells (lanes 7 and 15), or infected, mock-transfected (inf) cells (lanes 8 and 16), was harvested at 24 h post-mock-infection and postinfection; respectively. (C) Bar graphs illustrating the amounts of BCoV DI RNA accumulated relative to amounts accumulated with BCoV helper virus as measured in (A) and (B). (D) Bar graphs illustrating the amounts of helper virus mRNA 7 relative to amounts of BCoV mRNA 7 as measured in (A) and (B) (E). Sequence analysis of the leader fusion region of DI RNA progeny from cells infected with HCoV-OC43 helper virus and transfected with reporter-containing BCoV DI RNA. The HCoV-OC43-specific bases G60 and A95 and the BCoV-specific base C133 show that the progeny DI RNA molecule possesses a leader derived from the HCoV-OC43 helper virus and that the recombination site exists somewhere between nts 95 and 133.
Table 1.
Oligonucleotides used in this study
| Oligonucleotidea | Polarity | Sequence (5′→3′) | Nt binding region |
|---|---|---|---|
| 3′UTRPSEU(+) | − | CTGCAAGTCATCCATTCTGATAGAGAGTG | nts 63–91 in BCoV 3′ UTRb |
| 5RAAP(−)c | (+) | GGCCACGCGTCGACTAGTACGGGGGGGGGG | 3′ poly(C) on cDNA |
| Ada3(+) | − | GGAATTCTCGAGCTCAAGCTTTTTTTTTTTTTTTT | 3′ poly(A) tail |
| DI3′(+) | − | CGGGATCCGTCGACACGCGTTTTTTTTTTTTTTTTTTTT | 3′ poly (A) tail |
| DI420(−) | + | GTTGTGTGCAGTCTAGCCTAATAC | nts 419–442 in pDrep1 |
| HpaI(+) | − | GGCTGAAAGCTGTTAACAGCAGAAATG | nts 140–166 in pDrep1 |
| Ia1096(+) | − | CGCACAACGTGCCATGCCAC | nts 1096–1215 in BCoV genomed |
| Leader(−) | + | GAGCGATTTGCGTGCGTGCATCCCGC | nts 7–32 in pDrep1 |
| NdeI(+) | − | CCTCCAAATCATATGGACGTGTATTC | nts 456–481 in pDrep1 |
| Pst(−) | + | GAGAGTTGACGCAGAGGAC | nts 1799–1818 in pDrep1 |
| TGEV8(+) | − | CATGGCACCATCCTTGGCAACCCAGA | nts 1098–1123 in pDrep1 |
The positive and negative symbols in the oligonucleotide names indicate the polarity of the nucleic acid to which the oligonucleotide anneals.
The sequence in nts 63–91 in BCoV is common to all the group 2 coronaviruses examined in this study (note Fig. 3).
Supplied with the RACE kit (Invitrogen).
From BCoV-Mebus genome sequence as determined by K. Nixon and D. Brian (GenBank Accession No. U00735).
In previous studies on the regulatory effects of BCoV genomic 5′ UTR sequence on translation (Senanayake and Brian, 1999) it was learned that whereas BCoV replicated in murine OST-7 cells [a T7 RNA polymerase-expressing mouse L cell-derived line (Elroy-Stein and Moss, 1990)] nearly as well as in human rectal tumor cells [HRT-18 cells used for BCoV propagation (Chang et al., 1994)] the replication of transfected pDrep1 transcripts could not be observed at all with BCoV helper virus in the murine cells. An attempt was therefore made to test for the helper effect of MHV in HRT cells. For this we used the MHV-H2 strain that had been selected as a host range mutant of a MHV-A59/JHM recombinant on persistently infected BHK cells now able to grow on human cells (Baric et al., 1997). In MHV-H2-infected HRT cells, pDrep1 RNA accumulated as well as in HCoV-OC43 and ECoV-NC99 helper virus infected cells through 48 h posttransfection (20–40% of BCoV helper virus levels vs 15–40%, respectively; Fig. 5B, lanes 1–4), but not as well after virus passage 1 (5% of BCoV helper virus levels vs 40–100%: Fig. 5B, lanes 5 and 6, and Fig. 5C). These results indicate that MHV can serve as a helper virus for the replication of BCoV DI RNA and thus there are 5′-proximal as well as 3′-proximal cis-acting replicating signaling elements in BCoV DI RNA recognized by the MHV RNA replication apparatus. RNA replication signals are therefore likely to be common between the two viruses. The relatively low abundance of BCoV RNA in VP1 and VP2 of MHV-infected cells (Fig. 5B, lane 5) suggests, furthermore, that MHV does not package the BCoV DI RNA as well as do the BCV-like helper viruses. A difference in migration rates between marker DI RNA (Fig. 5B, lane 1) and intracellular DI RNA (Fig. 5B, lanes 2–6) has been previously observed Chang et al., 1994, Senanayake and Brian, 1999 and apparently represents an in-gel displacement of intracellular DI RNA by abundant comigrating cellular RNA species.
Leader switching from the HCoV-OC43 helper virus to BCoV DI RNA
Leader switching was first described between an MHV-JHM-derived DI RNA and MHV-A59 (Makino and Lai, 1989). Leader switching is thought to result from a high-frequency recombination step that is similar, but not necessarily identical to, the RdRp jump by which leader is acquired on coronavirus subgenomic mRNAs Brian and Spaan, 1997, Chang et al., 1996, Makino and Lai, 1989, Zhang and Lai, 1996. It was therefore anticipated that if replication of BCoV DI RNA resulted from the use of a heterologous group 2 coronavirus that the progeny DI RNA would acquire the leader of the helper virus. This possibility was sought on DI RNA progeny in cells infected with all helper viruses except HECoV, which has a leader sequence indistinguishable from that of BCoV. Leader switching was observed for HCoV-OC43 (four of four cDNA clones in VP1 RNA), ECoV (three of four clones), and HEV (three of four clones), but not for MHV (zero of eight cDNA clones examined and data not shown). In the sequence data from one of the VP1 cDNA clones from HCoV-OC43 helper virus (Fig. 5E) it can be seen that the recombination site occurred somewhere between HCoV-OC43 marker nt A95 and BCoV marker nt C133, slightly downstream of a high-frequency leader switching window (nts 70–93) previously identified in BCoV (Chang et al., 1996). These results confirm and extend our understanding of the high-frequency nature of leader switching by strains of group 2 coronaviruses and document for the first time a recombination event between human and animal coronavirus molecules.
Discussion
The results of this study show first a distinct subdivision of group 2 coronaviruses into two subgroups, those that are BCoV-like and those that are MHV-like, with regard to sequence patterns in the 5′ and 3′ UTRs (Fig. 2, Fig. 3). The significance of this divergence, however, is not clear since the replication apparatus of one virus will replicate the DI RNA of another, at least in the one-way tests employed here. Although we cannot judge the relative efficiencies of the helper viruses used since replication rates differed in the HRT cells, we think it quite likely that UTR sequence differences reflect a fine-tuning of structural interactions between each virus genome and its cognate replication apparatus for optimal replication. Since previous studies had shown that the 3′ terminus is interchangeable between the BCoV and MHV 3′ UTRs (Hsue and Masters, 1997; S. Ku, G.D. Williams, and D.A. Brian, unpublished data), the new information derived from this study is that there are also shared signals in the genome 5′ terminus. It remains to be determined what explains the lower rate of accumulation and packaging for BCoV DI RNA with MHV-H2 helper virus as compared to the BCoV-like helper viruses. It also remains to be determined what precise signaling elements are shared among the members of the group.
It has been found in a number of coronavirus DI RNA systems that DI RNA upon replication takes on the leader of the helper virus. The same was found here for the BCoV DI RNA and all helper viruses with a distinguishable leader sequence except for MHV. The absence of leader switching with MHV may reflect the relatively poor replication of MHV-H2 in BCoV DI RNA-transfected cells or it may possibly reflect other mechanistically incompatible features. The data showing leader switching with HCoV-OC43 are, to our knowledge, the first to document an in vivo recombination event between molecules of a human and animal coronavirus. Coronavirus leader switching has been compared mechanistically to RdRp strand switching that gives rise to leader-containing subgenomic transcripts Chang et al., 1996, Makino and Lai, 1989, Zhang and Lai, 1996. A demonstration of leader switching therefore raises the question of whether the heterologous helper viruses can support synthesis of subgenomic mRNAs from the BCoV DI RNA template as has been previously observed in the BCoV Krishnan et al., 1996, Ozdarendeli et al., 2001 and MHV Makino et al., 1991, van der Most et al., 1994 homologous systems. Preliminary experiments indicate that they do (H.-Y. Wu and D.A. Brian, unpublished data), but that the patterns of transcription differ among the helper viruses, suggesting a different RdRp mechanism and hence probably different signaling processes are at play between the processes of transcription and replication (H.-Y. Wu, A. Ozdarendeli, and D. Brian, unpublished data). A mechanistic separation of transcription and replication has been noted in arteriviruses (van Dinten et al., 1997), a family placed along with coronaviruses in the order Nidovirales (Cavanagh et al., 1997).
The results shown here establish that despite sequence differences in the 5′ and 3′ UTRs and in the first 289 nts within ORF 1 of group 2 coronaviruses, the replication apparatuses of these viruses recognize and replicate the BCoV DI RNA, indicating the recognition of a common set of replication signals within the group. The results suggest, furthermore, that as a consequence of common replication signals there exists a high potential for recombination among members of group 2. To what extent common replication signals exist among the coronavirus groups, including the recently described SARS coronavirus Marra, et al., 2003, Rota, et al., 2003, remains to be determined. Based on the results presented here, such information would seem useful in predicting the recombination potential among coronaviruses.
Materials and methods
Cells and viruses
The human adenocarcinoma (human rectal tumor) cell line HRT-18 (Tompkins et al., 1974) was used as previously described for BCoV Lapps et al., 1987, Chang et al., 1994 for the growth of the BCoV-like and MHV-H2 viruses. BCoV-Mebus was plaque purified three times (Lapps et al., 1987) and a DI RNA-free stock (Williams et al., 1999) was used. HCoV-OC43 (Hogue and Brian, 1986) obtained from S.R. Weiss (Univ. of Pennsylvania, Philadelphia, PA), HECoV-4408 (Zhang et al., 1994), obtained from X.M. Zhang (Univ. of Arkansas, Little Rock, AR), HEV-TN11 obtained from John Black (C.E. Kord Animal Disease Laboratory, Nashville, TN), and ECoV-NC99 (Guy et al., 2000) were each plaque purified three times and used to prepare stocks at concentrations of >107 PFU/ml. MHV-H2, a strain adapted for growth on human epithelial cells (Baric et al., 1997), was obtained from R.S. Baric (Univ. of North Carolina, Chapel Hill, NC) and used directly to prepare a stock at a concentration of >107 PFU/ml. L2 (Percy) cells were used to grow SDAV-681 as previously described (Yoo et al., 2000) and mouse L cells were used to grow PV as previously described (Klausegger et al., 1999).
Oligonucleotides
The oligonucleotides used in this study are described in Table 1.
Genome sequence determination
To determine the 5′ proximal genome sequences of HCoV-OC43, HECoV-4408, HEV-TN11, and ECoV-NC99, RNA was extracted with Trizol (Life Technologies) from infected cells at 24 hpi and cDNAs were synthesized from RNA using RT and oligonucleotide Ia1096(+). cDNAs were amplified by PCR using oligonucleotides Ia1096(+) and leader(−); the products were cloned into the TOPO XL vector (Invitrogen), and resulting plasmid DNAs were sequenced by automated dideoxynucleotide sequencing using oligonucleotides leader(−), DI420(−), and Ia1096(+) as sequencing primers. To determine the 5′ terminal genome sequences, RACE was used as recommended by the manufacturer of the RACE kit (Invitrogen). For this, first-strand cDNA was made with oligonucleotide NdeI(+), and the product was C-tailed and then PCR amplified with oligonucleotides HpaI(+) and 5RAAP(−), the bridge anchor primer supplied with the kit. The products were cloned into the TOPO XL vector and the resulting plasmids were sequenced by automated sequencing using oligonucleotide HPaI(+) and as sequencing primer.
To determine the 3′-terminal genome sequences of the BCoV-like viruses, cDNA was made by using oligonucleotide DI3′(+), which binds to the 3′-poly(A) tail and provides BamHI, SalI, AccI, HincI, and MluI restriction endonuclease cloning sites, amplified by PCR with oligonucleotides DI3′(+) and Pst(−), and the PCR products were cloned into TOPO XL cloning vector (Invitrogen). For PV, purified genomic RNA was reverse transcribed employing oligonucleotide Ada3(+). The second strand was synthesized and cloned as described previously (Klausegger et al, 1999). The resulting plasmids were sequenced by automated DNA sequencing.
Phylogenetic analyses
Nucleotide sequences for 3′ and 5′ UTRs and amino acid sequences for the partial 5′ ORF were aligned manually by using the Genetic Data Environment (GDE) (Smith et al., 1994). Nucleotides and amino acid residues were aligned in a manner that minimized the number of gaps, and nucleotide positions between the first identical nucleotide in all positions and the gaps were omitted from phylogenetic calculations to minimize ambiguity. Sufficiency of the phylogenetic “signal” in each aligned dataset was assessed by the random tree-length distribution method implemented in PAUP* (Version 4.0b10; Hillis and Hulsenbeck, 1992, Swofford, 1996. The sequence alignments contained sufficient information for valid phylogenetic analyses (g1 skew statistics −0.61, −0.55, and −1.033 for 5′ UTR, 3′ UTR, and 5′ partial ORF, respectively). Phylogenetic frameworks were inferred from nucleotide sequences by maximum likelihood in PAUP* with settings that correspond to the HKY85 + G model (two-substitution types, four rate categories following a γ-distribution with no invariant sites and nucleotide frequencies, transition/transversion ratio, and the γ-shape parameter estimated by maximum likelihood). Bootstrap proportions were calculated from 100 resampled datasets by using neighbor-joining (Kimura two-parameter model, transition/transversion ratio of 2) and maximum parsimony, also in PAUP*. Phylogenetic relationships among amino acid sequence translations for the partial 5′ ORF were inferred by neighbor-joining from distances calculated by using the PAM matrix of amino acid substitution frequencies, as implemented in PHYLIP (Felsenstein, 1993). Bootstrap proportions were inferred from 100 resampled datasets by using PHYLIP's PAM distance/neighbor joining and protein parsimony methods.
Plasmid constructs
Construction of pGEM3Zf(−) (Promega)-based pDrep 1 (Fig. 1) has been described (Chang et al., 1994).
Northern assay for DI RNA and virus replication
The Northern assay for detecting reporter-containing DI RNA was performed essentially as previously described Chang et al., 1994, Chang and Brian, 1996. Briefly, cells at 80% confluency (∼1 × 106 cells/dish) in 35-mm-diameter dishes were infected with the indicated helper virus at a multiplicity of approximately 10 PFU per cell and transfected with 600 ng of pDrep1 transcript in Lipofectin (Invitrogen) at 1 hpi. For passage of progeny virus, supernatant fluids were harvested at 48 hpi and 500 μl was used to directly infect freshly confluent cells in a 35-mm dish. RNA extracted with Trizol (yielding approximately 10 μg per plate) was stored at −20°C as an ethanol precipitate. For Northern analysis, precisely one-fourth of the total RNA preparation (approximately 2.5 μg per lane) was used for electrophoresis in a formaldehyde-agarose gel. Approximately 1 ng of transcript was loaded per lane when used as a size marker. RNA was transferred from the gel to Nytran membrane by vacuum blotting and blots were probed with oligonucleotide TGEV8 5′-end labeled with 32P to specific activities of 1.5 × 105 to 3.5 × 105 cpm/pmol (Cerenkov counts). For quantitating viral RNA replication, the blots used earlier for DI RNA detection on which radioactivity was no longer detectible were stripped and probed with 32P-radiolabeled oligonucleotide 3′UTRPSEV(+), a probe complementary to a common 3′ UTR sequence in all helper viruses (Table 1). Probed blots were read with a Packard InstantImager Autoradiography System for quantitation and exposed to Kodak XAR-5 film for 6 to 48 h at −80°C for imaging. Images were recorded by digital scanning of films.
Sequence of progeny DI RNAs
To determine the 5′-terminal leader sequence of progeny DI RNA, TGEV8(+) and RNA from cells infected with VP1 virus were used for RACE analysis as described above.
Accession numbers
GenBank Accession Nos. for the sequences studied here are as follows: U00735 for BCoV-Mebus, AF523843 and AF523847 for HCoV-OC43, AF523844 and AF523848 for HECoV-4408, AF523845 and AF523849 for HEV-TN11, AF523846 and AF523850 for ECoV-NC99, NC001846 for MHV-A59, AF201929 for MHV-2, M55148 and X00990 for MHV-JHM, AJ544718 for PV, and AF124990 and AF207551 for SDAV.14 32
Acknowledgements
We thank Cary Gay Brown, Kim Nixon, Aykut Ozdarendeli, Sharmila Raman, and Gwyn Williams for helpful discussions. This work was supported by Grant AI14367 from the National Institutes of Health, and by the University of Tennessee College of Veterinary Medicine Center of Excellence Program for Livestock Diseases and Human Health (H.-Y.W. and D.A.B.) and by Grant P-14104 from the Austrian Science Fund (R.V.).
References
- Baric R.S, Yount B, Hensley L, Peel S.A, Chen W. Episodic evolution mediates interspecies transfer of a murine coronavirus. J. Virol. 1997;71:1946–1955. doi: 10.1128/jvi.71.3.1946-1955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A, Spaan W.J.M. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D, Brian D.A, Briton P, Enjuanes L, Horzinek M.C, Lai M.M.C, Laude H, Plagemann P.G.W, Siddell S, Spaan W, Talbot P.J. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;1452:629–635. [PubMed] [Google Scholar]
- Chang R.-Y, Brian D.A. cis requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J. Virol. 1996;70:2201–2207. doi: 10.1128/jvi.70.4.2201-2207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.-Y, Hofmann M.A, Sethna P.B, Brian D.A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.-Y, Krishnan R, Brian D.A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci., USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L, Brian D.A, Cavanagh D, Holmes K, Lai M.M.C, Laude H, Masters P, Rottier P.J.M, Siddell S.G, Spaan W.J.M, Taguchi F, Talbot P. Family Coronaviridae. In: van Regenmortel M.H.V, Fauquet C.M, Bishop D.H.L, editors. Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press; New York: 2000. pp. 834–849. [Google Scholar]
- Felsenstein J. University of Washington; Seattle, WA: 1993. PHYLIP (Phylogeny Inference Package) v. 3.5c. [Google Scholar]
- Guy J.D. Turkey coronavirus is more closely related to avian infectious bronchitis virus than to mammalian coronaviruses: a review. Avian Pathol. 2000;29:207–212. doi: 10.1080/03079450050045459. [DOI] [PubMed] [Google Scholar]
- Guy J.S, Breslin J.J, Breuhaus B, Vivrette S, Smith L.G. Characterization of a coronavirus isolated from a diarrheic foal. J. Clin. Microbiol. 2000;38:4523–4526. doi: 10.1128/jcm.38.12.4523-4526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A, I. Smeenk I, Horzinek M.C, Rottier P.J, de Groot R.J. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis D.M, Hulsenbeck J.P. Signal, noise, and reliability in molecular phylogenetic analyses. J. Hered. 1992;83:189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- Hofmann M.A, Chang R.-Y, Ku S, Brian D.A. Leader-mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology. 1993;196:163–171. doi: 10.1006/viro.1993.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue B.G, Brian D.A. Structural proteins of the human coronavirus OC43. Virus Res. 1986;5:131–144. doi: 10.1016/0168-1702(86)90013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B, Hartshorne T, Masters P.S. Characterization of an essential RNA secondary structure in the 3′ untranslated region of murine coronavirus genome. J. Virol. 2000;74:6911–6921. doi: 10.1128/jvi.74.15.6911-6921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B, Masters P.S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Karaca K, Parrish C.R, Naqi S.A. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck J.G, Matsushima G.K, Makino S, Fleming J.O, Vannier D.M, Stohlman S.A, Lai M.M. In vivo RNA-RNA recombination of coronavirus in mouse brain. J. Virol. 1988;62:1810–1813. doi: 10.1128/jvi.62.5.1810-1813.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.N, Song Y.S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausegger A, Strobl B, Regl G, Kaser A, Luytjes W, Vlasak R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J. Virol. 1999;73:3737–3743. doi: 10.1128/jvi.73.5.3737-3743.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottier S.A, Cavanagh D, Britton P. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology. 1995;213:569–580. doi: 10.1006/viro.1995.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Chang R.-Y, Brian D.A. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology. 1996;218:400–405. doi: 10.1006/viro.1996.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L, Godeke G.-J, Raamsman M.J.B, Masters P.S, Rottier P.J.M. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–14006. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C, Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapps W, Hogue B.G, Brian D.A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.J, Liao C.L, Lai M.M.C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Johnson R.F, Leibowitz J.L. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J. Virol. 2001;75:12105–12113. doi: 10.1128/JVI.75.24.12105-12113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Joo M, Makino J.K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 1991;65:3304–3311. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Lai M.M.C. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J. Virol. 1989;63:5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengeling W.L, Boothe A.D, Ritche A.E. Characteristics of a coronavirus (strain 67N) of Pigs. Am. J. Vet. Res. 1972;33:297–308. [PubMed] [Google Scholar]
- Marra, M. A., Jones, S. J. M., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S. N., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., Cloutier, A., Coughlin, S. M., Freeman, D., Gim, N., Griffith, O. L., Leach, S. R., Mayo, M., McDonald, H., Montgomery, S. B., Pandoh, P. K., Petrescu, A. S., Robertson, A. G., Schein, J. E., Siddiqui, A., Smailus, D. E., Stott, J. M., Yang, G. S., Plummer, F., Andonov, A., Artsob, H., Bastien, N., Bernard, K., Booth, T. F., Bowness, D., Drebot, M., Fernando, L., Flick, R., Garbutt, M., Gray, M., Grolla, A., Jones, S., Feldmann, H., Meyers, A., Kabani, A., Li, Y., Normand, S., Stroher, U., Tipples, G. A., Tyler, S., Vogrig, R., Ward, D., Watson, B., Brunham, R. C., Krajden, M., Petric, M., Skowronski, D. M., Upton, C., Roper, R. L. 2003. The genome sequence of the SARS-associated coronavirus. Published online May 1 2003; 10.1126/science.1085953 (Science Express Research Articles) [DOI] [PubMed]
- Ozdarendeli A, Ku S, Rochat S, Williams G.D, Senanayake S.D, Brian D.A. Downstream sequences influence the choice between a naturally-occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 2001;75:7362–7374. doi: 10.1128/JVI.75.16.7362-7374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Bouma P, Williams G.D, Brian D.A. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003;77:6720–6730. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Peñaranda, S., Bankamp, B., Maher, K., Chen, M., Tong, S., Tamin, A., Lowe, L., Frace, M., DeRisi, J. L., Chen, Q., Wang, D., Erdman, D. D., Peret, T. C. T., Burns, C., Ksiazek, T. G., Rollin, P. E., Sanchez, A., Liffick, S., Holloway, B., Limor, L., McCaustland, K., Olsen-Rassmussen, M., Fouchier, R., Günther, S., Osterhaus, A. D. M. E., Drosten, C., Pallansch, M. A., Anderson, L. A., Bellini, W. J. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Published online May 1 2003; 10.1126/science.1085952 (Science Express Research Articles) [DOI] [PubMed]
- Senanayake S.D, Brian D.A. Translation from the 5′ untranslated region (UTR) of mRNA 1 is repressed, but that from the 5′ UTR of mRNA 7 is stimulated in coronavirus-infected cells. J. Virol. 1999;73:8003–8009. doi: 10.1128/jvi.73.10.8003-8009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S.G. The Coronaviridae, An Introduction. In: Siddell S.G, editor. The Coronaviridae. Plenum Press; New York, London: 1995. pp. 1–10. [Google Scholar]
- Smith S.W, Overbeek R, Woese C.R, Gilbert W, Gillevet P. The Genetice Data Environment: an expandable GUI for multiple sequence analysis. Comp. Appl. Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- Spagnolo J.F, Hogue B.J. Host protein interactions with the 3′ end of the bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sutherland, MA: 1996. PAUP*: phylogenetic analysis using parsimony (and other methods). v. 4.0. [Google Scholar]
- Tompkins W.A.F, Watrach A.M, Schmale J.D, Schulze R.M, Harris J.A. Cultural and antigenic properties of newly established cell strains derived from adenocarcinomas of the human colon and rectum. J. Natl. Cancer Inst. 1974;52:1101–1106. doi: 10.1093/jnci/52.4.1101. [DOI] [PubMed] [Google Scholar]
- van der Most R.G, de Groot R.J, Spaan W.J.M. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 1994;65:3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinten L.C, den Boon J.A, Wassenaar A.L.M, Spaan W.J.M, Snijder E.J. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D, Pei Y, Christie N, Cooper M. Primary structure of the sialodacryoadenitis virus genome: sequence of the structural-protein region and its application for differential diagnosis. Clin. Diagn. Lab. Immunol. 2000;7:568–573. doi: 10.1128/cdli.7.4.568-573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.D, Chang R.-Y, Brian D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzer W.J, Obojes K, Vlasak R. The sialate-4-O-acetylesterases of coronaviruses related to mouse hepatitis virus: a proposal to reorganize group 2 Coronaviridae. J. Gen. Virol. 2002;83:395–402. doi: 10.1099/0022-1317-83-2-395. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lai M.M.C. A 5′-proximal RNA sequence of murine coronavirus as a potential initiation site for genomic-length mRNA transcription. J. Virol. 1996;70:705–711. doi: 10.1128/jvi.70.2.705-711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M, Herbst W, Kousoulas K.G, Stortz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]


