Abstract
The 72 nt 3′ non-translated region (NTR) of potato virus X (PVX) RNA is identical in all sequenced PVX strains and contains sequences that are conserved among all potexviruses. Computer folding of the 3′ NTR sequence predicted three stem-loop structures (SL1, SL2, and SL3 in the 3′ to 5′ direction), which generally were supported by solution structure analyses. The importance of these sequence and/or structural elements to PVX RNA accumulation was further analyzed by inoculation of Nicotiana tabacum (NT-1) protoplasts with PVX transcripts containing mutations in the 3′ NTR. Analyses of RNA accumulation by S1 nuclease protection indicated that multiple sequence elements throughout the 3′ NTR were important for minus-strand RNA accumulation. Formation of SL3 was required for accumulation of minus-strand RNA, whereas SL1 and SL2 formation were less important. However, sequences within all of these predicted structures were required for minus-strand RNA accumulation, including a conserved hexanucleotide sequence element in the loop of SL3, and the CU nucleotide in a U-rich sequence within SL2. In contrast, 13 nucleotides that were predicted to reside in SL1 could be deleted without any significant reduction in minus or plus-strand RNA levels. Potential polyadenylation signals (near upstream elements; NUEs) in the 3′ NTR of PVX RNA were more important for plus-strand RNA accumulation than for minus-strand RNA accumulation. In addition, one of these NUEs overlapped with other sequence required for optimal minus-strand RNA levels. These data indicate that the PVX 3′ NTR contains multiple, overlapping elements that influence accumulation of both minus and plus-strand RNA.
Keywords: potato virus X, RNA replication, RNA structure, RNA virus, stem-loop
Abbreviations: NTR, non-translated region; PVX, potato virus X; SL, stem-loop; NT, Nicotiana tabacum; NUE, near upstream element; ORF, open reading frame; sgRNA, subgenomic RNA; DMS, dimethyl sulfate; CMCT, N-cyclohexyl-N′-[2-(N-methyl-4-morpholinio)-ethyl]-carboiimide p-toluenesulfonate
1. Introduction
The replication of viral RNA, for both eucaryotic and procaryotic plus-strand RNA viruses, initiates with the production of minus-strand RNA. This process involves recognition of cis-acting elements within the viral genome by the replication machinery. Although many of these elements include sequences and/or structures that reside in the 3′ non-translated regions (NTR) of viral RNAs,1., 2., 3. some cis-acting elements have been found to reside upstream and in the 5′ NTR.4 In addition, a long-distance interaction between elements near the 3′ terminus and 1200 nt upstream has been shown to be important for bacteriophage Qβ replication minus-strand RNA synthesis in vitro.5 Analyses of the roles of cis-acting elements in the synthesis of minus-strand RNA are complicated because 3′-terminal sequences may be required for other aspects of virus replication such as assembly, translation and/or RNA stabilization.6., 7., 8., 9., 10., 11., 12., 13.
Cis-acting elements required for minus-strand RNA accumulation in vivo or RNA synthesis in vitro have been well studied for the plant plus-strand RNA viruses that contain tRNA-like structures in the 3′ NTR2 such as brome mosaic virus (BMV),14., 15., 16. cucumber mosaic virus (CMV),17 tobacco mosaic virus (TMV)18 and turnip yellow mosaic virus (TYMV),19., 20. and for several other viruses such as alfalfa mosaic virus (AlMV),21., 22. barley stripe mosaic virus (BSMV),23 cymbidium ringspot virus (CymRSV),24 red clover necrotic mosaic virus (RCNMV)25 and turnip crinkle virus (TCV).26., 27. Much less is known about the roles of 3′ NTR cis-acting elements during minus-strand RNA synthesis of polyadenylated viruses. Although sequence and structural features in the 3′ NTR have been shown to be important for replication of beet necrotic yellow vein virus (BNYVV),28., 29. clover yellow vein virus (ClYVV),30 cowpea mosaic virus (CPMV),31 and the potexviruses, bamboo mosaic virus (BaMV),32., 33., 34. clover yellow mosaic virus (CYMV)35 and white clover mosaic virus (WClMV),36 only Sriskanda et al.37 have specifically shown that an element in the 3′ NTR affects accumulation of minus-strand RNA.
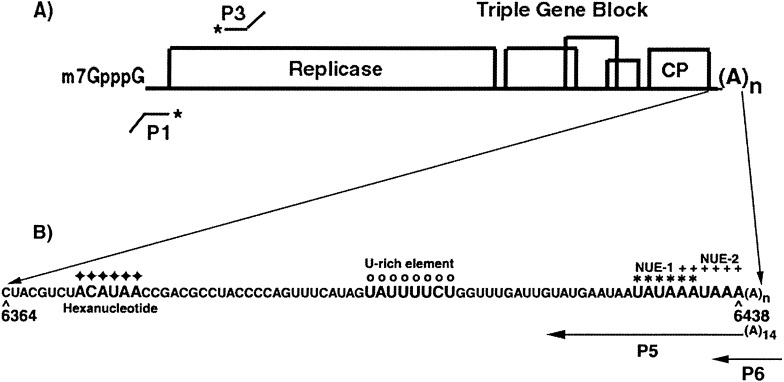
One potexvirus that has proven to be useful as a model system for analyses of viral RNA replication is potato virus X.38 This flexuous rod-shaped virus, which contains a 6.4 kb RNA genome that is capped and polyadenylated, is the type member of the potexvirus family.39., 40. The genome (Figure 1(A)) has an 84 nt 5′ NTR, five open reading frames (ORFs) and a 72 nt 3′ NTR.41., 42. ORF1 encodes the viral replicase, which is the only viral protein absolutely required for replication. The overlapping ORFs 2–4, collectively called the triple gene block, encode for proteins required for viral cell-to-cell and long-distance movement.43., 44., 45. ORF 5 encodes the coat protein, which is also required for viral movement.45., 46., 47. Our previous studies have shown that there are sequence and structural elements present in the 5′ NTR of PVX RNA that affect genomic and subgenomic (sgRNA) plus-strand RNA accumulation.48., 49., 50. These analyses revealed that although the plus-strand RNA accumulation was affected by mutations in the 5′ NTR, there was no significant effect on minus-strand RNA accumulation. Both local elements upstream of the sgRNAs, as well as long-distance interactions with complementary sequence at the terminus, are important for PVX plus-strand RNA accumulation. None of these elements in the 5′ NTR or upstream of the sgRNAs affected PVX minus-strand RNA accumulation. In contrast, Sriskanda et al.37 showed that deletion of 38 nt in the 3′ NTR of PVX RNA affected minus-strand RNA accumulation. Moreover, an eight nucleotide U-rich element within this region was shown to be important for host protein binding and replication.
Figure 1.
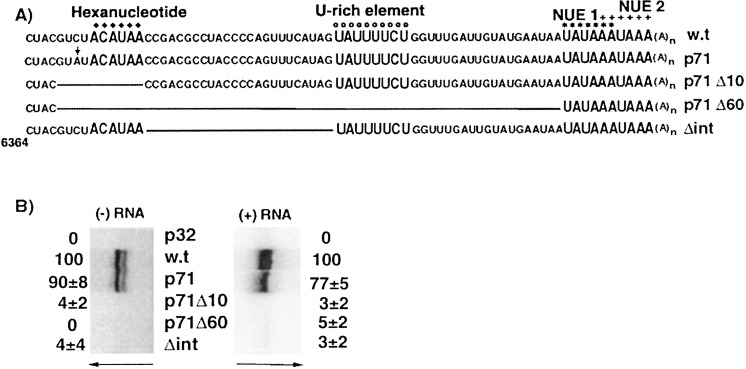
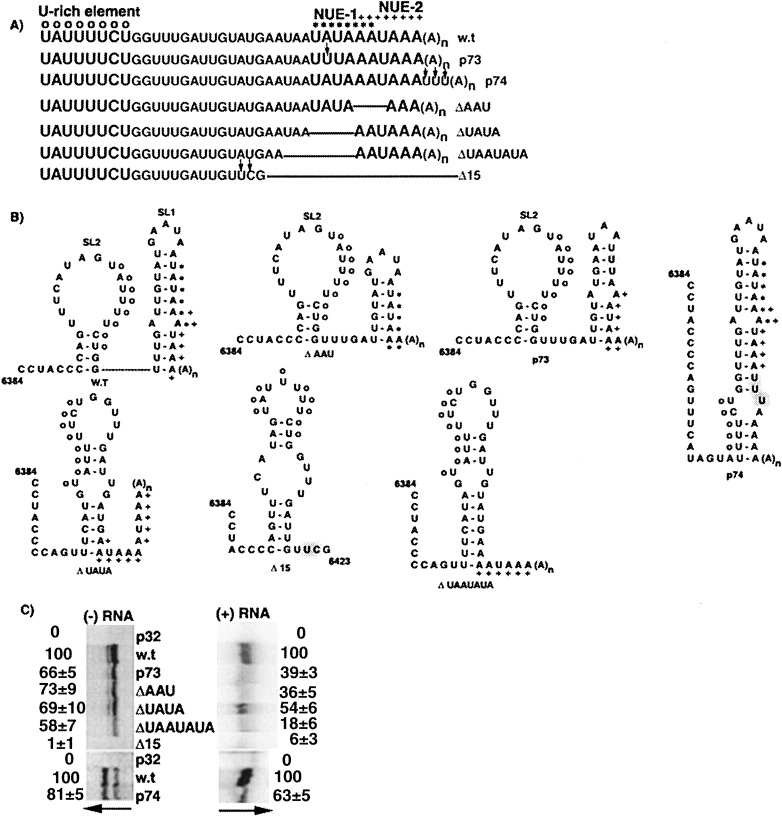
A representation of the PVX genome. (A) The five ORFs of the PVX genome are denoted by open boxes. The relative positions of the plus-strand probe (P1) and the minus-strand probe (P3) used for S1 nuclease protection assays are indicated below and above the genome, respectively, with asterisks (∗) denoting the 32P-labeled positions. (B) Within the 3′ NTR sequence, symbols are used to depict the hexanucleotide sequence (filled diamonds), the U-rich sequence (open circles), the first potential near-upstream element (NUE1; asterisk), and the second potential NUE (NUE2; plus sign). The relative positions of the primers P5 and P6 used for RNA sequencing and primer extension experiments are depicted below the 3′ NTR sequence.
In this study, we have investigated the importance of sequences and structures in the PVX 3′ NTR for accumulation of viral RNAs. Both minus and plus-strand viral RNA species were measured to determine the extent to which elements in the 3′ NTR affect minus-strand RNA levels and if this region contains signals that may regulate plus-strand RNA accumulation. We have identified multiple sequence elements throughout the 3′ NTR that were required for minus-strand RNA accumulation, as well as a short region near the 3′ terminus that was dispensable. In contrast, only one stem-loop structure was critical for RNA accumulation. We have localized elements important for optimal plus-strand RNA accumulation, indicating that the 3′ NTR is required for multiple aspects of virus replication.
2. Results
The role of the 3′ NTR cis-acting sequences and/or structures in PVX RNA accumulation was studied by solution structure probing and by introducing wild-type (w.t.) and mutant transcripts into tobacco protoplasts for analysis of minus and plus-strand RNA accumulation by S1 nuclease protection assays.
2.1. Solution structure analyses indicate multiple stem-loop structures in the 3′ NTR of PVX RNA
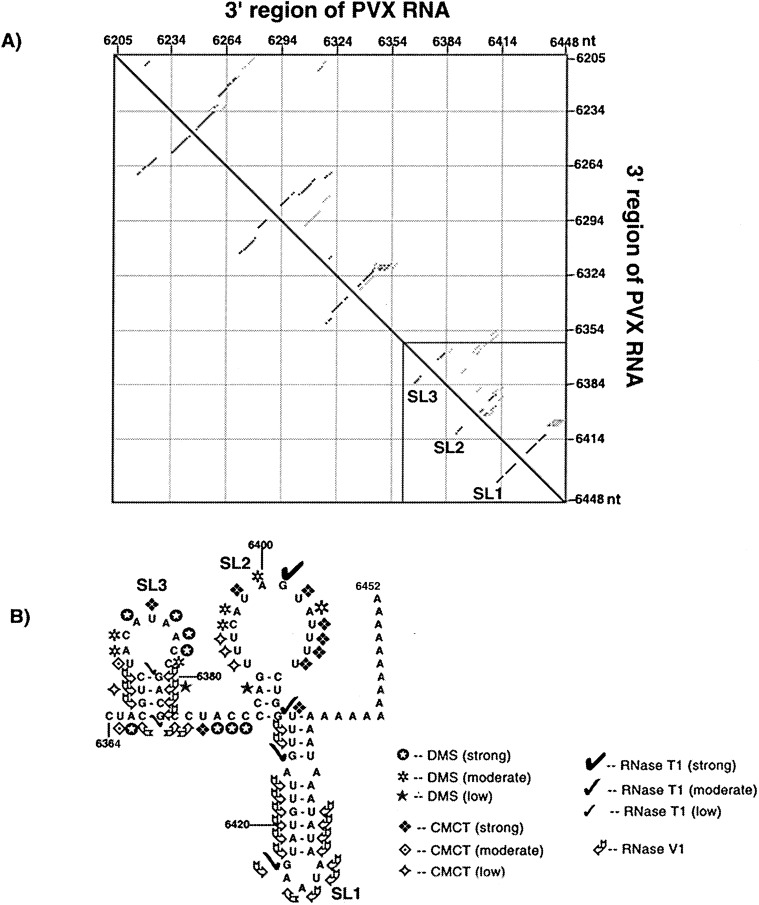
The predicted secondary structure for the 3′ 250 nt region of PVX RNA was generated using the Zuker Mfold program (Figure 2(B)). The thermodynamic energy dot plot shown in Figure 2(A) represents all secondary structures for the 250 nt sequence within 10% of the minimal energy. The triangle below the diagonal represents the most optimal folding of the minimum energy structure, with the rows of dots corresponding to nucleotide positions involved in pairing and the gaps within the rows indicating internal loops or bulges. The triangle above the diagonal represents sub-optimal foldings within 4 kcal (1 cal=4.184 J) of minimum free energy of the optimal folding. The dark dots represent optimal pairings and alternative folding patterns are represented by progressively lighter dots, with the lightest dots representing the least-favored pairing. Regions that are scattered with differently shaded dots exhibit multiple folding possibilities. The 3′ NTR (nucleotides 6364–6435 plus 17 A residues) of PVX RNA was predicted to contain three stem-loop structures, denoted SL1, SL2 and SL3 in the 3′ to 5′ direction (Figure 2(A) and (B)). As seen in Figure 2(A), SL3 was strongly predicted, but SL1 had some scatter near the base of the stem. The region containing SL2 and the region between SL2 and SL3 contained considerable scatter, suggesting multiple pairing possibilities. Similar structures were obtained when foldings were done with additional residues from the SpeI site. The secondary structure predicted by STAR program strongly supported the existence of SL3. There was no stable pseudoknot predicted in the 3′ region of PVX RNA.
Figure 2.
Summary of structural analyses of the 3′ NTR of PVX RNA. (A) An energy dot plot was derived for secondary structures in the 250 nt 3′region of PVX RNA predicted by the Zuker program (http://www.bioinfo.rpi.edu/applications/mfold/). The axes are labeled to denote the position of each nucleotide in the 3′ region of PVX RNA. The three stem-loop structures predicted to form in the 3′ NTR are boxed and denoted as SL1, SL2 and SL3. (B) A summary of the solution structure probing experiments is superimposed on the optimal predicted secondary structures within the 3′ NTR. The positions of RNase T1 cleavage are indicated by check marks, with larger check marks indicating more reactivity. RNase V1 cleavage positions are marked by curved arrows. The DMS modification positions are identified by asterisks with intensities of low ( ), moderate (
), moderate ( ), or strong (
), or strong ( ). The intensities of uridine modification by CMCT are denoted as low (
). The intensities of uridine modification by CMCT are denoted as low ( ), moderate (
), moderate ( ), or strong (
), or strong ( ). Filled circles represent ambiguous CMCT reactivity.
). Filled circles represent ambiguous CMCT reactivity.
Chemical and enzymatic probing experiments were conducted to analyze the solution structure of the 3′ NTR of PVX RNA. For chemical modification experiments, dimethyl sulfate (DMS) was used to modify unpaired A (at N-1) and C (at N-3) residues and N-cyclohexyl-N′-[2-(N-methyl-4-morpholinio)-ethyl]-carboiimide p-toluenesulfonate (CMCT) was used to modify unpaired U residues (at N-3). Samples were analyzed using Rnase T1 and Rnase V1 to detect unpaired G residues and base-paired or stacked helical regions, respectively. Reaction conditions for both the chemical modification and the RNase digestion were optimized such that less than one nucleotide per RNA molecule was modified. The extent of modification and cleavage was analyzed by primer extension of the products and subsequent gel electrophoresis. A particular nucleotide was considered reactive if the intensity of the extended product signal was stronger in the modified (+) lane than that in the unmodified (−) lane. Transcripts derived from clone p10, containing 185 nt of the PVX RNA 3′ region were used for the enzymatic cleavage experiments and full-length PVX transcripts derived from clone pMON8453 were utilized for the chemical modification experiments. Comparison of these transcripts and viral RNA resulted in similar modification patterns (data not shown).
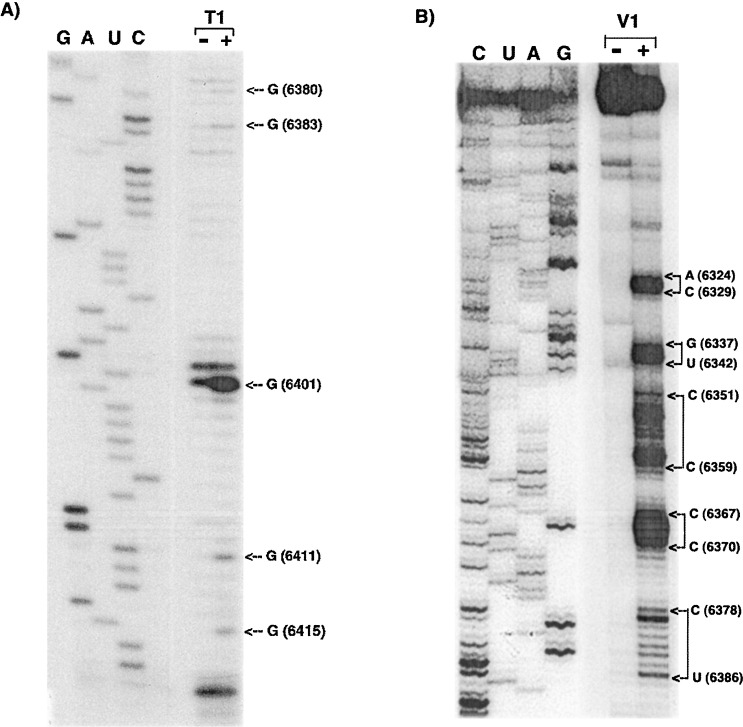
A summary of the data obtained from these experiments was superimposed on the most stable 3′ NTR structure (ΔG—55.6 Kcal/mole) predicted by the Zuker program (Figure 2(B)). Autoradiographs corresponding to some of these data are shown in Figure 3, Figure 4 . RNase V1 cleaved many of the nucleotides in the stem of SL1 and none was reactive to DMS, indicating base-pairing or stacking in this region. Lack of modification in the 3′ end of the lower region of SL1 was due to protection by primers used in the reverse transcription reactions. Exceptions to the predicted pairing were indicated by susceptibility of G6415 to Rnase T1 (Figure 2, Figure 4) and U6412 to CMCT (Figure 3(B)). Although the nucleotides in the predicted mismatch were not modified by DMS, it was not susceptible to RNase V1. The terminal loop residue G6423 was cleaved by RNase T1, suggesting that it was not paired, but none of the other terminal loop residues was accessible to DMS or CMCT. In contrast, all of the loop residues were accessible to RNase V1. Thus, it appears that the upper stem region of SL1 may be base-paired as predicted, and that the terminal loop residues of SL1 may be stacked or be involved in pairing with nucleotides located elsewhere. The lower region of the predicted stem region appears to be more flexible and may form alternative structures, which is consistent with the thermodynamic energy dot plot (Figure 2(A)).
Figure 3.
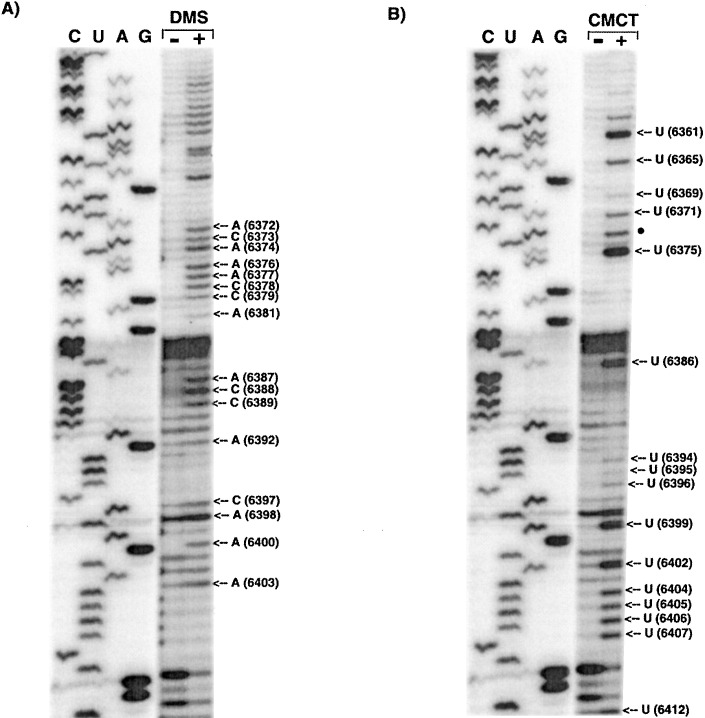
Chemical modification patterns of PVX transcripts. Full length transcripts from pMON8453 were incubated either (A) with (+) or without (−) DMS or (B) CMCT. Samples then were subjected to primer extension with primer P5, and run on denaturing 8% polyacrylamide gels. Lanes marked C, U, A, G correspond to PVX RNA sequenced using the same primer noted above. The positions of the modified nucleotides are noted on the right side of the autoradiogram. A filled circle marks an ambiguous CMCT reactivity where there is a signal in the modified lane, but it corresponds to a C rather than a U residue.
Figure 4.
Enzymatic cleavage of PVX transcripts. Transcripts from p10 corresponding to nucleotides 6265–6435 (plus 17 A residues and 4 nt of the SpeI site) from the 3′ end of PVX RNA were incubated either (A) with (+) or without (−) RNase T1 or (B) RNase V1, and were subjected to primer extension and gel electrophoresis. Consecutively modified nucleotides are denoted as connected arrows to the right of the autoradiogram. All other labeling is similar to that described in the legend to Figure 3.
Although SL2 is not strongly predicted and has considerable scatter compared to SL3 and SL1 (Figure 2(A)), the solution structure probing does suggest that the predicted structure is somewhat maintained (Figure 2(B)). Of the loop residues, U6399, U6402 and U6404–7 were all strongly modified by CMCT (Figure 3(B)); A6403, A6400, A6398 and C6397 were modified by DMS (Figure 3(A)). Similarly, G6401 in this loop had strong reaction with Rnase T1 (Figure 4(A)). In contrast, U6394–96 exhibited low-level modification by CMCT, as seen in Figure 3(B). Low reactivity of A6392 to DMS (Figure 3(A)) and G6411 to RNase T1 (Figure 4(A)), combined with weak or absent reactivity of some loop residues, may indicate that alternative structures can form. Since SL1 is predicted to be immediately adjacent to predicted SL2, it is possible that these two stems are co-axially stacked. The low level of modification of the residue G6411 at the base of SL2, and CMCT reaction of the contiguous U6412 at the base of SL1 (Figure 4, Figure 3, respectively), suggests that the bases of these stems may have been in alternative conformations.
Most of the predicted single-stranded residues between SL2 and SL3 were modified strongly by DMS (A6387–C6389) and CMCT (U6386) (Figure 3(A) and (B), respectively). As seen in Figure 4(B), C6384 and C6385 were cleaved by RNase V1, suggesting that they may be stacked.
The solution structure analyses of SL3 indicated that all the residues in the loop of SL3 were accessible, as evidenced by either moderate or strong reactivity to DMS (A6372–A6374, A6376–C6379) or CMCT (U6371 and U6375), as shown in Figure 3(A) and (B), respectively. RNase V1 cleaved residues C6367–C6370 and G6380–G6383 (Figure 4(B)), indicating that they are stacked or paired as predicted by the computer folding in Figure 2. Some of the residues in the stem of SL3 (U6369, A6381, G6380, G6383) were modified to a very small extent. Thus, both the digestion and modification data fit fairly well with the Mfold-predicted SL3 structure.
2.2. Multiple sequence elements important for PVX RNA accumulation reside within the 3′ NTR
The importance of elements in the upstream region of the PVX 3′ NTR for RNA accumulation in protoplasts was analyzed by directed deletions using exonuclease III (Figure 5(A)). First, an SnaBI site was created in pMON8453 by changing C6370 to A. The resulting clone, p71, was digested with SnaBI and then exonuclease III to generate mutants p71Δ10 and p71Δ60 that lack ten and 60 nucleotides, respectively. The smaller deletion, p71Δ10, extends through the hexanucleotide sequence. The larger deletion, p71Δ60, lacks most of the 3′ NTR, except for four nucleotides at the 5′ region of the 3′ NTR and the putative NUE signals. Deletion of the sequence between the hexanucleotide and eight nucleotide U-rich region in the Δint mutant, was created by site-directed mutagenesis.
Figure 5.
Effects of deletions extending from the 5′ region of the PVX 3′ NTR. (A) The sequence of the wild-type, pMON8453 3′ NTR is aligned with corresponding sequences from deletion mutants p71Δ10, p71Δ60 and Δint. The vertical arrow indicates the nucleotide mutated in pMON8453 to generate p71. Deleted sequences in p71Δ10, p71Δ60 and Δint are noted by lines. Different elements are illustrated above the wild-type sequence with symbols as described in the legend to Figure 1. (B) Protoplasts inoculated with replication-defective transcripts derived from p32 (p32), wild-type transcripts derived from pMON8453 (w.t.), and mutant transcripts (p71, p71Δ10, p71Δ60 and Δint) were analyzed at 40 hours post-inoculation by digestion with S1 nuclease. The protected products were separated on denaturing 8% polyacrylamide gels; arrows indicate the direction of electrophoresis. The relative average values (±standard error) for minus-strand (−RNA) and plus-strand (+RNA) accumulation are noted at the outer edges of the autoradiograms.
As shown in Figure 5(B), protoplasts inoculated with p71 accumulated near wild-type levels of minus-strand and plus-strand RNA. In contrast, protoplasts inoculated with the three deletion mutants (p71Δ10, p71Δ60, Δint) contained negligible levels of minus or plus-strand RNAs. The signals for minus-strand RNA frequently appeared as doublets. These are likely due to breathing in the AUAUC sequence immediately adjacent to the non-specific portion of the probe, which resulted in two S1 protection patterns. The intensity of each band in the doublet varies from experiment to experiment, and shows no correlation to a particular transcript used as inoculum. Coat protein accumulation (data not shown) mirrored the plus-RNA accumulation, suggesting that sub-genomic RNA accumulation was affected similarly by the deletions. These data indicate that deletion of 10 nt, including the hexanucleotide element near the 5′ region of the 3′ NTR, resulted in inhibition of minus-strand RNA accumulation. As expected, the longer deletion of most of the 3′ NTR in p71Δ60 also was unable to initiate minus-strand RNA synthesis. Although the Δint mutant contains the hexanucleotide element, the U-rich element and other sequences downstream, deletion of the sequence between these elements was detrimental to minus-strand RNA accumulation. Either the sequence in this region, the importance of this sequence to structure, and/or the spacing between the hexanucleotide and U-rich elements are necessary for RNA synthesis. Because minus-strand RNA synthesis was inhibited for all of these deletion mutants, plus-strand RNA levels were reduced similarly. These data demonstrate that multiple elements within the 3′ NTR are required for minus-strand, and hence plus-strand, RNA accumulation.
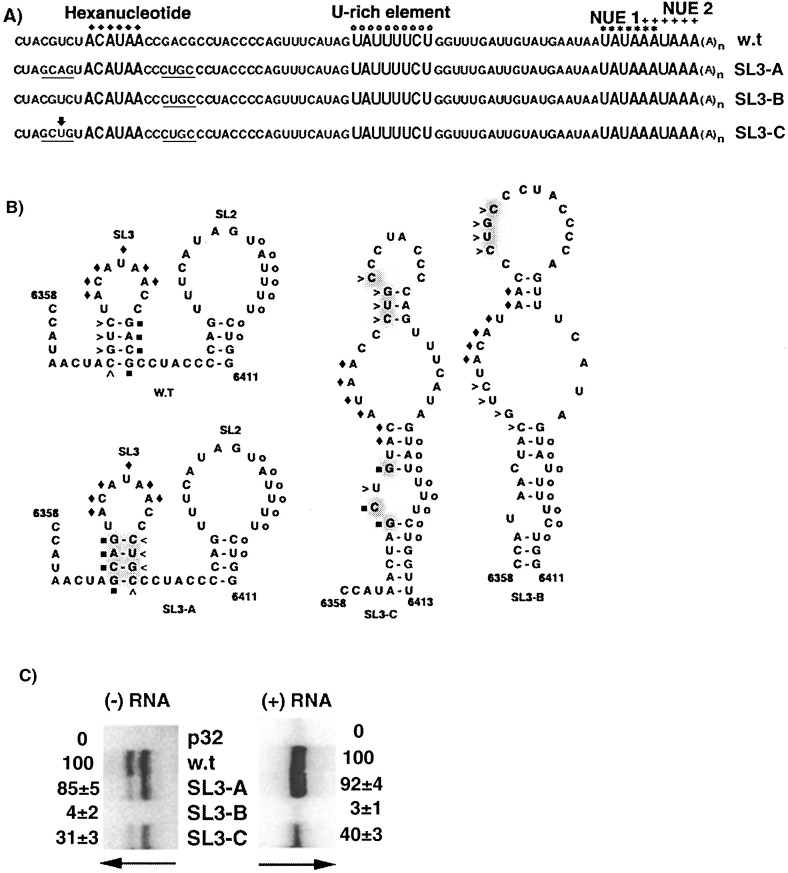
2.3. Pairing, and not sequence in the stem of SL3, is important for minus-strand RNA accumulation
The importance of SL3 structure in minus and plus-strand RNA accumulation was examined by creating mutants SL3-A, SL3-B, and SL3-C (Figure 6(A)). As shown in Figure 6(B), inversion of the sequence on the two sides of the stem in mutant SL3-A was not predicted to alter the structure. In contrast, mutant SL3-C, which differs from mutant SL3-A by only one nucleotide (A6369 to U), was predicted to form an alternative structure in the SL3 and SL2 regions. Rearrangement of structure was predicted for mutant SL3-B, which had sequence altered only on the 3′ side of SL3.
Figure 6.
Analyses of mutations predicted to affect SL3. (A) The 3′ NTR sequences of the wild-type and mutant transcripts are noted, with altered nucleotides underlined. (B) The predicted optimal secondary structures are depicted for wild-type and mutant RNAs between nucleotides 6358 and 6411. The wild-type sequence (CUGC) on the 5′ side of the SL3 stem is marked by arrowheads, and the wild-type sequence (GACG) on the 3′ side of the SL3 stem is indicated by filled squares. Alterations of these sequences are depicted by shading in the mutant structures. The hexanucleotide and U-rich elements are marked as in Figure 1. (C) Protoplasts inoculated with replication-defective transcripts (p32), wild-type (w.t.), and mutant transcripts (SL3-A, SL3-B and SL3-C) were analyzed for RNA accumulation as described in the legend to Figure 5(B).
Minus and plus-strand RNA accumulation in protoplasts inoculated with SL3-A transcripts was comparable to wild-type levels (Figure 6(C)). In contrast, protoplasts inoculated with either SL3-B or SL3-C transcripts exhibited negligible or significantly reduced levels of minus and plus-strand RNA accumulation, respectively. These data suggest that base-pairing, and not the specific sequence of the stem in SL3, is important for minus and plus-strand RNA accumulation.
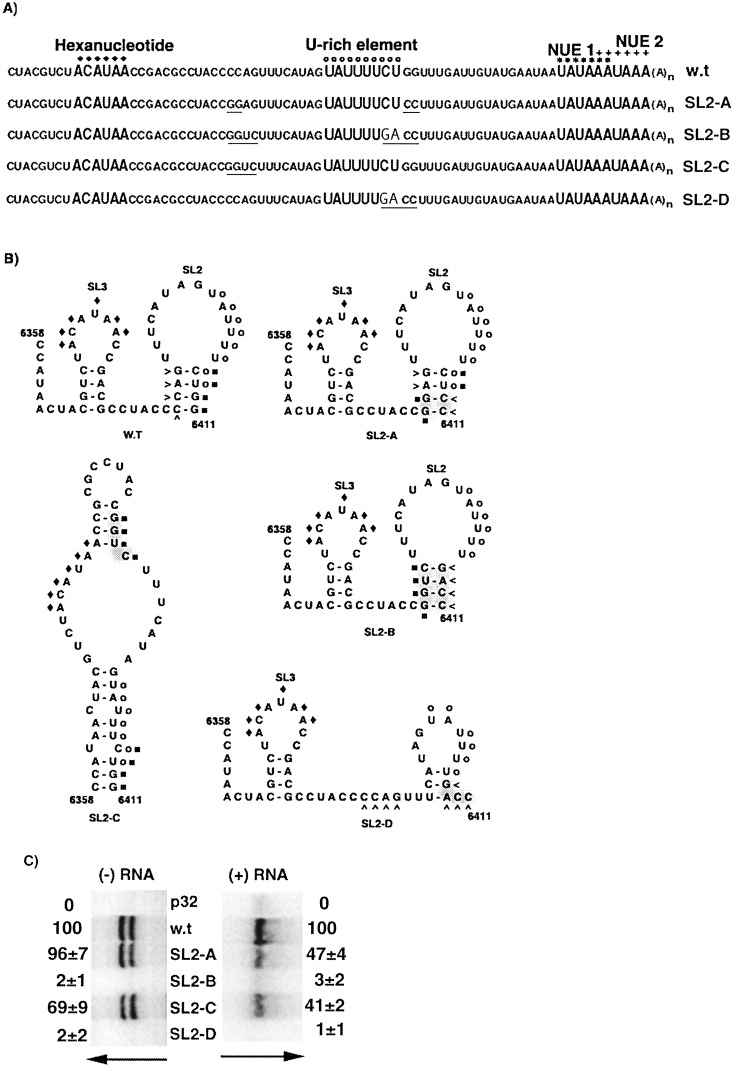
2.4. Sequence elements in the stem of SL2 are important for minus and/or plus-strand RNA accumulation
The importance of the predicted structure SL2 to PVX RNA accumulation was examined by creating mutants SL2-A, SL2-B, SL2-C and SL2-D (Figure 7(A)). As the last two nucleotides of the U-rich sequence are present in the stem of SL2, several sequence modifications were made to determine the relative importance of these nucleotides in the U-rich element and the pairing in the stem.
Figure 7.
Effects of mutations in U-rich sequence and predicted SL2. (A) The 3′ NTR sequences of the wild-type and mutant transcripts are noted, with. altered nucleotides underlined. (B) The predicted secondary structures of wild-type and mutant RNA between nucleotides 6358 and 6411 are illustrated, with designations as described in the legend to Figure 6(B). (C) Analysis of RNA accumulation in protoplasts inoculated with transcripts derived from p32 (p32), pMON8453 (w.t.), and mutant (SL2-A, SL2-B, SL3-C and SL2-D) transcripts was carried out as described in the legend to Figure 5(B).
Only the two base-pairs at the base of SL2 were reversed in mutant SL2-A, keeping the U-rich element and predicted SL2 intact (Figure 7(B)). Protoplasts inoculated with SL2-A transcripts had wild-type levels of minus-strand RNA accumulation, but the plus-strand RNA levels were reduced to approximately 40% of those observed with wild-type transcripts. These data suggest that the sequence in the lower part of the stem in SL2 was not necessary for minus-strand RNA synthesis, but was necessary for optimal accumulation of plus-strand RNA. In contrast, mutant SL2-C was created to retain the U-rich element within an altered structure by changing only the 5′ side of the stem (C6390CAG to GGUC). This resulted in disruption of both SL3 and SL2, and predicted formation of a single, large stem-loop structure (Figure 7(B)). Inoculation of protoplasts with mutant SL2-C resulted in reduced levels of both minus and plus-strand RNA (Figure 7(C)). These data are similar to those observed for mutant SL3-C (Figure 6(C)), which also was predicted to have a combined SL3/SL2 structure. Thus, disruption of SL3 structure in theSL2-C mutant caused a reduction in minus-strand RNA synthesis and consequently lowers levels of plus-strand RNA.
Both mutants SL2-B and SL2-D contain alterations of the U-rich element. In mutant SL2-B, the two sides of the stem were swapped to maintain the SL2 structure, but resulting in alteration of the 3′ two nucleotides of the U-rich sequence (Figure 7(B)). Similarly, in mutant SL2-D, these two nucleotides were changed because the 3′ side of SL2 was mutated (C6408UGG to GACC) to disrupt base-pairing. When either SL2-B or SL2-D transcripts were inoculated into protoplasts, both minus and plus-strand RNA levels were negligible (Figure 7(C)). These data indicate that the 3′-terminal CU nucleotides of the U-rich sequence are important for minus-strand RNA synthesis.
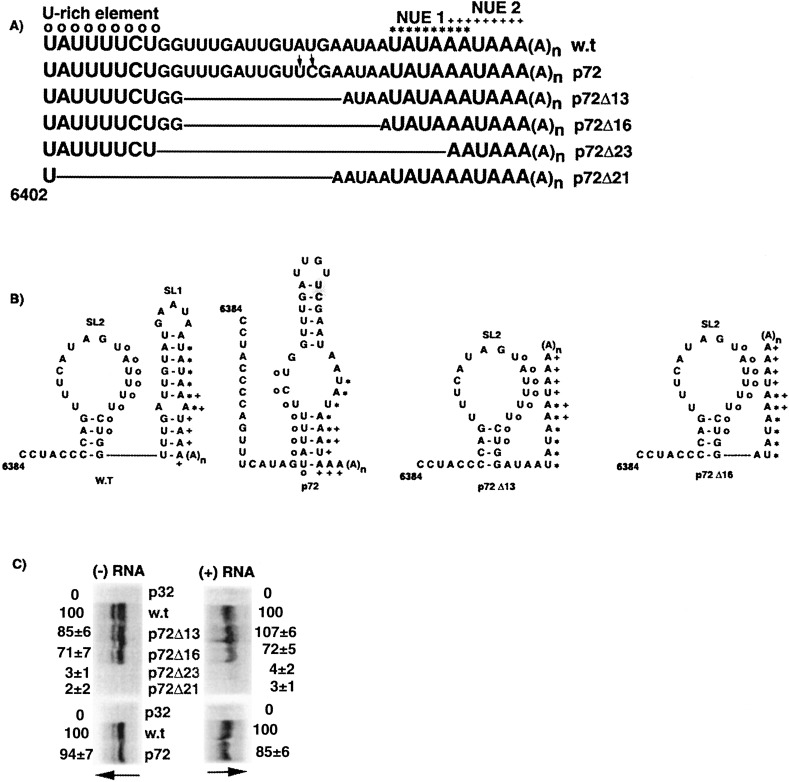
2.5. Sequences 3′ to the U-rich region contain signals for minus-strand RNA synthesis
The importance of sequence between the eight nucleotide U-rich region and the potential NUEs for PVX replication was examined using mutants listed in Figure 8(A) . A Bst BI site was created in the mutant p72 by site-directed mutagenesis to change nucleotides A6421–U6422 to UC. All the deletion mutants were created by digestion of p72 with Bst BI and exonuclease III, except for p72Δ23, which was created by site-directed mutagenesis of p72Δ16.
Figure 8.
Deletion analyses of sequences between the U-rich element and the potential NUEs. (A) Sequences between nucleotides 6402 and 6438 for p72, p72Δ13, p72Δ16, p72Δ23 and p72Δ21 are aligned, with deleted regions marked as lines. (B) Predicted secondary structures for wild-type and some of the mutant RNAs between nucleotides 6384 and 6438 are illustrated and marked as described in the legend to Figure 1. Although not shown, SL3 is predicted to be unchanged in all transcripts illustrated in A and B. (C) Minus and plus-strand RNA accumulation at 40 h.p.i. in protoplasts inoculated with transcripts derived from p32, pMON8453 (w.t), and mutants p72Δ13, p72Δ16, p72Δ23 and p72Δ21 were analyzed as described in the legend to Figure 5(B).
Mutant p72 and all the deletion mutants were predicted to contain a wild-type SL3 structure (not shown in Figure 8(B)). The predicted stem-loop structure downstream of SL3 for p72 is different from wild-type and has part of the U-rich sequence pairing with parts of both the potential NUEs (Figure 8(B)). Both mutant p72Δ13 (nucleotides 6412–6424 deleted) and p72Δ16 (nucleotides 6412–6427 deleted) were predicted to form wild-type SL3 and SL2, but were lacking SL1. Interestingly, protoplasts inoculated with p72 or with p72Δ13 transcripts exhibited minusand plus-strand RNA accumulation levels similar to wild-type (Figure 8(C)). These data indicate that removal of nucleotides 6412–6424 (p72Δ13) between the U-rich element and the potential NUEs did not affect RNA accumulation. In contrast, deletion of sequence beyond nucleotide 6424 in p72Δ16 resulted in reduction in minus and plus-strand RNA accumulation to 70% of wild-type levels. Thus, although p72Δ13 and p72Δ16 have similar structures downstream of SL3, the removal of three extra nucleotides from p72Δ16 affected RNA accumulation.
In mutant p72Δ23, the sequence between the U-rich element and the second NUE was deleted (G6410–A6432) and the RNA was predicted to be unstructured downstream of SL3 (data not shown). This deletion extends through G6410 and G6411, which are included in mutants p72Δ13a and p72Δ16. When the p72Δ23 transcripts were inoculated into protoplasts, both minus and plus-strand RNA levels were reduced substantially. This reduction likely can be attributed to continued deletion in the 3′ direction (removal of the second NUE), and not to the two G residues (nucleotides 6410–6411) because they were previously shown not to be important for minus-strand RNA accumulation when they were changed to two C residues in mutant SL2-A (Figure 7(B)).
As expected, deletion of the U-rich sequence in mutant p72Δ21 transcripts resulted in negligible amounts of minus and plus-strand RNA in protoplasts. This confirms that the U-rich sequence is necessary for minus-strand RNA synthesis. Overall, these data indicate that elements for minus-strand RNA synthesis reside in nucleotides 6425–6432, between the U-rich region and poly(A) tail, but that nucleotides 6412–6424 downstream of the U-rich sequence are not necessary for RNA accumulation.
2.6. Alteration of potential NUEs affects minus and plus-strand RNA accumulation differentially
The significance of the potential NUEs was studied with mutants p73, p74, ΔAAU, ΔUAUA, ΔUAAUAUA and Δ15 (Figure 9(A)). In mutant p73, A6340 was changed to U, disrupting the first NUE and leaving the second NUE intact. The predicted secondary structure for this mutant contains wild-type SL3 and SL2 and another structure downstream that is different from SL1 (Figure 9(B)). In mutant ΔAAU, the AAU sequence of the second NUE was deleted, keeping the first NUE intact. The secondary structure for ΔAAU also contains wild-type SL3 and SL2, and an alternative structure instead of SL1. Protoplasts inoculated with transcripts of mutants p73 and ΔAAU had minus-strand RNA levels reduced to approximately 70% that of wild-type. Surprisingly, plus-strand RNA accumulation in protoplasts inoculated with these mutants was further reduced to approximately 35% of wild-type (Figure 9(C)).
Figure 9.
RNA accumulation in response to modification of potential NUEs. (A) Sequences between nucleotides 6402 and 6438 of wild-type and mutant RNAs are compared. Transcripts derived from all mutants, except Δ15, have the poly(A) tail. Nucleotides mutated in p73, p74 and Δ15 are denoted by vertical arrows and other designations are as described in the legend to Figure 1. (B) The predicted secondary structures from nucleotides 6384–6438 of wild-type and mutant RNAs are illustrated; the predicted structure for Δ15 contains nucleotides 6384–6423. All of these RNA transcripts are predicted to have a wild-type SL3 structure. All markings are as denoted in the legends to Figure 1, Figure 3. (C) Detection of minus and plus-strand RNA accumulation in protoplasts inoculated with p32, w.t, and mutant (p73, p74, ΔAAU, ΔUAUA, ΔUAAUAUA and Δ15) transcripts as described in the legend to Figure 5(B).
This differential minus and plus-strand RNA accumulation was more pronounced in protoplasts inoculated with the mutant ΔUAAUAUA, which has a deletion of U6426–A6432. The predicted structure for this mutant contains wild-type SL3 and one altered stem-loop structure downstream of SL3 (Figure 9(B)). Protoplasts inoculated with ΔUAAUAUA transcripts have minus-strand RNA accumulation reduced to approximately 60% of wild-type, whereas plus-strand RNA accumulation was further reduced to approximately 20% of wild-type (Figure 9(C)). A similar result was observed in protoplasts inoculated with mutant p74, where minus-strand RNA accumulated to almost wild-type levels, while plus-strand RNA was reduced to 65% of wild-type. Mutant p74 had the sequence UUU inserted after the second NUE to keep both the NUEs intact, but increase their distance from the poly(A) tail. The secondary structure of p74 was predicted to include a wild-type SL3 and one stem-loop replacing SL2 and SL1. These data suggest that the potential NUEs and/or the sequences surrounding them may affect plus-strand RNA stability and/or accumulation, and have less of an effect on minus-strand RNA accumulation.
The importance of these potential NUEs can be be seen in the mutant ΔUAUA, which has a wild-type SL3 and another structure downstream. Deletion of U6429–A6432 in this mutant resulted in the removal of the first NUE and formation of two overlapping AAUAAA sequences (nucleotides 6424–6429 and 6429–6434). After inoculation of ΔUAUA transcripts into protoplasts, minus and plus-strand RNA levels were reduced equally to approximately 60% of wild-type. The absence of differential accumulation may indicate that the two overlapping AAUAAA sequences that are formed are sufficient for plus-strand RNA accumulation, at least to levels equal to minus-strand RNA accumulation. This was similar to the reduction in minus and plus-strand RNA in protoplasts inoculated with mutant p72Δ16, which still has the two potential NUE sequences (Figure 8). Consequently, the consecutive NUEs may need to be present for optimal PVX plus-strand RNA stability and accumulation.
Mutant Δ15, which lacks G6423–A6438 and the poly(A) tail, was predicted to contain wild-type SL3 and an altered stem-loop structure downstream of SL3. Protoplasts inoculated with Δ15 transcripts accumulated negligible amounts of minus and plus-strand RNA (Figure 9(C)). Similar data were seen with protoplasts inoculated with mutant p72Δ23 (Figure 8(C)), which lacks G6410–A6432, confirming that elements downstream of the U-rich region are important for minus-strand RNA synthesis. These elements probably overlap with the potential NUEs, and may be why all the mutants disrupting either of the potential NUEs (p73, ΔAAU, ΔUAAUAUA and ΔUAUA) have reduced minus-strand RNA levels (around 60–70% of wild-type). On the other hand, mutant p74, which has both the NUEs intact has near-wild-type levels of minus-strand RNA accumulation, but reduced plus-strand RNA accumulation. Thus, the distance of the potential NUEs from the poly(A) tail and/or the requirement of the second potential NUE to be continuous with the poly(A) tail may be important for optimal plus-strand RNA accumulation.
3. Discussion
For several plant and animal plus-strand RNA viruses, both sequence and structural elements in the 3′ NTR are required for minus-strand RNA accumulation.1., 2., 3. Similarly, our deletion and site-directed mutational analyses of the cis-acting elements in the 3′ NTR of PVX RNA identified multiple sequence and structural elements necessary for minus-strand RNA accumulation. Surprisingly, our data also indicated that additional sequences in the 3′ NTR differentially regulate plus-strand rather than minus-strand PVX RNA levels and overlap with sequences important for minus-strand RNA accumulation.
3.1. Only one of three predicted stem-loop structures is important for PVX RNA accumulation
The Mfold computer prediction for PVX 3′ NTR secondary structure indicated three SL structures (SL1, SL2 and SL3), with SL1 and SL3 being the most strongly predicted structures in the thermodynamic dot plot. Because all of the PVX strains have identical 3′ NTR sequences, no covariation data exist for these predicted structures. For another potexvirus, BaMV, the STAR program was utilized to predict that the 3′ NTR contained a pseudoknot involving some of the poly(A) tail residues.34 The existence of this pseudoknot configuration for the BaMV 3′ NTR was supported by solution structure studies and functional analyses.34 Modeling of the PVX 3′ NTR with the STAR program did predict a stem-loop similar to the SL3 predicted by the Mfold program, but did not predict any stable pseudoknot configurations or stem-loop structures identical with SL1 or SL251. Overall, the chemical modification and enzymatic digestion analyses of the PVX 3′ NTR fit the Mfold model better than the model predicted by STAR.
Although SL1, SL2 and SL3 were predicted by computer folding and these structures were generally supported by solution structure analyses, only SL3 was shown to be required for PVX minus-strand RNA accumulation in protoplasts. Alteration of predicted structures for SL2 and SL1 was not critical for minus-strand accumulation as seen with mutants p72 and p72Δ13. In contrast, alteration of predicted SL3 structure (mutant SL3-B) reduced minus-strand RNA accumulation in protoplasts. However, when compensatory changes were made to restore the SL3 structure (mutant SL3-A), minus-strand RNA levels were restored to near-wild-type levels, indicating that structure rather than sequence in the stem was the most important factor for accumulation of minus-strand RNA. In contrast, replication of BaMV was dependent on both pairing and sequence in stem D of the 3′ NTR,33 but both the BaMV stem D and PVX SL3 have terminal loops containing conserved hexanucleotide motifs that are required for replication.
3.2. Placement of the conserved hexanucleotide motif in a terminal loop is important for PVX minus-strand RNA accumulation
The requirement for formation of SL3 may be to present the conserved hexanucleotide sequence in the terminal loop. Bancroft et al.52 showed that the hexanucleotide sequence was conserved in all potexviral RNAs near the 3′ terminus and upstream of the TGB and CP coding regions. Such an element located in the 3′ NTR of CYMV D-RNA was required for accumulation of the genomic CYMV D-RNA,35 and positions 2 and 4 were most important. Chiu et al.32 recently reported that positions 4–6 of the hexanucleotide sequence were absolutely required for genomic and subgenomic BaMV RNA accumulation. Our data indicate that deletion of a region containing the hexanucleotide motif in PVX RNA (mutant p71Δ10) or disruption of the SL3 structure (mutants SL3-B, SL3-C) resulted in negligible to reduced minus and plus-strand RNA accumulation, suggesting that the hexanucleotide sequence and its inclusion in a stem-loop region are required for minus-strand RNA synthesis.
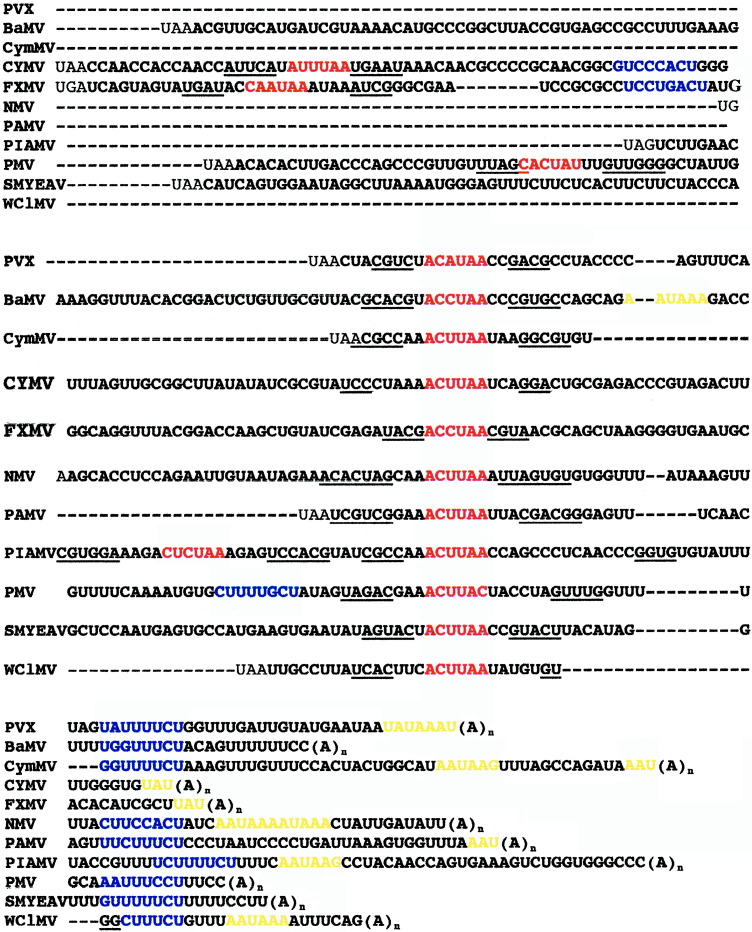
The significance of a paired structure including the hexanucleotide motif can also be modeled for other potexviruses.32 We have found that hand-alignment of the 3′ NTR sequences of 11 different potexviruses (Figure 10), supports conservation of the hexanucleotide (in red) within the context of palindromic sequences (underlined). When the 3′ NTR sequences of these potexviruses were folded using the Mfold, eight out of the 11 folded with the hexanucleotide sequence presented in terminal loop of a stem and the stems contained the palindromic sequences underlined in Figure 10 (data not shown).
Figure 10.
Alignments of the 3′ NTR sequences of different potexviruses. Alignments were done by hand and were centered around hexanucleotide elements (in red) conserved among potexviruses. Additional potential hexanucleotide elements found upstream in CYMV, FXMV, PIAMV, and PMV sequences are also noted in red. The sequences aligned include our PVX strain,49 bamboo mosaic virus (BaMV),84 cymbidium mosaic virus (CyMV),85 clover yellow mosaic virus (CYMV),86 foxtail mosaic virus (FXMV),52 narcissus mosaic virus (NMV),87 potato aucuba mosaic virus (PAMV),88Plantago asiatica mosaic virus (PIAMV),89 papaya mosaic virus (PMV),90 strawberry mild yellow edge-associated virus (SMYEAV)91 and white clover mosaic virus (WClMV).92 Palindromic sequences are underlined on either side of the hexanucleotide elements. Similarities in the U-rich (blue) and putative NUE (yellow) sequences among the potexvirus RNAs are indicated.
The presentation of the hexanucleotide in a loop may be required for RdRp and/or host protein binding during replication. Although the hexanucleotide sequence was not required for host protein binding to the PVX 3′ NTR,37 Huang et al.53 found that an Escherichia coli expressed RdRp domain of BaMV binds to the hexanucleotide loop, part of the surrounding stem, and the poly(A) tail in the BaMV 3′ NTR. Alternatively, the presentation of the hexanucleotide sequence in a loop in the 3′ NTR may allow for interaction with complementary sequences upstream of the sg RNAs. A sequence complementary to the hexanucleotide element, located near the 5′ terminus of PVX plus-strand RNA (or 3′ terminus of minus-strand RNA), was important for a long-distance interaction required for plus-strand RNA accumulation in protoplasts.50 Such a long-distance interaction may be required for PVX minus-strand RNA synthesis in vivo, similar to that required for PVX plus-strand RNA synthesis.50
Our data also showed that deletion of sequence between the PVX hexanucleotide and U-rich elements (mutant Δint) abolished minus and plus-strand RNA accumulation in protoplasts. This may be because this deletion is predicted to disrupt SL3 and SL2, resulting in a combined SL structure with the two elements adjacent to each other in a large loop (data not shown). Thus, the predicted disruption of SL3 could cause the reduction in RNA accumulation similar to what we have observed for the SL3 mutants. Alternatively, the spacing or the sequence between these elements may be important for replication.
3.3. The CU nucleotides at the 3′ end of the U-rich element contribute to minus-strand RNA accumulation
Sriskanda et al. 37 showed that a 38 nucleotide region in the PVX 3′ NTR containing the U-rich sequence (UAUUUUCU) was important for host protein binding, and deletion of this region reduced minus-strand RNA accumulation in protoplasts. Substitutions for the four consecutive U residues affected both host protein binding in vitro and replication in protoplasts. Our data indicate that deletions of the U-rich sequence (Δ60 and Δ21) from the PVX 3′ NTR specifically affected minus-strand RNA accumulation. Moreover, in this study we have further defined this region by showing that mutation of the CU residues at the 3′ end of the U-rich sequence (mutants SL2-B and SL2-D) is sufficient to abolish minus-strand RNA accumulation.
Similar U-rich elements can be found in the 3′ NTR of other potexviruses. As shown in the alignment in Figure 10, UC-rich regions (blue text) are found downstream of the hexanucleotide motif in nine out of 11 potexviruses and all of these have 3′ CU residues. Additional experiments will be required to determine if the UC-rich regions in other potexviruses are important for minus-strand RNA accumulation and/or host protein binding. Furthermore, it will be important to determine if the CU residues of the U-rich element in the PVX 3′ NTR are important for host protein binding. Sriskanda et al.37 postulated that host protein binding to the PVX U-rich element may be important for viral RNA synthesis, RNA stability, and/or translation. Similar results have been shown for the coronavirus, MHV, which has an 11 nucleotide sequence (UGAAUGAAGUU) at the 3′ end of the genome that is conserved among coronaviruses. This sequence was required for host protein binding and MHV viral RNA synthesis, with both minus and plus-strand replication being affected by mutations in this region.54
3.4. Sequence between the U-rich element and potential NUEs is dispensable for minus-strand RNA accumulation
Surprisingly, the sequence immediately downstream of the U-rich sequence in the PVX 3′ NTR (nucleotides 6412–6424) could be deleted (p72Δ13) without affecting minus-strand RNA accumulation. Because the p72Δ13 mutant is predicted to have wild-type SL3 and SL2, but no structure downstream of SL2, these data confirm that the SL1 structure is not required for minus-strand RNA accumulation. However, further deletion of three nucleotides downstream in mutant p72Δ16 was partially detrimental to minus-strand RNA levels. Although p72Δ13 and p72Δ16 have identical structures, they differ in only three nucleotides and this sequence difference reduces the minus-strand RNA accumulation from wild-type levels in p72Δ13 to 70% of wild-type in p72Δ16. When the deletion was extended two nucleotides in the upstream direction and five nucleotides in the downstream direction (mutant p72Δ23), minus-strand RNA accumulation in protoplasts dropped to 5% of wild-type. Thus, it was not surprising that deletion of the 3′- terminal 15 nt plus the poly(A) tail in the mutant Δ15 resulted in no minus-strand RNA accumulation. It is unlikely that the deletion of GG at the upstream end of the deletion in p72Δ23 was solely responsible for the reduction in minus-strand RNA accumulation because these two nucleotides were mutated in SL2-2A, which exhibited minus-strand RNA accumulation similar to wild-type levels. Thus, nucleotides from A6425–A6432 are required for minus-strand RNA accumulation, whereas at least U6412–A6424 and potentially G6410–A6424 are not necessary. Similarly, other groups have identified dispensable sequences in the 3′ NTRs of several plant33., 55., 56., 57. and animal58., 59. viral RNAs.
3.5. The PVX 3′ NTR contains sequences that affect both plus-strand and minus-strand RNA accumulation
Two overlapping AU-rich sequences, UAUAAA and AAUAAA, at the PVX 3′ terminus were chosen for mutagenesis because of their similarity to near upstream elements (NUEs) used as polyadenylation signals for plant mRNAs,60., 61. plant pararetroviruses62., 63. and cytoplasmic polyadenylation in maturing Xenopus and mouse oocytes.64 Nuclear polyadenylation can also be affected by secondary and/or tertiary structures of the viral RNA. For example, part of the AAUAAA sequence in the retrovirus, HIV-1, is present in a stem-loop structure; the stability of this structure regulates the use of the AAUAAA sequence with preference given to less stable structure.65
Our data show that when either of the potential NUEs in the PVX 3′ NTR are modified (mutants p73, ΔAAU, ΔUAUA and ΔUAAUAUA), minus-strand RNA accumulation is somewhat reduced compared to wild-type, but plus-strand RNA is reduced to a greater extent. Comparison of the 3′ NTR sequences of other potexviruses in Figure 10 indicates that sequences for eight out of 11 viruses contain one or more potential NUE (yellow sequences). Other sequences similar to the canonical AAUAAA were colored because several other sequences have been found to function as polyadenylation signals for plant mRNAs (i.e. UAUAAA, AAUAAG).60 When the AAUAAA signal in the 3′ NTR of the potexvirus, white clover mosaic virus (WClMV), was mutated by one nucleotide, no replication was observed.36 Additional experiments will be required to determine if these potential NUEs regulate PVX and other potexvirus plus-strand RNA accumulation by affecting polyadenylation efficiency and subsequent plus-strand RNA stability.
Although changes in the potential NUEs in the PVX 3′ NTR (mutants p73, ΔAAU, ΔUAUA, and Δ UAAUAUA), were more detrimental to plus-strand RNA accumulation, minus-strand RNA accumulation was also somewhat reduced. Thus, this region of the PVX 3′ NTR appears to have a dual role in minus-strand RNA synthesis and plus-strand RNA stability and/or polyadenylation. It is possible that correctly polyadenylated templates are important for PVX minus-strand RNA synthesis. When WClMV transcripts containing no poly(A) tail were inoculated into pea protoplasts, replication proceeded very slowly.36 The importance of poly(A) length was observed also for BaMV, where no or short poly(A) tails did not support genomic and subgenomic plus-strand RNA accumulation.34 In addition, it appears that BaMV minus-strand RNA initiates at multiple sites within the 5′ most 15 A residues of the poly(A) tail.66 The importance of a poly(A) tail in virus RNA accumulation was observed for coronaviruses. The 3′ NTR and the poly(A) tail of MHV RNA are required for minus-strand synthesis,59 and a poly(A) tail of at least five A residues was required for efficient replication of bovine coronavirus (BCV) RNA.67 Our data show that deletions in the regions of the PVX NUEs (mutants p72Δ23 and Δ15) resulted in negligible levels of minus-strand RNA, suggesting that elements necessary for minus-strand RNA accumulation and those needed for efficient polyadenylation and/or stability overlap near the 3′ end of the PVX 3′ NTR.
3.6. Relative roles of the 3′ NTR elements during PVX replication
Mutations to the multiple regulatory elements localized throughout the PVX 3′ NTR exhibited four phenotypes: (I) those resulting in negligible levels of minus-strand and plus-strand RNAs (mutants p71Δ10, p71Δ60, Δint, SL3-B, SL2-B, SL2-D, p72Δ23, p72Δ21, and Δ15); (II) those with detectable, but significantly reduced minus and plus-strand RNA levels (SL3-C, and p72Δ16); (III) those with greater reduction of plus than minus-strand RNA (mutants SL2-A, p73, ΔAAU, ΔUAUA, and ΔUAAUAU); and (IV) one (mutant SL2-A) that showed reduction in plus-strand with no reduction in minus-strand RNA.
Many of the elements in t he first two categories, which include the U-rich element and others upstream such as the hexanucleotide motif and SL3, are likely to regulate minus-strand RNA levels at the level of RNA synthesis. Alternatively, some of these elements may affect minus or plus-strand RNA stability, leading to reductions in minus-strand RNA levels. However, modeling of the minus-strand RNA 5′ terminus of these mutants using Mfold did not indicate any significant disruptions to predicted structures and/or stabilities (data not shown). In addition, we previously found that mutations to a stem-loop element in the 5′ NTR disrupted accumulation of plus-strand RNA, but did not affect minus-strand RNA levels.48., 49. It is possible that the reduced minus-strand RNA levels reflect decreased replicase translation, given that 3′-terminal elements have been found to be important for translation in several other systems.11., 12., 68., 69., 70., 71., 72., 73., 74., 75., 76.
The role(s) of the elements in the third and fourth categories, which are near the 3′ end of PVX RNA, may involve stabilization of the plus-strand RNA, either directly or as a result of disruption of efficient polyadenylation. Because some of the sequences in the third category significantly affect minus-strand RNA levels, they may function during minus-strand RNA synthesis or be important for optimal translation of the PVX replicase. Studies are underway to distinguish among these possibilities.
4. Materials and Methods
RNase T1, N-cyclohexyl-N′-[2-(N-methyl-4-morpholinio)ethyl]carboiimide p-toluenesulfonate (CMCT) and RNase A were purchased from Sigma. RNase V1 and dimethyl sulfate (DMS) was obtained from Pierce and Fluka, respectively. Trizol reagent was purchased from Life Technologies. Nuclease S1 was purchased from Roche Molecular Biochemicals. Cellulase “Onozuka” R-10 was purchased from Yakult Honsha Co. Ltd. and pectolyase Y23 is a product of Karlan Research Products. RNasin and RQ1 DNase were obtained from Promega, and all other enzymes and cap analogue were purchased from New England BioLabs.
4.1. Construction of site-directed and deletion mutants
Site-directed mutations were introduced into the wild-type (w.t.) PVX cDNA construct, pMON845377 according to method of Kunkel et al.78 to make the following constructs: SL3-A, SL3-B, SL3-C, SL2-A, SL2-B, SL2-C, SL2-D, ΔAAU, ΔUAUA, ΔUAAUAUA, Δint, p73, p74, p71 with an SnaBI site, and p72 with a BstBI site. Regions containing mutations were sequenced and resected back into the wild-type pMON8453 construct. For larger nested deletions, p71 and p72 were digested with SnaBI and BstBI, respectively, and then digested with exonuclease III. The resulting deletion mutants p71Δ10, p71Δ60, p72Δ13, p72Δ16 and p72Δ21 were digested with BsaAI and resected into p71 digested with BsaAI and SnaBI or p72 digested with BsaAI and BstBI (followed by mung bean nuclease digestion to remove 3′ overhangs). All resected clones were sequenced to confirm the extent of deletions. Mutant p72Δ23 was made by site-directed mutagenesis of the mutant p72Δ16 and then resected back into the wild-type pMON8453 construct.
4.2. Preparation of transcripts
The wild-type PVX cDNA construct, pMON8453, all mutant plasmids and p10 construct containing PVX 3′ 185 nucleotide were linearized with SpeI and transcribed in vitro, as described previously.48 The resulting transcript has an authentic 3′ end, 4 nt from the SpeI site, and 17 A residues at the 3′ end. The quality and concentration of the transcripts were verified on 1% agarose gels and visualized by staining with ethidium bromide.
4.3. Preparation and inoculation of protoplasts
Nicotiana tabaccum (NT-1) cells were maintained in a rapidly growing suspension culture with constant shaking at 25 °C in a 14 hours light/ten hour dark cycle. Protoplasts were prepared from this suspension as described by Miller et al.49 For protoplast inoculation, 10 μg of capped transcripts was diluted to 800 μl in electroporation buffer (0.8% (w/v) NaCl, 0.02% (w/v) KCl, 0.02% (w/v) KH2PO4, 0.11% (w/v) Na2HPO4, 0.4 M mannitol, pH 6.5), and added to 800 μl of protoplasts (4×106 cells) by electroporation. After 15 minutes incubation on ice, inoculated protoplasts were plated in 15 ml of NT-1 protoplast culture medium (0.43% (w/v) MS salts, 88 mM sucrose, 55 mM myo-inositol, 0.3 mM thiamine, 440 mM KH2PO4, 0.4 mg/ml of 2,4-dichlorophenoxyacetic acid, 0.4 M mannitol) containing 20 μg/ml of RNase A and incubated for 40 hours at 25 °C with a 14 hours light/ten hours dark cycle.
4.4. Analysis of RNA accumulation in protoplasts
At 40 hours post inoculation (hpi), protoplasts were washed and re-incubated for one hour in 1 ml of NT-1 protoplast culture medium supplemented with 20 μg/ml of RNase A to remove the remaining input transcripts. Total RNA was extracted using Trizol reagent (Life Technologies) and the final RNA pellets were resuspended in 100 μl of diethyl pyrocarbonate (DEPC)-treated water and stored at −80 °C. Both probes used for S1 nuclease protection assays were labeled with 32P using kinase as described.48
For plus-strand RNA detection, 10 μl of the total RNA isolated from protoplasts was hybridized with >10,000 cpm of 32P-labeled P1 probe (Figure 1(A)) and digested with S1 nuclease as described by Kim & Hemenway.48 For analysis of minus-strand RNA accumulation, 40 μl of total RNA was pre-annealed and treated with 0.1 μg of RNase A to remove cellular RNAs and excess plus-strand RNA, followed by addition of proteinase K and SDS to final concentrations of 30 μg/μl and 0.4 %, respectively.48., 79. This treated RNA was hybridized to >10,000 cpm of 32P-labeled P3 probe (Figure 1(A)) for S1 nuclease analysis.
Protected fragments resulting from S1 nuclease digestion were separated on 8% (w/v) polyacrylamide sequencing gels, visualized by using a Molecular Dynamics phosphorimager, and quantified with the ImageQuant 1.0 program. At least five individual experiments were conducted for each mutant and wild-type transcript. The RNA levels in protoplasts inoculated with wild-type pMON8453 transcripts or replication-defective p32 transcripts were set at 100% and 0%, respectively. Consequently, values for RNA levels in protoplasts inoculated with mutants represent relative values plus or minus the standard error.
4.5. Secondary structure prediction of the 3′ 250 nucleotides
The secondary structure predicted for the PVX 3′ 250 nucleotides was obtained using the Mfold program, version 2.3 on the Zuker website† . The STAR program,80., 81., 82. obtained from C. W. A. Pleij (Leiden University, Netherlands), was also used to predict the secondary structure for the 250 nucleotides.
4.6. Chemical and enzymatic analyses of the 3′ NTR structure
The transcripts used for the solution structure analyses were derived from a wild-type PVX cDNA construct, pMON8453, and p10, a construct that contains only 188 nt of the 3′ region of PVX RNA. For modification experiments, 5 μg of transcripts was incubated in a final volume of 50 μl containing either CMK buffer (80 mM potassium cacodylate (pH 7.2), 300 mM KCl, 20 mM magnesium acetate) for DMS reactions, or BMK buffer (50 mM potassium borate (pH 8.1), 50 mM KCl, 20 mM magnesium acetate, 0.5 mM DTT) for CMCT reactions, respectively.83 Either 0.3 M DMS was added to the reaction for incubation at 37 °C for 20 minutes or 12.5 μl of CMCT (42 mg/ml in BMK buffer) was added to the reaction for incubation at 25 °C for 30 minutes. Unmodified control reactions were treated similarly. DMS reactions were stopped by addition of 12.5 μl of stop solution (1 M Tris-acetate (pH 7.5), 1.5 M sodium acetate, 0.1 mM EDTA, 1.0 M 2-mercaptoethanol), incubated on ice for ten minutes, and precipitated in ethanol. CMCT reactions were precipitated directly with ethanol. Pellets from both the reactions were vacuum-dried, resuspended in water, and subjected to extraction withphenol/chloroform, followed by another extraction with chloroform. After extraction, samples were precipitated with ethanol, washed with cold 80% (v/v) ethanol, air-dried and resuspended in 12 μl of water. Enzymatic cleavage experiments were carried out as described,49 except that the final concentrations of RNase T1 and RNase V1 were 0.03 unit/μl and 5×10−4 units/μl, respectively.
4.7. Primer extension analyses of modified RNA
The modified RNA samples were analyzed by primer extension as described.49., 83. Two primers, P4 (TTTTTTTTTTTTTTTTTATTTATATTATTCATAC complementary to G6419–A6452) and P5 (CTAGTTTTTTTTTTTTTTTTT complementary to the 17 A residues of the poly(A) tail and the four non-viral nucleotides) were utilized to evaluate the extension products. The positions of these primers are indicated below the genome in Figure 1(A). The primers were first end-labeled49 and then combined with 5 μl of modified RNAs. Samples were dried and resuspended in 1 μl of R-loop buffer (80% (v/v) formamide, 400 mM NaCl, 40 mM Pipes (pH 6.8), 1 mM EDTA), and incubated for 15 minutes at 45 °C. After hybridization, 20 μl of reverse transcription mix (25 mM Tris–HCl (pH 8.3), 50 mM KCl, 2.0 mM DTT, 5 mM MgCl2, 1.0 mM each dNTP, 1.0 unit/μl of RNasin, 0.4 unit/μl of AMV reverse transcriptase) was added to the samples and incubated at 45 °C for another ten minutes. This extension reaction was stopped by addition of EDTA and SDS to final concentrations of 30 mM and 0.1% (w/v), respectively. This was followed by one extraction with phenol/chloroform, a second extraction with chloroform alone, and then precipitation in ethanol. The pellets were washed with 80% ethanol, air-dried and resuspended in 3 μl of water and 9 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% (w/v) xylene cyanol FF, 0.05% (w/v) bromophenol blue). After boiling samples at 90 °C for five minutes, followed by cooling on ice for a minute, samples (3 μl) were loaded onto an sequencing 8% polyacrylamide gel.
Acknowledgements
We are grateful to Dr Shigeo Yoshinari, D. Jeff Batten and Bin Hu for critical reading of the manuscript. This research was supported by Public Health Service grant GM4984 to C. L. Hemenway.
Edited by D. E. Draper
Footnotes
References
- 1.Buck K.W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Advan. Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreher T.W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- 3.Hemenway C.L., Lommel S.A. Manipulating plant viral RNA transcription signals. Genet. Eng. (NY) 2000;22:171–195. doi: 10.1007/978-1-4615-4199-8_10. [DOI] [PubMed] [Google Scholar]
- 4.Barton D.J., O'Donnell B.J., Flanegan J.B. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klovins J., van Duin J. A long-range pseudoknot in Qβ RNA is essential for replication. J. Mol. Biol. 1999;294:875–884. doi: 10.1006/jmbi.1999.3274. [DOI] [PubMed] [Google Scholar]
- 6.Danthinne X., Seurinck J., Meulewaeter F., Van Montagu M., Cornelissen M. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol. Cell. Biol. 1993;13:3340–3349. doi: 10.1128/mcb.13.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timmer R.T., Benkowski L.A., Schodin D., Lax S.R., Metz A.M., Ravel J.M., Browning K.S. The 5′ and 3′ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′ cap structure. J. Biol. Chem. 1993;268:9504–9510. [PubMed] [Google Scholar]
- 8.Reusken C.B., Neeleman L., Bol J.F. The 3′-untranslated region of alfalfa mosaic virus RNA 3 contains at least two independent binding sites for viral coat protein. Nucl. Acids Res. 1994;22:1346–1353. doi: 10.1093/nar/22.8.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S., Miller W.A. A sequence located 4.5–5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J. Biol. Chem. 1995;270:13446–13452. doi: 10.1074/jbc.270.22.13446. [DOI] [PubMed] [Google Scholar]
- 10.Gallie D.R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 11.Wu B., White K.A. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol. 1999;73:8982–8988. doi: 10.1128/jvi.73.11.8982-8988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergamini G., Preiss T., Hentze M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalton K., Casais R., Shaw K., Stirrups K., Evans S., Britton P. Cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J. Virol. 2001;75:125–133. doi: 10.1128/JVI.75.1.125-133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller W.A., Bujarski J.J., Dreher T.W., Hall T.C. Minus-strand initiation by brome mosaic virus replicase within the 3′ t-RNA-like structure of native and modified RNA templates. J. Mol. Biol. 1986;187:537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- 15.Dreher T.W., Hall T.C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J. Mol. Biol. 1988;201:31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- 16.Chapman M.R., Kao C.C. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 17.Sivakumaran K., Bao Y., Roossinck M.J., Kao C.C. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of Brome mosaic virus and Cucumber mosaic virus. J. Virol. 2000;74:10323–10331. doi: 10.1128/jvi.74.22.10323-10331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buck K.W. Replication of tobacco mosaic virus RNA. Philos. Trans. R. Soc. ser. B. 1999;354:613–627. doi: 10.1098/rstb.1999.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skuzeski J.M., Bozarth C.S., Dreher T.W. The turnip yellow mosaic virus tRNA-like structure cannot be replaced by generic tRNA-like elements or by heterologous 3′ untranslated regions known to enhance mRNA expression and stability. J. Virol. 1996;70:2107–2115. doi: 10.1128/jvi.70.4.2107-2115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshinari S., Nagy P.D., Simon A.E., Dreher T.W. CCA initiation boxes without unique promoter elements support in vitro transcription by three viral RNA-dependent RNA polymerases. RNA. 2000;6:698–707. doi: 10.1017/s1355838200992410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Kuyl A.C., Neeleman L., Bol J.F. Deletion analysis of cis and trans-acting elements involved in replication of alfalfa mosaic virus RNA 3 in vivo. Virology. 1991;183:687–694. doi: 10.1016/0042-6822(91)90997-p. [DOI] [PubMed] [Google Scholar]
- 22.van Rossum C.M., Reusken C.B., Brederode F.T., Bol J.F. The 3′ untranslated region of alfalfa mosaic virus RNA3 contains a core promoter for minus-strand RNA synthesis and an enhancer element. J. Genet. Virol. 1997;78:3045–3049. doi: 10.1099/0022-1317-78-11-3045. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H., Jackson A.O. Analysis of cis-acting elements required for replication of barley stripe mosaic virus RNAs. Virology. 1996;219:150–160. doi: 10.1006/viro.1996.0232. [DOI] [PubMed] [Google Scholar]
- 24.Havelda Z., Burgyan J. 3′ terminal putative stem-loop structure required for the accumulation of cymbidium ringspot viral RNA. Virology. 1995;214:269–272. doi: 10.1006/viro.1995.9929. [DOI] [PubMed] [Google Scholar]
- 25.Turner R.L., Buck K.W. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of red clover necrotic mosaic virus. Virology. 1999;253:115–124. doi: 10.1006/viro.1998.9495. [DOI] [PubMed] [Google Scholar]
- 26.Song C., Simon A.E. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 27.Carpenter C.D., Simon A.E. Analysis of sequences and predicted structures required for viral satellite RNA accumulation by in vivo genetic seletion. Nucl. Acids Res. 1998;26:2426–2432. doi: 10.1093/nar/26.10.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jupin I., Bouzoubaa S., Richards K., Jonard G., Guilley H. Multiplication of beet necrotic yellow vein virus RNA 3 lacking 3′ poly(A) tail is accompanied by reappearance of the poly(A) tail and a novel short U-rich tract preceding it. Virology. 1990;178:281–284. doi: 10.1016/0042-6822(90)90404-f. [DOI] [PubMed] [Google Scholar]
- 29.Lauber E., Guilley H., Richards K., Jonard G., Gilmer D. Conformation of the 3′-end of beet necrotic yellow vein benevirus RNA 3 analysed by chemical and enzymatic probing and mutagenesis. Nucl. Acids Res. 1997;25:4723–4729. doi: 10.1093/nar/25.23.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tacahashi Y., Uyeda I. Restoration of the 3′ end of potyvirus RNA derived from Poly(A)-deficient infectious cDNA clones. Virology. 1999;265:147–152. doi: 10.1006/viro.1999.0027. [DOI] [PubMed] [Google Scholar]
- 31.Eggen R., Verver J., Wellink J., Pleij K., Van Kammen A., Goldbach R. Analysis of sequences involved in cowpea mosaic virus RNA replication using site-specific mutants. Virology. 1989;173:456–464. doi: 10.1016/0042-6822(89)90558-8. [DOI] [PubMed] [Google Scholar]
- 32.Chiu W.W., Hsu Y.H., Tsai C.H. Specificity analysis of the conserved hexanucleotides for the replication of bamboo mosaic potexvirus RNA. Virus Res. 2002;83:159–167. doi: 10.1016/s0168-1702(01)00433-6. [DOI] [PubMed] [Google Scholar]
- 33.Cheng C.-P., Tsai C.-H. Structural and functional analysis of the 3′ untranslated region of bamboo mosaic potexvirus genomic RNA. J. Mol. Biol. 1999;288:555–565. doi: 10.1006/jmbi.1999.2716. [DOI] [PubMed] [Google Scholar]
- 34.Tsai C.H., Cheng C.P., Peng C.W., Lin B.Y., Lin N.S., Hsu Y.H. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. J. Virol. 1999;73:2703–2709. doi: 10.1128/jvi.73.4.2703-2709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White K.A., Bancroft J.B., Mackie G.A. Mutagenesis of a hexanucleotide sequence conserved in potexvirus RNAs. Virology. 1992;189:817–820. doi: 10.1016/0042-6822(92)90614-u. [DOI] [PubMed] [Google Scholar]
- 36.Guilford P.J., Beck D.L., Forster R.L. Influence of the poly(A) tail and putative polyadenylation signal on the infectivity of while clover mosaic potexvirus. Virology. 1991;182:61–67. doi: 10.1016/0042-6822(91)90648-u. [DOI] [PubMed] [Google Scholar]
- 37.Sriskanda V.S., Pruss G., Ge X., Vance V.B. An eight-nucleotide sequence in the potato virus X 3′ untranslated region is required for both host protein binding and viral multiplication. J. Virol. 1996;70:5266–5271. doi: 10.1128/jvi.70.8.5266-5271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batten J.S., Yoshinari S., Hemenway C. Potato virus X: a model system for virus replication, movement and gene expression. Mini-review Mol. Plant Pathology. 2003;4:2–8. doi: 10.1046/j.1364-3703.2003.00156.x. [DOI] [PubMed] [Google Scholar]
- 39.Milne R.G. The Plant Viruses, Vol. 4, Flimanetous Plant Viruses. Plenum Press; New York: 1985. [Google Scholar]
- 40.Brunt A.A., Foster G.D., Morozov S.Y., Zavriev S.K. Genus potexvirus. In: van Regenmortel M.H.V., editor. Seventh Report of International Committee on Taxonomy of Viruses. Academic Press; San Diego: 2000. pp. 975–981. [Google Scholar]
- 41.Huisman M.J., Linthorst H.J.M., Bol J.F., Cornelissen B.J.C. The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus-stranded RNA viruses. J. Genet. Virol. 1988;69:1789–1798. doi: 10.1099/0022-1317-69-8-1789. [DOI] [PubMed] [Google Scholar]
- 42.Skryabin K.G., Kraev A.S., Morozov S.Y., Rozanov M.N., Chernov B.K., Lukasheva L.I., Atabekov J.G. The nucleotide sequence of potato virus X RNA. Nucl. Acids Res. 1988;16:10929–10930. doi: 10.1093/nar/16.22.10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angell S.M., Davies C., Baulcombe D.C. Cell-to-cell movement of potato virus X is associated with a change in the size-exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii. Virology. 1996;216:197–201. doi: 10.1006/viro.1996.0046. [DOI] [PubMed] [Google Scholar]
- 44.Tamai A., Meshi T. Cell-to-cell movement of Potato virus X: the role of p12 and p8 encoded by the second and third open reading frames of the triple gene block. Mol. Plant Microbe Interact. 2001;14:1158–1167. doi: 10.1094/MPMI.2001.14.10.1158. [DOI] [PubMed] [Google Scholar]
- 45.Cruz S.S., Roberts A.G., Prior D.A., Chapman S., Oparka K.J. Cell-to-cell and phloem-mediated transport of potato virus X. The role of virions. Plant Cell. 1998;10:495–510. doi: 10.1105/tpc.10.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cruz S.S., Chapman S., Roberts A.G., Roberts I.M., Prior D.A., Oparka K.J. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl Acad. Sci. USA. 1996;93:6286–6290. doi: 10.1073/pnas.93.13.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baulcombe D.C., Chapman S., Santa Cruz S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- 48.Kim K.-H., Hemenway C. The 5′ nontranslated region of potato virus X RNA affects both genomic and subgenomic RNA synthesis. J. Virol. 1996;70:5533–5540. doi: 10.1128/jvi.70.8.5533-5540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller E.D., Plante C.A., Kim K.-H., Hemenway C. Stem-loop structure in the 5′ region of potato virus X genome required for plus-strand RNA accumulation. J. Mol. Biol. 1998;284:591–608. doi: 10.1006/jmbi.1998.2174. [DOI] [PubMed] [Google Scholar]
- 50.Kim K.H., Hemenway C.L. Long-distance RNA–RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA. 1999;5:636–645. doi: 10.1017/s1355838299982006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pillai-Nair, N. (2002). Cis-acting elements important for PVX replication. Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, pp. 38–78.
- 52.Bancroft J.B., Rouleau M., Johnston R., Prins L., Mackie G.A. The entire nucleotide sequence of foxtail mosaic virus RNA. J. Genet. Virol. 1991;72:2173–2181. doi: 10.1099/0022-1317-72-9-2173. [DOI] [PubMed] [Google Scholar]
- 53.Huang C.Y., Huang Y.L., Meng M., Hsu Y.H., Tsai C.H. Sequences at the 3′ untranslated region of bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. J. Virol. 2001;75:2818–2824. doi: 10.1128/JVI.75.6.2818-2824.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu W., Leibowitz J.L. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology. 1995;214:128–138. doi: 10.1006/viro.1995.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandrika R., Rabindran S., Lewandowski D.J., Manjunath K.L., Dawson W.O. Full-length tobacco mosaic virus RNAs and defective RNAs have different 3′ replication signals. Virology. 2000;273:198–209. doi: 10.1006/viro.2000.0414. [DOI] [PubMed] [Google Scholar]
- 56.Deiman B.A., Kortlever R.M., Pleij C.W. The role of the pseudoknot at the 3′ end of turnip yellow mosaic virus RNA in minus-strand synthesis by the viral RNA-dependent RNA polymerase. J. Virol. 1997;71:5990–5996. doi: 10.1128/jvi.71.8.5990-5996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh R.N., Dreher T.W. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology. 1997;233:430–439. doi: 10.1006/viro.1997.8621. [DOI] [PubMed] [Google Scholar]
- 58.Kuhn R.J., Hong Z., Strauss J.H. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J. Virol. 1990;64:1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin Y.J., Liao C.L., Lai M.M. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rothnie H.M., Reid J., Hohn T. The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′-end formation in plants. EMBO J. 1994;13:2200–2210. doi: 10.1002/j.1460-2075.1994.tb06497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rothnie H.M. Plant mRNA 3′ end formation. Plant Mol. Biol. 1996;32:43–61. doi: 10.1007/BF00039376. [DOI] [PubMed] [Google Scholar]
- 62.Sanfacon H. Analysis of figwort mosaic virus (plant pararetrovirus) polyadenylation signal. Virology. 1994;198:39–49. doi: 10.1006/viro.1994.1006. [DOI] [PubMed] [Google Scholar]
- 63.Rothnie H.M., Reid J., Hohn T. The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′-end formation in plants. EMBO J. 1994;13:2200–221065. doi: 10.1002/j.1460-2075.1994.tb06497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fox C.A., Sheets M.D., Wickens M.P. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- 65.Klasens B.I., Thiesen M., Virtanen A., Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucl. Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng J.H., Peng C.W., Hsu Y.H., Tsai C.H. The synthesis of minus-strand RNA of bamboo mosaic potexvirus initiates from multiple sites within the poly(A) tail. J. Virol. 2002;76:6114–6120. doi: 10.1128/JVI.76.12.6114-6120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spagnolo J.F., Hogue B.G. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gallie D.R., Walbot V. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev. 1990;4:1149–1157. doi: 10.1101/gad.4.7.1149. [DOI] [PubMed] [Google Scholar]
- 69.Leathers V., Tanguay R., Kobayashi M., Gallie D.R. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol. Cell. Biol. 1993;13:5331–5347. doi: 10.1128/mcb.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gallie D.R., Kobayashi M. The role of the 3′ untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene. 1994;142:159–165. doi: 10.1016/0378-1119(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 71.Gallie D.R., Tanguay R.L., Leathers V. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene. 1995;165:233–238. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- 72.Hann L.E., Webb A.C., Cai J.M., Gehrke L. Identification of a competitive translation determinant in the 3′ untranslated region of alfalfa mosaic virus coat protein mRNA. Mol. Cell. Biol. 1997;17:2005–2013. doi: 10.1128/mcb.17.4.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang S., Guo L., Allen E., Miller W.A. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA. 1999;5:728–738. doi: 10.1017/s1355838299981979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo L., Allen E., Miller W.A. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 2000;6:1808–1820. doi: 10.1017/s1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo L., Allen E.M., Miller W.A. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- 76.Neeleman L., Olsthoorn R.C., Linthorst H.J., Bol J.F. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc. Natl Acad. Sci. USA. 2001;98:14286–14291. doi: 10.1073/pnas.251542798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hemenway C., Weiss J., O'Connel K., Tumer N.E. Characterization of infectious transcripts from a potato virus X cDNA clone. Virology. 1990;175:365–371. doi: 10.1016/0042-6822(90)90421-m. [DOI] [PubMed] [Google Scholar]
- 78.Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ishikawa M., Meshi T., Ohno T., Okada Y. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J. Virol. 1991;65:861–868. doi: 10.1128/jvi.65.2.861-868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abrahams J.P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucl. Acids Res. 1990;18:3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gultyaev A.P. The computer simulation of RNA folding involving pseudoknot formation. Nucl. Acids Res. 1991;19:2489–2494. doi: 10.1093/nar/19.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Batenburg F.H., Gultyaev A.P., Pleij C.W. An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J. Theor. Biol. 1995;174:269–280. doi: 10.1006/jtbi.1995.0098. [DOI] [PubMed] [Google Scholar]
- 83.Moazed D., Stern S., Noller H.F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 84.Lin N.S., Lin B.Y., Lo N.W., Hu C.C., Chow T.Y., Hsu Y.H. Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Genet. Virol. 1994;75:2513–2518. doi: 10.1099/0022-1317-75-9-2513. [DOI] [PubMed] [Google Scholar]
- 85.Chia T.F., Chan Y.S., Chua N.H. Characterization of cymbidium mosaic virus coat protein gene and its expression in transgenic tobacco plants. Plant Mol. Biol. 1992;18:1091–1099. doi: 10.1007/BF00047712. [DOI] [PubMed] [Google Scholar]
- 86.Sit T.L., White K.A., Holy S., Padmanabhan U., Eweida M., Hiebert M., Mackie G.A., AbouHaidar M.G. Complete nucleotide sequence of clover yellow mosaic virus RNA. J. Genet. Virol. 1990;71:1913–1920. doi: 10.1099/0022-1317-71-9-1913. [DOI] [PubMed] [Google Scholar]
- 87.Zuidema D., Linthorst H.J., Huisman M.J., Asjes C.J., Bol J.F. Nucleotide sequence of narcissus mosaic virus RNA. J. Genet. Virol. 1989;70:267–276. doi: 10.1099/0022-1317-70-2-267. [DOI] [PubMed] [Google Scholar]
- 88.Xu H., Leclerc D., Leung B., AbouHaidar M.G. The entire nucleotide sequence and genomic organization of potato aucuba mosaic potexvirus. Arch. Virol. 1994;135:461–469. doi: 10.1007/BF01310031. [DOI] [PubMed] [Google Scholar]
- 89.Solovyev A.G., Novikov V.K., Merits A., Savenkov E.I., Zelenina D.A., Tyulkina L.G., Morozov S. Genome characterization and taxonomy of Plantago asiatica mosaic potexvirus. J. Genet. Virol. 1994:259–267. doi: 10.1099/0022-1317-75-2-259. [DOI] [PubMed] [Google Scholar]
- 90.Sit T.L., Abouhaidar M.G., Holy S. Nucleotide sequence of papaya mosaic virus RNA. J. Genet. Virol. 1989;70:2325–2331. doi: 10.1099/0022-1317-70-9-2325. [DOI] [PubMed] [Google Scholar]
- 91.Jelkmann W., Maiss E., Martin R.R. The nucleotide sequence and genome organization of strawberry mild yellow edge-associated potexvirus. J. Genet. Virol. 1992;73:475–479. doi: 10.1099/0022-1317-73-2-475. [DOI] [PubMed] [Google Scholar]
- 92.Forster R.L., Bevan M.W., Harbison S.A., Gardner R.C. The complete nucleotide sequence of the potexvirus white clover mosaic virus. Nucl. Acids Res. 1988;16:291–303. doi: 10.1093/nar/16.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]