Abstract
The global risk of viral disease outbreaks emphasizes the need for rapid, accurate, and sensitive detection techniques to speed up diagnostics allowing early intervention. An emerging field of microfluidics also known as the lab-on-a-chip (LOC) or micro total analysis system includes a wide range of diagnostic devices. This review briefly covers both conventional and microfluidics-based techniques for rapid viral detection. We first describe conventional detection methods such as cell culturing, immunofluorescence or enzyme-linked immunosorbent assay (ELISA), or reverse transcription polymerase chain reaction (RT-PCR). These methods often have limited speed, sensitivity, or specificity and are performed with typically bulky equipment. Here, we discuss some of the LOC technologies that can overcome these demerits, highlighting the latest advances in LOC devices for viral disease diagnosis. We also discuss the fabrication of LOC systems to produce devices for performing either individual steps or virus detection in samples with the sample to answer method. The complete system consists of sample preparation, and ELISA and RT-PCR for viral-antibody and nucleic acid detection, respectively. Finally, we formulate our opinions on these areas for the future development of LOC systems for viral diagnostics.
Keywords: LOC, Microfluidic, Viral detection, Immunoassays, Nucleic acid amplification, Commercialization
Graphical abstract
Highlights
-
•
Conventional methods for viral detection are time consuming and labor intensive.
-
•
Newly emerged microfluidic devices can perform practically any conventional laboratory technique.
-
•
Here we concentrated on microfluidics-based viral detection by immunoassays as well as by nucleic acid amplification.
-
•
We used a few examples of successfully commercialized rapid devices.
-
•
We discuss perspectives for technology of viral detection in near future.
1. Introduction
Reducing infectious disease mortality is a challenging and critical task despite the tremendous efforts and recent significant advances in public healthcare. Several viruses such as Lassa mammarenavirus, the dengue fever virus (DENV), the Ebola virus, human immunodeficiency virus (HIV) causing acquired immunodeficiency syndrome (AIDS), the Zika virus (ZIKV), West Nile virus, measles morbillivirus, severe acute respiratory syndrome coronavirus (SARS-CoV), avian and other influenza viruses posed the risk of worldwide outbreaks, potentially resulting in enormous economic and social burdens on affected countries. We chose them as typical representatives of viral diseases (summarized in Table 1 ) having a potential to cause worldwide problem including pandemic shown by spreading of HIV/AIDS as well as by SARS and Ebola outbreak.
Table 1.
Summary of emerging viruses with abbreviations: molecular (mol), serological (ser), reverse transcription loop-mediated isothermal amplification (RT-LAMP), immunoglobulin type M (IgM), lateral flow assay (LFA), nonstructural protein (NS1), immunoglobulin type G (IgG), loop mediated isothermal amplification (LAMP), digital PCR (dPCR) enzyme-immunoassay (EIA) and IgM antibody capture ELISA (MAC-ELISA), double antibody sandwich ELISA (DAS-ELISA), enzyme linked immunospotting (ELISPOT), data not available (N.A.), number of copies (c).
|
Target disease |
Detection method |
LOD |
Test time (min) |
Ref | |||
|---|---|---|---|---|---|---|---|
| mol | Ser | mol | ser | mol | ser | ||
| Lassa fever | RT-PCR; RT-LAMP | IgM ELISA | 4 c·μL−1 | 230 PFU μL−1 | 60 | 180 | (Fukuma et al., 2011; Satterly et al., 2017) |
| Dengue fever | RT-PCR | ELISA, NS1 IgG ELISA, ELISPOT | 10 c·μL−1 | 5.2 ng μL−1 | 90 | 60 | (Linares et al., 2013; Santiago et al., 2018) |
| Ebola hemorrhagic fever | RT-PCR | immunofluorescence, IgM/IgG ELISA, LFA | 10 c·μL−1 | 6.8 PFU μL−1 | 30–50 | N.A. | (Fernández-Carballo et al., 2018; Satterly et al., 2017) |
| AIDS | dPCR; RT-PCR | flow cytometry, ELISA | 0.05 c·μL−1 | 1 ng mL−1 | 90 | N.A. | (Gan et al., 2012; Tanriverdi et al., 2010) |
| Zika fever | RT-PCR | IgM antibody capture MAC-ELISA, DAS-ELISA | 10 c·μL−1 | 0.1 ng μL−1 | 90 | 60 | (Lee and Zeng, 2017; Santiago et al., 2018) |
| West Nile fever | PCR; RT-PCR | IgM-capture, indirect ELISA | 100 c·μL−1 | 195 PFU μL−1 | 90 | 180 | (Herrmann et al., 2007; Wang et al., 2016) |
| Measles | RT-PCR; RT-LAMP | ELISA | 0.4 c·μL−1 | N.A. | 90 | N.A. | Hummel et al. (2006) |
| SARS | RT-PCR; PCR | ELISA, immunofluorescence | 5 c·μL−1 | 50 pg mL−1 | 50–90 | 180 | (Huang et al., 2010; Xiao-Yan et al., 2004) |
| Influenza | RT-PCR; RT-LAMP | ELISA, immunosensor | 0.4 c·μL−1 | 10 PFU μL−1 | 40 | 180 | (Hewa et al., 2009; Jung et al., 2015a) |
Effective laboratory techniques enabling early and affordable detection of these disease-causing viruses would play a critical role in tackling their outbreaks (Broadhurst et al., 2016; Kluge et al., 2018). Currently, the standard technique for diagnosis of viral disease and viral quantification is based on cell culture methods requiring two days of culturing in a well-controlled laboratory environment (Otter et al., 2016; van Doremalen et al., 2013). Other viral disease detection methods include enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR), both with high sensitivity and specificity (Lee and Zeng, 2017b; Reperant and Osterhaus, 2017), as well as flow cytometry and microscopy (Yang et al., 2017; Zarei, 2018). However, all these methods require expensive equipment and reagents, well-trained operators, and labor-intensive processing, and they are also typically time-consuming (Nasseri et al., 2018) (Table 1). Nevertheless, the ELISA is currently considered a gold standard for protein detection.
Recently, lab-on-a-chip (LOC) technologies advanced from original devices that can conduct a single task to integrated systems capable of performing complex jobs (Haeberle and Zengerle, 2007; Schumacher et al., 2012). Each integrated LOC platform typically contains sets of microfluidic elements, each of which are dedicated to single operations such as reagent storage, fluid transport, fluid mixing, product detection, and possibly collection. Currently, the systems consist of complex devices with interconnected fluidic microchannel networks, valves, mixers, pumps, reaction chambers, and detectors, and they are able to perform many laborious benchtop protocols without human intervention (Jung et al., 2015b). These abilities make LOCs suitable for applications in clinical diagnostics as well as near-patient or point-of-care (POC) testing (Wang et al., 2017), potentially enabling low-cost mass production.
LOC-based techniques are widely used for viral detection, and there have been reviews published in the past such as comprehensive description of pathogen detection using microfluidic systems (Mairhofer et al., 2009). Other reviews covering the topic of viral infection in conjunction with microfluidics are devoted to single virus diagnoses including sample preparation, and detection methods such as Ebola (Coarsey et al., 2017), dengue fever (DENV) (Darwish et al., 2018), hepatitis (Duchesne and Lacombe, 2018) and HIV (Mauk et al., 2017).
In this review, we provide an overview of conventional as well as LOC-based techniques to diagnose diseases caused by either ribonucleic acid (RNA) or deoxyribonucleic (DNA) viruses. We also review the fabrication methods suitable for variable substrate materials as well as detection methods based on immunoassays and nucleic acid (NA) amplification.
2. Conventional techniques for detection of viral diseases
Conventional techniques for the diagnosis of viral diseases are typically based on virus-infected cell cultures or the detection of viral antigens, antibodies, and nucleic acids (NA), which are labor-intensive and time-consuming techniques. Methods based on electron microscopy, cytology, or histology (Storch, 2000) are less common. Currently, serological testing is the mainstay of viral disease diagnostics in clinical laboratories. It enables the monitoring of the immune system's antibody response to viral antigen exposure, both the IgM response representing acute infection, and IgG as a consequence of primary infection or as the response to the acute phase of secondary infections. In this chapter we wrote a brief overview of the selected detection and quantification methods of viruses.
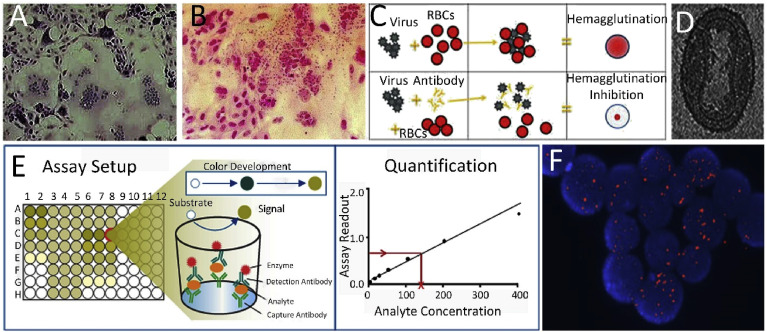
The culture methods are based on the virus growth and isolation from cell lines according to the specific viruses. The viruses are subsequently detected by cytopathic effects, eliciting specific characteristics and alterations in host cells caused by viral invasion (Agol, 2012) (Fig. 1 A), followed by immunofluorescence staining to its identification. The alternative method based on cell cultures is haemadsorption where the erythrocytes adsorb to the plasma membrane of the virus-infected cell monolayer (Fig. 1B). Cell culturing is a standard technique, but one of its drawbacks is that it takes approximately four weeks, which can be too long for tackling a potential viral disease outbreak (Papafragkou et al., 2013). Therefore, the rapid shell vial culture method was developed (Jayakeerthi et al., 2006). It is based on inoculation of the specimen on to the cell monolayer grown on a cover slip, followed by low-speed centrifugation and detection of the virus by immunofluorescence. However, this rapid culture test cannot target the required range of viruses and it does not possess sufficient sensitivity. Another simple and inexpensive technique is hemagglutination assay, which is based on the detection of the virus via agglutination of erythrocytes (Fig. 1C, upper). This test later led to the hemagglutination inhibition assay (Hierholzer et al., 1969) (Fig. 1C, bottom), a serological approach that detects specific antibodies against viral antigens (Hierholzer et al., 1969). The complement fixation test also introduces a convenient and easy method based on the reaction of the complement with an antigen-antibody complex (Adone et al., 2016).
Fig. 1.
Conventional methods for viral detection. (A) Cytopathic effect of measles virus infection of the human HeLa cell line. (B) Haemadsorption (Al-Shammari et al., 2014). (C) Hemagglutination inhibition test (Hierholzer et al., 1969). (D) Cryo-electro micrograph of vaccinia virus (Cyrklaff et al., 2005). (E) ELISA test principle (Niikura et al., 2001). (F) DNA of the Epstein-Barr virus (in red) within lymphoma cells detected by fluorescent imaging after in-situ hybridization (Leenman et al., 2004). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
A rather expensive and especially time-consuming method of electron microscopy (EM) (Fig. 1D) (Cyrklaff et al., 2005) has for a long time been considered an efficient tool for viral particle identification, given its characteristic morphology and quantification by counting the viral particles (Richert-Pöggeler et al., 2019). Despite some limitations, such as the need of technical skills and expertise, EM has been combined with culture-based methods and serological detection of antibodies against viruses in clinical laboratories.
However, the most conventional techniques mentioned above were later replaced with more sensitive and quantitative immunoassays capable of detecting the virus antigen or antibodies in clinical specimens (Weinberg and Walker, 2005). The high specificity and binding affinity between antigen and antibody generated a number of immunomethods with different detection approaches such as radio-immunoassay (RIA), enzyme-linked techniques (EIA), and ELISA. The most commonly used one is RIA, utilizing a radioisotope–labeled antibody to detect antigens or vice versa. As an alternative to danger radioactive labels, the utilization of enzymatic labels such as peroxidase or alkaline phosphatase led to the development of EIA. Currently, EIA variants such as ELISA (Niikura et al., 2001) (Fig. 1E) and chemiluminescent (Azar and Landry, 2018) or microparticle enzyme immunoassay (van den Berk et al., 2003) are widely used due to their high sensitivity. Especially ELISA, due to its dual specificity, became a gold standard for protein detection. Nevertheless, the co-circulation of closely related flaviviruses such as ZIKV and DENV in number of regions makes the conventional detection of a specific virus using immunoassays a complicated task (Wen and Shresta, 2019). Also there is a window period between the viral infection and antibody production resulting in false negative results using immunoassays, which can be up as long as 35–45 days for first generation HIV testing (Cornett and Kirn, 2013). The increase sensitivity acquired in fourth generation of immunoassays led to shortening this window to 10–15 days (Branson and Stekler, 2011).
NA-based detection methods have revolutionized virus-related diagnostics (Roy et al., 2017) having the false negative window period HIV between 10 and 15 days (Branson and Stekler, 2011). Several techniques can directly detect specific viral DNA or RNA via in-situ hybridization (Pfankuche et al., 2018) (Fig. 1F), dot-blot (Zhang et al., 2018a), or Southern blotting (Cai et al., 2013), but their sensitivity is insufficient. More sensitive techniques are based on NA amplification and its subsequent detection. Apart from endpoint PCR, the widely used variants are quantitative PCR (qPCR) for DNA (Edin et al., 2015) and RT-PCR for RNA (Zhang et al., 2018b), and both are becoming benchmarks in viral load assessment. Currently, dPCR is gaining popularity due to its ability to detect either DNA or RNA, with absolute gene quantification being more immune to background noise than conventional qPCR (Martinez-Hernandez et al., 2019). A number of alternative NA techniques have been developed including NA sequence-based amplification (Lanciotti and Kerst, 2001), strand displacement amplification (Shi et al., 2014), or branched DNA probes (Zhang et al., 2018a).
Since each of these established methods has several limitations, such as poor reproducibility and being labor-intensive and time-consuming, improved techniques of virus identification and quantification, such as mass spectrometry (He et al., 2014) and next-generation sequencing (Barzon et al., 2011), have been explored to overcome these limitations. However, the trend of miniaturization, cost-effectiveness and rapid viral monitoring via diagnostic methods based on LOC is undeniably a global public health ambition.
3. Technology for LOC fabrication
LOC and microfluidic devices for viral detection are being fabricated by several conventional and unconventional techniques as well as by rapid prototyping methods. In this chapter we summarize currently utilized techniques and materials for LOC fabrication. First, we start with planar technology used for silicon and glass substrate, computer numerical control (CNC) and laser ablation (LA) used for plastics. The printed circuit boards (PCBs), soft lithography used for popular material such as polydimethyl-siloxane (PDMS), 3D printing and paper-based structures are being also discussed.
Microfluidic devices are typically made of materials including silicon, glass, plastics, such as cyclic olefin copolymer (COC) (Levkin et al., 2008), poly(methyl methacrylate) (PMMA) (Levkin et al., 2008), polycarbonate (PC), PDMS (McDonald et al., 2000), polyimide (Levkin et al., 2008); and metals (Nguyen et al., 2012). Conventional methods include CNC micromachining, LA, and micro-electro-mechanical systems (MEMS) techniques. More recently, novel technologies have been developed for the fast, low-cost fabrication of LOC devices, such as soft lithography, printed circuit boards (PCB)-based methods, xurography, and paper-based methods.
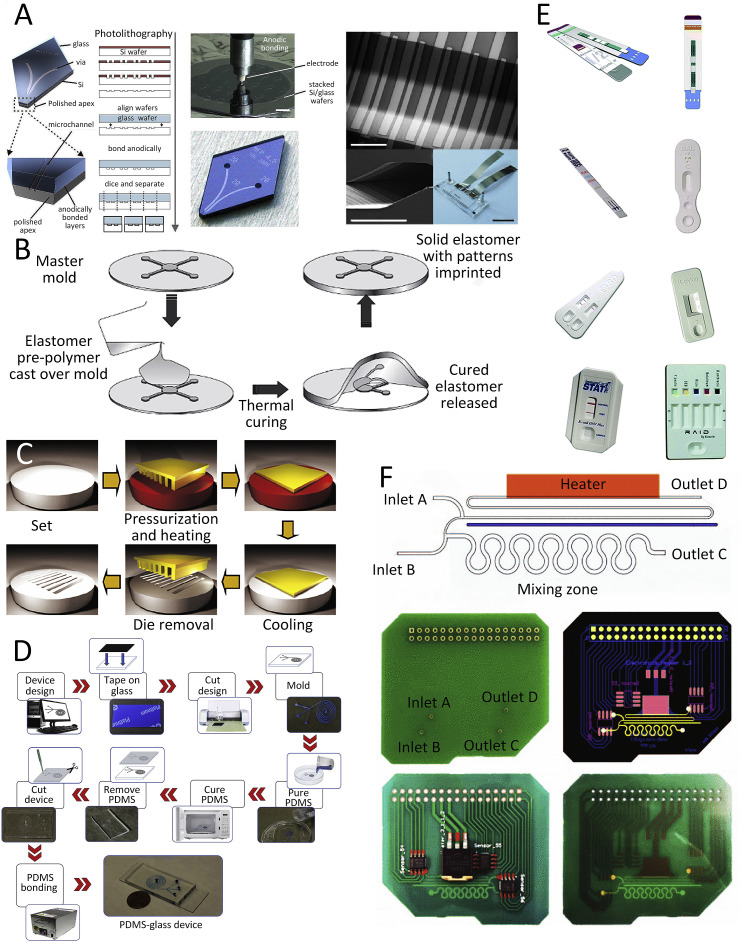
CNC and LA machining are traditional techniques for macroscopic material fabrication which is also suitable for microfluidic devices based on plastic substrates, such as PMMA and PC. But these methods are limited by the fabrication accuracy and size. The MEMS technique is widely used for silicon and glass substrate chip fabrication. Lithography is a general MEMS technology used in microfabrication to transform microstructures from masks to a substrate with a limited width of a few nanometers. This technology is utilized to fabricate silicon and glass based microfluidic chips or masters for microstructures, followed by substrate etching and wafer bonding. One of the earliest widespread MEMS technology for LOC and microfluidic devices is anodic bonding, wherein micromachined silicon is capped by glass to produce the microfluidic device (Fig. 2 A) (Qi et al., 2018). This fabrication method is relatively simple, cost-effective, and has a high yield of glass/silicon chips with excellent transparency of glass and practically no self-induced fluorescence (Iliescu et al., 2012).
Fig. 2.
Fabrication methods for LOC and microfluidic devices for viral detection. (A) An example of permanently sealed microfluidics produced by anodically bonding a silicon wafer to a glass wafer (Temiz et al., 2015). (B) Schematic diagram of a micromolding process involving elastomers such as PDMS (Feldman, 2014). (C) Schematic view of the hot embossing process (Sahli et al., 2013). (D) Schematic of xurography fabrication process (Speller et al., 2019). (E) Examples of commercial rapid paper-based microfluidic devices (Yetisen et al., 2013). (F) Lab-on-PCB integrating active control diluter (Moschou and Tserepi, 2017). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Soft lithography represents a conceptually different approach to rapid prototyping of various types of both microscale and nanoscale structures, and devices on variable materials and complex microstructures. A large number of patterning techniques are developed for the microfabrication of microfluidic devices, such as replica molding, microcontact printing, micromolding in capillaries, hot embossing, and microtransfer molding. For example, the micromolding technique was developed with two approaches involving either liquid or solid materials. A monomer of an elastomer such as PDMS (Karthik et al., 2015; Xu et al., 2012) is poured over a three-dimensional (3D) mask used as the mold and left to cross-link in a vacuum either at room temperature or at an elevated temperature. The mold is typically made of materials such as silicon, metal, or plastic, often produced by lithography (Fig. 2B). Then, the PDMS is separated from the mold, its surface is activated by O2 plasma, and then it is bonded to the glass substrate (Haubert et al., 2006). Another popular technique for fabricating microfluidic chips, called hot embossing (Fig. 2C), involves the use of hard materials. It can be wafer-based using a silicon wafer with a 3D surface pattern or a roll-to-roll using nickel or steel patterns to press against the substrate such as COC, PC, or PMMA. This roll-to-roll hot embossing technique is suitable for the large-scale production of microfluidic chips with a throughput of over 250,000 chips per day per line (Peng et al., 2016).
In the last decade, researchers adopted rather unconventional rapid prototyping methods to fabricate microfluidic devices such as xurography (Fig. 2D) (Speller et al., 2019) and paper-based methods (Fig. 2E) (Channon et al., 2018; Zhao and Liu, 2016). Xurography is a technique for creating microstructures with computer-controlled cutting from various thin-film polymer materials without requiring cleanroom facilities. The cutters are often capable of fabricating structures with feature sizes as small as ≈ 20 μm. Unfortunately, in spite of its potential, xurography has not yet been fully utilized to fabricate microfluidic devices for virus detection.
PCB recently has emerged as an LOC platform (lab-on-PCB) as its utilization requires minimal capital investments. The lab-on-PCB system can eliminate most of the obstacles in the commercialization of microfluidic devices based on other platforms: standardization, and system-level integration at a minimal cost. The PCB industry is well established all around the world for the mass production of electronic circuits, with standardized fabrication facilities and processes (Fig. 2F) (Moschou and Tserepi, 2017).
Paper-based methods (Govindaraju et al., 2019) are promising as the fabricated systems are disposable, portable, and biodegradable. The methods for paper-based device fabrication are fully developed forming channels originally performed by printing (Carrilho et al., 2009). The paper surface properties can be modified by formation of hydrophilic patterns on a hydrophobic membrane with plasma (Li et al., 2008) or laser treated surface (Chitnis et al., 2011). Another technique was employed to form superhydrophobic properties using polyhydroxybutyrate (Obeso et al., 2013). The paper-based microfluidics can be also direct cut with a laser or cutting plotter, printing with wax or biomolecules forming microchannel and test zones, mask-based fabrication, such as photolithography and wet etching, and screen-printing. Those methods are utilized to fabricate single layer paper devices, also allowed to form 3D devices by stacking papers such as origami (Liu and Crooks, 2011), even in easily reconfigurable configuration (Kong et al., 2017). As of now, paper-based devices do not have sufficient sensitivity, so further research and development in this area as well as in the implementation of suitable functionalization and immobilization of biomolecules (Yamada et al., 2017) is required. Paper-based detection, such as lateral flow strips (LFS), serve as qualitative diagnosis caused by the low precision of colorimetric device both for nucleic acids and proteins. The results are influenced by different lighting conditions and the variation in the color perception of users. In order to achieve quantitative analysis in paper-based diagnostics, commercial strip readers and color sensors have been used to record the color intensity, followed by the use of image-processing and analyzing software.
4. LOC-based microfluidic immunoassays
In this chapter we discuss sample preparation for immunoassays as well as on-chip immunoassays methods to detect antibodies related to the viral infection and problems related with these methods.
4.1. Sample preparation
Viral infections can be confirmed by a multitude of laboratory methods and a variety of samples can be used for virological testing. Thus, steps of separation and concentration are needed to exclude unwanted cell types and matrix fluids and to enrich the target cells when handling complex matrices such as blood, urine, and saliva (Lee et al., 2010). Microfluidics offers different techniques for viral particle separation. In the case of filtration with a charge-based membrane, the membrane is applied within the microfluidic cassette for HIV particle isolation, concentration, and detection in suspended raw saliva. HIV particles can be isolated from whole blood using an LOC filter-based chip with pore sizes between 1 μm and 2 μm from HIV-spiked samples at recovery efficiencies of (89.9 ± 5.0)%, (80.5 ± 4.3)%, and (78.2 ± 3.8)% for viral loads of 1000, 10,000, and 100,000 copies·mL−1, respectively (Reinholt and Baeumner, 2014; Wang et al., 2012).
A hand-held microfluidic device was integrated with vertically aligned carbon nanotube (VACNT) nanostructures to achieve high throughput virus capture using size-based filtration. The VACNT were synthetized at channel walls using chemical vapor deposition forming a VACNT forest with 97% porosity able to capture lentivirus measuring ≈128 nm in diameter, with an efficiency of 97% (Yeh J.T. 2018).
Another microfluidic device was integrated with a porous silicon nanowires (pSiNWs) forest for efficient virus isolation and their subsequent release. The device represents a continuous flow design with cross-flow filtration configuration to achieve large volume and high processing speed. Approximately 50% of influenza viruses were physically isolated in ≈30 min in the pSiNWs forest. The viruses could be released through degradation on the pSiNWs forest in ≈24 h (Wu et al., 2017). A sample preparation platform was developed that used a spiral inertial microfluidic device with continuous circulation to separate host cells from viral particles and free NAs. Removal with this device led to the reduction in human (host) background reads in metagenomic sequencing for viral detection and sequencing (Choi et al., 2018). Another approach for separating the sample is dielectrophoresis (DEP). It is implemented in microfluidic chips and used to capture DENV. The mechanism of detection involves the use of the DEP force to capture the modified beads in the microfluidic chip and the DENV modified with the fluorescence label, as the detection target can be captured on the modified beads by immunoreaction (Iswardy et al., 2017). Aptamers and antibodies are used for heterogeneous cell separation (Lin et al., 2016). An aptamer-based sandwich system was developed to capture and detect rare cells on a chip. An acoustic microfluidic device was developed for DENV cell separation from mammalian cells (Fong et al., 2014), with an efficiency of ≈70% using sample flow rate of ≈100 mL min−1.
4.2. On-chip immunoassay
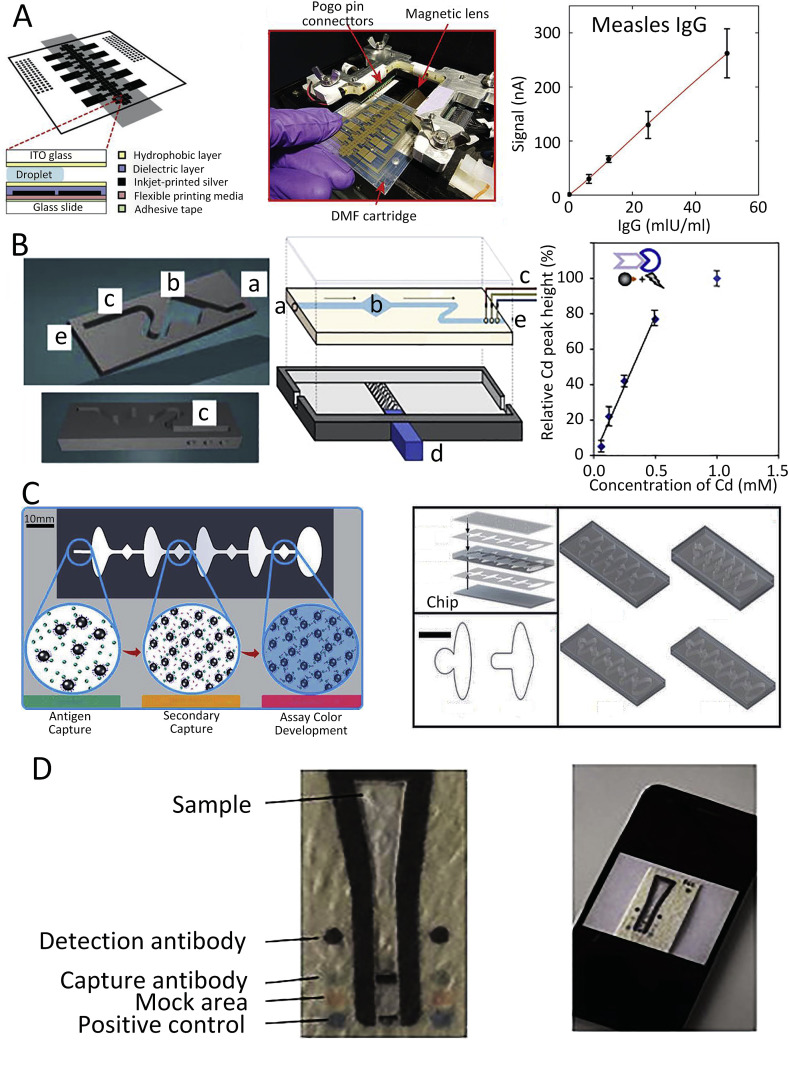
Miniaturization of immunoassays leads to reduced test time, increased test sensitivity, reduced sample volume, parallel processing, and portability. Inkjet-printed digital microfluidic cartridges were integrated with an instrument to perform ELISA and combined with a portable control system. The system was used for the detection of measles virus- and rubella virus-specific IgG in human blood samples (Fig. 3 A) (Ng et al., 2018). Detection can be enhanced by using quantum dots (QDs) that provide an alternative to the dye-based system with multiplexed detection of many colors.
Fig. 3.
Different approaches for chip-based immunoassay. (A) Digital microfluidic cartridge and ELISA used for measles detection (Ng et al., 2018). (B) 3D bead-based microfluidic chip for infection disease detection (Krejcova et al., 2014). (C) A flow-free magnetic actuation platform for microfluidic ELISA test (Coarsey et al., 2019). (D) Paper-based microfluidic test for ZIKV NS1 and DENV detection (Bedin et al., 2017).
A bead-based microfluidic chip was fabricated using 3D printing technology for detection of influenza hemagglutinin. The hemagglutinin labeled with QDs made of CdS was separated from the sample using streptavidin-modified paramagnetic beads, and the hemagglutinin concentration was subsequently indirectly detected by the sensitive anodic-stripping voltammetry of Cd2+ (Fig. 3B) (Krejcova et al., 2014).
Bead-based immunofluorescence assay on microfluidic chips is widely being used for virus detection (Zhang et al., 2013). The DEP module was integrated into a microfluidic chip to detect DENV with fluorescence immunosensing (Iswardy et al., 2017). The platform provided rapid on-chip detection in ≈5 min with a small sample volume of ≈15 μL and life-time reusability of >50 × . Also, an automated magnetic bead-based microfluidic ELISA method for detection of HIV-1 based on p24 capsid antigen detection was developed (Fig. 3C) (Coarsey et al., 2019). The developed platform provided a free-flow design, with no syringe pumps, and accepted target values as low as ≈ 20 pg mL−1 and middle range target values of ≈60 pg mL−1 (Fig. 3D) (Chunduri et al., 2016). Typically, the reagent flow is induced by an external force and controlled by either integrated or external valves. Microfluidic paper-based analytical devices (μPADs) have microporous structures that allow efficient absorption and sample flow without any peripheral force. Therefore, they provide an attractive alternative to traditional laboratory-based assays and allow affordable and rapid disease testing. Recently, μPADs for immunoassay were developed for the detection of the DENV and ZIKV via viral nonstructural glycoprotein 1 (NS1) in ≈7 min from samples containing ≈ 10 ng mL−1 of viral protein in the blood and plasma. The device consists of a large area at the top for sample collection either directly from blood or from plasma, followed by coating of the area with specific antibodies against both the DENV and ZIKV's NS1 protein (Bedin et al., 2017) using a custom-made application on a smartphone for results reading. A paper-based microfluidic dot ELISA chip was fabricated to detect influenza A (Wu et al., 2017). The chip consists of two modules: the first one is the reagent storage and dispensing module, and the second one is the reaction module with an absorbent pad and functionalized nitrocellulose membrane for influenza A detection.
5. NA amplification in microfluidic systems
In this chapter we discuss NA amplification testing (NAAT) methods (Niemz et al., 2011). The NAAT methods typically consist of three parts: sample preparation, NA amplification, and result detection; each part can be integrated in a microfluidic chip or independently operated off-chip. as well as sample preparations techniques. PCR based on thermal cycling and isothermal amplification such as LAMP have been widely used to diagnose infectious diseases. Compared with bulky benchtop equipment, microfluidic devices possess the advantages of portability and low sample and time consumption, indicating potential application in POC systems.
5.1. Sample preparation
Sample preparations, such as cell lysis (Heiniger et al., 2016), NA extraction, and purification, are typically accomplished with benchtop equipment in central laboratories. Sample preparations from clinical specimens could be integrated and coupled with amplification and detection in an inexpensive, automatic, miniaturized, and closed microfluidic NAAT system. A series of procedures of sample preparation, such as lysis, extraction, and purification of DNA or RNA have been extensively developed to obtain high-quality NAs. All the required steps of sample preparation such as lysis, either chemical, thermal, or electrical, NA extraction, and purification were utilized in microfluidic devices, followed by amplification and product detection.
5.1.1. On-chip cell lysis
In order to extract NAs from samples, cell lysis is applied first to release lysate inside cells followed by extraction and purification. Cell lysis methods utilize physical-chemical properties to destroy biological membranes, such as with chemical reagents, high temperature, electrical field and mechanical force. These methods require reagents and a specific environment for cell lysis, which is achievable on a chip through microstructure design and additional integrated parts.
Chemical lysis utilizing reagents (Maharjan and Ferenci, 2003) to disrupt cell membranes by solubilizing their phospholipids is a widely used method either in laboratories or in LOC systems. An example is a self-contained microfluidic chip utilized to detect the RNA of the hemagglutinin 3 and neuraminidase 2 (H3N2) influenza A virus, consisting of sample lysis, NA extraction, and RT-PCR (Stumpf et al., 2016). The sample lysis was performed in the lysis chamber with a lysis buffer for 10 min in a shake mode mixing controlled by frequency.
Thermal lysis of cells is conducted by exposing them to an elevated temperature of 80°C or more (Shetty et al., 2017) to rupture their membranes and release the NAs. This method can be used to direct the release of DNA from the spores (Přibylka et al., 2013), which is normally a difficult or time consuming process. Elevated temperatures at a level of ≈95°C utilized by PCR for DNA denaturation can be used for cell lysis. Thermal cell lysis was performed in the same chamber for RT-PCR at ≈ 95°C for ≈5 min on a single chip to detect DENV and enteroviruses (Lien et al., 2007). This method can be one step in an isothermal NAAT by adding a high temperature step. An immunoassay-based RT-LAMP assay for rapid detection of avian influenza virus subtype H5N1 in whole blood samples requires the viral RNA to be released from samples incubated at ≈ 95°C for ≈5 min (Tang et al., 2016).
Another method is electric cell lysis where the cell transmembrane potential is set over its critical value causing the cell membrane to break and release the NAs. The cell membrane was perforated by pulses above the electroporation threshold, resulting in pores for lysosome to diffuse into the cell and inter-membrane space and digest the cell wall (De Lange et al., 2016). The whole process was performed in a microfluidic droplet in the electrified channel to induce cell lysis.
Mechanical cell lysis is used to crush the cell membranes by mechanical force with, for example, shear forces, cell compression, the collision of cells with sharp objects, or other methods (Belgrader et al., 1999). Mechanical lysis is more effective than other methods, as other reagents are not introduced, and the DNA/RNA is not damaged by elevated temperatures. Porous polymers were utilized in the microfluidic chip fabrication, providing a mechanical shear force for cell lysis (Mahalanabis et al., 2009). The pore size of the polymer material was normally smaller than 1 μm causing a mechanical shearing force with a certain flow rate.
5.1.2. On-chip NA extraction and purification
After cell lysis, NA extraction and purification steps are required to separate them from NA amplification inhibitors such as proteins, polysaccharides, and fat molecules. There are two popular methods for utilizing magnetic beads or paper.
Magnetic beads are formed by coating a core of magnetic material such as Fe3O4 with an active group to bind the NAs. Magnetic bead-based separation can be combined with NA elution by temperature, pH, or salt concentration. Monodisperse Fe3O4 magnetic nanoparticles with a mean particle diameter of ≈300 nm covered by ≈ 20 nm of silicon matrix functionalized for RNA binding were developed for rapid NA extraction from clinical samples (Wang et al., 2018b). HIV-infected cells were loaded into a reaction chamber that was pre-loaded with 10 mL suspension of these magnetic beads. Then, the sample was exposed to a temperature of ≈95°C for ≈5 min to lyse the cells. The temperature was then decreased to ≈60°C and the sample was kept for ≈10 min to enable the designed probes to hybridize with the proviral DNA in the cells of the HIV-infected Jurkat T cell line. Then, the researchers employed a magnetic field with an amplitude of ≈4300 G to isolate the magnetic bead complexes from the unbound materials during the washing process with a micropump (Wang et al., 2013).
Paper-based devices, such as commercial filter paper (Gan et al., 2014) FTATM or Fusion 5, are used in the filtration isolation of NA and paper origami-based extraction to extract NA from various biological samples. NA extraction and subsequent purification of RNA from H1N1 influenza A virus was performed in a polyethersulfone matrix-based chip directly with human clinical nasopharyngeal specimens (Rodriguez et al., 2015). Hepatitis B virus DNA from clinical blood samples was extracted by a Fusion 5 paper-based system, which includes buffer storage and microvalves and channels of different lengths to direct the reagent and sample (Tang et al., 2017). Furthermore, helicase-dependent isothermal amplification and lateral flow assay detection were integrated with extraction, resulting in a fully disposable paper-based sample-in-answer-out device for NAAT (Tang et al., 2017).
5.2. NA amplification
NAAT is a key step in infectious disease diagnosis as it increases the concentration of NAs from the pathogens, making them detectable. Thermal cycling amplification methods, such as PCR as well as RT-PCR, and isothermal amplification methods have been developed to amplify and detect the NAs. The PCR system requires thermal cycling resulting in a simplified molecular biology protocol. Isothermal amplification techniques require simpler hardware as there is no thermal cycling, but the molecular biology protocol is more complex.
5.2.1. Thermal cycling amplification
PCR was invented by Kary Mullis (Mullis et al., 1986) over 30 years ago, and it quickly became a standard clinical diagnostic technique. A PCR protocol typically consists of ≈40 thermal cycles effectively doubling the number of DNA molecules in each cycle. This process can theoretically achieve multiplication of the original number of DNA copies (Valasek and Repa, 2005) by a factor of 240. The original end point detection PCR was modified by adding either intercalating dye or a probe forming real-time PCR (Higuchi et al., 1993). Parameter called critical threshold is used for relative DNA quantification by comparison with standard PCR curve, thus the method is also called quantitative PCR (qPCR). A new method of digital PCR (dPCR) was developed (Vogelstein and Kinzler, 1999) with sample compartmentalized (digitized) into thousands sub samples to get either one or none DNA copy per sample. Once the PCR was completed the automated system determined number of sub samples with amplified DNA resulting in absolute DNA quantification in the sample. The dPCR method is more complex than the qPCR, but the copy number resolution is far superior. The system also allows performing the PCR multiplexing with large difference in copy number of concurrently amplified DNAs (Ottesen et al., 2006).
PCR-based microfluidic devices can be prepared by using the time-domain or space-domain heating/cooling style.
5.2.1.1. Time-domain PCR
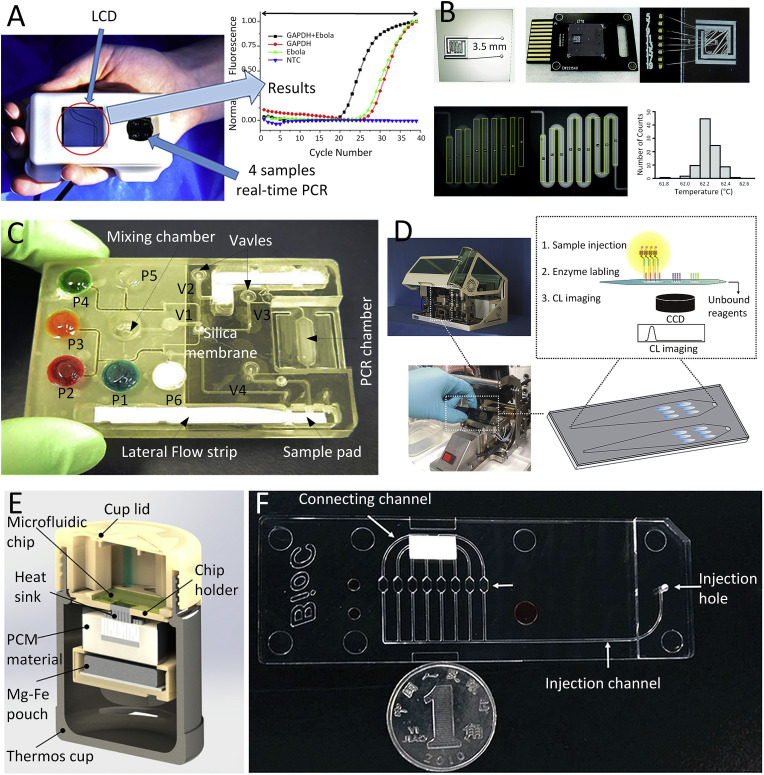
The thermal cycling of a time-domain PCR device is accomplished by temperature changes in the reaction chamber with stationary sample(s) resulting in a simple design. The original Mr. Cycler (Bartlett and Stirling, 2003) used by the Mullis at Cetus corporation as well as the first micromachined PCR chip were both time-domain systems (Northrup, 1993). An integrated handheld real-time PCR device was utilized to detect the Ebola virus RNA using single-step RT-PCR (Ahrberg et al., 2016b). The device (Fig. 4 A) concurrently performed four PCRs, each with a sample volume of ≈0.1 μL covered with ≈2 μL of mineral oil. The entire process consisted of an RT step to convert the RNA to complementary DNA (cDNA), PCR, and melting curve analysis, all conducted in <37 min. Although it is a relatively fast device, the sample preparation was not integrated with the real-time PCR system. Nevertheless, this system was demonstrated to be capable of performing qPCR multiplexing using a single fluorescent channel, either by combining an FAM probe and an intercalating dye (Ahrberg and Neužil, 2015) or using only an intercalating dye (Ahrberg et al., 2015).
Fig. 4.
Microfluidic NA amplification testing in different methods: (A) An integrated real-time PCR device for detection of Ebola virus (Ahrberg et al., 2016b). (B) Detail of a fluorescence image-based RT-PCR for detection of hepatitis C virus (HCV), HIV, ZIKV, and human papilloma virus (HPV) (Powell et al., 2018). (C) An integrating sample pretreatment RT-PCR chip for detection of HIV virus (Chen et al., 2010). (D) A real-time LAMP system for detection of respiratory infection virus (Wang et al., 2018b). (E) Fully integrated RT-LAMP system for detection of ZIKV (Song et al., 2016). (F) A completed recombinase polymerase amplification (RPA) system for detection of human adenovirus (Kunze et al., 2015).
Other microfluidic devices (Fig. 4B) were fabricated from silicon performing qPCR for detection of viral RNA and DNA, including the hepatitis C virus (HCV), HIV, ZIKV, and the human papilloma virus (HPV) (Powell et al., 2018). Another system had integrated sample preparation consisting of a cell (virus) lysis, NA isolation, amplification, and detection (Fig. 4C) (Chen et al., 2010). All the liquids and dry reagents needed for the reactions were pre-stored in the cassette with liquid reagents located in flexible pouches on the chip surface. The functionality of the device was demonstrated by detecting bacterial B. Cereus and HIV in saliva samples.
5.2.1.2. Space-domain PCR
Thermal cycling in the space-domain PCR device is performed by moving the sample through different temperature zones as seen in the PCR systems in the early days of PCR development (Bartlett and Stirling, 2003) as well as the first microfluidic device performing PCR (Kopp et al., 1998). The design of the microfluidic device typically predetermines the number of temperature cycling steps. Their duration can be then adjusted by variation of the flow rate or sample motion. Sample circulating systems offer more flexibility in cycle duration, number, and ratio (Pipper et al., 2008; Sun et al., 2007). A system incorporated with a disposable microfluidic chip can be produced in large volumes using cost-effective roll-to-roll embossing methods such as the one for detection of the Ebola virus (Fernández-Carballo et al., 2018). The chip has a long microfluidic channel that directs the PCR solution through areas heated to different temperatures. The solution first enters the zone with temperatures between ≈50°C and ≈57°C where the RNA is converted to cDNA and subsequently amplified by PCR while the amplification product is detected in real time by an optical fluorescence sensing system.
5.2.2. Isothermal amplification
Isothermal NA amplification methods (Zanoli and Spoto, 2013; Zhang et al., 2019) are carried out at a constant temperature, typically between ≈37°C and ≈65°C, making the isothermal amplification system hardware simpler than the one used for PCR. Many isothermal amplification methods have been reported and can be grouped based on the reaction principle. The most popular systems of isothermal DNA amplification are LAMP, recombinase polymerase amplification (RPA), and helicase-dependent amplification (HDA).
LAMP is based on primer annealing followed by auto-cycling strand displacement, typically at temperatures of ≈65°C. This amplification method requires six specific oligonucleotide sequences in the initial step and four sequences during the amplification, elongation, and recycling steps. An integrated microsystem (Fig. 4D) based on real-time LAMP was developed for diagnosing multiple respiratory viruses, including cDNA from influenza A virus subtypes of H1N1, H3N2, H5N1, and H7N9; influenza B virus; and human adenoviruses (Wang et al., 2018b). The researchers employed magnetic beads for NA extraction in an eight-channel microfluidic array chip integrated with a LAMP system for POC screening of respiratory viruses. Another isothermal amplification device was used to detect ZIKV with a single-step RT-LAMP assay (Fig. 4E) (Song et al., 2016). The elevated temperature required to perform RT-LAMP was achieved by heating the system using an exothermic chemical reaction, eliminating the requirement for external electrical power supply. Amplification products were detected by the naked eye using a leucocrystal violet dye, eliminating additional instrumentation moving towards a lab-on-a-chip system.
RPA operates at a relatively low temperature range from ≈37°C to ≈42°C. The target DNA sequence can be amplified using recombinase, single-strand binding proteins, and strand displacement polymerase. An RPA-based microfluidic system (Fig. 4F) was used to spatially separate the amplification reaction of DNA from two viruses: the human adenovirus and phage (Kunze et al., 2015) of Phi X 174.
HDA is a reaction based on natural DNA replication mechanisms working optimally at ≈ 65°C, utilizing a DNA helicase enzyme to separate dsDNA into an ssDNA template. Microfluidic devices based on HDA technology can amplify a fragment of the cDNA of the SARS virus with a single-step manual sample-pipetting step (Ramalingam et al., 2009). The sample was incubated at ≈ 62°C for ≈30 min using the real-time NA amplification benchtop system. Fluorescence amplitude of the NA product using EvaGreen intercalating dye was subsequently detected.
5.3. Detection method for LOC
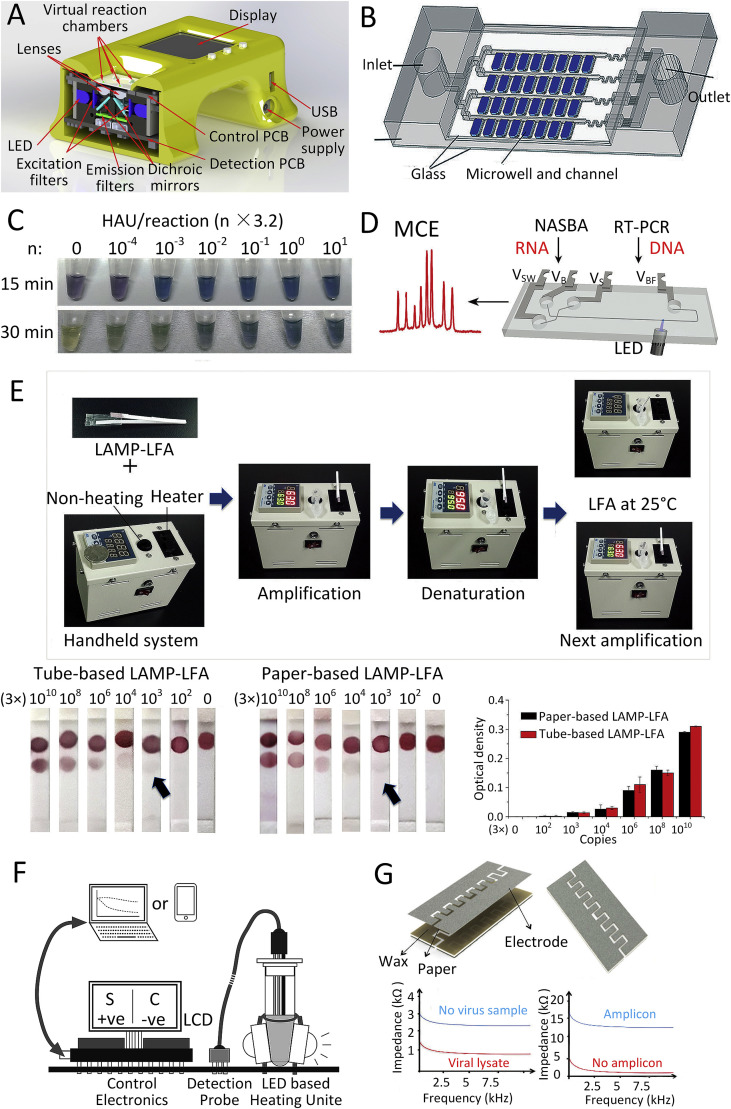
NA amplification can be either analyzed after the reaction is completed (endpoint detection) or while the reaction is progressing (real-time detection). Endpoint detection requires less complex instrumentation and provides simpler outputs for interpretation. Real-time methods integrate amplification with detection and are superior for quantitative analyte detection having a larger dynamic range. Alternative methods are developed for NA amplification detection, such as optical, mechanical, chemical, electrochemical, and nanomaterials-based detection. Fluorescence detection is the most popular optical detection strategy in biosensing caused by the advantages of sensitivity and selectivity, was developed in an integrated low cost unit (Novak et al., 2007). Fluorescence detection performed by fluorescent probes or beacons, and intercalating fluorescence dye, could be utilized for both real-time and endpoint detection. An integrated real-time fluorescence hand-held system was first developed to perform a detection of RNA of H5N1 avian influenza virus using real-time RT-PCR (Neuzil et al., 2010), later on upgraded into a palm-size device employed for detect the RNA of Ebola virus (Ahrberg et al., 2016b). Both systems utilized light-emitting diodes with a principal wavelength of ≈490 nm to excite the fluorophore and photodiodes to detect the emission. The output signal from the photodiodes was processed by a lock-in amplifier to increase the signal-to-noise ratio. This handheld device (Fig. 5 A) was further applied to provide an internet of things platform for infection disease diagnosis verified by DENV (Zhu et al., 2020). A similar platform was also developed as a modular system (Neuzil et al., 2014) capable to perform both time and space domain PCR for NA, localized surface plasmon resonance for protein detection (Neuzil and Reboud, 2008), electrochemical sensing using an array of two electrode systems and also using nanowires as biosensors.
Fig. 5.
Technique of NA amplification detection. (A) Optic design of integrated real-time fluorescence detection (Zhu et al., 2020). (B) Digital NASBA chip for endpoint detection by fluorescence imaging (Wang et al., 2018a). (C) A series of reaction tubes showing the color change when running the LAMP of H1N1 virus (Ma et al., 2019). (D) Microchip-based CE for NASBA product detection (Liu et al., 2015). (E) LFS-based NA amplification detection (Choi et al., 2016). (F) Schematic of real-time RT-LAMP detection by electrochemical hydrogen ion sensor (Jogezai and Shabbir, 2018). (G) Integrated paper-based chip for LAMP products detection by electrical conductivity measurement (Safavieh et al., 2017). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The endpoint fluorescence detection is mostly utilized in a continuous-flow as well as in digital PCR (or LAMP) systems. The results are only captured and analyzed once the sample amplification process is complete. A digital microfluidic NA sequence-based amplification (NASBA) chip (Fig. 5B) with a self-digitization sample loaded in vacuum was used to quantify HIV RNA (Wang et al., 2018a). The detection of the amplification result was performed by capturing and analyzing the fluorescence images with calcein added in the NASBA mixture to generate a fluorescent signal.
Visual detection, also named colorimetric detection, based on color changes under ambient light during amplification causing by chemical additives, such as hydroxynaphthol blue (HNB), calcein, and spatial nanoparticles, is the most promising detection means for POC applications. HNB is one of several metal ion indicators serving as a visual dye for the LAMP technique and changing from its original violet to sky blue after the LAMP reaction caused by a decrease in the magnesium ion concentration. A mixed dye of HNB and calcein was utilized to show the color change during LAMP for H1N1 virus (Fig. 5C) (Ma et al., 2019). The amplification result with the mixed dye could be detected by integrated real-time color sensors with higher precision. A system utilizes the sediment of DNA amplicons by spermine and flocculation of a mix of charcoal and diatomaceous earth particles in suspension for visual detection (Mason and Botella, 2019). The initial charcoal particles suspension is a black, non-transparent colloid solution without any precipitation. In positive samples in which DNA amplification has occurred, the particles flocculate and settle, leaving a transparent liquid phase distinguishable with the naked eye.
The separation-based detection of an NA amplification product is always an endpoint type. A typical example is an electrophoresis, which was for years the most popular PCR detection technique. Its capillary (CE) version conducted in a commercially available microfluidic chip is suitable for NA separation and detection due to sufficient resolution and operation convenience. A microchip-based CE (Fig. 5D) was integrated with NASBA for HPV detection (Liu et al., 2015). The sample was loaded and separated into individual NAs based on their lengths by applied a sequence of specific voltages delivered to the solution by four Pt electrodes. This CE integration with the amplification chip has a high throughput and a short analysis time, but it requires external high voltage sources with an amplitude of 1500 V or more.
The detection performed on an LFS is based on NA hybridization and antigen-antibody reactions, providing rapid and sensitive detection for multiplex detection of DNA and antigens/antibodies. The nanoparticles with ssDNA detector probe are typically prepared and assembled on the LFS membrane. The target DNAs bind to the immobilized probe producing a color change observable with the naked eye. A fully integrated paper-based LAMP device with an LFS layer (Fig. 5E) was utilized to detect DENV (Choi et al., 2016). They immobilized gold nanoparticles and detect probe conjugates on a nitrocellulose membrane for the LFS layer to capture target amplicons. After amplification, the nanoparticles-probe conjugates would then hybridize with the LAMP products in the test zone forming a visible red signal expending 15 min at room temperature.
Electrochemical detection for NA amplification product is based on hydrogen ions released during nucleic acid replication detected in real-time by an ion sensor, which also resulted in a pH value change of the solution (Zhang et al., 2014). The pH value change is not obviously caused by the solution condition and small volume. A designed detection probe was utilized in a 200 μL reaction tube to detect HCV using RT-LAMP by monitoring hydrogen ions concentration change (Fig. 5F). The probe comprises an ion sensitive field effect transistor, a micro-capillary based Ag/AgCl/KCl reference electrode and a temperature sensor (Jogezai and Shabbir, 2018). The electrical conductivity of the solution is also one parameter for the NA amplification test of the electrochemical method. The sample electrical conductivity is influenced by accumulate non-precipitated magnesium pyrophosphate and protons due to the consumption of primers and dNTPs during amplification process. A cellulose paper with printed graphene-modified silver electrodes (Fig. 5G) was used for the electrical conductivity measurement integrated with LAMP for HIV detection (Safavieh et al., 2017). The reduction in the sample electrical conductivity is correlated to the concentration of the target nucleic acid resulting in both qualitative and quantitative detection.
6. Commercial rapid devices for viral detection
Here we discuss commercialized integrated systems for viral detection based either on antibody detection or NAAT as well as fundamental problem associated with POC applications for highly contentious viruses such as SARS.
Recently emerging molecular diagnostics for the detection of NAs meet the requirements for speed, cost efficiency, and user friendliness for POCs (Niemz et al., 2011). A number of commercial systems were developed, to name a few, such as the Cobas® influenza A/B & RSV (Fig. 6 A) (Chen et al., 2015), Simplexa flu A/B, RSV direct (Fig. 6B), GenXpert to diagnose HIV in flu tests (Fig. 6C), and other tests for the rapid detection of HIV (Nash et al., 2017).
Fig. 6.
Commercial rapid devices for viral detection. (A) Cobas® influenza A/B & RSV assay (Chen et al., 2015), (B) Simplexa flu A/B RSV assay, (C) GeneXpert MTB/RIF machine (Nash et al., 2017). (D) Veredus Laboratories Pte. Ltd. system (Tan et al., 2014).
Rapid tests to detect viral antigens in small sets of samples using either a cassette or a dipstick are sensitive methods taking only a few minutes (Anderson et al., 2019). These systems were commercialized in the form of POC devices to detect different viruses and represent a user-friendly solution. The RIDA®QUICK rapid test based on LFA was developed to detect the presence of norovirus, rotavirus and adenovirus (Kim et al., 2014). The Binaxnow® influenza A & B test kit is another example of an in-vitro immunochromatographic assay for qualitative detection of influenza A and B (Mitamura et al., 2013). A system using immunochromatographic membrane assay to detect the antigen of a fusion protein secreted by the respiratory syncytial virus (RSV) was also commercialized (Thuy Tien et al., 2018). Several rapid tests are also available for diagnosis of DENV via its IgM and IgG antibodies and NS1 antigens (Granger et al., 2017). Also, a variety of rapid tests for the diagnosis of HIV have been developed in the form of POC systems, such as the OraQuick Rapid HIV-1/2 antibody test and the fourth-generation HIV tests detecting both the antigen and antibodies, such as the ARCHITECT HIV Ag/Ab combo and the Alere Determine HIV 1/2 Ag/Ab combo (Moshgabadi et al., 2015).
Another example of a POC for viral diagnostic is an endpoint detection system made by Veredus Laboratories Pte. Ltd. (Fig. 6D) (Tan, 2005). The device has two small chambers of a triangular profile embedded inside a Si chip integrated with heater and temperature sensor mounted on a PCB (Fuchs et al., 2002). An external electronic control system for temperature cycling required to conduct PCR. The sample is loaded by a vacuum inside the chambers, the RT-PCR is conducted and then the product is loaded onto an area with hybridization probes. The results are read by a florescence camera system and the diagnosed RNA/DNA is determined from the captured fluorescent pattern. The company CEO's philosophy was to conduct only RT-PCR as it can amplify both RNA as well as the DNA and there is only one protocol, making the system operation simpler. Based on this philosophy, various RNA/DNA kits were developed (Tan et al., 2014). Each device has its own bar code linked to the system database for device history tracking.
A major driver for POC development is the ability to diagnose infectious diseases at sites with limited infrastructure without the requirement to transport the clinical sample to a centralized facility. We expect POCs to be able to tackle the spread of infectious diseases. This is necessary for all countries regardless of their wealth. During the SARS outbreak in 2002, the only facility equipped with diagnostic equipment in Singapore was Tan Tock Seng hospital and people were sent there, thus increasing the chance of spreading this disease during their transportation.
7. Summary and conclusion
In this review, we summarized advances of LOC technologies employed for viral diagnostics. We listed typical representatives of viral infections causing potential problems and pandemic in last 20 year (Table 1). First, we described conventional techniques, their advantages and shortcomings as well as advantages of LOC type methods. Then we discussed different technologies for LOC fabrications starting with conventional using planar technologies. An emphasis was given to modern techniques based on 3D printing and paper-based microfluidics. Next chapter was dedicated to immunoassay techniques including sample preparation. Large part of this review describes nucleic acid amplification methods again starting with sample preparation, then different types of PCR, isothermal amplifications and detection methods of amplified product. Final chapter of this review is used to describe commercial devices for rapid viral diagnostics.
8. Future perspectives
Tackling the outbreak of viral disease requires rapid action locally. The conventional diagnostic systems for viral disease are expensive, bulky, and their utilization is labor intensive requiring skilled personnel to operate them, thus they are typically located in centralized laboratories. LOC systems offer tremendous advantages in infectious disease diagnostics especially in combination with newly developed technologies such as 3D printing (Krejcova et al., 2014), suitable for rapid prototyping and eventually even manufacturing. These new technologies can come together with new design tools such as the Nanolithography Toolbox (Balram et al., 2016) and simplify the complex microfluidic chip design.
We can also predict the further development of existing simple microfluidic systems with the electrochemical detection of proteins (Karon et al., 2007) and expansion of their capabilities for public health. This user-friendly system can be modified to perform ELISA with fluorescent optical detection, bringing the cost per test down to a reasonable level. These diagnostic kits will be available in a ready-to-use format and not do it yourself. Palm-sized RT-PCR systems for RNA detention have already been developed (Ahrberg et al. 2016a, 2016b) as well as complex stationary sample-answer systems based on RT-PCR (Nash et al., 2017). We envision the miniaturization of sample preparation followed by RT-PCR for viral RNA diagnosis. However, the sample volume needed to be processed for diseases with a low viral load, such as HIV, prevents the sample from being directly processed by LOC. This problem can be solved by adding a sample pre-concentration step (Pipper et al., 2007) where the viruses or the RNA from a volume of tens or hundreds of μL will be concentrated to a few μL and then processed by the LOC system. This would be ideally performed in a touch-free manner to avoid possible contamination of the person handling the blood, which would be irrelevant if the infected person were handling his/her own sample. Nevertheless, minimal human intervention is essential, and thus the development should lead to a sample-to-answer system.
Smartphone platforms will be heavily used as they have high resolution cameras and test results can be directly sent to the medical personnel or heath centers monitoring the infectious disease outbreak. Connecting all these diagnostic systems with centers and large data-processing facilities will form a true IoT (Zhu et al., 2020) for healthcare authorities. User-friendly software for data acquisition and analysis can be developed using Android or different operating systems to increase public safety. The LOC technology for POC assays is moving towards speed and efficiency in the diagnosis of viral diseases. Portability, cost, automation, speed, efficiency, and connectivity are critical technical parameters for the future generation of LOCs in viral disease diagnosis.
CRediT author contributions
Hanliang Zhu: NA amplification in microfluidic systems section, manuscript editing. Zdenka Fohlerova: Conventional techniques for detection of viral diseases and commercial rapid devices for viral detection section. Jan Pekarek: Technology for LOC fabrication section, Evgenia Basova: LOC-based microfluidic immunoassays section. Pavel Neuzil: Introduction, Perspectives section and manuscript editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
H.Z. and P.N. were financially supported by the grant from State Administration of Foreign Experts Affairs number W099109. The authors would also like to acknowledge support by a grant from the Czech Science Foundation number 17-20716S.
References
- Adone R., Sali M., Francia M., Iatarola M., Donatiello A., Fasanella A. Front. Microbiol. 2016;7(19):1–7. doi: 10.3389/fmicb.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agol V.I. Trends Microbiol. 2012;20(12):570–576. doi: 10.1016/j.tim.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrberg C.D., Ilic B.R., Manz A., Neužil P. Lab Chip. 2016;16(3):586–592. doi: 10.1039/c5lc01415h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrberg C.D., Manz A., Neuzil P. Sci. Rep. 2015;5:1–7. doi: 10.1038/srep12588. 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrberg C.D., Manz A., Neuzil P. Anal. Chem. 2016;88(9):4803–4807. doi: 10.1021/acs.analchem.6b00278. [DOI] [PubMed] [Google Scholar]
- Ahrberg C.D., Neužil P. Sci. Rep. 2015;5:1–9. doi: 10.1038/srep12595. 12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shammari A.M., Ismaeel F.E., Salih S.M., Yaseen N.Y. Oncolytic Virother. 2014;3:57–68. doi: 10.2147/OV.S59037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.E., Buser J.R., Fleming A.M., Strauch E.-M., Ladd P.D., Englund J., Baker D., Yager P. Lab Chip. 2019;19(5):885–896. doi: 10.1039/c8lc00691a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azar M.M., Landry M.L. J. Clin. Microbiol. 2018;56(7) doi: 10.1128/JCM.00367-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balram K.C., Westly D.A., Davanco M., Grutter K.E., Li Q., Michels T., Ray C.H., Yu L., Kasica R., Wallin C.B. J. Res. Natl. Inst. Stand. Technol. 2016;121:1–12. doi: 10.6028/jres.121.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J.M., Stirling D. Springer; 2003. A Short History of the Polymerase Chain Reaction. PCR Protocols; pp. 3–6. [DOI] [PubMed] [Google Scholar]
- Barzon L., Militello V., Lavezzo E., Franchin E., Peta E., Squarzon L., Trevisan M., Pagni S., Dal Bello F., Toppo S., Palù G. J. Clin. Virol. 2011;52(2):93–97. doi: 10.1016/j.jcv.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Bedin F., Boulet L., Voilin E., Theillet G., Rubens A., Rozand C. J. Med. Virol. 2017;89(9):1520–1527. doi: 10.1002/jmv.24806. [DOI] [PubMed] [Google Scholar]
- Belgrader P., Hansford D., Kovacs G.T.A., Venkateswaran K., Mariella R., Milanovich F., Nasarabadi S., Okuzumi M., Pourahmadi F., Northrup M.A. Anal. Chem. 1999;71(19):4232–4236. doi: 10.1021/ac990347o. [DOI] [PubMed] [Google Scholar]
- Branson B.M., Stekler J.D. Oxford University Press; 2011. Detection of Acute HIV Infection: We Can't Close the Window. [DOI] [PubMed] [Google Scholar]
- Broadhurst M.J., Brooks T.J.G., Pollock N.R. Clin. Microbiol. Rev. 2016;29(4):773–793. doi: 10.1128/CMR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Nie H., Yan R., Guo J.-T., Block T.M., Guo H. Methods Mol. Biol. 2013;1030:151–161. doi: 10.1007/978-1-62703-484-5_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrilho E., Martinez A.W., Whitesides G.M. Anal. Chem. 2009;81(16):7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- Channon R.B., Yang Y., Feibelman K.M., Geiss B.J., Dandy D.S., Henry C.S. Anal. Chem. 2018;90(12):7777–7783. doi: 10.1021/acs.analchem.8b02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Mauk M., Qiu X., Liu C., Kim J., Ramprasad S., Ongagna S., Abrams W.R., Malamud D., Corstjens P.L.A.M. Biomed. Microdevices. 2010;12(4):705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Tian Y., Chen S., Liesenfeld O. Eur. J. Microbiol. Immunol. 2015;5(4):236–245. doi: 10.1556/1886.2015.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis G., Ding Z., Chang C.-L., Savran C.A., Ziaie B. Lab Chip. 2011;11(6):1161–1165. doi: 10.1039/c0lc00512f. [DOI] [PubMed] [Google Scholar]
- Choi J.R., Hu J., Gong Y., Feng S., Abas W.A.B.W., Pingguan-Murphy B., Xu F. Analyst. 2016;141(10):2930–2939. doi: 10.1039/c5an02532j. [DOI] [PubMed] [Google Scholar]
- Choi K., Ryu H., Siddle K.J., Piantadosi A., Freimark L., Park D.J., Sabeti P., Han J. Anal. Chem. 2018;90(7):4657–4662. doi: 10.1021/acs.analchem.7b05200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunduri L.A.A., Haleyurgirisetty M.K., Patnaik S., Bulagonda P.E., Kurdekar A., Liu J., Hewlett I.K., Kamisetti V. Microfluid. Nanofluidics. 2016;20(12):1–10. 167. [Google Scholar]
- Coarsey C., Coleman B., Kabir M.A., Sher M., Asghar W. RSC Adv. 2019;9(15):8159–8168. doi: 10.1039/c8ra07607c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coarsey C.T., Esiobu N., Narayanan R., Pavlovic M., Shafiee H., Asghar W. Crit. Rev. Microbiol. 2017;43(6):779–798. doi: 10.1080/1040841X.2017.1313814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett J.K., Kirn T.J. Clin. Infect. Dis. 2013;57(5):712–718. doi: 10.1093/cid/cit281. [DOI] [PubMed] [Google Scholar]
- Cyrklaff M., Risco C., Fernández J.J., Jiménez M.V., Estéban M., Baumeister W., Carrascosa J.L. Proc. Natl. Acad. Sci. U. S. A. 2005;102(8):2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwish N.T., Sekaran S.D., Khor S.M. Sensor. Actuator. B Chem. 2018;255:3316–3331. [Google Scholar]
- De Lange N., Tran T.M., Abate A.R. Biomicrofluidics. 2016;10(2):1–8. doi: 10.1063/1.4944742. 024114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesne L., Lacombe K. J. Viral Hepat. 2018;25(2):108–117. doi: 10.1111/jvh.12827. [DOI] [PubMed] [Google Scholar]
- Edin A., Granholm S., Koskiniemi S., Allard A., Sjöstedt A., Johansson A. J. Mol. Diagn. 2015;17(3):315–324. doi: 10.1016/j.jmoldx.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M. Elsevier Science; 2014. Nanolithography: the Art of Fabricating Nanoelectronic and Nanophotonic Devices and Systems. [Google Scholar]
- Fernández-Carballo B.L., McBeth C., McGuiness I., Kalashnikov M., Baum C., Borrós S., Sharon A., Sauer-Budge A.F. Anal. Bioanal. Chem. 2018;410(1):33–43. doi: 10.1007/s00216-017-0689-8. [DOI] [PubMed] [Google Scholar]
- Fong E.J., Johnston A.C., Notton T., Jung S.Y., Rose K.A., Weinberger L.S., Shusteff M. Analyst. 2014;139(5):1192–1200. doi: 10.1039/c4an00034j. [DOI] [PubMed] [Google Scholar]
- Fuchs A., Jeanson H., Claustre P., Gruss J., Revol-Cavalier F., Caillat P., Mastromatteo U., Scurati M., Villa F., Barlocchi G. 2nd Annual International IEEE-EMBS Special Topic Conference on Microtechnologies in Medicine and Biology. Proceedings (Cat. No. 02EX578) IEEE; 2002. A silicon lab-on-chip for integrated sample preparation by PCR and DNA analysis by hybridization; pp. 227–231. [Google Scholar]
- Fukuma A., Kurosaki Y., Morikawa Y., Grolla A., Feldmann H., Yasuda J. Microbiol. Immunol. 2011;55(1):44–50. doi: 10.1111/j.1348-0421.2010.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan N., Bo S., Liu Q., Jia L., Li B., Zheng L., Wang Q. Molecules. 2012;17(5):5988–6000. doi: 10.3390/molecules17055988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W., Zhuang B., Zhang P., Han J., Li C.X., Liu P. Lab Chip. 2014;14(19):3719–3728. doi: 10.1039/c4lc00686k. [DOI] [PubMed] [Google Scholar]
- Govindaraju S., Lee M.H., Yun K. Elsevier; 2019. Recent Trends in the Development of Paper-Based Diagnostic Chips for the Detection of Human Viruses. Recent Developments in Applied Microbiology and Biochemistry; pp. 349–361. [Google Scholar]
- Granger D., Leo Y.S., Lee L.K., Theel E.S. Diagn. Microbiol. Infect. Dis. 2017;88(2):120–124. doi: 10.1016/j.diagmicrobio.2017.03.015. [DOI] [PubMed] [Google Scholar]
- Haeberle S., Zengerle R. Lab Chip. 2007;7(9):1094–1110. doi: 10.1039/b706364b. [DOI] [PubMed] [Google Scholar]
- Haubert K., Drier T., Beebe D. Lab Chip. 2006;6(12):1548–1549. doi: 10.1039/b610567j. [DOI] [PubMed] [Google Scholar]
- He Q., Zhu Z., Jin L., Peng L., Guo W., Hu S. J. Anal. Atomic Spectrom. 2014;29(8):1477–1482. [Google Scholar]
- Heiniger E.K., Buser J.R., Mireles L., Zhang X., Ladd P.D., Lutz B.R., Yager P. J. Microbiol. Methods. 2016;128:80–87. doi: 10.1016/j.mimet.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Herrmann S., Leshem B., Lobel L., Bin H., Mendelson E., Ben-Nathan D., Dussart P., Porgador A., Rager-Zisman B., Marks R.S. J. Virol. Methods. 2007;141(2):133–140. doi: 10.1016/j.jviromet.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Hewa T.M.P., Tannock G.A., Mainwaring D.E., Harrison S., Fecondo J.V. J. Virol. Methods. 2009;162(1):14–21. doi: 10.1016/j.jviromet.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer J.C., Suggs M.T., Hall E.C. Appl. Microbiol. 1969;18(5):824–833. doi: 10.1128/am.18.5.824-833.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Fockler C., Dollinger G., Watson R. Biotechnology. 1993;11(9):1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- Huang J.L., Wang Y.K., Liu H.W., Lin C.S. J. Med. Virol. 2010;77(2):151–158. doi: 10.1002/jmv.20432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel K.B., Lowe L., Bellini W.J., Rota P.A. J. Virol. Methods. 2006;132(1):166–173. doi: 10.1016/j.jviromet.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Iliescu C., Taylor H., Avram M., Miao J., Franssila S. Biomicrofluidics. 2012;6(1):1–16. doi: 10.1063/1.3689939. 016505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iswardy E., Tsai T.C., Cheng I.F., Ho T.C., Perng G.C., Chang H.C. Biosens. Bioelectron. 2017;95:174–180. doi: 10.1016/j.bios.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Jayakeerthi R.S., Potula R.V., Srinivasan S., Badrinath S. Virol. J. 2006;3:1–7. doi: 10.1186/1743-422X-3-2. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogezai N., Shabbir M.I. Anal. Methods. 2018;10(35):4233–4241. [Google Scholar]
- Jung J.H., Oh S.J., Kim Y.T., Kim S.Y., Kim W.J., Jung J., Seo T.S. Anal. Chim. Acta. 2015;853:541–547. doi: 10.1016/j.aca.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W., Han J., Choi J.-W., Ahn C.H. Microelectron. Eng. 2015;132:46–57. [Google Scholar]
- Karon B.S., Gandhi G.Y., Nuttall G.A., Bryant S.C., Schaff H.V., McMahon M.M., Santrach P.J. Am. J. Clin. Pathol. 2007;127(6):919–926. doi: 10.1309/6RFQCKAAJGKWB8M4. [DOI] [PubMed] [Google Scholar]
- Karthik A., Margulis K., Ren K., Zare R.N., Leung L.W. Nanoscale. 2015;7(45):18998–19003. doi: 10.1039/c5nr06114h. [DOI] [PubMed] [Google Scholar]
- Kim J., Kim H.S., Kim H.-S., Kim J.-S., Song W., Lee K.M., Lee S., Park K.U., Lee W., Hong Y.J. Ann. Lab Med. 2014;34(3):216–222. doi: 10.3343/alm.2014.34.3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge H., Martin-Moreno J.M., Emiroglu N., Rodier G., Kelley E., Vujnovic M., Permanand G. BMJ Global Health. 2018;3:1–7. doi: 10.1136/bmjgh-2017-000656. e000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong T., Flanigan S., Weinstein M., Kalwa U., Legner C., Pandey S. Lab Chip. 2017;17(21):3621–3633. doi: 10.1039/c7lc00620a. [DOI] [PubMed] [Google Scholar]
- Kopp M.U., Mello A.J.d., Manz A. Science. 1998;280(5366):1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- Krejcova L., Nejdl L., Rodrigo M.A.M., Zurek M., Matousek M., Hynek D., Zitka O., Kopel P., Adam V., Kizek R. Biosens. Bioelectron. 2014;54:421–427. doi: 10.1016/j.bios.2013.10.031. [DOI] [PubMed] [Google Scholar]
- Kunze A., Dilcher M., Abd El Wahed A., Hufert F., Niessner R., Seidel M. Anal. Chem. 2015;88(1):898–905. doi: 10.1021/acs.analchem.5b03540. [DOI] [PubMed] [Google Scholar]
- Lanciotti R.S., Kerst A.J. J. Clin. Microbiol. 2001;39(12):4506–4513. doi: 10.1128/JCM.39.12.4506-4513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Zeng H.Q. Anal. Chem. 2017;89(23):12743–12748. doi: 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
- Lee W.G., Kim Y.G., Chung B.G., Demirci U., Khademhosseini A. Adv. Drug Deliv. Rev. 2010;62(4–5):449–457. doi: 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenman E.E., Panzer-Grümayer R.E., Fischer S., Leitch H.A., Horsman D.E., Lion T., Gadner H., Ambros P.F., Lestou V.S. Mod. Pathol. 2004;17:1564–1572. doi: 10.1038/modpathol.3800228. [DOI] [PubMed] [Google Scholar]
- Levkin P.A., Eeltink S., Stratton T.R., Brennen R., Robotti K., Yin H., Killeen K., Svec F., Fréchet J.M.J. J. Chromatogr. 2008;1200(1):55–61. doi: 10.1016/j.chroma.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Tian J., Nguyen T., Shen W. Anal. Chem. 2008;80(23):9131–9134. doi: 10.1021/ac801729t. [DOI] [PubMed] [Google Scholar]
- Lien K.-Y., Lee W.-C., Lei H.-Y., Lee G.-B. Biosens. Bioelectron. 2007;22(8):1739–1748. doi: 10.1016/j.bios.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Lin X.X., Sun X.Y., Luo S.D., Liu B., Yang C.X. Trends Anal. Chem. 2016;80:132–148. [Google Scholar]
- Linares E.M., Pannuti C.S., Kubota L.T., Thalhammer S. Biosens. Bioelectron. 2013;41(1):180–185. doi: 10.1016/j.bios.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Liu H., Crooks R.M. J. Am. Chem. Soc. 2011;133(44):17564–17566. doi: 10.1021/ja2071779. [DOI] [PubMed] [Google Scholar]
- Liu Q., Lin X., Lin L., Yi L., Li H., Lin J.-M. Analyst. 2015;140(19):6736–6741. doi: 10.1039/c5an00944h. [DOI] [PubMed] [Google Scholar]
- Ma Y.-D., Li K.-H., Chen Y.-H., Lee Y.-M., Chou S.-T., Lai Y.-Y., Huang P.-C., Ma H.-P., Lee G.-B. Lab Chip. 2019;19(22):3804–3814. doi: 10.1039/c9lc00797k. [DOI] [PubMed] [Google Scholar]
- Mahalanabis M., Al-Muayad H., Kulinski M.D., Altman D., Klapperich C.M. Lab Chip. 2009;9(19):2811–2817. doi: 10.1039/b905065p. [DOI] [PubMed] [Google Scholar]
- Maharjan R.P., Ferenci T. Anal. Biochem. 2003;313(1):145–154. doi: 10.1016/s0003-2697(02)00536-5. [DOI] [PubMed] [Google Scholar]
- Mairhofer J., Roppert K., Ertl P. Sensors. 2009;9(6):4804–4823. doi: 10.3390/s90604804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hernandez F., Garcia-Heredia I., Lluesma Gomez M., Maestre-Carballa L., Martínez Martínez J., Martinez-Garcia M. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01226. 1226-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Botella J.R. RSC Adv. 2019;9(42):24440–24450. doi: 10.1039/c9ra04725e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauk M., Song J., Bau H.H., Gross R., Bushman F.D., Collman R.G., Liu C. Lab Chip. 2017;17(3):382–394. doi: 10.1039/c6lc01239f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.C., Duffy D.C., Anderson J.R., Chiu D.T., Wu H., Schueller O.J.A., Whitesides G.M. Electrophoresis. 2000;21(1):27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Mitamura K., Kawakami C., Shimizu H., Abe T., Konomi Y., Yasumi Y., Yamazaki M., Ichikawa M., Sugaya N. J. Infect. Chemother. 2013;19(4):633–638. doi: 10.1007/s10156-012-0533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou D., Tserepi A. Lab Chip. 2017;17(8):1388–1405. doi: 10.1039/c7lc00121e. [DOI] [PubMed] [Google Scholar]
- Moshgabadi N., Galli R.A., Daly A.C., Ko S.M.S., Westgard T.E., Bulpitt A.F., Shackleton C.R. J. Clin. Virol. 2015;71:67–72. doi: 10.1016/j.jcv.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Mullis K., Faloona F., Scharf S., Saiki R., Horn G., Erlich H. Cold Spring Harbor Symp. Quant. Biol. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- Nash M., Ramapuram J., Kaiya R., Huddart S., Pai M., Baliga S. Lancet Glob. Health. 2017;5(8):e754–e755. doi: 10.1016/S2214-109X(17)30247-4. [DOI] [PubMed] [Google Scholar]
- Nasseri B., Soleimani N., Rabiee N., Kalbasi A., Karimi M., Hamblin M.R. Biosens. Bioelectron. 2018;117:112–128. doi: 10.1016/j.bios.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuzil P., Campos C.D.M., Wong C.C., Soon J.B.W., Reboud J., Manz A. Lab Chip. 2014;14(13):2168–2176. doi: 10.1039/c4lc00310a. [DOI] [PubMed] [Google Scholar]
- Neuzil P., Novak L., Pipper J., Lee S., Ng L.F.P., Zhang C. Lab Chip. 2010;10(19):2632–2634. doi: 10.1039/c004921b. [DOI] [PubMed] [Google Scholar]
- Neuzil P., Reboud J. Anal. Chem. 2008;80(15):6100–6103. doi: 10.1021/ac800335q. [DOI] [PubMed] [Google Scholar]
- Ng A.H.C., Fobel R., Fobel C., Lamanna J., Rackus D.G., Summers A., Dixon C., Dryden M.D.M., Lam C., Ho M., Mufti N.S., Lee V., Asri M.A.M., Sykes E.A., Chamberlain M.D., Joseph R., Ope M., Scobie H.M., Knipes A., Rota P.A., Marano N., Chege P.M., Njuguna M., Nzunza R., Kisangau N., Kiogora J., Karuingi M., Burton J.W., Borus P., Lam E., Wheeler A.R. Sci. Transl. Med. 2018;10(438):1–13. doi: 10.1126/scitranslmed.aar6076. eaar6076. [DOI] [PubMed] [Google Scholar]
- Nguyen T.D., Markova S., Liu W., Gow J.M., Baldwin R.M., Habashian M., Relling M.V., Ratain M.J., Kroetz D.L. Pharmacogenomics J. 2012;13(5):396–402. doi: 10.1038/tpj.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemz A., Ferguson T.M., Boyle D.S. Trends Biotechnol. 2011;29(5):240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura M., Ikegami T., Saijo M., Kurane I., Miranda M.E., Morikawa S. J. Clin. Microbiol. 2001;39(9):3267–3271. doi: 10.1128/JCM.39.9.3267-3271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrup M.A. Technical digest of 7th Intl. Conf.on Solid-State Sensors and Actuators. 1993:924–926. [Google Scholar]
- Novak L., Neuzil P., Pipper J., Zhang Y., Lee S. Lab Chip. 2007;7(1):27–29. doi: 10.1039/b611745g. [DOI] [PubMed] [Google Scholar]
- Obeso C.G., Sousa M.P., Song W., Rodriguez-Pérez M.A., Bhushan B., Mano J.F. Colloid. Surface. 2013;416:51–55. [Google Scholar]
- Otter J.A., Donskey C., Yezli S., Douthwaite S., Goldenberg S.D., Weber D.J. J. Hosp. Infect. 2016;92(3):235–250. doi: 10.1016/j.jhin.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen E.A., Hong J.W., Quake S.R., Leadbetter J.R. Science. 2006;314(5804):1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- Papafragkou E., Hewitt J., Park G.W., Greening G., Vinjé J. PloS One. 2013;8(6) doi: 10.1371/journal.pone.0063485. e63485-e63485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.F., Wu H., Shu Y.Y., Yi P.Y., Deng Y.J., Lai X.M. Rev. Sci. Instrum. 2016;87(10):1–13. doi: 10.1063/1.4963907. 105120. [DOI] [PubMed] [Google Scholar]
- Pfankuche V.M., Hahn K., Bodewes R., Hansmann F., Habierski A., Haverkamp A.-K., Pfaender S., Walter S., Baechlein C., Postel A., Steinmann E., Becher P., Osterhaus A., Baumgärtner W., Puff C. Viruses. 2018;10(7):1–16. doi: 10.3390/v10070384. 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipper J., Inoue M., Ng L.F.P., Neuzil P., Zhang Y., Novak L. Nat. Med. 2007;13(10):1259–1263. doi: 10.1038/nm1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipper J., Zhang Y., Neuzil P., Hsieh T.-M. Angew. Chem., Int. Ed. Engl. 2008;47(21):3900–3904. doi: 10.1002/anie.200705016. [DOI] [PubMed] [Google Scholar]
- Powell L., Wiederkehr R.S., Damascus P., Fauvart M., Buja F., Stakenborg T., Ray S.C., Fiorini P., Osburn W.O. Analyst. 2018;143(11):2596–2603. doi: 10.1039/c8an00552d. [DOI] [PubMed] [Google Scholar]
- Přibylka A., Almeida A.V., Altmeyer M.O., Petr J., Ševčík J., Manz A., Neužil P. Lab Chip. 2013;13(9):1695–1698. doi: 10.1039/c3lc41305e. [DOI] [PubMed] [Google Scholar]
- Qi Z.B., Xu L.N., Xu Y., Zhong J.J., Abedini A., Cheng X., Sinton D. Lab Chip. 2018;18(24):3872–3880. doi: 10.1039/c8lc01109e. [DOI] [PubMed] [Google Scholar]
- Ramalingam N., San T.C., Kai T.J., Mak M.Y.M., Gong H.-Q. Microfluid. Nanofluidics. 2009;7(3):325–336. doi: 10.1007/s10404-008-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholt S.J., Baeumner A.J. Angew. Chem., Int. Ed. Engl. 2014;53(51):13988–14001. doi: 10.1002/anie.201309580. [DOI] [PubMed] [Google Scholar]
- Reperant L.A., Osterhaus A. Vaccine. 2017;35(35):4470–4474. doi: 10.1016/j.vaccine.2017.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert-Pöggeler K.R., Franzke K., Hipp K., Kleespies R.G. Front. Microbiol. 2019;9 doi: 10.3389/fmicb.2018.03255. 3255-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N.M., Linnes J.C., Andy F., Ellenson C.K., Pollock N.R., Klapperich C.M. Anal. Chem. 2015;87(15):7872–7879. doi: 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy F., Mendoza L., Hiebert J., McNall R.J., Bankamp B., Connolly S., Lüdde A., Friedrich N., Mankertz A., Rota P.A. J. Clin. Microbiol. 2017;55(3):735–743. doi: 10.1128/JCM.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavieh M., Kaul V., Khetani S., Singh A., Dhingra K., Kanakasabapathy M.K., Draz M.S., Memic A., Kuritzkes D.R., Shafiee H. Nanoscale. 2017;9(5):1852–1861. doi: 10.1039/c6nr06417e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahli M., Millot C., Gelin J.C., Barriere T. J. Mater. Process. Technol. 2013;213(6):913–925. [Google Scholar]
- Santiago G.A., Vázquez J., Courtney S., Matías K.Y., Andersen L.E., Colón C., Butler A.E., Roulo R., Bowzard J., Villanueva J.M. Nat. Commun. 2018;9(1):1–10. doi: 10.1038/s41467-018-03772-1. 1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterly N.G., Voorhees M.A., Ames A.D., Schoepp R.J. J. Clin. Microbiol. 2017;55(1):68–78. doi: 10.1128/JCM.01693-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher S., Nestler J., Otto T., Wegener M., Ehrentreich-Förster E., Michel D., Wunderlich K., Palzer S., Sohn K., Weber A. Lab Chip. 2012;12(3):464–473. doi: 10.1039/c1lc20693a. [DOI] [PubMed] [Google Scholar]
- Shetty P., Ghosh D., Paul D. J. Microbiol. Methods. 2017;143:1–5. doi: 10.1016/j.mimet.2017.09.013. [DOI] [PubMed] [Google Scholar]
- Shi C., Liu Q., Ma C., Zhong W. Anal. Chem. 2014;86(1):336–339. doi: 10.1021/ac4038043. [DOI] [PubMed] [Google Scholar]
- Song J., Mauk M.G., Hackett B.A., Cherry S., Bau H.H., Liu C. Anal. Chem. 2016;88(14):7289–7294. doi: 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speller N.C., Morbioli G.G., Cato M.E., Cantrell T.P., Leydon E.M., Schmidt B.E., Stockton A.M. Sensor. Actuator. B Chem. 2019;291:250–256. [Google Scholar]
- Storch G.A. Clin. Infect. Dis. 2000;31(3):739–751. doi: 10.1086/314015. [DOI] [PubMed] [Google Scholar]
- Stumpf F., Schwemmer F., Hutzenlaub T., Baumann D., Strohmeier O., Dingemanns G., Simons G., Sager C., Plobner L., Von Stetten F. Lab Chip. 2016;16(1):199–207. doi: 10.1039/c5lc00871a. [DOI] [PubMed] [Google Scholar]
- Sun Y., Kwok Y.C., Nguyen N.T. Lab Chip. 2007;7(8):1012–1017. doi: 10.1039/b700575j. [DOI] [PubMed] [Google Scholar]
- Tan J.J., Capozzoli M., Sato M., Watthanaworawit W., Ling C.L., Mauduit M., Malleret B., Grüner A.-C., Tan R., Nosten F.H. PLoS Negl. Trop. Dis. 2014;8(7):1–14. doi: 10.1371/journal.pntd.0003043. e3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. Asia-Pacific Biotech News. 2005;9(24):1316–1318. [Google Scholar]
- Tang R., Yang H., Gong Y., You M.L., Liu Z., Choi J.R., Wen T., Qu Z., Mei Q., Xu F. Lab Chip. 2017;17(7):1270–1279. doi: 10.1039/c6lc01586g. [DOI] [PubMed] [Google Scholar]
- Tang Y., Yu X., Chen H., Diao Y. Biosens. Bioelectron. 2016;86:255–261. doi: 10.1016/j.bios.2016.06.063. [DOI] [PubMed] [Google Scholar]
- Tanriverdi S., Chen L., Chen S. J. Infect. Dis. 2010;201(1):S52–S58. doi: 10.1086/650387. Suppl 1(8) [DOI] [PubMed] [Google Scholar]
- Temiz Y., Lovchik R.D., Kaigala G.V., Delamarche E. Microelectron. Eng. 2015;132:156–175. [Google Scholar]
- Thuy Tien T.T., Park H., Tuong H.T., Yu S.-T., Choi D.-Y., Yeo S.-J. Int. J. Mol. Sci. 2018;19(10):1–16. doi: 10.3390/ijms19103013. 3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek M.A., Repa J.J. Adv. Physiol. Educ. 2005;29(3):151–159. doi: 10.1152/advan.00019.2005. [DOI] [PubMed] [Google Scholar]
- van den Berk G.E.L., Frissen P.H.J., Regez R.M., Rietra P.J.G.M. J. Clin. Microbiol. 2003;41(8):3868–3869. doi: 10.1128/JCM.41.8.3868-3869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Munster V.J. Euro Surveill. 2013;18(38):7–10. doi: 10.2807/1560-7917.es2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Kinzler K.W. Proc. Natl. Acad. Sci. U. S. A. 1999;96(16):9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-H., Cheng L., Wang C.-H., Ling W.-S., Wang S.-W., Lee G.-B. Biosens. Bioelectron. 2013;41:484–491. doi: 10.1016/j.bios.2012.09.011. [DOI] [PubMed] [Google Scholar]
- Wang J., Kreutz J.E., Thompson A.M., Qin Y., Sheen A.M., Wang J., Wu L., Xu S., Chang M., Raugi D.N. Lab Chip. 2018;18(22):3501–3506. doi: 10.1039/c8lc00956b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.S., Maw M.M., Yu X.M., Dai B.W., Wang G., Jiang Z. Microfluid. Nanofluidics. 2017;21(3):1–16. 39. [Google Scholar]
- Wang R., Zhao R., Li Y., Kong W., Guo X., Yang Y., Wu F., Liu W., Song H., Hao R. Lab Chip. 2018;18(22):3507–3515. doi: 10.1039/c8lc00841h. [DOI] [PubMed] [Google Scholar]
- Wang S.Q., Sarenac D., Chen M.H., Huang S.H., Giguel F.F., Kuritzkes D.R., Demirci U. Int. J. Nanomed. 2012;7:5019–5028. doi: 10.2147/IJN.S32579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ostlund E.N., Jun Y., Nie F.-p., Li Y.-g., Johnson D.J., Lin R., Li Z.-g. Vet. J. 2016;212:27–35. doi: 10.1016/j.tvjl.2015.10.040. [DOI] [PubMed] [Google Scholar]
- Weinberg A., Walker M.L. Clin. Diagn. Lab. Immunol. 2005;12(3):367–370. doi: 10.1128/CDLI.12.3.367-370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Shresta S. Curr. Opin. Immunol. 2019;59:1–8. doi: 10.1016/j.coi.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Zhang J.H., Xu F.H., Wen X., Li P.F., Zhang X.L., Qiao S., Ge S.X., Xia N.S., Qian S.Z., Qiu X.B. Microfluid. Nanofluidics. 2017;21(3):1–9. 43. [Google Scholar]
- Xiao-Yan C., Li-Wen Q., Yu-Xian P., Kun W., Wei H., Li-Ya Z., Ya-Di W., Zhi-Yong L., Xu H., Cheng V.C.C. J. Clin. Microbiol. 2004;42(6):2629–2635. doi: 10.1128/JCM.42.6.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Zhang Z.-F., Wang L., Gao B., Pang D.-W., Wang H.-Z., Zhang Z.-L. Biomicrofluidics. 2012;6(3) doi: 10.1063/1.4756793. 34122-34122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Shibata H., Suzuki K., Citterio D. Lab Chip. 2017;17(7):1206–1249. doi: 10.1039/c6lc01577h. [DOI] [PubMed] [Google Scholar]
- Yang Y.L., Han Q.J., Hou Z.H., Zhang C., Tian Z.G., Zhang J. Cell. Mol. Immunol. 2017;14(5):465–475. doi: 10.1038/cmi.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh J.T., L H., Zheng S., Maldonado M.T. Vol. 4. 2018. pp. 6072–6075. (2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)). [DOI] [PubMed] [Google Scholar]
- Yetisen A.K., Akram M.S., Lowe C.R. Lab Chip. 2013;13(12):2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- Zanoli L., Spoto G. Biosens. 2013;3(1):18–43. doi: 10.3390/bios3010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei M. Biosens. Bioelectron. 2018;106:193–203. doi: 10.1016/j.bios.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang F., Wu J., Wang R., Wang L., Ying Y. Chem. Commun. 2014;50(61):8416–8419. doi: 10.1039/c4cc03011g. [DOI] [PubMed] [Google Scholar]
- Zhang H., Xu Y., Fohlerova Z., Chang H., Iliescu C., Neuzil P. Trends Anal. Chem. 2019;113:44–53. doi: 10.1016/j.trac.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Dai Y., Chen J., Hong L., Liu Y., Ke Q., Chen Y., Cai C., Liu X., Chen Z. J. Med. Virol. 2018;90(1):177–183. doi: 10.1002/jmv.24931. [DOI] [PubMed] [Google Scholar]
- Zhang R., Yao D., Chen J., Ye W., Ou X., Chen T., Sun B. J. Virol. Methods. 2018;257:79–84. doi: 10.1016/j.jviromet.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Zhang R.Q., Liu S.L., Zhao W., Zhang W.P., Yu X., Li Y., Li A.J., Pang D.W., Zhang Z.L. Anal. Chem. 2013;85(5):2645–2651. doi: 10.1021/ac302903p. [DOI] [PubMed] [Google Scholar]
- Zhao C., Liu X. Biomicrofluidics. 2016;10(2) doi: 10.1063/1.4945311. 024119-024119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Podesva P., Liu X., Zhang H., Teply T., Xu Y., Chang H., Qian A., Lei Y., Li Y. Sensor. Actuator. B Chem. 2020;303:1–7. doi: 10.1016/j.snb.2019.127098. [DOI] [PMC free article] [PubMed] [Google Scholar]