Abstract
Angiotensin-converting enzyme (ACE) plays diverse roles in the animal kingdom. However, whether ACE plays an immune function against viral infection in vector insects is unclear. In this study, an ACE gene (LsACE) from the small brown planthopper was found to respond to Rice stripe virus (RSV) infection. The enzymatic activities of LsACE were characterized at different pH and temperature. Twenty planthopper proteins were found to interact with LsACE. RSV infection significantly upregulated LsACE expression in the testicle and fat body. When the expression of LsACE in viruliferous planthoppers was inhibited, the RNA level of the RSV SP gene was upregulated 2-fold in planthoppers, and all RSV genes showed higher RNA levels in the rice plants consumed by these planthoppers, leading to a higher viral infection rate and disease rating index. These results indicate that LsACE plays a role in the immune response against RSV transmission by planthoppers.
Keywords: Rice stripe virus, Small brown planthopper, Angiotensin-converting enzyme, Enzyme activity, Immune function
Graphical abstract
1. Introduction
Angiotensin-converting enzyme (ACE, EC 3.4.15.1), also known as dipeptidyl carboxypeptidase, is well studied in the animal kingdom. The mammalian ACE is best known for its role in the renin–angiotensin system, which is involved in the regulation of blood pressure homeostasis (Bernstein et al., 2013). Angiotensin I is converted to the vasoconstrictor angiotensin II by ACE through the cleavage of a C-terminal dipeptide of angiotensin I. Apart from this, ACE takes part in thermoregulation (Schwimmer et al., 2004), hematopoiesis (Hubert et al., 2006), and immune responses (Bernstein et al., 2013). The mammalian ACE has two distinct isoforms, i.e., a double-domain somatic form and a single-domain testicular form (Bernstein et al., 2013).
Insects have an open circulatory hemolymph system that is considered to lack all components of the renin–angiotensin system except ACE homologs (Fournier et al., 2012). Insect ACE only has one isoform, which resembles the testicular form of the mammalian ACE (Lemeire et al., 2008). The biological functions of ACEs in insects are diversified. They are involved in larval growth and development (Isaac et al., 2007), metabolic inactivation and biosynthesis of neuropeptides (Isaac et al., 2009), digestion (Lemeire et al., 2008), regulation of male and female fecundity (Isaac et al., 1999; Ekbote et al., 2003; Sun et al., 2018), modulation of insect-plant interactions as effector proteins (Wang et al., 2015), and immune response (Aguilar et al., 2005; Macours et al., 2003; Duressa and Huybrechts, 2016). Regarding the roles of ACE in the innate immune system of insects, upregulation of ACE transcripts was observed in defense against both gram-negative and gram-positive bacteria but not against fungal infection in Locusta migratoria and Anopheles gambiae (Aguilar et al., 2005; Macours et al., 2003; Duressa and Huybrechts, 2016).
Rice stripe virus (RSV), a single-stranded RNA virus of the genus Tenuivirus, is one of the most destructive pathogens to rice and has caused serious rice yield loss in many East Asian countries (Toriyama, 1986). This virus is efficiently transmitted by the small brown planthopper Laodelphax striatellus in a persistent-propagative manner (Falk and Tsai, 1998). The genome of RSV contains four RNA segments. RNA1 is negative-sense and encodes the RNA-dependent RNA polymerase (RdRp). The other three segments are ambisense and encode NS2, NSvc2 (putative membrane glycoprotein), NS3 (gene silencing suppressor), CP (nucleocapsid protein), SP (disease-specific protein), and NSvc4 (movement protein) (Cho et al., 2013). As a persistent-propagative plant virus, RSV has a limited replication level in the vector insect under the surveillance of the insect immune system, such as pattern recognition molecules, immune-responsive effectors, reactive oxygen species, and the Toll pathway (Zhao et al., 2016, 2019a). In addition to these well-known immune pathways, the ACE gene of L. striatellus (LsACE) was found to response; its transcript level was upregulated in viruliferous planthoppers (Zhang et al., 2010). However, whether this ACE gene plays an immune response function against RSV infection in the vector insect is unclear.
The aim of this study was to determine the effect of LsACE on RSV infection in the vector insect. The enzymatic dynamics of in vitro expressed LsACE were characterized. The planthopper proteins that may interact with LsACE were identified using yeast two-hybrid screening. The temporal and spatial expression patterns of LsACE in nonviruliferous and viruliferous planthoppers were revealed. Finally, RNAi-based knockdown of LsACE was performed to determine the effect of LsACE on RSV infection in planthoppers and rice plants.
2. Materials and methods
2.1. Small brown planthoppers and rice plants
The viruliferous and nonviruliferous small brown planthopper strains used in this study were established from a field population collected in Hai'an, Jiangsu Province, China. The two strains were reared separately in the laboratory on 2-cm to 3-cm seedlings of rice, Oryza sativa Huangjinqing, in glass incubators at 25 °C with 16 h of light daily as described previously (Zhao et al., 2016). To maintain the RSV-carrying frequency of the viruliferous strain at no less than 90%, nonviruliferous individuals were identified and eliminated via dot-ELISA using the monoclonal anti-CP antibody every three months (Zhao et al., 2016).
2.2. RNA isolation and cDNA synthesis
Total RNA was isolated from planthoppers, six tissues (brain, salivary gland, gut, fat body, ovary, and testicle), or rice leaves following the standard TRIzol reagent protocol (Invitrogen, Carlsbad, CA, USA). The concentration and quality of total RNA were determined using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and by gel electrophoresis. RNA was treated using DNase I (Qiagen, Valencia, CA, USA) to remove genomic DNA contamination before being used for cDNA synthesis. RNA (1 μg) was reverse transcribed to cDNA using MLV reverse transcriptase (Promega, Madison, WI, USA) and random primers or oligo dT primers following the manufacturer's instructions.
2.3. Gene cloning and sequence analysis
Based on the 254 bp fragment of ACE from the L. striatellus transcriptome (Zhang et al., 2010), specific primers 5′-RACE-in-p, 5′-RACE-ex-p, 3′-RACE-in-p, and 3′-RACE-ex-p (Table S1) were designed to obtain the 5′-terminal and 3′-terminal sequences by 5′ RACE and 3′ RACE using SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA) according to the manufacturer's instructions. Based on the RACE results, the open reading frame (ORF) of LsACE was amplified from the cDNA library of L. striatellus with primers LsACE-ORF-F/LsACE-ORF-R (Table S1). The PCR product was subcloned into the pGEM-T easy vector (Promega) and transfected into DH5α cells for sequencing. The protein sequence was determined from the sequenced ORF. Possible secretory signal peptides and transmembrane helices were predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) and TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). The theoretical molecular weight and isoelectric point of mature LsACE were computed in ExPASy (http://web.expasy.org/compute_pi/). Possible glycosylation sites were predicted in NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/). The protein sequence of LsACE was aligned with the homologs of Drosophila melanogaster using ClustalW at EBI (http://www.ebi.ac.uk/Tools/msa/clustalw2/). An unrooted tree was constructed with the neighbor-joining method using pairwise deletion and the p-distance model in Mega 7.0 software. Bootstrap analysis with 1000 replicates was performed to evaluate the internal support of the tree topology.
2.4. Protein expression and purification
A 2052 bp LsACE fragment from amino acid residues 50 to 716 was amplified using the primers LsACE-EXPS-F/LsACE-EXPS-R and then subcloned into the pET28a vector to generate His-tag recombinant plasmids. The recombinant plasmids were transformed to Escherichia coli strain Rosetta for expression. After 4 h induction with 0.8 mM isopropyl β-D-thiogalactoside (IPTG) at 37 °C, the cells were pelleted by centrifugation and sonicated for 30 min in ice water. The expressed recombinant LsACE was purified from the supernatant using Ni Sepharose (GE Healthcare, Little Chalfont, Buckinghamshire, UK) with different concentrations of imidazole following the manufacturer's instructions. The purified LsACE was dissolved in 80 mM HEPES buffer using Amicon Ultra Centrifugal Filters (10 kDa cut-off) (Millipore, Burlington, MA, USA) after centrifugation at 3000 rpm for 10 min at 4 °C. The empty pET28a vector was used in the same procedure as the negative control.
2.5. ACE activity assay
The enzyme activity of in vitro expressed LsACE was evaluated by quantifying the hydrolysis of N-[3-(2-furyl)acryloyl]-L-phenylalanyl-glycylglycine (FAPGG), which is one of the most widely used ACE substrates, to N-(3-[2-furyl]acryloyl)-L-phenylalanine (FAP) and glycylglycine (GG) using a modified method described by Murray et al. (2004). The standard curves of FAPGG (Sigma-Aldrich, Santa Clara, CA, USA) and FAP + GG (equal molar concentration) (Sigma-Aldrich) were constructed with a series of concentrations from 0 to 1.0 mM in a buffer containing 80 mM HEPES, 300 mM NaCl and 0.1 mM ZnCl2, pH 8.0. Three replicates for each concentration were prepared. The linear regression equation, along with the coefficient of determination (R2), was used to evaluate the standard curves. The change in the extinction coefficient (Δε), i.e., the maximum absorbance change at 340 nm due to the complete hydrolysis of 1 mM FAPGG to FAP and GG, was obtained from the two standard curves. The absorbance at 340 nm was recorded on a SpectraMax Paradigm Multi-Mode Microplate Reader (Molecular Devices, San Jose, CA, USA).
The purified LsACE protein was dissolved in 80 mM HEPES buffer to yield the LsACE solution at a final concentration of 65 μg/mL. The substrate solution contained 80 mM HEPES, 300 mM NaCl, 0.1 mM ZnCl2 and 1 mM FAPGG. The reaction was carried out in one well of a 96-well plate, where 30 μL of LsACE solution was mixed with 200 μL of substrate solution. After incubation for 30 min, the reaction was terminated by the addition of 20 μL of 100 mM ethylene diamine tetraacetic acid (Merck, Kenilworth, NJ, USA). A pH range from 4.0 to 10 at 25 °C and a series of temperatures, 20, 25, 30, 35, 40, and 45 °C, at pH 8.0 were tested to determine the optimal pH and temperature for LsACE activity. Five replicates for each pH and temperature were performed. The activity of LsACE was computed using Equation (1):
| ACE activity (units/L)=(Vt×1000)/(Δε×Vs×d)×ΔA/min | (1) |
where Vt = the final assay volume (250 μL); V s = the volume of ACE solution (30 μL); d = light path (cm); Δε = the maximum absorbance change at 340 nm due to the complete hydrolysis of 1 mM FAPGG to FAP and GG; and ΔA/min = decrease in absorbance at 340 nm per minute. Differences were statistically evaluated using one-way ANOVA followed by Tukey's test for multiple comparisons in SPSS 17.0 software.
An appropriate amount of LsACE was incubated with a range of concentrations of FAPGG solutions at 25 °C and pH 6.0. The concentration of FAPGG was quantified using the standard curve for the theoretical breakdown of FAPGG to FAP and GG. Five replicates were carried out for each substrate concentration. The kinetic constants V max and K m were computed with HYPER software according to the Lineweaver-Burk method and presented as the mean ± SE.
2.6. Yeast two-hybrid analysis
Yeast two-hybrid screening was performed with the Matchmaker™ GAL4 Two-Hybrid System 3 & Libraries (Clontech) according to the manufacturer's protocol. A whole-body cDNA library of the small brown planthopper was constructed using the SMART™ cDNA Library Construction Kit (Clontech), normalized with the Trimmer-Direct cDNA Normalization Kit (Evrogen, Moscow, Russia), and then subcloned into the vector pGADT7 (Clontech) to create prey plasmids. The mature LsACE without secretory signal peptide was subcloned into the vector pGBKT7 (Clontech) to create the pGBKT7-LsACE bait plasmid. MaV203 yeast cells were cotransformed with pGBKT7-LsACE and the pGADT7 library. Positive clones were selected on triple dropout medium (SD/-Leu/-Trp/-His) and quadruple dropout medium (SD/-Leu/-Trp/-His/-Ade). Prey plasmids were isolated from these clones for sequencing. To confirm the interaction of bait and prey proteins, we cotransformed the two plasmids into the yeast strain AH109 and repeated the selection on triple and quadruple dropout media. The prey sequences were used in a BLAST search of the gene set of the small brown planthopper (Zhu et al., 2017) and annotated by National Center for Biotechnology Information (NCBI).
2.7. Real-time quantitative PCR
A 168 bp fragment of LsACE, 142 bp fragment of RdRp, 142 bp fragment of NS2, 148 bp fragment of NSvc2, 129 bp fragment of NS3, 60 bp fragment of CP, 152 bp fragment of SP, and 128 bp fragment of NSvc4 were amplified using real-time quantitative PCR (qPCR) to quantify the relative RNA levels of respective genes in planthoppers and rice plants. A 64 bp fragment of the planthopper elongation factor 2 gene (EF2, Wang et al., 2017) and a 185 bp fragment of the rice ubiquitin 5 gene (AK061988) were quantified to normalize the cDNA templates of insect and plant samples, respectively. The primers are listed in Table S1 qPCR was performed on a Light Cycler 480 II (Roche, Basel, Switzerland). The thermal cycling conditions were 95 °C for 2 min, 40 cycles of 95 °C for 20 s, 58 °C for 20 s and 68 °C for 20 s, followed by one cycle of 95 °C for 30 s, 58 °C for 30 s and 95 °C for 10 s to collect the melting curve. The relative RNA level of each gene to that of EF2 or ubiquitin 5 was reported as the mean ± SE. Differences were statistically evaluated either by Student's t-test to compare two means or one-way ANOVA followed by Tukey's test for multiple comparisons in SPSS 17.0 software.
2.8. Quantification of LsACE expression in various tissues and developmental stages of viruliferous and nonviruliferous planthoppers
Brain, salivary gland, gut, fat body, ovary, and testicle were dissected from 20 to 50 viruliferous and nonviruliferous adults of planthoppers, and 6 biological replicates for each tissue were prepared for RNA extraction. Five or ten individuals from each nymphal stage and female and male adults of viruliferous and nonviruliferous insects were prepared for RNA extraction, and 6 biological replicates were performed. qPCR was performed to quantify and compare the transcript levels of LsACE in viruliferous and nonviruliferous samples.
2.9. Double-stranded RNA synthesis and delivery
PCR primers with T7 promoter sequences, LsACE-dsRNA-F and LsACE-dsRNA-R, were used to prepare 559 bp double-stranded RNA (dsRNA) of LsACE (Table S1). A 420 bp dsRNA for green fluorescent protein (GFP) was amplified using primers GFP-dsRNA-F and GFP-dsRNA-R as negative controls (Table S1). dsRNA was generated using T7 RiboMAX Express RNAi System (Promega) and purified using Wizard SV Gel and PCR Clean-Up System (Promega) following the manufacturer's protocol. 23 nL of dsRNAs at 6 μg/μL was infected into the fourth instar of nymphs. The dsRNAs were delivered into the hemolymph in the ventral thorax by microinjection through a glass needle using Nanoliter 2000 (World Precision Instruments, Sarasota, Florida, USA).
2.10. Effects of LsACE on RSV infection in planthoppers and rice plants
After injection of dsLsACE-RNA for 5 d in viruliferous planthoppers, the RNA levels of seven RSV genes and LsACE were measured using qPCR. Insects injected with dsGFP-RNA were used as controls. Twelve biological replicates and 5 insects per replicate were prepared for treatment or control groups. On the fifth day after injection of dsLsACE-RNA or dsGFP-RNA, 10 viruliferous planthoppers were transferred to 5 rice seedlings for 4 d of feeding and then removed from the plants. One batch of plants was collected for measurement of the RNA levels of seven RSV genes using qPCR. Twenty biological replicates and 5 seedlings per replicate were prepared. Another batch of plants was cultured continually for disease symptom observation. The infection rate (IR) and the disease rating index (DRI) were calculated at 10 d, 13 d, and 16 d after viral inoculation (DAI). The IR was the ratio of the number of plants showing RSV symptoms to the total number of inoculated plants. The DRI was scored based on the severity of disease symptoms according to the definition described by Washio et al. (1967). Three biological replicates with 20–42 seedlings per replicate were used to calculate IRs and DRIs.
2.11. Western blot assay
The variation of viral proteins in planthoppers and rice plants was checked with Western blot assay after injection of dsLsACE-RNA in viruliferous planthoppers as described in 2.10. Total protein was extracted from planthoppers using T-PER® Tissue Protein Extraction Reagent (Thermo Fisher Scientific) and from plants using Plant Protein Extraction Reagent (CWBIO, Beijing, China) containing a protease inhibitor cocktail (CWBIO). The amounts of NS3, CP, SP, and NSvc4 were measured using homemade monoclonal anti-CP, anti-SP, anti-NS3 antibodies and polyclonal anti-NSvc4 antibody (Zhao et al., 2019b). Beta-tubulin of the small brown planthoppers and beta-actin of the rice were applied as the internal controls against the monoclonal anti-beta-tubulin antibody (CWBIO) and monoclonal anti-beta-actin antibody (Sungene Biotech, Tianjin, China). The immune signal was visualized using Image Station 4000 MM ProCFL (Carestream, Rochester, NY, USA). The density of viral proteins was quantified with image analysis software ImageJ and normalized to that of beta-tubulin or beta-actin. Differences were statistically evaluated by Student's t-test in SPSS 17.0 software.
3. Results
3.1. LsACE cDNA and amino acid sequence
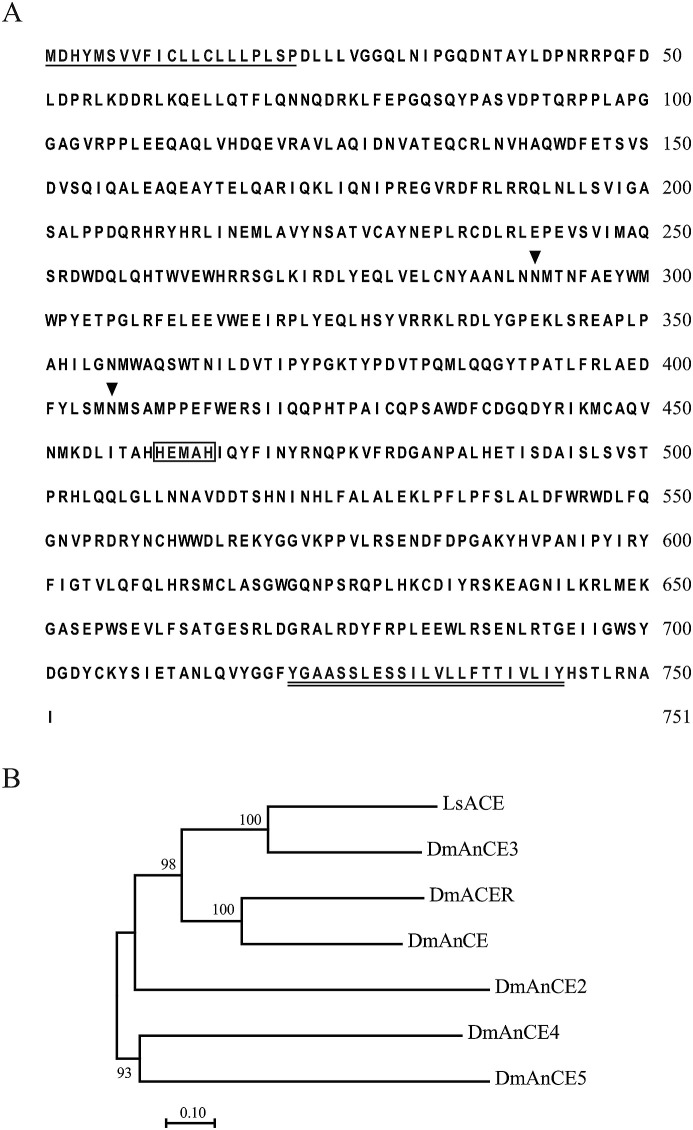
The full-length transcript of LsACE (MK387339 in GenBank) was obtained using the RACE technique. The sequence contained a 174 bp and 273 bp untranslated region at the 5′ and 3’ terminals, respectively. The 2256 bp open reading frame encoded a protein of 751 amino acid residues. A 21-amino-acid secretory signal peptide at the N-terminal and a 23-amino-acid transmembrane helix (position 726 to 748) at the C-terminal were predicted, indicating that LsACE was probably a membrane protein. Two putative N-glycosylation sites were identified at Asn291 and Asn406. The activity site motif “HEMAH” (“HEXXH”, position 460 to 464) was identified (Fig. 1 A). The mature LsACE without the secretory signal peptide had a theoretical molecular mass of 84.6 kDa and an isoelectric point of 5.84. Phylogenetic analysis showed that LsACE was most similar to D. melanogaster ACE3 (DmAnCE3) among the six D. melanogaster homologs (Fig. 1B).
Fig. 1.
Sequence characteristics of LsACE. (A) The amino acid sequence of LsACE. The secretory signal peptide is underlined. N-glycosylation sites are marked with triangles. Active site sequences are boxed. The transmembrane region at the C-terminal is underlined with two lines. (B) The phylogenetic tree of LsACE and six homologs of Drosophila melanogaster constructed with the neighbor-joining method using pairwise deletion and the p-distance model. Bootstrap analysis with 1000 replicates was performed. The accession numbers for D. melanogaster ACE homologs are NP_477195.1 for DmACER, NP_477046.1 for DmAnCE, NP_788042.1 for DmAnCE2, NP_001033904.1 for DmAnCE3, NP_610442.2 for DmAnCE4, and NP_573392.2 for DmAnCE5.
3.2. Enzymatic activity of LsACE
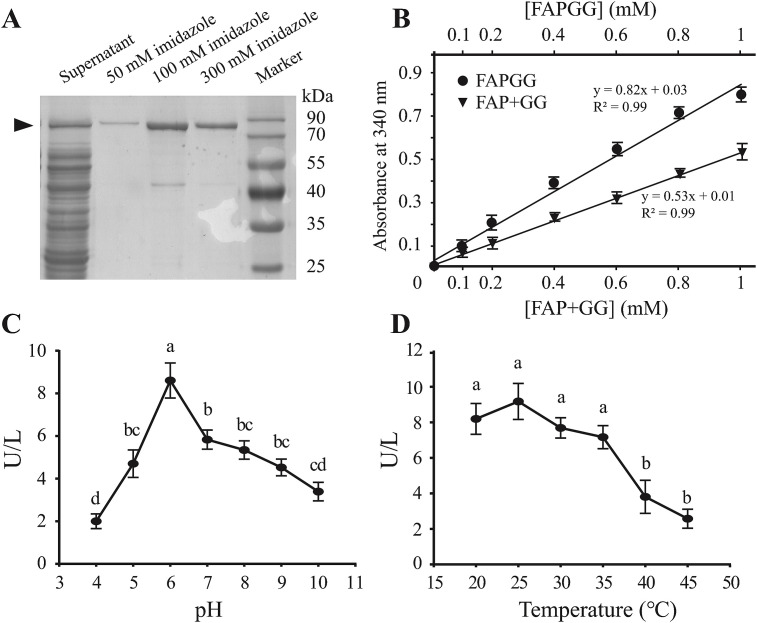
To characterize the enzymatic activity of LsACE, the fragment from amino acid residues 50 to 716 (without the secretory signal peptide and transmembrane helix) was expressed in E. coli cells and purified. The molecular weight of the His-tagged recombinant protein was 79.5 kDa (Fig. 2 A). The hydrolysis activity of LsACE from FAPGG to FAP and GG was measured based on standard curves (Fig. 2B). The coefficient of determination (R2) was over 0.98 for each standard curve. In the pH range from 4.0 to 12, the enzyme activity increased from 2.0 U/L at pH 4.0–8.6 U/L at pH 6.0 and then dropped at pH 7 (Fig. 2C). LsACE maintained a high activity between 7.2 U/L and 9.2 U/L in a range of temperatures from 20 °C to 35 °C, while the activity remarkably decreased at 40 or 45 °C (Fig. 2D). Thus, the optimal pH and temperature for LsACE activity were pH 6.0 and from 20 °C to 35 °C, with the best activity at 25 °C. The kinetic constant V max was 38.5 ± 3.3 μmol min−1 mg−1, and K m was 36.1 ± 1.4 μM at pH 6.0 and 25 °C.
Fig. 2.
Enzymatic activity of LsACE. (A) SDS-PAGE of the recombinantly expressed and purified LsACE fragment from amino acid residues 50 to 716. The arrow indicates the 79.5 kDa His-tagged recombinant protein that was eluted from Ni Sepharose using different concentrations of imidazole. (B) Standard curves of N-[3-(2-furyl)acryloyl]-L-phenylalanyl-glycylglycine (FAPGG) and a mixture of equal molar N-(3-[2-furyl]acryloyl)-L-phenylalanine (FAP) and glycylglycine (GG) measured at 340 nm. Points represent the means ± SE of three replicates. The linear regression equation and coefficient of determination (R2) for each curve are presented. (C) Enzymatic activity of LsACE at different pH values. (D) Enzymatic activity of LsACE at different temperatures. Points represent means ± SE of five replicates. Differences were statistically evaluated using one-way ANOVA followed by Tukey's test for multiple comparisons. Significant differences are indicated by different lowercase letters.
3.3. Potential proteins that interact with LsACE in planthoppers
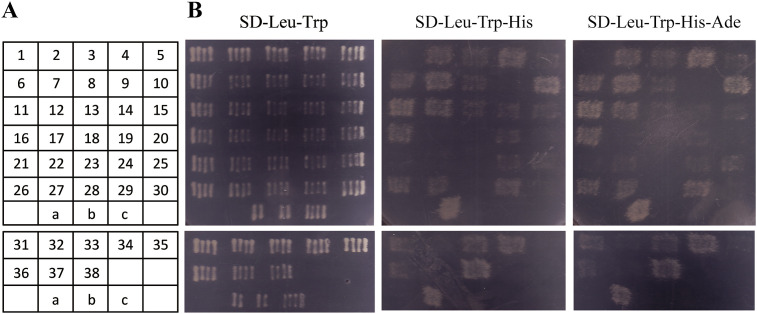
A whole-body cDNA library of the small brown planthopper was constructed and expressed in yeast. The potential interactive proteins were screened using LsACE as bait in yeast two-hybrid analysis. Forty positive clones were selected on triple dropout medium (SD/-Leu/-Trp/-His) or quadruple dropout medium (SD/-Leu/-Trp/-His/-Ade), among which 38 clones were successfully sequenced. The interaction between LsACE and the 38 clones was further verified with the yeast two-hybrid system, and only 29 clones were confirmed to interact with LsACE (Fig. 3 ). After alignment with the gene set of the small brown planthopper, 28 clones encoding 20 different proteins were found in the gene set (Table 1 ). Five clones encoded an unknown protein. AMP deaminase 2, DnaJ-like protein subfamily A member 2, NADH-ubiquinone oxidoreductase 49 kDa subunit, and an unknown protein each were retrieved from two clones. Other known proteins included 2-oxoisovalerate dehydrogenase subunit beta, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-like, aspartate-tRNA ligase, cob(I)yrinic acid a,c-diamide adenosyltransferase, kynurenine-oxoglutarate transaminase 3, endocuticle structural glycoprotein SgAbd-2, RR1 cuticle protein 2 precursor, macrophage migration inhibitory factor, putative proteasome inhibitor, calcium-binding protein, integrator complex subunit 9, and clustered mitochondria protein.
Fig. 3.
Yeast two-hybrid assay to screen planthopper proteins that putatively interact with LsACE. (A) Numbers and positions of the 38 clones that were further verified with the yeast two-hybrid system after screening the cDNA library of a small brown planthopper using LsACE as bait. The clones with red numbers are deemed to interact with LsACE in Fig. 3B a, pGBKT7-53 + pGADT7-T, positive control; b, pGBKT7-Lam + pGADT7-T, negative control; c, pGBKT7-LsACE + pGADT7, self-activation. (B) Verification of the interaction between LsACE and the 38 clones on triple dropout medium (SD/-Leu/-Trp/-His) and quadruple dropout medium (SD/-Leu/-Trp/-His/-Ade) in the yeast two-hybrid system. Only 29 clones were determined to interact with LsACE on the quadruple dropout medium and encoded 20 different proteins (Table 1).
Table 1.
Annotations of the Laodelphax striatellus proteins that putatively interact with LsACE screened by the yeast two-hybrid assay.
| No. of clones∗ | Genome ID# | Annotation |
|---|---|---|
| 2 | evm.model.Contig112.94 | Calcium-binding protein |
| 3 | evm.model.Contig 30.97 | Kynurenine-oxoglutarate transaminase 3 |
| 4,25 | evm.model.Contig 33.13 | DnaJ-like protein subfamily A member 2 |
| 5 | evm.model.Contig 1409.2 | Aspartate-tRNA ligase, cytoplasmic |
| 6,29 | evm.model.Contig 84.28 | NADH-ubiquinone oxidoreductase 49 kDa subunit |
| 7 | evm.model.Contig 47.45 | Unknown protein |
| 10 | evm.model.Contig 348.76 | Cob(I)yrinic acid a,c-diamide adenosyltransferase, mitochondrial |
| 11,38 | evm.model.Contig 40.65 | Unknown protein |
| 12 | evm.model.Contig 374.60 | Endocuticle structural glycoprotein SgAbd-2 |
| 13 | evm.model.Contig 1058.2 | Integrator complex subunit 9 |
| 14,15,19,24,26 | evm.model.Contig 309.3 | Unknown protein |
| 16,27 | evm.model.Contig 323.46 | AMP deaminase 2 |
| 20 | evm.model.Contig 78.125 | 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial |
| 21 | evm.model.Contig 42.39 | Unknown protein |
| 22 | evm.model.Contig 0.394 | Macrophage migration inhibitory factor homolog |
| 31 | evm.model.Contig 92.72 | Putative proteasome inhibitor |
| 32 | evm.model.Contig 18.302 | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-like |
| 33 | evm.model.Contig112.100 | RR1 cuticle protein 2 precursor |
| 34 | evm.model.Contig 440.4 | Clustered mitochondria protein homolog |
| 36 | evm.model.Contig 192.23 | Uncharacterized protein LOC105272240 |
*The numbers of clones are same as in Fig. 3A.
3.4. Temporal and spatial expression of LsACE in nonviruliferous and viruliferous planthoppers
The transcript levels of LsACE in various tissues and developmental stages were quantified and compared between nonviruliferous and viruliferous planthoppers using qPCR. LsACE was mainly expressed in the brain, salivary gland, and testicle, while it had a lower expression level in the fat body, gut, and ovary (Fig. 4 A). RSV infection increased LsACE expression in the testicle and fat body (Fig. 4A). At different developmental stages, LsACE showed a higher expression level in male than in female adult and nymphal stages (Fig. 4B). However, RSV infection significantly upregulated LsACE expression in nymphal stages but not in adult stages (Fig. 4B).
Fig. 4.
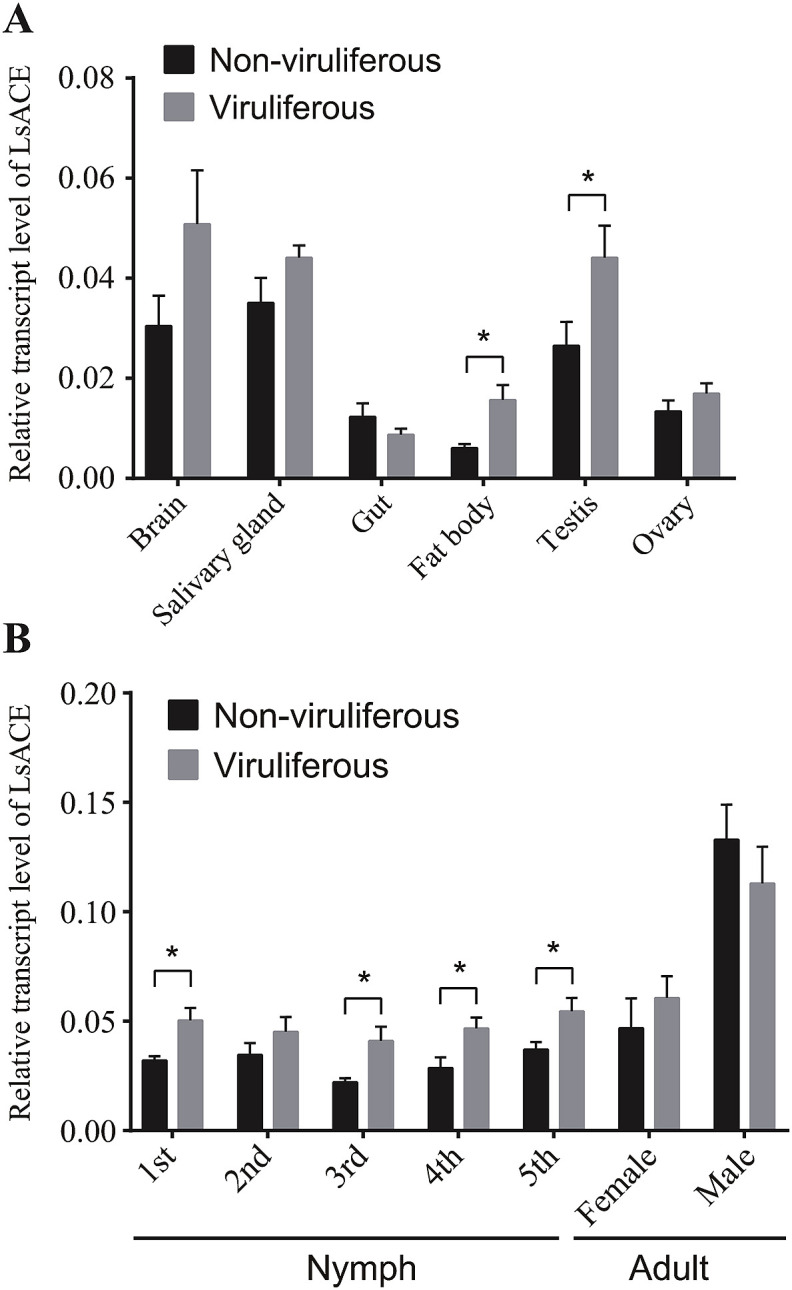
Temporal and spatial expression of LsACE in viruliferous and nonviruliferous planthoppers. (A) Relative transcript levels of LsACE in the brain, salivary glands, gut, fat body, testis and ovary. (B) Relative transcript levels of LsACE in different developmental stages. The transcript level of LsACE was normalized to that of the elongation factor 2 transcript and was represented as the mean ± SE. Differences were statistically evaluated by Student's t-test to compare two means. *, P < 0.05.
3.5. Effect of LsACE on RSV infection in planthoppers and rice plants
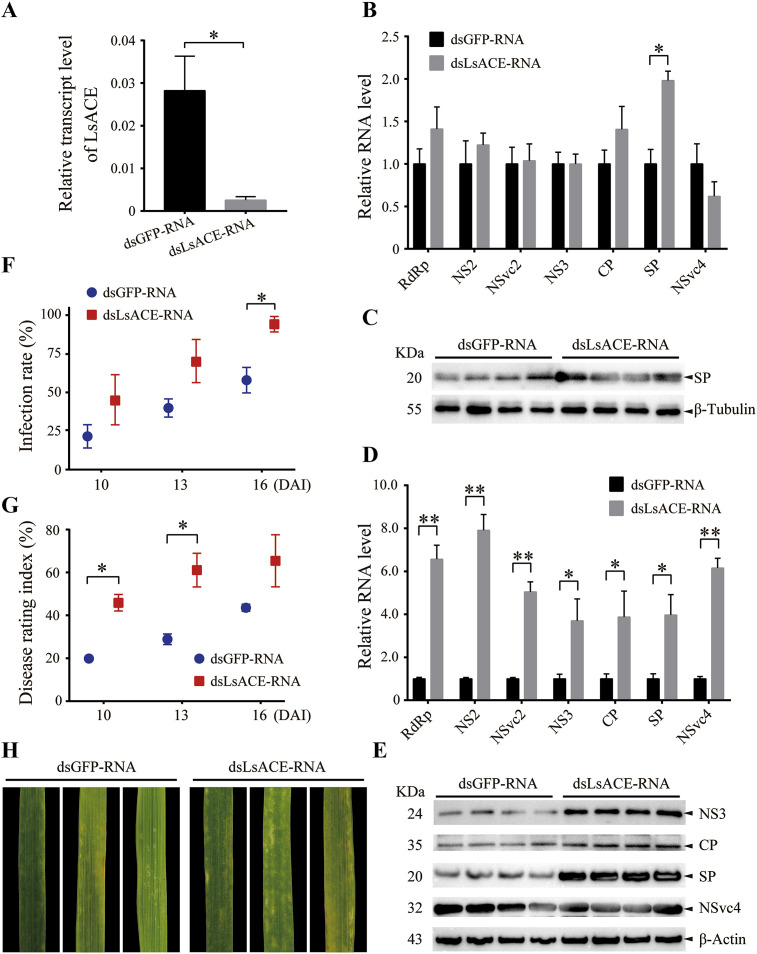
To explore the influence of LsACE on RSV infection in planthoppers, the expression of LsACE in viruliferous planthoppers was inhibited by injection of dsLsACE-RNA. After 5 d, the RNA levels of seven RSV genes and LsACE in planthoppers were measured using qPCR. Compared to the control group, which was injected with dsGFP-RNA, the transcript level of LsACE decreased by 80% (Fig. 5 A). The RNA level of the SP gene was significantly upregulated by 2-fold, while other RSV genes did not show changes when the expression of LsACE was inhibited (Fig. 5B). The protein level of SP only had an increasing tendency without statistical significance (Fig. 5C, Supplemental Fig. 1A).
Fig. 5.
Effect of LsACE on RSV infection in planthoppers and rice plants. (A) Relative transcript level of LsACE to that of elongation factor 2 5 d after dsLsACE-RNA or dsGFP-RNA injection in planthoppers. (B) Relative RNA levels of seven viral genes to that of elongation factor 2 5 d after dsLsACE-RNA or dsGFP-RNA injection in viruliferous planthoppers. (C) Western blot to show the protein levels of SP in viruliferous planthoppers after dsLsACE-RNA or dsGFP-RNA injection using monoclonal anti-SP antibodies. Beta-tubulin of the small brown planthoppers was applied as the internal control against the monoclonal anti-beta-tubulin antibody. (D) Relative RNA levels of seven viral genes to that of ubiquitin 5 in rice plants 4 d after consumed by dsLsACE-RNA or dsGFP-RNA-injected viruliferous planthoppers. (E) Western blot to show the protein levels of NS3, CP, SP, and NSvc4 in rice plants 4 d after consumed by dsLsACE-RNA or dsGFP-RNA-injected viruliferous planthoppers using monoclonal anti-CP, anti-SP, anti-NS3 antibodies and polyclonal anti-NSvc4 antibody. Beta-actin of the rice was applied as the internal control against the monoclonal anti-beta-actin antibody. (F) Infection rates of the rice plants consumed by dsLsACE-RNA or dsGFP-RNA-injected viruliferous planthoppers at 10, 13, and 16 d after viral inoculation (DAI). (G) Disease rating index of the rice plants consumed by dsLsACE-RNA or dsGFP-RNA-injected viruliferous planthoppers at 10, 13, and 16 DAI. (H) Disease symptoms of the rice leaves consumed by dsLsACE-RNA or dsGFP-RNA-injected viruliferous planthoppers at 16 DAI. Values represent means ± SE. Differences were statistically evaluated by Student's t-test to compare two means. *, P < 0.05; **, P < 0.01.
To test whether LsACE affected RSV infection in rice plants, the RNA levels of the seven RSV genes from the rice plants consumed by dsLsACE-RNA-injected viruliferous planthoppers were measured using qPCR and compared to those consumed by dsGFP-RNA-injected planthoppers at 4 DAI. All RSV genes showed higher RNA levels in the rice consumed by dsLsACE-RNA-injected planthoppers, indicating a higher viral activity (Fig. 5D). The protein levels of NS3, CP, SP, and NSvc4 were also checked with Western blot assay. Only NS3 and SP proteins showed a significantly elevated level in the rice plants consumed by these planthoppers (Fig. 5E, Supplemental Fig. 1B). The reaction of rice to RSV infection was also evaluated. The infection rate (IR) and the disease rating index (DRI) were calculated at 10, 13, and 16 DAI. A higher IR at 16 DAI and a higher DRI at 10 and 13 DAI were observed in the rice consumed by dsLsACE-RNA-injected planthoppers compared to the control group (Fig. 5F and G), reflecting a higher number of plants with more severe disease symptoms (Fig. 5H).
From these data, we conclude that the lower LsACE expression promoted the expression of the RSV SP gene in planthoppers and thus facilitated RSV infection in rice plants. Therefore, LsACE plays a role in immune response against RSV in planthoppers.
4. Discussion
In this study, we presented evidence to propose a possible immune function of an ACE of the small brown planthopper during the transmission of RSV. RSV infection upregulated the expression of LsACE in the fat body, an immune organ of insects. RNAi-based knockdown of LsACE led to a significant upregulation of viral SP gene expression in planthoppers and a more severe viral infection in rice. This finding expands our knowledge of the physiological functions of ACE in insects and is the first report of a relationship between ACE and plant virus in vector insects.
The immune function of LsACE in RSV infection of vector insects is different from that of the mammalian renin–angiotensin system in the pathogenesis of various human viruses. The expression of LsACE is activated by RSV, and the increase in LsACE is not in favor of the viral infection, as shown by inhibited viral SP expression. Cytoplasmic viral inclusions comprise SP that directly interacts with the ribonucleoprotein particles of RSV; the knockdown of SP expression significantly limited the spread of RSV in the infected insects (Wu et al., 2014). In our study, overexpression of SP due to the knockdown of LsACE in planthoppers led to a more serious disease incidence in rice plants, suggesting a higher viral transmission efficiency from vector insects to host plants. Therefore, the immune function of LsACE may be embodied in blocking the RSV spread in vector insects. In contrast, the SARS coronavirus and avian influenza viruses H5N1 and H7N9 share a common regulation on the mammalian renin–angiotensin system, namely, the elevation of angiotensin II levels in parallel with reduced ACE2 and sometimes enhanced ACE activities (Kuba et al., 2005; Wösten-van Asperen et al., 2013; Zou et al., 2014; Huang et al., 2014). The plasma level of angiotensin II is strongly correlated with the disease severity of acute respiratory distress syndrome. Angiotensin I is cleaved by ACE to produce angiotensin II and is further converted to angiotensin 1-7 by ACE2 (Imai et al., 2010). Therefore, these two enzymes have opposite functions in the mammalian renin–angiotensin system. Activation of the renin–angiotensin system in viral acute respiratory distress syndrome and in RSV-infected vector insects produces a different effect on viral infection. This is probably due to differences in the substrates that are catalyzed by the two enzymes in mammals and insects.
LsACE may have extensive protein interactions. As a member of the M2 metalloprotease family, ACE is usually involved in the digestion of small hormonal peptides and neuropeptides, such as leucokinin, locustatachykinin, and allatostatin in insects (Lamango et al., 1997). We identified 20 proteins that putatively interact with LsACE. Of these proteins, several were associated with immune and inflammatory reactions. Kynurenine-oxoglutarate transaminase 3 (EC 2.6.1.7), also known as kynurenine aminotransferase III, is a component of the kynurenine pathway that is partially involved in neuroactive and immunomodulatory metabolites (Asp et al., 2011). The DnaJ-like protein subfamily A member 2 is generally considered a potential target of the immune response in rheumatoid arthritis (Kotlarz et al., 2013). AMP deaminase 2 regulates the accumulation of extracellular ATP, which provokes pro-inflammation and immune effects in systemic lupus erythematosus (Guo et al., 2018). The RR1 cuticle protein 2 precursor may take part in viral transmission because the homologous cuticle proteins in the small brown planthopper (CPR1) and in the aphid Schizaphis graminum were found to be involved in the viral transmission process (Liu et al., 2015; Cilia et al., 2011). The macrophage migration inhibitory factor has a central role as a regulator of innate immune and inflammatory responses in human sepsis and other inflammatory diseases (Baumann et al., 2003). Considering that these proteins are not small peptides, we suggest that LsACE directly binds these proteins to affect their functions instead of cleaving them. One or more immune pathways could be targeted by LsACE.
LsACE shows typical dipeptidyl carboxypeptidase activity. LsACE is most closely related in sequence to D. melanogaster AnCE3. Although both ACEs have the HExAH active site motif, recombinantly expressed D. melanogaster AnCE3 does not show enzymatic activities using Hip-His-Leu or Locustatachykinin-1 as substrates (Isaac et al., 2007), while LsACE is able to cleave the C-terminal dipeptide of FAPGG. The K m value of LsACE towards FAPGG is 36.1 μM, reflecting a high affinity to this substrate, compared to the K m of 2200 μM with the substrate Hip-His-leu and 420 μM with the substrate Hip-Phe-Arg for the mammalian somatic ACEs (Cheung et al., 1980). The optimal pH for LsACE activity is 6.0, an acidic condition, while the optimal pH for an ACE homolog of Musca domestica is 8.2, an alkaline condition (Lamango and Isaac, 1994). LsACE maintains high activity in a broad temperature range from 20 °C to 35 °C, which adapts to the seasonal temperature variations in temperate and subtropical East Asia where the small brown planthoppers distribute.
In conclusion, we presented basic evidence for the immune function of an ACE acting in a vector insect against the transmission of a plant virus. How RSV regulates the expression of LsACE and what immune pathways are targeted by LsACE require further investigation.
Author contributions
X.W., W.W., W.Z., and J.L. performed the experiments. F.C. and L.Q. designed the experiments. W.W. and F.C. wrote the manuscript.
Conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was funded by the Natural Science Foundation of China (No. 31802017), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB11040200), and the grant from the State Key Laboratory of Integrated Management of Pest Insects and Rodents (No. Chinese IPM1901).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2019.05.007.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
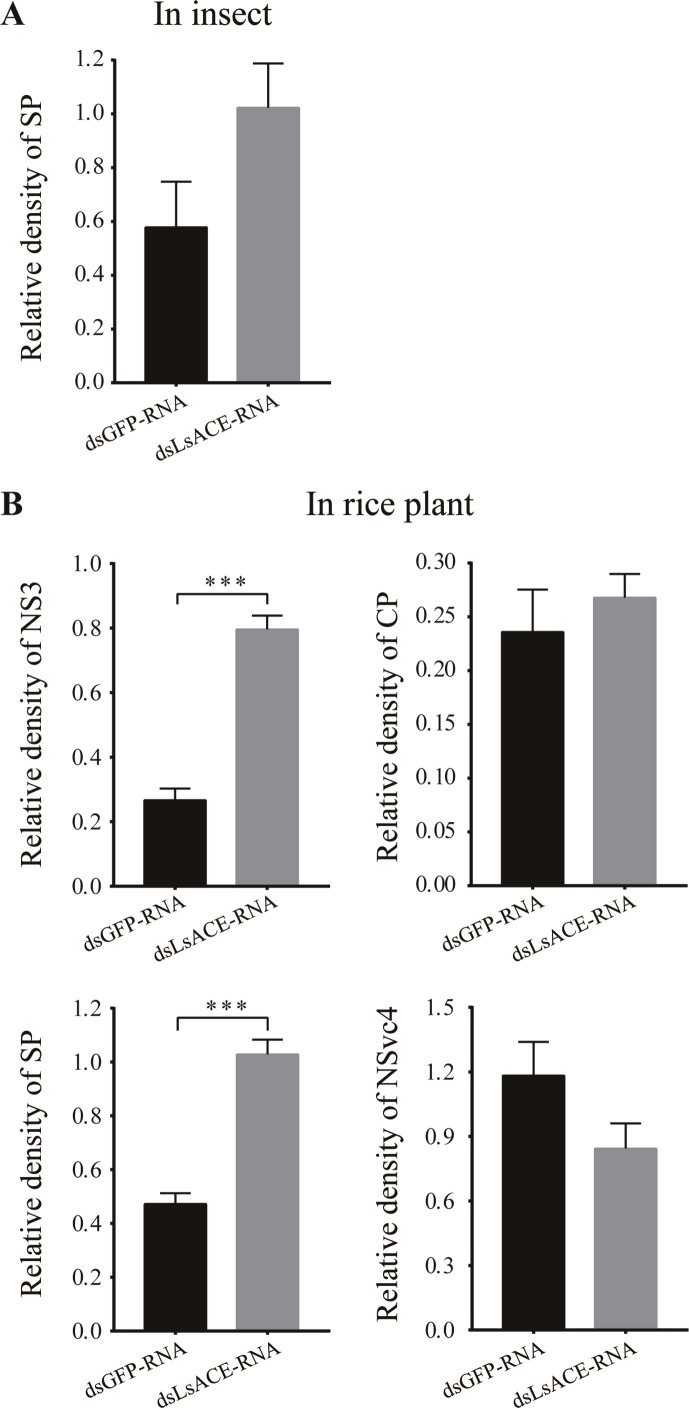
Fig. S1.
Densitometry analysis for the viral protein image bands from Fig. 5C and E. (A) The relative densities of SP in planthoppers from Fig. 5C were normalized with those of beta-tubulin and presented as mean ± SE. (B) The relative densities of NS3, CP, SP, and NSvc4 in rice plants from Fig. 5E were normalized with those of beta-actin and presented as mean ± SE. ***, P < 0.001.
References
- Aguilar R., Jedlicka A.E., Mintz M., Mahairaki V., Scott A.L., Dimopoulos G. Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem. Mol. Biol. 2005;35:709–719. doi: 10.1016/j.ibmb.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Asp L., Johansson A.-S., Mann A., Owe-Larsson B., Urbanska E.M., Kocki T., Kegel M., Engberg G., Lundkvist G.B., Karlsson H. Effects of pro-inflammatory cytokines on expression of kynurenine pathway enzymes in human dermal fibroblasts. J. Inflamm. 2011;8:25. doi: 10.1186/1476-9255-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann R., Casaulta C., Simon D., Conus S., Yousefi S., Simon H.-U. Macrophage migration inhibitory factor delays apoptosis in neutrophils by inhibiting the mitochondria-dependent death pathway. FASEB J. 2003;17:2221–2230. doi: 10.1096/fj.03-0110com. [DOI] [PubMed] [Google Scholar]
- Bernstein K.E., Ong F.S., Blackwell W.L.B., Shah K.H., Giani J.F., Gonzalez-Villalobos R.A., Shen X.Z., Fuchs S. A modern understanding of the traditional and nontraditional biological functions of angiotensin-converting enzyme. Pharmacol. Rev. 2013;65:1–46. doi: 10.1124/pr.112.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung H.S., Wang F.L., Ondetti M.A., Sabo E.F., Cushman D.W. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH-terminal dipeptide sequence. J. Biol. Chem. 1980;255:401–407. [PubMed] [Google Scholar]
- Cho W.K., Lian S., Kim S.M., Park S.H., Kim K.H. Current insights into research on Rice stripe virus. Plant Pathol. J. 2013;29:223. doi: 10.5423/PPJ.RW.10.2012.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia M., Tamborindeguy C., Fish T., Howe K., Thannhauser T.W., Gray S. Genetics coupled to quantitative intact proteomics links heritable aphid and endosymbiont protein expression to circulative polerovirus transmission. J. Virol. 2011;85:2148–2166. doi: 10.1128/JVI.01504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duressa T.F., Huybrechts R. Development of primary cell cultures using hemocytes and phagocytic tissue cells of Locusta migratoria: an application for locust immunity studies. In Vitro Cell. Dev. Biol. Anim. 2016;52:100–106. doi: 10.1007/s11626-015-9952-5. [DOI] [PubMed] [Google Scholar]
- Ekbote U., Looker M., Isaac R.E. ACE inhibitors reduce fecundity in the mosquito, Anopheles stephensi. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003;134:593–598. doi: 10.1016/s1096-4959(03)00019-8. [DOI] [PubMed] [Google Scholar]
- Falk B.W., Tsai J.H. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 1998;36:26. doi: 10.1146/annurev.phyto.36.1.139. [DOI] [PubMed] [Google Scholar]
- Fournier D., Luft F.C., Bader M., Ganten D., Andrade-Navarro M.A. Emergence and evolution of the renin-angiotensin-aldosterone system. J. Mol. Med. 2012;90:495–508. doi: 10.1007/s00109-012-0894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Wang H., Shi X., Ye L., Wu K., Lin K., Ye S., Li B., Zhang H., Lin Q., Ye S., Xue X., Chen C. NovelmiRNA-25 inhibits AMPD2 in peripheral blood mononuclear cells of patients with systemic lupus erythematosus and represents a promising novel biomarker. J. Transl. Med. 2018;16:370. doi: 10.1186/s12967-018-1739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Guo J., Zou Z., Liu J., Cao B., Zhang S., Li H., Wang W., Sheng M., Liu S., Pan Ji, Bao C., Zeng M., Xiao H., Qian G., Hu X., Chen Y., Chen Y., Zhao Y., Liu Q., Zhou H., Zhu J., Gao H., Yang S., Liu X., Zheng S., Yang J., Diao H., Cao H., Wu Y., Zhao M., Tan S., Guo D., Zhao X., Ye Y., Wu W., Xu Y., Penninger J.M., Li D., Gao G.F., Jiang C., Li L. Angiotensin II plasma levels are linked to disease severity and predict fatal outcomes in H7N9-infected patients. Nat. Commun. 2014;5:3595. doi: 10.1038/ncomms4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert C., Savary K., Gasc J.M., Corvol P. The hematopoietic system: a new niche for the renin-angiotensin system. Nat. Clin. Pract. Cardiovasc. Med. 2006;3:80–85. doi: 10.1038/ncpcardio0449. [DOI] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Ohto-Nakanishi T., Penninger J.M. Angiotensinconverting enzyme 2 (ACE2) in disease pathogenesis. Circ. J. 2010;74:405–410. doi: 10.1253/circj.cj-10-0045. [DOI] [PubMed] [Google Scholar]
- Isaac R.E., Bland N.D., Shirras A.D. Neuropeptidases and the metabolic inactivation of insect neuropeptides. Gen. Comp. Endocrinol. 2009;162:8–17. doi: 10.1016/j.ygcen.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Isaac R.E., Ekbote U.M.A., Coates D., Shirras A.D. Insect angiotensin-converting enzyme: a processing enzyme with broad substrate specificity and a role in reproduction. Ann. N. Y. Acad. Sci. 1999;897:342–347. doi: 10.1111/j.1749-6632.1999.tb07904.x. [DOI] [PubMed] [Google Scholar]
- Isaac R.E., Lamango N.S., Ekbote U., Taylor C.A., Hurst D., Weaver R.J., Carhan A., Burnham S., Shirras A.D. Angiotensin-converting enzyme as a target for the development of novel insect growth regulators. Peptides. 2007;28:153–162. doi: 10.1016/j.peptides.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Kotlarz A., Tukaj S., Krzewski K., Brycka E., Lipinska B. Human Hsp40 proteins, DNAJA1 and DNAJA2, as potential targets of the immune response triggered by bacterial DnaJ in rheumatoid arthritis. Cell Stress Chaperones. 2013;18:653–659. doi: 10.1007/s12192-013-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamango N.S., Isaac R.E. Identification and properties of a peptidyl dipeptidase in the housefly, musca-domestica, that resembles mammalian angiotensin-converting enzyme. Biochem. J. 1994;299:651–657. doi: 10.1042/bj2990651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamango N.S., Nachman R.J., Hayes T.K., Strey A., Isaac R.E. Hydrolysis of insect neuropeptides by an angiotensin-converting enzyme from the housefly, Musca domestica. Peptides. 1997;18:47–52. doi: 10.1016/s0196-9781(96)00232-x. [DOI] [PubMed] [Google Scholar]
- Lemeire E., Vanholme B., Van Leeuwen T., Van Camp J., Smagghe G. Angiotensin-converting enzyme in Spodoptera littoralis: molecular characterization, expression and activity profile during development. Insect Biochem. Mol. Biol. 2008;38:166–175. doi: 10.1016/j.ibmb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Liu W., Gray S., Huo Y., Li L., Wei T., Wang X. Proteomic analysis of interaction between a plant virus and its vector insect reveals new functions of hemipteran cuticular protein. Mol. Cell. Proteom. 2015;14:2229–2242. doi: 10.1074/mcp.M114.046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macours N., Hens K., Francis C., Loof A.D., Huybrechts R. Molecular evidence for the expression of angiotensin converting enzyme in hemocytes of Locusta migratoria: stimulation by bacterial lipopolysaccharide challenge. J. Insect Physiol. 2003;49:739–746. doi: 10.1016/s0022-1910(03)00110-0. [DOI] [PubMed] [Google Scholar]
- Murray B.A., Walsh D.J., FitzGerald R.J. Modification of the furanacryloyl-L-phenylalanylglycylglycine assay for determination of angiotensin-I-converting enzyme inhibitory activity. J. Biochem. Biophys. Methods. 2004;59:127–137. doi: 10.1016/j.jbbm.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Schwimmer H., Gerstberger R., Horowitz M. Nitric oxide and angiotensin II: neuromodulation of thermoregulation during combined heat and hypohydration stress. Brain Res. 2004;1006:177–189. doi: 10.1016/j.brainres.2004.01.064. [DOI] [PubMed] [Google Scholar]
- Sun Z., Shi Q., Xu C., Wang R., Wang H., Song Y., Zeng R. Regulation of NlE74A on vitellogenin may be mediated by angiotensin converting enzyme through a fecundity-related SNP in the brown planthopper, Nilaparvata lugens. Comp. Biochem. Physiol. Mol. Integr. Physiol. 2018;225:26–32. doi: 10.1016/j.cbpa.2018.06.009. [DOI] [PubMed] [Google Scholar]
- Toriyama S. Rice stripe virus: prototype of a new group of viruses that replicate in plants and insects. Microbiology. 1986;3:347–351. [PubMed] [Google Scholar]
- Wang W., Luo L., Lu H., Chen S., Kang L., Cui F. Angiotensin-converting enzymes modulate aphid-plant interactions. Sci. Rep. 2015;5:8885. doi: 10.1038/srep08885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhao W., Li J., Luo L., Kang L., Cui F. The c-Jun N-terminal kinase pathway of a vector insect is activated by virus capsid protein and promotes viral replication. Elife. 2017;6 doi: 10.7554/eLife.26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio O., Ezuka A., Sakurai Y., Toriyama K. Studies on the breeding of rice varieties resistant to stripe disease: I. Varietal difference in resistance to stripe disease. Breed Sci. 1967;17:91–98. [Google Scholar]
- Wösten-van Asperen R.M., Bos A.P., Bem R.A., Dierdorp B.S., Dekker T., van Goor H., Kamilic J., van der Loos C.M., van den Berg E., Bruijn M., van Woensel J.B., Lutter R. Imbalance between pulmonary angiotensin-converting enzyme and angiotensin-converting enzyme 2 activity in acute respiratory distress syndrome. Pediatr. Crit. Care Med. 2013;14:e438–e441. doi: 10.1097/PCC.0b013e3182a55735. [DOI] [PubMed] [Google Scholar]
- Wu W., Zheng L., Chen H., Jia D., Li F., Wei T. Nonstructural protein NS4 of Rice Stripe Virus plays a critical role in viral spread in the body of vector insects. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.J., Guo H.Y., Zheng H.J., Zhou T., Zhou Y.J., Wang S.Y., Fang R.X., Qian W., Chen X.Y. Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV) BMC Genomics. 2010;11:303. doi: 10.1186/1471-2164-11-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Lu L., Yang P., Cui N., Le Kang, Cui F. Organ-specific transcriptome response of the small brown planthopper toward rice stripe virus. Insect Biochem. Mol. Biol. 2016;70:60–72. doi: 10.1016/j.ibmb.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Zhao W., Wang Q., Xu Z., Liu R., Cui F. Immune responses induced by different genotypes of the disease-specific protein of Rice stripe virus in the vector insect. Virology. 2019;527:122–131. doi: 10.1016/j.virol.2018.11.011. [DOI] [PubMed] [Google Scholar]
- Zhao W., Wang Q., Xu Z., Liu R., Cui F. Distinct replication and gene expression strategies of the Rice stripe virus in vector insects and host plants. J. Gen. Virol. 2019 doi: 10.1099/jgv.0.001255. (in press) [DOI] [PubMed] [Google Scholar]
- Zhu J., Jiang F., Wang X., Yang P., Bao Y., Zhao W., Wang W., Lu H., Wang Q., Cui N., Li J., Chen X., Luo L., Yu J., Kang L., Cui F. Genome sequence of the small brown planthopper, Laodelphax striatellus. GigaScience. 2017;6:1–12. doi: 10.1093/gigascience/gix109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Yan Y., Shu Y., Gao R., Sun Y., Li X., Ju X., Liang Z., Liu Q., Zhao Y., Guo F., Bai T., Han Z., Zhu J., Zhou H., Huang F., Li C., Lu H., Li Ni, Li D., Jin N., Penninger J.M., Jiang C. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat. Commun. 2014;5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.