Abstract
Background & Aims
The liver is an organ with paradoxic immunologic properties and is known for its tolerant microenvironment, which holds important implications for hepatic diseases. The molecular basis for this local immune suppression, however, is poorly understood. In this study, we aimed to determine the role of liver sinusoidal endothelial cell lectin (LSECtin), a recently identified member of the dendritic cell-specific ICAM-3 grabbing nonintegrin (DC-SIGN) family, in the regulation of hepatic T-cell immune response.
Methods
The regulation of T-cell effector function by LSECtin was determined by co-stimulated T cells with anti-CD3/CD28 monoclonal antibody and LSECtin protein, or co-culture of T-cell receptor transgenic T cells with mouse LSECs in vitro. We generated LSECtin knockout mice and prepared recombinant LSECtin protein and complementary DNA plasmids to analyze the role of LSECtin in hepatic T-cell immune regulation in vivo.
Results
We showed that LSECtin specifically recognized activated T cells and negatively regulated their immune responses. In mice with T-cell–mediated acute liver injury, the lack of LSECtin accelerated the disease owing to an increased T-cell immune response, whereas the exogenous administration of recombinant LSECtin protein or plasmid ameliorated the disease via down-regulation of T-cell immunity.
Conclusions
Our results reveal that LSECtin is a novel regulator of T cells and expose a crucial mechanism for hepatic T-cell immune suppression, perhaps opening up a new approach for treatment of inflammatory diseases in the liver.
Abbreviations used in this paper: CLR, C-type lectin receptors; CRD, carbohydrate recognition domain; DC, dendritic cell; DC-SIGN, dendritic cell-specific ICAM-3 grabbing nonintegrin; EGTA, ethylene glycol-bis(b-aminoethyl ether)-N,N,N′,N′-tetraacetic acid; IFN, interferon; IL, interleukin; L-SIGN, liver/lymph node specific ICAM-3 grabbing nonintegrin; LSEC, liver sinusoidal endothelial cell; mAb, monoclonal antibody; MGl, macrophage galactose-type C-type lectin; NF-κΒ, nuclear factor-κΒ; OVA, ovalbumin; PBL, peripheral blood lymphocyte; PBMC, peripheral blood mononuclear cell; PCR, polymerase chain reaction; PGK, phosphoglycerate kinase; PMA, phorbol-12-myristate-13-acetate; Q-PCR, quantitative PCR; SARS, severe acute respiratory syndrome; siRNA, small interfering; TCR, T-cell receptor; TNF, tumor necrosis factor; wt, wildtype.
Accumulating evidence indicates that the liver is an important immune organ that favors the induction of tolerance rather than immunity,1, 2, 3 which is critical for maintaining immunologic silence in response to harmless antigenic material present in food. The immune tolerance of the liver also might account for the survival of liver allografts and diverse hepatic inflammatory diseases,2 which highlights the importance of understanding the nature of hepatic immune regulation. Two crucial mechanisms have been proposed for liver tolerance. First, the liver is unlike any other organ in the T-cell biology, as best illustrated by inactivation, tolerance, and apoptosis.4 Second, liver sinusoidal endothelial cells (LSECs) and Kupffer cells play significant roles in the distinctive constraints on hepatic T cells.5, 6, 7, 8 The exact molecular mechanisms underlying the hepatic T-cell immune regulation, however, have not been elucidated.
T-cell immune regulation is regulated precisely through a variety of mechanisms, and cell-surface regulators such as co-signaling molecules have been identified as crucial components. Even so, the understanding of cell-surface regulators for T-cell immunity has been greatly expanded by the introduction of the lectin superfamily.9 For example, the C-type lectin macrophage galactose-type C-type lectin (MGL) on dendritic cells (DCs) negatively regulates effector T cells via interaction with CD45.10 A genome-wide association study identified the predicted C-type lectin KIAA0350 as a type 1 diabetes gene, suggesting its regulatory role in the activation of cytotoxic T cells.11 It is clear that lectins are crucial players in immune tolerance and homeostasis, and are involved in diseases of the immune system. It is noteworthy that several lectins also are localized to the liver, although no lectins have been confirmed to be T-cell regulators in the liver thus far.
We identified LSECtin from the liver, which belongs to the C-type lectin subfamily including CD23, dendritic cell-specific ICAM-3 grabbing nonintegrin (DC-SIGN), and liver/lymph node-specific ICAM-3 grabbing nonintegrin (L-SIGN).12 Accumulating evidence suggests that this family is vital in the regulation of immune effector cells: DC-SIGN is essential for DC-induced T-cell proliferation13; L-SIGN has a similar function; and CD23 is involved in the up-regulation or down-regulation of immunoglobulin (Ig)E via interaction with CD21 on B cells or IgE, respectively.14 It was reported that LSECtin functions as an attachment factor for Ebola virus and severe acute respiratory syndrome (SARS).15 Until now, the function of LSECtin in immune regulation has remained unclear. The specific localization of LSECtin in the LSECs and Kupffer cells12, 16 inspired us to investigate whether LSECtin interacts with immune cells and contributes to hepatic immune regulation.
In the present study, we found that LSECtin specifically recognizes activated T cells and negatively regulates T-cell receptor–mediated signaling and the T-cell immune response in vitro, characterized by decreased proliferation and activation, as well as down-regulation of T-cell cytokines. We generated LSECtin knockout (KO) mice and observed an increased accumulation of T cells in liver in the absence of LSECtin. In mice with T-cell–mediated acute liver injury, the lack of LSECtin accelerated the disease owing to an increased T-cell immune response, whereas the exogenous administration of recombinant LSECtin protein or plasmid ameliorated the disease by down-regulating T-cell immunity. Thus, our results reveal a crucial mechanism by which hepatic T-cell immunity is controlled by the C-type lectin LSECtin.
Materials and Methods
Cell Lines and Cell Preparation
The cell lines HL-60, U937, Daudi, Molt-4, CEM, Jurkat, CHO, and MCF7 were from American Type Culture Collection. Human LSECs were from Sciencell Research Laboratories (Carlsbad, CA). CD4+ and CD8+ subsets or total T cells were purified using the MACS beads system (Miltenyi Biotech, Bergisch Gladbach, Germany). Mouse LSECs were prepared as reported previously.8
Monoclonal Antibodies, Reagents, Immunoglobulin Fusion Proteins, and Plasmids
The monoclonal antibody (mAb) against CD3 (h: HIT3a; m: 500A2), anti-CD4 (RPA-T4), anti-CD8 (RPA-T8), anti-CD69 (FN50), and anti-CD25 (M-A251) were from BD Pharmingen (San Jose, CA). The mAb against CD28 (ANC28.1/5D10) was from Calbiochem (San Diego, CA). Anti-mouse interleukin (IL)-2 (JES6-5H4), tumor necrosis factor (TNF)-α (MP6-XT22), and interferon (IFN)-γ (AN18.17.24) were from MACS. Anti-mouse CD3 (145-2C11), anti-mouse CD4 (L3T4), anti-mouse CD8 (Ly-2), anti-mouse CD44 (IM7), anti-DO11.10 T-cell receptor (TCR) (KJ1-26), and the mAb against CD28 (37.51) were from eBioscience (San Diego, CA). Anti-mouse CD62L (MEL-14) was from BioLegend (San Diego, CA). All phospho-specific antibodies were from Cell Signaling Technology (Danvers, MA). hLSECtin–Fc protein was prepared as described.12 The complementary DNA (cDNA) coding full-length and soluble mLSECtin were subcloned into the expression vector pcDNA3.1a (Invitrogen, Carlsbad, CA).
Cell Adhesion Assay
Cells were incubated with LSECtin-Fc, mAb against LSECtin, and then incubated with fluorescein isothiocyanate–conjugated goat anti-mouse IgG, and analyzed on a FACScalibur (BD Biosciences, Franklin Lakes, NJ). Activated peripheral blood lymphocyte (PBLs) labeled with PKH26 were incubated with the cells stably expressing pIRES2–EGFP LSECtin with or without blocking antibodies against LSECtin. Adherent cells were lysed and fluorescence was quantified on a fluostar spectrofluorometer (BMG Labtech, Offenburg, Germany).
T-Cell Proliferation and Cytokine Assays
PBLs were seeded onto CHO or CHO–LSECtin cells and then were stimulated with soluble anti-CD3 or were seeded onto plates coated with LSECtin-Fc and anti-CD3 mAb, followed by 72 hours of culture. In some cases anti-CD28 was added to the cells. DO11.10 CD4+ T cells were cultured in 96-well plates with spleen cells (3 × 105, irradiated with 2500 rads), along with the ovalbumin (OVA) (323–339) peptide and precoated LSECtin-Fc, or plated directly onto LSECs in the presence of OVA peptide. Cell proliferation was estimated by incorporation of bromodeoxyuridine. Supernatants were collected and the cytokines were determined by sandwich enzyme-linked immunosorbent assay kits.
Generation of LSECtin KO Mice
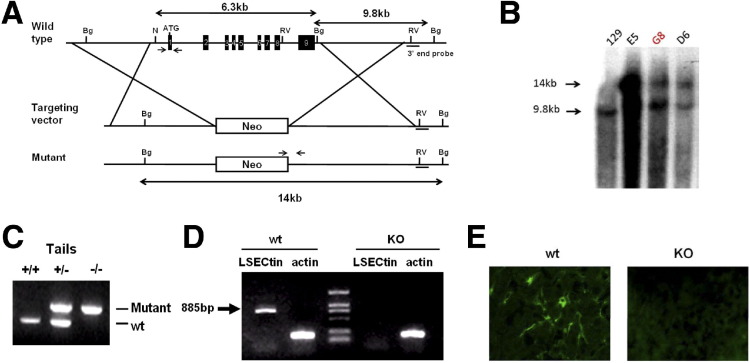
A genomic mouse LSECtin clone was isolated from a 129/SvJ library and was used to construct a targeting vector that was electroporated into 129 R1 embryonic stem cells. Targeted clones that had undergone homologous recombination were screened by polymerase chain reaction (PCR) and verified by Southern blot analysis, and these were injected into C57BL/6 blastocysts to generate chimeric mice. To remove the neo-loxP cassette carrying the phosphoglycerate kinase (PGK) promoter-driven neo gene flanked by lox sequences, agouti F1 heterozygous progeny were crossed with E2a-Cre transgenic mice, and the resultant mice were back-crossed to C57BL/6 strains for 6 generations. Germ line transmission was confirmed by PCR of tail DNA.
Model of Experimental Acute Inflammatory Liver Injury
Male Balb/C mice were injected with an optimum dose (18 mg/kg) of Con A (Sigma–Aldrich, St. Louis, MO) via the tail vein. A reduced dose (8 mg/kg) of Con A was used in KO mice. The small interfering RNAs (siRNAs) or the mLSECtin cDNA plasmid were delivered in vivo as described.17, 18 Mice were administered LSECtin–Fc or control–Fc via tail vein. Serum alanine aminotransferase (ALT)/aspartate aminotransferase (AST) levels were estimated using a detection kit.
Statistical Analyses
A Student t test and 1-way analysis of variance with the Tukey post hoc test were used for statistical analysis. Results with a P value of less than .05 were considered as statistically significant.
Results
LSECtin Binds to Activated T Cells Via a Protein–Glycan Interaction
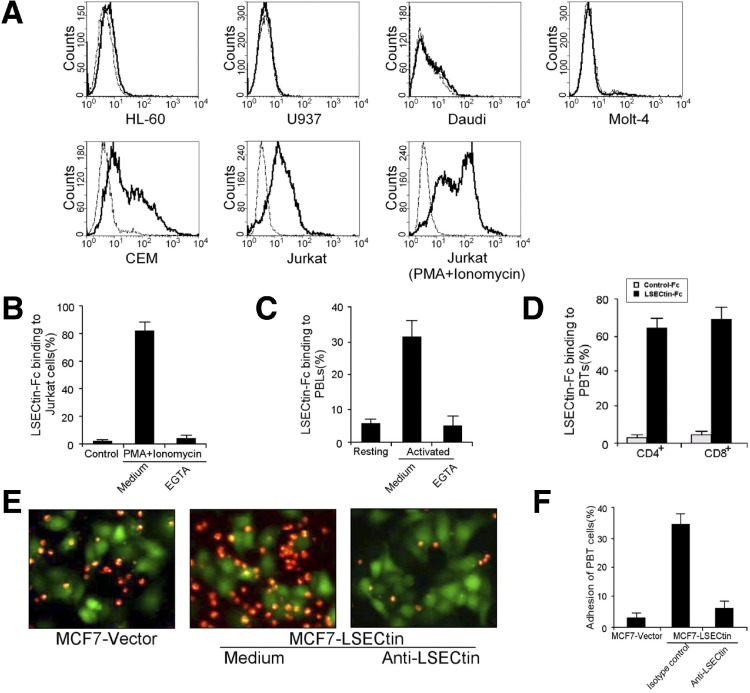
Characterization of LSECtin family members such as DC-SIGN suggest that LSECtin might act as a cell adhesion molecule in the immune system. To test this hypothesis we screened a series of cell lines to identify the LSECtin ligand by flow cytometry. We found that LSECtin–Fc interacted with the CEM and Jurkat T cell lines (Figure 1 A). Moreover, when tested after stimulation with phorbol-12-myristate-13-acetate (PMA) combination with ionomycin, Jurkat T cells showed markedly increased binding of LSECtin, and this binding required the presence of Ca2+ because it was inhibited by addition of the calcium chelator ethylene glycol-bis(b-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) (Figure 1 B). We then sought to determine whether LSECtin receptor is expressed naturally on T cells in vivo. By using flow cytometry analysis, we found that LSECtin–Fc specifically bound peripheral blood mononuclear cells (PBMCs) activated by PMA and ionomycin, and this interaction could be inhibited completely by the addition of EGTA (Figure 1 C). However, LSECtin did not bind nonactivated PBMCs. To verify T-cell binding, we purified either CD4+ or CD8+ cells from PBMCs by positive selection using antibody-coated magnetic beads. LSECtin showed strong binding to activated CD4+ and CD8+ T cells (Figure 1 D). To confirm the interaction of LSECtin and T cells at a cellular level, cell–cell adhesion experiments were performed to assess the adhesion of peripheral blood T cells to LSECtin transfectants. Activated peripheral blood T cells specifically adhered to MCF7 cells expressing LSECtin compared with background binding to parental MCF7 cells. This binding was inhibited by a blocking antibody to LSECtin (Figure 1 E and F), confirming the specificity of the interaction.
Figure 1.
LSECtin recognizes activated T cells. (A) Adhesion of LSECtin–Fc to various human hematopoietic cell lines: promyelocytic leukemia cells (HL-60), monoblastic leukemia cells (U937), Burkitt's lymphoma cells (Daudi), and T-cell leukemia cells (Molt-4, CEM, and Jurkat). (B–D) Adhesion of LSECtin–Fc to activated T cells. (B) Jurkat cells, (C) PBMCs, or purified (D) CD4+ and CD8+ subsets treated with PMA and ionomycin were stained with LSECtin–Fc or human IgG control and analyzed by fluorescence-activated cell sorter. (E) PKH-26–labeled activated peripheral blood T cells were incubated with MCF7 cells transfected with pIRES2-EGFP-LSECtin or vector, and anti-LSECtin was used to block LSECtin-dependent adhesion. Cell–cell adhesion was visualized by fluorescence microscopy and representative pictures were taken. (F) The percentage of activated peripheral blood T cells adherent to MCF7 or MCF7–LSECtin cells was measured using a fluorstar spectrofluorimeter.
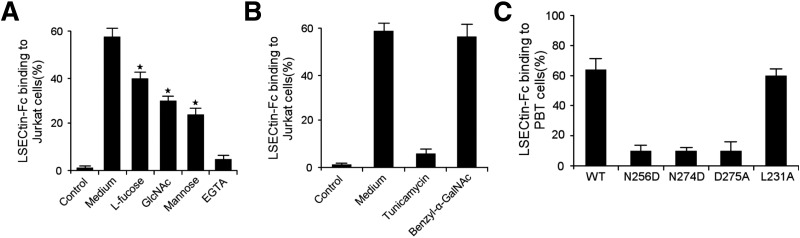
LSECtin has a typical carbohydrate recognition domain (CRD) and binds mannose, GlcNAc, and fucose in a Ca2+-dependent manner.12 Therefore, we sought to determine whether LSECtin binds to activated T cells through a protein–glycan interaction, The binding of LSECtin to activated T cells actually was prevented by EGTA and saccharides such as D-mannose, N-GlcNAc, and L-fucose (Figure 2 A). To assess the contribution of N- or O-linked glycosylation in LSECtin adhesion, activated T cells were cultured in the presence of tunicamycin or benzyl-α-GalNAc, which could block the formation of proteins N- or O-glycosidic linkages, respectively. Tunicamycin abrogated the adhesion of T cells to LSECtin, whereas benzyl-α-GalNAc did not (Figure 2 B). Amino acid sequence alignment of the CRD of LSECtin with that of other C-type lectins indicates that 4 amino acids, Glu254, Asn256, Asn 274, and Asp275, interact with Ca2+ through their carbonyl groups, and Glu254/Asn256 exist in a conserved EPN motif, which is important for carbohydrate recognition.12, 19 Thus, we generated a series of LSECtin point mutants, including N256D, N274D, and D275A, in the soluble isoform. Alteration of each residue completely abolished the binding of LSECtin–Fc to T cells (Figure 2 C), compared with the control substitution L231A. These results indicate that LSECtin binds to activated T cells through a protein–glycan interaction.
Figure 2.
Protein–glycan interaction regulates LSECtin binding to T cells. (A) Binding inhibition of LSECtin to activated Jurkat cells by D-mannose, GlcNAc, L-fucose, and EGTA. (B) Binding inhibition of LSECtin to activated Jurkat cells by tunicamycin or benzyl-α-GalNAc. (C) Binding of wt or mutant LSECtin–Fc to peripheral blood T (pbt) cells, in which amino acids within the CRD domain are mutated. Mutation: N256D, Asn256Asp; N274D, Asn274Asp; D275A, Asp275Ala; and L231A, Leu231Ala.
LSECtin Inhibits T-Cell Immune Response In Vitro
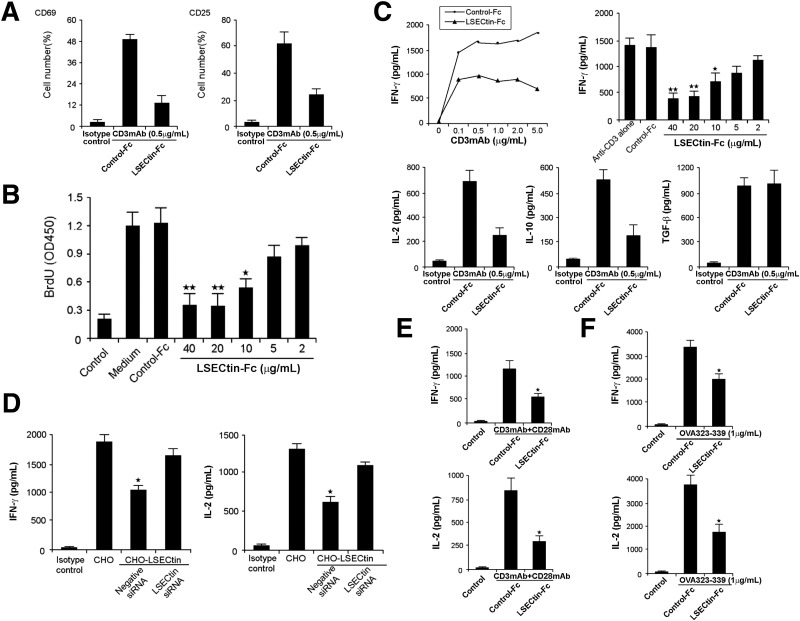
Considering that LSECtin recognizes activated T cells, we investigated whether LSECtin regulates T-cell immune response. We cultured peripheral blood T cells with anti-CD3 mAb and LSECtin–Fc for 72 hours and found that the up-regulation of cell surface activation molecules, including CD69 and CD25, was inhibited by LSECtin (Figure 3 A). Proliferation was estimated by incorporation of bromodeoxyuridine, and it was found that LSECtin significantly decreased T-cell proliferation stimulated by anti-CD3 (Figure 3 B). To examine the effects of soluble LSECtin ligation on the production of cytokines, culture supernatants from T-cell proliferation assays, as shown previously, were harvested at 48 hours. Secretion of IFN-γ, IL-2, and IL-10 was inhibited significantly by LSECtin–Fc, although the production of transforming growth factor-β was not affected (Figure 3 C). To further test whether LSECtin on the cell surface inhibits T-cell responses, we established a stably expressing CHO cell line of LSECtin (CHO–LSECtin). PBMCs were co-cultured with irradiated CHO–LSECtin cells or mock CHO cells and soluble anti-CD3 mAb for 48 hours. Membrane-bound LSECtin decreased the secretion of IFN-γ and IL-2 in the culture supernatants of PBMCs; on the other hand, its siRNA reversed this effect (Figure 3 D). The negative effect of LSECtin also was observed in the presence of anti-CD28 mAb (Figure 3 E), suggesting that the suppression cannot be overcome by co-stimulation. To study antigen-specific signals, we examined the effects of LSECtin on T cells that were expressing an OVA-specific TCR transgene. The secretion of IFN-γ and IL-2 was shown to be inhibited significantly by LSECtin–Fc (Figure 3 F).
Figure 3.
LSECtin inhibits T-cell immune responses in vitro. (A) Inhibition of T-cell activation by LSECtin–Fc. Peripheral blood T cells were stimulated with immobilized T-cell–specific mitogen anti-CD3 antibody plus 10 μg/mL or indicated concentration of LSECtin–Fc or human IgG. Cell activation was detected by staining with antibodies against cell surface activation molecules, including CD69 and CD25. (B) Inhibition of T-cell proliferation by LSECtin–Fc. T-cell proliferation was assayed by pulsing with bromodeoxyuridine (brdu). (C) Inhibition of cytokine production by LSECtin–Fc. IFN-γ, IL-2, IL-10, and transforming growth factor (tgf)-β concentrations in the supernatants was measured by enzyme-linked immunosorbent assay. (D) Inhibition of cytokine production by membrane-bound LSECtin. CHO cells stably expressing LSECtin were transfected with control siRNA or LSECtin siRNA and irradiated after 48 hours. PBMCs were co-cultured with the CHO cells together with the soluble anti-CD3 mAb for 48 hours. Cytokine expression in the supernatants was determined by enzyme-linked immunosorbent assay. (E) Inhibition of cytokine production by LSECtin–Fc in the presence of anti-CD28 antibody. (F) LSECtin–Fc inhibited cytokine production by DO11.10 CD4 T cells stimulated with OVA peptide 323-339. ★P < .05; ★★P < .01.
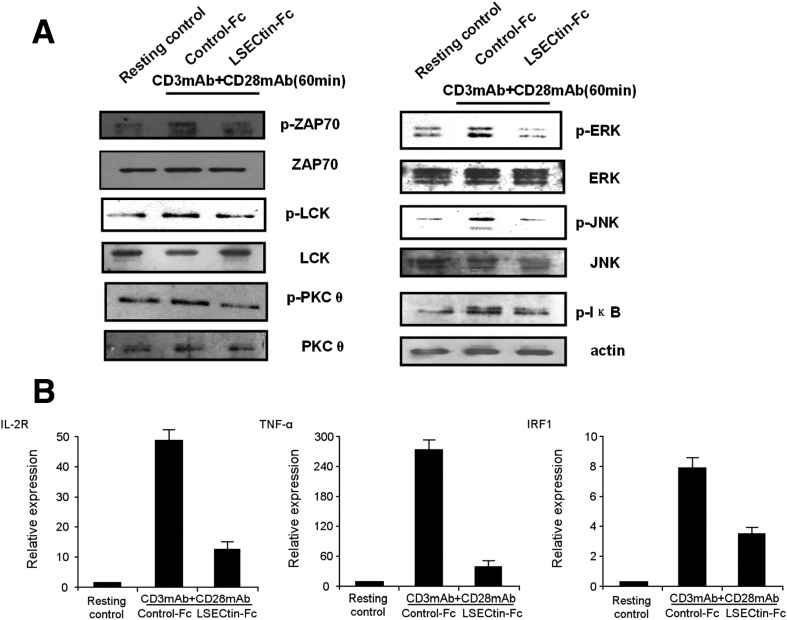
To illustrate the mechanism of LSECtin-mediated T-cell inhibition, we first examined tyrosine phosphorylation of proteins after stimulation via the TCR for 60 minutes. We examined TCR-mediated phosphorylation of ZAP-70, LCK, and PKCθ tyrosine kinase and found that the phosphorylation of these proteins was decreased (Figure 4 A). ERK and JNK are 2 important downstream targets of TCR activation in response to TCR signals. The phosphorylation of ERK and JNK was up-regulated in activated T cells, whereas it was down-regulated by LSECtin engagement (Figure 4 A). We also examined the TCR-mediated nuclear factor-κB (NF-κB) pathway. The phosphorylation of IκB was decreased, which is important for its proteasomal degradation and for the activation of NF-κB. Moreover, the expression of NF-κB target genes IL-2R, TNF-α, and IRF1 obviously was decreased using quantitative PCR (Q-PCR) assay (Figure 4 A and B).
Figure 4.
LSECtin inhibits TCR-mediated signaling. (A) Tyrosine phosphorylation of proteins. T cells were incubated with anti-CD3 and anti-CD28 in the presence of LSECtin–Fc for 60 minutes before lysis in RIPA buffer. Cell lysates were analyzed by immunoblotting with phospho-specific, or pan antibodies against ZAP-70, LCK, PKCθ, Erk, JNK, and anti-pIκB. (B) Q-PCR analysis of changes in the expression of the NF-κB target genes IL-2R, TNF-α, and IRF1. The data shown are from 1 representative of 3 experiments.
Generation of LSECtin-Deficient Mice
To determine the in vivo function of LSECtin, we generated LSECtin-deficient mice. Our gene targeting vector replaced the entire coding region (including all exons) of the endogenous LSECtin allele with a PGK-neo cassette, thereby deleting all functional domains of LSECtin (Figure 5 A). Among 96 G418-resistant ES clones, 4 (4.1%) positive clones were obtained via Southern blot verification (Figure 5 B). Chimeric male mice were derived from these ES clones by standard procedures. They were crossed to C57BL/6 females, and heterozygous mutant mice were established from targeted ES clone G8. Heterozygous and homozygous LSECtin mutant mice then were identified by PCR analysis of genomic DNA isolated from tail biopsies (Figure 5 C). To confirm the absence of LSECtin in these mice, the expression of LSECtin in the liver was estimated by reverse-transcription PCR analysis. As expected, LSECtin was detected in wild-type (wt) but not in KO liver (Figure 5 D). The loss of LSECtin protein in liver also was confirmed by immunofluorescein staining (Figure 5 E). These mice initially were back-crossed to the C57BL/6 background for at least 6 generations before they were used in the studies described.
Figure 5.
Generation of LSECtin KO mice. (A) The targeting map of the LSECtin genomic locus. The whole coding region was replaced with PGK-neo cassette. RE sites: N, Nco I; Bg, Bgl II; RV, EcoR V. The short black bar indicates the external southern probe, and short arrows are genotyping primers. (B and C) Southern blot and PCR detection of heterozygous and homozygous LSECtin mutants in genomic DNA from targeted (B) embryonic stem (es) cells and (C) mouse tails. (D and E) The expression of LSECtin in liver from wt or KO mice estimated by (D) reverse-transcription PCR analysis and (E) immunofluorescein staining.
LSECtin Limits the Hepatic T-Cell Immune Response In Vivo
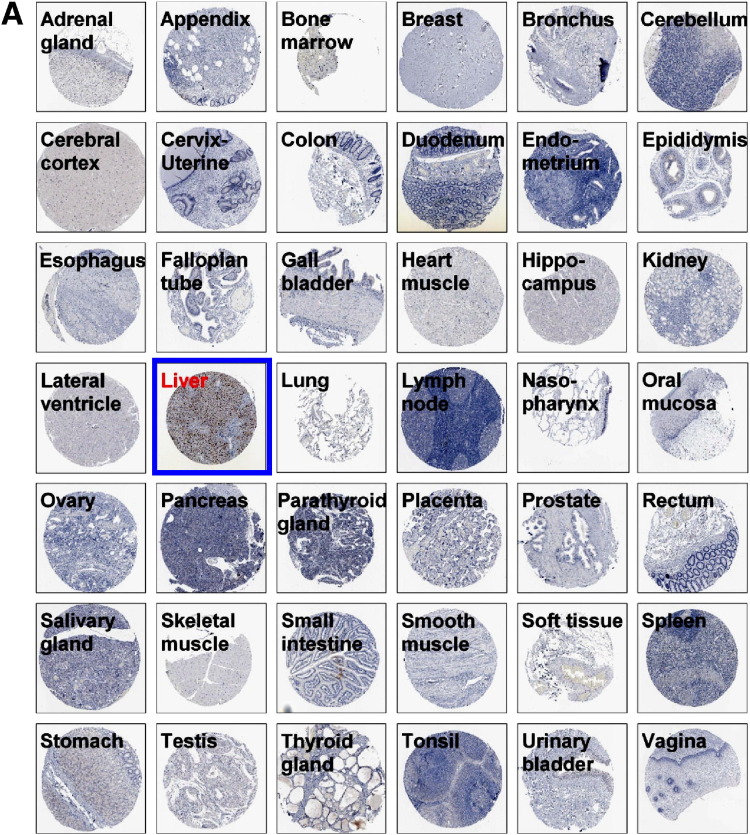
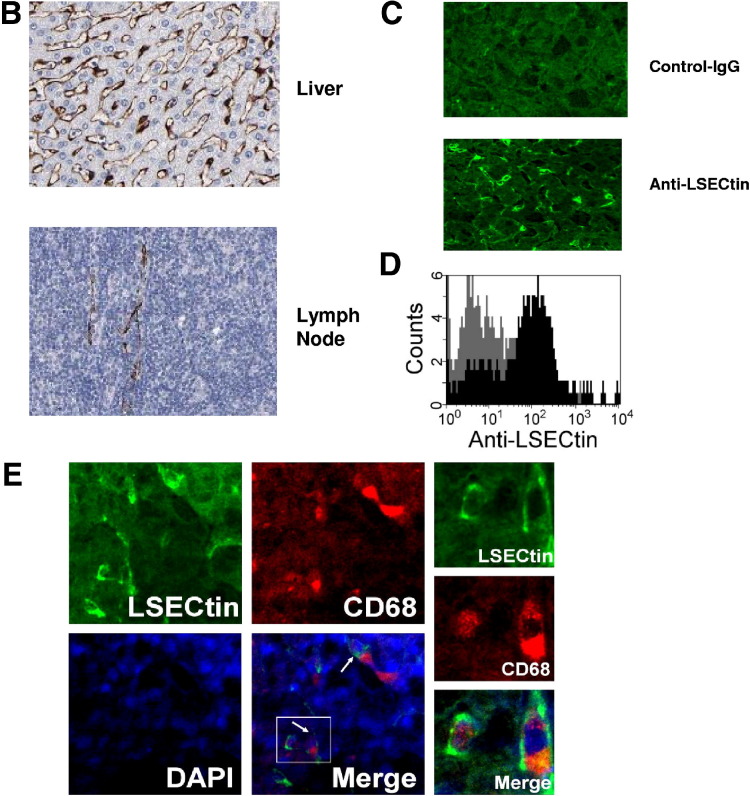
Our previous work showed that LSECtin messenger RNA is expressed in liver sinusoidal endothelial cells.12 Recent studies have indicated that LSECtin also is present in Kupffer cells,16 which was confirmed in our study (Supplementary Figure 1, Supplementary Figure 2). LSECtin limited the activation of T cells in vitro, and the specific localization of LSECtin in the LSECs and Kupffer cells provided a very strong clue for its important function in the regulation of T-cell immune responses in the liver.
Supplementary Figure 1.
LSECtin specifically expresses in LSECs and Kupffer cells (A) Tissue array of LSECtin expression in 42 normal human tissues. All immunohistochemically stained sections were scanned using an automated slide-scanning system at 20× magnification. (B) Highlight of LSECtin expression in liver or lymph node from tissue array. (C) Immunofluorescein staining of normal liver sections was processed using antibody against LSECtin. (D) Expression of LSECtin in LSECs. Purified LSECs were stained with anti-LSECtin mAb. (E) Expression of LSECtin in Kupffers. Normal liver sections were stained with the antibodies against LSECtin (green) and CD68 (red).
Supplementary Figure 2.
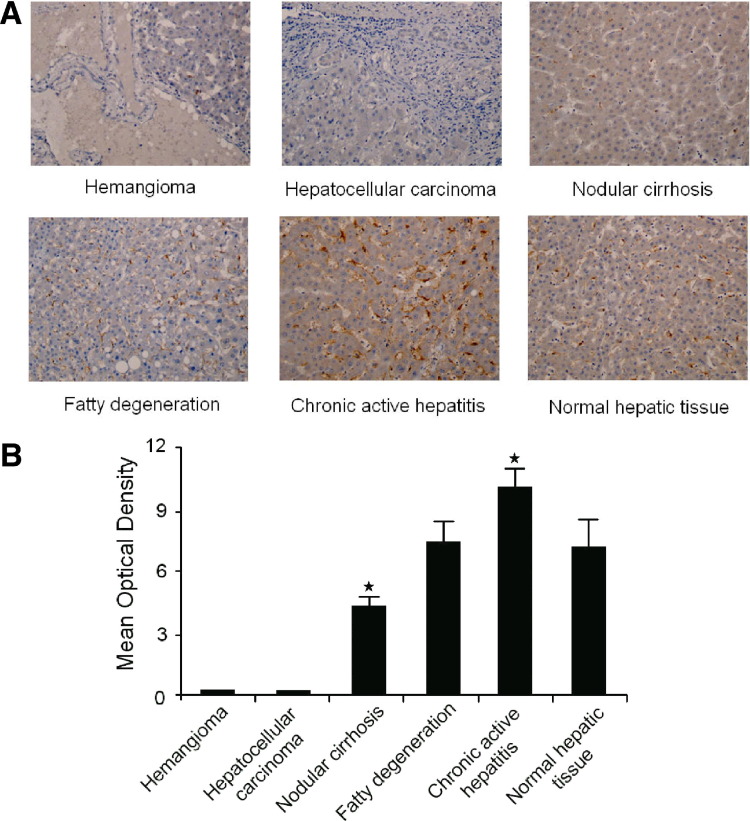
LSECtin expression in liver tissue from patients with various liver diseases (A) Tissue array of LSECtin expression in various liver diseases. All immunohistochemically stained sections were scanned using an automated slide-scanning system at 200× magnification. (B) LSECtin expression in various liver diseases evaluated by mean optical density (hemangioma, n = 10; nodular cirrhosis, n = 30; fatty degeneration, n = 15; chronic active hepatitis, n = 22; hepatocellular carcinoma, n = 25; normal hepatic tissue, n = 14).
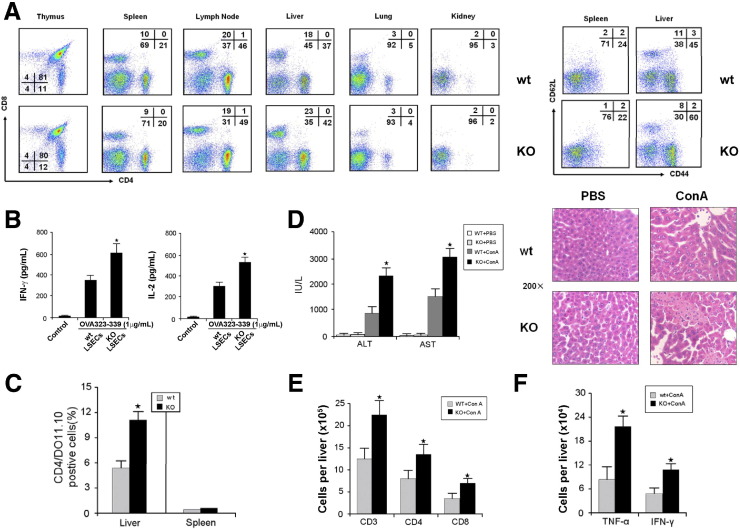
LSECtin KO mice displayed normal numbers and ratios of CD4+CD8+, CD4+, and CD8+ T cells in the thymus. The T-cell populations in the spleen and lymph nodes also appeared to show normal numbers (Figure 6 A). In contrast, an increase in CD4+ (42% in KO mice vs 37% in wt mice) and CD8+ (23% in KO mice vs 18% in wt mice) T cells were found in the liver, but no effect was detected in the lung and kidney. To determine the phenotype of accumulated T cells in the liver of LSECtin KO mice, we examined the expression of CD62L and CD44 on intrahepatic T cells. A fraction of intrahepatic T cells in LSECtin KO mice were CD62LloCD44Hi in comparison with normal mice (60% vs 45%), although no difference was found in the spleen of the same mice (Figure 6 A), suggesting that a large number of these T cells were activated. These results implicated that LSECtin could play a key role in the regulation of hepatic T-cell immunity.
Figure 6.
LSECtin inhibits T-cell immune response in vivo. (A) Lymphocytes were isolated from lymphoid organs and nonlymphoid organs of wt and KO mice (8–12 wk), and subsequently analyzed by CD4 and CD8 expression using specific mAbs. The numbers represent the percentage of CD4+, CD8+, and CD4+CD8+ subsets. Lymphocytes isolated from the spleen or liver of the same mice were stained with anti-CD62L–phycoerythrin and anti-CD44–fluorescein isothiocyanate and subjected to flow cytometry analysis. (B) DO11.10 CD4+ T cells were activated with 1 μg/mL of OVA peptide presented by LSECs from wt or KO mice for 48 hours. Supernatants were collected and cytokines were measured by enzyme-linked immunosorbent assay. (C) Naive DO11.10 T cells were transferred into wt or KO mice via intravenous infusion on day 0. OVA peptide was injected intraperitoneally daily for 3 days from day 1. On day 4 after the first OVA peptide injection, lymphocytes from the liver and spleen were isolated and stained with anti-CD4 and DO11.10. The results represent the average percentage of CD4+/DO11.10. T cells from 2–3 mice at each time point from 3 experiments. (D) Acceleration of acute liver inflammatory injury in LSECtin KO mice. Wt or KO mice were administered Con A. After 24 hours, plasma ALT/AST levels were measured, and liver samples were collected for H&E staining. (E) Liver infiltrating mononuclear cells were collected and lymphocyte populations were analyzed using anti-CD3, anti-CD4, and anti-CD8 antibodies. (F) Total cell numbers of TNF-α– and IFN-γ–producing T cells were counted by intracellular cytokine staining. ★P < .05; n = 5.
Given that the amount of LSECtin present in the in vitro models could well have been nonphysiologic, we performed analyses using co-culture TCR transgenic T cells with KO or wt LSECs. The T cells co-cultured with KO LSECs showed higher responses than those cultured with wt LSECs, indicating that LSECtin on LSECs down-regulates T-cell immune responses (Figure 6 B). The role of LSECtin in the regulation of T cells in liver was evaluated further in a DO11.10 TCR transgenic T-cell transfer system in which wt DO11.10 CD4+ T cells were transferred into either wt or LSECtin KO mice. The mice subsequently were challenged with OVA peptide. We observed a significant increase in DO11.10 CD4+ T cells in the liver of KO mice compared with wt mice on day 4 post-OVA peptide injection (Figure 6 C).
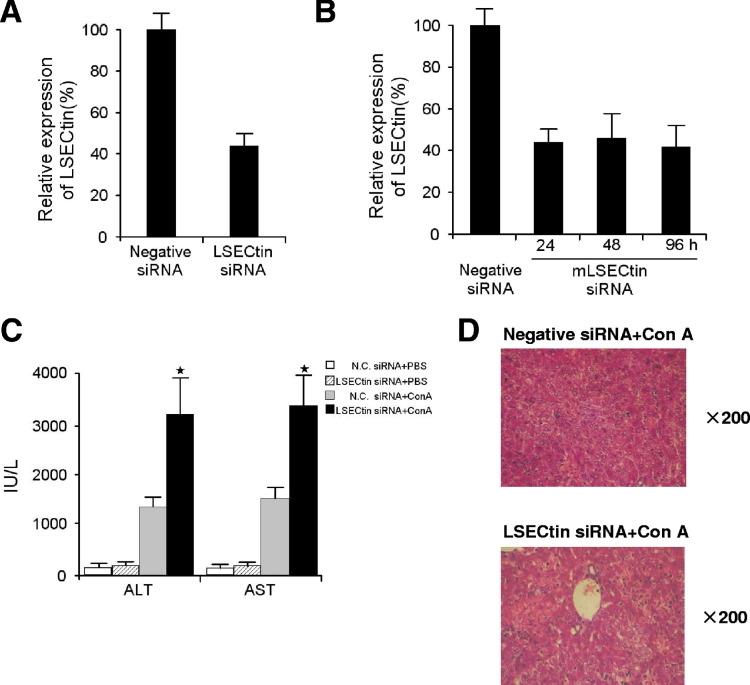
We also induced experimental acute hepatitis in mice using Con A, which is an activated T-cell–mediated liver disease model.20 We then analyzed the effect of LSECtin on activated T-cell–mediated acute liver injury induced via Con A using LSECtin KO mice. Inflammatory liver injury was exacerbated with increased serum ALT/AST, and more serious liver histologic damage was observed in the LSECtin KO compared with the wt mice (Figure 6 D). The total number of lymphocytes in the liver was significantly higher in LSECtin KO mice than in wt mice (Figure 6 E). Moreover, the hepatic T cells of LSECtin KO mice were found to show higher levels of TNF-α and IFN-γ compared with wt mice (Figure 6 F). We also investigated the in vivo silencing effect of siRNA duplexes targeting the LSECtin gene by delivering synthetic siRNA duplexes into the mouse liver by hydrodynamic tail vein injection. In concordance with data from the LSECtin KO mice, the siRNA knock-down mice showed higher levels of plasma ALT and AST, serious liver histologic damage, and higher activity of hepatic T cells compared with wt mice (Supplementary Figure 3). Combining the results from in vitro and in vivo studies, it is reasonable to conclude that LSECtin is involved in constraining T-cell immunity in the liver.
Supplementary Figure 3.
Knockdown of LSECtin accelerates acute inflammatory liver injury in mice. (A) LSECtin was down-regulated in the liver by synthetic siRNA duplexes injection. Expression of LSECtin was estimated by Q-PCR analysis. (B) The expression of LSECtin in liver at 24h, 48h and 96h after siRNA injection. Expression of LSECtin was estimated by Q-PCR analysis. (C, D) Acceleration of acute liver inflammatory injury by the knockdown of LSECtin in siRNA delivered mice. Mice were injected with control siRNA or LSECtin siRNA and then administered with Con A. After 24h, plasma ALT/AST levels were measured (C), and liver samples were collected for hematoxylin-eosin staining (D).
Administration of LSECtin Protein or cDNA Plasmids In Vivo Ameliorates Acute Liver Inflammatory Injury
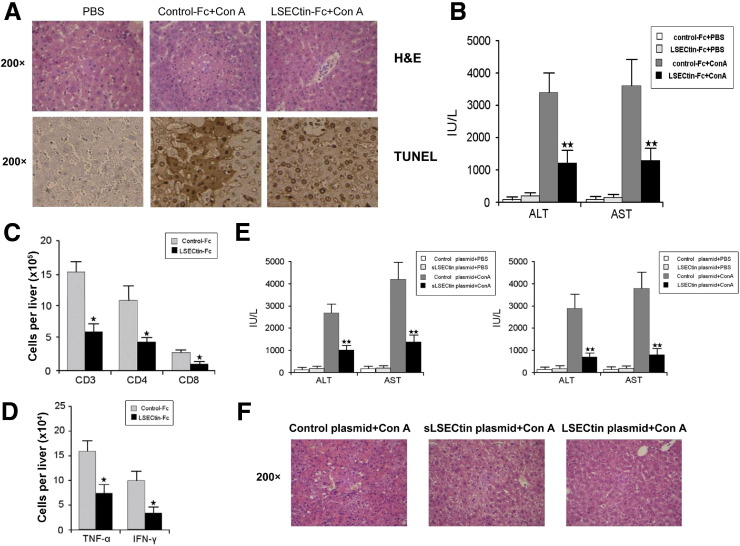
To show the therapeutic effect of LSECtin on Con A-induced hepatitis, 2 approaches were used. We first tried a protein therapeutic approach in which we directly administered recombinant soluble LSECtin before Con A via the tail. It was found that LSECtin protein significantly reduced disease severity, as illustrated by H&E and terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling staining, and remarkably decreased serum ALT/AST levels (Figure 7 A and B). However, when given after Con A, LSECtin could not reverse established inflammation (Supplementary Figure 4). To confirm whether the protective effect was caused by suppression of T-cell activity, we determined the infiltrating lymphocyte subpopulations in the liver. A significant decrease in T-cell infiltration was detected in the livers of LSECtin–Fc–treated mice (Figure 7 C). Moreover, LSECtin–Fc treatment decreased the numbers of TNF-α– and IFN-γ–producing T cells in liver (Figure 7 D).
Figure 7.
Therapeutic effect of recombinant LSECtin protein or LSECtin cDNA on acute inflammatory liver injury in mice via down-regulation of T-cell immunity. Mice were treated with control–Fc or LSECtin–Fc 30 minutes before Con A injection. After 24 hours, liver samples were collected for (A) H&E staining and terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (tunel) staining and (B) plasma ALT/AST levels were measured. (C) Liver infiltrating mononuclear cells were collected and lymphocyte populations were analyzed using anti-CD3, anti-CD4, and anti-CD8 antibodies; (D) liver infiltrating mononuclear cells were restimulated with PMA and ionomycin and the numbers of TNF-α– and IFN-γ–producing T cells were counted by intracellular cytokine staining. Mice were injected with soluble or full-length LSECtin cDNA plasmids 12 hours before Con A injection. After 24 hours, (E) plasma ALT/AST levels were measured and (F) liver samples were collected for H&E staining. ★P < .05; ★★P < .01; n = 5.
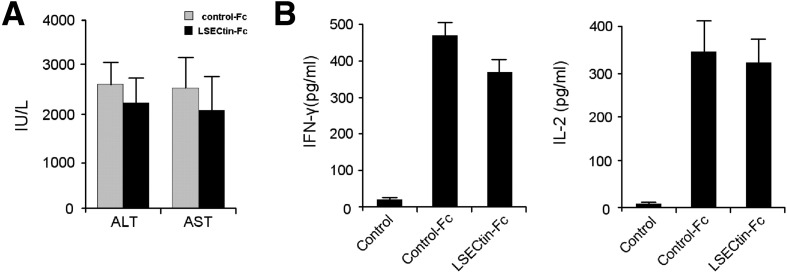
Supplementary Figure 4.
LSECtin did not reverse established inflammation induced by Con A. (A) Mice were treated with control-Fc or LSECtin-Fc 3 hours after Con A injection. After 24 hours, plasma ALT/AST levels were measured. (B) After 6-hour treatment with Con A, the T cells in mouse liver were separated and stimulated with immobilized anti-CD3 antibody plus LSECtin-Fc. Cytokine expression in the supernatants was determined by ELISA.
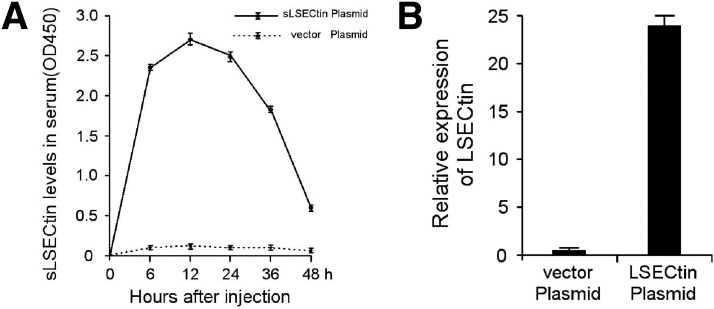
A soluble LSECtin cDNA plasmid was transfected into mice by hydrodynamic tail vein injection. To rule out the possibility that the soluble LSECtin protein blocks the interaction between LSECs and activated T cells, a full-length LSECtin cDNA encoding the membrane-bound protein also was transfected into mice by hydrodynamic tail vein injection. The expression of each in plasma and liver was confirmed by enzyme-linked immunosorbent assay and Q-PCR, respectively (Supplementary Figure 5). Considerable protective effects were observed in the transfected mice, which showed both a tremendous decrease in plasma ALT and AST levels (Figure 7 E) and a clear amelioration of the histologic damage of hepatitis (Figure 7 F). Thus, it could be concluded that LSECtin is not only a significant negative checkpoint in modulating hepatic T-cell activation, but also a new promising therapeutic target for acute liver inflammatory injury.
Supplementary Figure 5.
The expression of soluble and full-length LSECtin cDNA plasmid mice were injected with soluble or full-length LSECtin cDNA plasmids. Plasma LSECtin protein was measured by ELISA-based assay at 6, 12, 24, 36, and 48 h (A). LSECtin mRNA in liver was estimated by Q-PCR analysis at 12 h (B).
Discussion
The liver is a unique immunologic organ, functioning either as a site amenable to effective immune responses to pathogenic microorganisms or in the generation of tolerance to portally derived food and probiotic antigens. The dual functions of the liver require a delicate balance between immunity and tolerance.3, 21 The mechanisms behind the unique immune tolerance of liver have been elucidated in the past few years. One of the principal findings is that the T-cell biology in the liver is unlike that in any other organ, highlighted by the remarkable preference for the inactivation, tolerance, and apoptosis of hepatic T cells.4 In TCR transgenic mice, activated CD8+ T cells dispersed from the lymph nodes and spleen, and an increased number of activated CD8+ T cells simultaneously were found undergoing apoptosis in the liver.22 This phenomenon appears to be specific and is evident in a variety of in vivo model systems, suggesting that the liver is a “graveyard” or “killing field” for activated T cells.2 In a mouse liver transplant model, CD4+ and CD8+ T cells were found to infiltrate significantly and then undergo apoptosis in liver allografts.23 Moreover, many clinical investigations have shown that intrahepatic hepatitis C virus–specific CD8+ T cells have functional alterations, including weak IFN-γ production, impaired proliferation, and enhanced apoptosis, which is associated with viral persistence and the progression of the disease to a chronic phase.24, 25 Obviously, the unique T-cell immune response contributes greatly to specific hepatic tolerance. The exact molecular mechanisms underlying hepatic immune regulation, however, have not been elucidated clearly.
The investigation of B7-H1 and its contribution to hepatic T-cell immune regulation is noteworthy. Iwai et al5 found that expression of B7-H1 in liver sinusoidal endothelial cells and Kupffer cells could inhibit the function of activated T cells via a PD-1–dependent mechanism. Dong et al26 discovered an important role for B7-H1 in controlling the deletion of activated intrahepatic CD8+ T cells in the liver via a PD-1–independent mechanism.26 It is highly likely that hepatic immune regulation is multifactorial. Furthermore, the broad expression of B7-H1 in vivo makes it difficult to solely explain the unique characteristics of hepatic immunity. It is reasonable to speculate about other novel and specific molecular mechanisms identified in the control of hepatic immunity.
LSECtin is expressed specifically in LSECs and Kupffer cells. Its specific localization and roles in the negative regulation of T cells give rise to its important function in hepatic immunity and tolerance induction. Indeed, we showed that LSECtin negatively regulates T-cell receptor–mediated signaling and T-cell immune response stimulated by plate-bound anti-CD3 mAb; this suppression could not be overcome by co-stimulation. Moreover, the role of LSECtin in the antigen-specific activation of T cells was analyzed using T cells from TCR transgenic mice and murine LSECs with physiologic levels of LSECtin expression. The lack of LSECtin in mice resulted in increased accumulation of T cells in the liver both in normal mice and in DO11.10 TCR transgenic T-cell transfer mice. In mice with T-cell–mediated acute liver injury, KO of LSECtin accelerated the disease caused by an increased T-cell immune response, whereas exogenous administration of recombinant LSECtin protein or plasmid ameliorated the disease via down-regulation of T-cell immunity. Thus, our study provides important insight into the molecular mechanism of hepatic T-cell constraint based on investigation of the liver-specific C-type lectin LSECtin.
Recently, the importance of C-type lectin receptors (CLRs) was highlighted by the fact that they have a crucial immune regulatory role. First, CLRs on the surface of DCs could sense the extracellular environment and modulate cellular responses, especially the TLR-dependent inflammatory response.27 Second, CLRs reversibly could modulate T-cell receptor signaling and exert effects on those immune cells.10 This strongly implies that CLRs are important players in immune activation, tolerance, and homeostasis, and are involved in diseases of the immune system. It is noteworthy that several lectins also are localized to the liver, including galectin-1,7 DC-SIGN, and L-SIGN.28 None of the lectins, however, has been confirmed to be a T-cell regulator in liver thus far. In the present study, we showed that the liver-specific C-type lectin LSECtin recognized activated T cells and negatively regulated T-cell immune response in the liver. Our study not only expands the knowledge about C-type lectins in T-cell immune regulation, but also reveals a mechanism by which the T-cell homeostasis and tolerance of the liver is regulated by C-type lectin.
Studies of the mechanisms involved in liver diseases have provided unequivocal evidence that the pathogenesis of virtually all inflammatory liver diseases involves the innate and/or adaptive immune responses. Undoubtedly, research on the mechanisms of hepatic immune regulation will provide new perspectives on liver physiology, and this knowledge will be translated rapidly into more effective therapies in the near future,29 similar to the noticeable progress with the T-cell regulator B7-H1 in liver diseases and other inflammatory diseases.30 In our study, we uncovered a novel and crucial molecular mechanism for hepatic T-cell constraint, and successfully used LSECtin protein and cDNA to treat acute liver injury. This hepatic immune regulation mechanism may provide important insights into hepatic physiology and pathophysiology and could be promising for the treatment of hepatic diseases.
Acknowledgments
Li Tang and Juntao Yang were responsible for the acquisition of data, analysis and interpretation of data, and drafting of the manuscript; Wanli Liu, Xiaoming Tang, Jie Chen, Dianyuan Zhao, Min Wang, Feng Xu, Yantao Lu, Biao Liu, Qihong Sun, and Lingqiang Zhang provided technical and material support; and Fuchu He was responsible for study concept and design, study supervision, and revision of the manuscript.
Footnotes
L.T. and J.Y. contributed equally to this work.
Cell lines used were HL-60 (ATCC number: CCL-240), U937 (ATCC number: CRL-1593.2), Daudi (ATCC number: CCL-213), Molt-4 (ATCC number: CRL-1582), CEM (ATCC number: CCL-119), Jurkat (ATCC number: TIB-152), MCF7 (ATCC number: HTB-22TM), CHO: (ATCC number: CCL-61), and hLSEC (Sciencell 5000, Carlsbad, CA).
Complementary DNA clones used were LSECtin (GenBank accession NM_198492) and mLSECtin (GenBank accession NM_029465).
Antibodies used were from BD Pharmingen: the monoclonal antibody against CD3 (h: HIT3a; m: 500A2), anti-CD4 (RPA-T4), anti-CD8 (RPA-T8), anti-CD69 (FN50), and anti-CD25 (M-A251); Calbiochem: the monoclonal antibody against CD28 (ANC28.1/5D10); MACS: anti-mouse interleukin-2 (JES6-5H4), tumor necrosis factor-α (MP6-XT22), and interferon-γ (AN18.17.24); eBioscience: anti-mouse CD3 (145-2C11), anti-mouse CD4 (L3T4), anti-mouse CD8 (Ly-2), anti-mouse CD44 (IM7), anti-DO11.10 T-cell receptor (KJ1-26), and CD28 (37.51); and BioLegend: anti-mouse CD62L (MEL-14).
Unique animals used were DO11.10 mice (stock number: 003303; The Jackson Laboratory).
Conflicts of interest The authors disclose no conflicts.
FundingThis study was supported by Chinese National Natural Science Foundation Projects (30400398, 30730050, 30621063, and 30772030), the Chinese State Key Program in Basic Research (2009CB52250, 2006CB910802, and 2006CB910800), and the Chinese National High-tech Program (2006AA02Z165).
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi:10.1053/j.gastro.2009.07.051.
Supplementary data
References
- 1.Crispe I.N. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 2.Crispe I.N., Dao T., Klugewitz K., et al. The liver as a site of T-cell apoptosis: graveyard, or killing field? Immunol Rev. 2000;174:47–62. doi: 10.1034/j.1600-0528.2002.017412.x. [DOI] [PubMed] [Google Scholar]
- 3.Knolle P.A., Gerken G. Local control of the immune response in the liver. Immunol Rev. 2000;174:21–34. doi: 10.1034/j.1600-0528.2002.017408.x. [DOI] [PubMed] [Google Scholar]
- 4.Crispe I.N. Hepatic T cells and liver tolerance. Nat Rev Immunol. 2003;3:51–62. doi: 10.1038/nri981. [DOI] [PubMed] [Google Scholar]
- 5.Iwai Y., Terawaki S., Ikegawa M., et al. PD-1 inhibits antiviral immunity at the effector phase in the liver. J Exp Med. 2003;198:39–50. doi: 10.1084/jem.20022235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuniyasu Y., Marfani S.M., Inayat I.B., et al. Kupffer cells required for high affinity peptide-induced deletion, not retention, of activated CD8+ T cells by mouse liver. Hepatology. 2004;39:1017–1027. doi: 10.1002/hep.20153. [DOI] [PubMed] [Google Scholar]
- 7.Karrar A., Broomé U., Uzunel M., et al. Human liver sinusoidal endothelial cells induce apoptosis in activated T cells: a role in tolerance induction. Gut. 2007;56:243–252. doi: 10.1136/gut.2006.093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knolle P.A., Schmitt E., Jin S., et al. Induction of cytokine production in naive CD4+ T cells by antigen-presenting murine liver sinusoidal endothelial cells but failure to induce differentiation toward Th1 cells. Gastroenterology. 1999;116:1428–1440. doi: 10.1016/s0016-5085(99)70508-1. [DOI] [PubMed] [Google Scholar]
- 9.van Kooyk Y., Rabinovich G.A. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 10.van Vliet S.J., Gringhuis S.I., Geijtenbeek T.B., et al. Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat Immunol. 2006;7:1200–1208. doi: 10.1038/ni1390. [DOI] [PubMed] [Google Scholar]
- 11.Hakonarson H., Grant S.F., Bradfield J.P., et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;2:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 12.Liu W., Tang L., Zhang G., et al. Characterization of a novel C-type lectin-like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J Biol Chem. 2004;279:18748–18758. doi: 10.1074/jbc.M311227200. [DOI] [PubMed] [Google Scholar]
- 13.Geijtenbeek T.B., Torensma R., van Vliet S.J., et al. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 14.Hibbert R.G., Teriete P., Grundy G.J., et al. The structure of human CD23 and its interactions with IgE and CD21. J Exp Med. 2005;20:751–760. doi: 10.1084/jem.20050811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gramberg T., Hofmann H., Moller P., et al. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 2005;340:224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domínguez-Soto A., Aragoneses-Fenoll L., Gómez-Aguado F., et al. The pathogen receptor liver and lymph node sinusoidal endothelial cell C-type lectin is expressed in human Kupffer cells and regulated by PU.1. Hepatology. 2009;49:287–296. doi: 10.1002/hep.22678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song E., Lee S.K., Wang J., et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 18.Zhang G., Budker V., Wolff J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 19.Geijtenbeek T.B., van Duijnhoven G.C., van Vliet S.J., et al. Identification of different binding sites in the dendritic cell-specific receptor DC-SIGN for intercellular adhesion molecule 3 and HIV-1. J Biol Chem. 2002;277:11314–11320. doi: 10.1074/jbc.M111532200. [DOI] [PubMed] [Google Scholar]
- 20.Tiegs G., Hentschel J., Wendel A. A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J Clin Invest. 1992;90:196–203. doi: 10.1172/JCI115836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowen D.G., McCaughan G.W., Bertolino P. Intrahepatic immunity: a tale of two sites? Trends Immunol. 2005;26:512–517. doi: 10.1016/j.it.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Huang L., Soldevila G., Leeker M., et al. The liver eliminates T cells undergoing antigen-triggered apoptosis in vivo. Immunity. 1994;1:741–749. doi: 10.1016/s1074-7613(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 23.Qian S., Lu L., Fu F., et al. Apoptosis within spontaneously accepted mouse liver allografts: evidence for deletion of cytotoxic T cell and implications for tolerance induction. J Immunol. 1997;158:4654–4661. [PMC free article] [PubMed] [Google Scholar]
- 24.Toubi E., Kessel A., Goldstein L., et al. Enhanced peripheral T-cell apoptosis in chronic hepatitis C virus infection: association with liver disease severity. J Hepatol. 2001;35:774–780. doi: 10.1016/s0168-8278(01)00207-0. [DOI] [PubMed] [Google Scholar]
- 25.Spangenberg H.C., Viazov S., Kersting N., et al. Intrahepatic CD8+ T-cell failure during chronic hepatitis C virus infection. Hepatology. 2005;42:828–837. doi: 10.1002/hep.20856. [DOI] [PubMed] [Google Scholar]
- 26.Dong H., Zhu G., Tamada K., et al. B7-H1 determines accumulation and deletion of intrahepatic CD8+ T lymphocytes. Immunity. 2004;20:327–336. doi: 10.1016/s1074-7613(04)00050-0. [DOI] [PubMed] [Google Scholar]
- 27.Gringhuis S.I., den Dunnen J., Litjens M., et al. C-type lectin DC-SIGN modulates toll-like receptor signaling via Raf-1 kinase-dependent acetylation of transcription factor NF-kappa B. Immunity. 2007;26:605–616. doi: 10.1016/j.immuni.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Lai W.K., Sun P.J., Zhang J., et al. Expression of DC-SIGN and DC-SIGNR on human sinusoidal endothelium: a role for capturing hepatitis C virus particles. Am J Pathol. 2006;69:200–208. doi: 10.2353/ajpath.2006.051191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gershwin M.E., Vierling J.M., Manns M.P. Humana Press; Totowa, NJ: 2007. Liver immunology: principles and practice. [Google Scholar]
- 30.Sharpe A.H., Wherry E.J., Ahmed R., et al. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol. 2007;8:239–245. doi: 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]