Abstract
In domain-swapping, two or more identical protein monomers exchange structural elements and fold into dimers or multimers whose units are structurally similar to the original monomer. Domain-swapping is of biotechnological interest because inhibiting domain-swapping can reduce disease-causing fibrillar protein aggregation. To achieve such inhibition, it is important to understand both the energetics that stabilize the domain-swapped structure and the protein dynamics that enable the swapping. Structure-based models (SBMs) encode the folded structure of the protein in their potential energy functions. SBMs have been successfully used to understand diverse aspects of monomer folding. Symmetrized SBMs model interactions between two identical protein chains using only intra-monomer interactions. Molecular dynamics simulations of such symmetrized SBMs have been used to correctly predict the domain-swapped structure and to understand the mechanism of domain-swapping. Here, we review such models and illustrate that monomer topology determines key aspects of domain-swapping. However, in some proteins, specifics of local energetic interactions modulate domain-swapping and these need to be added to the symmetrized SBMs. We then summarize some general principles of the mechanism of domain-swapping that emerge from the symmetrized SBM simulations. Finally, using our own results, we explore how symmetrized SBMs could be used to design domain-swapping in proteins.
Keywords: Mechanism of domain swapping, Protein topology, Symmetrized structure-based models, Molecular dynamics simulations
1. Introduction
Domain-swapping is a process in which two or more identical protein monomers exchange structural elements and fold into dimers or multimers (Bennett et al., 1994a, Liu and Eisenberg, 2002). The individual units of such oligomers are structurally similar to the original monomer (Fig. 1 ). The earliest evidence for domain-swapping was seen in RNase A (Crestfield et al., 1962) and the first determination of a domain-swapped structure was that of the dimeric diphtheria toxin (Bennett et al., 1994a, Bennett et al., 1994b). Since then, it has become clear that domain-swapping is common, and several domain-swapped proteins have been crystallized (Shameer et al., 2011). Recent data indicates that RNA structures may also domain-swap (Suslov et al., 2015). In this section, we first define several terms used in the study of domain-swapping (Liu and Eisenberg, 2002, Gronenborn, 2009, Rousseau et al., 2012) and then summarize the biological consequences of domain-swapping (Rousseau et al., 2012, Wodak et al., 2015).
Fig. 1.
The elements of domain swapping. (A) Two unfolded monomer chains shown in green and orange. These can either fold to two monomers (B) or exchange structural elements and fold to a domain-swapped dimer (C). The hinge is the peptide connector between the two domains in a single protein and is shown in (B). The hinge is in different conformations in the monomer (B) and the swapped structures (C, D). (D) Alternate outcomes of domain swapping which could lead to larger aggregates.
1.1. Domain-swapping nomenclature
In order for a protein to be considered domain-swapped the structures of both the monomer and the domain-swapped oligomer need to be present. If one of the structures is that of a homologous protein then the protein is said to be quasi-domain swapped. If only the domain-swapped structure is available then the protein is considered a candidate for domain-swapping. Several inter-protein interfaces are formed upon domain-swapping. The interface whose inter-protein interactions mimic those present in the monomer is called the primary interface. However, new interfaces which are not present in the monomer can also be formed and such interfaces are called secondary interfaces. Many proteins swap only a single secondary structural element and in such cases this “domain” can be thought of as being swapped. However, there are proteins where the size of the “swapped” region is similar to that of the other region. To account for both cases, in this review, we call both parts of the monomer swapped domains. So, a domain-swapping protein usually consists of two swapping-domains connected by a peptide segment (of about 4–5 residues) called the hinge (Fig. 1B and C). The hinge undergoes a conformational change upon swapping, usually being a loop or a turn in the monomer (Fig. 1B) and adopting an extended conformation after domain-swapping (Fig. 1C). There are also examples where two different hinges induce two different modes of domain swapping in the same protein (Liu et al., 1998, Liu et al., 2001, Chen et al., 2010). In some proteins (Nilsson et al., 2004), one swapped domain is inserted into the sequence of the other domain and there are two peptide segments connecting the two domains. Upon swapping, both segments undergo a conformational change. Given the diversity of structural hinge types, there have been efforts to understand the sequence determinants of hinges. Early efforts concentrated on the proline composition of hinges but although prolines contribute to domain-swapping in some proteins (Bergdoll et al., 1997, Rousseau et al., 2001, Miller et al., 2010) this is not a universal effect (Barrientos et al., 2002, Cho et al., 2005). A recent study analyses the distribution of all amino acids in the hinge regions of domain swapping proteins and finds that the amino acid distribution of hinge residues is similar to that of other loop regions. However, there are some amino acids such as valine which are more likely to be found in hinges (Shingate and Sowdhamini, 2012).
1.2. Biological consequences of domain swapping
1.2.1. Aggregation
Open-ended domain-swapping (Fig. 1D) can lead to the formation of large protein aggregates (Esposito et al., 2010). Such aggregation can lead to the loss of protein function and cause disease (Law et al., 2006). Domain-swapping has also been implicated in the formation of disease-causing fibrillar aggregates (Chiti and Dobson, 2006, Herczenik and Gebbink, 2008, Bennett et al., 1995, Bennett et al., 2006, Janowski et al., 2005). For instance, mutants of cystatin-C with reduced domain-swapping also have reduced fibrillar aggregation (Nilsson et al., 2004). The question that arises, then, is, “Why are the amino acids that cause increased domain-swapping retained through evolution?” In a recent study, we showed that in stefin-B, residues that increase the propensity of domain-swapping are part of the protease binding-site and thus, domain-swapping can be a by-product of the need to conserve protein function (Mascarenhas and Gosavi, 2016). In some proteins, domain-swapping is itself involved in protein function and we discuss this next.
1.2.2. Modulation of protein function
Domain-swapping has been implicated in the regulation of protein function (Saint-Jean et al., 1998, Vitagliano et al., 1999, Baxter et al., 2012). The addition of glutathione converted the Pseudomonas putida glyoxylase I from an active domain-swapped dimer with two metal binding sites to a metastable less active monomer with one metal binding site (Saint-Jean et al., 1998). Domain-swapping was also observed to induce allosteric communication in the N-terminal swapped bovine seminal ribonuclease A (Vitagliano et al., 1999). Additionally, domain swapping has been suggested as a mechanism for the evolution of larger complex folds when it is followed by gene duplication and fusion (Bennett et al., 1995). Further support for this hypothesis was found in directed evolution experiments on NF-κB p50, where domain-swapping was able to rescue both structure and function from destabilizing mutations (Chirgadze et al., 2004). Finally, domain-swapping has been seen to induce protein self-assembly (Baker et al., 2016).
In order to understand the consequences of domain-swapping, it is important not only to be able to predict the final domain-swapped structure but also to understand the mechanism of domain-swapping. Here, we review a class of computational protein models called Gō (Gō and Taketomi, 1979) or structure-based models (SBMs) (Clementi et al., 2000, Whitford et al., 2009). SBMs use information derived only from the monomeric protein structure to model domain-swapping (Ding et al., 2002, Yang et al., 2004). Molecular dynamics (MD) simulations of such SBMs have been able to correctly predict hinge regions and the structure of the domain-swapped dimers (Ding et al., 2002, Yang et al., 2004, Cho et al., 2005, Ding et al., 2006). They have also been used to understand whether swapping starts from a transition-state ensemble, an intermediate ensemble or an unfolded ensemble (Yang et al., 2004, Cho et al., 2005, Ono et al., 2015). We summarize insights about the mechanism of domain swapping gained from SBM simulations and then briefly describe how this information might be used to design domain-swapping in proteins.
2. Structure-based models (SBMs)
2.1. SBMs and protein folding
Despite having a vast conformational space, proteins fold on a biologically reasonable timescale because of a funnel shaped energy landscape (Bryngelson et al., 1995). This funnel shape arises because the interactions present in the native or the folded structure of the protein are stabilizing while non-native interactions, which would need to be broken before further folding, are mostly destabilizing. SBMs (Clementi et al., 2000, Hyeon and Thirumalai, 2011, Whitford et al., 2012) use this funnel shape of the energy landscape to simplify the potential energy function and make it easier to sample the conformational space. The potential energy functions of SBMs encode the structure of the protein and in doing so, ignore all non-native interactions and assume that only native interactions contribute to folding. Further, solvation is taken into account implicitly. Both these simplifications speed up computation and make it possible to reach folding timescales in a reasonable amount of computer time. Molecular dynamics (MD) simulations of SBMs of proteins have been successfully used to describe various aspects of protein folding such as folding mechanisms, trends in folding barrier heights, structures of folding intermediate ensembles, etc (Hyeon and Thirumalai, 2011, Whitford et al., 2012).
SBMs have been reviewed in detail elsewhere (Hyeon and Thirumalai, 2011, Whitford et al., 2012, Yadahalli et al., 2014). Here, we summarize a few general features of the models. The protein structure is encoded into different flavours of SBMs in different ways. For instance, some SBMs are coarse-grained and represent each residue by only its Cα (see for example: Clementi et al., 2000) or its Cα and its Cβ atoms (see for example: Cheung et al., 2005). In general, in all SBMs, constraints derived from chemical bonding such as bond distances (Fig. 2 A), angles (Fig. 2B) or improper dihedrals maintaining chirality are encoded using strong harmonic potentials which do not allow these terms to deviate much from their values in the folded structure. Some SBMs encode a cosine-like dihedral potential which mimics secondary structure formation (Fig. 2C). The energetics of tertiary structure is encoded through Lennard-Jones-like potentials between atoms which are in “contact” in the folded structure of the protein (Fig. 2D). The strength of this contact potential is such that contacts can break and form (the distance between the atoms in contact is similar to that in the folded state) allowing the protein to unfold and fold. Atom pairs which are in contact and have an attractive interaction potential between them are usually calculated using one of two recipes. In the first, a contact is said to be present between two atoms if the second atom is within a pre-set cut-off radius (say 4.5 Å) of the first atom (see for example: Karanicolas and Brooks, 2002, Chen and Chan, 2015). Such contact maps are called cut-off contact maps. In the second recipe, an additional screening term is imposed which removes contacts where the contact is occluded by a third atom. Such screened contact maps (e.g. shadow (Noel et al., 2012) and CSU (Sobolev et al., 1999)), are advantageous because they reduce the sensitivity of contact lists to the cut-off radius. In several coarse-grained SBMs, contacts are calculated using the atomistic representation of the protein and then projected onto the coarse-grained representation (Clementi et al., 2000, Karanicolas and Brooks, 2002). Some coarse-grained SBMs weight the strengths of contacts (see Yadahalli and Gosavi, 2016 for a detailed description of several such SBMs) or change the shape of the contact potential (Azia and Levy, 2009, Chen and Chan, 2015) to account for sequence specific effects. Such heterogeneous contacts can be used to represent salt bridges (Azia and Levy, 2009), disulphide bonds (Cho et al., 2005), single effective contacts between large amino acids (Yadahalli and Gosavi, 2016), etc. Finally, excluded volume interactions keep atoms which are not in contact from passing through each other.
Fig. 2.
A summary of the symmetrized SBM interactions. The two protein chains which interact to form the domain-swapped dimer are shown in orange and green. (A-E) show intra-monomer interactions which are present both in the monomer SBM and the symmetrized SBM. (A, B) These are strong harmonic interactions between bonds and angles respectively. They preserve the chemical connectivity of the monomer while it folds or domain-swaps. (C) There are weaker cosine dihedral potentials between four consecutive beads. (D) Lennard-Jones-like interaction potentials are present between all beads which are in contact in the folded state of the protein. Here such interactions are represented by dashed lines. (E) Intra-protein interactions in the second monomer are exactly the same as those in the first monomer. (F) Inter-protein interactions present in the symmetrized SBM are shown as dashed lines. These are calculated based only on the monomer contacts.
2.2. Symmetrized SBMs and domain-swapping
SBMs are computationally efficient and can be used to simulate long timescales for multiple protein chains. Thus, SBMs can be used to study domain-swapping. However, SBMs used for protein folding use only native-like interactions in the potential energy function. Although such SBMs can be used to understand the mechanism of domain-swapping using the domain-swapped structure, they cannot be used for predicting the domain-swapped structure or for understanding the causes of domain-swapping. In order to address these issues, symmetrized SBMs, which use information only from the monomer structure, were invented (Ding et al., 2002, Yang et al., 2004). The potential energy functions of symmetrized SBMs are constructed as follows: The energy function of each identical protein chain (say A or B) is derived from the monomer structure in the same manner as in the folding SBMs. A weak harmonic restraint of the form k(x-x0)2 is usually applied between the centres of mass of the two protein chains (A and B) to maintain a given protein concentration. Additionally, for every pair of intra-molecular contacts between atoms iA and jA of chain A and iB and jB of chain B present in the monomer SBMs, a pair of inter-molecular contacts (iA,jB) and (iB,jA) are introduced into the symmetrized SBM. Thus, atoms can form the same set of contacts either within their own chain, which leads to a pair of monomers or with atoms of another chain, which can lead to a diversity of domain-swapped structures. However, it was observed that the largest population of dimers seen in symmetrized SBM simulations had structures which were similar to the domain-swapped dimer crystal structure (Yang et al., 2004, Cho et al., 2005).
In some proteins, specific features of the amino acid sequence promote or repress domain-swapping. Such sequence effects can either be present (as stress) in the hinges (e.g. isomerization of hinge prolines (Gronenborn, 2009) or the hydrophobic effect driving the dihedral angle of valine into disallowed regions of the Ramachandran plot (Mascarenhas and Gosavi, 2016)) or elsewhere in the protein (e.g. energetic heterogeneity created by the presence of disulphide bonds (Cho et al., 2005)). Symmetrized SBMs modified to account for such effects have been able to correctly predict the structure of the domain-swapped dimers (Mascarenhas and Gosavi, 2016, Cho et al., 2005).
3. Insights gained about domain-swapping from symmetrized SBM simulations
3.1. Monomer topology determines the domain-swapped structure
For several proteins, even symmetrized SBMs which include only a coarse-grained representation can correctly predict the structure of the domain-swapped dimers (Yang et al., 2004, Cho et al., 2005, Ding et al., 2006). Further, the physical understanding of domain-swapping from symmetrized SBMs has also led to a method for predicting the unfolding hotspots and hinge regions using only the monomer structure (Ding et al., 2006). This method calculates the difference in contact energies (ΔE) between the folded structure and an “open” (or domain-swapped) structure of the monomer which assumes that a particular hinge contributes to the domain-swapping. The gain in entropy (ΔS) upon going from the folded structure to the open structure is modelled using polymer theory. A hinge is predicted to be the likely cause of domain-swapping if its ΔE/ΔS is the lowest in the protein. This method was able to correctly predict the hinges for several proteins including the two different hinges present in RNase-A which give rise to two different domain-swapped dimers. It was also able to predict an entirely new domain-swapped structure for the focal adhesion targeting domain which was subsequently experimentally confirmed. Thus, in general, the structure of the domain-swapped dimer is determined by the overall topology of the monomer.
3.2. Mechanistic principles that emerge from MD simulations of domain swapping
We next discuss potential protein fates that have been seen in symmetrized SBM simulations and how these fates are determined by the energetics of the protein structure. At low protein concentrations (a low inter-protein interaction force constant, k, and a high inter-protein mean interaction distance, x0 in symmetrized SBM simulations), proteins interact little and mainly fold to monomers. At higher protein concentrations (a larger inter-protein interaction force constant, k and a small mean interaction distance, x0), there is a competition between folding to a monomer structure and domain-swapping (Yang et al., 2005). In simulations of proteins which do not domain-swap and have a high barrier to folding (e.g. WT CI2 (Yang et al., 2004, Cho et al., 2005)), a high protein concentration promotes domain-swapping but induces a diversity of swapped structures with no particular structure being given a preference (Yang et al., 2004, Cho et al., 2005). In high concentration simulations of proteins which domain-swap, the correct domain-swapped structure is predominant (Yang et al., 2004, Cho et al., 2005, Ding et al., 2006). It has also been observed that “hotspots” of unfolding are present around the hinge region (Ding et al., 2006, Dehouck et al., 2003). Such hotspots can promote domain-swapping from the unfolded state by remaining unfolded while the rest of the protein folds (Yang et al., 2004, Ding et al., 2006, Liu and Huang, 2013).
In some proteins, the free energy barrier to folding is low potentially due to the formation of folding intermediates (Tsytlonok and Itzhaki, 2013). In such cases, partial monomer folding can still occur even at high protein concentrations. The structure of the intermediate (or possibly the folding transition state (Moschen and Tollinger, 2014)) then determines the structure of the domain-swapped dimers with the intermediate (or the transition state) being mostly identical to one of swapped regions in the domain-swapped structure (O'Neill et al., 2001, Kim et al., 2000, Moschen and Tollinger, 2014, Rousseau et al., 2012, Mascarenhas and Gosavi, 2016). Further, we find that if the energetics of the inter-protein interactions are weak and cannot overcome the loss of entropy that accompanies protein dimerization, then domain-swapping is inhibited despite the accumulation of an intermediate that can potentially domain-swap (Mascarenhas and Gosavi, 2016). We note in passing that this effect is not accounted for in the algorithm to predict domain-swapping hotspots (Ding et al., 2006) described in the previous section. Domain-swapping can also occur from the folded state where unfolding hotspots in the structure promote local unfolding in the folded protein. Such local unfolding events can give rise to near-native intermediates (Straub and Thirumalai, 2011, Das et al., 2011) whose role in domain swapping has recently been demonstrated using SBM simulations of the SH3 domain (Zhuravlev et al., 2014).
An altogether different mechanism of domain swapping has been proposed for a quadruple mutant of GB1 using simulations of non-structure based forcefields (Malevanets et al., 2008). Since natural GB1 is not known to domain swap, it is possible that the hydrophobic core mutations that promote domain-swapping (Byeon et al., 2003) alter the funnel-shaped energy landscape of natural GB1. So, the domain-swapping mechanism of mutant GB1 may not be generally applicable to the domain-swapping of natural proteins and we do not elaborate on it here. Finally, more complex mechanisms which may involve non-native interactions (interactions not present in the folded monomer structure) have been proposed for the C-terminal domain of the SARS coronavirus main protease (Kang et al., 2012). We do not comment on the underlying energetics of such mechanisms here because this protein is yet to be studied using SBMs. We next demonstrate how an understanding of the mechanism of domain-swapping might help in the design of domain-swapping.
4. Introducing domain-swapping into proteins that do not swap
Design of domain-swapping is likely to have applications in protein assembly (see for example Ogihara et al., 2001) and functional regulation (Ha et al., 2015). Several protein-specific methods have been used to design domain-swapping (Green et al., 1995, Murray et al., 1998, Pica et al., 2013, Rousseau et al., 2001, Ogihara et al., 2001, Reis et al., 2014, Kuhlman et al., 2001, Orlikowska et al., 2011, Picone et al., 2005). More recently, insertion of a lever protein (ubiquitin) into a non-domain swapping protein (ribose binding protein: RBP) was used to force RBP open and enable domain swapping. This method is likely to be generalizable to other proteins. Using this RBP construct it was found that a primary hinge existed in the protein and if the entropic constraints of intra-protein interactions were lifted then domain-swapping occurred at this hinge (Ha et al., 2015).
Symmetrized-SBM simulations of non-domain swapping proteins such as CI2 at high protein concentrations have shown that such proteins lead to diverse groups of domain-swapped structures each of which uses a different loop for swapping (Yang et al., 2004). Here, we define the primary hinge as that loop which leads to the domain-swapped structure with the highest population. It follows that it should be easiest to destabilize the loop conformation of this hinge to engineer domain-swapping into the protein. In fact, it was seen in simulations that experimental mutations that led to a domain-swapped version of the protein CI2 were made in the loop that was the primary hinge of CI2 (Yang et al., 2004, Cho et al., 2005). To understand how simulations can guide design, we discuss domain-swapping simulations of the symmetrized SBMs of two proteins of the cystatin fold, one of which domain-swaps and the other does not.
4.1. Proteins of the monellin-cystatin fold
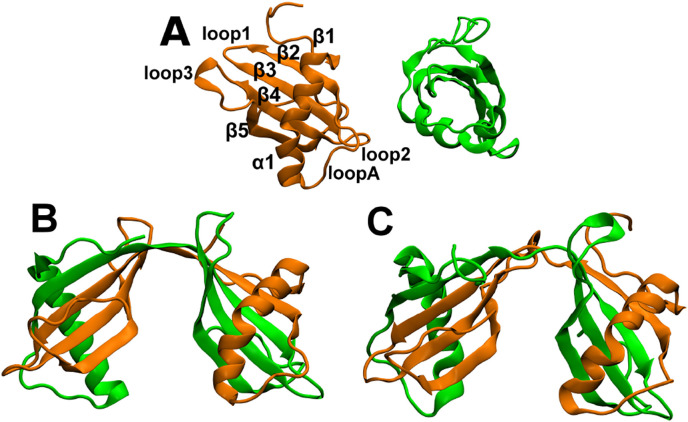
Proteins of the monellin-cystatin fold are composed of five antiparallel strands docked to a single helix with an overall topology of β1-α1-β2-β3-β4-β5 (Fig. 3 A). Here, we compare domain-swapping simulations of the cysteine protease inhibitor, stefin-B (Turk and Bode, 1991), which readily domain swaps (Jenko Kokalj et al., 2007), and a single chain version of the sweet tasting protein monellin (scMn) (Tancredi et al., 1992), where domain-swapping has not been observed in experiment (Patra and Udgaonkar, 2007, Malhotra and Udgaonkar, 2014, Malhotra and Udgaonkar, 2015). We recently showed using the simulations of symmetrized SBMs which encode all heavy atoms of the protein that the primary cause for domain-swapping in stefin-B is the presence of a frustrated loop whose composition is QVVAG (Mascarenhas and Gosavi, 2016). The backbone dihedrals of the second Val in this loop lie in the disallowed region of the Ramachandran plot (Ramachandran et al., 1963). Domain-swapping is favoured because it eases the destabilization created by this disallowed dihedral. We also found that the energetics of this destabilization could only be described correctly if structural information from the dimer structure was encoded to simulate this hinge (QVVAG). In particular, when the hinge dihedrals were extracted from the dimer structure and encoded at ten times the strength of all other dihedrals, we observed that over 60% of the quenched structures folded to the correct domain-swapped dimer structure (Fig. 3B and C) (Mascarenhas and Gosavi, 2016). Here, we show results from the domain-swapping simulations of wild-type monellin (scMn) and a variant which encodes the energetics (and the structure) of the stefin-B loop (QVVAG).
Fig. 3.
Representative structures from symmetrized-SBM simulations of stefin-B (PDB ID:4N6Vchain 0). (A) Monomer structures seen in simulations. The secondary structural elements (β1-α1-loopA-β2-loop1-β3-loop2-β4-loop3-β5) are labeled. (B) Crystal structure of the domain swapped dimer (PDB ID: 2OCT). (C) A domain-swapped structure seen in simulations. It can be seen that the simulated structure is very similar to the crystal structure.
4.2. Domain-swapping simulations of monellin variants
As before (Mascarenhas and Gosavi, 2016), we used symmetrized SBMs which encode all the heavy atoms of the protein (Whitford et al., 2009) for these simulations. Two unfolded chains (with different structures in each replicate) were used as starting structures in the simulations. These were then simulated for 1 × 107 simulation steps at ∼0.9Tf, where Tf is that temperature below which the protein is folded. The chains were restrained to be within interacting distance of each other by a weak harmonic restraint as explained in the section on symmetrized SBMs. Simulations were performed using the GROMACS 5.0 program suite (Abraham et al., 2015) with the leap frog stochastic dynamics integrator and a time step of 0.0005 in reduced units. The basic energy scale of the simulations, ε, is set to 1.0 kJ/mol, the basic length scale to 1 nm and the basic time scale to 1 ps, units which are used in GROMACS. We performed 100 replicate simulations and visually analysed the final structures from these simulations.
The scMn mutant was created by deleting residues 47–56 of scMn and replacing them by a loop with residues QVVA. This loop was built using I-TASSER (Yang et al., 2015). The 57th residue of scMn, a Gly, is not deleted and thus, the mutant scMn has the QVVAG segment from stefin-B in its loop1 (Fig. 3). The resulting sequence has 90 residues instead of the 96 of scMn. Further, the minima of the backbone dihedrals of the QVVAG sequence are encoded from the dimer structure of stefin-B (PDB ID: 2OCT). The potential energies of these dihedrals are also strengthened to 10 times those of the normal dihedrals (Mascarenhas and Gosavi, 2016).
Table 1 summarizes the analysis of final structures from the simulations of both wild-type and mutant proteins. As expected, the simulations of scMn show a diversity of domain-swapped structures with the highest populations (Table 1) being those with loopA swapped (Fig. 4 A) and loop1 swapped (Fig. 4B). At a lower protein concentration (here a lower value of the strength of the harmonic potential, k), the population of the loop1 swapped mutant reduces while that of the two monomers increases. Thus, loopA is likely to be the primary hinge in scMn. However, loopA is a long loop and it is not apparent what mutations will destabilize the monomer but stabilize the dimer. The primary hinge in stefin-B is loop1 which is a β-turn and we introduce this into scMn. This loop mutation almost completely suppresses the population of all domain-swapped structures except that of the loop1 swapped-dimer (Table 1). Interestingly, this mutation suppresses the formation of two monomers even at lower protein concentrations (Table 1) and thus, promotes domain-swapping quite significantly. Experiments with the loop1 mutant of scMn can be easily performed to test our predictions.
Table 1.
Summary of the results from domain swapping simulations. A total of 100 quenching runs were performed for each protein and the final snapshots of each simulation were categorized by visual inspection. The underlined numbers give the populations of the most commonly occurring structures for each protein.
| loop1-hinge | loop1-hinge (one) | two monomers | monomers (one) | loopA-hinge | loopA-hinge (one) | loop2-hinge | loop3-hinge | uncategorized |
|---|---|---|---|---|---|---|---|---|
| scMn | ||||||||
| (i) k = 1.0; x0 = 0.5 | ||||||||
| 20 | 8 | 23 | 10 | 21 | 11 | 2 | 0 | 5 |
| (ii) k = 0.5; x0 = 0.5 | ||||||||
| 7 | 4 | 44 | 10 | 18 | 9 | 0 | 1 | 7 |
| scMn with stefin-B loop1 | ||||||||
| (i) k = 1.0; x0 = 0.5 | ||||||||
| 64 | 11 | 1 | 2 | 1 | 0 | 0 | 0 | 21 |
| (ii) k = 0.5; x0 = 0.5 | ||||||||
| 42 | 23 | 0 | 8 | 0 | 1 | 0 | 0 | 26 |
All structural elements have been marked in Fig. 3A Loop1 is the loop between β2-β3 and loopA is the loop between α1-β2.
loop1-hinge (one) or loopA-hinge (one): loop1 (or loopA) is the hinge, but only one subunit of the domain-swapped dimer is formed while the other is unfolded.
Monomers (one): Only one of the monomers is folded.
K is the strength of the harmonic potential in units of ε/nm2 and x0 is the equilibrium distance between the centres of mass of the two chains.
Fig. 4.
Representative structures from symmetrized-SBM simulations of monellin (PDB ID:1IV7). Monellin is not known to domain-swap and diverse domain-swapped structures are populated in the WT monellin simulations (See Table 1). (A) Dimer with loopA swapped. This structure is rarely observed in stefin-B simulations because the β1-β2 interactions are weak in stefin-B and this reduces the likelihood that they are formed between proteins. (B) Dimer with loop1 swapped. Similar to Fig. 3C. In simulations, where the monellin loop1 is replaced by the stefin-B loop1, this is the predominant structure observed. (C) Other complex swapped-dimers are also observed. A structure with β1-α1-loopA swapped between proteins is shown. The hinge is the peptide segment between loopA and β2.
4.3. Using symmetrized SBMs to design domain-swapping in proteins
A general feature of swapping proteins is that the hinge region is stressed in the monomer structure and this stress is relieved upon domain swapping. Several structurally homologous pairs of proteins exist where only one of the proteins is known to domain-swap (Shingate and Sowdhamini, 2012). A detailed understanding of the mechanism of domain-swapping made possible by symmetrized-SBMs can be used to predict whether hinge-swap mutations between such pairs of proteins can induce domain-swapping in the protein which does not domain-swap. The domain-swapping hinges could also be used to create a library of loops which introduce different kinds and degrees of destabilization into the monomer structure. For example, the QVVAG creates destabilization due to the tradeoff between the hydrophobic effect and excluded volume while adding prolines might create destabilization due to the population of a proline isomer in the monomer form which is less stable than the isomer populated in the domain-swapped structure (Bergdoll et al., 1997, Rousseau et al., 2001, Kuhlman et al., 2001, Miller et al., 2010). Finally, changes in (both increases and decreases in) the length of hinges can also promote swapping (Green et al., 1995, Murray et al., 1998, Picone et al., 2005). Such a library could then be used along with symmetrized-SBM simulations to understand the effect of hinge-swap mutations of any given loop in a protein and whether such mutations are likely to promote domain-swapping and to what degree. This loop library could then be used to guide mutation experiments designed to introduce domain-swapping into proteins that do not domain-swap.
5. Conclusions
Protein domain-swapping can have both beneficial (protein evolution, functional regulation, etc.) and detrimental (fibrillar and amorphous aggregation and loss of function) effects. We review symmetrized-SBMs and their simulations, which can both predict the domain-swapped structure and identify the mechanism of domain-swapping. Such simulations have been useful for understanding the consequences of domain-swapping in specific proteins. The design of domain-swapping has applications in protein-assembly and in the regulation of protein function. In this review, we also outline how symmetrized-SBMs could be used to tune the energetics of domain-swapping in order to achieve the required dimer-structure and the degree of domain-swapping.
Acknowledgements
NMM is supported by the Govt of India-SERB Young Scientist scheme (No. SB/FT/LS-267/2012). SG is supported by core funding from the Govt of India-DAE.
References
- Abraham M.J., Murtolad T., Schulz R., Pálla S., Smith J.C., Berk Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- Azia A., Levy Y. Nonnative electrostatic interactions can modulate protein folding: molecular dynamics with a grain of salt. J. Mol. Biol. 2009;393:527–542. doi: 10.1016/j.jmb.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Baker M.A., Hynson R.M., Ganuelas L.A., Mohammadi N.S., Liew C.W., Rey A.A., Duff A.P., Whitten A.E., Jeffries C.M., Delalez N.J., Morimoto Y.V., Stock D., Armitage J.P., Turberfield A.J., Namba K., Berry R.M., Lee L.K. Domain-swap polymerization drives the self-assembly of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 2016;23:197–203. doi: 10.1038/nsmb.3172. [DOI] [PubMed] [Google Scholar]
- Barrientos L.G., Louis J.M., Botos I., Mori T., Han Z., O'Keefe B.R., Boyd M.R., Wlodawer A., Gronenborn A.M. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures. Structure. 2002;10:673–686. doi: 10.1016/s0969-2126(02)00758-x. [DOI] [PubMed] [Google Scholar]
- Baxter E.L., Jennings P.A., Onuchic J.N. Strand swapping regulates the iron-sulfur cluster in the diabetes drug target mitoNEET. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1955–1960. doi: 10.1073/pnas.1116369109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.J., Choe S., Eisenberg D. Domain swapping: entangling alliances between proteins. Proc. Natl. Acad. Sci. U. S. A. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.J., Choe S., Eisenberg D. Refined structure of dimeric diphtheria toxin at 2.0 A resolution. Protein Sci. 1994;3:1444–1463. doi: 10.1002/pro.5560030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.J., Sawaya M.R., Eisenberg D. Deposition diseases and 3D domain swapping. Structure. 2006;14:811–824. doi: 10.1016/j.str.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Bennett M.J., Schlunegger M.P., Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergdoll M., Remy M.H., Cagnon C., Masson J.M., Dumas P. Proline-dependent oligomerization with arm exchange. Structure. 1997;5:391–401. doi: 10.1016/s0969-2126(97)00196-2. [DOI] [PubMed] [Google Scholar]
- Bryngelson J.D., Onuchic J.N., Socci N.D., Wolynes P.G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- Byeon I.-J.L., Louis J.M., Gronenborn A.M. A protein contortionist: core mutations of GB1 that induce dimerization and domain swapping. J. Mol. Biol. 2003;333:141–152. doi: 10.1016/s0022-2836(03)00928-8. [DOI] [PubMed] [Google Scholar]
- Chen T., Chan H.S. Native contact density and nonnative hydrophobic effects in the folding of bacterial immunity proteins. PLoS Comput. Biol. 2015;11:e1004260. doi: 10.1371/journal.pcbi.1004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M.S., Klimov D., Thirumalai D. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgadze D.Y., Demydchuk M., Becker M., Moran S., Paoli M. Snapshot of protein structure evolution reveals conservation of functional dimerization through intertwined folding. Structure. 2004;12:1489–1494. doi: 10.1016/j.str.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Cho S.S., Levy Y., Onuchic J.N., Wolynes P.G. Overcoming residual frustration in domain-swapping: the roles of disulfide bonds in dimerization and aggregation. Phys. Biol. 2005;2:S44–S55. doi: 10.1088/1478-3975/2/2/S05. [DOI] [PubMed] [Google Scholar]
- Clementi C., Nymeyer H., Onuchic J.N. Topological and energetic factors: what determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J. Mol. Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- Crestfield A.M., Stein W.H., Moore S. On the aggregation of bovine pancreatic ribonuclease. Arch. Biochem. Biophys. Suppl. 1962;1:217–222. [PubMed] [Google Scholar]
- Das P., King J.A., Zhou R. Aggregation of γ-crystallins associated with human cataracts via domain swapping at the C-terminal β-strands. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10514–10519. doi: 10.1073/pnas.1019152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehouck Y., Biot C., Gilis D., Kwasigroch J.M., Rooman M. Sequence-structure signals of 3D domain swapping in proteins. J. Mol. Biol. 2003;330:1215–1225. doi: 10.1016/s0022-2836(03)00614-4. [DOI] [PubMed] [Google Scholar]
- Ding F., Dokholyan N.V., Buldyrev S.V., Stanley H.E., Shakhnovich E.I. Molecular dynamics simulation of the SH3 domain aggregation suggests a generic amyloidogenesis mechanism. J. Mol. Biol. 2002;324:851–857. doi: 10.1016/s0022-2836(02)01112-9. [DOI] [PubMed] [Google Scholar]
- Ding F., Prutzman K.C., Campbell S.L., Dokholyan N.V. Topological determinants of protein domain swapping. Structure. 2006;14:5–14. doi: 10.1016/j.str.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Esposito V., Guglielmi F., Martin S.R., Pauwels K., Pastore A., Piccoli R., Temussi P.A. Aggregation mechanisms of cystatins: a comparative study of monellin and oryzacystatin. Biochemistry. 2010;49:2805–2810. doi: 10.1021/bi902039s. [DOI] [PubMed] [Google Scholar]
- Gō N., Taketomi H. Studies on protein folding, unfolding and fluctuations by computer simulation. IV. Hydrophobic interactions. Int. J. Pept. Protein Res. 1979;13:447–461. doi: 10.1111/j.1399-3011.1979.tb01907.x. [DOI] [PubMed] [Google Scholar]
- Green S.M., Gittis A.G., Meeker A.K., Lattman E.E. One-step evolution of a dimer from a monomeric protein. Nat. Struct. Biol. 1995;2:746–751. doi: 10.1038/nsb0995-746. [DOI] [PubMed] [Google Scholar]
- Gronenborn A.M. Protein acrobatics in pairs–dimerization via domain swapping. Curr. Opin. Struct. Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J.-H., Karchin J.M., Walker-Kopp N., Castañeda C.A., Loh S.N. Engineered domain swapping as an on/off switch for protein function. Chem Biol. 2015;22:1384–1393. doi: 10.1016/j.chembiol.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herczenik E., Gebbink M.F.B.G. Molecular and cellular aspects of protein misfolding and disease. FASEB J. 2008;22:2115–2133. doi: 10.1096/fj.07-099671. [DOI] [PubMed] [Google Scholar]
- Hyeon C., Thirumalai D. Capturing the essence of folding and functions of biomolecules using coarse-grained models. Nat. Comm. 2011;2:487. doi: 10.1038/ncomms1481. [DOI] [PubMed] [Google Scholar]
- Janowski R., Kozak M., Abrahamson M., Grubb A., Jaskolski M. 3D domain-swapped human cystatin C with amyloidlike intermolecular beta-sheets. Proteins. 2005;61:570–578. doi: 10.1002/prot.20633. [DOI] [PubMed] [Google Scholar]
- Jenko Kokalj S., Guncar G., Stern I., Morgan G., Rabzelj S., Kenig M., Staniforth R.A., Waltho J.P., Zerovnik E., Turk D. Essential role of proline isomerization in stefin B tetramer formation. J. Mol. Biol. 2007;366:1569–1579. doi: 10.1016/j.jmb.2006.12.025. [DOI] [PubMed] [Google Scholar]
- Kang X., Zhong N., Zou P., Zhang S., Jin C., Xia B. Foldon unfolding mediates the interconversion between M(pro)-C monomer and 3D domain-swapped dimer. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14900–14905. doi: 10.1073/pnas.1205241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanicolas J., Brooks C. The origins of asymmetry in the folding transition states of protein L and protein G. Protein Sci. 2002;11:2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.E., Fisher C., Baker D. A breakdown of symmetry in the folding transition state of protein L. J. Mol. Biol. 2000;298:971–984. doi: 10.1006/jmbi.2000.3701. [DOI] [PubMed] [Google Scholar]
- Kuhlman B., O'Neill J.W., Kim D.E., Zhang K.Y.J., Baker D. Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10687–10691. doi: 10.1073/pnas.181354398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law R.H.P., Zhang Q., McGowan S., Buckle A.M., Silverman G.A., Wong W., Rosado C.J., Langendorf C.G., Pike R.N., Bird P.I., Whisstock J.C. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gotte G., Libonati M., Eisenberg D. A domain-swapped RNase A dimer with implications for amyloid formation. Nat. Struct. Biol. 2001;8:211–214. doi: 10.1038/84941. [DOI] [PubMed] [Google Scholar]
- Liu Y., Hart P.J., Schlunegger M.P., Eisenberg D. The crystal structure of a 3D domain-swapped dimer of RNase A at a 2.1-A resolution. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3437–3442. doi: 10.1073/pnas.95.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Huang Y. Evidences unfolding Mech. three-dimensional domain swapping Prot Sci. 2013;22:280–286. doi: 10.1002/pro.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malevanets A., Sirota F.L., Wodak S.J. Mechanism and energy landscape of domain swapping in the B1 domain of protein G. J. Mol. Biol. 2008;382:223–235. doi: 10.1016/j.jmb.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Malhotra P., Udgaonkar J.B. Tuning cooperativity on the free energy landscape of protein folding. Biochemistry. 2015;54:3431–3441. doi: 10.1021/acs.biochem.5b00247. [DOI] [PubMed] [Google Scholar]
- Malhotra P., Udgaonkar J.B. High-energy intermediates in protein unfolding characterized by thiol labeling under nativelike conditions. Biochemistry. 2014;53:3608–3620. doi: 10.1021/bi401493t. [DOI] [PubMed] [Google Scholar]
- Mascarenhas N.M., Gosavi S. Protein domain swapping can Be a consequence of functional residues. J. Phys. Chem. B. 2016;120:6929–6938. doi: 10.1021/acs.jpcb.6b03968. [DOI] [PubMed] [Google Scholar]
- Miller K.H., Karr J.R., Marqusee S. A hinge region cis-proline in ribonuclease A acts as a conformational gatekeeper for C-terminal domain swapping. J. Mol. Biol. 2010;400:567–578. doi: 10.1016/j.jmb.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschen T., Tollinger M. A kinetic study of domain swapping of Protein L. Phys. Chem. Chem. Phys. 2014;16:6383–6390. doi: 10.1039/c3cp54126f. [DOI] [PubMed] [Google Scholar]
- Murray A.J., Head J.G., Barker J.J., Brady R.L. Engineering an intertwined form of CD2 for stability and assembly. Nat. Struct. Biol. 1998;5:778–782. doi: 10.1038/1816. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Wang X., Rodziewicz-Motowidlo S., Janowski R., Lindström V., Onnerfjord P., Westermark G., Grzonka Z., Jaskolski M., Grubb A. Prevention of domain swapping inhibits dimerization and amyloid fibril formation of cystatin C: use of engineered disulfide bridges, antibodies, and carboxymethylpapain to stabilize the monomeric form of cystatin C. J. Biol. Chem. 2004;279:24236–24245. doi: 10.1074/jbc.M402621200. [DOI] [PubMed] [Google Scholar]
- Noel J.K., Whitford P.C., Onuchic J.N. The shadow map: a general contact definition for capturing the dynamics of biomolecular folding and function. J. Phys. Chem. B. 2012;116:8692–8702. doi: 10.1021/jp300852d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara N.L., Ghirlanda G., Bryson J.W., Gingery M., DeGrado W.F., Eisenberg D. Design of three-dimensional domain-swapped dimers and fibrous oligomers. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5596–5601. doi: 10.1073/pnas.98.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill J.W., Kim D.E., Johnsen K., Baker D., Zhang K.Y. Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus. Structure. 2001;9:1017–1027. doi: 10.1016/s0969-2126(01)00667-0. [DOI] [PubMed] [Google Scholar]
- Ono K., Ito M., Hirota S., Takada S. Dimer domain swapping versus monomer folding in apo-myoglobin studied by molecular simulations. Phys. Chem. Chem. Phys. 2015;17:5006–5013. doi: 10.1039/c4cp05203j. [DOI] [PubMed] [Google Scholar]
- Orlikowska M., Jankowska E., Kołodziejczyk R., Jaskólski M., Szymańska A. Hinge-loop mutation can Be used to control 3D domain swapping and amyloidogenesis of human cystatin C. J. Struct. Biol. 2011;173:406–413. doi: 10.1016/j.jsb.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Patra A.K., Udgaonkar J.B. Characterization of the folding and unfolding reactions of single-chain monellin: evidence for multiple intermediates and competing pathways. Biochemistry. 2007;46:11727–11743. doi: 10.1021/bi701142a. [DOI] [PubMed] [Google Scholar]
- Pica A., Merlino A., Buell A.K., Knowles T.P., Pizzo E., D'Alessio G., Sica F., Mazzarella L. Three-dimensional domain swapping and supramolecular protein assembly: insights from the X-ray structure of a dimeric swapped variant of human pancreatic RNase. Acta Crystallogr. D. Biol. Crystallogr. 2013;69:2116–2123. doi: 10.1107/S0907444913020507. [DOI] [PubMed] [Google Scholar]
- Picone D., Di Fiore A., Ercole C., Franzese M., Sica F., Tomaselli S., Mazzarella L. The role of the hinge loop in domain swapping. The special case of bovine seminal ribonuclease. J. Biol. Chem. 2005;280:13771–13778. doi: 10.1074/jbc.M413157200. [DOI] [PubMed] [Google Scholar]
- Ramachandran G.N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Reis J.M., Burns D.C., Woolley G.A. Optical control of Protein−Protein interactions via blue light-induced domain swapping. Biochemistry. 2014;53:5008–5016. doi: 10.1021/bi500622x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F., Schymkowitz J., Itzhaki L.S. Implications of 3D domain swapping for protein folding, misfolding and function. Adv. Exp. Med. Biol. 2012;747:137–152. doi: 10.1007/978-1-4614-3229-6_9. [DOI] [PubMed] [Google Scholar]
- Rousseau F., Schymkowitz J.W., Wilkinson H.R., Itzhaki L.S. Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5596–5601. doi: 10.1073/pnas.101542098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jean A.P., Phillips K.R., Creighton D.J., Stone M.J. Active monomeric and dimeric forms of Pseudomonas putida glyoxalase I: evidence for 3D domain swapping. Biochemistry. 1998;37:10345–10353. doi: 10.1021/bi980868q. [DOI] [PubMed] [Google Scholar]
- Shameer K., Shingate P.N., Manjunath S.C.P., Karthika M., Pugalenthi G., Sowdhamini R. 3DSwap: curated knowledgebase of proteins involved in 3D domain swapping. Database Oxf. 2011;2011 doi: 10.1093/database/bar042. bar042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingate P., Sowdhamini R. Analysis of domain-swapped oligomers reveals local sequence preferences and structural imprints at the linker regions and swapped interfaces. PLoS One. 2012;7:e39305. doi: 10.1371/journal.pone.0039305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V., Sorokine A., Prilusky J., Abola E.E., Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- Straub J.E., Thirumalai D. Toward a molecular theory of early and late events in monomer to amyloid fibril formation. Annu. Rev. Phys. Chem. 2011;62:437–463. doi: 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslov N.B., DasGupta S., Huang H., Fuller J.R., Lilley D.M.J., Rice P.A., Piccirilli J.A. Crystal structure of the Varkud satellite ribozyme. Nat. Chem. Biol. 2015;11:840–846. doi: 10.1038/nchembio.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancredi T., Iijima H., Saviano G., Amodeo P., Temussi P.A. Structural determination of the active site of a sweet protein. A 1H NMR investigation of pMNEI. FEBS Lett. 1992;310:27–30. doi: 10.1016/0014-5793(92)81138-c. [DOI] [PubMed] [Google Scholar]
- Tsytlonok M., Itzhaki L.S. The how's and why's of protein folding intermediates. Arch. Biochem. Biophys. 2013;531:14–23. doi: 10.1016/j.abb.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Turk V., Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285:213–219. doi: 10.1016/0014-5793(91)80804-c. [DOI] [PubMed] [Google Scholar]
- Vitagliano L., Adinolfi S., Sica F., Merlino A., Zagari A., Mazzarella L. A potential allosteric subsite generated by domain swapping in bovine seminal ribonuclease. J. Mol. Biol. 1999;293:569–577. doi: 10.1006/jmbi.1999.3158. [DOI] [PubMed] [Google Scholar]
- Whitford P.C., Noel J.K., Gosavi S., Schug A., Sanbonmatsu K.Y., Onuchic J.N. An all-atom structure-based potential for proteins: bridging minimal models with all-atom empirical forcefields. Proteins. 2009;75:430–441. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford P.C., Sanbonmatsu K.Y., Onuchic J.N. Biomolecular dynamics: order–disorder transitions and energy landscapes. Rep. Prog. Phys. 2012;75:076601. doi: 10.1088/0034-4885/75/7/076601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodak S.J., Malevanets A., MacKinnon S.S. The landscape of intertwined associations in Homo-oligomeric proteins. Biophys. J. 2015;109:1087–1100. doi: 10.1016/j.bpj.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadahalli S., Giri Rao V.V.H., Gosavi S. Modeling non-native interactions in designed proteins. Isr. J. Chem. 2014;54:1230–1240. [Google Scholar]
- Yadahalli S., Gosavi S. Functionally relevant specific packing can determine protein folding routes. J. Mol. Biol. 2016;428:509–521. doi: 10.1016/j.jmb.2015.12.014. [DOI] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Meth. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Cho S.S., Levy Y., Cheung M.S., Levine H., Wolynes P.G., Onuchic J.N. Domain swapping is a consequence of minimal frustration. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13786–13791. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Levine H., Onuchic J.N. Protein oligomerization through domain swapping: role of inter-molecular interactions and protein concentration. J. Mol. Biol. 2005;352:202–211. doi: 10.1016/j.jmb.2005.06.062. [DOI] [PubMed] [Google Scholar]
- Zhuravlev P.I., Reddy G., Straub J.E., Thirumalai D. Propensity to form amyloid fibrils is encoded as excitations in the free energy landscape of monomeric proteins. J. Mol. Biol. 2014;426:2653–2666. doi: 10.1016/j.jmb.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]