Abstract
Tuberculosis causes serious health problem for the world population. Antigenic peptides selected by pathogen-specific cytotoxic T lymphocytes (CTLs) are presented by major histocompatibility complex (MHC; or human leukocyte antigen [HLA] in humans) molecules, and HLA-A restricted responses may be of interest for vaccine development and the understanding of cellular immunity. A series of peptides derived from the 10-KDa culture filtrate protein (CFP10) and the 6 kDa early secretory antigenic target (ESAT-6) in the Mycobacterium tuberculosis (Mtb) have been screened and a CTL epitope restricted by the human leukocyte antigen HLA-A24, a common HLA allele in Asian people, has been identified. In this study, we studied a panel of CFP10 and ESAT-6-derived peptides to identify those with binding motifs for HLA-A24 molecules. The antigenicity of candidate peptides was assessed with in vitro refolding tests and an enzyme-linked immunospot (ELISPOT) assay, and by tetramer staining to determine the capacity to stimulate CTLs from peripheral blood mononuclear cells (PBMCs) of HLA-A24-positive TB Patients. We report that one novel candidate peptide at positions 5–14 of ESAT-6 of Mtb could induce peptide-specific CTLs from PBMCs of HLA-A24-positive patients, but not from HLA-A24-negative patients and HLA-A24-positive healthy controls. Identified epitope is a weak binder for HLA-A24 molecule in a mini MHC refolding assay. Since the peptide is presented by a common HLA class I molecule, it may be useful for immunotherapy against Mtb infection and vaccine development in the large population of Mtb-infected patients.
Keywords: Mycobacterium tuberculosis, HLA-A24, Epitope, ESAT-6
Abbreviations: Mtb, Mycobacterium tuberculosis; CTLs, cytotoxic T lymphocytes; IFN-γ, interferon gamma; PBMCs, peripheral blood mononuclear cells; CFP10, 10-KDa culture filtrate protein; ESAT-6, 6 kDa early secretory antigenic target; MHC, major histocompatibility complex; HLA, human leukocyte antigen; TB, tuberculosis; β2m, β2 microglobulin; SFCs, spot-forming cells; HPLC, high performance liquid chromatography; PHA, Phytohaemagglutinin; rhIL-2, recombinant human interleukine-2; rhIL-7, recombinant human interleukine-7; AFB, acid-fast bacilli; PE, phycoerythrin; BCG, bacillus Calmette-Guérin; LTBI, latent TB infection
Highlights
► We study a panel of Mtb-derived peptides to identify binders for HLA-A24. ► Peptide P5 significantly elicits IFN-producing CTLs from HLA-A24+ TB donors. ► Peptide P5 is a weak binder for HLA-A24 molecule in a mini refolding assay. ► The identified epitope may be useful for immunotherapy and vaccine development.
1. Introduction
Currently, tuberculosis remains one of the top three fatal infectious diseases, together with acquired immune deficiency syndrome (AIDS) and malaria. In China, it is estimated that 550 million (about 44.5% of the population) individuals latently have been infected with Mycobacterium tuberculosis (Mtb), and 2 million are with smear-positive or culture-positive pulmonary tuberculosis (TB) patients [1].
Although the mechanisms of protection against tuberculosis are not completely understood, many lines of evidence have shown that antigen-specific cytotoxic T lymphocytes (CTLs) might be effective in limiting the spread of Mtb and clearing Mtb during infection. To control mycobacterial growth, MHC class I-restricted CD8+ CTLs are expected to recognize and react to target epitopes presented by antigen presenting cells during the course of infection. Moreover, the cytokine interferon gamma (IFN-γ) produced by T cells has a critical role in the protective immunological response against primary infection. However, because the antigen specificity of the human T cell response is known to be strongly disciplined by HLA polymorphism [2], the immunogenic potential of candidate vaccines needs to be elucidated in the context of major HLA polymorphisms. Therefore, the identification of specific CTL epitopes in the context of major HLA alleles remains a critical step in both understanding mycobacterial control mechanisms and evaluating TB vaccines.
The antigenicity of TB seems to be largely dependent upon two important secreted proteins: the 6-KDa early secretory antigenic target (ESAT-6) and the 10-KDa culture filtrate protein (CFP10). It has been previously demonstrated that ESAT-6 and CFP10 are able to stimulate T cells to produce IFN-γ and display cytolytic CTL activity in humans infected with Mtb, suggesting them as potential candidates for application in an anti-tuberculosis subunit vaccine [3], [4]. A series of epitopes for CD8+ T cells, restricted by certain MHC molecules, have been identified [4], [5], [6], [7]. According to the report of the frequency of HLA alleles in humans, HLA-A24 is one of the most common HLA-A alleles worldwide, especially in Asian populations [8]. Nevertheless, relatively few epitopes presented by this molecule have been defined for human CD8 T cells in Mtb infection. In the present study, we adapted an approach utilizing a panel of synthetic peptides containing the HLA-A24-binding motifs for identification of CTL epitopes in ESAT-6 and CFP10 proteins.
2. Material and methods
2.1. Subjects
Seven TB patients (nos. 1–7) and two healthy controls were examined for the induction of TB-specific CTLs (Table 1 ). All patients were recruited from Nanjing Chest Hospital and The People's Hospital of Luhe, Nanjing. The diagnosis of tuberculosis was confirmed in all cases by one of the diagnostic standards developed by the Ministry of Health of the People's Republic of China: (1) two positive acid-fast bacilli (AFB) smears, or positive sputum culture for M. tuberculosis, and chest X-rays showing radiographical abnormalities characteristic of pulmonary tuberculosis; or (2) at least two sputum specimen negative for AFB and culture and chest X-rays showing signs of active pulmonary tuberculosis, accompanied with symptoms such as cough, cough with blood-tinged sputum, coughing up blood, chest distress, chest pain, and tiredness. Four patients (nos. 1–4) were HLA-A24-positive, and all other patients (nos. 5–7) were HLA-A24-negative. Both of the two healthy controls were HLA-A24-positive and had no history of tuberculosis and no history of contact with patients with tuberculosis. All patients were HIV negative. The TB patient group included three women and four men. The purpose and performance of the study were fully explained to all participants from Nanjing Chest Hospital and The People's Hospital of Luhe. Collection of peripheral blood mononuclear cell (PBMC) samples was authorized by the Hospital Ethics Review Committees.
Table 1.
HLA alleles of subjects used for inducing CTLs specific for the HLA-A24 synthetic peptides.
| Subjects | Age (yr) | Sex | HLA allele |
|---|---|---|---|
| TB patients | |||
| 1 | 17 | Male | A24 A31 |
| 2 | 24 | Male | A24 A11 |
| 3 | 23 | Female | A24 |
| 4 | 20 | Male | A24 A2 |
| 5 | 32 | Female | A2 A30 |
| 6 | 21 | Male | A2 |
| 7 | 51 | Female | A2 A11 |
| Healthy donors | |||
| 8 | 55 | Female | A24 A2 |
| 9 | 49 | Male | A24 A33 |
Subjects 1–4 are HLA-A24-positive TB patients; subjects 5–7 are HLA-A24-negative TB patients; subjects 8 and 9 are HLA-A24-positive healthy controls.
Fifteen milliliters of peripheral blood was obtained from each subject, and the PBMCs were prepared by Ficoll–Conray density gradient centrifugation. All of the samples were cryopreserved in liquid nitrogen until use in the experiments. The expression of HLA-A molecules on the PBMCs was determined as follows:
2.2. HLA typing
HLA typing of the subjects was done using fresh PBMCs. DNA was extracted from PBMCs using RelaxGene Blood (Tiangen, Beijing, China). Low resolution HLA typing was performed using PCR with sequence-specific primers (PROTRANS, Germany). MHC alleles were identified with the 20090122 version of SCORE Virtual Sequencing software. The results of the HLA typing of the subjects are listed in Table 1.
2.3. Peptides
Peptides were purchased from SBS Genetech (Beijing, China). Precise information regarding these peptides is provided in Table 2 . The purities of all the peptides used here were >90%, as determined by high performance liquid chromatography (HPLC). All peptides were dissolved in dimethylsulfoxide (Sigma, St. Louis, MO, USA) at a concentration of 10 mg/ml and stored at −70 °C.
Table 2.
Predicted HLA-A*24-restricted peptides for TB epitopes.
| No. | Sequence | Position | Scorea | HLA-A24 bindingb |
|---|---|---|---|---|
| P1 | Q Q A L S S Q M G F | CFP10, 91–100 | 187.3 | High |
| P2 | Q W R G A A G T A A | CFP10, 42–51 | 316.5 | Intermediate |
| P3 | E L N N A L Q N L | ESAT-6, 64–72 | 1015.9 | Intermediate |
| P4 | A Y Q G V Q Q K W | ESAT-6, 50–58 | 609.6 | No |
| P5 | Q W N F A G I E A A | ESAT-6, 5–14 | 411.2 | Intermediate |
| P6 | Q Y S R A D E E Q Q | CFP10, 82–91 | 635.8 | No |
| P8 | V T S I H S L L D | ESAT-6, 22–30 | 631.4 | High |
| P9 | E M K T D A A T L | CFP10, 3–11 | 306.4 | No |
In the evaluation of HLA-A24-restricted peptide prediction, the predicted binding score in the computer algorithm is reported as the IC50 value. IC50 refers to the concentration of peptide required to inhibit 50% of reporter peptide-MHC binding. The source of the peptide binding prediction was a web site: www.immuneepitope.org. (see Ref. [15]).
The binding of peptides to the RMA-S/hβ2m-A24 cell line was demonstrated by an increase in the fluorescence index MFI (see Ref. [15]). Peptides with an MFI ratio >1.5 defines a high-affinity binding, and intermediate binders have an MFI ratio of 1.25–1.5. The other peptides showed no binding affinity.
2.4. Cytokines
Human recombinant IL-2 and human recombinant IL-7 were purchased from R&D Systems (Minneapolis, MN, USA).
2.5. Refolding of candidate peptides with HLA-A24 heavy chain and β2m
Refolding was performed as described previously with minor modifications [9]. Briefly, the HLA-A24 heavy chain and β2m were expressed in a prokaryotic expression system. HLA-A24 heavy chain, β2m and peptide were subsequently added into refolding buffer (100 mM Tris/HCl, 400 mM l-arginine-HCl, 2 mM sodium EDTA, 0.5 mM oxidized glutathione, 5 mM reduced glutathione, 1 mM PMSF, 1 mg/ml pepstatin, and 1 mg/ml leupeptin). After stirring at 4 °C for 48 h, the soluble refolded portion was concentrated and then purified by Superdex G200 gel filtration chromatography (GE Healthcare Bio-Sciences AB) to indicate the correctly refolded peptide–HLA complex.
2.6. ELISPOT assay
A 96-well plate membrane was pre-coated with 10 μg/ml anti-IFN-γ mAb (Epigen, Beijing, China) overnight at 4 °C. PBMCs (2 × 105 cells per well) were added in duplicate wells. Candidate peptides were used at concentrations of 10 μg/ml to stimulate the effector cells. As a positive control, 5 μg/ml Phytohaemagglutinin (PHA) was also added to pairs of ELISPOT wells. TB-stimulants was provided by Epigen. After incubation (37 °C/5% CO2), the medium was discarded, and the wells were washed prior to the addition of anti-human biotinylated interferon-γ (Epigen, Beijing, China) at 2 μg/ml per well. The plates were incubated at room temperature for 2 h and then washed, followed by the addition of streptavidin–alkaline phosphatase and then the chromogenic substrate NBT/BCIP (Epigen, Beijing, China). After development, SFCs were counted using an ELISPOT reader (Autoimmun Diagnostika GmbH, Strasberg, Germany). Typically, the negative control will have less than 5 spots. In this case, a positive or reactive sample will have at least 6 spots more than the negative control. If the negative control has 6 or more spots, a response was considered positive if the well contained more than two times the mean SFCs of the negative controls.
2.7. In vitro stimulation of IFN-γ-producing CTLs with selected peptides
PBMCs were separated from the whole blood of two HLA-A24+ and one HLA-A24− TB patients, as well as one HLA-A24+ healthy control. PBMCs (2 × 106/ml) were cultured with P5 peptide at a concentration of 10 μM in 1 ml of medium consisting of RPMI 1640 supplemented with 10% fetal calf serum, amino acids, pyruvate, antibiotics, 20 ng/ml rhIL-7, and 20 units/ml rhIL-2 (EuroCetus, Amsterdam, Netherlands) in 24-well tissue culture plates. Half of the medium was then changed every 3 days with supplementation of rhIL-2 at a final concentration of 20 U/ml. At day 10, the cells were harvested and tested for the presence of peptide-specific CD8+ T cells by an IFN-release ELISPOT assay.
2.8. Tetramer preparing and staining
Tetrameric HLA-A24–peptide complex–peptide molecules containing candidate peptides were constructed using the previously described method [10]. The cells were incubated at 37 °C for 20 min with the PE-labeled tetrameric complex and washed once with PBS prior to incubation at 4 °C with FITC-labeled anti-CD8 mAb (eBioscience, San Diego, CA, USA). The samples were detected by flow cytometry (Guava easycyte). Tetramer-positive cells gated from CD8+ T lymphocytes were counted as epitope-specific CTLs.
2.9. Statistical evaluation
Statistical analysis was performed using the unpaired two-tailed Student's t-test to compare cytokine release. Differences were considered significant when the P was <0.05.
3. Results
3.1. An HLA-A24-restricted epitope was identified by ex vivo color IFN-γ ELISPOT assay and tetramer staining with freshly isolated PBMCs
To identify peptides recognized by TB-specific CTLs in the context of HLA-A24 molecules, the IFN-γ ELISPOT assay was performed using freshly isolated PBMCs. PBMC samples obtained from 20 donors who had recovered from TB were typed for HLA-A24 expression. The samples of 4 donors were HLA-A24-positive. Each of the 8 candidate peptides from TB-ESAT-6 and CFP10 were tested for their capacity to stimulate IFN-secretion in the PBMCs from the four HLA-A24+ TB donors ex vivo.
The induction of IFN-secretion was revealed by a 24-h direct ELISPOT assay with freshly isolated PBMCs. As shown in Fig. 1 , P5 significantly elicited specific IFN-producing CTLs from the PBMCs of all HLA-A24+ TB donors compared to the negative controls (40.6 ± 17.8 vs. 10.7 ± 3.7 spot-forming cells (SFCs)/2 × 105 PBMCs, p < 0.05). Such a T cell response was not observed from the PBMCs stimulated by the other tested peptides except P8 (20.1 ± 17.9 vs. 10.7 ± 3.7). However, there was no statistical difference in IFN-producing cell numbers between the P8 group and negative control wells. Both HLA-A24− TB donors and HLA-A24+ healthy donors had similar levels of SFCs in the P5 group as in the negative controls.
Fig. 1.
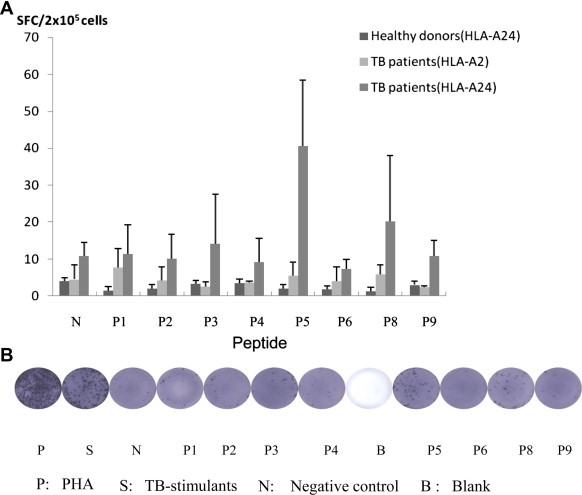
Enumeration of SFCs in a 24-h direct ELISPOT assay performed with freshly isolated PBMCs from HLA-A24+ TB donors, HLA-A24− TB donors, or HLA-A2 TB donors. (A) Results are means ± SD from three separate experiments. The spots are a measure of IFN-γ-producing cells from PBMCs stimulated with various candidate peptides. (B) Results are from one representative experiment. 5 μg/ml PHA and TB-stimulants were used as positive controls. No peptide was added in the negative control.
Consequently, an HLA-A24/P5 tetramer was prepared and used to confirm the existence of P5-specific CTLs. Fresh PBMCs from HLA-A24+ healthy donors, HLA-A24+ TB donors, and HLA-A24− TB donors were stained with HLA-A24/P5 tetramer, and the proportion of P5-specific CD8+ T cells was detected by flow cytometry. The results showed that the percentages of P5-specific CTLs detected from all HLA-A24+ TB donors were an average of three times higher than those in the PBMCs of all tested HLA-A24+ healthy controls and HLA-A24− TB donors (Fig. 2 ).
Fig. 2.
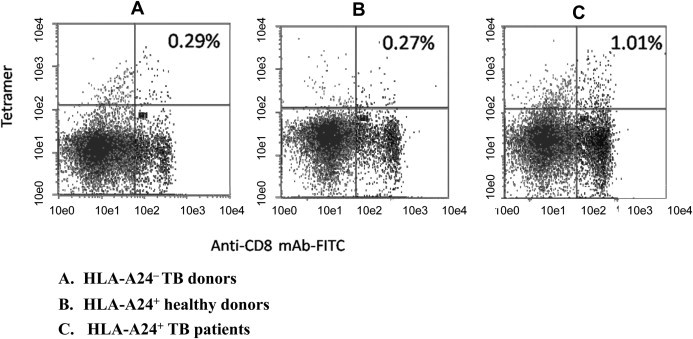
Measurement of P5-specific CTLs by tetramer staining. P5-specific CTLs were measured from fresh PBMCs of (A) HLA-A24− TB donors; (B) HLA-A24+ healthy donors; and (C) HLA-A24+ TB patients, using PE-labeled HLA-A24/P5 tetramer along with FITC-labeled anti-CD8 mAbs for cell staining. The numbers shown represent the percentage of tetramer+ cells within CD3 + CD8+ lymphocytes. The results are representative of three independent experiments.
3.2. Identified peptide could stimulate bulk peptide-specific CTL expansion from HLA-A24-positive TB patients with ELISPOT assay
The PBMCs were stimulated for 10 days in the presence of P5 peptide and 20 U/ml recombinant human interleukine-2(rhIL-2) and 20 ng/ml recombinant human interleukine-7(rhIL-7). The induction of IFN-secretion was revealed by the ELISPOT assay in the presence of P5. We adopted the assay because it can test many samples simultaneously in a few plates using relatively small numbers of CTLs. As shown in Table 3 , the polyclonal TB-specific CD8+ T cells established from PBMCs of the HLA-A24+ TB donor produced significant numbers of IFN-γ spots when incubated with P5 peptide, while the responders generated from the controls produced small numbers of spots. Peptide P5 significantly elicited specific IFN-producing CTLs from the PBMCs of all HLA-A24+ TB donors, compared to the HLA-A24+ healthy controls or HLA-A2+ TB donors.
Table 3.
Induction of P5-specific CTLs in PBMC cultures from TB-infected patients.
| IFN-γspot-forming unit | ||
|---|---|---|
| Group |
Ex vivo |
Cells cultured for 10 days in the presence of P5 peptide |
| Per 2 × 105 cells | Per 5 × 104 cells | |
| A24 TB patient 1 | ||
| Positive control (+) | 223 ± 77 | 583 ± 151 |
| Negative control (−) | 3 ± 3 | 11 ± 9 |
| Sample | 22 ± 20 | 213 ± 54 |
| A24 TB patient 2 | ||
| Positive control (+) | 226 ± 37 | 639 ± 67 |
| Negative control (−) | 8 ± 2 | 6 ± 4 |
| Sample | 36 ± 9 | 193 ± 48 |
| A24 healthy donor | ||
| Positive control (+) | 233 ± 42 | 548 ± 80 |
| Negative control (−) | 7 ± 4 | 8 ± 6 |
| Sample | 7 ± 5 | 48 ± 24 |
| A2 TB patient | ||
| Positive control (+) | 179 ± 35 | 519 ± 29 |
| Negative control (−) | 5 ± 4 | 6 ± 2 |
| Sample | 4 ± 4 | 58 ± 8a |
Mean ± SD from three independent experiments. Values shown are the mean number of IFN-γ+ cells, based on duplicated wells.
3.3. Identified epitope is a weak binder for HLA-A24 confirmed by a mini MHC refolding assays
To test the binding affinity of the candidate peptides to HLA-A24 molecules, we adopted a peptide-induced stabilization assay of the HLA-A24 heavy chain and light chain in vitro. All the 8 predicted peptides could refold with HLA-A24 H chain and β2m molecules with varying avidities (Fig. 3 ). Among these, P1, P3, P4, and P6 had stronger avidities. Compared to the positive control peptide and those peptides with higher avidities, P5 had a lower binding efficiency to HLA-A24 molecules. Peptide N1(QFKDNVILL) acted as the positive control and showed a high binding capacity to HLA-A24 molecules. HLA-A24 heavy chain and light chain could not form the MHC complexes in the presence of the HLA-A2-restricted peptide GILGFVFTL.
Fig. 3.
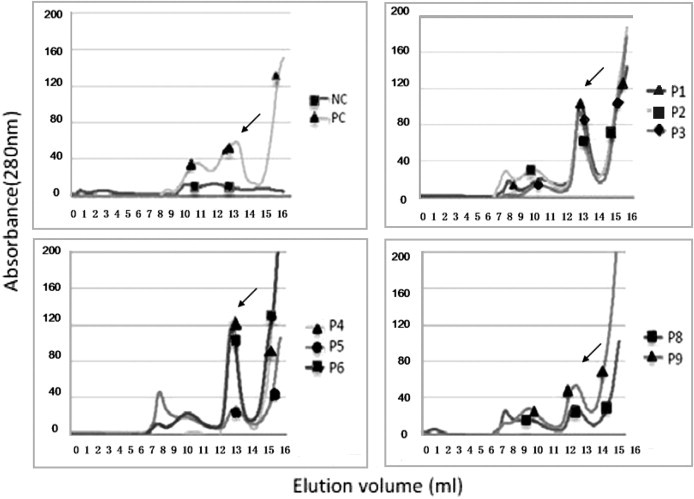
In vitro refolding of the HLA-A24 heavy chain and β2m molecules with candidate peptides of ESAT-6 and CFP10 proteins. The refolded complexes were tested by fast protein liquid chromatography Superdex G75 gel filtration. The positive control (PC) peptide was a HLA-A24-restricted severe acute respiratory syndrome coronavirus (SARS-CoV) nucleocapsid (N) protein-derived CTL epitope, N1 (QFKDNVILL), and the negative control (NC) peptide was a HLA-A2-restricted influenza A matrix peptide 58-66 (GILGFVFTL). All the peptide could refold with the HLA-A24 heavy chain and light chain. The results are representative of three independent experiments.
4. Discussion
M. tuberculosis, which is the leading cause of death from a single bacterial species, severely impacts global health [11]. Vaccination with Mycobacterium bovis BCG (bacillus Calmette-Guérin) reduces the severity of tuberculosis in children; however, there is limited evidence of the efficacy of BCG vaccination when applied to adults [12]. Furthermore, vaccination may cause severe disease in immunocompromised patients, such as those infected with HIV or suffering from tumors [13]. T cells play a vital role in fighting against tuberculosis. It has been proven that CD4+ T cells are essential for immunity and that CD8 T cells are indispensable parts of the anti-bacterial immunity [5]. Considering that BCG is a powerful stimulator of CD4 T cells and a poor stimulator of CD8 T cells, further work should focus on eliciting both potent CD8 T cell and CD4 T cell responses to achieve better immunity [14].
In this study, we used an approach starting with a computer motif prediction. The predicted binding affinity in the computer algorithm is reported as the IC50 value. A panel of 9- and 10-mer peptides derived from TB ESAT-6 and CFP10 proteins were selected for screening based on computer algorithms [15]. All the peptides could bind with the H and L chains of HLA-A24 molecules with various avidities in vitro, as determined by dilution refolding. The candidate peptides were previously tested for their capacity to bind to HLA-A24 molecules on the cell surface [15]. We observed that the affinities of these peptides were not entirely consistent when measured by the two assays. Peptides P4, P6 and P9 could refold with the HLA-A24 heavy chain and light chain; however, no binding affinity was observed with the three peptides in the cell surface stabilization assay [15]. It has been suggested that factors such as temperature or the conformation of the heavy chain of MHC class I may affect the formation of MHC-I molecular complexes, which may account for this discrepancy [16]. To verify the results, we assayed PBMCs from TB donors to determine the CD8+ T cell-specific response to the candidate peptides. Only peptide P5-specific CTLs from HLA-A24+ TB donors were significantly detected by stimulated ELISPOT assays and tetramer staining. Our study demonstrates that, although in vitro refolding is an effective approach to identify HLA class I-restricted T cell epitopes, the peptides which have refolding capacity with the corresponding HLA allele, for example P4, may not react well with T cells in further assays, such as the ELISPOT assay and tetramer staining. While the peptides which present lower binding capacity with the target HLA allele, for example P5, may distinctly induced peptide-specific CTLs. We assume that some potential T cell epitopes may be missed if we use the refolding test solely to identify peptides because there may be some peptides that bind HLA with relatively low affinity but still bind TCR with high affinity. Therefore, establishing that an immunodominant HLA-A24-restricted, TB-specific CD8+ T cell epitope exists would require screening by an IFN-γ ELISPOT assay and tetramer staining.
This study demonstrates the presence of HLA-A24-restricted CD8 CTL specific for an M. tuberculosis protein antigen in infected individuals. Different levels of epitope-specific CD8 T cells may be found between subjects with active Mtb infection and those who contain infection (LTBI individuals). During active disease, in which antigen load is high, large numbers of CD8 T cells are indeed sequestered in the lung, and not only in peripheral blood [17]. The results obtained with PBMC in humans may not necessarily reflect the local immune response to the infection. The CTL epitope discovered could be further investigated for the subjects with latent TB where either elevated T cell frequencies could be the major force for the containment of infection, or the consequence of the persistence of pathogens inside macrophages for a long time [18].
In conclusion, we provide evidence that ESAT-6-derived peptide P5 (QWNFAGIEAA) may be a novel, naturally processed HLA-A24-restricted CTL epitope, which adequately sensitized the target for recognition by CTL in the Mtb-infected patients carrying HLA-A24. Peptide P5 was a weak binder for HLA-A24 molecule and light chain in the refolding assay, and showed medium affinity in the MHC stabilization assay [15]. This sequence, encompassing residues QWNFAGIEAA, is located in the N-terminal side of the ESAT-6 protein, whose corresponding gene (esat-6) is located in RD1, which is a 10-kb DNA region deleted in the attenuated tuberculosis vaccine BCG [19]. It is well known that ESAT-6 is a dominant target for cell-mediated immunity and is frequently recognized during early TB infection [20]. In this regard, the HLA-A24-restricted CTL epitope identified in this study may be useful to enhance the CTL activity specific for Mtb protein to prevent the spread of mycobacteria in a large population of Mtb-infected patients.
Conflicts of interest
None of the authors has a conflict of interest related to this study.
Acknowledgments
The authors acknowledge the following supports: The Key Project supported by the Medical Science and Technology Development Foundation, Nanjing Department of Health (2007, No. 18); Scientific Program for Key Infectious Diseases, The Ministry of Science and Technology (2008ZX10003-012, 2009ZX10601 and 2009ZX10004-305); CAS Grant (KSCX2-YW-R-164) and Science Grant from Nanjing Health Bureau (ZKX08020).
References
- 1.National Technical Steering Group of the Epidemiological Sampling Survey for Tuberculosis Report on fourth national epidemiological sampling survey of tuberculosis. Chin J Tuberc Respir Dis (Chin) 2002;25:3–7. [PubMed] [Google Scholar]
- 2.Geluk A., Taneja V., van Meijgaarden K.E., Zanelli E., Abou-Zeid C., Thole J.E.R. Identification of HLA class II-restricted determinants of M. tuberculosis-derived proteins using HLA-transgenic, class II deficient mice. Proc Natl Acad Sci U S A. 1998;95:10797. doi: 10.1073/pnas.95.18.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arend S.M., Geluk A., van Meijgaarden K.E., van Dissel J.T., Theisen M., Andersen P. Antigenic equivalence of human T-cell responses to Mycobacterium tuberculosis-specific RD1-encoded protein antigens ESAT-6 and culture filtrate protein 10 and to mixtures of synthetic peptides. Infect Immun. 2000;68:3314. doi: 10.1128/iai.68.6.3314-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalvani A., Brookes R., Wilkinson R., Malin A., Pathan A., Andersen P. Human cytolytic and interferon γ-secreting CD8+T lymphocytes specific for Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1998;95:270. doi: 10.1073/pnas.95.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shams H., Klucar P., Weis S.E., Lalvani A., Moonan P.K., Safi H. Characterization of a Mycobacterium tuberculosis peptide that is recognized by human CD4+ and CD8+ T cells in the context of multiple HLA alleles. J Immunol. 2004;173:1966–1977. doi: 10.4049/jimmunol.173.3.1966. [DOI] [PubMed] [Google Scholar]
- 6.Geluk A., van Meijgaarden K.E., Franken K.L., Drijfhout J.W., D'Souza S., Necker A. Identification of major epitopes of Mycobacterium tuberculosis AG85B that are recognized by HLA-A*0201-restricted CD8+T cells in HLA-transgenic mice and humans. J Immunol. 2000;165:6463–6471. doi: 10.4049/jimmunol.165.11.6463. [DOI] [PubMed] [Google Scholar]
- 7.Billeskov R., Vingsbo-Lundberg C., Andersen P., Dietrich J. Induction of CD8 T cells against a novel epitope in TB10.4:correlation with mycobacterial virulence and the presence of a functional region of difference-1. J Immunol. 2007;179(6):3973–3981. doi: 10.4049/jimmunol.179.6.3973. [DOI] [PubMed] [Google Scholar]
- 8.Kondo A., Sidney J., Southwood S., del Guercio M.F., Appella E., Sakamoto H. Prominent roles of secondary anchor residues in peptide binding to HLA-A24 human class I molecules. J Immunol. 1995;155:4307–4312. [PubMed] [Google Scholar]
- 9.Garboczi D.N., Hung D.T., Wiley D.C. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci U S A. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman J.D., Moss P.A.H., Goulder P.J.R., Barouch D.H., McHeyzer-Williams M.G., Bell J.I. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 11.Coker R.J. Review: multidrug-resistant tuberculosis: public health challenges. Trop Med Int Health. 2004;9:25–40. doi: 10.1046/j.1365-3156.2003.01156.x. [DOI] [PubMed] [Google Scholar]
- 12.Chan E.D., Iseman M.D. Current medical treatment for tuberculosis. BMJ. 2002;325(7375):1282–1286. doi: 10.1136/bmj.325.7375.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sterling T.R., Brehm W.T., Moore R.D., Chaisson R.E. Tuberculosis vaccination versus isoniazid preventive therapy: a decision analysis to determine the preferred strategy of tuberculosis prevention in HIV-infected adults in the developing world. Int J Tuberc Lung Dis. 1999;3:248. [PubMed] [Google Scholar]
- 14.Wang J.L., Zhang H.M., Wang H.H. Analysis of predicted CD8+ T cell epitopes from proteins encoded by the specific RD regions of Mycobacterium tuberculosis for vaccine development and specific diagnosis. Mol Biol Rep. 2010;37:1793–1799. doi: 10.1007/s11033-009-9613-4. [DOI] [PubMed] [Google Scholar]
- 15.Ding J., Wang Y., Cheng T.T., Chen X.W., Gao B. Identification of HLA-A24-binding peptides of Mycobacterium tuberculosis derived proteins with beta 2m linked HLA-A24 single chain expressing cells. Immunol Invest. 2010;39:103–113. doi: 10.3109/08820130903496777. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M.H., Xu D.P., Li X.J., Li H.T., Shan M., Tang J.R. Screening and Identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- 17.Caccamo N., Guggino G., Meraviglia S., Gelsomino G., Di Carlo P., Titone L. Analysis of Mycobacterium tuberculosis-specific CD8 T cells in patients with active tuberculosis and in individuals with latent infection. PLoS ONE. 2009;4(5):e5528. doi: 10.1371/journal.pone.0005528. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Tully G., Kortsik C., Höhn H., Zehbe I., Hitzler W.E., Neukirch C. Highly focused T cell responses in latent human pulmonary Mycobacterium tuberculosis infection. J Immunol. 2005;174:2174–2184. doi: 10.4049/jimmunol.174.4.2174. [DOI] [PubMed] [Google Scholar]
- 19.Berthet F.X., Rasmussen P.B., Rosenkrands I., Andersen P., Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10) Microbiology. 1998;144(Pt 11):3195–3203. doi: 10.1099/00221287-144-11-3195. [DOI] [PubMed] [Google Scholar]
- 20.Ravn P., Demissie A., Eguale T., Wondwosson H., Lein D., Amoudy H.A. Human T cell responses to the ESAT-6 antigen from Mycobacterium tuberculosis. J Infec Dis. 1999;179(3):637–645. doi: 10.1086/314640. [DOI] [PubMed] [Google Scholar]


