Abstract
The causative agent for the most fatal form of malaria, Plasmodium falciparum, has developed insecticide and drug resistance with time. Therefore combating this disease is becoming increasingly difficult and this calls for finding alternate ways to control malaria. One of the feasible ways could be to find out inhibitors/drugs specific for the indispensable enzymes of malaria parasite such as helicases. These helicases, which contain intrinsic nucleic acid-dependent ATPase activity, are capable of enzymatically unwinding energetically stable duplex nucleic acids into single-stranded templates and are required for all the nucleic acid transactions. Most of the helicases contain a set of nine extremely conserved amino acid sequences, which are called ‘helicase motifs’. Due to the presence of the DEAD (Asp–Glu–Ala–Asp) in one of the conserved motifs, this family is also known as the ‘DEAD-box’ family. In this review, using bioinformatic approach, we describe the ‘DEAD-box’ helicases of malaria parasite P. falciparum. An in depth analysis shows that the parasite contains 22 full-length genes, some of which are homologues of well-characterized helicases of this family from other organisms. Recently we have cloned and characterized the first member of this family, which is a homologue of p68 and is expressed during the schizont stage of the development of the parasite [Pradhan, A., Chauhan, V.S., Tuteja, R., 2005a. A novel ‘DEAD-box’ DNA helicase from Plasmodium falciparum is homologous to p68. Mol. Biochem. Parasitol. 140, 55–60.; Pradhan A., Chauhan V.S., Tuteja R., 2005b. Plasmodium falciparum DNA helicase 60 is a schizont stage specific, bipolar and dual helicase stimulated by PKC phosphorylation. Mol. Biochem. Parasitol. 144, 133–141.]. It will be really interesting to clone and characterize other members of the ‘DEAD-box’ family and understand their role in the replication and transmission of the parasite. These detailed studies may help to identify a parasite-specific enzyme, which could be a potential drug target to treat malaria. The various steps at which this probable drug can act are also discussed.
Abbreviations: ATP, adenosine triphosphate; DEAD, single letter code for amino acids Asp–Glu–Ala–Asp; eIF-4A, eukaryotic initiation factor 4A; NTP, nucleoside triphosphate; p68, protein 68 kDa; SF, super-family
Keywords: Helicase, Malaria parasite, Motor proteins, Plasmodium falciparum, Unwinding activity
1. Introduction
1.1. Malaria and the genome of P. falciparum
Malaria caused by Plasmodium, particularly P. falciparum, is the most serious and widespread parasitic disease of humans. Each year approximately 300–500 million people become infected with malaria and 2–3 million die as a result. The malaria parasite has a four-stage life cycle as it passes from humans to mosquitoes and back again. The parasite is very efficient at evading the human immune response. A malaria vaccine would be the ultimate weapon to fight this deadly disease but unfortunately despite encouraging advances a vaccine is not likely soon. Anti-malarial drugs are the last certain defense but due to the rapid spread of resistance of both the parasite and the mosquito vector to currently available anti-malarial drugs and insecticides, control of malaria is becoming increasingly difficult. There is a need to bring new classes of drugs, both alone and in combination to meet the future challenges of drug resistance and other areas of medical needs. There are a limited number of drugs in widespread use for treatment of P. falciparum malaria and a lack of new affordable drugs (Winstanley, 2000). P. falciparum has developed resistance to nearly all the available anti-malarial drugs (White, 1998, Hyde, 2005). The rational development of novel and affordable drugs for the treatment of malaria and the identification of new drug targets is an important goal. The recent completion of Malaria Genome Project and availability of new technologies for genome wide comparison of genomes will help to identify key targets in biochemical pathways that are parasite specific and can be interrupted without deleterious consequences for the host. The genome of P. falciparum has 14 chromosomes (which encode about 5400 genes), a circular plastid-like genome and a linear mitochondrial genome (Bozdech et al., 2003).
The completed genome of P. falciparum has opened new avenues for research. It has been reported that homologues for a number of genes are present in the genome of the parasite (Gardner et al., 2002). On the other hand the studies report that of the 5268 predicted proteins, about 60% did not contain adequate resemblance to proteins reported in other organisms. This may be due to the A + T richness of the Plasmodium genome. It is interesting to note that the overall (A + T) composition of P. falciparum genome is ∼ 81% and it increases to ∼ 90% in introns and intergenic regions (Gardner et al., 2002). Studies have indicated that overall the proteins from P. falciparum are consistently larger than their homologous counterparts from other species (Pizzi and Frontali, 2001). This size difference can normally be attributed to the presence of long insertions, which separate the well-conserved regions that are flanking in the homologous proteins (Pizzi and Frontali, 2000, Pizzi and Frontali, 2001). Previous studies have indicated that the genes for DNA metabolism such as DNA replication, repair and recombination are present in the genome of P. falciparum and the apicoplast of P. falciparum also contains a gene encoding contiguous DNA polymerase, DNA primase and DNA helicase components (Gardner et al., 2002, Seow et al., 2005).
1.2. Nucleic acid metabolism and helicases
DNA recombination, repair and replication are necessary for the maintenance and the reliable transmission of genetic information to the offspring. Each of these processes requires opening of the double helix to provide a single-stranded template. Helicases are enzymes, which catalyze the unwinding of nucleic acid duplexes in an NTP-dependent manner. Therefore all the helicases contain a distinct nucleic acid binding site and an NTP-binding site. These enzymes are also called “molecular motors” that use the energy of hydrolysis of NTP to separate energetically stable duplex momentarily into single strands. These are important enzymes, which play essential roles in all aspects of nucleic acid metabolism. The helicase genes have been found in a variety of organisms ranging from bacteria to eukaryotes (Tuteja and Tuteja, 2004a). It is interesting to note that in the genome of the yeast Saccharomyces cerevisiae there are 134 open reading frames, which code for helicase-like proteins (Shiratori et al., 1999).
1.3. Conserved helicase core
Primary structure sequence analysis and comparisons of a number of helicases from a variety of organisms have revealed the presence of seven to nine short highly conserved amino acid sequence motifs or fingerprints which are known as ‘helicase signature motifs’ and are designated as Q, I, Ia, Ib, II, III, IV, V and VI (Hodgman, 1988, Gorbalenya et al., 1988, Gorbalenya and Koonin, 1993, Tanner, 2003, Tuteja and Tuteja, 2004b). Various structural studies have shown that in three-dimensional structure of the protein these motifs are generally clustered in the middle of the protein to form an ATP-hydrolyzing pocket, which is capable of providing energy for the unwinding activity (Hall and Matson, 1999). Due to the presence of DEAD, DEAH or DEXH in motif II, this family of helicases is also known as the ‘DEAD-box’ protein family (Gorbalenya et al., 1988, Gorbalenya et al., 1989, Linder et al., 1989). Similar to helicases this family of proteins is also present ubiquitously ranging from bacteria to mammals (Tuteja and Tuteja, 2004a). Some partial genes of this family have also been reported from the malaria parasites P. falciparum and P. cynomolgi (Song et al., 1999). It has been reported that humans have 36 members and S. cerevisiae has 26 members of the ‘DEAD-box’ family of helicases (de la Cruz et al., 1999, Abdelhaleem et al., 2003). The structural studies of various RNA and DNA helicases have shown that there is a close association between the conserved motifs and the three-dimensional structures of the enzymatic cores (Tanner and Linder, 2001). These observations suggest that although various helicases have different biological activities, their catalytic cores are almost identical (Hall and Matson, 1999, Tanner and Linder, 2001). ‘DEAD-box’ helicases also have roles in ribosome biogenesis and act by way of regulation of small ribosomal and nucleolar RNAs. Two members of this ‘DEAD-box’ family, eIF-4A and p68 have been well characterized and shown to contain ATP-dependent RNA helicase activity (Hirling et al., 1989, Rozen et al., 1990). Mutational analysis of eIF-4A and p68 indicate that these are required for cell growth and translation in yeast and other systems (Schmid and Linder, 1991, Wassarman and Steitz, 1991). It has been suggested that in humans the helicases of the ‘DEAD-box’ family have roles in differentiation and carcinogenesis (Abdelhaleem et al., 2003).
The discovery of ‘helicase signature motifs’ resulted in the classification of these important proteins into three distinct super-families (SF) such as SF1, SF2 and SF3. This classification is based on the extent of similarity and organization of the helicase signature motifs (Gorbalenya and Koonin, 1993). SF1 and 2 represent the major and closely related super-families, which contain at least seven conserved motifs such as I, Ia, II–VI. The members of these families contain enzymes from a variety of organisms such as bacteria to humans. On the other hand SF3 is very small and contains the viral Rep-like helicases. It has been observed that the conserved motifs of SF1 and SF2 are generally clustered in the central 200–700 amino acids known as the ‘core-region’ of the protein. The crystal structure comparison studies between SF1 and SF2 have revealed that the helicases are highly conserved at the structural level throughout evolution (Hall and Matson, 1999). The stretches of low sequence but high length conservation separate the various conserved motifs. Contrary to this, the N-terminal and C-terminal regions are characterized by a high degree of sequence as well as length variability. It is well documented now that the highly conserved motifs are responsible for ATP-binding and hydrolysis or binding and unwinding of nucleic acid duplexes. However the divergent N- and C-terminal regions have been suggested to be responsible for individual protein functions (Hall and Matson, 1999). It has also been postulated that the non-conserved or subsidiary part of the various helicase proteins may contain special domains responsible for protein–protein interaction, cellular localization and oligomerization (Hall and Matson, 1999). These domains can also directly bind to the specific nucleic acid substrate and help to deliver the helicase enzyme to its place of action (Silverman et al., 2003). The ‘Q motif’, which was discovered recently, has been shown to be involved in ATP-binding and hydrolysis (Tanner, 2003, Tanner et al., 2003).
2. Bioinformatics of ‘DEAD-box’ proteins of P. falciparum
The completion of P. falciparum genome prompted us to search for the presence of members of putative ‘DEAD-box’ protein family in the parasite. The database of the genome of P. falciparum PlasmoDB (Bahl et al., 2003) was investigated using keyword ‘helicase’ as a query. There were a total of 60 positive hits after this query. The results of this search are presented in Table 1 . This list includes all of the genes containing putative helicase in their gene name. Each positive hit was analyzed further in detail and the genes containing the ‘DEAD-box’ motif were selected for further studies. Out of all the ‘DEAD-box’ genes, only the genes, which contained the complete open reading frame with the presence of all the conserved boxes (shown in bold in Table 1) were considered and further studied in detail.
Table 1.
List of all helicases in Plasmodium falciparum
| Gene ID | Gene names |
|---|---|
| 1. MAL6P1.192 | ATP-dependent DEAD-box helicase, putative |
| 2. MAL6P1.24 | ATP-dependent RNA helicase, putative |
| 3. MAL6P1.49 | DNA helicase, putative |
| 4. MAL6P1.119 | DEAD/DEAH-box ATP-dependent RNA helicase, putative |
| 5. PFB0445c | Helicase, putative |
| 6. PFB0730w | DNA helicase, putative |
| 7. PFB0860c | RNA helicase, putative |
| 8. PFC0440c | Helicase, putative |
| 9. PFC0915w | ATP-dependent RNA helicase, putative |
| 10. PFD1070w | Eukaryotic initiation factor, putative |
| 11. PFC0955w | ATP-dependent RNA helicase |
| 12. PFD0565c | RNA helicase, putative |
| 13. PFE0205w | ATP-dependent helicase, putative |
| 14. PFE0215w | ATP-dependent helicase, putative |
| 15. PFE0430w | ATP-dependent RNA helicase, putative |
| 16. PFE0705c | Helicase, belonging to UvrD family, putative |
| 17. PFE1085w | DEAD-box subfamily ATP-dependant helicase, putative |
| 18. PFE1390w | RNA helicase-1 |
| 19. PFF0100w | Putative ATP-dependent RNA helicase |
| 20. PFF0225w | DNA helicase, putative |
| 21. PFF1140c | ATP-dependent DEAD-box helicase, putative |
| 22. PFF1500c | DEAD/DEAH-box ATP-dependent RNA helicase, putative |
| 23. MAL7P1.113 | DEAD-box helicase, putative |
| 24. PF08_0100 | RuvB-like DNA helicase, putative |
| 25. PF08_0096 | RNA helicase, putative |
| 26. PF08_0048 | ATP-dependant helicase, putative |
| 27. PF08_0042 | ATP-dependent RNA helicase prh1, putative |
| 28. PFI0165c | DEAD/DEAH-box helicase, putative |
| 29. PFI0480w | Helicase with Zn-finger motif, putative |
| 30. PFI0860c | ATP-dependant RNA helicase, putative |
| 31. PFI0910w | DNA helicase, putative |
| 32. PFI1650w | DNA excision-repair helicase, putative |
| 33. PF10_0209 | RNA helicase, putative |
| 34. PF10_0294 | RNA helicase, putative |
| 35. PF10_0369 | Helicase, putative |
| 36. PF11_0071 | RuvB DNA helicase, putative |
| 37. PFL0100c | ATP-dependent RNA helicase, putative |
| 38. PFL1310c | ATP-dependent RNA helicase, putative |
| 39. PFL1525c | Pre-mRNA splicing factor RNA helicase, putative |
| 40. PFL2010c | DEAD/DEAH-box helicase, putative |
| 41. PFL2475w | DEAD/DEAH-box helicase, putative |
| 42. MAL13P1.14 | ATP-dependent DEAD-box helicase, putative |
| 43. PF13_0037 | DEAD-box helicase, putative |
| 44. PF13_0077 | DEAD-box helicase, putative |
| 45. MAL13P1.134 | Helicase, putative |
| 46. MAL13P1.166 | Helicase, putative |
| 47. PF13_0177 | ATP-dependent RNA helicase, putative |
| 48. MAL13P1.216 | DNA helicase, putative |
| 49. PF13_0308 | DNA helicase |
| 50. PF13_0330 | ATP-dependent DNA helicase, putative |
| 51. PF14_0081 | DNA repair helicase, putative |
| 52. PF14_0183 | RNA helicase, putative |
| 53. PF14_0185 | ATP-dependent RNA helicase, putative |
| 54. PF14_0278 | ATP-dependent DNA helicase, putative |
| 55. PF14_0370 | RNA helicase, putative |
| 56. PF14_0429 | RNA helicase, putative |
| 57. PF14_0436 | Helicase, truncated, putative |
| 58. PF14_0437 | Helicase, truncated, putative |
| 59. PF14_0563 | DEAD-box RNA helicase, putative |
| 60. PF14_0655 | RNA helicase-1, putative |
Keyword helicase in gene name.
Table 2 shows the ‘PlasmoDB’ gene number (systematic name), chromosomal location, gene and protein size and the status of exons/introns of these putative helicases. It is interesting to note that these 22 full-length ‘DEAD-box’ genes are unequally distributed on each chromosome. The chromosome number 3, 4, 6, 7, 8 and 10 each contain one gene, chromosome number 2 and 13 each contain 2 genes, chromosome number 12 contains 3 genes, chromosome number 5 has 4 genes and chromosome number 14 has 5 genes. As described for the majority of the P. falciparum genes, it is noteworthy that most of these helicase genes (fourteen) are intron-less. Only eight genes contain introns and the number of introns ranges from 1 to 3 (Table 2). The evolution of intron/exon structures of ‘DEAD-box’ helicase family genes has been studied and it was reported that the mean number of introns per gene is seven in Arabidopsis thaliana, six in Caenorhabditis elegans and three in Drosophila melanogaster (Boudet et al., 2001). The size of proteins encoded by P. falciparum ‘DEAD-box’ helicase genes ranges from 44 to 164 kDa (Table 2). The most probable functions of these genes based on the studies of their homologues in other systems and the sequence similarity to the highest similar homologues are shown in Table 3 . The various characteristics of individual important motifs of all of these putative members are described in detail in the following sections.
Table 2.
‘DEAD-box’ helicases of Plasmodium falciparum
| S. no. | PlasmoDB no. | Location (chr. no./position) | Size (gene/protein) kb/kDa | Exon (no.) | Intron (no.) |
|---|---|---|---|---|---|
| 1. | PFB0445c | 2 comp. (404281..406355) | 1374/50 | 2 | 1 |
| 2. | PFB0860c | 2 comp. (750048..751736) | 1689/62 | 1 | 0 |
| 3. | PFC0915w | 3 (867487..869012) | 1302/48 | 2 | 1 |
| 4. | PFD1070w | 4 (1044910..1046082) | 1173/45 | 1 | 0 |
| 5. | PFE 0215w | 5 (172150..174417) | 2268/83 | 1 | 0 |
| 6. | PFE0430w | 5 (358261..362733) | 4473/164 | 1 | 0 |
| 7. | PFE1085w | 5 (882373..884898) | 2526/93 | 1 | 0 |
| 8. | PFE1390w | 5 (1153670..1155667) | 1998/73 | 1 | 0 |
| 9. | PFF 1500c | 6 comp. (1291038..1292975) | 1806/66 | 2 | 1 |
| 10. | MAL7P1.113 | 7 (977663..980362) | 2700/99 | 1 | 0 |
| 11. | PF08_0096 | 8 (543092..545917) | 2826/104 | 1 | 0 |
| 12. | PF10_0209 | 10 comp. (867633..869675) | 2043/75 | 1 | 0 |
| 13. | PFL1310c | 12 comp. (1093533..1095761) | 2229/82 | 1 | 0 |
| 14. | PFL2010c | 12 comp. (1779911..1782635) | 2685/99 | 2 | 1 |
| 15. | PFL2475w | 12 (2099837..2102082) | 2154/79 | 2 | 1 |
| 16. | PF13_0037 | 13 (333800..335692) | 1893/69 | 1 | 0 |
| 17. | PF13_0177 | 13 comp. (1345004..1346476) | 1473/54 | 1 | 0 |
| 18. | PF14_0183 | 14 (782104..785818) | 2841/104 | 3 | 2 |
| 19. | PF14_0185 | 14 (793751..796876) | 3126/115 | 1 | 0 |
| 20. | PF14_0429 | 14 (1856643..1859251) | 2391/88 | 3 | 2 |
| 21. | PF14_0563 | 14 (2420700..2422925) | 2226/82 | 1 | 0 |
| 22. | PF14_0655 | 14 (2825306..2826854) | 1197/44 | 4 | 3 |
Table 3.
Plasmodium falciparum helicases: homologues, functions and similarity
| Gene ID | Homologues/functions | Similarity (%) |
|---|---|---|
| 1. PFB0445c | Sub2p like SF2 helicase involved in SnRNP biogenesis, spliceosome RNA helicase BAT1 (UAP56). | C. parvum (78) |
| 2. PFB0860c | RNA helicase, Rrp3p, eIF-4A family, SF2 helicase. | C. parvum (72) |
| 3. PFC0915w | RNA helicase, stimulates RNA decapping, coordinates distinct steps in mRNA function and decay, interacts with both the decapping and deadenylase complexes, may have a role in mRNA transport and translation, Dhh1p. | S. cerevisiae (82) |
| 4. PFD1070w | eIF-4A from the following sources: Theileria, Xenopus, Oryza, Nicotiana, Humans, Mouse, Neurospora etc. | T. parva (87) |
| 5. PFE0215w | RNA helicase. | A. thaliana (57) |
| 6. PFE0430w | RNA helicase, pre-mRNA splicing (processing) factor. | C. neoformans (62) |
| 7. PFE1085w | ATP-dependent RNA helicase, DDX1 homologue. | H. sapiens (50) |
| 8. PFE1390w | RNA helicase. | H. sapiens (75) |
| 9. PFF1500c | RNA helicase. | T. parva (78) |
| 10. MAL7P1.113 | DEAD-box helicase, Dbp7p, eIF-4A family, SF2 helicase. | C. parvum (55) |
| 11. PF08_0096 | Dbp7p, eIF-4A homologue. | C. parvum (70) |
| 12. PF10_0209 | RNA helicase, Rok1p, eIF-4A homologue, required for 18s rRNA synthesis. | C. parvum (46) |
| 13. PFL1310c | p68 homologue (PfDH60). | C. neoformans (70) |
| 14. PFL2010c | Dbp9p, eIF-4A family SF2 helicase involved in biogenesis of 60s ribosomal subunit. Probable nucleolar RNA helicase. | C. parvum (46) |
| 15. PFL2475w | Drs1p, eIF-4A family SF2 helicase, required for ribosome assembly and function, including synthesis of 60s ribosomal subunits, constituent of 66s pre-ribosomal particles. DDX27 homologue. | C. parvum (58) |
| 16. PF13_0037 | myc regulated DEAH/D-box RNA helicase. | O. sativa (59) |
| 17. PF13_0177 | Rrp3p, RNA SF2 helicase involved in rRNA processing; required for maturation of 35s primary transcript of pre-rRNA and for cleavage leading to mature 18s rRNA; eIF-4A DEAD-box RNA-dependent ATPase with helicase activity. | S. cerevisiae (54) |
| 18. PF14_0183 | RNA helicase, Ddx10 Mouse/Humans, Hca4p helicase, Dbp4, eIF-4A-1 family RNA SF2 helicase. | H. sapiens (43) |
| 19. PF14_0185 | RNA helicase, homologous to DDX55, DDX47 and DDX31. | M. musculus (45) |
| 20. PF14_0429 | Dbp4 like eIF-4A-1 family SF2 RNA helicase involved in rRNA processing, Mak5 pre-mRNA splicing, DDX31, Hca4p. | C. parvum (45) |
| 21. PF14_0563 | Dbp5p like eIF-4A-1 family SF2 RNA helicase, cytoplasmic helicase involved in mRNA export from the nucleus, Zinc response protein (Rat). Zd10A gonadotropin regulated testicular helicase (Rat or Humans). DDX19 (Humans). | C. parvum (67) |
| 22. PF14_0655 | RNA helicase shows high homology with eIF-4A from the following sources: C. parvum, Theileria, Toxoplasma, Xenopus, Oryza, Arabidopsis, Zea mays, Nicotiana, etc. | T. gondii (91) |
2.1. ‘Q motif’ of P. falciparum helicases
The ‘Q motif’ is a newly discovered motif, which contains 9 amino acids and is distinctive to and characteristic of the ‘DEAD-box’ family of helicases. The consensus sequence for ‘Q motif’ is Gly (G)–Phe (F)–c–c–Pro (P)–Thr (T)–Pro (P)–Ile (I)–Gln (Q), where c is a charged side chain residue (Tanner, 2003). It has been shown in recent studies that this ‘Q motif’ is part of a highly conserved structure, which consists of a loop–helix–loop, which in turn is capable of interactions with motif I and a bound ATP (Cordin et al., 2004). The studies involving site-specific mutants in yeast have shown that this motif controls the helicase activity by regulating the ATP-binding and hydrolysis (Cordin et al., 2004). The importance of ‘Q motif’ is shown in a recent study where it has been shown that phosphorylation by cdc2 protein kinase at Thr (T) 204 of ‘Q motif’ of hamster DDX3 causes its loss of function (Sekiguchi and Fukumura, 2005).
Fig. 2 shows the schematic representation of various conserved motifs of the 22 putative ‘DEAD-box’ helicases of P. falciparum. It is noteworthy that this recently discovered ‘Q motif’ is present in all the ‘DEAD-box’ family members of P. falciparum. In all the members a conserved Phe (F) also occurred in a range of 23 to 31 amino acids from the highly conserved Gln (Q) of the ‘Q motif’ (Fig. 2). The consensus of ‘Q motif’ is shown in the box in Fig. 1A . It also shows the specific distribution and frequency of amino acids at respective positions in the ‘Q motif’ of all the ‘DEAD-box’ helicases of P. falciparum. It is interesting to note that the amino acid Gln (Q) is present at position 9 of the ‘Q motif’ in all the members. The positions 6 and 8 of the ‘Q motif’ are least variable, positions 1–5 are moderately variable and position 7 is highly variable (Fig. 1A).
Fig. 2.
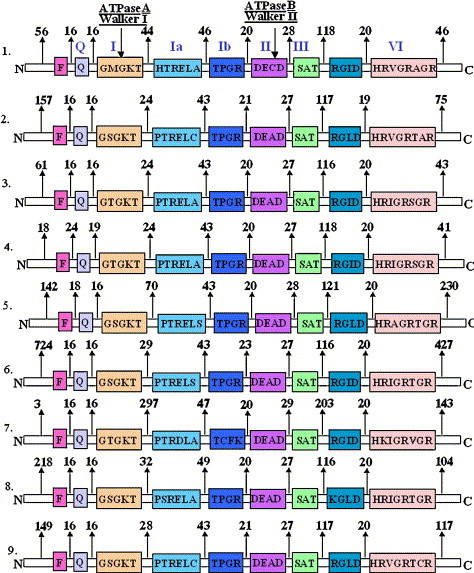
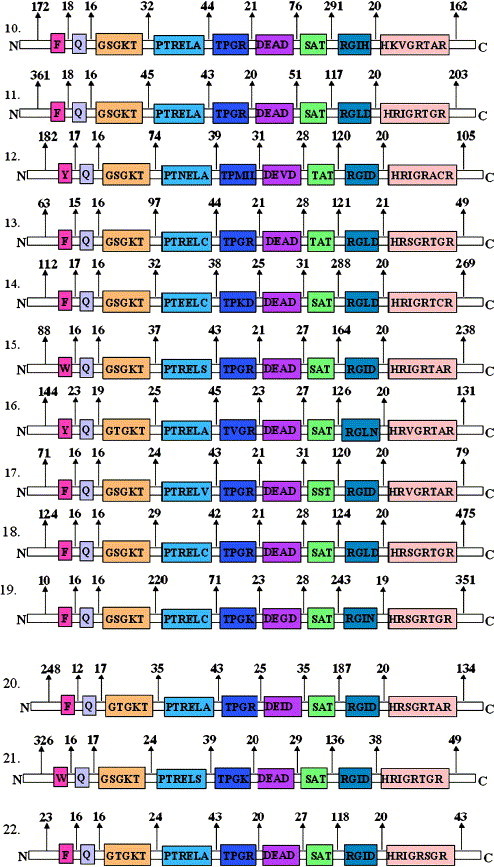
Schematic diagram showing the various conserved motifs of members of ‘DEAD-box’ helicases of Plasmodium falciparum. Open boxes represent the conserved helicase motifs and the amino acid sequence of each motif of each member is written by the single letter code inside the box. Labels above the boxes in number 1 (Q, I, Ia etc.) are the names assigned to these motifs. The number between the motif and above the arrow is the number of amino acids separating the various motifs.
Fig. 1.
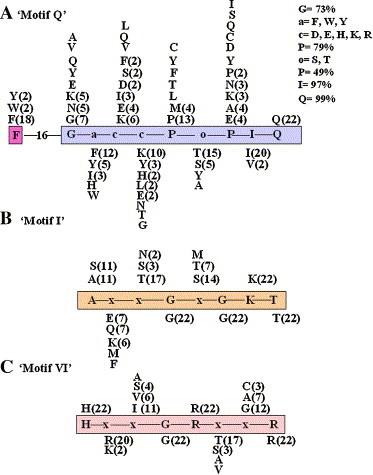
Distribution of amino acids at various positions in a. ‘Q motif; b. ‘Motif I’ and c. ‘Motif VI’ of ‘DEAD-box’ helicases of Plasmodium falciparum. In panel ‘a’ the variation in occurrence of amino acid upstream of ‘Q motif’ has also been shown. The numbers in parenthesis indicate the frequency of occurrence of each amino acid at respective position. The amino acids without any number means that they have occurred only once. Single letter code for amino acids has been used.
2.2. ‘Motif I’ of P. falciparum helicases
This motif is also known as ‘A’ motif of ATPase or ‘Walker motif A’. This motif is present in many nucleotide-binding proteins also and forms a phosphate binding loop or ‘P loop’ (Tanner and Linder, 2001). The consensus sequence of this motif for RNA helicases of SF1 is Ala (A)–x–x–Gly (G)–x–Gly (G)–Lys (K)–Thr (T) and for a few DNA helicases, ATPases and GTPases belonging to SF2, this sequence is Gly (G)–x–x–x–x–Gly (G)–Lys (K)–Thr (T) (Gorbalenya et al., 1989). In the classical version of this motif either Gly (G) or Ala (A) is present at first position and in the short version G is present at fourth position of this motif (Gorbalenya and Koonin, 1989). It is well established that the last three residues ‘GKT’ are required for the interaction of the protein with Mg2+ and ATP (Walker et al., 1982). More precisely the amino group of Lys (K) of GKT interacts with the β and γ phosphates of the ATP molecule (Walker et al., 1982). The role of conserved Gly (G) is structural and it helps to maintain the flexible loop conformation (Gorbalenya and Koonin, 1989). In a separate study it was shown that the mutation of K of ‘GKT’ to uncharged amino acid Asn (N) in mammalian translation initiation factor eIF-4A eliminated the binding of ATP to the protein (Pause and Sonenberg, 1992).
The consensus of ‘A motif’ is shown in the box in Fig. 1B. The figure also shows the specific distribution and frequency of amino acids at respective positions in the ‘A motif’ of all the ‘DEAD-box’ helicases of P. falciparum. As expected the Gly (G) at fourth position and the Gly Lys Thr (GKT) at the last three positions are highly conserved in all the members. It is interesting to note that the first position is equally shared by amino acid Ala (A) and Ser (S). It is a well known fact that Ser is the most frequent substituent for Ala in homologous proteins and it can replace Ala without alteration of the protein conformation. The position two is highly variable and is occupied by Glu (E), Gln (Q), Lys (K), Met (M) or Phe (F) (Fig. 1B). The positions three and five are mainly occupied by Thr (T) or Ser (S) and in a few cases by Asn (N) or Met (M) (Fig. 1B).
2.3. ‘Motif Ia’ of P. falciparum helicases
This motif was discovered as one more conserved sequence motif in helicases (Gorbalenya and Koonin, 1988). The consensus sequence for this motif is Pro (P)–Thr (T)–Arg (R)–Glu (E)–Leu (L)–Ala (A) and its secondary structure prediction suggests that it may form an element of the β/α core of a number of proteins of the ‘DEAD-box’ family (Gorbalenya et al., 1988, Gorbalenya and Koonin, 1988). Based on the structural information on SF1 and SF2 helicases, it has been suggested that this motif is involved in single-stranded DNA binding and it forms the groove for facilitating this binding (Velankar et al., 1999). All the members of the ‘DEAD-box family’ in P. falciparum contain the conserved Pro (P) at position one except one, which contains a His (H) and on position two also all the members contain Thr (T) except one, which contains a Ser (S) (Fig. 2 ). Conserved Arg (R) occupies position three in all except two members, where it is Glu (E) or Asn (N) respectively. Similarly conserved Glu (E) occupies position four in all except one member, where it is Asp (D). Position five is extremely conserved and Leu (L) is present in all the members, whereas the last position six is highly variable and occupied by Ala (A), Cys (C), Ser (S) or Val (V) (Fig. 2). The variability in this motif at various positions falls within the permissible limits.
2.4. ‘Motif II’ of P. falciparum helicases
This motif is also known as ‘B’ motif of ATPase or ‘Walker motif B’ (Walker et al., 1982). The consensus sequence for this motif is Asp (D)–Glu (E)–Ala (A)–Asp (D), therefore the proteins containing this motif are also known as ‘DEAD-box’ proteins (Linder et al., 1989). Various studies have shown that the first highly conserved residue Asp (D) of this motif interacts with the Mg2+ cation, which is required for NTP binding (Gorbalenya et al., 1989). Mutations in this motif have been shown to affect the ATPase and coupled helicase activities of a number of proteins such as Ded1p, eIF4 A, Dbp8p and a number of other putative helicases (Blum et al., 1992, Pause and Sonenberg, 1992, Iost et al., 1999, Daugeron and Linder, 2001). In yeast eIF-4A the mutation of the first Asp (D) of this motif to a Glu (E) resulted in the inactivation of this protein (Schmid and Linder, 1991). In another study involving mammalian eIF-4A, mutation of second Glu (E) to Gln (Q) severely affected ATP hydrolysis (Pause and Sonenberg, 1992). It has been suggested that this replacement may cause steric hindrance and this may be the reason, which explains the presence of this conserved ATPase B motif in the ‘DEAD-box’ family of helicases and some ATPases (Hodgman, 1988, Schmid and Linder, 1991). These studies also emphasize that negative charges at the first two positions of this motif are required for Mg2+-coordinated ATP hydrolysis. It is interesting to note that there is an enormously extensive range of variation in the distances separating the ‘Walker A’ and ‘B’ motifs. Since the two domains are believed to come together to form the NTP-binding center, this means that large domains can be looped out and allow this interaction.
In all the members of the ‘DEAD-box’ family in P. falciparum, the ‘Walker B’ motif is present and the first two amino acids Asp (D) and Glu (E) and the last amino acid Asp (D) are extremely conserved but there is variation at the third position. Although in most of the members Ala (A) occupies the third position, in some it is occupied by Cys (C), Gly (G), Ile (I) or Val (V) (Fig. 2). The distance between Walker motif A and B ranges from 83 to 368 amino acids in the ‘DEAD-box’ helicases of P. falciparum (Fig. 2).
2.5. ‘Motif III’ of P. falciparum helicases
The consensus sequence for this motif is Ser (S)–Ala (A)–Thr (T) and various studies have shown that this motif is involved in the unwinding reaction (Gorbalenya et al., 1989, Tuteja and Tuteja, 2004a). The crystallographic structural studies of the amino-terminal domain of yeast eIF-4A reveal specific interactions between amino acid residues of the phosphate binding loop, the DEAD-motif and the SAT motif (Johnson and McKay, 1999). Double mutations in this domain (SAT to AAA) of yeast eIF-4A lead to the loss of RNA helicase activity while the ATP-binding, hydrolysis and RNA binding activities remained intact (Pause and Sonenberg, 1992). The studies using mutants in motif III of yeast Prp22p helicase revealed that this motif is required for coupling of NTPase and helicase activities because these mutants were able to hydrolyze ATP but were defective in unwinding duplex RNA (Schwer and Meszaros, 2000). It has been shown recently that the mutations of the first Ser (S) to Ala (A) and the third Thr (T) to Ala (A) of the yeast ‘DEAD-box’ protein Has1p partially dissociated ATPase and helicase activities (Rocak et al., 2005). These mutational data collectively suggest the importance of the interactions between the amino acid residues of various domains, which may be responsible for yet unknown mechanism of coupling the NTPase activity with the RNA unwinding. It has also been suggested that the SAT residues might be involved in the transmission of energy derived from ATP hydrolysis to unwind the duplex RNA.
In all the members of the ‘DEAD-box’ family in P. falciparum, the motif III is present and the amino acid Ser (S) is present at first position in all of the members except two where it is Thr (T) (Fig. 2). This is a conserved substitution and has been found in other putative helicases also (Gorbalenya et al., 1989). Ala (A) occupies the second position of this domain in all the members except one where it is Ser (S) but the last position is highly conserved and Thr (T) is present in all the members in P. falciparum (Fig. 2).
2.6. ‘Motif VI’ of P. falciparum helicases
The consensus sequence for this motif is His (H)–x–x–Gly (G)–Arg (R)–x–x–R and it is the third most conserved segment in the proteins of this family (Gorbalenya et al., 1989). This motif has been suggested to be involved in interaction with RNA. It has been proposed that this motif is a part of the ATP-binding cleft and is also involved in coupling between helicase and NTPase activities of the protein. In mammalian eIF-4A, it has been shown that mutants having first His (H) changed to Gln (Q) of motif VI had reduced ATP-binding and ATPase activity but no helicase activity (Pause and Sonenberg, 1992). In a recent study it has been shown that the substitution of first His (H) to Glu (E) in motif VI of the yeast DEAD-box protein Has1p significantly decreased helicase but not the ATPase activity and this mutation was lethal in vivo (Rocak et al., 2005). The mutation of last Arg (R) of this motif in hepatitis C virus full-length NS3 protein resulted in loss of both the NTPase and helicase activities (Wardell et al., 1999). Similarly in UL9, the replacement of Arg (R) in this domain with Lys (K) resulted in reduction of the ssDNA-dependent intrinsic ATPase activity and complete elimination of the helicase activity (Marintcheva and Weller, 2001). The studies with DEN-2 NS3 protein with mutation of Arg (R) of this motif to Ala (A) also showed no detectable RNA unwinding activity but it retained the ATPase activity (Matusan et al., 2001). These studies suggest that motif VI mediates ligand-induced conformational changes, which are required for the helicase to move along the nucleic acid substrate. The crystal structure of NS3 has shown that the arginines of this domain make contact with the α and γ phosphates of ATP and facilitate this interaction (Kim et al., 1998).
In all the members of the ‘DEAD-box’ family in P. falciparum, the motif VI is present and the first, fourth, fifth and eighth position of this domain are occupied by highly conserved amino acids His (H), Gly (G), Arg (R) and Arg (R) respectively (Fig. 1C). The position two is least variable and mainly occupied by Arg (R) or in some cases Lys (K) also. The positions third, sixth and seventh are highly variable and occupied mainly by Ile (I), Thr (T) and Gly (G) respectively followed by Val (V), Ser (S), Cys (C) or Ala (A) (Fig. 1C).
2.7. ‘Other motifs’ and homorepeats of P. falciparum helicases
Besides the motifs described in above sections, there are some additional conserved motifs in the ‘DEAD-box’ family of helicases, which are not very well studied. There are two highly conserved Gly (G) residues in between the motif Ia and Ib, which may be required for proper bending of the protein molecule so that the domains can come together for interaction. The motif V with conserved sequence Arg (R)–Gly (G)–x–Asp (D) is also present in all the members of DEAD-box family in P. falciparum (Fig. 2). It has been suggested that this motif is capable of forming an extended network of interactions (Caruthers and McKay, 2002). Structural studies have shown that motifs V and VI “link” the carboxy-terminal domain to the amino-terminal domain through interactions with ATP and the ‘DEAD-motif’ providing a mechanism for the coupling of ATP-binding and hydrolysis with conformational changes, which modulate the binding of the nucleic acid substrate (Caruthers et al., 2000, Tuteja et al., 2005). With structural studies of yeast eIF-4A it has been shown that first Arg (R) of this motif could interact with the ‘DEAD-motif’ and this interaction plays a role in coupling ATP-binding and hydrolysis to RNA binding or it induces a conformational change (Caruthers et al., 2000). In a recent study the first Arg (R) of motif V of human p68 was mutated to Leu (L) and it was observed that both the RNA-dependent ATPase and RNA unwinding activity were abolished in the mutant (Lin et al., 2005). These observations collectively suggest that although the conserved motifs possibly act in synchronization with one another to carry out the basic helicase action, their distinct mechanistic roles might be susceptible to some extent of variation to correspond to the specific function of the particular enzyme.
Another interesting feature of the ‘DEAD-box’ family in P. falciparum is the presence of homorepeats i.e. the presence of repeated stretches of a single amino acid. These regions are referred to as hydrophilic low complexity regions and are due to the presence of amino acid Asn (N), Lys (K), Glu (E) or Asp (D), which are mainly coded by AT-rich codons (Pizzi and Frontali, 2000, Pizzi and Frontali, 2001). The frequency and presence of these homorepeats are highly variable and they are mainly present in most of the ‘DEAD-box’ helicases of P. falciparum at the N-terminal or C-terminal regions. In some of the ‘DEAD-box’ helicases these homorepeats are also present in between the various conserved domains. It is interesting to note that although a stretch of amino acid Asn (N) is present in between the motif I and Ia in the ‘DEAD-box’ DNA helicase PfDH60 from P. falciparum it still shows the nucleic acid-dependent ATPase, DNA and RNA helicase and ATP-binding activities (Pradhan et al., 2005a, Pradhan et al., 2005b). Therefore it can be postulated that in most of the proteins the presence of these homorepeats may not have any effect on their biological activity. It has been suggested that these regions encode non-globular domains of unidentified function, which are extruded from the proteins core and therefore have no effect on the functional folding of the protein (Pizzi and Frontali, 2001).
3. Structure of helicases
It was proposed initially that due to the presence of conserved motifs helicases might possibly be organized into a modular structure consisting of a minimum of DNA- and NTP-binding domains. Till to date, the crystal structure of a total of 24 helicases has been solved (Tuteja et al., 2005). These data suggest that all the conserved helicase motifs are clustered together at the level of tertiary structure.
The eIF-4A is the best-studied member of the DEAD-box family and it has been shown that this group of protein contains ATP-dependent nucleic acid unwinding activity. The crystallographic structure of the amino-terminal domain (residues 1–223) of yeast eIF-4A has been solved (Johnson and McKay, 1999). This structure shows that there are precise interactions between the amino acid residues of the Walker A motif, the ‘DEAD-motif’ and the SAT motif. On the other hand, the structure of full-length eIF-4A reveals that it is a dumbbell consisting of two compact domains that are connected by an extended linker (Caruthers et al., 2000) (Fig. 3 ). By using the structure of yeast eIF-4A, a model for the structure of eIF-4A homologue of P. falciparum has been created (Fig. 3). As is clear from the modeled structure, the amino- and carboxy-terminal domains are connected via an extended linker. It is also noticeable that motifs V and VI ‘link’ the carboxy-terminal domain to the amino-terminal domain through interactions with the ‘DEAD-motif’ and ATP, which provide a means for coupling the ATP-binding and hydrolysis with conformational changes. These results collectively suggest that there might be minor variations in the length and sequence between the conserved domains but overall for the enzyme action all the domains are clustered and act in co-ordination with each other (Fig. 3).
Fig. 3.
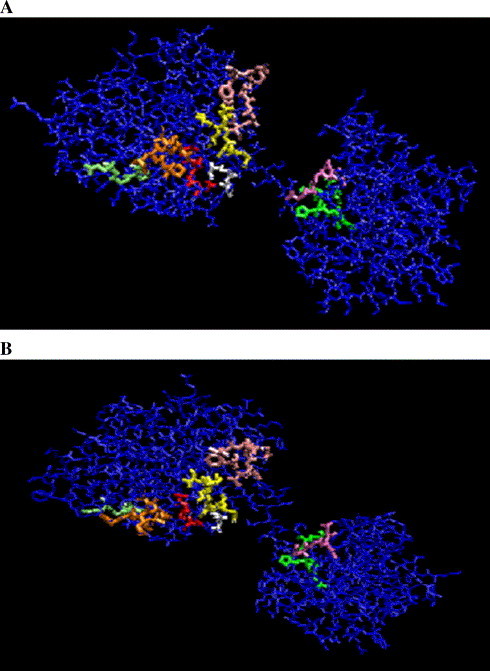
Crystal structure of eIF-4A homologue (PF14_0655) of Plasmodium falciparum based on the solved structure of eIF-4A of yeast (Saccharomyces cerevisiae). The structural model of eIF-4A homologue of P. falciparum (PF14_0655) with ID no. Q8IKFO was retrieved from the modbase database (www.modbase.compbio.ucsf.edu). This shows ∼ 61% sequence identity with its template, which is yeast eIF-4A (1fuuB) and was retrieved from the RCSB protein databank (www.pdb.org). The conserved helicase motifs of both of these proteins are indicated in different colors using molecular visualization program for displaying, animating and analyzing large biomolecule systems using 3-dimensional graphics and built-in scripting (VMD software www.ks.uiuc.edu). The colors used for various motifs are: Q motif — pink; motif I — yellow; motif Ia — orange; motif Ib — lime; motif II — red; motif III — white; motif V — mauve and motif VI — green.
4. Helicase as drug target
As discussed in the preceding sections and various other studies it has been shown that the ‘DEAD-box’ helicases are involved in a wide variety of cellular processes and studies in yeast have shown that each DEAD-box gene is independently necessary because loss of one ‘DEAD-box’ gene cannot be complemented by the over-expression of another family member (Kessler et al., 1997). Recent studies have shown that some helicases are also essential for the viability of a variety of bacteria and viruses and the helicases can be used as antiviral targets (Borowski et al., 2002, Coen and Schaffer, 2003, Kwong et al., 2005). These data suggest that the ‘DEAD-box’ proteins are essential components of the cellular machinery and the modulation of their activity by inhibitors can have grave consequences. Therefore these enzymes represent a potential drug target for anti-malarial therapy.
Any helicase reaction is a result of three tightly coupled consecutive steps, which are binding of the nucleic acid substrate, ATP-binding and hydrolysis, and ATP-hydrolysis-dependent unwinding of the nucleic acid duplex (Tuteja and Tuteja, 2004a). Hence, the compounds, which can inhibit any of these steps, can act as potential inhibitors. The compounds, which can bind at the nucleic acid binding pocket of the helicase, can also inhibit the reaction by preventing the interaction between the enzyme and the substrate or cause a block for the movement of the enzyme on the substrate. These compounds may inhibit the reaction either by interfering with ATP-binding, inhibiting nucleic acid-dependent ATPase activity or by uncoupling the ATP-hydrolysis-dependent unwinding reaction. The compounds, which diminish the accessibility of ATP for ATP-binding site, can also be considered as significant inhibitors of helicase and potential anti-malarial agents. A number of previous studies including ours have shown that the DNA-interacting agents are well-known anti-helicase agents and these act at the level of the substrate (Tuteja et al., 1997, Tuteja et al., 2003, Borowski et al., 2002). It has been shown that out of various compounds tested, only cisplatin, 4,′6′-diamidino-2-phenylindole, daunorubicin and nogalamycin were inhibitory to the unwinding activity of a P. cynomolgi helicase (Tuteja et al., 2003).
On the other hand the nucleoside analogues inhibit the enzyme activity by blocking the ATP-binding site of helicase resulting in uncoupling the ATPase and helicase activities and therefore operate via interaction with the enzyme. These compounds have been used in previous studies to block the helicase activity of hepatitis C virus (Borowski et al., 1999, Borowski et al., 2000). It has also been suggested that poxviruses, papovaviruses, some RNA viruses such as West Nile virus, severe acute respiratory syndrome coronavirus and dengue virus encoded viral helicases can be probable antiviral drug targets (Kwong et al., 2005). All these data support the notion that gradually helicases are becoming an attractive and promising drug target but still a lot of work needs to be done especially with regard to the helicases of P. falciparum.
5. Conclusion and future prospects
Helicases, the important enzymatic tools of cells, are capable of unwinding energetically stable duplex nucleic acids into single strands and can be considered as “screw driver” of the cellular machinery. With the completion of genome sequences of a number of organisms, it is interesting to note that each genome contains a large number of putative helicases. These putative helicases are classified as helicases due to the presence of conserved sequence motifs in them. Even though a number of these ‘DEAD-box’ proteins have been documented and predicted as ‘putative helicases’ from a variety of organisms, only few have been characterized biochemically. Previously we have cloned and characterized an eIF-4A homologue belonging to the ‘DEAD-box’ family from the malaria parasite P. cynomolgi (Tuteja et al., 2002, Tuteja et al., 2003). Recently we have cloned and characterized the first ‘DEAD-box’ helicase, which is homologous to p68 from P. falciparum (Pradhan et al., 2005a, Pradhan et al., 2005b). Therefore it is safe to state that based only on the presence of these conserved motifs it is preliminary to classify a protein as a helicase and it should be characterized biochemically before it is considered as a bonafide helicase.
DNA helicases have been shown to be associated with a number of human genetic diseases and RNA helicases are required for growth and proliferation of bacteria and viruses. Hence these enzymes are excellent targets for antibacterial and antiviral drugs. Studies in the past have shown that the inhibitors of the herpes simplex virus helicase-primase have potent anti-herpes activity and recently some other viral helicases have emerged as novel targets for the treatment of viral infections (Kleymann et al., 2002, Kwong et al., 2005). It has also been suggested that these important enzymes can be used as targets for anti-cancer drugs (Sharma et al., 2005). The studies with PfDH60 (a ‘DEAD-box’ helicase from P. falciparum) have shown that DNA-binding compounds can inhibit its unwinding and ATPase activities (Tuteja and Pradhan, unpublished observation). The detailed studies including cloning and characterization of helicases of malaria parasite P. falciparum may help to identify a specific enzyme, which is inhibited by a compound that has no effect on the enzymes of the host and consequently could be used as the potential drug to treat malaria. These studies should also make a significant contribution to our better understanding of the nucleic acid transaction in the parasite.
Acknowledgements
This work was supported by Defence Research and Development Organization grant (to R. T.). Infrastructural support from the Department of Biotechnology, Government of India is gratefully acknowledged. The authors sincerely thank the editor and referees for helpful comments.
Received by A.J. van Wijnen
References
- Abdelhaleem M., Maltais L., Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2003;81:618–622. doi: 10.1016/s0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- Bahl A. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum S. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7664–7668. doi: 10.1073/pnas.89.16.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski P., Kuehl R., Mueller O., Hwang L.H., Schulze Zur Wiesch J., Schmitz H. Biochemical properties of a minimal functional domain with ATP-binding activity of the NTPase/helicase of hepatitis C virus. Eur. J. Biochem. 1999;266:715–723. doi: 10.1046/j.1432-1327.1999.00854.x. [DOI] [PubMed] [Google Scholar]
- Borowski P. ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy. Acta Biochim. Pol. 2000;47:173–180. [PubMed] [Google Scholar]
- Borowski P. Characterization of imidazo[4,5-d]pyridazine nucleosides as modulators of unwinding reaction mediated by West Nile virus nucleoside triphosphatase/helicase: evidence for activity on the level of substrate and/or enzyme. Antimicrob. Agents Chemother. 2002;46:1231–1239. doi: 10.1128/AAC.46.5.1231-1239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet N., Aubourg S., Toffano-Nioche C., Kreis M., Lecharny A. Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res. 2001;11:2101–2114. doi: 10.1101/gr.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z., Llinas M., Pulliam B.L., Wong E.D., Zhu J., DeRisi J.L. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struck. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- Caruthers J.M., Johnson E.R., McKay D.B. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D.M., Schaffer P.A. Antiherpesvirus drugs: a promising spectrum of new drugs and drug targets. Nat. Rev. Drug Discov. 2003;2:278–288. doi: 10.1038/nrd1065. [DOI] [PubMed] [Google Scholar]
- Cordin O., Tanner N.K., Doere M., Linder P., Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron M.C., Linder P. Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis. Nucleic Acids Res. 2001;29:1144–1155. doi: 10.1093/nar/29.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Kressler D., Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box protein and related families. Trends Biochem. Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Gardner M.J. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V. One more conserved sequence motif in helicases. Nucleic Acids Res. 1988;16:7734. doi: 10.1093/nar/16.15.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 1989;17:8413–8440. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure–function relationship. Curr. Opin. Struck. Biol. 1993;3:419–429. [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS. Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M.C., Matson S.W. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989;339:562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- Hodgman T.C. A new superfamily of replicative proteins. Nature (London) 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- Hyde J.E. Drug resistant malaria. Trends Parasitol. 2005;21:494–498. doi: 10.1016/j.pt.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I., Dreyfus M., Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- Johnson E.R., McKay D.B. Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase. RNA. 1999;5:1526–1534. doi: 10.1017/s1355838299991410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M.M. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.L. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- Kleymann G. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- Kwong A.D., Rao B.G., Jeang K.T. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Yang L., Yang J.J., Huang Y., Liu Z.R. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol. Cell. Biol. 2005;25:7484–7493. doi: 10.1128/MCB.25.17.7484-7493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Marintcheva B., Weller S.K. Residues within the conserved helicase motifs of UL9, the origin-binding protein of herpes simplex virus-1, are essential for helicase activity but not for dimerization or origin binding activity. J. Biol. Chem. 2001;276:6605–6615. doi: 10.1074/jbc.M007743200. [DOI] [PubMed] [Google Scholar]
- Matusan A.E., Pryor M.J., Davidson A.D., Wright P.J. Mutagenesis of the Dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J. Virol. 2001;75:9633–9643. doi: 10.1128/JVI.75.20.9633-9643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzi E., Frontali C. Divergence of noncoding sequences and of insertions encoding nonglobular domains at a genomic region well conserved in plasmodia. J. Mol. Evol. 2000;50:474–480. doi: 10.1007/s002390010050. [DOI] [PubMed] [Google Scholar]
- Pizzi E., Frontali C. Low-complexity regions in Plasmodium falciparum proteins. Genome Res. 2001;11:218–229. doi: 10.1101/gr.152201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A., Chauhan V.S., Tuteja R. A novel ‘DEAD-box’ DNA helicase from Plasmodium falciparum is homologous to p68. Mol. Biochem. Parasitol. 2005;140:55–60. doi: 10.1016/j.molbiopara.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pradhan A., Chauhan V.S., Tuteja R. Plasmodium falciparum DNA helicase 60 is a schizont stage specific, bipolar and dual helicase stimulated by PKC phosphorylation. Mol. Biochem. Parasitol. 2005;144:133–141. doi: 10.1016/j.molbiopara.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Rocak S., Emery B., Tanner N.K., Linder P. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 2005;33:999–1009. doi: 10.1093/nar/gki244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T.E., Merrick W.C., Sonenberg N. Bidirectional RNA helicase activity of eukaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S.R., Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T., Fukumura J. Phosphorylation of Dead-box RNA helicase DDX3 by mitotic cyclin B/CDC2, but not cyclin A/CDK2. J. Biol. Chem. 2005 doi: 10.1074/jbc.M505018200. (Nov 9; Electronic publication ahead of print) [DOI] [PubMed] [Google Scholar]
- Seow F. The plastidic DNA replication enzyme complex of Plasmodium falciparum. Mol. Biochem. Parasitol. 2005;141:145–153. doi: 10.1016/j.molbiopara.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Sharma S., Doherty K.M., Brosh R.M., Jr. DNA helicases as targets for anti-cancer drugs. Curr. Med. Chem. Anti-Canc. Agents. 2005;5:183–199. doi: 10.2174/1568011053765985. [DOI] [PubMed] [Google Scholar]
- Shiratori A., Shibata T., Arisawa M., Hanaoka F., Murakami Y., Eki T. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast. 1999;15:219–253. doi: 10.1002/(SICI)1097-0061(199902)15:3<219::AID-YEA349>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Silverman E., Edwalds-Gilbert G., Lin R.J. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- Song P., Malhotra P., Tuteja N., Chauhan V.S. RNA helicase-related genes of Plasmodium falciparum and Plasmodium cynomolgi. Biochem. Biophys. Res. Commun. 1999;255:312–316. doi: 10.1006/bbrc.1999.0204. [DOI] [PubMed] [Google Scholar]
- Tanner N.K. The newly identified Q motif of DEAD box helicases is involved in adenine recognition. Cell Cycle. 2003;2:18–19. doi: 10.4161/cc.2.1.296. [DOI] [PubMed] [Google Scholar]
- Tanner N.K., Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Tanner N.K., Cordin O., Banroques J., Doere M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- Tuteja N., Tuteja R. Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery. Eur. J. Biochem. 2004;271:1835–1848. doi: 10.1111/j.1432-1033.2004.04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N., Tuteja R. Unraveling DNA helicases. Motif, structure, mechanism and function. Eur. J. Biochem. 2004;271:1849–1863. doi: 10.1111/j.1432-1033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- Tuteja N., Phan T.N., Tuteja R., Falaschi A. Inhibition of DNA unwinding and ATPase activities of human DNA helicase II by chemotherapeutic agents. Biochem. Biophys. Res. Commun. 1997;236:636–640. doi: 10.1006/bbrc.1997.7021. [DOI] [PubMed] [Google Scholar]
- Tuteja R., Malhotra P., Song P., Tuteja N., Chauhan V.S. Isolation and characterization of an eIF-4A homologue from Plasmodium cynomolgi. Mol. Biochem. Parasitol. 2002;124:79–83. [PubMed] [Google Scholar]
- Tuteja R., Tuteja N., Malhotra P., Chauhan V.S. Replication fork stimulated eIF-4A from Plasmodium cynomolgi unwinds DNA in the 3′ to 5′ direction and is inhibited by DNA-interacting compounds. Arch. Biochem. Biophys. 2003;414:108–114. doi: 10.1016/s0003-9861(03)00176-0. [DOI] [PubMed] [Google Scholar]
- Tuteja N., Vashisht A.A., Tuteja R. Structural and mechanistic aspect of DEAD-box helicases. Natl. Acad. Sci. Lett. 2005;28:313–323. [Google Scholar]
- Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardell A.D., Errington W., Ciaramella G., Merson J., McGarvey M.J. Characterization and mutational analysis of the helicase and NTPase activities of hepatitis C virus full-length NS3 protein. J. Gen. Virol. 1999;80:701–709. doi: 10.1099/0022-1317-80-3-701. [DOI] [PubMed] [Google Scholar]
- Wassarman D.A., Steitz J.A. Alive with dead proteins. Nature. 1991;349:463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- White N.J. Drug resistance in malaria. Br. Med. Bull. 1998;54:703–715. doi: 10.1093/oxfordjournals.bmb.a011721. [DOI] [PubMed] [Google Scholar]
- Winstanley P.A. Chemotherapy for falciparum malaria: the armoury, the problems and the prospects. Parasitol. Today. 2000;16:146–153. doi: 10.1016/s0169-4758(99)01622-1. [DOI] [PubMed] [Google Scholar]


