Abstract
In September 2012, a novel coronavirus was isolated from a patient who died in Saudi Arabia after presenting with acute respiratory distress and acute kidney injury. Analysis revealed the disease to be due to a novel virus which was named Middle East Respiratory Coronavirus (MERS-CoV). There have been several MERS-CoV hospital outbreaks in KSA, continuing to the present day, and the disease has a mortality rate in excess of 35%. Since 2012, the World Health Organization has been informed of 2220 laboratory-confirmed cases resulting in at least 790 deaths. Cases have since arisen in 27 countries, including an outbreak in the Republic of Korea in 2015 in which 36 people died, but more than 80% of cases have occurred in Saudi Arabia.. Human-to-human transmission of MERS-CoV, particularly in healthcare settings, initially caused a ‘media panic’, however human-to-human transmission appears to require close contact and thus far the virus has not achieved epidemic potential. Zoonotic transmission is of significant importance and evidence is growing implicating the dromedary camel as the major animal host in spread of disease to humans. MERS-CoV is now included on the WHO list of priority blueprint diseases for which there which is an urgent need for accelerated research and development as they have the potential to cause a public health emergency while there is an absence of efficacious drugs and/or vaccines. In this review we highlight epidemiological, clinical, and infection control aspects of MERS-CoV as informed by the Saudi experience. Attention is given to recommended treatments and progress towards vaccine development.
Keywords: Coronavirus, MERS, Respiratory, Infection, Transmission, Saudi Arabia, Middle East
Highlights
-
•
2220 laboratory-confirmed cases of MERS-CoV resulting in at least 790 deaths since 2012
-
•
MERS-CoV is on the WHO list of priority blueprint diseases
-
•
Zoonotic and human-to-human transmission modes need further clarification.
-
•
No specific therapy has yet been approved.
-
•
There is a need for well-controlled clinical trials on potential direct therapies.
1. Introduction
Middle East Respiratory Syndrome (MERS) arises from infection with the MERS-coronavirus (MERS-CoV), a beta coronavirus. Since the first confirmed case in June 2012, the World Health Organization (WHO) have been informed of 2220 laboratory-confirmed cases resulting in at least 790 deaths (Middle East respiratory syndrome coronavirus (MERS-CoV), 2018; Zaki et al., 2012). Although cases have arisen in 27 countries to date, including a major outbreak in the Republic of Korea in 2015, the overwhelming burden of infection has occurred in the Middle East and most particularly in the Kingdom of Saudi Arabia (KSA), where more than 80% of cases have occurred according to WHO estimates (Middle East respiratory syndrome coronavirus (MERS-CoV), 2018; Middle East respiratory syndrome coronavirus (MERS-CoV). Fact sheet, 2018; Nishiura et al., 2016a, Nishiura et al., 2016b, Park et al., 2015). In this review, we consider current knowledge of MERS-CoV virology, molecular biology, immunology, epidemiology, diagnosis, transmission, therapy and vaccinology with special reference to the impact on the Middle East and KSA in particular.
2. Epidemiology
The first confirmed case of Middle East Respiratory Syndrome (MERS) was in June 2012. A previously healthy 60-year old Saudi male was hospitalized on 10th June 2012 in Bisha in the Kingdom of Saudi Arabia (KSA) with acute community-acquired pneumonia and was subsequently transferred to a private hospital in Jeddah on 13th June 2012, where he died on 24th June due to respiratory and renal failure (Zaki et al., 2012). Indirect immunofluorescence assays on day 1 sputum samples were negative for influenza A and B, parainfluenza 1 to 3, respiratory syncytial virus and adenovirus, however cytopathic changes in LLC-MK2 and Vero cells inoculated with the patient's sputum indicated the likelihood of viral replication (Zaki et al., 2012). PCR testing was negative for adenovirus, enterovirus, metapneumovirus, herpesviruses, and paramyxoviruses but positive for detection of coronaviruses (Zaki et al., 2012). Sequencing of the PCR products confirmed the identification of a new virus belonging in lineage C of the betacoronavirus genus and initially named human coronavirus EMC (HCoV-EMC) (Zaki et al., 2012). In September 2012, the same virus was identified in a 49-year-old man who had been transferred from a hospital in Qatar to London with an unexplained, severe respiratory illness which required intubation and ventilation (Bermingham et al., 2012). Importantly, this man had a history of travel in KSA, where he had experienced a mild undiagnosed respiratory illness in August 2012 (Bermingham et al., 2012). The first cluster of human cases was retrospectively confirmed from a group of 13 people who had become ill with an unexplained respiratory illness in a public hospital in Zarqa city in Jordan in April 2012 (Hijawi et al., 2013).
Since then, most outbreaks have occurred in KSA. These include a cluster of 25 cases in Al-Hasa between April 1st and May 23rd 2013 (Assiri et al., 2013a), 255 laboratory-confirmed cases in Jeddah between January 1st and May 16th, 2014 (Drosten et al., 2015, Oboho et al., 2015), 45 cases in King Fahad Medical City in Riyadh between March 29th and May 21st, 2014, with contemporaneous outbreaks in other Riyadh hospitals between March and April 2014 (Almekhlafi et al., 2016, Fagbo et al., 2015), and 130 cases at King Abulaziz Medical City in Riyadh during late June–late August 2015 (Balkhy et al., 2016a). An exception was the major outbreak that occurred in in the Republic of Korea between 20 May and 27 July 2015 (Nishiura et al., 2016a, Nishiura et al., 2016b, Park et al., 2015). This outbreak encompassed 186 MERS-CoV cases, and resulted in 36 deaths (Middle East respiratory syndrome coronavirus (MERS-CoV) – Republic of Korea, 2015; Nishiura et al., 2016a, Nishiura et al., 2016b). However, in common with cases that have arisen in other countries outside the Middle East, the Korean outbreak began with a man with a preceding travel history to Middle Eastern countries. According to reports made to WHO and the Centers of Disease Control and Prevention (CDC), laboratory-confirmed cases of MERS have occurred in Middle Eastern countries including KSA, Bahrain, Iran, Jordan, Kuwait, Lebanon, Oman, Qatar, United Arab Emirates (UAE), and Yemen, as well as in countries outside the Middle East including Algeria, Austria, China, Egypt, France, Germany, Greece, Italy, Malaysia, Netherlands, Philippines, Republic of Korea, Thailand, Tunisia, Turkey, United Kingdom (UK), and United States of America (USA), but associated with individuals with a travel history in the Middle East.
MERS-CoV is now included on the WHO list of priority blueprint diseases for which there which is an urgent need for accelerated research and development as they have the potential to cause a public health emergency while there is an absence of efficacious drugs and/or vaccines (List of Blueprint priority diseases, 2018). Cases continue to arise in KSA and exact a high mortality rate, including 20 cases from 11 areas of the country reported to WHO by the National IHR Focal Point between December 2017 and 17 January 2018, resulting in 9 deaths (Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia, 2018). Frequent small outbreaks include 2 clusters of cases in the Al Jawf Region of KSA, i.e. a cluster of 13 cases in a hospital between 2nd and 11th August 2017, among them 8 healthcare workers (HCWs), and 7 cases in Dawmet Aljandal City between 24th and 31st August 2017 (Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia, 2017a, Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia, 2017b). In 3 clusters in 3 Riyadh hospitals in June 2017, 2 of which were related, 49 individuals were infected of whom 10 died (Coronavirus infections, 2017).
Clearly, MERS-CoV is a serious public health issue in KSA. Extended outbreaks of the disease have been focused on healthcare facilities, with transmission apparently dependent on close human-to-human contact (Almekhlafi et al., 2016, Assiri et al., 2013a, Balkhy et al., 2016a, Drosten et al., 2015, Fagbo et al., 2015, Oboho et al., 2015). The emergence of this disease has therefore had a profound impact on infection control and prevention procedures in KSA as outbreaks in healthcare facilities have been associated with defective or inadequate infection prevention and control measures (Balkhy et al., 2016b, Butt et al., 2016, Cotten et al., 2014, Hastings et al., 2016).
3. Infection prevention and control measures in Saudi Arabia
Public health authorities in KSA worked with WHO in identifying shortcomings in infection and control procedures in healthcare facilities which contributed to MERS-CoV transmission (Middle East respiratory syndrome coronavirus (MERS-CoV), 2018). Problems which were identified included emergency room overcrowding and neglect of basic infection and prevention control measures such as handwashing (Middle East respiratory syndrome coronavirus (MERS-CoV), 2018). The KSA Ministry of Health updated guidelines for infection prevention and control in line with WHO recommendations (Butt et al., 2016, Hastings et al., 2016). The Ministry of Health now specifies that “Standard Precautions” should be adhered to in all patient interactions within hospitals, and that these should be further supplemented with the specific precautions for suspected or confirmed MERS-CoV cases (Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia, n.d.). Infection prevention and control measures include comprehensive basic procedures such as hand hygiene, including application of ‘my 5 moments for hand hygiene’ (About SAVE LIVES: Clean Your Hands, 2018), respiratory precautions, contacts control, and use of personal protective equipment (PPE), which comprises surgical or correctly fitted and sealed N95 mask, gloves and gown, and goggles/face shield where indicated, and prevention of overcrowding in emergency rooms. More advanced precautions for care of patients with acute respiratory infections include use of effective triage, droplet and airborne precautions, safe patient transport and continuous training and education of healthcare workers. Frequent and thorough cleaning of MERS patient rooms with special attention to frequently touched surfaces, preferably by designated, well-trained housekeeping staff and with a clearly defined scope for cleaning of patient-care equipment, is also recommended (Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia, n.d.). Ministry of Health guidelines should also be followed for cleaning and disinfection after MERS patient discharge, handling of textiles, use of disposable dishes and eating utensils for MERS-CoV patients and diposal of medical waste (Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia, n.d.). Detailed guidelines are included on management of contacts of MERS-CoV patients, including household contacts, healthcare workers and patients; contact monitoring for 14 days after date of exposure is recommended (Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia, n.d.). Home isolation procedures and duration of isolation precautions should be based on laboratory testing if available to assure absence of viral shedding; appropriate duration of isolation is an area that is still being researched (Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia, n.d.). Handling of bodies in the mortuary, as well as guidelines for extracorporeal membrane oxygenation (ECMO), which is available in designated MERS-CoV centers in Riyadh, Jeddah and Dammam, but which is of uncertain benefit for MERS-CoV treatment, are also detailed (Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia, n.d.).
Implementation of these infection prevention and control guidelines for MERS-CoV in line with most up-to-date case definition and surveillance guidance have resulted in a decline in cases in KSA (Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia, n.d.). However, diligence in needed in maintaining surveillance standards and furthering understanding of transmission patterns within KSA and elsewhere. Comparison of epidemiology of KSA outbreaks to that of the Republic of Korea 2015 outbreak suggests that while there are similarities in terms of mean age of infected individuals (51 and 54 y respectively) and the higher risk of infection or death for older males (≥70 y), nevertheless there is evidence that transmission patterns and risk factors are different in KSA (Chen et al., 2017). While in Korea the transmission pattern was almost exclusively nosocomial, in KSA zoonotic transmission, human-to-human transmission and unknown pathways were all present in addition to nosocomial infection (Chen et al., 2017). In some 59.9% of cases in KSA outbreaks, exposure risk was unknown (Chen et al., 2017). Thus in addition to the infection prevention and control guidelines for healthcare facilities, WHO has also issued guidance on potential zoonotic transmission in the community, in particular with respect to dromedary camels which are recognized as a major MERS-CoV host reservoir and animal source for human infection (Middle East respiratory syndrome coronavirus (MERS-CoV). Fact sheet, 2018). In KSA it is recommended that people visiting places where dromedary camels are present should practice general hygiene measures and avoid contact with sick animals. Furthermore, consumption of raw or uncooked meat, milk or urine from dromedaries is discouraged, with pasteurization, cooking, or other heat treatments recommended for rendering these products fit for consumption (Middle East respiratory syndrome coronavirus (MERS-CoV). Fact sheet, 2018). Immunocompromised people and other vulnerable groups such as people with diabetes, renal failure or chronic lung disease are advised to avoid contact with dromedaries in general and not to consume camel food products that have not been pasteurized or adequately cooked (Middle East respiratory syndrome coronavirus (MERS-CoV). Fact sheet, 2018). Recent studies, including those based on serological evidence, support the role of dromedary camels as important zoonotic sources of human MERS-CoV infection. MERS-CoV antibodies are present in more than 90% of dromedary camels tested in the Middle East and in many African countries (Ali et al., 2017a, Chu et al., 2015, Farag et al., 2015, Hemida et al., 2013, Hemida et al., 2014, Hemida et al., 2017a, Müller et al., 2015, Reusken et al., 2014). Dromedary camel exposure within 2 weeks of illness onset has been identified as a significant risk factor in a study examining MERS-CoV infection cases documented between May and November 2014 in KSA (Alraddadi et al., 2016a). Changes in dromedary camel production and farming practices, including intensification and location close to cities, may have contributed to zoonotic transmission in KSA (Gossner et al., 2016). Thus, in KSA the emergence of MERS-CoV has had an impact on the agricultural, animal husbandry, food production and veterinary fields, as well as infection and prevention control procedures in healthcare settings (Hemida et al., 2017b). For example, the association between the calving season and MERS-CoV infection in dromedary camels and the highest risk of MERS-CoV infection in calves compared to adult cows, has led to suggestions that weaning of calves could be delayed to reduce the opportunity for human exposure to calves (Hemida et al., 2014, Hemida et al., 2017a, Hemida et al., 2017b). Furthermore, there is a need to increase understanding of the implications in terms of MERS-CoV transmission and spread, as well as viral exchange, amplification and dissemination, of the economically important bidirectional movement of camels between African countries and the Middle East, including KSA (Hemida et al., 2017b).
Meanwhile, when a case of MERS-CoV is suspected, effective identification is achieved by molecular methods. The currently WHO-recommended methods used in KSA are based on polymerase chain reaction (PCR) targeting of a number of MERS-CoV genes, which has been made possible by development in understanding of MERS-CoV classification and genomics.
4. General virology
4.1. Classification
In the 1960s, the first human respiratory illness-causing coronaviruses, (HCoVs) 229E and HCoV-OC43, were discovered (Becker et al., 1967, Hamre and Procknow, 1966). In 2003, a new CoV named Severe Acute Respiratory Syndrome (SARS)-CoV SARS was involved in a series of international outbreaks causing close to 800 deaths (Marra et al., 2003, Peiris et al., 2003, Rota et al., 2003). The NL63 and HKU1 human coronaviruses were discovered in 2004, both of which also cause human respiratory illness (Fouchier et al., 2004, van der Hoek et al., 2004). MERS-CoV was first isolated in September 2012, and initially named human coronavirus EMC (Zaki et al., 2012).The coronavirus study group later renamed this novel virus as the Middle East respiratory syndrome coronavirus (MERS-CoV), reflecting its origin (de Groot et al., 2013).
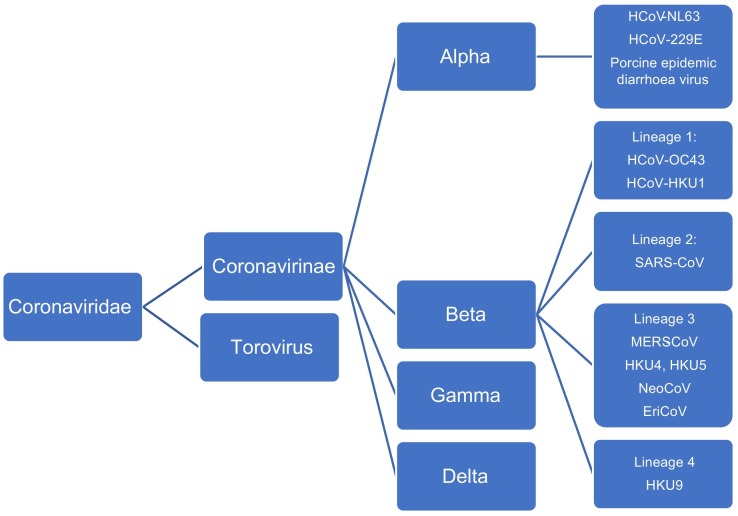
Coronaviruses (CoVs) are members of the Coronavirinae subfamily of the Coronaviridae family. CoVs infect humans as well as other species. The subfamily is comprised of 4 genera, alpha CoVs, beta CoVs, gamma CoVs, and delta CoVs (Fig. 1 ) CoVs are enveloped single-stranded, positive-sense RNA viruses with genomes of 25 to 32 kilobases (kb). HCoV-NL63 and HCoV-229E are alphaCoVs, while SARS-CoV, MERS-CoV, HCoV-HKU1 and HCoV-OC43 are beta coronoaviruses (Fig. 1). The beta coronavirses can be further subdivided into 4 lineages. MERS-CoV is unique among CoVs infecting humans in belonging to lineage C (lineage 3) of the beta CoVs (Fig. 1) (Chan et al., 2015a, Corman et al., 2014a, de Groot et al., 2012).
Fig. 1.
Taxonomy of the Coronaviridae family.
Bats are potentially the main MERS-CoV mammalian reservoir, as with other coronaviruses (Drexler et al., 2014). Closely related lineage 3 viruses include the bat viruses NeoCoV, isolated from a Neoromicia zuluensis bat in South Africa, and the prototypic lineage c betacoronaviruses, Tylonycteris bat virus HKU4 and Pipistrellus bat HKU5 virus (Fig. 1) (Middle East respiratory syndrome coronavirus (MERS-CoV) Fact sheet, 2017; Corman et al., 2014b, Ithete et al., 2013). Studies on the phylogeny of lineage C betacoronaviruses suggest that evolution of MERS-CoV in camels occurred prior to that in humans and that there was exchange of genetic elements among ancestral viruses either in bats, or within the camel genetic ‘mixing vessel’, leading to MERS-CoV emergence (Corman et al., 2014b). Other potentially important mammalian hosts are members of the Eulipotyphla taxon, the closest sister taxon to bats which includes hedgehogs (Corman et al., 2014a). EriCoV, another lineage C virus which is closely related to both MERS-CoV and the bat lineage C coronaviruses, was found to be present in approximately 59% of European hedgehog (Erinaceus europaeus) fecal samples in a study in Germany (Corman et al., 2014a).
4.2. Genomics
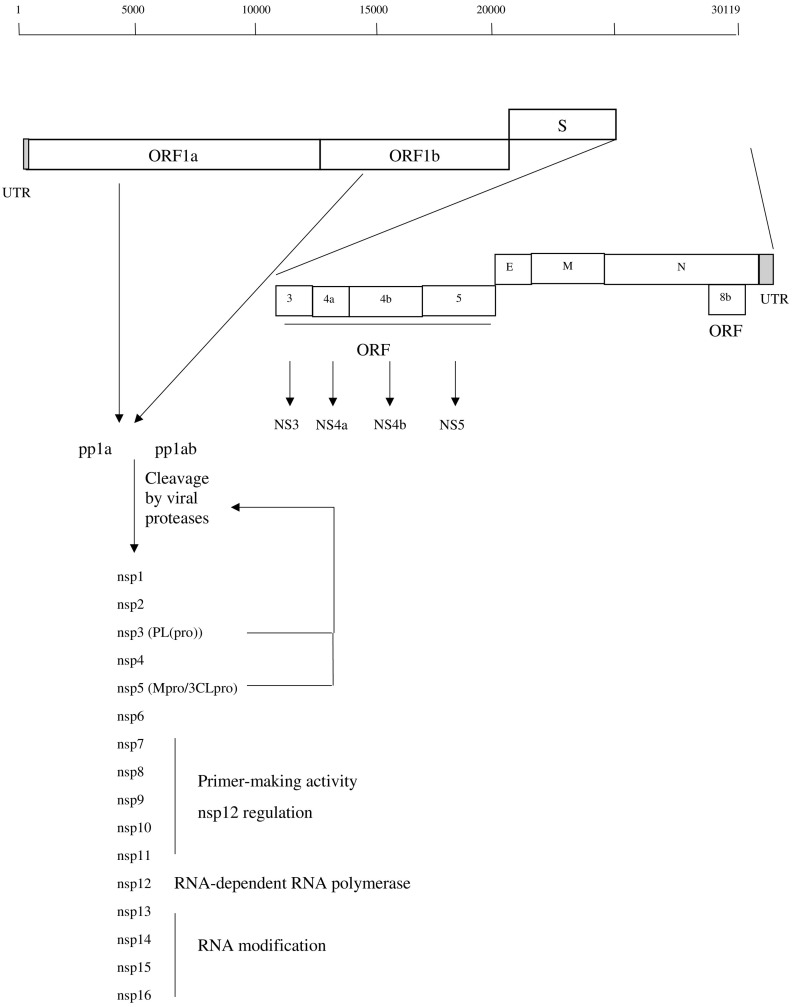
The MERS-CoV has a genome of 30,119 nucleotides comprising 7 predicted open reading frames (ORFs) (1a, 1b, 3, 4a, 4b, 5, 8b) and 4 structural genes encoding the spike (S), nucleocapsid (N), membrane (M) and envelope (E) proteins (Fig. 2 ) (Forni et al., 2016, Mackay and Arden, 2015, Zhang et al., 2016a). The overlapping ORF1a and 1b are located at the 5′ end of the single stranded positive RNA alongside a 278 nucleotide un-translated region (UTR) (Fig. 2). ORF1a and ORF1b comprise the majority of the MERS-CoV genome and are translated into polyproteins pp1a and pp1ab, which are then cleaved by viral proteases to give 16 non-structural proteins termed nsp1 to nsp16 (Fig. 2). These form the replication-transcription complex (RTC) of the virus. Individual nsp proteins have different roles in viral replication. For example, nsp3 has a papain-like protease (PLpro) activity which mediates the initial processing of pp1a (Forni et al., 2016, Hagemeijer et al., 2012, Neuman et al., 2014). Nsp3 also works with nsp4 and nsp6 to anchor the viral RTC to intracellular membranes and form a reticulovesicular membranous network where the viral RNA can replicate. Meanwhile nsp5 also has a protease activity, 3C-like protease (3CLpro), which also mediates pp1a and pp1ab cleavage into nsp 1–16. Nsp7 to nsp11 medate primer-making activity and regulate nsp12, which is the main viral RNA-dependent RNA polymerase (RdRp) (Forni et al., 2016, Hagemeijer et al., 2012, Neuman et al., 2014). Nsp13 to 16 are involved in viral RNA modification (Forni et al., 2016, Hagemeijer et al., 2012, Neuman et al., 2014).
Fig. 2.
Genomic Mapping of MERS-CoV.
The genes for the S, E, M and N proteins are downstream of ORF1 (Fig. 2). The S protein is vital in MERS-CoV transmission and host cell infection, determining tropism of the virus and host cell entry. The S protein is a trimeric, envelope protein which can be cleaved by host proteases into S1 (N-terminal) and S2 (C-terminal) subunits (Lu et al., 2015). The S1 subunit contains a receptor binding domain (RBD), which mediates binding of S protein to the host cell human dipeptidyl peptidase 4 (DPP4; CD26) receptor (Raj et al., 2013, Wang et al., 2013). Once the MERS-CoV binds to DPP4 via the S1 RBD, endocytosis occurs. Cleavage at the S1/S2 junction then occurs, mediated by host proteases including the serine protease TMPRSS2, the endosomal cathepsin L, and furin protease (Millet and Whittaker, 2014, Qian et al., 2013, Shirato et al., 2013, Yang et al., 2015a, Zhang et al., 2016a), followed by viral fusion with the host cell membrane mediated by the S2 subunit. The S2 subunit contains a fusion peptide, 2 heptad repeat domains HR1 and HR2, and a transmembrane (TM) domain (Durai et al., 2015). Fusion is facilitated by rearrangement of S2 into a 6-helix bundle (6HB) fusion core, centred on a trimer of the HR1 and HR2 dimer. This folding of H1/H2 allows exposure of the fusion peptide and insertion into host cell membrane, and hence fusion (Durai et al., 2015, Zhang et al., 2016a).
4.3. DPP4 receptor
The MERS-CoV S protein DPP4 receptor is widely expressed in human cells including lower respiratory tract non ciliated bronchial epithelium, kidney epithelial cells, small intestine cells, T lymphocytes and macrophages (Al-Qahtani et al., 2017, Boonacker and Van Noorden, 2003, Tang et al., 2017, Widagdo et al., 2016). There is limited expression of DPP4 in the upper respiratory tract epithelium in humans when compared to dromedary camels, which may contribute to the limited replication of MERS-CoV in the human upper respiratory tract and to restriction of human-to-human transmission (Widagdo et al., 2016). Infection of macrophages by lentiviral particles pseudotyped with MERS-CoV S protein resulted in attenuation of macrophage responses via expression of IRAK-M, a negative regulator of Toll-like receptor (TLR) signaling, and of the transcriptional repressor PPARγ (Al-Qahtani et al., 2017). Use of the DPP4 inhibitor sitagliptin or DPP4-siRNA reduced the effects of MERS-CoV S protein on IRAK-M, PPARγ and IL-10, indicating that the suppression of macrophage immune responses by MERS-CoV is mediated via DPP4 (Al-Qahtani et al., 2017). Mathematical modeling suggests that reducing the rate of DPP4 expression would reduce MERS-CoV spread (Tang et al., 2017). Indeed, levels of DPP4 mRNA and protein are higher in lung tissues of smokers and individuals with chronic obstructive pulmonary disease (COPD) compared to never-smokers (Seys et al., 2018); both smoking and COPD are associated with increased susceptibility to MERS-CoV infection. Host species restriction of MERS-CoV infection has been linked to 13 DPP4 residues which are key in interacting with the S protein RBD (Lu et al., 2015, Peck et al., 2015, van Doremalen et al., 2014). Phylogenetic analyses have shown that these residues are either conserved or differ by only one or 2 residues in DPP4 of species that are permissive either in vitro or in vivo, including camel, macaque, marmoset, goat, pig, civet, and horse (Lu et al., 2015), but to have multiple variations in non-permissive species including mouse, hamster and ferret (Peck et al., 2015).
Other host cell mediators may also be involved along with DPP4 in MERS-CoV S protein binding and viral infection. In a recent virus overlay protein binding assay (VOPBA) study, the carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) was identified as a another MERS-CoV cell surface binding target which interacts with the S protein in cell culture (Chan et al., 2016). While over-expression of CEACAM5 could not independently support MERS-CoV entry into non-permissive cells, it did enhance viral attachment, while in permissive cells CEACAM5 over-expression enhanced viral entry in conjunction with DPP4 (Chan et al., 2016). MERS-CoV has also been shown to bind with high specificity but low affinity to sialic acid (Sia) in a hemagglutination assay with human erythrocytes and intact virus (Li et al., 2017). The S1 domain or its S1A subdomain expressed on nanoparticles could bind Sia-dependently to human erythrocytes or mucin, while Sia depletion on the surface of Calu-3 human airway cells reduced MERS-CoV viral entry (Li et al., 2017). Thus in addition to DPP4 expression, Sia may also contribute to MERS-CoV host range and tissue tropism.
5. Pathogenesis and immunity
5.1. Infection routes
The human respiratory tract is the primary target for infection by MERS-CoV (Muller et al., 2012, Zielecki et al., 2013). DPP4-expressing bronchial epithelial cells, bronchiolar epithelial cells, alveolar epithelial cells and the endothelial cells of pulmonary vessels have all been found to be infected by the virus in ex vivo human lung tissue (Hocke et al., 2013, Mackay and Arden, 2015, Muller et al., 2012, Raj et al., 2013, Seys et al., 2018, Tang et al., 2017, Widagdo et al., 2016, Zielecki et al., 2013). The human intestinal tract has been recently proposed to be an alternative route for MERS-CoV infection (Zhou et al., 2017). Human primary intestinal epithelial cells, small intestine explants, and intestinal organoids have all been shown to be susceptible to MERS-CoV infection and replication, while enteric MERS-CoV has been identified in clinical patient stool samples (Zhou et al., 2017). In DPP4-transgenic mice, direct intragastri inoculation with MERS-CoV resulted in lethal infection while histology demonstrated the presence of enteric infection in all inoculated mice, with development of sequential respiratory infection (Zhou et al., 2017). MERS-CoV can target both the innate and adaptive human immune responses in a number of direct and indirect ways. A feature of MERS-CoV infection spread is the occurrence of nosocomial outbreaks. In a recent outbreak which occurred in May/June, 2017, there were 44 reported MERS-CoV cases from 3 simultaneous clusters in 3 different healthcare facilities in Riyadh; 11 cases were fatal (Amer et al., 2018). This outbreak highlights the need to develop rapid point-of-care testing to enable emergency room healthcare staff to rapidly identify MERS-CoV cases as the outbreak was the result of delay in diagnosis of MERS-CoV in a patient who presented with acute renal failure and who directly exposed 120 contacts including healthcare workers and other patients during 14 hours spent in the open area of the emergency department and 2 hemodialysis sessions (Amer et al., 2018).Hospital outbreaks, the fact that up to 50% of MERS-CoV cases in Saudi Arabia have been classified as due to human-to-human transmission through contact with asymptomatic or symptomatic individuals and the difficulty inherent in distinguishing the clinical features of MERS-CoV infection from other respiratory tract infections further highlights the importance of specific point-of-care testing and high degree of clinical awareness among clinical staff in Saudi Arabia (Hui et al., 2018).
5.2. Innate immune response: interferon
Detection of positive-stranded RNA viruses such as MERS-CoV by the host innate immune system depends on recognition of pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (PRRs) such as toll-like receptors (TLRs) (tenOever, 2016). An important PAMP relevant to MERS-CoV is viral double-stranded (ds)-RNA. The host cell innate immune response to ds-RNA involves induction of type I interferon (IFN) expression via the RIG-1-like helicases including Rig-1 and MDA-5, as well as other activities including activation of protein kinase R (PKR), which reduces translation in the infected host cell, and activation of the 2′,5′-oligoadenylate synthetase (OAS)/RNaseL pathway, which can degrade viral RNA (Schneider et al., 2014, tenOever, 2016). Importantly, while MERS-CoV is significantly more susceptible to type I interferon (IFN)- mediated innate immune responses than SARS-CoV, it also has strategies for evading these responses. In common with other coronaviruses, the MERS-CoV nsp3 PLpro removes ubiquitin (Ub) (deubiquitination; DUB), and interferon-stimulated gene 15 (ISG15) (deISGylation) from host cell proteins, which in turn blocks production of IFN-β and hence reduces type-1 interferon responses in cell line studies (Báez-Santos et al., 2014, Daczkowski et al., 2017). MERS-CoV nsp15, which contains an endonuclease (EndoU) activity, has also been recently shown in primary cell lines and in macrophages to reduce early innate immune responses by inhibition of MDA-5, PKR and OAS responses and IFN activation (Kindler et al., 2017). Nsp16, a viral 2’O-methyltransferase (2’O-MTase), has also been recently implicated in viral pathogenesis and type I- IFN inhibition in both primary human airway cell cultures and in vivo mouse models (Menachery et al., 2017a).
Meanwhile non-structural protein NS3, NS4a, NS4b and NS5, as well as the structural M protein, have been implicated in IFN antagonism and inhibition of the innate immune response in cell culture studies (Canton et al., 2018, Matthews et al., 2014, Menachery et al., 2017b, Rabouw et al., 2016, Siu et al., 2014, Thornbrough et al., 2016, Yang et al., 2013, Yang et al., 2015b). Lack of homology between MERS-CoV and SARS-CoV in their accessory ORF-3, 4a, 4b and 5 genes highlights the fact that immune defense mechanisms may differ between the viruses. Deletion of MERS-CoV ORF-3 to 5 has been shown both in vitro and in vivo mouse models to impact on viral replication and pathogenesis via dysregulation of host cell responses, including increased activation of the type-1 IFN pathway and induction of inflammatory responses (Menachery et al., 2017b). ORF5 has been shown to partially modulate the inflammation-associated NF-κB transcription factor (Menachery et al., 2017b). The ORF4b-encoded NS4b protein has been shown in cell culture studies to inhibit IFN- and NF-κB- mediated signaling, IFN-β production and the (OAS)/RNaseL pathway (Matthews et al., 2014, Thornbrough et al., 2016, Yang et al., 2015b). Presence of NS4b in MERS-CoVinfected cells results in tethering of NF-κB in the cytoplasm while NS4a is located in the nucleus (Canton et al., 2018). However in the absence of NS4b, or in the presence of mutant NS4b lacking a nuclear localization signal (NLS), NF-κB can translocate to the nucleus and induce pro-inflammatory cytokine expression (Canton et al., 2018). NS4b-induced NF-κB translocation inhibition appears to be mediated by its binding to karyopherin-α4 (KPNA4), a protein essential for NF-κB nuclear translocation (Canton et al., 2018). Binding of NS4b to KPNA4 during infection inhibited its interaction with the NF-κB-p65 subunit. NS4a is potentially particularly potent in IFN-inhibition as it targets both IFN-β production and signaling via interferon-sensitive response element (ISRE) promoter elements (Yang et al., 2013). NS4a-mediated inhibition of IFN production has been linked in vitro to its binding to the host cell ds-RNA-binding protein, interferon-inducible double-stranded RNA-dependent protein kinase activator A (PACT), which is a critical innate immune mediator responsible for activation of Rig-1 and MDA-5 and hence type 1-IFN in response to coronavirus infection (Siu et al., 2014). This is linked to NS4a-mediated inhibition of the PKR-induced stress response, as PACT is a PKR-associated protein (Rabouw et al., 2016). NS4a is a ds-RNA binding protein and hence can effectively mask the viral ds-RNA PAMP from the host innate immune response (Batool et al., 2017).
5.3. Innate immune response: cellular targeting
MERS-CoV virus infects and replicates in human macrophages -including alveolar macrophages- and can induce pro-inflammatory and chemotactic cytokines and chemokines expression from the infected macrophages (Zhou et al., 2014, Zhou et al., 2015a). Binding and infection of MERS-CoV is supported by expression of DPP4 receptor on alveolar macrophages (Meyerholz et al., 2016). Levels of DPP4 are higher on alveolar macrophages, as well as on alveolar epithelial cells, in individuals with pre-existing pulmonary disease such as cystic fibrosis or chronic obstructive pulmonary disease, which could predispose them to MERS-CoV morbidity and mortality (Meyerholz et al., 2016). In human monocyte-derived macrophages (MDMs), MERS-CoV productive infection did not induce expression of antiviral IFN-α or IFN-β, but induced similar levels of interleukin (IL)-6 and tumor necrosis factor (TNF)-α to SARS-CoV, and significantly higher levels of other proinflammatory cytokines including IL-12 and IFN-γ, and chemokines including IP-10/CXCL-10, MCP-1/CCL-2, MIP-1α/CCL-3, RANTES/CCL-5, and IL-8 (Zhou et al., 2014). This could contribute to the level of pulmonary inflammation and tissue damage associated with MERS-CoV induced progressive pneumonia. On the other hand, recent studies in differentiated THP-1 macrophages infected with lentiviral particles pseudotyped with MERS-CoV S protein suggested that macrophage responses including IL-6 and TNF-α production were reduced, while LPS-induced production of the immunosuppressive IL-10 was increased (Al-Qahtani et al., 2017). This increase in IL-10 production was mediated by DPP4 binding and activation of IRAK-M, a negative regulator of TLR signaling and the transcriptional repressor PPARγ (Al-Qahtani et al., 2017). These results suggest that MERS-CoV may employ IRAK-M and PPARγ to evade destruction by macrophages.
In vitro studies on antigen-presenting cells (APCs) have shown that human plasmacytoid dendritic cells (pDCs) could be infected by MERS-CoV and that unlike B cells, macrophages, or monocyte-derived dendritic cells (MDDCs) they secreted type I- and type III- IFNs upon MERS-CoV infection (Scheuplein et al., 2015). This was accompanied by initial steps of viral infection and replication, evidenced by increased N protein RNA in infected cells, but not by productive replication or viral amplification (Scheuplein et al., 2015). Recent studies suggested that while mature MDDCs did not seem to be permissive to MERS-CoV infection, immature MDDCs were permissive but, unlike with macrophages, infection in vitro did not result in up-regulation of proinflammatory cytokine and chemokine production (Cong et al., 2018). As dendritic cells enter peripheral tissues and carry antigens to lymphoid tissues, it has been suggested that they may contribute to MERS-CoV dissemination by acting as vehicles, possibly explaining the isolation of MERS-CoV from specimens other than respiratory tract samples such as blood, stool, and urine from MERS-CoV infected patients (Drosten et al., 2013, Guery et al., 2013).
5.4. Adaptive immune response
In one cell culture study, MERS-CoV but not SARS-CoV could efficiently infect human primary T cells, including cells from peripheral blood, spleen and tonsils (Chu et al., 2016). CD4 T cells appeared to be more susceptible than CD8 T cells, and infection resulted in DPP4 receptor down-regulation and in T cell apoptosis by both extrinsic and intrinsic pathways (Chu et al., 2016). Spleen and tonsil cells were apparently vulnerable to a higher degree of infection and apoptosis than peripheral blood cells (Chu et al., 2016). Infection of common marmosets with MERS-CoV resulted in dissemination of virus to the spleen and infection of T cells in vivo ( Chu et al., 2016). Results of a recent study on a transgenic mouse model expressing human DPP4 (hDPP4) suggested that depletion of CD8 T cells could actually protect from MERS-CoV-induced pathology and symptoms, whereas depletion of macrophages exacerbated the pathology and symptoms (Coleman et al., 2017). Meanwhile recent in vitro studies suggested that, in common with H5N1-VN1203 influenza virus, MERS-CoV can attempt to evade the adaptive immune response by down-regulation of antigen-presentation gene expression, mediated by epigenetic mechanisms (Menachery et al., 2018). Down-regulated genes in the human airway epithelial cell line Calu3 included HLA-A, -B, or –C, whose expression was increased in the presence of SARS-CoV infection, as well as transcription factors (CTIIA) and genes expressing elements of the antigen processing machinery (TAP2 and PDIA3). HLA-A, -B, or –C peptides were also decreased by MERS-CoV infection, although H5N1-VN1203 reduced only HLA-A and-C peptides. In the case of MERS-CoV the major epigenetic mechanism appeared to be DNA methylation whereas H5N1-VN1203 employed a number of mechanisms (Menachery et al., 2018). Results from use of mutant viruses suggested that both host and viral processes were involved in the antigen presentation down-regulation, although this conclusion awaits definitive data (Menachery et al., 2018). In terms of humoral responses to MERS-CoV, the S protein has been shown to be the most immunogenic MERS-CoV antigen and is central to neutralizing antibody and T cell responses to MERS-CoV (Zhang et al., 2014). As a result, the S protein is the target of a number of proposed MERS-CoV vaccines, which we have recently extensively reviewed (Rabaan et al., 2017) and which are considered in more detail below.
5.5. Case definition
In the light of the pathogenicity of MERS-CoV, its ability to potentially evade the immune system, and its high mortality rate, it is vital that accurate case definition criteria are established and updated as knowledge of the virus expands. This is of particular concern in KSA, which remains the site of the greatest number of cases. The WHO and the CDC regularly update case definitions in order to help healthcare professionals in recognition and classification of cases. Case definitions categorize patients into either confirmed or probable cases.
5.6. Confirmed Case
Both WHO and the CDC define a confirmed case as a patient with a laboratory confirmation of MERS-CoV regardless of clinical presentation (Centers for Disease Control and Prevention (CDC), 2017, World Health Organization (WHO), n.d, World Health Organization (WHO), 2018). Laboratory confirmation as currently defined can be via detection of viral nucleic acid or serology. The bases for WHO and CDC definitions are shown in Table 1 . Viral nucleic acid confirmation can be either by positive results for nucleic acid amplification assays (NAAT), for example reverse transcription polymerase chain reaction (RT-PCR) directed against a minimum of 2 specific genomic targets (either upstream of the E protein gene (upE) and ORF1a, ORF1b or N gene), or against a single positive target with sequencing of a second target, preferably the RNA-dependent RNA polymerase (RdRp; nsp12) or N genes (Al Johani and Hajeer, 2016, Centers for Disease Control and Prevention (CDC), 2017, Corman et al., 2012a, Corman et al., 2014c, World Health Organization (WHO), n.d, World Health Organization (WHO), 2018). In the USA, an Emergency Use Authorization (EUA) was issued by the FDA to authorize the use of the WHO-approved RealStar® MERS-CoV RT-PCR Kit, as there is currently no FDA-cleared/approved test available for MERS-CoV testing in the USA (Centers for Disease Control and Prevention (CDC), 2017, Corman et al., 2012a, Food and Drug Administration, 2016, Lu et al., 2014a). For serology, WHO case confirmation requires demonstration of sero-conversion in 2 samples, ideally taken at least 14 days apart, by a screening test including enzyme-linked immunosorbent assay (ELISA) or immunofluorescence assay (IFA) and a neutralization assay for confirmation (World Health Organization (WHO), n.d, World Health Organization (WHO), 2018). For the CDC, a 2-phase approach is also adopted, involving one screening test (ELISA) and 2 confirmatory tests (IFA, microneutralization) to detect MERS-CoV antibodies (Table 1) (Centers for Disease Control and Prevention, 2017). The CDC specifies that serology tests are for surveillance or investigational purposes rather than for diagnosis.
Table 1.
WHO and CDC case definitions for MERS-CoV.
| Case definition | WHO | CDC |
|---|---|---|
| Confirmed |
Nucleic acid testing RT-PCR: 2 specific genomic targets • upE31 • ORF1a, ORF1b or N gene OR RT-PCR: one specific genomic target and: Sequencing of a second target: nsp12 or M gene |
Nucleic acid testing RT-PCR: 2 specific genomic targets • upE31 • ORF1a, ORF1b or N gene OR RT-PCR: one specific genomic target and: Sequencing of a second target: nsp12 or M gene |
|
Serology Screening test: • ELISA • IFA Confirmation test: • Neutralization |
Serology Screening test: • ELISA Confirmation test: • IFA • Microneutralization Surveillance, investigation Not diagnosis |
|
| Probable (WHO) or Patient under investigation (PUI) (CDC) |
1. Febrile acute respiratory illness with clinical, radiological, or histopathological evidence of pulmonary parenchymal disease AND Direct epidemiologic link with a laboratory-confirmed MERS-CoV case AND Testing for MERS-CoV is unavailable, negative on a single inadequate specimen or inconclusive |
1. Fever AND pneumonia or acute respiratory distress syndrome AND EITHER: history of travel from countries in or near the Arabian Peninsula within 14 days before symptom onset, OR close contact with a symptomatic traveler who developed fever and acute respiratory illness (not necessarily pneumonia) within 14 days after traveling from countries in or near the Arabian Peninsula, OR a member of a cluster of patients with severe acute respiratory illness of unknown etiology in which MERS-CoV is being evaluated, in consultation with state and local health departments |
| 2. Febrile acute respiratory illness with clinical, radiological, or histopathological evidence of pulmonary parenchymal disease that cannot be explained fully by any other etiology AND Resides or traveled in the Middle East, or in countries where MERS-CoV is known to be circulating in dromedary camels or where human infections have recently occurred AND Testing for MERS-CoV is inconclusive |
2. Fever AND symptoms of respiratory illness (not necessarily pneumonia) AND being in a healthcare facility (as a patient, worker, or visitor) within 14 days before symptom onset in a country or territory in or near the Arabian Peninsula in which recent healthcare-associated cases of MERS have been identified. | |
| 3. Acute febrile respiratory illness of any severity AND Direct epidemiologic link with a confirmed MERS-CoV case AND Testing for MERS-CoV is inconclusive |
3. Fever OR symptoms of respiratory illness (not necessarily pneumonia) AND close contact with a confirmed MERS case while the case was ill. |
5.7. Probable case
There are different possible definitions of probable cases according to WHO criteria, all of which involve a febrile patient with respiratory disease, either with evidence of pulmonary parenchymal disease (e.g. pneumonia or Acute Respiratory Distress Syndrome (ARDS)); or of any severity, along with other criteria shown in Table 1, including residence or recent travel in the Middle East, or a direct epidemiological link to a laboratory-confirmed case (World Health Organization, n.d.). The CDC criteria for a probable case or person under investigation (PUI) are also shown in Table 1, and again involve patients who are febrile and/or have evidence of respiratory illness (acute or otherwise), along with criteria including recent travel, or being in a healthcare facility, in or near the Arabian Peninsula, or close contact with a laboratory-confirmed case (Centers for Disease Control and Prevention, 2017).
6. Diagnosis
6.1. Detection of viral RNA
Several assays for detection of MERS-CoV RNA have been developed using real-time PCR. Corman and colleagues introduced assays that target the region upstream of the E protein (upE), ORF1b, and ORF1a (Fig. 2) (Corman et al., 2012a, Corman et al., 2012b, Corman et al., 2014c). The high sensitivity of RT-PCR (upE) and RT-PCR (ORF1a) compared to ORF1b rendered them valuable options for screening of MERS-CoV RNA (Corman et al., 2012a, Corman et al., 2012b, Corman et al., 2014c, World Health Organization (WHO), 2018). The CDC validated a suggested alternate testing strategy based on screening of one N gene signature sequence (N2) combined with upE testing for enhanced sensitivity, and a second N gene signature (N3) for confirmation of positive tests (Lu et al., 2014a, World Health Organization (WHO), 2018). Corman and colleagues ultimately developed the upE and ORF1a real-time PCRs into the RealStar® MERS-CoV RT-PCR Kit, which was clinically validated using samples of a German MERS-CoV case and respiratory samples from other respiratory disease patients (Corman et al., 2014c). The RealStar® MERS-CoV RT-PCR Kit has been WHO approved and granted an FDA Emergency Use Authorization (EUA) in the United States (Food and Drug Administration, 2016, World Health Organization (WHO), n.d). The same study group introduced RT-PCR assays for sequencing in RdRp gene (RdRpSeq assay) and in the N gene (NSeq assay) now recommended by the WHO as confirmatory tests (Corman et al., 2012b).
Although the RealStar® MERS-CoV RT-PCR Kit is the only upE and ORF1a-detecting kit approved by WHO and the Conformité Européenne (CE), and permitted FDA Emergency Use Authorization (EUA), several MERS-CoV RNA detection kits have been developed. For example, in a recent study 6 commercially available real-time RT-PCR MERS-CoV RNA detection kits based on upE and ORF1a were analyzed and clinically validated on nasopharyngeal swabs taken during the 2015 outbreak in Korea (Kim et al., 2016a). Results suggested that sensitivity and specificity of all of these assay systems would be sufficient for confirmation of MERS-CoV infection, although use of appropriate internal controls would be important in specimens where PCR inhibition is an issue (Kim et al., 2016a). In another recent study, a MERS-CoV r-gene ® 32 rRT-PCR assay 33 (bioMérieux, France), targeting the S protein gene, was shown to have comparable accuracy to the WHO recommended in-house rRT-PCR assay targeting upE and ORF1a in a set of 130 respiratory samples (Lee et al., 2017). Loop-mediated isothermal amplification RT PCR assays (RT-LAMP) have also been developed for field use given their rapid results with high sensitivity profiles. They require minimal instrumentation, thus they can also be used for portable point-of-care testing (Bhadra et al., 2015). Other assays targeting small RNA molecules (leader sequences) have good sensitivity profiles (Chan et al., 2015b).
Respiratory specimens - nasopharyngeal swabs, sputum, tracheal aspirate and bronchoalveolar lavage (BAL) are commonly used for detection of viral RNA. Results of tests on patients from KSA and elsewhere comparing the viral load and genomic fraction yield among respiratory specimens obtained from different sites have shown that lower respiratory samples (e.g. tracheal aspirate and bronchoalveolar lavage) yield significantly higher viral loads and genomic fractions compared with upper respiratory tract samples (Drosten et al., 2013, Guery et al., 2013, Memish et al., 2014a). In the context of MERS-CoV testing in RSA and elsewhere, this means that WHO recommends that lower respiratory tract specimens should be collected whenever possible (World Health Organization, 2018). However, a case series from KSA also showed that there is value in collecting and testing upper respiratory tract specimens such as nasopharyngeal/oropharyngeal swabs so long as nasopharyngeal swabs are taken from the nasopharynx following WHO guidelines, not just from the nostril, and that nasopharyngeal and oropharyngeal swabs are placed in the same tube (Assiri et al., 2013b, World Health Organization (WHO), 2006, World Health Organization (WHO), 2018). Thus WHO recommends that when it is possible, both upper and lower respiratory tract specimens should be collected, while specimens from sites outside the respiratory tract should not be used for routine diagnostic testing (World Health Organization, 2018). WHO further recommends that samples should be collected for symptomatic patients for NAAT testing on presentation, followed by repeat sequential sampling every 2–4 days, until results are negative on 2 sequential samples to confirm viral clearance (World Health Organization, 2018).
6.2. Antigen detecting tests
For diagnosis of MERS-CoV in camels, which is highly relevant in the KSA context, molecular testing based on NAAT is not always a feasible option largely due to the expense and impracticality of carrying out large numbers of tests on animal herds in a timely manner. Recognition of the need for a relatively affordable test for use in diagnosis in camels which would also be sensitive and specific led to the development of an immunochromatographic assay (ICA) for the rapid and direct qualitative detection of MERS-CoV antigen (Song et al., 2015). The test was based on use of monoclonal antibodies for detection of N protein at room temperature and was 93.9% and 100% sensitive and specific respectively in relation to UpE and ORF1a real-time RT-PCR-based detection in a study on 571 camel nasal swabs (Song et al., 2015). Another N protein antigen-detection test, this time capture enzyme-linked immunosorbent assay (ELISA) based on 2 N protein-specific monoclonal antibodies (MAbs) has also been developed and shown to be 100% specific in testing of a series of 129 nasopharyngeal aspirates known to be positive for various respiratory viruses (Chen et al., 2015). Such a sensitive and specific ELISA test would be feasible for MERS-CoV detection both in clinical samples, in particular for point-of-care testing, and in dromedaries and other animals, and may have particular utility in field studies in KSA and elsewhere in the Middle East and in mass gathering contexts such as Hajj (Chan et al., 2017, Chen et al., 2015). The relative affordability and lower resource-intensiveness would give it an advantage over RT-PCR based methods in these types of contexts. These antigen-detection tests require further refinement as they have not yet been completely validated for use in human samples and are usually not as sensitive as NAAT, which has limited their use to date (Chan et al., 2017).
6.3. Detecting human immune response
Several serological assays have been developed for detection of anti MERS-CoV antibodies, notably against N protein or S protein. While NAAT-based testing is the gold standard for MERS-CoV diagnosis, serological assays have some advantages such as a less restricted time frame for antibody versus viral RNA detection, easier application in the field during an outbreak situation, and more economical application in animal testing (Fukushi et al., 2018, Meyer et al., 2014a, Trivedi et al., 2018). However, potential pitfalls of serological testing were exposed during the SARS-CoV outbreak, including the possibility of cross-reactivity to antigens from other coronaviruses (Meyer et al., 2014a). A recent assessment of the utility of ELISA-based detection of MERS-CoV S1 IgG compared to viral RNA detection was carried out on nasopharyngeal.
swab specimens from 174 patients in a hospital in Riyadh, between January 2016 and December 2016, during which a MERS-CoV.
outbreak occurred (Alhetheel et al., 2017). While MERS-CoV RNA was detected in 30 patient samples, only 6 samples were positive by serological testing, including 4 who were recently MERS-CoV RNA-positive and 2 who were MERS-CoV RNA-negative. This lack of correlation between NAAT and serological results suggested that MERS-CoV-IgG testing may not be appropriate for diagnosis of acute infection, estimation of outbreak prevalence, or determination of disease severity (Alhetheel et al., 2017). Nevertheless, serological testing remains one of the approved methods for MERS-CoV case confirmation by both WHO and CDC (World Health Organization (WHO), n.d, World Health Organization (WHO), 2018, Centers for Disease Control and Prevention (CDC), 2017). One recent validation study suggested that combination of indirect MERS-CoV N and S ELISAs in combination with confirmation by microneutralization assay can improve overall detection sensitivity and specificity (Trivedi et al., 2018). Another recent innovation suggests the possibility of using competitive ELISA rather than IgG/IgM ELISAs that rely on a species-specific secondary antibody (Fukushi et al., 2018). In this case, labeled monoclonal antibodies (MAb) against MERS-CoV S protein were developed and used to compete with test serum antibodies for target epitopes, allowing detection of antibodies in a species-independent manner (Fukushi et al., 2018). The competitive ELISA successfully detected MERS-CoV-specific antibodies in sera from infected rats and rabbits immunized with MERS-CoV S protein, and the test was also validated on sera from 66 Ethiopian dromedary camels in comparison to a neutralization test, giving sensitivity and specificity of 98% and 100%, respectively. These results suggest that competitive ELISA might be a useful serological test in epidemiological investigations in KSA and elsewhere in the Middle East (Fukushi et al., 2018). WHO recommends that for serology testing in symptomatic patients, paired samples should be collected within the first week of illness and the second ideally 3 to 4 weeks later (World Health Organization, 2018).
7. Clinical manifestations of MERS-CoV
7.1. Incubation period
Variable incubation periods for MERS-CoV have been calculated in studies from different countries (Assiri et al., 2013b, Mailles et al., 2013, Oh et al., 2015). A median of 5.2 days (95% CI 1.9–14.7 days) (range 2–13 days) was reported in one study of 47 laboratory confirmed MERS-CoV cases in KSA (Assiri et al., 2013b). Investigators in France reported a longer incubation period of between 9 and 12 days (Mailles et al., 2013). Early during the 2015 Korean outbreak, the median incubation period of MER-CoV was found to be 6.3 days (Oh et al., 2015). Accommodating the range of these observations, it is currently recommended that people who have contact with confirmed cases must be evaluated for a full 14 days from day of contact for any symptoms or signs suggestive of MERS-CoV.
7.2. Clinical features
The clinical spectrum of MERS-CoV infection ranges from mild respiratory illness to severe disease with severe acute respiratory distress syndrome, septic shock and multi-organ failure (Memish et al., 2013a, Memish et al., 2013b). Most reported cases do run a severe clinical course. Fever and cough are the predominant symptoms in symptomatic cases. Early in the history of MERS-CoV, analyzing the clinical presentation among 47 confirmed cases in KSA showed fever with temperature above 38 °C in almost 98% of the patients (Assiri et al., 2013b). Fever was also found to be a predictive factor for progression of pneumonia in a study following up the clinical course of 5 confirmed MERS-CoV cases during the Korean outbreak. The progression of pneumonia appeared to slow or even stop after fever subsided (Rhee et al., 2016).
Cough was present in 83% of infected individuals in the KSA study of 47 cases, while gastrointestinal (GI) symptoms including abdominal pain, vomiting and diarrhea were also reported in a significant number of patients included in this study (Assiri et al., 2013b). GI symptoms were also reported in 12.9% of the 186 cases involved in the South Korean MERS-CoV outbreak (Korea Centers for Disease Control, and Prevention, 2015). Arabi et al. reported the clinical manifestation in 12 cases from 2 hospitals in KSA, showing that symptoms could be attributed to the lower respiratory tract (Arabi et al., 2014). Upper respiratory tract symptoms, such as rhinorrhea and sore throat, were found to be uncommon (Arabi et al., 2014). Renal complications are well known to occur in MERS-CoV infection. The first ever reported case suffered from acute kidney injury (Zaki et al., 2012). Proteinuria, hematuria and acute kidney injury (AKI) were noted in a retrospective study of 30 MERS-CoV cases in South Korea, in which diabetes, AKI, and the application of a continuous renal replacement therapy (CRRT) were observed to be risk factors for MERS-CoV-related mortality (Cha et al., 2015). Seizures, DIC, and rhabdomyolysis were also reported as complications related to MERS-CoV infection in a study of seventy patients in a single centre in KSA (Shalhoub et al., 2015).
About 75% of confirmed MERS-CoV infections occur in patients with comorbid disease. Frequent comorbid conditions seen in patients with MERS-CoV infection are diabetes mellitus, obesity, chronic kidney disease, cardiac diseases, and hypertension, as well as respiratory diseases including asthma and COPD (Ahmed, 2018, Arabi et al., 2014, Assiri et al., 2013b, Badawi and Ryoo, 2016, Banik et al., 2016, Cha et al., 2015, Korea Centers for Disease Control, and Prevention, 2015, Matsuyama et al., 2016, Park et al., 2018, Saad et al., 2014, Shalhoub et al., 2015). Disease severity and mortality risk is impacted by comorbidities and age. For example, in one study age >50 years and diabetes were significantly associated with mortality and all patients in this series requiring renal replacement therapy died (Shalhoub et al., 2015). Age >65 years was significantly associated with mortality in another single centre study in KSA (Saad et al., 2014). A study analyzing publicly available data from KSA reported that pre-existing lung disease appeared not to be a significant risk factor for severity and mortality, however this study did not use multivariate risk modeling (Ahmed, 2018, Banik et al., 2016). Other case–control and retrospective observational studies from both KSA and Korea have suggested that smoking and/or comorbid respiratory diseases are significant risk factors for MERS-CoV-related mortality (Alraddadi et al., 2016b, Choi et al., 2016, Korea Centers for Disease Control, and Prevention, 2015, Matsuyama et al., 2016, Park et al., 2018). Higher levels of DPP4 mRNA and protein in lung tissues of smokers and COPD patients compared to never-smokers may predispose these individuals to MERS-CoV infection (Seys et al., 2018). Systematic review and meta-analysis has shown that obesity is present in 16% of MERS-CoV cases and may influence disease severity as with other respiratory conditions (Badawi and Ryoo, 2016). Asymptomatic MERS-CoV infection also occurs in household contacts, healthcare workers and people who have contact with dromedary camel (Memish et al., 2013a, Memish et al., 2013b).
7.3. Children
Although older age has been confirmed as a risk factor for MERS-CoV infection and mortality, it is not only a disease of adults but also occurs in children, albeit rarely (Al-Tawfiq et al., 2016, Memish et al., 2014b, Thabet et al., 2015). 80.6% of the 31 pediatric cases reported between June 2012 and April 2016 were from KSA, with a mean age of 9.8 ± 5.4 years, and they were most commonly infected due to household contacts (Al-Tawfiq et al., 2016). Mortality is lower in children than in adults and is commonly associated with underlying comorbid conditions In one study from KSA, MERS-CoV was detected in 11 pediatric patients ranging in age from 2 to 16 years (Memish et al., 2014b). While 9 of the 11 were asymptomatic and were detected during a contacts investigation on older patients, 2 symptomatic patients had underlying conditions and one died (Memish et al., 2014b). Meanwhile a 9-month-old infant with infantile nephrotic syndrome being treated with prednisolone died in the PICU of a Riyadh hospital as result of MERS-CoV infection and his clinical course was complicated by acute renal failure (Thabet et al., 2015).
7.4. Pregnancy
Information is limited on the impact of MERS-CoV in pregnancy, but in common with other severe respiratory viral infections the impact appears to be severe both maternally and perinatally. In one study on 5 pregnant women in KSA infected with MERS-CoV, all 5 needed ICU admission (Assiri et al., 2016). While 2 recovered and went on to deliver healthy infants, one of the mothers died due to multiple organ failure related to her infection after delivering a healthy infant at 38 weeks gestation, another died due to complications of her infection a few weeks after her infant was surgically delivered at 24 weeks and died after 4 hours of life, and one infant was stillborn at 34 weeks (Assiri et al., 2016). One case of a second trimester stillbirth during a MERS-CoV outbreak in Jordan was attributed to MERS-CoV on the basis of maternal exposure history and serological testing (Payne et al., 2014). In another case a woman at 32 weeks gestation died due to MERS-CoV-related complications including ARDS and septic shock after delivering a healthy infant by caesarean section (Malik et al., 2016).
7.5. Laboratory and radiological manifestation
In a study of 47 cases of MERS-CoV infection in KSA, 14% had leukopenia, 34% had lymphopenia and 11% had lymphocytosis, while thrombocytopenia was present in 36% of cases (Assiri et al., 2013b). Lymphocytopenia and thrombocytopenia have also been detected in other studies, including among members of a KSA MERS-CoV family cluster (Memish et al., 2013a). Impaired liver function findings are a feature of MERS-CoV infection, including the 47-case study which revealed raised concentrations of lactate dehydrogenase (49% of patients), alanine aminotransferase (11% of patients) and aspartate amino transferase (15% of patients), although other liver function tests were normal (Assiri et al., 2013b) and in a retrospective study of 29 confirmed cases of MERS-CoV infections from March to May 2014 at 2 hospitals in the Al-Madinah region of KSA, in which elevated liver enzymes were observed in 50% of cases (Sherbini et al., 2017). Elevation of urea and creatinine levels indicating renal impairment has also been widely observed, including in case series from KSA (Arabi et al., 2014, Sherbini et al., 2017). Animal studies on human DPP4 (hDPP4)-expressing transgenic mouse models infected with MERS-CoV, while not entirely reflective of disease in humans, have shown multi-organ damage, including to liver and kidney as well as brain and spleen (Zhao et al., 2015). However, other studies on a hDPP4 transgenic mouse models have suggested that while infection with 10 LD50 of MERS-CoV resulted in persistent inflammatory infiltrates in the lungs and brain stems 2 and 4 days post-infection respectively, and focal infiltrates in the liver, there was no definite pathology in other organs (Tao et al., 2016). Recently, post-mortem histopathological findings on a 33-year-old male T lymphoma patient who contracted MERS-CoV were reported (Alsaad et al., 2018). Histopathological examination of tissue needle biopsies from multiple sites including brain, heart, lung, liver, kidney and skeletal muscle showed evidence of virally induced pulmonary and extrapulmonary pathological changes. These included necrotising pneumonia, pulmonary diffuse alveolar damage, acute kidney injury, hepatitis and myositis with muscle atrophic changes, however there were no notable findings for brain and heart. For the first time, ultrastructural viral particles were shown in renal cells, as well in pneumocytes, pulmonary macrophages and macrophages infiltrating the skeletal muscles (Alsaad et al., 2018). A wide range of radiological features have been shown on chest X-rays of MERS-CoV infected patients including ground glass opacification, consolidation (either patchy or confluent), reticular opacities, nodular opacities and reticulo-nodular infiltrates (Assiri et al., 2013b, Das et al., 2015, Das et al., 2016). Use of serial chest radiographs can be used to classify disease progression into 4 types ranging from type 1, in which initial radiographic deterioration is followed by improvement, all the way up to type 4, where there is progressive radiographic deterioration (Das et al., 2016). Importantly, in a study of 55 adult patients with acute MERS-CoV infection, chest radiographic score was shown to be an independent predictor of mortality, with mean chest radiographic score significantly higher in patients who died than in those who survived. Pneumothorax, bilateral pleural effusion and type 4 radiographic progression were all significantly higher in patients who died (Das et al., 2015). Bilateral pleural effusion has also been identified as an independent predictor of short-term mortality for community-acquired pneumonia but not SARS (Hasley et al., 1996, Wong et al., 2003). Similar to the radiographic findings, the more sensitive computed tomography (CT) scans also showed ground glass opacity (53% of patients), or consolidation (20% of patients), or both together (33% of patients), as well as pleural effusion (33%) and interlobular thickening (26%) within a week of infection (Das et al., 2015, Das et al., 2016). As disease progressed, bronchial abnormalities and organizing pneumonia emerged on CT scans (Das et al., 2015, Das et al., 2016).
8. Source and transmission
8.1. Bats
As mentioned above, it has been assumed that bats are the likely main MERS-CoV mammalian source reservoir, as with other coronaviruses, because sequences related to the MERS-CoV were found in samples taken from different bat species (Fig. 1) (Drexler et al., 2014, Memish et al., 2013c). The HKU4 bat coronavirus RBD in the S protein shares high sequence identity to MERS-CoV and pseudotyped viruses embedding HKU4 S protein can bind human DPP4 and enter cells in vitro (Wang et al., 2014). HKU4 S protein binds human DPP4 with only low affinity, however introduction of 2 mutations, N762A and S746R, into the bat S gene enabled HKU4 to bind with higher affinity and more efficiently enter human cells (Yang et al., 2015a). These mutations are part of human protease motifs in the S1/S2 junction in MERS-CoV and thus facilitate S protein cleavage and human cell infection and may have been instrumental in transmission of MERS-CoV from bats to humans (Yang et al., 2015a). However, positing that bats are a direct source of MERS-CoV human infections is difficult given the infrequent contact of human with bats. In a study in KSA, it was found that samples from only one bat found near the home of a MERS-CoV infected patient among 823 samples collected from different bat species had total nucleotide identity with MERS-CoV sequence obtained from the patient (Memish et al., 2013c).
8.2. Camels
There is growing evidence that dromedary camels act as the source of MERS-CoV. Dromedary camels' sera from different parts of the world –especially from the Middle East and broad areas of Africa, including Nigeria, Tunisia, Egypt and Ethiopia – have tested positive for anti-MERS-CoV antibodies (Ali et al., 2017b, Chu et al., 2015, Farag et al., 2015, Meyer et al., 2014b, Müller et al., 2015, Reusken et al., 2013a, Reusken et al., 2014, Saqib et al., 2017). Serological studies on camels in Africa and the Middle East within the last 30 years suggest that MERS-CoV was circulating among camels for decades before it was first documented in human beings in 2012 (Meyer et al., 2014b, Reusken et al., 2014). All Canary Islands dromedary camels which have positive serological evidence of MERS-CoV infection were originally imported from Africa 20 years ago or more (Gutiérrez et al., 2015). However, there are lower than expected levels of MERS-CoV human infection in Africa, which suggests there may be under-reporting of human cases, possibly related to limited resources for testing. Extension of sero-surveys among the human population would help in furthering understanding of the extent of levels of MERS-CoV infection in Africa. In one study use of ELISA, IFA and ppNT showed that there was evidence for unrecorded cases of human MERS-CoV in Kenya, similar to previous reports in KSA (Liljander et al., 2016, Müller et al., 2015).
There is some genetic evidence to suggest transmission of MERS-CoV occurs from camels to humans. During one outbreak in Qatar, MERS-CoV sequences obtained from nasopharyngeal swabs from 2 infected human cases residing on a farm and from 3 seropositive camels within the same farm were found to be identical (Haagmans et al., 2014). In another case in Jeddah in KSA, a shared unique single nucleotide polymorphism (SNP) signature was found in both a MERS CoV patient and a MERS-CoV-carrying dromedary camel for which he had been caring (Azhar et al., 2014, Memish et al., 2014c). Comparison of the sequence of the full genome of the MERS-CoV variant associated with the Korean outbreak showed 99.96–99.98% similarity with the full genome of CoVs obtained from a camel in Riyadh, Saudi Arabia (Sabir et al., 2016). In this study RT-PCR testing was carried out on nasal swab samples from 1309 camels. Coronaviruses were identified in 25.3% of samples and 3 different lineages of coronaviruses, including MERS-CoV, betacoronavirus 1 (betacoronavirus, group A); and human CoV 229E (alphacoronavirus) were found to be circulating among dromedary camels (Sabir et al., 2016). The study showed camels aged less than 1 year have the highest rate of infection with coronaviruses compared to older camels (Sabir et al., 2016). The identification of camels as the probable natural zoonotic source for human infection with MERS-CoV has economic implications for countries of the Middle East, including KSA, given the importance of the camel trade between the Middle East and Africa (Younan et al., 2016).
8.3. Other animals
There was no evidence of MERS CoV upon testing of other animals such as sheep, goats, cattle, or water buffalo, although results of one study suggests alpaca may be a possible viral reservoir (Perera et al., 2013, Reusken et al., 2013b, Reusken et al., 2016). Detection of MERS-CoV in this New World camelid raises the possibility of zoonotic spread of MERS CoV to areas where alpacas are farmed, including South America and the United States (Reusken et al., 2016).
8.4. Human-to-human transmission
Strong evidence of human to human transmission was obtained from epidemiological and genomic studies investigating clustering of cases in hospitals and among household contacts (Assiri et al., 2013a, Memish et al., 2013a, Müller et al., 2015). Investigating a hospital outbreak in the city of Al-Ahsa in the Eastern Province of KSA revealed that all isolates of MERS-CoV infecting the 23 patients were from one monophyletic lineage and 91.3% of cases occurred as a result of person-to-person contact (Assiri et al., 2013a). Human-to-human transmission was also responsible for most of the MERS-CoV cases reported during the outbreak that occurred in Jeddah in 2014 (Oboho et al., 2015). The majority of cases were attributable to contact with a health care facility, other patients, or both, highlighting the role of healthcare facilities in human-to-human transmission that also arose in subsequent outbreaks, including hospital outbreaks in Riyadh and the 2015 outbreak in Korea (Oboho et al., 2015, Drosten et al., 2015, Fagbo et al., 2015, Almekhlafi et al., 2016, Balkhy et al., 2016a; Middle East respiratory syndrome coronavirus (MERS-CoV) – Republic of Korea, 2015). As explained above, healthcare facility human-to-human transmission has been associated with defective or inadequate infection prevention and control measures (Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia, 2018, Balkhy et al., 2016b, Butt et al., 2016, Coronavirus infections, 2017, Cotten et al., 2014, Hastings et al., 2016). The infection tends to be milder in secondary cases, in which a patient is infected as a result of close contact with a primary source, and can even be asymptomatic. The number of cases who get infected from confirmed cases is low; the rate of transmission among household contacts has been calculated to be around 5% in one study done in KSA in 2014 (Memish et al., 2014d). However, epidemiological analysis of the Korean hospital outbreak in 2015 showed that the fatality rate was not significantly different between primary cases and subsequent generations (Kim, 2015). This outbreak highlighted the danger posed by a combination of circumstances including a primary source traveling from the Middle East, infection among secondary and tertiary contacts due to movement of infected individuals between healthcare facilities, and inadequate infection prevention and control measures (Nishiura et al., 2016a, Nishiura et al., 2016b, Park et al., 2015).
8.5. Epidemic potential
From the data available to date, MERS-CoV has failed to demonstrate the potential to result in an epidemic. A study based on Bayesian analysis was carried out to estimate the basic MERS-CoV reproduction number (R0), which represents the number of secondary cases for each index case in a fully susceptible population (Breban et al., 2013). Epidemic potential is achieved when R0 is above 1,. R0 for MERS-CoV was estimated to be between 0·60 and 0·69, however these calculations were based on data obtained in June 2013 in advance of many of the important outbreaks and so may be underestimated (Breban et al., 2013). There is in any case no room for complacency, as the potential is always present for viral mutations that could increase zoonotic or human-to-human transmissibility. Thus development of effective directed therapies remains a top priority.
9. Vaccination and therapy
9.1. Current and potential treatments
In 2015, Public Health England (PHE) and the WHO International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC-WHO) published a position paper on MERS-CoV therapies (Treatment of MERS-CoV; information for clinicians, 2015). They concluded that there was a positive benefit versus risk balance for convalescent plasma, lopinavir/ritonavir, interferons and monoclonal/polyclonal antibodies, but a negative balance for ribavirin monotherapy or corticosteroids (Treatment of MERS-CoV; information for clinicians, 2015). It was deemed that there was insufficient information available for interferon/ribavirin combination therapy, nitazoxanide and chloroquine (Treatment of MERS-CoV; information for clinicians, 2015). Currently, no specific evidence-based therapy or vaccine for MERS-CoV is available. We have recently extensively reviewed candidate MERS-CoV therapies and vaccines (Rabaan et al., 2017). Table 2 shows a summary of current and proposed therapies and vaccines, including targets, advantages and disadvantages, updated to include some potential agents that have emerged since the publication of our review (Alharbi et al., 2017, Jung et al., 2018, Langenmayer et al., 2018, Li et al., 2018, Liu et al., 2018, Niu et al., 2018, Rabaan et al., 2017, Sun et al., 2017). Development of a targeted anti- MERS-CoV therapy and availability of effective vaccines would require coordinated efforts to carry out properly controlled and organized clinical trials. This would be of particular importance for KSA, given the relatively major impact of MERS-CoV there; availability of reliable directed therapies and the possibility of either a prophylactic vaccine programme or a vaccine that could be rapidly available in the event of a major outbreak would be a major advantage in effectively tackling this disease.
Table 2.
Summary of potential MERS-CoV therapies and vaccines.
| Therapeutic target | Type of therapy | Therapy/ Vaccine name |
Study type | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|---|
| S1/DPP4 binding | Antibody (mouse): S1 RBD | Mersmab | In vitro | (Du et al., 2014) | ||
| Antibody (human): S1 RBD |
m336, m337, m338 |
In vitro In vivo (Mouse, rabbit- m336) |
(Agrawal et al., 2016, Houser et al., 2016, Ying et al., 2014) | |||
| Antibody (human): S1 RBD |
MERS-4, MERS-27 | In vitro | (Jiang et al., 2014, Yu et al., 2015) | |||
| Antibody (mouse- humanized): S1 RBD | 4C2 |
In vitro In vivo (Mouse) |
Prophylactic and therapeutic | (Li et al., 2015) | ||
| Antibody (mouse- humanized): S1 RBD | hMS-1 |
In vitro In vivo (Mouse) |
(Qiu et al., 2016) | |||
| Antibody (human): S1 RBD |
LCA60 |
In vitro In vivo (Mouse) |
Targets both NTD and RBD; stable CHO cell line; prophylactic and therapeutic | (Corti et al., 2016) | ||
| Antibody (human): S1 RBD |
3B11-N |
In vitro In vivo (rhesus monkeys) |
Prophylactic | (Johnson et al., 2016) | ||
| Antibody (human): S1 RBD |
MERS-GD27 MERS-GD33 |
In vitro | Synergistic effect; Different epitopes; MERS-GD27 overlaps receptor binding site | (Niu et al., 2018) | ||
| Antibody (human- anti-DPP4) |
2F9, 1F7, YS110 | In vitro | (Ohnuma et al., 2013) | |||
| RBD-IgG fusion vaccine candidate | RBD s377–588- Fc IgG fusion |
In vitro In vivo (Mouse) |
Humoral response in mice; potential intranasal administration; improved by adjuvant; divergent strains/ escape mutants; CHO cell line |
(Du et al., 2013, Ma et al., 2014, Nyon et al., 2018, Tai et al., 2017, Zhang et al., 2015, Zhang et al., 2016b) | ||
| Nanoparticles vehicle (vaccine candidate) | Full-length S protein proprietary nanoparticles |
In vitro In vivo (Mouse) |
Use of adjuvants improves humoral response | Stable expression of abundant full-length S protein difficult | (Coleman et al., 2014) | |
| Nanoparticles and virus vehicle (vaccine candidate) | Full-length S protein: Ad5/MERS and S protein nanoparticles | Heterologous prime-boost: | In vivo (Mouse) | T cell and neutralizing antibody responses; potentially prophylactic | (Jung et al., 2018) | |
| Virus vehicle (vaccine candidate) | MVA expressing full-length S protein | MVA-MERS-S |
In vitro In vivo (Mouse, camel) |
T cell and neutralizing antibody responses; entering human clinical trials; potential for veterinary use- | (Langenmayer et al., 2018, Volz et al., 2015) | |
| ad5 or ad41 adenovirus expressing full-length S |
In vitro In vivo (Mouse) |
T cell and neutralizing antibody responses | (Guo et al., 2015) | |||
| Measles virus expressing full-length S |
In vitro In vivo (Mouse) |
T cell and neutralizing antibody responses | (Malczyk et al., 2015) | |||
| Chimeric vesicular stomatitis virus (VSV) expressing full-length S |
In vitro In vivo (Rhesus monkeys) |
T cell and neutralizing antibody responses | (Liu et al., 2018) | |||
| Chimpanzee adenovirus (ChAdOx1) expressing full-length S |
In vitro In vivo (mouse) |
T cell and neutralizing antibody responses; entering human clinical trials; potential for veterinary use | (Alharbi et al., 2017) | |||
| Plasmid vaccine | GLS-5300 |
In vitro In vivo (Mouse, camels, and macaques) Human clinical trials |
T cell and neutralizing antibody responses; in phase I clinical trial | (Inovio, 2016, Muthumani et al., 2015) | ||
| Viral S2-host membrane fusion | Anti-HR2 viral peptide | HR2P | In vitro | (Lu et al., 2014b) | ||
| Anti-HR2 viral peptide | HR2P-M2 |
In vitro In vivo (Mouse) |
Blocks 6HB bundle formation; enhances IFN-β effect; potential intranasal treatments | (Bosch et al., 2004, Channappanavar et al., 2015, Liu et al., 2004) | ||
| Three HR1 and two HR2 protein | MERS-5HB | In vitro | Inhibits fusion and entry | (Sun et al., 2017) | ||
| Immune evasion response | IFN-α2b and ribavirin |
In vitro In vivo (Macaque) |
Combination therapy- reduced dose of each; non-human primate model; 10 different gene pathways | (Falzarano et al., 2013a, Falzarano et al., 2013b, Zheng and Wang, 2016) | ||
| IFN-β1b and lopinavir |
In vitro In vivo (Marmoset) |
Combination therapy- reduced dose of each | (Chan et al., 2015c) | |||
| IFN combination therapy (ribavirin and/or lopinavir | Case studies (human) | Only prophylactic or early use; insufficient evidence of clinical efficacy as yet | (Khalid et al., 2015, Kim et al., 2016b, Spanakis et al., 2014, Strayer et al., 2014) | |||
| IFN combination therapy (ribavirin) | Retrospective cohort studies (human) | Probable benefit of early use in less vulnerable patients; safety and efficacy established for other viral illnesses | Only prophylactic or early use; insufficient evidence of clinical efficacy as yet | (Al-Tawfiq et al., 2014, de Wilde et al., 2013, Khalid et al., 2014, Lau et al., 2013, Omrani et al., 2014, Shalhoub et al., 2015) | ||
| IFN combination therapy (cyclosporine) |
In vitro Human ex-vivo explant |
Synergistic effect; safety and efficacy established for other viral illnesses | (Li et al., 2018) | |||
| S protein host proteases | TMPRSS2 inhibitor | Camostat | In vivo- mouse, SARS-CoV | Already in clinical use | (Zhou et al., 2015b) | |
| TMPRSS2 inhibitor | Nafamostat | Split-protein-based cell–cell fusion assay | Already in clinical use | (Yamamoto et al., 2016) | ||
| Cathepsin L inhibitor | Teicoplanin dalbavancin oritavancin telavancin | High-throughput screening | Already in clinical use | (Zhou et al., 2016) | ||
| Viral proteases | PL(pro) inhibitor | 6-mercaptopurine (6MP) 6-thioguanine (6TG) |
In vitro | Potential for more MERS-specific agents | (Cheng et al., 2015) | |
| PL(pro) inhibitor | F2124–0890 | In vitro | May lose potency in physiological reducing environments | (Clasman et al., 2017) | ||
| Mpro | Lopinavir |
In vitro In vivo (marmosets) |
High activity at low micromolar range in vitro; better outcomes, in marmosets | Clinical efficacy not fully established in humans | (Chan et al., 2015, de Wilde et al., 2014, Rambaut, 2014) |
The S protein and its binding to DPP4 is the target of many proposed direct MERS-CoV therapies, including a large number of antibodies which target the interaction either from the viral or the host side (Agrawal et al., 2016, Corti et al., 2016, Du et al., 2014, Houser et al., 2016, Jiang et al., 2014, Johnson et al., 2016, Li et al., 2015, Niu et al., 2018, Ohnuma et al., 2013, Qiu et al., 2016, Ying et al., 2014, Yu et al., 2015). Monoclonal antibodies against the RBD of the S1 region have particularly strong neutralizing capacity, although full-length S or S1 targeting antibodies may have greater potential in a vaccine context given their larger number of target epitopes and the reduced chance of escape mutations (Agrawal et al., 2016, Corti et al., 2016, Du et al., 2014, Houser et al., 2016, Jiang et al., 2014, Johnson et al., 2016, Li et al., 2015, Niu et al., 2018, Ohnuma et al., 2013, Qiu et al., 2016, Ying et al., 2014, Yu et al., 2015). A fusion product in which truncated RBD (residues 377–588) was joined to the Fc portion of human IgG could bind human DPP4 and inhibit MERS-CoV infection in vitro in cell culture and in vivo in infected mice (Table 2) (Du et al., 2013). In vivo studies on mice have also indicated that intranasal administration of this fusion product induced comparable sustained IgG humoral responses to subcutaneous injection, and superior cellular immune responses and local mucosal responses in lungs (Ma et al., 2014, Zhang et al., 2015). Use of an adjuvant, particularly MF59 or AddaVax, significantly improved both the humoral and T cell responses in subcutaneously immunized mice (Zhang et al., 2016b). Recently, a high-yield CHO cell line capable of large-scale production of this S1 RBD-Fc fusion product was described, strengthening the possibility of sustainable manufacture and human testing for this potential vaccine antigen (Nyon et al., 2018). Another recent study showed that 5 recombinant RBDs incorporating mutations which arose in different MERS-CoV outbreaks or in camel strains could induce neutralizing antibody responses against several MERS-CoV pseudoviruses (Tai et al., 2017).
A particularly promising antibody candidate for MERS-CoV therapy is the human antibody LCA60, as it targets both the N-terminal domain (NTD) and the RBD of S1 (Table 2) (Corti et al., 2016). LCA60 was isolated from B cells of a MERS-CoV-infected human donor, and has been used to establish a stable CHO cell line from which clinical grade antibody is reliably available (Corti et al., 2016); this type of ready availability would be of particular benefit in KSA for outbreak situations. It had both prophylactic and therapeutic activities against MERS-CoV infection in 2 transgenic mouse models, Ad5/hDPP4 and type I interferon receptor (IFNAR)- KO (Corti et al., 2016). Another human anti-RBD antibody, 3B11-N, has shown promising results in a non-human primate model, i.e. rhesus monkeys infected with MERS-CoV, in which it prophylactically reduced lung pathology (Johnson et al., 2016). Recently a suite of potent MERS-CoV-neutralizing anti-S protein antibodies were derived from B cells of an infected patient, specifically from the first imported case in China (Niu et al., 2018). Two of the antibodies in particular, MERS-GD27 and MERS-GD33, had potent and synergistic neutralizing in vitro activity against both pseudotyped and live MERS-CoV (Table 2) (Niu et al., 2018). The 2 antibodies targeted different epitopes, with the MERS-GD27 epitope almost entirely overlapping the receptor binding site (Niu et al., 2018). Thus there is a wide range of S protein directed antibodies and fusion products available for potential passive immunization strategies, but thus far they have not entered human clinical trials. Availability of monoclonal antibodies may be of particular use in outbreak situations, which continue to arise in KSA. Other potential S protein-targeting vaccine candidates include nanoparticles expressing full-length S protein (Coleman et al., 2014) and active immunization strategies using vectors including modified vaccinia, adenoviruses or measles viruses or plasmids expressing full-length S protein as potential vaccine candidates, discussed in more detail in the next section (Alharbi et al., 2017, Guo et al., 2015, Inovio, 2016, Jung et al., 2018, Langenmayer et al., 2018, Liu et al., 2018, Malczyk et al., 2015, Muthumani et al., 2015, Volz et al., 2015).
Antiviral peptides that target the HR2 regions of the S protein and hence virus-host cell fusion have also been shown to have potential therapeutic activities in cell culture and transgenic animal studies (Bosch et al., 2004, Channappanavar et al., 2015, Liu et al., 2004, Lu et al., 2014b). The HR2 peptide HR2P, covering residues 1251–1286, reduced viral replication and fusion in vitro (Lu et al., 2014b) while its analogue, HR2P-M2, blocked fusion even more potently in vitro, and inhibited pseudovirus infection by blocking 6HB bundle formation (Bosch et al., 2004, Channappanavar et al., 2015, Liu et al., 2004). Convenient intranasal administration of HR2P-M2 in vivo to Ad5/hDPP4 transgenic mice protected them from MERS-CoV infection, which was enhanced by co-administration of IFN-β (Bosch et al., 2004). Recently a synthetic protein named MERS-5-helix bundle (MERS-5HB) was derived from the 6HB bundle involved in MERS-CoV fusion and was shown to bind strongly to HR2P and to effectively inhibit pseudotyped MERS-CoV fusion and entry in in vitro studies (Table 2) (Sun et al., 2017). This represents another potentially useful directed MERS-CoV therapeutic candidate.
At present, combined antiviral therapies tend to be used in patients who develop respiratory illness, based on experience with SARS-CoV therapy, for example pegylated interferon (IFN)-α, ribavirin, and/or lopinavir/ritonavir (Al-Tawfiq et al., 2014, de Wilde et al., 2013, Khalid et al., 2014, Khalid et al., 2015, Kim et al., 2016b, Lau et al., 2013, Omrani et al., 2014, Shalhoub et al., 2015, Spanakis et al., 2014, Strayer et al., 2014). While in vitro and animal studies suggested their potential efficacy, in vivo and clinical evidence is less well-established (Al-Tawfiq et al., 2014, Chan et al., 2015, de Wilde et al., 2013, Falzarano et al., 2013a, Falzarano et al., 2013b, Khalid et al., 2014, Khalid et al., 2015, Kim et al., 2016b, Lau et al., 2013, Omrani et al., 2014, Shalhoub et al., 2015, Spanakis et al., 2014, Strayer et al., 2014, Zheng and Wang, 2016). Clinical studies have been mainly confined to case studies and case series, and retrospective analyses (Al-Tawfiq et al., 2014, de Wilde et al., 2013, Khalid et al., 2014, Khalid et al., 2015, Kim et al., 2016b, Lau et al., 2013, Omrani et al., 2014, Spanakis et al., 2014, Strayer et al., 2014). Thus there is a need for properly controlled clinical trials of IFN combination therapy in MERS-CoV, preferably early in the illness when it seems to be most effective. These types of therapies function essentially by challenging the immune evasion tactics employed by the virus (Al-Tawfiq et al., 2014, Chan et al., 2015, de Wilde et al., 2013, Falzarano et al., 2013a, Falzarano et al., 2013b, Khalid et al., 2014, Khalid et al., 2015, Kim et al., 2016b, Lau et al., 2013, Omrani et al., 2014, Shalhoub et al., 2015, Spanakis et al., 2014, Strayer et al., 2014, Zheng and Wang, 2016). Recently, results of a study using in vitro and human ex vivo explant cultures suggested that a combination of IFN-α and cyclosporine had a synergistic effect on reduction of MERS-CoV replication, based on immunomodulation and induction of IFN-stimulated gene expression, suggesting that clinical trials may be warranted (Table 2) (Li et al., 2018). Corticosteroid treatment is also commonly used in treatment of critically ill MERS-CoV patients, despite the findings of the ISARIC-WHO group that its risks may outweigh benefits (Treatment of MERS-CoV; information for clinicians, 2015). A recent marginal structural modeling study was carried out on data from 309 critically ill ICU patients with MERS-CoV, of whom 151 received corticosteroids, from 14 KSA health facilities between September 2012 and October 2015 (Arabi et al., 2018). The results indicated that corticosteroid therapy was not associated with significantly different mortality outcomes when time-varying confounding effects such as worsening condition of the patient were considered, but that it was associated with delayed clearance of viral RNA. These findings suggest that bias in determining potentially harmful effects of therapies can emerge in observational studies if only the baseline characteristics rather than time-variant characteristics of the patients are considered and further highlight the need for properly controlled clinical trial data.
Another useful tactic would be to make use of therapies that have already been clinically approved for other purposes and for which there is a sound scientific rationale for possible use in MERS-CoV therapy. An example would be camostat, which is an inhibitor of TMPRSS2 (Shirato et al., 2013, Zhou et al., 2015b). Camostat has been shown to block infection, viral spread and pathogenesis in a pathogenic mouse model of SARS-CoV and would be likely to have a similar inhibitory effect on MERS-CoV (Zhou et al., 2015b). Camostat is already used clinically for treatment of chronic pancreatitis, and is thus a potentially safe and effective therapeutic option. Another TMPRSS2 inhibitor, nafamostat, has also been identified in vitro as a potent inhibitor of MERS-CoV S protein-mediated host-viral membrane fusion and is also already in clinical use as an FDA-approved anticoagulant (Yamamoto et al., 2016). In a screen of FDA-approved drugs, an inhibitor of cathepsin L called teicoplanin has been shown to block cytoplasmic entry of MERS-CoV, SARS-CoV and Ebola pseudoviruses (Qian et al., 2013, Zhou et al., 2016). Teicoplanin is in current clinical use as an antibiotic for serious Gram-positive bacterial infections and its derivatives, including dalbavancin, oritavancin, and telavancin, also block cytoplasmic viral entry. While these therapies all target host proteases, another possibility is targeting of viral proteases. The nsp3 encoded PL(pro) activity, which mediates the initial processing of pp1a (Fig. 2) (Forni et al., 2016, Hagemeijer et al., 2012, Neuman et al., 2014), can be inhibited in vitro by the SARS-CoV PL(pro) inhibitors, 6-mercaptopurine (6MP) and 6-thioguanine (6TG) and by a commercial compound F2124–0890 (Life Chemicals) (Cheng et al., 2015, Clasman et al., 2017). The main MERS-CoV protease Mpro/3CLpro, encoded by nsp5 (Fig. 2) can be targeted in vitro and in vivo by lopinavir, a protease inhibitor with activity against the SARS-CoV Mpro and which emerged in a screen of a library of 348 FDA-approved drugs as one of 4 compounds that inhibited MERS-CoV viral activity in a low micromolar range (Chan et al., 2015, de Wilde et al., 2014, Rambaut, 2014). However, lopinavir clinical efficacy has not been convincingly established in MERS-CoV treatment as it has generally been used clinically in combination with IFN and data is only available from case studies and series. In marmosets infected with MERS-CoV, it gave favorable clinical outcomes and reduced mortality in combination with ritonavir (Chan et al., 2015).
Early treatment (within 4–5 days of symptoms onset) with convalescent plasma (or hyperimmune IV immunoglobulin (HVIG) from plasma of convalescent donors) has been associated with decreased viral load and reduced mortality for influenza and SARS-CoV infection, although the quality of studies for SARS-CoV has been uneven and there have been few adequate clinical trials (Hui and Lee, 2013, Hung et al., 2013, Mair-Jenkins et al., 2015, Stockman et al., 2006). The PHE and ISARIC-WHO position paper identified convalescent plasma as a potential treatment for MERS-CoV infection, however no clinical trial have yet been completed (Treatment of MERS-CoV; information for clinicians, 2015). A clinical trial in KSA on safety and efficacy of convalescent plasma treatment for critically ill MERS-CoV patients was initiated in May 2014 and is still listed as active but not recruiting [247; ClinicalTrials.gov Identifier: NCT02190799]. It was due to report in June 2017 but in common with many convalescent plasma trials it has been affected by logistical and technical issues, such as availability both of sufficient donors and sufficient levels of MERS-CoV antibodies in the plasma that is collected (Arabi et al., 2015, Modjarrad, 2016). Clinical data is sparse on use of convalescent plasma in treatment of MERS-CoV and is confined to 2 case reports in which its role in patient recovery was unclear (Arabi et al., 2014, Kapoor et al., 2014). Use in marmosets infected with MERS-CoV in a recent study indicated that while convalescent plasma treatment reduced signs of clinical disease, including reduced respiratory tract viral load, it did not induce a decrease in gross pathology (van Doremalen et al., 2017). Thus while convalescent plasma is a possible candidate MERS-CoV therapy, technical and logistical difficulties with its collection and preparation and uncertainty over the extent of its protective effects may undermine its potential usefulness.
9.2. Vaccines
Studies from KSA have suggested that while patients who survived MERS-CoV produced anti-MERS-CoV IgG and neutralizing antibodies, these antibody levels only weakly inversely correlated with lower respiratory tract (LRT) viral load and would be insufficient to eliminate LRT virus (Corman et al., 2015). T cell responses to MERS-CoV infection are not yet well-understood, but in a recent study on 21 survivors of MERS-CoV in KSA, both CD4 and CD8 T cell responses developed in all of them (Zhao et al., 2017). MERS-CoV specific neutralizing antibody responses along with memory CD4 T cell but not CD8 T cell responses were shown to correlate with disease severity, while virus-specific CD8 T cell responses were observed in all MERS-CoV survivors, even when serological responses were not observed (Zhao et al., 2017). Robust CD8 T cell responses might therefore be important in early clearance of viral infection and hence antibody and CD4 T cell responses may not develop so strongly. Measurement of T cell responses along with antibodies may also give a more accurate estimate of disease prevalence. In vitro studies have shown that MERS-CoV infection down-regulates MHC and antigen presentation molecules via a methylation-based mechanism, which could have implications for both T cell and humoral adaptive immune responses [old ref. (Menachery et al., 2018)]. The combination of the apparent inadequacy of the humoral adaptive immune response to clear MERS-CoV and the high mortality rate associated with the disease point up the importance of vaccine development, particularly for the Middle East and KSA in particular. Induction of both antibody and T cell responses would be an important feature of a useful vaccine. WHO have issued guidelines on proposed MERS-CoV vaccines; they will consider prospective vaccines on a case-by-case basis (WHO target product profiles for MERS-CoV vaccines, 2017). WHO distinguished between vaccine types to be aimed at 3 different defined target populations, all of which have direct relevance in the Middle East. The 3 types are: dromedary camel vaccines designed to prevent camel-camel and camel-human transmission; prophylactic human vaccines for individuals who may be at long-term risk, for example healthcare workers and people working with potentially infected animals; and finally human vaccines which would be suitable for use in outbreaks (WHO target product profiles for MERS-CoV vaccines, 2017). WHO have defined preferred and minimally acceptable criteria for each vaccine type. These WHO guidelines are particularly welcome in the context of the difficulties that have prevailed in defining populations who should be targeted in a MERS-CoV vaccination program and/or in randomized clinical trials, especially given the current relatively low incidence of disease in humans, and the difficulties in developing suitable small animal models, depending on transduced or transgenic human DPP4-expressing mouse models (Cho et al., 2018).
As with therapy development, the S protein is the focus of many candidate vaccines (Alharbi et al., 2017, Coleman et al., 2014, Guo et al., 2015, Inovio, 2016, Jung et al., 2018, Langenmayer et al., 2018, Liu et al., 2018, Malczyk et al., 2015, Muthumani et al., 2015, Volz et al., 2015). Vectors including modified vaccinia virus Ankara (MVA), ad5 or ad41-type adenoviruses, measles virus, chimeric vesicular stomatitis virus (VSV) and chimpanzee adenovirus (ChAdOx1) have been successfully used to express MERS-CoV S protein and induce neutralizing antibodies in mice and in other animal models including camels and rhesus monkeys (Table 2) (Alharbi et al., 2017, Coleman et al., 2014, Guo et al., 2015, Inovio, 2016, Jung et al., 2018, Langenmayer et al., 2018, Liu et al., 2018, Malczyk et al., 2015, Muthumani et al., 2015, Volz et al., 2015). These virus vectors have the advantage of good safety profiles in humans. Production of candidate vaccines with potential for veterinary use in dromedary camels in order to reduce cross-species transmission is a welcome development in keeping with the WHO guidelines. These include an S protein-expressing MVA-based vaccine (MVA-MERS-S) which can a strong neutralizing antibody and cytotoxic T lymphocyte response and reduction of viral replication in transduced mice and induce mucosal immunity in MERS-CoV-infected dromedary camels (Langenmayer et al., 2018, Volz et al., 2015). These viruses are due to enter human clinical trials soon as a candidate prophylactic MERS-CoV vaccine. Another potential vaccine due to be evaluated in camels and to enter human clinical trials is a ChAdOx1 MERS vaccine (Table 2) (Alharbi et al., 2017). In mouse studies, a single dose of ChAdOx1 MERS with the leader sequence of the human tissue plasminogen activator gene (tPA) induced an equivalent humoral response to 2 doses of an MVA-based vaccine (Alharbi et al., 2017). Another potentially efficient prophylactic vaccination strategy recently tested in mice involved heterologous prime–boost vaccination regimens using Ad5/MERS in combination with S protein nanoparticles (Jung et al., 2018). Heterologous prime-boost elicited both anti-MERS-CoV neutralizing antibodies and simultaneous Th1 and Th2 responses, while homologous prime–boost regimens did not induce simultaneous Th1 and Th2 responses. Homologous Ad5/MERS also did not induce neutralizing antibody responses, while immunization schedules involving Ad5/MERS did induce Th1 cell activation and those including only S protein nanoparticles did not. Thus overall, heterologous prime–boost schedules gave superior results and are likely to induce more effective and sustained immune responses against MERS-CoV (Jung et al., 2018). This type of vaccine would again be in keeping with WHO guidelines. A DNA-plasmid-based vaccine called GLS-5300 which encodes MERS-CoV S protein and was co-developed by Inovio, GeneOne Life Science Inc. and the Walter Reed Army Institute of Research, is meanwhile the first potential MERS-CoV vaccine to be tested in clinical trials in humans (Inovio, 2016). A phase I clinical trial in healthy volunteers is ongoing to evaluate its safety and its ability to generate sustained humoral and cellular immune responses over a 1 year period (Inovio, 2016). Pre-clinical trials were performed in mice, camels, and macaques, in which the vaccine induced robust immune responses which were effective in preventing viral infection (Muthumani et al., 2015).
10. Conclusions
Since its initial description in 2012, MERS-CoV has exacted a high mortality rate particularly in KSA. While epidemic potential has not been evident thus far, the potential exists for viral mutation that could increase zoonotic and/or human-to-human transmission. Outbreaks have tended to occur in healthcare facility settings and infection rates in KSA have been reduced by stringent efforts to improve infection control and prevention standards. However, the inclusion by WHO of MERS-CoV on its list of priority blueprint diseases is a timely reminder of the urgent need for accelerated research and development as this disease has the potential to cause a public health emergency and there are currently no directly efficacious drugs and/or vaccines available (List of Blueprint priority diseases, 2018). The virus clinical spectrum varies from asymptomatic, to mild–moderate disease and potential for severe disease with a high case fatality rate. The impact of asymptomatic cases, including healthcare workers, on transmission is not yet fully understood. Multiple studies have suggested that dromedary camels are the likely main zoonotic source of MERS-CoV infection in humans, and this has major implications for the valuable camel trade between the Middle East and Africa. The apparently lower than expected numbers of human cases in Africa may be attributable to inadequacies in surveillance systems that should be addressed. The role of other animals such as bats and hedgehogs also needs further clarification, and the possible emergence of alpacas as a potential zoonotic source deserves attention. While NAAT detection systems are highly sensitive and specific, further attention is needed to the most effective and feasible detection systems that can be employed in the field. A major ongoing issue is the lack of any accepted specific treatment for MERS-CoV infection. Current treatment guidelines are too much based on experience with SARS-CoV therapy, despite numerous key differences between these coronaviruses, and there is an urgent need to move from in vitro and in vivo models and clinical case studies to properly managed randomized control trials on some of the numerous direct therapeutic and vaccine candidates that have been identified. Further clarification of issues such as duration of isolation of patients with MERS-CoV infection is also needed. Thus in our view, priorities include further clarification of transmission modes, for example the role of asymptomatic individuals in disease spread, ongoing vigilance in monitoring possible cross-species transmission, the ongoing need for well-validated human and animal sera panels, and the need to add some urgency to the clinical response progress, including advancement of possible direct therapies to human clinical trials. While progress has undoubtedly been made in our understanding of MERS-CoV, much remains to be done to reduce the impact of this disease, particularly in KSA, and to ensure that any future outbreaks can be effectively contained.
Compliance with Ethics Guidelines
This manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee. Special thanks to research center, Dr. Sulaiman Al-Habib Medical Group for supporting this research project.
References
- My 5 Moments for Hand Hygiene. World Health Organization (WHO); 2018. About SAVE LIVES: Clean Your Hands.http://www.who.int/gpsc/5may/background/5moments/en/ [cited 2018 September 27]. Available from. [Google Scholar]
- Agrawal A.S., Ying T., Tao X. Passive transfer of a germline-like neutralizing human monoclonal antibody protects transgenic mice against lethal Middle East respiratory syndrome coronavirus infection. Sci Rep. 2016;6:31629. doi: 10.1038/srep31629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A.E. Estimating survival rates in MERS-CoV patients 14 and 45 days after experiencing symptoms and determining the differences in survival rates by demographic data, disease characteristics and regions: a worldwide study. Epidemiol Infect. 2018;146(4):489–495. doi: 10.1017/S095026881700293X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Johani S., Hajeer A.H. MERS-CoV diagnosis: an update. J Infect Public Health. 2016;9:216–219. doi: 10.1016/j.jiph.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alharbi N.K., Padron-Regalado E., Thompson C.P., Kupke A., Wells D., Sloan M.A. ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine. 2017;35(30):3780–3788. doi: 10.1016/j.vaccine.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhetheel A., Altalhi H., Albarrag A., Shakoor Z., Mohamed D., El-Hazmi M. Assessing the detection of Middle East respiratory syndrome coronavirus IgG in suspected and proven cases of Middle East respiratory syndrome coronavirus infection. Viral Immunol. 2017;30(9):649–653. doi: 10.1089/vim.2017.0091. [DOI] [PubMed] [Google Scholar]
- Ali M., El-Shesheny R., Kandeil A., Shehata M., Elsokary B., Gomaa M. Cross-sectional surveillance of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels and other mammals in Egypt, August 2015 to January 2016. Euro Surveill. 2017;22(11):1560–7917. doi: 10.2807/1560-7917.ES.2017.22.11.30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M., El-Shesheny R., Kandeil A. Cross-sectional surveillance of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels and other mammals in Egypt, August 2015 to January 2016. Euro Surveill. 2017;22(11):1560–7917. doi: 10.2807/1560-7917.ES.2017.22.11.30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almekhlafi G.A., Albarrak M.M., Mandourah Y., Hassan S., Alwan A., Abudayah A. Presentation and outcome of Middle East respiratory syndrome in Saudi intensive care unit patients. Crit Care. 2016;20:123. doi: 10.1186/s13054-016-1303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Qahtani A., Lyroni K., Aznaourova M., Tseliou M., Al-Anazi M., Al-Ahdal M. Middle East respiratory syndrome coronavirus spike glycoprotein suppresses macrophage responses via DPP4-mediated induction of IRAK-M and PPARγ. Oncotarget. 2017;8(6):9053–9066. doi: 10.18632/oncotarget.14754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alraddadi B., Bawareth N., Omar H. Patient characteristics infected with Middle East respiratory syndrome coronavirus infection in a tertiary hospital. Ann Thorac Med. 2016;11(2):128–131. doi: 10.4103/1817-1737.180027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alraddadi B.M., Watson J.T., Almarashi A., Abedi G.R., Turkistani A., Sadran M. Risk factors for primary Middle East respiratory syndrome coronavirus illness in humans, Saudi Arabia, 2014. Emerg Infect Dis. 2016;22(1):49–55. doi: 10.3201/eid2201.151340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaad K.O., Hajeer A.H., Al Balwi M., Al Moaiqel M., Al Oudah N., Al Ajlan A. Histopathology of Middle East respiratory syndrome coronovirus (MERS-CoV) infection - clinicopathological and ultrastructural study. Histopathology. 2018;72(3):516–524. doi: 10.1111/his.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J., Kattan R.F., Memish Z.A. Middle East respiratory syndrome coronavirus disease is rare in children: an update from Saudi Arabia. World J Clin Pediatr. 2016;5(4):391–396. doi: 10.5409/wjcp.v5.i4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J.A., Momattin H., Dib J., Memish Z.A. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int J Infect Dis. 2014;20:42–46. doi: 10.1016/j.ijid.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer H., Alqahtani A.S., Alzoman H., Aljerian N., Memish Z.A. Unusual presentation of Middle East respiratory syndrome coronavirus leading to a large outbreak in Riyadh during 2017. Am J Infect Control. 2018;46(9):1022–1025. doi: 10.1016/j.ajic.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y., Balkhy H., Hajeer A.H., Bouchama A., Hayden F.G., Al-Omari A. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: a study protocol. SpringerPlus. 2015;4:709–716. doi: 10.1186/s40064-015-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., Arifi A.A., Balkhy H.H., Najm H., Aldawood A.S., Ghabashi A. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med. 2014;160(6):389–397. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am J Respir Crit Care Med. 2018;197(6):757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- Assiri A., Abedi G.R., Al Masri M., Bin Saeed A., Gerber S.I., Watson J.T. Middle East respiratory syndrome coronavirus infection during pregnancy: a report of 5 cases from Saudi Arabia. Clin Infect Dis. 2016;63(7):951–953. doi: 10.1093/cid/ciw412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., Al-Tawfiq J., Al-Rabeeah A., Al-Rabiah F., Al-Hajjar S., Al-Barrak A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A.T. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M. Evidence for camel-to-human transmission of MERS coronavirus. NEJM. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Badawi A., Ryoo S.G. Prevalence of comorbidities in the Middle East respiratory syndrome coronavirus (MERS-CoV): a systematic review and meta-analysis. Int J Infect Dis. 2016;49:129–133. doi: 10.1016/j.ijid.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y.M., Mielech A.M., Deng X., Baker S., Mesecar A.D. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J Virol. 2014;88(21):12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkhy H.H., Alenazi T.H., Alshamrani M.M., Baffoe-Bonnie H., Al-Abdely H.M., El-Saed A. Notes from the Field: Nosocomial Outbreak of Middle East Respiratory Syndrome in a Large Tertiary Care Hospital--Riyadh, Saudi Arabia, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:163–164. doi: 10.15585/mmwr.mm6506a5. [DOI] [PubMed] [Google Scholar]
- Balkhy H.H., Perl T.M., Arabi Y.M. Preventing healthcare-associated transmission of the Middle East Respiratory Syndrome (MERS): our Achilles heel. J Infect Public Health. 2016;9:208–212. doi: 10.1016/j.jiph.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik G.R., Alqahtani A.S., Booy R., Rashid H. Risk factors for severity and mortality in patients with MERS-CoV: Analysis of publicly available data from Saudi Arabia. Virol Sin. 2016;31(1):81–84. doi: 10.1007/s12250-015-3679-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batool M., Shah M., Patra M.C., Yesudhas D., Choi S. Structural insights into the Middle East respiratory syndrome coronavirus 4a protein and its dsRNA binding mechanism. Sci Rep. 2017;7(1):11362. doi: 10.1038/s41598-017-11736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W.B., McIntosh K., Dees J.H., Chanock R.M. Morphogenesis of avian infectious bronchitis virus and a related human virus (strain 229E) J Virol. 1967;1:1019–1027. doi: 10.1128/jvi.1.5.1019-1027.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17(40):20290. [PubMed] [Google Scholar]
- Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PloS One. 2015;10 doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonacker E., Van Noorden C.J.F. The multifunctional or moonlighting protein CD26/DPPIV. Eur J Cell Biol. 2003;82(2):53–73. doi: 10.1078/0171-9335-00302. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., Martina B.E., van der Zee R., Lepault J., Haijema B.J., Versluis C. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. PNAS. 2004;101(22):8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breban R., Riou J., Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382(9893):694–699. doi: 10.1016/S0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt T.S., Koutlakis-Barron I., AlJumaah S., AlThawadi S., AlMofada S. Infection control and prevention practices implemented to reduce transmission risk of Middle East respiratory syndrome-coronavirus in a tertiary care institution in Saudi Arabia. Am J Infect Control. 2016;44(5):605–611. doi: 10.1016/j.ajic.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton J., Fehr A.R., Fernandez-Delgado R., Gutierrez-Alvarez F., Sanchez-Aparicio M., García-Sastre A. MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog. 2018;14(1):1–25. doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control & Prevention (CDC) CDC Laboratory Testing for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) 2017. https://www.cdc.gov/coronavirus/mers/lab/lab-testing.html Available from:
- Cha R., Joh J., Jeong I., Lee J.Y., Shin H., Kim G. Renal complications and their prognosis in Korean patients with Middle East respiratory syndrome-coronavirus from the central MERS-CoV designated hospital. J Korean Med Sci. 2015;30(12):1807–1814. doi: 10.3346/jkms.2015.30.12.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C., Chu H., Wang Y., Wong B.H., Zhao X., Zhou J. Carcinoembryonic antigen-related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of Middle East respiratory syndrome coronavirus. J Virol. 2016;90(20):9114–9127. doi: 10.1128/JVI.01133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Choi G.K., Tsang A.K., Tee K.M., Lam H.Y., Yip C.C. Development and evaluation of novel real-time reverse transcription-PCR assays with locked nucleic acid probes targeting leader sequences of human-pathogenic coronaviruses. J Clin Microbiol. 2015;53:2722–2726. doi: 10.1128/JCM.01224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yao Y., Yeung M., Deng W., Bao L., Jia L. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212(12):1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yuen K. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Sridhar S., Yip C.C.Y., Lau S.K.P., Woo P.C.Y. The role of laboratory diagnostics in emerging viral infections: the example of the Middle East respiratory syndrome epidemic. J Microbiol. 2017;55(3):172–182. doi: 10.1007/s12275-017-7026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Lu L., Xia S., Du L., Meyerholz D.K., Perlman S. Protective effect of intranasal regimens containing peptidic Middle East respiratory syndrome coronavirus fusion inhibitor against MERS-CoV infection. J Infect Dis. 2015;212(12):1894–1903. doi: 10.1093/infdis/jiv325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chughtai A.A., Dyda A., MacIntyre C.R. Comparative epidemiology of Middle East respiratory syndrome coronavirus (MERS-CoV) in Saudi Arabia and South Korea. Emerg Microbes Infect. 2017;6(6) doi: 10.1038/emi.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chan K., Kang Y., Chen H., Luk H.K.H., Poon R.W.S. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerg Microbes Infect. 2015;4(4):1–5. doi: 10.1038/emi.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K., Cheng S., Chen W., Lin M.H., Chuang S.J., Cheng I.H. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antiviral Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Excler J., Kim J.H., Yoon I. Development of Middle East respiratory syndrome coronavirus vaccines – advances and challenges. Hum Vaccin Immunother. 2018;14(2):304–313. doi: 10.1080/21645515.2017.1389362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.S., Kang C., Kim Y., Choi J., Joh J.S., Shin H. Clinical presentation and outcomes of Middle East respiratory syndrome in the Republic of Korea. Infect Chemother. 2016;48(2):118–126. doi: 10.3947/ic.2016.48.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Oladipo J.O., Perera R., Kuranga S.A., Chan S.M., Poon L.L. Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Nigeria, 2015. Euro Surveill. 2015;20(49) doi: 10.2807/1560-7917.ES.2015.20.49.30086. [DOI] [PubMed] [Google Scholar]
- Chu H., Zhou J., Wong B.H., Li C., Chan J.F., Cheng Z. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis. 2016;213(6):904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasman J.R., Báez-Santos Y.M., Mettelman R.C., O'Brien A., Baker S.C., Mesecar A.D. X-ray structure and enzymatic activity profile of a core papain-like protease of MERS coronavirus with utility for structure-based drug design. Sci Rep. 2017;7:40292. doi: 10.1038/srep40292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M., Liu Y.V., Mu H., Taylor J.K., Massare M., Flyer D.C. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32(26):3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M., Sisk J.M., Halasz G., Zhong J., Beck S.E., Matthews K.L. CD8+ T Cells and macrophages regulate pathogenesis in a mouse model of Middle East respiratory syndrome. J Virol. 2017;91(1):1825-16. doi: 10.1128/JVI.01825-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Hart B.J., Gross R., Zhou H., Frieman M., Bollinger L. MERS-CoV pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells. PLoS One. 2018;13(3):1–17. doi: 10.1371/journal.pone.0194868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M. Viral shedding and antibody response in 37 patients with MERS-coronavirus infection. Clin Infect Dis. 2015;62(4):477–483. doi: 10.1093/cid/civ951. civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17(39):1–6. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Ithete N.L., Richards L.R., Schoeman M.C., Preiser W., Drosten C. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014;88(19):11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Kallies R., Philipps H., Göpner G., Müller M.A., Eckerle I. Characterization of a novel betacoronavirus related to Middle East respiratory syndrome coronavirus in European hedgehogs. J Virol. 2014;88(1):717–724. doi: 10.1128/JVI.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Müller M.A., Costabel U., Timm J., Binger T., Meyer B. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveill. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Ölschläger S., Wendtner C.M., Drexler J.F., Hess M., Drosten C. Performance and clinical validation of the RealStar MERS-CoV kit for detection of Middle East respiratory syndrome coronavirus RNA. J Clin Virol. 2014;60:168–171. doi: 10.1016/j.jcv.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disease outbreak news. World Health Organization (WHO); Geneva: 2017. Coronavirus infections.http://www.who.int/csr/don/archive/disease/coronavirus_infections/en/ [cited 2018 April 4]. Available from. [Google Scholar]
- Corti D., Passini N., Lanzavecchia A., Zambon M. Rapid generation of a human monoclonal antibody to combat Middle East respiratory syndrome. J Infect Public Health. 2016;9(3):231–235. doi: 10.1016/j.jiph.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Watson S.J., Zumla A.I., Makhdoom H.Q., Palser A.L., Ong S.H. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio. 2014;5(1):e01062-13. doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daczkowski C.M., Dzimianski J.V., Clasman J.R., Goodwin O., Mesecar A.D., Pegan S.D. Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J Mol Biol. 2017;429(11):1661–1683. doi: 10.1016/j.jmb.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K.M., Lee E.Y., Al Jawder S.E., Enani M.A., Singh R., Skakni L. Acute Middle East respiratory syndrome coronavirus: temporal lung changes observed on the chest radiographs of 55 patients. Am J Roentgenol. 2015;205(3):W267–W274. doi: 10.2214/AJR.15.14445. [DOI] [PubMed] [Google Scholar]
- Das K.M., Lee E.Y., Langer R.D., Larsson S.G. Middle East respiratory syndrome coronavirus: what does a radiologist need to know? Am J Roentgenol. 2016;206(6):1193–1201. doi: 10.2214/AJR.15.15363. [DOI] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Falzarano D., Ying T., de Wit E., Bushmaker T., Feldmann F. Efficacy of antibody-based therapies against Middle East respiratory syndrome coronavirus (MERS-CoV) in common marmosets. Antiviral Res. 2017;143:30–37. doi: 10.1016/j.antiviral.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Miazgowicz K.L., Milne-Price S., Bushmaker T., Robertson S., Scott D. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J Virol. 2014;88(16):9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Muth D., Corman V.M., Hussain R., Al Masri M., HajOmar W. An observational, laboratory-based study of outbreaks of Middle East respiratory syndrome coronavirus in Jeddah and Riyadh, kingdom of Saudi Arabia, 2014. Clin Infect Dis. 2015;60:369–377. doi: 10.1093/cid/ciu812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G., Sack S. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Yang Y., Qiu H., Wang L., Kou Z. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol. 2014;88(12):7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai P., Batool M., Shah M., Choi S. Middle east respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med. 2015;47(8) doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbo S.F., Skakni L., Chu D.K.W., Garbati M.A., Joseph M., Peiris M. Molecular epidemiology of hospital outbreak of Middle East respiratory syndrome, Riyadh, Saudi Arabia, 2014. Emerg Infect Dis. 2015;21:1981–1988. doi: 10.3201/eid2111.150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P. Treatment with interferon-[alpha] 2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19(10):1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag E.A.B.A., Reusken C.B.E.M., Haagmans B.L., Mohran K.A., Stalin Raj V., Pas S.D. High proportion of MERS-CoV shedding dromedaries at slaughterhouse with a potential epidemiological link to human cases, Qatar 2014. Infect Ecol Epidemiol. 2015;5 doi: 10.3402/iee.v5.28305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration Fact sheet for health care providers: interpreting RealStar® MERS-CoV RT-PCR Kit U.S. Test Results. 2016. https://www.fda.gov/downloads/MedicalDevices/Safety/EmergencySituations/UCM455349.pdf Available from:
- Forni D., Cagliani R., Mozzi A., Pozzoli U., Al-Daghri N., Clerici M. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J Virol. 2016;90(7):3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H. A previously undescribed coronavirus associated with respiratory disease in humans. PNAS. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi S., Fukuma A., Kurosu T., Watanabe S., Shimojima M., Shirato K. Characterization of novel monoclonal antibodies against the MERS-coronavirus spike protein and their application in species-independent antibody detection by competitive ELISA. J Virol Methods. 2018;251:22–29. doi: 10.1016/j.jviromet.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossner C., Danielson N., Gervelmeyer A., Berthe F., Faye B., Kaasik Aaslav K. Human-dromedary camel interactions and the risk of acquiring zoonotic Middle East respiratory syndrome coronavirus infection. Zoonoses Public Health. 2016;63(1):1–9. doi: 10.1111/zph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R. Family Coronaviridae. In: AMQ King, Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus taxonomy: classification and nomenclature of viruses. Academic Press; London (UK): 2012. pp. 806–820. (Ninth report of the International Committee on Taxonomy of Viruses). [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guery B., Poissy J., el Mansouf L., Séjourné C., Ettahar N., Lemaire X. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Deng Y., Chen H., Lan J., Wang W., Zou X. Systemic and mucosal immunity in mice elicited by a single immunization with human adenovirus type 5 or 41 vector-based vaccines carrying the spike protein of Middle East respiratory syndrome coronavirus. Immunology. 2015;145(4):476–484. doi: 10.1111/imm.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez C., Tejedor-Junco M., González M., Lattwein E., Renneker S. Presence of antibodies but no evidence for circulation of MERS-CoV in dromedaries on the Canary Islands, 2015. Euro Surveill. 2015;20(37):1560–7917. doi: 10.2807/1560-7917.ES.2015.20.37.30019. [DOI] [PubMed] [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H.S., Reusken C.B.E.M., Raj V.S., Galiano M., Myers R. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Rottier P.J., de Haan C.A. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4:3245–3269. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Hasley P.B., Albaum M.N., Li Y.H., Fuhrman C.R., Britton C.A., Marrie T.J. Do pulmonary radiographic findings at presentation predict mortality in patients with community-acquired pneumonia? Arch Intern Med. 1996;156(19):2206–2212. [PubMed] [Google Scholar]
- Hastings D.L., Tokars J.I., Abdel Aziz I.Z., Alkhaldi K.Z., Bensadek A.T., Alraddadi B.M. Outbreak of Middle East respiratory syndrome at tertiary care hospital, Jeddah, Saudi Arabia, 2014. Emerg Infect Dis. 2016;22(5):794–801. doi: 10.3201/eid2205.151797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Alnaeem A., Chu D.K., Perera R.A., Chan S.M., Almathen F. Longitudinal study of Middle East respiratory syndrome coronavirus infection in dromedary camel herds in Saudi Arabia, 2014-2015. Emerg Microbes Infect. 2017;6(6):1. doi: 10.1038/emi.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Chu D.K.W., Poon L.L.M., Perera R.A.P.M., Alhammadi M.A., Ng H. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis. 2014;20(7):1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Elmoslemany A., Al-Hizab F., Alnaeem A., Almathen F., Faye B. Dromedary camels and the transmission of Middle East respiratory syndrome coronavirus (MERS-CoV) Transbound Emerg Dis. 2017;64(2):344–353. doi: 10.1111/tbed.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Perera R.A., Wang P., Alhammadi M.A., Siu L.Y., Li M. Middle East respiratory syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill. 2013;18(50):20659. doi: 10.2807/1560-7917.es2013.18.50.20659. [DOI] [PubMed] [Google Scholar]
- Hijawi B., Abdallat M., Sayaydeh A., Alqasrawi S., Haddadin A., Jaarour N. Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation/Infections par le nouveau coronavirus en Jordanie, avril 2012: résultats épidémiologiques d'une étude rétrospective. East Mediterr Health J. 2013;19(Suppl. 1):S12–S18. [PubMed] [Google Scholar]
- Hocke A.C., Becher A., Knepper J., Peter A., Holland G., Tönnies M. Emerging human Middle East respiratory syndrome coronavirus causes widespread infection and alveolar damage in human lungs. Am J Respir Crit Care Med. 2013;188:882–886. doi: 10.1164/rccm.201305-0954LE. [DOI] [PubMed] [Google Scholar]
- Houser K.V., Gretebeck L., Ying T., Wang Y., Vogel L., Lamirande E.W. Prophylaxis with a Middle East respiratory syndrome coronavirus (MERS-CoV)-specific human monoclonal antibody protects rabbits from MERS-CoV infection. J Infect Dis. 2016;213(10):1557–1561. doi: 10.1093/infdis/jiw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., Azhar E.I., Kim Y., Memish Z.A., Oh M., Zumla A. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect Dis. 2018;18(8):e217–e227. doi: 10.1016/S1473-3099(18)30127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S.C., Lee N. Adjunctive therapies and immunomodulating agents for severe influenza. Influenza Other Respi Viruses. 2013;7(Suppl. 3):52–59. doi: 10.1111/irv.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.N., To K.K.W., Lee C., Lee K.L., Yan W.W., Chan K. Hyperimmune IV immunoglobulin treatment: a multicenter double-blind randomized controlled trial for patients with severe 2009 influenza a(H1N1) infection. Chest. 2013;144(2):464–473. doi: 10.1378/chest.12-2907. [DOI] [PubMed] [Google Scholar]
- Inovio GLS-5300 SynCon® immunotherapy targeting Middle East Respiratory Syndrome. 2016. http://www.inovio.com/products/infectious-disease-vaccines/mers/ Available from.
- Ithete N.L., Stoffberg S., Corman V.M., Cottontail V.M., Richards L.R., Schoeman M.C. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis. 2013;19(10):1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang N., Zuo T., Shi X., Poon K.M., Wu Y. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med. 2014;6(234) doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- Johnson R.F., Bagci U., Keith L., Tang X., Mollura D.J., Zeitlin L. 3B11-N, a monoclonal antibody against MERS-CoV, reduces lung pathology in rhesus monkeys following intratracheal inoculation of MERS-CoV Jordan-n3/2012. Virology. 2016;490:49–58. doi: 10.1016/j.virol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Kang K.W., Lee E., Seo D., Kim H., Kim H. Heterologous prime-boost vaccination with adenoviral vector and protein nanoparticles induces both Th1 and Th2 responses against Middle East respiratory syndrome coronavirus. Vaccine. 2018;36(24):3468–3476. doi: 10.1016/j.vaccine.2018.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M., Pringle K., Kumar A., Dearth S., Liu L., Lovchik J. Clinical and laboratory findings of the first imported case of Middle East respiratory syndrome coronavirus to the United States. Clin Infect Dis. 2014;59(11):1511–1518. doi: 10.1093/cid/ciu635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid M., Al Rabiah F., Khan B., Al Mobeireek A., Butt T.S., Al Mutairy E. Ribavirin and interferon-α2b as primary and preventive treatment for Middle East respiratory syndrome coronavirus: a preliminary report of two cases. Antivir Ther. 2015;20(1):87–91. doi: 10.3851/IMP2792. [DOI] [PubMed] [Google Scholar]
- Khalid M., Khan B., Rabiah F.A., Alismaili R., Saleemi S., Rehan-Khaliq A.M. Middle eastern respiratory syndrome coronavirus (MERS CoV): case reports from a tertiary care hospital in Saudi Arabia. Ann Saudi Med. 2014;34(5):396–400. doi: 10.5144/0256-4947.2014.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S. Lessons learned from new emerging infectious disease, Middle East respiratory syndrome coronavirus outbreak in Korea. Epidemiol Health. 2015;37:1–3. doi: 10.4178/epih/e2015051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.N., Ko Y.J., Seong M.W., Kim J.S., Shin B.M., Sung H. Analytical and clinical validation of six commercial Middle East respiratory syndrome coronavirus RNA detection kits based on real-time reverse-transcription PCR. Ann Lab Med. 2016;36(5):450–456. doi: 10.3343/alm.2016.36.5.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U.J., Won E., Kee S., Jung S.I., Jang H.C. Combination therapy with lopinavir/ritonavir, ribavirin and interferon-α for Middle East respiratory syndrome. Antivir Ther. 2016;21(5):455–459. doi: 10.3851/IMP3002. [DOI] [PubMed] [Google Scholar]
- Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13(2):1–26. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korea Centers for Disease Control, and Prevention Middle East Respiratory Syndrome Coronavirus Outbreak in the Republic of Korea, 2015. Osong Public Health Res Perspect. 2015;6(4):269–278. doi: 10.1016/j.phrp.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenmayer M.C., Lülf-Averhoff A., Adam-Neumair S., Fux R., Sutter G., Volz A. Distribution and absence of generalized lesions in mice following single dose intramuscular inoculation of the vaccine candidate MVA-MERS-S. Biologicals. 2018;54:58–62. doi: 10.1016/j.biologicals.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Lau C.C.Y., Chan K., Li C.P., Chen H., Jin D.Y. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- Lee J., Ahn J.S., Yu B.S., Cho S.I., Kim M.J., Choi J.M. Evaluation of a real-time reverse transcription-PCR (RT-PCR) assay for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) in clinical samples from an outbreak in South Korea in 2015. J Clin Microbiol. 2017;55(8):2554–2555. doi: 10.1128/JCM.00667-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.S., Kuok D.I.T., Cheung M.C., Ng M.M.T., Ng K.C., Hui K.P.Y. Effect of interferon alpha and cyclosporine treatment separately and in combination on Middle East respiratory syndrome coronavirus (MERS-CoV) replication in a human in-vitro and ex-vivo culture model. Antiviral Res. 2018;155:89–96. doi: 10.1016/j.antiviral.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A. 2017;114(40):E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wan Y., Liu P., Zhao J., Lu G., Qi J. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015;25(11):1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljander A., Meyer B., Jores J., Müller M.A., Lattwein E., Njeru I. MERS-CoV antibodies in humans, Africa, 2013–2014. Emerg Infect Dis. 2016;22(6):1086–1089. doi: 10.3201/eid2206.160064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2018 annual review of the Blueprint list of priority diseases. World Health Organization (WHO); Geneva, Switzerland: 2018. List of Blueprint priority diseases.http://www.who.int/blueprint/priority-diseases/en/ [cited 2018 April 4]. Available from: [Google Scholar]
- Liu R., Wang J., Shao Y., Wang X., Zhang H., Shuai L. A recombinant VSV-vectored MERS-CoV vaccine induces neutralizing antibody and T cell responses in rhesus monkeys after single dose immunization. Antiviral Res. 2018;150:30–38. doi: 10.1016/j.antiviral.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363(9413):938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23(8):468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5 doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Whitaker B., Sakthivel S.K.K., Kamili S., Rose L.E., Lowe L. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014;52(1):67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Li Y., Wang L., Zhao G., Tao X., Tseng C.T. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: implication for designing novel mucosal MERS vaccines. Vaccine. 2014;32(18):2100–2108. doi: 10.1016/j.vaccine.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I.M., Arden K.E. MERS coronavirus: diagnostics, epidemiology and transmission. Virol J. 2015;12:222. doi: 10.1186/s12985-015-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailles A., Blanckaert K., Chaud P., van dW, Lina B., Caro V. First cases of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infections in France, investigations and implications for the prevention of human-to-human transmission, France, May 2013. Euro Surveill. 2013;18(24):20502–20505. [PubMed] [Google Scholar]
- Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.M., Lim W.S. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211(1):80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malczyk A.H., Kupke A., Prüfer S., Scheuplein V.A., Hutzler S., Kreuz D. A highly immunogenic and protective Middle East respiratory syndrome coronavirus vaccine based on a recombinant measles virus vaccine platform. J Virol. 2015;89(22):11654–11667. doi: 10.1128/JVI.01815-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A., El Masry K.M., Ravi M., Sayed F. Middle East respiratory syndrome coronavirus during pregnancy, Abu Dhabi, United Arab Emirates, 2013. Emerg Infect Dis. 2016;22(3):515–517. doi: 10.3201/eid2203.151049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Matsuyama R., Nishiura H., Kutsuna S., Hayakawa K., Ohmagari N. Clinical determinants of the severity of Middle East respiratory syndrome (MERS): a systematic review and meta-analysis. BMC Public Health. 2016;16(1):1203. doi: 10.1186/s12889-016-3881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.L., Coleman C.M., van dM, Snijder E.J., Frieman M.B. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Al-Tawfiq J., Assiri A., AlRabiah F.A., Al Hajjar S., Albarrak A. Middle East respiratory syndrome coronavirus disease in children. Pediatr Infect Dis J. 2014;33(9):904–906. doi: 10.1097/INF.0000000000000325. [DOI] [PubMed] [Google Scholar]
- Memish Z.A., Al-Tawfiq J., Makhdoom H.Q., Al-Rabeeah A., Assiri A., Alhakeem R.F. Screening for Middle East respiratory syndrome coronavirus infection in hospital patients and their healthcare worker and family contacts: a prospective descriptive study. Clin Microbiol Infect. 2014;20(5):469–474. doi: 10.1111/1469-0691.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Al-Tawfiq J., Makhdoom H.Q., Assiri A., Alhakeem R.F., Albarrak A. Respiratory tract samples, viral load, and genome fraction yield in patients with Middle East respiratory syndrome. J Infect Dis. 2014;210(10):1590–1594. doi: 10.1093/infdis/jiu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Cotten M., Meyer B., Watson S.J., Alsahafi A.J., Al Rabeeah A.A. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014;20(6):1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Mishra N., Olival K.J., Fagbo S.F., Kapoor V., Epstein J.H. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19(11):1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Zumla A.I., Al-Hakeem R., Al-Rabeeah A., Stephens G.M. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;368(26):2487–2494. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- Memish Z.A., Zumla A.I., Assiri A. Middle East respiratory syndrome coronavirus infections in health care workers. N Engl J Med. 2013;369(9):884–886. doi: 10.1056/NEJMc1308698. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.H., Leist S.R., Yount B.L. Middle East respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere. 2017;2(6):1–12. doi: 10.1128/mSphere.00346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Mitchell H.D., Cockrell A.S., Gralinski L.E., Yount B.L., Graham R.L. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio. 2017;8(4):665–678. doi: 10.1128/mBio.00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Schäfer A., Burnum-Johnson K., Mitchell H.D., Eisfeld A.J., Walters K.B. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc Natl Acad Sci U S A. 2018;115(5):E1012–E1021. doi: 10.1073/pnas.1706928115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Müller M.A., Corman V.M., Reusken C.B.E.M., Ritz D., Godeke G. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis. 2014;20(4):552–559. doi: 10.3201/eid2004.131746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz D.K., Lambertz A.M., McCray P.B. Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East respiratory syndrome. Am J Pathol. 2016;186(1):78–86. doi: 10.1016/j.ajpath.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middle East respiratory syndrome coronavirus (MERS-CoV) World Health Organization (WHO); Geneva, Switzerland: 2018. http://www.who.int/emergencies/mers-cov/en/ [cited 2018 March 23]. Available from: [Google Scholar]
- Middle East respiratory syndrome coronavirus (MERS-CoV) – Republic of Korea. Disease outbreak news. World Health Organization (WHO); Geneva: 2015. http://www.who.int/csr/don/25-october-2015-mers-korea/en/ October 2015. [cited 2018 April 4]. Available from. [Internet] [Google Scholar]
- Disease outbreak news. 17 August 2017. World Health Organization (WHO); Geneva, Switzerland: 2017. Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia.http://www.who.int/csr/don/17-august-2017-mers-saudi-arabia/en/ [cited 2018 April 4]. Available from: [Google Scholar]
- Disease outbreak news. 9 October 2017. World Health Organization (WHO); Geneva, Switzerland: 2017. Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia.http://www.who.int/csr/don/09-october-2017-mers-saudi-arabia/en/ [cited 2018 April 4]. Available from: [Google Scholar]
- Disease outbreak news. 26 January 2018. World Health Organization (WHO); Geneva, Switzerland: 2018. Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia.http://www.who.int/csr/don/26-january-2018-mers-saudi-arabia/en/ [cited 2018 April 4]. Available from: [Google Scholar]
- Middle East respiratory syndrome coronavirus (MERS-CoV) Fact sheet. World Health Organization (WHO); Geneva: 2017. http://www.who.int/mediacentre/factsheets/mers-cov/en/# May 2017. [cited 2018 April 19]. Available from. [Google Scholar]
- Middle East respiratory syndrome coronavirus (MERS-CoV). Fact sheet. World Health Organization (WHO); Geneva, Switzerland: 2018. http://www.who.int/mediacentre/factsheets/mers-cov/en/ [cited 2018 March 23]. Available from: [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. PNAS. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modjarrad K. Treatment strategies for Middle East respiratory syndrome coronavirus. J Virus Erad. 2016;2(1):1–4. doi: 10.1016/S2055-6640(20)30696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.A., Meyer B., Corman V.M., Al-Masri M., Turkestani A., Ritz D. Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infect Dis. 2015;15(5):559–564. doi: 10.1016/S1473-3099(15)70090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., Raj V.S., Muth D., Meyer B., Kallies S., Smits S.L. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio. 2012;3:515–519. doi: 10.1128/mBio.00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthumani K., Falzarano D., Reuschel E.L., Tingey C., Flingai S., Villarreal D.O. A synthetic consensus anti–spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med. 2015;7(301) doi: 10.1126/scitranslmed.aac7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Chamberlain P., Bowden F., Joseph J. Atlas of coronavirus replicase structure. Virus Res. 2014;194:49–66. doi: 10.1016/j.virusres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Endo A., Saitoh M., Kinoshita R., Kinoshita R., Ueno R. Identifying determinants of heterogeneous transmission dynamics of the Middle East respiratory syndrome (MERS) outbreak in the Republic of Korea, 2015: a retrospective epidemiological analysis. BMJ Open. 2016;6(2) doi: 10.1136/bmjopen-2015-009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Miyamatsu Y., Mizumoto K. Objective determination of end of MERS outbreak, South Korea, 2015. Emerg Infect Dis. 2016;22(1):146–148. doi: 10.3201/eid2201.151383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu P., Zhang S., Zhou P., Huang B., Deng Y., Qin K. Ultra-potent human neutralizing antibody repertoires against MERS-CoV from a recovered patient. J Infect Dis. 2018;218:1249–1260. doi: 10.1093/infdis/jiy311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever B.R. The evolution of antiviral defense systems. Cell Host Microbe. 2016;19(2):142–149. doi: 10.1016/j.chom.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Nyon M.P., Du L., Tseng C.K., Seid C.A., Pollet J., Naceanceno K.S. Engineering a stable CHO cell line for the expression of a MERS-coronavirus vaccine antigen. Vaccine. 2018;36(14):1853–1862. doi: 10.1016/j.vaccine.2018.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oboho I.K., Tomczyk S.M., Al-Asmari A.M., Banjar A.A., Al-Mugti H., Aloraini M.S. 2014 MERS-CoV outbreak in Jeddah--a link to health care facilities. N Engl J Med. 2015;372:846–854. doi: 10.1056/NEJMoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M., Choe P.G., Oh H.S., Park W.B., Lee S., Park J. Middle East respiratory syndrome coronavirus superspreading event involving 81 persons, Korea 2015. J Korean Med Sci. 2015;30(11):1701–1705. doi: 10.3346/jkms.2015.30.11.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma K., Haagmans B.L., Hatano R., Raj V.S., Mou H., Iwata S. Inhibition of Middle East respiratory syndrome coronavirus infection by anti-CD26 monoclonal antibody. J Virol. 2013;87(24):13892–13899. doi: 10.1128/JVI.02448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrani A.S., Saad M.M., Baig K., Bahloul A., Abdul-Matin M., Alaidaroos A.Y. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis. 2014;14(11):1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.Y., Lee E.J., Ryu Y.A., Kim Y., Kim H., Lee H. Epidemiological investigation of MERS-CoV spread in a single hospital in South Korea, may to June 2015. Euro Surveill. 2015;20(25):21169. doi: 10.2807/1560-7917.es2015.20.25.21169. [DOI] [PubMed] [Google Scholar]
- Park J., Jung S., Kim A. MERS transmission and risk factors: a systematic review. BMC Public Health. 2018;18(1):574. doi: 10.1186/s12889-018-5484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D.C., Iblan I., Alqasrawi S., Al Nsour M., Rha B., Tohme R.A. Stillbirth during infection with Middle East respiratory syndrome coronavirus. J Infect Dis. 2014;209(12):1870–1872. doi: 10.1093/infdis/jiu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck K.M., Burch C.L., Heise M.T., Baric R.S. Coronavirus host range expansion and Middle East respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Ann Rev Virol. 2015;2(1):95–117. doi: 10.1146/annurev-virology-100114-055029. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A., Wang P., Gomaa M.R., El-Shesheny R., Kandeil A., Bagato O. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18(36):20574. doi: 10.2807/1560-7917.es2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- Qian Z., Dominguez S.R., Holmes K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Sun S., Xiao H., Feng J., Guo Y., Tai W. Single-dose treatment with a humanized neutralizing antibody affords full protection of a human transgenic mouse model from lethal Middle East respiratory syndrome (MERS)-coronavirus infection. Antiviral Res. 2016;132:141–148. doi: 10.1016/j.antiviral.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan A.A., Alahmed S.H., Bazzi A.M., Alhani H.M. A review of candidate therapies for Middle East respiratory syndrome from a molecular perspective. J Med Microbiol. 2017;66(9):1261–1274. doi: 10.1099/jmm.0.000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouw H.H., Langereis M.A., Knaap R.C.M., Dalebout T.J., Canton J., Sola I. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog. 2016;12(10):1371–1398. doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. MERS-coronavirus molecular epidemiology and genetic analysis - origin and evolution. 2014. http://epidemic.bio.ed.ac.uk/coronavirus_analysis Available at.
- Reusken C.B., Ababneh M., Raj V.S., Meyer B., Eljarah A., Abutarbush S. Middle East respiratory syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.50.20662. [DOI] [PubMed] [Google Scholar]
- Reusken C.B., Chrispijn Schilp V., De Bruin E., Kohl R.H., Farag E.A., Haagmans B.L. MERS-CoV infection of alpaca in a region where MERS-CoV is endemic. Emerg Infect Dis. 2016;22(6):1129–1131. doi: 10.3201/eid2206.152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Messadi L., Feyisa A., Ularamu H., Godeke G.J., Danmarwa A. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg Infect Dis. 2014;20(8):1370–1374. doi: 10.3201/eid2008.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B.E.M., Haagmans B.L., Müller M.A., Gutierrez C., Godeke G., Meyer B. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13(10):859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J., Hong G., Ryu K.M. Clinical implications of 5 cases of Middle East respiratory syndrome coronavirus infection in a south Korean outbreak. Jpn J Infect Dis. 2016;69(5):361–366. doi: 10.7883/yoken.JJID.2015.445. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saad M., Omrani A.S., Baig K., Bahloul A., Elzein F., Matin M.A. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single-center experience in Saudi Arabia. Int J Infect Dis. 2014;29:301–306. doi: 10.1016/j.ijid.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir J.S.M., Lam T.T., Ahmed M.M.M., Li L., Shen Y., Abo-Aba S. C-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351(6268):81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- Saqib M., Sieberg A., Hussain M.H., Mansoor M.K., Zohaib A., Lattwein E. Serologic evidence for MERS-CoV infection in dromedary camels, Punjab, Pakistan, 2012–2015. Emerg Infect Dis. 2017;23(3):550–551. doi: 10.3201/eid2303.161285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuplein V.A., Seifried J., Malczyk A.H., Miller L., Höcker L., Vergara-Alert J. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J Virol. 2015;89(7):3859–3869. doi: 10.1128/JVI.03607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia . 4th. (KSA): Ministry of Health; Kingdom of Saudi Arabia: 2018. Scientific Advisory Board, Ministry of Health, Kingdom of Saudi Arabia. Infection prevention and control guidelines for the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infection. [Google Scholar]
- Seys L.J.M., Widagdo W., Verhamme F.M., Kleinjan A., Janssens W., Joos G.F. DPP4, the Middle East respiratory syndrome coronavirus receptor, is upregulated in lungs of smokers and chronic obstructive pulmonary disease patients. Clin Infect Dis. 2018;66(1):45–53. doi: 10.1093/cid/cix741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalhoub S., Farahat F., Al-Jiffri A., Simhairi R., Shamma O., Siddiqi N. IFN-α2a or IFN-β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother. 2015;70(7):2129–2132. doi: 10.1093/jac/dkv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherbini N., Iskandrani A., Kharaba A., Khalid G., Abduljawad M., Al-Jahdali H. Middle East respiratory syndrome coronavirus in Al-Madinah City, Saudi Arabia: demographic, clinical and survival data. J Epidemiol Glob Health. 2017;7(1):29–36. doi: 10.1016/j.jegh.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013;87(23):12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K., Yeung M.L., Kok K., Yuen K., Kew C., Lui P. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol. 2014;88(9):4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Song D., Ha G., Serhan W., Eltahir Y., Yusof M., Hashem F. Development and validation of a rapid immunochromatographic assay for detection of Middle East respiratory syndrome coronavirus antigen in dromedary camels. J Clin Microbiol. 2015;53(4):1178–1182. doi: 10.1128/JCM.03096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanakis N., Tsiodras S., Haagmans B.L., Raj V.S., Pontikis K., Koutsoukou A. Virological and serological analysis of a recent Middle East respiratory syndrome coronavirus infection case on a triple combination antiviral regimen. Int J Antimicrob Agents. 2014;44(6):528–532. doi: 10.1016/j.ijantimicag.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3(9):1525–1531. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer D.R., Dickey R., Carter W.A. Sensitivity of SARS/MERS CoV to interferons and other drugs based on achievable serum concentrations in humans. Infect Disord Drug Targets. 2014;14(1):37–43. doi: 10.2174/1871526514666140713152858. [DOI] [PubMed] [Google Scholar]
- Sun Y., Zhang H., Shi J., Zhang Z., Gong R. Identification of a novel inhibitor against Middle East respiratory syndrome coronavirus. Viruses. 2017;9(9):1–12. doi: 10.3390/v9090255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Wang Y., Fett C.A., Zhao G., Li F., Perlman S. Recombinant receptor-binding domains of multiple Middle East respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J Virol. 2017;91(1):e01651-16. doi: 10.1128/JVI.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Ma W., Bai P. A novel dynamic model describing the spread of the MERS-CoV and the expression of dipeptidyl peptidase 4. Comput Math Methods Med. 2017;2017:5285810. doi: 10.1155/2017/5285810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., Garron T., Agrawal A.S., Algaissi A., Peng B., Wakamiya M. Characterization and demonstration of the value of a lethal mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol. 2016;90(1):57–67. doi: 10.1128/JVI.02009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thabet F., Chehab M., Bafaqih H., Al Mohaimeed S. Middle East respiratory syndrome coronavirus in children. Saudi Med J. 2015;36(4):484–486. doi: 10.15537/smj.2015.4.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbrough J.M., Jha B.K., Yount B., Goldstein S.A., Li Y., Elliott R. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio. 2016;7(2):1–12. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical decision-making support for treatment of MERS-CoV patients. Public Health England/ISARIC; Oxford (UK): 2015. Treatment of MERS-CoV; information for clinicians.https://www.google.ie/?gws_rd=ssl#q=public+health+england+treatment+mers-cov September 2015. [cited 2018 June 06]. Available from. [Internet] [Google Scholar]
- Trivedi S., Miao C., Al-Abdallat M., Haddadin A., Alqasrawi S., Iblan I. Inclusion of MERS-spike protein ELISA in algorithm to determine serologic evidence of MERS-CoV infection. J Med Virol. 2018;90(2):367–371. doi: 10.1002/jmv.24948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz A., Kupke A., Song F., Jany S., Fux R., Shams-Eldin H. Protective efficacy of recombinant modified vaccinia virus Ankara delivering Middle East respiratory syndrome coronavirus spike glycoprotein. J Virol. 2015;89(16):8651–8656. doi: 10.1128/JVI.00614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16(3):328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO target product profiles for MERS-CoV vaccines. World Health Organization; Geneva: 2017. http://www.who.int/emergencies/mers-cov/en/ [cited 2018 June 13]. Available from. [Internet] [Google Scholar]
- Widagdo W., Raj V.S., Schipper D., Kolijn K., van Leenders G.J.L.H., Bosch B.J. Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. J Virol. 2016;90(9):4838–4842. doi: 10.1128/JVI.02994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58(8):4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin a or interferon-α treatment. J Gen Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.T., Antonio G.E., Hui D.S.C., Lee N., Yuen E.H.Y., Wu A. Severe acute respiratory syndrome: radiographic appearances and pattern of progression in 138 patients. Radiology. 2003;228(2):401–406. doi: 10.1148/radiol.2282030593. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) Collecting, preserving and shipping specimens for the diagnosis of avian influenza A(H5N1) virus infection. Guide for field operations. 2006. http://www.who.int/csr/resources/publications/surveillance/CDS_EPR_ARO_2006_1.pdf Available from:
- World Health Organization (WHO) Laboratory testing for Middle East respiratory syndrome coronavirus interim guidance (revised) 2018. http://apps.who.int/iris/bitstream/handle/10665/259952/WHO-MERS-LAB-15.1-Rev1-2018-eng.pdf?sequence=1 Available from:
- World Health Organization (WHO) Middle East respiratory syndrome coronavirus: case definition for reporting to WHO. Interim case definition as of 26 July 2017. http://www.who.int/csr/disease/coronavirus_infections/case_definition/en/ Available from:
- Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60(11):6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liu C., Du L., Jiang S., Shi Z., Baric R.S. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J Virol. 2015;89(17):9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4(12):951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C., Lu L. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol. 2014;88(14):7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younan M., Bornstein S., Gluecks I.V. MERS and the dromedary camel trade between Africa and the Middle East. Tropl Anim Health Prod. 2016;48(6):1277–1282. doi: 10.1007/s11250-016-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zhang S., Jiang L., Cui Y., Li D., Wang D. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci Rep. 2015;5:13133. doi: 10.1038/srep13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang N., Channappanavar R., Ma C., Wang L., Tang J., Garron T. Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against Middle East respiratory syndrome coronavirus. Cell Mol Immunol. 2016;13(2):180–190. doi: 10.1038/cmi.2015.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Jiang S., Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev Vaccines. 2014;13(6):761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Tang J., Lu L., Jiang S., Du L. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res. 2015;202:151–159. doi: 10.1016/j.virusres.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Shen L., Gu X. Evolutionary dynamics of MERS-CoV: potential recombination, positive selection and transmission. Sci Rep. 2016;6:25049. doi: 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Jiang Y., Qiu H., Gao T., Zeng Y., Guo Y. Multi-organ damage in human dipeptidyl peptidase 4 transgenic mice infected with Middle East respiratory syndrome-coronavirus. PLoS One. 2015;10(12):1–14. doi: 10.1371/journal.pone.0145561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci Immunol. 2017;2(14) doi: 10.1126/sciimmunol.aan5393. [pii: eaan5393] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wang Q.Y. Bioinformatics analysis on molecular mechanism of ribavirin and interferon-α in treating MERS-CoV. Chung-Hua Liu Hsing Ping Hsueh Tsa Chih. 2016;37(2):291–293. doi: 10.3760/cma.j.issn.0254-6450.2016.02.028. [DOI] [PubMed] [Google Scholar]
- Zhou J., Chu H., Chan J.F., Yuen K. Middle East respiratory syndrome coronavirus infection: virus-host cell interactions and implications on pathogenesis. Virol J. 2015;12:218. doi: 10.1186/s12985-015-0446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Chu H., Li C., Wong B.H., Cheng Z., Poon V.K. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. 2014;209(9):1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Zhao G., Chu H., Wang D., Yan H.H. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv. 2017;3(11):1–12. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Pan T., Zhang J., Li Q., Zhang X., Bai C. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J Biol Chem. 2016;291(17):9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Jr., Nunneley J.W. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A.M., Matrosovich M. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013;87(9):5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]