Abstract
Cytometry by time-of-flight (CyTOF) simultaneously measures multiple cellular proteins at the single-cell level and is used to assess inter- and intra-tumor heterogeneity. This approach may be used to investigate the variability of individual tumor responses to treatments. Herein, we stratified lung tumor subpopulations based on AXL signaling as a potential targeting strategy. Integrative transcriptome analyses were used to investigate how TP-0903, an AXL kinase inhibitor, influences redundant oncogenic pathways in metastatic lung cancer cells. CyTOF profiling revealed that AXL inhibition suppressed SMAD4/TGF-β signaling and induced JAK1-STAT3 signaling to compensate for the loss of AXL. Interestingly, high JAK1-STAT3 was associated with increased levels of AXL in treatment-naïve tumors. Tumors with high AXL, TGF-β and JAK1 signaling concomitantly displayed CD133-mediated cancer stemness and hybrid EMT features in advanced stage patients, suggesting greater potential for distant dissemination. Diffusion pseudotime analysis revealed cell-fate trajectories among four different categories that were linked to clinicopathologic features for each patient. Patient-derived organoids (PDOs) obtained from tumors with high AXL and JAK1 were sensitive to TP-0903 and ruxolitinib (JAK inhibitor) treatments supporting the CyTOF findings. This study shows that single-cell proteomic profiling of treatment-naïve lung tumors, coupled with ex vivo testing of PDOs, identifies continuous AXL, TGF-β and JAK1-STAT3 signal activation in select tumors that may be targeted by combined AXL-JAK1 inhibition.
Introduction
AXL, a member of Tyro3-AXL-Mer (TAM) receptor tyrosine kinases (RTKs), is a promising therapeutic target in lung cancer (1,2). Frequently overexpressed in metastatic tumors, AXL is associated with drug resistance and poor survival outcomes (3–7). The oncogenic action is achieved primarily through AXL dimerization or hetero-dimerization with other RTKs, which activates TAM kinases in a ligand-dependent or -independent manner for downstream oncogenic networks, promoting cancer stemness and epithelial-to-mesenchymal transition (EMT) (8,9). Upon acquiring an EMT phenotype, lung cancer cells show loss of cell-to-cell contacts and escape from primary sites into the circulation and lymphatic channels (10–14). These invasive cells then revert back to an epithelial state during tumor implantation on vital organs. It is also believed that hybrid EMT states of invasive cells contribute to immune evasion and distant colonization (10–14). Other major pathways known to regulate mesenchymal/epithelial plasticity for advanced tumor phenotypes include transforming growth factor β (TGF-β), epidermal growth factor, hepatocyte growth factor, and the WNT/β-catenin and NOTCH pathways (10,11,13,15,16). Elucidation of those complex pathways and their partnership with AXL is critically important for developing combination treatment strategies in lung cancer. TP-0903 is a small molecule inhibitor of AXL kinase and have 80% inhibition of other two TAM family currently being investigated in patients with refractory lung cancer and solid tumors (17,18). Despite the advance of AXL inhibitors in the clinic, little is known about resistance mechanisms of these treatments in lung cancer. We speculate that oncogenic signaling crosstalk and bypass mechanisms orchestrated by deregulated AXL in vitro is similarly observed in treatment-naïve tumors.
Knowledge of diverse tumor subpopulations during lung cancer progression is essential for understanding differential responses to AXL treatment. In this regard, cytometry by time-of-flight (CyTOF) is a single-cell detection technology that allows for measurement of 30–45 protein markers in diverse cell subpopulations of a tumor (19–21). This high-dimensional analysis has been described as a “single-cell atlas” of tumor ecosystem, which can link a tumor’s cellular landscape with its clinicopathologic features. For example, CyTOF is being used to profile the immune ecosystem in early-stage lung adenocarcinoma for optimal design of immunotherapies (22,23). In this way, CyTOF is becoming integrated in the drug screening process and can detect intracellular signaling perturbations to short-term drug exposure for prediction of long-term response (24–26). CyTOF also provides opportunities for studying cellular dynamic processes that can be modeled using a trajectory inference method, also called pseudotime analysis, to predict tumor cell progression and lineage branching (27).
In this study, we first conducted transcriptomic analysis of metastatic lung cancer cells to probe key pathways perturbed by TP-0903. The profiling revealed previously uncharacterized AXL-associated signaling pathways that contribute to diversified treatment responses of lung tumor subpopulations. From the in silico analysis, we designed a CyTOF panel of 21 antibodies to recognize AXL, SMAD4/TGF-β and JAK1-STAT3 signaling components, characteristics of cancer stemness and EMT. The CyTOF panel was used to assess intra- and inter-tumor heterogeneity and stratify tumor subpopulations based on their AXL expression and signaling networks as a potential targeting strategy. Computational modeling with pseudotime analysis further ordered tumor cells along a trajectory based on similarities in their CyTOF expression patterns and comparisons made based on clinicopathologic features of patients. We also determined the feasibility of using tumor CyTOF data to identify patient-derived organoids (PDOs) suitable for combined AXL-JAK1 targeting. We suggest this work as a step toward a broader strategy will ultimately account for tumor heterogeneity at the single-cell level to optimize combination treatments in lung cancer patients.
Materials and Methods
Patient samples
Fresh lung tumors were obtained from treatment naïve patients (n=11) with non-small cell lung cancer at the time of surgery (Supplementary Table S1). Peripheral blood mononuclear cells (PBMCs) were isolated from two blood samples of a patient before and after surgery (Detail in the supplementary method). The protocol was approved by the University of Texas Health Science Center Institution Review Board. All patients were enrolled at the University of Texas Health Science Center at San Antonio between October 2018 and July 2019. Written informed consent was obtained from all patients in compliance with the Declaration of Helsinki, Belmont Report, US Common Rule following the US Department of Health and Human Services and the FDA regulations and GCP (Good Clinical Practice) guidelines. No patients received any prior treatment, and the site from which specimens were obtained had not been previously treated with radiotherapy. For CyTOF assays, tumor samples were digested into single-cell suspensions as described (28).
Cell lines
A549 and H2009 cell lines were obtained from and authenticated by the American Type Culture Collection, and routinely maintained in RPMI-1640 medium supplemented 10% FBS, penicillin (100 units/mL) and streptomycin (100μg/mL) in aired with 5% CO2 at 37 °C. The absence of Mycoplasma contamination was validated using DAPI staining. These cells were treated with TP-0903 and/or ruxolitinib (SelleckChem) at appropriate doses over 72 hr. The CellTiter-Glo Luminescent Cell Viability assay was used to determine cell responsiveness. shRNA knockdown was performed in A549 cells by using lentiviral delivery of short-hairpin AXL or vehicle plasmid pLKO.1 puro in two biological repeats (Addgene; Supplementary Table S2) (29).
Patient-derived organoids (PDOs)
Tumor tissues were minced on ice into ≤1 mm3 small pieces. Tumor pieces (~20 μl in volume) were resuspended in 200 μl Matrigel and seeded into 24 well plates for 15 min until gel solidify, followed by culture in advanced DMEM/F12 medium supplemented with B27 and N2 (Thermo Fisher Scientific), 0.01% BSA (Roche), 100 units/mL penicillin-streptomycin (Thermo Fisher Scientific), and others (Supplementary Table S3) for 4–8 weeks to grow organoids (30). Organoids were digested into single-cell suspensions and treated with 1) TP-0903, 20 nmol/L; 2) ruxolitinib, 15 μmol/L; 3) TP-0903 plus ruxolitinib; and 4) DMSO control for 72 hr in 5 replicates per treatment with 200 cells per replicate. The CellTiter-Glo Luminescent Cell Viability assay was used to determine drug responsiveness.
Cytometry by time-of-flight (CyTOF)
Antibodies were conjugated in-house according to the manufacturer’s instructions or purchased in pre-conjugated forms from the supplier (Fluidigm; Supplementary Table S4). Single cells from cell lines, tumors, or PBMCs were harvested and stained with cisplatin and metal-conjugated surface antibodies sequentially for viability and surface staining. After fixation and permeabilization, cells were stained with metal-conjugated antibodies. The cells were then labeled with an iridium-containing DNA intercalator (191Ir+ or 193Ir+) for identification of cell events before analysis on a Helios mass cytometer. Signals were bead-normalized using EQ Four Element Calibration Beads.
Signals of samples were normalized using CyTOF software (Version 6.7.1014, Fluidigm). The generated files underwent signal cleanup and filtering for single cells using Cytobank (https://www.cytobank.org/). The gated Flow Cytometry Standard (FCS) file were downloaded for further analysis using Cytofkit. The PhenoGraph clustering algorithm in Cytofkit was implemented in R from the Bioconductor website (https://bioconductor.org/packages/cytofkit/). CyTOF data were clustered and visualized using t-distributed stochastic neighbor embedding (t-SNE) algorithm based on normalized expression levels (Z-score) of 21 markers (AXL, JAK1, pSTAT3, SMAD2, SMAD4, TGFBRII, OCT3/4, NANOG, CD133, CD44, ALDH1A1, SNAIL, TWIST, Vimentin, N-cadherin, Fibronectin, β-catenin, ZO-2, PECAM, EpCAM, and CK8/18) and plotted on a continuum of protein expression with phenotypically related cells clustered together (31,32). Violin plots and scatter plots were generated by R package ggplot2 based on Z-score from the results of Cytofkit. Epithelial and mesenchymal indices were calculated based on the average Z-score of epithelial and mesenchymal markers. Pseudotime analysis was performed with the destiny package in R using expression levels of oncogenic signaling markers from normalized CyTOF data from individual patients to calculate dimensionality of data (DC1 and DC2) and diffusion pseudotime (DPT) (33). Diffusion maps were plotted based on dimensionality of data and DPT using R package ggplot2.
Atomic force microscopy (AFM)
AFM was performed to determine response of mechanical properties of lung cancer cells to TP-0903 treatment (34). Briefly, live cells cultured in 60 mm dishes were imaged with a Nanoscope Catalyst AFM (Bruker) mounted on a Nikon Ti inverted epifluorescent microscope. The cells were treated with 40 nmol/L TP-0903 or DMSO (control) for 24 hr. To collect the nanomechanical phenotypes of single cells immersed in culture media, we captured 30 × 30 μm images with a resolution of 256 × 256 pixels using the Peak Force Quantitative Nanomechanical Mapping (QNM) AFM (Bruker). For imaging, SCANASYST-AIR probes were used with the nominal spring constant 0.4 N/m. Following the Sneddon model and the Sokolov’s rules (35), nanomechanical parameters were calculated with Nanoscope Analysis software v.1.7 using retrace images.
Statistical analysis
Statistical significance was determined in GraphPad Prism by using Student t test (unpaired 2-tailed) and Duncan multiple range test for comparing pre- and post-treated cell lines among groups.
Results
Compensatory activation of JAK1-STAT3 following anti-AXL treatment
AXL overexpression in primary lung tumors is a single negative predictor of survival outcomes and represents a potential drug target (Supplementary Fig. S1A–C) (5). Accordingly, we first tested the effects of AXL inhibition on the growth of two metastatic lung cancer cell lines A549 and H2009 (Supplementary Fig. S1D). Proliferation rates decreased with increasing concentrations of TP-0903 (AXL inhibitor) in both cell lines and with shAXL knockdown in A549 cells (Supplementary Fig. S1E and S1F). Growth inhibition was confirmed in an A549-derived mouse xenograft model (Supplementary Fig. S1G). To determine the effect of AXL inhibition on gene expression, RNA-seq was conducted in A549 cells treated with TP-0903 (40 nmol/L) or shAXL knockdown and vehicle control cells (Supplementary Fig. S2A and S2B). Pathway enrichment analysis of differentially expressed genes showed that TGF-β signaling axis was attenuated by AXL inhibition, but JAK1-STAT3 signaling was upregulated likely due to a bypass mechanism (Supplementary Fig. S2C–E). Transcriptomic alterations in cancer stemness and EMT programs were also observed in TP-0903-treated cells (Supplementary Fig. 2F and 2G). Capillary WES protein analysis confirmed the downstream influence of AXL on TGF-β signaling, but the TP-0903 treatment had minor effects on suppressing the oncoproteins well-known for AXL-associated pathways (Supplementary Fig. S3 and S4) (9).
Effective targeting of AXL and JAK1 in metastatic cancer cells
To further probe AXL and JAK1-STAT3 signaling in different tumor populations, we analyzed the CyTOF data to identify common cellular communities among both cell lines untreated and treated with TP-0903 (n=4) and untreated lung tumors (n=11) (36). A total of 21 antibodies for CyTOF were selected for subpopulation analysis: 1) oncogenic signaling components of AXL, JAK1-STAT3 and TGF-β; 2) markers for cancer stemness; and 3) EMT (Fig. 1A) (37,38). Markers for immune, stromal and endothelial cells were initially used to segregate non-epithelial components in lung tumors and PBMCs. Leukocyte common antigen (CD45)-negative epithelial cell subpopulations were manually gated based on the expression of CK8/18 and EpCAM (Fig. 1B). Second, tSNE was used to cluster single cells based on shared protein expression collectively to identify metaclusters common across all samples. A total of 92,798 CD45−/CK8+/18+/EPCAM+ single cells were categorized into 27 subpopulations (Fig. 1C). Diverse expression profiles of oncogenic signaling, stemness and EMT were observed among these subpopulations from all samples (Fig 1D–G). There was also extensive inter-patient variability (Fig.1 H–L; Supplementary Fig. S5–S14).
Figure 1.
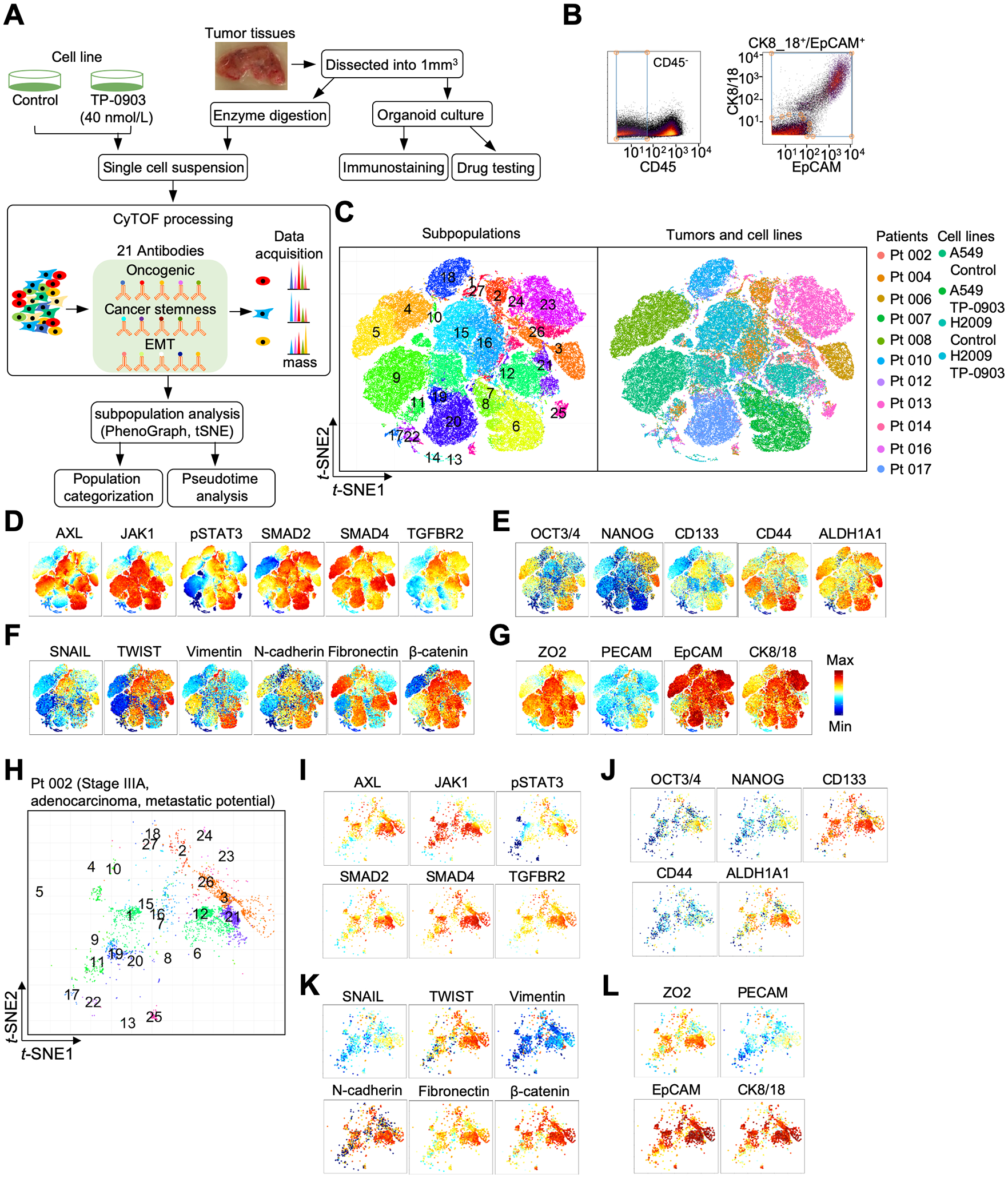
Cytometry by mass-of-flight (CyTOF) profiling of oncogenic signaling, cancer stemness, and epithelial-mesenchymal transition (EMT) in lung tumors and cell lines. A, A flow chart was drawn to illustrate the CyTOF and organoid processing. B, Tumor epithelial cells were identified based on CD45−/CK8+/18+/EpCAM+ profiles. C, t-distributed stochastic neighbor embedding (t-SNE) scatter plots stratified 27 subpopulations derived from different lung tumors and cell lines. D-G, t-SNE scatter plots were utilized to display expression levels of oncogenic signaling components and markers for cancer stemness and epithelial-mesenchymal transition (EMT). H, t-SNE scatter plot of subpopulations in a patient (Pt 002). See profiles of other patients in Supplementary Fig S5–S14. I-L, t-SNE scatter plots showed expression levels of oncogenic signaling components, markers for cancer stemness and EMT in Pt 002.
In general, subpopulations from cell lines displayed less variability than lung tumors based on CyTOF profiling of the aforementioned markers (Fig. 2A). In untreated A549 cells, there was one dominant subpopulation (#9) with high AXL expression. Following TP-0903 treatment at 40 nmol/L, three new subpopulations emerged in A549 cells with #6 and 7 displaying high levels of AXL and #8 exhibiting attenuated AXL (Fig. 2B, left panel). High JAK1-STAT3 signaling activities were observed in these subpopulations, supporting the original RNA-seq findings that JAK1-STAT3 might serve as a bypass mechanism leading to drug resistance (Supplementary Fig. S2E). Specifically, phosphorylated STAT3 (pSTAT3) levels were dramatically increased in A549-treated cells while JAK1 stably maintained high activities even in the presence of TP-0903 (Fig. 2B). Consistent with the capillary WES protein analysis (Supplementary Fig. S3 and S4), this treatment suppressed SMAD4 in the three main subpopulations of A549 cells (#6–8) (Fig. 2B). The upregulation of SMAD2 might be promoted via increasing pSTAT3 (Fig. 2B) (39). The second cell line H2009 was less responsive to AXL inhibition based on CyTOF data, confirming prior observations by capillary WES (Supplementary Fig. S3 and S4). In this cell line, subpopulations #12a and 12b displaying high AXL levels were observed prior to the TP-0903 treatment. Two main tumor subpopulations (#15 and 16) emerged following the treatment and had amplified JAK1-pSTAT3 expression, implicating a drug resistant phenotype. Compared to JAK1 signaling, AXL and SMAD4 had lower expression suggesting drug influence on these signaling pathways (Fig. 2C). Taken together, this CyTOF analysis of metastatic cell lines identified signaling components of JAK1-STAT3 that can be either extrinsically induced by AXL inhibition or intrinsically present as a bypass mechanism for cell survival and invasion.
Figure 2.

Single-cell profiling was performed using lung cancer cells treated with TP-0903 by cytometry by mass-of-flight (CyTOF). A, t-distributed stochastic neighbor embedding (t-SNE) scatter plots of subpopulations in A549 and H2009 cells treated with and without 40 nmol/L TP-0903. B-C, t-SNE scatter plots displaying expression levels of oncogenic signaling components in TP-0903-treated and untreated lung cancer cells. D, The bar graph of cell viability at 72 hr in TP-0903 and/or ruxolitinib treated A549 and H2009 cells (Duncan multiple range test; ***, P < 0.001). See the detailed description of treatment protocols in the Materials and Methods section.
The above in vitro study indicated that a single-target drug treatment is not effective in repressing lung cancer progression. To verify JAK1 as a potential bypass mechanism of AXL inhibition, short-term (72 hr) testing of TP-0903 and/or ruxolitinib (JAK inhibitor) was pursued in A549 and H2009 cells (Fig. 2D). Compared with the H2009 line, A549 cells were again more sensitive to TP-0903 treatments at 20, 30 and 40 nmol/L. However, the cell killing effect became more apparent in both cell lines when ruxolitinib (15 and 20 μmol/L) was additionally included in the treatment (P < 0.001). To confirm the finding of CyTOF and drug combination effect, western blots revealed upregulation of oncogenic signaling markers (JAK, STAT3, pSTAT3 and pAKT), increasing cancer stemness (CD133 and ALDH1A1), and upregulation of epithelial (EpCAM and CK8/18) and mesenchymal (Vimentin and N-cadherin) markers in TP-0903-treated A549 cells (Supplementary Fig. S15). The level of pSTAT3 and pAKT was additionally reduced in treated H2009 cells. Combination treatment with TP-0903 (20 nmol/L) and ruxolitinib (15 μmol/L) greatly attenuated JAK1, pSTAT3, pAKT, CD133, Vimentin, and EpCAM compared with single agent TP-0903 in both cell lines (Supplementary Fig. S15). Together, this result suggests that the combined therapy may be effective in suppressing lung cancer cells with activated AXL and JAK1-STAT3 and supports the CyTOF and RNA-seq findings.
Increased JAK1-STAT3 and TGF-β in AXL-overexpressing cell subpopulations
To explore intra-tumor and inter-patient heterogeneity of AXL-related oncogenic signaling activities in lung tumors, we classified the aforementioned 27 subpopulations into four categories (i.e., I, II, III, and IV) on the basis of 1) AXL expression levels, 2) JAK (JAK1 and pSTAT3) and TGF-β (SMAD2, SMAD4, and TGFBRII) signaling components, and 3) subpopulation sizes (Fig. 3A and 3B). Violin plot analysis further supported this subpopulation categorization: I) low expression of AXL, JAK1 and TGF-β signaling components; II) intermediate expression of AXL and high expression of JAK1 and TGF-β signaling components; III) high expression of AXL and TGF-β and intermediate expression of JAK1 signaling components; and IV) High expression of all five signaling components, including AXL (Fig. 3C). Collectively, cell lines demonstrated less heterogeneity than lung tumors. The majority (57–98%) of subpopulations in cell lines assigned to Category IV exhibited concomitant upregulation of AXL, JAK1, and TGF-β signaling (Fig. 3A, 3B, and 3D). As redundant mechanisms, these signaling components had already existed in some subpopulations or could be induced through in vitro inhibition of AXL. The remaining subpopulations were assigned to Category I-III with intermediate signaling activities.
Figure 3.

Four categories among different subpopulations of lung cancer cell lines and lung tumors ordered by AXL expression levels. A, Subpopulations were aligned according to increasing AXL levels (violin plots). Expression heat maps of JAK1, pSTAT3, SMAD2, SAMD4 and TGFBR2 of each subpopulation were arranged accordingly. B, Sizes of each subpopulation in cell lines and lung tumors were indicated. C, Violin plots were employed to illustrate the six signaling components in cell lines and lung tumors. D, Percentage of four categories in patients and cell lines.
Compared to cell lines, lung tumor subpopulations were more diverse, spanning all four categories (Fig. 3A and 3B). For example, tumor subpopulations of patient (Pt) 008 and 010 belonged to Category I and II (Fig. 3C and 3D). Pt 004, 014 and 017 had predominant Category II subpopulations (Fig. 3C and 3D). Pt 006 had 67% tumor cells in Category III (Fig. 3C and 3D). Subpopulations of Pt 002, 007, 009, 012, and 016 were preferentially assigned to Category IV (Fig. 3C and 3D). This inter-patient variability spanning all four categories underscores the need for tailored treatments based on a tumor’s predominant phenotype. Category II and IV subpopulations cells were present in every patient to varying degrees, suggesting pre-existing and redundant signaling pathways in treatment-naïve lung tumors (Fig. 3D). Furthermore, 464 circulating tumor cells (CTCs) derived from PBMC of Pt 006 belonged exclusively to Category IV, confirming a greater potential of these cells to disseminate to vital organs of the patient through the blood circulation (Supplementary Fig. S16).
Increased cancer stemness and hybrid EMT in AXL-overexpressing cell subpopulations
AXL and JAK1 signaling are well-established in cancer stemness regulation (8,40). Therefore, we included cancer stemness markers OCT3/4, NANOG, CD133, CD44 and ALDH1A1 in our CyTOF analysis. Generally speaking, the highest expression of cancer stemness markers was observed in Category III/IV subpopulations of cell lines and lung tumors (Fig. 4A and 4B). Furthermore, TP-0903 treatment preferentially gave rise to subpopulations with elevated CD133, a self-renewal regulator for metastasis and therapeutic resistance (Fig. 4C) (37,41). Moreover, higher levels of CD133 relative to other markers were frequently observed in Category IV subpopulations and aggressive stages of lung cancer, suggesting their innate resistance to TP-0903 and other treatments (Fig. 4B and 4D). Generally speaking, high expression levels of cancer stemness markers were observed in advanced stage patients. In only two cases, early-stage lung tumors of Pt 007 and Pt 016 with mixed histologies demonstrated high stemness markers, suggesting a more aggressive phenotype (Supplementary Table S1 and Fig 4D).
Figure 4.

Features of cancer stemness in cancer cell lines and lung tumors. A, Expression heat maps of OCT3/4, NANOG, CD133, CD44 and ALDH1A1 of each subpopulation were aligned at an increasing AXL level in individual subpopulations. B, Violin plots were employed to highlight the five cancer stemness markers in four categories of cell lines and lung tumors. C, Expression of five cancer stemness markers in cell lines before and after 40 nmol/L TP-0903 treatment was compared in violin plots (Student t test; ***, P < 0.001). D, Expression of five cancer stemness markers in early- and advanced- stage patients shown as violin plots.
Increased CD133 expression is a classic signature marker of EMT (37,41). For this reason, we conducted CyTOF analysis of 10 EMT markers (SNAIL, TWIST, Vimentin, N-cadherin, Fibronectin, β-catenin, ZO2, PECAM, EPCAM, and CK8/18) across 27 cell subpopulations. The levels of mesenchymal markers corresponded with high AXL levels while epithelial markers were more dominant in Categories II and IV (Fig. 5A). Based on epithelial (E) and mesenchymal (M) index values, Category IV subpopulations displayed the highest EMT hybrid states (Fig. 5B). Furthermore, TP-0903 treatment engendered higher E and M index values of these subpopulations, allowing greater mesenchymal/epithelial plasticity for metastasis (Fig. 5C) (11). To confirm this hybrid state, we applied AFM to probe biophysical properties - stiffness, deformation, and adhesion in TP-0903-treated and untreated cells (Fig. 5D–F). Stiffness is expressed in units of pressure as the Young’s modulus, whereas deformation is presented in units of length and assesses the depth of cell indentation at a selected point by a preset force (35,42,43). Adhesion is measured in units of force (Newtons) and quantifies a cell’s ability to stick to another cell or to base membranes (34,44). Overall, TP-0903-treated cells became more epithelial-like with increased stiffness and adhesion and attenuated deformity, relative to untreated cells (Fig. 5F). A549 cells responded to TP-0903 treatment with a 3-fold increase in stiffness, decreased deformation (25%) and increased adhesion (50%). The response of H2009 cells was moderate with only 61% increase of stiffness and 35% increase of adhesion noted (Fig. 5F). In general, early-stage tumors demonstrated lower E and M index values while advanced-stage tumors displayed higher E and M index values (Fig. 5G and 5H). However, tumors from early-stage patients, Pt 007 and Pt 016, showed high E and M index values, suggesting more aggressive phenotypes (Fig. 5G and 5H). Our finding implicates that the acquisition of a hybrid EMT phenotype allows invasive cells to simultaneously retain epithelial and mesenchymal traits for distant metastasis (45).
Figure 5.
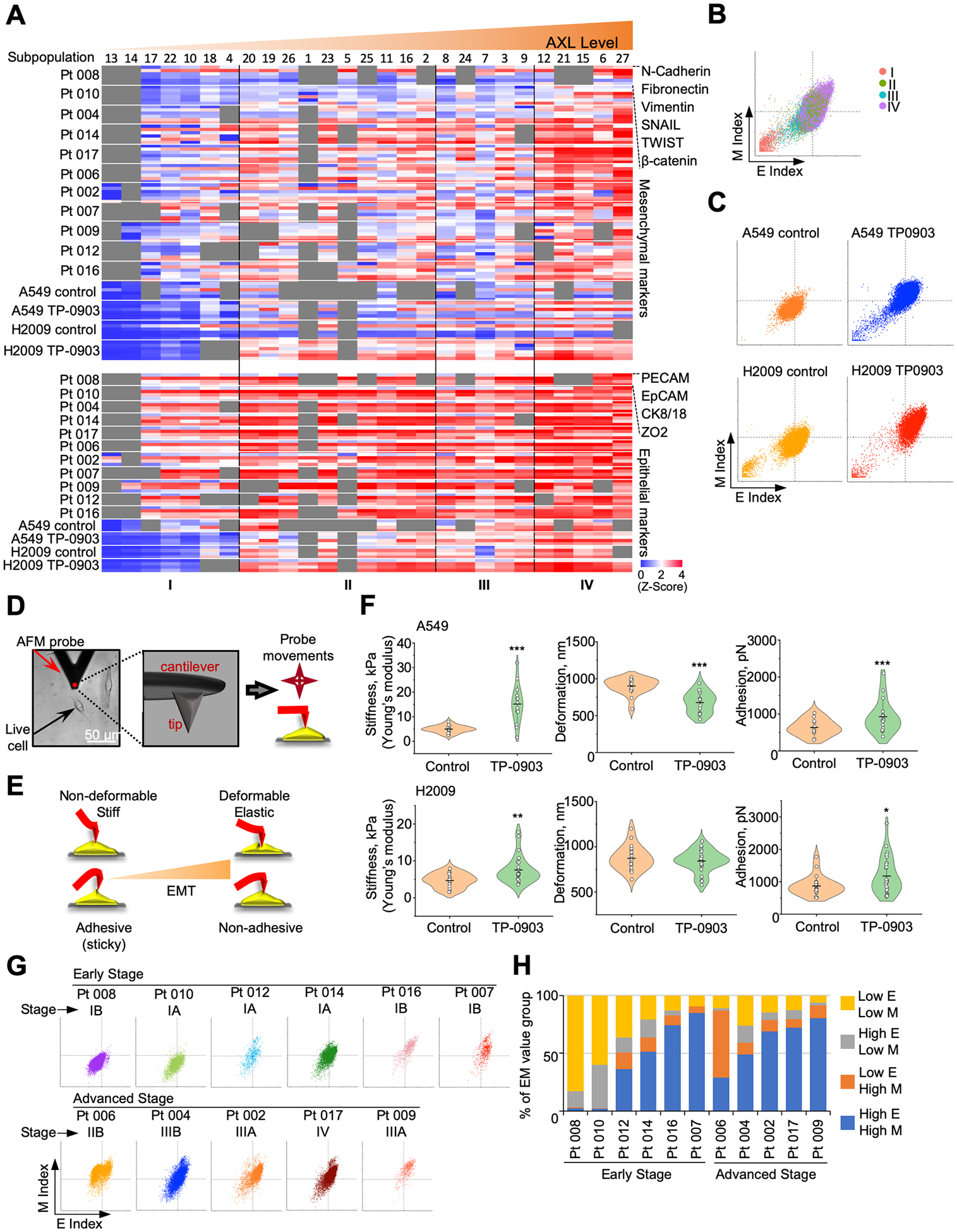
Profiles of epithelial-mesenchymal transition (EMT) in lung cancer cell lines and lung tumors. A, Expression heat maps of mesenchymal (M) and epithelial (E) markers of each subpopulation were aligned in order of increasing AXL levels accordingly. B-C, E and M index values in each subpopulation category of A549 and H2009 cells treated with and without TP-0903 were compared by scatter plots. D, A bright field image of H2009 cells probed with atomic force microscopy (AFM) is shown. A black triangle represents an AFM cantilever equipped with a scanning tip perpendicularly positioned (red dot). The 3D rendering of an AFM probe showed probe tip location. E, A schematic representation of AFM image formation is illustrated. F, Biophysical profiles (i.e., stiffness, deformation, and adhesion) were compared in A549 and H2009 cells with and without 40 nmol/L TP-0903 treatment. Each symbol represents a single-cell data point. Long vertical lines represent the mean and short vertical lines represent ±SD. (Student’s T-test; *, P < 0.05; **, P < 0.01; ***, P < 0.001) G, Scatter plots were plotted for E and M index values in each subpopulation category among patients’ cells. H, Percentages of different E/M groups were compared among early- and advanced-stage patients.
Diverse progression and regression patterns in lung tumors
Pseudotime analysis was performed to model cellular transition states among the four categories. Developmental trajectories of the 11 lung tumors were reminiscent of linear or punctuated models of evolution (Fig. 6 and Supplementary Table S1) (46). Lung tumors from six patients displayed a conventional trajectory, transitioning seamlessly from Category I to IV. Tumor specimen of Pt 008, for example, had early Stage IB invasive adenocarcinoma with papillary features and cell fate shifted from Category I to II, displaying the least invasive phenotype (Fig. 6A). Pt 010, on the other hand, transitioned from Category I to IV suggesting a more invasive phenotype. Interestingly, this patient had Stage IA moderately differentiated adenocarcinoma with additional micropapillary and acinar features on histopathologic examination. These features are often associated with stromal invasion and poorer outcomes than invasive adenocarcinoma without these features (Fig. 6A) (47). Analogously, Pt 014 had early Stage IA invasive adenocarcinoma (acinar predominant) with cell fates transitioning abruptly from Category II to IV (omitting Category I) (Fig. 6A). Both Pt 002 (metastatic paratracheal lymph node) and Pt 009 (moderate differentiated carcinoma with a mixed histology of lepidic, solid and glandular patterns) had aggressive Stage IIIA adenocarcinoma with highest metastatic potential and tumor-cell fates leading with Category II and culminating to Category IV subpopulations (Fig. 6A). Tumor subpopulations from these patients likely came from a common origin and progressively diverged into more advanced categories.
Figure 6.

Pseudotime analysis and organoid testing of lung tumors. For patients’ clinicopathological information, see Supplementary Table S1. A, Diffusion maps of linear model. B, Diffusion maps of punctuated model. C, Diffusion maps of punctuated regression model. D, Flow chart of a short-term drug treatment process in patient-derived organoids (PDOs). E, Bright view images of organoid morphology (Scale bar = 500 μm). F, Examples of Immunofluorescence images of DAPI (blue), CD45 (red), pan-cytokeratin (green), and EpCAM (purple) in PDOs (Scale bar = 40 μm). G, Bar graph of cell viability at 72 hr in 20 nmol/L TP-0903 and/or 15 μmol/L ruxolitinib treated PDOs (Duncan multiple range test; *, P < 0.05; **, P < 0.01; ***, P < 0.001). Doses were selected based on in vitro testing of lung cancer cell lines (see Fig. 2D).
Pt 004 and 016 revealed tumor cell fates that transitioned to high risk Category IV, but unlike the others, the intermediate stages reverted from III→II then jumped to IV (Fig. 6A). This dichotomy can be partially explained by their distinct histopathologic findings. Pt 004 had moderately differentiated Stage IIIB adenosquamous lung cancer; the two synchronous tumor components might explain the abrupt transition from low to high metastatic potential. Even more striking was the fact that this patient had a separate tumor nodule of invasive carcinoma in the same right upper lung lobe, indicating a higher metastatic potential than other patients in this category. By contrast, Pt 016 had early Stage IB invasive adenocarcinoma with a papillary predominant growth pattern and focal stromal invasion. This less aggressive histologic pattern may account for this instability of abrupt transition from Category II to IV through III/II intermediate stage.
The pseudotime analysis of the remaining lung tumors lacked intermediate stages and cell fates evolved nonlinearly in short bursts. Pt 007 had Stage IB adenocarcinoma with acinar predominant histology, which could explain the punctuated tumor model (Fig. 6B). Acinar adenocarcinomas have intermediate prognosis and notoriously display stromal invasion (bundles of broken elastic fibers) with desmoplastic tumor stroma and asymmetrical glands (47). Pt 012 had early Stage IA lung adenocarcinoma with acinar predominance and micropapillary features that may explain the branched tumor patterns (high-risk Category III→IV and Category II→IV progression) (Fig. 6B). Micropapillary-predominant adenocarcinoma has the poorest survival outcomes compared with acinar-predominant tumor. This tumor type is often associated with advanced lymph node staging (47). Lymph node involvement by tumor could not be assessed for Pt 012 who underwent a limited wedge resection.
The punctuated regression models with tumor subpopulations transitioning from a high-risk to lower-risk category were unique to Pt 006, 009 and 017 (Fig. 6C). Pt 006 presented with Stage IIB poorly differentiated adenocarcinoma with subpopulations assigned high-risk Category III/IV (Fig. 6C) and CTCs belonging to Category IV (Supplementary Fig. S16). Strikingly, pseudotime analysis of tumor specimen of Pt 006 exposed diverse clonal lineages: 1) tumor progression from Category III to IV; 2) tumor regression from category III to II; and 3) stasis (Category III) (Fig. 6C). Pt 009 had advanced stage IIIB moderately differentiated, invasive adenocarcinoma with a 2.1 cm tumor with mixed histology (lepidic, solid and glandular patterns), pleural and lymphovascular invasion and lymph node involvement (3 out of 13). Lepidic-predominant adenocarcinomas invade with a predominant lepidic growth pattern and have a favorable prognosis, while solid predominant adenocarcinoma presents with tumor necrosis, invasion of lymphovascular spaces and visceral pleura, and have a poor prognosis. Tumor specimen 009 revealed multiple clonal lineages indicative of tumor progression (Category III→II→IV and III→IV) and tumor regression (Category III→II), which can be explained by advanced disease stage and mixed histology (Fig. 6C). Pt 017 presented with Stage IV invasive adenocarcinoma (well to moderately differentiated). Tumor specimen of this patient originated from pleural metastasis, and pseudotime analysis represents a punctuated model consisting of spontaneous regression with tumor cell subpopulations transitioning to lower risk category (Category II→I) and higher risk categories (Category II→IV) (Fig. 6C). Fitting into this punctuated model, cell subpopulations for all these tumors might be pre-programmed in earlier stage to become metastatic or resistant to therapy (41).
Optimized targeting of AXL and JAK1 recapitulated in patient-derived organoids
PDOs are three-dimensional cultures of cancer and related cells that can be established from tumor specimens for drug testing (Fig. 6D–F). Short-term treatments of PDOs were pursued to examine the overall effect of AXL and/or JAK inhibitors on tumor cell subpopulations of Category I through IV. We hypothesized that tumors expressing moderate to high AXL and JAK-related proteins (Category III and IV) are most responsive to these therapies, whereas tumors belonging to Category I (lowest AXL and JAK1-STAT3 expression) may not respond. Based on the aforementioned in vitro testing (see Fig. 2D), the doses of TP-0903 (20 nmol/L) and ruxolitinib (15 μmol/L) were chosen for PDO testing. In a short-term drug treatment design (Fig. 6G), PDOs of Pt 008 and 010 with Category I/II tumor cell subpopulations did not respond robustly to either TP-0903 (20 nmol/L) or ruxolitinib (15 μmol/L) (Fig. 6G; Supplementary Fig. S8 and S10). In contrast, PDO of Pt 016 had 59% tumor subpopulations that belonged to Category IV (high expression of AXL and JAK1-STAT3 signaling components) responded robustly to 15 μmol/L ruxolitinib alone, but the synergy with 20 nmol/L TP-0903 was less apparent at 72 hr after combined treatment (Fig. 6G; Supplementary Fig. S13). PDOs of Pt 014 and 017 belonged to Category II (moderate levels of AXL and JAK1-STAT3 expression) and each responded to TP-0903 or ruxolitinib treatment alone with 10–20% reduction in cell viability (Fig. 6G; Supplementary Fig. S12 and S14). These preliminary results suggest that CyTOF profiling of lung tumors can provide predictive information for optimal testing of anti-AXL and -JAK1 agents in corresponding organoids, which will support the personalization of treatment for lung cancer patients.
Discussion
Small molecule inhibitors of AXL, like TP-0903, have entered clinical trials (48). However, the successful development of these drugs will depend on predictive markers for patient stratification. In this regard, CyTOF offers valuable knowledge of single-cell alterations of intracellular and surface markers in response to drug treatments, providing a powerful tool for rational design of AXL targeting strategies (49). To identify potential predictive markers, we started with the transcriptomic analysis of lung cancer cells treated with AXL inhibitor (TP-0903 which revealed AXL-TGF-β crosstalk, as well as upregulation of JAK1-STAT3 signaling as a bypass mechanism. With this in mind, we designed a CyTOF panel of 21 markers for AXL-related pathways, cancer stemness and EMT markers as a drug targeting strategy. This single-cell proteomic analysis revealed that tumor subpopulations with increasing AXL activities also intrinsically express higher levels of TGF-β and JAK1 signaling components, suggesting progression towards higher grade malignancies with enhanced cancer stemness and hybrid EMT features (45). TP-0903 treatment induced hybrid EMT and changed the nanomechanical properties of LAC cells. It is well-established that AFM can characterize the biophysical properties of cancer cells and corresponds to tumor cell invasion and EMT progression (50,51). Both pharmacologic and genetic targeting of AXL increased stiffness of lung cancer cells. Accordingly, it was found that stress fiber formation was stimulated following AXL knockdown (52). This finding was further supported by the presence of Category IV subpopulations with the highest AXL/TGF-β/JAK1 expressions. As expected, we found that CTCs of Pt 006 analyzed by CyTOF belonged solely to Category IV, representing tumor cells with the highest metastatic potential. The concordant upregulation of AXL, TGF-β and JAK1 suggests that these redundant networks promote tumor growth and metastatic spread.
Pseudotime analysis was conducted to predict tumor-cell fates based on subpopulation categorization. The three trajectories identified from this analysis resemble linear, punctuate and regression models (46). The assimilation of pseudotime results with patients’ histomorphologic patterns provides additional prognostic information based on the assumption that functional phenotypes reflect an underlying genotype. For example, punctate models seem to correlate with advanced tumor stages and/or high-risk histopathologic features (e.g., micropapillary, papillary, and acinar histologies). Another interesting discovery with pseudotime is tumor regression where tumor subpopulations could revert to low-risk phenotype. Spontaneous tumor regression occurs in primary tumors and metastatic niches and have been attributed to apoptosis, immunity and tumor microenvironment conditions (53). Tumor specimen 017, for example, originated from pleural metastasis and demonstrated a punctuated regression pattern with cell fate transitioning from Category II/III→I. Future analysis that links CyTOF to histopathology in a larger patient cohort may prove useful for adjuvant treatment strategies with curative intent.
The inter-patient variability and tumor subpopulations traversing all four categories underscore the need for tailored and personalized treatments based on a tumor’s predominant phenotype. Ex vivo drug testing of PDOs recapitulate tumor growth and can more accurately predict individual treatment responses to anti-AXL and -JAK combinations compared to other preclinical models (30). While patients with Category I tumor subpopulations might not benefit from these targeted agents due to low AXL and JAK activities, PDOs belonging to advanced categories exhibited sensitivity to single-agent inhibition, particularly with ruxolitinib (i.e., JAK inhibitor). Synergy of TP-0903 and ruxolitinib combination was not apparent in the present study. One explanation is that JAK inhibition can attenuate AXL signaling, and further exploration of crosstalk between AXL and JAK1 signaling is warranted. Another explanation could be the shorter drug exposure time (i.e., 72 hr) used to treat organoids. Most targeted therapies are given at lower doses when used in combination, which significantly reduces adverse events (54). For this reason, lower doses of ruxolitinib should be pursued in organoids, which may prove to be synergistic when combined with TP-0903. As we used organoids derived from “curative-intent” surgical resection samples without parallel patient treatment, future prospective studies will be required to establish definitive correlations between organoid and patient. Additional profiling of tumor ecosystem will be our next step in determining how non-tumor cells (e.g., immune, stromal, and endothelial cells) support expansions of residual tumor subpopulations after drug treatments. One major advantage to combining single-cell profiling of tumors and drug testing of corresponding organoids is that they can be realistically performed within a one-week time frame that is clinically relevant for making treatment decisions for cancer patients.
The CyTOF panel used in this study may prove useful in identifying lung cancer patients who should be considered for investigational agents, like TP-0903 or ruxolitinib. Similarly, the subpopulation categorization and trajectory modeling may predict which patients are at higher risk for tumor recurrence following their lung tumor resections. The proposed protein markers would be readily available for validation and can be implemented in clinical trials using liquid and/or tumor biopsies. If validated, these or similar markers will serve as surrogates for patient classification and can be used for treatment decisions.
Supplementary Material
Significance:
Single-cell proteomic profiling of clinical samples may facilitate the optimal selection of novel drug targets, interpretation of early-phase clinical trial data and development of predictive biomarkers valuable for patient stratification.
Acknowledgment
We are grateful for the contribution made from patients to this study. We also thank the staff of the BioAnalytics and Single-Cell Core (BASiC) and the Next-Generation Sequencing Shared Resource at the University of Texas Health Science Center at San Antonio for technical support in transcriptomic analyses. We thank Dr. Patrick Sung for critical comments of the manuscript. This work was supported by NIH grants U54CA217297, NCI Cancer Center Support Grant (P30CA54174) and the Ministry of Science and Technology of Taiwan, MOST 106-2917-I-029-001 (C.N.H.). The project described was also supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant KL2 TR001118 and KL2 TR002646.
Footnotes
Disclosure of Potential Conflicts of Interest: J.A.T. serves has no financial disclosures to report. M.W. and L.M. serve as directors of Biomarker Drug Discovery for Tolero Pharmaceuticals. S.W. is principal scientist at Tolero Pharmaceuticals. D.J.B. is the CEO of Tolero Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Zhang G, Wang M, Zhao H, Cui W. Function of Axl receptor tyrosine kinase in non-small cell lung cancer. Oncol Lett 2018;15:2726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linger RM, Keating AK, Earp HS, Graham DK. Taking aim at Mer and Axl receptor tyrosine kinases as novel therapeutic targets in solid tumors. Expert Opin Ther Targets 2010;14:1073–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae SY, Hong JY, Lee HJ, Park HJ, Lee SK. Targeting the degradation of AXL receptor tyrosine kinase to overcome resistance in gefitinib-resistant non-small cell lung cancer. Oncotarget 2015;6:10146–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa M, Sonobe M, Nakayama E, Kobayashi M, Kikuchi R, Kitamura J, et al. Higher expression of receptor tyrosine kinase Axl, and differential expression of its ligand, Gas6, predict poor survival in lung adenocarcinoma patients. Ann Surg Oncol 2013;20 Suppl 3:S467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F, Li J, Jang C, Wang J, Xiong J. The role of Axl in drug resistance and epithelial-to-mesenchymal transition of non-small cell lung carcinoma. Int J Clin Exp Pathol 2014;7:6653–61. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gay CM, Balaji K, Byers LA. Giving AXL the axe: targeting AXL in human malignancy. Br J Cancer 2017;116:415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scaltriti M, Elkabets M, Baselga J. Molecular Pathways: AXL, a membrane receptor mediator of resistance to therapy. Clin Cancer Res 2016;22:1313–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer 2018;18:128–34. [DOI] [PubMed] [Google Scholar]
- 11.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev 2016;35:645–54. [DOI] [PubMed] [Google Scholar]
- 12.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol 2015;25:675–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francart ME, Lambert J, Vanwynsberghe AM, Thompson EW, Bourcy M, Polette M, et al. Epithelial-mesenchymal plasticity and circulating tumor cells: Travel companions to metastases. Dev Dyn 2018;247:432–50. [DOI] [PubMed] [Google Scholar]
- 14.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer 2010;10:415–24. [DOI] [PubMed] [Google Scholar]
- 16.Espinoza I, Miele L. Deadly crosstalk: Notch signaling at the intersection of EMT and cancer stem cells. Cancer Lett 2013;341:41–5. [DOI] [PubMed] [Google Scholar]
- 17.Pharmaceuticals T A phase 1a/1b, first-in-human, open-label, dose-escalation, safety, pharmacokinetic, and pharmacodynamic study of oral TP-0903 administered daily for 21 days to patients with advanced solid tumors. (CTMS# 16–0092). 2018:1–108. [Google Scholar]
- 18.Mollard A, Warner SL, Call LT, Wade ML, Bearss JJ, Verma A, et al. Design, synthesis and biological evaluation of a series of novel Axl kinase inhibitors. ACS Med Chem Lett 2011;2:907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnaswamy S, Spitzer MH, Mingueneau M, Bendall SC, Litvin O, Stone E, et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science 2014;346:1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol 2012;30:858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anchang B, Davis KL, Fienberg HG, Williamson BD, Bendall SC, Karacosta LG, et al. DRUG-NEM: Optimizing drug combinations using single-cell perturbation response to account for intratumoral heterogeneity. Proc Natl Acad Sci U S A 2018;115:E4294–E303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, et al. A Single-Cell Atlas of the tumor and immune ecosystem of human breast cancer. Cell 2019;177:1330–45 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, et al. An immune atlas of clear cell renal cell carcinoma. Cell 2017;169:736–49 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niepel M, Hafner M, Pace EA, Chung M, Chai DH, Zhou L, et al. Profiles of basal and stimulated receptor signaling networks predict drug response in breast cancer lines. Sci Signal 2013;6:ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo LH, Bougen-Zhukov NM, Tan WC. Early spatiotemporal-specific changes in intermediate signals are predictive of cytotoxic sensitivity to TNFalpha and co-treatments. Sci Rep 2017;7:43541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider A, Klingmuller U, Schilling M. Short-term information processing, long-term responses: Insights by mathematical modeling of signal transduction. Early activation dynamics of key signaling mediators can be predictive for cell fate decisions. Bioessays 2012;34:542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods 2016;13:845–8. [DOI] [PubMed] [Google Scholar]
- 28.Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 2018;24:1277–89. [DOI] [PubMed] [Google Scholar]
- 29.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 2003;9:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018;359:920–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Der Maaten L Accelerating t-SNE using tree-based algorithms. J Mach Learn Res 2015;15:3221–45. [Google Scholar]
- 32.Van Der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579–625. [Google Scholar]
- 33.Angerer P, Haghverdi L, Buttner M, Theis FJ, Marr C, Buettner F. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 2016;32:1241–3. [DOI] [PubMed] [Google Scholar]
- 34.Hsu YT, Osmulski P, Wang Y, Huang YW, Liu L, Ruan J, et al. EpCAM-regulated transcription exerts influences on nanomechanical properties of endometrial cancer cells that promote epithelial-to-mesenchymal transition. Cancer Res 2016;76:6171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dokukin ME, Guz NV, Sokolov I. Quantitative study of the elastic modulus of loosely attached cells in AFM indentation experiments. Biophys J 2013;104:2123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 2015;162:184–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weidenfeld K, Barkan D. EMT and stemness in tumor dormancy and outgrowth: Are they intertwined processes? Front Oncol 2018;8:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polak KL, Chernosky NM, Smigiel JM, Tamagno I, Jackson MW. Balancing STAT activity as a therapeutic strategy. Cancers (Basel) 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen F JNK-induced apoptosis, compensatory growth, and cancer stem cells. Cancer Res 2012;72:379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liou GY. CD133 as a regulator of cancer metastasis through the cancer stem cells. Int J Biochem Cell Biol 2019;106:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokolov I, Dokukin ME. AFM indentation analysis of cells to study cell mechanics and pericellular coat. Methods Mol Biol 2018;1814:449–68. [DOI] [PubMed] [Google Scholar]
- 43.Sokolov I, Dokukin ME, Guz NV. Method for quantitative measurements of the elastic modulus of biological cells in AFM indentation experiments. Methods 2013;60:202–13. [DOI] [PubMed] [Google Scholar]
- 44.Cross SE, Jin YS, Tondre J, Wong R, Rao J, Gimzewski JK. AFM-based analysis of human metastatic cancer cells. Nanotechnology 2008;19:384003. [DOI] [PubMed] [Google Scholar]
- 45.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature 2018;556:463–8 [DOI] [PubMed] [Google Scholar]
- 46.Davis A, Gao R, Navin N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim Biophys Acta Rev Cancer 2017;1867:151–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell PA, Wainer Z, Wright GM, Daniels M, Conron M, Williams RA. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 2011;6:1496–504. [DOI] [PubMed] [Google Scholar]
- 48.Shen Y, Chen X, He J, Liao D, Zu X. Axl inhibitors as novel cancer therapeutic agents. Life Sci 2018;198:99–111. [DOI] [PubMed] [Google Scholar]
- 49.Bouzekri A, Esch A, Ornatsky O. Multidimensional profiling of drug-treated cells by Imaging Mass Cytometry. FEBS Open Bio 2019;9:1652–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zemla J, Danilkiewicz J, Orzechowska B, Pabijan J, Seweryn S, Lekka M. Atomic force microscopy as a tool for assessing the cellular elasticity and adhesiveness to identify cancer cells and tissues. Semin Cell Dev Biol 2018;73:115–24. [DOI] [PubMed] [Google Scholar]
- 51.Lekka M Discrimination Between Normal and Cancerous Cells Using AFM. Bionanoscience 2016;6:65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iida K, Sakai R, Yokoyama S, Kobayashi N, Togo S, Yoshikawa HY, et al. Cell softening in malignant progression of human lung cancer cells by activation of receptor tyrosine kinase AXL. Sci Rep 2017;7:17770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ricci SB, Cerchiari U. Spontaneous regression of malignant tumors: Importance of the immune system and other factors (Review). Oncol Lett 2010;1:941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sachs JR, Mayawala K, Gadamsetty S, Kang SP, de Alwis DP. Optimal dosing for targeted therapies in oncology: Drug development cases leading by example. Clin Cancer Res 2016;22:1318–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


