Abstract
By using a sensitive search program based on hidden Markov models (HMM), we identified 74 viruses carrying frameshift sites among 1500 fully sequenced virus genomes. These viruses are clustered in specific families or genera. Sequence analysis of the frameshift sites identified here, along with previously characterized sites, identified a strong bias toward the two nucleotides 5′ of the shifty heptamer signal. Functional analysis in the yeast Saccharomyces cerevisiae demonstrated that high frameshifting efficiency is correlated with the presence of a Ψ39 modification in the tRNA present in the E site of the ribosome at the time of frameshifting. These results demonstrate that an extended signal is involved in eukaryotic frameshifting and suggest additional interactions between tRNAs and the ribosome during decoding.
Introduction
The universal rules of genetic translation have long been known. However, the present situation is more complex. Firstly, the genetic code is not universal. In several organelles and protists, all genes are decoded by a variant code in which a stop codon reads as a sense codon. Moreover, the genetic code can be locally extended by alteration of standard rules in an individually specific manner for given mRNAs. Such extensions of the genetic code are termed reprogrammed genetic decoding or recoding events. Recoding is determined by particular sequences that force the ribosome to escape standard translation. In vivo, recoding is often in competition with standard decoding and permits the synthesis of an elongated polypeptide. Only a defined proportion of the ribosomes translating a given recoded mRNA actually use reprogrammed genetic decoding. Recoding extends the possibilities of increased diversity in gene expression or regulation.
During programmed −1 frameshifting, ribosomes switch to an alternative frame at a specific shift site, and classical triplet decoding follows. Efficient −1 frameshifting necessitates specific signals on the mRNA. Basically, the model proposed by Jacks and Varmus identified two components of frameshift signals: a slippery heptamer sequence, X XXY YYZ (the frame of the initiator AUG is indicated by a space), and a downstream structural element, often a pseudoknot or a stem-loop (Jacks et al., 1988). The secondary structure induces a ribosomal pause at the slippery site, and the heptamer sequence allows the slippage of ribosome bound A and P site tRNAs by one nucleotide in the 5′ direction. The pause increases the probability of ribosomal movement in the 5′ direction, although pausing seems necessary, but not sufficient, for stimulation (Kontos et al., 2001). More recently, a new sequence element—the spacer region located between the heptamer and the secondary structure—has been shown to modulate frameshift efficiency both in prokaryotes and eukaryotes Bekaert et al. 2003, Bertrand et al. 2002.
Most −1 frameshift events have been reported in viruses and transposons, where they serve in synthesis of replicase activities. This mode of expression allows both a very precise control of the Gag-Pol/Gag ratio and a means of incorporating the enzymes necessary for the replication cycle into the viral particle. Enhancement or reduction of the efficiency of this mechanism can influence virus viability Dinman and Wickner 1992, Shehu-Xhilaga et al. 2001. Very few cellular genes with −1 frameshifting are presently known: the dnaX gene of Escherichia coli (Tsuchihashi and Kornberg, 1990), the cdd gene of Bacillus subtilis (Mejlhede et al., 1999), and the Edr gene of mouse (Shigemoto et al., 2001). Even though the identification of novel genes regulated by −1 frameshifting constitutes one of the postgenomic challenges, there is presently no general method to do so (Namy et al., 2004). We recently began a computational approach to model frameshifting sites. The rationale was to observe more elements of the sequence in order to obtain a more precise description of characterized frameshift signals, identify pertinent characteristics, and develop appropriate algorithms. This approach previously allowed us to identify a strong bias in the spacer sequence of eukaryotic viral frameshift signals. This bias was shown to be functionally relevant for the frameshifting mechanism (Bekaert et al., 2003). However, only few frameshift signals have been functionally characterized, although several other sites are suspected to possess such signals from sequence analysis. The original goal of this study was to identify new −1 frameshift sites in viruses, so as to enrich our collection of sites in the modeling process. We first quantified frameshifting efficiency directed by 20 viruses carrying a putative or characterized frameshift site. From these results, we were able to develop a sensitive search based on HMM. This algorithm enabled us to detect −1 frameshift sites in 74 viruses among the available, fully sequenced viruses. Sequence alignment of these virus sites identified a strong composition bias just upstream of the shifty heptanucleotide site. We demonstrate that the sequence of this region actually affects frameshift efficiency in Saccharomyces cerevisiae. These results point to the possible involvement of pseudouridine at position 39 of the tRNA present in the E site of the host cell ribosome in modulating frameshifting efficiency. By using PUS3 yeast mutants lacking Ψ38/39 modifications, we show that frameshifting efficiency is modulated by the modification status of the E site tRNA. Overall, our results propose an extended model for −1 frameshift sites.
Results and Discussion
Characterization of Viral −1 Frameshift Sites
The RECODE database resource (http://recode.genetics.utah.edu, Baranov et al., 2001) describes 35 viruses suspected or demonstrated to carry a frameshift site. Few −1 frameshift signals are fully documented. The genomes of these 35 viruses are entirely sequenced, and only five sites are precisely characterized, including the structure of the stimulatory pseudoknot (BWYV, HIV-1, MMTV, PEMV-1, and SRV-1). For the remaining sites, 13 have been analyzed by extensive directed mutagenesis coupled with quantification of frameshifting efficiency, but the others are only partially characterized. Most of the sites are therefore putative; i.e., they carry the typical heptamer and secondary structure but have never been proven to be functional.
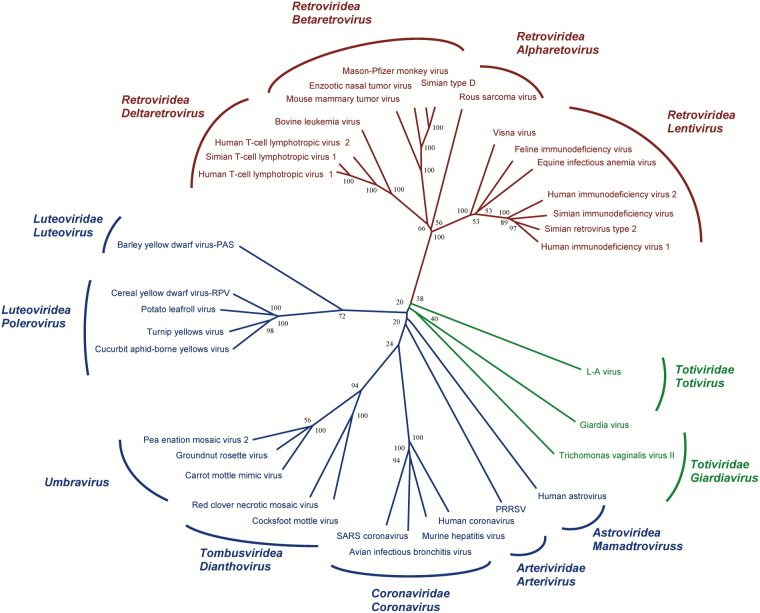
Initially, we functionally characterized a larger number of viruses containing a putative frameshifting site. To explore the widest viral diversity possible (order, family, and genus), we deduced a neighbor-joining tree from the 35 viruses, based on the multiple alignment of the polymerase protein sequence (Figure 1) . From this tree, we selected a subset of 20 viruses representative of the global viral diversity. To assay the frameshift competence of each putative site, we cloned the entire viral −1 frameshift region of the 20 representative viruses in a dual-reporter vector and estimated in vivo frameshifting efficiency in yeast (see Experimental Procedures). Frameshift sites from different eukaryotic species have been shown to function in yeast Bekaert et al. 2003, Stahl et al. 1995. The existence of a functional frameshift signal was demonstrated for all candidates (Table 1) except the Sugarcane yellow leaf virus (ScYLV). It is unlikely that the low level of expression of ScYLV is due to the use of a heterologous host cell, because ten frameshift sites from other plant viruses are functional in our assay. This site might be nonfunctional or carry polymorphic variations. The −1 frameshifting frequencies varied between 8% and 31%, compatible with those previously obtained with in vitro or in vivo assays (e.g., EAV 15%–20%, den Boon et al. [1991]; BWYV 5%, Kim et al. [1999]; HCV 20%–30%, Herold and Siddell [1993]; MMTV 20%, Chamorro et al. [1992]; and RSV 4%, Marczinke et al. [1998]). Although our assay does not provide exact frameshift rates in natural host cells (except for L-A and LB-C viruses, which are natural in S. cerevisiae), these results strongly suggest that most of the frameshift sites identified purely by sequence analysis are in fact functional.
Figure 1.
Unrooted Neighbor-Joining Phylogeny Based on the Amino Acid Sequence of the Pol Protein
Labels at branch tips represent taxa as provided by the Universal Virus Database, ICTVdB (http://phene.cpmc.columbia.edu; Büchen-Osmond, 2003). Numbers at branch nodes indicate the percentage of bootstrap support for that node based on 1000 replications. Color codes are according to the clade, with the following scheme: ssRNA-RT viruses (red), positive-sense ssRNA viruses (blue), and dsRNA viruses (green). Abbreviations: PRRSV, Porcine reproductive and respiratory syndrome virus.
Table 1.
Nucleic Acid Alignment of Heptameric Sequence
| Virus Acronym | FS-1 | Slippery Region | Genbank | ||
|---|---|---|---|---|---|
| BChV | 15.8% ± 2% | cacaucugcC | GGgAAAu | gGacuGaGcG | NC_002766 |
| BLV gag/pro | 8.1% ± 1% | cccucaaaUC | aaAAAAC | UaauAGaGGG | NC_001414 |
| BWYV | 12.0% ± 1% | ccaagagcUC | GGgAAAC | gGGagaGcGG | NC_004756 |
| BYDV | 12.2% ± 1% | uugacucugu | GGguuuu | UaGagGGGcu | NC_002160 |
| CABYV | 17.5% ± 1% | aauacgagUC | GGgAAAC | gGGcAGGcGG | NC_003688 |
| EIAV | 7.0% ± 1% | gaaguguucC | aaAAAAC | gGGagcaaGG | NC_001450 |
| FIV | 9.0% ± 1% | gaaagaauUC | GGgAAAC | UGGaAGGcGG | NC_001482 |
| HIV1 | 6.0% ± 1% | gacaggcuaa | uuuuuua | gGGaAGaucu | NC_001802 |
| IBV | 19.3% ± 1% | auaagaauUa | uuuAAAC | gGGuAcGGGG | NC_001451 |
| L-A | 10.0% ± 1% | guacucagca | GGguuua | gGaguGGuaG | NC_003745 |
| L-BC | 13.0% ± 2% | cugagaagUu | GGauuuu | cGuguaGcaG | NC_001641 |
| LDV | 13.1% ± 1% | aggcaucggC | uuuAAAC | UGcuAGccac | NC_002534 |
| MMTV gag/pro | 20.2% ± 2% | cugaaaauUC | aaAAAAC | UuGuAaaGGG | NC_001503 |
| PEMV1 | 31.0% ± 2% | ccagacgcUC | GGgAAAC | gGauuauucc | NC_003629 |
| PLRV | 19.0% ± 1% | caaacaagcC | GGgAAAu | gGGcAaGcGG | NC_001747 |
| PLRV-W | 17.8% ± 2% | caaacaagcC | uuuAAAu | gGGcgaGcGG | Y07496 |
| PRRSV | 15.7% ± 1% | aggagcagUg | uuuAAAC | UGcuAGccGc | NC_001961 |
| SARS | 10.3% ± 1% | caucaacgUu | uuuAAAC | gGGuuuGcGG | NC_004718 |
| ScYLV | 0.7% ± 0% | cuccagacca | GGgAAAu | gaGccaaGuG | NC_000874 |
| SRV1 gag/pro | 13.0% ± 2% | caccccauca | GGgAAAC | gGacuGaGGG | NC_001551 |
|
Pseudoconsensus |
xxxxxxxxUC |
GGAAAAC |
UGGxAGGGGG |
||
Nucleotides in agreement with the functional pseudoconsensus inferred from the HMM profile are in uppercase. Acronyms are as follows: BChV, Beet chlorosis virus; BLV, Bovine leukemia virus (gag/pro junction); BWYV, Beet western yellows virus; BYDV, Barley yellow dwarf virus; CABYV, Cucurbit aphid-borne yellows virus; EIAV, Equine infectious anemia virus; FIV, Feline immunodeficiency virus; HIV1, Human immunodeficiency virus 1; IBV, Avian infectious bronchitis virus; L-A, Saccharomyces cerevisiae virus L-A; L-BC, Saccharomyces cerevisiae virus L-BC; LDV, Lactate dehydrogenase-elevating virus; MMTV, Mouse mammary tumor virus (gag/pro junction); PEMV1, Pea enation mosaic virus 1; PLRV, Potato leafroll virus; PLRV-W, Potato leafroll virus, Germany strain (Wageningen); PRRSV, Porcine reproductive and respiratory syndrome virus; SARS, SARS coronavirus; ScYLV, Sugarcane yellow leaf virus; and SRV1, Simian type D virus 1 (gag/pro junction).
We then aligned the newly characterized sites with sites already identified. Strikingly, we observed an important bias not only at the slippery heptamer but also in the spacer region and just upstream of the heptamer. The upstream bias was never before observed, and its detailed analysis is presented below.
Sensitive Search of Viral Frameshift Sites
We established a HMM profile of efficient viral −1 frameshift signals with the alignment of the slippery regions from the 20 viruses that we had functionally characterized (see Experimental Procedures) and used it to search the GenBank viral genome database (release 02/10/2004). 285 motifs were identified and subsequently manually inspected to eliminate false positives. We checked (1) that the first nucleotide of the heptamer is in frame with the AUG of the upstream coding region, (2) for a protein motif associated to the upstream and downstream coding regions, and (3) for the presence of a potential secondary structure downstream of the heptamer. By this procedure, we identified 74 frameshift sites in viral genomes. Most false positives exhibited no secondary structure after the shifty site and were found in the large and highly complex herpesvirus, papillomavirus, and nucleopolyhedrovirus genomes. We consider this assessment to be accurate, because it depends not only on in silico methods but also on the biological assay of the HMM learning set. It is noteworthy that this method is very efficient, even though we did not take into account the stimulatory secondary structures when we defined the profile. RNA folding algorithms are time consuming and cannot be restrained to a defined window in the vicinity of the heptamer. Moreover, the theoretical evaluation of thermodynamic stability of secondary structures is not accurate for pseudoknots (Walter et al., 1994).
With the HMM profile based on only 20 sites, we were able to find all known frameshifting viruses and 39 that are new or uncharacterized (Table 2) . The list of the viruses with the position of their frameshift signals is in Supplemental Table S1 available online at http://www.molecule.org/cgi/content/full/17/1/61/DC1/. Ten putative frameshift sites were never previously annotated and are associated to an upstream and a downstream coding region. 12 frameshifting structures were already annotated as such in the RECODE database, and eight were only annotated in the sequence field of GenBank or in relation to a publication that did not mention any evidence of frameshift. For the remaining 44 sequences, a site was suspected, but it was not precisely localized between two coding regions. For those, we were able to propose a precise position for the frameshift event and in some cases, a more accurate annotation. For example, for the Ovine astrovirus (ssRNA+, Astroviridae family), a putative −1 frameshift event sequence was previously reported but without data on the position of the frameshift site (Jonassen et al., 1998). By adding the newly identified virus to the initial set, we established an enhanced profile HMM of viral −1 frameshift sites (see Experimental Procedures), available as Supplemental Data.
Table 2.
Detailed Distributions of the HMM Profile Results from GenBank Compared to the Initial Data Set (RECODE database)
| Status | GenBank (1500 virus) | RECODE (35 viruses) |
|---|---|---|
| New viruses | 10 | – |
| New annotations | 32 | 15 |
| Frameshift localized | 12 | 8 |
| Already annotated | 20 | 12 |
| 74 | 35 | |
Among 82 viral families, only seven are involved in −1 frameshifting: Astroviridae, Arteriviridae, Coronaviridae, Luteoviridae, Retroviridae, Tombusviridae, and Totiviridae. Within each family, only a few subfamilies/genera were capable of −1 frameshifting (see Supplemental Table S1 for details). However, in this latter case, all members of the genus submitted to HMM analyses appear capable of −1 frameshifting: they carry not only the HMM profile but also secondary structures as a canonical frameshift signal (Supplemental Table S1). For example, manual checking of the Poleroviruses found by using HMM successfully identifies a pseudoknot three to nine nucleotides downstream from the heptamer site. In the Totiviridae family (dsRNA virus), only the Totivirus genus, represented by the L-A and LB-C viruses (two yeast viruses), has a −1 frameshift signal. Moreover, all viruses from the Totivirus and the Giardiavirus genera appear to exhibit such sites. The only exception is the Ustilago maydis virus H1 (Totivirus genus): it shows a perfectly conserved canonical slippery sequence and a strong pseudoknot, despite an in-frame gag-pol gene. This could be due to either a sequencing/annotation error or to an unusual configuration where canonical decoding would be responsible for the synthesis of the Gag-Pol protein, whereas the frameshift would lead to the production of only the Gag domain, reminiscent of the control of the dnaX gene expression in E. coli (Tsuchihashi and Kornberg, 1990). Another ambiguous case concerns the Helminthosporium victiriae virus 190S, where previous experimental investigation of gene expression concluded that translation of the second ORF is initiated on its own internal AUG codon (Huang and Ghabrial, 1996). However, this does not exclude the possibility that both mechanisms are at play to express different polypeptides involved in polymerase activity, as observed in some bacterial transposons (Fayet et al., 1990).
Upstream Bias
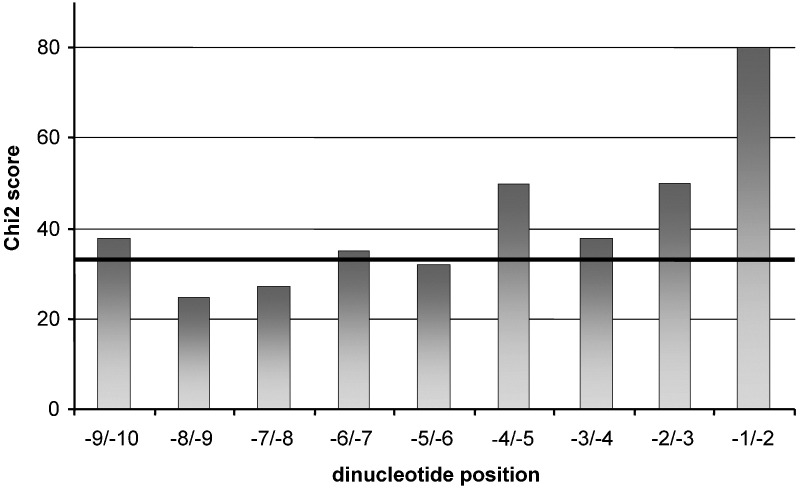
As mentioned above, the alignment of the initial set of 20 viruses −1 frameshift signal sequences revealed that the base composition around the slippery sequence follows a preferential use of nucleotides. Composition bias in the spacer has been previously reported for first nucleotides Bekaert et al. 2003, Bertrand et al. 2002; the second part of the bias has been reported in relation with the first stem composition bias (ten Dam et al., 1990). Bias in upstream sequences of the slippery regions was accurately detected in the larger scale data derived from the 74 virus sequences identified through an order 1 HMM search where the probability of a given nucleotide is dependent on the identity of the previous nucleotide. Accordingly, Figure 2 shows the bias of dinucleotide distribution. The χ2 score for the last dinucleotide position before the heptamer is 80 with 15 degrees of freedom, which makes it significant for a p value of 6.4 × 10−11. The −4/−5 position also seems biased, but this has not been analyzed further.
Figure 2.
Estimation of Dinucleotide Usage Bias
χ2 values were calculated at each position upstream of the slippery heptamer. Black line represents the threshold score for a p value of 1% with 15 degrees of freedom. Higher values are more indicative of a significant bias. The first dinucleotide is strongly significant (p value = 6.4 10−11).
To determine the role of this dinucleotide in frameshifting, we constructed dual-reporter vectors with the 16 possible sequences within the context of the wild-type (wt) frameshift signal of the Avian infectious bronchitis virus (IBV), because it has been extensively used as a model virus for −1 frameshifting studies Brierley et al. 1991, Brierley et al. 1992. Table 3 shows that a 3.3-fold variation was found between the frameshifting efficiencies directed by these 16 IBV variant sites. Compared to the wt sequence, the frameshifting level is significantly reduced (p value < 10−4) in ten of the mutants.
Table 3.
Effect of the Upstream Dinucleotide on −1 Frameshifting Efficiency
| Plasmids | Modified Sequence | tRNA (Anticodon loop) | Frameshift |
|---|---|---|---|
| pAC.5.AA | aau AAuuua aac | ACU GUU t6 AAΨ | 18.0% ± 2% |
| pAC.5.AC | aau ACuuua aac | Am5CU IGU t6 AAΨ | 22.0% ± 1% |
| pAC.5.UA | aau UAuuua aac | ACU GΨA i6 AAΨ | 19.3% ± 1% |
| pAC.5.UC | aau UCuuua aac | AΨU IGA i6 AAΨ | 22.1% ± 2% |
| pAC.5.UG | aau UGuuua aac | AΨU GCA i6 AAΨ | 19.0% ± 2% |
| pAC.5.UU | aau UUuuua aac | ACmU GmAA YAΨ | 21.0% ± 1% |
| pAC.5.AG | aau AGuuua aac | CCU GCU AAG | 9.3% ± 1% |
| pAC.5.AU | aau AUuuua aac | GCU IAU t6 AAC | 7.0% ± 1% |
| pAC.5.CA | aau CAuuua aac | GΨU GUG m1 GCC | 9.0% ± 1% |
| pAC.5.CC | aau CCuuua aac | CUU AGG GUG | 9.0% ± 1% |
| pAC.5.CG | aau CGuuua aac | GCU ICG AAC | 7.5% ± 1% |
| pAC.5.CU | aau CUuuua aac | GUC GAG GUC | 10.0% ± 2% |
| pAC.5.GA | aau GAuuua aac | CΨU GUC m1GCG | 9.4% ± 1% |
| pAC.5.GC | aau GCuuua aac | CUU IGC m1IΨG | 8.0% ± 1% |
| pAC.5.GG | aau GGuuua aac | GΨU GCC AΨC | 8.5% ± 1% |
| pAC.5.GU | aau GUuuua aac | CΨU IAC ACG | 6.7% ± 1% |
The FY strain was transformed with one of the plasmids harboring the test sequence as indicated (from 5′ to 3′). Frameshifting efficiencies were measured at 30°C, and the data are expressed as percentages. Codons including the dinucleotides are underlined and anticodon of tRNA anticodon loops are in bold (Lecointe, 2002); heptamers are in bold and dinucleotides in uppercase.
Role of Base 39 of tRNA
The dinucleotide situated 5′ of the heptamer corresponds to the first two nucleotides of the preceding codon; its impact can thus be interpreted as an effect either of the amino acid, the codon, or the decoding tRNA. Because it was previously shown that tRNA modification can affect recoding efficiency Lecointe et al. 2002, Licznar et al. 2003, we looked for a bias in modifications of tRNA involved in decoding high and low frameshifting constructs. A correlation between the presence of pseudouridine at position 39 (Ψ39) of the tRNA anticodon domain was observed (Table 3): all constructs that exhibited a high-frameshifting level use a cognate (or near-cognate) tRNA carrying the Ψ39 modification. Conversely, the sequences that do not involve a codon decoded by a tRNA with the Ψ39 modification direct low-frameshifting efficiency. This observation prompted us to investigate the effect of the mutation of the PUS3 gene, whose product is specifically responsible for the Ψ39 modification (Lecointe et al., 1998). If Ψ39 is actually involved, inactivation of PUS3 should result in a lower frameshift efficiency.
Two low- and two high-frameshift rate constructs were tested in modification mutants (Table 4) . With the low-frameshifting rate subset (frameshift efficiency lower than 10%), pus3Δ mutants show no significant effect (Table 4). In contrast, with the high-frameshifting rate subset (frameshift efficiency higher than 18%), which involves decoding by a Ψ39 modified tRNA, a reduced frameshifting frequency was observed in pus3Δ mutants. This frequency was similar to that directed by the low-frameshifting rate subset, indicating that most of the effect was reversed in the mutant. We verified that the effect is actually due to the modifying activity of Pus3p and not to a possible chaperone-like activity by using the pus3[D151A] mutant, which harbors a mutation in the active site of the PUS3 protein. In this mutant, the high-frameshifting constructs yield lower frameshifting efficiency, as in a pus3Δ mutant context (Table 4).
Table 4.
Effect of PUS3 Gene Deletion on −1 Frameshifting Efficiency
| Plasmids | Wt | pus3Δ | pus3Δ + pRS315 | pus3Δ + PUS3 | pus3Δ + pus3[D151A] |
|---|---|---|---|---|---|
| pAC.5.CG | 5.3% ± 1% | 5.9% ± 1% (1.1) | 5.8% ± 0% (1.1) | 5.5% ± 1% (1.0) | 6.0% ± 1% (1.1) |
| pAC.5.GA | 7.6% ± 1% | 8.0% ± 1% (1.1) | 8.1% ± 1% (1.1) | 7.2% ± 0% (0.9) | 7.3% ± 1% (1.0) |
| pAC.5.UA | 21.7% ± 1% | 12.8% ± 1% (0.6) | 11.5% ± 1% (0.5) | 19.3% ± 1% (0.9) | 11.8% ± 1% (0.5) |
| pAC.5.UC | 19.5% ± 1% | 10.3% ± 1% (0.5) | 10.2% ± 1% (0.5) | 18.7% ± 2% (1.0) | 12.7% ± 1% (0.7) |
Wild-type (wt) and pus3Δ mutants of 74-D694 strains were transformed with the test plasmids. The 74-D694 pus3Δ strain was also transformed with empty pRS315 or the same plasmid containing the PUS3 gene or the mutant pus3[D151A] gene, as indicated. −1 frameshifting efficiencies were measured at 30°C, and the data are expressed as percentages. Numbers in parentheses correspond to ratios of recoding efficiency in the wt strain over the recoding efficiency in the pus3Δ derivative strain. No significant difference can be expected by a Mann-Whitney statistical test, except between pAC.5.UA/UC in wt or pus3Δ + PUS3 compared to other transformed strains (p value < 0.005).
The effect of the dinucleotide upstream of the heptamer suggests that the three ribosomal site tRNAs are involved in the mechanism of −1 frameshifting. However, although the mechanism of frameshifting in eukaryotes is thought to involve mostly tandem slippage of the tRNAs occupying the A and P sites, single slippage at the P site has been reported to occur (Jacks et al., 1988). If this is the case in the experimental system used here, there is no tRNA in the A site at the time of slippage Baranov et al. 2004, Leger et al. 2004. To test the occurrence of single slippage, we used a mutant site in which the UUUAAAC heptamer was mutated to UUUAUAC. In this case, tandem slippage should be inefficient due to the presence of two mismatches after repairing of the A site tRNA in the −1 frame, but single slippage would not be affected. The frameshifting efficiency obtained with this construct was <0.1%, similar to the background level. This result demonstrates that in these experiments, frameshifting actually occurred through a tandem tRNA slippage mechanism. This implies that the three sites are involved in ribosomal frameshifting (see below).
The Ψ39 modification is conserved over the tree of life; its role on −1 frameshifting could thus be similar in a broad spectrum of organisms. This is consistent with the fact that the bias at the two positions upstream of the heptamer was deduced from a wide variety of viruses of different origins. However, each host cell, like the yeast strains used here, carries a specific tRNA pool that differs from one organism to another. This could explain the different dinucleotide usage observed between viruses; however, not enough sequence data are available to assess this point. In any case, the existence of a bias indicates an important role of tRNA modification on −1 frameshifting in eukaryotes. A role of tRNA modifications on +1 frameshifting has been previously described both in E. coli and in S. cerevisiae Bjork et al. 1989, Lecointe et al. 2002, Urbonavicius et al. 2001. For −1 frameshifting, a few examples have been reported in E. coli Brierley et al. 1997, Licznar et al. 2003, but not in eukaryotes. In these cases, the tRNAs involved were acting at the A or P site.
Overall, these results demonstrate that the effect of the upstream context of the heptamer is directed by the modification status of the tRNA decoding the −1 codon.
Conclusions
Viral Frameshifting Signals
It is striking that all members of a genus (or family, in some cases) use a frameshifting event to produce their Pol protein but that phylogenetic analyses of frameshift sequences give rise to patterns inconsistent with accepted trees (data not shown). Inconsistency of frameshifting patterns with accepted phylogenetic trees is not surprising taking into account the recombinant nature of many viruses; functional requirements probably account for both this complete conservation and the variability of the frameshifting site sequences. Indeed, in the Retroviridae family, the Alpharetrovirus genus is exceptional because some members exhibit frameshift signals but others do not. In fact, this genus is subdivided in two categories: replication-competent viruses, which possess the pol gene, and defective viruses, which do not. Logically, frameshift signals are found only in the latter category. It is even more interesting that despite their position among the Totiviridae, the Leishmaniavirus genus members do not carry −1 frameshift sites but, rather, use +1 frameshifting to express their polymerase domain. This suggests that strong biological constraints are at play in the selection of a recoding event in the life cycle of these viruses, possibly related to the incorporation of the polymerase as a fusion protein in the viral particle.
Role of E Site in Frameshifting
An interesting feature of the results presented here is the involvement of an extended nonanucleotide signal in ribosomal frameshifting. As demonstrated above, no single slippage is observed in the experimental system used here; this nonanucleotide-directed frameshifting thus involves classical tandem slippage where both A and P site tRNAs slip by one nucleotide upstream. This implies that the three ribosomal sites are involved in −1 frameshifting. Two hypotheses can be proposed to account for the role of the E site tRNA in frameshifting. Firstly, frameshifting might be enhanced by the absence of a tRNA in the E site. In this case, the Ψ39 modification would destabilize the tRNA:E site interaction. Secondly, Ψ39 might interfere directly or indirectly with the interaction of the P site tRNA with the mRNA, decreasing pairing stability.
The first hypothesis is supported by recent results in which premature release of the E site tRNA from the ribosome has been shown to be coupled with high-level +1 frameshifting at the prfB gene, encoding the prokaryotic termination factor RF2 (Marquez et al., 2004). Likewise, in eukaryotes, Ψ39 may induce an unusual E site conformation. If this is the case, one would predict that the Ψ39 modification induces a higher frequency of release of the tRNA from the E site. If E-tRNA normally helps prevent tRNA slippage in the P site, this could explain the different susceptibilities of a given heptamer to slippage. Probably the E-tRNA is released during the accommodation step of the A-tRNA and not during the preceding decoding reaction Nierhaus 1990, Noller et al. 2002. However, in the case of a −1 frameshift event, E-tRNA release at the decoding step would facilitate the slippery event of A and P site tRNAs, and this precisely might be the effect of Ψ39. Biochemical experiments will be required to clarify this point.
The second hypothesis is supported by structural data on the prokaryotic ribosome Ramakrishnan and Moore 2001, Yusupov et al. 2001 and inferred cryo-EM reconstruction of the yeast ribosome (Spahn et al., 2001) that strongly suggest that the E-tRNA interacts with several partners. The closest distance between the anticodon stem backbones of the P- and E-tRNAs is about 6 Å, which is closer than the distance separating the A- and P-tRNAs. The two tRNAs are not in direct contact but are linked by the 16S rRNA helices H24, H28, and H29, and loops 690 and 790, both of which they directly interact with through their anticodon loops (Yusupov et al., 2001). Another link between E and P sites is through the mRNA. A single possible contact was noted between the mRNA and E-tRNA in the crystal structure, but the latter was noncognate. Even this noncognate E site anticodon was close enough to the codon, such that cognate interaction would be structurally plausible; moreover, there is biochemical evidence for codon-anticodon specificity in the E site Lill and Wintermeyer 1987, Rheinberger et al. 1986. E site tRNA is thus sufficiently connected to the P site to suggest that it very likely plays a role in promoting the stability of P site codon-anticodon pairing. Ψ39 modification can be expected to improperly fill the E site during the slippage-prone state, probably resulting in an unstable P site codon-anticon interaction and enhanced −1 frameshifting. This is reminiscent of the role played by a particular context of a bacterial tmRNA resume codon. In this case, an unusual E site conformation destabilizes the P site codon-anticodon interaction and induces frameshifting (Trimble et al., 2004).
The results presented here demonstrate that the slippery component of −1 frameshift signals, at least in yeast, is more complex than previously anticipated. Compared to the initial model of Jacks et al. (1988), sequence elements of both the 3′ and 5′ heptamer elements are now shown to participate in frameshiting efficiency through interactions between tRNA, mRNA, and the ribosome. Similarly, downstream secondary structures can directly or indirectly influence frameshifting. A combinatorial use of upstream codons, heptamer sequences, downstream codons, and stimulatory secondary structures permit a given frameshifting efficiency for a given virus in a given host. Whether or not these different sequence elements act independently remains to be established.
Experimental Procedures
Polymerase Tree
A ClustalW 1.83 (Thompson et al., 1994) alignment of viral polymerase amino acid sequences retrieved from GenBank was used. It was employed to deduce a neighbor-joining tree with 1000 bootstrap replications (Saitou and Nei, 1987) by using Mega 2.1 package (Kumar et al., 2001), which provides a graphical representation repartition of selected viruses. Pairwise distances were calculated as mean observed substitutions per site. The unrooted tree is shown in Figure 1 and is color coded to mark each clade.
Profile Construction
Frameshift sites—the heptamers surrounded by ten nucleotides on both sides—from the selected viruses were used to construct and calibrate an HMM by using the HMMER package 2.3.2 (Eddy, 1998). Each sequence was aligned on the shifty heptamer and the HMM established. Sequences from viruses not selected as representative subset but reported as frameshifting viruses were used to validate our profile. All sites were found (data not shown).
Searches with Profile
With this HMM profile, searches against the publicly available viral genome database (GenBank, downloaded February 10th, 2004) were carried out. All searches against nucleotide databases were performed with the HMMER 2.3.2 package. As threshold, we assigned a minimal e value of 0.5. Subsequently, an enhanced HMM profile was established with the HMMER package 2.3.2. The frameshift sites found—the heptamers surrounded by ten nucleotides on either side—were used to construct and calibrate the enhanced HMM profile.
Bias
Dinucleotide biases were estimated by counting each dinucleotide of the 74 sequences and comparing the distribution with an equiprobability model. Because we compared different viruses from different hosts, we should use the lesser bias model where the frequency of each dinucleotide is 1/16. This estimation used a χ2 probability with 15 degrees of freedom.
Yeast Strains and Media
The S. cerevisiae strains used were FY1679-18B (Mat α his3-Δ200, trp1-Δ63, ura3-52, and leu2-Δ1), 74-D694 (Mat a, ade1-14, trp1-289, his3Δ200, leu2-3, 112, and ura3-52), and its derivative pus3Δ (Mat a, ade1-14, trp1-289, his3Δ200, leu2-3, 112, ura3-52, and pus3Δ::KAN). Strains were grown in minimal media (0.67% yeast nitrogen base, 2% glucose) supplemented with the appropriate amino acids to allow maintenance of the different plasmids under standard growth conditions. Yeast transformations were performed by the lithium acetate method (Ito et al., 1983).
Plasmids and Molecular Biology Methods
pAC99 derivatives were constructed by cloning the synthetic oligonucleotides of interest at the unique MscI site of pAC99 (Bidou et al., 2000). For viral frameshift sites, heptamer, spacer, and secondary structure surrounded by ten nucleotides on each side were inserted. For mutants of the upstream region of the heptamer, the IBV frameshift site was used and the wt sequence (UA) was changed to the 15 other possible sequences (see Supplemental Table S2). Plasmids containing the PUS3 gene or the mutant pus3[D151A] gene were from Lecointe et al. (2002). All constructs were verified by sequencing the region of interest.
Quantification of −1 Frameshifting Efficiency
Luciferase and β-galactosidase activities were assayed in the same crude extract as previously described (Stahl et al., 1995). The assays were carried out at least five times by using two independent transformants grown in the same conditions. The luciferase β-galactosidase ratio obtained with test constructs was normalized to the ratio obtained with the in-frame control and expresses frameshift efficiency.
Acknowledgements
We would like to thank Dominique Fourmy, Henri Grosjean, and Olivier Namy for helpful discussions and suggestions and François Lecointe for providing us with the pus3Δ stains and plasmids. We thank members of the Génétique Moléculaire de la Traduction laboratory and the “frameshift team” for numerous stimulating discussions. We are especially grateful to Anne-Lise Haenni for critically reading the manuscript. This work was supported by the Association pour la Recherche sur le Cancer (contract 4699).
Published: January 6, 2005
Supplementary Material
References
- Baranov P.V., Gurvich O.L., Fayet O., Prere M.F., Miller W.A., Gesteland R.F., Atkins J.F., Giddings M.C. Recode: a database of frameshifting, bypassing and codon redefinition utilized for gene expression. Nucleic Acids Res. 2001;29:264–267. doi: 10.1093/nar/29.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Gesteland R.F., Atkins J.F. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M., Bidou L., Denise A., Duchateau-Nguyen G., Forest J.P., Froidevaux C., Hatin I., Rousset J.P., Termier M. Towards a computational model for −1 eukaryotic frameshifting sites. Bioinformatics. 2003;19:327–335. doi: 10.1093/bioinformatics/btf868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand C., Prere M.F., Gesteland R.F., Atkins J.F., Fayet O. Influence of the stacking potential of the base 3′ of tandem shift codons on −1 ribosomal frameshifting used for gene expression. RNA. 2002;8:16–28. doi: 10.1017/s1355838202012086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidou L., Stahl G., Hatin I., Namy O., Rousset J.P., Farabaugh P.J. Nonsense-mediated decay mutants do not affect programmed −1 frameshifting. RNA. 2000;6:952–961. doi: 10.1017/s1355838200000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork G.R., Wikstrom P.M., Bystrom A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- Brierley I., Rolley N.J., Jenner A.J., Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Meredith M.R., Bloys A.J., Hagervall T.G. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J. Mol. Biol. 1997;270:360–373. doi: 10.1006/jmbi.1997.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchen-Osmond C. The universal virus database ICTVdB. Comput. Sci. Eng. 2003;5:16–25. [Google Scholar]
- Chamorro M., Parkin N., Varmus H.E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl. Acad. Sci. USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D., Wickner R.B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Fayet O., Ramond P., Polard P., Prere M.F., Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol. Microbiol. 1990;4:1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- Herold J., Siddell S.G. An `elaborated' pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 1993;21:5838–5842. doi: 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Ghabrial S.A. Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc. Natl. Acad. Sci. USA. 1996;93:12541–12546. doi: 10.1073/pnas.93.22.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen C.M., Jonassen T.O., Grinde B. A common RNA motif in the 3' end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J. Gen. Virol. 1998;79:715–718. doi: 10.1099/0022-1317-79-4-715. [DOI] [PubMed] [Google Scholar]
- Kim Y.G., Su L., Maas S., O'Neill A., Rich A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl. Acad. Sci. USA. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos H., Napthine S., Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lecointe F. Etude d'Enzymes de Modification de Nucléotides des ARNt et Leurs Fonctions dans le Métabolisme Cellulaire Chez Saccharomyces cerevisiae. Université Paris XI; Orsay, France: 2002. [Google Scholar]
- Lecointe F., Simos G., Sauer A., Hurt E.C., Motorin Y., Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J. Biol. Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- Lecointe F., Namy O., Hatin I., Simos G., Rousset J.P., Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 2002;277:30445–30453. doi: 10.1074/jbc.M203456200. [DOI] [PubMed] [Google Scholar]
- Leger M., Sidani S., Brakier-Gingras L. A reassessment of the response of the bacterial ribosome to the frameshift stimulatory signal of the human immunodeficiency virus type 1. RNA. 2004;10:1225–1235. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznar P., Mejlhede N., Prere M.F., Wills N., Gesteland R.F., Atkins J.F., Fayet O. Programmed translational −1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 2003;22:4770–4778. doi: 10.1093/emboj/cdg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Wintermeyer W. Destabilization of codon-anticodon interaction in the ribosomal exit site. J. Mol. Biol. 1987;196:137–148. doi: 10.1016/0022-2836(87)90516-x. [DOI] [PubMed] [Google Scholar]
- Marczinke B., Fisher R., Vidakovic M., Bloys A.J., Brierley I. Secondary structure and mutational analysis of the ribosomal frameshift signal of rous sarcoma virus. J. Mol. Biol. 1998;284:205–225. doi: 10.1006/jmbi.1998.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez V., Wilson D.N., Tate W.P., Triana-Alonso F., Nierhaus K.H. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Mejlhede N., Atkins J.F., Neuhard J. Ribosomal −1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J. Bacteriol. 1999;181:2930–2937. doi: 10.1128/jb.181.9.2930-2937.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Rousset J.P., Napthine S., Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- Nierhaus K.H. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- Noller H.F., Yusupov M.M., Yusupova G.Z., Baucom A., Cate J.H. Translocation of tRNA during protein synthesis. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V., Moore P.B. Atomic structures at last: the ribosome in 2000. Curr. Opin. Struct. Biol. 2001;11:144–154. doi: 10.1016/s0959-440x(00)00184-6. [DOI] [PubMed] [Google Scholar]
- Rheinberger H.J., Sternbach H., Nierhaus K.H. Codon-anticodon interaction at the ribosomal E site. J. Biol. Chem. 1986;261:9140–9143. [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shehu-Xhilaga M., Crowe S.M., Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto K., Brennan J., Walls E., Watson C.J., Stott D., Rigby P.W., Reith A.D. Identification and characterisation of a developmentally regulated mammalian gene that utilises −1 programmed ribosomal frameshifting. Nucleic Acids Res. 2001;29:4079–4088. doi: 10.1093/nar/29.19.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Stahl G., Bidou L., Rousset J.P., Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E.B., Pleij C.W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. Clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble M.J., Minnicus A., Williams K.P. tRNA slippage at the tmRNA resume codon. RNA. 2004;10:805–812. doi: 10.1261/rna.7010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z., Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. USA. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J., Qian Q., Durand J.M., Hagervall T.G., Bjork G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A.E., Turner D.H., Kim J., Lyttle M.H., Muller P., Mathews D.H., Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.