Abstract
Background: Surveillance of winter respiratory viral illness has been carried out for nearly 30 years using a clinical diagnosis by general practitioners as part of the Scottish Sentinel General Practice (SSGP) network. Contemparaneous laboratory diagnosis has not been available previously. Objectives: To assess the proportion of influenza-like illness (ILI) attributable to influenza, respiratory syncytial virus (RSV) and picornavirus infection during the winter season. To compare the influenza PCR data with serology of paired blood samples. Study design: Combined nose and throat swabs, from patients with ILI attending 15 general practices across Scotland, were submitted to the laboratory in virus PCR sample solution (VPSS). The extracted nucleic acid was tested using a multiplex reverse-transcription polymerase chain reaction (RT-PCR) assay. Serological analysis was performed on paired serum samples using complement fixation assays. The rate of influenza virus positivity was compared with reports of ILI obtained from the SSGP network. Results: Of 240 samples received at the laboratory, 132 (55%) were PCR positive for influenza A virus. There were nine (3.8%) picornavirus and three (1.2%) RSV PCR positives, two (0.8%) were dual influenza A/picornavirus infections. Ninety four (39.2%) were negative for all viruses tested. Results on paired sera from 89 patients showed a rising titre to influenza A in 48 of the 57 PCR positive samples (84.2%). One PCR negative patient displayed a significant rising titre to influenza A. Virological data paralleled the SSGP data but was available at least a week earlier. Conclusions: Influenza A infection was detected in the majority of patients with ILI; picornavirus infection was also shown to be an important cause of illness. PCR is a rapid and sensitive method for respiratory virus surveillance. Serology is slow, insensitive and difficult to interpret at low titres.
Abbreviations: SSGP, Scottish Sentinel General Practice; ILI, influenza-like illness; RT-PCR, reverse-transcription polymerase chain reaction; VPSS, viral PCR sample solution; CFT, complement fixation test
Keywords: Multiplex PCR, Respiratory viruses, Surveillance
1. Introduction
Influenza virus infection is an important cause of respiratory infection in the population over the winter season (Makela et al., 1998, Nicolson et al., 1997). All age groups are affected by influenza infection, but serious complications occur much more frequently in young children (<1-year-old) and the elderly (>65 years old), (Sczucs, 1999). In young children, influenza infection leads to significantly increased antibiotic prescription, outpatient visits and hospitalisation (Neuzil et al., 2000), while elderly nursing home patients with culture proven influenza A or B infection show significantly increased mortality (Drinka et al., 1999). Other at risk groups also suffer considerable morbidity and significant mortality (Neuzil et al., 1999, Nicholson, 1996).
The neuraminidase inhibitor class of anti-influenza drugs, i.e. zanamivir and oseltamivir (Fiddian, 2000), are most effective if administered early in infection, so modernisation of the monitoring of the community influenza outbreak is required. Rapid diagnosis of influenza will be required to enable early treatment. Reverse-transcription polymerase chain reaction (RT-PCR) is a more sensitive and specific method for the detection of influenza infection than virus isolation or direct immunofluorescence and can be used for rapid diagnosis (Magnard et al., 1999, Pregliasco et al., 1998, Stockton et al., 1998, Wallace et al., 1999). Our data (unpublished) demonstrate that RT-PCR can detect up to twice as many influenza positive samples than virus culture and direct immunofluorescence combined. Multiplex PCR has been used to monitor the influenza outbreak in England and Wales (Ellis et al., 1997). As respiratory syncytial virus (RSV) was shown to be a significant cause of influenza-like illness (ILI), in a wide age range of patients, during the winter respiratory season (Zambon et al., 2001), it is therefore essential to make a virological diagnosis to effect appropriate treatment.
In a pilot study, performed during the 1998–1999 influenza season, RT-PCR for influenza A and B was optimised (Wallace et al., 1999) and used to monitor the community influenza outbreak in Scotland as PCR results were available within 24 h of the sample arriving in the laboratory (Carman et al., 2000). In this paper, we report the extension of virological surveillance to include the detection of respiratory syncytial virus and picornavirus infection in patients reporting with influenza-like illness at general practices in Scotland during the 1999/2000 season.
2. Patients and methods
2.1. Sample collection
Patients presenting with influenza-like illness at 15 general practices across Scotland from October 1999 until March 2000, were asked to provide a throat swab and a high nasal swab. Both swabs were swirled vigorously in a single vial containing 2 ml of a guanidinium-based lysis buffer, (virus PCR sample solution, VPSS), provided with the High Pure Viral RNA Extraction Kit™ (Roche Diagnostics GmbH, Penzberg, Germany). The swabs were discarded after use and the VPSS sample was posted to the laboratory. Blood samples for serological analysis were taken at presentation and at least 3 weeks later (range 15–59 days, mean 29.5 days) from consenting patients at two sites (Fort William and Dingwall). A total of 240 combined swab samples and 89 paired blood samples were received.
The clinical protocols were approved by the relevant local area medical ethics committees.
2.2. RT-PCR
A multiplex RT-PCR was performed to identify influenza A or B, respiratory syncytial virus and picornavirus. RNA was extracted from samples using an adapted protocol for the High Pure Viral RNA Extraction Kit™ (Roche Diagnostics GmbH, Penzberg, Germany); 400 μl of the VPSS sample was loaded directly onto the spin-column and then the manufacturer’s instructions were followed. RNA was eluted in a volume of 50 μl. cDNA was synthesised by reverse-transcription as previously described (Wallace et al., 1999). Nested PCR was performed using primers which amplified regions within the genes for: (i) the influenza A matrix protein (Zhang and Evans, 1991); (ii) the influenza B haemagglutinin (Stockton et al., 1998); (iii) the N and P regions of the RSV genome (Stockton et al., 1998) and the FI subunit of fusion F glycoprotein (Henkel et al., 1997); (iv) the picornavirus 5′NCR (Andeweg et al., 1999). TaqStart™ antibody (Clontech, Palo Alto, USA) was used to increase sensitivity and product specificity in both rounds of PCR (Wallace et al., 1999). cDNA (20 μl) was amplified in a reaction volume of 100 μl containing each of the four primer pairs at 5 pmol/μl and 3.5 mM MgCl2. PCR was performed in thin walled 0.2 ml tubes on a PE9700 thermal cycler (PE Applied Biosystems, Warrington, Cheshire, UK). After an initial denaturation step at 95 °C, 35 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 45 s were followed by a final extension of 2 min at 72 °C. The second round of amplification was performed using an increased annealing temperature of 60 °C with nested primers (at 25 pmol/μl) in a final volume of 50 μl containing 2 μl of first round product. PCR products were analysed on 2% (3:1 Nusieve® agarose, FMC Bioproducts, USA) gels containing 0.2 μg/ml ethidium bromide. Product sizes of 767, 401, 334, 290 and 183 bp were expected for influenza B, influenza A, RSV A, picornavirus and RSV B, respectively.
2.3. Serology
Specific complement fixation assays to detect an elevated antibody titre consistent with influenza A or influenza B virus infection were performed on paired serum samples from 89 patients from the Fort William and Dingwall practices. A four-fold rise in the antibody titre of the second sample when tested in parallel with the first was defined as significant and diagnostic of a current or recent infection with influenza A or B. Any first sample taken late (>7–10 days since onset) with a titre of 64 or greater was also considered significant.
3. Results
3.1. Patients
A total of 240 swab samples were received. Patients were aged 3.3 to 95.3 years (mean 52.7 years), with 141 females and 99 males, of whom 58 were smokers and 72 were defined as ‘at risk’ according to the UK national guidelines for influenza vaccination. Forty three patients had received influenza vaccination.
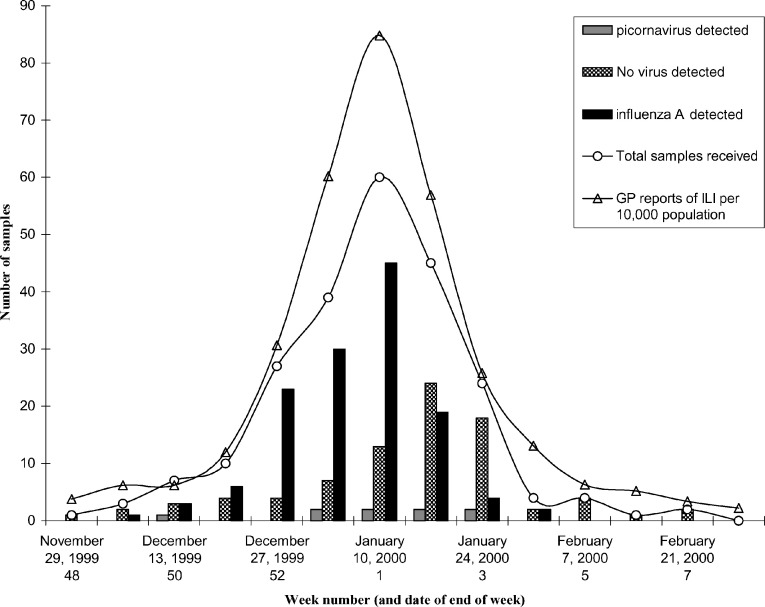
Samples were taken between 1 and 21 days after the perceived onset of ILI (mean 5.1 days), with 80% of samples taken within 7 days. The relative proportion of samples taken each week correlated well with the figures from the Scottish Sentinel GP network (Fig. 1 ).
Fig. 1.
PCR detection of influenza A and picornavirus during the 1999–2000 winter season. The number of samples received at the laboratory each week during the winter respiratory season is shown in comparison to the number of consultations (per 10,000 population) for influenza-like illness reported to The Scottish Centre for Infection and Environmental Health (SCIEH) by the general practitioner sentinel network. The number of samples which were influenza A positive, picornavirus positive or negative for influenza A and B, picornavirus and RSV by nested PCR are shown for each week. Key: (○) total samples received; (▵) sentinel GP reports of influenza-like illness; (■) influenza A PCR positive; ( ) picornavirus PCR positive; (
) picornavirus PCR positive; ( ) PCR negative samples.
) PCR negative samples.
3.2. PCR results
Of the 240 samples collected, 132 (55%) were PCR positive for influenza A only, 9 (3.8%) were picornavirus PCR positive only, 3 (of 77 samples) were RSV PCR positive only, 2 (0.8%) samples were PCR positive for both influenza A and picornavirus and 94 (39.2%) were negative for all four viruses. No influenza B virus was detected by PCR in this study population.
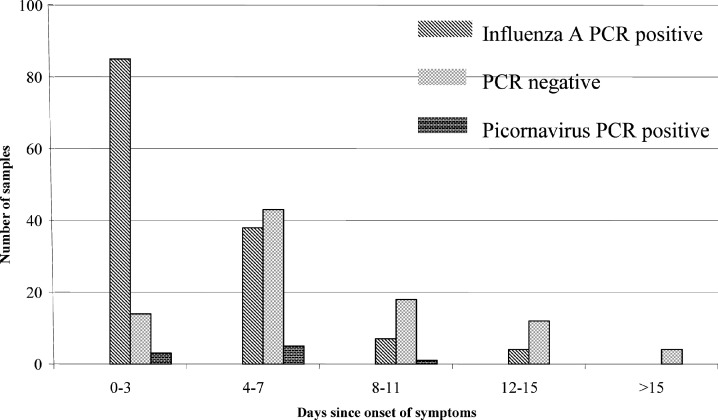
The rate of influenza A PCR positivity in this study fell significantly in samples taken longer after onset of ILI (83.3%, days 0–3; 45.2%, days 4–7 and 23.9% after 8 or more days (Fig. 2 )).
Fig. 2.
The number of PCR positive samples decreases as the number of days between the onset of symptoms and the delay in patients reporting to the GP increases. Key: ( ) influenza A PCR positive samples; (
) influenza A PCR positive samples; ( ) PCR negative samples; (
) PCR negative samples; ( ) picornavirus PCR positive samples.
) picornavirus PCR positive samples.
3.3. Serology and PCR positives
A total of 89 paired blood samples were obtained for influenza A/B complement fixation test (CFT). Fifty-seven of these blood samples had matching throat/nose swab samples which were PCR positive for influenza A. Fourty-eight (84.2%) of these influenza A positive patients displayed a significant (four-fold or greater) increase in antibody titre to influenza A virus by CFT (Table 1 ). Of the nine remaining influenza A PCR positive patients, two (3.5%) had a static high titre (64 or above) to influenza A virus (Table 1, footnotes h and n), five (8.7%) displayed a two-fold rise in CFT titre (Table 1, footnotes c and j) and two, who had been vaccinated, had falling titres. (Table 1, footnote l).
Table 1.
Complement fixation assay titres for influenza A virus compared to PCR results
| Titre; |
Fold increase | Number of samples with titres; |
||
|---|---|---|---|---|
| Sample 1 | Sample 2 | Influenza A PCR positive | Influenza A PCR negative | |
| <8 | <8 | 0 | 0 | 4 |
| <8 | 16 | 4 | 2 | 0 |
| <8 | 32 | 8 | 7 | 0 |
| <8 | 64 | 16 | 2 | 0 |
| <8 | 128 | 32 | 5 | 1a, b |
| <8 | 256 | 64 | 1 | 0 |
| <8 | 512 | 128 | 1 | 0 |
| 8 | <8 | – | 0 | 1 |
| 8 | 8 | 0 | 0 | 6 |
| 8 | 16 | 2 | 0 | 1 |
| 8 | 32 | 4 | 2 | 0 |
| 8 | 64 | 8 | 4 | 0 |
| 8 | 128 | 16 | 2 | 0 |
| 8 | 512 | 64 | 1 | 0 |
| 16 | 16 | 0 | 0 | 8 |
| 16 | 32 | 2 | 3a, c | 0 |
| 16 | 64 | 4 | 13 | 0 |
| 16 | 128 | 8 | 5 | 0 |
| 16 | 256 | 16 | 1 | 0 |
| 32 | 16 | – | 0 | 2d |
| 32 | 32 | 0 | 0 | 2e |
| 32 | 64 | 2 | 0 | 1f |
| 32 | 128 | 4 | 1 | 0 |
| 64 | 32 | – | 0 | 2g |
| 64 | 64 | 0 | 1a, h | 1i |
| 64 | 128 | 2 | 2a, j | 1k |
| 64 | 256 | 4 | 1 | 0 |
| 128 | 64 | – | 2a, l | 1m |
| 128 | 128 | 1 | 1a, n | 0 |
| 128 | 256 | 2 | 0 | 1o |
Results that do not correspond, i.e. (i) influenza A positive PCR samples with CFT titre rises less than four-fold; (ii) PCR negative samples with a significant CFT increase.
Sample taken 8 days since onset of symptoms. Viral nucleic acid may no longer have been present at this time.
Samples taken 1 day, 1 day since (vaccinated subject), and 3 days since onset, respectively.
Samples taken 1 day since (a vaccinated subject) and 11 days since onset. Viral nucleic acid may no longer have been present after 11 days.
Samples taken 9 and 13 days since onset. Viral nucleic acid may no longer have been present.
Samples taken 11 days since onset. Viral nucleic acid may no longer have been present after 11 days.
Samples taken 1 and 2 days since onset.
Sample taken 8 days since onset. Antibodies may have risen already in the first sample.
Samples taken 4 days since onset.
Samples taken 9 and 14 days since onset, respectively. Antibodies may have risen already in the first sample.
Samples taken 9 days since onset. Viral nucleic acid may no longer have been present after 9 days.
Samples taken 2 and 13 days since onset, both from vaccinated subjects.
This is a second sample from a patient who presented to their GP twice. The initial sample was PCR and serology positive for influenza A infection.
Sample taken 7 days since onset of symptoms. Antibodies may have risen already in the first sample.
Samples taken 10 days since onset. Viral nucleic acid may no longer have been present after 10 days.
There were 32 influenza PCR negative samples with matched serology samples; 20 (62.5%) showed no evidence of influenza A infection by CFT, with titres in the second sample of 16 or lower and no significant increase in titre between the samples. One PCR negative patient displayed a 32-fold (<8 to 128) increase in CFT titre to influenza A, consistent with recent infection (Table 1, footnote b). The swab sample from this patient was taken 8 days since onset of symptoms. Eleven patients had raised titres (32 or above) in both samples; three of these showed less than significant (two-fold) rises in titre, and the remaining eight patients had static or falling titres. The two-fold rise was noted in three patients whose swab samples were taken >9 days since onset (Table 1, footnotes f, k and o). Static or falling titres were noted in: one vaccinated patient (Table 1, footnote d); a patient who had recently been diagnosed with laboratory confirmed influenza A and who re-presented to the GP 20 days later (Table 1, footnote m); three patients whose swab samples had been taken >9 days since onset (Table 1, footnotes d and e); and three whose swabs had been taken apparently early in infection at Days 1, 2 and 4 (Table 1, footnotes g and i).
Five of the seven picornavirus PCR positive patients for whom serology results were available displayed no significant titre to either influenza A or B. The two other samples were dual picornavirus/influenza A infections and consequently had significantly elevated titres to influenza A virus.
4. Discussion
We have previously demonstrated that influenza PCR is rapid, sensitive and a user friendly assay. (Wallace et al., 1999). Here, we have expanded the capability of the RT-PCR assay to include RSV and picornavirus infection in addition to influenza A and B viruses and have employed the assay as a rapid test to monitor circulating respiratory viruses in the community. This is an improvement on our previously reported capacity to provide real-time surveillance to inform public health and policy makers and further confirms that there is no practical alternative to nucleic acid testing.
Over the course of the winter season, the majority (55%) of patients reporting to their GPs who were diagnosed as having ILI were infected with influenza A virus. At the 2-week peak of the influenza season (weeks 52/1999 to 2/2000 inclusive (Fig. 1)), the proportion of patients who were influenza A PCR positive rose to 68%, with 85% of samples positive in week 52/1999. Picornavirus infection was the cause of 3.8% of respiratory illness in patients who reported to their GP with ILI during the winter season. We failed to detect influenza B infection in this study population. This is not a lack of sensitivity as influenza B was detected in one of a set of samples received in the laboratory from a separate study taking place in England. Initially all study samples were RSV PCR negative, although it was clear that RSV was diagnosed with greater sensitivity than either virus culture or direct immunofluorescence throughout the same season in our routine diagnostic samples (data not shown). However, a different set of RSV primers were introduced into the laboratory after this study was completed and three of the 77 remaining, stored negative samples retested positive for RSV.
Therefore, nearly 60% of all samples were positive for one of the viruses tested. It is noteworthy that 83% of the samples taken within 3 days of the onset of symptoms were influenza A positive and there was a 50% or more decrease in positivity in samples taken after 3 days when viral nucleic acid may no longer be present in a proportion of patients presenting later in the course of infection. The severity of the symptoms of influenza infection may encourage patients who actually have an influenza infection to report to their GP earlier than those patients who do not have influenza infection.
Both the rate of sampling in our study (which was dependent on patients feeling ill and reporting to their GP), and the number of influenza A positive samples, correlated with the reports of ILI per 100,000 population from GP sentinel practices collated by SCIEH (Christie et al., 2000); however, as our results were available within 24 h, at least a week earlier than the published surveillance figures, we were able to monitor community influenza infection. This is especially important when there is circulation of a non-vaccine strain in the community which adds extra burden to health care providers. Also, additional information on the cause of illness is available when multiplex PCR is performed; picornavirus infection was identified as the cause of around 4% of influenza-like illness over the season. A proportion of patients presenting with ILI do not have an influenza infection; 15% of patients were influenza A PCR negative even at the height of the influenza season. In a community-based study, Zambon et al. (2001) have shown that RSV is an important cause of ILI in all age groups, however this was not found in the sample group in this study. PCR assays are being developed for other respiratory pathogens, including parainfluenza, coronavirus, including the virus related to severe acute respiratory syndrome (SARS, Drosten et al., 2003), human metapneumovirus (van den Hoogen et al., 2001), Mycoplasma and Chlamydia species, to assess the contribution of these agents to community influenza-like illness.
One disadvantage of using PCR alone to monitor the community influenza outbreak is the lack of viable virus for culture and epidemiological analysis. Although PCR can be used to a limited extent to subtype influenza A virus, e.g. by the use of heamagglutinin-specific primers, a proportion of samples should continue to be submitted in a manner that facilitates more detailed analysis.
The surveillance samples were submitted in VPSS which is a guanidium-based buffer used in the first phase of RNA extraction. In previous studies it has been shown to increase our positivity rate for detection of these RNA viruses by 10% (Carman et al., 2000) and is ideal for posting samples to the laboratory if there is no requirement for live virus.
Serological results are difficult to interpret and even more difficult when matched to PCR results. Only one sample was positive by serology and PCR negative. This sample was taken at 8 days since the onset of symptoms when a 50% decrease in sensitivity of detection has been noted (Fig. 2). Fig. 2 also shows that samples can be PCR positive up to 15 days after onset; this reflects either good quality sampling or a longer perceived length of illness. There were nine non-diagnostic serology results from influenza A PCR positive patients. Further analysis of these nine influenza A PCR positive patients showed that five (including one who was vaccinated) had had their first sample taken 7 days or more since onset (Table 1, footnotes h, j, l and n) and all had a titre of 64 and above in the first sample which may explain the lack of a significant rise.
There were 11 PCR negative patients with suggestive serology results (all had initial titres of 32 or above). Six of the patients with higher than background serology titres (greater or equal to 32) did not report to their GP until 9 or more days after the onset of symptoms (three of them showed a two-fold rise); they may have been PCR positive if they had been sampled earlier in the course of their infection. Three other patients (Table 1, footnotes g and i) were sampled early in infection; two had titres of 64 falling to 32, and the other was static at 64, as there is no PCR evidence of influenza infection in these patients, the elevated CFT titre may be due to infection in a previous year or an asymptomatic infection earlier in the season.
In all, except two cases (Table 1, footnotes m and o), the initial titres in the PCR negative group are too low, in practice, to be considered diagnostic of infection, even those who were sampled after 8 days. In addition, the second serum in the majority of the samples which are both PCR and serology positive would not be considered diagnostic of infection unless part of a pair.
Although a titre of 128 is considered diagnostic by many virology laboratories, only 21 of 57 PCR positive samples achieved this level or higher. An additional 22 patients had a titre of 64 and 12 had a titre of 32 in the second, convalescent sample. Twenty-one of 32 PCR negative patients had a titre of 16 or less. It may be that during the influenza season, titres of 64 and perhaps even 32 should be considered indicative of recent infection. The difficulty in interpreting serology results meaningfully has led us to favour PCR as the frontline assay.
The serology results obtained suggest that most patients who were PCR negative had not been infected with influenza A virus, supporting the hypothesis that true influenza infection encourages earlier presentation at the GP surgery.
In conclusion, the differential diagnosis of influenza by serological methods requires two blood samples over 3 weeks and our results show that it is not always easy to interpret. Blood samples are an inappropriate sample to take from patients with acute respiratory illness; however the increasing use of nose and throat swabs in the GP setting and a rapid PCR result turnaround time allows the viruses contributing to the annual winter respiratory season to be monitored in a timely and useful manner. The development of a set of multiplex PCRs to achieve this is the future for surveillance and diagnosis of respiratory viral infection.
Acknowledgements
The authors would like to thank the staff at the participating general practices for their help in patient recruitment and sample collection. This study was supported by a grant from The Chief Scientists Office, The Scottish Office, UK.
References
- Andeweg A.C., Bestebroer T.M., Huybreghs M., Kimman T.G., deJong J.C. Improved detection of rhinoviruses in clinical samples by using a newly developed nested reverse-transcription-PCR assay. J. Clin Microbiol. 1999;37:524–530. doi: 10.1128/jcm.37.3.524-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman W.F., Wallace L.A., Walker J., McIntyre S., Noone A., Christie P. Rapid virological monitoring of the community influenza outbreak. Br. Med. J. 2000;321:736–737. doi: 10.1136/bmj.321.7263.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P., Mooney J., Johnston F., Brownlie S. Surveillance report: respiratory infections. SCIEH Wkly. Rep. 2000;34:70–71. [Google Scholar]
- Drinka P.J., Gravenstein S., Langer E., Krause P., Shult P. Mortality following isolation of various respiratory viruses in nursing home residents. Infect. Control Hosp. Epidemiol. 1999;20:812–815. doi: 10.1086/501589. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., Van der Wer F.S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Ellis J.S., Fleming D.M., Zambon M.C. Multiplex reverse transcription PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J. Clin. Microbiol. 1997;35:2076–2082. doi: 10.1128/jcm.35.8.2076-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddian A.P. Progress with neuraminidase inhibitors. Rev. Med. Virol. 2000;10:135–137. doi: 10.1002/(sici)1099-1654(200003/04)10:2<135::aid-rmv276>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Henkel J.H., Aberle S.W., Kundi M., Popow-Kraupp T. Improved detection of respiratory synctial virus in nasal aspirates by semi-nested RT-PCR. J. Med. Virol. 1997;53:366–371. [PubMed] [Google Scholar]
- Magnard C., Valette M., Aymard M., Lina B. Comparison of two nested PCR, cell culture and antigen detection for the diagnosis of upper respiratory tract infections due to influenza virus. J. Med. Virol. 1999;59:215–220. doi: 10.1002/(sici)1096-9071(199910)59:2<215::aid-jmv15>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Makela M.J., Puhakka T., Ruuskanen O., Leinonen M., Saikku P., Kimpimaki M. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuzil K.M., Reed G.W., Mitchel E.F., Jr., Griffin M.R. Influenza-associated morbidity and mortality in young and middle-aged women. JAMA. 1999;281:901–907. doi: 10.1001/jama.281.10.901. [DOI] [PubMed] [Google Scholar]
- Neuzil K.M., Mellen B.G., Wright P.F., Mitchel E.F., Griffin M.R. The effect of influenza on hospitalisations, outpatient visits, and courses of antibiotics in children. N. Engl. J. Med. 2000;342:225–231. doi: 10.1056/NEJM200001273420401. [DOI] [PubMed] [Google Scholar]
- Nicholson K.G. Impact of influenza and respiratory syncytial virus on mortality in England and Wales from January 1975 to December 1990. Epidemiol. Infect. 1996;116:51–63. doi: 10.1017/s0950268800058957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson K.G., Kent J., Hammersley V., Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population-based study of disease burden. Br. Med. J. 1997;315:1060–1064. doi: 10.1136/bmj.315.7115.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregliasco F., Mensi C., Camorali L., Anselmi G. Comparison of RT-PCR with other diagnostic assays for rapid detection of influenza viruses. J. Med. Virol. 1998;56:168–173. doi: 10.1002/(sici)1096-9071(199810)56:2<168::aid-jmv11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Sczucs T.D. Role of burden of illness research. Pharmacoeconomics. 1999;116:27–32. doi: 10.2165/00019053-199916001-00004. [DOI] [PubMed] [Google Scholar]
- Stockton J., Ellis J.S., Saville M., Clewley J.P., Zambon M.C. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J. Clin. Microbiol. 1998;36:2990–2995. doi: 10.1128/jcm.36.10.2990-2995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A.M. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nature Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace L.A., McAulay K.A., Douglas J.D.M., Elder A.G., Stott D.J., Carman W.F. Influenza diagnosis: from dark isolation into the molecular light. J. Infection. 1999;39:221–226. doi: 10.1016/s0163-4453(99)90053-1. [DOI] [PubMed] [Google Scholar]
- Zambon M.C., Stockton J.D., Clewley J.P., Fleming D.M. Contribution of influenza and respiratory syncytial virus to community cases of influenza-like illness: an observational study. Lancet. 2001;358:1410–1416. doi: 10.1016/s0140-6736(01)06528-x. [DOI] [PubMed] [Google Scholar]
- Zhang W., Evans D. Detection and identification of human influenza viruses by the polymerase chain reaction. J. Virol. Methods. 1991;33:165–189. doi: 10.1016/0166-0934(91)90017-t. [DOI] [PubMed] [Google Scholar]