Abstract
The mechanisms and factors involved in the replication of positive stranded RNA viruses are still unclear. Using poliovirus as a model, we show that a long-range interaction between ribonucleoprotein (RNP) complexes formed at the ends of the viral genome is necessary for RNA replication. Initiation of negative strand RNA synthesis requires a 3′ poly(A) tail. Strikingly, it also requires a cloverleaf-like RNA structure located at the other end of the genome. An RNP complex formed around the 5′ cloverleaf RNA structure interacts with the poly(A) binding protein bound to the 3′ poly(A) tail, thus linking the ends of the viral RNA and effectively circularizing it. Formation of this circular RNP complex is required for initiation of negative strand RNA synthesis. RNA circularization may be a general replication mechanism for positive stranded RNA viruses.
Introduction
Positive stranded RNA viruses amplify their genomes from one single molecule to tens of thousands of copies in only a few hours. Efficiency and speed are important characteristics of the viral replication machinery. In addition, they must also be highly specific to discriminate between the viral RNA and cellular RNAs in the cytoplasm of the infected cell.
The genomic RNA initially directs the synthesis of viral proteins, and then, once the viral RNA–dependent RNA polymerase and other essential proteins are synthesized, the viral RNA is copied, starting from the 3′ end, to generate a complementary, negative stranded RNA that, in turn, is transcribed into new molecules of positive stranded genomic RNA. Thus, due to its double role, the genomic RNA must be recognized by both the cellular translation machinery and the viral replication apparatus. Recent evidence indicates that, besides the virally encoded factors, host cell proteins play an essential role in the replication of positive stranded RNA viruses Gamarnik and Andino 1996, Diez et al. 2000.
To examine the role of intracellular determinants of viral replication, we have used poliovirus as a model, because both in vitro and in vivo systems are available to dissect the viral replication cycle Molla et al. 1991, Barton et al. 1995, Gamarnik and Andino 1996. The poliovirus genome consists of ∼7500 nucleotides and contains a poly(A) tail at the 3′ end and a small peptide, VPg, covalently linked to the 5′ end Yogo and Wimmer 1972, Flanegan et al. 1977, Lee et al. 1977, Racaniello and Baltimore 1981. The highly structured 5′ untranslated region (UTR) regulates both translation and RNA replication (Rohll et al., 1994). Two functional regions have been described within the 5′ UTR: a long element involved in cap-independent initiation of translation (IRES) Pelletier et al. 1988, Trono et al. 1988 and a shorter 5′-terminal structure (the cloverleaf RNA) involved in viral RNA replication (Andino et al., 1990a).
The cloverleaf RNA forms a ternary complex with two proteins: the uncleaved viral protease polymerase precursor 3CD Andino et al. 1990a, Andino et al. 1993, Silvera et al. 1999 and the poly(rC) binding protein (PCBP, also known as hnRNP E or α-CP) Gamarnik and Andino 1997, Parsley et al. 1997. PCBP is a host protein that regulates the stability and expression of several cellular mRNAs (reviewed by Ostareck-Lederer et al., 1998). The molecular mechanism by which PCBP interacts with the translation apparatus remains poorly understood, but it has been shown to interact with other RNA binding proteins, including poly(A) binding protein 1 (PABP1) (Wang et al., 1999). Evidence suggests that the poliovirus 5′ cloverleaf ternary complex has a bifunctional role, participating in both viral translation and RNA replication Simoes and Sarnow 1991, Gamarnik and Andino 1998. The binding of the cellular protein PCBP to the cloverleaf RNA enhances viral translation. In contrast, the binding of the viral polymerase precursor, 3CD, to the cloverleaf represses viral translation and promotes the synthesis of negative stranded RNA Gamarnik and Andino 1998, Barton et al. 1999.
An open question concerning the replication of RNA viruses is how the same viral replication machinery can initiate RNA synthesis from both positive and negative strands, since they carry very different cis-acting elements. In this regard, it also is intriguing that specific binding of the viral polymerase precursor to the 5′ end of the RNA genome is required for negative strand RNA synthesis (Gamarnik and Andino, 1998), given that initiation of negative strand RNA synthesis takes place at the opposite end of the genomic RNA. Clearly, successful initiation of negative strand RNA synthesis depends on the specific recognition of the viral RNA as a template, as well as the replication start site. Poliovirus negative strand RNA synthesis initiates within the poly(A) tail of the genomic RNA Larsen et al. 1980, Herold and Andino 2000. The poliovirus poly(A) tail is an important cis-acting element for RNA replication since removal or shortening of the poly(A) tail results in a defect in RNA replication Spector et al. 1975, Sarnow 1989, Barton et al. 1996. Furthermore, purified 3Dpol is able to uridylate the putative primer for RNA synthesis, VPg, in vitro using poly(A) as a template (Paul et al., 1998). However, the poly(A) tail cannot be the primary cis-acting element that specifies replication of the viral RNA, as all cellular mRNAs contain poly(A) at their 3′ ends. One proposal is that the short 3′ UTR that precedes the poly(A) tail is involved in determining template specificity (oriR) (Pilipenko et al., 1996). However, recombinant polioviruses with deleted 3′ UTRs are viable, although they replicate with slower kinetics than the wild-type virus (Todd et al., 1997). Furthermore, the poliovirus 3′ UTR is interchangeable with the 3′ UTR of human rhinovirus 14, which is predicted to fold into a completely different structure (Rohll et al., 1995). These results suggest that the 3′ UTR plays a regulatory role rather than acting as the origin of replication for negative strand RNA synthesis.
Here, we examined cis- and trans-acting elements involved in the initiation of negative strand RNA. We discover that PABP1 interacts with both the poly(A) tail and the cloverleaf ribonucleoprotein (RNP) complex. Our results explain the requirement of a long poly(A) tail for RNA replication and uncover a direct role for the requirement of 3CD binding to the cloverleaf in negative strand RNA synthesis. The viral polymerase precursor 3CD binds to the 5′ end of the genomic RNA and reaches its site of action within the poly(A) tail of the genome via circularization of the genomic RNA using an RNA–protein–protein–RNA bridge that involves at least two cellular factors, PCBP and PABP1. We propose that the formation of a circular genomic structure by interaction of the 5′ and 3′ ends may be a general mechanism by which positive stranded RNA viruses initiate replication.
Results
Sequences at the 5′ End of the Genomic RNA Are Directly Involved in the Initiation of Negative Strand Synthesis
It has been previously shown that binding of the viral RNA polymerase precursor 3CD to the cloverleaf is essential for initiation of negative strand RNA synthesis (Gamarnik and Andino, 1998). This phenotype could result from the inability of these mutants to shut down translation, a prerequisite in order to initiate RNA synthesis. However, it is also possible that the cloverleaf participates in both repression of translation and initiation of RNA synthesis.
To examine these possibilities, we used a cell-free system that supports complete poliovirus replication Molla et al. 1991, Barton et al. 1995. A crude cytoplasmic extract can be programmed with in vitro synthesized viral RNA. The extract contains all of the cellular components required for authentic initiation of RNA synthesis. Thus, each of the poliovirus replication intermediates and final products is correctly produced. The reaction is carried out in two steps. First, viral RNA is used to direct the synthesis of viral proteins. This reaction is performed in the presence of 2 mM guanidinium hydrochloride (GuHCl), a potent inhibitor of poliovirus replication that allows viral proteins to accumulate but no RNA to be synthesized. It has been proposed that structures (called preinitiation complexes) form during this step of the reaction, which, in turn, later participate in the RNA replication step. The second part is the RNA replication reaction, whereby the GuHCl is removed from the reaction mixture and radiolabeled nucleotides are used to monitor the synthesis of RNA. Since translation is uncoupled from RNA synthesis in this two-step system, the requirement for each of these steps can be examined independently.
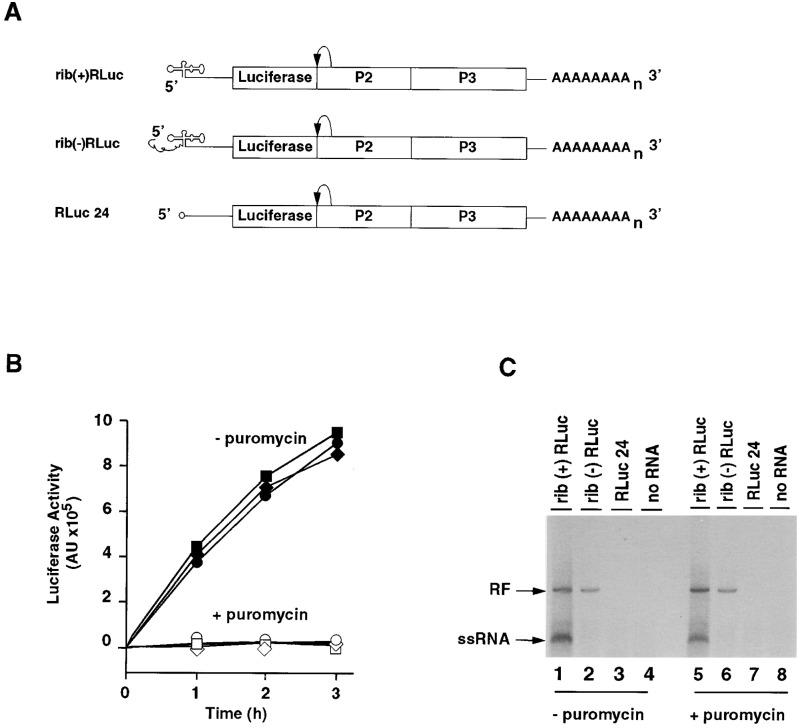
Two mutants were used (Figure 1A) , one in which the cloverleaf structure was deleted (RLuc24) and another that carries an extension of ∼50 nucleotides at the 5′ end (rib[−]Rluc). The extra sequences present at the 5′ end of rib(−)RLuc abolish synthesis of positive strand RNA without affecting negative strand RNA synthesis (Herold and Andino, 2000). As a positive control, we used a transcript in which extra sequences at the 5′ end were removed by a cis-active hammerhead ribozyme and therefore containing the authentic poliovirus 5′ end (rib[+]Rluc) (Herold and Andino, 2000). All three constructs carried a 3′ poly(A) tail of 80 nucleotides.
Figure 1.
The Cloverleaf Is an Important cis Active Element for Negative Strand RNA Synthesis
(A) Schematic representation of three replicons used, in which the structural protein coding region was replaced by the luciferase gene. Luciferase is released from the poliovirus polyprotein by the proteolytic activity of 2Apro. Replicon rib(+)RLuc RNA carries authentic 5′ ends; in contrast, the rib(−)RLuc replicon contains an extension of 50 nucleotides that prevents positive strand RNA synthesis in vitro (Herold and Andino, 2000). RLuc24 carries a deletion of the entire 5′ cloverleaf RNA. This construct contains a 5′-terminal cap structure (o-). All three constructs had a poly(A) tail of 80 adenylates.
(B) Translational activity of the replicon RNAs in vitro. HeLa extracts were programmed with the replicon RNAs (rib[+]RLuc, squares; rib[−]RLuc, diamonds; RLuc24, circles). Translation was efficient in the absence of puromycin (black symbols). However, luciferase activity was not observed in the presence of puromycin (negative control, white symbols).
(C) RNA synthesis in the absence and presence of puromycin. Translation/replication extracts were programmed with the replicon RNAs. After isolation of preinitiation complexes, the reactions were divided into two aliquots, and RNA synthesis reaction was performed either in the absence (lanes 1–4) or the presence (lanes 5–8) of 100 μg/ml puromycin. The products of the reaction were analyzed by native 1% agarose gel electrophoresis
We first studied the ability of these mutants to direct translation in the cell-free replication system. The extracts were programmed with in vitro synthesized RNAs in the presence of GuHCl to inhibit viral replication. The amount of luciferase activity produced by the mutants during the first 3 hr was similar to that of the positive control, rib(+)RLuc (Figure 1B). These results demonstrated that, under the experimental conditions used, the cloverleaf RNA is not necessary for efficient translation or for the stability of the viral RNA.
We next examined RNA synthesis in both rib(+)RLuc and mutants. When extracts were programmed with rib(+)RLuc RNA, we observed the accumulation of both radiolabeled replicative form RNA (RF) and single strand RNA (ssRNA) (Figure 1C, lane 1). The labeled RF RNA observed is composed of the unlabeled input positive stranded and newly synthesized 33P-labeled negative stranded RNA; thus, RF can be taken as a direct measure of negative strand RNA synthesis. As expected, the mutant containing extra nucleotides at the 5′ end was able to synthesize RF but not ssRNA (lanes 2). More importantly, the cloverleaf deleted mutant was unable to synthesize either RF or ssRNA (lanes 3), indicating that the cloverleaf RNA is essential for negative strand RNA synthesis.
As discussed above, downregulation of translation via 3CD–cloverleaf interaction is necessary to generate a ribosome-free template capable of negative strand RNA synthesis. To release the ribosomes from the viral RNA in the absence of a 5′ cloverleaf, we added puromycin to the labeling reaction. Puromycin induces translation chain termination and inhibits protein synthesis by releasing ribosomes from their RNA template. RNA synthesis was not substantially altered by puromycin when cloverleaf-containing replicon RNAs were used (Figure 1C, lanes 5 and 6). Importantly, the release of ribosomes was not sufficient to stimulate synthesis of negative strand in the cloverleaf-less RNA (Figure 1C, lane 7), although the amount of puromycin added was sufficient to completely prevent translation (Figure 1B). In addition, cloverleaf RNA added in trans did not restore replication of a cloverleaf-less replicon (data not shown). Thus, we conclude that the cloverleaf is an essential cis-acting sequence that, in addition to its function in translational control, directly participates in the initiation of negative strand RNA synthesis.
The Poly(A) Tail at the 3′ End of the Poliovirus Genome Is an Important cis-Acting Element for Negative Strand RNA Synthesis
The observation that the 5′ cloverleaf directly participates in negative strand synthesis led us to examine in more detail the requirements for this reaction. Since negative strand synthesis initiates at the other end of the genome, 7500 nucleotides downstream from the cloverleaf RNA, we focused on the 3′ end and its potential communication with the 5′ end.
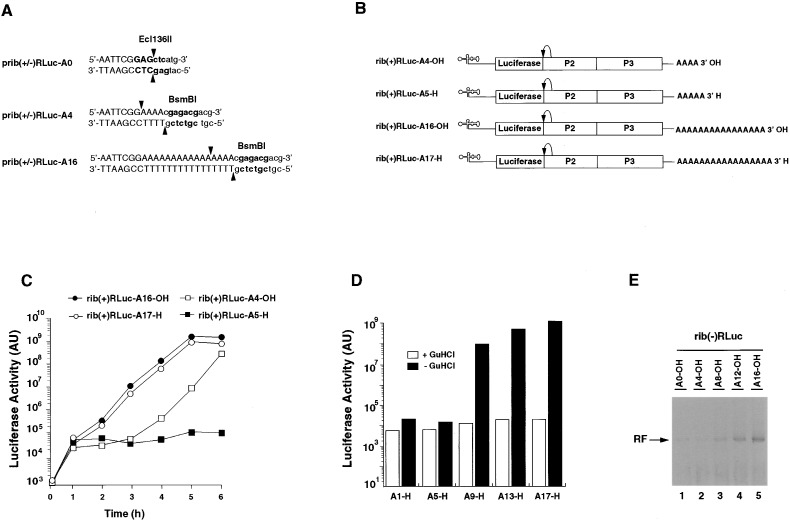
Previous results suggested that the poly(A) tail may be involved in the initiation of negative strand RNA synthesis (Barton et al., 1996). Transcripts with only 12 adenylates at their 3′ ends are less efficient at producing negative stranded RNA in vitro than those with 80. To confirm and expand these results, we engineered poliovirus replicon RNAs containing a defined number of adenylate residues—A0, A4, A8, A12, or A16—at their 3′ ends (rib[+]RLuc-A0-OH to rib[+]RLuc-A16-OH, schematized in Figures 2A and 2B). It is possible that during the course of replication, transcripts could regain long poly(A) tails by addition of adenylates at the 3′ OH. To prevent this possibility, we also generated transcripts rib(+)RLuc-A1-H to rib(+)RLuc-A17-H with blocked 3′ ends via incorporation of a single 3′ deoxyadenosine (3′-dA), which cannot accept additional nucleotides. Transcripts containing a long poly(A) tail (A16-OH or A17-H) replicated very efficiently when transfected into 293 cells and had similar kinetics regardless of whether or not the 3′ end was blocked with 3′-dA. In contrast, RNAs with short poly(A) tails with 3′ OH ends (A4-OH) displayed a delay in replication of ∼1–2 hr, but by 6 hr posttransfection, long poly(A) and short poly(A) constructs produced similar amounts of luciferase (Figure 2C). Total RNA was extracted 7 hr posttransfection and the 3′ ends were analyzed. Irrespective of the length of the poly(A) of the input RNA, all RNAs contained long poly(A) tails by 7 hr posttransfection (data not shown). Thus, the delay observed in RLuc-A4-OH presumably represents the time required to increase the length of the poly(A) before replication can start. More importantly, rib(+)RLuc-A5-H with a short poly(A) and blocked 3′ end was unable to replicate (Figure 2C).
Figure 2.
A Minimum Poly(A) Tail Is Necessary for RNA Replication In Vivo and In Vitro
(A) Restriction endonuclease recognition sites used for linearization of replicon-encoding cDNAs to generate transcripts with defined 3′ ends. Recognition sites are shown in bold, cleavage sites are indicated by arrows. Transcribed regions are shown in uppercase letters.
(B) Schematic presentation of the replicon RNAs with modified 3′ ends. Rib(+)Rluc RNA with 4 or 16 adenylates at their 3′ ends. In rib(+)RLuc-A5-H and -A17-H, the 3′ end were blocked with a 3′ deoxyadenylate.
(C) Transcripts with blocked 3′ ends and short poly(A) tails do not replicate in vivo. Replicon transcripts were transfected into 293 cells and incubated at 37°C. Luciferase activity in the transfected cells was determined at the indicated time points.
(D) A minimum poly(A) tail containing 8 adenylate residues is necessary for replication. Luciferase expressing replicon RNAs with 0, 4, 8, 12, or 16 adenylate residues and a single 3′-dA at their 3′ ends were transfected into 293 cells. The cells were incubated at 37°C in the absence or presence of 2 mM GuHCl, and luciferase activity was determined at 5 hr posttransfection.
(E) A minimum poly(A) tail containing 8 adenylate residues is necessary for efficient negative strand RNA synthesis in vitro. Replicon rib(−)RLuc RNAs with 0, 4, 8, 12, or 16 adenylate residues at their 3′ ends were used to program an in vitro translation/replication extract. The radiolabeled products of the reaction were analyzed on native 0.8% agarose gels and visualized by autoradiography
Next, we determined the precise length of the poly(A) tail required for replication in vivo. Five constructs containing poly(A) tails ranging from a single A to 17 As (A1-H, A5-H, A9-H, A13-H, and A17-H) were tested for their ability to replicate in 293 cells. All constructs contained 3′-dA at their 3′ end to block potential extension of the poly(A) tail. After transfection, cells were incubated at 37°C in the presence or absence of 2 mM GuHCl to distinguish between direct translation of the input RNA and translation of amplified RNA. Luciferase activity was determined at 4 hr posttransfection. In the presence of GuHCl, luciferase activity produced by RNAs with shorter poly(A) tails (A1-H, A5-H) was only two to three times lower than that produced by long poly(A) tail constructs (A9-H, A13-H, and A17-H) (Figure 2D). In the absence of GuHCl, only those constructs with more than 5 As at their 3′ ends (A9-H, A12-H, and A17-H) were able to replicate. However, replicons with a 3′ A9 produced five times less luciferase activity at the 4 hr time point than those constructs with longer poly(A) tails (A13-H and A17-H), suggesting that, within this range of poly(A) length, the ability of a particular construct to replicate closely correlates with the number of A residues present at the 3′ end.
To confirm that the reduced levels of luciferase produced by RNA replicons with short poly(A) tails indeed reflected a defect in RNA synthesis, we examined the ability of these replicons to synthesize negative stranded RNA in vitro. The rib(−)RLuc replicon RNAs, which are only able to produce negative stranded RNA (Herold and Andino, 2000), were synthesized with 0, 4, 8, 12, or 16 3′-terminal As. These RNAs were then examined using the cell-free translation/replication system. Constructs with no poly(A) tail or with only four 3′ adenylates produced almost undetectable levels of double stranded RF RNA (Figure 2E, lanes 1 and 2). In contrast, constructs with poly(A) tails comprising 8–16 adenylates were able to synthesize negative strand RNA. Interestingly, as the poly(A) tails increased in length, we observed a progressive increase in the efficiency of negative strand synthesis (Figure 2E, lanes 3, 4, and 5).
These results indicate that the 3′ poly(A) tail of the poliovirus genomic RNA is an important cis-acting element for negative strand RNA synthesis in vitro and in vivo. A minimum length of 8 to 12 adenylate residues is sufficient to support efficient initiation of RNA synthesis.
Interactions of PABP1 with the Poliovirus Poly(A) Tail
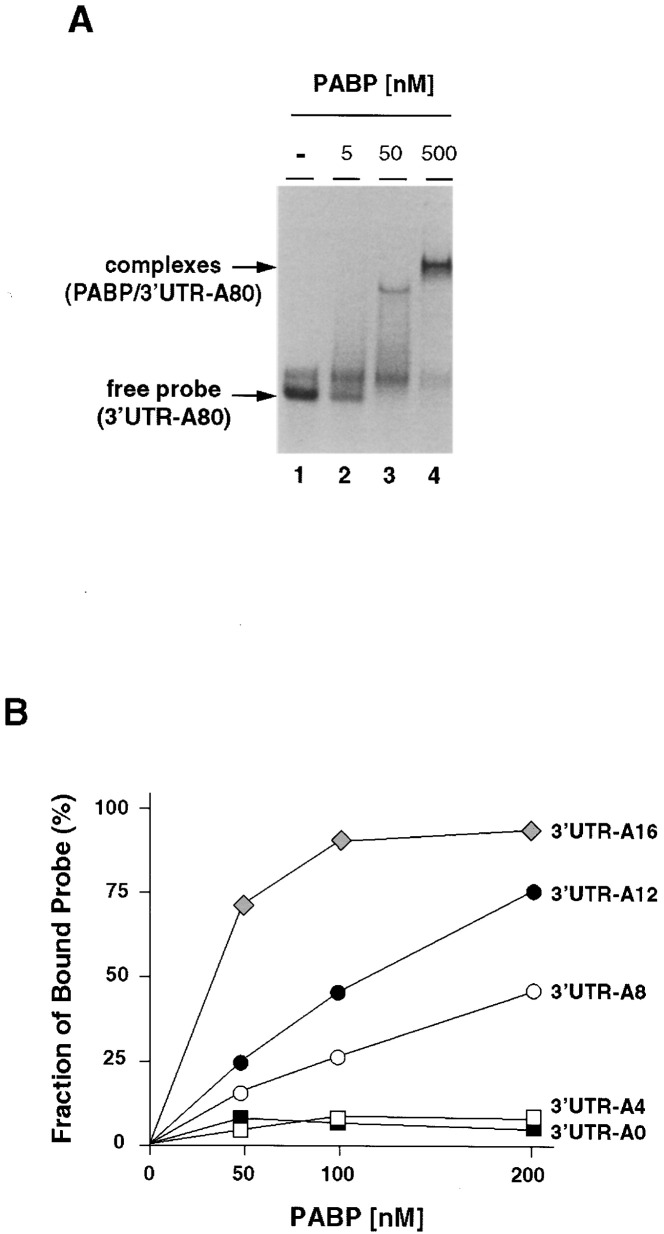
Since both the 5′ end cloverleaf and the 3′ end poly(A) tail are required for efficient negative strand synthesis Figure 1, Figure 2, we next studied the proteins that associate with these structures. Using poly(A) homopolymers, it has been shown that ∼12 nucleotides is the minimal length for high-affinity binding to RRM1/2, two of the RNA binding domains of mammalian PABP1 (Sachs et al., 1987). Also, it has been shown that a single RRM domain binds to poly(A) in a fashion similar to that of the entire protein. A single RRM domain and the full-length protein interact with a similar sized segment of the poly(A) and with comparable affinity. To examine the interaction of PABP1 with poly(A) in the context of the poliovirus 3′ UTR, we performed electrophoretic mobility shift assays (EMSA) using bacterial expressed PABP1 (PABP1-H6). We first determined the affinity of recombinant PABP1-H6 for poly(A) using a poliovirus 3′ UTR with a poly(A) tail of 80 nucleotides (Figure 3A) . The apparent Kd was ∼40 nM, which is higher than reported for PABP1 purified from eukaryotic cells (Gorlach et al., 1994) but closely matches what has been reported for the bacterially expressed RNA binding domains Sachs et al. 1987, Deo et al. 1999. Next, we determined the apparent Kd (see Experimental Procedures) of PABP1-H6 using poliovirus 3′ UTRs with 0-, 4-, 8-, 12-, and 16 nucleotide poly(A) tails (3′ UTR-A0, 3′ UTR-A4, 3′ UTR-A8, 3′ UTR-A12, and 3′ UTR-A16, respectively). As expected, PABP1 was unable to interact with 3′ UTR-A0 and 3′ UTR-A4 probes (Figure 3B). In contrast, PABP1-H6 interacted with 3′ UTR-8, 3′ UTR-12, and 3′ UTR-16 with high affinities. Apparent Kd values were ∼200, ∼100, and ∼40 nM, respectively (Figure 3B), indicating that a minimum of eight nucleotides are required for binding and that the affinity increases as a function of the poly(A) tail length. These results are consistent with previously published observations indicating that the affinity of yeast PABP for poly(A) increases with increasing length of oligo(A) up to 12 residues (Sachs et al., 1987).
Figure 3.
A Poly(A) Tail with a Defined Length Is Necessary for Efficient PABP1 Binding
(A) Recombinant human PABP1-H6 interacts with the poliovirus 3′ UTR. PABP1-H6 (5–500 nM) was incubated with a radiolabeled probe representing the poliovirus 3′ UTR containing 80 adenylate residues followed by electrophoresis through a native polyacrylamide gel.
(B) The interaction of PABP1 with the poliovirus 3′ UTR requires a minimum poly(A) tail with 8 adenylate residues. Recombinant PABP1 was incubated at the indicated concentrations with a radiolabeled probe representing the poliovirus 3′ UTR containing 0, 4, 8, 12, and 16 adenylate residues at the 3′ end and analyzed as in (A)
In conclusion, we observed a close correlation between the affinity of PABP1-H6 for the poly(A) tail and the initiation of negative strand RNA synthesis in vitro; constructs with longer poly(A) tails (A12 and A16) interact with PABP1-H6 with higher affinity and initiate negative strand synthesis more efficiently (compare Figure 2, Figure 3) than do replicons with shorter poly(A) tails.
PABP1 Interacts with PCBP and 3CD In Vitro
As mentioned before, it has been shown that the C-terminal part of PABP1 interacts with PCBP (Wang et al., 1999). Since PCBP is a known RNA binding protein that binds to the cloverleaf domain of the poliovirus 5′ UTR, we reasoned that the 5′ and 3′ ends of the viral genome might interact with each other through a PABP1–PCBP interaction.
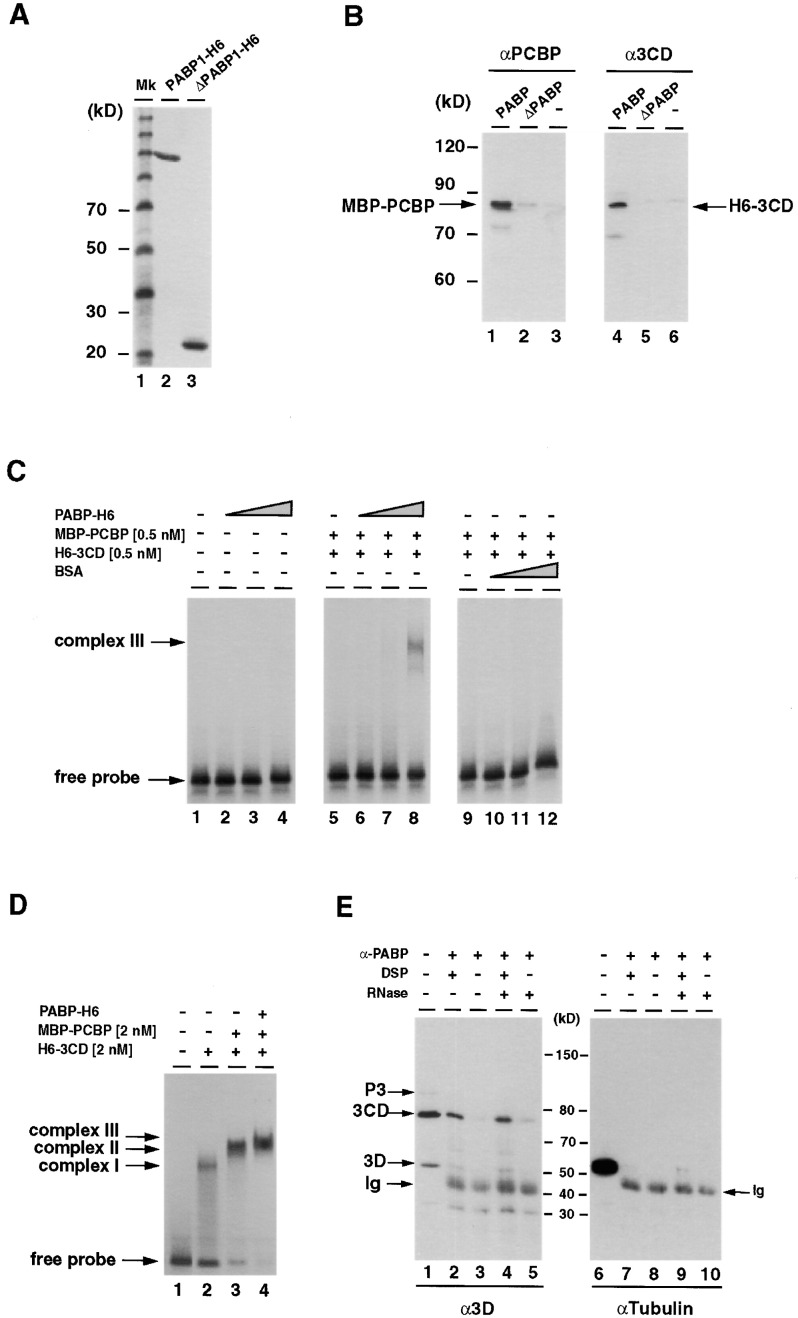
Full-length PABP1-H6 and the N-terminal region of PABP, ΔPABP-H6 (residues 1–190, containing RRM1/2), were expressed in E. coli and purified by Ni-NTA chromatography. As expected (Kuhn and Pieler, 1996), both recombinant proteins were able to bind to a poly(A)-Sepharose column (Figure 4A , lanes 2 and 3). To test whether proteins that bind to the poliovirus cloverleaf RNA can interact with PABP1, we incubated recombinant MBP-PCBP (PCBP2 fused to the maltose binding protein) or H6–3CD (His-tagged 3CD) with PABP1-H6 bound to poly(A)-Sepharose. Both MBP-PCBP and H6-3CD interacted with full-length PABP1-H6 (Figure 4B, lanes 1 and 4) but did not interact with ΔPABP-H6 or poly(A)-Sepharose alone (Figure 4B, lanes 2, 3, 5, and 6). Thus, PCBP and 3CD have the potential to associate with PABP1, presumably through interaction with the C-terminal part of PABP1.
Figure 4.
PABP1 Interacts with PCBP and 3CD In Vitro and in Poliovirus-Infected Cells
(A) Recombinant full-length PABP1-H6 and ΔPABP1-H6 (containing RRM1 and -2 domains) were loaded on to poly(A)-Sepharose column. After extensive washing, bound proteins were eluted using protein sample buffer, separated on SDS-polyacrylamide gel, and stained with Coomassie blue.
(B) Recombinant 3CD and PCBP interact specifically with PABP1. Bacterial expressed MBP-PCBP (lanes 1–3) or 3CD-H6 (lanes 4–6) was incubated with either PABP1-H6 bound to poly(A)-Sepharose (lanes 1 and 4), ΔPABP-H6 bound to poly(A)-Sepharose (lanes 2 and 5), or poly(A)-Sepharose alone (lanes 3 and 6). Bound proteins were eluted in protein sample buffer, separated on SDS-polyacrylamide gel, and detected by Western blotting using specific antisera.
(C) PABP1 stimulates ternary complex formation at the 5′ end of the genomic RNA. Radiolabeled cloverleaf RNA was incubated with various concentrations of MBP-PCBP, H6–3CD, PABP1-H6, or BSA. In lanes 2 through 4, PABP1 was incubated alone with cloverleaf RNA at 1, 10, and 100 nM (lanes 2, 3, and 4). In lanes 5 through 12, the concentrations of MBP-PCBP and H6-3CD were kept constant at 0.5 nM. Increasing amounts of either PABP1-H6 (lanes 5–8) or BSA (lanes 9–12) were added to each binding reaction (1 nM in lanes 6 and 7, 10 nM in lanes 7 and 11, and 100 nM in lanes 8 and 12). Complexes were analyzed by native polyacrylamide gel electrophoresis.
(D) Comparison of electrophoretic mobility of complexes containing 3CD (lane 2), 3CD + MBP-PCBP (lane 3), or 3CD + MBP-PCBP + PABP (lane 4).
(E) PABP1 interacts with 3CD-containing polypeptides in poliovirus-infected cells. Five hours postinfection, the cells were harvested, treated with the cross-linker DSP for 30 min (lanes 2, 4, 7, and 9), or left untreated (lanes 3, 5, 8, and 10). Cytoplasmic extracts were immunoprecipitated using anti-PABP1 polyclonal antibodies. Samples were analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting using anti-3Dpol monoclonal antibodies (lanes 1–5). The blot was stripped and reprobed using anti-tubulin antibodies (lanes 6–10). In lanes 1 and 6, total lysates are shown
To provide further evidence for the interaction of PABP1 with PCBP and 3CD, we performed mobility shift assays using the 5′-terminal cloverleaf RNA as a probe. Interactions between the cloverleaf, PCBP, and 3CD control the regulation of both viral translation and RNA synthesis. Experiments have demonstrated that PCBP and 3CD specifically interact with different stem–loops of the poliovirus cloverleaf structure. The cloverleaf RNA has been shown to form low-affinity complexes with either PCBP or 3CD alone. However, PCBP and 3CD together form a stable ternary RNP complex Andino et al. 1993, Parsley et al. 1997, Silvera et al. 1999. The estimated dissociation constant for the ternary complex was calculated to be ∼1 nM. As expected, the formation of the ternary complex was not detected at low concentrations (0.5 nM) of MBP-PCBP and H6–3CD (Figure 4C, lane 5). However, a high-affinity complex was observed by the inclusion of PABP1-H6 in the binding reaction (lane 8). PABP1-H6 by itself does not interact with the cloverleaf RNA (Figure 4C, lanes 1–4), and an excess of bovine serum albumin (BSA) did not induce the formation of the high-affinity complex (Figure 4C, lanes 9–12). Furthermore, the complex formed by only 3CD and MBP-PCBP runs slightly faster than the complex formed by 3CD, MBP-PCBP, and PABP (Figure 4D). In summary, these results indicate that PABP1 directly interacts with PCBP and 3CD, that the proteins can interact with each other while bound to their cognate RNAs, and that PABP1 can induce the formation of a stable complex at the 5′ end of the genomic RNA.
PABP1 Interacts In Vivo with 3CD
Next, we determined whether PABP1 interacts with 3CD in intact cells. HeLa cells were infected with poliovirus and incubated for 5 hr at 37°C. Cells were harvested and treated with dithiobis(succinimidylpropionate) (DSP), a reversible, cell membrane–permeable cross-linker that induces ester formation of the primary amines of proteins within a range of 12 Å. After washing, cells were lysed, and cytoplasmic extracts were subjected to immunoprecipitation using antibodies directed against PABP1. Immunoprecipitates were heated to 100°C in the presence of dithiothreitol (DTT) to reverse the crosslinking and analyzed by western blotting using 3D-specific monoclonal antibodies. As shown in Figure 4, Figure 3CD coimmunoprecipitated with PABP1 (Figure 4D, lane 2). Coimmunoprecipitation of 3CD was not mediated by an RNA bridge but rather through a direct PABP1–3CD protein–protein interaction because RNase A treatment did not reduce the amount of 3CD coimmunoprecipitated (Figure 4D, lane 4). In the absence of DSP, we were able to detect small amounts of coimmunoprecipitated 3CD, suggesting that the interaction of PABP1 with 3CD is relatively stable (Figure 4D, lanes 3 and 5). The cross-linking of PABP1 with 3CD was specific because after DSP treatment, the abundant cellular protein α-tubulin did not coimmunoprecipitate with PABP1 (Figure 4D, lanes 7–10). Thus, PABP1 interacts with 3CD both in vitro and in vivo in intact poliovirus-infected cells.
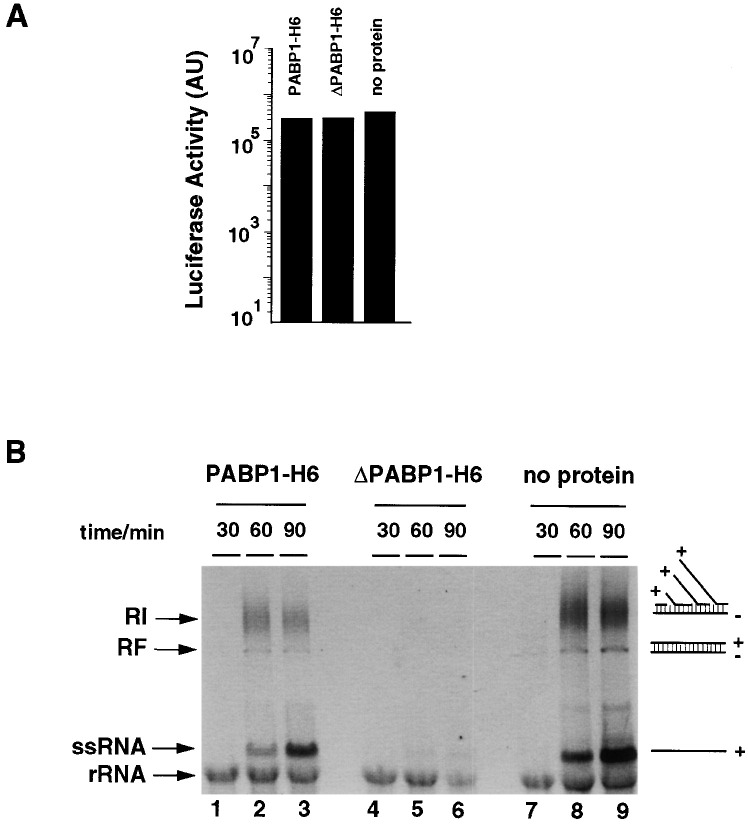
ΔPABP1 Inhibits Negative Strand Synthesis In Vitro
As shown above, ΔPABP1 with a large C-terminal deletion is able to bind efficiently to poly(A) but does not interact with 3CD and PCBP. To investigate whether the interaction of PABP1 with 3CD and PCBP is necessary for RNA synthesis, we examined the effect of recombinant ΔPABP-H6 on translation and RNA synthesis using the cell-free replication system. A replicon RNA with a long poly(A) tail (rib[+]RLuc-A80) was incubated with 500 nM of purified recombinant ΔPABP1-H6 or full-length recombinant PABP1 for 5 min and then used to program the translation/replication cell-free system. While the recombinant proteins had no detrimental effect on translation (Figure 5A) , the addition of ΔPABP1-H6 completely abrogated RNA replication (Figure 5B, lanes 4–6). In contrast, the addition of recombinant PABP1-H6 did not block replication (Figure 5B, lanes 1–3). Taken together, these results strongly suggest that ΔPABP acts as a dominant-negative mutant by disrupting the interaction of PABP1 with PCBP and/or 3CD and that this interaction is required for initiation of poliovirus RNA synthesis.
Figure 5.
ΔPABP1 Inhibits Negative Strand RNA Synthesis In Vitro
(A) Recombinant PABP1-H6 and ΔPABP1-H6 do not effect translation in vitro. Replicon RNA was preincubated with 500 nM of recombinant protein for 5 min and then used to program a translation/replication extract. Luciferase activity was measured at 3 hr.
(B) Recombinant ΔPABP1-H6 inhibits negative strand RNA synthesis. Preinitiation complexes were isolated from the reactions described above, and RNA synthesis was monitored by the incorporation of radiolabel in newly synthesized RNA. Samples were taken after 30, 60, and 90 min, and the RNA was isolated and analyzed on a 1% native agarose gel
It has been reported that PABP1 is proteolytically cleaved during picornavirus infection by viral proteases Joachims et al. 1999, Kerekatte et al. 1999. We have not yet determined whether proteolytically cleaved PABP1 is capable of supporting the initiation of negative strand RNA synthesis. However, cleavage of PABP1 is not detectable until 4 hr postinfection, around the peak of maximum RNA synthesis, and at 8 hr postinfection, when RNA synthesis has ceased, more than 30% of PABP1 remains uncleaved in the infected cell (our unpublished data). These results are in accordance with previous findings (Joachims et al., 1999) and demonstrate that cleavage of PABP1 occurs only late in the RNA replication cycle.
Discussion
Negative stranded RNA synthesis is initiated at the 3′ end of the genome of positive stranded RNA viruses. Depending on the virus group, two distinct RNA structures located at the 3′ ends of these viruses have been proposed to participate in RNA replication: a poly(A) tail (e.g., in the picorna-, α, corona-, or potexviruses) or a stable, often t-RNA-like secondary structure (e.g., in the flavi- and bromoviruses; reviewed by Buck, 1996). It has been shown for a few viruses that the length of the poly(A) tail is critical for efficient replication Sarnow 1989, Raju et al. 1999, Tsai et al. 1999, Spagnolo and Hogue 2000. However, if the poly(A) tail directly was the sole major determinant of initiation of negative strand RNA synthesis, it is difficult to understand how the viral replication apparatus discriminates between cellular mRNAs and the viral genomic RNA.
In this study, we have provided evidence that the poly(A) tail is indeed critical for organizing proteins around the start site of negative strand RNA synthesis. However, part of the initiation complex assembles at the 5′ end of the genome, 7500 nucleotides upstream from the start site. We also demonstrated that RNP complexes formed at the 5′ and 3′ ends of the genomic RNA are able to interact with each other via a protein bridge that could effectively circularize the genome, and we propose that the formation of this supercomplex is an essential step for the initiation of negative strand RNA synthesis.
The Ribonucleoprotein Complex Formed around the Cloverleaf RNA and Its Role in Negative Strand RNA Synthesis
The cloverleaf RNA appears to be involved in the synthesis of both positive and negative stranded RNA. Previously, we have shown that several point mutations within the cloverleaf RNA reduced both positive and negative strand RNA synthesis, with negative strand RNA synthesis being less affected, resulting in a 5- to 10-fold decrease in the ratio of plus to minus strands (Andino et al., 1990a). These results implied that mutations within the cloverleaf abrogated positive strand RNA synthesis and/or stability (Andino et al., 1990a). Recently, we have shown that the 3CD–cloverleaf interaction downregulates translation to yield ribosome-free templates that could be replicated, thus implying that the cloverleaf played a role in coordinating the use of the genomic RNA for translation or RNA replication (Gamarnik and Andino, 1998). The experiments presented here suggest that the cloverleaf also plays an important direct role in negative strand RNA synthesis, as we have now shown that deletion of the entire cloverleaf results in complete inhibition of negative strand RNA synthesis. More importantly, inhibition of translation with puromycin does not promote negative strand RNA synthesis of a cloverleaf-deficient genome (Figure 1C).
Since 3CD binds to the RNA through the 3C proteinase RNA binding domain (Andino et al., 1990b), an additional role of 3Cpro may be analogous to that of transcription factors which recruit PolII RNA polymerase to the cellular transcription promoters. The cloverleaf is a cis active promoter element necessary for initiation of negative strand RNA synthesis to which the transcription factor binds and recruits the RNA polymerase. Thus, the cloverleaf RNA could play a central role in regulating the usage of the genome; it facilitates or inhibits translation depending on which proteins it interacts with, and it directs the synthesis of both negative and positive strand RNA (Figure 6) . Interestingly, it has been previously shown that an RNA structure at the 5′ end of BVDV (bovine viral diarrhea virus), a pestivirus, modulates translation and replication of the positive stranded genome (Yu et al., 2000). Thus, this structure could play a role homologous to that of the cloverleaf RNA in BVDV replication.
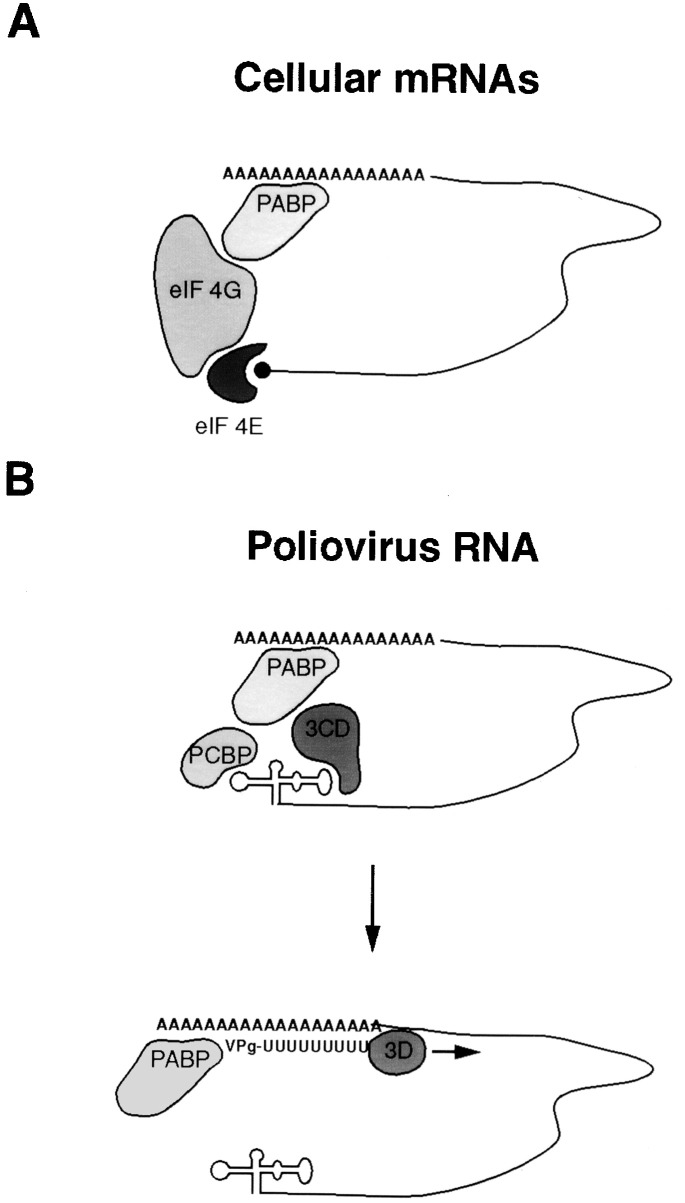
Figure 6.
Circularization of Cellular and Viral RNAs
(A) Circularization of eukaryotic mRNA during translation. PABP1 and eIF4E bind to the 3′ and 5′ ends of a capped and polyadenylated RNA, respectively (Wells et al., 1998).
(B) A model for initiation of negative strand RNA synthesis during poliovirus replication. After translation of the poliovirus polyprotein, the viral polymerase containing polypeptide 3CD binds, together with the cellular factor PCBP, to the 5′ cloverleaf, thus downregulating translation. Interactions between 3CD, PCBP, and PABP1 hold the 5′ and the 3′ end of the poliovirus RNA in a noncovalent juxtaposition that leads to the circularization of the genomic RNA. These interactions bring the viral polymerase in close proximity of the 3′ poly(A) tail and allow for the initiation of negative strand RNA synthesis
The Function of the Poly(A) Tail and PABP1
Our results suggest that, in order to reach their site of action during initiation of negative strand RNA synthesis, proteins bound to the 5′ cloverleaf interact with PABP1 bound to the 3′ poly(A) tail, thus circularizing the genomic RNA. It has been previously shown that enzymatic removal of the poly(A) tail from virion RNA renders replication of these RNAs incompetent (Spector et al., 1975), whereas translation was not affected. Indeed, our analysis of the minimal poly(A) length required for viral replication suggests that the high-affinity interaction of the poly(A) tail with PABP1 is more critical for efficient RNA replication than for IRES-dependent translation (although minor effects on translation have been observed in vivo; Figure 2D). PABP1 is an important regulatory factor involved in the control of cellular mRNA stability and translation (reviewed by Sachs and Wahle 1993, Sachs et al. 1997). It has been shown that PABP1 interacts with the translation initiation factor eIF4G (reviewed by Gingras et al., 1999), and because eIF4G forms a complex with the cap-recognizing eIF4E, this interaction probably leads to the effective circularization of cellular mRNAs (Figure 6A). Using purified homologous yeast proteins, this circularization has been recently visualized with atomic force microscopy (Wells et al., 1998).
For poliovirus RNA, circularization also appears to be mediated by PABP1, but in this case PABP1 interacts with PCBP and viral 3CD bound to the 5′ end of the RNA (Figure 6B). PCBP is a known component of an RNP complex that forms within the 3′ UTR of the human α-globin mRNA and determines its stability (Kiledjian et al., 1995). Within that complex, PCBP has been shown to interact with the C-terminal part of PABP1 (Wang et al., 1999). Thus, poliovirus uses assistance from established intracellular protein–protein and protein–RNA interactions to circularize its genomic RNA.
Circularization in Other Positive Stranded RNA Viruses?
The replication machinery of the positive stranded RNA bacteriophage Qβ appears also to utilize a circularization mechanism, but in this case circularization is accomplished through RNA–RNA interactions Klovins et al. 1998, Klovins and van Duin 1999. Flaviviruses also appear to circularize their genome utilizing RNA–RNA interactions (Hahn et al., 1987). For dengue virus, a member of the flavivirus family, initiation of negative strand RNA synthesis depends on these complementary sequences (You and Padmanabhan, 1999). Flaviviruses might employ a strategy similar to that described here for poliovirus, in which interactions between the 3′ and 5′ end cyclization elements bring the initiation complex close to the start site of negative stranded RNA synthesis.
For coronaviruses, intriguing data are beginning to accumulate that point to a similar mechanism. Because coronavirus genomes contain a 5′-terminal cap structure and a poly(A) tail at their 3′ ends, circularization in this case could be mediated by a PABP1–eIF4G–eIF4E interaction like that used by cellular mRNAs for efficient initiation of translation. Using coronavirus replicons, it was recently shown that a minimum length of the 3′ poly(A) tail is required for efficient replication. As with poliovirus, the minimal poly(A) tail length required for replication correlates with the length required for efficient PABP1 binding (Spagnolo and Hogue, 2000).
Recently, a host protein, Lsm1p, was identified that is required for efficient template selection during Brome mosaic virus replication in yeast (Diez et al., 2000). Interestingly, the function of this protein could be replaced by addition of a poly(A) tail to the otherwise unpolyadenylated genomic RNA. The authors propose that factors as yet unidentified binding to the 5′ and 3′ ends of the RNA communicate with each other in order to allow RNA replication.
Genome circularization and interaction of the replicase with the 5′ end may be a common feature of many, if not all, positive stranded RNA viruses, but the specific details of circularization are likely to vary from family to family (e.g., RNA–RNA interaction for flaviviruses, RNA–protein–protein–RNA interaction for picornaviruses, or cap–eIF4E–eIF4G–PABP1–poly(A) interaction for coronaviruses and α viruses). Genome circularization may provide several advantages for viral replication, including the coordination of translation and RNA synthesis, the localization of the viral polymerase at the appropriate start site, and a control mechanism for the integrity of the viral genome.
Experimental Procedures
Plasmids
The following plasmids were used to transcribe luciferase-expressing replicon RNA in vitro. Prib(+)RLuc, prib(−)RLuc, and pRLuc31 have been described previously Andino et al. 1993, Herold and Andino 2000. pRLuc24, a derivative of pRLuc31 lacking the sequences that encode for the 5′-terminal cloverleaf, was constructed using standard PCR procedures. Again, standard cloning techniques were used to construct luciferase expressing with defined poly(A) tail length. The sequences and restriction enzyme recognition sites introduced are outlined in Figure 2A.
To express proteins in bacteria, the following plasmids were used: pET3b-PABP-His, encoding for a C terminally His-tagged version of the human PABP1 protein (PABP1-H6), was kindly provided by N. Sonenberg. pET3b-ΔPABP-His, encoding for ΔPABP-H6, the first two RNA recognition motifs (RRM 1/2, amino acids 1–186) of human PABP1 fused to six consecutive histidine residues, was obtained by PCR mutagenesis as described previously (Herold et al., 1999). The construction of the plasmid pMal-PCBP2, encoding for a maltose binding protein–PCBP2 fusion protein (MBP-PCBP), has been described previously (Gamarnik and Andino, 1997). The His-tagged recombinant 3CD protein used in this study contains a catalytic site mutation (H40E) that abolishes proteolysis (Andino et al., 1993).
Blocking the 3′ End of In Vitro Transcripts with Cordycepin-5′-Triphosphate
Cordycepin (3′-desoxyadenosine, Sigma Chemical, St. Louis, MO) was added to the 3′ end of in vitro transcribed RNA using yeast poly(A) polymerase (United States Biochemical, Cleveland, OH) as recommended by the manufacturer.
The incorporation efficiency was tested using [α-P32]Co-TP (NEN, Boston, MA). Co-TP-treated (100 ng) and untreated RNA were incubated for 60 min at 37°C with 10 μCi cordycepin-5′-triphosphate (10 μCi/μl) and analyzed by TCA precipitation. It was determined that at least 99% of the 3′ ends were blocked.
RNA Transfection
Human 293 cells were electroporated as described previously (Herold and Andino, 2000). GuHCl (Sigma Chemical) was added to the medium to a final concentration of 2 mM when indicated. At different time points, cells were lysed, and luciferase activity was determined as recommended (Promega, Madison, WI).
To isolate and quantify polyadenylated viral transcripts, 5 × 106 cells were transfected with 20 μg of in vitro synthesized viral RNA with or without poly(A) tails using the conditions described above. At 7 hr posttransfection, viral RNA was isolated using oligo dT25 DynaBeads (Oslo, Norway) and analyzed on a nondenaturing 1.5% agarose gel in 1 × TAE in the presence of ethidium bromide. Control experiments indicated that only those RNAs with poly(A) tails longer than 17 nucleotides are effectively isolated by this method. Importantly, if the transfected poliovirus RNAs were blocked at the 3′ end via incorporation of a single 3′ deoxyadenylate, we failed to detect polyadenylated poliovirus genomes.
Translation/Replication in Cell-Free Extracts
Preparation of HeLa cell S10 extracts and translation initiation factors has been described in detail (Barton et al., 1995). Negative and positive strand RNA synthesis were analyzed as previously described (Herold and Andino, 2000).
Protein Expression and Purification
pET3b-PABP-His, pET3b-ΔPABP-His, and H6–3CD were transformed into BL21(DE3) bacteria. The bacteria growth, lysis, and protein Ni-NTA superflow column purification were preformed as recommended by the manufacturer (Qiagen, Valencia, CA). MBP-PCBP has been expressed and purified as described previously (Silvera et al., 1999). After purification, proteins were dialysed twice against 500 volumes of 40 mM HEPES (pH 8.0), 120 mM potassium acetate, 5.5 mM magnesium acetate, 10 mM potassium chloride, 6 mM DTT, 10% glycerol (PABP1-H6, Δ-PABP1, MBP-PCBP) or 40 mM HEPES (pH 8.0), 120 mM potassium acetate, 5.5 mM magnesium acetate, 400 mM potassium chloride, 6 mM DTT, 10% glycerol (H6–3CD) and frozen in aliquots at 80°C.
Mobility Shift Assays
The generation of the 5′-terminal cloverleaf RNA probe has been described earlier (Andino et al., 1990a). PCR products encoding a T7 RNA polymerase promotor, the complete poliovirus 3′ UTR, and a variable amount of adenylate residue at the 3′ end were used as templates to generate 3′ end–specific RNA probes. The RNA–protein complexes were analyzed as previously described (Andino et al., 1990a). The radioactivity in the complexes was quantified using a STORM 860 system and ImageQuaNT software (Molecular Dynamics, Sunnyvale, CA).
Dissociation constants were determined by quantifying the fraction of RNA bound (θ) with a PhosphoImager (Molecular Dynamics). The data were fitted by nonlinear least squares as a function of total PABP1-H6 concentration: (θ) = [PABP]/[PABP] Kd.
Protein Interaction In Vitro
Poly(A)-Sepharose slurry (100 μl; Amersham Pharmacia, Uppsala, Sweden) was incubated for 1 hr at 4°C with 2 μg of recombinant PABP1-H6, ΔPABP1-H6, or MPB-PCBP. The resin was then washed with buffer D (5 mM HEPES [pH 7.0], 75 mM potassium chloride, 2 mM magnesium chloride, 0.5% NP-40, 5% glycerol, 2 μg/μl tRNA). For 3CD, 100 μl of poly(A)-Sepharose slurry was incubated with recombinant H6–3CD for 10 min at 4°C in 100 μl buffer E (5 mM HEPES [pH 7.0], 150 mM potassium chloride, 2 mM magnesium chloride, 0.5% NP-40, 5% glycerol, 2 μg/μl tRNA). After three washes with 10 volumes of the respective buffer, proteins were eluted from the slurry using SDS-containing protein sample buffer. Eluted proteins were subjected to SDS-polyacrylamide gel electrophoresis and subsequent Western blotting with PCBP- or 3CD-specific antisera.
In Vivo Cross-Linking
Cross-linking of proteins in intact cells using DSP (Lomant's reagent; Pierce, Rockford, IL) has been done essentially as described (Grundhoff et al., 1999). Briefly, 107 HeLa cells were infected with poliovirus at an MOI of 10 and incubated at 37°C. At 5 hr p.i., the cells were washed twice with phosphate-buffered saline (PBS). The cells were then incubated at 4°C in either PBS or PBS/1 mM DSP for 30 min. Cells were then washed twice with PBS and lysed with 1% NP-40, and the cytoplasmic fraction was subjected to immunoprecipitation using PABP1-specific antiserum in the presence or absence of RNase A (1 μg/μl). Proteins were eluted from the beads, and cross-linking was reversed by heating in protein sample buffer containing 100 mM DTT to 100°C for 5 min and analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting with 3CD or α-tubulin (T5168, Sigma) antibodies.
Acknowledgements
We are grateful to Shane Crotty and Judith Frydman for useful comments on the manuscript and to N. Sonenberg for the gift of pET-PABP1-H6. J. H. is supported by a grant from the Deutsche Akademie der Naturforscher Leopoldina (BMBF-LPD 9801-2). This work was also supported by a Public Health Service grant (AI 40085) to R. A.
References
- Andino R., Rieckhof G.E., Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. a. [DOI] [PubMed] [Google Scholar]
- Andino R., Rieckhof G.E., Trono D., Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5′ noncoding region. J. Virol. 1990;64:607–612. doi: 10.1128/jvi.64.2.607-612.1990. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Rieckhof G.E., Achacoso P.L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′- end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D.J., Black E.P., Flanegan J.B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D.J., Morasco B.J., Flanegan J.B. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 1996;275:35–57. doi: 10.1016/s0076-6879(96)75005-x. [DOI] [PubMed] [Google Scholar]
- Barton D.J., Morasco B.J., Flanegan J.B. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 1999;73:10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck K.W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo R.C., Bonanno J.B., Sonenberg N., Burley S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- Diez J., Ishikawa M., Kaido M., Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J.B., Petterson R.F., Ambros V., Hewlett N.J., Baltimore D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc. Natl. Acad. Sci. USA. 1977;74:961–965. doi: 10.1073/pnas.74.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gorlach M., Burd C.G., Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA- binding specificity. Exp. Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- Grundhoff A.T., Kremmer E., Tureci O., Glieden A., Gindorf C., Atz J., Mueller-Lantzsch N., Schubach W.H., Grasser F.A. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 1999;274:19136–19144. doi: 10.1074/jbc.274.27.19136. [DOI] [PubMed] [Google Scholar]
- Hahn C.S., Hahn Y.S., Rice C.M., Lee E., Dalgarno L., Strauss E.G., Strauss J.H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Herold J., Andino R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Siddell S.G., Gorbalenya A.E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+- binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims M., Van Breugel P.C., Lloyd R.E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerekatte V., Keiper B.D., Badorff C., Cai A., Knowlton K.U., Rhoads R.E. Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M., Wang X., Liebhaber S.A. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovins J., van Duin J. A long-range pseudoknot in Qbeta RNA is essential for replication. J. Mol. Biol. 1999;294:875–884. doi: 10.1006/jmbi.1999.3274. [DOI] [PubMed] [Google Scholar]
- Klovins J., Berzins V., van Duin J. A long-range interaction in Qbeta RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA. 1998;4:948–957. doi: 10.1017/s1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U., Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein–protein interaction. J. Mol. Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- Larsen G.R., Dorner A.J., Harris T.J., Wimmer E. The structure of poliovirus replicative form. Nucleic Acids Res. 1980;8:1217–1229. doi: 10.1093/nar/8.6.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.F., Nomoto A., Detjen B.M., Wimmer E. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A., Paul A.V., Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A., Ostareck D.H., Hentze M.W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- Parsley T.B., Towner J.S., Blyn L.B., Ehrenfeld E., Semler B.L. Poly (rC) binding protein 2 forms a ternary complex with the 5′- terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Paul A.V., van Boom J.H., Filippov D., Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Kaplan G., Racaniello V.R., Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E.V., Poperechny K.V., Maslova S.V., Melchers W.J., Slot H.J., Agol V.I. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (`kissing') interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- Racaniello V.R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc. Natl. Acad. Sci. USA. 1981;78:4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R., Hajjou M., Hill K.R., Botta V., Botta S. In vivo addition of poly(A) tail and AU-rich sequences to the 3′ terminus of the Sindbis virus RNA genome: a novel 3′-end repair pathway. J. Virol. 1999;73:2410–2419. doi: 10.1128/jvi.73.3.2410-2419.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohll J.B., Percy N., Ley R., Evans D.J., Almond J.W., Barclay W.S. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohll J.B., Moon D.H., Evans D.J., Almond J.W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A., Wahle E. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- Sachs A., Davis R.W., Kornberg R.D. A similar domain of yeast poly (A)–binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow P., Hentze M.W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D., Gamarnik A.V., Andino R. The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′-untranslated region and acts as an inhibitor of viral translation. J. Biol. Chem. 1999;274:38163–38170. doi: 10.1074/jbc.274.53.38163. [DOI] [PubMed] [Google Scholar]
- Simoes E.A., Sarnow P. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J. Virol. 1991;65:913–921. doi: 10.1128/jvi.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo J.F., Hogue B.G. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.H., Villa-Komaroff L., Baltimore D. Studies on the function of polyadenylic acid on poliovirus RNA. Cell. 1975;6:41–44. doi: 10.1016/0092-8674(75)90071-9. [DOI] [PubMed] [Google Scholar]
- Todd S., Towner J.S., Brown D.M., Semler B.L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D., Pelletier J., Sonenberg N., Baltimore D. Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science. 1988;241:445–448. doi: 10.1126/science.2839901. [DOI] [PubMed] [Google Scholar]
- Tsai C.H., Cheng C.P., Peng C.W., Lin B.Y., Lin N.S., Hsu Y.H. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. J. Virol. 1999;73:2703–2709. doi: 10.1128/jvi.73.4.2703-2709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Day N., Trifillis P., Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Yogo Y., Wimmer E. Polyadenylic acid at the 3′-terminus of poliovirus RNA. Proc. Natl. Acad. Sci. USA. 1972;69:1877–1882. doi: 10.1073/pnas.69.7.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S., Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999;274:33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- Yu H., Isken O., Grassmann C.W., Behrens S.E. A stem-loop motif formed by the immediate 5′ terminus of the bovine viral diarrhea virus genome modulates translation as well as replication of the viral RNA. J. Virol. 2000;74:5825–5835. doi: 10.1128/jvi.74.13.5825-5835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]