Highlights
-
•
Development of a tool to track viral RNA in living cells.
-
•
Tick-borne encephalitis virus as a model system to study Flaviviral RNA dynamics.
-
•
FRAP and FLIP to measure the dynamic properties of viral RNA.
Keywords: Flavivirus, RNA, Replication, Innate immunity, Live imaging, FRAP, FLIP
Abstract
Flaviviruses include a wide range of important human pathogens delivered by insects or ticks. These viruses have a positive-stranded RNA genome that is replicated in the cytoplasm of the infected cell. The viral RNA genome is the template for transcription by the virally encoded RNA polymerase and for translation of the viral proteins. Furthermore, the double-stranded RNA intermediates of viral replication are believed to trigger the innate immune response through interaction with cytoplasmic cellular sensors. Therefore, understanding the subcellular distribution and dynamics of Flavivirus RNAs is of paramount importance to understand the interaction of the virus with its cellular host, which could be of insect, tick or mammalian, including human, origin. Recent advances on the visualization of Flavivirus RNA in living cells together with the development of methods to measure the dynamic properties of viral RNA are reviewed and discussed in this essay. In particular the application of bleaching techniques such as fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) are analysed in the context of tick-borne encephalitis virus replication. Conclusions driven by this approached are discussed in the wider context Flavivirus infection.
1. Introduction
Flaviviridae is a family of viruses causing severe diseases and mortality in humans and animals. The Flavivirus genus is the largest in the family and includes among the most important emerging viruses known to man, which are mostly transmitted by mosquitoes or ticks (arthropod-borne viruses), such as yellow fever virus (YFV), the dengue viruses (DV), Japanese encephalitis virus (JEV), West Nile virus (WNV) and tick-borne encephalitis virus (TBEV). Hepatitis C virus (HCV, genus Hepacivirus) is the unique blood-borne virus in the family [1]. HCV is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma (HCC) worldwide. Although these viruses belong to different genera with different biological properties, the members of this family share similarity in terms of virion morphology, genome organization and replication strategies [2], [3], [4], [5].
Flaviviruses are icosahedral enveloped 50 nm viruses with a RNA genome packaged by the viral capsid protein (C). The spherical nucleocapsid core of about 30 nm is covered by a host-derived lipid bilayer with two surface glycoproteins, membrane (M, which is expressed as prM, the precursor to M) and envelope (E), that have double-membrane anchors at the C-terminus. The Flavivirus genome is an 11-kilobase single-stranded RNA molecule of positive polarity that encodes a single long open reading frame (ORF). The ORF of all Flaviviruses is flanked by 5′ (about 100 nucleotides) and 3′ (400–700 nucleotides) untranslated regions (UTR) carrying RNA sequence motifs and secondary structures that function as cis-acting regulatory elements for genome amplification, translation or packaging [3]. Translation of the genome by the host cell machinery produces a long polyprotein precursor that is co- and post-translationally cleaved into at least 10 proteins. The N-terminal end of this polyprotein encodes the structural proteins (C-prM-E) followed by seven non-structural (NS) proteins (i.e. for TBEV: NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) each of which is an essential component of the viral replication complex (RC). Processing of the polyprotein is carried out by both host proteases and the viral protease NS3/NS4B in the lumen of the ER.
Flavivirus infection is initiated when mature viral particles attach to the target cell surface through interaction of the large glycoprotein E with cellular receptors. After attachment, the virus is internalized by receptor-mediated endocytosis and delivered to the endosome. The low pH in the endosomal compartment induces a conformational change in the surface protein E that triggers the fusion of the viral and host cell membrane [6], [7]. This process results in the release of the nucleocapsid and viral RNA into the cell cytoplasm. At this stage, the uncoated genome can be translated by the cell machinery to generate the polyprotein precursor. Processing of the polyprotein by host and viral proteases then leads to the generation of the individual viral proteins and to the initiation of genome replication. The viral RNA-dependent RNA polymerase (RdRp) NS5 copies complementary minus-strand RNA from genomic RNA, which then serves as template for the synthesis of new positive strand viral RNAs. Viral RNA synthesis is asymmetric, with the plus-sense RNA synthesized in excess over minus-sense RNA [8], [9], [10]. The newly synthesized plus-sense RNA is subsequently: (i) used for translation of further viral proteins; (ii) used for the synthesis of additional minus-sense RNA; (iii) incorporated as genomic viral RNA into new viral particles. Hence, the viral RNA has at least three different functions (translation, replication, and association with nascent viral particles), which need to be tightly regulated and coordinated during the viral replication cycle. More recently, an abundant non-coding RNA derived from incomplete degradation of the viral 3′UTR by the cellular 5′-3′ exonuclease Xrn1 and produced by all Flaviviruses (termed sfRNA for subgenomic flaviviral RNA) was reported to be required for viral pathogenicity, possibly by regulating the interferon response (see below) [11], [12], [13].
Viruses are obligate intracellular parasites, therefore they must exploit the host cell machinery to efficiently replicate and produce infectious progeny. Flaviviruses usurp and modify cytoplasmic membranes in order to build functional sites of protein translation, processing and RNA replication [5], [14], [15], [16], [17], [18], [19]. These sites, enriched in cellular membranes, viral RNA and virus- and host-encoded proteins, are generally defined as replication complexes. Membrane wrapping of the RC is thought to provide a physical framework in which RNA synthesis can occur and to ensure protection from host-response proteins recognizing the viral RNA. Recently, elegant three-dimensional EM tomography studies have shown that Flaviviruses such as DENV, TBEV and WNV share the property of forming vesicles of approximately 80 nm of diameter as invaginations towards the lumen of the ER bearing necked connections to the cytoplasm [19], [20], [21]. At variance, HCV induces the formation of double-membrane extrusions from the ER membrane, probably with the same purpose of protecting the viral RC [22], [23].
Mammalian cells have evolved a variety of defence mechanisms to detect, contain and clear viral infections. There are two fundamentally different types of responses to invading pathogens: the innate and the acquired immune response. The innate immune response offers the first early protection against foreign invaders and is mediated by a limited number of pattern-recognition receptors (PRRs). In contrast, acquired immunity is implicated in pathogens clearance during the late phase of the infection and long-term protection, which involves lymphocytes (T and B cells) clonally expressing a large repertoire of rearranged antigen-specific receptors. An effective innate immune response to viral infection involves two phases: an early phase of interferon production, triggered by the recognition of conserved “non-self” signatures, also known as pathogen-associated molecular patterns (PAMPs), by host PRRs, and a later phase of IFN signalling and interferon stimulated genes (ISGs) expression [24]. Indeed, upon recognition, PRRs initiate signalling cascades that result in the activation of transcription factors critical for type I interferons (INFα and IFNβ) expression. Thereafter, as secreted factors, type I IFNs can regulate a variety of immune responses through interaction with the type I IFN receptor. These responses include induction of a protective antiviral state in the infected and neighbouring cells as well as initiation of the acquired immunity. To date, two distinct families of sensors have been characterized as key players in the detection of RNA viruses: the Toll-like receptor (TLR) and the RIG-I (retinoic acid inducible gene-I)-like receptor (RLR) families [25], [26], [27], [28], [29], [30]. RNA viruses are specifically sensed by the intracellular TLRs such as TLR3, TLR7 and TLR8, which recognise dsRNA and ssRNA [31], [32]. Whereas TLRs detect viruses-derived nucleic acids within intracellular compartments of specific cell types, such as dendritic cells and macrophages, RLRs sense viral components that are present in the cytoplasm of most infected cells. The RLR family is composed of three members: RIG-I, melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) [33], [34]. RIG-I and MDA5 are DExD/H-containing RNA helicases with two caspase activation and recruitment domains (CARD), which are essential for the interaction with the IFNβ promoter stimulator (IPS)-1 adaptor protein (also known as mitochondrial anti-viral signalling protein (MAVS)). Interestingly, IPS-1 is localized on specialized ER membranes associated to mitochondria or peroxisomes, suggesting a critical function of these organelles as a signalling platform for antiviral innate immunity [35], [36], [37], [38]. IPS1 activation then associates with tumour necrosis factor (TNF) receptor-associated factor (TRAF) 3 leading to TBK1 and inhibitor of kB kinase (IkB) ε (IKKε) activation and subsequent phosphorylation of the IRF3 transcription factor. Alternatively, IPS-1 recruits the adaptor Fas-associated death domain (FADD) and the kinases receptor- interacting protein 1 (RIP1) in order to trigger the NF-kB pathway. Upon activation, IRF3 and NF-kB translocate to the nucleus to drive type I IFN transcription and subsequent induction of the antiviral state [39].
Originally, both RIG-I and MDA5 were thought to sense cytoplasmic dsRNA during viral infection [40]. However, through numerous studies, it has been clearly demonstrated that, despite their structural and functional similarities, the two sensors are not redundant in their ability to recognize non-self RNA [41]. Poly (I:C) as well as chemically synthesized RNA oligonucleotides annealed to a complementary strand trigger RIG-I [42], [43]. RNAs carrying a 5′-triphosphate (5′-PPP) moiety, generally produced during infection by influenza and other negative-strand RNA viruses, are RIG-I agonists as well [44], [45]. Long, possibly branched dsRNAs found for example in picornaviruses [45] and mRNAs lacking 2′-O-methylation at their 5′ cap structure [46], [47] are MDA5 agonists. Total RNA extracted from virally infected cells can also stimulate specific RLRs. However, the form of viral RNA that is recognized depends on the specific virus. Concerning Flaviviridae, while RIG-I knockout mice demonstrated increased susceptibility to JEV infection compared to control mice, MDA5 deficient mice responded normally to infection [41]. Furthermore, siRNA-mediated knock out RIG-I, but not MDA5, affected TBEV-mediated induction of interferon [48]. In contrast, WNV and DV were shown to induce both RLRs-dependent pathways of PAMP recognition. Consistently, cells lacking RIG-I or MDA5 were not able to properly counteract viral infection [49], [50], [51]. Interestingly, temporal regulation of PRR appears to take place in WNV, with early activation of RIG-I and a later role for MDA5 [52]. Altogether, these studies suggest that RIG-I plays a critical early role in establishing effective immune responses to all Flaviviruses, whereas MDA5 role appear to be virus-dependent and acting at later stages. However, the real Flavivirus RNA structure that is recognized by cellular PRRs has not been identified so far and only few reports investigated this topic in the context of the whole cell [53].
As mammalian hosts have evolved several sensors for viral infection, viruses, on the other hand, adapted multiple tricks to escape or at least counteract innate immune response. These can be distinguished in those that target PRR signalling, thus delaying the first induction of IFNs, and those that target IFNs signalling, thus limiting their antiviral potential [54]. Typically, evasion of IFN induction is accomplished by viral proteins that directly inhibit the function of PRRs. For example, the NS1 viral protein is the main IFN antagonist of influenza A viruses [55]. NS1 acts both by preventing the nucleation of the IFN enhanceosome [56], [57], [58] and by targeting RIG-I activity [59]. The HCV NS3/4A protease cleaves IPS-1, the adaptor molecule that bridges RLRs to downstream effectors, affecting type I IFN production in infected cells [60], [61], [62], [63]. In a similar way, DV protease was shown to cleave and degrade STING and, as mentioned before, DV sfRNA binds TRIM25 to inhibit interferon induction [13], [64]. Passive mechanisms of immune evasion have also been proposed. Concerning Flaviviruses, the main mechanism appears to be related to the sequestration of the replication intermediates in vesicles of the ER to avoid PRR activation and delay interferon induction [19], [20], [21], [48], [65], [66]. These vesicles are believed to be able to extrude newly replicated viral RNA to the cytosol, but are impenetrable to cellular PRRs until the content gets eventually exposed later during the infection. Such delay of interferon activation allows these viruses to replicate to sufficient levels before the antiviral pathway is activated.
2. Overview of the method
2.1. Visualization of RNA in living cells
Labelling either by fusion with a fluorescent protein or with a fluorescent-dye moiety allows the dynamic study of a protein of interest in live cells. Imaging of nucleic acids in living cells is instead a less common technique since the DNA or RNA targets are typically not accessible and only recently non-invasive labelling approaches became available. So far, much of our knowledge about intracellular RNA dynamics came from either in situ hybridization of fixed cells or from biochemical fractionation of sub-cellular components. However, these methods can only provide a static picture at the time of fixation or fractionation of the sample and are certainly not compatible for the real-time monitoring of nucleic acids in live cells. Labelling of chromosomes in living cells can be achieved by fluorescent tagging of chromosome binding factors. Usage of the lac operator (lacO) DNA sequence that is inserted into the DNA of interest provides an elegant way to label specific regions of nuclear chromatin, which could be visualized by the expression of a fluorescently tagged lac repressor (lacI) fusion protein, which specifically binds to lacO sequence [67]. This approach has been used to study chromatin organization, chromosome dynamics and even genomes from DNA viruses [68], [69], [70], [71]. More recently, in addition to their applications in genome editing, programmable DNA recognition systems, such as CRISPR/Cas9 and TALEN, have been exploited for the visualization of endogenous genomic elements in living cells [72].
RNA can be conjugated to fluorescent dyes and directly microinjected into the cells. However, this is a rather invasive procedure [73]. Santangelo et al. provided an interesting method for short-term imaging of viral RNA in live cells [74]. To this end they delivered labelled RNA probes conjugated to streptavidin into cells reversibly permeabilized with streptolysin to detect non-engineered human respiratory syncytial virus RNA. Several other techniques have been developed which include the use of fluorescently labelled probes, the use of molecular “beacons” which are active only when bound to target RNA or the use of fluorescently tagged RNA-binding proteins [75], [76], [77]. However, few of those methods provide information about the behaviour of a single RNA molecule in real-time in living cells. Alternatively, researchers described several approaches based on the insertion of a series of RNA aptamers–stem–loops into the transcript of interest and detection by high affinity specific interaction of RNA stem-loops with a fluorescently labelled RNA binding protein [78], [79], [80], [81]. One of the most studied and versatile RNA reporter is based on the MS2 bacteriophage coat protein. MS2 binds specifically and avidly to an RNA stem–loop structure consisting of 19 nucleotides. The MS2-based system has been effectively used for tracking specific viral, cytoplasmic and nuclear mRNA in yeast, plants, flies and mammalian cells [71], [82], [83], [84], [85], [86], [87], [88], [89]. So far, RNA detection by the MS2 system coupled with live-cell imaging technologies represents the most powerful approach for RNA analysis in single cells in real time. However, novel methods of tracking RNA in living cells are continuously being developed. These include the ‘spinach’ approach [90], recent advances in the CRISPR/Cas9 for RNA targeting [91] and possibly click-chemistry approaches, which have proven successful for Coronavirus RNA detection [92]. Therefore, the possibility to track viral genomes by these methods is of great importance for better understanding virus-host interaction and exploring how RNA dynamics is correlated with protein or organelle dynamics during viral infection.
In our laboratory we successfully applied the MS2 tagging method to the study of HIV-1 transcriptional and post- transcriptional processes [85], [86], [87], [88], [93], [94]. Concerning Flaviviridae, we were able to visualize TBEV RNA in cells expressing MS2-EYFP transfected with a sub-genomic replicon carrying MS2 repeats [21], [48], [95], [96] (see below). The MS2 repeats were cloned in the 3′-NCR, within a region which has shown to be dispensable for virus replication in cell culture and upstream of the TBEV sfRNA [97], [98]. A minimum of 12× MS2 repeats was required to detect viral RNA. The choice of using replicons instead of full genomes was dictated principally by the requirement of a biosafety level 3 containment (BSL-3) for the manipulation of TBEV (BSL-4 in the US), thus limiting their use in live cell microscopy. Despite the success with TBEV, we failed to extend this technology to HCV. HCV sub-genomic replicons carrying MS2 repeats of various lengths in different positions of the untranslated genome were capable of replicating, but none could be visualized following expression of MS2-EYFP (Cevik RE and Marcello A unpublished observations). The most likely explanation is that MS2-EYFP binding to viral RNA disrupts the secondary structures required for viral replication resulting in cells that either express MS2-EYFP or replicate HCV RNA, but never both. Alternatively, levels of HCV genomic RNA could be lower than those of acute viruses such as TBEV, below the visualization threshold. Hence, not all Flaviviridae are amenable to the MS2-tagging method and the correct positioning of the MS2 repeats within the viral genome is critical for a successful outcome. Nevertheless, successful tracking of TBEV RNA should allow conclusions to be extrapolated at least to the Flavivirus genus in general.
2.2. Tools for imaging intracellular dynamics of Flavivirus RNA in living cells
The discovery and development of fluorescent proteins as molecular tags [99] combined with technical advances in the field of live-cell imaging has provided profound insight into how molecules are organized in cells and how they interact with each other and the cellular environment [100]. Cells that express proteins or nucleic acids tagged with these fluorescent proteins can be imaged with low light intensity over many hours and can provide useful information about changes in protein localization and steady-state level over time. Time-lapse imaging alone, however, cannot reveal the dynamic properties of a molecule, for example, whether the protein is immobilized to a scaffold, free to diffuse, or undergoing constant exchange between compartments. To obtain such information the combination of time-lapse imaging with photobleaching and photoactivation techniques is required. Photobleaching occurs when a fluorophore permanently loses its ability to fluoresce due to photon-induced chemical damage and covalent modification. This can occur through either repeated excitation/emission cycles or by exposure to a high intensity light from a laser beam. Photoactivation, on the other hand, works by converting molecules to a fluorescence state by using a brief pulse of high-intensity irradiation [100]. Both these techniques distinguish a selected pool of fluorescent proteins from other fluorescent proteins in the cells in order to monitor how they re-equilibrate. Results from fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) experiments can provide important quantitative information about protein diffusion rates, binding kinetics, and movements between cellular compartments.
2.2.1. Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) is a technique developed in the mid-1970s to study the diffusive properties of molecules in living cells [101]. Irreversible, or almost irreversible, photochemical bleaching of a fluorophore in a specific portion of the living cell, which contains mobile fluorescent molecules, is obtained by exposure to a brief and intense focused laser beam. A quantitative measure of fluorophore mobility in the region is obtained by the kinetic of recovery of fluorescence, which depends on the diffusion of fluorescent molecules from unirradiated parts of the system into the area of bleach and the diffusion of irradiated non-florescent molecules out of the area of bleach.
As already mentioned, analysis of fluorescence recovery can be used to determine the kinetic parameters of a protein, including its diffusion constant, mobile fraction, transport rate or binding/dissociation rate from other proteins [102]. When binding interactions are present, they retard a FRAP recovery in relation to what would be observed if only diffusion occurred. Indeed a bleached protein has to dissociate from its binding site before being free to leave the bleached area.
FRAP has now been adopted as a common technique for studying protein dynamics in the cytoplasm, nucleus, organelle lumens and membranes of living cells. Unconjugated GFP is considered a good standard marker of mobility in the absence of binding. It has been calculated that soluble GFP in the cytoplasm and in the nucleoplasm is highly mobile with diffusion rates of about 20 μm2/s [103], [104]. Diffusion of GFP within the ER lumen appeared instead to be three-to sixfold slower than GFP in the cytoplasm indicating that a greater viscosity characterizes this compartment [105]. FRAP has also unveiled important features of GFP-tagged membrane proteins. Many trans-membrane proteins localized in ER and Golgi compartments appeared to be highly mobile, with diffusion rates ranging from 0.2 to 0.5 μm2/s, which is near the theoretical limit for protein diffusion in a lipid bilayer [106], [107]. This suggested that these proteins are not immobilized but may instead retain lateral mobility in the membrane of these compartments. Consistently, ER associated HCV NS4B and NS5A proteins exhibited a high degree of mobility when expressed alone from a plasmid in the absence of viral replication [108], [109]. However, the mobility of the same proteins expressed from a sub-genomic replicon or analyzed within membrane associated foci (MAFs) appeared dramatically slower. Therefore, most likely, once viral proteins get incorporated into active replication foci they became restricted in their movements by interacting with other viral as well as host factors that are anchored to intracellular membranes. In keeping with these findings, large NS5A-GFP labelled structures, representing HCV-induced membranous webs, showed restricted mobility and displayed a static internal architecture with limited exchange of viral proteins within and between neighbouring RCs [110].
2.2.2. Fluorescence loss in photobleaching
Fluorescence loss in photobleaching (FLIP) is a complementary approach to FRAP most often employed when the continuity of a cell compartment or the mobility of a molecule within the whole compartment is examined. FLIP experiments are similar to FRAP experiments in that a region of the cell is subjected to photobleaching from intense laser light, but in the FLIP approach, the bleaching is repeated in between imaging scans. Over time this will result in the loss of the fluorescent signal in any compartment that is contiguous with the bleaching region or in any compartment in which the protein is freely mobile [104]. As example, when unconjugated GFP, which localizes both in the nucleus and in the cytoplasm of cells, is continuously bleached in the cytoplasm, a fast and uniform decay of fluorescence is observed in this compartment. To the contrary at the same time the nucleus loses its fluorescence much more slowly since the nuclear membrane acts as a diffusion barrier for GFP [111]. Therefore, if labelled molecules in certain subcellular compartments are not bleached, the clear implication is that these compartments do not exchange with the compartment being bleached.
3. Detailed protocols
3.1. Cells
Baby hamster kidney (BHK-21) and human osteosarcoma (U2OS) cell lines are grown under standard conditions in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% foetal bovine serum. For imaging of living cells it is essential that the cells are healthy, routinely checked for mycoplasma contamination and not passaged too long.
3.2. Lentiviral vector transductions
EYFP-MS2nls, EYFP-MS2, Cherry-MS2nls expression cassettes are inserted in lentiviral vectors (WPI-BLR obtained from Volker Lohman at the University of Heidelberg). Lentiviral vectors are obtained by cotransfection of the WPI-BLR based vectors with packaging constructs pCMVR8.91 and pMD.G (Addgene) in HEK 293T cells following standard procedures. The cell culture medium 48 h post-transfection is collected, pelleted at low speed, filtered (0.4 μm) and kept in aliquots in the freezer. Target cells are incubated with lentiviral vector preparations and selection medium for 48 h before analysis. This procedure allows a good level of signal in most cells and avoids the perturbations of cell transfection. It is recommended not to maintain the polyclonal population of transduced cells too long in culture, but rather to transduce fresh cells for each experiment.
3.3. Electroporation of TBEV sub-genomic replicons
The capped viral RNA is introduced into cells by electroporation of a subgenomic RNA replicon of the TBEV Western subtype prototypic strain Neudoerfl. Construct pTNd/ΔME_24 × MS2 has been generated by replacing the variable region of the 3′ non-coding region with 24 repeats of a stem–loop RNA structure (19 nucleotide each) specifically recognized by the MS2 bacteriophage coat protein, as described previously [96]. Capped RNA is synthesized with the m7GpppG Cap analogue and reagents from the T7 Megascript kit (Ambion). Template DNA (1 μg) is degraded by incubation with DNase for 15 min at 37 °C and the RNA is purified by phenol–chloroform extraction, precipitated with ethanol and washed with 70% ethanol. The pellet is then resuspended in RNase-free water. The correct size and integrity of the RNA is checked on denaturing agarose gels. RNA concentrations are measured spectrophotometrically. For transfection, subconfluent cells are collected with trypsin, washed once in growth medium and once in ice-cold PBS buffer. Aliquots of 5 × 106 cells are then resuspended in 500 μl ice cold PBS and mixed in a 0.4 cm cuvette with 10 μg of RNA and 5–10 μg of MS2-encoding plasmids together with other constructs such as GFP-RIG-I (kind gift from Takashi Fujita, Kyoto University). GFP-RIG-I was functional when co-transfected together with a reporter luciferase under the control of the IFNβ promoter (data not shown). The cells are electroporated with a Bio-Rad Gene Pulser apparatus applying two subsequent pulses (setup values of 0.25 kV and 500 μF) usually resulting in a time constant of 16 ms. After electroporation, cells are washed three times in complete growth medium without antibiotics and seeded in the same medium.
3.4. Live imaging, FRAP and FLIP
For the visualization of the viral RNA, BHK-21 or U2OS cells are transduced either with CherryMS2nls, expressing a hybrid protein composed by the core protein of the MS2 bacteriophage fused to Cherry and to a nuclear localization signal (nls), or with the MS2-EYFP construct with or without the nls. Choice of localization of the tagged MS2 depends on the kind of experiment in mind. Since TBEV RNA localizes in the cytoplasm associated to ER compartments, when monitoring RNA mobility also outside ER compartments it is suggested to use MS2 without the nls, while, when monitoring viral RNA within the ER compartments, MS2-nls allows detection against a dark background with a higher signal-to-noise ratio. Subsequently, cells are electroporated with the TBEV replicons’ RNA and plated on glass-bottom plates (MatTek, Ashland, MA, USA or similar) as described above. At the appropriate time point post-transfection, cells are transferred on a humidified and CO2-controlled on-stage incubator (PeCon GmbH, Erbach, Germany) at 37 °C in complete DMEM medium without phenol red for live cell imaging.
FRAP and FLIP analysis are performed by using the inverted META LSM510 confocal microscope (Zeiss, Jena, Germany) with an oil immersion 63 × objective (NA 1.4, Zeiss). For FRAP experiments images of 512 × 512 pixels (29.25 × 29.25 μm for BHK-21 cells; 48.75 × 48.75 μm for U2OS cells) and optical thickness of 1 μm are acquired using 1% or less of the power of the 514 nm laser line. EYFP and GFP are bleached at 514 nm and 488 nm respectively (Argon laser, maximum output 500 MilliWatt) in a circle of 30 pixels of diameter, at full laser power, for 10 passages. For FLIP measurements images are acquired as described above, but bleaching is repeated at every acquisition. EYFP is less photostable and is bleached more efficiently than GFP, making it more suitable for techniques such as FRAP and FLIP [112]. This was experimentally tested on fixed cells exposed in different conditions (data not shown).
Images are analyzed with ImageJ (Rasband, W.S., ImageJ, National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/). For the mathematical analysis of the data experimental recovery curves (for FRAP) or loss of fluorescence curves (for FLIP) from at least 10 cells are normalized and averaged [88], [113].
4. Experimental examples
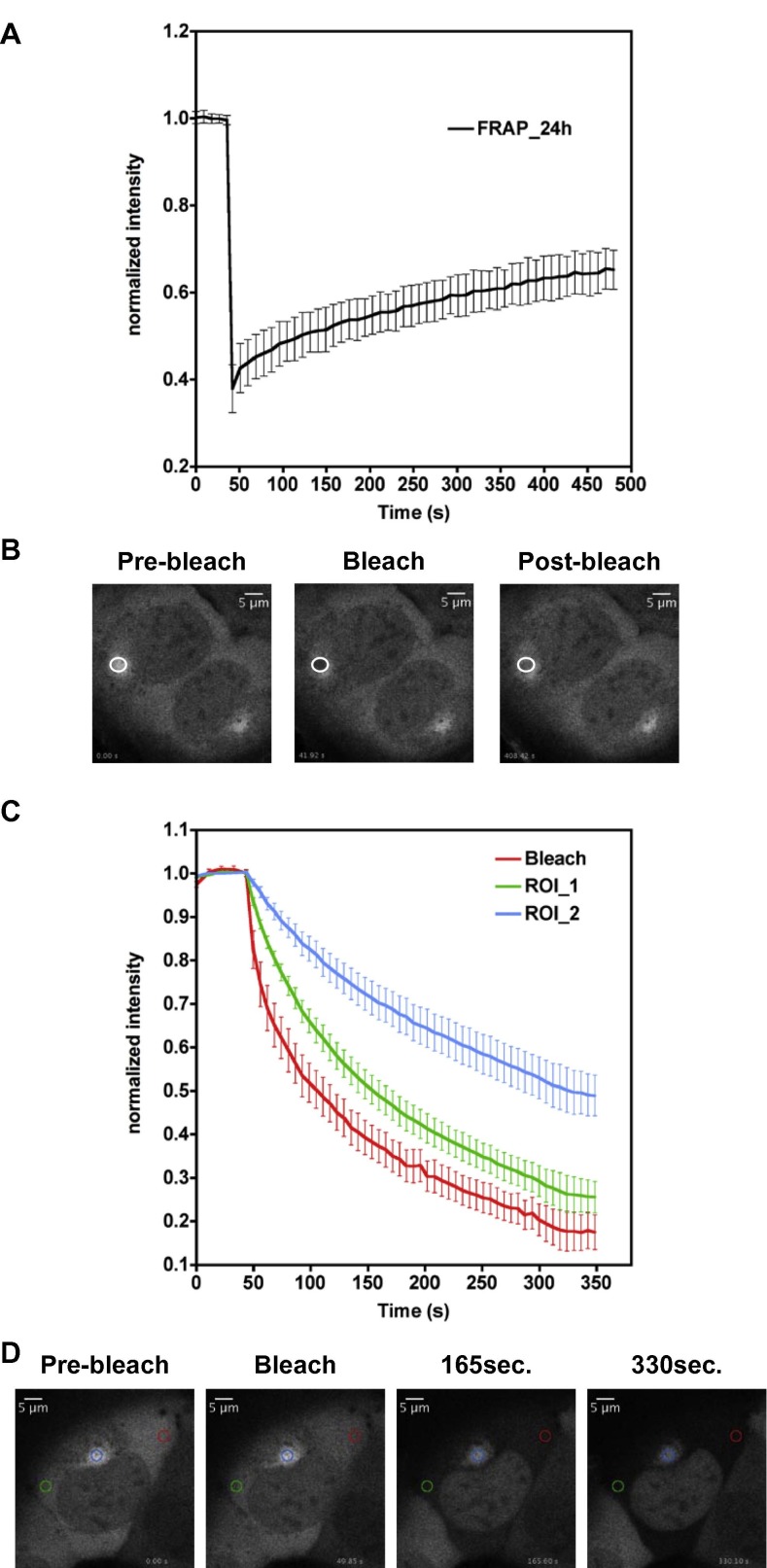
The RNA of Flaviviruses can be found within a cell in four states: (+)RNA, (−)RNA, dsRNA and sfRNA as well as associated to viral proteins during entry, uncoating, replication and packaging or bound to cellular proteins that regulate viral processes or sense exogenous RNA to trigger an innate immune response. Localization varies in time following infection: from the incoming virion to the ER vesicles and extra-vesicular space, to the newly formed virions. The ability of visualizing specific viral RNAs and their partners and their sub-cellular localization in living cells replicating a Flavivirus has been the aim of our studies. To this end we tagged sub-genomic TBEV replicons with the MS2 method and tracked viral RNA in living cells. By positioning the MS2 RNA binding sites in antisense orientation we originally hoped to be able to track also (−)RNA. However, we were not able to visualize such constructs, but only (+)RNA, which localized to the perinuclear ER. Since the (−)RNA template is much less abundant than the replicated (+)RNA, the MS2-EYFP signal would be either too small to be visible above background or not accessible to MS2, either being protected by the replication complex or being present only in a double-stranded conformation. Replication-dependent production of TBEV (+)RNA rapidly (within hours post replicon transfection) captures the MS2 tagged with EYFP present in the cytoplasm to form clusters of EYFP signal above background. The viral RNA is continuously synthesized by the viral polymerase and should be able to diffuse in the cytoplasm in complex with EYFP-MS2 unless bound to an immobile structure or limited in its movements by physical barriers. To test this hypothesis we took advantage of FRAP. As shown in Fig.1 A and B, U2OS cells expressing the TNd/ΔME_24 × MS2 replicon RNA and EYFP-MS2 were subjected to FRAP analysis at 24 h post-electroporation. The curve shows an initial recovery that quickly tails off and never reaches pre-bleach values, which is indicative of the presence of a relevant immobile fraction, as it has been observed for BHK21 cells [21]. The initial portion of the recovery curve represents the unbound EYFP-MS2 that remains free to diffuse in the bleached area. We could conclude that during TBEV replication, the amount of freely mobile EYFP-MS2 is reduced, being sequestered by the increasing amount of replicated RNA. To test this hypothesis we performed the FLIP experiment shown in Fig.1C and D. We bleached continuously an area distant from the MS2-enriched perinuclear compartment and measured loss of fluorescence at different sites: (i) in the area of bleach, (ii) in a different location in the cytoplasm as well as (iii) within perinuclear regions of EYFP-MS2 accumulation. After each bleaching, the intensity of fluorescence was measured in the locations mentioned above. According to the principle of FLIP, each mobile fluorescent molecule that diffuses at the bleaching ROI will be irreversibly bleached. Therefore, areas of where the fluorescent proteins are freely mobile will be losing fluorescence quickly, whereas regions where these proteins are not mobile, either because irreversibly bound or secluded into membrane compartments, will resist bleaching. This kind of measurements clearly showed that fluorescent TBEV RNA-bound EYFP-MS2 is depleted less efficiently than freely diffusible EYFP-MS2. These FRAP and FLIP data are only selected examples of the experiments that helped us demonstrate that replicated TBEV (+)RNA is maintained within an extra-vesicular compartment of the ER, as confirmed by electron-microscopy studies [21]. Progeny viral (+)RNAs are synthesized within the lumen of virus-induced vesicles and are then extruded through the pore into the cytoplasmic extravesicular space. Once released into this area, viral RNAs are available for downstream assembly into new viral particles that bud back into the lumen of ER cisternae of infected cells. Alternatively, these RNAs may be engaged in further rounds of translation and replication.
Fig. 1.
FRAP and FLIP analysis of TBEV RNA dynamics in U2OS cells. (A) U2OS cells transduced with MS2-EYFP were electroporated with TNd/ΔME_24 × MS2 sub-genomic RNA. After 24 h the fluorescence recovery of the MS2-EYFP protein in the area of bleaching was analyzed (white circles). The graph shows values of fluorescence intensity normalized to the pre-bleach values and corrected for the loss of fluorescence due to the imaging procedure. Data represent the average of acquisitions from 10 cells ± standard deviation. (B) Image sequence from the FRAP experiment described in (A) (48,75 × 48,75 μm). Times were collected before bleaching (pre-bleach, 0 s.), immediately after the bleaching (bleach, 42 s.) and at 408 s. after the bleaching event (post-bleach). (C) U2OS cells were electroporated as described in (A) and after 24 h viral RNA release from the replication compartment was monitored by FLIP. The graph compares the loss in fluorescence intensity within the three different ROIs (Bleach; red circle in (D), ROI_1; green circle in (D), and ROI_2; blue circle in (D)) analyzed. Data are normalized as described in (A) and represent the average of acquisitions from 10 cells ± standard deviation. (D) Image sequence from the FLIP experiment described in (C) (48,75 × 48,75 μm). The region for bleaching (red circle) was chosen in the cytoplasm away from the clustered TBEV RNA. Loss of fluorescence was measured in the cytoplasm both within (blue circle, ROI_2) and outside (green circle, ROI_1) the replication compartment.
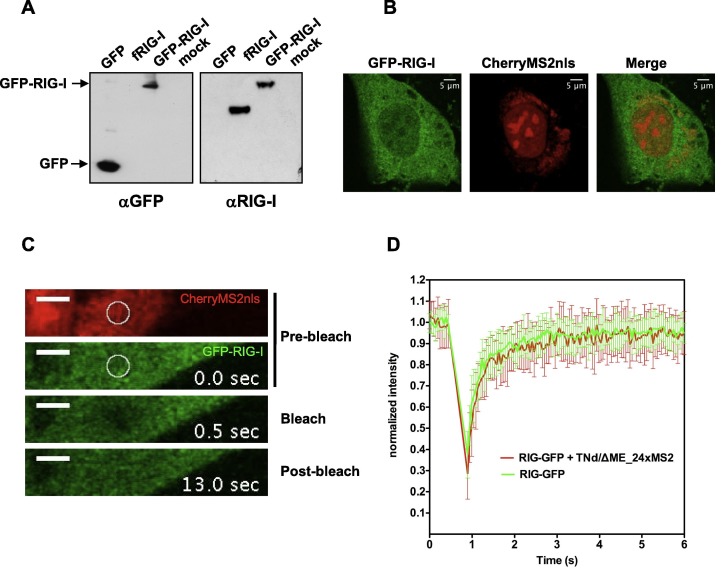
Some cellular proteins are certainly able to access this extra-vesicular space to bind the viral (+)RNA as we demonstrated for the TIA-1/TIAR proteins, which shuttle between stress-granules (SG) and viral RNA [114]. However, we became intrigued by the accessibility of this compartment to a PRR like RIG-I. Therefore we performed a FRAP experiment using RIG-I fused to GFP (Fig. 2 ). To be sure that we were tracking the full-length fusion protein, and not only GFP from a degradation product, we immunoblotted extracts from cells with antibodies against GFP or RIG-I. As shown in Fig.2A, GFP-RIG-I is expressed as a full-length protein product. Localization of GFP-RIG-I didn’t seem to be affected by TBEV replication (Fig.2B). Mobility of GFP-RIG-I within the compartment, defined by Cherry-MS2nls in TNd/ΔME_24 × MS2 transfected BHK21 cells (Fig.2C), didn’t differ significantly from the free mobility of the protein in the cytoplasm of mock-transfected cells (compare red and green curves in Fig.2D). Interestingly, we also couldn’t observe any increase of the immobile fraction in the recovery of GFP-RIG-I in cells transfected with TNd/ΔME_24 × MS2 at 24 hpe. At this time point IRF3 is already being translocated to the nucleus and IFNβ is activated [48]. Therefore, either the fraction of activated GFP-RIG-I is too low to be significantly detected in the FRAP experiment, or RIG-I is activated in another cellular location, or activation doesn’t affect mobility of the protein significantly. This latter point would be in contrast to the formation of a protein complex with IPS-1. Further experiments are needed to clarify these points by conjugating live imaging with in situ hybridization and biochemical assays.
Fig. 2.
FRAP of RIG-I in TBEV replication compartments. (A) Immunoblot of whole cell extracts expressing GFP-RIG-I. Antibodies where against GFP (left panel) or RIG-I (right panel) position of full-length GFP-RIG-I and GFP are indicated. Flag-tagged RIG-I was used as control (fRIG-I) because endogenous RIG-I is not sufficiently expressed in non-induced cells. (B) Representative image of BHK-21 cells transfected with GFP-RIG-I and CherryMS2nls in the presence of the TBEV replicating RNA. (C) Image sequence from the FRAP experiment described in (D) (36,56 × 7,14 μm). Top, pre-bleach stacks in both channels (0 s.). The circle indicates the area of bleach chosen in the RC marked by CherryMS2nls. Middle, time point immediately after the bleaching event (0,5 s.); bottom, post-bleach stack (13 s.). (D) Analysis of GFP-RIG-I mobility within the replication compartment. BHK-21 cells were electroporated with CherryMS2nls and with a GFP-RIG-I expressing plasmid both in the presence (red line, GFP-RIG-I + TNd/ΔME_24 × MS2) and in the absence (green line, GFP-RIG-I) of the TBEV replicon RNA. The choice of transfecting tagged MS2 instead of using lentivectors was dictated by the necessity of co-expressing both MS2 and RIG-I at the same time in the same cells. After 24 h GFP kinetics was investigated by FRAP. In the graph the recovery curves of the GFP protein in the two different experimental conditions are compared. The values of fluorescence intensity are normalized to the pre-bleach values and corrected for the loss of fluorescence due to the imaging procedure. Data represent the average of acquisitions from 10 cells ± standard deviation.
5. Concluding remarks
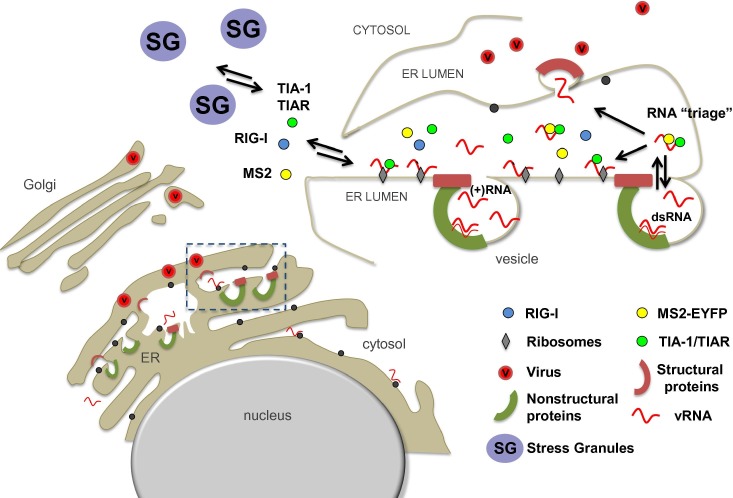
Subcellular localization and dynamics of viral RNAs is critical both for the successful replication of Flaviviruses and for the induction of innate immunity. By exploiting the MS2-tagged replicon system, we could combine structural information with dynamic data of newly replicated TBEV-RNA within functional intracellular compartments, providing new insights into the spatiotemporal organization of Flavivirus replication compartments (see Fig. 3 ). This approach could be extended to other members of the Flaviviridae and to RNA viruses in general.
Fig. 3.
Schematic model of the dynamic exchange of proteins and viral RNA between replication compartments and the cytosol. Upon infection, the viral (+)RNA genome associates with the rough ER (ribosomes are indicated in dark grey) and the viral polyprotein, composed of structural (pink) and non-structural (green) proteins. Invaginations of the ER membrane are induced, leading to the formation of 80 nm vesicles that are connected with the cytosol by a pore [21]. Inside vesicles the viral RNA is replicated through a dsRNA intermediate. Newly generated (+)RNA is extruded from the vesicles into an extra-vesicular compartment where it is either translated, or further replicated or packaged (RNA “triage”). Packaging occurs on the ER membrane and new virus buds into the ER lumen. The extra-vesicular compartment is connected to the cytosol and accessible to proteins such as MS2-EYFP or TIA-1/TIAR, which bind the viral RNA to regulate viral translation and shuttle between the extra-vesicular compartment and stress granules (SG) [114]. Also RIG-I is freely diffusible into this compartment as shown in Fig. 2. This diagram is not drawn to scale.
Acknowledgments
Work on Flaviviridae in AM lab is supported by Beneficentia Stiftung and FLAVIPOC, a Regional grant (Regione FVG, Italy).
References
- 1.Choo Q.L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay S., Kuhn R.J., Rossmann M.G. Nat. Rev. Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 3.Lindenbach B.D., Thiel H.J., Rice C.M. In: Fields Virology, Lippincott. Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., editors. Williams & Wilkins; Philadelphia: 2007. pp. 1101–1152. [Google Scholar]
- 4.Gould E.A., Solomon T. Lancet. 2008;371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Garcia M.D., Mazzon M., Jacobs M., Amara A. Cell Host Microbe. 2009;5:318–328. doi: 10.1016/j.chom.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Heinz F.X., Allison S.L. Adv. Virus Res. 2003;59:63–97. doi: 10.1016/s0065-3527(03)59003-0. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan M.N., Sukumaran B., Pal U., Agaisse H., Murray J.L., Hodge T.W., Fikrig E. J. Virol. 2007;81:4881–4885. doi: 10.1128/JVI.02210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu P.W., Westaway E.G. Virology. 1985;140:68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 9.Cleaves G.R., Ryan T.E., Schlesinger R.W. Virology. 1981;111:73–83. doi: 10.1016/0042-6822(81)90654-1. [DOI] [PubMed] [Google Scholar]
- 10.Wengler G., Gross H.J. Virology. 1978;89:423–437. doi: 10.1016/0042-6822(78)90185-x. [DOI] [PubMed] [Google Scholar]
- 11.Pijlman G.P., Funk A., Kondratieva N., Leung J., Torres S., van der Aa L., Liu W.J., Palmenberg A.C., Shi P.Y., Hall R.A., Khromykh A.A. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Bidet K., Dadlani D., Garcia-Blanco M.A. PLoS Pathog. 2014;10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manokaran G., Finol E., Wang C., Gunaratne J., Bahl J., Ong E.Z., Tan H.C., Sessions O.M., Ward A.M., Gubler D.J., Harris E., Garcia-Blanco M.A., Ooi E.E. Science. 2015 doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackenzie J. Traffic. 2005;6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.den Boon J.A., Diaz A., Ahlquist P. Cell Host Microbe. 2010;8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller S., Krijnse-Locker J. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahlquist P. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novoa R.R., Calderita G., Arranz R., Fontana J., Granzow H., Risco C. Biol. Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillespie L.K., Hoenen A., Morgan G., Mackenzie J.M. J. Virol. 2010 doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miorin L., Romero-Brey I., Maiuri P., Hoppe S., Krijnse-Locker J., Bartenschlager R., Marcello A. J. Virol. 2013;87:6469–6481. doi: 10.1128/JVI.03456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero-Brey I., Bartenschlager R. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero-Brey I., Merz A., Chiramel A., Lee J.Y., Chlanda P., Haselman U., Santarella-Mellwig R., Habermann A., Hoppe S., Kallis S., Walther P., Antony C., Krijnse-Locker J., Bartenschlager R. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goubau D., Deddouche S., e Sousa C. Reis. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai T., Akira S. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Lo M.K., Tilgner M., Bernard K.A., Shi P.Y. J. Virol. 2003;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel J.R., Garcia-Sastre A. eLife. 2014;3:e02369. doi: 10.7554/eLife.02369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel J.R., Garcia-Sastre A. Cytokine Growth Factor Rev. 2014;25:513–523. doi: 10.1016/j.cytogfr.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Nazmi A., Dutta K., Hazra B., Basu A. Virus Res. 2014;185:32–40. doi: 10.1016/j.virusres.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Suthar M.S., Aguirre S., Fernandez-Sesma A. PLoS Pathog. 2013;9:e1003541. doi: 10.1371/journal.ppat.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akira S., Uematsu S., Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Akira S., Takeda K. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 33.Pichlmair A., e Sousa C. Reis. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Loo Y.M., Gale M., Jr. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixit E., Boulant S., Zhang Y., Lee A.S., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M., Nibert M.L., Superti-Furga G., Kagan J.C. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seth R.B., Sun L., Ea C.K., Chen Z.J. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., Chen Z.J. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 39.Yoneyama M., Fujita T. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 40.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 41.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., Yamaguchi O., Otsu K., Tsujimura T., Koh C.S., e Sousa C. Reis, Matsuura Y., Fujita T., Akira S. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 42.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. Mol. Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 44.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., Endres S., Hartmann G. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 45.Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., e Sousa C. Reis. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Jr., Shi P.Y., Diamond M.S. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miorin L., Albornoz A., Baba M.M., D’Agaro P., Marcello A. Virus Res. 2012;163:660–666. doi: 10.1016/j.virusres.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 49.Fredericksen B.L., Gale M., Jr. J. Virol. 2006;80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fredericksen B.L., Keller B.C., Fornek J., Katze M.G., Gale M., Jr. J. Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., Garcia-Sastre A., Katze M.G., Gale M., Jr. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Errett J.S., Suthar M.S., McMillan A., Diamond M.S., Gale M., Jr. J. Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Triantafilou K., Vakakis E., Kar S., Richer E., Evans G.L., Triantafilou M. J. Cell Sci. 2012;125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 54.Suthar M.S., Gale M., Jr., Owen D.M. Cell. Microbiol. 2009;11:880–888. doi: 10.1111/j.1462-5822.2009.01311.x. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Sastre A., Egorov A., Matassov D., Brandt S., Levy D.E., Durbin J.E., Palese P., Muster T. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 56.Ludwig S., Wang X., Ehrhardt C., Zheng H., Donelan N., Planz O., Pleschka S., Garcia-Sastre A., Heins G., Wolff T. J. Virol. 2002;76:11166–11171. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talon J., Horvath C.M., Polley R., Basler C.F., Muster T., Palese P., Garcia-Sastre A. J. Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X., Li M., Zheng H., Muster T., Palese P., Beg A.A., Garcia-Sastre A. J. Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gack M.U., Albrecht R.A., Urano T., Inn K.S., Huang I.C., Carnero E., Farzan M., Inoue S., Jung J.U., Garcia-Sastre A. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin R., Lacoste J., Nakhaei P., Sun Q., Yang L., Paz S., Wilkinson P., Julkunen I., Vitour D., Meurs E., Hiscott J. J. Virol. 2006;80:6072–6083. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loo Y.M., Owen D.M., Li K., Erickson A.K., Johnson C.L., Fish P.M., Carney D.S., Wang T., Ishida H., Yoneyama M., Fujita T., Saito T., Lee W.M., Hagedorn C.H., Lau D.T., Weinman S.A., Lemon S.M., Gale M., Jr. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 64.Aguirre S., Maestre A.M., Pagni S., Patel J.R., Savage T., Gutman D., Maringer K., Bernal-Rubio D., Shabman R.S., Simon V., Rodriguez-Madoz J.R., Mulder L.C., Barber G.N., Fernandez-Sesma A. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Overby A.K., Popov V.L., Niedrig M., Weber F. J. Virol. 2010;84:8470–8483. doi: 10.1128/JVI.00176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uchida L., Espada-Murao L.A., Takamatsu Y., Okamoto K., Hayasaka D., Yu F., Nabeshima T., Buerano C.C., Morita K. Sci. Rep. 2014;4:7395. doi: 10.1038/srep07395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tumbar T., Sudlow G., Belmont A.S. J. Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukamoto T., Hashiguchi N., Janicki S.M., Tumbar T., Belmont A.S., Spector D.L. Nat. Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- 70.Fraefel C., Bittermann A.G., Bueler H., Heid I., Bachi T., Ackermann M. J. Virol. 2004;78:389–398. doi: 10.1128/JVI.78.1.389-398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., Spector D.L. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen B., Huang B. Methods Enzymol. 2014;546:337–354. doi: 10.1016/B978-0-12-801185-0.00016-7. [DOI] [PubMed] [Google Scholar]
- 73.Ainger K., Avossa D., Morgan F., Hill S.J., Barry C., Barbarese E., Carson J.H. J. Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santangelo P.J., Lifland A.W., Curt P., Sasaki Y., Bassell G.J., Lindquist M.E., Crowe J.E., Jr. Nat. Methods. 2009;6:347–349. doi: 10.1038/nmeth.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tyagi S., Kramer F.R. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 76.Politz J.C., Browne E.S., Wolf D.E., Pederson T. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6043–6048. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui Z.Q., Zhang Z.P., Zhang X.E., Wen J.K., Zhou Y.F., Xie W.H. Nucleic Acids Res. 2005;33:3245–3252. doi: 10.1093/nar/gki629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 79.Daigle N., Ellenberg J. Nat. Methods. 2007;4:633–636. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- 80.Lange S., Katayama Y., Schmid M., Burkacky O., Brauchle C., Lamb D.C., Jansen R.P. Traffic. 2008;9:1256–1267. doi: 10.1111/j.1600-0854.2008.00763.x. [DOI] [PubMed] [Google Scholar]
- 81.Chen J., Grunwald D., Sardo L., Galli A., Plisov S., Nikolaitchik O.A., Chen D., Lockett S., Larson D.R., Pathak V.K., Hu W.S. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E5205–E5213. doi: 10.1073/pnas.1413169111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fusco D., Accornero N., Lavoie B., Shenoy S.M., Blanchard J.M., Singer R.H., Bertrand E. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shav-Tal Y., Darzacq X., Shenoy S.M., Fusco D., Janicki S.M., Spector D.L., Singer R.H. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darzacq X., Shav-Tal Y., de Turris V., Brody Y., Shenoy S.M., Phair R.D., Singer R.H. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Molle D., Maiuri P., Boireau S., Bertrand E., Knezevich A., Marcello A., Basyuk E. Retrovirology. 2007;4:36. doi: 10.1186/1742-4690-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boireau S., Maiuri P., Basyuk E., de la Mata M., Knezevich A., Pradet-Balade B., Backer V., Kornblihtt A., Marcello A., Bertrand E. J. Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Marco A., Dans P.D., Knezevich A., Maiuri P., Pantano S., Marcello A. Amino Acids. 2010;38:1583–1593. doi: 10.1007/s00726-009-0378-9. [DOI] [PubMed] [Google Scholar]
- 88.Maiuri P., Knezevich A., Bertrand E., Marcello A. Methods. 2011;53:62–67. doi: 10.1016/j.ymeth.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 89.Maiuri P., Knezevich A., De Marco A., Mazza D., Kula A., McNally J.G., Marcello A. EMBO Rep. 2011;12:1280–1285. doi: 10.1038/embor.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Strack R.L., Jaffrey S.R. Methods Enzymol. 2015;550:129–146. doi: 10.1016/bs.mie.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Connell M.R., Oakes B.L., Sternberg S.H., East-Seletsky A., Kaplan M., Doudna J.A. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hagemeijer M.C., Vonk A.M., Monastyrska I., Rottier P.J., de Haan C.A. J. Virol. 2012;86:5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Marco A., Biancotto C., Knezevich A., Maiuri P., Vardabasso C., Marcello A. Retrovirology. 2008;5:98. doi: 10.1186/1742-4690-5-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dieudonne M., Maiuri P., Biancotto C., Knezevich A., Kula A., Lusic M., Marcello A. EMBO J. 2009;28:2231–2243. doi: 10.1038/emboj.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoenninger V.M., Rouha H., Orlinger K.K., Miorin L., Marcello A., Kofler R.M., Mandl C.W. Virology. 2008;377:419–430. doi: 10.1016/j.virol.2008.04.035. [DOI] [PubMed] [Google Scholar]
- 96.Miorin L., Maiuri P., Hoenninger V.M., Mandl C.W., Marcello A. Virology. 2008;379:64–77. doi: 10.1016/j.virol.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 97.Gehrke R., Heinz F.X., Davis N.L., Mandl C.W. J. Gen. Virol. 2005;86:1045–1053. doi: 10.1099/vir.0.80677-0. [DOI] [PubMed] [Google Scholar]
- 98.Rouha H., Thurner C., Mandl C.W. Nucl. Acids Res. 2010 doi: 10.1093/nar/gkq681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsien R.Y. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 100.Lippincott-Schwartz J., Patterson G.H. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 101.Axelrod D., Koppel D.E., Schlessinger J., Elson E., Webb W.W. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lippincott-Schwartz J., Altan-Bonnet N., Patterson G.H. Nat. Cell Biol. 2003;(Suppl.):S7–S14. [PubMed] [Google Scholar]
- 103.Swaminathan R., Hoang C.P., Verkman A.S. Biophys. J. 1997;72:1900–1907. doi: 10.1016/S0006-3495(97)78835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lippincott-Schwartz J., Snapp E., Kenworthy A. Nat. Rev. Mol. Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 105.Dayel M.J., Hom E.F., Verkman A.S. Biophys. J. 1999;76:2843–2851. doi: 10.1016/S0006-3495(99)77438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cole N.B., Smith C.L., Sciaky N., Terasaki M., Edidin M., Lippincott-Schwartz J. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- 107.Nehls S., Snapp E.L., Cole N.B., Zaal K.J., Kenworthy A.K., Roberts T.H., Ellenberg J., Presley J.F., Siggia E., Lippincott-Schwartz J. Nat. Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- 108.Gretton S.N., Taylor A.I., McLauchlan J. J. Gen. Virol. 2005;86:1415–1421. doi: 10.1099/vir.0.80768-0. [DOI] [PubMed] [Google Scholar]
- 109.Jones D.M., Gretton S.N., McLauchlan J., Targett-Adams P. J. Gen. Virol. 2007;88:470–475. doi: 10.1099/vir.0.82363-0. [DOI] [PubMed] [Google Scholar]
- 110.Wolk B., Buchele B., Moradpour D., Rice C.M. J. Virol. 2008;82:10519–10531. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wustner D., Solanko L.M., Lund F.W., Sage D., Schroll H.J., Lomholt M.A. BMC Bioinformatics. 2012;13:296. doi: 10.1186/1471-2105-13-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shaner N.C., Steinbach P.A., Tsien R.Y. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 113.Phair R.D., Misteli T. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 114.Albornoz A., Carletti T., Corazza G., Marcello A. J. Virol. 2014;88:6611–6622. doi: 10.1128/JVI.03736-13. [DOI] [PMC free article] [PubMed] [Google Scholar]