Significance
Life dissipates energy far from thermodynamic equilibrium via electron transport systems that are coupled to external sources of oxidants and reductants. Biological electron transport, in turn, is catalyzed by a suite of enzymes that comprise the superfamily of oxidoreductases. The origin of oxidoreductases is enigmatic. Comparing protein topology and the sequence of modern oxidoreductases, we deduce a putative common ancestor that may have existed at the earliest stages of metabolism. Through duplication, recruitment of other proteins, and diversification, this ancestral protein may have evolved to facilitate electron transfer and redox catalysis at a very early stage in the origin of metabolism.
Keywords: electron transfer, biological networks, ferredoxin, flavodoxin, Rossmann fold
Abstract
Life on Earth is driven by electron transfer reactions catalyzed by a suite of enzymes that comprise the superfamily of oxidoreductases (Enzyme Classification EC1). Most modern oxidoreductases are complex in their structure and chemistry and must have evolved from a small set of ancient folds. Ancient oxidoreductases from the Archean Eon between ca. 3.5 and 2.5 billion years ago have been long extinct, making it challenging to retrace evolution by sequence-based phylogeny or ancestral sequence reconstruction. However, three-dimensional topologies of proteins change more slowly than sequences. Using comparative structure and sequence profile-profile alignments, we quantify the similarity between proximal cofactor-binding folds and show that they are derived from a common ancestor. We discovered that two recurring folds were central to the origin of metabolism: ferredoxin and Rossmann-like folds. In turn, these two folds likely shared a common ancestor that, through duplication, recruitment, and diversification, evolved to facilitate electron transfer and catalysis at a very early stage in the origin of metabolism.
All life on Earth is driven by electron transfer (i.e., redox) reactions. Indeed, biological redox reactions unify metabolisms across the tree of life (1). These reactions are catalyzed by a suite of enzymes that comprise the superfamily of oxidoreductases (Enzyme Classification EC1). However, the origin(s) and evolution of these proteins remain enigmatic. Most modern oxidoreductases are complex in their structure and chemistry. Logically, the extant oxidoreductases must have evolved from a small set of ancient folds, which became increasingly complex via repeated gene duplication, recruitment, and diversification events (2–4) either from a universal common ancestor or from several independent origins.
Ancient oxidoreductases from the Archean Eon between ca. 4.0 and 2.5 billion years ago have been long extinct. Their antiquity makes it challenging to retrace evolution by sequence-based phylogeny or ancestral sequence reconstruction. However, three-dimensional (3D) topologies of proteins are relatively robust to variations in their amino acid sequence (5), providing an opportunity to reconstruct the origins of oxidoreductases in deep time. Metabolism of the last universal common ancestor (LUCA) has been proposed to include enzymes with both transition metals and organic cofactors (6, 7). As proteins diverge over time, the catalytic centers and metal coordination sites evolve more slowly than the rest of the fold (8, 9), making structural analyses of these regions particularly attractive for reconstructing their origins.
Previously, we published a network analysis of over 30,000 metal-coordination sites in high-resolution protein structures and discovered a small number of metal-binding modules that were recurrent across many proteins (10). Modules that bound transition metals often occurred in pairs or larger chains, making paths for electron transfer through the protein matrix. Connecting modules that putatively function in electron-transfer couples produced a single network that comprised nearly all metal-containing oxidoreductases. This network provided an insight into the evolution of protein structures underlying redox reactions in metabolism across the tree of life (11). The topology of this network strongly suggests that nearly all extant metal-binding modules arose from a small number of early building blocks and that functional connections between modules potentially indicate evolutionary history. In effect, the network approach strongly suggests that protein structural topology can be used to infer metabolic phylogeny and, hence, potentially the origin(s) of the engines of life.
For practical reasons, our previous analysis excluded the microenvironments of nontransition metal cofactors such as flavins within oxidoreductases; it was unclear whether including them would significantly influence the topology of the protein-wiring diagram. Nontransition metal cofactor sites are included in the current work, more than doubling the number of protein sites considered. Furthermore, we had yet to develop phylogenetic methods to test whether evolutionary inferences based on network topology were valid.
Here, using comparative structure and sequence profile-profile alignments, we quantify the evolutionary significance of similarity between proximal cofactor-binding folds and generate a more complete picture of electron transfer. While many features of the network are unchanged relative to the previous metal-only version, the addition of organic cofactors shows a transition from reducing modules at the center of the network to oxidizing modules at the periphery, strongly suggesting that the metabolic network across the tree of life has coevolved through Earth’s history with the changing redox conditions of the planet (12). At the center of the network are two recurring folds central to the origin of metabolism: ferredoxin (Fd) and Rossmann-like (flavodoxin) folds. In turn, these two folds potentially shared a common ancestor that, through duplication, recruitment, and diversification, evolved to facilitate electron transfer and catalysis at a very early stage in the origin of metabolism.
Results and Discussion
Identifying Oxidoreductase Modules.
To identify the building blocks of oxidoreductases, we applied a comparative structural alignment approach described previously (10) to a larger dataset of proteins encompassing both metal-containing and organic redox cofactors. Briefly, we isolated local cofactor-binding protein motifs (microenvironments) from deposited high-resolution structures from the Protein Data Bank (PDB) at http://wwpdb.org (13). Using the cofactor as a fiducial marker, we defined a spherical microenvironment within a 15-Å radius from the metal center (8) (SI Appendix, Fig. S1 and Dataset S1). For those cofactors without a transition metal, the center was chosen based on the geometric center of mass of the redox-active component of the prosthetic group. A simpler metric for comparative structural alignments was used relative to the previous work. Here, pairs of microenvironments were aligned and clustered based on a backbone structural similarity of <0.1 Å rmsd per residue and the overlap of cofactors in the alignment. Each cluster of microenvironments, referred to as a “module,” represent a putative ancestral cofactor-binding fold.
The modified threshold for similarity and the addition of nonmetal cofactor-containing microenvironments to the analysis did not greatly redistribute the classification of structures into modules and instead resulted in more modules overall. Most modules contained either transition-metal or organic cofactors; only two, type 7 and type 105, included both cofactors. In many of the nucleoside-based cofactors, such as NAD(P) and FAD, the binding site was distinct and notably more conserved than the electron transfer site of the cofactor (SI Appendix, Fig. S2). Thus, we first analyzed both sites separately and subsequently merged the resulting clusters. The nucleoside-base module (type 7 or Rm) contained the most microenvironments: Rossmann-like folds (Rm) consist of alternating beta (β) strands and alpha (α) helices wherein the β‐strands form a parallel β‐sheet (14).
Constructing the Electron Transfer Network.
In the next step, a network of electron transfer pathways was constructed by connecting modules based on their spatial proximity using the same approach as previously applied to metal-only cofactor sites (10). Proximity in the Spatial-Adjacency Network (SpAN) (Fig. 1) was defined as a cofactor edge-to-edge distance of <14 Å (10), a distance that permits electron transfer on biologically relevant timescales (15). The addition of organic cofactor modules did not significantly alter the topology of the metal-only SpAN, but rather added additional connections to subgraphs consisting largely of the new organic cofactor modules. One notable difference was the rubredoxin module, which previously bridged the iron-symmerythrin and iron-sulfur modules. With the revised similarity threshold, this group split into the monovalent iron-containing rubredoxins and Reiske FeS-containing rubredoxin modules (modules 100 and 538, respectively). However, connectivity in the SpAN was largely preserved with both modules connected to symmerythrin and through the Rm module to ferredoxin. Type 538 connected with multiple FeS modules.
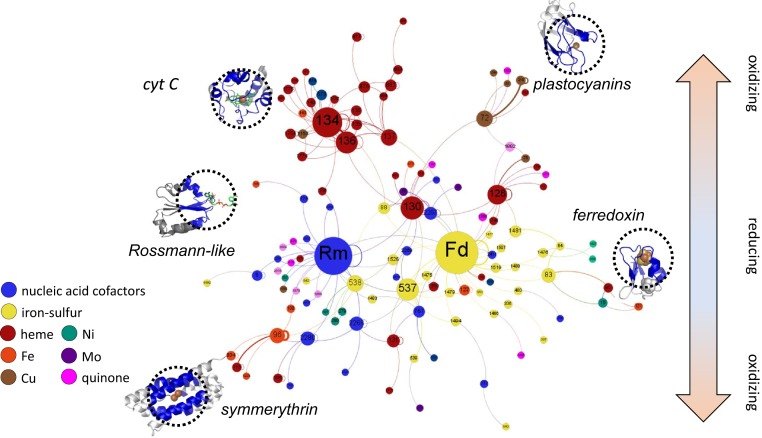
Fig. 1.
SpAN of the EC.1 proteins showing putative electron transfer paths in proteins. Nodes correspond to distinct modules based on structural similarity of microenvironments while the lines connecting nodes (or edges) represent adjacency in the protein. Node size corresponds to the number of electron-transfer–competent connections to other modules. Edge thickness reflects the number of connections in the dataset. Node color represents cofactor type. Examples of protein structures of key modules in the SpAN are shown with the cofactor microenvironment circled. See Dataset S2 for module composition of the SpAN. Rm, module 7; Fd, module 85.
Evolutionary Inference from the SpAN.
Signatures of metabolic pathway evolution found in the earlier SpAN of metalloproteins were also present in the current network containing both metal and organic cofactors. For example, self-connecting loops were likely a result of tandem module duplications resulting in an extended electron transport chain. Moreover, modules with similar cofactors colocalized in a connected subgraph within the SpAN. These features suggest a model of duplication and diversification in the evolution of oxidoreductases (10). Evident in the current SpAN were clear vectoral paths for electron transfer from lower potential ferredoxins and flavodoxins at the center to higher potential cytochromes and copper and iron enzymes at the periphery. This observation is consistent with a model of metabolic network evolution that parallels transitions from a mildly reducing to a more oxidizing planetary environment over geologic time (12, 16).
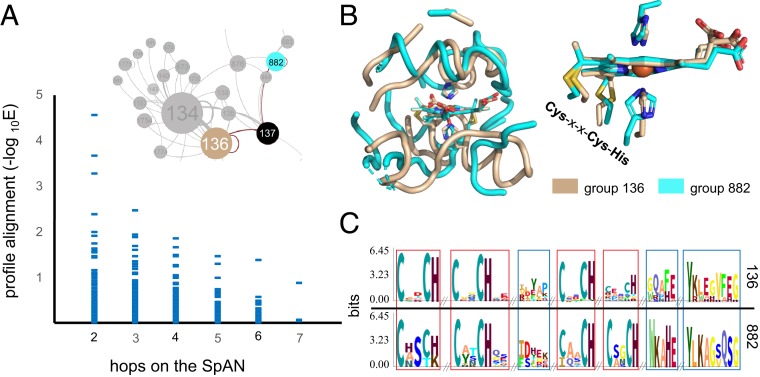
To test the evolutionary significance of connections within the SpAN, we analyzed sequence profile-profile alignments of modules (17). Profiles represent the weighted frequency of amino acids at each position rather than the sequences themselves and allow for more sensitive detection of distantly related proteins (17). Analysis of profile alignments of adjacent modules (i.e., one “hop” on the SpAN) was complicated by the physical overlap resulting from 15-Å radius microenvironments separated by an electron transfer cofactor cutoff of 14 Å or less. Thus, while single-hop profile alignment scores were very high (SI Appendix, Fig. S3A), we focused the analysis on only profile-profile alignments that were two hops or more on the SpAN. The closer the two modules were, the more likely they were to have a significant profile alignment (Fig. 2A). In the stringent case of modules separated by two or three hops, a statistically significant increase in profile similarity was observed (SI Appendix, Fig. S3B). For example, the structural alignment of microenvironments from modules type 136 and type 882 separated by two hops shows weak structural homology other than the first-shell ligands (Fig. 2B). In contrast, a profile alignment of the same two domains shows notable similarity beyond the first shell (Fig. 2C). Connections on the SpAN can be inferred as evidence of diversification of ancestral folds upon duplication, where sequence and structural similarity are no longer evident.
Fig. 2.
(A) Pairwise module profile alignment E-values grouped by shortest distance in the SpAN. HemeC-binding module groups 136 and 882 alignment E-value of 5.7E-4. Seven percent of all profile alignments, and 13% of two-hop alignments have a −log E > 1. (B) Structural superposition of cofactor microenvironments from group 136 Desulfovibrio cytochrome C3 (1GYO [cofactor shown: HEC 113E]) and group 882 cell-surface cytochrome MtrF from Shewanella (3PMQ [cofactor shown: HEC 670B]) show modest structural overlap beyond the central cofactor and first shell ligands. (C) Profiles of the two groups aligned by HHBlits (17) show profile similarity beyond the first shell ligands. Red boxes highlight regions of the alignment with HHBlits confidence scores of ≥2, and blue boxes highlight regions where the confidence score is <2 (see SI Appendix, Fig. S8, for more information).
Two Ancient Modules.
The SpAN exhibits many features consistent with that of a scale-free network, a signature of natural networks where the distribution of the degree (i.e., number of connections) of nodes follows a power law (SI Appendix, Fig. S4). The structure of a scale-free network is driven by two behaviors. The first is a growing network (18), where the highest-degree nodes are among the earliest members. From these early proteins, the SpAN grew in size and complexity with the evolution of novel metabolisms taking advantage of evolving electron sources and sinks in the geosphere. The second feature is preferential attachment (18); nodes have unequal probabilities to form new connections. Hubs in metabolic networks such as the SpAN are proposed to be older (19, 20). Furthermore, highly connected protein-cofactor modules are particularly useful for electron transfer and thus recur in multiple oxidoreductases.
The two modules with the highest node degree were the following: type 85, the bacterial Fd, which coordinates iron-sulfur cofactors, and type 7, the Rm, containing mostly flavins, Co-A, NAD(P), and related cofactors. Both folds are ancient and thought to have been present in LUCA (14, 21, 22). The age of folds can be constrained by combining functional annotations of oxidoreductases with the emergence of metabolic pathways inferred from the geologic record (21) (SI Appendix, Fig. S5A). Fd and other iron-sulfur modules as well as Rm modules are replete in oxidoreductases that make up the earliest posited metabolic pathways (23) including acetogenesis (e.g., the Wood–Ljundhal pathway) and methanogenesis (SI Appendix, Fig. S5B).
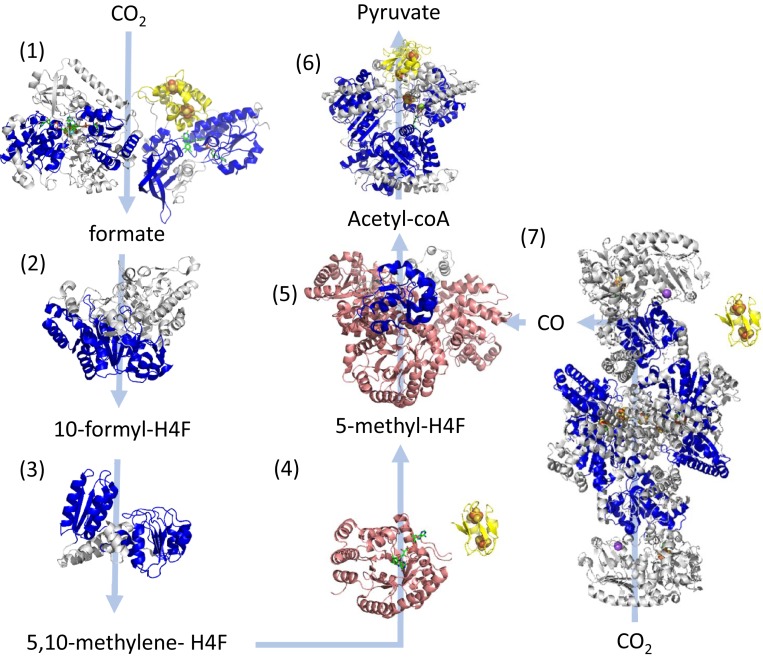
If Rm and Fd folds are ancient, they should be replete in early metabolic pathways. An examination of key oxidoreductases in acetogenesis revealed that enzymes in nearly every step were composed of these modules (Fig. 3). Furthermore, key enzymes in this pathway were composed of (β/α)8-barrels, which have been proposed to share ancestry with Rm-related folds (24). Ancient microorganisms which inhabited the anoxic Archean oceans likely used H2 as their electron source and reduced CO2 by the Wood–Ljundhal pathway, the most ancient extant CO2 fixation pathway (25–27). Anaerobic acetogens and methanogens that obtained carbon sources by autotrophically reducing CO2 via the electrons acquired from H2 are proposed to be living examples of the earliest forms of life (6, 26). For example, these organisms reduce NAD(P)+ using the electrons that are extracted from H2 by a soluble ferredoxin (7). The NADH is then used to reduce CO2 to formic acid in first step in the Wood–Ljungdhal pathway (28). The modules comprising the central enzymes of this pathway would have emerged early in Earth’s history, consistent with the placement of their microenvironments near the center of the SpAN.
Fig. 3.
Structurally determined oxidoreductases in the Wood–Ljundhal carbon-fixation pathway from CO2 to pyruvate performed by early acetogens (26). Modules within these proteins are colored as follows: blue, Rm folds; yellow, Fd folds; and pink, β/α barrels. Gray domains designate other modules or unassigned structural motifs. Where possible, the locations of cofactors are included in the structure seen as spheres for metals or as green sticks for organic cofactors. Light blue arrows indicate substrate-product transformations catalyzed by each enzyme. Numbers designate each of the reactions and the structures of enzymes catalyzing these reactions are the following: reaction 1—NADPH formate dehydrogenase (PDB ID codes 1FDO.A and 4YRY.B); reaction 2—10-formyl-H4 folate synthetase (PDB ID code 4IOJ); reaction 3—bifunctional protein FolD including methylene tetrahydrofolate dehydrogenase and methenyl tetrahydrofolate cyclohydrolase (PDB ID 5A5O); reaction 4—5,10-methylene-H4 folate reductase (PDB ID code 3APT); reaction 5—corrinoid iron-sulfur protein (CFeSP) (PDB ID code 4DJD); reaction 6—pyruvate synthase (PDB ID code 6CIN); and reaction 7—CO dehydrogenase/acetyl-CoA synthase (PDB ID code 3I01). The enzymes shown in groups 4 and 7 utilize the electrons from a soluble ferredoxin, which is not in the experimental structure and is illustrated using the bacterial ferredoxin structure PDB ID code 1FDN.
Divergence of Fd and Rm.
Although no detectable sequence profile similarity was observed across Fd and Rm modules, connectivity between the two modules on SpAN suggested potential common ancestry. A total of 59 connections were observed from 18 proteins, including enzymes from putative ancient metabolic pathways. Examples included adenylsulfate reductases from dissimilatory sulfate-reducing metabolisms of the hyperthermophiles Archeaoglobus fulgidis and Desulfovibrio gigas. Archeaoglobus members are found in deep-sea hydrothermal vents and other high-temperature environments (29). Connections were also observed in NADH-dependent ferredoxin oxidoreductases in Pyrococcus furiosus, which plays a central role in maintaining redox balance within the cell (30). This enzyme utilizes flavin-based electron bifurcation, which has been proposed as an energetic strategy utilized by early oxidoreductases (27).
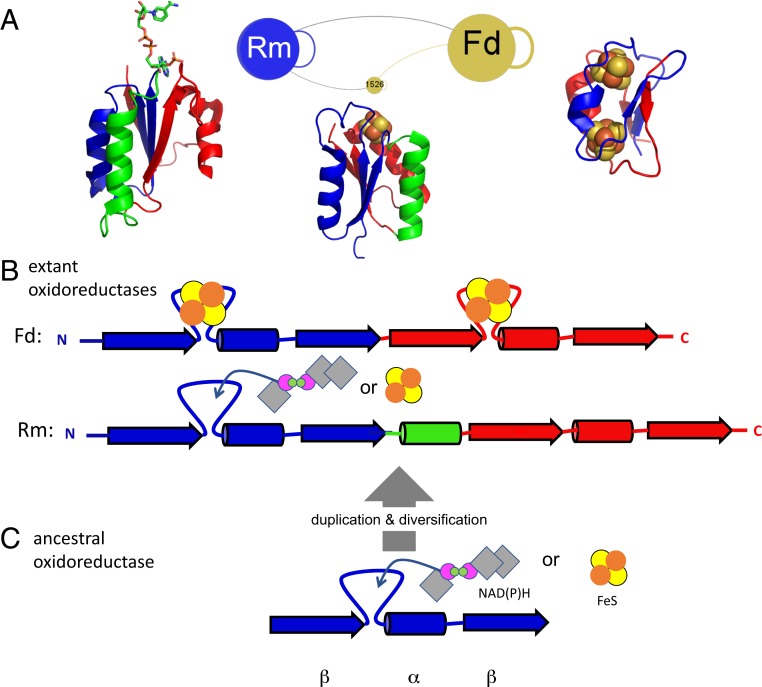
Beyond proximity in electron transfer pathways, the two modules show synteny of secondary structural elements (α-helices and β-strands) (Fig. 4). Both contain repeats of a β-α-β motif with a ligand-binding loop between the first β-strand and the α-helix that coordinates either an FeS center or nucleoside phosphate moiety. In phosphate-binding sites, these features are referred to as P loops or Walker motifs (31, 32), whereas, for FeS-binding sites, they are a variation of Cys-x-x-Cys motifs (33). Both loops adopt unusual combinations of α-left/α-right structures, forming what Watson and Milner-White refer to as “cationic nests” (34), where backbone amides coordinate and stabilize bound anions and, for FeS sites, support redox reversibility (35–38).
Fig. 4.
(A) Portion of SpAN with ferredoxin module (Fd), Rossmann-like module (Rm), and module 1526 (similar to Rm fold with an 4Fe4S bound). Examples of structures shown are the following: ferredoxin—PDB ID code 1FDN; heterodisulfide reductase (group 1526) —PDB ID code 5ODC; ferredoxin/NADPH oxidoreductase—PDB ID code 2VNH. (B) Secondary structure synteny of the primordial β-α-β duplication between Fd and the Rm modules (N-terminal β-α-β element in blue and C-terminal element in red; colors match those shown in A). One α-helix (green) differentiates the two folds. Loops that either bind iron-sulfur or nucleoside-based cofactors are shown between the first β-strand and α-helix. FeS cofactors are shown as yellow/orange circles and NAD(P)H as gray diamonds with purple/green circles. (C) Deduced primordial β-α-β peptide capable of binding either cofactor may have been a common precursor to both Fd and Rm modules.
There are other modules that share aspects of structure and/or chemistry with Rm and Fd. Several modules have Rossmann-like topologies but coordinate iron-sulfur clusters instead of organic cofactors. One such module (type 1526), found in nickel-containing hydrogenases in extant Archaea, bridges Fd and nucleoside-binding Rm modules in the SpAN. This module might be a contemporary descendant of an ancestral molecule that diverged, giving rise to Fd and Rm oxidoreductases. The Rm fold also shares ancestry with another enzyme fold class, the (β/α)8 barrel (24), suggesting that a common ancestor roots a significant fraction of modern proteins.
The duplication of an ancestral β-α-β fold in Fd is plausible (39, 40), and recent demonstrations of symmetric Fd designs that function in vivo support this hypothesis (41); however, the path between Fd and Rm folds is less obvious. The synteny of secondary structural elements between Fd and Rm modules is not matched by homology at the tertiary structure level—hence the division into separate modules. Supposing that the two modules diverged from a common ancestor, then an early insertion/deletion (indel) event could have produced the additional α-helix in the Rm fold (green helix in Fig. 4 A and B). Such an indel would require reorganization of the antiparallel β-sheet in Fd folds to the parallel one characteristic of Rm and related folds, which is a significant change in the fold topology. While most indels in proteins are very small, comprising a few amino acids in surface-facing loops (42), much larger structural arrangements are occasionally observed, with sequence and structural similarity manifesting at a local rather than global fold scale (43, 44). Models for larger structural rearrangements often describe a process of structural drift through multiple intermediates (45). In the best understood examples of fold changes, intermediates are anchored by a stable hydrophobic core that remains largely constant across the transition pathway. In the case of smaller folds with a modest hydrophobic core, a bound FeS or nucleoside phosphate could impose structural specificity, facilitating structural drift between Fd and Rm folds. The original Rm and Fd molecules and any putative intermediates, if they existed, are now extinct, but their traces may be inferred from the SpAN.
Supposing the existence of a primordial peptide oxidoreductase ancestral to both the Fd and Rm, its topology may have been that of a half-ferredoxin, namely a β-α-β fold, with a cationic nest at the N-terminal end of the central α-helix (Fig. 4C). Synthetic miniproteins with a parallel β-hairpin and a central helix have been designed using disulfide bridges to produce highly stable molecules (46). While solvent-exposed parallel β-sheets are statistically rare in natural proteins, this may be due to the abundance of larger β−α folds such as the Rm and (β−α)8 barrels. Smaller proteins such as Ig binder G(β1) contain exposed parallel β-hairpins that are tolerant to mutations (47). Even simpler β-(P-loop)-α designs that bind nucleic acids developed by Romero Romero and colleagues (48) suggest a possible Rm-derived ancestor.
Conclusions
The electronic circuits that sustain life on this planet are expressed in the SpAN as a metabolic network expanding over geologic timescales from adaptations to increasing potentials across cathodes and anodes in the geosphere (11, 12, 49). At the SpAN center are simple protein folds that coordinate iron-sulfur clusters and flavins, matching the reducing potential of early earth. Through duplication, diversification, and recruitment, the SpAN expanded its repertoire of electron transport chains toward more oxidizing domains. Tracing this network to its root suggests that oxidoreductases may have arisen from two simple protein folds that in turn shared an ancestral β-α-β redox cofactor-binding peptide.
There are important caveats to be considered. The first is a lack of quantitative measures of parsimony for phylogenetic models inferred from structural distances. Evolutionary inferences of homology over analogy based on topology must be considered with caution (50, 51). Combining structure with sequence or network-based analyses can be used to constrain evolutionary models (16, 24, 52). In this work, these issues are mitigated by the use of cofactor placement to filter structure alignments (8, 9) and the use of profile alignments of modules in the SpAN to examine how adjacency in electron transfer pathways supports homology. The second caveat is the challenge associated with using phylogenetic models to explain the growth of biological networks as multiple network dynamics models can lead to the same outcome (53). In this case, associating centrality and age of ferredoxin and Rossmann-like folds with phylogenetic age is supported by our understanding of redox evolution of the geosphere and the corresponding expansion of metabolic networks (12, 49).
Nevertheless, in the realm of deep-time evolutionary inference, we are necessarily limited to deducing what could have happened, rather than proving what did happen. As such, the next step will be to design and study plausible primordial peptides for redox activity. Engineered minimal forms of ferredoxin and Rossmann-like folds are currently being pursued (37, 41, 48, 54), testing the plausibility that modern complex metabolism had its origins in a few ancient peptide oxidoreductases.
Methods
Compiling Organic and Inorganic Microenvironments.
Three-dimensional structures of natural proteins from the PDB (as of August 2018) that contained transition metals cofactors (Fe, Cu, Mn, Ni, Mo, Co, V, and W) or electron-transfer–related organic cofactors were used as the database for this work. We extracted a sphere (microenvironment) of the residues within a 15-Å geometric center of each cofactor (8). While metal-containing cofactors consisted of only a few metal atoms, the organic cofactors varied considerably in length and shape. Therefore, in cases of a large organic cofactor, we focused only on the 15-Å sphere around the active moiety of the cofactors, where the electron transfer is most likely centered. The new database, which was composed of both metal and organic cofactors, included a total of 74,340 microenvironments and included the ∼30-K microenvironments from the previous study (10). The list of included organic cofactors used for this database was taken from The CoFactor database (55). Dataset S3 presents a full list of the cofactors that we considered, as well as the atoms we used to calculate the center and the cofactor-cofactor distance. Many of the organic cofactors were nucleoside-based cofactors (e.g., FAD, NAD, CoA). In addition to their electron transfer activity, these cofactors are known for the unique conserved protein fold which binds the nucleoside base (21, 56, 57). To allow for detection of the nucleoside-base–binding site, we generated a separate microenvironment around the center of the nucleoside base (SI Appendix, Fig. S2A). These microenvironments clustered together due to the conserved binding site of the nucleotide base despite having a more variable electron-transfer-site microenvironment. We did not include chlorophyll microenvironments in this work, as chlorophyll tends to form a dense bundle of porphyrin with very little protein structure. These microenvironments were not suitable for this protein-structure-centric approach and will be considered in future work.
Clustering Similar Microenvironments.
The microenvironments were clustered into various groups based on their structural similarity. All collected microenvironments of the same cofactor were individually subjected to a comparative pairwise alignment using the “align” package in the PyMOL Molecular Graphics System (Version 1.8, Schrödinger). In addition, a nonredundant list of all collected microenvironments [<90% sequence similarity (58)] were subjected to the same comparative pairwise alignment. Poor alignments (i.e., <25 amino acids, >4 Å rmsd) were not included. Aligned microenvironments from the nonredundant dataset that met a threshold of 0.1 Å rmsd per alignment length and had the cofactors’ center distance ≤2 Å apart were grouped into a cluster (SI Appendix, Fig. S6). In addition, if the cofactor contained a nucleoside base, the nucleoside-base microenvironment was combined with the active-site microenvironment, resulting in a merged microenvironment. This was done so that every cofactor, despite size, was analyzed in the context of both electron transfer and binding. The resulting modules varied in size and sequence diversity (SI Appendix, Fig. S7).
Generating SpAN.
The SpAN was generated using previously published methods (10). Briefly, a network was constructed by treating each module (a cluster of microenvironments) as a node, where nodes were connected by edges if at least one microenvironment from each module was located in the same protein structure and cofactors were in electron-transfer distance range (i.e., ≤14 Å). Maximal electron-transfer distance was determined by the observed maximum possible distance for electron tunneling in natural proteins (15). In cases of two similar cofactors, we used a minimum distance of 4.5 Å in order to ignore self-connections in multi-ion microenvironments. The total number of edges connecting the same modules were used as the weight of the edge. This produced a total of 15,688 pairs of microenvironments, which were used to construct a SpAN containing 1,850 connected components. We focus our discussion on the giant component of this network which represents the electron-transfer network of oxidoreductases (SpAN). We calculate functional diversity of each node using the same method as previously described (10). The scripts used to generate SpAN have been deposited in https://github.com/hraanan/SpAN_scripts (59).
Sequence Profile-Profile Alignment.
The residues that fell into each microenvironment were individually extracted. It is important to note that these include fragments of positions in the sequence that falls into the 15-Å sphere. In many cases, interstitial sequences outside the designated sphere were large enough to incorporate much, if not all of, the adjacent microenvironments, confounding profile definitions. In addition, large indels also increased the level of nonspecific alignments in profile generation. Hence, to identify motifs that were associated with a given metal or cofactor, we considered the collection of fragments within a microenvironment as continuous elements. Identical sequences were removed from the module when creating profiles. The extracted sequences from each microenvironment in each cluster were individually aligned using Clustal Omega (60). The aligned sequences for each cluster were used to generate a sequence profile using the HHblits package (17). Following the generation of profiles for each cluster, the sequences were subjected to profile-profile alignment using the HHblits package.
Analysis of profile-profile alignments of adjacent modules on the SpAN was complicated by the physical overlap resulting from a 15-Å radius microenvironment separation and an electron-transfer-cofactor separation cutoff of 14 Å or less. Thus, we considered only profile-profile alignments that were two hops or more from each other on the SpAN. The average and SE of the profile-profile alignments score for each hop were calculated.
Three-Dimensional Structures of the Wood–Ljungdhal Pathway.
We used the enzymes of the Wood-Ljungdhal pathway in the acetogen Moorella thermoacetica as described by Fuchs (26). For enzymes where an experimentally determined structure had not been deposited in the PDB, we used the protein sequence to search for the best existing 3D template structure and generated a homology model using the homology modeling tool SWISS-MODEL (61, 62). SI Appendix, Table S1, summarizes the sequence similarity and coverage of each structural model to the original sequence.
Data Availability Statement.
Key data files are included as supplemental material and code has been uploaded to https://github.com/hraanan/SpAN_scripts.
Supplementary Material
Acknowledgments
This work was supported by the Gordon and Betty Moore Foundation on “Design and Construction of Life’s Transistors” Grant GBMF-4742 (to V.N. and P.G.F.) and by NASA Grant 80NSSC18M0093 from the NASA Astrobiology Institute. S.P. acknowledges support from the Rutgers University Institute of Earth, Ocean, and Atmospheric Science Postdoctoral Fellowship Program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data depositon: The scripts used to generate SpAN have been deposited in GitHub, https://github.com/hraanan/SpAN_scripts.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914982117/-/DCSupplemental.
References
- 1.Goldford J. E., Segre D., Modern views of ancient metabolism. Curr. Opin. Syst. Biol. 8, 117–124 (2018). [Google Scholar]
- 2.Smith E., Morowitz H., The Origin and Nature of Life on Earth: The Emergence of the Fourth Geosphere (Cambridge University Press, 2016). [Google Scholar]
- 3.Lynch M., Genomics. Gene duplication and evolution. Science 297, 945–947 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Baymann F., et al. , The redox protein construction kit: Pre-last universal common ancestor evolution of energy-conserving enzymes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 267–274 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lupas A. N., Ponting C. P., Russell R. B., On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J. Struct. Biol. 134, 191–203 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Weiss M. C., et al. , The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Lane N., Martin W. F., The origin of membrane bioenergetics. Cell 151, 1406–1416 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Senn S., Nanda V., Falkowski P., Bromberg Y., Function-based assessment of structural similarity measurements using metal co-factor orientation. Proteins 82, 648–656 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Catazaro J., Caprez A., Swanson D., Powers R., Functional evolution of proteins. Proteins 87, 492–501 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raanan H., Pike D. H., Moore E. K., Falkowski P. G., Nanda V., Modular origins of biological electron transfer chains. Proc. Natl. Acad. Sci. U.S.A. 115, 1280–1285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkowski P. G., Fenchel T., Delong E. F., The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Moore E. K., Jelen B. I., Giovannelli D., Raanan H., Falkowski P. G., Metal availability and the expanding network of microbial metabolisms in the Archaean eon. Nat. Geosci. 10, 629–636 (2017). [Google Scholar]
- 13.Berman H., Henrick K., Nakamura H., Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10, 980 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Rao S. T., Rossmann M. G., Comparison of super-secondary structures in proteins. J. Mol. Biol. 76, 241–256 (1973). [DOI] [PubMed] [Google Scholar]
- 15.Page C. C., Moser C. C., Chen X., Dutton P. L., Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Raymond J., Segrè D., The effect of oxygen on biochemical networks and the evolution of complex life. Science 311, 1764–1767 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Remmert M., Biegert A., Hauser A., Söding J., HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Barabasi A.-L., Linked: The New Science of Networks (Perseus, 2002). [Google Scholar]
- 19.Fell D. A., Wagner A., The small world of metabolism. Nat. Biotechnol. 18, 1121–1122 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Wagner A., The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes. Mol. Biol. Evol. 18, 1283–1292 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Laurino P., et al. , An ancient fingerprint indicates the common ancestry of Rossmann-fold enzymes utilizing different ribose-based cofactors. PLoS Biol. 14, e1002396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma B. G., et al. , Characters of very ancient proteins. Biochem. Biophys. Res. Commun. 366, 607–611 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Braakman R., Smith E., The emergence and early evolution of biological carbon-fixation. PLoS Comput. Biol. 8, e1002455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farías-Rico J. A., Schmidt S., Höcker B., Evolutionary relationship of two ancient protein superfolds. Nat. Chem. Biol. 10, 710–715 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Braakman R., Smith E., The compositional and evolutionary logic of metabolism. Phys. Biol. 10, 011001 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Fuchs G., Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu. Rev. Microbiol. 65, 631–658 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Martin W. F., Sousa F. L., Lane N., Evolution. Energy at life’s origin. Science 344, 1092–1093 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Ragsdale S. W., Pierce E., Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim. Biophys. Acta 1784, 1873–1898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poudel S., et al. , Origin and evolution of flavin-based electron bifurcating enzymes. Front. Microbiol. 9, 1762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen D. M. N., et al. , Two functionally distinct NADP+-dependent ferredoxin oxidoreductases maintain the primary redox balance of Pyrococcus furiosus. J. Biol. Chem. 292, 14603–14616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saraste M., Sibbald P. R., Wittinghofer A., The P-loop: A common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Walker J. E., Saraste M., Runswick M. J., Gay N. J., Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J. D., Rodriguez-Granillo A., Case D. A., Nanda V., Falkowski P. G., Energetic selection of topology in ferredoxins. PLoS Comput. Biol. 8, e1002463 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson J. D., Milner-White E. J., A novel main-chain anion-binding site in proteins: The nest. A particular combination of phi,psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol. 315, 171–182 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Adman E., Watenpaugh K. D., Jensen L. H., NH—S hydrogen bonds in Peptococcus aerogenes ferredoxin, Clostridium pasteurianum rubredoxin, and Chromatium high potential iron protein. Proc. Natl. Acad. Sci. U.S.A. 72, 4854–4858 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blake P. R., et al. , Quantitative measurement of small through-hydrogen-bond and ‘through-space’ 1H-113Cd and 1H-199Hg J couplings in metal-substituted rubredoxin from Pyrococcus furiosus. J. Biomol. NMR 2, 527–533 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Kim J. D., et al. , Minimal heterochiral de novo designed 4Fe-4S binding peptide capable of robust electron transfer. J. Am. Chem. Soc. 140, 11210–11213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nanda V., et al. , De novo design of a redox-active minimal rubredoxin mimic. J. Am. Chem. Soc. 127, 5804–5805 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Romero Romero M. L., Rabin A., Tawfik D. S., Functional proteins from short peptides: Dayhoff’s hypothesis turns 50. Angew. Chem. Int. Ed. Engl. 55, 15966–15971 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Davis B. K., Molecular evolution before the origin of species. Prog. Biophys. Mol. Biol. 79, 77–133 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Mutter A. C., et al. , De novo design of symmetric ferredoxins that shuttle electrons in vivo. Proc. Natl. Acad. Sci. U.S.A. 116, 14557–14562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascarella S., Argos P., Analysis of insertions/deletions in protein structures. J. Mol. Biol. 224, 461–471 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Grishin N. V., Fold change in evolution of protein structures. J. Struct. Biol. 134, 167–185 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Zhang D., Iyer L. M., Burroughs A. M., Aravind L., Resilience of biochemical activity in protein domains in the face of structural divergence. Curr. Opin. Struct. Biol. 26, 92–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishna S. S., Grishin N. V., Structural drift: A possible path to protein fold change. Bioinformatics 21, 1308–1310 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Bhardwaj G., et al. , Accurate de novo design of hyperstable constrained peptides. Nature 538, 329–335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merkel J. S., Sturtevant J. M., Regan L., Sidechain interactions in parallel beta sheets: The energetics of cross-strand pairings. Structure 7, 1333–1343 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Romero Romero M. L., et al. , Simple yet functional phosphate-loop proteins. Proc. Natl. Acad. Sci. U.S.A. 115, E11943–E11950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dupont C. L., Butcher A., Valas R. E., Bourne P. E., Caetano-Anollés G., History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc. Natl. Acad. Sci. U.S.A. 107, 10567–10572 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krishna S. S., Sadreyev R. I., Grishin N. V., A tale of two ferredoxins: Sequence similarity and structural differences. BMC Struct. Biol. 6, 8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janin J., Shared structural motif in proteins. Nature 365, 21 (1993). [DOI] [PubMed] [Google Scholar]
- 52.Franklin M. W., et al. , Evolutionary pathways of repeat protein topology in bacterial outer membrane proteins. eLife 7, e40308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lima-Mendez G., van Helden J., The powerful law of the power law and other myths in network biology. Mol. Biosyst. 5, 1482–1493 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Scintilla S., et al. , Duplications of an iron-sulphur tripeptide leads to the formation of a protoferredoxin. Chem. Commun. (Camb.) 52, 13456–13459 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Fischer J. D., Holliday G. L., Thornton J. M., The CoFactor database: Organic cofactors in enzyme catalysis. Bioinformatics 26, 2496–2497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ojha S., Meng E. C., Babbitt P. C., Evolution of function in the “two dinucleotide binding domains” flavoproteins. PLoS Comput. Biol. 3, e121 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesk A. M., NAD-binding domains of dehydrogenases. Curr. Opin. Struct. Biol. 5, 775–783 (1995). [DOI] [PubMed] [Google Scholar]
- 58.Wang G., Dunbrack R. L. Jr, PISCES: Recent improvements to a PDB sequence culling server. Nucleic Acids Res. 33, W94–W98 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raanan H., Poudel S., Pike D. H., Nanda V., Falkowski P. G., SpAN scripts. GitHub. https://github.com/hraanan/SpAN_scripts. Deposited 14 June 2019.
- 60.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guex N., Peitsch M. C., Schwede T., Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30 (suppl. 1), S162–S173 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Key data files are included as supplemental material and code has been uploaded to https://github.com/hraanan/SpAN_scripts.