Abstract
The mechanism of tumor-selective replication of oncolytic measles virus (MV) is poorly understood. Using a stepwise model of cellular transformation, in which oncogenic hits were additively expressed in human bone marrow-derived mesenchymal stromal cells, we show that MV-induced oncolysis increased progressively with transformation. The type 1 interferon (IFN) response to MV infection was significantly reduced and delayed, in accordance with the level of transformation. Consistently, we observed delayed and reduced signal transducer and activator of transcription (STAT1) phosphorylation in the fully transformed cells. Pre-treatment with IFNβ restored resistance to MV-mediated oncolysis. Gene expression profiling to identify the genetic correlates of susceptibility to MV oncolysis revealed a dampened basal level of immune-related genes in the fully transformed cells compared to their normal counterparts. IFN-induced transmembrane protein 1 (IFITM1) was the foremost basally downregulated immune gene. Stable IFITM1 overexpression in MV-susceptible cells resulted in a 50% increase in cell viability and a significant reduction in viral replication at 24 h after MV infection. Overall, our data indicate that the basal reduction in functions of the type 1 IFN pathway is a major contributor to the oncolytic selectivity of MV. In particular, we have identified IFITM1 as a restriction factor for oncolytic MV, acting at early stages of infection.
Keywords: innate immune response, type 1 interferon, measles virus, MV, interferon-induced transmembrane protein 1, IFITM1, interferon-stimulated gene, ISG, oncolytic measles virus
Using a well-characterized, stepwise cellular transformation model to determine the mechanism of selectivity of oncolytic measles virus (MV), Aref et al. demonstrated the key contribution of the type 1 interferon pathway in resistance to MV-mediated oncolysis. They also showed that IFITM1 acts as a restriction factor for oncolytic MV strains at early stages of infection.
Introduction
During the last two decades, oncolytic viruses (OVs) have emerged as promising cancer therapeutics by preferentially infecting tumor cells, thereby mediating cell killing and eliciting an anti-tumor immune response.1 Recombinant derivatives of the measles virus (MV) Edmonston B vaccine strain (MV-Edm) are efficacious against a number of human malignancies2 and are currently being evaluated in phase I/II clinical trials for the treatment of glioblastoma (ClinicalTrials.gov: NCT00450814), ovarian cancer (ClinicalTrials.gov: NCT02364713), breast cancer (ClinicalTrials.gov: NCT01846091), mesothelioma (ClinicalTrials.gov: NCT01503177), multiple myeloma (ClinicalTrials.gov: NCT01503177), head and neck cancer (ClinicalTrials.gov: NCT01846091), and malignant peripheral nerve sheath tumors (ClinicalTrials.gov: NCT02700230).
MV is an enveloped morbillivirus from the paramyxoviruses (family: Paramyxoviridae). It has a non-segmented negative single-stranded RNA genome encoding eight viral proteins, of which two envelope glycoproteins hemagglutinin (H) and fusion (F) proteins mediate attachment and entry into infected host cells, respectively.3 Cellular entry of MV is facilitated by three known receptors, that is, CD46,4,5 SLAM,6 and nectin-4,7,8 with CD46 being the preferential receptor for vaccine strains. Upon infection, MV induces a strong cytopathic effect, which results in multi-nucleated syncytia.3,9
The mechanism underlying the tumor selectivity of oncolytic MV is unclear. The selective tropism of oncolytic MV has been attributed to CD46 overexpression by tumor cells.10 However, SLAM-dependent entry of oncolytic MV has been documented in mantle cell lymphoma cells and xenografts, which was not correlated with CD46 expression levels.11 Defective type 1 interferon (IFN) signaling, first shown by Stojdl et al.12 to facilitate oncolytic vesicular stomatitis virus replication, also represents a potential mechanism of action for oncolytic MV. In normal cells, sensing of MV RNA by pathogen recognition receptors RIG-I and MDA-5 results in the production of type 1 IFN (IFNα/β).13, 14, 15 The binding of IFNs to their cognate receptor (IFNAR) on infected and neighboring uninfected cells activates the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway, resulting in the phosphorylation of STAT1 and STAT2. Phosphorylated STAT1 and STAT2 bind to IRF9 to form the ISGF3 transcription factor complex, which then binds to IFN-stimulated response elements (ISREs) in promoters of IFN-stimulated genes (ISGs) to induce the transcription of hundreds of ISGs. These ISGs collectively establish an antiviral state in the infected cell by inhibiting multiple stages of viral infection.16,17 Indeed, constitutive IFN pathway activation was identified as the key determinant for MV replication in MV-permissive and MV-resistant glioblastoma xenografts.18
We set out to probe the precise mechanisms of oncolytic MV specificity using an established stepwise model of cellular transformation, in which progressive oncogenic hits were stably and additively expressed in human bone marrow-derived mesenchymal stromal cells (BM-MSCs).19 We showed that progressive MV-mediated cell killing is closely correlated with the degree of transformation, and that lower basal expression of genes within the type 1 IFN pathway play an important role in facilitating MV infection. In particular, we implicate the ISG IFITM1 in the tumor-selective replication and killing mediated by oncolytic MV.
Results
Susceptibility to MV-Mediated Oncolysis Is Positively Correlated with Progressive Transformation of MSCs
To investigate MV replication kinetics in the model of progressively transformed human BM-MSCs, cells at all the transformational stages, as shown in Figure 1, were infected with MV-NSe at a multiplicity of infection (MOI) of 1.0. Figure 2A shows MV-specific cell death at three time points per cell type and demonstrates a progressive increase in MV-specific cell death with increasing transformation. MV-mediated cell killing was minimal in hTERT cells, where even at 72 h post-infection (hpi), fewer than 5% of the cells had died, compared to more than 60% cell death in 4+V and 5H at the same time point. Figure 2B shows fluorescence microscopy at the same time points after infection with MV-green fluorescent protein (GFP); MV-induced syncytia were small and barely evident in hTERT but copious and large in transformed 3H, 4+V, and 5H MSCs following MV infection.
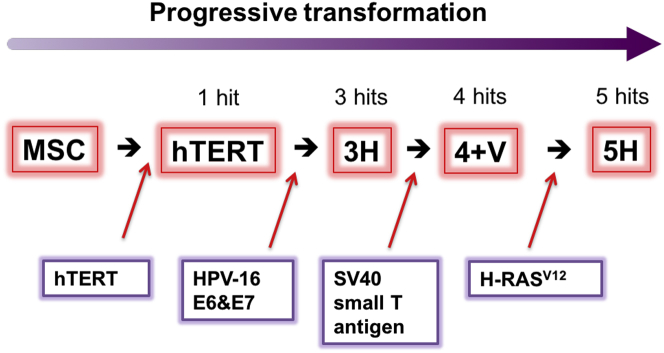
Figure 1.
Model of Stepwise Transformation of Human Bone Marrow-Derived Mesenchymal Stromal Cells
Schematic diagram of MSC stepwise transformation (adapted from Funes et al.19). MSCs were named according to the number of oncogenes inserted by retroviral transduction. hTERT encodes the catalytic subunit of human telomerase and confers the cells extended lifespan in vitro. Human papilloma virus (HPV-16) E6 and E7 genes abrogate the functions of p53 and pRb tumor suppressors, respectively. SV40 small T antigen leads to the stabilization of c-Myc by inactivating protein phosphatase 2A. Finally, the insertion of an oncogenic allele of Ras (H-RasV12) provides the acquisition of a constitutive mitogenic signal.
Figure 2.
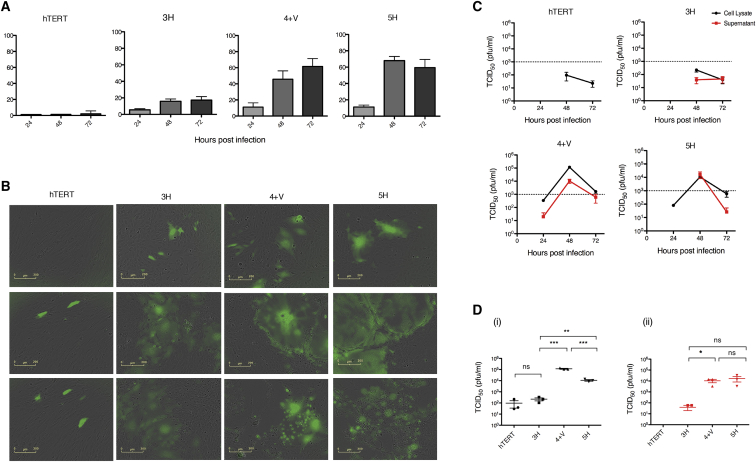
Susceptibility to MV-Mediated Oncolysis Is Positively Correlated with Progressive Transformation of MSCs
(A) MSCs were infected with MV-NSe (MOI of 1.0) and cell viability was assessed by trypan blue exclusion at 24, 48 and 72 h post-infection (hpi). Data are expressed as a percentage of cell killing by MV relative to uninfected control cells (n = 3). (B) Representative GFP and bright-field (BF) microscopy images of MSCs at 24, 48, and 72 hpi with MV-NSe-GFP (MOI of 1.0). Scale bars, 200 μm. (C) Tissue culture supernatants (red lines) and cell lysates (black lines) were harvested at the indicated time points after infection. (D) Viral titres at 48 hpi are shown for (i) cell lysates and (ii) supernatants. Viral titrations were performed on Vero cells and are calculated as TCID50 (plaque-forming units [PFU]/mL) (n = 3). Data are expressed as mean ± SEM. All results shown are representative of three independent experiments (unpaired t test, *p < 0.05, **p < 0.01, ***p < 0.001). MV, measles virus; NS, not significant.
Consistent with this, representative one-step viral growth curves (Figure 2C) showed that MV replication and release correlated with progressive transformation. In Figure 2D, a comparison of the peak titers of cell-associated and supernatant MV (shown as mean and SEM of three independent replicates) shows a 2–3 log difference between hTERT/3H and 4+V/5H cells. We found no significant difference in MV receptor (SLAM, CD46, or Nectin-4) expression across the stages of transformation, ruling this out as potential mechanism for variable MV permissiveness (Figure S2). Taken together, clear differences in MV replication that were unrelated to density of receptor expression suggest that the model of stepwise transformation of human BM-MSCs is a good model for further elucidating mechanisms of MV-mediated oncolysis.
Differential Production of Type 1 IFN by MSCs in Response to Oncolytic MV
Production of type 1 IFN is essential for the induction of normal anti-viral immune responses, and it results in the expression of a series of ISGs. To determine whether differential IFN responses to oncolytic MV infection play a role in MV susceptibility, we quantified IFNα and IFNβ from tissue culture supernatants after infection. Only primary, unmodified normal human MSC control cells produced any IFNα in response to MV infection (Figure 3A). In comparison, IFNβ was produced by all MSCs following infection, but the level was inversely correlated with the transformational stage. While primary MSCs (683 pg/mL) and hTERT cells (560 pg/mL) produced IFNβ at 24 hpi, the more transformed MSCs only started to produce any IFNβ at 48 hpi, with a maximum of 98, 34, and 42 pg/mL in 3H, 4+V, and 5H cells, respectively. This suggests that although all the MSCs were capable of mounting a type 1 IFN response, the magnitude and timing of the response is very different in the MV-resistant MSCs compared to their malignant counterparts.
Figure 3.
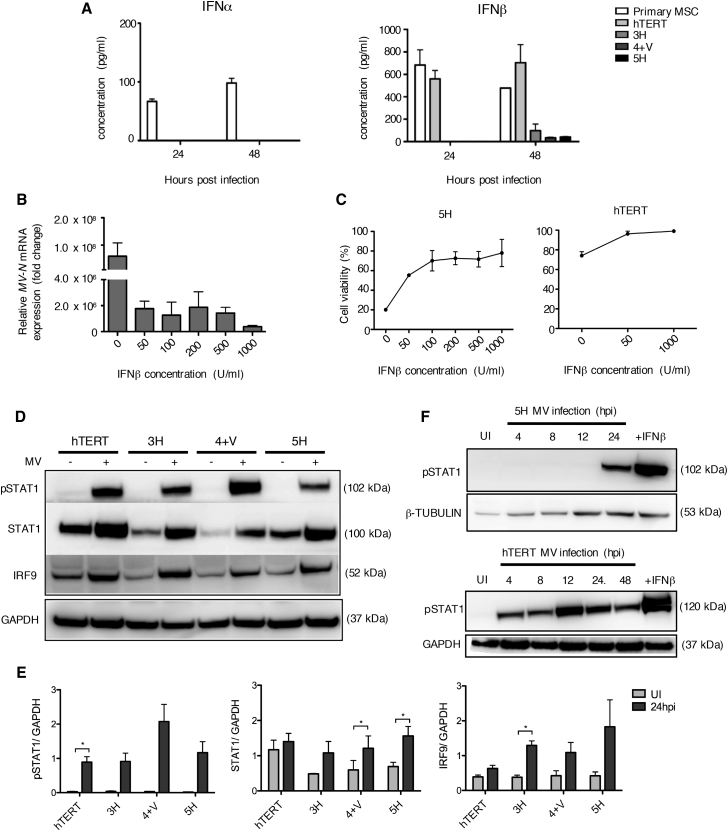
Differential Production of Type 1 IFN by MSCs in Response to Oncolytic MV
(A) IFNα and IFNβ production levels as assessed by ELISA using tissue culture supernatants collected from all MSCs, including primary patient-derived MSCS, at 24 and 48 hpi. Data are expressed as mean ± SEM of two independent experiments (n = 2) with samples measured in duplicates. (B) MV-N mRNA expression levels as assessed by qRT-PCR for 5H cells pre-treated with different concentrations of exogenous IFNβ for 16 h prior to MV infection. Data shown are relative to housekeeping gene GAPDH and normalized to uninfected control cells. Data are expressed as mean ± SEM (n = 3). (C) Cell viability of 5H cells following pre-treatment with IFNβ, and MV infection was assessed by trypan blue exclusion at 30 hpi. hTERT cells were used as a control. Results are reported as a the percentage of cell viability relative to uninfected control cells. Data are expressed as mean ± SEM (n = 3). (D) Immunoblotting of total STAT1, phosphorylated STAT1 (pSTAT1), and IRF9 using cell lysate of uninfected and MV-infected MSCs collected at 24 hpi. β-Tubulin or GAPDH was used as a loading control (n = 3). (E) Densitometry analysis of blot in (D) performed by ImageJ. (F) Immunoblot analysis of pSTAT1 during the time course of MV infection in 5H cells and hTERT cells using cell lysates collected at the indicated time points. Cells stimulated with IFNβ (1,000 U/mL) for 1 h were used as a positive control. Expression of β-tubulin and GAPDH was used as loading controls as indicated.
In order to determine whether the deficiency of IFNβ production could account for the increased susceptibility to MV infection and MV-mediated oncolysis, we asked whether it was possible to “rescue” the highly MV-susceptible cells with exogenous IFN by adding IFNβ 16 h prior to infection, followed by quantification of viral replication by qRT-PCR for the MV-N gene and assessment of cell viability at 30 hpi. As shown in Figures 3B and 3C, IFNβ pretreatment reduced viral replication and increased cell viability in 5H cells in a dose-dependent manner, with the steepest increase at the lowest dose of IFNβ. The impact on viability in hTERT control cells was very modest.
In order to confirm downstream IFN pathway activation, we investigated the phosphorylation status of STAT1 and the expression of total STAT1 and IRF9 proteins by immunoblotting using cell lysates collected at 24 hpi and compared to control, uninfected cells. STAT1 phosphorylation occurred in all MSCs in response to MV infection, more strongly in hTERT as shown in Figures 3D (representative blot) and 3E (densitometry from n = 3 blots). While endogenous STAT1 was expressed at higher basal levels in hTERT compared to the other cells, MV infection significantly increased STAT1 expression in 4+V and 5H cells. Similarly, IRF9 protein expression was induced in all MSCs following MV infection, with a statistically significant increase observed for 3H cells (Figures 3D and 3E). The time course of MV-stimulated STAT1 phosphorylation was determined in the most MV-susceptible 5H cells and compared with the most MV-resistant hTERT cells (Figure 3F). STAT1 phosphorylation was not detected until 24 hpi in 5H compared to a brisk response beginning at 4 hpi in hTERT cells. The delayed STAT1 phosphorylation in 5H compared to hTERT cells parallels the pattern observed for IFNβ production.
RNA Sequencing and qRT-PCR Reveal Differential Baseline Expression Levels of Genes Involved in the Type 1 IFN Pathway Response in MV-Resistant and MV-Susceptible MSCs
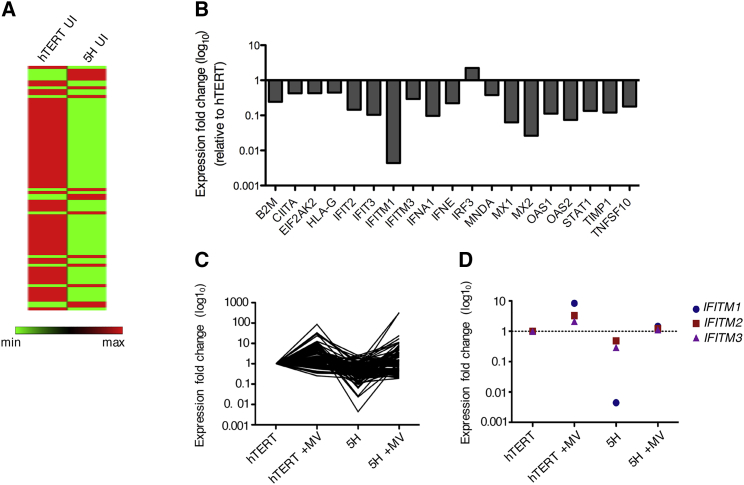
To gain a more global understanding of host responses and identify candidate genes, which could potentially be associated with increased susceptibility to oncolytic MV infection in this model, gene expression profiling by RNA sequencing (RNA-seq) was performed using total RNA extracted from uninfected and MV-infected hTERT and 5H cells. We reasoned that the optimal time point was 24 hpi, given that the MV-stimulated immune response was clearly underway by this time in both cell types, yet the MV-specific cell death was still minimal. Gene selection criteria were based on log2 fold change >1 and padjusted <0.05. At baseline, a total of 4,902 differentially expressed genes (DEGs) were identified between hTERT and 5H cells with 2,438 upregulated genes and 2,464 downregulated genes relative to hTERT cells, as shown in the volcano plot in Figure 4A. Reactome pathway enrichment analysis of genes significantly downregulated by more than 3-fold in 5H compared to hTERT cells revealed a number of enriched pathways, out of which type 1 IFN signaling was among the top 10, indicating significant baseline variability in immune-related functions (Figure 4B). Upon MV infection, a total of 795 DEGs (155 downregulated and 640 upregulated) were identified in MV-infected hTERT compared to uninfected controls. In 5H cells, 1,109 DEGs were identified (75 downregulated and 1034 upregulated) in MV-infected compared to uninfected 5H cells as shown in the volcano plots in Figure 4C. We selected the DEGs that were commonly upregulated in both cell lines following MV infection (394 genes) and analyzed them for pathway enrichment. (Figure 4D). The pathways most affected by MV infection were, not surprisingly, mostly related to the immune system and included antigen presentation and IFN signaling. Given our prior finding of the clear relevance of the type 1 IFN pathway in oncolytic MV susceptibility and noting that RNA-seq also identified it as one of the most differentially activated pathways, we validated RNA-seq data in separate experiments, by quantifying the expression of 84 genes involved in the type 1 IFN signaling pathway in uninfected hTERT and 5H cells using the RT2 Profiler PCR array, done as n = 3 independent experiments. The results are presented as a heatmap in Figures 5A and S3. In the absence of infection, 18 of the 84 genes analyzed were significantly differentially expressed between hTERT and 5H at baseline (fold change > 2, p < 0.05) The largest downregulation was found in the gene encoding IFITM1 (225.62-fold regulation, p = 0.045), followed by MX2 (−38.09-fold regulation, p = 0.002) and MX1 (−15.76-fold regulation, p = 0.049) (Figure 5B; Table S1). Figure 5C shows the changes in the 84 genes at 24 hpi; genes repressed in 5H cells relative to hTERT are induced to levels equivalent to, or greater than, those observed in hTERT cells at baseline, suggesting that the differential susceptibility to MV infection is due to baseline differences in type 1 IFN pathway gene expression, rather than the inability of 5H cells to deploy that pathway upon challenge. The IFITM genes seemed to show the greatest changes. The expression profiles of these genes before and after MV infection are plotted separately (see Figure 5C). IFTIM1 was greatly and very significantly downregulated in 5H compared to hTERT (−225.6-fold regulation) and was induced upon MV infection by 8.37-fold and 134.92-fold in hTERT and 5H cells, respectively (Figure 5D).
Figure 4.
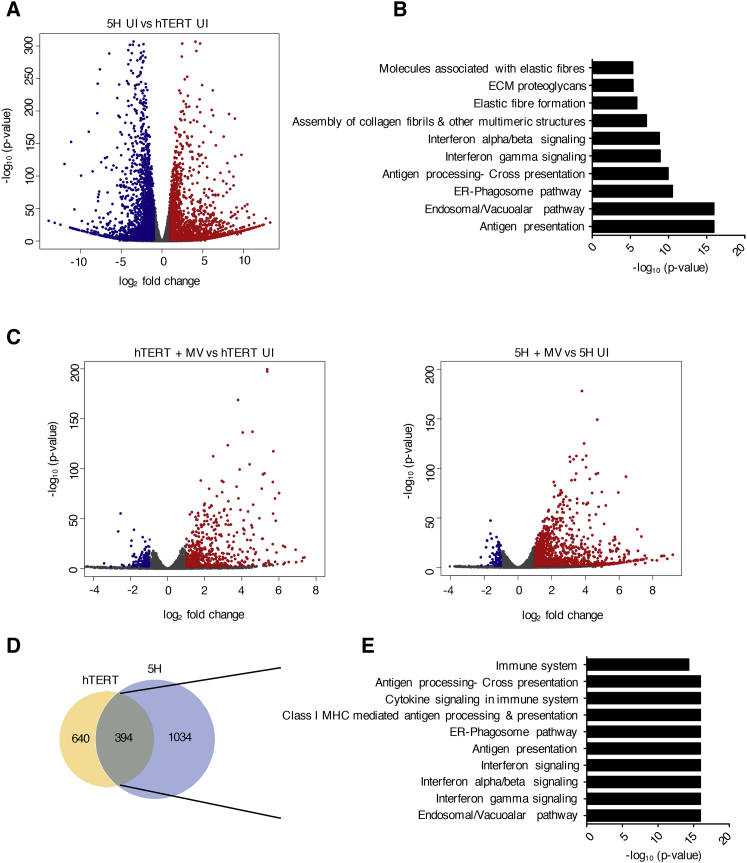
RNA Sequencing Reveals Differential Baseline Gene Expression in MV-Resistant and MV-Susceptible MSCs
RNA-seq was performed on total RNA that was extracted from uninfected and MV-infected hTERT and 5H cells at 24 hpi. (A) Volcano plot of differentially expressed genes (DEGs) in 5H cells compared to hTERT cells at baseline levels (n = 3). Cut-off criteria for DEGs are absolute log2 fold change >1 and padjusted value <0.05. The y axis displays the log10 p value for each gene, while the x axis displays the log2 fold change for that gene relative to hTERT. Red dots indicate upregulated genes, green dots indicate downregulated genes, and gray dots indicate non-significant relative to hTERT. (B) Genes downregulated (more than 3-fold downregulation, p value < 0.05) in 5H compared to hTERT cells were selected and analyzed using the functional annotation tool in REACTOME. The top 12 enriched pathways (p value < 0.001) are shown. (C) Volcano plots depicting DEGs after infection of (i) hTERT cells and (ii) 5H cells (compared to mock-infected control cells). (D) Venn diagram illustrating the overlap of genes found to be upregulated in response to MV infection in both hTERT and 5H cells (absolute log2 fold change [FC] > 1, padjusted value < 0.05). (E) REACTOME pathway enrichment analysis of the 394 upregulated genes. The top 10 pathways are represented in the bar plot (p value < 0.001).
Figure 5.
Genes Involved in the Type 1 IFN Signaling Pathway Are Repressed in 5H Compared to hTERT Cells and Induced by MV Infection
Gene expression of 84 genes was validated by a custom type 1 IFN RT2 Profiler PCR array (n = 3). (A) Heatmap representation of array genes in uninfected hTERT and 5H cells. Green represents low expression levels, and red represents high expression levels. (B) Gene expression profile showing differentially expressed genes in 5H compared to hTERT (absolute log2 fold change [FC] > 1 and p < 0.05). Gene names are shown on the x axis. Data are expressed as mean ± SEM. (C) Fold change in gene expression of the 84 analyzed genes at basal levels and upon induction by MV infection. (D) Differential expression of IFITM genes (IFITM1, IFITM2, and IFITM3) before and after MV infection of hTERT and 5H cells. *p = 0.0338 for IFITM1, hTERT versus 5H.
IFITM1 Expression Is Correlated with Progressive MSC Transformation
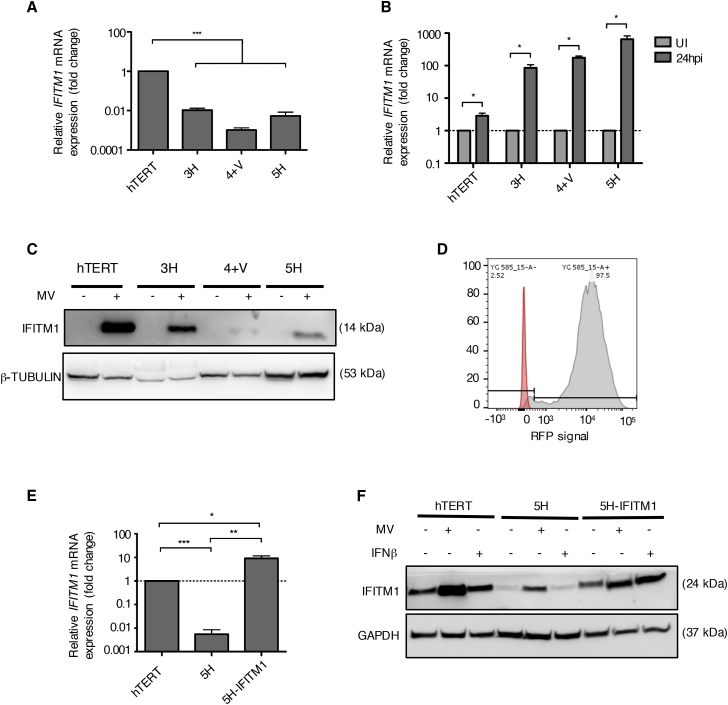
The viral-restricting properties of IFITM1 proteins have been documented for a number of viruses but not specifically for MV. Due to its prominence in our findings, we focused on IFITM1 as a candidate gene for potentially restricting MV infection. We analyzed our full set of sequentially transformed MSCs for basal expression of IFITM1 in the absence of MV infection by qRT-PCR (Figure 6A). IFITM1 expression was significantly lower in the more transformed MSCs compared to hTERT cells, with 4+V cells exhibiting the largest downregulation (−988.18-fold). IFITM1 expression was significantly induced in all MSCs at 24 h after MV infection to levels correlated with the MSCs transformational stage, with hTERT cells exhibiting the lowest induction (2.88-fold) and 5H showing the highest induction (771.4-fold) (Figure 6B). IFITM1 protein expression was evaluated at immunoblotting using cell lysates collected at 24 hpi. Interestingly, results did not follow the gene expression results; MV infection strongly induced IFITM1 protein expression in hTERT, and to lesser extent in 3H and 5H cells. IFITM1 expression was almost undetectable in 4+V cells (Figure 6C).
Figure 6.
IFITM1 Expression Is Correlated with Progressive MSC Transformation
(A) IFITM1 mRNA expression in all MSCs as assessed by qRT-PCR. Data shown are relative to housekeeping gene GAPDH and normalized to hTERT cells (n = 3). (B) IFITM1 mRNA expression measured by qRT-PCR at 24 hpi of all MSCs. Data shown are relative to housekeeping gene GAPDH and normalized to uninfected control cells (n = 3). (C) Immunoblot showing IFITM1 protein expression at 24 hpi in uninfected and MV-infected MSCs. β-Tubulin is used as a loading control. (D) Histograms of FACS analysis of the expression levels of red fluorescent protein (RFP), which was co-expressed with IFITM1, in transduced 5H cells. (E) IFITM1 mRNA expression levels in mock-infected and MV-infected hTERT, 5H, and 5H-IFITM1 cells at 24 hpi. Data shown are relative to housekeeping gene GAPDH and normalized to uninfected control cells (n = 3). (F) Immunoblots confirming the overexpression of IFITM1 in transduced 5H cells. GAPDH is used as a loading control. Data are expressed as mean ± SEM. All results shown are representative of three independent experiments (unpaired t test, *p < 0.05, **p < 0.01, ***p < 0.001). NS, not significant.
We hypothesized that if IFITM1 was an important factor restricting MV oncolysis in non-transformed cells, overexpression in transformed 5H cells would restrict the ability oncolytic capacity of MV. Hence, we stably overexpressed IFITM1 in the highly MV-permissive 5H cells (termed 5H-IFITM1) by retroviral vector transduction. Cells were sorted via fluorescence-activated cell sorting (FACS) by red fluorescent protein (RFP) expression (Figure 6D). Quantification of IFITM1 gene expression by qRT-PCR in transduced 5H cells showed that more than a 10,000-fold overexpression was obtained compared to control non-transduced 5H cells. Compared to hTERT cells, 5H-IFITM1 showed a 9.2-fold upregulation in IFITM1 expression (Figure 6E). Overexpression of IFITM1 in uninfected, non-MV-infected 5H cells was confirmed by western blotting (Figure 6F).
Restoration of IFITM1 Expression Restricts Oncolytic MV Infection in Transformed Cells
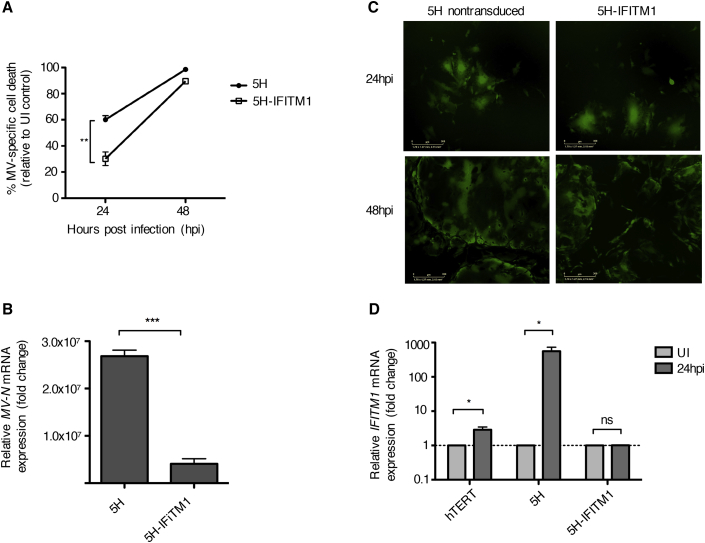
In order to determine the extent to which restoration of IFITM1 could restrict MV infection in transformed cells, 5H-IFITM1 and control 5H cells were infected with MV-NSe (MOI of 1.0) and assessed for cell viability, syncytia formation, and viral replication. As shown in Figure 7A, IFITM1 overexpression increased cell viability after MV infection by 50% at 24 hpi. By 48 hpi, most of the 5H and 5H-IFITM1 cells had succumbed to MV oncolysis. MV-N expression was significantly reduced in 5H-IFITM1 compared to 5H at 24 h (Figure 7B). MV-induced syncytia formation was only modestly affected by IFITM1 overexpression at 24 h and not at all by 48 h (Figure 7C). We next measured the expression of IFITM1 following MV infection in non-transduced 5H cells, 5H-IFITM1 cells and hTERT control cells. Both hTERT and 5H showed an increase of IFITM1 expression at 24 hpi, with hTERT cells exhibiting a smaller induction. However, the level of IFITM1 expression in 5H-IFITM1 cells remained unchanged following MV infection, suggesting that baseline IFITM1 expression is the most appropriate correlate of resistance to oncolytic MV infection in non-transformed cells (Figure 7D). Interestingly, despite finding no difference in the relative expression of IFITM1 mRNA after MV infection of 5H-IFITM1 cells, protein expression clearly increased as shown in Figure 6F. There are presumably a number of reasons for this reported in the literature, including post-transcriptional modifications, degradation of the mRNA, and the “translation on demand” where mRNA is preferentially translated under stressful conditions to ensure that the protein is rapidly available in response to signals.20,21 Taken together, using a stromal cell model of sequential transformation, our data indicate that the lack of an adequate IFN response is a key reason for the oncolytic selectivity of the vaccine strain of MV. In particular, we have identified IFITM1 as a restriction factor for oncolytic MV.
Figure 7.
IFITM1 Has Anti-Viral Properties and Partly Restricts MV Infection
(A) Cell viability of 5H cells and IFITM1-overexpressing 5H cells was assessed by trypan blue exclusion at 24 and 48 hpi with MV-NSe (MOI of 1.0). Data are expressed as a percentage of cell killing by MV relative to mock-infected control cells (n = 3). (B) MV-N gene expression measured by qRT-PCR at 24 hpi. Results shown are relative to GAPDH and normalized to mock-infected control cells (n = 3). (C) Representative fluorescence microscopy images of MV-GFP-infected non-transduced and 5H-IFITM1 cells. Scale bars, 300 μm. (D) IFITM1 mRNA expression was measured by qRT-PCR at 24 hpi. Results shown are relative to GAPDH and normalized to mock-infected control cells (n = 3). All data are expressed as mean ± SEM (unpaired t test, *p < 0.05, **p < 0.01, ***p < 0.001). MV, measles virus; UI, uninfected; NS, not significant.
Discussion
The mechanisms by which attenuated, vaccine strain MV exerts its oncolytic effects remain to be fully elucidated. In a well-characterized model of stepwise cellular transformation of BM-MSCs, we can clearly show that productive infection leading to higher levels of MV-mediated cell killing occurred only in more highly transformed cells. We were able to use this model to probe the contribution of the type 1 IFN pathway at different stages of transformation. While other groups have studied MV oncolysis in MV-permissive and non-permissive cancer cell lines or xenografts, our study allows for the direct comparison between transformed cells and their normal healthy counterparts. However, we have broadly similar findings. Berchtold et al.22 showed that in sarcoma cell lines, the upregulation of RIG-I and IFIT1, as well as STAT1 phosphorylation in response to MV infection, was correlated with their resistant phenotype. Furthermore, Achard et al.23 showed that replication of oncolytic MV in malignant pleural mesothelioma cell lines was restricted in cells with intact IFN signaling. Upon examining antiviral responses in MV-resistant and MV-permissive patient-derived glioblastoma xenografts (PDXs) using genome expression profiling, Kurokawa et al.18 showed that constitutive activation of the IFN pathway was critical for MV replication. Our data show that type 1 IFN production was closely correlated with progressive transformation, with cells at late stages of transformation producing lower levels of IFNβ in response to oncolytic MV infection compared to primary and hTERT MSCs. The observation that pre-treating MV-susceptible 5H cells with exogenous IFNβ restored resistance to MV-mediated oncolysis suggested that this was indeed a biologically plausible mechanism. Consistently, we observed a muted but also very delayed type 1 IFN response with increasing transformation, which was mirrored by the time course of STAT1 phosphorylation, which was already maximal at 4 hpi in hTERT cells but only began at 24 h in the fully transformed 5H cells.
Transcriptomic evaluation allowed us to gain a more global view of the impact of oncolytic MV in transformed and non-transformed cells. 5H cells expressed significantly lower basal levels of genes involved in the IFNγ pathway, the IFNα/β pathway, and the antigen presentation and processing pathways than did hTERT cells. However, oncolytic MV infection triggered the upregulation of genes involved in immune responses, including antigen presentation and IFN signaling in both cell types. In both the RNA-seq and the RT2 Profiler validation experiments, genes that were basally repressed in 5H cells, relative to hTERT cells, were induced by MV infection to levels equivalent to, or greater than, those that had been observed in hTERT cells at baseline. However, despite the induction of an immune response in 5H cells, the viral replication outpaced the antiviral response, suggesting that the differential susceptibility to MV infection is due to significant baseline differences in type 1 IFN pathway gene expression leading to a delayed response, rather than the inability of 5H cells to deploy those pathways upon challenge.
Unsurprisingly, given the importance of the IFN pathway, a number of ISGs such as OAS2, PKR, and MX1/2 have been reported as viral restriction factors.24 However, most ISGs remain to be functionally characterized, and it is of interest to know whether they might serve as predictive biomarkers for identifying patients who are more likely to respond to oncolytic MV virotherapy. Recently, Kurokawa et al.25 identified the ISG RSAD2 as an inhibitor of oncolytic MV replication by blocking viral release in both 293T cells and SR-B2, an ovarian cancer cell line. Our RNA-seq results showed that RSAD2 was also downregulated in 5H compared to hTERT cells; however, this was not statistically significant, suggesting that different ISGs may function in a cell type-specific manner. In contrast, our data highlighted IFN-inducible transmembrane protein 1 (IFITM1) as the foremost downregulated ISG in 5H transformed cells compared to non-transformed hTERT cells at baseline. Following oncolytic MV infection, IFITM1 expression increased only modestly in hTERT cells compared to a much higher, significant 771-fold increase in 5H cells, suggesting it may play a role in the selectivity of MV-mediated oncolysis in transformed cells. Interestingly, even though MV infection resulted in increased IFITM1 transcription in transformed MSCs, IFITM1 protein expression was not detected in the same cells, strongly implying the existence of post-transcriptional modifications. This observation is possibly a compensatory mechanism that contributes to the MV-susceptible phenotype of these cells. In support of this notion, mircoRNA-mediated regulations were reported to inhibit IFITM1 protein expression following infection with Kaposi’s sarcoma-associated herpesvirus26 and hepatitis C virus.27
The antiviral activities of IFITM family members IFITM1, IFITM2, and IFITM3 were demonstrated by a number of studies. IFITM1 particularly restricted enveloped RNA viruses, including hepatitis C virus,28, 29, 30 HIV,31 influenza virus A H1N1,32 Zika virus,33 respiratory syncytial virus (RSV),34,35 mumps virus, Newcastle disease virus (NDV), human metapneumovirus, and parainfluenza virus.34 The viral restriction properties of IFITM1 on a number of viruses that enter cells via the plasma membrane were recently investigated by Smith et al.34 A small but significant effect of IFITM1 overexpression was observed on the replication of strains of MV that are currently not used as cancer therapies, namely the Edmonston-Zagreb strain36 and a recombinant MV strain with a replaced P gene37 that has been shown to be competent for immune evasion.38 A proposed mechanism of action for the IFITM proteins in this regard is that the fluidity of cellular membranes can be altered, preventing fusion with the infecting virus envelope. Consistent with this, our own data do show fewer and smaller syncytia in hTERT versus 5H cells whereas virus release does not appear to be impacted, as we observed viral titers in the supernatants that were consistent with titers in the lysates at all of the transformational stages.
Consistent with a block to entry, our studies showed a 50% increase in cell viability after 24 h of infection in IFITM1-overexpressing 5H cells relative to control 5H cells. Moreover, IFITM1 overexpression altered the ability to establish an infection, as monitored by the significant decrease in expression of MV-N viral transcripts. However, by 48 hpi, multinucleated syncytia were readily apparent and cell killing reached levels similar to those in control 5H cells, suggesting that the antiviral effects of IFITM1 were indeed limited to early stages of infection.
Taken together, we have used a model of sequential transformation to show that a reduction in function of the type 1 IFN pathway is a major contributor to the selectivity of MV-mediated oncolysis for transformed cells. Our data have also identified IFITM1 as an oncolytic MV restriction factor. The mechanisms of action of IFITM1 remain to be determined. Co-factors that may be acting in a combined manner with IFITM1 may also be identified from our data and those of others. Mechanistic insights into MV oncolysis will assist with candidate tumor selection, trial design, and also help direct potential therapeutic combinations.
Materials and Methods
Cell Culture
Vero cells (African green monkey kidney cells, ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 5% fetal bovine serum (FBS; Gibco), l-glutamine (2 mM; Gibco), and 100 U/mL penicillin-streptomycin (Gibco). Primary MSCs were obtained from healthy BM samples after written consent. BM samples were filtered through a 40-μm cell strainer (Becton Dickinson), and BM mononuclear cells were isolated by density gradient centrifugation using Ficoll-Paque (Amersham Biosciences). All MSCs were cultured in MesenCult MSC basal medium with MesenCult stimulatory supplements (STEMCELL Technologies), 5% FBS, l-glutamine, penicillin-streptomycin, and 1 ng/mL recombinant basic human fibroblast growth factor (FGF) (R&D Systems). Phoenix-AMPHO cells (ATCC) were grown in DMEM supplemented with 10% FBS, penicillin-streptomycin, and l-glutamine. All cells were maintained in a 37°C humidified 5% CO2 incubator.
Model of Sequentially Transformed BM-MSCs
The transformed human BM-MSCs were developed and characterized elsewhere by Funes et al.19 Briefly, human BM-MSCs were sequentially infected with retroviral particles carrying the following expression vectors: pBABE-puro-EST2 (hTERT expression), pLXSN-neo-E6E7 (inactivation of p53 and Rb) to generate 3H cells, pBABE-zeo-ST (introduction of SV40 small T antigen) to generate 4+V cells, and pWZL-hygro-RasV12 (expression of oncogenic H-RasV12) to generate 5H cells. Following serial retroviral infections, drug selection with puromycin (100 μg/mL), neomycin (300 μg/mL), Zeocin (50 μg/mL), and hygromycin (100 μg/mL), respectively, were used to purify cell populations. The validity of this model was demonstrated by anchorage-independent growth, tumor growth in immunodeficient mice, and gene expression profiles consistent with transformed cells.19
Cell Counting and Viability Assays
Cell viability was measured using trypan blue (Sigma-Aldrich, Poole, UK) dye exclusion. Viability after MV infection was expressed as a percentage of non-infected cell viability.
Propagation and Titration of MV
Live-attenuated vaccine strain of MV (MV-NSe) and MV expressing recombinant GFP (MVNSe-GFP) were propagated on Vero cells. Vero cells were plated in 14-cm plates and infected at an MOI of 0.01 in Opti-MEM (Gibco). When 80%–90% cytopathic effect was observed, the virus was harvested by scraping the cells and subjected to two freeze-thaw cycles to release cell-associated viral particles. This was followed by high-speed centrifugation (4,000 rpm for 5 min at 4°C) to remove cell debris. Aliquots were stored at −80°C. Viral titers were determined by the end-point infectivity assay. The 50% tissue culture infected dose (TCID50) of MV stock was calculated using the modified Kärber39 formula.
MV Infections
For MV infection of adherent Vero cells and MSCs, cells were washed once with PBS and then inoculated with virus in Opti-MEM (Gibco) at an MOI of 1.0 in all experiments. Mock-infected cells were washed once and inoculated with Opti-MEM only. Cells were incubated for 2 h at 37°C before removal of the inoculum and replacement with fresh media. In conditions where cells were cultured in the presence of exogenous IFNβ, cells were pre-treated with concentrations ranging from 50 to 1,000 U/mL recombinant IFNβ (Millipore, UK, IF014) for 16 h before infection with MV-NSe at an MOI of 1.0.
MV Receptor Expression by Flow Cytometry
A total of 1–10 × 105 cells per aliquot were incubated with a FITC-labeled anti-human CD46 antibody (R&D Systems), or phycoerythrin (PE)-labeled anti-human CD105 antibody (BD Biosciences), or PE-labeled anti-human nectin-4 (R&D Systems) at 4°C in the dark for 30 min. Cells were then washed and resuspended in PBS. Samples were acquired on a BD FACSAria or LSR II flow cytometer (Becton Dickinson, Oxford, UK) with 5,000–10,000 events being recorded and analyzed with FlowJo (Tree Star). Results are expressed as mean fluorescence intensity (MFI). As a positive control for CD46 surface expression, Raji cells were used. Isotype-stained cells were used as negative controls.
Enzyme-Linked Immunosorbent Assay
MSCs were plated at a density of 5 × 105 per well in six-well plates and infected with MV-NSe. At 24 and 48 hpi, tissue culture supernatants were collected and stored at −80°C. Quantification of IFNα and IFNβ was performed using VeriKine human IFNα multi-subtype ELISA (PBL Assay Science, Piscataway, NJ, USA) and VerKine human IFNβ (PBL Assay Science, Piscataway, NJ, USA) kits, respectively, according to the manufacturer’s protocol.
qRT-PCR
MSCs were infected in six-well plates at 3 × 105 cells per well. Total cell RNA was extracted using the TRIzol (Invitrogen) method at 24 and 48 hpi. Quantification of RNA sample concentrations and purity was achieved by measuring UV-light absorbance at 260 (A260) and 280 nm (A280) using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). For each sample, 0.4–1 μg of total RNA was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol, and 60 ng of the synthesized cDNA product was used in triplicate reactions of quantitative PCR (qPCR). qPCR reactions were performed by mixing cDNA with 12.5 μL of 2× TaqMan universal PCR master mix (Applied Biosystems) and 1.25 μL of 20× TaqMan gene-specific primer and a probe mix (TaqMan gene expression assay, catalog nos. 4331182 and 4331348; Applied Biosystems) to a final reaction volume of 25 μL. The TaqMan assays used were as follows: IFITM1 (Hs00705137_s1), GAPDH (Hs02786624_g1), and custom MV-N assay (ID AIY896X) (all from Applied Biosystems). PCR reactions were carried out on an ABI 7500 fast real-time PCR system (Applied Biosystems) according to the following conditions: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. GAPDH was used as a housekeeping gene for all assays. Samples were run in triplicate for each gene, and non-template controls (NTCs) were included for each primer set. PCR cycle number at threshold is represented as Ct. Relative expression level of genes of interest was calculated using the comparative 2−ΔΔCt formula and expressed in fold change as compared to corresponding uninfected control cells.
RT2 Profiler PCR Array
Total RNA was analyzed using the human type 1 IFN response RT2 Profiler PCR array (catalog no. PAHS-016Z; QIAGEN), which profiles the expression of 84 gene transcripts that are known to be involved in the type 1 IFN response, as well as the expression of five housekeeping genes (ACTB, B2M, GAPDH, HPRT1, and RPLP0). In addition, one well contains a genomic DNA control, three wells contain reverse transcription controls, and three wells contain a positive PCR control. For each sample, 0.5 μg of RNA was reverse transcribed into cDNA using the RT2 first-strand kit (QIAGEN, UK). The cDNA was then mixed with the RT2 SYBR Green Mastermix (QIAGEN, UK) and nuclease-free water. Next, 25 μL of the PCR mix was added to each well of the 96-well plate. All steps were done according to the manufacturer’s instructions. The qPCR reaction was run on an ABI 7500 (fast block) according to the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 minute. Data analysis was conducted using a software-based tool (QIAGEN’s GeneGlobe Data Analysis Center). The exported Ct values for each well were uploaded into the software, which performed fold-change calculations based on the 2−ΔΔCt method. Expression levels were quantified relative to the values obtained for housekeeping genes.
Immunoblotting
MSCs were infected in six-well plates at a density of 3 × 105 cells per well. At specific time points after infection, cells were washed once with cold PBS before adding 100 μL of radioimmunoprecipitation assay (RIPA) cell lysis buffer (Sigma-Aldrich) with complete mini protease inhibitor cocktail tablets (Sigma-Aldrich, Roche) and harvested. After incubation on ice for 30 min, cell lysates were centrifuged at 14,000 rpm for 5 min at 4°C to pellet the cell debris. The supernatant was aliquoted and stored at −80°C until use. Protein concentrations were determined using the BCA (bicinchoninic acid) protein assay kit (Sigma-Aldrich). Equal amounts of protein (30μg-50 μg) were mixed with loading dye, incubated at 70°C for 10 min to denature the proteins, and then loaded onto 4%–12% Bolt Bis-Tris Plus gels (Thermo Fisher Scientific). Gels were subject to electrophoresis at 200 V for 22 min. After SDS-PAGE, the proteins were transferred to nitrocellulose membranes using the iBlot 2 gel transfer device (Thermo Fisher Scientific). Membranes were blocked in 3% non-fat dry milk in Tris-buffered saline (TBS) with Tween 20 (TBS-T) for 1 h at room temperature. This was followed by incubation with the following primary antibodies: rabbit anti-STAT1 (Cell Signaling Technology [CST], #9172), rabbit anti-phospho-STAT1 (Tyr701, CST, #7649), rabbit anti-IRF9 (CST, #28492), mouse anti-GAPDH (CST, #97166), rabbit anti-IFITM1 (Proteintech, 11727-3-AP; CST, #13126), and mouse β-tubulin (Thermo Fisher Scientific, BT7R). Primary antibodies were diluted 1:1,000 in 3% BSA or 3% milk in TBS-T at 4°C overnight. Membranes were washed three times for 5 min with TBS-T followed by incubating with anti-mouse immunoglobulin G (IgG) or anti-rabbit horseradish peroxidase-conjugated IgG secondary antibodies (Promega) for 1 h at room temperature. All secondary antibodies were diluted 1:50,000 in 3% milk in TBS-T. Membranes were washed again at this point with TBS-T to remove any unbound antibodies. Immunodetection was performed with Amersham ECL (enhanced chemiluminescence) prime western blotting detection reagent (GE Healthcare Life Sciences) using the ImageQuant Las 4000 system (GE Healthcare). Densitometry of bands was carried out using ImageJ and was normalized to the expression level of the relevant loading control.
RNA Sequencing and Differential Expression Analysis
TRIzol reagent (Invitrogen) was used to extract total RNA from uninfected or MV-infected hTERT and 5H MSCs at 24 hpi. Samples were processed using the KAPA mRNA library prep hyper kit and sequenced on the Illumina NextSeq 500 by the UCL Genomics facility (London, UK). RNA-seq data were analyzed for differential gene expression using the DESeq240,41 and SARTools (developed at PF2- Institut Pasteur) R packages. DESeq2 provides statistical methods for determining differential expression data using a model based on the negative binomial distribution. Volcano plots were generated by R (R Core Team, 2014) to display differential gene expression results between two comparison groups. Each point represents the average value of one transcript in three replicate experiments. DEGs were selected based on an absolute log2 fold change >1 and an adjusted p value <0.05. Red and green dots represent upregulated and downregulated genes compared to control, respectively. Gray dots represent non-significantly regulated genes. Pathway enrichment analysis was performed on DEGs using Reactome (https://www.reactome.org).42 RNA-seq data have been deposited in the GEO repository under the accession number GSE131840.
Construction of the IFITM1-Expressing Vector
The full-length IFITM1 cDNA was PCR amplified from the pCMV-HA-IFITM1 plasmid (a gift from H. Hang and J. Yount, Addgene, plasmid #58399), using primers tailed with BamHI and XhoI linker restriction sites at the 5′ and 3′ ends, respectively (Sigma-Aldrich): forward primer, 5′-tctgatGGATCCtctgatATGCACAAGGAGGAACAT-3′; reverse primer, 5′- GAAAAACGGGGTTACTAGtctgatCTCGAGtctgat-3′. The amplified PCR fragment (378 bp) was cloned into a pCR II-TOPO vector using the TOPO TA cloning kit (Invitrogen) according to the manufacturer’s protocol and sequenced for confirmation (Eurofins) before being cloned into the retroviral vector MSCV-IRES-mRFP (a gift from Charles G. Mullighan, St. Jude Children’s Research Hospital, Memphis, TN, USA; Figure S1). To obtain the overexpressing construct, pCR II-TOPO-IFITM1 and MSCV-IK6-IRES-mRFP plasmids were both digested using BamHI high-fidelity and XhoI restriction enzymes (New England Biolabs [NEB]). The digested vectors were separated by electrophoresis using 2% agarose gels, and the target IFITM1 insert and MSCV backbone were purified using the QIAex II gel purification kit (QIAGEN). The purified fragments were ligated using T4 DNA ligase (NEB) and transformed into One Shot DH5α-T1 E. coli competent cells (Invitrogen). The recombinant plasmid was isolated from bacterial pellets using the HiSpeed Plasmid Midi kit (QIAGEN). Agarose gel electrophoresis confirmed the presence of the correct IFITM1 insert, and the sequence was verified (Eurofins Genomics, UK). A schematic representation of the construction of the MSCV-IRES-RFP-IFITM1 plasmid is illustrated in Figure S1.
Retroviral Transduction of 5H MSCs
For the preparation of retroviral vector particles, Phoenix-AMPHO cells were co-transfected with the appropriate plasmids using a FuGENE high-density transfection reagent (Roche) method. Briefly, 2 × 106 Phoenix-AMPHO cells were plated into 10-cm Petri dishes in 8 mL of fresh media and incubated overnight. On the following day, the transfection mix was prepared by first adding 10 μL of FuGENE and 150 μL of Opti-MEM, which represents solution A. Solution B was prepared by adding 1.5 μg of pCL-ampho retrovirus packaging vector (Imgenex), 2.6 μg of MSVC-IFITM1-IRES-mRFP plasmid, and adjusting the volume to 50 μL with distilled H2O. The DNA mix was then added to the FuGENE solution, mixed by gentle pipetting, and incubated at room temperature for 15–20 min. The liposomal complexes were added directly to the Phoenix-AMPHO cells and then returned to the incubator. Three days after transfection, medium was replaced with 5 mL of fresh DMEM media. Retroviral supernatants from the transfected packaging cells were ready to be used for transduction on the following day. Collected supernatants were spun at 1,500 rpm for 5 min to remove remaining cells and cell debris and used for infection of 5H MSCs. FACS analysis was performed for sorting RFP-positive cells.
Statistical Analysis
Statistical analysis and graph plotting were performed using GraphPad Prism 5.0. Graph data are represented as mean ± SEM. For comparisons involving two groups, paired or unpaired Student’s t tests (two-tailed) were used for statistical analysis. A p value less than 0.05 was considered to be statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001). The number of independent experiments performed is indicated by “n” in the figure legends.
Author Contributions
S.A. and A.Z.C. designed experiments, performed experiments, and interpreted data. A.K.F. and S.A. wrote the paper. D.L. and S.A. performed RNA-seq analysis. A.D., R.B., D.O., K.B., and R.J.M. provided technical assistance and contributed to the interpretation of the results. A.K.F. conceived the project, critically evaluated experiments, and supervised the study.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank all of the members of the Fielding laboratory for their feedback on this project. We thank David Gullinghan (St. Jude Children’s Research Hospital) for providing the MSCV-IK6 plasmid. We thank Juan M. Funes and Chris Boshoff for the transformed MSC model. We thank UCL Genomics for the RNA sequencing. This work was funded by a grant to S.A. from the King Abdullah International Medical Research Center (KAIMRC; Riyadh, Saudi Arabia).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.01.027.
Supplemental Information
References
- 1.Prestwich R.J., Harrington K.J., Pandha H.S., Vile R.G., Melcher A.A., Errington F. Oncolytic viruses: a novel form of immunotherapy. Expert Rev. Anticancer Ther. 2008;8:1581–1588. doi: 10.1586/14737140.8.10.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Msaouel P., Opyrchal M., Dispenzieri A., Peng K.W., Federspiel M.J., Russell S.J., Galanis E. Clinical trials with oncolytic measles virus: current status and future prospects. Curr. Cancer Drug Targets. 2018;18:177–187. doi: 10.2174/1568009617666170222125035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moss W.J., Griffin D.E. Global measles elimination. Nat. Rev. Microbiol. 2006;4:900–908. doi: 10.1038/nrmicro1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naniche D., Varior-Krishnan G., Cervoni F., Wild T.F., Rossi B., Rabourdin-Combe C., Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dörig R.E., Marcil A., Chopra A., Richardson C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 6.Tatsuo H., Ono N., Tanaka K., Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 7.Noyce R.S., Bondre D.G., Ha M.N., Lin L.T., Sisson G., Tsao M.S., Richardson C.D. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mühlebach M.D., Mateo M., Sinn P.L., Prüfer S., Uhlig K.M., Leonard V.H., Navaratnarajah C.K., Frenzke M., Wong X.X., Sawatsky B. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navaratnarajah C.K., Oezguen N., Rupp L., Kay L., Leonard V.H., Braun W., Cattaneo R. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat. Struct. Mol. Biol. 2011;18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson B.D., Nakamura T., Russell S.J., Peng K.W. High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus. Cancer Res. 2004;64:4919–4926. doi: 10.1158/0008-5472.CAN-04-0884. [DOI] [PubMed] [Google Scholar]
- 11.Miest T.S., Frenzke M., Cattaneo R. Measles virus entry through the signaling lymphocyte activation molecule governs efficacy of mantle cell lymphoma radiovirotherapy. Mol. Ther. 2013;21:2019–2031. doi: 10.1038/mt.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stojdl D.F., Lichty B., Knowles S., Marius R., Atkins H., Sonenberg N., Bell J.C. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 2000;6:821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 13.Plumet S., Herschke F., Bourhis J.M., Valentin H., Longhi S., Gerlier D. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE. 2007;2:e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikegame S., Takeda M., Ohno S., Nakatsu Y., Nakanishi Y., Yanagi Y. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J. Virol. 2010;84:372–379. doi: 10.1128/JVI.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berghäll H., Sirén J., Sarkar D., Julkunen I., Fisher P.B., Vainionpää R., Matikainen S. The interferon-inducible RNA helicase, mda-5, is involved in measles virus-induced expression of antiviral cytokines. Microbes Infect. 2006;8:2138–2144. doi: 10.1016/j.micinf.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurokawa C., Iankov I.D., Anderson S.K., Aderca I., Leontovich A.A., Maurer M.J., Oberg A.L., Schroeder M.A., Giannini C., Greiner S.M. Constitutive interferon pathway activation in tumors as an efficacy determinant following oncolytic virotherapy. J. Natl. Cancer Inst. 2018;110:1123–1132. doi: 10.1093/jnci/djy033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funes J.M., Quintero M., Henderson S., Martinez D., Qureshi U., Westwood C., Clements M.O., Bourboulia D., Pedley R.B., Moncada S., Boshoff C. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc. Natl. Acad. Sci. USA. 2007;104:6223–6228. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenbaum D., Colangelo C., Williams K., Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Berchtold S., Lampe J., Weiland T., Smirnow I., Schleicher S., Handgretinger R., Kopp H.G., Reiser J., Stubenrauch F., Mayer N. Innate immune defense defines susceptibility of sarcoma cells to measles vaccine virus-based oncolysis. J. Virol. 2013;87:3484–3501. doi: 10.1128/JVI.02106-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achard C., Boisgerault N., Delaunay T., Roulois D., Nedellec S., Royer P.J., Pain M., Combredet C., Mesel-Lemoine M., Cellerin L. Sensitivity of human pleural mesothelioma to oncolytic measles virus depends on defects of the type I interferon response. Oncotarget. 2015;6:44892–44904. doi: 10.18632/oncotarget.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurokawa C., Iankov I.D., Galanis E. A key anti-viral protein, RSAD2/VIPERIN, restricts the release of measles virus from infected cells. Virus Res. 2019;263:145–150. doi: 10.1016/j.virusres.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussein H.A.M., Akula S.M. miRNA-36 inhibits KSHV, EBV, HSV-2 infection of cells via stifling expression of interferon induced transmembrane protein 1 (IFITM1) Sci. Rep. 2017;7:17972. doi: 10.1038/s41598-017-18225-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhanja Chowdhury J., Shrivastava S., Steele R., Di Bisceglie A.M., Ray R., Ray R.B. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J. Virol. 2012;86:10221–10225. doi: 10.1128/JVI.00882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkins C., Woodward J., Lau D.T., Barnes A., Joyce M., McFarlane N., McKeating J.A., Tyrrell D.L., Gale M., Jr. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology. 2013;57:461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raychoudhuri A., Shrivastava S., Steele R., Kim H., Ray R., Ray R.B. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayana S.K., Helbig K.J., McCartney E.M., Eyre N.S., Bull R.A., Eltahla A., Lloyd A.R., Beard M.R. The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J. Biol. Chem. 2015;290:25946–25959. doi: 10.1074/jbc.M115.657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J., Pan Q., Rong L., He W., Liu S.L., Liang C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011;85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savidis G., Perreira J.M., Portmann J.M., Meraner P., Guo Z., Green S., Brass A.L. The IFITMs inhibit Zika virus replication. Cell Rep. 2016;15:2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- 34.Smith S.E., Busse D.C., Binter S., Weston S., Diaz Soria C., Laksono B.M., Clare S., Van Nieuwkoop S., Van den Hoogen B.G., Clement M. Interferon-induced transmembrane protein 1 restricts replication of viruses that enter cells via the plasma membrane. J. Virol. 2019;93:e02003-18. doi: 10.1128/JVI.02003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W., Zhang L., Zan Y., Du N., Yang Y., Tien P. Human respiratory syncytial virus infection is inhibited by IFN-induced transmembrane proteins. J. Gen. Virol. 2015;96:170–182. doi: 10.1099/vir.0.066555-0. [DOI] [PubMed] [Google Scholar]
- 36.Rennick L.J., de Vries R.D., Carsillo T.J., Lemon K., van Amerongen G., Ludlow M., Nguyen D.T., Yüksel S., Verburgh R.J., Haddock P. Live-attenuated measles virus vaccine targets dendritic cells and macrophages in muscle of nonhuman primates. J. Virol. 2015;89:2192–2200. doi: 10.1128/JVI.02924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Vries R.D., Lemon K., Ludlow M., McQuaid S., Yüksel S., van Amerongen G., Rennick L.J., Rima B.K., Osterhaus A.D., de Swart R.L., Duprex W.P. In vivo tropism of attenuated and pathogenic measles virus expressing green fluorescent protein in macaques. J. Virol. 2010;84:4714–4724. doi: 10.1128/JVI.02633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devaux P., von Messling V., Songsungthong W., Springfeld C., Cattaneo R. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology. 2007;360:72–83. doi: 10.1016/j.virol.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 39.Kärber G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol. 1931;162:480–483. [Google Scholar]
- 40.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabregat A., Sidiropoulos K., Viteri G., Forner O., Marin-Garcia P., Arnau V., D’Eustachio P., Stein L., Hermjakob H. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18:142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.