Abstract
Spinal muscular atrophy (SMA) is a severe neuromuscular disease affecting infants caused by alterations of the survival motor neuron gene, which results in progressive degeneration of motor neurons (MNs). Although an effective treatment for SMA patients has been recently developed, the molecular pathway involved in selective MN degeneration has not been yet elucidated. In particular, miR-206 has been demonstrated to play a relevant role in the regeneration of neuromuscular junction in several MN diseases, and particularly it is upregulated in the quadriceps, tibialis anterior, spinal cord, and serum of SMA mice. In the present paper, we demonstrated that miR-206 was transiently upregulated also in the brainstem of the mouse model of SMA, SMAΔ7, in the early phase of the disease paralleling MN degeneration and was down-regulated in the late symptomatic phase. To prevent this downregulation, we intracerebroventricularly injected miR-206 in SMA pups, demonstrating that miR-206 reduced the severity of SMA pathology, slowing down disease progression, increasing survival rate, and improving behavioral performance of mice. Interestingly, exogenous miRNA-206-induced upregulation caused a reduction of the predicted target sodium calcium exchanger isoform 2, NCX2, one of the main regulators of intracellular [Ca2+] and [Na+]. Therefore, we hypothesized that miR-206 might exert part of its neuroprotective effect modulating NCX2 expression in SMA disease.
Keywords: spinal muscular atrophy, motor neuron disease, miR-206, NCX2, Brainstem
Motor neuron degeneration occurs very early in the facial nuclei of SMA mice, paralleling a transient upregulation of miR-206. The prolonged increase of miR-206 induced by i.c.v. administration reduces motor neuron loss, ameliorates behavioral performance, and increases mouse lifespan, also through the modulation of the plasma membrane Na+/Ca2+ exchanger 2, NCX2.
Introduction
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disease, most frequent in infants, characterized by selective degeneration of lower motor neurons (MNs), resulting in progressive muscle weakness, atrophy, paralysis and death, due to respiratory failure.1 The disease is caused by homozygous deletions or mutations in the survival motor neuron gene 1, SMN1, leading to the deficiency of the ubiquitous SMN protein.2 The human genome contains the SMN2 gene, that is an SMN1 paralog, and it differs only in few nucleotides. However, a C-to-T transition occurring 6 bp inside SMN2 exon 7 profoundly influences mRNA splicing, leading to an unstable, rapidly degradable protein, lacking exon 7, SMNΔ7.1,3 Almost 10% of SMN2-encoded protein is a full-length functional SMN (fl-SMN) protein; indeed, the severity of the disease is inversely proportionate to SMN2 copy number.4,5 Clinical features range from the most severe form SMAI to the mildest SMAIV, according to the age of onset and motor milestones achieved by patients.4 Although several therapeutic options have been recently available for SMA patients, the molecular pathway involved in selective MN degeneration has not been yet elucidated. It is known that SMN1 encodes a 38-kDa protein6 that forms a large multiprotein complex containing SMN, Gemin2,7 Gemin5,8 Gemin6,9 and Gemin7.10 This multiprotein complex has its primary and most prominent role in spliceosomal assembly, pre-mRNA maturation, and assembly of microRNA (miRNA)-ribonucleoprotein complexes (miRNPs) and transcriptosomes.11 In addition, the importance of some miRNA families in SMA pathophysiology has been recently recognized. In fact, miRNAs are evolutionarily conserved non-coding RNA of 18–22 nucleotides that post-transcriptionally regulate the expression of most mammalian genes and play a major role in a variety of biological processes such as differentiation, proliferation, apoptosis, and development. miRNA dysregulation has been related to several disorders, including muscular dystrophy,12 diabetes,13 and cancer.14,15 In particular, several pieces of evidence showed a close relationship between miRNA alteration and SMA. In fact, it has been reported that mice that specifically lack the miRNA-processing enzyme Dicer in MNs display hallmarks of SMA,16 and that SMN protein is able to alter miRNA expression and distribution in neurons.17,18 In particular, miR-206, a miRNA highly conserved in Homo sapiens, Rattus norvegicus, and Mus musculus, is involved in the proper formation and regeneration of the mature neuromuscular junction (NMJ).19 In fact, miR-206 acts as a key regulator of the interplay between MNs and muscle fibers, sensing MN damage and promoting the regeneration of functional synapses in the muscle.19 In particular, Williams and colleagues,20 demonstrated that healthy mice that are genetically deficient in miR-206 form normal neuromuscular synapses during development, but knocking out of miR-206 in the amyotrophic lateral sclerosis (ALS) mouse model SOD1-G93A accelerates disease progression, thus demonstrating that miR-206 is required for efficient regeneration of neuromuscular synapses. On the other hand, it has been previously demonstrated that miR-206 is upregulated in the quadriceps of SMAΔ7, a mouse model of SMA,21 and in the spinal cord of SMAI,22 only at post-natal day 10 (P10), a few days before animal death, suggesting that this miRNA upregulation occurs too late to revert disease progression.
In addition, with the help of in silico analysis, we found that among the numerous putative targets of miR-206, there is the SLC8A2 gene coding for the plasma membrane protein sodium calcium exchanger isoform 2, NCX2. This protein belongs to the Na+/Ca2+ exchanger (NCX) family, a group of three NCX1, NCX2, and NCX3 transmembrane proteins that play a relevant role in the maintenance of Ca2+ and Na+ homeostasis in the central nervous system (CNS) cells, and it is involved in several pathophysiological conditions such as brain ischemia,23, 24, 25, 26 multiple sclerosis (MS),27 Alzheimer’s disease (AD),28 and ALS.29,30 In this regard, it should be underlined that Ca2+ homeostasis deregulation seems to be a key factor in triggering SMA toxicity of MNs, motor nerve terminals, and astrocytes.31, 32, 33
In the light of these premises, we were prompted to investigate whether (1) miR-206 might play a role as a sensor of MN degeneration in brain regions of CNS, (2) the administration of exogenous miR-206 in a very early phase might represent a strategy to attenuate MN degeneration, and (3) miR-206 modulates NCX2 expression, thus interfering with Ca2+ homeostasis.
Results
miR-206 Is Upregulated in the Brainstem of Early Symptomatic SMA Mice
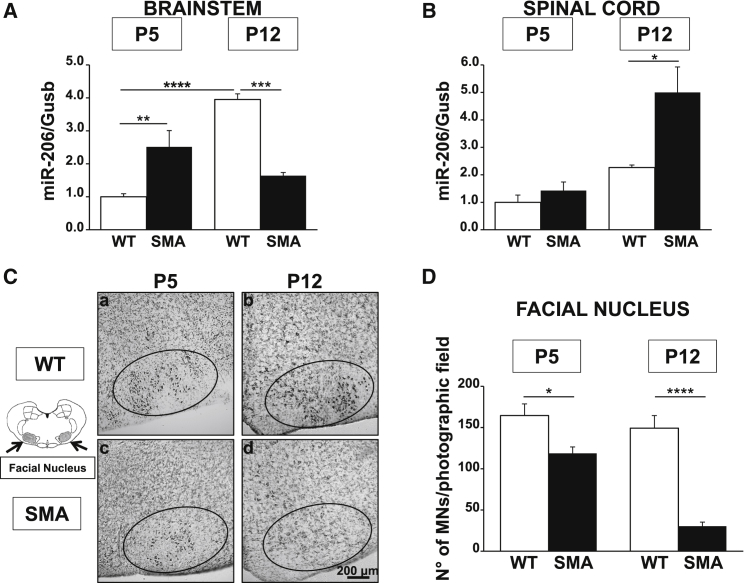
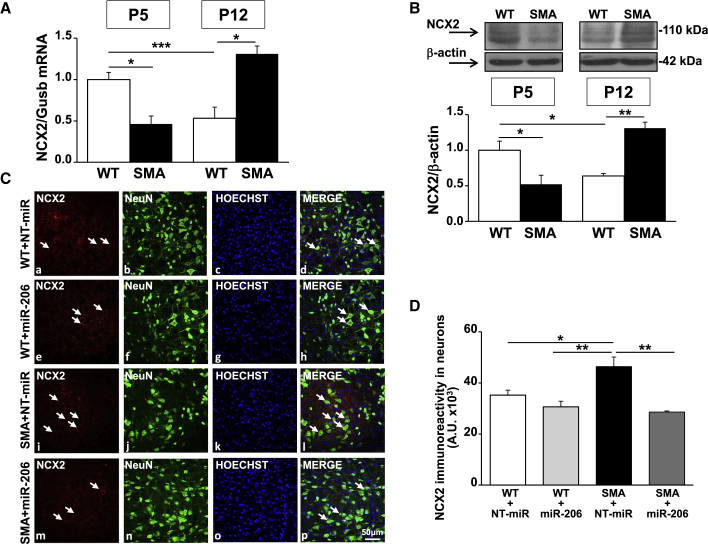
To investigate whether miR-206 may play a role in SMA disease, we measured miR-206 levels in the brainstem and in the spinal cord of early (P5) and late (P12) symptomatic smn−/−; SMN2+/+; SMNΔ7+/+ (SMA) mice and compared these values to those of age-matched healthy controls (smn+/+; SMN2+/+; SMNΔ7+/+; wild-type [WT]) (Figures 1A and 1B). This mouse model, SMNΔ7, is one of the most common used to study SMA at the preclinical level. Mice at birth are smaller than normal littermates and have a lifespan of about 13 days and impairment of motor behavior clearly detectable 4–5 days after birth (P4–P5).34 In particular, our data revealed that a statistically significant 2.5-fold increase in miR-206 expression level was observed in the brainstem of early symptomatic SMA mice compared to WT littermate animals. This effect was completely abolished in the late phase of the disease (Figure 1A). Indeed, a 60% reduction in miR-206 levels in the brainstem of SMA pups at P12, compared to age-matched WT animals, was observed (Figure 1A). Interestingly, in WT animals, a physiological increase in miR-206 expression during growth occurred, while in SMA mice this upregulation did not take place. In contrast, in the spinal cord of SMA mice, a significant increase (2.2-fold) in miR-206 level was observed in SMA pups compared to WT only in the late phase of the disease (P12) (Figure 1B). On the other hand, miR-206 levels in the spinal cord of SMA animals at P5 were not statistically different from WT mice, while the latter did not show a statistically significant increase of miR-206 levels during growth (Figure 1B). Moreover, the number of MNs in the facial nuclei of SMA mice at different ages was evaluated by Nissl staining and compared to age-matched WT pups (Figures 1C and 1D). Interestingly, a significant 25% reduction in the amount of MNs was already detected at P5 (WT, 164 ± 14 MNs/mm2; versus SMA, 119 ± 8 MNs/mm2); this phenomenon was more evident at P12 (WT, 149 ± 15 MNs/mm2; versus SMA, 30 ± 5 MNs/mm2) (Figure 1D). In fact, in the facial nuclei of SMA mice, an 80% loss of MNs compared to WT pups of the same age was detected (Figure 1D).
Figure 1.
miR-206 Was Upregulated in the Brainstem of SMA Mice Parallel to MN Loss in the Facial Nuclei in the Early Phase of the Disease
(A and B) Real-time PCR of miR-206 for the evaluation of its expression levels in the brainstem (A) and in the spinal cord (B) of WT (white columns) and SMA (black) mice, five (P5) and 12 (P12) days after birth showing miR-206 upregulation at P5 in the brainstem and at P12 in the spinal cord. (C) Nissl-stained sections (20 μm) of the brainstem of WT (a and b) and SMA (c and d) mice at P5 (a and c) and P12 (b and d) showing MN reduction in the facial nuclei (ellipse area) of SMA mice. Scale bars 200 μm. (D) Quantification of MN number normalized for photographic field in the facial nuclei of WT and SMA mice at P5 and P12. *p < 0.05; **p < 0.005; ***p < 0.001; ****p < 0.0001 by one-way ANOVA analysis followed by Bonferroni test. Each column represents the mean ± SEM (n = 3/6).
Intracerebroventricular Administration of miR-206 Reduced MN Loss in the Facial Nuclei of the Brainstem and Extended Survival of SMA Mice
In order to assess the protective effect of miR-206, it was intracerebroventricularly (i.c.v.) administered to WT and SMA pups three times at P3, P6, and P10 (10 pmol/1 μL per mouse), randomly dividing them into four experimental groups: WT+NT-miR, WT+miR-206, SMA+NT-miR, and SMA+miR-206. To evaluate whether miR-206 could effectively reach the brainstem and the spinal cord regions, its levels were measured at P12 in WT animals i.c.v. injected at P3, P6, and P10 with NT-miR or miR-206 molecules (Figure S1). Interestingly, a 50% and 85% increase in miR-206 levels was detected in the brainstem (Figure S1A) and in the spinal cord (Figure S1B) of WT pups treated with miR-206 (Figure S1).
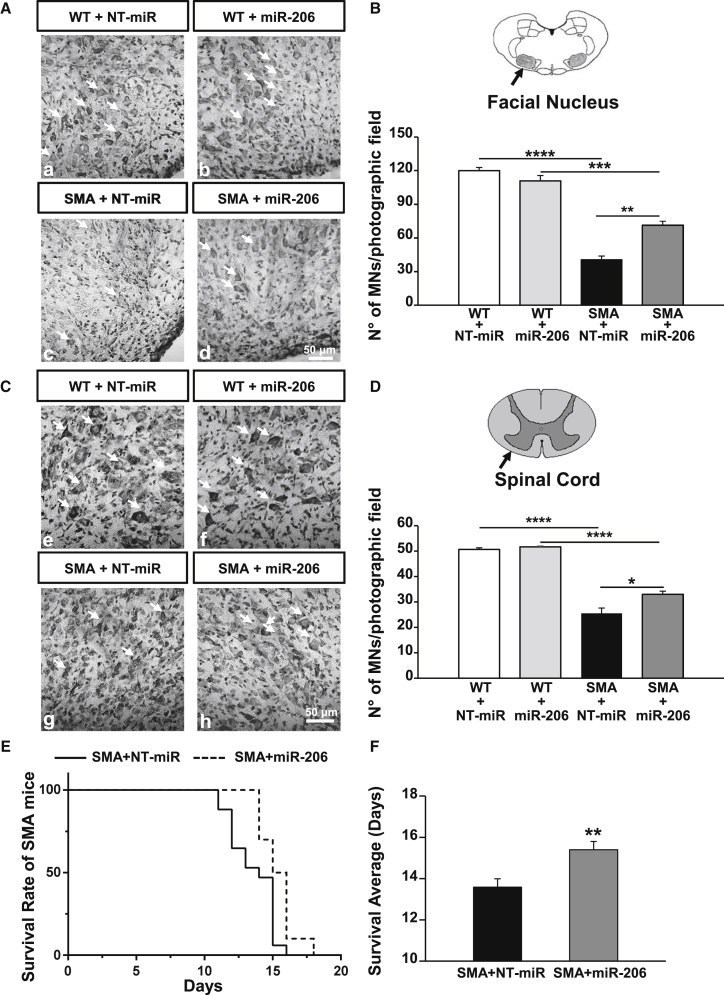
The brainstem (Figures 2A and 2B) and spinal cord sections (Figures 2C and 2D) of WT and SMA pups at P12, treated three times with miR-206 or with NT-miR, were stained with Nissl, and MNs were counted. miR-206 i.c.v. administration preserved large MNs in the facial nuclei of SMA mice (Figures 2A and 2B). In fact, a 65% reduction of MNs was observed in this region of SMA mice treated with NT-miR (WT+NT-miR, 120 ± 3 MNs/mm2; versus SMA+NT-miR, 40 ± 3 MNs/mm2), while in SMA mice treated with miR-206, MN loss was limited to 40% (WT+ miR-206, 111 ± 5 MNs/mm2; versus SMA+miR-206, 71 ± 3 MNs/mm2) (Figure 2B). More importantly, miR-206 i.c.v. administration reduced MN loss of approximatively 25% in the facial nuclei of SMA animals. Notably, miR-206 administration did not alter the number of MNs in the facial nuclei of WT mice (Figure 2B). Furthermore, miR-206 i.c.v. administration reduced MN death also in the spinal cord of SMA mice (Figures 2C and 2D). In particular, a 50% reduction in MN number was observed in SMA mice treated with NT-miR, compared to age-matched WT+NT-miR pups (WT+NT-miR, 50.7 ± 0.7 MNs/mm2; versus SMA+NT-miR, 26 ± 2 MNs/mm2) (Figure 2D). Interestingly, MN loss in the spinal cord of SMA animals treated with miR-206 was limited to 35% (WT+miR-206, 51.7 ± 0.4 MNs/mm2; versus SMA+miR-206, 33.0 ± 1.3 MNs/mm2) (Figure 2D).
Figure 2.
miR-206 Reduced MN Loss in the Facial Nucleus and in the Spinal Cord and Extended Survival of SMA Mice
(A and C) Nissl-stained sections (20 μm) of the brainstem (A) and the spinal cord (C) of WT (a, b, e, and f) and SMA (c, d, g, and h) mice at P12, i.c.v. treated with 1 μL (10 μM) of a non-targeting microRNA (NT-miR; a, c, e, and g) or with miR-206 (b, d, f, and h) showing MNs loss in the facial nuclei (ellipse area) and in the spinal cord of miR-206 treated SMA mice (MNs are indicated by arrows). Scale bars, 50 μm. (B and D) Quantification of MN number normalized for the photographic field in the facial nuclei (B) and in the spinal cord (D) of WT+NT-miR (white columns); WT+miR-206 (light gray); SMA+NT-miR (black); SMA+miR-206 (dark gray) mice at P12. *p < 0.05; **p < 0.005; ***p < 0.001; ****p < 0.0001 by one-way ANOVA analysis followed by Bonferroni test. Each column represents the mean ± SEM (n = 3). (E and F) Kaplan-Meier survival analysis, expressed as a percentage (E) or in days (F), of SMA+NT-miR (continuous line and black column; n = 17) and SMA+miR-206 (dashed line and dark gray column, n = 11) indicating that i.c.v. miR-206 administration can significantly extend SMA lifespan. *p < 0.05 by log rank statistical test for (E) and **p < 0.005 by Student’s t test for (F).
Finally, miR-206 extended the lifespan of SMA pups (Figure 2E). In fact, the average lifespan of SMA mice treated with NT-miR was 13.6 ± 0.4 days, while that of SMA mice treated with miR-206 was significantly longer, being 15.4 ± 0.4 days (Figure 2F).
i.c.v. Administration of miR-206 Reduced the Severity of Motor Deficits in SMA Mice
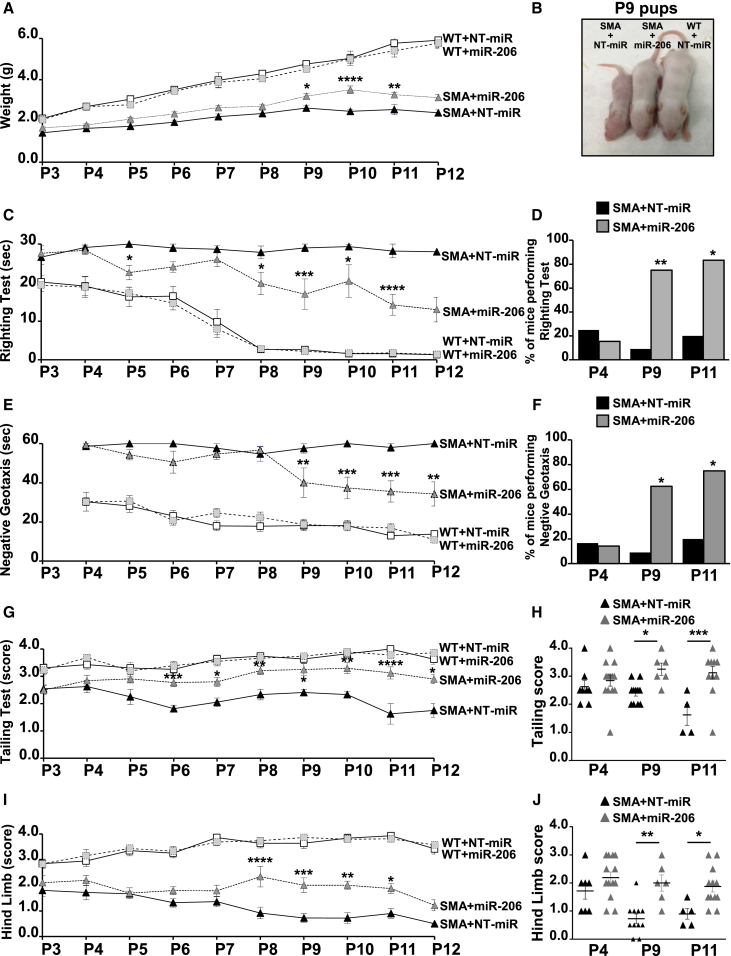
To investigate whether the reduced loss of MNs observed in miR-206-treated SMA pups could ameliorate the motor performance of these mice, a battery of behavioral tests was daily performed on SMA pups for 10 days (P3–P12), starting from the third day after birth (P3), when symptoms and i.c.v. injection of miR-206 began. The animals, randomly divided in four experimental groups, WT+NT-miR, WT+miR-206, SMA+NT-miR, and SMA+miR-206, were weighed and subjected to righting, negative geotaxis, tailing, and hind limb suspension tests, the most adequate behavioral tests for SMA mice of this age35 (Figure 3).
Figure 3.
miR-206 Ameliorated Growth Curve and Motor Behavioral Performances of SMA Mice
(A) Growth curves of WT+NT-miR (white squares, continuous line); WT+miR-206 (light gray squares, dashed line); SMA+NT-miR (black triangles, continuous line); SMA+miR-206 (dark gray triangles, dashed line) mice. (B) A representative picture of P9 littermates: SMA+NT-miR; SMA+miR-206; WT+NT-miR. (C, E, and G–J) Righting (C), negative geotaxis (E), tailing suspension (G and H), and hind limb suspension (score; I and J) tests of WT+NT-miR; WT+miR-206; SMA+NT-miR; SMA+miR-206 pups. (D and F) Contingency table for righting (D) and negative geotaxis (F) tests for SMA+NT-miR (black column) and SMA+miR-206 (gray) mice. (H and J) Scattered graph plots of tailing (H) and hind limb (J) scores of each SMA+NT-miR (black triangle) and SMA+miR-206 (gray) animal at P4, P9, and P11. *p < 0.05; **p < 0.005; ***p < 0.001; ****p < 0.0001 SMA+miR-206 versus SMA+NT-miR by 2-ways ANOVA analysis (A, C, E, G, and I), by one-way ANOVA analysis (H and J) followed by Bonferroni test and by Fisher exact test for (D) and (F).
Body Weight
miR-206-treated SMA mice showed higher weight than NT-miR-treated ones from P9 (Figures 3A and 3B). Indeed, at P9, a statistically significant higher value of approximately 22% was reached, compared to NT-miR-treated mice (Figures 3A and 3B). 10 and 11 days after birth, the weight of SMA+miR-206 mice was 42% and 28% higher compared to age-matched SMA+NT-miR, respectively (Figures 3A). No differences were observed between WT pups treated with miR-206 and WT mice treated with the non-targeting miRNA.
Righting Test
Motor coordination of WT and SMA pups treated with miR-206 or with NT-miR was evaluated measuring the time the pups required, positioned upside down, to reposition themselves dorsal side up (30 s was considered the cutoff time). WT animals could complete the test in 20 s at P3 and within a few seconds from P8 (Figure 3C). Also in this case, differences between WT pups treated with miR-206 or WT mice treated with NT-miR were not detected. On the other hand, approximately 25% of SMA+NT-miR pups completed the test with an average time of 29.1, 27.8, and 28.2 s at P4, P8, and P11, respectively (Figures 3C and 3D). Interestingly, the 75% of SMA mice (P8) treated with miR-206 was able to pass the test in an average time of 19.8 s, with a significant improvement in behavioral performance also at P9 (SMA+miR-206, 17.0 ± 3.9 s; versus SMA+NT-miR, 29.0 ± 1.0 s), P10 (SMA+miR-206, 20.4 ± 4.3 s; versus SMA+NT-miR, 29.3 ± 0.5 s), and P11 (SMA+miR-206, 14.2 ± 2.7 s; versus SMA+NT-miR, 28.2 ± 1.8 s) when the 80% of mice were able to complete the test (Figure 3D).
Negative Geotaxis Test
To evaluate both motor coordination and vestibular sensitivity, the mice were positioned on an inclined grid (approximately 35°) with the head facing down, and the time the mice spent to turn around with the head facing up was measured (the cutoff time was 60 s) (Figure 3E). At P4, only 16.7% of SMA+NT-miR pups were able to perform the test with no significant differences with miR-206-treated mice, taking a time of 58.7 and 59.4 s, respectively (Figures 3E and 3F). Interestingly, at P9, the 60% of SMA+miR-206 pups were able to complete the test with an average time of 40.1 s, compared with the 9.1% of age-matched SMA+NT-miR mice that completed the test in 57.0 s (Figures 3E and 3F). Significant improvements upon miR-206 i.c.v. injection were measured also at P10 (SMA+miR-206, 37.4 ± 5.5 s; versus SMA+NT-miR, 60.0 ± 0.0 s), P11 (SMA+miR-206, 35.6 ± 5.4 s; versus SMA+NT-miR, 58.0 ± 2.0 s), and P12 (SMA+miR-206, 34.3 ± 6.2 s; versus SMA+NT-miR, 60.0 ± 0.0 s), when the time necessary for SMA+miR-206 mice to complete the test was significantly shorter compared to pups treated with the non-targeting miRNA (Figure 3E). The contingency table analysis also confirmed a significant increase in the number of SMA mice treated with miR-206 that successfully completed the test at P9 and P11 (Figure 3F).
Tailing-Suspension Test
This test assigns a score based on hindlimb posture when mice are suspended by the tail for 15 s. The highest value, 4, indicates a normal spreading of hind paws that decreases with self-clasping events that are usually observed in SMA animals. In fact, WT mice showed a score between 3.3 and 4 at all ages considered, with no differences between NT-miR or miR-206 treated groups (Figure 3G). On the other hand, SMA+NT-miR pups showed a score ranging from 2.6 at P3 to 1.8 at P12, indicating hindlimbs often close together. Interestingly, SMA+miR-206 mice showed a significant increase in hindlimb posture at P6 with a score of 2.8 ± 0.1, compared with age-matched SMA+NT-miR pups (1.8 ± 0.1) and reached the highest values at P10 (SMA+miR-206, 3.3 ± 0.2; versus SMA+NT-miR, 2.3 ± 0.1). At P11 and P12, the SMA+miR-206 mice score was still significantly higher compared to SMA+NT-miR, but it started to decline (Figure 3G). The scattered graph plot of each SMA+NT-miR and SMA+miR-206 animal evaluated at P4, P9, and P11 were also reported in Figure 3H.
Hindlimb-Suspension Test
This test evaluates hindlimb muscle strength in rodent neonates. In particular, the position of legs and tail was evaluated by assigning a score ranging from 4 (normal hindlimb separation with tail raised) to 0 (constant clasping and tail lowered). Interestingly, WT animals treated with miR-206 or with the non-targeting molecule showed an improvement in hindlimb score during growth (Figure 3I). By contrast, SMA+NT-miR animals showed a score level of 1.8 at P3 that decreased day by day to a score level close to zero at P12 (Figure 3I). Furthermore, miR-206-treated pups showed a slower decrease in hindlimb score level that was approximately 2 from P3 to P11, then it declined to values close to 1. In particular, a significantly higher score for SMA+miR-206 pups was observed at P8 (SMA+miR-206, 2.3 ± 0.4; versus SMA+NT-miR, 0.9 ± 0.2), P9 (SMA+miR-206, 2.0 ± 0.3; versus SMA+NT-miR, 0.73 ± 0.17), P10 (SMA+miR-206, 2.00 ± 0.16; versus SMA+NT-miR, 0.7 ± 0.2), and P11 (SMA+miR-206, 1.9 ± 0.2; versus SMA+NT-miR, 0.90 ± 0.19), compared to age-matched SMA+NT-miR mice. The scattered graph plot of each SMA+NT-miR and SMA+miR-206 animal evaluated at P4, P9, and P11 were also reported in Figure 3J.
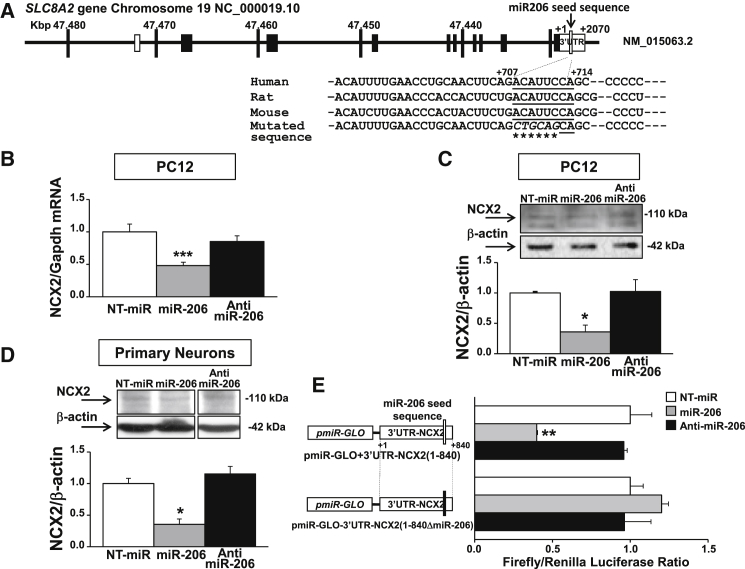
SLC8A2 Gene Is a Novel Target of miR-206
In order to shed light on the neuroprotective pathway activated by miR-206, we investigated among the list of 637 predictable targets generated by McCarthy36 those involved in calcium homeostasis, considering that Ca2+ deregulation is a key factor in triggering SMA toxicity in several cellular compartments.31, 32, 33 Interestingly, a miR-206 putative consensus sequence highly conserved among H. sapiens, R. norvegicus, and M. musculus were identified on the 3′ UTR of the SLC8A2 gene that codes for the plasma membrane transporter NCX2 (Figure 4A). Therefore, in order to investigate whether the SLC8A2 gene can be a novel target gene for miR-206, rat pheochromocytoma cells (PC12) were transfected with 50 nM of NT-miR, miR-206, and anti-miR-206, an antagonist molecule for miR-206. Forty-eight hours after transfection, NCX2 mRNA and protein levels were measured. A significant reduction in NCX2 transcript (∼50%) and protein (∼64%) was observed in cells transfected with miR-206 (Figures 4B and 4C). A significant 65% reduction of NCX2 protein was also observed in primary rat cortical neurons 48 h after transfection with 50 nM miR-206 (Figure 4D).
Figure 4.
SLC8A2 Gene Is a New Target of miR-206
(A) Schematic representation of human SLC8A2 gene structure on chromosome 19. The rectangles represent coding (black) and non-coding (white) exons. Sequence alignment among human, rat, and mouse miR-206 seeds (underlined), showing a high percentage of conservation, was zoomed in. The nucleotides mutated in pmiR-GLO+3′UTR-NCX2(1-840ΔmiR-206) were indicated with a star. (B) Real-time PCR for NCX2 mRNA in PC12 cells transfected with 50 nM of NT-miR (white bars), miR-206 (gray), or anti-miR-206 (black) showing NCX2 mRNA reduction after miR-206 transfection. (C and D) Western blot analysis of NCX2 protein in PC12 cells (C) and primary neurons (D) transfected with 50 nM of NT-miR, miR-206, or anti-miR-206 showing NCX2 protein reduction after miR-206 transfection. (E) Luciferase assay in PC12 cells co-transfect with pmiR-GLO+3′UTR-NCX2(1-840) or pmiR-GLO+3′UTR-NCX2(1-840ΔmiR-206) plus NT-miR, miR-206, or anti-miR-206 showing luciferase decrease in cells co-transfected with the wild-type construct and miR-206 molecules. *p < 0.05; **p < 0.005; ***p < 0.001 versus NT-miR by one-way ANOVA analysis followed by Bonferroni test. Each column represents the mean ± SEM (n = 3/6).
Furthermore, to confirm that the SLC8A2 gene is a target of miR-206, an 840-bp region of the 3′ UTR of this gene, starting from the end of the open reading frame (ORF; indicated as +1) that includes a miR-206 putative binding site at +707/714 bp (Figure 4A), was cloned downstream of the firefly luciferase gene in the pmiR-GLO reporter vector. PC12 cells were then co-transfected with the pmiR-GLO+3′UTR-NCX2(1-840) construct and with NT-miR, miR-206, or anti-miR-206 molecules. Interestingly, when cells were co-transfected with miR-206, the luciferase assay showed a significant 60% reduction compared to NT-miR-transfected cells (Figure 4E). On the other hand, the co-transfection of miR-206 and a new mutated construct pmiR-GLO-3′UTR-NCX2(1-840ΔmiR-206), in which the core sequence of miR-206 seed was mutated by site-directed mutagenesis (as shown in Figure 4A), did not show any reduction in luciferase assay, thus demonstrating that miR-206 was able (1) to bind a specific sequence on the 3′ UTR of NCX2 gene and (2) to modulate its expression.
NCX2 Transcript and Protein Levels Are Inversely Correlated to miR-206 Expression in the Brainstem of SMA Mice
NCX2 expression was measured in the brainstem of WT and SMA mice at different ages with the aim to investigate whether miR-206 expression evaluated in SMA mice correlates with that of the putative target NCX2. Interestingly, a significant downregulation of both NCX2 mRNA and protein expression of approximately 50% was observed in the early phase of the disease (Figures 5A and 5B). On the contrary, NCX2 mRNA and protein levels were upregulated by 2- and 2.5-fold, respectively, in P12 SMA mice (Figures 5A and 5B). This pattern of expression inversely correlated with the expression of miR-206 observed in SMA mice at P5 and P12 (Figure 1A). Furthermore, while NCX2, both at transcript and protein levels, showed a physiological downregulation during growth in WT animals, SMA mice showed an upregulation of NCX2 during disease progression. On the other hand, in the spinal cord of SMA mice, NCX2 expression did not change at P5, while it showed a slightly significant increase at P12 compared to WT (data not shown), even though we observed an upregulation in miR-206 levels in this region in a late phase of the disease (Figure 1B). Interestingly, NCX2 protein expression was downregulated both in the brainstem and in the spinal cord of WT mice i.c.v. injected with miR-206 compared to WT mice treated with NT-miR by 45% and 30%, respectively (Figures S1C and S1D). miR-206 i.c.v. did not affect the other two NCX gene products expressed in the CNS, NCX1, and NCX3. Indeed, their expression did not change in animals i.c.v. injected with miR-206 (Figure S2).
Figure 5.
In Brainstem Facial Nucleus NCX2 Transcript and Protein Levels Were Downregulated in the Early Phase of SMA Pathology, While They Were Upregulated in the Late Phase of the Disease, Particularly in NeuN-Positive Cells
(A and B) Real-time PCR (A) and western blot analysis (B) for NCX2 mRNA and protein in the brainstem of WT (white columns) and SMA (black) mice, five (P5) and 12 (P12) days after birth showing that NCX2 expression is inversely correlated to miR-206 modulation. (C) Confocal images showing the double labeling of NCX2 (red) and NeuN (green) in the facial nuclei of brainstem slices (40 μm) of WT+NT-miR (a–d); WT+miR-206 (e–h); SMA+NT-miR (i–l), and SMA+miR-206 (m–p) animals at P12. Nuclei were labeled with Hoechst (blue). The merge panels (d, h, l, and p) showed the co-localization of NCX2 in neurons as indicated by arrows. Scale bar, 50 μm. (D) Quantification of NCX2 fluorescence in neurons of WT+NT-miR (white column), WT+miR-206 (light gray), SMA+NT-miR (black), and SMA+miR-206 (dark gray) animals at P12. *p < 0.05; **p < 0.005; ***p < 0.001 by one-way ANOVA analysis followed by Bonferroni test. Each column represents the mean ± SEM (n = 3/5).
NCX2 Protein Is Downregulated in Brainstem Neurons of miR-206-Injected SMA Mice
Finally, NCX2 cellular localization was evaluated by confocal microscopy experiments in the facial nuclei of WT and SMA mice injected with miR-206. Brainstem sections of WT+NT-miR, WT+miR-206, SMA+NT-miR, and SMA+miR-206 animals at P12 were double-labeled with anti-NCX2 antibody in combination with the neuronal marker anti-NeuN antibody (Figure 5C). In particular, in the facial nuclei of WT+NT-miR and WT+miR-206 mice, NCX2 immunostaining was mainly localized to neurons, as indicated by arrows in the merge panels (Figures 5Cd and 5Ch). Interestingly, in the whole brainstem of SMA+NT-miR mice at P12, a 30% increase in the immunofluorescence signal of NCX2 was observed in NeuN-positive cells, compared to the other experimental groups (Figure 5D), in agreement with the increase of NCX2 mRNA and protein measured by real-time PCR and WB analysis (Figures 5A and 5B). Moreover, SMA+miR-206 animals showed a reduced immunolabelling of NCX2 in neurons within the facial nucleus (Figure 5Cp) that was comparable to WT+NT-miR and WT+miR-206 groups (Figure 5D).
Discussion
In the present paper, taking advantage of a SMNΔ7 mouse model, we demonstrated for the first time that (1) MN degeneration occurs in the facial nuclei of SMA mice in an early phase of the disease; (2) a parallel upregulation of miR-206 levels takes place in the brainstem; (3) the i.c.v. administration of miR-206 in SMA mice reduces MN loss in the facial nucleus of the brainstem by 25% and in the spinal cord by 15%, ameliorates behavioral performance, and improves SMA mouse survival; and (4) miR-206 might exert its protective effect in SMA, also modulating the expression of the newly identified target gene NCX2, particularly in neurons of the facial nuclei.
An important finding of our work is represented by the demonstration that MN degeneration in the facial nuclei occurs at an early stage of the disease (P5) and may represent an early sign of SMA. On the other hand, MN loss in a late phase of the disease has been described in the spinal cord and in cortical layer V of SMAΔ7 mice,37 thus suggesting that MN degeneration occurs mainly in the facial nuclei of the brainstem in an early phase of the disease and then it proceeds to spinal cord and cerebral cortex in a subsequent, most severe, stage of the disease.
Interestingly, we observed that in an early phase of the disease, MN loss in the facial nuclei of SMA mice was paralleled by the upregulation of miR-206, a member of the myomiR family, linked to SMA pathophysiology,38 which also includes miR-1, miR-133, and miR-208a/b, among few others.39 Therefore, we hypothesized that miR-206 could represent a sensor of MN death, early activated in the brainstem of SMA mice. Indeed, an upregulation of miR-206 levels was also observed in the spinal cord of SMA mice in a late phase of the disease, in agreement with previous studies carried out in two other transgenic mice modeling SMA, SMAI, and SMAIII.22 By contrast, in a late phase of the disease, when MN loss in the facial nucleus was approximatively 80% compared to age-matched WT animals, miR-206 levels were strongly reduced in the brainstem. Therefore, we hypothesize that miR-206 early overexpressed in the brainstem could be widespread to other tissues, thus explaining its downregulation in a later phase of the disease. This hypothesis is supported by Catapano and colleague’s22 findings, that reported an early altered levels of miR-206 in the serum of SMAI mice at P7, followed by a late change in miR-206 expression in the spinal cord of SMAI mice at P10. Moreover, the mRNA precursor of the mature miRNA, the pri-miR-206 molecule, decreased in the spinal cord of SMA mice at P12 (data not shown), suggesting a distant origin of the mature miRNA in this region. Another possible explanation could be represented by the possibility that other mechanisms intervene in the neurodegenerative process during disease progression that could specifically counteract miR-206 upregulation in the brainstem at P12.
Notably, we demonstrated for the first time that miR-206 may represent a new therapeutic option for SMA disease, in agreement with previous reports demonstrating an important role for miR-206 in other CNS disorders.40,41 Interestingly, miR-206 i.c.v. injection in SMA mice ameliorated behavioral performance and improved survival rate. In particular, the increased animal growth induced by the treatment of SMA mice with miR-206 can be attributed to the preservation of approximatively the 25% of MNs in the facial nuclei. Indeed, the facial nucleus is the main nucleus of the facial nerve, which projects MNs to upper and lower facial muscles, mainly the auricular, orbicularis oculi, buccinator, and the nasolabialis muscle groups,42 all of them involved in the action of chewing. Hence, miR-206, by preserving MNs in the facial nuclei, may be responsible for the greater weight observed in SMA animals. On the other hand, improvement in motor performance could be ascribed to the higher number of surviving spinal cord MNs (approximatively the 15%) that project directly to muscles or, alternatively, to greater muscle strength due to easier nutrition. Furthermore, miR-206 administration increased the mouse lifespan by 2 days, thus increasing the life expectancy by almost 15%. It should be underlined that although an increase in lifespan of only 2 days might appear short, this time interval should be evaluated considering the short lifespan of approximately 13 days of SMA pups. On the other hand, other promising therapeutic strategies such as autophagosome inhibition, increasing agrin function and JNK inhibition described in this genetic model of SMA showed similar effects on lifespan increase.43, 44, 45
In order to improve the translatability of our results, the pharmacological treatment was started 3 days after birth. Indeed, several reports demonstrated that the therapeutic window for SMA therapy is quite narrow. In particular, Foust and colleagues46 demonstrated that intravenous injection of self-complementary adeno-associated virus 9 to delivery SMN (scAAV9-SMN) in SMAII mice rescues motor function and extends lifespan up to 250 days only when given at P1 or P2. On the contrary, the therapeutic effects were more modest when the treatment started at P5 and totally ineffective when started at P10.46 Analogously, trichostatin A (TSA) administration, by increasing SMN and SMNΔ7 proteins, is able to exert its protective effect only when administered at P5, but not at P10.47 Moreover, clinical trials on SMA patients with nusinersen, the first treatment approved for SMA by the US Food and Drug Administration (FDA) in 2016 and by the European Medicines Agency (EMA) in 2017, demonstrated that early initiation of treatment within 13 weeks after the onset of the disease may maximize its efficacy in SMAI children.48,49
Finally, among the list of 637 predictable targets of miR-206 generated by McCarthy compiling data from TargetScan (http://www.targetscan.org), PicTar (https://pictar.mdc-berlin.de), and Miranda (https://microrna.sanger.ac.uk) databases,36 we investigated the putative miR-206 seed sequence on the 3′ UTR of the SLC8A2 gene, which codes for the plasma membrane transporter NCX2. Notably, we demonstrated that miR-206 can directly downregulate NCX2 expression in cellular cultures and in the brainstem of SMA mice. NCX2 is a “low-affinity high-capacity” transporter, belonging to the Na+/Ca2+ exchanger family, that can rapidly respond to transient changes in [Ca2+]i23,50 and hence represent one of the master regulators of Na+ and Ca2+ homeostasis.51,52 Therefore, the modulation of NCX2 expression represents a reasonable mechanism to preserve MN function and survival. In particular, our data revealed that in the brainstem, NCX2 expression decreases physiologically during the first weeks of age in WT animals, while in SMA mice, NCX2 levels increased during development, thus suggesting that NCX2 upregulation may be linked to disease progression. In agreement with this hypothesis, i.c.v. injection of miR-206 prevented NCX2 upregulation, particularly in the neuronal compartment of the facial nuclei of SMA mice, inducing a neuroprotective effect on MN degeneration. In particular, as indicated by double immunofluorescence experiments. in the facial nucleus of WT mice, NCX2 expression was mainly localized in the neuronal compartment. Interestingly, SMA mice neurons of the facial nucleus showed a 30% upregulation of NCX2 levels that was prevented by miR-206 administration. However, besides neurons, we cannot exclude NCX2 modulation in other cell types, i.e., glial cells, that could promote MN survival. On the other hand, in a late phase of the disease, we reported an upregulation of miR-206 in the spinal cord of SMA mice, but in this anatomical region, NCX2 protein levels were not reduced (data not shown). This discrepancy could be due to a late activation of miR-206 in this region and/or to the concomitant activation of alternative biochemical pathways that counteracted miR-206 action.
Concerning the protective effect exerted by NCX2 downregulation induced by miR-206, a possible explanation is that during the first 2 weeks of mouse life, γ-aminobutyric acid (GABA) switches from an excitatory to an inhibitory function (GABA switch).53 Indeed, at birth, it has been reported that there is an increased activity of Na+-K+-2Cl− co-transporter (NKCC1) that causes an intracellular increase of Cl−, Na+, and K+ ions. Regarding Cl− ion accumulation, GABA, by activating its GABAA receptor, differently from what occurs in mature CNS cells, promotes Cl− exit. Concerning the increased levels of [Na+]i and the mechanism involved in its reduction, we can suggest that NCX2, working in the Na+ efflux manner, takes part in this process.53 However, the activation of NCX2 in the reverse mode causes an overload of intracellular Ca2+ ions that has been already observed in SMN-knocked-down neurons54 and that can trigger MN degeneration.
Overall, in the present paper, we contributed to shedding light on SMA pathology, indicating the facial nucleus in the brainstem as one of the first areas in which MN degeneration occurs. We supported the hypothesis that miR-206 could represent an effective strategy in SMA since, by modulating NCX2 gene expression and hence Na+ and Ca2+ homeostasis, it could act in a synergistic way with the currently available therapies. Indeed, the concomitant targeting of different pathological mechanisms could be more effective than individual treatment in this multifactorial non-autonomous disease.
Materials and Methods
Animal Model
The original breeding smn+/−; hSMN2+/+; hSMNΔ7+/+ mice, heterozygous and healthy carriers for smn gene mutation, were purchased from Jackson Laboratory (stock number 005025; Jackson Laboratories) and were bred to obtain smn+/+; hSMN2+/+; hSMNΔ7+/+ (WT) animals and smn−/−; hSMN2+/+; hSMNΔ7+/+ (SMA) animals. The colony was maintained by interbreeding carrier mice, and the offspring were genotyped by PCR assays on tail DNA according to the protocols provided by Jackson Laboratory, as previously reported.45 Mice were sacrificed at P5 and P12, considering P0 as the day of birth. Overall, 80 WT mice and 74 SMA mice, housed under diurnal lighting conditions (12 h darkness/light) were used. Experiments were performed according to the international guidelines for animal research and approved by the Animal Care Committee of “Federico II” University of Naples, Italy, and Ministry of Health, Italy. All efforts were made to minimize animal suffering and to reduce the number of animals used.
Real-Time PCR Analysis
Mice were deeply anaesthetized with 3% isoflurane vaporized in O2/N2O 50:50 and sacrificed. Brainstem and spinal cord were rapidly removed and immediately frozen on dry ice and stored at −80°C until use. Total RNA was extracted following supplier’s instructions (Life Technologies), and cDNA was synthesized using 0.5 or 2 μg of total RNA to obtain miRNA-specific cDNA or total cDNA, respectively, with the high-capacity transcription kit following supplier’s instruction (Life Technologies, Monza, Italy). Quantitative real-time PCR was performed with TaqMan assays in a 7500 real-time PCR system (Life Technologies). Changes in mRNA levels were determined as the difference in threshold cycle (2ˆ−ΔΔCt) between the target gene (SLC8A2 TaqMan ID, Mm00455836_m1) and the reference gene: beta-glucuronidase (Gusb) for mouse tissues (ID: Mm00446953_m1) and glyceraldehyde-3-phosphate dehydrogenase (Gapdh) for cell tissues (ID: Mm99999915_g1). Changes in miRNA levels were determined as the difference in threshold cycle between miR-206 (ID: 00510) and Gusb as previously reported.21,55 U6 small nuclear RNA (snRNA) (ID: 001973) was also used as endogenous control for miR-206, giving results fully overlapping to the one obtained using Gusb as housekeeping gene.
Western Blot
Total extracts were obtained as previously described.56,57 To detect the proteins of interest, specific antibodies were used: anti-NCX1 (rabbit polyclonal antibody, 1:1,000; Swant), anti-NCX2 (rabbit polyclonal antibody, 1:1,000; Alpha Diagnostic Intl., TX, USA), and mouse monoclonals (Sigma-Aldrich, Milan, Italy) anti β-actin (1:1,000) and anti-tubulin (1:5,000). Immunoreaction was revealed using anti-mouse and anti-rabbit immunoglobulin G conjugated to peroxidase 1:10,000 (GE Healthcare, Norwalk, CT, USA) by the enhanced chemiluminescence (ECL) reagent (GE Healthcare). The optical density of the bands was determined by Chemi Doc Imaging System (Bio-Rad, Milan, Italy) and normalized to the optical density of β-actin or tubulin, used as internal controls.
Stereotaxic Surgery and Administration of miRNAs
i.c.v. administration of 1 μL of locked lucleic acid-modified miR-206 (miRCURY LNA microRNA mimic, Exiqon, Denmark) or of a non-targeting miRNA (NT-miR negative control miRCURY LNA miRNA mimic, Exiqon) at the concentration of 10 μM at P3, P6, and P10 was performed as previously described.58 In brief, neonatal mice were anesthetized by hypothermia (total duration 2′), and their heads were immobilized on a custom neonatal stereotaxic apparatus at 4°C during surgery. miR-206 or NT-miR molecules were injected at stereotaxic coordinates of 0 mm from bregma, 0.8 mm lateral to sagittal sinus, and 1.5 mm deep with a Hamilton microsyringe. The pups were then placed on a heat pad, quickly revitalized, and subsequently returned to their mother.
Behavioral Assessment
In order to analyze the progression of motor symptoms in WT and SMA mice treated with miR-206 or with NT-miR, the animals were divided into four experimental groups, WT+NT-miR (n = 25), WT+miR-206 (n = 26), SMA+NT-miR (n = 14), and SMA+miR-206 (n = 13), body weight was checked daily, and behavioral tests were performed, starting at P3 as previously reported.35
For the righting reflex test, pups were placed on their backs on a flat surface. The time employed to right themselves was recorded (cutoff 30 s). For the tail-suspension test (self-clasping test), pups were suspended by the tail for 15 s, and their hindlimb posture was scored. The scores were as follows: 4, hindlimbs spread open; 3, hindlimbs not completely spread; 2, hindlimbs often close together; 1, hindlimbs always close together; 0, hindlimbs always close together with additional postural abnormalities (clasping). For the hindlimb-suspension test (tube test), the mouse was placed head down, hanging by its hindlimbs, in a plastic 50-mL tube with a cotton ball cushion at the bottom to protect the animal in case of fall. We considered two parameters: the latency to fall (time) from the edge of the tube and the hindlimb score. The score was divided into four criteria: 4, normal hindlimb separation with tail raised; 3, apparent weakness and hindlimbs close together without touching each other; 2, closer hindlimbs, almost touching; 1, hindlimbs always clasping with the tail raised; and 0, constant clasping of the hindlimbs with the tail lowered. This test, developed at PsychoGenics, evaluated the proximal hindlimb fatigue, weakness, and muscle strength in pups. For the negative geotaxis from P4 text, the animals were placed on an inclined grid (at approximately 35° inclination) with the mouse head facing down. We measured the time needed for mice to orient themselves (cutoff 60 s). This test is useful to evaluate both motor coordination and vestibular sensitivity.
Immunofluorescence
Immunostaining and confocal immunofluorescence procedures were performed as previously described.59 In brief, animals were anaesthetized and transcardially perfused with a saline solution containing 0.01 mL heparin, followed by 4% paraformaldehyde in 0.1 mol/L PBS saline solution. Brains were rapidly removed on ice and post-fixed overnight at +4°C and cryoprotected in 30% sucrose in 0.1 M phosphate buffer (PB) with sodium azide 0.02% for 24 h at 4°C. Next, brains were sectioned frozen on a sliding cryostat at 40 μm thickness, in rostrum-caudal direction. Afterward, free-floating serial sections were incubated with PB Triton X 0.3% and blocking solution (0.5% milk, 10% fetal bovine serum [FBS], 1% BSA) for 1 h and 30 min. The sections were incubated overnight at +4°C with the following primary antibodies: anti-NeuN (mouse monoclonal antibody; 1:1,000; Millipore, Milan, Italy), anti-NCX2 (1:500; Alpha Diagnostic). The sections were then incubated with the corresponding florescent-labeled secondary antibodies, Alexa 488/Alexa 594 conjugated anti-mouse/anti-rabbit immunoglobulin Gs (Molecular Probes, Invitrogen, Milan, Italy). Nuclei were counterstained with Hoechst (Sigma-Aldrich). Images were observed using a Zeiss LSM700 META/laser scanning confocal microscope (Zeiss, Oberkochen, Germany). Single images were taken with an optical thickness of 0.7 m and a resolution of 1,024 × 1,024. In double-labeled sections, the pattern of immune reactivity for both antigens was identical to that seen in single-stained material. Standard Nissl staining was employed on coronal step serial sections from the brainstem and spinal cord.
Fluorescence Intensity and MN Counting Analysis
NCX2 fluorescence intensity in neurons was quantified in terms of pixel intensity value by using the NIH image software, as described previously.60 In brief, digital images were taken with ×40 or ×10 objectives, and identical laser power settings and exposure times were applied to all the photographs from each experimental set. Images from the same areas of each brain region were compared.
To obtain an indirect measure of the amount of NCX2 in neurons, image analysis of NeuN was performed by NIH image software by measuring the intensity of fluorescent NCX2 immunolabeling in 20 NeuN-positive neurons for each group. Results were expressed in arbitrary units. Three sections from each mouse were analyzed, with n = 3 mice per treatment group.
MNs were counted in the facial nucleus in the brainstem. Sections of each area were analyzed as previously described.30 Frozen brain tissues were sectioned on a sliding cryostat at 20 μm in rostrum-caudal direction. Three mice for each group and three slides from every mouse were analyzed. Analyses were performed using ImageJ software in polygonal-shaped neurons larger than 20 μm with a well-defined cytoplasm, nucleus, and nucleolus.
Cloning of the Human SLC8A2 3′ UTR
PCR amplification was performed using PrimeSTAR GLX DNA polymerase (Takara, Kusatsu, Japan) on human cDNA, priming from NCX2 stop codon at +3,042 bp to +3,930 bp (GenBank: NM_015063.2), inserting an NheI and a XhoI restriction site within the forward and the reverse primers, respectively. The PCR product was purified using StrataPrep DNA gel extraction kits (Agilent, Milan, Italy) and cloned into multiple cloning sites of pmirGLO dual-luciferase miRNA target expression vector (Promega, Milan, Italy) downstream of the firefly luciferase gene (luc2). The primer sequences flanked by NheI and XhoI sites used for the amplification were as follows: NCX2-3′ UTR fwd, 5′-atagctagctagggcctcgcgcagagactcg-3′, and NCX2-3′ UTR rev, 5′-tatctcgagcccttttcccaagacatgcaccc-3′. The fidelity of the constructs was confirmed by DNA sequencing. Site-directed mutations were carried out in the miR-206-3p binding seed sequence using the quick change site-directed mutagenesis kit (Agilent) according to the manufacturer’s instructions as previously reported.61 Desired mutations were confirmed by sequencing. The primers used for site-directed mutagenesis were 5′-gaacctgcaacttcagctgcagcagctcccccactcgc-3′ and NCX2-3′ UTR mut rev 5′-gcgagtgggggagctgctgcagctgaagttgcaggttc-3′.
Cell Cultures, Transient Transfection, and Reporter Assay
Primary cortical neurons were obtained as previously reported26 according to the experimental protocols approved by the Ethics Committee of “Federico II” University of Naples. They were prepared from 17-day-old Wistar rat embryos (Charles River, Milan, Italy) and used after 7 days. In brief, dissection and dissociation were performed in Ca2+/Mg2+-free PBS containing glucose (30 mM). Tissues were incubated with papain for 10 min at 37°C and dissociated by trituration in Earl’s Balanced Salt Solution containing DNase (0.16 U/mL), BSA (10 mg/mL), and ovomucoid (10 mg/mL). Neurons, plated in plastic Petri dishes (Falcon Becton Dickinson) precoated with poly-D-lysine (20 μg/mL), were grown in MEM/F12 containing glucose, 5% deactivated FBS, 5% HS, 2 mM glutamine, and 1% penicillin-streptomycin (5 U/mL) (Invitrogen). Cytosine arabinoside (2.5 μM) was added on the second day to reduce glial contamination.
PC12 cells were maintained in culture in complete RPMI 1640 medium containing 10% heat-inactivated HS, 5% FBS, 1% L-glutamine, 1% penicillin-streptomycin and were plated 24 h before transfection.
PC12 cells and cortical neurons were transfected with 50 nM of miR-206-3p mimic (Mimic; Ambion, Milan, Italy) or the negative control (NT-miR; Ambion) or specific miRNA inhibitor (anti-miR-206; Ambion) with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Forty-eight hours after transfection, cells were harvested.
For dual luciferase assays, PC12 cells were transfected with 200 ng of each plasmid in combination with 50 nM of miR-206-3p mimic or the NT-miR or anti-miR-206. Forty-eight hours after transfection, cells were harvested following the supplier’s instructions (Promega), and the light emitted was measured with a manual luminometer (Glomax 20/20, Promega). The effect of 3′ UTR activity on the reporter gene was calculated as firefly-to-renilla ratio and normalized for the luciferase of the empty vector.
Statistical Analysis
Data were evaluated as means ± SEM. Statistically significant differences among means were determined by one-way ANOVA followed by Bonferroni post hoc test for western blotting, cell counting, real-time PCR and luciferase analysis. Two-way ANOVA followed by Bonferroni post hoc was used for motor performances test and body weight analysis. The Kaplan-Meier plot was used to evaluate survival. Student’s t test was used for two-group comparison. Statistical significance was accepted at the 95% confidence level (*p < 0.05; **p < 0.005; ***p < 0.001; ****p < 0.0001). Statistical analyses were performed using GraphPad Prism 5.0 (La Jolla, CA, USA). All experiments were carried out in a blinded manner.
Author Contributions
V.V. conceived and designed the project in collaboration with G.P. and L.A.; V.V., S.A., A.S., G.L., and P.B. performed the experiments; O.C. and N.G. contributed with animal experiments; V.V., G.P., and L.A. wrote the manuscript.
Acknowledgments
This work was supported by the following grants: Programma Operativo Nazionale (PON_01602 and PON03PE_00146_1) from MIUR to L.A.; PRIN 2015 2015783N45-CUP: E62F1600136001 to G.P.; and the Italian Ministry of Economic Development (MISE), Nanomirnictus, F/050139/01,02/X32 to L.A.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.01.013.
Contributor Information
Valeria Valsecchi, Email: valeria.valsecchi@unina.it.
Giuseppe Pignataro, Email: giuseppe.pignataro@unina.it.
Lucio Annunziato, Email: lannunzi@unina.it.
Supplemental Information
References
- 1.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 5.Feldkötter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burghes A.H., Beattie C.E. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 8.Meister G., Bühler D., Laggerbauer B., Zobawa M., Lottspeich F., Fischer U. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- 9.Pellizzoni L., Baccon J., Rappsilber J., Mann M., Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 10.Baccon J., Pellizzoni L., Rappsilber J., Mann M., Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- 11.Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 12.Novák J., Kružliak P., Bienertová-Vašků J., Slabý O., Novák M. MicroRNA-206: a promising theranostic marker. Theranostics. 2014;4:119–133. doi: 10.7150/thno.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zampetaki A., Kiechl S., Drozdov I., Willeit P., Mayr U., Prokopi M., Mayr A., Weger S., Oberhollenzer F., Bonora E. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 14.Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catalanotto C., Cogoni C., Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016;17:E1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haramati S., Chapnik E., Sztainberg Y., Eilam R., Zwang R., Gershoni N., McGlinn E., Heiser P.W., Wills A.M., Wirguin I. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. USA. 2010;107:13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kye M.J., Gonçalves I.d.C.G. The role of miRNA in motor neuron disease. Front. Cell. Neurosci. 2014;8:15. doi: 10.3389/fncel.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L.T., Chiou S.S., Liao Y.M., Jong Y.J., Hsu S.H. Survival of motor neuron protein downregulates miR-9 expression in patients with spinal muscular atrophy. Kaohsiung J. Med. Sci. 2014;30:229–234. doi: 10.1016/j.kjms.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Ma G., Wang Y., Li Y., Cui L., Zhao Y., Zhao B., Li K. MiR-206, a key modulator of skeletal muscle development and disease. Int. J. Biol. Sci. 2015;11:345–352. doi: 10.7150/ijbs.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams A.H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J.L., Bassel-Duby R., Sanes J.R., Olson E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valsecchi V., Boido M., De Amicis E., Piras A., Vercelli A. Expression of Muscle-Specific MiRNA 206 in the Progression of Disease in a Murine SMA Model. PLoS ONE. 2015;10:e0128560. doi: 10.1371/journal.pone.0128560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catapano F., Zaharieva I., Scoto M., Marrosu E., Morgan J., Muntoni F., Zhou H. Altered Levels of MicroRNA-9, -206, and -132 in Spinal Muscular Atrophy and Their Response to Antisense Oligonucleotide Therapy. Mol. Ther. Nucleic Acids. 2016;5:e331. doi: 10.1038/mtna.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Annunziato L., Pignataro G., Boscia F., Sirabella R., Formisano L., Saggese M., Cuomo O., Gala R., Secondo A., Viggiano D. ncx1, ncx2, and ncx3 gene product expression and function in neuronal anoxia and brain ischemia. Ann. N Y Acad. Sci. 2007;1099:413–426. doi: 10.1196/annals.1387.050. [DOI] [PubMed] [Google Scholar]
- 24.Valsecchi V., Pignataro G., Sirabella R., Matrone C., Boscia F., Scorziello A., Sisalli M.J., Esposito E., Zambrano N., Cataldi M. Transcriptional regulation of ncx1 gene in the brain. Adv. Exp. Med. Biol. 2013;961:137–145. doi: 10.1007/978-1-4614-4756-6_12. [DOI] [PubMed] [Google Scholar]
- 25.Formisano L., Guida N., Valsecchi V., Pignataro G., Vinciguerra A., Pannaccione A., Secondo A., Boscia F., Molinaro P., Sisalli M.J. NCX1 is a new rest target gene: role in cerebral ischemia. Neurobiol. Dis. 2013;50:76–85. doi: 10.1016/j.nbd.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Formisano L., Guida N., Valsecchi V., Cantile M., Cuomo O., Vinciguerra A., Laudati G., Pignataro G., Sirabella R., Di Renzo G., Annunziato L. Sp3/REST/HDAC1/HDAC2 Complex Represses and Sp1/HIF-1/p300 Complex Activates ncx1 Gene Transcription, in Brain Ischemia and in Ischemic Brain Preconditioning, by Epigenetic Mechanism. J. Neurosci. 2015;35:7332–7348. doi: 10.1523/JNEUROSCI.2174-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casamassa A., La Rocca C., Sokolow S., Herchuelz A., Matarese G., Annunziato L., Boscia F. Ncx3 gene ablation impairs oligodendrocyte precursor response and increases susceptibility to experimental autoimmune encephalomyelitis. Glia. 2016;64:1124–1137. doi: 10.1002/glia.22985. [DOI] [PubMed] [Google Scholar]
- 28.Pannaccione A., Secondo A., Molinaro P., D’Avanzo C., Cantile M., Esposito A., Boscia F., Scorziello A., Sirabella R., Sokolow S. A new concept: Aβ1-42 generates a hyperfunctional proteolytic NCX3 fragment that delays caspase-12 activation and neuronal death. J. Neurosci. 2012;32:10609–10617. doi: 10.1523/JNEUROSCI.6429-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirabella R., Valsecchi V., Anzilotti S., Cuomo O., Vinciguerra A., Cepparulo P., Brancaccio P., Guida N., Blondeau N., Canzoniero L.M.T. Ionic Homeostasis Maintenance in ALS: Focus on New Therapeutic Targets. Front. Neurosci. 2018;12:510. doi: 10.3389/fnins.2018.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anzilotti S., Brancaccio P., Simeone G., Valsecchi V., Vinciguerra A., Secondo A., Petrozziello T., Guida N., Sirabella R., Cuomo O. Preconditioning, induced by sub-toxic dose of the neurotoxin L-BMAA, delays ALS progression in mice and prevents Na+/Ca2+ exchanger 3 downregulation. Cell Death Dis. 2018;9:206. doi: 10.1038/s41419-017-0227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jablonka S., Beck M., Lechner B.D., Mayer C., Sendtner M. Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J. Cell Biol. 2007;179:139–149. doi: 10.1083/jcb.200703187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz R., Casañas J.J., Torres-Benito L., Cano R., Tabares L. Altered intracellular Ca2+ homeostasis in nerve terminals of severe spinal muscular atrophy mice. J. Neurosci. 2010;30:849–857. doi: 10.1523/JNEUROSCI.4496-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGivern J.V., Patitucci T.N., Nord J.A., Barabas M.A., Stucky C.L., Ebert A.D. Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production. Glia. 2013;61:1418–1428. doi: 10.1002/glia.22522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 35.El-Khodor B.F., Edgar N., Chen A., Winberg M.L., Joyce C., Brunner D., Suárez-Fariñas M., Heyes M.P. Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy. Exp. Neurol. 2008;212:29–43. doi: 10.1016/j.expneurol.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy J.J. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.d’Errico P., Boido M., Piras A., Valsecchi V., De Amicis E., Locatelli D., Capra S., Vagni F., Vercelli A., Battaglia G. Selective vulnerability of spinal and cortical motor neuron subpopulations in delta7 SMA mice. PLoS ONE. 2013;8:e82654. doi: 10.1371/journal.pone.0082654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawley Z.C.E., Campos-Melo D., Droppelmann C.A., Strong M.J. MotomiRs: miRNAs in Motor Neuron Function and Disease. Front. Mol. Neurosci. 2017;10:127. doi: 10.3389/fnmol.2017.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horak M., Novak J., Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016;410:1–13. doi: 10.1016/j.ydbio.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Sun W., Zhang L., Li R. Overexpression of miR-206 ameliorates chronic constriction injury-induced neuropathic pain in rats via the MEK/ERK pathway by targeting brain-derived neurotrophic factor. Neurosci. Lett. 2017;646:68–74. doi: 10.1016/j.neulet.2016.12.047. [DOI] [PubMed] [Google Scholar]
- 41.Lee S.T., Chu K., Jung K.H., Kim J.H., Huh J.Y., Yoon H., Park D.K., Lim J.Y., Kim J.M., Jeon D. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012;72:269–277. doi: 10.1002/ana.23588. [DOI] [PubMed] [Google Scholar]
- 42.Watson C., Paxinos G., Puelles L. Academic Press; 2012. The Mouse Nervous System. [Google Scholar]
- 43.Piras A., Schiaffino L., Boido M., Valsecchi V., Guglielmotto M., De Amicis E., Puyal J., Garcera A., Tamagno E., Soler R.M., Vercelli A. Inhibition of autophagy delays motoneuron degeneration and extends lifespan in a mouse model of spinal muscular atrophy. Cell Death Dis. 2017;8:3223. doi: 10.1038/s41419-017-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schellino R., Boido M., Borsello T., Vercelli A. Pharmacological c-Jun NH2-Terminal Kinase (JNK) Pathway Inhibition Reduces Severity of Spinal Muscular Atrophy Disease in Mice. Front. Mol. Neurosci. 2018;11:308. doi: 10.3389/fnmol.2018.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boido M., De Amicis E., Valsecchi V., Trevisan M., Ala U., Ruegg M.A., Hettwer S., Vercelli A. Increasing Agrin Function Antagonizes Muscle Atrophy and Motor Impairment in Spinal Muscular Atrophy. Front. Cell. Neurosci. 2018;12:17. doi: 10.3389/fncel.2018.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H., Kaspar B.K. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Avila A.M., Burnett B.G., Taye A.A., Gabanella F., Knight M.A., Hartenstein P., Cizman Z., Di Prospero N.A., Pellizzoni L., Fischbeck K.H., Sumner C.J. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., ENDEAR Study Group Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 49.Parente V., Corti S. Advances in spinal muscular atrophy therapeutics. Ther. Adv. Neurol. Disorder. 2018;11 doi: 10.1177/1756285618754501. 1756285618754501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khananshvili D. Sodium-calcium exchangers (NCX): molecular hallmarks underlying the tissue-specific and systemic functions. Pflugers Arch. 2014;466:43–60. doi: 10.1007/s00424-013-1405-y. [DOI] [PubMed] [Google Scholar]
- 51.Pignataro G., Sirabella R., Anzilotti S., Di Renzo G., Annunziato L. Does Na+/Ca2+ exchanger, NCX, represent a new druggable target in stroke intervention? Transl. Stroke Res. 2014;5:145–155. doi: 10.1007/s12975-013-0308-8. [DOI] [PubMed] [Google Scholar]
- 52.Pignataro G., Esposito E., Sirabella R., Vinciguerra A., Cuomo O., Di Renzo G., Annunziato L. nNOS and p-ERK involvement in the neuroprotection exerted by remote postconditioning in rats subjected to transient middle cerebral artery occlusion. Neurobiol. Dis. 2013;54:105–114. doi: 10.1016/j.nbd.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 53.Rivera C., Voipio J., Payne J.A., Ruusuvuori E., Lahtinen H., Lamsa K., Pirvola U., Saarma M., Kaila K. The K+/Cl- co-transporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature. 1999;397:251–255. doi: 10.1038/16697. [DOI] [PubMed] [Google Scholar]
- 54.Acsadi G., Lee I., Li X., Khaidakov M., Pecinova A., Parker G.C., Hüttemann M. Mitochondrial dysfunction in a neural cell model of spinal muscular atrophy. J. Neurosci. Res. 2009;87:2748–2756. doi: 10.1002/jnr.22106. [DOI] [PubMed] [Google Scholar]
- 55.Boido M., Piras A., Valsecchi V., Spigolon G., Mareschi K., Ferrero I., Vizzini A., Temi S., Mazzini L., Fagioli F., Vercelli A. Human mesenchymal stromal cell transplantation modulates neuroinflammatory milieu in a mouse model of amyotrophic lateral sclerosis. Cytotherapy. 2014;16:1059–1072. doi: 10.1016/j.jcyt.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Guglielmotto M., Monteleone D., Piras A., Valsecchi V., Tropiano M., Ariano S., Fornaro M., Vercelli A., Puyal J., Arancio O. Aβ1-42 monomers or oligomers have different effects on autophagy and apoptosis. Autophagy. 2014;10:1827–1843. doi: 10.4161/auto.30001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guida N., Valsecchi V., Laudati G., Serani A., Mascolo L., Molinaro P., Montuori P., Di Renzo G., Canzoniero L.M., Formisano L. The miR206-JunD Circuit Mediates the Neurotoxic Effect of Methylmercury in Cortical Neurons. Toxicol. Sci. 2018;163:569–578. doi: 10.1093/toxsci/kfy051. [DOI] [PubMed] [Google Scholar]
- 58.Guida N., Laudati G., Mascolo L., Valsecchi V., Sirabella R., Selleri C., Di Renzo G., Canzoniero L.M., Formisano L. p38/Sp1/Sp4/HDAC4/BDNF Axis Is a Novel Molecular Pathway of the Neurotoxic Effect of the Methylmercury. Front. Neurosci. 2017;11:8. doi: 10.3389/fnins.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anzilotti S., Tornincasa M., Gerlini R., Conte A., Brancaccio P., Cuomo O., Bianco G., Fusco A., Annunziato L., Pignataro G., Pierantoni G.M. Genetic ablation of homeodomain-interacting protein kinase 2 selectively induces apoptosis of cerebellar Purkinje cells during adulthood and generates an ataxic-like phenotype. Cell Death Dis. 2015;6:e2004. doi: 10.1038/cddis.2015.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gargiulo S., Anzilotti S., Coda A.R., Gramanzini M., Greco A., Panico M., Vinciguerra A., Zannetti A., Vicidomini C., Dollé F. Imaging of brain TSPO expression in a mouse model of amyotrophic lateral sclerosis with (18)F-DPA-714 and micro-PET/CT. Eur. J. Nucl. Med. Mol. Imaging. 2016;43:1348–1359. doi: 10.1007/s00259-016-3311-y. [DOI] [PubMed] [Google Scholar]
- 61.Valsecchi V., Pignataro G., Del Prete A., Sirabella R., Matrone C., Boscia F., Scorziello A., Sisalli M.J., Esposito E., Zambrano N. NCX1 is a novel target gene for hypoxia-inducible factor-1 in ischemic brain preconditioning. Stroke. 2011;42:754–763. doi: 10.1161/STROKEAHA.110.597583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.